
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 December 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.613704
Bacteria face diverse stresses in the environment and, sometimes, respond by forming multi-cellular structures, e.g., biofilms. Here, we report a novel macroscopic and multi-cellular structure formed by Salmonella Typhimurium, which resembles small strings. These string-like structures, ∼1 cm long, are induced under some stress conditions: iron deprivation by 2,2-Bipyridyl or low amounts of antibiotics or ethanol in minimal media. However, cells in strings revert back to planktonic growth upon return to nutrient rich media. Compared to planktonic cells, strings are more resistant to antibiotics and oxidative stress. Also, strains lacking csgD or rpoS, which are defective in the classical rdar biofilm formation, form strings. Furthermore, some biofilm inducing conditions do not result in strings and vice-versa, demonstrating that strings are not related to classical CsgD-dependent biofilms. Cells in a string are held together by cellulose and a strain lacking bcsA, which is defective in cellulose production, does not form strings. In addition, reductive stress conditions such as dithiothreitol (DTT) or mutations in the Disulfide bonding system (DSB) also give rise to strings. The amounts of c-di-GMP are increased upon string formation and studies with single and double deletion strains of the diguanylate cyclases, yedQ (STM1987) primarily and yfiN (STM2672) partly, revealed their importance for string formation. This is the first study showcasing the ability of Salmonella to produce high amounts of cellulose in liquid culture, instead of an interface, in a CsgD-independent manner. The relevance and possible applications of strings in the production of bacterial cellulose and bioremediation are discussed.
Bacteria constantly assess their environment for nutrient availability, presence of chemical messengers, stress-inducing chemicals etc. These cues dictate bacterial responses, resulting in changes in morphology at a single cell level or forming multi-cellular structures. (Galperin, 2018). An array of bacterial multi-cellular structures has been described, including those made by filamentous bacteria e.g., Candidatus, Mycobacterial cording in detergent free media, aggregates of Zymomonas in minimal media, clumping behavior by certain Rhodobacter mutants, to the most well studied ones like biofilms which could be composed of a single or multiple species (Puskas et al., 1997; Shapiro, 1998; Rudolph et al., 2001; Bernut et al., 2014; Lyons and Kolter, 2015; Jones-Burrage et al., 2019). Multi-cellular structures, the most well studied being biofilms, can be induced by a myriad of stresses including differences in temperature, osmolarity, nutrient availability etc. (Stanley and Lazazzera, 2004; Ray et al., 2020). Sub-Minimum Inhibitory Concentration (MIC) levels of antibiotics are also known to induce biofilms. Biofilms are known to be more resistant to antibiotics (up to a 1000-fold) and other stresses over planktonic cells by forming a barrier, reducing cellular activity, increasing expression of multiple drug resistance pumps, expression of glycans that trap antibiotics etc. (Mah and O’Toole, 2001; Mah et al., 2003; Høiby et al., 2010; Kaplan, 2011).
Salmonella Typhimurium, an intracellular pathogen, is known to form robust biofilms at interfaces/surfaces in lab settings as well as inside the host gall bladder, where it resides during the carrier state (Mireles et al., 2001; Crawford et al., 2010; Pontes et al., 2015). Cells in the Salmonella biofilms are held together by various proteinaceous components like curli fimbriae, exopolysaccharides like cellulose, and others like colanic acid, enterobacterial common antigen, O-antigen capsule, extracellular DNA (Pontes et al., 2015). Also, S. Typhimurium biofilm formation is controlled by the bacterial secondary messenger c-di-GMP and the transcription factor CsgD, which control production of various extracellular polymers as well as quorum sensing molecules (Xavier and Bassler, 2003; Parsek and Greenberg, 2005; Ahmad et al., 2017). However, a CsgD-independent Type 1 fimbriae mediated biofilm formation has also been reported (Monteiro et al., 2012). The two major extracellular components of Salmonella biofilms are curli fimbriae and cellulose. The production of cellulose depends on the cellulose synthase complex, consisting of members of the bcs operon and c-di-GMP which allows the cellulose synthase complex to become active (Römling and Galperin, 2015). Levels of c-di-GMP are in turn controlled by GGDEF-domain containing diguanylate cyclases (DGCs) which produce c-di-GMP and EAL or HD-GYP domain containing phosphodiesterases (PDEs) which degrade c-di-GMP (Simm et al., 2014). Salmonella has been reported to produce cellulose in multiple conditions such as low osmolarity in rich medium, in nutrient deficient medium such as ATM and inside macrophages (Römling, 2002; Stanley and Lazazzera, 2004; Pontes et al., 2015; Römling and Galperin, 2015). Recent studies in E. coli and Salmonella have, however, shown that cellulose production may occur in a manner independent of transcription factor CsgD via DGC YedQ or under reductive stresses via YfiN (Da Re and Ghigo, 2006; Grantcharova et al., 2010; Hufnagel et al., 2014; Römling and Galperin, 2015; Ahmad et al., 2016). Notably, cellulose production represses bacterial virulence (Pontes et al., 2015; Römling and Galperin, 2015). Cellulose produced by E. coli and Salmonella Typhimurium is a modified form, wherein alternate glycosyl residues in the cellulose carry a phosphoethanoloamine group (Galperin and Shalaeva, 2018; Thongsomboon et al., 2018). Interestingly, bacterial cellulose is a material of great biomedical significance and cellulose produced by Gluconacetobacter hanseni is presently being used for burn wound dressings. Bacterial cellulose is suitable for this due to properties such as high-water retention capacity, low toxicity, low immunogenicity and ease of purification compared to plant cellulose (Fu et al., 2013; Portela et al., 2019).
Apart from swarming motility and classical biofilms, group behavior/structures in Salmonella have not been reported extensively. Our lab has studied the various genetic and environmental factors that affect biofilm formation and stress responses by Salmonella (Kumar and Nandi, 2007; Bhosale et al., 2013; Ray et al., 2019; Ray et al., 2020; Thakur et al., 2020). While studying some stress responses, we observed macroscopic string-like structures. In this study, we report this novel string-like multi-cellular structure dependent on cellulose in Salmonella Typhimurium that is induced under certain stress conditions, e.g., iron deprivation by 2,2 –Bipyridyl (Bipd), Dithiothreitol (DTT), sub-MIC amounts of antibiotics in minimal media. We find that this macroscopic structure is not related to classical CsgD-dependent biofilms and confers increased resistance to various stresses compared to planktonic cells. We also report that high amounts of c-di-GMP are found in strings and the diguanylate cyclases, yedQ primarily and yfiN partly, are important for formation of strings.
The bacterial strains used in this study are listed in Table 1. All cultures were grown in Luria-Bertani (LB) medium consisting of 10 g of tryptone (Himedia, Mumbai, India), 10 g of NaCl (Merck, Mumbai, India) and 5 g of yeast extract (Himedia) per liter at 37°C with constant shaking at 180 rpm. Single colony cultures grown for 8 h-10 h served as pre-inoculum cultures for all experiments. Antibiotics were used at 30 μg/ml chloramphenicol, 50 μg/ml of streptomycin (Sigma, United States), 100 μg/ml ampicillin (Himedia). Single gene knockouts were generated using one step chromosomal gene inactivation as described using plasmids and primers in Tables 1 and 2 (Datsenko and Wanner, 2000). The antibiotic resistance cassette was then removed using pCP20 before being used.
Single and double gene knockouts were generated using one step chromosomal gene inactivation as previously described using plasmids and primers in Tables 1 and 2 (Datsenko and Wanner, 2000). Double knockouts were constructed after the removal of the antibiotic resistance cassette from the single gene knockouts (Ray et al., 2019). The antibiotic resistance cassette was then removed using pCP20 before being used.
M9 minimal medium was made as per the Cold Spring Harbor Laboratory protocol: 5X salt solution (per liter: 33.92 g Na2HPO4, 15 g KH2PO4, 5 g NH4Cl, all from Fisher Scientific, Mumbai, India) 2.5 g NaCl (Merck), 2 mM MgSO4 (Fisher Scientific), 100 μM CaCl2 (Merck), 4 g Glucose (Himedia). To induce strings unless mentioned 200 μM of 2,2-Bipyridyl (Bipd) (Sigma) was added to M9 minimal medium. Strings were harvested using sterile cut-tips after 10 hours at 37 °C with constant shaking at 180 rpm and processed appropriately. Planktonic growth was measured as optical density at 600 nm.
Strings were washed thoroughly twice with sterile PBS to remove planktonic cells and were stained using 50 μl of 2.5 μM of SYTO 9 (Molecular Probes, OR, United States), followed by incubation at room temperature at 90 rpm for 3 hours. The fluorescence intensity was measured with 488 nm excitation and 528 nm emission wavelength (Römling and Galperin, 2015) using Tecan Infinite PRO 200 (Austria). Due to fluctuations in laser intensity during some experiments involving M9 + Bipd, fluorescence values in those experiments were normalized such that the average WT fluorescence in M9 + Bipd was set at 10000 a.u.
Both strings and planktonic cells were harvested at 10 h and washed with PBS. They were then placed on a glass slide and 100 μl of 100 μg/ml Fluorescent Brightener 28 solution (pH 10) (Sigma) was added and incubated for 5 min followed by washing to remove excess stain before confocal imaging. For Calcofluor staining on LB minus NaCl agar plates, 20 μg/ml of Calcofluor white was added (Fluka, Canada).
Strings approximately 0.5 cm were isolated at 6-hour time point from M9 + 200 μM 2,2- Bipyridyl and washed with PBS once. Strings were stained with 2.5 μM of CFSE (Sigma) in PBS and kept for 90 minutes at 37°C and 180 rpm. The excess stain was washed using PBS. The washed strings were inoculated in fresh M9 + 200 μM 2,2- Bipd + Glu for specified time points and in M9- Glu + 200 μM 2,2- Bipd for the remaining time that would add up to 18 hours at 37°C and 180 rpm. Strings were harvested after 18 hours and prepared for confocal imaging.
Planktonic cells and strings at 10-hour time points were taken for the experiment. 50 μl of 2 OD culture or strings roughly 1 cm in length were added to M9 medium for 3 h at 37°C and 180 rpm. The viability of planktonic cells was determined using CFU enumeration on SS Agar plates after plating appropriate dilutions. CFU/ml was then compared among various conditions to estimate survival. CFU was used for planktonic cells, since single cells would most likely get lysed due to ampicillin and kanamycin treatments.
The viability of cells in a string was determined using Propidium Iodide staining. Strings were washed twice with PBS and stained with 15 μM of PI for 20 min at 37°C and 180 rpm. The excess PI was removed by gentle washing.
To calculate the viability of strings (VT) following treatment ‘T’, the following equation was used,
where, FT is Mean Fluorescence Intensity (MFI) of PI stained strings post treatment T, FD is MFI of dead strings and FM9 for strings kept in M9 alone.
To obtain the formula, we assumed that the all cells in the strings kept in M9 alone were viable, leading to FM9 being the background signal due to basal auto-fluorescence or small amount of residual stain after washing. We have also taken FD to be the maximum MFI, since all cells in a string treated with Isopropanol were dead (by observing no growth after adding washed isopropanol treated strings into fresh LB, data not shown). Thus, FD-FM9 represents the difference in MFI when all cells are either dead or alive. Any intermediate survival reflected by FT can be translated to percentage viability by removing the fluorescence due dead cells shown by FT.
E.coli (MG1655) was transformed with (pProEX HTb mCherry) and S. Typhimurium with (pProEX HTb GFP). Pre-inoculums were made with LB medium containing 100 μg/ml ampicillin and 400 μM IPTG. The bacteria were mixed in 1:1 ratio amounting to a total of 10^8 cells in total.
P.aeruginosa was stained with 10 mM CFSE (Sigma) for 3 h and then washed twice to remove excess stain. These cells and WT S. Typhimurium were mixed in 1:1 ratio amounting to a total of 10^8 cells.
Strings post staining and washing were placed on a glass coverslip and fixed using 4% PFA for 30 min at room temperature. Excess PFA was removed by gentle washing with PBS. 1% DABCO was added as the anti-fade. Imaging was done using Leica SP8 confocal microscope and images were analyzed using Fiji (Schindelin et al., 2012).
Strings isolated at 10 h were washed three times with sterile MilliQ water to remove any residual media components. The strings were then placed onto a poly-L-lysine coated glass coverslip. The samples were then air-dried at room temperature in a laminar flow hood for 1 h. The non-adherent bacteria were removed by washing the coverslips once with MilliQ water. These were again air-dried for 1 h. AFM imaging was performed as previously reported (Bhaskarla et al., 2016; Verma et al., 2018).
bcsA was cloned under trc promoter in pTrc99a plasmid. The gene encoding bcsA was amplified from WT 14028s genomic DNA using Phusion polymerase using appropriate primers (Table 2). The amplified DNA was digested using Xba1 and Sac1 and ligated into the pTrc99a vector digested with the same enzymes to yield the plasmid pbcsA and verified using sequencing.
The protocol for c-di-GMP extraction and quantitation was the one previously reported (Petrova and Sauer, 2017). 1 ml of 2 OD cells and strings isolated from 20 ml of culture were used for this assay. Post extraction the c-di-GMP containing pellet was resuspended in 200 μl of nanopure water just before use (stored at −80°C). 20 μl was injected and analyzed using HPLC (Thermo Finnigan Surveyor HPLC) and compared to standard plots generated with sodium salt of c-di-GMP (HPLC grade, Sigma) at concentrations 1, 2, 5, 10 and 20 pmol. The values of c-di-GMP detected were then normalized for protein content.
Biofilms were generated in 12 well plastic plates (Tarsons, India) in LB minus NaCl, M9, M9 + 200 μM 2,2- Bipyridyl. 50 μl of 2 OD cells were inoculated in 1.5 ml media and kept at 30°C for 5 days in static condition. Biofilms were quantitated using 1% (w/v) crystal violet staining and 33% (v/v) acetic acid extraction and the absorbance was measured at 570 nm (Ray et al., 2020; Thakur et al., 2020).
Total RNA was extracted from WT cells and strings after 10 h using TRIzol reagent (Sigma, St. Louis, MO, United States), according to manufacturer’s instructions. The pellet was dried and dissolved in 25 μL Nuclease free water (NEB, MA, United States). RNA concentration and purity were quantified using NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA, United States). This was followed by DNase treatment of 1 μg of RNA using a Turbo DNA free kit (Invitrogen, United States) as per manufacturer’s instructions. Subsequently, 500ng of RNA was reverse transcribed to cDNA using RevertAid (Thermo Scientific).
cDNA was diluted 1:20 and analyzed using BioRad CFX-Connect Real Time PCR Detection system (BioRad, CA, United States) with SYBR Green I detection. Each sample was measured in triplicate in a 96-well plate (BioRad) in a reaction mixture (10 μL final volume) containing 2X SYBR iQ SYBR Green supermix and 10 μM primer mix (Supplementary Table 2). The threshold cycles (Ct) were calculated using the iQ5 Optical system software, and fold changes normalized to the reference control rrlC were determined using the 2–ΔΔCt method (Ray et al., 2020).
Initially, strings formed by Salmonella Typhimurium 14028s were observed when 200 μM of 2,2-Bipyridyl (Bipd) was added to M9 medium with 0.4% (w/v) glucose as carbon source. We observed around 1-3 strings per 5 ml of media. A protocol was then designed, after optimizing dose and time points, to quantify strings using SYTO9 a nucleic acid stain which binds DNA (Figures 1A,B, Supplementary Figures 1, 2). SYTO9 stains both live and dead Salmonella Typhimurium and has been used for biofilm quantitation (Berney et al., 2007; Peeters et al., 2008). Extensive cell-cell contact due to the rotating motion of the vessel containing the culture was required for string formation and a larger proportion of cells entered the string with increasing RPM as shown by the decreasing normalized OD with increasing RPM (Supplementary Figures 2C,D). After calibrating SYTO9 fluorescence with known number of single cells, we found that a single string on average contains nearly a billion cells (Supplementary Figure 3). These structures were, however, not formed upon addition of Bipd to LB (Figures 1A,B). Strings were also found to have an average length of ∼ 1 cm (Figure 1C). Confocal imaging of strings formed by GFP positive S. Typhimurium demonstrated that strings were composed of cells, but they were not packed in any discernible pattern (Figure 1D) and confirmed by AFM imaging (Figure 1E). To rule out strain specific effects, string formation was studied in the S. Typhimurium SL1344 strain in M9 containing 200 μM Bipd (M9 plus Bipd). S. Typhimurium SL1344 also formed strings in similar amounts to the 14028s strain, demonstrating that string formation was not strain specific (Supplementary Figure 4).
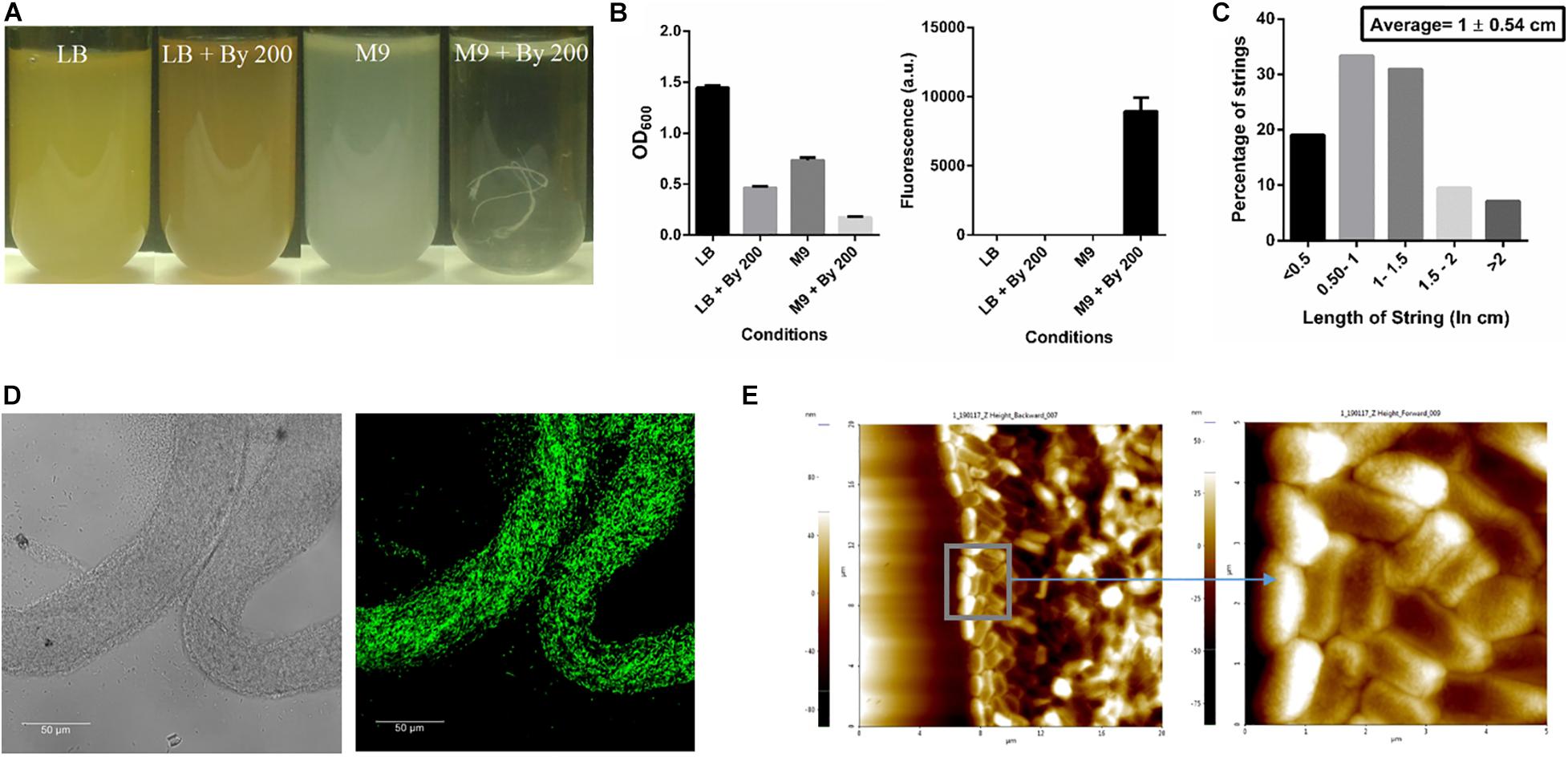
Figure 1. SalmonellaTyphimurium under Bipyridyl stress in minimal medium produces string-like structures. (A) Representative images of test tubes of Salmonella Typhimurium after 10 h at 37°C under 180 rpm in different growth conditions: Nutrient rich media (LB), Nutrient rich media with By 200 (200 μM 2,2 Bipyridyl, Fe2+ chelator) (LB + By 200), Minimal media (M9) and Minimal media with By 200 (M9 + By 200/M9 + Bipd) containing 0.4% (w/v) glucose as the carbon source, with white string like structures clearly visible (B) Quantification of strings formed using SYTO9 (nucleic acid stain, staining both live and dead Salmonella cells) after through washing to remove planktonic cells and planktonic growth under various conditions (n = 3) (C) Histogram depicting distribution of string length with average ± SD (n > 40) under M9 + By 200 conditions (D) Confocal Images of a strings formed by cells harboring GFP expressing plasmid at 63X (scale bar 50 μm) (E) Atomic Force Microscopy (AFM) imaging of strings to study arrangement of cells with 4X magnification of inset (20 μm and 5 μm squares). Data is represented as mean ± SEM.
We studied if other bacterial species could also form strings. E. coli MG1655 did not form macroscopic strings under Bipd stress although the planktonic growth was similar to S. Typhimurium (Supplementary Figure 4). We then studied whether multi-species strings could be formed like multi-species biofilms (Galperin, 2018). When both E. coli and S. Typhimurium were inoculated in 1:1, multi-species could be formed, suggesting E. coli could become a part of Salmonella strings (Supplementary Figure 5A). Although Pseudomonas aeruginosa did not grow well in M9 alone itself (data not shown), it could also become part of multi-species strings when P. aeruginosa and S. Typhimurium were inoculated in a 1:1 ratio (Supplementary Figure 5B).
Next, we studied whether string formation would help in evasion of the inducing stress. Bipd has been shown to exert a bacteriostatic effect due to chelation of Fe2+, an essential micronutrient. (Jurado, 1997; Abdul-Tehrani et al., 1999). First, we verified that the Bipd effect was dependent on Fe2+ chelation: the addition of fresh FeSO4 to minimal media containing Bipd restored planktonic growth to levels observed in M9 alone (Figure 2A). We also observed a reduction in string formation with the addition of FeSO4, we could not quantify it due to conversion of soluble Fe2+ to insoluble Fe3+ salt precipitates which could not be washed off from strings. We then verified that Bipd exerted a bacteriostatic effect on planktonic cells by observing no significant increase in growth from 4h to 24h (Figure 2B). If strings could evade the bacteriostatic effect, then cells inside the string would divide. Carboxyfluorescein succinimidyl ester (CFSE) is a protein staining dye, exhibiting fluorescence properties similar to GFP, that is routinely used to study cell division in various immune cells and has been used for bacteria as well (Lyons, 2000; Vander Top et al., 2006). The method is based on initially prestaining cells with CFSE which is non-toxic. As cells grow, the protein content increases and gets divided into the daughter cells. Therefore, the CFSE dye bound to the proteins would also get divided between the daughter cells (Lyons, 2000). Thus, dilution of CFSE dye acts as a proxy for measuring cell growth and division. To study if strings could evade Bipd’s bacteriostatic effect, we stained strings (6h old formed in M9 + Bipd) with CFSE and then transferred them to fresh M9 + Bipd in the absence or presence of 0.4% (w/v) glucose. M9 + Bipd not containing glucose acted as a control to ensure reduction in CFSE fluorescence was not due to bleaching or decay of the dye. The CFSE fluorescence intensity in M9 + Bipd containing glucose was significantly lower in both 9 hr and 18 hr time points compared to the control, suggesting that cells in the strings divided and were able to evade the bacteriostatic effects of Bipd (Figures 2C,D). To verify whether string formation was a stress response, we inoculated washed strings into fresh nutrient rich LB media and investigated whether cells in strings could revert to a planktonic state upon reversion to favorable conditions. We reasoned that if cells came together to form strings to evade stress, they would be able to revert back upon removal of stress inducing conditions. We observed that cells in the strings did revert to a planktonic state, since the OD data reached comparable levels of the inoculum of ∼1 billion planktonic cells (grown in M9) by 6 h (Figure 2E).
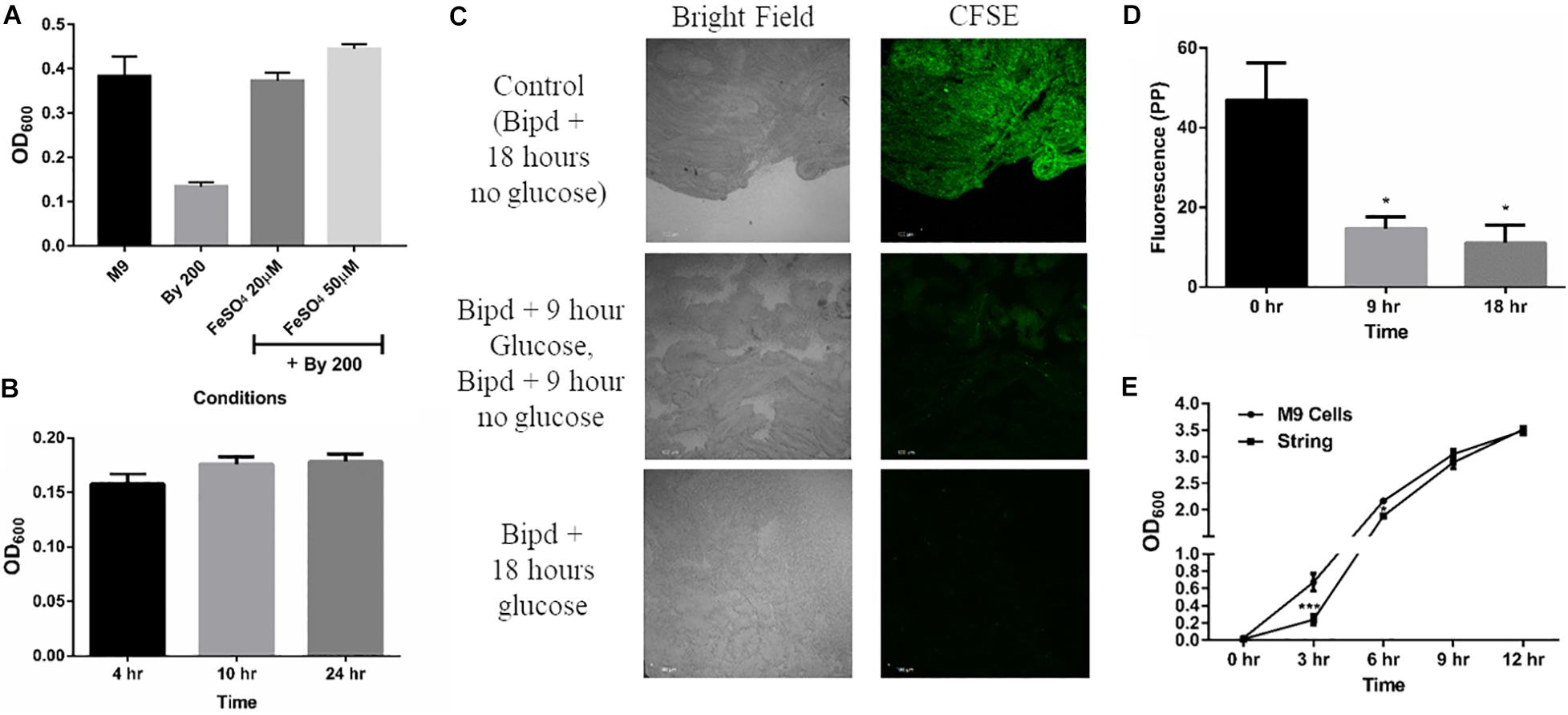
Figure 2. String-like structure help in the evasion of Bipyridyl induced bacteriostasis. (A) Planktonic growth in M9 + 200 μM 2,2-Bipyridyl upon supplementation with Fe2+ by adding fresh FeSO4 at indicated concentrations (n = 3). (B) Planktonic growth of cells in M9 + Bipd at the varying time points (n = 3). (C) Imaging of Strings pre-stained using CFSE (Carboxyfluorescein succinimidyl ester, stains cell proteins) after 18 h in M9 + Bipd with or without C source (glucose at 0.4% w/v) for indicated time points to visualize cell division via reduction of CFSE fluorescence. Control of strings in media with no glucose for 18 h to account for reduction in CFSE fluorescence due to bleaching. Images at 20X (scale bar 100 μm) (n = 3). (D) Quantitation of CFSE fluorescence to detect protein synthesis and cell division * = p < 0.05, One-way ANOVA from images in 2C (E) Planktonic growth emanating from strings harvested from M9 + Bipd and control planktonic M9 cells (Starting at 10^7 CFU/ml) upon removal of Bipd stress and entering nutrient rich media (LB) at varying time points. * = p < 0.05, *** = p < 0.005 Two-way ANOVA. Data is represented as mean ± SEM.
Next, we studied whether strings, like biofilms, were equipped to resist higher doses of antibiotics like ampicillin and kanamycin and other stresses such as oxidative stress, compared to planktonic cells (Mah and O’Toole, 2001; Høiby et al., 2010; Kaplan, 2011) To study the death induced by various stresses in strings, we used Propidium Iodide (PI) staining and then measured relative fluorescence intensity of each treatment group between untreated strings kept in fresh M9 and strings killed with 60% isopropanol to quantify the percentage of dead cells in strings. Planktonic cells subjected to similar stresses were plated on SS agar plates and CFU/ml was calculated and compared to the same controls. PI staining was not used for the planktonic cells since both ampicillin and kanamycin would lyse the planktonic cells (Heller and Spence, 2019). Strings were more resistant to higher doses of both bactericidal antibiotics tested, e.g., Ampicillin and Kanamycin (> 100 fold) compared to planktonic cells (Figures 3A,B). In oxidative stress, strings were more resilient and showed > 1000-fold survival compared to planktonic cells (Figure 3A).
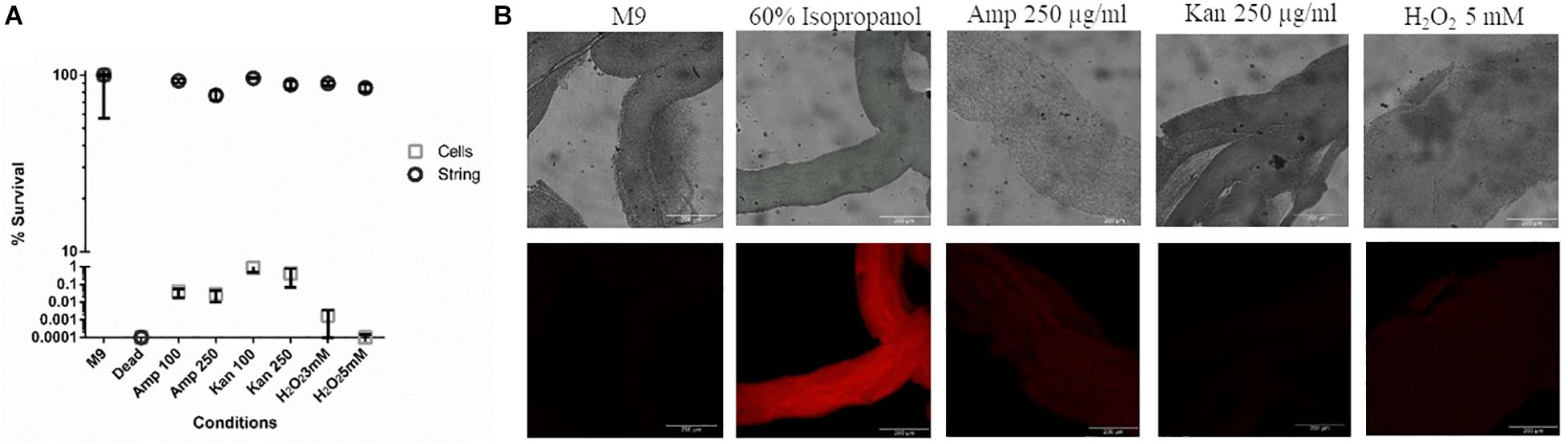
Figure 3. Strings confer protection against high doses of antibiotics and oxidative stress (A) The percentage survival of washed strings formed in M9 + Bipd and planktonic cells in M9 after 3 h of treatment under 8-20 fold MBC (Minimum Bactericidal Concentration) of ampicillin, kanamycin or oxidative stress individually at multiple doses in fresh M9. Viability of strings determined by PI staining and planktonic cells by plating (instead of PI due to single cell lysis). (60% Isopropanol treatment kills all cells in the string, Error for dead (strings) is 7.53%, kanamycin (Kan) and ampicillin (Amp) in μg/ml) (n = 3). (B) Confocal images of PI stained strings post varying stresses for 3 h taken at 20X (scale bar 200 μm) (n = 3). The data is represented as mean ± SEM.
Given that S. Typhimurium forms robust biofilms at surfaces/interfaces, it was important to address if strings are related to classical CsgD-dependent biofilms. The approach was to determine whether conditions that induce these biofilms could also induce strings and vice-versa. While low osmolarity (LB minus NaCl) condition produced robust biofilms, it did not give rise to strings. Also, ethanol which also led to strings in M9, did not induce biofilm formation while giving robust string formation (Supplementary Figures 6B,C). Further verification was performed using mutants known to be defective in classical CsgD-dependent biofilm formation, e.g., strains lacking CsgD or RpoS. As expected, the ΔcsgD and ΔrpoS strains were defective in biofilm formation in the LB minus NaCl, M9 and M9 plus Bipd conditions compared to WT (Grantcharova et al., 2010; Mika and Hengge, 2014; Figure 4A). However, string formation was similar in all three strains (Figures 4B,C). These two observations together demonstrate that mechanisms involved in classical CsgD-dependent biofilm formation and string formation in Salmonella are distinct.
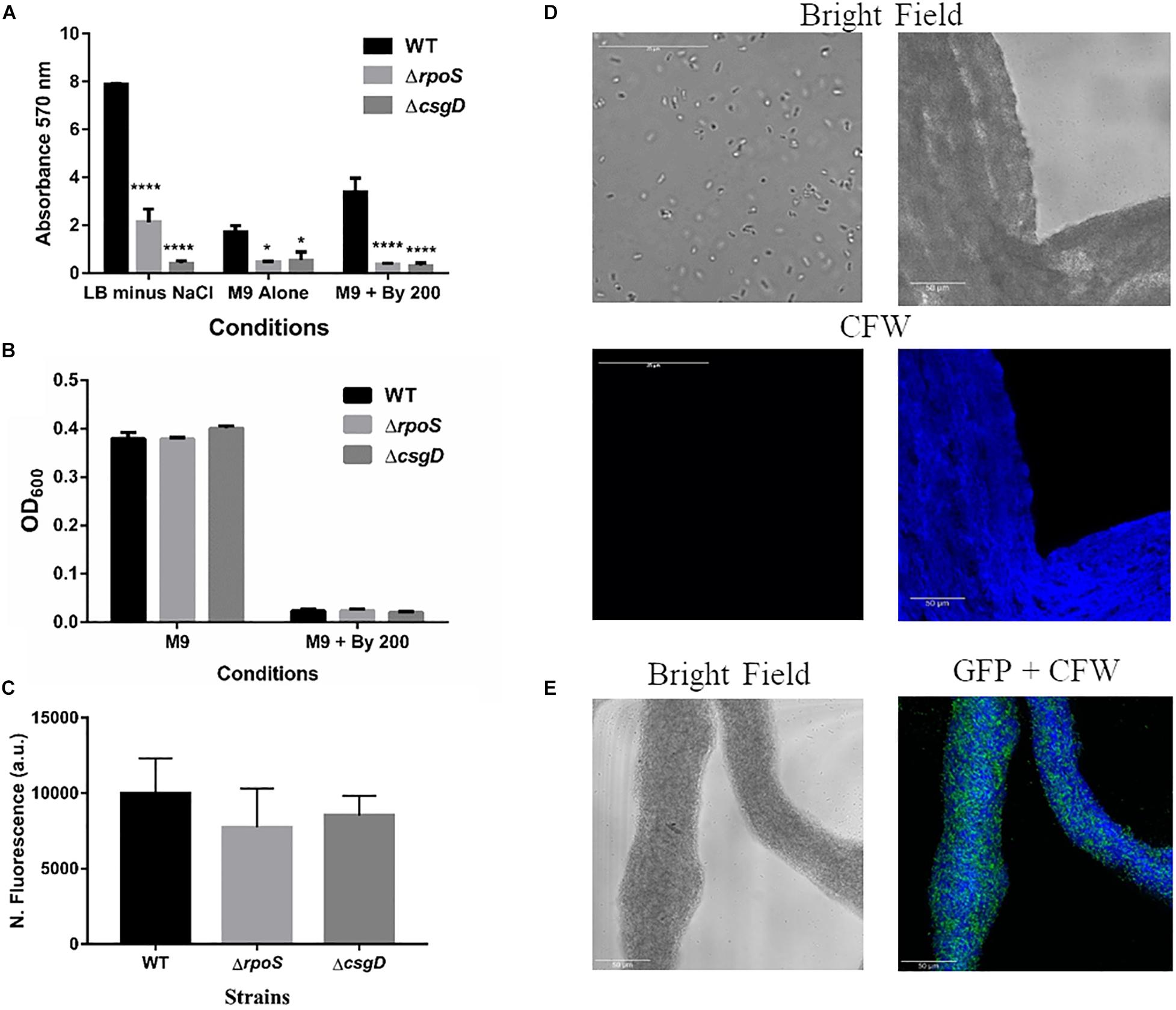
Figure 4. String formation is independent of key molecules required for biofilms but show increased cellulose. (A) Biofilm formation by WT, ΔrpoS and ΔcsgD under various conditions at day 5 time point in 30°C without shaking. * = p < 0.05, **** = p < 0.001, Two-way ANOVA, (B) Planktonic growth in WT, ΔrpoS and ΔcsgD strains in M9 or M9 + Bipd (n = 4). (C) String formation in M9 + Bipd by WT, ΔrpoS and ΔcsgD. (D) Images of cells from M9 and strings formed in M9 + Bipd (63X) after Calcofluor White (CFW) staining to detect cellulose. Bright field on upper half and after UV excitation in the lower half. (E) Represents CFW stained string formed by GFP positive cells to show cellulose encasement of cells. The scale bar is 25 μm for planktonic cells and 50 μm for strings. The data represented as mean ± SEM.
It was important to identify the matrix involved in packing of cells within strings. An extracellular matrix comprising of cellulose could encase cells and lead to the formation of strings. Staining with Calcoflour white, a cellulose binding dye, demonstrated higher fluorescence for strings compared to planktonic cells grown in M9 (Figure 4D). Imaging strings formed by GFP positive S. Typhimurium after Calcoflour staining, clearly showed that cells were encased in a cellulose matrix (Figure 4E). To further confirm the roles of cellulose, a deletion strain of a cellulose synthase complex component bcsA was generated. The ΔbcsA strain did not form strings suggesting that cellulose was critical for string formation (Figures 5A,B), although the ΔbcsA strain was not defective in biofilm formation (Supplementary Figure 6A). Further, complementing the ΔbcsA strain with WT bcsA, but not the empty vector control, restored its ability to form strings. Notably, the planktonic growth remained similar in all four strains (Figure 6A). These results clearly demonstrate a key role of cellulose in string formation.
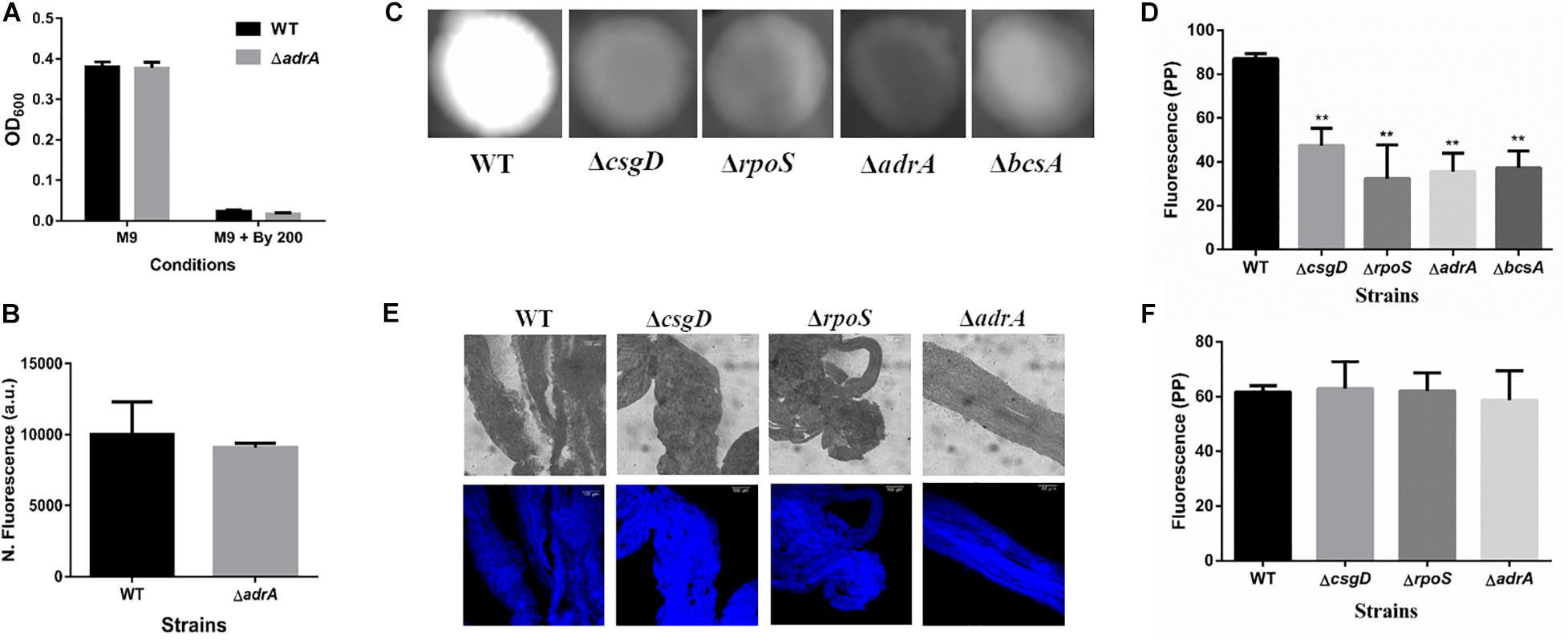
Figure 5. Cellulose production in strings is independent of CsgD. (A) Planktonic growth in WT and ΔadrA strains in M9 or M9 + Bipd (n = 4). (B) String formation in M9 + Bipd by WT and ΔadrA (n = 4). (C) Cellulose production in WT, ΔbcsA, ΔcsgD and ΔrpoS mutants in LB minus NaCl agar determined by Calcoflour staining after 48 h at 30°C. (D) Quantitation of fluorescence as a measure of cellulose production in colonies, ** = p < 0.01, *** = p < 0.005, One-way ANOVA. (E) Calcofluor White staining of strings formed by various mutants (Images at 20X), scale bar 100 μm. (F) Quantitation of fluorescence as a measure of cellulose production in strings. The data represented as mean ± SEM.
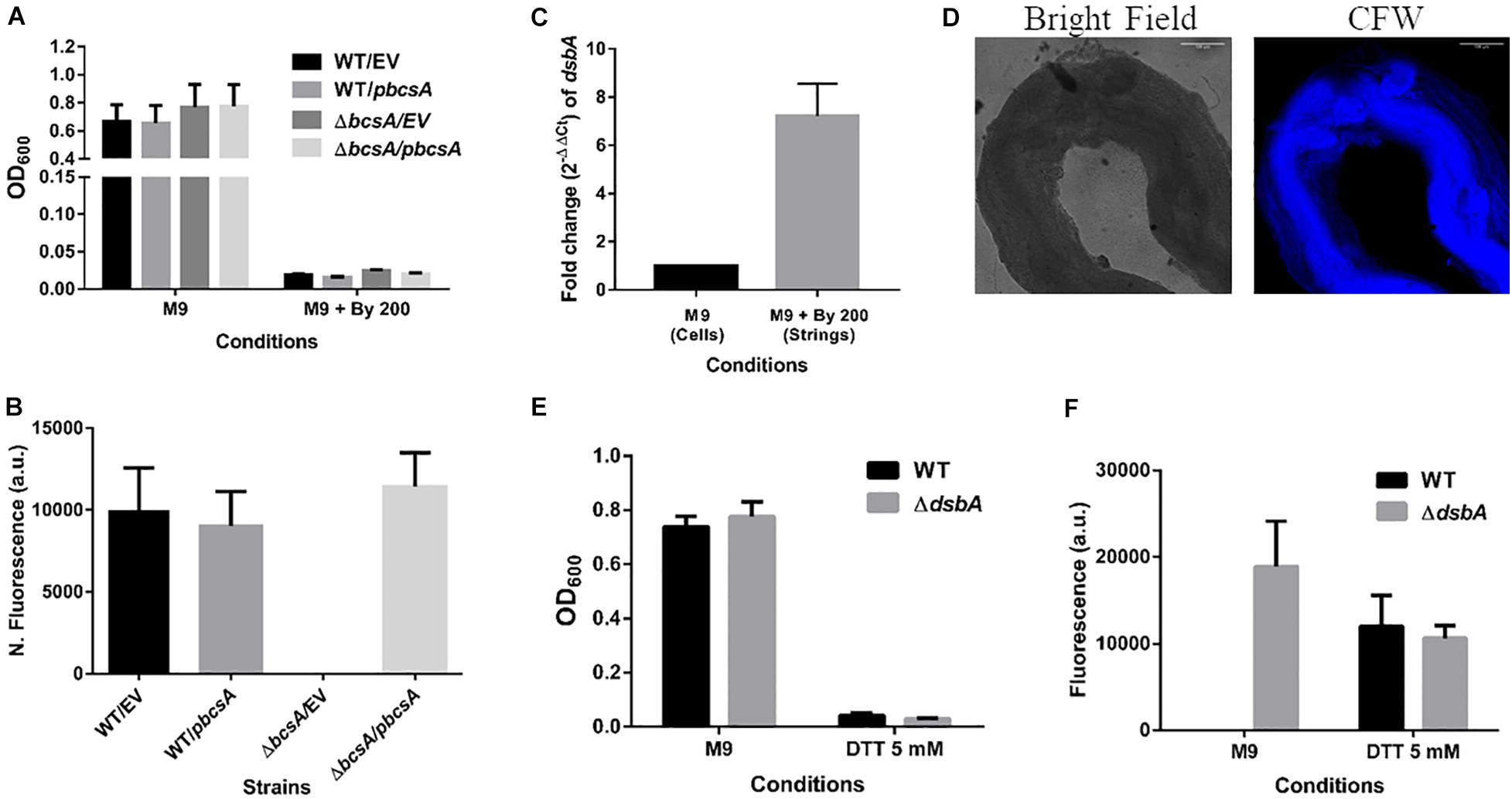
Figure 6. Cellulose is critical for string formation and reductive stress is sufficient to activate string formation. (A) Planktonic growth of WT and ΔbcsA strains complemented with Empty Vector (pTrc99a) or pbcsA. (n = 4). (B) Quantitation of string formation in the four strains in M9 + Bipd. (C) The fold difference in transcript levels for dsbA in cells from M9 and strings formed under M9 + Bipd at 10 h time point (n = 4). (D) Images of Calcofluor White stained strings formed under dithiothreitol (DTT) 5 mM stress (20X) in bright field and after UV excitation to detect cellulose production. The scale bar is 100 μm. (E) Planktonic growth in M9 and 5 mM DTT of WT and ΔdsbA mutant. (F) Quantitation of string formation in WT and ΔdsbA mutant in M9 and 5 mM DTT stress. (n = 4). The data represented as mean ± SEM.
Next, we studied if strings formed by the mutants ΔcsgD,ΔrpoS and SL1344 strain were also dependent on cellulose. As previously reported (García et al., 2004; Mika and Hengge, 2014; Römling and Galperin, 2015), these strains were defective in cellulose production in LB minus NaCl agar model as determined by Calcoflour staining and was comparable to the ΔbcsA strain. (Figures 5C,D, Supplementary Figures 7A,B). However, we observed that strings formed by all these strains produced equal amounts of cellulose based on Calcoflour staining, further demonstrating that cellulose is critical for string formation. Importantly, this observation also suggested a mode of regulation of cellulose production independent of the classical CsgD-dependent biofilm pathway (Figures 5E,F, Supplementary Figures 7C,D). Cellulose production independent of CsgD is known in both E. coli and Salmonella (Da Re and Ghigo, 2006; Hufnagel et al., 2014; Simm et al., 2014). The SL1344 strain is defective in cellulose production due to a deficiency in MlrA which controls the expression of CsgD (García et al., 2004; Römling and Galperin, 2015) and further confirms our results that cellulose production in strings is independent of CsgD.
Next, we investigated whether other stress could also induce string formation. Interestingly, we found that other stresses such as chloramphenicol at sub- optimal doses (5 μg/ml) and ethanol [2–5% (v/v)] also induced string formation (Supplementary Figure 8A). However, others like oxidative stress (Supplementary Figure 8A), pH (4–10), bile (1–5% w/v), and detergents like SDS (0.2– 1% w/v), Tween 20 (0.5–2% v/v) induced stress did not form strings in both M9 and LB (data not shown). Taken together, these results suggest that string formation is induced by some, but not all, stress conditions. We further investigated whether string formation in other stresses were also dependent on cellulose production. Interestingly, strings formed under the stresses of chloramphenicol and ethanol, also stained positive for cellulose while cells exposed to oxidative stress that did not form strings stained negative (Supplementary Figure 8B). Next, we studied whether the ΔbcsA strain could give rise to strings in these conditions. We observed that ΔbcsA strain was defective in string formation in both these stresses and upon complementation with WT bcsA string formation was restored, proving that cellulose is critical for string formation across varying conditions (Supplementary Figure 8C).
CsgD-independent cellulose production in both E. coli and Salmonella has been shown to be activated by reductive stress conditions (Hufnagel et al., 2014; Simm et al., 2014). To test whether Bipd in minimal media induced reductive stress via chelation of Fe2+ that can contribute to ROS via Fenton reaction, we studied dsbA transcript levels in the cells. The disulfide bonding (DSB) system is involved in oxidizing reduced proteins, and an increase in dsbA and dsbB is expected when cells face reductive environments (Gutteridge et al., 1990; Bardwell et al., 1993). Compared to M9, M9 + Bipd cells had 6-fold higher levels of dsbA suggesting that Bipd does induce reductive stress (Figure 6C). To directly determine the role of reductive stress, we tested whether DTT alone in minimal media would also lead to string formation in a cellulose dependent manner. We observed that DTT did induce string formation in a cellulose-dependent manner. (Figures 6D–F). Next, we confirmed the role of reductive stress by observing string formation in M9 alone for the ΔdsbA strain, which would intrinsically have a higher reduced state (Bardwell et al., 1993; Hufnagel et al., 2014) (Figures 5E,F). Together, these studies unequivocally demonstrate that reductive stresses in minimal media are important for string formation.
Cellulose production through the cellulose synthase complex is controlled by c-di-GMP (Römling, 2002; Römling and Galperin, 2015). Therefore, we performed an HPLC analysis to quantify c-di-GMP levels in strings compared to M9 planktonic cells. We observed a 6-fold increase in c-di-GMP levels in strings under Bipd stress compared to planktonic cells in M9 (Figure 7A). To confirm the roles of c-di-GMP, we utilized Coumarin, a compound that has multiple effects but also importantly reduces intracellular c-di-GMP amounts (Zhang et al., 2018; Thakur et al., 2020). Treatment with Coumarin decreased string formation in M9 plus Bipd extensively but not planktonic growth, suggesting a potential role for c-di-GMP during string formation (Figures 7B,C).
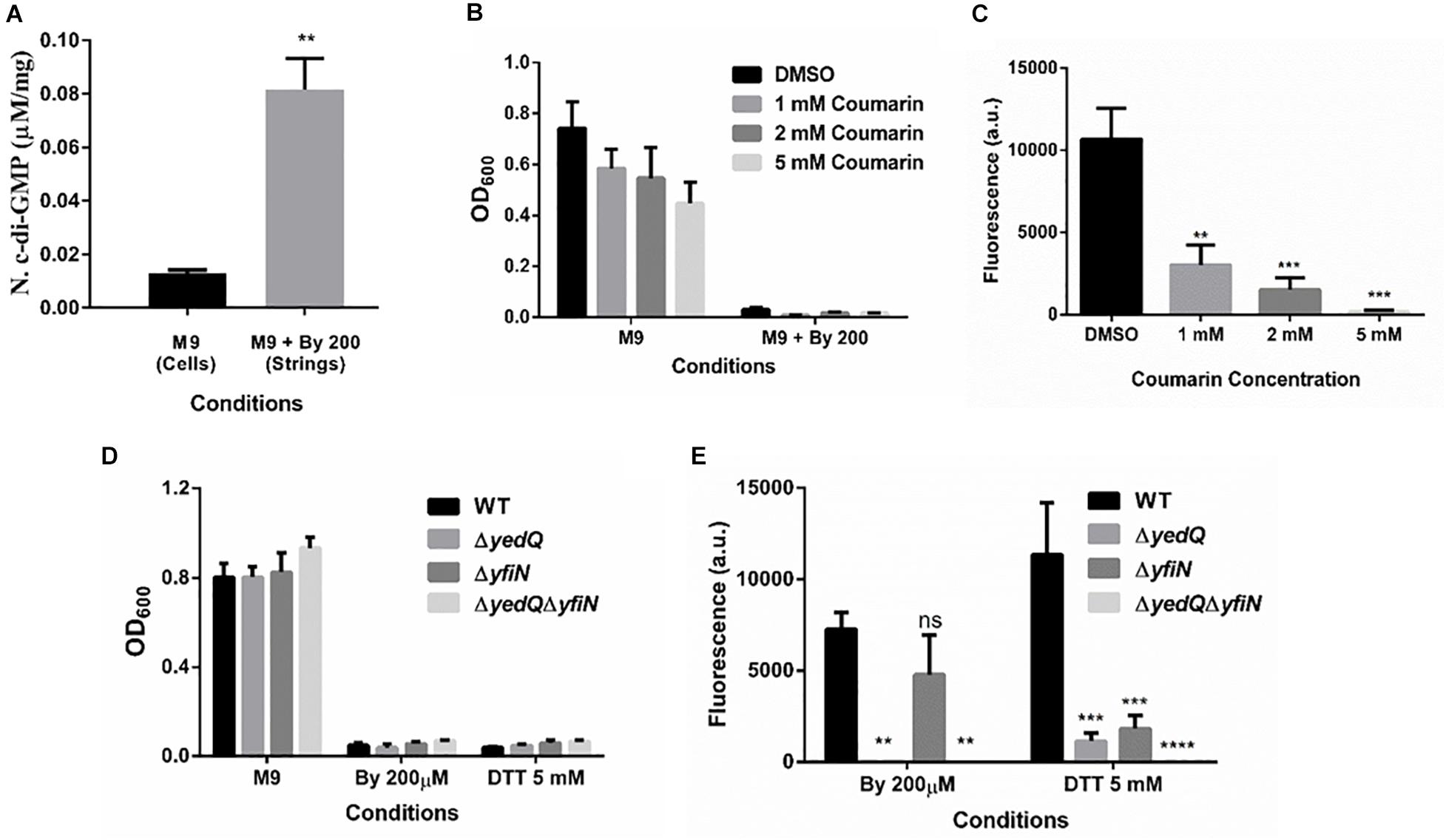
Figure 7. Increase in levels of bacterial secondary messenger c-di-GMP via diguanylate cyclases YedQ primarily and YfiN is critical for string formation. (A) c-di-GMP levels in cells in M9 vs strings formed in M9 + Bipd at 10 hr measured by using HPLC (n = 3) and normalized using protein levels determined by Bradford assay. ** = p < 0.01, Students T-test (B) Effect of Coumarin a molecule known to reduce c-di-GMP levels in cells, on planktonic growth in M9 and M9 + Bipd. (C) Effect of Coumarin on String formation in M9 + Bipd. (D) Planktonic growth of WT, ΔyedQ, ΔyfiN and ΔyedQΔyfiN mutants under various conditions. (E) String formation of WT, ΔyedQ, ΔyfiN and ΔyedQΔyfiN mutants under various conditions ** = p < 0.01, *** = p < 0.005 **** = p < 0.001, Two-way ANOVA. The data is represented as mean ± SEM.
c-di-GMP production in Salmonella is mediated by the major DGCs, AdrA and YedQ; therefore, we tested whether mutants for either would abrogate string formation (Römling and Galperin, 2015). We found the adrA deletion had no effect on string formation (Figures 5A,B). The ΔadrA strain, however, did reduce cellulose levels in the LB minus NaCl growth conditions, while strings of the ΔadrA strain contained cellulose amounts similar to WT strings (Figure 5). String formation was, however, completely abrogated in the ΔyedQ mutant in M9 + Bipd conditions while in M9 + DTT the mutant was significantly less efficient compared to WT. (Figures 7D,E). Given that YfiN also contributes to c-di-GMP production in reductive environments in E coli and had been shown to substitute YedQ in ATM media, we constructed a ΔyfiN single and ΔyfinΔyedQ double mutant to test their roles (Solano et al., 2009; Hufnagel et al., 2014). The single yfiN deletion lowered string formation in M9 + DTT to the same extent as yedQ but the effect in M9 + Bipd was only modest. (Figures 7D,E). However, the ΔyfinΔyedQ mutant did not form strings in both M9 + Bipd and M9 + DTT conditions. (Figures 7D,E). This data demonstrates that string formation is mediated primarily by YedQ and partly by YfiN.
Filamentous forms of bacteria, small aggregates like biofilms, multi-cellular magneto-tactic prokaryotes, Mycobacterial cording, Rhodobacter clumping, Zymomonas aggregates are the main multi-cellular structures previously reported in bacteria (Puskas et al., 1997; Shapiro, 1998; Rudolph et al., 2001; Bernut et al., 2014; Lyons and Kolter, 2015; Jones-Burrage et al., 2019). In this study, we report a novel macroscopic multi-cellular structure formed by Salmonella Typhimurium referred to as “strings” which are ∼ 1 cm in size and contain around a billion tightly packed cells. (Figure 1, Supplementary Figure 3). In addition, we show that strings and biofilms are unrelated, and that string formation requires high amounts of cellulose production in liquid culture. This high cellulose production is mediated by increased c-di-GMP production via DGCs, YedQ primarily and YfiN partly. Extensive cell-cell contact brought by shaking conditions were required to form strings. This suggested that upon cellulose production by individual cells, they must come in contact and the shear force/drag drives cells to form structures as large as a string (Supplementary Figure 2D). In the absence of shaking conditions, cells may clump after settling or form smaller aggregates similar to cords formed by Mycobacterium. Lower planktonic growth under some stress conditions correlated with string formation suggesting that cells become part of the growing strings instead of remaining as single cells (Supplementary Figure 2C). In addition, we show that multi-species strings can be formed by S. Typhimurium with E. coli or P. aeruginosa (Supplementary Figure 5). Further studies are required to identify the ability of other bacteria, especially pathogenic bacteria, to form strings and characterize them under different conditions. How the host environment may induce formation of strings in pathogenic bacteria and their ability in resisting antibiotics or immune responses in vivo is an area that need to be further investigated.
To investigate the relationship between strings and classical CsgD-dependent biofilms, we studied whether conditions inducing strings induce biofilms and vice-versa. We observed that conditions inducing biofilms do not necessarily result in strings in shaking conditions and vice-versa, which suggested that biofilm and string formation are distinct (Supplementary Figure 6). Interestingly, some of the key genes like csgD and rpoS which are involved in biofilm formation (Mika and Hengge, 2014; Simm et al., 2014) do not play critical roles in string formation (Figure 4). The observation that the ΔcsgD strain forms strings suggests that major extracellular polymers like curli fimbriae, O antigen capsule etc, whose transcription is directly regulated by CsgD may not be important for string formation (Mika and Hengge, 2014). The fact that ΔbcsA also formed biofilms by day 5 (Supplementary Figure 6A) is supported by the fact that curli is the major component of biofilms and that lack of bcsA only affects initial biofilm formation (Grantcharova et al., 2010). All these observations together suggest that string formation is independent of the classical CsgD-dependent biofilms.
Previous reports of CsgD-independent cellulose production pointed to reductive stresses playing a role (Hufnagel et al., 2014). To investigate whether reductive stresses led to an increase in string formation, we added DTT into minimal media and scored for string formation. As seen in Figures 6D–F, DTT-induced reductive stress led to string formation in a cellulose-dependent manner. Addition of Bipd leading to a 6-fold increase in dsbA levels suggested that Bipd also induces a reducing stress possibly via Fe2+ chelation (Figure 6C) (Gutteridge et al., 1990; Bardwell et al., 1993). We also observed that just deleting dsbA led to string formation in minimal media alone under no other stress (Figures 6E,F). Overall, these results demonstrate that reductive stress along with nutritional stress is sufficient to induce string formation (Figure 6).
The basic mechanism for string formation appears to be increased production of the bacterial secondary messenger c-di-GMP which regulates cellulose production (Römling, 2002; Römling and Galperin, 2015; Figure 8). Coumarin a molecule known to reduce c-di-GMP levels at the concentrations used, lowered planktonic growth marginally while greatly reducing string formation (Figures 7B,C). This suggests that Coumarin reduced string formation by lowering c-di-GMP levels in strings. However, coumarin could also have other targets like quorum sensing, type III secretion system as well, raising the possibility that other factors may also be involved in this reduction (Zhang et al., 2018; Thakur et al., 2020). Strings stained positive for cellulose as judged by calcofluor staining but not planktonic single cells in M9. The role of cellulose was confirmed by constructing a ΔbcsA strain which did not form strings in M9 + Bipd and upon complementing with WT bcsA restored string formation (Figure 6). String formation under all other conditions tested relied on cellulose production whereas cells exposed to oxidative stress which did not stain positive for cellulose and did not induce strings (Supplementary Figure 8). The critical role of cellulose could explain why E. coli MG1655 did not form strings, since it is defective in cellulose production (Zogaj et al., 2001; Da Re and Ghigo, 2006). Thus, string formation seems to occur due to increased cellulose production during reductive stresses.
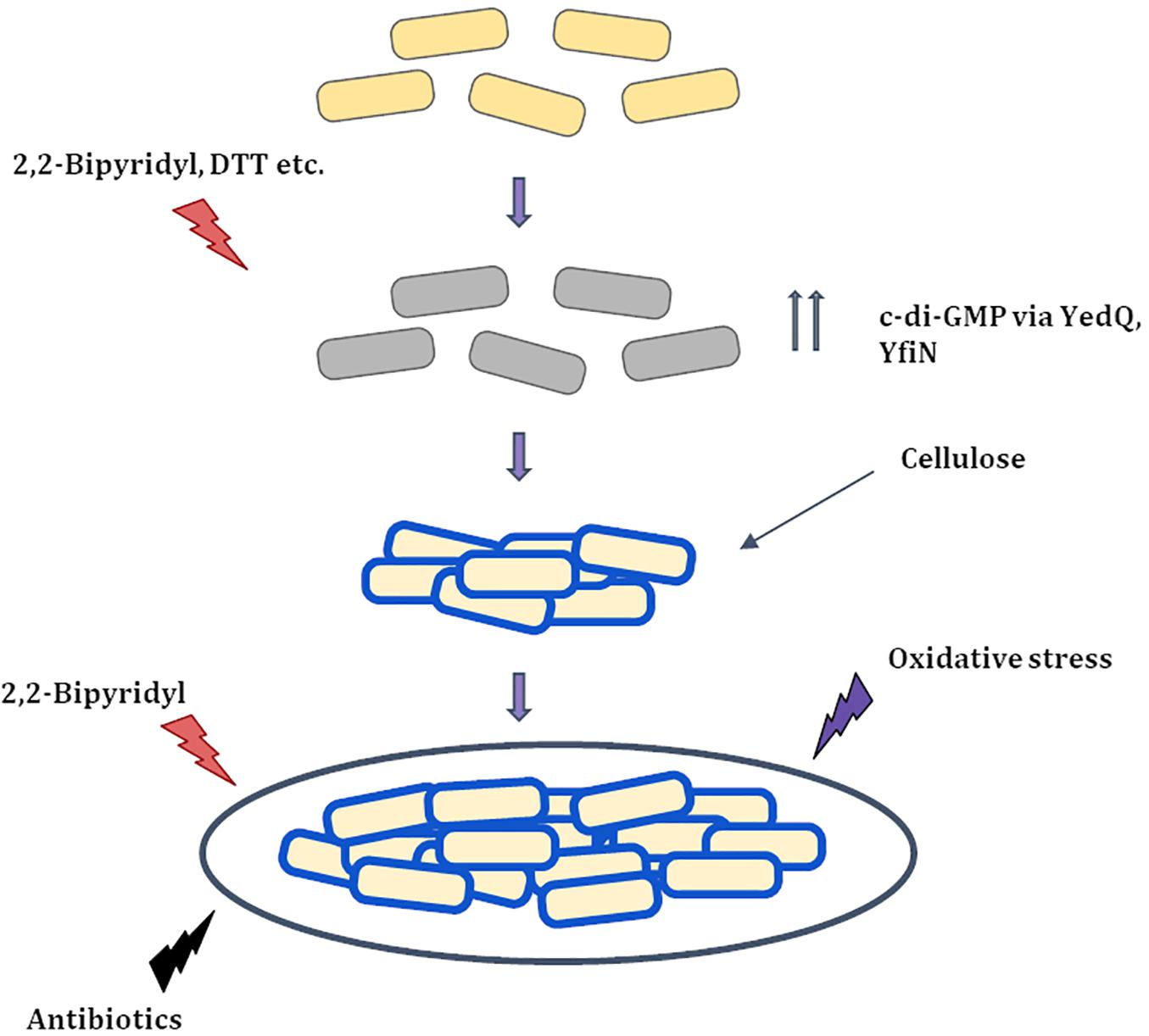
Figure 8. Proposed Model for string formation (i) Cells upon exposure to 2,2-Bipyridyl or Dithiothreitol (DTT) increase the production of c-di-GMP via DGC’s YedQ and YfiN to produce cellulose. (ii) The cells surface gets coated by cellulose and the shaking condition causes them to cluster and form strings. (iii) String formation and cellulose coating leads to greater resistance to stress conditions that induced them and others like antibiotics and oxidative stress.  Healthy cells
Healthy cells  Stressed cells
Stressed cells  Partially recovered cells with cellulose coating (
Partially recovered cells with cellulose coating ( ).
).
In Salmonella c-di-GMP production is mediated mainly by DGCs: adrA (in a CsgD-dependent manner) or yedQ (in a CsgD-independent manner) (Solano et al., 2009; Römling and Galperin, 2015). Given that string formation was independent of CsgD, we found that the ΔadrA strain could still form strings. (Figure 5A). Interestingly, mutants of adrA, csgD, rpoS and the SL1344 strain all, as expected, produced lesser cellulose compared to WT (Kader et al., 2006; Mika and Hengge, 2014; Römling and Galperin, 2015) and similar to the ΔbcsA strain in the LB minus NaCl plate but produce similar amounts of cellulose in strings (Figure 5, Supplementary Figure 7). The SL1344 strain forming strings had indeed suggested that the cellulose production is independent of CsgD, since the strain is deficient in MlrA, leading to reduced CsgD and AdrA levels (García et al., 2004). Since the adrA mutant could still form strings, we generated a ΔyedQ deletion strain. We saw that the ΔyedQ mutant’s ability to form strings was abrogated in M9 + Bipd and greatly reduced in M9 + DTT, suggesting that YedQ plays a critical role in string formation. These results are consistent with our observation that string formation is independent of CsgD regulated AdrA, as suggested by previous studies delineating YedQ and AdrA dependent cellulose production pathways (Kader et al., 2006). The DGC YfiN has also been shown to play a role in producing c-di-GMP under reductive stresses specifically in E. coli (Hufnagel et al., 2014). We saw that the yfiN single deletion greatly reduced string formation in M9 + DTT while modestly reduced string formation in M9 + Bipd. However, string formation was completely abrogated in the double mutant in both conditions, suggesting that YedQ is the primary c-di-GMP producer and YfiN partially contributes to string formation. YedQ and YfiN being the two DGC’s important for string formation in minimal media agrees with previous reports showing them as the only two DGCs important in minimal media (Solano et al., 2009).
There are reports by Solano et al. (1998) and García et al. (2004) of aggregates by Salmonella in ATM (another minimal media). Strings are unlikely to be similar to those aggregates: strings are formed only during additional stress in minimal media, are not predominantly attached to a surface and require much lower amounts of glucose (4 g/L compared to 20 g/L). Importantly, no aggregates where formed by the WT SL1344 strain in ATM (García et al., 2004); however, the WT SL1344 strain clearly formed strings in the presence of Bipd in M9 (Supplementary Figure 7). Most likely, the increased cellulose production leading to string formation is a bacterial response to evade stress, e.g., the bacteriostatic effect of Bipd or sub-optimal amounts of antibiotics on planktonic cells. Notably, cells within a string divided under these stress conditions (Figures 2C,D). Strings are also more resistant to various bactericidal stresses compared to planktonic cells (Figure 3). The reasons for increased resistance by cells within strings could be cellulose coating, tight packing of cells and possible lower metabolic rate, similar to biofilms (Mah and O’Toole, 2001; Høiby et al., 2010; Kaplan, 2011).
The link between minimal media and cellulose production also is quite interesting as recently reported aggregation of Zymomonas mobilis in minimal media, but not nutrient rich media, is dependent on cellulose production (Jones-Burrage et al., 2019). Our data along with previous reports further demonstrate that the CsgD-independent pathway for cellulose production is more prominent in minimal media (Solano et al., 2009). Additionally, we also provide more insights into the CsgD-independent pathway for cellulose production and how two DGCs, YedQ and YfiN, contribute to formation of strings (Figure 8). Furthermore, we observed that the ΔdsbA strain formed strings in minimal media but not in nutritionally rich media LB, demonstrating that reductive conditions in combination with nutritional stress are required for string formation. This is the first report to the best of our knowledge where Salmonella produces high amounts of cellulose in a liquid culture instead of an interface such as liquid air or solid liquid.
Strings could have at least two potential applications. First, the high amounts of cellulose and the ease as well as their production rate suggest that strings can be used to mass produce bacterial cellulose. This material has significant biomedical applications for making artificial skin, tissues like blood vessels as well as drug delivery (Portela et al., 2019). Current methods to produce bacterial cellulose mainly utilize G. hansenii which takes around 7–8 days and hence is time consuming (Shoda and Sugano, 2005; Costa et al., 2017). Strings could potentially solve this problem as mature strings can be formed within 10 h under shaking conditions. Given that both E. coli and Salmonella produce modified cellulose having phosphoethanoloamine groups, these modifications may have to be removed before use, or genetic manipulations have to be done to prevent the modification (Galperin and Shalaeva, 2018; Thongsomboon et al., 2018). Secondly, multi-species biofilms are being explored for bioremediation since the microbes effective in clearing the contaminants do not tolerate the harsh environment. Being a component of biofilms helps these cells resist these harsh conditions (Singh et al., 2006; Edwards and Kjellerup, 2013). As multi-species strings can also form and strings, like biofilms, can evade various harsh conditions, strings can also be used for bioremediation (Figure 3).
Overall, strings are a novel macroscopic structure made by Salmonella which must be explored further to determine their relevance and possibly realize their potential applications. Further studies are required to investigate whether other bacteria form strings and understand the reasons as to why only certain stresses induce string formation and if there are other pathways for string formation. Additional investigations into string formation in vivo, identification of other factors that play a role in string formation, stability, function etc. also need to be studied in future.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author DN (bmFuZGlAaWlzYy5hYy5pbg==).
AV and DN designed the research and analyzed the data. AV, SR, and TV were involved in performing the experiments and AV did majority of experiments. All authors contributed in writing and editing the draft.
The study was possible through funding by the DBT-IISc partnership program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Judith Armitage (Oxford University, United Kingdom) for suggestions and critical comments to improve the quality of the study. We appreciate the kind gift of plasmids and reagents from Deepak Saini (MRDG, IISc), Neha Bahl (UG, IISc) and encouragements from all members of the DpN lab. The facilities in the Biological Sciences Building deserve a special mention: Confocal imaging, AFM and HPLC.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.613704/full#supplementary-material
Abdul-Tehrani, H., Hudson, A. J., Chang, Y. S., Timms, A. R., Hawkins, C., Williams, J. M., et al. (1999). Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181, 1415–1428. doi: 10.1128/JB.181.5.1415-1428.1999
Ahmad, I., Cimdins, A., Beske, T., and Römling, U. (2017). Detailed analysis of c-di-GMP mediated regulation of csgD expression in Salmonella typhimurium. BMC Microbiol. 17:27. doi: 10.1186/s12866-017-0934-5
Ahmad, I., Rouf, S. F., Sun, L., Cimdins, A., Shafeeq, S., Le Guyon, S., et al. (2016). BcsZ inhibits biofilm phenotypes and promotes virulence by blocking cellulose production in Salmonella enterica serovar Typhimurium. Microb. Cell Fact. 15:177. doi: 10.1186/s12934-016-0576-6
Bardwell, J. C., Lee, J. O., Jander, G., Martin, N., Belin, D., and Beckwith, J. (1993). A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. U S A. 90, 1038–1042. doi: 10.1073/pnas.90.3.1038
Berney, M., Hammes, F., Bosshard, F., Weilenmann, H. U., and Egli, T. (2007). Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl. Environ. Microbiol. 73, 3283–3290. doi: 10.1128/AEM.02750-06
Bernut, A., Herrmann, J. L., Kissa, K., Dubremetz, J. F., Gaillard, J. L., Lutfalla, G., et al. (2014). Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc. Natl. Acad. Sci. U S A. 111, E943–E952. doi: 10.1073/pnas.1321390111
Bhaskarla, C., Das, M., Verma, T., Kumar, A., Mahadevan, S., and Nandi, D. (2016). Roles of Lon protease and its substrate MarA during sodium salicylate-mediated growth reduction and antibiotic resistance in Escherichia coli. Microbiology 162, 764–776. doi: 10.1099/mic.0.000271
Bhosale, M., Kumar, A., Das, M., Bhaskarla, C., Agarwal, V., and Nandi, D. (2013). Catalytic activity of Peptidase N is required for adaptation of Escherichia coli to nutritional downshift and high temperature stress. Microbiol. Res. 168, 56–64. doi: 10.1016/j.micres.2012.06.003
Costa, A. F. S., Almeida, F. C. G., Vinhas, G. M., and Sarubbo, L. A. (2017). Production of bacterial cellulose by Gluconacetobacter hansenii using corn steep liquor as nutrient sources. Front. Microbiol. 8:2027. doi: 10.3389/fmicb.2017.02027
Crawford, R. W., Rosales-Reyes, R., Ramírez-Aguilar Mde, L., Chapa-Azuela, O., Alpuche-Aranda, C., and Gunn, J. S. (2010). Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. U S A. 107, 4353–4358. doi: 10.1073/pnas.1000862107
Da Re, S., and Ghigo, J. M. (2006). A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188, 3073–3087. doi: 10.1128/JB.188.8.3073-3087.2006
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U S A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Edwards, S. J., and Kjellerup, B. V. (2013). Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl. Microbiol. Biotechnol. 97, 9909–9921. doi: 10.1007/s00253-013-5216-z
Fu, L., Zhang, J., and Yang, G. (2013). Present status and applications of bacterial cellulose-based materials for skin tissue repair. Carbohydr. Polym. 92, 1432–1442. doi: 10.1016/j.carbpol.2012.10.071
Galperin, M. Y. (2018). What bacteria want. Environ. Microbiol. 20, 4221–4229. doi: 10.1111/1462-2920.14398
Galperin, M. Y., and Shalaeva, D. N. (2018). A bacterial coat that is not pure cotton. Science 359, 276–277. doi: 10.1126/science.aar5253
García, B., Latasa, C., Solano, C., García-del Portillo, F., Gamazo, C., and Lasa, I. (2004). Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54, 264–277. doi: 10.1111/j.1365-2958.2004.04269.x
Grantcharova, N., Peters, V., Monteiro, C., Zakikhany, K., and Römling, U. (2010). Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. J. Bacteriol. 192, 456–466. doi: 10.1128/JB.01826-08
Gutteridge, J. M., Maidt, L., and Poyer, L. (1990). Superoxide dismutase and Fenton chemistry. Reaction of ferric-EDTA complex and ferric-bipyridyl complex with hydrogen peroxide without the apparent formation of iron(II). Biochem. J. 269, 169–174. doi: 10.1042/bj2690169
Heller, A. A., and Spence, D. M. (2019). A rapid method for post-antibiotic bacterial susceptibility testing. PLoS One 14:e0210534. doi: 10.1371/journal.pone.0210534
Høiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Holloway, B. W., and Morgan, A. F. (1986). Genome organization in Pseudomonas. Annu. Rev. Microbiol. 40, 79–105. doi: 10.1146/annurev.mi.40.100186.000455
Hufnagel, D. A., DePas, W. H., and Chapman, M. R. (2014). The disulfide bonding system suppresses CsgD-independent cellulose production in Escherichia coli. J. Bacteriol. 196, 3690–3699. doi: 10.1128/JB.02019-14
Jones-Burrage, S. E., Kremer, T. A., and McKinlay, J. B. (2019). Cell aggregation and aerobic respiration are important for Zymomonas mobilis ZM4 survival in an aerobic minimal medium. Appl. Environ. Microbiol. 85:e00193-19. doi: 10.1128/AEM.00193-19
Jurado, R. L. (1997). Iron, infections, and anemia of inflammation. Clin. Infect. Dis. 25, 888–895. doi: 10.1086/515549
Kader, A., Simm, R., Gerstel, U., Morr, M., and Römling, U. (2006). Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60, 602–616. doi: 10.1111/j.1365-2958.2006.05123.x
Kaplan, J. B. (2011). Antibiotic-induced biofilm formation. Int. J. Artif. Organs 34, 737–751. doi: 10.5301/ijao.5000027
Kumar, A., and Nandi, D. (2007). Characterization and role of peptidase N from Salmonella enterica serovar Typhimurium. Biochem. Biophys. Res. Commun. 353, 706–712. doi: 10.1016/j.bbrc.2006.12.073
Lyons, A. B. (2000). Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J. Immunol. Methods 243, 147–154. doi: 10.1016/s0022-1759(00)00231-3
Lyons, N. A., and Kolter, R. (2015). On the evolution of bacterial multicellularity. Curr. Opin. Microbiol. 24, 21–28. doi: 10.1016/j.mib.2014.12.007
Mah, T. F., and O’Toole, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39. doi: 10.1016/s0966-842x(00)01913-2
Mah, T. F., Pitts, B., Pellock, B., Walker, G. C., Stewart, P. S., and O’Toole, G. A. (2003). A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426, 306–310. doi: 10.1038/nature02122
Mika, F., and Hengge, R. (2014). Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 11, 494–507. doi: 10.4161/rna.2886
Mireles, J. R. II, Toguchi, A., and Harshey, R. M. (2001). Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183, 5848–5854. doi: 10.1128/JB.183.20.5848-5854.2001
Monteiro, C., Papenfort, K., Hentrich, K., Ahmad, I., Le Guyon, S., Reimann, R., et al. (2012). Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 489–502. doi: 10.4161/rna.19682
Parsek, M. R., and Greenberg, E. P. (2005). Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13, 27–33. doi: 10.1016/j.tim.2004.11.007
Peeters, E., Nelis, H. J., and Coenye, T. (2008). Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 72, 157–165. doi: 10.1016/j.mimet.2007.11.010
Petrova, O. E., and Sauer, K. (2017). High-Performance liquid chromatography (HPLC)-based detection and quantitation of cellular c-di-GMP. Methods Mol. Biol. 1657, 33–43. doi: 10.1007/978-1-4939-7240-1_4
Pontes, M. H., Lee, E. J., Choi, J., and Groisman, E. A. (2015). Salmonella promotes virulence by repressing cellulose production. Proc. Natl. Acad. Sci. U S A. 112, 5183–5188. doi: 10.1073/pnas.1500989112
Portela, R., Leal, C. R., Almeida, P. L., and Sobral, R. G. (2019). Bacterial cellulose: a versatile biopolymer for wound dressing applications. Microb. Biotechnol. 12, 586–610. doi: 10.1111/1751-7915.13392
Puskas, A., Greenberg, E. P., Kaplan, S., and Schaefer, A. L. (1997). A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 179, 7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997
Ray, S., Da Costa, R., Das, M., and Nandi, D. (2019). Interplay of cold shock protein E with an uncharacterized protein, YciF, lowers porin expression and enhances bile resistance in Salmonella Typhimurium. J. Biol. Chem. 294, 9084–9099. doi: 10.1074/jbc.RA119.008209
Ray, S., Da Costa, R., Thakur, S., and Nandi, D. (2020). Salmonella Typhimurium encoded cold shock protein E is essential for motility and biofilm formation. Microbiology 166, 460–473. doi: 10.1099/mic.0.000900
Römling, U. (2002). Molecular biology of cellulose production in bacteria. Res. Microbiol. 153, 205–212. doi: 10.1016/s0923-2508(02)01316-5
Römling, U., and Galperin, M. Y. (2015). Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 23, 545–557. doi: 10.1016/j.tim.2015.05.005
Rudolph, C., Wanner, G., and Huber, R. (2001). Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology. Appl. Environ. Microbiol. 67, 2336–2344. doi: 10.1128/AEM.67.5.2336-2344.2001
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Shapiro, J. A. (1998). Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104. doi: 10.1146/annurev.micro.52.1.81
Shoda, M., and Sugano, Y. (2005). Recent advances in bacterial cellulose production. Biotechnol. Bioprocess. Eng. 10, 1–8. doi: 10.1007/BF02931175
Simm, R., Ahmad, I., Rhen, M., Le Guyon, S., and Römling, U. (2014). Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 9, 1261–1282. doi: 10.2217/fmb.14.88
Singh, R., Paul, D., and Jain, R. K. (2006). Biofilms: implications in bioremediation. Trends Microbiol. 14, 389–397. doi: 10.1016/j.tim.2006.07.001
Solano, C., García, B., Latasa, C., Toledo-Arana, A., Zorraquino, V., Valle, J., et al. (2009). Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc. Natl. Acad. Sci. U S A. 106, 7997–8002. doi: 10.1073/pnas.0812573106
Solano, C., Sesma, B., Alvarez, M., Humphrey, T. J., Thorns, C. J., and Gamazo, C. (1998). Discrimination of strains of Salmonella enteritidis with differing levels of virulence by an in vitro glass adherence test. J. Clin. Microbiol. 36, 674–678. doi: 10.1128/JCM.36.3.674-678.1998
Stanley, N. R., and Lazazzera, B. A. (2004). Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52, 917–924. doi: 10.1111/j.1365-2958.2004.04036.x
Subramanian, N., and Qadri, A. (2006). Lysophospholipid sensing triggers secretion of flagellin from pathogenic salmonella. Nat. Immunol. 7, 583–589. doi: 10.1038/ni1336
Thakur, S., Ray, S., Jhunjhunwala, S., and Nandi, D. (2020). Insights into coumarin-mediated inhibition of biofilm formation in Salmonella Typhimurium. Biofouling 36, 479–491. doi: 10.1080/08927014.2020.1773447
Thongsomboon, W., Serra, D. O., Possling, A., Hadjineophytou, C., Hengge, R., and Cegelski, L. (2018). Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose. Science 359, 334–338. doi: 10.1126/science.aao4096
Vander Top, E. A., Perry, G. A., and Gentry-Nielsen, M. J. (2006). A novel flow cytometric assay for measurement of in vivo pulmonary neutrophil phagocytosis. BMC Microbiol. 6:61. doi: 10.1186/1471-2180-6-61
Verma, T., Bhaskarla, C., Sadhir, I., Sreedharan, S., and Nandi, D. (2018). Non-steroidal anti-inflammatory drugs, acetaminophen and ibuprofen, induce phenotypic antibiotic resistance in Escherichia coli: roles of marA and acrB. FEMS Microbiol. Lett. 365:fny251. doi: 10.1093/femsle/fny251
Xavier, K. B., and Bassler, B. L. (2003). LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6, 191–197. doi: 10.1016/s1369-5274(03)00028-6
Zhang, Y., Sass, A., Van Acker, H., Wille, J., Verhasselt, B., Van Nieuwerburgh, F., et al. (2018). Coumarin reduces virulence and biofilm formation in Pseudomonas aeruginosa by Affecting quorum sensing, type iii secretion and C-di-GMP levels. Front. Microbiol. 9:1952. doi: 10.3389/fmicb.2018.01952
Keywords: Salmonella, multi-cellularity, cellulose, c-di-GMP, stress responses
Citation: Varghese A, Ray S, Verma T and Nandi D (2020) Multicellular String-Like Structure Formation by Salmonella Typhimurium Depends on Cellulose Production: Roles of Diguanylate Cyclases, YedQ and YfiN. Front. Microbiol. 11:613704. doi: 10.3389/fmicb.2020.613704
Received: 06 October 2020; Accepted: 24 November 2020;
Published: 14 December 2020.
Edited by:
Catherine Ann Wakeman, Texas Tech University, United StatesReviewed by:
Irfan Ahmad, University of Health Sciences, PakistanCopyright © 2020 Varghese, Ray, Verma and Nandi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipankar Nandi, bmFuZGlAaWlzYy5hYy5pbg==; bmFuZGlAYmlvY2hlbS5paXNjLmVybmV0Lmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.