- 1Department of Microbiological Diagnostics and Infectious Immunology, Medical University of Białystok, Białystok, Poland
- 2Institute of Biological Chemistry, Biophysics and Bioengineering, Heriot-Watt University, Edinburgh, United Kingdom
- 3Department of Microbiology, Institute of Biology, University of Białystok, Białystok, Poland
- 4Regional Centre for Transfusion Medicine, Białystok, Poland
- 5Second Department of Nephrology and Hypertension with Dialysis Unit, Medical University of Białystok, Białystok, Poland
- 6Second Department of General and Gastroenterological Surgery, Medical University of Białystok, Białystok, Poland
- 7Department of Neurology, Medical University of Białystok, Białystok, Poland
- 8Department of Vascular Surgery and Transplantation, Medical University of Białystok, Białystok, Poland
- 9Department of Clinical Molecular Biology, Medical University of Białystok, Białystok, Poland
- 10Department of Anesthesiology and Intensive Care, Medical University of Białystok, Białystok, Poland
- 11Department of Hematology, Medical University of Białystok, Białystok, Poland
Objectives: The growing incidence of multidrug-resistant (MDR) bacteria is an inexorable and fatal challenge in modern medicine. Colistin is a cationic polypeptide considered a “last-resort” antimicrobial for treating infections caused by MDR Gram-negative bacterial pathogens. Plasmid-borne mcr colistin resistance emerged recently, and could potentially lead to essentially untreatable infections, particularly in hospital and veterinary (livestock farming) settings. In this study, we sought to establish the molecular basis of colistin-resistance in six extraintestinal Escherichia coli strains.
Methods: Molecular investigation of colistin-resistance was performed in six extraintestinal E. coli strains isolated from patients hospitalized in Medical University Hospital, Bialystok, Poland. Complete structures of bacterial chromosomes and plasmids were recovered with use of both short- and long-read sequencing technologies and Unicycler hybrid assembly. Moreover, an electrotransformation assay was performed in order to confirm IncX4 plasmid influence on colistin-resistance phenotype in clinical E. coli strains.
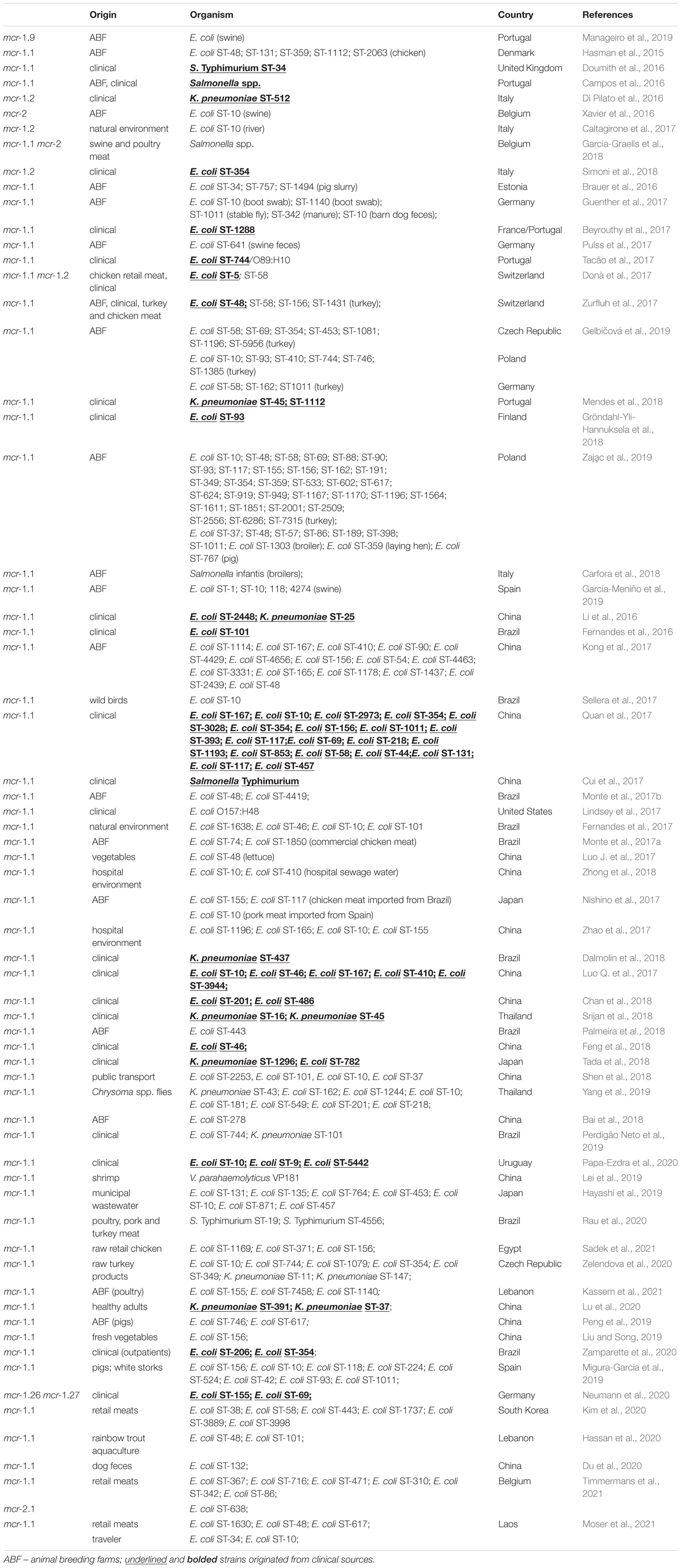
Results: Here we report on the emergence of six mcr-1.1-producing extraintestinal E. coli isolates with a number of virulence factors. Mobile pEtN transferase-encoding gene, mcr-1.1, has been proved to be encoded within a type IV secretion system (T4SS)-containing 33.3 kbp IncX4 plasmid pMUB-MCR, next to the PAP2-like membrane-associated lipid phosphatase gene.
Conclusion: IncX4 mcr-containing plasmids are reported as increasingly disseminated among E. coli isolates, making it an “epidemic” plasmid, responsible for (i) dissemination of colistin-resistance determinants between different E. coli clones, and (ii) circulation between environmental, industrial, and clinical settings. Great effort needs to be taken to avoid further dissemination of plasmid-mediated colistin resistance among clinically relevant Gram-negative bacterial pathogens.
Introduction
The growing incidence of multidrug-resistant (MDR) bacteria is an unavoidable challenge in modern medicine. Constant selection of MDR bacteria significantly contributes to the reduction of available therapeutic options. Colistin, also referred to as polymyxin E, is a cationic polypeptide considered a “last-resort” antimicrobial for treating infections caused by MDR Gram-negative bacterial pathogens, along with carbapenems and tigecycline (Livermore et al., 2011). Colistin was originally introduced in the 1950s for the treatment of infections caused by Gram-negative bacteria; however, polymyxins fell out of favor in the middle of the 1970s due to high rates of nephro- and neurotoxicity coupled with the advent of less toxic antibacterial agents. Nevertheless, by the mid-1990s (El-Sayed Ahmed et al., 2020), polymyxins were reintroduced into clinical practice due to the emergence of extensively drug-resistant (XDR) Gram-negative bacteria, and currently serve a critical role in the antimicrobial armamentarium (Kaye et al., 2016). Moreover, colistin often stands as the last antimicrobial agent retaining activity against carbapenem-resistant Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii (Olaitan et al., 2014). Unfortunately, bacterial resistance to polymyxin E emerged rapidly, and could potentially lead to essentially untreatable infections, particularly in the hospital setting where aforementioned XDR microorganisms frequently cause life-threatening infections in the most vulnerable patient populations (Kaye et al., 2016).
Colistin is capable of interacting with lipid A moiety of the lipopolysaccharide (LPS), thereby expelling Ca2+ and Mg2+ ions from phosphate groups and resulting in disruption of the negatively charged outer membrane (OM) of Gram-negative bacteria. Therefore, the ability of bacteria to resist killing by antimicrobial cationic polypeptides often entails modification of the OM (LPS modification resulting in reduced OM net negative charge). Polymyxin resistance has increased gradually within the last few years, and knowledge on a wide variety of possible chromosomal or acquired resistance mechanisms is still expanding (Olaitan et al., 2014). Most mechanisms conferring polymyxin resistance are directed at modifications of the lipid A moiety of the LPS, which is the primary target of colistin. Most genetic alterations, either chromosomal or acquired, entail a common lipid A modification pathway with 4-amino-4-deoxy-L-arabinose (L-Ara4N) and/or phosphoethanolamine (pEtN) addition. The most important target of L-Ara4N is the 4′-phosphate group of lipid A, but it can also be added to the 1-phosphate group or 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) (Baron et al., 2016). Substitution of the phosphate groups by L-Ara4N is followed by significant reduction of net negative charge of lipid A to 0, while pEtN modifications are associated with a net charge decrease from −1,5 to −1 (Nikaido, 2003). Therefore, L-Ara4N modification seems to be the most effective, owing to the nature of the OM charge modification (Olaitan et al., 2014). In Enterobacteriaceae, the aforementioned modifications of lipid A can result from mutation in the two-component systems (TCSs) such as PhoPQ, BasSR (PmrAB), small feedback-inhibition peptide MgrB, as well as from plasmid-mediated determinants (i.e., mcr gene encoding pEtN transferase) (Zeng et al., 2016; Litrup et al., 2017; Roer et al., 2017; Torpdahl et al., 2017; Osei Sekyere, 2019). The first plasmid-mediated polymyxin resistance gene, termed as mcr-1 (currently mcr-1.1) was identified in China in November 2015, and was subsequently reported all over the world, in both retrospective and prospective studies (Liu et al., 2016). The earliest, so far described, mcr-producing strains date back to the end of the previous century, in the 1980s (Shen et al., 2016). The earliest mcr-producing bacterial strain of clinical origin was a Shigella sonnei isolated from a pediatric patient in Vietnam, in 2008 (Pham Thanh et al., 2016). The various mcr variants (mcr-1 to mcr-10) have been so far, identified in various species of Gram-negative pathogens originating from animals, meat, food products, environmental, and human sources (Partridge et al., 2018; Nang et al., 2019). The emergence of plasmid-mediated pEtN transferase-encoding genes is a matter of serious concern due to the potential for rapid dissemination via horizontal gene transfer. Broad distribution of mcr genes in multidrug-resistant hospital strains would be especially dangerous in clinical settings, and could possibly result in wide dissemination of pandrug-resistant bacteria and untreatable infections. Here, we report on the emergence of mcr-1.1-harboring IncX4 plasmid in six extraintestinal Escherichia coli strains of clinical origin isolated in University Hospital of Bialystok, Poland between 2016 and 2018.
Materials and Methods
Clinical Isolates Used in the Study
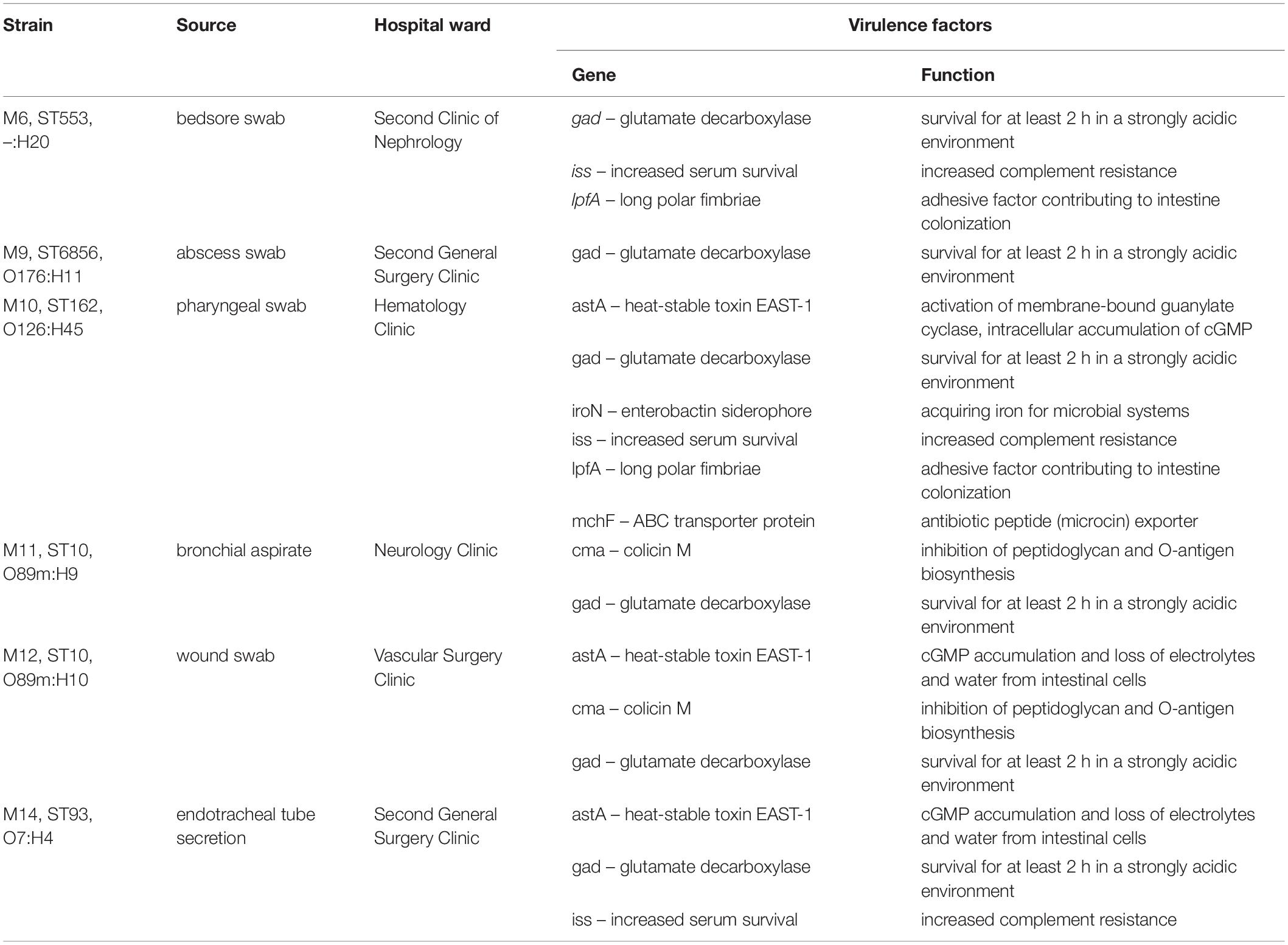
Colistin-resistant E. coli isolates were obtained during microbiological screening of infected patients hospitalized in Medical University Hospital in Bialystok, between 2016 and 2018. Extraintestinal solates originated from postoperative wound swab, bedsore swab, perianal abscess swab, pharyngeal swab, bronchial aspirate, and endotracheal tube secretion. Clinical characteristics of the patients colonized by extraintestinal mcr-1.1-producing E. coli strains are presented in Table 1.
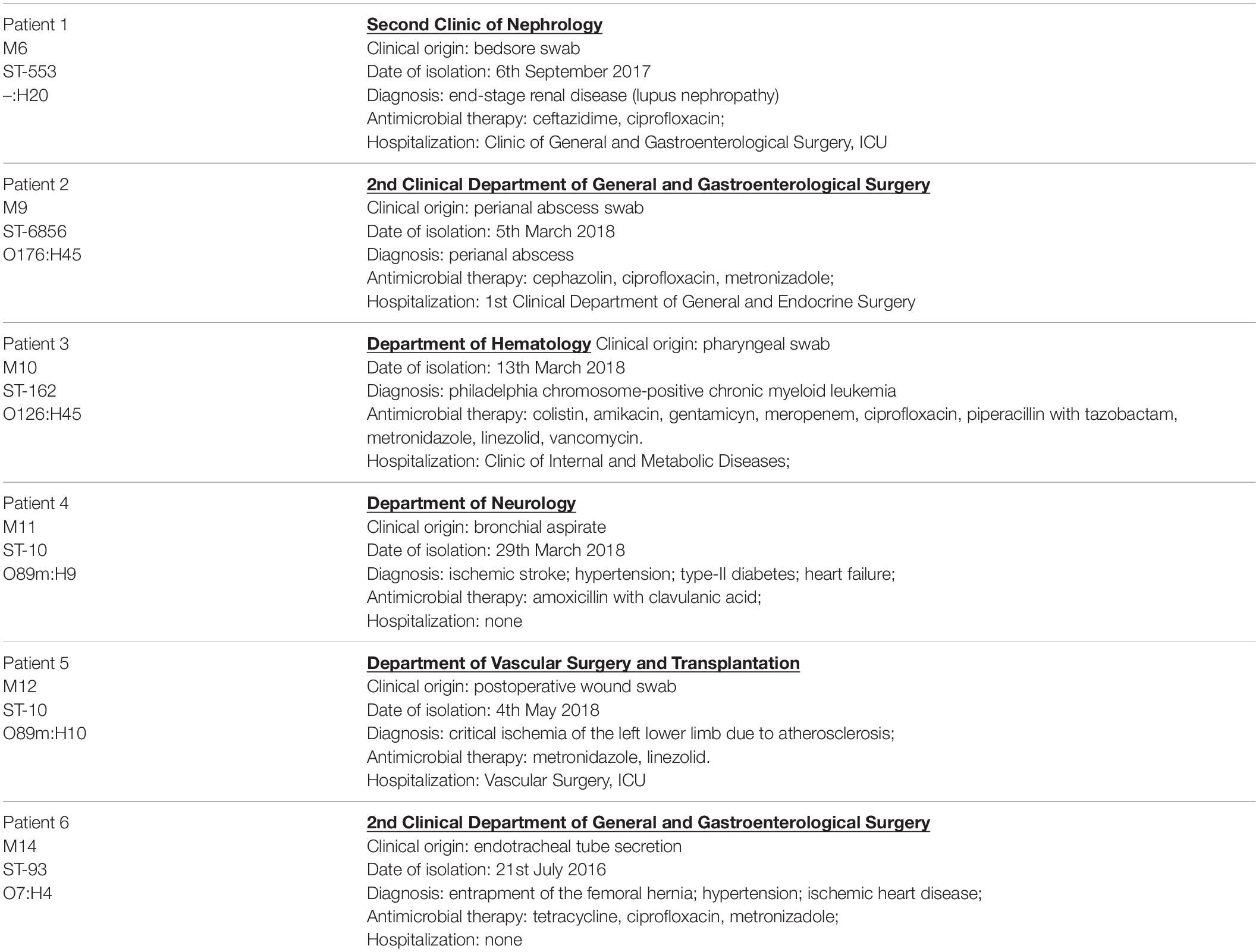
Table 1. Clinical characteristics of the patients colonized by extraintestinal mcr-1.1-producing E. coli strains.
Bacterial Identification and Antimicrobial Susceptibility Testing
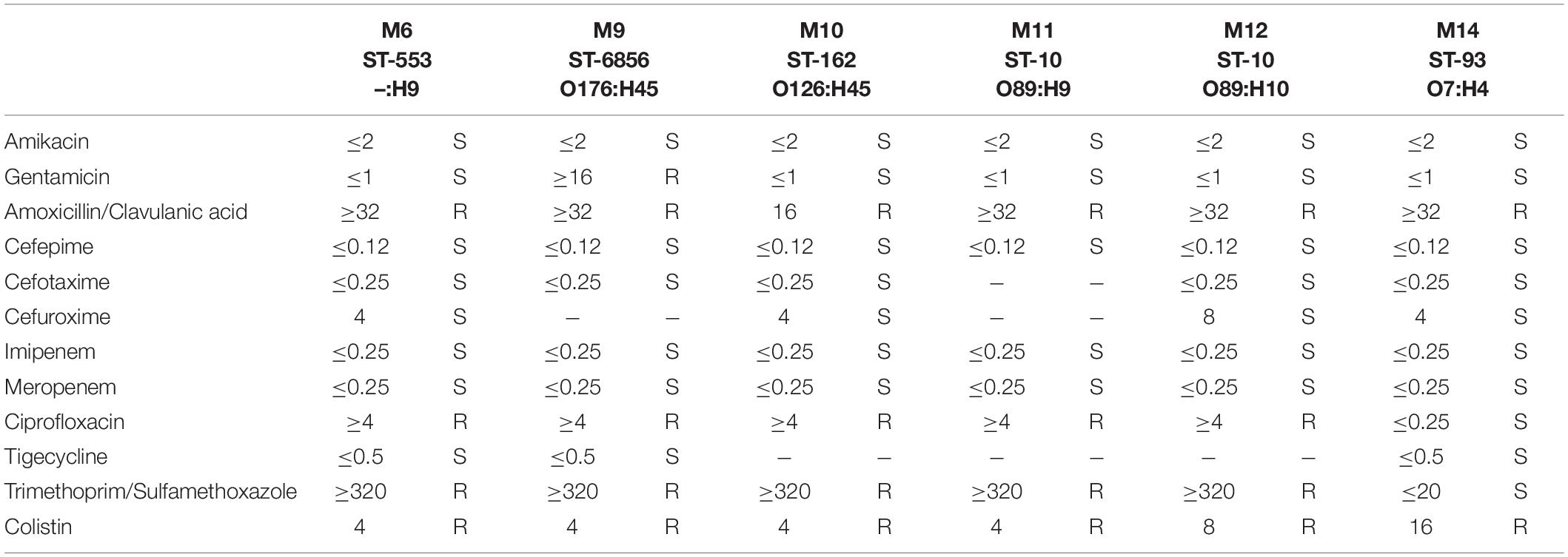
Bacterial isolates identification was performed with VITEK-MS (bioMérieux, Marcy l’Etoile, France); with subsequent antimicrobial susceptibility testing (AST) using the VITEK 2 system (bioMérieux, Marcy l’Etoile, France), SensiTest Colistin broth microdilution method (Liofilchem, Roseto degli Abruzzi, Italy), and MIC Test Strips (Liofilchem, Roseto degli Abruzzi, Italy) following manufacturer guidelines. AST results were interpreted in accordance with European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria.
Whole Bacterial DNA Extraction and Sequencing
Whole bacterial DNA from six clinical extraintestinal E. coli was isolated from Luria Broth overnight cultures with use of silica column-based Genomic Mini AX Bacteria kit (A&A Biotechnology). Purified bacterial DNA was sequenced using both short- and long-read methodologies (Illumina and Oxford Nanopore Technology).
In the first step of molecular analysis, Nextera XT library preparation kit and Nextera XT Indexes (Illumina) were used for previously quantified bacterial DNA, which was simultaneously fragmented and tagged with sequencing adapters in a single-tube enzymatic reaction. Quality and quantity of libraries were assessed by fluorometry (Qubit, Thermo Fisher Scientific) and chip electrophoresis (2100 Bioanalyzer, Agilent). FASTQ reads were generated with the use of MiSeq Reagent Kit v3 (600 cycles) and MiSeq analyzer (Illumina).
In the next step, DNA libraries were prepared with the use of a Ligation Sequencing Kit (SQK-LSK109) with Native Barcoding Expansion (EXP-NBD104). Quality and quantity of libraries were assessed by fluorometry (Qubit, Thermo Fisher Scientific) and chip electrophoresis (2100 Bioanalyzer, Agilent). FASTQ reads were generated with the use of Spot-ON Flow Cell (FLO-MIN106D R9 Version) and MinION Mk1b analyzer (Oxford Nanopore Technology).
Raw Data Quality Assessment and Downstream Bioinformatics
After quality assessment and quality filtering, reads were trimmed (Trimmomatic in case of Illumina reads), and demultiplexed with Porechop in case of long ONT reads (Bolger et al., 2014). Full structures of bacterial chromosomes and plasmids were recovered using Unicycler hybrid assembler which utilizes spades.py, racon, makeblastdb, tblastn, bowtie2, samtools, bcftools, and pilon (Wick et al., 2017). Alignment and mapping of nucleotide sequences were performed using Geneious 10.0.9 software (Biomatters Ltd., Auckland, New Zealand). A RAST (Rapid Annotation using Subsystem Technology)-annotated genomes were subjected to subsequent in silico analyses with use of PlasmidFinder, Resfinder, Virulence Finder (Carattoli et al., 2014; Brettin et al., 2015; Bortolaia et al., 2020).
Strain Phylogenomics
The final assembled genome sequence data were uploaded to the Type (Strain) Genome Server (TYGS), a free bioinformatics platform available under https://tygs.dsmz.de, for a whole genome-based taxonomic analysis (Meier-Kolthoff and Göker, 2019).
For the phylogenomic inference, all pairwise comparisons among the set of genomes were conducted using the Genome BLAST Distance Phylogeny approach (GBDP) and accurate intergenomic distances inferred under the algorithm ‘trimming’ and distance formula d5 (Meier-Kolthoff et al., 2013). Phylogenomic tree inferred with FastME 2.1.6.1 (Lefort et al., 2015) from GBDP distances calculated from genome sequences.
IncX4 Plasmids Phylogenomics
For the purpose of IncX4 plasmid phylogenomic inference, 100 similar sequences from the BLAST database were aligned and analyzed with the use of Clustal Omega (clustalo 1.2.4). Resulting phylogenetic tree was visualized using iTOL v61 (Letunic and Bork, 2021). Structural comparison between colistin-conferring plasmids harbored by studied extraintestinal E. coli isolates and IncX4 plasmid sequences deposited in NCBI was prepared using BLAST Ring Image Generator (BRIG) – default parameters with 90/70 as upper/lower threshold (Alikhan et al., 2011).
Transconjugation Assays
Electrotransformation of the IncX4 plasmid into the recipient E. coli strain was performed in order to confirm its influence on colistin-resistance phenotype in clinical E. coli isolates. To determine whether mcr-1.1 gene was located on pMUB-MCR 33.3 kbp IncX4 plasmid, transconjugation experiments were performed, with plasmid profiles preparation using Plasmid Mini AX kit (A&A Biotechnology), and subsequent electrotransformation into plasmid-free and colistin-sensitive E. coli TOP10 strain. Electrotransformation with subsequent selection of the transformants on the Luria-Bertani medium containing 1 mg/L colistin was conducted for the E. coli TOP10 strain.
Results
Extraintestinal E. coli isolates incorporated into the described study presented a relatively similar antimicrobial resistance pattern (66.66%; 4 of 6). E. coli MIN6 ST-553, MIN10 ST-162, MIN11 ST-10, and MIN12 ST-10 were found to be resistant to amoxicillin/clavulanic acid (MIC > 32 mg/L), ciprofloxacin (MIC ≥ 4 mg/L), trimethoprim/sulfamethoxazole (MIC ≥ 320 mg/L), and colistin (MIC = 4 mg/L, except of strain MIN12 – MIC = 8 mg/L). E. coli MIN9 ST-6856 was found to be resistant to amoxicillin/clavulanic acid (MIC > 32 mg/L), gentamicin (MIC ≥ 16 mg/L), ciprofloxacin (MIC ≥ 4 mg/L), trimethoprim/sulfamethoxazole (MIC ≥ 320 mg/L), and colistin (MIC = 4 mg/L). Furthermore, E. coli MIN14 strain was resistant to amoxicillin/clavulanic acid (MIC > 32 mg/L), and presented the highest colistin MIC = 16 mg/L. Detailed antimicrobial susceptibility testing results of six extraintestinal E. coli strains are presented in Table 2.
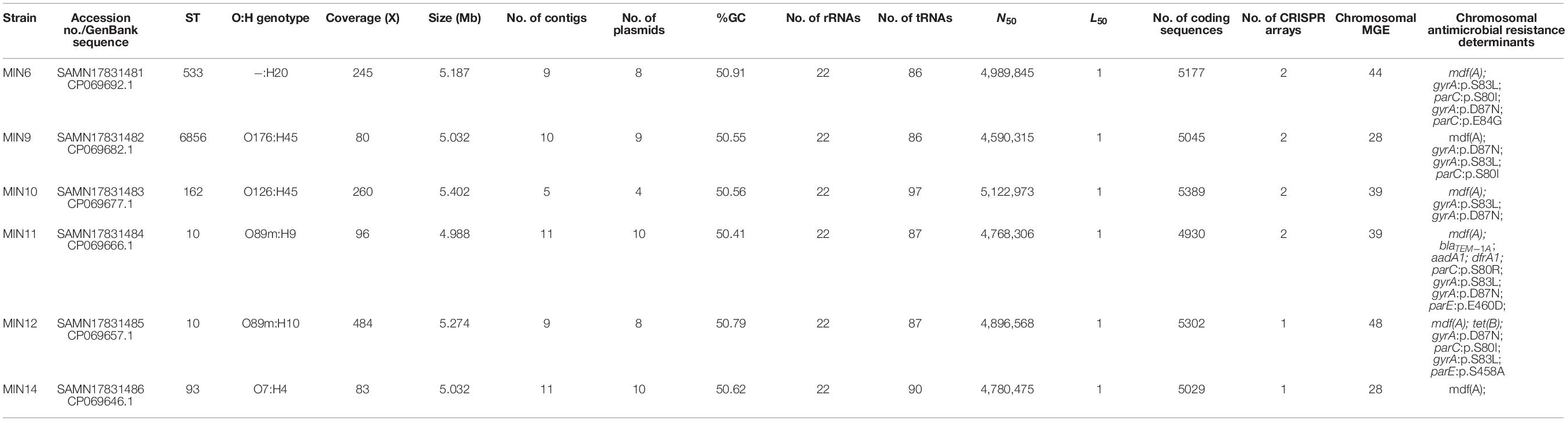
Whole-genome sequencing with use of a hybrid assembly approach allowed us to recover full genome and mobilome structures of tested extraintestinal colistin-resistant E. coli strains. Detailed characteristics of the sequenced genomes are presented in Table 3.
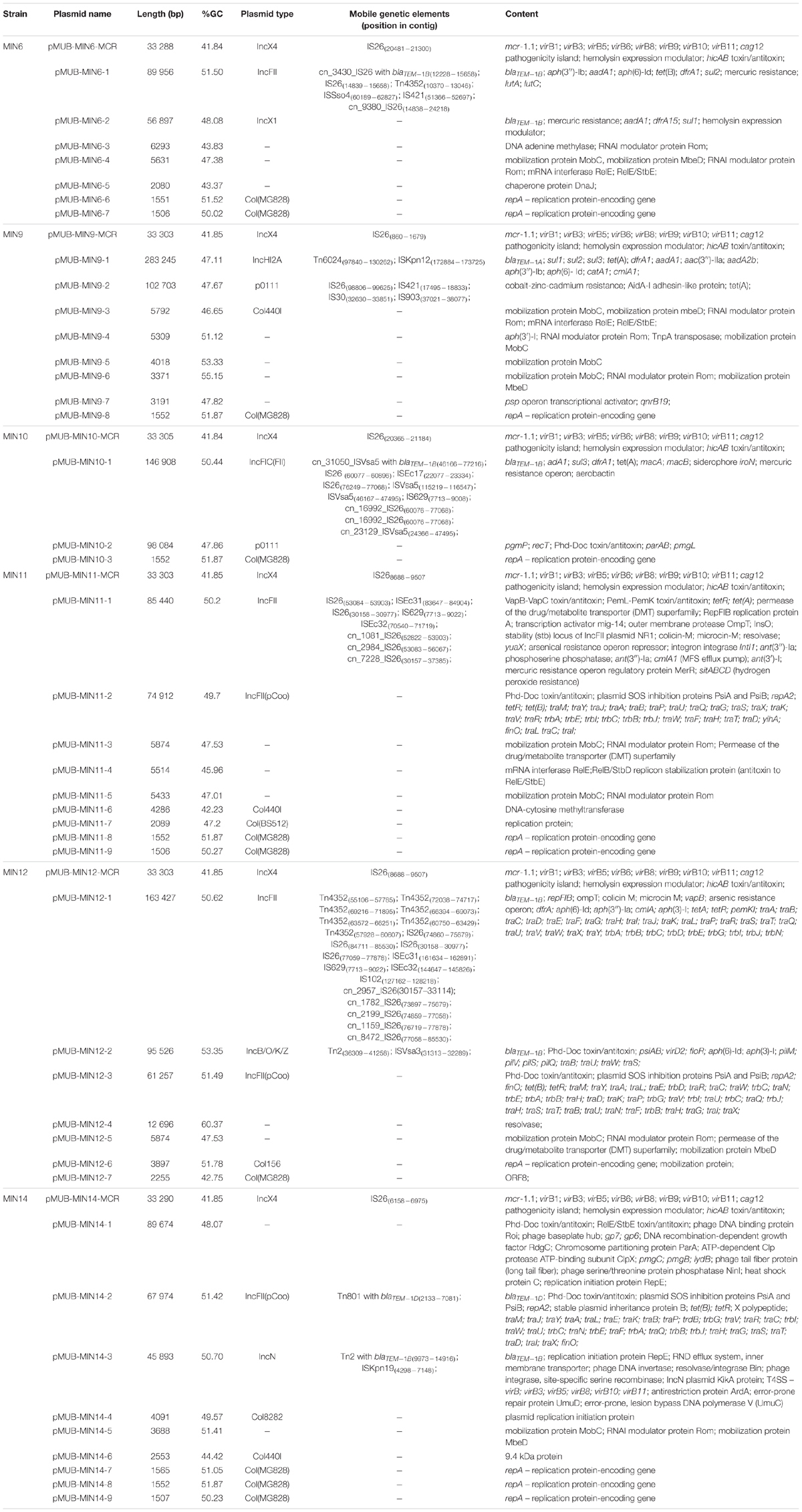
Moreover, all six colistin-resistant extraintestinal E. coli strains were equipped with a relatively rich plasmidic panel (Table 4). Interestingly, all of the tested strains harbored a IncX4 33.3 kbp plasmid pMUB-MCR, with the mobile pEtN transferase-encoding gene, mcr-1.1, which has been proved to be encoded within a type IV secretion system (T4SS), next to the PAP2-like membrane-associated lipid phosphatase gene (Figures 1, 2). The biological consequences of pMUB-MCR IncX4 plasmid possession were evaluated with the use of a transconjugation assay. The E. coli TOP10 electrotransformants carrying the 33.3 kbp IncX4 plasmid showed a MIC of colistin of 2 mg/L, which corresponded to a 16-fold increase as compared to the recipient E. coli TOP10 strain. These data confirmed that 33.3 kbp IncX4 pMUB-MCR conjugative plasmid is responsible for colistin-resistance in six extraintestinal clinical E. coli strains isolated from patients hospitalized in Medical University Hospital in Bialystok, Poland. Moreover, phylogenomics of IncX4 plasmids bearing mobile pEtN transferase-encoding genes is presented in Figure 3.
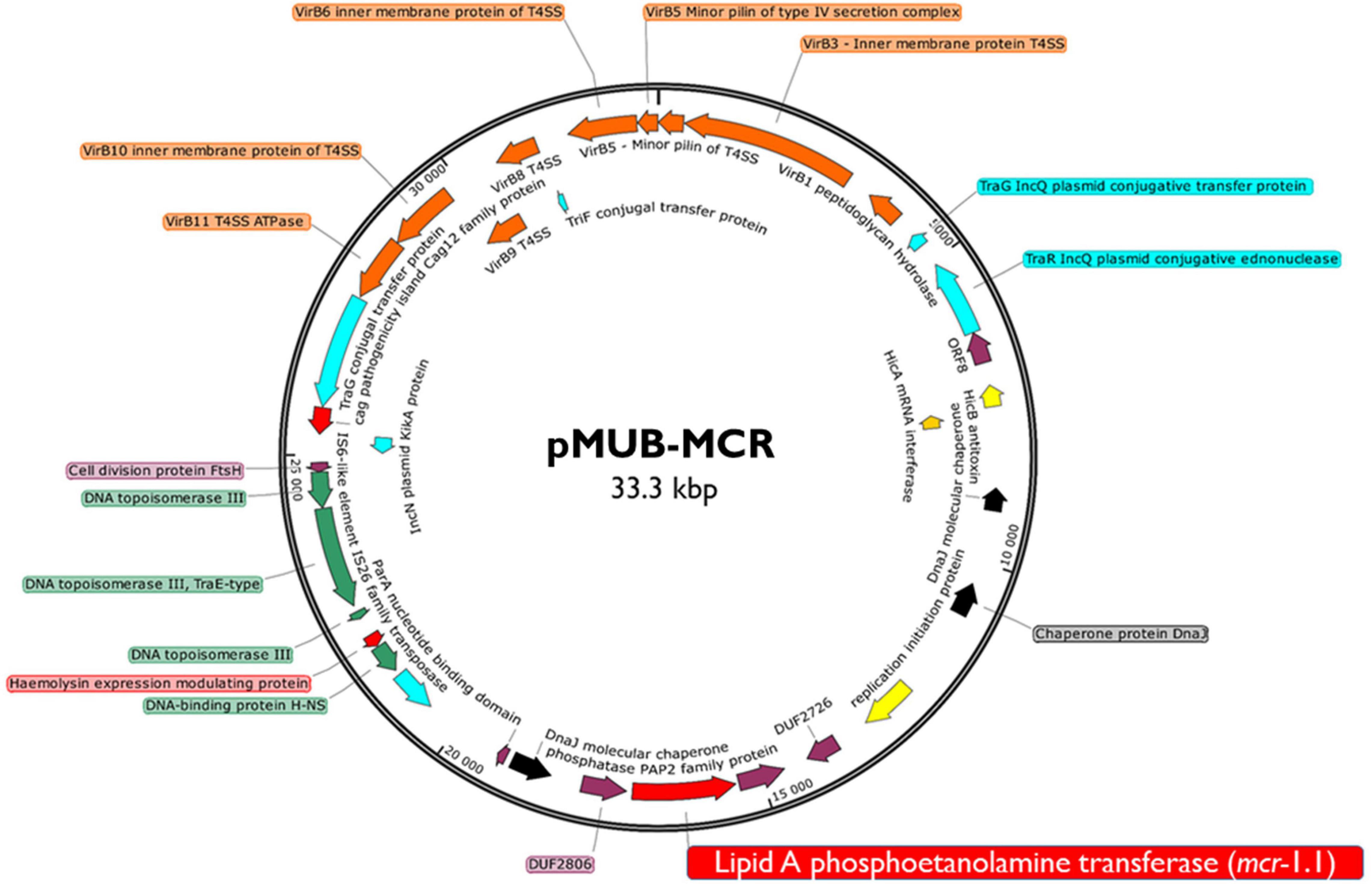
Figure 1. Genetic structure of mcr-1.1 harboring IncX4 plasmid. Figure represents a RAST (Rapid Annotation using Subsystem Technology)-annotated plasmid produced by the E. coli MIN6 strain. Arrows on the diagram represents annotated functional genes present within the structure of 33.3 kb IncX4 plasmid.
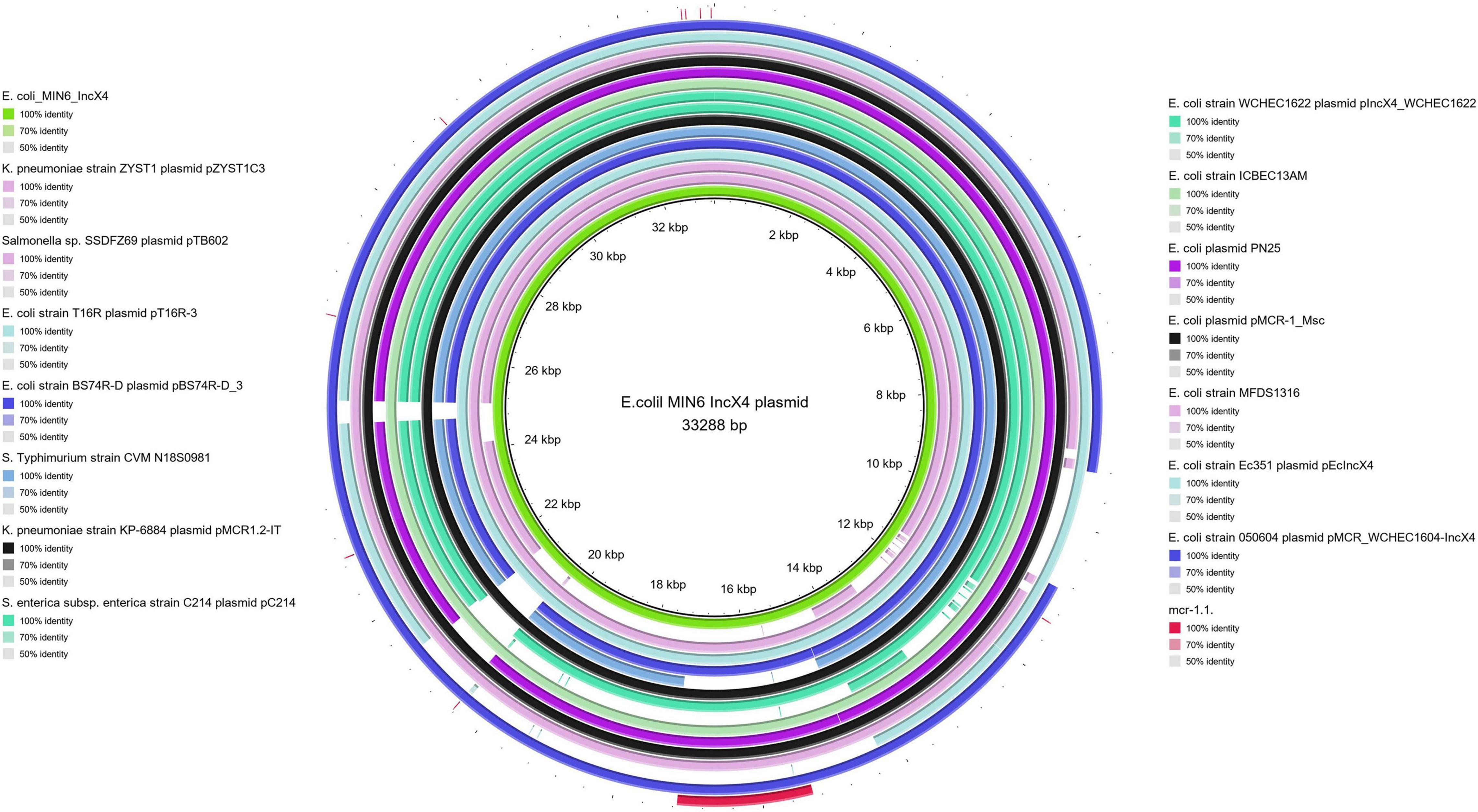
Figure 2. BRIG alignment of IncX4 plasmids recovered from different Enterobacterales. Structural comparison between colistin-conferring plasmids harbored by studied extraintestinal E. coli isolates and IncX4 plasmid sequences deposited in NCBI was prepared using BLAST Ring Image Generator (BRIG). The alignment includes the mcr-1.1. sequence (red fragment), and the one IncX4 mcr-1–bearing plasmid harbored by E. coli MIN6 described in our study (green inner circle).
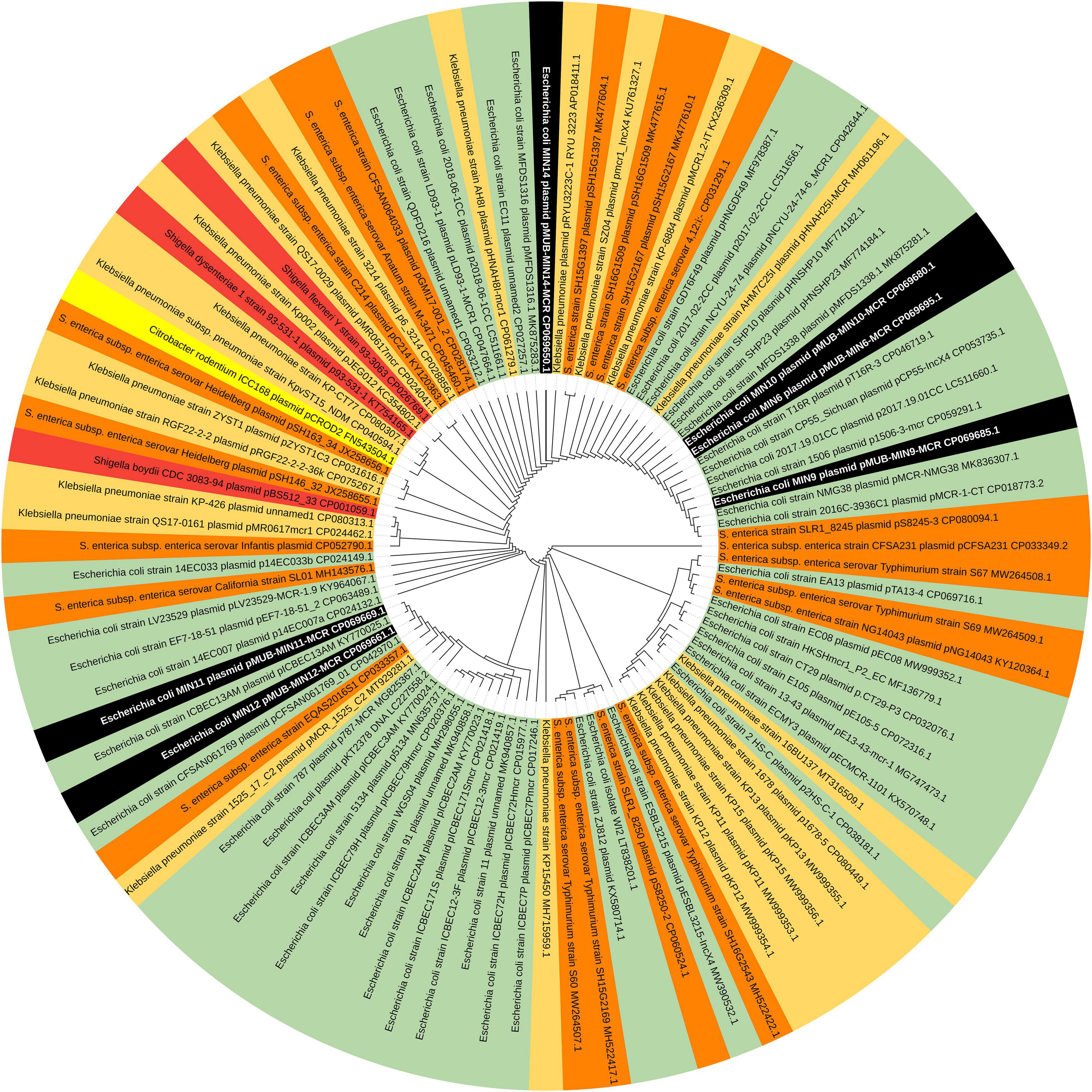
Figure 3. Phylogenomics of IncX4 plasmids bearing mobile pEtN transferase-encoding genes. Colored segments represents bacterial genera harboring IncX4 plasmids – black segments – E. coli strains MIN6, MIN9, MIN10, MIN11,MIN12, MIN14; green segments – E. coli; light orange segments – K. pneumoniae; orange segments – Salmonella spp.; red segments – Shigella spp., yellow segments – Citrobacter spp.
In addition to the IncX4 33.3 kbp plasmid pMUB-MCR, the tested E. coli strains were equipped with relatively rich plasmid panels. E. coli MIN6 possessed seven plasmids, two of which constituted a vehicle for antimicrobial resistance determinants, namely pMUB-MIN6-1 (IncFII plasmid), and pMUB-MIN-6-2 (IncX1 plasmid). E. coli MIN9 harbored eight plasmids, four of which constituted a vehicle for antimicrobial resistance determinants, namely pMUB-MIN9-1 (IncHI2A), pMUB-MIN9-2 (p0111), pMUB-MIN9-4, and pMUB-MIN9-7. E. coli MIN10 possessed three plasmids, one of which constituted a vehicle for antimicrobial resistance determinants, namely pMUB-MIN10-1 – IncFIC(FII). Among nine plasmids harbored by E. coli MIN11, two were responsible for antimicrobial resistance determinants carriage, namely pMUB-MIN-11-1 (IncFII), and pMUB-MIN-11-2 [IncFII(pCoo)]. E. coli MIN12 harbored seven plasmids, three of which possessed antimicrobial resistance determinants, namely pMUB-MIN12-1 (IncFII), pMUB-MIN12-2 (IncB/O/K/Z), and pMUB-MIN12-3 [IncFII(pCoo)]. Furthermore, among nine plasmids possessed by E. coli MIN14, two harbored antimicrobial resistance genes, namely pMUB-MIN14-2 [IncFII(pCoo)], and pMUB-MIN14-3 (IncN). Detailed features of plasmids harbored by studied E. coli strains are presented in Table 4.
The MLST approach was utilized in order to evaluate the molecular relatedness of tested extraintestinal colistin-resistant E. coli strains. Among 6 tested clinical strains, two were proven to be clonally related, namely E. coli MIN11 and MIN12, which belonged to ST-10. Those two strains were isolated within an interval of 36 days, in the Department of Neurology (MIN11) and Department of Vascular Surgery and Transplantation (MIN12). However, those two strains were easily distinguished by O:H genotype (MIN11 – O89m:H9 vs., MIN12 – O89m:H10), plasmid content (MIN11 – 10 plasmids vs. MIN12 – 8 plasmids), number of chromosomal mobile genetic elements (MIN11 – 39 vs. MIN12 – 48), virulence factors (MIN-12 was distinguished by the presence of heat-stable toxin EAST-1) and content of chromosomal resistance determinants. The remaining extraintestinal colistin-resistant E. coli strains belonged to ST-533 (E. coli MIN6), ST-6856 (E. coli MIN9), ST-162 (E. coli MIN10), and ST-93 (E. coli MIN14). Moreover, whole-genome sequence-based phylogenomics of colistin-resistant E. coli strains is presented in Figure 4.
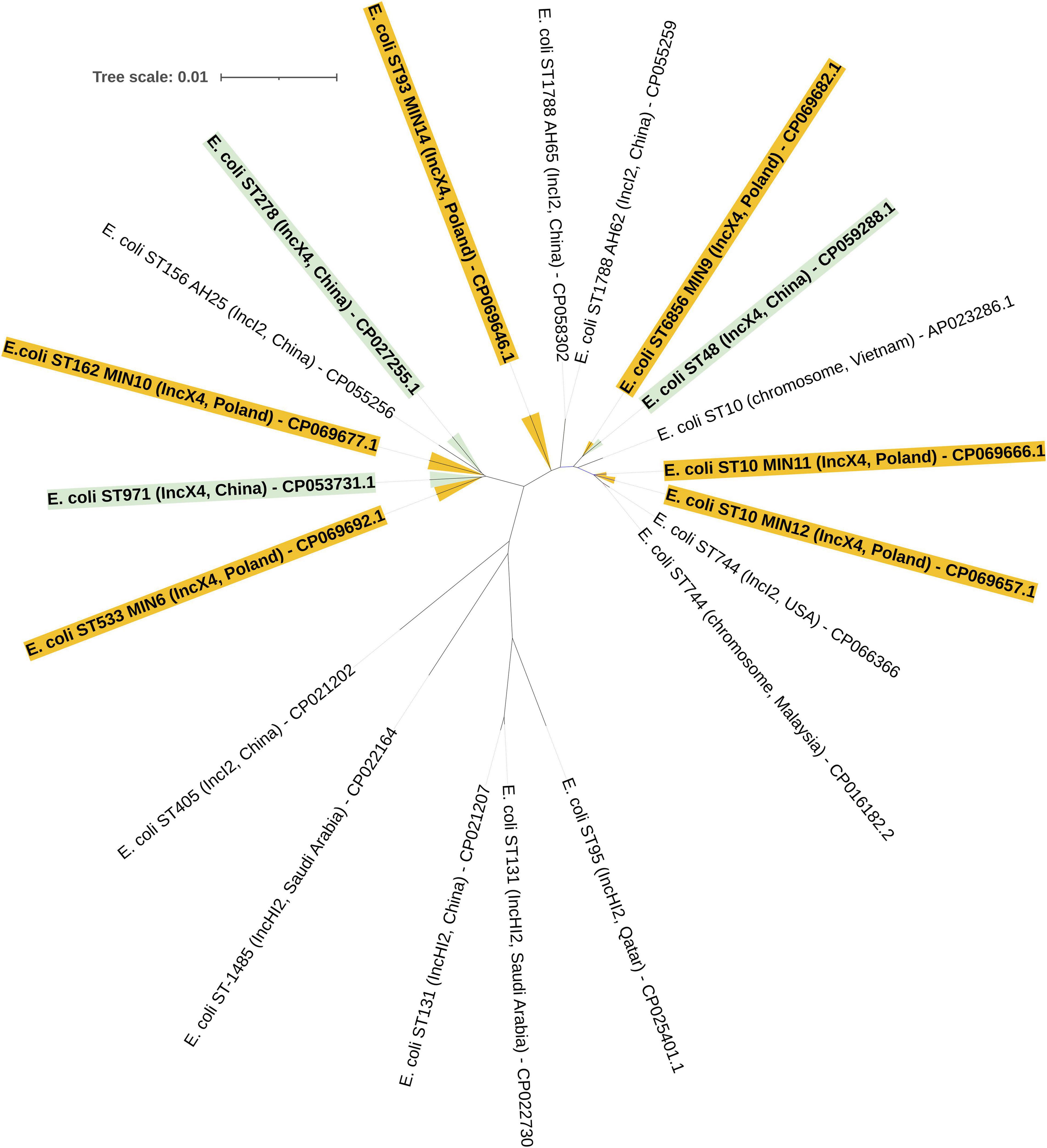
Figure 4. Whole-genome sequence-based GBDP tree of mcr-producing E. coli complete genomes available at NCBI. Tree inferred with FastME 2.1.6.1 [6 (Lefort et al., 2015)] from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5.
Beta-Lactam Resistance
In the present study, all tested strains presented isolated resistance to amoxicillin-clavulanate. Three strains (MIN6, MIN12, MIN14) were equipped with multiple blaTEM–1 genes carried by various plasmids. E. coli MIN6 and MIN12 strain harbored duplicate blaTEM–1B genes, within two distinct mobile vectors, namely pMUB-MIN6-1 (89 956 bp), and MUB-MIN6-2 (56 897 bp) in E. coli MIN6, and pMUB-MIN12-1 (163 427 bp), and pMUB-MIN12-2 (95 526 bp) in E. coli MIN12. E. coli MIN14 possessed two TEM variants, namely blaTEM–1D and blaTEM–1B harbored by pMUB-MIN-14-2 (67 974 bp), and pMUB-MIN-14-3 (45 893 bp), respectively. Moreover, E. coli MIN9 possessed a single blaTEM–1a gene within the pMUB-MIN9-1 plasmid (283 245 bp). Interestingly, in case of E. coli MIN11, blaTEM–1B gene was present within the bacterial chromosome structure.
Fluoroquinolone Resistance
All tested E. coli strains, except MIN14, were ciprofloxacin-resistant due to multiple mutations of gyrA (p.S83L – 5/5 strains; p.D87N – 5/5 strains), parC (p.S80I – 3/5 strains; p.E84G – 1/5 strain; p.S80R – 1/5 strain), and parE (p.E460D – 1/5 strain; p.S458A – 1/5 strain) genes, coupled with additional acquired fluoroquinolone resistance gene, qnrB19, in case of MIN9 strain (pMUB-MIN9-7). The only ciprofloxacin-susceptible strain MIN14, possessed a plasmid-borne qnrS1 gene, which could be associated with low ciprofloxacin MICs.
Aminoglycoside Resistance
All tested E. coli strains were amikacin-susceptible, while E. coli MIN9 was the only tested strain that presented phenotypic resistance against gentamicin (MIC ≥ 16 mg/L), due to presence of aac(3″)-IIa (gene conferring resistance to gentamicin, apramycin, tobramycin, dibekacin, netilmicin, sisomicin) within pMUB-MIN9-1 (InCHI2A). Furthermore, all tested strains except MIN14, produced various aminoglycoside-resistance factors conferring resistance to spectinomycin, streptomycin [aadA1; aadA2b; aph(6)-Id; aph(3″)-Ib]; neomycin, kanamycin, lividomycin, paromomycin, ribostamycin (aph(3′)-Ia).
Folate Pathway Antagonist Resistance
All tested E. coli strains, except MIN14, presented trimethoprim/sulfamethoxazole-resistance, in accordance with WGS data screening for antimicrobial resistance determinants. E. coli MIN11 possesses the chromosomal trimethoprim-resistance gene, dfrA1, coupled with plasmidic sulfamethoxazole-resistance gene sul3. Moreover, remaining strains harbor plasmidic resistance genes, such as, dfrA1, dfrA14, dfrA15, sul1, sul2, and sul3.
Phenicol Resistance
Escherichia coli MIN9, MIN11, and MIN12 were also equipped with acquired genes conferring resistance to phenicols, namely chloramphenicol acetyltransferase gene catA1 (pMUB-MIN9-1), and MFS transporters cmlA1 (pMUB-MIN9-1; pMUB-MIN11-1; pMUB-MIN12-1) and floR (pMUB-MIN12-2).
Extraintestinal E. coli strains also possess a number of virulence factors, such as long polar fimbriae, heat-stable toxin EAST-1 or enterobactin siderophore. All genes encoding virulence factors are listed in Table 5.
Discussion
In this paper we sought to investigate the mechanism of colistin-resistance in six extraintestinal E. coli strains isolated from patients hospitalized in Medical University Hospital, Bialystok, Poland. Full structures of bacterial chromosomes and plasmids were recovered with use of both short- and long-read sequencing technologies and Unicycler hybrid assembly. Results of antimicrobial resistance testing were in accordance with genomic and mobilome screening for antimicrobial resistance determinants. All tested extraintestinal E. coli strains harbored an IncX4 33.3 kbp plasmid pMUB-MCR, with the mobile pEtN transferase-encoding gene, mcr-1.1. Moreover, its influence on colistin-resistance phenotype was confirmed by transconjugation assays.
In the present study, we report the first detailed description of mcr-containing IncX4 plasmid harbored by clinical E. coli strains in Poland. In six extraintestinal E. coli strains, mcr-1.1 was found within a type IV secretion system (T4SS) contained within a 33.3 kbp IncX4 plasmid that is known to be involved in the disseminating of multiple mcr variants. It is widely accepted that some type IV secretion systems (T4SSs) in pathogenic Gram-negative bacteria are utilized in order to translocate virulence factors into the host cell, mediate downregulation of the hosts innate immune response genes and an increase bacterial uptake and survival within macrophages and epithelial cells (Gokulan et al., 2013). Moreover, T4SSs could be also responsible for horizontal gene transfer (Juhas et al., 2008), thus contributing to genome plasticity and the evolution of pathogens through dissemination of antibiotic resistance and virulence genes (Juhas et al., 2008). Conjugative T4SSs are often encoded on self-transmissible plasmids coupled with genes that provide selective advantages for the cell such as antibiotic resistance, virulence factors or other metabolic functions that enhance survival (Wallden et al., 2010). The 33 kbp IncX4 plasmid was proven to be highly transmissible, showing 102–105-fold higher transfer frequencies relative to epidemic IncFII plasmid (Lo et al., 2014; Xavier et al., 2016). Moreover, Lo and colleagues proved that 33 kbp IncX4 plasmid carriage is associated with relatively low fitness cost, which makes it a highly effective vehicle for drug resistance determinants (Wu et al., 2018). Interestingly, it has been also recently reported that IncX4 plasmid can be relatively easily and stably maintained in host bacteria (Beyrouthy et al., 2017). In fact, IncX4 plasmids have been recently shown to harbor multiple mcr variants, CTX-M extended spectrum β-lactamase, as well as the 33.3 kbp IncX4 vehicles without any drug-resistance determinants (Lo et al., 2014; Chen et al., 2019). Similar IncX4 mcr-containing plasmids are reported as increasingly disseminated mainly among E. coli isolates (Table 6), suggesting that it is becoming an “epidemic” plasmid, responsible for (i) disseminating colistin-resistance determinants between different E. coli clones, and (ii) circulating between environmental, industrial, and clinical settings. The phylogenomics of IncX4 plasmids bearing mobile pEtN transferase-encoding genes is presented in Figure 3.
The first European environmental mcr-producing E. coli strain was obtained from Italian diarrhoeic veal calves in 2005 (Haenni et al., 2016), whereas the first European strain of clinical origin harboring plasmid-mediated colistin-resistance gene, was described in Denmark in a Salmonella Typhimurium ST34 strain with an mcr-3 variant (Litrup et al., 2017).
Here, we describe mobile pEtN transferase, mcr-1.1, encoded within a T4SS-containing 33.3 kbp IncX4 plasmid, pMUB-MCR. Similar IncX4 plasmids harboring different mcr variants were recently reported all over the world, with particular reference to animal breeding farms and environmental settings. Global dissemination of similar mcr-harboring IncX4 plasmids in Enterobacterales is presented in Table 6. So far, IncX4 mcr-harboring strains have been reported mainly in animal breeding farms, meat industry, and natural environments. Incidence of IncX4 mcr-harboring strains originating from clinical sources has been recently reported, in Switzerland (E. coli ST-5, ST-48), Portugal (K. pneumoniae ST-45, ST-1112, Salmonella spp.), Italy (E. coli ST-354; K. pneumoniae ST-512), France (E. coli ST-1288), Germany (E. coli ST-155; E. coli ST-69), Finland (E. coli ST-93), China (i. a. E. coli ST-2448; E. coli ST-167; E. coli ST-10), Brazil (E. coli ST-101; K. pneumoniae ST-437), United States of America (E. coli O157:H48), Thailand (K. pneumoniae ST-16; K. pneumoniae ST-45), Japan (K. pneumoniae ST-1296; E. coli ST-782), and United Kingdom (S. Typhimurium ST-34). Moreover, according to the current state of knowledge, E. coli is the major IncX4 clinical producer present in natural environments, animal breeding farms, as well as, in hospital settings. Recent research study performed by Zaja̧c et al. (2019) highlighted the importance of poultry farming, with particular emphasis on turkey, providing important reservoirs of mcr-1.1-carrying E. coli strains in Poland. The authors showed a wide diversity of IncX4 harboring strains, including 32 distinct sequence types (Table 6; Zaja̧c et al., 2019). Furthermore, the first clinical occurrence of a mcr-producing pathogen in Poland was reported by Izdebski et al. (2016). Clinical E. coli ST-617 strain, a member of ST-10 complex, possesses ∼250 kbp plasmid carrying mcr-1.1 and blaCMY–2-containing IncA/C2 plasmids (∼160 kbp).
In this study, we described the following strains – two ST-10, ST-93, ST-162, ST-553, and ST-6856. Interestingly, ST-553 and ST-162 strains have been recently described as mcr-producers in turkeys from animal breeding farms in Poland and Germany, while ST-93 have been already reported in clinical settings in Finland. Moreover, colistin-resistant IncX4-producing E. coli ST-10 seems to be widely distributed globally, and were already reported in Belgium (swine), Italy (river), Germany (barn dog feces), Poland (turkeys), Spain (swine), Czech Republic (raw turkey products), Brazil (wild birds; natural environment), Thailand (Chrysoma spp. flies), China (hospital setting; public transport), Uruguay (clinical source), and Japan (retail meat; municipal wastewater). This is in accordance with a recent report published by Matamoros et al. (2017), suggesting that E. coli ST-10 lineage, a sequence type known for its ubiquity in human fecal samples and in food samples, may function as an important reservoir of the mcr-1.1 gene (Matamoros et al., 2017).
The enormous genome plasticity of Gram-negative bacteria enables the accumulation of many different mechanisms of resistance to various antimicrobial agents. As a result, the increased emergence of MDR or XDR pathogens considerably reduces the opportunities for effective treatments against these bacteria (Livermore et al., 2011; Ojdana et al., 2015). A number of recent reports highlights the importance of mcr dissemination in clinical MDR bacteria, especially among subpopulations of pathogens persisting in hospital environments. Co-occurrence of mcr and ESBLs (CTX-M-15), different carbapenemases (KPC-type, OXA-181), and other antimicrobial resistance determinants may possibly lead to formation of pandrug-resistant bacterial lineages (Brauer et al., 2016; Di Pilato et al., 2016; Haenni et al., 2016; Caltagirone et al., 2017; Pulss et al., 2017; Tacão et al., 2017; Dalmolin et al., 2018; Mendes et al., 2018; Manageiro et al., 2019). In this study mcr coexisted with determinants of resistance to aminoglycosides [aph(3″)-Ib; aph(3″)-Ia; aph(6)-Id; aadA1; aac(3″)-IIa; aadA2b], chloramphenicol (catA1, cmlA1), β-lactams (blaTEM–1A; blaTEM–1B; blaTEM–1D), quinolones (qnrB19, qnrS1), sulfonamides (sul1, sul2, sul3), and trimethoprim (dfrA1, dfrA14, dfrA15). Interestingly, in case of E. coli MIN11, blaTEM–1B gene was present within the bacterial chromosome structure. Di Conza et al. (2014) proved that amoxicillin-clavulanate resistance with retained second-and third-generation cephalosporins susceptibility may be linked with blaTEM–1 overproduction (Di Conza et al., 2014). Furthermore, Salverda et al. (2010) have recently proved that several amino acid substitutions were also identified as factors involved in increased resistance to β-lactam-clavulanate. Interestingly, in the case of tested extraintestinal E. coli subpopulation, the only ciprofloxacin-susceptible strain MIN14, possessed a plasmid-borne qnrS1 gene, which could be associated with low ciprofloxacin MICs. Allou et al. (2009) showed that qnrS1-possesing E. coli transconjugants showed low-level resistance to fluoroquinolones, with ciprofloxacin MIC ranging from 0.25 to 0.5 mg/L (Allou et al., 2009). In our study, qnrS1-producing E. coli MIN14 strain was classified as ciprofloxacin susceptible, with MIC ≤ 0.25 mg/L.
In conclusion, the increasing prevalence of plasmids responsible for colistin-resistance, often carrying other determinants of drug resistance, may possibly lead to formation of pandrug-resistant bacterial lineages. Great effort needs to be taken to avoid further dissemination of plasmid-mediated colistin resistance among clinically relevant Gram-negative pathogens.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository under BioProject number PRJNA700422 and accession numbers SAMN17831481 (E. coli MIN6—CP069692.1 for chromosome and CP069693.1–CP069700.1 for plasmids); SAMN17831482 (E. coli MIN9—CP069682.1 for chromosome and CP069683.1–CP069691.1 for plasmids); SAMN17831483 (E. coli MIN10—CP069677.1 for chromosome and CP069678.1–CP069681.1 for plasmids); SAMN17831484 (E. coli MIN11—CP069666.1 for chromosome and CP069667.1–CP069676.1 for plasmids); SAMN17831485 (E. coli MIN12—CP069657.1 for chromosome and CP069658.1–CP069665.1 for plasmids); and SAMN17831486 (E. coli MIN14—CP069646.1 for chromosome and CP069647.1–CP069656.1 for plasmids).
Ethics Statement
This molecular investigation uses strains obtained from collection of strains deposited in Department of Microbiological Diagnostics and Infectious Immunology, Medical University of Bialystok, Poland. The Bioethics Commission of the Medical University in Bialystok did not require the study to be reviewed or approved by an ethics committee because apart from the strains from the Department’s collection, no data enabling patient identification was used in the study.
Author Contributions
PiM, DS, JN, and ET substantially contributed to the conception of the submitted research manuscript, designing and validation of the experiments, and data acquisition and interpretation (antimicrobial susceptibility testing, short-read sequencing, long-read sequencing, and preparation of the figures and tables). PaM, AG, AS, and DS wrote the main manuscript. PiM was responsible for library preparation, WGS, and bioinformatics. PaM, DG, DI, PS, AZ, PR, PW, THa, IS, KM, RC, and JD contributed to the validation of the designed experiments and data acquisition and interpretation. THr, BK, JK, JG, SC, and PR were responsible for the medical care of the patient. All authors reviewed the manuscript.
Funding
This work was financed by the MINIATURA research project (2017/01/X/NZ6/01852 - National Science Center, Poland), and supported by the Medical University of Bialystok statutory subsidy (SUB/1/DN/19/005/2222).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Steven J. Snodgrass for editorial assistance.
Footnotes
References
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Allou, N., Cambau, E., Massias, L., Chau, F., and Fantin, B. (2009). Impact of low-level resistance to fluoroquinolones due to qnrA1 and qnrS1 genes or a gyrA mutation on ciprofloxacin bactericidal activity in a murine model of Escherichia coli urinary tract infection. Antimicrob. Agents Chemother. 53, 4292–4297. doi: 10.1128/AAC.01664-08
Bai, F., Li, X., Niu, B., Zhang, Z., Malakar, P. K., Liu, H., et al. (2018). A mcr-1-carrying conjugative IncX4 plasmid in colistin-resistant Escherichia coli ST278 strain isolated from dairy cow feces in Shanghai, China. Front. Microbiol. 9:2833. doi: 10.3389/fmicb.2018.02833
Baron, S., Hadjadj, L., Rolain, J.-M., and Olaitan, A. O. (2016). Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents 48, 583–591. doi: 10.1016/j.ijantimicag.2016.06.023
Beyrouthy, R., Robin, F., Lessene, A., Lacombat, I., Dortet, L., Naas, T., et al. (2017). MCR-1 and OXA-48 In Vivo acquisition in KPC-producing Escherichia coli after colistin treatment. Antimicrob. Agents Chemother. 61:e02540-16. doi: 10.1128/AAC.02540-16
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Brauer, A., Telling, K., Laht, M., Kalmus, P., Lutsar, I., Remm, M., et al. (2016). Plasmid with colistin resistance gene mcr-1 in extended-spectrum-β-lactamase-producing Escherichia coli strains isolated from pig slurry in Estonia. Antimicrob. Agents Chemother. 60, 6933–6936. doi: 10.1128/AAC.00443-16
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Caltagirone, M., Nucleo, E., Spalla, M., Zara, F., Novazzi, F., Marchetti, V. M., et al. (2017). Occurrence of extended spectrum β-lactamases, KPC-Type, and MCR-1.2-producing Enterobacteriaceae from wells, River water, and wastewater treatment plants in Oltrepò Pavese Area, Northern Italy. Front. Microbiol. 8:2232. doi: 10.3389/fmicb.2017.02232
Campos, J., Cristino, L., Peixe, L., and Antunes, P. (2016). MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. 21:30270. doi: 10.2807/1560-7917.ES.2016.21.26.30270
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carfora, V., Alba, P., Leekitcharoenphon, P., Ballarò, D., Cordaro, G., Di Matteo, P., et al. (2018). Colistin resistance mediated by mcr-1 in ESBL-producing, multidrug Resistant Salmonella infantis in broiler chicken industry, Italy (2016-2017). Front. Microbiol. 9:1880. doi: 10.3389/fmicb.2018.01880
Chan, W.-S., Au, C.-H., Ho, D. N., Chan, T.-L., Ma, E. S.-K., and Tang, B. S.-F. (2018). Prospective study on human fecal carriage of Enterobacteriaceae possessing mcr-1 and mcr-2 genes in a regional hospital in Hong Kong. BMC Infect. Dis. 18:81. doi: 10.1186/s12879-018-2987-y
Chen, F., Zhang, W., Schwarz, S., Zhu, Y., Li, R., Hua, X., et al. (2019). Genetic characterization of an MDR/virulence genomic element carrying two T6SS gene clusters in a clinical Klebsiella pneumoniae isolate of swine origin. J. Antimicrob. Chemother. 74, 1539–1544. doi: 10.1093/jac/dkz093
Cui, M., Zhang, J., Gu, Z., Li, R., Chan, E. W.-C., Yan, M., et al. (2017). Prevalence and molecular characterization of mcr-1-positive Salmonella strains recovered from clinical specimens in China. Antimicrob. Agents Chemother. 61:e02471-16. doi: 10.1128/AAC.02471-16
Dalmolin, T. V., Martins, A. F., Zavascki, A. P., de Lima-Morales, D., and Barth, A. L. (2018). Acquisition of the mcr-1 gene by a high-risk clone of KPC-2-producing Klebsiella pneumoniae ST437/CC258, Brazil. Diagn. Microbiol. Infect. Dis. 90, 132–133. doi: 10.1016/j.diagmicrobio.2017.09.016
Di Conza, J. A., Badaracco, A., Ayala, J., Rodríguez, C., Famiglietti, A., and Gutkind, G. O. (2014). β-lactamases produced by amoxicillin-clavulanate-resistant enterobacteria isolated in Buenos Aires, Argentina: a new blaTEM gene. Rev. Argent. Microbiol. 46, 210–217. doi: 10.1016/S0325-7541(14)70075-6
Di Pilato, V., Arena, F., Tascini, C., Cannatelli, A., Henrici De Angelis, L., Fortunato, S., et al. (2016). mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-Resistant KPC Carbapenemase-producing Klebsiella pneumoniae strain of sequence Type 512. Antimicrob. Agents Chemother. 60, 5612–5615. doi: 10.1128/AAC.01075-16
Donà, V., Bernasconi, O. J., Pires, J., Collaud, A., Overesch, G., Ramette, A., et al. (2017). Heterogeneous genetic location of mcr-1 in colistin-resistant Escherichia coli isolates from humans and retail chicken meat in Switzerland: emergence of mcr-1-carrying IncK2 plasmids. Antimicrob. Agents Chemother. 61:e01245-17. doi: 10.1128/AAC.01245-17
Doumith, M., Godbole, G., Ashton, P., Larkin, L., Dallman, T., Day, M., et al. (2016). Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 71, 2300–2305. doi: 10.1093/jac/dkw093
Du, C., Feng, Y., Wang, G., Zhang, Z., Hu, H., Yu, Y., et al. (2020). Co-occurrence of the mcr-1.1 and mcr-3.7 Genes in a multidrug-resistant Escherichia coli Isolate from China. Infect. Drug Resist. 13, 3649–3655. doi: 10.2147/IDR.S268787
El-Sayed Ahmed, M. A. E.-G., Zhong, L.-L., Shen, C., Yang, Y., Doi, Y., and Tian, G.-B. (2020). Colistin and its role in the Era of antibiotic resistance: an extended review (2000-2019). Emerg. Microbes Infect. 9, 868–885. doi: 10.1080/22221751.2020.1754133
Feng, S., Shen, C., Chen, H., Zheng, X., Xia, Y., Zhong, L.-L., et al. (2018). Co-production of MCR-1 and NDM-5 in Escherichia coli isolated from a colonization case of inpatient. Infect. Drug Resist. 11, 1157–1161. doi: 10.2147/IDR.S171164
Fernandes, M. R., McCulloch, J. A., Vianello, M. A., Moura, Q., Pérez-Chaparro, P. J., Esposito, F., et al. (2016). First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence Type 101 isolate from a human infection in Brazil. Antimicrob. Agents Chemother. 60, 6415–6417. doi: 10.1128/AAC.01325-16
Fernandes, M. R., Sellera, F. P., Esposito, F., Sabino, C. P., Cerdeira, L., and Lincopan, N. (2017). Colistin-Resistant mcr-1-Positive Escherichia coli on public beaches, an infectious threat emerging in recreational waters. Antimicrob. Agents Chemother. 61:e00234-17. doi: 10.1128/AAC.00234-17
Garcia-Graells, C., De Keersmaecker, S. C. J., Vanneste, K., Pochet, B., Vermeersch, K., Roosens, N., et al. (2018). Detection of plasmid-mediated colistin resistance, mcr-1 and mcr-2 genes, in Salmonella spp. Isolated from food at retail in Belgium from 2012 to 2015. Foodborne Pathog. Dis. 15, 114–117. doi: 10.1089/fpd.2017.2329
García-Meniño, I., Díaz-Jiménez, D., García, V., de Toro, M., Flament-Simon, S. C., Blanco, J., et al. (2019). Genomic characterization of prevalent mcr-1, mcr-4, and mcr-5 Escherichia coli within swine enteric Colibacillosis in Spain. Front. Microbiol. 10:2469. doi: 10.3389/fmicb.2019.02469
Gelbíčová, T., Baráková, A., Florianová, M., Jamborová, I., Zelendová, M., Pospíšilová, L., et al. (2019). Dissemination and comparison of genetic determinants of mcr-mediated Colistin resistance in Enterobacteriaceae via retailed raw meat products. Front. Microbiol. 10:2824. doi: 10.3389/fmicb.2019.02824
Gokulan, K., Khare, S., Rooney, A. W., Han, J., Lynne, A. M., and Foley, S. L. (2013). Impact of plasmids, including those encodingVirB4/D4 type IV secretion systems, on Salmonella enterica serovar Heidelberg virulence in macrophages and epithelial cells. PLoS One 8:e77866. doi: 10.1371/journal.pone.0077866
Gröndahl-Yli-Hannuksela, K., Lönnqvist, E., Kallonen, T., Lindholm, L., Jalava, J., Rantakokko-Jalava, K., et al. (2018). The first human report of mobile colistin resistance gene, mcr-1, in Finland. APMIS 126, 413–417. doi: 10.1111/apm.12834
Guenther, S., Falgenhauer, L., Semmler, T., Imirzalioglu, C., Chakraborty, T., Roesler, U., et al. (2017). Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 72, 1289–1292. doi: 10.1093/jac/dkw585
Haenni, M., Poirel, L., Kieffer, N., Châtre, P., Saras, E., Métayer, V., et al. (2016). Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect. Dis. 16, 281–282. doi: 10.1016/S1473-3099(16)00007-4
Hasman, H., Hammerum, A. M., Hansen, F., Hendriksen, R. S., Olesen, B., Agersø, Y., et al. (2015). Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 20:30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085
Hassan, J., Eddine, R. Z., Mann, D., Li, S., Deng, X., Saoud, I. P., et al. (2020). The mobile colistin resistance gene, mcr-1.1, is carried on IncX4 plasmids in multidrug resistant E. coli isolated from rainbow trout aquaculture. Microorganisms 8:1636. doi: 10.3390/microorganisms8111636
Hayashi, W., Tanaka, H., Taniguchi, Y., Iimura, M., Soga, E., Kubo, R., et al. (2019). Acquisition of mcr-1 and cocarriage of virulence genes in avian pathogenic Escherichia coli isolates from municipal wastewater influents in Japan. Appl. Environ. Microbiol. 85:e001661-19. doi: 10.1128/AEM.01661-19
Izdebski, R., Baraniak, A., Bojarska, K., Urbanowicz, P., Fiett, J., Pomorska-Wesołowska, M., et al. (2016). Mobile MCR-1-associated resistance to colistin in Poland. J. Antimicrob. Chemother. 71, 2331–2333. doi: 10.1093/jac/dkw261
Juhas, M., Crook, D. W., and Hood, D. W. (2008). Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10, 2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x
Kassem, I. I., Mann, D., Li, S., and Deng, X. (2021). Draft genome sequences and resistome analysis of multidrug-resistant mcr-1-harbouring Escherichia coli isolated from pre-harvest poultry in Lebanon. J. Glob. Antimicrob. Resist. 25, 114–116. doi: 10.1016/j.jgar.2021.03.001
Kaye, K. S., Pogue, J. M., Tran, T. B., Nation, R. L., and Li, J. (2016). Agents of last resort: polymyxin resistance. Infect. Dis. Clin. North Am. 30, 391–414. doi: 10.1016/j.idc.2016.02.005
Kim, S., Kim, H., Kang, H.-S., Kim, Y., Kim, M., Kwak, H., et al. (2020). Prevalence and genetic characterization of mcr-1-positive Escherichia coli isolated from retail meats in South Korea. J. Microbiol. Biotechnol. 30, 1862–1869. doi: 10.4014/jmb.2007.07008
Kong, L.-H., Lei, C.-W., Ma, S.-Z., Jiang, W., Liu, B.-H., Wang, Y.-X., et al. (2017). Various sequence types of Escherichia coli isolates coharboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob. Agents Chemother. 61:e02167-16. doi: 10.1128/AAC.02167-16
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Lei, T., Zhang, J., Jiang, F., He, M., Zeng, H., Chen, M., et al. (2019). First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int. J. Food Microbiol. 308:108290. doi: 10.1016/j.ijfoodmicro.2019.108290
Letunic, I., and Bork, P. (2021). Interactive tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, A., Yang, Y., Miao, M., Chavda, K. D., Mediavilla, J. R., Xie, X., et al. (2016). Complete sequences of mcr-1-harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 60, 4351–4354. doi: 10.1128/AAC.00550-16
Lindsey, R. L., Batra, D., Rowe, L., Loparev, V. N., Stripling, D., Garcia-Toledo, L., et al. (2017). High-quality genome sequence of an Escherichia coli O157 strain carrying an mcr-1 resistance gene isolated from a patient in the United States. Genome Announc. 5:e01725-16. doi: 10.1128/genomeA.01725-16
Litrup, E., Kiil, K., Hammerum, A. M., Roer, L., Nielsen, E. M., and Torpdahl, M. (2017). Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009-17. Euro Surveill. 22:30587. doi: 10.2807/1560-7917.ES.2017.22.31.30587
Liu, B.-T., and Song, F.-J. (2019). Emergence of two Escherichia coli strains co-harboring mcr-1 and blaNDM in fresh vegetables from China. Infect. Drug Resist. 12, 2627–2635. doi: 10.2147/IDR.S211746
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Livermore, D. M., Warner, M., Mushtaq, S., Doumith, M., Zhang, J., and Woodford, N. (2011). What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int. J. Antimicrob. Agents 37, 415–419. doi: 10.1016/j.ijantimicag.2011.01.012
Lo, W.-U., Chow, K.-H., Law, P. Y., Ng, K.-Y., Cheung, Y.-Y., Lai, E. L., et al. (2014). Highly conjugative IncX4 plasmids carrying blaCTX-M in Escherichia coli from humans and food animals. J. Med. Microbiol. 63, 835–840. doi: 10.1099/jmm.0.074021-0
Lu, J., Dong, N., Liu, C., Zeng, Y., Sun, Q., Zhou, H., et al. (2020). Prevalence and molecular epidemiology of mcr-1-positive Klebsiella pneumoniae in healthy adults from China. J. Antimicrob. Chemother. 75, 2485–2494. doi: 10.1093/jac/dkaa210
Luo, J., Yao, X., Lv, L., Doi, Y., Huang, X., Huang, S., et al. (2017). Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob. Agents Chemother. 61:e01139-17. doi: 10.1128/AAC.01139-17
Luo, Q., Yu, W., Zhou, K., Guo, L., Shen, P., Lu, H., et al. (2017). Molecular epidemiology and colistin resistant mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front. Microbiol. 8:2262. doi: 10.3389/fmicb.2017.02262
Manageiro, V., Clemente, L., Romão, R., Silva, C., Vieira, L., Ferreira, E., et al. (2019). IncX4 plasmid carrying the new mcr-1.9 gene variant in a CTX-M-8-producing Escherichia coli isolate recovered from swine. Front. Microbiol. 10:367. doi: 10.3389/fmicb.2019.00367
Matamoros, S., van Hattem, J. M., Arcilla, M. S., Willemse, N., Melles, D. C., Penders, J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. doi: 10.1038/s41598-017-15539-7
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Mendes, A. C., Novais, Â, Campos, J., Rodrigues, C., Santos, C., Antunes, P., et al. (2018). mcr-1 in carbapenemase-producing Klebsiella pneumoniae with hospitalized patients, Portugal, 2016-2017. Emerging Infect. Dis. 24, 762–766. doi: 10.3201/eid2404.171787
Migura-Garcia, L., González-López, J. J., Martinez-Urtaza, J., Aguirre Sánchez, J. R., Moreno-Mingorance, A., Perez de Rozas, A., et al. (2019). mcr-Colistin resistance genes mobilized by IncX4, IncHI2, and IncI2 plasmids in Escherichia coli of pigs and white stork in Spain. Front. Microbiol. 10:3072. doi: 10.3389/fmicb.2019.03072
Monte, D. F., Fernandes, M. R., Cerdeira, L., de Souza, T. A., Mem, A., Franco, B. D. G. M., et al. (2017a). Draft genome sequences of colistin-resistant MCR-1-producing Escherichia coli ST1850 and ST74 strains isolated from commercial chicken meat. Genome Announc. 5:e00329-17. doi: 10.1128/genomeA.00329-17
Monte, D. F., Mem, A., Fernandes, M. R., Cerdeira, L., Esposito, F., Galvão, J. A., et al. (2017b). Chicken meat as a reservoir of colistin-Resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 61:e02718-16. doi: 10.1128/AAC.02718-16
Moser, A. I., Kuenzli, E., Campos-Madueno, E. I., Büdel, T., Rattanavong, S., Vongsouvath, M., et al. (2021). Antimicrobial-resistant Escherichia coli strains and their plasmids in people, poultry, and chicken meat in laos. Front. Microbiol. 12:708182. doi: 10.3389/fmicb.2021.708182
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841X.2018.1492902
Neumann, B., Rackwitz, W., Hunfeld, K.-P., Fuchs, S., Werner, G., and Pfeifer, Y. (2020). Genome sequences of two clinical Escherichia coli isolates harboring the novel colistin-resistance gene variants mcr-1.26 and mcr-1.27. Gut Pathog. 12:40. doi: 10.1186/s13099-020-00375-4
Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656. doi: 10.1128/mmbr.67.4.593-656.2003
Nishino, Y., Shimojima, Y., Suzuki, Y., Ida, M., Fukui, R., Kuroda, S., et al. (2017). Detection of the mcr-1 gene in colistin-resistant Escherichia coli from retail meat in Japan. Microbiol. Immunol. 61, 554–557. doi: 10.1111/1348-0421.12549
Ojdana, D., Sacha, P., Olszańska, D., Majewski, P., Wieczorek, P., Jaworowska, J., et al. (2015). First report of Klebsiella pneumoniae-carbapenemase-3-producing Escherichia coli ST479 in Poland. Biomed Res. Int. 2015:256028. doi: 10.1155/2015/256028
Olaitan, A. O., Morand, S., and Rolain, J.-M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Osei Sekyere, J. (2019). Mcr colistin resistance gene: a systematic review of current diagnostics and detection methods. Microbiologyopen 8:e00682. doi: 10.1002/mbo3.682
Palmeira, J. D., Ferreira, H., Madec, J.-Y., and Haenni, M. (2018). Draft genome of a ST443 mcr-1- and blaCTX-M-2-carrying Escherichia coli from cattle in Brazil. J. Glob. Antimicrob. Resist. 13, 269–270. doi: 10.1016/j.jgar.2018.05.010
Papa-Ezdra, R., Grill Diaz, F., Vieytes, M., García-Fulgueiras, V., Caiata, L., Ávila, P., et al. (2020). First three Escherichia coli isolates harbouring mcr-1 in Uruguay. J. Glob. Antimicrob. Resist. 20, 187–190. doi: 10.1016/j.jgar.2019.07.016
Partridge, S. R., Di Pilato, V., Doi, Y., Feldgarden, M., Haft, D. H., Klimke, W., et al. (2018). Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother. 73, 2625–2630. doi: 10.1093/jac/dky262
Peng, Z., Li, X., Hu, Z., Li, Z., Lv, Y., Lei, M., et al. (2019). Characteristics of carbapenem-resistant and colistin-resistant Escherichia coli Co-producing NDM-1 and MCR-1 from pig farms in China. Microorganisms 7:482. doi: 10.3390/microorganisms7110482
Perdigão Neto, L. V., Corscadden, L., Martins, R. C. R., Nagano, D. S., Cunha, M. P. V., Neves, P. R., et al. (2019). Simultaneous colonization by Escherichia coli and Klebsiella pneumoniae harboring mcr-1 in Brazil. Infection 47, 661–664. doi: 10.1007/s15010-019-01309-2
Pham Thanh, D., Thanh Tuyen, H., Nguyen Thi Nguyen, T., Chung The, H., Wick, R. R., Thwaites, G. E., et al. (2016). Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J. Antimicrob. Chemother. 71, 2314–2317. doi: 10.1093/jac/dkw173
Pulss, S., Semmler, T., Prenger-Berninghoff, E., Bauerfeind, R., and Ewers, C. (2017). First report of an Escherichia coli strain from swine carrying an OXA-181 carbapenemase and the colistin resistance determinant MCR-1. Int. J. Antimicrob. Agents 50, 232–236. doi: 10.1016/j.ijantimicag.2017.03.014
Quan, J., Li, X., Chen, Y., Jiang, Y., Zhou, Z., Zhang, H., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17, 400–410. doi: 10.1016/S1473-3099(16)30528-X
Rau, R. B., de Lima-Morales, D., Wink, P. L., Ribeiro, A. R., and Barth, A. L. (2020). Salmonella enterica mcr-1 positive from food in Brazil: detection and characterization. Foodborne Pathog. Dis. 17, 202–208. doi: 10.1089/fpd.2019.2700
Roer, L., Hansen, F., Stegger, M., Sönksen, U. W., Hasman, H., and Hammerum, A. M. (2017). Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 22:30584.
Sadek, M., Ortiz de la Rosa, J. M., Abdelfattah Maky, M., Korashe Dandrawy, M., Nordmann, P., and Poirel, L. (2021). Genomic features of MCR-1 and extended-spectrum β-lactamase-producing Enterobacterales from retail raw chicken in Egypt. Microorganisms 9:195. doi: 10.3390/microorganisms9010195
Salverda, M. L. M., De Visser, J. A. G. M., and Barlow, M. (2010). Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol. Rev. 34, 1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x
Sellera, F. P., Fernandes, M. R., Sartori, L., Carvalho, M. P. N., Esposito, F., Nascimento, C. L., et al. (2017). Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J. Antimicrob. Chemother. 72, 1255–1256. doi: 10.1093/jac/dkw543
Shen, C., Feng, S., Chen, H., Dai, M., Paterson, D. L., Zheng, X., et al. (2018). Transmission of mcr-1-producing multidrug-resistant Enterobacteriaceae in Public transportation in Guangzhou, China. Clin. Infect. Dis. 67, S217–S224. doi: 10.1093/cid/ciy661
Shen, Z., Wang, Y., Shen, Y., Shen, J., and Wu, C. (2016). Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 16:293. doi: 10.1016/S1473-3099(16)00061-X
Simoni, S., Morroni, G., Brenciani, A., Vincenzi, C., Cirioni, O., Castelletti, S., et al. (2018). Spread of colistin resistance gene mcr-1 in Italy: characterization of the mcr-1.2 allelic variant in a colistin-resistant blood isolate of Escherichia coli. Diagn. Microbiol. Infect. Dis. 91, 66–68. doi: 10.1016/j.diagmicrobio.2017.12.015
Srijan, A., Margulieux, K. R., Ruekit, S., Snesrud, E., Maybank, R., Serichantalergs, O., et al. (2018). Genomic characterization of nonclonal mcr-1-positive multidrug-resistant Klebsiella pneumoniae from clinical samples in Thailand. Microb. Drug Resist. 24, 403–410. doi: 10.1089/mdr.2017.0400
Tacão, M., Tavares, R. D. S., Teixeira, P., Roxo, I., Ramalheira, E., Ferreira, S., et al. (2017). mcr-1 and blaKPC-3 in Escherichia coli sequence Type 744 after Meropenem and Colistin therapy. Portugal. Emerging Infect. Dis. 23, 1419–1421. doi: 10.3201/eid2308.170162
Tada, T., Uechi, K., Nakasone, I., Nakamatsu, M., Satou, K., Hirano, T., et al. (2018). Emergence of IncX4 plasmids encoding mcr-1 in a clinical isolate of Klebsiella pneumoniae in Japan. Int. J. Infect. Dis. 75, 98–100. doi: 10.1016/j.ijid.2018.08.011
Timmermans, M., Wattiau, P., Denis, O., and Boland, C. (2021). Colistin resistance genes mcr-1 to mcr-5, including a case of triple occurrence (mcr-1, -3 and -5), in Escherichia coli isolates from faeces of healthy pigs, cattle and poultry in Belgium, 2012-2016. Int. J. Antimicrob. Agents 57:106350. doi: 10.1016/j.ijantimicag.2021.106350
Torpdahl, M., Hasman, H., Litrup, E., Skov, R. L., Nielsen, E. M., and Hammerum, A. M. (2017). Detection of mcr-1-encoding plasmid-mediated colistin-resistant Salmonella isolates from human infection in Denmark. Int. J. Antimicrob. Agents 49, 261–262. doi: 10.1016/j.ijantimicag.2016.11.010
Wallden, K., Rivera-Calzada, A., and Waksman, G. (2010). Type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 12, 1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, R., Yi, L.-X., Yu, L.-F., Wang, J., Liu, Y., Chen, X., et al. (2018). Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in Vitro. Front. Microbiol. 9:331. doi: 10.3389/fmicb.2018.00331
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Yang, Q. E., Tansawai, U., Andrey, D. O., Wang, S., Wang, Y., Sands, K., et al. (2019). Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): an increasing public health risk. Environ. Int. 122, 281–290. doi: 10.1016/j.envint.2018.11.021
Zaja̧c, M., Sztromwasser, P., Bortolaia, V., Leekitcharoenphon, P., Cavaco, L. M., Ziȩtek-Barszcz, A., et al. (2019). Corrigendum: occurrence and characterization of mcr-1-positive Escherichia coli isolated from food-producing animals in Poland, 2011-2016. Front. Microbiol. 10:2816. doi: 10.3389/fmicb.2019.02816
Zamparette, C. P., Schorner, M., Campos, E., Moura, Q., Cerdeira, L., Tartari, D. C., et al. (2020). IncX4 plasmid-mediated mcr-1.1 in polymyxin-resistant Escherichia coli from outpatients in Santa Catarina, Southern Brazil. Microb. Drug Resist. 26, 1326–1333. doi: 10.1089/mdr.2019.0203
Zelendova, M., Papagiannitsis, C. C., Valcek, A., Medvecky, M., Bitar, I., Hrabak, J., et al. (2020). Characterization of the complete nucleotide sequences of mcr-1-encoding plasmids from Enterobacterales isolates in retailed raw meat products from the Czech Republic. Front. Microbiol. 11:604067. doi: 10.3389/fmicb.2020.604067
Zeng, K.-J., Doi, Y., Patil, S., Huang, X., and Tian, G.-B. (2016). Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob. Agents Chemother. 60, 3862–3863. doi: 10.1128/AAC.00345-16
Zhao, F., Feng, Y., Lü, X., McNally, A., and Zong, Z. (2017). Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of Two New mcr-1 variants. Front. Microbiol. 8:2094. doi: 10.3389/fmicb.2017.02094
Zhong, L.-L., Phan, H. T. T., Shen, C., Vihta, K.-D., Sheppard, A. E., Huang, X., et al. (2018). High rates of human fecal carriage of mcr-1-positive multidrug-resistant Enterobacteriaceae emerge in China in association with successful plasmid families. Clin. Infect. Dis. 66, 676–685. doi: 10.1093/cid/cix885
Keywords: colistin-resistance, IncX4 plasmid, mcr-1.1, extraintestinal E. coli, plasmid
Citation: Majewski P, Gutowska A, Smith DGE, Hauschild T, Majewska P, Hryszko T, Gizycka D, Kedra B, Kochanowicz J, Glowiński J, Drewnowska J, Swiecicka I, Sacha PT, Wieczorek P, Iwaniuk D, Sulewska A, Charkiewicz R, Makarewicz K, Zebrowska A, Czaban S, Radziwon P, Niklinski J and Tryniszewska EA (2021) Plasmid Mediated mcr-1.1 Colistin-Resistance in Clinical Extraintestinal Escherichia coli Strains Isolated in Poland. Front. Microbiol. 12:547020. doi: 10.3389/fmicb.2021.547020
Received: 30 March 2020; Accepted: 02 November 2021;
Published: 10 December 2021.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Gerald Larrouy-Maumus, Imperial College London, United KingdomJohn Osei Sekyere, University of Pretoria, South Africa
Copyright © 2021 Majewski, Gutowska, Smith, Hauschild, Majewska, Hryszko, Gizycka, Kedra, Kochanowicz, Glowiński, Drewnowska, Swiecicka, Sacha, Wieczorek, Iwaniuk, Sulewska, Charkiewicz, Makarewicz, Zebrowska, Czaban, Radziwon, Niklinski and Tryniszewska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Piotr Majewski, cGlvdHIubWFqZXdza2lAdW1iLmVkdS5wbA==
 Piotr Majewski
Piotr Majewski Anna Gutowska1
Anna Gutowska1 David G. E. Smith
David G. E. Smith Tomasz Hryszko
Tomasz Hryszko Justyna Drewnowska
Justyna Drewnowska Izabela Swiecicka
Izabela Swiecicka Pawel T. Sacha
Pawel T. Sacha Piotr Radziwon
Piotr Radziwon