
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 February 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.625488
Adult rumen fluid inoculations have been considered to facilitate the establishment of rumen microbiota of pre-weaned dairy calves. However, the sustained effects of the inoculations remain to be explored. In our previous study, 20 pre-weaned dairy calves had been dosed with four types of adult rumen inoculums [autoclaved rumen fluid, bacterial-enriched rumen fluid (BE), protozoal-enriched (PE), and BE + PE] weekly at 3 to 6 weeks of age. To verify the sustained effect of adult rumen inoculation, the rumen bacterial communities, fermentation characteristics, and animal performance measurements were measured after sacrifice from 20 post-weaned dairy bull calves (9 weeks of age). Ruminal pH tended to be lower in BE treated calves (n = 10). All PE treated calves had rumen ciliates (>104 cells per ml of rumen fluid). PE treated calves had greater VFA concentrations (P = 0.052), lower molar proportions of isobutyrate (P = 0.073), and butyrate (P = 0.019) compared to those of control calves. No treatment differences were found in all animal performance measurements. Both PE and BE inocula increased bacterial species richness, Faith’s phylogenetic diversity, and Shannon’s index in rumen liquid fractions. However, the relative proportion of those bacterial taxa possibly transferred from the donor’s rumen was minor. Microbial network analysis showed different co-occurrence and mutually exclusive interactions between treatments of microbial inoculations. Collectively, adult rumen inoculations in pre-weaned dairy calves slightly altered the rumen bacteriome of post-weaned calves without changing fermentation and animal performance.
The rumen microbial community of an adult dairy cow consists of anaerobic bacteria, protozoa, fungi, archaea and harmless viruses. The bacteria, fungi, and protozoa convert nutrients into volatile fatty acids (VFA) as the main source of energy for the host, while the archaea utilize fermentation by-products to produce methane. Dairy calves are born with a sterile, undeveloped rumen, and the initial colostrum and subsequent milk they consume, bypasses the rumen via the esophageal groove to the abomasum (Church, 1976; Rey et al., 2014; Meale et al., 2016). At and after birth, dairy calves acquire fibrolytic bacteria and methanogenic archaea in their gastrointestinal tracts through contact with the vaginal, mammary gland, colostrum, and milk microbial communities (Vi et al., 2004; Guzman et al., 2015). With the onset of solid feed intake, increases in ruminal mass, papillae growth, and fermentation and further establishment of the rumen microbial ecology occur (Vi et al., 2004; Roth et al., 2009; Dill-Mcfarland et al., 2017).
Several studies have demonstrated the resiliency and host-specificity of the rumen microbial ecology of adult dairy cows, suggesting that the established rumen microbial environment is resistant to manipulative strategies (Jewell et al., 2015; Weimer, 2015; Malmuthuge and Guan, 2017). In contrast, the period of early ruminal development shows potential to be a favorable time to manipulate and direct the rumen environment (Yáñez-Ruiz et al., 2015). Direct-fed microbials, early nutrition, and artificial dosing with ruminal contents from adults have been utilized as strategies to direct the microbial ecology of the gastrointestinal tract of young ruminants and subsequent production performance (Schönhusen et al., 2003; Ishaq et al., 2015; Fouladgar et al., 2016; Zhang et al., 2017; Li et al., 2019). Li and co-authors found that separate ruminal wall microbial communities formed between dairy calves dosed with and without adult-derived rumen fluid (Li et al., 2019). These results were interpreted to support the manipulation of early, artificial dosing on the microbial community in ruminal wall. Additionally, Ishaq et al. demonstrated that moose-derived fibrolytic bacteria inoculation increased dietary efficiency of lambs (Ishaq et al., 2015).
In our previous study investigating the effect of adult-derived rumen fluid inoculation, two different microbial inocula (bacterial- or protozoal-enriched; BE or PE, respectively) were dosed in pre-weaned dairy calves to evaluate the effect of specific rumen microbiota on host growth responses, ruminal fermentation, and bacterial community composition (Cersosimo et al., 2019). We found that inoculation resulted in minor changes in some bacterial abundances and were accompanied with altered ruminal ammonia and butyrate concentrations, although calf health and growth were unaffected (Cersosimo et al., 2019). However, the effect of altered microbial community by inoculation might be accentuated after accelerated solid feeding that occurs after weaning. Furthermore, in our previous study, only 60% of the dairy calves treated with PE inoculum had a consistent rumen protozoa population pre-weaning (Cersosimo et al., 2019). Successful establishment of the protozoal population was hypothesized to induce shifts in the rumen microbiome and fermentation profile in post-weaned calves, though currently there is limited information about a potential carry-over effect. With the post-weaning period as the focus of this study, our objective was to investigate if the BE and PE microbial inoculations administered at 3–6 weeks of age during the pre-weaning period (Cersosimo et al., 2019) would affect rumen fermentation characteristics, rumen tissue (e.g., papillae length), and bacterial community structures of post-weaned, dairy bull calves (sacrificed at 9 weeks).
This work is a continuation of our previously published work (Cersosimo et al., 2019). All animal procedures were approved by The University of Wisconsin’s Institutional Animal Care and Use Committee under protocol A005829. Holstein bull calves (n = 20) were enrolled into the study at birth over a 4-week period from July to August 2017. Calves were removed from their dam at birth, received colostrum within 4 h after birth, and were housed in individual calf hutches at the US Dairy Forage Research Farm in Prairie du Sac, WI. Calves were randomly assigned to a 2 × 2 factorial design with 4 different types of rumen inocula. Briefly, treatments previously described in Cersosimo et al., 2019, included 50 mL autoclaved, clarified rumen fluid (RF), PE, BE or 50 mL of each BE and PE. Five of the dairy calves were assigned to each treatment. Calves received 2.5 L pasteurized, antibiotic-free waste milk 3 × daily and were offered Vita Plus BSF 18 texturized calf starter (Vita Plus Corp., Madison, WI) at 6 d of age that contained shell corn, soybean meal, cottonseed hulls, kibbled corn, cane molasses, and heat processed soybeans. As-fed starter and refusals were measured on a daily basis from d 6 to 9 weeks of age. Calves were weaned at 7 weeks of age, thereafter their diet was only comprised of the texturized calf starter. Pre-weaned calves were orally dosed once per week with treatment inocula at 3 to 6 weeks of age. Dose administration was followed by 50 mL 0.7% sterile saline to clear the tube of residual inocula. Specific tubes were designated by inoculum type to avoid cross contamination of treatments.
The stomach compartments (reticulorumen, omasum, and abomasum) were harvested after euthanasia by penetrating captive bolt followed by exsanguination at 9 weeks of age at the University of Wisconsin Meat Science Laboratory. Each stomach compartment was isolated from each other with zip ties and weighed. Ruminal pH was measured in situ and whole rumen contents were collected from each rumen compartment and squeezed through four layers of cheesecloth to obtain a total of 50 mL of fluid for volatile fatty acid, ammonia, protozoal identification, and bacterial community analyses. After squeezing whole rumen contents through cheesecloth, 20–30 g of rumen solids were collected and stored at -80°C for bacterial community analyses. Complete ruminal contents were collected to measure rumen dry matter contents. Each rumen was washed to remove sand and any remaining feed particles before tissue sampling. A total of three 1 × 1 cm tissue samples were collected from the caudal ventral region of the rumen. The samples were stored in 10% formalin before measuring papillae length and number, and rumen wall width. Papillae length and rumen wall width were measured with digital calipers.
Individual calf body length, weight, paunch and cardiac girths, wither and hip heights were measured once per week, from 1–9 weeks of age. Calf body measurements from 1–6 weeks of age were previously reported in Cersosimo et al., 2019.
A total of 5 mL of strained rumen fluid fixed in 50% formalin (v/v) at room temperature was collected for protozoal quantification, 5 mL for DNA extractions, and 5 mL with 0.1 mL 50% H2SO4 immediately frozen at -80°C for NH3 and VFA analyses, with the remaining 35 mL frozen and saved as extra rumen fluid at -80°C. A total of 4 mL of acidified rumen fluid was centrifuged at 30,000 × g at 4°C for 30 min and the supernatant was subsequently collected and stored at -20°C for VFA and NH3 analyses. Gas-liquid chromatography and Lachat methods, as previously described by Paula et al. (2018), were used to analyze VFA and NH3, respectively. Protozoa were detected using the microscopy methods outlined by Dehority (1984, 1993) and modified by Cersosimo et al. (2019).
Rumen fluid, BE and PE inocula were thawed and centrifuged at 10,000 × g for 30 min at 4°C to retain the microbial pellet for the DNA extraction method (Yu and Morrison, 2004). For the rumen solids, a total of 30 mL sterile 0.9% NaCl (w/v) kept cold at 4°C was added to 10 g of rumen solids in a sterile Stomacher bag with a strainer to remove solid material (feed, hair, sand from bedding) and obtain microbial cells. The bag was placed and homogenized in a Stomacher (Seward, West Sussex, BN) on full speed for 2 min. and a total of 20 mL of the strained liquid was collected into a sterile conical tube and kept cold on ice. The liquid obtained from the solid sample was centrifuged at 500 × g for 15 min at 4°C to remove residual particles. The supernatant was centrifuged at 10,000 × g for 30 min at 4°C to obtain the microbial pellet for DNA extraction.
DNA was extracted from the microbial pellets obtained from each rumen fluid and solid fractions, and microbial inocula. DNA concentrations were quantified using the Broad Range kit for the Qubit ®Fluorometer (Invitrogen, San Diego, CA, United States). The methodologies and universal primers outlined previously (Kozich et al., 2013) were used to target the V4 hypervariable region of the bacterial 16S rRNA gene. PCR reaction details, thermal cycler conditions, and gel extraction information were previously described by Cersosimo et al., 2019. The pooled DNA library was sequenced, in-house, with the MiSeq 2 × 250 kit with 500 cycles (Illumina, San Diego, CA, United States) on an Illumina MiSeq. Note that the week 9 post-weaned samples were sequenced in the same sequencing run as samples collected from weeks 3–6 of age. The demultiplexed paired-end reads were processed using QIIME2 plugins (version 2019.10) (Bolyen et al., 2018). Quality filtering (Q-score ≥ 25), denoising, read merging, and chimeric sequence removal were sequentially done using the DADA2 plugin (Callahan et al., 2016). The resulting amplicon sequencing variants (ASVs) were taxonomically classified using the naïve Bayes taxonomy classifier pre-trained on Silva 16S databases (NR 132 version; clustered at 99% similarity). ASVs identified as unassigned, mitochondria, chloroplasts, or archaea were filtered out before downstream analysis. Major classified taxa, which were detected in over 50% of the samples at least one of the treatments, were discussed in this study.
Alpha- and beta-diversity analyses were performed with the rarefied ASV table using the lowest sequence count (14,893 ASVs). Richness (observed ASVs and Chao1 estimates), Evenness, Faith’s phylogenetic diversity, Shannon’s index, and Simpson’s index were calculated from the rarefied ASV table. Beta-diversity shaped by different microbial inoculations was visualized using principal coordinates analysis (PCoA) plots based on the unweighted and weighted UniFrac distances using the QIIME2 emperor plugin (Vázquez-Baeza et al., 2013). Microbial metabolic functions were predicted from 16S ASVs using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) (Douglas et al., 2019). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were reconstructed from predicted KEGG ortholog profiles using the python script implemented in PICRUSt2. The effects on the overall functional profiles were examined based on the relative abundance of KEGG ortholog annotations and then principal components analysis (PCA) plots based on the Bray-Curtis similarity index was generated. The PCA plots were visualized using the R package ggfortify (Tang et al., 2016).
Microbial inoculation effects on the specific bacterial co-occurrence and mutual exclusion network were determined based on the compositional data of major bacterial genera, which were present in over 50% of the samples in at least one of the inoculated treatments. Correlations among major bacterial genera were determined using Sparse Co-occurrence Network Investigation for Compositional data (SCNIC) (https://github.com/shafferm/SCNIC) by computing Spearman correlation coefficients. Among the significant correlations (P < 0.05), specific microbial interactions at each inoculated treatment were defined according to their commonalities calculated using the R package, Co-expression Differential Network Analysis (CoDiNA) (Gysi et al., 2018). Additionally, the number of shared and exclusively found microbial taxa either in rumen liquid or solid between treated and control calves at phylum and genus levels were visualized using Venn diagrams.
Treatment groups were denoted as follows; with or without BE inoculation: BE(+) or BE(−), respectively, and with or without PE inoculation: PE(+) or PE(−), respectively. To compare alpha-diversity measurements between different ruminal fractions, PROC MIXED in SAS 9.3 (SAS Institute Inc., Cary, NC, United States) was used with the fixed effect of ages or ruminal fractions and random effect of calf. Animal performance measurements, fermentation characteristics, protozoal counts, and alpha diversity measurements were statistically analyzed using GLIMMIX procedure of SAS 9.3 with BE, PE and the interaction between BE and PE as fixed effects.
Relative abundance of both the classified microbial taxa and the predicted KEGG pathways were statistically analyzed by non-parametric Kruskal-Wallis test in R (v3.5.0). A PERMANOVA test implemented in PAST3 (Hammer et al., 2001) with 9,999 random permutations was used to check the significance of beta-diversity differences by ruminal fraction types (solid vs. liquid) and microbial inoculation types. Pearson correlation coefficients (correlation coefficient, | r| ≥ 0.5, P ≤ 0.05) between animal performance measurements, stomach compartment weights, fermentation characteristics, and the relative abundance of differentially abundant genera were determined using the PROC CORR procedure in SAS 9.3 and subsequently visualized using the corrplot package in R. Within each exclusive network selected by CoDiNA, the network statistics including the measurements of centrality (i.e., eigenvector centrality and authority) were calculated using the built-in plugins in Gephi (Bastian et al., 2009). Significance was declared at P ≤ 0.05 and trends at 0.05 < P ≤ 0.1.
Stomach compartment weights, ruminal dry matter%, ammonia, and total free amino acids concentrations did not differ by microbial inoculum type (Table 1). Likewise, body measurements and rumen papillae characteristics did not differ by microbial inoculum type (Supplementary Table 1). Omasum wet weights tended to be greater in PE(+) calves than PE(−) (P = 0.094). Ruminal pH tended to be lower in BE(+) calves than BE(−) (P = 0.079; Table 1). Proportions of ruminal butyrate (% of total VFA) were lower in PE(+) calves (5.57 ± 0.29%) than with PE(−) (7.95 ± 0.83%; P = 0.019). Calves treated with PE tended to have lower concentrations of isobutyrate than PE(−) (Table 1). Total ruminal VFA concentrations tended to be higher in PE(+) calves (73.71 ± 4.04 mM) than PE(−) (58.76 ± 5.76 mM; P = 0.052). No differences in total or individual VFA were observed between BE(+) and BE(−) calves. Only PE(+) treated calves had established rumen protozoal populations and mean protozoal counts were 4.34 ± 0.05 log10 counts per ml of rumen fluid. One calf treated with the PE inoculum had 1,000 holotrichs per ml of rumen fluid and entodiniomorphs, while the other 9 calves had entodiniomorphs exclusively.
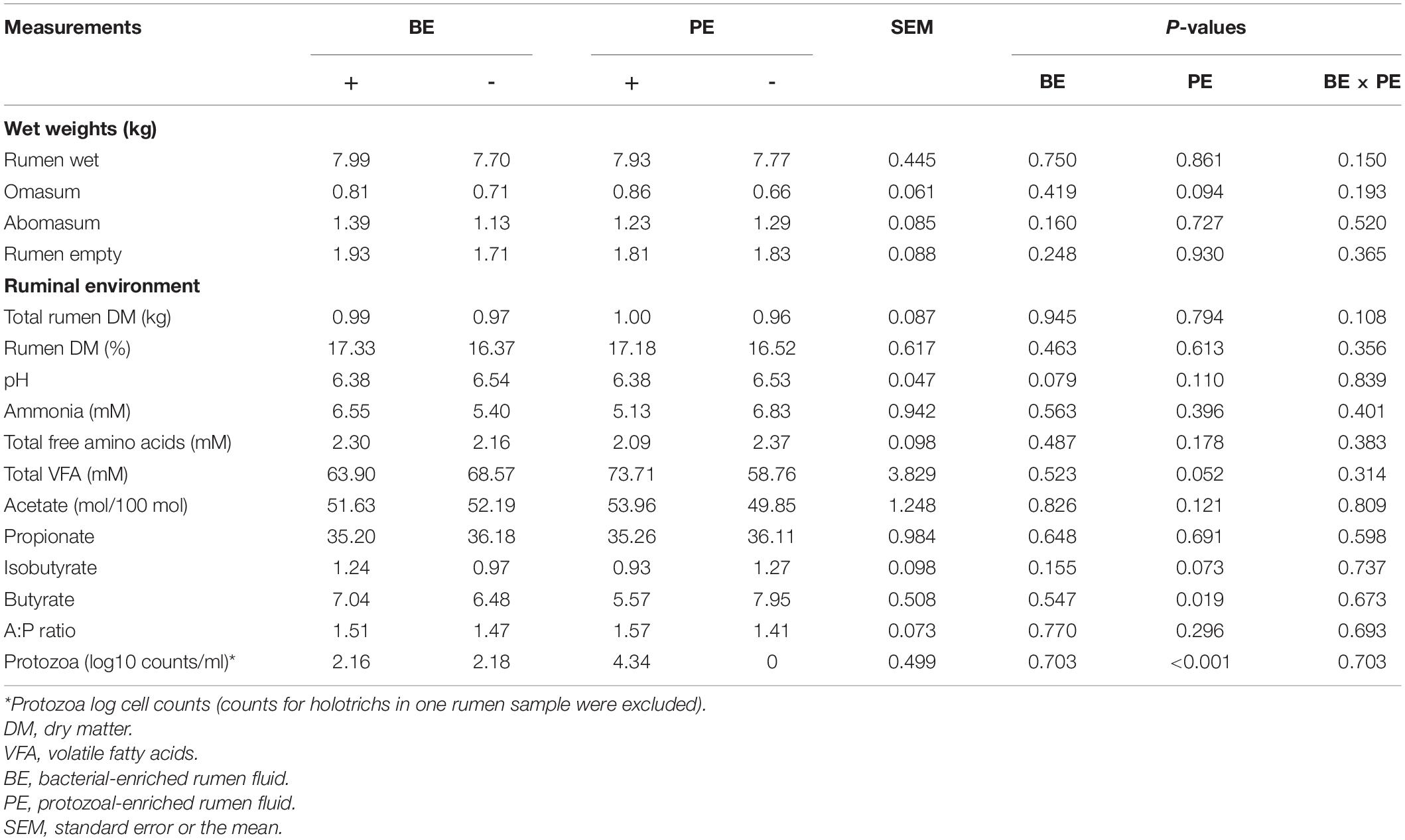
Table 1. The main effects of microbial inoculations on the gut size, ruminal fermentation characteristics, and protozoal counts.
Amplicon sequencing of 40 rumen samples from both the ruminal fractions resulted in a mean of 26,616 sequences per sample (Supplementary Table 2). High-quality ASVs averaging 25,053 per sample were obtained after quality- and taxa-filtering (ranging from 14,893 to 51,348 ASVs per sample) (Supplementary Table 2). Alpha- and beta-diversity analyses were done using a rarefied BIOM table at 14,893 ASVs per sample. Good’s coverage of all the samples was higher than 99.8%. In rumen fluid, species richness, phylogenetic diversity, and Shannon’s index were significantly higher in calves treated with both the BE(+) and PE(+) compared to their controls (P < 0.05) (Table 2). Furthermore, in rumen fluid, PE(+) calves tended to have more evenness (P < 0.1) and had significantly greater Simpson’s index than PE(−) (P < 0.05). In rumen solids, tendencies of greater species richness and phylogenetic diversity were observed between BE(+) and BE(−) calves (P < 0.1), while no alpha-diversity measurements were different between PE(+) and PE(−) calves. Alpha diversity measurements did not differ by fraction type except the evenness which was greater in the solid fraction (Supplementary Table 3). A greater number of ASVs were found in the PE inoculum compared to that of the BE inoculum (P < 0.05) accompanied with the tendency of greater Chao1 estimates in PE inoculum (P = 0.056; Supplementary Table 4).
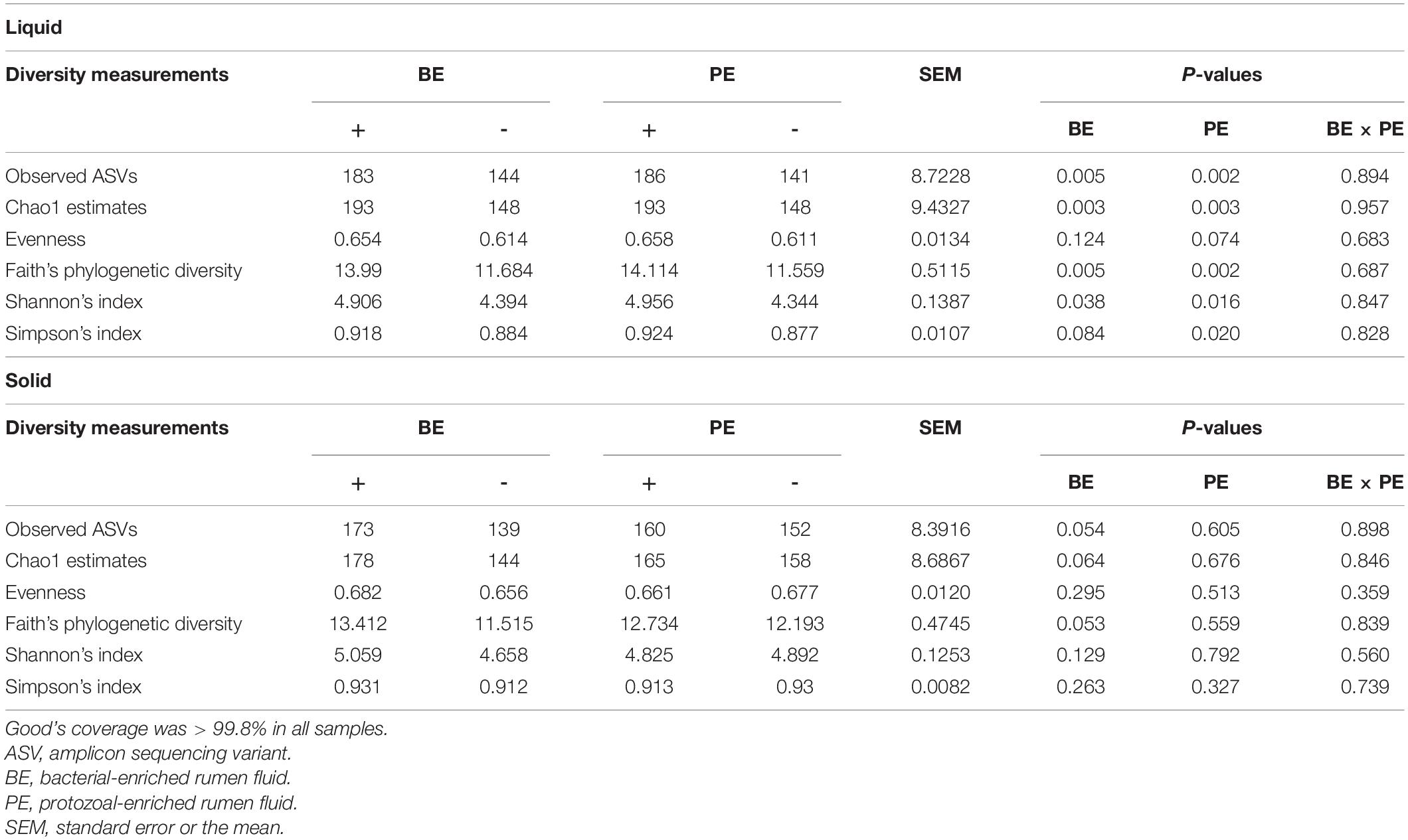
Table 2. The main effects of microbial inoculations on alpha diversity measurements in different ruminal fractions.
Based on the unweighted (qualitative) and weighted (quantitative) Unifrac distance matrices, including the impact of phylogeny, BE and PE affected overall bacterial community qualitatively in rumen fluids, but not quantitatively (P < 0.1; Figure 1A). In rumen solids, PE altered the overall bacterial community (P = 0.023 for unweighted UniFrac; P = 0.063 for weighted UniFrac) while BE had no significant impact (Figure 1B). Two different ruminal fractions had significantly different overall bacterial community, qualitatively and quantitatively (P < 0.01; Supplementary Figure 1).
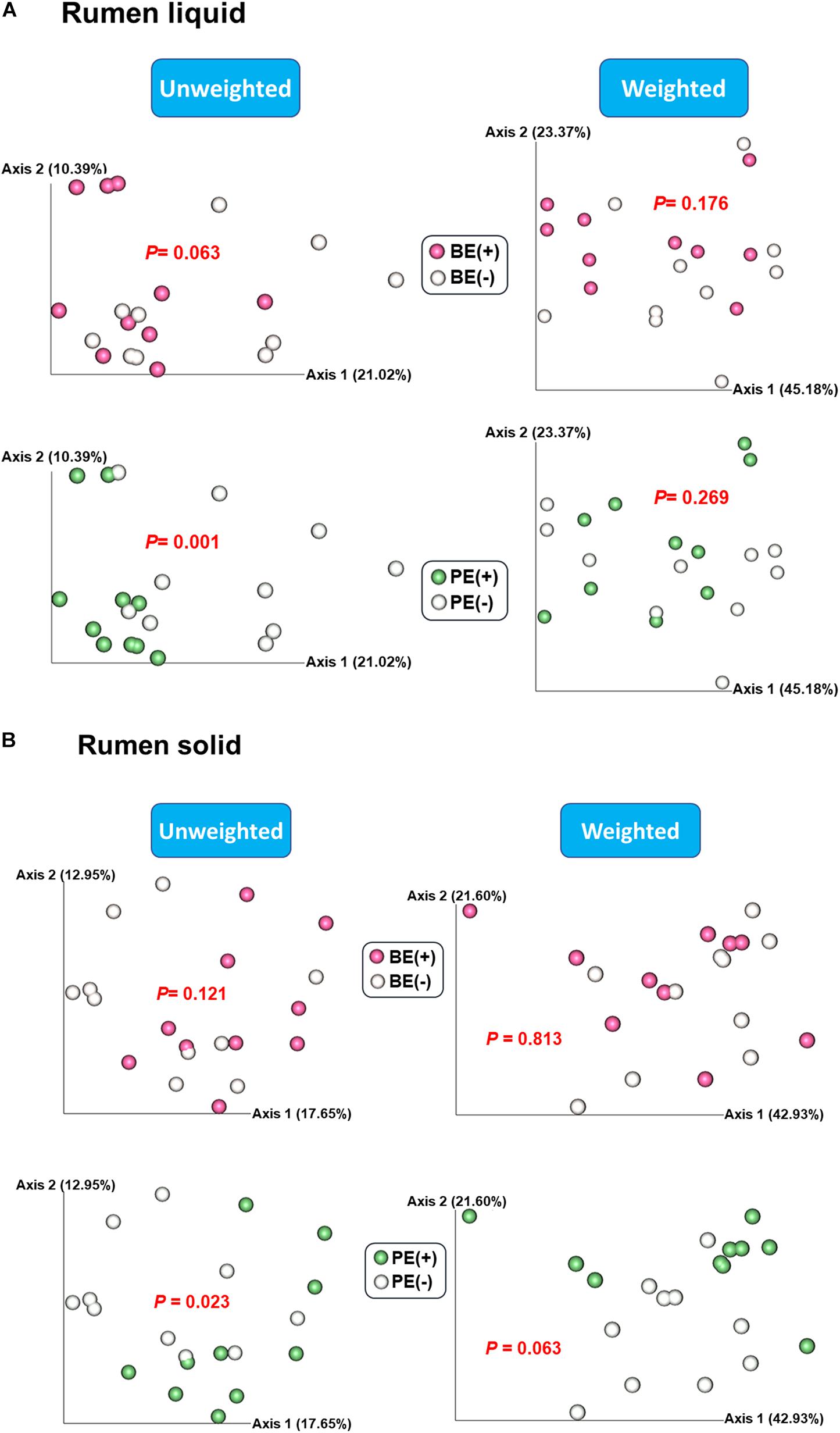
Figure 1. Principal coordinates analysis (PCoA) plot based on unweighted and weighted UniFrac distance matrices showing overall rumen bacterial microbiota distributions in (A) liquid and (B) solid fraction of dairy calves differed by microbial inoculations with two types of inoculums (BE and PE).
The Venn diagram showed shared bacterial taxa between donor inoculum and two treatment groups by either BE or PE inoculation (Figure 2). In rumen liquid, 10 phyla were shared among the respective inoculum sample, treated- and non-treated calves in both the BE and PE inoculations. In rumen solid, nine phyla were shared among these three groups (i.e., inoculum, treated- and non-treated) in both the BE and PE inoculations. Two phyla were shared only between inoculum and inoculum-treated calves regardless of the ruminal fractions and inoculum types, and Chloroflexi, Lentisphaerae, Kiritimatiellaeota, and Patescibacteria were the phyla belonging to that category. The phylum Verrucomicrobia was exclusively found in one, non-treated calf. At the genus level, over 90 genera were shared by three groups in both the BE and PE inoculations. Invariably, calves receiving either the PE or BE shared more phyla and genera than control groups. More than 80 genera were exclusively found in inocula, but those genera comprised only 4.44 to 5.88% of overall abundance in both the inocula. Furthermore, exclusively found genera by microbial inoculation treatments occupied a minor portion regardless of the ruminal fractions and inoculum types (< 1% of average relative abundance). Erysipelotrichaceae UCG-007 and Family XIII AD3011 group were the only major genera (present in at least 50% of the samples) exclusively found in the solid fraction of BE treatments and in the liquid fraction of PE treatments, respectively. The number of detected taxa between rumen liquid and solid fractions was also shown in Supplementary Figure 2.
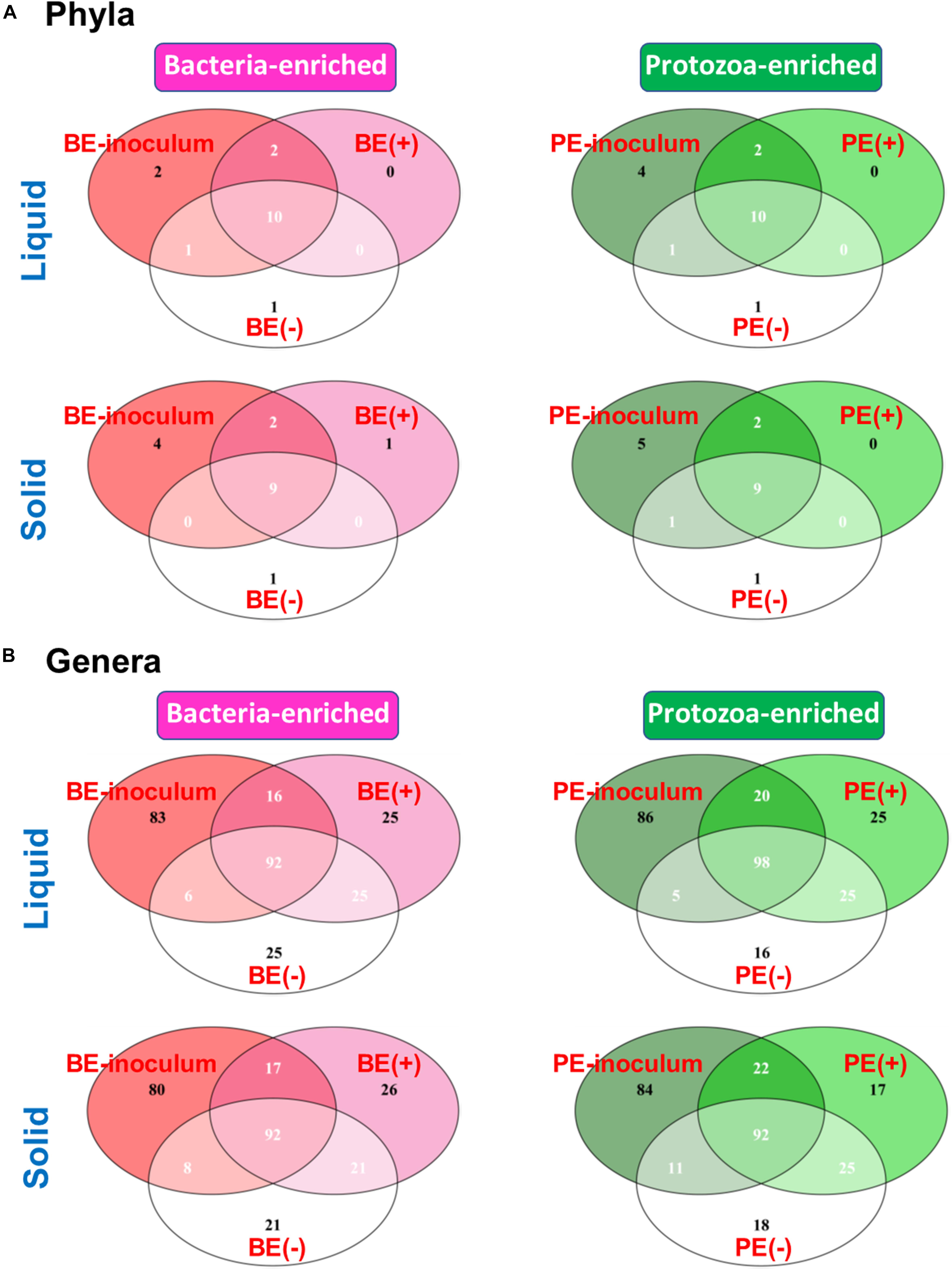
Figure 2. Venn Diagram depicting the number of shared and exclusive bacterial (A) phyla or (B) genera by microbial inoculations at either rumen liquid or solid fractions.
An opposite trend of the relative abundance of the phyla Fibrobacteres and Proteobacteria in rumen fluid was observed by BE. The bacterial phylum Fibrobacteres was dominant in BE(+) calves [BE(+): 0.21 ± 0.08% and BE(−): 0.06 ± 0.03%, P = 0.02], whereas the relative abundance of Proteobacteria was higher in BE(−) calves [BE(+): 13.90 ± 3.63% and BE(−): 28.29 ± 4.94%, P = 0.03; Table 3A]. The same trend for both phyla was observed in rumen solids by PE [for Fibrobacteres, PE(+): 3.42 ± 0.94% and PE(−): 0.61 ± 0.24%, P = 0.02; for Proteobacteria, PE(+): 3.20 ± 1.31% and PE(−): 10.03 ± 2.40%, P = 0.03; Table 3B].
By PE, the abundance of Fibrobacter and Lachnospiraceae UCG-008 in the solid fraction was also dominant in PE(+) calves compared to that of PE(−) calves as shown in the liquid fraction. In the solid fraction, the abundance of Bacteroidales bacterium Bact_22 and Eubacterium xylanophilum group was greater in PE(+) calves compared to that of PE(−) calves, while the opposite distribution was found in the abundance of five genera (unclassified genus of Eggerthellaceae, Prevotella 9, Roseburia, Ruminococcaceae UCG-004, and unclassified genus of Succinivibrionaceae) by PE.
To show the preferential environmental niches of major bacterial phyla and genera, a comparison of their relative abundance between liquid and solid fractions was also analyzed (Supplementary Table 5). The bacterial phylum Proteobacteria and 14 genera were dominant in the liquid fraction (P < 0.05). A greater abundance of the three phyla (Bacteroidetes, Fibrobacteres, and Spirochaetes) and 14 genera were detected in the solid fraction compared to that of the liquid fraction (P < 0.05; Supplementary Table 5).
In the liquid fraction, two uncultured genera within the phylum Bacteroidetes (F082 uncultured rumen bacterium and Rikenellaceae U29-B03), Fibrobacter, and four genera within Firmicutes (Lachnoclostridium 1, Lachnospira, Lachnospiraceae UCG-010, and Erysipelotrichaceae UCG-007) were differentially abundant in BE(+) calves, whereas unclassified genus of Succinivibrionaceae was the only genus abundant in BE(−) calves compared to their counterparts. Comparing by PE, 15 genera belonging to four different phyla were differentially abundant in PE(+) calves, but there were no taxa significantly abundant in PE(−) calves were found (Table 3C). In the solid fraction, the relative abundance of F082 uncultured rumen bacterium, Rikenellaceae U29-B03, and Erysipelotrichaceae UCG-007 showed same distribution between BE(+) calves and BE(−) calves as determined in the liquid fraction (Table 3D).
Differential network analysis identified 36 and 52 nodes with 35 and 136 significant interactions among the major genera in rumen liquid between BE(+) and BE(−) calves, respectively (Figure 3). A total of 56 and 31 microbial nodes with 116 and 47 significant interactions were exclusively identified in rumen liquid bacteria of PE(+) and PE(−) calves, respectively. While a total of 31 and 22 nodes with 51 and 25 significant interactions from rumen solids consisted of the exclusive networks between BE(+) and BE(−) calves. A total of 28 and 10 nodes with 59 and 8 significant interactions from rumen solid fraction of PE inoculated calves were exclusively found either in microbial networks of PE(+) and PE(−) calves, respectively. Based on the centrality measurements, four bacterial genera in the rumen liquid [Prevotella 9 for BE(+), Olsenella for BE(−), F082 uncultured rumen bacterium for PE(+), Alloprevotella for PE(−)] and five bacterial genera in the rumen solid [Eubacterium xylanophilum group for BE(+), Megasphaera for BE(−), Eubacterium coprostanoligenes for PE(+), Rikenellaceae RC9 gut group and Roseburia for PE(−)] were denoted as keystone genera in the exclusive microbial networks of each microbial inoculation treatment (Table 4). Among those keystone microbial nodes at each specific network, the genera Megasphaera and Eubacterium coprostanoligenes group were liquid-dominant, while three bacterial genera (F082 uncultured rumen bacterium, Eubacterium xylanophilum group, and Rikenellaceae RC9 gut group) were solid-dominant (Supplementary Table 5).
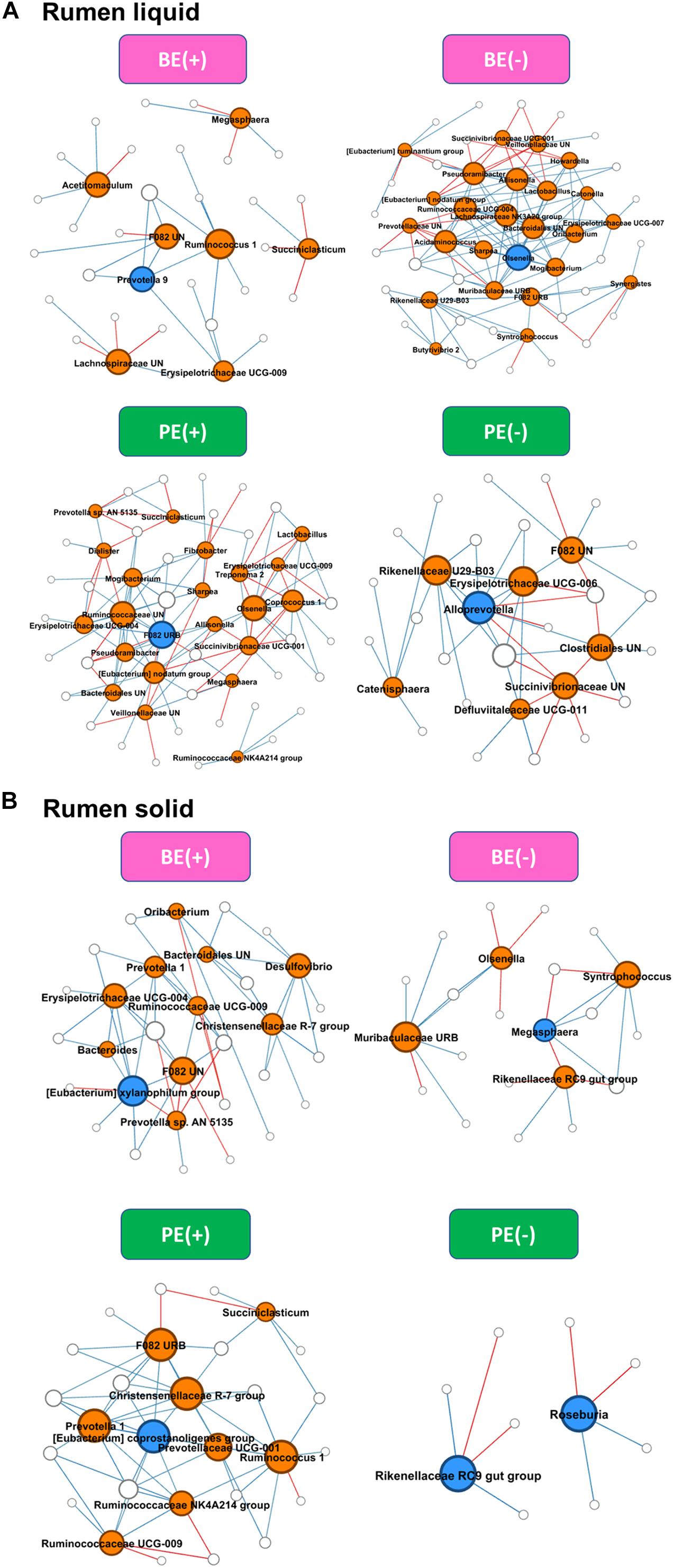
Figure 3. Exclusive co-occurrence and mutual exclusion microbial network either at (A) rumen liquid and (B) rumen solid. Node color represents exclusive microbial nodes (orange) and keystone taxa (blue) selected based on the betweenness centrality, authority, and eigenvector centrality measurements within each exclusive network. Edge color represents co-occurrence (blue) or mutual exclusive (red) interactions. Edge thickness was adjusted based on the absolute value of the correlation coefficients of each interaction.
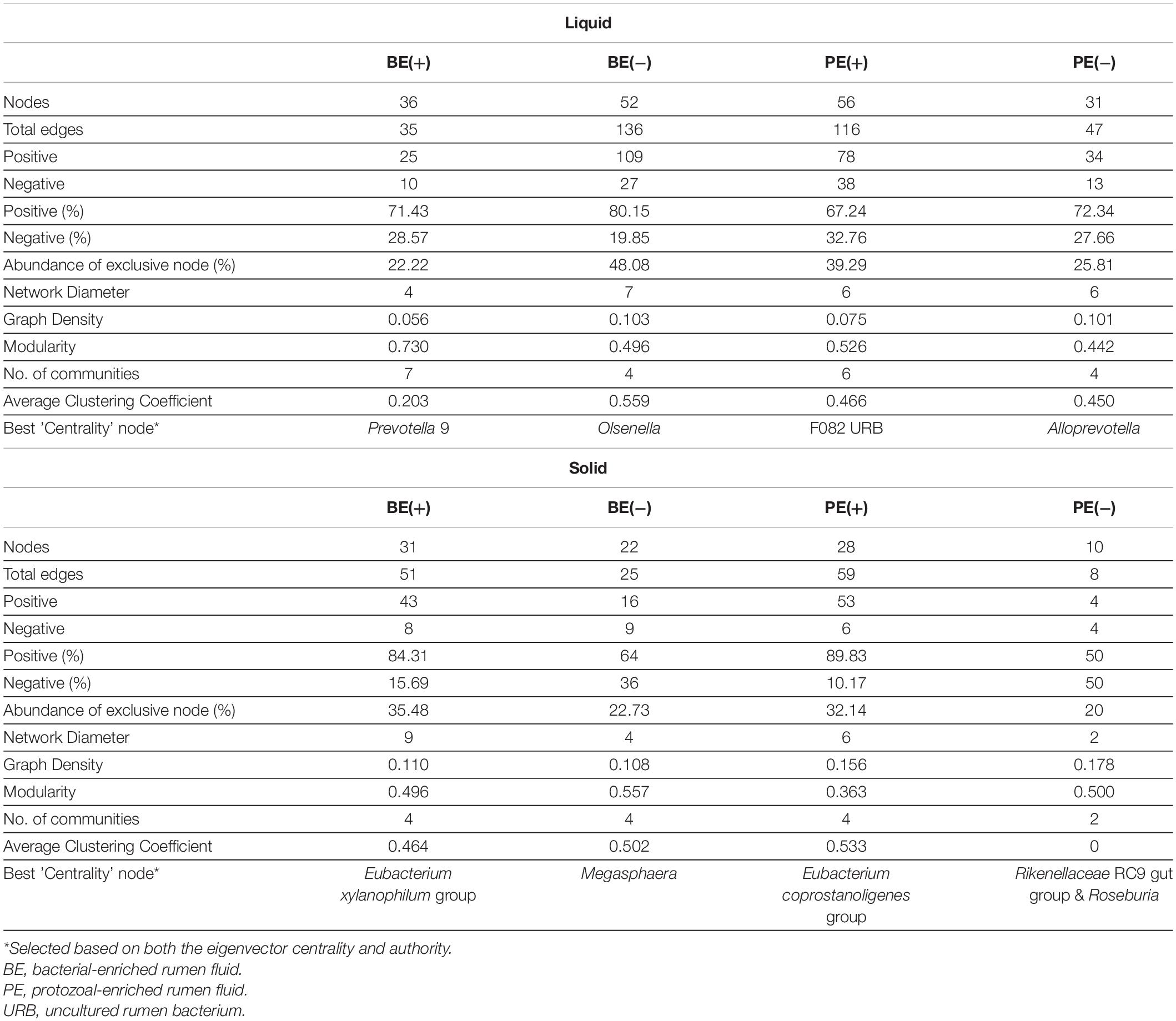
Table 4. Exclusive network statistics by microbial inoculations either in rumen liquid or solid fraction.
The overall distribution of microbial functions represented by KEGG ortholog profiles tended to be affected by PE in rumen liquid (P = 0.089; Figure 4). Two ruminal fractions had significantly different overall distribution of microbial functions from one another (P < 0.001; Supplementary Figure 3). Among the major KEGG pathways, which have over 0.1% average relative abundance either in treated or non-treated calves, eight pathways [“Glycolysis/Gluconeogenesis (ko00010),” “Citrate cycle (TCA cycle) (ko00240),” “Valine, leucine and isoleucine degradation (ko00280),” “Lysine degradation (k00310)”, “D-Alanine metabolism (ko00473),” “Amino sugar and nucleotide sugar metabolism (ko00520),” “Aminoacyl-tRNA biosynthesis (ko00970),” and “Nucleotide excision repair (ko03420)”] were enriched in rumen liquid of BE(+) calves and five pathways [“Ascorbate and aldarate metabolism (ko00053),” “Tyrosine metabolism (ko00350),” “Phenylalanine metabolism (ko00360,” “C5-Branched dibasic acid metabolism (ko00660),” and “Bacterial secretion system (ko03070)”] were enriched in rumen liquid of BE(−) calves (Table 5). In the rumen solid fraction, a total of three pathways [“Selenocompound metabolism (ko00450),” “Glycerophospholipid metabolism (ko00564),” and “Aminoacyl-tRNA biosynthesis (ko00970)” in BE(+) calves] were differentially abundant by BE. In both the rumen fractions, PE(+) calves were more enriched with the “Fatty acid degradation (ko00071)” pathway, while PE(−) calves had a greater abundance of “Pantothenate and CoA biosynthesis (ko00770)”, “Sulfur metabolism (ko00920)”, and “Biofilm formation - Vibrio cholerae (ko05111)” pathways (Table 6). The solid fraction of PE(+) calves exclusively had seven more metabolism-related pathways [“Lysine degradation (ko00310),” “N-Glycan biosynthesis (ko00510),” “Other glycan degradation (ko00511),” “Streptomycin biosynthesis (ko00521),” “Inositol phosphate metabolism (ko00562),” “Methane metabolism (ko00680),” and “Biosynthesis of vancomycin group antibiotics (ko01055)”], whereas two pathways in rumen liquid [“Vitamin B6 metabolism (ko00750)” and “RNA degradation (ko03018)”] and four additional pathways in rumen solids [“Fatty acid biosynthesis (ko00061),” “Secondary bile acid biosynthesis (ko00121),” “Lysine biosynthesis (ko00300),” and “Phosphotransferase system (PTS) (ko02060)”] were differentially abundant in PE(−) treated calves. A total of 19 and 32 major pathways were enriched in rumen liquid and solid, respectively (Supplementary Table 6).
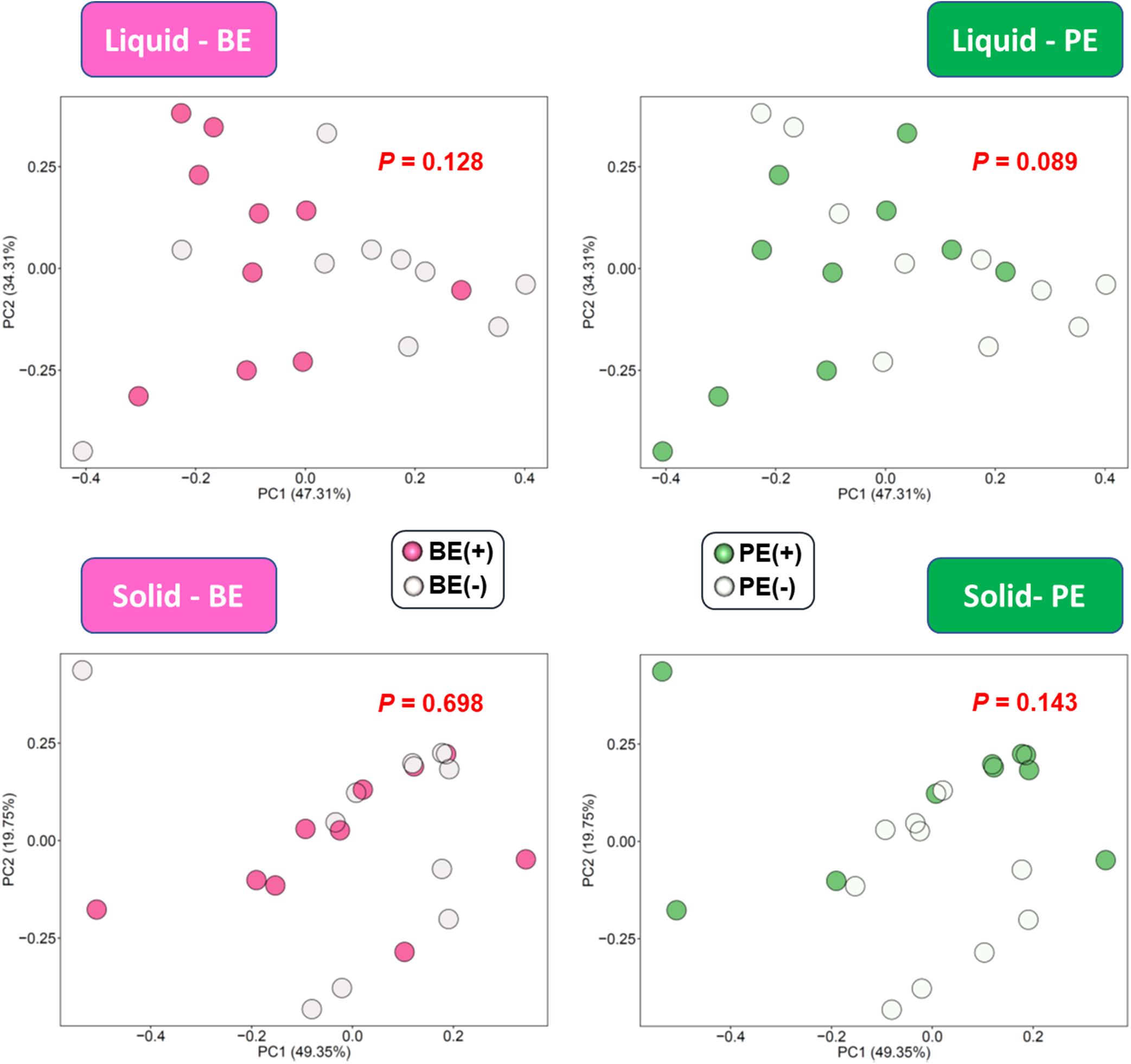
Figure 4. PCA plot showing the effect of microbial inoculations on the overall functional distribution at rumen liquid and solid fractions. Overall functional distribution was computed based on the relative abundance of KEGG orthologs predicted by PICRUSt2.
Correlation analysis identified various differentially abundant bacterial genera correlated (correlation coefficient, |r| ≥ 0.5; P ≤ 0.05) with either rumen (Figure 5A) or animal performance measurements (Figure 5B). Unclassified genus of Coriobacteriales and Syntrophococcus, which were abundant in PE(+) calves in the liquid fraction, were positively correlated with total VFA concentration, while a negative correlation was found with Prevotella 9 enriched in PE(−) calves in the solid fraction. No bacterial taxa showed significant correlations with butyrate molar proportions, however, F082 URB and Ruminococcaceae UCG-004 showed significant positive correlation with isobutyrate molar proportions whose dominance was found in rumen solid fractions of calves treated with autoclaved rumen fluid. Particularly with log10-transformed counts of rumen protozoa, eight genera enriched by PE showed significant positive correlations. Three genera dominant in PE(−) calves in the solid fraction (Unclassified genus of Eggerthellaceae, Ruminococcaceae UCG-004, and unclassified genus of Succinivibrionaceae) were negatively correlated with protozoal counts. Bacteroidales bacterium Bact_22 was positively correlated with rumen papillae length, while the bacterial genus Lachnospira was negatively correlated with papillae thickness.
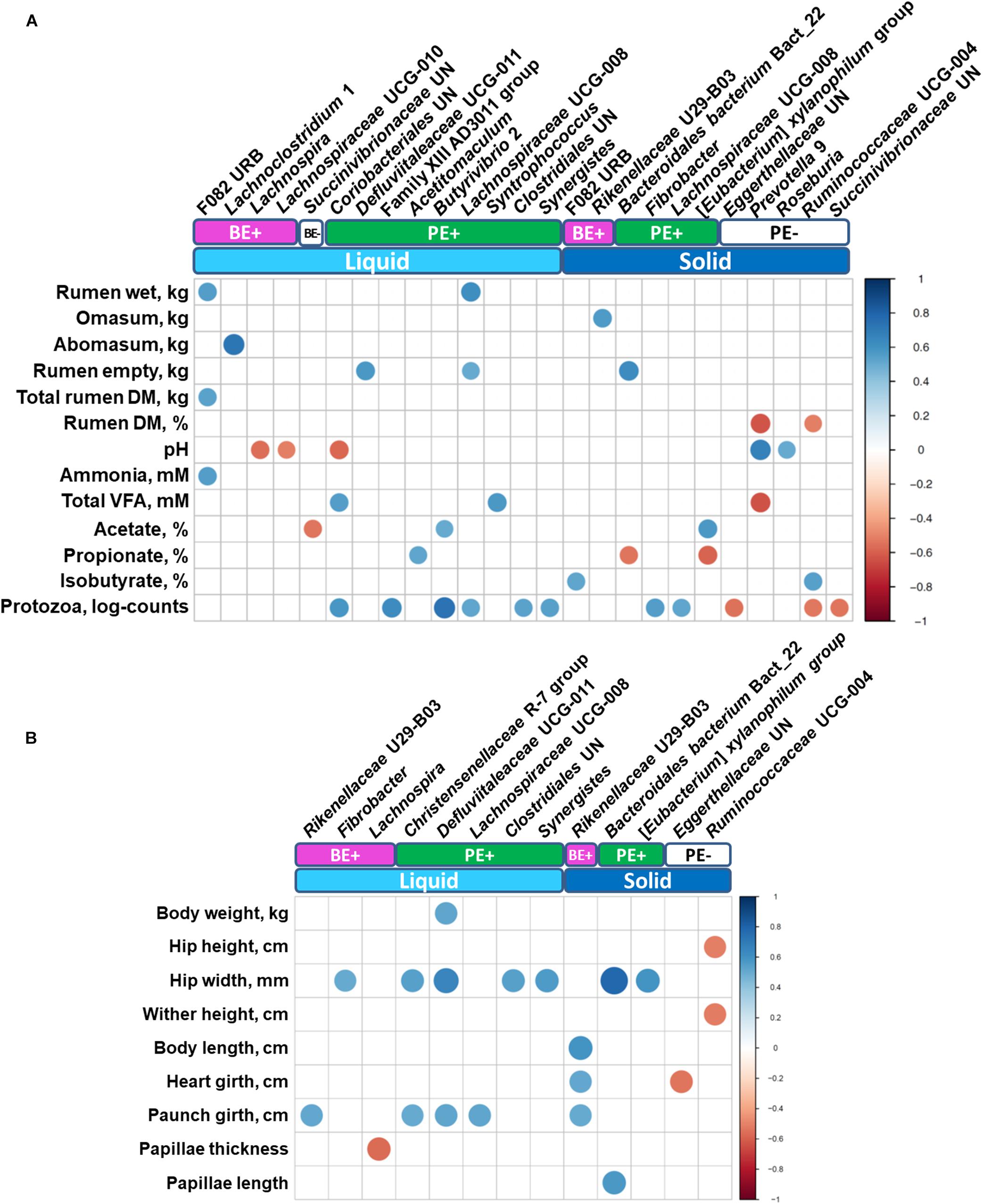
Figure 5. Correlations between (A) rumen measurements (stomach compartments, fermentation, and protozoal counts) and (B) animal performance measurements with differentially abundant bacterial genera. Only strong significant correlation coefficients (| r| ≥ 0.5, P ≤ 0.05) were shown on the plot.
Rumen fluid inoculation was applied to direct the early establishment of the rumen microbial community of dairy calves. Dairy calves have much simpler rumen microbiota before weaning than that of post-weaning, but its composition has shown rapid changes by the onset of solid feeding (Jami et al., 2013; Rey et al., 2014). Other researchers have administered microbial inocula during a similar feeding period to enable young ruminants to establish a more diverse microbial community (Belanche et al., 2020; Bu et al., 2020; Yu et al., 2020), which is suggested to facilitate the utilization of complex lignocellulosic contents in the rumen (Ribeiro et al., 2017). In our previous study with pre-weaned dairy calves (3–6 weeks of age), we confirmed that repeated rumen fluid inoculation moderately affected ruminal fermentation parameters, bacterial diversity measurements and composition (Cersosimo et al., 2019). However, sustainability of the inoculation effects on the ruminal microbial community post weaning has not been well demonstrated. This study analyzed rumen bacterial community structures and the performance of post-weaned dairy calves, which were treated with BE and PE inocula before weaning to determine if the previous inoculation effects are detectable.
The increased alpha-diversity measurements observed in post-weaned calves treated with the combined PE/BE inoculum in the present study, particularly in rumen liquid fraction, was not observed in our previous study (Cersosimo et al., 2019). This is possibly due to the growth of transferred microbial taxa in the post-weaned dairy calves fed increased amounts of solids. The immediate establishment of transferred microbiota was not likely (Bryant and Small, 1960) and the inoculation effect conferred by newly transferred microbial taxa could be delayed. The BE increased species richness and phylogenetic diversity in both the solid and liquid fractions, whereas the effect of PE on alpha-diversity measurements was more pronounced in the liquid fraction of treated calves. This might be due to the colonization of the first protozoal group, small entodinia, which are known to prey on bacteria and prefer starch in the rumen (Williams and Coleman, 1992; Newbold et al., 2015). However, the increased alpha-diversity measurements did not result in the significant differences on overall bacterial community structure when the abundance of bacterial taxa were combined. In addition to the different bacterial community structure by quantitative assessment between PE(+) and PE(−) calves in the solid fraction (P = 0.063), more distinguishable changes of beta-diversity were observed by qualitative assessment in both of the fractions by PE (P < 0.05). This might be due to the transfer of protozoa-associated microbiota which contributed to greater species richness and phylogenetic diversity in PE-inoculum than those of BE-inoculum.
Although not considered as major taxa in this study, the phyla and genera exclusively detected in both of the inoculum and inoculum-treated calves were possibly established by inoculations. Kiritimatiellaeota was recently diverged from the phylum Verrucomicrobia (Spring et al., 2016). Microbiota from this phylum were previously detected in anaerobic conditions, like the animal gut, and could potentially utilize xylose, but no other types of sugars (Spring et al., 2016). The presence of this phylum solely in the solid fraction could indicate an increase in hemicellulose digestion in the rumen of dairy calves. Patescibacteria was observed in a variety of anoxic environments (Herrmann et al., 2019), but because this metagenomically identified phylum has not been well characterized in the rumen, their role in the early rumen is still unknown. In our previous study with the same calves at earlier ages (from 3–6 weeks of age), the abundance of Erysipelotrichaceae unclassified genus linearly increased by age (Cersosimo et al., 2019). Some species belonging to the family Erysipelotrichaceae are saccharolytic and produce lactate by fermenting a variety of sugars (Deusch et al., 2017), which in turn could lower the ruminal pH of inoculum treated calves, especially in BE(+) (P < 0.1). While Family XIII AD3011 group was exclusively found in rumen liquid of PE(+) calves as a major bacterial genus, not much information is known for this candidate genus. The establishment of those two tentative genera was previously demonstrated by repeated rumen fluid inoculation of lambs (Yu et al., 2020).
The increased diversity and the qualitatively altered rumen liquid bacterial community structure by PE tended to affect overall functional differences based on the KEGG orthologs profile.
Fatty acid degradation (ko00071) consists of two modules related to beta-oxidation. This pathway was enriched by PE(+) in both of the rumen fractions, so that increasing the yield of acetyl-CoA might be further used to produce energy for cellular biosynthesis in PE treated calves (Goepfert and Poirier, 2007).
Monophyletic Fibrobacteres and Proteobacteria showed opposite distributions by microbial inoculations in the liquid fraction by BE and in the solid fraction by PE. Increased abundance of the genus Fibrobacter was observed after faunation of defaunated Holstein bull calves (Ozutsumi et al., 2006). This solid-enriched bacteria, represented over 3% of relative abundance in the solid fraction of PE(+) calves and was suspected to contribute to an increase in fiber digestion resulting in tendency for higher VFA concentration in PE(+) than PE (−) calves (P = 0.052). The abundance of the phylum Proteobacteria decreased with age in dairy calves especially after the onset of solid feeding (Jami et al., 2013). Inoculation of dairy calves with adult-derived rumen fluid enriched with Bacteroidetes and Firmicutes in pre-weaned dairy calves might facilitate this transition by lowering the abundance of Proteobacteria.
The unclassified genus of the bacterial family Succinivibrionaceae was a representative genus within Proteobacteria whose abundance was more enriched in the liquid fraction of BE(−) and in the solid fraction of PE(−) calves compared to their respective controls. A 4-fold decrease in the relative abundance of the genus Succinivibrio was previously observed between 2 month and 6 month old dairy calves (Jami et al., 2013). The unclassified genus of Rikenellaceae, which was differentially abundant by BE in both of the fractions, is generally considered as primary saccharolytic in many ecosystems and utilizes a variety of fermentable sugars which could have resulted in slightly lowered ruminal pH in BE(+) calves. The known butyrate producers including Butyrivibrio 2, Coprococcus 1, and Lachnospiraceae UCG-008 were differentially abundant in PE(+) calves in the liquid fraction while only Roseburia, as a butyrate-producing bacterium (Vos et al., 2011), was abundant in PE(−) calves in the solid fraction. The lower butyrate proportions in PE(+) calves might be explained by two possible scenarios: 1. a greater abundance of acetogenic bacteria (Acetitomaculum and Syntrophococcus), or 2. a greater abundance of fibrolytic bacteria, which produce more VFA but not butyrate, inversely lowered the proportion of butyrate in PE(+) calves. Collectively, microbial inoculations facilitated the transition from the Proteobacteria-enriched microbiota to the microbiota occupied by a greater number of bacterial species which can enhance fiber digestion in the rumen. However, opposite distribution of known butyrate producers in between PE(+) and PE(−) calves (e.g., Butyrivibrio 2, Coprococcus 1, and Lachnospiraceae UCG-008) calls into question these explanations of the significantly different butyrate molar proportions by PE. Analyzing active microbiota using transcriptomic approaches could help fill the gaps between rumen fermentation and these microbial compositions.
Rumen microbiota produce B vitamins, including pantothenic acid for the host or other microorganisms (Schlau et al., 2012). Several vitamin metabolism enriched pathways were previously reported to be related to high residual feed intake in beef cattle (Li and Guan, 2017) and might be associated with the enrichment of the lactate-utilizers, which require vitamins for their growth (Ogunade et al., 2019). The abundance of unclassified genus of Succinivibrionaceae in the solid fraction of PE(−) calves may also be associated with increased vitamin metabolism. A relatively greater abundance of the biofilm formation pathway in both rumen fractions of PE(−) calves might be related to the absence of rumen protozoa which affect overall composition and activities of rumen microflora by their bacterial predation and close metabolic association particularly with symbionts (Belanche et al., 2014; Park and Yu, 2018).
Exclusive keystone microbial nodes were defined by two centrality measurements. Among those, the genus Roseburia was the only node whose abundance corresponded to both the microbial inoculations and ruminal fractions. Roseburia is a known butyrate producer that consumes acetate during the fermentation of carbohydrates (Vos et al., 2011), which might contribute to the difference of butyrate molar proportions by PE inoculation. On the other hand, Roseburia only exclusively interacts with four microbial nodes which are not considered as butyrate producers (data not shown).
The single largest classified genus in the rumen, Prevotella, was also defined as an important exclusive microbial node in the liquid fraction of BE(+) calves. This genus consists of several uncultured Prevotella spp. which need further investigation of their metabolism. Prevotella usually are considered as multi-functional, utilizing a variety of substrates produced by the rumen microbiome (Bekele et al., 2010). Thus, they might be associated with other microbes whose metabolic functions are diverse. Many exclusive co-occurrence interactions were found with Eubacterium xylanophilum group in the solid fraction of BE(+) calves, which might contribute to xylan degradation (Van Gylswyk and Van Der Toorn, 1985). This genus was defined as an important node in the solid fraction of PE(+) calves in this study. Interestingly, the relative abundance of another Eubacterium genus, E. coprostanoligenes group was increased in lambs inoculated with adult sheep rumen fluid during weaning (Yu et al., 2020).
Within their specific microbial networks altered by microbial inoculations, those bacterial genera might be associated differently with other microbes particularly in different environmental niches. However, since those exclusive nodes only represent 20 to 48.1% of overall bacterial community, the ruminal fermentation and animal phenotypic differences might be derived from the undefined or shared microbial networks occurred in each microbial inoculation treatment.
There were ten and three bacterial genera exhibiting positive and negative correlations with animal performance measurements, respectively. Among those taxa, Bacteroidales bacterium Bact_22 showed positive correlation with papillae length, which could have contributed to the nutrient absorption through the rumen wall (Dehority, 2003; Millen et al., 2016). The order Bacteroidales was previously associated with a greater feed efficiency in steers (Hernandez-Sanabria et al., 2012; Mccann et al., 2014; Myer et al., 2015). Bacterial genera exhibiting significant positive or negative correlations with protozoal log counts might have physical or metabolic associations with rumen protozoa but little is known about any specific relationship. Lachnospiraceae UCG-008, a known butyrate producer, showed positive correlation with the weights of the wet and empty rumen and circumference of paunch girth in addition to the potential association with rumen protozoa in both the ruminal fractions. Future research is required to verify those potential relationships between bacterial taxa and both animal development and ruminal fermentation.
Compared to our previous results with these calves prior to weaning, a greater number of taxa were shared between treated and non-treated groups. This was also accompanied with increased species richness in groups treated with the microbial inoculations. In PE treated calves, adult-derived rumen inoculations successfully triggered the establishment of some of microbial taxa including entodiniomorphs. In this study, the post-weaned dairy calves, regardless of the microbial inoculations, shared most of the core-bacterial taxa, which predominate in the rumen. Observed differentially abundant taxa and functional pathways by microbial inoculations were associated with a disparity in butyrate proportions by PE, but didn’t result in significant variation in other fermentation measurements or animal development measurements. Lastly, unique microbial networks in each treatment group within different ruminal fractions might contribute to the differential microbial compositions and metabolic shifts as a result of the inoculations.
The 16S amplicon sequences generated for this study can be found in NCBI Sequence Read Archive, PRJNA454463.
The animal study was reviewed and approved by University of Wisconsin’s Institutional Animal Care and Use Committee. The protocol was approved by the University of Wisconsin’s Institutional Animal Care and Use Committee under the protocol A005829.
LC and GZ conceived and designed the study. LC, WR, and GZ conducted the experiment and analyzed the animal data. TP, LC, and WL did the metataxonomic analysis. TP and LC drafted the manuscript. GZ and WL reviewed and edited the manuscript. All the authors read and approved the manuscript.
This research was supported by funding from the USDA, Agricultural Research Service under National Program 101 Food Animal Production. This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 0500-00093-001-00-D. Mention of any trademark or proprietary product in this manuscript does not constitute a guarantee or warranty of the product by the USDA or the Agricultural Research Service and does not imply its approval to the exclusion of other products that also may be suitable. USDA is an equal opportunity provider and employer.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge the excellent work of Dairy Forage Research Center farm staff for animal care and Rebecca Shilts and Ali Pelletier for assisting with sample collection. We also thank Dr. Garret Suen, Madison Cox, and Joseph Skarlupka of the Suen Lab for their assistance. We thank Robby Weyker and personnel at the Meat Science and Muscle Biology Laboratory for assistance with animal sacrifice and processing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.625488/full#supplementary-material
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: an open source software for exploring and manipulating networks,” in Proceedings of the Third International Aaai Conference on Weblogs and Social Media, San Jose, CA.
Bekele, A. Z., Koike, S., and Kobayashi, Y. (2010). Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 305, 49–57. doi: 10.1111/j.1574-6968.2010.01911.x
Belanche, A., De La Fuente, G., and Newbold, C. J. (2014). Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol. Ecol. 90, 663–677. doi: 10.1111/1574-6941.12423
Belanche, A., Palma-Hidalgo, J., Nejjam, I., Jiménez, E., Martín-García, A., and Yáñez-Ruiz, D. (2020). Inoculation with rumen fluid in early life as a strategy to optimize the weaning process in intensive dairy goat systems. J. Dairy Sci. 103, 5047–5060. doi: 10.3168/jds.2019-18002
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C., Al-Ghalith, G. A., et al. (2018). QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ [Preprints] doi: 10.7287/peerj.preprints.27295v1
Bryant, M., and Small, N. (1960). Observations on the ruminal microorganisms of isolated and inoculated calves. J. Dairy Sci. 43, 654–667. doi: 10.3168/jds.S0022-0302(60)90216-2
Bu, D., Zhang, X., Ma, L., Park, T., Wang, L., Wang, M., et al. (2020). Repeated inoculation of young calves with rumen microbiota does not significantly modulate the rumen prokaryotic microbiota consistently but decreases diarrhea. Front. Microbiol. 11:1403. doi: 10.3389/fmicb.2020.01403
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cersosimo, L. M., Radloff, W., and Zanton, G. I. (2019). Microbial inoculum composition and pre-weaned dairy calf age alter the developing rumen microbial environment. Front. Microbiol. 10:1651. doi: 10.3389/fmicb.2019.01651
Dehority, B. A. (1984). Evaluation of subsampling and fixation procedures used for counting rumen protozoa. Appl. Environ. Microbiol. 48, 182–185. doi: 10.1128/Aem.48.1.182-185.1984
Dehority, B. A. (1993). Laboratory Manual for Classification and Morphology of Rumen Ciliate Protozoa. Boca Raton, FL: CRC Press.
Deusch, S., Camarinha-Silva, A., Conrad, J., Beifuss, U., Rodehutscord, M., and Seifert, J. (2017). A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments. Front. Microbiol. 8:1605. doi: 10.3389/fmicb.2017.01605
Dill-Mcfarland, K. A., Breaker, J. D., and Suen, G. (2017). Microbial succession in the gastrointestinal tract of dairy cows from 2 weeks to first lactation. Sci. Rep. 7:40864. doi: 10.1038/srep40864
Douglas, G. M., Maffei, V. J., Zaneveld, J., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2019). PICRUSt2: an improved and extensible approach for metagenome inference. bioRxiv [Preprint] 672295. doi: 10.1101/672295
Fouladgar, S., Shahraki, A. F., Ghalamkari, G., Khani, M., Ahmadi, F., and Erickson, P. S. (2016). Performance of Holstein calves fed whole milk with or without kefir. J. Dairy Sci. 99, 8081–8089. doi: 10.3168/jds.2016-10921
Goepfert, S., and Poirier, Y. (2007). β-Oxidation in fatty acid degradation and beyond. Curr. Opin. Plant Biol. 10, 245–251. doi: 10.1016/j.pbi.2007.04.007
Guzman, C. E., Bereza-Malcolm, L. T., De Groef, B., and Franks, A. E. (2015). Presence of selected methanogens, fibrolytic bacteria, and proteobacteria in the gastrointestinal tract of neonatal dairy calves from birth to 72 hours. PLoS One 10:e0133048. doi: 10.1371/journal.pone.0133048
Gysi, D. M., Fragoso, T. M., Buskamp, V., Almaas, E., and Nowick, K. (2018). Comparing multiple networks using the Co-expression differential network analysis (CoDiNA). arXiv [Preprint] arXiv:1802.00828
Hammer, Ø, Harper, D. A., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9.
Hernandez-Sanabria, E., Goonewardene, L. A., Wang, Z., Durunna, O. N., and Moore, S. S. (2012). Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 78, 1203–1214. doi: 10.1128/AEM.05114-11
Herrmann, M., Wegner, C.-E., Taubert, M., Geesink, P., Lehmann, K., Yan, L., et al. (2019). Predominance of Cand. Patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front. Microbiol. 10:1407. doi: 10.3389/fmicb.2019.01407
Ishaq, S. L., Kim, C. J., Reis, D., and Wright, A.-D. G. (2015). Fibrolytic bacteria isolated from the rumen of North American moose (Alces alces) and their use as a probiotic in neonatal lambs. PLoS One 10:e0144804. doi: 10.1371/journal.pone.0144804
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Jewell, K. A., Mccormick, C. A., Odt, C. L., Weimer, P. J., and Suen, G. (2015). Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl. Environ. Microbiol. 81, 4697–4710. doi: 10.1128/Aem.00720-15
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Li, F., and Guan, L. L. (2017). Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl. Environ. Microbiol. 83:e00061-17. doi: 10.1128/AEM.00061-17
Li, W., Edwards, A., Riehle, C., Cox, M., Raabis, S., Skarlupka, J., et al. (2019). Transcriptomics analysis of host liver and meta-transcriptome analysis of rumen epimural microbial community in young calves treated with artificial dosing of rumen content from adult donor cow. Sci. Rep. 9:790.
Malmuthuge, N., and Guan, L. L. (2017). Understanding host-microbial interactions in rumen: searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 8:8. doi: 10.1186/s40104-016-0135-3
Mccann, J. C., Wiley, L. M., Forbes, T. D., Rouquette, F. M. Jr., and Tedeschi, L. O. (2014). Relationship between the rumen microbiome and residual feed intake-efficiency of Brahman bulls stocked on bermudagrass pastures. PLoS One 9:e91864. doi: 10.1371/journal.pone.0091864
Meale, S. J., Li, S., Azevedo, P., Derakhshani, H., Plaizier, J. C., Khafipour, E., et al. (2016). Development of ruminal and fecal microbiomes are affected by weaning but not weaning strategy in dairy calves. Front. Microbiol. 7:582. doi: 10.3389/fmicb.2016.00582
Myer, P. R., Smith, T. P., Wells, J. E., Kuehn, L. A., and Freetly, H. C. (2015). Rumen microbiome from steers differing in feed efficiency. PLoS One 10:e0129174. doi: 10.1371/journal.pone.0129174
Newbold, C. J., De La Fuente, G., Belanche, A., Ramos-Morales, E., and Mcewan, N. R. (2015). The role of ciliate protozoa in the rumen. Front. Microbiol. 6:1313. doi: 10.3389/fmicb.2015.01313
Ogunade, I., Pech-Cervantes, A., and Schweickart, H. (2019). Metatranscriptomic analysis of sub-acute ruminal acidosis in beef cattle. Animals 9:232. doi: 10.3390/ani9050232
Ozutsumi, Y., Tajima, K., Takenaka, A., and Itabashi, H. (2006). Real-time PCR detection of the effects of protozoa on rumen bacteria in cattle. Curr. Microbiol. 52, 158–162. doi: 10.1007/s00284-005-0266-9
Park, T., and Yu, Z. (2018). Do ruminal ciliates select their preys and prokaryotic symbionts? Front. Microbiol. 9:1710. doi: 10.3389/fmicb.2018.01710
Paula, E., Broderick, G., Danes, M., Lobos, N., Zanton, G., and Faciola, A. (2018). Effects of replacing soybean meal with canola meal or treated canola meal on ruminal digestion, omasal nutrient flow, and performance in lactating dairy cows. J. Dairy Sci. 101, 328–339. doi: 10.3168/jds.2017-13392
Rey, M., Enjalbert, F., Combes, S., Cauquil, L., Bouchez, O., and Monteils, V. (2014). Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential. J. Appl. Microbiol. 116, 245–257. doi: 10.1111/jam.12405
Ribeiro, G. O., Oss, D. B., He, Z., Gruninger, R. J., Elekwachi, C., Forster, R. J., et al. (2017). Repeated inoculation of cattle rumen with bison rumen contents alters the rumen microbiome and improves nitrogen digestibility in cattle. Sci. Rep. 7:1276. doi: 10.1038/s41598-017-01269-3
Roth, B., Keil, N., Gygax, L., and Hillmann, E. (2009). Influence of weaning method on health status and rumen development in dairy calves. J. Dairy Sci. 92, 645–656. doi: 10.3168/jds.2008-1153
Schlau, N., Guan, L., and Oba, M. (2012). The relationship between rumen acidosis resistance and expression of genes involved in regulation of intracellular pH and butyrate metabolism of ruminal epithelial cells in steers. J. Dairy Sci. 95, 5866–5875. doi: 10.3168/jds.2011-5167
Schönhusen, U., Zitnan, R., Kuhla, S., Jentsch, W., Derno, M., and Voigt, J. (2003). Effects of protozoa on methane production in rumen and hindgut of calves around time of weaning. Arch. Anim. Nutr. 57, 279–295. doi: 10.1080/00039420310001594423
Spring, S., Bunk, B., Spröer, C., Schumann, P., Rohde, M., Tindall, B. J., et al. (2016). Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. 10, 2801–2816. doi: 10.1038/ismej.2016.84
Tang, Y., Horikoshi, M., and Li, W. (2016). ggfortify: unified interface to visualize statistical results of popular R packages. R J. 8, 474–489. doi: 10.32614/rj-2016-060
Van Gylswyk, N., and Van Der Toorn, J. (1985). Eubacterium uniforme sp. nov. and Eubacterium xylanophilum sp. nov., fiber-digesting bacteria from the rumina of sheep fed corn stover. Int. J. Syst. Evol. Microbiol. 35, 323–326. doi: 10.1099/00207713-35-3-323
Vázquez-Baeza, Y., Pirrung, M., Gonzalez, A., and Knight, R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. doi: 10.1186/2047-217x-2-16
Vi, R. B., Mcleod, K., Klotz, J., and Heitmann, R. (2004). Rumen development, intestinal growth and hepatic metabolism in the pre-and postweaning ruminant. J. Dairy Sci. 87, E55–E65. doi: 10.3168/jds.S0022-0302(04)70061-2
Vos, P., Garrity, G., Jones, D., Krieg, N. R., Ludwig, W., Rainey, F. A., et al. (2011). Bergey’s Manual of Systematic Bacteriology: The Firmicutes, Vol. 3. New York, NY: Springer.
Weimer, P. J. (2015). Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front. Microbiol. 6:296. doi: 10.3389/fmicb.2015.00296
Williams, A., and Coleman, G. (1992). “The Rumen Protozoa”. Springer Series in Contemporary Bioscience. New York, NY: Springer-Verlag.
Yáñez-Ruiz, D. R., Abecia, L., and Newbold, C. J. (2015). Manipulating rumen microbiome and fermentation through interventions during early life: a review. Front. Microbiol. 6:1133. doi: 10.3389/fmicb.2015.01133
Yu, S., Zhang, G., Liu, Z., Wu, P., Yu, Z., and Wang, J. (2020). Repeated inoculation with fresh rumen fluid before or during weaning modulates the microbiota composition and co-occurrence of the rumen and colon of lambs. BMC Microbiol. 20:29. doi: 10.1186/s12866-020-1716-z
Yu, Z., and Morrison, M. (2004). Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 36, 808–812. doi: 10.2144/04365ST04
Keywords: adult rumen inoculation, rumen bacterial communities, protozoa, dairy calves, microbial network
Citation: Park T, Cersosimo LM, Li W, Radloff W and Zanton GI (2021) Pre-weaning Ruminal Administration of Differentially-Enriched, Rumen-Derived Inocula Shaped Rumen Bacterial Communities and Co-occurrence Networks of Post-weaned Dairy Calves. Front. Microbiol. 12:625488. doi: 10.3389/fmicb.2021.625488
Received: 03 November 2020; Accepted: 01 February 2021;
Published: 26 February 2021.
Edited by:
Francesca Turroni, University of Parma, ItalyReviewed by:
Robert J. Gruninger, Lethbridge Research and Development Centre, Agriculture and Agri-Food Canada, CanadaCopyright © 2021 Park, Cersosimo, Li, Radloff and Zanton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Geoffrey I. Zanton, Z2VvZmZyZXkuemFudG9uQHVzZGEuZ292; Tansol Park, VGFuc29sLlBhcmtAdXNkYS5nb3Y=; dGFuc29sMTcxOUBnbWFpbC5jb20=
†These authors have contributed equally to this work
‡Present address: Tansol Park, Department of Animal Science and Technology, Chung-Ang University, Anseong, South Korea
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.