
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 June 2021
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.645353
This article is part of the Research Topic Genomic and Evolutionary Analysis of Microsporidian Parasites View all 7 articles
 Qiang Huang1,2*†
Qiang Huang1,2*† Zhi Hao Wu1,2†
Zhi Hao Wu1,2† Wen Feng Li3
Wen Feng Li3 Rui Guo4
Rui Guo4 Jin Shan Xu5
Jin Shan Xu5 Xiao Qun Dang5
Xiao Qun Dang5 Zheng Gang Ma5
Zheng Gang Ma5 Yan Ping Chen6
Yan Ping Chen6 Jay D. Evans6
Jay D. Evans6Microsporidia comprise a phylum of single cell, intracellular parasites and represent the earliest diverging branch in the fungal kingdom. The microsporidian parasite Nosema ceranae primarily infects honey bee gut epithelial cells, leading to impaired memory, suppressed host immune responses and colony collapse under certain circumstances. As the genome of N. ceranae is challenging to assembly due to very high genetic diversity and repetitive region, the genome was re-sequenced using long reads. We present a robust 8.8 Mbp genome assembly of 2,280 protein coding genes, including a high number of genes involved in transporting nutrients and energy, as well as drug resistance when compared with sister species Nosema apis. We also describe the loss of the critical protein Dicer in approximately half of the microsporidian species, giving new insights into the availability of RNA interference pathway in this group. Our results provided new insights into the pathogenesis of N. ceranae and a blueprint for treatment strategies that target this parasite without harming honey bees. The unique infectious apparatus polar filament and transportation pathway members can help to identify treatments to control this parasite.
As the earliest branch from the fungal kingdom, microsporidia comprise a large and widespread group of obligate intracellular animal parasites (Keeling and Fast, 2002; Williams, 2009; Capella-Gutiérrez et al., 2012). In humans, microsporidia are opportunistic parasites that infect immuno-compromised patients (Didier, 2005). Microsporidia also showed substantial damage to the silkworm and fisheries industries and are a driving factor for honey bee colony losses which seriously threaten the agricultural economy and global food security (Higes et al., 2008; Aizen et al., 2009; Freeman and Sommerville, 2011; Stentiford et al., 2016; Santhoshkumar et al., 2017; Meng et al., 2018). In response to their intracellular parasitic life cycle, microsporidia have undergone massive reductions in gene content, including decayed glycolytic pathways, leading to extremely compact eukaryotic genomes (Pelin et al., 2015, 2016; Ndikumana et al., 2017; Wiredu Boakye et al., 2017). Strikingly, mitochondrial genes were lost completely, leaving only a mitochondrion-related organelle called the mitosome (Burri et al., 2006). As a result, energy and resources needed for the proliferation of the parasite are acquired directly from the host, causing energetic stress (Mayack and Naug, 2009; Martin-Hernandez et al., 2011).
Nosema ceranae is a microsporidian parasite which infects honey bee mid-gut epithelial cells (Fries et al., 1996). As with all microsporidian parasites, N. ceranae produce infectious spores. The spore wall, which is comprised of an electron-dense proteinaceous exospore and an electron-transparent endospore protects the parasite from environmental stressors, allowing spores to remain infective for long term (Li et al., 2003; Fayer, 2004). N. ceranae infection starts from the ingestion of spores contaminated nectar and transmitted through oral-fecal and oral-oral routes. The spores germinate in the gut lumen and extrude a polar tube which ejects the sporoplasm into the host cytoplasm (Klee et al., 2007). This leads to parasite proliferation in the subsequent 4 days, resulting in a huge number of offspring spores. The life cycle destroys the gut membrane matrix and epithelial cell integrity (Higes et al., 2006, 2007). Infected honey bees showed suppressed immune responses, impaired memory, and energetic stress (Antunez et al., 2009; Mayack and Naug, 2009; Higes et al., 2013; Gage et al., 2018).
This paper aims to improve the N. ceranae genome assembly using long-read sequencing technology, reducing redundancies and improving the integrity of the assembled genome. We also more fully analyzed the transporters, spore wall, and polar tube proteins, essential proteins that fuel proliferation and determine the success of infections. These analyses improve our understanding of parasite evolution but also provide targets to treat an important bee disease.
N. ceranae spores were collected from the midgut tissues of heavily infected honey bee colonies. As the genetic diversity of N. ceranae is higher within a colony than among colonies, the impacts of multi-colony spores on the quality of the assembled genome was minor (Gómez-Moracho et al., 2014, 2015). The spores were purified using a Percoll gradient procedure and genomic DNA was extracted using the CTAB protocol (Chen Y. et al., 2013). The species status of N. ceranae was confirmed by species-specific PCR (Fries et al., 2013). A library was prepared and sequenced following the Oxford Nanopore protocol using MinION cell. Long reads were self-corrected and assembled using Mecat (version 1.0) with default parameters (Xiao et al., 2017). Redundant contigs were collapsed using redundans (version 0.13c) with default parameters (Pryszcz and Gabaldón, 2016). The assembly was aligned against the NCBI bacteria database and honey bee genome by BLASTN to remove contamination. The long reads were re-aligned to the assembled genome to determine structural variations (SVs) using the Ngmlr and Sniffles pipline (Sedlazeck et al., 2018). The assembled genome has been deposited in GenBank with assembly accession number GCA_004919615.1. The raw reads were deposited to NCBI BioProject PRJNA514060.
Previously, we quantified N. ceranae gene expression profiles at various proliferation stages with RNA-seq (Huang et al., 2019). To improve the gene annotation, we re-mapped those reads to the current N. ceranae genome assembly and retrieved the aligned reads using Hisat2 with default parameters (Kim et al., 2013). Both the assembled contigs (Supplementary File 1) and RNA-seq reads were imported into GenSAS, a free online gene features annotation pipeline (Humann et al., 2019). Briefly, the genomes were first masked using RepeatMasker and RepeatMolder (Smit et al., 2015a,b). Next, the genes were predicted using GeneMarkES and Augustus (Lomsadze et al., 2005; Stanke et al., 2008). After that, RNA-seq reads were assembled to longer transcripts using Spades, and these reads were used to polish the annotated gene features using PASA (Haas et al., 2008; Bankevich et al., 2012). In order to infer biological function, the predicted protein sequences were aligned by BLASTN to the Pfam, Uniprot, and NCBI non-redundant databases. The completeness of the assembly was gauged using BUSCO (version 4) against the microsporidia_odb10 dataset (Simao et al., 2015; Seppey et al., 2019).
The protein sequences of other 19 microsporidian parasites (Encephalitozoon romaleae, Encephalitozoon hellem, Encephalitozoon intestinalis, Encephalitozoon cuniculi, Ordospora colligata, Nosema apis, Nosema bombycis, Enterocytozoon bieneusi, Enterospora canceri, Enterocytozoon hepatopenae, Vittaforma corneae, Trachipleistophora hominis, Vavraia culicis, Pseudoloma neurophilia, Edhazardia aedis, Anncaliia algerae, and Nematocida parisii) with assembled genomes were retrieved from NCBI and MicrosporidianDB1,2 (Katinka et al., 2001; Corradi et al., 2007, 2009, 2010; Cuomo et al., 2012; Heinz et al., 2012; Pombert et al., 2012, 2013, 2015; Campbell et al., 2013; Chen Y. et al., 2013; Pan et al., 2013; Haag et al., 2014; Desjardins et al., 2015; Ndikumana et al., 2017; Reinke et al., 2017; Wiredu Boakye et al., 2017). These protein sequences were all used to query the BUSCO gene set microsporidian_odb 10 (Simao et al., 2015; Seppey et al., 2019). The 48 shared BUSCO groups among all 20 microsporidian species were aligned using Muscle with default parameters (Edgar, 2004). Resulting alignments were trimmed with trimAI (-w 3 -gt 0.95 -st 0.01) and then concatenated for phylogenetic analyses with Mrbayes (nchain = 4, aamodelpr = mixed, ngen = 1,000,000) (Ronquist and Huelsenbeck, 2003). The tree was then viewed and edited using FigTree3. The species M. daphniae was used to root the tree.
A paired synteny analysis among N. ceranae, N. apis, and N. bombycis was performed and viewed using SyMAP (V5.0.5) (Soderlund et al., 2011). In order to further analyze the selection of the gene Dicer, protein sequences were retrieved from 11 microsporidian species that have maintained the gene encoding Dicer. These Dicer orthologs have been described previously (Ndikumana et al., 2017). The sequences were aligned with Muscle, and a phylogenic tree was constructed using Mrbayes (nchain = 4, aamodelpr = mixed, ngen = 1,000,000) (Ronquist and Huelsenbeck, 2003; Edgar, 2004). Additionally, as the intergenic regions were fragmented, the maximal length of nucleotides up-(1 Kbp) and down-(2 Kbp) stream of the gene for Dicer were retrieved and aligned with Muscle (Edgar, 2004). These aligned sequences were then concatenated for the phylogenetic calculation with Mrbayes (nchain = 4, rates = invgamma, ngen = 1,000,000) (Ronquist and Huelsenbeck, 2003). The species M. daphniae, A. algerae, and N. apis were excluded, as the gene for Dicer was located at either the start or end of contigs or at a gap, where the nucleotides up- or down-stream of the gene for Dicer were not long enough to perform phylogenic analysis.
In order to further characterize the PTP genes and SWP genes, protein and CDS sequences encoded SWP were downloaded from NCBI and used as a library. The genome sequences of N. ceranae were used to query this library. Through blast with an E-value cutoff of ≤1e−5, the best aligned sequence was used for further analysis. To identify PTPs in N. ceranae, about 10 kb sequences of upstream and downstream of PTPs in E. cuniculi and N. bombycis were obtained, then candidate sequences were retrieved by BLAST (E-value ≤ 1e−10). Synteny blocks were identified manually from BLAST coordinates.
The components of the Fe-S cluster assembly machinery have been preliminarily identified for the microsporidians E. cuniculi and T. hominis (Goldberg et al., 2008). To identify the Fe-S cluster assembly proteins for N. ceranae, the amino acid sequences of E. cuniculi and T. hominis Fe-S cluster assembly genes Isu1, Nfs1, and Hsp70 gene were aligned to N. ceranae protein set using BLASTP. Putative Fe-S cluster assembly genes for N. ceranae were designated by E-value ≤ 1e−20 and query coverage ≥ 95%, and only one record for each gene was obtained. Sequences were aligned using Muscle (Edgar, 2004). The phylogenetic trees were built up using the PhyML program with the WAG model under maximum likelihood (Anisimova and Gascuel, 2006). The TreeDyn program was applied to visualize the trees (Chevenet et al., 2006). All the above analyses from the sequence alignment to tree reconstruction were carried out on the phylogeny.fr platform (Dereeper et al., 2008). Outputs in Newick format from this platform were downloaded and further used as input in the iTOL program to generate an unrooted, circular phylogenetic tree (Letunic and Bork, 2007).
The protein sequences of 22 fungal species were downloaded from NCBI, including 20 microsporidian species (E. romaleae, E. hellem, E. intestinalis, E. cuniculi, O. colligata, N. apis, N. bombycis, E. bieneusi, E. canceri, Enterocytozoon hepatopenae, V. corneae, T. hominis, V. culicis, P. neurophilia, E. aedis, A. algerae, Nematocida spERTm5, Nematocida sp1, N. parisii, M. daphniae) and two yeast species (Schizosaccharomyces pombe and Saccharomyces cerevisiae). The sequences of fungal ABC transporters were obtained from previous studies, which were used as seed sequences to query the downloaded protein sequence sets using BLAST with cutoff P < 0.05 (Paumi et al., 2009; Kovalchuk and Driessen, 2010). Sequences were examined manually to remove apparently incomplete sequences against query seeds. All candidate ABC amino acids were then aligned with MAFFT (Katoh et al., 2005). Amino acid substitution models for the ABC family were selected based on Prottest3 (Darriba et al., 2011). The phylogenetic tree was generated using FastTree based on the Jones-Taylor-Thornton (JTT) model with 1,000 bootstraps (Price et al., 2009). The final tree was viewed with MEGA7 (Kumar et al., 2016).
Additionally, protein sequences of ATP/ADP carriers for S. cerevisiae, E. cuniculi and E. bieneusi were downloaded from NCBI and then used to query against the microsporidian protein sequences using BLAST with P < 0.05 as cutoff. Sequences were examined manually to remove apparently incomplete sequences against query seeds. All candidate protein sequences were then aligned with MAFFT (Katoh et al., 2005). Amino acid substitution models for ATP/ADP carriers were selected based on Prottest3 (Darriba et al., 2011). A phylogenetic tree was generated using PhyML based on the Jones-Taylor-Thornton (JTT) model with 1,000 bootstraps (Guindon and Gascuel, 2003). The final tree was viewed with MEGA7 (Kumar et al., 2016).
The genomes of microsporidian parasites are generally compact due to their intracellular parasitic life history, ranging from 2–15 Mbp (Ndikumana et al., 2017). However, an exceptionally large genome of 51 Mpb has also been observed (Desjardins et al., 2015). Within the microsporidian parasites, the species maintained RNAi genes also showed a number of transposable elements, which might contribute to the observed larger genome sizes (Ndikumana et al., 2017). The genome of N. ceranae is notoriously difficult to assemble due to an extremely high level of within-colony genetic diversity (Gómez-Moracho et al., 2014) and the inability to produce pure cultures outside of honey bee hosts. In our study, 2,186,226 reads were generated, resulting in an 8.8 Mbp assembled genome (1,141× genome coverage). The genome was composed of 110 contigs, a substantial improvement over the previous assembly. In total, 2,280 genes were predicted with an average length of 1,057 nucleotides per gene, all supported by transcriptomic reads (Table 1 and Supplementary File 1). Alternative splicing has not been found, but 3′ UTR were identified, which enhanced miRNA-targeted gene prediction. By aligning the long reads back to the assembly, 97.2% of reads can be aligned. The majority of structural variations (SVs) were inversion duplications, and the number of SVs were positively correlated with contig length (Figure 1). The impacts of SVs on any phenotypic effects in microsporidia remain unclear, even though SVs contribute to genomic diversity (Borneman et al., 2011). In the budding yeast, SVs were suggested to be involved in tolerance toward stressors (Zhang et al., 2016). Also, SVs in fission yeasts showed strong impacts on quantitative traits and reproductive isolation (Jeffares et al., 2017). In our study, 1,785 genes were found within SVs. By aligning the protein sequences to the KEGG database, the distribution of genes among the six functional categories was not significantly different across the genome (Pearson’s Chi-squared test, P > 0.05). By aligning the RNA-seq reads back to the assembly, all the predicted protein-coding genes were expressed, suggesting these protein-coding genes were functional. Out of 600 conserved BUSCO groups, 541 complete, 6 fragmented and 53 missing BUSCOs were identified, indicating the assembly is nearly complete (Figure 2). As the BUSCO gene sets for microsporidia were primarily based on Encephalitozoon species, genes could be lost during the divergence, such as RNAi genes, which might partly explain the missing BUSCO genes in microsporidian species. Overall, the genome-based phylogenetic tree is consistent with previous ones (Ndikumana et al., 2017).
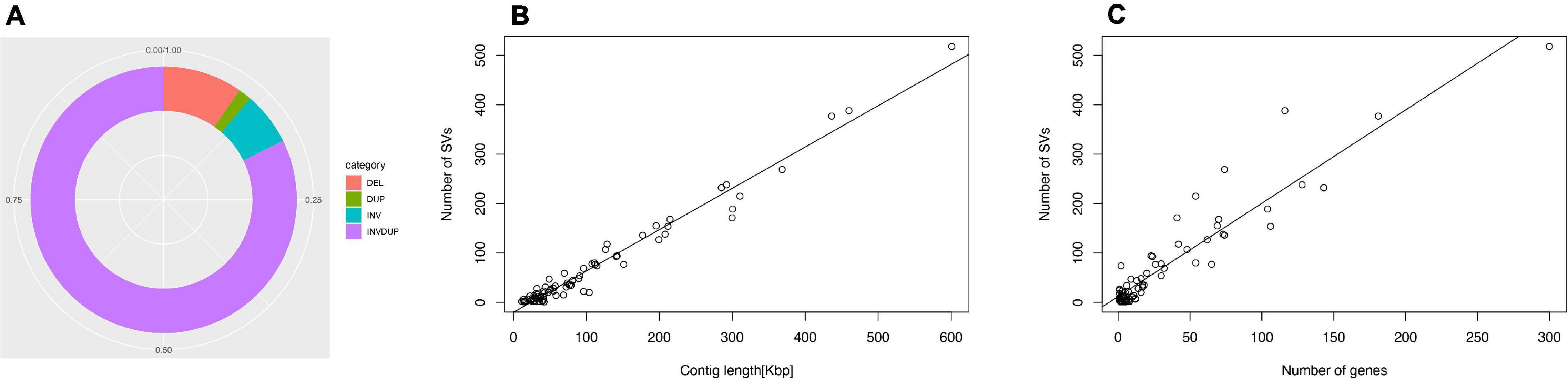
Figure 1. Structural variation of the N. ceranae genome. (A) In total, 8209 SV events were identified. The inversion duplication dominated the SV events, followed by indels, inversion and duplication. (B) The number of SV events is positively correlated with the contig length. (C) The number of SVs within genes.
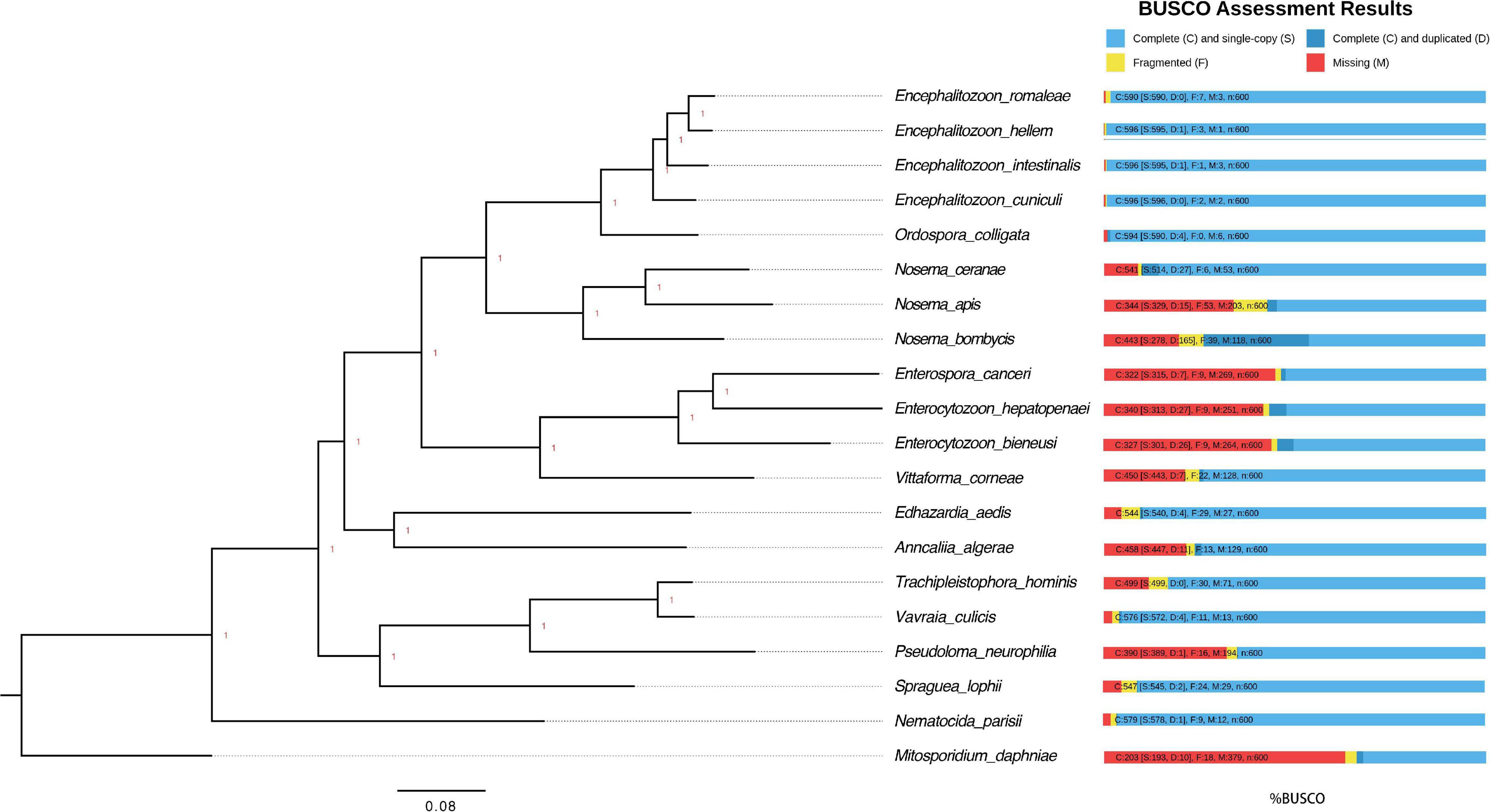
Figure 2. Phylogenetic tree and estimated completeness of the genome of 20 microsporidian species. The phylogenic tree was constructed based on 48 BUSCO genes shared among the microsporidian species and M. daphniae is used to root the tree. All the branches were 100% supported. Overall, 90.2% of microsporidian BUSCOs (V10) were identified from N. ceranae. The studied species N. ceranae has been highlighted. The RNAi genes have been lost twice without reversal, which have been highlighted.
RNAi is a mechanism to regulate gene expression at the post-transcriptional level, which is crucial for the development and defenses of organisms (Zhao and Srivastava, 2007; Obbard et al., 2009). Based on the assembled genomes, a few microsporidian species have lost RNAi genes (Ndikumana et al., 2017). However, the evolutionary forces driving the loss of RNAi genes in microsporidian species remain unclear. Out of 20 selected microsporidian species, a subset of 11 species has maintained RNAi genes Dicer and Argonaute, including M. daphniae, S. lophii, P. neurophilia, V. culicis, T. hominis, A. algerae, E. aedis, V. corneae, N. bombycis, N. apis, and N. ceranae. We did not find evidence suggesting that the loss of RNAi genes was associated with host specificity, either between insect and non-insect, or between vertebrate and invertebrate (Huang, 2018). As the flanking sequences and Dicer were selected as a unit, the topology of the phylogenic tree suggests that the loss of RNAi occurred late in the divergence of the microsporidian species twice without reversal. The events of RNAi maintenance significantly deviated from random (Fisher’s Exact test, P < 0.05; Figure 2). As N. ceranae, N. apis and N. bombycis were the most closely related sister species, and all three species maintained the gene Dicer, a synteny block with the gene Dicer is expected among the three species. By pair-wise analyses, 28 synteny blocks were identified between N. bombycis and N. ceranae, including a synteny block containing the gene Dicer (Figure 3). Additionally, 34 synteny blocks were identified between N. apis and N. ceranae. However, a synteny block containing the gene Dicer was not found between these two species, which might be due to a genome rearrangement. Alternatively, the orthologous anchor genes might have been fragmented during the assembly processes of other species (Liu et al., 2018). Therefore, two additional phylogenic analyses were further performed using protein sequences of the gene Dicer, as well as the up and downstream nucleotides of the gene Dicer. The topology of the phylogenetic tree for Dicer was consistent with the proposed divergence of microsporidian species (Figure 4). It is then reasonable to expect signals of genetic hitchhiking around the gene Dicer (Barton, 2000). Therefore, 1 Kbp up-stream and 2 Kbp down-stream the gene Dicer were extracted to construct a phylogenic tree. The topologies of the trees of both datasets were highly congruent, suggesting the upstream and downstream of the gene Dicer were under selection in parallel with the gene Dicer without insertion or recombination, at least within the studied clades due to hitchhiking.
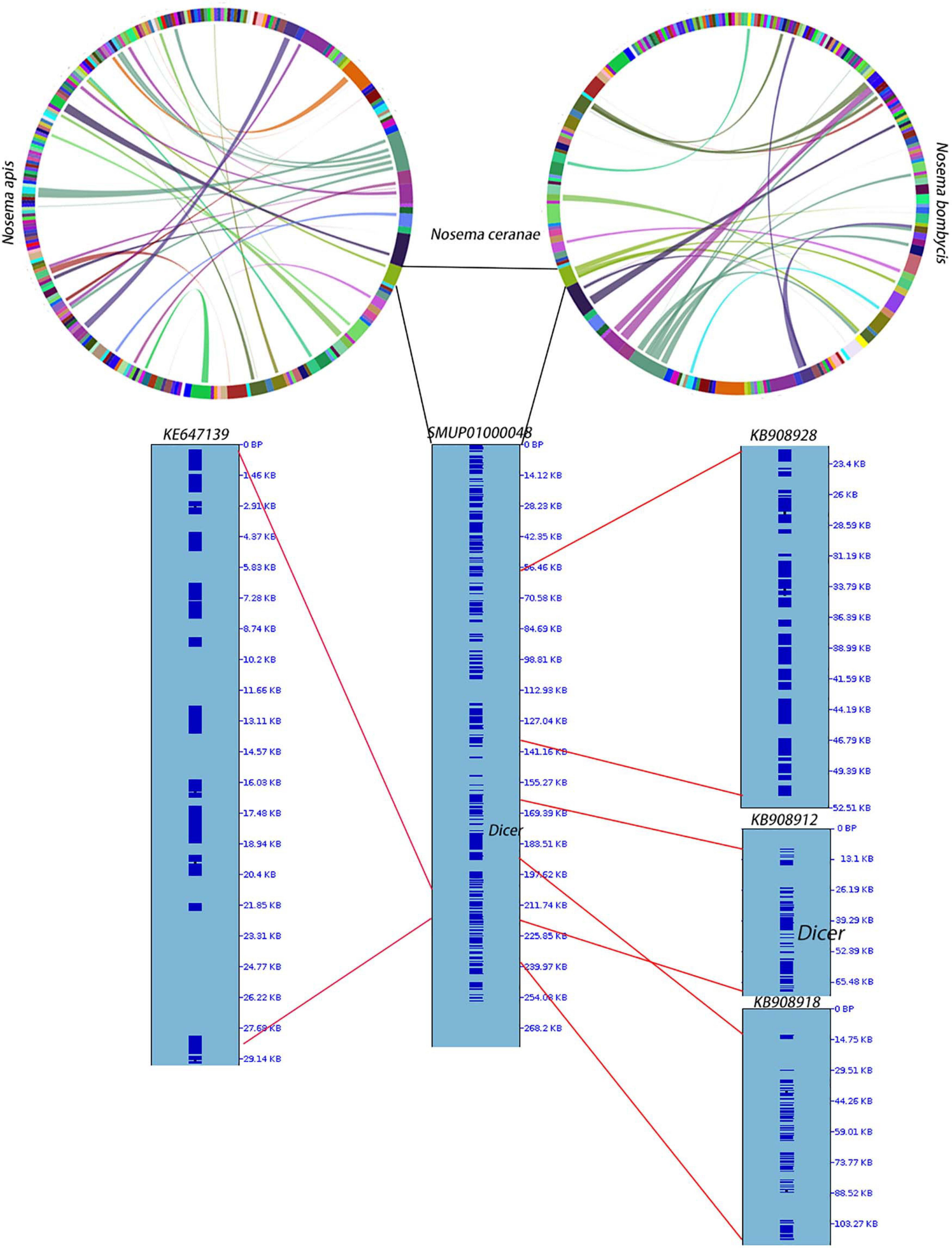
Figure 3. Synteny among N. apis, N. ceranae and N. bombycis. At the genome level, 28 synteny blocks were identified between N. bombycis and N. ceranae. Additionally, 34 synteny blocks were identified between N. apis and N. ceranae. Surprisingly, a synteny block with the gene Dicer is not shared among the three species. The gene Dicer was predicted in N. ceranae contig (SMUP01000048). The paired synteny block between N. ceranae and N. apis, as well as between N. ceranae and N. bombycis were further shown in this contig. The corresponding region are shown on the right of the contig and the region with red lines indicates a synteny block. A synteny block with the gene Dicer was identified between N. ceranae and N. bombycis.
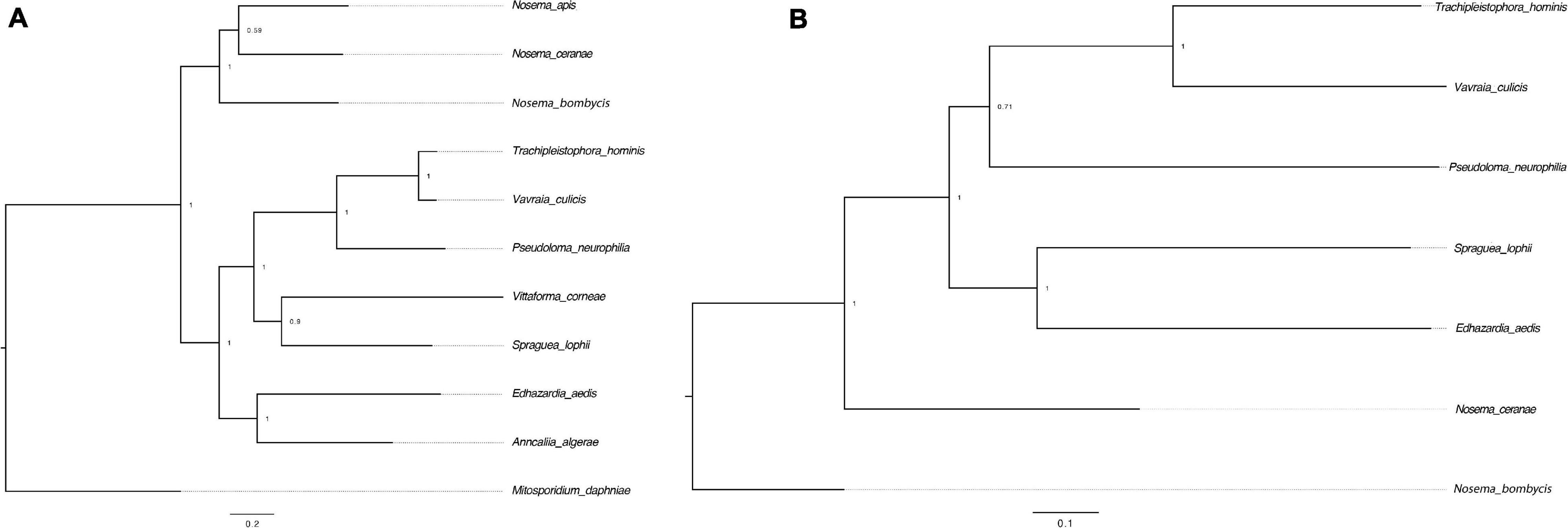
Figure 4. Phylogenetic trees of the gene Dicer and surrounding regions. (A) The tree was constructed based on the protein sequences of the gene Dicer. M. daphniae was used to root the tree. (B) The tree was constructed based on the nucleotides up and down stream of the gene Dicer. N. bombycis was selected to root the tree. The congruence of the two trees suggests a hitchhiking effect of Dicer during microsporidian divergence.
Microsporidia are highly obligate intracellular parasites of animals with extremely compact genomes and decreased cellular and biochemical reactions (Katinka et al., 2001; Keeling and Fast, 2002; Corradi et al., 2010). The microsporidian phylum lacks typical mitochondria but contains a mitosome, a tiny mitochondrial remnant (Williams et al., 2002). Although mitosomes have lost canonical mitochondrial functions like aerobic respiration and biosynthesis, their ability to generate Fe-S proteins essential for the maturation of proteins of diverse functions is maintained (Goldberg et al., 2008). Several components are required for the Fe-S cluster assembly machinery of mitosomes. First, the scaffold proteins Isu1 and Isu2 are involved in the de novo biosynthesis of a transiently bound Fe-S cluster. In this step, iron and sulfur are donated by frataxin (Yfh1) and the cysteine desulphurase complex Nfs1–Isd11, respectively. The electrons are delivered by ferredoxin (Yah1). Then, the Fe–S cluster pass from Isu1 and Isu2 to target apoproteins with the support from a Hsp70 (Ssq1) protein, co-chaperone Jac1, and the monothiol glutaredoxin Grx5.
Isu1, Nfs1, and Hsp70 are key components of Fe-S cluster assembly machinery. Orthologs for these proteins were retrieved to build the phylogeny trees crossing eukaryotic and prokaryotic organisms. By comparing the amino sequences, one significant hit was identified from the N. ceranae genome for Isu1 (Nn.00g008470, Figure 5), Nfs1 (Nn.00g019510, Figure 6), and Hsp70 (Nn.00g001540, Figure 7), respectively. The homologs of all three genes are inter-kingdom conserved, suggesting the conservation and importance of the Fe-S cluster assembly machinery. Generally, the genes from microsporidia (including N. ceranae) were relatively close and clustered. However, significant intra-microsporidian divergence was observed for Isu1 and Nfs1. Interestingly, all three phylogenetic trees suggest that the microsporidian sequences are branched relatively early from other species, indicating microsporidia may contain specific features of the Fe-S cluster assembly, supported by the Siddal and Whiting method (Supplementary Figure 1). A follow-up study will be to investigate the biosynthetic function of Fe-S proteins in the microsporidian mitosomes.
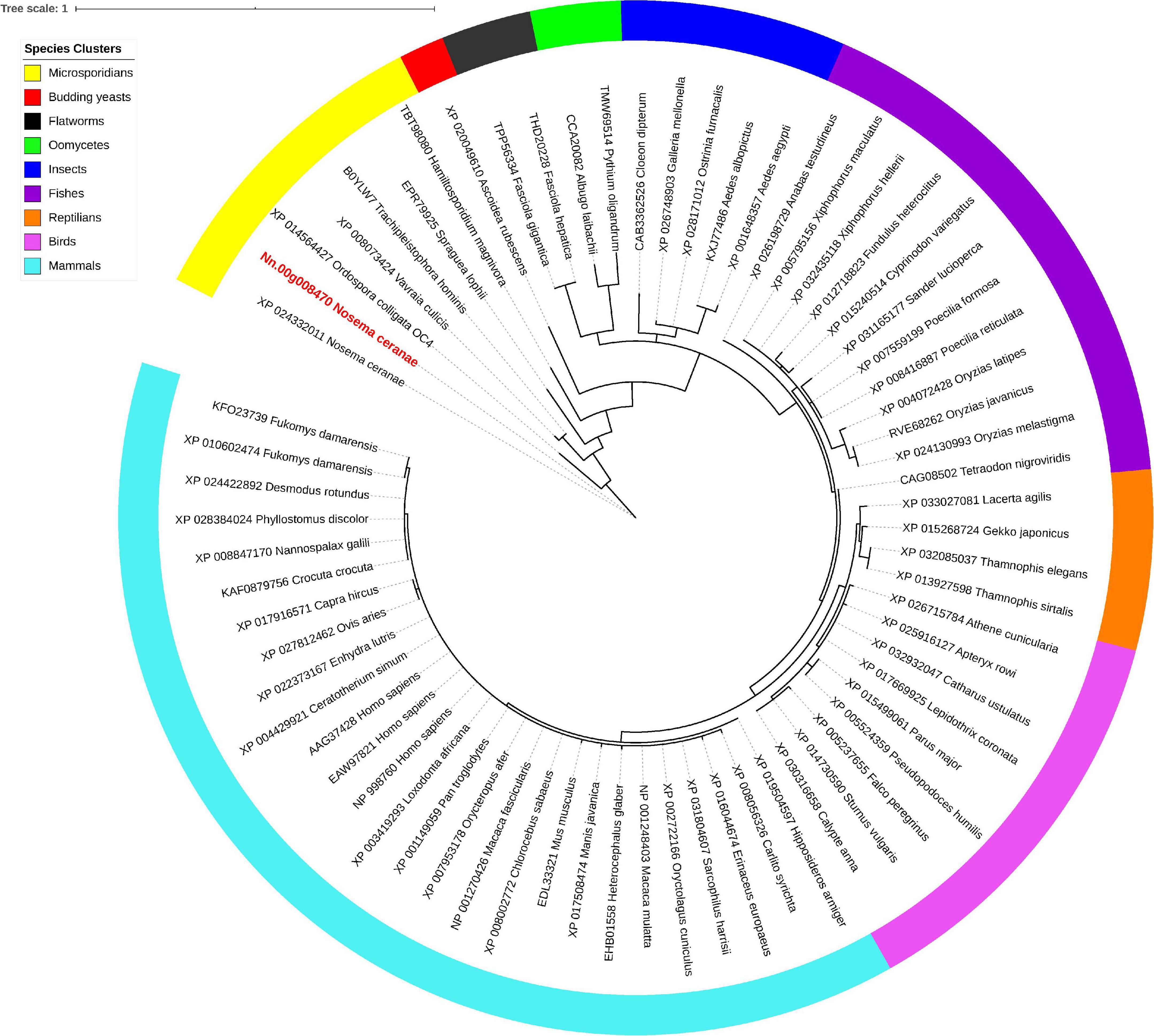
Figure 5. Maximum likelihood phylogenetic tree of Isu1 proteins. Branches are labeled as GenBank accession numbers followed by species names. Branches with bootstrap values (1,000 replicates) less than 0.5 were discarded. The genetic distance is drawn to scale. The current Nosema ceranae record is highlighted with red color. The definitions of the species clusters are adopted from the NCBI Taxonomy database.
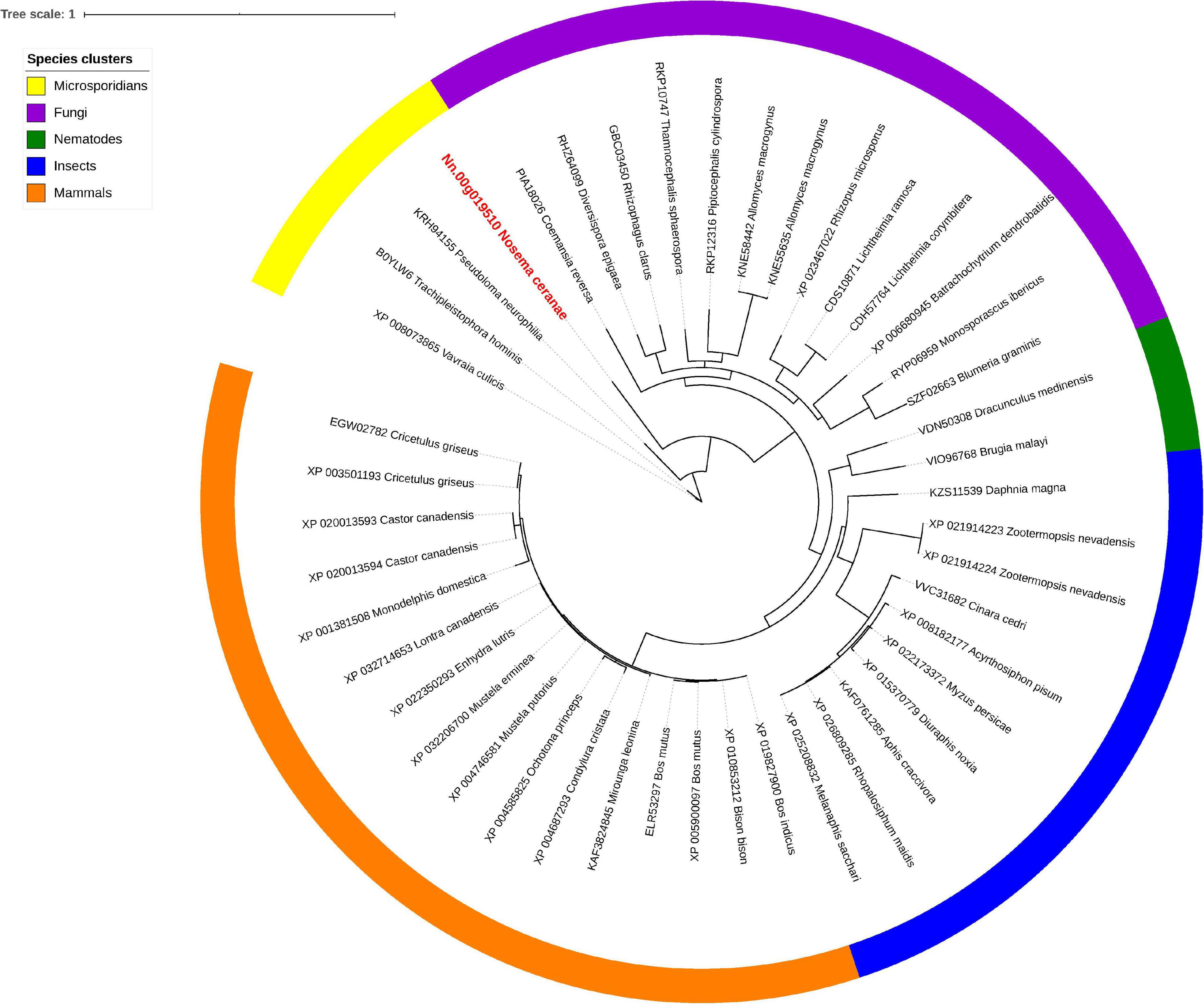
Figure 6. Maximum likelihood phylogenetic tree of Nfs1 proteins. Branches are labeled as GenBank accession numbers followed by species names. Branches with bootstrap values (1,000 replicates) less than 0.5 were discarded. The genetic distance is drawn to scale. The current Nosema ceranae record is highlighted with red color. The definitions of the species clusters are adopted from NCBI Taxonomy database.
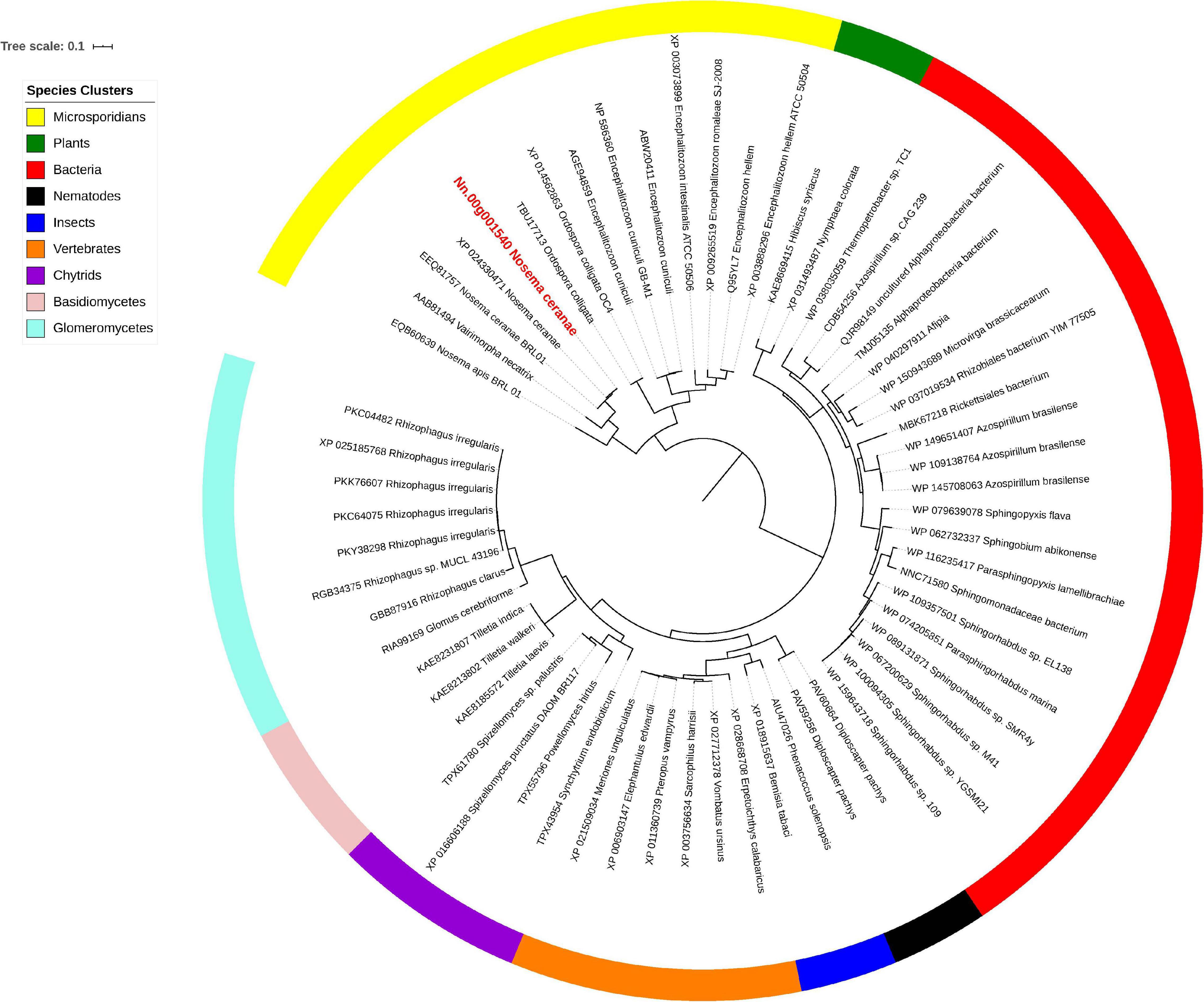
Figure 7. Maximum likelihood phylogenetic tree of Hsp70 proteins. Branches are labeled as GenBank accession numbers followed by species names. Branches with bootstrap values (1,000 replicates) less than 0.5 were discarded. The genetic distance is drawn to scale. The current Nosema ceranae record is highlighted with red color. The definitions of the species clusters are adopted from NCBI Taxonomy database.
ABC transporters constitute one of the largest superfamilies found in all living organisms, with the number of known members exceeding more than 10,000 species (Dassa and Bouige, 2001). ABC transporters contain a pair of nucleotide-binding domains (NBDs) that hydrolyze ATP and facilitate conformational alterations in the associated transmembrane domains (TMDs), thus permitting substrates to cross the membrane lipid bilayer and either be exported out of or imported into the cytoplasm (Locher, 2016). ABC transporter proteins are engaged in the ATP-dependent transport of extensive substrates across biological membranes, as well as receptors, ion channels, mRNA translation, and ribosome biogenesis (Kovalchuk and Driessen, 2010). Importantly, ABC transporters have been found to contribute to multidrug resistance in microbial pathogens and tumor cells (Piddock, 2006; Lubelski et al., 2007; Wu et al., 2019). According to the Human Genome Organization (HUGO) approved scheme, all eukaryotic ABC transporter proteins are categorized into eight major subfamilies (A to H) (Dean et al., 2001). In the fungal kingdom, the ABC transporter proteins have been well described in the budding yeast S. cerevisiae and the fission yeast S. pombe (Iwaki et al., 2006). However, knowledge of microsporidian ABC transporters remains limited. With the rapid development of genome sequencing projects in the past two decades, a diversity of ABC transporter proteins in genomes of microsporidian species has been uncovered, which allows for the comparative survey of ABC transporters within this group of organisms.
In the current research, five of eight subfamilies (ABC-B, ABC-C, ABC-E, ABC-F, and ABC-G) were present in genomes of 21 microsporidian species, and among these ABC subfamilies, members of ABC-G proteins were the most abundant, followed by ABC-B subfamily (Table 2). ABC-A, ABC-D, ABC-H subfamilies seem to be lost in most or all microsporidia. ABC-C only presents in the basal microsporidian M. daphniae. ABC-B proteins represent a large category of ABC transporters, which are widely distributed among eukaryotes, including fungi. Their diverse functions are associated with the export of mitochondrial peptides, biogenesis of iron-sulfur (Fe-S) cluster proteins, multidrug resistance, and antigen processing (Kovalchuk and Driessen, 2010). Genomic investigations demonstrate that reduction in metabolic capabilities is the dominating feature of microsporidian genome evolution, which has been supported by expanding transporter gene families to compensate for pathway loss (Heinz et al., 2012; Nakjang et al., 2013; Freibert et al., 2017). In our study, ABC-B members were identified in all microsporidian genomes though the number varies among different species, suggestive of the importance of ABC-B family in microsporidia (Table 2 and Figure 8). The number of ABC-B members was conserved in Encephalitozoon species, suggesting an essential function in this group of microsporidia. However, the number of ABC-B has diverged within Nosema family, where Nosema ceranae maintained the highest number of ABC-B paralogs, indicative of a lineage-specific duplication of this subfamily, which can also be seen in the evolutionary tree (Figure 8). Alternatively, the lower number of ABC-B might be due to incompletely assembled genomes in other Nosema species. Considering that N. ceranae has become a globally predominant microsporidian species in honey bees, it’s of great interest to perform additional studies to explore the relationship between the high number of ABC-B and the high virulence of N. ceranae.
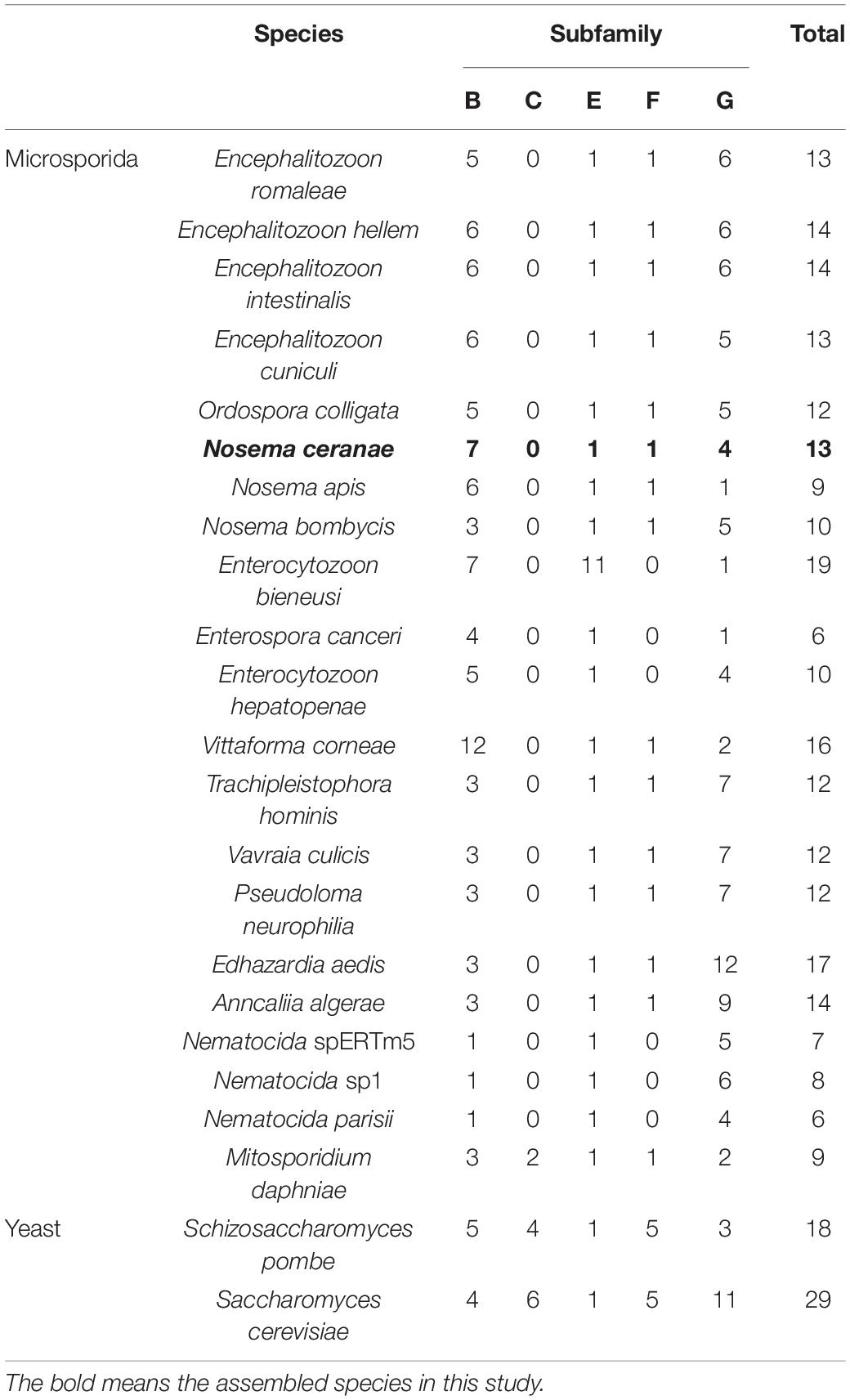
Table 2. Numbers of ABC transporter genes in 22 microsporidian species and two yeast species, in each of five subfamilies.
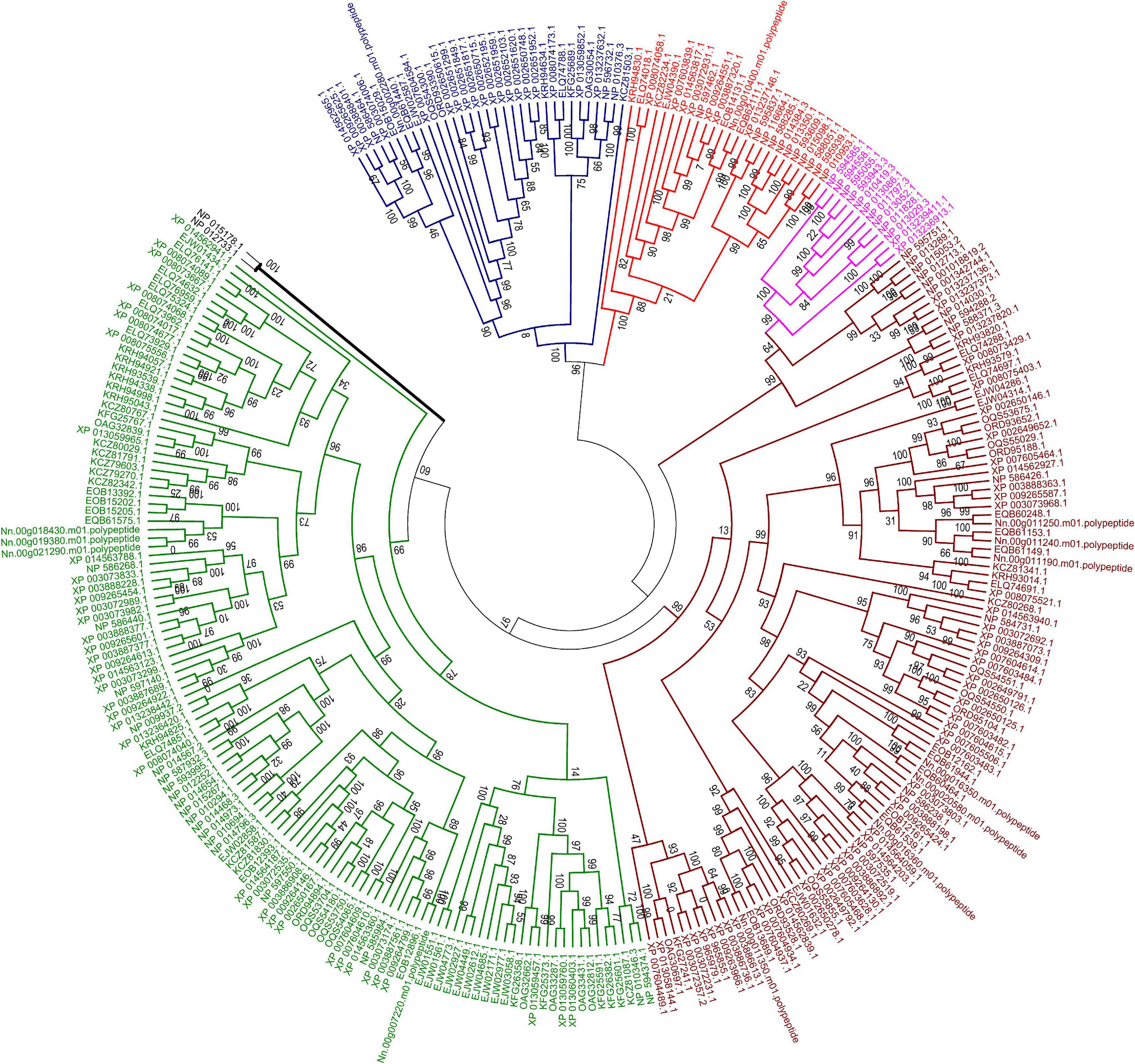
Figure 8. Maximum likelihood phylogenetic tree of ABC transporter proteins of 23 fungal genomes. The tree was based on a total of 293 ABC transporter protein sequences. The unrooted tree was calculated with FASTTREE based on JTT mode with 1,000 replications. The color code associated with each protein indicated the subfamily, with brown for subfamily B, purple for subfamily C, blue for subfamily E, red for subfamily F and green for subfamily G. Two ABC proteins from subfamily D in S. cerevisiae were labeled with black line.
ABC-G transporters in fungal species were often reported to be engaged in drug resistance and lipid translocation (Smriti et al., 2002; Coleman and Mylonakis, 2009). The number of ABC-G in the investigated microsporidian species was diverse, from only a single copy in N. apis, E. bieneusi, E. canceri, and V. culicis to 12 copies in E. aedis (Table 2). There might be some host- or environment-driven pressures to explain the huge birth and death rates of genes in this essential protein family, suggesting that this subfamily has undergone lineage-specific divergence during microsporidian evolution. N. ceranae (four ABC-G) showed higher copy number of ABC-G than that of its most closely related species N. apis (one ABC-G). However, the other three ABC-G orthologs were clustered into a single group (Figure 8), which indicates a recent gene duplication in ABC-G of N. ceranae. In another closely related species, N. bombycis, ABC-G was constantly expressed over the entire proliferation stages and was a key player in substrate transportation from ions to proteins (He et al., 2019). It is then interesting to decipher the detailed function of ABC-G during N. ceranae proliferation.
As the result of an intracellular lifestyle, the microsporidian parasites have lost canonical mitochondria and oxidative phosphorylation pathway; hence glycolysis is the mean to produce ATP (Timofeev et al., 2020). To satisfy their energy demands, microsporidia acquired the capability to import ATP directly from the host cell cytoplasm during proliferation (Tsaousis et al., 2008). Indeed, microsporidia have frequently been detected to be surrounded by host mitochondria (Han et al., 2019). ATP/ADP carriers, which were gained via HGT from intracellular bacteria, play a pivotal part in the transportation of ATP from infected cells to E. cuniculi (Tsaousis et al., 2008; Heinz et al., 2014). Additional efforts are required to conclude whether this is a common strategy during the evolution of microsporidian species.
Here, a phylogenetic tree of 21 microsporidian species and two yeast species S. cerevisiae and S. pombe was built based on the ATP/ADP carriers (Figure 9). There were three ATP/ADP carriers in P. neurophilia and E. aedis, similar to the number of ATP/ADP carriers identified in S. cerevisiae and S. pombe. Two members of the ATP/ADP carrier family were identified in A. algerae, T. hominis, Nematocida sp1, Nematocida spERTm5, and N. parisii. Only one ATP/ADP carrier was found in M. daphnia, which has a microsporidia-like morphology and is regarded as a basal microsporidia species (Bass et al., 2018). The different number of ATP/ADP carriers among various microsporidian parasites demonstrated that the selection of ATP/ADP carrier genes occurs during the lineage divergence. Species of the genus Encephalitozoon are ubiquitous vertebrate pathogens except for E. romaleae, which has been isolated from a grasshopper (Corradi, 2015). These species are well-known for their miniature genomes (ranging from 2.3 to 2.9 Mb) with the smallest coding capacity (Pombert et al., 2012). We observed that the numbers of ATP/ADP carrier members in Encephalitozoon species are highly conserved, and E. cuniculi, E. bieneusi, E. hepatopenaei, E. intestinalis, E. hellem, and E. romaleae all have four ATP/ADP carrier proteins. Similar to Encephalitozoon species, N. ceranae has four ATP/ADP carriers, which was higher than N. apis (two ATP/ADP carriers) and N. bombycis (one ATP/ADP carrier). N. ceranae Nn.00g020520 and Nn.00g004970 are, respectively, homologous to N. apis EQB60298.1 and EQB60147.1, while N. ceranae Nn.00g004860 is homologous to N. bombycis EOB13854.1 (Figure 9). Additionally, N. ceranae Nn.00g006340 has no homolog in N. apis or N. bombycis. This is suggestive of a recent lineage-specific gene expansion of ATP/ADP carrier family in N. ceranae. N. ceranae and A. mellifera have evolved together over just a short period, and arguably this imbalance explains why N. ceranae exerts more energetic stress on the bee host (Martin-Hernandez et al., 2011).
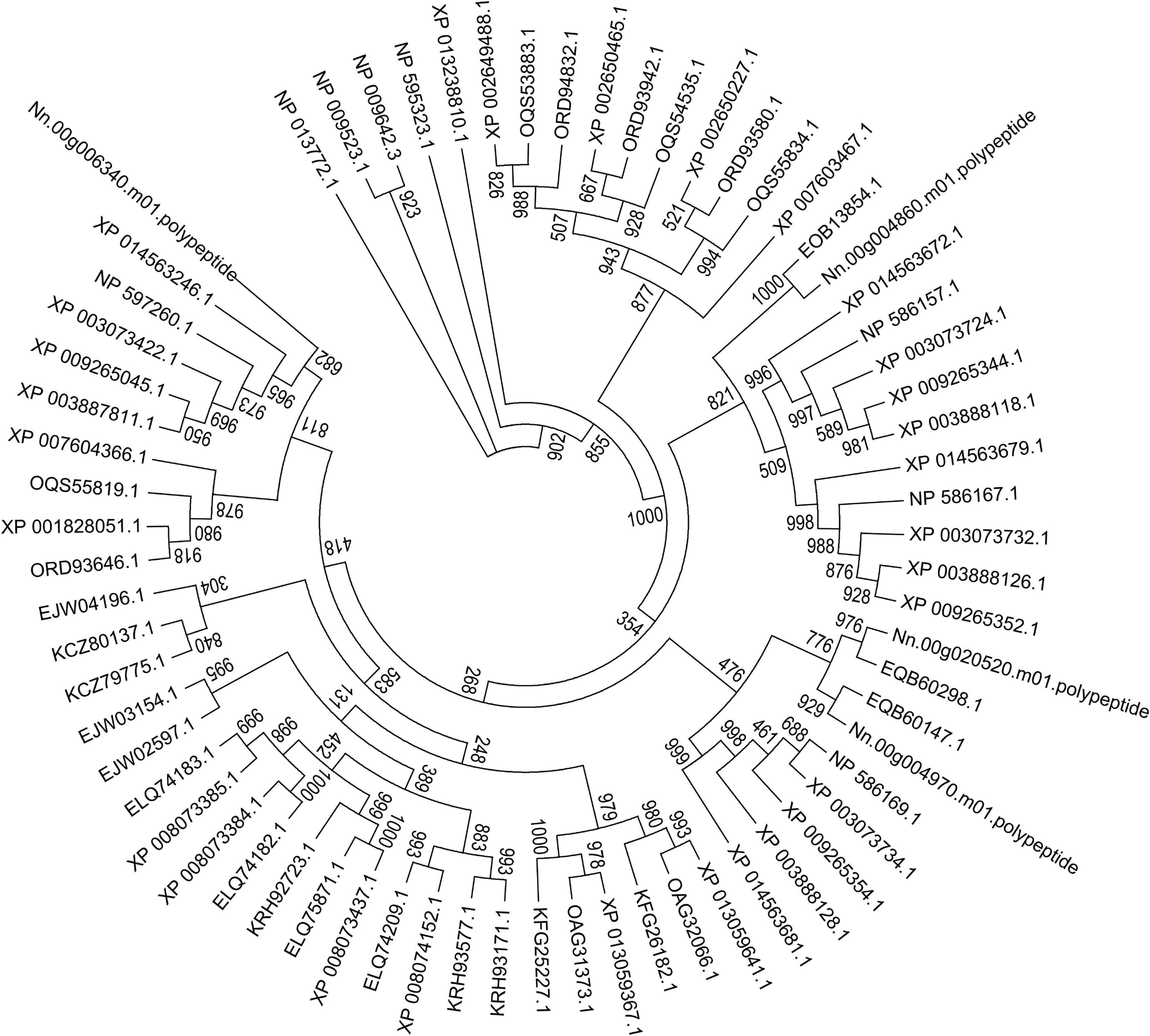
Figure 9. Maximum-likelihood phylogenetic tree generated based on ATP/ADP carrier proteins in 23 fungal genomes. Based on the total aligned 68 ATP/ADP carrier proteins, the tree, rooted with ATP/ADP carrier in S. cerevisiae and S. pombe, was calculated with PhyML based on JTT mode with 1,000 replications. Bootstrap values are indicated at the base of each clade.
Due to a lack of well-established tools for genetic manipulation, functional study on ATP/ADP carriers in microsporidian is extremely limited, especially in Nosema species. Given the severe influence of N. ceranae on the worldwide beekeeping industry, it’s necessary and significant to carry out experimental work on the function of identified members of ATP/ADP carrier family in N. ceranae adopting molecular approach such as RNAi, which has been proved to be efficient in knockdown of several N. ceranae genes (Paldi et al., 2010; Li et al., 2016; Rodriguez-Garcia et al., 2018; Huang et al., 2019). We previously used purified spores of N. bombycis to infect Bombyx mori BmN cells, followed by transfection with a non-transposon vector pIZT/V5-His vector and the exogenous gfp gene was successfully inserted into the N. bombycis genome (Guo et al., 2016). Additional studies reported the successful establishment of the gypsy moth (Porthetria dispar) IPL-LD-65Y cell-based system of N. ceranae infection (Gisder et al., 2011; Gisder and Genersch, 2015), which provides an excellent opportunity to conduct functional exploration of ATP/ADP carrier genes of N. ceranae.
Microsporidia infects host cells by employing a unique, highly specialized invasion device including the spore wall (SW) and polar tube. After germination, the polar tube protein PTP1 can interact with lectin receptors on the host cell surface (Xu et al., 2003, 2004). PTP2 with a basic lysine-rich core was clustered closely with PTP1 on a contig. PTP3 was up-regulated during sporogony at the transcriptional level (Peuvel et al., 2002). Polar tube protein 4 (PTP4) has been demonstrated to have a specific epitope on the tip of the PT, and this epitope was shown to interact with the host cell transferrin receptor (TfR1) (Han et al., 2017). The five known genes encoding the polar tube protein were all present in the current genome assembly, which include PTP1, PTP2, PTP3, PTP4, PTP5 genes. After determining the homologous gene loci in the genome, one syntenic block harboring the PTP1 and PTP2 was identified between N. bombycis, N. ceranae, E. cuniculi and E. intestinalis (Figure 10). Remarkably, the PTP1 and PTP2 were arrayed conversely both in N. ceranae and N. bombycis compared with E. cuniculi and E. intestinalis (Supplementary Figure 2). Similarly, another syntenic block, including the PTP4 and PTP5 genes, was identified (Figure 10). According to the composition of the spore wall, there are multiple SWPs in both the exospore and endospore (Han et al., 2020). In the current assembly, seven genes encoding the spore wall protein were identified based on homologous searches with BLASTP (Table 3). Protein domain prediction showed that NcSWP12 contains BAR/IMD domain which served as sensors of membrane curvature (Quinones et al., 2010). In N. bombycis, NbSHWP12 with BAR/IMD domain is localized to the spore wall and can adhere to the deproteinized chitin coats (Chen J. et al., 2013). The homologous gene to NcHSWP7, NbSWP7, is localized in the exospore, endospore and polar tube of the mature N. bombycis spores where it mediates adherence to host cells (Yang et al., 2015). NcHSWP1, which is the ortholog of EnP1, was found to contain the heparin-binding motif (HBM), which mediates interactions between spores and glycosaminoglycan from the surface of host cells (Southern et al., 2007). The low number of conserved spore wall genes might be due to high divergence of these components among microsporidian species.
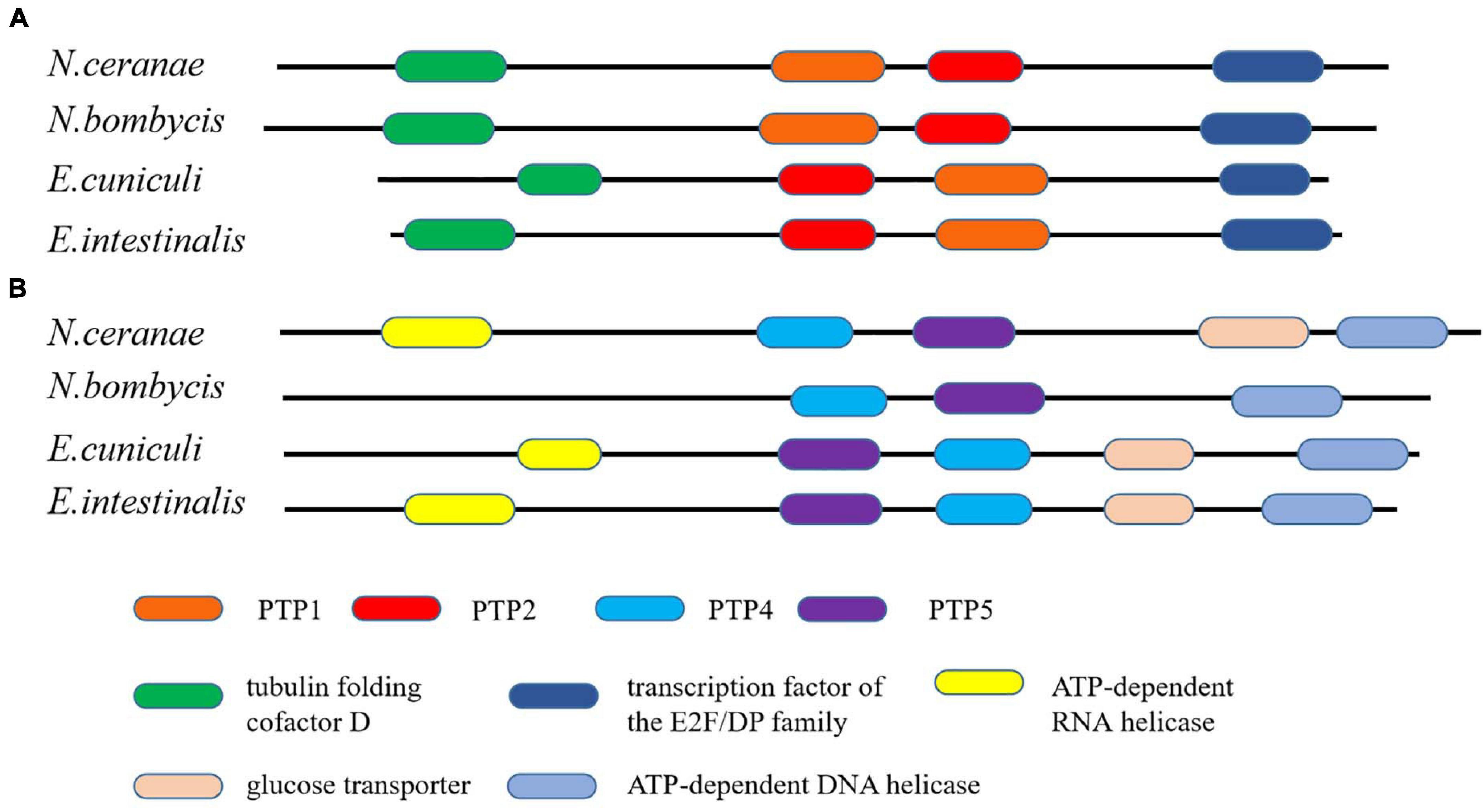
Figure 10. The conservation of gene order among N. ceranae and three other related microsporidian species. (A) The distribution of PTP1 and PTP2 genes in the synteny block. (B) The distribution of PTP4 and PTP5 genes in the synteny block. The colored oblong represents genes with functional annotations.
N. ceranae destroys the gut integrity of honey bees, leading to impaired flying and memory abilities, which can lead to the loss of colonies. We anticipate that the updated genome resource and comparative analyses provided here lead to novel methods to control this parasite without harming honey bees. The provided genome also reveals numerous evolutionary features compared with other microsporidian parasites, which may help to clarify the evolution of virulence and co-evolution with hosts.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR8536193.
The apiaries for bee sample collection are the property of the USDA-ARS Bee Research Laboratory, Beltsville, MD, United States. No specific permits are required for the described studies. Studies involved the European honey bee (Apis mellifera), which is neither an endangered nor a protected species.
QH and JE designed the investigation. QH assembled the genome. WL performed Fe-S phylogenetic analysis. RG performed ABC and ATP/ADP carriers’ phylogenetic analysis. JX, XD, and ZM performed SW and Polar tube protein phylogenetic analysis. QH, ZW, WL, RG, JX, XD, ZM, YC, and JE wrote the manuscript. All authors contributed to the article and approved the submitted version.
The project was supported by the initiation package of Jiangxi Agricultural University 050014/923230722 (QH) and The National Natural Science Foundation of China 32060778 (QH). The publication fee was funded by USDA-ARS Bee Research Laboratory Beltsville (JE). WL was supported by the Guangzhou Science and Technology Plan Project 201904010115. The funders had no role in experimental design and manuscript preparation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We appreciate Competence Centre in Bioinformatics and Computational Biology, SWISS Institute of Bioinformatic (Vital-IT) for bioinformatics support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.645353/full#supplementary-material
Supplementary File 1 | Additional results.
Supplementary File 2 | De novo transcriptome assembly.
Supplementary File 3 | Genes in SVs.
Supplementary File 4 | Sequences for microsporidian phylogenetic tree of Figure 2.
Supplementary File 5 | Sequences for Dicer phylogenetic tree of Figure 4A.
Supplementary File 6 | Sequences for Dicer up and down stream phylogenetic tree of Figure 4B.
Supplementary File 7 | Sequences for ISU1 phylogenetic tree of Figure 5.
Supplementary File 8 | Sequences for NFS1 phylogenetic tree of Figure 6.
Supplementary File 9 | Sequences for HSP1 phylogenetic tree of Figure 7.
Supplementary File 10 | Sequences for ABC phylogenetic tree of Figure 8.
Supplementary File 11 | Sequences for ATP phylogenetic tree of Figure 9.
Supplementary File 12 | Sequences for PTP synteny of Figure 10.
Aizen, M. A., Garibaldi, L. A., Cunningham, S. A., and Klein, A. M. (2009). How much does agriculture depend on pollinators? Lessons from long-term trends in crop production. Ann. Bot. 103, 1579–1588. doi: 10.1093/aob/mcp076
Anisimova, M., and Gascuel, O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. doi: 10.1080/10635150600755453
Antunez, K., Martin-Hernandez, R., Prieto, L., Meana, A., Zunino, P., and Higes, M. (2009). Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 11, 2284–2290. doi: 10.1111/j.1462-2920.2009.01953.x
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barton, N. H. (2000). Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 355, 1553–1562.
Bass, D., Czech, L., Williams, B. A. P., Berney, C., Dunthorn, M., Mahé, F., et al. (2018). Clarifying the relationships between microsporidia and cryptomycota. J. Eukaryot. Microbiol. 65, 773–782. doi: 10.1111/jeu.12519
Borneman, A. R., Desany, B. A., Riches, D., Affourtit, J. P., Forgan, A. H., Pretorius, I. S., et al. (2011). Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 7:e1001287. doi: 10.1371/journal.pgen.1001287
Burri, L., Williams, B. A. P., Bursac, D., Lithgow, T., and Keeling, P. J. (2006). Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc. Natl. Acad. Sci. U.S.A. 103, 15916–15920. doi: 10.1073/pnas.0604109103
Campbell, S. E., Williams, T. A., Yousuf, A., Soanes, D. M., Paszkiewicz, K. H., and Williams, B. A. P. (2013). The genome of spraguea lophii and the basis of host-microsporidian interactions. PLoS Genet. 9:e1003676. doi: 10.1371/journal.pgen.1003676
Capella-Gutiérrez, S., Marcet-Houben, M., and Gabaldón, T. (2012). Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol. 10:47. doi: 10.1186/1741-7007-10-47
Chen, J., Geng, L., Long, M., Li, T., Li, Z., Yang, D., et al. (2013). Identification of a novel chitin-binding spore wall protein (NbSWP12) with a BAR-2 domain from Nosema bombycis (microsporidia). Parasitology 140, 1394–1402. doi: 10.1017/S0031182013000875
Chen, Y., Pettis, J. S., Zhao, Y., Liu, X., Tallon, L. J., Sadzewicz, L. D., et al. (2013). Genome sequencing and comparative genomics of honey bee microsporidia, Nosema apis reveal novel insights into host-parasite interactions. BMC Genomics 14:451. doi: 10.1186/1471-2164-14-451
Chevenet, F., Brun, C., Bañuls, A.-L., Jacq, B., and Christen, R. (2006). TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439.
Coleman, J. J., and Mylonakis, E. (2009). Efflux in fungi: la pièce de résistance. PLoS Pathog. 5:e1000486. doi: 10.1371/journal.ppat.1000486
Corradi, N. (2015). Microsporidia: eukaryotic intracellular parasites shaped by gene loss and horizontal gene transfers. Annu. Rev. Microbiol. 69, 167–183. doi: 10.1146/annurev-micro-091014-104136
Corradi, N., Akiyoshi, D. E., Morrison, H. G., Feng, X., Weiss, L. M., Tzipori, S., et al. (2007). Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi. PLoS One 2:e1277. doi: 10.1371/journal.pone.0001277
Corradi, N., Haag, K. L., Pombert, J. F., Ebert, D., and Keeling, P. J. (2009). Draft genome sequence of the Daphnia pathogen Octosporea bayeri: insights into the gene content of a large microsporidian genome and a model for host-parasite interactions. Genome Biol. 10:R10. doi: 10.1186/gb-2009-10-10-r106
Corradi, N., Pombert, J. F., Farinelli, L., Didier, E. S., and Keeling, P. J. (2010). The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat. Commun. 1:77. doi: 10.1038/ncomms1082
Cuomo, C. A., Desjardins, C. A., Bakowski, M. A., Goldberg, J., Ma, A. T., Becnel, J. J., et al. (2012). Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 22, 2478–2488. doi: 10.1101/gr.142802.112
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Dassa, E., and Bouige, P. (2001). The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152, 211–229. doi: 10.1016/s0923-2508(01)01194-9
Dean, M., Hamon, Y., and Chimini, G. (2001). The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 42, 1007–1017. doi: 10.1016/s0022-2275(20)31588-1
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Desjardins, C. A., Sanscrainte, N. D., Goldberg, J. M., Heiman, D., Young, S., Zeng, Q., et al. (2015). Contrasting host-pathogen interactions and genome evolution in two generalist and specialist microsporidian pathogens of mosquitoes. Nat. Commun. 6:7121.
Didier, E. S. (2005). Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. 94, 61–76. doi: 10.1016/j.actatropica.2005.01.010
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fayer, R. (2004). Infectivity of microsporidia spores stored in seawater at environmental temperatures. J. Parasitol. 90, 654–657. doi: 10.1645/ge-3335rn
Freeman, M. A., and Sommerville, C. (2011). Original observations of Desmozoon lepeophtherii, a microsporidian hyperparasite infecting the salmon louse Lepeophtheirus salmonis, and its subsequent detection by other researchers. Parasit. Vectors 4:231.
Freibert, S.-A., Goldberg, A. V., Hacker, C., Molik, S., Dean, P., Williams, T. A., et al. (2017). Evolutionary conservation and in vitro reconstitution of microsporidian iron-sulfur cluster biosynthesis. Nat. Commun. 8:13932.
Fries, I., Chauzat, M.-P., Chen, Y.-P., Doublet, V., Genersch, E., Gisder, S., et al. (2013). Standard methods for Nosema research. J. Apic. Res. 52, 1–28. doi: 10.1007/978-3-319-23534-9_1
Fries, I., Feng, F., da Silva, A., Slemenda, S. B., and Pieniazek, N. J. (1996). Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 32, 356–365. doi: 10.1016/s0932-4739(96)80059-9
Gage, S. L., Kramer, C., Calle, S., Carroll, M., Heien, M., and DeGrandi-Hoffman, G. (2018). Nosema ceranae parasitism impacts olfactory learning and memory and neurochemistry in honey bees (Apis mellifera). J. Exp. Biol. 221:jeb161489. doi: 10.1242/jeb.161489
Gisder, S., and Genersch, E. (2015). Identification of candidate agents active against N. ceranae infection in honey bees: establishment of a medium throughput screening assay based on N. ceranae infected cultured cells. PLoS One 10:e0117200. doi: 10.1371/journal.pone.0117200
Gisder, S., Mockel, N., Linde, A., and Genersch, E. (2011). A cell culture model for Nosema ceranae and Nosema apis allows new insights into the life cycle of these important honey bee-pathogenic microsporidia. Environ. Microbiol. 13, 404–413. doi: 10.1111/j.1462-2920.2010.02346.x
Goldberg, A. V., Molik, S., Tsaousis, A. D., Neumann, K., Kuhnke, G., Delbac, F., et al. (2008). Localization and functionality of microsporidian iron–sulphur cluster assembly proteins. Nature 452, 624–628. doi: 10.1038/nature06606
Gómez-Moracho, T., Bartolomé, C., Bello, X., Martín-Hernández, R., Higes, M., and Maside, X. (2015). Recent worldwide expansion of Nosema ceranae (Microsporidia) in Apis mellifera populations inferred from multilocus patterns of genetic variation. Infect. Genet. Evol. 31, 87–94. doi: 10.1016/j.meegid.2015.01.002
Gómez-Moracho, T., Maside, X., Martin-Hernandez, R., Higes, M., and Bartolome, C. (2014). High levels of genetic diversity in Nosema ceranae within Apis mellifera colonies. Parasitology 141, 475–481.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Guo, R., Cao, G., Lu, Y., Xue, R., Kumar, D., Hu, X., et al. (2016). Exogenous gene can be integrated into Nosema bombycis genome by mediating with a non-transposon vector. Parasitol. Res. 115, 3093–3098. doi: 10.1007/s00436-016-5064-8
Haag, K. L., James, T. Y., Pombert, J.-F., Larsson, R., Schaer, T. M. M., Refardt, D., et al. (2014). Evolution of a morphological novelty occurred before genome compaction in a lineage of extreme parasites. Proc. Natl. Acad. Sci. U.S.A. 111, 15480–15485. doi: 10.1073/pnas.1410442111
Haas, B. J., Salzberg, S. L., Zhu, W., Pertea, M., Allen, J. E., Orvis, J., et al. (2008). Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9:R7.
Han, B., Ma, Y., Tu, V., Tomita, T., Mayoral, J., Williams, T., et al. (2019). Microsporidia interact with host cell mitochondria via voltage-dependent anion channels using sporoplasm surface protein 1. mBio 10:e01944-19.
Han, B., Polonais, V., Sugi, T., Yakubu, R., Takvorian, P. M., Cali, A., et al. (2017). The role of microsporidian polar tube protein 4 (PTP4) in host cell infection. PLoS Pathog. 13:e1006341. doi: 10.1371/journal.ppat.1006341
Han, B., Takvorian, P. M., and Weiss, L. M. (2020). Invasion of host cells by microsporidia. Front. Microbiol. 11:172.
He, Q., Vossbrinck, C. R., Yang, Q., Meng, X.-Z., Luo, J., Pan, G.-Q., et al. (2019). Evolutionary and functional studies on microsporidian ATP-binding cassettes: insights into the adaptation of microsporidia to obligated intracellular parasitism. Infect. Genet. Evol. 68, 136–144. doi: 10.1016/j.meegid.2018.12.022
Heinz, E., Hacker, C., Dean, P., Mifsud, J., Goldberg, A. V., Williams, T. A., et al. (2014). Plasma membrane-located purine nucleotide transport proteins are key components for host exploitation by microsporidian intracellular parasites. PLoS Pathog. 10:e1004547. doi: 10.1371/journal.ppat.1004547
Heinz, E., Williams, T. A., Nakjang, S., Noel, C. J., Swan, D. C., Goldberg, A. V., et al. (2012). The genome of the obligate intracellular parasite trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS Pathog. 8:e1002979. doi: 10.1371/journal.ppat.1002979
Higes, M., Garcia-Palencia, P., Martin-Hernandez, R., and Meana, A. (2007). Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 94, 211–217. doi: 10.1016/j.jip.2006.11.001
Higes, M., Juarranz, A., Dias-Almeida, J., Lucena, S., Botias, C., Meana, A., et al. (2013). Apoptosis in the pathogenesis of Nosema ceranae (Microsporidia: Nosematidae) in honey bees (Apis mellifera). Environ. Microbiol. Rep. 5, 530–536.
Higes, M., Martin, R., and Meana, A. (2006). Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 92, 93–95. doi: 10.1016/j.jip.2006.02.005
Higes, M., Martín-Hernández, R., Botías, C., Bailón, E. G., González-Porto, A. V., Barrios, L., et al. (2008). How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10, 2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x
Huang, Q. (2018). Evolution of Dicer and Argonaute orthologs in microsporidian parasites. Infect. Genet. Evol. 65, 329–332. doi: 10.1016/j.meegid.2018.08.011
Huang, Q., Li, W., Chen, Y., Retschnig-Tanner, G., Yanez, O., Neumann, P., et al. (2019). Dicer regulates Nosema ceranae proliferation in honey bees. Insect Mol. Biol. 28, 74–85. doi: 10.1111/imb.12534
Humann, J. L., Lee, T., Ficklin, S., and Main, D. (2019). “Structural and functional annotation of eukaryotic genomes with GenSAS BT - gene prediction: methods and protocols,” in Gene Prediction. Methods in Molecular Biology ed. M. Kollmar (New York, NY: Springer), 29–51. doi: 10.1007/978-1-4939-9173-0_3
Iwaki, T., Giga-Hama, Y., and Takegawa, K. (2006). A survey of all 11 ABC transporters in fission yeast: two novel ABC transporters are required for red pigment accumulation in a Schizosaccharomyces pombe adenine biosynthetic mutant. Microbiology 152, 2309–2321. doi: 10.1099/mic.0.28952-0
Jeffares, D. C., Jolly, C., Hoti, M., Speed, D., Shaw, L., Rallis, C., et al. (2017). Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 8:14061. doi: 10.1038/ncomms14061
Katinka, M. D., Duprat, S., Cornillot, E., Méténier, G., Thomarat, F., Prensier, G., et al. (2001). Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450–453. doi: 10.1038/35106579
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Keeling, P. J., and Fast, N. M. (2002). Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 56, 93–116. doi: 10.1146/annurev.micro.56.012302.160854
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36.
Klee, J., Besana, A. M., Genersch, E., Gisder, S., Nanetti, A., Tam, D. Q., et al. (2007). Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 96, 1–10. doi: 10.1016/j.jip.2007.02.014
Kovalchuk, A., and Driessen, A. J. M. (2010). Phylogenetic analysis of fungal ABC transporters. BMC Genomics 11:177. doi: 10.1186/1471-2164-11-177
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Letunic, I., and Bork, P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Li, W., Evans, J. D., Huang, Q., Rodríguez-García, C., Liu, J., Hamilton, M., et al. (2016). Silencing the Honey Bee (Apis mellifera) Naked Cuticle Gene (n.d.) improves host immune function and reduces Nosema ceranae infections. Appl. Environ. Microbiol. 82, 6779–6787. doi: 10.1128/AEM.02105-16
Li, X., Palmer, R., Trout, J. M., and Fayer, R. (2003). Infectivity of microsporidia spores stored in water at environmental temperatures. J. Parasitol. 89, 185–188. doi: 10.1645/0022-3395(2003)089[0185:iomssi]2.0.co;2
Liu, D., Hunt, M., and Tsai, I. J. (2018). Inferring synteny between genome assemblies: a systematic evaluation. BMC Bioinformatics 19:26.
Locher, K. P. (2016). Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493. doi: 10.1038/nsmb.3216
Lomsadze, A., Ter-Hovhannisyan, V., Chernoff, Y. O., and Borodovsky, M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33, 6494–6506. doi: 10.1093/nar/gki937
Lubelski, J., Konings, W. N., and Driessen, A. J. M. (2007). Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 71, 463–476. doi: 10.1128/mmbr.00001-07
Martin-Hernandez, R., Botias, C., Barrios, L., Martinez-Salvador, A., Meana, A., Mayack, C., et al. (2011). Comparison of the energetic stress associated with experimental Nosema ceranae and Nosema apis infection of honeybees (Apis mellifera). Parasitol. Res. 109, 605–612. doi: 10.1007/s00436-011-2292-9
Mayack, C., and Naug, D. (2009). Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J. Invertebr. Pathol. 100, 185–188. doi: 10.1016/j.jip.2008.12.001
Meng, X.-Z., Luo, B., Tang, X.-Y., He, Q., Xiong, T.-R., Fang, Z.-Y., et al. (2018). Pathological analysis of silkworm infected by two microsporidia Nosema bombycis CQ1 and Vairimorpha necatrix BM. J. Invertebr. Pathol. 153, 75–84. doi: 10.1016/j.jip.2017.12.005
Nakjang, S., Williams, T. A., Heinz, E., Watson, A. K., Foster, P. G., Sendra, K. M., et al. (2013). Reduction and expansion in microsporidian genome evolution: new insights from comparative genomics. Genome Biol. Evol. 5, 2285–2303. doi: 10.1093/gbe/evt184
Ndikumana, S., Pelin, A., Williot, A., Sanders, J. L., Kent, M., and Corradi, N. (2017). Genome analysis of pseudoloma neurophilia: a microsporidian parasite of zebrafish (Danio rerio). J. Eukaryot. Microbiol. 64, 18–30. doi: 10.1111/jeu.12331
Obbard, D. J., Gordon, K. H. J., Buck, A. H., and Jiggins, F. M. (2009). The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. B Biol. Sci. 364, 99–115. doi: 10.1098/rstb.2008.0168
Paldi, N., Glick, E., Oliva, M., Zilberberg, Y., Aubin, L., Pettis, J., et al. (2010). Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl. Environ. Microbiol. 76, 5960–5964. doi: 10.1128/aem.01067-10
Pan, G., Xu, J., Li, T., Xia, Q., Liu, S.-L., Zhang, G., et al. (2013). Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation. BMC Genomics 14:186. doi: 10.1186/1471-2164-14-186
Paumi, C. M., Chuk, M., Snider, J., Stagljar, I., and Michaelis, S. (2009). ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol. Mol. Biol. Rev. 73, 577–593. doi: 10.1128/mmbr.00020-09
Pelin, A., Moteshareie, H., Sak, B., Selman, M., Naor, A., Eyahpaise, M.-E., et al. (2016). The genome of an Encephalitozoon cuniculi type III strain reveals insights into the genetic diversity and mode of reproduction of a ubiquitous vertebrate pathogen. Heredity (Edinb) 116, 458–465. doi: 10.1038/hdy.2016.4
Pelin, A., Selman, M., Aris-Brosou, S., Farinelli, L., and Corradi, N. (2015). Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ. Microbiol. 17, 4443–4458. doi: 10.1111/1462-2920.12883
Peuvel, I., Peyret, P., Méténier, G., Vivarès, C. P., and Delbac, F. (2002). The microsporidian polar tube: evidence for a third polar tube protein (PTP3) in Encephalitozoon cuniculi. Mol. Biochem. Parasitol. 122, 69–80. doi: 10.1016/s0166-6851(02)00073-7
Piddock, L. J. V. (2006). Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 4, 629–636. doi: 10.1038/nrmicro1464
Pombert, J.-F., Haag, K. L., Beidas, S., Ebert, D., and Keeling, P. J. (2015). The Ordospora colligata genome: evolution of extreme reduction in microsporidia and host-to-parasite horizontal gene transfer. mBio 6:e02400-14. doi: 10.1128/mBio.02400-14
Pombert, J. F., Selman, M., Burki, F., Bardell, F. T., Farinelli, L., Solter, L. F., et al. (2012). Gain and loss of multiple functionally related, horizontally transferred genes in the reduced genomes of two microsporidian parasites. Proc. Natl. Acad. Sci. U.S.A. 109, 12638–12643. doi: 10.1073/pnas.1205020109
Pombert, J.-F., Xu, J., Smith, D. R., Heiman, D., Young, S., Cuomo, C. A., et al. (2013). Complete genome sequences from three genetically distinct strains reveal high intraspecies genetic diversity in the microsporidian Encephalitozoon cuniculi. Eukaryot. Cell 12, 503–511. doi: 10.1128/EC.00312-12
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Pryszcz, L. P., and Gabaldón, T. (2016). Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 44, e113. doi: 10.1093/nar/gkw294
Quinones, G. A., Jin, J., and Oro, A. E. (2010). I-BAR protein antagonism of endocytosis mediates directional sensing during guided cell migration. J. Cell Biol. 189, 353–367. doi: 10.1083/jcb.200910136
Reinke, A. W., Balla, K. M., Bennett, E. J., and Troemel, E. R. (2017). Identification of microsporidia host-exposed proteins reveals a repertoire of rapidly evolving proteins. Nat. Commun. 8:14023.
Rodriguez-Garcia, C., Evans, J. D., Li, W., Branchiccela, B., Li, J. H., Heerman, M. C., et al. (2018). Nosemosis control in European honey bees Apis mellifera by silencing the gene encoding Nosema ceranae polar tube protein 3. J. Exp. Biol. 221(Pt 19):jeb184606. doi: 10.1242/jeb.184606
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Santhoshkumar, S., Sivakumar, S., Vimal, S., Abdul Majeed, S., Taju, G., Haribabu, P., et al. (2017). Biochemical changes and tissue distribution of Enterocytozoon hepatopenaei (EHP) in naturally and experimentally EHP-infected whiteleg shrimp, Litopenaeus vannamei (Boone, 1931), in India. J. Fish Dis. 40, 529–539. doi: 10.1111/jfd.12530
Sedlazeck, F. J., Rescheneder, P., Smolka, M., Fang, H., Nattestad, M., von Haeseler, A., et al. (2018). Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468. doi: 10.1038/s41592-018-0001-7
Seppey, M., Manni, M., and Zdobnov, E. M. (2019). “BUSCO: assessing genome assembly and annotation completeness BT - gene prediction: methods and protocols,” in Bioinformatics, ed. M. Kollmar (New York, NY: Springer), 227–245. doi: 10.1007/978-1-4939-9173-0_14
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Smit, A., Hubley, R., and Green, P. (2015a). RepeatMasker. Open-4.0. Available online at: http://www.repeatmasker.org
Smit, A., Hubley, R., and Green, P. (2015b). Repeatmodeler. Open-1.0. Available online at: http://www.repeatmasker.org
Smriti, Krishnamurthy, S., Dixit, B. L., Gupta, C. M., Milewski, S., and Prasad, R. (2002). ABC transporters Cdr1p, Cdr2p and Cdr3p of a human pathogen Candida albicans are general phospholipid translocators. Yeast 19, 303–318. doi: 10.1002/yea.818
Soderlund, C., Bomhoff, M., and Nelson, W. M. (2011). SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res. 39:e68. doi: 10.1093/nar/gkr123
Southern, T. R., Jolly, C. E., Lester, M. E., and Hayman, J. R. (2007). EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryot. Cell 6, 1354–1362. doi: 10.1128/EC.00113-07
Stanke, M., Diekhans, M., Baertsch, R., and Haussler, D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644. doi: 10.1093/bioinformatics/btn013
Stentiford, G. D., Becnel, J. J., Weiss, L. M., Keeling, P. J., Didier, E. S., Williams, B.-A. P., et al. (2016). Microsporidia–emergent pathogens in the global food chain. Trends Parasitol 32, 336–348. doi: 10.1016/j.pt.2015.12.004
Timofeev, S., Tokarev, Y., and Dolgikh, V. (2020). Energy metabolism and its evolution in Microsporidia and allied taxa. Parasitol. Res. 119, 1433–1441. doi: 10.1007/s00436-020-06657-9
Tsaousis, A. D., Kunji, E. R. S., Goldberg, A. V., Lucocq, J. M., Hirt, R. P., and Embley, T. M. (2008). A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453, 553–556. doi: 10.1038/nature06903
Williams, B. A. P. (2009). Unique physiology of host-parasite interactions in microsporidia infections. Cell. Microbiol. 11, 1551–1560. doi: 10.1111/j.1462-5822.2009.01362.x
Williams, B. A. P., Hirt, R. P., Lucocq, J. M., and Embley, T. M. (2002). A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418, 865–869. doi: 10.1038/nature00949
Wiredu Boakye, D., Jaroenlak, P., Prachumwat, A., Williams, T. A., Bateman, K. S., Itsathitphaisarn, O., et al. (2017). Decay of the glycolytic pathway and adaptation to intranuclear parasitism within Enterocytozoonidae microsporidia. Environ. Microbiol. 19, 2077–2089. doi: 10.1111/1462-2920.13734
Wu, C., Chakrabarty, S., Jin, M., Liu, K., and Xiao, Y. (2019). Insect ATP-binding cassette (ABC) transporters: roles in xenobiotic detoxification and Bt insecticidal activity. Int. J. Mol. Sci. 20:2829. doi: 10.3390/ijms20112829
Xiao, C.-L., Chen, Y., Xie, S.-Q., Chen, K.-N., Wang, Y., Han, Y., et al. (2017). MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat. Methods 14, 1072–1074. doi: 10.1038/nmeth.4432
Xu, Y., Takvorian, P., Cali, A. N. N., and Weiss, L. M. (2003). Lectin binding of the major polar tube protein (PTPl) and its role in invasion. J. Eukaryot. Microbiol. 50, 600–601. doi: 10.1111/j.1550-7408.2003.tb00644.x
Xu, Y., Takvorian, P. M., Cali, A., Orr, G., and Weiss, L. M. (2004). Glycosylation of the major polar tube protein of Encephalitozoon hellem, a microsporidian parasite that infects humans. Infect. Immun. 72, 6341–6350. doi: 10.1128/iai.72.11.6341-6350.2004
Yang, D., Pan, G., Dang, X., Shi, Y., Li, C., Peng, P., et al. (2015). Interaction and assembly of two novel proteins in the spore wall of the microsporidian species Nosema bombycis and their roles in adherence to and infection of host cells. Infect. Immun. 83, 1715–1731. doi: 10.1128/IAI.03155-14
Zhang, K., Zhang, L.-J., Fang, Y.-H., Jin, X.-N., Qi, L., Wu, X.-C., et al. (2016). Genomic structural variation contributes to phenotypic change of industrial bioethanol yeast Saccharomyces cerevisiae. FEMS Yeast Res. 16:fov118. doi: 10.1093/femsyr/fov118
Keywords: honey bee, parasite, microsporidia, phylogeny, transporter, long reads
Citation: Huang Q, Wu ZH, Li WF, Guo R, Xu JS, Dang XQ, Ma ZG, Chen YP and Evans JD (2021) Genome and Evolutionary Analysis of Nosema ceranae: A Microsporidian Parasite of Honey Bees. Front. Microbiol. 12:645353. doi: 10.3389/fmicb.2021.645353
Received: 23 December 2020; Accepted: 29 April 2021;
Published: 02 June 2021.
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesReviewed by:
Paul Dean, Teesside University, United KingdomCopyright © 2021 Huang, Wu, Li, Guo, Xu, Dang, Ma, Chen and Evans. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Huang, cWlhbmctaHVhbmdAbGl2ZS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.