- 1Institute of Molecular Biotechnology, Graz University of Technology, NAWI Graz, BioTechMed-Graz, Graz, Austria
- 2acib—Austrian Centre of Industrial Biotechnology, Graz, Austria
Komagataella phaffii (Pichia pastoris) is one of the most extensively applied yeast species in pharmaceutical and biotechnological industries, and, therefore, also called the biotech yeast. However, thanks to more advanced strain engineering techniques, it recently started to gain attention as model organism in fundamental research. So far, the most studied model yeast is its distant cousin, Saccharomyces cerevisiae. While these data are of great importance, they limit our knowledge to one organism only. Since the divergence of the two species 250 million years ago, K. phaffii appears to have evolved less rapidly than S. cerevisiae, which is why it remains more characteristic of the common ancient yeast ancestors and shares more features with metazoan cells. This makes K. phaffii a valuable model organism for research on eukaryotic molecular cell biology, a potential we are only beginning to fully exploit. As methylotrophic yeast, K. phaffii has the intriguing property of being able to efficiently assimilate methanol as a sole source of carbon and energy. Therefore, major efforts have been made using K. phaffii as model organism to study methanol assimilation, peroxisome biogenesis and pexophagy. Other research topics covered in this review range from yeast genetics including mating and sporulation behavior to other cellular processes such as protein secretion, lipid biosynthesis and cell wall biogenesis. In this review article, we compare data obtained from K. phaffii with S. cerevisiae and other yeasts whenever relevant, elucidate major differences, and, most importantly, highlight the big potential of using K. phaffii in fundamental research.
Introduction: Why Use K. phaffii As Model Organism?
Most of the species that enabled the majority of biological discoveries during the last century did not start out as genetic model organisms, but rather matured into their roles. By being abundant human commensals (e.g., Escherichia coli, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, and Mus musculus), they were easily available, and in addition found to be amenable to genetic modification. Among those model organisms, yeasts pose a special role. As unicellular organisms, they possess many of the advantages that made E. coli the first model organism for molecular biology, e.g., fast growth, cheap and easy cultivation conditions, as well as well-established and precise genetic modification strategies. The use of single celled organisms allows one to work with large numbers of individuals, e.g., to discover rare phenotypes and the genes involved in particular biological processes. The composition of the growth medium and the growth conditions can be varied quickly, whereas high-throughput screening assays can be applied easily. As simple eukaryotes, yeasts can be used to study processes that are conserved from yeast to humans, such as cell organelle biogenesis, cell cycle progression, cytoskeletal organization, DNA replication, and protein secretion. S. cerevisiae (commonly known as baker’s yeast, brewer’s yeast, or simply yeast) has long dominated yeast-based research. S. cerevisiae was the first eukaryotic organism to have its genome sequenced (Goffeau et al., 1996), and genetic manipulation techniques were established early on. Additionally, it has the ability to grow as a stable haploid or diploid, which facilitates the analysis of mating behavior and the characterization of essential genes. However, new advances in genetic modification techniques (e.g., CRISPR/Cas9) allow many more species to enter the privileged circle of “model organisms,” and other yeasts started to gain attention, e.g., Kluyveromyces lactis, Kluyveromyces marxianus, Scheffersomyces stipitis, Yarrowia lipolytica, Arxula adeninivorans, Ogataea (Hansenula) polymorpha, and, as will be highlighted in this review article, Komagataella phaffii (Löbs et al., 2017; Gündüz Ergün et al., 2019; Sibirny, 2019). While non-conventional yeasts have hitherto widely been used for a range of biotechnological applications, they are now starting to gain attention as valuable model systems in fundamental research.
K. phaffii (often still referred to by its obsolete name Pichia pastoris) is an obligate aerobic and methylotrophic yeast, which means that it can metabolize methanol as sole carbon and energy source. The first enzyme in the methanol assimilation pathway is alcohol oxidase (AOX). In K. phaffii, alcohol oxidase (AOX) is encoded by two genes, AOX1 and AOX2 (Cregg et al., 1989). The availability of the extremely strong AOX1 promoter was one of the major reasons why K. phaffii gained attention as expression host for recombinant proteins in the first place. One of the most common strategies for (methanol-induced) gene expression is to integrate a recombinant gene into the AOX1 region (Invitrogen, 2014). While it was proven to be an easy-to-target locus for gene insertions, the knockout of AOX1 also creates a MutS strain. MutS strains still grow slowly on methanol due to the expression of AOX2, but direct the force of the AOX1 promoter mainly toward recombinant protein production.
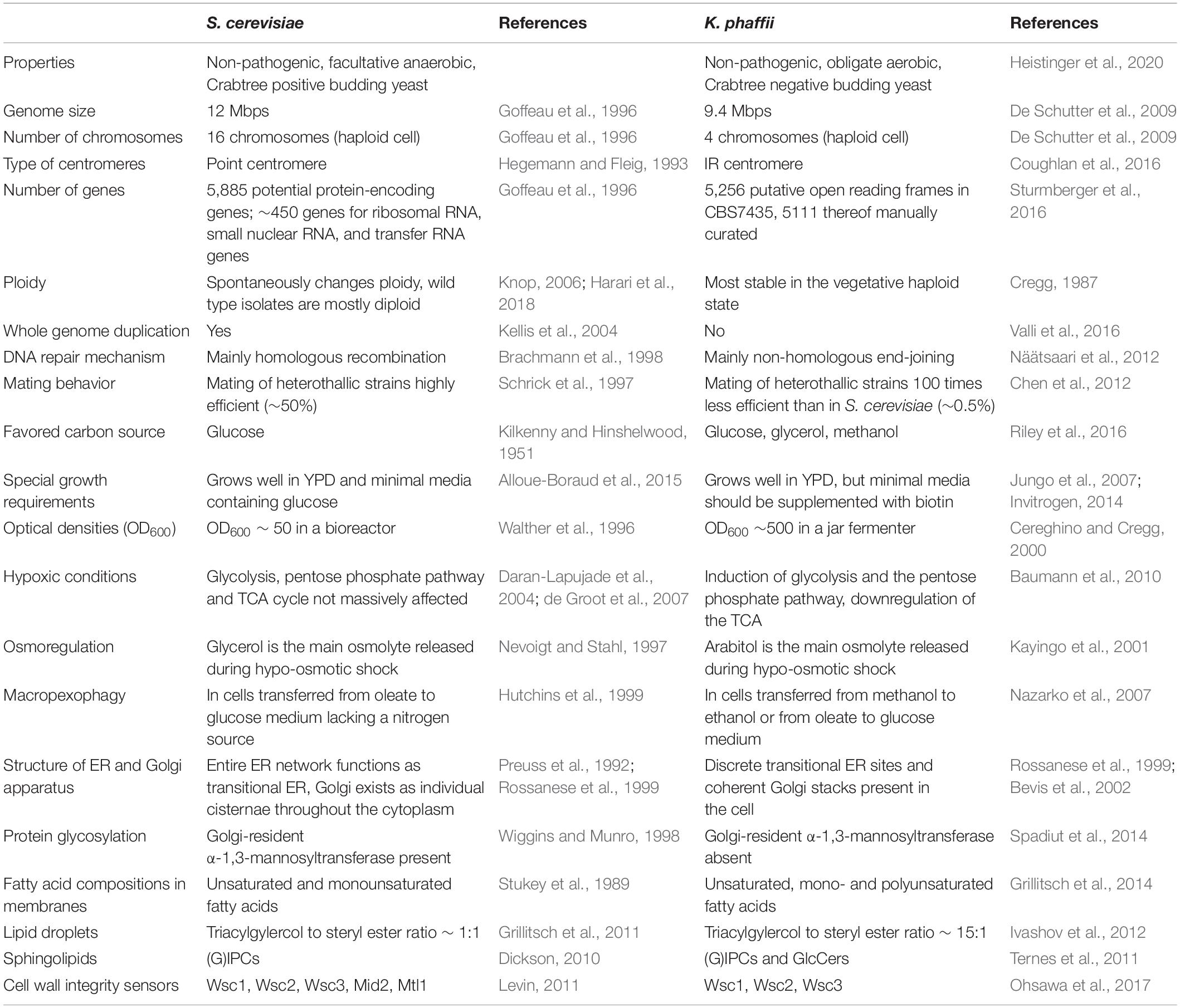
Since K. phaffii became such a popular production host for the pharmaceutical, feed and food industries, it is also called the biotech yeast. K. phaffii has several advantages over S. cerevisiae, including better thermo- and osmo-tolerance, respiratory growth to extremely high cell densities, effective protein secretion, and the ability to express recombinant proteins at high levels from strong constitutive and inducible promoters (Karbalaei et al., 2020). Furthermore, secretory mammalian protein production has been facilitated upon extensive engineering of glycosylation pathways (Hamilton et al., 2003; Hamilton and Gerngross, 2007; Jacobs et al., 2009). Bioprocesses also benefit from K. phaffii’s ability to efficiently produce membrane proteins (Jahic et al., 2006; Byrne, 2015). While most of these advances served the primary purpose to improve heterologous protein production, they also created an explosion in the knowledge based on the system as described in numerous publications. This, in turn, inspired a significant number of research labs to start using K. phaffii for fundamental studies. As methylotrophic yeast, K. phaffii exhibits methanol assimilation pathways, which cannot be studied in yeasts that either did not evolve or lost this feature in the course of evolution (Riley et al., 2016). Phylogenetically, Komagataella species are members of the methylotrophic yeasts clade (family Phaffomycetaceae) and are only distantly related to better-known yeasts such as S. cerevisiae, S. pombe, and Candida albicans (Riley et al., 2016; Shen et al., 2018; Figure 1). This fact adds great value to K. phaffii as emerging model yeast. If a process is found to be conserved among K. phaffii, S. cerevisiae, and other yeasts, it is likely to be more widely conserved. Vice versa, mechanistic differences found between different yeasts may indicate functional diversity among higher eukaryotes (Table 1).
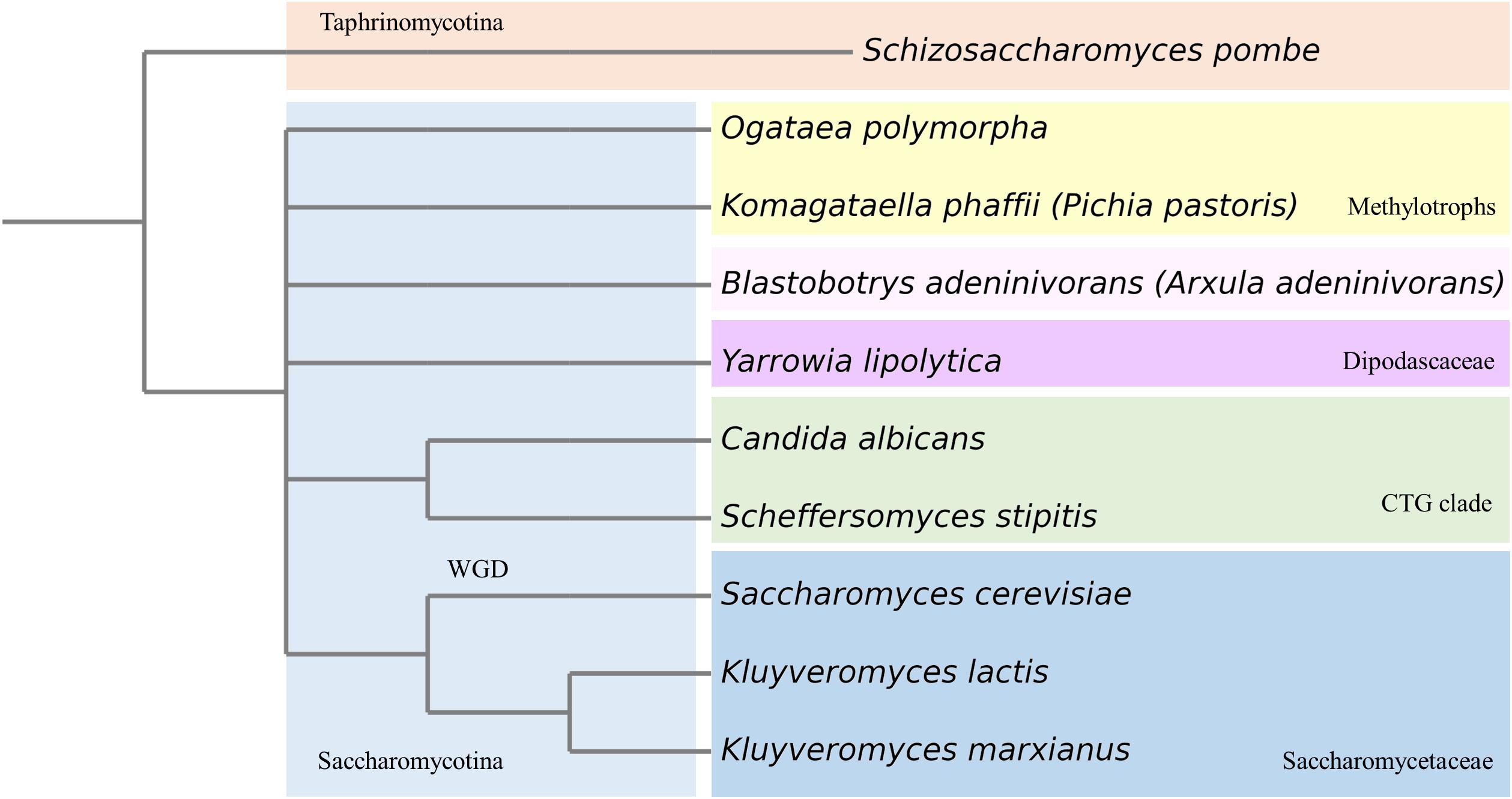
Figure 1. Phylogenetic tree of most prominent yeasts used in (fundamental) research. WGD, whole genome duplication.
Origins of K. phaffii Research
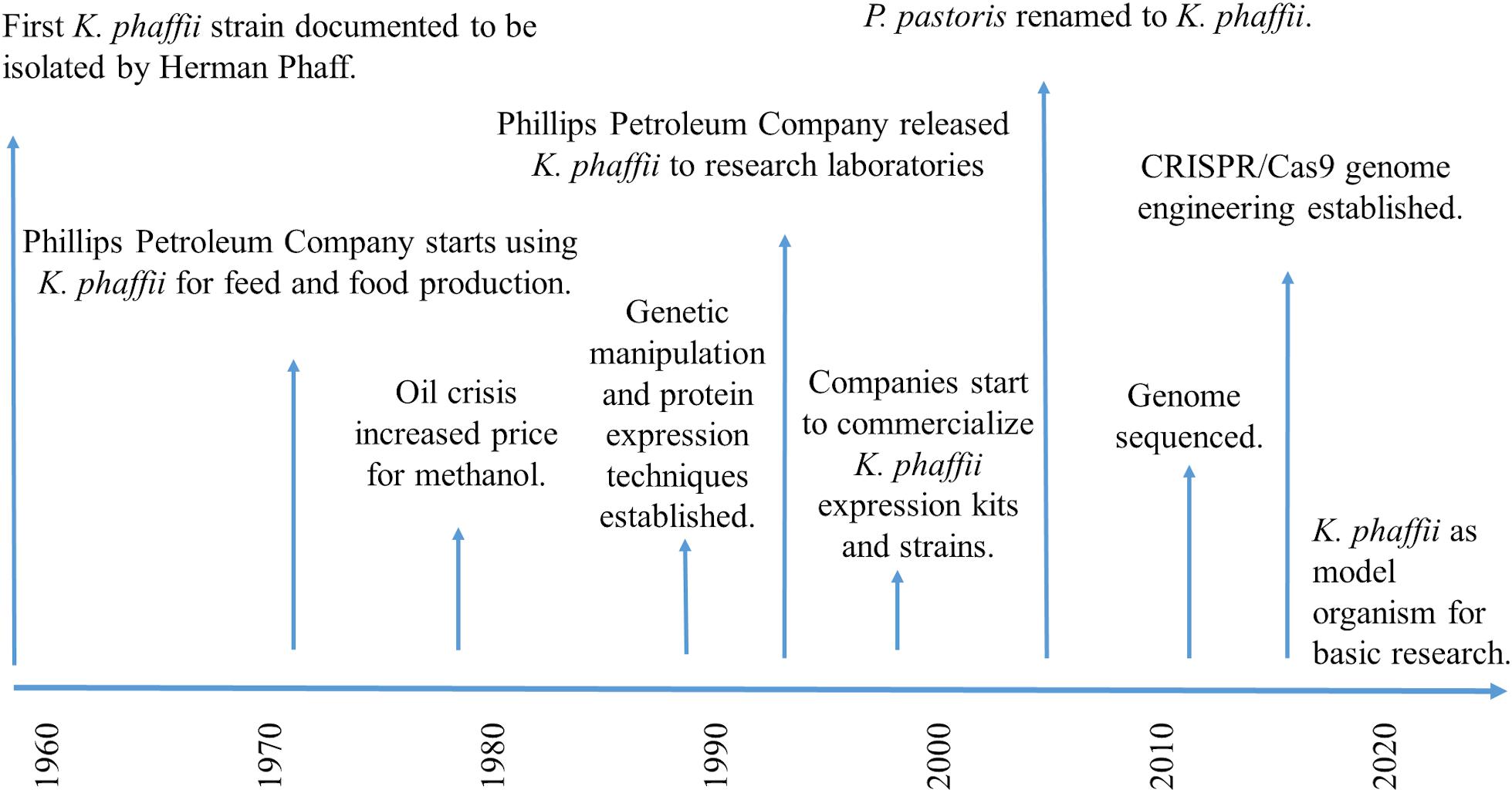
For decades, K. phaffii has been thought to belong to the genus Pichia, and therefore, mistakenly been called Pichia pastoris. The first strain ever assigned to the species P. pastoris was isolated by Alexandre Guilliermond from the exudate of a French chestnut tree in 1919, and was originally named Zygosaccharomyces pastori (species type strain CBS704, NRRL Y-1603) (Guilliermond, 1920; Figure 2). In the 1950s, Herman Phaff isolated further strains from black oak trees in California, United States, and renamed the species to Pichia pastoris (Phaff et al., 1956). However, in 1995, new insights generated by sequencing of ribosomal RNA caused all P. pastoris strains to be moved to a new genus, Komagataella (Yamada et al., 1995), and later separated into two species (Kurtzman, 2005): Komagataella pastoris (which includes the French strain) and K. phaffii (which includes the American isolates). The members of the genus Komagataella are phenotypically too similar to be distinguished from one another by routine tests usually employed in yeast taxonomy, which is why classification is mainly based on sequence alignments of a limited number of genes. The genomes of K. pastoris and K. phaffii differ by approximately 10% DNA sequence divergence and two reciprocal translocations (Love et al., 2016), and both strains are in use for recombinant protein production under the name P. pastoris. This seems quite confusing, but exact strain identifications, such as CBS2612 (NRRL Y-7556) or CBS7435, usually help allocate which (wild type) strain has been used. At this point it should be noted, that applied sciences still preferentially use the name Pichia pastoris (often as a synonym for all Komagataella species), while fundamental studies rather shifted to using the correct name, K. phaffii.
On a commercial basis, K. phaffii initially gained attention for the production of biomass and single-cell protein from methanol, a back then comparably cheap carbon source. In the 1970s, Phillips Petroleum Company (Bartlesville, Okla, United States), [and continued by Research Corporation Technologies (RCT)] started to develop and improve K. phaffii (CBS7435) fermentations for industrial production of high protein animal feed and biomass (Wegner, 1983, 1990). However, the world-wide oil crisis increased the price for methanol, and the economics of biomass production became unfavorable compared to proteins extracted from soybean and other sources. Phillips Petroleum Company decided to release the expression system to research laboratories in 1993 (BioWorldTM, 1993), and in the following years, diverse academic groups and biotech companies started to shift their interest toward the exploitation of K. phaffii as potential host for recombinant protein production (Wegner and Harder, 1987; Cereghino and Cregg, 2000). In the 1990s, a diverse set of expression vectors and genetic strain modification techniques started to be developed, published and commercialized (reviewed by Ahmad et al., 2014). Invitrogen (now Thermo Fisher Scientific, Waltham, Massachusetts, United States), for example, put their well-known “Pichia expression Kit” to market, containing two expression strains (GS115 and KM71), two different expression vectors, either with (pPIC9) or without (pHIL-S1) a secretion signal, for stable and selectable insertion of any gene of interest into the AOX1 gene locus, as well as extensive and easy to follow method descriptions (Invitrogen, 2014). The “Pichia Expression Kit” has been used by many laboratories world-wide to get started with protein expression in K. phaffii. Initially in collaboration with the Glieder group, VTU Technology (now Validogen GmbH, Graz, Austria), generated a set of methanol-inducible and methanol-free AOX1 promoter libraries with varying strengths, as well as diverse platform strains, which they now commercialize under the name “UNLOCK PICHIA”1. Simultaneously, many renowned laboratories published excellent “freedom to operate” studies on expression systems and genome modification tools, e.g., the Cregg, Glieder, Matthanovich, Schwab, Lin-Cereghino, Callewaert, and Ferrer groups. In this regard, we would like to highlight that close collaboration of research groups like the Mattanovich, Callewaert, and the Glieder group with industrial partners had a strong impact on the industrial application and commercialization of K. phaffii. Additionally, many of these research labs and companies (e.g., Bisy GmbH, Biogrammatics Inc., Bioingenium, Research Corporation Technologies (RCT) and Validogen GmbH) provide a diverse set of engineered K. phaffii strains, e.g., displaying improved protein expression, secretion, and glycosylation. On a technical basis, hands-on workshops have proven to be helpful to experience in person how to handle and genetically modify K. phaffii (e.g., https://www.hands-on-pichia.com/).
Even though the many strategies for recombinant protein expression in K. phaffii are highly interesting and constantly evolve (Ahmad et al., 2019), they are beyond the focus of this review, which is why we would like to refer the reader to some of the many excellent review articles summarizing and describing the progress made in this field (Ahmad et al., 2014; Karbalaei et al., 2020).
K. phaffii Genome Content and Chromosome Organization
Even though many K. phaffii strains have been isolated from methanol enriched media since the 1960s (Ogata et al., 1969; Kato et al., 1974), only four distinct natural isolates of K. phaffii exist in public yeast culture collections (Braun-Galleani et al., 2019). To date, almost all laboratory work on K. phaffii utilizes strains derived from the same natural isolate, CBS7435 (Braun-Galleani et al., 2019). For example, K. phaffii CBS7435 is the parental strain of the broadly commercialized GS115, which was developed by RCT/Philips petroleum by chemical mutagenesis and selected for histidine auxotrophy (US Patent 4,879,231 A). One of the biggest milestones in K. phaffii related research was the whole-genome sequencing of important K. phaffii strains GS115 (De Schutter et al., 2009) and CBS7435 (Küberl et al., 2011; Sturmberger et al., 2016), and K. pastoris strain DSMZ 70382 (Mattanovich et al., 2009). The genome sequences provide a robust platform for the ongoing annotation of features and functional annotations of those features to the genome content. To facilitate communication, the genetic nomenclature used for S. cerevisiae has been applied to K. phaffii (Valli et al., 2016). For example, the HIS4 gene in both S. cerevisiae and K. phaffii encodes histidinol dehydrogenase. There is also cross-complementation between gene products in both S. cerevisiae and K. phaffii. Several wild type genes from K. phaffii complement comparable mutant genes (e.g., HIS4, ARG4, TRP1, URA3, and ADE1) in S. cerevisiae (Cosano et al., 1998; Lin Cereghino et al., 2001). Gradually, a more complete picture of K. phaffii is emerging as new data sets are collected from a diverse range of experiments to refine the annotated gene structures and assign functional data to them. For example, in order to identify essential genes in K. phaffii, a combination of transposon mutagenesis and high-throughput sequencing has been applied (Zhu et al., 2018), and based on the genome sequencing of K. phaffii, genome-scale metabolic networks could be reconstructed to model the yeasts’ metabolism from a systems perspective (reviewed by Chung et al., 2013).
The K. phaffii genome size is 9.4 Mbp, compared to the 12 Mbp genome of S. cerevisiae. Despite similar genome sizes, K. phaffii has only four relatively large chromosomes of 2.9, 2.4, 2.3, and 1.8 Mbp in contrast to 16 smaller chromosomes in S. cerevisiae. With an average size of 2.5 Mbp, K. phaffii chromosomes are almost twice the length of the largest S. cerevisiae chromosome (chromosome I), which is 1.5 Mbp. Recently, Coughlan et al. structurally defined the centromeres of K. phaffii by transcriptome sequencing (RNA-seq) and chromatin immunoprecipitation with high-throughput sequencing (ChIP-seq) by binding the histone H3-like centromere protein Cse4 (Coughlan et al., 2016). This study demonstrates that K. phaffii has large modular centromeres, more reminiscent of those of higher organisms than of the 125 bp element sufficient for centromere function in S. cerevisiae (Fitzgerald-Hayes et al., 1982). Its four centromeres are unrelated in sequence and consist of a 1 kb central core (mid) region flanked by a 2 kb inverted repeat (IR). The histone H3 variant CenH3 (Cse4) binds strongly to the mid region, and gradually less strongly along the IRs. In principle, the organization and structure of K. phaffii centromeres resembles those of S. pombe (Wood et al., 2002), but they are smaller in size and lack the extensive flanking heterochromatic outer repeats. Due to recombination in the IRs, different isolates of K. phaffii show polymorphism for the orientation of the mid regions. The general conservation of centromere features, including size, structure, and multilayered organization, highlights K. phaffii as a valuable model for the study of eukaryotic chromatin remodeling and centromere function.
Growth Behavior: Carbon Sources, Methanol Assimilation, and Auxotrophies
In principle, K. phaffii can be grown on the same media as S. cerevisiae, e.g., YPD (yeast extract, peptone dextrose/glucose) and minimal media containing a nitrogen source and glucose as main carbon source. In these media, the doubling time of a log phase wild-type strain is ∼2 h. However, it has been shown that K. phaffii has a special requirement for biotin, which is why there have been efforts to optimize biotin concentrations in bioreactor cultivations (Jungo et al., 2007), or to engineer strains to become prototrophic for biotin (Gasser et al., 2010). Based on transciptomics, the Love lab created a specially designed rich medium for large-scale cultivations (Matthews et al., 2018). K. phaffii can utilize glucose, glycerol, sorbitol, methanol, ethanol, L-rhamnose, and acetate as carbon source (Riley et al., 2016). A comprehensive overview of the carbon sources and their diauxism is given by Zepeda et al. (2018).
As most yeasts, K. phaffii is “Crabtree-negative,” which means that it produces energy mostly via respiration. Oxygen limitation forces a cell to readjust its metabolic fluxes from cellular respiration to fermentation, which causes massive energy deprivation in a cell due to strongly reduced availability of ATP. Therefore, K. phaffii is more sensitive to the availability of oxygen than the “Crabtree-positive” yeast S. cerevisiae (Pfeiffer and Morley, 2014). There are several discussions, why and how a small set of yeasts evolved Crabtree positive phenotypes. Clearly, it gives them an evolutionary advantage of consuming glucose faster and producing ethanol to outcompete other microorganisms in sugar rich environments. The Mattanovich lab recently managed to convert K. phaffii into a Crabtree positive yeast through overexpression of a single Gal4-like transcription factor, which provides novel insights into the evolution of the Crabtree effect (Ata et al., 2018). As obligate aerobe, K. phaffii displays exclusively respiratory metabolism and naturally does not switch to an anaerobic metabolism that would lead to toxic metabolite accumulation under oxygen limited conditions. This is the main reason why such high cell density fermentations can be achieved with this organism under dissolved oxygen controlled processes. Another interesting engineering approach recently published converted K. phaffii into an autotroph that grows on CO2. In this study, the insertion of eight heterologous genes and deletion of three native genes generated a CO2-fixation pathway resembling the Calvin–Benson–Bassham cycle, the predominant natural CO2-fixation pathway (Gassler et al., 2020). Adaptive laboratory evolution could improve the growth rate from 0.008 to 0.018 h–1.
As methylotrophic yeast, K phaffii is able to grow on methanol as sole carbon source. This makes it one of approximately a dozen yeast species representing four different genera capable of assimilating methanol. The other genera include Candida, Ogataea, and Torulopsis. The methanol utilization (MUT) metabolic pathway appears to be the same in all these yeasts and involves a unique set of pathway enzymes—including alcohol oxidase (AOX), dihydroxyacetone synthase (DAS), and formate dehydrogenase (FDH) (Hartner and Glieder, 2006; Yurimoto and Sakai, 2009). In K. phaffii, the MUT pathway has been studied extensively, which is summarized in some excellent review articles (Daly and Hearn, 2005; Hartner and Glieder, 2006). Therefore, we will just outline the most important key aspects of methanol utilization. The first step of the MUT pathway is the oxidation of methanol to formaldehyde and hydrogen peroxide in the peroxisomes by alcohol oxidase (AOX). In K. phaffii, AOX is encoded by two genes, AOX1 and AOX2 (Cregg et al., 1989). Methanol assimilation is subject to carbon-source-dependent repression, derepression, and induction mechanisms. The presence of glucose, glycerol, and ethanol represses expression of both AOX genes, whereas the presence of methanol strongly induces expression at non-growth-limiting conditions (reviewed by Vogl and Glieder, 2013). Certain growth conditions can also trigger co-assimilation of a multicarbon source and methanol (Egli et al., 1982). Although glycerol does not derepress expression of AOX1, other non-repressing carbon sources like sorbitol, mannitol, trehalose, and alanine do so (Thorpe et al., 1999; Inan and Meagher, 2001). Aox1 has a much higher cellular abundance than Aox2. Thus, knockout of AOX2 results in a wild type like phenotype on methanol (Mut+), while an AOX1 knockout exhibits extremely slow growth on methanol (MutS, methanol utilization slow). Double knockout strains are unable to grow on methanol (Mut–). After the oxidation of methanol to formaldehyde and hydrogen peroxide, catalase breaks down the toxic hydrogen peroxide to water and oxygen. Formaldehyde can either undergo a dissimilation pathway by two subsequent dehydrogenase reactions or an assimilation pathway by condensation with xylulose 5-phosphate (Xu5P). A dihydroxyacetone synthase (DAS) catalyzes the latter peroxisomal condensation reactions of Xu5P and formaldehyde into the C3-compounds dihydroxyacetone (DHA) and glyceraldehyde 3-phosphate (GAP), which are further metabolized in the cytosol. Again, all of these steps are explained in much detail by Hartner and Glieder (2006).
In the last decades, metabolic studies and the generation of genome-scale metabolic models have been successfully employed to characterize the cellular physiology of K. phaffii and improve metabolic engineering (reviewed by Chung et al., 2013). In general, comparative genomics enabled the direct comparison of different yeast species and shows their differences, e.g., growth behavior under certain conditions on a broad basis (Solà et al., 2007; Dragosits et al., 2009). Chung et al. (2010) highlight and compare three genome-scale metabolic models available for K. phaffii and Pichia stipitis (Chung et al., 2010; Sohn et al., 2010; Caspeta et al., 2012). Overall, diverse metabolic pathways have been constructed and the metabolites and reactions assigned to subcellular compartments including the cytosol, endoplasmic reticulum, extracellular fluid, Golgi apparatus, mitochondria, nucleus, peroxisome, and vacuole. Growth predictions made on the basis of these models came reasonably close to the experimental data reported in earlier studies. In another interesting, recent study, organelles were isolated from K. phaffii cells grown on glucose or methanol, the proteome quantified and compared to S. cerevisiae (Valli et al., 2020). Differences in protein localization were found mostly for cytosolic, mitochondrial, and peroxisomal proteins, and the extensive analysis of the carbon-source dependent, organelle specific proteome gives insight in protein localization on a very broad basis. Both strategies, genome-scale metabolic models and the subcellular proteome atlas, provide powerful tools for targeted strain engineering strategies.
Genetic Manipulation Techniques
Based on the vast amount of excellent reviews on this topic (Cereghino and Cregg, 2000; Ahmad et al., 2014; Baghban et al., 2018, to name a few), we will keep this chapter rather short, and only focus on the most relevant aspects for the present review article.
Autonomously Replicating Plasmids
In many yeasts, as for example in S. cerevisiae and S. pombe, gene expression can be driven from small extrachromosomal plasmids (Siam et al., 2004; Gnügge and Rudolf, 2017). The use of plasmids has the big advantage that they can be cloned and transformed easily, but also get rid of, if not required any longer. Depending on the type of origin of replication, cells can either contain one (CEN/ARS origin) or several (two micron circle replication origin) copies of the plasmid, which can help modulate protein production levels. Autonomously replicating sequences (ARSs), which generally serve as the origins of DNA replication during mitosis, constitute the primary origins utilized to replicate plasmids in yeast host cells. A K. phaffii-specific ARS (PARS1) was identified over 30 years ago (Cregg et al., 1985). Similar to the S. cerevisiae-specific ARS, PARS1 enabled the high-efficiency transformation of K. phaffii with circular plasmids (Weninger et al., 2016; Pan et al., 2018). Recently, a 452 bp ARS element (panARS) was identified in K. lactis and shown to facilitate transformation in a wide range of yeast species, including K. phaffii (Liachko and Dunham, 2014; Camattari et al., 2016), and a mitochondrial DNA (mtDNA) fragment was discovered to function as a novel ARS in K. phaffii (Schwarzhans et al., 2017). All plasmids bearing these ARS sequences were found to be poorly stable for replication and segregation in K. phaffii, which restricted their use in many applications. Plasmid instabilities are not only detrimental for recombinant protein expression, but may also lead to unwanted random integration of the plasmid DNA into the yeast genome, especially if selection pressure is applied. As discussed earlier, recent advances in identifying putative centromeres on each of the four K. phaffii chromosomes (Coughlan et al., 2016; Sturmberger et al., 2016) now enabled the construction of more stably replicating episomal plasmids containing the entire chromosome two centromere DNA sequence (Nakamura et al., 2018).
However, for some applications it is beneficial to apply plasmids with low stability. For example, in CRISPR/Cas9 applications, it is important for cells to lose Cas9 activity after cell engineering to avoid unspecific mutations in the genome. To that effect, Weninger et al. (2016, 2018) successfully used PARS1 in their CRISPR/Cas9 plasmids, whereas a more recent study from Gu et al. (2019) compared different ARS sequences in CRISPR/Cas9 approaches and found very good efficiencies with plasmids containing panARS. In order to get rid of the plasmid after genome engineering, cells simply need to be streaked out on plates without selection pressure. Single colonies grown on these plates have usually already lost the plasmid.
DNA Recombination
Genomic integration of expression cassettes and DNA fragments can occur via two distinct DNA repair mechanisms in eukaryotic cells: homologous recombination (HR) and non-homologous end joining (NHEJ). HR is a highly accurate repair mechanism mediated through base pairing of rather long stretches of homologous DNA sequences and catalyzed by proteins encoded by genes in the RAD52 epistasis group (Pastwa and Blasiak, 2003). In contrast, NHEJ requires no sequence homology to operate (Krejci et al., 2012). The heterodimer Ku70/80 binds to free ends of a DNA strand and recruits DNA protein kinases (DNA-PKCs) to initiate a quick and unspecific repair of the DNA double strand break (Pastwa and Blasiak, 2003; Dudásová et al., 2004). Both mechanisms play overlapping roles in yeasts, but are used to a different extent depending on the yeast species. In S. cerevisiae, HR represents the dominant repair mechanism, whereas NHEJ occurs very rarely. In order to achieve targeted gene knockouts and knockins, short homologous flanking regions of 40 bp were shown to be sufficient (Brachmann et al., 1998), and integration events usually occur with > 70% efficiency at the correct locus (Gueldener et al., 2002). This highly efficient mechanism offers another intriguing possibility; a process known as in vivo ligation, which promotes the direct self-assembly of multiple recombinant DNA fragments in the nucleus of S. cerevisiae and fully eliminates the cloning process for plasmid assembly (Oldenburg et al., 1997; Juhas and Ajioka, 2017). In vivo recombination could also be observed in K. phaffii when a library of Rhizopus chinensis lipase mutants was assembled directly by the host and integrated into the targeted genomic locus (Yu et al., 2012). Overlapping ends as short as 50 nucleotides were reported to be sufficient to promote assembly at a relatively high efficiency. However, this seems rather surprising, since recombination and targeted gene insertions were shown to be difficult to achieve in K. phaffii because of the high ratio of NHEJ-to-HR activity (Näätsaari et al., 2012; Tsakraklides et al., 2015; Schwarzhans et al., 2016). For example, homologous targeting sequences of < 500 bp only led to < 0.1% of positive targeting events indicating a very low efficiency of gene replacement. This efficiency could be increased up to 1.5-fold by extending the homologous regions up to 1 kb at each side (Näätsaari et al., 2012). Due to this low efficiency, several strategies were developed to improve HR activity in K. phaffii, e.g., hydroxyurea-mediated cell cycle arrest (Tsakraklides et al., 2015) and providing extra copies of the gene to be deleted on a helper plasmid (Chen et al., 2013). Deletion of KU70 also significantly improved the NHEJ-to-HR ratio (Näätsaari et al., 2012). Deletion of KU70 can, however, also negatively affect cellular fitness (Näätsaari et al., 2012). This has also been demonstrated indirectly in a study where a CBS7435 ku70Δ was engineered toward production of different terpenoids (Wriessnegger et al., 2015). RNAseq data revealed that RAD52, a protein involved in DNA repair, was highly upregulated in all of the ku70 deletion strains. Methanol-induced overexpression of RAD52 drastically improved terpenoid production, which was most probably due to rescuing the loss of Ku70 function. In contrast, RAD52 overexpression had no effect on a S. cerevisiae strains producing terpenoids.
HR frequency can also be increased by several orders of magnitude (up to 4,000-fold in S. cerevisiae) through targeted single and double strand break induced DNA repair (Storici et al., 2003; Caldecott, 2008), which is why precise mechanisms introducing double-strand breaks in the genome are of great interest. The currently most prominent system that mediates targeted genome engineering in various pro- and eukaryotic hosts is CRISPR/Cas9 (Jakočiunas et al., 2016; Stovicek et al., 2017; Adli, 2018). The CRISPR gene-editing technology is composed of an endonuclease protein (Cas9), whose DNA-targeting specificity and cutting activity can be programmed by a short guide RNA (sgRNA). Optimal conditions required for efficient CRISPR/Cas9 function are very narrow in K. phaffii. Weninger et al. (2016) systematically tested more than 90 constructs containing different codon optimized DNA sequences of CAS9, various sgRNA sequences, several RNA Pol III, and RNA Pol II promoters (in combination with ribozymes) for the expression of the sgRNAs and different RNA Pol II promoters for the expression of CAS9 and sgRNAs. Only ∼6% (6/95) of all tested constructs mediated efficient CRISPR/Cas9 targeting, namely those bearing RNA Pol II promoters, ribozymes, and a human codon optimized CAS9 sequence. Multiplexity could be proven upon expressing different sgRNAs targeting multiple loci from one plasmid (Weninger et al., 2016; Liu et al., 2019). Follow-up studies have shown that deletion of KU70 drastically improved the efficiency of CRISPR/Cas9 engineering approaches due to the downregulation of NHEJ (Weninger et al., 2018; Liu et al., 2019).
K phaffii’s preference for NHEJ can also be advantageous for the construction of random gene knockout or knockin libraries. In S. cerevisiae, which prefers HR over NHEJ, this can only be achieved by labor intense implementation of transposon insertion strategies (Xu et al., 2011) or yeast oligo-mediated genome engineering (YOGE, a recombineering strategy) (Dicarlo et al., 2013). Recently, we also published a broad set of auxotrophic K. phaffii strains in the CBS7435 strain background, including strains auxotrophic for arginine and lysine, but also novel strains, e.g., auxotrophic for proline (Ahmad et al., 2019). These strains broaden the spectrum of possible markers to be used and can be highly beneficial for future strain modification strategies.
Mating, Sporulation, and Tetrad Dissection
The first step of sexual reproduction and mating in yeasts involves the mutual recognition of haploid cells of opposite mating types (MATa and MATα). The expression of the respective MAT gene hence determines the mating type of a cell, and the process causing DNA rearrangements in sexually differentiating cells is called mating type switching (Hanson and Wolfe, 2017). In S. cerevisiae, mating type switching is enabled through the presence of silent copies of both MAT variants, HMLα and HMRa. During mating type switching, HO endonucleases create double strand breaks at the MAT loci and genes at the active locus are replaced with a silent copy of the opposing mating type through synthesis dependent strand annealing (Haber, 2012). By definition, primary homothallic species usually express both MAT genes allowing them to mate with one other. Heterothallic species have individuals that reside in different mating types, and only cells of opposite mating types can mate. The same is true for secondary homothallic species, but cells can switch their mating types and mate with cells of the same strain. Since wild type S. cerevisiae strains are self-fertile, they are classified as secondary homothallic. Laboratory strains, which carry non-functional HO endonucleases are classified as heterothallic.
Mating type switching greatly differs between different yeast species, but has similar mechanisms in the methylotrophic yeasts O. polymorpha (Maekawa and Kaneko, 2014) and K. phaffii (Hanson et al., 2014). Both yeasts contain one copy each of MATa and MATα. The loci are flanked by sequences containing inverted repeats that are orthologous to regions generally found in Saccharomycetaceae. In both yeasts, one of the MAT loci is located close to a heterochromatin region (a centromere in case of O. polymorpha, and a telomere in case of K. phaffii), which causes transcriptional repression of the respective MAT locus. The inverted repeat sequences flanking the MAT loci are essential for mating type switching. Mating type switching occurs by homologous recombination of these sequences, which inverts the entire genomic region between the MATa and MATα locus (19 kb in O. polymorpha, 138 kb in K. phaffii). This means that once mating type switching is induced, active and repressed positions of MAT genes are swapped, without the involvement of any major synthesis or degradation of DNA. By contrast, in S. cerevisiae and S. pombe exonucleases degrade the exchanged MAT genes that have been replaced by newly synthesized DNA copied from the silent locus.
In contrast to S. cerevisiae, K. phaffii is most stable in the vegetative haploid state and (like S. pombe and K. lactis) remains haploid unless forced to mate, under certain conditions such as nitrogen limited-starvation (Cregg, 1987). Nitrogen limitation and other nutritional stresses also cause mated diploid K. phaffii cells to efficiently undergo meiosis, sporulation, and rapidly switch back to the haploid state. Upon mating two K. phaffii strains expressing the heavy and the light chain of the anti-HER2 antibody, it was shown that the mating efficiency of wild-type haploid K. phaffii strain (NRRL-Y11430) is about 0.1–1% (1 diploid per 1,000–100 haploid cells) (Chen et al., 2012). S. cerevisiae, in contrast, exhibits mating efficiencies of about 50% (Schrick et al., 1997).
The Mattanovich group recently published their studies on how to produce heterothallic K. phaffii strains with defined mating types (Heistinger et al., 2017, 2018). Even though tetrads appeared to be significantly less stable than described for S. cerevisiae, strains of opposite mating types were shown to efficiently mate and sporulate. The generation of heterothallic S. cerevisiae strains was one of the reasons for the establishment of this yeast as the most common yeast model organism. Hence, the establishment of heterothallic K. phaffii strains can be a powerful tool in K. phaffii related research. Heterothallic (or haploid) yeast cells are especially useful when screening for mutant alleles that produce a desired phenotype. Moreover, mating of S. cerevisiae has been successfully employed in diverse research applications such as yeast two-hybrid libraries (Golemis and Khazak, 1997). Also, the use of haploid cells of opposite mating types could greatly benefit the identification of synthetic lethality and facilitate complex strain construction, a task still difficult to achieve with K. phaffii.
Peroxisome Studies: Proliferation and Pexophagy
Peroxisomes are essential, subcellular organelles that are ubiquitously present in all eukaryotic cells. They have the intriguing ability to sequester specific enzymes that metabolize a variety of substrates, which often enables an organism to survive in unique environments. For example, in K. phaffii and other methylotrophic yeasts, the metabolism of methanol takes place within the peroxisome in order to separate toxic hydrogen peroxide, one of the major catabolites of methanol utilization, from the rest of the cell (van der Klei et al., 2006). In methylotrophic yeasts, peroxisomal proliferation can easily be stimulated upon shifting cells to methanol-rich media (Yurimoto et al., 2011). Vice versa, the transfer of cells from methanol-rich to methanol-deplete media induces pexophagy, the selective autophagy of peroxisomes. This feature has made methylotrophic yeasts like K. phaffii an attractive model system to dissect the molecular mechanisms controlling peroxisome biogenesis, proliferation and degradation.
Peroxisomes proliferate either by growth and division of pre-existing peroxisomes or arise de novo. This knowledge has been substantiated by studies using yeast peroxisome-deficient mutants (pex mutants). The endoplasmic reticulum (ER) was shown to provide most of the lipids and proteins needed for the formation of peroxisomes. This has been indicated upon studying the trafficking of diverse peroxisomal membrane proteins (PMPs) in S. cerevisiae, K. phaffii, and O. polymorpha, e.g., Pex3, Pex19, and Pex22, which are delivered to the peroxisome through the ER (reviewed by Agrawal and Subramani, 2016; Yuan et al., 2016). PEX genes whose deletion results in abnormal peroxisome number and size are usually less well studied, because mutants defective in these genes often do not exhibit defective peroxisome function or sorting of PMPs. Interestingly, the phenotype of such mutants can vary from organism to organism. For example, the deletion of PEX30 results in fewer and clustered peroxisomes in K. phaffii, while in S. cerevisiae, an increase in normal-sized peroxisomes has been reported (Yuan et al., 2016).
Upon glucose- or ethanol-induced catabolite inactivation, peroxisomes are rapidly and selectively degraded within the vacuole by a process called pexophagy, an autophagy-like process. In K. phaffii, pexophagy can proceed by micro or macro events (extensively reviewed by Dunn et al., 2005; Oku and Sakai, 2016). Micropexophagy is induced when methanol-grown cells are adapting to glucose, whereas macropexophagy is induced when cells are shifted from methanol to ethanol. During micropexophagy, an autophagic membrane structure called the micropexophagy-specific apparatus (MIPA) emerges on the peroxisome surface and fuses with arm-like extensions of the vacuole, which eventually engulfs the target peroxisome cluster (Mukaiyama et al., 2004). Although this mode of pexophagy has been extensively characterized in K. phaffii, similar dynamics of vacuole engulfment of the peroxisome were detected in oleate-grown S. cerevisiae cells when replenished with glucose (Chiang et al., 1996). During macropexophagy, the peroxisome is completely entrapped by a double-membrane structure, termed the macropexophagosome (MPP). The outer membrane of the MPP fuses with the vacuolar membrane and releases the captured peroxisome into the vacuolar lumen, a process that shows high similarity to macroautophagy. Macropexophagy has also been discovered in other yeast species, including O. polymorpha (when transferred from methanol to various other media) (Monastryska et al., 2004) and S. cerevisiae (when transferred from oleate medium to glucose medium lacking a nitrogen source) (Hutchins et al., 1999). Due to the vast amount of literature covering pexophagy in K. phaffii, we would like to refer the interested reader to excellent review articles published by the Veenhuis, Suzuki, and Sakai labs (Mukaiyama et al., 2004; Dunn et al., 2005; Oku and Sakai, 2016).
Protein Secretion and Unfolded Protein Response
Yeast has always been an important model organism to study secretory events because of the high conservation of the secretory machinery from lower to higher eukaryotes. The yeast secretory pathway is a very complex process that involves about 160 proteins responsible for different post translational processes, such as glycosylation and folding (Nielsen, 2013). Among the 550 proteins, which carry signal peptides and are processed by the secretory pathway, only very few are secreted to the extracellular matrix. The majority is targeted to the endoplasmic reticulum (ER), Golgi, vacuole, and cytoplasm. All of these secretory proteins must be folded correctly in the ER, and any accumulation of misfolded proteins causes ER stress that induces unfolded protein response (UPR). Activation of the UPR results in transcriptional change of about 400 genes (Tyo et al., 2012) and upregulation of chaperones and foldases, as well as ER associated degradation (ERAD), whereas many of these processes are under the regulation of the Hac1p transcription factor. In contrast to S. cerevisiae, K. phaffii has discrete transitional ER sites (Bevis et al., 2002) and coherent Golgi stacks (Rossanese et al., 1999), which makes this yeast an ideal host for studying the organization of ER subdomains and the early secretory pathway. Similar to the studies done with S. cerevisiae by the Schekman lab (Novick et al., 1981), the Glick lab investigated K. phaffii homologs of the S. cerevisiae SEC12, SEC13, SEC17, SEC18, and SAR1 genes and showed that they complement the corresponding S. cerevisiae mutants (Payne et al., 2000). Comparison of these proteins revealed differences in intron structures and conserved domains.
One of the biggest advantages of K. phaffii as industrial production host is the extraordinary efficiency of its protein secretion machinery (Damasceno et al., 2012; Puxbaum et al., 2015). Because K. phaffii secretes few of its own proteins, the secreted recombinant protein is usually the major polypeptide species found in the extracellular medium. The most extensively applied system is secretory expression of recombinant genes fused to the α-factor prepro leader sequence originating from S. cerevisiae (Fuller et al., 1988). Fusion proteins efficiently enter the secretory pathway, and mature in the late Golgi upon being processed by the K. phaffii endogenous Ca2+ dependent serine endoprotease Kex2. Several studies have proven that Kex2 activity is a rate limiting step in α-factor prepro leader sequence driven protein secretion in S. cerevisiae (Bevan et al., 1998) and K. phaffii (Yang et al., 2013; Sun et al., 2019).
The endeavor to optimize protein secretion also led to extensive studies of the UPR in K. phaffii (Gasser et al., 2007; Graf et al., 2008; Lin et al., 2013). In order to induce UPR, either HAC1 was overexpressed or cells were treated with DTT, and microarray analysis was performed. Results were allocated to GO groups and compared to studies using S. cerevisiae. K. phaffii reacted to DTT treatment mainly by the regulation of genes related to chemical stimulus, electron transport, and respiration. In contrast, overexpression of HAC1 induced many genes involved in translation, ribosome biogenesis, and organelle biosynthesis. This indicates that the cellular reactions to DTT treatment only slightly overlap with the reactions to overexpression of HAC1. In another study, the secretion of Fap fragments was examined under different oxygen conditions (Baumann et al., 2010). Gene expression profiling was combined with proteomic analyses and the 13C isotope labeling based experimental determination of metabolic fluxes in the central carbon metabolism. Since the protein expression machinery is a multistep metabolic process that requires ATP, it is not surprising that a shift to fermentative metabolism negatively impacted protein synthesis and secretion. However, more importantly, differences in the regulation of the core metabolism regulated by oxygen availability between K. phaffii and S. cerevisiae could be investigated. In contrast to studies using S. cerevisiae (de Groot et al., 2007), a strong transcriptional induction of glycolysis and the non-oxidative pentose phosphate pathways, as well as downregulation of the TCA cycle could be observed in K. phaffii under hypoxic conditions. Upon testing the influence of high osmolarity stress on protein secretion it has been demonstrated that processes such as protein folding, ribosome biogenesis and cell wall organization were affected (Dragosits et al., 2010). In contrast to S. cerevisiae, the main osmolyte released during hypo-osmotic stress was arabitol rather than glycerol, a response mainly observed in osmotolerant yeasts (Kayingo et al., 2001). Also, in contrast to S. cerevisiae, no transcriptional activation of the high osmolarity glycerol (HOG) pathway was observed at steady state conditions.
Another big, commercial research field is glycoengineering of K. phaffii for secretory expression of humanized glycoproteins. Due to its endogenous glycosylation machinery lacking specific terminal mannose glycopeptides known to be clinically incompatible for human administration (Hamilton and Gerngross, 2007), studies of antibody production in yeast have predominantly been conducted in K. phaffii (Spadiut et al., 2014). Several glycoengineering approaches provided valuable insights into the glycosylation machinery of K. phaffii and enabled the production of proteins with human linked glycans causing only mild to low antigenicity in humans (reviewed by Ferreira et al., 2019).
Biological Membranes
Biological membranes play many essential roles. Most importantly, they serve as intracellular boundaries, which compartmentalize organelles and shield the interior of the cell from the exterior, but they also create an indispensable environment for membrane-linked enzymes and biosynthetic processes. Among other yeasts, K. phaffii is an interesting model to investigate diverse aspects of cellular membranes, e.g., lipid profiling, lipid storage and cellular responses caused by lipid modifications (reviewed by Pichler and Emmerstorfer-Augustin, 2018).
The Daum lab for example optimized the isolation of diverse cellular compartments, and then thoroughly analyzed the lipid profiles, e.g., of the plasma membrane (PM) (Grillitsch et al., 2014), peroxisomes (Wriessnegger et al., 2007), mitochondria (Wriessnegger et al., 2009), lipid droplets (Ivashov et al., 2012), and the endoplasmic reticulum (Klug et al., 2014). In all studies it was concluded that cellular membranes from K. phaffii are more similar to the membranes found in multicellular eukaryotes than those found in S. cerevisiae. For example, lipids found in K. phaffii harbor higher amounts of polyunsaturated fatty acids, while S. cerevisiae can only produce unsaturated and monounsaturated fatty acids. The presence of polyunsaturated fatty acids in the PM was linked to improved protein secretion (Wei et al., 2010), which may explain the pole position of K. phaffii as yeast protein secretion host. As expected, the PM of K. phaffii was shown to be enriched in ergosterol, although not to the same extend as in S. cerevisiae (Zinser et al., 1993). Plasma membranes from K. phaffii were also enriched in complex sphingolipids (glycosyl), inositol phosphorylceramides [(G)IPCs], over neutral sphingolipids, which accumulated in internal membranes (Grillitsch et al., 2014).
Due to their low abundance and highly diverse molecular structures, the analysis of sphingolipids is more complex than that of other lipids. Studies on S. cerevisiae gave important insights into sphingolipid metabolism and functions (Dickson, 2010), but S. cerevisiae is an exception among eukaryotic organisms because it contains only (G)IPCs. Most other fungi contain both (G)IPCs and glycosylceramides (GlcCers). Hence, K. phaffii is an attractive alternative model organism to investigate the biosynthesis of GlcCer, because it produces both sphingolipid classes (Michaelson et al., 2009; Ternes et al., 2011).
Lipid profiles of peroxisomes isolated from K. phaffii cells grown on methanol and oleic acid revealed that independent from the carbon source, phosphatidylcholine, and phosphatidylethanolamine were the major peroxisomal phospholipids (Wriessnegger et al., 2007). Cardiolipin was present in peroxisomal membranes at a substantial amount. Interestingly, growth on oleic acid changed fatty acid compositions of phospholipids extracted from total cells and peroxysomes. Phospholipids were shown to majorly incorporate oleic acid indicating that the fatty acid is not only utilized as carbon source but also as a direct building block for complex membrane lipids.
K. phaffii is also an exceptional well-suited host for the examination of lipid droplets. In general, the synthesis of non-polar lipids has been investigated in K. phaffii and contributing proteins have been identified (Ivashov et al., 2012). Different from S. cerevisiae and similar to Y. lipolytica, K. phaffii lipid droplets have a very high triacylgylercol to steryl ester ratio (Koch et al., 2014; Table 1).
The Cell Wall
Cell walls comprise some 20–30% of the cell dry weight and are essential for the survival of fungal cells. Hence, cell wall perturbations and changes in cell wall morphology are efficiently transmitted to, and regulated by, the cell wall integrity (CWI) pathway (Levin, 2011). In S. cerevisiae, the response to cell wall stress signals is triggered by a family of cell-surface sensors, Wsc1, Wsc2, Wsc3, Mtl1, and Mid2, who activate the Pkc1-Mpk1 MAP kinase pathway. These sensors have highly conserved structural domains and consist of extracellular hyperglycosylated regions that function as mechanosensors, short single transmembrane domains, and flexible, cytosolic tails responsible for signal transduction and protein turnover (reviewed by Kock et al., 2015). Once cell wall sensors activate Rho1, which in turn activates protein kinase C (Pkc1), the signaling cascade is fired efficiently, resulting in signal transduction via several MAP kinases, and, finally, in hyperphosphorylation and nuclear localization of the terminal MAPK Slt2.
So far, several homologous proteins of the CWI pathway have been identified in K. phaffii. In 2001, the K. phaffii homolog of Slt2 was characterized and named Pim1 (P. pastoris cell integrity MAPK) (Cosano et al., 2001). Similar to what has been published for ScSlt2, Pim1/Slt2 is dually phosphorylated, and thereby activated in response to heat stress, caffeine and agents that affect the integrity of the fungal cell wall. However, no link to any upstream signaling components has been examined in this study. A more recent publication characterized and investigated CWI sensors Wsc1, Wsc2, and Wsc3 in K. phaffii (Ohsawa et al., 2017). The study revealed a novel function of Wsc1 and Wsc3 in sensing and responding to external methanol levels. Additionally, the binding to Rom2, the down-stream Rho1 GDP-GTP nucleotide exchange factor, has been investigated. In another study, a methanol-induced upregulation of Pim1/Slt2 has been reported (Zhang et al., 2020). Cell wall integrity also explained the majority of variations among different K. phaffii strains (e.g., CBS7435 and GS115), impacting transformation efficiency, growth, methanol metabolism, and secretion of heterologous proteins (Brady et al., 2020).
In a screening assay searching for a super secretor strain, a mutation of BGS13 (Beta-galactosidase supersecretion) was found to improve extracellular levels of a variety of secreted reporter proteins (Larsen et al., 2013). A BLAST search revealed that Bgs13 is the K. phaffii homolog of S. cerevisiae Pkc1. The role of Bgs13/Pkc1 has been further investigated by analyzing strains expressing the truncated variant of this essential protein (Naranjo et al., 2019). The mutant strain showed an abnormal localization of the Bgs13 variant and its cell wall suffered from inherent structural problems, which most probably promoted improved protein secretion.
Vive La (Bio)Diversity: Concluding Remarks
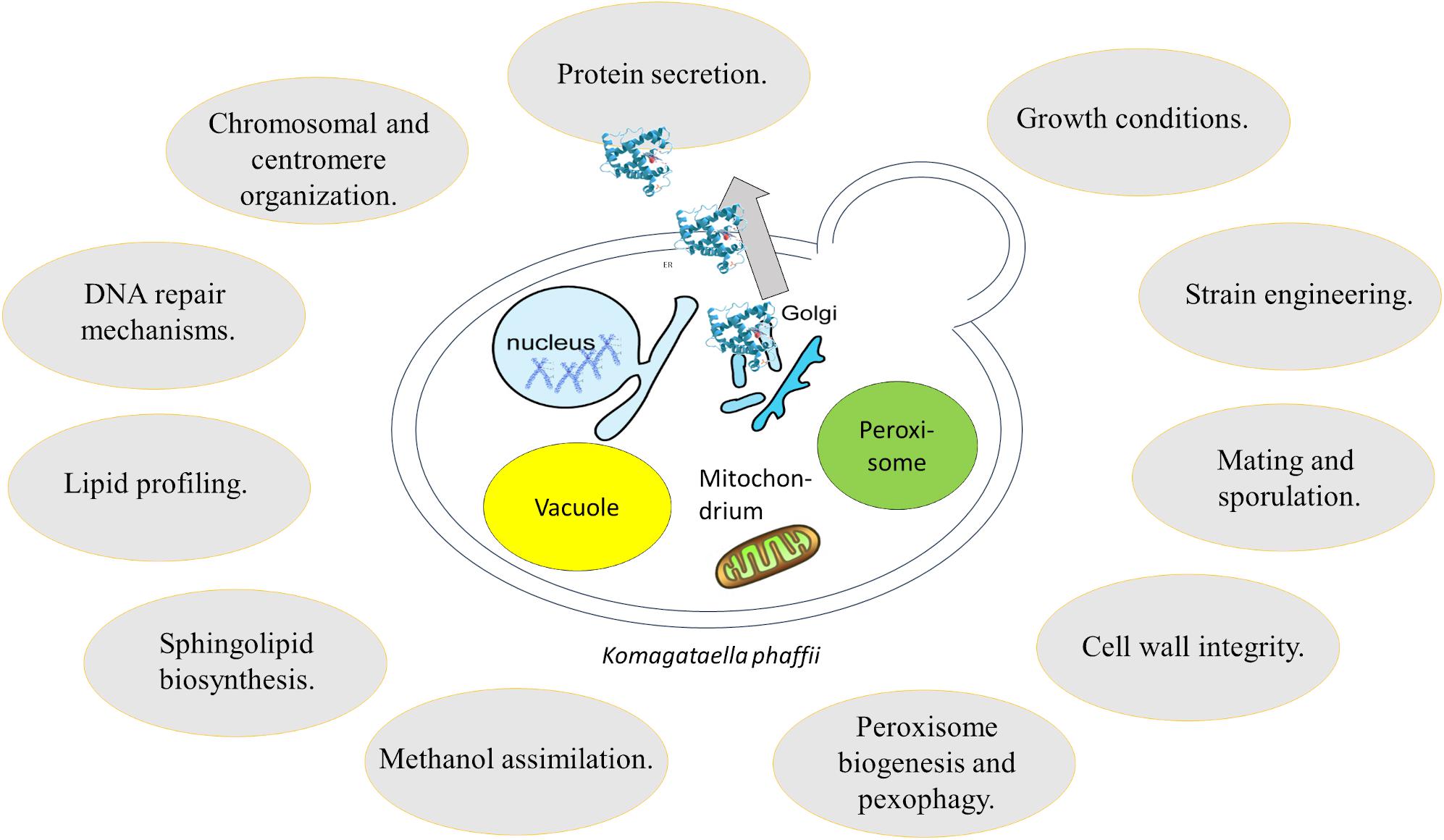
As summarized aptly by Matthews and Vosshall, there are 10 essential steps to build a genetic model organism, including “learning how to work with the organism in the lab” and “develop precise mutagenesis for tagged mutants and gene replacement” (Matthews and Vosshall, 2020). K. phaffii, which has a strong track record as expression host in biotechnological applications, has mostly been in the focus of straightforward pathway engineering in order to leverage advantageous phenotypes. However, data obtained from next generation sequencing and system wide—omics studies in combination with advanced strain modification tools allow a better understanding of the unique physiology and metabolism of this yeast. Thereby, K. phaffii easily passed the first nine steps toward becoming a model system, and is currently tackling the last step, “grow a field of interesting questions using your new model organism.” One may ask now: why should we add additional species to the already existing, most common model systems? The answer is fairly simple. Evolution greatly diversified life on our planet, with many secrets yet to be revealed and many mechanisms yet to be discovered. This review highlights that the study of specialist species, such as K. phaffii gives new and important insights in diverse areas of biology (Figure 3). As shown numerous times, results produced with K. phaffii significantly differ from those obtained using S. cerevisiae (Table 1). Even though S. cerevisiae still is an essential key player in molecular biology and genetics, it is not a very good representative of yeast, since it has a rather atypical physiology compared to others. To summarize a few specifics, S. cerevisiae is a Crabtree-positive yeast, it prefers HR over NHEJ as DNA repair mechanism, and it did undergo whole genome duplication. Without any doubt, S. cerevisiae still offers many advantages including tools such as yeast two-hybrid screening assays (Ito et al., 2001), yeast oligo-mediated genome engineering (YOGE, a recombineering strategy) (Dicarlo et al., 2013) and the yeast deletion collection (Giaever and Nislow, 2014). While most of these tools are not feasible in K. phaffii, some alternative strategies are on the rise to fill this niche. Strategies like REMI which cause random integration and knockout of genes, will allow the performance of genome-wide loss or gain of function studies (Larsen et al., 2013). Another emerging strategy for the application of genome-wide screens is the application of CRISPR/Cas9 or CRISPRi. While the strategy has already widely been used and validated in mammalian studies, an application in K. phaffii is still missing. CRISPR-Cas9 can be used to randomly introduce an insertion or deletion in the genome, which can facilitate the screening for, and analysis of desirable phenotypes. Taken together, future research and engineering efforts will further leverage K. phaffii as powerful model organism to address a critical lack of fundamental biochemical information, maximize desired phenotypes, and increase productivity to reach industrially relevant production yields of new products.
Author Contributions
LB, AR, LL, and AE-A drafted the manuscript. AE-A worked on all figures and tables and carefully reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Austrian Science Fund (FWF) project J3787 (AE-A) and BioTechMed-Graz (Young Researcher Group Program) (LB, AR, LL, AE-A).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We want to thank Assoc.-Prof. Dr. Christoph Augustin for help with the phylogenetic tree (Figure 1). Also, we want to thank Prof. Harald Pichler for useful discussions.
Footnotes
References
Adli, M. (2018). The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9:1911. doi: 10.1038/s41467-018-04252-4252
Agrawal, G., and Subramani, S. (2016). De novo peroxisome biogenesis: evolving concepts and conundrums. Biochim. Biophys. Acta Mol. Cell Res. 1863, 892–901. doi: 10.1016/j.bbamcr.2015.09.014
Ahmad, M., Hirz, M., Pichler, H., and Schwab, H. (2014). Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 98, 5301–5317. doi: 10.1007/s00253-014-5732-5735
Ahmad, M., Winkler, C. M., Kolmbauer, M., Pichler, H., Schwab, H., and Emmerstorfer-Augustin, A. (2019). Pichia pastoris protease-deficient and auxotrophic strains generated by a novel, user-friendly vector toolbox for gene deletion. Yeast 36, 557–570. doi: 10.1002/yea.3426
Alloue-Boraud, W. A. M., N’Guessan, K. F., Djeni, N. T., Hiligsmann, S., Djè, K. M., and Delvigne, F. (2015). Fermentation profile of Saccharomyces cerevisiae and Candida tropicalis as starter cultures on barley malt medium. J. Food Sci. Technol. 52, 5236–5242. doi: 10.1007/s13197-014-1526-1520
Ata, Ö, Rebnegger, C., Tatto, N. E., Valli, M., Mairinger, T., Hann, S., et al. (2018). A single Gal4-like transcription factor activates the Crabtree effect in Komagataella phaffii. Nat. Commun. 9:4911. doi: 10.1038/s41467-018-07430-7434
Baghban, R., Farajnia, S., Ghasemi, Y., Mortazavi, M., Zarghami, N., and Samadi, N. (2018). New developments in pichia pastoris expression system, review and update. Curr. Pharm. Biotechnol. 19, 451–467. doi: 10.2174/1389201019666180718093037
Baumann, K., Carnicer, M., Dragosits, M., Graf, A. B., Stadlmann, J., Jouhten, P., et al. (2010). A multi-level study of recombinant Pichia pastoris in different oxygen conditions. BMC Syst. Biol. 4:141. doi: 10.1186/1752-0509-4-141
Bevan, A., Brenner, C., and Fuller, R. S. (1998). Quantitative assessment of enzyme specificity in vivo: P2 recognition by Kex2 protease defined in a genetic system. Proc. Natl. Acad. Sci. U. S. A. 95, 10384–10389. doi: 10.1073/pnas.95.18.10384
Bevis, B. J., Hammond, A. T., Reinke, C. A., and Glick, B. S. (2002). De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 4, 750–756. doi: 10.1038/ncb852
BioWorldTM (1993). RCT Buys Pichia Expression System from Pillips. Available online at: https://www.bioworld.com/articles/361392-rct-buys-pichia-expression-system-from-phillips?v=preview (accessed October 4, 1993)
Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., et al. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. doi: 10.1002/(sici)1097-0061(19980130)14:2<115::aid-yea204>3.0.co;2-2
Brady, J. R., Whittaker, C. A., Tan, M. C., Kristensen, D. L., Ma, D., Dalvie, N. C., et al. (2020). Comparative genome-scale analysis of Pichia pastoris variants informs selection of an optimal base strain. Biotechnol. Bioeng. 117, 543–555. doi: 10.1002/bit.27209
Braun-Galleani, S., Dias, J. A., Coughlan, A. Y., Ryan, A. P., Byrne, K. P., and Wolfe, K. H. (2019). Genomic diversity and meiotic recombination among isolates of the biotech yeast Komagataella phaffii (Pichia pastoris). Microb. Cell Fact. 18:211. doi: 10.1186/s12934-019-1260-1264
Byrne, B. (2015). Pichia pastoris as an expression host for membrane protein structural biology. Curr. Opin. Struct. Biol. 32, 9–17. doi: 10.1016/j.sbi.2015.01.005
Caldecott, K. W. (2008). Single-strand break repair and genetic disease. Nat. Rev. Genet. 9, 619–631. doi: 10.1038/nrg2380
Camattari, A., Goh, A., Yip, L. Y., Meng Tan, A. H., Ng, S. W., Tran, A., et al. (2016). Characterization of a panARS-based episomal vector in the methylotrophic yeast Pichia pastoris for recombinant protein production and synthetic biology applications. Microb. Cell Fact. 15:139. doi: 10.1186/s12934-016-0540-545
Caspeta, L., Shoaie, S., Agren, R., Nookaew, I., and Nielsen, J. (2012). Genome-scale metabolic reconstructions of Pichia stipitis and Pichia pastoris and in silico evaluation of their potentials. BMC Syst. Biol. 6:24. doi: 10.1186/1752-0509-6-24
Cereghino, J. L., and Cregg, J. M. (2000). Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 24, 45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x
Chen, M. T., Lin, S., Shandil, I., Andrews, D., Stadheim, T. A., and Choi, B. K. (2012). Generation of diploid Pichia pastoris strains by mating and their application for recombinant protein production. Microb. Cell Fact. 11:91. doi: 10.1186/1475-2859-11-91
Chen, Z., Sun, H., Li, P., He, N., Zhu, T., and Li, Y. (2013). Enhancement of the gene targeting efficiency of non-conventional yeasts by increasing genetic redundancy. PLoS One 8:e57952. doi: 10.1371/journal.pone.0057952
Chiang, H. L., Schekman, R., and Hamamoto, S. (1996). Selective uptake of cytosolic, peroxisomal, and plasma membrane proteins into the yeast lysosome for degradation. J. Biol. Chem. 271, 9934–9941. doi: 10.1074/jbc.271.17.9934
Chung, B. K. S., Lakshmanan, M., Klement, M., Ching, C. B., and Lee, D. Y. (2013). Metabolic reconstruction and flux analysis of industrial Pichia yeasts. Appl. Microbiol. Biotechnol. 97, 1865–1873. doi: 10.1007/s00253-013-4702-4707
Chung, B. K. S., Selvarasu, S., Andrea, C., Ryu, J., Lee, H., Ahn, J., et al. (2010). Genome-scale metabolic reconstruction and in silico analysis of methylotrophic yeast Pichia pastoris for strain improvement. Microb. Cell Fact. 9:50. doi: 10.1186/1475-2859-9-50
Cosano, I. C., Álvarez, P., Molina, M., and Nombela, C. (1998). Cloning and sequence analysis of the Pichia pastoris TRP1, IPP1 and HIS3 genes. Yeast 14, 861–867. doi: 10.1002/(sici)1097-0061(19980630)14:9<861::aid-yea276>3.0.co;2-n
Cosano, I. C., Martín, H., Flández, M., Nombela, C., and Molina, M. (2001). Pim1, a MAP kinase involved in cell wall integrity in Pichia pastoris. Mol. Genet. Genomics 265, 604–614. doi: 10.1007/s004380100452
Coughlan, A. Y., Hanson, S. J., Byrne, K. P., and Wolfe, K. H. (2016). Centromeres of the yeast Komagataella phaffii (Pichia pastoris) have a simple inverted-repeat structure. Genome Biol. Evol. 8, 2482–2492. doi: 10.1093/gbe/evw178
Cregg, J. M. (1987). “Genetics of Methylotrophic yeasts,” in Microbial Growth on C1 Compounds, eds H. W. Verseveld and J. A. Duine (Berlin: Springer), 158–167. doi: 10.1007/978-94-009-3539-6_20
Cregg, J. M., Barringer, K. J., Hessler, A. Y., and Madden, K. R. (1985). Pichia pastoris as a host system for transformations. Mol. Cell Biol. 5, 3376–3385. doi: 10.1128/mcb.5.12.3376
Cregg, J. M., Madden, K. R., Barringer, K. J., Thill, G. P., and Stillman, C. A. (1989). Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris. Mol. Cell. Biol. 9, 1316–1323. doi: 10.1128/mcb.9.3.1316
Daly, R., and Hearn, M. T. W. (2005). Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J. Mol. Recognit. 18, 119–138. doi: 10.1002/jmr.687
Damasceno, L. M., Huang, C. J., and Batt, C. A. (2012). Protein secretion in Pichia pastoris and advances in protein production. Appl. Microbiol. Biotechnol. 93, 31–39. doi: 10.1007/s00253-011-3654-z
Daran-Lapujade, P., Jansen, M. L. A., Daran, J. M., Van Gulik, W., De Winde, J. H., and Pronk, J. T. (2004). Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae: a chemostat culture study. J. Biol. Chem. 279, 9125–9138. doi: 10.1074/jbc.M309578200
de Groot, M. J. L., Daran-Lapujade, P., van Breukelen, B., Knijnenburg, T. A., de Hulster, E. A. F., Reinders, M. J. T., et al. (2007). Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 153, 3864–3878. doi: 10.1099/mic.0.2007/009969-9960
De Schutter, K., Lin, Y.-C., Tiels, P., Van Hecke, A., Glinka, S., Weber-Lehmann, J., et al. (2009). Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 27, 561–566. doi: 10.1038/nbt.1544
Dicarlo, J. E., Conley, A. J., Penttilä, M., Jäntti, J., Wang, H. H., and Church, G. M. (2013). Yeast oligo-mediated genome engineering (YOGE). ACS Synth. Biol. 2, 741–749. doi: 10.1021/sb400117c
Dickson, R. C. (2010). Roles for sphingolipids in Saccharomyces cerevisiae. Adv. Exp. Med. Biol. 688, 217–231. doi: 10.1007/978-1-4419-6741-1_15
Dragosits, M., Stadlmann, J., Albiol, J., Baumann, K., Maurer, M., Gasser, B., et al. (2009). The effect of temperature on the proteome of recombinant Pichia pastoris. J. Proteome Res. 8, 1380–1392. doi: 10.1021/pr8007623
Dragosits, M., Stadlmann, J., Graf, A., Gasser, B., Maurer, M., Sauer, M., et al. (2010). The response to unfolded protein is involved in osmotolerance of Pichia pastoris. BMC Genomics 11:207. doi: 10.1186/1471-2164-11-207
Dudásová, Z., Dudás, A., Chovanec, M., Dudášová, Z., Dudáš, A., and Chovanec, M. (2004). Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 28, 581–601. doi: 10.1016/j.femsre.2004.06.001
Dunn, W. A., Cregg, J. M., Kiel, J. A. K. W., van der Klei, I. J., Oku, M., Sakai, Y., et al. (2005). Pexophagy: the selective autophagy of peroxisomes. Autophagy 1, 75–83. doi: 10.4161/auto.1.2.1737
Egli, T., Käppeli, O., and Fiechter, A. (1982). Mixed substrate growth of methylotrophic yeasts in chemostat culture: influence of the dilution rate on the utilisation of a mixture of glucose and methanol. Arch. Microbiol. 131, 8–13. doi: 10.1007/BF00451491
Ferreira, R., Limeta, A., and Nielsen, J. (2019). Tackling cancer with yeast-based technologies. Trends Biotechnol. 37, 592–603. doi: 10.1016/j.tibtech.2018.11.013
Fitzgerald-Hayes, M., Clarke, L., and Carbon, J. (1982). Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29, 235–244. doi: 10.1016/0092-8674(82)90108-90108
Fuller, R. S., Sterne, R. E., and Thorner, J. (1988). Enzymes required for yeast prohormone processing. Annu. Rev. Physiol. 50, 345–362. doi: 10.1146/annurev.ph.50.030188.002021
Gasser, B., Dragosits, M., and Mattanovich, D. (2010). Engineering of biotin-prototrophy in Pichia pastoris for robust production processes. Metab. Eng. 12, 573–580. doi: 10.1016/j.ymben.2010.07.002
Gasser, B., Maurer, M., Rautio, J., Sauer, M., Bhattacharyya, A., Saloheimo, M., et al. (2007). Monitoring of transcriptional regulation in Pichia pastoris under protein production conditions. BMC Genomics 8:179. doi: 10.1186/1471-2164-8-179
Gassler, T., Sauer, M., Gasser, B., Egermeier, M., Troyer, C., Causon, T., et al. (2020). The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 38, 210–216. doi: 10.1038/s41587-019-0363-360
Giaever, G., and Nislow, C. (2014). The yeast deletion collection: a decade of functional genomics. Genetics 197, 451–465. doi: 10.1534/genetics.114.161620
Gnügge, R., and Rudolf, F. (2017). Saccharomyces cerevisiae Shuttle vectors. Yeast 34, 205–221. doi: 10.1002/yea.3228
Goffeau, A., Barrell, G., Bussey, H., Davis, R. W., Dujon, B., Feldmann, H., et al. (1996). Life with 6000 genes. Science 274, 546–567. doi: 10.1126/science.274.5287.546
Golemis, E. A., and Khazak, V. (1997). Alternative yeast two-hybrid systems. the interaction trap and interaction mating. Methods Mol. Biol. 63, 197–218. doi: 10.1385/0-89603-481-X:197
Graf, A., Gasser, B., Dragosits, M., Sauer, M., Leparc, G. G., Tüchler, T., et al. (2008). Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays. BMC Genomics 9:390. doi: 10.1186/1471-2164-9-390
Grillitsch, K., Connerth, M., Köfeler, H., Arrey, T. N., Rietschel, B., Wagner, B., et al. (2011). Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: Lipidome meets Proteome. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1811, 1165–1176. doi: 10.1016/j.bbalip.2011.07.015
Grillitsch, K., Tarazona, P., Klug, L., Wriessnegger, T., Zellnig, G., Leitner, E., et al. (2014). Isolation and characterization of the plasma membrane from the yeast Pichia pastoris. Biochim. Biophys. Acta - Biomembr. 1838, 1889–1897. doi: 10.1016/j.bbamem.2014.03.012
Gu, Y., Gao, J., Cao, M., Dong, C., Lian, J., Huang, L., et al. (2019). Construction of a series of episomal plasmids and their application in the development of an efficient CRISPR/Cas9 system in Pichia pastoris. World J. Microbiol. Biotechnol. 35:79. doi: 10.1007/s11274-019-2654-2655
Gueldener, U., Heinisch, J., Koehler, G. J., Voss, D., and Hegemann, J. H. (2002). A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23.
Guilliermond, A. (1920). Zygosaccharomyces Pastori, Nouvelle Espèce de Levures à Copulation Hétérogamique. France: Bulletin de la Société Mycologique de France.
Gündüz Ergün, B., Hüccetoğulları, D., Öztürk, S., Çelik, E., and Çalık, P. (2019). Established and upcoming yeast expression systems. Methods Mol. Biol. 1923, 1–74. doi: 10.1007/978-1-4939-9024-5_1
Haber, J. E. (2012). Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191, 33–64. doi: 10.1534/genetics.111.134577
Hamilton, S. R., and Gerngross, T. U. (2007). Glycosylation engineering in yeast: the advent of fully humanized yeast. Curr. Opin. Biotechnol. 18, 387–392. doi: 10.1016/j.copbio.2007.09.001
Hamilton, S. R., Bobrowicz, P., Bobrowicz, B., Davidson, R. C., Li, H., Mitchell, T., et al. (2003). Production of complex human glycoproteins in yeast. Science 301, 1244–1246. doi: 10.1126/science.1088166
Hanson, S. J., and Wolfe, K. H. (2017). An evolutionary perspective on yeast mating-type switching. Genetics 206, 9–32. doi: 10.1534/genetics.117.202036
Hanson, S. J., Byrne, K. P., and Wolfe, K. H. (2014). Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc. Natl. Acad. Sci. U. S. A. 111, E4851–E4858. doi: 10.1073/pnas.1416014111
Harari, Y., Ram, Y., Rappoport, N., Hadany, L., and Kupiec, M. (2018). Spontaneous changes in ploidy are common in yeast. Curr. Biol. 28, 825–835.e4. doi: 10.1016/j.cub.2018.01.062
Hartner, F. S., and Glieder, A. (2006). Regulation of methanol utilisation pathway genes in yeasts. Microb. Cell Fact. 5:39. doi: 10.1186/1475-2859-5-39
Hegemann, J. H., and Fleig, U. N. (1993). The centromere of budding yeast. BioEssays 15, 451–460. doi: 10.1002/bies.950150704
Heistinger, L., Gasser, B., and Mattanovich, D. (2017). Creation of stable heterothallic strains of komagataella phaffii enables dissection of mating gene regulation. Mol. Cell. Biol. 38:e00398–17. doi: 10.1128/mcb.00398-317
Heistinger, L., Gasser, B., and Mattanovich, D. (2020). Microbe profile: Komagataella phaffii: a methanol devouring biotech yeast formerly known as Pichia pastoris. Microbiology 166, 614–616. doi: 10.1099/mic.0.000958
Heistinger, L., Moser, J., Tatto, N. E., Valli, M., Gasser, B., and Mattanovich, D. (2018). Identification and characterization of the Komagataella phaffii mating pheromone genes. FEMS Yeast Res. 18:foyo51. doi: 10.1093/femsyr/foy051
Hutchins, M. U., Veenhuis, M., and Klionsky, D. J. (1999). Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112, 4079–4087.
Inan, M., and Meagher, M. M. (2001). Non-repressing carbon sources for alcohol oxidase (AOX1) promoter of Pichia pastoris. J. Biosci. Bioeng. 92, 585–589. doi: 10.1263/jbb.92.585
Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U. S. A. 98, 4569–4574. doi: 10.1073/pnas.061034498
Ivashov, V. A., Grillitsch, K., Koefeler, H., Leitner, E., Baeumlisberger, D., Karas, M., et al. (2012). Lipidome and proteome of lipid droplets from the methylotrophic yeast Pichia pastoris. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1831, 282–290. doi: 10.1016/j.bbalip.2012.09.017
Jacobs, P. P., Geysens, S., Vervecken, W., Contreras, R., and Callewaert, N. (2009). Engineering complex-type N-glycosylation in Pichia pastoris using GlycoSwitch technology. Nat. Protoc. 4, 58–70. doi: 10.1038/nprot.2008.213
Jahic, M., Veide, A., Charoenrat, T., Teeri, T., and Enfors, S. O. (2006). Process technology for production and recovery of heterologous proteins with Pichia pastoris. Biotechnol. Prog. 22, 1465–1473. doi: 10.1021/bp060171t
Jakočiunas, T., Jensen, M. K., and Keasling, J. D. (2016). CRISPR/Cas9 advances engineering of microbial cell factories. Metab. Eng. 34, 44–59. doi: 10.1016/j.ymben.2015.12.003
Juhas, M., and Ajioka, J. W. (2017). High molecular weight DNA assembly in vivo for synthetic biology applications. Crit. Rev. Biotechnol. 37, 277–286. doi: 10.3109/07388551.2016.1141394
Jungo, C., Urfer, J., Zocchi, A., Marison, I., and von Stockar, U. (2007). Optimisation of culture conditions with respect to biotin requirement for the production of recombinant avidin in Pichia pastoris. J. Biotechnol. 127, 703–715. doi: 10.1016/j.jbiotec.2006.08.001
Karbalaei, M., Rezaee, S. A., and Farsiani, H. (2020). Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 235, 5867–5881. doi: 10.1002/jcp.29583
Kato, K., Kurimura, Y., Makiguchi, N., and Asai, Y. (1974). Determination of methanol strongly assimilating yeasts. J. Gen. Appl. Microbiol. 20, 123–127. doi: 10.2323/jgam.20.123
Kayingo, G., Kilian, S. G., and Prior, B. A. (2001). Conservation and release of osmolytes by yeasts during hypo-osmotic stress. Arch. Microbiol. 177, 29–35. doi: 10.1007/s00203-001-0358-352
Kellis, M., Birren, B. W., and Lander, E. S. (2004). Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624. doi: 10.1038/nature02424
Kilkenny, B. C., and Hinshelwood, C. N. (1951). The utilization of carbon sources by certain yeast strains. Proc. R. Soc. London. Ser. B Biol. Sci. 138, 375–385. doi: 10.1098/rspb.1951.0030
Klug, L., Tarazona, P., Gruber, C., Grillitsch, K., Gasser, B., Trötzmüller, M., et al. (2014). The lipidome and proteome of microsomes from the methylotrophic yeast Pichia pastoris. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1841, 215–226. doi: 10.1016/j.bbalip.2013.11.005
Knop, M. (2006). Evolution of thehemiascomycete yeasts: on life styles and the importance of inbreeding. BioEssays 28, 696–708. doi: 10.1002/bies.20435
Koch, B., Schmidt, C., and Daum, G. (2014). Storage lipids of yeasts: a survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol. Rev. 38, 892–915. doi: 10.1111/1574-6976.12069
Kock, C., Dufrêne, Y. F., and Heinisch, J. J. (2015). Up against the wall: is yeast cell wall integrity ensured by mechanosensing in plasma membrane microdomains? Appl. Environ. Microbiol. 81, 806–811. doi: 10.1128/AEM.03273-3214
Krejci, L., Altmannova, V., Spirek, M., and Zhao, X. (2012). Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795–5818. doi: 10.1093/nar/gks270
Küberl, A., Schneider, J., Thallinger, G. G., Anderl, I., Wibberg, D., Hajek, T., et al. (2011). High-quality genome sequence of Pichia pastoris CBS7435. J. Biotechnol. 154, 312–320. doi: 10.1016/j.jbiotec.2011.04.014
Kurtzman, C. P. (2005). Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella. Int. J. Syst. Evol. Microbiol. 55, 973–976. doi: 10.1099/ijs.0.63491-63490
Larsen, S., Weaver, J., de Sa Campos, K., Bulahan, R., Nguyen, J., Grove, H., et al. (2013). Mutant strains of Pichia pastoris with enhanced secretion of recombinant proteins. Biotechnol. Lett. 35, 1925–1935. doi: 10.1007/s10529-013-1290-1297
Levin, D. E. (2011). Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189, 1145–1175. doi: 10.1534/genetics.111.128264
Liachko, I., and Dunham, M. J. (2014). An autonomously replicating sequence for use in a wide range of budding yeasts. FEMS Yeast Res. 14, 364–367. doi: 10.1111/1567-1364.12123
Lin Cereghino, G. P., Lin Cereghino, J., Jay Sunga, A., Johnson, M. A., Lim, M., Gleeson, M. A. G., et al. (2001). New selectable marker/auxotrophic host strain combinations for molecular genetic manipulation of Pichia pastoris. Gene 263, 159–169. doi: 10.1016/S0378-1119(00)00576-X
Lin, X. Q., Liang, S. L., Han, S. Y., Zheng, S. P., Ye, Y. R., and Lin, Y. (2013). Quantitative iTRAQ LC-MS/MS proteomics reveals the cellular response to heterologous protein overexpression and the regulation of HAC1 in Pichia pastoris. J. Proteomics 91, 58–72. doi: 10.1016/j.jprot.2013.06.031
Liu, Q., Shi, X., Song, L., Liu, H., Zhou, X., Wang, Q., et al. (2019). CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Fact. 18:144. doi: 10.1186/s12934-019-1194-x
Löbs, A. K., Schwartz, C., and Wheeldon, I. (2017). Genome and metabolic engineering in non-conventional yeasts: current advances and applications. Synth. Syst. Biotechnol. 2, 198–207. doi: 10.1016/j.synbio.2017.08.002
Love, K. R., Shah, K. A., Whittaker, C. A., Wu, J., Bartlett, M. C., Ma, D., et al. (2016). Comparative genomics and transcriptomics of Pichia pastoris. BMC Genomics 17:550. doi: 10.1186/s12864-016-2876-y
Maekawa, H., and Kaneko, Y. (2014). Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha. PLoS Genet. 10:e1004796. doi: 10.1371/journal.pgen.1004796
Mattanovich, D., Callewaert, N., Rouzé, P., Lin, Y. C., Graf, A., Redl, A., et al. (2009). Open access to sequence: browsing the Pichia pastoris genome. Microb. Cell Fact. 8:53. doi: 10.1186/1475-2859-8-53
Matthews, B. J., and Vosshall, L. B. (2020). How to turn an organism into a model organism in 10 ‘easy’ steps. J. Exp. Biol. 223:jeb218198. doi: 10.1242/jeb.218198
Matthews, C. B., Kuo, A., Love, K. R., and Love, J. C. (2018). Development of a general defined medium for Pichia pastoris. Biotechnol. Bioeng. 115, 103–113. doi: 10.1002/bit.26440
Michaelson, L. V., Zäuner, S., Markham, J. E., Haslam, R. P., Desikan, R., Mugford, S., et al. (2009). Functional characterization of a higher plant sphingolipid Δ4-desaturase: defining the role of sphingosine and sphingosine-1- phosphate in arabidopsis. Plant Physiol. 149, 487–498. doi: 10.1104/pp.108.129411
Monastryska, I., Sjollema, K., Van Der Klei, I. J., Kiel, J. A. K. W., and Veenhuis, M. (2004). Microautophagy and macropexophagy may occur simultaneously in Hansenula polymorpha. FEBS Lett. 568, 135–138. doi: 10.1016/j.febslet.2004.05.018
Mukaiyama, H., Baba, M., Osumi, M., Aoyagi, S., Kato, N., Ohsumi, Y., et al. (2004). Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 15, 58–70. doi: 10.1091/mbc.E03-05-0340
Näätsaari, L., Mistlberger, B., Ruth, C., Hajek, T., Hartner, F. S., and Glieder, A. (2012). Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One 7:e39720. doi: 10.1371/journal.pone.0039720
Nakamura, Y., Nishi, T., Noguchi, R., Ito, Y., Watanabe, T., Nishiyama, T., et al. (2018). A stable, autonomously replicating plasmid vector containing Pichia pastoris centromeric DNA. Appl. Environ. Microbiol. 84:e02882-17. doi: 10.1128/AEM.02882-2817
Naranjo, C. A., Jivan, A. D., Vo, M. N., de Sa Campos, K. H., Deyarmin, J. S., Hekman, R. M., et al. (2019). Role of BGS13 in the secretory mechanism of Pichia pastoris. Appl. Environ. Microbiol. 85:e001615-19. doi: 10.1128/AEM.01615-1619
Nazarko, T. Y., Polupanov, A. S., Manjithaya, R. R., Subramani, S., and Sibirny, A. A. (2007). The requirement of sterol glucoside for pexophagy in yeast is dependent on the species and nature of peroxisome inducers. Mol. Biol. Cell 18, 106–118. doi: 10.1091/mbc.E06-06-0554
Nevoigt, E., and Stahl, U. (1997). Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 21, 231–241. doi: 10.1111/j.1574-6976.1997.tb00352.x
Nielsen, J. (2013). Production of biopharmaceutical proteins by yeast: advances through metabolic engineering. Bioengineered 4, 207–211. doi: 10.4161/bioe.22856
Novick, P., Ferro, S., and Schekman, R. (1981). Order of events in the yeast secretory pathway. Cell 25, 461–469. doi: 10.1016/0092-8674(81)90064-90067
Ogata, K., Nishikawa, H., and Ohsugi, M. (1969). A yeast capable of utilizing methanol. Agric. Biol. Chem. 33, 1519–1520. doi: 10.1271/bbb1961.33.1519
Ohsawa, S., Yurimoto, H., and Sakai, Y. (2017). Novel function of Wsc proteins as a methanol-sensing machinery in the yeast Pichia pastoris. Mol. Microbiol. 104, 349–363. doi: 10.1111/mmi.13631
Oku, M., and Sakai, Y. (2016). Pexophagy in yeasts. Biochim. Biophys. Acta Mol. Cell Res. 1863, 992–998. doi: 10.1016/j.bbamcr.2015.09.023
Oldenburg, K. R., Vo, K. T., Michaelis, S., and Paddon, C. (1997). Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25, 451–452. doi: 10.1093/nar/25.2.451
Pan, Y., Wu, D., and Wu, J. (2018). Improving the expression of recombinant xylanase in Pichia pastoris with episomal expression plasmid. Shengwu Gongcheng Xuebao/Chinese. J. Biotechnol. 34, 712–721. doi: 10.13345/j.cjb.170409
Payne, W. E., Kaiser, C. A., Bevis, B. J., Soderhalm, J., Fu, D., Sears, I. B., et al. (2000). Isolation of Pichia pastoris genes involved in ER-to-Golgi transport. Yeast 16, 979–993. doi: 10.1002/1097-0061(200008)16:11<979::aid-yea594>3.0.co;2-c
Pfeiffer, T., and Morley, A. (2014). An evolutionary perspective on the Crabtree effect. Front. Mol. Biosci. 1:17. doi: 10.3389/fmolb.2014.00017
Phaff, H. J., Miller, M. W., and Shifrine, M. (1956). The taxonomy of yeasts isolated from drosophila in the yosemite region of California. Antonie Van Leeuwenhoek 22, 145–161. doi: 10.1007/BF02538322
Pichler, H., and Emmerstorfer-Augustin, A. (2018). Modification of membrane lipid compositions in single-celled organisms – from basics to applications. Methods 147, 50–65. doi: 10.1016/j.ymeth.2018.06.009
Preuss, D., Mulholland, J., Franzusoff, A., Segev, N., and Botstein, D. (1992). Characterization of the saccharomyces golgi complex through the cell cycle by immunoelectron microscopy. Mol. Biol. Cell 3, 789–803. doi: 10.1091/mbc.3.7.789
Puxbaum, V., Mattanovich, D., and Gasser, B. (2015). Quo vadis? the challenges of recombinant protein folding and secretion in Pichia pastoris. Appl. Microbiol. Biotechnol. 99, 2925–2938. doi: 10.1007/s00253-015-6470-z
Riley, R., Haridas, S., Wolfe, K. H., Lopes, M. R., Hittinger, C. T., Göker, M., et al. (2016). Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. U. S. A. 113, 9882–9887. doi: 10.1073/pnas.1603941113
Rossanese, O. W., Soderholm, J., Bevis, B. J., Sears, I. B., O’Connor, J., Williamson, E. K., et al. (1999). Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell Biol. 145, 69–81. doi: 10.1083/jcb.145.1.69
Schrick, K., Garvik, B., and Hartwell, L. H. (1997). Mating in Saccharomyces cerevisiae: the role of the pheromone signal transduction pathway in the chemotropic response to pheromone. Genetics 147, 19–32.
Schwarzhans, J. P., Luttermann, T., Wibberg, D., Winkler, A., Hübner, W., Huser, T., et al. (2017). A mitochondrial autonomously replicating sequence from Pichia pastoris for uniform high level recombinant protein production. Front. Microbiol. 8:780. doi: 10.3389/fmicb.2017.00780
Schwarzhans, J.-P., Wibberg, D., Winkler, A., Luttermann, T., Kalinowski, J., and Friehs, K. (2016). Non-canonical integration events in Pichia pastoris encountered during standard transformation analysed with genome sequencing. Sci. Rep. 6:38952. doi: 10.1038/srep38952
Shen, X. X., Opulente, D. A., Kominek, J., Zhou, X., Steenwyk, J. L., Buh, K. V., et al. (2018). Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175, 1533–1545.e20. doi: 10.1016/j.cell.2018.10.023
Siam, R., Dolan, W. P., and Forsburg, S. L. (2004). Choosing and using Schizosaccharomyces pombe plasmids. Methods 33, 189–198. doi: 10.1016/j.ymeth.2003.11.013
Sohn, S. B., Graf, A. B., Kim, T. Y., Gasser, B., Maurer, M., Ferrer, P., et al. (2010). Genome-scale metabolic model of methylotrophic yeast Pichia pastoris and its use for in silico analysis of heterologous protein production. Biotechnol. J. 5, 705–715. doi: 10.1002/biot.201000078
Solà, A., Jouhten, P., Maaheimo, H., Sánchez-Ferrando, F., Szyperski, T., and Ferrer, P. (2007). Metabolic flux profiling of Pichia pastoris grown on glycerol/methanol mixtures in chemostat cultures at low and high dilution rates. Microbiology 153, 281–290. doi: 10.1099/mic.0.29263-29260
Spadiut, O., Capone, S., Krainer, F., Glieder, A., and Herwig, C. (2014). Microbials for the production of monoclonal antibodies and antibody fragments. Trends Biotechnol. 32, 54–60. doi: 10.1016/j.tibtech.2013.10.002
Storici, F., Durham, C. L., Gordenin, D. A., and Resnick, M. A. (2003). Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl. Acad. Sci. U. S. A. 100, 14994–14999. doi: 10.1073/pnas.2036296100
Stovicek, V., Holkenbrink, C., and Borodina, I. (2017). CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res. 17:fox030. doi: 10.1093/FEMSYR/FOX030
Stukey, J. E., McDonough, V. M., and Martin, M. C. (1989). Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264, 16537–16544.
Sturmberger, L., Chappell, T., Geier, M., Krainer, F., Day, K. J., Vide, U., et al. (2016). Refined Pichia pastoris reference genome sequence. J. Biotechnol. 235, 121–131. doi: 10.1016/j.jbiotec.2016.04.023
Sun, J., Jiang, J., Zhai, X., Zhu, S., Qu, Z., Yuan, W., et al. (2019). Coexpression of Kex2 endoproteinase and hac1 transcription factor to improve the secretory expression of bovine lactoferrin in Pichia pastoris. Biotechnol. Bioprocess Eng. 24, 934–941. doi: 10.1007/s12257-019-0176-175
Ternes, P., Wobbe, T., Schwarz, M., Albrecht, S., Feussner, K., Riezman, I., et al. (2011). Two pathways of sphingolipid biosynthesis are separated in the yeast Pichia pastoris. J. Biol. Chem. 286, 11401–11414. doi: 10.1074/jbc.M110.193094
Thorpe, E. D., D’Anjou, M. C., and Daugulis, A. J. (1999). Sorbitol as a non-repressing carbon source for fed-batch fermentation of recombinant Pichia pastoris. Biotechnol. Lett. 21, 669–672. doi: 10.1023/A:1005585407601
Tsakraklides, V., Brevnova, E., Stephanopoulos, G., and Shaw, A. J. (2015). Improved gene targeting through cell cycle synchronization. PLoS One 10:e0133434. doi: 10.1371/journal.pone.0133434
Tyo, K. E. J., Liu, Z., Petranovic, D., and Nielsen, J. (2012). Imbalance of heterologous protein folding and disulfide bond formation rates yields runaway oxidative stress. BMC Biol. 10:16. doi: 10.1186/1741-7007-10-16
Valli, M., Grillitsch, K., Grünwald-Gruber, C., Tatto, N. E., Hrobath, B., Klug, L., et al. (2020). A subcellular proteome atlas of the yeast Komagataella phaffii. FEMS Yeast Res. 20:foaa001. doi: 10.1093/femsyr/foaa001
Valli, M., Tatto, N. E., Peymann, A., Gruber, C., Landes, N., Ekker, H., et al. (2016). Curation of the genome annotation of Pichia pastoris (Komagataella phaffii) CBS7435 from gene level to protein function. FEMS Yeast Res. 16:fow051. doi: 10.1093/femsyr/fow051
van der Klei, I. J., Yurimoto, H., Sakai, Y., and Veenhuis, M. (2006). The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim. Biophys. Acta Mol. Cell Res. 1763, 1453–1462. doi: 10.1016/j.bbamcr.2006.07.016
Vogl, T., and Glieder, A. (2013). Regulation of Pichia pastoris promoters and its consequences for protein production. N. Biotechnol. 30, 385–404. doi: 10.1016/j.nbt.2012.11.010
Walther, I., Bechler, B., Müller, O., Hunzinger, E., and Cogoli, A. (1996). Cultivation of Saccharomyces cerevisiae in a bioreactor in microgravity. J. Biotechnol. 47, 113–127. doi: 10.1016/0168-1656(96)01375-1372
Wegner, E. H. (1983). Biochemical conversions by yeast fermentation at high cell densities. United States Pat. 4, 329–414.
Wegner, G. H. (1990). Emerging applications of the methylotrophic yeasts. FEMS Microbiol. Lett. 87, 279–284. doi: 10.1111/j.1574-6968.1990.tb04925.x
Wegner, G. H., and Harder, W. (1987). Methylotrophic yeasts - 1986. Antonie Van Leeuwenhoek 53, 29–36. doi: 10.1007/BF00422632
Wei, D., Li, J., Shen, M., Jia, W., Chen, N., Chen, T., et al. (2010). Cellular production of n-3 PUFAs and reduction of n-6 -to-n-3 ratios in the pancreatic β-cells and islets enhance insulin secretion and confer protection against cytokine-induced cell death. Diabetes Metab. Res. Rev. 59, 471–478. doi: 10.2337/db09-0284
Weninger, A., Fischer, J. E., Raschmanová, H., Kniely, C., Vogl, T., and Glieder, A. (2018). Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J. Cell. Biochem. 119, 3183–3198. doi: 10.1002/jcb.26474
Weninger, A., Hatzl, A. M., Schmid, C., Vogl, T., and Glieder, A. (2016). Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol. 235, 139–149. doi: 10.1016/j.jbiotec.2016.03.027
Wiggins, C. A. R., and Munro, S. (1998). Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc. Natl. Acad. Sci. U. S. A. 95, 7945–7950. doi: 10.1073/pnas.95.14.7945
Wood, V., Gwilliam, R., Rajandream, M. A., Lyne, M., Lyne, R., Stewart, A., et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880. doi: 10.1038/nature724
Wriessnegger, T., Gübitz, G., Leitner, E., Ingolic, E., Cregg, J., de la Cruz, B. J., et al. (2007). Lipid composition of peroxisomes from the yeast Pichia pastoris grown on different carbon sources. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1771, 455–461. doi: 10.1016/j.bbalip.2007.01.004
Wriessnegger, T., Leitner, E., Belegratis, M. R., Ingolic, E., and Daum, G. (2009). Lipid analysis of mitochondrial membranes from the yeast Pichia pastoris. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1791, 166–172. doi: 10.1016/j.bbalip.2008.12.017
Wriessnegger, T., Moser, S., Emmerstorfer-Augustin, A., Leitner, E., Müller, M., Kaluzna, I., et al. (2015). Enhancing cytochrome P450-mediated conversions in P. pastoris through RAD52 over-expression and optimizing the cultivation conditions. Fungal Genet. Biol. 89, 114–125. doi: 10.1016/j.fgb.2016.02.004
Xu, T., Bharucha, N., and Kumar, A. (2011). Genome-wide transposon mutagenesis in Saccharomyces cerevisiae and Candida albicans. Methods Mol. Biol. 765, 207–224. doi: 10.1007/978-1-61779-197-0_13
Yamada, Y., Matsuda, M., Maeda, K., and Mikata, K. (1995). The phylogenetic relationships of methanol-assimilating yeasts based on the partial sequences of 18S and 26S ribosomal RNAs: the proposal of Komagataella Gen. Nov. (Saccharomycetaceae). Biosci. Biotechnol. Biochem. 59, 439–444. doi: 10.1271/bbb.59.439
Yang, S., Kuang, Y., Li, H., Liu, Y., Hui, X., Li, P., et al. (2013). Enhanced production of recombinant secretory proteins in Pichia pastoris by optimizing Kex2 P1’ site. PLoS One 8:e75347. doi: 10.1371/journal.pone.0075347
Yu, X. W., Wang, R., Zhang, M., Xu, Y., and Xiao, R. (2012). Enhanced thermostability of a Rhizopus chinensis lipase by in vivo recombination in Pichia pastoris. Microb. Cell Fact. 11:102. doi: 10.1186/1475-2859-11-102
Yuan, W., Veenhuis, M., and van der Klei, I. J. (2016). The birth of yeast peroxisomes. Biochim. Biophys. Acta Mol. Cell Res. 1863, 902–910. doi: 10.1016/j.bbamcr.2015.09.008
Yurimoto, H., and Sakai, Y. (2009). Methanol-inducible gene expression and heterologous protein production in the methylotrophic yeast Candida boidinii. Biotechnol. Appl. Biochem. 53, 85–92. doi: 10.1042/BA20090030
Yurimoto, H., Oku, M., and Sakai, Y. (2011). Yeast methylotrophy: metabolism, gene regulation and peroxisome homeostasis. Int. J. Microbiol. 2011:101298. doi: 10.1155/2011/101298
Zepeda, A. B., Pessoa, A., and Farías, J. G. (2018). Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast. Braz. J. Microbiol. 49, 119–127. doi: 10.1016/j.bjm.2018.03.010
Zhang, C., Ma, Y., Miao, H., Tang, X., Xu, B., Wu, Q., et al. (2020). Transcriptomic analysis of Pichia pastoris (Komagataella phaffii) GS115 during heterologous protein production using a high-cell-density fed-batch cultivation strategy. Front. Microbiol. 11:463. doi: 10.3389/fmicb.2020.00463
Zhu, J., Gong, R., Zhu, Q., He, Q., Xu, N., Xu, Y., et al. (2018). Genome-wide determination of gene essentiality by transposon insertion sequencing in yeast Pichia pastoris. Sci. Rep. 8:10223. doi: 10.1038/s41598-018-28217-z
Keywords: Pichia pastoris, Komagataella phaffii, methylotrophic yeast, yeast genetics, mating, CRISPR/Cas9, chromosome structure, pexophagy
Citation: Bernauer L, Radkohl A, Lehmayer LGK and Emmerstorfer-Augustin A (2021) Komagataella phaffii as Emerging Model Organism in Fundamental Research. Front. Microbiol. 11:607028. doi: 10.3389/fmicb.2020.607028
Received: 16 September 2020; Accepted: 14 December 2020;
Published: 11 January 2021.
Edited by:
Junbiao Dai, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Alexander D. Frey, Aalto University, FinlandSara Hanson, Colorado College, United States
Copyright © 2021 Bernauer, Radkohl, Lehmayer and Emmerstorfer-Augustin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anita Emmerstorfer-Augustin, ZW1tZXJzdG9yZmVyLWF1Z3VzdGluQHR1Z3Jhei5hdA==
†These authors have contributed equally to this work
 Lukas Bernauer1†
Lukas Bernauer1† Anita Emmerstorfer-Augustin
Anita Emmerstorfer-Augustin