- 1Guangzhou Marine Geological Survey, China Geological Survey, Guangzhou, China
- 2Gas Hydrate Engineering Technology Center, China Geological Survey, Guangzhou, China
- 3Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 4Department of Ocean Science and Engineering, Southern University of Science and Technology, Shenzhen, China
- 5Shenzhen Key Laboratory of Marine Archaea Geo-Omics, Southern University of Science and Technology, Shenzhen, China
- 6Earth and Planetary Sciences, University of Tennessee, Knoxville, Knoxville, TN, United States
Cold seep ecosystems are developed from methane-rich fluids in organic rich continental slopes, which are the source of various dense microbial and faunal populations. Extensive studies have been conducted on microbial populations in this unique environment; most of them were based on DNA, which could not resolve the activity of extant organisms. In this study, RNA and DNA analyses were performed to evaluate the active archaeal and bacterial communities and their network correlations, particularly those participating in the methane cycle at three sites of newly developed cold seeps in the northern South China Sea (nSCS). The results showed that both archaeal and bacterial communities were significantly different at the RNA and DNA levels, revealing a higher abundance of methane-metabolizing archaea and sulfate-reducing bacteria in RNA sequencing libraries. Site ROV07-01, which exhibited extensive accumulation of deceased Calyptogena clam shells, was highly developed, and showed diverse and active anaerobic archaeal methanotrophs (ANME)-2a/b and sulfate-reducing bacteria from RNA libraries. Site ROV07-02, located near carbonate crusts with few clam shell debris, appeared to be poorly developed, less anaerobic and less active. Site ROV05-02, colonized by living Calyptogena clams, could likely be intermediary between ROV07-01 and ROV07-02, showing abundant ANME-2dI and sulfate-reducing bacteria in RNA libraries. The high-proportions of ANME-2dI, with respect to ANME-2dII in the site ROV07-01 was the first report from nSCS, which could be associated with recently developed cold seeps. Both ANME-2dI and ANME-2a/b showed close networked relationships with sulfate-reducing bacteria; however, they were not associated with the same microbial operational taxonomic units (OTUs). Based on the geochemical gradients and the megafaunal settlements as well as the niche specificities and syntrophic relationships, ANMEs appeared to change in community structure with the evolution of cold seeps, which may be associated with the heterogeneity of their geochemical processes. This study enriched our understanding of more active sulfate-dependent anaerobic oxidation of methane (AOM) in poorly developed and active cold seep sediments by contrasting DNA- and RNA-derived community structure and activity indicators.
Introduction
Methane is a potent greenhouse gas. It is 28 times more efficient than CO2 and contributes more than 20% to global warming (IPCC, 2014). From 2014 to the end of 2018 atmospheric methane increased at twice the rate observed in 2007 (Fletcher and Schaefer, 2019). The exact reason for the increasing methane in the atmosphere remains unclear. Therefore, studying global methane oxidation process is becoming increasingly important.
Continental margins account for only 11% of the ocean area (Levin and Sibuet, 2012); however, their subsurface seabed contains large reservoirs of methane in the dissolved state, gaseous state, and in the form of solid natural gas hydrates. Driven by a variety of unstable geological factors, low-temperature and methane-rich fluids emit to the seabed surface along the seabed channels, forming a unique deep-sea ecosystem—cold seep (Boetius and Wenzhöfer, 2013). There are thousands of active cold-seep systems distributed on the continental margins around the world. Although they emit 0.01–0.05 Gt of carbon up to the atmosphere annually (Milkov et al., 2003; Kvenvolden and Rogers, 2005; Reeburgh, 2007; Judd and Hovland, 2009), large amounts of methane have been consumed by microbial oxidation during upward migration in sediments (Reeburgh, 2007; Regnier et al., 2011). Therefore, methane oxidation by microorganisms in sediments plays an important role in preventing cold seep methane from methane from entering the atmosphere (Knittel and Boetius, 2009; Boetius and Wenzhöfer, 2013).
Anaerobic archaeal methanotrophs (ANMEs) compose the major microbial groups in cold seep sediments, with a relative abundance up to 80–90% in total archaea (Vigneron et al., 2013; Cui et al., 2019). They can be divided into three distinct methanotrophic groups, namely ANME-1, ANME-2, and ANME-3. The large phylogenetic distance between ANME groups represents their internal sequence similarity of 75–92% according to 16S rRNA genes (Knittel and Boetius, 2009). ANME-1 is further divided into two subgroups, designated ANME-1a and ANME-1b (Knittel et al., 2005). ANME-2 is the most widely distributed and highly diverse group, which is divided into four distinct subgroups, ANME-2a, ANME-2b, ANME-2c, and ANME-2d (Orphan et al., 2001; Mills et al., 2003). ANME-2a/b is highly abundant in cold seep sediments (Knittel et al., 2005; Niu et al., 2017), and ANME-2c can be found in both hydrothermal and cold seep sediments (Vigneron et al., 2013; McKay et al., 2016). ANME-2d is also named “GOM Arc I” (Lloyd et al., 2006) due to its late discovery and distant relationship with other subgroups of ANME-2. “Candidatus Methanoperedens nitroreducens” has also been used to refer to ANME-2d due to its capability of performing AOM with nitrate as the ultimate electron acceptor (Haroon et al., 2013).
ANMEs frequently form consortia with sulfate-reducing bacteria to conduct anaerobic oxidation of methane (AOM). ANME-1 and ANME-2 are connected with Desulfosarcina–Desulfococcus (DSS) as sulfate-reducing bacteria (Orphan et al., 2002), while ANME-3 are connected with Desulfobulbus (DBB) (Niemann et al., 2006). ANME-1, as an anaerobic methanotroph ecotype, often forms monospecific chains without attached bacterial partner (Maignien et al., 2012). The ANME-2-dominated community presents significantly higher AOM rates in anoxic incubation than that of ANME-1 (Nauhaus et al., 2005).
It has been estimated that about 75% of migratory methane emitted from subsurface reservoirs is microbially consumed by ANMEs during AOM (Boetius and Wenzhöfer, 2013). In addition, AOM associated with sulfate reduction is responsible for increasing sulfide and dissolved inorganic carbon, promoting the precipitation of authigenic carbonates (Liang et al., 2017), and creating a favorable habitat for benthic megafaunal and microbial communities. For instance, the biomass of benthic communities at cold seep sites is up to tens of kilograms per square meter, exceeding by orders of magnitude the biomass in nearby seabed sediments without seeps (Zhang et al., 2002; Zhang C. L. et al., 2003; Levin, 2005). The benthic communities at cold seep sites include motile sulfide-oxidizing bacteria (Sahling et al., 2002; Zhang et al., 2005; Niemann et al., 2006), bivalve Calyptogena clams, and tubeworms (Cordes et al., 2005; Niemann et al., 2006). Therefore, the AOM process not only reduces methane leakage but also plays an important role in changing marine ecological habitats.
Recently, ANME communities were investigated using high-throughput sequencing technology in the sediments of the South China Sea (SCS) (Niu et al., 2017; Cui et al., 2019; Zhuang et al., 2019). However, these studies only focused on the overall microbial communities at the DNA level, without distinguishing among active, dormant, and dead microbes. DNA molecules are much more resistant to degradation and can be detected in dead cells (Lorenz and Wackernagel, 1987; Karl and Bailiff, 1989). Therefore, DNA-based abundance estimates could include both the extant and necromass of microbial communities. In contrast, high-throughput sequencing technologies focusing on RNA can be used to evaluate relative metabolic activities due to RNA relatedness to enzyme production and their sensitivity to environmental changes (Charvet et al., 2014; Pawlowski et al., 2014). Several studies have successfully determined the microbial composition in the water column of the SCS using both DNA and RNA investigations (Xu et al., 2017; Wu and Liu, 2018). However, the survey of sedimentary microbial communities conducted by employing DNA and RNA molecules is currently lacking, especially in SCS cold seep unities.
Various cold seeps have been identified on the northern continental slope of the SCS (Han et al., 2008; Feng and Chen, 2015; Liang et al., 2017; Hu et al., 2019). Among them, Formosa Ridge (site F) and Haima cold seeps are known as active seeps (Feng and Chen, 2015; Liang et al., 2017). Observations using a remotely operated vehicle (ROV) confirmed the existence of massive authigenic carbonate crusts and various benthic faunal assemblages, such as dead bivalves (Calyptogena sp.), living tubeworms (Paraescarpia echinospica), and living mussels (Bathymodiolus plantifrons) (Feng and Chen, 2015; Liang et al., 2017). Large numbers of ANMEs have been found in sediments from Haima cold seeps through DNA investigation, including ANME-1a, ANME-1b, ANME-2a/b, and ANME-2c (Niu et al., 2017; Guan et al., 2018; Cui et al., 2019; Zhuang et al., 2019). In 2018, two weak seeps (ROV05 and ROV07, 5.22 km apart) were newly discovered using the Haima ROV. They are located about 110 km to the northeast of the Haima cold seeps below 1,721–1,753 m of water depth (Feng et al., 2020). These two seeps are obviously different from Haima cold seeps since they belong to different stages of cold seep evolution and harbor simple megafauna (mainly including Calyptogena) communities. Large amounts of methane gas hydrates have been revealed in the area where the two seeps occur, with hydrate depths of 15–160 m and 8–174 m below sediment surface at sites ROV05 and ROV07, respectively (Liang et al., 2019; Wei et al., 2019; Ye et al., 2019).
This comparative study was conducted at the DNA and RNA levels in combination with network analysis to evaluate the changes in microbial (archaeal and bacterial) communities in these two new seeps, aiming to (1) determine and contrast the active microbial communities involved in AOM on the northern slope of the SCS; (2) investigate the network correlations and partnerships between ANMEs and other microbes at these differing sites; and (3) explore the variation of ANMEs in different stages of cold seep evolution.
Materials and Methods
Site Description and Sampling
Sediment samples were collected with the Haima ROV from the new weak seeps (ROV05 and ROV07) during the cruise HYLH2018-01 (Figure 1). ROV05 was observed to show a small mud mound (diameter: 10 m) colonized by dense populations of living clams (Calyptogena sp., Supplementary Figure 1). One push core was carried out near the clam patch and was named ROV05-02. Another surface sediment sample with living clams was collected by an ROV gripper and was named LC (living clams). ROV07 was observed to show that the shells of dead clams (Calyptogena) and carbonate crusts were scattered over a few 100 m. Two push cores were obtained from the sediments near the shells of dead clams and the sediments near the carbonate crusts and were named ROV07-01 and ROV07-02, respectively. In addition, another sediment sample was collected in dead clam shells near the push core ROV07-01 by an ROV gripper and was named CS (Clam shells). In brief, the site of ROV05-02 was colonized by living Calyptogena clams with 1,721 m of water depth (Supplementary Figure 1), the site of ROV07-01 accumulated deceased Calyptogena clam shells with 1,753 m of water depth, and the site of ROV07-02 was located near carbonate crusts with few clam shell debris (1,751 m of water depth). The site of ROV05-02 is far away from ROV07-01 (5.22 km apart), while site ROV07-01 is 20 m away from ROV07-02.
Figure 1. Locations of the cold seeps ROV05 and ROV07 in the northern part of the SCS. ROV05-02 samples were collected from ROV05, and ROV07-01 and ROV07-02 samples from ROV07.
Each of the sediment cores was cut into halves on board, whereby one half was subsampled for molecular analysis and the other half was collected for geochemical analysis of pore water. All sediment samples were collected aseptically from 5cm-thick layers using sterile spoons, and were then frozen at −20°C until further processing. The dissolved inorganic carbon (DIC), δ13CDIC values, total alkalinity (TA), and cation-anion concentrations (, , Cl−, Mg2+, Ca2+, K+, and Na+) were determined as previously described (Feng et al., 2020). Particle size was measured using a Malvern Mastersizer2000 (Malvern Panalytical, UK).
DNA and RNA Extraction and PCR and Quantitative PCR Amplification
The total DNA and RNA of the sediments were extracted and purified using the E.Z.N.A.® soil DNA Kit and the E.Z.N.A.® soil RNA Mini Kit (Omega Bio-tek, Norcross, GA, U.S.), respectively. Aliquots of rRNA were reversely transcribed using HiScript® Q RT SuperMix (Vazyme Biotech Co., Ltd.) according to the manufacturer's protocols after purification. DNA concentration and quality were determined using a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA) and 1% agarose gel electrophoresis, respectively.
The DNA amplification were performed by a thermocycler PCR system (GeneAmp 9700, ABI, USA) through the following process: 5 min of denaturation at 95°C; 35 cycles of sub-process constituting 30 s at 95°C, 30 s for annealing at appropriate temperature according to targeting genes (listed in Supplementary Table 1), and 1 min for elongation at 72°C successively; a final extension at 72°C for 10 min. PCR reactions were performed in triplicate 20 μL mixture containing 10 μL of 2X Taq Plus Master Mix, 0.8 μL of each primer (5 μM), 7.4 μL of ddH2O, and 1 μL of template DNA. Primer-pairs were used to amplify the different microbial communities as shown in Supplementary Table 1.
Quantitative PCR (qPCR) was performed using an ABI7500 Real-Time system (Applied Biosystem, U.S.A.). The qPCR reaction was performed according to the following process: 5 min of denaturation at 95°C, followed by 40 cycles of sub-process consisting of 5 s at 95°C, 30 s for annealing at appropriate temperature according to targeting genes (listed in Supplementary Table 1), and 40 s for elongation at 72°C. The qPCR reactions were performed in triplicate 20 μL mixture containing 10 μL of ChamQ SYBR Color qPCR Master Mix, 0.8 μL of each primer (5 μM), and 1 μL of template DNA. Standard templates were generated using a dilution series of purified plasmids with R2 values greater than 0.99.
Illumina MiSeq Sequencing
16S rDNA and rRNA sequencing of target microbial (archaeal and bacterial, Supplementary Table 1) communities were conducted with an Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols of Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Low-quality sequences were demultiplexed, filtered by Trimmomatic, and merged by FLASH (Bolger et al., 2014). After removing chimeric sequences using UCHIME (Edgar et al., 2011), high-quality sequences were clustered into operational taxonomic units (OTUs) at similarity levels of 97% using UPARSE (version 7.1, http://drive5.com/uparse/). The taxonomy was assigned to representative sequences by RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU132) 16S rRNA database using a confidence threshold of 70% (Quast et al., 2013).
Statistical Analysis
The differences of archaeal and bacterial structure between DNA and RNA sequences were analyzed in R language using Bray Curtis method. Principal Component Analysis (PCA) was performed to evaluate the variations at different sites based on RNA level. Archaeal sequences were aligned with BLAST hits from GenBank using the MEGA software (Hall, 2013), followed by manual adjustments. Phylogenic tree was generated based on maximum likelihood analysis. The robustness of inferred topology was measured by bootstrap resampling (1,000). The tree was then drawn in the web-based interactive tree of life (iTOL) (Letunic and Bork, 2016).
The co-occurrence network was analyzed in R language (V.3.2.4) and visualized by using Cytoscape (Shannon et al., 2003). The archaeal OTUs (the top 50, representing 87.2–94.6% of total archaeal reads) and bacterial OTUs with the highest absolute abundance (the top 200, representing 61.8–74.2% of total bacterial reads) were firstly selected to be analyzed based on Pearson correlation matrix in R language. The absolute abundance was based on their population and equal to their relative abundance in DNA sequence libraries multiplied by archaeal and bacterial DNA quantification, respectively. The significant correlations (coefficient >0.9 or < −0.9, P ≤ 0.01) between two OTUs were secondarily selected and visualized using Cytoscape.
Redundancy analysis (RDA) was performed using PRIMER V6.1.16 & PERMANOVA+ V1.0.6. The distance matrix of archaeal communities serves as response variables and was calculated by the method of Bray Curtis similarity. Meanwhile, predictor variables of environmental factors were selected by the step-wise method through R2 criterion and further tested by Monte-Carlo significance test through 999 permutations.
Results
Geochemical Characterization
Comparing and contrasting data revealed similarities and contrasts between the three sample locations (Figure 2). Seawater impacts on shallow sediments were common across sites with sulfate ~25 mM, DIC ~5 mM and δ13CDIC ~ −20‰. Overall, it appeared that seawater impacts dominated in 0–20 cm depths. Methane concentration was likely undervalued due to outgassing during sampling, however, the methane range of the push cores clearly differed. Site ROV07-02 exhibited very low methane concentrations (0.19–0.54 μM) throughout the 60 cm profile as well as DIC and similar to overlying waters. Below ~30 cm there was a slight increase in DIC with concomitant decrease in δ13CDIC. Meanwhile, the sulfate concentrations remained constant at 24–25 mM in the sediments of ROV07-02 indicative of insignificant sulfate reduction. Combined with the lack of sulfide smell, these findings suggest low biological activity in the 60 cm core ROV07-02. Core ROV05-02 appeared similar to ROV07-02 except for higher methane concentrations (0.21–3.71 μM). These results suggested a significant methane energy source available in the sediments of ROV05-02 and perhaps some methane production or at least accumulation, suggestive of biological activity.
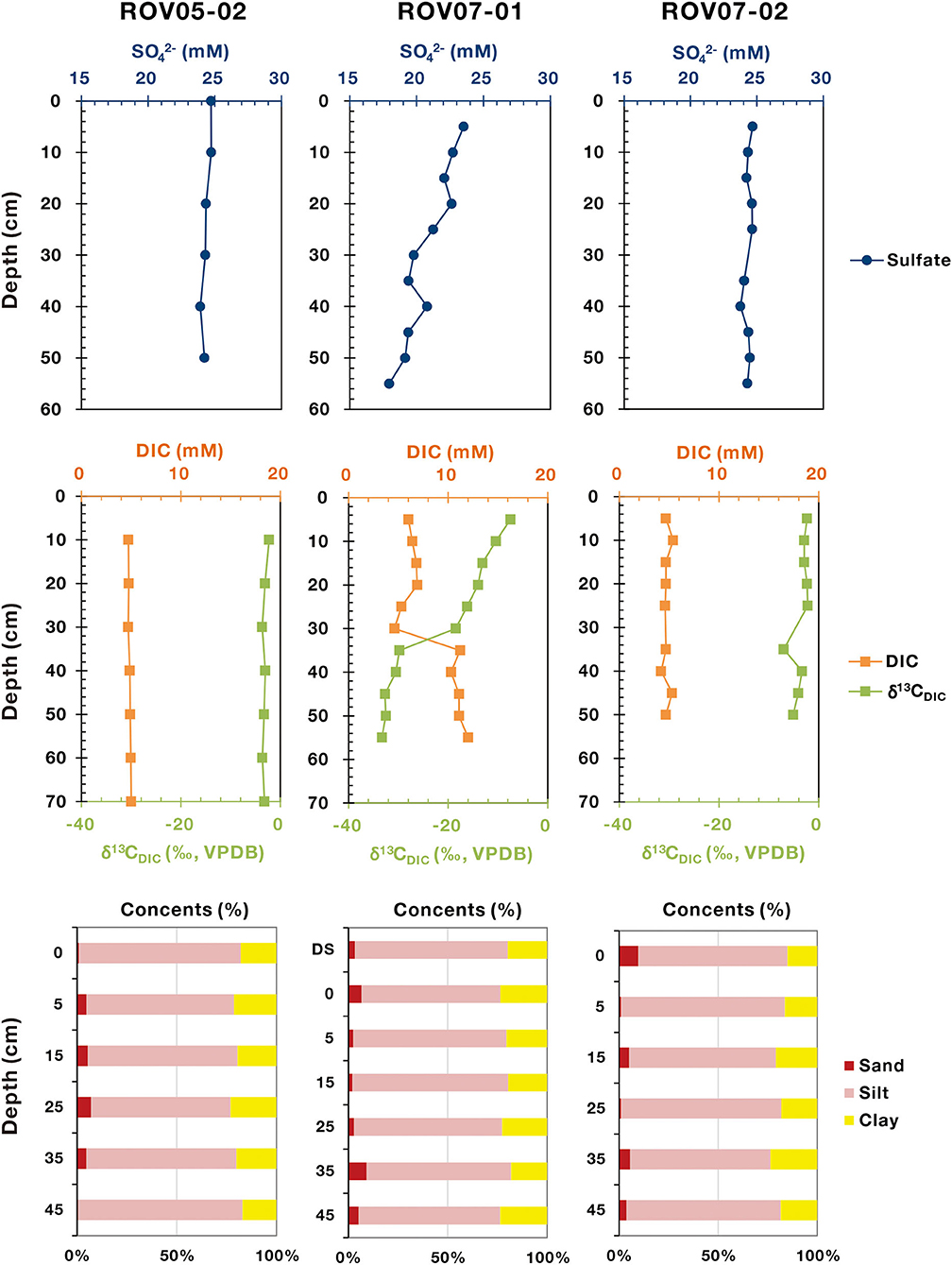
Figure 2. Geochemical profiles of sulfate, DIC, δ13CDIC, and particle sizes in the sediments of cold seeps. The sulfate, DIC, and δ13CDIC data are quoted from Feng et al. (2020).
In stark contrast were sediments from site ROV07-01. The sulfate concentrations decreased rapidly from 23.52 to 17.95 mM within 55 cm depth. Unlike sediments from ROV07-02 and ROV05-02, the ROV07-01 sediments exhibited a strong smell of sulfide, indicative of a highly reducing environment. Concomitantly, methane was presented at a higher level in the ROV07-01 sediments rising from 0.49 μM to 7.03 μM. Further evidence of enhanced microbial activities in ROV07-01 sediments was increased DIC at depth. The DIC content was ~5 mM at a depth of 0–30 cm. Then it greatly increased to 11.2 mM at a depth of 35 cm, and finally remained constant again at a depth of 35–55 cm. While DIC at the sediment water interface was likely in equilibrium with the overlying waters, the DIC at 60 cm depth was 2 times higher than that of the overlying waters. In parallel, the δ13C value of the DIC in the sediments of ROV07-01 presented an opposite trend. It was slightly negative at a depth of 0–30 cm (−7.5~-18.5‰), similar to that of ROV05-02 and ROV07-02, then became largely depleted at a depth of 35 cm, and finally remained constant (~-32‰) at a depth of 35–55 cm. The depletion of δ13CDIC is characteristic of biological oxidation of reduced carbon. Presence of sulfide, removal of sulfate in the upper 5+ cm depths, and increased DIC with concomitant decreasing δ13CDIC provided multiple lines of evidence for significant anaerobic activities and likely extending below the 60 cm core in ROV07-01 sediments.
Profiles of particle size of the three sediment cores were similar, with a high abundance of silt content ranging from 69.7 to 82.6%. As for the sand content (0.5–9.6%), it showed a high abundance in middle layers in core ROV05-02 (up to 7.0%), with the highest abundance occurring at 35 cm depth in core ROV07-01.
Quantification of Archaea, Bacteria and ANME Subgroups
Bacteria were nearly three times more abundant than archaea in the three sites, with abundance ranging from 1.71 × 107 to 5.24 × 106 copies/g (wet weight), respectively (Figure 3). Sediments of ROV05-02 and ROV07-01 exhibited significant abundance of the apsA functional gene ranging from 3.84 × 106 to 5.74 × 106 copies/g wet weight sediments on average, with the presumably active ROV07-01 sediments evidencing 1.65 times more than ROV05-02 sediments. The functional gene mcrA for methane metabolism was retrieved from ROV05-02 and ROV07-01, with abundance ranging from 1.64 × 105 to 3.71 × 105 copies/g (wet weight), respectively. Again, the higher abundances were noted in the more active and diverse ROV07-01 samples. Unsurprisingly, mcrA gene was not detected in the presumably lower activity sediments of ROV07-02.
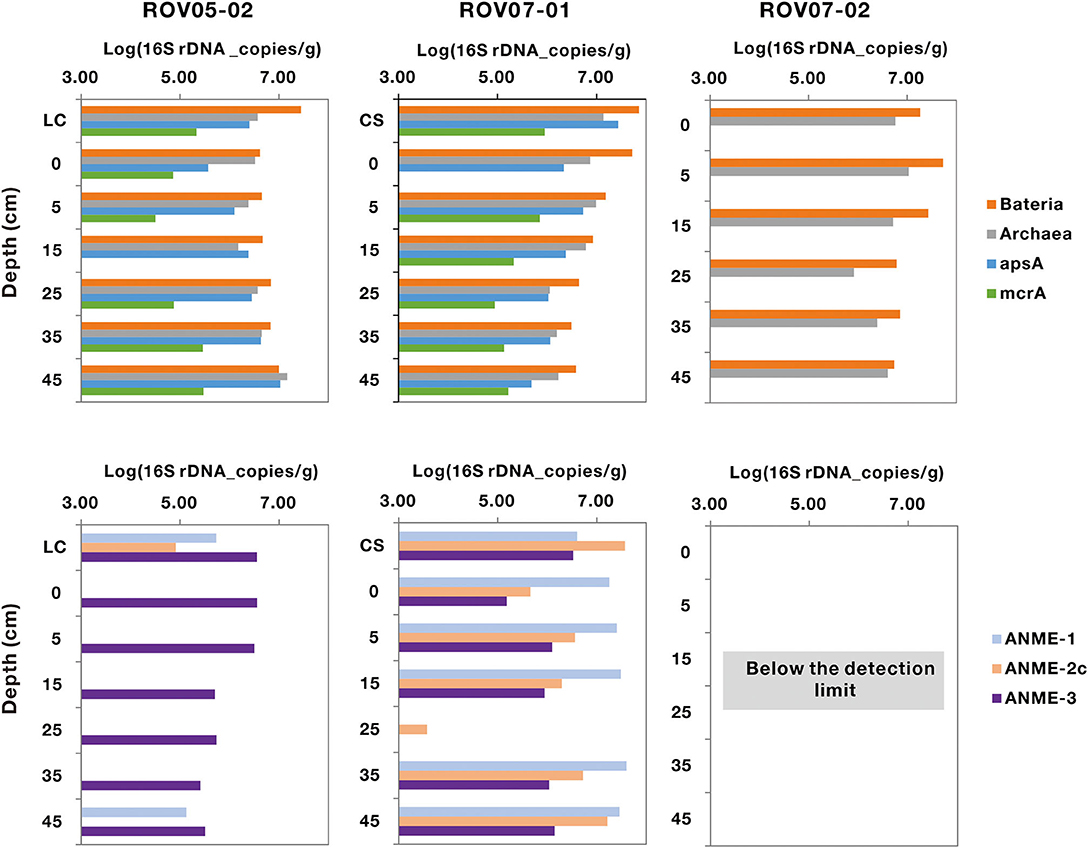
Figure 3. Quantification of the populations of archaeal, bacterial, apsA, mcrA, ANME-1, ANME-2c, and ANME-3 genes in cold seep sediments. LC and CS represent the sediment samples with living clams and die-off clam shells at surface sediment, respectively. The function gene apsA was not measured in core ROV07-02; the genes of mcrA, ANME-1, ANME-2c, and ANME-3 were below the detection limit.
The abundance of ANME-1, ANME-2c, and ANME-3 could only be quantified in the sediments of ROV05-02 and ROV07-01 by Q-PCR. All ANME groups were below detectable limits in each depth in the lower activity ROV07-02 core. ANME-1 and ANME-2c genes were more abundant in the sediments of ROV07-01, with an average of 2.44 × 107 and 9.25 × 106 copies/g wet weight sediments, respectively. However, they were hardly detected from most of the sediments of ROV05-02. ANME-3 gene was detectable from the sediments of ROV05-02 and ROV07-01, averaging 1.71 × 106 and 1.35 × 106 copies/g wet weight sediments, respectively.
Phylogenetic Diversity of Metabolically Active Archaea
The differences in archaeal and bacterial community structures were shown between DNA and RNA sequence libraries. Significant differences in archaeal structural patterns were shown in sediment cores ROV07-01 (R = 0.68, Sig. = 0.003) and ROV07-02 (R = 0.63, Sig. = 0.003) (Supplementary Table 2). Significant differences in bacterial structural pattern were observed in the sediment cores ROV05-02 (R = 0.50, Sig. = 0.003) and ROV07-01 (R = 0.72, Sig. =0.001; bacterial RNA communities were not detected). In general, differences between DNA and RNA sequence libraries of the sediment core ROV07-01 were greater than that of the sediment core ROV05-02.
A total of 367,333 16S rRNA sequences of archaea were obtained from 20 sediment samples, and were assigned to 919 archaeal OTUs (Supplementary Table 3). In general, both DNA and RNA sequences retrieved were mostly affiliated with methane-metabolizing archaea (ANMEs and methanogens, Figure 4), with the exception of sediments of ROV07-02, which consistently appeared low in both methane-metabolizing biomass and likely activity. Site ROV05-02 exhibited considerable abundance of ANMEs by DNA sequencing and likely activity as exhibited by RNA sequencing. In addition, the live clam area (ROV05-02) at the surface sediment exhibited significant ANME-3 and methanogenic DNA and RNA, at depth ANME-2d were in higher preponderance than other archaea. Site ROV07-01 revealed a diverse community of methanogens and ANMEs with near corresponding proportions of RNA suggesting diverse activity as well.
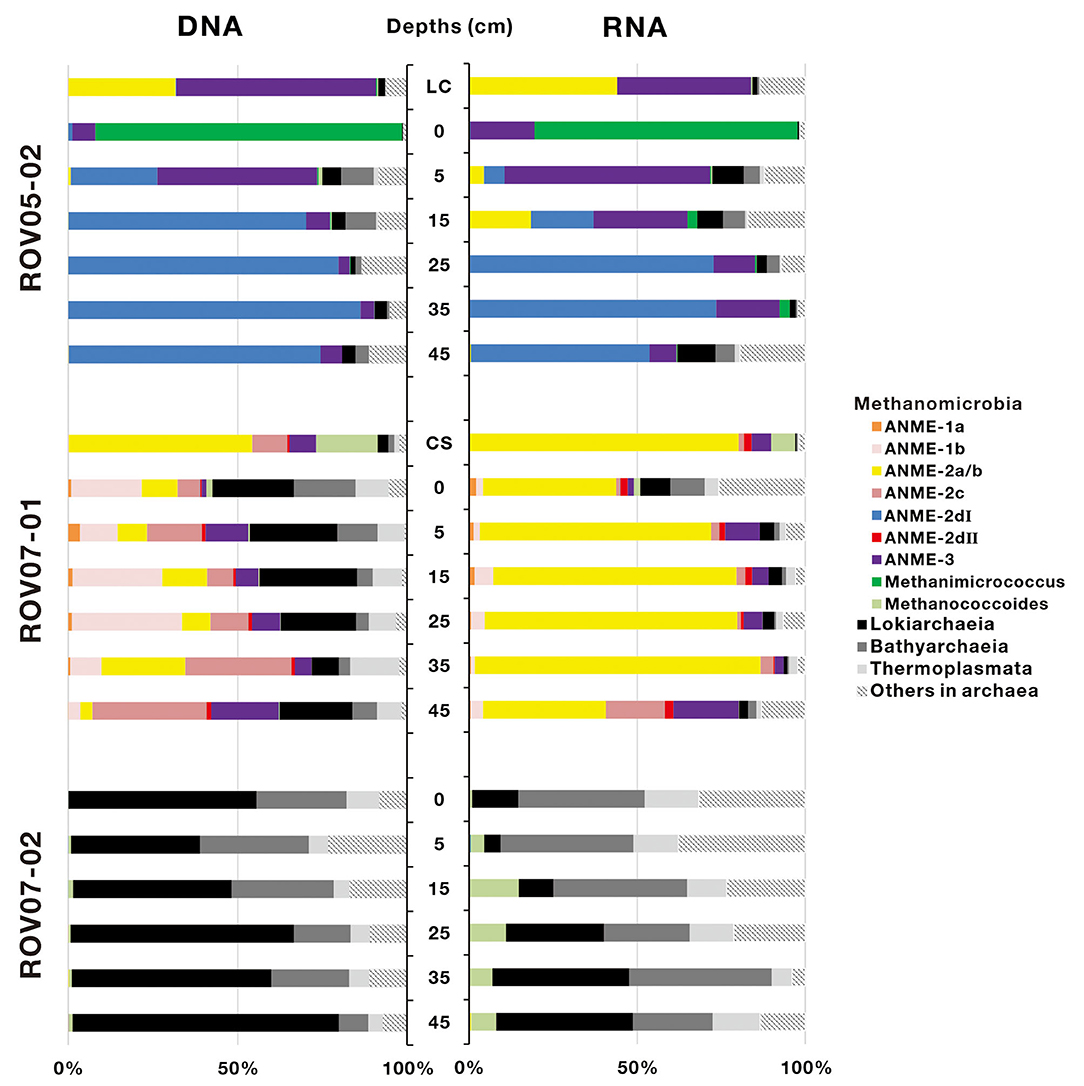
Figure 4. Archaeal community composition based on 16S rDNA and 16S rRNA-derived sequences in cold seep sediments. LC and CS represent the sediment samples with living clams and die-off clam shells at surface sediment, respectively.
ANME-2d increased proportionally with depth and became predominant in deep sediment layers in both DNA and RNA sequence libraries at the ROV05-02 site. In contrast, ANME-2d accounted for a small proportion of archaea at the ROV07-01 site and was not observed at ROV07-02. ANME-2d was further divided into two clades, namely ANME-2dI and ANME-2dII, with a similarity of 83.88–87.91% (Supplementary Figure 2). The two clades were respectively derived from the sediments of ROV05-02 and ROV07-01. The RNA sequences of ANME-2a/b were significantly predominant in all sediment layers (averaging 65.4% of archaea) at ROV07-01. The DNA and RNA sequences of ANME-2a/b were detected in shallow sediment layers but were present at low percentage in archaea at the site of ROV05-02. Interestingly, ANME-2a/b was dominant in sediments with living Calyptogena clam or dead clam shells. ANME-3 was dominant in shallow sediment layers at site ROV05-02. In contrast, it showed a low relative abundance of archaea in both DNA and RNA sequence libraries at site ROV07-01. The sequences affiliated to ANME-1a, ANME-1b, and ANME-2c were only obtained from the sediments of ROV07-01. Methanogen sequences belonging to Methanimicrococcus were only predominant in the surface sediment layer at site ROV05-02, and Methanococcoides occurred predominantly in the sediments with dead clam shells. In addition, Methanococcoides were dominant in all sediment layers at site ROV07-02 in RNA libraries, although few ANME communities were detected at this site. The amplification of Lokiarchaeia, Bathyarchaeia, and Thermoplasmata also occurred as major groups at site ROV07-02.
PCA analysis was performed to examine variations of the active archaeal communities at the RNA levels across the three different cores, and two optimal PCA dimensions (37.1% by first axis and 22.1% by second axis) were presented in Supplementary Figure 3. The results confirmed that archaeal communities in most samples collected from each push core were clustered together, substantiating the different communities and activities in each site. In detail, ANME-2dI and Methanimicrococcus showed a preference for the sediments of ROV05-02; ANME-2a/b, ANME-2dII, ANME-1a, and ANME-1b were mainly detected in the sediments of ROV07-01; Methanococcoides, Lokiarchaeia, Bathyarchaeia, and Thermoplasmata were mostly located in the sediments of ROV07-02. The lack of ANMEs in ROV07-02 was in contrast to the diverse and apparently active ANMEs in ROV07-01.
Phylogenetic Diversity of Metabolically Active Bacteria
A total of 674,266 16S rRNA sequences of bacteria were obtained from 16 sediment samples (Site ROV05-02 and ROV07-01), and were assigned to 7,083 bacterial OTUs (Supplementary Table 3). The bacterial phyla mainly consisted of Proteobacteria, Chloroflexi, Acidobacteria, Bacteroidetes, Atribacteria, Actinobacteria, and Epsilonbacteraeota (Figure 5). Among them, Proteobacteria were predominant in both DNA and RNA sequence libraries (especially in RNA sequence libraries) at the site of ROV07-01 (averaging 69.8%). According to RNA sequence libraries of the sediments of ROV05-02 and ROV07-01, Deltaproteobacteria was the most important group of Proteobacteria. Most of the sequences belonging to Deltaproteobacteria were affiliated with sulfate-reducing bacteria from families of Desulfobacteraceae, Desulfobulbaceae, Desulfarculaceae, and Desulfuromonadaceae. More abundant sulfate-reducing bacteria in ROV07-01 even further substantiated the likely highly anaerobic conditions in ROV07-01 sediments and likely low redox condition in ROV05-02 sediments. Aerobic methanotrophs of Gammaproteobacteria that are related to type I methanotrophs occurred in RNA sequences and showed more abundance in shallow sediment layers. However, aerobic methanotrophs of Alphaproteobacteria related to Type II methanotrophs occurred in most sediment samples in DNA sequence libraries and were barely detected in RNA sequence libraries. Giant sulfide-oxidizing bacteria of Gammaproteobacteria represented by Beggiatoa showed relatively higher abundance at the site of ROV05-02, especially in the surface layer. However, sulfide-oxidizing bacteria of Ectothiorhodospiraceae and Chromatiaceae within Gammaproteobacteria showed a high abundance in the sediments of ROV07-01. Geobacteraceae consisting almost entirely of Geothermobacter were abundant in the sediments of ROV05-02; however, they were rarely detected at the sites of other push cores. Thermodesulfovibrionia affiliated to Nitrospirae was also found from the sediments of ROV05-02. Sequences that belong to Chloroflexi were dominant throughout all sediment cores in DNA sequence libraries; other sequences affiliated to Acidobacteria, Bacteroidetes, Atribacteria, and Actinobacteria were found in a low proportion in all sediments. Active bacteria were not ascertained at site ROV07-02 due to their low abundance.
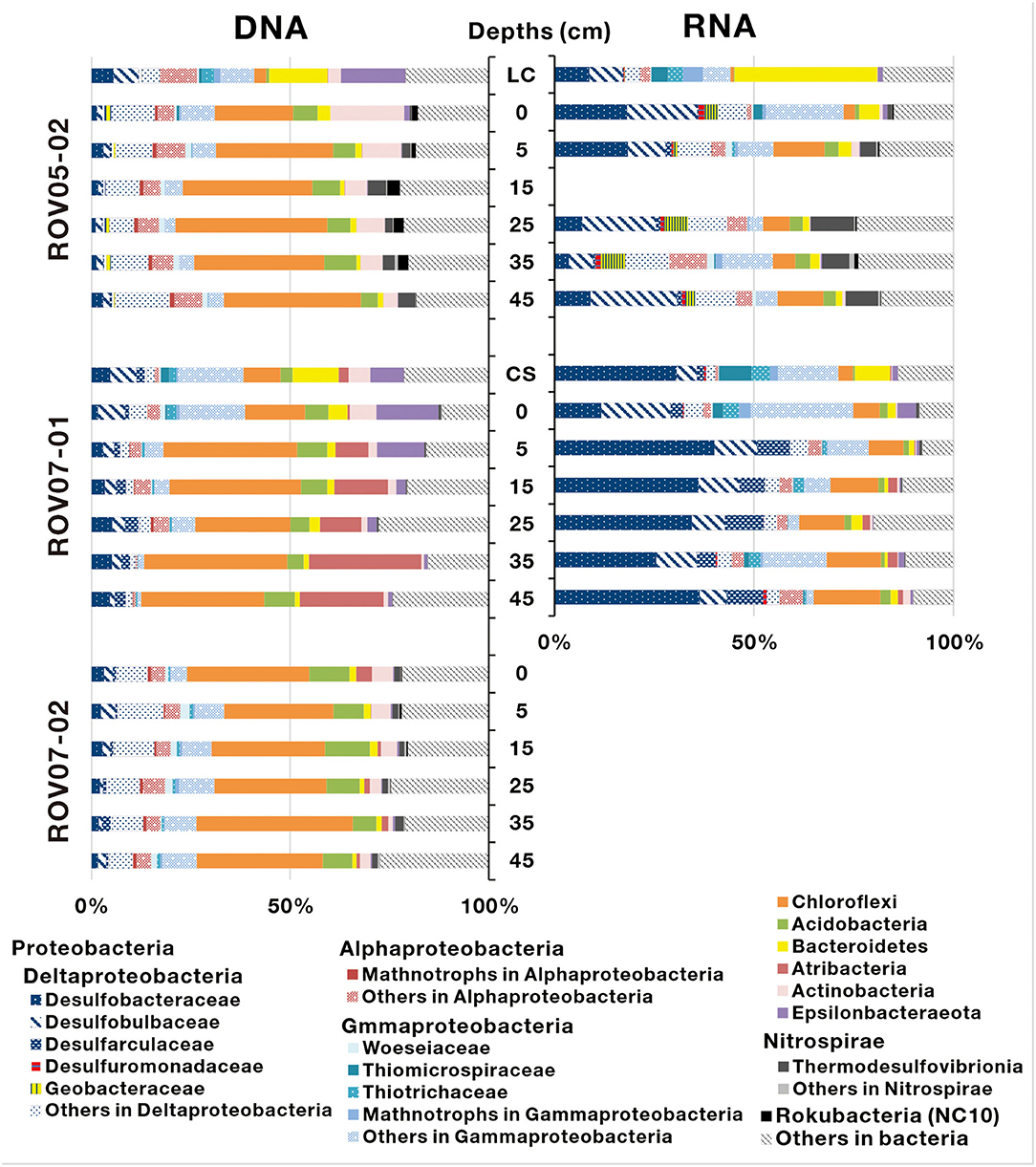
Figure 5. Bacterial community composition determined based on 16S rDNA and 16S rRNA-derived sequences in the sediments of cold seeps. LC and CS indicate the sediment samples with living clams and die-off clam shells at surface sediment, respectively.
Network Co-occurrence Analysis Regarding Archaeal Anaerobic Methanotrophs
To gain insights into potential biotic interplays, where ANMEs exhibited their distinct metabolic capabilities within communities and strong correlations with their syntrophic partnerships, network co-occurrence patterns of archaeal and bacterial populations were determined based on Pearson correlation analysis (Supplementary Figure 4). In order to focus on methane-metabolizing archaea, the sub-networks regarding relationships between the methane-metabolizing archaea with other microbial taxa were selected (Figure 6).
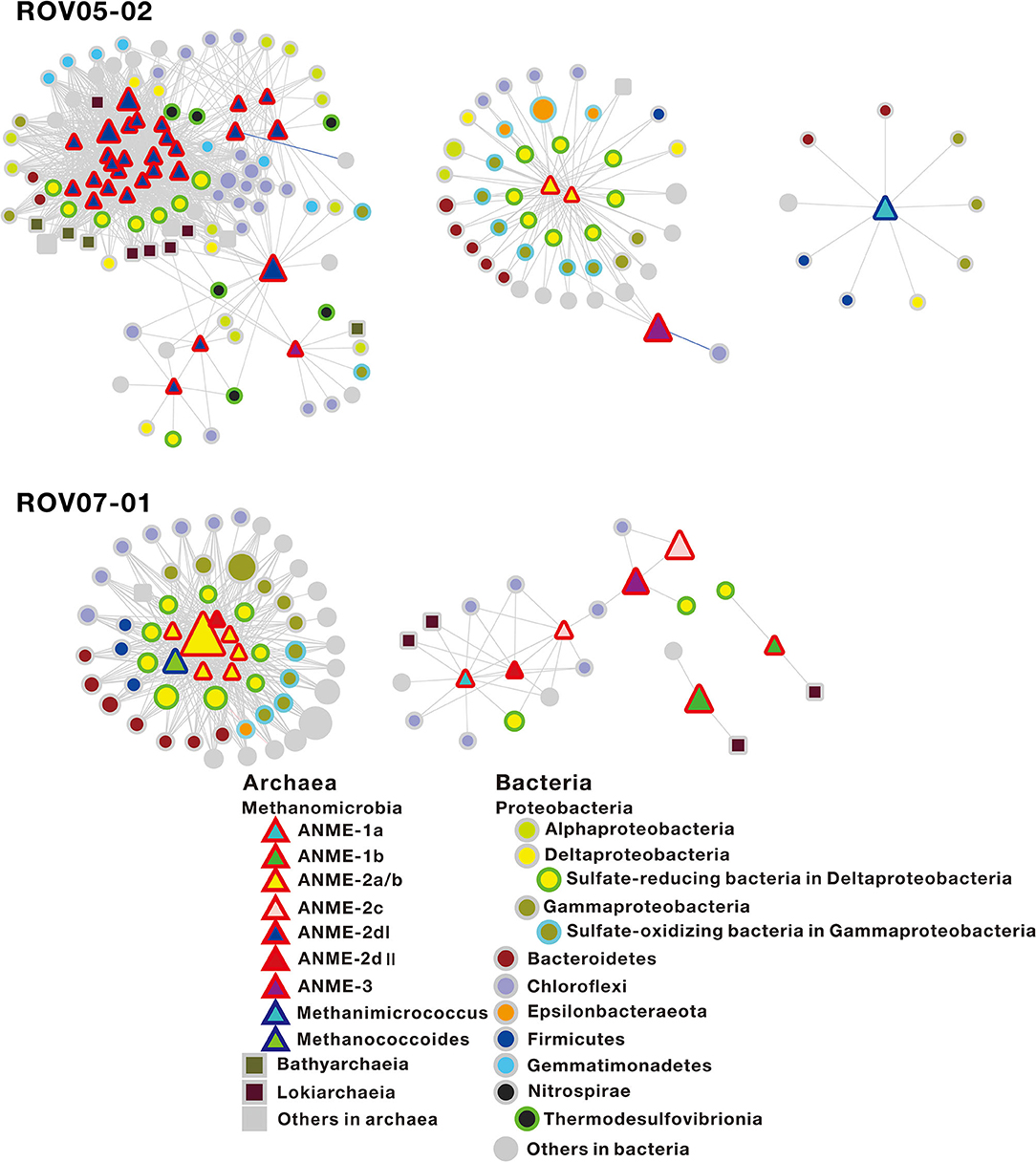
Figure 6. Sub-network co-occurrence patterns regarding the relationship of the methane-metabolizing archaea with other microbial taxa in the sediments of ROV05-02 and ROV07-01 in the cold seeps. The sub-network of the sediments of ROV07-02 is not presented, as only one node belongs to methane-metabolizing archaea. The red-sided triangles, blue-sided triangles, squares, and circles in the network represent ANMEs, archaeal methanotrophs, other archaea, and bacteria, respectively. Node size indicates the relative abundance of OTUs. Edges represent co-occurrence relationships between nodes (coefficient >0.9 or < −0.9, P ≤ 0.01). Gray edges and blue edges represent positive and negative relationships, respectively. The specific OTUs correlated with ANME-2dI and ANME-2a/b are shown in Supplementary Table 5.
At the site of ROV05-02, ANME-2a/b (2 nodes), ANME-2dI (28 nodes), ANME-3 (3 nodes), and Methanimicrococcus (1 node) were represented in the co-occurrence network, with ANME-2a/b and ANME-2d being present in the centers of different modules. In general, ANME-2dI tended to be connected with sulfate-reducing bacteria, such as Desulfatiglans (Desulfarculaceae), Sva0081 sediment group (Desulfobacteraceae), unclassified groups (Desulfobulbaceae), and Thermodesulfovibrionia (Nitrospirae) (Supplementary Table 5). In addition, ANME-2dI most likely had indirect trophic relationships with other microorganisms, including Dehalococcoidia (Chloroflexi), JG30-KF-CM66 (Chloroflexi), Gemmatimonadetes, Bathyarchaeia, and Lokiarchaeia. ANME-2a/b tended to be connected with multiple sulfate-reducing bacteria and sulfide-oxidizing bacteria, with the former including SEEP-SRB1 (Desulfobulbaceae), SEEP-SRB4 (Desulfobulbaceae), Desulfocapsa (Desulfobulbaceae), and the latter including Gammaproteobacteria and Epsilonbacteraeota (such as Woeseiaceae, Thiomicrospiraceae, Thiotrichaceae, Sulfurovum, and Sulfurimonas) (Figure 6). In addition, ANME-3 was present on the margin of ANME-2a/b and ANME-2d modules or isolated alone. The network analyses were insightful in delineating distinct relationships between consortia such as ANME-2dI/Deltaproteobacteria. These were distinct from the ANME-2a/b/Deltaproteobacteria and ANME-2a/b/Gammaproteobacteria networks. At site ROV07-01, ANME-1a (1 node), ANME-1b (2 nodes), ANME-2a/b (6 nodes), ANME-2c (2 nodes), ANME-2dII (2 nodes), ANME-3 (1 node), and Methanococcoides (1 node) were represented in the co-occurrence network. ANME-2a/b was still in the center of the subnetwork module (Figure 6), where the connected taxa were similar to those at site ROV05-02. In addition, two nodes belonging to ANME-2dII and Methanococcoides were also present in the module center. For site ROV07-02, the sub-network was not presented since nodes belonging to methanogens and ANMEs were lacking (Supplementary Figure 4). Chloroflexi were connected with other microbial nodes. In Archaea, Bathyarchaeia preferred connections with other groups; however, Lokiarchaeia preferred connections with themselves.
The Control of Environmental Geochemical Factors on Archaeal Community Distribution
The environmental geochemical factors shaping the distribution of archaeal communities at the RNA level were analyzed by redundancy analysis (RDA) to gain a better understanding of the ecological niche of methane-metabolizing archaea. The predictor variables of methane, sulfate, DIC and δ13CDIC, TA, particle size, and Ca2+, Mg2+, Na+, K+, and Cl− were selected for RDA (Figure 7, Supplementary Table 4). The results indicated that methane, sulfate, DIC and δ13CDIC, TA, particle size, Ca2+ were significant factors, which explained 73.7% (43.4% by first axis and 30.4% by second axis) of the variation in archaeal community distribution. Site ROV07-01 was associated with ANME-1 and−2, while site ROV05-02 was with ANME-2I and−3. For site ROV07-01 the loss of sulfate and diminishing 13CDIC were obvious explanations of the variables in concert with the ANMEs. Methane and TA were predominantly associated with the site ROV05-02 and ROV07-01 in agreement with previous data presented that the site ROV07-02 showed little and sparse evidence of reducing conditions, methanogenesis, or methane oxidation.
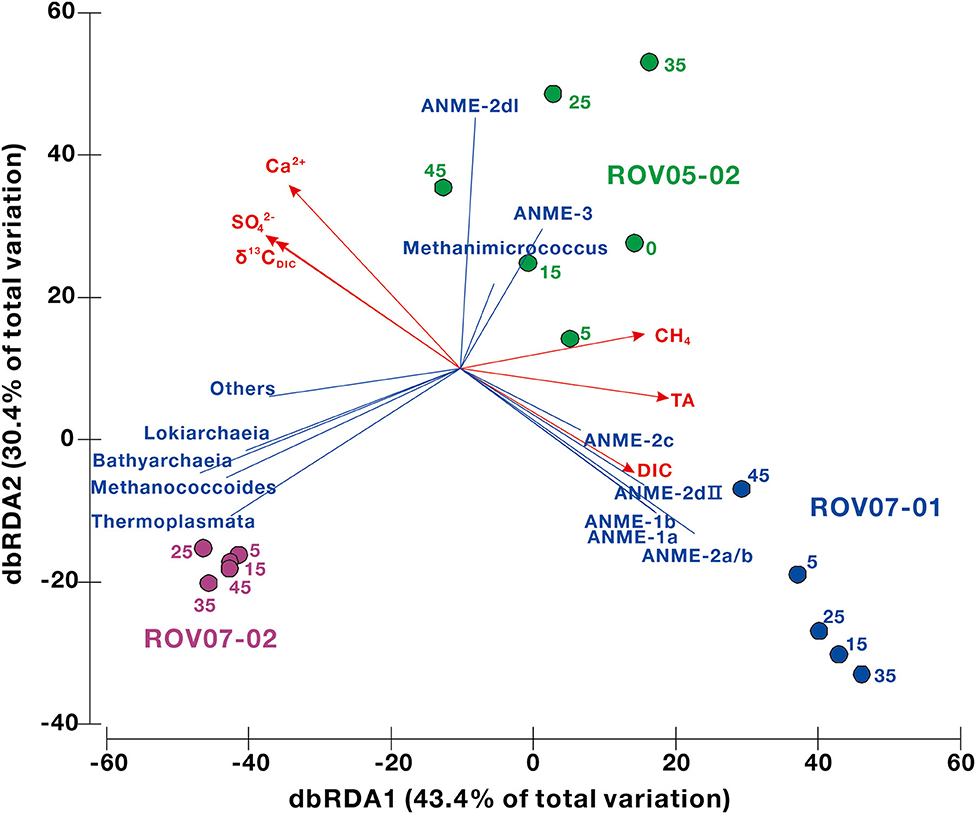
Figure 7. The relationships between the archaeal community structure and the environmental parameters in cold seep sediments determined using the redundancy analysis (RDA). Only significant factors are shown by red vectors. The number next to each node represents sample depth. The proportional contributions of different environmental factors are shown in Supplementary Table 4.
Discussion
Multiple studies have shown that ANMEs are dominant in cold seeps in the SCS (Niu et al., 2017; Cui et al., 2019; Zhuang et al., 2019). However, previous results were based on the DNA level, and thus inactive microorganisms may have been included (Luna et al., 2002; Ostle et al., 2003). The application of the experimental method at the RNA level can more accurately and authentically reflect the changes in the microbial community structure (Ostle et al., 2003; Allison et al., 2010). In this study, the structural patterns of both archaeal and bacterial communities differed substantially between DNA and RNA libraries. In RNA libraries, a high abundance of methane-metabolizing archaea and sulfate-reducing bacteria were present in archaeal and bacterial communities.
Active Archaeal Anaerobic Methanotrophs in Cold Seeps
Active ANMEs appeared to have different ecological preferences in different sediment cores (Supplementary Figure 3). ANME-2a/b is widely distributed in global sediments, and especially dominates in cold seep environments (Knittel et al., 2005; Biddle et al., 2012). A large amount of ANME-2a/b has been detected in cold seeps in the SCS (Niu et al., 2017; Cui et al., 2019; Zhuang et al., 2019). ANME-2a/b was found throughout the entire sediment core ROV07-01 in this study. A large number of dead Calyptogena clam shells and carbonate precipitates accumulated in the ROV07 region (Supplementary Figure 1), suggesting that the cold seep system has developed for quite a long time. The chemical parameters exhibited sulfate removal, DIC increase and δ13CDIC distinction with depth in site ROV07-01 (Figure 2), suggesting this site was highly developed, active and strictly anaerobic.
A large reduction or cessation of seepage activities may have occurred at one time in Haima cold seeps as reported (Liang et al., 2017), causing the covering of Calyptogena clam deaths. Therefore, benthic organisms did not include Bathymodiolin mussels and tubeworms, which are representative of later stage cold seeps evolution (Bowden et al., 2013). Presently, methane continues to seep, supporting archaeal anaerobic methanotrophs in ROV07-01 sediments. Compared to DNA sequences, ANME-2a/b was more predominant and the abundance of ANME-1a and ANME-1b was relatively lower at the RNA level (Figure 4).
The obvious changes in archaea community structure detected by these two methods may be related to the change in methane leakage flux in the ROV07 region. Since ANME-2 had a preference for sediments with high methane flux and showed significantly higher AOM rates while ANME-1 exhibited an opposite trend, it is speculated that an increase in methane flux may have occurred in recent times. Moreover, the abundance of sulfate-reducing bacteria significantly increased at the RNA level, suggesting that sulfate-dependent methane oxidation occurred in the sediments of ROV07-01. The abundant sulfide-oxidizing bacteria of Ectothiorhodospiraceae and Chromatiaceae (Meyer et al., 2007) within Gammaproteobacteria suggested the sufficient source of sulfide at this site. In addition, dense populations of ANME-2a/b discovered in the sediments with living Calyptogena clams (LC) or die-off clam shells (CS) might be linked to seawater sulfate and substrate availability to Calyptogena clams.
All ANME-2d in the samples were of mud volcano and hydrothermal origin (Pachiadaki et al., 2011; Schauer et al., 2011; Chang et al., 2012; Li et al., 2013), and were not derived from terrestrial origins (Supplementary Figure 2). The ANME-2d in the samples was further divided into sub-clades according to phylogenetic analysis, namely ANME-2dI and ANME-2dII, with an internal similarity of approximately 83.88–87.91%. The majority of ANME-2d sequences fell into ANME-2dI, which was dominant in ROV05-02 sediments (up to 73.5% in total archaea at the RNA level). ANME-2d was previously found in marine sediments such as a deep-sea gas chimney (Schrenk et al., 2003), hydrothermal vents (Schauer et al., 2011; Li et al., 2013), mud volcanos (Pachiadaki et al., 2011), and cold seep sediments (Reed et al., 2009; Vigneron et al., 2013). However, its abundance remained in extremely low proportion. Although a high abundance of ANME-2d was detected from deep submarine permafrost, the majority of sequences were derived from terrestrial areas (Winkel et al., 2018).
In this study, a high abundance of ANME-2dI was found in the sediments of ROV05-02, which might be related to the special habitat of the cold seep ROV05. It covered a single benthic fauna—living Calyptogena clams, which are early colonizers of short-lived (years to decades) and sulfide-rich surficial sediment patches (Bowden et al., 2013). The existence of Calyptogena suggested that ROV05-02 site possibly contained a small amount of sulfide, even though we did not smell it. The occurrence of giant sulfide-oxidizing bacteria of Gammaproteobacteria represented by Beggiatoa (Grünke et al., 2011) further substantiate the presence of sulfide. The chemical parameters evidencing a lack of sulfate removal, DIC increase and δ13CDIC distinction in site ROV05-02 indicated this site was less developed and less anaerobic than ROV07-01 (Figure 2). The sediments with living Calyptogena (LC) contained higher proportions of Type I methanotrophs, which were often identified as symbionts in marine invertebrates, and they had higher energy efficiency over type II methanotrophs (Petersen and Dubilier, 2009). Meanwhile, the existence of living Calyptogena suggested that the sediments of ROV05-02 originated from the early evolutionary stage of cold seeps, which has been rarely discovered so far in nSCS.
Most of the cold seeps are hundreds or thousands of years old and are rich in diversified fauna and carbonate rocks (Vigneron et al., 2013; Feng and Chen, 2015; Liang et al., 2017). Organic matter accumulating in megafaunal tissues can be released into underlying sediments to be utilized by microbial communities (Sørensen and Glob, 1987). The high abundance of ANME-2dI in deep layers might be linked to upper Calyptogena clams, which could release organic matter to support ANME-2d.
ANME-3 was detected in cold seeps on the northern slope of the SCS, while previously found in the Sonora Margin cold seeps (Vigneron et al., 2013) and submarine mud volcano in the Barents Sea (Losekann et al., 2007). ANME-3 was found to be distributed in both ROV05-02 and ROV07-01 push cores and had a high abundance in shallow layers at 5 cm depth of ROV05-02 (Figure 4). It was previously reported that the ANME-3 habitat was restricted to a rather narrow horizon below surface layers affected by surface communities (Losekann et al., 2007; Vigneron et al., 2013).
Methanococcoides were detected in ROV07-02 sediments as unique methane-metabolizing archaea (Figure 4), or they could be involved in methane generation through reverse methanogenesis as previously suggested (Vigneron et al., 2013; Timmers et al., 2017). The relative abundance of ANMEs in the sediments of ROV07-02 significantly decreased, which was similar to that in the non-cold seep sediments on the northern slope of the SCS (Niu et al., 2017). The most abundant taxa in DNA libraries belonged to Chloroflexi (Figure 5). Some groups in Chloroflexi (such as Anaerolineae) are able to utilize polysaccharides and cellulose in strictly anaerobic environments (Podosokorskaya et al., 2013; Xia et al., 2016). Other sequences affiliated to Acidobacteria, Bacteroidetes, Atribacteria, and Actinobacteria that are capable of degrading organic matter (Steger et al., 2007; Hanreich et al., 2013; Nobu et al., 2016; Wegner and Liesack, 2017) were found in a low proportion in all sediments. Due to the lack of methane, the microbial communities and geochemical factors were like the background in nSCS. Considering the lack of sulfide smell or sulfate removal, little DIC or δ13CDIC distinction, site ROV07-02 was likely poorly developed, less anaerobic, possessed less methane metabolic communities and exhibited less activity with respect to methane oxidation.
Network Relationship Among ANMEs
Network co-occurrence analysis can be used to elucidate relationships such as syntrophic partnerships and explore the potential ecological roles of ANMEs. According to the analysis, large numbers of OTUs belonging to ANME-2d and ANME-2a/b were in the center of the sub-networks of ROV05-02 and ROV07-01 sediments, respectively (Figure 6). This suggests that ANME-2d and ANME-2a/b may interact strongly with surrounding microorganisms, directly or indirectly affecting their activity. ANME-2dII was connected with ANME-2a/b, while ANME-2dI in different modules existed separately from ANME-2a/b. Both ANME-2dI and ANME-2a/b were connected with sulfate-reducing bacteria in Deltaproteobacteria, suggesting that they could conduct sulfate-dependent AOM. However, ANME-2dI and ANME-2a/b were not sharing the OTUs of microbes, including the sulfate-reducing bacteria. ANME-2a/b can form syntrophic aggregates with bacterial partners, such as Desulfosarcina (Knittel and Boetius, 2009).
As shown by the network analysis in this study, ANME-2a/b was associated with multiple sulfate-reducing bacteria, including SEEP-SRB1 (Desulfobulbaceae), SEEP-SRB4 (Desulfobulbaceae), and Desulfocapsa (Desulfobulbaceae) (Figure 6). AOM associated with sulfate reduction performed by ANME-2a/b was also observed in incubations at high sulfate levels (Timmers et al., 2015). ANME-2a/b could be distinguished from ANME-2dI by their relationship with OTUs affiliated with Gammaproteobateira and Epsilonbacteraeota in networks. Meanwhile, most of the related OTUs belong to sulfur-oxidizing bacteria, such as Woeseiaceae (Marques et al., 2019), Thiomicrospiraceae (Eberhard et al., 1995), Thiotrichaceae (Fossing et al., 1995), Sulfurovum (Mori et al., 2018), and Sulfurimonas (Takai et al., 2006). This suggests that ANME-2a/b was indirectly associated with strong sulfur oxidation while coupling with sulfate reduction. In addition, ANME-2a/b was associated with a variety of other groups of microorganisms, but their specific metabolic processes require further scrutiny. Interactions between ANME-2a/b and their symbiotic bacteria might play an important role in the anaerobic environment and promote the high development of site ROV07-01.
The OTUs connected with ANME-2dI belonged to Desulfatiglans (Desulfarculaceae), Sva0081 sediment group (Desulfobacteraceae), and unclassified groups (Desulfobulbaceae) (Supplementary Table 5). ANME-2dI was also connected to the OTUs of Thermodesulfovibrionia (Nitrospirae). Some of the Thermodesulfovibrio were isolated from hydrothermal-related freshwater environments (Henry et al., 1994) and can perform sulfate reduction with a limited range of electron donors (Sekiguchi et al., 2008). In addition, ANME-2dI most likely had an indirect trophic relationship with other microorganisms, including Dehalococcoidia (Chloroflexi), JG30-KF-CM66 (Chloroflexi), Gemmatimonadetes, Bathyarchaeia, and Lokiarchaeia. Both Dehalococcoidia and JG30-KF-CM66 are affiliated to Chloroflexi. Meanwhile, the former contains the genes associated with degradation of various organic matter such as fatty acids and aromatic compounds (Wasmund et al., 2014) and the latter contains genes related to nitrite oxidoreduction (Mori et al., 2019). Gemmatimonadetes can be involved in polyphosphate accumulation and carotenoid production in soil (Zhang H. et al., 2003; Takaichi et al., 2010). However, their ecological function in marine sediments remains unclear. Bathyarchaeia is capable of utilizing recalcitrant organic matter, and it is involved in methane cycling in sediments (Biddle et al., 2006; He et al., 2016; Lazar et al., 2016). Lokiarchaeia are strictly anaerobic and hydrogen-dependent archaea (Sousa et al., 2016) and can degrade substances and syntrophically transfer hydrogen and electrons to bacterial partners (Spang et al., 2019; Imachi et al., 2020).
The metabolic flexibility of ANME-2d is seen in terrestrial environments, facilitating AOM coupled to multiple terminal electron acceptors (Leu et al., 2020) such as nitrate (Haroon et al., 2013), iron, or manganese oxides (Ettwig et al., 2016; Weber et al., 2017). “Ca. Methylomirabilis oxyfera” (NC10 bacteria) and Geobacteraceae have been observed to be the syntrophic bacteria of ANME-2d (Holmes et al., 2002; Arshad et al., 2015). NC10 bacteria perform denitrification coupled to methane oxidation by ANME-2d (Vaksmaa et al., 2017). In detail, the nitrite produced through AOM by ANME-2d will be continually reduced to N2 or ammonia by NC10 (Raghoebarsing et al., 2006; Hu et al., 2011). Geobacteraceae can perform iron reduction coupled to AOM (He et al., 2018). Geothermobacter, one genus of Geobacteraceae, is a representative of iron-reducing microbes that transfer electrons extracellularly to iron oxides (Kashefi et al., 2003; Li et al., 2020).
In this study, the secondary pore water sampling analysis (in May 2020) found that the sediments of ROV05-02 contained extremely low nitrate concentration but high bivalent iron concentration (up to 33.68 μmol/L, unpublished data). The higher relative abundance of Geothermobacter in RNA sequence libraries probably indicates that Geothermobacter was involved in iron reduction in the sediments of ROV05-02. The lack of correlation between ANME-2dI and Geothermobacter suggests that Geothermobacter may not be a syntrophic partner of ANME-2dI. However, the correlation analysis might have been influenced by some unknown factors. In general, the network of ANME-2dI with their relative microbes in site ROV05-02 was significantly different from that of ANME-2a/b, possibly caused by unique metabolic characteristics of ANME-2dI or the lower redox condition than site ROV07-01.
Molecular data at the DNA level highlighted the high-proportion presence of ANME-1, ANME-2c, and ANME-3 relatives in the sediments of ROV05-02 and ROV07-01. However, they were associated with bacteria at a low level. At site ROV07-02, the lack of methanogens and ANMEs associated with other OTUs indicated that the effect of methane-metabolizing archaea on the whole community structure is extremely small, further substantiating that site ROV07-02 was poorly developed and low activity with respect to methane metabolism.
Control of Geochemical Factors Over ANME Distribution
The distribution and metabolism of ANMEs can also be linked to geochemical composition, such as methane, sulfate, and DIC (Timmers et al., 2015; Bowles et al., 2016). As shown by the RDA analysis in this study, methane, sulfate, DIC, and δ13CDIC, TA, and calcium could be the significant factors relating to the archaeal distribution at the RNA level (Figure 7). These environmental factors were dominantly associated with ANMEs in ROV07-01 sediments. Due to predominance of ANME-2a/b in RNA libraries of site ROV07-01 and its high correlation with sulfate-reducing and sulfide-oxidizing bacteria, the majority of AOM could be mediated by ANME-2a/b, utilizing sulfate and producing DIC and TA as revealed by the depleted δ13CDIC (Whiticar, 1999). This could have led to the precipitation of authigenic carbonates and induced the decrease in calcium (Hu et al., 2019). ANME-2a/b have been observed growing in incubation experiments with high sulfate and AOM activity (Timmers et al., 2015). The decrease in sulfate with depth in our study indicated that the effective AOM was likely performed by ANME-2a/b. The DIC and δ13CDIC profile changed sharply at a depth of 30–35 cm in ROV07-01 sediments, while the archaeal community and quantification of mcrA genes did not show any corresponding change. The increasing sand content at this depth could explain the fact that porosity might improve AOM efficiency (Figure 2) (Treude et al., 2005). Thus, the site of ROV07-01 was highly developed, diverse, anaerobic, and active in methane oxidation, providing an environment for ANME-2a/b occurrence.
Dense archaeal populations (such as Lokiarchaeia, Bathyarchaeia, Thermoplasmata) at site ROV07-02 did not show similar correlations with these significant geochemical factors. This result could also be deduced from the lack of methane, sulfate removal, DIC or δ13CDIC distinction with depth. Site ROV07-02 could be less anaerobic and less active with respect to methanogenesis, methane oxidation and related network relationships. Dominant and active ANMEs (such as ANME-2dI and ANME-3) occurred at the ROV05-02 site, suggesting the activity of methane oxidation was present. However, the data of sulfate, DIC, and δ13CDIC with depth was not as convincing as for the site of ROV07-01. This was probably due to the weak AOM performed by ANME-2dI and ANME-3. Meanwhile, the advective import of seawater and impacts by Calyptogena clams (Wallmann et al., 1997; Bertics et al., 2007; Fischer et al., 2012) could also explain the unchanging environmental factors. Findings suggest the ROV05-02 site could be anaerobic in spots and in micro-niches and likely intermediary between the highly diverse and active ROV07-01 site and the poorly developed low activity ROV07-02 site.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA664582.
Author Contributions
TZ and JZ conceived the study and designed the experiments. ZC provided the samples and the picture of seafloor observations. XX determined the physicochemical parameters. TZ, XX, and SC analyzed the data and wrote the manuscript. JF, JZ, QL, TP, and CZ edited and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (Nos. 41806074 and 41730528); the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. GML2019ZD0307); the Guangdong Basic and Applied Basic Research Foundation (No. 2019B030302004); and the Marine Geological Survey Program of China Geological Survey (No. DD20190218). CZ's contribution in this project was supported by the Shenzhen Key Laboratory of Marine Archaea Geo-Omics, Southern University of Science and Technology (No. ZDSYS 20180208184349083), and the Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. K19313901).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all the scientists and crews, in particular ROV operators for their assistance with sampling during cruise. Sincerely thanks to Hongpeng Cui (China University of Geosciences, Beijing) for providing methane data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.612135/full#supplementary-material
References
Allison, S. D., Wallenstein, M. D., and Bradford, M. A. (2010). Soil-carbon response to warming dependent on microbial physiology. Nat. Geosci. 3, 336–340. doi: 10.1038/ngeo846
Arshad, A., Speth, D. R., de Graaf, R. M., Op den Camp, H. J., Jetten, M. S., and Welte, C. U. (2015). A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front. Microbiol. 6:1423. doi: 10.3389/fmicb.2015.01423
Bertics, V., Treude, T., and Ziebis, W. (2007). “Vesicomyid clams alter biogeochemical processes at pacific methane seeps,” in AGU Fall Meeting Abstracts (San Francisco, CA).
Biddle, J. F., Cardman, Z., Mendlovitz, H. P., Albert, D. B., Lloyd, K. G., Boetius, A., et al. (2012). Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 6, 1018–1031. doi: 10.1038/ismej.2011.164
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K. G., Sorensen, K. B., Anderson, R., et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851. doi: 10.1073/pnas.0600035103
Boetius, A., and Wenzhöfer, F. (2013). Seafloor oxygen consumption fuelled by methane from cold seeps. Nat. Geosci. 6:725. doi: 10.1038/ngeo1926
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowden, D. A., Rowden, A. A., Thurber, A. R., Baco, A. R., Levin, L. A., and Smith, C. R. (2013). Cold seep epifaunal communities on the hikurangi margin, New Zealand: composition, succession, and vulnerability to human activities. PLoS ONE 8:e76869. doi: 10.1371/journal.pone.0076869
Bowles, M., Hunter, K. S., Samarkin, V., and Joye, S. (2016). Patterns and variability in geochemical signatures and microbial activity within and between diverse cold seep habitats along the lower continental slope, Northern Gulf of Mexico. Deep Sea Res. II Top. Stud. Oceanogr. 129, 31–40. doi: 10.1016/j.dsr2.2016.02.011
Chang, Y. H., Cheng, T. W., Lai, W. J., Tsai, W. Y., Sun, C. H., Lin, L. H., et al. (2012). Microbial methane cycling in a terrestrial mud volcano in eastern Taiwan. Environ. Microbiol. 14, 895–908. doi: 10.1111/j.1462-2920.2011.02658.x
Charvet, S., Vincent, W. F., and Lovejoy, C. (2014). Effects of light and prey availability on Arctic freshwater protist communities examined by high-throughput DNA and RNA sequencing. FEMS Microbiol. Ecol. 88, 550–564. doi: 10.1111/1574-6941.12324
Cordes, E. E., Arthur, M. A., Shea, K., Arvidson, R. S., and Fisher, C. R. (2005). Modeling the mutualistic interactions between tubeworms and microbial consortia. PLoS Biol. 3:e77. doi: 10.1371/journal.pbio.0030077
Cui, H., Su, X., Chen, F., Holland, M., Yang, S., Liang, J., et al. (2019). Microbial diversity of two cold seep systems in gas hydrate-bearing sediments in the South China Sea. Mar. Environ. Res. 144, 230–239. doi: 10.1016/j.marenvres.2019.01.009
Eberhard, C., Wirsen, C. O., and Jannasch, H. W. (1995). Oxidation of polymetal sulfides by chemolithoautotrophic bacteria from deep-sea hydrothermal vents. Geomicrobiol. J. 13, 145–164. doi: 10.1080/01490459509378014
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Ettwig, K. F., Zhu, B., Speth, D., Keltjens, J. T., Jetten, M. S., and Kartal, B. (2016). Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. U.S.A. 113, 12792–12796. doi: 10.1073/pnas.1609534113
Feng, D., and Chen, D. (2015). Authigenic carbonates from an active cold seep of the northern South China Sea: new insights into fluid sources and past seepage activity. Deep Sea Res. II Top. Stud. Oceanogr. 122, 74–83. doi: 10.1016/j.dsr2.2015.02.003
Feng, J., Li, N., Luo, M., Liang, J., Yang, S., Wang, H., et al. (2020). A quantitative assessment of methane-derived carbon cycling at the cold seeps in the Northwestern South China Sea. Minerals 10:256. doi: 10.3390/min10030256
Fischer, D., Sahling, H., Nöthen, K., Bohrmann, G., Zabel, M., and Kasten, S. (2012). Interaction between hydrocarbon seepage, chemosynthetic communities, and bottom water redox at cold seeps of the Makran accretionary prism: insights from habitat-specific pore water sampling and modeling. Biogeosciences 9, 2013–2031. doi: 10.5194/bg-9-2013-2012
Fletcher, S. E. M., and Schaefer, H. (2019). Rising methane: a new climate challenge. Science 364, 932–933. doi: 10.1126/science.aax1828
Fossing, H., Gallardo, V. A., Jorgensen, B. B., Huttel, M., Nielsen, L. P., Schulz, H. N., et al. (1995). Concentration and transport of nitrate by the mat-forming sulphur bacterium Thioploca. Nature 374, 713–715. doi: 10.1038/374713a0
Grünke, S., Felden, J., Lichtschlag, A., Girnth, A. C., de Beer, D., Wenzhöfer, F., et al. (2011). Niche differentiation among mat-forming, sulfide-oxidizing bacteria at cold seeps of the Nile Deep Sea Fan (Eastern Mediterranean Sea). Geobiology 9, 330–348. doi: 10.1111/j.1472-4669.2011.00281.x
Guan, H., Birgel, D., Peckmann, J. L., Liang, Q., Feng, D., Yang, S., et al. (2018). Lipid biomarker patterns of authigenic carbonates reveal fluid composition and seepage intensity at Haima cold seeps, South China Sea. J. Asian Earth Sci. 168, 163–172. doi: 10.1016/j.jseaes.2018.04.035
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
Han, X., Suess, E., Huang, Y., Wu, N., Bohrmann, G., Su, X., et al. (2008). Jiulong methane reef: microbial mediation of seep carbonates in the South China Sea. Mar. Geol. 249, 243–256. doi: 10.1016/j.margeo.2007.11.012
Hanreich, A., Schimpf, U., Zakrzewski, M., Schlüter, A., Benndorf, D., Heyer, R., et al. (2013). Metagenome and metaproteome analyses of microbial communities in mesophilic biogas-producing anaerobic batch fermentations indicate concerted plant carbohydrate degradation. Syst. Appl. Microbiol. 36, 330–338. doi: 10.1016/j.syapm.2013.03.006
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
He, Y., Li, M., Perumal, V., Feng, X., Fang, J., Xie, J., et al. (2016). Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 1:16035. doi: 10.1038/nmicrobiol.2016.35
He, Z., Zhang, Q., Feng, Y., Luo, H., Pan, X., and Gadd, G. M. (2018). Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane. Sci. Total Environ. 610–611, 759–768. doi: 10.1016/j.scitotenv.2017.08.140
Henry, E. A., Devereux, R., Maki, J. S., Gilmour, C. C., Woese, C. R., Mandelco, L., et al. (1994). Characterization of a new thermophilic sulfate-reducing bacterium thermodesulfovibrio yellowstonii, gen. nov. and sp. nov.: its phylogenetic relationship to thermodesulfobacterium commune and their origins deep within the bacterial domain. Arch. Microbiol. 161, 62–69. doi: 10.1007/s002030050022
Holmes, D. E., Finneran, K. T., O'neil, R. A., and Lovley, D. R. (2002). Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68, 2300–2306. doi: 10.1128/AEM.68.5.2300-2306.2002
Hu, S., Zeng, R. J., Keller, J., Lant, P. A., and Yuan, Z. (2011). Effect of nitrate and nitrite on the selection of microorganisms in the denitrifying anaerobic methane oxidation process. Environ. Microbiol. Rep. 3, 315–319. doi: 10.1111/j.1758-2229.2010.00227.x
Hu, Y., Luo, M., Liang, Q., Chen, L., Feng, D., Yang, S., et al. (2019). Pore fluid compositions and inferred fluid flow patterns at the Haima cold seeps of the South China Sea. Mar. Petroleum Geol. 103, 29–40. doi: 10.1016/j.marpetgeo.2019.01.007
Imachi, H., Nobu, M. K., Nakahara, N., Morono, Y., Ogawara, M., Takaki, Y., et al. (2020). Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519–525. doi: 10.1038/s41586-019-1916-6
IPCC (2014). Climate Change 2014: Mitigation of Climate Change. Contribution of Working Group III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, UK: Cambridge University Press.
Judd, A., and Hovland, M. (2009). Seabed Fluid Flow: the Impact on Geology, Biology and the Marine Environment. Cambridge, UK: Cambridge University Press.
Karl, D. M., and Bailiff, M. D. (1989). The measurement and distribution of dissolved nucleic acids in aquatic environments. Limnol. Oceanogr. 34, 543–558. doi: 10.4319/lo.1989.34.3.0543
Kashefi, K., Holmes, D. E., Baross, J. A., and Lovley, D. R. (2003). Thermophily in the geobacteraceae: geothermobacter ehrlichii gen. nov., sp. nov., a novel thermophilic member of the geobacteraceae from the “Bag City” hydrothermal vent. Appl. Environ. Microbiol. 69, 2985–2993. doi: 10.1128/AEM.69.5.2985-2993.2003
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Knittel, K., Lösekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479. doi: 10.1128/AEM.71.1.467-479.2005
Kvenvolden, K. A., and Rogers, B. W. (2005). Gaia's breath—global methane exhalations. Mar. Petroleum Geol. 22, 579–590. doi: 10.1016/j.marpetgeo.2004.08.004
Lazar, C. S., Baker, B. J., Seitz, K., Hyde, A. S., Dick, G. J., Hinrichs, K. U., et al. (2016). Genomic evidence for distinct carbon substrate preferences and ecological niches of bathyarchaeota in estuarine sediments. Environ. Microbiol. 18, 1200–1211. doi: 10.1111/1462-2920.13142
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucl. Acids Res. 44, W242–5. doi: 10.1093/nar/gkw290
Leu, A. O., McIlroy, S. J., Ye, J., Parks, D. H., Orphan, V. J., and Tyson, G. W. (2020). Lateral gene transfer drives metabolic flexibility in the anaerobic methane oxidising archaeal family methanoperedenaceae. MBio 11:e01325. doi: 10.1128/mBio.01325-20
Levin, L. A. (2005). “Ecology of cold seep sediments: interactions of fauna with flow, chemistry and microbes,” in Oceanography and Marine Biology: An Annual Review, Vol. 43, eds R. N. Gibson, R. J. A. Atkinson, and J. D. M. Gordon (Taylor & Francis), 1–46.
Levin, L. A., and Sibuet, M. (2012). Understanding continental margin biodiversity: a new imperative. Ann. Rev. Mar. Sci. 4, 79–112. doi: 10.1146/annurev-marine-120709-142714
Li, J., Peng, X., Zhou, H., Li, J., and Sun, Z. (2013). Molecular evidence for microorganisms participating in Fe, Mn, and S biogeochemical cycling in two low-temperature hydrothermal fields at the Southwest Indian Ridge. J. Geophys. Res. Biogeosci. 118, 665–679. doi: 10.1002/jgrg.20057
Li, X., Ding, L., Li, X., and Zhu, Y. (2020). Abundance, diversity, and structure of geobacteraceae community in paddy soil under long-term fertilization practices. Appl. Soil Ecol. 153:103577. doi: 10.1016/j.apsoil.2020.103577
Liang, J., Zhang, W., Lu, J. A., Wei, J., Kuang, Z., and He, Y. (2019). Geological occurrence and accumulation mechanism of natural gas hydrates in the eastern qiongdongnan basin of the South China Sea: insights from site GMGS5-W9-2018. Mar. Geol. 418:106042. doi: 10.1016/j.margeo.2019.106042
Liang, Q., Hu, Y., Feng, D., Peckmann, J., Chen, L., Yang, S., et al. (2017). Authigenic carbonates from newly discovered active cold seeps on the northwestern slope of the South China Sea: Constraints on fluid sources, formation environments, and seepage dynamics. Deep Sea Res. I Oceanogr. Res. Papers 124, 31–41. doi: 10.1016/j.dsr.2017.04.015
Lloyd, K. G., Lapham, L., and Teske, A. (2006). An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl. Environ. Microbiol. 72, 7218–7230. doi: 10.1128/AEM.00886-06
Lorenz, M. G., and Wackernagel, W. (1987). Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl. Environ. Microbiol. 53, 2948–2952. doi: 10.1128/AEM.53.12.2948-2952.1987
Losekann, T., Knittel, K., Nadalig, T., Fuchs, B. M., Niemann, H., Boetius, A., et al. (2007). Diversity and abundance of aerobic and anaerobic methane oxidizers at the haakon mosby mud volcano, barents sea. Appl. Environ. Microbiol. 73, 3348–3362. doi: 10.1128/AEM.00016-07
Luna, G. M., Manini, E., and Danovaro, R. (2002). Large fraction of dead and inactive bacteria in coastal marine sediments: comparison of protocols for determination and ecological significance. Appl. Environ. Microbiol. 68, 3509–3513. doi: 10.1128/AEM.68.7.3509-3513.2002
Maignien, L., Parkes, R. J., Cragg, B., Niemann, H., Knittel, K., Coulon, S., et al. (2012). Anaerobic oxidation of methane in hypersaline cold seep sediments. FEMS Microbiol. Ecol. 83, 214–231. doi: 10.1111/j.1574-6941.2012.01466.x
Marques, E. L. S., Dias, J. C. T., Gross, E., de Cerqueira e Silva, A. B., Moura, S. R., Rezende, R. P., et al. (2019). Purple sulfur bacteria dominate microbial community in brazilian limestone cave. Microorganisms 7:29. doi: 10.3390/microorganisms7020029
McKay, L., Klokman, V. W., Mendlovitz, H. P., LaRowe, D. E., Hoer, D. R., Albert, D., et al. (2016). Thermal and geochemical influences on microbial biogeography in the hydrothermal sediments of G uaymas B asin, G ulf of C alifornia. Environ. Microbiol. Rep. 8, 150–161. doi: 10.1111/1758-2229.12365
Meyer, B., Imhoff, J. F., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria–evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9, 2957–2977. doi: 10.1111/j.1462-2920.2007.01407.x
Milkov, A. V., Sassen, R., Apanasovich, T. V., and Dadashev, F. G. (2003). Global gas flux from mud volcanoes: a significant source of fossil methane in the atmosphere and the ocean. Geophys. Res. Lett. 30:1037. doi: 10.1029/2002GL016358
Mills, H. J., Cassie, H., Kristin, W., Macdonald, I. R., and Sobecky, P. A. (2003). Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46, 39–52. doi: 10.1016/S0168-6496(03)00191-0
Mori, J. F., Chen, L., Jessen, G. L., Rudderham, S. B., Mcbeth, J. M., Lindsay, M. B. J., et al. (2019). Putative mixotrophic nitrifying-denitrifying gammaproteobacteria implicated in nitrogen cycling within the ammonia/oxygen transition zone of an oil sands pit lake. Front. Microbiol. 10:2435. doi: 10.3389/fmicb.2019.02435
Mori, K., Yamaguchi, K., and Hanada, S. (2018). Sulfurovum denitrificans sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing epsilonproteobacterium isolated from a hydrothermal field. Int. J. Syst. Evol. Microbiol. 68, 2183–2187. doi: 10.1099/ijsem.0.002803
Nauhaus, K., Treude, T., Boetius, A., and Krüger, M. (2005). Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 7, 98–106. doi: 10.1111/j.1462-2920.2004.00669.x
Niemann, H., Lösekann, T., De Beer, D., Elvert, M., Nadalig, T., Knittel, K., et al. (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858. doi: 10.1038/nature05227
Niu, M., Fan, X., Zhuang, G., Liang, Q., and Wang, F. (2017). Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea. FEMS Microbiol. Ecol. 93, 1–13. doi: 10.1093/femsec/fix101
Nobu, M. K., Dodsworth, J. A., Murugapiran, S. K., Rinke, C., Gies, E. A., Webster, G., et al. (2016). Phylogeny and physiology of candidate phylum ‘Atribacteria’(OP9/JS1) inferred from cultivation-independent genomics. ISME J. 10, 273–286. doi: 10.1038/ismej.2015.97
Orphan, V., Hinrichs, K.-U., Ussler, W., Paull, C. K., Taylor, L., Sylva, S. P., et al. (2001). Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67, 1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001
Orphan, V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D., and DeLong, E. F. (2002). Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U.S.A. 99, 7663–7668. doi: 10.1073/pnas.072210299
Ostle, N. J., Whiteley, A. S., Bailey, M. J., Sleep, D., Ineson, P., and Manefield, M. (2003). Active microbial RNA turnover in a grassland soil estimated using a 13CO2 spike. Soil Biol. Biochem. 35, 877–885. doi: 10.1016/S0038-0717(03)00117-2
Pachiadaki, M. G., Kallionaki, A., Dählmann, A., De Lange, G. J., and Kormas, K. A. (2011). Diversity and spatial distribution of prokaryotic communities along a sediment vertical profile of a deep-sea mud volcano. Microb. Ecol. 62, 655–668. doi: 10.1007/s00248-011-9855-2
Pawlowski, J., Esling, P., Lejzerowicz, F., Cedhagen, T., and Wilding, T. A. (2014). Environmental monitoring through protist next-generation sequencing metabarcoding: assessing the impact of fish farming on benthic foraminifera communities. Mol. Ecol. Resour. 14, 1129–1140. doi: 10.1111/1755-0998.12261
Petersen, J. M., and Dubilier, N. (2009). Methanotrophic symbioses in marine invertebrates. Environ. Microbiol. Rep. 1, 319–335. doi: 10.1111/j.1758-2229.2009.00081.x
Podosokorskaya, O., Bonch-Osmolovskaya, E., Novikov, A., Kolganova, T., and Kublanov, I. (2013). Ornatilinea apprima gen. nov., sp. nov., a cellulolytic representative of the class anaerolineae. Int. J. Syst. Evol. Microbiol. 63, 86–92. doi: 10.1099/ijs.0.041012-0
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Raghoebarsing, A. A., Pol, A., van de Pas-Schoonen, K. T., Smolders, A. J., Ettwig, K. F., Rijpstra, W. I., et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921. doi: 10.1038/nature04617
Reeburgh, W. S. (2007). Oceanic methane biogeochemistry. Chem. Rev. 107, 486–513. doi: 10.1021/cr050362v
Reed, A. J., Dorn, R., Van Dover, C. L., Lutz, R. A., and Vetriani, C. (2009). Phylogenetic diversity of methanogenic, sulfate-reducing and methanotrophic prokaryotes from deep-sea hydrothermal vents and cold seeps. Deep Sea Res. II Top. Stud. Oceanogr. 56, 1665–1674. doi: 10.1016/j.dsr2.2009.05.012
Regnier, P., Dale, A. W., Arndt, S., LaRowe, D., Mogollón, J., and Van Cappellen, P. (2011). Quantitative analysis of anaerobic oxidation of methane (AOM) in marine sediments: a modeling perspective. Earth Sci. Rev. 106, 105–130. doi: 10.1016/j.earscirev.2011.01.002
Sahling, H., Rickert, D., Lee, R. W., Linke, P., and Suess, E. (2002). Macrofaunal community structure and sulfide flux at gas hydrate deposits from the Cascadia convergent margin, NE Pacific. Mar. Ecol. Prog. Ser. 231, 121–138. doi: 10.3354/meps231121
Schauer, R., Røy, H., Augustin, N., Gennerich, H. H., Peters, M., Wenzhoefer, F., et al. (2011). Bacterial sulfur cycling shapes microbial communities in surface sediments of an ultramafic hydrothermal vent field. Environ. Microbiol. 13, 2633–2648. doi: 10.1111/j.1462-2920.2011.02530.x
Schrenk, M. O., Kelley, D. S., Delaney, J. R., and Baross, J. A. (2003). Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol. 69, 3580–3592. doi: 10.1128/AEM.69.6.3580-3592.2003
Sekiguchi, Y., Muramatsu, M., Imachi, H., Narihiro, T., Ohashi, A., Harada, H., et al. (2008). Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic, thermophilic, sulfate-reducing bacteria isolated from thermophilic methanogenic sludge, and emended description of the genus thermodesulfovibrio. Int. J. Syst. Evol. Microbiol. 58, 2541–2548. doi: 10.1099/ijs.0.2008/000893-0
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sørensen, J., and Glob, E. (1987). Influence of benthic fauna on trimethylamine concentrations in coastal marine sediments. Mar. Ecol. Progr. Ser. 39, 15–21. doi: 10.3354/meps039015
Sousa, F. L., Neukirchen, S., Allen, J. F., Lane, N., and Martin, W. (2016). Lokiarchaeon is hydrogen dependent. Nat. Microbiol. 1:16034. doi: 10.1038/nmicrobiol.2016.34
Spang, A., Stairs, C. W., Dombrowski, N., Eme, L., Lombard, J., Caceres, E. F., et al. (2019). Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat. Microbiol. 4, 1138–1148. doi: 10.1038/s41564-019-0406-9
Steger, K., Jarvis, Å., Vasara, T., Romantschuk, M., and Sundh, I. (2007). Effects of differing temperature management on development of actinobacteria populations during composting. Res. Microbiol. 158, 617–624. doi: 10.1016/j.resmic.2007.05.006
Takai, K., Suzuki, M., Nakagawa, S., Miyazaki, M., Suzuki, Y., Inagaki, F., et al. (2006). Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 56, 1725–1733. doi: 10.1099/ijs.0.64255-0
Takaichi, S., Maoka, T., Takasaki, K., and Hanada, S. (2010). Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2′-dirhamnoside. Microbiology 156, 757–763. doi: 10.1099/mic.0.034249-0
Timmers, P. H., Welte, C. U., Koehorst, J. J., Plugge, C. M., Jetten, M. S., and Stams, A. J. (2017). Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237
Timmers, P. H., Widjaja-Greefkes, H., Ramiro-Garcia, J., Plugge, C. M., and Stams, A. J. (2015). Growth and activity of ANME clades with different sulfate and sulfide concentrations in the presence of methane. Front. Microbiol. 6:988. doi: 10.3389/fmicb.2015.00988
Treude, T., Niggemann, J., Kallmeyer, J., Wintersteller, P., Schubert, C., Boetius, A., et al. (2005). Anaerobic oxidation of methane and sulfate reduction along the chilean continental margin. Geochim. Cosmochim. Acta 69, 2767–2779. doi: 10.1016/j.gca.2005.01.002
Vaksmaa, A., Guerrero-Cruz, S., van Alen, T. A., Cremers, G., Ettwig, K. F., Luke, C., et al. (2017). Enrichment of anaerobic nitrate-dependent methanotrophic 'Candidatus methanoperedens nitroreducens' archaea from an Italian paddy field soil. Appl. Microbiol. Biotechnol. 101, 7075–7084. doi: 10.1007/s00253-017-8416-0
Vigneron, A., Cruaud, P., Pignet, P., Caprais, J. C., Cambon Bonavita, M. A., Godfroy, A., et al. (2013). Archaeal and anaerobic methane oxidizer communities in the sonora margin cold seeps, guaymas basin (Gulf of California). ISME J. 7, 1595–608. doi: 10.1038/ismej.2013.18
Wallmann, K., Linke, P., Suess, E., Bohrmann, G., Sahling, H., Schlüter, M., et al. (1997). Quantifying fluid flow, solute mixing, and biogeochemical turnover at cold vents of the eastern aleutian subduction zone. Geochim. Cosmochim. Acta 61, 5209–5219. doi: 10.1016/S0016-7037(97)00306-2
Wasmund, K., Schreiber, L., Lloyd, K. G., Petersen, D. G., Schramm, A., Stepanauskas, R., et al. (2014). Genome sequencing of a single cell of the widely distributed marine subsurface dehalococcoidia, phylum chloroflexi. ISME J. 8, 383–397. doi: 10.1038/ismej.2013.143
Weber, H. S., Habicht, K. S., and Thamdrup, B. (2017). Anaerobic methanotrophic archaea of the ANME-2d cluster are active in a low-sulfate, iron-rich freshwater sediment. Front. Microbiol. 8:619. doi: 10.3389/fmicb.2017.00619
Wegner, C.-E., and Liesack, W. (2017). Unexpected dominance of elusive Acidobacteria in early industrial soft coal slags. Front. Microbiol. 8:1023. doi: 10.3389/fmicb.2017.01023
Wei, J., Liang, J., Lu, J., Zhang, W., and He, Y. (2019). Characteristics and dynamics of gas hydrate systems in the northwestern South China Sea - results of the fifth gas hydrate drilling expedition. Mar. Petroleum Geol. 110, 287–298. doi: 10.1016/j.marpetgeo.2019.07.028
Whiticar, M. J. (1999). Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 161, 291–314. doi: 10.1016/S0009-2541(99)00092-3
Winkel, M., Mitzscherling, J., Overduin, P. P., Horn, F., Winterfeld, M., Rijkers, R., et al. (2018). Anaerobic methanotrophic communities thrive in deep submarine permafrost. Sci. Rep. 8:1291. doi: 10.1038/s41598-018-19505-9
Wu, W., and Liu, H. (2018). Disentangling protist communities identified from DNA and RNA surveys in the Pearl River-South China Sea continuum during the wet and dry seasons. Mol. Ecol. 27, 4627–4640. doi: 10.1111/mec.14867
Xia, Y., Wang, Y., Wang, Y., Chin, F. Y., and Zhang, T. (2016). Cellular adhesiveness and cellulolytic capacity in anaerolineae revealed by omics-based genome interpretation. Biotechnol. Biofuels 9:111. doi: 10.1186/s13068-016-0524-z
Xu, D., Li, R., Hu, C., Sun, P., Jiao, N., and Warren, A. (2017). Microbial eukaryote diversity and activity in the water column of the south china sea based on DNA and RNA high throughput sequencing. Front. Microbiol. 8:1121. doi: 10.3389/fmicb.2017.01121
Ye, J., Wei, J., Liang, J., Lu, J., Lu, H., and Zhang, W. (2019). Complex gas hydrate system in a gas chimney, South China Sea. Mar. Petroleum Geol. 104, 29–39. doi: 10.1016/j.marpetgeo.2019.03.023
Zhang, C. L., Huang, Z., Cantu, J., Pancost, R. D., Brigmon, R. L., Lyons, T. W., et al. (2005). Lipid biomarkers and carbon isotope signatures of a microbial (Beggiatoa) mat associated with gas hydrates in the Gulf of Mexico. Appl. Environ. Microbiol. 71, 2106–2112. doi: 10.1128/AEM.71.4.2106-2112.2005
Zhang, C. L., Li, Y., Wall, J. D., Larsen, L., Sassen, R., Huang, Y., et al. (2002). Lipid and carbon isotopic evidence of methane-oxidizing and sulfate-reducing bacteria in association with gas hydrates from the Gulf of Mexico. Geology 30, 239–242. doi: 10.1130/0091-7613(2002)030<0239:LACIEO>2.0.CO;2
Zhang, C. L., Pancost, R. D., Sassen, R., Qian, Y., and Macko, S. A. (2003). Archaeal lipid biomarkers and isotopic evidence of anaerobic methane oxidation associated with gas hydrates in the Gulf of Mexico. Org. Geochem. 34, 827–836. doi: 10.1016/S0146-6380(03)00003-2
Zhang, H., Sekiguchi, Y., Hanada, S., Hugenholtz, P., Kim, H., Kamagata, Y., et al. (2003). Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate- accumulating micro-organism, the first cultured representative of the new bacterial phylum gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53, 1155–1163. doi: 10.1099/ijs.0.02520-0
Keywords: anaerobic methanotrophs, methane, anaerobic oxidation of methane, cold seeps, calyptogena
Citation: Zhang T, Xiao X, Chen S, Zhao J, Chen Z, Feng J, Liang Q, Phelps TJ and Zhang C (2020) Active Anaerobic Archaeal Methanotrophs in Recently Emerged Cold Seeps of Northern South China Sea. Front. Microbiol. 11:612135. doi: 10.3389/fmicb.2020.612135
Received: 30 September 2020; Accepted: 27 November 2020;
Published: 16 December 2020.
Edited by:
Qunxin She, Shandong University, ChinaReviewed by:
Changyi Zhang, University of Illinois at Urbana–Champaign, United StatesShiQi Ji, Shandong University, China
Copyright © 2020 Zhang, Xiao, Chen, Zhao, Chen, Feng, Liang, Phelps and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Zhao, emhhb2ppbmdfMDMxNUAxNjMuY29t
†These authors have contributed equally to this work
 Tingting Zhang
Tingting Zhang Xi Xiao1,2,3†
Xi Xiao1,2,3† Songze Chen
Songze Chen Junxi Feng
Junxi Feng Qianyong Liang
Qianyong Liang Tommy J. Phelps
Tommy J. Phelps Chuanlun Zhang
Chuanlun Zhang