- 1Australian Infectious Diseases Research Centre, School of Chemistry and Molecular Biosciences, The University of Queensland, St. Lucia, QLD, Australia
- 2Institute for Molecular Bioscience, The University of Queensland, St. Lucia, QLD, Australia
Mononuclear molybdoenzymes are highly versatile catalysts that occur in organisms in all domains of life, where they mediate essential cellular functions such as energy generation and detoxification reactions. Molybdoenzymes are particularly abundant in bacteria, where over 50 distinct types of enzymes have been identified to date. In bacterial pathogens, all aspects of molybdoenzyme biology such as molybdate uptake, cofactor biosynthesis, and function of the enzymes themselves, have been shown to affect fitness in the host as well as virulence. Although current studies are mostly focused on a few key pathogens such as Escherichia coli, Salmonella enterica, Campylobacter jejuni, and Mycobacterium tuberculosis, some common themes for the function and adaptation of the molybdoenzymes to pathogen environmental niches are emerging. Firstly, for many of these enzymes, their role is in supporting bacterial energy generation; and the corresponding pathogen fitness and virulence defects appear to arise from a suboptimally poised metabolic network. Secondly, all substrates converted by virulence-relevant bacterial Mo enzymes belong to classes known to be generated in the host either during inflammation or as part of the host signaling network, with some enzyme groups showing adaptation to the increased conversion of such substrates. Lastly, a specific adaptation to bacterial in-host survival is an emerging link between the regulation of molybdoenzyme expression in bacterial pathogens and the presence of immune system-generated reactive oxygen species. The prevalence of molybdoenzymes in key bacterial pathogens including ESKAPE pathogens, paired with the mounting evidence of their central roles in bacterial fitness during infection, suggest that they could be important future drug targets.
Introduction
Molybdenum and tungsten are redox-active transition metals and, when incorporated into enzymes, are able to mediate a variety of reactions including the oxidation of substrates using oxygen from water (Murray et al., 1966; Hille, 2005; Schwarz et al., 2009; Hille et al., 2014; Hagen, 2017). Currently, the so-called mononuclear Mo/W enzymes, which contain a single Mo or W atom complexed by an organic pyranopterin (PPT) cofactor at their active site, are the most abundant group of Mo/W-containing enzymes (Magalon et al., 2011; Iobbi-Nivol and Leimkühler, 2013; Hille et al., 2014; Rothery and Weiner, 2015; Hagen, 2017). Mo/W enzymes are evolutionarily old and likely already existed in the last universal common ancestor (Lebrun et al., 2003; Zhang and Gladyshev, 2008; Schoepp-Cothenet et al., 2012; Nitschke and Russell, 2013; Mayr et al., 2020; Wells et al., 2020). As a result, they are found in all domains of life and are particularly abundant in bacteria, where more than 50 distinct types of enzymes have been identified (Leimkühler and Iobbi-Nivol, 2016; Maia et al., 2017), and physiological functions range from sulfur compound oxidation, degradation of aromatic compounds to supporting anaerobic respiration in bacteria (Hille et al., 2014; Maia et al., 2017).
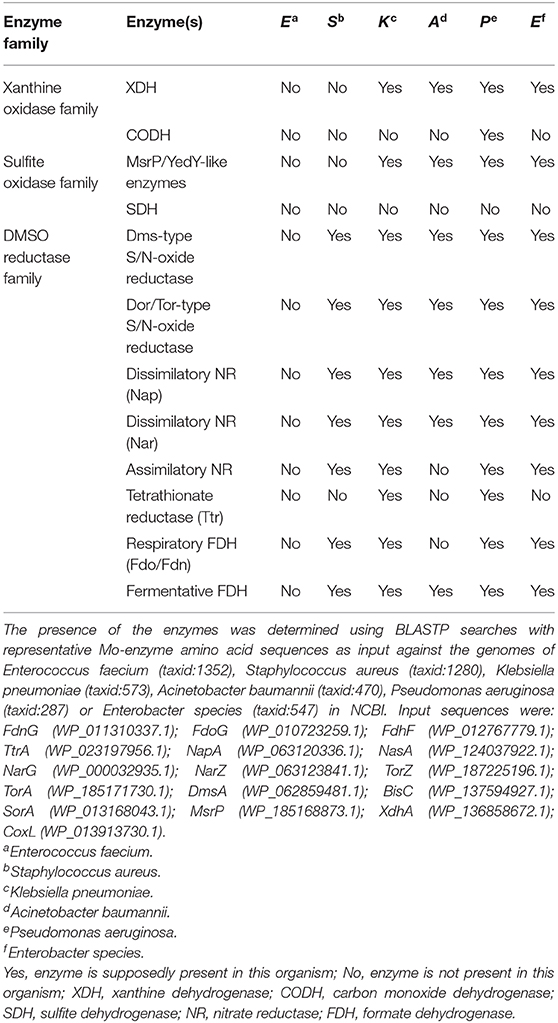
Bacterial Mo/W enzymes are found in each of the three major Mo/W enzyme families, the xanthine oxidase (XO) and sulfite oxidase (SO) families, which also contain enzymes from eukaryotic organisms, and the dimethyl sulfoxide (DMSO) reductase (DMSOR) enzyme family, which exclusively contains prokaryotic enzymes. Very few bacteria lack mononuclear Mo/W enzymes, and this is usually associated with highly adapted lifestyles that are found in Chlamydiae and certain types of Firmicutes (Zhang and Gladyshev, 2008).
While mononuclear Mo/W enzymes have been studied for over five decades, the role of these enzymes in pathogenic bacteria and their influence on bacterial fitness and virulence has only recently become an area of focus. Various studies have linked in-host survival of prevalent pathogenic bacteria such as Mycobacterium tuberculosis (Mtb), Escherichia coli, and Salmonella enterica to the presence of functional molybdoenzymes (Winter et al., 2010, 2013b; Denkel et al., 2013; Williams et al., 2014). Here, we have collated current data on the roles of Mo enzymes as well as enzymes involved in molybdenum uptake and PPT cofactor biosynthesis in bacterial virulence, revealing that Mo enzymes are essential for both body niche specific and systemic infections (Figure 1). Since tungstoenzymes are predominantly found in extremophiles (Andreesen and Makdessi, 2008; Hagen, 2017; Crichton, 2019; Niks and Hille, 2019; Seelmann et al., 2020), the focus of this review will be on molybdenum-containing enzymes.
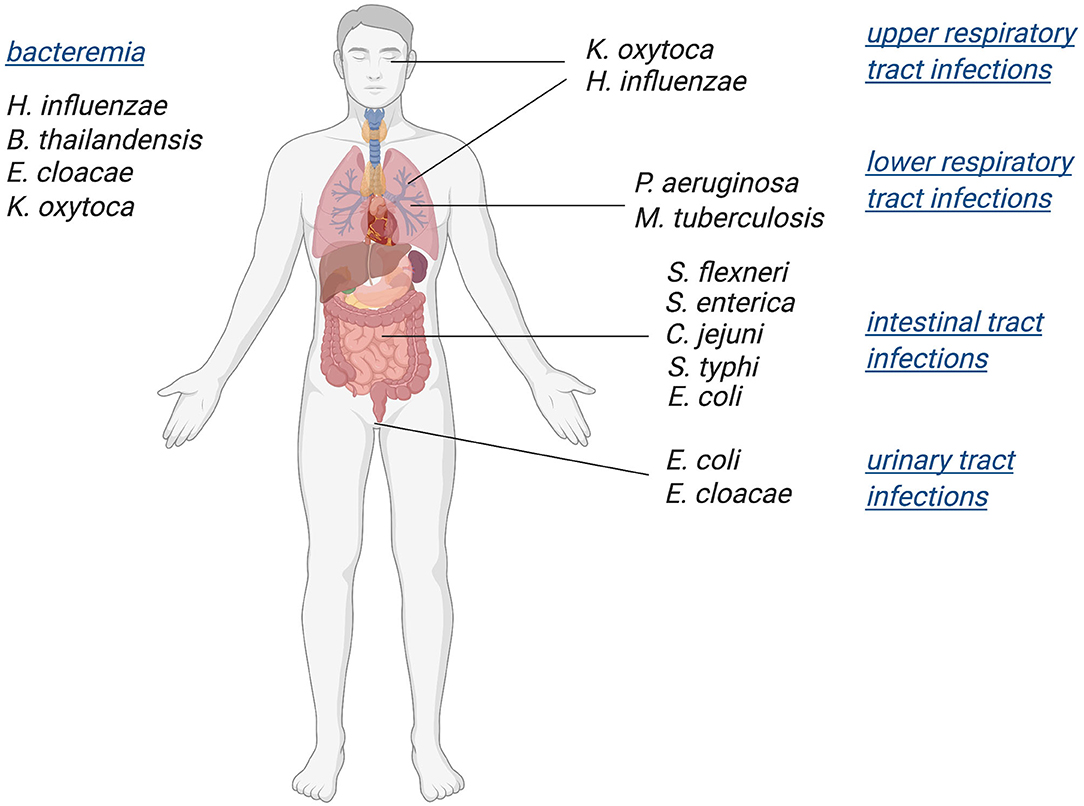
Figure 1. Schematic overview of bacterial pathogens in which Mo-enzymes have been shown to support virulence. Infections with these pathogens affect a variety of body niches, as well as systemic infections.
Molybdenum Uptake, Cofactor Biosynthesis, and Bacterial Virulence
Two processes are required for the synthesis of Mo-PPT cofactors, molybdate uptake via a transport protein and the actual cofactor biosynthesis pathway, and defects in either process have profound impacts on Mo enzyme activity.
Molybdenum Uptake
In most bacteria, the ATP-binding-cassette (ABC)-type molybdate transport system, ModABC, mediates high affinity uptake of molybdate from the environment (Rech et al., 1995; Hu et al., 1997; Imperial et al., 1998; Lawson et al., 1998; Self et al., 2001; Gisin et al., 2010; Perinet et al., 2016; Xia et al., 2018). The substrate-binding protein of this transporter, ModA, is a molybdate-specific, type II periplasmic-binding-protein (PBP2, cl21456) that interacts with the ModB and ModC proteins that form the channel for molybdate import and hydrolyze ATP to provide the energy required for transport, respectively (Hu et al., 1997; Lawson et al., 1998; Self et al., 2001; Santacruz et al., 2006; Hollenstein et al., 2007; Davidson et al., 2008). In the absence of ModABC, non-specific (e.g., sulfate transporters) or lower affinity Mo uptake systems such as the MolBCA transporter found in some strains of Haemophilus influenzae may enable uptake of small amounts of molybdate (Tirado-Lee et al., 2011; Rice et al., 2014). Defects in the Mo uptake system will lower the availability of molybdate for incorporation into Mo enzymes and result in a reduction or even loss of Mo enzyme activities, which can reduce bacterial environmental fitness. High affinity uptake of molybdate is particularly important for pathogenic bacteria, as they may be competing with the host for molybdate. Molybdate concentrations are known to be <1 μg of molybdenum per 1 g of wet organ/ash in many body niches affected by pathogenic bacteria such as the GI tract, lung, bladder, and uterus (Schroeder et al., 1970).
ModABC has been identified as essential for host interactions in Salmonella typhi, Mtb, and Pseudomonas aeruginosa, where strains defective in molybdate uptake were attenuated in different models of infection such as macrophages and mice (Table 1) (Contreras et al., 1997; Camacho et al., 1999; Perinet et al., 2016). A P. aeruginosa PAO1 ΔmodA mutant also showed significantly reduced biofilm formation as a result of an unidentified physiological change (Pederick et al., 2014), however, biofilm phenotypes were not reported for ΔmodA mutants in other bacteria. Loss of high affinity Mo uptake also caused reduced nitrate reductase activity or impaired growth of E. coli, P. aeruginosa, S. typhi, and Campylobacter jejuni on nitrate-containing media (Glaser and DeMoss, 1971; Contreras et al., 1997; Taveirne et al., 2009; Pederick et al., 2014), which likely resulted from the reduced availability of molybdate for incorporation into the molybdenum-dependent nitrate reductase active site.
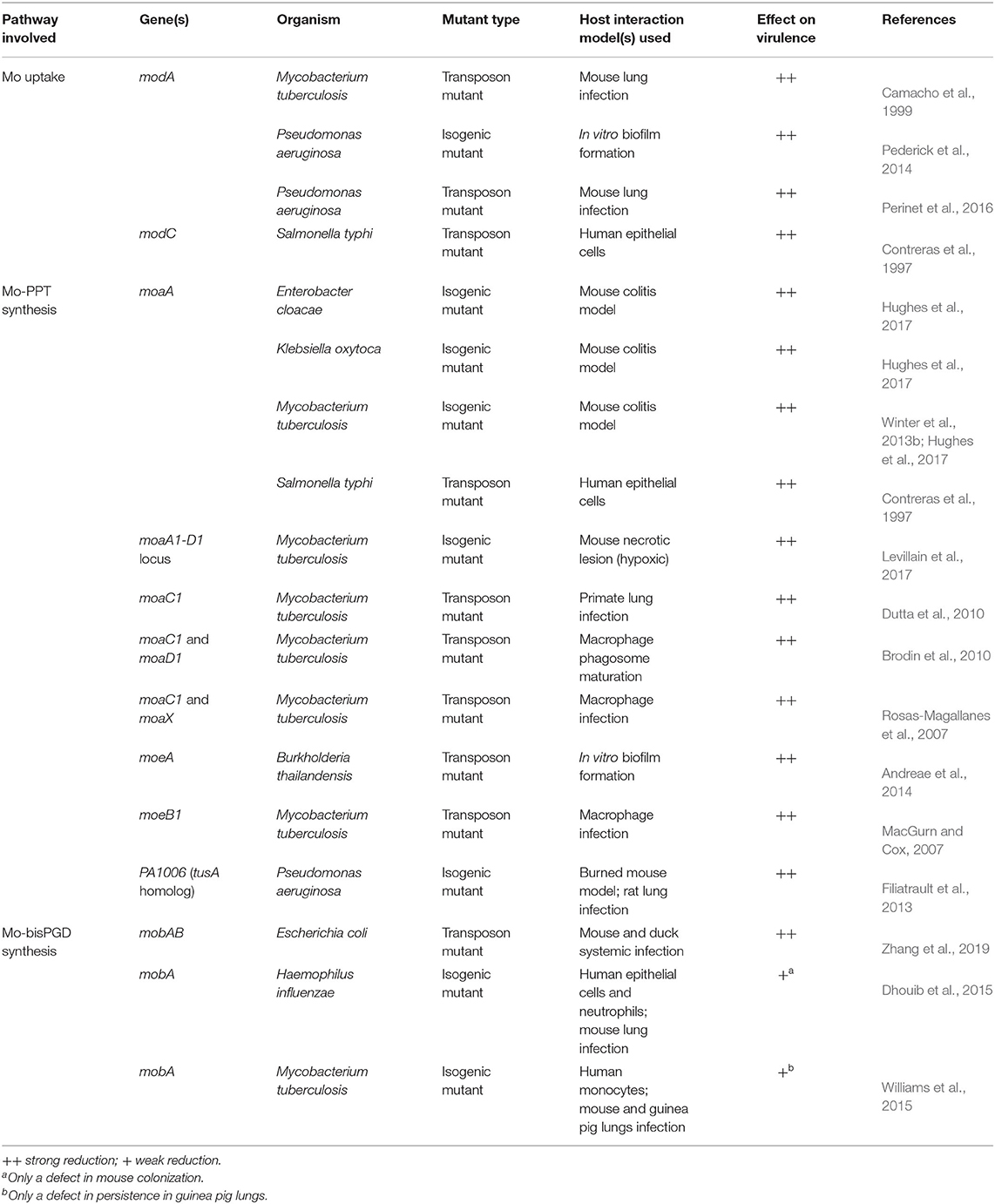
Table 1. Defects in molybdate uptake or Mo-PPT biosynthesis genes and their effects on bacterial virulence.
The significant reduction in bacterial virulence in the absence of effective Mo uptake has led to the molybdate-binding protein ModA being proposed as a promising drug target. Alignments of known ModA amino acid sequences, however, show that there is a relatively high divergence between species (sequence identities: 20–64%), and additionally there is some variability in the molybdate binding residues, which could make it difficult to find drugs that bind effectively to ModA from a variety of bacteria (Hu et al., 1997; Lawson et al., 1998; Santacruz et al., 2006; Hollenstein et al., 2007).
The Molybdenum-Pyranopterin Cofactor and Its Biosynthesis
The tricyclic pyranopterin cofactor (PPT, previously referred to as molybdopterin, MPT) that complexes the Mo atom is required for activity of all mononuclear molybdoenzymes. The Mo-PPT cofactor is synthesized from GTP in three major steps that involve six highly conserved enzymes, and at least two accessory enzyme complexes (Leimkühler et al., 2011; Mendel, 2013; Hille et al., 2014; Leimkühler, 2014; 2020; Mayr et al., 2020) (Figure 2). Depending on the molybdoenzyme family an enzyme belongs to, the basic Mo-PPT may contain different modifications (Magalon et al., 2011; Hille et al., 2014; Leimkühler, 2020).
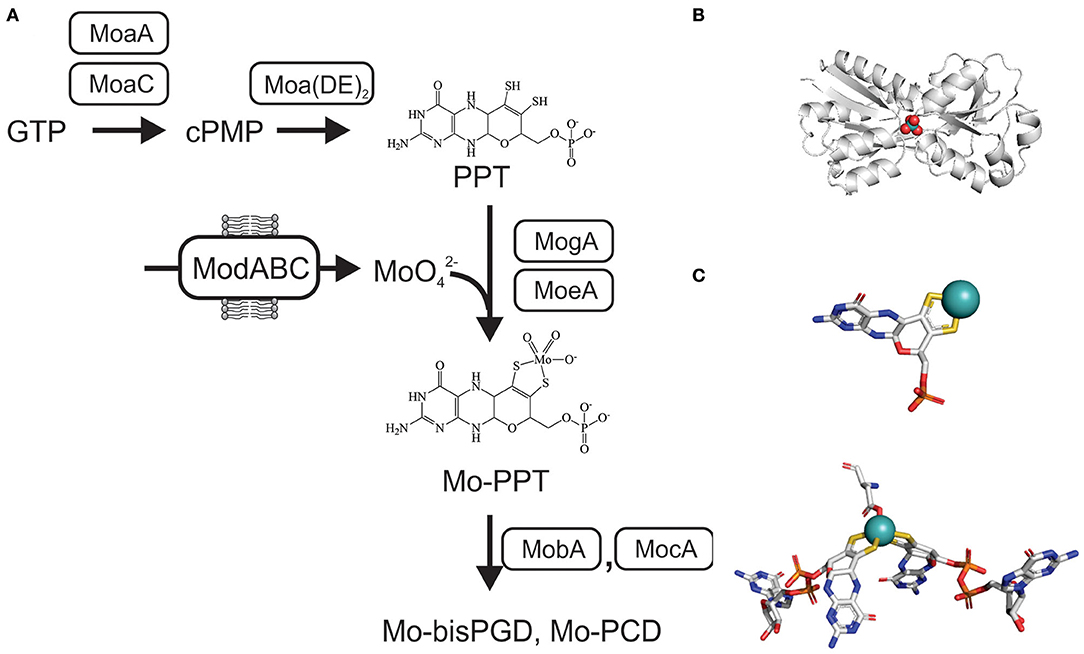
Figure 2. Schematic representation of the biosynthesis of Mo-PPT and Mo-PPT-dinucleotide forms from GTP/CTP in bacteria. cPMP, cyclic pyranopterin monophosphate; PPT, pyranopterin; PGD/PCD, pyranopterin guanine/cytosine dinucleotide; (MoaDE)2, PPT synthase. (A) Overview over molybdate uptake and Mo-PPT biosynthesis (B) Crystal structure of ModA from Escherichia (PDB: 1AMF) with a molybdate ion () bound, bluewhite- protein backbone, teal– molybdenum, red– oxygen. (C) 3D conformations of Mo-PPT and Mo-bisPGD in the enzyme-bound form, Mo-PPT is from MsrP (PDB: 1XDQ), Mo-bisPGD – NarGH (PDB: 1R27). Colors: white – carbon, blue – nitrogen, orange – phosphor, red – oxygen, yellow – sulfur, light teal– molybdenum.
Initially, cyclization of GTP via carbon rearrangement, catalyzed by MoaA and MoaC, leads to the formation of a cyclic pyranopterin monophosphate (cPMP) (Wuebbens et al., 2000; Hanzelmann and Schindelin, 2004; Hover et al., 2013, 2015; Srivastava et al., 2013; Yokoyama and Lilla, 2018; Pang et al., 2020). Sulfuration of cPMP, the second step, is carried out by PPT synthase, a heterotetrametric (α2β2) complex consisting of the MoaD and MoaE proteins, and leads to the formation of a PPT molecule (Pitterle et al., 1993; Daniels et al., 2008) (Figure 2). The PPT synthase reaction is supported by three accessory enzymes, MoeB, IscS, and TusA, which resulfurate the MoaD subunit of PPT synthase after each reaction cycle using cysteine as the sulfur donor (Leimkühler and Rajagopalan, 2001; Leimkühler et al., 2001; Zhang et al., 2010; Dahl et al., 2013; Kozmin et al., 2013; Mayr et al., 2020) (Figure 2).
The third and final step of Mo-PPT biosynthesis is the insertion of the Mo atom. This requires activation of the PPT by adenylation, which is catalyzed by MogA (Liu et al., 2000), and is followed by MoeA-mediated Mo incorporation (Schrag et al., 2001; Xiang et al., 2001; Nichols and Rajagopalan, 2005) (Figure 2). While Mo-PPT is found in most plant and mammalian Mo enzymes, many bacterial Mo enzymes require a modified, dinucleotide version of the Mo-PPT, such as pyranopterin guanine/cytosine dinucleotide (PGD/PCD) and, less frequently, also an adenine dinucleotide (Borner et al., 1991; Rajagopalan and Johnson, 1992; Guse et al., 2003; Neumann et al., 2009b). Formation of these Mo-PPT derivatives is catalyzed by the MocA and MobA proteins, with MobB likely to contribute to the process (Palmer et al., 1994, 1996; Guse et al., 2003; Neumann et al., 2009b; Leimkühler, 2014) (Figure 2). Pathways for the incorporation of the cofactors into individual molybdoenzymes follow enzyme-specific pathways that often require maturation chaperones (Leimkühler and Klipp, 1999; Oresnik et al., 2001; Turner et al., 2004; Iobbi-Nivol and Leimkühler, 2013; Cherak and Turner, 2017). Further information about Mo-PPT biosynthesis can be found in several excellent review papers (Leimkühler et al., 2011; Magalon et al., 2011; Iobbi-Nivol and Leimkühler, 2013; Mendel, 2013; Hille et al., 2014; Leimkühler and Iobbi-Nivol, 2016; Leimkühler, 2020; Mayr et al., 2020).
Mo-PPT biosynthesis enzymes have been shown to be crucial for virulence in several pathogenic bacteria, including E. coli and Mtb (Table 1) (MacGurn and Cox, 2007; Rosas-Magallanes et al., 2007; Brodin et al., 2010; Dutta et al., 2010; Winter et al., 2013b; Williams et al., 2014, 2015; Hughes et al., 2017; Levillain et al., 2017). The essential role of many of these Mo-PPT synthesis defects in bacterial pathogens was not identified by targeted mutagenesis, but instead during global transposon mutagenesis aimed at identifying genes essential for host-pathogen interactions (Contreras et al., 1997; Camacho et al., 1999; MacGurn and Cox, 2007; Rosas-Magallanes et al., 2007; Brodin et al., 2010; Dutta et al., 2010; Perinet et al., 2016; Zhang et al., 2019). As a result, mechanistic insights into the physiological effects of these defects are often unavailable.
As a general rule, mutations in the Mo-PPT synthesis pathway completely abolish cofactor formation, leading to a loss of activity for all Mo enzymes present and creating a pleiotropic phenotype. This is clearly demonstrated by studies in Mtb and pathogens of the family Enterobacteriaceae, where regardless of which Mo-PPT-biosynthetic enzyme was lost, the mutant strains displayed significantly reduced fitness and/or virulence in different host-pathogen interaction models (Table 1) (Contreras et al., 1997; Rosas-Magallanes et al., 2007; Brodin et al., 2010; Dutta et al., 2010; Winter et al., 2013b; Hughes et al., 2017; Levillain et al., 2017).
Virulence attenuation was also associated with defects in accessory proteins of Mo-PPT synthesis, such as the PPT synthase resulfuration complex in Mtb and P. aeruginosa (Table 1) (MacGurn and Cox, 2007; Filiatrault et al., 2013). A P. aeruginosa strain lacking the TusA-related PA1006 protein, which is part of the resulfurase complex (Kozmin et al., 2013), exhibited an ~10-fold reduction in cytoplasmic and membrane fraction molybdate concentrations, as well as impaired biofilm formation and maturation in vitro (Filiatrault et al., 2013; Tombline et al., 2013). PA1006 was also shown to interact with almost all Mo-PPT biosynthesis proteins (Tombline et al., 2013), providing a possible reason for the observed defects and introducing a new layer of complexity in the Mo-PPT cofactor biosynthesis pathway.
By contrast, mutations in enzymes catalyzing the formation of Mo-PPT dinucleotide forms have less striking effect on bacterial virulence (Table 1), possibly because they only affect enzymes requiring specific types of PPT cofactors. Mutations in mobA, which encodes the enzyme responsible for Mo-bisPGD formation (Lake et al., 2000), decreased the activities of all DMSOR family molybdoenzymes in P. aeruginosa, Mtb, and H. influenzae (Noriega et al., 2005; Tombline et al., 2013; Dhouib et al., 2015; Williams et al., 2015), but only resulted in a mild reduction of bacterial fitness in the Mtb and H. influenzae mutants in vivo (Dhouib et al., 2015; Williams et al., 2015). However, an E. coli carrying mutations in both mobA and mobB genes [the product of latter was proposed to have a non-essential role in Mo-PPT cofactor synthesis (Iobbi-Nivol et al., 1995; Leimkühler, 2014, 2020)], showed clearly reduced virulence in both mouse and duck systemic infection models (Zhang et al., 2019). This suggests that further investigations are needed to elucidate role of MobA-MobB complexes in the maturation of Mo enzymes from different enzyme families.
Xanthine Oxidase Family Enzymes and Their Roles in Bacterial Pathogens
XO family enzymes typically catalyze conversions of nucleobases and aldehydes, with typical representatives being xanthine oxidoreductases (XORs, include both XOs and xanthine dehydrogenases, XDHs), aldehyde oxidases (AOX), and carbon monoxide dehydrogenases (CODHs) (Hille et al., 2014; Nishino et al., 2017). The majority of enzymes found in this family are cytoplasmic proteins, and only some, such as particular bacterial aldehyde oxidases (Neumann et al., 2009a), are located in the bacterial periplasm (Figure 3).
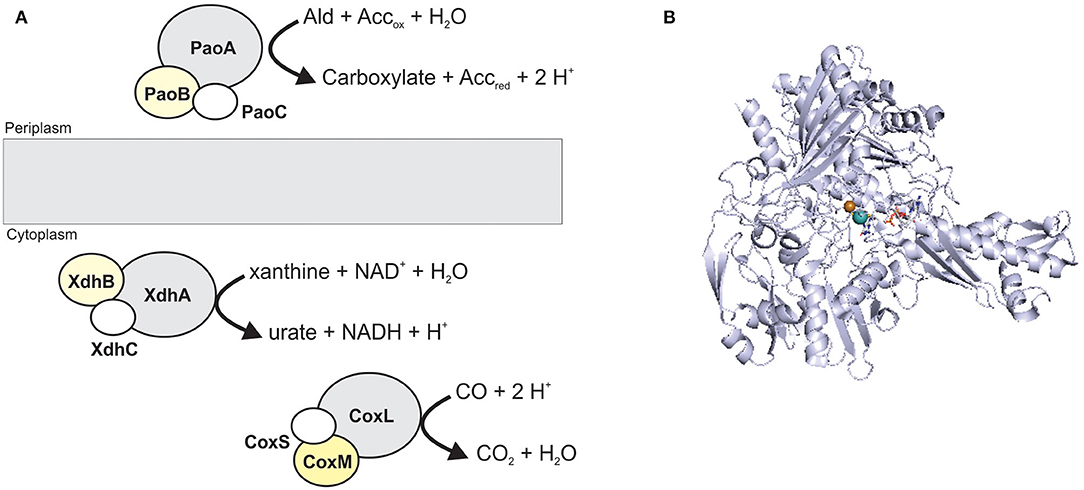
Figure 3. The structure and location of typical XO family enzymes. (A) schematic representation of enzyme structure. Enzymes of this family most commonly have a heterotrimeric (αβγ) structure containing a Mo-binding catalytic subunit (gray); an iron-sulfur protein (white); a FAD-binding subunit (yellow). CoxMSL is a carbon monoxide dehydrogenase, XdhABC is a xanthine dehydrogenase and PaoABC is an aldehyde oxidoreductase. (B) Crystal structure of CoxL, the catalytic subunit of the carbon monoxide dehydrogenase from Oligotropha carboxidovorans (PDB: 1N63) with the CuMo-PCD cofactor shown in stick form. Colors: white – carbon, blue – nitrogen, orange – phosphor, red – oxygen, yellow – sulfur, light teal– molybdenum, brown– copper, protein backbone - bluewhite.
AOXs oxidize numerous aromatic and non-aromatic aldehydes to their respective carboxylic acids, and are responsible for metabolizing various xenobiotics and drugs in mammals (Romão et al., 2017; Terao et al., 2020), while the bacterial aldehyde oxidoreductases are likely involved in metabolizing aromatic aldehydes for detoxification (Correia et al., 2016).
XO family enzymes typically contain not only a molybdenum redox center, but also at least one iron-sulfur cluster, while flavin adenine dinucleotide (FAD) is not always present (Bray et al., 1996; Nishino et al., 2017). In eukaryotic as well as some bacterial enzymes, two or more cofactor binding domains may be fused to form a single polypeptide, and enzymes with between one and three subunits have been identified (Figure 3) (Gremer and Meyer, 1996; Leimkühler et al., 1998; Enroth et al., 2000; Neumann et al., 2009a; Coelho et al., 2012; Otrelo-Cardoso et al., 2014). The active site of XO enzymes is typically pentacoordinated, with two Mo-PPT dithiolene sulfurs, an oxo-, a hydroxo/hydro-, and a sulfido-ligand coordinating the Mo atom. The sulfido-group can be replaced by selenium or another oxo-group in some XO enzymes, and interestingly, there is no amino acid ligand to the Mo center (Boyington et al., 1997; Schräder et al., 1999; Hille et al., 2014; Nishino et al., 2017). All mammalian enzymes of this family contain Mo-PPT, while Mo-PCD is present in most bacterial enzymes (Gremer and Meyer, 1996; Leimkühler et al., 1998; Magalon et al., 2011; Hille et al., 2014; Leimkühler, 2020; Terao et al., 2020).
A large number of studies have reported properties of XO family enzymes in bacteria, where they support the use of CO or purines as carbon, and in the latter case, also nitrogen sources as well as the detoxification of aldehydes (Leimkühler et al., 1998; Santiago et al., 1999; Xi et al., 2000; Park et al., 2003; Neumann et al., 2009a), however, only limited insights are available on their role in bacterial pathogens. The reactions catalyzed by XO family enzymes are relevant during infection as potential substrate molecules occur in the host, and XO family enzymes are found in human pathogens, including several opportunistic pathogens and species on the WHO top priority pathogen list such as Acinetobacter baumannii, Mtb, and Enterobacteriaceae family species (World Health Organization, 2017).
Xanthine Dehydrogenases
XORs catalyze the conversion of the purine base hypoxanthine to xanthine, and xanthine to uric acid, which is part of purine catabolism (Schwarz and Belaidi, 2013; Wang et al., 2016; Nishino et al., 2017). Human XOR deficiency leads to xanthinuria, which is characterized by elevated blood xanthine levels, and, if untreated, can eventually result in critical conditions such as urinary tract calculi or renal failure (Schwarz and Belaidi, 2013; Battelli et al., 2016). Human XOR is highly expressed in the liver and small intestine (Sarnesto et al., 1996), implying that xanthine is present in these body niches. In bacteria, XDHs are often required for growth on purine compounds as nitrogen and/or possibly also carbon sources, again indicating a role in purine catabolism (Christiansen et al., 1997; Xi et al., 2000).
XDH occurs in bacteria present in the human gut microbiome, and there is evidence that XDH may contribute to the overall fitness of these bacteria by mediating access to (hypo-)xanthine as an additional nitrogen or carbon source (Christiansen et al., 1997; Xi et al., 2000). B. subtilis growth with xanthine or hypoxanthine as the sole nitrogen source was XDH-dependent and could be inhibited by allopurinol (Christiansen et al., 1997). In contrast, growth of E. coli on a (hypo-)xanthine-supplemented medium required an additional nitrogen source such as aspartate, suggesting that purines might be used as a carbon source in E. coli (Xi et al., 2000). However, to the best of our knowledge, there is so far no direct evidence for XDHs or AOXs having a role in bacterial virulence.
Carbon Monoxide (CO) Dehydrogenases
Carbon monoxide dehydrogenases (CODH, CoxMSL) (Figure 3) catalyze the oxidation of CO to CO2 and enable growth with CO as the sole energy and carbon source in carboxydobacteria (Santiago et al., 1999; King and Weber, 2007; Robb and Techtmann, 2018). CODHs contain a special Mo-Cu binuclear active site (Dobbek et al., 2002; Gnida et al., 2003) and have been found in several pathogenic bacteria including P. aeruginosa, Burkholderia spp., and Mycobacterium spp. (King and Weber, 2007).
In the human body, CO is produced by the action of heme oxygenases (Hmox1) and can act as a signaling molecule in general physiological responses such as blood pressure adjustment, and in immune cells such as macrophages, where it activates the NALP3 inflammasome for bacterial clearance (Chung et al., 2008; Chin and Otterbein, 2009; Wegiel et al., 2014). CO also acts directly as an antimicrobial, attributable to its ability to bind metalloproteins and alter their biological functions (Nobre et al., 2007; Zacharia and Shiloh, 2012).
Resistance of Mtb to CO produced by macrophages has been attributed to the presence of the CutBCA CODH, an enzyme that is also present in many other Mycobacterium spp. (Shiloh et al., 2008; Song et al., 2010; Zacharia and Shiloh, 2012). CO triggers expression of the Mtb dormancy regulon (Kumar et al., 2008; Shiloh et al., 2008), and high levels (20.5 U/mg) of CODH activity were present in Mtb cultures following in vitro growth in the presence of CO (Park et al., 2007). CutBCA not only allows Mtb to use CO as a carbon source, but also possesses nitric oxide dehydrogenase activity (Park et al., 2007), which is thought to be unique to mycobacterial CODHs as these enzymes are phylogenetically distinct from other bacterial MoCu-CODHs (Song et al., 2010). In keeping with this, purified mycobacterial CODH was able to protect E. coli from nitrosative stress (Park et al., 2007), and reduced transcription of the cut operon in Mycobacterium sp. strain JC1 increased susceptibility to NO stress (Lee et al., 2017). However, direct evidence for an in vivo role of the Mtb CODH in host-association is still needed.
Sulfite Oxidase Family Enzymes in Pathogens
Sulfite oxidase (SO) family enzymes are found in all domains of life and, in addition to prokaryotic and eukaryotic sulfite-oxidizing enzymes (SOEs), this enzyme family also includes plant assimilatory nitrate reductases (aNRs), as well as bacterial sulfur dehydrogenases, sulfoxide reductases and a group of uncharacterized, single-subunit, cytoplasmic enzymes (Kappler, 2008; Workun et al., 2008; Kappler and Enemark, 2015). A new class of human Mo enzymes, the mitochondrial amidoxime reducing components (mARCs), have also been tentatively assigned to this enzyme family in preliminary investigations (Wahl et al., 2010; Llamas et al., 2017; Kubitza et al., 2018).
SO family enzymes typically contain a Mo-PPT cofactor bound to a protein with a characteristic sulfite oxidase (SUOX)-fold (Figure 4). The Mo atom is complexed by the two dithiolene sulfurs of the PPT cofactor as well as a cysteine residue and two oxo ligands, one of which is active in catalysis (Workun et al., 2008; Kappler and Enemark, 2015). Unlike the XO family enzymes that contain Fe/S clusters and, frequently, also FAD as redox cofactors, SO family enzymes often contain only a Mo center, or a combination of the Mo redox center with one or more heme groups, while FAD is only present in plant aNRs (Crawford et al., 1988; Kappler and Schwarz, 2017).
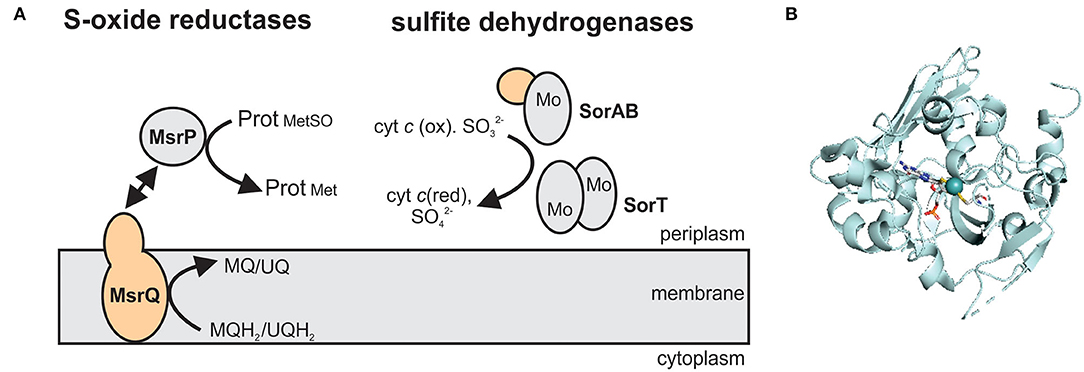
Figure 4. The structure and subcellular location of SO family enzymes associated with bacterial virulence. (A) Overview of enzyme structures. SorAB and SorT are two types of sulfite dehydrogenases that oxidize sulfite to sulfate, while the MsrP protein reduces protein-bound methionine sulfoxides with electrons transferred from the membrane integral cytochrome MsrQ. Gray – Mo-binding subunit; orange – heme-binding subunits. MQ – menaquinone; MQH2 - menaquinol; UQ – ubiquinone; UQH2 – ubiquinol. (B) Crystal structure of the methionine sulfoxide reductase MsrP from Escherichia coli (PDB: 1XDQ) with the Mo-PPT cofactor shown in stick form. Colors: white – carbon, blue – nitrogen, orange – phosphor, red – oxygen, yellow – sulfur, light teal – molybdenum, protein backbone – pale-cyan.
At least three enzyme subgroups that differ in the structure and size of the catalytic Mo subunit and the amino acid residues surrounding the active site have been identified in this enzyme family (Figure 4) (Kappler, 2008), namely (1) YedY and YedY-like enzymes that are prevalent in bacterial pathogens, (2) the main SOE group that also contains plant aNRs and bacterial sulfur dehydrogenases, and (3) uncharacterized, single subunit enzymes from environmental bacteria and archaea (Figure 4). For further information on the structural and catalytic properties of this family of enzymes, the reader is referred to recent review articles (Kappler, 2008, 2011; Hille et al., 2014; Kappler and Enemark, 2015; Kappler and Schwarz, 2017).
YedY and YedY-Like Enzymes
Genes encoding the group 1 SO family enzymes are predominantly found in bacterial pathogens and form two subgroups, the YedY (group 1A) and YedY-like (group 1B) enzymes (Kappler, 2008). The two subgroups differ in the size of the Mo-binding domain (1A: 35 kDa, 1B: 30 kDa) and the cytochrome they interact with (Kappler, 2008). The membrane-bound cytochrome subunits these enzymes interact with provide electrons from the quinone pool to the catalytic subunit of the enzyme (Loschi et al., 2004; Brokx et al., 2005) (Figure 4).
Group 1 SO family enzymes likely all act as reductases: the currently best characterized enzyme of this group is the E. coli MsrP (formerly YedY) methionine sulfoxide reductase that belongs to group 1A which also contains sequences from Yersinia spp. and C. jejuni (Kappler, 2008). MsrP/YedY was initially characterized as a weak DMSO reductase, before further investigations revealed that this enzyme is a methionine sulfoxide reductase, able to repair both free methionine sulfoxide (MetSO) and MetSO residues in oxidatively damaged periplasmic proteins (Loschi et al., 2004; Gennaris et al., 2015). Two E. coli cell envelope proteins, SurA, an outer membrane protein chaperone, and Pal, a lipoprotein that mediates outer membrane integrity, have been identified as physiological substrates of MsrP (Gennaris et al., 2015). At present, MsrP is the only Mo-containing sulfoxide reductase identified outside of the DMSOR enzyme family, to which these enzymes normally belong (Gennaris et al., 2015). Interestingly, expression of the msrPQ genes is induced by hypochlorite, an antimicrobial compound produced by neutrophils during infection, and regulated by the YedY/W two-component system (Gennaris et al., 2015), which clearly links this enzyme to host-associated environmental conditions; however, infection studies confirming this have not been reported so far.
While an E. coli ΔmsrP strain has not been tested in infection models, an MsrP-related protein from C. jejuni encoded by the cj0379c gene (NCBI Gene ID: 904702) was shown to be required for successful colonization of the chicken gut (Hitchcock et al., 2010). Chickens are the natural host of C. jejuni, a bacterium that causes gastroenteritis in humans when ingested.
The Cj0379 protein is also a group 1A enzyme and, like E. coli MsrP, displays poor S-oxide reductase activity with standard substrates (Hitchcock et al., 2010). However, a Δcj0379c strain had a higher sensitivity to nitrosative stress but not superoxides, and a slower NO degradation rate in vitro (Hitchcock et al., 2010), suggesting that Cj0379 has a role in nitrosative stress resistance in C. jejuni. To fully understand the role of this enzyme in vivo, a further characterization of its catalytic properties would contribute to identifying the mechanism by which this enzyme supports C. jejuni virulence.
Sulfite Dehydrogenases and Related Enzymes
Although better known as an antimicrobial preservative in food and beverages, sulfite is also present in the human body as a byproduct of amino-acid degradation that takes place mostly in the liver, while in serum, sulfite exists at micromolar levels in a protein-bound form that is less toxic (Ji et al., 1995; Lester, 1995; Mitsuhashi et al., 2005). Elevated serum sulfite levels have been observed in an LPS-stimulated rat model (Mitsuhashi et al., 2005) and also in patients with pneumonia or chronic renal failure (Kajiyama et al., 2000; Mitsuhashi et al., 2004), suggesting a pro-inflammatory activity of this molecule. Sulfite can also act as a signaling molecule in human cells, where it has been shown to promote proliferation of lymphocytes, as well as upregulation of reactive oxygen species (ROS) production in neutrophils, either directly or indirectly, via activation of alveolar macrophages to produce factors that stimulate neutrophils (Pettit et al., 1991; Beck-Speier et al., 1993; Mitsuhashi et al., 1998, 2005).
Interestingly, in response to LPS treatment, human neutrophils can also produce sulfite, at estimated local concentrations of 5–6 μM, likely to trigger an inflammatory response (Mitsuhashi et al., 2005). However, these physiological concentrations are unlikely to have significant bactericidal effects, as 0.96 mM sulfite was required to inhibit growth of Helicobacter pylori in vitro (Hawrylik et al., 1994), and at least 2 mM sulfite was needed to inhibit growth of Lactobacillus spp. and Streptococcus spp. (Irwin et al., 2017).
Despite the relatively low physiological concentrations of sulfite in at least the human host, a C. jejuni enzyme homologous to characterized SOEs (Figure 4) was shown to be required for successful infection of the Caco-2 human intestinal cell line. The C. jejuni sulfite dehydrogenase (SDH) has two components, the catalytic protein Cj0005, and a c-type cytochrome, Cj0004 (Myers and Kelly, 2005; Tareen et al., 2011). A Δcj0004c strain showed reduced respiration on sulfite, while a Δcj0005c strain exhibited a growth defect on sulfite-containing medium (Myers and Kelly, 2005; Tareen et al., 2011), indicating that these proteins are genuinely involved in C. jejuni sulfite oxidation. The Δcj0005c mutant also showed decreased motility, as well as down-regulation of flagellar biosynthesis- and autoagglutination-associated genes that are important for virulence (Tareen et al., 2011), but the mechanisms underlying these additional physiological effects are unknown. At present, this appears to be the only direct evidence that SOEs can support host-cell colonization.
DMSO Reductase Family Enzymes in Pathogens
Unlike the XO and SO family enzymes, enzymes of the DMSOR family occur uniquely in prokaryotes. They are functionally versatile and able to convert a diverse range of sulfur and nitrogen compounds such as DMSO, tetrathionate, trimethylamine N-oxide (TMAO), or nitrate, making many of them directly relevant to global sulfur and nitrogen cycles (McDevitt et al., 2002; Kappler and Schäfer, 2014; Leimkühler and Iobbi-Nivol, 2016). Additionally, enzymes of this family contribute to the detoxification of arsenic and selenium compounds, and degradation of aromatic compounds such as ethylbenzene (Hille et al., 2014).
DMSOR family enzymes very commonly have a heterotrimeric structure, consisting of a Mo-containing catalytic subunit, an iron-sulfur protein that transfers electrons to the catalytic subunit, and a membrane anchor subunit that connects the other redox centers to the cellular quinone pool (Figure 5) (Rothery et al., 2008; Grimaldi et al., 2013; Magalon et al., 2017). These heterotrimeric complexes can face either the bacterial cytoplasm or the periplasm (Grimaldi et al., 2013; Magalon et al., 2017). In some cases, two-subunit enzymes are also found and typically consist of a Mo-binding catalytic protein, or a multi-subunit catalytic protein complex (e.g., NapAB nitrate reductase), that interacts transiently with a membrane-bound, electron transfer subunit (Figure 5) (Arnoux et al., 2003; Grimaldi et al., 2013). All DMSOR family enzymes contain a Mo-bisPGD cofactor (Figure 5), and the four dithiolene sulfur atoms of the two cofactor molecules coordinate the Mo atom, together with an oxo- or a sulfido-group and an amino acid ligand (Johnson et al., 1990; Leimkühler et al., 2011; Leimkühler and Iobbi-Nivol, 2016; Kaufmann et al., 2018; Leimkühler, 2020). Different amino acid ligands including serine, cysteine, selenocysteine, and aspartate have been identified in DMSOR enzymes and their distribution largely coincides with three major phylogenetic groups identified in various studies: type I enzymes (e.g., NapAB nitrate reductase and FdnGHI formate dehydrogenase) contain a cysteine or selenocysteine ligand; type II enzymes, such as the heterotrimeric NarGHI nitrate reductase, the DmsABC DMSO reductase and the EbdABC ethylbenzene dehydrogenase, have an aspartate amino acid ligand; and the type III Dor/Tor-type S- and N-oxide reductases typically have a serine amino acid ligand (McDevitt et al., 2002; Grimaldi et al., 2013; Kappler and Schäfer, 2014; Wells et al., 2020). The AioAB arsenite oxidase is an outlier in this classification system, because of the absence of an amino acid ligand to the Mo; however, the AioAB amino acid sequence is most closely related to type I enzymes (McEwan et al., 2002; Warelow et al., 2013). Crystal structures as well as functional and spectroscopic properties are available for many of these enzymes and have been reviewed previously (Rothery et al., 2008; Magalon et al., 2011, 2017; Gonzalez et al., 2013; Grimaldi et al., 2013; Hille et al., 2014; Wells et al., 2020).
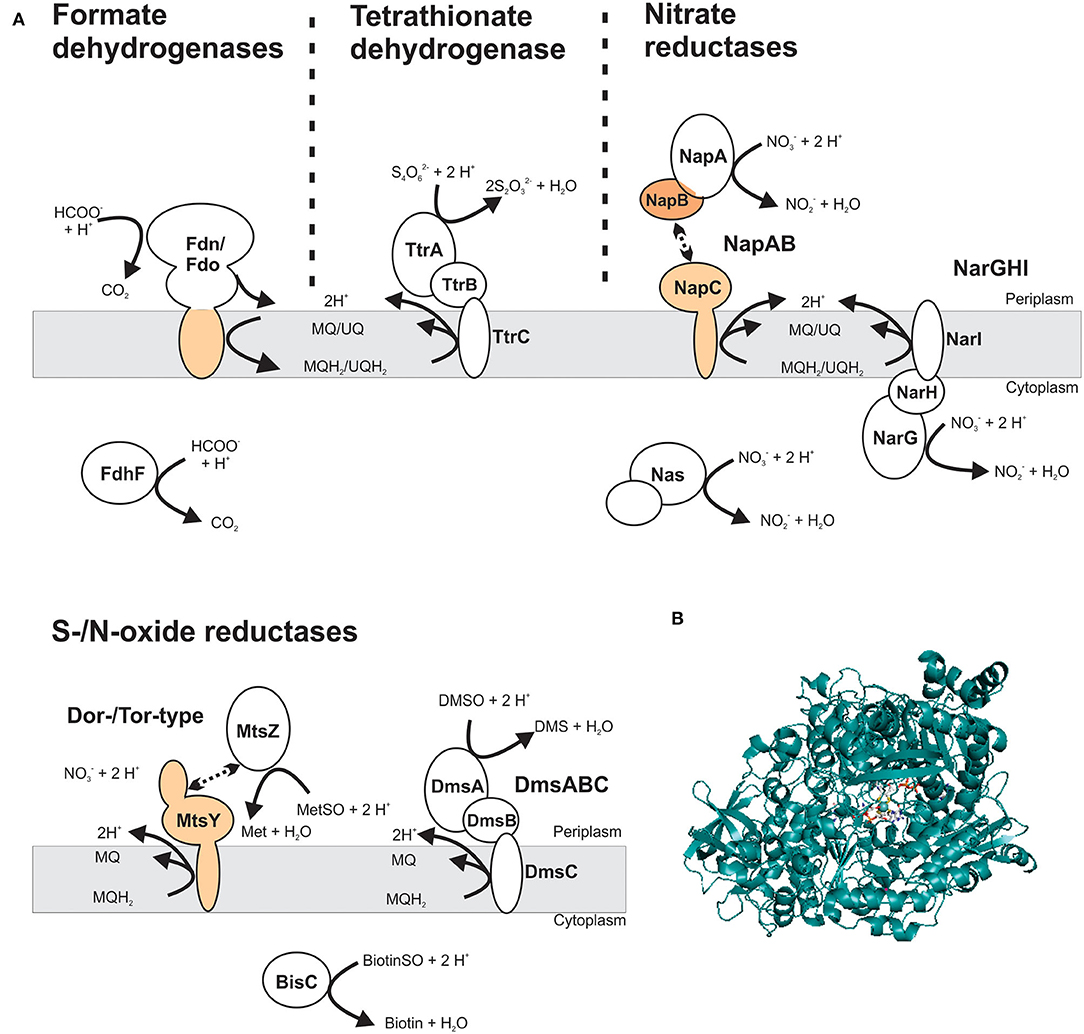
Figure 5. Structure and subcellular location of DMSOR family enzyme that support virulence in different bacterial pathogens. (A) Schematic representation of enzyme structures, showing both membrane-bound and soluble Mo-enzyme complexes. The most common structure for DMSOR family enzymes is heterotrimeric (αβγ), consisting of a Mo-containing subunit; an iron-sulfur protein and a membrane-integral subunit that may or may not bind hemes. Additionally, some enzymes that consist of a soluble Mo-binding subunit or a protein complex that includes the catalytic subunit that interacts transiently with a heme-binding membrane-integral cytochrome. Gray – Mo-binding subunit; orange – heme-binding subunits. MQ – menaquinone; MQH2 – menaquinol; UQ – ubiquinone; UQH2 – ubiquinol. (B) Crystal structure of NarG, the catalytic subunit of the respiratory nitrate reductase from Escherichia coli (PDB: 1R27) with the Mo-bisPGD cofactor shown in stick form. Colors: white – carbon, blue – nitrogen, orange – phosphor, red – oxygen, yellow – sulfur, light teal– molybdenum, protein backbone – teal.
Many DMSOR family enzymes play an essential role in the bacterial respiratory chain as alternative terminal reductases that support anaerobic energy generation, or as dehydrogenases that donate electrons to the respiratory chain (Unden et al., 2014). Anaerobic respiration provides a fitness advantage to bacteria in low oxygen environments, including during interactions with a host, where oxygen depletion is common during host tissue inflammation (Winter et al., 2013a,b; Hughes et al., 2017).
S- and/or N-oxide Reductases
S- and/or N-oxide reductases are a common type of DMSOR enzymes and occur in the structurally distinct type II and type III enzyme groups (Grimaldi et al., 2013). Both types of reductases have been implicated in supporting bacterial virulence (Baltes et al., 2003; Denkel et al., 2013; Dhouib et al., 2016). The heterotrimeric, membrane-bound DmsABC S/N-oxide reductase is a type II enzyme, and evidence for its involvement in bacterial virulence is currently mixed. DmsABC has been shown to be required for virulence in the pig pathogen Actinobacillus pleuropneumoniae (Baltes et al., 2003), but no mechanistic insights were provided beyond evidence of reduced survival of a ΔdmsA strain in a model of porcine pleuropneumonia. In contrast, in E. coli, DmsABC did not appear to be essential for colonization of the mouse gut (Jones et al., 2011; Hughes et al., 2017).
By contrast, for type III S-/N-oxide reductases several studies have detailed a role in bacterial fitness and virulence (Denkel et al., 2013; Dhouib et al., 2016). Phylogenetically, the Dor/Tor enzymes form at least two groups (Figure 6) (Dhouib et al., 2016): the first group comprises the DorA DMSORs (Mouncey et al., 1997; Shaw et al., 1999) and the TorA TMAO reductases (Mejean et al., 1994; Dos Santos et al., 1998), both of which enable anaerobic respiration with their respective substrates; representatives of the other lineage are the BisC biotin sulfoxide reductase (Ezraty et al., 2005) and TorZ S/N-oxide reductase (Gon et al., 2000) from E. coli and the MtsZ methionine sulfoxide reductase from H. influenzae (Dhouib et al., 2016) (Figures 5, 6). The proposed substrates of BisC and MtsZ, biotin sulfoxide (BSO) and MetSO, can form following exposure of biotin or methionine to reactive oxygen species (ROS) produced by immune cells such as neutrophils and macrophages at sites of infections (Pattison and Davies, 2001; Ray et al., 2012; Kim et al., 2014). The ability to reverse sulfoxidation of amino acids and vitamins is important for bacterial survival, as the oxidative damage renders these molecules biologically inactive (Levine et al., 2000; Zempleni et al., 2009).
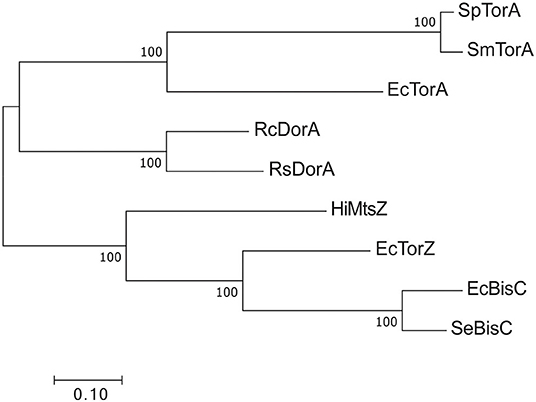
Figure 6. Phylogenetic relationships of Dor/Tor-type S- and N-oxide reductases of the DMSOR family. Sequences were aligned using MUSCLE in MEGA7 (Kumar et al., 2016) and phylogenetic relationships determined using the maximum-likelihood method. Robustness testing used bootstrapping (500 cycles), bootstrap values above 80 are shown near relevant nodes. All sequences were downloaded from the NCBI protein database. Grouping was well-supported by the bootstrap values. Sp TorA, Shewanella putrefaciens (WP_086903686.1); SmTorA, Shewanella massilia (WP_115334087.1); RcDorA, Rhodobacter capsulatus (WP_013068552.1); RsDorA, Rhodobacter sphaeroides (WP_011338998.1); HiMtsZ, Haemophilus influenzae (WP_0 108 69026.1); Se BisC, Salmonella enterica (WP_183053592.1); EcTorA, Escherichia coli (WP_001063164.1); EcTorZ (WP_000176781.1); EcBisC (WP_000013974.1).
Only one enzyme of the DorA/TorA lineage, the TorA TMAO reductase from E. coli, has been studied for its relevance during infection, and it was dispensable for mouse intestinal colonization (Jones et al., 2011). In contrast, enzymes of the second lineage within the Dor/Tor enzyme group, such as the BisC biotin sulfoxide reductase from S. enterica serovar Typhimurium and the MtsZ methionine sulfoxide reductase from H. influenzae, have been shown to be important for bacterial virulence (Denkel et al., 2013; Dhouib et al., 2016).
BisC was originally described as a biotin sulfoxide reductase (del Campillo-Campbell and Campbell, 1982), but later studies in both S. Typhimurium and E. coli showed that this enzyme also has MetSO reductase activity and is essential for MetSO- and BSO-dependent growth in methionine or biotin auxotrophic strains (Ezraty et al., 2005; Denkel et al., 2013). In S. Typhimurium, inactivation of bisC resulted in growth defects in the presence of H2O2 and attenuation of virulence in macrophages and during intraperitoneal infections in mice (Denkel et al., 2013). Interestingly, the increased in vitro sensitivity to H2O2 of the S. Typhimurium ΔbisC strain was linked to the MetSO reductase activity of BisC, while attenuation of virulence in the infection models was caused by the loss of BSO reductase activity. The MetSO reductase activity of BisC was not essential for this function (Denkel et al., 2013). This may be due to the important role of biotin as a cofactor for enzymes with essential cellular functions such as acetyl-CoA carboxylase that plays a key role in fatty acid biosynthesis and is a known antimicrobial target (Freiberg et al., 2004). BisC is an unusual DMSOR family enzyme, as it is not encoded in an operon with electron-donating proteins, and the physiological electron donor for BisC is still unknown (Ezraty et al., 2005), but will likely provide further insights into mechanistic aspects and physiological functions of this enzyme.
While the MetSO reductase activity of BisC is dispensable for virulence in S. Typhimurium, a methionine sulfoxide reductase, the periplasmic MtsZ enzyme from the respiratory pathogen H. influenzae, was shown to be important for bacterial virulence (Dhouib et al., 2016). Within the BisC/TorZ/MtsZ lineage, MtsZ and related enzymes form a distinct phylogenetic clade (Dhouib et al., 2016), suggesting that MtsZ may have a different physiological role to the E. coli TorZ or BisC. In keeping with this, kinetic data showed that the KM of MtsZ for BSO was in the millimolar range, making this activity unlikely to have physiological relevance, while that for MetSO was in the micromolar range (Dhouib et al., 2016). Like BisC, MtsZ only reduced free MetSO and was unable to repair protein-bound MetSO, but an obvious difference between the two enzymes is that MtsZ is linked to the H. influenzae respiratory chain, connecting it to redox balancing and energy generation in oxygen-limited niches (Dhouib et al., 2016). In H. influenzae, loss of MtsZ reduced biofilm formation and in-biofilm survival, and survival of the MtsZ mutant strains was attenuated in both human tissue cells and a mouse lung infection model (Dhouib et al., 2016). MtsZ was proposed to mediate redox balancing by reducing MetSO to methionine, which at the same time increases the availability of this amino acid as a nutrient.
However, not all enzymes in the BisC/TorZ/MtsZ lineage appear to affect in-host survival of bacterial pathogens. In C. jejuni, mutation in cj0264, which encodes an enzyme related to the BisC/TorZ/MtsZ group of S/N-oxide reductases, did not lead to a defect in a chicken caecum colonization model (Weingarten et al., 2008), and similarly, a torZ mutation in E. coli did not alter its ability to colonize the mouse intestine (Jones et al., 2011). As the physiological roles of these enzymes are currently unknown, it may be possible that, under other conditions, these enzymes are relevant for bacterial fitness, which may include other infection models. What is striking is the modulation of the substrate spectrum in enzymes of the MtsZ/BisC/TorZ group toward S-oxides found in the host; which has been demonstrated to correlate with specific and differential physiological functions during infection especially for BisC from S. Typhimurium.
Nitrate Reductases
Nitrate is another compound that is readily converted by molybdoenzymes. It occurs naturally in terrestrial and aqueous environments and is used as a food additive to prevent bacterial spoilage (Gassara et al., 2016; Ma et al., 2018). At sites of infection, nitrate forms following reaction of nitric oxide with oxygen, with nitrite and peroxynitrite being other NO oxidation products, that together cause nitrosative stress in cells (Lundberg et al., 1994; Nathan and Shiloh, 2000; Bogdan, 2001; Enocksson et al., 2004). Nitrate can be used by many bacteria, including pathogens as an alternative terminal electron acceptor for anaerobic respiration, and in several cases, this mode of anaerobic energy generation has been linked to bacterial virulence and fitness in the host (Jones et al., 2007; Lopez et al., 2012, 2015; Spees et al., 2013; Winter et al., 2013b).
There are three types of prokaryotic nitrate reductases (NRs): the membrane-bound, cytoplasm-facing NarGHI respiratory NR and the periplasmic NapAB NR both enable nitrate-dependent anaerobic respiration, while the assimilatory NasA NR enables the use of nitrate as a nitrogen source (Goldman et al., 1994; Moreno-Vivián et al., 1999; Potter et al., 1999; Richardson et al., 2001; Sparacino-Watkins et al., 2014) (Figure 5). In addition to enabling anaerobic respiration with nitrate, NapAB has also been proposed to have roles in redox balancing and growth under nitrate-limited conditions (Potter et al., 1999). In E. coli, expression of both nar and nap operons is strongly induced during anaerobiosis, and nar expression is further enhanced when nitrate is present (Stewart, 1982; Grove et al., 1996; Wang et al., 1999). In contrast, oxygen status does not seem to affect expression of the nas operon; instead, and in keeping with its role as an assimilatory enzyme, its expression is induced by nitrate and repressed by ammonium (Ramos et al., 1993; Goldman et al., 1994).
While all three types of NRs are present in pathogenic bacteria (Jeter et al., 1984; Cali et al., 1989), roles in bacterial virulence have only been identified for Nar- and Nap-type NRs (Spector et al., 1999; Weber et al., 2000; Kohler et al., 2002; Jones et al., 2007; Van Alst et al., 2007; Weingarten et al., 2008; Tan et al., 2010; Kassem et al., 2012; Winter et al., 2013b; Lopez et al., 2015; Martín-Rodríguez et al., 2020).
Dissimilatory NRs in the enteric pathogens E. coli and S. Typhimurium are particularly well studied (Lundberg et al., 1994; Enocksson et al., 2004). Both E. coli and S. Typhimurium contain two Nar-type NRs, NarGHI and NarZYW, and a Nap-type NR, NapAB. Of the three NRs, NarG is the major contributor to dissimilatory nitrate reduction, although NapA also plays a part in nitrate reduction (Potter et al., 1999, 2000; Stewart et al., 2002; Gates et al., 2003; Martín-Rodríguez et al., 2020). Interestingly, NarZ seemed to contribute little to nitrate reduction and anaerobic growth on nitrate, and is repressed by anaerobiosis, even in the presence of nitrate (Iobbi-Nivol et al., 1987, 1990; Chang et al., 1999; Spector et al., 1999; Stewart et al., 2002).
In E. coli, NarG is the only nitrate reductase required for successful colonization of the mouse intestine, while NapA and NarZ are non-essential (Jones et al., 2007). However, some synergy between the different NR systems appears to exist, as the colonization defect observed for the ΔnarG strain was more pronounced in both a ΔnarG ΔnarZ double mutant and a ΔnarG ΔnapA ΔnarZ triple mutant (Jones et al., 2007). The E. coli triple NR mutant was strongly outcompeted by the WT strain during colonization of healthy and inflamed mouse gut environments (Jones et al., 2007; Spees et al., 2013; Winter et al., 2013b), and in a rat urinary tract infection model (Martín-Rodríguez et al., 2020). This documents that nitrate reduction confers a growth advantage to pathogenic E. coli strains and supports effective host colonization.
NarG is also the more important dissimilatory NR for virulence in the cystic-fibrosis-associated, opportunistic pathogen P. aeruginosa, where NarGHI, NapAB, and NasA NRs have been identified. Mutations in P. aeruginosa narG led to growth defects in cystic fibrosis sputum medium, reduced swarming ability in vitro and decreased virulence in C. elegans infections (Palmer et al., 2007; Van Alst et al., 2007), while a ΔnapA mutant displayed wild-type phenotypes (Van Alst et al., 2007). The swarming defect was suggested to be attributable to a reduced level of rhamnolipid production, a swarming-facilitating biosurfactant that is only produced when exogenous nitrate is depleted (Van Alst et al., 2007). A loss of NarG also reduced biofilm formation and in-biofilm survival of P. aeruginosa (Van Alst et al., 2007). In the brucellosis-causing opportunistic human pathogen Brucella suis, where NarGHI is the only respiratory NR, a ΔnarG strain was attenuated in human macrophages (Kohler et al., 2002), but NarGHI appeared to be dispensable for mouse infections (Loisel-Meyer et al., 2006).
However, NarG is not always a major virulence determinant, and in S. Typhimurium it was not required for virulence in a mouse model of colitis (Lopez et al., 2015). Instead, NapA was necessary for wild type-level virulence of S. Typhimurium in the mouse colitis model, and a ΔnarZ mutant also showed somewhat reduced fitness in this infection model (Lopez et al., 2012, 2015). NarZ appears to have additional roles in S. Typhimurium, where narZ expression was induced upon interaction with tissue cells, and a ΔnarZ mutant also showed significantly reduced virulence in a mouse oral infection model (Spector et al., 1999).
In C. jejuni, where NapA is the only NR encoded in the genome, NapA also appeared to be essential for in vitro resistance to H2O2 and colonization of the chicken caecum (Weingarten et al., 2008; Kassem et al., 2012; Liu et al., 2012).
NRs are also found in prominent non-Gram-negative pathogens. Both Mtb and Mycobacterium bovis contain two NR homologs, NarG and NarX, of which only NarG has nitrate reductase activity while the cellular function of NarX, a “fused-domain NR” with amino acid sequence similarity to the subunits of NarGHI, remains unclear (Hutter and Dick, 1999; Sohaskey and Wayne, 2003).
NarG-mediated nitrate reductase activity in Mtb increases during hypoxic conditions (Sohaskey and Wayne, 2003), and has been proposed as a marker for the transition from aerobic growth to non-replicating persistence of Mtb e.g., in granulomas (Wayne and Hayes, 1998). In vitro, exogenous nitrate, which would serve as a substrate for NRs, protected Mtb from radical nitrogen species and acid stress (Tan et al., 2010). Exogenous nitrate also enhanced survival of wild-type Mtb during sudden anaerobiosis in vitro, but not when oxygen was gradually depleted (Sohaskey, 2008), which suggests that the NarG NR plays a role in maintaining redox balance. Despite showing a loss of nitrate-dependent, anaerobic respiration and reduced survival in vitro (Aly et al., 2006; Malm et al., 2009), an Mtb ΔnarG mutant exhibited wild type-level survival in a mouse lung infection model and primary human macrophages under low oxygen tension (Aly et al., 2006; Cunningham-Bussel et al., 2013), but was cleared more readily within infected macrophages by the anti-tuberculosis drug, isoniazid (Cunningham-Bussel et al., 2013). In contrast, NarG was essential for M. bovis infection of SCID mice and a loss of NarG also reduced persistence in BALB/c mice (Weber et al., 2000; Fritz et al., 2002).
Although studies of Mtb ΔnarX mutant strains have not been reported, in Mtb clinical isolates, narX expression was observed in granuloma samples, and both narX and narG were induced in pericavity and distant lung samples, indicating a role for both enzymes during infection (Fenhalls et al., 2002; Rachman et al., 2006). This observation matches the elevated levels of NarX and NarG detected in the proteome of Mtb in a guinea pig infection model (Kruh et al., 2010).
In summary, dissimilatory nitrate reduction underpins fitness and virulence in many bacterial pathogens; however, the exact physiological roles and enzymes involved in nitrate reduction differ significantly between species, and various NR homologs exist in bacteria for which the exact physiological functions still need to be determined.
Tetrathionate Reductases
In mammalian host organisms, the sulfur compound tetrathionate (S4) forms in inflamed gut environments, where H2S is produced by the microbiota and converted to thiosulfate (S2), by the action of mitochondria (Hildebrandt and Grieshaber, 2008). Thiosulfate can then be oxidized to tetrathionate by reactive oxygen species (Winter et al., 2010). Several different types of enzymes catalyze the reduction of tetrathionate, such as the membrane-bound, heterotrimeric TtrABC tetrathionate reductase (Hinojosa-Leon et al., 1986; Hensel et al., 1999), the octaheme tetrathionate reductases found in Shewanella spp. (Mowat et al., 2004), or thiosulfate dehydrogenases in, for example, C. jejuni (Liu et al., 2013). Of these three enzymes, only TtrABC is a molybdoenzyme (Hinojosa-Leon et al., 1986) (Figure 5), and the role of this enzyme in tetrathionate reduction was first discovered in S. enterica serovar Typhimurium (Knox et al., 1943). Tetrathionate reduction has since been shown to occur in other bacterial pathogens such as P. aeruginosa, S. typhi, and Citrobacter freundii, while E. coli and Klebsiella pneumoniae lack this activity (Barrett and Clark, 1987).
In S. Typhimurium, expression of the ttr operon is induced by anaerobiosis and the presence of tetrathionate (Hensel et al., 1999), but repressed by nitrate (Lopez et al., 2012), with the TtrSR two-component system as a major regulator of ttr expression (Hensel et al., 1999). A functional TtrABC complex was required for growth in tetrathionate-containing medium (Hensel et al., 1999), as well as for Aer-mediated energy taxis toward tetrathionate, likely because TtrABC modulates the redox state of S. Typhimurium (Rivera-Chávez et al., 2013).
An S. Typhimurium ΔttrA mutant strain was outcompeted by the wild-type strain in both mouse and bovine gut colitis infection models, but not during infection of a healthy gut (Winter et al., 2010). In fact, the inflamed gut was the only body niche where the ΔttrA strain showed reduced virulence compared to the wild-type strain, possibly due to the absence of tetrathionate in other body niches (Winter et al., 2010).
Unlike several of the other Mo enzymes discussed above, TtrABC thus confers a highly niche-specific fitness advantage to bacteria that is associated specifically with an inflamed gut. Confirming this advantage in other bacteria that harbor this enzyme, especially those that infect other body niches such as P. aeruginosa, would add further insights into the physiological roles of tetrathionate reductases.
Formate Dehydrogenases
Formate dehydrogenases convert formate to CO2, and unlike the enzymes discussed previously in this section, they feed electrons into the cellular quinone pool rather than re-oxidizing them. The substrate of these enzymes, formate, is a product of serine catabolism in mammalian cells, but can also result from microbial fermentation in the GI tract (Pietzke et al., 2020). Once produced, formate can enter the circulation and is used in various key metabolic pathways, including nucleotide synthesis (Pietzke et al., 2020). Formate may also play a role during infections, as a 2-fold increase of formate concentration (at millimolar level) correlated with increased inflammation in the mouse gut (Hughes et al., 2017).
In bacteria, formate is converted to CO2 by one of three types of molybdenum-containing formate dehydrogenases (FDHs) (Figure 5). The two membrane-integral, periplasm-facing formate dehydrogenases, FDH-O (encoded by the fdo operon), and FDH-N (encoded by the fdn operon) (Figure 5), contribute to both aerobic and anaerobic respiration with formate as the electron donor (Ruiz-Herrera and DeMoss, 1969; Yamamoto and Ishimoto, 1977; Unden et al., 2014). Expression of fdnG is induced predominantly during anaerobiosis, when nitrate is present (Darwin et al., 1997; Wang and Gunsalus, 2003), while fdoG is expressed constitutively at low levels regardless of oxygen status (Abaibou et al., 1995; Benoit et al., 1998). The cytoplasmic FDH-H (fdhF) is a component of the formate hydrogen lyase complex that disproportionates formate into CO2 and H2 and is involved in redox balancing during fermentative growth (McDowall et al., 2014; Pinske and Sawers, 2016). In keeping with this role, expression of fdhF in E. coli is induced under anaerobic condition in the presence of formate, while oxygen and nitrate act as repressors (Birkmann and Bock, 1989; Rossmann et al., 1991; Wang and Gunsalus, 2003). Interestingly, some bacterial FDHs have no strict functional requirement for Mo as the catalytic metal center: the C. jejuni FDH, a distant relative of the respiratory FDH-N and FDH-O enzymes, is active with either a Mo- or W-bound cofactor; however, the same is not true for the E. coli FDHs, which are strictly molybdenum-dependent (Enoch and Lester, 1972; Andreesen and Ljungdahl, 1973; Taveirne et al., 2009; Niks and Hille, 2019).
Similar to what was reported above for the TtrABC tetrathionate reductase, and in keeping with increased formate production in the inflamed mouse gut, E. coli FDHs enhance bacterial survival in an inflamed gut, but are not required in the absence of inflammation (Hughes et al., 2017). Expression of both fdnG and fdoG in E. coli was induced during colonization of an inflamed mouse gut (Hughes et al., 2017). Single-gene knockout mutants revealed that FDH-N is a major contributor to E. coli fitness in a mouse colitis model, while FDH-O played a minor role (Hughes et al., 2017). This observation matches the increased expression of fdnG during anaerobiosis and may also be connected to the fact that nitrate, generated during inflammation, induces fdnG expression (Darwin et al., 1997; Wang and Gunsalus, 2003). However, the cellular roles of FDH enzymes vary, and in Yersinia pestis, a FDH-H mutation did not attenuate virulence in a mouse infection model (Pradel et al., 2014).
In contrast, FdhA, an enzyme distantly related to FDH-N from E. coli, supported virulence of C. jejuni in human and chicken cell line infections and chicken caecum colonization (Weerakoon et al., 2009; Kassem et al., 2012; Pryjma et al., 2012). In addition to respiratory chain-linked FDHs, there is also evidence that the cytoplasmic FDH-H can provide a fitness advantage, for example, to the human pathobiont Aggregatibacter actinomycetemcomitans in a murine abscess infection model (Jorth et al., 2013).
However, the role of FDHs in bacterial virulence is complex, and in Shigella flexneri ΔfdnG and ΔfdoG mutant strains even showed increased virulence in infections of a human intestinal cell line. This was attributed to the increase in formate concentrations in S. flexneri cells lacking functional FDHs, and the fact that in this bacterium, formate controls virulence gene expression (Koestler et al., 2018). Thus, in S. flexneri, FDHs are negative modulators of bacterial virulence, while in E. coli and C. jejuni, FDHs are positive modulators of bacterial fitness in the host (Koestler et al., 2018). Formate-based regulation of virulence factor expression was also observed in S. enterica, where formate upregulated the expressions of sipC, a gene encoding a type III secretion system effector protein (Lara-Tejero and Galán, 2009), and two major regulators in the Salmonella pathogenicity island 1, leading to enhanced virulence in the mouse gut (Huang et al., 2008). While infection data for S. enterica FDH mutant strains has not been reported, formate degradation might also be a negative regulator of virulence gene expression in S. enterica, but this needs to be investigated.
It thus appears that there are two distinct roles for FDHs in supporting or modulating bacterial virulence. The first role is in supporting preferred modes of energy generation, either by respiration or during fermentative growth, while the second appears to consist in controlling of the concentrations of formate in bacteria where formate acts as molecular signal that enhances expression of virulence-associated genes.
Outlook
Mo enzymes from all three major families clearly have the potential to play important roles in supporting virulence and bacterial fitness in human pathogens, despite the fact that phylogenetic analyses indicate that these enzymes are not required in selected groups of bacterial pathogens that have undergone extreme adaptation of life in the host environment, such as the Chlamydiae.
While enzymes from the xanthine oxidase family are the least studied in this regard, and only CO dehydrogenases appear to have a possible role in bacterial virulence, for both the sulfite oxidase family and DMSO reductase family enzymes, there is clear evidence for the involvement of sulfite dehydrogenases, S/N-oxides reductases, nitrate reductases and formate dehydrogenases in supporting virulence in bacteria as diverse as E. coli, S. enterica, C. jejuni, Mtb, and H. influenzae.
In most cases, the physiological roles of these enzymes appear to be very similar to those already identified for many molybdoenzymes, namely, in facilitating adaptation of the bacterial pathogen to its environment (Figure 7), by supporting energy generation and enabling the use or detoxification of compounds present in the host during inflammation. In this way, nitrate and tetrathionate reductases support anaerobic respiration in E. coli, C. jejuni, or S. Typhimurium (Jones et al., 2007; Weingarten et al., 2008; Winter et al., 2010, 2013b; Kassem et al., 2012; Liu et al., 2012; Lopez et al., 2012, 2015; Spees et al., 2013), while other enzymes such as FDH-H can be important for optimal fermentative growth (Jorth et al., 2013). While this physiological role is not surprising, the resulting fitness defects generally manifest themselves in a reduced ability of the pathogen to survive in the host environment, which highlights the importance of an optimally poised energy generating system for successful bacterial pathogenesis. The specific physiological roles of the Mo enzymes appear to be closely linked to the preferred growth modes of each bacterial pathogen, and in some cases, enzymes that are essential for in-host fitness of one pathogen are expendable in another organism, where the metabolic network has different functional requirements (Figure 7).
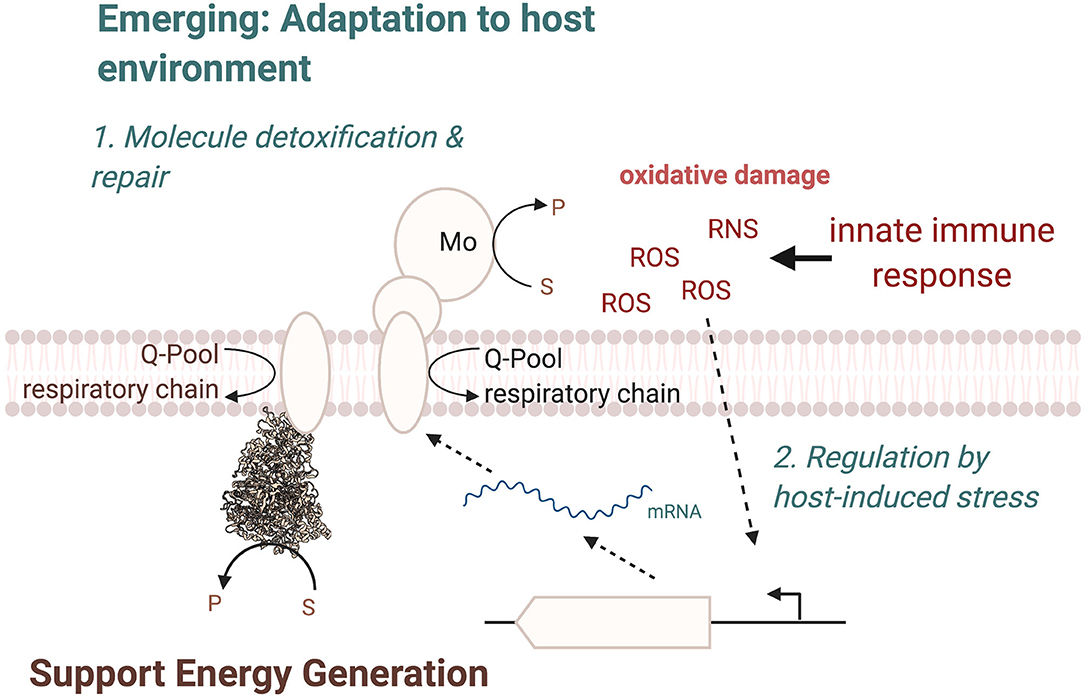
Figure 7. Overview of cellular functions of Mo-enzymes in bacterial pathogens. The three main functions are in energy generation, enabling optimal use of the metabolic network, and the two emerging functions of converting substrates that only occur in the human host and changes in the enzyme regulatory circuits, where Mo-enzyme expression is induced by molecules produced by the host organisms during infection. ROS, Reactive Oxygen Species; RNS, Reactive Nitrogen Species.
A second key consideration are the substrates that are converted by the Mo enzymes involved in bacterial virulence. Where the type of substrate has not changed during evolution, as is the case e.g., for nitrate or carbon monoxide, the source of the compound has changed and nitrate, for example, is predominantly produced from NO, while CO or sulfite are present because they act as signaling molecules or antimicrobials in the host (Figure 7). In other cases, notably the S- and N-oxide reductases, while the general reaction catalyzed is again similar to what has been identified in environmental bacteria such as the phototrophic Rhodobacter sp., the type of preferred substrate has changed. Instead of DMSO, which can occur in lake and marine environments (Kappler and Schäfer, 2014), the enzymes now preferentially convert sulfoxides and possible also N-oxides that are found in the human body, such as MetSO and BSO, both of which have been shown to be substrates converted by enzymes that are required for wild type-level virulence in different bacteria.
Thus, while S- and N-oxide reductases still have roles in supporting redox balancing and supporting anaerobic respiration, a subtle change in substrate preference appears to be occurring as a result of adaptation of bacteria to the host environment. To identify additional Mo enzymes that have the potential to support in-host fitness of bacterial pathogens, the presence of suitable substrates at sites of infection and in body niches that are colonized should be considered, as well as the possibility that some adjustment to the documented enzyme substrate preferences may have occurred.
A final point that should be considered when investigating the role of Mo enzymes in bacterial pathogens is the regulation of these enzymes in the pathogens (Figure 7). A well-established pattern of regulation for Mo enzymes is induction of expression in the presence of their substrate, which is often paired with increased expression in the absence of oxygen, which is in keeping with the known oxygen sensitivity of the Mo-PPT cofactor and the roles of many enzymes in anaerobic energy generation; this has been observed for nitrate and tetrathionate reductases as well as formate dehydrogenases. However, in some instances, a specific adaptation of the regulation of expression to the host environment seems to have occurred. As an example, expression of the MsrP methionine sulfoxide reductase is induced in response to exposure of the bacteria to HOCl and possibly other ROS, which are produced by the host during infection, rather than by the presence of the enzyme's substrate (Gennaris et al., 2015). This is a pathogen-specific adaptation in the way expression of MsrP is regulated and it seems likely that similar adaptations may have occurred for other Mo enzymes in pathogenic bacteria.
Finally, while there are many studies on Mo enzymes found in pathogenic E. coli, S. enterica, C. jejuni, and even Mtb, how Mo enzymes support survival of priority pathogens such as the ESKAPE pathogens or WHO priority pathogens is unclear at present. Many of these pathogens contain several Mo enzymes, including many of those that already have been shown to support bacterial virulence (Table 2), making them interesting targets for further research. Additionally, the majority of Mo enzymes that are relevant to bacterial pathogenicity belong to groups that are unique to prokaryotes, and thus could be promising targets for the development of new antimicrobial agents.
Author Contributions
QZ carried out the literature review, drafting, editing of all manuscript sections, carried out data analyses, and created tables and figures. BK provided critical analysis of the manuscript and help with the preparation of figures. UK provided help with the initial conception, structuring of the manuscript, contributed to manuscript writing, and to the generation of figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Health and Medical Research Council Grant GNT1158451 to UK and GNT1180826 to BK. BK is an Australian Research Council Laureate Fellow (FL180100109).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Figures were prepared using BioRender, Pymol, and CorelDraw.
References
Abaibou, H., Pommier, J., Benoit, S., Giordano, G., and Mandrand-Berthelot, M. A. (1995). Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J. Bacteriol. 177, 7141–7149. doi: 10.1128/JB.177.24.7141-7149.1995
Aly, S., Wagner, K., Keller, C., Malm, S., Malzan, A., Brandau, S., et al. (2006). Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol. 210, 298–305. doi: 10.1002/path.2055
Andreae, C. A., Titball, R. W., and Butler, C. S. (2014). Influence of the molybdenum cofactor biosynthesis on anaerobic respiration, biofilm formation and motility in Burkholderia thailandensis. Res. Microbiol. 165, 41–49. doi: 10.1016/j.resmic.2013.10.009
Andreesen, J. R., and Ljungdahl, L. G. (1973). Formate dehydrogenase of Clostridium thermoaceticum: incorporation of selenium-75, and the effects of selenite, molybdate, and tungstate on the enzyme. J. Bacteriol. 116, 867–873. doi: 10.1128/JB.116.2.867-873.1973
Andreesen, J. R., and Makdessi, K. (2008). Tungsten, the surprisingly positively acting heavy metal element for prokaryotes. Ann. N. Y. Acad. Sci. 1125, 215–229. doi: 10.1196/annals.1419.003
Arnoux, P., Sabaty, M., Alric, J., Frangioni, B., Guigliarelli, B., Adriano, J.-M., et al. (2003). Structural and redox plasticity in the heterodimeric periplasmic nitrate reductase. Nat. Struct. Mol. Biol. 10, 928–934. doi: 10.1038/nsb994
Baltes, N., Hennig-Pauka, I., Jacobsen, I., Gruber, A. D., and Gerlach, G. F. (2003). Identification of dimethyl sulfoxide reductase in Actinobacillus pleuropneumoniae and its role in infection. Infect. Immun. 71, 6784–6792. doi: 10.1128/IAI.71.12.6784-6792.2003
Barrett, E. L., and Clark, M. A. (1987). Tetrathionate reduction and production of hydrogen sulfide from thiosulfate. Microbiol. Rev. 51, 192–205. doi: 10.1128/MMBR.51.2.192-205.1987
Battelli, M. G., Polito, L., Bortolotti, M., and Bolognesi, A. (2016). Xanthine oxidoreductase-derived reactive species: physiological and pathological effects. Oxid. Med. Cell. Longev. 2016:3527579. doi: 10.1155/2016/3527579
Beck-Speier, I., Liese, J. G., Belohradsky, B. H., and Godleski, J. J. (1993). Sulfite stimulates NADPH oxidase of human neutrophils to produce active oxygen radicals via protein kinase C and Ca2+/calmodulin pathways. Free Radic. Biol. Med. 14, 661–668. doi: 10.1016/0891-5849(93)90148-N
Benoit, S., Abaibou, H., and Mandrand-Berthelot, M. A. (1998). Topological analysis of the aerobic membrane-bound formate dehydrogenase of Escherichia coli. J. Bacteriol. 180, 6625–6634. doi: 10.1128/JB.180.24.6625-6634.1998
Birkmann, A., and Bock, A. (1989). Characterization of a cis regulatory DNA element necessary for formate induction of the formate dehydrogenase gene (fdhF) of Escherichia coli. Mol. Microbiol. 3, 187–195. doi: 10.1111/j.1365-2958.1989.tb01807.x
Bogdan, C. (2001). Nitric oxide and the immune response. Nat. Immunol. 2, 907–916. doi: 10.1038/ni1001-907
Borner, G., Karrasch, M., and Thauer, R. K. (1991). Molybdopterin adenine dinucleotide and molybdopterin hypoxanthine dinucleotide in formylmethanofuran dehydrogenase from Methanobacterium thermoautotrophicum (Marburg). FEBS Lett. 290, 31–34. doi: 10.1016/0014-5793(91)81218-W
Boyington, J. C., Gladyshev, V. N., Khangulov, S. V., Stadtman, T. C., and Sun, P. D. (1997). Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275, 1305–1308. doi: 10.1126/science.275.5304.1305
Bray, R. C., Bennett, B., Burke, J. F., Chovnick, A., Doyle, W. A., Howes, B. D., et al. (1996). Recent studies on xanthine oxidase and related enzymes. Biochem. Soc. Trans. 24, 99–105. doi: 10.1042/bst0240099
Brodin, P., Poquet, Y., Levillain, F., Peguillet, I., Larrouy-Maumus, G., Gilleron, M., et al. (2010). High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog. 6:e1001100. doi: 10.1371/journal.ppat.1001100
Brokx, S. J., Rothery, R. A., Zhang, G., Ng, D. P., and Weiner, J. H. (2005). Characterization of an Escherichia coli sulfite oxidase homologue reveals the role of a conserved active site cysteine in assembly and function. Biochemistry 44, 10339–10348. doi: 10.1021/bi050621a
Cali, B. M., Micca, J. L., and Stewart, V. (1989). Genetic regulation of nitrate assimilation in Klebsiella pneumoniae M5al. J. Bacteriol. 171, 2666–2672. doi: 10.1128/JB.171.5.2666-2672.1989
Camacho, L. R., Ensergueix, D., Perez, E., Gicquel, B., and Guilhot, C. (1999). Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34, 257–267. doi: 10.1046/j.1365-2958.1999.01593.x
Chang, L., Wei, L. I., Audia, J. P., Morton, R. A., and Schellhorn, H. E. (1999). Expression of the Escherichia coli NRZ nitrate reductase is highly growth phase dependent and is controlled by RpoS, the alternative vegetative sigma factor. Mol. Microbiol. 34, 756–766. doi: 10.1046/j.1365-2958.1999.01637.x
Cherak, S. J., and Turner, R. J. (2017). Assembly pathway of a bacterial complex iron sulfur molybdoenzyme. Biomol. Concepts 8, 155–167. doi: 10.1515/bmc-2017-0011
Chin, B. Y., and Otterbein, L. E. (2009). Carbon monoxide is a poison to microbes! CO as a bactericidal molecule. Curr. Opin. Pharmacol. 9, 490–500. doi: 10.1016/j.coph.2009.06.025
Christiansen, L. C., Schou, S., Nygaard, P., and Saxild, H. H. (1997). Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J. Bacteriol. 179, 2540–2550. doi: 10.1128/JB.179.8.2540-2550.1997
Chung, S. W., Liu, X., Macias, A. A., Baron, R. M., and Perrella, M. A. (2008). Heme oxygenase-1-derived carbon monoxide enhances the host defense response to microbial sepsis in mice. J. Clin. Investig. 118, 239–247. doi: 10.1172/JCI32730
Coelho, C., Mahro, M., Trincao, J., Carvalho, A. T., Ramos, M. J., Terao, M., et al. (2012). The first mammalian aldehyde oxidase crystal structure: insights into substrate specificity. J. Biol. Chem. 287, 40690–40702. doi: 10.1074/jbc.M112.390419
Contreras, I., Toro, C. S., Troncoso, G., and Mora, G. C. (1997). Salmonella typhi mutants defective in anaerobic respiration are impaired in their ability to replicate within epithelial cells. Microbiology 143, 2665–2672. doi: 10.1099/00221287-143-8-2665
Correia, M. A., Otrelo-Cardoso, A. R., Schwuchow, V., Sigfridsson Clauss, K. G., Haumann, M., Romão, M. J., et al. (2016). The Escherichia coli periplasmic aldehyde oxidoreductase is an exceptional member of the xanthine oxidase family of molybdoenzymes. ACS Chem. Biol. 11, 2923–2935. doi: 10.1021/acschembio.6b00572
Crawford, N. M., Smith, M., Bellissimo, D., and Davis, R. W. (1988). Sequence and nitrate regulation of the Arabidopsis thaliana mRNA encoding nitrate reductase, a metalloflavoprotein with three functional domains. Proc. Natl. Acad. Sci. U. S. A. 85, 5006–5010. doi: 10.1073/pnas.85.14.5006
Crichton, R. (2019). “Chapter 17 - Molybdenum, tungsten, vanadium and chromium,” in Biological Inorganic Chemistry, 3rd Edn, ed R. Crichton (London: Academic Press), 475-498. doi: 10.1016/B978-0-12-811741-5.00017-5
Cunningham-Bussel, A., Bange, F. C., and Nathan, C. F. (2013). Nitrite impacts the survival of Mycobacterium tuberculosis in response to isoniazid and hydrogen peroxide. MicrobiologyOpen 2, 901–911. doi: 10.1002/mbo3.126
Dahl, J. U., Radon, C., Bühning, M., Nimtz, M., Leichert, L. I., Denis, Y., et al. (2013). The sulfur carrier protein TusA has a pleiotropic role in Escherichia coli that also affects molybdenum cofactor biosynthesis. J. Biol. Chem. 288, 5426–5442. doi: 10.1074/jbc.M112.431569
Daniels, J. N., Wuebbens, M. M., Rajagopalan, K. V., and Schindelin, H. (2008). Crystal structure of a molybdopterin synthase-precursor Z complex: insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency. Biochemistry 47, 615–626. doi: 10.1021/bi701734g
Darwin, A. J., Tyson, K. L., Busby, S. J., and Stewart, V. (1997). Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol. Microbiol. 25, 583–595. doi: 10.1046/j.1365-2958.1997.4971855.x
Davidson, A. L., Dassa, E., Orelle, C., and Chen, J. (2008). Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364. doi: 10.1128/MMBR.00031-07
del Campillo-Campbell, A., and Campbell, A. (1982). Molybdenum cofactor requirement for biotin sulfoxide reduction in Escherichia coli. J. Bacteriol. 149, 469–478. doi: 10.1128/JB.149.2.469-478.1982
Denkel, L. A., Rhen, M., and Bange, F. C. (2013). Biotin sulfoxide reductase contributes to oxidative stress tolerance and virulence in Salmonella enterica serovar typhimurium. Microbiology 159, 1447–1458. doi: 10.1099/mic.0.067256-0
Dhouib, R., Othman, D. S., Lin, V., Lai, X. J., Wijesinghe, H. G., Essilfie, A. T., et al. (2016). A novel, molybdenum-containing methionine sulfoxide reductase supports survival of Haemophilus influenzae in an in vivo model of infection. Front. Microbiol. 7:1743. doi: 10.3389/fmicb.2016.01743
Dhouib, R., Pg Othman, D. S., Essilfie, A. T., Hansbro, P. M., Hanson, J. O., McEwan, A. G., et al. (2015). Maturation of molybdoenzymes and its influence on the pathogenesis of non-typeable Haemophilus influenzae. Front. Microbiol. 6:1219. doi: 10.3389/fmicb.2015.01219
Dobbek, H., Gremer, L., Kiefersauer, R., Huber, R., and Meyer, O. (2002). Catalysis at a dinuclear [CuSMoOH] cluster in a CO dehydrogenase resolved at 1.1-Å resolution. Proc. Natl. Acad. Sci. U. S. A. 99, 15971–6. doi: 10.1073/pnas.212640899
Dos Santos, J. P., Iobbi-Nivol, C., Couillault, C., Giordano, G., and Méjean, V. (1998). Molecular analysis of the trimethylamine N-oxide (TMAO) reductase respiratory system from a Shewanella species. J. Mol. Biol. 284, 421–433. doi: 10.1006/jmbi.1998.2155
Dutta, N. K., Mehra, S., Didier, P. J., Roy, C. J., Doyle, L. A., Alvarez, X., et al. (2010). Genetic requirements for the survival of tubercle bacilli in primates. J. Infect. Dis. 201, 1743–1752. doi: 10.1086/652497
Enoch, H. G., and Lester, R. L. (1972). Effects of molybdate, tungstate, and selenium compounds on formate dehydrogenase and other enzyme systems in Escherichia coli. J. Bacteriol. 110, 1032–1040. doi: 10.1128/JB.110.3.1032-1040.1972
Enocksson, A., Lundberg, J., Weitzberg, E., Norrby-Teglund, A., and Svenungsson, B. (2004). Rectal nitric oxide gas and stool cytokine levels during the course of infectious gastroenteritis. Clin. Diagn. Lab. Immunol. 11, 250–254. doi: 10.1128/CDLI.11.2.250-254.2004
Enroth, C., Eger, B. T., Okamoto, K., Nishino, T., Nishino, T., and Pai, E. F. (2000). Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion. Proc. Natl. Acad. Sci. U. S. A. 97, 10723–10728. doi: 10.1073/pnas.97.20.10723
Ezraty, B., Bos, J., Barras, F., and Aussel, L. (2005). Methionine sulfoxide reduction and assimilation in Escherichia coli: new role for the biotin sulfoxide reductase BisC. J. Bacteriol. 187, 231–237. doi: 10.1128/JB.187.1.231-237.2005
Fenhalls, G., Stevens, L., Moses, L., Bezuidenhout, J., Betts, J. C., Helden Pv, P., et al. (2002). In situ detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect. Immun. 70, 6330–6338. doi: 10.1128/IAI.70.11.6330-6338.2002
Filiatrault, M. J., Tombline, G., Wagner, V. E., Van Alst, N., Rumbaugh, K., Sokol, P., et al. (2013). Pseudomonas aeruginosa PA1006, which plays a role in molybdenum homeostasis, is required for nitrate utilization, biofilm formation, and virulence. PLoS ONE 8:e55594. doi: 10.1371/journal.pone.0055594
Freiberg, C., Brunner, N. A., Schiffer, G., Lampe, T., Pohlmann, J., Brands, M., et al. (2004). Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors with antibacterial activity. J. Biol. Chem. 279, 26066–26073. doi: 10.1074/jbc.M402989200
Fritz, C., Maass, S., Kreft, A., and Bange, F.-C. (2002). Dependence of Mycobacterium bovis BCG on anaerobic nitrate reductase for persistence is tissue specific. Infect. Immun. 70, 286–91. doi: 10.1128/IAI.70.1.286-291.2002
Gassara, F., Kouassi, A. P., Brar, S. K., and Belkacemi, K. (2016). Green alternatives to nitrates and nitrites in meat-based products—a review. Crit. Rev. Food Sci. Nutr. 56, 2133–2148. doi: 10.1080/10408398.2013.812610
Gates, A. J., Hughes, R. O., Sharp, S. R., Millington, P. D., Nilavongse, A., Cole, J. A., et al. (2003). Properties of the periplasmic nitrate reductases from Paracoccus pantotrophus and Escherichia coli after growth in tungsten-supplemented media. FEMS Microbiol. Lett. 220, 261–269. doi: 10.1016/S0378-1097(03)00122-8
Gennaris, A., Ezraty, B., Henry, C., Agrebi, R., Vergnes, A., Oheix, E., et al. (2015). Repairing oxidized proteins in the bacterial envelope using respiratory chain electrons. Nature 528, 409–412. doi: 10.1038/nature15764
Gisin, J., Müller, A., Pfänder, Y., Leimkühler, S., Narberhaus, F., and Masepohl, B. (2010). A Rhodobacter capsulatus member of a universal permease family imports molybdate and other oxyanions. J. Bacteriol. 192, 5943–5952. doi: 10.1128/JB.00742-10
Glaser, J. H., and DeMoss, J. A. (1971). Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J. Bacteriol. 108, 854–860. doi: 10.1128/JB.108.2.854-860.1971
Gnida, M., Ferner, R., Gremer, L., Meyer, O., and Meyer-Klaucke, W. (2003). A novel binuclear [CuSMo] cluster at the active site of carbon monoxide dehydrogenase: characterization by X-ray absorption spectroscopy. Biochemistry 42, 222–230. doi: 10.1021/bi026514n
Goldman, B. S., Lin, J. T., and Stewart, V. (1994). Identification and structure of the nasR gene encoding a nitrate- and nitrite-responsive positive regulator of nasFEDCBA (nitrate assimilation) operon expression in Klebsiella pneumoniae M5al. J. Bacteriol. 176, 5077–5085. doi: 10.1128/JB.176.16.5077-5085.1994
Gon, S., Patte, J. C., Méjean, V., and Iobbi-Nivol, C. (2000). The torYZ (yecK bisZ) operon encodes a third respiratory trimethylamine N-oxide reductase in Escherichia coli. J. Bacteriol. 182, 5779–5786. doi: 10.1128/JB.182.20.5779-5786.2000
Gonzalez, P. J., Rivas, M. G., Mota, C. S., Brondino, C. D., Moura, I., and Moura, J. J. G. (2013). Periplasmic nitrate reductases and formate dehydrogenases: biological control of the chemical properties of Mo and W for fine tuning of reactivity, substrate specificity and metabolic role. Coord. Chem. Rev. 257, 315–331. doi: 10.1016/j.ccr.2012.05.020
Gremer, L., and Meyer, O. (1996). Characterization of xanthine dehydrogenase from the anaerobic bacterium Veillonella atypica and identification of a molybdopterin-cytosine-dinucleotide-containing molybdenum cofactor. Eur. J. Biochem. 238, 862–866. doi: 10.1111/j.1432-1033.1996.0862w.x
Grimaldi, S., Schoepp-Cothenet, B., Ceccaldi, P., Guigliarelli, B., and Magalon, A. (2013). The prokaryotic Mo/W-bisPGD enzymes family: a catalytic workhorse in bioenergetic. Biochim. Biophys. Acta 1827, 1048–1085. doi: 10.1016/j.bbabio.2013.01.011
Grove, J., Tanapongpipat, S., Thomas, G., Griffiths, L., Crooke, H., and Cole, J. (1996). Escherichia coli K-12 genes essential for the synthesis of c-type cytochromes and a third nitrate reductase located in the periplasm. Mol. Microbiol. 19, 467–481. doi: 10.1046/j.1365-2958.1996.383914.x
Guse, A., Stevenson, C. E., Kuper, J., Buchanan, G., Schwarz, G., Giordano, G., et al. (2003). Biochemical and structural analysis of the molybdenum cofactor biosynthesis protein MobA. J. Biol. Chem. 278, 25302–25307. doi: 10.1074/jbc.M302639200
Hagen, W. R. (2017). “Chapter 10 Tungsten-containing enzymes,” in Molybdenum and Tungsten Enzymes: Biochemistry. (The Royal Society of Chemistry), 313–342. doi: 10.1039/9781782623915-00313
Hanzelmann, P., and Schindelin, H. (2004). Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl. Acad. Sci. U. S. A. 101, 12870–12875. doi: 10.1073/pnas.0404624101
Hawrylik, S. J., Wasilko, D. J., Haskell, S. L., Gootz, T. D., and Lee, S. E. (1994). Bisulfite or sulfite inhibits growth of Helicobacter pylori. J. Clin. Microbiol. 32, 790–792. doi: 10.1128/JCM.32.3.790-792.1994
Hensel, M., Hinsley, A. P., Nikolaus, T., Sawers, G., and Berks, B. C. (1999). The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32, 275–287. doi: 10.1046/j.1365-2958.1999.01345.x
Hildebrandt, T. M., and Grieshaber, M. K. (2008). Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 275, 3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x
Hille, R. (2005). Molybdenum-containing hydroxylases. Arch. Biochem. Biophys. 433, 107–116. doi: 10.1016/j.abb.2004.08.012
Hille, R., Hall, J., and Basu, P. (2014). The mononuclear molybdenum enzymes. Chem. Rev. 114, 3963–4038. doi: 10.1021/cr400443z
Hinojosa-Leon, M., Dubourdieu, M., Sanchez-Crispin, J. A., and Chippaux, M. (1986). Tetrathionate reductase of Salmonella thyphimurium: a molybdenum containing enzyme. Biochem. Biophys. Res. Commun. 136, 577–581. doi: 10.1016/0006-291X(86)90479-1
Hitchcock, A., Hall, S. J., Myers, J. D., Mulholland, F., Jones, M. A., and Kelly, D. J. (2010). Roles of the twin-arginine translocase and associated chaperones in the biogenesis of the electron transport chains of the human pathogen Campylobacter jejuni. Microbiology 156, 2994–3010. doi: 10.1099/mic.0.042788-0
Hollenstein, K., Frei, D. C., and Locher, K. P. (2007). Structure of an ABC transporter in complex with its binding protein. Nature 446, 213–216. doi: 10.1038/nature05626
Hover, B. M., Loksztejn, A., Ribeiro, A. A., and Yokoyama, K. (2013). Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J. Am. Chem. Soc. 135, 7019–7032. doi: 10.1021/ja401781t
Hover, B. M., Tonthat, N. K., Schumacher, M. A., and Yokoyama, K. (2015). Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 112, 6347–6352. doi: 10.1073/pnas.1500697112
Hu, Y., Rech, S., Gunsalus, R. P., and Rees, D. C. (1997). Crystal structure of the molybdate binding protein ModA. Nat. Struct. Biol. 4, 703–707. doi: 10.1038/nsb0997-703
Huang, Y., Suyemoto, M., Garner, C. D., Cicconi, K. M., and Altier, C. (2008). Formate acts as a diffusible signal to induce Salmonella invasion. J. Bacteriol. 190, 4233–41. doi: 10.1128/JB.00205-08
Hughes, E. R., Winter, M. G., Duerkop, B. A., Spiga, L., Furtado de Carvalho, T., Zhu, W., et al. (2017). Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe 21, 208–219. doi: 10.1016/j.chom.2017.01.005
Hutter, B., and Dick, T. (1999). Up-regulation of narX, encoding a putative 'fused nitrate reductase' in anaerobic dormant Mycobacterium bovis BCG. FEMS Microbiol. Lett. 178, 63–69. doi: 10.1111/j.1574-6968.1999.tb13760.x
Imperial, J., Hadi, M., and Amy, N. K. (1998). Molybdate binding by ModA, the periplasmic component of the Escherichia coli mod molybdate transport system. Biochim. Biophys. Acta 1370, 337–346. doi: 10.1016/S0005-2736(98)00003-0
Iobbi-Nivol, C., and Leimkühler, S. (2013). Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1827, 1086–1101. doi: 10.1016/j.bbabio.2012.11.007
Iobbi-Nivol, C., Palmer, T., Whitty, P. W., McNairn, E., and Boxer, D. H. (1995). The mob locus of Escherichia coli K12 required for molybdenum cofactor biosynthesis is expressed at very low levels. Microbiology 141, 1663–1671. doi: 10.1099/13500872-141-7-1663
Iobbi-Nivol, C., Santini, C.-L., Blasco, F., and Giordano, G. (1990). Purification and further characterization of the second nitrate reductase of Escherichia coli K12. Eur. J. Biochem. 188, 679–687. doi: 10.1111/j.1432-1033.1990.tb15450.x
Iobbi-Nivol, C., Santini, C. L., Bonnefoy, V., and Giordano, G. (1987). Biochemical and immunological evidence for a second nitrate reductase in Escherichia coli K12. Eur. J. Biochem. 168, 451–459. doi: 10.1111/j.1432-1033.1987.tb13438.x
Irwin, S. V., Fisher, P., Graham, E., Malek, A., and Robidoux, A. (2017). Sulfites inhibit the growth of four species of beneficial gut bacteria at concentrations regarded as safe for food. PLoS ONE 12:e0186629. doi: 10.1371/journal.pone.0186629
Jeter, R. M., Sias, S. R., and Ingraham, J. L. (1984). Chromosomal location and function of genes affecting Pseudomonas aeruginosa nitrate assimilation. J. Bacteriol. 157, 673–677. doi: 10.1128/JB.157.2.673-677.1984
Ji, A. J., Savon, S. R., and Jacobsen, D. W. (1995). Determination of total serum sulfite by HPLC with fluorescence detection. Clin. Chem. 41, 897–903. doi: 10.1093/clinchem/41.6.897
Johnson, J. L., Bastian, N. R., and Rajagopalan, K. V. (1990). Molybdopterin guanine dinucleotide: a modified form of molybdopterin identified in the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides forma specialis denitrificans. Proc. Natl. Acad. Sci. U. S. A. 87, 3190–3194. doi: 10.1073/pnas.87.8.3190
Jones, S. A., Chowdhury, F. Z., Fabich, A. J., Anderson, A., Schreiner, D. M., House, A. L., et al. (2007). Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75, 4891–4899. doi: 10.1128/IAI.00484-07
Jones, S. A., Gibson, T., Maltby, R. C., Chowdhury, F. Z., Stewart, V., Cohen, P. S., et al. (2011). Anaerobic respiration of Escherichia coli in the mouse intestine. Infect. Immun. 79, 4218–4226. doi: 10.1128/IAI.05395-11
Jorth, P., Trivedi, U., Rumbaugh, K., and Whiteley, M. (2013). Probing bacterial metabolism during infection using high-resolution transcriptomics. J. Bacteriol. 195, 4991–4998. doi: 10.1128/JB.00875-13
Kajiyama, H., Nojima, Y., Mitsuhashi, H., Ueki, K., Tamura, S., Sekihara, T., et al. (2000). Elevated levels of serum sulfite in patients with chronic renal failure. J. Am. Soc. Nephrol. 11:923–927.
Kappler, U. (2008). “Bacterial sulfite-oxidizing enzymes – enzymes for chemolithotrophs only?,” in Microbial Sulfur Metabolism, eds. C. Dahl and C. G. Friedrich (Berlin, Heidelberg: Springer Berlin Heidelberg), 151–169. doi: 10.1007/978-3-540-72682-1_13
Kappler, U. (2011). Bacterial sulfite-oxidizing enzymes. Biochim. Biophys. Acta 1807, 1–10. doi: 10.1016/j.bbabio.2010.09.004
Kappler, U., and Enemark, J. H. (2015). Sulfite-oxidizing enzymes. J. Biol. Inorg. Chem. 20, 253–264. doi: 10.1007/s00775-014-1197-3
Kappler, U., and Schäfer, H. (2014). Transformations of dimethylsulfide. Met. Ions Life Sci. 14, 279–313. doi: 10.1007/978-94-017-9269-1_11
Kappler, U., and Schwarz, G. (2017). “Chapter 7 - The sulfite oxidase family of molybdenum enzymes,” in Molybdenum and Tungsten Enzymes: Biochemistry. (The Royal Society of Chemistry), 240–273. doi: 10.1039/9781782623915-00240
Kassem, I. I., Khatri, M., Esseili, M. A., Sanad, Y. M., Saif, Y. M., Olson, J. W., et al. (2012). Respiratory proteins contribute differentially to Campylobacter jejuni's survival and in vitro interaction with hosts' intestinal cells. BMC Microbiol. 12:258. doi: 10.1186/1471-2180-12-258
Kaufmann, P., Duffus, B. R., Mitrova, B., Iobbi-Nivol, C., Teutloff, C., Nimtz, M., et al. (2018). Modulating the molybdenum coordination sphere of Escherichia coli trimethylamine N-oxide reductase. Biochemistry 57, 1130–1143. doi: 10.1021/acs.biochem.7b01108
Kim, G., Weiss, S. J., and Levine, R. L. (2014). Methionine oxidation and reduction in proteins. Biochim. Biophys. Acta 1840, 901–905. doi: 10.1016/j.bbagen.2013.04.038
King, G. M., and Weber, C. F. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 107–118. doi: 10.1038/nrmicro1595
Knox, R., Gell, P. G., and Pollock, M. R. (1943). The selective action of tetrathionate in bacteriological media: a report to the medical research council. J. Hyg. 43, 147–158.141. doi: 10.1017/S0022172400012766
Koestler, B. J., Fisher, C. R., and Payne, S. M. (2018). Formate promotes Shigella intercellular spread and virulence gene expression. mBio 9, e01777–e01718. doi: 10.1128/mBio.01777-18
Kohler, S., Foulongne, V., Ouahrani-Bettache, S., Bourg, G., Teyssier, J., Ramuz, M., et al. (2002). The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. U. S. A. 99, 15711–15716. doi: 10.1073/pnas.232454299
Kozmin, S. G., Stepchenkova, E. I., and Schaaper, R. M. (2013). TusA (YhhP) and IscS are required for molybdenum cofactor-dependent base-analog detoxification. MicrobiologyOpen 2, 743–755. doi: 10.1002/mbo3.108
Kruh, N. A., Troudt, J., Izzo, A., Prenni, J., and Dobos, K. M. (2010). Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PLoS ONE 5:e13938. doi: 10.1371/journal.pone.0013938
Kubitza, C., Bittner, F., Ginsel, C., Havemeyer, A., Clement, B., and Scheidig, A. J. (2018). Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes. Proc. Natl. Acad. Sci. U. S. A. 115, 11958–11963. doi: 10.1073/pnas.1808576115
Kumar, A., Deshane, J. S., Crossman, D. K., Bolisetty, S., Yan, B.-S., Kramnik, I., et al. (2008). Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283, 18032–18039. doi: 10.1074/jbc.M802274200
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lake, M. W., Temple, C. A., Rajagopalan, K. V., and Schindelin, H. (2000). The crystal structure of the Escherichia coli MobA protein provides insight into molybdopterin guanine dinucleotide biosynthesis. J. Biol. Chem. 275, 40211–40217. doi: 10.1074/jbc.M007406200
Lara-Tejero, M., and Galán, J. E. (2009). Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 77, 2635–2642. doi: 10.1128/IAI.00077-09
Lawson, D. M., Williams, C. E., Mitchenall, L. A., and Pau, R. N. (1998). Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA. Structure 6, 1529–1539. doi: 10.1016/S0969-2126(98)00151-8
Lebrun, E., Brugna, M., Baymann, F., Muller, D., Lièvremont, D., Lett, M.-C., et al. (2003). Arsenite oxidase, an ancient bioenergetic enzyme. Mol. Biol. Evol. 20, 686–693. doi: 10.1093/molbev/msg071
Lee, J. H., Park, S. W., Kim, Y. M., and Oh, J. I. (2017). Functional characterization of the cutI gene for the transcription of carbon monoxide dehydrogenase genes in Mycobacterium sp. strain JC1 DSM 3803. J. Microbiol. 55, 31–36. doi: 10.1007/s12275-017-6572-7
Leimkühler, S. (2014). The biosynthesis of the molybdenum cofactor in Escherichia coli and its connection to FeS cluster assembly and the thiolation of tRNA. Adv. Biol. 2014:808569. doi: 10.1155/2014/808569
Leimkühler, S. (2020). The biosynthesis of the molybdenum cofactors in Escherichia coli. Environ. Microbiol. 22, 2007–2026. doi: 10.1111/1462-2920.15003
Leimkühler, S., and Iobbi-Nivol, C. (2016). Bacterial molybdoenzymes: old enzymes for new purposes. FEMS Microbiol. Rev. 40, 1–18. doi: 10.1093/femsre/fuv043
Leimkühler, S., Kern, M., Solomon, P. S., McEwan, A. G., Schwarz, G., Mendel, R. R., et al. (1998). Xanthine dehydrogenase from the phototrophic purple bacterium Rhodobacter capsulatus is more similar to its eukaryotic counterparts than to prokaryotic molybdenum enzymes. Mol. Microbiol. 27, 853–869. doi: 10.1046/j.1365-2958.1998.00733.x
Leimkühler, S., and Klipp, W. (1999). Role of XdhC in molybdenum cofactor insertion into xanthine dehydrogenase of Rhodobacter capsulatus. J. Bacteriol. 181, 2745–2751. doi: 10.1128/JB.181.9.2745-2751.1999
Leimkühler, S., and Rajagopalan, K. V. (2001). A sulfurtransferase is required in the transfer of cysteine sulfur in the in vitro synthesis of molybdopterin from precursor Z in Escherichia coli. J. Biol. Chem. 276, 22024–22031. doi: 10.1074/jbc.M102072200
Leimkühler, S., Wuebbens, M. M., and Rajagopalan, K. V. (2001). Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J. Biol. Chem. 276, 34695–34701. doi: 10.1074/jbc.M102787200
Leimkühler, S., Wuebbens, M. M., and Rajagopalan, K. V. (2011). The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coord. Chem. Rev. 255, 1129–1144. doi: 10.1016/j.ccr.2010.12.003
Lester, M. R. (1995). Sulfite sensitivity: significance in human health. J. Am. Coll. Nutr. 14, 229–232. doi: 10.1080/07315724.1995.10718500
Levillain, F., Poquet, Y., Mallet, L., Mazeres, S., Marceau, M., Brosch, R., et al. (2017). Horizontal acquisition of a hypoxia-responsive molybdenum cofactor biosynthesis pathway contributed to Mycobacterium tuberculosis pathoadaptation. PLoS Path. 13:e6752. doi: 10.1371/journal.ppat.1006752
Levine, R. L., Moskovitz, J., and Stadtman, E. R. (2000). Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life 50, 301–307. doi: 10.1080/15216540051081056
Liu, M. T., Wuebbens, M. M., Rajagopalan, K. V., and Schindelin, H. (2000). Crystal structure of the gephyrin-related molybdenum cofactor biosynthesis protein MogA from Escherichia coli. J. Biol. Chem. 275, 1814–1822. doi: 10.1074/jbc.275.3.1814
Liu, X., Gao, B., Novik, V., and Galán, J. E. (2012). Quantitative proteomics of intracellular Campylobacter jejuni reveals metabolic reprogramming. PLoS Path. 8:e1002562. doi: 10.1371/journal.ppat.1002562
Liu, Y. W., Denkmann, K., Kosciow, K., Dahl, C., and Kelly, D. J. (2013). Tetrathionate stimulated growth of Campylobacter jejuni identifies a new type of bi-functional tetrathionate reductase (TsdA) that is widely distributed in bacteria. Mol. Microbiol. 88, 173–188. doi: 10.1111/mmi.12176
Llamas, A., Chamizo-Ampudia, A., Tejada-Jimenez, M., Galvan, A., and Fernandez, E. (2017). The molybdenum cofactor enzyme mARC: moonlighting or promiscuous enzyme? BioFactors 43, 486–494. doi: 10.1002/biof.1362
Loisel-Meyer, S., Jiménez de Bagüés, M. P., Bassères, E., Dornand, J., Köhler, S., Liautard, J.-P., et al. (2006). Requirement of norD for Brucella suis virulence in a murine model of in vitro and in vivo infection. Infect. Immun. 74, 1973–1976. doi: 10.1128/IAI.74.3.1973-1976.2006
Lopez, C. A., Rivera-Chávez, F., Byndloss, M. X., and Bäumler, A. J. (2015). The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella enterica serovar Typhimurium during colitis. Infect. Immun. 83, 3470–3478. doi: 10.1128/IAI.00351-15
Lopez, C. A., Winter, S. E., Rivera-Chávez, F., Xavier, M. N., Poon, V., Nuccio, S.-P., et al. (2012). Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. Mbio 3, e00143–e00112. doi: 10.1128/mBio.00143-12
Loschi, L., Brokx, S. J., Hills, T. L., Zhang, G., Bertero, M. G., Lovering, A. L., et al. (2004). Structural and biochemical identification of a novel bacterial oxidoreductase. J. Biol. Chem. 279, 50391–50400. doi: 10.1074/jbc.M408876200
Lundberg, J. O., Hellström, P. M., Lundberg, J. M., and Alving, K. (1994). Greatly increased luminal nitric oxide in ulcerative colitis. Lancet 344, 1673–1674. doi: 10.1016/S0140-6736(94)90460-X
Ma, L., Hu, L., Feng, X., and Wang, S. (2018). Nitrate and nitrite in health and disease. Aging Dis. 9, 938–945. doi: 10.14336/AD.2017.1207
MacGurn, J. A., and Cox, J. S. (2007). A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect. Immun. 75, 2668–2678. doi: 10.1128/IAI.01872-06
Magalon, A., Fedor, J. G., Walburger, A., and Weiner, J. H. (2011). Molybdenum enzymes in bacteria and their maturation. Coord. Chem. Rev. 255, 1159–1178. doi: 10.1016/j.ccr.2010.12.031
Magalon, A., Ceccaldi, P., and Schoepp-Cothenet, B. (2017). “Chapter 5 the prokaryotic Mo/W-bisPGD enzymes family,” in Molybdenum and Tungsten Enzymes: Biochemistry. (The Royal Society of Chemistry), 143–191. doi: 10.1039/9781782623915-00143
Maia, L. B., Moura, I., and Moura, J. J. G. (2017). “Chapter 1 Molybdenum and tungsten-containing enzymes: an overview,” in Molybdenum and Tungsten Enzymes: Biochemistry. (The Royal Society of Chemistry), 1–80. doi: 10.1039/9781782623915-00001
Malm, S., Tiffert, Y., Micklinghoff, J., Schultze, S., Joost, I., Weber, I., et al. (2009). The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155, 1332–1339. doi: 10.1099/mic.0.023275-0
Martín-Rodríguez, A. J., Rhen, M., Melican, K., and Richter-Dahlfors, A. (2020). Nitrate metabolism modulates biosynthesis of biofilm components in uropathogenic Escherichia coli and acts as a fitness factor during experimental urinary tract infection. Front. Microbiol. 11:26. doi: 10.3389/fmicb.2020.00026
Mayr, S. J., Mendel, R. R., and Schwarz, G. (2020). Molybdenum cofactor biology, evolution and deficiency. Biochim. Biophys. Acta Mol. Cell Res. 1868:118883. doi: 10.1016/j.bbamcr.2020.118883
McDevitt, C. A., Hugenholtz, P., Hanson, G. R., and McEwan, A. G. (2002). Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes. Mol. Microbiol. 44, 1575–1587. doi: 10.1046/j.1365-2958.2002.02978.x
McDowall, J. S., Murphy, B. J., Haumann, M., Palmer, T., Armstrong, F. A., and Sargent, F. (2014). Bacterial formate hydrogenlyase complex. Proc. Natl. Acad. Sci. U. S. A. 111, E3948–E3956. doi: 10.1073/pnas.1407927111
McEwan, A. G., Ridge, J. P., McDevitt, C. A., and Hugenholtz, P. (2002). The DMSO reductase family of microbial molybdenum enzymes; molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 19, 3–21. doi: 10.1080/014904502317246138
Mejean, V., Iobbi-Nivol, C., Lepelletier, M., Giordano, G., Chippaux, M., and Pascal, M. C. (1994). TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol. Microbiol. 11, 1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x
Mendel, R. R. (2013). The molybdenum cofactor. J. Biol. Chem. 288, 13165–13172. doi: 10.1074/jbc.R113.455311
Mitsuhashi, H., Ikeuchi, H., Yamashita, S., Kuroiwa, T., Kaneko, Y., Hiromura, K., et al. (2004). Increased levels of serum sulfite in patients with acute pneumonia. Shock 21, 99–102. doi: 10.1097/01.shk.0000105501.75189.85
Mitsuhashi, H., Nojima, Y., Tanaka, T., Ueki, K., Maezawa, A., Yano, S., et al. (1998). Sulfite is released by human neutrophils in response to stimulation with lipopolysaccharide. J. Leukoc. Biol. 64, 595–599. doi: 10.1002/jlb.64.5.595
Mitsuhashi, H., Yamashita, S., Ikeuchi, H., Kuroiwa, T., Kaneko, Y., Hiromura, K., et al. (2005). Oxidative stress-dependent conversion of hydrogen sulfide to sulfite by activated neutrophils. Shock 24, 529–534. doi: 10.1097/01.shk.0000183393.83272.de
Moreno-Vivián, C., Cabello, P., Martínez-Luque, M., Blasco, R., and Castillo, F. (1999). Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 181, 6573–6584. doi: 10.1128/JB.181.21.6573-6584.1999
Mouncey, N. J., Choudhary, M., and Kaplan, S. (1997). Characterization of genes encoding dimethyl sulfoxide reductase of Rhodobacter sphaeroides 2.4.1T: an essential metabolic gene function encoded on chromosome II. J. Bacteriol. 179, 7617–7624. doi: 10.1128/JB.179.24.7617-7624.1997
Mowat, C. G., Rothery, E., Miles, C. S., McIver, L., Doherty, M. K., Drewette, K., et al. (2004). Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat. Struct. Mol. Biol. 11, 1023–1024. doi: 10.1038/nsmb827
Murray, K. N., Watson, J. G., and Chaykin, S. (1966). Catalysis of the direct transfer of oxygen from nicotinamide N-oxide to xanthine by xanthine oxidase. J. Biol. Chem. 241, 4798–4801.
Myers, J. D., and Kelly, D. J. (2005). A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology 151, 233–242. doi: 10.1099/mic.0.27573-0
Nathan, C., and Shiloh, M. U. (2000). Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 97, 8841–8848. doi: 10.1073/pnas.97.16.8841
Neumann, M., Mittelstadt, G., Iobbi-Nivol, C., Saggu, M., Lendzian, F., Hildebrandt, P., et al. (2009a). A periplasmic aldehyde oxidoreductase represents the first molybdopterin cytosine dinucleotide cofactor containing molybdo-flavoenzyme from Escherichia coli. FEBS J. 276, 2762–2774. doi: 10.1111/j.1742-4658.2009.07000.x
Neumann, M., Mittelstadt, G., Seduk, F., Iobbi-Nivol, C., and Leimkühler, S. (2009b). MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in Escherichia coli. J. Biol. Chem. 284, 21891–21898. doi: 10.1074/jbc.M109.008565
Nichols, J. D., and Rajagopalan, K. V. (2005). In vitro molybdenum ligation to molybdopterin using purified components. J. Biol. Chem. 280, 7817–7822. doi: 10.1074/jbc.M413783200
Niks, D., and Hille, R. (2019). Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: structure, mechanism, and cofactor insertion. Protein Sci. 28, 111–122. doi: 10.1002/pro.3498
Nishino, T., Okamoto, K., and Leimkühler, S. (2017). “Chapter 6 Enzymes of the xanthine oxidase family,” in Molybdenum and Tungsten Enzymes: Biochemistry. (The Royal Society of Chemistry), 192–239. doi: 10.1039/9781782623915-00192
Nitschke, W., and Russell, M. J. (2013). Beating the acetyl coenzyme A-pathway to the origin of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120258. doi: 10.1098/rstb.2012.0258
Nobre, L. S., Seixas, J. D., Romão, C. C., and Saraiva, L. M. (2007). Antimicrobial action of carbon monoxide-releasing compounds. Antimicrob. Agents Chemother. 51, 4303–4307. doi: 10.1128/AAC.00802-07
Noriega, C., Hassett, D. J., and Rowe, J. J. (2005). The mobA gene is required for assimilatory and respiratory nitrate reduction but not xanthine dehydrogenase activity in Pseudomonas aeruginosa. Curr. Microbiol. 51, 419–424. doi: 10.1007/s00284-005-0125-8
Oresnik, I. J., Ladner, C. L., and Turner, R. J. (2001). Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 40, 323–331. doi: 10.1046/j.1365-2958.2001.02391.x
Otrelo-Cardoso, A. R., da Silva Correia, M. A., Schwuchow, V., Svergun, D. I., Romão, M. J., Leimkühler, S., et al. (2014). Structural data on the periplasmic aldehyde oxidoreductase PaoABC from Escherichia coli: SAXS and preliminary X-ray crystallography analysis. Int. J. Mol. Sci. 15, 2223–2236. doi: 10.3390/ijms15022223
Palmer, K. L., Brown, S. A., and Whiteley, M. (2007). Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 189, 4449–4455. doi: 10.1128/JB.00162-07
Palmer, T., Santini, C. L., Iobbi Nivol, C., Eaves, D. J., Boxer, D. H., and Giordando, G. (1996). Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli Mol. Microbiol. 20, 875–884. doi: 10.1111/j.1365-2958.1996.tb02525.x
Palmer, T., Vasishta, A., Whitty, P. W., and Boxer, D. H. (1994). Isolation of protein FA, a product of the mob locus required for molybdenum cofactor biosynthesis in Escherichia coli. Eur. J. Biochem. 222, 687–692. doi: 10.1111/j.1432-1033.1994.tb18913.x
Pang, H., Lilla, E. A., Zhang, P., Zhang, D., Shields, T. P., Scott, L. G., et al. (2020). Mechanism of rate acceleration of radical C-C bond formation reaction by a radical SAM GTP 3',8-cyclase. J. Am. Chem. Soc. 142, 9314–9326. doi: 10.1021/jacs.0c01200
Park, S. W., Hwang, E. H., Park, H., Kim, J. A., Heo, J., Lee, K. H., et al. (2003). Growth of mycobacteria on carbon monoxide and methanol. J. Bacteriol. 185, 142–147. doi: 10.1128/JB.185.1.142-147.2003
Park, S. W., Song, T., Kim, S. Y., Kim, E., Oh, J. I., Eom, C. Y., et al. (2007). Carbon monoxide dehydrogenase in mycobacteria possesses a nitric oxide dehydrogenase activity. Biochem. Biophys. Res. Commun. 362, 449–453. doi: 10.1016/j.bbrc.2007.08.011
Pattison, D. I., and Davies, M. J. (2001). Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem. Res. Toxicol. 14, 1453–1464. doi: 10.1021/tx0155451
Pederick, V. G., Eijkelkamp, B. A., Ween, M. P., Begg, S. L., Paton, J. C., and McDevitt, C. A. (2014). Acquisition and role of molybdate in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 80, 6843–6852. doi: 10.1128/AEM.02465-14
Perinet, S., Jeukens, J., Kukavica-Ibrulj, I., Ouellet, M. M., Charette, S. J., and Levesque, R. C. (2016). Molybdate transporter ModABC is important for Pseudomonas aeruginosa chronic lung infection. BMC Res. Notes 9:23. doi: 10.1186/s13104-016-1840-x
Pettit, F. H., Lyon, D., Brown, J. R., and Shive, W. (1991). Evidence for sulfite as an essential metabolite for human peripheral lymphocytes. Biochem. Biophys. Res. Commun. 179, 611–614. doi: 10.1016/0006-291X(91)91415-9
Pietzke, M., Meiser, J., and Vazquez, A. (2020). Formate metabolism in health and disease. Mol. Metab. 33, 23–37. doi: 10.1016/j.molmet.2019.05.012
Pinske, C., and Sawers, R. G. (2016). Anaerobic formate and hydrogen metabolism. EcoSal Plus 7. doi: 10.1128/ecosalplus.ESP-0011-2016
Pitterle, D. M., Johnson, J. L., and Rajagopalan, K. V. (1993). In vitro synthesis of molybdopterin from precursor Z using purified converting factor. Role of protein-bound sulfur in formation of the dithiolene. J. Biol. Chem. 268, 13506–13509.
Potter, L. C., Millington, P., Griffiths, L., Thomas, G. H., and Cole, J. A. (1999). Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem. J. 344, 77–84. doi: 10.1042/bj3440077
Potter, L. C., Millington, P. D., Thomas, G. H., Rothery, R. A., Giordano, G., and Cole, J. A. (2000). Novel growth characteristics and high rates of nitrate reduction of an Escherichia coli strain, LCB2048, that expresses only a periplasmic nitrate reductase. FEMS Microbiol. Lett. 185, 51–57. doi: 10.1111/j.1574-6968.2000.tb09039.x
Pradel, E., Lemaître, N., Merchez, M., Ricard, I., Reboul, A., Dewitte, A., et al. (2014). New insights into how Yersinia pestis adapts to its mammalian host during bubonic plague. PLoS Path. 10:e1004029. doi: 10.1371/journal.ppat.1004029
Pryjma, M., Apel, D., Huynh, S., Parker, C. T., and Gaynor, E. C. (2012). FdhTU-modulated formate dehydrogenase expression and electron donor availability enhance recovery of Campylobacter jejuni following host cell infection. J. Bacteriol. 194, 3803–3813. doi: 10.1128/JB.06665-11
Rachman, H., Strong, M., Ulrichs, T., Grode, L., Schuchhardt, J., Mollenkopf, H., et al. (2006). Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect. Immun. 74, 1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006
Rajagopalan, K. V., and Johnson, J. L. (1992). The pterin molybdenum cofactors. J. Biol. Chem. 267, 10199–10202.
Ramos, F., Blanco, G., Gutiérrez, J. C., Luque, F., and Tortolero, M. (1993). Identification of an operon involved in the assimilatory nitrate-reducing system of Azotobacter vinelandii. Mol. Microbiol. 8, 1145–1153. doi: 10.1111/j.1365-2958.1993.tb01659.x
Ray, P. D., Huang, B. W., and Tsuji, Y. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 24, 981–990. doi: 10.1016/j.cellsig.2012.01.008
Rech, S., Deppenmeier, U., and Gunsalus, R. P. (1995). Regulation of the molybdate transport operon, modABCD, of Escherichia coli in response to molybdate availability. J. Bacteriol. 177, 1023–1029. doi: 10.1128/JB.177.4.1023-1029.1995
Rice, A. J., Harrison, A., Alvarez, F. J. D., Davidson, A. L., and Pinkett, H. W. (2014). Small substrate transport and mechanism of a molybdate atp binding cassette transporter in a lipid environment. J. Biol. Chem. 289, 15005–15013. doi: 10.1074/jbc.M114.563783
Richardson, D. J., Berks, B. C., Russell, D. A., Spiro, S., and Taylor, C. J. (2001). Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell. Mol. Life Sci. 58, 165–178. doi: 10.1007/PL00000845
Rivera-Chávez, F., Winter, S. E., Lopez, C. A., Xavier, M. N., Winter, M. G., Nuccio, S.-P., et al. (2013). Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Path. 9:e1003267. doi: 10.1371/journal.ppat.1003267
Robb, F. T., and Techtmann, S. M. (2018). Life on the fringe: microbial adaptation to growth on carbon monoxide. F1000Res 7:F1000 Faculty Rev-1981. doi: 10.12688/f1000research.16059.1
Romão, M. J., Coelho, C., Santos-Silva, T., Foti, A., Terao, M., Garattini, E., et al. (2017). Structural basis for the role of mammalian aldehyde oxidases in the metabolism of drugs and xenobiotics. Curr. Opin. Chem. Biol. 37, 39–47. doi: 10.1016/j.cbpa.2017.01.005
Rosas-Magallanes, V., Stadthagen-Gomez, G., Rauzier, J., Barreiro, L. B., Tailleux, L., Boudou, F., et al. (2007). Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect. Immun. 75, 504–7. doi: 10.1128/IAI.00058-06
Rossmann, R., Sawers, G., and Böck, A. (1991). Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol. Microbiol. 5, 2807–2814. doi: 10.1111/j.1365-2958.1991.tb01989.x
Rothery, R. A., and Weiner, J. H. (2015). Shifting the metallocentric molybdoenzyme paradigm: the importance of pyranopterin coordination. J. Biol. Inorg. Chem. 20, 349–372. doi: 10.1007/s00775-014-1194-6
Rothery, R. A., Workun, G. J., and Weiner, J. H. (2008). The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta Biomembr. 1778, 1897–1929. doi: 10.1016/j.bbamem.2007.09.002
Ruiz-Herrera, J., and DeMoss, J. A. (1969). Nitrate reductase complex of Escherichia coli K-12: participation of specific formate dehydrogenase and cytochrome b1 components in nitrate reduction. J. Bacteriol. 99, 720–729. doi: 10.1128/JB.99.3.720-729.1969
Santacruz, C. P., Balan, A., Ferreira, L. C. S., and Barbosa, J. A. R. G. (2006). Crystallization, data collection and phasing of the molybdate-binding protein of the phytopathogen Xanthomonas axonopodis pv. citri. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 289–291. doi: 10.1107/S1744309106003812
Santiago, B., Schübel, U., Egelseer, C., and Meyer, O. (1999). Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236, 115–124. doi: 10.1016/S0378-1119(99)00245-0
Sarnesto, A., Linder, N., and Raivio, K. O. (1996). Organ distribution and molecular forms of human xanthine dehydrogenase/xanthine oxidase protein. Lab. Invest. 74, 48–56.
Schoepp-Cothenet, B., van Lis, R., Philippot, P., Magalon, A., Russell, M. J., and Nitschke, W. (2012). The ineluctable requirement for the trans-iron elements molybdenum and/or tungsten in the origin of life. Sci. Rep. 2:263. doi: 10.1038/srep00263
Schräder, T., Rienhöfer, A., and Andreesen, J. R. (1999). Selenium-containing xanthine dehydrogenase from Eubacterium barkeri. Eur. J. Biochem. 264, 862–871. doi: 10.1046/j.1432-1327.1999.00678.x
Schrag, J. D., Huang, W., Sivaraman, J., Smith, C., Plamondon, J., Larocque, R., et al. (2001). The crystal structure of Escherichia coli MoeA, a protein from the molybdopterin synthesis pathway. J. Mol. Biol. 310, 419–431. doi: 10.1006/jmbi.2001.4771
Schroeder, H. A., Balassa, J. J., and Tipton, I. H. (1970). Essential trace metals in man: molybdenum. J. Chronic Dis. 23, 481–499. doi: 10.1016/0021-9681(70)90056-1
Schwarz, G., and Belaidi, A. A. (2013). Molybdenum in human health and disease. Met. Ions Life Sci. 13, 415–450. doi: 10.1007/978-94-007-7500-8_13
Schwarz, G., Mendel, R. R., and Ribbe, M. W. (2009). Molybdenum cofactors, enzymes and pathways. Nature 460, 839–847. doi: 10.1038/nature08302
Seelmann, C. S., Willistein, M., Heider, J., and Boll, M. (2020). Tungstoenzymes: occurrence, catalytic diversity and cofactor synthesis. Inorganics 8:44. doi: 10.3390/inorganics8080044
Self, W. T., Grunden, A. M., Hasona, A., and Shanmugam, K. T. (2001). Molybdate transport. Res. Microbiol. 152, 311–321. doi: 10.1016/S0923-2508(01)01202-5
Shaw, A. L., Leimkühler, S., Klipp, W., Hanson, G. R., and McEwan, A. G. (1999). Mutational analysis of the dimethylsulfoxide respiratory (dor) operon of Rhodobacter capsulatus. Microbiology 145, 1409–1420. doi: 10.1099/13500872-145-6-1409
Shiloh, M. U., Manzanillo, P., and Cox, J. S. (2008). Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3, 323–330. doi: 10.1016/j.chom.2008.03.007
Sohaskey, C. D. (2008). Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J. Bacteriol. 190, 2981–2986. doi: 10.1128/JB.01857-07
Sohaskey, C. D., and Wayne, L. G. (2003). Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J. Bacteriol. 185, 7247–7256. doi: 10.1128/JB.185.24.7247-7256.2003
Song, T., Park, S. W., Park, S. J., Kim, J. H., Yu, J. Y., Oh, J. I., et al. (2010). Cloning and expression analysis of the duplicated genes for carbon monoxide dehydrogenase of Mycobacterium sp strain JC1 DSM 3803. Microbiology 156, 999–1008. doi: 10.1099/mic.0.034769-0
Sparacino-Watkins, C., Stolz, J. F., and Basu, P. (2014). Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 43, 676–706. doi: 10.1039/C3CS60249D
Spector, M. P., Garcia Del Portillo, F., Bearson, S. M. D., Mahmud, A., Magut, M., Finlay, B. B., et al. (1999). The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvation-inducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiology 145, 3035–3045. doi: 10.1099/00221287-145-11-3035
Spees, A. M., Wangdi, T., Lopez, C. A., Kingsbury, D. D., Xavier, M. N., Winter, S. E., et al. (2013). Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. Mbio 4, e00430–e00413. doi: 10.1128/mBio.00430-13
Srivastava, V. K., Srivastava, S., Arora, A., and Pratap, J. V. (2013). Structural insights into putative molybdenum cofactor biosynthesis protein C (MoaC2) from Mycobacterium tuberculosis H37Rv. PLoS ONE 8:e58333. doi: 10.1371/annotation/49d4d96b-fafa-43cc-8c98-6146816656ed
Stewart, V. (1982). Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J. Bacteriol. 151, 1320–1325. doi: 10.1128/JB.151.3.1320-1325.1982
Stewart, V., Lu, Y., and Darwin, A. J. (2002). Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184, 1314–1323. doi: 10.1128/JB.184.5.1314-1323.2002
Tan, M. P., Sequeira, P., Lin, W. W., Phong, W. Y., Cliff, P., Ng, S. H., et al. (2010). Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS ONE 5:e13356. doi: 10.1371/journal.pone.0013356
Tareen, A. M., Dasti, J. I., Zautner, A. E., Gro,ß, U., and Lugert, R. (2011). Sulphite : cytochrome c oxidoreductase deficiency in Campylobacter jejuni reduces motility, host cell adherence and invasion. Microbiology 157, 1776–1785. doi: 10.1099/mic.0.045567-0
Taveirne, M. E., Sikes, M. L., and Olson, J. W. (2009). Molybdenum and tungsten in Campylobacter jejuni: their physiological role and identification of separate transporters regulated by a single ModE-like protein. Mol. Microbiol. 74, 758–771. doi: 10.1111/j.1365-2958.2009.06901.x
Terao, M., Garattini, E., Romão, M. J., and Leimkühler, S. (2020). Evolution, expression, and substrate specificities of aldehyde oxidase enzymes in eukaryotes. J. Biol. Chem. 195, 5377–5389. doi: 10.1074/jbc.REV119.007741
Tirado-Lee, L., Lee, A., Rees, D. C., and Pinkett, H. W. (2011). Classification of a Haemophilus influenzae ABC transporter HI1470/71 through its cognate molybdate periplasmic binding protein, MolA. Structure 19, 1701–1710. doi: 10.1016/j.str.2011.10.004
Tombline, G., Schwingel, J. M., Lapek, J. D. Jr., Friedman, A. E., Darrah, T., Maguire, M., et al. (2013). Pseudomonas aeruginosa PA1006 is a persulfide-modified protein that is critical for molybdenum homeostasis. PLoS ONE 8:e55593. doi: 10.1371/journal.pone.0055593
Turner, R. J., Papish, A. L., and Sargent, F. (2004). Sequence analysis of bacterial redox enzyme maturation proteins (REMPs). Can. J. Microbiol. 50, 225–238. doi: 10.1139/w03-117
Unden, G., Steinmetz, P. A., and Degreif-Dünnwald, P. (2014). The aerobic and anaerobic respiratory chain of Escherichia coli and Salmonella enterica: enzymes and energetics. EcoSal Plus 6. doi: 10.1128/ecosalplus.ESP-0005-2013
Van Alst, N. E., Picardo, K. F., Iglewski, B. H., and Haidaris, C. G. (2007). Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect. Immun. 75, 3780–3790. doi: 10.1128/IAI.00201-07
Wahl, B., Reichmann, D., Niks, D., Krompholz, N., Havemeyer, A., Clement, B., et al. (2010). Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J. Biol. Chem. 285, 37847–37859. doi: 10.1074/jbc.M110.169532
Wang, C. H., Zhang, C., and Xing, X. H. (2016). Xanthine dehydrogenase: an old enzyme with new knowledge and prospects. Bioengineered 7, 395–405. doi: 10.1080/21655979.2016.1206168
Wang, H., and Gunsalus, R. P. (2003). Coordinate regulation of the Escherichia coli formate dehydrogenase fdnGHI and fdhF genes in response to nitrate, nitrite, and formate: roles for NarL and NarP. J. Bacteriol. 185, 5076–5085. doi: 10.1128/JB.185.17.5076-5085.2003
Wang, H., Tseng, C. P., and Gunsalus, R. P. (1999). The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J. Bacteriol. 181, 5303–5308. doi: 10.1128/JB.181.17.5303-5308.1999
Warelow, T. P., Oke, M., Schoepp-Cothenet, B., Dahl, J. U., Bruselat, N., Sivalingam, G. N., et al. (2013). The respiratory arsenite oxidase: structure and the role of residues surrounding the rieske cluster. PLoS ONE 8:e72535. doi: 10.1371/journal.pone.0072535
Wayne, L. G., and Hayes, L. G. (1998). Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber. Lung Dis. 79, 127–132. doi: 10.1054/tuld.1998.0015
Weber, I., Fritz, C., Ruttkowski, S., Kreft, A., and Bange, F.-C. (2000). Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35, 1017–1025. doi: 10.1046/j.1365-2958.2000.01794.x
Weerakoon, D. R., Borden, N. J., Goodson, C. M., Grimes, J., and Olson, J. W. (2009). The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb. Pathog. 47, 8–15. doi: 10.1016/j.micpath.2009.04.009
Wegiel, B., Larsen, R., Gallo, D., Chin, B. Y., Harris, C., Mannam, P., et al. (2014). Macrophages sense and kill bacteria through carbon monoxide–dependent inflammasome activation. J. Clin. Investig. 124, 4926–4940. doi: 10.1172/JCI72853
Weingarten, R. A., Grimes, J. L., and Olson, J. W. (2008). Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 74, 1367–1375. doi: 10.1128/AEM.02261-07
Wells, M., Kanmanii, N. J., Al Zadjali, A. M., Janecka, J. E., Basu, P., Oremland, R. S., et al. (2020). Methane, arsenic, selenium and the origins of the DMSO reductase family. Sci. Rep. 10:10946. doi: 10.1038/s41598-020-67892-9
Williams, M., Mizrahi, V., and Kana, B. D. (2014). Molybdenum cofactor: a key component of Mycobacterium tuberculosis pathogenesis? Crit. Rev. Microbiol. 40, 18–29. doi: 10.3109/1040841X.2012.749211
Williams, M. J., Shanley, C. A., Zilavy, A., Peixoto, B., Manca, C., Kaplan, G., et al. (2015). Bis-Molybdopterin guanine dinucleotide is required for persistence of Mycobacterium tuberculosis in guinea pigs. Infect. Immun. 83, 544–550. doi: 10.1128/IAI.02722-14
Winter, S. E., Lopez, C. A., and Bäumler, A. J. (2013a). The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 14, 319–327. doi: 10.1038/embor.2013.27
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Winter, S. E., Winter, M. G., Xavier, M. N., Thiennimitr, P., Poon, V., Keestra, A. M., et al. (2013b). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711. doi: 10.1126/science.1232467
Workun, G. J., Moquin, K., Rothery, R. A., and Weiner, J. H. (2008). Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold. Microbiol. Mol. Biol. Rev. 72, 228–248. doi: 10.1128/MMBR.00041-07
World Health Organization (2017). Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. Geneva: WHO. Available online at: https://www.who.int/medicines/areas/rational_use/prioritizationof-pathogens/en/
Wuebbens, M. M., Liu, M. T., Rajagopalan, K., and Schindelin, H. (2000). Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC. Structure 8, 709–718. doi: 10.1016/S0969-2126(00)00157-X
Xi, H., Schneider, B. L., and Reitzer, L. (2000). Purine catabolism in Escherichia coli and function of xanthine dehydrogenase in purine salvage. J. Bacteriol. 182, 5332–5341. doi: 10.1128/JB.182.19.5332-5341.2000
Xia, Z., Lei, L., Zhang, H.-Y., and Wei, H.-L. (2018). Characterization of the ModABC molybdate transport system of Pseudomonas putida in nicotine degradation. Front. Microbiol. 9:3030. doi: 10.3389/fmicb.2018.03030
Xiang, S., Nichols, J., Rajagopalan, K. V., and Schindelin, H. (2001). The crystal structure of Escherichia coli MoeA and its relationship to the multifunctional protein gephyrin. Structure 9, 299–310. doi: 10.1016/S0969-2126(01)00588-3
Yamamoto, I., and Ishimoto, M. (1977). Anaerobic growth of Escherichia coli on formate by reduction of nitrate, fumarate, and trimethylamine N-oxide. Z. Allg. Mikrobiol. 17, 235–242. doi: 10.1002/jobm.19770170309
Yokoyama, K., and Lilla, E. A. (2018). C-C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products. Nat. Prod. Rep. 35, 660–694. doi: 10.1039/C8NP00006A
Zacharia, V. M., and Shiloh, M. U. (2012). Effect of carbon monoxide on Mycobacterium tuberculosis pathogenesis. Med. Gas Res. 2, 30–30. doi: 10.1186/2045-9912-2-30
Zempleni, J., Wijeratne, S. S. K., and Hassan, Y. I. (2009). Biotin. BioFactors 35, 36–46. doi: 10.1002/biof.8
Zhang, H. B., Chen, X. H., Nolan, L. K., Zhang, W., and Li, G. W. (2019). Identification of host adaptation genes in extraintestinal pathogenic Escherichia coli during infection in different hosts. Infect. Immun. 87:19. doi: 10.1128/IAI.00666-19
Zhang, W., Urban, A., Mihara, H., Leimkühler, S., Kurihara, T., and Esaki, N. (2010). IscS functions as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in Escherichia coli. J. Biol. Chem. 285, 2302–2308. doi: 10.1074/jbc.M109.082172
Keywords: infectious disease, metabolic pathways, pyranopterin molybdenum enzymes, bacterial pathogen, molybdenum enzymes, virulence, energy generation, metalloenzyme
Citation: Zhong Q, Kobe B and Kappler U (2020) Molybdenum Enzymes and How They Support Virulence in Pathogenic Bacteria. Front. Microbiol. 11:615860. doi: 10.3389/fmicb.2020.615860
Received: 10 October 2020; Accepted: 23 November 2020;
Published: 11 December 2020.
Edited by:
Jai Justin Tree, University of New South Wales, AustraliaReviewed by:
Silke Leimkuehler, University of Potsdam, GermanyRalf R. Mendel, Technical University of Braunschweig, Germany
Copyright © 2020 Zhong, Kobe and Kappler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulrike Kappler, dS5rYXBwbGVyJiN4MDAwNDA7dXEuZWR1LmF1
 Qifeng Zhong
Qifeng Zhong Bostjan Kobe
Bostjan Kobe Ulrike Kappler
Ulrike Kappler