
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 December 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.622785
This article is part of the Research Topic Bioactive Compounds with Potential Medicinal Properties Derived from Fungi: Recent and Future Developments in Microbial Biotechnology View all 15 articles
Transcription regulation caused by global regulators exerts important effects on fungal secondary metabolism. By overexpression of the global regulator Talae1 in a Ficus elastica-associated fungus Trichoderma afroharzianum, two structurally new polyketides (1 and 2) that were newly produced in the transformant were isolated and identified. Their structures, including the absolute configurations, were elucidated through a combination of high resolution mass spectrometer (HRMS), NMR, and electronic circular dichroism (ECD) calculations. The growth inhibitory activities of compounds 1 and 2 were evaluated against four bacteria and six plant-pathogenic fungi. Compound 1 showed the highest antifungal activity against Botrytis cinerea and Fusarium oxysporum f. sp. nicotianae with MIC of 8 μg/ml. To the best of our knowledge, this is the first study to report on the application of the global regulator in T. afroharzianum to activate the biosynthesis of bioactive secondary metabolites.
The genus Trichoderma is a ubiquitous fungal group comprising of more than 340 species (Zeng and Zhuang, 2019). Some Trichoderma species are widely used in agriculture as biological control agents to prevent the growth of other phytopathogenic fungi and to promote the development of crop plants (Benítez et al., 2004; Reino et al., 2008). The genus Trichoderma has been reported to be an excellent producer of bioactivity secondary metabolites (SMs). Several hundreds of SMs, including polyketides, non-ribosomal peptides, terpenoids, and alkaloids, have been identified and isolated in Trichoderma species and strains from different environments (Reino et al., 2008; Degenkolb et al., 2015; Li et al., 2019). Some of the SMs are considered to play a significant role in stimulating plant growth or providing defense against plant pathogens (Vinale et al., 2008).
Recent genome sequencing of many fungal species has revealed that most of the biosynthetic gene clusters (BGCs) are cryptic or lowly expressed under general culture conditions (Rutledge and Challis, 2015; Wu et al., 2020). Similarly, although diverse metabolites have been discovered from the genus Trichoderma, genome sequencing has revealed that there are more BGCs than we have discovered (Mukherjee et al., 2013; Zeilinger et al., 2016), suggesting that the potential of Trichoderma fungi to produce more undetected metabolites has hidden and needs to be activated.
To activate the cryptic biosynthetic potential and reveal more SMs, several strategies, such as ribosome engineering, transcriptional regulation, epigenetic perturbation, and heterologous expression, have been developed (Wiemann and Keller, 2014; Wu et al., 2016). Besides, the manipulation of global transcriptional regulators is also reported to be a feasible strategy for the activation of cryptic BGCs. The LaeA protein was first discovered as a global regulator of secondary metabolism in Aspergillus nidulans (Bok and Keller, 2004). Because of sequence similarity to histone methyltransferases, LaeA may achieve global transcriptional regulation by influencing chromatin modification (Brakhage, 2013). Reyes-Dominguez et al. (2010) reported that LaeA reversed the repressive chromatin structure caused by some negative regulators, resulting in the activation of BGCs in A. nidulans. Furthermore, LaeA is conserved in filamentous fungi, and thus its gene homologs are found in other filamentous fungi including Aspergillus carbonarius (Linde et al., 2016), Aspergillus fumisynnematus (Hong et al., 2015), Alternaria alternata (Estiarte et al., 2016), Fusarium verticillioides (Butchko et al., 2012), Monascus ruber (Liu et al., 2016), and Penicillium chrysogenum (Kosalková et al., 2009) and Penicillium brocae (Wang et al., 2020). Numerous studies have also determined that the presence of LaeA facilitates the expression of multiple BGCs. For instance, Jiang et al. (2016) reported that overexpression of laeA in Chaetomium globosum upregulated the expression of chaetoglobosin BGC and led to the discovery of a new cytochalasan. Yu et al. (2019) reported the discovery of a series of sorbicillinoids including two new ones by overexpression of laeA in Penicillium dipodomyis. Thus, overexpression of LaeA is considered as a useful strategy in activating silent biosynthetic pathways and promoting the discovery of novel SMs in fungi.
Endophytic fungi are increasingly recognized as a significant reservoir of bioactive metabolites (Chandra, 2012; Wu et al., 2016). Due to the absence of a simulated ecological environment, the BGCs of endophytic fungi are not effectively expressed under standard laboratory conditions, thus reducing the probability of detecting new SMs (Mao et al., 2015). In our previous research work, the endophytic fungus Trichoderma afroharzianum Fes1712 isolated from rubber tree Ficus elastica was shown to produce novel isocoumarin analogues (Ding et al., 2019). To further activate its chemical potential, we constructed a laeA-like gene (talae1) overexpression transformant of T. afroharzianum and investigated the effect of laeA-like gene overexpression on secondary metabolic profile. Chemical analysis of SMs produced by the overexpression strain led to the discovery of two new polyketides (1, 2). In this study, structural elucidation and biological evaluation of the two new compounds are presented.
DNA restriction enzymes were purchased from Transgen Biotech Co., LTD (Beijing, China). Polymerase chain reaction (PCR) was performed using TransStart® Fastpfu Fly DNA Polymerase (Transgen Biotech, Beijing, China). Optical rotations were measured on a P-1020 digital polarimeter (JASCO Corporation, Tokyo, Japan). ECD spectra were recorded on a JASCO J-815 spectropolarimeter (JASCO Corporation). UV spectra were recorded on Waters 2487 (Waters Corporation, Milford, MA, United States). HRESIMS and ESIMS spectra were measured on Thermo Scientific LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). NMR spectra were recorded on Agilent 500 MHz DD2 spectrometer (Agilent Technologies Inc., Santa Clara, CA, United States). Semi-preparative HPLC was performed using a YMC Pack ODS-A column (250 × 10 mm, 5 μm, 3 ml/min, YMC Co., Ltd., Kyoto, Japan). Column chromatography was performed on silica gel (200–300 mesh, Qingdao Marine Chemical Inc., Qingdao, China) and Sephadex LH-20 (GE Healthcare, Uppsala, Sweden).
The fungal wild strain Fes1712 (WT) was originally isolated from fresh leaves of F. elastica collected from Liaocheng University Arboretum, Liaocheng, Shandong Province of China and identified as T. afroharzianum based on tef1 and rpb2 sequences (Chaverri et al., 2015; Supplementary Figure S1).
Because the genomic sequence of T. afroharzianum Fes1712 is currently unknown, the genomic sequence of Trichoderma harzianum CBS 226.95 was used for gene selection and initial primer design. A laeA-like gene was identified using Local-Blast with A. nidulans LaeA gene (AN0807) as the query. The two pairs of specific primers, including two inner primers (Ta85012iF/R) with terminal XbaI and EcoRV restriction sites, were used as primers of the nested-PCR amplification of talae1 from genomic DNA of strain Fes1712. The PCR product of the talae1 gene was digested using endonucleases and inserted into the same restriction site of the pZeo vector (Tang et al., 2017) to create pZeo-talae1 (Supplementary Figure S2). The talae1 gene from strain Fes1712 was sequenced by Sangon Biotech (Shanghai, China) and deposited in the GenBank Database under the accession numbers MT313929. The overexpression vector pZeo-talae1 comprised of a continuous expression cassette, including the PgpdA promoter, talae1 gene, and TtrpC terminator, and two selection markers (ampicillin resistance gene for selection of Escherichia coli strain and bleomycin resistance gene for the Fes1712 transformant). The recombinant vector was transformed into E. coli strain Trans1-T1 to extract plasmids for transformation. The extracted plasmids were then transferred into WT yielding a transformant with overexpressed talae1 gene (OE::Talae1) by PEG-mediated protoplast transformation (Tang et al., 2017), while the control transformant was also generated using a vacant pZeo. The transformants were selected and purified in the presence of bleomycin. The genotype of the overexpression transformant was confirmed by diagnostic PCR (Supplementary Figure S3). The oligonucleotide sequences for PCR primers are presented in Supplementary Table S1.
For SMs production, the overexpression transformant and control transformant were cultured on PDA medium (PDA, 20% potato, 2% dextrose, and 1.5% agar) at 28°C for 5 days. They were then inoculated into 1 L Erlenmeyer flasks, each containing 80 g of rice and 120 ml distilled water and incubated at 28°C under static conditions. After 15 days, the fermented substrate in each flask was broken using a disperser (T18, IKA, Germany) and extracted three times with 200 ml MeOH. The liquid layers were collected and evaporated to remove MeOH. The residual extract was suspended in H2O and extracted three times with an equal volume of EtOAc. The EtOAc layers were separated and evaporated under reduced pressure to yield the solid extract.
The extract from the overexpression transformant (6.7 g) was applied on silica gel using a step gradient elution with petroleum ether−EtOAc (v/v 10:1, 5:1, 2:1, 1:1, 1:3) and EtOAc−MeOH (v/v 10:1, 3:1, 1:1, 0:1) to give nine fractions (Fr. 1–9). Fraction 7 was fractionated on MPLC (60–100% MeOH/H2O, 45 min) to give five fractions (Fr. 7.1–7.5). Fraction 7.3 and Fraction 7.4 were further purified by semipreparative HPLC eluting with 70% MeOH−H2O to obtain compound 1 (9.0 mg) and 2 (4.5 mg).
Compound 1: white amorphous powder; [α]20D − 36.1 (c 0.10, MeOH); ECD (MeOH) λmax (∆ε) 304 (+2.01), 233 (−10.55) nm; UV (MeOH) λmax (log ε) 299 (2.01), 210 (3.14) nm; positive HR-ESI-MS (m/z): 263.1282 [M + H]+ (calcd. for C15H19O4, 263.1283; Supplementary Figure S5); 1H and 13C NMR data, see Table 1 and Supplementary Figures S6–S11.
Compound 2: white amorphous powder; [α]20D − 44.2 (c 0.10, MeOH); ECD (MeOH) λmax (∆ε) 304 (+2.52), 235 (−9.74) nm; UV (MeOH) λmax (log ε) 300 (2.10), 210 (3.15) nm; positive HR-ESI-MS (m/z): 277.1435 [M + H]+ (calcd. for C16H21O4, 277.1439; Supplementary Figure S12); 1H and 13C NMR data, see Table 1 and Supplementary Figures S13–S18.
Antimicrobial activities of the isolated compounds were evaluated against four bacteria (Bacillus subtilis CMCC 63501, E. coli CMCC 44102, Pseudomonas aeruginosa CMCC 10104, and Staphylococcus aureus CMCC 26003) and six plant-pathogenic fungi (A. alternata ACCC 36110, Botrytis cinerea ACCC 36028, Colletotrichum lagenarium ACCC 30016, Fusarium oxysporum f. sp. nicotianae TRICAAS 0101, Gaeumannomyces graminis var. graminis TRICAAS 0191, and Thielaviopsis basicola TRICAAS 0207) using the 96-well plate microdilution method (Zhang et al., 2019). The phytopathogenic fungi were provided and deposited by the Tobacco Research Institute of the Chinese Academy of Agricultural Sciences, Qingdao, China. The tested compounds were prepared as 2-fold dilutions with DMSO and added (10 μl) to each well, containing the spore suspension (106 CFU/ml, 10 μl) and culture medium (180 μl, Luria-Bertani medium for bacteria, potato dextrose medium for fungi), to obtain a final concentration of between 512 μg/ml to 1 μg/ml. After adequate mixing, the assay plates were incubated in the dark for 24–72 h at 30°C. The minimal inhibitory concentration (MIC) was determined as the lowest concentration at which no growth of pathogen was observed. Chloramphenicol and prochloraz were used as positive controls for antibacterial and antifungal assays, respectively. All tests were performed in triplicate.
A laeA-like gene was identified in the genomic sequence of T. harzianum CBS 226.95 using Local-blast. In the genomic sequence of T. harzianum CBS 226.95, the laeA-like gene is designated as M431DRAFT_85012 (DOE Joint Genome Institute). Subsequently, we designed special primers to clone talae1 ORF from the genomic DNA of strain Fes1712 via nested-PCR. The obtained PCR fragment was 1,228 bp in size, and the predicted coding sequence was 999 bp, encoding a 332-residue polypeptide. Sequence analysis via InterProScan indicated that talae1 protein was an S-adenosyl-L-methionine-dependent methyltransferase, which is consistent with the function of LaeA. BLAST analysis indicated that Talae1 protein had 73.9, 68.1, 57.1, and 56.8% sequence identity to Lae1 (XP_006966726.1) of Trichoderma reesei (Seiboth et al., 2012), Lae1 (XP_013938600.1) of Trichoderma atroviride (Aghcheh et al., 2013), LaeA (XP_018233436.1) of F. oxysporum (López-Berges et al., 2014), and Lae1 (XP_018742755.1) of F. verticillioides (Butchko et al., 2012), respectively (Figure 1).
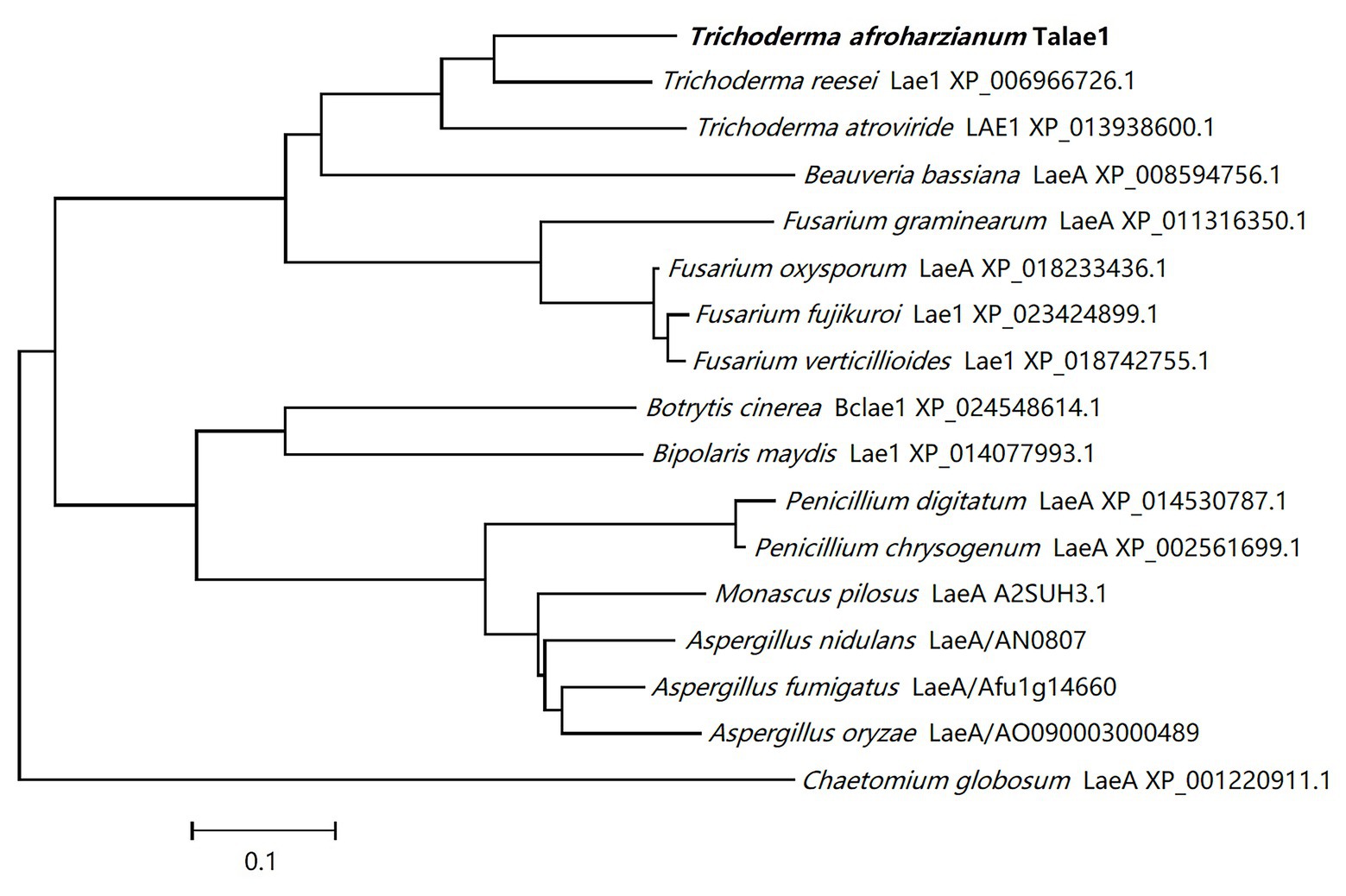
Figure 1. Phylogenetic tree analyses of Talae1 and its homologs from different species. Branch lengths are in proportion to distance.
To examine the effect of talae1 gene overexpression on secondary metabolism in strain Fes1712, OE::Talae1 transformant and control transformant were constructed and cultured on rice medium at 28°C for 15 days under static conditions. Real time PCR was performed to determine the difference in the transcriptional levels of talae1 gene between the OE::Talae1 and control transformants. Results indicated that the transcriptional level of talae1 gene was 7.7-fold upregulated in OE::Talae1 transformant (Supplementary Figure S4). The fermentation products were extracted and analyzed using HPLC. OE::Talae1 transformant showed an obvious change in the secondary metabolic profile, evidenced by the emergence of several new peaks (Figure 2), compared with that of the control transformant. Purification of the new peaks led to the isolation of two relatively high-yield compounds 1 and 2 (Figure 3).
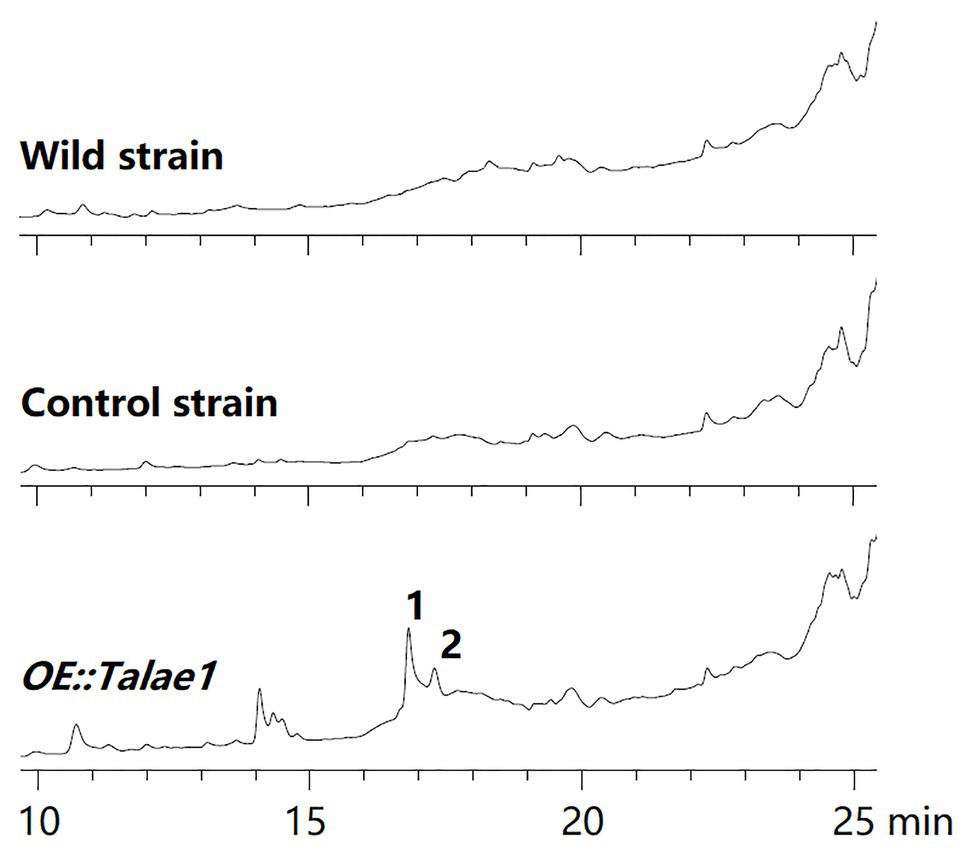
Figure 2. Comparative analysis of secondary metabolite HPLC profiles of OE::Talae1 and control transformants. The numbers represent the compounds isolated from the OE::Talae1 transformant. Detection was carried out at 210 nm.
Compound 1 was obtained as a white amorphous powder. The molecular formula C15H18O4 was determined by the positive HRESIMS m/z 263.1282 ([M + H]+, calcd. 263.1283), indicating 7 degrees of unsaturation. The 1H NMR spectrum of 1 revealed signals for five olefinic protons (δH 7.24, 6.29, 6.25, 6.21, and 6.12), an oxymethine (δH 5.33), and three methyl groups (δH 2.16, 2.06, and 1.81). While the 13C NMR, DEPT, and HSQC spectra exhibited the presence of 15 carbon resonance signals, including one conjugated ketone carbonyl (δC 199.1), nine olefinic or aromatic carbons with five protonated, one oxymethine (δC/H 76.6/5.33), and three methyls (δC/H 17.4/1.81, 10.2/2.16, and 7.6/2.06; Table 1). Analysis of the 2D NMR data of 1 revealed contiguous COSY correlations extending from H-9 to H-13, as well as heteronuclear multiple bond correlation (HMBC) correlation from H-10, H-9, and the oxymethine proton H-7 to C-8 carbonyl, indicating the presence of a 1-hydroxyhepta-3,5-dien-2-one side chain (Figure 4). The large J values of H-9/H-10 (J = 15.3 Hz) and H-11/H-12 (J = 15.5 Hz) revealed the E-configuration of the ∆10 and ∆12 double bonds (Figure 4). HMBC correlations from H3-15 to C-5, C-6, and the hydroxylated aromatic carbon C-4, from H3-14 to C-3 and the two hydroxylated aromatic carbons C-4 and C-2, and from the aromatic proton H-1 to C-3 and C-5 collectively led to the construction of the penta-substituted benzene core structure of compound 1 as shown in Figure 4. The presence of C-6/C-7 linkage between the penta-substituted benzene core structure and the 1-hydroxyhepta-3,5-dien-2-one side chain was demonstrated by HMBC correlations from H-7 to C-1, C-5, and C-6. To determine the absolute configuration of C-7 in 1, the ECD spectrum of 1 was calculated using the time-dependent density functional theory (TD-DFT) at the B3LYP/6-31 + G (d) level using the Gaussian 09 program (Frisch et al., 2009). The result showed that the experimental ECD spectrum of 1 was in good agreement with the calculated ECD spectrum of (7R)-1 (Figure 5), designating the absolute configuration of C-7 as R. Thus, compound 1 was determined to be (R,3E,5E)-1-(3,5-dihydroxy-2,4-dimethylphenyl)-1-hydroxyhepta-3,5-dien-2-one.
Compound 2 was isolated as a white amorphous powder with the molecular formula C16H20O4 determined based on HRESIMS (m/z 277.1413 [M + H]+), with an additional CH3 group compared to 1. The 1D and 2D NMR data of compound 2 showed close similarity to those of 1, with the only distinction attributable to the presence of a methoxy signal at δH/C 3.30/55.6 in 2 (Table 1). The key HMBC correlation from H3-16 (δ 3.67) to the oxymethine carbon C-7 (δ 172.1) indicated that the hydroxyl group at C-7 in 1 was methylated in 2. The absolute configuration of C-7 in 2 was determined to be similar to that of 1 based on ECD spectra (Figure 5). Therefore, the structure of compound 2 was determined to be (R,3E,5E)-1-(3,5-dihydroxy-2,4-dimethylphenyl)-1-methoxyhepta-3,5-dien-2-one.
A plausible biosynthetic pathway for 1 and 2 is proposed as shown in Figure 6. The polyketide chain primed with one acetyl-CoA starter unit and six malonyl-CoA extender units was selectively enolized, hydroxylated, methylated, cyclized, and finally released as the intermediate a (Wu et al., 2019). Compounds 1 and 2 are proposed to be generated by further decarboxylation and O-methylation (Shaw et al., 2015; Lebe and Cox, 2019).
The newly produced compounds 1 and 2 were evaluated for antifungal activity against four bacteria and six plant-pathogenic fungi were tested. As shown in Table 2, compounds 1 and 2 showed selective antifungal activity with MIC values ranging from 8 to 32 μg/ml. Compound 1 displayed the highest growth inhibitory activity with MIC values of 8, 8, and 32 toward B. cinerea, F. oxysporum f. sp. nicotianae, and C. lagenarium, respectively. The genus Trichoderma has been shown to produce numerous polyketides with various bioactivities (Reino et al., 2008; Harned and Volp, 2011). For instance, fungal species of the Trichoderma genus are known to be the main producers of sorbicillin family (Harned and Volp, 2011; Derntl et al., 2017), which are structurally similar to the compounds 1 and 2. Reátegui et al. (2006) have reported antifungal activity of sorbicillin analogues against Aspergillus flavus and Fusarium verticillioides. Besides, the compounds 1 and 2 also showed weak antibacterial activity against Gram-positive bacteria S. aureus.
In summary, overexpression of the global regulator Talae1 upregulated the production of SMs in an endophytic fungus T. afroharzianum, and two new polyketides (1 and 2) were isolated and identified from overexpression transformant. These results indicate that the global regulator Talae1 is involved in secondary metabolic regulation of T. afroharzianum and affects the biosynthesis of a series of antifungal polyketides. This study also demonstrates that genetic manipulation of the global regulator presents a promising approach for activating new SMs and improving the metabolic potential of biocontrol fungi.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
ZD conceived and designed the experiments, prepared the manuscript, and was involved in isolation of compounds. XW and H-MH performed genetic manipulation, strain fermentation, and extraction. F-DK contributed to determinate the structures of isolated compounds. Y-NZ and ML contributed to bioactivity assay. Z-PW and JH supervised the work and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by the Natural Science Foundation of Shandong Province (ZR2017BB077 and ZR2018BH043) and Tai-Shan Scholar Foundation of Shandong Province (319190201).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.622785/full#supplementary-material
Aghcheh, R. K., Druzhinina, I. S., and Kubicek, C. P. (2013). The putative protein methyltransferase LAE1 of Trichoderma atroviride is a key regulator of asexual development and mycoparasitism. PLoS One 8:e67144. doi: 10.1371/journal.pone.0067144
Benítez, T., Rincón, A. M., Limón, M. C., and Codón, A. C. (2004). Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 7, 249–260. doi: 10.2436/im.v7i4.9480
Bok, J. W., and Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3, 527–535. doi: 10.1128/ec.3.2.527-535.2004
Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. doi: 10.1038/nrmicro2916
Butchko, R. A. E., Brown, D. W., Busman, M., Tudzynski, B., and Wiemann, P. (2012). Lae1 regulates expression of multiple secondary metabolite gene clusters in Fusarium verticillioides. Fungal Genet. Biol. 49, 602–612. doi: 10.1016/j.fgb.2012.06.003
Chandra, S. (2012). Endophytic fungi: novel sources of anticancer lead molecules. Appl. Microbiol. Biotechnol. 95, 47–59. doi: 10.1007/s00253-012-4128-7
Chaverri, P., Branco-Rocha, F., Jaklitsch, W., Gazis, R., Degenkolb, T., and Samuels, G. J. (2015). Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 107, 558–590. doi: 10.3852/14-147
Degenkolb, T., Nielsen, K. F., Dieckmann, R., Branco-Rocha, F., Chaverri, P., Samuels, G. J., et al. (2015). Peptaibol, secondary-metabolite, and hydrophobin pattern of commercial biocontrol agents formulated with species of the Trichoderma harzianum complex. Chem. Biodivers. 12, 662–684. doi: 10.1002/cbdv.201400300
Derntl, C., Guzmán-Chávez, F., Mello-de-Sousa, T. M., Busse, H. -J., Driessen, A. J. M., Mach, R. L., et al. (2017). In vivo study of the sorbicillinoid gene cluster in Trichoderma reesei. Front. Microbiol. 8:2037. doi: 10.3389/fmicb.2017.02037
Ding, Z., Tao, T., Wang, L., Zhao, Y., Huang, H., Zhang, D., et al. (2019). Bioprospecting of novel and bioactive metabolites from endophytic fungi isolated from rubber tree Ficus elastica leaves. J. Microbiol. Biotechnol. 29, 731–738. doi: 10.4014/jmb.1901.01015
Estiarte, N., Lawrence, C. B., Sanchis, V., Ramos, A. J., and Crespo-Sempere, A. (2016). LaeA and VeA are involved in growth morphology, asexual development, and mycotoxin production in Alternaria alternata. Int. J. Food Microbiol. 238, 153–164. doi: 10.1016/j.ijfoodmicro.2016.09.003
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2009). Gaussian 09, Revision A.1, Gaussian, Inc., Wallingford, CT.
Harned, A. M., and Volp, K. A. (2011). The sorbicillinoid family of natural products: isolation, biosynthesis, and synthetic studies. Nat. Prod. Rep. 28, 1790–1810. doi: 10.1039/c1np00039j
Hong, E. J., Kim, N. K., Lee, D., Kim, W. G., and Lee, I. (2015). Overexpression of the laeA gene leads to increased production of cyclopiazonic acid in Aspergillus fumisynnematus. Fungal Biol. 119, 973–983. doi: 10.1016/j.funbio.2015.06.006
Jiang, T., Wang, M., Li, L., Si, J., Song, B., Zhou, C., et al. (2016). Overexpression of the global regulator LaeA in Chaetomium globosum leads to the biosynthesis of chaetoglobosin Z. J. Nat. Prod. 79, 2487–2494. doi: 10.1021/acs.jnatprod.6b00333
Kosalková, K., García-Estrada, C., Ullán, R. V., Godio, R. P., Feltrer, R., Teijeira, F., et al. (2009). The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 91, 214–225. doi: 10.1016/j.biochi.2008.09.004
Lebe, K. E., and Cox, R. J. (2019). O-methylation steps during strobilurin and bolineol biosynthesis. RSC Adv. 9, 31527–31531. doi: 10.1039/c9ra06412e
Li, M. F., Li, G. H., and Zhang, K. Q. (2019). Non-volatile metabolites from Trichoderma spp. Metabolites 9:58. doi: 10.3390/metabo9030058
Linde, T., Zoglowek, M., Lübeck, M., Frisvad, J. C., and Lübeck, P. S. (2016). The global regulator LaeA controls production of citric acidand endoglucanases in Aspergillus carbonarius. J. Ind. Microbiol. Biotechnol. 43, 1139–1147. doi: 10.1007/s10295-016-1781-3
Liu, Q., Cai, L., Shao, Y., Zhou, Y., Li, M., Wang, X., et al. (2016). Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biol. 120, 297–305. doi: 10.1016/j.funbio.2015.10.008
López-Berges, M. S., Schäefer, K., Hera, C., and Pietro, A. D. (2014). Combinatorial function of velvet and AreA in transcriptional regulation of nitrate utilization and secondary metabolism. Fungal Genet. Biol. 62, 78–84. doi: 10.1016/j.fgb.2013.11.002
Mao, X. M., Xu, W., Li, D., Yin, W. B., Chooi, Y. H., Li, Y. Q., et al. (2015). Epigenetic genome mining of an endophytic fungus leads to the pleiotropic biosynthesis of natural products. Angew. Chem. Int. Ed. 54, 7592–7596. doi: 10.1002/anie.201502452
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M., and Kenerley, C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129. doi: 10.1146/annurev-phyto-082712-102353
Reátegui, R. F., Wicklow, D. T., and Gloer, J. B. (2006). Phaeofurans and sorbicillin analogues from a fungicolous Phaeoacremonium species (NRRL 32148). J. Nat. Prod. 69, 113–117. doi: 10.1021/np0504199
Reino, J. L., Guerrero, R. F., Hernández-Galán, R., and Collado, I. G. (2008). Secondary metabolites from species of the biocontrol agent Trichoderma. Phytochem. Rev. 7, 89–123. doi: 10.1007/s11101-006-9032-2
Reyes-Dominguez, Y., Bok, J. W., Berger, H., Shwab, E. K., Basheer, A., Gallmetzer, A., et al. (2010). Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 76, 1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x
Rutledge, P. J., and Challis, G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523. doi: 10.1038/nrmicro3496
Seiboth, B., Karimi, R. A., Phatale, P. A., Linke, R., Hartl, L., Sauer, D. G., et al. (2012). The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol. Microbiol. 84, 1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x
Shaw, J. J., Spakowicz, D. J., Dalal, R. S., Davis, J. H., Lehr, N. A., Dunican, B. F., et al. (2015). Biosynthesis and genomic analysis of medium-chain hydrocarbon production by the endophytic fungal isolate Nigrograna mackinnonii E5202H. Appl. Microbiol. Biotechnol. 99, 3715–3728. doi: 10.1007/s00253-014-6206-5
Tang, M. C., Cui, X., He, X., Ding, Z., Zhu, T., Tang, Y., et al. (2017). Late-stage terpene cyclization by an integral membrane cyclase in the biosynthesis of isoprenoid epoxycyclohexenone natural products. Org. Lett. 19, 5376–5379. doi: 10.1021/acs.orglett.7b02653
Vinale, F., Sivasithamparam, K., Ghisalberti, E. L., Marra, R., Barbetti, M. J., Li, H., et al. (2008). A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 72, 80–86. doi: 10.1016/j.pmpp.2008.05.005
Wang, L., Zhang, X., Zhang, K., Zhang, X., Zhu, T., Che, Q., et al. (2020). Overexpression of global regulator PbrlaeA leads to the discovery of new polyketide in fungus Penicillium brocae HDN-12-143. Front. Chem. 8:270. doi: 10.3389/fchem.2020.00270
Wiemann, P., and Keller, N. P. (2014). Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 41, 301–313. doi: 10.1007/s10295-013-1366-3
Wu, J. S., Shi, X. H., Zhang, Y. H., Yu, J. Y., Fu, X. M., Li, X., et al. (2019). Co-cultivation with 5-azacytidine induced new metabolites from the zoanthid-derived fungus Cochliobolus lunatus. Front. Chem. 7:763. doi: 10.3389/fchem.2019.00763
Wu, J. S., Yao, G. S., Shi, X. H., Rehman, S. U., Xu, Y., Fu, X. M., et al. (2020). Epigenetic agents trigger the production of bioactive nucleoside derivatives and bisabolane sesquiterpenes from the marine-derived fungus Aspergillus versicolor. Front. Microbiol. 11:85. doi: 10.3389/fmicb.2020.00085
Wu, G., Zhou, H., Zhang, P., Wang, X., Li, W., Zhang, W., et al. (2016). Polyketide production of pestaloficiols and macrodiolide ficiolides revealed by manipulations of epigenetic regulators in an endophytic fungus. Org. Lett. 18, 1832–1835. doi: 10.1021/acs.orglett.6b00562
Yu, J., Han, H., Zhang, X., Ma, C., Sun, C., Che, Q., et al. (2019). Discovery of two new sorbicillinoids by overexpression of the global regulator LaeA in a marine-derived fungus Penicillium dipodomyis YJ-11. Mar. Drugs 17:446. doi: 10.3390/md17080446
Zeilinger, S., Gruber, S., Bansal, R., and Mukherjee, P. K. (2016). Secondary metabolism in Trichoderma—chemistry meets genomics. Fungal Biol. Rev. 30, 74–90. doi: 10.1016/j.fbr.2016.05.001
Zeng, Z. Q., and Zhuang, W. Y. (2019). Two new species and a new Chinese record of Hypocreaceae as evidenced by morphological and molecular data. Mycobiology 47, 280–291. doi: 10.1080/12298093.2019.1641062
Keywords: global regulator, LaeA, polyketides, antifungal activity, endophytic fungi, Trichoderma afroharzianum
Citation: Ding Z, Wang X, Kong F-D, Huang H-M, Zhao Y-N, Liu M, Wang Z-P and Han J (2020) Overexpression of Global Regulator Talae1 Leads to the Discovery of New Antifungal Polyketides From Endophytic Fungus Trichoderma afroharzianum. Front. Microbiol. 11:622785. doi: 10.3389/fmicb.2020.622785
Received: 29 October 2020; Accepted: 04 December 2020;
Published: 23 December 2020.
Edited by:
Paola Angelini, University of Perugia, ItalyReviewed by:
Weaam Ebrahim, Mansoura University, EgyptCopyright © 2020 Ding, Wang, Kong, Huang, Zhao, Liu, Wang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuang Ding, ZGluZ3podWFuZ0BsY3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.