
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 February 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.609840
This article is part of the Research Topic Zoonotic Microorganisms and Spread of Acquired Polymyxin Resistance Determinants View all 15 articles
Background: The resistance to colistin and carbapenems in Klebsiella pneumoniae infections have been associated with increased morbidity and mortality worldwide. A retrospective observational study was conducted to determine the prevalence and molecular events contributing to colistin resistance.
Methods: Clinical samples were screened for colistin resistance and underlying mechanisms were studied by PCR-based amplification and sequence analysis of genes of two-component regulatory system (phoPQ and pmrAB), regulatory transmembrane protein-coding mgrB, and mobilized colistin resistance genes (mcr-1-8). Gene expression of pmrC and pmrK was analyzed by qRT-PCR, and the genetic relationship was assessed by MLST. The putative effect of amino-acid substitutions was predicted by a combination of bioinformatics tools.
Results: Of 335 Klebsiella spp. screened, 11 (3.2%) were identified as colistin-resistant (MIC range, 8 to >128 μg/ml). K. pneumoniae isolates belonged to clonal complex-11 (CC11) with sequence types (STs): 14, 16, 43, 54, 147 and 395, whereby four isolates conferred three novel STs (3986, 3987 and 3988) profiles. Sequence analysis revealed non-synonymous potentially deleterious mutations in phoP (T151A), phoQ (del87–90, del263–264, L30Q, and A351D), pmrA (G53S), pmrB (D150V, T157P, L237R, G250C, A252G, R315P, and Q331H), and mgrB (C28G) genes. The mgrB gene in three strains was disrupted by insertion sequences encoding IS1-like and IS5/IS1182 family-like transposase genes. All 11 isolates showed an elevation in the transcription level of pmrC gene. Mobilized colistin-resistance (mcr) genes were not detected. All but one of the colistin-resistant isolates was also resistant to carbapenems; β-lactamase genes blaNDM-1-like, blaOXA-48-like, and blaCTX-M-like were detected in eight, five, and nine isolates, respectively.
Conclusion: All the studied colistin- and carbapenem-resistant K. pneumoniae isolates were genetically distinct, and various mechanisms of colistin resistance were detected, indicating its spontaneous emergence in this bacterial species.
Klebsiella pneumoniae, a nosocomial pathogen, accounts for one-third of worldwide reported Gram-negative infections (Navon-Venezia et al., 2017). Multidrug-resistant (MDR) K. pneumoniae are resistant to extended-spectrum cephalosporins, carbapenems, aminoglycosides, and fluoroquinolones and present a significant challenge to the clinicians. Available data showcase a significant increase in K. pneumoniae in India’s carbapenem-resistant isolates from 29% in 2008 to 57% in 2016, limiting the treatment of life-threatening infections (Gandra et al., 2016; Dixit et al., 2019). The unavailability of new antimicrobial agents to combat carbapenem-resistant K. pneumoniae infections has revived the use of polymyxins (colistin and polymyxin B). However, the indiscriminate use of polymyxins in animals, aquaculture, and agriculture in the last decades has compounded the issue of drug resistance (Nguyen et al., 2016; Zhang et al., 2019).
Colistin-resistant K. pneumoniae exhibits a high degree of genetic plasticity where a point mutation and/or genetic disruption in two-component regulatory systems (TCRS), i.e., pmrAB and phoPQ, are known to confer polymyxin resistance (Cannatelli et al., 2014a; Jayol et al., 2014). Besides, polymyxin resistance is often observed by the inactivation of mgrB, a regulatory trans-membrane protein that controls the kinase activity of phoQ in phoPQ TCRS, by point mutations, indels, or insertion sequences (IS5-like, IS1F, ISKpn13, ISKpn14, IS10R; Cannatelli et al., 2015; Aires et al., 2016). The spread of colistin resistance has accelerated by acquiring plasmid-encoded mobile colistin resistance (mcr)- genes. After the first report in China (November 2015), mcr-genes were detected in E. coli, K. pneumoniae, Shigella sonnei, Salmonella enterica, and many other bacterial strains from colonized and infected humans, food (meat and vegetables), farm and wild animals, and aquatic environments (Liu et al., 2016; Carroll et al., 2019; Zhang et al., 2019). Selective colistin pressure in different sectors has induced constant emergence and evolution of mcr-genes, and mcr-1 to mcr-10 genes with multiple variants have been identified in colistin-resistant bacteria isolated from several sources (Gharaibeh and Shatnawi, 2019; Wang et al., 2020).
Increasing incidences of colistin resistance among nosocomial K. pneumoniae isolates have been reported from Europe, Asia, North America, South America, and Africa (Rojas et al., 2017; Lomonaco et al., 2018; Boszczowski et al., 2019; Zafer et al., 2019). Many isolated case reports and outbreaks of MDR K. pneumoniae infections were reported from different parts of India (Goel et al., 2014; Bhaskar et al., 2017; Kaur et al., 2017; Mohanty et al., 2017; Pragasam et al., 2017; Aggarwal et al., 2018). High mortality rate (approximately 69%) in bloodstream infections due to carbapenem- and colistin-resistant K. pneumoniae was also noted among Indian patients (Kaur et al., 2017; Manohar et al., 2017; Pragasam et al., 2017; Jajoo et al., 2018; Palani et al., 2020).
The accretion in clinical isolates of colistin- and carbapenem-resistant K. pneumoniae warrants further investigation into the epidemiology and underlying molecular mechanisms. Detection of colistin resistance is a challenge in clinical diagnosis, and evolving breakpoints have complicated the problem. Furthermore, horizontal gene transfer and the spread of clones with resistance traits confront the therapeutic control in nosocomial settings (Diene and Rolain, 2014). This study aimed to determine the prevalence of colistin-resistant K. pneumoniae in a tertiary care hospital in India; their clonal relationship and the molecular events contributing to colistin resistance.
Clinical samples collected at Safdarjung Hospital and directed to the Department of microbiology for routine identification from August 2017 to January 2018 were included in the study. Klebsiella spp. were identified and screened for colistin resistance following standard broth microdilution method using cation-adjusted Mueller Hinton Broth (HiMedia, India) with EUCAST interpretation (S: ≤2 mg/L, R: >2 mg/L) guidelines (2018). Isolates were identified by a VITEK 2 GN card and confirmed by 16S rRNA gene sequence analysis (Azam et al., 2016). Ethics clearance was obtained from the institutional ethics committee: (i) ICMR-National Institute of Pathology, New Delhi, India (IEC No: NIP-IEC/2/3/17/06) and (ii) VMMC and Safdarjung Hospital, New Delhi, India (IEC No: IEC/VMMC/SJH/Project/1028). The demographic and clinical details of the patients were obtained from electronic medical records available in the hospital intranet.
Colistin-resistant isolates were subjected to antibiotic susceptibility testing by Kirby-Bauer disc diffusion method on Mueller Hinton Agar plates (HiMedia, India) against the ampicillin, ampicillin/clavulanic acid, amikacin, azithromycin, cefotaxime, chloramphenicol, ciprofloxacin, gentamicin, ertapenem, imipenem, tazobactam/piperacillin, tetracycline, and trimethoprim.
Minimum inhibitory concentrations (MICs) were determined by E-test (trimethoprim-sulfamethoxazole, amoxicillin-clavulanic acid, and imipenem) and broth microdilution (chloramphenicol, ciprofloxacin, colistin, gentamicin, kanamycin, polymyxin B, rifampicin, nalidixic acid, tigecycline, and tetracycline). E. coli ATCC 25922 (antibiotic-susceptible), K. pneumoniae ATCC 700603 (ESBL-producing), and E. coli NCTC 13846 (colistin-resistant) isolates were used as quality control strains. Results were interpreted as per CLSI (2018) guidelines and for tigecycline, EUCAST (2018) guidelines were followed. The susceptibility profile of K. pneumoniae isolates was determined using the WHONET (v20.8.21) database software.
Multilocus sequence typing (MLST) for seven housekeeping genes, rpoB, gapA, mdh, pgi, phoE, infB, and tonB, was performed using the Pasteur institute MLST scheme (Diancourt et al., 2005). A total of 3,550 MLST profiles of K. pneumoniae available till 2nd March 2019, were extracted from Pasteur institute MLST database.1 Sequence types (STs) were determined by comparing the sequence of isolates against K. pneumoniae MLST database (Feil et al., 2004). Novel ST Profiles were submitted to the curator for assignment of new sequence type. Clustering and comparative analysis of related STs were performed using default conservative definition of sharing six of the seven loci of eBURST version 3 software.
Lipid A modifying genes, i.e., phoP, phoQ, pmrA, pmrB, and mgrB, associated with colistin resistance were amplified and sequenced using gene-specific primers designed using the external region of the gene sequence (Supplementary Table S1). To determine the presence of plasmid-encoded colistin resistance, gene-specific primers were used to amplify mcr-1-8 genes. K. pneumoniae isolates were also analyzed for the presence of the ESBL gene (blaCTX-M-like) and carbapenemase genes (blaKPC-like, blaNDM-1-like, and blaOXA-48-like), conferring resistance to a broad range of β-lactam antibiotics (primer sequence information used for the amplification of specific genes is given in Supplementary Table S1). Genomic DNA from K. pneumoniae ATCC 700603 (colistin-susceptible), E. coli NCTC 13846 (mcr-1 positive), K. pneumoniae ATCC BAA1705 (blaKPC positive), K. pneumoniae ATCC BAA2156 (blaNDM positive), and E. coli MRE2 (blaCTX-M positive, GenBank accession # KM873162) were used as control strains.
The expression level of pmrK (encoding L-Ara4N transferase) and pmrC (encoding PEtN transferase) genes were analyzed using gene-specific primers by qRT-PCR in colistin treated and untreated samples. Colistin-resistant isolates and K. pneumoniae ATCC 700603 were grown to the mid-log phase in cation-adjusted Mueller Hinton Broth supplemented with and without colistin sulphate (4 μg/ml; Sigma). Bacterial cells were harvested, and total RNA was extracted using RNeasy kit (Qiagen) and treated with DNaseI. cDNA synthesized from 1 μg of total RNA using RevertAid first-strand cDNA synthesis kit (Thermo Scientific) was subjected to Real-time PCR amplification in 25 μl reactions (in triplicates) containing 10 pmoL forward and reverse primers and 1x Fast SYBR green PCR master mix (Applied BioSystems) under the following conditions: 1 cycle of 95°C for 20 s, 40 cycles of 95°C for 3 s, and 60°C for 30 s. After each run, melt curve analysis was performed to ensure single amplicon production, under the conditions 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Relative gene expression levels were calculated using the 2ΔΔCT formula, and ≥2 fold change in the expression (relative to wild type) was considered the increase in expression and below ≤0.5 as repression. The rpsL gene (encoding ribosomal protein) was used as an internal control. Colistin-susceptible K. pneumoniae ATCC 700603 was used for normalization. The student’s paired “t” test was performed for calculating the significance of differences observed in the expression levels under colistin untreated and treated conditions with the help of GraphPad Prism software version 8.0 (GraphPad software inc. CA, United States).
The amino acid substitutions in the genes (mgrB, phoP, phoQ, pmrA, and pmrB) associated with the colistin resistance phenotype in clinical isolates were defined by comparing the query sequence against K. pneumoniae ATCC 700603. To predict the phenotypic effect of these substitutions on protein structure and function, these were analyzed by three different bioinformatics tools: sorting intolerant from tolerant (SIFT), polymorphism phenotyping (PolyPhen-2), and protein variation effect analyzer (PROVEAN) with default parameters (Ng and Henikoff, 2003; Adzhubei et al., 2010; Sim et al., 2012; Choi and Chan, 2015). The mutations found to be deleterious by at least two of the three analysis software were considered potentially deleterious, while mutations with low confidence intervals were treated as neutral.
The nucleotide sequences of colistin-resistant K. pneumoniae isolates under study were deposited at a GenBank nucleotide sequence database under the following accession numbers: 16S rRNA gene (MH411220, MH410611, MH411072–MH411080), phoP gene (MH424384–MH424392, MH450205, MH450206), phoQ gene (MH424393–MH424402, MH450207), pmrA gene (MH450188–MH450197), and pmrB gene (MH450198–MH450204, MH688167–MH688170). Nucleotide sequences of the mgrB genes have been deposited under GenBank accession numbers: wild type mgrB gene (MH424403–MH424409, MH671333) and mutated/disrupted mgrB gene (MK625062–MK625064).
A total of 335 K. pneumoniae were isolated from urine (n = 153), blood (n = 20), pus (n = 85), tracheal aspirate (n = 9), wound (n = 7), and other specimens (n = 61) within a period of 6 months (2017–2018) in the hospital. Of the total, 11 (3.2%) Klebsiella isolates representing one isolate per patient from the clinical samples viz. urine (5), pus (3), tissue (1), wound swab (1), and blood (1) were identified to be colistin-resistant. All 11 patients had a hospital stay for more than 10 days and encountered invasive devices like intravenous lines, ventilators, or catheters. These patients received colistin during or before their sample collection for microbiology culture analysis. Molecular identification confirmed the isolates as K. pneumoniae and MLST analysis revealed the sequence types ST14 (n = 1), ST16 (n = 1), ST43 (n = 1), ST54 (n = 1), ST147 (n = 1), and ST395 (n = 2; Table 1). Four of the isolates belonged to three novel ST profiles and were assigned new sequence type numbers as ST3986 (MRK1), ST3987 (MRK5 and MRK6), and ST3988 (MRK10). Comparative and clustering analysis by eBURST v3 of query dataset with a total of 3,550 already reported MLST profiles of K. pneumoniae suggested that all 11 isolates of the query dataset belonged to the most prominent and diverse clonal complex 11 (CC11). The novel sequence types ST3986, ST3987, and ST3988 had evolutionarily related sub-founders as ST16, ST43, ST39, respectively (Figure 1). The reference strain K. pneumoniae ATCC 700603 used in this study belonged to ST489 and was designated as a singleton by eBURST v3.
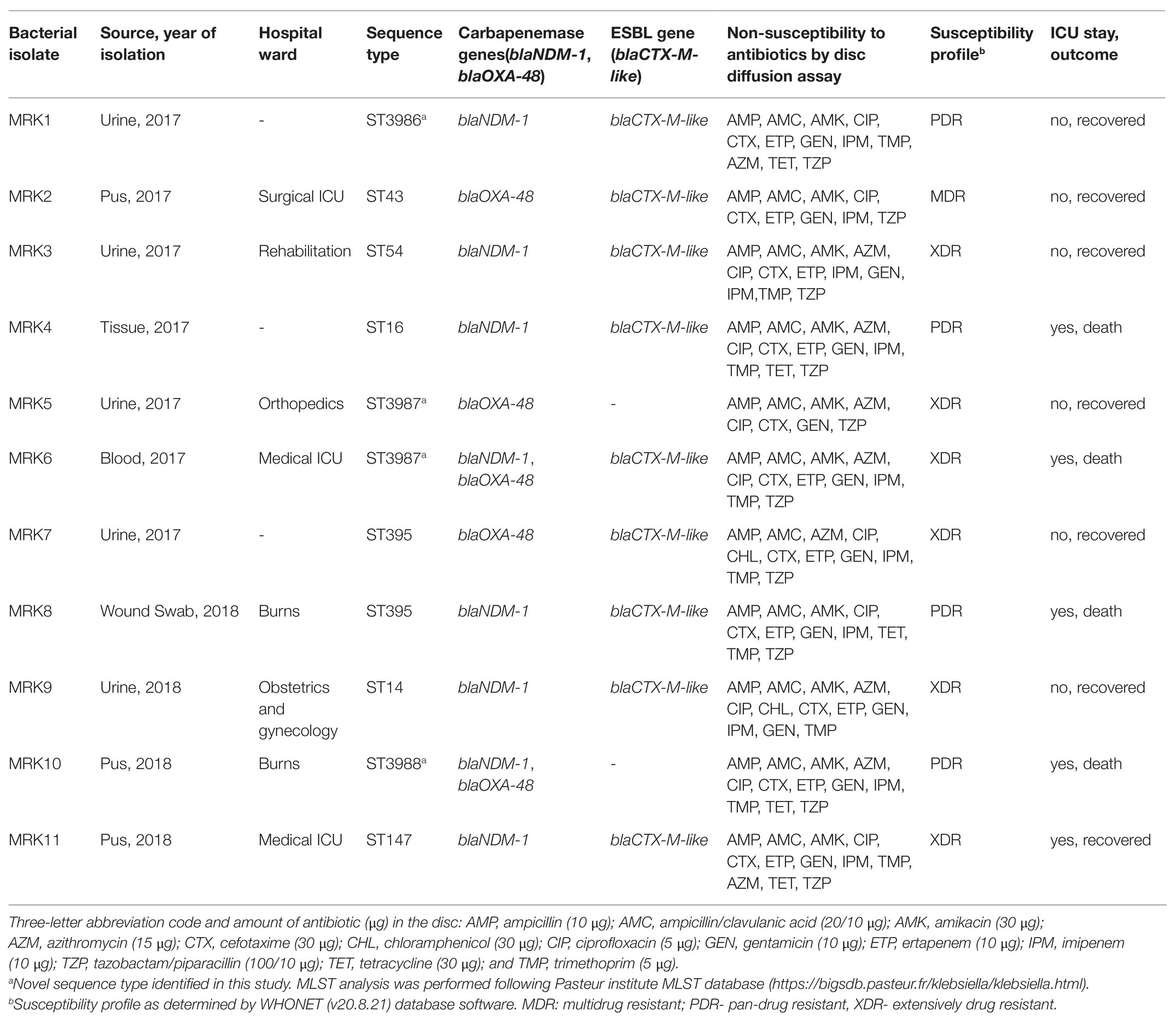
Table 1. Demographic, clinical, and microbiological features of 11 colistin-resistant K. pneumoniae isolates.
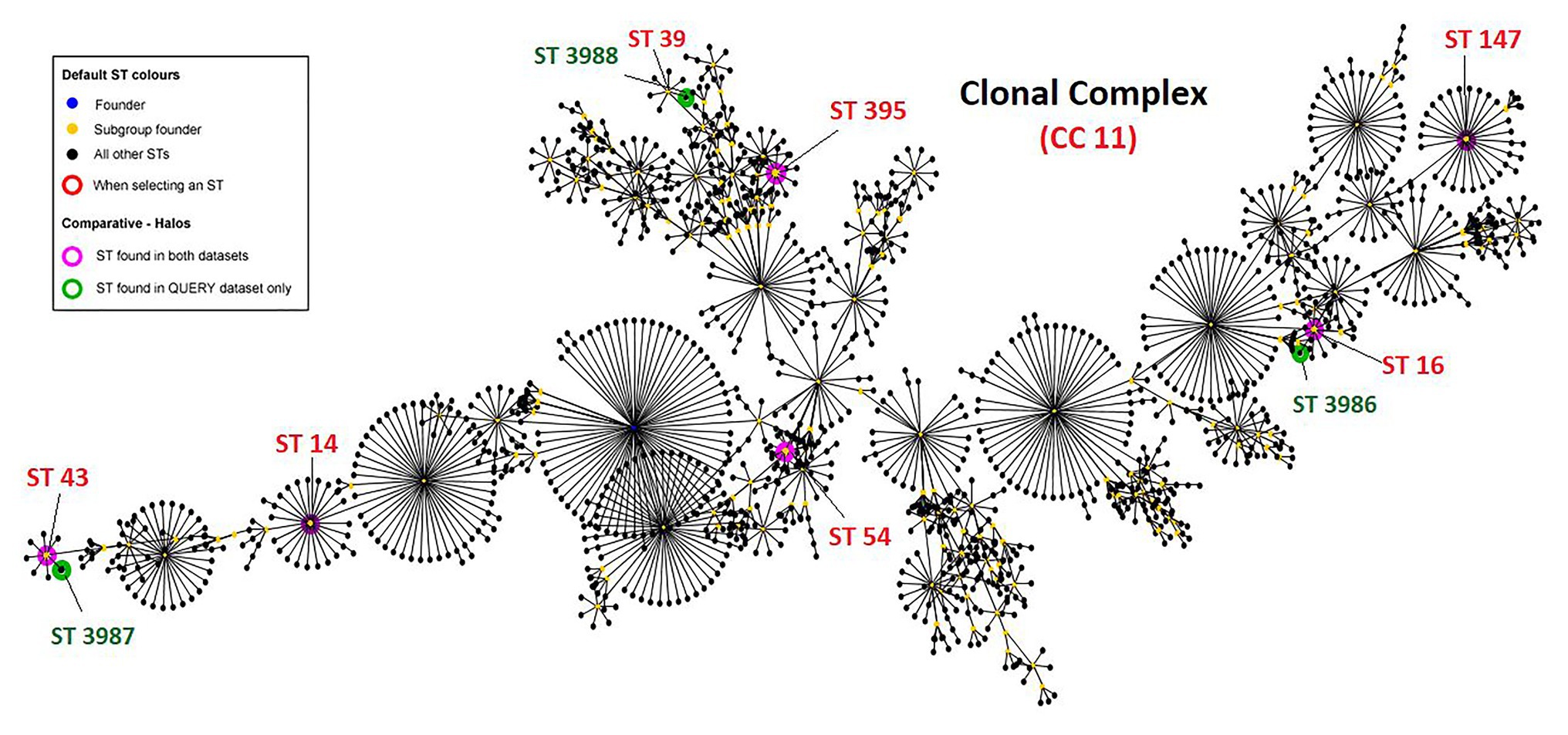
Figure 1. Clonal complex CC11 with 1,562 sequence types. Green halos represents novel sequence types found in this study and submitted to curator of Klebsiella pneumoniae MLST database Pasteur institute (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html), pink halos represent isolates with defined sequence types. K. pneumoniae ATCC 700603 is ST489 and designated as singleton by eBURST v3 and hence not shown here.
All 11 tested K. pneumoniae isolates were resistant to cephalosporins, carbapenems, fluoroquinolones, aminoglycosides, macrolides, and trimethoprim by disc diffusion method (Table 1). Three K. pneumoniae isolates (MRK1, MRK9, and MRK10) were resistant to all antibiotics tested by disc diffusion assay except with intermediate resistant phenotype to chloramphenicol (MRK1 and MRK10) and tetracycline (MRK9), as per CLSI standards. MIC-values for colistin and polymyxin B ranged from 8 to >128 μg/ml and 16 to >64 μg/ml, respectively. Unexpectedly, MICs of 5 (45.5%) isolates to colistin were ≥128 μg/ml (Table 2). MICs of amoxicillin/clavulanic acid and imipenem were >256 μg/ml and ≥8 μg/ml, respectively, for the 11 tested isolates except for MRK5 (4 μg/ml and 2 μg/ml). High MIC values were noted for kanamycin (≥256 μg/ml; n = 10), gentamycin (≥256 μg/ml; n = 6), co-trimoxazole (>32 μg/ml; n = 9), and tetracycline (>128 μg/ml; n = 5) among colistin-resistant K. pneumoniae.
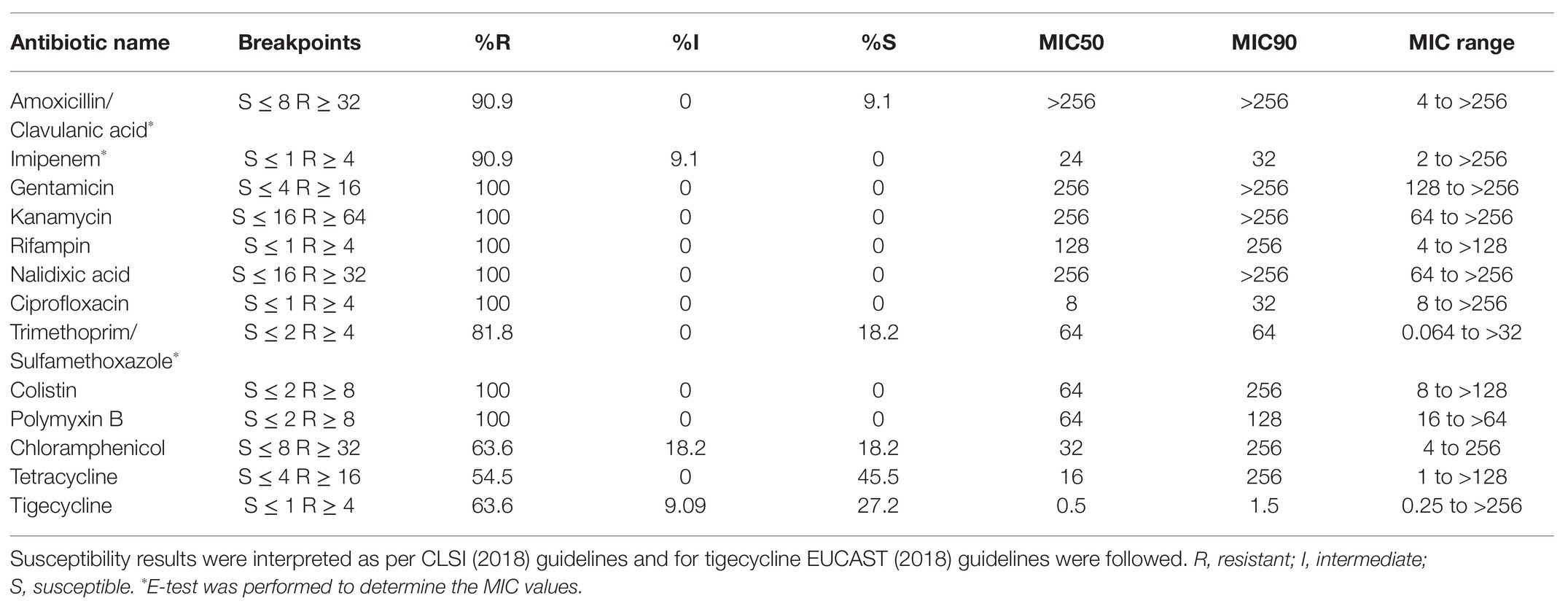
Table 2. Minimum inhibitory concentration (MIC) in μg/ml of different antibiotics tested by E-test and broth microdilution method for 11 K. pneumoniae isolates.
Sequence analysis of Lipid A modifying genes of all the 11 isolates of K. pneumoniae revealed non-synonymous nucleotide mutations at two different positions of the phoP gene, 18 positions of the phoQ gene, nine positions of the pmrA gene, 37 positions of the pmrB gene, and two positions of the mgrB gene. Of these mutations, one in phoP, four in phoQ, one in pmrA, eight in pmrB, and one in mgrB were found to be potentially deleterious (Table 3). The protein sequence of phoP genes of all colistin-resistant K. pneumoniae isolates showed no difference with the wild type strain (K. pneumoniae ATCC 700603) except for MRK9 strain that exhibited T151A substitution. phoQ gene was mutated in two strains, i.e., MRK8 (L30Q) and MRK11(A351D), whereas, MRK5 and MRK6 isolates exhibited deletion of four and two amino acids at 87–90 and 267–268 positions, respectively. All other isolates exhibited neutral changes compared to sensitive strain at phoQ locus.
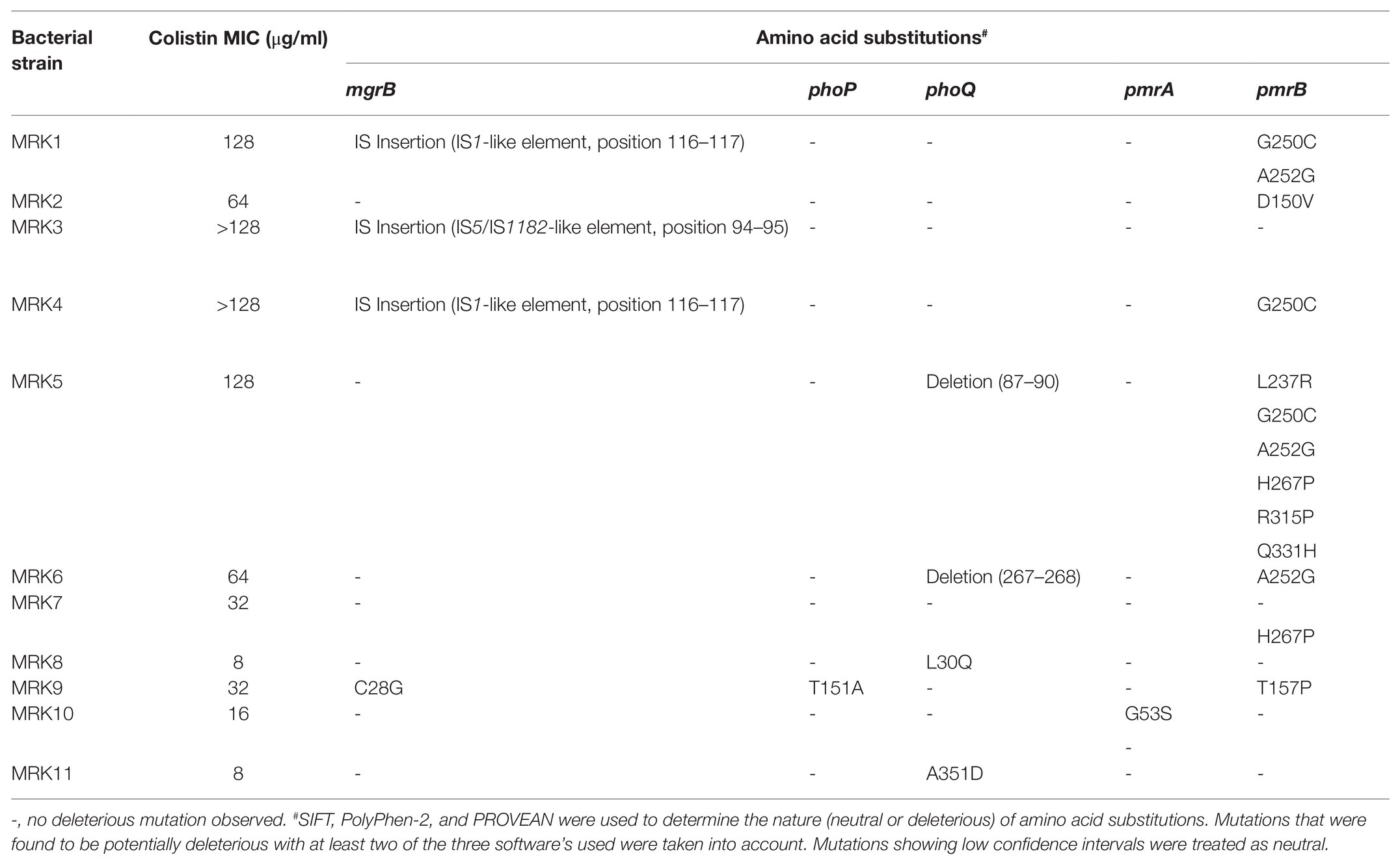
Table 3. List of amino acid substitutions or disruptions among mgrB, phoP, phoQ, pmrA, and pmrB proteins among 11 colistin-resistant K. pneumoniae isolates.
The insertional inactivation of the mgrB gene was observed in three isolates, i.e., MRK1, MRK3, and MRK4. IS1 family transposase was observed in K. pneumoniae MRK1 strain. In K. pneumoniae MRK3, mgrB gene was interrupted with a 1,066 bp fragment composed of the IS5/IS1182 family transposase gene, non-coding sequence, and nine base pair inverted repeats (ACCAGGATG). An insertion sequence of 768 base pairs comprising IS1 family transposase and a non-coding fragment was observed interrupting the mgrB gene of K. pneumoniae MRK4 strain. A base change T to G at position 82, leading to an amino acid substitution of C28G being observed in the mgrB gene of K. pneumoniae MRK9, which was found to be deleterious.
Analysis of pmrA/pmrB genes regulating the expression of arnBCADTEF operon decorating LPS showed amino acid changes at five positions in pmrA gene of four different isolates, none being deleterious except for MRK-10 exhibiting G53S. The pmrB gene was mutated in most isolates with a varying number of potentially deleterious mutations ranging from one mutation in MRK2, MRK6 and MRK9 to six deleterious mutations in MRK5 strain. In total, 23 neutral changes and eight potentially deleterious changes were observed in the pmrB gene. L30Q substitution of phoQ and D150V substitution of pmrB were observed to be highly intolerant under PROVEAN, SIFT, and PolyPhen-2 scoring criteria. Plasmid-encoded colistin resistance genes (mcr-1-8) did not amplify in any of the tested colistin-resistant K. pneumoniae isolates.
Colistin-resistant K. pneumoniae isolates resistant to carbapenems showed positive amplification of blaNDM-1-like and blaOXA-48-like gene in eight and five isolates, respectively. Furthermore, ESBL encoding blaCTX-M-like gene was found in six isolates, and one of the isolates, MRK6, showed positive amplification for three β-lactamase (blaNDM-1-like, blaOXA-48-like, and blaCTX-M-like) genes (Table 1).
The fold change in the expression level of pmrC and pmrK genes is given in Figure 2. All the colistin-resistant K. pneumoniae isolates exhibited increased transcript levels of pmrC gene both in the presence or absence of colistin sulphate. The fold change in pmrC expression ranged from 2.7 ± 1.29 to 226.37 ± 118.63 when bacteria was subcultured with 4 μg/ml of colistin sulphate and 13.76 ± 4.091 to 404.71 ± 116.25-fold under untreated conditions. MRK4, MRK8, and MRK10 exhibited a comparable level of pmrC expression in both treated and untreated conditions (p = >0.05) whereas two isolates (MRK1 and MRK2) showed more than 2-fold increase in expression (p < 0.05) upon colistin treatment, and the remaining six isolates exhibited a decrease in pmrC expression from 1.5- to 5-fold (p < 0.05) upon colistin treatment. The expression level of pmrK gene under untreated condition was comparable to colistin-susceptible isolate in seven isolates (MRK1, MRK2, MRK3, MRK5, MRK6, MRK7, and MRK8), high (3.42 ± 1.171 to 21.67 ± 6.87 fold) in three isolates and down in isolate MRK11. Upon colistin treatment, the pmrK expression increased by 1.63- to 36.9-fold in seven isolates (MRK1, MRK2, MRK6, MRK7, MRK9, MRK10, and MRK11; p < 0.05) and was comparable in three isolates except for MRK4 where it decreased approximately 3-fold.
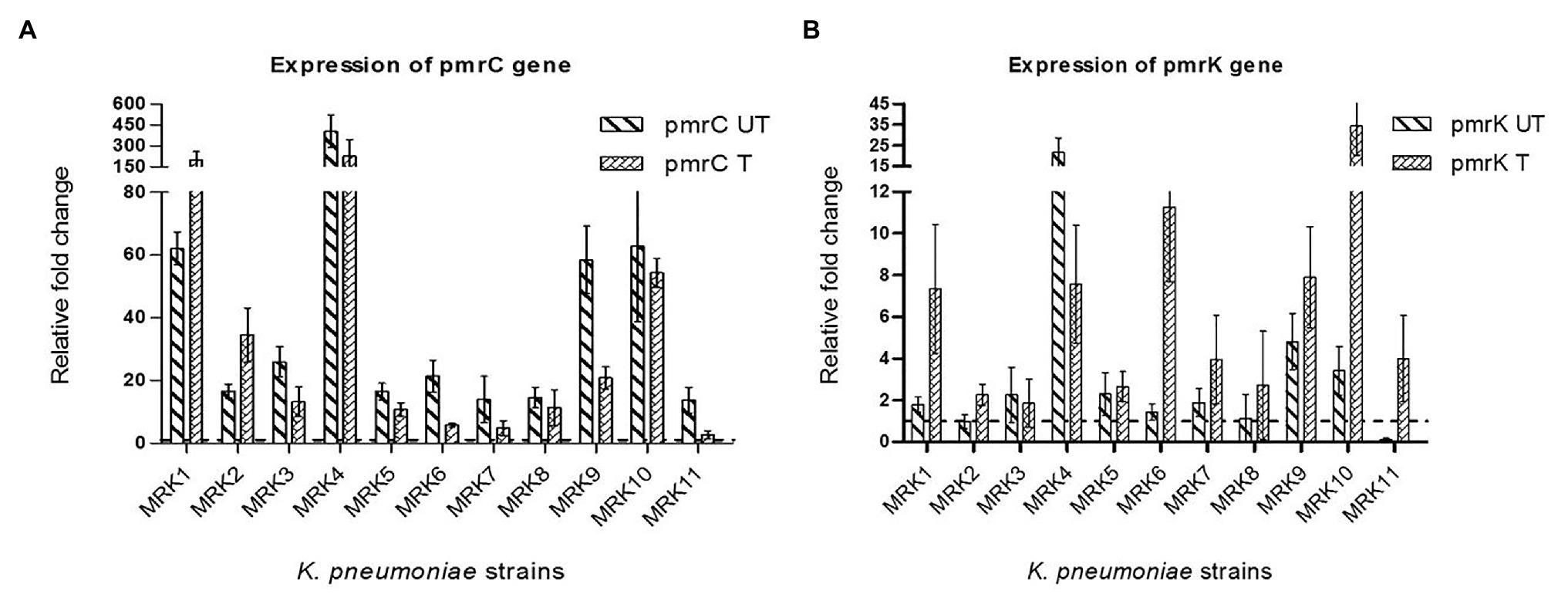
Figure 2. Fold changes ±SD in the expression of pmrC and pmrK genes among colistin-resistant K. pneumoniae isolates (n = 11) with respect to colistin-susceptible K. pneumoniae ATCC 700603 isolate are represented here. The rpsl gene was used as internal control. All the reactions (both under colistin treated and untreated conditions) were normalized using colistin-susceptible K. pneumoniae ATCC 700603 (colistin untreated- wild type). (A) relative fold-change of pmrC gene expression under colistin treated (pmrC T) and untreated conditions (pmrC UT); (B) relative fold-change of pmrK gene expression under colistin treated (pmrK T) and untreated conditions (pmrK UT). Values given are mean ± SD of three different experiments with qRT-PCR reactions performed in triplicate.
The rise in carbapenem-resistant K. pneumoniae isolates has enforced the increased application of polymyxin based therapies (mono and combination), ensuing in the development of colistin- and carbapenem-resistant K. pneumoniae strains. Reports of colistin resistance are emerging from different countries (Monaco et al., 2014; Rojas et al., 2017; Boszczowski et al., 2019). Our data showed 3.2% (11/335) of K. pneumoniae as colistin-resistant with minimal clonal relatedness. All 11 K. pneumoniae isolates belonged to same most prominent and diverse clonal complex-11 with sequence types (STs): 14, 16, 43, 54, 147, and 395, whereby four isolates conferred three novel ST (3986, 3987, and 3988) profiles. Although isolates MRK5 and MRK6 belonged to ST3987 and isolates MRK7 and MRK8 belonged to ST395, all four were isolated from patients of different wards of the hospital. The PubMed search for published literature identified limited studies describing the molecular mechanism of colistin resistance among K. pneumoniae isolates in India and abroad (Table 4). A thorough analysis showed that compared to other global studies, K. pneumoniae of very different STs conferred colistin resistance in India. However, Mavroidi et al. (2016) demonstrated colistin resistance due to clonal spread of KPC-producing K. pneumoniae belonging to clonal complex CC258 in Greece. Moubareck et al. (2018) showed 59% of colistin resistance in carbapenem-resistant K. pneumoniae was associated with locally prevalent ST14 clone in Dubai. High MIC values for colistin and carbapenems in K. pneumoniae causing bloodstream infections were observed in different studies from India (Ramesh et al., 2017; Aggarwal et al., 2018). K. pneumoniae resistant to both colistin and carbapenems have also been reported to be associated with increased risk of in-hospital mortality from India (Kaur et al., 2017; Aggarwal et al., 2018) and other parts of the world (Giacobbe et al., 2015; Rojas et al., 2017).

Table 4. Studies performed in India and abroad to understand the molecular mechanisms of colistin resistance among K. pneumoniae isolates.
In this study, in vitro antimicrobial susceptibility profiling and WHONET analysis categorized four K. pneumoniae isolate as pan drug-resistant and six as extensively drug-resistant (XDR) as per the definition by Magiorakos et al. (2012). Additionally, these isolates fall to the category of difficult-to-treat Gram-negative infections, where resistance to aminoglycosides emphasizes the difficulty of choosing salvage antibiotics for clinical containment (Kadri et al., 2018). The mortality in the present study was 36.4%, where only 7 out of 11 patients could recover. Three of the four patients deceased were infected with pan-drug resistant (PDR) K. pneumoniae. Among the 11 study isolates, positive amplification of blaNDM-1 and blaOXA-48 carbapenemase genes was found in 8 (73%) and 5 (45%) of the isolates, and two isolates were positive for both blaNDM-1 and blaOXA-48. Colistin-resistant K. pneumoniae with PDR and XDR phenotype co-producing blaNDM-1 and blaOXA-48 carbapenamases have been reported to cause severe nosocomial infections in several countries (Guducuoglu et al., 2018; Moubareck et al., 2018; Haller et al., 2019).
In K. pneumoniae, the positively charged groups 4-amino-4-deoxy-L-arabinose (L-Ara4N) and phosphoethanolamine (PEtN) mediate the covalent modifications of lipid A moiety of LPS reducing the net negative charge and subsequently binding affinity of colistin (Tamayo et al., 2005a). Mutations in TCRSs and/or genetic alteration in the mgrB gene, the negative regulator of phoP/phoQ TCRS can cause constitutive expression of the pmrHFIJKLM and pmrCAB operons transferring the L-Ara4N and PEtN respectively, to lipid A of the cell membrane (Tamayo et al., 2005b). Along with six potentially deleterious mutations in pmrB gene, a deletion of 12 nucleotides resulting in the omission of four amino acids in the phospho-transfer domain of phoQ gene was observed in K. pneumoniae MRK5 strain tolerating 128 μg/ml of colistin. Marina et al. also reported similar deletions in the phoQ gene reducing the colistin susceptibility (Marina et al., 2001). MRK7 strain also demonstrated a MIC of 128 μg/ml for colistin having potentially deleterious substitution (H267P) in pmrB gene, underlining its significance. L30Q substitution in phoQ gene of MRK8 was similar to the earlier report (Cheng et al., 2015). The L30Q substitution in the hydrophobic domain (constituting 17–44 amino acid residues) of phoQ influences the protein conformation and oligomer stability potentially changing the phosphate transfer and the phosphatase ability (Goldberg et al., 2010). In two of K. pneumoniae isolates (MRK1 and MRK4), point mutation A170G was detected in the pmrA gene resulting in amino acid change (E57G) potentially deleterious while R256G in pmrB of MRK7 predicted not to cause any functional change in the protein function (Pragasam et al., 2017). The novel G250C potentially deleterious substitution in the pmrB gene was observed in three of K. pneumoniae isolates (MRK1, MRK4, and MRK5) that exhibited high MIC (≥128 μg/ml) for colistin. Several other deleterious substitutions in the pmrB (D150V, A252G, L237R, H267P, R315P, and Q331H) genes with their scores in the intolerant and potentially intolerant range are being reported here for the first time. These amino acid substitutions (L237R, A252G, H267P, R315P, Q331H, except D150V) were observed in a single isolate (MRK5) along with a four amino acid deletion in phoQ gene, emphasizing the need for further evaluation for their role in colistin resistance. Predicted deleterious substitution pmrA-Q140L (found in all 11 test isolates) and pmrB R256G (present in MRK7) have been previously reported in colistin-resistant as well as susceptible isolates negating its decisive role in colistin resistance (Pitt et al., 2018). However, the other mutations observed here, L30Q and A351D substitutions in the phoQ gene of MRK8 and MRK11 strains, respectively, and G53S in the pmrA gene of MRK10 and D150V and H267P in the pmrB gene of the MRK2 and MRK7 strain, respectively, may impart resistance in the absence of any other contributing factor.
The mgrB gene-mediated inactivation of the phoP/phoQ TCRS has been extensively reported to play a prominent role in polymyxin resistance in K. pneumoniae (Cannatelli et al., 2014b; Poirel et al., 2017). Three of 11 K. pneumoniae isolates MRK1, MRK3 and MRK4 had insertion inactivation of mgrB gene disrupting the translation of a functional protein, along with mutations in pmrA and pmrB genes showing high tolerance to colistin (MIC ≥128 μg/ml). The role of IS elements (IS5-like, IS1F and ISKpn14, ISKpn13, and IS10R) in mgrB inactivation that inhibits phoQ phosphorylation resulting in increased expression of pmrHFIJKLM mRNA and leading to reduced colistin susceptibility in K. pneumoniae have been extensively demonstrated (Cannatelli et al., 2014a; Poirel et al., 2017; Zhu et al., 2019). K. pneumoniae MRK1 and MRK4 were interrupted by an IS1-like transposase elements. The inactivation of mgrB protein with a similar IS1-like element interrupting at the same nucleotide position (117–118) was observed in K. pneumoniae recovered from a dead broiler from Iran (Pishnian et al., 2019). The mgrB gene in K. pneumoniae MRK3 isolate was disrupted by 969 bp long IS5/IS1182-like element at a different position. IS5/IS1182 fragment is present in various plasmids isolated from different Gram-negative bacteria (Hala et al., 2019). Cannatelli et al. (2013) and Poirel et al. (2015) have reported insertional inactivation of mgrB gene with a similar IS5-like insertion sequence at 74–75 nucleotide position; however, the IS5-like element reported in MRK3 isolate of this study showed a different interruption of the nucleotide sequence at position 86–87. To the best of our knowledge, this insertional inactivation of mgrB gene by the IS5/IS1182 fragment has not been reported before. Isolate MRK7 had M23R substitution having SIFT score of 0 with low confidence, PROVEAN score of −3.121 depicting intolerance and is found to be neutral with PolyPhen-2 hence treated as neutral while MRK9 had C28G substitution also observed by Shankar et al. (2019b), which shows highly intolerant tendency (Table 4). The remaining seven isolates had an intact wild type mgrB gene. Mutations observed in this study in the pmrB (T157P) and mgrB (C28Y) gene have been reported in previous studies to be linked with reduced colistin susceptibility in K. pneumoniae (Cannatelli et al., 2014a, 2015; Jayol et al., 2014; Lomonaco et al., 2018; Mathur et al., 2018). Isolates harboring ISKpn26-like and other IS elements disrupting mgrB also exhibited mutations in pmrA and pmrB genes (Table 4).
To establish a correlation between lipid A modifying operon (pmrCAB and pmrHFIJKLM) expression pattern and colistin resistance among the test isolates, the expression level of pmrK and pmrC genes was evaluated. All isolates showed an over-expressed pmrC gene in comparison to the pmrK gene, implying that PEtN-mediated LPS modification plays a significant role in conferring colistin resistance in tested isolates. Among colistin-resistant K. pneumoniae isolates, fold-change increase in expression of the pmrC gene was higher in 6/11 (54%) isolates that decreased significantly upon colistin treatment (p < 0.05), though overall in both the conditions that level of expression of pmrC was higher compared to colistin sensitive isolate. For three isolates, the expression in both colistin treated and untreated conditions did not differ significantly, while the expression of pmrC increased significantly upon colistin treatment in two isolates.
The basal expression level of pmrK gene in 6/11 (54%) colistin-resistant K. pneumoniae isolates was similar to colistin sensitive isolate and showed significantly higher expression in three isolates. The expression of pmrK increased significantly upon colistin treatment in 7/11 (63%) isolates, remained unaltered in 3/11 (27%) isolates and repressed in one isolate MRK4. Isolate MRK11 exhibited repressed expression of pmrK in untreated condition exhibited >30 fold increase in the expression of pmrK gene upon colistin treatment. This isolate harbored deleterious change in phoQ gene that upon activation may increase phosphorylation of phoP activating transcription of pmrHFIJKLM operon resulting in increased pmrK expression. A similar increase in the expression of pmrC gene as operon regulators has been reported previously (Cheng et al., 2015; Haeili et al., 2017). T157P substitution in pmrB (observed in MRK9 isolate) has been shown to contribute to the overexpression of pmrCAB and pmrHFIJKLM operons with the pmrC being highly overexpressed (170-fold) compared to pmrK (40-fold; Jayol et al., 2014). The majority of the tested isolates (8/11) harbored pmrB gene with several mutations possibly contributing to constitutive expression of pmrA gene that may preferentially activate pmrCAB promoter compared to pmrHFIJKLM resulting in increased expression of the pmrC gene in these isolates. Similar to previous reports, no direct correlation between colistin exposure and gene upregulation was observed (Can et al., 2018). A single mechanism may not explain the role of pmrC and pmrK in mediating colistin resistance in K. pneumoniae, as we observed diverse strain-specific mutations that may affect the expression of these operons differentially. Our data indicates discrete emergence of colistin resistance in clinical isolates through strain-specific pathways where multiple mechanisms might be involved in resistance development.
All 11 colistin and carbapenem-resistant K. pneumoniae isolates under study were distinct (with nine different ST types) and present the emergence of discrete colistin resistance mechanisms. The role of mgrB gene as a hot-spot for insertion inactivation and its functional loss associated with colistin resistance was observed in three isolates. Multiple mutations in regulatory genes (phoP, phoQ, pmrA, pmrB, and mgrB) and their association in the expression pattern of LPS decorating operons raises the colistin MIC values. The 11 test isolates were negative for the mobilized colistin resistance genes mcr-1-8, however, the occurrence of other mcr gene variants not investigated here could not be excluded as a contributory factor towards increased colistin MIC values. The clinical and public health concerns in a background where the pipeline for new antibiotics is limited, demand implementation of antimicrobial stewardship and infection control measures to prevent the spread of resistant bacteria in the health care settings. The upshot of the exorbitant and lavish use of antibiotics has developed severe concern regarding multidrug-resistant bacteria, especially nosocomial pathogens that necessitates the rational use of colistin as a last-resort antibiotic.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by ICMR-National Institute of Pathology, New Delhi, India (IEC No: NIP-IEC/2/3/17/06) and VMMC and Safdarjung Hospital, New Delhi, India (IEC No: IEC/VMMC/SJH/Project/January/2018/1028). The patients/participants provided their written informed consent to participate in this study.
Conceptualization, methodology, writing – review and editing: MA, RS, and RG. Data curation: MA, GY, AS, and KU. Formal analysis: MA, GY, AS, KU, and MJ. Funding acquisition: MA and RS. Investigation: RS and RG. Validation: RS, RG, and MJ. Writing – original draft: MA and GY. All authors contributed to the article and approved the submitted version.
The authors acknowledge the funding by the Indian Council of Medical Research (ICMR), New Delhi, India. MA received SERB-National Post-Doctoral Fellowship (Grant no. PDF/2016/001961) from The Science &Engineering Research Board (SERB), Department of Science &Technology, Government of India. GY is grateful to the University Grant Commission (UGC), New Delhi, India, for providing the Junior Research Fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.609840/full#supplementary-material
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., and Bork, P. (2010). Catalog of somatic mutations in cancer and bioinformatics mutation-prediction tools PolyPhen2. Nat. Methods 7, 248–249. doi: 10.1038/nmeth0410-248
Aggarwal, R., Rastogi, N., Mathur, P., Soni, K. D., Kumar, S., Gupta, A., et al. (2018). Colistin-resistant Klebsiella pneumoniae in surgical polytrauma intensive care unit of level-1 trauma center: first case series from trauma patients in India. Indian J. Crit. Care Med. 22:103. doi: 10.4103/ijccm.IJCCM_430_17
Aires, C. A. M., Pereira, P. S., Asensi, M. D., and Carvalho-Assef, A. P. D. A. (2016). mgrB mutations mediating polymyxin B resistance in Klebsiella pneumoniae isolates from rectal surveillance swabs in Brazil. Antimicrob. Agents Chemother. 60, 6969–6972. doi: 10.1128/AAC.01456-16
Azam, M., Jan, A. T., and Haq, Q. M. R. (2016). blaCTX-M-152, a novel variant of CTX-M-group-25, identified in a study performed on the prevalence of multidrug resistance among natural inhabitants of river Yamuna, India. Front. Microbiol. 7:176. doi: 10.3389/fmicb.2016.00176
Bhaskar, B. H., Mulki, S. S., Joshi, S., Adhikari, R., and Venkatesh, B. M. (2017). Colistin resistance in carbapenem-resistant Klebsiella pneumoniae strains. Asian J. Pharm. Clin. Res. 10, 70–73. doi: 10.22159/ajpcr.2017.v10i9.18369
Boszczowski, I., Salomão, M. C., Moura, M. L., Freire, M. P., Guimarães, T., Cury, A. P., et al. (2019). Multidrug-resistant Klebsiella pneumoniae: genetic diversity, mechanisms of resistance to polymyxins and clinical outcomes in a tertiary teaching hospital in Brazil. Rev. Inst. Med. Trop. Sao Paulo 61:e29. doi: 10.1590/s1678-9946201961029
Cain, A. K., Boinett, C. J., Barquist, L., Dordel, J., Fookes, M., Mayho, M., et al. (2018). Morphological, genomic and transcriptomic responses of Klebsiella pneumoniae to the last-line antibiotic colistin. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-28199-y
Can, F., Menekse, S., Ispir, P., Atac, N., Albayrak, O., Demir, T., et al. (2018). Impact of the ST101 clone on fatality among patients with colistin-resistant Klebsiella pneumoniae infection. J. Antimicrob. Chemother. 73, 1235–1241. doi: 10.1093/jac/dkx532
Cannatelli, A., D’Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Cannatelli, A., Di Pilato, V., Giani, T., Arena, F., Ambretti, S., Gaibani, P., et al. (2014a). In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob. Agents Chemother. 58, 4399–4403. doi: 10.1128/AAC.02555-14
Cannatelli, A., Giani, T., D’Andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014b). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/AAC.03110-14
Cannatelli, A., Santos-Lopez, A., Giani, T., Gonzalez-Zorn, B., and Rossolini, G. M. (2015). Polymyxin resistance caused by mgrB inactivation is not associated with significant biological cost in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 59, 2898–2900. doi: 10.1128/AAC.04998-14
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10, e00853–e00919. doi: 10.1128/mBio.00853-19
Cheng, Y. H., Lin, T. L., Pan, Y. J., Wang, Y. P., Lin, Y. T., and Wang, J. T. (2015). Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob. Agents Chemother. 59, 2909–2913. doi: 10.1128/AAC.04763-14
Cheong, H. S., Kim, S. Y., Wi, Y. M., Peck, K. R., and Ko, K. S. (2019). Colistin heteroresistance in Klebsiella pneumoniae isolates and diverse mutations of PmrAB and PhoPQ in resistant subpopulations. J. Clin. Med. 8:1444. doi: 10.3390/jcm8091444
Choi, Y., and Chan, A. P. (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. doi: 10.1093/bioinformatics/btv195
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Diene, S. M., and Rolain, J. M. (2014). Carbapenemase genes and genetic platforms in gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 20, 831–838. doi: 10.1111/1469-0691.12655
Dixit, A., Kumar, N., Kumar, S., and Trigun, V. (2019). Antimicrobial resistance: progress in the decade since emergence of New Delhi Metallo-β-lactamase in India. Indian J. Community Med. 44, 4–8. doi: 10.4103/ijcm.IJCM_217_18
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Gandra, S., Mojica, N., Klein, E. Y., Ashok, A., Nerurkar, V., Kumari, M., et al. (2016). Trends in antibiotic resistance among major bacterial pathogens isolated from blood cultures tested at a large private laboratory network in India, 2008–2014. Int. J. Infect. Dis. 50, 75–82. doi: 10.1016/j.ijid.2016.08.002
Gharaibeh, M. H., and Shatnawi, S. Q. (2019). An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: a review. Vet. World 12:1735. doi: 10.14202/vetworld.2019.1735-1746
Giacobbe, D. R., Del Bono, V., Trecarichi, E. M., De Rosa, F. G., Giannella, M., Bassetti, M., et al. (2015). Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case-control-control study. Clin. Microbiol. Infect. x21, 1106.e1-8. doi: 10.1016/j.cmi.2015.08.001
Goel, G., Hmar, L., De, M. S., Bhattacharya, S., and Chandy, M. (2014). Colistin-resistant Klebsiella pneumoniae: report of a cluster of 24 cases from a new oncology center in eastern India. Infect. Control Hosp. Epidemiol. 35, 1076–1077. doi: 10.1086/677170
Goldberg, S. D., Clinthorne, G. D., Goulian, M., and DeGrado, W. F. (2010). Transmembrane polar interactions are required for signaling in the Escherichia coli sensor kinase PhoQ. Proc. Natl. Acad. Sci. U. S. A. 107, 8141–8146. doi: 10.1073/pnas.1003166107
Guducuoglu, H., Gursoy, N. C., Yakupogullari, Y., Parlak, M., Karasin, G., Sunnetcioglu, M., et al. (2018). Hospital outbreak of a colistin-resistant, NDM-1-and OXA-48-producing Klebsiella pneumoniae: high mortality from pandrug resistance. Microb. Drug Resist. 24, 966–972. doi: 10.1089/mdr.2017.0173
Haeili, M., Javani, A., Moradi, J., Jafari, Z., Feizabadi, M. M., and Babaei, E. (2017). MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front. Microbiol. 8:2470. doi: 10.3389/fmicb.2017.02470
Hala, S., Antony, C. P., Alshehri, M., Althaqafi, A. O., Alsaedi, A., Mufti, A., et al. (2019). First report of Klebsiella quasipneumoniae harboring blaKPC-2 in Saudi Arabia. Antimicrob. Resist. Infect. Control 8:203. doi: 10.1186/s13756-019-0653-9
Halaby, T., Kucukkose, E., Janssen, A. B., Rogers, M. R., Doorduijn, D. J., van der Zanden, A. G., et al. (2016). Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob. Agents Chemother. 60, 6837–6843. doi: 10.1128/AAC.01344-16
Haller, S., Kramer, R., Becker, K., Bohnert, J. A., Eckmanns, T., Hans, J. B., et al. (2019). Extensively drug-resistant Klebsiella pneumoniae ST307 outbreak, North-Eastern Germany, June to October 2019. Eurosurveillance 24:1900734. doi: 10.2807/1560-7917.ES.2019.24.50.1900734
Jaidane, N., Bonnin, R. A., Mansour, W., Girlich, D., Creton, E., Cotellon, G., et al. (2018). Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob. Agents Chemother. 62, e01601–e01617. doi: 10.1128/AAC.01601-17
Jajoo, M., Manchanda, V., Chaurasia, S., Sankar, M. J., Gautam, H., Agarwal, R., et al. (2018). Alarming rates of antimicrobial resistance and fungal sepsis in outborn neonates in North India. PLoS One 13:e0180705. doi: 10.1371/journal.pone.0180705
Jayol, A., Nordmann, P., Brink, A., and Poirel, L. (2015). Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 59, 2780–2784. doi: 10.1128/AAC.05055-14
Jayol, A., Poirel, L., Brink, A., Villegas, M. V., Yilmaz, M., and Nordmann, P. (2014). Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 58, 4762–4766. doi: 10.1128/AAC.00084-14
Kadri, S. S., Adjemian, J., Lai, Y. L., Spaulding, A. B., Ricotta, E., Prevots, D. R., et al. (2018). Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin. Infect. Dis. 67, 1803–1814. doi: 10.1093/cid/ciy378
Kaur, A., Gandra, S., Gupta, P., Mehta, Y., Laxminarayan, R., and Sengupta, S. (2017). Clinical outcome of dual colistin- and carbapenem-resistant Klebsiella pneumoniae bloodstream infections: a single-center retrospective study of 75 cases in India. Am. J. Infect. Control 45, 1289–1291. doi: 10.1016/j.ajic.2017.06.028
Leung, L. M., Cooper, V. S., Rasko, D. A., Guo, Q., Pacey, M. P., McElheny, C. L., et al. (2017). Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 72, 3035–3042. doi: 10.1093/jac/dkx234
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lomonaco, S., Crawford, M. A., Lascols, C., Timme, R. E., Anderson, K., Hodge, D. R., et al. (2018). Resistome of carbapenem-and colistin-resistant Klebsiella pneumoniae clinical isolates. PLoS One 13:e0198526. doi: 10.1371/journal.pone.0198526
Lu, Y., Feng, Y., McNally, A., and Zong, Z. (2018). The occurence of colistin-resistant hypervirulent Klebsiellapneumoniae in China. Front. Microbiol. 9:2568. doi: 10.3389/fmicb.2018.02568
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Manohar, P., Shanthini, T., Ayyanar, R., Bozdogan, B., Wilson, A., Tamhankar, A. J., et al. (2017). The distribution of carbapenem-and colistin-resistance in gram-negative bacteria from the Tamil Nadu region in India. J. Med. Microbiol. 66, 874–883. doi: 10.1099/jmm.0.000508
Marina, A., Mott, C., Auyzenberg, A., Hendrickson, W. A., and Waldburger, C. D. (2001). Structural and mutational analysis of the PhoQ histidine kinase catalytic domain insight into the reaction mechanism. J. Biol. Chem. 276, 41182–41190. doi: 10.1074/jbc.M106080200
Mathur, P., Veeraraghavan, B., Devanga Ragupathi, N. K., Inbanathan, F. Y., Khurana, S., Bhardwaj, N., et al. (2018). Multiple mutations in lipid-a modification pathway &novel fosA variants in colistin-resistant Klebsiella pneumoniae. Future Sci. OA 4:FSO319. doi: 10.4155/fsoa-2018-0011
Mavroidi, A., Katsiari, M., Likousi, S., Palla, E., Roussou, Z., Nikolaou, C., et al. (2016). Characterization of ST258 colistin-resistant, Bla KPC-producing Klebsiella pneumoniae in a Greek hospital. Microb. Drug Resist. 22, 392–398. doi: 10.1089/mdr.2015.0282
Mohanty, S., Gajanand, M., and Gaind, R. (2017). Identification of carbapenemase-mediated resistance among Enterobacteriaceae bloodstream isolates: a molecular study from India. Indian J. Med. Microbiol. 35:421. doi: 10.4103/ijmm.IJMM_16_386
Monaco, M., Giani, T., Raffone, M., Arena, F., Garcia-Fernandez, A., Pollini, S., et al. (2014). Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Eurosurveillance 19:20939. doi: 10.2807/1560-7917.ES2014.19.42.20939
Moubareck, C. A., Mouftah, S. F., Pál, T., Ghazawi, A., Halat, D. H., Nabi, A., et al. (2018). Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int. J. Antimicrob. Agents 52, 90–95. doi: 10.1016/j.ijantimicag.2018.03.003
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Ng, P. C., and Henikoff, S. (2003). SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814. doi: 10.1093/nar/gkg509
Nguyen, N. T., Nguyen, H. M., Nguyen, C. V., Nguyen, T. V., Nguyen, M. T., Thai, H. Q., et al. (2016). Use of colistin and other critical antimicrobials on pig and chicken farms in southern Vietnam and its association with resistance in commensal Escherichia coli bacteria. Appl. Environ. Microbiol. 82, 3727–3735. doi: 10.1128/AEM.00337-16
Palani, G. S., Ghafur, A., Krishnan, P., Rayvathy, B., and Thirunarayan, M. A. (2020). Intestinal carriage of colistin resistant Enterobacteriaceae in hospitalized patients from an Indian center. Diagn. Microbiol. Infect. Dis. 97:114998. doi: 10.1016/j.diagmicrobio.2020.114998
Pishnian, Z., Haeili, M., and Feizi, A. (2019). Prevalence and molecular determinants of colistin resistance among commensal Enterobacteriaceae isolated from poultry in northwest of Iran. Gut Pathog. 11:2. doi: 10.1186/s13099-019-0282-0
Pitt, M. E., Elliott, A. G., Cao, M. D., Ganesamoorthy, D., Karaiskos, I., Giamarellou, H., et al. (2018). Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae. Microb. Genom. 4:e000158. doi: 10.1099/mgen.0.000158
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. microbiol. rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Poirel, L., Jayol, A., Bontron, S., Villegas, M. V., Ozdamar, M., Türkoglu, S., et al. (2015). The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. doi: 10.1093/jac/dku323
Pragasam, A. K., Shankar, C., Veeraraghavan, B., Biswas, I., Nabarro, L. E., Inbanathan, F. Y., et al. (2017). Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India—a first report. Front. Microbiol. 7:2135. doi: 10.3389/fmicb.2016.02135
Ramesh, N., Prasanth, M., Ramkumar, S., Suresh, M., Tamhankar, A. J., Gothandam, K. M., et al. (2017). Colistin susceptibility of gram-negative clinical isolates from Tamil Nadu, India. Asian Biomed. 10, 35–39. doi: 10.5372/1905-7415.1001.462
Rojas, L. J., Salim, M., Cober, E., Richter, S. S., Perez, F., Salata, R. A., et al. (2017). Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin. Infect. Dis. 64, 711–718. doi: 10.1093/cid/ciw805
Shankar, C., Karunasree, S., Manesh, A., and Veeraraghavan, B. (2019a). First report of whole-genome sequence of colistin-resistant Klebsiella quasipneumoniae subsp. similipneumoniae producing KPC-9 in India. Microb. Drug Resist. 25, 489–493. doi: 10.1089/mdr.2018.0116
Shankar, C., Venkatesan, M., Rajan, R., Mani, D., Lal, B., Prakash, J. A. J., et al. (2019b). Molecular characterization of colistin-resistant Klebsiella pneumoniae &its clonal relationship among Indian isolates. Indian J. Med. Res. 149:199. doi: 10.4103/ijmr.IJMR_2087_17
Sim, N. L., Kumar, P., Hu, J., Henikoff, S., Schneider, G., and Ng, P. C. (2012). SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457. doi: 10.1093/nar/gks539
Tamayo, R., Choudhury, B., Septer, A., Merighi, M., Carlson, R., and Gunn, J. S. (2005a). Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar typhimurium lipopolysaccharide core. J. Bacteriol. 187, 3391–3399. doi: 10.1128/JB.187.10.3391-3399.2005
Tamayo, R., Prouty, A. M., and Gunn, J. S. (2005b). Identification and functional analysis of Salmonella enterica serovar Typhimurium PmrA-regulated genes. FEMS Immunol. Med. Microbiol. 43, 249–258. doi: 10.1016/j.femsim.2004.08.007
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Zafer, M. M., El-Mahallawy, H. A., Abdulhak, A., Amin, M. A., Al-Agamy, M. H., and Radwan, H. H. (2019). Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann. Clin. Microbiol. Antimicrob. 18:40. doi: 10.1186/s12941-019-0339-4
Zhang, H., Hou, M., Xu, Y., Srinivas, S., Huang, M., Liu, L., et al. (2019). Action and mechanism of the colistin resistance enzyme MCR-4. Commun. Biol. 2, 1–14. doi: 10.1038/s42003-018-0278-1
Zhu, Y., Galani, I., Karaiskos, I., Lu, J., Aye, S. M., Huang, J., et al. (2019). Multifaceted mechanisms of colistin resistance revealed by genomic analysis of multidrug-resistant Klebsiella pneumoniae isolates from individual patients before and after colistin treatment. J. Inf. Secur. 79, 312–321. doi: 10.1016/j.jinf.2019.07.009
Keywords: colistin resistance, Klebsiella pneumoniae, phoPQ, pmrAB, carbapenem resistance
Citation: Azam M, Gaind R, Yadav G, Sharma A, Upmanyu K, Jain M and Singh R (2021) Colistin Resistance Among Multiple Sequence Types of Klebsiella pneumoniae Is Associated With Diverse Resistance Mechanisms: A Report From India. Front. Microbiol. 12:609840. doi: 10.3389/fmicb.2021.609840
Received: 24 September 2020; Accepted: 22 January 2021;
Published: 22 February 2021.
Edited by:
Alberto Quesada, University of Extremadura, SpainReviewed by:
Yvonne Pfeifer, Robert Koch Institute (RKI), GermanyCopyright © 2021 Azam, Gaind, Yadav, Sharma, Upmanyu, Jain and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruchi Singh, cnVjaGlzcEBnbWFpbC5jb20=; cnVjaGlzaW5naC5uaXBAZ292Lmlu; Rajni Gaind, cmdhaW5kNUBob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.