- 1School of Life Sciences, Liaoning Normal University, Dalian, China
- 2Liaoning Provincial Key Laboratory of Biotechnology and Drug Discovery, Liaoning Normal University, Dalian, China
- 3Clinical Laboratory Department of the First Affiliated Hospital, Dalian Medical University, Dalian, China
Pseudomonas aeruginosa uses quorum sensing (QS) to control virulence, biofilm formation and antibiotic efflux pump expression. The development of effective small molecules targeting the QS system and biofilm formation represents a novel attractive strategy. In this present study, the effects of a series of Trp-containing peptides on the QS-regulated virulence and biofilm development of multidrug-resistant P. aeruginosa, as well as their synergistic antibacterial activity with three classes of traditional chemical antibiotics were investigated. The results showed that Trp-containing peptides at low concentrations reduced the production of QS-regulated virulence factors by downregulating the gene expression of both the las and rhl systems in the strain MRPA0108. Biofilm formation was inhibited in a concentration-dependent manner, which was associated with extracellular polysaccharide production inhibition by downregulating pelA, algD, and pslA transcription. These changes correlated with alterations in the extracellular production of pseudomonal virulence factors and swarming motility. In addition, the combination of Trp-containing peptides at low concentration with the antibiotics ceftazidime and piperacillin provided synergistic effects. Notably, L11W and L12W showed the highest synergy with ceftazidime and piperacillin. A mechanistic study demonstrated that the Trp-containing peptides, especially L12W, significantly decreased β-lactamase activity and expression of efflux pump genes OprM, MexX, and MexA, resulting in a reduction in antibiotic efflux from MRPA0108 cells and thus increasing the antibacterial activity of these antibiotics against MRPA0108.
Introduction
The gram-negative bacterium Pseudomonas aeruginosa is an opportunistic human pathogen found in hospital-acquired infections, such as cystic fibrosis (CF) and burn and wound infections, particularly in immunocompromised patients (Lyczak et al., 2000; Breidenstein et al., 2011). With the indiscriminate use of antibiotics to treat P. aeruginosa-caused infections, there is a widespread appearance of the multidrug resistant (MDR) strains, leading to a significantly increased incidence of antibiotic resistance. In addition to innate resistance mechanisms and acquired resistance to antibiotics (Strateva and Yordanov, 2009), P. aeruginosa contributes antibiotic resistance through its ubiquitous ability to thrive in different environmental conditions by virtue of its highly complex regulatory networks and ability to form communities called biofilms. The quorum sensing (QS) regulates the population density of bacteria by a highly organized cell-to-cell signaling communication system in which constitutively expressed QS signaling molecules regulate virulence factors production to respond to changes in P. aeruginosa population density (Lee and Zhang, 2015; Pattnaik et al., 2018). When the bacterial population densities are low, virulence gene expression is not activated to avoid pathogen detection and immune stimulation against virulence factors. Once a threshold level of signal molecules has been reached, coordinated gene expression is induced. This system gives bacteria ample time for colonization and establishment in the host. Such virulence factors can affect biofilm formation and swarming motility by the extracellular polymeric substance (EPS), oxygen availability and matrix density (Bjarnsholt et al., 2010; Coenye, 2010; Brackman and Coenye, 2015).
P. aeruginosa QS depends on 3 well-defined and highly characterized signaling systems, i.e., the lasI/lasR, rhlI/rhlR, and pqs/mvfR systems, for the communal response to extracellular signaling autoinducer molecules, such as 3OC12-homoserine lactone (C12-HSL) and N-butyryl-L-homoserine lactone (C4-HSL) (Pattnaik et al., 2018). In the las system, the transcription factor lasR is first activates by the autoinducer C12-HSL, and then lasR drives lasI expression and triggers the production of exotoxin A, the LasA protease and the LasB elastase. Similarly, in the rhl system, the autoinducer C4-HSL increases rhlI expression by the binding and interaction of C4-HSL with rhlR and induces controlled production of the LasB elastase, rhamnolipids, pyocyanin, and cytotoxic lectins that play crucial roles in virulence and biofilm formation and development. The rhl system is also regulated by the lasR/C12-HSL complex at the transcriptional and posttranscriptional levels. As present, QS has not been found in humans, so it will be a specific antibacterial target. Therefore, it seems to be highly appropriate and significant to develop novel antimicrobial drugs for use as potent antiinfectives targeting QS systems and associated virulence factors.
Antimicrobial peptides (AMPs) are composed of short, positively charged amino acid residues, which represent one of the most promising alternative agents for conventional antibiotics because they quickly kill a broad spectrum of microorganisms, including species resistant to other antimicrobials (Fjell et al., 2011; Abd-El-Aziz et al., 2015). Although currently available AMPs exhibit limitations, such as protease instability, high production costs and inactivity under physiological conditions, it is important to develop more effective AMPs as a novel class of antibiotics (Pasupuleti et al., 2011). Many AMPs, especially cationic AMPs with an amphipathic arrangement of cationic and hydrophobic residues, can directly cover bacterial membranes, alter membrane permeability, and create pores across the membrane, leading to cell content leakage and bacterial death (Yeaman and Yount, 2003). However, a previous study reported that colistin, a cationic AMP, altered the expression of 30 genes associated with virulence and bacterial colonization in chronic P. aeruginosa infection (Cummins et al., 2009). Clearly, this finding points to a non-membrane disruption mechanism of AMPs in combating MDR bacteria.
Tryptophan (Trp) residues are often found in naturally occurring AMPs, and their aromatic side chain can form hydrogen bonds with a dipole moment of ∼21 D in an interfacial membrane region (Khandelia and Kaznessis, 2007). This special feature makes Trp-containing peptides partition into the bilayer interface, and Trp-containing peptides been proven effective as antibacterial agents (Chan et al., 2006; Rydberg et al., 2012). A series of Trp-containing peptides has been designed and synthesized by replacing isoleucine residues or leucine residues with Trp residues at different positions of L-K6, a AMP derived from the frog skin peptide temporin-1CEb in our lab (Shang et al., 2012; Bi et al., 2013, 2014). I1W and I4W exhibited stronger antibacterial activity against both gram-positive and gram-negative bacteria than the other peptides, but L11W and L12W had only moderate or low antibacterial activity. I1WL5W and I4WL5W had a broad-spectrum antibacterial activity. We have also determined that the position of Trp residues rather than the number of Trp residues in these peptides play a key role in the antibacterial activity (Bi et al., 2013, 2014). The present study focused on the effects of Trp-containing AMPs on virulence factors and biofilm formation and development regulated by QS in MDR P. aeruginosa (MRPA0108) and aimed to provide an improved prospect for developing more effective antiinfective drugs targeting bacterial virulence.
Materials and Methods
Materials
Peptides were purchased from GL Biochemistry Inc. (Shanghai, China). The purity of peptides was ≥5%. Amino acid sequences and physical characteristics for peptides are shown in Supplementary Table S1. TRIzol reagent and Super ScriptTM III kits were purchased from TaKaRa Inc. (Dalian, China). Ceftazidime, piperacillin and levofloxacin were purchased from Solarbio Inc. (Beijing, China). C4-HSL standards is purchased from Sigma (Sigma, China). All other reagents were of analytical grade. The MDR P. aeruginosa (MRPA) strain 0108 (MRPA0108) was obtained from the Clinical Laboratory Department, the First Affiliated Hospital of Dalian Medical University. MRPA0108 exhibits resistance to gentamicin, kanamycin, ampicillin, cefepime, streptomycin, tetracycline, amoxicillin, and ciprofloxacin. Identification and antimicrobial susceptibility testing (AST) of MRPA0108 were performed by an automated MicroScan WalkAway 96 Plus system (Siemens Ltd., Germany) at the First Affiliated Hospital of Dalian Medical University.
Real-Time Quantitative PCR Analysis
Mid-logarithmic (mid-log) growth-phase MRPA0108 cells (1 × 108 CFU/ml) were incubated with the different concentrations of peptide at 37°C for 18 h. The bacterial cells were collected by centrifugation at 13,000×g and digested in lysis buffer containing 10 mg/ml lysozyme. Total RNA was isolated by using a TRIzol® MaxTM Kit (Invitrogen, Shanghai, China) according to the manufacturer’s instructions and subsequently reverse transcribed into cDNA using a Prime Script RT Master Mix Kit (Takara, Dalian, China). Real-time PCR was performed in an ABI QuantStudioTM 5 sequence detection system (Applied Biosystems, United States) using 2 × SYBR Premix Ex Taq II (Solarbio Science and Technology Co., Ltd.). The PCR cycling conditions were as follows: 95°C for 30 s and 40 cycles of 5 s at 95°C, 30 s at 60°C, and 20 s at 72°C. The fold changes calculated according to the comparative cycle threshold (Ct) method were used for quantifying gene expression, normalizing to the expression of the housekeeping gene rpsL. PCR primers, including those for lasA, lasB, rhlA, rhlB, pelA, algD, psl, oprD, oprM, mexA, and mexB, were synthesized by Sangon Biotech (Shanghai, China) and are presented in Supplementary Table S2.
Determination of QS-Regulated Virulence Factor Production
Mid-log growth-phase MRPA0108 cells (1 × 105 CFU/ml) were incubated in the presence or absence of peptide at 1/4 × the minimal inhibitory concentration (MIC) at 37°C in a shaking culture for 24 h. The culture supernatant was obtained after centrifugation at 12,000×g for 10 min. (1) LasB elastase activity was measured using elastin-Congo red (ECR) as the substrate, as described by Pattnaik et al. (2018). Briefly, 100 μl of the culture supernatant was added to 900 μl of ECR buffer (100 mM Tris-HCl, 1 mM CaCl2, pH 7.5) containing 10 mg of ECR and then incubated with shaking at 37°C for 18 h. Then, 100 μl of 0.12 M EDTA was added to stop the reaction. The insoluble ECR was removed by centrifugation, and the supernatant absorbance was measured at 495 nm. The optical density (OD) 495 nm/OD 600 nm represents the elastase activity. (2) LasA protease activity was measured as follows (Pattnaik et al., 2018): first, the culture supernatant was filtered to remove the bacterial cells, and then 150 μl of sterile-filtered supernatant was added to 2% azocasein (prepared in 50 mM Tris-HCl) and incubated for 4 h at 37°C. Trichloroacetic acid (TCA, 10%) was added to precipitate the undigested substrate. After centrifugation at 10,000 rpm for 10 min, 1 M NaOH was added to the supernatant, and the relative protease activity was measured at OD 440 nm. (3) The pyocyanin assay was based on the absorbance of excreted pyocyanin at 520 nm in an acidic solution (Kong et al., 2005). Three milliliters of culture supernatant were mixed with 2 ml of chloroform for 20 min, and then the pyocyanin from the chloroform phase was reextracted using 0.2 M HCl for 15 min to obtain a pink-colored pyocyanin solution. The OD 520 nm was measured to evaluate the change in pyocyanin content. (4) Rhamnolipid detection was carried out as previously described (Wittgens et al., 2011). Briefly, 500 μl of culture supernatant was mixed with 500 μl of ethyl acetate and vortexed vigorously for 15 s to extract rhamnolipid. After centrifugation at 10,000×g for 5 min at 4°C, the organic phase containing rhamnolipids was obtained and evaporated at room temperature overnight. The rhamnolipid extract dissolved in ultrapure water was mixed with orcinol reagent (0.19% orcinol in 53% H2SO4) and incubated at 80°C for 30 min. The absorbance was measured at 420 nm. (5) The level of C4-HSL was quantified by GC-MS as previously described (Guo et al., 2012).
Swarming Motility Assay
The swarming motility of MRPA0108 was assessed as described by O’May and Tufenkji (2011), with slight modifications. Briefly, swarming agar medium (1% tryptone, 0.5% NaCl, and 0.5% agar) was prepared in the presence or absence of peptide at a final concentration of 1/4 × MIC. MRPA0108 cells from an overnight culture grown on LB agar plates were placed directly on the agar surface (center) with a sterile toothpick so that the motility within the semisolid agar could be evaluated. After 24 h of incubation at 37°C, the diameters of swarming motility were observed and measured. LB liquid medium and erythromycin were used as negative control and positive controls, respectively. All experiments were repeated three times.
Observation of Bacterial Biofilms Using Confocal Laser Microscopy
Biofilms of MRPA0108 were visualized by confocal microscopy. Mid-log growth-phase bacteria were diluted to 1 × 107 CFU/ml. One milliliter of diluted culture and peptide (at a final concentration of 1 × MIC or 2 × MIC) were incubated in 12-well plates with chambered coverglass at 37°C for 24 h. LB liquid medium and erythromycin were used as negative and positive controls, respectively. After incubation, the supernatant was removed, and the slides were washed and stained in a dark environment for 30 min using a LIVE/DEAD BacLightTM Bacterial Viability Kit (Invitrogen, Life Technologies, Shanghai, China). The slides were washed three times and imaged with a confocal laser scanning microscope (LSM 710, Carl Zeiss, Germany) with excitation/emission wavelengths of 480/500 and 490/635 nm for SYTO9 and propidium iodide (PI), respectively. SYTO9 was used to indicate live bacteria, and PI was used to mark dead bacteria.
Biofilm Formation Assay
The biofilm formation of the MRPA0108 strain was assessed as described previously (Shang et al., 2017). Briefly, 50 μl of a mid-log growth-phase bacterial solution (1 × 105 CFU/ml) was placed into a 96-well microtiter plate and incubated overnight with 50 μl of peptide at a final concentration of 1/8 × MIC, 1/4 × MIC, 1/2 × MIC, or 1 × MIC. LB liquid medium and erythromycin were used as negative and positive controls, respectively. After 24 h of incubation at 37°C, planktonic bacteria were carefully removed, and the biofilms were fixed with formaldehyde for 15 min and stained with 0.1% (w/v) crystal violet (CV) dye for 5 min. Before being quantified by reading the absorbance at 590 nm in a microtiter plate reader (Varioskan Flash Microplate Reader, Thermo Fisher Scientific Co., Beijing), biofilms were washed and solubilized with 95% ethanol. The OD 590 nm was determined as a measure of biofilm biomass.
Degradation of 1-day-old biofilms was measured as described previously (Breij et al., 2018). First, biofilms were grown by a 24 h incubation of mid-log growth-phase culture (1 × 105 CFU/ml) at 37°C. Next, planktonic bacteria were removed by washing with PBS. The biofilms were incubated with peptides at a final concentration of 1/8 × MIC, 1/4 × MIC, 1/2 × MIC, or 1 × MIC at 37°C for 24 h and then fixed, stained and quantified as described above. All experiments were repeated three times. LB liquid medium and erythromycin were used as negative control and positive controls, respectively.
Determination of the Polysaccharides in Biofilms
First, mid-log growth-phase bacteria (1 × 105 CFU/ml) were incubated with peptide at a final concentration of 1/8 × MIC, 1/4 × MIC, 1/2 × MIC, or 1 × MIC at 37°C overnight. Exopolysaccharides were determined using the phenol-H2SO4 method. Briefly, after incubation, the cultures were centrifuged at 10,000 rpm for 15 min, and the pellets were resuspended in high-salt buffer (10 mM KPO4, 5 mM NaCl, 2.5 mM MgSO4, pH 7.0), followed by recentrifugation at 10,000 rpm for 30 min. The resulting supernatant was mixed with three volumes of chilled ethanol (100%) and incubated overnight at 4°C. The precipitated exopolysaccharide was added to a mixture containing cold phenol and H2SO4, and the absorbance of the solution was measured at 490 nm. Alginate content was determined using the H2SO4-carbazole method. Briefly, after incubation, the precooled cultures were mixed with an H2SO4 solution (4:1) and vortexed in ice water, and then 0.2% carbazole dissolved in ethanol was added. The mixture was revortexed and incubated at 55°C for 30 min, and the OD 530 nm was measured.
Psl content was detected using the liquid Congo red (CR, Sigma-Aldrich, Shanghai, China) method described by Ma et al. (2006). Briefly, mid-log growth-phase bacteria (1 × 105 CFU/ml) were incubated in no-salt LB liquid medium containing 40 μg/ml CR in the presence or absence of peptide at a final concentration of 1/8 × MIC, 1/4 × MIC, 1/2 × MIC, or 1 × MIC at 37°C overnight. After the absorbance was measured at 600 nm, the cultures were centrifuged at 14,000 rpm for 5 min, and then the absorbance of the supernatant was measured at 490 nm to determine the binding of CR and bacterial cells.
Checkerboard Assay
The synergistic effect of peptides and antibiotics was evaluated using the checkerboard method, in which the fractional inhibitory concentration index (FICI) was used to analyze the type of interaction between compounds (Berditsch et al., 2015). A 100 μl inoculum of 105 CFU/ml planktonic bacteria was added to the respective wells of 96-well microtiter plates, and then serial dilutions of peptide and antibiotic at 100 μl volumes were added to each well and incubated at 37°C for 24 h. The OD 600 nm value was measured with a microplate plate reader (Varioskan Flash Microplate Reader, Thermo Fisher Scientific Co., Beijing) as an indicator of growth inhibition. The resulting checkerboard contained each combination of peptide and antibiotic in 9 doubly increasing concentrations, with wells containing the highest concentration of each antibiotic at opposite corners. The concentrations tested for every combination of peptide-antibiotic were their respective MICs. MH medium and the peptide/antibiotic combination served as negative and positive controls, respectively. The FICI is calculated as FICa plus FICb, where FICa is the MIC of the peptide in the presence of antibiotics/the MIC of the peptide alone, and FICb is the MIC of the antibiotic in the presence of peptide/the MIC of the antibiotic alone. Drug interactions were defined as follows: synergy, FICI ≤ 0.5; additive effect, 0.5 < FICI ≤ 1; and indifference, 1 < FICI ≤ 2 (Hollander et al., 1998; Yoon et al., 2004). The results were recorded in at least three independent experiments, and the median FICI values were used in the analysis.
Intracellular Antibiotic Assay by HPLC
Mid-log growth-phase bacteria were diluted to 1 × 108 CFU/ml and treated with I1W or L12W at concentrations ranging from 3.13 to 50 μM for 12 h at 37°C. After induction, the cultures were harvested by centrifugation at 12,000 rpm for 2 min, washed three times with PBS buffer (pH 7.2), and then incubated with lysozyme (20 mg/ml) at 37°C for 1 h in a water bath. After centrifugation at 12,000 rpm for 5 min, the supernatant of the cell lysates was collected, added to methanol (1:3) and vortexed to precipitate proteins. After recentrifugation at 12,000 rpm for 5 min, the supernatant was measured by using HPLC with a mobile phase of 0.1% trifluoroacetic acid/methanol (92:8, v/v) and a flow rate of 0.7 ml/min.
β-Lactamase Activity Assay
β-lactamase activity was measured by using a β-lactamase assay kit (YIASEN Biotech Co., Ltd., Shanghai, China) according to the manufacturer’s instructions. Briefly, mid-log growth-phase bacteria were diluted to 1 × 108 CFU/ml and treated with I1W or L12W at concentrations ranging from 3.13 to 50 μM for 12 h at 37°C. After induction, the cultures were harvested by centrifugation at 5,000 rpm at 4°C, washed three times with PBS buffer (pH 7.2), and then resuspended in the same buffer. The cells were sonicated on ice for 15 min, and the cell lysates containing β-lactamase were collected by centrifugation at 12,000 rpm for 30 min at 4°C. Fifty microliters of the prepared β-lactamase-containing lysates was mixed with 50 μl of assay buffer containing nitrocefin and reacted at room temperature in a dark environment for 20 min. Nitrocefin hydrolysis was measured spectrophotometrically at 490 nm. Concurrently, the total protein content was determined with a Bradford assay using the same supernatant. One milliunit of β-lactamase is defined as 1 nanomole of nitrocefin hydrolyzed per minute per microgram of protein.
Statistical Analysis
All the experiments were performed in triplicate. The results are generally expressed as the means and standard errors. The paired Student’s t-test was used to test for significance. The significance is indicated as ∗ for p < 0.05 and ∗∗ for p < 0.01.
Results
A previous study showed that Trp-containing peptides had antibacterial activity against the MRPA0108 strain, with MIC values ranging from 6.25 to 25 μM (Supplementary Table S1; Han et al., 2020). Among them, I1W and I4W exhibited higher antibacterial activity than the other peptides, especially L11W and L12W with fourfold higher MICs, and L5W, I1WL5W, and I4WL5W showed moderate antibacterial activity. As previously reported, neither of the peptides showed hemolytic activity at the tested concentrations of 500 μM (Supplementary Table S1; Bi et al., 2013).
Effect of the Trp-Containing Peptides on QS
The relative expression of key virulence and QS genes were investigated in MRPA0108 after Trp-containing peptide treatment. The RT-qPCR results revealed that four virulence factor regulatory genes in the MRPA0108 QS system were significantly downregulated upon peptide exposure, namely, lasA, lasB, rhlA, and rhlB. In particular, the rhlA and rhlB relative expression levels were extremely low after peptide treatment (Figure 1). Trp-containing peptides at 1/8 × MIC inhibited the gene expression of lasA 10.7–47.8% and lasB 7.1–55.6%. As the concentration of peptide increased to 1/4 × MIC, the expression of the lasA and lasB genes was downregulated by 20.3–68.8 and 31.0–81.5%, respectively (Figures 1A,B). Among the peptides, I1W exhibited the highest inhibitory effect. In the same way, the gene expression of rhlA and rhlB was downregulated 24.8–50.7 and 34.1–55.5%, respectively, in MRPA0108 cells upon Trp-containing peptide treatment at 1/8 × MIC and downregulated 51.1–82.1 and 49.1–82.0%, respectively, at a peptide concentration of 1/4 × MIC (Figures 1C,D).
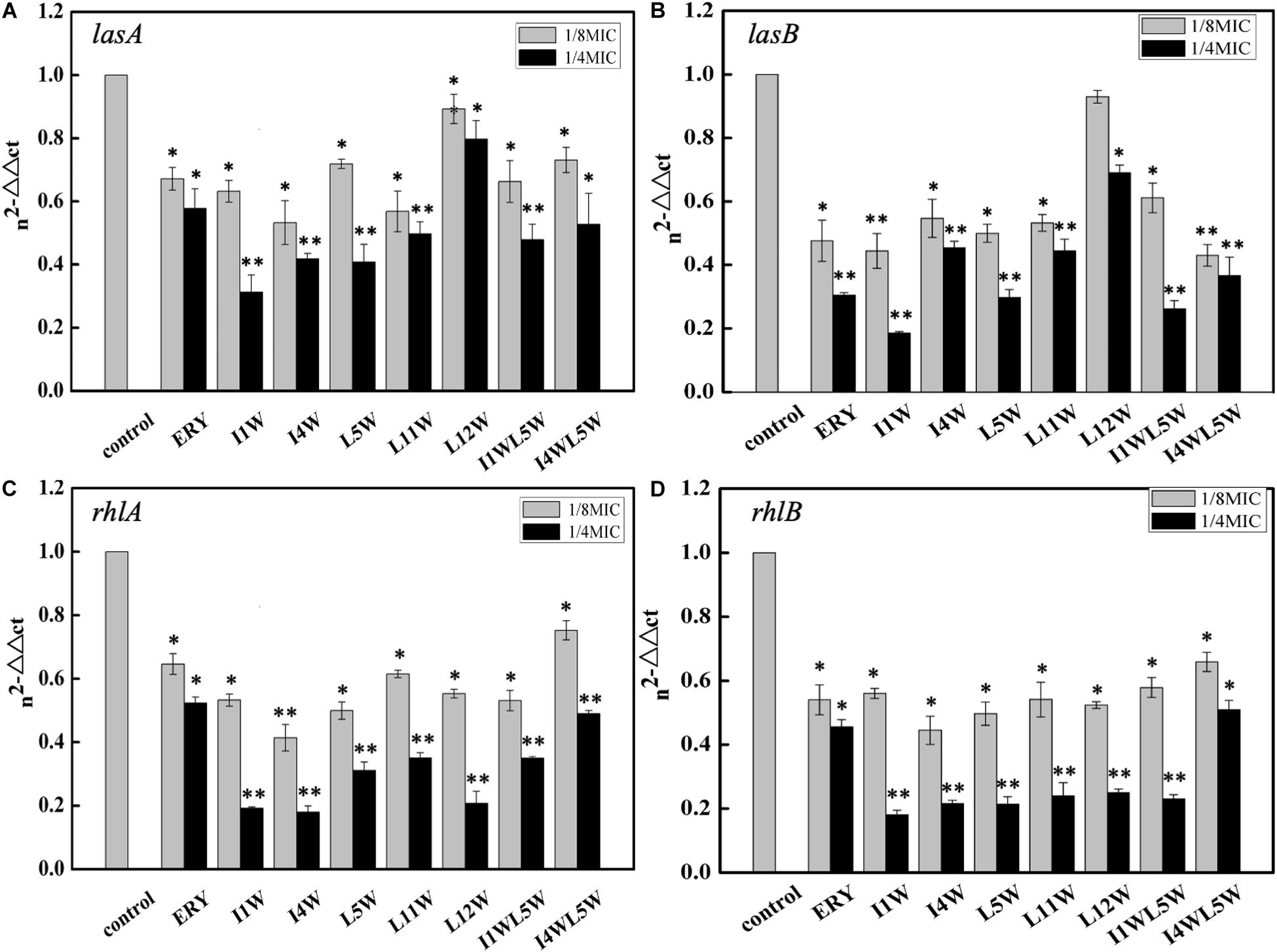
Figure 1. Relative expression of quorum-sensing genes measured by real-time qPCR. (A) lasA; (B) lasB; (C) rhlA; (D) rhlB. The MRPA0108 cells were treated in the absence (control) and presence of 1/4 × MIC and 1/8 × MIC of the Trp-containing peptides for 24 h. The fold changes of QS gene expression were normalized to the level of rpsL gene expression and then quantitated relative to gene expression of control was normalized to 1 using the comparative Ct method. Results represent the means and SD from triplicate experiments. *P < 0.05 and **P < 0.01 indicate statistically significant differences between peptide and control.
The effect of the Trp-containing peptides on the activity of elastase and alkaline protease, virulence factors regulated by the las QS system in MRPA0108, was examined by using the ECR method and azocasein method, respectively. The LasB elastase activity of MRPA0108 after exposure to the Trp-containing peptides is shown in Figure 2A. Compared to that of the control, the LasB elastase activity of MRPA0108 treated with I1W, I4W, and L5W at 1/4 × MIC decreased by 21.0, 22.3, and 24.1%, respectively, and decreased less than 20% with the other Trp-containing peptides. The activity of alkaline protease also decreased by 18–44% in MRPA0108 treated with Trp-containing peptides at 1/4 × MIC compared to that in the control (Figure 2B). These results suggested that the Trp-containing peptides reduced the production of the two virulence factors regulated by the las system and that the inhibitory effect increased as the peptide concentration increased.
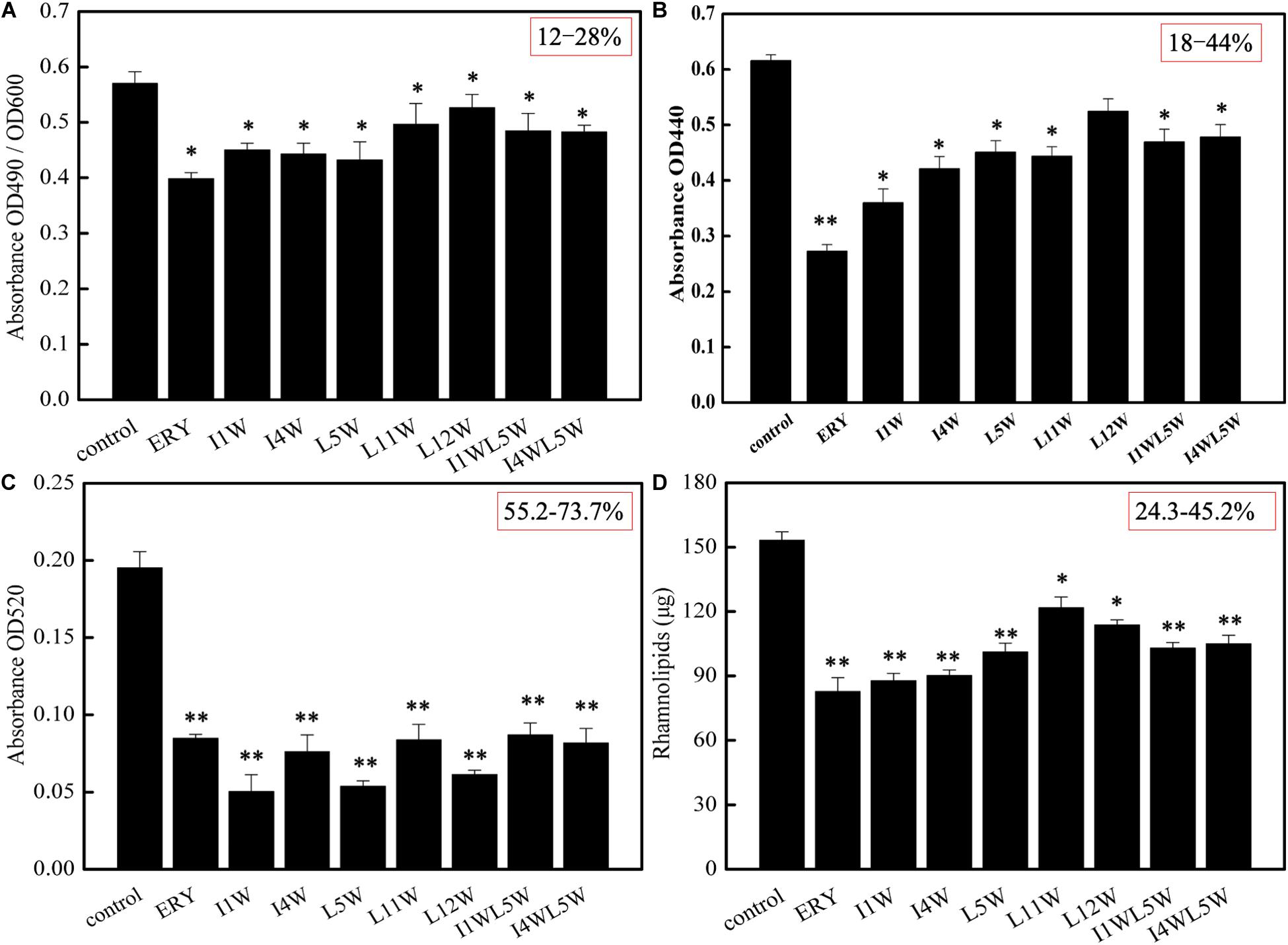
Figure 2. The production of virulence factors regulated by QS system in MRPA0108. (A) lasB elastase; (B) lasA protease; (C) pyocyanin; (D) rhamnolipid. The MRPA0108 cells were treated in the absence (control) and presence of 1/4 × MIC of the Trp-containing peptides for 24 h. The production of virulence factors was examined, respectively. Results represent the means and SD from triplicate experiments. *P < 0.05 and **P < 0.01 indicate statistically significant differences between peptide and control.
Pyocyanin and rhamnolipid are important virulence factors regulated by the rhl QS system in MRPA0108. Pyocyanin induces neutrophil apoptosis and damages neutrophil-mediated host defense, resulting in severe toxic effects in humans (Fothergill et al., 2007). The results showed that the Trp-containing peptides significantly inhibited pyocyanin production in a dose-dependent manner. Pyocyanin production was reduced maximally to 73.7, 69.4, and 63.1% over that in the untreated control after the I1W, L5W, and L12W treatments at 1/4 × MIC, respectively. The other peptides produced approximately 55% inhibition of pyocyanin production (Figure 2C). Similar concentration-dependent results were observed with the positive control. Rhamnolipid production was examined by using the orcinol method. As shown in Figure 2D, I1W and I4W inhibited rhamnolipid production by 45.2 and 43.9%, respectively, and 40.7, 38.8, 37.5, and 24.3% inhibition was observed with L5W, I1WL5W, I4WL5W, and L11W, respectively. This result is consistent with the RT-qPCR results, suggesting that the inhibitory effect of the Trp-containing peptides on virulence factor production regulated by the rhl system is better than that on virulence factor production regulated by the las system.
Effect of the Trp-Containing Peptides on MRPA 0108 Biofilms
The Trp-Containing Peptides Reduced MRPA 0108 Biofilm Formation
QS controls biofilm formation in addition to modulating virulence and pathogenicity. Biofilms greatly enhance the microbial capacity to resist antimicrobial agents (Sergey et al., 2018). Confocal laser scanning microscopy was used to observe biofilms by using SYTO9 and PI, membrane permeability-sensitive DNA-binding dyes as markers. As shown in Figure 3, the biofilm of the MRPA0108 strain had a dense highly aggregated intercellular structure (Figure 3A), but it became sparse with the addition of I1W, I4W, or L5W (Figures 3C–H), suggesting that the peptides inhibited MRPA0108 biofilm formation in a concentration-dependent manner. The average thickness of the 1-day old MRPA0108 biofilm was 6 μm (Figure 3a), but an average biofilm thickness of 1.5–2.5 μm (Figures 3c–h) was observed after treatment with low concentrations of the Trp-containing peptides I1W, I4W, and L5W. The percentage of dead/live cells was 8.7% for control, 68.9% for erythromycin treatment, 29.9 and 47.9% for I1W at a concentration of 1 × MIC and 2 × MIC, 31.2 and 41.7% for I4W at a concentration of 1 × MIC and 2 × MIC, and 21.4 and 32.5% for L5W at a concentration of 1 × MIC and 2 × MIC, respectively.
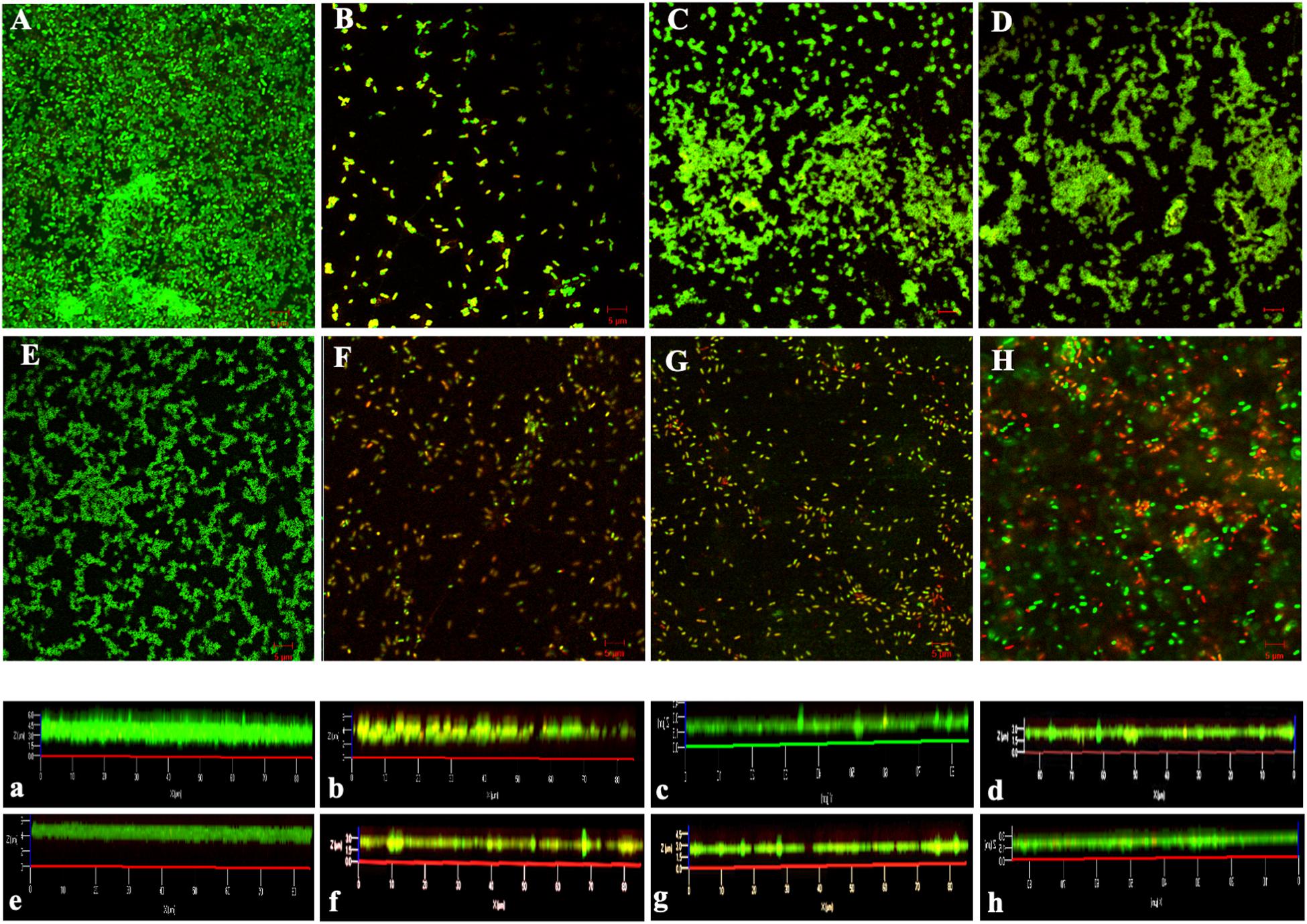
Figure 3. Confocal laser scanning microscopy images of MRPA0108 biofilms. The MRPA0108 cells were incubated at 37°C for 24 h in the absence (control) and presence of the Trp-containing peptides, respectively, and then stained using a LIVE/DEAD BacLight TM bacterial viability kit. Dead cells are stained red, whereas live cells are stained green. (A) Control; (B) erythromycin; (C–E) 1 × MIC of I1W, I4W, and L5W; (F–H) 2 × MIC of I1W, I4W, and L5W. (a–h) Illustrate an average thickness of biofilms. The micrographs are representative of two independent experiments.
Next, the biofilm-forming capability of the MRPA0108 strain was observed after incubation with Trp-containing peptides at concentrations ranging from 1/8 × MIC to 1 × MIC for 24 h. The results showed that the peptides significantly decreased biofilm formation in a dose-dependent manner. A maximum reduction of 53% occurred with the treatment with I1W at 1 × MIC. I1W and I4W at a concentration of 1/8 × MIC inhibited 30 and 33% of biofilm formation, respectively. As peptide concentration increased onefold to eightfold (1/4 × MIC, 1/2 × MIC and 1 × MIC), inhibition of biofilm formation increased 33–53 and 40–48% compared to the control, respectively. Similar observations were recorded with the other peptides. Erythromycin, the positive control, exhibited inhibition similar to that by L5W, L11W and L12W (Figure 4A). For 1-day mature biofilms, the Trp-containing peptides at concentrations of 1 × MIC reduced the attached biofilm biomass by 26–31%, but the mature biofilm was only reduced by 11–15% when the peptide concentration decreased to 1/8 × MIC (Figure 4B), indicating that the Trp-containing peptides at a low concentration had no effect on the dispersal of the preformed biofilms compared with the control except for I1W and I4W.
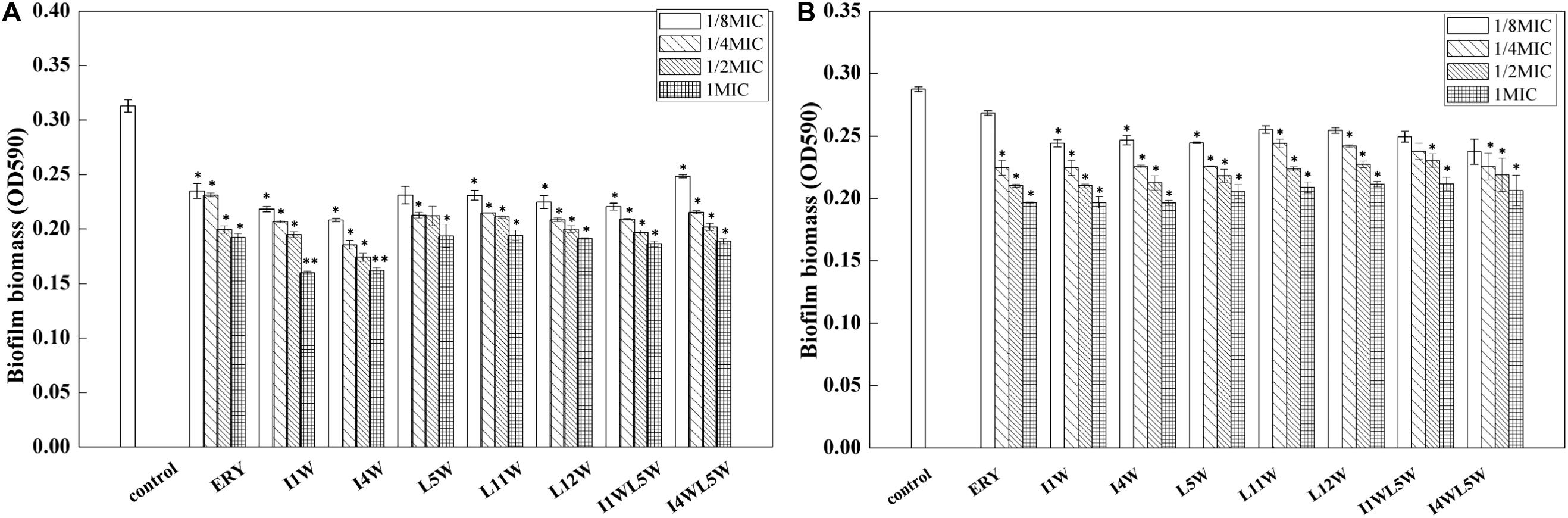
Figure 4. Effect of the Trp-containing peptides on the MRPA0108 biofilm. (A) Effect of the peptides on biofilm formation: the MRPA0108 cells were incubated at 37°C for 24 h in the absence (control) and presence of a series concentration of the peptides, respectively. Biofilm biomass was quantified by crystal violet dye (CV) staining. (B) Effect of the peptides on 24 h mature biofilm: Biofilms of MRPA 0108 cells were grown for 24 h in the absence of the peptide, and then planktonic bacteria were removed and the 24 h mature biofilm was treated with a series concentration of the peptides at 37°C for 24 h. Biofilm biomass was quantified as described above. Results represent the means and SD from triplicate experiments. *P < 0.05 and **P < 0.01 indicate statistically significant differences between peptide and control.
The Trp-Containing Peptides Inhibited the Swarming Motility of MRPA0108 Biofilms
Swarming motility is vital in the initial attachment of bacterial cells to the matrix during biofilm formation. In the present study, the swarming motility of MRPA0108 cells was tested by measuring the diameter of colony growth in a semisolid medium. The MRPA0108 cells exhibited significantly reduced pilus-mediated swarming upon treatment with the Trp-containing peptides at 1/4 × MIC, with an inhibition of 90% for I1W, 85% for I4W, and 80% for other peptides compared to that of the control (Supplementary Figure S1). The positive control erythromycin also induced a significant reduction of 85% in swarming motility for MRPA0108 compared to that of the control.
The Trp-Containing Peptides Reduced the Polysaccharide Content of MRPA0108 Biofilms
Exopolysaccharide is an important component of biofilms to protect them from antimicrobial agents. It was hypothesized that the Trp-containing peptides at subinhibitory concentrations would decrease the forming of MRPA0108 biofilm. As shown in Figure 5A, 1/4 × MICs of I1W and I4W interfered with the exopolysaccharide production in MRPA0108 by 26.3 and 25.0%, respectively. As the peptide concentration increased onefold, the exopolysaccharide production was reduced by 42.6 and 42.4%, respectively, and reduced by 67.3 and 65.2% when the peptide concentration increased to 1 × MIC, respectively.
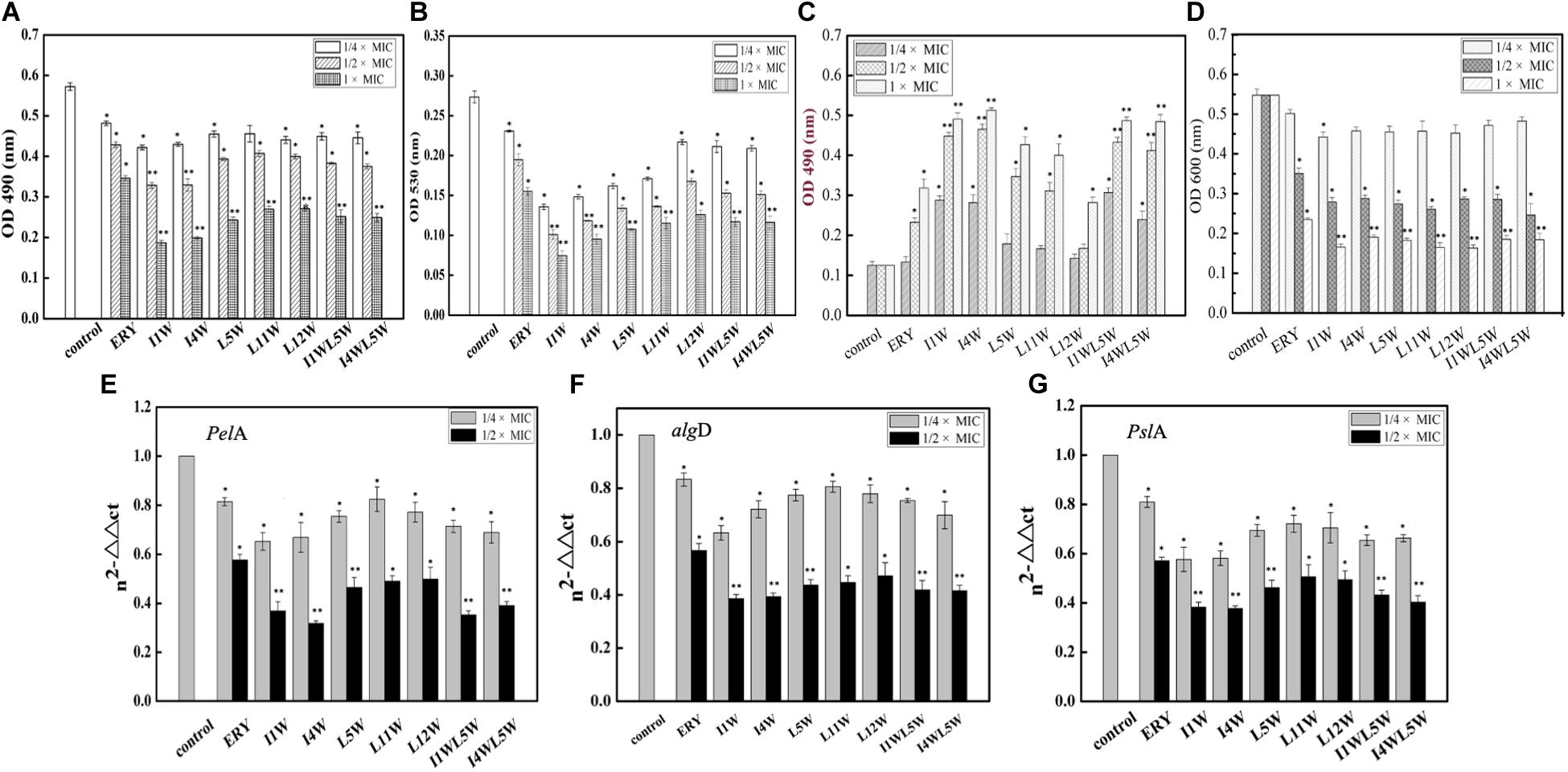
Figure 5. Effect of the Trp-containing peptides on polysaccharide biosynthesis of MRPA0108 biofilm: extracellular polysaccharide (A), alg-(B), and Psl polysaccharide (C,D) was assessed as described in the “Materials and Methods” section. (E–G) Expressions of biofilm polysaccharide-associated genes PelA (E), algD (F), and PslA (G) was assessed by real-time qPCR. Fold changes of gene expression were normalized to the level of rpsL gene and quantitated relative to gene expression of control was normalized to 1 using the comparative Ct method. Results represent the means and SD from triplicate experiments. *P < 0.05 and **P < 0.01 indicate statistically significant differences between peptide and control.
Exopolysaccharide includes at least three polysaccharides, such as alginate, Pel, and Psl, to attach to the surface of the planktonic cells of P. aeruginosa (Ryder et al., 2007; Ghafoor et al., 2011). Alginate is a polyuronide that enhances bacterial cell adhesion to the surface of the matrix. The results showed that the Trp-containing peptides significantly reduced alginate production in a concentration-dependent manner (Figure 5B). I1W and I4W at 1/4 × MIC inhibited alginate production by 53 and 47% of and inhibited alginate production by 74 and 68% when the peptide concentration increased to 1 × MIC. L5W, L11W, L12W, I1WL5W, and I4WL5W exhibited lower inhibition than I1W and I4W. Psl polysaccharide synthesis in MRPA0108 cells was determined by the CR binding assay (Ma et al., 2006). As more Psl containing either -1,3- or -1,4-glucopyranosyl units in the culture supernatant binds to the liquid Congo red, the A 490 nm value decreases, but the A600 nm value increases. As shown in Figures 5C,D, the A600 and A490 nm values were slightly decreased and increased, respectively, in MRPA0108 treated with the low concentration of the Trp-containing peptides (1/4 × MIC) compared to those in the control. When the peptide concentration increased to 1 × MIC, the A490 nm value increased threefold to fivefold compared to those in the control, and the A600 nm value decreased twofold to fourfold, suggesting that the Trp-containing peptides inhibited Psl synthesis in MRPA0108 cells in a concentration-dependent manner.
The Trp-Containing Peptides Downregulated the Expression of Polysaccharide Synthesis-Related Genes in MRPA0108 Biofilms
The gene expression of three biofilm polysaccharide-associated genes, pslA, alginate (algD), and pelA, was quantified using real-time qPCR. The results showed that the Trp-containing peptides significantly reduced the transcription level of the three genes in MRPA0108 cells in a concentration-dependent manner (Figures 5E–G). Expression of pelA, algD and PslA genes decreased by 28–41, 21–39, and 20–38% compared with the control levels, respectively, after MRPA0108 cells were incubated with the 1/4 × MIC Trp-containing peptides for 24 h. In addition, the gene expression levels of pelA, algD and PslA s were inhibited by 50–61, 53–62, and 52–65%, respectively, compared with the control levels when the peptide concentration increased to 1/2 × MIC. I1W and I4W exhibited higher inhibition effect than other peptides and the positive control erythromycin.
The Trp-Containing Peptides Exhibit Synergistic Action in Combination With Antibiotics
The Trp-Containing Peptides Exhibited Synergy With Antibiotics
We investigated the synergistic effect of the Trp-containing peptides in combination with commercially available antibiotics, including ceftazidime, piperacillin and levofloxacin by using the checkerboard method. These antibiotics in combination with a low peptide concentration equivalent to 1/4 × MIC showed noticeably improved antimicrobial activity, with 8–32-fold reduced MIC values for ceftazidime and 2–11-fold reduced MIC values for piperacillin, but the antimicrobial activity of levofloxacin exhibted no significant improvement, with only 0–4-fold reduced MIC values (Figure 6). Ceftazidime and piperacillin showed better antibacterial activity against the MRPA0108 strain in combination with 1/4 × MIC L12W, whose MIC values decreased from 30 to 0.94 μM and from 500 to 187 μM, respectively. The FICI defining synergy between the antibacterial agents is shown in Table 1. According to the previous studys (Hollander et al., 1998; Yoon et al., 2004), FICI ≦ 0.5 were defined as synergy, and 0.5 < FICI ≦ 1.0 and 1.0 < FICI ≦ 2.0 were defined as addition and indifference, respectively. The antibacterial activities of ceftazidime were synergistic with all of the tested Trp-containing peptides, with FICI values of 0.23–0.43, and L5W, L11W, and L12W exhibited the highest synergistic effects. Piperacillin had a synergistic effect with I1W, I4W, L5W, L11W, and L12W, with FICI values of 0.28–0.49, but additive action with I1WL5W and I4W5W, with FICI values of 0.59 and 0.68, respectively. Levofloxacin exhibited only additive action with the tested Trp-containing peptides, with FICI values of 0.51–0.87, except I4W5W, with which levofloxacin had an indifferent effect (FICI value: 1.12). Notably, although L11W and L12W alone had the lowest antibacterial activity against the MRPA0108 strain because of their low membrane disruption ability, they showed the highest synergy with ceftazidime and piperacillin.
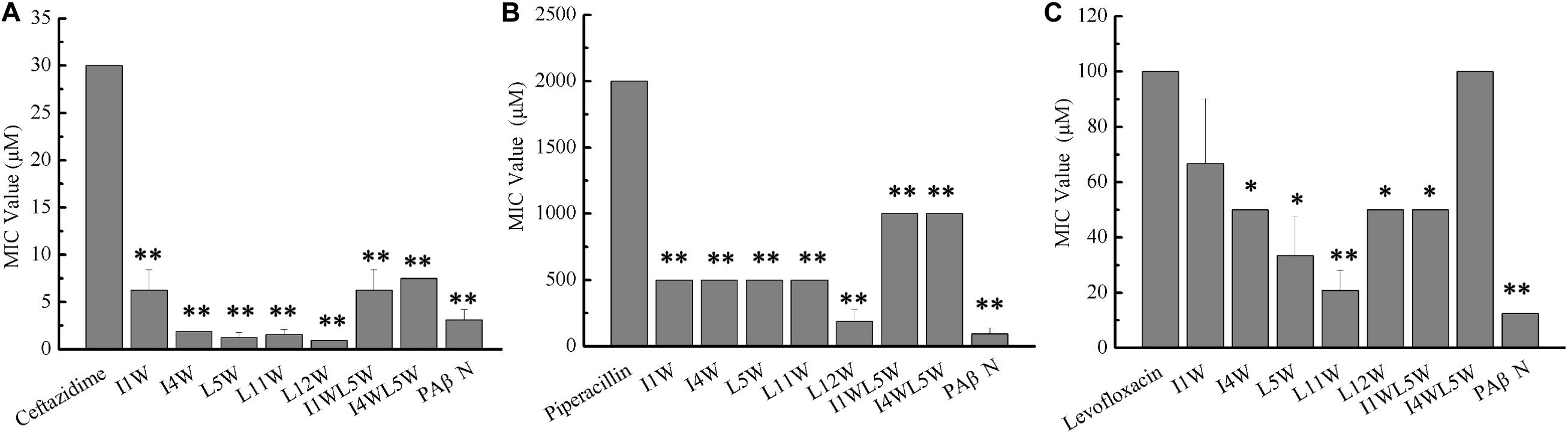
Figure 6. Antibacterial activity of antibiotics in combination with the Trp-containing peptides. Bacterial cultures were treated with a series of concentration of antibiotics ceftazidime (A), piperacillin (B), and levofloxacin (C) in the absence (control) and presence of a 1/4 × MIC peptide concentration at 37C overnight. The OD600 was recorded using a microtiter plate reader. The MIC was defined as the lowest antibiotics concentration that inhibited 95% of the bacterial growth. Phenylalanyl arginyl β-naphthylamide (PAβN), a broad-spectrum inhibitor for efflux pump, was used as a positive control. Each data point represents an average of six independent experiments. *P < 0.05 and **P < 0.01 indicate statistically significant differences between antibiotics alone and antibiotics in combination with peptide.
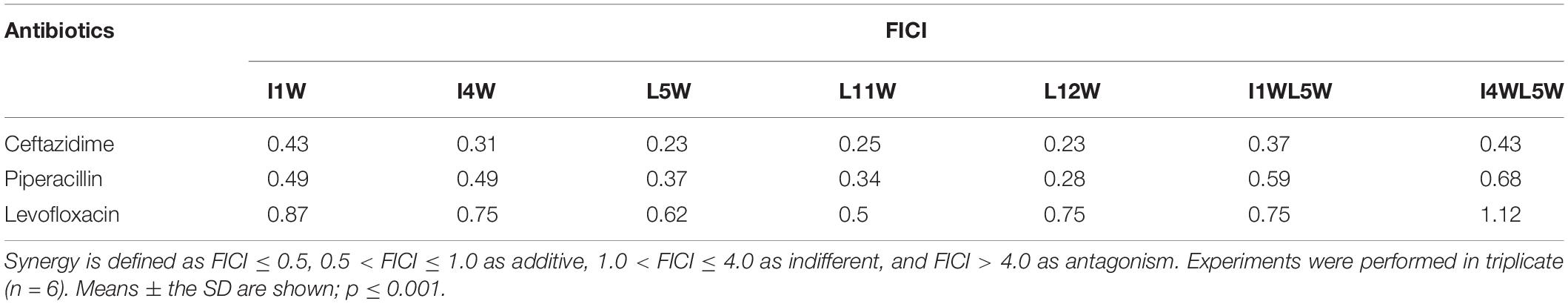
Table 1. The fractional inhibitory concentration index (FICI) of the Trp-containing peptides in combination with antibiotics against MRPA0108.
The Trp-Containing Peptides Increased the Antibacterial Activity of Antibiotics by Inhibiting β-Lactamase Activity in MRPA0108 Cells
Both ceftazidime and piperacillin are β-lactam antibiotics. To understand the synergistic mechanism of I1W and L12W in combination with ceftazidime and piperacillin, β-lactamase activity in MRPA0108 cells was detected. The results showed that the β-lactamase activity increased fourfold and 1.8-fold in MRPA0108 cells treated with ceftazidime and piperacillin alone compared to that in the control, respectively (Figure 7). I1W and L12W alone at concentrations greater than 12.5 μM reduced β-lactamase activity compared to that in the control, but at low concentrations, they had no effect on β-lactamase activity. The combination of I1W and L12W with ceftazidime or piperacillin decreased β-lactamase activity in a concentration-dependent manner compared to that with the antibiotics alone. Specifically, the combination of ceftazidime or piperacillin with L12W even at low concentrations (3.13 μM), significantly decreased β-lactamase activity by 15.8 and 23.5%, respectively. L12W at 25 μM combined with ceftazidime and piperacillin caused a reduction of 31.6 and 58.8% in β-lactamase activity, respectively. These results suggested that the ceftazidime/piperacillin-peptide combination displayed highly synergistic activity on β-lactamase production.
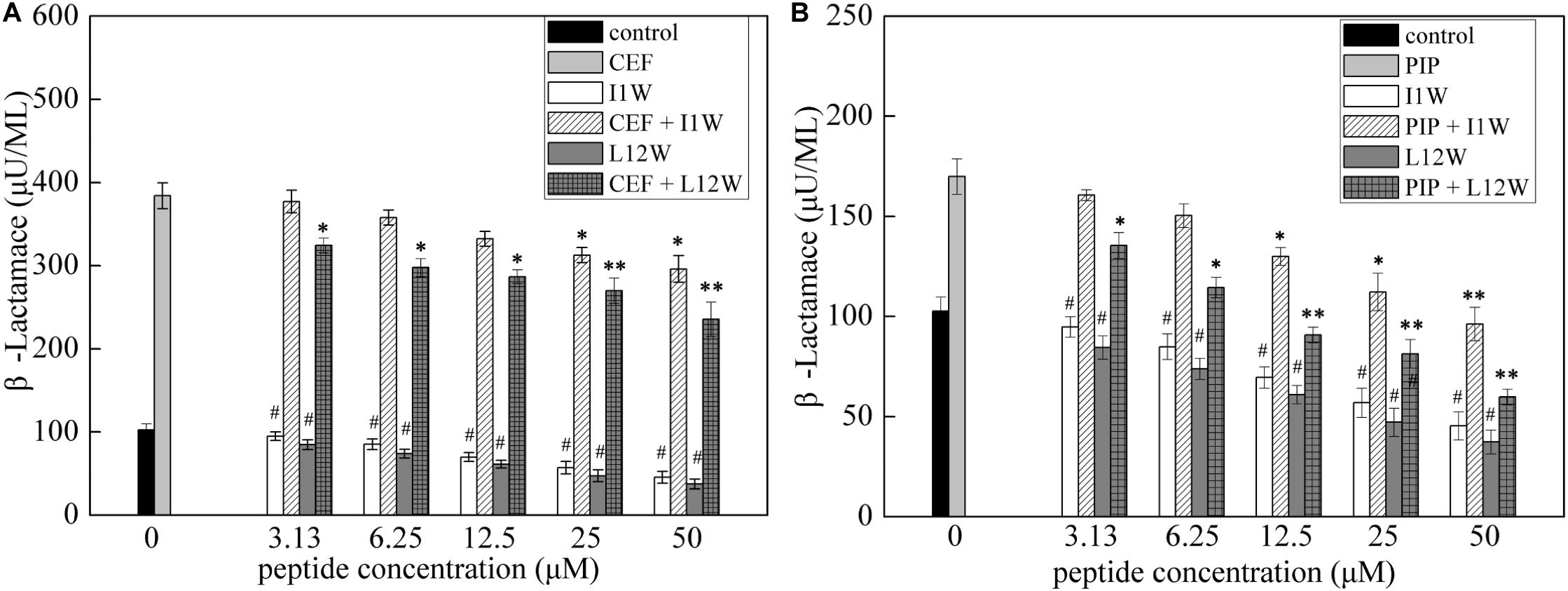
Figure 7. β-lactamase activity in the MRPA0108 cells treated with the Trp-containing peptides in combination with β-lactama antibiotics ceftazidime (A) and piperacillin (B). Bacteria were treated with a series of concentration of I1W and L12W for 12 h at 37°C. β-lactamase activity in the prepared β-lactamase-containing lysates was measured by using a β-lactamase assay kit. *P < 0.05 and **P < 0.01 indicate statistically significant differences between antibiotics alone and antibiotics in combination with peptide, and #P < 0.01 between peptide alone and antibiotics alone, respectively. CEF, ceftazidime; PIP, piperacillin.
The Trp-Containing Peptides Inhibit the Gene Expression of Efflux Pumps, Resulting in an Increase in Antibiotics in MRPA0108 Cells
Previous studies have clearly shown that QS systems are involved in antimicrobial resistance regulation via efflux pump genes, including MexAB-OprM and MexXY/OprM (Evans et al., 1998; Minagawa et al., 2012; Rasamiravaka and Jaziri, 2016). Many of the clinical strains of P. aeruginosa with high expression of the lasI and lasR genes exhibit elevated MexA/B gene expression, resulting in high resistance to currently used broad-spectrum drugs (Pourmand et al., 2013). In our study, relative expression of the MexA, MexX, and OprM/D genes was detected using RT-qPCR. The results showed that the transcription level of the OprD gene significantly increased and that the expression of the OprM, MexX, and MexA genes was reduced upon Trp-containing peptide addition at 25 μM in MRPA0108 cells (Figures 8A–D). The peptides increased the gene expression of OprD 1.3–2.5-fold compared to that in the control and decreased the expression of the OprM, MexX and MexA genes by 21–68, 40–75, and 70–90%, respectively. However, I1W at its MIC (6.25 μM) reduced the OprD/M gene expression by 27 and 22.3%, respectively, and L12W at its MIC (25 μM) reduced the OprD/M gene expression by 72 and 41.3%, respectively (Supplementary Figure S2). The results suggested that the Trp-containing peptides were able to increase antibiotic content in cells by inhibiting the expression of efflux pump genes.
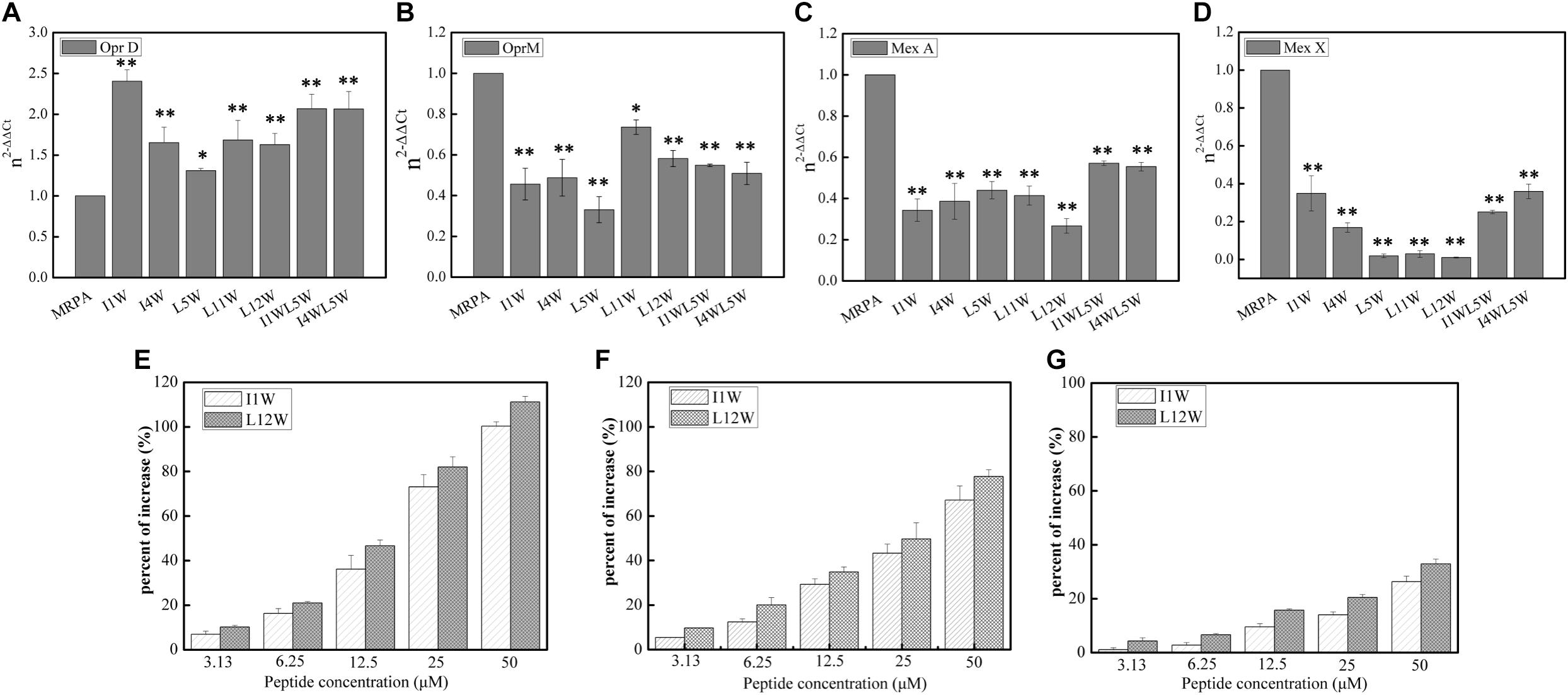
Figure 8. Effect of the Trp-containing peptides on the antibiotics content in MRPA0108 cells. (A–D) Relative expression of porin gene OprD (A), efflux pump gene OprM (B), MexA (C), and MexX (D) was measured by real-time qPCR. Fold changes of gene expression were normalized to the level of rpsL gene and quantitated relative to gene expression of control was normalized to 1 using the comparative Ct method. (E–G) The content of antibiotics ceftazidime (E), piperacillin (F), and levofloxacin (G) in MRPA0108 cells treated with the Trp-containing peptides was measured by HPLC. Results represent the average of at least four independent experiments SD (*P < 0.05 and **P < 0.01).
Subsequently, we detected the contents of the antibiotic ceftazidime, piperacillin, and levofloxacin in MRPA0108 cells treated with or without the Trp-containing peptides I1W and L12W by RP-HPLC. As shown in Figures 8E–G, I1W and L12W increased the three antibiotics contents inside the MRPA0108 cells in a concentration-dependent manner. I1W caused an increase of 7.5-100% for ceftazidime, 8–80% for piperacillin and 0.01–22% for levofloxacin. L12W caused an increase of 8–100% for ceftazidime, 5–62% for piperacillin and 2.1–38% for levofloxacin. Similarly, I1W at its MIC increased the content of ceftazidime by 19%, piperacillin by 17% and levofloxacin by 1% in MRPA0108 cells, but L12W at its MIC increased the content of ceftazidime by 81%, piperacillin by 58%, and levofloxacin by 23%. These results indicated that the Trp-containing peptides reduced antibiotic efflux from MRPA0108 cells by inhibiting efflux pump gene expression, thus increasing the antibacterial activity of these antibiotics against the MRPA0108 strain.
Discussion
QS systems in P. aeruginosa play a role in the production of virulence factors, bacterial swarming motility and development of biofilm and are a potential target as a new therapeutic approach in bacterial infection and biofilm formation (Rasamiravaka and Jaziri, 2016). The present study focused on the QS attenuation potential of Trp-containing peptides and the role of the peptides on the virulence and biofilm formation regulated by quorum sensing of MRPA. In P. aeruginosa, QS activates virulence factors proteases and elastase regulated by the las system, and pyocyanin and rhamnolipids regulated by the rhl system (Pattnaik et al., 2018). This study confirms that Trp-containing peptides affect global regulatory QS signaling in MRPA0108. In this study, four virulence factor regulatory genes in the QS system of MRPA0108 were significantly downregulated in a concentration-dependent manner upon Trp-containing peptide exposure, namely, lasA, lasB, rhlA, and rhlB. The peptides at low concentrations (1/4 × MIC or 1/8 × MIC) strongly decreased the relative expression levels of the rhlA and rhlB genes. Consistent with these results, the Trp-containing peptides significantly inhibited the production of the LasB elastase and LasA protease regulated by the las system, with the highest inhibition being 24 and 44% at peptide concentrations of 1/4 × MIC, respectively. The production of pyocyanin and rhamnolipids regulated by the rhl system was reduced maximally to 73 and 44% over that of the untreated control after the peptide treatments at 1/4 × MIC, respectively. Compared with the effect on elastase and total protease production, the Trp-containing peptides showed more significant inhibitory effect on the production of pyocyanin and rhamnolipid, suggesting that the inhibitory effect of the peptides on the rhl system is better than that on the las system.
Davies et al. (1998) reported that the lasI mutant produced a flat and densely packed multilayer biofilm that was different from the large-aggregate wild-type biofilms and was sensitive to detergent treatment. Subsequent studies revealed that QS plays an important role in biofilm formation in P. aeruginosa, in which the lasI/lasR and rhlR/rhlI systems are expressed during the initial stage of biofilm formation and the maturation stage of P. aeruginosa biofilm development (Bjarnsholt et al., 2010; Wrafy et al., 2016). Among virulence factors produced by P. aeruginosa, pyocyanin, and rhamnolipids regulated by the rhl system are responsible for the deposition of extracellular DNA (eDNA), which is a major biofilm component of the EPS, microcolony formation and structural development in the early and late biofilm development stages, respectively (Pamp and Nielsen, 2007; Brackman and Coenye, 2015; Qadri et al., 2016; Silva et al., 2017). Thus, they are key mediators of bacterial persistence in P. aeruginosa. Rhamnolipid-deficient P. aeruginosa cannot maintain open spaces to aid nutrient diffusion through densely populated mature biofilms (Davey et al., 2003). The ability of the Trp-containing peptides to modify QS signaling and reduce the production of pyocyanin, proteases and rhamnolipids in MRPA0108 may influence critical downstream functions, such as biofilm formation. Confocal laser scanning microscopy and CV dye assays demonstrated that peptide-treated biofilms had a concentration-dependent decrease in biomass. A maximum reduction of 53% occurred with the treatment with I1W at 1 × MIC, and an average biofilm thickness of 6 μm decreased to 1.5 μm after treatment with a low concentration of I1W. For 1 day mature biofilms, Trp-containing peptides at concentrations ranging from 1/8 × MIC to 1 × MIC reduced the existing-biofilm biomass, but they were less effective in the degradation of 1-day mature biofilms and reduced 1-day-old MRPA0108 strain biofilms by only 15–30%. Although QS regulation of biofilms is complex and involves numerous intrinsic and environmental factors, such as the matrix density, EPS composition, cell number, and oxygen availability, rhamnolipids, proteases, and pyocyanin participate in optimal QS signaling of biofilm (Furiga et al., 2016).
At least three polysaccharides, Psl, Pel, and alginate are produced to stabilize the biofilm structure in P. aeruginosa (Ryder et al., 2007; Ghafoor et al., 2011). Our data also showed that the synthesis of alginate and Psl was significantly inhibited in MRPA0108 cells treated with the Trp-containing peptides for 24 h, and the transcription levels of the pelA, algD, and pslA genes were significantly downregulated. As a result, alginate and Psl production showed a significant reduction, with maximum inhibition of 53–74% and 12–74%, after treatment with Trp-containing peptides at 1 × MIC, respectively. Psl, a major biofilm matrix polysaccharide, can act as a signal to stimulate its own production and that of other biofilm components (alginate and Pel) via the intracellular secondary messenger molecule c-di-GMP, which is regulated by two diguanylate cyclases (Irie et al., 2012). On the other hand, Gilbert et al. found that the QS regulator LasR can regulate psl expression by binding itself to the promoter region of the psl operon (Gilbert et al., 2009). Here, peptide-driven downregulation of polysaccharide biosynthesis plays an important role in biofilm information, in which the Trp-containing peptides reduce the amounts of EPSs to decrease the biofilm structural stability and interfere with MRPA0108 biofilm formation.
The bacterial swarming motility assay further supported a significant effect of the peptides on bacterial flagellum-and pilus-mediated chemotaxis. QS regulates swarming motility in P. aeruginosa to affect both biofilm formation and established biofilm disruption (Overhage et al., 2007). Motility plays an important role in colonization and exploitation of new niches of P. aeruginosa. Swarming motility is a complex behavior influenced by a large number of different genes and has been linked to increased antibiotic resistance (Overhage et al., 2008). Rhamnolipids can modulate the intricate P. aeruginosa swarming motility patterns (Caiazza et al., 2005). Therefore, it was perhaps unsurprising that Trp-containing peptides affected the MRPA0108 swarming motility. The study of Overhage et al. (2007) demonstrated that a series of “swarming-negative” P. aeruginosa mutants obtained via the insertion of mini-Tn5 exhibited impaired biofilm formation, indicating the link between the two phenotypes.
Previous studies have demonstrated that combination of antibiotics with some cationic AMPs have synergistic effects (Maisetta et al., 2009; Kajal et al., 2015; Regmi et al., 2017). Such synergy increases the antibacterial activity of antibiotic and peptide and helps prevent bacterial resistance. Most cationic AMPs, such as magainin II, ranalexin, cecropin P1, indolicidin, and buforin II, adopt an amphiphilic α-helical confirmation and show membrane-disrupting activity (Giacometti et al., 2000; He et al., 2015). The increase of membrane permeability leads to easy cellular uptake of small molecule antibiotics and exerts synergistic effects. Ranalexin, a 20-residue AMP from the skin secretion of Rana catesbiana Shaw, was demonstrated to act in a synergistic manner with clarithromycin, polymyxin E, and doxycycline against Staphylococcus aureus because of its membrane-disrupting activity, leading to entry of hydrophobic compounds into the cell (Viljanen et al., 1991; Zhou and Peng, 2013). Similarly, magainin II was observed to be synergistic with the clinically used antibiotics amoxicillin, ceftriaxone, ceftazidime, piperacillin, and meropenem, but buforin II, cecropin P1 and indolicidin did not show synergistic effects in combinations with these antibiotics against S. aureus (Falla and Hancock, 1997; Zhou and Peng, 2013). Our previous study showed that the combination of penicillin, ampicillin and erythromycin with the Trp-containing peptides L11W, L12W, I1WL5W, and I4WL5W had a distinct synergistic effect against MDR Staphylococcus epidermidis (MRSE) in vitro and in vivo, but ceftazidime in combination with the peptides exhibited additive activity (Shang et al., 2019). Interestingly, the present study demonstrated that the combination of all of the tested Trp-containing peptides with ceftazidime provided a synergistic effect against MRPA0108, with FICI values of 0.23–0.43. Piperacillin had a synergistic effect with I1W, I4W, L5W, L11W, and L12W, with FICI values of 0.28–0.49, but additive action with I1WL5W and I4W5W, with FICI values of 0.59 and 0.68, respectively. Levofloxacin exhibited only additive action with the tested Trp-containing peptides, with FICI values of 0.51–0.87, except I4W5W, which had an indifferent effect (FICI value: 1.12). In combination with a low peptide concentration equivalent to 1/4 × MIC, ceftazidime and piperacillin showed improved antimicrobial activity, with 32-fold- and 11-fold-reduced MIC values for ceftazidime and piperacillin, respectively, but only fourfold reduced MIC values for levofloxacin. Notably, although L11W and L12W alone had the lowest antibacterial activities against the MRPA0108 strain because of their low membrane disruption ability, they showed the highest synergy with ceftazidime and piperacillin. In addition, Reffuveille et al. (2014) demonstrated that the combinations of a broad-spectrum antibiofilm peptide at a subinhibitory levels and the antibiotics ceftazidime and ciprofloxacin prevented biofilm formation in P. aeruginosa and triggered cell death in mature P. aeruginosa biofilms, indicating a novel strategy to potentiate antibiotic activity against biofilms formed by multidrug-resistant pathogens.
In Pseudomonas, β-lactamases production is the most important mechanism of antibiotic resistance to penicillin, monobactams, and cephalosporins (Berrazeg et al., 2015; Bush and Bradford, 2016). At present, in addition to developing novel antibiotics by modifying the structure of β-lactams, the coadministration of a β-lactam antibiotic together with other compounds or biomolecules to inhibit β-lactamase is another relevant strategy (Peppoloni et al., 2020). Our results showed that β-lactamase activity increased fourfold and 1.8-fold in MRPA 0108 cells treated with ceftazidime or piperacillin alone, respectively, but the tested Trp-containing peptides I1W and L12W in combination with ceftazidime or piperacillin significantly decreased β-lactamase activity. Specifically, the β-lactamase activity was reduced by 31.6 and 58.8% in MRPA0108 treated with L12W along with ceftazidime or piperacillin, respectively. These results suggested that the ceftazidime/piperacillin-peptide combination displayed highly synergistic activity on β-lactamase production. Both the ceftazidime and piperacillin used in the present study are β-lactam antibiotics. In P. aeruginosa, AmpC encoding β-lactamase is regulated by AmpR, the AmpC β-lactamase regulator. The previous studys found that AmpR altered the production of β-lactamase by upregulating AmpC gene expression. AmpR, the LysR-type transcriptional regulator, not only regulates AmpC transcription by changing its conformation to modulate RNA polymerase activity (Ito et al., 2018; Torrens et al., 2019), but also controls the expression of 313 genes including metabolic, antibiotic-resistance and virulence genes (Kong et al., 2005). AmpR plays an important role in multiple virulence mechanisms by positively regulating the gene expression of lasB and rhlR and downstream genes (Lister et al., 2009; Balasubramanian et al., 2012). Moreover, mutation of AmpR results in a high expression of β-lactamases and higher levels of the virulence factors extracellular proteases and pyocyanin through quorum sensing (Kong et al., 2005). However, little is known about how QS signal molecules impact chromosomal AmpC β-lactamases. Our present results suggest a positive correlation between β-lactamase production inhibition and las/rhl system downregulation by the tested Trp-containing peptides. However, whether inhibition of β-lactamase production by the peptides results from its regulation of the QS system remains to be fully elucidated. Notably, further studies about the effect of the peptides on β-lactamases expression modulated by AmpC/AmpR will be helpful for further understanding the complex molecular mechanisms.
In addition to the production of β-lactamase, the expression of antibiotic multidrug efflux pumps, such as the MexAB/OprM, MexCD/OprJ, MexEF/OprN, and MexXY/OprM systems, intrinsically high levels of antibiotic resistance occur through the extrusion of a broad range of substrates, including antibiotics and chemotherapeutic agents in P. aeruginosa (Pourmand et al., 2013; Rasamiravaka and Jaziri, 2016). MexB, MexD, MexF, and MexY, which are located in the inner membrane, act in conjunction with the outer membrane proteins OprM, OprJ, and OprN through the periplasmic proteins MexA, MexC, MexE, and MexX, respectively. This study showed that the Trp-containing peptides I1W and L12W significantly inhibited the relative expression of the OprM, MexX and MexA genes but increased the level of transcription of the OprD gene. I1W at its MIC (6.25 μM) reduced OprD/M expression by 27 and 22.3%, and L12W at its MIC (25 μM) reduced OprD/M expression by 72 and 41.3%, respectively, suggesting that L12W had a better effect than I1W on inhibiting molecule efflux. Our results also confirm that the Trp-containing peptides I1W and L12W increased the contents of the three antibiotics inside the MRPA0108 cells by RP-HPLC. I1W at its MIC increased the content of ceftazidime by 19%, piperacillin by 17%, and levofloxacin by 1% in MRPA0108 cells, but L12W at its MIC increased the content of ceftazidime by 81%, piperacillin by 58%, and levofloxacin by 23%. These results indicated that the Trp-containing peptides reduced antibiotic efflux from MRPA0108 cells by inhibiting efflux pump gene expression, thus increasing the antibacterial activity of these antibiotics against MRPA0108. The biochemical targets of ceftazidime, piperacillin and levofloxacin are located intracellularly, and the mechanism of inhibiting β-lactamase activity and antibiotic multidrug efflux pump expression could have assisted the antibiotics in inhibiting their molecular targets.
Previous studies have clearly shown that the efflux pump system is associated with QS systems in P. aeruginosa (Evans et al., 1998; Minagawa et al., 2012). Many of the clinical MRPA strains overexpressing lasI and lasR exhibit elevated MexAB gene expression, resulting in high levels of resistance to currently used broad-spectrum antibiotics (Jeannot et al., 2008; Stickland et al., 2010). Similarly, a positive correlation between mexXY/oprM expression with the las system has also been found in some clinical MRPA strains (Pourmand et al., 2013). Furthermore, the exogenous addition of the signaling autoinducer C4-HSL enhanced mexAB/oprM operon expression in P. aeruginosa, indicating that C4-HSL accumulation in the medium directly induced mexAB-oprM operon expression in the stationary phase (Maseda et al., 2004). However, Evans et al. reported that the overexpressing mexAB/oprM in P. aeruginosa mutant strains lowered the expression of lasI, resulting in the decrease in the production of C12-HSL and virulence factors pyocyanin, elastase, and casein protease compared to wild-type strains (Evans et al., 1998; Rico et al., 2018). In agreement with the findings of Pourmand et al. (2013), the Trp-containing peptides inhibited MexAB/OprM expression and decreased virulence factor production regulated by las and rhl, indicating a positive correlation between mexAB/oprM expression and the las and rhl systems in the clinical strain MRPA 0108, in which decreased las and rhl gene expression result in reduced mexA, mexB, and oprM gene expression. This phenomenon may not be related simply to QS inhibition observed here with the peptides in P. aeruginosa, and the underlying reasons for the correlation between QS and the efflux pump mexAB/oprM remain to be elucidated.
Conclusion
Assessing the effects of Trp-containing AMPs on virulence factors and biofilm formation and development regulated by QS in MRPA is essential for the development of potential drug targets. In the present study, the Trp-containing peptides exhibited significant QS attenuation potential in downregulating QS-regulated virulence factor production in MRPA. In addition, the Trp-containing peptides showed significant antibiofilm potential, suggesting their role in altering bacterial tolerance to conventional antibiotics. The present study will provide a new perspective for developing novel and highly effective antiinfective drugs targeting bacterial virulence.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
This study uses strains obtained from clinical specimens submitted to The First Affiliated Hospital of Dalian Medical University. The ethics committee of The First Affiliated Hospital of Dalian Medical University did not require the study to be reviewed or approved by an ethics committee because the samples were primarily isolated for clinical diagnosis, rather than for scientific research.
Author Contributions
DS conceived and designed the experiments and wrote the manuscript. XH, WD, and ZK performed the experiments. FJ performed the isolation, identification, and antimicrobial susceptibility testing (AST) of MRPA0108. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Nature Natural Science Foundation of China (31672289).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.611009/full#supplementary-material
References
Abd-El-Aziz, A. S., Agatemor, C., Etkin, N., Overy, D. P., Lanteigne, M., McQuillan, K., et al. (2015). Antimicrobial organ ometallic dendrimers with tunable activity against multidrug-resistant bacteria. Biomacromolecules 16, 3694–3703. doi: 10.1021/acs.biomac.5b01207
Balasubramanian, D., Schneper, L., Merighi, M., Smith, R., Narasimhan, G., Lory, S., et al. (2012). The regulatory repertoire of Pseudomonas aeruginosa AmpC ß-lactamase regulator AmpR includes virulence genes. PLoS One 7:e34067. doi: 10.1371/journal.pone.0034067
Berditsch, M., Jäger, T., Strempel, N., Schwartz, T., Overhage, J., and Ulricha, A. S. (2015). Synergistic effect of membrane-active peptides polymyxin B and gramicidins on multidrug-resistant strains and biofilms of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 259, 5288–5296. doi: 10.1128/aac.00682-15
Berrazeg, M., Jeannot, K., Ntsogo Enguéné, V. Y., Broutin, I., Loeffert, S., Fournier, D., et al. (2015). Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob. Agents Chemother. 59, 6248–6255. doi: 10.1128/aac.00825-15
Bi, X., Wang, C., Dong, W., Zhu, W., and Shang, D. (2014). Antimicrobial properties and interaction of two Trp-substituted cationic antimicrobial peptides with a lipid bilayer. J. Antibiot. 67, 361–368. doi: 10.1038/ja.2014.4
Bi, X., Wang, C., Ma, L., Sun, Y., and Shang, D. (2013). Investigation of the role of tryptophan residues in cationic antimicrobial peptides to determine the mechanism of antimicrobial action. J. Appl. Microbiol. 115, 663–672. doi: 10.1111/jam.12262
Bjarnsholt, T., Tolker-Nielsen, T., Høiby, N., and Givskov, M. (2010). Interference of Pseudomonas aeruginosa signalling and biofilm formation for infection control. Expert. Rev. Mol. Med. 12:e11.
Brackman, G., and Coenye, T. (2015). Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 21, 5–11. doi: 10.2174/1381612820666140905114627
Breidenstein, E. B. M., Fuente-Núñez, C., and Hancock, R. E. W. (2011). Pseudomonas aeruginosa: all roads lead to resistance. Trend Microbiol. 19, 419–426. doi: 10.1016/j.tim.2011.04.005
Breij, A., Riool, M., Cordfunke, R. A., Malanovic, N., Boer, L., Koning, R. I., et al. (2018). The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 10:eaan4044. doi: 10.1126/scitranslmed.aan4044
Bush, K., and Bradford, P. A. (2016). β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb. Perspect. Med. 6, 295–306.
Caiazza, N. C., Shanks, R. M., and O’Toole, G. A. (2005). Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187, 7351–7361. doi: 10.1128/jb.187.21.7351-7361.2005
Carver, T., and Bleasby, A. (2003). The design of Jemboss: a graphical user interface to EMBOSS. Bioinformatics 19, 1837–1843. doi: 10.1093/bioinformatics/btg251
Chan, D. I., Prenner, E. J., and Vogel, H. J. (2006). Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim. Biophys. Acta-Biomembr. 1758, 1184–1202. doi: 10.1016/j.bbamem.2006.04.006
Coenye, T. (2010). Social interactions in the Burkholderia cepacia complex: biofilms and quorum sensing. Future Microbiol. 10, 87–99.
Cummins, J., Reen, F. J., Baysse, C., Mooij, M. J., and O’Gara, F. (2009). Subinhibitory concentrations of the cationic antimicrobial peptide colistin induce the pseudomonas quinolone signal in Pseudomonas aeruginosa. Microbiology 155, 2826–2837. doi: 10.1099/mic.0.025643-0
Davey, M. E., Caiazza, N. C., and O’Toole, G. A. (2003). Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas Aeruginosa PAO1. J. Bacteriol. 185, 1027–1036. doi: 10.1128/jb.185.3.1027-1036.2003
Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W., and Greenberg, E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298. doi: 10.1126/science.280.5361.295
Eisenberg, D., Weiss, R. M., and Terwilliger, T. C. (1982). The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299, 371–374. doi: 10.1038/299371a0
Evans, K., Passador, L., Srikumar, R., Tsang, E., Nezezon, J., and Poole, K. (1998). Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180, 5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998
Falla, T., and Hancock, R. E. (1997). Improved activity of a synthetic indolicidin analog. Antimicrob. Agents Chemother. 41, 771–775. doi: 10.1128/aac.41.4.771
Fjell, C. D., Hiss, J. A., Hancock, R. E. W., and Schneider, G. (2011). Designing antimicrobial peptides: from follows function. Nat. Rev. Drug Discov. 11, 37–51. doi: 10.1038/nrd3591
Fothergill, J. L., Panagea, S., Hart, C. A., Walshaw, M. J., Pitt, T. L., and Winstanley, C. (2007). Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 7:45. doi: 10.1186/1471-2180-7-45
Furiga, A., Lajoie, B., El Hage, S., Baziard, G., and Roques, C. (2016). Impairment of Pseudomonas aeruginosa biofilm resistance to antibiotics by combining the drugs with a new quorum-sensing inhibitor. Antimicrob. Agents Chemother. 60, 1676–1686. doi: 10.1128/aac.02533-15
Ghafoor, A., Hay, I. D., and Rehm, B. H. A. (2011). Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77, 5238–5246. doi: 10.1128/aem.00637-11
Giacometti, A., Cirioni, O., Del Prete, M. S., Barchiesi, F., Fortuna, M., Drenaggi, D., et al. (2000). In vitro activities of membrane-active peptides alone and in combination with clinically used antimicrobial agents against Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 44, 1716–1719. doi: 10.1128/aac.44.6.1716-1719.2000
Gilbert, K. B., Kim, T. H., Gupta, R., Greenberg, E. P., and Schuster, M. (2009). Global position analysis of the pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 73, 1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x
Guo, X., Zheng, L., Zhang, K., Xu, L., and Wang, X. (2012). Determination of N-Acyl-homoserine lactones signal molecules by gas chromatography-mass spectrometry. J. Inst. Anal. 31, 347–350.
Han, X., Kou, Z., Jiang, F., Sun, X., and Shang, D. (2020). Interactions of designed trp-containing antimicrobial peptides with DNA of multidrug-resistant Pseudomonas aeruginosa. DNA Cell Biol. doi: 10.1089/dna.2019.4874
CrossRef Full Text [Epub ahead of print], | PubMed Abstract | Google Scholar
He, J., Starr, C. G., and Wimley, W. C. (2015). A lack of synergy between membrane-permeabilizing cationic antimicrobial peptides and conventional antibiotics. Biochim. Biophys. Acta 1848, 8–15. doi: 10.1016/j.bbamem.2014.09.010
Hollander, J. G., Mouton, J. W., and Verbrugh, H. A. (1998). Use of pharmacodynamic parameters to predict efficacy of combination therapy by using fractional inhibitory concentration kinetics. Antimicrob. Agents Chemother. 42, 744–748. doi: 10.1128/aac.42.4.744
Irie, Y., Borlee, B. R., O’Connor, J. R., Hill, P. J., Harwood, C. S., Wozniak, D. J., et al. (2012). Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. PNAS 109, 20632–20636. doi: 10.1073/pnas.1217993109
Ito, A., Nishikawa, T., Ota, M., Ito-Horiyama, T., Ishibashi, N., Sato, T., et al. (2018). Stability and low induction propensity of cefiderocol against chromosomal AmpC b-lactamases of Pseudomonas aeruginosa and Enterobacter cloacae. J. Antimicrob. Chemother. 73, 3049–3052. doi: 10.1093/jac/dky317
Jeannot, K., Elsen, S., Köhler, T., Attree, I., van Delden, C., and Plésiat, P. (2008). Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob. Agents Chemother. 52, 2455–2462. doi: 10.1128/aac.01107-07
Kajal, G., Singh, S., and Monique, L. H. (2015). Short, synthetic cationic peptides have antibacterial activity against Mycobacterium smegmatis by forming pores in membrane and synergizing with antibiotics. Antibiotics 4, 358–378. doi: 10.3390/antibiotics4030358
Khandelia, H., and Kaznessis, Y. N. (2007). Cation–π interactions stabilize the structure of the antimicrobial peptide indolicidin near membranes: molecular dynamics simulations. J. Phys. Chem. B. 111, 242–250. doi: 10.1021/jp064776j
Kong, K. F., Jayawardena, S. R., Indulkar, S. D., Del Puerto, A., and Koh, C. L. (2005). Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 49, 4567–4575. doi: 10.1128/aac.49.11.4567-4575.2005
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein. Cell. 6, 26–41. doi: 10.1007/s13238-014-0100-x
Lister, P. D., Wolter, D. J., and Hanson, N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610. doi: 10.1128/cmr.00040-09
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2, 1051–1060. doi: 10.1016/s1286-4579(00)01259-4
Ma, L., Jackson, K. D., Landry, R. M., Parsek, M. R., and Wozniak, D. J. (2006). Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure post attachment. J. Bacteriol. 188, 8213–8221. doi: 10.1128/jb.01202-06
Maisetta, G., Mangoni, M. L., Esin, S., Pichierri, G., Capria, A. L., Brancatisano, F. L., et al. (2009). In vitro bactericidal activity of the N-terminal fragment of the frog peptide esculentin-1b (Esc 1-18) in combination with conventional antibiotics against Stenotrophomonas maltophilia. Peptides 30, 1622–1626. doi: 10.1016/j.peptides.2009.06.004
Mant, C. T., Kovacs, J. M., Kim, H. M., Pollock, D. D., and Hodges, R. S. (2009). Intrinsic amino acid side-chain hydrophilicity/hydrophobicity coefficients determined by reversed-phase high-performance liquid chromatography of model peptides: comparison with other hydrophilicity/hydrophobicity scales. Biopolymers 92, 573–595. doi: 10.1002/bip.21316
Maseda, H., Sawada, I., Saito, K., Uchiyama, H., Nakae, T., and Nomura, N. (2004). Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. AgentsChemother. 48, 1320–1328. doi: 10.1128/aac.48.4.1320-1328.2004
Minagawa, S., Inami, H., Kato, T., Sawada, S., Yasuki, T., and Miyairi, S. (2012). RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl- homoserine lactones, for cell-to-cell communication. BMC Microbiol. 12:70. doi: 10.1186/1471-2180-12-70
O’May, C., and Tufenkji, N. (2011). The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl. Environ. Microbiol. 77, 3061–3067. doi: 10.1128/aem.02677-10
Overhage, J., Campisano, A., Bains, M., Torfs, E. C. W., Rehm, B. H. A., and Hancock, R. E. W. (2008). Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76, 4176–4182. doi: 10.1128/iai.00318-08
Overhage, J., Lewenza, S., Marr, A. K., and Hancock, R. E. W. (2007). Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 189, 2164–2169. doi: 10.1128/jb.01623-06
Pamp, S. J., and Nielsen, T. T. (2007). Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 189, 2531–2539. doi: 10.1128/jb.01515-06
Pasupuleti, M., Malmsten, M., and Schmidtchen, A. (2011). Antimicrobial peptides: a key component of innate immunity. Crit. Rev. Biotechnol. 32, 143–171.
Pattnaik, S. S., Ranganathan, S. K., Ampasala, D. R., Syed, A., Ameen, F., and Busi, S. (2018). Attenuation of quorum sensing regulated virulence and biofilm development in Pseudomonas aeruginosa PAO1 by Diaporthe phaseolorum SSP12. Microb. Pathogenesis 118, 177–189. doi: 10.1016/j.micpath.2018.03.031
Peppoloni, S., Pericolini, E., Colombari, B., Pinetti, D., Cermelli, C., Fini, F., et al. (2020). The β-lactamase inhibitor boronic acid derivative SM23 as a new anti-Pseudomonas aeruginosa biofilm. Front. Microbiol. 11:35. doi: 10.3389/fmicb.2020.00035
Pourmand, M. R., Sadighian, H., and Naderi, M. (2013). Relation between Expression of the las quorum-sensing system in clinical isolates of Pseudomonas aeruginosa and expression of efflux pump and ampC. J. Med. Bacteriol. 2, 32–40.
Qadri, S. M., Donkor, D. A., Bhakta, V., Eltringham-Smith, L. J., Dwivedi, D. J., Moore, J. C., et al. (2016). Phosphatidylserine externalization and procoagulant activation of erythrocytes induced by Pseudomonas aeruginosa virulence factor pyocyanin. J. Cell Mol. Med. 20, 710–720. doi: 10.1111/jcmm.12778
Rasamiravaka, T., and Jaziri, M. E. (2016). Quorum-sensing mechanisms and bacterial response to antibiotics in P. aeruginosa. Curr. Microbiol. 73, 747–753. doi: 10.1007/s00284-016-1101-1
Reffuveille, F., Fuente-Núñez, C., Mansour, S., and Hancock, R. E. W. (2014). A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 58, 5363–5371. doi: 10.1128/aac.03163-14
Regmi, S., Choi, Y. S., Choi, Y. H., Kim, Y. K., Cho, S. S., Yoo, J. C., et al. (2017). Antimicrobial peptide from Bacillus subtilis CSB138: characterization, killing kinetics, and synergistic potency. Int. Microbiol. 20, 43–53.
Rico, M. A., Pacheco, J. O., Ortega, C. A., Cámara, M., and Martiìnez, J. L. (2018). Role of the multidrug resistance efflux pump MexCD-OprJ in the Pseudomonas aeruginosa quorum sensing response. Front. Microbiol. 9:2752. doi: 10.3389/fmicb.2018.02752
Rydberg, H. A., Matson, M., Åmand, H. L., Esbjörner, E. K., and Nordén, B. (2012). Effects of tryptophan content and backbone spacing on the uptake efficiency of cell- penetrating peptides. Biochemistry 51, 5531–5539. doi: 10.1021/bi300454k
Ryder, C., Byrd, M., and Wozniak, D. J. (2007). Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10, 644–648. doi: 10.1016/j.mib.2007.09.010
Sergey, C., Natalia, G., Dmitry, T., and Andrey, Y. (2018). Biofilm infections between Scylla and Charybdis: interplay of host antimicrobial peptides and antibiotics. Infect. Drug Resist. 11, 501–514. doi: 10.2147/idr.s157847
Shang, D., Li, X., Sun, Y., Wang, C., Sun, L., and Wei, S. (2012). Design of potent, non-toxic antimicrobial agents based upon the structure of the frog skin peptide, temporin-1CEb from Chinese brown frog, Rana chensinensis. Chem. Biol. Drug Des. 79, 653–662. doi: 10.1111/j.1747-0285.2012.01363.x
Shang, D., Liu, Y., Jiang, F., Ji, F., Wang, H., and Han, X. (2019). Synergistic antibacterial activity of designed Trp-containing antibacterial peptides in combination with antibiotics against multidrug-resistant Staphylococcus epidermidis. Front. Microbiol. 10:2719. doi: 10.3389/fmicb.2019.02719
Shang, D., Meng, X., Zhang, D., and Kou, Z. (2017). Antibacterial activity of chensinin-1b, a peptide with a random coil conformation, against multiple-drug-resistant Pseudomonas aeruginosa. Biochem. Pharmacol. 143, 65–78. doi: 10.1016/j.bcp.2017.07.017
Silva, D. P., Schofield, M., Parsek, M. R., and Tseng, B. (2017). An update on the sociomicrobiology of quorum sensing in gram-negative biofilm development. Pathogens. 6:51. doi: 10.3390/pathogens6040051
Stickland, H. G., Davenport, P. W., Lilley, K. S., Griffin, J. L., and Welch, M. (2010). Mutation of nfxB causes global changes in the physiology and metabolism of Pseudomonas aeruginosa. J. Proteome Res. 9, 2957–2967.
Strateva, T., and Yordanov, D. (2009). Pseudomonas aeruginosa – a phenomenon of bacterial resistance. J. Med. Microbiol. 58, 1133–1148. doi: 10.1099/jmm.0.009142-0
Torrens, G., Hernaìndez, S. B., Ayala, J. A., Moya, B., Juan, C., Cava, F., et al. (2019). Regulation of AmpC-Driven β-Lactam Resistance in Pseudomonas aeruginosa: different pathways, different signaling. mSystems 4, e00524–19 doi: 10.1128/mSystems.00524-19
Viljanen, P., Matsunaga, H., Kimura, Y., and Vaara, M. (1991). The outer membrane permeability-increasing action of deacylpolymyxins. J. Antibiot. 44, 517–523. doi: 10.7164/antibiotics.44.517
Wittgens, A., Tiso, T., Arndt, T. T., Wenk, P., Hemmerich, J., and Muller, C. (2011). Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb. Cell Fact. 10:80. doi: 10.1186/1475-2859-10-80
Wrafy, F. A., Brzozowska, E., Górska, S., and Gamian, A. (2016). Pathogenic factors of Pseudomonas aeruginosa-the role of biofilm in pathogenicity and as a target for phage therapy. Postepy Hig. Med. Dosw (online) 70, 78–91. doi: 10.5604/01.3001.0010.3792
Yeaman, M. R., and Yount, N. Y. (2003). Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55. doi: 10.1124/pr.55.1.2
Yoon, J., Urban, C., Terzian, C., Mariano, N., and Rahal, J. J. (2004). In vitro double and triple synergistic activities of Polymyxin B, imipenem, and rifampin against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 48, 753–757. doi: 10.1128/aac.48.3.753-757.2004
Keywords: multidrug-resistant Pseudomonas aeruginosa, Trp-containing peptide, quorum sensing, virulence factor, biofilm, synergy
Citation: Shang D, Han X, Du W, Kou Z and Jiang F (2021) Trp-Containing Antibacterial Peptides Impair Quorum Sensing and Biofilm Development in Multidrug-Resistant Pseudomonas aeruginosa and Exhibit Synergistic Effects With Antibiotics. Front. Microbiol. 12:611009. doi: 10.3389/fmicb.2021.611009
Received: 28 September 2020; Accepted: 20 January 2021;
Published: 11 February 2021.
Edited by:
Younes Smani, Institute of Biomedicine of Seville (IBIS), SpainReviewed by:
Cesar de la Fuente-Nunez, University of Pennsylvania, United StatesJorge Andres Olivares Pacheco, Pontificia Universidad Católica de Valparaíso, Chile
Copyright © 2021 Shang, Han, Du, Kou and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dejing Shang, ZGpzaGFuZ0Bsbm51LmVkdS5jbg==
 Dejing Shang
Dejing Shang Xue Han1
Xue Han1