
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 26 November 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.604458
This article is part of the Research TopicRNA Biology of MicroorganismsView all 12 articles
6S RNA is a kind of high-abundance non-coding RNA that globally regulates bacterial transcription by interacting with RNA polymerase holoenzyme. Through bioinformatics analysis, we found that there are two tandem 6S RNA-encoding genes in the genomes of Bacillus cereus group bacteria. Using Bacillus thuringiensis BMB171 as the starting strain, we have explored the physiological functions of 6S RNAs, and found that the genes ssrSA and ssrSB encoding 6S-1 and 6S-2 RNAs were located in the same operon and are co-transcribed as a precursor that might be processed by specific ribonucleases to form mature 6S-1 and 6S-2 RNAs. We also constructed two single-gene deletion mutant strains ΔssrSA and ΔssrSB and a double-gene deletion mutant strain ΔssrSAB by means of the markerless gene knockout method. Our data show that deletion of 6S-1 RNA inhibited the growth of B. thuringiensis in the stationary phase, leading to lysis of some bacterial cells. Furthermore, deletion of 6S-1 RNA also significantly reduced the spore number and parasporal crystal content. Our work reveals that B. thuringiensis 6S RNA played an important regulatory role in ensuring the sporulation and parasporal crystal formation.
In 1967, Hindley (1967) first discovered a highly abundant non-coding RNA in Escherichia coli. Because it exhibits a sedimentation coefficient of 6S, it was thus named 6S RNA, with its nucleotide sequence further determined by Brownlee (1971) afterwards. However, the study of 6S RNA remains silent for a long time until 2000, when Wassarman and Storz (2000) found that E. coli 6S RNA exists in high abundance throughout all bacterial growth phases, and exhibits a concentration as high as 10,000 molecules/cell in E. coli cells in the stationary phase. High abundant 6S RNA is found to regulate gene transcription in the stationary phase by combining with σ70-denpendent RNA polymerase holoenzyme, thus leading to the response regulation of bacterial to stresses such as starvation. Infact, all previous studies also seem to indicate that 6S RNA is a global regulatory factor (Wassarman and Storz, 2000; Cavanagh and Wassarman, 2014; Steuten et al., 2014; Wassarman, 2018). Later on, 6S RNA is found to exist in even more bacteria. Wehner et al. (2014) identified 1,750 6S RNA-encoding genes in 1,611 bacterial genomes, and found that the sizes of 6S RNA-encoding genes ranging from 153 to 237 base pairs (with an average of 183 bp). While most bacteria seem to contain only one 6S RNA-encoding gene in their genomes, some bacteria, such as Bacillus genus in Firmicutes phylum, do contain two 6S RNA-encoding genes. Interestingly, even four such 6S RNA-encoding genes are found in the genome of Magnetococcus sp. MC-1 of α-Proteobacteria. In fact, 6S RNA-encoding genes are now found to be widely distributed in various bacteria, even in bacteria from the phyla Chloroflexi and Aquificae at the root of the phylogenetic tree (Wehner et al., 2014), indicating that 6S RNAs are originated quite early and are widely distributed.
Besides, Trotochaud and Wassarman (2004) also reveal that the survival ability of E. coli is distinctively reduced in the stationary phase when the 6S RNA-encoding gene ssrS1 is deleted, indicating that 6S RNA is beneficial to the normal growth and survival of E. coli in the stationary phase. Indeed, more and more regulatory functions of 6S RNA are gradually unveiled after many more in-depth researches. For example, it can now delay the sporulation of Bacillus subtilis (Cavanagh and Wassarman, 2013), promote the photosynthesis of Synechocystis (Heilmann et al., 2017), enhance the gene expression of pathogenicity island in Salmonella enterica serovar Typhimurium (Ren et al., 2017), increase the synthesis of antibiotics in Streptomyces coelicolor (Mikulík et al., 2014), optimize the symbiosis of Bradyrhizobium with leguminous plants (Madhugiri et al., 2012), and so on.
B. subtilis contains two kinds of 6S RNAs, called 6S-1 and 6S-2 RNAs, with their encoding genes bsrA and bsrB located in different regions of the genome. While deletion of 6S-1 RNA inhibits the growth of B. subtilis in the stationary phase (Hoch et al., 2015) and promotes earlier sporulation initiation by accelerating the utilization of nutrients (Cavanagh and Wassarman, 2013), the lack of 6S-2 RNA does not, however, seem to affect the growth and sporulation in the stationary phase (Cavanagh and Wassarman, 2013; Hoch et al., 2015). Until now, its physiological function remains unclear.
The Bacillus cereus group is a taxonomic group comprising closely related species of the Bacillus genus that includes more than 20 different species (Miller et al., 2016; Liu et al., 2017). Among them, the insect pathogen Bacillus thuringiensis, the anthracnose pathogen Bacillus anthracis, and the food-borne opportunistic pathogen B. cereus (Wang et al., 2019), have attracted extensive attention. Different from B. subtilis that harbors two 6S RNA-encoding genes located in different regions, B. cereus group bacteria contain the 6S RNA-encoding genes arranged in tandem in their genomes (Wehner et al., 2014). In this study, taking the B. thuringiensis BMB171 as a model to study the physiological function of 6S RNAs in B. cereus group bacteria, we found that the encoding genes ssrSA and ssrSB of 6S-1 and 6S-2 RNAs in BMB171 were located in the same operon and were co-transcribed as a precursor, which might be processed by ribonucleases to form mature 6S-1 and 6S-2 RNAs. Furthermore, deletion of the ssrSA gene inhibited the growth of B. thuringiensis in the stationary phase and decreased the sporulation and parasporal crystal formation. Besides, we found that diminished sporulation is primarily due to the decreased growth rate of the ssrSA deletion mutant in the stationary phase.
The plasmids and strains used in this study are listed in Tables 1, 2, with primers used listed in Supplementary Table S1, respectively. Escherichia coli DH5α was used for cloning experiment and was cultured at 37°C in lysogeny broth (LB) medium (g/L: tryptone, 10; yeast extract, 5; and NaCl, 10). The medium was adjusted to pH 7.0 before autoclaving at 121°C for 15 min. Bacillus thuringiensis BMB171 and its derivative strains were cultured at 28°C in the GYS medium (g/L: glucose, 1.00; yeast extract, 2.00; K2HPO4·3H2O, 0.66; (NH4)2SO4, 2.00; MgSO4·7H2O, 0.04; MnSO4·H2O, 0.04; and CaCl2, 0.08), with the medium autoclaved at 115°C for 30 min after pH value being adjusted to 7.8. When necessary, relevant antibiotics were added to the cultures with the following final concentration: 50 μg/ml kanamycin, 25 μg/ml erythromycin, 100 μg/ml ampicillin, 300 μg/ml spectinomycin, or 60 U polymyxin (Wang et al., 2019). For the decoyinine-added experiment, cells were grown to an OD600 of 0.5 in GYS medium, followed by the addition of decoyinine to 0.4 mg/ml as previously described (Mitani et al., 1977; Ikehara et al., 1982; Cavanagh and Wassarman, 2013).
The samples of 30 ml each from BMB171 and its derivative strains were cultured in GYS medium for 11 h before being centrifuged, followed by total RNA extraction and RT-qPCR experiments as previously described (Zheng et al., 2015, 2020; Fu et al., 2018). In these experiments, the gapdh gene was used as an internal control.
The 5'-rapid amplification of complementary DNA (cDNA) ends (5'-RACE) experiment was performed to identity the trancription start site (TSS) as described previously with some modifications (Ali et al., 2017). RNA was first extracted from BMB171 cells that were grown in GYS, followed by reverse transcription to cDNA. The 3'-end of cDNA was then labeled by poly(dA) using terminal deoxynucleotidyl transferase (Takara, Japan). The cDNA was then PCR amplified using primers of Primer-8 and ssrSB-R as listed in Supplementary Table S1. The PCR products were then cloned to the pMD19-T vector (Takara, Japan) and sequenced (Wang et al., 2019).
BMB171/pHT1K, BMB171/P1-lacZ, and BMB171/P2-lacZ strains were grown at 28°C in a shaking incubator at 200 rpm in 100 ml GYS with 25 μg/ml erythromycin. Two milliliters of each culture was separately collected at indicated time and assayed for β-galactosidase activity as described previously (Zhou et al., 2017; Wang et al., 2019).
BMB171 and its single deletion 6S-1 RNA mutant ΔssrSA, single deletion 6S-2 RNA mutant ΔssrSB, and the double deletion mutant ΔssrSAB were cultured in GYS medium at 28°C for 24 h, which were then heated to 65°C for 30 min, followed by gradient dilution (10 times) with M9 minimum medium. Around 100 μl of each diluent was then spread onto LB plates. The colony-forming units (CFUs) per ml were then counted (Wang et al., 2016).
BMB171 and its derivative strains were cultured at 28°C in GYS medium. To observe the morphology of vegetative cells and spores, 5 μl of each cell sample was collected at indicated time points, spotted onto the center of a glass slide, and covered with a coverslip. Spores were then observed with a phase-contrast microscope (Olympus, Japan; Zheng et al., 2020).
To observe the morphology of BMB171 cells and its derivative strains, 4 ml of each sample was harvested by centrifugation at 17 h, with the cell pellets resuspended in 2.5% glutaraldehyde, and stored at 4°C overnight. Ultra-thin sections were finally prepared and stained as described (Craig et al., 1997). A Hitachi H-7000 FA transmission electron microscope (Hitachi, Japan) was then used for observation.
The markerless gene deletion mediated by homing endonuclease I-SceI was performed in B. thuringiensis as previously reported (Zheng et al., 2015; Tang et al., 2016; Wang et al., 2016). Intermediate plasmids (pRP1028-ssrSA-UD, pRP1028-ssrSB-UD, and pRP1028-ssrSAB-UD) used in this study for gene deletion experiments were listed in Table 1. The helper plasmid pSS1827 for conjugational transfer and B. thuringiensis-E. coli shuttle plasmid pRP1028 carrying an I-SceI recognition site and pSS4332 carrying I-SceI restriction enzyme encoding gene was listed in Table 1.
Crystalliferous strains of BMB171-cry, ΔssrSA-cry, ΔssrSB-cry, and ΔssrSAB-cry were obtained by transformation of the cry1Ac10 gene with its original promoter in the plasmid pBMB43-304 (Qi et al., 2015) into BMB171, ΔssrSA, ΔssrSB, and ΔssrSAB strains, respectively. To observe parasporal crystals, the crystalliferous strains were grown at 28°C and 200 rpm for 24 h in GYS medium supplemented with 25 mg/ml erythromycin. One drop from each culture was spotted onto the center of a glass slide, and covered with a coverslip. Parasporal crystals were then observed with a phase-contrast microscope (Olympus, Japan). To extract Cry1Ac10 protein, each culture was collected separately by centrifugation at 6,000 g for 15 min (AG Eppendorf, Hamburg, Germany). Procedure for the separation of Cry1Ac10 protein was carried out according to a previous study (Wang et al., 2013b). Finally, the Cry1Ac10 protein was visualized by SDS-PAGE, with its concentration measured by the Bradford method (Wang et al., 2016).
The BMB171 strain was found to possess two 6S RNAs, with their encoding genes ssrSA (BMB171_RS29145) and ssrSB (BMB171_RS291506) located in tandem in the genome (Figure 1A). To explore whether the ssrSA and ssrSB genes are within the same operon, we first verified the co-transcription of the ssrSA and ssrSB genes by the semi-quantitative reverse transcription PCR (SqRT-PCR; Supplementary Figure S1), and found they were indeed co-transcribed. To further confirm that the ssrSB gene is not transcribed individually, we carried out the β-galactosidase assays to detect the promoter activities of the upstream regions of ssrSA and ssrSB genes in the BMB171 genome, respectively. We found that the upstream sequence of the ssrSA gene (P1 region) exhibited strong transcription initiation activities in the logarithmic phase (5 h), transition phase (11 h), and stationary phase (17 h), while those of the ssrSB gene (P2 region) basically exhibited no such transcription initiation activities at all when compared to the no promoter control (Figure 1B). These data indicate that the ssrSB gene was co-transcribed along, but not individually, with ssrSA. Finally, through the 5’-RACE experiment (Figure 2A), we identified a TSS as well as the canonical −35 and −10 regions (Figure 2B) upstream of the ssrSA gene, but found no independent TSS upstream of the ssrSB gene. These results thus clearly confirmed that the 6S RNA-encoding genes ssrSA and ssrSB were co-transcribed in BMB171.
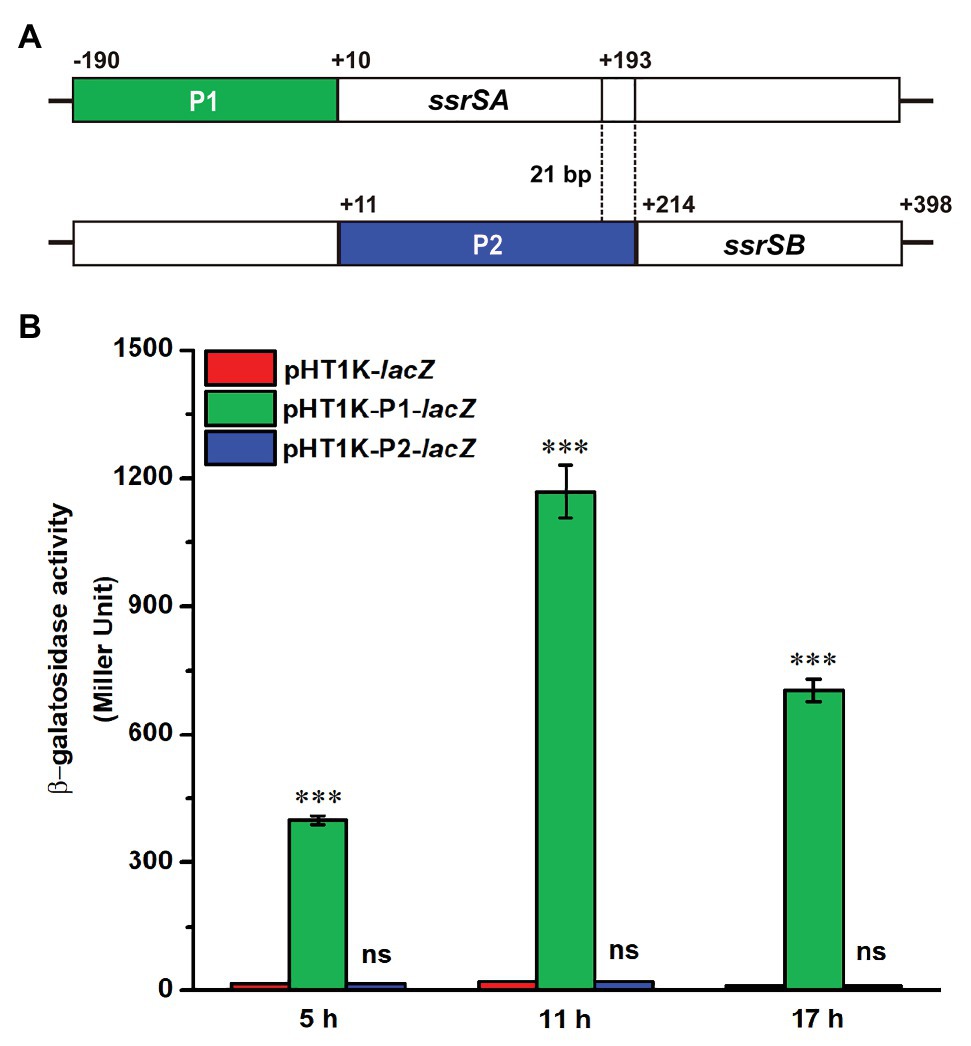
Figure 1. Promoter activity assays of the ssrSA and ssrSB genes in BMB171. (A) Schematic diagram of ssrSA and ssrSB genes and their putative promoter regions of P1 (marked in green) and P2 (marked in blue). The P1 and P2 regions were each fused with a β-galactosidase gene (lacZ) and transformed into the plasmid pHT1K, respectively. (B) The β-galactosidase activity assay of strains BMB171/P1-lacZ and BMB171/P2-lacZ harboring plasmids pHT1K-P1-lacZ and pHT1K-P2-lacZ with the BMB171/-lacZ harboring pHT1K carrying a promoter-less lacZ as the control. The strains were cultured at 28°C in GYS medium and were determined in the logarithmic phase (5 h), transition phase (11 h), and stationary phase (17 h). The values were means ± SDs for triplicate assays. Significances of differences by Student’s t-test are indicated. ***p < 0.001; **p < 0.01; *p < 0.05; ns, p > 0.05.
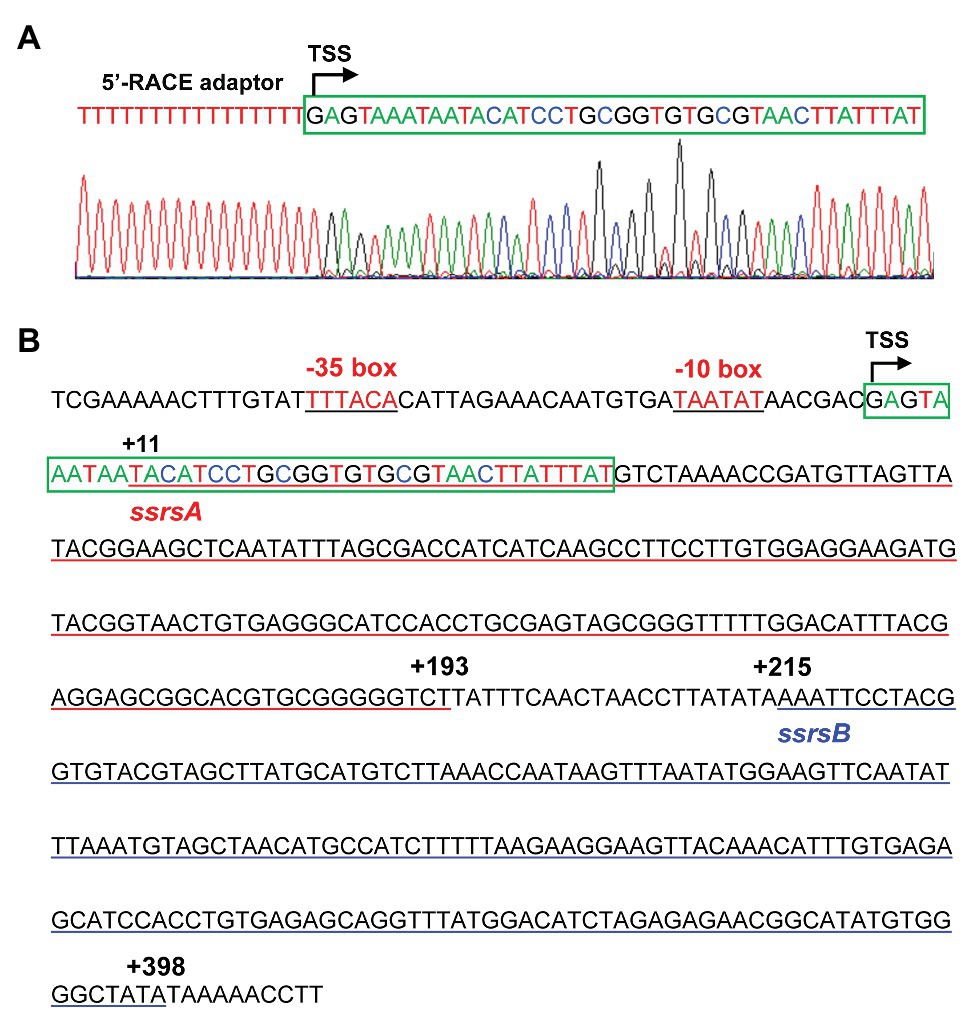
Figure 2. Identification of the transcription start site (TSS) and promoter regions of the ssrSAB operon. (A) TSS is identified by 5'-rapid amplification of complementary DNA (cDNA) ends (5'-RACE), with the 5'-RACE aptamer sequence (a tandem of 16 thymine nucleotides) shown in red, and the TSS marked with a curved arrow. The 5'-end 40 bp sequence of the ssrSAB operon was highlighted with a green box, with the colored nucleotides consistently as those in the sequencing diagram (below). (B) The promoter (−51 to −1) and encoding region (+1 to +398) sequences of the ssrSAB operon. The −35 and −10 regions were underlined in black, and the ssrSA (+1 to +193) and ssrSB (+215 to +398) encoding regions underlined in red and blue, respectively. The TSS was marked with a curved arrow with the 5'-end 40 bp sequence enclosed by a green box and colored as those in (A).
It has been reported that deletion of 6S-1 RNA results in inhibition of the growth of B. subtilis in the stationary phase (Hoch et al., 2015). We thus wonder whether deletion of 6S RNAs also exhibits the similar effect on the growth of B. thuringiensis. We have thus used the markerless gene knockout technology to precisely delete the two 6S RNA-encoding genes in the starting strain BMB171, and constructed the single deletion 6S-1 RNA mutant ΔssrSA, single deletion 6S-2 RNA mutant ΔssrSB, and the double deletion mutant ΔssrSAB. We first determined the growth curves of BMB171 and its mutants, and found that the growth rates of mutants ΔssrSA and ΔssrSAB declined much more rapidly than that of starting strain BMB171 in the stationary phase, while ΔssrSB exhibited no significant change compared to BMB171 (Figure 3A). Next, we checked the cell morphologies of BMB171, ΔssrSA, ΔssrSB, and ΔssrSAB at 17 h with transmission electron microscope, and found that deletion of 6S-1 RNA caused more cells to lyse (Figure 3B), while the complemented strains ΔssrSA/P1-ssrSA and ΔssrSAB/P1-ssrSAB showed no significant differences in bacterial growth and morphology (Supplementary Figure S2) compared to the control strain BMB171/pHT1K. Taken together, these results indicate that deletion of 6S-1 RNA but not 6S-2 RNA inhibited the growth of B. thuringiensis in the stationary phase.
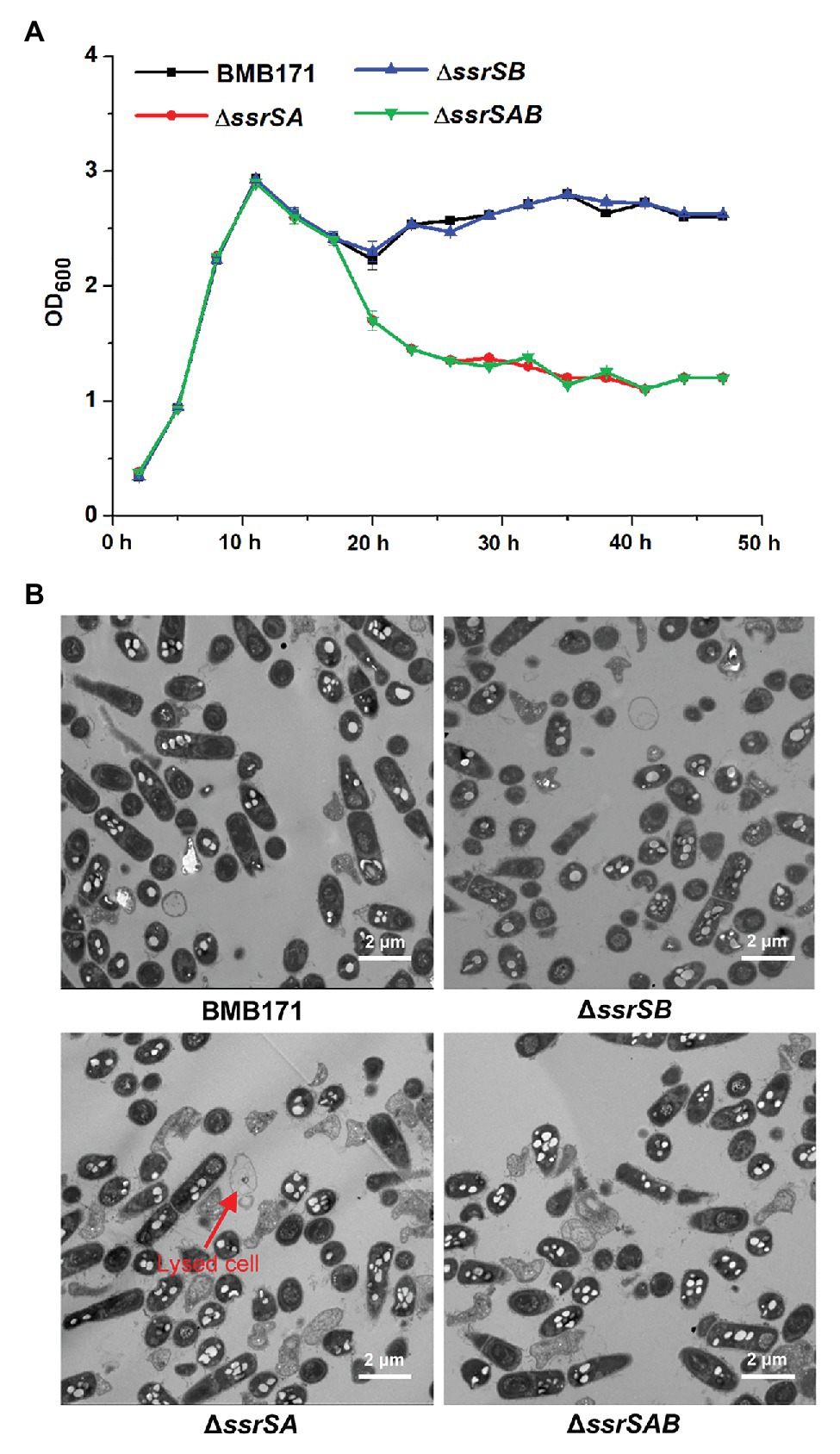
Figure 3. The effect of deleting of 6S RNA-encoding genes on the growths of Bacillus thuringiensis. (A) Determination of the growth curves of BMB171 and its 6S RNA-encoding genes deletion mutants ΔssrSA, ΔssrSB, and ΔssrSAB. (B) Transmission electron microscope images of the cell morphologies of BMB171 and its deletion mutants ΔssrSA, ΔssrSB, and ΔssrSAB at 17 h (stationary phase). Lysed cells are indicated by red arrows. The above-mentioned strains were cultured at 28°C in GYS medium. The values were means ± SDs for triplicate assays.
To investigate the roles of 6S-1 and 6S-2 RNAs in the process of sporulation, we first checked the spores of BMB171 and its mutants in the stationary phase through a phase-contrast microscope, and found that the spores formed by mutants ΔssrSA and ΔssrSAB were much fewer than those of the starting strain BMB171. However, the spore amount of ΔssrSB was similar to that of BMB171 (Figure 4A). Second, we counted the spores by spread-plate method, and found that the spore numbers formed by ΔssrSA and ΔssrSAB were much fewer than those of BMB171, whereas the spore number in ΔssrSB did not differ much from that in BMB171 (Figure 4B). Meanwhile, we also complemented back the ssrSA and ssrSAB genes into the mutants ΔssrSA and ΔssrSAB, respectively. Phase-contrast microscopy observation and spore count did show that the sporulation capabilities of the complemented strains ΔssrSA/P1-ssrSA and ΔssrSAB/P1-ssrSAB have returned to the original level of the control strain BMB171/pHT1K (Supplementary Figure S3). These results indicate that deletion of the ssrSA gene, but not ssrSB, inhibited sporulation; namely, the presence of 6S-1 RNA was required for the normal sporulation.
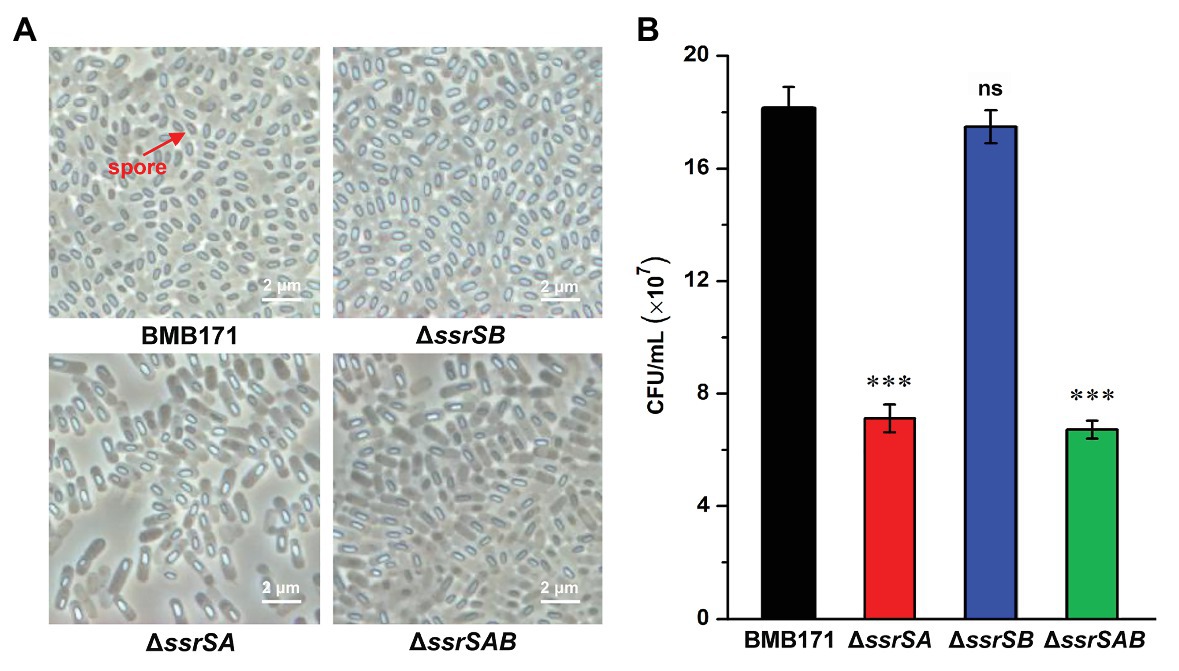
Figure 4. The effect of deleting of 6S RNA-encoding genes on the B. thuringiensis sporulation. (A) Observation of the spore formed by BMB171 and its deletion mutants ΔssrSA, ΔssrSB, and ΔssrSAB at 24 h (stationary phase) using a phase contrast microscope. Oval-shaped spores are indicated by red arrows. (B) The spore counts of BMB171 and its mutants ΔssrSA, ΔssrSB, and ΔssrSAB at 24 h (stationary phase). The strains were cultured at 28°C in GYS medium. The values were means ± SDs for triplicate assays. Significances of differences by Student’s t-test are indicated. ***p < 0.001; **p < 0.01; *p < 0.05; ns, p > 0.05.
BMB171 is an acrystalliferous mutant of wild-type YBT-1463 (Li et al., 2000; Qi et al., 2015). To explore whether 6S RNAs affect the parasporal crystal formation, we introduced a pBMB43-304 plasmid containing the parasporal crystal protein encoding gene cry1Ac10 and its original promoter sequence (Qi et al., 2015) into the BMB171 and its mutant strains ΔssrSA, ΔssrSB, and ΔssrSAB, respectively, to obtain the crystalliferous strains of BMB171-cry, ΔssrSA-cry, ΔssrSB-cry, and ΔssrSAB-cry. We first checked the parasporal crystal formation of BMB171 and its mutants in the stationary phase through a phase-contrast microscope, and found that the amounts of parasporal crystals formed by the ΔssrSA-cry and ΔssrSAB-cry strains were far less than that of BMB171-cry, while the amount of crystals in the ΔssrSB-cry was similar to that of BMB171-cry (Figure 5A). Subsequently, we measured the content of parasporal crystal protein Cry1Ac10, and found that the concentrations of Cry1Ac10 in ΔssrSA-cry and ΔssrSAB-cry were remarkably reduced compared to BMB171-cry, while the ΔssrSB-cry exhibited no substantial difference (Figures 5B,C). These experiments show that deletion of 6S-1 RNA not only inhibited the sporulation of bacterial cells, but also repressed the parasporal crystal formation.
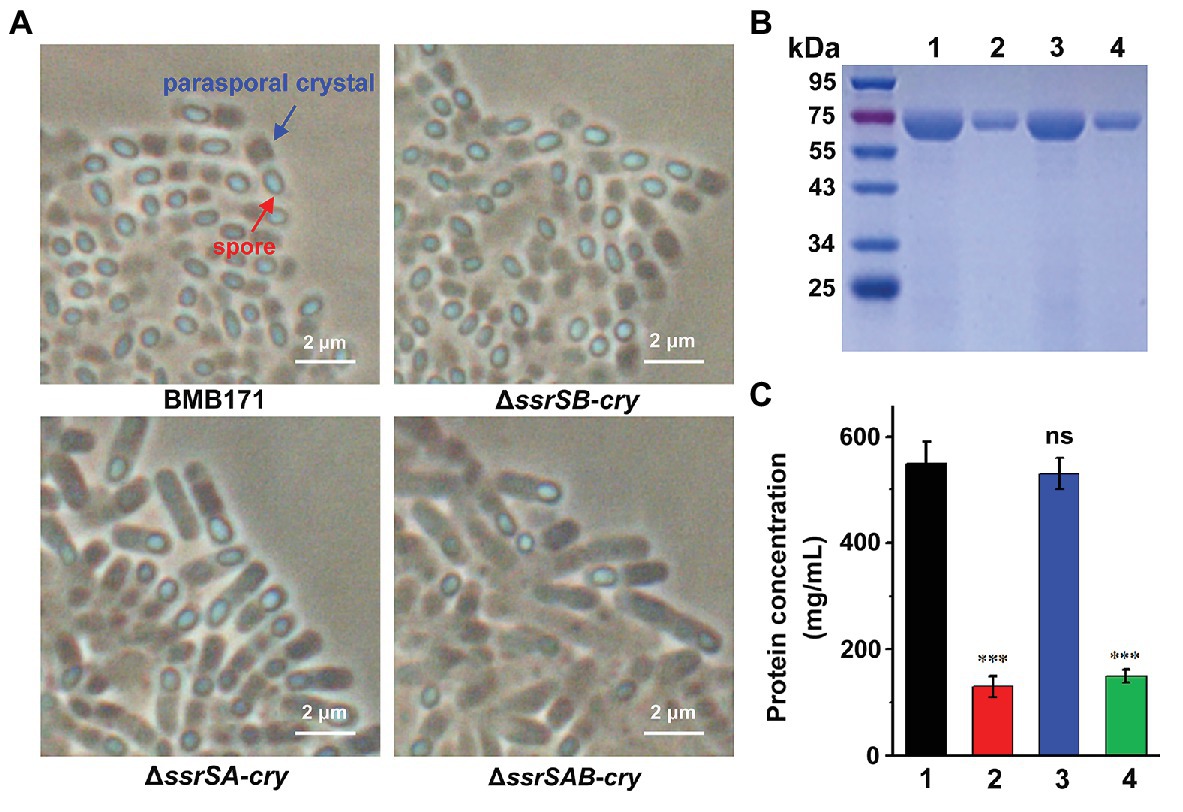
Figure 5. The effect of deleting 6S RNA-encoding genes on the parasporal crystal formation of B. thuringiensis (A) Phase contrast microscopy image of the parasporal crystal formation in crystalliferous strains BMB171-cry, ΔssrSA-cry, ΔssrSB-cry, and ΔssrSAB-cry at 24 h (stationary phase). Cells with parasporal crystals and spores are indicated by blue and red arrows, respectively. (B) Separation of Cry1Ac10 from the four strains by SDS-PAGE. Lines 1–4 represent BMB171-cry, ΔssrSA-cry, ΔssrSB-cry, and ΔssrSAB-cry, respectively. (C) The concentrations of Cry1Ac10 in BMB171-cry (1), ΔssrSA-cry (2), ΔssrSB-cry (3), and ΔssrSAB-cry (4) at 24 h (stationary phase). The above-mentioned strains were cultured at 28°C in GYS medium. The values were means ± SDs for triplicate assays. Significances of differences by Student’s t-test are indicated. ***p < 0.001; **p < 0.01; *p < 0.05; ns, p > 0.05.
To figure out the reasons accounting for inhibition of sporulation in BMB171 after 6S-1 RNA deletion, we used the real-time quantitative PCR (RT-qPCR) to examine the transcription levels of several key early sporulation-related genes in BMB171 and its mutant’s strains ΔssrSA, ΔssrSB, and ΔssrSAB. These relevant genes include spo0A, which encodes an essential transcriptional regulatory factor in the initial stage of sporulation (Lopez et al., 2009); kinA, which encodes a phosphorylation kinase that is mainly responsible for the phosphorylation of Spo0A (Jiang et al., 2000a); and spo0H, which encodes σH to promote the transcription of spo0A and kinA (Predich et al., 1992). Taking gene gapdh encoding glyceraldehyde-3-phosphate dehydrogenase as an internal control, we demonstrated that the transcription levels of spo0A, kinA, and spo0H from mutants ΔssrSA, ΔssrSB, and ΔssrSAB at 0.5, 1.0, 1.5, and 2.0 h showed almost no difference to those of BMB171 (Supplementary Figure S4). Besides, we further determined the transcriptomes of BMB171 and ΔssrSAB in the stationary phase and found that the transcription levels of sporulation-related genes did not change significantly between BMB171 and ΔssrSAB (Supplementary Table S2). These data demonstrate that deletion of 6S-1 RNA did not change the start time of sporulation at the molecular level.
Since deletion of 6S-1 RNA did inhibit the growth of B. thuringiensis in the stationary phase, we wonder whether such inhibition also affects their sporulation? Because BMB171 and its various mutants ΔssrSA, ΔssrSB, and ΔssrSAB exhibited no obvious difference in the growth curves in the logarithmic phase, we wonder whether there is difference in sporulation efficacy when these strains were induced to produce spores at this time? Given that decoyinine can induce Bacillus cells to produce spores in advance (Cavanagh and Wassarman, 2013; Wang et al., 2016; Zhou et al., 2017), we thus conducted an experiment by adding decoyinine to the GYS medium in the logarithmic phase. The results showed that the spore numbers formed by ΔssrSA, ΔssrSB, and ΔssrSAB did not differ much from that of BMB171 (Figure 6). This experiment proves that the reduced sporulation efficacy was mainly caused by inhibition of the growth of B. thuringiensis in the stationary phase, not in the logarithmic phase. Further, we found that the transcription levels of genes involved in carbohydrate transport and metabolism, nucleotide transport and metabolism, protein translation, and energy production and conversion decreased significantly in ΔssrSAB (Supplementary Table S3). Therefore, the decreased growth rate of the ssrSA deletion mutant in the stationary phase is the main reason for the diminished sporulation of B. thuringiensis.
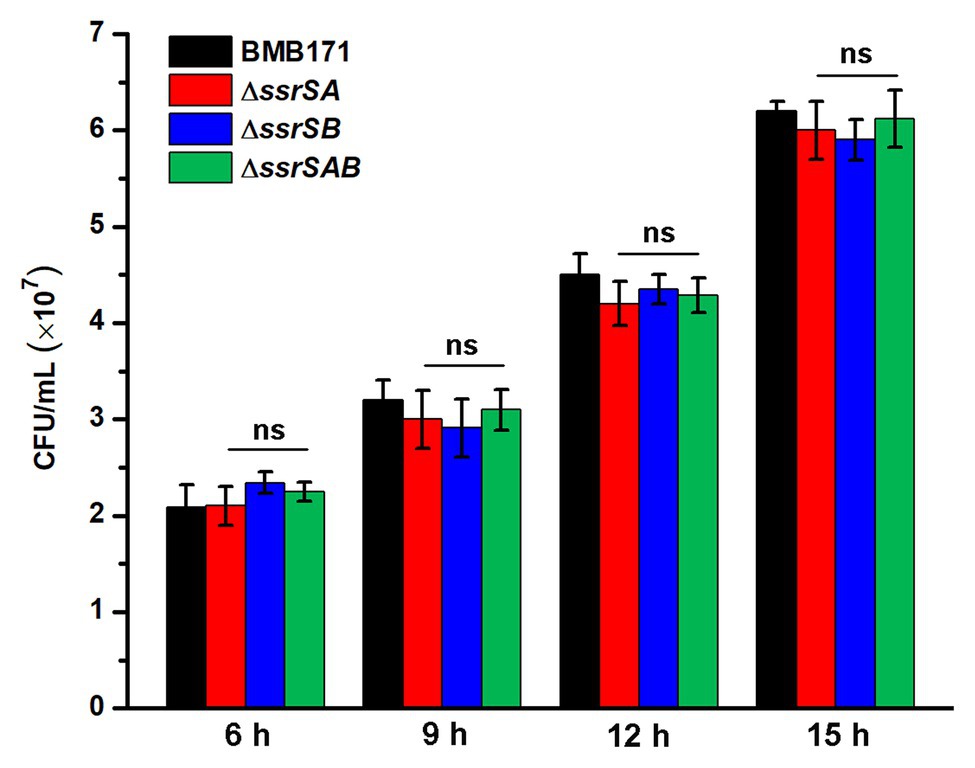
Figure 6. The effect of deleting 6S RNA-encoding genes on the sporulation of BMB171 and its mutants in the logarithmic phase. After adding the sporulation inducer decoyinine (0.4 mg/ml) to the GYS cultures of above-mentioned strains in the logarithmic phase (3 h, OD600 ≈ 0.5), the spore numbers were counted at 6, 9, 12, and 15 h after induction. The strains were cultured at 28°C and the numbers are revealed as means ± SDs in triplicate assays. Significances of differences by Student’s t-test are indicated. ***p < 0.001; **p < 0.01; *p < 0.05; ns, p > 0.05.
In this study, we verified that the 6S-1 and 6S-2 RNAs-encoding genes ssrSA and ssrSB were located in the same operon and co-transcribed in B. thuringiensis. Through the phenotypic study of BMB171 and its deletion mutants of ΔssrSA, ΔssrSB, and ΔssrSAB, we found that deletion of 6S-1 RNA not only inhibited the growth of B. thuringiensis in the stationary phase, but also decreased the sporulation and parasporal crystal formation. We further confirmed that inhibition of the growth in the stationary phase is likely the primary reason for the reduced sporulation efficacy.
In BMB171, we validated that the 6S-1 and 6S-2 RNAs-encoding genes ssrSA and ssrSB were arranged in tandem and co-transcribed, and verified that the upstream sequence of the ssrSA gene exhibited the transcription initiation activity, but not the upstream sequence of the ssrSB gene (Figure 1). These results indicate that the transcriptions of ssrSA and ssrSB genes were initiated through the upstream promoter region of the ssrSA gene, and were jointly transcribed into a long RNA precursor. In E. coli, the 6S RNA-encoding gene is first transcribed into a long RNA precursor, which is then cleaved by RNase E and RNase G into mature 6S RNA (Kim and Lee, 2004). Given the fact that RNase E and RNase G prefer to cut the regions enriched in nucleotides A and U on single-stranded RNA (Mackie, 1998; Jiang et al., 2000b; Tock et al., 2000), so we analyzed the RNA precursor sequence in BMB171 and found that, similar to E. coli, the 5'- and 3'-ends of the precursor RNA of 6S-1 RNA and 6S-2 RNA in BMB171 are also rich in A and U (Supplementary Figure S5). Moreover, transcriptomic data showed that 6S RNAs existed mainly as two separate mature 6S-1 and 6S-2 RNAs (Supplementary Figure S6). Combining the above results (Supplementary Figures S1, S5, S6), we suggest that a long precursor 6S RNA is first transcribed, followed by processing with RNase E, RNase G, or other ribonucleases to form mature and functional 6S-1 and 6S-2 RNAs.
In the present manuscript, we have analyzed the 6S RNA-encoding genes of Bacillus genus bacteria and their gene organization. Bacillus genus bacteria usually contain two different 6S RNA-encoding genes of ssrSA and ssrSB (Supplementary Tables S4–S6). However, unlike other Bacillus genus bacteria that have these two genes located in different regions of the genome, the two 6S RNA-encoding genes of B. cereus group bacteria were located in tandem positions. Furthermore, we also verified that they were in the same operon and are transcribed together. The ssrSA and ssrSB genes of B. cereus group bacteria are located between the genes encoding thiocyanase and glutathione spermidine synthase. The former is related to the detoxification of exogenous toxins while the latter involved in the resistance to stress such as oxidation (Supplementary Table S4). Other than the B. cereus group, there is also a B. subtilis group bacteria composed of more than 20 important bacteria species, such as, B. subtilis, Bacillus licheniformis, Bacillus pumilus, Bacillus velezensis, and Bacillus amyloliquefaciens in Bacillus genus (Caulier et al., 2019; Fayad et al., 2019). Unlike the B. cereus group bacteria, the ssrSA gene of B. subtilis group bacteria was mainly located between the FMN-dependent NADH-azoreductase and DNA helicase RecQ encoding genes. The former is related to the detoxification of exogenous toxins, while the latter is linked with DNA repair, recombination, and replication. The ssrSB gene, on the other hand, was mainly located between the aspartate-tRNA ligase and tRNA threonylcarbamoyladenosine dehydratase, both of which were associated with protein translation (Supplementary Table S5). Among Bacillus genus bacteria excluding B. cereus group and B. subtilis group, only the aspartate-tRNA ligase encoding gene locus upstream of the ssrSB gene was relatively conservative (Supplementary Table S6).
The 6S RNAs in B. cereus group bacteria were quite different to those of B. subtilis group bacteria regarding their gene organization, which might lead to their differences in sporulation. For example, in B. subtilis, deletion of 6S-1 RNA accelerates the utilization of nutrients, leading to the early arrival of nutrient deprivation conditions, which in turn induces the earlier expression of key early sporulation-related genes, ultimately promoting the earlier sporulation (Wassarman, 2018), while deletion of 6S-1 RNA in B. thuringiensis resulted in suppression of sporulation. In order to explore its possible regulatory mechanism, we have determined the transcriptomes of BMB171 and ΔssrSAB in the stationary phase. After analyses, we found that the transcription levels of sporulation-related genes did not change significantly between BMB171 and ΔssrSAB (Supplementary Table S2), indicating that 6S RNA does not affect sporulation by directly regulating the temporal expression of sporulation-related genes. Moreover, the experiments of decoyinine-induced sporulation in the logarithmic phase (Figure 6) and RT-qPCR detection of the transcription levels of key early sporulation-related genes (Supplementary Figure S4) further proved this conclusion. Since there is no available transcriptomic data from the 6S-1 RNA deletion in B. subtilis, we are currently unable to compare the 6S-1 RNA regulatory mechanisms between the two bacteria in a more comprehensive way. Altogether, the 6S RNAs of B. subtilis group and B. cereus group bacteria exhibited different gene organization and physiological functions, indicating that 6S RNAs might regulate the biological functions of B. subtilis group and B. cereus group bacteria via different mechanisms.
Then, how does deletion of 6S-1 RNA inhibit sporulation of B. thuringiensis? After further analyzing the transcriptomic data, we found that the transcription levels of genes involved in carbohydrate transport and metabolism, nucleotide transport and metabolism, protein translation, and energy production and conversion decreased significantly in ΔssrSAB (Supplementary Table S3). According to our previous report that sporulation and parasporal crystal formation do require a lot of material and energy supply in B. thuringiensis (Wang et al., 2013c), we therefore speculate that the insufficient supply of material and energy in ΔssrSAB was the main reason for inhibition of sporulation.
How does deletion of 6S-1 RNA inhibits the parasporal crystal formation of B. thuringiensis? In crystalliferous strain BMB171-cry, cry1Ac10 with the original promoter was regulated by a sporulation-specific sigma factor SigE. In the transcriptomic data, we did not find a significant expression difference of sigE between BMB171 and ΔssrSAB (Supplementary Table S2). Meanwhile, RT-qPCR assays confirmed that cry1Ac10 had no expression difference between BMB171 and ΔssrSAB (Supplementary Figure S7). However, the content of parasporal crystals was significantly reduced in ΔssrSAB (Figure 5B). This indicates that deletion of 6S RNA may inhibit the translation of Cry1Ac10, further reducing the parasporal crystal formation.
Like 6S RNA, CsrA, CarD, and (p)ppGpp are all global regulatory factors that can respond to starvation stress in the stationary phase (Kalia et al., 2013; Romeo et al., 2013; Flentie et al., 2016). CsrA is believed to inhibit bacterial translation by interacting with conserved sequences on target mRNA under carbon starvation conditions (Romeo et al., 2013). Both CarD and (p)ppGpp, on the other hand, seem to regulate downstream gene transcription by interacting with RNA polymerase; in addition, the binding of CarD with RNA polymerase has been found to stabilize the transcription initiation complex to initiate the transcription of downstream genes (Flentie et al., 2016). Under starvation conditions, high levels of intracellular (p)ppGpp can also inhibit translation, which not only regulates the transcription of tRNA, rRNA, and ribosomal protein genes by binding to RNA polymerase, but also directly inhibits the bacterial translation activity through combining translation initiation factor IF2 with the translation elongation factors EF-Tu and EF-G (Kalia et al., 2013). Like CarD and (p)ppGpp, 6S RNA is also a global regulatory factor that can regulate gene transcription by binding to RNA polymerase. The consistent function of 6S RNA, CsrA, CarD, and (p)ppGpp may be to maintain and optimize the survival rates of bacteria under different stress conditions, which may be manifested in rather complex metabolic regulation networks. Currently we cannot figure out what are the specific roles 6S RNA plays in these networks and also how they co-regulate the bacterial response to starvation stress in the stationary phase, which are issues deserve further exploration.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
JH and ZL designed the experiments. ZL, LZ, ZY, and LL did the experiments. JH, S-HC, JW, and ZL wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This paper is supported by the National Natural Science Foundation of China (grants 31770087 and 31970074), and the Cultivation fund project of Fujian Academy of Agricultural Sciences (AGP2018-2).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.604458/full#supplementary-material
Ali, M. K., Li, X. F., Tang, Q., Liu, X. Y., Chen, F., Xiao, J. F., et al. (2017). Regulation of inducible potassium transporter KdpFABC by the KdpD/KdpE two-component system in Mycobacterium smegmatis. Front. Microbiol. 8:570. doi: 10.3389/fmicb.2017.00570
Brownlee, G. G. (1971). Sequence of 6S RNA of E. coli. Nat. New Biol. 229, 147–149. doi: 10.1038/newbio229147a0
Caulier, S. C., Nannan, C., Gillis, A., Licciard, F., Bragard, C., and Mahillon, J. (2019). Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 10:302. doi: 10.3389/fmicb.2019.00302
Cavanagh, A. T., and Wassarman, K. M. (2013). 6S-1 RNA function leads to a delay in sporulation in Bacillus subtilis. J. Bacteriol. 195, 2079–2086. doi: 10.1128/JB.00050-13
Cavanagh, A. T., and Wassarman, K. M. (2014). 6S RNA, a global regulator of transcription in Escherichia coli, Bacillus subtilis, and beyond. Annu. Rev. Microbiol. 68, 45–60. doi: 10.1146/annurev-micro-092611-150135
Craig, J. E., Ford, M. J., Blaydon, D. C., and Sonenshein, A. L. (1997). A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependentgene expression. J. Bacteriol. 179, 7351–7359. doi: 10.1128/jb.179.23.7351-7359.1997
Fayad, N., Awad, M. K., and Mahillon, J. (2019). Diversity of Bacillus cereus sensu lato mobilome. BMC Genomics 20:436. doi: 10.1186/s12864-019-5764-4
Flentie, K., Garner, A. L., and Stallings, C. L. (2016). Mycobacterium tuberculosis transcription machinery: ready to respond to host attacks. J. Bacteriol. 198, 1360–1373. doi: 10.1128/JB.00935-15
Fu, Y., Yu, Z. Q., Liu, S., Chen, B., Zhu, L., Li, Z., et al. (2018). C-di-GMP regulates various phenotypes and insecticidal activity of gram-positive Bacillus thuringiensis. Front. Microbiol. 9:45. doi: 10.3389/fmicb.2018.00045
He, J., Shao, X. H., Zheng, H. J., Li, M. S., Wang, J. P., Zhang, Q. Y., et al. (2010). Complete genome sequence of Bacillus thuringiensis mutant strain BMB171. J. Bacteriol. 192, 4074–4075. doi: 10.1128/JB.00562-10
Heilmann, B., Hakkila, K., Georg, J., Tyystjärvi, T., Hess, W. R., Axmann, I. M., et al. (2017). 6S RNA plays a role in recovery from nitrogen depletion in Synechocystis sp. PCC 6803. BMC Microbiol. 17:229. doi: 10.1186/s12866-017-1137-9
Hindley, J. (1967). Fractionation of 32P-labelled ribonucleic acids on polyacrylamide gels and their characterization by fingerprinting. J. Mol. Biol. 30, 125–136. doi: 10.1016/0022-2836(67)90248-3
Hoch, P. G., Burenina, O. Y., Weber, M. H. W., Elkina, D. A., Nesterchuk, M. V., Sergiev, P. V., et al. (2015). Phenotypic characterization and complementation analysis of Bacillus subtilis 6S RNA single and double deletion mutants. Biochimie 117, 87–99. doi: 10.1016/j.biochi.2014.12.019
Ikehara, K., Okamoto, M., and Sugae, K. (1982). Induction of Bacillus subtilis sporulation by decoyinine and the concomitant disappearance of ppGpp in vegetative cells. J. Biochem. 91, 1089–1092. doi: 10.1093/oxfordjournals.jbchem.a133759
Janes, B. K., and Stibitz, S. (2006). Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 74, 1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006
Jiang, X., Diwa, A., and Belasco, J. G. (2000b). Regions of RNase E important for 5'-end-dependent RNA cleavage and autoregulated synthesis. J. Bacteriol. 182, 2468–2475. doi: 10.1128/jb.182.9.2468-2475.2000
Jiang, M., Shao, W., Perego, M., and Hoch, J. A. (2000a). Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38, 535–542. doi: 10.1046/j.1365-2958.2000.02148.x
Kalia, D., Merey, G., Nakayama, S., Zheng, Y., Zhou, J., Luo, Y. L., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341. doi: 10.1039/c2cs35206k
Kim, K. S., and Lee, Y. (2004). Regulation of 6S RNA biogenesis by switching utilization of both sigma factors and endoribonucleases. Nucleic Acids Res. 32, 6057–6068. doi: 10.1093/nar/gkh939
Li, L., Yang, C., Liu, Z., Li, F., and Yu, Z. (2000). Screening of acrystalliferous mutants from Bacillus thuringiensis and their transformation properties. Wei Sheng Wu Xue Bao 40, 85–90.
Liu, Y., Du, J., Lai, Q. L., Zeng, R. Y., Ye, D. Z., Xu, J., et al. (2017). Proposal of nine novel species of the Bacillus cereus group. Int. J. Syst. Evol. Microbiol. 67, 2499–2508. doi: 10.1099/ijsem.0.001821
Lopez, D., Vlamakis, H., and Kolter, R. (2009). Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33, 152–163. doi: 10.1111/j.1574-6976.2008.00148.x
Mackie, G. A. (1998). Ribonuclease E is a 5'-end-dependent endonuclease. Nature 395, 720–723. doi: 10.1038/27246
Madhugiri, R., Pessi, G., Voss, B., Hahn, J., Sharma, C. M., Reinhardt, R., et al. (2012). Small RNAs of the Bradyrhizobium/Rhodopseudomonas lineage and their analysis. RNA Biol. 9, 47–58. doi: 10.4161/rna.9.1.18008
Mikulík, K., Bobek, J., Zídková, J., and Felsberg, J. (2014). 6S RNA modulates growth and antibiotic production in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 98, 7185–7197. doi: 10.1007/s00253-014-5806-4
Miller, R. A., Beno, S. M., Kent, D. J., Carroll, L. M., Martin, N. H., Boor, K. J., et al. (2016). Bacillus wiedmannii sp. nov., a psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and dairy environments. Int. J. Syst. Evol. Microbiol. 66, 4744–4753. doi: 10.1099/ijsem.0.001421
Mitani, T., Heinze, J. E., and Freese, E. (1977). Induction of sporulation in Bacillus subtilis by decoyinine or hadacidin. Biochem. Biophys. Res. Commun. 77, 1118–1125. doi: 10.1016/s0006-291x(77)80094-6.
Predich, M., Nair, G., and Smith, I. (1992). Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing sigma H. J. Bacteriol. 174, 2771–2778. doi: 10.1128/JB.174.9.2771-2778.1992
Qi, M. X., Mei, F., Wang, H., Sun, M., Wang, G. J., Yu, Z. N., et al. (2015). Function of global regulator CodY in Bacillus thuringiensis BMB171 by comparative proteomic analysis. J. Microbiol. Biotechnol. 25, 152–161. doi: 10.4014/jmb.1406.06036
Ren, J., Sang, Y., Qin, R., Cui, Z. L., and Yao, Y. F. (2017). 6S RNA is involved in acid resistance and invasion of epithelial cells in Salmonella enterica serovar Typhimurium. Future Microbiol. 12, 1045–1057. doi: 10.2217/fmb-2017-0055
Romeo, T., Vakulskas, C. A., and Babitzke, P. (2013). Posttranscriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15, 313–324. doi: 10.1111/j.1462-2920.2012.02794.x
Steuten, B., Schneider, S., and Wagner, R. (2014). 6S RNA: recent answers-future questions. Mol. Microbiol. 91, 641–648. doi: 10.1111/mmi.12484
Tang, Q., Yin, K., Qian, H. L., Zhao, Y. W., Wang, W., Chou, S. H., et al. (2016). Cyclic di-GMP contributes to adaption and virulence of Bacillus thuringiensis through a riboswitch-regulated collagen adhesion protein. Sci. Rep. 6:28807. doi: 10.1038/srep28807
Tock, M. R., Walsh, A. P., Carroll, G., and McDowall, K. J. (2000). The CafA protein required for the 5'-maturation of 16S rRNA is a 5'-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 275, 8726–8732. doi: 10.1074/jbc.275.12.8726
Trotochaud, A. E., and Wassarman, K. M. (2004). 6S RNA function enhances long-term cell survival. J. Bacteriol. 186, 4978–4985. doi: 10.1128/JB.186.15.4978-4985.2004
Wang, J. P., Ai, X. L., Mei, H., Fu, Y., Chen, B., Yu, Z. N., et al. (2013a). High-throughput identification of promoters and screening of highly active promoter-5'-UTR DNA region with different characteristics from Bacillus thuringiensis. PLoS One 8:e62960. doi: 10.1371/journal.pone.0062960
Wang, X., Cai, X., Ma, H. D., Yin, W., Zhu, L., Li, X. F., et al. (2019). A c-di-AMP riboswitch controlling kdpFABC operon transcription regulates the potassium transporter system in Bacillus thuringiensis. Commun. Biol. 2:151. doi: 10.1038/s42003-019-0414-6
Wang, X., Li, Z., Li, X., Qian, H. L., Cai, X., Li, X. F., et al. (2016). Poly-β-hydroxybutyrate metabolism is unrelated to the sporulation and parasporal crystal protein formation in Bacillus thuringiensis. Front. Microbiol. 7:836. doi: 10.3389/fmicb.2016.00836
Wang, J. P., Mei, H., Qian, H. L., Tang, Q., Liu, X. C., Yu, Z. N., et al. (2013b). Expression profile and regulation of spore and parasporal crystal formation-associated genes in Bacillus thuringiensis. J. Proteome Res. 12, 5487–5501. doi: 10.1021/pr4003728
Wang, J., Mei, H., Zheng, C., Qian, H. L., Cui, C., Fu, Y., et al. (2013c). The metabolic regulation of sporulation and par-asporal crystal formation in Bacillus thuringiensis revealed by transcriptomics and proteomics. Mol. Cell. Proteomics 12, 1363–1376. doi: 10.1074/mcp.M112.023986
Wassarman, K. M. (2018). 6S RNA, a global regulator of transcription. Microbiol. Spectr. 6:RWR-0019-2018. doi: 10.1128/microbiolspec.RWR-0019-2018
Wassarman, K. M., and Storz, G. (2000). 6S RNA regulates E. coli RNA polymerase activity. Cell 101, 613–623. doi: 10.1016/s0092-8674(00)80873-9
Wehner, S., Damm, K., Hartmann, R. K., and Marz, M. (2014). Dissemination of 6S RNA among bacteria. RNA Biol. 11, 1467–1478. doi: 10.4161/rna.29894
Zheng, C., Ma, Y., Wang, X., Xie, Y. Q., Ali, M. K., and He, J. (2015). Functional analysis of the sporulation-specific diadenylate cyclase CdaS in Bacillus thuringiensis. Front. Microbiol. 6:908. doi: 10.3389/fmicb.2015.00908
Zheng, C., Yu, Z. Q., Du, C. Y., Gong, Y. J., Yin, W., Li, X. F., et al. (2020). 2-Methylcitrate cycle: a well-regulated controller of Bacillus sporulation. Environ. Microbiol. 22, 1125–1140. doi: 10.1111/1462-2920.14901
Keywords: non-coding RNA, 6S RNA, Bacillus thuringiensis, sporulation, parasporal crystal formation
Citation: Li Z, Zhu L, Yu Z, Liu L, Chou S-H, Wang J and He J (2020) 6S-1 RNA Contributes to Sporulation and Parasporal Crystal Formation in Bacillus thuringiensis. Front. Microbiol. 11:604458. doi: 10.3389/fmicb.2020.604458
Received: 09 September 2020; Accepted: 04 November 2020;
Published: 26 November 2020.
Edited by:
Jiqiang Ling, University of Maryland, College Park, United StatesReviewed by:
Ma. Cristina Del Rincón-Castro, University of Guanajuato, MexicoCopyright © 2020 Li, Zhu, Yu, Liu, Chou, Wang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin He, aGVqaW5AbWFpbC5oemF1LmVkdS5jbg==; Jieping Wang, d2FuZ2pwZmFhc0Bmb3htYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.