
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 October 2020
Sec. Virology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.604618
To counteract host-encoded restriction systems, bacteriophages (phages) incorporate modified bases in their genomes. For example, phages carry in their genomes modified pyrimidines such as 5-hydroxymethyl-cytosine (5hmC) in T4gt deficient in α- and β-glycosyltransferases, glucosylated-5-hydroxymethylcytosine (5gmC) in T4, 5-methylcytosine (5mC) in Xp12, and 5-hydroxymethyldeoxyuridine (5hmdU) in SP8. In this work we sequenced phage Xp12 and SP8 genomes and examined Type II restriction of T4gt, T4, Xp12, and SP8 phage DNAs. T4gt, T4, and Xp12 genomes showed resistance to 81.9% (186 out of 227 enzymes tested), 94.3% (214 out of 227 enzymes tested), and 89.9% (196 out of 218 enzymes tested), respectively, commercially available Type II restriction endonucleases (REases). The SP8 genome, however, was resistant to only ∼8.3% of these enzymes (17 out of 204 enzymes tested). SP8 DNA could be further modified by adenine DNA methyltransferases (MTases) such as M.Dam and M.EcoGII as well as a number of cytosine DNA MTases, such as CpG methylase. The 5hmdU base in SP8 DNA was phosphorylated by treatment with a 5hmdU DNA kinase to achieve ∼20% phosphorylated 5hmdU, resulting resistance or partially resistant to more Type II restriction. This work provides a convenient reference for molecular biologists working with modified pyrimidines and using REases. The genomic sequences of phage Xp12 and SP8 lay the foundation for further studies on genetic pathways for 5mC and 5hmdU DNA base modifications and for comparative phage genomics.
Type II restriction and modification (R-M) systems encoded by bacteria and archaebacteria restrict foreign DNA from bacteriophages (phages) and mobile genetic elements (Smith, 1979). The companion methyltransferase (MTase) usually modifies the host genome rendering it refractory to the endonuclease and thereby preventing self-restriction (reviewed in Pingoud and Jeltsch, 2001; Pingoud et al., 2016). To counteract host restriction systems, many phages synthesize heavily modified (aka hypermodified) bases in their genomes by a combination of biosynthetic activities acting before and/or after DNA replication (Gold and Schweiger, 1969; Weigele and Raleigh, 2016; Lee et al., 2018). Additionally, some phage or prophage genomes encode multi-specificity C5 methyltransferases (Schumann et al., 1996; Xu et al., 2012), Dam methylase (Hattman et al., 1985), or frequent adenine MTase (Drozdz et al., 2012) to similarly protect phage genome from Type II restriction. Coliphage T4 synthesizes the hypermodified base, glucosylated 5-hydroxymethylcytosine completely replacing cytosine in its genomic DNA and as a result, can infect Escherichia coli (E. coli) strains encoding McrBC and McrA, which are modification-dependent restriction systems that attack 5hmC/5mC-containing DNA (Lehman and Pratt, 1960; Fleischman et al., 1976; Raleigh et al., 1989). Phage T4gt is a mutant deficient in DNA α- and β-glucosyltransferases and as a result contains 5-hydroxymethycytosines (5hmC) in its genome (Miller et al., 2003) and thus is restricted by the host McrBC and McrA systems. Phage Xp12 (natural host Xanthomonas oryzae) contains 5-methylcytosines replacing all cytosines in its genome (Kuo et al., 1968a,b). The Bacillus phage SP8, like the phages SPO1 and ϕe, contains 5-hydroxymethyl-2′-deoxyuridine (5hmdU) in its genome (Hoet et al., 1992).
It has been known for many years since the discovery of Type II R-M systems that these modified phage genomes are somewhat resistant to Type II restriction in vitro (Miller et al., 1985). But only limited information is available for a small number of restriction endonucleases (REases) on heavily modified phage genomes (Huang et al., 1982). The goal of this work is to test a vast array of commercially available REases (Roberts et al., 2015) on the four phage DNAs (T4gt, T4, Xp12, and SP8) and compile a reference list for molecular biologists who use REases to create recombinant DNA. As part of this goal, we sequenced phages Xp12 and SP8 genomes and deposited the sequences in GenBank. In addition, we confirmed the modified base compositions in these four phage genomes by LC-MS. We examined several adenine and cytosine methyltransferase activities on SP8 DNA. We also tested Type II restriction on SP8 DNA after its phosphorylation by treatment with 5hmdU DNA kinase. The results detailed herein comprise a comprehensive reference for the in vitro activity of Type II R-M enzymes on hypermodified DNA.
Bacteriophages Xp12 and SP8 were obtained from ATCC (#35934-B1 and #15563-B1, respectively). Bacteriophages T4 and T4gt were kindly provided by Dr. Elisabeth Raleigh (NEB). T4GT7 DNA was provided by Dr. Geoff Wilson (NEB). The bacteriophages supplying the genomic DNAs used in this study were cultured by infection of host cells at early log phase in liquid medium/broth cultured until a significant drop in optical density (OD) occurred indicating lysis of the majority of the cells in the culture. Phage particles were precipitated from centrifugated clarified lysates by addition of PEG8000 to 10% weight-by-volume (w/v) and 1 M NaCl to the phage lysates and collected by centrifugation. Phages were further purified by cesium chloride density gradient centrifugation and dialyzed against three changes of phage buffer (50 mM Tris–HCl, pH 7.5, 75 mM NaCl, 10 mM MgCl2). DNA was extracted from the phage by phenol-CHCl3 extraction, and ethanol precipitation (Sambrook et al., 1989).
REases, MTases, 5hmdU DNA kinase, and Proteinase K were provided by New England Biolabs, Inc., (NEB). NEBcutter v2.1 software (Vincze et al., 2003) was used to generate restriction patterns of phage DNA with the assumption of no base modification. We used excess of REases in restriction digestions (5 to 40 U to cleave 0.25 to 0.5 μg phage DNA) in 50 μl total volume incubated at the recommended temperature for 1 h (e.g., 5 μl of REases for low concentration enzyme supplied at 1,000 U/ml, 2 μl of REase for high concentration REase supplied at 20,000 U/ml). Four general restriction buffers (NEBuffer 1.1 (low salt), 2.1 (medium salt), 3.1 (high salt) and CutSmart buffer were used except those unique buffers recommended by the enzyme supplier. Digested DNAs were analyzed by agarose gel electrophoresis (0.8–1% gel). The DNA cleavage patterns were compared to NEBcutter-generated restriction patterns to determine digestion results as complete (C), partial (P), very partial (VP), or resistant (X) to digestions. To test phosphorylation of 5hmdU base in SP8 viral DNA, the DNA was first treated with 5hmdU DNA kinase for 2 h at 37°C in the presence of ATP (1 mM) and subsequently purified by spin column purification (NEB Monarch DNA clean up kit) before being subjected to nucleoside analysis (see below).
SP8 phage DNA was methylated by treatment with excess DNA MTase and methyl-donor SAM in the recommended buffer for 2 h. After heat inactivation of the MTase (65°C for 30 min), the methylated DNA was digested by cognate or non-cognate REases to evaluate the degree of resistance to restriction.
Modified or unmodified phage DNA was precipitated in ethanol, dried, and stored at −20°C. DNA samples (5 μg) were digested to nucleosides by treatment with the Nucleoside Digestion Mix (NEB, M0649S) overnight at 37°C. Nucleoside analysis was performed on an Agilent LC/MS System 1200 Series instrument equipped with a G1315D diode array detector and a 6120 Single Quadrupole Mass Detector operating in positive (+ESI) and negative (−ESI) electrospray ionization modes. LC was carried out on a Waters Atlantis T3 column (4.6 mm × 150 mm, 3 μm) with a gradient mobile phase consisting of 10 mM aqueous ammonium acetate (pH 4.5) and methanol. MS data acquisition was recorded in total ion chromatogram (TIC) mode. Each nucleoside was identified as follows: dC [M + H]+ 228.1 and [M − H]– 226.2; dG [M + H]+ 268.1 and [M − H]– 266.1; dT [M + H]+ 243.1 and [M − H]– 241.1; dA [M + H]+ 252.1 and [M − H]– 250.1; 5mdC [M + H]+ 242.1 and [M − H]– 240.2; 6mdA [M + H]+ 266.1 and [M − H]– 264.1; 5hmdC [M + H]+ 258.1 and [M − H]– 256.1; α- and β-5hmdC [M + H]+ 420.2 and [M − H]– 418.1. The relative abundance of each nucleoside was determined by dividing the UV absorbance by the corresponding extinction coefficient at 260 nm. To estimate the ratio of phosphorylation of 5-hydroxymethyluridine, kinase-treated SP8 gDNA was digested to nucleotides by treatment with Nuclease P1 (NEB, M0660S) overnight at 37°C. Each nucleotide was identified as follows: dCMP [M + H]+ 308.0 and [M − H]– 306.0; 5hmdUMP [M − H]– 337.0; dGMP [M + H]+ 348.0 and [M − H]– 346.0; dAMP [M + H]+ 332.0 and [M − H]– 330.0.
Samples of genomic DNA extracted from bacteriophages Xp12 and SP8 were sheared to ∼5 kb average fragment length using the Covaris gTube (Covaris Inc., Woburn, MA) according to manufacturer’s instructions. A total of 5 μg of sheared genomic DNA was used to prepare libraries for Pacific Biosciences (PacBio) Single Molecule Real-Time (SMRT) sequencing on the RSII model sequencer using P6-C4 chemistry and a flow-cell for each phage library. Following sequencing, reads were de novo assembled using the HGAP2 algorithm each yielding a single contig with average 200-fold coverage. Open reading frames and some gene assignments were performed by Rapid Annotation of Subsystems Technology (RAST) via web server1 (Aziz et al., 2008). The phage genome sequences for Xp12 and SP8 have been deposited in GenBank.
Phage DNAs were extracted from phage particles, purified by the CsCl gradient centrifugation and had their base composition analyzed by the LC-MS as described previously (Flodman et al., 2019). Table 1 shows the base composition of seven phage genomes. T4GT7, a mutant deficient in 5hmC synthesis (containing only canonical cytosines in its genome) (Snyder et al., 1976), and phage λ were used as controls for host-encoded M.Dam and M.Dcm transient methylation. Figure 1 shows the modified bases 5mC, 5hmC, 5gmC, and 5hmdU found in four phage genomes.
In recent years, stable levels 5hmC were discovered in human and mouse stems cells and brain cells. This base derives from an active DNA demethylation pathway involving the oxidation of 5mC by Ten-eleven-translocation (TET; 5-methylcytosine dioxygenase) enzyme: 5mC, 5hmC, 5fC (5-formylcytosine), 5caC (5-carboxycytosine) and subsequent removal by the thymine DNA glycosylase (TDG) repair enzyme (Tahiliani et al., 2009; He et al., 2011; Parker et al., 2019). Thus, there is a practical need in knowing how REases perform on 5mC or 5hmC modified DNA. We examined Type II restriction enzyme activity (enzymes commercially available from NEB) on four modified phage gDNAs. Phage DNA sensitivity or resistance to Type II restriction is summarized in Table 1, and all restriction data is presented in Supplementary Tables S1–S4. T4gt, T4, and Xp12 genome show 81.9, 94.3, and 89.9% resistance to all Type II REases, respectively (Table 1 and Supplementary Figure S1. See below). The SP8 genome (5hmdU), however, was resistant to only ∼8.3% of all Type II restrictions. The 5hmdU bases can be further modified as in phages ViI and ΦW-14 to provide higher resistance (Flodman et al., 2019). In addition, a simple phosphorylation step by treatment with 5hmdU DNA kinase can also increase DNA resistance to Type II restriction (see below).
The DNA of phage T4gt, a mutant T4 strain having diminished glucosyltransferase activity, contains 5hmC replacing all C in its genome. Due to the presence of 5hmC, the gDNA was previously reported by others to be resistant to some restriction digestions in vitro (NEB catalog 2019/20), but an extensive list of all commercially available REases is lacking. We purified phage T4gt DNA and analyzed its base composition. T4gt genomic DNA contained ∼83% of 5hmC, and 10% of α-5gmC and 7% of β-5gmC among all cytosine bases (Supplementary Figure S2A). The residual amount of 5gmC is probably a result of low activity of the mutated α- and β-glucosyltransferases. Null mutant in α- and β-glucosyltransferase genes would be required to completely substitute all 5gmC bases with 5hmC. A small amount of modified adenine 6mA (0.5%) was also detected in the T4gt DNA, which may originate from either the host Dam or phage-encoded Dam methyltransferase (AF158101). T4gt DNA can also be used as a substrate for modification-dependent endonucleases (see below). Despite the presence of residual 5gmC, we decided to use the T4gt DNA for testing Type II restriction. Examples of restriction digestion are shown in Figure 2. ApoI (RAATTY), AseI (ATTAAT), and a Type IIS enzyme BtsCI (GGATGN2↓) completely digested the DNA. The DNA is nearly resistant to digestion by PflFI (GACN3GTC), and completely resistant to digestion by NaeI (GCCGGC), PstI (CTGCAG), or ScaI (AGTACT). While BccI (CCATC), MlyI (GAGTC), and MboII (GAAGA) partially digested the substrate. Further restriction results are presented in Supplementary Table S1. Overall, phage T4gt was resistant to 81.9% of all Type II restriction enzymes tested here (186 out of 227 enzymes).
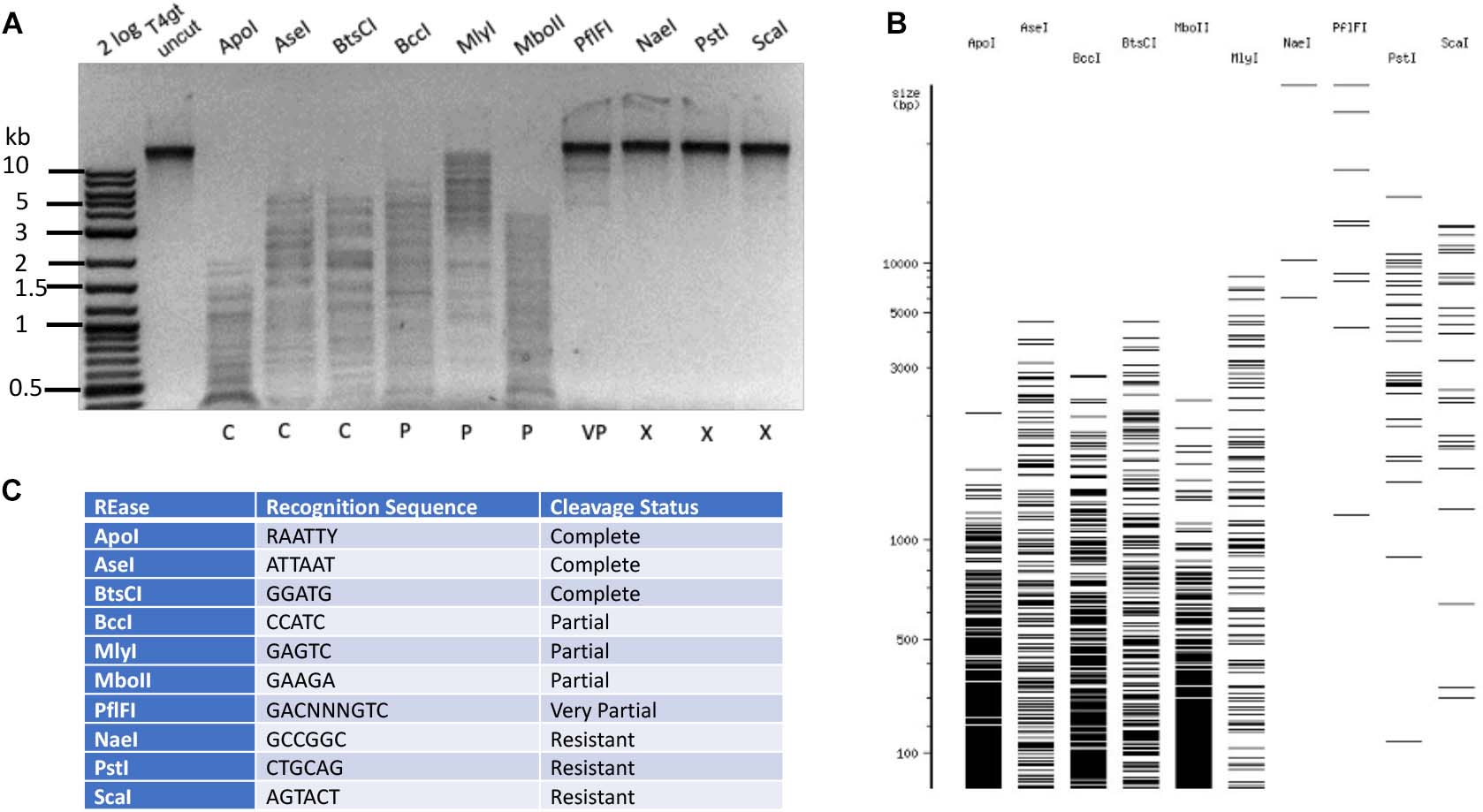
Figure 2. Type II restriction of phage T4gt DNA. Phage T4gt gDNA was digested by 10 REases and analyzed by agarose gel electrophoresis (A). 2 log, DNA size marker in 100 bp to 10 kb (NEB). The predicted digestion patterns by NEBcutter are shown in (B). The target recognition sequence and digestion results are summarized in (C). C = complete digestion, P = partial digestion, VP = very partial digestion (only a few weak bands), X = resistant to restriction.
The base composition of phage T4 DNA was confirmed by LC-MS analysis to carry 60% α-5gmC and 40% β-5gmC (Supplementary Figure S2B). Based on cleavage patterns obtained for phage T4gt DNA, it is reasonable to assume that negative restriction on T4gt would be mirrored on T4 DNA. To confirm this hypothesis, we tested twenty enzymes and found that all of them were negative on both T4gt and T4 (data not shown). Next, we selected and tested a subset of REases that could either completely or partially cleave T4gt. Examples of Type II restriction of phage T4 DNA are shown in Figure 3. MluCI (AATT), MseI (TTAA), and NdeI (CATATG) digested T4 DNA completely. But AseI (ATTAAT), SspI (AATATT), and SwaI (ATTTAAAT) did not show complete digestion, even though they all recognize sites containing A/T sequence and they do not overlap with dam methylation site. This partial digestion was reproducible (data not shown) and suggested an indirect inhibitory effect of 5gmC modifications on REases having only A/T nucleotides in their targets. T4 DNA was nearly resistant to HpyCH4III (ACNGT) restriction, and completely resistant to restriction by MboII (GAAGA), NsiI (ATGCAT), or SalI (GTCGAC), all of which containing 2–4 modified cytosines in their target sites. Further Type II restriction data are presented in Supplementary Table S2. Phage T4 DNA was resistant to 94.3% of all Type II restrictions examined here (214 out of 227 enzymes), indicating the highly resistant nature of 5gmC modified DNA. In the arms race between phage and host, bacteria had developed modification-dependent restriction systems to restrict such hypermodified genomes (see below).
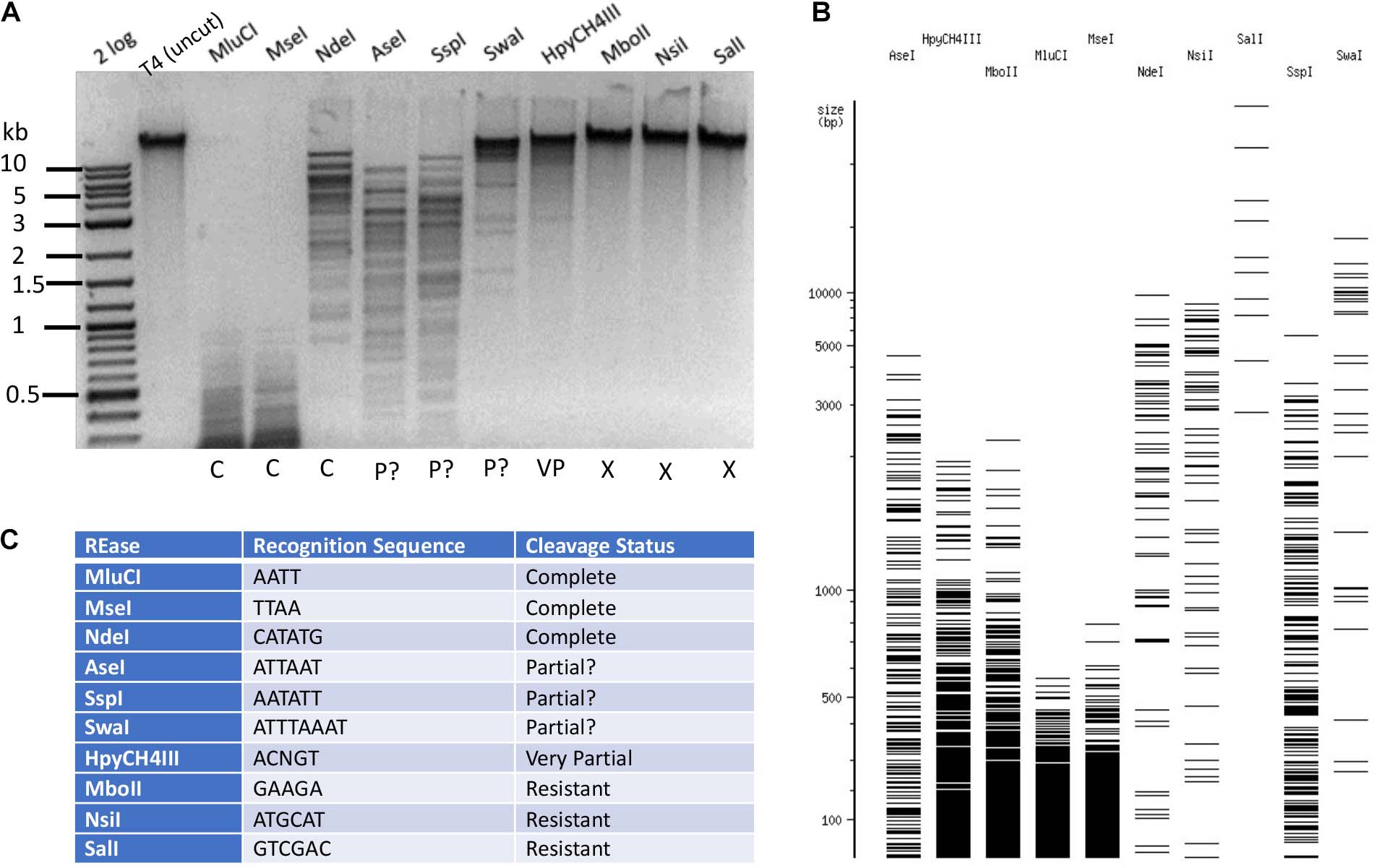
Figure 3. Type II restriction of phage T4 DNA. Phage T4 DNA was digested by 10 REases and analyzed by agarose gel electrophoresis (A). The predicted digestion patterns in the absence of modifications by NEBcutter are shown in (B). The recognition sequence and results are shown in (C). The question marks in “P?” indicate possible partial digestions (the presence of a few large DNA bands not seen in NEBcutter patterns).
Hydroxymethylation-deficient T4 GT7 and λ DNA were used as controls. Only low levels of modified bases 5mC (0.3%) and 6mA (0.5%) were found in phage T4 GT7 (Supplementary Figure S3). The 5mC and 6mA bases are most likely transiently modified by the bacterial host enzymes M.Dcm and M.Dam. Similarly, low levels of modified bases 5mC and 6mA were found in Dcm+ Dam+ phage λ DNA (Supplementary Figure S4A). As could be expected, Dam– phage λ DNA displayed a much lower level of 6mA (<0.05%) (Supplementary Figure S4B). Type II restriction data of phage T4 GT7 DNA are presented in Supplementary Table S4 (spread sheet in the Supplementary).
The phage Xp12 genomic DNA was sequenced using PacBio sequencing kit. The viral genome contains 63,783 bp (GenBank accession number MT664984). The detailed analysis of the Xp12 genome and 5mC modification pathway will be published elsewhere (PW). Examples of Type II RE cleavage of phage Xp12 DNA (5mC) are shown in Figure 4. The DNA was resistant to restrictions by AfeI (AGCGCT), ApaI (GGGCCC), and ApaLI (GTGCAC). It was partially resistant to Type IIS enzymes MlyI (GAGTC) and MseI (TTAA) (a 3 kb MseI fragment was missing). AhdI, BstNI, KpnI, MboII, and TspA15I completely digested Xp12 DNA, even though their target sites contain 2–5 modified cytosines. It is known that M. AhdI, M. KpnI, and M1. MboII (GGAGG) are N6-adenine MTases and M2. MboII (TCTTC) is an N4-cytosine MTase (Roberts et al., 2015). The non-cognate C5 methylations did not have inhibitory effect on these REases. Further Type II restriction data are presented in Supplementary Table S3. Phage Xp12 DNA was nearly resistant to 90% of all Type II restrictions tested here (196 out of 218 enzymes).
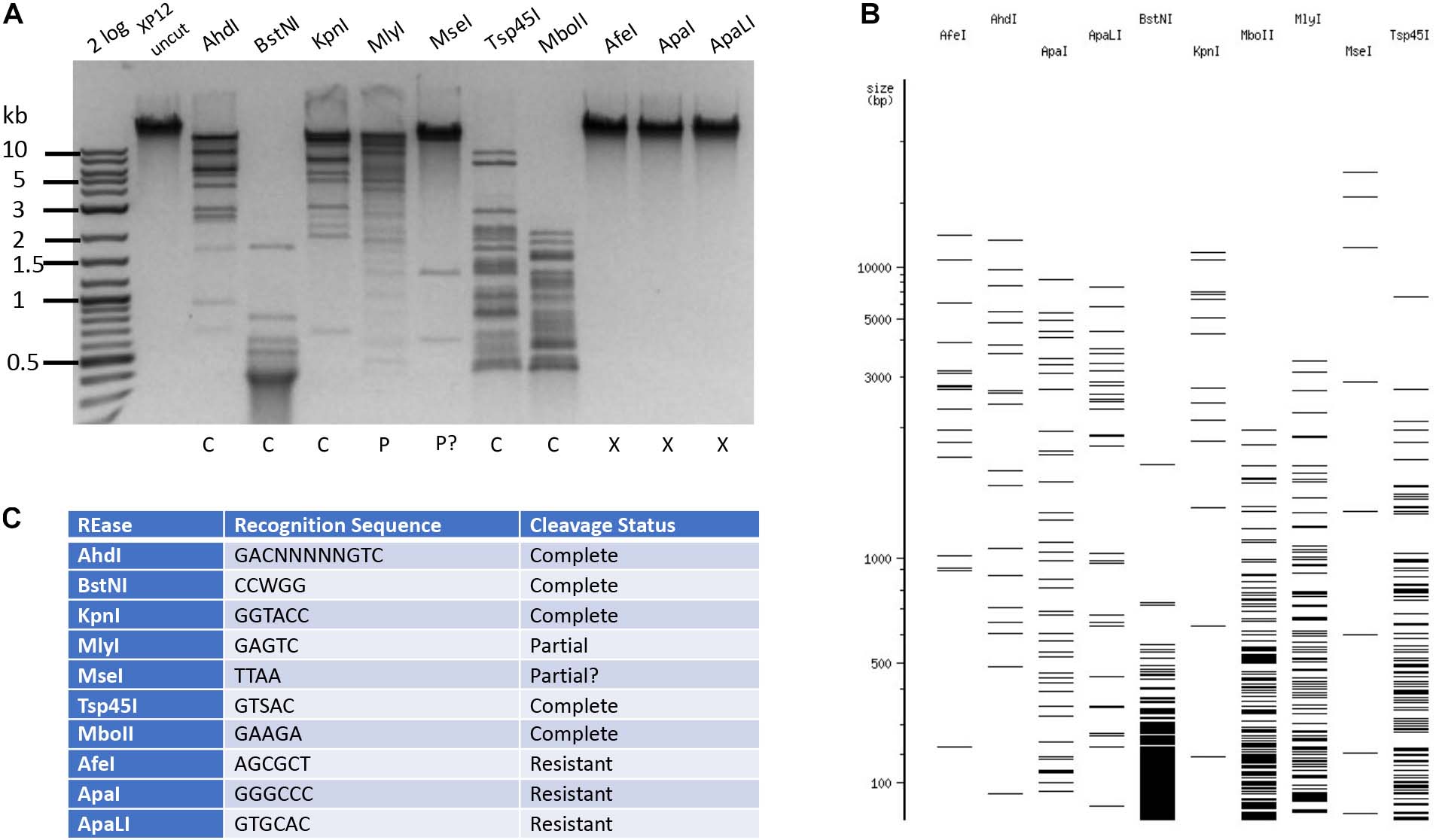
Figure 4. Restriction of phage Xp12 DNA. Example of 10 restriction digestions analyzed in 1% agarose gel (A). NEBcutter predicted cleavage patterns in the absence of cytosine modification (B). Enzyme recognition sequences and cleavage result (C).
The phage SP8 genomic DNA was sequenced by PacBio sequencing platform. The viral genome contains 138,741 bp (GenBank accession number MW001214). The Bacillus phage SP8 contains 100% 5hmdU replacing all T in its genome (Hoet et al., 1992). LC-MS analysis of SP8 DNA confirmed its predicted base composition (Table 1 and Supplementary Figure S5). In the time since the publication of a study examining cleavage of 5hmdU DNA by a small set of REases (Huang et al., 1982), many more Type II REases have become commercially available. We decided to test all currently available Type II REases on SP8 DNA. Examples of Type II restriction of phage SP8 DNA are shown in Figure 5. Phage SP8 DNA was partially resistant to restriction by ApaLI (GTGCAC) and AseI (ATTAAT), and completely resistant to restriction by BsmI (GAATGC with three 5hmdU), BspMI (ACCTGC), and EcoRI (GAATTC). Five REases (AccI, AciI, AcuI, AlwI, and EcoRV) completely digested the substrate DNA. While both EcoRI and EcoRV sites contain four 5hmdU replacing T in SP8 DNA, its sensitivity to restriction was completely different: SP8 DNA was resistant to EcoRI digestion (24 EcoRI sites in the genome) but sensitive to EcoRV restriction. The genomic DNA can be digested by MluCI (AATT). Therefore, it is difficult to predict which REase will completely digest SP8 DNA, even with the presence of up to six 5hmdUs in their target recognition sequences. The SP8 genome was resistant to only a small fraction (∼8.3%) of all Type II restrictions tested here (17 out of 204 enzymes, Supplementary Table S5). Type II restrictions of four phage genomes are summarized in Table 2.
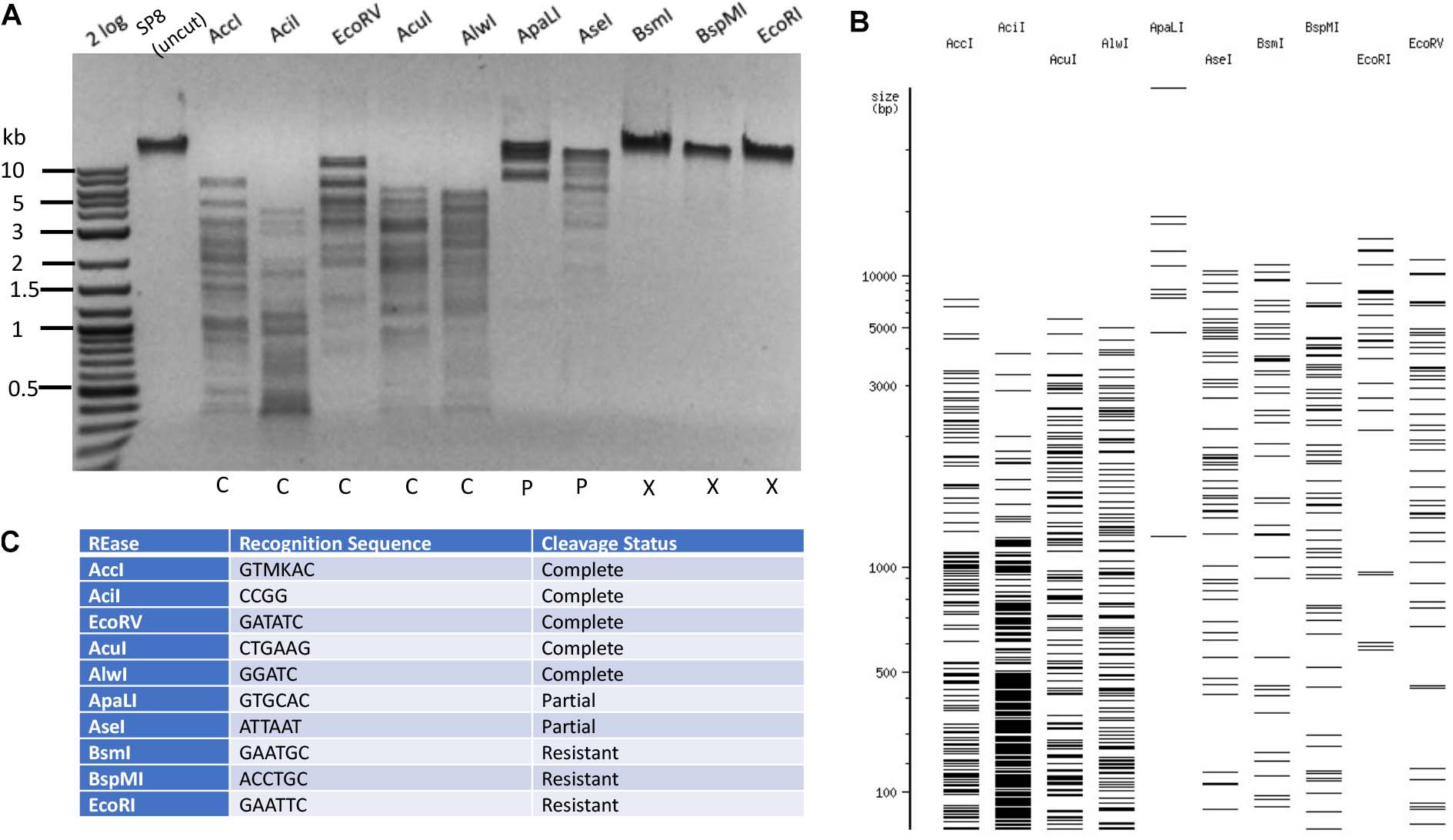
Figure 5. Restriction of phage SP8 (5hmdU) DNA. Example of 10 restriction digestions analyzed in 1% agarose gel (A). NEBcutter predicted cleavage patterns in the absence of T modification (B). Enzyme recognition sequences and restriction result (C).
Next, we tested a number of DNA methyltransferases on phage SP8 gDNA to see whether 5hmdU could potentially interfere with adenine methylation. After DNA methylation, the modified DNA was subjected to either cognate or non-cognate restriction enzymes. SP8 DNA was partially modified by M.Dam (GATC) or M.EcoGII (frequent adenine methyltransferase). The modified DNA became partially resistant to restriction by MboI (GATC, restriction blocked by 6mA modification). SP8 DNA, however, was a poor substrate for M. TaqI (Supplementary Figure S5). In a control experiment, M. TaqI was able to modify λ DNA and rendered the DNA resistant to TaqI restriction (data not shown). M. EcoRI was able to modify λ DNA and protect it from EcoRI restriction. However, in the case of SP8 DNA, no conclusion was derived from the results because the substrate DNA itself was already resistant to EcoRI even before methylation (24 EcoRI sites present in the genome). We have not analyzed other adenine methyltransferase activities on phage SP8 DNA due to the lack of enzyme availability. Overall, phage SP8 DNA can be efficiently methylated by C5 MTases (M. HaeIII, M. MspI, M. HhaI, M. AluI, M. HpaII, CpG, and GpC methyltransferases) as one expected. These MTases could modify phage SP8 efficiently and render the DNA resistant to both cognate restriction and overlapping restriction (Supplementary Figure S6). The methylation results are summarized in Table 3.
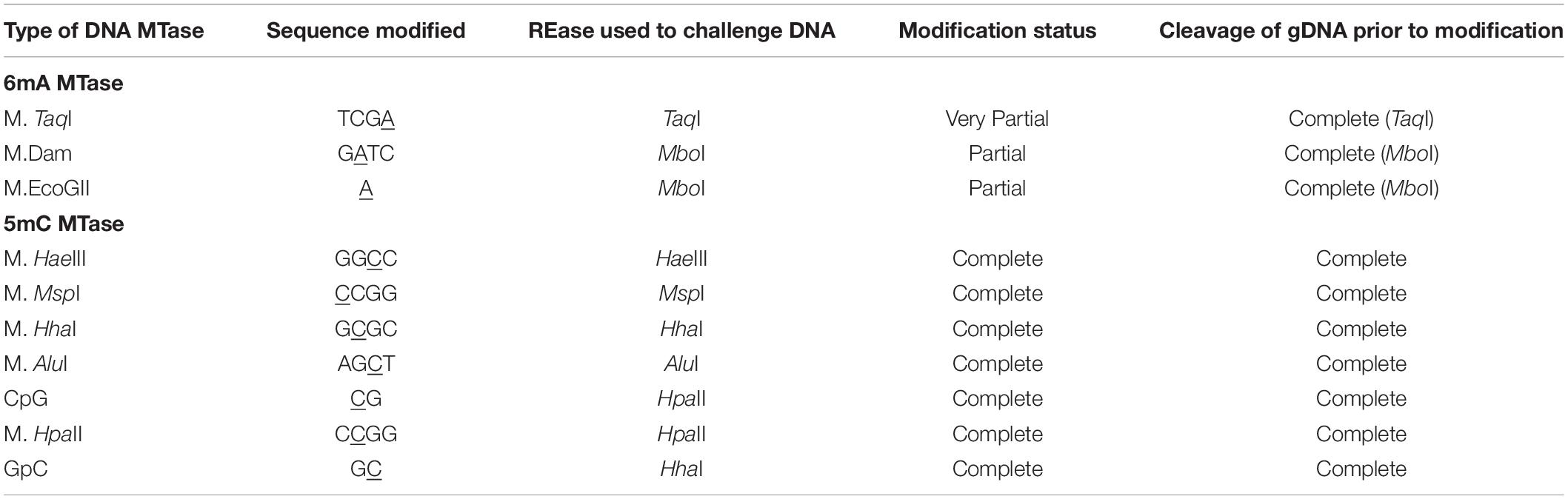
Table 3. DNA methylation by adenine or cytosine MTases followed by restriction challenge of the methylated phage SP8 DNA.
MDREs (Type IIM and Type IV) cleave modified DNA specifically (Roberts et al., 2003). These enzymes are thought to be evolved in bacterial hosts to attack modified phage genomes in the host-virus arms race (Bair and Black, 2007). We examined a few commercially available enzymes for activity on modified phage DNA. T4gt, T4, Xp12, and T4GT7 were digested with AbaSI, FspEI, LpnPI, MspJI, and McrBC. Table 4 summarizes the restriction results. All five enzymes digested T4gt DNA efficiently. Only AbaSI (a PvuRts1I homolog) was able to digest T4 and T4gt DNA (Wang et al., 2011). T4 DNA was resistant to restriction by FspEI, LpnPI, MspJI, and McrBC. The T4GT7 DNA was partially digested by FspEI and MspJI, probably due to partial M.Dcm methylation of the genomic DNA. AbaSI and McrBC were incapable of cleaving T4GT7 DNA. As expected, AbaSI was able to cleave 5hmC and 5gmC modified DNA, but not 5mC or unmodified DNA. FspEI, MspJI, and McrBC were able to cleave 5mC and 5hmC modified DNA, but not on 5gmC and unmodified DNA. It has been reported that GmrSD endonuclease is able to cleave both T4gt and T4 DNA in ATP/GTP dependent manner (He et al., 2015).
5hmdU DNA kinase can phosphorylate the 5-hydroxymethyl group in the 5hmdU in a sequence-specific manner (Lee et al., 2018) making the base more negatively charged. The 5hmdU DNA kinase has been shown to block NcoI restriction after the kinase reaction2. We used the 5hmdU DNA kinase to phosphorylate phage SP8 DNA. After the kinase treatment, the DNA was purified by spin column and its base composition was analyzed by LC-MS. Supplementary Figure S7 shows that ∼20% of 5hmdU had been converted to its phosphorylated form in SP8 DNA.
Next, we set out to test more Type II restriction after phosphorylation. Phosphorylated phage SP8 DNA was tested with 20 restriction enzymes. Phosphorylated SP8 DNA was resistant to restriction by NcoI, AlwNI, NdeI, BbvCI, BccI, MslI, NlaIII, PvuII, PmlI, NspI, and NsiI, and partially resistant to restriction by EcoRV, XmnI, MlyI, MboI, and Hpy188I (Figure 6). Inspection of the resistant and partially resistant sites indicated that these sites contain TG (or NG) and TC dinucleotides in their recognition sequences, confirming previous findings with phage M6, ViI, and ϕW-14 DNAs (Flodman et al., 2019). This type of base modification (phosphorylation) is a useful method to render the site resistant to Type II restriction when a DNA MTase is not yet available to modify the same sequence.
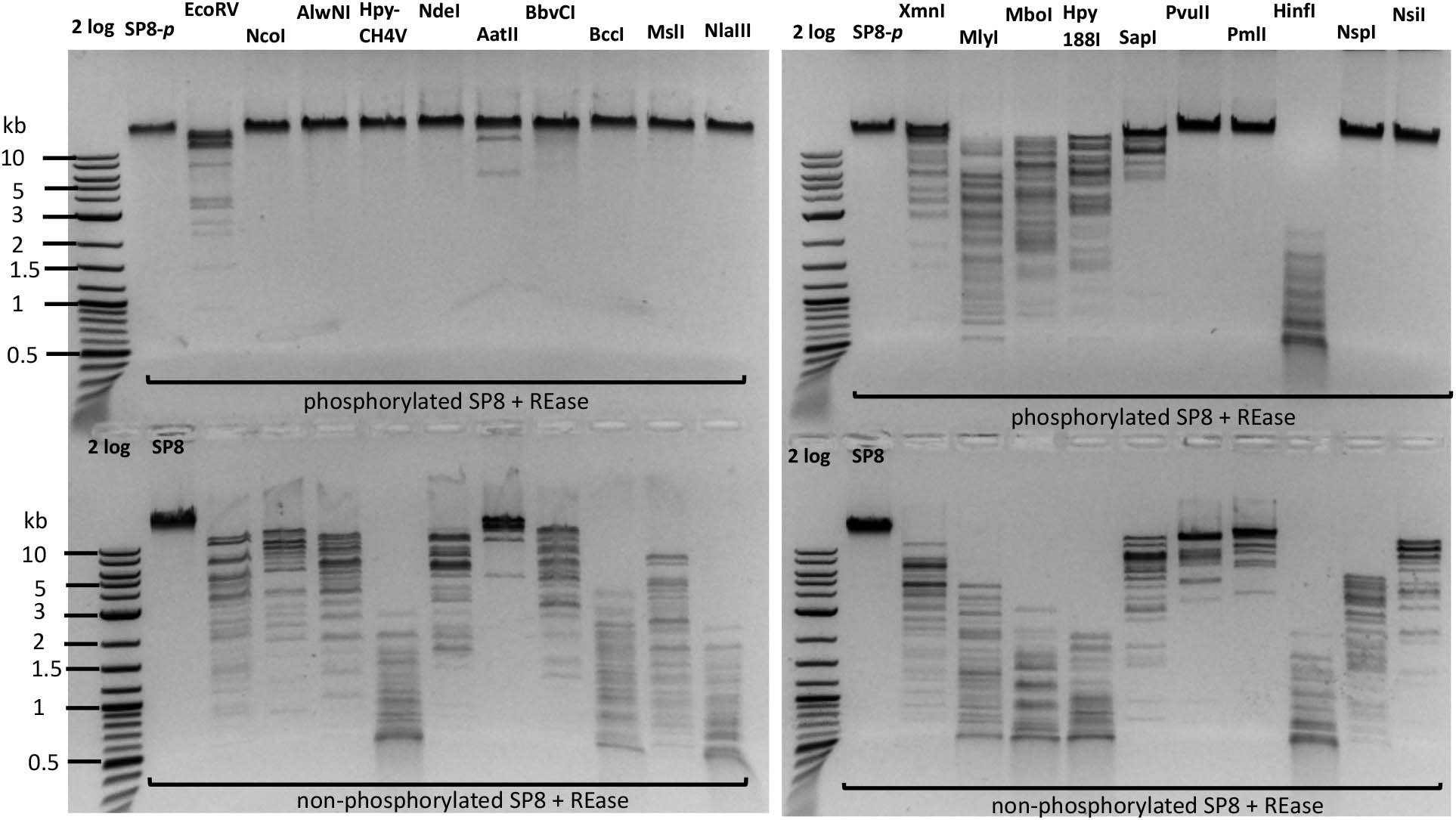
Figure 6. Restriction digestion of SP8 gDNA following treatment with 5hmdU DNA kinase. Top panel, restriction of phosphorylated SP8 DNA (SP8-p). Bottom panel, control: restriction of non-p phage SP8 DNA. The phosphorylated DNA appeared largely resistant to restrictions by NcoI (CCATGG), AlwNI (CAGN3CTG), HpyCH4V (ACGTn), NdeI (CATATG), BbvCI (CCTCAGC), BccI (CTATC), MslI (CAYN4RTG), NlaIII (CATG), PvuII (CAGCTG), PmlI (CACGTG), NspI (RCATGY), and NsiI (ATGCATn) digestion; and partially resistant to EcoRV (nGATATCn), MlyI (nGAGTCn), MboI (nGATCn), Hpy188I (TCNGA), SapI (nGCTCTTC) digestion. Phosphorylation of 5hmdU had no inhibitory effect on AatII (nGACGTC) and HinfI (nGANTC) digestions. The TG, TC, nG, and Tn dinucleotides in the restriction sites are underlined.
In this work, we analyzed Type II restriction of modified phage genomes T4gt (5hmC), T4 (5gmC), Xp12 (5mC), and SP8 (5hmdU). We found T4gt, T4, and Xp12 genomes are highly resistant to Type II restriction; while SP8 DNA is only modestly resistant to Type II restriction.
A few REases that recognize only A/T target sites partially digested phage T4 and Xp12 DNA. It is not clear whether this observation is an artifact or true indirect effect of base modification on neighboring sequences. In any event, practitioners using REases to digest hypermodified phage genomes should be aware of this partial inhibition.
The MDREs (AbaSI, FspEI, LpnPI, MspJI, and McrBC) tested here digested phage T4gt DNA efficiently. No undigested phage DNA was detected. It is important to note that LC-MS analysis of T4gt DNA indicated that only 83% of cytosines are modified as 5hmC, with the remaining cytosines found in the form of 5gmC. We have observed a slight variation in phage T4gt base composition (83 to 90% of 5hmC vs. 10 to 17% of 5gmC) across independent batches of phage lysates. The reason for this variation is unknown, but it may be related to suppressor efficiency of different E. coli strains growing T4gt α- and β- glucosyltransferase amber mutants.
Phage T4 DNA shows highest resistance to bacteria-encoded Type II restriction. However, bacteria also evolved new restriction systems that can target 5gmC-modified DNA. Examples of these restriction systems include PvuRts1I-like enzymes (Janosi et al., 1994), GmrSD-like enzymes (He et al., 2015), and EVE-HNH endonucleases (Lutz et al., 2019). Many more Type IV restriction systems have been found to restrict 5mC and 5hmC modified DNA (Loenen and Raleigh, 2014).
We demonstrated here that M.Dam and M.EcoGII could efficiently modify phage SP8 DNA. However, M. TaqI modified the viral DNA poorly. The cytosine MTases tested here mostly recognize G/C sequence (except M. AluI) and they methylated SP8 DNA efficiently, rendering it resistant to cognate restriction or overlapping restriction. In addition, we tested Type II restriction of phosphorylated (p) SP8 DNA following treatment with 5hmdU DNA kinase. The kinase-treated DNA became more resistant to Type II restriction only if the targets sites contained TG (NG) or TC dinucleotides. A few enzymes (AatII and HinfI) were not affected by base phosphorylation. 5hmdU-containing plasmid or phage DNA could be potentially grown in an engineered E. coli strain to achieve ∼75% maximum incorporation (Mehta et al., 2016). Alternatively, PCR amplification could be used to incorporate 5hmdU using 5hmdUTP in the dNTP pool, depending on the efficiency of 5hmdUTP incorporation by a PCR DNA polymerase. We anticipate that 5hmdU DNA kinase will be a useful tool to manipulate DNA, making it resistant to certain restriction digestions, especially where DNA MTases are not commercially available.
In addition to DNA methylation and 5hmdU phosphorylation, researchers also utilize dUMP derivatives for photocaging which can be reversed by UV and light treatment. For example, 5-[(2-Nitrobenzyl)oxymethyl]-2′-deoxyuridine 5′-O-triphosphate can be incorporated into DNA in PCR reaction and such photocaged DNA has been shown to be resistant to cleavage by AflII (CTTAAG), KpnI (GGTACC), PvuII (CAGCTG), and RsaI (GTAC) endonucleases. Deprotection of the photocaged DNA by UV light treatment (355–425 nm) converted it to 5hmdU-containing DNA which could be cleaved by the REases (Vanikova and Hocek, 2014; Bohacova et al., 2018a). Our work confirmed that phage SP8 DNA with natural 5hmdU modified bases can be cleaved by the four REases mentioned above. It has been shown that 5hmdU phosphorylation of PCR DNA also negatively impacted in vitro transcription efficiency by E. coli RNA polymerase and restriction by AluI (AGCT) and RsaI (GTAC) (Vanikova et al., 2019). AluI site contains TN dinucleotides in both strands. But RsaI site does not contain TG or TC dinucleotides. How the 5hmdU DNA kinase is able to phosphorylate the RsaI site is not clear. Similarly, photocaged dCTP derivatives (dCNBTP and dCNPTP, NB, 2-nitrobenzyl; NP, 2-nitropiperonyl) can be incorporated into PCR DNA, which becomes resistant to RsaI restriction. Photochemical release of the protected bases by UV or visible light treatment converted it to 5hmC-containing DNA, which is sensitive to RsaI restriction (Bohacova et al., 2018b). We found that RsaI partially digested T4gt DNA. The variation of RsaI digestion results may result from the high density of 5hmC modification in the phage DNA vs. the synthetic PCR DNA described by others. The photocaged nucleotides provided a convenient and reversible way to control enzyme activities on modified DNA in vitro.
Phage Xp12 and SP8 genome sequences have been deposited in GenBank and assigned the accession numbers MT664984 and MW001214.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT664984 and MW001214.
KF, S-yX, IC, PW, and ND generated and analyzed experimental data. S-yX, KF, IC, and PW wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by New England Biolabs, Inc. KF was supported by NEB summer internship program.
S-yX, IC, ND, and PW are employed by New England Biolabs, Inc.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Andy Gardner, Tom Evans, and Richard Roberts for support and advice. We are grateful to Y. J. Lee for his kind assistance with this project. We thank the reviewers and editor for suggestions to improve the manuscript. The open access publication fee is provided by New England Biolabs, Inc.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.604618/full#supplementary-material
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bair, C. L., and Black, L. W. (2007). A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J. Mol. Biol. 366, 768–778. doi: 10.1016/j.jmb.2006.11.051
Bohacova, S., Ludvikova, L., Postova Slavetinska, L., Vanikova, Z., Klan, P., and Hocek, M. (2018a). Protected 5-(hydroxymethyl)uracil nucleotides bearing visible-light photocleavable groups as building blocks for polymerase synthesis of photocaged DNA. Org. Biomol. Chem. 16, 1527–1535. doi: 10.1039/C8OB00160J
Bohacova, S., Vanikova, Z., Postova Slavetinska, L., and Hocek, M. (2018b). Protected 2′-deoxyribonucleoside triphosphate building blocks for the photocaging of epigenetic 5-(hydroxymethyl)cytosine in DNA. Org. Biomol. Chem. 16, 5427–5432. doi: 10.1039/C8OB01106K
Drozdz, M., Piekarowicz, A., Bujnicki, J. M., and Radlinska, M. (2012). Novel non-specific DNA adenine methyltransferases. Nucleic Acids Res. 40, 2119–2130. doi: 10.1093/nar/gkr1039
Fleischman, R. A., Cambell, J. L., and Richardson, C. C. (1976). Modification and restriction of T-even bacteriophages. In vitro degradation of deoxyribonucleic acid containing 5-hydroxymethylctosine. J. Biol. Chem. 251, 1561–1570.
Flodman, K., Tsai, R., Xu, M. Y., Correa, I. R. Jr., Copelas, A., Lee, Y. J., et al. (2019). Type II restriction of bacteriophage DNA With 5hmdU-derived base modifications. Front. Microbiol. 10:584. doi: 10.3389/fmicb.2019.00584
Gold, L. M., and Schweiger, M. (1969). Synthesis of phage-specific alpha- and beta-glucosyl transferases directed by T-even DNA in vitro. Proc. Natl. Acad. Sci. U.S.A. 62, 892–898. doi: 10.1073/pnas.62.3.892
Hattman, S., Wilkinson, J., Swinton, D., Schlagman, S., MacDonald, P. M., and Mosig, G. (1985). Common evolutionary origin of the Phage T4 dam and host Escherichia Coli dam DNA-adenine methyltransferase genes. J. Bacteriol. 164, 932–937. doi: 10.1128/JB.164.2.932-937.1985
He, X., Hull, V., Thomas, J. A., Fu, X., Gidwani, S., Gupta, Y. K., et al. (2015). Expression and purification of a single-chain Type IV restriction enzyme Eco94GmrSD and determination of its substrate preference. Sci. Rep. 5:9747. doi: 10.1038/srep09747
He, Y. F., Li, B. Z., Li, Z., Liu, P., Wang, Y., Tang, Q., et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. doi: 10.1126/science.1210944
Hoet, P. P., Coene, M. M., and Cocito, C. G. (1992). Replication cycle of Bacillus subtilis hydroxymethyluracil-containing phages. Annu. Rev. Microbiol. 46, 95–116. doi: 10.1146/annurev.mi.46.100192.000523
Huang, L. H., Farnet, C. M., Ehrlich, K. C., and Ehrlich, M. (1982). Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 10, 1579–1591. doi: 10.1093/nar/10.5.1579
Janosi, L., Yonemitsu, H., Hong, H., and Kaji, A. (1994). Molecular cloning and expression of a novel hydroxymethylcytosine-specific restriction enzyme (PvuRts1I) modulated by glucosylation of DNA. J. Mol. Biol. 242, 45–61. doi: 10.1006/jmbi.1994.1556
Kuo, T. T., Huang, T. C., and Teng, M. H. (1968a). 5-Methylcytosine replacing cytosine in the deoxyribonucleic acid of a bacteriophage for Xanthomonas Oryzae. J. Mol. Biol. 34, 373–375. doi: 10.1016/0022-2836(68)90263-5
Kuo, T. T., Huang, T. C., Wu, R. Y., and Chen, C. P. (1968b). Phage Xp12 of Xanthomonas oryzae (Uyeda et Ishiyama) Dowson. Can. J. Microbiol. 14, 1139–1142. doi: 10.1139/m68-190
Lee, Y. J., Dai, N., Walsh, S. E., Muller, S., Fraser, M. E., Kauffman, K. M., et al. (2018). Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc. Natl. Acad. Sci. U.S.A. 115, E3116–E3125. doi: 10.1073/pnas.1714812115
Lehman, I. R., and Pratt, E. A. (1960). On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J. Biol. Chem. 235, 3254–3259.
Loenen, W. A., and Raleigh, E. A. (2014). The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 42, 56–69. doi: 10.1093/nar/gkt747
Lutz, T., Flodman, K., Copelas, A., Czapinska, H., Mabuchi, M., Fomenkov, A., et al. (2019). A protein architecture guided screen for modification dependent restriction endonucleases. Nucleic Acids Res. 47, 9761–9776. doi: 10.1093/nar/gkz755
Mehta, A. P., Li, H., Reed, S. A., Supekova, L., Javahishvili, T., and Schultz, P. G. (2016). Replacement of thymidine by a modified base in the Escherichia Coli genome. J. Am. Chem. Soc. 138, 7272–7275. doi: 10.1021/jacs.6b03904
Miller, E. S., Kutter, E., Mosig, G., Arisaka, F., Kunisawa, T., and Ruger, W. (2003). Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67, 86–156. doi: 10.1128/MMBR.67.1.86-156.2003
Miller, P. B., Wakarchuk, W. W., and Warren, R. A. J. (1985). Alpha-putrescinylthymine and the sensitivity of bacteriophage Phi W-14 DNA to restriction endonucleases. Nucleic Acids Res. 13, 2559–2568. doi: 10.1093/nar/13.7.2559
Parker, M. J., Weigele, P. R., and Saleh, L. (2019). Insights into the biochemistry, evolution, and biotechnological applications of the ten-eleven translocation (TET) enzymes. Biochemistry 58, 450–467. doi: 10.1021/acs.biochem.8b01185
Pingoud, A., and Jeltsch, A. (2001). Structure and function of type II restriction endonucleases. Nucleic Acids Res. 29, 3705–3727. doi: 10.1093/nar/29.18.3705
Pingoud, A., Wilson, G. G., and Wende, W. (2016). Type II restriction endonucleases - a historical perspective and more. Nucleic Acids Res. 44:8011. doi: 10.1093/nar/gkw513
Raleigh, E. A., Trimarchi, R., and Revel, H. (1989). Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia Coli K-12. Genetics 122, 279–296.
Roberts, R. J., Belfort, M., Bestor, T., Bhagwat, A. S., Bickle, T. A., Bitinaite, J., et al. (2003). A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31, 1805–1812. doi: 10.1093/nar/gkg274
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2015). REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299. doi: 10.1093/nar/gku1046
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning, A Laboratory Manual, 2 Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Schumann, J., Walter, J., Willert, J., Wild, C., Koch, D., and Trautner, T. A. (1996). M.Bsshii, A Multispecific Cytosine-C5-DNA-methyltransferase with unusual target recognizing properties. J. Mol. Biol. 257, 949–959. doi: 10.1006/jmbi.1996.0214
Smith, H. O. (1979). Nucleotide sequence specificity of restriction endonucleases. Science 205, 455–462. doi: 10.1126/science.377492
Snyder, L., Gold, L., and Kutter, E. (1976). A gene of bacteriophage T4 whose product prevents true late transcription on cytosine-containing T4 DNA. Proc. Natl. Acad. Sci. U.S.A. 73, 3098–3102. doi: 10.1073/pnas.73.9.3098
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Vanikova, Z., and Hocek, M. (2014). Polymerase synthesis of photocaged DNA resistant against cleavage by restriction endonucleases. Angew. Chem. Int. Ed. Engl. 53, 6734–6737. doi: 10.1002/anie.201402370
Vanikova, Z., Janouskova, M., Kambova, M., Krasny, L., and Hocek, M. (2019). Switching transcription with bacterial RNA polymerase through photocaging, photorelease and phosphorylation reactions in the major groove of DNA. Chem. Sci. 10, 3937–3942. doi: 10.1039/C9SC00205G
Vincze, T., Posfai, J., and Roberts, R. J. (2003). NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 31, 3688–3691. doi: 10.1093/nar/gkg526
Wang, H., Guan, S., Quimby, A., Cohen-Karni, D., Pradhan, S., Wilson, G., et al. (2011). Comparative characterization of the PvuRts1I family of restriction enzymes and their application in mapping genomic 5-hydroxymethylcytosine. Nucleic Acids Res. 39, 9294–9305. doi: 10.1093/nar/gkr607
Weigele, P., and Raleigh, E. A. (2016). Biosynthesis and function of modified bases in bacteria and their viruses. Chem. Rev. 116, 12655–12687. doi: 10.1021/acs.chemrev.6b00114
Keywords: Type II restriction, modified phage genome, phage SP8, phage Xp12 genome sequence, 5hmdU DNA kinase
Citation: Flodman K, Corrêa IR Jr, Dai N, Weigele P and Xu S-y (2020) In vitro Type II Restriction of Bacteriophage DNA With Modified Pyrimidines. Front. Microbiol. 11:604618. doi: 10.3389/fmicb.2020.604618
Received: 10 September 2020; Accepted: 05 October 2020;
Published: 27 October 2020.
Edited by:
Robert Czajkowski, University of Gdańsk, PolandReviewed by:
Michal Hocek, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic (ASCR), CzechiaCopyright © 2020 Flodman, Corrêa, Dai, Weigele and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuang-yong Xu, eHVzQG5lYi5jb20=; Peter Weigele, V2VpZ2VsZUBuZWIuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.