- 1College of Environmental Science and Engineering, China West Normal University, Nanchong, China
- 2Department of Environmental Engineering, College of Resources and Environmental Sciences, Nanjing Agricultural University, Nanjing, China
- 3State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
- 4College of Agricultural Engineering, Hohai University, Nanjing, China
- 5Key State Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, China
- 6Key Laboratory of Environment Remediation and Ecological Health, Ministry of Education, College of Environmental Resource Sciences, Zhejiang University, Hangzhou, China
- 7School of Geography and Ocean Science, Nanjing University, Nanjing, China
- 8Key Lab of Microbiology for Agricultural Environment, Ministry of Agriculture, College of Life Sciences, Nanjing Agricultural University, Nanjing, China
The discovery of complete ammonia oxidation (comammox), oxidizing ammonia to nitrate via nitrite in a single organism, has redefined the traditional recognition of the two-step nitrification driven by two functional groups (ammonia-oxidizing and nitrite-oxidizing microorganisms). However, the understanding of the distribution and niche differentiation of comammox Nitrospira in the estuarine mudflats and their reclaimed agricultural soils is still limited. Here, we investigated the abundance, diversity and community structures of comammox Nitrospira in the mudflats and the reclaimed agricultural soils in the northern Yangtze River estuary. Quantitative PCR showed the abundances of amoA genes of comammox were lower than that of ammonia-oxidizing bacteria (AOB) in nearly all samples. Amplicon sequencing of amoA genes revealed that the community structures of comammox Nitrospira were significantly (P < 0.001) different between the original mudflats and the reclaimed agricultural soils, indicating niche differentiation among comammox Nitrospira clades (clade A.1, clade A.2, and clade B). The clade A.1 was the dominant group of comammox Nitrospira in the mudflats, while clade B predominated in the agricultural soils. However, the members of clade A.2 could be clearly divided into two groups, the mudflat-preferred and agricultural soil-preferred groups, suggesting more complicated ecological preferences within this sub-clade. Furthermore, it was demonstrated that salinity, organic matter (OM) and NO3–-N had a significantly influence on the distribution of comammox Nitrospira in the estuarine environment. Clade A.1 and nearly half members of clade A.2 were positively correlated with salinity, and negatively correlated with the concentrations of OM and NO3–-N. In contrast, the clade B and the other half members of clade A.2 showed the exact opposite pattern: a negative correlation with salinity and positive correlation with OM and NO3–-N. The co-occurrence network demonstrated that the operational taxonomic units (OTUs) within the same (sub-)clade were mostly positively correlated, indicating the similar niche preferences among the members from the same (sub-)clade of comammox Nitrospira. Taken together, our results revealed the niche differentiation of comammox Nitrospira in estuarine ecosystems where salinity and OM were the primary factors responsible for the distinct ecological distribution patterns.
Introduction
Nitrification, the microbial conversion of ammonia to nitrate, is a central process in the global nitrogen cycle (Gruber and Galloway, 2008). For more than a century, nitrification was considered to be divided into two sequential step (Winogradsky, 1892), the ammonia oxidation (from ammonia to nitrite) performed by ammonia-oxidizing bacteria (AOB) and archaea (AOA) (Konneke et al., 2005), and the nitrite oxidation (from nitrite to nitrate) performed by nitrite-oxidizing bacteria (NOB). However, the recent discovery of complete ammonia oxidization (comammox) has broken this classical tenet (Daims et al., 2015; van Kessel et al., 2015). These novel comammox bacteria, affiliated to the genus Nitrospira (lineage II), contain all the necessary genes involved in the oxidation of ammonia and nitrite and thus can completely oxidize ammonia to nitrate in a single microorganism (Daims et al., 2015; van Kessel et al., 2015; Palomo et al., 2018). Phylogeny based on amoA gene encoding the alpha subunit of ammonia monooxygenase (AMO) revealed that comammox Nitrospira could be separated into clade A and clade B (Daims et al., 2015). Clade A could be further subdivided into two monophyletic groups (clade A.1 and A.2) (Xia et al., 2018). However, currently the ecological distribution of these phylotypes and their potential importance in the global nitrogen and carbon cycling remain poorly understood.
Metagenomic analysis and molecular surveys primarily based on amplicon sequencing of amoA genes demonstrated the widespread distribution of comammox Nitrospira in diverse habitats, including agricultural soils (Li et al., 2019; Zhao et al., 2019), groundwater-fed rapid sand filters (Palomo et al., 2016; Bartelme et al., 2017; Fowler et al., 2018), drinking water systems (Pinto et al., 2016; Wang et al., 2017), freshwater systems (Liu et al., 2020), wastewater treatment plants (WWTPs) (Pjevac et al., 2017; Annavajhala et al., 2018) and sediments (Yu et al., 2018), but not yet in marine ecosystems (Daims et al., 2015). The abundance of comammox Nitrospira was comparable to or even higher than those of AOA and AOB in some agricultural soils (Li et al., 2019) and engineered systems (Wang et al., 2018). Kinetic study indicates that the comammox species N. inopinata is more competitive in highly oligotrophic environments (Kits et al., 2017). Likewise, Xu Y. et al. (2020) recently found that eutrophication might inhibit the growth of comammox organisms in lake sediment. However, Wang J. et al. (2019) reported that the abundance of comammox bacteria was positively related to the amount of N fertilizer along a fertilization gradient in soil, and the two monophyletic clades (clade A and clade B) showed opposite responses to nutrient input. Recent studies also suggested the distinct distribution of clade A and clade B in different environments (Xia et al., 2018; Xu S. et al., 2020), which might be determined by multiple environmental factors (Hu and He, 2017; Shi et al., 2018; Roots et al., 2019; Xu Y. et al., 2020).
Estuary and coastal environments are the interface between land and sea serving as the hub of energy and material flows (Jickells, 1998). Over the past several decades, large-scale land reclamation of mudflats in estuaries has provided great economic benefits (Wang and Wall, 2010), but the nutrient input threatened the environment of estuarine ecosystems (Jickells, 1998; Chai et al., 2006). Nitrification coupling to denitrification or anaerobic ammonium oxidation is potentially an effective bioremediation pathway to remove excessive reactive nitrogen in these nitrogen-enriched environments (Rysgaard et al., 1996; Gruber and Galloway, 2008). Thus, the distribution and activity of ammonia-oxidizing microorganisms has become a subject of considerable interest (Wankel et al., 2011; Zheng et al., 2014). Previous studies have demonstrated that salinity, NH4+-N, organic carbon and other factors contribute to the niche differentiation of ammonia-oxidizing microorganisms (AOA and AOB) (Liu et al., 2013; Zhou et al., 2018). Recently, some studies suggested that comammox Nitrospira were quite abundant in the tidal flat sediments of estuaries (Sun et al., 2020), even as much so as AOA or AOB (Xia et al., 2018; Jiang et al., 2019). However, the distribution of comammox along a salinity gradient in estuarine continuum and the population dynamics in response to the land reclamation from mudflat to agricultural soils are still unknown.
The Yangtze River is the third-longest river of the world in terms of water volume, and carries a high biogeochemical flux of N nutrients (Zhu, 2005). In this study, quantitative PCR (qPCR) and Illumina HiSeq-based sequencing of amoA genes were employed to investigate the distribution and diversity of comammox Nitrospira in the original mudflat and the reclaimed agricultural soils along the north branch of Yangtze River estuary. Our aim is to reveal the niche differentiation of comammox Nitrospira phylotypes, and to explore potential factors driving the environmental distributions of comammox Nitrospira in estuarine ecosystems.
Materials and Methods
Soil Sampling and Physiochemical Analysis
The sampling sites were along the north branch of the Yangtze estuary, located in Jiangsu Province, China (Supplementary Figure S1). This region is characterized by a typical subtropical wet monsoon climate. The mean annual temperature in this region is 15.1°C and the mean annual rainfall is 1,040 mm. Since the 1990s, a large part of upper intertidal mudflats was reclaimed to agricultural fields with a rapeseed-rice rotation system. We sampled 22 locations including both mudflat sites (M1-M11) and the agricultural sites (A1-A11) in April 2017 (Supplementary Figure S1). Each mudflat site was ≤ 200 m apart from the paralleled agricultural site and located in the supratidal zone, which remain non-flooded condition at most time. At each sampling site, three replicates were collected, and each replicate contained five soil cores (0–5 cm). The soil samples were freeze-dried and passed through a 2 mm sieve and stored at −20°C for following DNA extraction and biochemical analysis.
Soil pH and salinity were measured using a soil-water suspension (1:2.5 soil/water) after shaking for 30 min using a Mettler-Toledo pH/EC meter (SevenMulti S40, Switzerland) (Wang et al., 2014). Organic matter (OM) in soils was determined using the K2Cr2O7 oxidation-reduction titration method. Nitrate (NO3–-N) and ammonium (NH4+-N) were extracted with 2 M KCl at a soil-to-solution ratio of 1:5 and measured with a continuous flow analyzer (Skalar Inc., Breda, Netherlands).
DNA Extraction and Quantitative PCR
Total soil DNA was extracted from each sample using the Fast DNA spin kit for soil (MP Biomedicals, Santa Ana, CA). DNA concentration was then quantified by a Nanodrop ND-1000 UV-vis Spectrophotometer (Thermo Scientific, Wilmington, DE, United States). The absolute abundances of comammox Nitrospira, AOA and AOB in each soil sample were determined by using a CFX96 Optical Real-Time Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, United States). The primer pairs of Ntsp-amoA 162F (5′-GGATTTCTGGNTSGATTGGA-3′)/Ntsp-amoA 359R (5′-WA GTTNGACCACCASTACCA-3′) (Fowler et al., 2018), Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′)/Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) (Francis et al., 2005), and amoA1F (5′-GGGGTTTCTACTGGTGGT-3′)/amoA2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (Rotthauwe et al., 1997) were used for the qPCR of comammox, AOA and AOB amoA genes, respectively. Each sample was amplified in triplicate 20 μl containing 10 μl of 2 × SYBR Premix Ex TaqTM (Takara, Dalian, China), 0.5 μl of each primer (10 μM) and 10 ng of template DNA. The qPCR reactions were performed using the following thermocycling protocol: 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 48°C (55°C for AOA/AOB amoA) for 30 s and extension at 72°C for 45 s, and melt curve (65–95°C) 0.2°C/s gradient. The qPCR standards were generated using serial dilutions (from 101 to 109 copies) of plasmid DNA from one representative clone containing amoA genes of comammox Nitrospira, AOA or AOB. The amplification efficiencies were determined as above 96% with R2 values of 0.995–0.999. Additionally, melting curve analyses and agarose gel electrophoresis were conducted to determine the specificity of the amplified products.
Amplicon Sequencing and Bioinformatic Analysis
For amplicon sequencing, the amoA gene of comammox Nitrospira was amplified using primers Ntsp-amoA 162F/Ntsp-amoA 359R, and the forward primers were modified to contain a unique barcode (10 bp) at the 5′ end for each sample. PCR reactions were performed in a 50 μl mixture containing 25 μl of 2 × Rapid Taq Master Mix (Vazyme, Nanjing, China), 2 μl of each primer (10 μM) and 20 ng of template DNA. The amplification conditions were as follows: 95°C for 3 min, and 30 cycles of 95°C for 15 s, 48°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min. Each sample was amplified in triplicate, and the PCR products were purified using AxyPrep gel extraction kit (AxyGen, Hangzhou, China) and quantified by Nano-Drop ND-1000 spectrophotometer. Purified amplicons were pooled together in equimolar ratios into one single tube and sent for paired-end sequencing (2 × 150 bp) on an Illumina HiSeq platform (Illumina, San Diego, United States) at Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China.
Raw data were processed using the Quantitative Insight into Microbial Ecology (QIIME) toolkit1 (Caporaso et al., 2010). Paired-end reads were first merged using the FLASH tool based on the matched overlapping regions (Magoc and Salzberg, 2011). Sequences were quality filtered (quality score > 20), then were split into libraries of samples by specific barcode sequences. Chimera detection and elimination was accomplished using uchime de novo function, which was based on self-sequences (Edgar et al., 2011). The remaining sequences sharing ≥ 95% identity were clustered into one operational taxonomic unit (OTU). The representative sequence of each OTU was blasted against NCBI-nr database, and only the candidates with the hits of amoA or pmoA genes (comammox amoA gene might be wrongly annotated as pmoA gene before the discovery of comammox in 2015; Daims et al., 2015; van Kessel et al., 2015) were collected and further confirmed by using phylogenetic analysis with the reference comammox amoA sequences (Lin et al., 2020). QIIME’s scripts alpha_rarefaction. py and beta_diversity_through_plots. py were used to compute alpha and beta diversity values, and all samples were uniformed to a certain number of sequences per sample. Phylogenetic tree was constructed using Maximum Likelihood method and calculated in MEGA 6.0 with 100 bootstrap replicates (Tamura et al., 2013).
Co-occurrence Network Analysis
To reduce rare OTUs in the data set, only the OTUs with relative abundance ≥ 0.01% and occurring in more than 50% of all samples were kept for co-occurrence network analysis (Ma et al., 2016; Delgado-Baquerizo et al., 2018). In order to reduce the spurious correlation associated with compositional bias, we calculated all possible correlation coefficients using SparCC method (Kurtz et al., 2015). We considered statistical significance with cutoff at P < 0.01 (adjusted with Benjamini-Hochberg false discovery rate (FDR) control procedure) (Yoav Benjamini et al., 2006) and correlations with cutoff at r ≥ 0.6 (Junker and Schreiber, 2008; Barberan et al., 2012). Network topological parameters were calculated with “igraph”2 and “WGCNA” package3. Network images were generated with Gephi 0.9.14 with the Fruchterman-Reingold layout. The nodes with the highest degree centrality (degree > 10) in were defined as hub nodes, and the other nodes were defined as peripheral nodes (Ma et al., 2018).
Statistical Analysis
ANOVA analysis was performed to evaluate differences in soil physiochemical properties, gene copies, diversity index among samples using SPSS Statistics 19.0 (IBM Corporation, Armonk, NY, United States). Spearman’s correlations were used to reveal significant correlations between the diversity, the relative abundance of each OTUs and soil physiochemical properties. Non-metric multidimensional scaling (NMDS) and pairwise comparisons (ADONIS, ANOSIM, and MRPP) were conducted to display differences in comammox Nitrospira community composition based on the Bray-Curtis dissimilarity matrix of detected OTUs using the “vegan” package in R3.5.4. Response Ratio (RR) (Luo et al., 2006) was performed to explain the changes in the relative abundance of OTU between niches. Mantel test (“vegan” package in R 3.5.4), Multiple Regression Tree analysis (MRT) (“mvpart” package in R 2.1.5) and Constrained Correspondence Analysis (CCA) (Canoco 5 v. 5.02) were conducted to analysis the relationships between environmental properties and community structure. Distance-based multivariate analysis (DistLM) (Mcardle and Anderson, 2001) was also performed to determine the effect of soil properties on community structure of comammox Nitrospira, the sequential and marginal tests were analyzed to determine the statistical significance and the relative contribution of each environmental variable.
NCBI Sequence Accession Numbers
Amplicon date was deposited in the Sequence Read Archive (SRA) at NCBI under the accession number PRJNA644509. The OTUs representative amoA genes were submitted to the NCBI GenBank under accession no. MT395049 to MT395142.
Results
Soil Physiochemical Properties
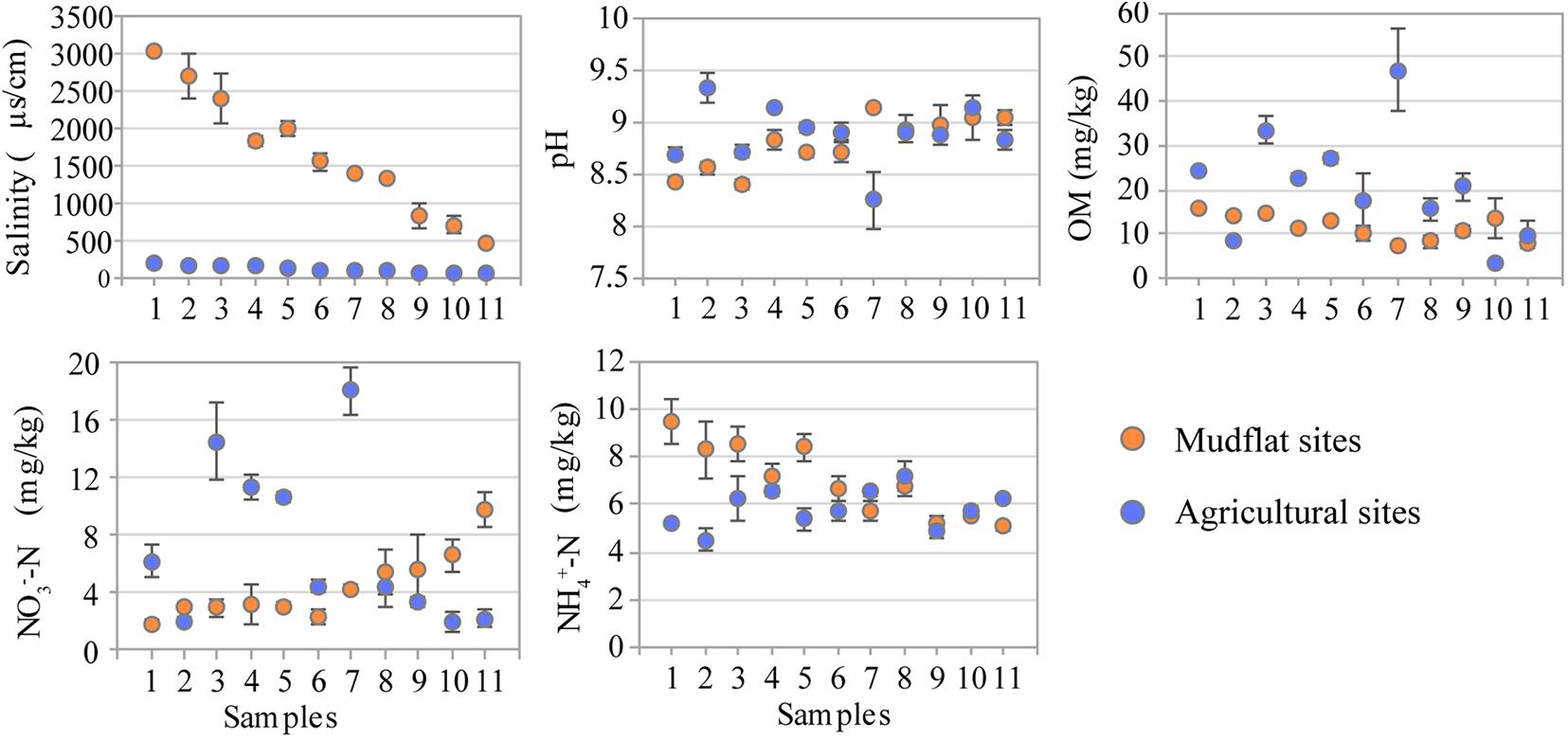
Soil salinity in the mudflats was typically high, showing a salinity gradient descending from the lower estuary (M1, 3040.0 μS/cm) to the upper estuary (M11, 391.0 μS/cm) (P < 0.01), which remarkably decreased (P < 0.01) in reclaimed agricultural soil (46.2–217.0 μS/cm) (Figure 1 and Supplementary Table S1). The soil pH was alkaline in all samples, ranging from 8.44 to 9.14 in mudflat and 7.88 to 9.59 in agricultural soils. The organic matter (OM) varied from 7.93 mg/kg to 15.93 mg/kg in mudflat sites with the mean value 11.65 mg/kg, which significantly increased in agricultural sites (range 8.26–46.97 mg/kg, mean 20.92 mg/kg). Similarly, the average content of NO3–-N in agricultural soils was also higher than that of mudflat (7.18 vs. 4.38 mg/kg). There was no significant difference in the concentration of NH4+-N between the mudflat (5.08–9.50 mg/kg) and agricultural sites (4.52–7.17 mg/kg) (Figure 1 and Supplementary Table S1).
Abundance of the amoA Genes of Ammonia Oxidizers
The amoA gene abundance of comammox Nitrospira, AOA and AOB in each sample were quantified using qPCR assays (Figure 2). In mudflat, the number of comammox Nitrospira amoA genes ranged from 1.93 × 106 to 3.00 × 107 copies g–1 soil, which were higher than or comparable to AOA amoA genes (1.45 × 106 copies − 9.05 × 106 copies g–1 soil) (Figure 2). The AOB amoA genes abundances were higher than counterparts of AOA and comammox Nitrospira, ranging from 1.58 × 107 to 1.15 × 108 copies g–1 soil, with the ratio of AOB/AOA amoA and AOB/comammox Nitrospira amoA varying from 1.75 to 45.59 and 1.08 to 34.73, respectively. In the agricultural soils, the comammox Nitrospira amoA gene abundances ranged from 9.76 × 105 to 7.74 × 107 copies g–1 soil (Figure 2). The number of AOA amoA genes reached to 7.38 × 106 − 5.74 × 108 copies g–1 soil. The AOB amoA genes ranged from 7.25 × 106 to 3.48 × 108 copies g–1 soil (Figure 2), and the ratio of AOB/AOA amoA and AOB/comammox Nitrospira amoA ranged from 0.03 to 1.05 (except site A1 was 5.22) and 1.02 to 36.09, respectively. Spearman correlation displayed that the amoA abundance of AOA and comammox Nitrospira were negatively correlated with salinity, and AOA amoA gene abundance also showed significant negative correlation with the concentration of NH4+-N (Figure 2). Additionally, the amoA gene abundances of all three groups of ammonia-oxidizing microorganisms exhibited positive relationship with OM and NO3–-N (Figure 2).
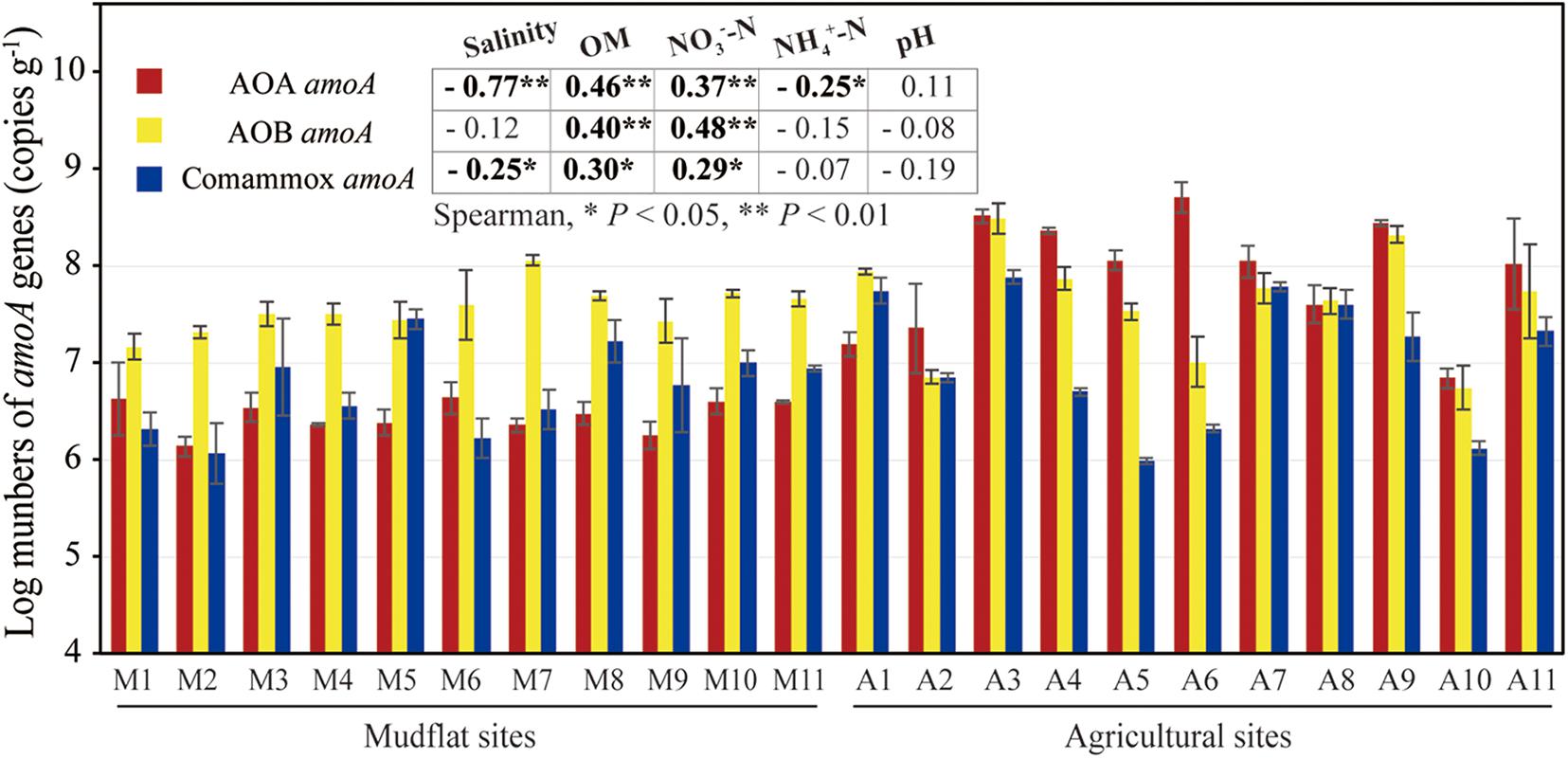
Figure 2. AOA, AOB and comammox Nitrospira amoA gene copy numbers in mudflat (M1–M11) and reclaimed agricultural soils (A1–A11). The mean values of amoA copy numbers were obtained from triplicate samples. Embedded chart shows the correlation between the amoA gene numbers and soil properties based on spearman rank (*P < 0.05, **P < 0.01).
Diversity and Community Structure of Comammox Nitrospira
After filtering, a total of 7,307,681 high-quality comammox Nitrospira amoA gene sequences was obtained from all the 66 soil samples, which were then clustered into 546 OTUs at 95% sequence identity. Rarefaction curve of each soil sample gradually reached a plateau after 5,000 sequences, indicating that the dataset was sufficient for the diversity analysis of comammox Nitrospira in the samples studied (Supplementary Figure S2). The sequences in each sample were rarified to 20,006 for subsequent analysis. The α diversity of comammox Nitrospira in the mudflat was higher than that in the agricultural soils, both in phylotype richness (Observed OTUs and Shannon) and phylogenetic diversity (PD) (P < 0.01) (Figure 3A). NMDS demonstrated that soil comammox Nitrospira communities varied between habitats (Figure 3B), which was further supported by non-parametric multivariate analysis (ADONIS, R2 = 0.3997, P < 0.001; ANOSIM, R2 = 0.3997, P < 0.001; MRPP, R2 = 0.2019, P < 0.001) (Supplementary Table S2). The within-habitat pairs of Bray-Curtis dissimilarity revealed that the community dissimilarity was greater in agricultural than that in mudflat (Figure 3C).
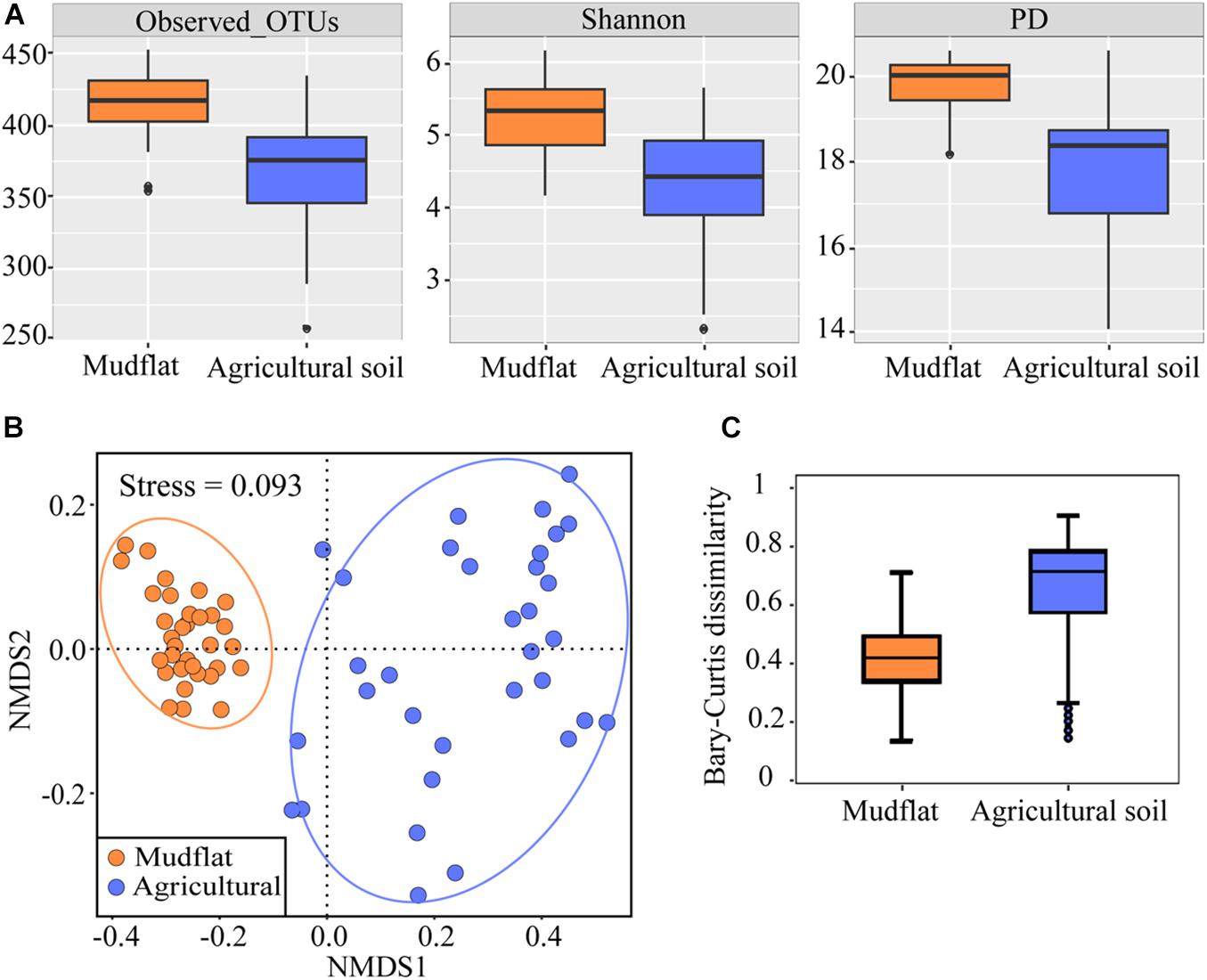
Figure 3. α-diversity and β-diversity of comammox Nitrospira amoA genes in mudflat and agricultural sites. (A) Number of observed OTUs, the Shannon index, and the PD values using OTU similarity thresholds of 95% for the amoA gene. (B) NMDS analysis based on Bray-Curtis dissimilarities of comammox Nitrospira communities. (C) Bray-Curtis dissimilarities between mudflat and agricultural soils, P < 0.01.
Phylogenetic analysis demonstrated that 90.8–99.2% of comammox Nitrospira amoA gene sequences in the mudflat fell into clade A, and 34.1–59.6% of the sequences belonged to clade A.1 and 38.9–59.9% belonged to clade A.2 (Figure 4A). Clade B only accounted for 0.8–9.2% of comammox Nitrospira community in the mudflat soils (Figure 4A). However, the relative abundance of clade A.1 sharply decreased in the reclaimed agricultural soils compared to the mudflat soils, and clade B was the most dominant lineage of comammox Nitrospira in most agricultural soils with the exception of sites A2, A10, and A11 (Figure 4A). Additionally, the relative abundance of clade A.2 was much higher than that of clade A.1 in the agricultural soils (Figure 4A). However, it did not mean that the relative abundance of each OTU from clade A.2 in the agricultural soils was higher than that in the mudflat. For instance, OTU1, 3, 10, 16, 24, 27, and 28 were primarily distributed in the mudflat with the relative abundance of 1.1–14.0% that were significantly higher (P < 0.05) than those in the agricultural soils, whereas OTU4, 7, 12, 18, 19, 22, 23, and 31 were much more abundant (P < 0.05) in the agricultural soils than in the mudflat soils (Figure 4B and Supplementary Table S3).

Figure 4. Community composition of comammox Nitrospira in different sites of this study. (A) Relative proportions of the different clades of comammox Nitrospira in 22 different sites samples. (B) Phylogenetic tree and relative abundance of comammox Nitrospira amoA genes sequences. Dominate OTUs with a relative abundance > 1% were included in the phylogenetic analysis, accounting for 74% of the total comammox Nitrospira amoA gene sequences. The log value of relative abundance (%) of individual taxa in each site was displayed as a heat map representation. The phylogenetic position of each representative OTU was shown in the phylogenetic tree on the left-hand side. OTUs labeled in red represent mudflat-preferred OTUs (MPOs), OTUs labeled in green represent agricultural soil-preferred OTUs (APOs).
Correlation Between Soil Properties and Diversity and Community Structure of Comammox Nitrospira
Spearman analysis demonstrated that the α-diversity (Observed OTUs, Shannon index and Phylogenetic diversity) of comammox Nitrospira communities was significantly positively correlated with salinity (P < 0.05), but was negatively correlated with content of OM and NO3–-N (P < 0.05) in the estuarine environments (Table 1). Mantel test and multivariate regression tree (MRT) analysis also suggested the significant influence of soil variables on comammox Nitrospira community structure in the estuary studied with the salinity at the top of the list, followed by OM and NO3–-N (Table 1 and Supplementary Figure S3). DistLM analysis further revealed that salinity, OM and NO3–-N could explain 27.43, 8.92, and 2.66% of the community variation of comammox in the estuarine environments (Supplementary Table S4).
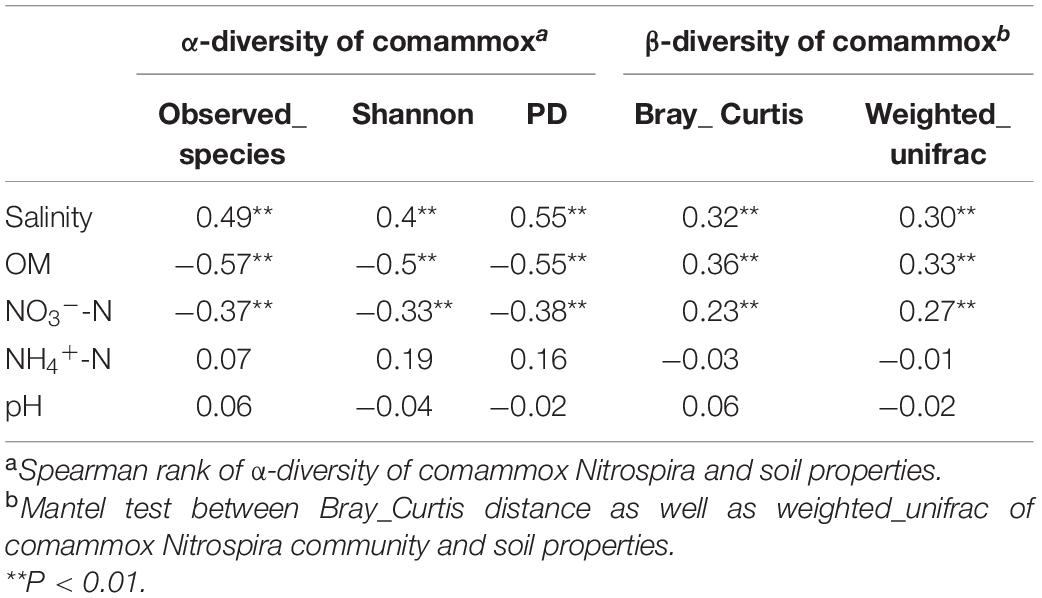
Table 1. Relationship between soil physiochemical properties and abundance and community diversity of comammox Nitrospira.
Linear regression analysis revealed that relative abundance of clade B showed significantly negative relationship with salinity (R2 = 0.448, P < 0.01), but positive relationship with OM (R2 = 0.475, P < 0.01) and NO3–-N (R2 = 0.310, P < 0.01) (Figure 5A). On the contrary, clade A.1 was positively correlated with salinity (R2 = 0.732, P < 0.01), but negatively correlated with OM (R2 = 0.169, P < 0.05) and NO3–-N (R2 = 0.142, P < 0.05). However, clade A.2 showed significant negative correlation with OM (R2 = 0.392, P < 0.01) and NO3–-N (R2 = 0.270, P < 0.01), but had no significant correlation with salinity on the whole sub-clade level (P > 0.05) (Figure 5A). Nonetheless, although there was no significant relationship between the distribution of clade A.2 and salinity on the whole sub-clade level, the OTUs of clade A.2 could be roughly divided into two groups according to their positive or negative relationship with salinity, respectively (Figure 5B). Taking the dominant OTUs for example, OTU 1, 3, 10, 16, 24, 27, and 28 were significantly positively related to salinity (P < 0.05), while OTU 4, 12, 18, and 22 showed negative relationship with salinity (P < 0.05) (Figure 5B). The similar patterns were also observed for most of the rare OTUs (0.1% < relative abundance <1%) within the clade A.2 (Supplementary Figure S4). All these results were also supported by the canonical correspondence analysis (CCA) (Supplementary Figure S5).
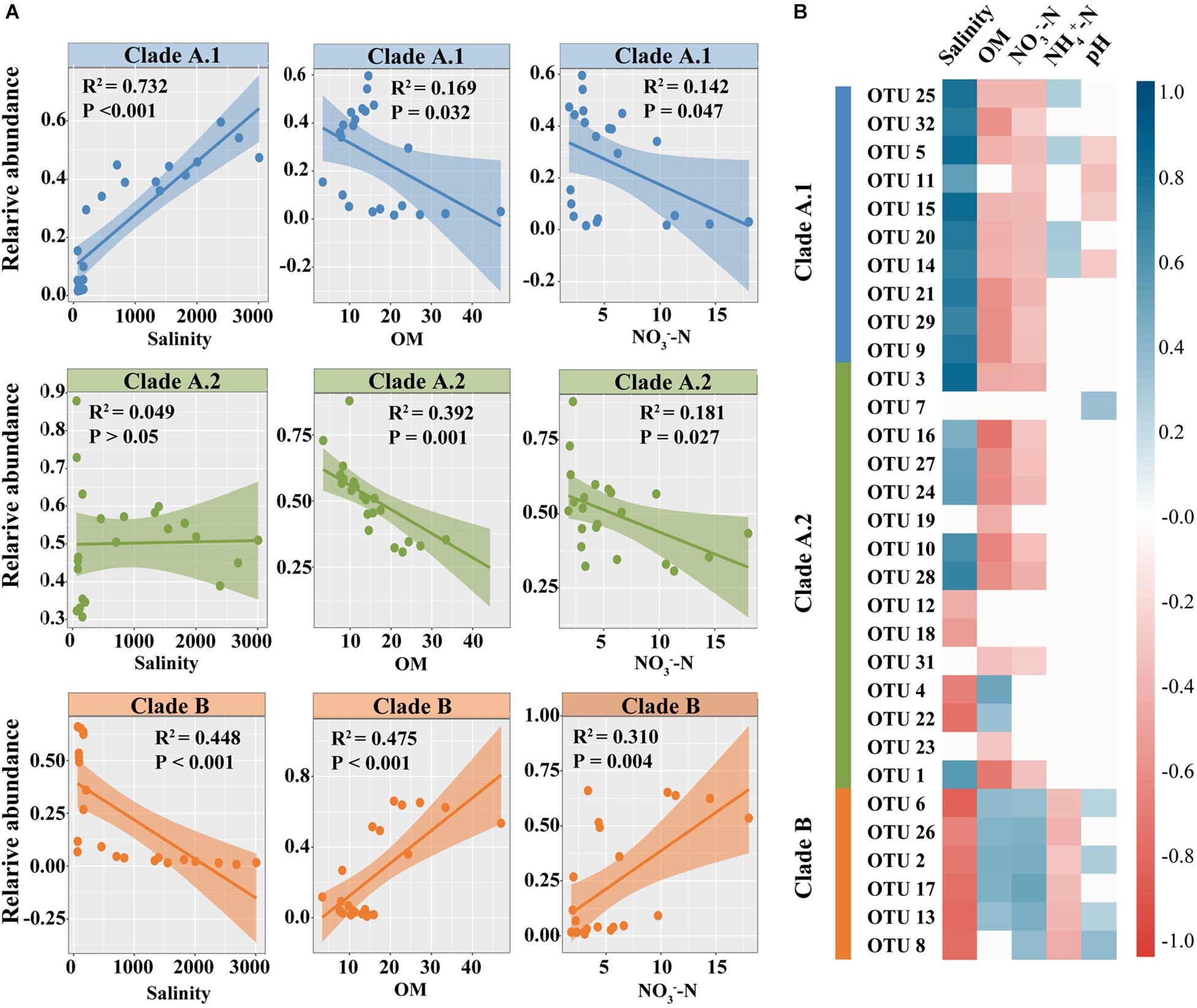
Figure 5. Relationships between relative abundance of comammox Nitrospira and soil properties. (A) Linear regression analysis between relative abundance of different comammox clades and environmental properties. (B) Spearman correlations of dominant OTUs (abundance > 1%) with soil properties in all samples, displayed as heatmaps. Color bar indicates correlation coefficients. Only significant correlations (P < 0.05) were shown.
Co-occurrence Network of Comammox Nitrospira Species
According to the statistically significant difference on the relative abundance of OTUs between the mudflat and agricultural soils (Response ratio, P < 0.05), we grouped the OTUs of comammox Nitrospira into three ecological clusters, mudflat-preferred OTUs (MPOs), agricultural soil-preferred OTUs (APOs), and a small part of ubiquitous OTUs (UBOs) (Supplementary Table S3). To identify the co-occurrence patterns of comammox Nitrospira species, we constructed a network based on significant (P < 0.01) correlations (r ≥ 0.6) among species pairs in mudflat and agricultural soils, respectively (Figure 6). The two empirical networks were verified as non-random and scale-free, based on topological comparison with random network and a power-law distribution pattern (P < 0.001) of degree (Supplementary Table S5 and Supplementary Figure S6).
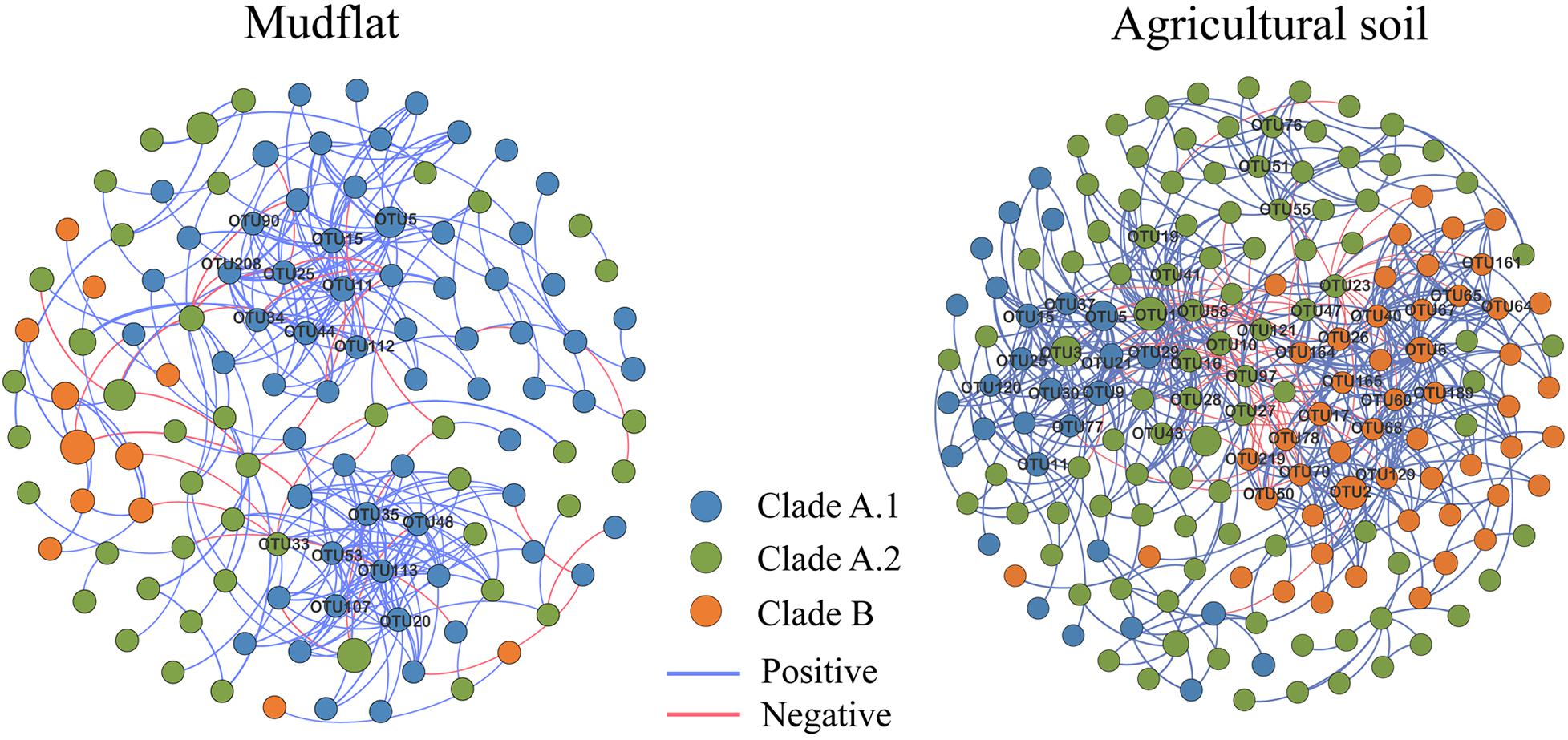
Figure 6. Co-occurrence network of comammox Nitrospira species. The nodes represent OTUs, the relationship of two OTUs in the network was shown by an edge (Coefficient r ≥ 0.6, significant P ≤ 0.01). Nodes in different colors represent different clades of comammox Nitrospira, and the size of the nodes was proportional to their relative abundance; the pink and blue edges represent the positive and negative correlations, respectively, and the thickness of each connection between two nodes is proportional to the value of correlation coefficient. Hub nodes of co-occurrence were labeled in networks.
The connection among the members in the network of agricultural soils seemed to be much tighter than that of mudflat soils, since the former one contained more nodes (i.e., OTUs), more links (i.e., correlation between two OTUs), higher average degree, and less average path length that those of the latter one (Figure 6 and Supplementary Table S5). In the network of mudflat, the members of clade A.1 had the highest average degree (6.39) compared to clade A.2 (2.95) and clade B (2.67), and most (15 in 16) of hub nodes (degree > 10) belonged to clade A.1 (Supplementary Table S6). However, in the network of agricultural soils, the average degree of clade B (8.98) were higher than those of clade A.1 (7.38) and clade A.2 (5.48), and a total of 47 nodes were determined as hub species, including 19 OTUs of clade B, 17 OTUs of clade A.2 and 11 OTUs of clade A.1 (Supplementary Table S6).
The connections among the members in both networks were mostly positive (90.4% for the mudflat network and 89.5% for the agricultural soil network) (Supplementary Table S7 and Figure 6). It was noteworthy that the connections mostly occurred among the members within each clade (Mudflat: 84.9%; Agricultural soil: 83.1%) rather than those between each clade (Mudflat: 15.1%; Agricultural soil: 16.9%) (Supplementary Table S7). Furthermore, 92 and 100% of connections among the members within MPO cluster in the two networks were positive, and 100 and 93% within APO cluster were positive, but more than 70% of connections among the members between MPO and APO cluster were negative (Supplementary Table S8).
Discussion
In this study, by using qPCR and high-throughput sequencing of comammox Nitrospira amoA genes, we determined the distribution of comammox Nitrospira in the mudflat and the reclaimed agricultural soils along the north branch of Yangtze river estuary, a typical land-sea transitional area. The abundance, diversity and community composition of comammox Nitrospira showed significant differences between the mudflat and agricultural soils, which was influenced by the various soil properties.
Previous studies demonstrated the ubiquitous distribution of comammox Nitrospira in terrestrial ecosystems (Daims et al., 2015; van Kessel et al., 2015; Lawson and Lücker, 2017), but not yet in marine environment. Here, we detected abundant comammox Nitrospira amoA genes in the mudflats and the reclaimed agricultural soils, around 106 to 107 copies g–1 soil, which was however generally lower than those of canonical ammonia oxidizers, AOB and AOA. Not surprisingly, similar results also detected in previous studies (Yu et al., 2018; Sun et al., 2020). Considering the increased salinity gradient from estuarine area to coastal water and then to the open sea coupled with the distribution patterns of comammox Nitrospira, high salinity may be one of major factors inhibiting occurrence of comammox Nitrospira in the open sea (see below).
The anthropogenic land transformation dramatically affected the community assembly of comammox Nitrospira in the estuarine environments. In this study, clade A was revealed as the predominant lineage of comammox Nitrospira in the original mudflats and was mostly characterized as MPOs cluster, while clade B and some members of clade A.2 combinedly dominated the comammox Nitrospira community in the reclaimed agricultural soils and termed as APOs. This result was consistent with the previous studies that estuarine sediment and coastal waters preferred the clade A comammox Nitrospira and most of them belonged to subclade A.1, but clade B was negligibly detected in these environments (Yu et al., 2018; Jiang et al., 2019). On the contrary, considerable abundant amoA genes of clade A.2 (Xu S. et al., 2020) and clade B (Wang Z.H. et al., 2019) were enriched in agricultural soils, and the 13CO2-DNA-stable isotope probing proved the activity of clade A.2 (Li et al., 2019) and clade B (Wang Z.H. et al., 2019) in the nitrification of agricultural soils. These results suggested high diversity and strong niche differentiation of comammox in diverse environments.
Salinity and OM were revealed as the major factors determining the distribution of comammox in the estuarine environments in our study. The abundance of comammox Nitrospira were significantly negatively correlated with salinity (Figure 2), indicating high salinity could depress the activity of most comammox Nitrospira members, especially of clade B. This may to some extent explain why the niche of comammox Nitrospira could expand to estuarine environment (Yu et al., 2018) and coastal water (Xia et al., 2018), but not yet to the open sea, in which salinity is as high as around 35‰ and much higher than those of estuarine and coastal waters. Moreover, we presumed that OM was also a key factor driving the distribution of APO members (clade B and a part of clade A.2), besides the salinity restriction. For instance, particularly, in two newly reclaimed (3 years) agricultural sites A10 and A11 with similar salinity with but lower OM than the rest of agricultural soils, the relative abundance of clade B were as few as that of the mudflat with low OM but high salinity (Figure 4A). Thus, OM might be the principal factor in governing niche adaptation of clade B in agricultural soils. Although currently, there is no direct evidence for the growth of clade B on organic carbon, the genomic analysis revealed the presence of enzymes and pathways involved in different carbon source degradation, suggesting potential mixotrophic lifestyle for clade B comammox Nitrospira as reported for other Nitrospira spp. (Watson et al., 1986; Daims et al., 2001; Spieck et al., 2006). Additionally, pH and NH4+-N had previously also been recognized as main factors impacting the diversity and activity of ammonia oxidizers in terrestrial environments (Prosser and Nicol, 2012; Shi et al., 2020; Xu Y. et al., 2020). However, we did not detect significant effect of pH and NH4+-N on the abundance and community of comammox Nitrospira in our study, that might due to the not wide range of pH (7.88–9.59) and NH4+-N (4.5–9.5 mg/kg) among our all samples (Supplementary Table S1). All these conclusions warrant the future physiological and biochemical study of pure cultures of comammox Nitrospira. However, it should be pointed out that we did not determine more comprehensive soil properties, such as distribution of particle size and concentration of metal ions, which could also influence the distribution of comammox Nitrospira.
Co-occurrence network analysis is a powerful tool in exploring the correlation or interaction among the members of microbial communities (Weiss et al., 2016), which has been performed to explore community patterns of bacteria and fungi (Mendes et al., 2014; Ma et al., 2016), as well as ammonia oxidizers (Shi et al., 2020; Sun et al., 2020). More nodes, more links and higher average degree as well as shorter average path length were found in the agricultural soil network, indicating a more complex co-occurrence pattern in agricultural soil. But a more complex co-occurrence pattern did not necessarily equal more ecological interactions among the members of comammox Nitrospira in the agricultural soils than the mudflat. In our study, positive correlations among comammox Nitrospira species predominated in the networks of both the mudflat and agricultural soils. As previously reported positive connection might be due to cross-feeding, co-aggregation, co-colonization, niche overlap or other reasons (Faust and Raes, 2012). It was hard to distinguish between true ecological interactions and other non-random processes (for example, cross-feeding vs. niche overlap) from most co-occurrence networks, but similar niche preference could be the reason why closely related species co-occur more often than those distantly related species (Chaffron et al., 2010; Faust and Raes, 2012). It was noteworthy that all comammox members depend on the energy generated from complete oxidation of ammonia to nitrate, and comammox Nitrospira within the same subclade were more connected. Therefore, we presumed that positive correlations among comammox Nitrospira of the MPOs or APOs groups largely due to the shared niche preference (Supplementary Table S8), but not biotic interaction as observed in macro-ecological co-occurrence networks (Freilich et al., 2018).
Taken together, this study revealed the ubiquitous distribution and significant niche differentiation among different (sub-)clade of comammox, clade A.1, clade A.2, and clade B, in the estuarine environment and reclaimed agricultural soils. Moreover, salinity and the concentration of OM were revealed as the major factors driving the distribution of each (sub-)clade in the estuarine environments, but more physiological and biochemical studies of pure cultures are needed to prove these conclusions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
BW proposed the idea. XW and BW designed the main idea of this work. XZ provided the resources of this study. XW and XT performed the experiments. XW, LL, and LK contributed to the interpretation of data for the work. XW drafted the manuscript. BW, LL, JA, JS, HL, HQ, and JC revised it critically for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 41977056, 41530857, 31971631, and 41701269) and State Key Laboratory of Subtropical Silviculture (No. KF2017-02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Michael Wagner from University of Vienna for the valuable discussion. We thank Dr. Yuntao Li for the assistance on data analysis and Dr. Petra Pjevac for sharing the comammox Nitrospira amoA gene sequences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.618287/full#supplementary-material
Footnotes
- ^ http://qiime.org/
- ^ https://igraph.org/
- ^ https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/
- ^ http://gephi.github.io/
References
Annavajhala, M. K., Kapoor, V., Santo-Domingo, J., and Chandran, K. (2018). Comammox functionality identified in diverse engineered biological wastewater treatment systems. Environ. Sci. Technol. Lett. 5, 110–116. doi: 10.1021/acs.estlett.7b00577
Barberan, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Bartelme, R. P., McLellan, S. L., and Newton, R. J. (2017). Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Front. Microbiol. 8:101. doi: 10.3389/fmicb.2017.00101
Benjamini, Y., Krieger, A. M., and Yekutieli, D. (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507. doi: 10.1093/biomet/93.3.491
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chaffron, S., Rehrauer, H., Pernthaler, J., and von Mering, C. (2010). A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 20, 947–959. doi: 10.1101/gr.104521.109
Chai, C., Yu, Z., Song, X., and Cao, X. (2006). The status and characteristics of eutrophication in the Yangtze River (Changjiang) estuary and the adjacent East China Sea, China. Hydrobiologia 563, 313–328.
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Daims, H., Nielsen, J. L., Nielsen, P. H., Schleifer, K.-H., and Wagner, M. (2001). In situ characterization of Nitrospira-Like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67, 5273–5284. doi: 10.1128/aem.67.11.5273-5284.2001
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-Gonzalez, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Fowler, S. J., Palomo, A., Dechesne, A., Mines, P. D., and Smets, B. F. (2018). Comammox Nitrospira are abundant ammonia oxidizers in diverse groundwater-fed rapid sand filter communities. Environ. Microbiol. 20, 1002–1015. doi: 10.1111/1462-2920.14033
Francis, C. A., Roberts, K. J., Michael Beman, J., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A 102, 14683–14688. doi: 10.1073/pnas.0506625102
Freilich, M. A., Wieters, E., Broitman, B. R., Marquet, P. A., and Navarrete, S. A. (2018). Species co-occurrence networks: can they reveal trophic and non-trophic interactions in ecological communities? Ecology 99, 690–699. doi: 10.1002/ecy.2142
Gruber, N., and Galloway, J. N. (2008). An Earth-system perspective of the global nitrogen cycle. Nature 451, 293–296. doi: 10.1038/nature06592
Hu, H. W., and He, J. Z. (2017). Comammox—a newly discovered nitrification process in the terrestrial nitrogen cycle. J. Soils Sediments 17, 2709–2717. doi: 10.1007/s11368-017-1851-9
Jiang, Q., Xia, F., Zhu, T., Wang, D., and Quan, Z. (2019). Distribution of comammox and canonical ammonia-oxidizing bacteria in tidal flat sediments of the Yangtze River estuary at different depths over four seasons. J. Appl. Microbiol. 127, 533–543. doi: 10.1111/jam.14337
Jickells, T. D. (1998). Nutrient biogeochemistry of the coastal zone. Science 281, 217–222. doi: 10.1126/science.281.5374.217
Junker, B. H., and Schreiber, F. (2008). “Correlation networks,” in Analysis of Biological Networks, eds B. H. Junker and F. Schreiber (Hoboken, NJ: Wiley-Interscience).
Kits, K. D., Sedlacek, C. J., Lebedeva, E. V., Han, P., and Wagner, M. (2017). Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 549, 269–272. doi: 10.1038/nature23679
Konneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Kurtz, Z. D., Muller, C. L., Miraldi, E. R., Littman, D. R., Blaser, M. J., and Bonneau, R. A. (2015). Sparse and compositionally robust inference of microbial ecological networks. PLoS. Comput. Biol. 11:e1004226. doi: 10.1371/journal.pcbi.1004226
Lawson, C. E., and Lücker, S. (2017). Complete ammonia oxidation: an important control on nitrification in engineered ecosystems? Curr. Opin. Biotechnol. 50, 158–165. doi: 10.1016/j.copbio.2018.01.015
Li, C., Hu, H. W., Chen, Q. L., Chen, D., and He, J. Z. (2019). Comammox Nitrospira play an active role in nitrification of agricultural soils amended with nitrogen fertilizers. Soil Biol. Biochem. 138:107609. doi: 10.1016/j.soilbio.2019.107609
Lin, C., Xu, H., Qin, W., Xu, S., Tang, X., Kuang, L., et al. (2020). Evaluation of two primer sets for amplification of comammox Nitrospira amoA genes in wetland soils. Front. Microbiol. 11:560942. doi: 10.3389/fmicb.2020.560942
Liu, S., Shen, L., Lou, L., Tian, G., Zheng, P., and Hu, B. (2013). Spatial distribution and factors shaping the niche segregation of ammonia-oxidizing microorganisms in the Qiantang River, China. Appl. Environ. Microbiol. 79, 4065–4071. doi: 10.1128/AEM.00543-13
Liu, S., Wang, H., Chen, L., Wang, J., Zheng, M., Liu, S., et al. (2020). Comammox Nitrospira within the Yangtze River continuum: community, biogeography, and ecological drivers. ISME J. 14, 2488–2504. doi: 10.1038/s41396-020-0701-8
Luo, Y., Hui, D., and Zhang, D. (2006). Elevated CO2 stimulates net accumulations of carbon and nitrogen in land ecosystems: a meta-analysis. Ecology 87, 53–63. doi: 10.1890/04-1724
Ma, B., Wang, H., Dsouza, M., Lou, J., He, Y., Dai, Z., et al. (2016). Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 10, 1891–1901. doi: 10.1038/ismej.2015.261
Ma, B., Zhao, K., Lv, X., Su, W., Dai, Z., Gilbert, J. A., et al. (2018). Genetic correlation network prediction of forest soil microbial functional organization. ISME J. 12, 2492–2505. doi: 10.1038/s41396-018-0232-8
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mcardle, B. H., and Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.2307/2680104
Mendes, L. W., Kuramae, E. E., Navarrete, A. A., van Veen, J. A., and Tsai, S. M. (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 8, 1577–1587. doi: 10.1038/ismej.2014.17
Palomo, A., Jane Fowler, S., Gulay, A., Rasmussen, S., Sicheritz-Ponten, T., and Smets, B. F. (2016). Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J. 10, 2569–2581. doi: 10.1038/ismej.2016.63
Palomo, A., Pedersen, A. G., Fowler, S. J., Dechesne, A., Sicheritz-Ponten, T., and Smets, B. F. (2018). Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 12, 1779–1793. doi: 10.1038/s41396-018-0083-3
Pinto, A. J., Marcus, D. N., Ijaz, U. Z., Bautista-de Lose Santos, Q. M., Dick, G. J., and Raskin, L. (2016). Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere 1:e00054–15. doi: 10.1128/mSphere.00054-15
Pjevac, P., Schauberger, C., Poghosyan, L., Herbold, C. W., van Kessel, M., Daebeler, A., et al. (2017). AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Front. Microbiol. 8:1508. doi: 10.3389/fmicb.2017.01508
Prosser, J. I., and Nicol, G. W. (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 20, 523–531. doi: 10.1016/j.tim.2012.08.001
Roots, P., Wang, Y., Rosenthal, A. F., Griffin, J. S., Sabba, F., Petrovich, M., et al. (2019). Comammox Nitrospira are the dominant ammonia oxidizers in a mainstream low dissolved oxygen nitrification reactor. Water Res. 157, 396–405. doi: 10.1016/j.watres.2019.03.060
Rotthauwe, J. H., Witzel, K. P., and Liesack, K. P. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712. doi: 10.1126/science.284.5411.63
Rysgaard, S., Risgaard-Petersen, N., and Sloth, N. P. (1996). Nitrification, denitrification, and nitrate ammonification in sediments of two coastal lagoons in Southern France. Hydrobiologia 329, 133–141. doi: 10.1007/bf00034553
Shi, X., Hu, H. W., Wang, J., He, J. Z., Zheng, C., Wan, X., et al. (2018). Niche separation of comammox Nitrospira and canonical ammonia oxidizers in an acidic subtropical forest soil under long-term nitrogen deposition. Soil Biol. Biochem. 126, 114–122. doi: 10.1016/j.soilbio.2018.09.004
Shi, Y., Jiang, Y., Wang, S., Wang, X., and Zhu, G. (2020). Biogeographic distribution of comammox bacteria in diverse terrestrial habitats. Sci. Total Environ 717:137257. doi: 10.1016/j.scitotenv.2020.137257
Spieck, E., Hartwig, C., McCormack, I., Maixner, F., Wagner, M., Lipski, A., et al. (2006). Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ. Microbiol. 8, 405–415. doi: 10.1111/j.1462-2920.2005.00905.x
Sun, D., Tang, X., Zhao, M., Zhang, Z., Hou, L., Liu, M., et al. (2020). Distribution and diversity of comammox Nitrospira in coastal wetlands of China. Front. Microbiol. 11:589268. doi: 10.3389/fmicb.2020.589268
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
van Kessel, M. A., Speth, D. R., Albertsen, M., Nielsen, P. H., Op den Camp, H. J., Kartal, B., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Wang, B., Zheng, Y., Huang, R., Zhou, X., Wang, D., and He, Y. (2014). Active ammonia oxidizers in an acidic soil are phylogenetically closely related to neutrophilic archaeon. Appl. Environ. Microbiol. 80, 1684–1691. doi: 10.1128/aem.03633-13
Wang, F., and Wall, G. (2010). Mudflat development in Jiangsu province, China: practices and experiences. Ocean Coast. Manage. 53, 691–699. doi: 10.1016/j.ocecoaman.2010.10.004
Wang, J., Wang, J., Rhodes, G., He, J. Z., and Ge, Y. (2019). Adaptive responses of comammox Nitrospira and canonical ammonia oxidizers to long-term fertilizations: implications for the relative contributions of different ammonia oxidizers to soil nitrogen cycling. Sci. Total Environ. 668, 224–233. doi: 10.1016/j.scitotenv.2019.02.427
Wang, M., Huang, G., Zhao, Z., Dang, C., Liu, W., and Zheng, M. (2018). Newly designed primer pair revealed dominant and diverse comammox amoA gene in full-scale wastewater treatment plants. Bioresour. Technol. 270, 580–587. doi: 10.1016/j.biortech.2018.09.089
Wang, Y., Ma, L., Mao, Y., Jiang, X., Xia, Y., Yu, K., et al. (2017). Comammox in drinking water systems. Water Res. 116, 332–341. doi: 10.1016/j.watres.2017.03.042
Wang, Z. H., Cao, Y. Q., Zhu-Barker, X., Nicol, G. W., Wright, A. L., Jia, Z. J., et al. (2019). Comammox Nitrospira clade B contributes to nitrification in soil. Soil Biol. Biochem. 135, 392–395. doi: 10.1016/j.soilbio.2019.06.004
Wankel, S. D., Mosier, A. C., Hansel, C. M., Paytan, A., and Francis, C. A. (2011). Spatial variability in nitrification rates and ammonia-oxidizing microbial communities in the agriculturally impacted elkhorn slough estuary, California. Appl.Environ. Microbiol. 77, 269–280. doi: 10.1128/aem.01318-10
Watson, S. W., Bock, E., Valois, F. W., Waterbury, J. B., and Schlosser, U. (1986). Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch. Microbiol. 144, 1–7. doi: 10.1007/BF00454947
Weiss, S., Van, T. W., Lozupone, C., Faust, K., Friedman, J., Deng, Y., et al. (2016). Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681. doi: 10.1038/ismej.2015.235
Winogradsky, S. (1892). Contributions a la morphologie des organisms de la nitrification. Arkhiv Biolog. Nauk (St. Petersbourg) 1, 87–137.
Xia, F., Wang, J. G., Zhu, T., Zou, B., Rhee, S. K., and Quan, Z. X. (2018). Ubiquity and diversity of complete ammonia oxidizers (comammox). Appl. Environ. Microbiol. 84:e1390–18. doi: 10.1128/AEM.01390-18
Xu, S., Wang, B., Li, Y., Jiang, D., Zhou, Y., Ding, A., et al. (2020). Ubiquity, diversity, and activity of comammox Nitrospira in agricultural soils. Sci. Total Environ. 706:135684. doi: 10.1016/j.scitotenv.2019.135684
Xu, Y., Lu, J., Wang, Y., Liu, G., Wan, X., Hua, Y., et al. (2020). Diversity and abundance of comammox bacteria in the sediments of an urban lake. J. Appl. Microbiol. 128, 1647–1657. doi: 10.1111/jam.14593
Yu, C., Hou, L., Zheng, Y., Liu, M., Yin, G., Gao, J., et al. (2018). Evidence for complete nitrification in enrichment culture of tidal sediments and diversity analysis of clade a comammox Nitrospira in natural environments. Appl. Microbiol. Biotechnol. 102, 9363–9377. doi: 10.1007/s00253-018-9274-0
Zhao, Z., Huang, G., He, S., Zhou, N., Wang, M., Dang, C., et al. (2019). Abundance and community composition of comammox bacteria in different ecosystems by a universal primer set. Sci. Total Environ. 691, 146–155. doi: 10.1016/j.scitotenv.2019.07.131
Zheng, Y., Hou, L., Newell, S., Liu, M., Zhou, J., Zhao, H., et al. (2014). Community dynamics and activity of ammonia-oxidizing prokaryotes in intertidal sediments of the Yangtze estuary. Appl. Environ. Microbiol. 80, 408–419. doi: 10.1128/AEM.03035-13
Zhou, X., Li, B., Guo, Z., Wang, Z., Luo, J., and Lu, C. (2018). Niche separation of ammonia oxidizers in mudflat and agricultural soils along the Yangtze River, China. Front. Microbiol. 9:3122. doi: 10.3389/fmicb.2018.03122
Keywords: complete ammonia oxidizer, estuary, amoA gene, community structure, niche differentiation, soil property, co-occurrence network
Citation: Wang X, Lu L, Zhou X, Tang X, Kuang L, Chen J, Shan J, Lu H, Qin H, Adams J and Wang B (2021) Niche Differentiation of Comammox Nitrospira in the Mudflat and Reclaimed Agricultural Soils Along the North Branch of Yangtze River Estuary. Front. Microbiol. 11:618287. doi: 10.3389/fmicb.2020.618287
Received: 16 October 2020; Accepted: 15 December 2020;
Published: 14 January 2021.
Edited by:
Yurong Liu, Huazhong Agricultural University, ChinaReviewed by:
Yongxin Lin, Fujian Normal University, ChinaXinda Lu, Massachusetts Institute of Technology, United States
Hirotsugu Fujitani, Chuo University, Japan
Copyright © 2021 Wang, Lu, Zhou, Tang, Kuang, Chen, Shan, Lu, Qin, Adams and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baozhan Wang, Ynp3YW5nQG5qYXUuZWR1LmNu; Jonathan Adams, Sm9uYWRhbXNAbmp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Xinxin Wang
Xinxin Wang Lu Lu1†
Lu Lu1† Xue Zhou
Xue Zhou Xiufeng Tang
Xiufeng Tang Junhui Chen
Junhui Chen Jun Shan
Jun Shan Huijie Lu
Huijie Lu Hua Qin
Hua Qin Baozhan Wang
Baozhan Wang