Abstract
Gamma-glutamyl transpeptidase (GGT) enzyme is ubiquitously present in all life forms and plays a variety of roles in diverse organisms. Higher eukaryotes mainly utilize GGT for glutathione degradation, and mammalian GGTs have implications in many physiological disorders also. GGTs from unicellular prokaryotes serve different physiological functions in Gram-positive and Gram-negative bacteria. In the present review, the physiological significance of bacterial GGTs has been discussed categorizing GGTs from Gram-negative bacteria like Escherichia coli as glutathione degraders and from pathogenic species like Helicobacter pylori as virulence factors. Gram-positive bacilli, however, are considered separately as poly-γ-glutamic acid (PGA) degraders. The structure–function relationship of the GGT is also discussed mainly focusing on the crystallization of bacterial GGTs along with functional characterization of conserved regions by site-directed mutagenesis that unravels molecular aspects of autoprocessing and catalysis. Only a few crystal structures have been deciphered so far. Further, different reports on heterologous expression of bacterial GGTs in E. coli and Bacillus subtilis as hosts have been presented in a table pointing toward the lack of fermentation studies for large-scale production. Physicochemical properties of bacterial GGTs have also been described, followed by a detailed discussion on various applications of bacterial GGTs in different biotechnological sectors. This review emphasizes the potential of bacterial GGTs as an industrial biocatalyst relevant to the current switch toward green chemistry.
Introduction
The enzyme γ-glutamyl transpeptidase (GGT; E.C.2.3.3.2) is conserved throughout all three domains of life, ranging from single-celled prokaryotes to multicellular higher eukaryotes (Rawlings et al., 2010). It belongs to the N-terminal nucleophile hydrolase superfamily and exists as a heterodimer of a large and a small subunit (Nash and Tate, 1984; Suzuki et al., 1989; Brannigan et al., 1995). It is a two-substrate enzyme catalyzing the transfer of γ-glutamyl moiety from donor substrates, such as glutathione/glutamine, to an acceptor substrate, which can be water (hydrolysis) or other amino acids/small peptides (transpeptidation) or the donor itself (autotranspeptidation) (Thompson and Meister, 1977; Tate and Meister, 1981). Here, the GGT is commonly referred to by its old name as “gamma-glutamyl transpeptidase;” however, according to the Nomenclature Committee of International Union of Biochemistry and Molecular biology, its accepted name is γ-glutamyltransferase (Balakrishna and Prabhune, 2014).
Gamma-glutamyl transpeptidase enzyme was first reported in mammals, such as sheep and rat (Hanes et al., 1952; Tate and Meister, 1976). It is worldwide recognized as a diagnostic marker for many physiological diseases in humans (Yamada et al., 1991; Whitfield, 2001; Grundy, 2007). Although its existence in the living world was known for a long time, only after its discovery in prokaryotes led to a profound understanding of its structure–function relationship and also suggested its application in the synthesis of various γ-glutamyl compounds. Presently, it finds biotechnological significance in the synthesis of various bioactive γ-glutamyl compounds for application in food and pharmaceutical sectors (Suzuki et al., 2004a; Wang et al., 2008; Shuai et al., 2010; Saini et al., 2017; Mu et al., 2019).
Mammalians GGTs have already been extensively reviewed (Tate and Meister, 1981; Taniguchi and Ikeda, 1998; Chikhi et al., 1999; Whitfield, 2001). Microbial GGTs, covering the structure–function relationship of the protein and their biotechnological significance, have been reviewed by Castellano and Merlino (2012) and subsequently by Balakrishna and Prabhune (2014).
The current review focuses mainly on bacterial GGTs with emphasis on their physiological significance, structural and molecular aspects, autoprocessing, and enzyme catalysis along with their relevance in biotechnology and biomedicine sectors.
Diversity of Bacterial GGTs
Gamma-glutamyl transpeptidases diversity ranging from microbes to mammals has been described by Verma et al. (2015) through a phylogenetic tree, including 47 diverse GGT sequences. This phylogenetic tree had two main branches and various clades (Verma et al., 2015). The tree also showed some interesting evolutionary relationships of prokaryotic and eukaryotic GGTs, as it grouped pathogenic bacterial GGTs including Escherichia coli GGT with eukaryotic ones, instead of their nonpathogenic bacterial homologues. This suggests high sequence similarity between pathogens and their hosts. Glutathione hydrolysis by E. coli and most of the pathogenic GGTs represents a functional similarity to mammalian GGTs. However, in Bacillus subtilis and Bacillus licheniformis, the presence of two types of GGT enzymes – the canonical GGT placed in close association to other Bacillus GGTs and GGT-like protein YwrD grouped with archaeal and extremophilic GGTs – suggest the horizontal transfer of YwrD protein to the mesophilic Bacillus strains from some other archaea or extremophiles. Thus, the diversification of GGT proteins gives intriguing insights into the evolutionary history of this ubiquitously conserved protein. Besides, GGT proteins were also classified into two distinct groups based on the presence or absence of a 12-residue long structural sequence known as lid loop. Accordingly, GGTs from eukaryotes and Gram-negative organisms have been termed as lid-loop positive and GGTs from Bacillus species, archaea, and extremophiles as lid-loop negative.
Physiological Significance of Bacterial GGT
Gamma-glutamyl transpeptidase protein shows widespread conservation, but its physiological role is not very clear yet (Balakrishna and Prabhune, 2014). Mammalian GGTs are involved in glutathione metabolism; on the other hand, prokaryotic GGTs are known to have diverse physiological functions stemming from the fact that the glutathione is not found among all prokaryotes (Table 1).
TABLE 1
| GGT group and associated organisms | Expression and localization | Putative Physiological role |
| 1. Glutathione degrading GGT – mainly present in glutathione producing Gram-negative bacteria | ||
| E. coli | Periplasmic expression | Glutathione utilization as a source of amino acid and nitrogen under nutrient limiting conditions |
| Proteus mirabilis | Localized on the outer cytoplasmic membrane facing the periplasm | |
| 2. GGT as virulence factor – mainly present in pathogenic Gram-negative bacteria | ||
| Helicobacter pylori | Periplasmic expression | Utilization of host’s glutathione and glutamine as a source of glutamate; Inhibition of T-cell proliferation in host |
| Campylobacter jejuni | ND# | |
| Fransicella tularensis | ND | Utilization of cytosolic glutathione and γ-glutamyl cysteine peptides present in host cytosol for cysteine acquisition |
| Neisseria meningitidis | Expressed on the inner cytoplasmic membrane facing cytoplasm | Utilization of cytosolic γ-glutamyl cysteine peptides for cysteine acquisition |
| 3. PGA hydrolyzing GGT – mainly present in PGA producing Gram-positive bacteria | ||
| Bacillus subtilis (natto) | Extracellular secretion | Degradation of PGA into glutamate as a source of nitrogen under nutrient-starved conditions |
| Bacillus licheniformis | Extracellular secretion | |
| Bacillus anthracis* | Membrane-bound expression | Covalent anchorage of PDGA to peptidoglycan layer to maintain capsule integrity; Depolymerization of PDGA inside the cytoplasm |
Classification of bacterial gamma-glutamyl transpeptidases (GGTs) into different physiological groups.
#Not determined.*Bacillus anthracis is also a pathogenic strain and GGT is involved in its virulence; however, it has been placed in the third group as it is a Gram-positive Bacillus with PDGA as a virulence factor.
Gamma-glutamyl transpeptidase was first reported in some Gram-negative proteobacteria, such as Proteus mirabilis and E. coli, as a periplasmic protein along with glutathione as the most abundant thiol (Fahey et al., 1978; Nakayama et al., 1984; Suzuki et al., 1986; Masip et al., 2006). In these organisms, deletion or inhibition of the GGT enzyme led to increased extracellular leakage of glutathione (Nakayama et al., 1984; Suzuki et al., 1986). Further studies indicated its possible role in the cleavage of glutathione in periplasmic space to provide cysteine and glycine in Gram-negative bacteria, similar to mammalian GGTs. Another report on E. coli GGT indicated its ability to uptake an exogenous supply of γ-glutamyl peptides and glutathione as the source of amino acids (Suzuki et al., 1993). For Pseudomonas species, periplasmic localization of GGT has been reported, but its involvement in glutathione metabolism could not be elucidated (Kushwaha and Srivastava, 2014).
In some Gram-negative pathogenic bacteria such as Helicobacter pylori, Fransicella tularensis, Neisseria meningitidis, and Campylobacter jejuni, the expression of GGT has been linked to their pathophysiology, and GGT has been ascribed as a formidable virulence factor (Ling, 2013). The pathogenicity mechanism in H. pylori, which causes gastritis, gastric ulcers, and cancers in humans, has been studied extensively. H. pylori cells colonize gastric mucosa of mammalian gut involving several virulence factors including GGT for the establishment of infection (Ling, 2013; Ricci et al., 2014). During colonization, constitutive periplasmic expression of H. pylori GGT (HpGGT) allowed metabolism of extracellular glutathione and glutamine present in the host cytosol as a source of glutamate by H. pylori cells (Chevalier et al., 1999; Shibayama et al., 2007). This has been suggested to be the key physiological role played by HpGGT resulting in profuse growth of the pathogen in the gastric mucosa (Gong and Ho, 2004). HpGGT has also been reported to induce apoptosis of gastric epithelial cells under oxidative stress aided by scavenging glutathione from the gastric environment (Shibayama et al., 2003; Gong et al., 2010; Flahou et al., 2011). It has also been demonstrated to have an immunosuppressive effect on the host, as it inhibits the proliferation of T cells resulting in persistent colonization and infection (Schmees et al., 2007). Higher activity of HpGGT from peptic ulcer patients corroborates its clinical relevance as a virulence factor in the diagnosis of gastric-related diseases caused by H. pylori (Gong et al., 2010; Franzini et al., 2014). A similar physiological function of GGT was reported in another pathogen C. jejuni, closely related to H. pylori. GGT from C. jejuni assisted in the inhibition of T-cell proliferation, thus promoting persistent colonization of gastric epithelial cells in mammals and avian gut (Barnes et al., 2007; Floch et al., 2014).
Fransicella tularensis, a facultative intracellular pathogen, causes tularemia by mainly infecting phagocytic macrophages (Clemens et al., 2004). The role of GGT in its pathogenesis was demonstrated by mutational disruption of ggt gene that resulted in drastic defects in its intracellular growth and its replication in macrophage cell lines and caused an attenuated virulence in mice. Here, GGT was reported to be responsible for utilization of glutathione and γ-glutamyl cysteine pools of host as a source of cysteine, essential in intracellular multiplication of pathogen (Alkhuder et al., 2009). In fact, ggt deletion mutant of a highly virulent F. tularensis SCHU S4 strain has been reported as a potential vaccine candidate; patent was granted in 2013 (United States 8,609,108B2) (Ireland et al., 2011; Le Butt et al., 2013). Another Gram-negative bacteria N. meningitidis, the causative agent of a deadly brain disease, meningitis, has also been reported to utilize GGT for cysteine acquisition from an extracellular pool of γ-glutamyl cysteine peptides (Takahashi et al., 2004, 2018). It was located in association with the inner membrane toward the cytoplasmic side, suggesting easy accessibility to extracellular peptides (Takahashi and Watanabe, 2004).
Gamma-glutamyl transpeptidase has been reported in other prokaryotes as well, such as in Gram-positive bacilli including B. subtilis, B. licheniformis, Bacillus anthracis (a pathogen), and Bacillus amyloliquefaciens, and in extremophiles such as Geobacillus thermodentrificans, Deinococcus radiodurans, and Thermus thermophilus and archaea like Picrophilus torridus. In Gram-positive bacilli, many studies on the putative physiological role of GGTs have ruled out their involvement in glutathione degradation for amino acid utilization, as these bacteria cannot synthesize glutathione (Fahey et al., 1978), although they can hydrolyze it under in vitro conditions (Minami et al., 2003b). In another report, when glutathione was supplemented externally as a nitrogen source, the cysteine and glycine auxotrophic mutants of B. subtilis could not grow owing to their inability in utilizing glutathione as a source of these amino acids (Xu and Strauch, 1996). However, B. subtilis GGT has shown efficacy in cleaving γ-glutamyl bond of glutathione to release cysteinylglycine as a sulfur source instead (Minami et al., 2004). GGT in Bacillus species is secreted extracellularly during the onset of the stationary phase, suggesting its role in stationary phase physiology (Xu and Strauch, 1996). Many Bacillus species produce poly-γ-glutamic acid (PGA) at the onset of the stationary phase, which is utilized as a nitrogen source during the late stationary phase under nutrient-starved conditions (Ashiuchi and Misono, 2002). The involvement of B. subtilis (natto) GGT in the hydrolysis of PGA was investigated for the first time by Kimura et al. (2004). Here, GGT knockout mutant was shown to produce an increased amount of medium-sized PGA fragments (1 × 105 Da) as compared to the wild-type strain, in which both the amount and the size of PGA fragments (5 × 103 Da) decreased considerably. They suggested that the large PGA fragments must have first got hydrolyzed to medium-sized fragments by an endopeptidase YwtD (now known as PgdS) (Suzuki and Tahara, 2003); B. subtilis GGT may have acted later as an exopeptidase catalyzing the cleavage of glutamate (both L-and D-forms) residue from amino-terminal of the medium-sized PGA fragments (Kimura et al., 2004). In another report, a double knockout of pgdS and ggt gene resulted in twofold enhanced production of PGA in B. subtilis 168 strain; however, single-gene knockout of pgdS or ggt did not improve PGA yields (Scoffone et al., 2013). Moreover, among the three mutants, the pgdS single mutant produced PGA with the highest molecular weight, which could be attributed to hampered endo-peptidase activity. Contrasting results were obtained in a recent report of pgdS and ggt knockout mutants of B. licheniformis RK14-46 strain. The single pgdS gene knockout, as well as the double knockout of pgdS and ggt gene, resulted in a drastic reduction in PGA production, while the single ggt gene knockout improved PGA production due to lesser degradation (Ojima et al., 2019). This suggests that GGT is crucial for PGA hydrolysis by Bacillus species, and these GGT-producing Gram-positive bacilli can be classified as PGA degraders.
In addition to GGT, Bacillus species like B. subtilis and B. licheniformis also code for another GGT-like protein named YwrD, expressed intracellularly and shares only 27% sequence similarity with well-described GGTs (Grundy and Henkin, 2001). However, this protein has not been assigned any role yet (Minami et al., 2004).
Another GGT-producing Gram-positive bacterium known to be a deadly pathogen, B. anthracis, produces capsular poly-γ-D-glutamic acid (PDGA) as a virulence factor to evade off immune response (Wu et al., 2009). It expresses a virulent protein named capsule depolymerase (CapD), which is located on the cellular surface in association with bacterial envelope (Candela and Fouet, 2005). CapD is considered to be a part of the GGT family, as it, respectively, shares 31 and 27% sequence similarity to E. coli and B. subtilis GGTs and exhibits similar autoprocessing steps (Uchida et al., 1993; Minami et al., 2004; Wu et al., 2009). CapD has been mainly reported to catalyze the covalent anchoring of PDGA to peptidoglycan for maintaining the integrity of the capsule (Richter et al., 2009). CapD minus B. anthracis mutant showed no capsule layer after heat and chemical treatment and was reported to have an attenuated virulence in mice models (Candela and Fouet, 2005). Apart from its anchoring function, CapD has also been suggested to carry out depolymerization of PDGA in the cell cytoplasm (Richter et al., 2009).
Recently, the role of B. subtilis GGT (BsGGT) as a novel virulence factor in the pathogenesis of bone resorption similar to mammalian GGT (Niida et al., 2004) has been demonstrated in a cell culture study (Kim et al., 2016). It was reported that the large subunit of BsGGT enhanced osteoclastogenesis activity, independent of enzymatic activity. Further molecular study in the presence of BsGGT large subunit showed that the induction of osteoclastogenesis was related to upregulation of an osteoclast differentiation factor receptor activator of nuclear factor kappa-B ligand (RANKL), which interacted with surface receptors of precursor osteoblast cells and promoted the formation of osteoclast cells. In addition, there was an enhancement in the messenger RNA (mRNA) expression of cyclooxygenase 2 (COX-2). Based on these findings, GGT has been hypothesized to act as a virulence factor in bone destruction, caused by periodontopathic bacteria (Kim et al., 2016).
The physiological significance of GGT in extremophilic microbes has not been elucidated yet. The reason could be the extreme cultural conditions required during cultivation and also the difficulty in the genetic manipulation of such strains.
Structural and Functional Aspects of Bacterial GGTs
Structural and Topological Features
Most microbial GGTs are synthesized as prepro-GGT with N-terminal signal peptide (Figure 1) sequence for extracellular secretion. After the signal is cleaved, the resulting unique precursor pro-GGT is modified through a self-proteolytic cleavage event termed autoprocessing, forming an active mature heterodimer of large and small subunits (Tate and Meister, 1981; Lo et al., 2007). Autoprocessing involves a conserved nucleophile threonine positioned subcentric inclined toward C-terminal (Balakrishna and Prabhune, 2014). Post autoprocessing, resulting C- and N-terminal, respectively, form large and small subunits; with conserved threonine as the first residue in smaller subunit to serve as a nucleophile for carrying out enzymatic reaction (Inoue et al., 2000; Figure 1).
FIGURE 1
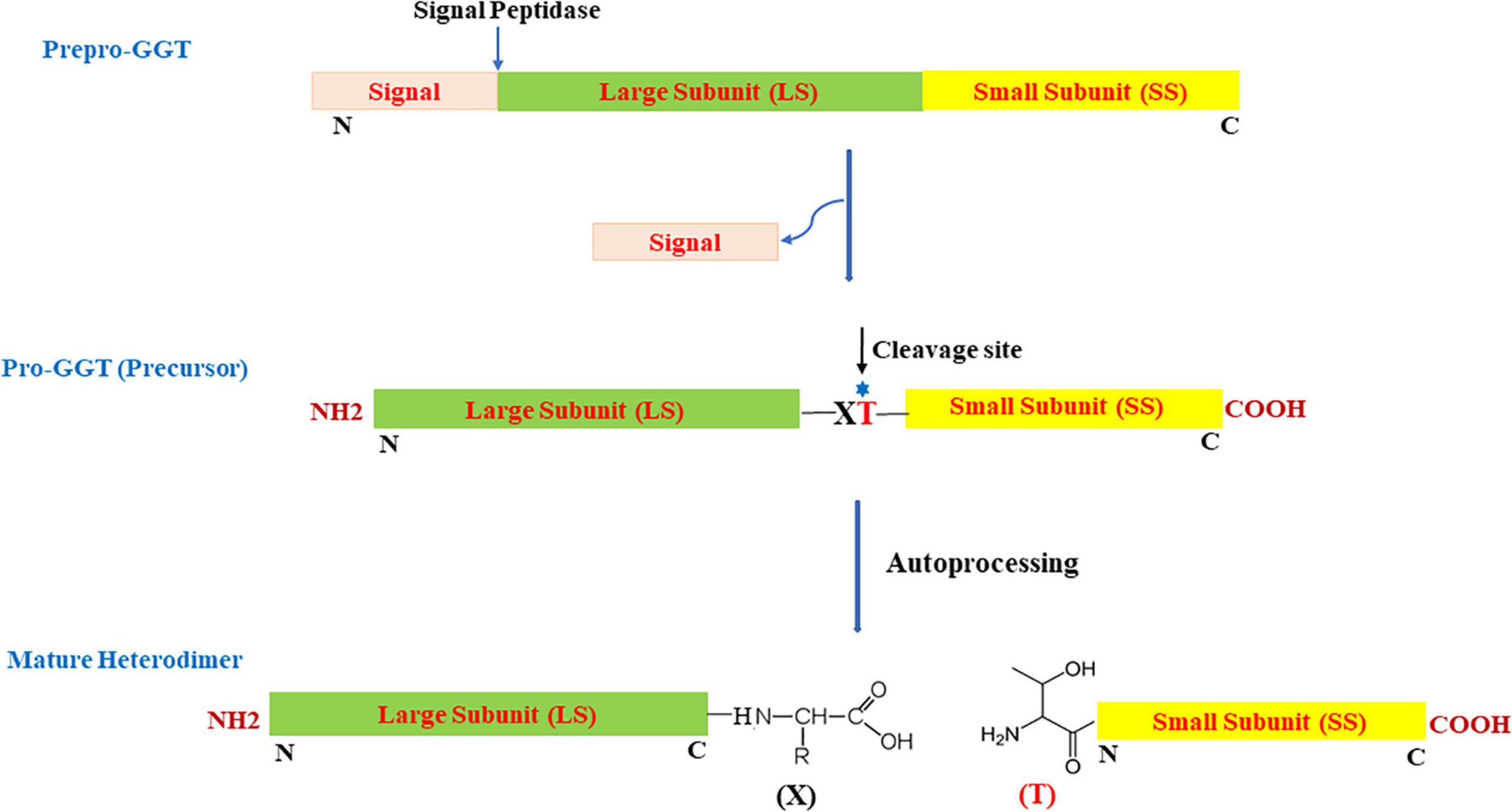
Schematic view of organization and two-step maturation of bacterial gamma-glutamyl transpeptidases (GGTs); the cleavage site represents the site of autoprocessing. T, conserved threonine; X, residue preceding the conserved threonine; N, N-terminus; C, C-terminus.
Primary structure analysis of various microbial GGTs has shown that the intact polypeptide is generally composed of 500 ± 100 amino acid residues (Supplementary Figure 1). GGTs from mesophilic organisms like E. coli (EcGGT), H. pylori (HpGGT), Pseudomonas, and Bacillus species contain around 24–35-residue long N-terminal signal peptide, while some extremophilic organisms like Geobacillus thermodenitrificans (GtGGT), Thermus hermophiles (TtGGT), D. radiodurans (DrGGT), etc., lack any signal peptide and are thus intracellular. Primary sequence comparison also reveals >25% sequence similarities among different species, including mammalian GGT from Homo sapiens (HsGGT). Further, putative cleavage site for autoprocessing occupied by threonine [T391 in EcGGT; T380 in HpGGT; T399 in B. licheniformis GGT (BlGGT)] and two glycine residues (G483/G484 in EcGGT; G481/G482 in BlGGT), constituting oxyanion hole for stabilizing tetrahedral enzyme intermediate during catalysis, is invariably conserved. Large subunit is less conserved and consists of 300–405 residues, while 170–195 amino acid long small subunit is relatively conserved and contains active site residues (Tate and Ross, 1977; Castonguay et al., 2003). These subunits are highly intertwined, mainly by noncovalent hydrophobic interactions and hydrogen bonding (Castellano and Merlino, 2012). The first crystal structure to be elucidated was of E. coli GGT (EcGGT) in 2006; in fact, three different structures of EcGGT (PDB IDs: 2DBU, 2D5G, 2DBX) were determined simultaneously (Table 2; Okada et al., 2006). Thereafter, GGT structures from a variety of microbes such as H. pylori (HpGGT), B. subtilis (BsGGT), B. licheniformis (BlGGT), B. anthracis (BanGGT; CapD), Bacillus halodurans (BhGGT), Pseudomonas nitoreducens (PnGGT), and Thermoplasma acidophilum (TaGGT) were determined (Boanca et al., 2007; Okada et al., 2007; Williams et al., 2009; Wu et al., 2009; Wada et al., 2010; Ida et al., 2014; Pica et al., 2016; Kumari et al., 2017; Table 2). Although the mammalian GGTs were the first to be described in detail, heavy glycosylation and membrane-bound localization made it difficult to determine their structure; the first crystal structure of human GGT was solved in 2013 (West et al., 2013).
TABLE 2
| Enzyme | PDB ID | Resolution | Structural form | GGT molecules in the asymmetric unit |
| EcGGT | 2DBU | 1.95 | Ligand free form | Heterotetramer – two heterodimeric molecules containing two large and two small subunits |
| 2DG5 | 1.60 | In complex with hydrolyzed glutathione (substrate) | ||
| 2DBX | 1.70 | In complex with glutamate (substrate) | ||
| 2DBW | 1.80 | Acyl-enzyme intermediate | ||
| 2E0Y | 2.02 | Samarium derivative | ||
| 2E0X | 1.95 | Monoclinic form (Se-Met GGT) | ||
| 2Z8K | 1.65 | In complex with acivicin (inhibitor) | ||
| 2Z8J | 1.65 | In complex with azaserine (inhibitor) | ||
| 2Z8I | 2.05 | In complex with azaserine in dark | ||
| 2E0W | 2.55 | T391A mutant | Dimer – two identical precursor molecules | |
| HpGGT | 2NQO | 1.90 | Ligand free form | Heterotetramer – two heterodimeric molecules containing two large and two small subunits |
| 2QMC* | 1.55 | T380A mutant in complex with S-(nitrobenzyl)glutathione | ||
| 2QM6 | 1.60 | In complex with glutamate | ||
| 3FMN | 1.70 | In complex with acivicin | ||
| 5BPK | 1.49 | − | ||
| BsGGT | 2V36 | 1.85 | Ligand free form | Heterotetramer – two heterodimeric molecules containing two large and two small subunits |
| 3A75 | 1.95 | In complex with glutamate | ||
| 3WHS | 1.80 | In complex with acivicin | Heterodimer – one large and small subunit | |
| 3WHQ | 1.58 | Soaked in acivicin for 0 min | ||
| 3WHR | 1.85 | Soaked in acivicin for 3 min | ||
| BlGGT | 4OTT | 2.98 | Ligand free mature form | Heterodimer – one large and small subunit |
| 4OTU# | 3.02 | In complex with glutamate | ||
| 5XLU | 1.45 | In complex with acivicin | ||
| 4Y23 | 2.89 | T399A precursor mutant | Monomer – one molecule of precursor | |
| PnGGT | 5ZJG | 1.70 | In complex with Gly–Gly | Heterotetramer – two heterodimeric molecules containing two large and two small subunits |
| BhGGT (cephalosporin acylase) | 2NLZ | 2.70 | Ligand free form | Heterotetramer – two heterodimeric molecules containing two large and two small subunits |
| BanGGT (CapD) | 3G9K | 1.79 | Ligand free form | Heterotetramer – two heterodimeric molecules containing two large and two small subunits |
| 3GA9 | 2.30 | Ligand free form | ||
| TaGGT | 2I3O | 2.03 | Ligand free form | Heterotetramer – two heterodimeric molecules containing two large and two small subunits |
List of crystal structures available for prokaryotic gamma-glutamyl transpeptidase (GGTs).
*Crystal of an inactive but processed T380A mutant using a bicistronic construct expressing large and small subunit separately.#Presence of Mg2+ ion crucial for crystal formation.
It has also been demonstrated that in various GGTs, the content of secondary structures formed of α-helices and β-sheets is nearly identical, resulting in similar tertiary and quaternary structural folding (Lin et al., 2014). A highly conserved tetralamellar α/ββ/α fold forms core of the enzyme appearing sandwich-like, made of two tightly packed antiparallel β-sheets, one from each subunit, surrounded by two α-helices (Okada et al., 2006). This fold is characteristic of the N-terminal nucleophile (Ntn) hydrolase superfamily (Brannigan et al., 1995). Most members of this family including penicillin G acylase (Hewitt et al., 2000), cephalosporin acylase (Kim et al., 2000), glycosylasparaginase (Guo et al., 1998), etc., are synthesized as inactive precursors, which undergo autocatalytic processing to form an active mature protein. The activated protein acquires either a nucleophile serine, threonine, or cysteine as its first residue at the newly formed N-terminus with suggested role in catalysis and autoprocessing. Similarly, in case of GGT enzyme, the mechanism of autoprocessing and catalysis also involves a conserved threonine; GGT is therefore considered to be a member of the Ntn hydrolase superfamily.
Molecular Mechanism of Autoprocessing
Through several mutational and crystallography studies, it was demonstrated for the first time in EcGGT that the processing was an autocatalytic event rather than a, earlier hypothesized, protease-dependent occurrence (Kuno et al., 1984). Suzuki and Kumagai (2002) proposed a molecular mechanism of autoprocessing of EcGGT based on its similarity to the Ntn hydrolase superfamily. They suggested that the hydroxyl group of Thr391 (in EcGGT) acts as a nucleophile for proteolytic cleavage of scissile peptide bond with the preceding residue Gln390; the presence of a base abstracts proton from the hydroxyl group of Thr391, generating a reactive oxyanion (Figure 2). The reactive oxygen at gamma position (OG) atom attacks the carbonyl carbon of Gln390 to form a transitional tetrahedral intermediate. The C–N bond later gets cleaved via protonation of nucleophile threonine’s amino group, leading to an ester intermediate (N–O acyl shift) which is hydrolyzed further into two subunits with Thr391 as the new N-terminal residue of small subunit (Suzuki and Kumagai, 2002).
FIGURE 2
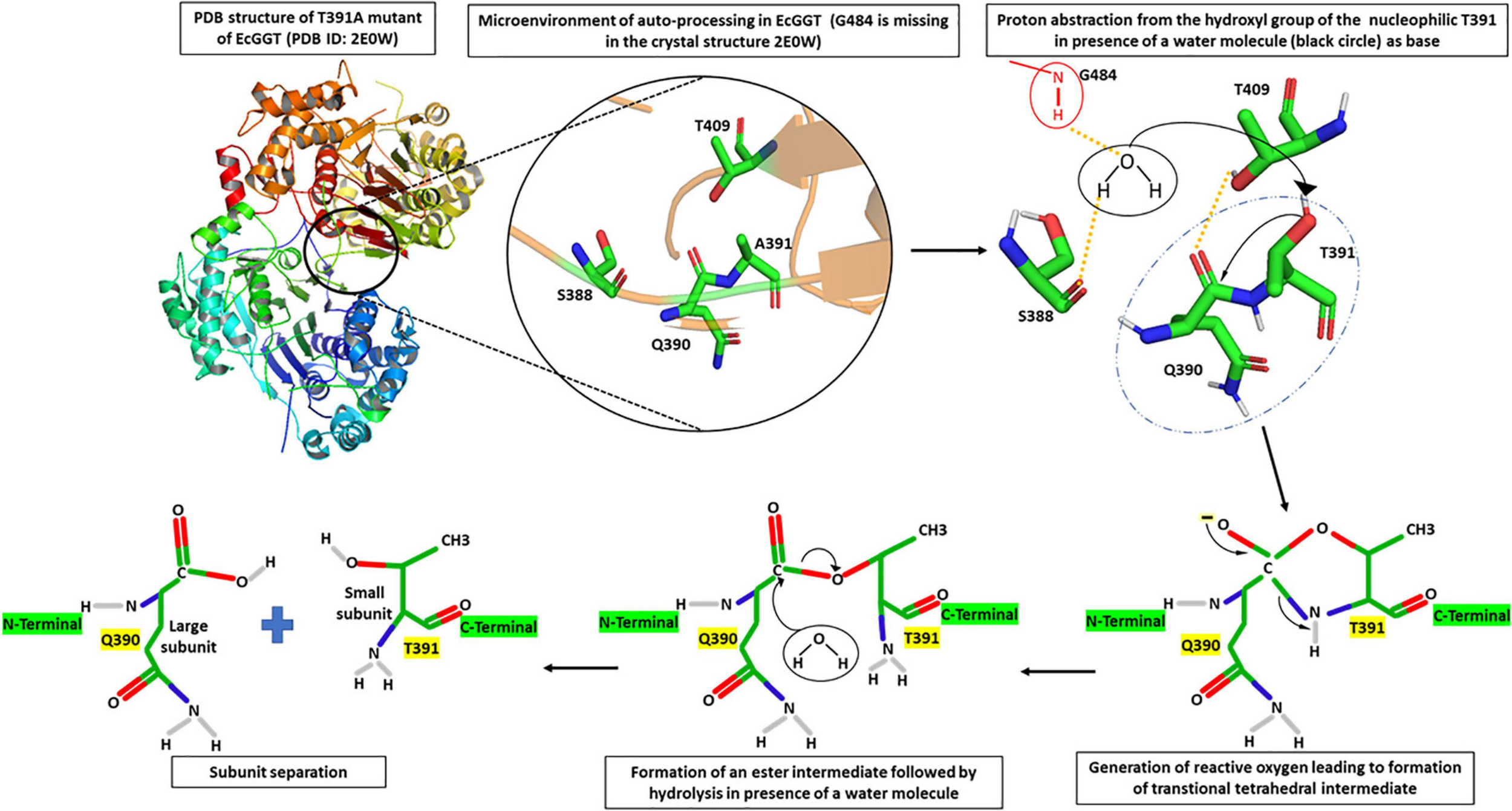
Microenvironment for autocatalytic processing of Escherichia coli gamma-glutamyl transpeptidase (EcGGT) and its detailed molecular mechanism. Crystal structure of T391A precursor mutant of EcGGT (PDB ID: 2E0W) has been used for studying autocatalytic environment. A391 in the structure has been replaced by T391 to show hydrogen bonding (yellow dashed lines) and nucleophilic attack (black arrow) by T391 during autoprocessing. A water molecule (black circle) and amide bond of G484 (red circle) have been drawn to show bonding. The structures are prepared using PyMOL2 software.
Further analysis of the crystal structure of mature E. coli GGT (PDB Id: 2DBU), as well as its mutated T391A precursor (PDB ID: 2E0W), provided more mechanistic details of autoprocessing. A water molecule in the close vicinity of Gln390 hydrogen bonded to the hydroxyl group of Ser388 and α-amino group of Gly484 has been reported to enhance nucleophilicity of OG atom of Thr391 that appears crucial in autoprocessing (Figure 2; Okada et al., 2006, 2007). Accordingly, it was reported that substituting conserved threonine in HpGGT (Thr380) and BlGGT (Thr399) with alanine led to complete inhibition of autoprocessing (Boanca et al., 2006; Lyu et al., 2009). Similarly, the cysteine mutant (T399C) of BlGGT remained unprocessed, indicating the importance of the hydroxyl group in cleaving peptide bond (Lyu et al., 2009). Serine mutants of both BlGGT (T399S) and HpGGT (T380S) showed a dramatic decrease in the processing rate; the mutants were, however, processed completely after prolonged incubation. For HpGGT, the role of the methyl group present in the side chain of nucleophile threonine has been suggested for properly positioning the hydroxyl group during the nucleophilic attack (Boanca et al., 2006).
Similar autoprocessing behavior was observed in GtGGT from an extremophilic organism G. thermodenitrificans where a homotetrameric precursor is initially produced, which underwent autocatalysis after incubation at 45°C for 48 h (Castellano et al., 2010). The alanine mutant (T353A) of its conserved threonine 353 was also expressed as an inactive unprocessed homotetramer (Castellano et al., 2010).
Analysis of the recently solved crystal structure of the T399A precursor mimic of BlGGT gave interesting insights; Pica et al. (2016) explored the microenvironment required for autocatalytic processing and proposed a slightly different mechanism of autoprocessing. They suggested that in BlGGT, Thr399 is activated by the OG atom of another Thr417 in lieu of a water molecule as is the case of EcGGT (Figure 3). This leads to a six-membered transition state involving two hydroxyl groups of Thr399 and Thr417 and one carbonyl group of Gln398. Following this, a transient tetrahedral intermediate is formed, and later, hydrolysis of ester bond yields two subunits. Contribution of some other residues, namely, His401, Thr415, Thr417, E419, and Arg571, reported to be important for proper positioning of Gln398 and Thr399, was also assessed by the same group (Figure 3). They generated alanine mutants of each of the five residues and characterized them concerning autoprocessing. Mutants E419A and R571A could process completely, while H401A and T417A showed 60–70% processing in a time-dependent fashion, and T415A mutant persisted as an inactive precursor. Similar maturational defects were observed in EcGGT when the corresponding residue Thr407 (Thr415 in BlGGT) was mutated to aspartate (T407D), lysine (T407K), and serine (T407S) (Lo et al., 2007). In another report from BlGGT, its T417S mutant could process considerably with time, while T417K mutant led to complete blockage of maturation, suggesting the importance of hydroxyl group of Thr417 for proper autoprocessing (Lyu et al., 2009). In HpGGT, mutation of the same threonine residue at position 398 to serine (T398S) and alanine (T398A) resulted in comparable processing rates with respect to native HpGGT (Boanca et al., 2007). Thus, despite being highly conserved among microbial as well as mammalian GGTs, the role of this threonine residue in autoprocessing is still controversial.
FIGURE 3
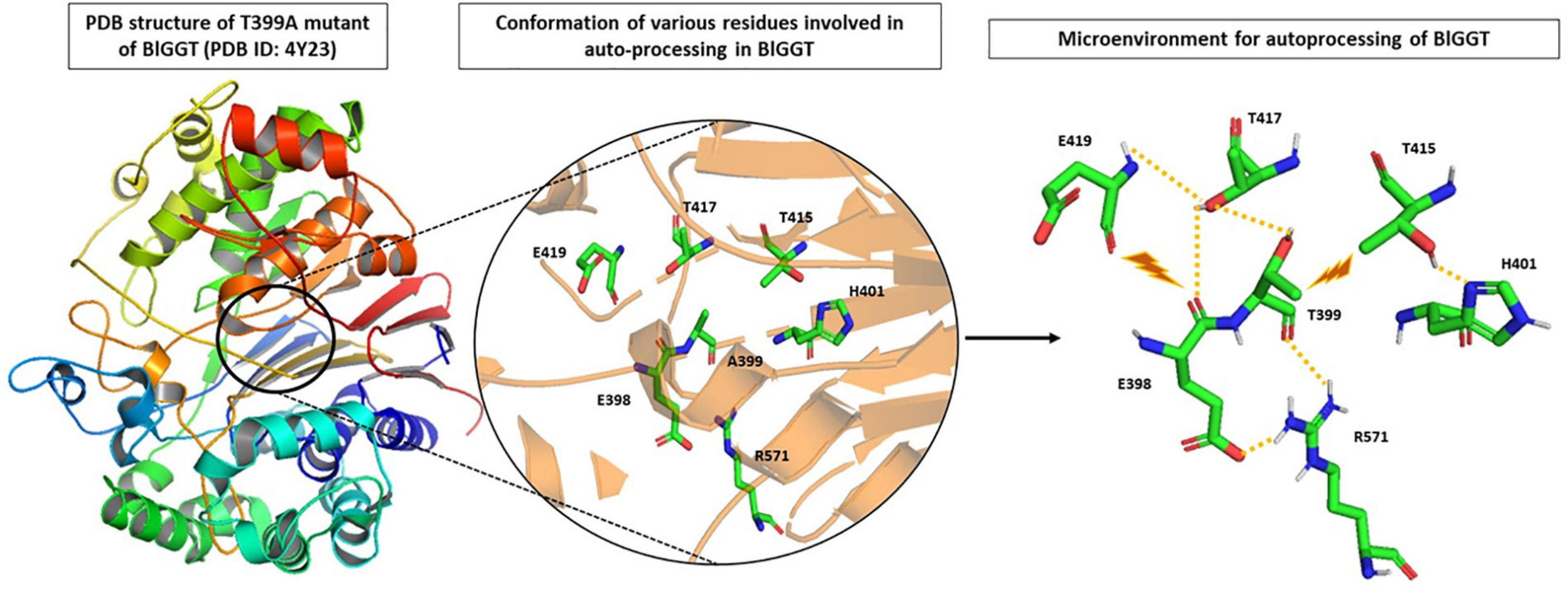
Microenvironment for autocatalytic processing of Bacillus licheniformis gamma-glutamyl transpeptidase (BlGGT). Crystal structure of T399A precursor mutant of BlGGT (PDB ID: 4Y23) has been used for studying autocatalytic environment. A399 is replaced by T399 to show hydrogen bonding (yellow dashed lines) and steric hindrance (brown lightning bolt) experienced by T399 for its proper positioning during autoprocessing. The structures are prepared using PyMOL2 software.
The role of residues present in the immediate vicinity of the cleavage site during autoprocessing was also explored. The residue preceding conserved nucleophile threonine was variable among microbial GGTs (Supplementary Figure 1). After autoprocessing, it acquired the last position of the newly formed C-terminus of the large subunit and was occupied by Gln390 in EcGGT and Glu398 in BlGGT. Analysis of four mutants, viz E398A, E398D, E398R, and E398Q of BlGGT, suggested that the C-terminal of the large subunit was indeed critical for autoprocessing of BlGGT (Chi et al., 2012). Among these, in three mutants, except E398Q, autoprocessing was hampered, which did not improve even after prolonged in vitro incubation. On the contrary, in EcGGT, replacement of the corresponding residue Q390 with alanine (Q390A) did not affect autoprocessing, indicating the insignificance of the large subunit C-terminus of EcGGT in autoprocessing (Hashimoto et al., 1995). The obtained contradictory results also possibly explain the low conservation of this residue among prokaryotic GGTs. Later, Chi et al. (2014) demonstrated the role of a 14-residue long extra sequence, exclusively present in Bacillus GGTs at the C-terminus of the large subunit, in autoprocessing, structural stability, and catalysis of BlGGT. They constructed six progressive deletion mutants within the extra sequence region with altered autoprocessing capabilities and variable catalytic activities (Chi et al., 2014). Further, the N-terminal of the small subunit, constituting a highly conserved TTH motif, was also investigated for its role in autoprocessing. The role of the first threonine residue, constituting the conserved nucleophile, in autoprocessing has been established well for many bacterial GGTs (Suzuki and Kumagai, 2002; Pica et al., 2012, 2016). In EcGGT, replacement of the second and third residue, respectively, with alanine (T392A) and glycine (H393G) impaired autoprocessing, suggesting that the small subunit N-terminus is more crucial than the large subunit C-terminus during autoprocessing (Hashimoto et al., 1995).
Apart from this, N- and C-termini of the intact polypeptide chain (prepro-GGT) of bacterial GGTs were also studied for their importance in autoprocessing. Truncation in the N-terminal region, including the signal peptide, of EcGGT and BlGGT demonstrated that signal peptide is critical for proper enzyme folding and processing (Lin et al., 2008; Lo et al., 2008). For EcGGT, truncation beyond 17 amino acids was found detrimental for maturation, indicating the crucial role of the last nine amino acids of the signal peptide in functional expression of the enzyme (Lo et al., 2008), while in BlGGT, all the truncated mutants except the native enzyme resulted in partial or complete loss of processing, emphasizing again on the importance of signal peptide in the enzyme’s folding and maturation (Lin et al., 2008). This is contradictory to the reports showing functional heterologous expression, with efficient autoprocessing, of EcGGT and BlGGT that lack signal peptide sequence (Lin et al., 2006; Yao et al., 2006). Similarly, N-terminal truncation in BpGGT up to 95 residues did not affect autoprocessing, but functional activity was lost with truncation beyond 63 residues (Murty et al., 2012). The importance of the C-terminus in autoprocessing has also been elucidated and was first demonstrated in EcGGT. Two arginyl residues present at the C-terminus of EcGGT were mutated with alanine (R513A) and glycine (R571G), resulting in a complete blockage of autocatalytic processing (Hashimoto et al., 1992). Further in HpGGT, the formation of salt bridges among four highly conserved residues, viz Glu515/Arg547 and Arg502/Asp562 at C-terminus, suggested their importance in enzyme processing. Disrupting salt bridges by mutating residue Glu515 (E515Q) and Arg502 (R502L) led to impaired autoprocessing and activity (Williams et al., 2009). C-terminal truncation in BlGGT revealed that deletion beyond V576, at ninth position from C-terminus, was detrimental for autoprocessing and activity (Chang et al., 2010).
Effect of Autoprocessing on Structure and Function of GGT
Structural analysis of EcGGT and HpGGT crystal structures showed that the new termini formed post autoprocessing are approximately 35 Å apart, which is quite distant, suggesting significant conformational changes during autoprocessing (Okada et al., 2006; Morrow et al., 2007). Further, comparing the overall structure of the mature and T391A precursor of EcGGT demonstrated that backbone atoms in the core regions of both the proteins remain unchanged, while significant conformational changes occurred proximal to the active site (Okada et al., 2007). Particularly after autoprocessing, the large subunit C-terminus (denoted as P-segment) flips out and is replaced by a flexible lid loop located in the smaller subunit. Adoption of an extended conformation by the P-segment has been suggested significant in forming binding pocket. However, these conformational changes do not occur in all microbial GGTs owing to the absence of lid loop as seen in Bacillus species. Analysis of the mature structures of BsGGT and BlGGT demonstrated that the C-terminus of large subunit possesses an extra sequence region located close to the N-terminus of the small subunit, suggesting no significant conformational changes upon autoprocessing (Wada et al., 2010; Lin et al., 2014). The extra sequence in BlGGT has been reported as important for autoprocessing and enzyme activity (Chi et al., 2014). Apart from structural implications, the correlation of autoprocessing with catalytic activity has also been well documented. Analysis of in vitro processing of native and mutant precursors of EcGGT and BlGGT, as a function of time, suggests that the extent of autoprocessing correlates well with increased enzyme activity (Suzuki and Kumagai, 2002; Lin et al., 2008). In GGTs like EcGGT, HpGGT, and BpGGT, uncoupling of autoprocessing and enzymatic activity has been attempted. Hashimoto et al. (1995) demonstrated that the coexpression of large and small subunit (using separate expression plasmid for each subunit) of EcGGT could result in reconstitution of two subunits under both in vivo and in vitro conditions. The retained activity was very less, indicating that only few molecules of the two subunits could fold properly and give rise to an active form (Hashimoto et al., 1995). In contrast, for HpGGT and BpGGT, coexpression of both the subunits in pET-DUET vector resulted in a fully active heterodimeric enzyme with comparable and enhanced activities (Boanca et al., 2006; Murty et al., 2012). This indicated that different bacterial GGTs can adopt different conformations; use of different expression systems can also effect both folding and active conformation of GGT, as uncoupling autoprocessing from enzymatic activity was successful for HpGGT and BpGGT but not for EcGGT.
Catalysis Mediated via Nucleophile Threonine
Gamma-glutamyl transpeptidase enzyme catalyzes a two-step reaction, involving cleavage of γ-glutamyl bond present in γ-glutamyl compounds like glutathione and glutamine subsequently, followed by transfer of γ-glutamyl moiety to a water molecule or another amino acid or short peptide (Tate and Meister, 1981). The first step is termed acylation, in which donor substrate donates γ-glutamyl moiety to the enzyme, forming an acyl-enzyme intermediate. In the second step termed deacylation, an acceptor substrate accepts γ-glutamyl moiety to form final end product (Figure 4). Based on the end product formation, GGT mediates three types of reactions.
FIGURE 4
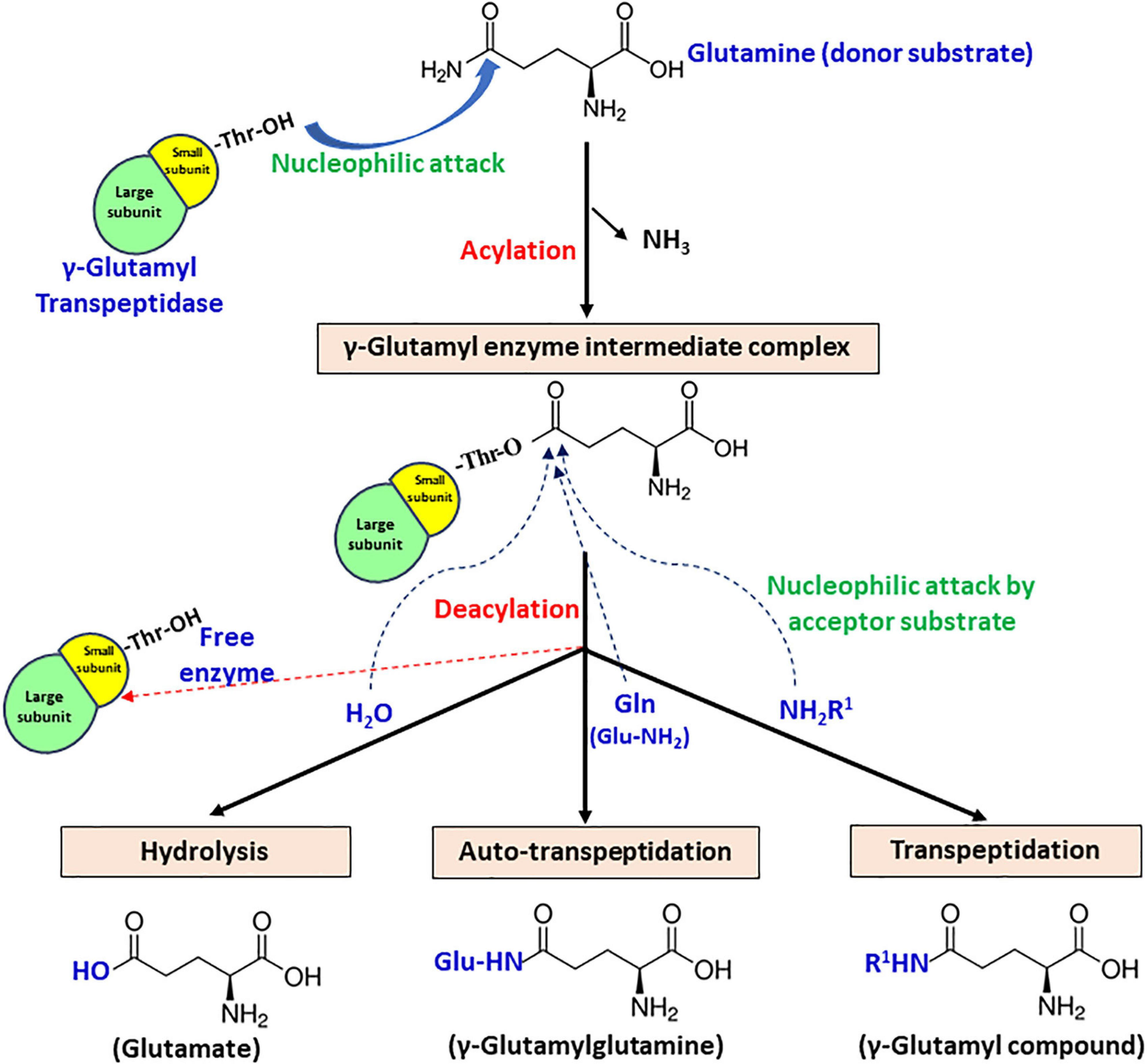
Schematic representation of the proposed catalytic mechanism of gamma-glutamyl transpeptidase (GGT) with glutamine as the γ-glutamyl moiety donor.
-
1.
Hydrolysis – acceptor is a water molecule, with end-product glutamate.
-
2.
Transpeptidation – acceptor is an amine group containing compound that can be any amino acid or short peptide, and the end-product formed is a γ-glutamyl compound.
-
3.
Autotranspeptidation – acceptor is the donor molecule itself, and the end-product is γ-glutamylated donor.
The formation of an acyl-enzyme intermediate during catalysis has been substantiated by both kinetic and crystallographic studies. Stop-flow kinetics performed under presteady-state conditions using rat kidney GGT revealed a biphasic pattern of enzyme catalysis, confirming its ping-pong mechanism (Keillor et al., 2004). Based on the observation, it has been interpreted that the acylation step is faster, while the deacylation step is rate limiting (Keillor et al., 2005). For bacterial GGTs, further examination of the X-ray crystal structure of EcGGT soaked in glutathione for varying time intervals also demonstrated the formation of γ-glutamyl enzyme intermediate, indicating that the second step is much slower than the first (Okada et al., 2006). Further analysis of EcGGT in complex with substrate analogs, acivicin and azaserine, revealed that they formed a tetrahedral adduct (structurally analogous to transient acyl-enzyme intermediate) with the enzyme (Wada et al., 2008).
Gamma-glutamyl transpeptidase-mediated reactions are known to be catalyzed by nucleophilic attack on the donor substrate. The N-terminal threonine residue of small subunit responsible for autoprocessing has also been reported as the relevant nucleophile in enzyme catalysis (Inoue et al., 2000). Inhibition of GGT activity by treatment with certain inhibitors like 6-diazo-5-oxo-L-norleucine (DON) and serine–borate complex suggested the involvement of the hydroxyl group of either a serine or a threonine residue (Tate and Meister, 1977, 1978). However, Inoue et al. (2000) were the first group to identify that the conserved Thr391 is indeed the catalytic nucleophile for EcGGT. They employed mechanism-based affinity labeling to modify the putative active site of GGT using 2-amino-4-(fluorophosphono) butanoic acid (1), a γ-phosphonic acid monofluoride derivative of glutamic acid. The compound phosphonylated the putative catalytic nucleophile, and subsequent liquid chromatography/mass spectrometry (LC/MS) and MS/MS analysis revealed that the small subunit N-terminal threonine is the catalytic nucleophile. Moreover, the structure of EcGGT in complex with glutathione clearly illustrates the formation of a covalent bond between the OG atom of Thr391 and carbonyl carbon of γ-glutamyl moiety of glutathione (Okada et al., 2006).
Analysis of the crystal structure of HpGGT also demonstrated the role of a second threonine (T398) residue in increasing nucleophilicity of the catalytic threonine (T380) (Boanca et al., 2007). It has been reported to be highly conserved among bacterial GGTs and suggested to interact with the hydroxyl group of the catalytic threonine, resulting in the formation of a TT dyad. Besides, the nucleophilicity of catalytic threonine has also been proposed to increase through interaction with its own α-amino group, which can act as a base to activate catalytic nucleophile threonine (Michalska et al., 2005; Boanca et al., 2007). The contribution of T398 in the activation of T380 in HpGGT was investigated by site-directed mutagenesis. Replacement of T398 with alanine (T398A) and serine (T398S) resulted in a complete and nearly fivefold reduction of enzymatic activity, respectively, with no maturational defects, suggesting the importance of TT dyad in efficient catalysis. A plausible explanation given for the same was the presence of a methyl group in T398, likely required for precise positioning of its side chain hydroxyl group for interaction with catalytic threonine (Boanca et al., 2007). Similarly, the replacement of the corresponding second threonine residue T417 with lysine, serine, and alanine in BlGGT also resulted in a dramatic decrease in activity as well as autoprocessing rate, indicating its functional significance in both autoprocessing and catalysis (Lyu et al., 2009).
Substrate Binding Pocket
The substrate-binding pocket of GGT can be divided into two parts – the donor binding site and the acceptor binding site. The donor binding site interacts with γ-glutamyl moiety in the donor molecule, while the presence of a discrete acceptor binding site is still ambiguous. In mammalian GGTs, these sites have been reported to be largely overlapping (Thompson and Meister, 1977). However, in bacterial GGTs, the donor binding site has been determined to be highly conserved, while the acceptor binding site is reported highly variable. Analysis of the crystal structures of EcGGT, HpGGT, BsGGT, and BlGGT in complex with glutamate demonstrated that the catalytic pocket is enclosed within the shallow groove at the interface of both large and small subunits (Okada et al., 2006; Morrow et al., 2007; Wada et al., 2010; Lin et al., 2014). The binding pocket resides the catalytic threonine at the bottom of the groove on one of the central β-sheets, and it further extends into the enzyme, appearing as a finger-like projection. The γ-glutamyl moiety of the donor molecule is bound to the active site at the bottom of the pocket by an extensive network of hydrogen bonds and salt bridges formed mainly with the residues present in the small subunit (Okada et al., 2006). In BsGGT, the α-carboxylate group of the bound glutamate interacts with Arg113, Ser464, and Ser465, and the α-amino group interacts with Glu423, Glu442, and Asp445 (Figure 5; Wada et al., 2010). The glutamate carbonyl carbon is covalently bonded to the OG atom of threonine, while the carbonyl oxygen is hydrogen bonded to the main chain amino group of two glycine residues, Gly485 and Gly486. Arg113 is reported to be the only residue from, respectively, the large subunit involved in binding pocket formation. Residues forming the binding-pocket of mesophilic GGTs are mostly conserved, except differences of one or two residues, suggesting rigidity of the binding pocket: in EcGGT, Asn411 and Gln430 while Glu423 and Glu442 in BsGGT. On the contrary, GGTs from extremophiles exhibit variability in their catalytic residues with respect to their mesophilic counterparts as has been revealed by multiple sequence alignment (Supplementary Figure 1). For example, the highly conserved arginine residue (R107 in BlGGT; R113 in BsGGT; R114 in EcGGT), present in the large subunit of mesophilic GGTs, is replaced by serine (S68 in PtGGT and S85 in GtGGT) in extremophiles. Likewise, the two consecutive serine residues (Ser462/Ser463 in EcGGT) of small subunit are replaced by histidine and threonine (414His/415Thr in GtGGT) in GGTs from extremophiles. The different nature of these residues is suggested to be responsible for the differences in catalytic properties of mesophilic and extremophilic GGTs (Castellano et al., 2011).
FIGURE 5
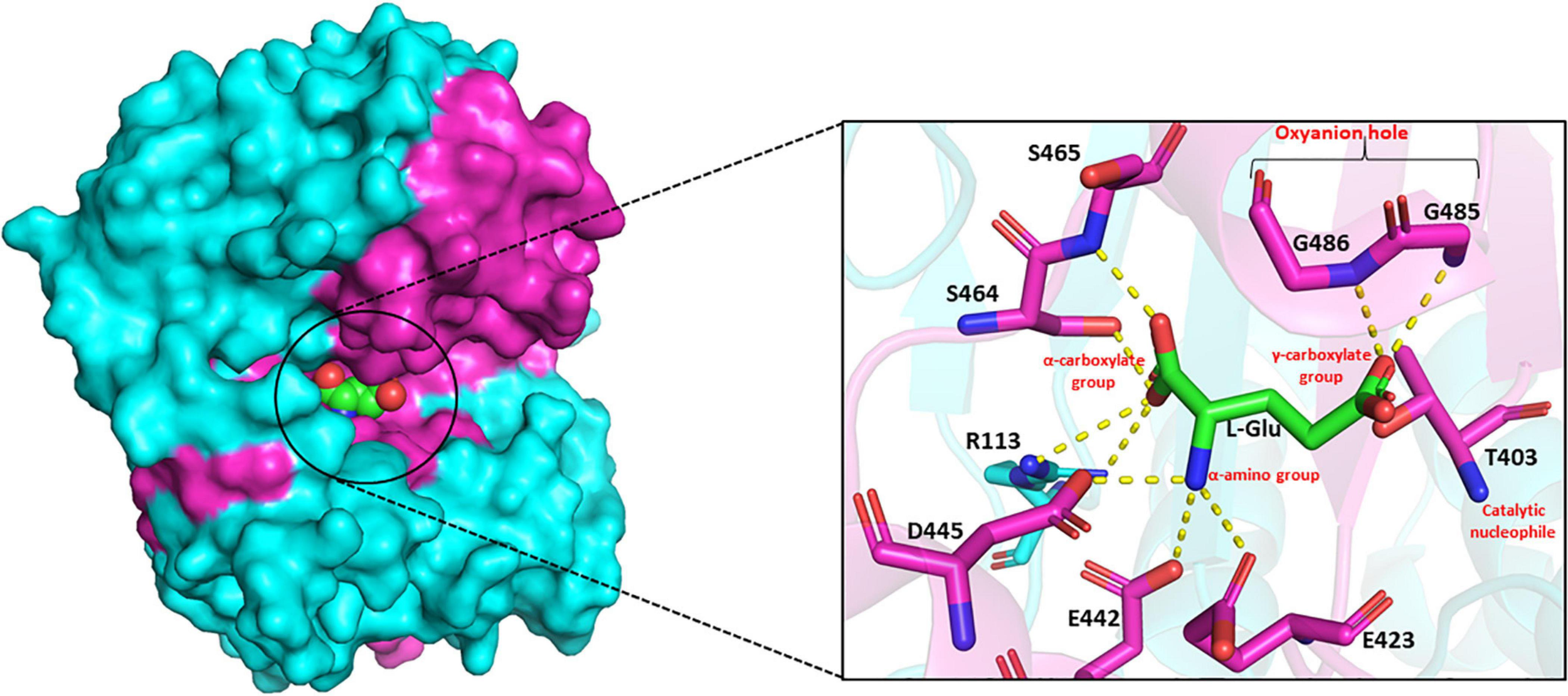
Side view of surface drawing of Bacillus subtilis gamma-glutamyl transpeptidase (BsGGT) (PDB ID: 3A75) showing the binding pocket groove and interaction of L-glutamic acid (L-Glu) with active site residues of BsGGT. Cyan color highlights large subunit, pink color highlights small subunit, and L-Glu is represented as a sphere in the surface drawing. Hydrogen bonding is represented as yellow dashed lines. The structures are prepared using PyMOL2 software.
Recent crystallographic studies on L-Glu-bound BlGGT demonstrated that its active site channel constitutes two extensive pockets at the intersubunit interface lined mainly by hydrophobic residues. In addition, substrate binding resulted in significant structural changes in BlGGT; particularly, there is a reordering of six residues at the C-terminus of the large subunit with suggested role in enzyme catalysis (Lin et al., 2014).
As GGT catalysis follows a ping-pong mechanism, it is assumed that it sequentially binds donor and acceptor molecules. It has been suggested that the acceptor molecule occupies the same site where initially the leaving group of the donor molecule binds, but nothing has been conclusively proven yet (Taniguchi and Ikeda, 1998). Hu et al. (2012) threw some light on the acceptor binding residues and their mode of interaction by computational and mutational studies performed using CapD protein from B. anthracis and HsGGT (Homo sapiens). Analysis of the structures of CapD and HsGGT suggested that the putative acceptor binding site is proximal to the donor binding site, located in the deep groove, and is highly variable and flexible as compared to the conserved and rigid donor binding site. Further, the putative acceptor binding site of HsGGT has been suggested to be formed by some polar residues like Asp46, Asn79, His81, Ser82, Tyr403, Gln476, and Lys562, while in CapD, Arg423 and Arg520 are substituted in place of Gln476 and Lys562. Both the residues Arg423 and Arg520 have been suggested to play a role in acceptor binding and imparting broader stereospecificity to CapD in accepting both L- and D-forms of amino acids as acceptors.
Interestingly, Pseudomonas nitroreducens GGT (PnGGT) exhibits higher hydrolytic activity than transpeptidase activity despite high sequence similarity with EcGGT (Imaoka et al., 2010). Recent crystallographic studies on PnGGT in complex with Gly–Gly acceptor demonstrated that the binding mode of Gly–Gly in the active site is probably the reason for its reduced activity as an acceptor (Hibi et al., 2019). The terminal amino group of Gly–Gly is oriented opposite to the nucleophilic active center. Moreover, tight packaging of three aromatic residues Trp385, Phe417, and Trp525 around the Gly–Gly binding pocket has been suggested to be advantageous for its binding; side chains of these residues are involved in the recognition of acceptor substrate. Phe417 is located in the lid loop, while Trp385 and Trp525 have been suggested to form side walls of the putative acceptor binding site. However, the hydrophobic pocket formed by the three aromatic residues has been suggested to shield the catalytic nucleophile from bulk solvent and activating hydrolysis. The functional significance of the putative acceptor site of PnGGT including Trp385, Phe417, and Trp525 has also been explored by mutational studies. Substitution of three aromatic residues with threonine (W385T), tyrosine (F417Y), and alanine (W525A), present at the corresponding positions in EcGGT, resulted in enhanced transpeptidase activity for all three mutants, while a 5–14% decrease in hydrolytic activity was observed. Likewise, in another report from Picrophilus torridus GGT (PtGGT), the replacement of an aromatic residue tyrosine at position 327 by asparagine (corresponding residue in EcGGT) introduced significant transpeptidase activity in PtGGT, while the native enzyme exhibited only hydrolytic activity (Rajput et al., 2013). Comparative docking of the acceptor ligand Gly–Gly in the structural model of PtGGT and its Y327N mutant identified some residues with suggested importance in acceptor recognition and binding. The acceptor Gly–Gly interacted with five residues (L87, E90, Y305, N327, S348) and perfectly docked in the binding pocket of Y327N.
Importance of Lid Loop in Catalysis
The presence of a unique lid-loop region in the small subunit of GGT of different bacterial and mammalian homologues as shown in multiple sequence alignment (Supplementary Figure 1) has structural and functional significance. Analysis of substrate-bound crystal structures of EcGGT and HpGGT demonstrated that the otherwise flexible lid loop acquires a well-defined position after binding of the substrate and extends over the binding site pocket, shielding it from the solvent molecule (Okada et al., 2006). The lid-loop spanning segment from Pro438 to Gly449 in EcGGT contains an aromatic residue tyrosine (Try444), which has been reported to form a hydrogen bond with conserved residue Asn411 by its side chain hydroxyl group (Figure 6). The hydrogen bond has been suggested to act as a gate, restricting entry of the solvent molecule into the active site and also regulating access of the substrate to the active site cleft that results in a rigid structural conformation around the binding pocket. A similar hydrogen bond has been reported between Tyr433 of the lid loop and Asn400 in HpGGT (Morrow et al., 2007). The shielding likely occludes water molecules from entering the active site, facilitating transpeptidation. However, human GGT exhibits higher transpeptidase activity, likely associated with higher mobility of its lid loop allowing rapid switch of its structural conformation from open to close resulting in faster product release (Hu et al., 2012). Analysis of recently determined crystal structure of human GGT also confirms high mobility of the lid loop (West et al., 2013). The probable explanation ascribed is the replacement of tyrosine with phenylalanine (Phe444), causing a lack of stabilizing hydrogen bond. In HpGGT, substitution of Try433 with Phe (Y433F) had no effect on the catalytic activity, but mutation with alanine (Y433A) resulted in a dramatic decrease in activity; substrate binding was, however, not affected (Morrow et al., 2007).
FIGURE 6
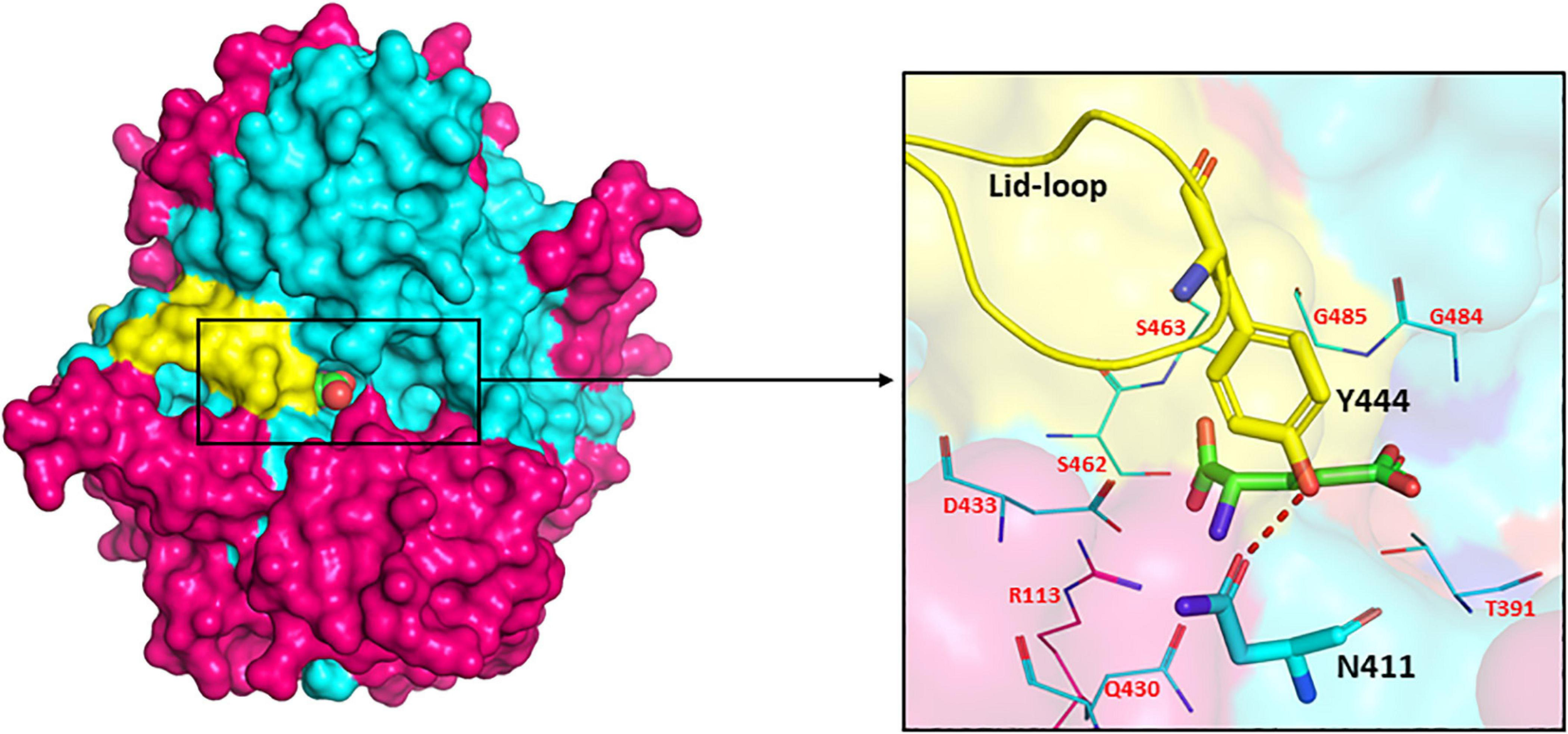
Surface drawing of Escherichia coli gamma-glutamyl transpeptidase (EcGGT) (PDB ID: 2DBX) showing the arrangement of lid-loop around substrate-binding pocket. Pink color highlights large subunit, cyan color highlights small subunit, and L-Glu is represented as a sphere in the surface drawing. Hydrogen bonding between residues Asn411 and Tyr444 is shown by the red dotted line. The active site residues surrounding the bound substrate L-Glu are pictorially shown as lines. The structures are prepared using PyMOL2 software.
Several microbial GGTs including BsGGT, BlGGT, GtGGT, TaGGT, PtGGT, DnGGT, and TtGGT lack the lid-loop segment. Structural analysis of BsGGT and BlGGT demonstrated that their binding pocket is solvent exposed, as they lack lid loop or any of that sort to cover the active site. They are thus suggested to have an open active site cleft to accommodate hydrolysis of large polymeric substrates like PGA (Wada et al., 2010; Hu et al., 2012; Lin et al., 2014). Based on this correlation between the absence of lid loop and hydrolysis of higher molecular weight substrates, it has been proposed that lid loop containing enzymes would not be able to utilize polymeric substrates due to closed catalytic pocket. In an interesting study by Calvio et al. (2018), the role of lid loop in substrate selection was investigated where they constructed a mutant BsGGT by inserting the lid loop from EcGGT. On comparing the activities of EcGGT (lid loop), BsGGT (no lid loop), and mutant BsGGT (lid loop), they concluded that entry of substrate into the active site was regulated by the lid loop depending on molecular weight. Both EcGGT and mutant BsGGT could efficiently utilize glutamine than bulky PGA. Moreover, the presence of the lid loop in mutant BsGGT enhanced its transpeptidation activity as compared to EcGGT and BsGGT (Calvio et al., 2018).
Mutations in Binding Pocket
Site-directed mutagenesis of the active site residues of microbial GGTs has contributed significantly in determining their functional role and also in the development of catalytically efficient variants (Table 3). As already mentioned, the only residue of the large subunit reported to participate in catalysis is the conserved arginine (Arg114 in EcGGT, Arg113 in BsGGT, Arg109 in BlGGT). This residue has been reported to form a weak interaction with the α-carboxylate group of donor substrate (Ikeda et al., 1993; Okada et al., 2006). However, the role of this residue has been elucidated in substrate recognition and binding rather than direct involvement in catalysis. In BlGGT, replacement of this arginine with another positively charged residue lysine (R109K) resulted in significant enhancement of activity (Bindal et al., 2017), whereas in EcGGT (R114K) and BsGGT (R113K), similar mutation resulted in 93 and 22% residual activities, respectively (Minami et al., 2003a; Ong et al., 2008). Substitutions with amino acids of different functional nature such as Leu, Met, Asp, Glu, Phe, etc., were detrimental (Table 3). R109M and R109L mutants of BlGGT exhibited only hydrolytic activity (Bindal et al., 2017).
TABLE 3
| Residue | Mutation | Effect | References | Suggested functional role |
| Arg113 in large subunit of EcGGT* | ||||
| Arg114 (EcGGT) | R114K | 93% activity retained | Ong et al., 2008 | Substrate recognition and binding |
| R114L | No detectable activity | |||
| R114D | No detectable activity | |||
| Arg113 (BsGGT) | R113K | Reduced activity by 78% | Minami et al., 2003a | |
| Arg109 (BlGGT) | R109K | 2-fold enhancement in activity | Bindal et al., 2017 | |
| R109S | 85% activity retained | |||
| R109L | Abolished transpeptidase activity; 5% hydrolytic activity retained | |||
| R109M | Abolished transpeptidase activity; 4% hydrolytic activity retained | |||
| R109F | No detectable activity | |||
| R109E | No detectable activity | |||
| Thr391 at small subunit N-terminal of EcGGT | ||||
| Thr391 (EcGGT) | T391A | No autoprocessing and detectable activity | Suzuki and Kumagai, 2002 | Catalytic nucleophile crucial for both autoprocessing and activity |
| T391S | Impaired autoprocessing and activity (values not mentioned) | |||
| T391C | Impaired autoprocessing and activity (values not mentioned) | |||
| Thr380 (HpGGT) | T380A | No autoprocessing and detectable activity | Boanca et al., 2006 | |
| T380S | Impaired autoprocessing; reduced activity by 92% | |||
| Thr399 (BlGGT) | T399A | No autoprocessing and detectable activity | Lyu et al., 2009 | |
| T399S | Impaired autoprocessing; reduced activity by 89% | |||
| T399C | No autoprocessing and detectable activity | |||
| Thr353 (GtGGT) | T353A | No autoprocessing; retained some activity (values not mentioned) | Castellano et al., 2010 | |
| Asp433 in small subunit of EcGGT | ||||
| Asp445 (BsGGT) | D445A | Abolished transpeptidase activity; 40% hydrolytic activity retained | Minami et al., 2003a | Substrate binding and its affinity |
| D445E | Reduced activity by 86% | |||
| D445N | Reduced activity by 90%; cephalosporin acylase activity increased by 55-fold | |||
| D445Y | Reduced activity by 91% | |||
| E423Y/E442Q/D445N | Cephalosporin acylase activity enhanced by 963-fold | Suzuki et al., 2010 | ||
| Asp433 (EcGGT) | D433N | Abolished transpeptidase activity 82% hydrolytic activity retained; introduced cephalosporin acylase activity | Suzuki et al., 2004c | |
| D433N/Y444A/G484A | Cephalosporin activity enhanced by 50-fold | Yamada et al., 2008 | ||
| Gly484 and Gly485 in small subunit of EcGGT | ||||
| Gly481 (BlGGT)# | G481A | 80% autoprocessed with 78% activity retained | Chi et al., 2018 | Stabilization of oxyanion hole during catalysis; crucial for both autoprocessing and catalysis |
| G481R | Complete loss of autoprocessing and activity | |||
| G481E | 60% autoprocessed with 13% activity retained | |||
| Gly482 (BlGGT)# | G482A | 95% processed; retained 70% activity | ||
| G482R | 20% processed; no detectable activity | |||
| G482E | 80% processed; significant loss in activity (4% left) | |||
| PLSSMXP motif in small subunit of EcGGT (Pro460, Leu461, Ser462, Ser463, Met464, Pro466) | ||||
| P458 (BlGGT) | P458A | 25% increase in activity | Chi et al., 2019b | Crucial for both autoprocessing and catalysis |
| L459 (BlGGT) | L459A | No effect on activity | ||
| S460 (BlGGT) | S460A | 2.1-fold enhancement in activity | ||
| S461 (BlGGT) | S461A | 2.4-fold enhancement in activity | ||
| S463 (EcGGT) | S463T | Reduced activity by 40% | Hsu et al., 2009 | |
| S463K | No autoprocessing and detectable activity | |||
| S463D | No autoprocessing and detectable activity | |||
| M462 (BlGGT) | M462A | 37% increase in activity | Chi et al., 2019b | |
| M464 (EcGGT) | M464E | No autoprocessing and detectable activity | Lo et al., 2007 | |
| M464K | No autoprocessing and detectable activity | |||
| M464L | Reduced activity by 50% | |||
| P464 (BlGGT) | P464A | Reduced activity by 53% | Chi et al., 2019b | |
| Deletion mutants (BlGGT) | △M462 | No autoprocessing and detectable activity | Chi et al., 2019b | |
| △S460-M462 | ||||
| △S461-M462 | ||||
| △P464 | ||||
| Asp452 residue in small subunit of EcGGT | ||||
| Asn450 (BlGGT)† | N450Q | 41% increase in transpeptidase activity | Lin et al., 2016 | Substrate binding and catalysis |
| N450A | 3.5-fold enhancement in transpeptidase activity | |||
| N450D | 3.6-fold enhancement in transpeptidase activity | |||
| N450K | Reduced activity by 27% | |||
List of mutations performed in conserved and catalytically important residues of bacterial gamma-glutamyl transpeptidases (GGTs) and their effect on autoprocessing and activity.
*The residues have been numbered according to their corresponding position in E. coli GGT for ease in reading the table. The activities mentioned in the table are transpeptidase activities measured in presence of both donor and acceptor; hydrolytic activity wherever mentioned is measured only with donor without the addition of any acceptor substrate.#The transpeptidase activity was recorded after an incubation of 21 days at 4°C.†Transpeptidase and hydrolytic activities were recorded after 10 days of incubation at 4°C.
Another active site residue of the small subunit (Asp445 in BsGGT) reported to interact with the α-amino group of donor substrate has also been suggested to have a putative role in substrate binding (Minami et al., 2003a). Replacement of Asp445 by alanine (D445A) resulted in a hydrolytic variant with 40% retained activity and complete abolishment of transpeptidase activity with respect to native BsGGT (Minami et al., 2003a). Likewise, substitution of the corresponding residue Asp433 with asparagine (D433N) in EcGGT abolished its transpeptidase activity, and the hydrolytic variant evolved retained almost 82% activity (Suzuki et al., 2004c). D433N mutant could catalyze deacylation of glutaryl-7-aminocephalosporanicacid (GL-7-ACA) producing 7-aminocephalosporanic acid, a starting material for the synthesis of semisynthetic cephalosporins. GGTs share 30% sequence homology with class IV cephalosporin acylases (CAs) that synthesize semisynthetic cephalosporins. The conserved Asp433 residue of EcGGT is replaced by asparagine in class IV CAs; point mutation (D433N) introduced CA activity in EcGGT, although the acylase activity was quite low; introduction of two random mutations of Y444A and G484A to D433N resulted in up to 50-fold increase in catalytic efficiency for GL-7-ACA substrate (Yamada et al., 2008). Similarly, a triple mutant E423Y/E442Q/D445N of BsGGT was also developed with enhanced acylase activity (Suzuki et al., 2010).
The functional role of two fully conserved tandem glycine repeats reported to form the oxyanion hole has also been explored (Chi et al., 2018). In BlGGT, substituting residues Gly481 and Gly482, respectively, with arginine (G481R) and lysine (G481K) led to impaired autoprocessing as well as catalytic activity, while alanine substitution was not detrimental (Table 3). It has been suggested that proper positioning of the glycine residues is critical for both autoprocessing and catalysis (Chi et al., 2018).
Recently, a highly conserved 458PLSSMXP464 motif present in the small subunit of BlGGT containing the two catalytic serine residues has been studied by deletion and alanine scanning mutagenesis (Chi et al., 2019b). All the deletion mutants showed complete loss of processing and activity. However, alanine substituent of four residues Pro458 (P458A), Ser460 (S460A), Ser461 (S461A), and Met462 (M462A) exhibited a significant increase in catalytic activity with no maturational defects (Table 3). Interestingly, S460A and S461A displayed about twofold enhancement in activity; the role of catalytic Ser463 in EcGGT corresponding to Ser461 in BlGGT was also investigated by mutating it to aspartate (S463D), lysine (S463K), and threonine (S463T). Variants S463D and S463K of EcGGT showed no detectable activities with considerable maturational defects (Hsu et al., 2009), while S463T mutant displayed 40% loss in activity without affecting autoprocessing, indicating that presence of hydroxyl group is required for autoprocessing and addition of charged residues at this position is detrimental for both processing and activity. The same phenomenon was observed for Met464 residue in EcGGT (Met462 in BlGGT) wherein M464E and M464K were maturationally blocked mutants, while M464L exhibited no loss in autoprocessing but had a 50% loss in activity (Lo et al., 2007).
Analysis of the L-Glu-bound crystal structure of BlGGT (PDB ID: 4OTU) suggested importance for another highly conserved residue, Asn450, located close to the binding pocket, in proper substrate orientation (Lin et al., 2016). Replacement of Asn450 by glutamine (N450Q), aspartate (N450D), and alanine (N450A) showed a significant increase in transpeptidase activity up to 3.6-fold enhancement and minor maturational defects, suggesting significant contribution of this residue in substrate binding and catalysis.
Based on mutations in catalytically conserved residues of bacterial GGTs, it was observed that contradictory results were obtained for similar mutations in different bacterial GGTs concerning autoprocessing and catalytic activities (Table 3). The possible reason for the same may be attributed to minor differences in the autoprocessing microenvironments and active structural conformations of different bacterial GGTs. For example, the presence of the lid loop in EcGGT can also influence its catalysis in comparison to Bacillus GGTs lacking the lid-loop region.
Physicochemical Properties of Bacterial GGTs
Wild strains like E. coli and Bacillus species produced very low titers of GGT, as it is a stress-related enzyme (Suzuki et al., 1987; Minami et al., 2003b). Further, cultivation of extremophilic organisms like G. thermodenitrificans, D. radiodurans, etc., is rather difficult; therefore, bacterial GGTs have been largely expressed heterologously in E. coli and B. subtilis via conventional methods for their biochemical and biophysical characterization (Table 4).
TABLE 4
| S. No. | Enzyme Source | Expression Host | Purification | Specific activity (U/mg) | Remarks | References |
| 1. | E. coli K-12 | GGT deficient E. coli K-12 | Two-step; ammonium sulfate precipitation and chromatofocusing | 3.0 | 37-fold enhanced expression; 4.4-fold purification with 42% yield | Suzuki et al., 1988 |
| 2. | E. coli Novablue | E. coli M15 | One-step; Ni-NTA affinity chromatography utilizing N-terminal His6 tag | 4.25 | 32.7-fold purification with 83% yield | Yao et al., 2006 |
| 3. | Bacillus licheniformis ATCC 27811 | E. coli M15 | One-step; Ni-NTA affinity chromatography utilizing N-terminal His6 tag | 185. 6 | 29-fold purification with 26 mg/L yield | Lin et al., 2006 |
| 4. | Geobacillus thermodenitrificans NG80-2 | E. coli BL21 (DE3) | One-step; Ni-NTA affinity chromatography utilizing C-terminal His6 tag | 0.36 (hydrolytic) | 10 mg/L purification yield; no transpeptidase activity | Castellano et al., 2010 |
| 5. | Pseudomonas nitroreducens IFO12694 | E. coli Rosetta-gami B (DE3) | Two-step; DEAE cellulofine and Butyl FF column chromatography | 30.2 (hydrolytic); 2.98 (transpeptidase) | 70-fold enhanced expression; high hydrolytic activity | Imaoka et al., 2010 |
| 6. | Thermus thermophilus | E. coli BL21 (DE3) | One-step; Ni-NTA affinity chromatography utilizing C-terminal His6 tag | 0.151 (hydrolytic) | No transpeptidase activity | Castellano et al., 2011 |
| 7. | Deinococcus radiodurans | E. coli BL21 (DE3) | One-step; Ni-NTA affinity chromatography utilizing C-terminal His6 tag | 0.060 (hydrolytic) | No transpeptidase activity | Castellano et al., 2011 |
| 8. | E. coli | E. coli BL21 (DE3) | One-step; Ni-NTA affinity chromatography utilizing N-terminal His6 tag | 51.41 (U/ml) | High transpeptidase activity; high purification yield of 62 mg/L | Wang et al., 2011 |
| 9. | Bacillus pumilus KS12 | E. coli BL21 (DE3) | One step; Ni-NTA affinity chromatography utilizing C-terminal His6 tag | 1.82 (hydrolytic); 4.35 (transpeptidase) | High hydrolytic activity | Murty et al., 2012 |
| 10. | Picrophilus torridus DSM 9790 | E. coli Rosetta | One-step; Ni-NTA affinity chromatography utilizing C-terminal His6 tag | 18.92 (hydrolytic) | Addition of 2% hexadecane enhanced production ∼10 times; no transpeptidase activity | Rajput et al., 2013 |
| 11. | Bacillus licheniformis ER-15 | E. coli BL21 (DE3) | Two-step; acetone precipitation and Q-sepharose anion exchange chromatography | 4.58 | 10.2-fold purification with 28.8 % yield | Bindal and Gupta, 2014 |
| 12. | Pseudomonas protegens Pf-5 | E. coli BL21 (DE3) | One-step; Ni-NTA affinity chromatography utilizing C-terminal His6 tag | 31.06 (hydrolytic); 4.28 (transpeptidase) | High hydrolytic activity | Kushwaha and Srivastava, 2014 |
| 13. | Pseudomonas fluorescence PfT-1 | E. coli BL21 (DE3) | One-step; Ni-NTA affinity chromatography utilizing C-terminal His6 tag | 36.67 (hydrolytic); 5.45 (transpeptidase) | High hydrolytic activity | Kushwaha and Srivastava, 2014 |
| 14. | Pseudomonas syringae | E. coli Rosetta-gami B (DE3) | Two-step; DEAE cellulofine and Butyl toyopearl column chromatography | 0.92 (transpeptidase) | Low purification yield (1.83%) due to protein instability | Prihanto et al., 2015a |
| 15. | Bacillus licheniformis | E. coli BL21 (DE3) | One-step; Ni-NTA affinity chromatography utilizing N-terminal His6 tag | − | High protein yield of 150–200 mg/2 L | Kumari et al., 2017 |
| 16. | Bacillus licheniformis ER-15 | E. coli BL21 (DE3) | − | 9000 U/L (extracellular activity); 3000 U/L (intracellular activity) | 75–80% extracellular translocation using native signal of the enzyme | Bindal et al., 2018 |
| 17. | Bacillus subtilis 168 | Sporulation-deficient B. subtilis 168 | Two-step; Gigapite and SOURCE 15Q PE column chromatography | 7.49 (hydrolytic); 100 (transpeptidase) | 15-fold higher expression; 30% purification yield | Minami et al., 2003b |
| 18. | Bacillus amyloliquefaciens SMB469 | B. subtilis KCTC 3135 | − | 24.7 (U/ml) | 23 times higher expression | Lee et al., 2017 |
| 19. | Bacillus amyloliquefaciens BH072 | B. subtilis 168 | One-step; Ni-NTA affinity chromatography utilizing His6 tag | 55 | Use of dual signal peptide enhanced secretion to 1.6 times; high purification yield of 90 mg/L | Mu et al., 2019 |
| 20. | Bacillus pumilus ML413 | B. subtilis 168 | Two-step; acetone precipitation and Ni-NTA affinity chromatography utilizing His6 tag | 18.65 (U/ml) | Addition of poly(T/A) tail to ggt mRNA and coexpression of PrsA lipoprotein resulted in 60% high GGT production and 2-fold enhanced extracellular expression | Yang T. et al., 2019 |
Heterologous expression of prokaryotic gamma-glutamyl transpeptidases (GGTs).
Bacterial GGTs are distantly related to plant and mammalian GGTs in terms of their biochemical characteristics (Castellano and Merlino, 2012), but large variations exist among bacterial homologues; for example, Bacillus GGTs are 20–35 times catalytically more efficient than EcGGT (Lyu et al., 2009). Extremophilic bacterial GGTs exhibit only hydrolytic activity suggesting them to be the ancient progenitors that evolved earliest with only hydrolytic activity, and later, transpeptidase activity was imparted to other mesophilic bacterial and eukaryotic GGTs (Castellano et al., 2010, 2011).
Biochemical properties of prokaryotic GGTs are not very diverse, and they optimally function at alkaline pH ranging from 8.0 to 11.0 with different pH optima for hydrolysis and transpeptidation reactions (Balakrishna and Prabhune, 2014; Morelli et al., 2014). Most bacterial GGTs are stable over a wide pH range (6.0–11.0) with maximum stability at alkaline pH range (Shuai et al., 2011; Murty et al., 2012; Bindal and Gupta, 2014; Mu et al., 2019); this is due to the alkaline pKa of GGT substrates, which is above pH 9.0. Moreover, the enzyme can be controlled to catalyze hydrolysis, transpeptidation, or autotranspeptidation simply by adjusting reaction pH (Morelli et al., 2014). Bacterial GGTs exhibit highly variable thermal stability (Castellano et al., 2010) and are reported to optimally catalyze reactions between 37 to 60°C (Castellano and Merlino, 2012). Inhibition in enzyme activity after addition of amino-acid-specific inhibitors like N-bromosuccinimide and phenylmethylsulfonyl fluoride suggested crucial involvement of a tryptophan and serine/threonine residue (Suzuki et al., 1986; Shuai et al., 2011; Bindal and Gupta, 2014). Cysteine-protease inhibitors like β-mercaptethanol, dithiothreitol, and iodoacetamide show only marginal inhibition due to lack of sulfhydryl groups (Moallic et al., 2006; Rajput et al., 2013). GGT-specific substrate analogs like DON and azaserine show complete inhibition (Bindal and Gupta, 2014; Kushwaha and Srivastava, 2014). Metal chelating agents ethylenediaminetetraacetic acid (EDTA) and ethylene glycol tetraacetic acid (EGTA) exhibit moderate inhibition, suggesting contribution of a metal ion in the active conformation of enzyme. The catalytic activity of some bacterial GGTs is reported to enhance with divalent cations such as Mg2++ and Ca2+; however, GGT is not considered to be a metallopeptidase and is sensitized by most of the heavy metal ions like Zn2+, Cd2+, Hg2+, and Pb2+ (Lin et al., 2006; Yao et al., 2006; Shuai et al., 2011; Kushwaha and Srivastava, 2014).
The determination of salt tolerance capacity of bacterial GGTs also characterizes them as halophilic and nonhalophilic. GtGGT is considered the most halotolerant, as it retains more than 90% of its hydrolytic activity in 4 M NaCl, followed by BsGGT retaining 86% of its hydrolytic activity at 3 M NaCl, while EcGGT lost 90% of its hydrolytic activity at the same salt concentration (Wada et al., 2010; Pica et al., 2013). BlGGT has also been reported to tolerate 4 M NaCl concentration without significant effect on its activity (Yang et al., 2011). The underlying mechanism for halotolerance has been reported to be associated with the presence of a relatively large number of negatively charged residues (aspartate and glutamate) on the solvent-exposed surfaces of GtGGT and BsGGT structures even at high salt concentrations resulting in increased water binding capacity, thus increasing solvation of the protein and preventing self-aggregation. EcGGT has also been reported to have acidic residues patches on its surface; however, they are not maintained at the surface under hypersaline conditions (Wada et al., 2010).
Bacterial GGTs exhibit broad substrate specificity for amino acids and other amine-group containing compounds like ethylamine, taurine, 3,4-dihydroxyphenylalanine (DOPA) as acceptors and can thus synthesize a variety of γ-glutamyl compounds utilized in food and pharmaceutical industries. Dipeptides like glycyl glycine and glycyl-L-alanine are fairly good acceptors for bacterial GGTs, with the former being the best (Ogawa et al., 1991; Moallic et al., 2006). Monomeric L-arginine and L-lysine seem to be good acceptors, while L-alanine, L-serine, L-glutamine, and L-glutamic acids have been reported to be poor acceptors (Lee et al., 2017); the propensity of hydrophobic and aromatic amino acids like L-methionine, L-phenylalanine, and L-DOPA is quite good compared to branched acceptors (Suzuki et al., 1986; Ogawa et al., 1991; Shuai et al., 2011). Interestingly, EcGGT poorly accepts D-amino acids, while Bacillus GGTs can accept both L- and D-isomers, thus showing relaxed stereoselectivity at the acceptor site (Ogawa et al., 1991; Hu et al., 2012).
Biophysical characterization of bacterial GGTs has been done to understand the molecular basis of enzyme stability. Temperature- and guanidine hydrochloride (GdnHCl)-induced unfolding of mature and precursor mimic (T399A mutant) forms of BlGGT using CD and emission fluorescence revealed that the mature form was structurally more stable and that both forms displayed an irreversible two-state pattern of thermal unfolding (Hung et al., 2011). A similar analysis showed that EcGGT was more sensitive toward thermal and chemical denaturation than BlGGT, which was more salt stable also (Yang et al., 2011). On the other hand, mature and precursor mutant (T353A) of GtGGT behaved differently; the thermal unfolding of both forms interestingly showed a three-state model including a stable intermediate species, and they exhibited remarkable temperature stability owing to strong electrostatic interactions but were sensitive toward GdnHCl (Pica et al., 2012). In another report, Ho et al. (2013) showed that heat-denatured EcGGT was renatured into active conformation upon incubation at 4°C, while BsGGT showed no such renaturation. They have suggested, based on biophysical characterization and native polyacrylamide gel electrophoresis (PAGE) analysis, that EcGGT dissociated into individual subunits upon heat treatment, which reassembled into active conformation at 4°C (Ho et al., 2013).
Immobilization of Bacterial GGTs
Immobilization of industrially relevant enzymes has been suggested to offset the cost of the process utilizing such enzymes and in circumventing problems of enzyme destabilization (Mohamad et al., 2015). Bacterial GGTs like EcGGT, BsGGT, and BlGGT have been extensively explored for their potential as industrial biocatalysts in the synthesis of a wide variety of γ-glutamyl compounds like L-theanine, γ-glutamyl taurine, γ-D-glutamyl-l-tryptophan, etc., with huge biotechnological applicability (Suzuki et al., 2002a, 2004a; Wang et al., 2008; Bindal and Gupta, 2014). Thus, immobilization has been mainly focused on these three enzymes using various techniques ranging from simple entrapment method to covalent linkage with a suitable support matrix (Table 5) to reduce enzyme production cost and to increase enzyme reusability and stability. However, there are only a few reports of GGT enzyme immobilization that might be attributed to the heterodimeric composition of the enzyme that makes it difficult to remain in the active conformation after immobilization.
TABLE 5
| S. No. | Enzyme | Immobilization matrix | Immobilization method | Cross-linking agent | Loading capacity/immobilization efficiency | Recyclability | Storage stability | Comments* | References |
| 1. | EcGGT | Ca-alginate-k-carrageenan beads | Entrapment | − | 1.5 mg enzyme/g of alginate | ∼55% activity retained after 6 cycles | 50% residual activity after 35 days of storage | Improve thermal stability at 40°C; improved storage stability | Hung et al., 2008 |
| 2. | BlGGT | Ca-alginate beads | Entrapment | − | 40–45% activity recovered; 0.02 U/mg of beads | >90% activity retained after three cycles | Not mentioned | Improve thermal stability at 60°C with >80% residual activity after 15 min | Bindal and Gupta, 2014 |
| 3. | BlGGT | Amino silane coated iron magnetic nanoparticles | Covalent linkage | Glutaraldehyde | 32.4 mg/g of support; 52.4% activity recovered | ∼36% activity retained after 10 cycles | 82% residual activity rafter 30 days of storage | Comparable thermal and storage stability | Chen et al., 2013 |
| 4. | BlGGT | HBPAA-modified magnetic nanoparticles | Covalent linkage | NHS/EDC | 16.2 mg/g of support; 47.4% activity recovered | ∼32% activity retained after 10 cycles | 98% residual activity after 63 days of storage | Slightly improved pH stability in alkaline pH range; comparable thermal and storage stability | Juang et al., 2014 |
| 5. | BsGGT | APES modified and unmodified TiO2 whiskers | Adsorption | − | 184 U/g of support (modified TiO2 whiskers) | 72 and 80% for unmodified and modified TiO2 whiskers, respectively, after 21 cycles | Stable for 60 days at 4°C | Improved pH stability at alkaline pH range (modified); | Wang et al., 2014 |
| 6. | BlGGT | Chitosan microspheres | Covalent linkage | Glutaraldehyde | 11.9 U/mg dry weight of support | 90% activity retained after 10 cycles | 85% residual activity after 32 days of storage | Active in broad pH range; improved thermal and salt stability | Bindal and Gupta, 2016 |
| 7. | BsGGT | Pharmalyte modified titania oxide whiskers | Adsorption | − | Not mentioned | ∼85% after 21 cycles | Stable for 60 days at 4°C | Active in broad pH range; 23 times improved pH stability under alkaline conditions | Ni et al., 2017 |
| 8. | BlGGT | Graphene oxide nanosheets | Covalent and noncovalent linkage | Glutaraldehyde | 34.7 mg/mg of support; 68.7% activity recovered | 46% activity retained after 5 cycles (noncovalent); 45% activity retained after 9 cycles (covalent) | 92% (noncovalent) and 97% (covalent) after 30 days of storage | Active in a broad pH range; improved thermal stability at 60°C | Lin et al., 2018 |
Immobilization of bacterial gamma-glutamyl transpeptidases (GGTs) on different matrices.
Heterologously expressed EcGGT and BlGGT have been immobilized successfully by entrapment methods using Ca-alginate-k-carrageenan and Ca-alginate, respectively, and have been used in theanine synthesis (Hung et al., 2008; Bindal and Gupta, 2014). Bacterial GGTs have also been immobilized on various nanoparticles like iron magnetic nanoparticles, titanium dioxide whiskers, and graphene oxide nanosheets to increase enzyme loading capacity as well as immobilization efficiency and recyclability. Chen et al. (2013) reported covalent immobilization of BlGGT on aldehyde-functionalized magnetic nanoparticles with an efficiency of around 52 and 36% activity retainment after 10 cycles (Chen et al., 2013). In another report, BlGGT was immobilized onto hyperbranched poly(amido acid) (HBPAA)-modified magnetic nanoparticles displaying high storage stability than free enzyme and recyclability up to 10 cycles with 32% activity retained (Juang et al., 2014). Recently, BlGGT has also been immobilized covalently on the highly biocompatible chitosan microsphere beads showing high recyclability with 90% activity retained even after 10 cycles along with improved thermal and salt stability (Bindal and Gupta, 2016).
Immobilization of BsGGT on unmodified and APES-modified mesoporous TiO2 whiskers (denoted as MTw and MTwA, respectively) improved its physicochemical properties. MTw-immobilized BsGGT displayed better thermal stability, while MTwA-immobilized BsGGT exhibited better pH stability in alkaline pH range beneficial for industrial use; as bacterial GGTs are reported to react optimally at alkaline pH (Wang et al., 2014). The modified MTwA BsGGT exhibited good operational stability with more than 80% activity retained after 21 cycles and storage of 60 days at 4°C as well as significantly higher loading capacity than unmodified MTw. In another study, BsGGT immobilized on mesoporous titania whiskers (MTWs) was further modified by adsorbing Pharmalyte on MWT (Ni et al., 2017). Pharmalyte with an isoelectric point of ∼9.2 formed a buffering layer around MTW-immobilized enzyme, thus protecting it from surrounding bulk liquid with significant improvement in pH stability of the immobilized enzyme under alkaline conditions. At an elevated pH 11.0, the pH stability of the immobilized enzyme was 23 times higher than the free enzyme.
Recently, Lin et al. (2018) reported a facile method of BlGGT immobilization on graphene oxide nanosheets by both covalent and noncovalent bonds. The noncovalently immobilized enzyme showed less operational stability with 46% activity remaining after five repeated cycles, while the covalently immobilized enzyme retained about 45% activity after nine cycles of usage with improved thermal stability (Lin et al., 2018). Further, the covalently immobilized enzyme was successfully used as a biocatalyst in synthesizing two γ-glutamyl peptides, vizγ-glutamyl-phenylalanine and γ-glutamyl-leucine.
Biotechnological Applications of Bacterial GGTs
Bacterial GGTs generate an array of compounds through enzymatic γ-glutamylation of amino acids (e.g., Cys, Glu, Phe, Leu, Val, Trp, and His) and other related amine compounds [e.g., 3,4-dihydroxyphenylalanine (L-DOPA) and ethylamine] exhibiting value-added properties like increased solubility, protease resistance, enhanced flavors, and therapeutic capabilities making such compounds significant in food and pharmaceutical industries. Recently, Hideyuki Suzuki (2019) has briefly reviewed the application of bacterial GGTs in food and medicine sectors also (Suzuki, 2019).
Increase in Solubility and Protease Resistance by γ-Glutamylation
Gamma-glutamylation of cysteine has been reported to increase its water-solubility by threefolds (Hara et al., 1992). In addition, γ-glutamylated glutamine exhibited high thermal stability as compared to glutamine, which gets readily converted to pyroglutamic acid in aqueous solutions, thus making the former suitable for nutritional and therapeutic use (Sayed et al., 2010).
Additionally, synthesis of prodrugs like γ-glutamyl 3,4-dihydroxyphenylalanine (L-DOPA) has been reported to have increased half-life due to resistance to cleavage by general serum proteases (Hara et al., 1992; Misicka et al., 1996). Gamma-glutamyl L-DOPA has been shown to increase organ-specific dopamine concentration in mouse models and is thus perceived a potential treatment for Parkinson’s disease (Ichinose et al., 1987). Moreover, γ-glutamyl L-DOPA has also been used as an assay substrate for clinical detection of GGT in human serum samples (Kiuchi et al., 1986). GGT from P. mirabilis and E. coli K-12 have been employed in enzymatic synthesis of γ-glutamyl L-DOPA with 6.7 and 79% conversion rates, respectively (Nakayama et al., 1985; Kumagai et al., 1988).
Gamma-Glutamylation for Synthesis of Flavor-Enhancing Peptides
Some essential amino acids like Phe, Leu, Val, His, etc., taste bitter when consumed orally. The addition of γ-glutamyl moiety to these amino acids using E. coli K-12 GGT has been reported to dramatically reduce their bitterness and improve their taste preference due to the introduction of a refreshing sour flavor, making them suitable for oral consumption as amino acid supplements (Suzuki et al., 2002b, 2004b).
Recently, the importance of γ-glutamyl peptides in imparting kokumi taste to various food items has also been highlighted (Yang J. et al., 2019). Many kokumi flavor-enhancing γ-glutamyl dipeptides, tripeptides, and sulfur-containing compounds like γ-glutamyl-S-alk(en)yl-L-cysteine have been reported to occur naturally in edible legumes, garlic, onion, and several fermented foods like cheese, fish, yeast extract, and soy sauce (Kuroda et al., 2012; Liao et al., 2013; Liu et al., 2015). Enzymatic synthesis of such kokumi peptides has also been successful employing bacterial GGTs as biocatalysts with L-glutamine as a donor and various amino acids as acceptors (Table 6). Large-scale production of kokumi peptides using bacterial GGTs has been suggested to be a cost-effective and suitable alternative for their commercial use, thus not depending on their natural sources, which involve agricultural and environmental limitations (Yang J. et al., 2019).
TABLE 6
| Enzyme source | Peptides | Application | References |
| E. coli GGT | γ-glutamyl phenylalanine; γ-glutamyl histidine; γ-glutamyl leucine; γ-glutamyl valine | Debittering of bitter amino acids | Suzuki et al., 2002b, 2004b |
| B. subtilis GGT | γ-glutamyl methionine; | Flavor-enhancing kokumi properties | Morelli et al., 2014 |
| B. licheniformis GGT | γ-glutamyl phenylalanine; γ-glutamyl leucine; γ-L-glutamyl-S-allyl-L-cysteine | Flavor-enhancing kokumi properties | Chen et al., 2015; Chi et al., 2017; Lee et al., 2018 |
| B. amyloliquefaciens GGT (L-glutaminase with transpeptidase activity) | γ-[Glu](1 ≦ n < 5)-phenylalanine γ-[Glu]n-valine; γ-[Glu]n-methionine γ-[Glu](n ≥ 1)-Gln | Short-chain peptides with kokumi flavor | Yang et al., 2017, 2018 |
| B. subtilis GGT (salt tolerant) | γ-glutamyl valyl glycine; γ-glutamyl valine | Flavor-enhancing kokumi properties | Ho and Suzuki, 2013 |
| B. subtilis GGT (salt tolerant) | Glutamic acid | Umami flavor to soy sauce | Kijima and Suzuki, 2007 |
List of enzymatically synthesized flavor-enhancing peptides.
The hydrolytic activity of bacterial GGTs has also been exploited in soy sauce fermentation for imparting a characteristic umami taste (Kijima and Suzuki, 2007). Traditionally, soy sauce is prepared by proteolytic digestion of soy protein by fungal proteases like those from Aspergillus oryzae/sojae (Ito et al., 2013). The umami flavor of soy sauce has been introduced by the conversion of free glutamine released during fermentation to glutamic acid by fungal glutaminases. However, inhibition of fungal glutaminases at high salt concentrations during fermentation leads to unwanted conversion of glutamine to pyroglutamic acid. Thus, a salt-tolerant glutaminase activity is crucial for soy sauce fermentation. For this, GGT from B. subtilis (Kijima and Suzuki, 2007) and G. thermodenitrificans (Castellano et al., 2010) have been discovered to be salt tolerant with more than 80 and 90% hydrolytic activity retained in the presence of 18% NaCl, respectively. As GGT from Bacillus subtilis shows both hydrolytic and transpeptidase activity, salt-tolerant GGT from extremophilic G. thermodenitrificans, harboring only hydrolytic activity, is a better alternative to glutaminases in soy sauce fermentation. However, salt-tolerant GGT from B. subtilis can be exploited in another Japanese traditional seasoning, meso, where both transpeptidase and hydrolytic activities are required in improving product taste (Ho and Suzuki, 2013).
Recently, Kalum et al. (United States 2020/0196617 A1) have filed a patent against the utilization of GGT enzyme from B. licheniformis/Bacillus horikoshii in dough relaxation (Kalum et al., 2020). They have claimed that the addition of bacterial GGT enzyme during dough making can result in better extensibility of dough, which would help in producing flattened dough during the manufacturing of products like bread, flatbread, crackers, pizzas, pasta, noodles, laminated baking products, biscuits, baguettes, and hamburgers.
Nutraceutical and Therapeutic γ-Glutamyl Compounds
In nature, γ-glutamyl peptides are ubiquitously present in bacteria, plants, and animals. They are the products and by-products of the glutathione cycle. Many naturally occurring γ-glutamyl peptides and their derivatives like theanine, γ-D-glutamyl aminomethylphosphonate, γ-D-glutamylglycine, γ-D-glutamyltaurine, γ-D-glutamyl-β-alanine, etc., have been reported to be beneficial for human consumption (Suzuki et al., 2007; Yang J. et al., 2019). Among them, peptides like L-theanine, γ-glutamyl taurine, and γ-D-glutamyl-L-tryptophan have been extensively studied and synthesized using bacterial GGTs. However, in the future, in-depth study and analysis of other naturally occurring γ-glutamyl peptides and their prospective use in pharmaceuticals would also demand their enzymatic synthesis, which eventually would increase the biotechnological demand of bacterial GGTs.
L-Theanine
L-Theanine (γ-L-glutamylethylamide) is a nonproteinogenic amino acid occurring naturally in tea. Besides imparting the characteristic umami flavor to tea, L-theanine has various neurophysiological benefits on human health like regulating high blood pressure (Yokogoshi et al., 1995), improving cognition power (Haskell et al., 2008), reducing stress and promoting relaxation (Lu et al., 2004; Kimura et al., 2007), strengthening the immune system (Miyagawa et al., 2008), and enhancing antitumor activity (Sadzuka et al., 2000; Liu et al., 2009) and antiobesity effects (Zheng et al., 2004).
Owing to such a wide array of health benefits, L-theanine has huge demand worldwide. However, due to shortcomings of natural extraction and chemical synthesis methods, scientists and industries have been moving toward the enzymatic synthesis of L-theanine (Mu et al., 2015). Bacterial enzymes like GGT, glutamine synthetase, L-glutaminase, and γ-glutamylmethylamide synthetase have been utilized in L-theanine synthesis. As compared to other enzymes, GGT has advantages like readily available enzyme source, convenient reaction set-up, short reaction time, and no ATP requirement (Mu et al., 2015).
There are numerous reports on the enzymatic synthesis of L-theanine using bacterial GGTs with variable conversion rates ranging from 27% to as high as 94% (Table 7). The percent conversion and yield of L-theanine obtained in different studies were observed to be highly influenced by the ratio of the donor (mainly L-glutamine) and acceptor (ethylamine) used in the reaction. In a recent report, N450A mutant of GGT from B. licheniformis showed a conversion rate of 94% as compared to 62% with wild type at pH 10.5 and 37°C after 4 h using substrate molar ratio of 1:2.4 corresponding to 250 mM L-glutamine and 600 mM ethylamine (Chi et al., 2019a). While in another report, a double mutant, V319A/S437G, of GGT from B. amyloliquefaciens resulted in an increased conversion rate for L-theanine synthesis from 58 to 83% at pH 10.5 and 37°C after 5 h using a substrate molar ratio of 1:10 (Li et al., 2020). Yang T. et al. (2019) demonstrated a fed-batch strategy to increase the yield of L-theanine employing GGT from B. pumilus overexpressed in B. subtilis 168 host. Addition of fixed amounts of glutamine and ethylamine after constant intervals resulted in a final yield of 53.5 g/L with a conversion rate of 63% after 16 h (Yang T. et al., 2019). Recently, the use of ultrasound power of 100 W has been reported to increase theanine yield by 2.61-fold using E. coli K-12 GGT with a conversion rate of 89.1% (Xu et al., 2020).
TABLE 7
| Biocatalyst | L-Glutamine concentration (donor; mM)* | Ethylamine concentration (acceptor; mM) | Enzyme concentration (U/ml) | Reaction conditions | Percent conversion/yield# | References |
| Pseudomonas nitroreducens IFO 12694 L-glutaminase | 700 | 1500 | 0.5 | pH 11.0; 30°C; 7 h | 39%; 38 g/L | Tachiki et al., 1998 |
| E. coli K-12 GGT | 200 | 1500 | 0.4 | pH 10.0; 37°C; 2 h | 60%; 120 mM | Suzuki et al., 2002a |
| E. coli Novablue GGT | 10 | 40 | 1.04 | pH 10.0; 37°C | 40% | Yao et al., 2006 |
| Ca-alginate-k-carrageenan immobilized E. coli Novablue GGT | 25 | 40 | 1.5 mg/g alginate | pH 10.0; 40°C; 12 h | 27% | Hung et al., 2008 |
| Sodium alginate immobilized E. coli K-12 cells expressing GGT | 300 (Glutamic acid γ-methyl ester) | 3000 | 0.1 g/ml of immobilized cells | pH 10.0; 45°C; 18 h | 69.3% | Zhang et al., 2010 |
| B. subtills SK 11.004 GGT | 20 | 50 | 0.06 | pH 9.0; 37°C; 4 h | 94%; 18.9 mM | Shuai et al., 2010 |
| Sumo-tag fused E. coli GGT | 267 | 2000 | 1.5 | pH 10.5; 45°C; 24 h | 80%; 41 g/L | Wang et al., 2011 |
| B. subtilis NX-2 GGT | 48 [L-glutamine-Zn(II) complex] | 1600 | 0.5 | pH 9.0; 37°C; 3 h | 63.8%; 61.3 mM | Wang et al., 2012 |
| E. coli GGT | 10 (γ-glutamyl-p-nitroanilide) | 200 | − | pH 9.0; 37°C; 6 h | 93% | Zhang et al., 2013 |
| P. nitroreducens DSM 14399 L-glutaminase | 300 | 1500 | 1.5 U/5 ml | pH 10.0; 37°C; 5 h | 40%; 28 g/L | Pu et al., 2013 |
| B. licheniformis ER 15 GGT | 80 | 600 | 1 | pH 9.0; 37°C; 4 h | >84% | Bindal and Gupta, 2014 |
| GGT from B. subtilis 168 | 200 | 2200 | − | pH 10.0; 37°C; 5 h | 78% | Chen et al., 2014 |
| B. amyloliquefaciens GGT | 20 | 100 | 0.5 | pH 9.0; 40°C; 6 h | 48% | Mu et al., 2019 |
| B. pumilus ML413 GGT | 250 | 2500 | 1 | pH 10.0; 30°C; 9 h (batch reaction) | 74%; 32 g/L | Yang T. et al., 2019 |
| B. pumilus ML413 GGT | 250 (25 g/L added after every 3 h) | 2500 (73 g/L added after every 3 h) | 1 | pH 10.0; 30°C; 16 h (fed batch reaction) | 63%; 53.5 g/L | Yang T. et al., 2019 |
| B. licheniformis N450A mutant GGT | 250 | 600 | 25 μg/ml | pH 10.5; 37°C; 4 h | 94% | Chi et al., 2019a |
| B. licheniformis N450D mutant GGT | 250 | 600 | 25 μg/ml | pH 10.5; 37°C; 4 h | 75% | Chi et al., 2019a |
| B. licheniformis GGT | 250 | 600 | 25 μg/ml | pH 10.5; 37°C; 4 h | 62% | Chi et al., 2019a |
| P. nitroreducens SP.001 L-glutaminase | 750 | 1000 | 1 | pH 9.5; 30°C; 11 h | 66.1%; 490 mM | Shuai et al., 2019 |
| Bacillus amyloliquefaciens GGT mutant V319A/S437G | 200 | 2000 | 20 μg/ml | pH 10.0; 37°C; 5 h | 83% | Li et al., 2020 |
| E. coli K-12 GGT | 120 | 480 | pH 9.0; 45°C; 5 h; 100 W ultrasonic waves | 89.1%; 18.5 g/L | Xu et al., 2020 |
Enzymatic synthesis of L-theanine using various bacterial gamma-glutamyl transpeptidases (GGTs) as biocatalysts.
*Donor substrate used other than glutamine mentioned in bracket.#Percent conversion calculated against initial concentration of L-glutamine donor.
Interestingly, in a recent report by Sun et al. (2019), endophytic bacteria present in Camellia sinensis has been shown to contribute toward the production of L-theanine. The endophyte has been identified as Luteibacter spp., and its GGT enzyme has been demonstrated to exhibit high activity for L-theanine synthesis using L-glutamine and ethylamine as substrates. Thus, high conversion rate and high yields obtained using bacterial GGTs suggest their potential in bio-catalytic synthesis of L-theanine at an industrial scale for commercial use.
Gamma-D-Glutamyl-L-Tryptophan
Gamma-D-glutamyl-L-tryptophan (also known as SCV-07 or Golotimod) is an immunomodulatory dipeptide with broad-spectrum stimulatory activities that boost immune response (Kolobov and Simbirtsev, 1999). It has been implicated as a potential medicine for the treatment of tuberculosis (Simbirtsev et al., 2003) and a potent antiviral therapeutic agent against genital HSV-2 recurrent disease (Rose et al., 2008). The role of this peptide in suppressing tumor growth and reducing severity and duration of oral mucositis induced by chemoradiation therapy has also been reported (Watkins et al., 2010; Alterovitz et al., 2011; Papkoff et al., 2011; Tuthill et al., 2011).
Enzymatic synthesis of SCV-07 can be a promising alternative to meet the future demand of this prospective drug. Bacterial GGTs can take both L- and D-forms of glutamine as donor and show high specificity toward aromatic amino acid L-tryptophan as acceptor and thus can be used as a biocatalyst. Bacterial GGTs from E. coli and Bacillus species have been employed successfully in enzymatic synthesis of SCV-07 (Suzuki et al., 2004a; Wang et al., 2008; Saini et al., 2017). GGT from E. coli could synthesize 33 mM product with a conversion rate of 66% using a 1:1 molar ratio of D-glutamine (donor; 50 mM) and L-tryptophan (acceptor; 50 mM) in 5 h at 37°C (Suzuki et al., 2004a). At similar reaction conditions, B. atrophaeus GGT led to 50% conversion within 6 h with a product yield of 25 mM (Saini et al., 2017). GGT from B. subtilis NX-2 catalyzed SCV-07 synthesis using a 5:7 molar ratio of D-glutamine and L-tryptophan with a conversion rate of 42% after 4 h at 40°C (Wang et al., 2008). However, B. subtilis NX-2 GGT also catalyzed irreversible hydrolysis of the product γ-D-glutamyl-L-tryptophan, which resulted in a lower conversion rate.
Gamma-Glutamyl-Taurine
Gamma-glutamyl taurine has been isolated from brains of different mammals and suggested to be synthesized by mammalian GGT in vivo (Török et al., 1981; Marnela et al., 1985). Many physiological effects of γ-glutamyl taurine on the central nervous system have been reported (Davies et al., 1982; Bittner et al., 2005). It has also been reported to show a positive ionotropic effect on the locust heart (Feuer and S.-Rózsa, 1981), on metamorphosis of amphibians (Feuer et al., 1978), and on the activity and concentration of plasma renin (Feuer and Gaál, 1979). This naturally occurring peptide with numerous physiological benefits has been synthesized enzymatically under in vitro conditions. As mammalian GGTs are difficult to purify owing to their membrane-bound location and heavy glycosylation, bacterial GGTs are being employed for enzymatic synthesis of γ-glutamyl taurine (Suzuki et al., 2002c). GGT from E. coli catalyzed the synthesis of the peptide using 200 mM L-glutamine and 200 mM taurine and a conversion rate of 22.5% corresponding to 45 mM γ-L-glutamyl taurine was achieved after 1 h of reaction at 37°C. Lower conversion rates were attributed to a poor affinity for taurine as an acceptor and the formation of by-products like γ-glutamyl glutamine and γ-glutamyl-γ-glutamyl-taurine (Suzuki et al., 2002c). Interestingly, γ-D-glutamyl taurine, the isomeric form of γ-L-glutamyl taurine, has also been reported to have similar physiological effects to some extent (Davies et al., 1982; Jones et al., 1984). E. coli GGT has been reported to exhibit poor specificity toward D-amino acids as γ-glutamyl acceptors (Suzuki et al., 1986). Thus, it has been employed to synthesize γ-D-glutamyl taurine with a conversion rate of 71% using D-glutamine as a donor (Suzuki et al., 2003).
Allied Applications of Microbial GGTs
In addition to transpeptidase and hydrolytic activities, a few microbial GGTs have also been reported to exhibit some unique activities like cephalosporin acylase (CA) and β-aspartyl transferase (BAT), as they can act on substrates like glutaryl-7-aminocephalosporanic acid and asparagine, respectively.
Cephalosporin acylases that catalyze the conversion of cephalosporin C to 7-aminocephalosporanic acid, a starting material for the synthesis of semisynthetic cephalosporins, exhibits a higher affinity for GGT substrate than CA substrate (Yamada et al., 2008). Thus, it has been speculated that class IV CAs are primarily GGTs with adventitious CA activity (Li et al., 1999). Microbial GGTs have also been explored for CA activity, and GGT from B. subtilis has been reported to possess an inherent CA activity, as it can catalyze deacylation of glutaryl-7-aminocephalosporanic acid (GL-7-ACA) and produce 7-aminocephalosporanic acid (Yamada et al., 2008). However, the activity reported was very low, which could be enhanced to 963-fold for a triple mutant E423Y/E442Q/D445N of BsGGT, developed by site-directed and random mutagenesis approach (Suzuki et al., 2010). In another study, a single-point mutation at residue 433 (D433N) introduced CA activity in E. coli GGT (EcGGT), which was initially absent (Suzuki et al., 2004c). Subsequent introduction of two random mutations, Y444A and G484A, to D433N EcGGT variant further enhanced catalytic efficiency for GL-7-ACA substrate by 50-fold (Yamada et al., 2008). Thus, microbial GGTs have the potential to be utilized in the pharmaceutical industry as CAs.
Gamma-glutamyl transpeptidase from Pseudomonas syringae (PsGGT) has been reported to utilize asparagine in addition to glutamine as a donor substrate and catalyze the transfer of β-aspartyl and γ-glutamyl moiety to hydroxylamine, respectively (Prihanto et al., 2015a). The transfer of β-aspartyl moiety was higher than the transfer of γ-glutamyl moiety, thus making it a novel GGT exhibiting both γ-glutamyl transferase and β-aspartyl transferase activities. Apart from PsGGT, only one enzyme from Mycobacterium tuberculosis, viz aspartotransferase, has been reported to exhibit β-aspartyl transferase activity (Reddy et al., 1969). Enzymes like asparaginase can only catalyze the hydrolysis of asparagine, but no transfer reaction has been reported (Hendriksen et al., 1969). The novel β-aspartyl transferase activity of PsGGT was applied for the synthesis of L-β-aspartyl hydroxamate (BAH) using 80 mM L-asparagine (donor substrate) and 40 mM hydroxylammonium chloride (acceptor substrate) with 0.106 mM product yield (Prihanto et al., 2015b).
In a recent report, degradation fragments of GGT from B. subtilis BU108 have been demonstrated to have antimicrobial activity against Streptomyces scabiei (Liu et al., 2020). Two antimicrobial peptides (AMPs), viz KT20 (KKGGNAIDAAVAIQFALNVT) and IF20 (IQKDLAKTFKLIRSNGTDAF), have been identified, which are the degradation fragments of GGT enzymes. Thus, B. subtilis GGT has been speculated to have an application in a potato common scab biocontrol.
Conclusion and Future Perspectives
Gamma-glutamyl transpeptidase enzyme has been studied extensively over the past few decades owing to its universal existence and high sequence similarity among the prokaryotes and eukaryotes, indicating evolutionary conservation, the reason for which is not clear yet. Just like mammals, GGTs from Gram-negative bacteria also degrade glutathione; Gram-positive ones, however, are recognized as PGA degraders and hence grouped separately. GGT has many physiological roles in Gram-positive and Gram-negative bacteria (Table 1). Recently, the involvement of H. pylori GGT (HpGGT) in pathogenesis has been well established, and HpGGT can be explored as a potential drug target in combating H. pylori infection in combination with antibiotics. Likewise, extensive research on GGTs from other bacterial pathogens is required in understanding their virulence mechanism for developing efficient drugs.
Besides their physiological and clinical relevance, crystallization of bacterial GGTs also unveiled molecular aspects of autoprocessing and catalysis, emphasizing the role of conserved threonine that acts as a nucleophile. However, the interplay between small and large subunit formed after autoprocessing is yet to be elucidated. There are few reports on in vitro association of large and small subunit with variable results, as the decoupling of autoprocessing was only successful in a few cases. Crystallization studies have provided details on donor binding site, while identification of residues of acceptor binding site is still nebulous. Thus, crystallization should be attempted in the presence of acceptor substrate to have information about acceptor site residues. Mutational studies have been done focusing mainly on catalytically important small subunit residues to develop variants with improved catalytic activity. However, crystallization of such mutants is also required for a better understanding of structure and function.
Simpler handling of bacterial cultures, their easy genetic manipulations for overexpression, simpler purification techniques, and utilization of glutamine as a cheaper donor source make them a potential candidate for industrial exploitation. Most work has been done on the enzymatic synthesis of theanine, a nutraceutical with huge market potential, and numerous patents have been granted for the same (Table 8); however, still, no product enzymatically synthesized by GGT has been commercialized to date. Lack of commercially available GGTs could be one of the major reasons along with meager studies on the bioactivity of other potential γ-glutamyl peptides like γ-D-glutamyl-L-tryptophan, γ-glutamyl taurine, and other kokumi peptides. Addressing these issues along with immobilization techniques would aid in increasing the market value of GGTs. Other allied applications of bacterial GGTs showing unique β-aspartyl transferase activity and as a biocontrol agent require in-depth exploration to widen their applicability.
TABLE 8
| S. No. | Patent No. | Title | Technology Organism/enzyme/substrates/conditions | Assignee/inventors | Filling date |
| 1. | United States 20200196617 A1 | Dough Relaxation Using Gamma Glutamyl Transpeptidase | Bacterial GGT is added to flour to make dough | Novozymes AS | 2018-06-20 |
| 2. | US9512177B2 | Method for producing γ-glutamyl-valyl-glycine crystal | Microbial GGT is used to convert valyl-glycine to γ-glutamyl-valyl-glycine | Ajinomoto Co Inc | 2015-02-10 |
| 3. | WO2006102722A1 | Process for the production of gamma-glutamylcysteine | Bovine GGT is used to synthesize gamma-glutamyl cysteine | Wallace John BridgeMartin Hani Zarka | 2006-03-31 |
| 4. | JP8919992A | Production of l-gamma-glutamyl-lower alkylamide | Purified Bacillus subtilis GGT; L-glutamine and ethylamine hydrochloride; pH 10.0, 37°C and 18 h | Daiwa Kasei Kk | 1992-04-10 |
| 5. | JP18431892A | Production of theanine | Immobilized Pseudomonas nitroreducens IFO 12694 cells; L-glutamine and ethylamine; pH 9.5 and 22 days | Taiyo Kagaku Co Ltd | 1992-05-30 |
| 6. | CN101270376A | Method for synthesizing L-theanine with enzyme | Gamma-glutamyltranspeptidase; L-glutamine-copper (II) complex and ethylamine; pH 8.0–11.0, 35–45°C, 2–5 h | Car Taiyo Kagaku Kabushiki Kaisha | 2008-05-14 |
| 7. | CN101343618A | Preparation method for natural theanine | GGT from Bacillus subtilis SK11.004 strain; glutamine and ethylamine; pH 9–10.5, 37°C and 2 h | Car Taiyo Kagaku Kabushiki Kaisha | 2008-08-19 |
| 8. | CN101445787A | Method for biosynthesizing theanine by immobilized gene-engineering strain | Immobilized E. coli cells harboring pET32a-GGT plasmid; glutamine and ethylamine hydrochloride; pH 9–10, 32°C | Tea Research Institute of Chinese Academy of Agricultural Sciences | 2008-12-04 |
| 9. | CN101445788A | Method for biosynthesizing theanine by gene-engineering strain | Recombinant E. coli GGT; glutamine and ethylamine hydrochloride; pH 9, 37°C | Tea Research Institute of Chinese Academy of Agricultural Sciences | 2008-12-04 |
| 10. | US20090169704A1 | Process for the enzymatic preparation of a gamma-glutamyl compound | GGT enzyme derived from a plant belonging to the Graminaceae or Leguminaceae family, or from Camellia sinensis; glutamine or glutamic acid; ethylamine or ethylamine hydrochloride; pH is < 8.0 | ConopcoInc | 2008-12-18 |
| 11. | CN101457241B | Method for preparing theanine by using species coupling ATP regenerative technology | GGT from immobilized Bacillus licheniformis cells and ATP regeneration from Bacillus stearothermophilus; glutamine (glutamic acid) and ethylamine; pH 10. 5, 30°C, 48 h | Tea Research Institute of Chinese Academy of Agricultural Sciences | 2009-01-04 |
| 12. | CN101560532A | L-theanine enzymatic transformation preparation method | GGT enzyme or Bacillus subtilis whole cells; L-glutamic acid-gamma-alkyl ester or L-glutamyl hydrazide and ethylamine; pH 9–11, 25-60°C | Tea Research Institute of Chinese Academy of Agricultural Sciences | 2009-05-25 |
| 13. | CN101643712B | Escherichia coli strain for efficiently converting glutamine to synthesize L-theanine and application thereof | E. coli of CCTCC No. M 209166; glutamine and ethylamine; 20–37°C, 40–48 h | 2009-09-15 | |
| 14. | CN103409475A | Method for synthesizing L-theanine through enzyme process | Recombinant GGT from Bacillus subtilis 168; glutamine and ethylamine; pH 9.5–10.5, 37–50°C, 2–5 h | 2013-07-18 | |
| 15. | CN104372046A | Method for producing L-theanine by supplementing material | GGT from Bacillus subtilis; glutamine and ethylamine hydrochloride; pH 9–11, 30–45°C, 10–20 h; substrates are supplemented after every 2 h | 2014-11-19 | |
| 16. | CN104404075A | Method for catalyzing to generate L-theanine by using recombinant Bacillus subtilis secreted gamma-glutamyltranspeptidase | Recombinant Bacillus subtilis GGT expressed in Bacillus subtilis expression host; glutamine and ethylamine hydrochloride; pH 10, 37°C, 5 h | 2014-12-09 | |
| 17. | CN104561160A | Method for preparing theanine by using biological method | Pseudomonas nitroreducens cells; glutamine and triethylamine hydrochloride; pH 5.5–6.8, 25–38°C, 4–6h; L-glutamine is supplemented 1–3 times. | 2014-12-22 | |
| 18. | CN104789538A | Supplementary strategy for improving catalytic synthesis of L-theanine from gamma-glutamyltranspeptidase | Recombinant Bacillus subtilis GGT expressed in Bacillus subtilis expression host; glutamine and ethylamine hydrochloride; pH 10, 37°C, 18 h; glutamine is supplemented after regular intervals. | 2015-03-30 |
List of patents on applications of bacterial gamma-glutamyl transpeptidases (GGTs).
Based on the present review, addressing bacterial GGTs and their relevance, it will be appropriate to put forth that detailed understanding of the structure–function relationship of bacterial GGTs utilized in synthesizing γ-glutamyl compounds has led to their emergence as the industrial biocatalyst in the current century and can be utilized in many other biotechnological sectors. It indicates that GGT is indeed an emerging biocatalyst, and in the coming decade, its commercial exploitation and new insights of its catalytic mechanism regarding acceptor binding can be visualized.
Statements
Author contributions
MS prepared the complete first draft along with conceptualization and review outline prepared in supervision of the corresponding author RG. AK contributed in finalization of a major portion of the review covering structural and functional aspects of bacterial GGTs and also contributed in finalizing bibliography. SB and KS contributed in finalizing parts of the manuscript covering biotechnological applications and physiological significance of bacterial GGTs. RG contributed during different stages of manuscript preparation starting from drawing the review outline to checking drafts at every stage for technical corrections and improving overall language for finalizing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We would like to acknowledge Indian Council of Medical Research (ICMR), Government of India for providing Senior Research Fellowship (Grant No. 3/1/3/JRF-2017/HRD-LS/55101/08).
Acknowledgments
The authors would like to acknowledge Shashi A. Verma, Editor (English) (Former) at Directorate of Knowledge Management, Indian Council of Agricultural Research for editing the language of the manuscript. The authors would like to acknowledge Indian Council of Medical Research (ICMR), Govt. of India for providing Senior Research Fellowship (Grant No. 3/1/3/JRF-2017/HRD-LS/55101/08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.641251/full#supplementary-material
References
1
Alkhuder K. Meibom K. L. Dubail I. Dupuis M. Charbit A. (2009). Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis.PLoS Pathog.5:e1000284. 10.1371/journal.ppat.1000284
2
Alterovitz G. Tuthill C. Rios I. Modelska K. Sonis S. (2011). Personalized medicine for mucositis: Bayesian networks identify unique gene clusters which predict the response to gamma-D-glutamyl-L-tryptophan (SCV-07) for the attenuation of chemoradiation-induced oral mucositis.Oral Oncol.47951–955. 10.1016/j.oraloncology.2011.07.006
3
Ashiuchi M. Misono H. (2002). Biochemistry and molecular genetics of poly-gamma-glutamate synthesis.Appl. Microbiol. Biotechnol.599–14. 10.1007/s00253-002-0984-x
4
Balakrishna S. Prabhune A. A. (2014). Gamma-glutamyl transferases: a structural, mechanistic and physiological perspective.Front. Biol.9:51–65. 10.1007/s11515-014-1288-0
5
Barnes I. H. A. Bagnall M. C. Browning D. D. Thompson S. A. Manning G. Newell D. G. (2007). γ-Glutamyl transpeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni.Microb. Pathog.43198–207. 10.1016/j.micpath.2007.05.007
6
Bindal S. Dagar V. K. Saini M. Khasa Y. P. Gupta R. (2018). High level extracellular production of recombinant γ-glutamyl transpeptidase from Bacillus licheniformis in Escherichia coli fed-batch culture.Enzyme Microb. Technol.11623–32. 10.1016/j.enzmictec.2018.05.004
7
Bindal S. Gupta R. (2014). L-Theanine synthesis using γ-glutamyl transpeptidase from Bacillus licheniformis ER-15.J. Agric. Food Chem.629151–9159. 10.1021/jf5022913
8
Bindal S. Gupta R. (2016). Thermo- and salt-tolerant chitosan cross-linked γ-glutamyl transpeptidase from Bacillus licheniformis ER15.Int. J. Biol. Macromol.91544–553. 10.1016/j.ijbiomac.2016.05.106
9
Bindal S. Sharma S. Singh T. P. Gupta R. (2017). Evolving transpeptidase and hydrolytic variants of γ-glutamyl transpeptidase from Bacillus licheniformis by targeted mutations of conserved residue Arg109 and their biotechnological relevance.J. Biotechnol.24982–90. 10.1016/j.jbiotec.2017.03.034
10
Bittner S. Win T. Gupta R. (2005). γ-L-glutamyltaurine.Amino Acids28343–356. 10.1007/s00726-005-0196-7
11
Boanca G. Sand A. Barycki J. J. (2006). Uncoupling the enzymatic and autoprocessing activities of Helicobacter pylori γ-glutamyltranspeptidase.J. Biol. Chem.28119029–19037. 10.1074/jbc.M603381200
12
Boanca G. Sand A. Okada T. Suzuki H. Kumagai H. Fukuyama K. et al (2007). Autoprocessing of Helicobacter pylori γ-glutamyltranspeptidase leads to the formation of a Threonine-Threonine catalytic dyad.J. Biol. Chem.282534–541. 10.1074/jbc.M607694200
13
Brannigan J. A. Dodson G. Duggleby H. J. Moody P. C. E. Smith J. L. Tomchick D. R. et al (1995). A protein catalytic framework with an N-terminal nucleophile is capable of self-activation.Nature378416–419. 10.1038/378416a0
14
Calvio C. Romagnuolo F. Vulcano F. Speranza G. Morelli C. F. (2018). Evidences on the role of the lid loop of γ-glutamyltransferases (GGT) in substrate selection.Enzyme Microb. Technol.11455–62. 10.1016/j.enzmictec.2018.04.001
15
Candela T. Fouet A. (2005). Bacillus anthracis CapD, belonging to the γ-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan.Mol. Microbiol.57717–726. 10.1111/j.1365-2958.2005.04718.x
16
Castellano I. Di Salle A. Merlino A. Rossi M. La Cara F. (2011). Gene cloning and protein expression of γ-glutamyltranspeptidases from Thermus thermophilus and Deinococcus radiodurans: comparison of molecular and structural properties with mesophilic counterparts.Extremophiles15259–270. 10.1007/s00792-011-0355-6
17
Castellano I. Merlino A. (2012). γ-Glutamyltranspeptidases: sequence, structure, biochemical properties, and biotechnological applications.Cell. Mol. Life Sci.693381–3394. 10.1007/s00018-012-0988-3
18
Castellano I. Merlino A. Rossi M. La Cara F. (2010). Biochemical and structural properties of gamma-glutamyl transpeptidase from Geobacillus thermodenitrificans: An enzyme specialized in hydrolase activity.Biochimie92464–474. 10.1016/j.biochi.2010.01.021
19
Castonguay R. Lherbet C. Keillor J. W. (2003). Kinetic studies of rat kidney γ-glutamyltranspeptidase deacylation reveal a general base-catalyzed mechanism.Biochemistry4211504–11513. 10.1021/bi035064b
20
Chang H.-P. Liang W.-C. Lyu R.-C. Chi M.-C. Wang T.-F. Su K.-L. et al (2010). Effects of C-terminal truncation on autocatalytic processing of Bacillus licheniformis γ-glutamyl transpeptidase.Biochemistry75919–929. 10.1134/S0006297910070151
21
Chen X. Su L. Wu D. Wu J. (2014). Application of recombinant Bacillus subtilis γ-glutamyltranspeptidase to the production of L-theanine.Process Biochem.491429–1439. 10.1016/j.procbio.2014.05.019
22
Chen Y.-Y. Lo H.-F. Wang T.-F. Lin M.-G. Lin L.-L. Chi M.-C. (2015). Enzymatic synthesis of γ-l-glutamyl-S-allyl-l-cysteine, a naturally occurring organosulfur compound from garlic, by Bacillus licheniformis γ-glutamyltranspeptidase.Enzyme Microb. Technol.718–24. 10.1016/j.enzmictec.2015.04.011
23
Chen Y.-Y. Tsai M.-G. Chi M.-C. Wang T.-F. Lin L.-L. (2013). Covalent immobilization of Bacillus licheniformisγ-glutamyl transpeptidase on aldehyde-functionalized magnetic nanoparticles.Int. J. Mol. Sci.144613–4628. 10.3390/ijms14034613
24
Chevalier C. Thiberge J. Ferrero R. L. Labigne A. (1999). Essential role of Helicobacter pylori γ-glutamyltranspeptidase for the colonization of the gastric mucosa of mice.Mol. Microbiol.311359–1372. 10.1046/j.1365-2958.1999.01271.x
25
Chi M.-C. Chen Y.-Y. Lo H.-F. Lin L.-L. (2012). Experimental evidence for the involvement of amino acid residue Glu398 in the autocatalytic processing of Bacillus licheniformis γ-glutamyltranspeptidase.FEBS Open Bio.2298–304. 10.1016/j.fob.2012.09.007
26
Chi M.-C. Lin M.-G. Chen Y.-Y. Lin L.-L. Wang T.-F. (2018). Functional role of the conserved glycine residues, Gly481 and Gly482, of the γ-glutamyltranspeptidase from Bacillus licheniformis.Int. J. Biol. Macromol.1091182–1188. 10.1016/j.ijbiomac.2017.11.116
27
Chi M.-C. Lin M.-G. Huang Y.-F. Chen Y.-Y. Wang T.-F. Lin L.-L. (2019a). Enzymatic synthesis of L-theanine from L-glutamine and ethylamine by Bacillus licheniformis γ-glutamyltranspeptidase and its mutants specialized in transpeptidase activity.Biocatal. Agric. Biotechnol.22:101393. 10.1016/j.bcab.2019.101393
28
Chi M.-C. Lo H.-F. Lin M.-G. Chen Y.-Y. Lin L.-L. Wang T.-F. (2017). Application of Bacillus licheniformis γ-glutamyltranspeptidase to the biocatalytic synthesis of γ-glutamyl-phenylalanine.Biocatal. Agric. Biotechnol.10278–284. 10.1016/j.bcab.2017.04.005
29
Chi M.-C. Lo H.-F. Lin M.-G. Chen Y.-Y. Wang T.-F. Lin L.-L. (2019b). Mutational analysis of a highly conserved PLSSMXP sequence in the small subunit of Bacillus licheniformis γ-glutamyltranspeptidase.Biomolecules9:508. 10.3390/biom9090508
30
Chi M.-C. Lo Y.-H. Chen Y.-Y. Lin L.-L. Merlino A. (2014). γ-Glutamyl transpeptidase architecture: Effect of extra sequence deletion on autoprocessing, structure and stability of the protein from Bacillus licheniformis.Biochim. Biophys. Acta Prot. Proteom.18442290–2297. 10.1016/j.bbapap.2014.09.001
31
Chikhi N. Holic N. Guellaen G. Laperche Y. (1999). Gamma-glutamyl transpeptidase gene organization and expression: a comparative analysis in rat, mouse, pig and human species.Comp. Biochem. Physiol. Part B Biochem. Mol. Biol.122367–380. 10.1016/S0305-0491(99)00013-9
32
Clemens D. L. Lee B.-Y. Horwitz M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages.Infect. Immun.723204–3217. 10.1128/IAI.72.6.3204-3217.2004
33
Davies J. Evans R. H. Jones A. W. Smith D. A. S. Watkins J. C. (1982). Differential activation and blockade of excitatory amino acid receptors in the mammalian and amphibian central nervous systems.Comp. Biochem. Physiol. Part C Comparat. Pharmacol.72211–224. 10.1016/0306-4492(82)90086-7
34
Fahey R. Brown W. Adams W. Worsham M. (1978). Occurrence of glutathione in bacteria.J. Bacteriol1331126–1129. 10.1128/jb.133.3.1126-1129.1978
35
Feuer L. Gaál K. (1979). Effect of glutaurine on plasma renin activity in the rat and the dog.Gen. Comp. Endocrinol.39330–335. 10.1016/0016-6480(79)90129-1
36
Feuer L. S.-Rózsa K. (1981). Effect of litoralon, taurine and other taurine derivatives on the heart muscle cell membrane of the locusta migratoria (migratoriodes R.F.).Comp. Biochem. Physiol. Part C Comparat. Pharmacol.69411–414. 10.1016/0306-4492(81)90162-3
37
Feuer L. Török L. Kapa E. Csaba G. (1978). The effect of gamma-L-glumatamyl-taurine (litoralon) on the amphibian metamorphosis.Comp. Biochem. Physiol. Part C Comparat. Pharmacol.6167–71. 10.1016/0306-4492(78)90113-2
38
Flahou B. Haesebrouck F. Chiers K. Van Deun K. De Smet L. Devreese B. et al (2011). Gastric epithelial cell death caused by Helicobacter suis and Helicobacter pylori γ-glutamyl transpeptidase is mainly glutathione degradation-dependent.Cell. Microbiol.131933–1955. 10.1111/j.1462-5822.2011.01682.x
39
Floch P. Pey V. Castroviejo M. Dupuy J. Bonneu M. de la Guardia A. et al (2014). Role of Campylobacter jejuni gamma-glutamyl transpeptidase on epithelial cell apoptosis and lymphocyte proliferation.Gut Pathog.6:20. 10.1186/1757-4749-6-20
40
Franzini M. Corti A. Fierabracci V. Pompella A. (2014). Helicobacter, gamma-glutamyltransferase and cancer: Further intriguing connections.World J. Gastroenterol.2018057–18058. 10.3748/wjg.v20.i47.18057
41
Gong M. Ho B. (2004). Prominent role of γ-glutamyl-transpeptidase on the growth of Helicobacter pylori.World J. Gastroenterol.102994–2996. 10.3748/wjg.v10.i20.2994
42
Gong M. Ling S. S. M. Lui S. Y. Yeoh K. G. Ho B. (2010). Helicobacter pylori γ-glutamyl transpeptidase is a pathogenic factor in the development of peptic ulcer disease.Gastroenterology139564–573. 10.1053/j.gastro.2010.03.050
43
Grundy F. J. Henkin T. M. (2001). “Synthesis of serine, glycine, cysteine, and methionine,” in Bacillus subtilis and its closest relatives: From genes to cells, edsSonensheinA. L.HochJ. A.LosickR. (Washington, DC: ASM Press), 245–254. 10.1128/9781555817992.ch18
44
Grundy S. M. (2007). Gamma-Glutamyl Transferase: another biomarker for metabolic syndrome and cardiovascular risk.Arterioscl. Thromb. Vasc. Biol.274–7. 10.1161/01.ATV.0000253905.13219.4b
45
Guo H.-C. Xu Q. Buckley D. Guan C. (1998). Crystal structures of Flavobacterium glycosylasparaginase.J. Biol. Chem.27320205–20212. 10.1074/jbc.273.32.20205
46
Hanes C. S. Hird F. J. R. Isherwood F. A. (1952). Enzymic transpeptidation reactions involving γ-glutamyl peptides and α-amino-acyl peptides.Biochem. J.5125–35. 10.1042/bj0510025
47
Hara T. Yokoo Y. Furukawa T. (1992). “Potential of γ-L-glutamyl-L-cystine and bis-γ-L-glutamyl-L-cystine as a cystine-containing peptide for parenteral nutrition,” in Frontiers and new Horizons in Amino Acid Research, ed.TakaiK. (Amsterdam: Elsevier), 607–611.
48
Hashimoto W. Suzuki H. Nohara S. Kumagai H. (1992). Escherichia coli γ-glutamyltranspeptidase mutants deficient in processing to subunits.Biochem. Biophy. Res. Commun.189173–178. 10.1016/0006-291X(92)91540-7
49
Hashimoto W. Suzuki H. Yamamoto K. Kumagai H. (1995). Effect of site-directed mutations on processing and activity of γ-glutamyltranspeptidase of Escherichia coli K-12.J. Biochem.11875–80. 10.1093/oxfordjournals.jbchem.a124894
50
Haskell C. F. Kennedy D. O. Milne A. L. Wesnes K. A. Scholey A. B. (2008). The effects of l-theanine, caffeine and their combination on cognition and mood.Biol. Psychol.77113–122. 10.1016/j.biopsycho.2007.09.008
51
Hendriksen H. V. Kornbrust B. A. Østergaard P. R. Stringer M. A. (2009). Evaluating the potential for enzymatic acrylamide mitigation in a range of food products using an asparaginase from aspergillus oryzae.J. Agric. Food Chem.57,4168–4176. 10.1021/jf900174q
52
Hewitt L. Kasche V. Lummer K. Lewis R. J. Murshudov G. N. Verma C. S. et al (2000). Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft.J. Mol. Biol.302887–898. 10.1006/jmbi.2000.4105
53
Hibi T. Imaoka M. Shimizu Y. Itoh T. Wakayama M. (2019). Crystal structure analysis and enzymatic characterization of γ-glutamyltranspeptidase from Pseudomonas nitroreducens.Biosci. Biotechnol. Biochem.83262–269. 10.1080/09168451.2018.1547104
54
Ho T. V. Kamei K. Wada K. Fukuyama K. Suzuki H. (2013). Thermal Denaturation and Renaturation of γ-Glutamyltranspeptidase of Escherichia coli.Biosci. Biotechnol. Biochem.77409–412. 10.1271/bbb.120780
55
Ho T. V. Suzuki H. (2013). Increase of “Umami” and “Kokumi” compounds in miso, fermented soybeans, by the addition of bacterial γ-glutamyltranspeptidase.Int. J. Food Stud239–47. 10.7455/ijfs/2.1.2013.a3
56
Hsu W.-H. Ong P.-L. Chen S.-C. Lin L.-L. (2009). Contribution of Ser463 residue to the enzymatic and autoprocessing activities of Escherichia coli gamma-glutamyltranspeptidase.Indian J. Biochem. Biophys.46281–288.
57
Hu X. Legler P. M. Khavrutskii I. Scorpio A. Compton J. R. Robertson K. L. et al (2012). Probing the donor and acceptor substrate specificity of the γ-Glutamyl transpeptidase.Biochemistry511199–1212. 10.1021/bi200987b
58
Hung C.-P. Lo H.-F. Hsu W.-H. Chen S.-C. Lin L.-L. (2008). Immobilization of Escherichia coli novablue γ-glutamyltranspeptidase in Ca-alginate-k-carrageenan beads.Appl. Biochem. Biotechnol.150157–170. 10.1007/s12010-008-8244-x
59
Hung C.-P. Yang J.-C. Chen J.-H. Chi M.-C. Lin L.-L. (2011). Unfolding analysis of the mature and unprocessed forms of Bacillus licheniformis γ-glutamyltranspeptidase.J. Biol. Phys.37463–475. 10.1007/s10867-011-9228-6
60
Ichinose H. Togari A. Suzuki H. Kumagai H. Nagatsu T. (1987). Increase of Catecholamines in Mouse Brain by Systemic Administration of γ-Glutamyl L-3,4-Dihydroxyphenylalanine.J. Neurochem.49928–932. 10.1111/j.1471-4159.1987.tb00982.x
61
Ida T. Suzuki H. Fukuyama K. Hiratake J. Wada K. (2014). Structure of Bacillus subtilis γ-glutamyltranspeptidase in complex with acivicin: diversity of the binding mode of a classical and electrophilic active-site-directed glutamate analogue.Acta Crystallogr. D. Biol. Crystallogr.70607–614. 10.1107/S1399004713031222
62
Ikeda Y. Fujii J. Taniguchi N. (1993). Significance of Arg-107 and Glu-108 in the catalytic mechanism of human gamma-glutamyl transpeptidase. Identification by site-directed mutagenesis.J. Biol. Chem.2683980–3985. 10.1016/S0021-9258(18)53567-5
63
Imaoka M. Yano S. Okumura M. Hibi T. Wakayama M. (2010). Molecular cloning and characterization of γ-glutamyltranspeptidase from Pseudomonas nitroreducens IFO12694.Biosci. Biotechnol. Biochem.741936–1939. 10.1271/bbb.100199
64
Inoue M. Hiratake J. Suzuki H. Kumagai H. Sakata K. (2000). Identification of catalytic nucleophile of Escherichia coli γ-glutamyltranspeptidase by γ-monofluorophosphono derivative of glutamic Acid: N-terminal Thr-391 in small subunit is the nucleophile.Biochemistry397764–7771. 10.1021/bi000220p
65
Ireland P. M. LeButt H. Thomas R. M. Oyston P. C. F. (2011). A Francisella tularensis SCHU S4 mutant deficient in γ-glutamyltransferase activity induces protective immunity: Characterization of an attenuated vaccine candidate.Microbiology1573172–3179. 10.1099/mic.0.052902-0
66
Ito K. Koyama Y. Hanya Y. (2013). Identification of the glutaminase genes of Aspergillus sojae involved in glutamate production during soy sauce fermentation.Biosci. Biotechnol. Biochem.771832–1840. 10.1271/bbb.130151
67
Jones A. W. Smith D. A. S. Watkins J. C. (1984). Structure-activity relations of dipeptide antagonists of excitatory amino acids.Neuroscience13573–581. 10.1016/0306-4522(84)90250-1
68
Juang T.-Y. Kan S.-J. Chen Y.-Y. Tsai Y.-L. Lin M.-G. Lin L.-L. (2014). Surface-functionalized hyperbranched poly(amido acid) magnetic nanocarriers for covalent immobilization of a bacterial γ-glutamyltranspeptidase.Molecules194997–5012. 10.3390/molecules19044997
69
Kalum L. Landvik S. M. Matveeva I. V. Joergensen S. T. Jensen K. (2020). Dough relaxation Using Gamma Glutamyl Transpeptidase.US Patent No. 2020/0196617A1.Alexandria, VA: U.S. Patent and Trademark Office.
70
Keillor J. W. Castonguay R. Lherbet C. (2005). Gamma−glutamyl transpeptidase substrate specificity and catalytic mechanism.Methods Enzymol.401449–467. 10.1016/S0076-6879(05)01027-X
71
Keillor J. W. Ménard A. Castonguay R. Lherbet C. Rivard C. (2004). Pre-steady-state kinetic studies of rat kidney γ-glutamyl transpeptidase confirm its ping-pong mechanism.J. Phys. Org. Chem.17529–536. 10.1002/poc.777
72
Kijima K. Suzuki H. (2007). Improving the umami taste of soy sauce by the addition of bacterial γ-glutamyltranspeptidase as a glutaminase to the fermentation mixture.Enzyme Microb. Technol.4180–84. 10.1016/j.enzmictec.2006.12.004
73
Kim J. Jang S. Kim A. Su H. Gunawardhana N. Jeon Y.-E. et al (2016). Role of bacterial γ-glutamyltranspeptidase as a novel virulence factor in bone-resorbing pathogenesis.J. Microbiol.54396–402. 10.1007/s12275-016-6137-1
74
Kim Y. Yoon K.-H. Khang Y. Turley S. Hol W. G. J. (2000). The 2.0 Å Crystal structure of cephalosporin acylase.Structure81059–1068. 10.1016/S0969-2126(00)00505-0
75
Kimura K. Ozeki M. Juneja L. R. Ohira H. (2007). L-Theanine reduces psychological and physiological stress responses.Biol. Psychol.7439–45. 10.1016/j.biopsycho.2006.06.006
76
Kimura K. Tran L.-S. P. Uchida I. Itoh Y. (2004). Characterization of Bacillus subtilis γ-glutamyltransferase and its involvement in the degradation of capsule poly-γ-glutamate.Microbiology1504115–4123. 10.1099/mic.0.27467-0
77
Kiuchi K. Kiuchi K. Nagatsu T. Togari A. Kumagai H. (1986). Highly sensitive assay for γ-glutamyltranspeptidase activity by high-performance liquid chromatography with electrochemical detection.J. Chromatogr. A357191–198. 10.1016/S0021-9673(01)95820-0
78
Kolobov A. A. Simbirtsev A. S. (1999). γ-Glutamyl and β-Aspartyl Containing Immunomodulator Compounds and Methods Therewith.US Patent No. 5,916,878. Alexandria, VA: U.S. Patent and Trademark Office.
79
Kumagai H. Echigo T. Suzuki H. Tochikura T. (1988). Synthesis of γ-glutamyl-DOPA from L-Glutamine and L-DOPA by γ-glutamyltranspeptidase of Escherichia coli K-12.Agr. Biol. Chem.521741–1745. 10.1080/00021369.1988.10868938
80
Kumari S. Pal R. K. Gupta R. Goel M. (2017). High resolution X-ray diffraction dataset for Bacillus licheniformis gamma glutamyl transpeptidase-acivicin complex: SUMO-tag renders high expression and solubility.Prot. J.367–16. 10.1007/s10930-017-9693-2
81
Kuno T. Matsuda Y. Katunuma N. (1984). Characterization of a processing protease that converts the precursor form of gamma-glutamyltranspeptidase to its subunits.Biochem. Int.8581–588.
82
Kuroda M. Kato Y. Yamazaki J. Kai Y. Mizukoshi T. Miyano H. et al (2012). Determination and quantification of γ-glutamyl-valyl-glycine in commercial fish sauces.J. Agric. Food Chem.607291–7296. 10.1021/jf3012932
83
Kushwaha N. Srivastava S. (2014). Gamma-glutamyl transpeptidase from two plant growth promoting rhizosphere fluorescent pseudomonads.Antonie Van Leeuwenhoek10545–56. 10.1007/s10482-013-0051-x
84
Le Butt H. Ireland P. M. Oyston P. C. F. (2013). Gamma-Glutamyl Transpeptidase Attenuated Francisella.US Patent No. 8,609,108B2. Alexandria, VA: U.S. Patent and Trademark Offic.
85
Lee J.-M. Lee J. Son B.-S. Jang M.-U. Lee S.-W. Hurh B.-S. et al (2017). Heterologous expression and enzymatic characterization of γ-glutamyltranspeptidase from Bacillus amyloliquefaciens.J. Microbiol.55147–152. 10.1007/s12275-017-6638-6
86
Lee Y.-C. Chi M.-C. Lin M.-G. Chen Y.-Y. Lin L.-L. Wang T.-F. (2018). Biocatalytic synthesis of γ-glutamyl-L-leucine, a kokumi-imparting dipeptide, by Bacillus licheniformis γ-glutamyltranspeptidase.Food Biotechnol.32130–147. 10.1080/08905436.2018.1444636
87
Li Y. Chen J. Jiang W. Mao X. Zhao G. Wang E. (1999). In vivo post−translational processing and subunit reconstitution of cephalosporin acylase from Pseudomonas sp. 130.Eur. J. Biochem.262713–719. 10.1046/j.1432-1327.1999.00417.x
88
Li Z. Zhu R. Liu Y. Li J. Gao H. Hu N. (2020). γ-Glutamyltranspeptidase from Bacillus amyloliquefaciens: transpeptidation activity enhancement and L-theanine production.Enzyme Microb. Technol.140:109644. 10.1016/j.enzmictec.2020.109644
89
Liao D. Cram D. Sharpe A. G. Marsolais F. (2013). Transcriptome profiling identifies candidate genes associated with the accumulation of distinct sulfur γ-glutamyl dipeptides in Phaseolus vulgaris and Vigna mungo seeds.Front. Plant Sci4:60. 10.3389/fpls.2013.00060
90
Lin L.-L. Chen Y.-Y. Chi M.-C. Merlino A. (2014). Low resolution X-ray structure of γ-glutamyltranspeptidase from Bacillus licheniformis: opened active site cleft and a cluster of acid residues potentially involved in the recognition of a metal ion.Biochim. Biophys. Acta, Proteins Proteomics18441523–1529. 10.1016/j.bbapap.2014.04.016
91
Lin L. L. Chi M. C. Lan Y. J. Lin M. G. Juang T.-Y. Wang T.-F. (2018). Facile immobilization of Bacillus licheniformis γ-glutamyltranspeptidase onto graphene oxide nanosheets and its application to the biocatalytic synthesis of γ-L-glutamyl peptides.Int. J. Biol. Macromol.1171326–1333. 10.1016/j.ijbiomac.2017.11.153
92
Lin L.-L. Chou P.-R. Hua Y. W. Hsu W. H. (2006). Overexpression, one-step purification, and biochemical characterization of a recombinant γ-glutamyltranspeptidase from Bacillus licheniformis.Appl. Microbiol. Biotechnol.73103–112. 10.1007/s00253-006-0440-4
93
Lin L. L. Yang L. Y. Hu H. Y. Lo H. F. (2008). Influence of N-Terminal truncations on the functional expression of Bacillus licheniformis γ-glutamyltranspeptidase in recombinant Escherichia coli.Curr. Microbiol.57603–608. 10.1007/s00284-008-9250-5
94
Lin M. G. Chi M. C. Chen Y. Y. Wang T. F. Lo H. F. Lin L. L. (2016). Site-directed mutagenesis of a conserved Asn450 residue of Bacillus licheniformis γ-glutamyltranspeptidase.Int. J. Biol. Macromol.91416–425. 10.1016/j.ijbiomac.2016.05.101
95
Ling S. S. M. (2013). Helicobacter pylori γ-glutamyl transpeptidase: a formidable virulence factor.World J. Gastroenterol.198203–8210. 10.3748/wjg.v19.i45.8203
96
Liu J. Song H. Liu Y. Li P. Yao J. Xiong J. (2015). Discovery of kokumi peptide from yeast extract by LC-Q-TOF-MS/MS and sensomics approach.J. Sci. Food Agric.953183–3194. 10.1002/jsfa.7058
97
Liu Q. Duan H. Luan J. Yagasaki K. Zhang G. (2009). Effects of theanine on growth of human lung cancer and leukemia cells as well as migration and invasion of human lung cancer cells.Cytotechnology59211–217. 10.1007/s10616-009-9223-y
98
Liu Q. Shen Y. R. Yin K. D. (2020). The degradation fragments of gamma-glutamyl transpeptidase from Bacillus subtilis BU108 have antimicrobial activity against Streptomyces scabiei.J. Plant Dis. Prot128279–285. 10.1007/s41348-020-00371-5
99
Lo H. Chou W. Chen P. Lin L. (2008). Influence of signal−peptide truncations on the functional expression of Escherichia coli γ−glutamyltranspeptidase.J. Basic Microbiol.48260–268. 10.1002/jobm.200700325
100
Lo H.-F. Lin L.-L. Chen P.-J. Chou W.-M. (2007). Site-directed mutagenesis of conserved Thr407, Asp433 and Met464 residues in small subunit of Escherichia coli gamma-glutamyltranspeptidase.Indian J. Biochem. Biophys.44197–203.
101
Lu K. Gray M. A. Oliver C. Liley D. T. Harrison B. J. Bartholomeusz C. F. et al (2004). The acute effects of L−theanine in comparison with alprazolam on anticipatory anxiety in humans.Hum. Psychopharmacol Clin. Exp.19457–465. 10.1002/hup.611
102
Lyu R. C. Hu H. Y. Kuo L. Y. Lo H.-F. Ong P. L. Chang H. P. et al (2009). Role of the conserved Thr399 and Thr417 residues of Bacillus licheniformis γ-glutamyltranspeptidase as evaluated by mutational analysis.Curr. Microbiol.59101–106. 10.1007/s00284-009-9403-1
103
Marnela K.-M. Morris H. R. Panico M. Timonen M. Lähdesmäki P. (1985). Glutamyl-taurine is the predominant synaptic taurine peptide.J. Neurochem.44752–754. 10.1111/j.1471-4159.1985.tb12878.x
104
Masip L. Veeravalli K. Georgiou G. (2006). The many faces of glutathione in bacteria.Antioxid. Redox Signal.8753–762. 10.1089/ars.2006.8.753
105
Michalska K. Brzezinski K. Jaskolski M. (2005). Crystal structure of isoaspartyl aminopeptidase in complex with L-Aspartate.J. Biol. Chem.28028484–28491. 10.1074/jbc.M504501200
106
Minami H. Suzuki H. Kumagai H. (2003a). A mutant Bacillus subtilis γ-glutamyltranspeptidase specialized in hydrolysis activity.FEMS Microbiol. Lett.224169–173. 10.1016/S0378-1097(03)00456-7
107
Minami H. Suzuki H. Kumagai H. (2003b). Salt-tolerant γ-glutamyltranspeptidase from Bacillus subtilis 168 with glutaminase activity.Enzyme Microb. Technol.32431–438. 10.1016/S0141-0229(02)00314-9
108
Minami H. Suzuki H. Kumagai H. (2004). γ-Glutamyltranspeptidase, but not YwrD, is important in utilization of extracellular glutathione as a sulfur source in Bacillus subtilis.J. Bacteriol.1861213–1214. 10.1128/JB.186.4.1213-1214.2004
109
Misicka A. Maszczynska I. Lipkowski A. W. Stropova D. Yamamura H. I. Hruby V. J. (1996). Synthesis and biological properties of gamma-glutamyl-dermorphin, a prodrug.Life Sci.58905–911. 10.1016/0024-3205(96)00033-1
110
Miyagawa K. Hayashi Y. Kurihara S. Maeda A. (2008). Co-administration of L-cystine and L-theanine enhances efficacy of influenza vaccination in elderly persons: nutritional status-dependent immunogenicity.Geriatr. Gerontol. Int.8243–250. 10.1111/j.1447-0594.2008.00483.x
111
Moallic C. Dabonne S. Colas B. Sine J. P. (2006). Identification and characterization of a gamma-glutamyl transpeptidase from a thermo-alcalophile strain of Bacillus pumilus.Prot. J.25:391. 10.1007/s10930-006-9025-4
112
Mohamad N. R. Marzuki N. H. C. Buang N. A. Huyop F. Wahab R. A. (2015). An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes.Biotechnol. Biotechnol. Equip.29205–220. 10.1080/13102818.2015.1008192
113
Morelli C. F. Calvio C. Biagiotti M. Speranza G. (2014). pH-Dependent hydrolase, glutaminase, transpeptidase and autotranspeptidase activities of Bacillus subtilis γ-glutamyltransferase.FEBS J.281232–245. 10.1111/febs.12591
114
Morrow A. L. Williams K. Sand A. Boanca G. Barycki J. J. (2007). Characterization of Helicobacter pylori γ-glutamyltranspeptidase reveals the molecular basis for substrate specificity and a critical role for the tyrosine 433-containing loop in catalysis.Biochemistry4613407–13414. 10.1021/bi701599e
115
Mu D. Li H. Chen Q. Zhu J. Wu X. Luo S. et al (2019). Secretion of Bacillus amyloliquefaciens γ-Glutamyltranspeptidase from Bacillus subtilis and its application in enzymatic synthesis of L-Theanine.J. Agric. Food Chem.6714129–14136. 10.1021/acs.jafc.9b06140
116
Mu W. Zhang T. Jiang B. (2015). An overview of biological production of L-theanine.Biotechnol. Adv.33335–342. 10.1016/j.biotechadv.2015.04.004
117
Murty N. A. R. Tiwary E. Sharma R. Nair N. Gupta R. (2012). γ-Glutamyl transpeptidase from Bacillus pumilus KS 12: decoupling autoprocessing from catalysis and molecular characterization of N-terminal region.Enzyme Microb. Technol.50159–164. 10.1016/j.enzmictec.2011.08.005
118
Nakayama R. Kumagai H. Akashi S. Sugiura H. Tochikura T. (1985). Synthesis of γ-glutamyl L-3, 4-dihydroxyphenylalanine by γ-glutamyltranspeptidase from Proteus mirabilis.Agr. Biol. Chem.491041–1046. 10.1080/00021369.1985.10866860
119
Nakayama R. Kumagai H. Tochikura T. (1984). Gamma-glutamyltranspeptidase from Proteus mirabilis: localization and activation by phospholipids.J. Bacteriol.1601031–1036. 10.1128/JB.160.3.1031-1036.1984
120
Nash B. Tate S. (1984). In vitro translation and processing of rat kidney gamma-glutamyl transpeptidase.J. Biol. Chem.259678–685. 10.1016/S0021-9258(17)43715-X
121
Ni F. Zhang F. Yao Z. Ye L. Sun Y. Wang H. et al (2017). Improving the catalytic properties and stability of immobilized γ-glutamyltranspeptidase by post-immobilization with Pharmalyte MT 8–10.5.Int. J. Biol. Macromol.1051581–1586. 10.1016/j.ijbiomac.2017.04.050
122
Niida A. Hiroko T. Kasai M. Furukawa Y. Nakamura Y. Suzuki Y. et al (2004). DKK1, a negative regulator of Wnt signaling, is a target of the β-catenin/TCF pathway.Oncogene238520–8526. 10.1038/sj.onc.1207892
123
Ogawa Y. Hosoyama H. Hamano M. Motai H. (1991). Purification and properties of γ-glutamyltranspeptidase from Bacillus subtilis (natto).Agr. Biol. Chem.552971–2977. 10.1080/00021369.1991.10857918
124
Ojima Y. Kobayashi J. Doi T. Azuma M. (2019). Knockout of pgdS and ggt gene changes poly-γ-glutamic acid production in Bacillus licheniformis RK14-46.J. Biotechnol.30457–62. 10.1016/j.jbiotec.2019.08.003
125
Okada T. Suzuki H. Wada K. Kumagai H. Fukuyama K. (2006). Crystal structures of γ-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.Proc. Natl. Acad. Sci. U.S.A.1036471–6476. 10.1073/pnas.0511020103
126
Okada T. Suzuki H. Wada K. Kumagai H. Fukuyama K. (2007). Crystal structure of the γ-glutamyltranspeptidase precursor protein from Escherichia coli: Structural changes upon autocatalytic processing and implications for the maturation mechanism.J. Biol. Chem.2822433–2439. 10.1074/jbc.M607490200
127
Ong P. L. Yao Y. F. Weng Y. M. Hsu W. H. Lin L. L. (2008). Residues Arg114 and Arg337 are critical for the proper function of Escherichia coli γ-glutamyltranspeptidase.Biochem. Biophys. Res. Commun.366294–300. 10.1016/j.bbrc.2007.11.063
128
Papkoff J. Lee H. Singh G. Singh S. Chitta S. Andrew K. et al (2011). Identification of signaling pathways involved in the mechanism of action of the immunomodulatory peptide gamma-D-glutamyl-L-tryptophan.J. Clin. Oncol.295591–5591. 10.1200/jco.2011.29.15_suppl.5591
129
Pica A. Chi M.-C. Chen Y.-Y. d’Ischia M. Lin L.-L. Merlino A. (2016). The maturation mechanism of γ-glutamyl transpeptidases: Insights from the crystal structure of a precursor mimic of the enzyme from Bacillus licheniformis and from site-directed mutagenesis studies.Biochim. Biophys. Acta, Proteins Proteomics1864195–203. 10.1016/j.bbapap.2015.10.006
130
Pica A. Russo Krauss I. Castellano I. Rossi M. La Cara F. Graziano G. et al (2012). Exploring the unfolding mechanism of γ-glutamyltranspeptidases: the case of the thermophilic enzyme from Geobacillus thermodenitrificans.Biochim. Biophys. Acta Prot. Proteom.1824571–577. 10.1016/j.bbapap.2012.01.014
131
Pica A. Russo Krauss I. Castellano I. La Cara F. Graziano G. Sica F. et al (2013). Effect of NaCl on the conformational stability of the thermophilic γ-glutamyltranspeptidase from geobacillus thermodenitrificans: Implication for globular protein halotolerance.Biochim. Biophys. Acta Prot. Proteom1834149–157. 10.1016/j.bbapap.2012.09.014
132
Prihanto A. A. Nonomura Y. Takagi K. Naohara R. Umekawa M. Wakayama M. (2015a). Novel properties of γ-glutamyltransferase from Pseudomonas syringae with β-aspartyltransferase activity.Biotechnol. Lett.372255–2263. 10.1007/s10529-015-1906-1
133
Prihanto A. A. Nonomura Y. Takagi K. Naohara R. Wakayama M. (2015b). “Synthesis optimization of L-Aspartic acid β-hydroxamate by a novel enzyme, β-Aspartyl-γ-glutamyl transferase,” in Proceeding of the 5th International Seminar on New Paradigm and Innovation on Natural Sciences and Its Application (5th ISNPINSA), Semarang.
134
Pu H. Wang Q. Zhu F. Cao X. Xin Y. Luo L. et al (2013). Cloning, expression of glutaminase from Pseudomonas nitroreducens and application to theanine synthesis.Biocatal. Biotransform.311–7. 10.3109/10242422.2012.749462
135
Rajput R. Verma V. V. Chaudhary V. Gupta R. (2013). A hydrolytic γ-glutamyl transpeptidase from thermo-acidophilic archaeon Picrophilus torridus: binding pocket mutagenesis and transpeptidation.Extremophiles1729–41. 10.1007/s00792-012-0490-8
136
Rawlings N. D. Barrett A. J. Bateman A. (2010). MEROPS: the peptidase database.Nucleic Acids Res.38D227–D233.
137
Reddy V. V. S. Jayaram H. N. Sirsi M. Ramakrishnan T. (1969). Inhibitory activity of l-asparaginase from Mycobcacterium tuberculosis on Yoshida ascites sarcoma in rats.Arch. Biochem. Biophys.132,262–267. 10.1016/0003-9861(69)90361-0
138
Ricci V. Giannouli M. Romano M. Zarrilli R. (2014). Helicobacter pylori gamma-glutamyl transpeptidase and its pathogenic role.World J. Gastroenterol.20:630. 10.3748/wjg.v20.i3.630
139
Richter S. Anderson V. J. Garufi G. Lu L. Budzik J. M. Joachimiak A. et al (2009). Capsule anchoring in Bacillus anthracis occurs by a transpeptidation reaction that is inhibited by capsidin.Mol. Microbiol.71404–420. 10.1111/j.1365-2958.2008.06533.x
140
Rose W. A. Tuthill C. Pyles R. B. (2008). An immunomodulating dipeptide, SCV-07, is a potential therapeutic for recurrent genital herpes simplex virus type 2 (HSV-2).Int. J. Antimicrob. Agents32262–266. 10.1016/j.ijantimicag.2008.04.010
141
Sadzuka Y. Sugiyama T. Sonobe T. (2000). Efficacies of tea components on doxorubicin induced antitumor activity and reversal of multidrug resistance.Toxicol. Lett.114155–162. 10.1016/S0378-4274(99)00290-8
142
Saini M. Bindal S. Gupta R. (2017). Heterologous expression of γ-glutamyl transpeptidase from Bacillus atrophaeus GS-16 and its application in the synthesis of γ-D-glutamyl-L-tryptophan, a known immunomodulatory peptide.Enzyme Microb. Technol9967–76. 10.1016/j.enzmictec.2017.01.003
143
Sayed A. S. A. F. E. Fujimoto S. Yamada C. Suzuki H. (2010). Enzymatic synthesis of γ-glutamylglutamine, a stable glutamine analogue, by γ-glutamyltranspeptidase from Escherichia coli K-12.Biotechnol. Lett.321877–1881. 10.1007/s10529-010-0364-z
144
Schmees C. Prinz C. Treptau T. Rad R. Hengst L. Voland P. et al (2007). Inhibition of T-Cell proliferation by Helicobacter pylori γ-glutamyl transpeptidase.Gastroenterology1321820–1833. 10.1053/j.gastro.2007.02.031
145
Scoffone V. Dondi D. Biino G. Borghese G. Pasini D. Galizzi A. et al (2013). Knockout of pgdS and ggt genes improves γ-PGA yield in B. subtilis.Biotechnol. Bioeng.1102006–2012. 10.1002/bit.24846
146
Shibayama K. Kamachi K. Nagata N. Yagi T. Nada T. Doi Y. et al (2003). A novel apoptosis-inducing protein from Helicobacter pylori.Mol. Microbiol.47443–451. 10.1046/j.1365-2958.2003.03305.x
147
Shibayama K. Wachino J. Arakawa Y. Saidijam M. Rutherford N. G. Henderson P. J. F. (2007). Metabolism of glutamine and glutathione via γ-glutamyltranspeptidase and glutamate transport in Helicobacter pylori: possible significance in the pathophysiology of the organism.Mol. Microbiol.64396–406. 10.1111/j.1365-2958.2007.05661.x
148
Shuai Y. Zhang T. Jiang B. Hua Y. Mu C.-F. (2019). An efficient method for the high-yield production of l-theanine using a newly isolated glutaminase-producing organism.Food Biosci.28164–169. 10.1016/j.fbio.2019.02.001
149
Shuai Y. Zhang T. Jiang B. Mu W. (2010). Development of efficient enzymatic production of theanine by γ-glutamyltranspeptidase from a newly isolated strain of Bacillus subtilis, SK11.004.J. Sci. Food Agric.902563–2567. 10.1002/jsfa.4120
150
Shuai Y. Zhang T. Mu W. Jiang B. (2011). Purification and characterization of γ-glutamyltranspeptidase from Bacillus subtilis SK11.004.J. Agric. Food Chem.596233–6238. 10.1021/jf2003249
151
Simbirtsev A. Kolobov A. Zabolotnych N. Pigareva N. Konusova V. Kotov A. et al (2003). Biological activity of peptide SCV-07 against murine tuberculosis.Russ. J. Immunol.811–22.
152
Sun J. Chang M. Li H. Zhang Z. Chen Q. Chen Y. et al (2019). Endophytic bacteria as contributors to theanine production in Camellia sinensis.J. Agri. Food Chem. 6710685–10693. 10.1021/acs.jafc.9b03946
153
Suzuki H. (2019). Bacterial gamma-glutamyltranspeptidase: Food and medicinal applications.Science Asia45503–508. 10.2306/scienceasia1513-1874.2019.45.503
154
Suzuki H. Hashimoto W. Kumagai H. (1993). Escherichia coli K-12 can utilize an exogenous gamma-glutamyl peptide as an amino acid source, for which gamma-glutamyltranspeptidase is essential.J. Bacteriol.1756038–6040. 10.1128/JB.175.18.6038-6040.1993
155
Suzuki H. Izuka S. Minami H. Miyakawa N. Ishihara S. Kumagai H. (2003). Use of bacterial γ-glutamyltranspeptidase for enzymatic synthesis of γ-D-Glutamyl Compounds.Appl. Environ. Microbiol.696399–6404. 10.1128/AEM.69.11.6399-6404.2003
156
Suzuki H. Izuka S. Miyakawa N. Kumagai H. (2002a). Enzymatic production of theanine, an “umami” component of tea, from glutamine and ethylamine with bacterial γ-glutamyltranspeptidase.Enzyme Microb. Technol.31884–889. 10.1016/S0141-0229(02)00213-2
157
Suzuki H. Kajimoto Y. Kumagai H. (2002b). Improvement of the bitter taste of amino acids through the transpeptidation reaction of bacterial γ-glutamyltranspeptidase.J. Agric. Food Chem.50313–318. 10.1021/jf010726u
158
Suzuki H. Kato K. Kumagai H. (2004a). Development of an efficient enzymatic production of γ-D-glutamyl-L-tryptophan (SCV-07), a prospective medicine for tuberculosis, with bacterial γ-glutamyltranspeptidase.J. Biotechnol.111291–295. 10.1016/j.jbiotec.2004.04.003
159
Suzuki H. Kato K. Kumagai H. (2004b). Enzymatic synthesis of γ-glutamylvaline to improve the bitter taste of valine.J. Agric. Food Chem.52577–580. 10.1021/jf0347564
160
Suzuki H. Kumagai H. (2002). Autocatalytic processing of γ-Glutamyltranspeptidase.J. Biol. Chem.27743536–43543. 10.1074/jbc.M207680200
161
Suzuki H. Kumagai H. Echigo T. Tochikura T. (1988). Molecular cloning of Escherichia coli K-12 ggt and rapid isolation of γ-glutamyltranspeptidase.Biochem. Biophys. Res. Commun.15033–38. 10.1016/0006-291X(88)90482-2
162
Suzuki H. Kumagai H. Echigo T. Tochikura T. (1989). DNA sequence of the Escherichia coli K-12 gamma-glutamyltranspeptidase gene, ggt.J. Bacteriol.1715169–5172. 10.1128/jb.171.9.5169-5172.1989
163
Suzuki H. Kumagai H. Tochikura T. (1986). gamma-Glutamyltranspeptidase from Escherichia coli K-12: formation and localization.J. Bacteriol.1681332–1335. 10.1128/JB.168.3.1332-1335.1986
164
Suzuki H. Kumagai H. Tochikura T. (1987). Isolation, genetic mapping, and characterization of Escherichia coli K-12 mutants lacking gamma-glutamyltranspeptidase.J. Bacteriol1693926–3931. 10.1128/JB.169.9.3926-3931.1987
165
Suzuki H. Miwa C. Ishihara S. Kumagai H. (2004c). A single amino acid substitution converts γ-glutamyltranspeptidase to a class IV cephalosporin acylase (glutaryl-7-aminocephalosporanic acid acylase).Appl. Environ. Microbiol.706324–6328. 10.1128/AEM.70.10.6324-6328.2004
166
Suzuki H. Miyakawa N. Kumagai H. (2002c). Enzymatic production of γ-L-glutamyltaurine through the transpeptidation reaction of γ-glutamyltranspeptidase from Escherichia coli K-12.Enzyme Microb. Technol.30883–888. 10.1016/S0141-0229(02)00038-8
167
Suzuki H. Yamada C. Kato K. (2007). γ-Glutamyl compounds and their enzymatic production using bacterial γ-glutamyltranspeptidase.Amino Acids32333–340. 10.1007/s00726-006-0416-9
168
Suzuki H. Yamada C. Kijima K. Ishihara S. Wada K. Fukuyama K. et al (2010). Enhancement of glutaryl−7−aminocephalosporanic acid acylase activity of γ−glutamyltranspeptidase of Bacillus subtilis.Biotechnol. J.5829–837. 10.1002/biot.201000015
169
Suzuki T. Tahara Y. (2003). Characterization of the Bacillus subtilis ywtD gene, whose product is involved in γ-polyglutamic acid degradation.J. Bacteriol.1852379–2382. 10.1128/JB.185.7.2379-2382.2003
170
Tachiki T. Yamada T. Mizuno K. Ueda M. Shiode J. Fukami H. (1998). γ-Glutamyl transfer reactions by glutaminase from Pseudomonas nitroreducens IFO 12694 and their application for the syntheses of theanine and γ-glutamylmethylamide.Biosci. Biotechnol. Biochem.621279–1283. 10.1271/bbb.62.1279
171
Takahashi H. Hirose K. Watanabe H. (2004). Necessity of meningococcal γ-glutamyl aminopeptidase for Neisseria meningitidis growth in rat cerebrospinal fluid (CSF) and CSF-Like medium.J. Bacteriol.186244–247. 10.1128/JB.186.1.244-247.2004
172
Takahashi H. Watanabe H. (2004). Post-translational processing of Neisseria meningitidis γ-glutamyl aminopeptidase and its association with inner membrane facing to the cytoplasmic space.FEMS Microbiol. Lett.23427–35. 10.1111/j.1574-6968.2004.tb09509.x
173
Takahashi H. Watanabe H. Kim K. S. Yokoyama S. Yanagisawa T. (2018). The meningococcal cysteine transport system plays a crucial role in Neisseria meningitidis survival in human brain microvascular endothelial cells.mBio9:e02332–18. 10.1128/mBio.02332-18
174
Taniguchi N. Ikeda Y. (1998). γ-Glutamyl transpeptidase: catalytic mechanism and gene expression.Adv. Enzymol. Relat. Areas Mol. Biol.72239–278. 10.1002/9780470123188.ch7
175
Tate S. Ross M. E. (1977). Human kidney gamma-glutamyl transpeptidase. Catalytic properties, subunit structure, and localization of the gamma-glutamyl binding site on the light subunit.J. Biol. Chem.2526042–6045. 10.1016/S0021-9258(17)40026-3
176
Tate S. S. Meister A. (1976). Subunit structure and isozymic forms of gamma-glutamyl transpeptidase.Proc. Natl. Acad. Sci. U.S.A.732599–2603. 10.1073/pnas.73.8.2599
177
Tate S. S. Meister A. (1977). Affinity labeling of gamma-glutamyl transpeptidase and location of the gamma-glutamyl binding site on the light subunit.Proc. Natl. Acad. Sci. U.S.A.74931–935. 10.1073/pnas.74.3.931
178
Tate S. S. Meister A. (1978). Serine-borate complex as a transition-state inhibitor of gamma-glutamyl transpeptidase.Proc. Natl. Acad. Sci. U.S.A.754806–4809. 10.1073/pnas.75.10.4806
179
Tate S. S. Meister A. (1981). “γ-Glutamyl transpeptidase: catalytic, structural and functional aspects,” in The Biological Effects of Glutamic Acid and Its Derivatives. Developments in Molecular and Cellular Biochemistry, Vol. 1ed.NajjarV. A. (Dordrecht: Springer).
180
Thompson G. Meister A. (1977). Interrelationships between the binding sites for amino acids, dipeptides, and gamma-glutamyl donors in gamma-glutamyl transpeptidase.J. Biol. Chem.2526792–6798. 10.1016/S0021-9258(17)39919-2
181
Török K. Varga V. Somogyi J. Feuer L. Gulyás J. (1981). Formation of γ-glutamyl-taurine in the rat brain.Neurosci. Lett.27145–149. 10.1016/0304-3940(81)90259-7
182
Tuthill C. Papkoff J. Watkins B. Sonis S. (2011). The effect of the immunomodulatory peptide gamma-D-glutamyl-L-tryptophan in leukemia, lymphoma, and head and neck cancer xenograft models.J. Clin. Oncol.295592–5592. 10.1200/jco.2011.29.15_suppl.5592
183
Uchida I. Makino S. Sasakawa C. Yoshikawa M. Sugimoto C. Terakado N. (1993). Identification of a novel gene, dep, associated with depolymerization of the capsular polymer in Bacillus anthracis.Mol. Microbiol.9487–496. 10.1111/j.1365-2958.1993.tb01710.x
184
Verma V. V. Gupta R. Goel M. (2015). Phylogenetic and evolutionary analysis of functional divergence among Gamma glutamyl transpeptidase (GGT) subfamilies.Biol. Direct.10:49. 10.1186/s13062-015-0080-7
185
Wada K. Hiratake J. Irie M. Okada T. Yamada C. Kumagai H. et al (2008). Crystal structures of Escherichia coli γ-glutamyltranspeptidase in complex with azaserine and acivicin: novel mechanistic implication for inhibition by glutamine antagonists.J. Mol. Biol.380361–372. 10.1016/j.jmb.2008.05.007
186
Wada K. Irie M. Suzuki H. Fukuyama K. (2010). Crystal structure of the halotolerant γ-glutamyltranspeptidase from Bacillus subtilis in complex with glutamate reveals a unique architecture of the solvent-exposed catalytic pocket.FEBS J.2771000–1009. 10.1111/j.1742-4658.2009.07543.x
187
Wang H. Q. Yao Z. Sun Y. Zhou Z. Xiong Q. Zhong Z. X. (2014). Immobilization of γ-glutamyltranspeptidase on silylated mesoporous TiO2 whiskers.Biotechnol. Bioproc. E.19304–310. 10.1007/s12257-013-0675-8
188
Wang H.-Q. Yao Z. Zhou Z. Sun Y. Wei P. Ouyang P. (2012). Enzymatic synthesis of theanine with L-glutamine-Zn(II) complexes.Biotechnol. Bioproc. E.171135–1139. 10.1007/s12257-012-0205-0
189
Wang Q. Min C. Zhu F. Xin Y. Zhang S. Luo L. et al (2011). Production of bioactive γ-glutamyl transpeptidase in Escherichia coli using SUMO fusion partner and application of the recombinant enzyme to L-Theanine synthesis.Curr. Microbiol.621535–1541. 10.1007/s00284-011-9891-7
190
Wang Q. Yao Z. Xun Z. Xu X. Xu H. Wei P. (2008). Properties and catalytic mechanism of γ-glutamyltranspeptidase from B. subtilis NX-2.Front. Chem. Eng. China2:456–461. 10.1007/s11705-008-0075-3
191
Watkins B. Pouliot K. Fey E. Tuthill C. Sonis S. (2010). Attenuation of radiation- and chemoradiation-induced mucositis using gamma-D-glutamyl-L-tryptophan (SCV-07).Oral Dis.16655–660. 10.1111/j.1601-0825.2010.01671.x
192
West M. B. Chen Y. Wickham S. Heroux A. Cahill K. Hanigan M. H. et al (2013). Novel insights into eukaryotic γ-glutamyltranspeptidase 1 from the crystal structure of the glutamate-bound human enzyme.J. Biol. Chem.28831902–31913. 10.1074/jbc.M113.498139
193
Whitfield J. B. (2001). Gamma Glutamyl transferase.Crit. Rev. Clin.38263–355. 10.1080/20014091084227
194
Williams K. Cullati S. Sand A. Biterova E. I. Barycki J. J. (2009). Crystal structure of acivicin-inhibited γ-glutamyltranspeptidase reveals critical roles for Its C-terminus in autoprocessing and catalysis.Biochemistry482459–2467. 10.1021/bi8014955
195
Wu R. Richter S. Zhang R. Anderson V. J. Missiakas D. Joachimiak A. (2009). Crystal structure of Bacillus anthracis transpeptidase enzyme CapD.J. Biol. Chem.28424406–24414. 10.1074/jbc.M109.019034
196
Xu K. Strauch M. A. (1996). Identification, sequence, and expression of the gene encoding gamma-glutamyltranspeptidase in Bacillus subtilis.J. Bacteriol.1784319–4322. 10.1128/JB.178.14.4319-4322.1996
197
Xu L. Han F. Zhang X. Yu Q. (2020). Ultrasound enhanced biosynthesis of L-theanine from L-glutamine and ethylamine by recombinant γ-glutamyltranspeptidase.Bioresour. Technol.307:123251. 10.1016/j.biortech.2020.123251
198
Yamada C. Kijima K. Ishihara S. Miwa C. Wada K. Okada T. et al (2008). Improvement of the glutaryl-7-aminocephalosporanic acid acylase activity of a bacterial γ-glutamyltranspeptidase.Appl. Environ. Microbiol.743400–3409. 10.1128/AEM.02693-07
199
Yamada Y. Ishizaki M. Kido T. Honda R. Tsuritani I. Ikai E. et al (1991). Alcohol, high blood pressure, and serum gamma-glutamyl transpeptidase level.Hypertension18819–826. 10.1161/01.HYP.18.6.819
200
Yang J. Bai W. Zeng X. Cui C. (2019). Gamma glutamyl peptides: the food source, enzymatic synthesis, kokumi-active and the potential functional properties – a review.Trends Food Sci. Techno91339–346. 10.1016/j.tifs.2019.07.022
201
Yang J. Sun-Waterhouse D. Cui C. Dong K. Wang W. (2017). Synthesis and sensory characteristics of kokumi γ-[Glu] n -Phe in the presence of Glutamine and Phenylalanine: Glutaminase from Bacillus amyloliquefaciens or Aspergillus oryzae as the Catalyst.J. Agric. Food Chem.658696–8703. 10.1021/acs.jafc.7b03419
202
Yang J. Sun-Waterhouse D. Xie J. Wang L. Chen H. Cui C. et al (2018). Comparison of kokumi γ -[Glu] (n>1) -Val and γ -[Glu] (n>1) -Met synthesized through transpeptidation catalyzed by glutaminase from Bacillus amyloliquefaciens.Food Chem.24789–97. 10.1016/j.foodchem.2017.11.096
203
Yang J.-C. Liang W.-C. Chen Y.-Y. Chi M.-C. Lo H.-F. Chen H.-L. et al (2011). Biophysical characterization of Bacillus licheniformis and Escherichia coli γ-glutamyltranspeptidases: a comparative analysis.Int. J. Biol. Macromol.48414–422. 10.1016/j.ijbiomac.2011.01.006
204
Yang T. Irene K. Liu H. Liu S. Zhang X. Xu M. et al (2019). Enhanced extracellular gamma glutamyl transpeptidase production by overexpressing of PrsA lipoproteins and improving its mRNA stability in Bacillus subtilis and application in biosynthesis of L-theanine.J. Biotechnol.30285–91. 10.1016/j.jbiotec.2019.06.302
205
Yao Y. Weng Y. Hu H. Lin L. (2006). Overexpression of a recombinant gamma-glutamyltranspeptidase from Escherichia coli Novablue.Indian J. Biochem. Biophys.43:345.
206
Yokogoshi H. Kato Y. Sagesaka Y. M. Takihara-Matsuura T. Kakuda T. Takeuchi N. (1995). Reduction effect of theanine on blood pressure and brain 5-hydroxyindoles in spontaneously hypertensive rats.Biosci. Biotechnol. Biochem.59615–618. 10.1271/bbb.59.615
207
Zhang F. Zheng Q.-Z. Jiao Q.-C. Liu J.-Z. Zhao G.-H. (2010). Enzymatic synthesis of theanine from glutamic acid γ-methyl ester and ethylamine by immobilized Escherichia coli cells with γ-glutamyltranspeptidase activity.Amino Acids391177–1182. 10.1007/s00726-010-0553-z
208
Zhang H. Zhang W. Wang Z. Zhan Y. Xu L. Liu J. et al (2013). Enzymatic synthesis of theanine with Escherichia coli γ-glutamyltranspeptidase from a series of γ-glutamyl anilide substrate analogues.Biotechnol. Bioproc. E.18358–364. 10.1007/s12257-012-0644-7
209
Zheng G. Sayama K. Okubo T. Juneja L. R. Oguni I. (2004). Anti-obesity effects of three major components of green tea, catechins, caffeine and theanine, in mice.In Vivo1855–62.
Summary
Keywords
autoprocessing, bacteria, catalytic mechanism, γ-glutamyl transpeptidase, γ-glutamyl peptides, glutathione, poly-γ-glutamic acid
Citation
Saini M, Kashyap A, Bindal S, Saini K and Gupta R (2021) Bacterial Gamma-Glutamyl Transpeptidase, an Emerging Biocatalyst: Insights Into Structure–Function Relationship and Its Biotechnological Applications. Front. Microbiol. 12:641251. doi: 10.3389/fmicb.2021.641251
Received
13 December 2020
Accepted
15 February 2021
Published
09 April 2021
Volume
12 - 2021
Edited by
Kian Mau Goh, University of Technology Malaysia, Malaysia
Reviewed by
Taowei Yang, Jiangnan University, China; Carlo Morelli, University of Milan, Italy; Jun Feng, University of Wisconsin-Madison, United States
Updates
Copyright
© 2021 Saini, Kashyap, Bindal, Saini and Gupta.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rani Gupta, rani.gupta@south.du.ac.in; ranigupta.udsc57@gmail.com; ranigupta15@rediffmail.com
†These authors have contributed equally to this work
This article was submitted to Microbiotechnology, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.