
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 05 May 2021
Sec. Microbiotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.638068
This article is part of the Research TopicNanomicrobiology: Emerging Trends in Microbial Synthesis of Nanomaterials and their ApplicationsView all 10 articles
In recent times, nanoparticles (NPs) have found increasing interest owing to their size, large surface areas, distinctive structures, and unique properties, making them suitable for various industrial and biomedical applications. Biogenic synthesis of NPs using microbes is a recent trend and a greener approach than physical and chemical methods of synthesis, which demand higher costs, greater energy consumption, and complex reaction conditions and ensue hazardous environmental impact. Several microorganisms are known to trap metals in situ and convert them into elemental NPs forms. They are found to accumulate inside and outside of the cell as well as in the periplasmic space. Despite the toxicity of NPs, the driving factor for the production of NPs inside microorganisms remains unelucidated. Several reports suggest that nanotization is a way of stress response and biodefense mechanism for the microbe, which involves metal excretion/accumulation across membranes, enzymatic action, efflux pump systems, binding at peptides, and precipitation. Moreover, genes also play an important role for microbial nanoparticle biosynthesis. The resistance of microbial cells to metal ions during inward and outward transportation leads to precipitation. Accordingly, it becomes pertinent to understand the interaction of the metal ions with proteins, DNA, organelles, membranes, and their subsequent cellular uptake. The elucidation of the mechanism also allows us to control the shape, size, and monodispersity of the NPs to develop large-scale production according to the required application. This article reviews different means in microbial synthesis of NPs focusing on understanding the cellular, biochemical, and molecular mechanisms of nanotization of metals.
Nanoparticles (NPs) are particles with dimensions between 1 and 100 nm that may have different chemical and physical properties relative to their bulk-metal counterpart in addition to the large surface-to-mass ratio (Sardar et al., 2014). The physical and chemical properties of NPs can be differing by their elemental composition, larger specific surface area, and composition of the coating agents (Ahmad et al., 2013a; Sadaf et al., 2020). NPs possess unique features such as mechanical properties (Mishra et al., 2015; Gao et al., 2020), antimicrobial capabilities (Ahmad et al., 2014a, 2015; Abdulla et al., 2021), drug delivery capacities (Ahmad et al., 2021), optical properties (Albrecht et al., 2018), and catalytic capabilities (Mishra et al., 2016; Liu and Liu, 2017; Perwez et al., 2017; Ghosh et al., 2018a, b) and act as artificial chaperones (Ahmad et al., 2014b; Ghosh et al., 2019).
There are various approaches to synthesize nanomaterials, namely, chemical, physical, and biological methods. In general, the chemical and physical methods are preferred routes by the industries, but their disadvantages overshadow their performance. They are usually expensive, consume enormous amounts of time and energy, have complicated procedures, and generate toxic by-products (Castro et al., 2014; Ngoepe et al., 2020). Biogenic NPs are therefore an alternative with the potential to be an eco-friendly and cost-effective method for NP synthesis (Mishra et al., 2013a; Khatoon et al., 2015; Mazumder et al., 2016; Kim et al., 2018; Jacob et al., 2020). In relation to the manufacture of NPs, the word “biogenic” encompasses a range of methodology used for the NP synthesis by either plant extracts or microbes by reduction of metal ions into NPs through their inherent NP manufacturing capabilities (Capeness et al., 2019; Castillo-Henriquez et al., 2020). Bacterial growth properties and genetic modifications render them potent for biogenic NP manufacture as well as industrial applications, particularly when combined with their ability to catch metals, such as those found as environmental pollutants (Nancharaiah et al., 2016).
Interactions between inorganic matter and biological entities have been responsible for geochemical cycles and the maintenance of life on this planet for millions of years. Many species during evolution rely on minerals of different sizes and shapes for a broad range of functions, including physical support, defense from foreign agents, and navigation (Wang and Nilsen-Hamilton, 2013). Biomineralization requires the absorption and modulation of ions from the atmosphere into highly ordered structures, activities that are subject to strict biological regulation (Mann, 2001). A solid-phase organic matrix comprising polysaccharides, phospholipids, and mostly proteins is needed to generate higher-order inorganic structures (Addadi and Weiner, 1985, 2014; Weiner and Dove, 2003). The processes of inorganic structure formation, nucleation, crystal growth, and developing minerals of particular size as well as shape are involved. Harsh conditions may be required to form minerals in the environment. To overcome these situations, organisms provide an offshore account in which specific biological systems help with specific membrane pumps to produce a saturating level of a specific ion (Weiner and Dove, 2003). During mineral formation, the organic matrix plays a major role. First of all, negatively charged residues pull and concentrate positive ions from solution. This contributes to ion saturation at particular points to initiate the nucleation. Nucleation involves a reduction in the free energy and formation of nano-size particles. As a result, the organic matrix decreases the free energy and stabilizes the ions to create a solid stable particle that will be growing into a crystal. Also, the organic matrix plays an important role in the crystal growth and formation of definite particle size. Earlier research has shown that diverse proteins can directly interact with the mineral surface for the formation of solid structure (Hunter, 1996; Mann, 2001; Addadi and Weiner, 2014; Nudelman et al., 2018). A specific organelle called magnetosome, which contains nano-magnetic particles synthesized by magnetotactic bacteria (MTB), is a good model to understand NP formation (Lower and Bazylinski, 2013). The magnetosome is composed of well-defined nano-magnetic particles in a chain or chains that are enclosed by a membrane layer and oriented as per the cell axis (Yamamoto et al., 2010). This magnetosome is often exploited to carry drugs or other loads as a delivery vehicle in cancer treatments (Vargas et al., 2018).
As is evident, several metals such as gold, silver, copper, cadmium, zinc, tellurium, platinum, titanium, and palladium are found to be synthesized through microbial routes, which are often harvested for industrial or biomedical applications. This mini review covers the microbial route to synthesize the various nanomaterials, focusing on the mechanistic aspects of nanotization.
Microorganisms such as bacteria, fungi, yeast, and algae are often favored for NP synthesis due to simpler cultivation, rapid growth rate, and their capacity to grow at atmospheric pH, temperature, and pressure conditions. Different biological agents behave differently with different metal solutions in order to form NPs (Fariq et al., 2017; Saravanan et al., 2020; Ghosh et al., 2021). Metal ions are initially trapped on the surface of the cell followed by the reduction of metal ions to NPs with the presence of enzymes synthesized by the microbes. The NP synthesis process is depicted in Figure 1. The microbial-mediated synthesis of NPs and their possible application is summarized in Table 1.
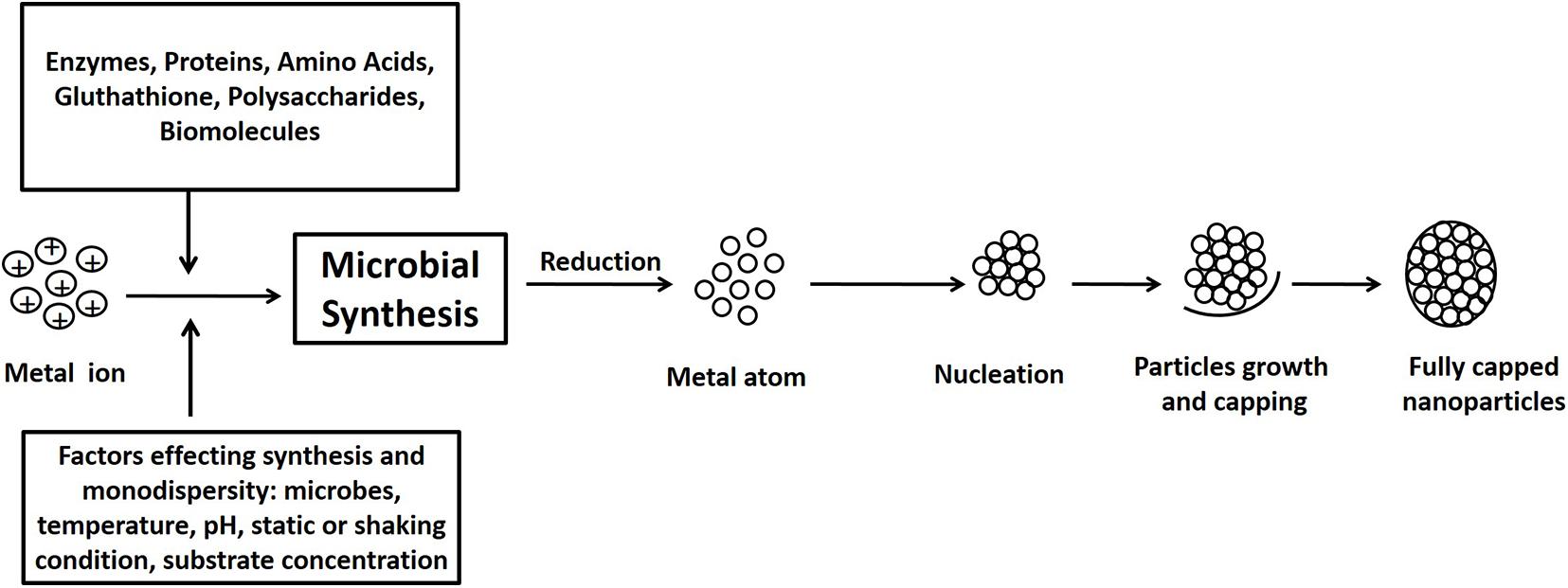
Figure 1. Schematic diagram of mechanisms of nanoparticle synthesis by microbes: the pathway of nanoparticle synthesis by microbes involves metal capture, enzymatic reduction, and capping. Several biomolecules such as proteins, amino acids, and polysaccharides found in the microbial extracts help in the stabilization of the nanoparticles.
Bacteria that reduce metals are found to be environmentally friendly catalysts for bioremediation as well as material synthesis. In general, through the microbial respiration processes, genus Shewanella has been found to synthesize diverse metal oxides (Kim et al., 2018). Electrons can be moved from reduced organic to oxidized inorganic compounds through microbial dissimilatory anaerobic respiration, thus promoting the formation of crystal along with bioremediation processes. It is well documented that the genus Shewanella is able to assist the oxidation of organic acids as electron donors and reduction of inorganic metals as electron acceptors (Heidelberg et al., 2002; Harris et al., 2018). Bacterial nanowires and flavins are extracellularly excreted by the genus Shewanella by bioreduction process (Marsili et al., 2008; El-Naggar et al., 2010; Beblawy et al., 2018). Few researchers have documented biosynthesis of copper NPs through biosorptive process with dead Rhodotorula mucilaginosa biomass. The synthesized NPs were spherical in form and were considered to be a good method for NP synthesis for simultaneous pollutant remediation. Another analysis involving Clostridium pasteurianum metallic molybdenum NP synthesis has also been published (Salvadori et al., 2014; Nordmeier et al., 2018). Ag NPs were synthesized by Shewanella oneidensis MR-1 with antibacterial activity (Suresh et al., 2010). Another report showed that Cu NPs were synthesized through bioreduction of Cu(II) by S. oneidensis MR-1 (Kimber et al., 2018). Shewanella loihica PV-4 and S. oneidensis MR-1 have the ability to produce Pd NPs with higher catalytic activities (Wang W. et al., 2018; Xiong et al., 2018). S. oneidensis MR-1 have properties to utilize toxic soluble tellurite as electron acceptor leading to Te nanorod formation. Also, needle-shaped crystalline Te nanorods were formed both intracellularly and extracellularly (Kim et al., 2012, 2013). Gold- and tellurite-containing nanostructures were biosynthesized in aerobic and anaerobic conditions by crude extracts from Enterobacter cloacae MF01 (Contreras et al., 2018). In another study, the investigators explored the formation biogenic selenium nanostructures by gram-negative bacteria under aerobic conditions (Piacenza et al., 2018). Studies have also revealed the importance of different volatile sulfur compounds (VSCs) in the biosynthesis of CdS quantum dots (QDs) by Pseudomonas fragi GC01 (Gallardo Benavente et al., 2019). Also, similar studies showed ruthenium and ruthenium-palladium NPs synthesized by Escherichia coli cells (Macaskie et al., 2019) and mercury NPs synthesized by Enterobacter sp. (Sinha and Khare, 2011).
Fungal NP synthesis is favored over other microbial synthesis methods due to the high resistance of fungal mycelial mesh to higher flow and agitation in bioreactors (Saravanan et al., 2020). Chitin was found to be the key ingredient in the fungal cell wall system that is involved in heavy metal complexation, resulting in synthesis of NPs (Wang L. et al., 2018). Due to several bioactive metabolites, strong aggregation, and increased efficiency, fungi are more resourceful than bacteria in the biosynthesis of NPs (Castro-Longoria et al., 2011; Alghuthaymi et al., 2015; Saravanan et al., 2020). In Au NP biosynthesis, many filamentous fungi have been reported to be capable. This research employed diverse approaches in order to biosynthesize Au NPs. The authors proposed that the NPs could be stabilized by fungal secreted compounds and media materials (Molnar et al., 2018; Guilger-Casagrande and De Lima, 2019). Formulation of silver NPs from Penicillium chrysogenum NG85 and Fusarium chlamydosporum NG30 was successful. Ag NPs were prepared using cell-free filtrate obtained from the autolyzed biomass. The involvement of enzymes and proteins in the cell-free filtrate was responsible for the formation of NPs in which the enzymes mediated the reduction of silver ions to silver atoms and the stabilization of silver atoms by capping materials was carried out by proteins (Khalil et al., 2019). Phomopsis liquidambaris extracellular filtrate isolated from healthy Salacia chinensis leaves contains proteins that could reduce and serve as a capping agent, thereby stabilizing the formulation of Ag NPs that demonstrated bactericidal activity against pathogens (Seetharaman et al., 2018). Filtered biomass extract of Aspergillus tamarii, Aspergillus niger, and Penicillium ochrochloron could act as a potential fungal nanofactories for the green and eco-friendly production (Devi and Joshi, 2015). An endophytic fungus Periconium sp. isolated from leaves of Balanites aegyptiaca mediated the synthesis of zinc NPs (Ganesan et al., 2020). The synthesis of copper oxide NPs from endogenous fungi has been accompanied by a similar method. Copper oxide NPs were synthesized using Trichoderma asperellum water extract and copper nitrate solution (Saravanakumar et al., 2019). P. chrysogenum provided a particular pigment that facilitated the synthesis of NPs (El-Sayyad et al., 2018).
Nanoparticles have been synthesized from many metals, including gold, silver, copper, nickel, cobalt, zinc, and titanium inside microorganisms. These metal and metal oxide nanomaterials comprise a large segment of the growing nanotechnology market. Increasing use of metallic nanomaterials is likely to result in the release of these particles into the aqueous environments, thus providing a path to enter the food chain and eventually disturbing the ecological balance. Toxicities associated with NPs in microorganisms are mainly related to their nano-size that causes membrane disorganization, generation of reactive oxygen species (ROS), and, in some cases, oxidative DNA damage and release of the toxic ions (Morones et al., 2005; Niazi and Gu, 2009).
The metal NPs cause adsorption on the cell wall leading to depolarization and increase in its permeability, followed by disintegration of membranes and further penetration into the membrane (Thill et al., 2006; McQuillan et al., 2012; Slavin et al., 2017). Inside microbes, these metallic NPs increase significant production of ROS, which target multiple sites simultaneously like conformational changes in protein, peroxidation of the lipids, and DNA damage, thus leading to membrane disintegration and ultimately may cause cell death (Madl et al., 2014; Slavin et al., 2017).
Ensuring monodispersity of NPs is important for biological studies and clinical translation. An important step in understanding the physical properties of NPs is purifying NPs into monodisperse fractions. NPs have been separated by size and form, and certain methods have been created to purify subpopulations of particles after their assembly. Size exclusion chromatography will separate both hard and soft NPs according to their size (Wei and Liu, 1999; Ahmad et al., 2013b; Satzer et al., 2014). Similarly, different density and mass-based methods can separate NPs according to their form (e.g., discoidal from spherical-like NPs) (Johansson et al., 2007; Robertson et al., 2016). There are a variety of techniques that manufacture uniformly appearing nanomaterials with desired properties. The assembly process can yield NPs with several properties simultaneously (Robertson et al., 2016). Characterization methods assess the form, scale, distribution, surface morphology, and surface area of the NPs. Methods to classify NPs are illustrated in Figure 2. The visual observation of color change is critical for NPs. Due to the variation in the surface plasmon resonance (SPR) measured by the NPs, color shift is observed (Zhang et al., 2016). SPR allows the presence of the metal to be monitored by UV–Vis spectroscopy, while X-ray diffraction (XRD) can be used to classify metal NPs. Analysis of XRD data will tell about the composition of materials. Apart from controlling the size of the dust, it is used for particle size determination (Reddy et al., 2016). Dynamic light scattering (DLS) is one of the widely used techniques for the determination of NP size. Hydrodynamic radii are determined using the time it takes light to pass around the particle. In colloidal suspensions, DLS helps ensure calculations are more precise (Kumari et al., 2019). Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) are other useful techniques for the determination of size, shape, morphology, and aggregation of NPs (Ahmad and Khare, 2018). Particle size, agglomeration, and shape can be determined by atomic force microscopy (AFM) topography imaging (Ahmad and Sardar, 2014). Biomolecules responsible for capping NPs can be observed by Fourier transform infra-red (FTIR) spectroscopy (Menon et al., 2017; Sardar et al., 2018). Inductively coupled plasma mass spectrometry (ICP-MS) can analyze NPs qualitatively and quantitatively. Single or combination methods are used to characterize and isolate monodisperse particles.
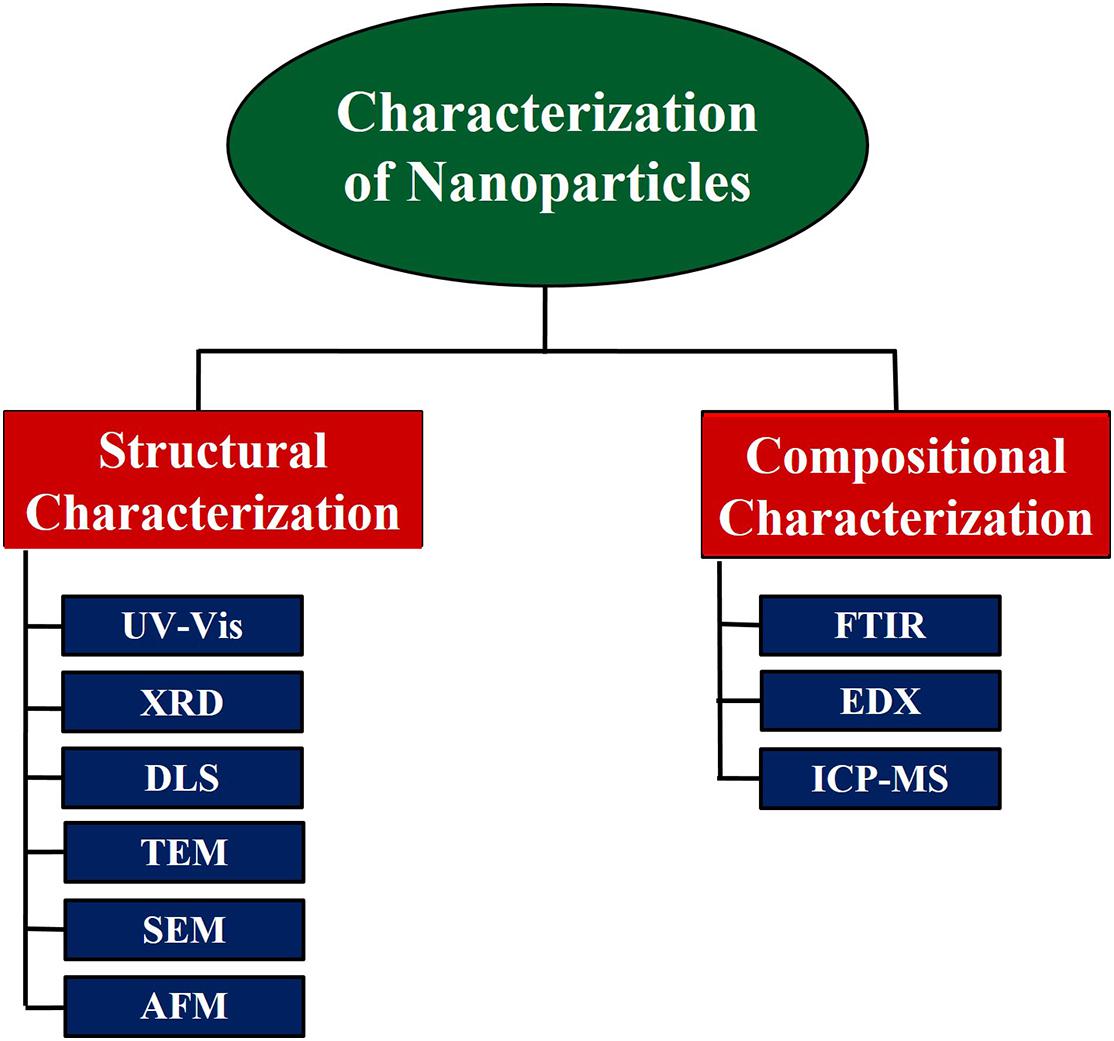
Figure 2. Characterization methods for nanoparticles: several analytical techniques are employed to isolate and characterize monodisperse nanoparticles. UV–Vis, ultraviolet–visible spectroscopy; XRD, X-ray powder diffraction; DLS, dynamic light scattering; TEM, transmission electron microscopy; SEM, scanning electron microscopy; AFM, atomic force microscopy; FTIR, Fourier transform infrared spectroscopy; EDX, energy-dispersive X-ray spectroscopy; ICP-MS, inductively coupled plasma mass spectrometry.
Why do microbes accumulate nanometals? Presence of metals in the environment entails their penetration into cells, disruption of membrane structure, inactivation of enzymes, and production of antimetabolites, which chelate with essential metabolites leading to toxicity or cell death for the microbial cells in the vicinity of the metal particles (Sobolev and Begonia, 2008). This property of metals has also been explored in case of antimicrobial activity against pathogenic strains. As such, microbes try their best to evade the toxicity of the metals by eliminating or reactively changing their properties or accumulating them in forms away for vital organelles. The nanotization of metals by microbes is an important part of geo-cycles and occurs mainly as a stress response resulting in adaptive or defense mechanism for the organism against the metal toxicity. Thus, the microorganisms capable of NP synthesis are often metal resistant in nature, and bacteria genera such as Pseudomonas are reported to grow in high metal concentrations (Iravani, 2014; Luo et al., 2015). The interaction of the anionic cell components with the cationic metal ions leads to the formation of metal, metal oxide, or metal sulfide NPs via reduction, chelation, or hydrolysis (Bansal et al., 2005). As detoxification measures, the organism can employ enzymatic reduction, dissimilatory oxidation, precipitation, complexation, or transport via efflux systems to remediate the metals from the cell (Arshad, 2017). The broad mechanisms of NP synthesis mainly involve either of the pathways involving enzymes, proteins, exopolysaccharides, and electron shuttle quinines (Gahlawat and Choudhury, 2019). Often external parameters such as temperature and pH play a definite role in NP synthesis with regard to properties such as size as well as concentration (Saklani et al., 2012).
Synthesis through microbial routes gives the distinct advantages of control of shape and size, in addition to being a greener alternative to chemical and physical processes. Compared with other biological synthesis methods such as plants, which may result in decrease in monodispersity due the presence of phytochemicals and yield difference due to seasonal variations, microbial means offer a monodisperse NP synthesis methods, which are reproducible in industrial conditions in bioreactors, without batch variations (Ahmad et al., 2021). Additionally, the small size and biocompatibility of biogenic NPs render them good candidates as drug delivery carriers, antimicrobials, anticancer drugs, and diagnostic agents. The disadvantages, though few, mainly refer to the high cost and technology involved in the development and production process of biogenic NPs. Moreover, some NPs may end up in the environment with polluting as well as disease-causing effects (Parveen et al., 2016). Thus, it is important to assess the toxicity and long-term effects of synthesized NPs before using them in any application.
Enzymes are believed to be the foremost entities involved in the reduction and capping of metals in microbes, through redox reactions occurring in either the intracellular or extracellular space, often acting as the nucleation sites. Broadly, the intracellular route involves metal capture, enzymatic reduction, and capping, while in the extracellular method, secreted or membrane-bound enzymes/proteins are involved. Additionally, shuttle quinones such as anthraquinones, naphthoquinones, and hydroquinones are involved in the process. Mostly NADH-dependent nitrate reductases are reported in fungi to be involved in the reduction process. Fungal species such as Penicillium brevicompactum, Cladosporium cladosporioides, Fusarium oxysporum, Fusarium semitectum, and Fusarium solani have been reported to use nitrate reductases to produce Ag NPs (Ovais et al., 2018; Meenakshi, 2020). NADH and NADH-dependent reductases in Pseudomonas aeruginosa JS-11 were reported for the biogenic reduction of SeO32– to insoluble Se0 NPs (Dwivedi et al., 2013). In a cell-free study, glutathione (GSH), NADPH, and GSH reductase have been utilized to quasi-biosynthesize Ag2Se QDs with tunable fluorescence (Gu et al., 2012). In Rhodopseudomonas capsulate, NADH and NADH-dependent enzymes aided in the conversion of Au ions into Au nanospheres through an electron shuttle mechanism (He et al., 2007). In case of palladium NPs, hydrogenases have been found to play a profound role in reducing Pd(II), with the deposit of Pd(0) NPs depending on the localization of hydrogenases in Desulfovibrio fructosivorans. The enzyme serves as the nucleation site, providing electrons to Pd(II) for its reduction (Mikheenko et al., 2008). Hydrogenases have also been reported in the reduction of several other metals such as selenium by C. pasteurianum, uranium by Micrococcus lactyliticus, and gold by Shewanella algae (Iravani, 2014). In case of metal sulfide NPs, the metal and sulfide moieties need to be present in soluble salt form as precursors, and furthermore, sulfide anions and metal cations react with each other to form the metal sulfide NPs, mediated by extracellular or intracellular enzymes. For intracellular reduction, the ions enter the cytoplasm through magnesium or manganese transport chains. For extracellular synthesis, secreted enzymes or those present on the cell membrane are involved (Hosseini and Sarvi, 2015; Fariq et al., 2017). In case of intracellular synthesis, cations are utilized in the capturing metallic ions from the exterior and subsequent reduction inside the cell, followed by accumulation in the cytoplasmic membrane, cell wall, or periplasmic space. Besides the commonly reported reductase, laccase and ligninase have also been reported for the intracellular synthesis route (Ovais et al., 2018).
In addition to the enzymes discussed in the above section, myriad proteins are also found to responsible for NP formation by microbes. Proteins and peptides are also involved in the capping and stabilization of the formed nanometals. Also, for intracellular entry of metals, transport proteins become essential. Especially in case of magnetic NPs, MTB employ magnetosome membrane proteins for biomineralization. Another protein termed MagA, isolated from the strain Magnetospirillum sp. AMB-1, was elemental in biogenic magnetic NP synthesis (Matsunaga and Takeyama, 1998). In another instance, for controlled size and shape, a small acidic protein Mms6 isolated from the same strain was utilized to precipitate uniform CoFe2O4 nanocrystals in vitro (Prozorov et al., 2007). In case of smaller peptides, dipeptides, and tripeptides having polar amino groups have shown mediation of assembly of Au NPs in the presence of HAuCl4 as a reducing agent. Gold NP-tripeptide was prepared using a novel tripeptide, harboring a C-terminus tyrosine residue that reduced Au3+ to Au NPs. Furthermore, the terminally located free amino group bound Au NPs resulting in stable colloidal Au (Ramesh et al., 2015). In a similar study, yeast strains of Schizosaccharomyces pombe and Candida glabrata accumulated CdS NPs inside the cell plasma coated with phytochelatins, a peptide known to arrest DNA disruption and cell cycle damage occurring in metal toxicity (Krumov et al., 2007).
As toxic metal ions accumulate around a microbial cell and further enter inside the cell membrane, efflux pumps try to eliminate and excrete them into the extracellular space, thus combating metal toxicity. The presence of efflux pump genes, including a multidrug resistant (MDR) one, is one of the main factors behind a metal-resistant phenotype and has an impact on the nanometal synthesis capacity of the organism. In case of silver, BaeSR, as a two-component signal transduction (TCS) system, is involved in the overexpression of efflux pumps related to metal and antibiotic resistance. Such proteins generally belong to the RND (resistance, nodulation, and cell division) family. Similarly, CusCFBA efflux system has been identified in the Escherichia coli, overexpressed in the presence of higher concentration of copper ions (Graves et al., 2015). A lower number of porins such as OmpF or OmpC present on the outer membrane of E. coli is also known to be responsible for the efflux of silver ions and hence the resistant phenotype (Salas-Orozco et al., 2019). These efflux pumps are also responsible for antibiotic resistance in bacteria, and one study explored vanillin-coated Au NPs as inhibitors of MexAB-OprM efflux pump components (Arya et al., 2019).
The interaction of metals with cells and subsequent nanosynthesis can be elucidated precisely by understanding how they interact with individual organelles and biomolecules. In fact, instead of whole cells, organelles and subcellular components in ex vivo conditions have been explored for their capability to synthesize nanometals with controlled dimensions. In one such instance, circular plasmid DNA molecules close to 4-kb size (acting as the reducing agent) were allowed to complex electrostatically with Ag ions, followed by UV irradiation, leading to the formation of Ag NPs, compared with the non-plasmid control (Liu et al., 2012). Isolated porins from the cell membrane of Mycobacterium smegmatis, termed as MspA, were docked onto the self-assembled organic thiosulfates and electrodeposited on Au plates. These self-assembled protein layers were further used to deposit Cu NPs with potential applications in transistors (Woerner et al., 2007). Separate classes of studies have also focused on protein self-assemblies into nanostructures. Genetically engineered Pichia pastoris has been used to synthesize protein nanostructures composed of blocks to coat DNA used in gene delivery applications (Hernandez-Garcia et al., 2012).
In the presence of elevated concentrations of heavy metals, bacteria usually respond by expressing specific heavy metal resistance genes (MRGs) for NP production. Since these genes are mostly present in plasmids or transposons, they are easily transferred to the neighboring microbial communities. Several research groups have studied these genes to elucidate the mechanisms behind nanotization. A group worked on estimating the amount of metals bioavailable for the microbial community using quantitative PCR and found that the gene czcA (a Cd/Zn/Co efflux pump) was responsible for Cd/Zn/Co bioavailability in microbes (Roosa et al., 2014). A study by yang and their group showed that some of the MRG such as pcoA, merA, silC, and arsA genes were present in higher frequencies in MDR blaNDM–1– and blaCTX–M–15– Enterobacteriaceae isolates (Yang et al., 2018). In Northern China near a copper tailing dam area, the following genes were found at the remedial site: copper resistance genes (copA, copB, pcoA, pcoC, and pcoD), other MRG (czcA, czcC, and czcD), arsenic resistance genes (arsB and arsC), nccA (for nickel), pbrT (for lead), and chrB (for chromium) (Chen et al., 2019). Some of the genes are present in operons such as silG gene in the sil operon for silver sequestration (Randall et al., 2015). A study has shown that arsRBCC operon and arsC gene in Desulfovibrio desulfuricans G20 helped in regulating arsenic in the microbial environment (Li and Krumholz, 2007). Studies have also found upregulated levels of efflux complexes in the presence of excess heavy metals, such as resistance-nodulation-cell division (RND) transporters, the P-type ATPases, efflux complexes made from membrane fusion protein (MFP) family, or the outer membrane factor (OMF) protein family.
The understanding of the genetics of microbe-mediated NP synthesis has been interestingly used to design genetically engineered microbes with metal-resistant phenotypes, which find applications in NP synthesis as well as remediating the metal-contaminated environment. Such an example was observed in Escherichia coli strains expressing PC synthetase of Schizosaccharomyces pombe, used for the synthesis of semiconductor CdS nanocrystals. The phytochelatins produced by the action of PC synthase act as a nucleation site for the nanocrystals and stabilize them against aggregation (Kang et al., 2008). Thus, several diverse gene families are reported to be involved in nanotization of metals by microbial strains.
Owing to their nanoscale sizes leading to an increase in the surface/volume ratio, NPs find applications in myriad industrial, environmental, and biomedical applications. TiO2 NPs from several microbial species such as Micrococcus lylae, Cellulosimicrobium sp., Micrococcus aloeverae, and Chlorella pyrenoidosa have been shown to show dye-degradation capability to remediate polluted waste waters (Fulekar et al., 2018). Similarly, Au NPs from Streptomyces griseoruber and ZnO NPs from Cordyceps have been specifically explored to degrade methylene blue dye via catalysis (Ranjitha and Rai, 2017; Li et al., 2019). Toxic compound remediation is also a major environmental application of biogenic NPs where PbS and Au NPs from bacterial and fungal strains have shown promise (Lok et al., 2006; Shen et al., 2017). In electronics, TiO2 NPs from Bacillus mycoides have been applied in the construction of green solar cells (Ordenes-Aenishanslins et al., 2014). Closer dimensions to biomolecules also render these biogenic NPs good candidates for biomedical applications. A major portion of microbial NPs such as Ag, Ag, Se, and Te NPs find applications as antibacterial, antifungal, antiviral, and antibiofilm agents (Abdeen et al., 2014; Zonaro et al., 2015; El-Sheekh et al., 2020; Gursoy, 2020). Biogenic NPs display good penetration across membranes, and blood–tissue and blood–brain barriers, thus finding applications as anticancer agents and drug delivery vehicles (Gonzalez-Ballesteros et al., 2017). An interesting application had utilized bacterial magnetosomes loaded with doxorubicin and tested on H22 tumor-bearing mice; they displayed higher tumor toxicity than only doxorubicin (Sun et al., 2009). These magnetosomes are also recognized as better MRI contrast agents with higher relaxivity than the conventional ones (Zhang Y. et al., 2018). In biosensing, CdSe QDs from C. pyrenoidosa have been exploited for imatinib sensing, while Fe2O3 NPs have been used to detect H2O2 and glucose (Mishra et al., 2016; Zhang Z. et al., 2018). Thus, it is evident that the applications of microbial NPs are diverse and multifaceted, holding great promise in several fields of research.
Several bacterial, viral, algal, fungal, and yeast species are known to trap metals in situ and convert them to elemental NP forms, while remediating their immediate environment in the process. This feature has been further exploited in industrial, environmental, and biomedical applications. However, most metals are toxic to microbial cells; thus, it is widely reported that the synthesis of metals into their elemental nano-forms results as a defense mechanism or stress response for the organism to eliminate, segregate from essential organelles, or reactively change the harmful nature of the metals. Mechanisms such as enzymatic reactions, precipitation, complexation, binding to peptides, and efflux pumps are involved in this process, which act independently or simultaneously for the metal remediation by the cell, resulting in a metal-resistant phenotype, with microbes harboring specific genes for this property. Even though a number of mechanisms have been reported for biogenic nanosynthesis, it is important to extend our studies to other reducing enzymes, catalytic proteins, and stabilizers along with their tandem action in the cell. The role of different classes of enzymes needs to be studied in detail. The precise understanding of pathways and mechanisms involved in the biogenic synthesis allows researchers to modulate existing microbes and engineer metabolic pathways for NP synthesis with controlled size and shapes for varied applications.
SG, RA, and KB conceptualized and prepared the manuscript. MFA and SR helped and addressing the review comments with inputs which were further included in the revised manuscript. All authors have critically reviewed the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors acknowledge the Indian Institute of Technology Delhi and Jamia Millia Islamia, New Delhi, for providing necessary facilities and resources. The authors also acknowledge the support of researchers supporting project under grant number RSP-2021/122, King Saud University, Riyadh, Saudi Arabia.
Abdeen, S., Geo, S., Praseetha, P. K., and Dhanya, R. P. (2014). Biosynthesis of silver nanoparticles from Actinomycetes for therapeutic applications. Int. J. Nano Dimen. 5, 155–162.
Abdel-Raouf, N., Al-Enazi, N. M., and Ibraheem, I. B. M. (2017). Green biosynthesis of gold nanoparticles using Galaxaura elongata and characterization of their antibacterial activity. Arab. J. Chem. 10, S3029–S3039.
Abdulla, N. K., Siddiqui, S. I., Fatima, B., Sultana, R., Tara, N., Hashmi, A. A., et al. (2021). Silver based hybrid nanocomposite: a novel antibacterial material for water cleansing. J. Clean. Product. 284, 124746. doi: 10.1016/j.jclepro.2020.124746
Acay, H. (2020). Utilization of Morchella esculenta-mediated green synthesis golden nanoparticles in biomedicine applications. Preparat. Biochem. Biotechnol. 51, 127–136. doi: 10.1080/10826068.2020.1799390
Acharya, D., Satapathy, S., Somu, P., Parida, U. K., and Mishra, G. (2020). Apoptotic effect and anticancer activity of biosynthesized silver nanoparticles from marine algae Chaetomorpha linum extract against human colon cancer cell HCT-116. Biol. Trace Elem. Res. 20, 1–11.
Addadi, L., and Weiner, S. (1985). Interactions between acidic proteins and crystals: stereochemical requirements in biomineralization. Proc. Natl. Acad. Sci. U.S.A. 82, 4110–4114. doi: 10.1073/pnas.82.12.4110
Addadi, L., and Weiner, S. (2014). Biomineralization: mineral formation by organisms. Phys. Script. 89:098003. doi: 10.1088/0031-8949/89/9/098003
Ahmad, R., and Khare, S. K. (2018). Immobilization of Aspergillus niger cellulase on multiwall carbon nanotubes for cellulose hydrolysis. Bioresour. Technol. 252, 72–75. doi: 10.1016/j.biortech.2017.12.082
Ahmad, R., and Sardar, M. (2014). Immobilization of cellulase on TiO2 nanoparticles by physical and covalent methods: a comparative study. Ind. J. Biochem. Biophys. 51, 314–320.
Ahmad, R., Khatoon, N., and Sardar, M. (2013a). Biosynthesis, characterization and application of TiO2 nanoparticles in biocatalysis and protein folding. J. Protein. Proteom. 4, 115–121. doi: 10.1385/1-59745-189-4:115
Ahmad, R., Khatoon, N., and Sardar, M. (2014a). Antibacterial effect of green synthesized TiO2 nanoparticles. Adv. Sci. Lett. 20, 1616–1620. doi: 10.1166/asl.2014.5563
Ahmad, R., Mishra, A., and Sardar, M. (2013b). Peroxidase-TiO2 nanobioconjugates for the removal of phenols and dyes from aqueous solutions. Adv. Sci. Eng. Med. 5, 1020–1025. doi: 10.1166/asem.2013.1387
Ahmad, R., Mishra, A., and Sardar, M. (2014b). Simultaneous immobilization and refolding of heat treated enzymes on TiO2 nanoparticles. Adv. Sci. Eng. Med. 6, 1264–1268. doi: 10.1166/asem.2014.1644
Ahmad, R., Mohsin, M., Ahmad, T., and Sardar, M. (2015). Alpha amylase assisted synthesis of TiO2 nanoparticles: structural characterization and application as antibacterial agents. J. Hazard. Mater. 283, 171–177. doi: 10.1016/j.jhazmat.2014.08.073
Ahmad, R., Srivastava, S., Ghosh, S., and Khare, S. K. (2021). Phytochemical delivery through nanocarriers: a review. Coll. Surf. B Biointerfac. 197:111389. doi: 10.1016/j.colsurfb.2020.111389
Ahmed, E., Kalathil, S., Shi, L., Alharbi, O., and Wang, P. (2018). Synthesis of ultra-small platinum, palladium and gold nanoparticles by Shewanella loihica PV-4 electrochemically active biofilms and their enhanced catalytic activities. J. Saud. Chem. Soc. 22, 919–929. doi: 10.1016/j.jscs.2018.02.002
Akter, S., and Huq, M. A. (2020). Biologically rapid synthesis of silver nanoparticles by Sphingobium sp. MAH-11T and their antibacterial activity and mechanisms investigation against drug-resistant pathogenic microbes. Artific. Cells Nanomed. Biotechnol. 48, 672–682. doi: 10.1080/21691401.2020.1730390
Albrecht, G., Ubl, M., Kaiser, S., Giessen, H., and Hentschel, M. (2018). Comprehensive study of plasmonic materials in the visible and near-infrared: linear, refractory, and nonlinear optical properties. Acs Photon. 5, 1058–1067. doi: 10.1021/acsphotonics.7b01346
Alghuthaymi, M. A., Almoammar, H., Rai, M., Said-Galiev, E., and Abd-Elsalam, K. A. (2015). Myconanoparticles: synthesis and their role in phytopathogens management. Biotechnol. Biotechnol. Equipm. 29, 221–236. doi: 10.1080/13102818.2015.1008194
Alsamhary, K. I. (2020). Eco-friendly synthesis of silver nanoparticles by Bacillus subtilis and their antibacterial activity. Saud. J. Biol. Sci. 27, 2185–2191. doi: 10.1016/j.sjbs.2020.04.026
Arshad, A. (2017). Bacterial synthesis and applications of nanoparticles. Nano Sci. Nano. Technol. 11:119.
Arya, S. S., Sharma, M. M., Das, R. K., Rookes, J., Cahill, D., and Lenka, S. K. (2019). Vanillin mediated green synthesis and application of gold nanoparticles for reversal of antimicrobial resistance in Pseudomonas aeruginosa clinical isolates. Heliyon 5:e02021. doi: 10.1016/j.heliyon.2019.e02021
Balagurunathan, R., Radhakrishnan, M., Rajendran, R. B., and Velmurugan, D. (2011). Biosynthesis of gold nanoparticles by actinomycete Streptomyces viridogens strain HM10. J. Biochem. Biophys. 48, 331–335.
Bansal, V., Rautaray, D., Bharde, A., Ahire, K., Sanyal, A., Ahmad, A., et al. (2005). Fungus-mediated biosynthesis of silica and titania particles. J. Mater. Chem. 15, 2583–2589. doi: 10.1039/b503008k
Beblawy, S., Bursac, T., Paquete, C., Louro, R., Clarke, T. A., and Gescher, J. (2018). Extracellular reduction of solid electron acceptors by Shewanella oneidensis. Mol. Microbiol. 109, 571–583. doi: 10.1111/mmi.14067
Bhuyar, P., Rahim, M. H. A., Sundararaju, S., Ramaraj, R., Maniam, G. P., and Govindan, N. (2020). Synthesis of silver nanoparticles using marine macroalgae Padina sp. and its antibacterial activity towards pathogenic bacteria. Beni Suef Univ. J. Basic Appl. Sci. 9, 1–15.
Borah, D., Das, N., Das, N., Bhattacharjee, A., Sarmah, P., Ghosh, K., et al. (2020). Alga-mediated facile green synthesis of silver nanoparticles: photophysical, catalytic and antibacterial activity. Appl. Organ. Chem. 34:e5597.
Capeness, M. J., Echavarri-Bravo, V., and Horsfall, L. E. (2019). Production of biogenic nanoparticles for the reduction of 4-nitrophenol and oxidative Laccase-Like reactions. Front. Microbiol. 10:997.
Castillo-Henriquez, L., Alfaro-Aguilar, K., Ugalde-Alvarez, J., Vega-Fernandez, L., Montes De Oca-Vasquez, G., and Vega-Baudrit, J. (2020). Green synthesis of gold and silver nanoparticles from plant extracts and their possible applications as antimicrobial agents in the agricultural area. Nanomaterials 10:1763. doi: 10.3390/nano10091763
Castro, L., Blazquez, M. L., Gonzalez, F. G., and Ballester, A. (2014). Mechanism and applications of metal nanoparticles prepared by bio-mediated process. Rev. Adv. Sci. Eng. 3, 199–216. doi: 10.1166/rase.2014.1064
Castro-Longoria, E., Vilchis-Nestor, A. R., and Avalos-Borja, M. (2011). Biosynthesis of silver, gold and bimetallic nanoparticles using the filamentous fungus Neurospora crassa. Coll. Surfac. B Biointerfac. 83, 42–48. doi: 10.1016/j.colsurfb.2010.10.035
Chen, J., Li, J., Zhang, H., Shi, W., and Liu, Y. (2019). Bacterial heavy-metal and antibiotic resistance genes in a copper Tailing Dam Area in Northern China. Front. Microbiol. 10:1916.
Contreras, F., Vargas, E., Jimenez, K., Munoz-Villagran, C., Figueroa, M., Vasquez, C., et al. (2018). Reduction of gold (III) and tellurium (IV) by Enterobacter cloacae MF01 results in nanostructure formation both in aerobic and anaerobic conditions. Front. Microbiol. 9:3118.
Costa, L. H., Hemmer, J. V., Wanderlind, E. H., Gerlach, O. M. S., Santos, A. L. H., Tamanaha, M. S., et al. (2020). Green synthesis of gold nanoparticles obtained from algae Sargassum cymosum: optimization, characterization and stability. BioNanoScience 20, 1–14.
Deljou, A., and Goudarzi, S. (2016). Green extracellular synthesis of the silver nanoparticles using thermophilic Bacillus sp. AZ1 and its antimicrobial activity against several human pathogenetic bacteria. Iran. J. Biotechnol. 14:25. doi: 10.15171/ijb.1259
Devi, L. S., and Joshi, S. R. (2015). Ultrastructures of silver nanoparticles biosynthesized using endophytic fungi. J. Microsc. Ultrastruct. 3, 29–37. doi: 10.1016/j.jmau.2014.10.004
Dwivedi, S., Alkhedhairy, A. A., Ahamed, M., and Musarrat, J. (2013). Biomimetic synthesis of selenium nanospheres by bacterial strain JS-11 and its role as a biosensor for nanotoxicity assessment: a novel Se-bioassay. PLoS One 8:e57404. doi: 10.1371/journal.pone.0057404
El-Naggar, M. Y., Wanger, G., Leung, K. M., Yuzvinsky, T. D., Southam, G., Yang, J., et al. (2010). Electrical transport along bacterial nanowires from Shewanella oneidensis MR-1. Proc. Natl. Acad. Sci. U.S.A. 107, 18127–18131. doi: 10.1073/pnas.1004880107
El-Sayyad, G. S., Mosallam, F. M., and El-Batal, A. I. (2018). One-pot green synthesis of magnesium oxide nanoparticles using Penicillium chrysogenum melanin pigment and gamma rays with antimicrobial activity against multidrug-resistant microbes. Adv. Powder Technol. 29, 2616–2625. doi: 10.1016/j.apt.2018.07.009
El-Sheekh, M. M., Shabaan, M. T., Hassan, L., and Morsi, H. H. (2020). Antiviral activity of algae biosynthesized silver and gold nanoparticles against Herps Simplex (HSV-1) virus in vitro using cell-line culture technique. Int. J. Environ. Health Res. 20, 1–12. doi: 10.1080/09603123.2020.1789946
Fariq, A., Khan, T., and Yasmin, A. (2017). Microbial synthesis of nanoparticles and their potential applications in biomedicine. J. Appl. Biomed. 15, 241–248. doi: 10.1016/j.jab.2017.03.004
Feroze, N., Arshad, B., Younas, M., Afridi, M. I., Saqib, S., and Ayaz, A. (2020). Fungal mediated synthesis of silver nanoparticles and evaluation of antibacterial activity. Microsc. Res. Techn. 83, 72–80.
Fulekar, J., Dutta, D. P., Pathak, B., and Fulekar, M. H. (2018). Novel microbial and root mediated green synthesis of TiO2 nanoparticles and its application in wastewater remediation. J. Chem. Technol. Biotechnol. 93, 736–743. doi: 10.1002/jctb.5423
Gahlawat, G., and Choudhury, A. R. (2019). A review on the biosynthesis of metal and metal salt nanoparticles by microbes. RSC Adv. 9, 12944–12967. doi: 10.1039/c8ra10483b
Gallardo Benavente, C. D., Carrión, O., Todd, J. D., Pieretti, J., Seabra, A., Duran, N., et al. (2019). Biosynthesis of cds quantum dots mediated by volatile sulfur compounds released by antarctic pseudomonas fragi. Front. Microbiol. 10:1866.
Ganesan, V., Hariram, M., Vivekanandhan, S., and Muthuramkumar, S. (2020). Periconium sp.(endophytic fungi) extract mediated sol-gel synthesis of ZnO nanoparticles for antimicrobial and antioxidant applications. Mater. Sci. Semiconduc. Proces. 105:104739. doi: 10.1016/j.mssp.2019.104739
Gao, L., Fan, K., and Yan, X. (2020). Iron oxide nanozyme: a multifunctional enzyme mimetics for biomedical application. Nanozymolog 2, 105–140. doi: 10.1007/978-981-15-1490-6_5
Ghosh, S., Ahmad, R., and Khare, S. K. (2018a). Immobilization of cholesterol oxidase: an overview. Open Biotechnol. J. 12, 176–188. doi: 10.2174/1874070701812010176
Ghosh, S., Ahmad, R., and Khare, S. K. (2019). Refolding of thermally denatured cholesterol oxidases by magnetic nanoparticles. Int. J. Biol. Macromol. 138, 958–965. doi: 10.1016/j.ijbiomac.2019.07.103
Ghosh, S., Ahmad, R., Zeyaullah, M., and Khare, S. K. (2021). Microbial nano-factories: synthesis and biomedical applications. Front. Chem. 9:626834. doi: 10.3389/fchem.2021.626834
Ghosh, S., Ahmad, R., Gautam, V. K., and Khare, S. K. (2018b). Cholesterol-oxidase-magnetic nanobioconjugates for the production of 4-cholesten-3-one and 4-cholesten-3, 7-dione. Bioresour. Technol. 254, 91–96. doi: 10.1016/j.biortech.2018.01.030
Gonzalez-Ballesteros, N., Prado-Lopez, S., Rodraguez-Gonzalez, J. B., Lastra, M., and Rodraguez-Arguelles, M. C. (2017). Green synthesis of gold nanoparticles using brown algae Cystoseira baccata: its activity in colon cancer cells. Coll. Surf. B Biointerfac. 153, 190–198. doi: 10.1016/j.colsurfb.2017.02.020
Graves, J. L., Tajkarimi, M., Cunningham, Q., Campbell, A., Nonga, H., Harrison, S. H., et al. (2015). Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front. Genet. 6:42.
Gu, Y.-P., Cui, R., Zhang, Z.-L., Xie, Z.-X., and Pang, D.-W. (2012). Ultrasmall near-infrared Ag2Se quantum dots with tunable fluorescence for in vivo imaging. J. Am. Chem. Soc. 134, 79–82.
Guilger-Casagrande, M., and De Lima, R. (2019). Synthesis of silver nanoparticles mediated by fungi: a review. Front. Bioeng. Biotechnol. 7:287. doi: 10.3389/fbioe.2019.00287
Gursoy, N. (2020). Fungus-mediated synthesis of silver nanoparticles (agnp) and inhibitory effect on Aspergillus spp. in combination with antifungal agent. Cumhur. Sci. J. 41, 311–318. doi: 10.17776/csj.653627
Harris, H. W., Sanchez-Andrea, I., Mclean, J. S., Salas, E. C., Tran, W., El-Naggar, M. Y., et al. (2018). Redox sensing within the genus Shewanella. Front. Microbiol. 8:2568.
He, S., Guo, Z., Zhang, Y., Zhang, S., Wang, J., and Gu, N. (2007). Biosynthesis of gold nanoparticles using the bacteria Rhodopseudomonas capsulata. Mater. Lett. 61, 3984–3987. doi: 10.1016/j.matlet.2007.01.018
Heidelberg, J. F., Paulsen, I. T., Nelson, K. E., Gaidos, E. J., Nelson, W. C., Read, T. D., et al. (2002). Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20, 1118–1123.
Hernandez-Garcia, A., Werten, M. W. T., Stuart, M. C., De Wolf, F. A., and De Vries, R. (2012). Coating of single DNA molecules by genetically engineered protein diblock copolymers. Small 8, 3491–3501. doi: 10.1002/smll.201200939
Hosseini, M. R., and Sarvi, M. N. (2015). Recent achievements in the microbial synthesis of semiconductor metal sulfide nanoparticles. Mater. Sci. Semiconduc. Proces. 40, 293–301. doi: 10.1016/j.mssp.2015.06.003
Hunter, G. K. (1996). Interfacial aspects of biomineralization. Curr. Opin. Solid State Mater. Sci. 1, 430–435. doi: 10.1016/s1359-0286(96)80036-2
Huq, M. (2020). Green synthesis of silver nanoparticles using Pseudoduganella eburnea MAHUQ-39 and their antimicrobial mechanisms investigation against drug resistant human pathogens. Int. J. Mol. Sci. 21:1510. doi: 10.3390/ijms21041510
Iravani, S. (2014). Bacteria in nanoparticle synthesis: current status and future prospects. Int. Schol. Res. Notic. 2014:1. doi: 10.1155/2014/359316
Jacob, J. M., Ravindran, R., Narayanan, M., Samuel, S. M., Pugazhendhi, A., and Kumar, G. (2020). Microalgae: a prospective low cost green alternative for nanoparticle synthesis. Curr. Opin. Environ. Sci. Health doi: 10.1016/j.coesh.2019.12.005
Jafari, M., Rokhbakhsh-Zamin, F., Shakibaie, M., Moshafi, M. H., Ameri, A., Rahimi, H. R., et al. (2018). Cytotoxic and antibacterial activities of biologically synthesized gold nanoparticles assisted by Micrococcus yunnanensis strain J2. Biocatalys. Agricult. Biotechnol. 15, 245–253. doi: 10.1016/j.bcab.2018.06.014
Jayaseelan, C., Rahuman, A. A., Kirthi, A. V., Marimuthu, S., Santhoshkumar, T., Bagavan, A., et al. (2012). Novel microbial route to synthesize ZnO nanoparticles using Aeromonas hydrophila and their activity against pathogenic bacteria and fungi. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 90, 78–84. doi: 10.1016/j.saa.2012.01.006
Johansson, E., Lundquist, A., Zuo, S., and Edwards, K. (2007). Nanosized bilayer disks: attractive model membranes for drug partition studies. Biochim. Biophys. Acta (BBA) Biomembr. 1768, 1518–1525. doi: 10.1016/j.bbamem.2007.03.006
Kalpana, V. N., Kataru, B. A. S., Sravani, N., Vigneshwari, T., Panneerselvam, A., and Rajeswari, V. D. (2018). Biosynthesis of zinc oxide nanoparticles using culture filtrates of Aspergillus niger: antimicrobial textiles and dye degradation studies. OpenNano 3, 48–55. doi: 10.1016/j.onano.2018.06.001
Kang, S. H., Bozhilov, K. N., Myung, N. V., Mulchandani, A., and Chen, W. (2008). Microbial synthesis of CdS nanocrystals in genetically engineered E. coli. Angew. Chem. 120, 5264–5267. doi: 10.1002/ange.200705806
Khalil, N. M., Abd El-Ghany, M. N., and Rodriguez-Couto, S. (2019). Antifungal and anti-mycotoxin efficacy of biogenic silver nanoparticles produced by Fusarium chlamydosporum and Penicillium chrysogenum at non-cytotoxic doses. Chemosphere 218, 477–486. doi: 10.1016/j.chemosphere.2018.11.129
Khatoon, N., Ahmad, R., and Sardar, M. (2015). Robust and fluorescent silver nanoparticles using Artemisia annua: biosynthesis, characterization and antibacterial activity. Biochem. Eng. J. 102, 91–97. doi: 10.1016/j.bej.2015.02.019
Kim, D.-H., Kanaly, R. A., and Hur, H.-G. (2012). Biological accumulation of tellurium nanorod structures via reduction of tellurite by Shewanella oneidensis MR-1. Bioresour. Technol. 125, 127–131. doi: 10.1016/j.biortech.2012.08.129
Kim, D.-H., Kim, M.-G., Jiang, S., Lee, J.-H., and Hur, H.-G. (2013). Promoted reduction of tellurite and formation of extracellular tellurium nanorods by concerted reaction between iron and Shewanella oneidensis MR-1. Environ. Sci. Technol. 47, 8709–8715.
Kim, T.-Y., Kim, M. G., Lee, J.-H., and Hur, H.-G. (2018). Biosynthesis of nanomaterials by Shewanella species for application in lithium ion batteries. Front. Microbiol. 9:2817.
Kimber, R. L., Lewis, E. A., Parmeggiani, F., Smith, K., Bagshaw, H., Starborg, T., et al. (2018). Biosynthesis and characterization of copper nanoparticles using Shewanella oneidensis: application for click chemistry. Small 14:1703145. doi: 10.1002/smll.201703145
Klaus, T., Joerger, R., Olsson, E., and Granqvist, C.-G. R. (1999). Silver-based crystalline nanoparticles, microbially fabricated. Proc. Natl. Acad. Sci. U.S.A. 96, 13611–13614. doi: 10.1073/pnas.96.24.13611
Krumov, N., Oder, S., Perner-Nochta, I., Angelov, A., and Posten, C. (2007). Accumulation of CdS nanoparticles by yeasts in a fed-batch bioprocess. J. Biotechnol. 132, 481–486. doi: 10.1016/j.jbiotec.2007.08.016
Kumar, V., Sowmya, B., Geetha, R., Karpagambigai, S., Rajeshkumar, S., and Lakshmi, T. (2019). Preparation of yeast mediated semiconductor nanoparticles by Candida albicans and its bactericidal potential against Salmonella typhi and Staphylococcus aureus. Int. J. Res. Pharmac. Sci. 10, 861–864. doi: 10.26452/ijrps.v10i2.262
Kumaresan, M., Anand, K. V., Govindaraju, K., Tamilselvan, S., and Kumar, V. G. (2018). Seaweed Sargassum wightii mediated preparation of zirconia (ZrO2) nanoparticles and their antibacterial activity against gram positive and gram negative bacteria. Microb. Pathog. 124, 311–315. doi: 10.1016/j.micpath.2018.08.060
Kumari, Y., Kaur, G., Kumar, R., Singh, S. K., Gulati, M., Khursheed, R., et al. (2019). Gold nanoparticles: new routes across old boundaries. Adv. Coll. Interface Sci. 274:102037. doi: 10.1016/j.cis.2019.102037
Li, J. F., Rupa, E. J., Hurh, J., Huo, Y., Chen, L., Han, Y., et al. (2019). Cordyceps militaris fungus mediated Zinc Oxide nanoparticles for the photocatalytic degradation of Methylene blue dye. Optik 183, 691–697. doi: 10.1016/j.ijleo.2019.02.081
Li, X., and Krumholz, L. R. (2007). Regulation of arsenate resistance in Desulfovibrio desulfuricans G20 by an arsRBCC operon and an arsC gene. J. Bacteriol. 189, 3705–3711. doi: 10.1128/jb.01913-06
Liu, B., and Liu, J. (2017). Surface modification of nanozymes. Nano Res. 10, 1125–1148. doi: 10.1007/s12274-017-1426-5
Liu, J., Zhang, X., Yu, M., Li, S., and Zhang, J. (2012). Photoinduced silver nanoparticles/nanorings on plasmid DNA scaffolds. Small 8, 310–316. doi: 10.1002/smll.201101423
Lok, C.-N., Ho, C.-M., Chen, R., He, Q.-Y., Yu, W.-Y., Sun, H., et al. (2006). Proteomic analysis of the mode of antibacterial action of silver nanoparticles. J. Proteom. Res. 5, 916–924. doi: 10.1021/pr0504079
Lower, B. H., and Bazylinski, D. A. (2013). The bacterial magnetosome: a unique prokaryotic organelle. J. Mol. Microbiol. Biotechnol. 23, 63–80. doi: 10.1159/000346543
Luo, C.-H., Shanmugam, V., and Yeh, C.-S. (2015). Nanoparticle biosynthesis using unicellular and subcellular supports. NPG Asia Mater. 7:e209. doi: 10.1038/am.2015.90
Lv, Q., Zhang, B., Xing, X., Zhao, Y., Cai, R., Wang, W., et al. (2018). Biosynthesis of copper nanoparticles using Shewanella loihica PV-4 with antibacterial activity: novel approach and mechanisms investigation. J. Hazard. Mater. 347, 141–149. doi: 10.1016/j.jhazmat.2017.12.070
Macaskie, L. E., Bolivar, J. G., Mikheenko, I., Orozco, R. L., Sharma, S., Banerjee, D., et al. (2019). Synthesis of Pd/Ru bimetallic nanoparticles by Escherichia coli and potential as a catalyst for upgrading 5-hydroxymethyl furfural into liquid fuel precursors. Front. Microbiol. 10:1276.
Madl, A. K., Plummer, L. E., Carosino, C., and Pinkerton, K. E. (2014). Nanoparticles, lung injury, and the role of oxidant stress. Annu. Rev. Physiol. 76, 447–465. doi: 10.1146/annurev-physiol-030212-183735
Mann, S. (2001). Biomineralization: Principles and Concepts in Bioinorganic Materials Chemistry. Oxford: Oxford University Press.
Marsili, E., Baron, D. B., Shikhare, I. D., Coursolle, D., Gralnick, J. A., and Bond, D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 3968–3973. doi: 10.1073/pnas.0710525105
Matsunaga, T., and Takeyama, H. (1998). Biomagnetic nanoparticle formation and application. Supramol. Sci. 5, 391–394. doi: 10.1016/s0968-5677(98)00037-6
Mazumder, J. A., Ahmad, R., and Sardar, M. (2016). Reusable magnetic nanobiocatalyst for synthesis of silver and gold nanoparticles. Int. J. Biol. Macromol. 93, 66–74. doi: 10.1016/j.ijbiomac.2016.08.073
McQuillan, J. S., Groenaga Infante, H., Stokes, E., and Shaw, A. M. (2012). Silver nanoparticle enhanced silver ion stress response in Escherichia coli K12. Nanotoxicology 6, 857–866. doi: 10.3109/17435390.2011.626532
Meenakshi, S. (2020). Bacterial cell–a bioreactor for the synthesis of nanoparticles. Chettinad Health City Med. J. 9, 130–136.
Menon, S., Rajeshkumar, S., and Kumar, V. (2017). A review on biogenic synthesis of gold nanoparticles, characterization, and its applications. Resour. Effic. Technol. 3, 516–527. doi: 10.1016/j.reffit.2017.08.002
Mikheenko, I. P., Rousset, M., Dementin, S., and Macaskie, L. E. (2008). Bioaccumulation of palladium by Desulfovibrio fructosivorans wild-type and hydrogenase-deficient strains. Appl. Environ. Microbiol. 74, 6144–6146. doi: 10.1128/aem.02538-07
Mishra, A., Ahmad, R., and Sardar, M. (2015). Biosynthesized iron oxide nanoparticles mimicking peroxidase activity: application for biocatalysis and biosensing. J. Nanoeng. Nanomanuf. 5, 37–42. doi: 10.1166/jnan.2015.1220
Mishra, A., Ahmad, R., Perwez, M., and Sardar, M. (2016). Reusable green synthesized biomimetic magnetic nanoparticles for glucose and H2O2 detection. BioNanoScience 6, 93–102. doi: 10.1007/s12668-016-0197-x
Mishra, A., Ahmad, R., Singh, V., Gupta, M. N., and Sardar, M. (2013a). Preparation, characterization and biocatalytic activity of a nanoconjugate of alpha amylase and silver nanoparticles. J. Nanosci. Nanotechnol. 13, 5028–5033. doi: 10.1166/jnn.2013.7593
Mishra, M., Paliwal, J. S., Singh, S. K., Selvarajan, E., Subathradevi, C., and Mohanasrinivasan, V. (2013b). Studies on the inhibitory activity of biologically synthesized and characterized zinc oxide nanoparticles using lactobacillus sporogens against Staphylococcus aureus. J. Pure Appl. Microbiol. 7, 1–6. doi: 10.1007/978-3-319-23534-9_1
Mohd Yusof, H., Rahman, A., Mohamad, R., and Zaidan, U. H. (2020). Microbial mediated synthesis of silver nanoparticles by Lactobacillus Plantarum TA4 and its antibacterial and antioxidant activity. Appl. Sci. 10:6973. doi: 10.3390/app10196973
Molnar, Z., Bodai, V., Szakacs, G., Erdelyi, B., Fogarassy, Z., Safran, G., et al. (2018). Green synthesis of gold nanoparticles by thermophilic filamentous fungi. Sci. Rep. 8, 1–12. doi: 10.1016/j.cplett.2015.12.019
Morones, J. R., Elechiguerra, J. L., Camacho, A., Holt, K., Kouri, J. B., Ramã-Rez, J. T., et al. (2005). The bactericidal effect of silver nanoparticles. Nanotechnology 16, 2346.
Mukherjee, P., Ahmad, A., Mandal, D., Senapati, S., Sainkar, S. R., Khan, M. I., et al. (2001). Fungus-mediated synthesis of silver nanoparticles and their immobilization in the mycelial matrix: a novel biological approach to nanoparticle synthesis. Nano Lett. 1, 515–519. doi: 10.1021/nl0155274
Munawer, U., Raghavendra, V. B., Ningaraju, S., Krishna, K. L., Ghosh, A. R., Melappa, G., et al. (2020). Biofabrication of gold nanoparticles mediated by the endophytic Cladosporium species: Photodegradation, in vitro anticancer activity and in vivo antitumor studies. Int. J. Pharmac. 588:119729. doi: 10.1016/j.ijpharm.2020.119729
Nancharaiah, Y. V., Mohan, S. V., and Lens, P. N. L. (2016). Biological and bioelectrochemical recovery of critical and scarce metals. Trends Biotechnol. 34, 137–155. doi: 10.1016/j.tibtech.2015.11.003
Nazari, N., and Kashi, F. J. (2021). A novel microbial synthesis of silver nanoparticles: its bioactivity, Ag/Ca-Alg beads as an effective catalyst for decolorization Disperse Blue 183 from textile industry effluent. Separat. Purificat. Technol. 259:118117. doi: 10.1016/j.seppur.2020.118117
Ngoepe, N. M., Hato, M. J., Modibane, K. D., and Hintsho-Mbita, N. C. (2020). Biogenic synthesis of metal oxide nanoparticle semiconductors for wastewater treatment. Photocatalys. Adv. Oxidat. Proces. Wastew. Treat. 20, 1–31. doi: 10.1002/9781119631422.ch1
Niazi, J. H., and Gu, M. B. (2009). Toxicity of metallic nanoparticles in microorganisms-a review. Atmosph. Biol. Environ. Monitor. 9, 193–206. doi: 10.1007/978-1-4020-9674-7_12
Nordmeier, A., Merwin, A., Roeper, D. F., and Chidambaram, D. (2018). Microbial synthesis of metallic molybdenum nanoparticles. Chemosphere 203, 521–525. doi: 10.1016/j.chemosphere.2018.02.079
Nudelman, H., Lee, Y.-Z., Hung, Y.-L., Kolusheva, S., Upcher, A., Chen, Y.-C., et al. (2018). Understanding the biomineralization role of Magnetite-Interacting Components (MICs) from magnetotactic bacteria. Front. Microbiol. 9:2480.
Ordenes-Aenishanslins, N. A., Saona, L. A., Duran-Toro, V. M., Monrãs, J. P., Bravo, D. M., and Perez-Donoso, J. M. (2014). Use of titanium dioxide nanoparticles biosynthesized by Bacillus mycoides in quantum dot sensitized solar cells. Microb. Cell Fact. 13:90.
Ovais, M., Khalil, A. T., Ayaz, M., Ahmad, I., Nethi, S. K., and Mukherjee, S. (2018). Biosynthesis of metal nanoparticles via microbial enzymes: a mechanistic approach. Int. J. Mol. Sci. 19:4100. doi: 10.3390/ijms19124100
Parveen, K., Banse, V., and Ledwani, L. (2016). “Green synthesis of nanoparticles: their advantages and disadvantages,” in AIP Conference Proceedings, (College Park, MD: AIP Publishing LLC), 020048.
Perwez, M., Ahmad, R., and Sardar, M. (2017). A reusable multipurpose magnetic nanobiocatalyst for industrial applications. Int. J. Biol. Macromol. 103, 16–24. doi: 10.1016/j.ijbiomac.2017.05.029
Piacenza, E., Presentato, A., Ambrosi, E., Speghini, A., Turner, R. J., Vallini, G., et al. (2018). Physical-chemical properties of biogenic selenium nanostructures produced by Stenotrophomonas maltophilia SeITE02 and Ochrobactrum sp. MPV1. Front. Microbiol. 9:3178.
Prozorov, T., Palo, P., Wang, L., Nilsen-Hamilton, M., Jones, D., Orr, D., et al. (2007). Cobalt ferrite nanocrystals: out-performing magnetotactic bacteria. ACS Nano 1, 228–233. doi: 10.1021/nn700194h
Pytlik, N., Kaden, J., Finger, M., Naumann, J., Wanke, S., Machill, S., et al. (2017). Biological synthesis of gold nanoparticles by the diatom Stephanopyxis turris and in vivo SERS analyses. Algal Res. 28, 9–15. doi: 10.1016/j.algal.2017.10.004
Rajakumar, G., Rahuman, A. A., Roopan, S. M., Khanna, V. G., Elango, G., Kamaraj, C., et al. (2012). Fungus-mediated biosynthesis and characterization of TiO2 nanoparticles and their activity against pathogenic bacteria. Spectrochim. Acta Part A Mol. Biomol. Spectros. 91, 23–29. doi: 10.1016/j.saa.2012.01.011
Ramesh, A., Sundari, M. T., and Thirugnanam, P. E. (2015). Microbial molecular mechanisms in biosynthesis of nanoparticles. Bio Nanopartic. Biosynth. Sustain. Biotechnol. Implicat. 15, 53–81. doi: 10.1002/9781118677629.ch3
Randall, C. P., Gupta, A., Jackson, N., Busse, D., and O’neill, A. J. (2015). Silver resistance in gram-negative bacteria: a dissection of endogenous and exogenous mechanisms. J. Antimicrob. Chemother. 70, 1037–1046.
Ranjitha, V. R., and Rai, V. R. (2017). Actinomycetes mediated synthesis of gold nanoparticles from the culture supernatant of Streptomyces griseoruber with special reference to catalytic activity. 3 Biotech 7:299.
Reddy, A. V. B., Yusop, Z., Jaafar, J., Reddy, Y. V. M., Aris, A. B., Majid, Z. A., et al. (2016). Recent progress on Fe-based nanoparticles: synthesis, properties, characterization and environmental applications. J. Environ. Chem. Eng. 4, 3537–3553. doi: 10.1016/j.jece.2016.07.035
Robertson, J. D., Rizzello, L., Avila-Olias, M., Gaitzsch, J., Contini, C., Magon, M. S., et al. (2016). Purification of nanoparticles by size and shape. Sci. Rep. 6, 1–9.
Roosa, S., Wattiez, R., Prygiel, E., Lesven, L., Billon, G., and Gillan, D. C. (2014). Bacterial metal resistance genes and metal bioavailability in contaminated sediments. Environ. Pollut. 189, 143–151. doi: 10.1016/j.envpol.2014.02.031
Sadaf, A., Ahmad, R., Ghorbal, A., Elfalleh, W., and Khare, S. K. (2020). Synthesis of cost-effective magnetic nano-biocomposites mimicking peroxidase activity for remediation of dyes. Environ. Sci. Pollut. Res. 27, 27211–27220. doi: 10.1007/s11356-019-05270-3
Saeed, S., Iqbal, A., and Ashraf, M. A. (2020). Bacterial-mediated synthesis of silver nanoparticles and their significant effect against pathogens. Environ. Sci. Pollut. Res. 2, 1–10.
Saklani, V., Suman, J. V. K., and Jain, K. (2012). Microbial synthesis of silver nanoparticles: a review. J. Biotechnol. Biomaterial. 12:13.
Salas-Orozco, M., Nino-Martinez, N., Martinez-Castanon, G.-A., Mendez, F. T., Jasso, M. E. C., and Ruiz, F. (2019). Mechanisms of resistance to silver nanoparticles in endodontic bacteria: a literature review. J. Nanomater. 2019, 1–12. doi: 10.1155/2019/7630316
Salvadori, M. R., Ando, R. A., Do Nascimento, C. A. O., and Correa, B. (2014). Intracellular biosynthesis and removal of copper nanoparticles by dead biomass of yeast isolated from the wastewater of a mine in the Brazilian Amazonia. PLoS One 9:e87968. doi: 10.1371/journal.pone.0087968
Saravanakumar, K., Shanmugam, S., Varukattu, N. B., Mubarakali, D., Kathiresan, K., and Wang, M.-H. (2019). Biosynthesis and characterization of copper oxide nanoparticles from indigenous fungi and its effect of photothermolysis on human lung carcinoma. J. Photochem. Photobiol. B Biol. 190, 103–109. doi: 10.1016/j.jphotobiol.2018.11.017
Saravanan, A., Kumar, P. S., Karishma, S., Vo, D.-V. N., Jeevanantham, S., Yaashikaa, P. R., et al. (2020). A review on biosynthesis of metal nanoparticles and its environmental applications. Chemosphere 2020: 128580. doi: 10.1016/j.chemosphere.2020.128580
Sardar, M., Mishra, A., and Ahmad, R. (2014). “Biosynthesis of metal nanoparticles and their applications,” in Biosensors and Nanotechnology, eds A. Tiwari and A. P. F. Turner (Beverly, MA: Scrivener Publishing), 239–266. doi: 10.1002/9781118773826.ch8
Sardar, M., Perwez, M., Ahmad, R., Mukherjee, J., and Gupta, M. N. (2018). “Immobilization of enzymes on magnetic nanoparticles,” in Encyclopedia of Nanoscience and Nanotechnology, ed. H. S. Nalwa (Los Angeles, CA: American Scientific Publishers).
Satzer, P., Wellhoefer, M., and Jungbauer, A. (2014). Continuous separation of protein loaded nanoparticles by simulated moving bed chromatography. J. Chromatogr. A 1349, 44–49. doi: 10.1016/j.chroma.2014.04.093
Sayadi, M. H., Salmani, N., Heidari, A., and Rezaei, M. R. (2018). Bio-synthesis of palladium nanoparticle using Spirulina platensis alga extract and its application as adsorbent. Surfac. Interfac. 10, 136–143. doi: 10.1016/j.surfin.2018.01.002
Seetharaman, P. K., Chandrasekaran, R., Gnanasekar, S., Chandrakasan, G., Gupta, M., Manikandan, D. B., et al. (2018). Antimicrobial and larvicidal activity of eco-friendly silver nanoparticles synthesized from endophytic fungi Phomopsis liquidambaris. Biocatalys. Agricult. Biotechnol. 16, 22–30. doi: 10.1016/j.bcab.2018.07.006
Senapati, S., Syed, A., Moeez, S., Kumar, A., and Ahmad, A. (2012). Intracellular synthesis of gold nanoparticles using alga Tetraselmis kochinensis. Mater. Lett. 79, 116–118. doi: 10.1016/j.matlet.2012.04.009
Shaker, M. A., and Shaaban, M. I. (2017). Synthesis of silver nanoparticles with antimicrobial and anti-adherence activities against multidrug-resistant isolates from Acinetobacter baumannii. J. Taib. Univ. Med. Sci. 12, 291–297. doi: 10.1016/j.jtumed.2017.02.008
Sharma, M., Behl, K., Nigam, S., and Joshi, M. (2018). TiO2-GO nanocomposite for photocatalysis and environmental applications: a green synthesis approach. Vacuum 156, 434–439. doi: 10.1016/j.vacuum.2018.08.009
Shen, W., Qu, Y., Pei, X., Li, S., You, S., Wang, J., et al. (2017). Catalytic reduction of 4-nitrophenol using gold nanoparticles biosynthesized by cell-free extracts of Aspergillus sp. WL-Au. J. Hazard. Mater. 321, 299–306. doi: 10.1016/j.jhazmat.2016.07.051
Sinha, A., and Khare, S. K. (2011). Mercury bioaccumulation and simultaneous nanoparticle synthesis by Enterobacter sp. cells. Bioresour. Technol. 102, 4281–4284. doi: 10.1016/j.biortech.2010.12.040
Slavin, Y. N., Asnis, J., Hafeli, U. O., and Bach, H. (2017). Metal nanoparticles: understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 15, 1–20.
Sobolev, D., and Begonia, M. (2008). Effects of heavy metal contamination upon soil microbes: lead-induced changes in general and denitrifying microbial communities as evidenced by molecular markers. Int. J. Environ. Res. Public Health 5, 450–456. doi: 10.3390/ijerph5050450
Srinath, B. S., Namratha, K., and Byrappa, K. (2018). Eco-friendly synthesis of gold nanoparticles by Bacillus subtilis and their environmental applications. Adv. Sci. Lett. 24, 5942–5946. doi: 10.1166/asl.2018.12224
Subramaniyan, S. A., Sheet, S., Vinothkannan, M., Yoo, D. J., Lee, Y. S., Belal, S. A., et al. (2018). One-pot facile synthesis of Pt nanoparticles using cultural filtrate of microgravity simulated grown P. chrysogenum and their activity on bacteria and cancer cells. J. Nanosci. Nanotechnol. 18, 3110–3125. doi: 10.1166/jnn.2018.14661
Sun, J.-B., Wang, Z.-L., Duan, J.-H., Ren, J., Yang, X.-D., Dai, S.-L., et al. (2009). Targeted distribution of bacterial magnetosomes isolated from Magnetospirillum gryphiswaldense MSR-1 in healthy Sprague-Dawley rats. J. Nanosci. Nanotechnol. 9, 1881–1885. doi: 10.1166/jnn.2009.410
Suresh, A. K., Pelletier, D. A., Wang, W., Moon, J.-W., Gu, B., Mortensen, N. P., et al. (2010). Silver nanocrystallites: biofabrication using Shewanella oneidensis, and an evaluation of their comparative toxicity on gram-negative and gram-positive bacteria. Environ. Sci. Technol. 44, 5210–5215. doi: 10.1021/es903684r
Thill, A., Zeyons, O., Spalla, O., Chauvat, F., Rose, J., Auffan, M., et al. (2006). Cytotoxicity of CeO2 nanoparticles for Escherichia coli Physico-chemical insight of the cytotoxicity mechanism. Environ. Sci. Technol. 40, 6151–6156.
Vargas, G., Cypriano, J., Correa, T., Leao, P., Bazylinski, D. A., and Abreu, F. (2018). Applications of magnetotactic bacteria, magnetosomes and magnetosome crystals in biotechnology and nanotechnology: mini-review. Molecules 23:2438. doi: 10.3390/molecules23102438
Wang, L., and Nilsen-Hamilton, M. (2013). Biomineralization proteins: from vertebrates to bacteria. Front. Biol. 8:234–246. doi: 10.1007/s11515-012-1205-3
Wang, L., Liu, X., Lee, D.-J., Tay, J.-H., Zhang, Y., Wan, C.-L., et al. (2018). Recent advances on biosorption by aerobic granular sludge. J. Hazard. Mater. 357, 253–270. doi: 10.1016/j.jhazmat.2018.06.010
Wang, W., Zhang, B., Liu, Q., Du, P., Liu, W., and He, Z. (2018). Biosynthesis of palladium nanoparticles using Shewanella loihica PV-4 for excellent catalytic reduction of chromium (VI). Environ. Sci. Nano 5, 730–739. doi: 10.1039/c7en01167a
Wei, G.-T., and Liu, F.-K. (1999). Separation of nanometer gold particles by size exclusion chromatography. J. Chromatogr. A 836, 253–260. doi: 10.1016/s0021-9673(99)00069-2
Weiner, S., and Dove, P. M. (2003). An overview of biomineralization processes and the problem of the vital effect. Rev. Mineral. Geochem. 54, 1–29. doi: 10.1515/9781501509346-006
Woerner, M., Lioubashevski, O., Basel, M. T., Niebler, S., Gogritchiani, E., Egner, N., et al. (2007). Characterization of nanostructured surfaces generated by reconstitution of the porin MspA from Mycobacterium smegmatis. Small 3, 1084–1097. doi: 10.1002/smll.200600559
Xiong, L., Zhang, X., Huang, Y.-X., Liu, W.-J., Chen, Y.-L., Yu, S.-S., et al. (2018). Biogenic synthesis of Pd-based nanoparticles with enhanced catalytic activity. ACS Appl. Nano Mater. 1, 1467–1475. doi: 10.1021/acsanm.7b00322
Yamamoto, D., Taoka, A., Uchihashi, T., Sasaki, H., Watanabe, H., Ando, T., et al. (2010). Visualization and structural analysis of the bacterial magnetic organelle magnetosome using atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 9382–9387. doi: 10.1073/pnas.1001870107
Yang, Q. E., Agouri, S. R., Tyrrell, J. M., and Walsh, T. R. (2018). Heavy metal resistance genes are associated with blaNDM-1-and blaCTX-M-15-carrying Enterobacteriaceae. Antimicrob. Agents Chemother. 62:e02642-17.
Zhang, X., Qu, Y., Shen, W., Wang, J., Li, H., Zhang, Z., et al. (2016). Biogenic synthesis of gold nanoparticles by yeast Magnusiomyces ingens LH-F1 for catalytic reduction of nitrophenols. Coll. Surfac. A Physicochem. Eng. Aspects 497, 280–285. doi: 10.1016/j.colsurfa.2016.02.033
Zhang, Y., Ni, Q., Xu, C., Wan, B., Geng, Y., Zheng, G., et al. (2018). Smart bacterial magnetic nanoparticles for tumor-targeting magnetic resonance imaging of HER2-positive Breast cancers. ACS Appl. Mater. Interfac. 11, 3654–3665. doi: 10.1021/acsami.8b15838
Zhang, Z., Chen, J., Yang, Q., Lan, K., Yan, Z., and Chen, J. (2018). Eco-friendly intracellular microalgae synthesis of fluorescent CdSe QDs as a sensitive nanoprobe for determination of imatinib. Sen. Actuat. B Chem. 263, 625–633. doi: 10.1016/j.snb.2018.02.169
Keywords: nanoparticles, synthesis mechanism, microbes, metal nanotization, biomaterials, therapeutic
Citation: Ghosh S, Ahmad R, Banerjee K, AlAjmi MF and Rahman S (2021) Mechanistic Aspects of Microbe-Mediated Nanoparticle Synthesis. Front. Microbiol. 12:638068. doi: 10.3389/fmicb.2021.638068
Received: 05 December 2020; Accepted: 25 March 2021;
Published: 05 May 2021.
Edited by:
Sougata Ghosh, RK University, IndiaReviewed by:
Ajay Kumar, Agricultural Research Organization (ARO), IsraelCopyright © 2021 Ghosh, Ahmad, Banerjee, AlAjmi and Rahman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shubhrima Ghosh, c2h1YmhyaW1hLmdob3NoQGdtYWlsLmNvbQ==; Razi Ahmad, cmF6aS5qbWlAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.