- 1Basic Medicine College, Hebei University of Chinese Medicine, Shijiazhuang, China
- 2Shijiazhuang Center for Disease Control and Prevention, Shijiazhuang, China
- 3College of Life Science, Hebei Normal University, Shijiazhuang, China
As an opportunistic pathogen worldwide, Staphylococcus aureus can cause food poisoning and human infections. This study investigated the sequence typing, the penicillin (blaZ) and methicillin (mec) resistance profiles of S. aureus from food samples and food poisoning outbreaks in Shijiazhuang City, and the staphylococcal enterotoxin (SE) types of the S. aureus isolates from food poisoning. A total of 138 foodborne S. aureus isolates were distributed into 8 clonal complexes (CCs) and 12 singletons. CC1, CC5, CC8, CC15, CC97, CC59, CC398, CC88, and CC7 were the predominant CCs of foodborne S. aureus isolates. Moreover, CC59, CC15, and CC5 were the most prevalent CCs in food poisoning outbreaks. SEE was the most commonly detected SE in food poisoning isolates. One hundred thirty-three S. aureus isolates harbored the penicillin-resistant gene blaZ, and nine isolates carried the mec gene. The present study further explained the relationship between S. aureus and foods and food poisoning and indicated the potential risk of S. aureus infection.
Introduction
As an important Gram-positive spherical pathogen, Staphylococcus aureus can release a variety of heat-stable staphylococcal enterotoxins (SEs) into food (Hennekinne et al., 2012; Wang et al., 2017; Oliveira et al., 2018) responsible for staphylococcal food poisoning (SFP) in human population. SFP is characterized by foodborne gastroenteritis (Argudín et al., 2010; Hennekinne et al., 2012; Lv et al., 2014), and it was recognized as one of the most prominent culprits in food poisoning outbreaks worldwide (Hennekinne et al., 2012; Wu et al., 2016). S. aureus can carry multiple SE genes and produce different SEs and SE-like toxins. The SEs (SEA to SEE; SEG to SEI; SEK; SEM to SET) and SE-like toxins (SElJ; SElL; SElU to SElZ) were reported as responsible agents for food poisoning outbreaks (Ono et al., 2008; Argudín et al., 2010; Hennekinne et al., 2012; Lv et al., 2014). SEA, SEB, SEC, SED, and SEE were the most common toxins implicated in SFP (Hennekinne et al., 2012; Kadariya et al., 2014).
Multilocus sequence typing (MLST) analysis of S. aureus is important for the prognosis of infection, and it provides means to trace epidemiologically related strains contributing to the tracking of the contamination source. The MLST types of S. aureus vary with regions and sources (Pereira et al., 2009; Cheng et al., 2013; Rao et al., 2015; Thapaliya et al., 2017; Htun et al., 2018; Kitti et al., 2018; Wu et al., 2018; Li et al., 2019).
The MLST genotype of S. aureus has been reported to influence the complications, severity, and mortality of infection; the strains of CC5 and CC30 exhibited a significant trend toward increased levels of hematogenous complications (Chen et al., 2010). The sequence type 121 of S. aureus (ST121) infected the patients who often required long hospitalization and prolonged antimicrobial therapy (Rao et al., 2015), CC398 were associated with high mortality (Bouiller et al., 2016).
Staphylococcus aureus can acquire the antibiotic resistance determinants that may confer resistance to many antibiotics. High resistance of S. aureus to penicillin was observed (Wu et al., 2018; Li et al., 2019; Dastmalchi Saei and Panahi, 2020); moreover, the resistance to most beta-lactams and the other antibiotics except ceftaroline and ceftobiprole was reported for methicillin-resistant S. aureus (MRSA) strains (Lakhundi and Zhang, 2018; Wu et al., 2018; Li et al., 2019). MRSA infections are associated with higher mortality rates than the infections caused by methicillin-susceptible strains. MRSA isolates carry a mobile genetic element of staphylococcal cassette chromosome mec (SCCmec), the determinant that encodes low-affinity penicillin-binding protein (PBP) similar to PBP2a or PBP2′. The mec genes share ≥70% nucleotide sequence identity with the mecA gene (Ito et al., 2012; Lakhundi and Zhang, 2018).
This study aimed to characterize S. aureus isolates from SFP outbreaks and the food samples isolated in Shijiazhuang, China. In particular, MLST genotypes and the presence of beta-lactam-resistant genes of blaZ and mec were determined on all isolates. In addition, the enterotoxigenic status of SFP isolates was explored.
Materials and Methods
Bacterial Isolates
One hundred and thirty-eight S. aureus isolates were used in this study. Eighty-two of the strains were isolated from food samples in 2011–2019, and 56 strains were isolated from 20 SFP outbreaks in 2009–2016. All the isolates were identified as S. aureus using conventional microbiological methods including Gram staining and catalase and coagulase tests and then stored in a Brain Heart Infusion (BHI) medium with 40% glycerine at −80°C. Species was confirmed by Matrix-Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) System (Bruker, Berlin, Germany) before further experiments.
Primers
All the primers used in the study were synthesized by TaKaRa (TaKaRa, Beijing, China) (Table 1). Three pairs of primers of mec, blaZ, and nucA were designed using the PrimerSelect program of the DNASTAR software according to the reported sequences from the Gene Databank (NCBI, NIH, Bethesda, MD, United States). The primers of MLST were available from the MLST website1.
Staphylococcus Enterotoxin Detection
Staphylococcal enterotoxins of the SPF isolates were detected by ELISA using a RIDASCREEN SET A, B, C, D, E assay kit (R-Biopharm, Pfungstadt, Germany). S. aureus isolates were pre-enriched in BHI broth, and the supernatant of 24-h cultures of one colony grown in BHI at 37°C was centrifuged at 6,000 × g for 10 min at 4°C. The supernatant was aseptically filtered by a 2.0-μm Millipore filter, and the filtered sample was added into an ELISA plate. The enterotoxins were tested according to the protocol recommended by the manufacturer’s instructions. The absorbance was measured at 450 nm in the Epoch ELISA reader (BioTek, Winooski, VT, United States).
DNA Extraction
Staphylococcus aureus isolates were grown overnight in BHI broth at 37°C. Genomic DNA was extracted using a bacterial genomic DNA extraction kit (DNeasy Blood and Tissue Kit, Qiagen Inc., Redwood City, CA, United States) according to the manufacturer’s instructions.
Detection of mec and blaZ
All the isolates underwent antimicrobial resistance gene typing. The presence of two antimicrobial resistance genes of blaZ and mec was assessed together with the reference gene of nucA, using a multiplex PCR assay. The three pairs of primers used are listed in Table 1. The PCR mixture contained 4 μl mec-F (2.5 μM), 4 μl mec-R (2.5 μM), 4 μl blaZ-F (2.5 μM), 4 μl blaZ-R (2.5 μM), 4 μl nucA-F (2.5 μM), 4 μl nucA-R (2.5 μM), 2.5 μl genome DNA template, 5 μl 10 × Ex Taq buffer (Mg2+ plus) (20 mM), 4 μl dNTP mixture (2.5 mM each), 0.5 μl TaKaRa Ex Taq DNA polymerase (TaKaRA, China), and 14 μl double-distilled water. The PCR conditions were as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 68°C for 60 s, extension at 72°C for 60 s, and a final extension at 72°C for 10 min. All the PCR products were analyzed by QIAxcel system (Qiagen, Germany). Separation was performed using the AM320 method (100 ng/μl sample was injected at the voltage of 5 kV for 10 s and separated at the voltage of 6 kV for 320 s). The samples with two fragments were considered positive to nucA and mec or blaZ according to the fragment size, and the samples with three fragments were positive to nucA, mec, and blaZ. The mec and blaZ PCR fragments were sequenced using Sanger dideoxy DNA sequencing (Sangon, Shanghai, China), and the sequences were blasted in NCBI BLAST web.
MLST of S. aureus Isolates
Seven housekeeping genes of S. aureus of arcC, aroE, glpF, gmk, pta, tpi, and yqiL were adapted for MLST. These seven housekeeping genes were amplified by PCR according to the original and alternative protocols available from the MLST website (see text footnote 2). The amplicons were sequenced using Sanger dideoxy DNA sequencing (Sangon, China). The sequences were compared with the known alleles in the MLST database to determine the allele number, ST types, and CCs. The isolates that did not match with the previously identified STs were submitted to the database and assigned as new STs. The phylogenetic tree was constructed using BioNumerics 7.6.2 (Applied Maths, Sint-Martens-Latem, Belgium).
Results
Presence of mec and blaZ
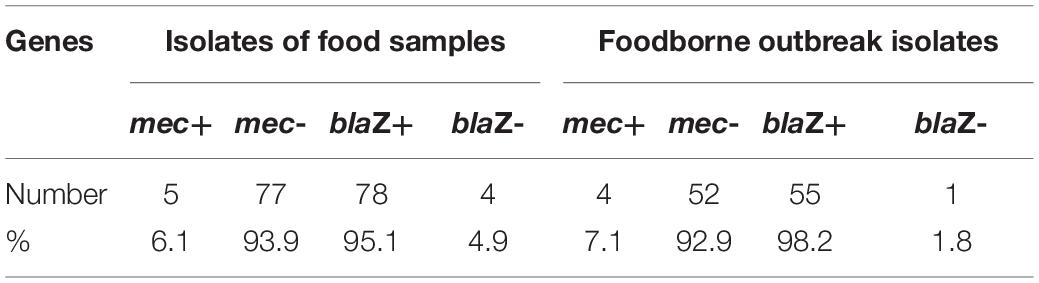
Of the 138 S. aureus isolates, 133 isolates harbored the penicillin-resistant gene of blaZ and nine isolates harbored the methicillin-resistant gene of mec (Table 2). Four blaZ-negative isolates were isolated from food samples, and one blaZ negative isolate was isolated from SFP outbreaks. The nine mec-positive isolates originated from four SFP outbreaks, and five mec-positive isolates originated from food samples (four cooked foods and one hamburger).
STs and CCs of S. aureus Isolates
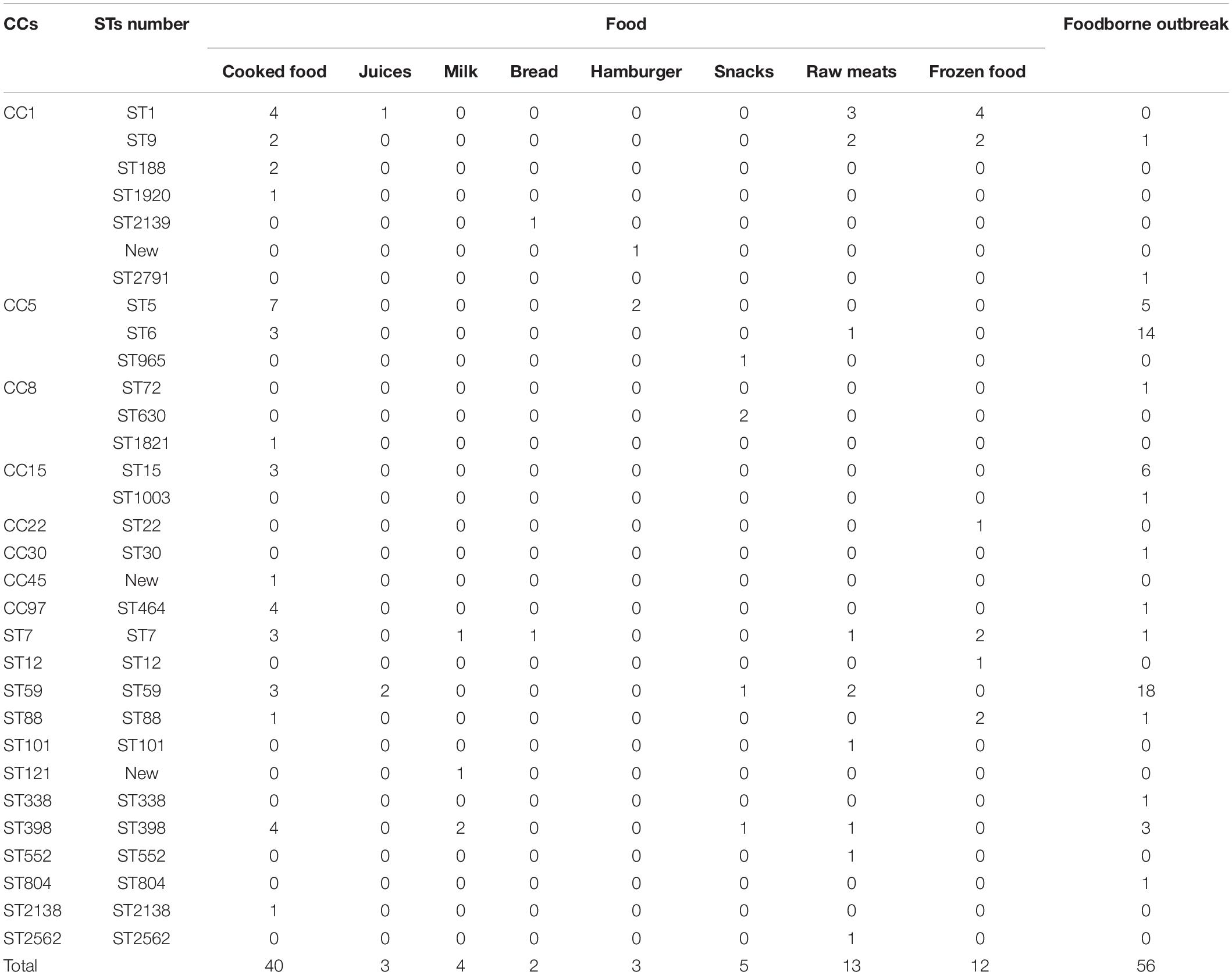
Multilocus sequence typing analysis allowed the distribution of the isolates in CCs, defined as those STs that matched the central genotype, and singletons that differed from all STs in the dataset and did not belong to any CC. The 138 foodborne S. aureus isolates, 82 isolates from food samples, and 56 isolates from food poisoning outbreak were respectively distributed into 31 distinct STs, 25 STs, and 15 STs. The 31 STs were divided into eight CCs by eBURST, and 12 STs were singletons. The minimum spanning network was built from the core genome MLST allelic profiles of 138 S. aureus isolates with CCs and singletons (Figure 1). The CCs of cladogram showed high diversity. The most prevalent foodborne S. aureus isolates were CC1, CC5, CC8, CC15, CC97, CC59, CC398, CC88, and CC7; other clonal complexes were isolated sporadically (Figure 1).
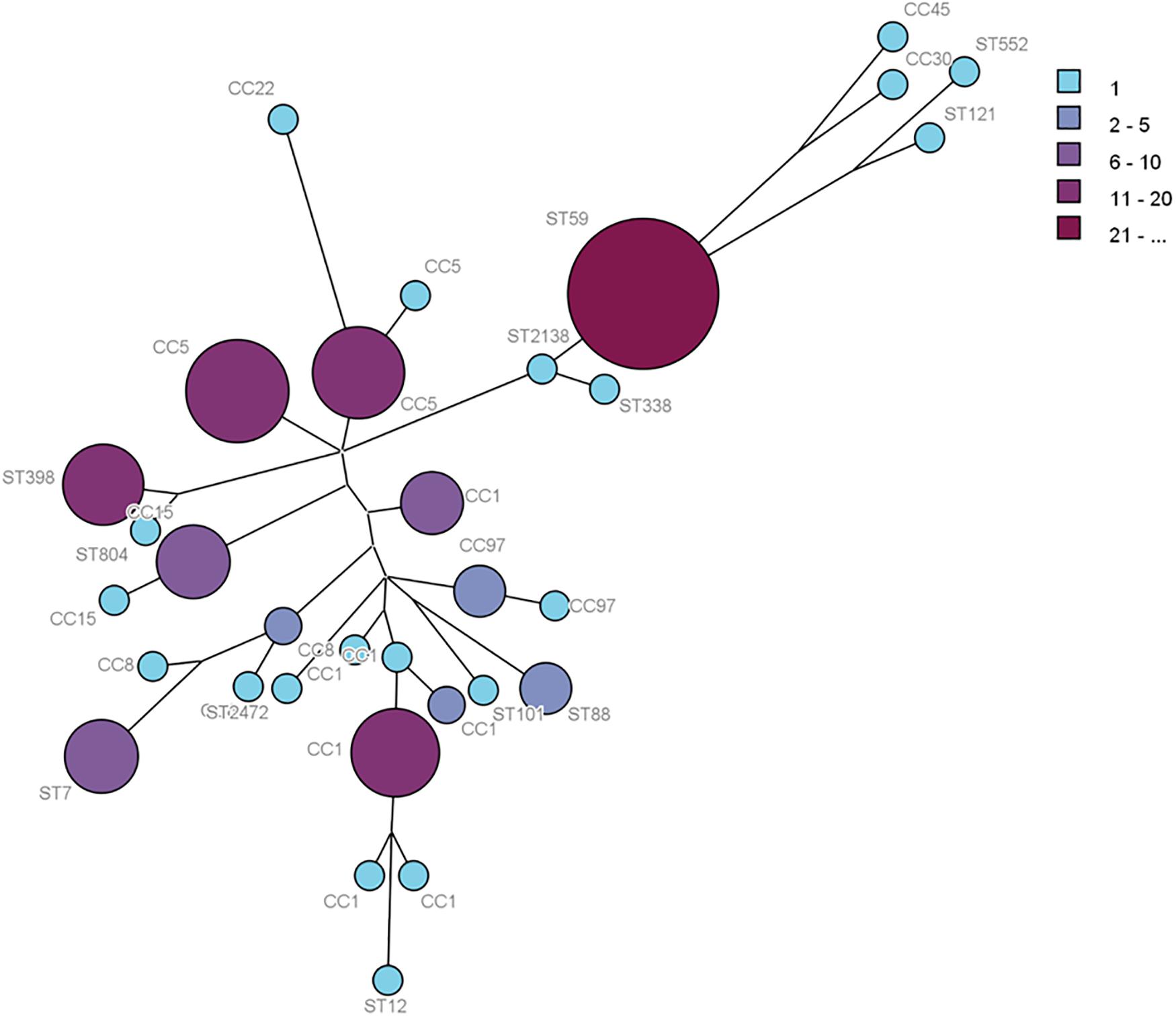
Figure 1. Phylogenetic tree of 138S. aureus isolates based on MLST allelic profiles. Each circle represents CC type or singleton, and the size and color depth of each circle correspond to the number of samples (1, 2–5, 6–10, 11–20, and ≥21).
In SFP isolates, ST59 was the most prevalent type (32.1%, n = 18), followed by ST6 (25.0%, n = 14), ST15 (10.7%, n = 6), ST5 (8.9%, n = 5), and ST398 (5.3%, n = 3). The other 10 STs, including ST9, ST2791, ST72, ST1003, ST30, ST464, ST7, ST88, ST338, and ST804, accounted for 1.8% (n = 1), respectively (Table 3).
Eighty-two S. aureus isolates were identified from eight kinds of food samples (Table 3). ST1 was the most prevalent isolate (14.6%, n = 12), followed by ST5 (11.0%, n = 9), ST59 (9.8%, n = 8), ST398 (9.8%, n = 8), ST7 (9.8%, n = 8), ST9 (7.3%, n = 6), ST6 (4.9%, n = 4), ST464 (4.9%, n = 4), ST15 (3.7%, n = 3), ST88 (3.7%, n = 3), and ST188 (2.4%, n = 2). Other STs were isolated sporadically (Table 3), including ST12, ST22, ST101, ST552, ST2138, ST2139, ST2462, ST1821, ST965, and ST1930 and three new STs. Twenty-five STs were divided into 7 CCs and 10 singletons (Table 3). Twelve ST1 isolates originated from four kinds of food samples (four from cooked food, four from frozen food, three from meat, and one from juice). Nine ST5 isolates were taken from two kinds of food samples (seven from cooked food and two from hamburgers). The distribution of the other STs is shown in Table 3.
Production of Enterotoxins in Isolates of SFP Outbreaks
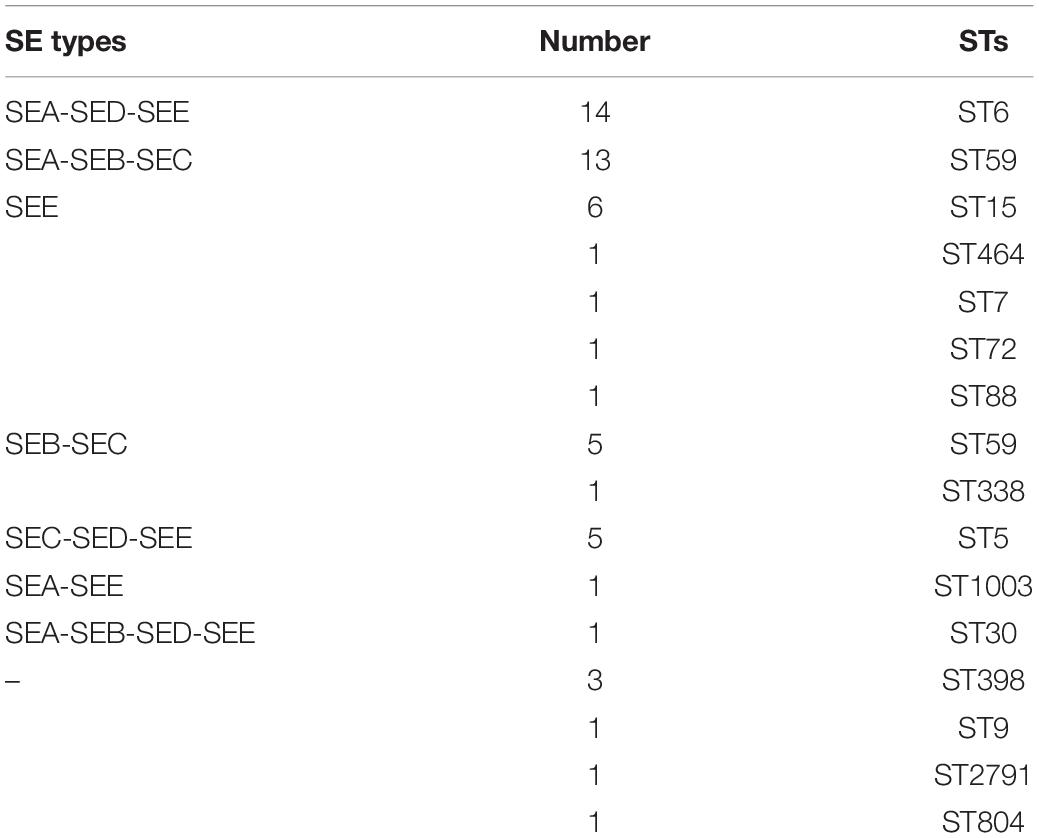
The production of SEA, SEB, SEC, SED, and SEE was evaluated for the 56 S. aureus isolates from food poisoning outbreaks, and eight distinct SE profiles were identified (Table 4). The most prevalent SEA-SED-SEE accounted for 25.0% (14/56), followed by SEA-SEB-SEC 23.2% (13/56), SEE 17.9% (10/56), SEB-SEC 10.7% (6/56), SEC-SED-SEE 8.9% (5/56), SEA-SEE 1.8% (1/56), and SEA-SEB-SED-SEE 1.8% (1/56). SEA, SEB, SEC, SED, and SEE were not detected in six strains.
Discussion
Staphylococcal food poisoning outbreaks are often caused by foodborne S. aureus. The CCs of SFP isolates have their distribution characteristics. In this study, the isolates were assigned to 12 CCs in SFP outbreaks, including CC5, CC59, CC15, CC30, CC8, CC1, CC97, CC38, CC398, CC7, CC804, and CC88. Among them, CC59, CC15, CC5, and CC398 were the predominant CCs. While in Tokyo, Japan, CC8, CC6, CC5, CC508, CC59, CC20, and CC30 were the prevalent CCs of the SFP isolates in the descending order (Suzuki et al., 2014). Some epidemic CCs of SFP isolates were also the prevalent CCs in foods and hospitals. CC1, CC5, CC8, CC15, CC97, CC7, CC59, and CC398 were the most prevalent clonal complexes in food samples in this study. CC5, CC8, CC188, CC59, CC7, and CC398 were the most prevalent CCs in the teaching hospitals of China (Li et al., 2018). The high prevalence of CC59, CC15, CC5, and CC398 was related with the SFP outbreaks. These CCs are common in China and often detected in foods, animals, and humans (Bi et al., 2018; Li et al., 2018; Peng et al., 2018; Wu et al., 2018). The S. aureus strains from humans and animals can contaminate foods; they are also a major cause of food contamination.
The SE produced by S. aureus is the direct cause of SFP. In this study, eight distinct SE types were identified from food poisoning isolates. SEE was the most commonly detected SE in SFP isolates, followed by SEA, SEC, SEB, and SED. SEA, SEB, SEC, SED, or SEE was not detected in several isolates, but other types of SEs could be produced by these strains, so they should be analyzed by more sensitive methods such as SE gene detection. Meanwhile, the assay kit should be improved to detect all types of SEs and SE-like toxins in the future, because many other SEs and SE-like toxins can also cause food poisoning outbreaks. In this study, there were some correlations between STs and SE types in the SFP isolates. The isolates of ST15, ST6, and ST5 were respectively of SEE, SEA-SED-SEE, and SEC-SED-SEE types. However, the ST59 isolates in food poisoning outbreaks were of SEA-SEB-SEC or SEB-SEC types (Table 4). The SE characteristics of the strain might be related with its region and source. So, more literature data is needed to prove the correlations.
The prevalence of foodborne S. aureus would increase the risk of causing human infection and food poisoning. The STs of epidemic S. aureus in foods were respectively ST1, ST5, ST59, ST398, ST7, ST9, ST6, ST464, ST15, and ST88 in descending order. ST1, ST5, ST6, ST7, ST9, ST15, ST59, and ST398 were the common STs of foodborne isolates in China (Wu et al., 2018; Li et al., 2021). They had different characteristics in SFP outbreaks in this study. As the low-risk SFP isolates, ST1, ST7, and ST9 are widely distributed in foods, but ST1 isolates were not involved in any SFP outbreaks, and ST7 and ST9 isolates rarely caused food poisoning. ST6 and ST15 were the highly frequent isolates in SFP outbreaks, whereas ST5, ST59, and ST398 isolates were the highly frequent isolates in SFP outbreaks and food contamination.
ST464 was the main isolate in cooked food, and it caused one SFP outbreak. The other dominant clones were ST5, ST398, ST1, ST15, and ST59. ST1, ST9, and ST59 were the dominant clones from raw meat in this study. With the exception of ST464, these STs were the prevalent isolates in retail meat and meat products of China (Wu et al., 2018). ST398, ST15, and ST59 were also the main STs from sushi samples in Beijing, China (Li et al., 2021). The ST5, ST15, and ST398 isolates were also the prevalent foodborne S. aureus in other countries (Thapaliya et al., 2017; Li et al., 2019). ST1, ST5, ST15, ST59, and ST398 were also included in hospitals, community, and animals (Li et al., 2017, 2018; Bi et al., 2018; Peng et al., 2018; Heaton et al., 2020). The pollution of these isolates was closely related to animal and human activities. The transmissions of these strains also increased the chance of acquiring drug resistance.
Nine MRSA isolates were obtained from foodborne S. aureus isolates. ST15, ST59, and ST338 were the ST types of four MRSA isolates from SFP outbreaks, and ST5, ST15, and ST59 were the ST types of the other five MRSA isolates from food samples. Two MRSA ST59 isolates were detected in sushi samples from Beijing, China (Li et al., 2021). ST5, ST15, and ST59 were also the ST types often detected among iatrogenic isolates. They were the prevalent isolates from healthcare-associated MRSA (HA-MRSA) and community-associated MRSA (CA-MRSA) of China (Li et al., 2018; Peng et al., 2018; Wu et al., 2018; Yuan et al., 2019). The contamination of these STs in food was associated with human activities. The proportion of MRSA isolates in foodborne S. aureus strains was 6.5% in this study. The low contamination rate of foodborne MRSA has been reported in China. MRSA was present in 7.9% of 1,150 S. aureus isolates from retail food and approximately 7.1% of S. aureus isolates from meat and meat products (Wang et al., 2017; Wu et al., 2018). The proportion of MRSA in hospitals and healthy human carriers was higher than that of the foodborne S. aureus in China. The MRSA accounted for 36.5% in teaching hospitals (Li et al., 2018), 44.7% in clinical S. aureus (Yuan et al., 2019), and 14.8% among the healthy adult carriers (Fan et al., 2016). The prevalence of foodborne MRSA is due to the human activity that spreads the bacteria through the food chain. It is possible that the transmissions between humans and foods are bidirectional.
In this study, 96.4% (133/138) of the foodborne S. aureus isolates contained the blaZ gene-encoding penicillin resistance. The resistance of S. aureus to penicillin was prevalent in China, which respectively accounted for 84.6% (735/868) in meat samples (Wu et al., 2018), 83.7% (963/1150) from retail foods (Wang et al., 2017), and 90.7% (98/108) among healthy adult carriers (Fan et al., 2016). The penicillin resistance rate of S. aureus in China was higher than that in other countries. Seventy-three percent (108/148) of the S. aureus strains from food products were resistant to penicillin in Portugal (Pereira et al., 2009) and 56.8% (50/88) in Denmark (Li et al., 2019). The prevalence of penicillin-resistant strains of foodborne S. aureus increases the risk of infection in humans.
Staphylococcus aureus can spread through the food chain. As the most prevalent foodborne ST, ST1 was also included in humans and animals and could cause infections at some time (Chen et al., 2017; Li et al., 2017; Peng et al., 2018; Wu et al., 2018). As the common foodborne STs, ST6 and ST7 were included in the transmission in communities and hospitals (Fan et al., 2016; Li et al., 2018; Peng et al., 2018). ST59 and ST5 were the main pathogenetic isolates and the predominant clonal lineages in community and healthcare in China (Bi et al., 2018; Li et al., 2018; Peng et al., 2018; Yuan et al., 2019). ST59 was the common food-related MRSA in China (Wu et al., 2018). ST398 and ST9 were the main livestock-associated S. aureus and the prevalent isolates to cause bovine mastitis in China (Yan et al., 2014; Li et al., 2017; Bi et al., 2018). ST398 was also common in communities and hospitals (Li et al., 2018; Peng et al., 2018). These epidemic isolates are widely distributed in the environment including humans, animals, and foods. Their transmissions in the environment have epidemiological correlation. It is important for public health to control the transmissions of foodborne S. aureus.
Conclusion
This study provides the epidemic characteristics of foodborne S. aureus isolates in Shijiazhuang, China. ST1 was the predominant isolate in food samples, followed by ST5, ST7, ST59, ST398, ST9, ST6, and ST464. Moreover, ST59 was the dominant ST in food poisoning outbreaks, followed by ST6, ST15, ST5, and ST398. The food poisoning isolates can produce one or more SEs. SEE was the most commonly detected SE in food poisoning isolates. Most of the foodborne S. aureus isolates contained the blaZ gene, and several isolates were MRSA. The prevalence and transmission of foodborne S. aureus will increase the risk of infection. These data could help us understand the contamination and transmission of this bacterium.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
GL and LQ performed the research. GL and RJ analyzed the data and wrote the manuscript. HaZ performed the blaZ and mec testing and analysis. RJ, LW, HoZ, and YP contributed to the MLST analysis. LL, WG, XW, and HD performed the detection of staphylococcus enterotoxins. All authors have approved the manuscript and agreed with submission to your esteemed journal.
Funding
This work was supported by the Science and Technology Program of Hebei Province (No. 17275506D).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank all the participants who took part in this study.
Footnotes
References
Argudín, M. A., Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel) 2, 1751–1773. doi: 10.3390/toxins2071751
Bi, Z., Sun, C., Börjesson, S., Chen, B., Ji, X., Berglund, B., et al. (2018). Identical genotypes of community-associated MRSA (ST59) and livestock-associated MRSA (ST9) in humans and pigs in rural China. Zoonoses Public Health 65, 367–371. doi: 10.1111/zph.12443
Bouiller, K., Gbaguidi-Haore, H., Hocquet, D., Cholley, P., Bertrand, X., and Chirouze, C. (2016). Clonal complex 398 methicillin-susceptible Staphylococcus aureus bloodstream infections are associated with high mortality. Clin. Microbiol. Infect. 22, 451–455. doi: 10.1016/j.cmi.2016.01.018
Chen, B. J., Xie, X. Y., Ni, L. J., Dai, X. L., Lu, Y., Wu, X. Q., et al. (2017). Factors associated with Staphylococcus aureus nasal carriage and molecular characteristics among the general population at a Medical College Campus in Guangzhou, South China. Ann. Clin. Microbiol. Antimicrob. 16:28. doi: 10.1186/s12941-017-0206-0
Chen, H., Liu, Y., Jiang, X., Chen, M., and Wang, H. (2010). Rapid change of methicillin resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob. Agents Chemother. 54, 1842–1847. doi: 10.1128/AAC.01563-09
Cheng, H., Yuan, W., Zeng, F., Hu, Q., Shang, W., Tang, D., et al. (2013). Molecular and phenotypic evidence for the spread of three major methicillin-resistant Staphylococcus aureus clones associated with two characteristic antimicrobial resistance profiles in China. J. Antimicrob. Chemother. 68, 2453–2457. doi: 10.1093/jac/dkt213
Dastmalchi Saei, H., and Panahi, M. (2020). Genotyping and Antimicrobial resistance of Staphylococcus aureus isolates from dairy ruminants: differences in the distribution of clonal types between cattle and small ruminants. Arch. Microbiol. 202, 115–125. doi: 10.1007/s00203-019-01722-z
Fan, Y., Wang, X., Li, L., Yao, Z., Chen, S., and Ye, X. (2016). Potential relationship between phenotypic and molecular characteristics in revealing livestock-associated Staphylococcus aureus in Chinese humans without occupational livestock contact. Front. Microbiol. 7:1517. doi: 10.3389/fmicb.2016.01517
Heaton, C. J., Gerbig, G. R., Sensius, L. D., Patel, V., and Smith, T. C. (2020). Staphylococcus aureus epidemiology in wildlife: a systematic review. Antibiotics (Basel) 9:89. doi: 10.3390/antibiotics9020089
Hennekinne, J. A., De Buyser, M. L., and Dragacci, S. (2012). Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 36, 815–836. doi: 10.1111/j.1574-6976.2011.00311.x
Htun, H. L., Kyaw, W. M., de Sessions, P. F., Low, L., Hibberd, M. L., and Chow, A. (2018). Methicillin-resistant Staphylococcus aureus colonisation: epidemiological and molecular characteristics in an acute-care tertiary hospital in Singapore. Epidemiol. Infect. 146, 1785–1792. doi: 10.1017/S0950268818001966
Ito, T., Hiramatsu, K., Tomasz, A., de Lencastre, H., Perreten, V., Holden, M. T., et al. (2012). Guidelines for reporting novel mecA gene homologues. Antimicrob. Agents Chemother. 56, 4997–4999. doi: 10.1128/AAC.01199-12
Kadariya, J., Smith, T. C., and Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. BioMed. Res. Int. 2014:827965. doi: 10.1155/2014/827965
Kitti, T., Seng, R., Saiprom, N., Thummeepak, R., Chantratita, N., Boonlao, C., et al. (2018). Molecular characteristics of methicillin-resistant staphylococci clinical isolates from a tertiary Hospital in Northern Thailand. Can. J. Infect. Dis. Med. Microbiol. 2018:8457012. doi: 10.1155/2018/8457012
Lakhundi, S., and Zhang, K. (2018). Methicillin-Resistant Staphylococcus aureus: molecular Characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31, e20–e18. doi: 10.1128/CMR.00020-18
Li, H., Andersen, P. S., Stegger, M., Sieber, R. N., Ingmer, H., Staubrand, N., et al. (2019). Antimicrobial resistance and virulence gene profiles of methicillin-resistant and susceptible Staphylococcus aureus from food products in Denmark. Front. Microbiol. 10:2681. doi: 10.3389/fmicb.2019.02681
Li, H., Tang, T., Stegger, M., Dalsgaard, A., Liu, T., and Leisner, J. J. (2021). Characterization of antimicrobial-resistant Staphylococcus aureus from retail foods in Beijing, China. Food Microbiol. 93:103603. doi: 10.1016/j.fm.2020.103603
Li, S., Sun, S., Yang, C., Chen, H., Yin, Y., Li, H., et al. (2018). The changing pattern of population structure of Staphylococcus aureus from bacteremia in China from 2013 to 2016: ST239-030-MRSA replaced by ST59-t437. Front. Microbiol. 9:332. doi: 10.3389/fmicb.2018.00332
Li, T., Lu, H., Wang, X., Gao, Q., Dai, Y., Shang, J., et al. (2017). Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front. Cell Infect. Microbiol. 7:127. doi: 10.3389/fcimb.2017.00127
Lv, G., Xu, B., Wei, P., Song, J., Zhang, H., Zhao, C., et al. (2014). Molecular characterization of foodborne-associated Staphylococcus aureus strains isolated in Shijiazhuang, China, from 2010 to 2012. Diagn Microbiol. Infect. Dis. 78, 462–468. doi: 10.1016/j.diagmicrobio.2013.12.006
Oliveira, D., Borges, A., and Simões, M. (2018). Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins(Basel) 10:252. doi: 10.3390/toxins10060252
Ono, H. K., Omoe, K., Imanishi, K., Iwakabe, Y., Hu, D. L., Kato, H., et al. (2008). Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 76, 4999–5005. doi: 10.1128/IAI.00045-08
Peng, H., Liu, D., Ma, Y., and Gao, W. (2018). Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus isolates at a Chinese tertiary hospital, 2012-2017. Sci. Rep. 8:17916. doi: 10.1038/s41598-018-36206-5
Pereira, V., Lopes, C., Castro, A., Silva, J., Gibbs, P., and Teixeira, P. (2009). Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 26, 278–282. doi: 10.1016/j.fm.2008.12.008
Rao, Q., Shang, W., Rao, X., and Rao, X. (2015). Staphylococcus aureus ST121: a globally disseminated hypervirulent clone. J. Med. Microbiol. 64, 1462–1473. doi: 10.1099/jmm.0.000185
Suzuki, Y., Omoe, K., Hu, D. L., Sato’o, Y., Ono, H. K., Monma, C., et al. (2014). Molecular epidemiological characterization of Staphylococcus aureus isolates originating from food poisoning outbreaks that occurred in Tokyo, Japan. Microbiol. Immunol. 58, 570–580. doi: 10.1111/1348-0421.12188
Thapaliya, D., Forshey, B. M., Kadariya, J., Quick, M. K., Farina, S., O’ Brien, A., et al. (2017). Prevalence and molecular characterization of Staphylococcus aureus in commercially available meat over a one-year period in Iowa, USA. Food Microbiol. 65, 122–129. doi: 10.1016/j.fm.2017.01.015
Wang, W., Baloch, Z., Jiang, T., Zhang, C., Peng, Z., Li, F., et al. (2017). Enterotoxigenicity and antimicrobial resistance of Staphylococcus aureus isolated from retail food in China. Front. Microbiol. 8:2256. doi: 10.3389/fmicb.2017.02256
Wu, S., Duan, N., Gu, H., Hao, L., Ye, H., and Gong, W. (2016). A review of the methods for detection of Staphylococcus aureus enterotoxins. Toxin(Basel) 8:176. doi: 10.3390/toxins8070176
Wu, S., Huang, J., Wu, Q., Zhang, J., Zhang, F., Yang, X., et al. (2018). Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front. Microbiol. 9:2767. doi: 10.3389/fmicb.2018.02767
Yan, X., Yu, X., Tao, X., Zhang, J., Zhang, B., Dong, R., et al. (2014). Staphylococcus aureus ST398 from slaughter pigs in northeast China. Int. J. Med. Microbiol. 304, 379–383. doi: 10.1016/j.ijmm.2013.12.003
Keywords: Staphylococcus aureus, food poisoning, MLST, antibiotics resistance, staphylococcal enterotoxin
Citation: Lv G, Jiang R, Zhang H, Wang L, Li L, Gao W, Zhang H, Pei Y, Wei X, Dong H and Qin L (2021) Molecular Characteristics of Staphylococcus aureus From Food Samples and Food Poisoning Outbreaks in Shijiazhuang, China. Front. Microbiol. 12:652276. doi: 10.3389/fmicb.2021.652276
Received: 12 January 2021; Accepted: 11 May 2021;
Published: 22 June 2021.
Edited by:
Dan Cristian Vodnar, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Daniela Bianchi, Istituto Zooprofilattico Sperimentale del Piemonte, Liguria e Valle d’Aosta, ItalyCalinoiu Florina Lavinia, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, Romania
Copyright © 2021 Lv, Jiang, Zhang, Wang, Li, Gao, Zhang, Pei, Wei, Dong and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoping Lv, bHZndW9waW5nMTE2QDE2My5jb20=; Liyun Qin, cWlubGl5dW4tMjAwN0AxNjMuY29t
†These authors have contributed equally to this work
 Guoping Lv
Guoping Lv Ruiping Jiang2†
Ruiping Jiang2† Hongyan Dong
Hongyan Dong