
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 06 April 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.630438
This article is part of the Research TopicMicrobiota: A Consequential Third Wheel in the Mosquito-Pathogen RelationshipView all 15 articles
 Paolo Gabrieli1†
Paolo Gabrieli1† Silvia Caccia2,3†
Silvia Caccia2,3† Ilaria Varotto-Boccazzi1
Ilaria Varotto-Boccazzi1 Irene Arnoldi4
Irene Arnoldi4 Giulia Barbieri4
Giulia Barbieri4 Francesco Comandatore5
Francesco Comandatore5 Sara Epis1*
Sara Epis1*In mosquitoes, the interaction between the gut microbiota, the immune system, and the pathogens that these insects transmit to humans and animals is regarded as a key component toward the development of control strategies, aimed at reducing the burden of severe diseases, such as malaria and dengue fever. Indeed, different microorganisms from the mosquito microbiota have been investigated for their ability to affect important traits of the biology of the host insect, related with its survival, development and reproduction. Furthermore, some microorganisms have been shown to modulate the immune response of mosquito females, significantly shaping their vector competence. Here, we will review current knowledge in this field, focusing on i) the complex interaction between the intestinal microbiota and mosquito females defenses, both in the gut and at humoral level; ii) how knowledge on these issues contributes to the development of novel and targeted strategies for the control of mosquito-borne diseases such as the use of paratransgenesis or taking advantage of the relationship between Wolbachia and mosquito hosts. We conclude by providing a brief overview of available knowledge on microbiota-immune system interplay in major insect vectors.
Bloodsucking insects are important vectors of pathogens that cause a variety of severe diseases worldwide, with a strong impact on human and animal health (Lee et al., 2018; Boulanger et al., 2019). Concern about vector-borne diseases has increased in the last decade, also because of the geographical spread of several insect vectors, caused by intense trade and climate changes (de La Rocque et al., 2011; Caminade et al., 2019).
In particular, mosquitoes are major vectors of pathogens, including protozoa (e.g., Plasmodium spp. which causes malaria), nematodes (e.g., filariae), and viruses (e.g., dengue, chikungunya, West Nile, and Zika). Over 3,500 species of mosquitoes have been described, but only a limited number of them can function as disease vectors, and varying levels of specificity are observed for different types of pathogens. Overall, mosquito-borne pathogens are estimated to cause around 500,000 deaths each year, with billions of people exposed to the risk of contracting these infectious agents1.
So far, the most effective preventive strategies to limit the impact of mosquito-borne diseases have focused on controlling mosquito vector populations heavily relying on the use of insecticides and personal preventive measures, such as insecticide-treated nets (ITN) (Wangdi et al., 2018; Carnevale and Gay, 2019). For example, massive use of LLINs (long-lasting insecticidal nets, ITN with longer duration of effectiveness due to the incorporation of the insecticide into fibers during the manufacturing process) has greatly contributed to combat malaria (Carnevale and Gay, 2019). However, the efficacy of these control measures is hampered by the selection and spread of resistance (Hemingway, 2018), which is a complex phenomenon that accounts for modifications of multiple biochemical processes in mosquitoes (Hemingway, 2018; Ingham et al., 2020) or, also, for alterations of the mosquito biting behavior (e.g., shifts from an indoor- to an out-door host-seeking behavior) (Moiroux et al., 2012; Kreppel et al., 2020; Perugini et al., 2020). The massive use of insecticides raises also concerns, in relation to the impact on non-target species and the environment (Mansouri et al., 2017). Furthermore, the spread of invasive mosquito species to new areas requires constant monitoring and availability of new and alternative control strategies, considering that the control methodologies applied in the area of origin of a given species are not always suitable to be used in different countries and environmental conditions (Bellini et al., 2020).
The improvement of integrated vector control strategies, and in particular the development of novel environment-friendly insecticides and control approaches, is therefore urgent. In this context, insect microbiota already inspired the development of innovative control tools, such as the use of “symbiotic control” to target insect pests and vectors.
In this review we will focus our attention on the interactions between the microbiota and the vector host, with particular emphasis on the immune response. We will describe how this interaction shapes, at least partially, the vectorial capacity of mosquitoes; we will then describe the microbiota- and symbiont-based strategies that are used to control mosquitoes and mosquito-borne diseases, or that have been proposed but not yet applied. Finally, we will provide an overview of the current knowledge about the interaction between microorganisms and the immune system in other bloodsucking insect vectors.
The vector competence of mosquitoes is a biological trait that is influenced by multiple factors (Azar and Weaver, 2019). It is shaped, in the first instance, by the genetic variability of the immune effectors of the mosquito; for example, thioester-containing protein 1 gene have multiple alleles that determine differences in susceptibility of Anopheles mosquitoes to the malaria infection (Le et al., 2012). The genomic variants of vectored pathogens or parasites can also play a major role, such as the case of the E1-226V variant of chikungunya virus that is preferentially transmitted by Aedes albopictus (Schuffenecker et al., 2006). Lastly, vector competence in mosquitoes can be also affected by the composition of the microbiota (Boissière et al., 2012).
Microorganisms, indeed, colonize different organs and tissues in mosquitoes, including gut, salivary glands and reproductive tissues (Segata et al., 2016; Scolari et al., 2019; Gao H. et al., 2020). They influence many aspects of the mosquito biology, including reproduction, development, adult survival and, overall, immunity (Coon et al., 2014). The main sites where cellular and humoral components of adult mosquito immunity exert their functions against invaders are the hemocoel with the circulating hemolymph, that contains the immune cells called hemocytes (Hillyer, 2010, 2016; Raddi et al., 2020), and the gut, which receives the sugar and blood meals and that hosts a major component of the insect microbiota (gut-associated microbiota).
For the purpose of this review, we will focus our attention on how bacteria interact with the gut of adult female mosquitoes and shape the immune responses after a blood meal (summarized in Figure 1). Blood meal, indeed, causes a proliferation of midgut microbiota (Gusmão et al., 2010; Kumar et al., 2010; Oliveira et al., 2011; Barletta et al., 2017) that, for instance, peaks at around 30 h after meal in Anopheles gambiae (Kumar et al., 2010).
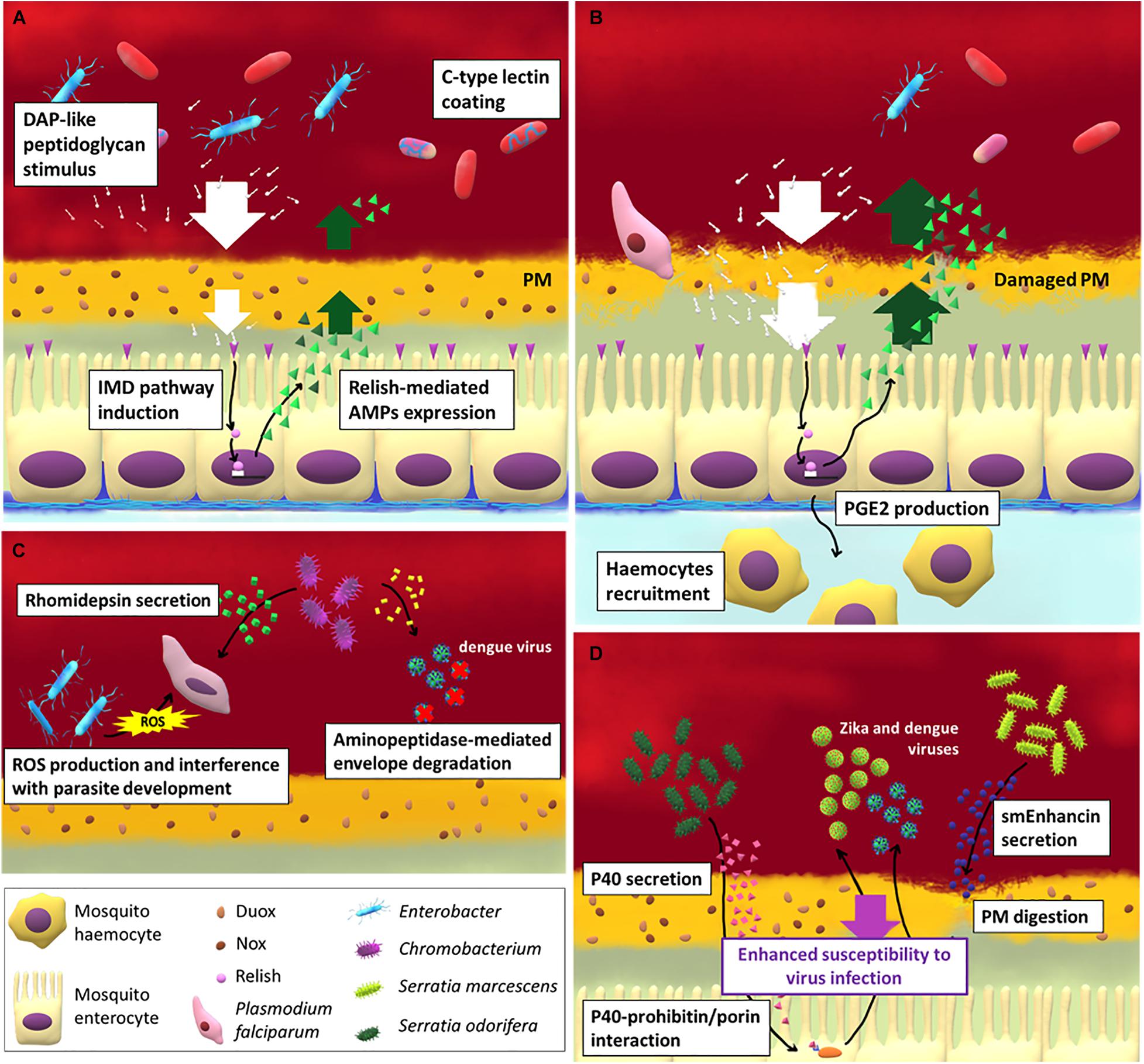
Figure 1. The interaction between gut immune response, microbiota and pathogens in mosquito females. (A) The strong increase of gut-associated microbial load after blood meal induces the activation of the IMD pathway in midgut epithelial cells and the release of antimicrobial peptides (AMPs). The contact between bacterial cells or bacterial-associated molecules (such as peptidoglycans) and epithelium is partially prevented by the peritrophic matrix (PM) which forms soon after a blood meal. Other mosquito-secreted molecules can be exploited by the bacteria as protection from AMPs, such as C-type lectins. When the PM integrity is impaired (B) by the action, for example, of the Plasmodium ookinetes, the IMD pathway is activated and hemocytes are recruited to the infection site at the base of the epithelium thanks to the release of prostaglandin E2 by midgut cells. Apart from activating vector immune response some bacterial species are able to directly limit (C) or to favor (D) pathogen and virus infection in mosquitoes.
Female mosquitoes acquire pathogens together with the blood meal and the microbes residing in the gut have a profound effect on the outcome of the infection (Cirimotich et al., 2011b; Dennison et al., 2014; Jupatanakul et al., 2014; Scolari et al., 2019).
For example, axenic An. gambiae mosquitoes are more susceptible to Plasmodium infection; conversely the co-feeding of a mixture of Escherichia coli, Staphylococcus aureus bacteria, and Plasmodium falciparum gametocytes decreases infection levels (Dong et al., 2009). Similarly, axenic Ae. aegypti have higher midgut dengue virus titers compared to normal septic mosquitoes (Xi et al., 2008) and some field-derived bacterial isolates affect dengue virus infection when introduced in axenic mosquitoes (Ramirez et al., 2012). Notably, the effect of microbiota on viral infection is specific and varies with the insect host and the virus: for example, it has been shown that axenic An. gambiae mosquitoes are less susceptible to o’nyong’nyong virus infection (Carissimo et al., 2015).
The protective role of the microbiota can be exerted by a specific class of microorganisms. It is the case of Enterobacteriaceae in Anopheles mosquitoes, which have a protective effect on Plasmodium infection (Cirimotich et al., 2011a; Boissière et al., 2012). In Ae. aegypti, different strains with different susceptibility to dengue infection harbor specific bacterial species that might be related to their vectorial capacity, with Pedobacter sp. and Janthinobacterium sp. identified only in resistant strains, while Bacillus sp. only in susceptible strains (Charan et al., 2013).
Physiological features and/or the genome variability of the mosquito vector can modulate vector competence in reason of their effect on the composition of gut bacteria community. The regulation of specific metabolic processes, as the branched chain amino acid degradation pathway, plays a role in the modulation of the microbial load of different Aedes aegypti strains (Short et al., 2017) that may in turn affect vector competence. Furthermore, genetic variation in immune genes encoding proteins with type III fibronectin domains (FN3D) in the gut correlates with interspecific variation of the load of Serratia marcescens, a common component of Anopheles gut Enterobacteriaceae (Stathopoulos et al., 2014). Indeed, silencing of three FN3D genes modulates S. marcescens load and alters the gut bacteria population favoring Enterobacteriaceae in Anopheles mosquitoes (Stathopoulos et al., 2014). This interaction, in turn, influences vector competence, since the abundance of Enterobacteriaceae in the mosquito midgut affect Plasmodium infection (Boissière et al., 2012).
The mosquito immune responses against infectious agents involves multiple pathways and effector molecules, which are summarized in Table 1.
The gut of mosquito females houses a wide spectrum of bacterial species, the most common of which are Gram-negative (Gendrin and Christophides, 2013; Scolari et al., 2019; Gao H. et al., 2020). Humoral responses against microbial pathogens have been deeply characterized in Drosophila and involve different pathways (Buchon et al., 2014; Mussabekova et al., 2017). Among them, the IMD pathway is conserved in mosquitoes (Christophides et al., 2002) and it appears to be functionally involved in antibacterial defense against both Gram-positive and Gram-negative bacteria (Meister et al., 2005; Cooper et al., 2009; Magalhaes et al., 2010; Barletta et al., 2017). In mosquito females, IMD pathway is activated in response to the proliferation of midgut microbiota that is triggered by the blood meal (Kumar et al., 2010; Barletta et al., 2017). The microbe-associated molecular pattern (MAMP) that triggers the activation of this pathway in Gram-negative bacteria is the diaminopimelic acid (DAP)-type peptidoglycan of the cell wall. In Drosophila, this molecule is recognized by two peptidoglycan recognition proteins (PGRP), i.e., the membrane-bound PGRP-LC in the anterior midgut and the intracellular PGRP-LE in the middle and posterior midgut (Kaneko et al., 2006; Buchon et al., 2014). Other pattern recognition proteins (PRRs) participate in the regulation of IMD pathway in a tissue specific manner: in the gut, it is positively regulated by PGRP-LA, while the amidases PGRP-LB and PGRP-SC, which cleave peptidoglycan into non-immunogenic fragments, negatively regulate the pathway (Zaidman-Rémy et al., 2006; Paredes et al., 2011; Gendrin et al., 2017). In mosquitoes, PGRP-LC is the main receptor that mediates immune response against Gram-positive and Gram-negative infections, with the isoform PGRP-LC3 recognized as key modulator of these responses at early stages of hemolymph colonization (Meister et al., 2009; Stathopoulos et al., 2014) and the isoform PGRP-LC1 having a main role in the midgut response (Rodgers et al., 2020). Similarly to Drosophila, PGRP-LC interacts with polymeric DAP-type peptidoglycan, while PGRP-LA and PGRP-LB positively and negatively regulate the pathway in Anopheles mosquitoes (Gendrin et al., 2017; Gao L. et al., 2020).
In Drosophila, the binding of the peptidoglycan ligand causes the dimerization of the receptor, activating an intracellular signaling cascade: the adaptor protein IMD is cleaved by the protease Dredd (Kim et al., 2014) and is rapidly ubiquitinated. This modification leads ultimately to the activation of the NF-κB transcription factor Relish, through the activity of Dredd and of the transforming growth factor β activated kinase-1 and the I-kappa B kinase complex (Paquette et al., 2010). Notably, the An. gambiae genome encodes two isoforms of the Relish homolog (i.e., REL-2); the short isoform, REL-2S, is involved in the response against Gram-negative bacteria, while the long isoform, REL-2F, against Gram-positives (Meister et al., 2005). It has been demonstrated that in Anopheles dirus REL-2F is involved in protection against both Gram-positive (with Lys-type peptidoglycan) and Gram-negative bacteria (with DAP-type peptidoglycan) (Khan et al., 2016). Relish, in turn, induces the expression of antimicrobial peptides (AMPs). These peptides have a highly conserved structure and they might exert their antimicrobial activity through peptide-lipid interaction or receptor-mediated recognition processes (Bulet et al., 1999). In mosquitoes, there are two classes of AMPs (defensins and cecropins) that have been found in many other insects, and one class, gambicins, that seems to be mosquito specific (Levashina, 2004).
Interestingly, it has been reported a direct interaction between PGRP-LD and gut-associated microbiota in Anopheles. Silencing of PGRP-LD, led to an over-activation of the immune response, leading to an over-expression of multiple AMP in An. stephensi prior blood feeding that causes a reduction of the bacterial load in the mosquito gut (Song et al., 2018).
A role of an immunomodulatory peroxidase (IMPer) and a dual oxidase (Duox) secreted by midgut cells in modulating gut-associated microbiota in Anopheles has also been described (Kajla et al., 2016) (see also section “The Interplay Between Physical Barriers Defenses in the Gut, Immune Responses, Microbiota and Implications for Vector Competence”). Indeed, when the peroxidase is silenced in Anopheles stephensi midgut, bacterial growth is significantly reduced by the overexpression of nitric oxide (NO) synthase gene (NOS), a final effector of the JAK/STAT pathway, while no significant recruitment of the classical immune pathways was observed (Kajla et al., 2016). Since NOS is a negative regulator of Plasmodium development (Oliveira et al., 2011), the authors suggested that the induction of the JAK/STAT pathway might be a strategy to modulate the vectorial capacity of Anopheles mosquitoes.
The expression of Duox is also regulated by a gut-membrane-associated protein, named Mesh, and the reduction of Duox activity lead to the increase of the microbiota load, suggesting that reactive oxygen species (ROS) might participate in controlling gut microbial homeostasis (Xiao et al., 2017). Notably, it has been also shown that blood meal-derived heme can decrease ROS levels in the mosquito midgut, allowing proliferation of bacteria (Oliveira et al., 2011).
The homeostatic balance governed by a tight control of both AMP transcripts and Duox expression is further confirmed by the effect of the mechanism exerted by the pathogenic fungus Beauveria bassiana: this fungus induces dysbiosis in the mosquito midgut by altering the expression of AMP transcripts and Duox with the secretion of the toxin oosporein, inducing bacterial growth, promoting the overgrowth of the opportunistic bacteria S. marcescens, which, once in the hemocoel, favors septicemia and thus the killing of mosquitoes (Wei et al., 2017).
On the other hand, the antimicrobial effect of AMPs produced by the mosquito against gut-associated microbiota is counteracted by multiple mechanisms: it has been demonstrated, for example, that the coating of bacteria with C-type lectins expressed in the mosquito midgut counteracts AMPs activity and favors gut microbiota homeostasis (Pang et al., 2016; Li et al., 2020).
The priming of the mosquito innate immune response by gut-associated microbiota can partially explain the effect of microbiota on pathogen virulence (Dong et al., 2009). In particular, some bacteria species are able to promote AMP genes expression in the gut, thus exerting a protective role against pathogens: this is the case of Proteus sp. in Ae. aegypti against dengue (Ramirez et al., 2012) and S. marcescens in An. stephensi against Plasmodium berghei (Bai et al., 2019).
An important immune role in the midgut of many insects is exerted by the peritrophic matrix (PM), a gel-like structure produced by midgut (Type I PM) or cardia region (Type II PM) cells (Hegedus et al., 2009). The PM is a non-cellular, selectively permeable layer composed by a scaffold of chitin fibrils associated with glycoproteins and proteoglycans that, among other functions, represents the first line of defense providing a physical barrier between the gut flora and the epithelium (Hegedus et al., 2009). In adult mosquitoes the PM is absent but in females the distension of the midgut induced by blood ingestion triggers the formation of a thick layer of Type I PM (around 20 μm) that surrounds the blood bolus (Shao et al., 2001).
As already mentioned, during blood meal, the load of gut-associated microbiota strongly increases and, interestingly, in Anopheles the synthesis and the integrity of PM appears to be microbiota dependent (Rodgers et al., 2017; Song et al., 2018) as already observed for other arthropod vectors (Weiss et al., 2013; Narasimhan et al., 2014). It is unclear which signaling pathway is responsible for this phenomenon, even though a potential role for the JAK/STAT pathway, which in mosquitoes has been implicated in antiviral response (Souza-Neto et al., 2009; Jupatanakul et al., 2017), has been suggested (Rodgers et al., 2017).
The structural integrity of PM is necessary for a proper response against pathogens: for example silencing of PGRP-LD in An. stephensi causes a dysbiosis, as a consequence of the altered expression of genes that codify for structural components of the PM and thus for its integrity (Song et al., 2018). Noteworthy, the fragmentation of the PM consequent to silencing increases the vectorial potential of the mosquito thanks to the enhanced susceptibility to P. berghei infections (Song et al., 2018).
In An. gambiae mosquitoes in addition to PM, the formation of a mucin-barrier lining the epithelium has been proposed (Kumar et al., 2010). In particular, upon the increase of microbiota load induced by blood meal, IMPer and Duox enzymes are secreted and their role in a process of crosslinking between mucins that may be secreted on cell surface is proposed. Although the presence of this mucin coat has to be demonstrated yet and the mechanism by which this coat should not interfere with physiological absorption/secretion processes at microvillar surface is still unknown, this mucin-barrier may regulate the access of immune elicitors secreted by bacteria to the epithelium and, vice versa, the access of immune effectors secreted by midgut cells into the endoperitrophic space where bacteria proliferate.
When PM integrity is disrupted by ookinete invasion in malaria-vectors, the direct contact between bacteria and midgut epithelial cells primes the immune cellular response in the hemocoel (Barletta et al., 2019). Hemocytes are recruited at the midgut basal surface by the prostaglandin E2 (PGE2) that is produced and secreted by the midgut cells. Hemocytes secrete an alpha macroglobulin with a structure similar to complement C3 protein in vertebrates, named thioester-containing protein 1 (TEP1) (Blandin et al., 2004; Baxter et al., 2007), which is involved in the lysis of pathogens, mainly Plasmodium ookinetes. In particular, TEP1 is a complement-like opsonin that upon binding to pathogens and parasites promote their recognition by hemocytes and thus promote their phagocytosis or lysis. The link between microbiota-induced immune priming and TEP1 expression has been further demonstrated in An. dirus (Wang Y. et al., 2013), showing that the microbiota participates in orchestrating the epithelial and complement-like immune responses. Hemocytes, in particular granulocytes, also participate in the phagocytosis of circulating microbes, while oenocytes are major players in the melanization response (Hillyer and Strand, 2014). The activation of this system heavily affect Plasmodium infection: the recruitment of hemocytes in proximity of the midgut basal surface (Barletta et al., 2019) and the production of NO (Kajla et al., 2016) leads to nitration of epithelial cells, which is required for a proper immune response against these parasites (Oliveira et al., 2012).
Some gut bacterial species can affect pathogen transmission directly, without influencing the mosquito immune response. Pseudomonas rhodesiae, Enterobacter ludwigii, and Vagococcus salmoninarium, isolated from the Ae. albopictus midgut, directly inhibit La Crosse virus infection, suggesting that they may produce anti-viral molecules (Joyce et al., 2011). Chromobacterium sp. Panama strain produces an aminopeptidase that degrades the dengue virus envelope protein, reducing dengue virus infection in Ae. aegypti (Ramirez et al., 2014; Saraiva et al., 2018a). The same species also produces an antiparasitic protein, named rhomidepsin, which restricts P. falciparium infection in An. gambiae (Saraiva et al., 2018b). An Enterobacter, isolated from wild Anopheles arabiensis mosquito populations in Zambia, has been demonstrated to generate ROS and to interfere with P. falciparum development before invasion of the midgut epithelium (Cirimotich et al., 2011a).
Bacteria may also enhance the infection of vectored pathogens. Serratia odorifera suppresses the immune response of the host by secreting a polypeptide, P40, that interacts with the mosquito prohibitin, similar to a cysteine rich protein present in some venoms, required for virus infection in mosquitoes (Londono-Renteria et al., 2015). As a result, susceptibility of Ae. aegypti to both dengue and chikungunya viruses infection is enhanced (Apte-Deshpande et al., 2012, 2014). Similarly, S. marcescens secretes smEnhancin, a protein that digests mucins associated with the PM, making mosquitoes more susceptible to virus infection (Wu et al., 2019).
The relationship between gut-microbiota and pathogens transmitted by mosquitoes is not only one way, but it is more and more clear that pathogens can shape the microbial load in the mosquito midgut and/or the composition of the bacterial population. For example, during the pre-invasive phase, Plasmodium vivax significantly decrease microbial load and 16S rRNA gene expression was not detectable before 36 h post meal, the time frame when ookinetes/early oocysts invaded the gut (Sharma et al., 2020). This suggests that Plasmodium can restrict bacterial growth minimizing the impact of microbiota on the mosquito immune response by out-competing the bacteria before ookinete invasion.
Finally, viral infection can shape the composition of the gut microbial community: Zika virus alters the microbiota profile in Ae. aegypti (Villegas et al., 2018), and chikungunya virus increases the abundance of Enterobacteriaceae in Ae. albopictus (Zouache et al., 2012).
The knowledge accumulated on the interaction between insects and resident microbiota inspired the development of new strategies for the control of vector-borne diseases, since the modulation or manipulation of microbiota may have a strong impact on the host fitness and its resistance to pathogens and parasites (Gendrin et al., 2013; Gupta and Nair, 2020). The main microbiota-mediated interventions for the control of vector-borne diseases include: i) the manipulation of the symbionts for the expression of effector molecules (i.e., paratransgenesis, Wang and Jacobs-Lorena, 2017), summarized in Figure 2; ii) the introduction of microorganisms (bacteria or fungi) into the insect in order to reduce vector competence (van Tol and Dimopoulos, 2016), also outlined in Figure 2.
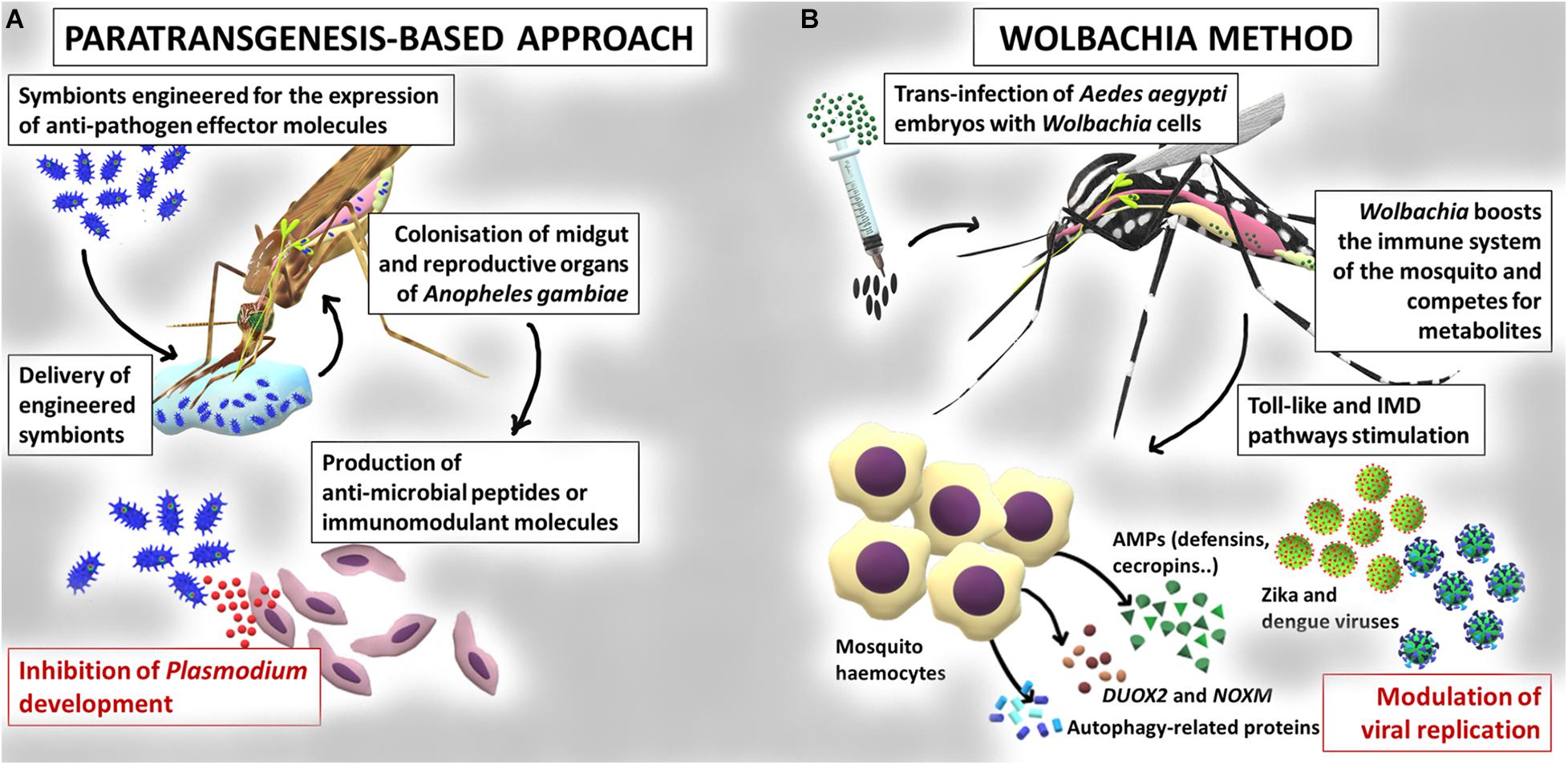
Figure 2. Paratransgenesis as a tool for the control of mosquito-borne diseases. (A) Example of application of a paratransgenesis based-approach for the control of mosquito vector competence. Engineered symbionts colonize midgut and reproductive organs of Anopheles gambiae mosquitoes and express anti-pathogen effector molecules, leading to the inhibition of Plasmodium parasite development. (B) Example of application of the Wolbachia based-approach. Wolbachia is artificially introduced into the Aedes aegypti mosquitoes; these bacteria can block development of viruses such as dengue and Zika, through priming the response of the insect immune system or competing for nutrients.
In arthropods, paratransgenesis is based on the genetic manipulation of symbionts for the production of effector molecules (e.g., antipathogens or immunomodulatory), followed by the re-introduction of the modified symbiont into the arthropod host, to reduce its vector competence (Ogaugwu and Durvasula, 2017; Wang and Jacobs-Lorena, 2017; Gao H. et al., 2020; Figure 2). The choice of a good candidate symbiont is crucial (Hoy, 2013). First, the symbiont should be stably associated with the insect vector, efficiently transmitted vertically and/or horizontally, and persist long enough to produce the effector molecules (Wilke and Marrelli, 2015). Second, the symbiont should be easily culturable and should be genetically manipulable (Wang and Jacobs-Lorena, 2017). Third, the engineered microorganism should have the same fitness of the wild type strain and should not affect the fitness of the host (van Tol and Dimopoulos, 2016). Finally, to better enhance the effect, the symbiont should secrete the antagonistic molecule to guarantee its interaction with the target pathogen (Wang and Jacobs-Lorena, 2017). Paratransgenesis was initially applied for the control of Chagas disease by exploiting the symbiont Rhodococcus rhodnii, engineered for the production of the AMP cecropin A in the host, the triatomine bug Rhodnius prolixus (Durvasula et al., 1997). Since then, several projects have explored paratrangenesis as a strategy to control malaria. In 2007, Riehle et al. (2007) engineered the bacterium Escherichia coli for the expression of the two anti-plasmodial molecules (i.e., salivary gland and midgut peptide 1 and the phospholipase-A2 PLA2). Although a significant inhibition of the parasite P. berghei development was detected, the persistence of the bacterium in the gut was very low and the expression of functional PLA2 was toxic to the bacterium (Riehle et al., 2007). The mosquito symbiotic bacteria belonging to the genera Pantoea, Serratia, and Asaia, have been regarded as very promising for paratransgenesis purposes. Pantoea agglomerans is a non-pathogenic bacterium, widespread in different mosquitoes belonging to the genus Anopheles and, differently from E. coli, can efficiently persist in the insect gut (Riehle et al., 2007). This bacterium has been engineered for the expression of five anti-Plasmodium factors which have determined a strong inhibition of the development of the parasite (Wang et al., 2012). Serratia colonizes male and female of An. stephensi mosquitoes with a very low fitness cost for the insect (Chiamaka et al., 2020). The release of five single effector molecules by this modified bacterium or their simultaneous expression efficiently inhibited P. falciparum infection in mosquitoes (Wang et al., 2017). Finally, the bacterium Asaia, commonly found in Anopheles and Aedes mosquitoes (Favia et al., 2007; Crotti et al., 2009) has been successfully engineered for the secretion of different effector proteins resulting in a significant inhibition of P. berghei development (Bongio and Lampe, 2015; Shane et al., 2018). In addition, more recently, a modified strain of the bacterium Asaia, able to stimulate the immune system of mosquitoes, has been proposed for the control of the heartworm Dirofilaria immitis (Epis et al., 2020). Examples of paratransgenic control approaches come also from the study of leishmaniases and trypanosomiasis. Engineered bacteria of the genus Bacillus, among others, are under study for their potential to reduce the capability of sand flies to transmit Leishmania (Wijerathna et al., 2020). In African trypanosomiasis, the symbiont of the genus Sodalis has been studied as a candidate vector to be exploited to block trypanosome transmission in the tsetse flies. Especially, attacin is a well characterized inducible immune peptide studied as an effector molecule for the engineering of Sodalis with specificity against some Gram-negative bacteria and protozoa (Aksoy et al., 2008).
In addition to bacteria, other microorganisms have been investigated for their potential to be exploited in paratransgenesis, in particular fungi and viruses. Metarhizium robertsii (previously named M. anisopliae), a fungus that infects several insects and proliferates in the hemolymph, was engineered to produce antimalaria effector proteins with encouraging results (Fang et al., 2011). As for viruses, densonucleosis viruses have been proposed as attractive agents for viral paratransgenesis in Aedes and Anopheles mosquitoes; Ren et al. (2008) described an efficient An. gambiae densovirus (AgDNV) which can be potentially used for the control of malaria by transduction of anti-Plasmodium peptides or insect-specific toxins. The same densovirus was proposed by Suzuki et al. (2014) as over-expression system for the malaria vector An. gambiae. Moreover, the pathogenic Aedes DNV (AeDNV) was manipulated to express the green fluorescent protein (Afanasiev et al., 1999) and the microRNAs that target host genes (Liu et al., 2016).
An important key point in the paratransgenic approach is the choice of the molecules with antagonistic activity against pathogens or parasites (Wang et al., 2017). While in the case of malaria parasites there are different effector molecules successfully studied and tested (Bisi and Lampe, 2011; Fang et al., 2011; Dehghan et al., 2017), in the case of viral infections the research is much more limited (Gao H. et al., 2020). Supplementary Table 1 highlights several effector molecules, including AMPs and specific single chain antibodies, currently investigated for their anti-parasite activities.
Before paratransgenesis is applied in large-scale in the field, an intermediate step is required to validate laboratory-based findings; recently, a semi-field study provided evidence for the potential capability of engineered Asaia bacteria to invade mosquito populations (Mancini et al., 2016). Many questions are still open about the introduction and maintenance of the engineered bacteria in mosquito populations; exploiting a bacterium that is naturally vertically and/or horizontally transmitted offers the possibility of a stably spreading the symbiont among target mosquito populations (van Tol and Dimopoulos, 2016). To date, one of the most important tools for the dissemination of engineered bacteria to mosquitoes is based on sugar baits (Lindh et al., 2006; Wang et al., 2012; Wang and Jacobs-Lorena, 2017). Furthermore, Bilgo et al. (2018) tested in a field study the attractivity and effectiveness of sugar baits as a delivery method for modified bacteria (Bilgo et al., 2018); in brief, they highlighted that Window entry trap (WET) attractive sugar bait stations are the most promising tool to introduce and spread engineered bacteria through the mosquito population. Despite these promising results and applications in semi-field condition or in the field, a real application of paratransgenesis has not yet been realized and possible disadvantages of this strategy are still to be investigated. Safety and risk assessments on humans and on non-target organisms, horizontal gene transfer, stability of the engineered symbionts in a natural habitat are some of the issues that will have to be addressed before the application (Coutinho-Abreu et al., 2010).
The second microbiota-mediated intervention exploits the introduction of non-modified microorganisms into the insects able to impair vector competence. The impairment may occur by different mechanisms such as resource competition with the vectored pathogen or parasite, stimulation of the host immune response, reduction of host lifespan (Cirimotich et al., 2011b; Dennison et al., 2014). Different bacteria isolated from the insect gut have been studied for their capability to affect pathogen transmission. Interestingly, a recent study showed that the bacterium S. marcescens, isolated from the midguts of field-collected mosquitoes, could negatively affect Plasmodium development in An. stephensi mosquitoes by activating immune response and in particular modulating effector genes such as TEP1 and fibrinogen immunolectin 9 (Bai et al., 2019). Moreover, Cappelli et al. (2019) described the interactions between the bacteria Asaia and the immune system of the mosquitoes An. stephensi; in particular, the introduction of Asaia triggers mosquito immune responses, eliciting an anti-Plasmodium response.
To date, the most promising microbiota-mediated intervention is based on the release of Ae. aegypti mosquitoes infected with a Wolbachia strain isolated from Drosophila melanogaster for the control of dengue virus (Hoffmann et al., 2011; Walker et al., 2011; O’Neill, 2018; see dedicated section).
Wolbachia is one of the most fascinating microorganisms associated with arthropods, due to its ability to influence the reproductive biology of the hosts, their metabolism, and immunity (Werren et al., 2008). The Wolbachia encompasses obligate intracellular bacteria, members of the order Rickettsiales, first observed in the mosquito Culex pipiens by Hertig and Wolbach (1924). Wolbachia is widespread in insect species and populations, but patchily distributed among them. In a seminal study, insects from 65% of the examined species tested positive for Wolbachia, with different prevalence rates within infected species, in some cases reaching fixation (Hilgenboecker et al., 2008). Among mosquitoes, Wolbachia has consistently been detected in species from the genera Culex, Aedes, Coquillettidia, Mansonia, and Uranotaenia (Huicong et al., 2020), where it is found both in reproductive organs and somatic tissues. These localizations are coherent with the effects that Wolbachia has on the hosts, i.e., with its capability to influence the mosquito survival and fertility. In general, the presence of these bacteria in insects determines reproductive alterations, such as feminization of genetic males, parthenogenesis and the killing of male embryos (sex-ratio distortions) and cytoplasmic incompatibility (CI). CI provides a reproductive advantage to Wolbachia infected females over uninfected ones, resulting in a rapid spread of Wolbachia into the host population (Jiggins, 2017). CI is caused by the sperm from infected males, which is capable of reducing the fertility of uninfected females. Briefly, the molecular mechanism at the basis of CI has been recently elucidated: CI displays as embryonic death when a male expressing prophage WO genes cifA and cifB mate with an uninfected female or a female infected by an incompatible Wolbachia strain. In mosquito females harboring a compatible cifA-expressing strain rescue the embryonic development (LePage et al., 2017; Shropshire et al., 2021). Wolbachia has recently been detected in Ae. aegypti and in some species of Anopheles mosquitoes, although its presence is in general variable, in terms of prevalence and abundance, from species to species (Baldini et al., 2014; Balaji et al., 2019). As for the presence of Wolbachia in Anopheles, a negative correlation between Wolbachia infection and Plasmodium was observed in An. gambiae, in which the presence of Wolbachia reduces malaria transmission with effects on sporozoites (Shaw et al., 2016; Gomes et al., 2017). More recently, the description of novel Wolbachia strains in Anopheles mosquitoes was reported on two large studies in Africa (Jeffries et al., 2018; Ayala et al., 2019); in these researches the authors proved that the Wolbachia prevalence varied among Anopheles species, suggesting that the sample size can be a key factor to detect the infection. Moreover, recent papers emphasized that the evidence for the infection of Wolbachia in Anopheles mosquitoes is largely molecular, which implies that active Wolbachia infections had not always been discriminated from the simple presence of “traces” of Wolbachia or its DNA (Chrostek and Gerth, 2019; Ross et al., 2020). However, another possible explanation for the limited presence of Wolbachia in several Anopheles mosquitoes can be the preponderant role of Asaia bacteria in these mosquitoes (Favia et al., 2007; Chouaia et al., 2012). In fact, Asaia symbionts had been shown to interfere with the vertical transmission of Wolbachia and to negatively correlate with Wolbachia in mosquito reproductive tissues (Hughes et al., 2014; Rossi et al., 2015).
Prior to the observation of naturally infected individuals of Ae. aegypti, stable and heritable Wolbachia infections had been generated in laboratory colonies of this species, by embryonic microinjection of Wolbachia from donor species (Xi et al., 2005; Figure 2). After the release of infected mosquitoes, Wolbachia was then able to spread into wild Ae. aegypti populations, by means of the CI mechanism (Xi et al., 2005; Hoffmann et al., 2011; Nazni et al., 2019). Wolbachia was also stably introduced into a colony of An. stephensi, where the bacteria increased host resistance to P. falciparum (Bian et al., 2013). A similar phenomenon was observed in Ae. aegypti where different Wolbachia strains have been shown to inhibit the infection by viruses of medical relevance, such as dengue (Moreira et al., 2009; Bian et al., 2010), chikungunya (Moreira et al., 2009), West Nile (Hussain et al., 2013), Zika (Aliota et al., 2016), and filarial worms (Kambris et al., 2009).
A stable infection of Wolbachia into a novel mosquito host implies that this symbiont must be able to cope with the host immune system. Thus, has Wolbachia evolved mechanisms to suppress or stimulate the immune system of the hosts?
Actually, when Wolbachia bacteria infect a new host, they are able to stimulate the mosquito immune system, including the Toll and IMD pathways. In detail, Pan et al. (2018), reported that the suppression of either the IMD pathway alone or both the Toll and IMD pathways reduced Wolbachia load in Ae. aegypti; on the other hand, the activation of these pathways increased Wolbachia load, suggesting that host innate immunity is utilized to establish and promote this new host-microbial symbiosis. Various studies indicated that Wolbachia-mediated interference with pathogens is associated with a boosted immunity in mosquitoes (Kambris et al., 2009, 2010; Moreira et al., 2009; Bian et al., 2010; Hughes et al., 2011). Overexpression of AMPs, such as defensins and cecropins, and of several Toll pathway genes, is induced by Wolbachia in Ae. aegypti, providing evidence that immune activation is crucial in the inhibition of dengue infection in these mosquitoes. Comparing the transcripts of Wolbachia-infected Ae. aegypti mosquitoes with wild type mosquitoes, Pan et al. (2012) described the up-regulation of genes in the midguts of Wolbachia-infected mosquitoes: defensin C, attacin, cecropin D, Copper superoxide dismutase, 13 cytochrome P450, two putative NADH dehydrogenase, and three heat-shock proteins, Gram-negative binding protein B1 (GNBPB1), Relish-like protein 1A (REL1A). Similarly, the components of the Toll pathway such as GNBPB1, Spaetzle 3, myeloid differentiation primary response 88 and REL1A were also up-regulated. Moreover, they demonstrated that Wolbachia infection leads to an up-regulation of genes encoding a NADPH oxidase and a dual oxidase (DUOX2), which are involved in the generation of ROS. Specifically, this increased ROS level is correlated with the activation of the Toll pathway, which contributes to the production of antioxidants, defensins and cecropins (Bian et al., 2010; Luplertlop et al., 2011; Pan et al., 2012).
A recent study provided evidence for the effect of a protein of Wolbachia in the activation of the immune response of Ae. aegypti and An. stephensi mosquitoes, consisting in the expression of genes coding for cecropin, TEPs, leucine-rich repeat protein and CLIP-domain serine protease, plus NADPH-oxidases and NO synthase. This priming of the immune response of mosquitoes was associated with the inhibition of the development of the heartworm parasite Dirofilaria immitis (Epis et al., 2020; Varotto-Boccazzi et al., 2020).
Additionally, Zug and Hammerstein (2015) proposed the hypothesis that newly introduced Wolbachia triggers the immune response and causes oxidative stress by upregulating the expression of several immune effectors such as AMPs, autophagy-related proteins, and ROS. In Drosophila, a native Wolbachia infection increases ROS level, leading to oxidative stress, which is involved in the resistance of these flies against viral infection and replication (Wong et al., 2015). On the contrary, in Ae. albopictus mosquitoes, which are naturally infected by Wolbachia, the presence of the bacteria is not associated with oxidative stress, but with balanced redox homeostasis.
In summary, although Wolbachia often determines an up-regulation of mosquito immunity in newly infected hosts, immune priming is not regarded as the sole mechanism involved in the inhibition of pathogen transmission. For example, it has been proposed that competition between viruses and Wolbachia for intracellular cholesterol and amino acids can result in metabolite depletion and cellular stress, thus reducing viral replication (Caragata et al., 2014; Lindsey et al., 2018).
Normally, when Wolbachia-free insects are artificially infected with the symbionts, it is expected that an anti-microbial immune response could be triggered leading to the elimination of Wolbachia itself. However, Wolbachia, through the evasion of the AMP-based immune response or the suppression of the autophagy-associated immune defense, are able to prevent their elimination (Zug and Hammerstein, 2015). In parallel, natural selection could favor the presence of the endosymbiont Wolbachia improving the fitness of the insect host; indeed, other studies suggest that Wolbachia provides an advantage to the host in the form of metabolic provisioning (Brownlie et al., 2009; Gerth and Bleidorn, 2016). In the long term, natural selection is also expected to favor a reduction in the immune stimulating property of Wolbachia, with a stabilization of the association (Dedeine et al., 2003).
The artificially infection of Aedes mosquitoes by Wolbachia affects the relative abundance of resident bacteria, but not species diversity (Audsley et al., 2018), and this effect may be related to an activation of immune pathways such as Toll and IMD (Rancès et al., 2012). Interestingly, in Anopheles mosquitoes, there are several bacterial species that negatively correlate with Wolbachia; for example, Hughes et al. (2014) demonstrated that native mosquito microbiota, in particular bacteria of the genus Asaia, is a major barrier for the transmission of Wolbachia. The same observation was reported in Rossi et al., 2015, in which, a mutual exclusion or a competition between Asaia and Wolbachia has been hypothesized in anophelines thus explaining the inability of Wolbachia to colonize the reproductive system.
Anyhow, due to the variable influence of Wolbachia on the composition of mosquito microbiota, e.g., in relation with the host species, developmental stage, sampling location (Muturi et al., 2016, 2017; Straub et al., 2020), an understanding of these factors is very important before Wolbachia is transinfected into a new mosquito species for the control of the pathogens.
Furthermore, another crucial aspect to be investigated is the long-term phenotypic stability of artificially infected Ae. aegypti mosquitoes in field conditions (O’Neill, 2018). As previously described, field application of Wolbachia-infected Ae. aegypti mosquitoes for the control of mosquito-borne viruses is relatively “new”; we can expect that this system (Wolbachia-Ae. aegypti) will evolve in the coming years (Dorigatti et al., 2018). Certainly, higher efficacy strains of Wolbachia must be investigated and the release of mosquitoes infected by two or more strains (“superinfected”) might be proposed as an alternative strategy to manage potential reductions of the efficiency of single Wolbachia to interfere with pathogen transmission (Joubert et al., 2016).
The role of microbiota in the modulation of vector immune responses and in the regulation of vector competence, has been also studied in tsetse flies (Diptera: Glossinidae), sand flies (Diptera: Psychodidae) and triatoma bugs (Hemiptera: Triatominae), major vectors of African trypanosomiasis, leishmaniases and American trypanosomiasis respectively (Cirimotich et al., 2011b; Weiss and Aksoy, 2011; Wang J. et al., 2013; Telleria et al., 2018). Indeed, the comprehension of the intimate relationship between these insect vectors and resident microbiota may be pivotal for the development of new tools to counteract the transmission and spread of diseases, such as paratransgenesis (Weiss and Aksoy, 2011).
Due to their reproduction and feeding habits, the life of the immature stages of tsetse flies is characterized by a relative sterility (Wang J. et al., 2013), since the larva develops inside the female uterus where it is fed by the maternal accessory gland (i.e., the milk gland) that produces a highly nutrient secretion. Once deposited, the larva immediately pupate, and adults, that are exclusively hematophagous, feed on sterile blood of different mammalian hosts including humans (Wang J. et al., 2013). The microbiota associated with tsetse flies is thus relatively simple compared to other insects and essentially constituted by three bacterial symbionts and a salivary-gland associated Hytrosavirus (Table 2). Moreover, the environment may marginally contribute to the establishment of gut microbiota through the ingestion of bacteria present on host skin during blood meals (Geiger et al., 2014). The obligate association with Wiggleworthia during larval stage is responsible for proper development of an adult functional immune system, in particular of the pathways mediating cellular responses. Wiggleworthia-free larvae develop into adults unable to counteract the septicemia induced by normally non-pathogenic E. coli due to a decrease in sessile and circulating immune cells and failure in melanization reaction (Weiss et al., 2011). Although a similar effect was observed in laboratory colonies of flies depleted of Sodalis and Wolbachia, field-flies that do not harbor these symbionts possess a functional immune system (Weiss et al., 2012). Interestingly, Wiggleworthia is able to trigger tsetse flies antibacterial immune responses against trypanosome by inducing the production of a peptidoglycan recognition protein (i.e., PGRP-LB) and, by the recruitment of the IMD pathway, of anti-trypanosome effector molecules (Wang et al., 2009). In addition, the competence of tsetse flies for trypanosomes has been linked to the capacity of Wiggleworthia to produce folate (vitamin B9) de novo, which thus seems to be a key metabolite for these parasites (Rio et al., 2019).

Table 2. Tsetse fly symbionts, main features of the association, and symbiont role in the modulation of host biology.
The knowledge about the interplay between microbiota and immune system in sand flies and triatome bugs is quite fragmented, although a role of intestinal microbiota in the maintenance of gut homeostasis and immune activation in these vectors has been reported (Araújo et al., 2006; Ursic-Bedoya and Lowenberger, 2007; Waniek et al., 2011; Castro et al., 2012; Diaz-Albiter et al., 2012; Vieira et al., 2015; Telleria et al., 2018).
Sand flies larvae acquire their gut microbiota from food, which is represented by soil organic matter and sand flies adults from carbohydrate-rich fluids (plant sap and aphid secretions). In addition, adult females feed on blood, principally from birds and mammals. Gut microbiota presence and composition has an impact on insect reproductive fitness (Telleria et al., 2018) and allows the activation of important immune pathways for the production of humoral effectors that allow the coexistence of insect and resident microbiota (Telleria et al., 2018). Moreover, studies on the sand fly Lutzomyia longipalpis have highlighted a key role of gut microbiota on vector competence for Leishmania (Sant’Anna et al., 2014; Kelly et al., 2017) and even that Leishmania protects L. longipalpis against bacterial infection (Diaz-Albiter et al., 2012; Sant’Anna et al., 2014). Intriguingly, recent work has demonstrated a remarkable role of Leishmania-infected sand fly microbiota. When regurgitated on the skin of the secondary host during bite, sand fly microbes are able to initiate an immune reaction at the bite site that positively impacts on the progression of infection (Dey et al., 2018).
The triatomine gut is a complex environment where microorganisms and parasites coexist and challenge each other in different ways (Díaz et al., 2016; de Fuentes-Vicente et al., 2018). This association has been well studied in R. prolixus, one of the vectors of the protozoa Trypanosoma cruzi (Azambuja et al., 2017). R. prolixus acquires enteric microbiota through horizontal transmission (i.e., by the consumption of feces of conspecifics or cannibalism, which allow the establishment of intestinal symbionts, such as R. rhodnii that provides vitamins to the bug) and through the skin of the animals during blood feeding, while infected blood is the source of T. cruzi (Azambuja et al., 2017). Although strain dependent, the capacity of the parasite to alter immune responses of the bug has been reported in different studies (Araújo et al., 2006; Ursic-Bedoya and Lowenberger, 2007; Waniek et al., 2011; Castro et al., 2012; Vieira et al., 2015). In particular, T. cruzi and Trypanosoma rangeli are able to trigger the production of immune effectors by the host (i.e., phenoloxidase and AMPs) that specifically reduce gut flora and, on the other hand, increase parasitemia (Araújo et al., 2006; Ursic-Bedoya and Lowenberger, 2007; Waniek et al., 2011; Castro et al., 2012; Vieira et al., 2015). In addition, the induction of a significant decrease of R. rhodnii load in the gut of R. prolixus infected with T. rangeli (but not with T. cruzi) has been observed (Eichler and Schaub, 2002).
The manipulation of the mosquito microbiota is an emerging strategy for the control of many deadly diseases, including malaria, dengue, chikungunya, and Zika. These strategies require a deep knowledge of the mosquito immunity and of the interactions occurring between the insect immune system and the microbiota. Three main applicative approaches are under study: i) development of microbial strains that express anti-parasitic or anti-viral effector molecules; ii) development of microbial strains expressing immune-priming molecules; iii) introduction of unmodified strains with immune-priming effects in mosquitoes and/or resource competitors that ultimately limit infections in the insects. The first two approaches require the release of genetically modified organisms in the field and, therefore, further studies are needed to understand the spread and the effect of these organisms in target and non-target species. The development of strategies for a safe removal of the organisms are necessary, in the case that adverse effects will be detected during releases in the field, as already suggested for transgenic mosquitoes (Zapletal et al., 2021). The development of these multiple tools in mosquito will foster the studies in other less-studied arthropod species, which anyhow can transmit a high number of human pathogens.
PG, SC, GB, and IA reviewed the mosquito immunity, the interaction with the mosquito gut microbiota, and the interactions of microbiota with other insect species. IV-B, FC, and SE reviewed the paratransgenesis and the applied application of the studies on microbiota interaction. All authors have made a direct and intellectual contribution to the work and approved the manuscript for publication.
This study was supported by the MIUR (Italian Minister of University and Research) PRIN Prot. 2017J8JR57 of SE and SC and by Fondazione Cariplo Prot. 2017-0798 to PG. None of the funding sources had roles in the analysis and interpretation of data or in the writing of the report.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Claudio Bandi for his valuable suggestions and revision.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.630438/full#supplementary-material
Afanasiev, B. N., Ward, T. W., Beaty, B. J., and Carlson, J. O. (1999). Transduction of Aedes aegypti mosquitoes with vectors derived from Aedes densovirus. Virology 257, 62–72. doi: 10.1006/viro.1999.9621
Aksoy, S., Weiss, B., and Attardo, G. (2008). “Paratransgenesis applied for control of tsetse transmitted sleeping sickness,” in Transgenesis and the Management of Vector-Borne Disease Adv Exp Med Biol, ed. S. Aksoy (New York, NY: Springer), 35–48.
Aliota, M. T., Peinado, S. A., Velez, I. D., and Osorio, J. E. (2016). The wMel strain of Wolbachia reduces transmission of Zika virus by Aedes aegypti. Sci. Rep. 6:28792. doi: 10.1038/srep28792
Apte-Deshpande, A., Paingankar, M., Gokhale, M. D., and Deobagkar, D. N. (2012). Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One 7:e40401. doi: 10.1371/journal.pone.0040401
Apte-Deshpande, A. D., Paingankar, M. S., Gokhale, M. D., and Deobagkar, D. N. (2014). Serratia odorifera mediated enhancement in susceptibility of Aedes aegypti for chikungunya virus. Indian J. Med. Res. 139, 762–768.
Araújo, C. A. C., Waniek, P. J., Stock, P., Mayer, C., Jansen, A. M., and Schaub, G. A. (2006). Sequence characterization and expression patterns of defensin and lysozyme encoding genes from the gut of the reduviid bug Triatoma brasiliensis. Insect Biochem. Mol. Biol. 36, 547–560. doi: 10.1016/j.ibmb.2006.04.003
Audsley, M. D., Seleznev, A., Joubert, D. A., Woolfit, M., O’Neill, S. L., and McGraw, E. A. (2018). Wolbachia infection alters the relative abundance of resident bacteria in adult Aedes aegypti mosquitoes, but not larvae. Mol. Ecol. 27, 297–309. doi: 10.1111/mec.14436
Ayala, D., Akone-Ella, O., Rahola, N., Kengne, P., Ngangue, M. F., Mezeme, F., et al. (2019). Natural Wolbachia infections are common in the major malaria vectors in Central Africa. Evol. Appl. 12, 1583–1594. doi: 10.1111/eva.12804
Azambuja, P., Garcia, E. S., Waniek, P. J., Vieira, C. S., Figueiredo, M. B., Gonzalez, M. S., et al. (2017). Rhodnius prolixus: from physiology by Wigglesworth to recent studies of immune system modulation by Trypanosoma cruzi and Trypanosoma rangeli. J. Insect. Physiol. 97, 45–65. doi: 10.1016/j.jinsphys.2016.11.006
Azar, S. R., and Weaver, S. C. (2019). Vector competence: what has Zika virus taught us? Viruses 11:867. doi: 10.3390/v11090867
Bai, L., Wang, L., Vega-Rodríguez, J., Wang, G., and Wang, S. (2019). A gut symbiotic bacterium Serratia marcescens renders mosquito resistance to Plasmodium infection through activation of mosquito immune responses. Front. Microbiol. 10:1580. doi: 10.3389/fmicb.2019.01580
Balaji, S., Jayachandran, S., and Prabagaran, S. R. (2019). Evidence for the natural occurrence of Wolbachia in Aedes aegypti mosquitoes. FEMS Microbiol. Lett. 366:fnz055. doi: 10.1093/femsle/fnz055
Baldini, F., Segata, N., Pompon, J., Marcenac, P., Shaw, W. R., Dabiré, R. K., et al. (2014). Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat. Commun. 5:3985. doi: 10.1038/ncomms4985
Barletta, A. B. F., Nascimento-Silva, M. C. L., Talyuli, O. A. C., Oliveira, J. H. M., Pereira, L. O. R., Oliveira, P. L., et al. (2017). Microbiota activates IMD pathway and limits Sindbis infection in Aedes aegypti. Parasit. Vectors 10:103. doi: 10.1186/s13071-017-2040-9
Barletta, A. B. F., Trisnadi, N., Ramirez, J. L., and Barillas-Mury, C. (2019). Mosquito midgut prostaglandin release establishes systemic immune priming. iScience 19, 54–62. doi: 10.1016/j.isci.2019.07.012
Baxter, R. H. G., Chang, C.-I., Chelliah, Y., Blandin, S., Levashina, E. A., and Deisenhofer, J. (2007). Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc. Natl. Acad. Sci. U.S.A. 104, 11615–11620. doi: 10.1073/pnas.0704967104
Bellini, R., Michaelakis, A., Petrić, D., Schaffner, F., Alten, B., Angelini, P., et al. (2020). Practical management plan for invasive mosquito species in Europe: I. Asian tiger mosquito (Aedes albopictus). Travel Med. Infect. Dis. 35:101691. doi: 10.1016/j.tmaid.2020.101691
Bian, G., Joshi, D., Dong, Y., Lu, P., Zhou, G., Pan, X., et al. (2013). Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science 340, 748–751. doi: 10.1126/science.1236192
Bian, G., Xu, Y., Lu, P., Xie, Y., and Xi, Z. (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 6:e1000833. doi: 10.1371/journal.ppat.1000833
Bilgo, E., Vantaux, A., Sanon, A., Ilboundo, S., and Dabirè, R. K. (2018). Field assessment of potential sugar feeding stations for disseminating bacteria in a paratransgenic approach to control malaria. Malar. J. 17:367. doi: 10.1186/s12936-018-2516-x
Bisi, D. C., and Lampe, D. J. (2011). Secretion of anti-Plasmodium effector proteins from a natural Pantoea agglomerans isolate by using PelB and HlyA secretion signals. Appl. Environ. Microbiol. 77, 4669–4675. doi: 10.1128/AEM.00514-11
Blandin, S., Shiao, S.-H., Moita, L. F., Janse, C. J., Waters, A. P., Kafatos, F. C., et al. (2004). Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670. doi: 10.1016/s0092-8674(04)00173-4
Boissière, A., Tchioffo, M. T., Bachar, D., Abate, L., Marie, A., Nsango, S. E., et al. (2012). Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 8:e1002742. doi: 10.1371/journal.ppat.1002742
Bongio, N. J., and Lampe, D. J. (2015). Inhibition of Plasmodium berghei development in mosquitoes by effector proteins secreted from asaia sp. bacteria using a novel native secretion signal. PLoS One 10:e0143541. doi: 10.1371/journal.pone.0143541
Boulanger, N., Boyer, P., Talagrand-Reboul, E., and Hansmann, Y. (2019). Ticks and tick-borne diseases. Med. Mal. Infect. 49, 87–97. doi: 10.1016/j.medmal.2019.01.007
Brownlie, J. C., Cass, B. N., Riegler, M., Witsenburg, J. J., Iturbe-Ormaetxe, I., McGraw, E. A., et al. (2009). Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 5:e1000368. doi: 10.1371/journal.ppat.1000368
Buchon, N., Silverman, N., and Cherry, S. (2014). Immunity in Drosophila melanogaster–from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810. doi: 10.1038/nri3763
Bulet, P., Hetru, C., Dimarcq, J. L., and Hoffmann, D. (1999). Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 23, 329–344. doi: 10.1016/s0145-305x(99)00015-4
Caminade, C., McIntyre, K. M., and Jones, A. E. (2019). Impact of recent and future climate change on vector-borne diseases. Ann. NY Acad. Sci. 1436, 157–173. doi: 10.1111/nyas.13950
Cappelli, A., Damiani, C., Mancini, M. V., Valzano, M., Rossi, P., Serrao, A., et al. (2019). Asaia activates immune genes in mosquito eliciting an anti-Plasmodium response: implications in malaria control. Front. Genet. 10:836. doi: 10.3389/fgene.2019.00836
Caragata, E. P., Rancès, E., O’Neill, S. L., and McGraw, E. A. (2014). Competition for amino acids between Wolbachia and the mosquito host, Aedes aegypti. Microb. Ecol. 67, 205–218. doi: 10.1007/s00248-013-0339-4
Carissimo, G., Pondeville, E., McFarlane, M., Dietrich, I., Mitri, C., Bischoff, E., et al. (2015). Antiviral immunity of Anopheles gambiae is highly compartmentalized, with distinct roles for RNA interference and gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 112, E176–E185. doi: 10.1073/pnas.1412984112
Carnevale, P., and Gay, F. (2019). Insecticide-treated mosquito nets. Methods Mol. Biol. 2013, 221–232. doi: 10.1007/978-1-4939-9550-9_16
Castro, D. P., Moraes, C. S., Gonzalez, M. S., Ratcliffe, N. A., Azambuja, P., and Garcia, E. S. (2012). Trypanosoma cruzi immune response modulation decreases microbiota in Rhodnius prolixus gut and is crucial for parasite survival and development. PLoS One 7:e36591. doi: 10.1371/journal.pone.0036591
Charan, S. S., Pawar, K. D., Severson, D. W., Patole, M. S., and Shouche, Y. S. (2013). Comparative analysis of midgut bacterial communities of Aedes aegypti mosquito strains varying in vector competence to dengue virus. Parasitol. Res. 112, 2627–2637. doi: 10.1007/s00436-013-3428-x
Chiamaka, L., Id, E., Abla, E., Id, A., Aboagye-Antwi, F., and Akorli, J. (2020). Mosquito midgut Enterobacter cloacae and Serratia marcescens affect the fitness of adult female Anopheles gambiae s.l. PLoS One 15:e0238931. doi: 10.1371/journal.pone.0238931
Chouaia, B., Rossi, P., Epis, S., Mosca, M., Ricci, I., Damiani, C., et al. (2012). Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol. 12(Suppl. 1):S2. doi: 10.1186/1471-2180-12-S1-S2
Christophides, G. K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., et al. (2002). Immunity-related genes and gene families in Anopheles gambiae. Science 298, 159–165. doi: 10.1126/science.1077136
Chrostek, E., and Gerth, M. (2019). Is Anopheles gambiae a natural host of Wolbachia? mBio 10:e00784-19. doi: 10.1128/mBio.00784-19
Cirimotich, C. M., Dong, Y., Clayton, A. M., Sandiford, S. L., Souza-Neto, J. A., Mulenga, M., et al. (2011a). Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332, 855–858. doi: 10.1126/science.1201618
Cirimotich, C. M., Dong, Y., Garver, L. S., Sim, S., and Dimopoulos, G. (2009). Mosquito immune defenses against Plasmodium infection. Dev. Comp. Immunol. 34, 387–395. doi: 10.1016/j.dci.2009.12.005
Cirimotich, C. M., Ramirez, J. L., and Dimopoulos, G. (2011b). Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe 10, 307–310. doi: 10.1016/j.chom.2011.09.006
Coon, K., Vogel, K., Brown, M., and Strand, M. (2014). Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 23, 2727–2739. doi: 10.1111/mec.12771
Cooper, D. M., Chamberlain, C. M., and Lowenberger, C. (2009). Aedes FADD: a novel death domain-containing protein required for antibacterial immunity in the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 39, 47–54. doi: 10.1016/j.ibmb.2008.09.011
Coutinho-Abreu, I. V., Zhu, K. Y., and Ramalho-Ortigao, M. (2010). Transgenesis and paratransgenesis to control insect-borne diseases: current status and future challenges. Parasitol. 59, 1–8. doi: 10.1016/j.parint.2009.10.002
Crotti, E., Damiani, C., Pajoro, M., Gonella, E., Rizzi, A., Ricci, I., et al. (2009). Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ. Microbiol. 11, 3252–3264. doi: 10.1111/j.1462-2920.2009.02048.x
de Fuentes-Vicente, J. A., Gutiérrez-Cabrera, A. E., Flores-Villegas, A. L., Lowenberger, C., Benelli, G., Salazar-Schettino, P. M., et al. (2018). What makes an effective Chagas disease vector? Factors underlying Trypanosoma cruzi-triatomine interactions. Acta Trop. 183, 23–31. doi: 10.1016/j.actatropica.2018.04.008
de La Rocque, S., Balenghien, T., Halos, L., Dietze, K., Claes, F., Ferrari, G., et al. (2011). A review of trends in the distribution of vector-borne diseases: is international trade contributing to their spread? Rev. Sci. Tech. 30, 119–130. doi: 10.20506/rst.30.1.2018
Dedeine, F., Bandi, C., Bouletreau, M., and Kramer, L. H. (2003). “Insights into Wolbachia obligatory symbiosis,” in Insect Symbiosis, eds K. Bourtzis and T. A. Miller (Boca Raton, FL: CRC Press), 267–282.
Dehghan, H., Oshaghi, M. A., Moosa-Kazemi, S. H., Yakhchali, B., Vatandoost, H., Maleki-Ravasan, N., et al. (2017). Dynamics of transgenic Enterobacter cloacae expressing green fluorescent protein defensin (GFP-D) in Anopheles stephensi under laboratory condition. J. Arthropod Borne Dis. 11, 515–532.
Dennison, N. J., Jupatanakul, N., and Dimopoulos, G. (2014). The mosquito microbiota influences vector competence for human pathogens. Curr. Opin. Insect. Sci. 3, 6–13. doi: 10.1016/j.cois.2014.07.004
Dey, R., Joshi, A. B., Oliveira, F., Pereira, L., Guimarães-Costa, A. B., Serafim, T. D., et al. (2018). Gut microbes egested during bites of infected sand flies augment severity of leishmaniasis via inflammasome-derived IL-1β. Cell Host Microbe 23, 134.e6–143.e6. doi: 10.1016/j.chom.2017.12.002
Díaz, S., Villavicencio, B., Correia, N., Costa, J., and Haag, K. L. (2016). Triatomine bugs, their microbiota and Trypanosoma cruzi: asymmetric responses of bacteria to an infected blood meal. Parasit. Vectors 9:636. doi: 10.1186/s13071-016-1926-2
Diaz-Albiter, H., Sant’Anna, M. R. V., Genta, F. A., and Dillon, R. J. (2012). Reactive oxygen species-mediated immunity against Leishmania mexicana and Serratia marcescens in the phlebotomine sand fly Lutzomyia longipalpis. J. Biol. Chem. 287, 23995–24003. doi: 10.1074/jbc.M112.376095
Dong, Y., Manfredini, F., and Dimopoulos, G. (2009). Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 5:e1000423. doi: 10.1371/journal.ppat.1000423
Dorigatti, I., McCormack, C., Nedjati-Gilani, G., and Ferguson, N. M. (2018). Using Wolbachia for dengue control: insights from modelling. Trends Parasitol. 34, 102–113. doi: 10.1016/j.pt.2017.11.002
Doudoumis, V., Alam, U., Aksoy, E., Abd-Alla, A. M. M., Tsiamis, G., Brelsfoard, C., et al. (2013). Tsetse-Wolbachia symbiosis: comes of age and has great potential for pest and disease control. J. Invertebr. Pathol. 112, S94–S103. doi: 10.1016/j.jip.2012.05.010
Durvasula, R. V., Gumbs, A., Panackal, A., Kruglov, O., Aksoy, S., Merrifield, R. B., et al. (1997). Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc. Natl. Acad. Sci. U.S.A. 94, 3274–3278. doi: 10.1073/pnas.94.7.3274
Eichler, S., and Schaub, G. A. (2002). Development of symbionts in triatomine bugs and the effects of infections with trypanosomatids. Exp. Parasitol. 100, 17–27. doi: 10.1006/expr.2001.4653
Epis, S., Varotto-Boccazzi, I., Crotti, E., Damiani, C., Giovati, L., Mandrioli, M., et al. (2020). Chimeric symbionts expressing a Wolbachia protein stimulate mosquito immunity and inhibit filarial parasite development. Commun. Biol. 3:105. doi: 10.1038/s42003-020-0835-2
Fang, W., Vega-Rodríguez, J., Ghosh, A. K., Jacobs-Lorena, M., Kang, A., and St Leger, R. J. (2011). Development of transgenic fungi that kill human malaria parasites in mosquitoes. Science 331, 1074–1077. doi: 10.1126/science.1199115
Favia, G., Ricci, I., Damiani, C., Raddadi, N., Crotti, E., Marzorati, M., et al. (2007). Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc. Natl. Acad. Sci. U.S.A. 104, 9047–9051. doi: 10.1073/pnas.0610451104
Gao, H., Cui, C., Wang, L., Jacobs-Lorena, M., and Wang, S. (2020). Mosquito microbiota and implications for disease control. Trends Parasitol. 36, 98–111. doi: 10.1016/j.pt.2019.12.001
Gao, L., Song, X., and Wang, J. (2020). Gut microbiota is essential in PGRP-LA regulated immune protection against Plasmodium berghei infection. Parasit. Vectors 13:3. doi: 10.1186/s13071-019-3876-y
Geiger, A., Ponton, F., and Simo, G. (2014). Adult blood-feeding tsetse flies, trypanosomes, microbiota and the fluctuating environment in sub-Saharan Africa. ISME J. 9, 1496–1507. doi: 10.1038/ismej.2014.236
Gendrin, M., and Christophides, G. (2013). “The Anopheles Mosquito Microbiota and Their Impact on Pathogen Transmission,” in Anopheles Mosquitoes - New Insights into Malaria Vectors. London: InTech.
Gendrin, M., Turlure, F., Rodgers, F. H., Cohuet, A., Morlais, I., and Christophides, G. K. (2017). The peptidoglycan recognition proteins PGRPLA and PGRPLB regulate Anopheles immunity to bacteria and affect infection by Plasmodium. J. Innate Immun. 9, 333–342. doi: 10.1159/000452797
Gendrin, M., Zaidman-Rémy, A., Broderick, N. A., Paredes, J., Poidevin, M., Roussel, A., et al. (2013). Functional analysis of PGRP-LA in Drosophila immunity. PLoS One 8:e69742. doi: 10.1371/journal.pone.0069742
Gerth, M., and Bleidorn, C. (2016). Comparative genomics provides a timeframe for Wolbachia evolution and exposes a recent biotin synthesis operon transfer. Nat. Microbiol. 2:16241. doi: 10.1038/nmicrobiol.2016.241
Gomes, F. M., Hixson, B. L., Tyner, M. D. W., Ramirez, J. L., Canepa, G. E., Alves, E., et al. (2017). Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. Proc. Natl. Acad. Sci. U.S.A. 114, 12566–12571. doi: 10.1073/pnas.1716181114
Gupta, A., and Nair, S. (2020). Dynamics of insect-microbiome interaction influence host and microbial symbiont. Front. Microbiol. 11:1357. doi: 10.3389/fmicb.2020.01357
Gusmão, D. S., Santos, A. V., Marini, D. C., Bacci, M., Berbert-Molina, M. A., and Lemos, F. J. A. (2010). Culture-dependent and culture-independent characterization of microorganisms associated with Aedes aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop. 115, 275–281. doi: 10.1016/j.actatropica.2010.04.011
Hegedus, D., Erlandson, M., Gillott, C., and Toprak, U. (2009). New insights into peritrophic matrix synthesis, architecture, and function. Annu. Rev. Entomol. 54, 285–302. doi: 10.1146/annurev.ento.54.110807.090559
Hemingway, J. (2018). Resistance: a problem without an easy solution. Pestic. Biochem. Physiol. 151, 73–75. doi: 10.1016/j.pestbp.2018.08.007
Hertig, M., and Wolbach, S. B. (1924). Studies on rickettsia-like micro-organisms in insects. J. Med. Res. 44, 329–374.7.
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia? – a statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Hillyer, J. F. (2010). Mosquito immunity. Adv. Exp. Med. Biol. 708, 218–238. doi: 10.1007/978-1-4419-8059-5_12
Hillyer, J. F. (2016). Insect immunology and hematopoiesis. Dev. Comp. Immunol. 58, 102–118. doi: 10.1016/j.dci.2015.12.006
Hillyer, J. F., and Strand, M. R. (2014). Mosquito hemocyte-mediated immune responses. Curr. Opin. Insect. Sci. 3, 14–21. doi: 10.1016/j.cois.2014.07.002
Hoffmann, A., Montgomery, B., Popovici, J., Iturbe-Ormaetxe, I., Johnson, P. H., Muzzi, F., et al. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476, 454–457. doi: 10.1038/nature10356
Hoy, M. A. (2013). “Chapter 14 - genetic modification of pest and beneficial insects for pest-management programs,” in Insect Molecular Genetics (Third Edition), ed. M. A. Hoy (San Diego: Academic Press), 661–736.
Hughes, G. L., Dodson, B. L., Johnson, R. M., Murdock, C. C., Tsujimoto, H., Suzuki, Y., et al. (2014). Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. PNAS 111, 12498–12503. doi: 10.1073/pnas.1408888111
Hughes, G. L., Koga, R., Xue, P., Fukatsu, T., and Rasgon, J. L. (2011). Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 7:e1002043. doi: 10.1371/journal.ppat.1002043
Huicong, D., Huiqing, Y., and Nalini, P. (2020). Wolbachia Infection in Wild Mosquitoes (Diptera: Culicidae): Implications for Transmission Modes and Host-Endosymbiont Associations. Available online at: https://www.researchsquare.com/article/rs-37816/v2 (Accessed October 28, 2020)
Hussain, M., Lu, G., Torres, S., Edmonds, J. H., Kay, B. H., Khromykh, A. A., et al. (2013). Effect of Wolbachia on replication of West Nile virus in a mosquito cell line and adult mosquitoes. J. Virol. 87, 851–858. doi: 10.1128/JVI.01837-12
Ingham, V. A., Anthousi, A., Douris, V., Harding, N. J., Lycett, G., Morris, M., et al. (2020). A sensory appendage protein protects malaria vectors from pyrethroids. Nature 577, 376–380. doi: 10.1038/s41586-019-1864-1
Jeffries, C., Lawrence, G., Golovko, G., Kristan, M., Orsborne, J., Spence, K., et al. (2018). Novel Wolbachia strains in Anopheles malaria vectors from Sub-Saharan Africa. Wellcome Open Res. 3:113. doi: 10.12688/wellcomeopenres.14765.1
Jiggins, F. M. (2017). The spread of Wolbachia through mosquito populations. PLoS Biol. 15:e2002780. doi: 10.1371/journal.pbio.2002780
Joubert, D. A., Walker, T., Carrington, L. B., De Bruyne, J. T., Kien, D. H. T., Hoang, N. L. T., et al. (2016). Establishment of a Wolbachia superinfection in Aedes aegypti mosquitoes as a potential approach for future resistance management. PLoS Pathog. 12:e1005434. doi: 10.1371/journal.ppat.1005434
Joyce, J. D., Nogueira, J. R., Bales, A. A., Pittman, K. E., and Anderson, J. R. (2011). Interactions between la crosse virus and bacteria isolated from the digestive tract of Aedes albopictus (Diptera: Culicidae). J. Med. Entomol. 48, 389–394. doi: 10.1603/ME09268
Jupatanakul, N., Sim, S., Angleró-Rodríguez, Y. I., Souza-Neto, J., Das, S., Poti, K. E., et al. (2017). Engineered Aedes aegypti JAK/STAT pathway-mediated immunity to dengue virus. PLoS Negl. Trop. Dis. 11:e0005187. doi: 10.1371/journal.pntd.0005187
Jupatanakul, N., Sim, S., and Dimopoulos, G. (2014). The insect microbiome modulates vector competence for arboviruses. Viruses 6, 4294–4313. doi: 10.3390/v6114294
Kajla, M., Choudhury, T. P., Kakani, P., Gupta, K., Dhawan, R., Gupta, L., et al. (2016). Silencing of Anopheles stephensi heme peroxidase HPX15 activates diverse immune pathways to regulate the growth of midgut bacteria. Front. Microbiol. 7:1351. doi: 10.3389/fmicb.2016.01351
Kambris, Z., Blagborough, A. M., Pinto, S. B., Blagrove, M. S. C., Godfray, H. C. J., Sinden, R. E., et al. (2010). Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 6:e1001143. doi: 10.1371/journal.ppat.1001143
Kambris, Z., Cook, P. E., Phuc, H. K., and Sinkins, S. P. (2009). Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326, 134–136. doi: 10.1126/science.1177531
Kaneko, T., Yano, T., Aggarwal, K., Lim, J.-H., Ueda, K., Oshima, Y., et al. (2006). PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to DAP-type peptidoglycan. Nat. Immunol. 7, 715–723. doi: 10.1038/ni1356
Kariithi, H. M., Boucias, D. G., Murungi, E. K., Meki, I. K., Demirbaş-Uzel, G., van Oers, M. M., et al. (2018). Coevolution of hytrosaviruses and host immune responses. BMC Microbiol. 18:183. doi: 10.1186/s12866-018-1296-3
Kelly, P. H., Bahr, S. M., Serafim, T. D., Ajami, N. J., Petrosino, J. F., Meneses, C., et al. (2017). The gut microbiome of the vector Lutzomyia longipalpis is essential for survival of Leishmania infantum. mBio 8:e01121-16. doi: 10.1128/mBio.01121-16
Khan, M. B., Liew, J. W. K., Leong, C. S., and Lau, Y.-L. (2016). Role of NF-kβ factor Rel2 during Plasmodium falciparum and bacterial infection in Anopheles dirus. Parasit. Vectors 9:525. doi: 10.1186/s13071-016-1810-0
Kim, C. H., Paik, D., Rus, F., and Silverman, N. (2014). The caspase-8 homolog Dredd cleaves Imd and Relish but is not inhibited by p35. J. Biol. Chem. 289, 20092–20101. doi: 10.1074/jbc.M113.544841
Kreppel, K. S., Viana, M., Main, B. J., Johnson, P. C. D., Govella, N. J., Lee, Y., et al. (2020). Emergence of behavioural avoidance strategies of malaria vectors in areas of high LLIN coverage in Tanzania. Sci. Rep. 10:14527. doi: 10.1038/s41598-020-71187-4
Kumar, A., Srivastava, P., Sirisena, P., Dubey, S. K., Kumar, R., Shrinet, J., et al. (2018). Mosquito innate immunity. Insects 9:95. doi: 10.3390/insects9030095
Kumar, S., Molina-Cruz, A., Gupta, L., Rodrigues, J., and Barillas-Mury, C. (2010). A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 327, 1644–1648. doi: 10.1126/science.1184008
Le, B. V., Williams, M., Logarajah, S., and Baxter, R. H. G. (2012). Molecular basis for genetic resistance of Anopheles gambiae to Plasmodium: structural analysis of TEP1 susceptible and resistant alleles. PLoS Pathog. 8:e1002958. doi: 10.1371/journal.ppat.1002958
Lee, H., Halverson, S., and Ezinwa, N. (2018). Mosquito-borne diseases. Prim. Care 45, 393–407. doi: 10.1016/j.pop.2018.05.001
LePage, D. P., Metcalf, J. A., Bordenstein, S. R., On, J., Perlmutter, J. I., Shropshire, J. D., et al. (2017). Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543, 243–247. doi: 10.1038/nature21391
Levashina, E. A. (2004). Immune responses in Anopheles gambiae. Insect Biochem. Mol. Biol. 34, 673–678. doi: 10.1016/j.ibmb.2004.03.020
Li, H.-H., Cai, Y., Li, J.-C., Su, M. P., Liu, W.-L., Cheng, L., et al. (2020). C-Type lectins link immunological and reproductive processes in Aedes aegypti. iScience 23:101486. doi: 10.1016/j.isci.2020.101486
Lindh, J. M., Terenius, O., Eriksson-Gonzales, K., Knols, B. G. J., and Faye, I. (2006). Re-introducing bacteria in mosquitoes-a method for determination of mosquito feeding preferences based on coloured sugar solutions. Acta Trop. 99, 173–183. doi: 10.1016/j.actatropica.2006.07.008
Lindsey, A. R. I., Bhattacharya, T., Newton, I. L. G., and Hardy, R. W. (2018). Conflict in the intracellular lives of endosymbionts and viruses: a mechanistic look at Wolbachia-mediated pathogen-blocking. Viruses 10:141. doi: 10.3390/v10040141
Liu, P., Li, X., Gu, J., Dong, Y., Liu, Y., Santhosh, P., et al. (2016). Development of non-defective recombinant densovirus vectors for microRNA delivery in the invasive vector mosquito, Aedes albopictus. Sci. Rep. 6:20979. doi: 10.1038/srep20979
Londono-Renteria, B., Troupin, A., Conway, M. J., Vesely, D., Ledizet, M., Roundy, C. M., et al. (2015). Dengue virus infection of Aedes aegypti requires a putative cysteine rich venom protein. PLoS Pathog. 11:e1005202. doi: 10.1371/journal.ppat.1005202
Luplertlop, N., Surasombatpattana, P., Patramool, S., Dumas, E., Wasinpiyamongkol, L., Saune, L., et al. (2011). Induction of a peptide with activity against a broad spectrum of pathogens in the Aedes aegypti salivary gland, following infection with dengue virus. PLoS Pathog. 7:e1001252. doi: 10.1371/journal.ppat.1001252
Magalhaes, T., Leandro, D. C., and Ayres, C. F. J. (2010). Knock-down of REL2, but not defensin A, augments Aedes aegypti susceptibility to Bacillus subtilis and Escherichia coli. Acta Trop. 113, 167–173. doi: 10.1016/j.actatropica.2009.10.013
Mancini, M. V., Spaccapelo, R., Damiani, C., Accoti, A., Tallarita, M., Petraglia, E., et al. (2016). Paratransgenesis to control malaria vectors: a semi-field pilot study. Parasit. Vectors 9:140. doi: 10.1186/s13071-016-1427-3
Mansouri, A., Cregut, M., Abbes, C., Durand, M.-J., Landoulsi, A., and Thouand, G. (2017). The environmental issues of DDT pollution and bioremediation: a multidisciplinary review. Appl. Biochem. Biotechnol. 181, 309–339. doi: 10.1007/s12010-016-2214-5
Meister, S., Agianian, B., Turlure, F., Relógio, A., Morlais, I., Kafatos, F. C., et al. (2009). Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 5:e1000542. doi: 10.1371/journal.ppat.1000542
Meister, S., Kanzok, S. M., Zheng, X., Luna, C., Li, T.-R., Hoa, N. T., et al. (2005). Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc. Natl. Acad. of Sci. U.S.A. 102, 11420–11425. doi: 10.1073/pnas.0504950102
Moiroux, N., Gomez, M. B., Pennetier, C., Elanga, E., Djènontin, A., Chandre, F., et al. (2012). Changes in Anopheles funestus biting behavior following universal coverage of long-lasting insecticidal nets in Benin. J. Infect. Dis. 206, 1622–1629. doi: 10.1093/infdis/jis565
Moreira, L. A., Iturbe-Ormaetxe, I., Jeffery, J. A., Lu, G., Pyke, A. T., Hedges, L. M., et al. (2009). A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139, 1268–1278. doi: 10.1016/j.cell.2009.11.042
Mukherjee, D., Das, S., Begum, F., Mal, S., and Ray, U. (2019). The mosquito immune system and the life of dengue virus: what we know and do not know. Pathogens 8:77. doi: 10.3390/pathogens8020077
Mussabekova, A., Daeffler, L., and Imler, J.-L. (2017). Innate and intrinsic antiviral immunity in Drosophila. Cell Mol. Life. Sci 74, 2039–2054. doi: 10.1007/s00018-017-2453-9
Muturi, E. J., Kim, C.-H., Bara, J., Bach, E. M., and Siddappaji, M. H. (2016). Culex pipiens and Culex restuans mosquitoes harbor distinct microbiota dominated by few bacterial taxa. Parasit Vectors 9:18. doi: 10.1186/s13071-016-1299-6
Muturi, E. J., Ramirez, J. L., Rooney, A. P., and Kim, C. H. (2017). Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl. Trop. Dis. 11:e0005377. doi: 10.1371/journal.pntd.0005377
Narasimhan, S., Rajeevan, N., Liu, L., Zhao, Y. O., Heisig, J., Pan, J., et al. (2014). Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe 15, 58–71. doi: 10.1016/j.chom.2013.12.001
Nazni, W. A., Hoffmann, A. A., NoorAfizah, A., Cheong, Y. L., Mancini, M. V., Golding, N., et al. (2019). Establishment of Wolbachia strain wAlbB in malaysian populations of Aedes aegypti for dengue control. Curr. Biol. 29, 4241.e3–4248.e3. doi: 10.1016/j.cub.2019.11.007
Ogaugwu, C. E., and Durvasula, R. V. (2017). “Developing the arsenal against pest and vector dipterans: inputs of transgenic and paratransgenic biotechnologies,” in Biological Control of Pest and Vector Insects, ed. V. D. C. Shields (Rijeka: InTech).
Oliveira, G. A., Lieberman, J., and Barillas-Mury, C. (2012). Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 335, 856–859. doi: 10.1126/science.1209678
Oliveira, J. H. M., Gonçalves, R. L. S., Lara, F. A., Dias, F. A., Gandara, A. C. P., Menna-Barreto, R. F. S., et al. (2011). Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog. 7:e1001320. doi: 10.1371/journal.ppat.1001320
O’Neill, S. L. (2018). “The use of Wolbachia by the World Mosquito Program to interrupt transmission of Aedes aegypti transmitted viruses,” in Dengue and Zika: Control and Antiviral Treatment Strategies, eds S. G. Vasudevan and R. Hilgenfeld (Berlin: Springer), 355–360.
Pan, X., Pike, A., Joshi, D., Bian, G., McFadden, M. J., Lu, P., et al. (2018). The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J. 12, 277–288. doi: 10.1038/ismej.2017.174
Pan, X., Zhou, G., Wu, J., Bian, G., Lu, P., Raikhel, A. S., et al. (2012). Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 109, E23–E31. doi: 10.1073/pnas.1116932108
Pang, X., Xiao, X., Liu, Y., Zhang, R., Liu, J., Liu, Q., et al. (2016). Mosquito C-type lectins maintain gut microbiome homeostasis. Nat. Microbiol. 1:16023. doi: 10.1038/nmicrobiol.2016.23
Paquette, N., Broemer, M., Aggarwal, K., Chen, L., Husson, M., Ertürk-Hasdemir, D., et al. (2010). Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-κB signaling. Mol. cell. 37, 172–182. doi: 10.1016/j.molcel.2009.12.036
Paredes, J. C., Welchman, D. P., Poidevin, M., and Lemaitre, B. (2011). Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35, 770–779. doi: 10.1016/j.immuni.2011.09.018
Perugini, E., Guelbeogo, W. M., Calzetta, M., Manzi, S., Virgillito, C., Caputo, B., et al. (2020). Behavioural plasticity of Anopheles coluzzii and Anopheles arabiensis undermines LLIN community protective effect in a Sudanese-savannah village in Burkina Faso. Parasit Vectors 13:277. doi: 10.1186/s13071-020-04142-x
Raddi, G., Barletta, A. B. F., Efremova, M., Ramirez, J. L., Cantera, R., Teichmann, S. A., et al. (2020). Mosquito cellular immunity at single-cell resolution. Science 369, 1128–1132. doi: 10.1126/science.abc0322
Ramirez, J. L., Short, S. M., Bahia, A. C., Saraiva, R. G., Dong, Y., Kang, S., et al. (2014). Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog. 10:e1004398. doi: 10.1371/journal.ppat.1004398
Ramirez, J. L., Souza-Neto, J., Torres Cosme, R., Rovira, J., Ortiz, A., Pascale, J. M., et al. (2012). Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 6:e1561. doi: 10.1371/journal.pntd.0001561
Rancès, E., Ye, Y. H., Woolfit, M., McGraw, E. A., and O’Neill, S. L. (2012). The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog. 8:e1002548. doi: 10.1371/journal.ppat.1002548
Ren, X., Hoiczyk, E., and Rasgon, J. L. (2008). Viral paratransgenesis in the malaria vector Anopheles gambiae. PLoS Pathog. 4:e1000135. doi: 10.1371/journal.ppat.1000135
Riehle, M. A., Moreira, C. K., Lampe, D., Lauzon, C., and Jacobs-Lorena, M. (2007). Using bacteria to express and display anti-Plasmodium molecules in the mosquito midgut. Int. J. Parasitol. 37, 595–603. doi: 10.1016/j.ijpara.2006.12.002
Rio, R. V. M., Jozwick, A. K. S., Savage, A. F., Sabet, A., Vigneron, A., Wu, Y., et al. (2019). Mutualist-provisioned resources impact vector competency. mBio 10:e00018-19. doi: 10.1128/mBio.00018-19
Rodgers, F. H., Cai, J. A., Pitaluga, A. N., Mengin-Lecreulx, D., Gendrin, M., and Christophides, G. K. (2020). Functional analysis of the three major PGRPLC isoforms in the midgut of the malaria mosquito Anopheles coluzzii. Insect Biochem. Mol. Biol. 118:103288. doi: 10.1016/j.ibmb.2019.103288
Rodgers, F. H., Gendrin, M., Wyer, C. A. S., and Christophides, G. K. (2017). Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes. PLoS Pathog. 13:e1006391. doi: 10.1371/journal.ppat.1006391
Ross, P. A., Callahan, A. G., Yang, Q., Jasper, M., Arif, M. A. K., Afizah, A. N., et al. (2020). An elusive endosymbiont: does Wolbachia occur naturally in Aedes aegypti? Ecol. Evol. 10, 1581–1591. doi: 10.1002/ece3.6012
Rossi, P., Ricci, I., Cappelli, A., Damiani, C., Ulissi, U., Mancini, M. V., et al. (2015). Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasites Vectors 8:278. doi: 10.1186/s13071-015-0888-0
Sant’Anna, M. R. V., Diaz-Albiter, H., Aguiar-Martins, K., Al Salem, W. S., Cavalcante, R. R., Dillon, V. M., et al. (2014). Colonisation resistance in the sand fly gut: Leishmania protects Lutzomyia longipalpis from bacterial infection. Parasit. Vectors 7:329. doi: 10.1186/1756-3305-7-329
Saraiva, R. G., Fang, J., Kang, S., Angleró-Rodríguez, Y. I., Dong, Y., and Dimopoulos, G. (2018a). Aminopeptidase secreted by Chromobacterium sp. Panama inhibits dengue virus infection by degrading the E protein. PLoS Negl. Trop. Dis. 12:e0006443. doi: 10.1371/journal.pntd.0006443
Saraiva, R. G., Huitt-Roehl, C. R., Tripathi, A., Cheng, Y.-Q., Bosch, J., Townsend, C. A., et al. (2018b). Chromobacterium spp. mediate their anti-Plasmodium activity through secretion of the histone deacetylase inhibitor romidepsin. Sci. Rep. 8:6176. doi: 10.1038/s41598-018-24296-0
Schuffenecker, I., Iteman, I., Michault, A., Murri, S., Frangeul, L., Vaney, M.-C., et al. (2006). Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3:e0030263. doi: 10.1371/journal.pmed.0030263
Scolari, F., Casiraghi, M., and Bonizzoni, M. (2019). Aedes spp. and their microbiota: a review. Front. Microbiol. 10:2036. doi: 10.3389/fmicb.2019.02036
Segata, N., Baldini, F., Pompon, J., Garrett, W. S., Truong, D. T., and Dabiré, R. K. (2016). The reproductive tracts of two malaria vectors are populated by a core microbiome and by gender-and swarm-enriched microbial biomarkers. Sci. Rep. 6:24207. doi: 10.1038/srep24207
Shane, J. L., Grogan, C. L., Cwalina, C., and Lampe, D. J. (2018). Blood meal-induced inhibition of vector-borne disease by transgenic microbiota. Nat. Commun. 9:4127. doi: 10.1038/s41467-018-06580-9
Shao, L., Devenport, M., and Jacobs-Lorena, M. (2001). The peritrophic matrix of hematophagous insects. Arch. Insect. Biochem. Physiol. 47, 119–125. doi: 10.1002/arch.1042
Sharma, P., Rani, J., Chauhan, C., Kumari, S., Tevatiya, S., Das De, T., et al. (2020). Altered gut microbiota and immunity defines Plasmodium vivax survival in Anopheles stephensi. Front. Immunol. 11:609. doi: 10.3389/fimmu.2020.00609
Shaw, W. R., Marcenac, P., Childs, L. M., Buckee, C. O., Baldini, F., Sawadogo, S. P., et al. (2016). Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat. Commun. 7:11772. doi: 10.1038/ncomms11772
Short, S. M., Mongodin, E. F., MacLeod, H. J., Talyuli, O. A., and Dimopoulos, G. (2017). Amino acid metabolic signaling influences Aedes aegypti midgut microbiome variability. PLoS Negl. Trop. Dis. 11:e0005677. doi: 10.1371/journal.pntd.0005677
Shropshire, J. D., Rosenberg, R., and Bordenstein, S. R. (2021). The impacts of cytoplasmic incompatibility factor (cifA and cifB) genetic variation on phenotypes. Genetics 217:iyaa007. doi: 10.1093/genetics/iyaa007
Sim, S., Jupatanakul, N., and Dimopoulos, G. (2014). Mosquito immunity against arboviruses. Viruses 6, 4479–4504. doi: 10.3390/v6114479
Song, X., Wang, M., Dong, L., Zhu, H., and Wang, J. (2018). PGRP-LD mediates A. stephensi vector competency by regulating homeostasis of microbiota-induced peritrophic matrix synthesis. PLoS Pathog. 14:e1006899. doi: 10.1371/journal.ppat.1006899
Souza-Neto, J. A., Sim, S., and Dimopoulos, G. (2009). An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U.S.A. 106, 17841–17846. doi: 10.1073/pnas.0905006106
Stathopoulos, S., Neafsey, D. E., Lawniczak, M. K., Muskavitch, M. A., and Christophides, G. K. (2014). Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens. PLoS Pathog. 10:e1003897. doi: 10.1371/journal.ppat.1003897
Straub, T. J., Shaw, W. R., Marcenac, P., Sawadogo, S. P., Dabiré, R. K., Diabaté, A., et al. (2020). The Anopheles coluzzii microbiome and its interaction with the intracellular parasite Wolbachia. Sci. Rep. 10:13847. doi: 10.1038/s41598-020-70745-0
Suzuki, Y., Niu, G., Hughes, G. L., and Rasgon, J. L. (2014). A viral over-expression system for the major malaria mosquito Anopheles gambiae. Sci. Rep. 4:5127. doi: 10.1038/srep05127
Telleria, E. L., Martins-da-Silva, A., Tempone, A. J., and Traub-Csekö, Y. M. (2018). Leishmania, microbiota and sand fly immunity. Parasitology 145, 1336–1353. doi: 10.1017/S0031182018001014
Tikhe, C. V., and Dimopoulos, G. (2021). Mosquito antiviral immune pathways. Dev. Comp. Immunol. 116:103964. doi: 10.1016/j.dci.2020.103964
Toh, H., Weiss, B. L., Perkin, S. A. H., Yamashita, A., Oshima, K., Hattori, M., et al. (2006). Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16, 149–156. doi: 10.1101/gr.4106106
Ursic-Bedoya, R. J., and Lowenberger, C. A. (2007). Rhodnius prolixus: identification of immune-related genes up-regulated in response to pathogens and parasites using suppressive subtractive hybridization. Dev. Comp. Immunol. 31, 109–120. doi: 10.1016/j.dci.2006.05.008
van Tol, S., and Dimopoulos, G. (2016). “Influences of the mosquito microbiota on vector competence,” in Progress in Mosquito Research, ed. A. Raikhel (London: Academic Press Inc), 249–291.
Varotto-Boccazzi, I., Epis, S., Arnoldi, I., Corbett, Y., Gabrieli, P., Paroni, M., et al. (2020). Boosting innate immunity: asaia bacteria expressing a protein from Wolbachia determine macrophage activation and killing. Leishmania Pharmacol. Res. 4:105288. doi: 10.1016/j.phrs.2020.105288
Vieira, C. S., Mattos, D. P., Waniek, P. J., Santangelo, J. M., Figueiredo, M. B., Gumiel, M., et al. (2015). Rhodnius prolixus interaction with Trypanosoma rangeli: modulation of the immune system and microbiota population. Parasit. Vectors 8:135. doi: 10.1186/s13071-015-0736-2
Villegas, L. E. M., Campolina, T. B., Barnabe, N. R., Orfano, A. S., Chaves, B. A., Norris, D. E., et al. (2018). Zika virus infection modulates the bacterial diversity associated with Aedes aegypti as revealed by metagenomic analysis. PLoS One 13:e0190352. doi: 10.1371/journal.pone.0190352
Walker, T., Johnson, P. H., Moreira, L. A., Iturbe-Ormaetxe, I., Frentiu, F. D., McMeniman, C. J., et al. (2011). The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476, 450–453. doi: 10.1038/nature10355
Wang, J., Weiss, B. L., and Aksoy, S. (2013). Tsetse fly microbiota: form and function. Front. Cell. Infect. Microbiol. 3:69. doi: 10.3389/fcimb.2013.00069
Wang, J., Wu, Y., Yang, G., and Aksoy, S. (2009). Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. U.S.A. 106, 12133–12138. doi: 10.1073/pnas.0901226106
Wang, S., and Jacobs-Lorena, M. (2017). Paratransgenesis applications: fighting malaria with engineered mosquito symbiotic bacteria. Arthropod. Vector Control. Dis. Trans. 1, 219–234. doi: 10.1016/B978-0-12-805350-8.00013-1
Wang, S., Dos-Santos, A. L. A., Huang, W., Liu, K. C., Oshaghi, M. A., Wei, G., et al. (2017). Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357, 1399–1402. doi: 10.1126/science.aan5478
Wang, S., Ghosh, A. K., Bongio, N., Stebbings, K. A., Lampe, D. J., and Jacobs-Lorena, M. (2012). Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 109, 12734–12739. doi: 10.1073/pnas.1204158109
Wang, Y., Wang, Y., Zhang, J., Xu, W., Zhang, J., and Huang, F. S. (2013). Ability of TEP1 in intestinal flora to modulate natural resistance of Anopheles dirus. Exp. Parasitol. 134, 460–465. doi: 10.1016/j.exppara.2013.04.003
Wangdi, K., Furuya-Kanamori, L., Clark, J., Barendregt, J. J., Gatton, M. L., Banwell, C., et al. (2018). Comparative effectiveness of malaria prevention measures: a systematic review and network meta-analysis. Parasites Vectors 11:210. doi: 10.1186/s13071-018-2783-y
Waniek, P. J., Jansen, A. M., and Araújo, C. A. C. (2011). Trypanosoma cruzi infection modulates the expression of Triatoma brasiliensis def1 in the midgut. Vector Borne Zoonotic Dis. 11, 845–847. doi: 10.1089/vbz.2010.0020
Wei, G., Lai, Y., Wang, G., Chen, H., Li, F., and Wang, S. (2017). Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. U.S.A. 114, 5994–5999. doi: 10.1073/pnas.1703546114
Weiss, B., and Aksoy, S. (2011). Microbiome influences on insect host vector competence. Trends Parasitol. 27, 514–522. doi: 10.1016/j.pt.2011.05.001
Weiss, B. L., Maltz, M., and Aksoy, S. (2012). Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 188, 3395–3403. doi: 10.4049/jimmunol.1103691
Weiss, B. L., Wang, J., and Aksoy, S. (2011). Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 9:e1000619. doi: 10.1371/journal.pbio.1000619
Weiss, B. L., Wang, J., Maltz, M. A., Wu, Y., and Aksoy, S. (2013). Trypanosome infection establishment in the tsetse fly gut is influenced by microbiome-regulated host immune barriers. PLoS Pathog. 9:e1003318. doi: 10.1371/journal.ppat.1003318
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Wijerathna, T., Gunathunga, S., and Gunathilaka, N. (2020). Recent developments and future directions in the paratransgenesis based control of Leishmania transmission. Biol. Control 145:104260. doi: 10.1016/j.biocontrol.2020.104260
Wilke, A. B. B., and Marrelli, M. T. (2015). Paratransgenesis: a promising new strategy for mosquito vector control. Parasit Vectors 8:342. doi: 10.1186/s13071-015-0959-2
Wong, Z. S., Brownlie, J. C., and Johnson, K. N. (2015). Oxidative stress correlates with Wolbachia-mediated antiviral protection in Wolbachia-Drosophila associations. Appl. Environ. Microbiol. 81, 3001–3005. doi: 10.1128/AEM.03847-14
Wu, P., Sun, P., Nie, K., Zhu, Y., Shi, M., Xiao, C., et al. (2019). A gut commensal bacterium promotes mosquito permissiveness to arboviruses. Cell Host Microbe 25, 101.e5–112.e5. doi: 10.1016/j.chom.2018.11.004
Xi, Z., Khoo, C. C. H., and Dobson, S. L. (2005). Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310, 326–328. doi: 10.1126/science.1117607
Xi, Z., Ramirez, J. L., and Dimopoulos, G. (2008). The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 4:e1000098. doi: 10.1371/journal.ppat.1000098
Xiao, X., Yang, L., Pang, X., Zhang, R., Zhu, Y., Wang, P., et al. (2017). A Mesh-Duox pathway regulates homeostasis in the insect gut. Nat. Microbiol. 2:17020. doi: 10.1038/nmicrobiol.2017.20
Zaidman-Rémy, A., Hervé, M., Poidevin, M., Pili-Floury, S., Kim, M.-S., Blanot, D., et al. (2006). The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473. doi: 10.1016/j.immuni.2006.02.012
Zapletal, J., Najmitabrizi, N., Erraguntla, M., Lawley, M. A., Myles, K. M., and Adelman, Z. N. (2021). Making gene drive biodegradable. Philos. Trans. R. Soc. Lond. B Biol. Sci. 376:20190804. doi: 10.1098/rstb.2019.0804
Zouache, K., Michelland, R. J., Failloux, A.-B., Grundmann, G. L., and Mavingui, P. (2012). Chikungunya virus impacts the diversity of symbiotic bacteria in mosquito vector. Mol. Ecol. 21, 2297–2309. doi: 10.1111/j.1365-294X.2012.05526.x
Keywords: Wolbachia, vector-borne diseases, control strategies, pathogens, insects
Citation: Gabrieli P, Caccia S, Varotto-Boccazzi I, Arnoldi I, Barbieri G, Comandatore F and Epis S (2021) Mosquito Trilogy: Microbiota, Immunity and Pathogens, and Their Implications for the Control of Disease Transmission. Front. Microbiol. 12:630438. doi: 10.3389/fmicb.2021.630438
Received: 17 November 2020; Accepted: 02 March 2021;
Published: 06 April 2021.
Edited by:
Jeremy Keith Herren, International Centre of Insect Physiology and Ecology (ICIPE), KenyaReviewed by:
Marco Pombi, Sapienza University of Rome, ItalyCopyright © 2021 Gabrieli, Caccia, Varotto-Boccazzi, Arnoldi, Barbieri, Comandatore and Epis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sara Epis, c2FyYS5lcGlzQHVuaW1pLml0
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.