
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Microbiol. , 28 May 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.650307
This article is part of the Research Topic Eukaryotic Soilborne Plant Pathogens: Interactions With Their Hosts and Biocontrol View all 6 articles
Clubroot is a devastating disease infecting roots of plants of the Brassicaceae family and responsible for 10–15% yield reduction on a global scale (Botero et al., 2019). The clubroot pathogen, Plasmodiophora brassicae, is an obligate intracellular biotroph that belongs to the class Phytomyxea within the eukaryote supergroup Rhizaria, one of the least studied groups of eukaryotes (Burki et al., 2010). The life cycle of P. brassicae starts in the soil, where resting spores, which can remain viable for up to 20 years, germinate in response to the presence of plant hosts and initiate primary infection. During secondary infection, susceptible plants develop galls that disrupt water and nutrient uptake, leading to wilting, stunting, and, in some instances, death of the infected plant (Dixon, 2006, 2009). The mechanisms by which the clubroot pathogen can escape plant immunity in the susceptible host and induce the root galls characteristics of the disease is still unknown (Pérez-López et al., 2018). Based on what we know about biotrophic pathogens from other evolutionary groups, effector proteins might play a key role in mediating P. brassicae pathogenicity (Jones and Dangl, 2006). Prediction of effectors based on conserved motifs has been very challenging for this pathogen, mainly because of unique features inherent to P. brassicae, including its inability to grow in axenic cultures (Chen et al., 2019; Pérez-López et al., 2020). The first P. brassicae effector characterized was Pro1, a serine protease that may play a key role in stimulating germination of P. brassicae resting spores (Feng et al., 2010). The methyltransferase PbBSMT was the second P. brassicae effector characterized (Ludwig-Müller et al., 2015), an effector that allows the pathogen to mimic regulation of salicylic acid within the susceptible host (Bulman et al., 2018; Djavaheri et al., 2019). The third and last P. brassicae effector characterized was the cysteine protease inhibitor SSPbP53, which plays a key role in protecting effectors from host apoplastic enzymes during secondary infection (Pérez-López et al., 2020, 2021). While some P. brassicae ubiquitin ligases have been functionally characterized, their role in pathogenicity has not been elucidated (Yu et al., 2019). This scarcity of information about P. brassicae effectors stresses the need to find other ways to assess more effector candidates to better understand the etiology of the clubroot pathogen. Luckily for us, the researchers aiming a better understanding of the clubroot pathogen, thale cress (Arabidopsis thaliana), the model plant in molecular biology, is susceptible to most of the P. brassicae pathotypes and its use can facilitate our search for new methodologies and the interpretation of the results.
Several strategies have been followed to study the effector repertoire of unculturable plant pathogens. Two of them are based on the heterologous expression of the effector candidates: (i) in the host plant or the model plant Arabidopsis thaliana, and (ii) by a related or unrelated cultured microorganism to evaluate the role of the effector in pathogenicity. The first strategy has allowed the characterization of several phytoplasma effectors such as SAP54, an effector that generates a short circuit between two key pathways of the host to alter plant development and to promote insect feeding of the infected plant to increase the propagation of the pathogen (MacLean et al., 2014; Tomkins et al., 2018). This strategy, although successful, is long and laborious and, for those effectors that do not produce a visible phenotype in the transgenic plant, as with SAP54, further analyses like RNAseq should be performed (dos Santos et al., 2020).
The use of a surrogate culturable microorganism represents an interesting approach to facilitate the understanding of the clubroot pathogen. It has been exploited (1) to characterize the flavescence doree phytoplasma variable membrane protein VmpA, using Spiroplasma citri (Renaudin et al., 2015) as surrogate, (2) to characterize a “Candidatus Liberibacter asiaticus” cysteine protease inhibitor, using Pseudomonas syringae pv. tomato (Clark et al., 2018), and (3) to characterize a “Candidatus Liberibacter asiaticus” glyoxalase using the culturable Liberibacter crescent (Jain et al., 2017), among other examples. In the previous studies, a culturable bacterium was used as a surrogate of an unculturable one, but evolutionary distant microorganisms have also been used. For instance, Pseudomonas syringae pv. tomato (PstDC3000) served as surrogate for the unculturable oomycete Hyaloperonospora arabidopsis (Hpa) (Fabro et al., 2011; Goritschnig et al., 2012), a breakthrough that allowed, for the first time, the identification and characterization of Hpa effectors suppressing plant immunity (Fabro et al., 2011) and eliciting effector-triggered immunity (ETI) (Goritschnig et al., 2012).
The use of a culturable surrogate to characterize P. brassicae proteins has been previously attempted. Recently, Magnaporthe oryzae was the used to identify the role of the cyclophilin PbCYP3 on pathogenicity (Singh et al., 2018). Although this is a sensible approach, a negative point of using M. oryzae as a P. brassicae surrogate is that it is a rice pathogen with a high degree of host specificity (Couch et al., 2005). This makes it unreliable to characterize P. brassicae proteins with a suspected target in plants.
Pseudomonas spp. has been exploited as a model organism allowing conceptual advances in understanding many key aspects of plant-microbe interactions. Recent technical advances such as the development of an image-based system to quantify plant immunity (Laflamme et al., 2016) and a high-throughput seedling screening of plant immunity (Martel et al., 2020), both using the Arabidopsis-Pseudomonas pathosystem, could facilitate the use of P. syringae as a surrogate to understand P. brassicae. Like P. brassicae, P. syringae is a biotrophic plant pathogen and can infect Arabidopsis, two more elements reinforcing Pseudomonas spp. as a good surrogate for the clubroot pathogen. Another milestone recently achieved in the study of P. syringae as a model organism was the identification of the P. syringae pan-genome effectors triggering ETI in Arabidopsis, expanding the number and protein families previously known to elicit ETI (Laflamme et al., 2020). Among those protein families able to elicit ETI during Arabidopsis-Pseudomonas interaction, proteases, lipases, carbohydrate esterases, ADP-ribosyltransferases, and GTPase-activating proteins were identified (Laflamme et al., 2020). Members of all those protein families are found in the P. brassicae genome, without transmembrane domains and functional signal peptide (Pérez-López et al., 2020), supporting the untested theory that resistance induced by ETI elicited by P. brassicae effectors and mediated by NLR receptors, is the base of the resistance observed for some canola cultivars (Hejna et al., 2019). Using Pseudomonas spp. as a surrogate would allow to deliver P. brassicae effectors individually and use them as probes to identify resistance proteins mediating ETI.
The use of a prokaryote as surrogate for an eukaryotic pathogen has however several disadvantages. These include incorrect protein folding, incorrect post-translational modifications, and the simple fact that some strategies used by eukaryotic plant pathogens are not used by prokaryotes (van den Burg et al., 2004; Macek et al., 2019). How can this problem be solved? A simple solution would be to explore another culturable surrogate. In a recent study, our group identified two effectors, PbChiB2 and PbChiB4, able to bind to chitin oligomers in vitro, and suppress chitin-triggered immunity (Muirhead and Pérez-López, 2020). These effectors were able to bind chitin through a CBM18 domain as previously described for the soil-borne pathogen Verticillium dahliae (Volk et al., 2020). Like P. brassicae, V. dahliae can affect Arabidopsis, with an hemibiotrophic lifestyle. Besides, V. dahliae can be transformed to express clubroot pathogen effectors (Gao et al., 2019). A common advantage with the use of P. syringae and V. dahliae is that the typical symptomatology induced by both plant pathogens is detected in the leaves, allowing the use of visual assessment to study the effect of P. brassicae candidate effectors on pathogenicity.
The complexity and uniqueness of the Brassicas-P. brassicae pathosystem do not allow us to select only one cultured surrogate for effectome studies. Is that complexity the reason why there is so little known about the clubroot pathogen biology and the mechanisms used to infect the plant and to suppress and/or escape plant immunity. For this reason, we believe that different approaches, including the use of versatile surrogates such as P. syringae and V. dahliae, can provide a multi-prong solution to further the characterization of P. brassicae effectors and their targets in plants (Figure 1), although questions like how the effectors are secreted to the plant cell are still to be answered. Through the application of such novel approaches, we can hope to make advancements toward the development of resistant germplasm that can mediate losses to this important pathogen.
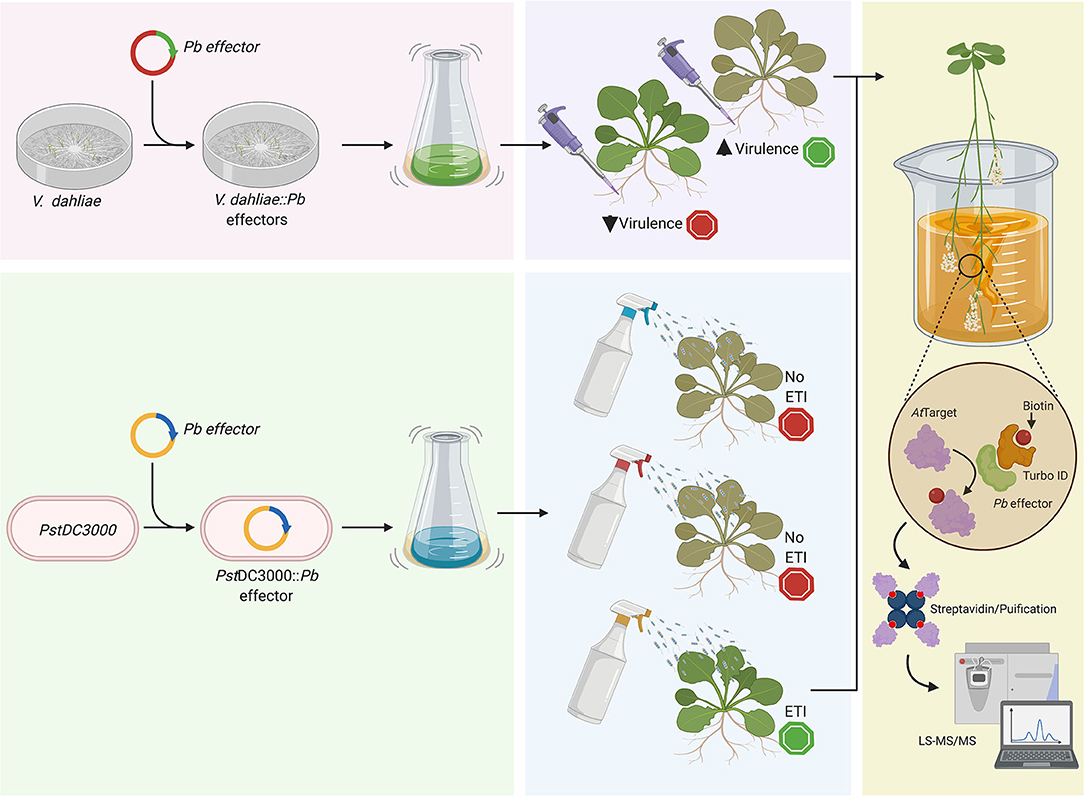
Figure 1. Schematic representation of the use of a cultured surrogate plant pathogen to study and characterized P. brassicae candidate effectors. Here we proposed the use of (i) V. dahliae to, for example, complement deleterious mutations like the case discussed in the main text about the chitin-binding effectors, and (ii) P. syringae pv. tomato (PstDC3000), which can be used to screen effectors eliciting effector-triggered immunity. After the initial step selecting candidate effectors with a role on pathogenicity through the assessment of foliar symptomatology, we could identify the target in the plant host using a protein-protein approach like BioID as previously described Khan et al. (2018). Created with BioRender.com.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by the Department of Phytologie, Université Laval. We also thank the Natural Sciences and Engineering Research Council of Canada for the support through the Discovery Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Richard Bélanger and Tim Dumonceaux for the critical review.
Botero, A., García, C., Gossen, B. D., Strelkov, S. E., Todd, C. D., Bonham-Smith, P. C., et al. (2019). Clubroot disease in Latin America: distribution and management strategies. Plant Pathol. 68, 827–833. doi: 10.1111/ppa.13013
Bulman, S., Richter, F., Marschollek, S., Benade, F., Jülke, S., and Ludwig-Müller, J. (2018). Arabidopsis thaliana expressing PbBSMT, a gene encoding a SABATH-type methyltransferase from the plant pathogenic protist Plasmodiophora brassicae, show leaf chlorosis and altered host susceptibility. Plant Biol. 21, 120–130. doi: 10.1111/plb.12728
Burki, F., Kudryavtsev, A., Matz, M. V., Aglyamova, G. V., Bulman, S., Fiers, M., et al. (2010). Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol. Biol. 10:377. doi: 10.1186/1471-2148-10-377
Chen, W., Li, Y., Yan, R., Xu, L., Ren, L., Liu, F., et al. (2019). Identification and characterization of Plasmodiophora brassicae primary infection effector candidates that suppress or induce cell death in host and nonhost plants. Phytopathol. 109, 1689–1697. doi: 10.1094/PHYTO-02-19-0039-R
Clark, K., Franco, J. Y., Schwizer, S., Pang, Z., Hawara, E., Liebrand, T. W. H., et al. (2018). An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Comm. 9:1718. doi: 10.1038/s41467-018-04140-9
Couch, B. C., Fudal, I., Lebrun, M.-H., Tharreau, D., Valent, B., van Kim, P., et al. (2005). Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 170, 613–630. doi: 10.1534/genetics.105.041780
Dixon, G. R. (2006). The biology of Plasmodiophora brassicae Wor.-a review of recent advances. Acta Hort. 706, 271–282. doi: 10.17660/ActaHortic.2006.706.32
Dixon, G. R. (2009). The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Reg. 28, 194–202. doi: 10.1007/s00344-009-9090-y
Djavaheri, M., Ma, L., Klessig, D. F., Mithöfer, A., Gropp, G., and Borhan, H. (2019). Mimicking the host regulation of salicylic acid: a virulence strategy by the clubroot pathogen Plasmodiophora brassicae. Mol. Plant Microbe Interact. 32, 296–305. doi: 10.1094/MPMI-07-18-0192-R
dos Santos, K. C. G., Pelletier, G., Séguin, A., Guillemette, F., Hawkes, J., Desgagné-Penix, I., et al. (2020). Differential alteration of plant functions by homologous fungal candidate effectors. bioRxiv [Preprint]. doi: 10.1101/2020.10.30.363010
Fabro, G., Steinbrenner, J., Coates, M., Ishaque, N., Baxter, L., Studholme, D. J., et al. (2011). Multiple candidate effectors from the oomycete pathogen Hyaloperonospora Arabidopsis suppress host plant immunity. PLoS Pathog. 7:e1002348. doi: 10.1371/journal.ppat.1002348
Feng, J. I. E., Hwang, R. U., Hwang, S. F., Strelkov, S. E., Gossen, B. D., Zhou, Q. X., et al. (2010). Molecular characterization of a serine protease Pro1 from Plasmodiophora brassicae that stimulates resting spore germination. Mol. Plant Pathol. 11, 503–512. doi: 10.1111/j.1364-3703.2010.00623.x
Gao, F., Zhang, B.-S., Zhao, Z.-H., Huang, J.-F., Jia, P.-S., Wang, S., et al. (2019). Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 5, 1167–1176. doi: 10.1038/s41477-019-0527-4
Goritschnig, S., Krasileva, K. V., Dahlbeck, D., and Staskawicz, B. J. (2012). Computational prediction and molecular characterization of an oomycete effector and the cognate Arabidopsis resistance gene. PLoS Pathog. 8:e1002502. doi: 10.1371/journal.pgen.1002502
Hejna, O., Havlickova, L., He, Z., Bancroft, I., and Curn, V. (2019). Analysing the genetic architecture of clubroot resistance variation in Brassica napus by associative transcriptomics. Mol. Breed. 39:112. doi: 10.1007/s11032-019-1021-4
Jain, K., Munoz-Bodnar, A., and Gabriel, D. W. (2017). Concomitant loss of the glyoxalase system and glycolysis makes the uncultured pathogen “Candidatus Liberibacter Asiaticus” an energy scavenger. Appl. Environ. Microbiol. 83:e01670-17. doi: 10.1128/AEM.01670-17
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Khan, M., Youn, J.-Y., Gingras, A.-C., Subramaniam, R., and Desveaux, D. (2018). In planta proximity dependent biotin identification (BioID). Sci. Rep. 8:9212. doi: 10.1038/s41598-018-27500-3
Laflamme, B., Dillon, M. M., Martel, A., Almeida, R. N. D., Desveaux, D., and Guttman, D. S. (2020). The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 376, 763–768. doi: 10.1126/science.aax4079
Laflamme, B., Middleton, M., Lo, T., Desveaux, D., and Guttman, D. S. (2016). Image-based quantification of plant immunity and disease. Mol. Plant Microbe Interact. 29, 919–924. doi: 10.1094/MPMI-07-16-0129-TA
Ludwig-Müller, J., Jülken, S., Geib, K., Richter, F., Mithöfer, A., Šola, I., et al. (2015). A novel methyltransferase from the intracellular pathogen Plasmodiophora brassicae methylates salicylic acid. Mol. Plant Pathol. 16, 349–364. doi: 10.1111/mpp.12185
Macek, B., Forchhammer, K., Hardouin, J., Weber-Ban, E., Grangeasse, C., and Mijakovic, I. (2019). Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 17, 651–664. doi: 10.1038/s41579-019-0243-0
MacLean, A. M., Orlovskis, Z., Kowitwanich, K., Zdziarska, A. M., Angenent, G. C., et al. (2014). Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 12:e1001835. doi: 10.1371/journal.pbio.1001835
Martel, A., Lo, T., Desveaux, D., and Guttman, D. S. (2020). A high-throughput, seedling screening for plant immunity. Mol. Plant Microbe Interact. 33, 394–401. doi: 10.1094/MPMI-10-19-0295-TA
Muirhead, K., and Pérez-López, E. (2020). Plasmodiophora brassicae chitin-binding effectors guard and mask spores during infection. bioRxiv [Preprint]. doi: 10.1101/2020.12.23.423615
Pérez-López, E., Hossain, M. M., Tu, J., Waldner, M., Todd, C. D., Kusalik, A. J., et al. (2020). Transcriptome analysis identifies Plasmodiophora brassicae secondary infection effector candidates. J. Eukaryot. Microbiol. 67:337–351. doi: 10.1111/jeu.12784
Pérez-López, E., Hossain, M. M., Wei, Y., Todd, C. D., and Bonham-Smith, P. C. (2021). A clubroot pathogen effector targets cruciferous cysteine proteases to suppress plant immunity. Mol. Plant Microbe Interact. [Epub ahead of print].
Pérez-López, E., Waldner, M., Hossein, M., Kusalik, A. J., Wei, Y., Bonham-Smith, P. C., et al. (2018). Identification of Plasmodiophora brassicae effectors – a challenging goal. Virulence 9, 1344–1353. doi: 10.1080/21505594.2018.1504560
Renaudin, J., Beven, L., Batailler, B., Duret, S., Desque, D., Arricau-Bouvery, N., et al. (2015). Heterologous expression and processing of the flavescence doree phytoplasma variable membrane protein VmpA in Spiroplasma citri. BMC Microbiol. 15:82. doi: 10.1186/s12866-015-0417-5
Singh, K., Tzelepis, G., Zouhar, M., Ryšánek, P., and Dixelius, C. (2018). The immunophilin repertoire of Plasmodiophora brassicae and functional analysis of PbCYP3 cyclophilin. Mol. Genet. Genom. 293, 381–390. doi: 10.1007/s00438-017-1395-0
Tomkins, M., Kliot, A., Marée, A. F. M., and Hogenhout, S. A. (2018). A multi-layered mechanistic modelling approach to understand how effector genes extend beyond phytoplasma to modulate plant hosts, insect vectors and the environment. Curr. Opin. Plants Biol. 44, 39–48. doi: 10.1016/j.pbi.2018.02.002
van den Burg, H. A., Spronk, C. A. E. M., Boeren, S., Kennedy, M. A., Vissers, J. P. C., Vuister, G., et al. (2004). Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein–protein interactions: the chitin-binding site of AVR4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin-binding domain. J. Biol. Chem. 279, 16786–16796. doi: 10.1074/jbc.M312594200
Volk, H., Marton, K., Flajšman, M., Radišek, S., Tian, H., Hein, I., et al. (2020). Chitin-binding protein of Verticillium nonalfalfae disguises fungus from plant chitinases and suppresses chitin-triggered host immunity. Mol. Plant Microbe Interact. 32, 1378–1390. doi: 10.1094/MPMI-03-19-0079-R
Yu, F., Wang, S., Zhang, W., Tang, J., Wang, H., Yu, L., et al. (2019). Genome-wide identification of genes encoding putative secreted E3 ubiquitin ligases and functional characterization of PbRING1 in the biotrophic protist Plasmodiophora brassicae. Curr. Genet. 65, 1355–1365. doi: 10.1007/s00294-019-00989-5
Keywords: clubroot, Plasmodiophora brassicae, Brassicas, surrogate, unculturable plant pathogen, effector biology, MPMI
Citation: González-García M and Pérez-López E (2021) Looking for a Cultured Surrogate for Effectome Studies of the Clubroot Pathogen. Front. Microbiol. 12:650307. doi: 10.3389/fmicb.2021.650307
Received: 06 January 2021; Accepted: 05 May 2021;
Published: 28 May 2021.
Edited by:
Georgios Tzelepis, Swedish University of Agricultural Sciences, SwedenReviewed by:
Jutta Ludwig-Müller, Technische Universität Dresden, GermanyCopyright © 2021 González-García and Pérez-López. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edel Pérez-López, ZWRlbC5wZXJlei1sb3Blei4xQHVsYXZhbC5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.