
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 08 January 2021
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.619618
This article is part of the Research Topic RNA Biology of Microorganisms View all 12 articles
Timely and accurate RNA synthesis depends on accessory proteins that instruct RNA polymerase (RNAP) where and when to start and stop transcription. Among thousands of transcription factors, NusG/Spt5 stand out as the only universally conserved family of regulators. These proteins interact with RNAP to promote uninterrupted RNA synthesis and with diverse cellular partners to couple transcription to RNA processing, modification or translation, or to trigger premature termination of aberrant transcription. NusG homologs are present in all cells that utilize bacterial-type RNAP, from endosymbionts to plants, underscoring their ancient and essential function. Yet, in stark contrast to other core RNAP components, NusG family is actively evolving: horizontal gene transfer and sub-functionalization drive emergence of NusG paralogs, such as bacterial LoaP, RfaH, and UpxY. These specialized regulators activate a few (or just one) operons required for expression of antibiotics, capsules, secretion systems, toxins, and other niche-specific macromolecules. Despite their common origin and binding site on the RNAP, NusG homologs differ in their target selection, interacting partners and effects on RNA synthesis. Even among housekeeping NusGs from diverse bacteria, some factors promote pause-free transcription while others slow the RNAP down. Here, we discuss structure, function, and evolution of NusG proteins, focusing on unique mechanisms that determine their effects on gene expression and enable bacterial adaptation to diverse ecological niches.
In every living cell, multi-subunit RNA polymerases (RNAPs) carry out the first step of gene expression, transcription of a DNA template into an RNA copy. Reflecting their common evolutionary origin in the last universal common ancestor (LUCA) and the basic mechanism of RNA synthesis, RNAPs share an overall architecture and structural elements that play key roles in the assembly of transcription complexes, substrate selection and catalysis, interactions with nucleic acids, etc. (Lane and Darst, 2010a,b). However, extant RNAPs differ greatly in subunit composition and sequence: core RNAPs are composed of 5–7 subunits in bacteria vs. 12+ subunits in archaea and eukaryotes, and even RNAPs from mesophilic bacteria Escherichia coli and Bacillus subtilis are only 50% identical. Differences in cellular transcriptional machinery are thought to reflect unique regulatory constraints imposed by diverse habitats. In support of this notion, even basal general transcription factors that assist RNAP during each step of the transcription cycle are not conserved between kingdoms. The sole exception to this trend is a transcription elongation factor NusG (Werner, 2012).
Bacterial Nus (N-utilization substance) proteins have been identified genetically based on their requirement for the coliphage λ development (Casjens and Hendrix, 2015). In E. coli and Salmonella, potentially harmful xenogenes are silenced by premature transcription termination by a hexameric RNA helicase Rho (Peters et al., 2012; Bossi et al., 2019). To escape silencing, bacteriophages have evolved antitermination mechanisms targeting Rho or RNAP (Santangelo and Artsimovitch, 2011). The immediate early gene N of phage λ is required for the expression of delayed-early genes. N nucleates the assembly of a large transcription antitermination complex (TAC) composed of RNAP and NusABEG proteins (Mason and Greenblatt, 1991; Krupp et al., 2019) and a similar TAC assembles during transcription of the E. coli ribosomal RNA operons (Squires et al., 1993; Huang et al., 2020). NusA and NusG are general transcription elongation factors, which are associated with RNAP transcribing all genes, at least in E. coli (Mooney et al., 2009a). NusE, a.k.a. the ribosomal protein S10, requires a binding partner NusB to remain soluble while not a part of the ribosome; NusB is selectively enriched on rRNA operons (Mooney et al., 2009a), consistent with its principal role in rRNA synthesis. Among the shared components of the TACs, NusG is the only factor that facilitates transcription elongation in vivo and in vitro (Burova et al., 1995; Burns et al., 1998; Zellars and Squires, 1999); by contrast, NusA increases RNAP pausing and intrinsic termination, whereas NusB/E have no effect (Belogurov and Artsimovitch, 2015).
All NusG-like proteins (NusG in bacteria; Spt5 in archaea and yeast, DSIF in mammals) bind to an evolutionary conserved site on the largest RNAP subunit (Klein et al., 2011; Martinez-Rucobo et al., 2011; Ehara et al., 2017; Kang et al., 2018; Vos et al., 2018). The NusG binding site is located on the tip of the RNAP clamp, a conserved flexible module that closes over the DNA binding channel. The clamp closes during the formation of a transcriptionally competent initiation complex, remains closed throughout elongation, and opens during termination (Belogurov and Artsimovitch, 2019); more subtle movements of the clamp have been proposed to accompany RNAP pausing, which serves as a prelude to termination (Kang et al., 2019). By keeping the clamp locked, NusG proteins are thought to promote continuous, pause-free RNA synthesis, an essential function given that the premature release of the RNA transcript is irreversible. The presence of a clamping factor in LUCA thus underscores the fundamental importance of transcription processivity, particularly on difficult templates (Werner, 2012).
The antipausing and, by inference, antitermination activity of NusG prompted its annotation as a transcription antiterminator. Likewise, many subsequently discovered bacterial NusG homologs have been shown to possess antitermination activity (Artsimovitch and Knauer, 2019). Nevertheless, this view has been challenged since the time of E. coli NusG discovery by the data in support of its role as a termination-promoting factor. NusG is essential in wild-type E. coli (Downing et al., 1990) and its depletion leads to defects in Rho-dependent termination (Sullivan and Gottesman, 1992). NusG aids Rho in silencing transcription of damaged and harmful RNAs genome-wide (Peters et al., 2012) and promotes efficient termination by Rho in vitro (Burns and Richardson, 1995). Indeed, the nusG gene can be deleted, albeit at a significant fitness cost, in an E. coli strain lacking the toxic rac prophage, which is silenced by Rho (Cardinale et al., 2008). Point mutations in nusG that lead to defects in transcription termination (Saxena and Gowrishankar, 2011) or interactions with the ribosome (Saxena et al., 2018) do not have significant fitness phenotypes.
Functional studies of NusG-like proteins from different bacteria support a picture in which these factors can mediate diverse effects on RNA synthesis (Figure 1). Through contacts to RNAP, nucleic acids, and auxiliary proteins, NusG homologs can suppress or promote transcriptional pausing and termination and bridge RNAP to other cellular machineries. Most unusually for a family of alternative transcription regulators, although binding to the same site on the transcribing RNAP, NusG-like proteins frequently have exactly opposite effects on the expression of some genes, most notably those encoding virulence determinants. Furthermore, even the housekeeping NusG proteins have seemingly opposite effects on RNA synthesis; for example, unlike its E. coli counterpart, B. subtilis NusG promotes RNAP pausing in vitro and in vivo (Yakhnin et al., 2016, 2020a). Below, we describe recent advances in our understanding of molecular mechanisms, evolution, and regulatory diversity of bacterial NusG-like proteins.
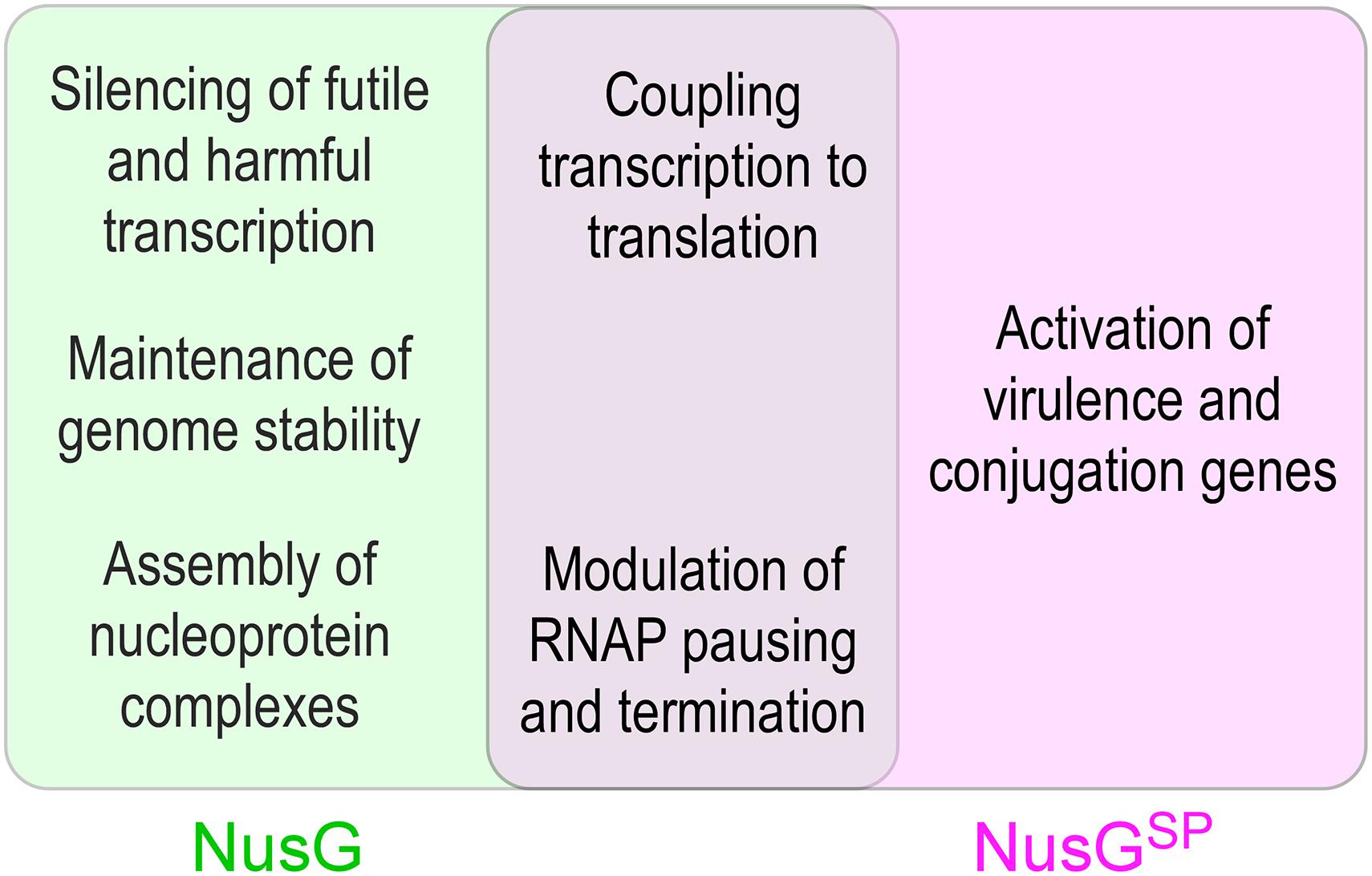
Figure 1. Unique and overlapping cellular functions of housekeeping E. coli NusG and its specialized paralogs.
NusG-like proteins have a similar structural core consisting of a NusG N-terminal domain (NGN) and a C-terminal domain with a 27-residue long Kyrpides-Ouzounis-Woese (KOW) motif common among RNA-binding proteins (Kyrpides et al., 1996; Ponting, 2002; Figure 2). Bacterial NusG alone can perform its function, while Spt5 has an obligatory partner—a small zinc finger protein Spt4 (called RpoE in archaea). Eukaryotic Spt5 contains several KOW domains, the first of which carries a large insertion, an N-terminal acidic region, and an unstructured C-terminal repeat (CTR) domain (Figure 2A); in metazoan DSIF, additional KOWs are present at the very C terminus of the protein (Decker, 2020). Apart from the KOW1 insertion, the NGN and KOW domains from all life have very similar topologies (Figure 2B).
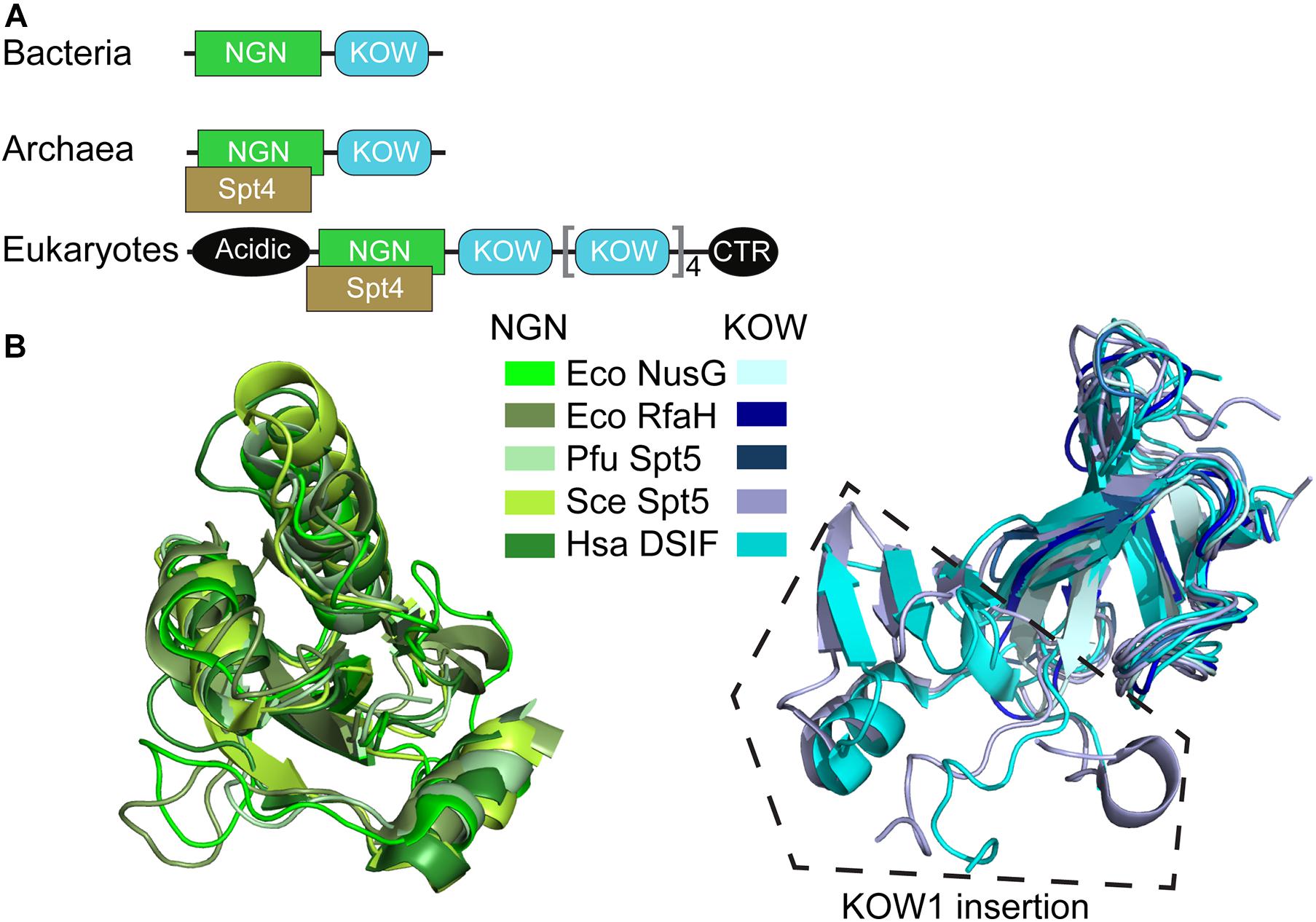
Figure 2. Structural conservation of NusG-like proteins. (A) Domain organization. (B) Superposition of NGN and KOW domains. PDB IDs: E. coli (Eco) NusG-NGN: 2K06; Eco NusG-KOW: 2KVQ; Eco RfaH-NGN: 2OUG; Eco RfaH-KOW: 2LCL; Pyrococcus furiosus (Pfu) Spt5-NGN/KOW: 3P8B; Saccharomyces cerevisiae (Sce) Spt5-NGN: 2EXU; Sce Spt5-KOW1: 4YTK; Sce Spt5-KOW2/3: 4YTL; Homo sapiens (Hsa) DSIF-NGN: 3H7H; Hsa DSIF-KOW1: 5OIK; Hsa DSIF-KOW2: 2E6Z; Hsa DSIF-KOW3: 2DO3; Hsa DSIF-KOW4: 5OHO; Hsa DSIF-KOW5: 2E70. Sce Spt5-KOW1/2/3 have similar structures and are shown in the same color, as are Hsa DSIF-KOW1/2/3/4/5 domains.
All NGN domains make very similar contacts to two conserved RNAP elements (Klein et al., 2011; Martinez-Rucobo et al., 2011; Ehara et al., 2017; Kang et al., 2018), the clamp helices (CH) in the largest RNAP subunit (β’ in Bacteria) and the gate loop in the second largest subunit (β in Bacteria).
In addition, some NGNs make sequence-specific contacts to the non-template DNA strand in the transcription bubble of the transcription elongation complex (TEC; see below). The NGN binding site on the TEC is structurally analogous to binding sites of transcription initiation factors in promoter complexes; e.g., bacterial σ factors recognize non-template DNA sequences and an adjacent region on the β’ CH during promoter-dependent initiation (Zhang et al., 2012). Consequently, NusG/Spt5 proteins compete with the cognate initiation factors for binding to RNAP, reducing pausing during transcription elongation and potentially facilitating promoter escape (Sevostyanova et al., 2008; Grohmann et al., 2011). Along with the housekeeping NusG present in every free-living cell, many species also contain NusG paralogs (Wang B. et al., 2020) that regulate expression of selected genes in a sequence- or condition-specific fashion.
While the “clamping” contacts between the NGN and TEC are sufficient for NusG/Spt5 effects on RNA synthesis (Mooney et al., 2009b; Hirtreiter et al., 2010), the KOW domains determine their regulatory properties. In E. coli NusG, interactions between the KOW domain and Rho facilitate termination (Lawson et al., 2018), whereas the KOW-ribosome interactions couple transcription to translation (Saxena et al., 2018). In eukaryotic Spt5, the presence of multiple KOWs and the CTR, which acts as a hub for recruitment of several RNA processing enzymes and other cellular factors (Decker, 2020), expands the range of regulatory interactions.
Accurate and timely execution of the gene expression program is essential for cell survival. By itself, RNAP is a passive interpreter of genetic information. Auxiliary proteins instruct RNAP to synthesize RNAs that are required for proper cellular function and prevent it from wasting resources on making useless or potentially harmful RNAs, such as antisense transcripts or mRNAs encoding toxic proteins. In E. coli, the housekeeping NusG travels with RNAP transcribing almost all genes (Mooney et al., 2009a), save a few controlled by its paralog RfaH (Belogurov et al., 2009), actively contributing to the transcriptome surveillance. First, NusG cooperates with Rho to silence transcription of aberrant RNAs; this is an essential function of E. coli NusG (Mitra et al., 2017). Second, NusG increases RNAP processivity by modifying properties of the TEC, a shared function of NusG proteins from all life. Third, NusG is an integral part of multi-component nucleoprotein complexes that promote facile synthesis and proper assembly of the ribosomal RNAs, and thus the ribosomes. Finally, NusG helps to protect translatable mRNAs from premature release by Rho by bridging the RNAP and the ribosome.
Rho is an ATP-dependent, RecA-type hexameric helicase that terminates transcription of a wide variety of genes in bacteria. Initially viewed as a sequence-specific terminator that requires a C-rich Rho utilization (rut) element for loading onto the nascent RNA and subsequent TEC dissociation, Rho has recently emerged as a global multi-functional regulator (Mitra et al., 2017). In addition to its canonical role, inducing termination at the end of some genes (Peters et al., 2012), Rho silences transcriptional noise and expression of horizontally acquired genes, reduces translational stress, and prevents replication-transcription collisions. Genome-wide studies demonstrate that E. coli Rho travels with the elongating RNAP, together with NusG and NusA (Mooney et al., 2009a), from the onset of elongation, and acts on numerous cellular targets that lack easily recognizable rut sequences (Peters et al., 2012).
To silence AT-rich xenogenes and trigger the release of antisense transcripts or low-quality mRNAs independently of their sequence, Rho relies on help from NusG, which has been implicated in Rho termination at suboptimal, C-less sites (Peters et al., 2012). In a binary system lacking RNAP, NusG activates Rho by promoting isomerization from an open-ring, RNA-loading state, to a closed-ring, translocation-competent state, the transition otherwise triggered by a perfect rut element in the RNA (Lawson et al., 2018). The NusG KOW interacts with the C-terminal translocase domain of Rho (Figure 3), inducing conformational changes that favor the ring closure even on RNAs devoid of C residues (Lawson et al., 2018). NusG-Rho contacts are mediated by the same KOW region that binds to the ribosomal protein S10 (Burmann et al., 2010), explaining why the translating pioneering ribosome protects the mRNA from a spurious attack by Rho. By contrast, the corresponding Rho-binding residues are missing in RfaH (Lawson et al., 2018), explaining why RfaH does not bind to Rho.
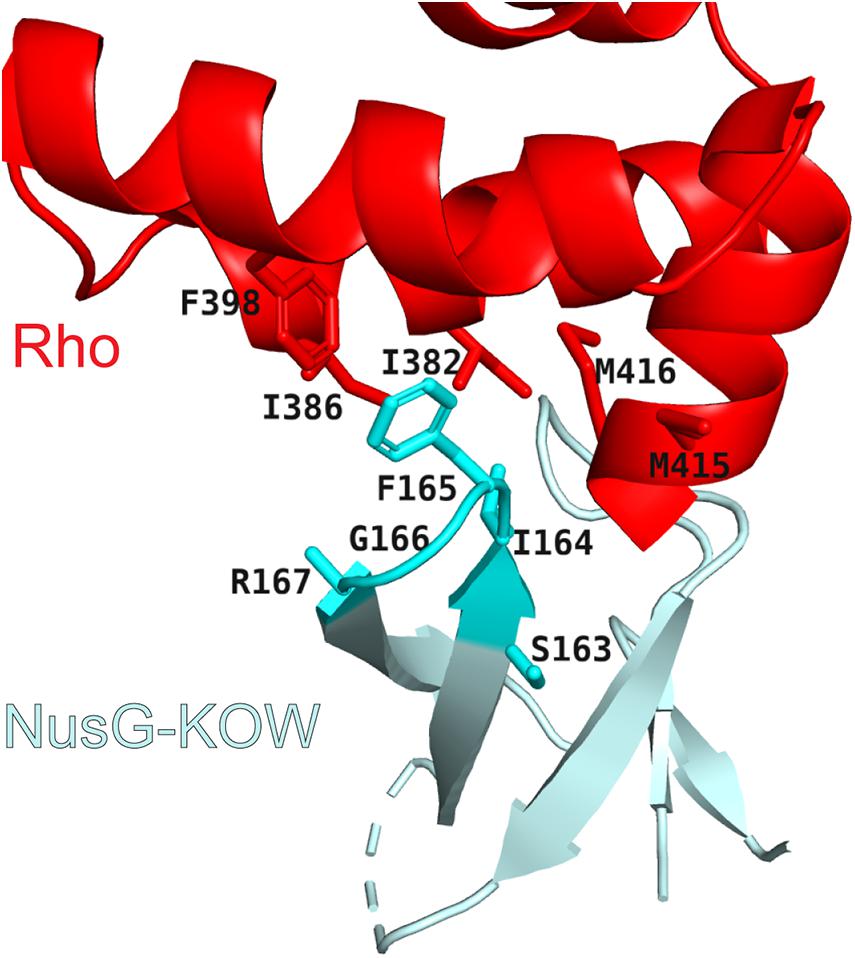
Figure 3. Rho/NusG-KOW interface. Rho residues that contact NusG are shown as red sticks. NusG KOW residues implicated in Rho and S10 binding are shown as cyan sticks; PDB ID: 6DUQ.
However, the ring closure activity of NusG may not be the main mechanism by which NusG stimulates Rho-dependent termination. Consistent with biochemical data (Schmidt and Chamberlin, 1984; Epshtein et al., 2010) and genome-wide mapping (Mooney et al., 2009a) that support persistent Rho-RNAP interactions, a recent cryo-EM analysis of the E. coli TEC under attack by Rho reveals seven complexes thought to represent sequential steps in the termination pathway (Said et al., 2020). During the initial binding to the TEC, Rho makes numerous contacts to the RNAP subunits, NusA and NusG NGN (Figure 4), but captures the nascent RNA transcript only later in the pathway. Once engaged, Rho induces dramatic conformational changes in RNAP and Nus factors, which ultimately trap a moribund TEC in which the clamp is wide open and the RNA 3′ end is dislodged from the RNAP active site (Said et al., 2020), a model initially proposed by Nudler and colleagues (Epshtein et al., 2010). In this structurally defined pathway, NusG NGN assists Rho loading onto the RNA and then dissociates to allow for Rho-mediated RNAP clamp opening, whereas NusG KOW is invisible. Remarkably, the Rho ring remains opens even in the moribund TEC, implying that the NusG-promoted Rho helicase activity is required to unwind the RNA:DNA hybrid only after RNAP inactivation; this model is supported by a report that the E. coli rho gene becomes dispensable in the presence of a heterologous RNA:DNA helicase (Leela et al., 2013). The allosteric model of termination explains how Rho selectively binds to RNAs that are still being made and reinforces the notion that, even in bacteria, transcriptional regulators act in the context of multi-protein complexes, rather than on RNAP alone.
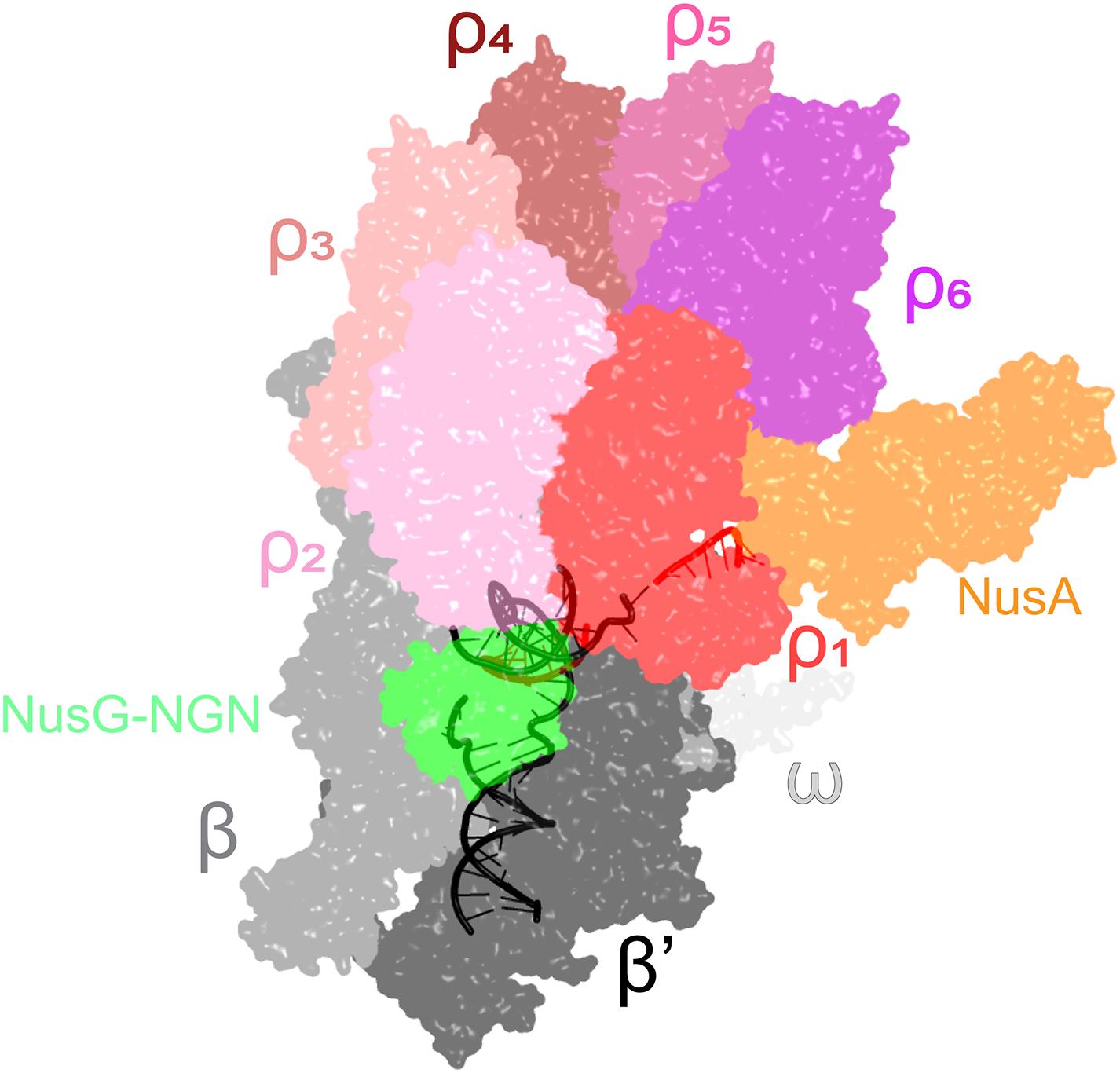
Figure 4. A cryo-EM structure of the Rho engagement complex, in which Rho hexamer makes initial contacts with the transcribing RNAP bound to NusA and NusG. The DNA is shown in black; the RNA—in red. PDB ID: 6Z9P.
Indeed, recent evidence suggests that Rho and NusG cooperate with the histone-like nucleoid-structuring (H-NS) protein, a prototypical xenogeneic silencer, to limit unwanted gene expression. In E. coli, Rho and H-NS co-localize on the chromosome (Chandraprakash and Seshasayee, 2014) and mutations in rho and hns lead to synergistic growth defects (Peters et al., 2012). In Salmonella, depletion of NusG leads to massive upregulation of H-NS silenced loci, which include pathogenicity islands and are devoid of rut sites; consistently, mutations that compromise Rho-rut contacts have no effect on NusG-mediated silencing (Bossi et al., 2019). While the molecular mechanism of this cooperation remains to be determined, it likely reflects RNAP stalling when running into nucleoprotein filaments assembled by H-NS and other nucleoid-associated proteins on the template DNA (Boudreau et al., 2018).
During transcription of cellular DNA, RNAP frequently encounters unfavorable sequences or obstacles, such as DNA-bound proteins or DNA lesions, that slow the enzyme down or induce arrest. Retrograde movement of the RNAP along the RNA and DNA chains, or backtracking, is a common mechanism of pausing and arrest (Nudler, 2012). Backtracked complexes are rendered inactive because the nascent RNA is extruded through the active site, blocking nucleotide addition (Figure 5). The arrested complexes are long-lived, blocking progression of other RNAPs and replisomes, and must be released or reactivated upon transcript cleavage. Cleavage of the backtracked RNA, which is mediated by the RNAP active site and is strongly enhanced by Gre cleavage factors (Sosunova et al., 2003), repositions the 3’ end of the RNA in the active site. By preventing backtracking, an activity well-documented in the case of NusG and RfaH (Svetlov et al., 2007; Herbert et al., 2010), NusG-like proteins facilitate processive transcription and promote genome stability. Recent functional and structural data suggest a molecular mechanism of enhanced RNAP processivity, in which the NGN domain loops out the non-template DNA, bringing the upstream and downstream DNA duplexes closer together (Turtola and Belogurov, 2016; Kang et al., 2018; Nedialkov et al., 2018), and establishes contacts to the upstream DNA duplex (Krupp et al., 2019; Said et al., 2020). Together, these interactions alter the upstream DNA trajectory (Figure 5) and stabilize the upstream edge of the transcription bubble, which must melt to allow backtracking, explaining how NusG and RfaH inhibit backtracking (Svetlov et al., 2007; Herbert et al., 2010). In addition, the NGN domain, at least in the case of RfaH (Kang et al., 2018), disfavors subtle conformational changes (termed swiveling) that accompany the formation of hairpin-stabilized paused TEC (Kang et al., 2019) and constrains the path of the non-template DNA, preventing it from assuming non-productive conformations (Nedialkov et al., 2018); a similar mechanism has been proposed for yeast Spt5 (Crickard et al., 2016). Together, the NGN-promoted changes in the TEC ensure pause-free RNA synthesis, preventing arrest and termination.
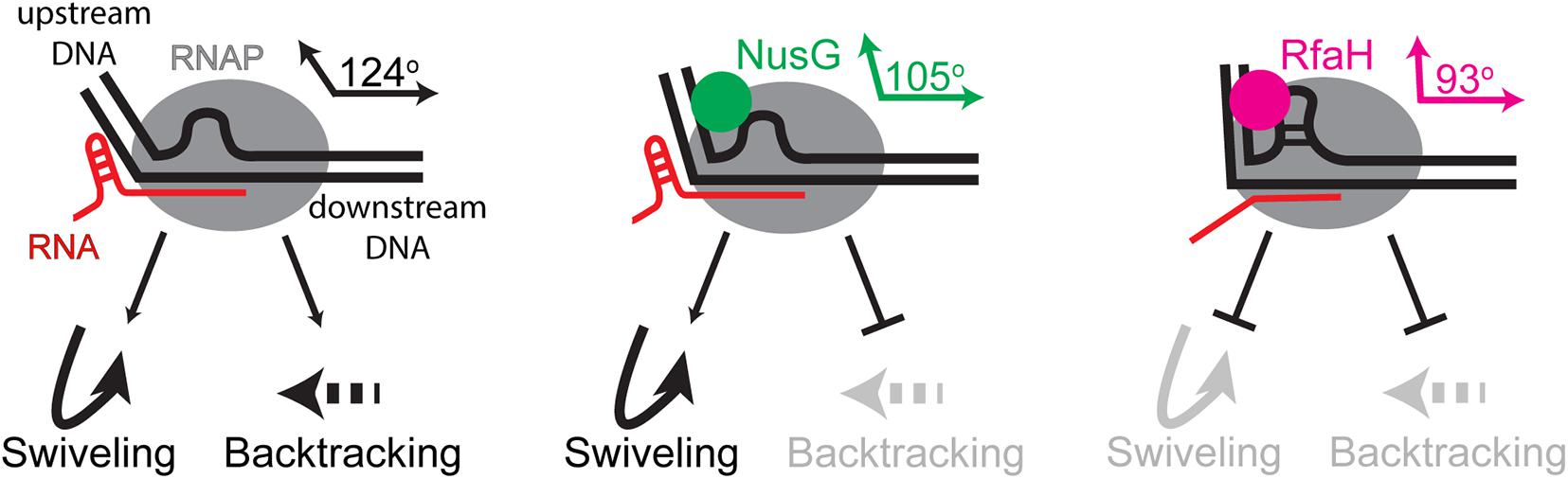
Figure 5. Antipausing activities of E. coli NusG and RfaH. Upon encountering a pause-inducing sequence, RNAP can either backtrack or undergo conformational changes termed swiveling; the latter are stabilized by formation of a pause hairpin in the nascent RNA. The NGN domains of both proteins bind near the upstream edge of the transcription bubble, promoting forward and thus inhibiting backward translocation. Transient (NusG) or stable (RfaH) interactions with the non-template DNA strand bring the upstream and downstream DNA duplexes closer together (indicated by angles between these duplexes), an effect that is more pronounced with RfaH. RfaH also binds to the β’ and β subunits with higher affinity, restricting the clamp movements to inhibit swiveling and hairpin-stabilized pausing. NusG lacks this activity.
To enact RNA surveillance, Rho travels with the elongating RNAP and probes the nascent RNA “translatability.” RNAs that contain premature stop codons or are poorly translated, e.g., under conditions of proteotoxic stress, are released by Rho (Richardson, 1991). Yet a very large fraction of cellular RNA is never translated, most notably the most abundant and absolutely essential rRNA which comprises ∼50% of the newly synthesized RNA during the exponential growth phase (Dennis et al., 2004). Thus, making rRNA rapidly while protecting it from Rho is key to the survival of cells. Similarly, phage replication is critically dependent on uninterrupted transcription of the phage genome, but Rho is known to broadly silence xenogenes, including phages (Mitra et al., 2017).
Protection of the phage λ early genes and E. coli rRNA operons (rrn) from Rho is conferred by multicomponent TACs. Recently solved cryo-EM structures of these TACs (Figure 6) revealed common and unique details of their action (Krupp et al., 2019; Huang et al., 2020). Both complexes assemble on boxA and boxB elements in the nascent RNA and share a set of NusABEG factors. Each complex also includes unique factors, N in the λN-TAC and an inositol monophosphatase SuhB dimer + the ribosomal protein S4 in the rrn-TAC.
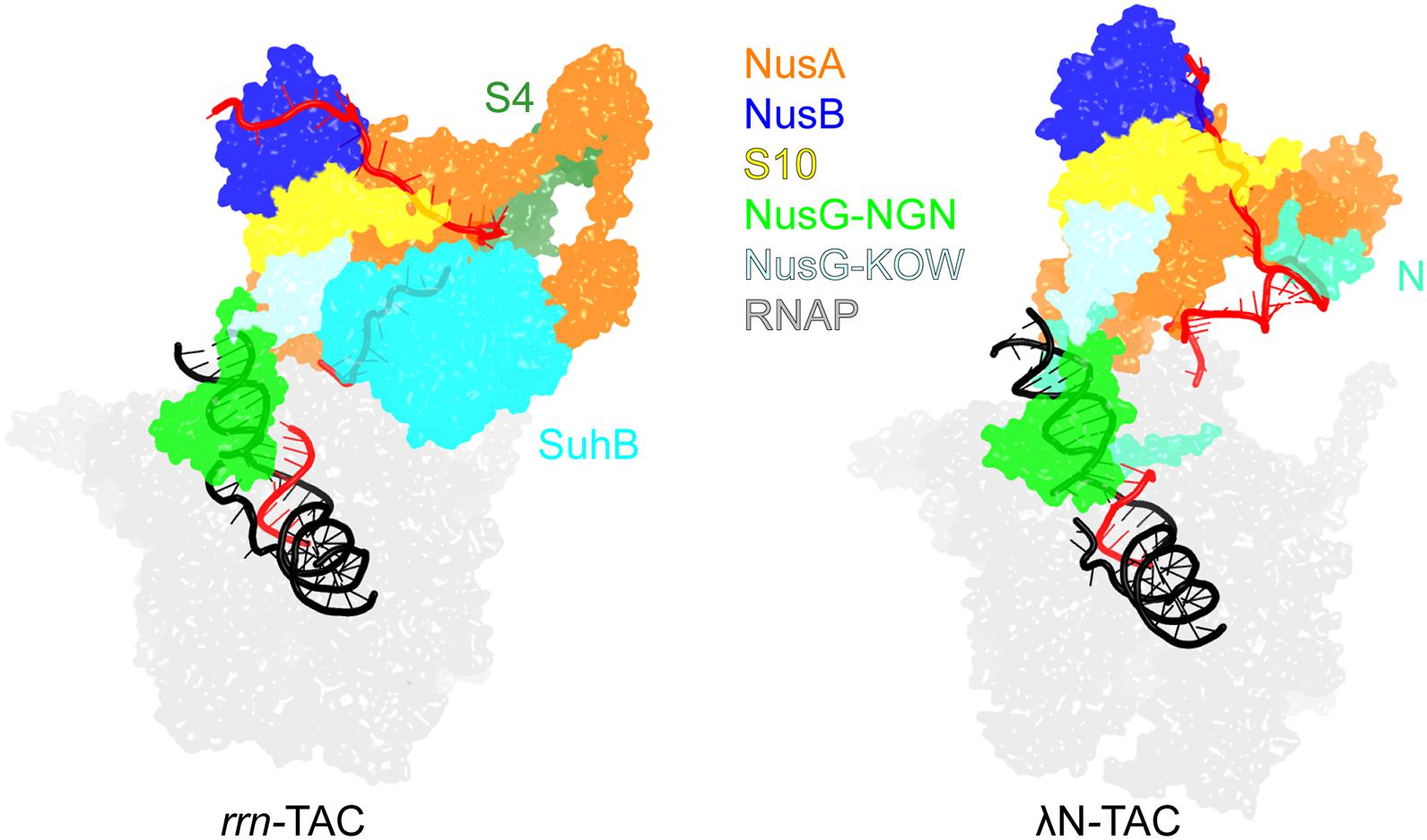
Figure 6. Transcription antitermination complexes (TAC). Left, rrn-TAC; right, phage λN-TAC. RNA is in red, DNA is in black. The unique proteins that play key roles in antitermination are shown on each complex; the shared components are indicated in the middle. PDB ID: rrn-TAC, 6TQO; λN-TAC, 6GOV.
The λN-TAC is resistant to pausing and termination elicited by hairpin signals and Rho. An intrinsically unstructured λN is the principal player which uses a range of mechanisms to modify the TEC (Krupp et al., 2019). λN snakes inside the RNAP, making contacts to multiple RNAP domains and repositioning others, and rearranges Nus factor interactions. λN stabilizes the elongation-competent state of RNAP, inhibiting the nascent RNA hairpin formation and its stabilization by NusA, supports the anti-backtracking and anti-swiveling action of the NusG NGN domain. In the λN-TAC, neither NusG domain can make contacts to Rho observed in the binary Rho-NusG complex (Lawson et al., 2018) and Rho-TEC (Said et al., 2020) structures. Consequently, in the λN-TAC, NusG anti-pausing activity is augmented while its termination-promoting activity is abolished.
Although the rrn-TAC has a different protein composition, analogous structural changes inhibit backtracking and NusA-stabilized hairpin pausing and sequester NusG from Rho, with a much larger, well-folded SuhB dimer playing a central role in restructuring of the TAC components instead of λN (Singh et al., 2016). Notably, in addition to promoting pause- and termination-free RNA synthesis, the rrn-TAC acts as a molecular chaperone that actively assists the folding and maturation of the nascent RNA (Huang et al., 2020). Similarly to the ribosome-associated chaperones, SuhB, S4 and Nus factors assemble into a ring around the RNA exit channel, extending the channel outward to accommodate a longer segment of the exiting RNA. The RNA is thus sequestered away from the upstream DNA, blocking formation of deleterious R-loops, and is held within a positively charged protein cage to promote folding of local secondary structures and annealing of distant segments, which is required for processing of rRNA precursors into mature forms (Young and Steitz, 1978).
NusG plays a supporting role in both TACs: e.g., λN alone has a short-range antitermination activity and requires the TAC assembly to act over long distances (Rees et al., 1996). By contrast, RfaH is a principal, self-sufficient antiterminator: RfaH acts over very long distances yet its activity is not affected by cellular factors, at least in vitro (Artsimovitch and Landick, 2002). Other NusGSP may similarly act alone.
In prokaryotic cells, the lack of a nuclear membrane provides an opportunity for direct physical interaction of the transcribing RNAP and the translating ribosome. The translation-coupled synthesis of the nascent mRNA is known as transcription-translation coupling. The coupling was directly observed by electron microscopy in 1970 in E. coli cells (Miller et al., 1970) and subsequently in archaeon Thermococcus kodakarensis (French et al., 2007). RNAP and ribosomes form a one-to-one complex with about 1 μM dissociation constant, which is already well within a physiologically relevant range, even in the absence of the nascent mRNA and accessory factors (Fan et al., 2017), resulting in factor-free coupling. Alternatively, the two complexes can be linked by bridging factors, e.g., via the NusG:S10 captured by NMR (Burmann et al., 2010). Substitutions at the E. coli NusG:S10 binding interface weakened NusG:S10 association in vivo and completely abolished it in vitro (Saxena et al., 2018).
The TEC-ribosome complexes, stabilized by general transcription factors, have been observed in vitro using cryo-EM (Wang C. et al., 2020; Webster et al., 2020) and analyzed inside cells using a combination of cross-linking mass spectrometry and cryo–electron tomography (O’Reilly et al., 2020). Evidence suggests that coupling may occur initially via direct RNAP:ribosome contacts and then is aided by accessory factors (Washburn et al., 2020). In the NusG/NusA coupled complex, the RNAP β’ subunit contacts the 30S subunit protein S3, NusA simultaneously binds to α/β subunits and S2/S5, and finally NusG binds to β/β’ and S10 (Figure 7). If the ribosome approaches the RNAP further, the collided state, in which the ribosome translocation and the factor-mediated coupling are no longer possible, forms (Wang C. et al., 2020; Webster et al., 2020). Preventing such unproductive collisions may be another function of NusA and NusG.
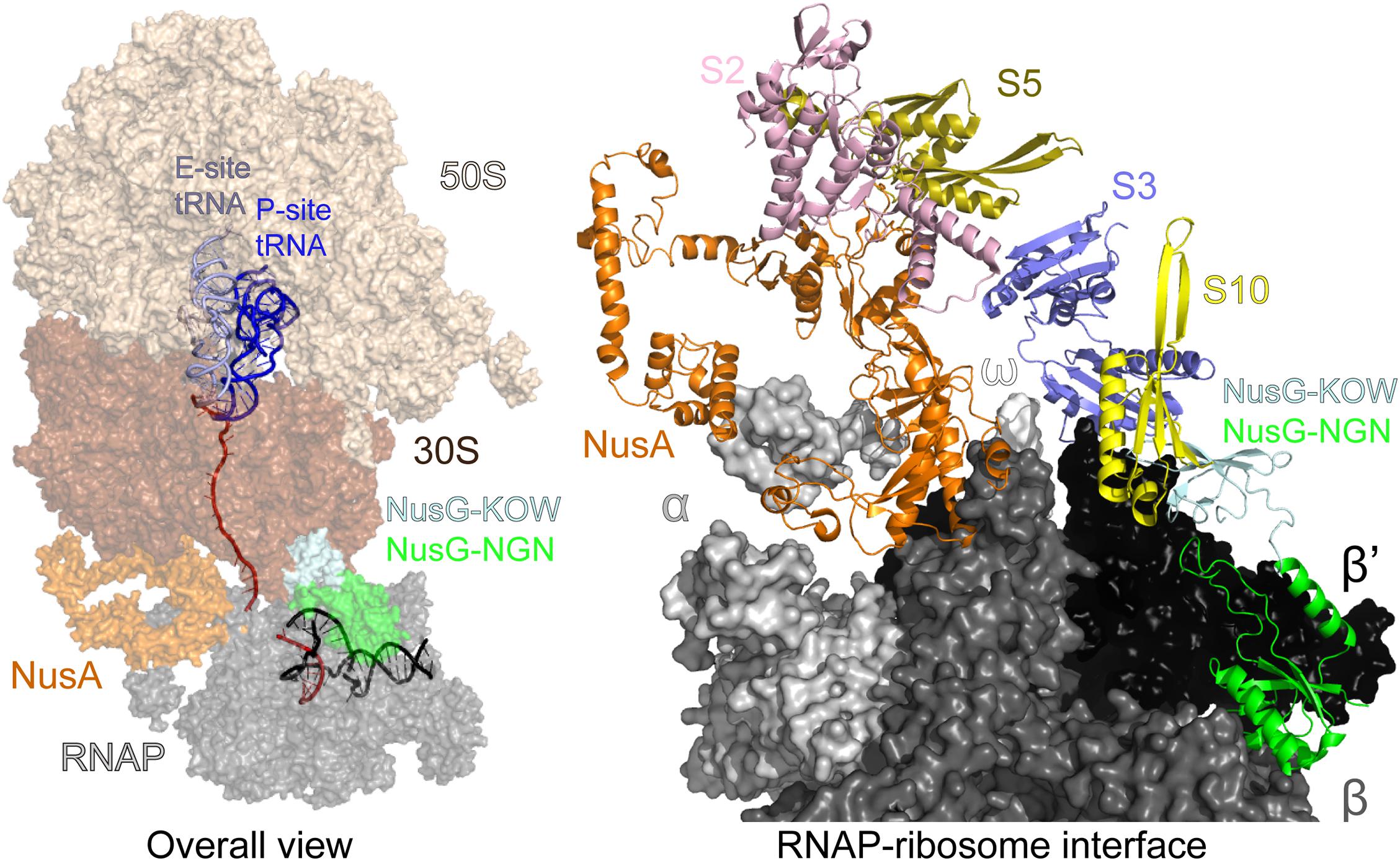
Figure 7. Transcription-translation coupling. Left—an overall view: mRNA is in red, DNA is in black. The interface between the ribosomal 30S subunit and RNAP is stabilized by NusA and NusG. Right—a view of the coupling interface; mRNA, DNA, the entire 50S and most of the 30S subunit have been removed for clarity. PDB ID: 6 × 7F.
Since RNAP might often transcribe without a linked ribosome (Chen and Fredrick, 2018), the coupling events must carry important regulatory information (McGary and Nudler, 2013). The closely coupled ribosome prevents the formation of R-loops and RNAP backtracking, thereby promoting genome stability (Gowrishankar and Harinarayanan, 2004; Proshkin et al., 2010; Stevenson-Jones et al., 2020) and inhibits factor-independent termination by blocking the formation of nascent RNA hairpins (Roland et al., 1988). The coupled ribosome also prevents mRNA degradation, by blocking the access of RNaseE (Iost and Dreyfus, 1995), or premature Rho termination, by sequestering NusG and shielding the nascent RNA (Washburn et al., 2020). When the coupling is broken, e.g., by the ribosome pausing or stalling, Rho releases the nascent RNA, a phenomenon known as polarity (Richardson, 1991). Transcription attenuation is another regulatory mechanism dependent on coupling between the RNAP and the trailing ribosome, wherein the formation of an RNA hairpin induces RNAP pausing and the trailing ribosome pushes the RNAP out of the pause (Turnbough, 2019). By stabilizing the RNAP-ribosome tandem or aiding Rho, NusG controls the fate of the nascent RNA, promoting its translation or release.
The universal conservation of the NusG structure and its binding site on the RNAP, as well as perceived common principles of gene expression control in bacteria, justified using the E. coli NusG as a paradigm. However, early and recent data suggest that, beyond occupying the same site on RNAP, even housekeeping NusGs, which are encoded within the conserved genomic locus, secE-nusG-rplK-rplA in evolutionary distant bacterial phyla (Wang B. et al., 2020), have relatively few common features. Comparison of NusG proteins from E. coli and B. subtilis, the best studied Gram-negative and Gram-positive model bacteria that grow very similarly in the lab, illustrates these differences.
In wild-type E. coli, nusG and rho genes are essential; their deletions can be obtained only in specially engineered strains (Leela et al., 2013) and confer significant growth defects. In contrast, neither gene is essential in B. subtilis (Ingham et al., 1999), in which Rho has limited effects on gene regulation (Nicolas et al., 2012), early stop codons do not induce polarity (Johnson et al., 2020), and most transcription termination is induced by hairpin signals (Mondal et al., 2016; Johnson et al., 2020). In contrast to E. coli, where NusG aids Rho in termination of rut-less RNAs (Lawson and Berger, 2019), Rho-dependent termination in B. subtilis is strongly linked to cis-encoded C-rich RNA elements (Johnson et al., 2020). Together, these results suggest that NusG is not involved in gene expression control by Rho in B. subtilis (and perhaps other related bacteria) and raise a possibility that an alternative mechanism of transcription noise silencing operates in these species.
Another key function of E. coli NusG is bridging the RNAP and the ribosome (Figure 7) to mediate transcription-translation coupling, which is thought to occur in all single-compartment cells (see above). In addition to preventing Rho-dependent termination, which may be irrelevant in B. subtilis, the coupled ribosome inhibits RNAP backtracking (Proshkin et al., 2010; Stevenson-Jones et al., 2020) and could disfavor the formation of deleterious R-loops (Gowrishankar et al., 2013). The pioneer round of translation may also prime the RNA for subsequent rounds of translation. Strikingly, a recent report demonstrates that transcription and translation are uncoupled in B. subtilis (Johnson et al., 2020), where RNAP moves along the template about twice as fast as the ribosome does. While in E. coli the coupled ribosome inhibits both intrinsic and Rho-dependent termination, termination in B. subtilis is unaffected by translation. The loss of coupling has a profound effect on operon structure: more than 70% of B. subtilis intrinsic terminators are positioned just downstream of the stop codon (Johnson et al., 2020), where they would be rendered inefficient by the trailing ribosome in E. coli (Roland et al., 1988). These findings are consistent with in vitro comparative analysis of B. subtilis and E. coli RNAP, which shows that B. subtilis enzyme transcribes faster and pauses less (Artsimovitch et al., 2000). In contrast, their ribosomes move at similar rates and are unable to catch up with the run-away B. subtilis RNAP (Johnson et al., 2020); even if B. subtilis NusG binds to the RNAP and the ribosome, it cannot bridge this gap.
In E. coli, RNAP pauses frequently and NusG facilitates RNA synthesis (Herbert et al., 2010). By contrast, B. subtilis RNAP rarely pauses and NusG stimulates pausing in vitro and in vivo (Yakhnin et al., 2020a). Unlike E. coli NusG, which is positioned next to the non-template DNA strand in the TEC but is not known to recognize any specific DNA elements (Kang et al., 2018), B. subtilis NusG specifically binds to T-rich DNA sequences and delays RNA chain elongation (Yakhnin et al., 2016). NusG-dependent RNAP pausing is required for regulation of several operons in B. subtilis (Yakhnin et al., 2020b); for example, NusG-dependent pausing in the trp and rib leader regions provides time for recruitment of an RNA-binding protein TRAP and for riboswitching by flavin mononucleotide, respectively. Sequence-specific pausing through non-template DNA contacts has been first shown for RfaH (Artsimovitch and Landick, 2002), which recognizes 12-nt ops elements in the E. coli genome (Belogurov et al., 2009); RfaH-induced RNAP delay is thought to facilitate the ribosome recruitment to the nascent RNA (see below) in a handful of leader regions. The ops sequence is a perfect match to the consensus pause sequence that induces pausing in E. coli (Larson et al., 2014; Vvedenskaya et al., 2014) but has additional recognition determinants for RfaH (Zuber et al., 2018).
By contrast, in B. subtilis, NusG recognizes a simpler consensus TTNTTT motif and stimulates pausing genome wide, favoring forward translocation of RNAP (Yakhnin et al., 2020a). Sequences that induce intrinsic, NusG-independent pausing of B. subtilis enzyme are also very different from the consensus pause elements documented in E. coli, and backtracking is not observed (Yakhnin et al., 2020a). Although the mechanism and regulation of pausing appear to be distinct, slowing RNAP is expected to be essential in both B. subtilis and E. coli. Pausing determines the overall rate of RNA chain synthesis, is an obligatory step in termination, and facilitates recruitment of regulatory factors (Kang et al., 2019). In both E. coli and B. subtilis, pausing has been implicated in attenuation control and co-transcriptional folding of riboswitches and catalytic RNAs (Landick et al., 1985; Pan et al., 1999; Perdrizet et al., 2012; Yakhnin et al., 2019), and contributes to coupling of transcription and translation in E. coli (McGary and Nudler, 2013). Pausing-defective E. coli RNAP variants do not support cell growth but can be rescued by small-molecule ligands that slow the RNAP down (Artsimovitch et al., 2003). In contrast to E. coli RNAP, which readily pauses at consensus sequences without the aid of accessory factors (Artsimovitch and Landick, 2000; Larson et al., 2014; Vvedenskaya et al., 2014), B. subtilis RNAP relies on NusG to slow it down (Yakhnin et al., 2016, 2020a). In this light, NusG can be viewed as a pause-promoting accessory subunit, a regulatory mechanism that could be widespread in bacteria (Yakhnin et al., 2020b). Indeed, Thermus thermophilus NusG reduces the RNA synthesis rate (Sevostyanova and Artsimovitch, 2010) and mycobacterial NusG promotes intrinsic termination (Czyz et al., 2014).
Is there any common function of NusG proteins? The conservation of the boxA and boxB RNA elements, all Nus factors, ribosomal proteins, and SuhB suggests that similar rrn-TACs may form in B. subtilis, a hypothesis supported by a report that rrn antitermination can be achieved in a heterologous E. coli/B. subtilis system (Arnvig et al., 2008). Observations that B. subtilis cells lacking NusG do not show defects in rRNA transcription argue that NusG is not required for rRNA synthesis (Yakhnin et al., 2020a). However, given that the principal role of the E. coli rrn-TACs appears to be in chaperoning of the nascent RNA (Huang et al., 2020), an analogous complex, with or without NusG, may be required to ensure the correct rRNA folding and processing in B. subtilis.
In addition to housekeeping NusG/Spt5 proteins present in all free-living cells, many genomes encode one or more NusG paralogs (Wang B. et al., 2020). While the primary sequences of these proteins are very diverse, the high conservation of residues that comprise the high-affinity RNAP binding site suggests that all of them bind to the TEC similarly. Indeed, E. coli NusG and RfaH, which are only 17% identical, make very similar contacts to that RNAP β’ subunit (Kang et al., 2018). However, in contrast to housekeeping NusG, which binds to RNAP and modulates transcription genome wide (Mooney et al., 2009a; Yakhnin et al., 2020a), these paralogs control expression of just a few target genes. Akin to alternative transcription initiation factors, these specialized NusGs (NusGSP) comprise a set of alternative transcription elongation factors that compete for the transcribing RNAP, an analogy further strengthened by their recruitment to the same site on RNAP (Sevostyanova et al., 2008).
However, this analogy does not extend to functions and mechanisms of gene-specific recruitment. Every σ factor activates transcription of its cognate promoters by recruiting RNAP and facilitating DNA melting; just the promoter sequences differ. In a stark contrast, NusGSP factors activate expression of genes that the housekeeping NusG silences (Figure 8). These genes can be a few in number, but critical for bacterial evolution and pathogenesis because they encode conjugation and virulence determinants (see below).
Furthermore, while σ factors bind to specific DNA sequences in static promoter complexes, NusG homologs are recruited to a moving RNAP. The available data suggest that these proteins use different recruitment mechanisms, only in some cases relying on specific protein-DNA interactions. Housekeeping NusGs are abundant proteins that can bind the TEC by chance, irrespective of the transcribed sequence; indeed, specific interactions would slow RNAP down, a regulatory feature used in B. subtilis (Yakhnin et al., 2020a) but not in E. coli, in which NusG is sequence blind. By contrast, the best characterized NusGSP, E. coli RfaH, uses a very complex mechanism to ensure efficient and selective recruitment to its targets (Zuber et al., 2018). RfaH is recruited to the TEC at operon polarity suppressor (ops; Figure 9A) sites (16 in E. coli MG1655 genome) which are present in leader regions of several operons silenced by NusG and Rho (Artsimovitch and Knauer, 2019). The ops element is a composite regulatory signal: it induces RNAP pausing and backtracking (Artsimovitch and Landick, 2000) and is directly recognized by RfaH (Artsimovitch and Landick, 2002). Pausing at ops is essential for RfaH recruitment (Zuber et al., 2018): it (i) provides additional time for RfaH, which is present in few copies/cell, to find its target; and (ii) presents the ops bases in a small hairpin, with a conserved T residue flipped out for specific recognition by RfaH (Kang et al., 2018; Zuber et al., 2018). This is a one-time opportunity because, once the RNAP moves past ops, the recruitment window is closed; thus, RfaH must bind to RNAP at ops and stay bound until the end of RNA synthesis. To fend off 100-fold more abundant NusG (Schmidt et al., 2016), RfaH binds RNAP much tighter (Kang et al., 2018), essentially becoming an RNAP subunit for one round of RNA synthesis. RfaH maintains the ability to trigger pausing at a downstream (engineered) ops site while traveling with RNAP but reduces pausing at any other sequence (Belogurov et al., 2009).
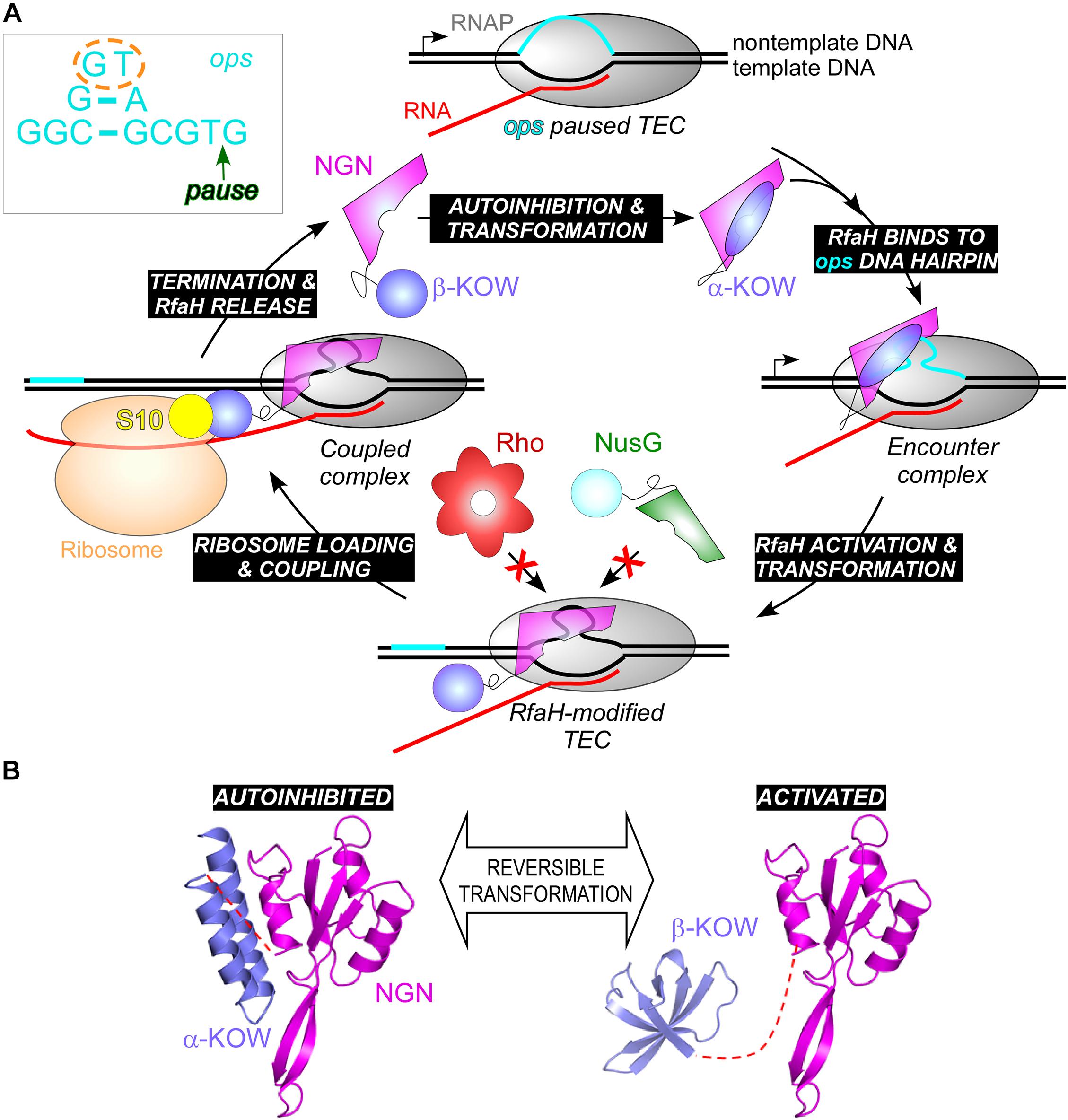
Figure 9. (A) A full cycle of RfaH; see text for details. The inset shows the ops DNA element, which forms a short hairpin on the TEC surface; ops bases that make most interactions with RfaH in the complex are circled; the pause position is indicated by an arrow. (B) RfaH domain dissociation and refolding. PDB IDs: autoinhibited RfaH, 5OND; activated RfaH, 6C6S.
If RfaH binds to RNAP very tightly during elongation, why does it need the ops signal in the first place? Unlike NusG, in which the RNAP-binding site on the NGN domain is exposed, this site is blocked by the KOW domain in free RfaH (Figure 9). Also unlike NusG, in which the KOW domain is in a β-barrel state (β-KOW; Figure 2), in this “autoinhibited” RfaH the KOW domain is folded as an α-helical hairpin (α-KOW; Figure 9B). To bind RNAP, RfaH must be “activated” by domain dissociation, which happens only in the presence of a complete ops-paused TEC (Zuber et al., 2019). The details of this process remain elusive, but the current model suggests that the NGN domain recognizes the ops hairpin via its exposed DNA-binding residues, forming a transient encounter complex and triggering the KOW dissociation (Artsimovitch and Knauer, 2019). It is possible that autoinhibition may be a common feature of NusG homologs. While in E. coli NusG the NGN and KOW domains move freely (Burmann et al., 2011), in NusG from a hyperthermophilic bacterium Thermotoga maritima, the two domains interact, masking the binding sites for RNAP, NusE, and Rho (Drögemüller et al., 2013). Domain dissociation enables T. maritima NusG-KOW binding to Rho and NusE, and these contacts may be stabilized by the NGN-RNAP contacts (Drögemüller et al., 2017).
RfaH recruitment relies on the multi-functional DNA element and elaborate structural rearrangements of the protein domains. Binding to a specific DNA element enables RfaH to control several operons scattered on the chromosome. But how is a wannabe NusGSP, which has just surfaced following gene duplication, targeted to a specific locus in the presence of overwhelming numbers of NusG molecules? An “ancestral” mechanism, in which NusGSP binds to the transcribing RNAP in cis has been proposed to explain this conundrum (Belogurov et al., 2009). This model is supported by bioinformatics analyses which reveal that the residues that mediate DNA contacts in RfaH arose late in evolution and that many NusGSP are encoded within long xenogeneic operons, in contrast to the standalone rfaH gene (Wang B. et al., 2020). However, observations that some of these cis-encoded regulators act in trans (Chatzidaki-Livanis et al., 2010) suggest that NusGSP recruitment strategies are multifaceted.
RfaH activation is not limited to the domain dissociation needed to expose the RNAP-binding site: the released α-KOW undergoes a dramatic transformation into a NusG-like β-KOW (Figure 9B) and binds to S10 similarly to NusG KOW (Burmann et al., 2012). The residues that make contacts with S10 are not available in the α-KOW domain, thus the free RfaH is autoinhibited with respect to both RNAP and ribosome binding, allowing RfaH to achieve high target specificity (Shi et al., 2017). The activated state persists until the TEC dissociates at a terminator and RfaH is released; the KOW then refolds into the α-helical hairpin and re-establishes contacts with the NGN, restoring autoinhibition (Figure 9A).
Interconversion between the alternative RfaH-KOW states is principally controlled by interdomain contacts: the KOW (re)folds into a β-barrel when expressed alone, separated from the NGN domain upon proteolytic cleavage of the linker, or as a result of interface-destabilizing substitutions (Burmann et al., 2012; Tomar et al., 2013; Shi et al., 2017). Deuteron incorporation reveals that the tip of the C-terminal α-hairpin is stably folded in the autoinhibited state, whereas the rest of the KOW is highly flexible, and its flexibility only decreases in the β-folded state (Galaz-Davison et al., 2020). The mechanism underlying this dramatic fold switch has been also pursued by computational approaches (Gc et al., 2014, 2015; Balasco et al., 2015; Ramírez-Sarmiento et al., 2015; Xiong and Liu, 2015; Xun et al., 2016). Although the β-barrel is a preferred state of the isolated RfaH-KOW, its free energy is only slightly lower than that of the α-helical conformation. The separation of the two alternative states is dependent on large energy barriers resulting from the main chain hydrogen bonds of the α-helical hairpin. An all-atom Monte Carlo simulations study suggests a possibility that the encounter complex between the autoinhibited RfaH and the ops-TEC is characterized by net attractive interactions with the NGN and net repulsive interactions with the KOW. The resulting opposing forces on the two domains, in combination with the peculiar mechanical rigidity profile of the autoinhibited RfaH, might help trigger domain separation (Seifi et al., 2020). The α→β rearrangement essentially depends on an unstructured state: upon dissolution of the α-helical hairpin, the KOW assumes a disordered state and then follows a step-wise assembly into the final five-stranded β-barrel (Bernhardt and Hansmann, 2018; Joseph et al., 2019).
Among NusG homologs, E. coli RfaH is the only known transformer protein. However, it is possible that other KOW domains are capable of transformation. In particular, an amazingly broad repertoire of known cellular targets of eukaryotic NusG homologs (Decker, 2020) could be due to metamorphic behavior of their KOWs.
RfaH-controlled genes encode toxins, adhesins, LPS and capsule biosynthesis enzymes, type IV secretion apparatus, etc. located in long horizontally acquired operons (Figure 10), which are silenced by Rho. RfaH abolishes Rho-dependent termination (Sevostyanova et al., 2011) and the ability to bind Rho appears to be lost early in RfaH evolution (Wang B. et al., 2020). RfaH elicits dramatic, 50 + fold activation of gene expression in vivo, an effect that was initially assumed to be mediated by its direct antitermination effects on RNAP (Artsimovitch and Landick, 2002). Surprisingly, RNAP modification by RfaH makes only a minor contribution in the cell (Sevostyanova et al., 2011). Instead, RfaH inhibits Rho-dependent termination by outcompeting NusG and activating translation.
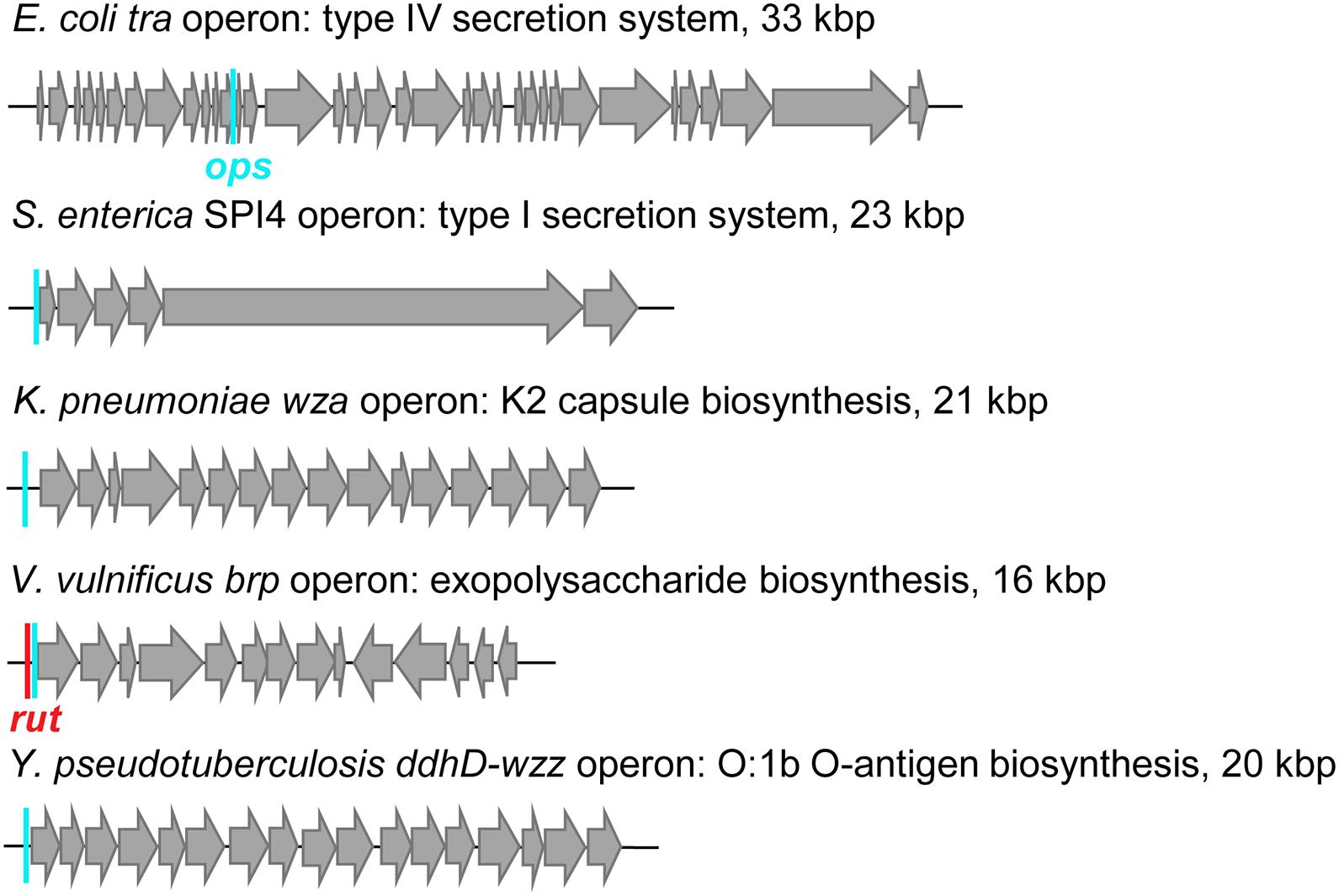
Figure 10. Examples of RfaH-controlled operons. Positions of ops sites (cyan bars) and a rut site (red bar) are indicated.
RfaH-controlled genes lack Shine-Dalgarno elements, which recruit the ribosome through RNA base-pairing with the 16S rRNA (Rodnina, 2018) and have many rare codons, limiting their translation and making them easy targets for Rho. Observations that the transformed β-KOW directly binds S10 (Burmann et al., 2012) prompted a hypothesis that RfaH recruits the ribosome via β-KOW/S10 contacts and then couples transcription to translation during elongation. In support of this model, expression of SD-less reporters is completely dependent on RfaH, and substitutions of residues that interact with S10 abolish expression (Burmann et al., 2012). In addition to the ribosome recruitment, by bridging the RNAP and the ribosome during elongation, RfaH may prevent uncoupling at rare codons; the ribosome stalling exposes mRNA to Rho (Elgamal et al., 2016). RfaH may be particularly important during synthesis of excessively long proteins such as Salmonella pathogenicity island IV giant 600 kDa adhesin (Figure 9B), which requires RfaH for expression (Main-Hester et al., 2008). Remarkably, the ops-RfaH module supports efficient expression of an SD-less reporter in vivo, ∼20% relative to that driven by a perfect SD element (Burmann et al., 2012).
Although RfaH and NusG make similar contacts to S10 (Burmann et al., 2012), their effects on translation are expected to be different. NusG binds to the RNAP transiently (Kang et al., 2018) and late in the operon, well after the first ORF (Mooney et al., 2009a). In contrast, RfaH binds to RNAP upstream of the first ORF and remains stably associated with the EC until termination (Belogurov et al., 2009). It is possible that RfaH recruits the ribosome to the ops-paused RNAP and promotes ribosome scanning for a downstream initiation codon. Future studies will reveal the details of translation activation by RfaH, but the available data suggest that this universally conserved transcription antiterminator may be acting primarily as an RNAP-tethered translation initiation/elongation factor and may employ the first protein-mediated ribosome recruitment mechanism outside of viruses.
Specialized NusG paralogs (Figure 11) are evolving in very different ecological niches but may have similar functions—to promote expression of long or silenced operons. Functional data implicate several NusGSP in transcription antitermination of very long gene clusters, whereas for others this function is inferred from their genomic associations. Bacillus amyloliquefaciens LoaP inhibits termination in two operons producing antibiotics difficidin and macrolactin (Goodson et al., 2017). Differently from RfaH, which is rather inefficient against intrinsic terminators (Artsimovitch and Landick, 2002; Carter et al., 2004), LoaP promotes readthrough of the hairpin termination signals (Goodson et al., 2017). Polyketide antibiotic TA made by Myxococcus xanthus inhibits bacterial cell wall synthesis and is produced by a 40 kb operon which is activated by NusGSP called TaA (Paitan et al., 1999) by an unknown mechanism. Human gut bacterium Bacteroides fragilis synthesizes eight capsular polysaccharides from separate operons, which are activated by UpxY family of NusGSP. UpxY proteins prevent premature transcriptional termination within the 5′ leaders upstream from the upxY gene (Chatzidaki-Livanis et al., 2009).
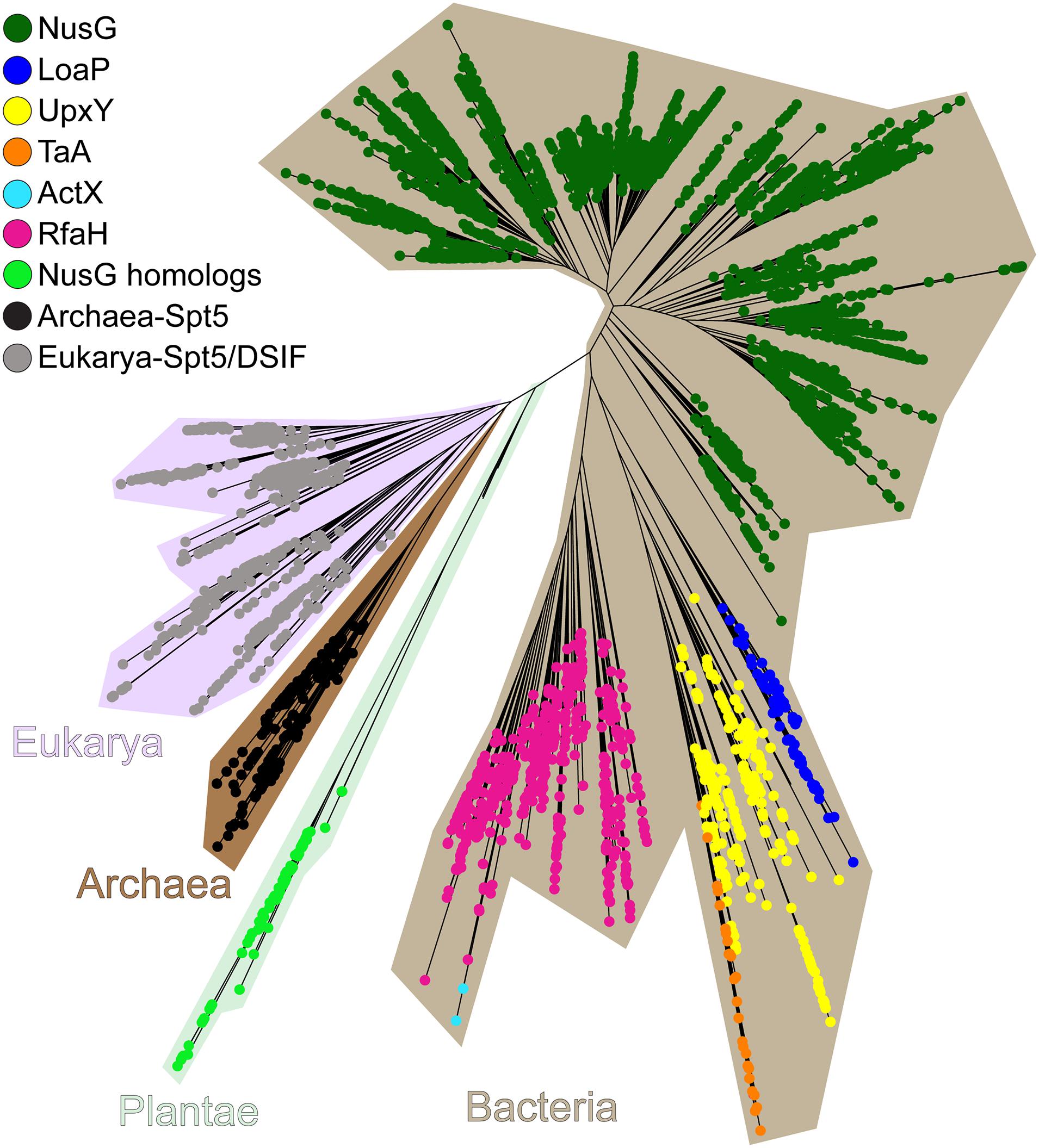
Figure 11. The maximum-likelihood phylogenetic tree of NusG-like proteins. Plantae is an artificial group used solely for brevity.
While functional data are available for just a few NusGSP, recent bioinformatics analysis suggests that these proteins fall into eight different clusters, which differ in their primary sequence signatures as well as regulatory contexts. Some NusGSP, such as RfaH, form one group and are encoded by single cistrons, whereas others (e.g., loaP, taA, and upxY) are adjacent to their target operons (Wang B. et al., 2020). ActX, which is closely related to RfaH (Figure 11), is encoded within pilus biosynthesis operons on antibiotic-resistant plasmids in E. coli and Klebsiella pneumoniae (Núñez et al., 1997), but its regulatory function remains unknown. Analysis of genomic contexts can be instrumental in predicting functional associations (Moreno-Hagelsieb and Santoyo, 2015). Gene neighbors of NusGSP (except for RfaH-like stand-alone genes) are enriched in genes involved in cell envelope biogenesis, with glycosyltransferases, nucleoside-diphosphate-sugar epimerases, and exopolysaccharide biosynthesis enzymes being the most common (Wang B. et al., 2020). However, notable differences exist among distinct clusters; for example, some NusGSP are adjacent to Tat protein secretion system, others are encoded near undecaprenyl pyrophosphate synthase and H-NS genes. A group of regulators from Shewanella are encoded within putative exopolysaccharide operons, an arrangement resembling B. fragilis operons controlled by UpxY proteins (Chatzidaki-Livanis et al., 2010). Future studies will be required to determine functional significance of these associations.
Extensive duplications, sub-functionalization, and horizontal transfer underpin the evolution of NusG paralogs. One NusG copy has gradually evolved into RfaH, starting from an “early” loss of binding to Rho terminator while tightening contacts to RNAP and culminating with the “late” acquisition of residues that interact with the ops DNA element and confer autoinhibition (Wang B. et al., 2020). While in most NusG homologs these changes do not alter the core domain structure, some factors acquired additional domains thought to promote adaptation to their unique niches. For example, in T. maritima NusG, an extra domain DII supports NusG recruitment to the TEC and stabilizes the NusG:RNAP complex, a necessary adaptation to high temperatures in the T. maritima natural habitat (Drögemüller et al., 2017).
In addition to Spt5, NusG homologs are also encoded in the genomes of all major land plant and algal lineages except for some green algal species (Wang B. et al., 2020). These bacterial regulators have recognizable chloroplast-localization signals and are presumably retained to assist the bacterial-type RNAPs that mediate chloroplast transcription. A NusG homolog of Arabidopsis thaliana has been identified as a component of the active transcriptional machinery in chloroplasts (Pfalz et al., 2006), and a Rho ortholog has been shown to terminate transcription by plastid-encoded RNAP (Yang et al., 2020).
Extensive functional studies have established RfaH as the paradigm for the regulation of transcription elongation, translation initiation, and protein folding. However, RfaH is also a key virulence factor. RfaH activates the expression of capsule, cell wall, toxins, adhesins, and pilus biosynthesis operons (Figure 9B), which are important for virulence and conjugal transfer in several Gram-negative pathogens including E. coli, K. pneumoniae, Vibrio vulnificus, Salmonella enterica, Yersinia pseudotuberculosis, and Yersinia pestis (Kong et al., 2011; Bachman et al., 2015; Garrett et al., 2016; Hoffman et al., 2017). RfaH effects on gene expression are very large (50+ fold); consequently, the loss of rfaH leads to dramatic defects in virulence, e.g., 104 decrease in K. pneumoniae survival in the lung (Bachman et al., 2015).
The first protein secretion process discovered in bacteria was the hemolysin A (HlyA) type 1 secretion system (T1SS), which is found in uropathogenic E. coli strains (Thomas et al., 2014). HlyA is a 107 kDa protein that induces hemolysis by creating pores in the erythrocyte membrane (Skals et al., 2009). RfaH, a.k.a. HlyT, has been identified genetically as an activator of the hly operon (Thomas et al., 2014). Inactivation of rfaH dramatically decreases virulence of uropathogenic E. coli strain in a murine model of urinary tract infection (Nagy et al., 2002). The capability to colonize the intestinal tract by efficiently competing with the commensal microbiota has been considered as a multifactorial virulence property. RfaH also plays a role in the infectious process during colonization of the intestinal tract: rfaH mutants are susceptible to bile salts and show reduced gut colonization capacity (Nagy et al., 2005).
Antibiotic-resistant K. pneumoniae is an urgent public health threat and a leading cause of pneumonia in hospitalized patients (David et al., 2019). Functional genomic profiling of four diverse serum-resistant K. pneumoniae strains reveals that the deletion of rfaH dramatically reduces resistance to serum complement system in all strains (Short et al., 2020). Vibrio vulnificus is another opportunistic human pathogen responsible for the majority of seafood-associated deaths worldwide, and antibiotic resistance has developed (Heng et al., 2017). Loss of rfaH also makes V. vulnificus highly sensitive to human serum (Garrett et al., 2016). Expression of the brp exopolysaccharide operon mediates surface adherence of V. vulnificus, and the presence of ops and rut sites in the leader region suggests RfaH-dependent antitermination (Chodur and Rowe-Magnus, 2018). S. enterica serovar Typhimurium is a primary enteric pathogen infecting both humans and animals and a major cause of diarrheal diseases, with antibiotic resistance on the rise (Fàbrega and Vila, 2013; Knodler and Elfenbein, 2019). Salmonella harbors five pathogenicity islands (SPI) required for infection in vertebrate hosts. Among them, SPI4 plays a role in the initial interaction with the intestinal epithelium and possibly contributes to long-term persistence (Gerlach et al., 2007). S. enterica RfaH is required for the expression of SPI4, which encodes a T1SS and its adhesin substrate (Main-Hester et al., 2008), as well as the expression of secreted and surface-associated polysaccharides (Lindberg and Hellerqvist, 1980; Bailey et al., 1997). Mutants of S. enterica serovar Typhimurium lacking rfaH are efficient as vaccines against salmonellosis and induce strong serum immune responses (Nagy et al., 2006; Liu et al., 2016). Given their association with capsular and TSS operons (Wang B. et al., 2020), other NusG paralogs likely play important roles during pathogenesis.
Antibiotic resistance determinants are frequently encoded on conjugative plasmids and can be rapidly transferred between bacteria (Wang et al., 2017). RfaH activates the F plasmid conjugation operon (Beutin and Achtman, 1979) and RfaH homologs are encoded on some clinical resistant plasmids (Wang B. et al., 2020), suggesting that they may contribute to plasmid transfer. A recent study showed that deletions of seven genes, including rfaH, prevented cefotaxime-induced up-regulation of traF and decreased the conjugative transfer of the resistance plasmid (Liu et al., 2019).
RfaH proteins from Vibrio, Yersinia, Salmonella, and Klebsiella bind to the E. coli TEC in vitro and complement the E. coli rfaH gene deletion (Carter et al., 2004). Small molecule inhibitors that block recruitment of E. coli and K. pneumoniae RfaH to RNAP (Svetlov et al., 2018) may have a potential to inhibit virulence and the spread of antibiotic resistance.
NusG homologs comprise the only universally conserved family of transcription factors, which includes housekeeping regulators and their specialized paralogs (Figure 11). Despite highly similar core domain architectures and interactions with RNAP, NusG-like proteins exert amazingly diverse, and frequently opposite, effects on gene expression. Bacterial NusG homologs can inhibit or stimulate transcription termination, accelerate RNA synthesis by suppressing RNAP backtracking or slow transcription down by halting RNAP at specific sequences, bridge the RNAP to the ribosome during translation elongation or recruit the ribosome to mRNAs that lack canonical ribosome binding sites, and likely perform other functions that remain to be discovered.
This regulatory plasticity depends on dynamic interactions of the NGN and KOW domains with each other, RNAP, single and double-stranded nucleic acids, and many auxiliary cellular proteins. While bound to the TEC through contacts mediated by highly conserved residues within RNAP and NGN, NusG homologs employ divergent residues in their NGN and KOW domains to enact a range of responses demanded by specific cellular circumstances. Some NusG paralogs augment their regulatory prowess by undergoing an unprecedented and reversible refolding of an entire KOW domain, during which the protein turns inside out. The presence of NusG in all free-living organisms, sometimes in several copies, confirms its unique place in gene expression control, from LUCA to present life forms.
BW prepared all original figures and wrote the first draft. IA revised and expanded the manuscript. Both authors prepared figures and edited the draft while preparing a revised manuscript.
Our research was supported by the National Institutes of Health (GM067153 to IA).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Arnvig, K. B., Zeng, S., Quan, S., Papageorge, A., Zhang, N., Villapakkam, A. C., et al. (2008). Evolutionary comparison of ribosomal operon antitermination function. J. Bacteriol. 190, 7251–7257. doi: 10.1128/jb.00760-08
Artsimovitch, I., Chu, C., Lynch, A. S., and Landick, R. (2003). A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science 302, 650–654. doi: 10.1126/science.1087526
Artsimovitch, I., and Knauer, S. H. (2019). Ancient transcription factors in the news. mBio 10:e01547-18.
Artsimovitch, I., and Landick, R. (2000). Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. U.S.A. 97, 7090–7095. doi: 10.1073/pnas.97.13.7090
Artsimovitch, I., and Landick, R. (2002). The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109, 193–203. doi: 10.1016/s0092-8674(02)00724-9
Artsimovitch, I., Svetlov, V., Anthony, L., Burgess, R. R., and Landick, R. (2000). RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J. Bacteriol. 182, 6027–6035. doi: 10.1128/jb.182.21.6027-6035.2000
Bachman, M. A., Breen, P., Deornellas, V., Mu, Q., Zhao, L., Wu, W., et al. (2015). Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6:e00775-15.
Bailey, M. J., Hughes, C., and Koronakis, V. (1997). RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26, 845–851. doi: 10.1046/j.1365-2958.1997.6432014.x
Balasco, N., Barone, D., and Vitagliano, L. (2015). Structural conversion of the transformer protein RfaH: new insights derived from protein structure prediction and molecular dynamics simulations. J. Biomol. Struct. Dyn. 33, 2173–2179. doi: 10.1080/07391102.2014.994188
Belogurov, G. A., and Artsimovitch, I. (2015). Regulation of transcript elongation. Annu. Rev. Microbiol. 69, 49–69. doi: 10.1146/annurev-micro-091014-104047
Belogurov, G. A., and Artsimovitch, I. (2019). The mechanisms of substrate selection, catalysis, and translocation by the elongating RNA polymerase. J. Mol. Biol. 431, 3975–4006. doi: 10.1016/j.jmb.2019.05.042
Belogurov, G. A., Mooney, R. A., Svetlov, V., Landick, R., and Artsimovitch, I. (2009). Functional specialization of transcription elongation factors. EMBO J. 28, 112–122. doi: 10.1038/emboj.2008.268
Bernhardt, N. A., and Hansmann, U. H. E. (2018). Multifunnel landscape of the fold-switching protein RfaH-CTD. J. Phys. Chem. B 122, 1600–1607. doi: 10.1021/acs.jpcb.7b11352
Beutin, L., and Achtman, M. (1979). Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J. Bacteriol. 139, 730–737. doi: 10.1128/JB.139.3.730-737.1979
Bossi, L., Ratel, M., Laurent, C., Kerboriou, P., Camilli, A., Eveno, E., et al. (2019). NusG prevents transcriptional invasion of H-NS-silenced genes. PLoS Genet. 15:e1008425. doi: 10.1371/journal.pgen.1008425
Boudreau, B. A., Hron, D. R., Qin, L., van der Valk, R. A., Kotlajich, M. V., Dame, R. T., et al. (2018). StpA and Hha stimulate pausing by RNA polymerase by promoting DNA-DNA bridging of H-NS filaments. Nucleic Acids Res. 46, 5525–5546. doi: 10.1093/nar/gky265
Burmann, B. M., Knauer, S. H., Sevostyanova, A., Schweimer, K., Mooney, R. A., Landick, R., et al. (2012). An α Helix to β Barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 150, 291–303. doi: 10.1016/j.cell.2012.05.042
Burmann, B. M., Scheckenhofer, U., Schweimer, K., and Rosch, P. (2011). Domain interactions of the transcription-translation coupling factor Escherichia coli NusG are intermolecular and transient. Biochem. J. 435, 783–789. doi: 10.1042/BJ20101679
Burmann, B. M., Schweimer, K., Luo, X., Wahl, M. C., Stitt, B. L., Gottesman, M. E., et al. (2010). A NusE:NusG complex links transcription and translation. Science 328, 501–504. doi: 10.1126/science.1184953
Burns, C. M., and Richardson, J. P. (1995). NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc. Natl. Acad. Sci. U.S.A. 92, 4738–4742. doi: 10.1073/pnas.92.11.4738
Burns, C. M., Richardson, L. V., and Richardson, J. P. (1998). Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J. Mol. Biol. 278, 307–316. doi: 10.1006/jmbi.1998.1691
Burova, E., Hung, S. C., Sagitov, V., Stitt, B. L., and Gottesman, M. E. (1995). Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J. Bacteriol. 177, 1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995
Cardinale, C. J., Washburn, R. S., Tadigotla, V. R., Brown, L. M., Gottesman, M. E., and Nudler, E. (2008). Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320, 935–938. doi: 10.1126/science.1152763
Carter, H. D., Svetlov, V., and Artsimovitch, I. (2004). Highly divergent RfaH orthologs from pathogenic proteobacteria can substitute for Escherichia coli RfaH both in vivo and in vitro. J. Bacteriol. 186, 2829–2840. doi: 10.1128/jb.186.9.2829-2840.2004
Casjens, S. R., and Hendrix, R. W. (2015). Bacteriophage lambda: early pioneer and still relevant. Virology 47, 310–330. doi: 10.1016/j.virol.2015.02.010
Chandraprakash, D., and Seshasayee, A. S. (2014). Inhibition of factor-dependent transcription termination in Escherichia coli might relieve xenogene silencing by abrogating H-NS-DNA interactions in vivo. J. Biosci. 39, 53–61. doi: 10.1007/s12038-014-9413-4
Chatzidaki-Livanis, M., Coyne, M. J., and Comstock, L. E. (2009). A family of transcriptional antitermination factors necessary for synthesis of the capsular polysaccharides of Bacteroides fragilis. J. Bacteriol. 191, 7288–7295. doi: 10.1128/jb.00500-09
Chatzidaki-Livanis, M., Weinacht, K. G., and Comstock, L. E. (2010). Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U.S.A. 107, 11976–11980. doi: 10.1073/pnas.1005039107
Chen, M., and Fredrick, K. (2018). Measures of single- versus multiple-round translation argue against a mechanism to ensure coupling of transcription and translation. Proc. Natl. Acad. Sci. U.S.A. 115, 10774–10779. doi: 10.1073/pnas.1812940115
Chodur, D. M., and Rowe-Magnus, D. A. (2018). Complex control of a genomic island governing biofilm and rugose colony development in Vibrio vulnificus. J. Bacteriol. 200:e00190-18.
Crickard, J. B., Fu, J., and Reese, J. C. (2016). Biochemical analysis of Yeast suppressor of Ty 4/5 (Spt4/5) reveals the importance of nucleic acid interactions in the prevention of RNA polymerase II arrest. J. Biol. Chem. 291, 9853–9870. doi: 10.1074/jbc.M116.716001
Czyz, A., Mooney, R. A., Iaconi, A., and Landick, R. (2014). Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and is aided by NusG at suboptimal terminators. mBio 5:e00931-14.
David, S., Reuter, S., Harris, S. R., Glasner, C., Feltwell, T., Argimon, S., et al. (2019). Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 4, 1919–1929. doi: 10.1038/s41564-019-0492-8
Decker, T.-M. (2020). Mechanisms of transcription elongation factor DSIF (Spt4–Spt5). J. Mol. Biol. doi: 10.1016/j.jmb.2020.09.016 [Epub ahead of print].
Dennis, P. P., Ehrenberg, M., and Bremer, H. (2004). Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68, 639–668. doi: 10.1128/mmbr.68.4.639-668.2004
Downing, W. L., Sullivan, S. L., Gottesman, M. E., and Dennis, P. P. (1990). Sequence and transcriptional pattern of the essential Escherichia coli secE-nusG operon. J. Bacteriol. 172, 1621–1627. doi: 10.1128/jb.172.3.1621-1627.1990
Drögemüller, J., Schneider, C., Schweimer, K., Strauß, M., Wöhrl, B. M., Rösch, P., et al. (2017). Thermotoga maritima NusG: domain interaction mediates autoinhibition and thermostability. Nucleic Acids Res. 45, 446–460. doi: 10.1093/nar/gkw1111
Drögemüller, J., Stegmann, C. M., Mandal, A., Steiner, T., Burmann, B. M., Gottesman, M. E., et al. (2013). An autoinhibited state in the structure of Thermotoga maritima NusG. Structure 21, 365–375. doi: 10.1016/j.str.2012.12.015
Ehara, H., Yokoyama, T., Shigematsu, H., Yokoyama, S., Shirouzu, M., and Sekine, S. I. (2017). Structure of the complete elongation complex of RNA polymerase II with basal factors. Science 357, 921–924. doi: 10.1126/science.aan8552
Elgamal, S., Artsimovitch, I., and Ibba, M. (2016). Maintenance of transcription-translation coupling by elongation factor P. mBio 7:e01373-16.
Epshtein, V., Dutta, D., Wade, J., and Nudler, E. (2010). An allosteric mechanism of Rho-dependent transcription termination. Nature 463, 245–249. doi: 10.1038/nature08669
Fàbrega, A., and Vila, J. (2013). Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341. doi: 10.1128/cmr.00066-12
Fan, H., Conn, A. B., Williams, P. B., Diggs, S., Hahm, J., Gamper, H. B., et al. (2017). Transcription–translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Res. 45, 11043–11055. doi: 10.1093/nar/gkx719
French, S. L., Santangelo, T. J., Beyer, A. L., and Reeve, J. N. (2007). Transcription and translation are coupled in Archaea. Mol. Biol. Evol. 24, 893–895. doi: 10.1093/molbev/msm007
Galaz-Davison, P., Molina, J. A., Silletti, S., Komives, E. A., Knauer, S. H., Artsimovitch, I., et al. (2020). Differential local stability governs the metamorphic fold switch of bacterial virulence factor RfaH. Biophys. J. 118, 96–104. doi: 10.1016/j.bpj.2019.11.014
Garrett, S. B., Garrison-Schilling, K. L., Cooke, J. T., and Pettis, G. S. (2016). Capsular polysaccharide production and serum survival of Vibrio vulnificus are dependent on antitermination control by RfaH. FEBS Lett. 590, 4564–4572. doi: 10.1002/1873-3468.12490
Gc, J. B., Bhandari, Y. R., Gerstman, B. S., and Chapagain, P. P. (2014). Molecular dynamics investigations of the α-Helix to β-Barrel conformational transformation in the RfaH transcription factor. J. Phys. Chem. B 118, 5101–5108. doi: 10.1021/jp502193v
Gc, J. B., Gerstman, B. S., and Chapagain, P. P. (2015). The role of the interdomain interactions on RfaH dynamics and conformational transformation. J. Phys. Chem. B 119, 12750–12759. doi: 10.1021/acs.jpcb.5b05681
Gerlach, R. G., Jäckel, D., Stecher, B., Wagner, C., Lupas, A., Hardt, W. D., et al. (2007). Salmonella pathogenicity Island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 9, 1834–1850. doi: 10.1111/j.1462-5822.2007.00919.x
Goodson, J. R., Klupt, S., Zhang, C., Straight, P., and Winkler, W. C. (2017). LoaP is a broadly conserved antiterminator protein that regulates antibiotic gene clusters in Bacillus amyloliquefaciens. Nat. Microbiol. 2:17003. doi: 10.1038/nmicrobiol.2017.3
Gowrishankar, J., and Harinarayanan, R. (2004). Why is transcription coupled to translation in bacteria? Mol. Microbiol. 54, 598–603. doi: 10.1111/j.1365-2958.2004.04289.x
Gowrishankar, J., Leela, J. K., and Anupama, K. (2013). R-loops in bacterial transcription: their causes and consequences. Transcription 4, 153–157. doi: 10.4161/trns.25101
Grohmann, D., Nagy, J., Chakraborty, A., Klose, D., Fielden, D., Ebright, R. H., et al. (2011). The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol. Cell 43, 263–274. doi: 10.1016/j.molcel.2011.05.030
Heng, S.-P., Letchumanan, V., Deng, C.-Y., Ab Mutalib, N.-S., Khan, T. M., Chuah, L.-H., et al. (2017). Vibrio vulnificus: an environmental and clinical burden. Front. Microbiol. 8:997. doi: 10.3389/fmicb.2017.00997
Herbert, K. M., Zhou, J., Mooney, R. A., Porta, A. L., Landick, R., and Block, S. M. (2010). E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J. Mol. Biol. 399, 17–30. doi: 10.1016/j.jmb.2010.03.051
Hirtreiter, A., Damsma, G. E., Cheung, A. C., Klose, D., Grohmann, D., Vojnic, E., et al. (2010). Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 38, 4040–4051. doi: 10.1093/nar/gkq135
Hoffman, J. M., Sullivan, S., Wu, E., Wilson, E., and Erickson, D. L. (2017). Differential impact of lipopolysaccharide defects caused by loss of RfaH in Yersinia pseudotuberculosis and Yersinia pestis. Sci. Rep. 7:10915.
Huang, Y. H., Hilal, T., Loll, B., Bürger, J., Mielke, T., Böttcher, C., et al. (2020). Structure-based mechanisms of a molecular RNA polymerase/chaperone machine required for ribosome biosynthesis. Mol. Cell 79, 1024–1036.e5. doi: 10.1016/j.molcel.2020.08.010
Ingham, C. J., Dennis, J., and Furneaux, P. A. (1999). Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol. Microbiol. 31, 651–663. doi: 10.1046/j.1365-2958.1999.01205.x
Iost, I., and Dreyfus, M. (1995). The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 14, 3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x
Johnson, G. E., Lalanne, J. B., Peters, M. L., and Li, G. W. (2020). Functionally uncoupled transcription-translation in Bacillus subtilis. Nature 585, 124–128. doi: 10.1038/s41586-020-2638-5
Joseph, J. A., Chakraborty, D., and Wales, D. J. (2019). Energy landscape for fold-switching in regulatory protein RfaH. J. Chem. Theory Comput. 15, 731–742. doi: 10.1021/acs.jctc.8b00912
Kang, J. Y., Mishanina, T. V., Landick, R., and Darst, S. A. (2019). Mechanisms of transcriptional pausing in Bacteria. J. Mol. Biol. 431, 4007–4029. doi: 10.1016/j.jmb.2019.07.017
Kang, J. Y., Mooney, R. A., Nedialkov, Y., Saba, J., Mishanina, T. V., Artsimovitch, I., et al. (2018). Structural basis for transcript elongation control by NusG family universal regulators. Cell 173, 1650–1662.e14. doi: 10.1016/j.cell.2018.05.017
Klein, B. J., Bose, D., Baker, K. J., Yusoff, Z. M., Zhang, X., and Murakami, K. S. (2011). RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc. Natl. Acad. Sci. U.S.A. 108, 546–550. doi: 10.1073/pnas.1013828108
Knodler, L. A., and Elfenbein, J. R. (2019). Salmonella enterica. Trends Microbiol. 27, 964–965. doi: 10.1016/j.tim.2019.05.002
Kong, Q., Yang, J., Liu, Q., Alamuri, P., Roland, K. L., and Curtiss, R. III, et al. (2011). Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infect. Immun. 79, 4227–4239. doi: 10.1128/iai.05398-11
Krupp, F., Said, N., Huang, Y.-H., Loll, B., Bürger, J., Mielke, T., et al. (2019). Structural basis for the action of an all-purpose transcription anti-termination factor. Mol. Cell 74, 143–157.e5. doi: 10.1016/j.molcel.2019.01.016
Kyrpides, N. C., Woese, C. R., and Ouzounis, C. A. (1996). KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21, 425–426. doi: 10.1016/s0968-0004(96)30036-4
Landick, R., Carey, J., and Yanofsky, C. (1985). Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc. Natl. Acad. Sci. U.S.A. 82, 4663–4667. doi: 10.1073/pnas.82.14.4663
Lane, W. J., and Darst, S. A. (2010a). Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 395, 671–685. doi: 10.1016/j.jmb.2009.10.062
Lane, W. J., and Darst, S. A. (2010b). Molecular evolution of multisubunit RNA polymerases: structural analysis. J. Mol. Biol. 395, 686–704. doi: 10.1016/j.jmb.2009.10.063
Larson, M. H., Mooney, R. A., Peters, J. M., Windgassen, T., Nayak, D., Gross, C. A., et al. (2014). A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1042–1047. doi: 10.1126/science.1251871
Lawson, M. R., and Berger, J. M. (2019). Tuning the sequence specificity of a transcription terminator. Curr. Genet. 65, 729–733. doi: 10.1007/s00294-019-00939-1
Lawson, M. R., Ma, W., Bellecourt, M. J., Artsimovitch, I., Martin, A., Landick, R., et al. (2018). Mechanism for the regulated control of bacterial transcription termination by a universal adaptor protein. Mol. Cell 71, 911–922.e4. doi: 10.1016/j.molcel.2018.07.014
Leela, J. K., Syeda, A. H., Anupama, K., and Gowrishankar, J. (2013). Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 258–263. doi: 10.1073/pnas.1213123110
Lindberg, A. A., and Hellerqvist, C. G. (1980). Rough mutants of Salmonella typhimurium: immunochemical and structural analysis of lipopolysaccharides from rfaH mutants. J. Gen. Microbiol. 116, 25–32. doi: 10.1099/00221287-116-1-25
Liu, G., Olsen, J. E., and Thomsen, L. E. (2019). Identification of genes essential for antibiotic-induced up-regulation of plasmid-transfer-genes in cephalosporin resistant Escherichia coli. Front. Microbiol. 10:2203. doi: 10.3389/fmicb.2019.02203
Liu, Q., Liu, Q., Yi, J., Liang, K., Liu, T., Roland, K. L., et al. (2016). Outer membrane vesicles derived from Salmonella typhimurium mutants with truncated LPS induce cross-protective immune responses against infection of Salmonella enterica serovars in the mouse model. Int. J. Med. Microbiol. 306, 697–706. doi: 10.1016/j.ijmm.2016.08.004
Main-Hester, K. L., Colpitts, K. M., Thomas, G. A., Fang, F. C., and Libby, S. J. (2008). Coordinate regulation of Salmonella pathogenicity island 1 (SPI1) and SPI4 in Salmonella enterica serovar Typhimurium. Infect. Immun. 76, 1024–1035. doi: 10.1128/iai.01224-07
Martinez-Rucobo, F. W., Sainsbury, S., Cheung, A. C., and Cramer, P. (2011). Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 30, 1302–1310. doi: 10.1038/emboj.2011.64
Mason, S. W., and Greenblatt, J. (1991). Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 5, 1504–1512. doi: 10.1101/gad.5.8.1504
McGary, K., and Nudler, E. (2013). RNA polymerase and the ribosome: the close relationship. Curr. Opin. Microbiol. 16, 112–117. doi: 10.1016/j.mib.2013.01.010
Miller, O. L., Hamkalo, B. A., and Thomas, C. A. (1970). Visualization of bacterial genes in action. Science 169, 392–395. doi: 10.1126/science.169.3943.392
Mitra, P., Ghosh, G., Hafeezunnisa, M., and Sen, R. (2017). Rho protein: roles and mechanisms. Annu. Rev. Microbiol. 71, 687–709. doi: 10.1146/annurev-micro-030117-020432
Mondal, S., Yakhnin, A. V., Sebastian, A., Albert, I., and Babitzke, P. (2016). NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol. 1:15007. doi: 10.1038/nmicrobiol.2015.7
Mooney, R. A., Davis, S. E., Peters, J. M., Rowland, J. L., Ansari, A. Z., and Landick, R. (2009a). Regulator trafficking on bacterial transcription units in vivo. Mol. Cell 33, 97–108. doi: 10.1016/j.molcel.2008.12.021
Mooney, R. A., Schweimer, K., Rosch, P., Gottesman, M., and Landick, R. (2009b). Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 391, 341–358. doi: 10.1016/j.jmb.2009.05.078
Moreno-Hagelsieb, G., and Santoyo, G. (2015). Predicting functional interactions among genes in prokaryotes by genomic context. Adv. Exp. Med. Biol. 883, 97–106. doi: 10.1007/978-3-319-23603-2_5
Nagy, G., Danino, V., Dobrindt, U., Pallen, M., Chaudhuri, R., Emödy, L., et al. (2006). Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 74, 5914–5925. doi: 10.1128/iai.00619-06
Nagy, G., Dobrindt, U., Grozdanov, L., Hacker, J., and Emõdy, L. (2005). Transcriptional regulation through RfaH contributes to intestinal colonization by Escherichia coli. FEMS Microbiol. Lett. 244, 173–180. doi: 10.1016/j.femsle.2005.01.038
Nagy, G., Dobrindt, U., Schneider, G., Khan, A. S., Hacker, J., and Emödy, L. (2002). Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect. Immun. 70, 4406–4413. doi: 10.1128/iai.70.8.4406-4413.2002
Nedialkov, Y., Svetlov, D., Belogurov, G. A., and Artsimovitch, I. (2018). Locking the nontemplate DNA to control transcription. Mol. Microbiol. 109, 445–457. doi: 10.1111/mmi.13983
Nicolas, P., Mader, U., Dervyn, E., Rochat, T., Leduc, A., Pigeonneau, N., et al. (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106. doi: 10.1126/science.1206848
Nudler, E. (2012). RNA polymerase backtracking in gene regulation and genome instability. Cell 149, 1438–1445. doi: 10.1016/j.cell.2012.06.003
Núñez, B., Avila, P., and De La Cruz, F. (1997). Genes involved in conjugative DNA processing of plasmid R6K. Mol. Microbiol. 24, 1157–1168. doi: 10.1046/j.1365-2958.1997.4111778.x
O’Reilly, F. J., Xue, L., Graziadei, A., Sinn, L., Lenz, S., Tegunov, D., et al. (2020). In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557. doi: 10.1126/science.abb3758
Paitan, Y., Orr, E., Ron, E. Z., and Rosenberg, E. (1999). A NusG-like transcription anti-terminator is involved in the biosynthesis of the polyketide antibiotic TA of Myxococcus xanthus. FEMS Microbiol. Lett. 170, 221–227. doi: 10.1111/j.1574-6968.1999.tb13377.x
Pan, T., Artsimovitch, I., Fang, X. W., Landick, R., and Sosnick, T. R. (1999). Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. U.S.A. 96, 9545–9550. doi: 10.1073/pnas.96.17.9545
Perdrizet, G. A. II, Artsimovitch, I., Furman, R., Sosnick, T. R., and Pan, T. (2012). Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl. Acad. Sci. U.S.A. 109, 3323–3328. doi: 10.1073/pnas.1113086109
Peters, J. M., Mooney, R. A., Grass, J. A., Jessen, E. D., Tran, F., and Landick, R. (2012). Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 26, 2621–2633. doi: 10.1101/gad.196741.112
Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K.-J., and Oelmüller, R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18, 176–197. doi: 10.1105/tpc.105.036392
Ponting, C. P. (2002). Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 30, 3643–3652. doi: 10.1093/nar/gkf498
Proshkin, S., Rahmouni, A. R., Mironov, A., and Nudler, E. (2010). Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508. doi: 10.1126/science.1184939
Ramírez-Sarmiento, C. A., Noel, J. K., Valenzuela, S. L., and Artsimovitch, I. (2015). Interdomain contacts control native state switching of RfaH on a dual-funneled landscape. PLoS Comput. Biol. 11:e1004379. doi: 10.1371/journal.pcbi.1004379
Rees, W. A., Weitzel, S. E., Yager, T. D., Das, A., and von Hippel, P. H. (1996). Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc. Natl. Acad. Sci. U.S.A. 93, 342–346. doi: 10.1073/pnas.93.1.342
Richardson, J. P. (1991). Preventing the synthesis of unused transcripts by Rho factor. Cell 64, 1047–1049. doi: 10.1016/0092-8674(91)90257-y
Rodnina, M. V. (2018). Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 10:a032664. doi: 10.1101/cshperspect.a032664
Roland, K. L., Liu, C. G., and Turnbough, C. L. Jr. (1988). Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 85, 7149–7153. doi: 10.1073/pnas.85.19.7149
Said, N., Hilal, T., Sunday, N. D., Khatri, A., Bürger, J., Mielke, T., et al. (2020). Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase ρ. Science doi: 10.1126/science.abd1673 [Epub ahead of print].
Santangelo, T. J., and Artsimovitch, I. (2011). Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 9, 319–329. doi: 10.1038/nrmicro2560
Saxena, S., and Gowrishankar, J. (2011). Compromised factor-dependent transcription termination in a nusA mutant of Escherichia coli: spectrum of termination efficiencies generated by perturbations of Rho, NusG, NusA, and H-NS family proteins. J. Bacteriol. 193, 3842–3850. doi: 10.1128/jb.00221-11
Saxena, S., Myka, K. K., Washburn, R., Costantino, N., Court, D. L., and Gottesman, M. E. (2018). Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol. Microbiol. 108, 495–504. doi: 10.1111/mmi.13953
Schmidt, A., Kochanowski, K., Vedelaar, S., Ahrné, E., Volkmer, B., Callipo, L., et al. (2016). The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110. doi: 10.1038/nbt.3418
Schmidt, M. C., and Chamberlin, M. J. (1984). Binding of rho factor to Escherichia coli RNA polymerase mediated by nusA protein. J. Biol. Chem. 259, 15000–15002.
Seifi, B., Aina, A., and Wallin, S. (2020). Structural fluctuations and mechanical stabilities of the metamorphic protein RfaH. Proteins doi: 10.1002/prot.26014 [Epub ahead of print].
Sevostyanova, A., and Artsimovitch, I. (2010). Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 38, 7432–7445. doi: 10.1093/nar/gkq623
Sevostyanova, A., Belogurov, G. A., Mooney, R. A., Landick, R., and Artsimovitch, I. (2011). The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol. Cell 43, 253–262. doi: 10.1016/j.molcel.2011.05.026
Sevostyanova, A., Svetlov, V., Vassylyev, D. G., and Artsimovitch, I. (2008). The elongation factor RfaH and the initiation factor sigma bind to the same site on the transcription elongation complex. Proc. Natl. Acad. Sci. U.S.A. 105, 865–870. doi: 10.1073/pnas.0708432105
Shi, D., Svetlov, D., Abagyan, R., and Artsimovitch, I. (2017). Flipping states: a few key residues decide the winning conformation of the only universally conserved transcription factor. Nucleic Acids Res. 45, 8835–8843. doi: 10.1093/nar/gkx523
Short, F. L., Di Sario, G., Reichmann, N. T., Kleanthous, C., Parkhill, J., and Taylor, P. W. (2020). Genomic profiling reveals distinct routes to complement resistance in Klebsiella pneumoniae. Infect. Immun. 88: e00043–20.
Singh, N., Bubunenko, M., Smith, C., Abbott, D. M., Stringer, A. M., Shi, R., et al. (2016). SuhB associates with Nus factors to facilitate 30S ribosome biogenesis in Escherichia coli. mBio 7:e00114-16.
Skals, M., Jorgensen, N. R., Leipziger, J., and Praetorius, H. A. (2009). α-Hemolysin from Escherichia coli uses endogenous amplification through P2X receptor activation to induce hemolysis. Proc. Natl. Acad. Sci. U.S.A. 106, 4030–4035. doi: 10.1073/pnas.0807044106
Sosunova, E., Sosunov, V., Kozlov, M., Nikiforov, V., Goldfarb, A., and Mustaev, A. (2003). Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc. Natl. Acad. Sci. U.S.A. 100, 15469–15474. doi: 10.1073/pnas.2536698100
Squires, C. L., Greenblatt, J., Li, J., Condon, C., and Squires, C. L. (1993). Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. U.S.A. 90, 970–974. doi: 10.1073/pnas.90.3.970
Stevenson-Jones, F., Woodgate, J., Castro-Roa, D., and Zenkin, N. (2020). Ribosome reactivates transcription by physically pushing RNA polymerase out of transcription arrest. Proc. Natl. Acad. Sci. U.S.A. 117, 8462–8467. doi: 10.1073/pnas.1919985117
Sullivan, S. L., and Gottesman, M. E. (1992). Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell 68, 989–994. doi: 10.1016/0092-8674(92)90041-a
Svetlov, D., Shi, D., Twentyman, J., Nedialkov, Y., Rosen, D. A., Abagyan, R., et al. (2018). In silico discovery of small molecules that inhibit RfaH recruitment to RNA polymerase. Mol. Microbiol. 110, 128–142. doi: 10.1111/mmi.14093
Svetlov, V., Belogurov, G. A., Shabrova, E., Vassylyev, D. G., and Artsimovitch, I. (2007). Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 35, 5694–5705. doi: 10.1093/nar/gkm600
Thomas, S., Holland, I. B., and Schmitt, L. (2014). The type 1 secretion pathway — the hemolysin system and beyond. Biochim. Biophys. Acta 1843, 1629–1641. doi: 10.1016/j.bbamcr.2013.09.017
Tomar, S. K., Knauer, S. H., Nandymazumdar, M., Rosch, P., and Artsimovitch, I. (2013). Interdomain contacts control folding of transcription factor RfaH. Nucleic Acids Res. 41, 10077–10085. doi: 10.1093/nar/gkt779
Turnbough, C. L. Jr. (2019). Regulation of bacterial gene expression by transcription attenuation. Microbiol. Mol. Biol. Rev. 83:e00019-19.
Turtola, M., and Belogurov, G. A. (2016). NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. eLife 5:e18096. doi: 10.7554/eLife.18096
Vos, S. M., Farnung, L., Urlaub, H., and Cramer, P. (2018). Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606. doi: 10.1038/s41586-018-0442-2
Vvedenskaya, I. O., Vahedian-Movahed, H., Bird, J. G., Knoblauch, J. G., Goldman, S. R., Zhang, Y., et al. (2014). Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 344, 1285–1289. doi: 10.1126/science.1253458
Wang, B., Gumerov, V. M., Andrianova, E. P., Zhulin, I. B., and Artsimovitch, I. (2020). Origins and molecular evolution of the NusG paralog RfaH. mBio 11:e02717-20.
Wang, C., Molodtsov, V., Firlar, E., Kaelber, J. T., Blaha, G., Su, M., et al. (2020). Structural basis of transcription-translation coupling. Science 369, 1359–1365. doi: 10.1126/science.abb5317
Wang, Y., Tian, G. B., Zhang, R., Shen, Y., Tyrrell, J. M., Huang, X., et al. (2017). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. doi: 10.1016/s1473-3099(16)30527-8
Washburn, R. S., Zuber, P. K., Sun, M., Hashem, Y., Shen, B., Li, W., et al. (2020). Escherichia coli NusG links the lead ribosome with the transcription elongation complex. iScience 23:101352. doi: 10.1016/j.isci.2020.101352
Webster, M. W., Takacs, M., Zhu, C., Vidmar, V., Eduljee, A., Abdelkareem, M., et al. (2020). Structural basis of transcription-translation coupling and collision in bacteria. Science 369, 1355–1359. doi: 10.1126/science.abb5036
Werner, F. (2012). A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 417, 13–27. doi: 10.1016/j.jmb.2012.01.031
Xiong, L., and Liu, Z. (2015). Molecular dynamics study on folding and allostery in RfaH. Proteins 83, 1582–1592. doi: 10.1002/prot.24839
Xun, S., Jiang, F., and Wu, Y. D. (2016). Intrinsically disordered regions stabilize the helical form of the C-terminal domain of RfaH: a molecular dynamics study. Bioorg. Med. Chem. 24, 4970–4977. doi: 10.1016/j.bmc.2016.08.012
Yakhnin, A. V., FitzGerald, P. C., McIntosh, C., Yakhnin, H., Kireeva, M., Turek-Herman, J., et al. (2020a). NusG controls transcription pausing and RNA polymerase translocation throughout the Bacillus subtilis genome. Proc. Natl. Acad. Sci. U.S.A. 117, 21628–21636. doi: 10.1073/pnas.2006873117
Yakhnin, A. V., Kashlev, M., and Babitzke, P. (2020b). NusG-dependent RNA polymerase pausing is a frequent function of this universally conserved transcription elongation factor. Crit. Rev. Biochem. Mol. Biol. 55, 716–728. doi: 10.1080/10409238.2020.1828261
Yakhnin, A. V., Murakami, K. S., and Babitzke, P. (2016). NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J. Biol. Chem. 291, 5299–5308. doi: 10.1074/jbc.M115.704189
Yakhnin, H., Yakhnin, A. V., Mouery, B. L., Mandell, Z. F., Karbasiafshar, C., Kashlev, M., et al. (2019). NusG-dependent RNA polymerase pausing and tylosin-dependent ribosome stalling are required for tylosin resistance by inducing 23S rRNA methylation in Bacillus subtilis. mBio 10:e02665-19.
Yang, Z., Li, M., and Sun, Q. (2020). RHON1 co-transcriptionally resolves R-Loops for Arabidopsis chloroplast genome maintenance. Cell Rep. 30, 243–256.e5. doi: 10.1016/j.celrep.2019.12.007
Young, R. A., and Steitz, J. A. (1978). Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc. Natl. Acad. Sci. U.S.A. 75, 3593–3597. doi: 10.1073/pnas.75.8.3593
Zellars, M., and Squires, C. L. (1999). Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol. 32, 1296–1304. doi: 10.1046/j.1365-2958.1999.01442.x
Zhang, Y., Feng, Y., Chatterjee, S., Tuske, S., Ho, M. X., Arnold, E., et al. (2012). Structural basis of transcription initiation. Science 338, 1076–1080. doi: 10.1126/science.1227786
Zuber, P. K., Artsimovitch, I., NandyMazumdar, M., Liu, Z., Nedialkov, Y., Schweimer, K., et al. (2018). The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. eLife 7:e36349. doi: 10.7554/eLife.36349
Keywords: antitermination, evolution, NusG, RfaH, transcriptional pausing, termination, virulence
Citation: Wang B and Artsimovitch I (2021) NusG, an Ancient Yet Rapidly Evolving Transcription Factor. Front. Microbiol. 11:619618. doi: 10.3389/fmicb.2020.619618
Received: 20 October 2020; Accepted: 07 December 2020;
Published: 08 January 2021.
Edited by:
Omar Orellana, University of Chile, ChileReviewed by:
Paul Babitzke, Pennsylvania State University (PSU), United StatesCopyright © 2021 Wang and Artsimovitch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Irina Artsimovitch, YXJ0c2ltb3ZpdGNoLjFAb3N1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.