- Department of Microbiology and Molecular Genetics, Faculty of Medicine, IMRIC, The Hebrew University of Jerusalem, Jerusalem, Israel
Coupled transcription-translation (CTT) is a hallmark of prokaryotic gene expression. CTT occurs when ribosomes associate with and initiate translation of mRNAs whose transcription has not yet concluded, therefore forming “RNAP.mRNA.ribosome” complexes. CTT is a well-documented phenomenon that is involved in important gene regulation processes, such as attenuation and operon polarity. Despite the progress in our understanding of the cellular signals that coordinate CTT, certain aspects of its molecular architecture remain controversial. Additionally, new information on the spatial segregation between the transcriptional and the translational machineries in certain species, and on the capability of certain mRNAs to localize translation-independently, questions the unanimous occurrence of CTT. Furthermore, studies where transcription and translation were artificially uncoupled showed that transcription elongation can proceed in a translation-independent manner. Here, we review studies supporting the occurrence of CTT and findings questioning its extent, as well as discuss mechanisms that may explain both coupling and uncoupling, e.g., chromosome relocation and the involvement of cis- or trans-acting elements, such as small RNAs and RNA-binding proteins. These mechanisms impact RNA localization, stability, and translation. Understanding the two options by which genes can be expressed and their consequences should shed light on a new layer of control of bacterial transcripts fate.
Coupled Transcription-Translation: A Hallmark Feature of Prokaryotic Gene Expression
Due to the scarcity of intracellular membrane-delimited compartmentalization, prokaryotic cells have historically been regarded as spatially unorganized. The lack of a nuclear membrane that physically separates the chromosomal DNA from the cytosolic environment led to the well-accepted notion that transcription and translation are spatiotemporally coupled in bacteria and archaea. Coupled transcription-translation (CTT) occurs when ribosomes bind and start to translate nascent mRNAs, whose transcription has not terminated yet, therefore forming an “RNAP·nascent mRNA·ribosome” complex (Figure 1).
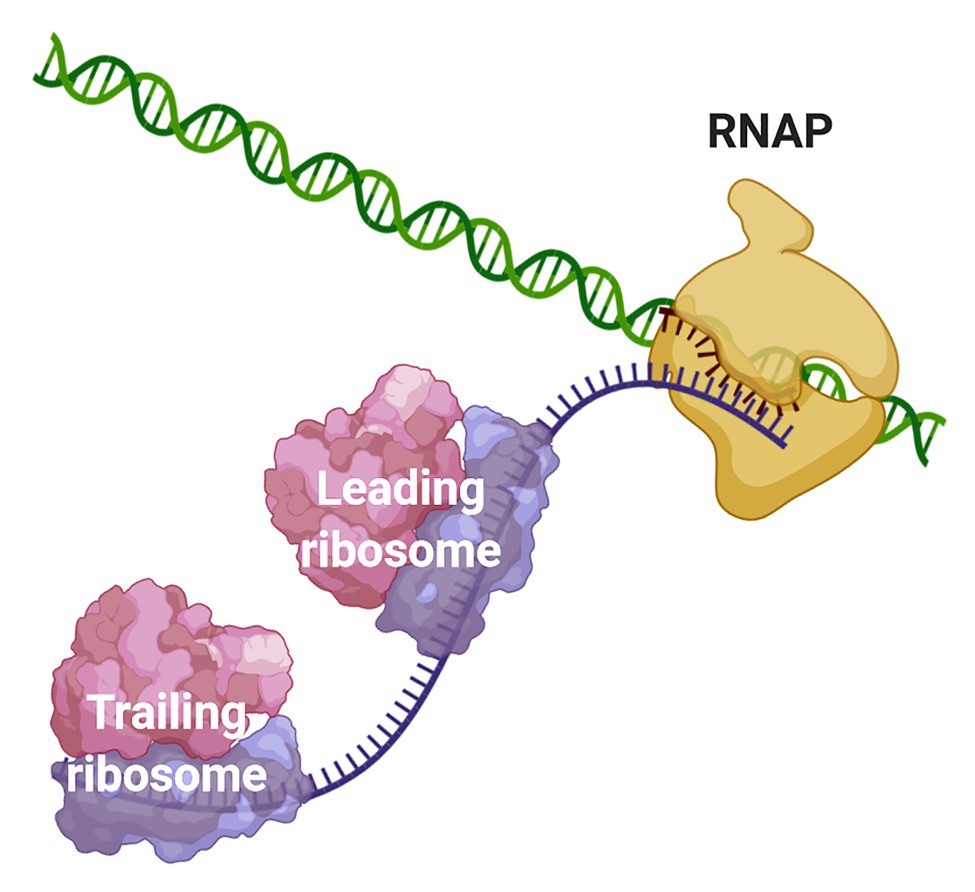
Figure 1. Coupling of transcription and translation in prokaryotes. When the nascent mRNA emerges from the RNAP, the transcript is bound by the leading ribosome forming a transcribing-translating complex. Additional ribosomes can associate with the nascent mRNA to form a convoy of trailing ribosomes on the transcript that is still bound to the transcription machinery. The leading ribosome can physically interact with the RNAP or the two machineries may be connected via the transcript.
The fact that transcription and translation could be coupled in prokaryotes was first proposed by Stent in the mid-1960s. He argued that due to the apparent inability to dissociate nascent transcripts from the chromosome in vitro, an active force exerted by translating ribosomes could be necessary to release the mRNAs from their templates. Indirectly, he implied that transcription and translation could be spatiotemporally coupled (Stent, 1964). Subsequent in vitro work from the Nirenberg lab demonstrated DNA·RNA·ribosome complexes (Byrne et al., 1964; Bladen et al., 1965). In the early 1970s, Miller et al. (1970) published an electron microscopy image, which showed ribosomes strongly associating with and translating nascent mRNA in a concatenated fashion forming polysomes. More recently, similar observations were reported in archaea (French et al., 2007), extending the occurrence of CTT to all prokaryotes. Miller’s micrographs have illustrated microbiology textbooks for decades and CTT is nowadays a widely accepted dogma of prokaryotic gene expression.
In this review, we will first highlight the biological significance of maintaining proper CTT and present the current understanding of how transcription and translation are coordinated under different growth conditions. We will then discuss the recent findings that shed light on the mechanistic and molecular details that mediate the physical coupling of these two processes. We will proceed by describing different models explaining where CTT takes place in the context of the prokaryotic cell. We will then introduce several types of evidence that challenge the CTT dogma and suggest that its occurrence may not be as general as currently assumed. Furthermore, we will extensively elaborate on molecular mechanisms that potentially promote the spatiotemporal uncoupling of transcription and translation. We will conclude by discussing open questions and principles emanating from this old but still exciting couple.
Coordination of CTT
In optimal growth conditions, Escherichia coli ribosomes translate 14–17 amino acids per second (Young and Bremer, 1976; Proshkin et al., 2010; Zhu et al., 2016), meaning that they translocate about 42–51 nucleotides per second (nt/s) along the mRNA being translated. On the other hand, RNAP synthesizes mRNA at a rate of 42–49 nt/s (Proshkin et al., 2010; Iyer et al., 2018). Thus, mRNA transcription and translation rates are well-matched. Of note, translation and transcription rates vary across different growth conditions, but the rates of both processes remain coordinated (Vogel and Jensen, 1994b; Proshkin et al., 2010; Iyer et al., 2018), suggesting CTT coordination is important.
Indeed, a balanced CTT regime is crucial for the proper function of E. coli cells, and the uncoupling of transcription and translation can lead to multiple conflicts that compromise cell viability. Many such conflicts are related to the intimate link between CTT and Rho-mediated premature transcription termination (PTT). It was assumed for years that, in the absence of ribosomes engaged in CTT, Rho utilization (rut) sites in the nascent transcripts become exposed and are readily recognized by Rho, which translocates towards RNAP and causes the disassembly of the transcription elongation complex (TEC; Chalissery et al., 2011; Lawson et al., 2018). Recent evidence, on the other hand, supports a different mechanism where, in the absence of a physically coupled ribosome, transcription termination factor Rho associates with RNAP early after transcription initiation (Mooney et al., 2009a) via protein-protein interactions with NusA and NusG, rendering the TEC into a moribund pre-termination complex (PTC). This state favors Rho recognition of the rut sites in the nascent mRNA, which is followed by the closure of the Rho ring and disassembly of the TEC (Epshtein et al., 2010; Hao et al., 2020; Said et al., 2020). These molecular events lead to PTT and operon polarity (Richardson, 1991). Ribosomes and Rho compete for the same binding interface of NusG and, thus, a coupled ribosome prevents Rho-mediated PTT (Burmann et al., 2010). Coupled ribosomes can additionally antiterminate intrinsic terminators (Li et al., 2015). Moreover, leading ribosomes push stalled RNAPs forward and facilitate elongation over transcriptional roadblocks (Proshkin et al., 2010; Stevenson-Jones et al., 2020). Beyond gene expression, this is especially important for avoiding clashes between replisomes and backtracked RNAPs, which cause double-strand breaks and lead to genome instability (Dutta et al., 2011). Furthermore, cotranscriptional translation prevents reannealing of the nascent RNA to the template DNA strand, which can give way to dangerous R-loops (Gowrishankar and Harinarayanan, 2004), and also protects nascent transcripts from ribonucleolytic attack (Makarova et al., 1995; Deana and Belasco, 2005). In agreement with this, the stability of lacZ transcripts was greatly reduced when transcribed by T7 RNAP (Iost and Dreyfus, 1995), which transcribes about 230 nt/s (Golomb and Chamberlin, 1974) and cannot be closely followed by translating ribosomes. Tight CTT could be of critical importance for the expression of genes whose decay initiates before the transcription of the full mRNA is completed (Chen et al., 2015). For this population of transcripts, post-transcriptional translation appears to be unfeasible and ribosomes should closely follow RNAP before ribonucleases trigger the degradation of the nascent mRNA. Additionally, it is speculated that physically associated ribosomes can perform a pioneering round of translation for mRNA quality control, a well-characterized phenomenon in eukaryotes (Maquat et al., 2010). On the other hand, controlled uncoupling of transcription and translation can also be utilized for gene regulation. A classical example of this is the operon for tryptophan biosynthesis (Yanofsky, 1981), where due to low tryptophan concentrations the leader ribosome lags behind RNAP and allows the formation of an antitermination secondary structure that precludes the formation of the attenuator structure.
rRNA transcription (90 nt/s; Vogel and Jensen, 1994b) exceeds by far the translation elongation rates in E. coli, implying that, when transcribing mRNA, RNAP does not work at its maximum biosynthetic capacity, and that mechanisms for equalizing mRNA transcription and translation exist. In this regard, physical CTT offers an elegant model to explain the transcription-translation correlation under different growth conditions. According to this model, translation elongation rates of the leading ribosome, dictate transcription elongation rates (Proshkin et al., 2010). This model is supported by the finding that in strains harboring ribosomal mutations that slow translation, transcription elongation rates decreased accordingly, and that transcription-translation rates of different genes correlated with their rare codon content, highlighting the role of translation in dictating transcription elongation (Proshkin et al., 2010). In this model, the leading ribosome equalizes transcription-translation rates by physically pushing forward the RNAP, which otherwise tends to spontaneous backtracking and/or pauses (Proshkin et al., 2010; Stevenson-Jones et al., 2020), so that CTT remains coordinated and futile transcription is prevented. Of note, this mode of CTT coordination does not necessarily imply persistent RNAP·ribosome interactions or the formation of a stable complex, and it could rather be driven by occasional ribosome-to-RNA pushing contacts at sites where transcription slows down.
Besides CTT coordinated by physical RNAP·ribosome contacts, recent evidence argues in favor of additional mechanisms for CTT coordination. In contrast to common thought, chloramphenicol does not inhibit translation by reducing ribosome elongation rates, but rather by diminishing the population of ribosomes engaged in translation (Dai et al., 2016). In contrast, cells challenged with fusidic acid showed slower translation elongation rates, but strikingly, no reduction in transcription elongation rates (Zhu et al., 2019), suggesting that transcription elongation can be modulated independently to translation. Also, in cells subjected to nitrogen starvation, which is characterized by a slowdown in translation elongation rates, transcription elongation slowed down correspondingly and no hints of PTT were observed (Iyer et al., 2018). Importantly, translation inhibition by chloramphenicol did not decrease transcription elongation rates under these experimental conditions (Iyer et al., 2018), indicating that the RNAP elongates independently of the leading ribosome. Possibly, under conditions where translation slows down below a certain threshold, transcriptional cooperation among RNAPs (see section The CTT Dogma has Been Challenged: Towards Uncoupled Transcription-Translation?) might suffice to maintain transcriptional elongation in a ribosome-independent manner. Additional studies support a model where the occurrence of CTT is a stochastic event that does not depend on the physical contact of both machineries (see section The CTT Dogma has Been Challenged: Towards Uncoupled Transcription-Translation?; Li et al., 2015; Chen and Fredrick, 2018, 2020). Thus, it is reasonable to argue that, besides CTT coordinated by physical contacts between the leading ribosome and RNAP, additional mechanisms ensure the coordination of CTT.
Notably, the alarmone (p)ppGpp has recently emerged as an important player in CTT coordination. Upon amino acid starvation, the ribosome-associated RelA detects uncharged tRNAs at the A-site of the ribosomes and triggers the rapid synthesis of (p)ppGpp (Wendrich et al., 2002), inducing stringent response. Traditionally (p)ppGpp has been linked to transcriptional regulation, primarily by binding to the interface between β and ω subunit of the RNAP (Mechold et al., 2013; Zuo et al., 2013) and negatively regulating rRNA transcription, as well as a transcription of a myriad of coding sequences (Potrykus and Cashel, 2008; Sanchez-Vazquez et al., 2019). In agreement with this, in cells treated with fusidic acid that maintained normal transcription (p)ppGpp accumulation caused a slowdown of transcription elongation in an alarmone dose-dependent manner (Zhu et al., 2019). Alarmone-dependent slowdown of transcription elongation was also observed in cells subjected to amino acid starvation (Vogel et al., 1992; Vogel and Jensen, 1994a) and nitrogen limitation (Iyer et al., 2018).
Recent publications have expanded the (p)ppGpp targetome (Zhang et al., 2018; Wang et al., 2019), and additional alarmone-mediated layers of regulation have been discovered (reviewed in Hauryliuk et al., 2015). For instance (p)ppGpp competes with GTP for binding translation initiation factor 2 and elongation factor G (EF-G; Milon et al., 2006; Mitkevich et al., 2010), inhibiting their activity. Interestingly, under nitrogen starvation, ribosome stalling at glutamine codons diminishes translation elongation, and alarmone-defective strains show even slower translational elongation rates compared to wild-type cells (Li et al., 2018). Apparently, bacteria prevent generalized translation initiation, which could deplete pools of charged tRNAs and lead to incomplete translational rounds, and instead ensure that basal translation of housekeeping and stress-responsive genes remains active (Dai et al., 2016). This phenomenon has been proven essential for survival under oxidative stress, which is also characterized by a dramatic slowdown of translation elongation rates (Zhu and Dai 2019, 2020). It was recently reported that (p)ppGpp-dependent translational selectivity depends on structural motifs formed by the mRNAs (Vinogradova et al., 2020). Thus, upon environmental challenges that reduce translation elongation rates, cells achieve CTT coordination by slowing down RNAP elongation and by prioritizing translation of a subset of essential genes that avoid alarmone-mediated translation inhibition. Highlighting the importance of this regulation is the finding that disrupting (p)ppGpp-mediated CTT coordination sensitizes cells to nitrogen starvation (Iyer et al., 2018).
Alarmones also coordinate transcription and translation less straightforwardly. It has recently been shown that (p)ppGpp binds inosine-guanosine kinase, which prevents the excessive accumulation of purine nucleotides and leads to pRpp synthesis, a precursor molecule for pyrimidine nucleotides, tryptophan, and histidine (Wang et al., 2020a). Thus, alarmones further coordinate CTT indirectly, by balancing nucleotide stocks required for transcription and ensuring tryptophan and histidine availability to maintain basal translation. Interestingly, the operons for the biosynthesis of these amino acids are regulated by transcriptional attenuation (Yanofsky, 1981), which links their regulation to sensing the CTT status.
The evidence presented above supports a scenario where CTT coordination is preserved by different, although complementary means, depending on the growth conditions (Figure 2). When transcription slows down below translation elongation rates, due to spontaneous backtracking or pausing, the leading ribosome physically pushes the RNAP forward in a way that translation rates dictate equal transcription elongation rates (Proshkin et al., 2010; Stevenson-Jones et al., 2020). Under conditions where ribosome elongation lags behind transcription, such as nitrogen limitation or oxidative stress, multilayered regulation via (p)ppGpp ensures that RNAP elongation rates decrease accordingly and maintains CTT coordination without the need for physical contacts of the transcription and translation machineries. Considering the ongoing discovery of (p)ppGpp-regulated processes and the tight relationship of alarmones with the metabolic status of the cell, we expect that additional pathways ensuring CTT coordination via (p)ppGpp will be discovered. These complementary mechanisms, i.e., physical contacts between RNAP and the leading ribosome and alarmone-mediated coordination, account for the observed correlation between transcription and translation elongation rates under different growth conditions.
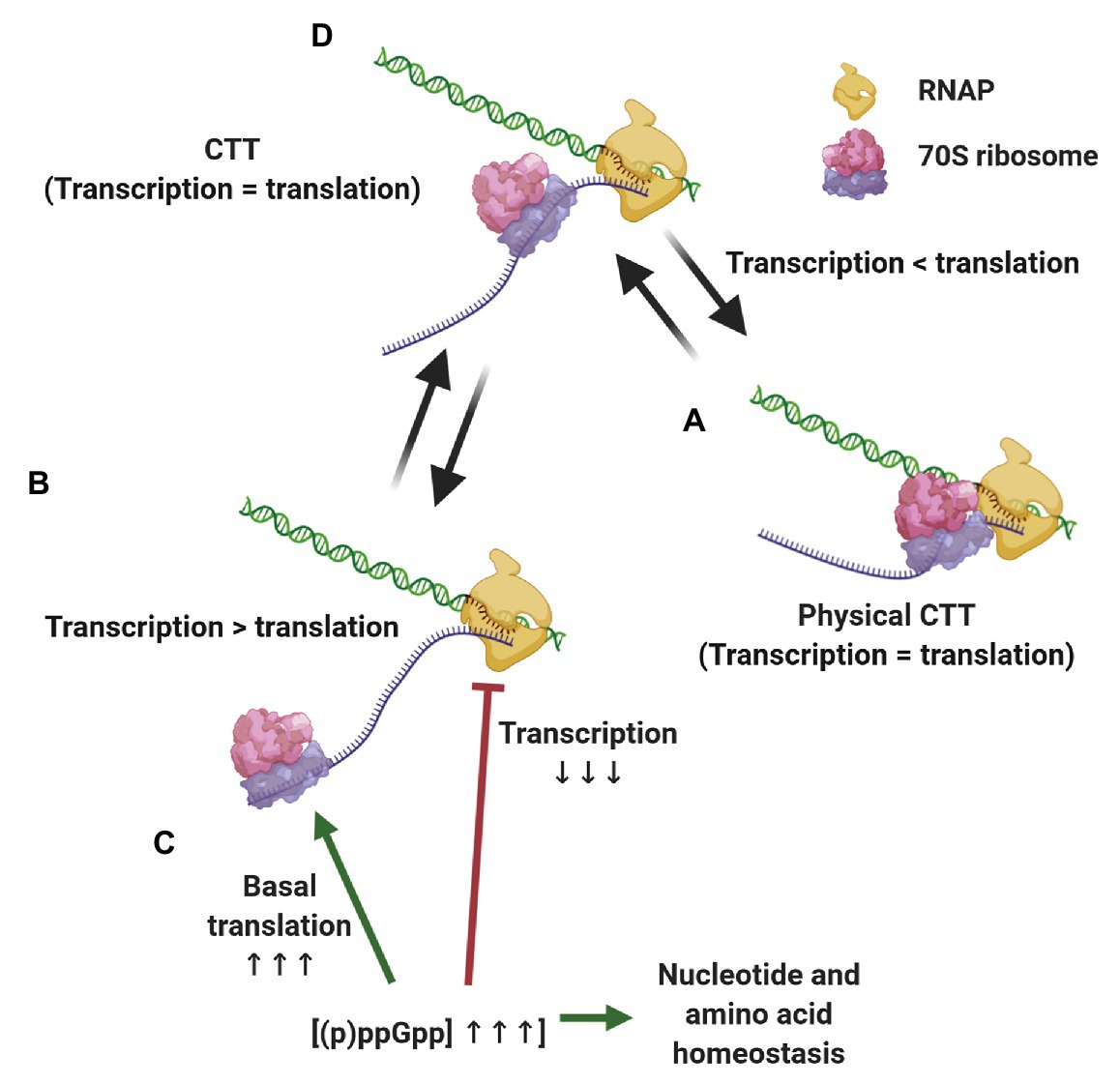
Figure 2. Strategies for maintaining coupled transcription-translation (CTT) coordination. Under conditions that slow down transcription elongation rates below translational rate, such as carbon limitation and RNAP backtracking (A), the leading ribosome catches up with and physically pushes the transcription elongation complex (TEC) forward, equalizing transcription and translation rates. When translation elongation lags behind transcription rate, e.g., upon nitrogen limitation and oxidative stress (B) (p)ppGpp concentrations increase, slowing down transcription elongation, promoting basal translation and balancing nucleotide and amino acid stocks (C), thus equalizing transcription and translation elongation rates (D).
The Molecular Architecture of CTT
How do the transcriptional and translational machineries intimately interact? Several recent publications have shed light on the molecular interactions that mediate the physical coupling of transcription and translation in E. coli, although their conclusions were not always in agreement (reviewed by Conn et al., 2019). CTT was initially proposed to be mediated by NusG. Specifically, nuclear magnetic resonance (NMR) experiments showed that the N-terminal domain (NTD) and C-terminal domain (CTD) of NusG interact with RNAP and NusE, which doubles as S10 protein in the ribosome (Mooney et al., 2009b; Burmann et al., 2010), respectively, thus, bridging the transcriptional and translational machineries. As Rho and the leading ribosome compete for NusG binding, this mode of coupling also explains why TECs engaged in CTT avoid Rho-mediated termination (Burmann et al., 2010). CTT of a subset of horizontally acquired operons was shown to be bridged by the NusG homolog RfaH in a similar fashion (Burmann et al., 2012; Zuber et al., 2019). The NusG-dependent CTT model was further supported by in vivo and in vitro experiments evidencing that NusG simultaneously interacts with the ribosome and RNAP (Saxena et al., 2018). A cryo-electron microscopy (cryo-EM) structure of a TEC·NusG complex did not show any defined density for the NusG CTD, suggesting that NusG mediates a flexible RNAP-ribosome association (Kang et al., 2018b). This was confirmed by another cryo-EM structure of a ribosome-bound NusG, that showed a defined density for the CTD but not for the NTD (Washburn et al., 2020). This latter publication also reported NMR results supporting the association of an RNAP·NusG complex with S10.
In parallel, a series of publications posited a model where transcription and translation are coupled independent of NusG, via direct RNAP·ribosome interactions. Chemical cross-linking experiments demonstrated multiple NusG-independent interactions between RNAP and both 30S and 50S ribosome subunits (Fan et al., 2017), and an RNAP·30S subunit cryo-EM structure (Demo et al., 2017) recapitulated several of the interactions detected by cross-linking. Simultaneously, a cryo-EM structure of a transcribing-translating RNAP·ribosome complex, named expressome, showed that direct interactions between both machineries can mediate the coupling (Kohler et al., 2017). Different from the previous works described above, this structure was produced by colliding a translating ribosome against a transcriptionally stalled RNAP. In this structure, the exit site of RNAP and the mRNA entry site of the ribosome were closely placed, which suggests continuous protection of the nascent mRNA from transcription to translation. This protection also excludes Rho from accessing nascent transcripts and from terminating transcription. Importantly, the NusG binding partners in the CTT complex, i.e., the β'-subunit of RNAP and the S10 protein of the ribosome, were located on opposite sites of the complex and the NusG linker was too short to bridge such distance. Thus, the collided expressome was not compatible with NusG-mediated CTT (Kohler et al., 2017).
How can the divergent views of NusG-dependent and -independent CTT, both emerging from structural studies, be reconciled? Two simultaneous studies presenting cryo-EM structures suggest that these views can co-exist. The first study by the Weixlbaumer lab describes the structures of transcribing-translating complexes assembled on mRNA scaffolds that allow different distances between the RNAP active site and the ribosomal P-site (Webster et al., 2020). One structure showed, for the first time, simultaneous binding of the NTD and CTD of NusG to RNAP and the leading ribosome, correspondingly. In this structure NusG forms a bridge between the ribosome and RNAP, thus stabilizing the interaction interface between the two machineries. Increasing the length of the intervening mRNA resulted in a similar structure, i.e., a NusG-coupled expressome. However, shortening of the intervening mRNA resulted in a structure in which RNAP is located closer to the ribosome entry tunnel. In this collided expressome, RNAP could still bind NusG, but the latter was not able to bridge the distance to S10. Hence, as shown previously (Kohler et al., 2017), the collided expressome is not compatible with NusG-mediated CTT. Of note, a structure nearly identical to this collided expressome was obtained when assembling expressomes on short scaffolds in the absence of NusG (Kohler et al., 2017). The conclusions that emerge from these structures are that coupling via NusG restrains RNAP motions, that the length of the intervening mRNA determines whether NusG can be involved, that expressome formation is strictly mRNA-dependent, although RNAP·30S complexes were previously observed (Demo et al., 2017), and that both the NusG-bridged and the collided expressomes are compatible with translation factor binding (Webster et al., 2020).
The second study, published by the Ebright lab, in addition to confirming many of the results presented by Weixlbaumer and coworkers and showing that collided expressomes are incompatible with ribosome·NusG binding, provides insights into the participation of NusA in CTT (Wang et al., 2020b). The researchers’ present high-resolution structures for expressomes that involve both NusG and NusA in CTT. In these structures, which accommodated spacer mRNAs of different lengths, NusA promoted expressome assembly by acting as a “coupling pantograph” between RNAP and the S2/S5 protein of the leading ribosome.
Based on molecular modeling, Ebright and co-workers point out several features of the collided expressome that question its capability to promote proper gene expression (Wang et al., 2020b). First, the collided expressome is sterically incompatible with NusA binding (Guo et al., 2018), with the formation of Q-dependent antitermination complexes (Shi et al., 2019; Yin et al., 2019) and with the formation of pause and termination RNA hairpins (Kang et al., 2018a; Roberts, 2019). Regarding translation, the collided expressome is not compatible with the swiveling of the 30S subunit head that takes place during ribosome translocation (Schuwirth et al., 2005; Ratje et al., 2010; Guo and Noller, 2012). Furthermore, collided expressomes lack densities corresponding to the RNAP ω subunit, which assists in TEC assembly and, by binding (p)ppGpp, mediates CTT coordination during stringent response (see section Coordination of CTT; Kurkela et al., 2020). Thus, it is highly unlikely that collided expressomes are responsible for general CTT and, instead, they could be specialized complexes in charge of CTT under specific conditions, or even anomalous complexes resulting from RNAP-ribosome clashes (Wang et al., 2020b). Supporting the latter, RNAP from a collided expressome was shown to reinitiate transcription elongation in vitro and detach from the ribosome that is purposely stalled (Stevenson-Jones et al., 2020), suggesting that it is not a stable complex with significant biological functions.
We propose a mechanistic model (Figure 3) where different expressomes could come into play sequentially during different stages of E. coli CTT. Translation initiation is a relatively lengthy process (median time of 15–30 s; Siwiak and Zielenkiewicz, 2013; Shaham and Tuller, 2018) compared to transcription elongation (49 nt/s; Iyer et al., 2018), so according to our model, RNAP can elongate several dozens, if not hundreds of nucleotides beyond the start codon translation-independently before the leader ribosome initiates elongating. Once the leading ribosome begins elongating, the two machineries are remotely connected by the nascent mRNA as an uncoupled expressome. The two machineries could remain kinetically coupled and the uncoupled expressome could conduct gene expression under conditions where the non-physical coupling is coordinated via (p)ppGpp (see section Coordination of CTT). In other cases, this kinetic coupling precedes the formation of a coupled expressome. Specifically, RNAP tends to pause within the first 100 nt after the start codon (Mooney et al., 2009a; Larson et al., 2014), which can allow the leading ribosome to catch up with the TEC. Several types of data indicate that ribosome translocation along the nascent mRNA to the proximity of the TEC may be a prerequisite for the formation of a physically coupled expressome. Firstly, NusG association with TECs, which occurs only after substantial transcription (Mooney et al., 2009a), is facilitated by cotranscriptional translation (Washburn et al., 2020). Additionally, the intranucleoidal ribosome concentration (2–8 μM; Bakshi et al., 2012; Sanamrad et al., 2014) is substantially lower than the NusG·ribosome dissociation constant (50 μM; Burmann et al., 2010), so the NusG·ribosome association is likely favored by the translocation of the leading ribosome towards the TEC along the nascent mRNA. Thus, the arrangement of the uncoupled expressome promotes interactions of NusG with both machineries and favors the formation of a coupled expressome. This complex could be further stabilized by NusA (Wang et al., 2020b), which associates with elongating TECs early after transcription initiation (Mooney et al., 2009a).
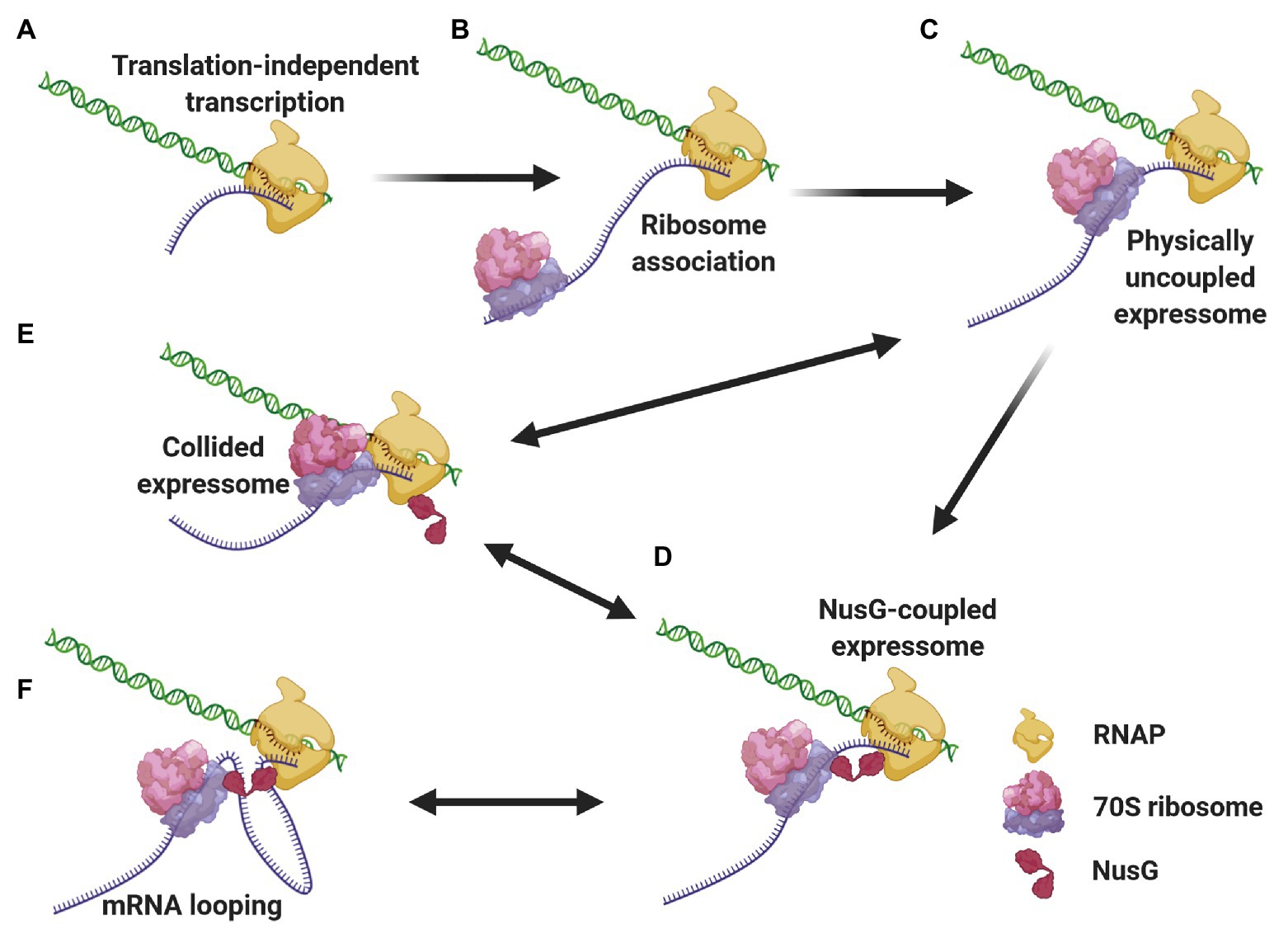
Figure 3. Different expressome configurations suggested to engage in gene expression. Transcription initiates over 5'-proximal coding sequences, eventually forming the TEC (A). After considerable elongation, the leading ribosome associates with the nascent transcript and initiates translation (B). RNAP pauses, allowing the leading ribosome to catch up with the TEC, forming a physically-uncoupled expressome (C). The proximity of the RNAP and the leading ribosome allows for NusG association with both machineries, forming a NusG-bridged coupled expressome (D). Transcriptional roadblocks can reduce the length of the connecting mRNA, giving way to a collided expressome, either with or without NusG (E). Conversely, translational slowdown lengthens the connecting mRNA, which can loop out the complex (F). The two latter events are reversible, giving way to a rearrangement of the coupled expressome.
The coupled expressome may proceed with CTT. Yet, transcriptional and translational elongation rates can respond to independent signals, resulting in varying lengths of the nascent mRNA connecting the RNAP and the leading ribosome (Conn et al., 2019). For instance, transcriptional pauses or backtracking reduce the distance between the leading ribosome and RNAP, which can give way to the formation of a collided expressome (Kohler et al., 2017; Wang et al., 2020b; Webster et al., 2020). As the leading ribosome pushes the stalled RNAP forward (Stevenson-Jones et al., 2020), the coupled expressome can be reconfigured. Such “collision-and-reconfiguration” events can also occur in the case of uncoupled expressomes. On the other hand, translational roadblocks will increase the distance between the leading ribosome and RNAP. As the mRNA connecting the two machineries in the coupled expressome is exposed to the solvent (Wang et al., 2020b; Webster et al., 2020), the longer nascent mRNA can be accommodated by looping out from the coupled expressome (Conn et al., 2019). Decreased transcription elongation and/or increased translation elongation reduce the length of the intervening mRNA and loop the protruding mRNA back into the complex.
The in-cell architectures of Mycoplasma pneumoniae expressomes published recently show similarities to the E. coli expressomes, as well as differences (O’Reilly et al., 2020). On one hand, structures obtained from pseudouridimycin-halted M. pneumoniae TECs, which likely represent collided expressomes, showed direct interactions between RNAP and the leading ribosome, similar to E. coli collided expressomes. On the other hand, M. pneumoniae elongating expressomes showed neither direct nor NusG-mediated interactions. Instead, the coupling between RNAP and the leading ribosome was mediated solely by NusA (O’Reilly et al., 2020). Thus, although direct RNAP·ribosome interactions, which characterize collided expressomes, seem a conserved phenomenon, the molecular actors and interactions that drive factor-mediated RNAP-ribosome coupling differ in evolutionarily unrelated species.
It should be emphasized that all E. coli expressome structures discussed in this section were obtained in vitro, and that their existence in vivo remains to be investigated. Also, the questions of whether RNAP and the lead ribosome are physically coupled on the nascent mRNA in vivo and whether there is a mechanism to ensure or promote this coupling remain open.
The Cell Biology of CTT
Where in the bacterial cell does CTT occur? The subcellular organization of the transcriptional and translational machineries offers clues to answer this question. RNAP spends most of its lifetime bound to DNA, either engaged in transcription or non-specifically searching promoters (Endesfelder et al., 2013; Stracy et al., 2015; Ladouceur et al., 2020), and very few RNAP molecules are observed outside the nucleoid region (Bakshi et al., 2012). In rich media, RNAPs form nucleolus-like clusters engaged in rRNA transcription, but cluster assembly is independent of ongoing transcription (Jin et al., 2013; Gaal et al., 2016; Weng et al., 2019). Recently, it has been shown that RNAPs nucleate and form biomolecular condensates by liquid-liquid phase separation (LLPS; Ladouceur et al., 2020).
Ribosome localization, on the other hand, differs considerably among species. In organisms with a high nucleocytoplasmic (NC) ratio (Gray et al., 2019), such as Caulobacter crescentus, the ribosomes, and the nucleoid are homogeneously mixed (Bowman et al., 2010; Montero Llopis et al., 2010; Bayas et al., 2018). This facilitates the encounter of the transcriptional and translational machineries, and the occurrence of CTT can be envisaged fairly intuitively in these organisms. Yet, in many other species with low NC ratio, including E. coli (Hobot et al., 1985; Azam et al., 2000; Valencia-Burton et al., 2007; Wang et al., 2011; Bakshi et al., 2012; Chai et al., 2014; Cougot et al., 2014; Mohapatra and Weisshaar, 2018; Zhu et al., 2020), Bacillus subtilis (Lewis et al., 2000; Mascarenhas et al., 2001), Bdellovibrio (Borgnia et al., 2008), and Pseudomonas putida (Kim et al., 2019a), ribosomes and nucleoids are strongly segregated. The nucleoid-excluded localization of E. coli factors engaged in translation, such as tRNAs (Plochowietz et al., 2016; Volkov et al., 2018) and translation EFs (Chai et al., 2014; Mohapatra et al., 2017; Mustafi and Weisshaar, 2018), also supports that in these species bulk translation takes place in spatial separation from the genetic material and the transcriptional machinery. Hence, in these species, the transcriptional and translational machineries rarely encounter each other and the occurrence of CTT is less intuitive.
Several biophysical forces cause the subcellular nucleoid-vs-ribosome segregation. By avoiding extensive contacts with the inner membrane, the DNA polymer maximizes its number of available conformational states, i.e., the conformational entropy, and by segregating from the nucleoid, the ribosomes optimize their freedom for motion and the translational entropy (Mondal et al., 2011; Bakshi et al., 2014). This segregation is further accentuated by volume exclusion forces mutually exerted by the nucleoid polymer against the bulky polysomes (Mondal et al., 2011; Bakshi et al., 2014; Castellana and Wingreen, 2016). Lastly, electrostatic repulsion forces between negatively charged nucleic acids, in this case, chromosomal DNA and RNA-rich ribosomes (Joyeux, 2016), and phase separation effects (Joyeux, 2018) could also account for the observed antilocalization of nucleoids and ribosomes.
Considering this subcellular segregation of RNAPs and ribosomes, how does CTT occur in these organisms? One possibility is that CTT occurs at the surface of the nucleoid, where both RNAPs and ribosomes may encounter each other (Figure 4A). Indeed, it was shown that highly transcribed gene loci migrate to the nucleoid periphery (Stracy et al., 2015; Weng et al., 2019; Yang et al., 2019). The association of several RNAPs into biomolecular condensates at highly transcribed loci (Ladouceur et al., 2020) could cause the exclusion of these condensates from the nucleoid surface by the biophysical forces described above. Supporting this hypothesis, the cotranscriptional association of bulky ribosomes amplifies loci migration to the nucleoid periphery (Yang et al., 2019).
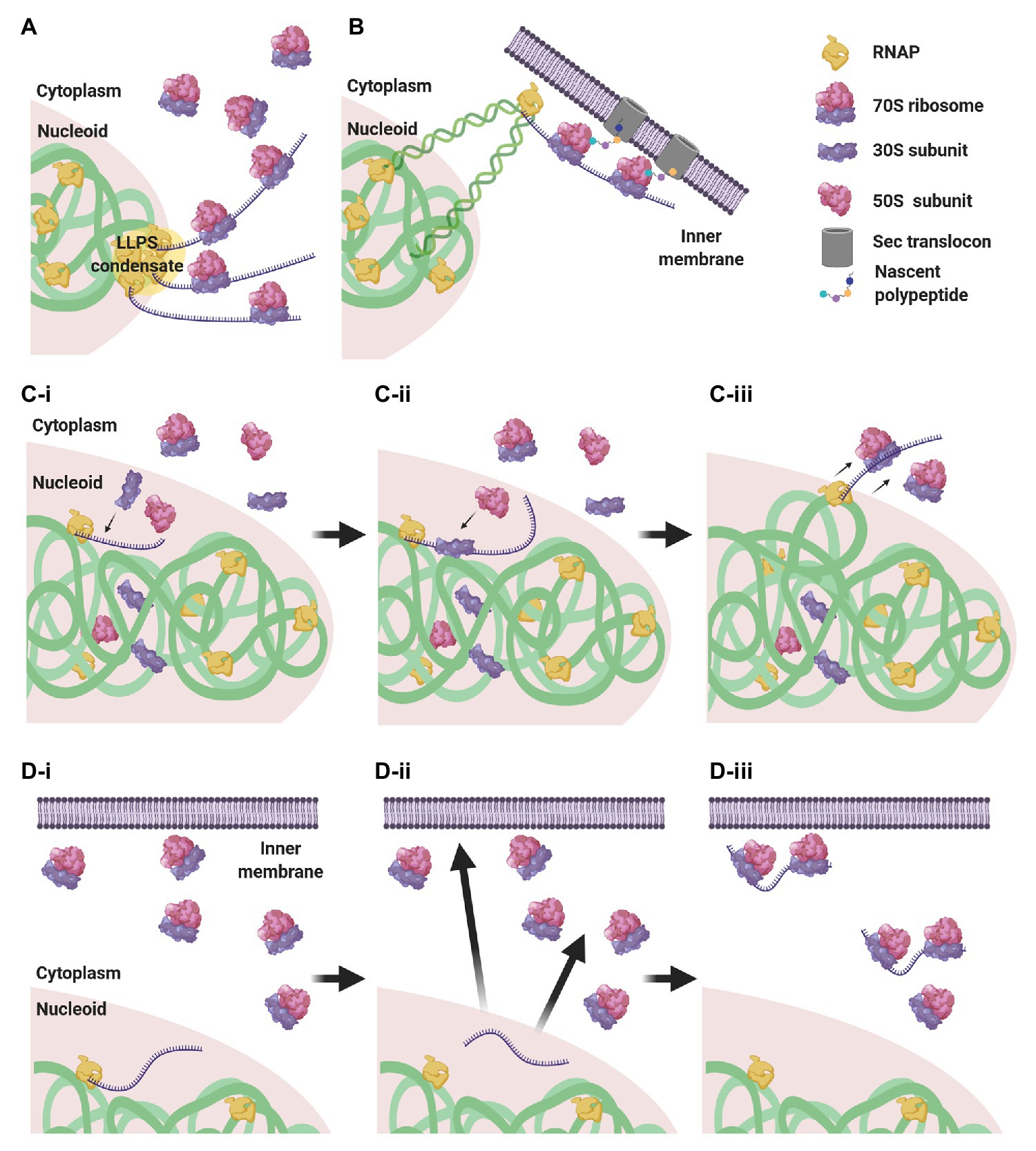
Figure 4. CTT scenarios in low nucleocytoplasmic (NC) ratio species. (A) RNAP clusters form condensates by liquid-liquid phase separation (LLPS) that are expelled to the nucleoid periphery, where they encounter ribosomes that engage in CTT. (B) Via transertion, gene loci encoding membrane proteins emerge from the nucleoid to the inner membrane, where CTT readily occurs. (C) Intranucleoidal translation initiation. Free ribosomal subunits penetrate the nucleoid and a 30S subunit associates with the SD sequence of the nascent transcript (C-i). A 50S subunit associates with the pre-initiation complex, forming a 70S ribosome (C-ii). The RNAP·mRNA·ribosome complex is expelled to the nucleoid periphery, where CTT proceeds (C-iii). (D) Uncoupled transcription-translation (UTT). Transcription takes place within the nucleoid translation-independently (D-i), and ribosome-free transcripts navigate the cell to their corresponding destination in the cytoplasm or in the membrane (D-ii), where they are locally translated (D-iii).
Gene loci encoding membrane proteins are thought to be expressed by coupled transcription-translation-membrane insertion, a mechanism known as transertion (Woldringh, 2002). This implies that certain gene loci migrate from the nucleoid mesh to the vicinity of the inner membrane and become exposed to ribosome-rich regions, where CTT could readily occur (Figure 4B). Although the occurrence of transertion still awaits direct experimental validation (Roggiani and Goulian, 2015), the notion is supported by the demonstration that a small population of ribosomes and RNAPs resides in the membrane vicinity (Herskovits and Bibi, 2000; Bakshi et al., 2012), and by the visualization of induction-dependent membrane relocation of several gene loci coding for membrane proteins (Libby et al., 2012; Kannaiah et al., 2019; Yang et al., 2019). Thus, localizing TECs to ribosome-rich regions via transertion may very well promote the encountering of RNAPs and ribosomes and facilitate CTT.
Alternatively, it was shown in E. coli that, although 70S ribosomes are segregated from the nucleoid, free 30S and 50S subunits can penetrate the nucleoid mesh (Sanamrad et al., 2014; Mohapatra and Weisshaar, 2018; Zhu et al., 2020). This implies that canonical translation initiation, which is conducted by a free 30S subunit that recognizes the Shine-Dalgarno (SD) sequence of the nascent transcript, could initiate within the nucleoid in a cotranscriptional manner. Upon assembly of the 70S monosome, the RNAP·nascent mRNA·ribosome complex would be pushed to the surface of the nucleoid by the biophysical forces mentioned above, where additional ribosomes could engage in CTT (Figure 4C).
An additional possibility is that transcription and translation are not compulsorily coupled. In this scenario, transcription takes place within the nucleoid, in spatiotemporal separation from translation, and the mRNAs then navigate the cytoplasm to their final destination where they are locally translated (Kannaiah and Amster-Choder, 2014; Irastortza-Olaziregi and Amster-Choder, 2020; Figure 4D). We will elaborate on this scenario in section The CTT Dogma has Been Challenged: Towards Uncoupled Transcription-Translation?
The CTT Dogma has Been Challenged: Towards Uncoupled Transcription-Translation?
Coupled transcription-translation is widely accepted by the microbiology community and supported by extensive work. Yet, most knowledge regarding CTT emanates either from in vitro studies or from experiments conducted with only a handful of genes. Importantly, the subcellular segregation of the transcriptional and translational machineries observed in some species raises the possibility that these processes could take place in spatiotemporal separation (see section The Cell Biology of CTT). Indeed, several lines of evidence have recently challenged the classical view that transcription is inherently coupled to translation, and the global occurrence of CTT has been questioned.
The Fredrick lab developed a hammerhead ribozyme-based reporter system that enables measuring and comparing protein synthesis carried out by limited vs. unlimited number of translation rounds (Chen and Fredrick, 2018). They applied this system to study the translation of six adjacent gene-pairs that are cotranscribed in the same operon. In a tight CTT scenario, limiting translation rounds should bring the relative protein amounts for each gene-pair close to 1:1, as protein synthesis of the two co-transcribed genes would presumably be carried out by a leading ribosome physically coupled to RNAP. However, for five out of six gene-pairs, they observed that the relative protein synthesis derived from limited translation rounds of the two genes was not close to 1:1. Rather, the ratio was similar to that measured for unlimited translation rounds. This indicates that the first translational rounds occur independently of transcription, i.e., they are not carried out by a ribosome that is physically bound to RNAP. Importantly, when these experiments were repeated with an RNAP mutant showing reduced transcription elongation rate, which, presumably, facilitates the physical coupling of the leading ribosome with RNAP (see section The Molecular Architecture of CTT), protein production after limited translation rounds was closer to 1:1 for one of the tested gene-pairs (Chen and Fredrick, 2018). These results support a model where the physical coupling of transcription and translation is a stochastic event, which depends on the rates of transcription and translation elongation. These observations indicate that physical association between the leading ribosomes and RNAPs, as well as coordinated elongation by the two machineries, is significantly less common than currently assumed, implying that RNAP often transcribes without a linked ribosome (Chen and Fredrick, 2020). Supporting this notion, pseudouridimycin, which stalls TECs, notably increased the percentage of ribosomes that are physically coupled to RNAP in M. pneumoniae (O’Reilly et al., 2020). This indicates that, in the absence of transcription-halting antibiotics, the majority of those ribosomes are engaged in CTT, but that they do not translate in physical association to RNAP.
Similar conclusions emanated from studying the relationship between termination efficiency (TE) at intrinsic terminators and their intragenic position (Li et al., 2015). TE of terminators located in the first 100 nt of ORFs was close to 100%, and it gradually decreased as the distance from the start codon increased. This position-dependent loss of TE was explained by the fact that ribosomes follow and catch up RNAPs closely enough to prevent the formation of terminators (Li et al., 2015). Indirectly, the dependence of TE on the distance from the start codon implies that transcription of the 5'-proximal coding sequences occurs translation-independently (see section The Molecular Architecture of CTT).
Whereas these studies argue against the idea of physical CTT, they are not incompatible with the existence of RNAP-ribosome complexes remotely connected by nascent mRNAs. Yet, further evidence supports the idea that transcription and translation in some cases are completely uncoupled. It is accepted that translation increases mRNA stability by ribosome shielding against ribonucleolytic attack (Iost and Dreyfus, 1995; Makarova et al., 1995; Deana and Belasco, 2005). A reassessment of RNA decay patterns unraveled that, contrary to the widespread idea that RNA decay is exponential, two-thirds of the analyzed transcripts followed a biphasic degradation pattern, with a very steep decay at short post-transcription times followed by an exponential decay at longer times (Deneke et al., 2013). These results suggest that most transcripts spend a minor fraction of their lifetime in a ribosome-free form, where they are highly vulnerable to ribonucleases until ribosomes engage in translation and slowdown mRNA decay. At the same time, similar to what happens in eukaryotic cells, other RNA-binding proteins could be responsible for the protection of the transcripts till they are translated. Whatever the reason is, these results imply that transcription is not tightly coupled to translation for most genes (Deneke et al., 2013).
Furthermore, previous work from our lab and others showed that transcripts can localize to sites overlapping with the localization of their encoded proteins in the cytoplasm, the membrane, or the poles of E. coli cells (Nevo-Dinur et al., 2011; Moffitt et al., 2016). Very importantly, these localization patterns were preserved when the translation of the tracked transcripts was inhibited by antibiotics, translational roadblocks, or mutations (Nevo-Dinur et al., 2011). Similarly, a transcript encoding a short membrane protein localized to the membrane even when the SD sequence of the mRNA was deleted (Steinberg et al., 2020). Likewise, translation-independent RNA localization has been observed in cyanobacteria, where transcripts encoding photosystem components localized to thylakoid membranes in the presence of puromycin concentrations that inhibit translation and detach ribosomes from mRNAs (Mahbub et al., 2020). These observations imply that bacterial mRNAs can skip tight CTT, navigate the cytosol as ribosome-free transcripts, and undergo local translation once they reach their corresponding destination (Nevo-Dinur et al., 2011; Kannaiah and Amster-Choder, 2014; Irastortza-Olaziregi and Amster-Choder, 2020). Another recent publication from our lab, which reports the distribution of E. coli RNAs between the membrane, cytoplasm, and poles by combining cell fractionation with deep-sequencing, showed that a significant fraction of the E. coli transcriptome localizes in a translation-independent manner and challenged the idea that CTT is a general mechanism for gene expression (Kannaiah et al., 2019). Collectively, this evidence supports the notion that CTT is not as predominant as currently assumed, and that spatiotemporally uncoupled transcription-translation (UTT) could be responsible for substantial gene expression.
UTT implies that processive transcription can occur independently of ribosomes that are coupled to RNAP by physical interaction, via protein factors or through mRNAs. For E. coli RNAPs, translation-independent transcription has been attributed only to TECs engaged in rRNA transcription, which are modified by the antitermination complex and show fast transcription elongation rates that outpace Rho and avoid termination (Squires and Zaporojets, 2000; Paul et al., 2004). For mRNA-transcribing TECs, the dogma that prevailed was that a coupled ribosome is required for transcription processivity (see section Coordination of CTT). Then again, early evidence already indicated that transcription can be tuned independently of translation. Although mRNA transcription-translation rates change and equalize each other at different growth rates, rRNA transcription rates also vary according to growth rates (Vogel and Jensen, 1994b), suggesting that ribosome-independent mechanisms exist in bacteria for determining transcription elongation rates. As discussed in section Coordination of CTT, the application of sublethal concentrations of fusidic acid, which slows down ribosome translocation, did not affect RNAP elongation rates (Zhu et al., 2019). Besides, when ribosomes stalled at proline-rich sequences of E. coli cells deleted for EF-P (Elgamal et al., 2016), or when (p)ppGpp-mediated CTT coordination was disrupted under nitrogen starvation (Iyer et al., 2018), RNAP was still able to elongate. In further agreement with ribosome-independent transcription, when backtracked TECs are pushed and reactivated by the leading ribosome, transcription elongation restarts even when translation elongation is inhibited (Stevenson-Jones et al., 2020). Likewise, in M. pneumoniae subjected to chloramphenicol treatment, the RNAP·ribosome association is lost (O’Reilly et al., 2020), which reinforces the idea that RNAP can detach from the expressome and transcription elongation proceeds independently to translation.
How can the transcription processivity of TECs that are engaged in mRNA transcription be maintained in the absence of a coupled ribosome? The trafficking of transcription elongation and termination factors can offer a partial explanation for this. NusG recruitment to E. coli TECs occurs after substantial transcription and is assisted by a translationally coupled ribosome (see section The Molecular Architecture of CTT). Although Rho is recruited to TECs early after transcription initiation (Mooney et al., 2009a), it still requires an RNAP-associated NusG for inducing the PTC before triggering termination (Epshtein et al., 2010; Hao et al., 2020; Said et al., 2020). Thus, translation-independent TECs could show less affinity for NusG and, consequently, may be less prone to Rho-mediated termination.
Cooperation among RNAPs (reviewed by Le and Wang, 2018) could further facilitate the processivity of ribosome-independent TECs. For instance, similar to a leading ribosome that pushes a backtracked RNAP forward (see section Coordination of CTT), trailing RNAPs facilitate transcription of the leading RNAP over DNA roadblocks and rescue backtracked TECs by physically pushing the preceding RNAP forward (Epshtein et al., 2003; Epshtein and Nudler, 2003). Effects related to DNA supercoiling offer alternative explanations for this transcriptional cooperation. A mathematical model predicts that the torque created by transcription elongation over the DNA double helix pushes and pulls TECs located in close proximity forward without the mediation of any physical contact among each other (Heberling et al., 2016). Recently, a publication from the Jacobs-Wagner lab evidenced that, as long as gene promoters remain induced and multiple RNAPs initiate transcription of the lacZ gene, elongation rates of ongoing transcription are maintained by the mutual cancelation of positive and negative DNA supercoiling upstream and downstream the TEC convoy (Kim et al., 2019b). Regardless of the actual underlying mechanism, we suggest that the cooperation between RNAPs could suffice for maintaining the transcription processivity required for UTT.
In vitro studies showed that the B. subtilis RNAP transcribes much faster than the E. coli RNAP (Artsimovitch et al., 2000) and, notably, a recent publication demonstrated that transcription elongation in B. subtilis outpaces ribosome translocation over nascent transcripts (Johnson et al., 2020). Although, as reported for E. coli, leading ribosomes could still physically push and rescue stalled or paused B. subtilis TECs (see section Coordination of CTT), this “runaway” transcription creates extensive distance between RNAP and the leading ribosome, supporting the idea that transcription and translation are mostly uncoupled in B. subtilis. Accordingly, transcription terminators in B. subtilis are located only a few nucleotides downstream of stop codons (Johnson et al., 2020), which would otherwise be masked by a ribosome physically associated with RNAP (Li et al., 2015). A bioinformatic exploration using this short distance between stop codons and intrinsic terminators as a proxy for runaway transcription suggested that this mode of gene expression could be a fairly widespread phenomenon in bacteria (Johnson et al., 2020), once again arguing against the universality of CTT (Wang and Artsimovitch, 2020).
Mechanisms Potentially Enabling UTT
Considering the evidence presented in section The CTT Dogma has Been Challenged: Towards Uncoupled Transcription-Translation, it is plausible that CTT may not be as universal as has been assumed for many years. In other words, transcription may occur translation-independently, and mRNAs could be transcribed and translated in spatiotemporal separation. Yet, for UTT to take place, bacteria would need to face three major challenges: (1) Prevent association between the leading ribosome and the nascent mRNA, once the SD sequence emerges from RNAP. (2) Protect the ribosome-free transcript from ribonucleolytic decay in the cytoplasm. (3) Carry out translation-uncoupled transcription without disruption by intrinsic and Rho-dependent terminators.
Dynamics of transcription elongation over 5'-proximal coding sequences are of special interest for UTT. In E. coli, RNAP shows pronounced transcriptional pausing at sites overlapping with SD sequences and start codons (Larson et al., 2014), thus opening a temporal window for regulation over the naked 5'-UTRs by RNA-Binding proteins (RBPs) or RNA folding events prior to ribosome association with the transcript. Furthermore, transcription of 5'-proximal coding sequences is anyhow expected to occur translation-independently (see section The Molecular Architecture of CTT), so why cannot this type of regulation continue to be exerted over the downstream ribosome-naked transcript before translation begins?
Below, we describe several molecular factors and mechanisms that, when acting in cooperation, potentially promote UTT in a way that the three challenges mentioned above would be satisfactorily resolved (Figure 5). Evidence supporting the capacity of these factors and mechanisms to act cotranscriptionally on nascent mRNAs is presented. Furthermore, formation of biomolecular condensates by LLPS and their putative implication in promoting UTT are briefly discussed.
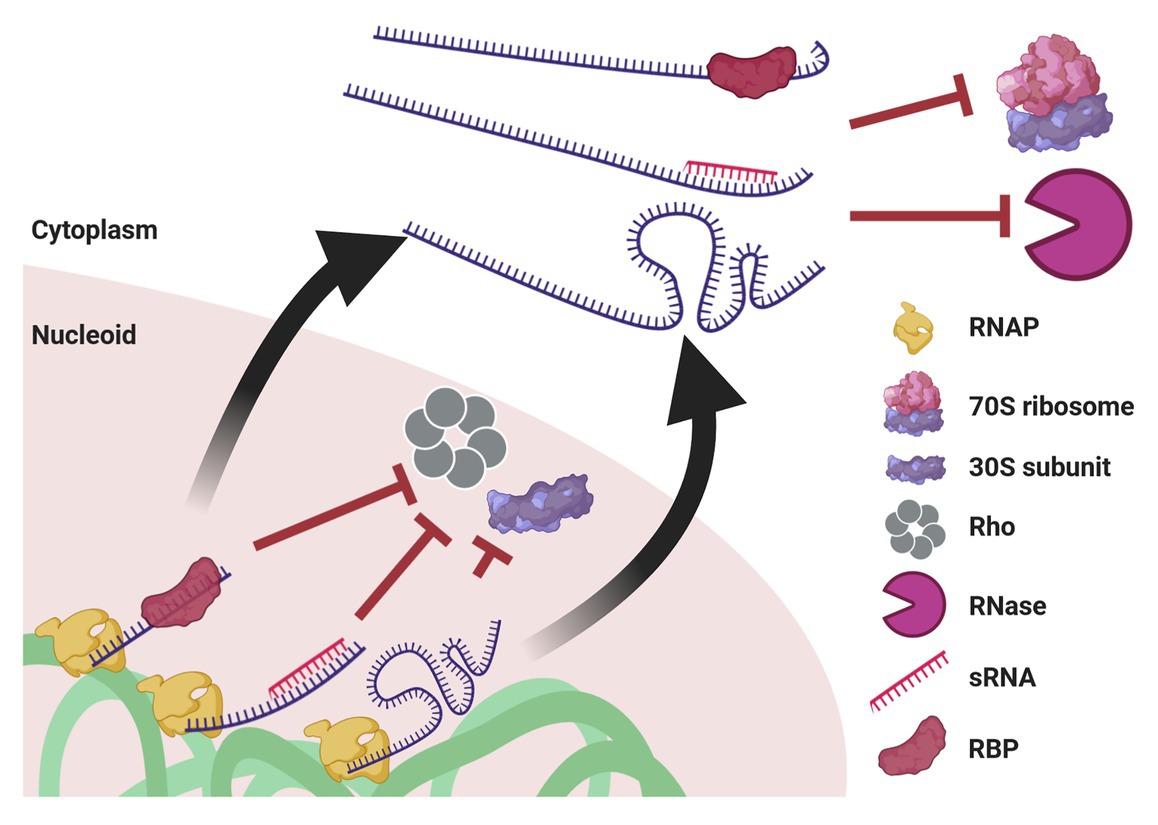
Figure 5. Mechanisms potentially enabling UTT. Cotranscriptional events, such as association with an RBP or with an sRNA, as well as riboswitch formation, prevent transcription termination by Rho and association with the leading ribosome. When ribosome-free transcripts are released to the cytoplasm, these transcript-protecting events counteract the activity of ribonucleases. Of note, although drawn linearly, transcripts supposedly acquire complex secondary and tertiary structures that confer further protection. This protection also prevents mRNA translation until they reach their final destination.
RNA-Binding Proteins
RBPs have gained major interest as a consequence of their capability to regulate transcript fate by affecting mRNA translation and stability (Holmqvist and Vogel, 2018; Richards and Belasco, 2019), and emerging evidence suggest they can promote UTT. For instance, Synechocystis RBPs Rbp2 and Rbp3 are required for the translation-independent thylakoid membrane localization of transcripts encoding photosystem proteins (Mahbub et al., 2020), implying that they act cotranscriptionally on mRNA targeting before translation initiates. Likewise, Grad-seq experiments performed in Salmonella and E. coli showed that several RBPs reviewed here co-sediment with RNAP (Smirnov et al., 2016; Hör et al., 2020), which could indicate a cotranscriptional association of RBPs with nascent RNAs. Considering their transcript fate-determining activities, these RBPs emerge as potential candidates for promoting UTT.
Cold Shock Proteins
Cold Shock Proteins (CSPs) belong to an evolutionarily widespread family of small, acidic proteins that were originally discovered as mediators of cold shock response, but it is currently understood that their function exceeds adaptation to cold (reviewed in Ermolenko and Makhatadze, 2002; Horn et al., 2007; Budkina et al., 2020). Notably, the presence of at least one csp gene in the cell is essential for viability in B. subtilis (Graumann et al., 1997). The E. coli genome encodes nine different CSP genes, from which only four, CspA, CspB, CspG, and CspI, are induced by cold shock (Etchegaray and Inouye, 1999; Wang et al., 1999), and two, CspC and CspE, are constitutively expressed at 37°C (Yamanaka et al., 1994; Bae et al., 1999). Of note, the cold shock domain (CSD) in CSPs, confers a capacity to bind single-stranded nucleic acids (Jiang et al., 1997; Lopez et al., 1999; Phadtare and Inouye, 1999) and melt secondary structures within them (Phadtare et al., 2002a; Phadtare and Severinov, 2005; Rennella et al., 2017). Hence, in addition to mediating UTT as described below, CSPs may aid CTT by ensuring the transfer of unfolded mRNA from the nucleoid to ribosomes (El-Sharoud and Graumann, 2007).
Bioinformatical explorations unraveled a sequence-level bias towards U-richness in transcripts encoding integral membrane proteins in E. coli (Prilusky and Bibi, 2009) and Lactococcus lactis (van Gijtenbeek et al., 2016). Information published by our lab confirmed that the U-richness in membrane-traversing domains is important for their membrane localization, as predicted bioinformatically (Kannaiah et al., 2019). Interestingly, E. coli CspC and CspE preferentially bind U-rich artificial transcripts and endogenous membrane mRNAs (Benhalevy et al., 2015). Moreover, CspE overexpression causes the accumulation of ribosome-free transcripts encoding membrane proteins in the cytoplasm and positively affects their translation in the membrane (Benhalevy et al., 2017). This suggests that, via CspE mediation, these transcripts avoid CTT until they reach the membrane, where they are locally translated.
Current evidence suggests that CSPs promotes UTT by protecting nascent transcripts from RNases. For instance, overexpression of CspC and CspE increases the stability of transcripts encoding stress response proteins RpoS and UspA (Phadtare and Inouye, 2001). Whereas CspE levels remain constant through different growth phases, CspC levels increase upon entry into stationary phase, which results in the stabilization of rpoS transcripts (Shenhar et al., 2012). These transcript-stabilizing capabilities can be explained by the tendency of CspE to bind poly(A) sequences, counteracting the ribonucleolytic activity of PNPase and RNase E (Feng et al., 2001). Considering that most transcripts undergo polyadenylation (Mohanty and Kushner, 2006), CspE could act as a global molecular shield against the concerted poly(A)-dependent exoribonucleolytic action of PNPase in the 3'-end and the endonucleolytic attack of RNase E in internal cleavage sites of nascent transcripts. Of note, cspE mRNA is downregulated in the absence of PNPase (Polissi et al., 2003), suggesting that a regulatory mechanism balances the ribonucleolytic activity of PNPase and the anti-RNase protection by CspE.
For these activities to promote UTT, CSPs should access nascent transcripts as soon as they emerge from RNAP. Indeed, CspE binds nascent RNAs and acts as an antiterminator by melting secondary structures of intrinsic terminators (Hanna and Liu, 1998; Bae et al., 2000; Phadtare et al., 2002a,b; Phadtare and Severinov, 2005). Collectively, these pieces of evidence indicate that CSPs can promote the uncoupling of transcription and translation by counteracting RNase activity over transcripts and promoting translation-independent transcription over intrinsic terminators.
Cold shock proteins show UTT-promoting activities in other species. In Salmonella, CspC and CspE bind about 20% of the transcriptome with important regulatory implications for its pathogenicity (Michaux et al., 2017). For example, the ecnB mRNA, which is bound by these CSPs, shows lower transcript levels and stability in a ΔcspCE background. Levels and stability of this transcript were restored in ΔcspCE cells expressing a temperature-sensitive RNase E mutant at the restrictive temperature. Thus, CSPs protect ecnB transcripts from RNase E-mediated decay (Michaux et al., 2017). Similarly, CspE binds and stabilizes yciF transcripts, conferring Salmonella increased resistance to bile salts by impermeabilizing the cell membrane (Ray et al., 2019b). Also, in B. subtilis, CspB shows extranucleoidal localization in a transcription-dependent manner (Mascarenhas et al., 2001; Weber et al., 2001), suggesting that CspB binds transcripts upon entry into the cytoplasm.
All in all, CSPs have emerged as RBPs with important roles in gene regulation and, potentially, promoting UTT. Their function resembles that of FRGY2, a CSD-containing protein that binds mRNAs in the nucleous of frog oocytes and protects transcripts from degradation and translation in the cytoplasm until they are released in a regulated manner during oocyte development (Bouvet and Wolffe, 1994). Their multifaceted functions as transcription antiterminators and antiribonucleolytic shields make CSPs promising subjects for future study regarding their putative role as UTT facilitators.
Ribosomal Proteins
In addition to multiple interactions with rRNAs within the ribosome, ribosomal proteins (RPs) perform extraribosomal moonlighting functions as free RBPs, often involved in gene regulation (reviewed in Bhavsar et al., 2010; Aseev and Boni, 2011). Several RPs bind their cognate mRNAs to negatively autoregulate their translation and to keep RP homeostasis. In other cases, RPs play functions that could enable UTT.
One example is the S1 RP in E. coli, which binds single-stranded AU-rich sequences located upstream of SD sequences within the 5'-UTRs (Boni et al., 1991; Mogridge and Greenblatt, 1998; Komarova et al., 2002), an activity shown to increase the stability of the lacZ mRNA (Komarova et al., 2005), most likely due to the overlap between S1 binding sites and RNase E cleavage sites (Kaberdin and Bläsi, 2006). In agreement with this hypothesis, S1 binds cspE and rpsO transcripts and protects them against RNase E attack (Delvillani et al., 2011). S1 was further shown to counteract PNPase-mediated mRNA decay (Briani et al., 2008). When overexpressed, S1 binds several mRNAs, including the pnp mRNA itself, and increases their stability against PNPase-mediated decay. In line with these results, depletion of S1 leads to the destabilization of these transcripts in vivo (Briani et al., 2008). Similar to CspE, S1 binds poly(A) sequences (Kalapos et al., 1997), but this binding does not protect transcripts against PNPase degradation in vitro (Feng et al., 2001). Whether S1 counteracts PNPase in vivo by promoting CspE-dependent transcript protection (see above) or through an alternative mechanism remains unknown. Additionally, S1 was shown to increase protein secretion that is mediated by the hemolysin signal peptide, and this was accompanied by the stabilization of the corresponding transcripts (Khosa et al., 2018). If S1 can promote UTT, these RNA-protecting activities should be implemented as soon as the nascent transcript emerges from RNAP. In this regard, S1 associates with RNAP and promotes its transcriptional processivity (Sukhodolets and Garges, 2003; Sukhodolets et al., 2006). Additionally, S1 binds RNAP indirectly forming a ribonucleoprotein (RNP) with IsrA sRNA (van Nues et al., 2015). Thus, it is plausible that S1 binds nascent transcripts cotranscriptionally and protects them from ribonucleolytic attack, thus favoring the occurrence of UTT.
Another example is the S4 RP, which negatively autoregulates its translation by binding its cognate cistron within the α-operon and creating a pseudoknot that leads to the entrapment of an inactive translation initiation complex (Spedding and Draper, 1993; Schlax et al., 2001). This autoregulation also represses the translation of several other RPs cotranscribed within the same operon. However, the regulation of the rpoA gene, which encodes the α-subunit of RNAP and is also cotranscribed within the α-operon, is not subjected to this negative regulation (Thomas et al., 1987). Although the mechanism of this exclusion remains unknown, it implies that the α-operon can be subjected to partial disruption of CTT; i.e., whereas the α-subunit of RNAP is cotranscriptionally translated, the genes encoding ribosomal proteins are cotranscribed but translationally repressed by S4. To exert such CTT disruption, S4 would need to act cotranscriptionally. Indeed, S4 can bind RNAP and antiterminate Rho-dependent terminators (Torres et al., 2001), thus protecting ribosome-free nascent transcripts from PTT. Therefore, S4 plays a double role in promoting UTT: disruption of the transcription-translation coupling in the α-operon and preventing Rho-mediated termination of TECs that are not physically coupled to a translating ribosome.
The L4 RP also shows multiple activities that could promote UTT. L4 binds the regulatory CTD of RNase E and inhibits its activity, leading to stabilization of a subset of stress-related transcripts crucial for cell survival (Singh et al., 2009). Alleviation of the ribonucleolytic pressure on these mRNAs could allow a less tight CTT regime for these transcripts. Among the L4-stabilized transcripts is that of the triptophanase (tna) operon, which is subjected to an additional layer of regulation by L4 that is degradosome-independent. Specifically, L4 overexpression increases the stability of the tnaCAB transcript but causes translational repression of the TnaA protein by binding to the spacer between tnaC-tnaA (Singh et al., 2020). This could lead to the partial disruption of CTT in this operon, i.e., tnaC can be expressed by CTT, whereas tnaA translation is repressed despite the increase in its mRNA levels. Interestingly, this translational repression of tnaA takes place at early stationary phase to prevent the degradation of tryptophan, which is required for long-term survival through deep stationary phase (Singh et al., 2020), highlighting that CTT disruptions can have important physiological implications. Importantly, L4 binds its cognate transcript to attenuate its transcription (Lindahl et al., 1983). Thus, L4 could act cotranscriptionally as an antitranslation agent also over other nascent transcripts. Further investigation of extraribosomal functions of RPs should lead to a better understanding of these activities that are potentially involved in UTT.
Hfq
Besides its highly studied role as an sRNA-mRNA matchmaker (see below), Hfq exerts post-transcriptional regulation in E. coli by directly binding to transcripts and affecting their fate (reviewed in Kavita et al., 2018). Hfq was shown to bind the 5'-UTRs of its cognate transcript (Večerek et al., 2005) and of cirA (Salvail et al., 2013) and mutS mRNAs (Chen and Gottesman, 2017), as well as the ribosome binding site (RBS) of the Tn10 transposase mRNA (Ellis et al., 2015). In all these cases Hfq binding leads to translational repression of the target transcripts. These activities can be explained by the observation that in enterohemorrhagic E. coli, Hfq shows a preference for binding ARN triplets located in the proximity of RBSs (Tree et al., 2014), which could preclude translation initiation. Likewise, translational repression by Hfq plays a central role in the catabolic repression of the opportunistic pathogen Pseudomonas aeruginosa. With the assistance of the catabolic repression control protein Crc, Hfq binds specific A-rich sequences in the proximity of translation initiation regions to suppress translation of catabolite repressed genes (Sonnleitner and Bläsi, 2014; Pei et al., 2019).
Hfq was also shown to preferentially bind intrinsic terminator sequences in Salmonella (Holmqvist et al., 2016). Correspondingly, Hfq binds PAP I and promotes the synthesis of poly(A) tails at intrinsic terminators in E. coli (Hajnsdorf and Régnier, 2000; Le Derout et al., 2003; Mohanty et al., 2004). Analogously to CspE (see above), Hfq binds to poly(A) sequences overlapping with RNase E cleavage sites and confers protection against exoribonucleolytic decay by PNPase and RNase II (Folichon et al., 2003; Moll et al., 2003; Zhang et al., 2003). Collectively, this evidence indicates that Hfq binds intrinsic termination sites in order to promote polyadenylation and, by binding to these poly(A) sequences, protects 3'-UTRs and upstream sequences from ribonucleolytic decay.
Importantly, Hfq interacts with RNAP to promote transcription (Sukhodolets and Garges, 2003). Furthermore, Hfq shares topological similarities with YaeO, the only so-far discovered protein that binds and inhibits Rho in E. coli (Pichoff et al., 1998; Gutiérrez et al., 2007) and Vibrio cholerae (Pal et al., 2019). Accordingly, Hfq suppresses Rho-dependent termination by simultaneously binding Rho and AU-rich sequences located upstream rut sites (Rabhi et al., 2011). Recently, it has been shown that Hfq pervasively binds nascent transcripts in E. coli (Persson et al., 2013; Sedlyarova et al., 2016) and P. aeruginosa (Kambara et al., 2018; Gebhardt et al., 2020). The helix-like localization of Hfq observed in E. coli under certain growth conditions (Taghbalout et al., 2014; Malabirade et al., 2017; Kannaiah et al., 2019) resembles the helical-ellipsoidal conformation of the nucleoid (Fisher et al., 2013), reinforcing the idea that Hfq could indirectly associate with the chromosome by binding nascent transcripts. Similar to S1, Hfq forms an RNP with IsrA sRNA that associates with RNAP (van Nues et al., 2015). Therefore, Hfq emerges as a potential UTT-promoting factor by antiterminating ribosome-free transcription that could otherwise be terminated by Rho. The fact that Hfq can interact with nascent transcripts implies that the antitranslation and antiribonuclease roles of Hfq described here could come into play cotranscriptionally to disrupt CTT and protect nascent mRNAs from RNases.
CsrA/RsmA
Although initially discovered as a regulator of glycogen biosynthesis upon entry into stationary phase (Romeo et al., 1993), CsrA has emerged as a global post-transcriptional regulator implicated in multiple cellular functions (reviewed in Vakulskas et al., 2015; Romeo and Babitzke, 2018). CsrA has been shown to bind hundreds of transcripts in E. coli (Edwards et al., 2011; Potts et al., 2017), Campylobacter (Dugar et al., 2016), Salmonella (Holmqvist et al., 2016), and Legionella (Sahr et al., 2017) affecting their post-transcriptional fate and potentially promoting UTT.
The main regulatory activity of CsrA is via translational repression (Dugar et al., 2016; Potts et al., 2017). For example, CsrA binds the SD sequence of the hfq mRNA and inhibits its translation (Baker et al., 2007). Similarly, CsrA binds at two sites in the 5'-UTR, including the SD sequence, of transcripts encoding the transcriptional regulator NhaR, which responses to high sodium concentrations and alkaline pH (Pannuri et al., 2012). Likewise, CsrA inhibits translation of the iron storage dsp mRNA by binding the 5'-UTR region of the transcript (Potts et al., 2017; Pourciau et al., 2019). In enteropathogenic E. coli (EPEC), CsrA represses translation of the virulence effector NleA (Katsowich et al., 2017). Upon contact with the host, EPEC injects effectors to the host cell through a type III secretion system with the assistance of the effector-bound chaperone CesT. After releasing the effector, free CesT binds and inhibits CsrA, leading to derepression of NleA and its subsequent translation and translocation to the host cell (Katsowich et al., 2017). The CsrA homolog RsmA binds the 5'-UTR of psl mRNA in charge of the biosynthesis of a structural polysaccharide in P. aeruginosa biofilms and inhibits its translation by refolding the transcript structure so that the SD sequence is not accessible to ribosomes (Irie et al., 2010). In all examples discussed here, translation inhibition was not accompanied by decreased transcript stability. Consequently, this can lead to the accumulation of ribosome-free transcripts in the cytoplasm, resembling the phenomenon observed upon CspE and S1 overexpression (see above). Interestingly, the CsrA/RsmA-regulated transcripts discussed here encode proteins responding to environmental stimuli. Thus, it is tempting to speculate that CsrA/RsmA promote UTT and the accumulation of untranslated pools of mRNAs whose expression can be rapidly derepressed upon environmental challenges without transcriptional delay.
Furthermore, CsrA can also act as an anti-RNase shield (Esquerré et al., 2016; Potts et al., 2017). For instance, CsrA was shown to be essential for E. coli motility by regulating the fhl operon, the master operon for flagellum biosynthesis, by increasing the stability of fhlDC transcripts (Wei et al., 2001), and this is achieved by protecting these mRNAs from RNase E cleavage (Yakhnin et al., 2013). Likewise, CsrA stabilizes the Legionella iron uptake regulator fur mRNA by binding a specific site in the proximity of a potential RNase E site (Sahr et al., 2017). CsrA and RsmA were also shown to increase the stability of the STM3611 transcript in Salmonella (Jonas et al., 2010) and of the hrpG transcript, the master regulator of T3SS genes in Xanthomonas, respectively (Andrade et al., 2014).
CsrA also displays transcription-related activities, e.g., binding to the gap operon transcript in Legionella cotranscriptionally to counteract Rho-dependent transcription termination (Sahr et al., 2017) and to RNAP as an RNP with IsrA sRNA in E. coli (van Nues et al., 2015). Furthermore, in P. aeruginosa, RsmA was shown to bind over 500 nascent transcripts cotranscriptionally (Gebhardt et al., 2020). Collectively, these publications indicate that CsrA/RsmA are major gene regulators that act over nascent transcripts. Thus, it is plausible that the antitranslation, anti-RNase, and antitermination activities of CsrA/RsmA favor the occurrence of UTT. Further investigating the cotranscriptional activities of CsrA/RsmA should shed light on the detailed involvement of CsrA/RsmA in promoting UTT.
ProQ
ProQ, a FinO-domain protein that acts as an sRNA-mRNA matchmaker (Smirnov et al., 2017; Westermann et al., 2019; Melamed et al., 2020), has recently emerged as an important RBP, which binds a substantial fraction of the E. coli and Salmonella transcriptomes (Smirnov et al., 2016; Holmqvist et al., 2018), suggesting that it may be involved in UTT implementation.
ProQ recognizes structural features present in sRNAs and 3'-UTRs of mRNAs and increases their stability upon binding (Smirnov et al., 2016; Holmqvist et al., 2018; Bauriedl et al., 2020; Stein et al., 2020). Specifically, ProQ was shown to stabilize cspE mRNA by binding its 3'-UTR and preventing RNase II-mediated exoribonucleolytic attack (Holmqvist et al., 2018). Stability of cspC, cspD, and ompD was also reduced in ΔproQ background (Holmqvist et al., 2018), but whether ProQ stabilizes these transcripts by antiribonuclease protection awaits experimental confirmation. Furthermore, about a third of ProQ binding events take place in sites overlapping with RNase E cleavage sites (Chao et al., 2017; Holmqvist et al., 2018). Collectively, these anti-RNase activities resemble those displayed by FinO, which recognizes and binds a similar structural feature of its only target FinP antisense RNA and exerts anti-RNase E protection (Jerome et al., 1999; Arthur et al., 2011). Importantly, ProQ associates with RNAP via an RNP formed with IsrA sRNA (van Nues et al., 2015). Thus, it is plausible that ProQ accesses and binds nascent transcripts cotranscriptionally, promoting UTT by protecting untranslated transcripts against ribonucleolytic decay.
Nus Factors
Nus factors regulate transcription elongation by affecting RNAP pausing and promoting transcription termination/antitermination (Santangelo and Artsimovitch, 2011; Sen et al., 2014). NusA is an essential component of the antitermination complex that, together with NusB, NusE, NusG, and several ribosomal proteins, enhances rRNA transcription rates (Squires and Zaporojets, 2000; Paul et al., 2004). Besides, NusA promotes RNAP pausing, which favors the formation, stability, and efficiency of intrinsic terminators (Farnham et al., 1982; Schmidt and Chamberlin, 1987; Toulokhonov et al., 2001). As discussed in section Coordination of CTT, NusA also mediates transcription termination by Rho (Epshtein et al., 2010; Hao et al., 2020; Said et al., 2020). Yet, NusA could possibly counteract Rho-dependent termination in certain instances. Specifically, NusA mutants with increased affinity for binding NusA utilization (nut) sites decreased Rho-dependent termination at specific cases where nut and rut sites overlap (Qayyum et al., 2016). Thus, regarding UTT, NusA could facilitate translation-independent transcription of mRNAs by interfering with certain Rho-dependent termination events on nascent ribosome-free transcripts.
Other Nus factors are involved in processive antitermination (PA) mechanisms. Opposite to dedicated antiterminators, PA factors associate with and modify TECs to promote transcriptional readthrough over multiple transcription terminators distally located in the operons under their regulation (Goodson and Winkler, 2018). For example, the NusG paralogue LoaP regulates transcription through termination sites located within two antibiotic biosynthesis operons in Firmicutes, Actinobacteria, and Spirochaetes (Goodson et al., 2017). Deletion of loaP led to a reduction in transcript levels of LoaP regulons. LoaP is thought to processively antiterminate intrinsic terminators, an activity that requires the 5' leader sequence of the transcripts under its regulation (Goodson et al., 2017). All in all, the PA activity of such Nus paralogs on their specific target operons could favor UTT by alleviating the need for a translationally coupled ribosome for counteracting intrinsic and Rho-dependent terminators.
Cis-Acting RNA Elements
Riboswitches are regulatory elements located in the 5'-UTR of mRNAs that are comprised of two modules: a structurally complex aptamer that binds a ligand and an expression platform that is regulated by the aptamer structural folding. Ligand binding causes structural refolding of the aptamer, which affects the expression of the downstream expression platform in an ON/OFF manner (Sherwood and Henkin, 2016). Some riboswitches act by translationally repressing the ORF under their regulation (Breaker, 2018). For example, in cobalamin riboswitches, the SD sequence is sequestered within the aptamer structure (Johnson et al., 2012), which becomes accessible for ribosomes only upon ligand binding. In thiamin pyrophosphate riboswitches, on the other hand, an anti-SD sequence within the aptamer structure anneals to and folds over the SD sequence located in the expression platform and inhibits translation initiation (Winkler et al., 2002). Again, ligand binding causes the refolding of the aptamer and liberates the SD sequence for ribosome binding. Similar to riboswitches, RNA thermometers are 5'-UTR elements that undergo structural refolding and affect downstream gene expression, but their refolding is caused by changes in temperature rather than ligand binding (Kortmann and Narberhaus, 2012). For example, the transcription of the cold-induced cspA gene takes place at all temperatures, but the cspA transcript is highly unstable at 37°C due to RNase E-mediated decay (Fang et al., 1997). Upon cold shock, the cspA transcript undergoes substantial refolding and is stabilized in a more RNase-resistant folding that allows translation of the protein (Giuliodori et al., 2010). Oppositely, the translation of rpoH transcript, coding for the heat shock sigma factor, is repressed at physiological temperatures by a secondary structure that sequesters the SD sequence. Upon temperature upshift, this structure refolds in a way that exposes the SD sequence for translation initiation (Morita et al., 1999). Likewise, several virulence factors are regulated by RNA thermometers that enable translation at 37°C upon entry in warm-blooded mammalian hosts (Johansson, 2009). Importantly, riboswitch structures are folded cotranscriptionally (Mironov et al., 2002; Frieda and Block, 2012; Watters et al., 2016; Uhm et al., 2017; Ray et al., 2019a). Considering that RNAP pauses at SD sequences and start codons (Larson et al., 2014), it is likely that riboswitch regulation comes into action shortly after they emerge from RNAP and before the translation initiation. Hence, it is reasonable to argue that riboswitches and RNA thermometers can cotranscriptionally block translation and protect transcripts from ribonucleolytic attack, which would promote UTT. Such inert transcripts would be activated upon environmental changes that induce their translation.
Similarly, cis-acting RNA elements act as translational repressors of type I toxin-antitoxin (TA) systems (Masachis and Darfeuille, 2018). In many type I TA systems the primary toxin transcript is translationally inert, and this is achieved by sequestering the SD sequence in a secondary structure formed with an anti-SD sequence located upstream in the transcript (Gultyaev et al., 1997; Darfeuille et al., 2007; Shokeen et al., 2008; Kristiansen et al., 2016; Wen et al., 2017). In other cases, the translation of the toxin is repressed by structures formed by interactions between the 5'- and 3'-ends of the full-length mRNA (Thisted et al., 1995; Franch and Gerdes, 1996; Gultyaev et al., 1997). The high structural complexity of toxin mRNAs is also thought to prevent the interaction of the nascent toxin transcript with the template DNA, which avoids the formation of deleterious R-loops, and to confer increased antiribonuclease resistance (Masachis and Darfeuille, 2018). Thus, structural features of toxin mRNAs ensure their translationally-inert transcription and protection against RNases until downstream activation events trigger post-transcriptional translation of these transcripts.
Some cis-acting RNA elements could facilitate UTT in a more indirect manner. For example, inhibitory RNA aptamers (iRAPs) interact with RNAP and facilitate Rho-dependent termination (Sedlyarova et al., 2017). Interestingly, many iRAPs map to the antisense strand and curb antisense transcription, which reduces transcriptional interference and favors sense transcription (Sedlyarova et al., 2017; Magán et al., 2019). Considering translation-independent TECs are less likely to reinitiate transcription after clashing with an antisense TEC (Hoffmann et al., 2019), diminishing antisense transcription by antisense iRAPs can alleviate the requirement of a leading ribosome that supports the TEC conducting sense transcription.
Other cis-acting RNA elements not only have the potential to attenuate CTT but enforce its disruption. For instance, the Salmonella virulence mgtCBR operon harbors a leader region that acts as a Rho-antagonizing RNA element (RARE) and a rut site that is necessary for Rho-mediated transcription termination of this operon (Sevostyanova and Groisman, 2015). The RARE counteracts termination by trapping Rho in a termination-defective conformation. Importantly, translation of the mgtCBR transcript sequesters the RARE in a stem-loop (Sevostyanova and Groisman, 2015). Thus, the only manner to express this operon is by UTT, in a way that the RARE inhibits Rho-mediated termination and allows translation-uncoupled transcription of the full mRNA, which would be translated post-transcriptionally. The corA operon of Salmonella is subjected to a very similar regulation and its leader region is highly conserved in enterobacteria (Kriner and Groisman, 2015), so such strictly UTT-dependent gene expression could be relatively widespread in these species.Lastly, besides elements that act against individual termination sites, other cis-acting RNA sequences are involved in PA (see above Goodson and Winkler, 2018). For example, in B. subtilis the eps operon, encoding exopolysaccharide biosynthesis proteins, is regulated by an eps-associated RNA (EAR) sequence, located in the intergenic region between the second and third genes of the operon (Irnov and Winkler, 2010). The EAR sequence is necessary for transcription of the entire operon, as it antiterminates several intrinsic terminators located in distal sites within the operon. The EAR sequence is also able to antiterminate heterologous terminators, supporting the hypothesis that it acts as a PA (Irnov and Winkler, 2010). Such cis-acting RNA elements with PA activities could promote UTT in the operons that they regulate since they would alleviate the requirement of a cotranscriptionally coupled ribosome to counteract terminators.
Trans-Acting RNA Elements
sRNAs are the archetypical example of trans-acting RNAs in bacterial gene regulation. They are typically 50–300 nt long transcripts that determine transcript fate by imperfect base-pairing with target mRNAs (Wagner and Romby, 2015). This process is often facilitated by the mRNA-sRNA matchmakers Hfq (Updegrove et al., 2016) and ProQ (Holmqvist et al., 2020). More recently, matchmaking activity has been reported for CsrA (Müller et al., 2019). sRNAs can affect the stability and/or translation of target mRNAs either positively or negatively, and these activities can potentially promote UTT.
In Salmonella, the glucose-responsive sRNA SgrS stabilizes the pdlB-yigL bicistronic transcript by preventing RNase E-mediated cleavage (Papenfort et al., 2013). Similarly, the sRNA RydC basepairs with the 5'-UTR of the cfa mRNA and stabilizes the longer isoform of this transcript by counteracting RNase E attack (Fröhlich et al., 2013). In B. subtilis, the sRNA RoxS binds the 5'-end of the yflS mRNA and prevents exoribonucleolytic decay by RNase J1 (Durand et al., 2017). Besides anti-RNase protection, the B. subtilis sRNA SR1 blocks translation binds in the 5'-UTR of its target ahrC mRNA and blocks its translation (Heidrich et al., 2006, 2007). Likewise, in Legionella pneumophila, the competence operon is translationally repressed by the sRNA RocR (Attaiech et al., 2016). sRNAs can further promote translational repression by recruiting Hfq to binding sites that prevent ribosomes association with SD sequences of their mRNA targets (Desnoyers and Massé, 2012; Azam and Vanderpool, 2018).
As sRNAs are relatively short and not translated by ribosomes they can penetrate the nucleoid mesh (Sheng et al., 2017) and putatively engage in cotranscriptional processes. Indeed, it was recently discovered that the sRNAs DsrA, ArcZ, and RprA, although induced by different stresses, bind the 5'-UTR of nascent rpoS transcripts to suppress Rho-dependent termination and allow expression of the stationary phase sigma factor (Sedlyarova et al., 2016). The expression of Rho itself is subjected to similar regulation, as the sRNA SraL basepairs with the 5'-UTR of the rho transcript to antiterminate its transcription (Silva et al., 2019). Thus, sRNAs could promote UTT by counteracting pervasive Rho-mediated transcription antitermination of ribosome-free mRNAs. The cotranscriptional association of sRNAs with mRNAs also suggests that they could exert their antitranslation and antiribonucleolytic regulation over nascent transcripts, facilitating the occurrence of UTT.
Interestingly, the E. coli sRNA IsrA associates with RNAP and forms RNPs together with important transcript fate-determining proteins Hfq, S1, CsrA, ProQ, and PNPase (van Nues et al., 2015). The association of these RNPs with RNAP and their corresponding regulatory outputs await further characterization. Yet, besides directly acting as UTT-inducing factors, sRNAs could promote UTT by serving as landing and integration platforms to enable cotranscriptional action of proteins with antitranslation, anti-RNase, and antitermination functions.
Bacterial RNP Bodies
Biomolecular condensates formed by multivalent interactions among proteins and nucleic acids have recently gained special interest. These condensates assemble and dissolve according to LLPS principles and they are involved in important cellular functions, including RNA metabolism (Banani et al., 2017). For example, processing bodies (P-bodies) and stress granules have been shown to sequester poorly translated transcripts for decay or storage in eukaryotes (Decker and Parker, 2012; Khong and Parker, 2020).
Biomolecular condensates show selective permeability and concentrate biomolecules and processes to discreet subcellular regions (Banani et al., 2017). Thus, they are attractive tools for prokaryotes to gain spatial complexity in their cytoplasms, which are generally devoid of membrane-bound organelles. Indeed, the existence of Bacterial RNP bodies (BR-bodies) in bacteria has been recently reported (Al-Husini et al., 2018; Muthunayake et al., 2020). In C. crescentus, RNase E assembles into BR-bodies together with the degradosome components and poorly translated RNAs, creating P-body-like condensates engaged in RNA degradation (Al-Husini et al., 2018, 2020). These RNase E condensates associate with the C. crescentus nucleoid in the proximity of rDNA loci (Bayas et al., 2018). Interestingly, the E. coli RNase E, which unlike in C. crescentus localizes to the inner membrane together with the other degradosome components, forms clusters that show RNA-dependent assembly and dynamics (Strahl et al., 2015), resembling the C. crescentus BR-bodies, thus suggesting that E. coli degradosomes may form condensates by LLPS in the inner membrane. Endoribonucleolytic attack by RNase E is believed to be the initial and the rate-limiting step of RNA degradation, so it is reasonable to argue that proximity to these RNase E condensates may increase the likelihood of a transcript to undergo decay. In agreement with this, E. coli membrane-localizing transcripts show a lower average stability, and artificially targeting cytoplasmic mRNAs to the membrane increases their degradation rate (Moffitt et al., 2016). Additionally, detaching RNase E from the membrane sensitized cytoplasmic ribosome-free transcripts (Hadjeras et al., 2019). Collectively, this evidence indicates that RNase activity is highly localized within bacterial cells by the mediation of BR-bodies, and that regions distant to RNA-degrading condensates may not be subjected to intensive ribonucleolytic pressure. This would certainly support UTT, especially in those organisms where the core ribonucleolytic machinery localizes in the membrane, as nascent ribosome-free transcripts would not be reachable for degradosomes and, hence, substantial antiribonucleolytic protection would not be required.
Furthermore, the bacterial cytoplasm shows glass-like properties and its fluidity varies according to the physiological state of the cell, where higher metabolic activity correlates with higher fluidity of the cytoplasm and vice versa (Parry et al., 2014). These biophysical properties may affect the function of biomolecular condensates, i.e., more fluid, liquid-like condensates allow higher motion and enzymatic activities, whereas more solid, aggregate-like entities specialize in storage (Banani et al., 2017). Bacteria undergo a notable metabolic slowdown upon entry into stationary phase or stress. This leads to the glassification of the cytoplasm (Parry et al., 2014), which could convert BR-bodies into RNA storage condensates rather than RNA processing bodies (Muthunayake et al., 2020). Such RNA-storing BR-bodies could further support UTT by accumulating and protecting untranslated transcripts until favorable conditions permit translation reinitiation.
Final Remarks
Although the individual processes leading to gene expression are subjected to extensive study, less is known about how these processes affect each other and how living cells spatiotemporally arrange the machineries that execute these functions. Yet, it is now acknowledged that these processes regulate each other and that their correct interplay is crucial for cell viability (Dahan et al., 2011). In this regard, prokaryotic CTT remains a paradigmatic example of such crosstalk. A proper CTT regime is of crucial importance for the overall cellular function, and is maintained over different growth conditions. Thus, upon environmental challenges that independently affect transcription or translation, bacteria quickly coordinate the kinetics of the unaffected process accordingly (see section Coordination of CTT). The radically different CTT regimes showed by different organisms (Johnson et al., 2020; O’Reilly et al., 2020; Wang et al., 2020b; Webster et al., 2020) reflect the underlying diversity in gene expression regulatory mechanisms among different species. Hence, further studying the physiological and molecular factors that mediate and regulate CTT is paramount. Of note, a transcriptome-wide assessment of the actual occurrence of CTT remains pending.
As a prokaryotic-specific phenomenon, CTT arises as an interesting target for the development of antimicrobial therapeutics and, for instance, targeting (p)ppGpp-mediated CTT regulation shows great potential. Alarmone-deficient strains are metabolically compromised but viable, so, unlike compounds that target essential targets, targeting (p)ppGpp metabolism may greatly reduce cell viability without exerting the selective pressure that leads to the arousal of antibiotic resistance (Hauryliuk et al., 2015; Syal et al., 2017).
Then again, studying evolutionarily shaped mechanisms that promote UTT (section Mechanisms Potentially Enabling UTT) can offer hints of how to purposely disrupt CTT under conditions that it is essential. Most likely, these mechanisms act in cooperation. In some cases, simultaneous action of UTT-promoting factors may be required, as suggested by the existence of RNA-mediated Hfq-CsrA complexes in E. coli (Caillet et al., 2019) and the partial overlap of Hfq and RsmA targetomes in P. aeruginosa (Gebhardt et al., 2020). Likewise, overexpression of S1 leads to the accumulation of ribosome-free cspE transcripts with increased stability, indicating that this population is subjected to simultaneous anti-translation and anti-RNase protection (Delvillani et al., 2011). RNPs that associate with RNAP, such as those mediated by IsrA sRNA (van Nues et al., 2015), could provide proper ground for such cooperation. Alternatively, UTT could be the result of tandem action of these factors, as in the case of the L. pneumophila RocC, a FinO-domain protein that protects the RocR sRNA from ribonucleolytic degradation, so that RocR subsequently represses translation of the competence operon (Attaiech et al., 2016). Similarly, cspE and cspC mRNAs are among the transcripts stabilized by the L4-mediated inhibition of RNase E (Singh et al., 2009), and cspE transcripts are also stabilized by S1 (Briani et al., 2008). Besides, ProQ also protects csp transcripts from RNases (Holmqvist et al., 2018). Thus, RPs and ProQ appear to be upstream activators of CSP-mediated UTT.
By now, the notion that prokaryotes have intricate intracellular organization is well-acknowledged (Rudner and Losick, 2010; Govindarajan and Amster-Choder, 2016; Surovtsev and Jacobs-Wagner, 2018). Consequently, prokaryotic UTT implies that the spatiotemporal separation between transcription and translation emerged earlier than the arousal of the eukaryotic cell. This is supported by the partial conservation of Rbp2 and Rbp3, RBPs mediating translation-independent RNA localization in cyanobacteria (Mahbub et al., 2020), in the eukaryotic algae Chlamydomonas, where they also bind and localize mRNAs to specific sites where local translation takes place (Uniacke and Zerges, 2009). Hence, it is likely that certain RNA-localizing pathways operating in eukaryotes originated from prokaryotic CTT-disrupting mechanisms. Further investigating these events in prokaryotes may unmask novel mechanisms operating at a suborganellar scale in eukaryotes.
In the past, researchers have drawn a clear line between transcriptional and post-transcriptional regulatory mechanisms in bacteria. In the light of the emerging evidence, we posit that many post-transcriptional regulatory mechanisms come into action on nascent transcripts and that their cotranscriptionality has so far been ignored. As already recognized in eukaryotes (Choder, 2011), factors acting cotranscriptionally can determine transcript fate in prokaryotes as well. Half a century after Miller’s famous micrographs (Miller et al., 1970), we foresee that the study of prokaryotic CTT and UTT will doubtlessly produce novel insights that will reshape our understanding of prokaryotic gene expression and subcellular organization.
Author Contributions
MIO and OAC wrote the original draft and edited the manuscript. OAC coordinated the work and acquired funding. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities (1274/19) and the Deutsch–Israeli Project Cooperation (AM 441/1-1 SO 568/1-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate helpful discussions with members of the OAC group. Figures were generated with BioRender.
References
Al-Husini, N., Tomares, D. T., Bitar, O., Childers, W. S., and Schrader, J. M. (2018). α-Proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell 71, 1027–1039.e14. doi: 10.1016/j.molcel.2018.08.003
Al-Husini, N., Tomares, D. T., Pfaffenberger, Z. J., Muthunayake, N. S., Samad, M. A., Zuo, T., et al. (2020). BR-bodies provide selectively permeable condensates that stimulate mRNA decay and prevent release of decay intermediates. Mol. Cell 78, 670–682.e8. doi: 10.1016/j.molcel.2020.04.001
Andrade, M. O., Farah, C. S., and Wang, N. (2014). The post-transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5' UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas. PLoS Pathog. 10:e1003945. doi: 10.1371/journal.ppat.1003945
Arthur, D. C., Edwards, R. A., Tsutakawa, S., Tainer, J. A., Frost, L. S., and Glover, J. N. M. (2011). Mapping interactions between the RNA chaperone FinO and its RNA targets. Nucleic Acids Res. 39, 4450–4463. doi: 10.1093/nar/gkr025
Artsimovitch, I., Svetlov, V., Anthony, L., Burgess, R. R., and Landick, R. (2000). RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J. Bacteriol. 182, 6027–6035. doi: 10.1128/JB.182.21.6027-6035.2000
Aseev, L. V., and Boni, I. V. (2011). Extraribosomal functions of bacterial ribosomal proteins. Mol. Biol. 45, 739–750. doi: 10.1134/S0026893311050025
Attaiech, L., Boughammoura, A., Brochier-Armanet, C., Allatif, O., Peillard-Fiorente, F., Edwards, R. A., et al. (2016). Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc. Natl. Acad. Sci. U. S. A. 113, 8813–8818. doi: 10.1073/pnas.1601626113
Azam, T. A., Hiraga, S., and Ishihama, A. (2000). Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells 5, 613–626. doi: 10.1046/j.1365-2443.2000.00350.x
Azam, M. S., and Vanderpool, C. K. (2018). Translational regulation by bacterial small RNAs via an unusual Hfq-dependent mechanism. Nucleic Acids Res. 46, 2585–2599. doi: 10.1093/nar/gkx1286
Bae, W., Phadtare, S., Severinov, K., and Inouye, M. (1999). Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol. Microbiol. 31, 1429–1441. doi: 10.1046/j.1365-2958.1999.01284.x
Bae, W., Xia, B., Inouye, M., and Severinov, K. (2000). Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. U. S. A. 97, 7784–7789. doi: 10.1073/pnas.97.14.7784
Baker, C. S., Eöry, L. A., Yakhnin, H., Mercante, J., Romeo, T., and Babitzke, P. (2007). CsrA inhibits translation initiation of Escherichia coli Hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol. 189, 5472–5481. doi: 10.1128/JB.00529-07
Bakshi, S., Choi, H., Mondal, J., and Weisshaar, J. C. (2014). Time-dependent effects of transcription- and translation-halting drugs on the spatial distributions of the Escherichia coli chromosome and ribosomes. Mol. Microbiol. 94, 871–887. doi: 10.1111/mmi.12805
Bakshi, S., Siryaporn, A., Goulian, M., and Weisshaar, J. C. (2012). Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 85, 21–38. doi: 10.1111/j.1365-2958.2012.08081.x
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. doi: 10.1038/nrm.2017.7
Bauriedl, S., Gerovac, M., Heidrich, N., Bischler, T., Barquist, L., Vogel, J., et al. (2020). The minimal meningococcal ProQ protein has an intrinsic capacity for structure-based global RNA recognition. Nat. Commun. 11:2823. doi: 10.1038/s41467-020-16650-6
Bayas, C. A., Wang, J., Lee, M. K., Schrader, J. M., Shapiro, L., and Moerner, W. E. (2018). Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 115, E3721–E3721. doi: 10.1073/pnas.1721648115
Benhalevy, D., Biran, I., Bochkareva, E. S., Sorek, R., and Bibi, E. (2017). Evidence for a cytoplasmic pool of ribosome-free mRNAs encoding inner membrane proteins in Escherichia coli. PLoS One 12:e0183862. doi: 10.1371/journal.pone.0183862
Benhalevy, D., Bochkareva, E. S., Biran, I., and Bibi, E. (2015). Model uracil-rich RNAs and membrane protein mRNAs interact specifically with cold shock proteins in Escherichia coli. PLoS One 10:e0134413. doi: 10.1371/journal.pone.0134413
Bhavsar, R. B., Makley, L. N., and Tsonis, P. A. (2010). The other lives of ribosomal proteins. Hum. Genomics 4, 327–344. doi: 10.1186/1479-7364-4-5-327
Bladen, H. A., Byrne, R., Levin, J. G., and Nirenberg, M. W. (1965). An electron microscopic study of a DNA-ribosome complex formed in vitro. J. Mol. Biol. 11, 78–83. doi: 10.1016/S0022-2836(65)80172-3
Boni, I. V., Lsaeva, D. M., Musychenko, M. L., and Tzareva, N. V. (1991). Ribosome-messenger recognition: MRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19, 155–162. doi: 10.1093/nar/19.1.155
Borgnia, M. J., Subramaniam, S., and Milne, J. L. S. (2008). Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J. Bacteriol. 190, 2588–2596. doi: 10.1128/JB.01538-07
Bouvet, P., and Wolffe, A. P. (1994). A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77, 931–941. doi: 10.1016/0092-8674(94)90141-4
Bowman, G. R., Comolli, L. R., Gaietta, G. M., Fero, M., Hong, S. H., Jones, Y., et al. (2010). Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol. Microbiol. 76, 173–189. doi: 10.1111/j.1365-2958.2010.07088.x
Breaker, R. R. (2018). Riboswitches and translation control. Cold Spring Harb. Perspect. Biol. 10:a032797. doi: 10.1101/cshperspect.a032797
Briani, F., Curti, S., Rossi, F., Carzaniga, T., Mauri, P., and Dehò, G. (2008). Polynucleotide phosphorylase hinders mRNA degradation upon ribosomal protein S1 overexpression in Escherichia coli. RNA 14, 2417–2429. doi: 10.1261/rna.1123908
Budkina, K. S., Zlobin, N. E., Kononova, S. V., Ovchinnikov, L. P., and Babakov, A. V. (2020). Cold shock domain proteins: structure and interaction with nucleic acids. Biochemistry (Mosc) 85, S1–S19. doi: 10.1134/S0006297920140011
Burmann, B. M., Knauer, S. H., Sevostyanova, A., Schweimer, K., Mooney, R. A., Landick, R., et al. (2012). An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 150, 291–303. doi: 10.1016/j.cell.2012.05.042
Burmann, B. M., Schweimer, K., Luo, X., Wahl, M. C., Stitt, B. L., Gottesman, M. E., et al. (2010). A NusE:NusG complex links transcription and translation. Science 328, 501–504. doi: 10.1126/science.1184953
Byrne, R., Levin, J. G., Bladen, H. A., and Nirenberg, M. W. (1964). The in vitro formation of a DNA-ribosome complex. Proc. Natl. Acad. Sci. U. S. A. 52, 140–148. doi: 10.1073/pnas.52.1.140
Caillet, J., Baron, B., Boni, I. V., Caillet-Saguy, C., and Hajnsdorf, E. (2019). Identification of protein-protein and ribonucleoprotein complexes containing Hfq. Sci. Rep. 9:14054. doi: 10.1038/s41598-019-50562-w
Castellana, M., and Wingreen, N. S. (2016). Spatial organization of bacterial transcription and translation. Proc. Natl. Acad. Sci. U. S. A. 113, 9286–9291. doi: 10.1073/pnas.1604995113
Chai, Q., Singh, B., Peisker, K., Metzendorf, N., Ge, X., Dasgupta, S., et al. (2014). Organization of ribosomes and nucleoids in Escherichia coli cells during growth and in quiescence. J. Biol. Chem. 289, 11342–11352. doi: 10.1074/jbc.M114.557348
Chalissery, J., Muteeb, G., Kalarickal, N. C., Mohan, S., Jisha, V., and Sen, R. (2011). Interaction surface of the transcription terminator rho required to form a complex with the C-terminal domain of the antiterminator NusG. J. Mol. Biol. 405, 49–64. doi: 10.1016/j.jmb.2010.10.044
Chao, Y., Li, L., Girodat, D., Förstner, K. U., Said, N., Corcoran, C., et al. (2017). In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell 65, 39–51. doi: 10.1016/j.molcel.2016.11.002
Chen, M., and Fredrick, K. (2018). Measures of single-versus multiple-round translation argue against a mechanism to ensure coupling of transcription and translation. Proc. Natl. Acad. Sci. U. S. A. 115, 10774–10779. doi: 10.1073/pnas.1812940115
Chen, M., and Fredrick, K. (2020). RNA polymerase’s relationship with the ribosome: not so physical, most of the time. J. Mol. Biol. 432, 3981–3986. doi: 10.1016/j.jmb.2020.03.018
Chen, J., and Gottesman, S. (2017). Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev. 31, 1382–1395. doi: 10.1101/gad.302547.117
Chen, H., Shiroguchi, K., Ge, H., and Xie, X. S. (2015). Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 11:781. doi: 10.15252/msb.20145794
Choder, M. (2011). mRNA imprinting: additional level in the regulation of gene expression. Cell Logist. 1, 37–40. doi: 10.4161/cl.1.1.14465
Conn, A. B., Diggs, S., Tam, T. K., and Blaha, G. M. (2019). Two old dogs, one new trick: a review of rna polymerase and ribosome interactions during transcription-translation coupling. Int. J. Mol. Sci. 20:2595. doi: 10.3390/ijms20102595
Cougot, N., Molza, A. E., Delesques, J., Giudice, E., Cavalier, A., Rolland, J. P., et al. (2014). Visualizing compaction of polysomes in bacteria. J. Mol. Biol. 426, 377–388. doi: 10.1016/j.jmb.2013.09.035
Dahan, O., Gingold, H., and Pilpel, Y. (2011). Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet. 27, 316–322. doi: 10.1016/j.tig.2011.05.008
Dai, X., Zhu, M., Warren, M., Balakrishnan, R., Patsalo, V., Okano, H., et al. (2016). Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2:16231. doi: 10.1038/nmicrobiol.2016.231
Darfeuille, F., Unoson, C., Vogel, J., and Wagner, E. G. H. (2007). An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell 26, 381–392. doi: 10.1016/j.molcel.2007.04.003
Deana, A., and Belasco, J. G. (2005). Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19, 2526–2533. doi: 10.1101/gad.1348805
Decker, C. J., and Parker, R. (2012). P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4:a012286. doi: 10.1101/cshperspect.a012286
Delvillani, F., Papiani, G., Dehò, G., and Briani, F. (2011). S1 ribosomal protein and the interplay between translation and mRNA decay. Nucleic Acids Res. 39, 7702–7715. doi: 10.1093/nar
Demo, G., Rasouly, A., Vasilyev, N., Svetlov, V., Loveland, A. B., Diaz-Avalos, R., et al. (2017). Structure of RNA polymerase bound to ribosomal 30S subunit. elife 6:e28560. doi: 10.7554/eLife.28560
Deneke, C., Lipowsky, R., and Valleriani, A. (2013). Effect of ribosome shielding on mRNA stability. Phys. Biol. 10:46008. doi: 10.1088/1478-3975/10/4/046008
Desnoyers, G., and Massé, E. (2012). Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev. 26, 726–739. doi: 10.1101/gad.182493.111
Dugar, G., Svensson, S. L., Bischler, T., Wäldchen, S., Reinhardt, R., Sauer, M., et al. (2016). The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat. Commun. 7:11667. doi: 10.1038/ncomms11667
Durand, S., Braun, F., Helfer, A. C., Romby, P., and Condon, C. (2017). sRNA-mediated activation of gene expression by inhibition of 5'-3' exonucleolytic mRNA degradation. elife 6:e23602. doi: 10.7554/eLife.23602
Dutta, D., Shatalin, K., Epshtein, V., Gottesman, M. E., and Nudler, E. (2011). Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146, 533–543. doi: 10.1016/j.cell.2011.07.034
Edwards, A. N., Patterson-Fortin, L. M., Vakulskas, C. A., Mercante, J. W., Potrykus, K., Vinella, D., et al. (2011). Circuitry linking the Csr and stringent response global regulatory systems. Mol. Microbiol. 80, 1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x
Elgamal, S., Artsimovitch, I., and Ibba, M. (2016). Maintenance of transcription-translation coupling by elongation factor P. mBio2 7:e01373-16. doi: 10.1101/gad.196741.112
Ellis, M. J., Trussler, R. S., and Haniford, D. B. (2015). Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Mol. Microbiol. 96, 633–650. doi: 10.1111/mmi.12961
El-Sharoud, W. M., and Graumann, P. L. (2007). Cold shock proteins aid coupling of transcription and translation in bacteria. Sci. Prog. 90, 15–27. doi: 10.3184/003685007780440549
Endesfelder, U., Finan, K., Holden, S. J., Cook, P. R., Kapanidis, A. N., and Heilemann, M. (2013). Multiscale spatial organization of RNA polymerase in Escherichia coli. Biophys. J. 105, 172–181. doi: 10.1016/j.bpj.2013.05.048
Epshtein, V., Dutta, D., Wade, J., and Nudler, E. (2010). An allosteric mechanism of rho-dependent transcription termination. Nature 463, 245–249. doi: 10.1038/nature08669
Epshtein, V., and Nudler, E. (2003). Cooperation between RNA polymerase molecules in transcription elongation. Science 300, 801–805. doi: 10.1126/science.1083219
Epshtein, V., Toulmé, F., Rachid Rahmouni, A., Borukhov, S., and Nudler, E. (2003). Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 22, 4719–4727. doi: 10.1093/emboj/cdg452
Ermolenko, D. N., and Makhatadze, G. I. (2002). Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59, 1902–1913. doi: 10.1007/PL00012513
Esquerré, T., Bouvier, M., Turlan, C., Carpousis, A. J., Girbal, L., and Cocaign-Bousquet, M. (2016). The Csr system regulates genome-wide mRNA stability and transcription and thus gene expression in Escherichia coli. Sci. Rep. 6:25057. doi: 10.1038/srep25057
Etchegaray, J. P., and Inouye, M. (1999). CspA, CspB, and CspG, major cold shock proteins of Escherichia coli, are induced at low temperature under conditions that completely block protein synthesis. J. Bacteriol. 181, 1827–1830. doi: 10.1128/jb.181.6.1827-1830.1999
Fan, H., Conn, A. B., Williams, P. B., Diggs, S., Hahm, J., Gamper, H. B., et al. (2017). Transcription–translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Res. 37, 5578–5588. doi: 10.1093/nar/gkx719
Fang, L., Jiang, W., Bae, W., and Inouye, M. (1997). Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol. Microbiol. 23, 355–364. doi: 10.1046/j.1365-2958.1997.2351592.x
Farnham, P. J., Greenblatt, J., and Platt, T. (1982). Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell 29, 945–951. doi: 10.1016/0092-8674(82)90457-3
Feng, Y., Huang, H., Liao, J., and Cohen, S. N. (2001). Escherichia coli poly(A)-binding proteins that interact with components of degradosomes or impede RNA decay mediated by polynucleotide phosphorylase and RNase E. J. Biol. Chem. 276, 31651–31656. doi: 10.1074/jbc.M102855200
Fisher, J. K., Bourniquel, A., Witz, G., Weiner, B., Prentiss, M., and Kleckner, N. (2013). Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153, 882–895. doi: 10.1016/j.cell.2013.04.006
Folichon, M., Arluison, V., Pellegrini, O., Huntzinger, E., Régnier, P., and Hajnsdorf, E. (2003). The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 31, 7302–7310. doi: 10.1093/nar/gkg915
Franch, T., and Gerdes, K. (1996). Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol. Microbiol. 21, 1049–1060. doi: 10.1046/j.1365-2958.1996.771431.x
French, S. L., Santangelo, T. J., Beyer, A. L., and Reeve, J. N. (2007). Transcription and translation are coupled in Archaea. Mol. Biol. Evol. 24, 893–895. doi: 10.1093/molbev/msm007
Frieda, K. L., and Block, S. M. (2012). Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338, 397–400. doi: 10.1126/science.1225722
Fröhlich, K. S., Papenfort, K., Fekete, A., and Vogel, J. (2013). A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 32, 2963–2979. doi: 10.1038/emboj.2013.222
Gaal, T., Bratton, B. P., Sanchez-Vazquez, P., Sliwicki, A., Sliwicki, K., Vegel, A., et al. (2016). Colocalization of distant chromosomal loci in space in E. coli: a bacterial nucleolus. Genes Dev. 30, 2272–2285. doi: 10.1101/gad.290312.116
Gebhardt, M. J., Kambara, T. K., Ramsey, K. M., and Dove, S. L. (2020). Widespread targeting of nascent transcripts by RsmA in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 117, 10520–10529. doi: 10.1073/pnas.1917587117
Giuliodori, A. M., Di Pietro, F., Marzi, S., Masquida, B., Wagner, R., Romby, P., et al. (2010). The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 37, 21–33. doi: 10.1016/j.molcel.2009.11.033
Golomb, M., and Chamberlin, M. (1974). Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J. Biol. Chem. 249, 2858–2863.
Goodson, J. R., Klupt, S., Zhang, C., Straight, P., and Winkler, W. C. (2017). LoaP is a broadly conserved antiterminator protein that regulates antibiotic gene clusters in Bacillus amyloliquefaciens. Nat. Microbiol. 2:17003. doi: 10.1038/nmicrobiol.2017.3
Goodson, J. R., and Winkler, W. C. (2018). Processive antitermination. Microbiol. Spectr. 6, 1–18. doi: 10.1128/microbiolspec.rwr-0031-2018
Govindarajan, S., and Amster-Choder, O. (2016). Where are things inside a bacterial cell? Curr. Opin. Microbiol. 33, 83–90. doi: 10.1016/j.mib.2016.07.003
Gowrishankar, J., and Harinarayanan, R. (2004). Why is transcription coupled to translation in bacteria? Mol. Microbiol. 54, 598–603. doi: 10.1111/j.1365-2958.2004.04289.x
Graumann, P., Wendrich, T. M., Weber, M. H. W., Schröder, K., and Marahiel, M. A. (1997). A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 25, 741–756. doi: 10.1046/j.1365-2958.1997.5121878.x
Gray, W. T., Govers, S. K., Xiang, Y., Parry, B. R., Campos, M., Kim, S., et al. (2019). Nucleoid size scaling and intracellular organization of translation across bacteria. Cell 177, 1632–1648.e20. doi: 10.1016/j.cell.2019.05.017
Gultyaev, A. P., Franch, T., and Gerdes, K. (1997). Programmed cell death by hok/sok of plasmid R1: coupled nucleotide covariations reveal a phylogenetically conserved folding pathway in the hok family of mRNAs. J. Mol. Biol. 273, 26–37. doi: 10.1006/jmbi.1997.1295
Guo, X., Myasnikov, A. G., Chen, J., Crucifix, C., Papai, G., Takacs, M., et al. (2018). Structural basis for NusA stabilized transcriptional pausing. Mol. Cell 69, 816–827.e4. doi: 10.1016/j.molcel.2018.02.008
Guo, Z., and Noller, H. F. (2012). Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc. Natl. Acad. Sci. U. S. A. 109, 20391–20394. doi: 10.1073/pnas.1218999109
Gutiérrez, P., Kozlov, G., Gabrielli, L., Elias, D., Osborne, M. J., Gallouzi, I. E., et al. (2007). Solution structure of YaeO, a rho-specific inhibitor of transcription termination. J. Biol. Chem. 282, 23348–23353. doi: 10.1074/jbc.M702010200
Hadjeras, L., Poljak, L., Bouvier, M., Morin-Ogier, Q., Canal, I., Cocaign-Bousquet, M., et al. (2019). Detachment of the RNA degradosome from the inner membrane of Escherichia coli results in a global slowdown of mRNA degradation, proteolysis of RNase E and increased turnover of ribosome-free transcripts. Mol. Microbiol. 111, 1715–1731. doi: 10.1111/mmi.14248
Hajnsdorf, E., and Régnier, P. (2000). Host factor Hfq of Escherichia coli stimulates elongation of poly(a) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. U. S. A. 97, 1501–1505. doi: 10.1073/pnas.040549897
Hanna, M. M., and Liu, K. (1998). Nascent RNA in transcription complexes interacts with CspE, a small protein in E. coli implicated in chromatin condensation. J. Mol. Biol. 282, 227–239. doi: 10.1006/jmbi.1998.2005
Hao, Z., Epshtein, V., Kim, K. H., Proshkin, S., Svetlov, V., Kamarthapu, V., et al. (2020). Pre-termination transcription complex: structure and function. Mol. Cell 81, 1–12. doi: 10.1016/j.molcel.2020.11.013
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
Heberling, T., Davis, L., Gedeon, J., Morgan, C., and Gedeon, T. (2016). A mechanistic model for cooperative behavior of co-transcribing RNA polymerases. PLoS Comput. Biol. 12:e1005069. doi: 10.1371/journal.pcbi.1005069
Heidrich, N., Chinali, A., Gerth, U., and Brantl, S. (2006). The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 62, 520–536. doi: 10.1111/j.1365-2958.2006.05384.x
Heidrich, N., Moll, I., and Brantl, S. (2007). In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 35, 4331–4346. doi: 10.1093/nar/gkm439
Herskovits, A. A., and Bibi, E. (2000). Association of Escherichia coli ribosomes with the inner membrane requires the signal recognition particle receptor but is independent of the signal recognition particle. Proc. Natl. Acad. Sci. U. S. A. 97, 4621–4626. doi: 10.1073/pnas.080077197
Hobot, J. A., Villiger, W., Escaig, J., Maeder, M., Ryter, A., and Kellenberger, E. (1985). Shape and fine structure of nucleoids observed on sections of ultrarapidly frozen and cryosubstituted bacteria. J. Bacteriol. 162, 960–971. doi: 10.1128/JB.162.3.960-971.1985
Hoffmann, S. A., Hao, N., Shearwin, K. E., and Arndt, K. M. (2019). Characterizing transcriptional interference between converging genes in bacteria. ACS Synth. Biol. 8, 466–473. doi: 10.1021/acssynbio.8b00477
Holmqvist, E., Berggren, S., and Rizvanovic, A. (2020). RNA-binding activity and regulatory functions of the emerging sRNA-binding protein ProQ. Biochim. Biophys. Acta – Gene Regul. Mech. 1863:194596. doi: 10.1016/j.bbagrm.2020.194596
Holmqvist, E., Li, L., Bischler, T., Barquist, L., and Vogel, J. (2018). Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3' ends. Mol. Cell 70, 971–982.e6. doi: 10.1016/j.molcel.2018.04.017
Holmqvist, E., and Vogel, J. (2018). RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 16, 601–615. doi: 10.1038/s41579-018-0049-5
Holmqvist, E., Wright, P. R., Li, L., Bischler, T., Barquist, L., Reinhardt, R., et al. (2016). Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011. doi: 10.15252/embj.201593360
Hör, J., Di Giorgio, S., Gerovac, M., Venturini, E., Förstner, K. U., and Vogel, J. (2020). Grad-seq shines light on unrecognized RNA and protein complexes in the model bacterium Escherichia coli. Nucleic Acids Res. 48, 9301–9319. doi: 10.1093/nar/gkaa676
Horn, G., Hofweber, R., Kremer, W., and Kalbitzer, H. R. (2007). Structure and function of bacterial cold shock proteins. Cell. Mol. Life Sci. 64, 1457–1470. doi: 10.1007/s00018-007-6388-4
Iost, I., and Dreyfus, M. (1995). The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 14, 3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x
Irastortza-Olaziregi, M., and Amster-Choder, O. (2020). RNA localization in prokaryotes: where, when, how, and why. Wiley Interdiscip. Rev. RNA, e1615. doi: 10.1002/wrna.1615
Irie, Y., Starkey, M., Edwards, A. N., Wozniak, D. J., Romeo, T., and Parsek, M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172. doi: 10.1111/j.1365-2958.2010.07320.x
Irnov, I., and Winkler, W. C. (2010). A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol. Microbiol. 76, 559–575. doi: 10.1111/j.1365-2958.2010.07131.x
Iyer, S., Le, D., Park, B. R., and Kim, M. (2018). Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli. Nat. Microbiol. 3, 741–748. doi: 10.1038/s41564-018-0161-3
Jerome, L. J., van Biesen, T., and Frost, L. S. (1999). Degradation of FinP antisense RNA from F-like plasmids: the RNA-binding protein, FinO, protects FinP from ribonuclease E. J. Mol. Biol. 285, 1457–1473. doi: 10.1006/jmbi.1998.2404
Jiang, W., Hou, Y., and Inouye, M. (1997). CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272, 196–202. doi: 10.1074/jbc.272.1.196
Jin, D. J., Cagliero, C., and Zhou, Y. N. (2013). Role of RNA polymerase and transcription in the organization of the bacterial nucleoid. Chem. Rev. 113, 8662–8682. doi: 10.1021/cr4001429
Johansson, J. (2009). RNA thermosensors in bacterial pathogens. Contrib. Microbiol. 16, 150–160. doi: 10.1159/000219378
Johnson, G. E., Lalanne, J. -B., Peters, M. L., and Li, G. -W. (2020). Functionally uncoupled transcription–translation in Bacillus subtilis. Nature 585, 124–128. doi: 10.1038/s41586-020-2638-5
Johnson, J. E., Reyes, F. E., Polaski, J. T., and Batey, R. T. (2012). B12 cofactors directly stabilize an mRNA regulatory switch. Nature 492, 133–137. doi: 10.1038/nature11607
Jonas, K., Edwards, A. N., Ahmad, I., Romeo, T., Römling, U., and Melefors, Ö. (2010). Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella typhimurium. Environ. Microbiol. 12, 524–540. doi: 10.1111/j.1462-2920.2009.02097.x
Joyeux, M. (2016). In vivo compaction dynamics of bacterial DNA: a fingerprint of DNA/RNA demixing? Curr. Opin. Colloid Interface Sci. 26, 17–27. doi: 10.1016/j.cocis.2016.08.005
Joyeux, M. (2018). A segregative phase separation scenario of the formation of the bacterial nucleoid. Soft Matter 14, 7368–7381. doi: 10.1039/c8sm01205a
Kaberdin, V. R., and Bläsi, U. (2006). Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 30, 967–979. doi: 10.1111/j.1574-6976.2006.00043.x
Kalapos, M. P., Paulus, H., and Sarkar, N. (1997). Identification of ribosomal protein S1 as a poly(a) binding protein in Escherichia coli. Biochimie 79, 493–502. doi: 10.1016/S0300-9084(97)82741-1
Kambara, T. K., Ramsey, K. M., and Dove, S. L. (2018). Pervasive targeting of nascent transcripts by Hfq. Cell Rep. 23, 1543–1552. doi: 10.1016/j.celrep.2018.03.134
Kang, J. Y., Mishanina, T. V., Bellecourt, M. J., Mooney, R. A., Darst, S. A., and Landick, R. (2018a). RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell 69, 802–815.e1. doi: 10.1016/j.molcel.2018.01.018
Kang, J. Y., Mooney, R. A., Nedialkov, Y., Saba, J., Mishanina, T. V., Artsimovitch, I., et al. (2018b). Structural basis for transcript elongation control by NusG family universal regulators. Cell 173, 1650–1662. doi: 10.1016/j.cell.2018.05.017
Kannaiah, S., and Amster-Choder, O. (2014). Protein targeting via mRNA in bacteria. Biochim. Biophys. Acta, Mol. Cell Res. 1843, 1457–1465. doi: 10.1016/j.bbamcr.2013.11.004
Kannaiah, S., Livny, J., and Amster-Choder, O. (2019). Spatiotemporal organization of the E. coli transcriptome: translation independence and engagement in regulation. Mol. Cell 76, 574–589.e7. doi: 10.1016/j.molcel.2019.08.013
Katsowich, N., Elbaz, N., Pal, R. R., Mills, E., Kobi, S., Kahan, T., et al. (2017). Host cell attachment elicits posttranscriptional regulation in infecting enteropathogenic bacteria. Science 355, 735–739. doi: 10.1126/science.aah4886
Kavita, K., de Mets, F., and Gottesman, S. (2018). New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 42, 53–61. doi: 10.1016/j.mib.2017.10.014
Khong, A., and Parker, R. (2020). The landscape of eukaryotic mRNPs. RNA 26, 229–239. doi: 10.1261/rna.073601.119
Khosa, S., Scholz, R., Schwarz, C., Trilling, M., Hengel, H., Jaeger, K. E., et al. (2018). An A/U-rich enhancer region is required for high-level protein secretion through the HlyA type I secretion system. Appl. Environ. Microbiol. 84:e01163-17. doi: 10.1128/AEM.01163-17
Kim, S., Beltran, B., Irnov, I., and Jacobs-Wagner, C. (2019b). Long-distance cooperative and antagonistic RNA polymerase dynamics via DNA supercoiling. Cell 179, 106–119.e16. doi: 10.1016/j.cell.2019.08.033
Kim, J., Goñi-Moreno, A., Calles, B., and de Lorenzo, V. (2019a). Spatial organization of the gene expression hardware in Pseudomonas putida. Environ. Microbiol. 21, 1645–1658. doi: 10.1111/1462-2920.14544
Kohler, R., Mooney, R. A., Mills, D. J., Landick, R., and Cramer, P. (2017). Architecture of a transcribing-translating expressome. Science 356, 194–197. doi: 10.1126/science.aal3059
Komarova, A. V., Tchufistova, L. S., Dreyfus, M., and Boni, I. V. (2005). AU-rich sequences within 5' untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 187, 1344–1349. doi: 10.1128/JB.187.4.1344-1349.2005
Komarova, A. V., Tchufistova, L. S., Supina, E. V., and Boni, I. V. (2002). Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA 8, 1137–1147. doi: 10.1017/S1355838202029990
Kortmann, J., and Narberhaus, F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265. doi: 10.1038/nrmicro2730
Kriner, M. A., and Groisman, E. A. (2015). The bacterial transcription termination factor rho coordinates Mg2+ homeostasis with translational signals. J. Mol. Biol. 427, 3834–3849. doi: 10.1016/j.jmb.2015.10.020
Kristiansen, K. I., Weel-Sneve, R., Booth, J. A., and Bjørås, M. (2016). Mutually exclusive RNA secondary structures regulate translation initiation of DinQ in Escherichia coli. RNA 22, 1739–1749. doi: 10.1261/rna.058461.116
Kurkela, J., Fredman, J., Salminen, T. A., and Tyystjärvi, T. (2020). Revealing secrets of the enigmatic omega subunit of bacterial RNA polymerase. Mol. Microbiol. mmi.14603. doi: 10.1111/mmi.14603
Ladouceur, A. -M., Parmar, B., Biedzinski, S., Wall, J., Tope, S. G., Cohn, D., et al. (2020). Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc. Natl. Acad. Sci. U. S. A. 117, 18540–18549. doi: 10.1101/2020.03.16.994491
Larson, M. H., Mooney, R. A., Peters, J. M., Windgassen, T., Nayak, D., Gross, C. A., et al. (2014). A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1042–1047. doi: 10.1126/science.1251871
Lawson, M. R., Ma, W., Bellecourt, M. J., Artsimovitch, I., Martin, A., Landick, R., et al. (2018). Mechanism for the regulated control of bacterial transcription termination by a universal adaptor protein. Mol. Cell 71, 911–922.e4. doi: 10.1016/j.molcel.2018.07.014
Le Derout, J., Folichon, M., Briani, F., Dehò, G., Régnier, P., and Hajnsdorf, E. (2003). Hfq affects the length and the frequency of short oligo(A) tails at the 3' end of Escherchia coli rpsO mRNAs. Nucleic Acids Res. 31, 4017–4023. doi: 10.1093/nar/gkg456
Le, T. T., and Wang, M. D. (2018). Molecular highways—navigating collisions of DNA motor proteins. J. Mol. Biol. 430, 4513–4524. doi: 10.1016/j.jmb.2018.08.006
Lewis, P. J., Thaker, S. D., and Errington, J. (2000). Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19, 710–718. doi: 10.1093/emboj/19.4.710
Li, S. H. J., Li, Z., Park, J. O., King, C. G., Rabinowitz, J. D., Wingreen, N. S., et al. (2018). Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 3, 939–947. doi: 10.1038/s41564-018-0199-2
Li, R., Zhang, Q., Li, J., and Shi, H. (2015). Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the rho-independent terminator. Nucleic Acids Res. 44, 2554–2563. doi: 10.1093/nar/gkv1285
Libby, E. A., Roggiani, M., and Goulian, M. (2012). Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc. Natl. Acad. Sci. U. S. A. 109, 7445–7450. doi: 10.1073/pnas.1109479109
Lindahl, L., Archer, R., and Zengel, J. M. (1983). Transcription of the s10 ribosomal protein operon is regulated by an attenuator in the leader. Cell 33, 241–248. doi: 10.1016/0092-8674(83)90353-7
Lopez, M. M., Yutani, K., and Makhatadze, G. I. (1999). Interactions of the major cold shock protein of Bacillus subtilis CspB with single-stranded DNA templates of different base composition. J. Biol. Chem. 274, 33601–33608. doi: 10.1074/jbc.274.47.33601
Magán, A., Amman, F., El-Isa, F., Hartl, N., Shamovsky, I., Nudler, E., et al. (2019). iRAPs curb antisense transcription in E. coli. Nucleic Acids Res. 47, 10894–10905. doi: 10.1093/nar/gkz791
Mahbub, M., Hemm, L., Yang, Y., Kaur, R., Carmen, H., Engl, C., et al. (2020). mRNA localization, reaction centre biogenesis and thylakoid membrane targeting in cyanobacteria. Nat. Plants 6, 1179–1191. doi: 10.1038/s41477-020-00764-2
Makarova, O. V., Makarov, E. M., Sousa, R., and Dreyfus, M. (1995). Transcribing of Escherichia coli genes with mutant T7 RNA polymerases: stability of lacZ mRNA inversely correlates with polymerase speed. Proc. Natl. Acad. Sci. U. S. A. 92, 12250–12254. doi: 10.1073/pnas.92.26.12250
Malabirade, A., Morgado-Brajones, J., Trépout, S., Wien, F., Marquez, I., Seguin, J., et al. (2017). Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep. 7:10724. doi: 10.1038/s41598-017-11157-5
Maquat, L. E., Tarn, W. Y., and Isken, O. (2010). The pioneer round of translation: features and functions. Cell 142, 368–374. doi: 10.1016/j.cell.2010.07.022
Masachis, S., and Darfeuille, F. (2018). Type I toxin-antitoxin systems: regulating toxin expression via Shine-Dalgarno sequence sequestration and small RNA binding. Microbiol. Spectr. 6, 1–18. doi: 10.1128/microbiolspec.rwr-0030-2018
Mascarenhas, J., Weber, M. H. W., and Graumann, P. L. (2001). Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2, 685–689. doi: 10.1093/embo-reports/kve160
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S., and Cashel, M. (2013). Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189. doi: 10.1093/nar/gkt302
Melamed, S., Adams, P. P., Zhang, A., Zhang, H., and Storz, G. (2020). RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol. Cell 77, 411–425.e7. doi: 10.1016/j.molcel.2019.10.022
Michaux, C., Holmqvist, E., Vasicek, E., Sharan, M., Barquist, L., Westermann, A. J., et al. (2017). RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc. Natl. Acad. Sci. U. S. A. 114, 6824–6829. doi: 10.1073/pnas.1620772114
Miller, O. L., Hamkalo, B. A., and Thomas, C. A. (1970). Visualization of bacterial genes in action. Science 169, 392–395. doi: 10.1126/science.169.3943.392
Milon, P., Tischenko, E., Tomšic, J., Caserta, E., Folkers, G., La Teana, A., et al. (2006). The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl. Acad. Sci. U. S. A. 103, 13962–13967. doi: 10.1073/pnas.0606384103
Mironov, A. S., Gusarov, I., Rafikov, R., Lopez, L. E., Shatalin, K., Kreneva, R. A., et al. (2002). Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756. doi: 10.1016/S0092-8674(02)01134-0
Mitkevich, V. A., Ermakov, A., Kulikova, A. A., Tankov, S., Shyp, V., Soosaar, A., et al. (2010). Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J. Mol. Biol. 402, 838–846. doi: 10.1016/j.jmb.2010.08.016
Moffitt, J. R., Pandey, S., Boettiger, A. N., Wang, S., and Zhuang, X. (2016). Spatial organization shapes the turnover of a bacterial transcriptome. elife 5:e13065. doi: 10.7554/eLife.13065
Mogridge, J., and Greenblatt, J. (1998). Specific binding of Escherichia coli ribosomal protein S1 to boxA transcriptional antiterminator RNA. J. Bacteriol. 180, 2248–2252. doi: 10.1128/jb.180.8.2248-2252.1998
Mohanty, B. K., and Kushner, S. R. (2006). The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 34, 5695–5704. doi: 10.1093/nar/gkl684
Mohanty, B. K., Maples, V. F., and Kushner, S. R. (2004). The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54, 905–920. doi: 10.1111/j.1365-2958.2004.04337.x
Mohapatra, S., Choi, H., Ge, X., Sanyal, S., and Weisshaar, J. C. (2017). Spatial distribution and ribosome-binding dynamics of EF-P in live Escherichia coli. MBio 8:e00300-17. doi: 10.1128/mbio.00300-17
Mohapatra, S., and Weisshaar, J. C. (2018). Functional mapping of the E. coli translational machinery using single-molecule tracking. Mol. Microbiol. 110, 262–282. doi: 10.1111/mmi.14103
Moll, I., Afonyushkin, T., Vytvytska, O., Kaberdin, V. R., and Bläsi, U. (2003). Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9, 1308–1314. doi: 10.1261/rna.5850703
Mondal, J., Bratton, B. P., Li, Y., Yethiraj, A., and Weisshaar, J. C. (2011). Entropy-based mechanism of ribosome-nucleoid segregation in E. coli cells. Biophys. J. 100, 2605–2613. doi: 10.1016/j.bpj.2011.04.030
Montero Llopis, P., Jackson, A. F., Sliusarenko, O., Surovtsev, I., Heinritz, J., Emonet, T., et al. (2010). Spatial organization of the flow of genetic information in bacteria. Nature 466, 77–81. doi: 10.1038/nature09152
Mooney, R. A., Davis, S. E., Peters, J. M., Rowland, J. L., Ansari, A. Z., and Landick, R. (2009a). Regulator trafficking on bacterial transcription units in vivo. Mol. Cell 33, 97–108. doi: 10.1016/j.molcel.2008.12.021
Mooney, R. A., Schweimer, K., Rösch, P., Gottesman, M., and Landick, R. (2009b). Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 391, 341–358. doi: 10.1016/j.jmb.2009.05.078
Morita, M. T., Tanaka, Y., Kodama, T. S., Kyogoku, Y., Yanagi, H., and Yura, T. (1999). Translational induction of heat shock transcription factor σ32: evidence for a built-in RNA thermosensor. Genes Dev. 13, 655–665. doi: 10.1101/gad.13.6.655
Müller, P., Gimpel, M., Wildenhain, T., and Brantl, S. (2019). A new role for CsrA: promotion of complex formation between an sRNA and its mRNA target in Bacillus subtilis. RNA Biol. 16, 972–987. doi: 10.1080/15476286.2019.1605811
Mustafi, M., and Weisshaar, J. C. (2018). Simultaneous binding of multiple EF-Tu copies to translating ribosomes in live Escherichia coli. MBio 9:e02143-17. doi: 10.1128/mBio.02143-17
Muthunayake, N. S., Tomares, D. T., Childers, W. S., and Schrader, J. M. (2020). Phase-separated bacterial ribonucleoprotein bodies organize mRNA decay. Wiley Interdiscip. Rev. RNA 11:e1599. doi: 10.1002/wrna.1599
Nevo-Dinur, K., Nussbaum-Shochat, A., Ben-Yehuda, S., and Amster-Choder, O. (2011). Translation-independent localization of mRNA in E. coli. Science 331, 1081–1084. doi: 10.1126/science.1195691
O’Reilly, F. J., Xue, L., Graziadei, A., Sinn, L., Lenz, S., Tegunov, D., et al. (2020). In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557. doi: 10.1101/2020.02.28.970111
Pal, K., Yadav, M., Jain, S., Ghosh, B., Sen, R., and Sen, U. (2019). Vibrio cholerae YaeO is a structural homologue of RNA chaperone Hfq that inhibits rho-dependent transcription termination by dissociating its hexameric state. J. Mol. Biol. 431, 4749–4766. doi: 10.1016/j.jmb.2019.09.019
Pannuri, A., Yakhnin, H., Vakulskas, C. A., Edwards, A. N., Babitzke, P., and Romeo, T. (2012). Translational repression of NhaR, a novel pathway for multi-tier regulation of biofilm circuitry by CsrA. J. Bacteriol. 194, 79–89. doi: 10.1128/JB.06209-11
Papenfort, K., Sun, Y., Miyakoshi, M., Vanderpool, C. K., and Vogel, J. (2013). Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153, 426–437. doi: 10.1016/j.cell.2013.03.003
Parry, B. R., Surovtsev, I. V., Cabeen, M. T., O’Hern, C. S., Dufresne, E. R., and Jacobs-Wagner, C. (2014). The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156, 183–194. doi: 10.1016/j.cell.2013.11.028
Paul, B. J., Ross, W., Gaal, T., and Gourse, R. L. (2004). rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38, 749–770. doi: 10.1146/annurev.genet.38.072902.091347
Pei, X. Y., Dendooven, T., Sonnleitner, E., Chen, S., Bläsi, U., and Luisi, B. F. (2019). Architectural principles for hfq/crc-mediated regulation of gene expression. elife 8:e43158. doi: 10.7554/eLife.43158
Persson, F., Lindén, M., Unoson, C., and Elf, J. (2013). Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat. Methods 10, 265–269. doi: 10.1038/nmeth.2367
Phadtare, S., and Inouye, M. (1999). Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 33, 1004–1014. doi: 10.1046/j.1365-2958.1999.01541.x
Phadtare, S., and Inouye, M. (2001). Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183, 1205–1214. doi: 10.1128/JB.183.4.1205-1214.2001
Phadtare, S., Inouye, M., and Severinov, K. (2002a). The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277, 7239–7245. doi: 10.1074/jbc.M111496200
Phadtare, S., and Severinov, K. (2005). Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 33, 5583–5590. doi: 10.1093/nar/gki859
Phadtare, S., Tyagi, S., Inouye, M., and Severinov, K. (2002b). Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277, 46706–46711. doi: 10.1074/jbc.M208118200
Pichoff, S., Alibaud, L., Guédant, A., Castanié, M. P., and Bouché, J. P. (1998). An Escherichia coli gene (yaeO) suppresses temperature-sensitive mutations in essential genes by modulating rho-dependent transcription termination. Mol. Microbiol. 29, 859–869. doi: 10.1046/j.1365-2958.1998.00981.x
Plochowietz, A., Farrell, I., Smilansky, Z., Cooperman, B. S., and Kapanidis, A. N. (2016). In vivo single-RNA tracking shows that most tRNA diffuses freely in live bacteria. Nucleic Acids Res. 45, 926–937. doi: 10.1093/nar/gkw787
Polissi, A., De Laurentis, W., Zangrossi, S., Briani, F., Longhi, V., Pesole, G., et al. (2003). Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154, 573–580. doi: 10.1016/S0923-2508(03)00167-0
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Potts, A. H., Vakulskas, C. A., Pannuri, A., Yakhnin, H., Babitzke, P., and Romeo, T. (2017). Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 8:1596. doi: 10.1038/s41467-017-01613-1
Pourciau, C., Pannuri, A., Potts, A., Yakhnin, H., Babitzke, P., and Romeo, T. (2019). Regulation of iron storage by CsrA supports exponential growth of Escherichia coli. MBio 10:e01034-19. doi: 10.1128/mBio.01034-19
Prilusky, J., and Bibi, E. (2009). Studying membrane proteins through the eyes of the genetic code revealed a strong uracil bias in their coding mRNAs. Proc. Natl. Acad. Sci. U. S. A. 106, 6662–6666. doi: 10.1073/pnas.0902029106
Proshkin, S., Rachid Rahmouni, A., Mironov, A., and Nudler, E. (2010). Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508. doi: 10.1126/science.1184939
Qayyum, M. Z., Dey, D., and Sen, R. (2016). Transcription elongation factor NusA is a general antagonist of rho-dependent termination in Escherichia coli. J. Biol. Chem. 291, 8090–8108. doi: 10.1074/jbc.M115.701268
Rabhi, M., Espéli, O., Schwartz, A., Cayrol, B., Rahmouni, A. R., Arluison, V., et al. (2011). The Sm-like RNA chaperone Hfq mediates transcription antitermination at rho-dependent terminators. EMBO J. 30, 2805–2816. doi: 10.1038/emboj.2011.192
Ratje, A. H., Loerke, J., Mikolajka, A., Brünner, M., Hildebrand, P. W., Starosta, A. L., et al. (2010). Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716. doi: 10.1038/nature09547
Ray, S., Chauvier, A., and Walter, N. G. (2019a). Kinetics coming into focus: single-molecule microscopy of riboswitch dynamics. RNA Biol. 16, 1077–1085. doi: 10.1080/15476286.2018.1536594
Ray, S., Costa, R.Da, Das, M., and Nandi, D. (2019b). Interplay of cold shock protein E with an uncharacterized protein, YciF, lowers porin expression and enhances bile resistance in Salmonella Typhimurium. J. Biol. Chem. 294, 9084–9099. doi: 10.1074/jbc.RA119.008209
Rennella, E., Sára, T., Juen, M., Wunderlich, C., Imbert, L., Solyom, Z., et al. (2017). RNA binding and chaperone activity of the E. coli cold-shock protein CspA. Nucleic Acids Res. 45, 4255–4268. doi: 10.1093/nar/gkx044
Richards, J., and Belasco, J. G. (2019). Obstacles to scanning by RNase E govern bacterial mRNA lifetimes by hindering access to distal cleavage sites. Mol. Cell 74, 284–295.e5. doi: 10.1016/j.molcel.2019.01.044
Richardson, J. P. (1991). Preventing the synthesis of unused transcripts by rho factor. Cell 64, 1047–1049. doi: 10.1016/0092-8674(91)90257-Y
Roberts, J. W. (2019). Mechanisms of bacterial transcription termination. J. Mol. Biol. 431, 4030–4039. doi: 10.1016/j.jmb.2019.04.003
Roggiani, M., and Goulian, M. (2015). Chromosome-membrane interactions in bacteria. Annu. Rev. Genet. 49, 115–129. doi: 10.1146/annurev-genet-112414-054958
Romeo, T., and Babitzke, P. (2018). Global regulation by CsrA and its RNA antagonists. Microbiol. Spectr. 6, 341–354. doi: 10.1128/microbiolspec.rwr-0009-2017
Romeo, T., Gong, M., Liu, M. Y., and Brun-Zinkernagel, A. M. (1993). Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175, 4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993
Rudner, D. Z., and Losick, R. (2010). Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2:a000307. doi: 10.1101/cshperspect.a000307
Sahr, T., Rusniok, C., Impens, F., Oliva, G., Sismeiro, O., Coppée, J. Y., et al. (2017). The Legionella pneumophila genome evolved to accommodate multiple regulatory mechanisms controlled by the CsrA-system. PLoS Genet. 13:e1006629. doi: 10.1371/journal.pgen.1006629
Said, N., Hilal, T., Sunday, N. D., Khatri, A., Bürger, J., Mielke, T., et al. (2020). Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase ρ. Science 371:eabd1673. doi: 10.1126/science.abd1673
Salvail, H., Caron, M. P., Bélanger, J., and Massé, E. (2013). Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J. 32, 2764–2778. doi: 10.1038/emboj.2013.205
Sanamrad, A., Persson, F., Lundius, E. G., Fange, D., Gynnå, A. H., and Elf, J. (2014). Single-particle tracking reveals that free ribosomal subunits are not excluded from the Escherichia coli nucleoid. Proc. Natl. Acad. Sci. U. S. A. 111, 11413–11418. doi: 10.1073/pnas.1411558111
Sanchez-Vazquez, P., Dewey, C. N., Kitten, N., Ross, W., and Gourse, R. L. (2019). Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 116, 8310–8319. doi: 10.1073/pnas.1819682116
Santangelo, T. J., and Artsimovitch, I. (2011). Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 9, 319–329. doi: 10.1038/nrmicro2560
Saxena, S., Myka, K. K., Washburn, R., Costantino, N., Court, D. L., and Gottesman, M. E. (2018). Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol. Microbiol. 108, 495–504. doi: 10.1111/mmi.13953
Schlax, P. J., Xavier, K. A., Gluick, T. C., and Draper, D. E. (2001). Translational repression of the Escherichia coli α operon mRNA. Importance of an mRNA conformational switch and a ternary entrapment complex. J. Biol. Chem. 276, 38494–38501. doi: 10.1074/jbc.M106934200
Schmidt, M. C., and Chamberlin, M. J. (1987). nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J. Mol. Biol. 195, 809–818. doi: 10.1016/0022-2836(87)90486-4
Schuwirth, B. S., Borovinskaya, M. A., Hau, C. W., Zhang, W., Vila-Sanjurjo, A., Holton, J. M., et al. (2005). Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834. doi: 10.1126/science.1117230
Sedlyarova, N., Rescheneder, P., Magán, A., Popitsch, N., Rziha, N., Bilusic, I., et al. (2017). Natural RNA polymerase aptamers regulate transcription in E. coli. Mol. Cell 67, 30–43.e6. doi: 10.1016/j.molcel.2017.05.025
Sedlyarova, N., Shamovsky, I., Bharati, B. K., Epshtein, V., Chen, J., Gottesman, S., et al. (2016). sRNA-mediated control of transcription termination in E. coli. Cell 167, 111–121.e13. doi: 10.1016/j.cell.2016.09.004
Sen, R., Chalissery, J., Qayyum, M. Z., Vishalini, V., and Muteeb, G. (2014). Nus factors of Escherichia coli. EcoSal Plus 6, 1–16. doi: 10.1128/ecosalplus.esp-0008-2013
Sevostyanova, A., and Groisman, E. A. (2015). An RNA motif advances transcription by preventing rho-dependent termination. Proc. Natl. Acad. Sci. U. S. A. 112, E6835–E6843. doi: 10.1073/pnas.1515383112
Shaham, G., and Tuller, T. (2018). Genome scale analysis of Escherichia coli with a comprehensive prokaryotic sequence-based biophysical model of translation initiation and elongation. DNA Res. 25, 195–205. doi: 10.1093/dnares/dsx049
Sheng, H., Stauffer, W. T., Hussein, R., Lin, C., and Lim, H. N. (2017). Nucleoid and cytoplasmic localization of small RNAs in Escherichia coli. Nucleic Acids Res. 45, 2919–2934. doi: 10.1093/nar/gkx023
Shenhar, Y., Biran, D., and Ron, E. Z. (2012). Resistance to environmental stress requires the RNA chaperones CspC and CspE. Environ. Microbiol. Rep. 4, 532–539. doi: 10.1111/j.1758-2229.2012.00358.x
Sherwood, A. V., and Henkin, T. M. (2016). Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374. doi: 10.1146/annurev-micro-091014-104306
Shi, J., Gao, X., Tian, T., Yu, Z., Gao, B., Wen, A., et al. (2019). Structural basis of Q-dependent transcription antitermination. Nat. Commun. 10:2925. doi: 10.1038/s41467-019-10958-8
Shokeen, S., Patel, S., Greenfield, T. J., Brinkman, C., and Weaver, K. E. (2008). Translational regulation by an intramolecular stem-loop is required for intermolecular RNA regulation of the par addiction module. J. Bacteriol. 190, 6076–6083. doi: 10.1128/JB.00660-08
Silva, I. J., Barahona, S., Eyraud, A., Lalaouna, D., Figueroa-Bossi, N., Massé, E., et al. (2019). SraL sRNA interaction regulates the terminator by preventing premature transcription termination of rho mRNA. Proc. Natl. Acad. Sci. U. S. A. 116, 3042–3051. doi: 10.1073/pnas.1811589116
Singh, D., Chang, S. J., Lin, P. H., Averina, O. V., Kaberdin, V. R., and Lin-Chao, S. (2009). Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106, 864–869. doi: 10.1073/pnas.0810205106
Singh, D., Murashko, O. N., and Lin-Chao, S. (2020). Posttranscriptional regulation of tnaA by protein-RNA interaction mediated by ribosomal protein L4 in Escherichia coli. J. Bacteriol. 202:e00799-19. doi: 10.1128/jb.00799-19
Siwiak, M., and Zielenkiewicz, P. (2013). Transimulation – protein biosynthesis web service. PLoS One 8:e73943. doi: 10.1371/journal.pone.0073943
Smirnov, A., Förstner, K. U., Holmqvist, E., Otto, A., Günster, R., Becher, D., et al. (2016). Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 113, 11591–11596. doi: 10.1073/pnas.1609981113
Smirnov, A., Wang, C., Drewry, L. L., and Vogel, J. (2017). Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 36, 1029–1045. doi: 10.15252/embj.201696127
Sonnleitner, E., and Bläsi, U. (2014). Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 10:e1004440. doi: 10.1371/journal.pgen.1004440
Spedding, G., and Draper, D. E. (1993). Allosteric mechanism for translational repression in the Escherichia coli α operon. Proc. Natl. Acad. Sci. U. S. A. 90, 4399–4403. doi: 10.1073/pnas.90.10.4399
Squires, C. L., and Zaporojets, D. (2000). Proteins shared by the transcription and translation machines. Annu. Rev. Microbiol. 54, 775–798. doi: 10.1146/annurev.micro.54.1.775
Stein, E. M., Kwiatkowska, J., Basczok, M. M., Gravel, C. M., Berry, K. E., and Olejniczak, M. (2020). Determinants of RNA recognition by the FinO domain of the Escherichia coli ProQ protein. Nucleic Acids Res. 48, 7502–7519. doi: 10.1093/nar/gkaa497
Steinberg, R., Origi, A., Natriashvili, A., Sarmah, P., Licheva, M., Walker, P. M., et al. (2020). Posttranslational insertion of small membrane proteins by the bacterial signal recognition particle. PLoS Biol. 18:e3000874. doi: 10.1371/journal.pbio.3000874
Stent, G. S. (1964). The operon: on its third anniversary – modulation of transfer RNA species can provide a workable model of an operator-less operon. Science 144, 816–820. doi: 10.1126/science.144.3620.816
Stevenson-Jones, F., Woodgate, J., Castro-Roa, D., and Zenkin, N. (2020). Ribosome reactivates transcription by physically pushing RNA polymerase out of transcription arrest. Proc. Natl. Acad. Sci. 117, 8462–8467. doi: 10.1073/pnas.1919985117
Stracy, M., Lesterlin, C., Garza de Leon, F., Uphoff, S., Zawadzki, P., and Kapanidis, A. N. (2015). Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc. Natl. Acad. Sci. U. S. A. 112, E4390–E4399. doi: 10.1073/pnas.1507592112
Strahl, H., Turlan, C., Khalid, S., Bond, P. J., Kebalo, J. M., Peyron, P., et al. (2015). Membrane recognition and dynamics of the RNA degradosome. PLoS Genet. 11:e1004961. doi: 10.1371/journal.pgen.1004961
Sukhodolets, M. V., and Garges, S. (2003). Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry 42, 8022–8034. doi: 10.1021/bi020638i
Sukhodolets, M. V., Garges, S., and Adhya, S. (2006). Ribosomal protein S1 promotes transcriptional cycling. RNA 12, 1505–1513. doi: 10.1261/rna.2321606
Surovtsev, I. V., and Jacobs-Wagner, C. (2018). Subcellular organization: a critical feature of bacterial cell replication. Cell 172, 1271–1293. doi: 10.1016/j.cell.2018.01.014
Syal, K., Flentie, K., Bhardwaj, N., Maiti, K., Jayaraman, N., Stallings, C. L., et al. (2017). Synthetic (p)ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob. Agents Chemother. 61:e00443-17. doi: 10.1128/AAC.00443-17
Taghbalout, A., Yang, Q., and Arluison, V. (2014). The Escherichia coli RNA processing and degradation machinery is compartmentalized within an organized cellular network. Biochem. J. 458, 11–22. doi: 10.1042/BJ20131287
Thisted, T., Sørensen, N. S., and Gerdes, K. (1995). Mechanism of post-segregational killing: secondary structure analysis of the entire Hok mRNA from plasmid R1 suggests a fold-back structure that prevents translation and antisense RNA binding. J. Mol. Biol. 247, 859–873. doi: 10.1006/jmbi.1995.0186
Thomas, M. S., Bedwell, D. M., and Nomura, M. (1987). Regulation of α operon gene expression in Escherichia coli. A novel form of translational coupling. J. Mol. Biol. 196, 333–345. doi: 10.1016/0022-2836(87)90694-2
Torres, M., Condon, C., Balada, J. -M., Squires, C., and Squires, C. L. (2001). Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 20, 3811–3820. doi: 10.1093/emboj
Toulokhonov, I., Artsimovitch, I., and Landick, R. (2001). Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science 292, 730–733. doi: 10.1126/science.1057738
Tree, J. J., Granneman, S., McAteer, S. P., Tollervey, D., and Gally, D. L. (2014). Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol. Cell 55, 199–213. doi: 10.1016/j.molcel.2014.05.006
Uhm, H., Kang, W., Ha, K. S., Kang, C., and Hohng, S. (2017). Single-molecule FRET studies on the cotranscriptional folding of a thiamine pyrophosphate riboswitch. Proc. Natl. Acad. Sci. U. S. A. 115, 331–336. doi: 10.1073/pnas.1712983115
Uniacke, J., and Zerges, W. (2009). Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc. Natl. Acad. Sci. U. S. A. 106, 1439–1444. doi: 10.1073/pnas.0811268106
Updegrove, T. B., Zhang, A., and Storz, G. (2016). Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138. doi: 10.1016/j.mib.2016.02.003
Vakulskas, C. A., Potts, A. H., Babitzke, P., Ahmer, B. M. M., and Romeo, T. (2015). Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79, 193–224. doi: 10.1128/mmbr.00052-14
Valencia-Burton, M., McCullough, R. M., Cantor, C. R., and Broude, N. E. (2007). RNA visualization in live bacterial cells using fluorescent protein complementation. Nat. Methods 4, 421–427. doi: 10.1038/nmeth1023
van Gijtenbeek, L. A., Robinson, A., van Oijen, A. M., Poolman, B., and Kok, J. (2016). On the spatial organization of mRNA, plasmids, and ribosomes in a bacterial host overexpressing membrane proteins. PLoS Genet. 12:e1006523. doi: 10.1371/journal.pgen.1006523
van Nues, R. W., Castro-Roa, D., Yuzenkova, Y., and Zenkin, N. (2015). Ribonucleoprotein particles of bacterial small non-coding RNA IsrA (IS61 or McaS) and its interaction with RNA polymerase core may link transcription to mRNA fate. Nucleic Acids Res. 44, 2577–2592. doi: 10.1093/nar/gkv1302
Večerek, B., Moll, I., and Bläsi, U. (2005). Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA 11, 976–984. doi: 10.1261/rna.2360205
Vinogradova, D. S., Zegarra, V., Maksimova, E., Nakamoto, J. A., Kasatsky, P., Paleskava, A., et al. (2020). How the initiating ribosome copes with ppGpp to translate mRNAs. PLoS Biol. 18:e3000593. doi: 10.1371/journal.pbio.3000593
Vogel, U., and Jensen, K. F. (1994a). Effects of guanosine 3',5'-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli. J. Biol. Chem. 269, 16236–16241.
Vogel, U., and Jensen, K. F. (1994b). The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 176, 2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994
Vogel, U., Sørensen, M., Pedersen, S., Jensen, K. F., and Kilstrup, M. (1992). Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol. Microbiol. 6, 2191–2200. doi: 10.1111/j.1365-2958.1992.tb01393.x
Volkov, I. L., Lindén, M., Aguirre Rivera, J., Ieong, K. -W., Metelev, M., Elf, J., et al. (2018). tRNA tracking for direct measurements of protein synthesis kinetics in live cells. Nat. Chem. Biol. 14, 618–626. doi: 10.1038/s41589-018-0063-y
Wagner, E. G. H., and Romby, P. (2015). Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208. doi: 10.1016/bs.adgen.2015.05.001
Wang, B., and Artsimovitch, I. (2020). A growing gap between the RNAP and the lead ribosome. Trends Microbiol. 29, 4–5. doi: 10.1016/j.tim.2020.09.011
Wang, B., Dai, P., Ding, D., Del Rosario, A., Grant, R. A., Pentelute, B. L., et al. (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150. doi: 10.1038/s41589-018-0183-4
Wang, B., Grant, R. A., and Laub, M. T. (2020a). ppGpp coordinates nucleotide and amino-acid synthesis in E. coli during starvation. Mol. Cell 80, 29–42.e10. doi: 10.1016/j.molcel.2020.08.005
Wang, W., Li, G. W., Chen, C., Xie, X. S., and Zhuang, X. (2011). Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333, 1445–1449. doi: 10.1126/science.1204697
Wang, C., Molodtsov, V., Firlar, E., Kaelber, J. T., Blaha, G., Su, M., et al. (2020b). Structural basis of transcription-translation coupling. Science 369, 1359–1365. doi: 10.1126/science.abb5317
Wang, N., Yamanaka, K., and Inouye, M. (1999). CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 181, 1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999
Washburn, R. S., Zuber, P. K., Sun, M., Hashem, Y., Shen, B., Li, W., et al. (2020). Escherichia coli NusG links the lead ribosome with the transcription elongation complex. iScience 23:101352. doi: 10.1016/j.isci.2020.101352
Watters, K. E., Strobel, E. J., Yu, A. M., Lis, J. T., and Lucks, J. B. (2016). Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat. Struct. Mol. Biol. 23, 1124–1131. doi: 10.1038/nsmb.3316
Weber, M. H. W., Volkov, A. V., Fricke, I., Marahiel, M. A., and Graumann, P. L. (2001). Localization of cold shock proteins to cytosolic spaces surrounding nucleoids in Bacillus subtilis depends on active transcription. J. Bacteriol. 183, 6435–6443. doi: 10.1128/JB.183.21.6435-6443.2001
Webster, M. W., Takacs, M., Zhu, C., Vidmar, V., Eduljee, A., Abdelkareem, M., et al. (2020). Structural basis of transcription-translation coupling and collision in bacteria. Science 369, 1355–1359. doi: 10.1126/science.abb5036
Wei, B. L., Brun-Zinkernagel, A. M., Simecka, J. W., Prüß, B. M., Babitzke, P., and Romeo, T. (2001). Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40, 245–256. doi: 10.1046/j.1365-2958.2001.02380.x
Wen, J., Harp, J. R., and Fozo, E. M. (2017). The 5', UTR of the type i toxin ZorO can both inhibit and enhance translation. Nucleic Acids Res. 45, 4006–4020. doi: 10.1093/nar/gkw1172
Wendrich, T. M., Blaha, G., Wilson, D. N., Marahiel, M. A., and Nierhaus, K. H. (2002). Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10, 779–788. doi: 10.1016/S1097-2765(02)00656-1
Weng, X., Bohrer, C. H., Bettridge, K., Lagda, A. C., Cagliero, C., Jin, D. J., et al. (2019). Spatial organization of RNA polymerase and its relationship with transcription in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 116, 20115–20123. doi: 10.1073/pnas.1903968116
Westermann, A. J., Venturini, E., Sellin, M. E., Förstner, K. U., Hardt, W. D., and Vogel, J. (2019). The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica serovar typhimurium. MBio 10:e02504-18. doi: 10.1128/mBio.02504-18
Winkler, W., Nahvi, A., and Breaker, R. R. (2002). Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956. doi: 10.1038/nature01145
Woldringh, C. L. (2002). The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 45, 17–29. doi: 10.1046/j.1365-2958.2002.02993.x
Yakhnin, A. V., Baker, C. S., Vakulskas, C. A., Yakhnin, H., Berezin, I., Romeo, T., et al. (2013). CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol. Microbiol. 87, 851–866. doi: 10.1111/mmi.12136
Yamanaka, K., Mitani, T., Ogura, T., Niki, H., and Hiraga, S. (1994). Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol. Microbiol. 13, 301–312. doi: 10.1111/j.1365-2958.1994.tb00424.x
Yang, S., Kim, S., Kim, D. K., Jeon An, H., Bae Son, J., Hedén Gynnå, A., et al. (2019). Transcription and translation contribute to gene locus relocation to the nucleoid periphery in E. coli. Nat. Commun. 10:5131. doi: 10.1038/s41467-019-13152-y
Yanofsky, C. (1981). Attenuation in the control of expression of bacterial operons. Nature 289, 751–758. doi: 10.1038/289751a0
Yin, Z., Kaelber, J. T., and Ebright, R. H. (2019). Structural basis of Q-dependent antitermination. Proc. Natl. Acad. Sci. U. S. A. 116, 18384–18390. doi: 10.1073/pnas.1909801116
Young, R., and Bremer, H. (1976). Polypeptide chain elongation rate in Escherichia coli B/r as a function of growth rate. Biochem. J. 160, 185–194. doi: 10.1042/bj1600185
Zhang, A., Wassarman, K. M., Rosenow, C., Tjaden, B. C., Storz, G., and Gottesman, S. (2003). Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50, 1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x
Zhang, Y., Zborníková, E., Rejman, D., and Gerdes, K. (2018). Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. MBio 9:e02188–17. doi: 10.1128/mBio.02188-17
Zhu, M., and Dai, X. (2019). Maintenance of translational elongation rate underlies the survival of Escherichia coli during oxidative stress. Nucleic Acids Res. 47, 7592–7604. doi: 10.1093/nar/gkz467
Zhu, M., and Dai, X. (2020). Bacterial stress defense: the crucial role of ribosome speed. Cell. Mol. Life Sci. 77, 853–858. doi: 10.1007/s00018-019-03304-0
Zhu, M., Dai, X., and Wang, Y. -P. (2016). Real time determination of bacterial in vivo ribosome translation elongation speed based on LacZα complementation system. Nucleic Acids Res. 44:e155. doi: 10.1093/nar/gkw698
Zhu, M., Mori, M., Hwa, T., and Dai, X. (2019). Disruption of transcription–translation coordination in Escherichia coli leads to premature transcriptional termination. Nat. Microbiol. 4, 2347–2356. doi: 10.1038/s41564-019-0543-1
Zhu, Y., Mustafi, M., and Weisshaar, J. C. (2020). Biophysical properties of Escherichia coli cytoplasm in stationary phase by superresolution fluorescence microscopy. MBio 11:e00143-20. doi: 10.1128/mBio.00143-20
Zuber, P. K., Schweimer, K., Rösch, P., Artsimovitch, I., and Knauer, S. H. (2019). Reversible fold-switching controls the functional cycle of the antitermination factor RfaH. Nat. Commun. 10:702. doi: 10.1038/s41467-019-08567-6
Keywords: coupled transcription-translation, uncoupled transcription-translation, subcellular organization of prokaryotes, translation-independent mRNA localization, local translation, expressome
Citation: Irastortza-Olaziregi M and Amster-Choder O (2021) Coupled Transcription-Translation in Prokaryotes: An Old Couple With New Surprises. Front. Microbiol. 11:624830. doi: 10.3389/fmicb.2020.624830
Edited by:
Hans-Georg Koch, University of Freiburg, GermanyReviewed by:
Evgeny Nudler, New York University, United StatesJared Schrader, Wayne State University, United States
Copyright © 2021 Irastortza-Olaziregi and Amster-Choder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mikel Irastortza-Olaziregi, bWlrZWwuaXJhc3RvcnphQG1haWwuaHVqaS5hYy5pbA==; Orna Amster-Choder, b3JuYWFtQGVrbWQuaHVqaS5hYy5pbA==
 Mikel Irastortza-Olaziregi
Mikel Irastortza-Olaziregi Orna Amster-Choder
Orna Amster-Choder