- 1State Key Laboratory of Crop Stress Biology for Arid Areas and College of Plant Protection, Northwest A&F University, Yangling, China
- 2Department of Botany and Plant Pathology and Center for Genome Research and Biocomputing, Oregon State University, Corvallis, OR, United States
Transcriptional plasticity enables oomycetes to rapidly adapt to environmental challenges including emerging host resistance. For example, the soybean pathogen Phytophthora sojae can overcome resistance conferred by the host resistance gene Rps1b through natural silencing of its corresponding effector gene, Avr1b-1. With the Phytophthora CRISPR/Cas9 genome editing system, it is possible to generate site-specific knock-out (KO) and knock-in (KI) mutants and to investigate the biological functions of target genes. In this study, the Avr1b-1 gene was deleted from the P. sojae genome using a homology-directed recombination strategy that replaced Avr1b-1 with a gene encoding the fluorescent protein mCherry. As expected, all selected KO transformants gained virulence on Rps1b plants, while infection of plants lacking Rps1b was not compromised. When a sgRNA-resistant version of Avr1b-1 was reintroduced into the Avr1b-1 locus of an Avr1b KO transformant, KI transformants with a well-transcribed Avr1b-1 gene were unable to infect Rps1b-containing soybeans. However, loss of expression of the incoming Avr1b-1 gene was frequently observed in KI transformants, which resulted in these transformants readily infecting Rps1b soybeans. A similar variability in the expression levels of the incoming gene was observed with AVI- or mCherry-tagged Avr1b-1 constructs. Our results suggest that Avr1b-1 may be unusually susceptible to transcriptional variation.
Introduction
Phytophthora sojae causes destructive root and stem rot diseases of soybeans and has been a model for molecular genetics research into oomycete plant pathogens (Tyler, 2007; Jiang and Tyler, 2012; Wang and Wang, 2018). The pathogenic mechanisms of Phytophthora pathogens have been extensively explored since the release of the P. sojae genome sequence (Tyler et al., 2006). In particular, hundreds of rapidly evolving effector proteins have been identified as playing a central role in manipulating host defenses and aiding pathogen infection and proliferation (Tyler and Gijzen, 2014; Wang Y. et al., 2019). Among those diverse infection-associated factors, a large superfamily of small secreted hydrophilic proteins with a conserved RXLR motif (RXLR effectors) has been characterized as key contributors to virulence by P. sojae and other oomycete plant pathogens (Dou et al., 2008a, b; Jiang et al., 2008; Jiang and Tyler, 2012; Wang and Wang, 2018; Wang Y. et al., 2019). The same superfamily includes most oomycete avirulence (Avr) determinants, which are proteins that enable recognition by host intracellular receptors encoded by major disease resistance (R) genes (Tyler and Gijzen, 2014). Recognition of avirulence determinants results in a vigorous defense response that often includes programmed cell death, called a hypersensitive response (HR) (Jones and Dangl, 2006). Most RXLR effector genes display transcriptional dynamics during host infection and other developmental stages (Wang et al., 2011; Ah-Fong et al., 2017).
The P. sojae RXLR effector Avr1b, encoded by the first cloned Avr gene of oomycete pathogens, Avr1b-1 (Shan et al., 2004), has been used as a probe in Phytophthora functional genomics (Dou et al., 2008a, b). Avr1b can be recognized by plants carrying the soybean resistance genes Rps1b or Rps1k in a gene-for-gene manner, triggering HR and leading to failure of infection (Song et al., 2013). Avr1b is capable of suppressing plant programmed cell death triggered by the mouse BAX protein (Dou et al., 2008b) and by other effector proteins (Wang et al., 2011). Constitutive expression of Avr1b-1 enhances the virulence of P. sojae (Dou et al., 2008b). Avr1b-mCherry fusion proteins mainly accumulated around haustoria during infection (Liu et al., 2014) and Avr1b can enter inside plant cells (Dou et al., 2008a; Kale et al., 2010; Tyler et al., 2013). Due to the existence of P. sojae natural isolates virulent on Rps1b plants, the Avr1b-1 gene was thought not to be essential for full virulence of P. sojae (Shan et al., 2004; Cui et al., 2012). One hypothesis is that paralogous RXLR genes may be functionally redundant with Avr1b-1. Another possibility is that numerous RXLR effectors quantitatively contribute to pathogenicity; thus, the variations in virulence from loss of one gene cannot be easily detected.
RXLR effector genes generally reside in gene-sparse regions of oomycete genomes (Tyler et al., 2006; Haas et al., 2009; Baxter et al., 2010). Diverse polymorphisms of those genes have been reported at both the DNA and RNA levels (Wang et al., 2011; Wang Q. et al., 2019). Besides various DNA sequence mutations, the loss of expression of RXLR effector genes encoding avirulence determinants has been described as a mechanism of P. sojae for overcoming a number of soybean resistance genes (Shan et al., 2004; Tyler and Gijzen, 2014; Chen et al., 2018; Wang Q. et al., 2019). For example, several virulence-conferring alleles of the P. sojae Avr1a, Avr1b, Avr1c, and Avr3a/5 genes displayed reduced transcript levels but had unchanged sequences relative to the respective avirulence-conferring alleles (Shan et al., 2004; Qutob et al., 2009, 2013; Dong et al., 2011). In the case of P. sojae Avr3a, the gene has been shown to undergo transgenerational gene silencing, resulting in loss of the avirulence-conferring phenotype (Qutob et al., 2013). Both histone methylation-induced transcriptional gene silencing (Wang et al., 2020) and double-stranded RNA-mediated posttranscriptional silencing (Wang Q. et al., 2019) have been proposed to be responsible for natural silencing of Avr1b-1 in the isolate P6497.
Recently, a Phytophthora-specific clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-mediated genome editing tool was developed for efficient gene knock-out (KO) and replacement (knock-in; KI) in P. sojae via either non-homologous end-joining (NHEJ) or homology-directed recombination (HDR) (Fang and Tyler, 2016). Soon after, an “all-in-one” system was developed by inserting the Cas9 and single guide RNA (sgRNA) cassette into one construct with a single neomycin phosphotransferase II (npt II) selection marker that substantially enhanced the efficiency of heritable genome modifications (Fang et al., 2017). As a result, knock-out and complementation of target genes have become basic requirements for functional genomics research in P. sojae. The tool has subsequently been deployed in other Phytophthora species including P. capsici (Chen et al., 2019; Wang W. et al., 2019), P. palmivora (Gumtow et al., 2018), P. parasitica (Zhang et al., 2020), and P. litchii (Situ et al., 2020), as well as in the animal pathogenic oomycete Aphanomyces invadans (Majeed et al., 2018).
In this study, we employed the Phytophthora-specific CRISPR/Cas9 system to knock out the RXLR effector gene Avr1b-1 of P. sojae and to evaluate the pathotype of Avr1b-1-deletion transformants using qualitative and quantitative inoculation assays. The results confirmed that Avr1b-1 is essential for avirulence of P. sojae on Rps1b soybean plants but not essential for full virulence. Reintroduction of sgRNA-resistant versions of Avr1b-1 was also completed using the same genome editing tool, in combination with a novel strategy for repeated reuse of the G418 resistance selection marker. All KI transformants carrying well-transcribed Avr1b-1 genes exhibited restored avirulence phenotypes on Rps1b plants. More interestingly, CRISPR-mediated gene replacements into both Avr1b-1 deletion transformants and wild-type isolate produced large variations in transcript levels of Avr1b-1, suggesting that the Avr1b-1 locus may be unusually susceptible to transcriptional polymorphisms.
Materials and Methods
P. sojae Isolates and Culture Conditions
P. sojae strains P6497 and P7063 were routinely cultured and maintained on clarified V8 agar plates (10% V8 juice with 0.01% w/v CaCO3 and 0.002% w/v β-sitosterol) at 25°C in the dark. Zoospores were induced and harvested from approximately 1 week-old cultures grown on clarified V8 agar or in clarified V8 broth, by flooding the plates or mycelia with sterile distilled water, followed by filtering through 50-mm nylon mesh to remove mycelial fragments (Fang and Tyler, 2016). P. sojae transformants were incubated in 12-well plates (Corning, NY, United States) containing V8 medium supplemented with 50 μg/ml G418 for 3–5 days before small-scale genomic DNA extraction. All transformants were routinely grown on clarified V8 plates with or without appropriate antibiotics for successive transfer.
Soybean Culture Conditions and Inoculation
Soybean plants were grown as described previously (Dou et al., 2008b) with some modifications. Seedlings were grown in a growth chamber with 16 h light at 28°C and 8 h darkness at 25°C. For inoculation assays, 1 week-old seedlings (used for hypocotyl inoculation) or unifoliate leaves of 9–12 day-old seedlings were inoculated with 10–20 μl P. sojae zoospore suspension (∼300 zoospores) and maintained in the growth chamber with 100% humidity. For hypocotyl inoculation assays, differences between numbers of surviving plants from Williams (rps1b) and L77-1863 (Rps1b) cultivars were compared using Fisher’s exact test with a P-value cutoff of 0.01. Strains showing significant differences between Williams (rps1b) and L77-1863 (Rps1b) cultivars were judged as avirulent. Quantitative virulence of P. sojae transformants was determined by measuring the lesion size. We analyzed 10 leaves within each treatment, and each treatment was repeated at least three times. Statistical analyses were performed by the Wilcoxon rank sum test with a cutoff of P < 0.001.
sgRNA Design and Plasmid Construction
sgRNAs were designed as described by Fang et al. (2017). In total, six synthesized DNA fragments encoding Avr1b-targeted and mCherry-targeted sgRNAs were individually inserted into the BsaI and NheI sites of “all-in-one” CRISPR/Cas9 vector pYF515 using annealing and ligation steps. The cassette harboring each sgRNA was verified by colony-PCR and Sanger sequencing. All HDR constructs contain an upstream flanking region, Avr1b or mutant’s sequence, and a downstream flanking region. Three fragments of interest were inserted into the EcoRI and HindIII restriction sites of pBluescript II KS via In-Fusion Cloning (TaKaRa, China). PCR amplification of the 1-kb upstream and downstream flanking ends was conducted with primer pairs Fl-Avr1b/Rl-Avr1b and Fr-Avr1b/Rr-Avr1b. PCR products of Avr1b or mCherry were obtained with primer pairs PsF/PsR and 1bmcF/1bmcR. The sgRNA-resistant version of Avr1b (Avr1bsgR) was synthesized from the company (GeneCreate, China) with multiple synonymous substitutions in three sgRNA targeting sites (Supplementary Figure S4A). The biotin-acceptor AVI-tag was fused to the C-terminus of Avr1bsgR by PCR using primer pairs PsF/Ps4WTR1/Ps4WTR2. The Avr1bsgR-mCherry fusion was obtained via fusion PCR using primer pairs PsF/1bmcR, 1bmcR/R-mcR, and PsF/R-mcR. The Avr1bsgR-AVI or Avr1bsgR-mCherry was used to replace Avr1b or mCherry in P. sojae transformation (Supplementary Figure S1). The vector for particle bombardment-mediated transient expression was as described by Dou et al. (2008b). Avr1b or Avr1bsgR was amplified with primer 1b-F/1b-R and was inserted into the SmaI and KpnI restriction sites of pUC1b via T4 DNA ligation (TaKaRa, China). High concentrations of plasmids for bombardment were prepared as described by Zhao et al. (2020). Standard molecular techniques were performed according to the instructions from kit manufacturers. The oligonucleotides used in this study are listed in Supplementary Table S3.
Transformation of P. sojae
Polyethylene glycol-mediated transformations were performed as reported previously (Fang et al., 2017). Specifically, young mycelia were collected by inoculating 30 ml of nutrient pea broth in 90 × 20 mm petri dishes with 30–40 thin agar disks (each 0.5 cm in diam.). Mycelial mats were rinsed twice with sterilized H2O and 0.8 M mannitol, followed by a 5 min wash in 0.8 M mannitol before enzyme digestion. Protoplasts were generated with a combination of 1% Trichoderma Lysing Enzymes (Sigma, China) and 0.1% CELLULYSIN® Cellulase (Merck, China) in Fry buffer (0.4 M mannitol, 20 mM KCl, 20 mM MES, pH 5.7, 10 mM CaCl2). The density of protoplasts was monitored to ensure that a minimum of 1 × 107 per ml was achieved. About 30–40 μg of plasmid DNA was used for transformation. Protoplasts were regenerated overnight in pea broth containing 0.4 M mannitol. Regenerated protoplasts were collected and distributed onto plates of regeneration media containing 50 μg/ml of G418 for 2–3 days. Half of the G418-insensitive colonies were transferred to 12-well plates containing V8 broth supplemented with 50 μg/ml of G418 and incubated for 3–4 days at 25°C before DNA extraction. The other half were routinely subcultured on V8 agar plates without antibiotics.
Zoospore Isolation
To obtain homokaryotic colonies, zoospore purification was routinely performed for selected transformants as described by Fang et al. (2017) with some modifications. Briefly, fresh cultures of PCR-verified transformants were flooded with sterile dH2O 6–8 times. Then, the mycelial colony (60 mm plate) was covered with 3 ml sterile dH2O, and zoospore release usually began after 3–4 h of room temperature incubation. Zoospore suspensions were diluted to 1 × 103 per milliliter. Zoospore drops (1 μl per drop) were loaded onto V8 plates and incubated at 25°C for no more than 24 h. With a light microscope, single zoospore-derived mycelial colonies were selected and transferred to a new V8 plate for further culturing.
DNA and RNA Isolation and cDNA Synthesis
Total gDNA was extracted as described by Fang and Tyler (2016). Total RNAs were extracted from zoospore-inoculated hypocotyls or unifoliate leaves with the RNeasy Plant Mini Kit (Qiagen, Germany). Both oligo-(dT)18 and random primer mix were used for first-strand cDNA synthesized by SuperScriptTM III (Thermo Fisher, China) according to the product instructions.
Semiquantitative and Quantitative PCR Analysis
Semiquantitative RT-PCR was carried out with 1 μl of fivefold diluted cDNA products in a 20 μl volume containing Taq PCR Mastermix (Tiangen, Beijing, China) and matching primers. PCR conditions consisted of one cycle of 95°C for 3 min, followed by 30 cycles of a three-step procedure (1 min at 95°C, 1 min at 56°C, and 1 min at 72°C) and a final step of 5 min at 72°C. Quantitative PCR of genomic DNA for biomass measurements and quantitative reverse transcription PCR (qRT-PCR) for transcript measurements were performed as previously described (Gu et al., 2020) using CFX connect (Bio-Rad, CA, United States). Genomic DNA levels or gene expression levels relative to the reference gene were calculated with the 2–ΔΔCT method. The Actin genes of P. sojae and soybean were chosen as the reference genes (Wang et al., 2011).
Detection and Validation of Targeted Mutagenesis
PCR amplifications using Taq DNA polymerase (TaKaRa, Japan) were conducted to detect the targeted mutations in transformants. Approximately 10 ng of gDNA was used as PCR template. To detect HDR events in individual transformants, primers located outside the Avr1b homology arms and within the Avr1b-1 locus were used. For screening homozygous transformants, PCR was performed with primers spanning from outside the homology arms to within the Avr1b-1 or mCherry coding regions.
Transient Expression by Particle Bombardment of Soybean Leaves
The unifoliate leaves of soybean seedlings were selected for bombardment using a double-barreled extension of the Bio-Rad He/1,000 particle delivery system (Kale and Tyler, 2011). Plasmid DNA was isolated with the EndoFree Maxi Plasmid Kit (Tiangen, China) and concentrated to 5–6 mg/ml in sterile H2O. Tungsten particles were prepared as reported previously (Dou et al., 2008a). Plasmids encoding beta-glucuronidase (GUS) mixed with a GFP control or a mixture of plasmids encoding Avr1b proteins and GUS were delivered into host cells side by side via the double barrel gene gun. Parameters were set as reported (Gu et al., 2011). After bombardment, the leaves were incubated for 2–3 days in darkness at 25°C and then stained for at least 4 h at 37°C in GUS staining solution. Blue spots were counted under a dissecting microscope at × 8– × 160 magnification. For each paired shot (GUS + Avr1b or Avr1bsgR vs. GUS + GFP), the logarithm of the ratio of the spot numbers with the fusion protein compared with that of the control was calculated, and then the log ratios obtained from the Rps1b and rps1b leaves were compared by the Wilcoxon rank sum test (Kale and Tyler, 2011).
Confocal Microscopy
A laser scanning confocal microscope FV3000 (Olympus, Japan) was used to examine the expression and subcellular localization of mCherry or mCherry-fused proteins in transformants upon infection. Living hyphae or zoospores were inoculated onto the hypocotyls of etiolated soybean seedlings grown for 7–10 days in darkness. Images were captured using a × 63 oil objective with excitation/emission settings of 561 nm/570–630 nm for mCherry. Calibrations of gain settings were performed with control hypocotyls as a range of background fluorescence to avoid capturing background fluorescence.
Results
Avr1b-1 KO Transformants Fail to Trigger HR on Rps1b Soybean Plants
To confirm that Avr1b-1 is essential for conferring HR on soybean plants carrying Rps1b, we employed CRISPR/Cas9-dependent mutagenesis to delete Avr1b-1 by gene replacement. The P. sojae isolate P7063 carries a single copy of Avr1b-1 and lacks the close paralog Avh1; this strain was used as the recipient strain for Avr1b-1 replacement. Based on in silico analysis, three sgRNAs matching different locations in the Avr1b-1 coding region were designed to guide Cas9 cleavage that would trigger homology direct repair (HDR) (Supplementary Figure S1 and Supplementary Table S1). One of the three sgRNAs, sg114R, produced the highest editing efficiency, with an average frequency of homozygous replacement events of 45%. In comparison, sg19 and sg285 yielded only a few homozygous transformants. Following PCR verification (Supplementary Figure S2A), two independent homozygous transformants Δ114-1-7 and Δ114-2-2, together with one non-replacement transformant Δ114-1-11f, were selected for further analysis. No abnormalities in morphology or fertility were observed in the selected mutants (Supplementary Table S2). All genes in the Avr1b-1 locus (either Avr1b-1 itself or mCherry) were normally transcribed during infection according to semiquantitative RT-PCR analysis (Supplementary Figure S2B). Next, the transformants, along with wild-type isolates P6497 and P7063, were subjected to avirulence analysis on soybean plants with or without the Rps1b gene. As shown in Figure 1, the Avr1b-1 KO transformants Δ114-1-7 and Δ114-2-2, plus P6497, which is naturally silenced at Avr1b-1, successfully infected the Rps1b soybean seedlings, suggesting that Rps1b-dependent disease resistance was not initiated. In contrast, the non-replacement transformant Δ114-1-11f and the control isolate P7063 failed to infect the Rps1b soybean plants. All wild-type isolates and transformants killed rps1b soybean seedlings (Figures 1A,B).
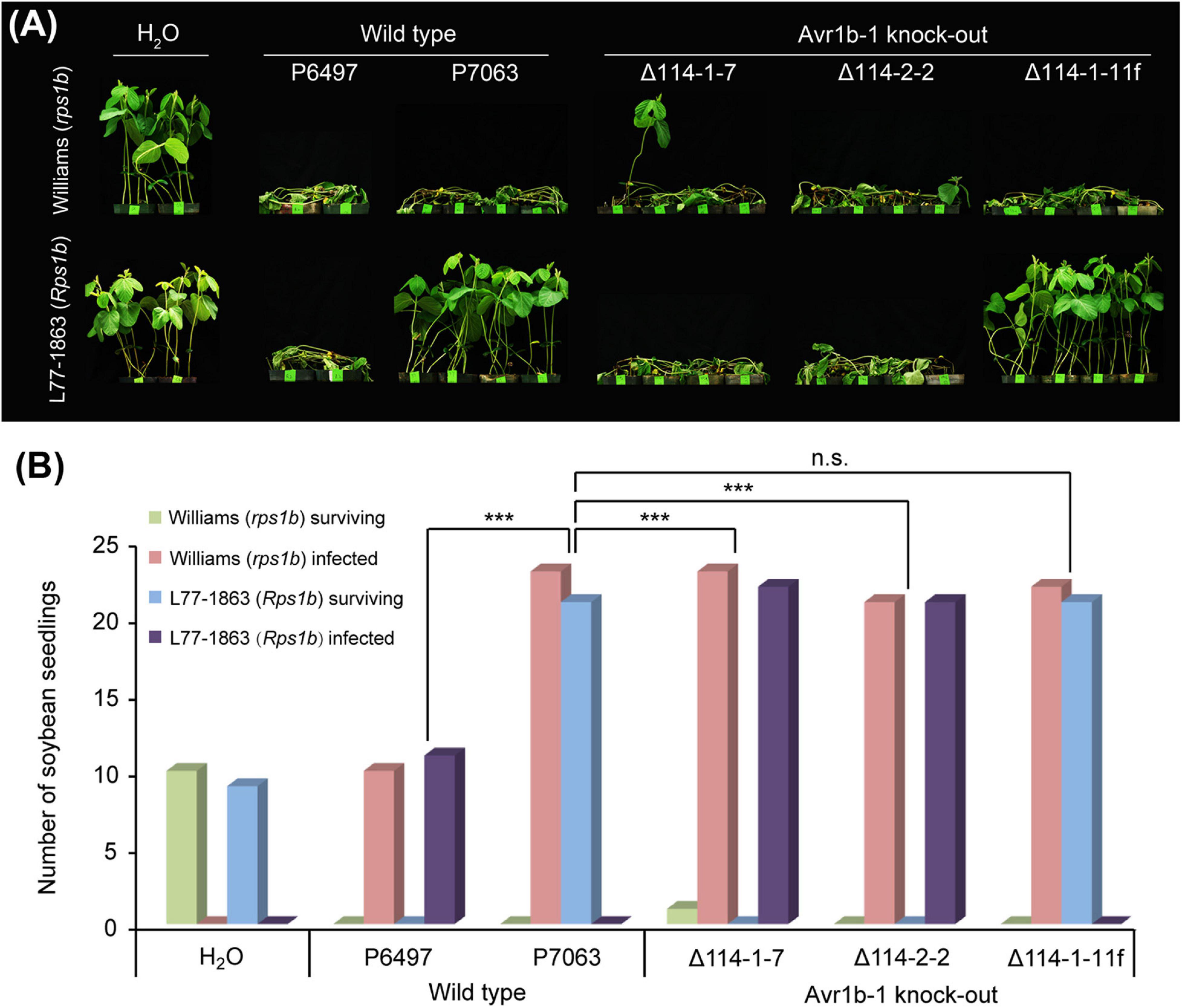
Figure 1. Avirulence phenotypes of Avr1b-1 knock-out transformants measured by hypocotyl inoculations. (A) Avr1b-1-silenced wild-type isolate P6497, Avr1b-1-expressing wild-type isolate P7063, and two homozygous Avr1b-1 deletion transformants Δ114-1-7 and Δ114-2-2, plus a non-replacement transformant Δ114-1-11f were inoculated onto the hypocotyls of 7 day-old seedlings of soybean cultivar Williams (rps1b) or L77-1863 (Rps1b). Pictures were taken 3 days after wound inoculation. (B) Numbers of infected and surviving seedlings after inoculation with the P. sojae strains. A strain was deemed to be avirulent if the number of inoculated Rps1b seedlings surviving was significantly higher than the number of surviving seedlings without the Rps1b gene, and the number was not significantly different from the number of surviving seedlings inoculated with wild-type P7063. At least eight seedlings were used for the inoculation of each control or transformant. The significance of differences was determined by Fisher’s exact test (P < 0.001). *** = significant; n.s. = not significant.
To quantitatively measure the resistance in L77-1863 (Rps1b) or Williams (rps1b) soybean cultivars, we inoculated soybean leaves with zoospores and monitored the lesion progression. The rate of lesion progression on the unifoliate leaves was measured, and no statistically significant differences were observed for any of the isolates and transformants infecting Williams (rps1b) soybean leaves (Figure 2). However, on L77-1863 leaves (containing Rps1b), strains P6497, Δ114-1-7, and Δ114-2-2 formed lesions that were significantly larger than the lesions produced by control strains P7063 and Δ114-1-11f (Figures 2A,B). Moreover, when the relative biomass of the pathogen was measured at 72 h post-inoculation (hpi) as genomic DNA ratios by qPCR, the biomasses of the Avr1b-1 KO mutants were about 45—110-fold greater on Rps1b soybean L77-1863 than those of the Avr1b-1-expressing transformant and P7063 control (Figure 2C). These results were consistent with the hypocotyl infection assay and confirmed that Avr1b-1 is necessary and sufficient for Rps1b recognition to trigger HR.
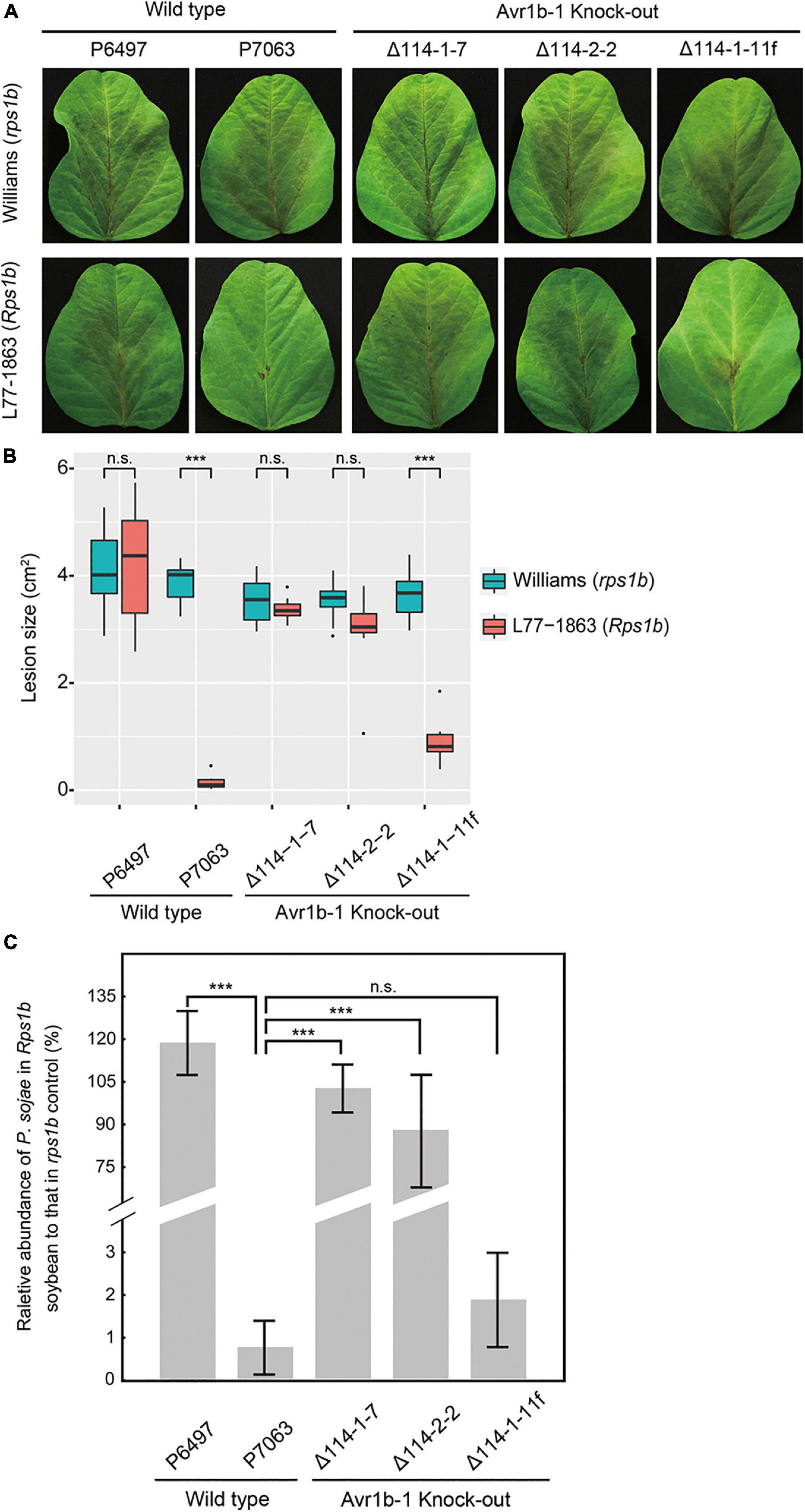
Figure 2. Avirulence phenotypes of Avr1b-1 knock-out transformants measured by detached leaf inoculations. (A) Soybean leaf lesions 3 days after zoospore inoculation of Williams (rps) or L77-1863 (Rps1b) unifoliate leaves. (B) Sizes of lesions after 3 days. Lesions on eight unifoliate leaves were measured in each of three independent experiments. The relative lesion size was measured from photographs with ImageJ software. Lesion sizes were compared between Williams (rps) and L77-1863 (Rps1b) using the Wilcoxon rank sum test. ***P < 0.001, n.s. P > 0.1. (C) Avirulence phenotypes of Avr1b-1 knock-out transformants measured by real-time PCR-based quantification of P. sojae genomic DNA in infected tissue. Proliferation of P. sojae was plotted as the ratio between the abundance of PsActin relative to GmActin DNA in infected L77-1863 leaves compared with that in infected Williams leaves, at 72 hpi. Error bars represent standard deviation of three technical replicates. This experiment was replicated three times. Error bars represent the mean ± standard deviation. P-values were determined by the Wilcoxon rank sum test; *** indicates P < 0.001.
Avr1b-1 Is Not Required for Full Virulence of P. sojae
Overexpression of Avr1b-1 in P. sojae confers increased virulence on soybean plants compared with the recipient strain (Dou et al., 2008b), but natural loss of Avr1b-1 transcripts does not compromise the virulence of P. sojae strain P6497 (Shan et al., 2004). To definitively confirm this result in a strain that normally expresses Avr1b-1, we measured the virulence of the P7063 KO mutants. P7063 and P6497, together with KO transformants Δ114-1-7, Δ114-1-8, Δ114-2-2, and non-replacement transformant Δ114-1-11f were assessed by zoospore inoculation of the leaves. Based on the lesion sizes, no significant virulence differences were observed among the six strains (Figures 3A,B). Furthermore, relative biomasses of the Avr1b-1 KO mutants measured on soybean plants at 72 hpi were similar to those produced by wild-type strains P7063 and P6497 and the non-replacement transformant (Figure 3C). These results demonstrated that the RXLR effector gene Avr1b-1 was not required for the full virulence of strain P7063 under the conditions of this assay.

Figure 3. Virulence phenotypes of Avr1b-1 knock-out transformants on Williams soybean leaves. (A) Avr1b-1-silenced isolate P6497; Avr1b-1-expressing isolate P7063; three homozygous Avr1b-1 deletion transformants Δ114-1-7, Δ114-1-8, and Δ114-2-2; plus non-HDR transformant Δ114-1-11f were inoculated onto unifoliate leaves of Williams (rps1b). Pictures were taken 3 days after inoculation. (B) Average lesion sizes determined from at least 10 inoculated unifoliate leaves in each of three independent experiments. The relative lesion sizes were measured by ImageJ software. Error bars represent the mean ± standard deviation. P-values were determined by the Wilcoxon rank sum test. (C) Virulence phenotypes of Avr1b-1 knock-out transformants measured by real-time PCR-based quantification of P. sojae genomic DNA in infected tissue. Error bars represent standard deviation of three technical replicates. This experiment was replicated three times. Error bars represent the mean ± standard deviation.
Repeated Use of Neomycin Phosphotransferase II (npt II) as a Selection Marker for Complementation
We noticed that successive subculture of the homozygous Avr1b-1 KO transformants in the absence of the antibiotic G418, used for the initial transformation, resulted in loss of G418 resistance. Most of the transformants, including Δ114-1-7, could not survive in media containing 50 μg/ml G418, while a smaller fraction of transformants, such as Δ114-2-2, retained weak growth in G418 media (Figure 4A). This observation raised the possibility that G418 could be used as a selection marker in complementation experiments using G418-sensitive transformants as recipients. The npt II genes in those G418-sensitive and semisensitive transformants were analyzed at both the DNA and RNA levels. As shown in Figure 4B, the npt II gene could be detected in genomic DNA from all transformants, but npt II was not detected in the control strain P7063 (with Actin as a positive control). When the npt II transcript levels were assessed, npt II could be detected in a transformant Δ114-1-8, which retained G418 resistance, and in Δ114-2-2, which retained weak G418 resistance. However, transcripts of npt II could not be detected in the G418-sensitive transformant Δ114-1-7 nor in the wild-type control, P7063. Thus, it appeared that the npt II gene had become partially or fully silenced in Δ114-2-2 and Δ114-1-7, respectively. To test if the npt II silencing in these two KO lines was stable enough for G418 selection following transformation and to test if the silencing of the ectopic npt II genes could be overcome by incoming plasmid DNA carrying npt II, we used both Δ114-1-7 and Δ114-2-2 as recipients to perform protoplast regeneration and transformation with a construct carrying mCherry-tagged Avr1b. Using the partially silenced Δ114-2-2 as the recipient resulted in numerous false-positive colonies on G418-containing plates during regeneration (Supplementary Figures S3A,B). In contrast, when Δ114-1-7 was used as a recipient, no G418-resistant colonies were observed in the absence of npt II-containing plasmid DNA. On the other hand, genuine transformants could be obtained using npt II-containing plasmids (Supplementary Figure S3B), indicating that silencing of the resident npt II gene did not affect the expression of incoming npt II genes. These results indicated that full loss of G418 resistance resulting from complete silencing of npt II in the recipient transformant was required for successful gene complementation when using G418 as a selection marker for the second time.

Figure 4. G418 sensitivity of Avr1b-1 knock-out transformants. (A) Wild-type P7063 and three selected Avr1b-1 deletion transformants cultured on clarified V8 medium with or without 50 μg/ml G418. Transformants Δ114-1-7 and Δ114-2-2 were isolated from G418 regeneration medium and then subcultured in V8 medium. Transformant Δ114-1-8 was maintained on V8 medium containing 50 μg/ml G418. (B) PCR detection of npt II sequences in genomic DNA and RNA of Avr1b-1 knock-out transformants and P7063 with primer pair qNptF/R. P. sojae Actin was used as the reference gene.
sgRNA-Resistant Version of Avr1b-1 for Complementation Experiments
As a prelude to conducting Avr1b-1 complementation experiments, we introduced synonymous substitutions at sgRNA binding sites in Avr1b-1 to prevent the potential cleavage of the incoming DNA by the previously introduced Cas9 protein in the transformants (Supplementary Figure S4A). To validate whether the sgRNA-resistant version of Avr1b-1 (Avr1bsgR) could be recognized by Rps1b, we used double-barreled particle bombardment (Dou et al., 2008a, b; Kale and Tyler, 2011) to co-express either Avr1b-1 or Avr1bsgR with a GUS reporter in rps1b and Rps1b soybean leaves. Compared with the GFP control, the number of surviving GUS-positive (blue) patches was reduced by about 80% in Rps1b soybean leaves in the presence of Avr1b-1 or Avr1bsgR, indicating that HR was activated in the soybean cells (Figure 5). In rps1b soybean leaves, Avr1b-1 and Avr1bsgR produced no reduction of GUS blue spots compared with GFP, indicating that no HR occurred (Figure 5B). These results confirmed that Avr1bsgR could produce a functional Avr1b gene product.
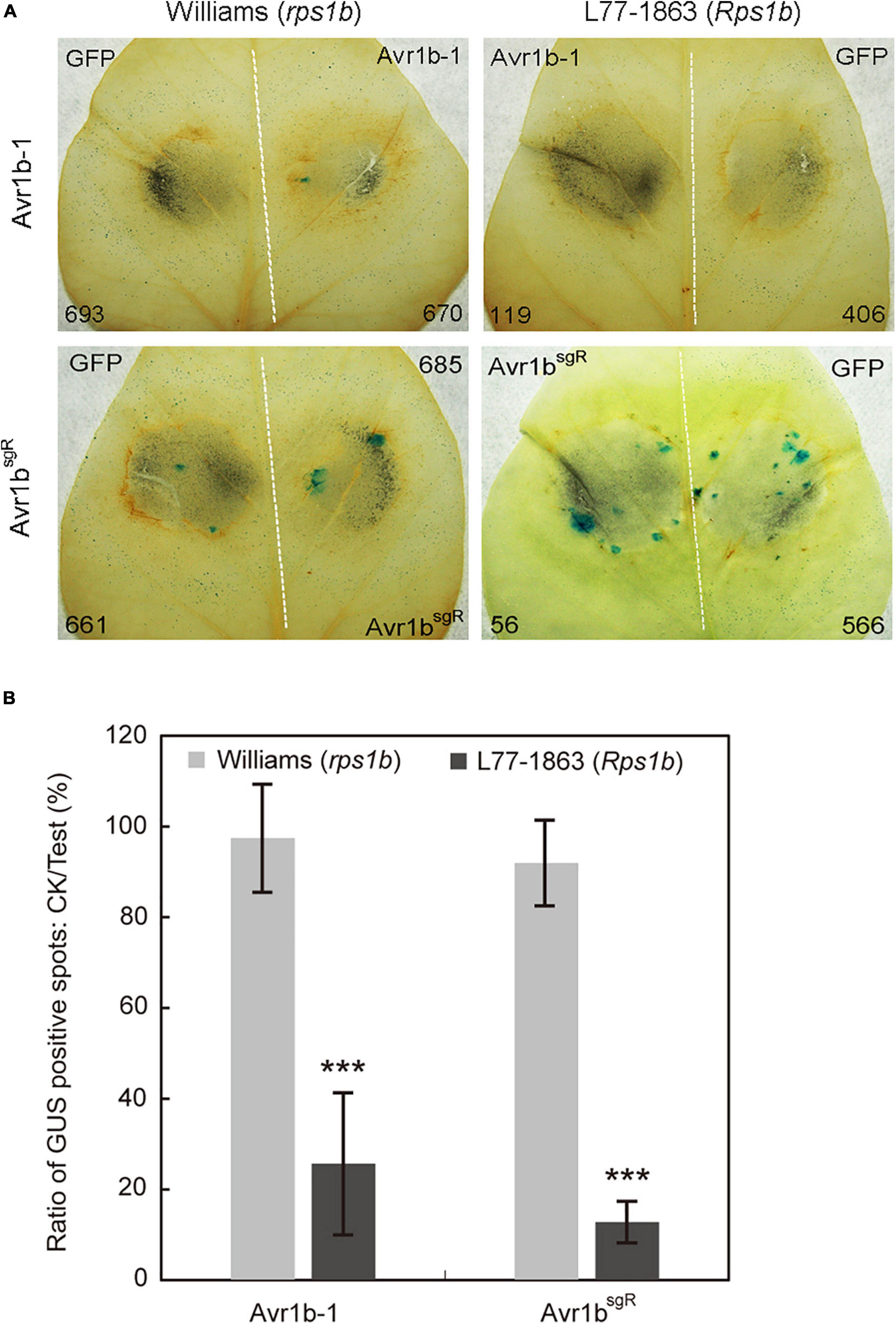
Figure 5. Transient expression of Avr1b-1sgR leads to cell death in Rps1b soybean leaves. (A) Soybean leaves from Williams (rps1b) and L77-1863 (Rps1b) were subjected to double-barreled particle bombardment with pairs of samples containing GUS reporter DNA plus either Avr1bsgR DNA or GFP control DNA. After staining, the numbers of blue spots on Williams (rps1b) and L77-1863 (Rps1b) leaves were counted. A functional Avr1b-1 gene is expected to significantly reduce the number of blue spots on L77-1863 (Rps1b) leaves, but not Williams (rps1b) leaves, due to triggering Rps1b-mediated cell death. The white dotted line separates the two bombardment sites. Numbers of GUS spots counted are indicated. (B) Ratios between the numbers of GUS spots produced in the presence of Avr1b-1 vs. control GFP DNA on each leaf, averaged from 10 soybean leaves, in a single experiment. Error bar represents the mean ± standard deviation. The significance of the difference between Williams and L77-1863 leaves was determined by the Wilcoxon rank sum test (Gu et al., 2011), ***P < 0.001.
Reintroduction of Avr1bsgR Into Deletion Transformants Restores Their Avirulence Phenotypes on Soybean Plants Containing Rps1b
In order to confirm that the deletion of Avr1b-1 was responsible for the loss of soybean resistance conferred by Rps1b, Avr1bsgR was introduced into Avr1b KO transformant Δ114-1-7 by replacement of the mCherry gene that was residing at the Avr1b-1 locus. Three sgRNAs, mc223, mc370, and mc391, targeted to different locations in mCherry, were designed to guide Cas9 cleavage and HDR (Supplementary Table S1 and Supplementary Figure S4B). Based on PCR assays (Supplementary Figure S5), three homozygous complementation transformants, C223-2, C223-3, and C391-10, and two non-replacement transformants, C223-5f and C391-12f, were identified. No morphological or fertility changes were observed in any of these P. sojae strains. The strains, together with Avr1b-1 non-replacement transformant Δ114-1-11f and wild-type strain P7063, were subjected to avirulence analysis on soybean plants with or without the Rps1b gene. Of the Avr1bsgR complementation transformants, C223-3 and C391-10 failed to infect Rps1b leaves but successfully infected Williams (rps1b) leaves, demonstrating that the sgRNA-resistant Avr1b gene could complement the Avr1b-1 KO (Figures 6A,B). The genomic biomass measurements were fully consistent with the lesion sizes on susceptible and resistant plants (Figure 6C). As expected, non-replacement transformants, C223-5f and C391-12f, were fully virulent on Rps1b as well as rps leaves. Surprisingly, however, homozygous complementation transformant C223-2 was virulent on both Rps1b and rps1b plants. RT-PCR analysis revealed that C223-2 failed to produce any detectable Avr1bsgR transcript, whereas Avr1bsgR transcripts were readily detected in C223-3 and C391-10 as well as in the controls P7063 and Δ114-1-11f (Figure 6D).
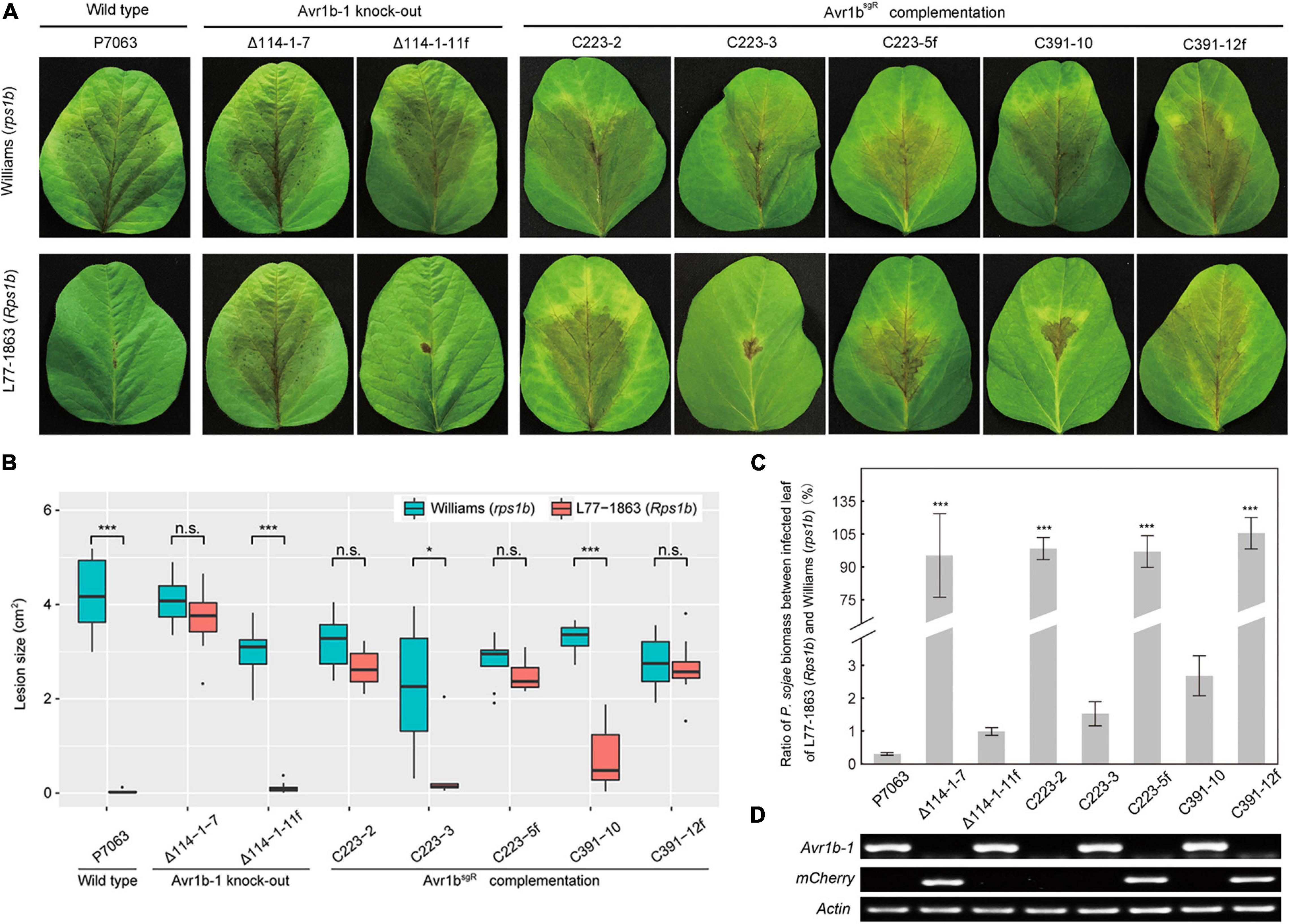
Figure 6. Evaluation of avirulence activity of Avr1bsgR knock-in transformants. (A) Avr1bsgR knock-in transformants (C223-3 and C391-10) and control strains (wild-type P7063, Δ114-1-7, Δ114-1-11f, C223-5f, and C391-12f) were tested for avirulence by zoospore inoculation onto unifoliate leaves of Williams (rps1b) and L77-1863 (Rps1b). Pictures were taken 3 days after inoculation. (B) Average lesion sizes on the leaves of Williams (rps1b) and L77-1863 (Rps1b) measured on 10 unifoliate leaves from each of three independent experiments. Sizes were measured with ImageJ software. Error bars represent the mean ± standard deviation. P-values were calculated with Wilcoxon rank sum test, * and *** indicate P-value < 0.05 and < 0.001, respectively. (C) Avirulence phenotypes of Avr1b-1 knock-in transformants measured by real-time PCR-based quantification of P. sojae genomic DNA in infected tissue. Proliferation of P. sojae is plotted as the ratio between abundance of PsActin relative to GmActin DNA in infected L77-1863 leaves compared with Williams leaves at 72 hpi. This experiment was replicated three times. Error bars represent the mean ± standard deviation. P-values were determined by the Wilcoxon rank sum test. ***P < 0.001. (D) RT-PCR analysis of Avr1b-1 and mCherry transcripts in Avr1b-1 knock-in transformants. P. sojae Actin was used as the reference gene.
Silencing of the Avr1b-1 Locus in Gene Replacement Transformants
In order to produce transformants expressing tagged versions of Avr1b from the Avr1b-1 locus, for Avr1b function experiments, genes encoding mCherry-tagged or AVI-tagged Avr1bsgR were used to replace Avr1b-1 in wild-type strain P7063 directly, using Avr1b-1 sgRNAs, sg114R. In each case, based on PCR screening, 20–45% of the candidates screened were homozygous replacement transformants (Supplementary Figure S6). Three homozygous replacement transformants and one non-replacement transformant were selected for each of Avr1bsgR-mCherry (C8W1, C8W2, C8W15, and C8W3f, respectively) and Avr1bsgR-AVI (C4W3, C4W6, C4W7, and C4W4f, respectively) and then subjected to virulence tests. Equal concentrations of zoospores produced by individual transformants or wild-type P7063 were inoculated onto both Williams (rps1b) and L77-1863 (Rps1b) soybean plants. Homozygous transformants C4W3 and C8W1 and the positive controls C4W4f, C8W3f, and P7063 exhibited the expected avirulence phenotype, failing to infect Rps1b plants and manifesting an HR (Figures 7A,B). Surprisingly, however, C4W6, C4W7, C8W2, and C8W15 infected Rps1b and rps leaves equally well, like the negative control, KO strain Δ114-1-7. The lesion size data were confirmed by the genomic biomass measurements on rps and Rps1b leaves (Figure 7C). Thus, these four strains had lost their avirulence phenotype.
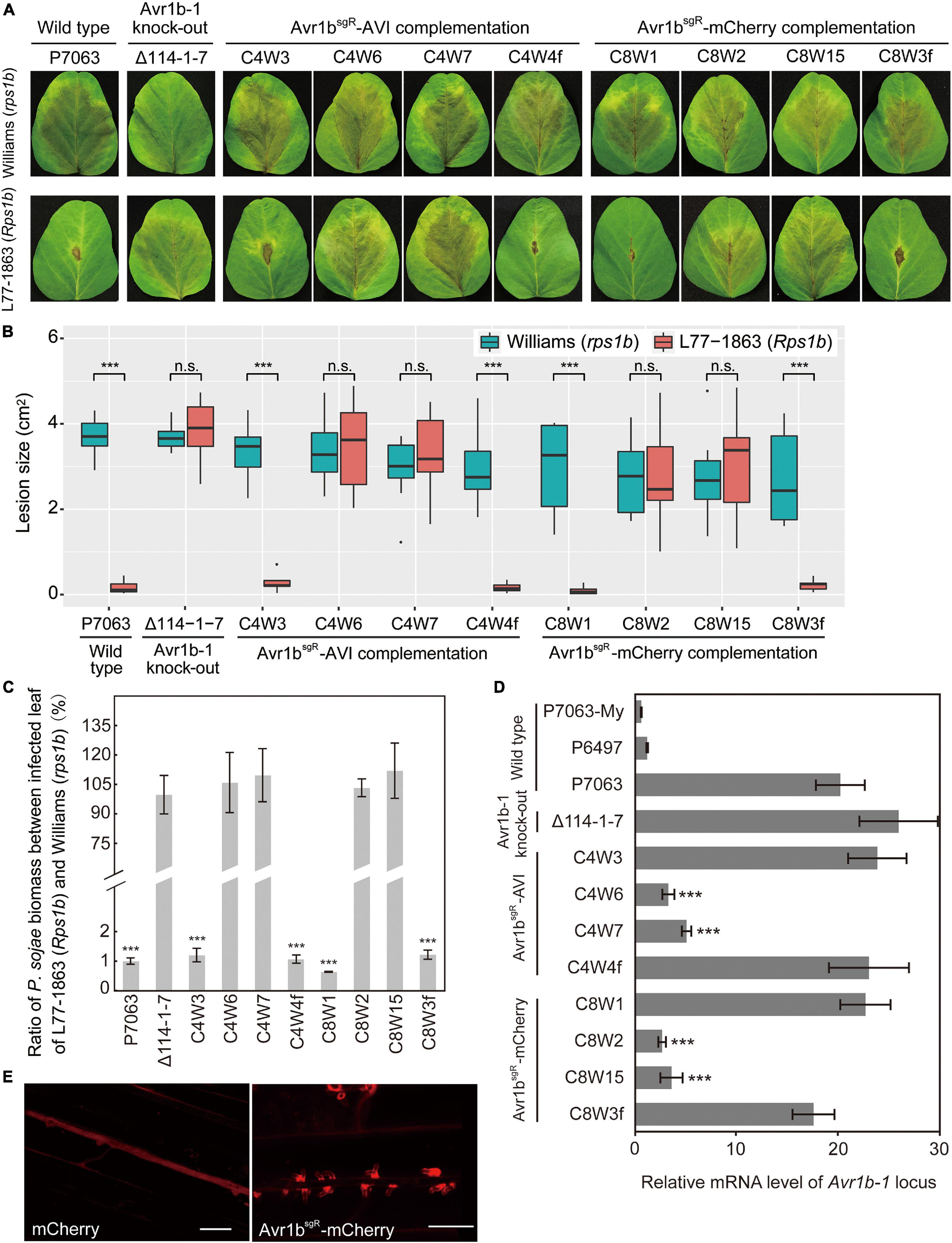
Figure 7. Characterization of Avr1b-1 knock-in transformants derived from Avr1b-1sgR fusion genes. (A) Lesions produced by transformants and control strains. Transformants were Avr1bsgR-AVI KI strains C4W3, C4W6, and C4W7, and Avr1bsgR-mCherry KI strains C8W1, C8W2, and C8W15. Control strains were wild-type P7063, Avr1b-1 deletion transformant Δ114-1-7, Avr1bsgR-AVI failed KI strain C4W4f, and Avr1bsgR-mCherry failed KI strain C8W3f. Zoospores from each strain were inoculated onto unifoliate leaves of soybean cultivar Williams (rps1b) or L77-1863 (Rps1b). Pictures were taken 3 days after zoospore inoculation. (B) Average lesion sizes on unifoliate soybean leaves inoculated with wild-type or transgenic P. sojae, measured from eight leaves in each of three independent experiments. Pictures were taken 3 days after inoculation, then relative lesion sizes were measured with ImageJ software. Error bars represent the mean ± standard deviation. P-values were calculated with the Wilcoxon rank sum test, ***P < 0.001. n.s. P > 0.1. (C) Avirulence phenotypes of Avr1b-1 knock-in transformants measured by real-time PCR-based quantification of P. sojae genomic DNA in infected tissue. Proliferation of P. sojae is plotted as the ratio between the relative abundance of PsActin DNA to GmActin DNA in infected L77-1863 leaves compared with Williams leaves at 72 hpi. This experiment was replicated three times. Error bars represent the mean ± standard deviation. P-values were determined with the Wilcoxon rank sum test. ***P < 0.001, n.s. P > 0.1. (D) Transcript levels produced by genes resident at the Avr1b-1 locus in wild-type and transgenic lines during soybean infection at 36 hpi. Levels were assayed by qRT-PCR using Avr1b-1 primers except for Δ114-1-7, for which mCherry primers were used (Supplementary Table S3). Transcript levels were normalized against P. sojae Actin. Error bars represent the mean ± standard deviation. P-values were calculated with the Wilcoxon rank sum test, ***P < 0.001, n.s. P > 0.1. RNA samples from the Avr1b-silenced isolate P6497 and in vitro-grown P7063 (P7063-My) were analyzed as controls. (E) Avr1b-mCherry fusion proteins preferentially accumulate in haustoria. Haustorial accumulation of Avr1b-mCherry during infection of etiolated hypocotyls by C8W1 (right panel) compared with uniform cytoplasmic distribution of mCherry in Δ114-1-7 (left panel). Images were obtained by confocal microscopy at 36 hpi. Scale bars = 20 μm.
To explore the reasons for loss of avirulence among the transformants, the relative transcript levels of the tagged Avr1b-1 genes during soybean infection were analyzed by qRT-PCR. As shown in Figure 7D, the transcript levels of Avr1b-1, Avr1bsgR-AVI, and Avr1bsgR-mCherry in C4W3 and C8W1 were comparable to those in the positive controls, C4W4f, C8W3f, and P7063, and the mCherry gene in Δ114-1-7. In contrast, the Avr1b-1 transcript levels in transformants C4W6, C4W7, C8W2, and C8W15 were 3–6-fold lower than in the wild type, P7063.
To further examine the expression of Avr1bsgR-mCherry in C8W1 during infection, zoospores from C8W1 were inoculated on the hypocotyls of Williams (rps1b) seedlings, then accumulation of the Avr1b-mCherry fusion protein was observed by confocal microscopy. The fusion protein could readily be observed to accumulate specifically around haustoria-like structures, as previously reported for ectopically expressed Avr1b-RFP (Liu et al., 2014). In contrast, when we observed the distribution of mCherry during infection by Δ114-1-7 which expresses free non-secreted mCherry from the Avr1b-1 locus, the fluorescent protein was uniformly distributed within the mycelium of the transformant (Figure 7E).
To extend the results described above, we tested additional Avr1bsgR-AVI and Avr1bsgR-mCherry gene replacement transformants from new independent transformations. Out of 13 homozygous Avr1bsgR-AVI KI transformants and 15 homozygous Avr1bsgR-mCherry KI transformants, only two and one transformants, respectively, induced HR on Rps1b soybean plants (Table 1), suggesting a very high frequency of silencing of the incoming transgene. The avirulence activities of the homozygous transformants were confirmed to be associated with normal transcript levels of either Avr1bsgR-AVI or Avr1bsgR-mCherry, respectively, whereas Avr1bsgR transcripts were not detected from the virulent transformants (Supplementary Figure S7). When the Avr1b-1 deletion mutant Δ114-1-7, which expresses mCherry at the Avr1b-1 locus, was used as a recipient for gene replacements with Avr1bsgR-AVI or Avr1bsgR-mCherry, failed expression was also observed at high frequency, based both on assays of avirulence (Table 1) and transcript presence (Supplementary Figure S7). Of the Avr1bsgR-mCherry homozygous replacement transformants, 9/11 showed failed expression based on inoculation of Rps1b soybean leaves and RT-PCR tests, while 9/9 Avr1bsgR-AVI homozygous replacement transformants exhibited failed expression (Table 1 and Supplementary Figure S7). These results suggest that the P. sojae Avr1b-1 locus is highly susceptible to loss of expression as a result of CRISPR/Cas9-mediated gene replacements.
Discussion
The diverse family of RXLR effectors are key virulence factors of Phytophthora and other oomycete plant pathogens. The Avr1b-1 RXLR gene of P. sojae, as well as its P. infestans homolog PiAvr3a, has served extensively as models for the study of RXLR effector functions, including impacts on host immunity and mechanisms of entry into host cells (Wang and Wang, 2018). Recently, Fang and Tyler (2016) developed a Phytophthora CRISPR/Cas9-mediated genome editing system using an oomycete-specific nuclear localization signal and oomycete-specific sgRNA transcription machinery. This Phytophthora genome editing tool allows detailed functional analysis of RXLR effector genes through knock-out, knock-in, and complementation experiments that avoid the uncertainties inherent in gene silencing and ectopic gene overexpression strategies. Here, we have conducted a series of knock-out and knock-in experiments with the Avr1b-1 gene of P. sojae. These experiments have confirmed previous conclusions about the function of Avr1b-1, have revealed some new features regarding its expression, and expanded the utility of the CRISPR/Cas9 system in P. sojae.
Previous characterization of Avr1b-1 had relied on transient expression in soybean leaves (Dou et al., 2008a, b), ectopic expression in P. sojae transformants (Dou et al., 2008a), gene silencing in P. sojae transformants (Song et al., 2013), assays of purified proteins (Shan et al., 2004; Kale et al., 2010), and phenotypes of P. sojae strains in which Avr1b-1 was naturally silenced or absent (Shan et al., 2004; Cui et al., 2012). Here, we used CRISPR/Cas9-mediated gene knock-outs and complementation experiments to unambiguously test the role of Avr1b-1 in producing an avirulence phenotype in the presence of the soybean Rps1b resistance gene. In these experiments, to avoid the potential off-targeting effects on a gene, Avh1, that is 97.6% identical to Avr1b-1, we selected P7063, which lacks Avh1, to perform CRISPR-mediated gene knock-out analysis. The results showed that the deletion of Avr1b-1 enabled mutants to infect soybean plants containing Rps1b, confirming that Avr1b-1 is essential for avirulence activity. The results also verified that the loss of Avr1b-1 did not compromise the pathogenicity of P. sojae transgenic strains.
The “all-in-one” CRISPR/Cas9 system (Fang et al., 2017) has been successfully applied in several different Phytophthora species except for P. infestans (van den Hoogen and Govers, 2018; Chen et al., 2019; Wang W. et al., 2019; Ochola et al., 2020; Zhang et al., 2020). In most cases, the efficiency of homozygous replacement events was low (<10%). Our results showed that the frequency of homozygous replacement events in the Avr1b-1 locus could reach 45%, but it varied among the sgRNAs. For this reason, when targeting a new gene, we typically try three different sgRNAs. For mCherry, we tested three different sgRNAs and all exhibited around 20% frequency of homozygous replacement events.
To conduct complementation following a knock-out, an alternative selection marker is needed for screening transformants. Besides G418, two other antibiotic markers, hygromycin B and streptomycin, have been used as selection markers in transgenic P. infestans (Ah-Fong and Judelson, 2011). In our hands, P. sojae isolates could maintain growth in V8 media containing up to 200 μg/ml hygromycin B. Therefore, it was not suitable for the screening of P. sojae transformants. Compared with G418, streptomycin is less effective as well. Although GFP has also been used as a selection marker for overexpression of spCas9 (Fang and Tyler, 2017), it is not suitable for the first round of screening due to the low transformation frequency. Oxathiapiprolin was described as an alternative selection marker in the genetic manipulation of Phytophthora pathogens (Wang W. et al., 2019), but the lack of commercial chemical products limits its application. Here, we found that G418 could be reused as a selection marker if the original KO transformant had lost its G418 resistance, even if the original gene was still present but silent. In P. sojae, G418 resistance is rapidly lost once G418 selection is removed, especially if the transformant is new. Therefore, reuse of G418 selection is feasible for routine complementation via a second round of transformation with the pYF515 vector. However, this strategy is only available if npt II was not used to replace the original target gene.
In order to conduct complementation, it was necessary to produce an Avr1b-1 gene that was resistant to the three sgRNAs used to target the native Avr1b-1 gene. This was needed in case the sgRNA genes were still being expressed in the original KO transformant and also to enable direct replacement of the native Avr1b-1 gene with modified Avr1b-1 genes. To produce Avr1bsgR, resistant to the three sgRNAs, we introduced multiple synonymous substitutions into the sgRNA target sites in Avr1b-1. The particle bombardment transient expression system was employed to verify that the expression of Avr1bsgR would produce a resistance response in Rps1b soybeans. The suitability of Avr1bsgR for this purpose was confirmed by the successful recovery of Avr1bsgR gene replacements from both KO and wild-type recipient strains, producing transformants avirulent on Rps1b plants.
Avr1b-1 is an infection-induced RXLR effector gene, and mCherry or Avr1b-mCherry genes introduced into the Avr1b-1 locus showed similar transcript levels during infection as that of Avr1b-1 in wild-type P7063 (except for those that had undergone silencing). Moreover, Avr1b-mCherry fusion proteins produced by the knock-in transformants specifically accumulated around haustoria during infection, whereas uniform distribution of mCherry proteins was observed in the Avr1b-1 KO transformant Δ114-1-7 which carried the Avr1b-1 promoter, but no signal peptide or effector sequences. Liu et al. (2014) reported that Avr1b-mRFP expressed from a constitutive promoter also specifically accumulated around haustoria during infection. These data suggest that the signal peptide and perhaps other targeting signals of Avr1b may be responsible for this localization pattern. In our experiments, we could not detect the effector-mCherry fusion inside haustorial host cells. Whisson et al. (2007) and Liu et al. (2014) reported similar results.
An unexpected feature of our results was the recovery of large numbers of knock-in transformants in which the incoming transgene had become silenced. We noticed that several different incoming genes, including mCherry, Avr1bsgR-AVI, and Avr1bsgR-mCherry, were subject to silencing in this way. Silencing occurred whether the transformation recipient was Δ114-1-7, carrying mCherry at the Avr1b-1 locus, or was the wild-type strain P7063. All incoming transgenes were embedded in the same upstream and downstream 1-kb flanking ends from the Avr1b-1 locus. Numerous studies have shown that sense as well as antisense constructs can readily induce silencing of both ectopic transgenes and endogenous genes in Phytophthora species (van West et al., 1999, 2008; Vu et al., 2019). However, our results are unusual in that only a single copy of the transgene is inserted into the genome, at the Avr1b-1 locus. Our results are also unusual because other studies have reported normal expression of incoming transgenes after gene replacements at other Phytophthora loci (Ma et al., 2017; Miao et al., 2020; Ochola et al., 2020).
Our observations raise the interesting question of whether the Avr1b-1 locus is unusually susceptible to silencing, due possibly to transcripts from the incoming plasmid DNA and/or epigenetic disturbances resulting from the replacement of genomic DNA with a segment of plasmid DNA. Among natural isolates of P. sojae, the frequency of Avr1b-1 silencing is also high. In a survey of 34 US isolates, 10 contained silent Avr1b-1 genes (Shan et al., 2004), while in a survey of 28 Chinese isolates, 5 contained silent Avr1b-1 alleles (Cui et al., 2012). Elevated levels of histone H3 Lysine27 (H3K27) trimethylation were observed at the Avr1b-1 locus in a naturally occurring Avr1b-silenced strain (P6497) but not in an Avr1b-expressing strain (Wang et al., 2020). Furthermore, mutations in a gene responsible for H3K27 methylation resulted in loss of silencing of Avr1b-1 (Wang et al., 2020). Thus, epigenetic changes likely underlie changes in the silencing state of Avr1b-1.
In summary, our findings extend the utility of CRISPR/Cas9-mediated genome editing for exploring gene functions in oomycetes, including transcriptional variability, while at the same time emphasizing the importance of confirming that incoming gene replacements are expressed normally.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
BG, XL, and BT conceived the research and wrote the manuscript with contributions from all authors. BG, GS, WG, and JM performed the experiments. BG, JM, and QW performed data analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (31301644), China Postdoctoral Science Foundation (2015M582716), The Changjiang Scholars Program of China (T2016106), grant #2018-67013-28438 to BT from the Agriculture and Food Research Initiative of the USDA NIFA, and Oregon State University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yufeng Fang (Greenlight Biosciences, NC, United States) for sharing the modified transformation protocol and CRISPR-mediated gene manipulation procedure in Phytophthora sojae and Kai Tao (Oregon Health & Science University, Portland, OR, United States) and Hua Zhao (SKLCSBAA, Northwest A&F University) for assistance in confocal experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.645331/full#supplementary-material
References
Ah-Fong, A. M. V., and Judelson, H. S. (2011). Vectors for fluorescent protein tagging in Phytophthora: tools for functional genomics and cell biology. Fungal Biol. 115, 882–890. doi: 10.1016/j.funbio.2011.07.001
Ah-Fong, A. M. V., Kim, K. S., and Judelson, H. S. (2017). RNA-seq of life stages of the oomycete Phytophthora infestans reveals dynamic changes in metabolic, signal transduction, and pathogenesis genes and a major role for calcium signaling in development. BMC Genomics 18:198. doi: 10.1186/s12864-017-3585-x
Baxter, L., Tripathy, S., Ishaque, N., Boot, N., Cabral, A., Kemen, E., et al. (2010). Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330, 1549–1551. doi: 10.1126/science.1195203
Chen, H., Shu, H., Wang, L., Zhang, F., Li, X., Ochola, S. O., et al. (2018). Phytophthora methylomes are modulated by 6mA methyltransferases and associated with adaptive genome regions. Genome Biol. 19:181. doi: 10.1186/s13059-018-1564-4
Chen, X., Zhang, Y., Li, H., Zhang, Z., Sheng, G., Li, Y., et al. (2019). The RXLR effector PcAvh1 is required for full virulence of Phytophthora capsici. Mol. Plant Microbe Interact. 32, 986–1000. doi: 10.1094/mpmi-09-18-0251-r
Cui, L., Yin, W., Dong, S., and Wang, Y. C. (2012). Analysis of polymorphism and transcription of the effector gene Avr1b in Phytophthora sojae isolates from China virulent to Rps1b. Mol. Plant Pathol. 13, 114–122. doi: 10.1111/j.1364-3703.2011.00733.x
Dong, S., Yu, D., Cui, L., Qutob, D., Tedman-Jones, J., Kale, S. D., et al. (2011). Sequence variants of the Phytophthora sojae RXLR effector Avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. PLoS One 6:e20172. doi: 10.1371/journal.pone.0020172
Dou, D., Kale, S. D., Wang, X., Chen, Y., Wang, Q., Wang, X., et al. (2008b). Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20, 1118–1133. doi: 10.1105/tpc.107.057067
Dou, D., Kale, S. D., Wang, X., Jiang, R. H. Y., Bruce, N. A., Arredondo, F. D., et al. (2008a). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20, 1930–1947. doi: 10.1105/tpc.107.056093
Fang, Y., Cui, L., Gu, B., Arredondo, F., and Tyler, B. M. (2017). Efficient genome editing in the oomycete Phytophthora sojae using CRISPR/Cas9. Curr. Protoc. Microbiol. 44, 21A.1.1–21A.1.26.
Fang, Y., and Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 17, 127–139. doi: 10.1111/mpp.12318
Fang, Y., and Tyler, B. M. (2017). Nuclear localization of a putative Phytophthora sojae bZIP1 transcription factor is mediated by multiple targeting motifs. Mol. Microbiol. 104, 621–635. doi: 10.1111/mmi.13652
Gu, B., Cao, X., Zhou, X., Chen, Z., Wang, Q., Liu, W., et al. (2020). The histological, effectoromic, and transcriptomic analyses of Solanum pinnatisectum reveal an upregulation of multiple NBS-LRR genes suppressing Phytophthora infestans infection. Int. J. Mol. Sci. 21:3211. doi: 10.3390/ijms21093211
Gu, B., Kale, S. D., Wang, Q., Wang, D., Pan, Q., Cao, H., et al. (2011). Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. PLoS One 6:e27217. doi: 10.1371/journal.pone.0027217
Gumtow, R., Wu, D., Uchida, J., and Tian, M. (2018). A Phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Mol. Plant Microbe Interact. 31, 363–373. doi: 10.1094/mpmi-06-17-0131-fi
Haas, B. J., Kamoun, S., Zody, M. C., Jiang, R. H. Y., Handsaker, R. E., Cano, L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398.
Jiang, R. H. Y., Tripathy, S., Govers, F., and Tyler, B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. U.S.A. 105, 4874–4879. doi: 10.1073/pnas.0709303105
Jiang, R. H. Y., and Tyler, B. M. (2012). Mechanisms and evolution of virulence in oomycetes. Annu. Rev. Phytopathol. 50, 295–318. doi: 10.1146/annurev-phyto-081211-172912
Kale, S. D., Gu, B., Capelluto, D. G. S., Dou, D., Feldman, E., Rumore, A., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142, 284–295. doi: 10.1016/j.cell.2010.06.008
Kale, S. D., and Tyler, B. M. (2011). “Assaying effector function in planta using double-barreled particle bombardment,” in Methods in Molecular Biology, Vol. 712, ed. J. M. McDowell (New York, NY: Humana Press), 153–172. doi: 10.1007/978-1-61737-998-7_13
Liu, T., Song, T., Zhang, X., Yuan, H., Su, L., Li, W., et al. (2014). Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5:4686. doi: 10.1038/ncomms5686
Ma, Z., Zhu, L., Song, T., Wang, Y., Zhang, Q., Xia, Y., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355, 710–714. doi: 10.1126/science.aai7919
Majeed, M., Soliman, H., Kumar, G., El-Matbouli, M., and Saleh, M. (2018). Editing the genome of Aphanomyces invadans using CRISPR/Cas9. Parasit. Vector 11:554. doi: 10.1186/s13071-018-3134-8
Miao, J., Liu, X., Li, G., Du, X., and Liu, X. (2020). Multiple point mutations in PsORP1 gene conferring different resistance levels to oxathiapiprolin confirmed using CRISPR-Cas9 in Phytophthora sojae. Pest Manag. Sci. 76, 2434–2440. doi: 10.1002/ps.5784
Ochola, S., Huang, J., Ali, H., Shu, H., Shen, D., Qiu, M., et al. (2020). Editing of an effector gene promoter sequence impacts plant-Phytophthora interaction. J. Integr. Plant Biol. 62, 378–392. doi: 10.1111/jipb.12883
Qutob, D., Chapman, B. P., and Gijzen, M. (2013). Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat. Commun. 4:1943. doi: 10.1038/ncomms2354
Qutob, D., Tedman-Jones, J., Dong, S., Kuflu, K., Pham, H., Wang, Y., et al. (2009). Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS One 4:e5066. doi: 10.1371/journal.pone.0005066
Shan, W., Cao, M., Dan, L., and Tyler, B. M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17, 394–403. doi: 10.1094/mpmi.2004.17.4.394
Situ, J., Jiang, L., Fan, X., Yang, W., Li, W., Xi, P., et al. (2020). An RXLR effector PlAvh142 from Peronophythora litchii triggers plant cell death and contributes to virulence. Mol. Plant Pathol. 21, 415–428. doi: 10.1111/mpp.12905
Song, T., Kale, S. D., Arredondo, F. D., Shen, D., Su, L., Liu, L., et al. (2013). Two RXLR avirulence genes in Phytophthora sojae determine soybean Rps1k-mediated disease resistance. Mol. Plant Microbe Interact. 26, 711–720. doi: 10.1094/mpmi-12-12-0289-r
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Tyler, B. M., and Gijzen, M. (2014). “The Phytophthora sojae genome sequence: foundation for a revolution,” in Genomics of Plant-Associated Fungi and Oomycetes: Dicot Pathogens, eds R. Dean, A. Lichens-Park, and C. Kole (Berlin: Springer Press), 2133–2157.
Tyler, B. M., Kale, S. D., Wang, Q., Tao, K., Clark, H. R., Drews, K., et al. (2013). Microbe-independent entry of oomycete RXLR effectors and fungal RXLR-like effectors into plant and animal cells is specific and reproducible. Mol. Plant Microbe Interact. 26, 611–616. doi: 10.1094/mpmi-02-13-0051-ia
Tyler, B. M., Tripathy, S., Zhang, X., Dehal, P., Jiang, R. H. Y., Aerts, A., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266. doi: 10.1126/science.1128796
van den Hoogen, J., and Govers, F. (2018). Attempts to implement CRISPR/Cas9 for genome editing in the oomycete Phytophthora infestans. BioRxiv [Preprint]. doi: 10.1101/274829
van West, P., Kamoun, S., van ’t Klooster, J. W., and Govers, F. (1999). Internuclear gene silencing in Phytophthora infestans. Mol. Cell 3, 339–348. doi: 10.1016/s1097-2765(00)80461-x
van West, P., Shepherd, S. J., Walker, C. A., Li, S., Appiah, A. A., Grenville-Briggs, L. J., et al. (2008). Internuclear gene silencing in Phytophthora infestans is established through chromatin remodelling. Microbiology 154, 1482–1490. doi: 10.1099/mic.0.2007/015545-0
Vu, A. L., Leesutthiphonchai, W., Ah-Fong, A. M. V., and Judelson, H. S. (2019). Defining transgene insertion sites and off-target effects of homology-based gene silencing informs the application of functional genomics tools in Phytophthora infestans. Mol. Plant Microbe Interact. 32, 915–927. doi: 10.1094/mpmi-09-18-0265-ta
Wang, L., Chen, H., Li, J., Shu, H., Zhang, X., Wang, Y., et al. (2020). Effector gene silencing mediated by histone methylation underpins host adaptation in an oomycete plant pathogen. Nucleic Acids Res. 48, 1790–1799. doi: 10.1093/nar/gkz1160
Wang, Q, Li, T., Zhong, C., Luo, S., Xu, K., Gu, B., et al. (2019). Small RNAs generated by bidirectional transcription mediate silencing of RXLR effector genes in the oomycete Phytophthora sojae. Phytopathol. Res. 1:18. doi: 10.1186/s42483-019-0026-6
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Wang, W., Xue, Z., Miao, J., Cai, M., Zhang, C., Li, T., et al. (2019). PcMuORP1, an oxathiapiprolin resistance gene, functions as a novel selection marker for Phytophthora transformation and CRISPR/Cas9 mediated genome editing. Front. Microbiol. 10:2402. doi: 10.3389/fmicb.2019.02402
Wang, Y., Tyler, B. M., and Wang, Y. (2019). Defense and counterdefense during plant-pathogenic oomycete infection. Annu. Rev. Microbiol. 73, 667–696. doi: 10.1146/annurev-micro-020518-120022
Wang, Y., and Wang, Y. C. (2018). Phytophthora sojae effectors orchestrate warfare with host immunity. Curr. Opin. Microbiol. 46, 7–13. doi: 10.1016/j.mib.2018.01.008
Whisson, S. C., Boevink, P. C., Moleleki, L., Avrova, A. O., Morales, J. G., Gilroy, E. M., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118. doi: 10.1038/nature06203
Zhang, Q., Li, W., Yang, J., Xu, J., Meng, Y., and Shan, W. (2020). Two Phytophthora parasitica cysteine protease genes, PpCys44 and PpCys45, trigger cell death in various Nicotiana spp. and act as virulence factors. Mol. Plant Pathol. 21, 541–554. doi: 10.1111/mpp.12915
Keywords: Phytophthora sojae, RXLR effector, clustered regularly interspaced short palindromic repeat, gene replacement, transcriptional variation
Citation: Gu B, Shao G, Gao W, Miao J, Wang Q, Liu X and Tyler BM (2021) Transcriptional Variability Associated With CRISPR-Mediated Gene Replacements at the Phytophthora sojae Avr1b-1 Locus. Front. Microbiol. 12:645331. doi: 10.3389/fmicb.2021.645331
Received: 23 December 2020; Accepted: 03 February 2021;
Published: 18 March 2021.
Edited by:
Tao Zhou, China Agricultural University, ChinaReviewed by:
Xiao-Ren Chen, Yangzhou University, ChinaQingshan Chen, Northeast Agricultural University, China
Copyright © 2021 Gu, Shao, Gao, Miao, Wang, Liu and Tyler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xili Liu, c2VlZGxpbmdAbndhZnUuZWR1LmNu; Brett M. Tyler, QnJldHQuVHlsZXJAb3JlZ29uc3RhdGUuZWR1
 Biao Gu
Biao Gu Guangda Shao1
Guangda Shao1 Qinhu Wang
Qinhu Wang Brett M. Tyler
Brett M. Tyler