- Department of Microbiology and Immunology, Weill Cornell Medical College, New York, NY, United States
Proper control of cell division in the intracellular pathogen Mycobacterium tuberculosis is central to its growth, survival, pathogenesis, and resistance to antibiotics. Nevertheless, the divisome components and mechanisms by which mycobacteria regulate their cell cycle are not entirely understood. Here we demonstrate that the previously uncharacterized Rv0954 protein localizes to the mid-cell during cell division and interacts with the division-related proteins LamA, PbpA, and PknH. Deletion of rv0954 did not result in alterations in cell morphology or sensitivity to cell wall-targeting antibiotics but transposon mutagenesis demonstrated genetic interactions with genes related to cell division. This work suggests that Rv0954 participates in cell division and reveals potential components of the mycobacterial divisome for future investigation.
Introduction
Our knowledge on how one bacterium becomes two has advanced significantly since Y. Hirota, A. Ryter, and F. Jacob first characterized a collection of thermosensitive Escherichia coli (E. coli) cell division mutants about half a century ago (Hirota et al., 1968). In the pathogenic mycobacterial species, Mycobacterium tuberculosis (Mtb) and Mycobacterium leprae, cell division control is an integral part of host-pathogen interactions and may contribute to disease outcomes (Kieser and Rubin, 2014). However, there are many remaining questions to answer and new components to discover in mycobacterial cell division.
Cell division in mycobacteria differs from model rod-shaped bacteria, E. coli and Bacillus subtilis, in several aspects (Kieser and Rubin, 2014; Baranowski et al., 2019). First, mycobacteria have a complex cell wall consisting of peptidoglycan, arabinogalactan, and mycolic acids (Baranowski et al., 2019). Splitting and synthesizing this highly sophisticated cell envelope at the division site poses a unique challenge for mycobacteria. Second, mycobacteria elongate at the poles in contrast to lateral elongation in the model organisms (Meniche et al., 2014; Cameron et al., 2015). As the division site matures into an elongation site, mycobacteria need to ensure both events are coordinated in time and space by tightly regulating the cell division complex (divisome) and the elongation machinery (Baranowski et al., 2019). Third, model bacteria divide precisely at mid-cell, creating two daughter cells of near-identical length; mycobacteria, on the other hand, grow and divide asymmetrically, leading to daughter cells of varying lengths (Aldridge et al., 2012; Santi et al., 2013). This heterogeneity is promoted by LamA and may be advantageous for the mycobacterial population to survive host stresses and antibiotics treatment (Rego et al., 2017). Mycobacteria’s unusual cell division features likely explain why they lack homologs of many divisome components found in model bacteria and instead encode species-specific factors (Kieser and Rubin, 2014; Baranowski et al., 2019).
Genes with related functions are often located in the same operon allowing co-regulation. The Mtb genome contains many such loci; for example, the rv0014c – rv0018c region encodes PknA (Rv0015c) and PknB (Rv0014c), two serine/threonine protein kinases that regulate cell division, and their cognate phosphatase, PstP (Rv0018c). The same region also contains pbpA (rv0016c) and rodA (rv0017c), which encode peptidoglycan synthases involved in septum formation during cell division (Sureka et al., 2010). Since rv0954 is in the same operon with perM, which encodes a division protein (Goodsmith et al., 2015; Wang et al., 2019), we hypothesized that Rv0954, a previously uncharacterized protein, may function in cell division or cell wall biosynthesis.
Like perM, rv0954 is one of 219 mycobacterial “core” genes conserved among mycobacterial species and without homologs in other bacteria (Marmiesse et al., 2004). Saturating transposon mutagenesis predicted rv0954 to be non-essential for growth of Mtb in rich medium (DeJesus et al., 2017), and topology prediction indicated that Rv0954 contains four transmembrane helixes (TMHs) flanked by cytoplasmic N- and C- termini (Supplementary Figure 1). The C- terminal half of Rv0954 (aa 155-303) is rich in proline (18%) and glutamine residues (15%), which often form extended and rigid structures (Williamson, 1994) and may allow Rv0954 to protrude into the cytoplasm for some distance. Here, we examined Rv0954’s localization during the cell cycle, identified its physical interaction partners, and discovered candidate genes that may encode proteins with overlapping functions with Rv0954. Our results support a role for Rv0954 in cell division.
Materials and Methods
Bacterial Culture Conditions
Mycobacterium smegmatis (Msm) mc2155 and derivative strains were cultured in Middlebrook 7H9 medium (BD Difco) containing 0.2% glycerol and 0.05% Tween 80 or Middlebrook 7H10 agar (BD Difco) containing 0.5% glycerol. Mtb H37Rv and derivative strains were cultured in Middlebrook 7H9 medium containing 0.2% glycerol, 0.2% dextrose, 0.5% BSA (Roche), 0.05% Tween 80 or tyloxapol, and 0.085% NaCl or Middlebrook 7H10 agar containing 10% OADC supplement (BD) and 0.5% glycerol. Strains bearing antibiotic resistance cassettes were cultured with hygromycin (50 μg/ml), zeocin (25 μg/ml), kanamycin (25 μg/ml) or streptomycin (20 μg/ml). Growth curves in acidic and low-Mg2+ conditions were performed as previously described (Goodsmith et al., 2015; Wang et al., 2019).
Mutant Construction
The Mtb Δrv0954 mutant was constructed by allelic exchange using a recombineering approach as previously described (Gee et al., 2012). The Δrv0954 strain was complemented by introducing a copy of rv0954 expressed from the hsp60 promoter into the Mtb genome. We generated the msmeg_5518 single deletion mutant (Δmsmeg_5518-perMmsm:perMmtb) by first integrating perMmtb to the attL5 site, then deleting the operon containing msmeg_5518 and perMmsm. The rv0954 phospho mutants were constructed by integrating a copy of phospho-ablative or phospho-mimetic Rv0954 fused with C-terminal GFP tags expressed from p38 promoters into the genome.
High-Resolution Microscopy
Microscopy imaging was performed using the same methods and equipment as described (Botella et al., 2017). The Mtb samples were fixed with 4% paraformaldehyde for 4 h before removal from BSL-3 containment. Single-cell suspensions were prepared by centrifugation at 800 rpm for 10 min. After spreading on a soft agar pad, the bacteria were visualized using a microscope with appropriate filter sets. For the time-lapse microscopy experiment, we place a drop of Msm single-cell culture (5∼10 μl) to a glass-bottom microwell dish (MatTek, 35 mm, 14 mm microwell). 1% low melting point agarose (UltraPure, Invitrogen) prepared in 7H9 broth was carefully added on top of the bacterial drop. Agarose was left at room temperature to allow solidification before microscopy. We captured snapshots every 15 min and analyzed images with ImageJ (Abramoff et al., 2003).
Co-immunoprecipitation
We harvested 150 ml of log-phase Mtb cultures and incubated whole-cell lysates with 1% DDM for 2 h before overnight incubation with anti-Flag affinity gel (Sigma) at 4°C. Beads were then washed with lysis buffer (50 mM Tris-HCl, 50 mM NaCl, pH 7.4) and eluted by incubation with 100 ng/μl Flag peptide with gentle rotation for 1 h at 4°C.
To confirm physical interactions, we harvested 30 ml Msm cultures for whole-cell protein lysates, which we incubated with 50 μl anti-GFP mAb-magnetic beads (MBL) overnight shaking at 4°C. After incubation, beads were collected using a DynaMag-Magnet 2 rack (Thermo Fisher Scientific) and washed three times with lysis buffer. Proteins bound to beads were eluted by boiling in Laemmli sample buffer for 10 min and analyzed by immunoblotting.
Antibiotic Susceptibility Assays
Mtb was grown to early log phase and diluted to an OD of 0.005 in regular 7H9 medium containing 0.2% glycerol, 0.2% dextrose, 0.5% BSA, 0.05% Tween 80, and 0.085% NaCl. Bacteria were then exposed to two-fold dilutions of compounds, and all wells contained 1% DMSO. We recorded MIC as the minimum concentration at which growth was inhibited by at least 90%, compared to controls after approximately 14 days.
Phosphorylation Assay and Site Identification
We incubated 20 ∼ 40 μg whole-cell protein lysates with 10 U of alkaline phosphatase (Fast AP, Thermo Scientific) for 1 h at 37°C. The same reactions without adding phosphatase served as controls. The samples were then boiled in Laemmli sample buffer and resolved on SDS-PAGE for western blotting. To identify Rv0954 phosphorylation sites, we enriched Rv0954-Flag from Mtb whole-cell lysates by pulldown. Eluates were resolved on SDS-PAGE, and gel pieces at the correct molecular weight range were collected for mass spectrometry identification.
Transposon Library Construction
The Mtb transposon libraries were constructed by himar1 mutagenesis as previously described (Abramoff et al., 2003). Briefly, approximately 5 × 1010 CFU of Mtb were incubated with 1011 PFU of MycoMarT7 phage for 4 h at 37°C. The cultures were collected, washed with 7H9 medium containing 0.05% Tween 80, and cultured on 7H9 agar (7H9 medium containing 1.5% bacto agar, 10% OADC, 0.5% glycerol, 0.05% Tween 80, and 25 μg/ml kanamycin) for about 20 days at 37°C. Both WT and Δrv0954 libraries have TA dinucleotide site coverages of around 50% by Illumina sequencing. Sequencing and data analysis were performed as previously described (Xu et al., 2017).
Results
Rv0954 Accumulates at the Mid-Cell Before Septum Formation and Persists Until Cell Constriction
We examined Rv0954’s localization by live-cell imaging using a fluorescent reporter Msm strain. Rv0954 localizes to the membrane in the pre-divisional cells, followed by its accumulation at the mid-cell that lasts until cell constriction (Figure 1). This localization pattern within the cell cycle suggests a role in cell division.
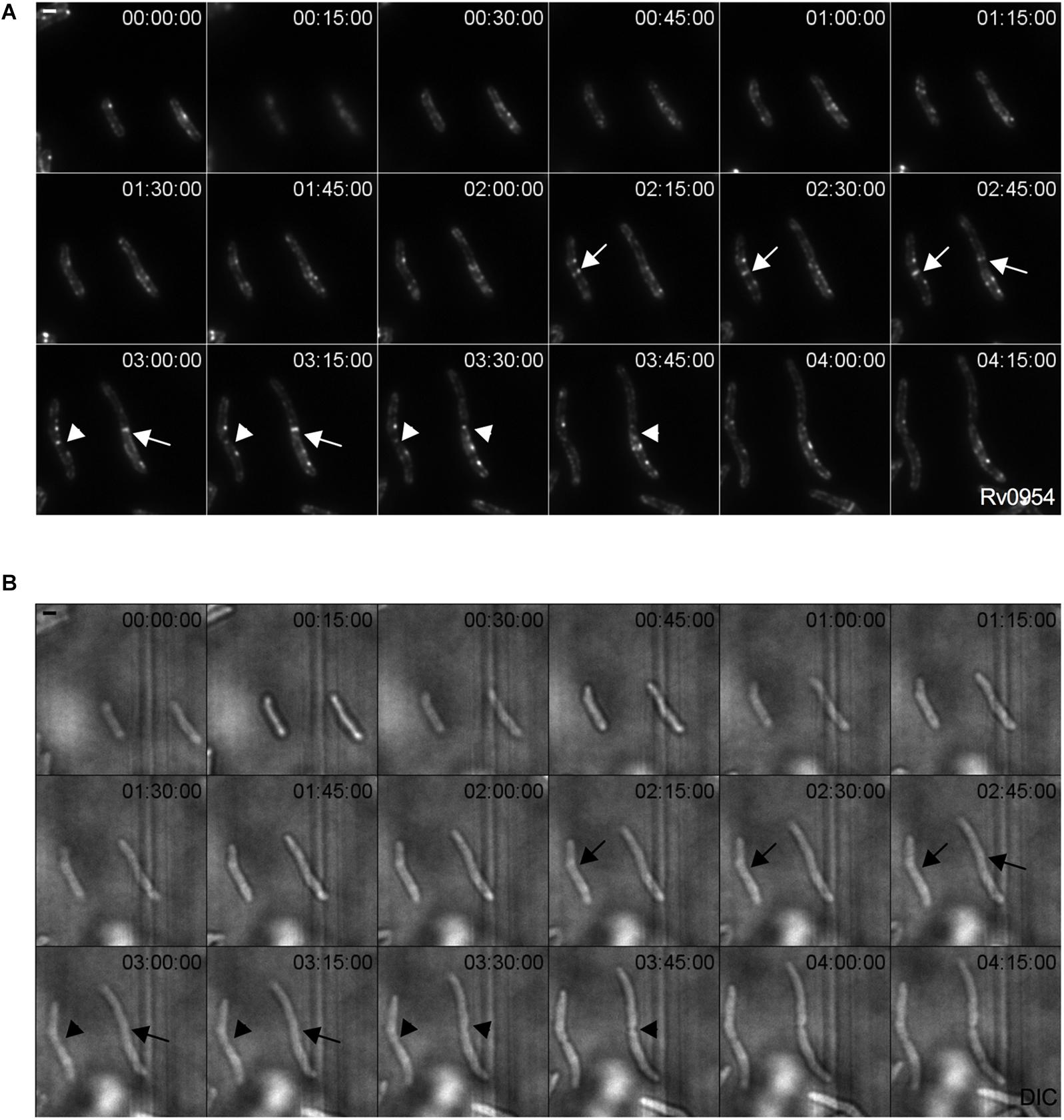
Figure 1. Rv0954 localizes to the mid-cell during cell division. Representative image series from a time-lapse movie of replicating Msm that constitutively express Rv0954-GFP over 4 h and 15 min. Arrows point to mid-cell localization of Rv0954 before cell constriction, and arrowheads point to Rv0954 in cells undergoing constriction. Numbers in the upper right corner indicate time. Scale bar, 1 μm. (A) GFP channel. (B) Transmitted-light snapshots.
We next compared the timing of Rv0954’s mid-cell localization to known markers of the cell cycle (Figure 2A). FtsZ is one of the earliest divisome components to assemble at the mid-cell, marking divisome formation (Kieser and Rubin, 2014). Snapshots and quantification of Msm cells containing septal FtsZ bands while lacking mid-cell Rv0954 signals indicates that Rv0954 localizes to the mid-cell after the initiation of divisome assembly (Figures 2B,E). The fluorescent dye FM5-95 stains the cytoplasmic membrane, and its mid-cell signal is indicative of septum formation. In 7.5% cells Rv0954 signals appeared at the mid-cell before septal membranes were visible (Figures 2C,E), indicating that Rv0954’s mid-cell localization precedes septum formation. The mid-cell localization of Wag31 coincides with septum closure, which results in the cytoplasmic compartmentalization of future daughter cells (Santi et al., 2013). Co-localization of Rv0954 and Wag31 at the mid-cell suggested that Rv0954 remains at the septum when cytokinesis is completed (Figures 2D,E). Of particular interest are the cells exhibiting ‘V-snapping’ morphology (Figure 2D, top two panels). Consistent with its role in organizing polar elongation, Wag31 associated with the newly formed poles of future daughter cells (Santi et al., 2013; Kieser and Rubin, 2014). In contrast, Rv0954 localized to the V shape’s inner edge, where the septum is not entirely resolved, perhaps to participate in the final steps of cell separation. Overall, these co-localization experiments demonstrate that the mid-cell localization of Rv0954 precedes septum formation and lasts until cell constriction.
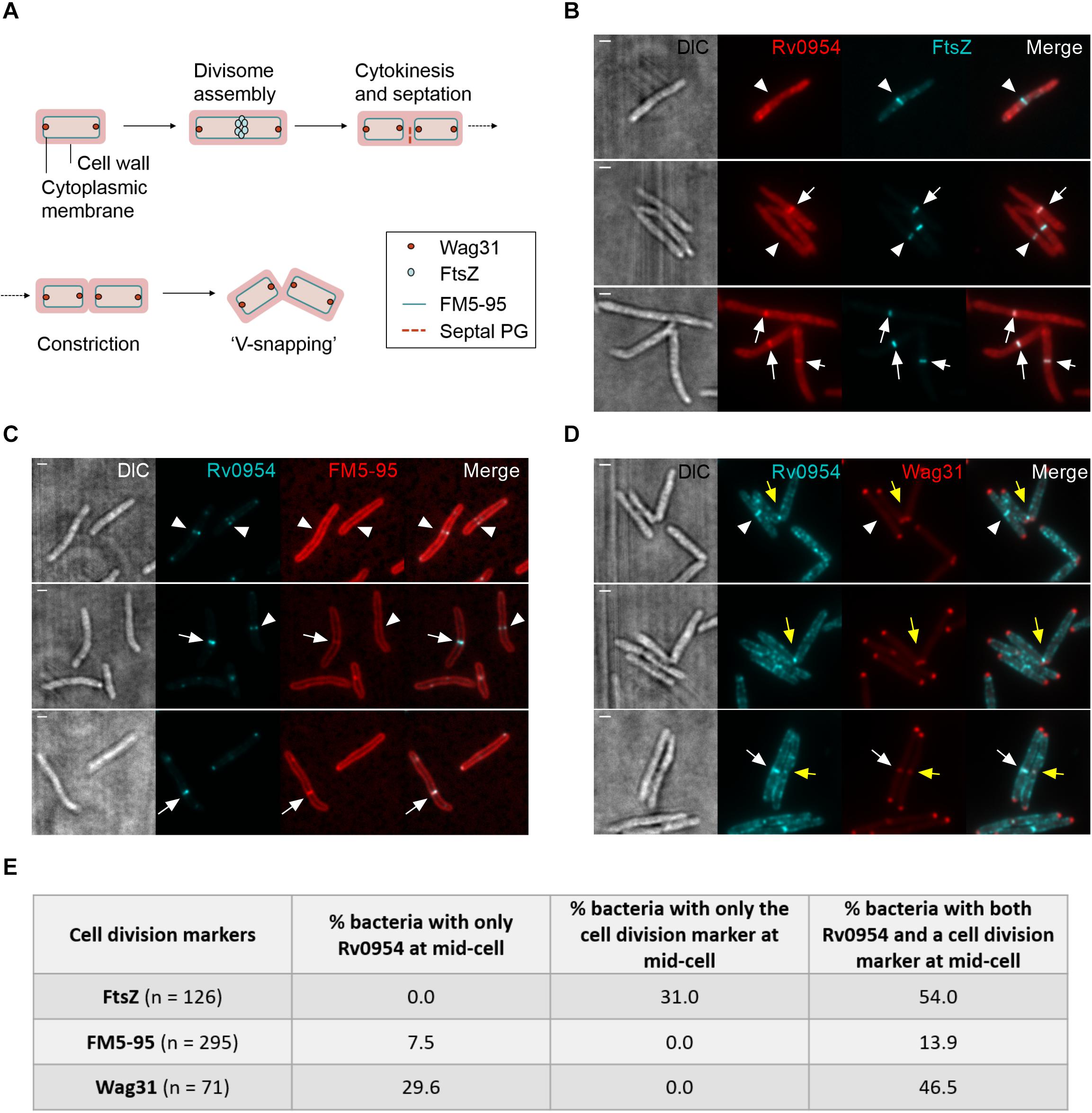
Figure 2. Mid-cell localization of Rv0954 relative to markers of cell division events. (A) Schematic of division markers in different stages of cell division. (B–D) Representative microscopy images of replicating Msm expressing (B) FtsZmsm-GFP and Rv0954-mCherry (C) Rv0954-GFP (D) Rv0954-GFP and Wag31mtb-mCherry constitutively. (B) Arrowheads point to the presence of FtsZ signals and the absence of Rv0954 signals at the mid-cell. Arrows point to the colocalization of FtsZ and Rv0954. (C) Replicating Msm stained with FM5-95 to label the cytoplasmic membrane. Arrowheads point to the presence of Rv0954 signals and the absence of septal FM5-95 staining. Arrows point to the overlapping of Rv0954 and FM5-95 signals at the mid-cell. (D) White arrowheads point to the presence of Rv0954 and the absence of Wag31mtb at the mid-cell. White arrows point to the mid-cell localization of both Rv0954 and Wag31mtb. Yellow arrows point to Msm undergoing constriction. Scale bar, 1 μm. (E) Quantification of bacteria with Rv0954 and respective cell division markers at mid-cell. N indicates the number of bacteria quantified.
Rv0954 Interacts With Several Proteins Involved in Cell Division and Cell Wall Biosynthesis
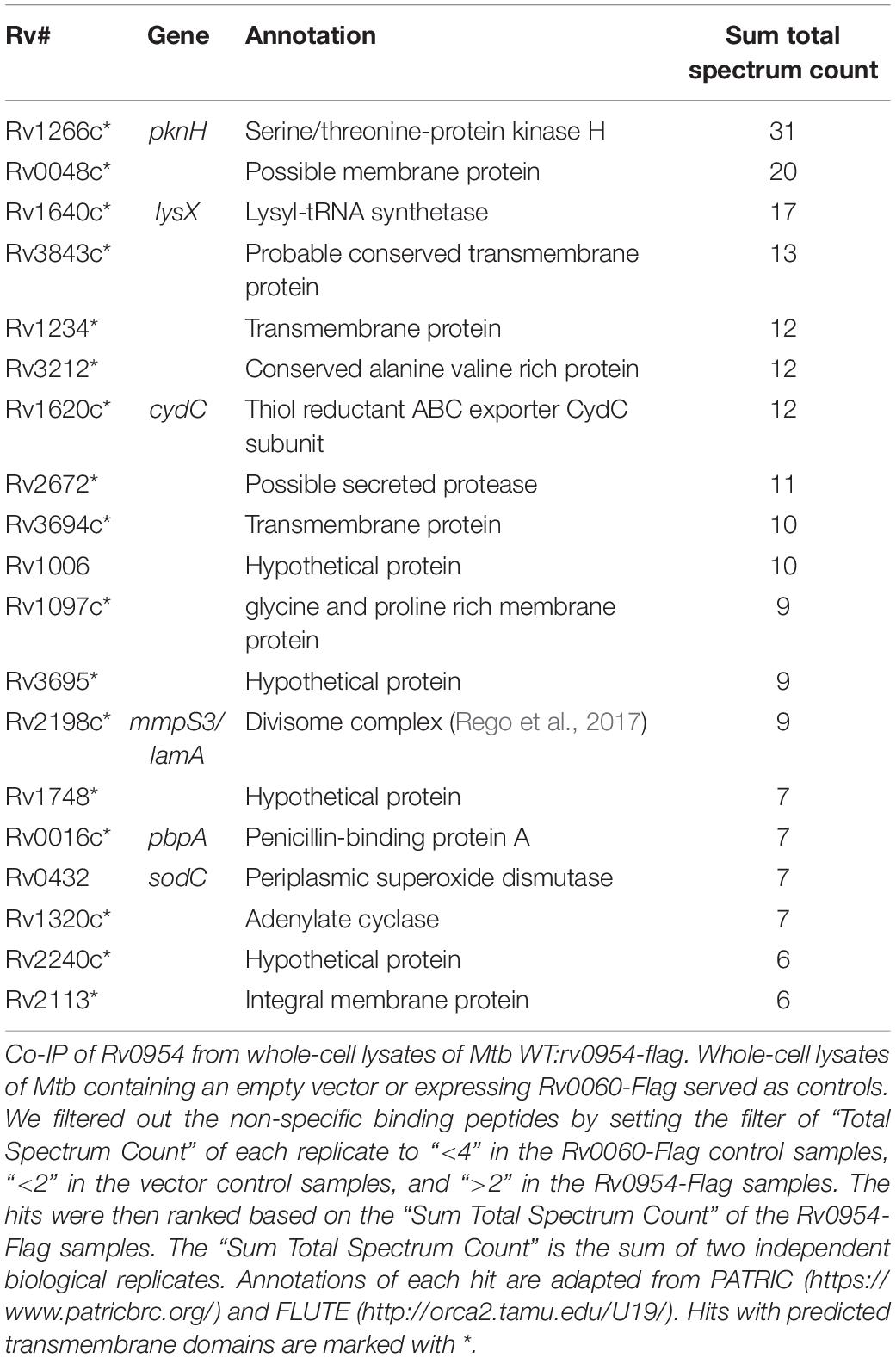
Mycobacterial cell division is mediated by a large protein complex termed ‘the divisome’ (Kieser and Rubin, 2014). To identify protein interactors, we immunoprecipitated Rv0954 from Mtb lysates, followed by mass spectrometry (Figure 3 and Table 1). The 19 identified interactors included a serine/threonine protein kinase involved in cell division regulation: PknH (Sharma et al., 2006; Zheng et al., 2007) and two other known divisome proteins: LamA (also known as MmpS3) and PbpA. LamA is a septal-localizing protein that inhibits cell elongation from the new pole, and PbpA is a transpeptidase that localizes to both the septum and poles (Rego et al., 2017; Viswanathan et al., 2017; Arora et al., 2018). We confirmed the interaction between Rv0954 and PknH in Msm by co-immunoprecipitation (Supplementary Figure 2). The interaction of Rv0954 with these three proteins further supports its involvement in cell division.
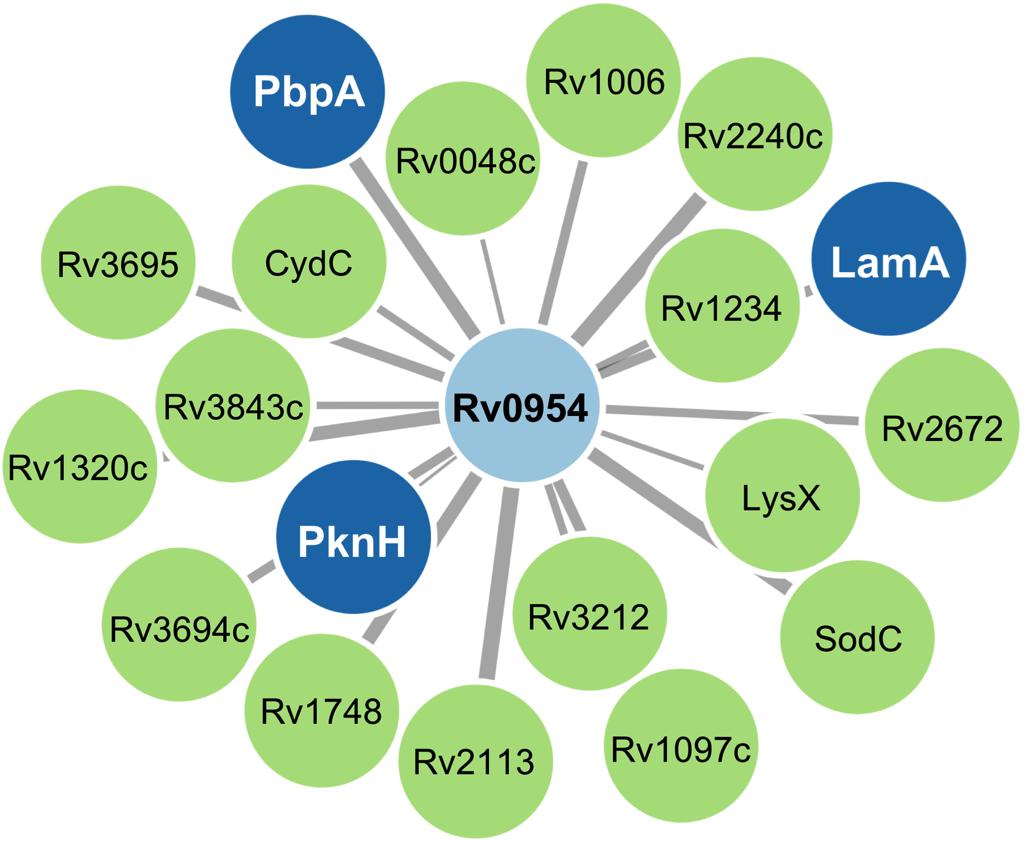
Figure 3. Network map of Rv0954 interacting partners. Cell division-related proteins are marked in blue.
Deletion of rv0954 in Mtb Did Not Result in Apparent Alterations in Cell Morphology, Growth, or Susceptibility to Cell Wall-Targeting Compounds
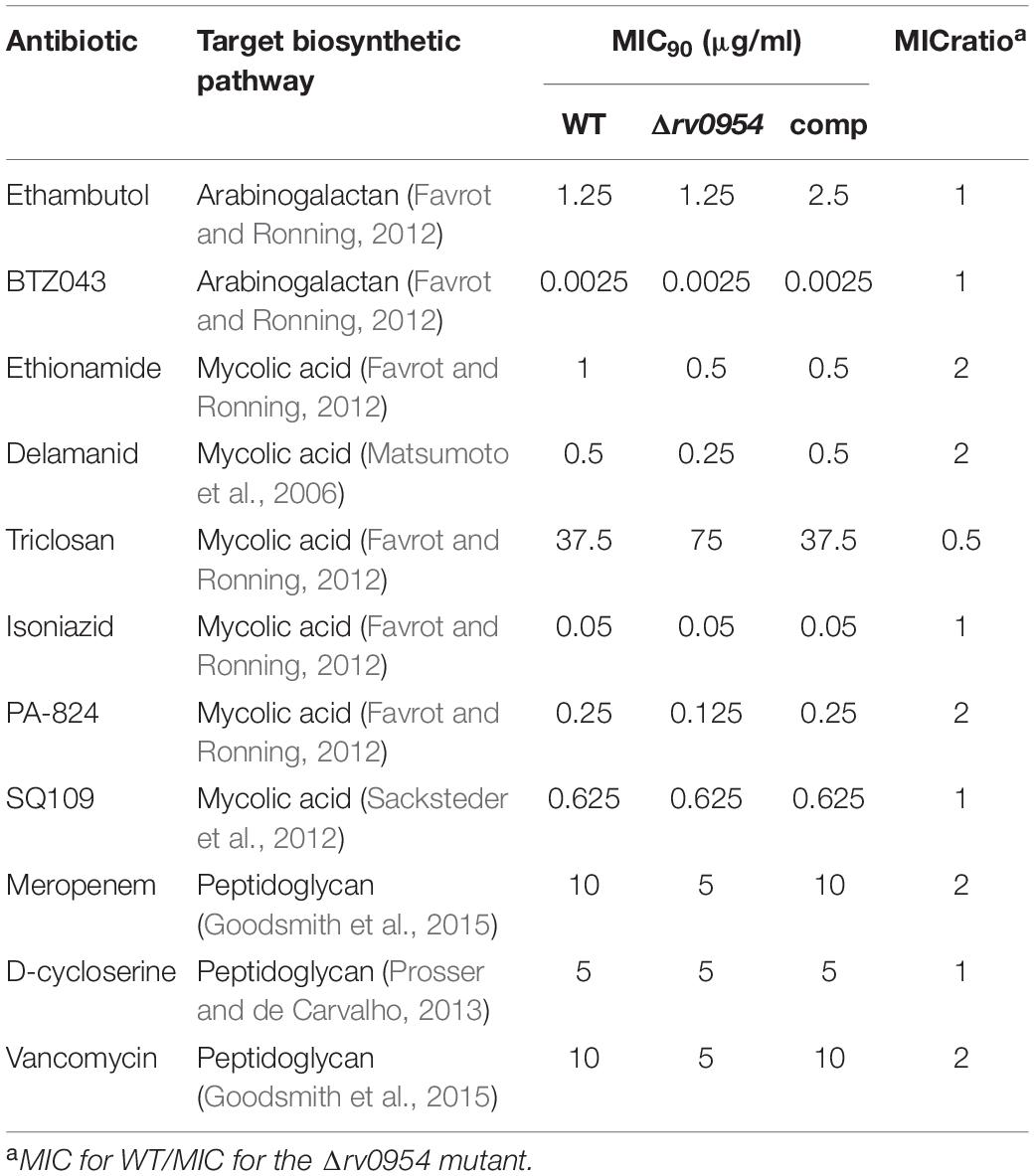
Rv0954 is predicted to be non-essential in Mtb (DeJesus et al., 2017); therefore, we generated a deletion mutant (Supplementary Figure 3). Cells lacking Rv0954 showed no apparent alterations in cell morphology or septum formation (Figure 4A). Deleting perM, which codes for a cell division component and is in an operon with rv0954, led to growth defects in acidic pH and in reduced Mg2+; thus, we tested Δrv0954 in the two conditions (Goodsmith et al., 2015; Wang et al., 2019). Acidification slowed Mtb growth, but the Δrv0954 mutant showed no growth defect relative to WT Mtb (Figure 4B). The Δrv0954 mutant also replicated comparably to WT in medium with reduced Mg2+ (Figure 4C). Moreover, Δrv0954 tolerated cell envelope targeting compounds similarly to WT (Table 2). These results indicate that the deletion of rv0954 does not result in significant defects in cell division or cell wall biosynthesis.
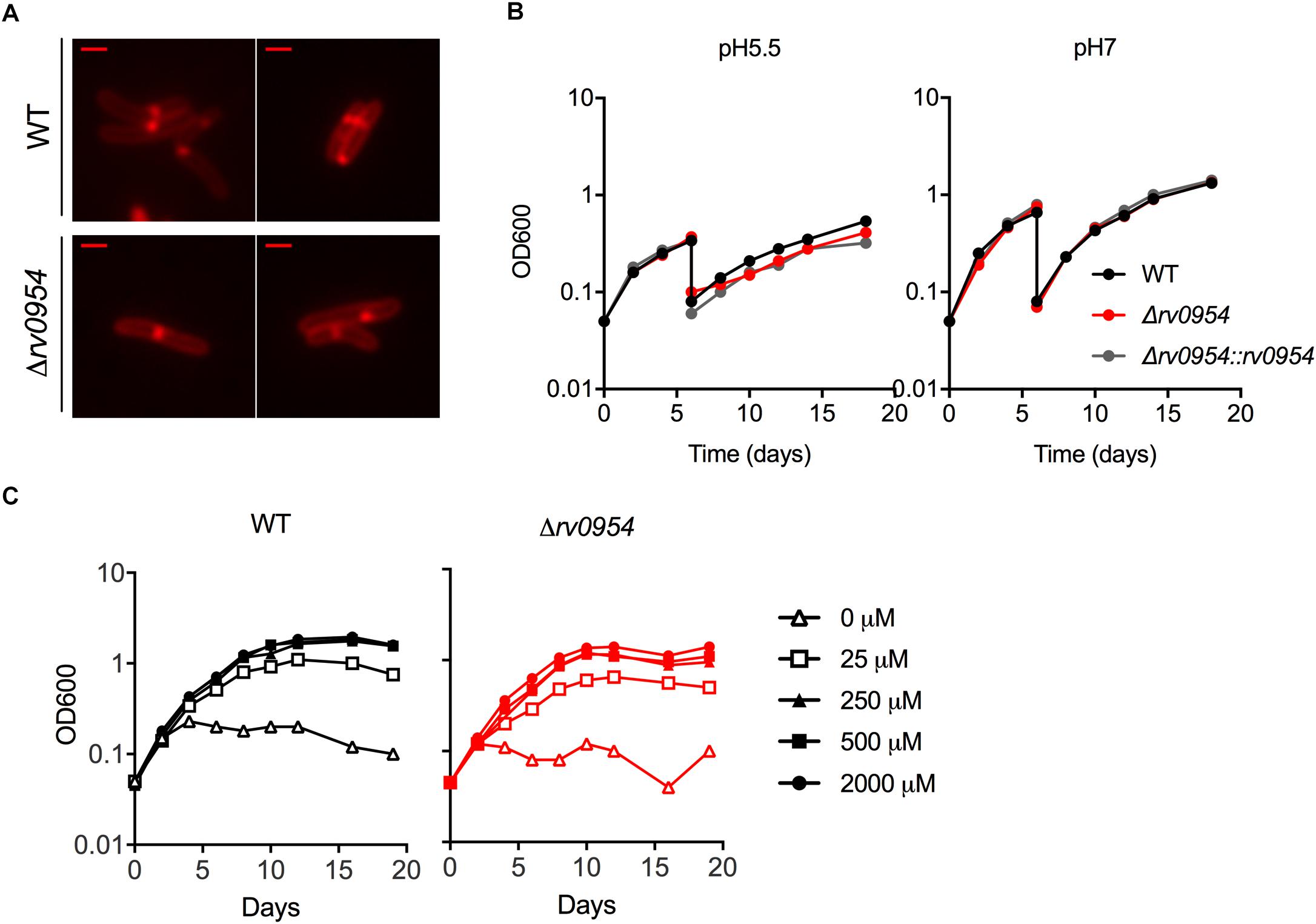
Figure 4. Characterization of Mtb Δrv0954. (A) Morphology of replicating WT and Δrv0954 cultured in 7H9 broth. The bacterial cells were collected and stained with FM5-95 before imaging. Scale bar, 1 μm. (B) Growth curves of Mtb strains sequentially cultured in 7H9 medium adjusted to pH5.5 or pH7. The cultures were diluted in the same medium on day 6. (C) Growth curves of Mtb incubated in chelated Sauton’s medium supplemented with Mg2+ at the indicated concentrations.
Transposon Sequencing (Tnseq) Experiments Revealed rv0954’s Genetic Interactions With Genes Encoding Integral Membrane Proteins
The lack of phenotypes associated with the deletion of rv0954 in Mtb suggested redundancy in the genome, as is the case for many other cell division components in bacteria, including mycobacteria (Kieser and Rubin, 2014). We expect genes of overlapping functions with rv0954 to be more critical for Mtb growth and survival when rv0954 is missing, and therefore transposon mutants of such genes should be underrepresented in the Δrv0954 mutant relative to WT Mtb.
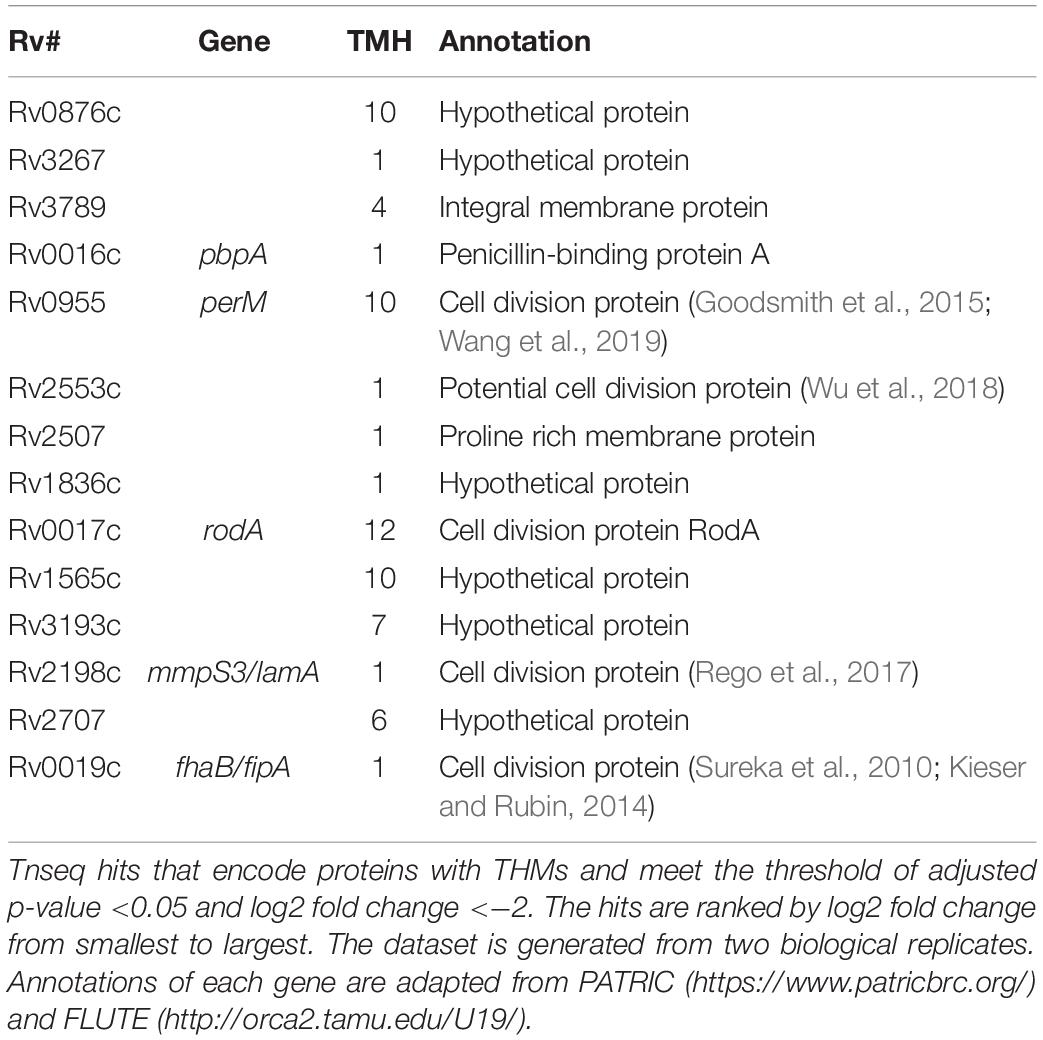
Four of the top 18 hits underrepresented in the Δrv0954 mutant (Table 3) were involved in either cell division or cell wall biosynthesis. PimE (encoded by rv1159) is a mannosyltransferase involved in the biosynthesis of phosphatidylinositol mannosides (PIMs), glycolipids in the cell wall of mycobacteria (Morita et al., 2006; Crellin et al., 2008). The deletion of pimE in Msm is associated with changes in the cell envelope’s structural integrity and increased sensitivity to toxic compounds, such as certain antibiotics and copper (Morita et al., 2006; Eagen et al., 2018). Rv0016c encodes PbpA, a transpeptidase involved in peptidoglycan crosslinking during cell division (Kieser and Rubin, 2014; Viswanathan et al., 2017), and PerM is a divisome component (Goodsmith et al., 2015; Wang et al., 2019). Rv2553 encodes a putative transglycosylase, and its Msm homolog localized to the septum and poles and interacted with FtsQ (Wu et al., 2018). We further analyzed underrepresented hits that encode integral membrane proteins (Figure 5 and Table 4). Six out of the 14 hits: pbpA, perM, rodA, rv2553c, mmpS3/lamA, and fhaB/fipA encode cell division-related proteins (Sureka et al., 2010; Kieser and Rubin, 2014; Goodsmith et al., 2015; den Blaauwen et al., 2017; Rego et al., 2017; Viswanathan et al., 2017; Arora et al., 2018; Wang et al., 2019). The increased requirements of these genes when Rv0954 is missing supports our hypothesis that Rv0954 is involved in cell division or cell wall biosynthesis.
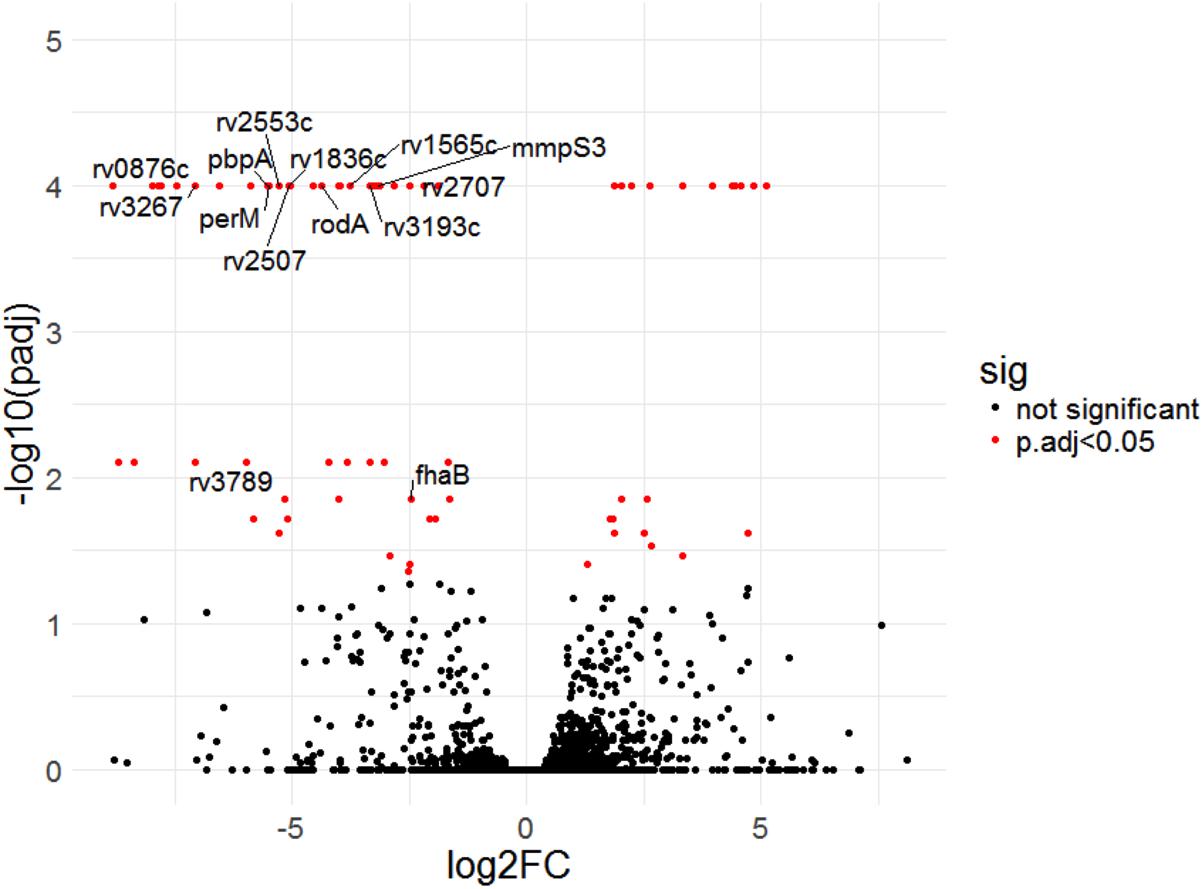
Figure 5. Transposon sequencing (Tnseq) experiments revealed rv0954’s genetic interactions with genes encoding integral membrane proteins. Log2 fold change of transposon insertions in WT and the Δrv0954 mutant (Δrv0954/WT) plotted against adjusted p-values for each genetic locus. Loci with an adjusted p-value of <0.05 are colored in red. The dataset was generated from biological duplicates. Annotations of each genetic locus are from publicly available databases PATRIC (https://www.patricbrc.org/) and FLUTE (http://orca2.tamu.edu/U19/). The significantly underrepresented hits encoding integral membrane proteins are indicated by name or gene number.
Rv0954 Is Phosphorylated in Mtb
The interaction between Rv0954 and PknH indicated that Rv0954 might undergo phosphorylation (Supplementary Figure 2). To test this hypothesis, we treated protein lysates with phosphatase and examined Rv0954’s migration pattern by western blotting (Figure 6A). Rv0954 migrated as double bands without phosphatase treatment; incubation with alkaline phosphatase down-shifted both bands, indicating that Rv0954 may be subject to phosphorylation and additional modifications, such as acetylation, as previously reported (Xie et al., 2015). The control protein Rv0060 showed no alteration in migration patterns after the same treatment.
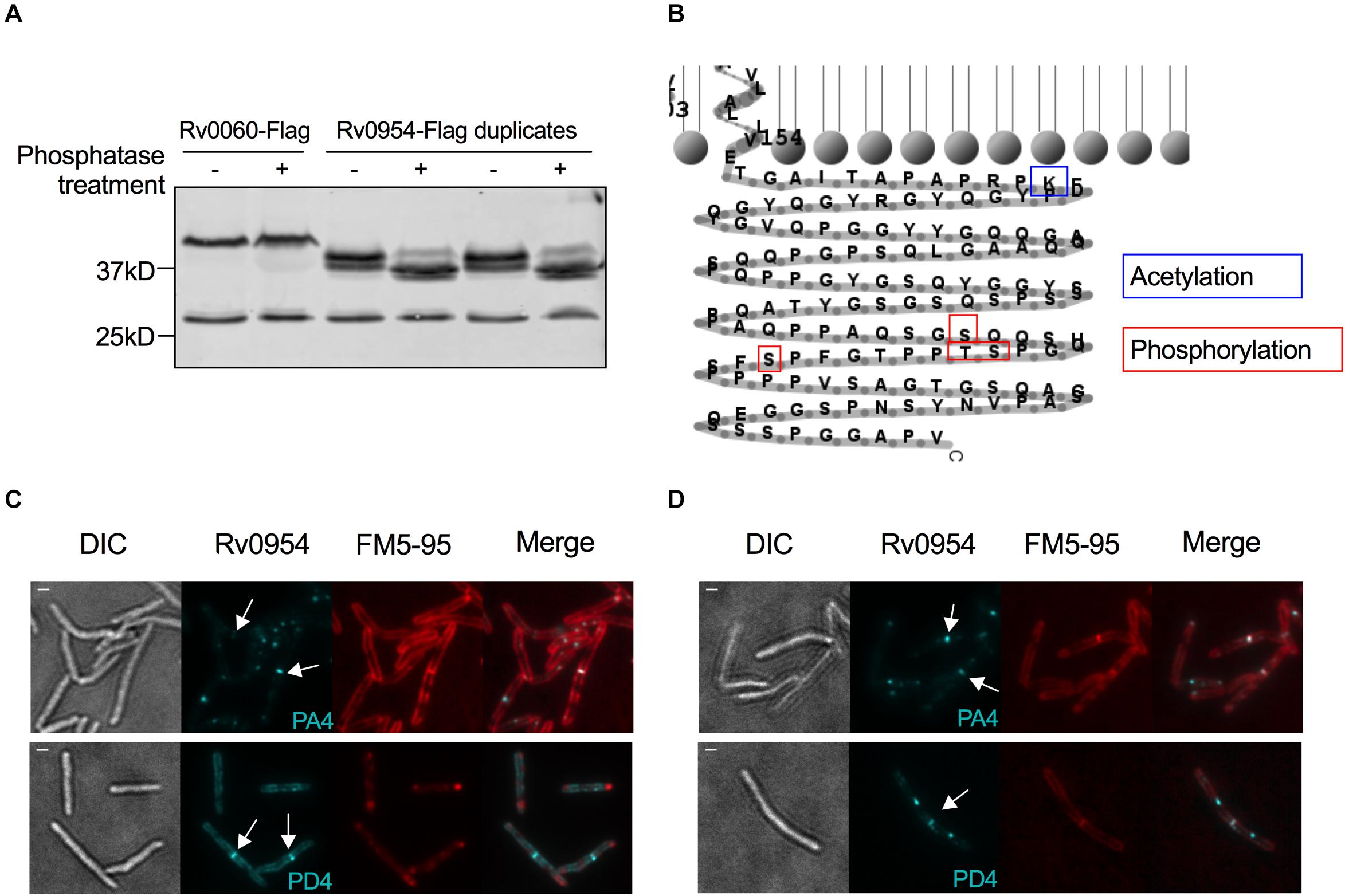
Figure 6. Rv0954 is phosphorylated. (A) Detection of Rv0060-Flag (42 kD) and Rv0954-Flag (34 kD) in Mtb whole-cell protein lysates after treatment or not with alkaline phosphatase. The western blot was probed with an anti-Flag antibody. Data are representative of two independent experiments. (B) Schematic of the previously reported acetylation (Xie et al., 2015) and newly discovered phosphorylation sites in the C-terminus of Rv0954. (C,D) Representative microscopy images of phosphoablative (PA4) or phosphomimetic (PD4) Rv0954 mutants expressed in panel (C). WT Msm, or (D) Msm ΔMSEMG_5518-perMmsm:perMmtb. Msm cells were collected and stained with membrane dye FM5-95 before imaging. Arrows point to the mid-cell localization of Rv0954. Scale bar, 1 μm.
We found four phosphorylation sites in the C-terminus of Rv0954 by mass spectrometry: T257, S248, S256, and S264 (Figure 6B). We mutated all four sites to generate phospho-ablative PA4 (Thr/Ser to Ala) or phospho-mimetic PD4 (Thr/Ser to Asp) mutants and evaluated the impact of phosphorylation on Rv0954. Both phospho mutants retained septal localization, either when expressed in addition to the WT MSMEG_5518 homologous allele (Figure 6C) or as the sole source of Rv0954 or MSMEG_5518 (Figure 6D). Our data indicate that Rv0954 phosphorylation does not affect cell division in standard growth conditions yet may play a role in regulating cell division in specific, to be determined physiological conditions.
Discussion
There are several possible roles that Rv0954 could play during cell division. With four predicted TMHs and a stretched C-terminal domain, Rv0954 could serve as a structural scaffold to recruit additional division proteins. Protein co-localization experiments (Figures 1, 2) showed that Rv0954 lingers at mid-cell from before cytokinesis to cell constriction, possibly recruiting late division proteins such as LamA, which localizes to the mid-cell after FtsZ (Rego et al., 2017), and PbpA. Another possible role of Rv0954 is to anchor intracellular divisome components to the inner cell membrane, similar to the role of ZipA in E. coli. In fact, like the proline/glutamine-rich intracellular tail of Rv0954, the cytoplasmic domain of ZipA contains a region that is made of 31% proline and 23% glutamine (Hale and de Boer, 1997). This region may form a long rigid linker to protrude into the cytoplasm for some distance and is proposed to facilitate interaction with FtsZ (Hale and de Boer, 1997). Mycobacteria lack homologs of ZipA, and Rv0954 is unlikely to fill the same role given that FtsZ localizes to the mid cell prior to Rv0954. However, it is plausible that a cytoplasmic division protein other than FtsZ needs membrane attachment via Rv0954.
Rv0954 is phosphorylated in Mtb, possibly by its interaction partner PknH (Figure 3, Table 1, and Supplementary Figure 2). Phosphorylation of division proteins is a major mechanism for mycobacteria to regulate cell division (Kieser and Rubin, 2014; Baranowski et al., 2019). Protein phosphorylation may alter its activity, localization, or interaction with other proteins. The Mtb phospho mutants did not display morphological abnormalities and Rv0954 accumulated at the mid-cell with and without phosphorylation (Figure 6), suggesting that phosphorylation may play an accessory role during normal growth. Alternative explanations include additional phosphorylation sites that exist within Rv0954 or phenotypes masked by proteins with overlapping functions.
It is common to find redundancies within the bacterial divisome (de Boer, 2010; Kieser and Rubin, 2014). E. coli, for instance, expresses ZipA and FtsA that have overlapping roles in membrane attachment of FtsZ; the Z-ring could form when either ZipA or FtsA was present, but not when both were inactivated (Pichoff and Lutkenhaus, 2002). Functional redundancy may explain Δrv0954’s lack of phenotypes in cell morphology, drug susceptibility, and growth in acidic pH or low magnesium. These results are in contrast to the phenotypes observed when perM, which is part of the same operon, was deleted (Goodsmith et al., 2015; Wang et al., 2019). Although we cannot exclude the possibility that rv0954 may have an essential role in vivo, our current data did not motivate infection experiments. Comparing transposon insertion profiles of the Δrv0954 mutant to WT Mtb could help reveal overlapping pathways. We detected a reduced number of insertions in several genes that encode integral membrane proteins (Table 4). These include six related to cell division and eight of unknown functions. Future studies of these hits might reveal additional mycobacterial division factors and unmask Rv0954’s functions.
Two cell division genes, pbpA and lamA, contained fewer transposon insertions in bacteria lacking Rv0954 than WT. PbpA and LamA also physically interacted with Rv0954 in the Co-IP experiment (Figure 3 and Table 1). PbpA is a transpeptidase that synthesizes the septal peptidoglycan; cells lacking PbpA were hypersusceptible to β-lactams (Arora et al., 2018). PbpA interacted with FhaA and CrgA, both are related to peptidoglycan biosynthesis and cell shape maintenance (Plocinski et al., 2011; Viswanathan et al., 2017). LamA is a highly conserved divisome component across mycobacterial species. It inhibits growth at the new cell poles and creates asymmetry. When treated with cell wall-targeting antibiotics such as rifampicin, cells lacking LamA were killed more uniformly and faster (Rego et al., 2017; Logsdon and Aldridge, 2018). Rv0954 mutant did not show increased sensitivity to cell wall-targeting compounds (Table 2), likely because of the functional overlap among divisome components and the complexity in cell division regulation. Future work on the mechanisms by which Rv0954 interacts with PbpA and LamA could help us understand how mycobacteria integrate growth and cell cycle and prioritize antitubercular targets.
The mycobacterial cell wall is the first barrier that protects bacteria from host stresses and antibiotics. Maintenance of cell wall integrity requires properly regulated cell division. Our genetic and biochemical study of a previously uncharacterized protein Rv0954, including its mid-cell localization, physical interactors, mutant phenotypes, and protein modifications, supported Rv0954 as a new component of the mycobacterial divisome.
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA768285; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA678285.
Author Contributions
RW designed and performed experiments, analyzed data, and drafted the manuscript. SE supervised the project, acquired funding, and reviewed and edited the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by NIH grants U19 AI111143 and P01 AI143575.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. G. Papavinasasundaram (University of Massachusetts) and J. Fernandez (Rockefeller University) for LC-MS/MS analyses. We thank A. North (Rockefeller University) for microscopy training and advice. We also thank M. Whitaker and S. Schrader for carefully editing the first draft. L. Chan and C. Harris helped with molecular cloning. Figure 3 was created with https://flourish.studio/.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.626461/full#supplementary-material
References
Abramoff, M., Magalhães, P., and Ram, S. J. (2003). Image Processing with ImageJ. Biophotonics Int. 11, 36–42.
Aldridge, B. B., Fernandez-Suarez, M., Heller, D., Ambravaneswaran, V., Irimia, D., Toner, M., et al. (2012). Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335, 100–104. doi: 10.1126/science.1216166
Arora, D., Chawla, Y., Malakar, B., Singh, A., and Nandicoori, V. K. (2018). The transpeptidase PbpA and noncanonical transglycosylase RodA of Mycobacterium tuberculosis play important roles in regulating bacterial cell lengths. J. Biol. Chem. 293, 6497–6516. doi: 10.1074/jbc.M117.811190
Baranowski, C., Rego, E. H., and Rubin, E. J. (2019). The Dream of a Mycobacterium. Microbiol. Spectr. 7, G3–G0008. doi: 10.1128/microbiolspec.GPP3-0008-2018
Botella, H., Yang, G., Ouerfelli, O., Ehrt, S., Nathan, C. F., and Vaubourgeix, J. (2017). Distinct Spatiotemporal Dynamics of Peptidoglycan Synthesis between Mycobacterium smegmatis and Mycobacterium tuberculosis. MBio 8, e01183–17. doi: 10.1128/mBio.01183-17
Cameron, T., Zupan, J., and Zambryski, P. (2015). The essential features and modes of bacterial polar growth. Trends Microbiol. 23, 347–353. doi: 10.1016/j.tim.2015.01.003
Crellin, P. K., Kovacevic, S., Martin, K. L., Brammananth, R., Morita, Y. S., Billman-Jacobe, H., et al. (2008). Mutations in pimE Restore Lipoarabinomannan Synthesis and Growth in a Mycobacterium smegmatis lpqW Mutant. J. Bacteriol. 190, 3690–3699. doi: 10.1128/JB.00200-08
de Boer, P. A. J. (2010). Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13, 730–737. doi: 10.1016/j.mib.2010.09.015
DeJesus, M. A., Gerrick, E. R., Xu, W., Park, S. W., Long, J. E., Boutte, C. C., et al. (2017). Comprehensive Essentiality Analysis of the Mycobacterium tuberculosis Genome via Saturating Transposon Mutagenesis. MBio 8, e02133–16. doi: 10.1128/mBio.02133-16
den Blaauwen, T., Hamoen, L. W., and Levin, P. A. (2017). The divisome at 25: the road ahead. Curr. Opin. Microbiol. 36, 85–94. doi: 10.1016/j.mib.2017.01.007
Eagen, W. J., Baumoel, L. R., Osman, S. H., Rahlwes, K. C., and Morita, Y. S. (2018). Deletion of PimE mannosyltransferase results in increased copper sensitivity in Mycobacterium smegmatis. FEMS Microbiol. Lett. 365:29390083. doi: 10.1093/femsle/fny025
Favrot, L., and Ronning, D. R. (2012). Targeting the mycobacterial envelope for tuberculosis drug development. Expert Rev. Anti Infec. Ther. 10, 1023–1036. doi: 10.1586/eri.12.91
Gee, C. L., Papavinasasundaram, K. G., Blair, S. R., Baer, C. E., Falick, A. M., King, D. S., et al. (2012). A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci. Signal. 5:ra7. doi: 10.1126/scisignal.2002525
Goodsmith, N., Guo, X. V., Vandal, O. H., Vaubourgeix, J., Wang, R., Botella, H., et al. (2015). Disruption of an M. tuberculosis Membrane Protein Causes a Magnesium-dependent Cell Division Defect and Failure to Persist in Mice. PLoS Pathog. 11:e1004645. doi: 10.1371/journal.ppat.1004645
Hale, C. A., and de Boer, P. A. (1997). Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185. doi: 10.1016/s0092-8674(00)81838-3
Hirota, Y., Ryter, A., and Jacob, F. (1968). Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb. Symp. Quant. Biol. 33, 677–693. doi: 10.1101/sqb.1968.033.01.077
Kieser, K. J., and Rubin, E. J. (2014). How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol. 12, 550–562. doi: 10.1038/nrmicro3299
Logsdon, M. M., and Aldridge, B. B. (2018). Stable Regulation of Cell Cycle Events in Mycobacteria: insights From Inherently Heterogeneous Bacterial Populations. Front. Microbiol. 9:514. doi: 10.3389/fmicb.2018.00514
Marmiesse, M., Brodin, P., Buchrieser, C., Gutierrez, C., Simoes, N., Vincent, V., et al. (2004). Macro-array and bioinformatic analyses reveal mycobacterial “core” genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150, 483–496. doi: 10.1099/mic.0.26662-0
Matsumoto, M., Hashizume, H., Tomishige, T., Kawasaki, M., Tsubouchi, H., Sasaki, H., et al. (2006). OPC-67683, a Nitro-Dihydro-Imidazooxazole Derivative with Promising Action against Tuberculosis In Vitro and In Mice. PLoS Med. 3:e466. doi: 10.1371/journal.pmed.0030466
Meniche, X., Otten, R., Siegrist, M. S., Baer, C. E., Murphy, K. C., Bertozzi, C. R., et al. (2014). Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Pro. Natl. Acad. Sci. U. S. A. 111, E3243–E3251. doi: 10.1073/pnas.1402158111
Morita, Y. S., Sena, C. B. C., Waller, R. F., Kurokawa, K., Sernee, M. F., Nakatani, F., et al. (2006). PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J. Biol. Chem. 281, 25143–25155. doi: 10.1074/jbc.M604214200
Pichoff, S., and Lutkenhaus, J. (2002). Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21, 685–693. doi: 10.1093/emboj/21.4.685
Plocinski, P., Ziolkiewicz, M., Kiran, M., Vadrevu, S. I., Nguyen, H. B., Hugonnet, J., et al. (2011). Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J. Bacteriol. 193, 3246–3256. doi: 10.1128/JB.00188-11
Prosser, G. A., and de Carvalho, L. P. S. (2013). Metabolomics Reveal d-Alanine:d-Alanine Ligase As the Target of d-Cycloserine in Mycobacterium tuberculosis. ACS Med. Chem. Lett. 4, 1233–1237. doi: 10.1021/ml400349n
Rego, E. H., Audette, R. E., and Rubin, E. J. (2017). Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 546, 153–157. doi: 10.1186/s40945-017-0033-9.Using
Sacksteder, K. A., Protopopova, M., Barry, C. E., Andries, K., and Nacy, C. A. (2012). Discovery and development of SQ109: a new antitubercular drug with a novel mechanism of action. Future Microbiol. 7, 823–837. doi: 10.2217/fmb.12.56
Santi, I., Dhar, N., Bousbaine, D., Wakamoto, Y., and McKinney, J. D. (2013). Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat. Commun. 4:2470. doi: 10.1038/ncomms3470
Sharma, K., Gupta, M., Pathak, M., Gupta, N., Koul, A., Sarangi, S., et al. (2006). Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 188, 2936–2944. doi: 10.1128/JB.188.8.2936-2944.2006
Sureka, K., Hossain, T., Mukherjee, P., Chatterjee, P., Datta, P., Kundu, M., et al. (2010). Novel Role of Phosphorylation-Dependent Interaction between FtsZ and FipA in Mycobacterial Cell Division. PLoS One 5:e8590. doi: 10.1371/journal.pone.0008590
Viswanathan, G., Yadav, S., Joshi, S. V., and Raghunand, T. R. (2017). Insights into the function of FhaA, a cell division-associated protein in mycobacteria. FEMS Microbiol. Lett. 364:28013239. doi: 10.1093/femsle/fnw294
Wang, R., Kreutzfeldt, K., Botella, H., Vaubourgeix, J., Schnappinger, D., and Ehrt, S. (2019). Persistent Mycobacterium tuberculosis infection in mice requires PerM for successful cell division. ELife 8:e49570. doi: 10.7554/eLife.49570
Williamson, M. P. (1994). The structure and function of proline-rich regions in proteins. Biochem. J. 297, 249–260. doi: 10.1042/bj2970249
Wu, K. J., Zhang, J., Baranowski, C., Leung, V., Rego, E. H., Morita, Y. S., et al. (2018). Characterization of Conserved and Novel Septal Factors in Mycobacterium smegmatis. J. Bacteriol. 200:29311277. doi: 10.1128/JB.00649-17
Xie, L., Wang, X., Zeng, J., Zhou, M., Duan, X., Li, Q., et al. (2015). Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int. J. Biochem. Cell Biol. 59, 193–202. doi: 10.1016/j.biocel.2014.11.010
Xu, W., DeJesus, M. A., Rücker, N., Engelhart, C. A., Wright, M. G., Healy, C., et al. (2017). Chemical Genetic Interaction Profiling Reveals Determinants of Intrinsic Antibiotic Resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 61, e01334–17. doi: 10.1128/AAC.01334-17
Keywords: Mycobacterium tuberculosis, cell division, divisome, phosphorylation, Tnseq
Citation: Wang R and Ehrt S (2021) Rv0954 Is a Member of the Mycobacterial Cell Division Complex. Front. Microbiol. 12:626461. doi: 10.3389/fmicb.2021.626461
Received: 05 November 2020; Accepted: 15 March 2021;
Published: 20 April 2021.
Edited by:
Hari S. Misra, Bhabha Atomic Research Centre (BARC), IndiaReviewed by:
Vinay Kumar Nandicoori, National Institute of Immunology (NII), IndiaMurty Madiraju, The University of Texas Health Science Center at Tyler, United States
Copyright © 2021 Wang and Ehrt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sabine Ehrt, c2FlMjAwNEBtZWQuY29ybmVsbC5lZHU=
†Present address: Ruojun Wang, Department of Molecular Biology, Princeton University, Princeton, NJ, United States
 Ruojun Wang
Ruojun Wang Sabine Ehrt
Sabine Ehrt