
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 February 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.601924
 Carl Basbas1
Carl Basbas1 Barbara A. Byrne2
Barbara A. Byrne2 Munashe Chigerwe3
Munashe Chigerwe3 Edlin D. Escobar4
Edlin D. Escobar4 Emir Hodzic4
Emir Hodzic4 Alda F. A. Pires1
Alda F. A. Pires1 Richard V. Pereira1*
Richard V. Pereira1*The objectives of this study were to evaluate the prevalence of extended spectrum β-lactamase (ESBL) genes, AmpC-type β-lactamase (ACBL) genes, and plasmid mediated quinolone resistance (PMQR) genes in Salmonella isolated at a Veterinary Medical Teaching Hospital microbiology laboratory, examine trends in presence of these resistance genes, and to explore the correlation between phenotypic resistance and presence of specific genes. The presence of ESBL, ACBL, and PMQR genes were detected using a single, novel multiplex qPCR. Only the genes blaCMY–2 and blaTEM were detected in the 110 Salmonella isolates tested. PMQR genes were not detected in isolates screened. Of 94 third-generation cephalosporin resistant isolates, representing eight serotypes, 48% (n = 45) were positive for blaCMY–2 only and 50% (n = 47) were simultaneously positive for blaCMY–2 and blaTEM. Two third-generation cephalosporin resistant isolates were tested negative for all β-lactamase genes in our qPCR assay and likely house ESBL genes not screened for by our qPCR assay. A logistic regression model revealed that for serotype Dublin isolates (n = 38) the odds ratio for testing positive for blaTEM when compared to all other serotypes was 51.6 (95% CI: 4.01–664.03, p = 0.0029). For serotype Typhimurium (n = 9) the odds ratio for testing positive for blaTEM when compared to all other serotypes was 43.3 (95% CI: 1.76–1000, p = 0.0216). Overall, our results suggest that the prevalence of resistance to cephalosporins and fluoroquinolones due to ESBLs, ACBLs, and PMQR genes present in bovine nontyphoidal Salmonella enterica isolates has remained relatively constant in the isolates screened over a 14-year period.
Globally in 2017, around 91 million cases of human gastrointestinal illness and diarrhea were believed to be caused by nontyphoidal Salmonella enterica (NTS) (Stanaway et al., 2019). In the United States alone, 1.35 million NTS infections amounted to an estimated $400 million in medical costs annually (Centers for Disease Control and Prevention, 2019). In humans, severe infections caused by Salmonella usually require treatment with specific recommended antimicrobials, including ciprofloxacin, azithromycin, and ceftriaxone (Centers for Disease Control and Prevention, 2019). With 3% and 7% of all human NTS infections in the U.S. classified as either ceftriaxone resistant or ciprofloxacin nonsusceptible, respectively, the U.S. Center for Disease Control and Prevention (CDC) has designated drug resistant NTS as a serious threat (Centers for Disease Control and Prevention, 2019). As resistance to third-generation cephalosporins and fluoroquinolones grows, increasing attention is being placed on extended spectrum β-lactamase (ESBL) genes, AmpC-type β-lactamase (ACBL) genes, and plasmid mediated quinolone resistance (PMQR) genes.
Extended spectrum β-lactamase (ESBL) genes encode for enzymes which are able to cleave the β-lactam ring of a wide range of β-lactam antimicrobials (e.g., penicillins and cephalosporins) (Dhillon and Clark, 2012). They confer β-lactam resistance to the bacteria that produce them, primarily Klebsiella pneumoniae and Escherichia coli. Worldwide, the most common ESBLs are the SHV, TEM, and CTX-M types. AmpC-type β-lactamase (ACBL) genes also encode for enzymes capable of degrading β-lactam antibiotics, including: extended spectrum cephalosporins (excluding cefepime and cefpirome), cephamycins, and ceftriaxone (Jacoby, 2009; Polsfuss et al., 2011). blaCMY–2 is the most common plasmid mediated ACBL gene globally (Jacoby, 2009).
Resistance to the quinolone and fluoroquinolone classes of antimicrobials has generally been attributed to chromosomal mutations in the bacterial enzymes targeted by these classes of antimicrobials: DNA gyrase and DNA topoisomerase IV (Strahilevitz et al., 2009). Additionally, three types of plasmid mediated quinolone resistance (PMQR) mechanisms have been identified: qnr genes protect DNA gyrase, the aac(6′)-lb-cr gene acetylates ciprofloxacin, norfloxacin, and certain other quinolones, and oqxAB and qepA genes produce efflux pumps (Rodriguez-Martinez et al., 2016; Karp et al., 2018).
Currently, a common method for identification of ESBL-producing bacteria is via culture-based phenotypic methods (Clinical and Laboratory Standards Institute, 2011; The European Committee on Antimicrobial Susceptibility Testing, 2013). Unfortunately, a definitive negative result may take 24–120 h (Engel et al., 2017). Furthermore, as these methods depend on the inhibition of ESBLs by clavulanic acid, the production of additional AmpC or metallo-β-lactamases (which are uninhibited by clavulanic acid) may decrease test sensitivity (Rawat and Nair, 2010). To identify particular genes responsible for ESBL production, reference laboratories use molecular analyses, primarily polymerase chain reaction (PCR) (Wittum et al., 2012).
Detection of ACBL-producing bacteria is generally done using phenotypic tests utilizing ACBL inhibitors such as boronic acid and cloxacillin, however, these tests are unable to distinguish between chromosomal or plasmid-mediated AmpC-type β-lactamases (Tamma et al., 2019). Detection of isolates carrying plasmid-mediated ACBL genes may be particularly important as these isolates may appear to be susceptible to cephalosporins in vitro, only to fail to respond to treatment (Thomson, 2001). Molecular approaches to identify plasmid-mediated ACBL genes are available, but are typically unavailable in clinical laboratories (Perez-Perez and Hanson, 2002). Additionally, there is currently no Clinical and Laboratory Standards Institute (CLSI) test for AmpC-type β-lactamases in clinical isolates (Clinical and Laboratory Standards Institute, 2020).
While there are multiplex PCR methods available for the detection of either ESBL, ACBL, or PMQR genes, there are few published multiplex PCR methods available for the combined detection of ESBL, ACBL, and PMQR genes relevant to NTS treatment (Ciesielczuk et al., 2013; Roschanski et al., 2014). The goal of this study was to identify trends in resistance of fecal Salmonella isolates to cephalosporins and fluoroquinolones due to the presence of ESBL, ACBL, and PMQR genes from Salmonella isolates obtained from cattle fecal samples isolated and tested in the University of California, Davis William R. Pritchard Veterinary Medical Teaching Hospital (VMTH) microbiology laboratory during a 14-year interval using a single, novel multiplex qPCR method.
A total of 110 Salmonella isolates were selected for qPCR analysis from a collection of 242 Salmonella isolates recovered from 9162 bovine fecal samples submitted to the University of California, Davis William R. Pritchard VMTH microbiology laboratory between January 1, 2002 and December 31, 2016 as detailed previously (Davidson et al., 2018). Sixty-eight isolates were recovered from dairy cattle exhibiting clinical signs of Salmonella infection, while 42 isolates were recovered from asymptomatic dairy cattle through the VMTH Infectious Disease Control (IDC) program. All isolates with phenotypic resistance to at least one of the following drugs, nalidixic acid, ceftiofur, and/or ceftriaxone, were included in the study (n = 94; Supplementary Table S1). None of the isolates were phenotypically resistant to ciprofloxacin (MIC ≥ 1.0 μg/ml). Only two isolates were resistant to nalidixic acid, and also presented simultaneous phenotypic resistance to ceftriaxone and ceftiofur. All isolates phenotypically resistant to ceftiofur were also resistant to ceftriaxone.
For each year a pan-susceptible Salmonella isolate, when available, was selected to serve as a control (n = 16) (Supplementary Table S2). For two years, namely 2002 and 2004, no pan-susceptible isolates were available, and an isolate resistant to streptomycin, and one isolate resistant to ampicillin, streptomycin, and tetracycline, respectively, were selected (Supplementary Table S2). These isolates were selected on the criteria that they were susceptible to quinolone and cephalosporin drugs and were the isolates resistant to the fewest number of antimicrobials for that year.
Data from phenotypic antimicrobial susceptibility testing conducted on the same isolates from a previous study were used (Davidson et al., 2018). Briefly, for that study all isolates were tested using the standardized National Antimicrobial Resistance Monitoring System (NARMS) (Thermo Fisher, Sensititre CMV3AGNF) for aerobic Gram-negative bacteria that included penicillins (ampicillin), β-lactam/ β-lactamase inhibitor combinations (amoxicillin/clavulanic acid), cephalosporins (ceftriaxone, ceftiofur, and cefoxitin), quinolones (ciprofloxacin and nalidixic acid), phenicols (chloramphenicol), sulfa-based drugs (sulfisoxazole and sulfamethoxazole/trimethoprim), tetracyclines (tetracycline), macrolides (azithromycin), and aminoglycosides (gentamicin and streptomycin). Plates were read using the Sensititre Vizion System® (Thermo Fisher) and minimum inhibitory concentrations (MIC) were interpreted using NARMS breakpoints (Centers for Disease Control and Prevention, 2014).
Frozen isolates were streak plated on blood agar plates and incubated overnight at 37°C to check for contamination. Visual inspection did not show any contamination therefore 1.5 ml of autoclaved BHI broth in a 2 ml micro centrifuge tube was inoculated from each isolate in a biological safety cabinet. The DNA was then extracted according to the manufacturer’s instructions for the DNeasy Blood and Tissue Kit (Qiagen N.V., Carlsbad, CA, United States). Two hundred microliter of DNA was eluted for each isolate into a sterile 2 ml micro centrifuge tube. The DNA samples were then stored at −80°C until further downstream processing.
In collaboration with the UC Davis Real-time PCR Research and Diagnostics Core Facility, a singleplex and several multiplex (duplex and triplex) qPCR assays were developed to facilitate rapid and sensitive analysis of samples. Isolates were analyzed for the presence of β-lactamase encoding genes (blaTEM, blaCTX–M, and blaCMY–2) and for presence of plasmid mediated quinolone resistance (PMQR) genes (oqxA, oqxB, qnrS, qnrB, and aac(6′)-Ib-cr). qnrA was not included in the qPCR assay because it has been rarely identified in Salmonella isolates with phenotypic resistance to fluoroquinolones. Cattle studies screening for qnr genes have more frequently detected qnrB and qnrS (Carroll et al., 2017). Furthermore, other recent studies screening Salmonella from isolates originating from broiler chicken and pork products for qnr genes did not detect qnrA genes (Tyson et al., 2017; Mahmud et al., 2018). Other recent studies screening human Salmonella isolates have also not detected qnrA, and noted it as infrequently detected when compared to qnrB and qnrS (Carroll et al., 2017; Karp et al., 2018).
Sequences from GenBank (blaTEM (LT985387), blaCTX–M (CP025146), blaCMY–2 (KY612500), oqxA (CP019074), oqxB (CP019074), qnrS (CP026578), qnrB (KP012539), and aac(6′)-Ib-cr (NG_056043)) were aligned using Sequence Analysis and Molecular Biology Data Management software Vector NTI AdvanceTM11 (Thermo Fisher Scientific, Carlsbad, CA, United States). The alignment was used to design primers specific to target for singleplex and multiplex qPCRs assays (Table 1). The specificity of the primers and probes was confirmed by BLAST searching against the non-redundant database of GenBank (NCBI). The primers for detecting blaTEM in our qPCR assay were designed to be more general and capable of annealing to both TEM non-extended spectrum β-lactamases and TEM-type ESBLs. This was done in the context of our assay being used as a screening tool.
Different fluorophores for each multiplex: blaTEM, blaCTX–M, aac(6′)-Ib-cr all used the fluorescent probe 6-carboxy-fluorescein (FAM); oqxA, blaCMY–2, and qnrB used the VIC probe; whereas, qnrS and oqxB used tetrachlorofluorescein (TET). All the probes utilized a 3′ minor groove binding quencher (Table 1). All qPCR assays were designed using Primer Express (Thermo Fisher Scientific) following the guidelines for multiplex qPCR assays. Amplicon lengths were ranging from 59 to 73 bp with each multiplex having similar lengths and GC percentage (Table 1). Primers and probes were synthesized by Life Technologies (Grand Island, NY, United States).
To test the efficiency of each primer/probe combination, singleplex mixes were prepared by combining 20 μL of 100 pmol/μl forward primer, 20 μl of 100 pmol/μl reverse primer, and 4 μl each of the 100 pmol/μl probes individually, in a final volume of 240 μl water. The qPCR multiplex primer/probe mix was prepared by mixing 40 μl for duplex and 60 μl for triplex of 100 pmol/μl forward primer, 40μl for duplex and 60 μl for triplex of 100 pmol/μl reverse primer, and 4 μl each of the two or three 100 pmol/μl probes in a final volume of 240 μl water.
The singleplex and multiplex qPCR for each target contained 0.42 μl water, 0.58 μl primer/probe mix (final concentration 400 nM of each primer and 80 nM probe), 6 μl of commercially available TaqManTM Universal Master Mix (UMM) (Thermo Fisher Scientific) for singleplex or Gene Expression Master Mix (Qiagen) for multiplex, and 5 μl of the DNA in a final volume of 12 μl.
All samples were placed in a 384-well plate and amplified in a 7900HT FAST Real-time PCR system (Thermo Fisher Scientific) using the manufacturer’s standard amplification conditions (2 min at 50°C, 10 min at 95°C, then 40 cycles of 15 s at 95°C and 60 s at 60°C). Fluorescent signals were collected during annealing and quantitative cycle (Cq) was calculated and exported with a threshold of 0.15 and a baseline of 3–10 for FAM labeled assays, 0.20 and a baseline of 3–10 for VIC assays, and 0.10 and a baseline of 3–15 for TET assays. The Cq was defined as the cycle in which there was a significant increase in reporter signal of the amount of PCR product detected during the exponential phase, above the threshold.
To assess and validate the efficiency of singleplex and multiplex qPCR assays for all assays, endpoint analysis of DNA using 10-fold dilutions was performed for each assay. In the singleplex qPCR mixtures (qnrS), only one target positive control was conducted using a nucleic acid template of known copy number. In multiplex qPCR mixtures, each of the two (blaTEM and oqxA, aac(6′)-Ib-cr, and qnrB) or three (blaCTX–M, blaCMY–2, and oqxB) target positive controls was combined in a single amplification tube. Standard curves were generated for each set of 10-fold serial dilutions of target. We calculated the amplification efficiency (E) of all assays from the slope (S) of the standard curves, using the formula E = 101/–s - 1 (Supplementary Table S3).
The multiplexes were very similar in sensitivity when all three targets were compared in the reaction. The sensitivity of each assay run as a single or multiplex was ∼10 or ∼100 gene copies. Such high similarity between the assays’ efficiency, sensitivity, amplicon length, and melting temperature assured the same competition efficiency during multiplex qPCR reaction.
Descriptive analysis for the distribution of Salmonella by year, phenotype, and resistance genes detected was conducted in JMP (SAS Institute Inc., Cary, NC, United States). To evaluate the reproducibility of Cq measurements and their associated error of the mean, a histogram was generated to visually evaluate the data (Figure 1) for each gene detected by qPCR, and mean and standard error for Cq values (Karlen et al., 2007). A 99% confidence interval of the standard error of the mean for Cq values was used for each gene to select the cut-off value for Cq values to classify an isolate as being positive for carrying that gene.

Figure 1. Histogram with cycle quantification (Cq) value distribution for Salmonella isolates screened for blaCMY–2 and blaTEM using multiplex qPCR. Blue represents isolates phenotypically resistant to ceftriaxone and orange represents isolates phenotypically susceptible to ceftriaxone. The red line indicates the cut-off Cq values for blaCMY–2 and blaTEM. (A) blaCMY–2 gene and blaTEM gene. Graph A depicts the Cq value distribution for blaCMY-2, and graph B depicts the Cq value distribution for blaTEM.
Logistic regression models using the GLIMMIX function in SAS using the logit link function were used to evaluate the population of blaCMY–2 positive, ceftriaxone resistant Salmonella isolates (n = 92) for the effect of explanatory variables sex, serotype, submission type (IDC vs. Suspect), and year group (calf vs. adult) on the risk of detecting an isolate positive for the gene blaTEM. Three models were constructed that differed by the presence of a binomial variable that allowed the evaluation of the effect of each of these three serotypes: S. Dublin, S. Typhimurium, and S. Newport when compared against any other serotype. This was a binomial variable that compared one of the three serotypes to all other serotypes combined (e.g., Dublin vs. Typhimurium, Newport, and any other serotype present in the study dataset). These three serotypes were selected because they represented the top three serotypes for isolates selected (92% of all isolates). Year group (2002–2009 vs. 2010–2016) when isolates were collected was included and maintained in all models as an independent variable to evaluate temporal changes on antimicrobial resistance. These two multi-year periods were chosen because they represent two halves of the time period evaluated. Additionally, these time periods were selected because of legislation related to antimicrobial use that occurred after 2009, such as changes on how ceftiofur could be used in an extra-label manner in livestock (Schmidt, 2012). Any explanatory variable that was not significant was removed from the model after evaluating if it negatively affected the model by evaluating the Akaike Information Criterion (AIC) value and overdispersion was evaluated using the Pearson chi-square divided by the degrees of freedom value. For all models, a P value of <0.05 was considered a significant difference.
Figure 1 contains two histograms of the Cq values for each of the two genes detected in our study: blaCMY–2 and blaTEM (Figures 1A,B). Only two genes were detected in the 110 Salmonella isolates tested, namely blaCMY–2 and blaTEM. The mean and 99% confidence interval for the Cq values of 23.0 (99% CI: 21.7–24.2) and 26.4 (99% CI: 24.9–27.7) were determined for blaCMY–2 and blaTEM, respectively. Figure 1A, which depicts the Cq value distribution for blaCMY–2, indicates that isolates below the cut-off of 23 were phenotypically resistant to ceftriaxone (Figure 1, blue color).
The opposite is primarily also true as most isolates above the cut-off of 23 are phenotypically susceptible to ceftriaxone (Figure 1, orange color). Results for logistic regression evaluating the effect of explanatory variables on the odds of isolating blaCMY–2-positive, ceftriaxone resistant Salmonella isolates also carrying blaTEM is depicted in Table 2.
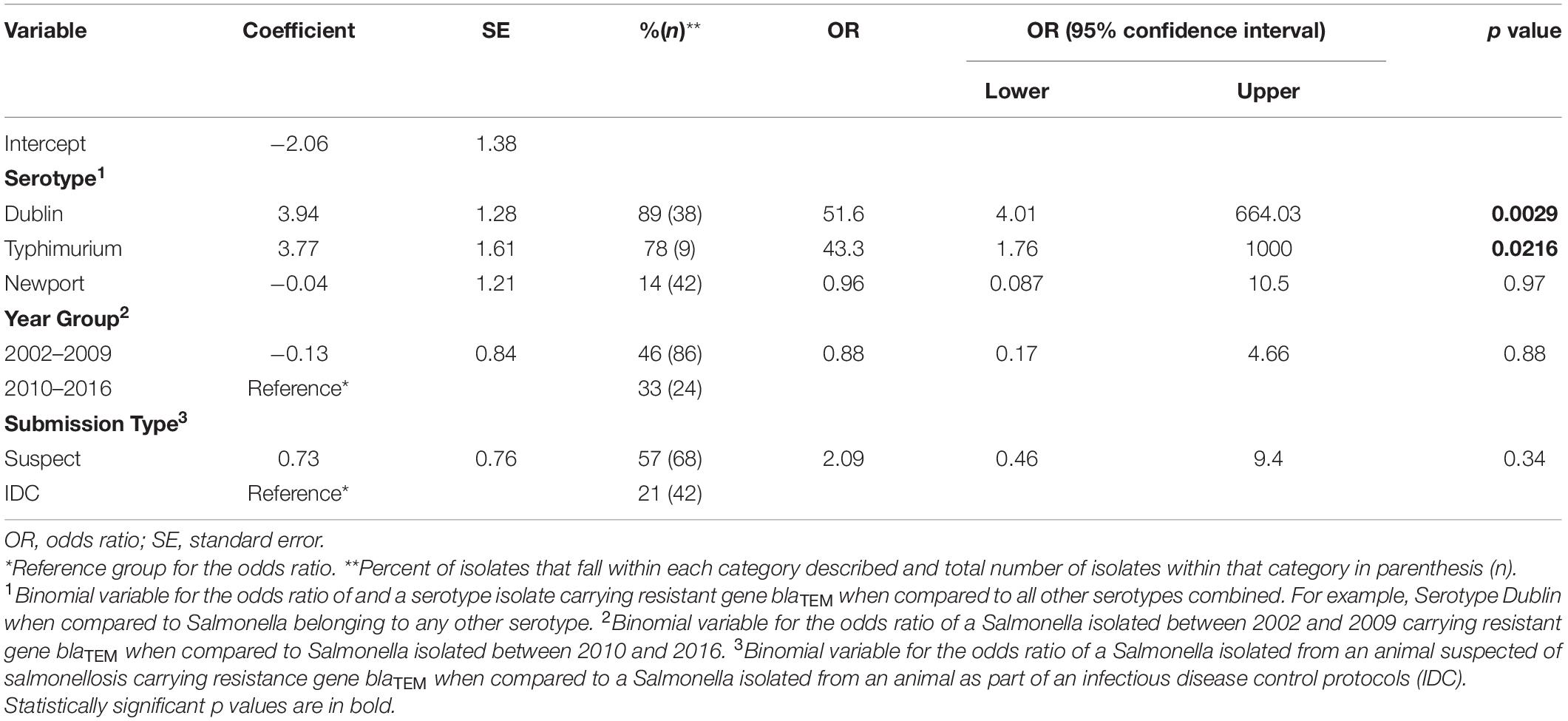
Table 2. Summary of the logistic regression model evaluating the effect of the explanatory variables serotype, year group, and submissionn type on the odds ratio of isolating a blaCMY–2–positive, ceftriaxone resistant Salmonella isolate also carrying blaTEM.
Isolates categorized as resistant to at least one third-generation cephalosporin (3GC) via MIC testing and were positive for either blaCMY–2 or blaTEM were analyzed by year (Table 3). Out of 242 Salmonella isolates, 94 (39%) were phenotypically resistant to at least one 3GC. Of these 94 3GC resistant isolates, 48% (n = 45) were positive for blaCMY–2 only and 50% (n = 47) were simultaneously positive for both blaCMY–2 and blaTEM. Two third-generation cephalosporin resistant isolates, which were negative for both blaCMY–2 and blaTEM, likely house ESBL genes not screened for by our qPCR assay.
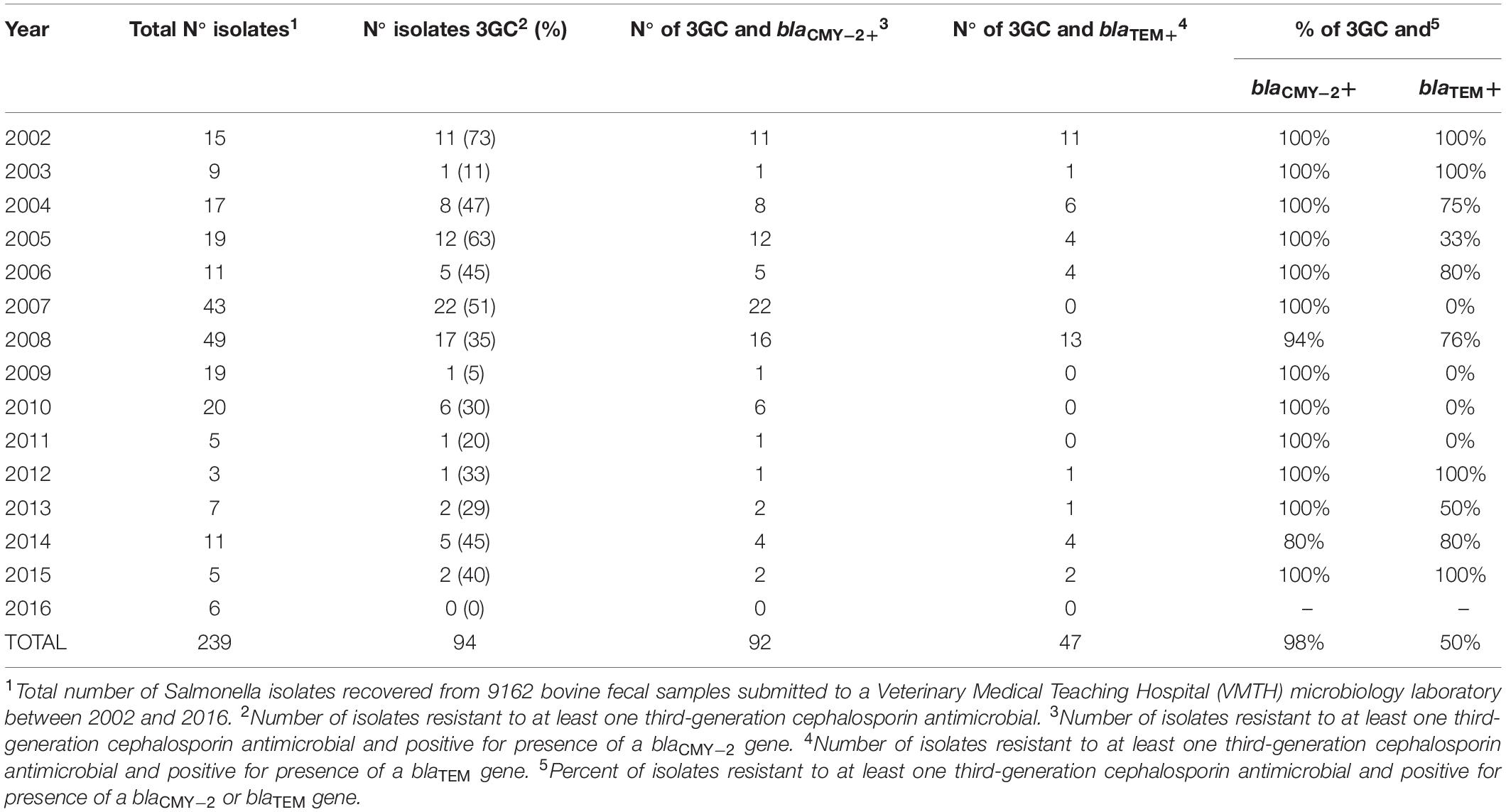
Table 3. Annual distribution of Salmonella isolates positive for phenotypic resistance to a third-generation cephalosporin (3GC) and for presence of antimicrobial resistance genes blaCMY–2 and blaTEM.
Despite fluctuations between years, there were no significant differences between individual years (Table 3). In addition, year group (2002–2009 vs. 2010–2016) was found to have no significant effect on the odds of isolating a blaCMY–2–positive, ceftriaxone resistant isolate positive for blaTEM (Table 2). For all years with isolates resistant to at least one 3GC, nearly all of these isolates were positive for blaCMY–2. Three years (2002, 2003, and 2012) reported 100% of third-generation cephalosporin (3GC) resistant isolates (Supplementary Table S1: Isolates 1–12, 85) were positive for both blaTEM and blaCMY–2.
A logistic regression model revealed that certain serotypes of Salmonella and submission type impacted the odds ratio of isolating a blaCMY–2 positive, ceftriaxone resistant isolate positive for blaTEM (Table 2). A similar analysis for calculating the odds ratio of isolating a ceftriaxone resistant isolate positive for only blaCMY–2 could not be conducted due to almost all isolates testing positive for blaCMY–2. For serotype Dublin, which accounted for 38% of all isolates, the odds ratio for testing positive for blaTEM when compared to all other serotypes was 51.6 (95% CI: 4.01–664.03, p = 0.0029). For serotype Typhimurium, which accounted for 9% of all isolates, the odds ratio for testing positive for blaTEM when compared to all other serotypes was 43.3 (95% CI: 1.76–1000, p = 0.0216). For serotype Newport, which accounted for 42% of all isolates, the odds ratio for testing positive for blaTEM when compared to all other serotypes was 0.96 (95% CI: 0.087–10.5, p = 0.97). For isolates in year group 2002–2009 (86% of total isolates) the odds ratio of carrying blaTEM was 0.88 when compared to Salmonella isolated between 2010 and 2016 (24% of total isolates) (95% CI: 0.17–4.66, p = 0.88). Although not significant, the odds ratio of having blaTEM for isolates collected from animals suspected of salmonellosis (68% of total isolates) was 2.09 when compared to isolates collected as part of an infectious disease control protocol (42% of total isolates) (95% CI: 0.46–9.7, p = 0.34).
While the three serotypes previously mentioned accounted for a majority of the 93 3GC resistant isolates, five other serotypes were also detected (Supplementary Table S1). These serotypes are Reading (Isolate 19), Meleagridis (Isolates 46, 50, and 52), Montevideo (Isolate 69), 9,12:nonmotile (Isolate 86), and Give (Isolate 93).
The ESBL gene blaCTX–M was not detected in any of the 110 NTS isolates screened. Additionally, none of the 110 NTS isolates, including two isolates phenotypically resistant to nalidixic acid, were positive for the PMQR genes screened in our assay (oqxA, oqxB, qnrS, qnrB, and aac(6′)-lb-cr). It should be noted that the methods used in our study did not allow for the delineation between aac(6′)-lb-cr and other highly similar variants like aac(6′)-lb. Likewise, the methods used in our study did not allow for differentiation between blaCTX–M groups.
Of the 242 NTS isolates tested, 39% of isolates (n = 94) were phenotypically resistant to a 3GC and 98% (n = 92) and 50% (n = 47) of these resistant isolates were PCR-positive for blaCMY–2 and blaTEM, respectively (Table 3). In the United States, ceftriaxone resistant NTS has primarily been observed to carry the gene blaCMY–2 encoding the AmpC-type β-lactamase (ACBL) CMY-2 (Centers for Disease Control and Prevention, 2014). The high prevalence of blaCMY–2 in 3GC resistant NTS in our study was similarly observed in a 2007 USDA study in which 81.6% of a subsample of ceftiofur resistant Salmonella isolates collected from 34,000 Salmonella isolates from the NARMS between 1999 and 2003 were positive for blaCMY–2 (Frye and Fedorka-Cray, 2007). More recently, a 2017 study which focused primarily on NTS isolated from beef cattle fecal samples detected blaCMY–2 in 8% of 571 isolates (Mollenkopf et al., 2017). Analysis of these CMY-2-positive isolates revealed 90% homology within serotypes, highlighting the clonal dissemination of blaCMY–2 within the cattle populations sampled in this study. Future work analyzing the homology within serotypes of the 92 isolates positive for blaCMY–2 in our study may be warranted given the number of isolates within the same serotype to be positive for blaCMY–2. blaCMY–2 is a very common resistance gene present when phenotypic resistance to ceftriaxone is observed, as shown in Table 3 and observed in other studies (Laurel, 2017), and represents a potential gene to focus future diagnostic approaches to classify an isolate as resistant to ceftriaxone without the need for use of phenotypic, culture-based methods. It should be noted that the methods used in our study did not allow for the delineation between blaCMY–2 and other highly similar variants like blaCMY–4.
The β-lactamase encoding gene blaTEM was present in 50% (n = 47) of 3GC resistant NTS isolates in our study and which were also simultaneously positive for blaCMY–2. TEM-1, discovered in 1965, is one of the most ubiquitous β-lactamases among Enterobacteriales (Paterson and Bonomo, 2005; Lachmayr et al., 2009). TEM-1 is not an ESBL and generally only degrades penicillins and the earliest developed cephalosporins. The first reported TEM-type ESBL, TEM-3, was discovered in 1989 (Sougakoff et al., 1988). With TEM variants now numbering greater than 200 and with many belonging to the ESBL subclass, a significant diversity exists within this resistance mechanism (Palzkill, 2018). A 2012 French study of 204 ESBL-producing E. coli isolates collected from sick cattle between 2006 and 2010 revealed only 7/204 (3.4%) expressed ESBL-type TEM-52 (Haenni et al., 2012). In the context of human medicine, the presence of TEM-type ESBLs in NTS in Bangladesh poses a public health concern (Ahmed et al., 2014). The 2014 Bangladesh study of 2120 Salmonella isolates from 128,000 human stool samples collected between 2005 and 2013 revealed that 88% (7/8) ceftriaxone resistant strains were positive for blaTEM. It should be noted that a limitation of our study was that all isolates positive for blaTEM were also positive for blaCMY–2 and that the methods used in our study did not allow for the delineation between the β-lactamase gene blaTEM–1 and other highly similar ESBL variants like blaTEM–52.
In the U.S., the first reported Enterobacteriales carrying blaCTX–M in dairy cattle was an E. coli strain in a study by Wittum et al. (2010) from Ohio in 2009. Identification of CTX-M-producing NTS in the U.S. has been relatively rare, but recent detection of such isolates in both livestock and retail chicken meat in the U.S. poses a potential threat to food safety (Wittum et al., 2012; Brown et al., 2018). None of the Salmonella isolates screened in our study were positive for blaCTX–M.
Our multiplex qPCR assay, while originally developed for use in an epidemiological or microbiological research setting, has potential advantages over traditional phenotypic testing common in a clinical setting. While research PCR assays tend to be low throughput and prioritize the ability to detect the lowest number of gene target copies, clinical PCR assays have additional requirements including high throughput and minimizing the chance of either false positive or negative results (Bustin et al., 2009). Frequently used in a clinical setting, phenotypic antimicrobial susceptibility testing (e.g., broth microdilution or Kirby–Bauer test) relies on multiple incubations of the microorganism and requires a minimum of 12 h (Doern, 2018). A multiplex qPCR assay could be clinically relevant when performing culture-based antibiotic resistance testing. A multiplex qPCR assay could serve as a complementary, rapid screening test for antimicrobial resistance genes while phenotypic tests are being conducted. Our qPCR assay (not including initial isolation) can be completed in about 6 h. Unlike phenotypic antimicrobial susceptibility tests, our assay requires DNA extraction and a qPCR run, but does not necessitate the bacteria to be incubated twice.
Only two serotypes were shown to significantly increase the odds ratio of isolating a blaCMY–2-positive, ceftriaxone resistant isolate also positive for blaTEM, namely Dublin and Typhimurium (Table 2). For our study, 98% of isolates resistant to ceftriaxone were also positive for blaCMY–2 (Table 3); because of that we cannot indicate causation of resistance to ceftriaxone as originating from blaTEM or blaCMY–2 gene (the latter being the most probable). A previous study conducted on colostrum fed to dairy calves screened cephalosporin resistant E. coli for β-lactamase resistance genes and observed, similarly to our study, that none of the isolates were positive for blaCTX–M; they also observed that 45% and 35% of these isolates were positive for blaCMY–2 and blaTEM, respectively (Awosile et al., 2017). The higher prevalence of blaTEM observed in our study compared to the colostrum study, in addition to increased odds for detection of blaTEM in S. Dublin isolates, is of critical importance as blaTEM has been linked to resistance to cephalosporins and various other β-lactam antibiotics; reducing the potential effective antimicrobial treatment options for infections caused by pathogens (Paterson and Bonomo, 2005).
While there is little research on the effect of serotype on the odds of isolating a ceftriaxone resistant and blaTEM-positive NTS isolate in cattle; a previous study has demonstrated both serotypes to possess high levels of ceftiofur (3GC) resistance, and the driver of resistance is most probably being driven by another antimicrobial gene (Otto et al., 2018). The most recent NARMS data of human NTS isolates revealed that 66.7% of serotype Dublin and 4.7% of serotype Typhimurium isolates were resistant to ceftriaxone. Despite being a cattle-adapted serotype, Dublin causes increased hospitalization and mortality in human infections when compared to other NTS serotypes (Harvey et al., 2017). Typhimurium is also one of the most common serotypes to cause human infection in both the US and globally (Gutema et al., 2019).
Our prior study evaluating phenotypic resistance of Salmonella isolates from cattle observed a 13.7 higher odds (p value = 0.0004) for isolating a multidrug resistant Salmonella from suspect clinical salmonellosis cases when compared to isolates originating from the VMTH IDC protocol sampling (Davidson et al., 2018). Our current study further evaluated specific resistance mechanisms for cephalosporin and fluoroquinolone resistance genes. We did not detect a significant difference in the odds ratio for isolating Salmonella from animals suspected of salmonellosis when compared to isolates originating from the IDC program for the resistance genes screened. This result could indicate that cephalosporin and fluoroquinolone resistance genes were not the main factors increasing the risk for MDR isolates between these two different sources of Salmonella isolates. Although antimicrobial resistance is not in itself a virulence factor, it is a key factor in development of infection, and may be considered a virulence-like factor in specific ecological niches which antibiotic resistant bacteria are able to colonize (Beceiro et al., 2013). This is especially consistent in a hospital environment where, if an opportunistic pathogen is drug resistant, it can cause disease more readily. Mutations increasing antimicrobial resistance have a range of effects on bacterial fitness during infection including decreased or increased pathogenic potential. Future studies should further elucidate the determinants of altered virulence potential in resistant pathogens and illuminate the mechanisms by which resistance traits modulate the outcome of disease in veterinary hospitals (Geisinger and Isberg, 2017). A limitation of our study was that the sample population were animals from a VMTH, and may not necessarily be extrapolated to other populations that may not be under similar circumstances and also explain a wider confidence interval for some of the variables evaluated in the model. Another limitation is that qnrA was not included as one of the PMQR genes screened in the qPCR assay; this was due to the very low risk of detecting qnrA in Salmonella of cattle origin (Carroll et al., 2017).
Out of 242 Salmonella isolates, 39% (n = 94) were resistant to at least one 3GC. Of these 3GC resistant isolates, 98% (n = 92) were positive for blaCMY–2 and 50% (n = 47) were positive for blaTEM and blaCMY–2. The consistently high prevalence of blaCMY–2 over time in isolates resistant to ceftriaxone suggests this gene may be a potential target for rapid molecular screening to identify isolates resistant to 3GC when compared to culture-based methods. The lack of isolates positive for blaCTX–M or PMQR genes screened suggest that the cattle population evaluated continued to be low risk group for carrier of these important resistance genes. There was also no significant association between the odds ratio of isolating a blaCMY–2-positive, ceftriaxone resistant isolate also positive for blaTEM and the year or year-group the isolates were collected. The higher odds for NTS serotype Dublin, ceftriaxone resistant isolate being positive for blaTEM highlight the need for continued monitoring of this important cattle host-adapted strain. Overall, our study suggests that the prevalence of resistance to cephalosporins due to ESBL and ACBL genes present in bovine NTS isolates has remained relatively constant in this hospital population in Northern California from 2002 to 2016.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
CB, BB, EE, EH, and RP conducted laboratory testing of samples. CB, BB, and RP performed data analysis and wrote the manuscript. CB, BB, AP, MC, and RP designed the study. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding was provided by USDA (Washington, DC) project number CA-V-PHR-4098-H. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the USDA. This work was partially funded by the Antimicrobial Use and Stewardship (AUS) through the California Department of Food and Agriculture (CDFA). Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the California Department of Food and Agriculture (CDFA). For her support to the project, the authors would like to thank Samantha Barnum.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.601924/full#supplementary-material
Ahmed, D., Ud-Din, A. I., Wahid, S. U., Mazumder, R., Nahar, K., and Hossain, A. (2014). Emergence of bla Tem type extended-spectrum beta -lactamase producing Salmonella spp. in the Urban Area of Bangladesh. Isrn Microbiol. 2014:715310. doi: 10.1155/2014/715310
Awosile, B. B., McClure, J. T., Sanchez, J., VanLeeuwen, J., Rodriguez-Lecompte, J. C., Keefe, G., et al. (2017). Short communication: extended-spectrum cephalosporin-resistant Escherichia coli in colostrum from New Brunswick, Canada, dairy cows harbor blaCMY-2 and blaTEM resistance genes. J. Dairy Sci. 100, 7901–7905. doi: 10.3168/jds.2017-12941
Beceiro, A., Tomas, M., and Bou, G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26, 185–230. doi: 10.1128/CMR.00059-12
Brown, A. C., Chen, J. C., Watkins, L. K. F., Campbell, D., Folster, J. P., Tate, H., et al. (2018). CTX-M-65 extended-spectrum beta-lactamase-producing Salmonella enterica Serotype Infantis, United States(1). Emerg. Infect. Dis. 24, 2284–2291. doi: 10.3201/eid2412.180500
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Carroll, L. M., Wiedmann, M., den Bakker, H., Siler, J., Warchocki, S., Kent, D., et al. (2017). Whole-Genome sequencing of drug-resistant Salmonella enterica isolates from dairy cattle and humans in New York and Washington States reveals source and geographic associations. Appl. Environ. Microbiol. 83, e00140–17. doi: 10.1128/AEM.00140-17
Centers for Disease Control and Prevention (2014). NARMS 2014 Human Isolates Surveillance Report. Atlanta, GA: Centers for Disease Control and Prevention.
Centers for Disease Control and Prevention (2019). Antibiotic Resistance Threats in the United States 2019. Atlanta, GA: US Department of Health and Human Services, CDC.
Ciesielczuk, H., Hornsey, M., Choi, V., Woodford, N., and Wareham, D. W. (2013). Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med. Microbiol. 62(Pt 12), 1823–1827. doi: 10.1099/jmm.0.064428-0
Clinical and Laboratory Standards Institute (2011). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-First Informational Supplement. CLSI Document M100-S21. Wayne, PA: Clinical and Laboratory Standards Institute.
Clinical and Laboratory Standards Institute (2020). Performance Standards for Antimicrobial Susceptibility Testing. M100, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Davidson, K. E., Byrne, B. A., Pires, A. F. A., Magdesian, K. G., and Pereira, R. V. (2018). Antimicrobial resistance trends in fecal Salmonella isolates from northern California dairy cattle admitted to a veterinary teaching hospital, 2002-2016. PLoS One 13:e0199928. doi: 10.1371/journal.pone.0199928
Dhillon, R. H., and Clark, J. (2012). ESBLs: a Clear and Present Danger? Crit. Care Res. Pract. 2012:625170. doi: 10.1155/2012/625170
Doern, C. D. (2018). The slow march toward rapid phenotypic antimicrobial susceptibility testing: are we there yet? J. Clin. Microbiol. 56:e1999–17. doi: 10.1128/JCM.01999-17
Engel, T., Slotboom, B. J., van Maarseveen, N., van Zwet, A. A., Nabuurs-Franssen, M. H., and Hagen, F. (2017). A multi-centre prospective evaluation of the Check-Direct ESBL Screen for BD MAX as a rapid molecular screening method for extended-spectrum beta-lactamase-producing Enterobacteriaceae rectal carriage. J. Hosp. Infect. 97, 247–253. doi: 10.1016/j.jhin.2017.07.017
Frye, J. G., and Fedorka-Cray, P. J. (2007). Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003. Int. J. Antimicrob. Agents 30, 134–142. doi: 10.1016/j.ijantimicag.2007.03.013
Geisinger, E., and Isberg, R. R. (2017). Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. J. Infect. Dis. 215(Suppl.1), S9–S17. doi: 10.1093/infdis/jiw402
Gutema, F. D., Agga, G. E., Abdi, R. D., De Zutter, L., Duchateau, L., and Gabriel, S. (2019). Prevalence and Serotype Diversity of Salmonella in apparently healthy cattle: systematic review and meta-analysis of published studies, 2000-2017. Front. Vet. Sci. 6:102. doi: 10.3389/fvets.2019.00102
Haenni, M., Saras, E., Métayer, V., Doublet, B., Cloeckaert, A., and Madec, J.-Y. (2012). Spread of the bla TEM-52 gene is mainly ensured by IncI1/ST36 plasmids in Escherichia coli isolated from cattle in France. J. Antimicrob. Chemother. 67, 2774–2776.
Harvey, R. R., Friedman, C. R., Crim, S. M., Judd, M., Barrett, K. A., Tolar, B., et al. (2017). Epidemiology of Salmonella enterica serotype dublin infections among Humans, United States, 1968-2013. Emerg. Infect. Dis. 23, 1493–1501. doi: 10.3201/eid2309.170136
Jacoby, G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Karlen, Y., McNair, A., Perseguers, S., Mazza, C., and Mermod, N. (2007). Statistical significance of quantitative PCR. BMC Bioinform. 8:131. doi: 10.1186/1471-2105-8-131
Karp, B. E., Campbell, D., Chen, J. C., Folster, J. P., and Friedman, C. R. (2018). Plasmid-mediated quinolone resistance in human non-typhoidal Salmonella infections: an emerging public health problem in the United States. Zoonoses Public Health 65, 838–849. doi: 10.1111/zph.12507
Lachmayr, K. L., Kerkhof, L. J., Dirienzo, A. G., Cavanaugh, C. M., and Ford, T. E. (2009). Quantifying nonspecific TEM beta-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 75, 203–211. doi: 10.1128/AEM.01254-08
Laurel, M. (2017). The National Antimicrobial Resistance Monitoring System: NARMS Integrated Report 2015. Silver Spring, MD: Food and Drug Administration.
Mahmud, S., Nazir, K., and Rahman, M. T. (2018). Prevalence and molecular detection of fluoroquinolone-resistant genes (qnrA and qnrS) in Escherichia coli isolated from healthy broiler chickens. Vet World 11, 1720–1724. doi: 10.14202/vetworld.2018.1720-1724
Mollenkopf, D. F., Mathys, D. A., Dargatz, D. A., Erdman, M. M., Habing, G. G., Daniels, J. B., et al. (2017). Genotypic and epidemiologic characterization of extended-spectrum cephalosporin resistant Salmonella enterica from US beef feedlots. Prev. Vet. Med. 146, 143–149. doi: 10.1016/j.prevetmed.2017.08.006
Otto, S. J. G., Ponich, K. L., Cassis, R., Goertz, C., Peters, D., and Checkley, S. L. (2018). Antimicrobial resistance of bovine Salmonella enterica ssp. enterica isolates from the Alberta Agriculture and Forestry Disease Investigation Program (2006-2014). Can. Vet. J. 59, 1195–1201.
Palzkill, T. (2018). Structural and mechanistic basis for extended-spectrum drug-resistance mutations in altering the specificity of TEM, CTX-M, and KPC β-lactamases. Front. Mol. Biosci. 5:16. doi: 10.3389/fmolb.2018.00016
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
Perez-Perez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40, 2153–2162. doi: 10.1128/jcm.40.6.2153-2162.2002
Polsfuss, S., Bloemberg, G. V., Giger, J., Meyer, V., Bottger, E. C., and Hombach, M. (2011). Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 49, 2798–2803. doi: 10.1128/JCM.00404-11
Rawat, D., and Nair, D. (2010). Extended-spectrum β-lactamases in gram negative bacteria. J. Global Infect. Dis. 2:263.
Rodriguez-Martinez, J. M., Machuca, J., Cano, M. E., Calvo, J., Martinez-Martinez, L., and Pascual, A. (2016). Plasmid-mediated quinolone resistance: two decades on. Drug Resist. Updat. 29, 13–29. doi: 10.1016/j.drup.2016.09.001
Roschanski, N., Fischer, J., Guerra, B., and Roesler, U. (2014). Development of a multiplex real-time PCR for the rapid detection of the predominant beta-lactamase genes CTX-M, SHV, TEM and CIT-type AmpCs in Enterobacteriaceae. PLoS One 9:e100956. doi: 10.1371/journal.pone.0100956
Schmidt, C. W. (2012). FDA proposes to ban cephalosporins from livestock feed. Environ. Health Perspect. 120:A106. doi: 10.1289/ehp.120-a106
Sougakoff, W., Goussard, S., and Courvalin, P. (1988). The TEM-3 β-lactamase, which hydrolyzes broad-spectrum cephalosporins, is derived from the TEM-2 penicillinase by two amino acid substitutions. FEMS Microbiol. Lett. 56, 343–348.
Stanaway, J. D., Parisi, A., Sarkar, K., Blacker, B. F., Reiner, R. C., Hay, S. I., et al. (2019). The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 19, 1312–1324.
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., and Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. doi: 10.1128/CMR.00016-09
Tamma, P. D., Doi, Y., Bonomo, R. A., Johnson, J. K., and Simner, P. J. (2019). Antibacterial resistance leadership G. A primer on AmpC beta-lactamases: necessary knowledge for an increasingly multidrug-resistant world. Clin. Infect. Dis. 69, 1446–1455. doi: 10.1093/cid/ciz173
The European Committee on Antimicrobial Susceptibility Testing (2013). EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Basel: EUCAST.
Thomson, K. S. (2001). Controversies about extended-spectrum and AmpC beta-lactamases. Emerg. Infect. Dis. 7, 333–336. doi: 10.3201/eid0702.010238
Tyson, G. H., Tate, H. P., Zhao, S., Li, C., Dessai, U., Simmons, M., et al. (2017). Identification of plasmid-mediated quinolone resistance in Salmonella Isolated from Swine Ceca and Retail Pork Chops in the United States. Antimicrob. Agents Chemother. 61, e1318–17. doi: 10.1128/AAC.01318-17
Wittum, T. E., Mollenkopf, D. F., Daniels, J. B., Parkinson, A. E., Mathews, J. L., Fry, P. R., et al. (2010). CTX-M-type extended-spectrum beta-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog. Dis. 7, 1575–1579. doi: 10.1089/fpd.2010.0615
Keywords: fluoroquinolone, cephalosporin, qPCR, antimicrobial resistance genes, Salmonella
Citation: Basbas C, Byrne BA, Chigerwe M, Escobar ED, Hodzic E, Pires AFA and Pereira RV (2021) Detection of Cephalosporin and Fluoroquinolone Resistance Genes via Novel Multiplex qPCR in Fecal Salmonella Isolates From Northern Californian Dairy Cattle, 2002–2016. Front. Microbiol. 12:601924. doi: 10.3389/fmicb.2021.601924
Received: 02 September 2020; Accepted: 20 January 2021;
Published: 15 February 2021.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Yvonne Pfeifer, Robert Koch Institute (RKI), GermanyCopyright © 2021 Basbas, Byrne, Chigerwe, Escobar, Hodzic, Pires and Pereira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard V. Pereira, cnZwZXJlaXJhQHVjZGF2aXMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.