
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 12 November 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.603652
Bacteria evolved multiple strategies to survive and develop optimal fitness in their ecological niche. They deployed protein secretion systems for robust and efficient delivery of antibacterial toxins into their target cells, therefore inhibiting their growth or killing them. To maximize antagonism, recipient factors on target cells can be recognized or hijacked to enhance the entry or toxicity of these toxins. To date, knowledge regarding recipient susceptibility (RS) factors and their mode of action is mostly originating from studies on the type Vb secretion system that is also known as the contact-dependent inhibition (CDI) system. Yet, recent studies on the type VI secretion system (T6SS), and the CDI by glycine-zipper protein (Cdz) system, also reported the emerging roles of RS factors in interbacterial competition. Here, we review these RS factors and their mechanistic impact in increasing susceptibility of recipient cells in response to CDI, T6SS, and Cdz. Past and future strategies for identifying novel RS factors are also discussed, which will help in understanding the interplay between attacker and prey upon secretion system-dependent competition. Understanding these mechanisms would also provide insights for developing novel antibacterial strategies to antagonize aggressive bacteria-killing pathogens.
Bacteria are one of the most abundant forms of life on earth, and they have developed multiple strategies to compete with each other and fight for limited resources and space (Foster and Bell, 2012; Ghoul and Mitri, 2016). An effective strategy in this war game is to deliver toxins into opponents in order to kill them or challenge their fitness (Costa et al., 2015; Filloux and Sagfors, 2015; Green and Mecsas, 2016; Coulthurst, 2019; Klein et al., 2020). These toxins are deadly when they destroy the cell membrane integrity (e.g., peptidoglycan hydrolase, amidase, lipase, or pore-forming protein) or degrade nucleic acid (DNase, RNase, or tRNase) (Willett et al., 2015b; Lien and Lai, 2017). The challenge is to deliver efficiently one or more toxins to the appropriate destination. Thus, sophisticated mechanisms are evolved to allow the toxins to transport across the membranes and outreaching their molecular targets of the recipient cells while avoiding self-intoxication or intoxication of kins. For the latter, it is most remarkable that each toxin is encoded together with a specific immunity protein that would prevent toxicity, usually through direct protein–protein interaction.
There are several protein secretion systems that have been designed by bacteria for robust and efficient delivery of protein from the cytosol across the cell envelope. Among the nine identified so far (reviewed in Filloux and Sagfors, 2015; Christie, 2019), some have a proven capability to deliver antibacterial toxins (Aoki et al., 2005; Hood et al., 2010; Souza et al., 2015; Cao et al., 2016; García-Bayona et al., 2017). These are the type I secretion system (T1SS), type IV secretion system (T4SS), type V secretion system (T5SS) and here more specially those called contact-dependent inhibition (CDI) system, type VI secretion system (T6SS), and type VII secretion system (T7SS) (Figure 1). There are also a number of other examples such as colicins whose delivery does not involve the assembly of a supramolecular secretion machine but relies upon cell lysis (Cascales et al., 2007).
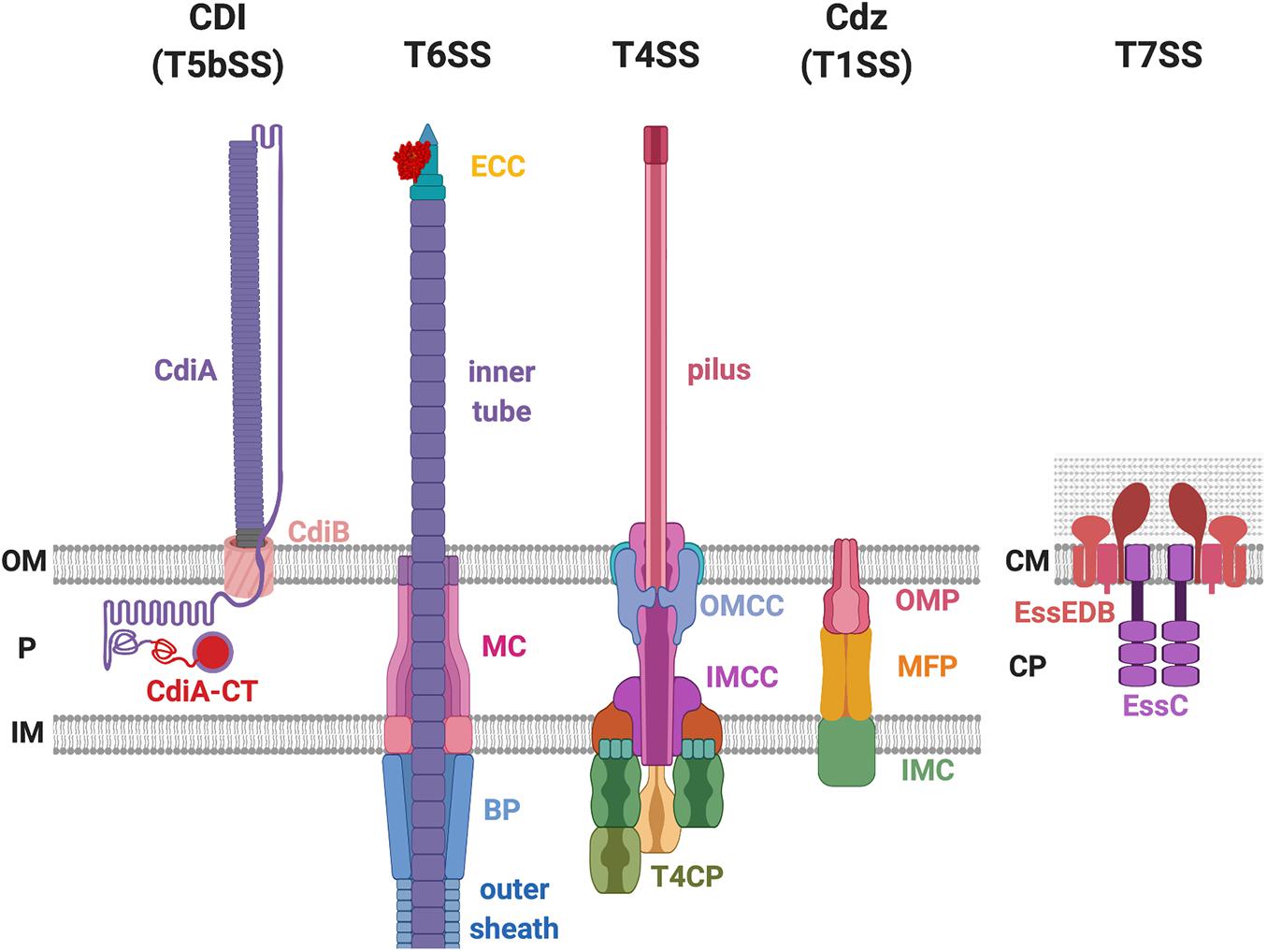
Figure 1. Bacterial secretion systems proven to deliver antibacterial toxins. The contact-dependent inhibition (CDI) system belongs to the type Vb secretion system (T5bSS), which is composed of outer membrane (OM) barrel CdiB and the surface-exposing CdiA protein. The type VI secretion system (T6SS) consists of the membrane complex (MC), baseplate (BP) complex, effector-containing complex (ECC), the outer sheath, and the inner tube. The type IV secretion system (T4SS) is composed of the outer membrane (OM) core complex (OMCC), inner membrane (IM) core complex (IMCC), type IV coupling protein (T4CP), and the pilus. The CDI by glycine-zipper protein (Cdz) belongs to the T1SS that consists of three proteins: the OM protein (OMP), the membrane fusion protein (MFP), and the IM component (IMC). The T7SS exists in the Gram-positive bacteria that only have one lipid bilayer. The T7SS is composed of the EssE, EssD, EssB, and the EssC protein. P, periplasm; CM, cell membrane; CP, cytoplasm.
To date, several papers have provided thorough overviews of the molecular mechanisms associated with these secretion systems, such as structural organization, regulatory networks, or the identity and mode of action of a repertoire of antibacterial toxins (Costa et al., 2015; Filloux and Sagfors, 2015; Green and Mecsas, 2016; Coulthurst, 2019; Klein et al., 2020). It is seldom considered what in the recipient cells might be required for an attack to be successful such as recipient susceptibility (RS) factors. To date, the best characterized RS factors are the ones recognized by the CDI system (Ruhe et al., 2020) and have been mostly identified by genetic screens (Aoki et al., 2008; Ruhe et al., 2014, 2017; Willett et al., 2015a; Jones et al., 2017). Recently, few other RS factors were identified in association with the T6SS or a novel CDI by glycine-zipper protein (Cdz) system, notably by screening resistant mutants or using knowledge-based approaches (Whitney et al., 2015; Mariano et al., 2018; García-Bayona et al., 2019; Lin et al., 2020). Our current knowledge suggests that CDI employs a receptor-based recognition mechanism for toxin delivery between close siblings at intraspecies levels, while T6SS uses mechanical force for toxin delivery into a wide range of recipient cells in a receptor-independent manner. The present review will focus on the CDI, T6SS, and Cdz by describing the secretion machine and their toxins with further highlights on the specific RS factors (e.g., membrane receptors and cytoplasmic proteins) that maximize delivery and activity of incoming toxins. In addition, we also discussed the current and potential strategies for identifying novel RS factors and proposed RS-mediated antibacterial strategies. The knowledge learned from these three systems may provide new insights to identify and investigate RS factors involved in regulating antibacterial activity from other systems, notably T4SS and T7SS.
Aoki et al. (2005) reported that wild-type Escherichia coli strain EC93 inhibits the growth of the laboratory strain MG1655 in a one-inhibits-many manner requiring direct cell-to-cell contact. Therefore, the authors defined this phenomenon as CDI. It was later on discovered that the CDI system is widely distributed in the α-, β-, and γ-proteobacteria (Aoki et al., 2010; Poole et al., 2011) and is functional in many species like E. coli, Burkholderia pseudomallei, Dickeya dadantii, Pseudomonas aeruginosa, and Acinetobacter baylyi (Aoki et al., 2010; Kiel et al., 2012; De Gregorio et al., 2018; Allen and Hauser, 2019).
The genes responsible for CDI in E. coli are cdiB, cdiA, and cdiI. The cdiI gene encodes an immunity protein that protects the attacker cell from self-intoxication (Aoki et al., 2005, 2010). The toxin domain is located at the C-terminal end of CdiA (termed CdiA-CT), which otherwise is a large protein (∼180–640 kDa) that forms a long filamentous structure with its N-terminus attached on the cell surface (Figure 1; Aoki et al., 2010; Willett et al., 2015b). CdiB is an outer-membrane beta-barrel protein that allows translocation and presentation of the CdiA toxin at the cell surface of the attacker cell (Figure 1). Both CdiB and CdiA are required to successfully inhibit the growth of the recipient cells (Aoki et al., 2005). CdiB and CdiA belong to a two-partner secretion (TPS) system also known as T5bSS, a subtype of the T5SS (Aoki et al., 2005; Filloux and Sagfors, 2015).
The domains of CdiA toxin include the N-terminal Sec-dependent signal peptide, the conserved TPS transport domain, the filamentous hemagglutinin adhesin domain 1 (FHA-1), the receptor binding domain (RBD), the Tyr/Pro-enriched (YP) domain, the second FHA domain (FHA-2), the pre-toxin domain (PD), and the C-terminal toxin domain (CdiA-CT) (Figure 2; Willett et al., 2015a; Ruhe et al., 2017, 2018). Both electron cryotomography and biochemical data support that the structure of CdiA resembles a U-shape hair clip and the RBD domain is at the bending point (Ruhe et al., 2018). One leg of the CdiA hair clip is composed of an elongated FHA-1 beta-helix whose filamentous structure extends out from the cell surface and another leg of CdiA is likely composed of the YP domain, which is required for cell surface presentation (Ruhe et al., 2017, 2018). The FHA-2 domain is required for toxin delivery into the recipient cell (Ruhe et al., 2018). The function of the PD domain is unclear, but it contains a VENN motif, which is highly conserved among different CdiA-harboring species and precisely precedes the N-terminal region of the toxin domain CdiA-CT (Aoki et al., 2010; Ruhe et al., 2018). The CdiA-CT consists of the N-terminal entry domain and the C-terminal toxin domain (Figure 2; Ruhe et al., 2018). The N-terminal entry domain is responsible for interacting with recipient’s inner membrane (IM) factor(s), and such interaction controls CdiA-CT toxin translocation into the recipient cytosol. Of note, the FHA-2 and the CdiA-CT reside in the attacker periplasm, and the delivery resumes only after the RBD domain binds to its specific recipient receptor. The FHA-2 domain is tightly associated with the recipient-cell outer membrane (OM), and this interaction is required for CdiA-CT translocation into the periplasm of target bacteria. The structure of FHA-2 is unknown but predicted to resemble an LptD lipopolysaccharide transporter consisting of a 26-stranded beta-barrel in the OM. These findings led to a proposed model that the FHA-2 domain may assemble into a transmembrane conduit for toxin translocation into the periplasm of the recipient cell (Figure 2; Ruhe et al., 2018).
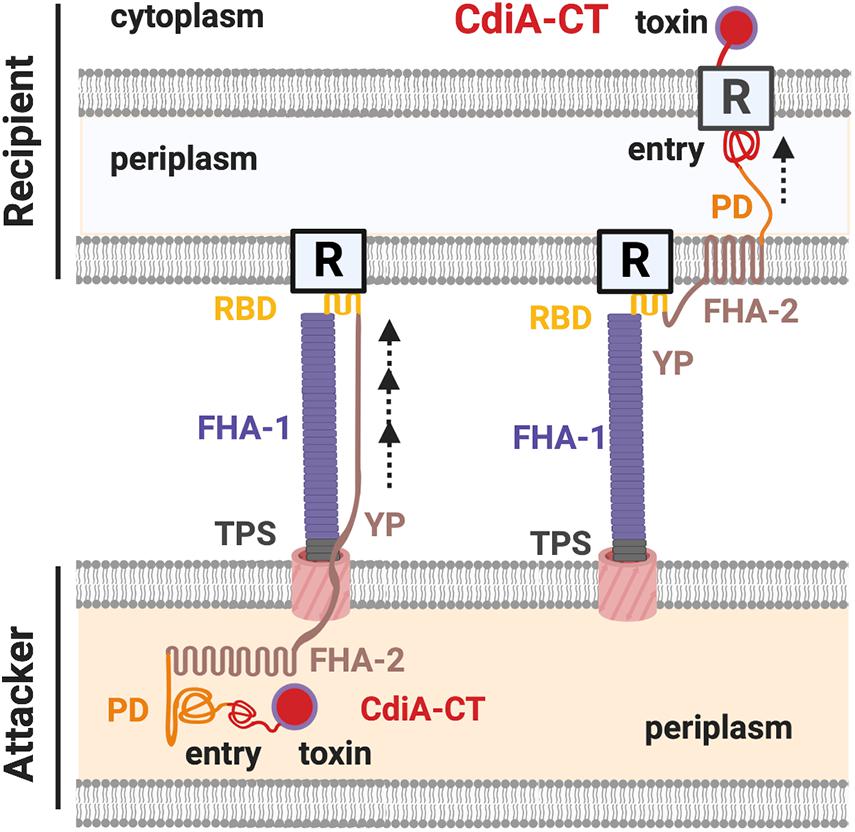
Figure 2. The CdiA domains and the contact-dependent inhibition (CDI) working model. The domains of a CdiA from the N-terminus are the conserved two-partner secretion (TPS) transport domain, the filamentous hemagglutinin domain 1 (FHA-1), the receptor-binding domain (RBD), the Tyr/Pro-enriched (YP) domain, the second FHA domain (FHA-2), the pre-toxin (PD) domain, and the C-terminus toxin domain (CdiA-CT). The CdiA-CT is further divided into the N-terminus entry subdomain and the C-terminus toxin domain. In the resting state, the FHA-2, PD, and the CdiA-CT remain in the attacker cell, while the TPS, FHA-1, RBD, and the YP domains are exposed in the extracellular milieu of the cell surface. Upon recognizing the outer membrane (OM) receptor (R) of a recipient cell by the RBD domain of CdiA, FHA-2 exposes and assembles into the recipient OM and translocates the CdiA-CT into the recipient periplasm. The entry domain then recognizes the IM receptor and translocates the toxin domain into the cytoplasm, where the toxin exerts its toxicity.
A contact-dependent process combined with the presence of CdiA at the cell surface of the attacker raised a question as to whether a cell surface receptor in the recipient cell is involved in docking/recognition of CdiA. If this was the case, then a variant of recipient cells for which the receptor is lacking or altered would become CDI-resistant (CDIR). Using CDIEC93 as a model, a transposon (Tn)-based mutagenesis screening led to the identification of such CDIR mutants that have Tn insertion in either acrB or the promoter region of bamA (Table 1; Aoki et al., 2008).
BamA is an OM protein at the core of the beta-barrel assembly machinery (BAM) complex and required for proper assembly/insertion of other beta-barrel proteins in the OM (Rigel and Silhavy, 2012). As bamA is an essential gene, the bamA-mutated CDIR mutant is not a null mutant but a knockdown mutant with five-fold less expression (Aoki et al., 2008). The biogenesis-inactive version of BamA, as well as other BAM complex variants, remains capable to mediate CDI, indicating that the presence of BamA but not the function of the BAM complex is required for CDI (Aoki et al., 2008). Treatment of bacterial cultures using anti-BamA antibody that recognizes the recipient BamA on the cell surface disrupted the attacker-recipient cell recognition and thus the CDI-mediated growth inhibition (Aoki et al., 2008). The results strongly support the idea that BamA is an OM receptor of the CdiAEC93.
Identification of the binding site between the CdiAEC93 toxin and BamA confirmed BamA as the receptor of CdiAEC93 (Figure 3). The RBD of CdiAEC93 (from Arg1358 to Phe1646) binds to BamA’s loop 6/loop 7 variable region that is identical in hundreds of other E. coli strains but shares low-sequence similarity among different CDI-encoding species (Ruhe et al., 2013, 2017). The results correlated well with the observation that CDIEC93 is unable to inhibit other CDI homologs-harboring species like Salmonella enterica serovar Typhimurium, Citrobacter freundii, Enterobacter aerogenes, Enterobacter cloacae, or Proteus mirabilis but was able to inhibit a variety of E. coli strains (Aoki et al., 2010; Ruhe et al., 2013). To summarize, the CdiAEC93 uses its RBD domain to bind specifically to the OM protein BamA of the E. coli recipient, demonstrating that CDI is restricted to intraspecies competition in a recipient receptor-dependent manner.
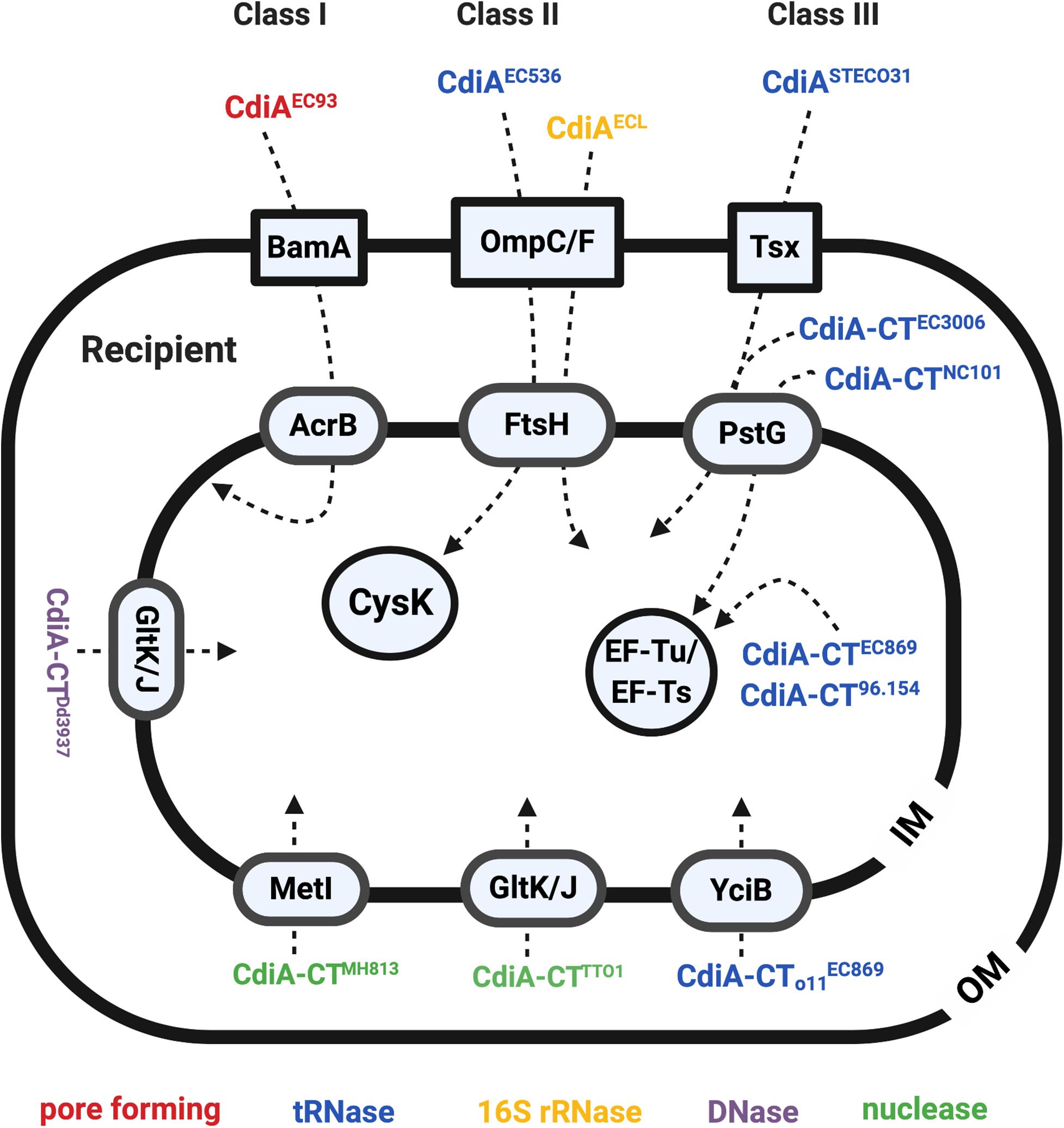
Figure 3. The recipient susceptibility factors that participate in exerting full toxicity of the CdiA. The CdiA toxins were classified into three different classes: the CdiAs that use BamA as outer membrane (OM) receptors are class I effector, the CdiAs that use the OmpC/F are the class II effector, and the ones that use Tsx are the class III effector. The OM, inner membrane (IM), and the cytosol proteins required for full toxicity of the CdiA are labeled in blue boxes in a square, oval, and circle, respectively. The CdiA functions in pore-forming toxins are labeled in red, tRNases are labeled in blue, 16S rRNases are labeled in yellow, DNases are labeled in purple, and the nuclease is labeled in green.
In contrast to CDIEC93, CdiA of the uropathogenic E. coli (UPEC) strain 536 (CdiAEC536) was shown to recognize the heterotrimeric OmpC-OmpF complex but not BamA (Figure 3; Beck et al., 2016). More specifically, the RBD region of the CdiAEC536 interacts with the extracellular loops L4 and L5 of OmpC (Beck et al., 2016; Ruhe et al., 2017). Unlike the binding region of BamA to CdiAEC93, which is highly conserved in protein sequence in hundreds of E. coli strains, the L4 and L5 of OmpC is highly diverse in protein sequence even among different E. coli strains (Aoki et al., 2008; Beck et al., 2016). Such OmpC polymorphism restricts the range of recipients for CDIEC536 (Beck et al., 2016). Although OmpF is strictly required for CDIEC536, using ompF alleles that are highly diverse from that of EC536 does not interfere with the CDIEC536 delivery process, consistent with the hypothesis that the recognition sites reside in OmpC. The data obtained on receptor preference or specificity of CDI from different E. coli strains suggest that E. coli may use the CDI systems to distinguish “self” from “non-self” cells and promote interactions between siblings.
After identifying the RBD region as the recipient OM receptor binding domain, the RBD region of all identifiable CdiA in the databae resoures of National Center for Biotechnology Information (NCBI) was compared to gain insights in binding specificity and selectivity toward either BamA or OmpC/OmpF (Ruhe et al., 2017). The results indicated that the CdiA RBD region can be divided into four main classes, instead of two, based on their amino acid sequences. Given the variability between the toxin classes, the CdiASTECO31 from E. coli STEC_O31 was used as a class III effector model to search for its CDIR mutants. The genetic screen of CdiASTECO31 CDIR mutants led to the discovery of a new receptor, namely, Tsx (Figure 3), an OM protein that functions as a monomeric nucleoside-specific porin (Bremer et al., 1990). Although the binding region of the Tsx remains elusive, the RBD region of the CdiASTECO31 lies in the Gln1385-Tyr1657 (Ruhe et al., 2017).
Besides identification of OM receptors, CDIR genetic screens also identified several genes encoding IM components (Figure 3; Aoki et al., 2008; Ruhe et al., 2014, 2017; Willett et al., 2015a). In the CDIR genetic screen using the attacker CDIEC93, acrB and bamA integrity in the prey cells were found mandatory for the attack to be effective. AcrB is an IM multidrug transport protein belonging to the multidrug/proton antiporter that is composed of AcrB, periplasmic protein AcrA, and OM protein TolC (Tikhonova and Zgurskaya, 2004). Intriguingly, only mutations in acrB but not acrA or tolC conferred resistance to the CDI, suggesting that the AcrB-mediated CDI is independent of its multidrug efflux pump function (Aoki et al., 2008). Of note, cells intoxicated with CdiAEC93 have reduced proton motive force and steady-state ATP levels, and their AcrB-containing multidrug/proton antiporter function is blocked (Aoki et al., 2009). These results suggested that CdiA-CTEC93 might interact with AcrB, thus resulting in dissipation of proton motive force (Aoki et al., 2009). Alternatively, AcrB could anchor the incoming CdiA-CTEC93 in the IM to activate the toxin that forms a pore (Jones et al., 2017).
The recipient’s IM factors required for CdiAEC536 is the filamenting temperature-sensitive H (FtsH) protein (Figure 3; Ruhe et al., 2014; Willett et al., 2015a). FtsH is an IM-anchored AAA+ protease, and its activity is stimulated by the proton motive force (Akiyama, 2002; Langklotz et al., 2012). As CdiA-CTEC536 is a well-defined tRNase that functions in the cytosol (Aoki et al., 2010; Ruhe et al., 2014), the role of FtsH is suggested to mediate toxin translocation across the IM. It is worth noting that CdiAEC536 and CdiAECL both require OmpC-OmpF heterotrimers and FtsH for toxicity (Willett et al., 2015a; Beck et al., 2016). However, the detailed mechanism of how FtsH is involved in CdiA toxicity remains elusive.
PtsG, the glucose-specific EIICB component of the sugar PTS (sugar phosphoenolpyruvate-dependent phosphotransferase) system, was found to be required for the toxicity of CdiA STECO31 (Gabor et al., 2011). As CdiA-CTSTECO31 encodes an EndoU anticodon nuclease that claves tRNAGlu in the cytosol (Michalska et al., 2018), the IM protein PtsG is believed to enable CdiA-CTSTECO31 translocation into the recipient’s cytosol. Further screening for CDIR mutants resisting intoxication for CdiA produced by a variety of different bacterial strains discovered that PtsG is also required for toxicity of CdiA-CTNC101 and CdiA-CT3006 (Willett et al., 2015a).
Additional recipient IM factors were also identified by screening for CDIR mutants resisting intoxication by CdiA produced by a variety of different bacterial strains (Willett et al., 2015a). The screening strategy was designed for identifying entry factors by using chimeric CdiA that harbors the N-terminus from E. coli strain EC93 and the C-terminal-containing toxin domain of other strains. The rationale is that the CdiA C-terminus (CdiA-CT) contains a variable domain that specifies the entry pathway into target bacteria and therefore recognizes and exploits specific proteins on the target cell for entry of the CdiA-CT toxin. Such screen has led to the discovery of six “permissive factors” conferring specific entry of different CDI toxins (Table 1 and Figure 3). Besides identifying known IM factor PtsG that is required for CdiA-CTNC101 and CdiA-CT3006, additional IM proteins including MetI for CdiA-CTMHI813, YciB for the orphan CdiA-CT of the EC869 (CdiA-CTo11EC869), GltK for Photorhabdus luminescens CdiA-CTTTO1, and RbsC for D. dadantii CdiA-CTDd3937 were uncovered (Figure 3 and Table 1; Willett et al., 2015a). Orphan cdiA-CTs encode toxins but have no translation initiation region and therefore are not translated unless grafted with a region encoding an N-terminal CdiA sequence (Poole et al., 2011).
It is worth mentioning that all the identified recipient proteins were IM protein, thus indicating that CdiA-CT is the region recognizing the recipient’s IM receptor but not the OM receptor. This finding is consistent with the evidence that the RBD but not the CdiA-CT region is responsible for binding to the OM receptor of CdiAEC93 (Figure 2; Ruhe et al., 2017). The authors also used chimeric CdiA-CTEC3006–EC869o11 to elucidate which part(s) of the CdiA-CT is responsible for recognition of the cognate IM receptor (Willett et al., 2015a). The CdiA-CTEC3006–EC869o11 consists of an N-terminal, CdiA-CT3006, and C-terminal fragments, CdiA-CTo11EC869, and requires PtsG but not YciB for growth inhibition. The results demonstrate that the IM receptor recognition domain of CdiA lies in the N-terminus of CdiA-CT and was thus designated as the entry domain, while the C-terminus of the CdiA-CT is the toxin domain itself (Willett et al., 2015a).
Cytoplasmic factors were also found to be required for effective CDI mechanism. In contrast to the roles of OM and IM factors involved in recognition and entry, cytoplasmic factors usually participate in enhancing toxin activity. The cytosolic factor of CdiAEC536 is the O-acetylserine sulfhydrylase A (CysK) (Figure 3; Diner et al., 2012; Beck et al., 2016). The requirement of CysK in antagonizing recipient growth stems from an unexpected result that CdiA-CTEC536 only displays tRNase activity in the presence of CysK both in vitro and in vivo (Diner et al., 2012). Crystal structure of the CysK/CdiA-CTEC536 complex revealed that CysK interacts with the C-terminal Gly-Tyr-Gly-Ile (GYGI) motif of CdiA-CTEC536, and this interaction increases the thermostability and tRNase activity of CdiA-CTEC536 (Johnson et al., 2016). Intriguingly, CysK also binds to and stabilizes the CdiA-CTEC536/CdiIEC536 complex in the attacker cell, and such binding reinforces protection against autoinhibition (Kaundal et al., 2016). The CysK/CdiA-CTEC536 interaction site mimics the binding site between CysK to its native substrate, CysE. Recent data demonstrated that CdiA-CTEC536 has a higher affinity to CysK even in the presence of excess CysE (Johnson et al., 2016; Jones et al., 2017). In brief, CdiA- CTEC536 utilizes CysK of the recipient cell to activate its tRNase activity once in the recipient cytosol.
Other recipient cytosolic factors required for CDI are the Elongation Factor Thermo-Unstable (EF-Tu) and the Elongation Factor Thermo-Stable (EF-Ts) (Figure 3; Jones et al., 2017; Michalska et al., 2017). The CdiA-CTEC869 toxin interacts with the EF-Tu/GTP/tRNA complex with high affinity. More importantly, the tRNase activity of CdiA-CTEC869 was only observed in the presence of this complex under in vitro conditions (Jones et al., 2017). Although EF-Ts was dispensable in activating CdiA-CTEC869 in vitro, it is required in vivo. The role of EF-Ts in vivo was proposed to be promoting the formation of the EF-Tu/GTP/tRNA complex. Aside from EC869, CdiA-CTs from strains NC101 and 96.154 also interact with EF-Tu, and both were unable to intoxicate a tsf mutant that lacks EF-Ts (Jones et al., 2017; Michalska et al., 2017). It is worth noting that CdiA-CTEC869, CdiA-CTNC101, and CdiA-CT96.154 share low-sequence similarity, suggesting that hijacking EF-Tu for activation may be a common strategy used by CDI toxins (Jones et al., 2017; Michalska et al., 2017).
As summarized above, the CDI system requires recipient membrane receptors and cytosolic activators to exert full toxicity. Exemplified by E. coli strains, a wide variety of the RS factors participates in recognition (OM receptor), translocation (IM proteins), and activity (cytoplasmic factors) of CDI toxins. Many other organisms harbor functional CDI, and they differ in gene organization, protein sequence, and cytotoxicity (Aoki et al., 2010; Kiel et al., 2012; De Gregorio et al., 2018; Allen and Hauser, 2019). As such, it is anticipated that novel CDI-dependent recipient receptors and activators would likely be discovered in future studies.
The T6SS was initially coined to be a virulence factor targeting eukaryotic hosts in many Gram-negative bacteria (Figure 1; Mougous et al., 2006; Pukatzki et al., 2006, 2007; Schell et al., 2007). Subsequent studies revealed that the T6SS could also target prokaryotic cells (Hood et al., 2010; MacIntyre et al., 2010; Schwarz et al., 2010). One of the first demonstrations came from P. aeruginosa on one of its T6SS substrate/effector Tse2 (Hood et al., 2010). Tse2 was toxic to E. coli and Burkholderia thailandensis when expressed ectopically, and this toxicity can be neutralized by the gene product encoded immediately downstream of tse2. The downstream gene was therefore named the tse2 immunity (tsi2). The authors also demonstrated that a P. aeruginosa strain lacking tse2-tsi2 lost fitness against its parental strain when the two strains were cocultured on solid but not liquid media and that this could be complemented by providing a plasmid-borne tsi2. The results demonstrated that T6SS uses the Tse2 toxin to gain fitness against a Tsi2-lacking sibling, and this occurred in a contact-dependent manner. In Vibrio cholerae, the T6SS-dependent antibacterial activity was shown against many Gram-negative bacteria, including Salmonella typhimurium, Citrobacter rodentium, and E. coli (MacIntyre et al., 2010). It has also been demonstrated that T6SS toxins can intoxicate a wide range of organisms including bacteria, archaea, fungi, and eukaryotic hosts (Coulthurst, 2019; Klein et al., 2020).
In contrast to CDI that employs a receptor-based recognition mechanism for toxin delivery at intraspecies levels, T6SS appears not to depend on a specific receptor for toxin delivery. T6SS’s action mold could explain it’s ability to target multiple organisms. The T6SS is composed of 13–14 core Type six secretion (Tss) proteins that are assembled in a structure highly similar to a contractile phage tail (Chang et al., 2017; Rapisarda et al., 2019; Wang et al., 2019). The current T6SS working model suggests that the formation of the membrane complex (MC) across the inner and outer membranes of the attacker cell is the first step in the assembly process (Figure 4). The membrane complex composed of (TssJ-)TssL-TssM (Ma et al., 2009; Rapisarda et al., 2019) functions as a scaffold for the recruitment of the baseplate (BP) complex and the effector-containing complex (ECC) for the initiation of the T6SS assembly (Brunet et al., 2015; Wang et al., 2019). The structure of the ECC resembles the tip of a spear that can puncture recipient cells (Basler and Mekalanos, 2012; Brunet et al., 2013). The BP serves as the docking site of the ECC and guides it to the MC. The BP is composed of TssE-TssF-TssG-TssK, and the ECC is composed of VgrG-(PAAR)-(adaptor)-effectors (Felisberto-Rodrigues et al., 2011; Brunet et al., 2015). The loading of the spear tip complex is believed to trigger polymerization of the spear handle that is composed of the Hcp inner tube and the TssB-TssC outer sheath (Figure 4; Mougous et al., 2006; Basler et al., 2012; Lossi et al., 2013; Zhang et al., 2013; Liang et al., 2019; Wu et al., 2020). When triggered, the outer sheath contracts and propels the inner tube and the “spear tip,” ECC, likely through the membrane complex scaffold to puncture the membrane of a recipient cell (Basler et al., 2012, 2013). The collective knowledge suggests that the toxin delivery to the recipient cell is through a mechanical force, rather than upon specific receptor binding (Figure 4).
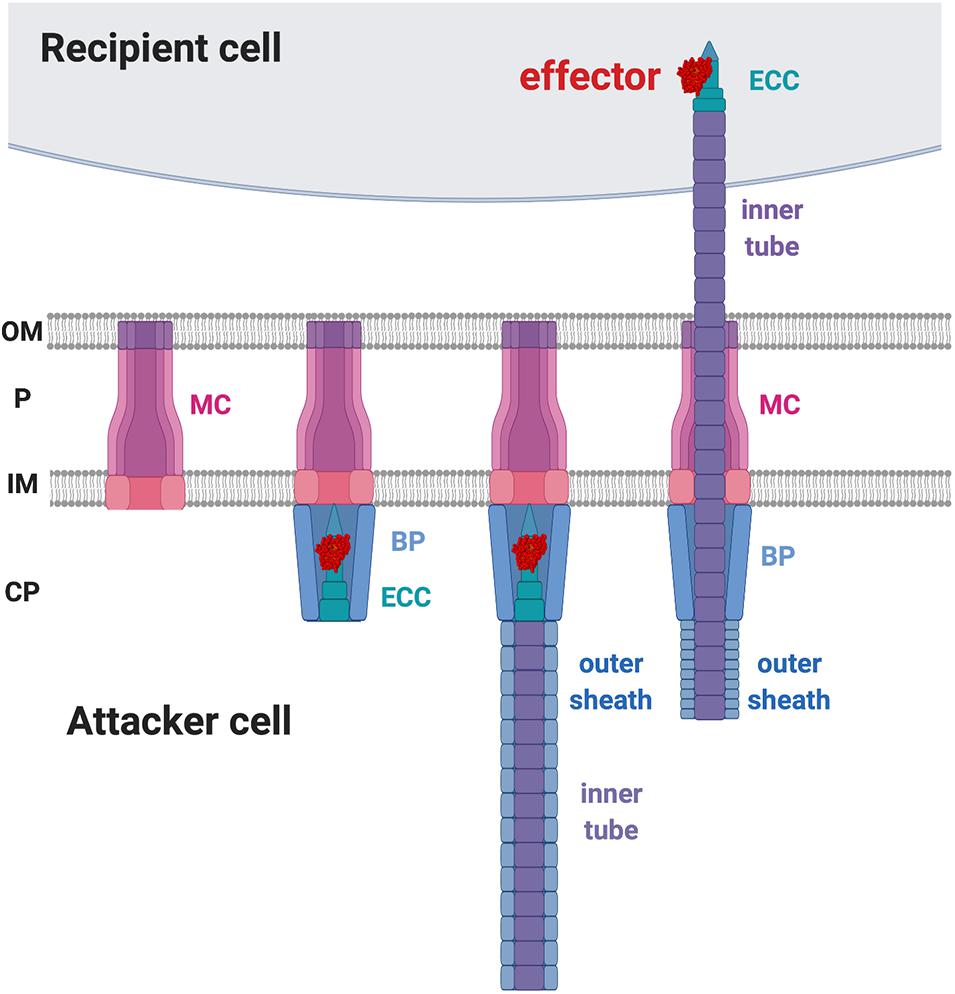
Figure 4. Type VI secretion system (T6SS) working model. The first step of T6SS assembly is the formation of the membrane complex (MC). The second step is the recruitment of the baseplate (BP) complex and the effector-containing complex (ECC) to the MC. The third step is the polymerization of the inner tube and the outer sheath inside the attacker cytosol. Upon trigger, the outer sheath contracts and propels the inner tube to penetrate the recipient membrane. OM, outer membrane; P, periplasm; IM, inner membrane; CP, cytoplasm.
The initial clues for recipient factors affecting the outcome of T6SS killing came from microscopic observations of T6SS firing events (Basler and Mekalanos, 2012; LeRoux et al., 2012). T6SS firing events were monitored by visualization of ClpV-GFP as ClpV is required for disassembly of contracted T6SS sheath, an event subsequent to T6SS firing (Bönemann et al., 2009; Pietrosiuk et al., 2011). The presence of ClpV-GFP foci thus indicates that T6SS firing has just happened (Mougous et al., 2006; Basler and Mekalanos, 2012). It was observed that P. aeruginosa ClpV-GFP foci occurred at the exact place where its neighboring sibling cells also had a ClpV-GFP foci, indicating that one of the activating signals for P. aeruginosa T6SS firing is the T6SS attack from a neighbor sibling cell (Basler and Mekalanos, 2012; LeRoux et al., 2012). This phenomenon was then demonstrated in an interspecies T6SS competition scenario. When punctured by the V. cholerae T6SS, P. aeruginosa fires back using its T6SS at the exact position where it was challenged (Basler et al., 2013). Similarly, Agrobacterium tumefaciens T6SS also triggers a P. aeruginosa counterattack, which led to higher killing of T6SS-active A. tumefaciens as compared to T6SS-inactive strains (Ma et al., 2014). Interestingly, the T6SS counterattack also occurs when sensing the pKM101 T4SS mating pair formation (Mpf) system of E. coli donor cells to resist T4SS-mediated gene transfer of foreign DNA (Ho et al., 2013). Because T6SS firing is also induced by membrane-disrupting compounds such as polymyxin B, the authors concluded that the T6SS counterattack results from Mpf-mediated membrane disruption. Recent studies further showed that the production of two adhesins (TraC and Pep), or the formation of a T4SS channel, but not assembly of conjugative pilus, is capable of activating a T6SS counterattack (Gordon et al., 2017; González-Rivera et al., 2019). Therefore, T6SS firing could be a defensive weapon in response to various assaults challenging membrane integrity (Figure 5).
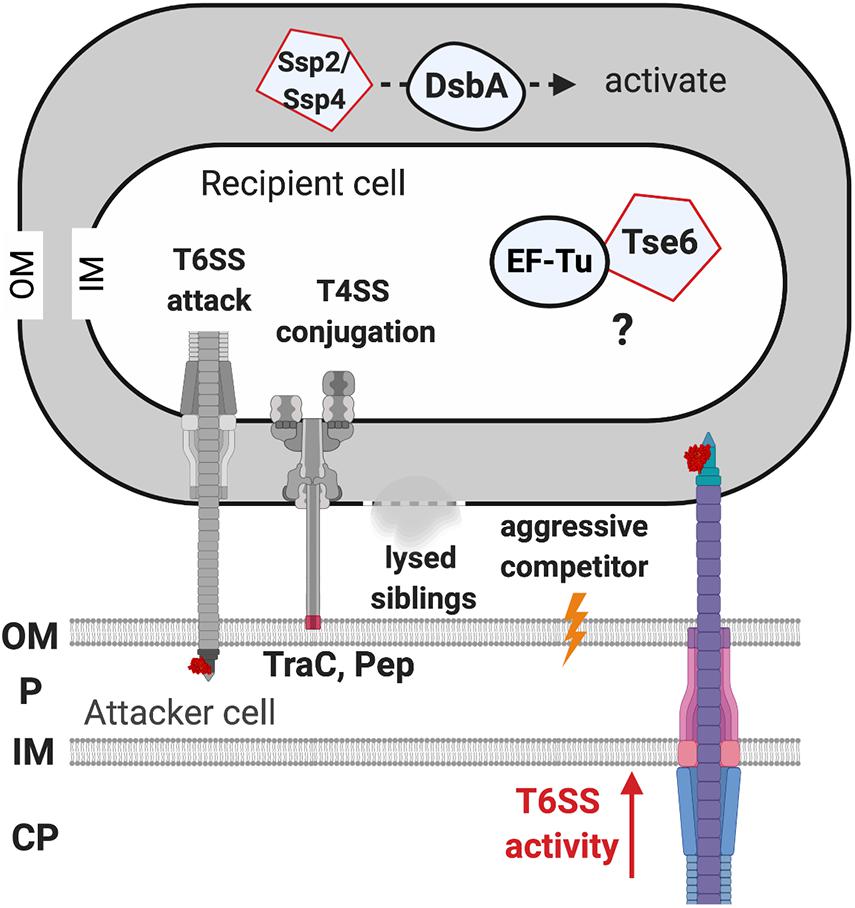
Figure 5. The recipient susceptibility factors enhancing the outcome of the type VI secretion system (T6SS) attack. The T6SS attack from the recipient cell triggers a T6SS counterattack. The T4SS-mediated membrane disruption that is caused by the adhesin TraC and Pep is required for activating the T6SS attack. Lysed siblings, aggressive competitors, and the non-self-competitors also trigger a T6SS attack. The elongation factor thermo-unstable (EF-Tu) of the recipient cell may be required for Tse6 to exert full toxicity, but the mechanism remains elusive. The DsbA of the recipient cell is required to activate the Ssp2 and Ssp4 periplasmic toxins. OM, outer membrane; P, periplasm; IM, inner membrane; CP, cytoplasm.
It has been demonstrated in multiple systems that the T6SS attack could be fine-tuned in response to different recipient cells (Ma et al., 2014; LeRoux et al., 2015; Lazzaro et al., 2017; Wu et al., 2019). For example, in P. aeruginosa, a non-self-recipient cell triggers a stronger T6SS attack than a susceptible sibling (LeRoux et al., 2015). Furthermore, the P. aeruginosa T6SS activity monitored by ClpV1-GFP was significantly elevated when cocultured with B. thailandensis as compared to a monoculture (LeRoux et al., 2012). The authors demonstrated that P. aeruginosa senses a “danger signal” released by lysed sibling cells and activates its T6SS to launch a counterattack (LeRoux et al., 2015). The enhanced T6SS susceptibility triggered by non-self-recipient cells was also demonstrated in A. tumefaciens (Ma et al., 2014). A. tumefaciens only exhibits antibacterial activity against E. coli but not against susceptible siblings in vitro (Ma et al., 2014). Furthermore, A. tumefaciens tends to antagonize other competitive A. tumefaciens strains from different genomospecies but not to the same degree to those within the same genomospecies in planta (Wu et al., 2019). In Serratia marcescens, the transcription level of T6SS is fine-tuned as the T6SS transcript level of S. marcescens varies when challenged by different competitors. Only basal levels of T6SS transcripts were detected when confronted with harmless recipient cells, while upregulation occurs at moderate or higher levels when confronted with contender or aggressive competitors (Lazzaro et al., 2017). Overall, these findings unveil the importance of kin recognition in determining the outcome of the T6SS attack, but future systematic analysis is required to identify the genetic features or determinants governing the fate of a competition (Figure 5).
The first evidence for the involvement of specific T6SS RS factors came from the characterization of the P. aeruginosa effector Tse6-loaded complex, which consists of Tse6, Tsi6 immunity protein, VgrG1, effector-associated gene with tse6 (EagT6), and EF-Tu (Whitney et al., 2015). The presence of EF-Tu in the Tse6-loaded complex was unexpected, and the authors addressed the role of EF-Tu by proposing four possibilities: EF-Tu may be required for (1) stabilizing Tse6, (2) activating Tse6, (3) facilitating Tse6 export from attacker cell, or (4) entering recipient cell. After ruling out the first three, the authors deduced that the interaction of Tse6 with EF-Tu might be required for entering the recipient cell. However, further study on the ability of the Tse6-loaded complex to translocate across membranes using liposome-based in vitro translocation assay showed that Tse6 translocation happened spontaneously in the absence of the inner-face EF-Tu (Quentin et al., 2018). Thus, EF-Tu may not play a role in entering recipient cells across the lipid bilayer, and the exact role of EF-Tu in the interbacterial competition is still to be elucidated.
Another example of RS factors affecting T6SS toxicity is DsbA that functions as a periplasmic disulfide bond-forming protein (Mariano et al., 2018). S. marcescens has two DsbAs, DsbA1 and DsbA2, which are functionally redundant for a proper T6SS functionality. Indeed, in S. marcescens-secreting cells, the presence of either DsbA1 or DsbA2 is sufficient for T6SS activity, but T6SS assembly and secretion levels are significantly compromised in the absence of DsbA1 and DsbA2. Strikingly, the peptidoglycan hydrolase Ssp2 and Ssp4 (English et al., 2012) are able to inhibit Ssp2- and Ssp4-susceptible S. marcescens strains, while a recipient lacking both DsbA1 and DsbA2 was entirely resistant against the activity of these periplasmic-acting effectors (Mariano et al., 2018). The requirement of DsbA for the toxicity of Ssp2 and Ssp4 was also confirmed by artificially expressing and targeting Ssp2 and Ssp4 to the E. coli periplasm, in which their toxicity is relieved if the E. coli strain lacks dsbA. Attacker cells expressing disulfide bond-lacking Ssp2 or Ssp4 did not show T6SS-mediated antibacterial activity. It is generally believed that T6SS delivers effectors from the attacker cell’s cytoplasm directly into the recipient cell, Ssp2 and Ssp4 effectors are unlikely to localize in the attacker cell’s periplasm to form a disulfide bond before its delivery. Thus, it remains unknown how DsbA influences T6SS activity in the attacker cell, but the contribution of DsbA or disulfide bond formation for activity of incoming periplasmic toxins in the recipient cell is likely a widespread mechanism (Figure 5; Mariano et al., 2018).
In A. tumefaciens, a high-throughput screening (HTS) aiming to identify RS factors that affect the T6SS killing outcome was performed (Lin et al., 2020). Using E. coli K12 strain BW25113 as the model recipient cell, several RS factors that enhance E. coli susceptibility to A. tumefaciens T6SS attack were identified. To date, the confirmed RS-encoding genes include clpA, clpP, gltA, ydhS, ydaE, and cbpA, all encoding cytosolic proteins. These results suggest that the RS factors affecting A. tumefaciens T6SS killing outcome are rather involved after injection of T6SS toxins into the recipient cells.
The clpP gene encoding ClpP protease is universal and highly conserved in both prokaryotes and eukaryotic organelles. Its activity depends on other adapter proteins such as ClpA or ClpX AAA+ ATPase for substrate recognition (Bhandari et al., 2018; Figaj et al., 2019). The authors showed that clpA but not clpX is required for enhancing susceptibility to A. tumefaciens T6SS killing, suggesting the involvement of ClpAP. ClpP variants deficient in ClpP protease activity or incapable of interacting with its adaptor protein could not restore T6SS effectiveness against a clpP knockout mutant, suggesting that ClpA–ClpP interaction and subsequent proteolysis are critical in enhancing susceptibility to T6SS killing. While the mode of action of recipient ClpAP complex involved in enhancing T6SS killing remains unknown, three hypotheses could be proposed for further testing. First, ClpAP complex may be used to enhance toxin activity, such as the Tde1 and/or Tde2 DNase activity, the major T6SS antibacterial weapons of A. tumefaciens strain C58 (Ma et al., 2014) used for the screen. Second, ClpAP complex could be hijacked by A. tumefaciens to trap or degrade an E. coli defense protein from inhibiting the activity of an incoming toxin. The third hypothesis is that the absence of a ClpAP system may result in substrate accumulation that interferes with T6SS firing or toxin activity of the attacker.
In summary, based on the broad spectrum of recipient cells that T6SS toxins act on, T6SS appears not to require recipient receptor for protein toxin entry. Current evidence suggests that specific RS factors may rather be used for the full activation of T6SS toxins once entering the recipient cells (Figure 5). However, future studies on the mode of action of identified RS factors and more comprehensive genetic screens are required to answer these questions. Besides RS factors, recent studies have revealed the presence of immunity-independent resistance in recipient cell that were nicely reviewed by Robitaille et al. (2020). These recipient defense factors or mechanisms include physical barriers such as exopolysaccharide (Toska et al., 2018), envelope stress responses (Hersch et al., 2020), or peptidoglycan editing (Le et al., 2020). Growing evidence of the involvement of recipient factors in either enhancing T6SS toxicity or defense against T6SS indicates an evolutionary arms race during interbacterial competition, which may play roles in shaping microbiome.
Recently, a novel Cdz system that requires the canonical T1SS proteins CdzA and CdzB has been described in Caulobacter crescentus (García-Bayona et al., 2017). The cdz operon consists of five genes encoding two T1SS components (CdzA IM component and CdzB membrane fusion protein) followed by two-peptide toxin (CdzC and CdzD) and the immunity protein CdzI (García-Bayona et al., 2017, 2019). The CdzC/CdzD two-peptide toxin kills the target cell by membrane depolarization, and its toxicity is neutralized by the immunity protein CdzI (García-Bayona et al., 2017). In contrast to log phase-specific CDI, the Cdz is stationary phase-specific. The transcript of the cdz operon and the gene products are highly induced in the stationary phase, while the Cdz protein levels are not detectable in the log phase.
The Cdz system is not species-dependent and can antagonize other closely related species. The Cdz of C. crescentus was able to inhibit a CdzI immunity protein-lacking sibling, Caulobacter segnis, and Brevundimonas subvibrioides sp. Poindexter. However, the C. crescentus Cdz was not able to antagonize Asticcacaulis excentricus, which also belongs to the Caulobacteraceae family but is more distantly related to C. crescentus, and other even more distantly related bacteria like A. tumefaciens or E. coli (García-Bayona et al., 2017). This implied that the T1SS-mediated growth inhibition by Cdz only occurs between close-related lineage but at broader scope than CDI. As the Cdz system can be found in Firmicutes, alpha-proteobacteria, beta-proteobacteria, and particularly widespread in gamma-proteobacteria, the Cdz is another common contact-dependent antibacterial strategy used by bacteria to thrive in the environment (García-Bayona et al., 2017).
A more recent study searching for recipient cells resistant to C. crescentus Cdz-killing led to the identification of a previously uncharacterized gene ccna_01968 (García-Bayona et al., 2019). The ccna_01968 was renamed as the pentapeptide envelope resistance A (perA) gene as it encodes a quadrilateral beta-helix protein. Biochemical data and microscopy observation demonstrated that PerA is a surface-exposed OM protein. The perA mutant strains resistant to Cdz were sensitized again by expressing perA in trans, suggesting that PerA may act as the receptor of CdzC/CdzD toxin (García-Bayona et al., 2019). The mode of action of PerA and whether additional RS factors in recipient cells are involved in Cdz-mediated antibacterial activity require further in-depth molecular studies and genetic screens.
The approaches used to identify RS factors were mostly by screening mutant libraries for resistant recipient cells. This led to the discovery of multiple RS genes involved in maximizing the toxicity of CDI, T6SS, or T1SS Cdz. Other RSs were identified through knowledge-based approaches such as on the basis of their association with the toxin either physically or biochemically. Here, we summarize the methods used and discuss potential strategies for the discovery of novel recipient factors.
Genetic screen is proven to be a powerful and non-biased method for identifying RS factors, which is applicable to any contact-dependent antibacterial system. As summarized in Table 2, the selection of resistant strains can be screened from identifying the survivors of a mutant library pool cocultured with attacker/recipient cells. The mutations responsible for the resistance phenotype can be later identified by complementation using a genomic library (Aoki et al., 2008; Ruhe et al., 2014, 2017; Willett et al., 2015a; Jones et al., 2017) or by whole-genome sequencing (García-Bayona et al., 2019). With the availability of the E. coli Keio library containing 3,909 knockout mutant strains (Baba et al., 2006), an HTS with the aid of pipetting robot and 96-well systems was established to screen E. coli recipient factors (Lin et al., 2020). Such screen can lead to the immediate identification of gene of interest without complementation by a genomic library and/or sequencing. However, the use of knockout mutant library cannot identify RS genes that are essential for bacterial growth. Thus, CRISPR interfering (CRISPRi) using a catalytic null mutant of the Cas9 endonuclease, dCas9, and guide RNA (gRNA) library (Cui et al., 2018) serves as an alternative and complementary method to screen for recipient factors that are not available in knockout or Tn-insertion mutant libraries. The availability of E. coli CRISPRi gRNA library (Addgene, Watertown, MA, United States) created by the Bikard lab enables such screen in E. coli and can be expanded to other bacterial species. A series of broad host range vectors that carry the dcas9 gene under control of the ptet promoter and the gRNA under control of a constitutive promoter are available for future applications in many Proteobacterial species (Depardieu and Bikard, 2020).
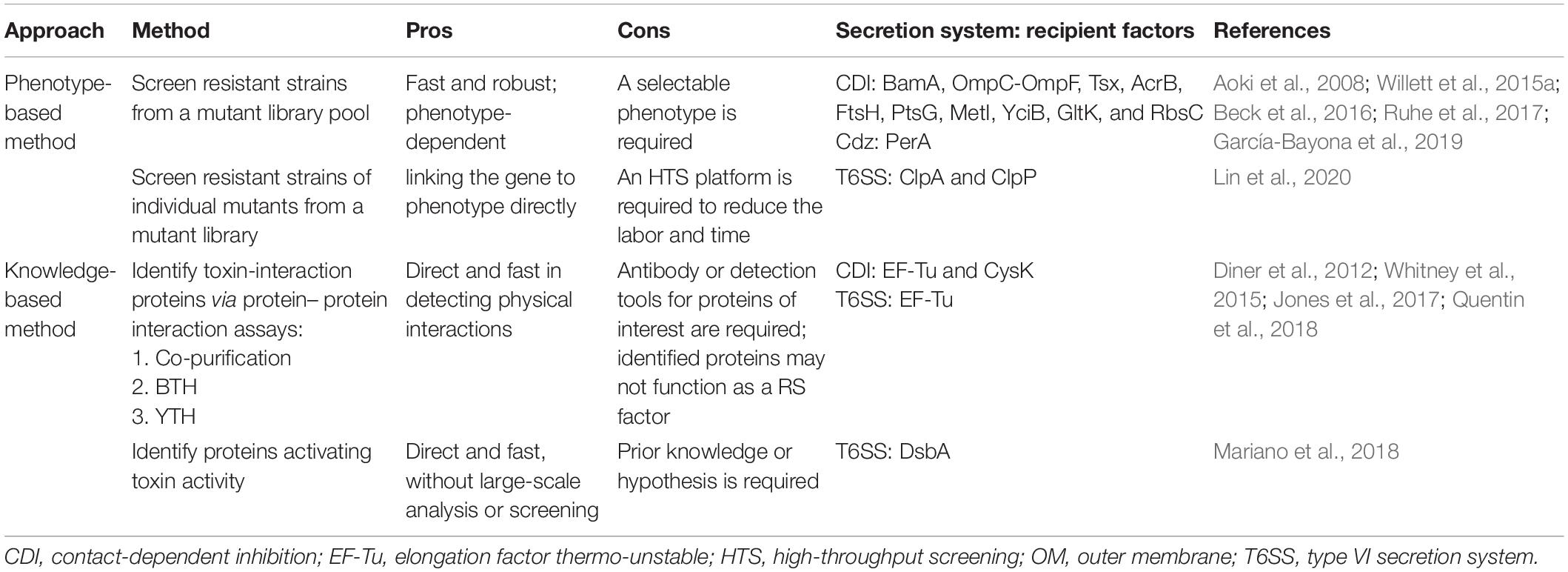
Table 2. Summary of current and potential methods for discovery of recipient susceptibility (RS) factors.
The major roles of RS factors are in recognition, entry, or activation of the toxins. Thus, an approach to identify recipient factors is to search for toxin-interacting proteins. Indeed, EF-Tu, the common RS factor involved in CDI and potentially for T6SS, was identified as one of the components taking part in the toxin–immunity protein complexes (Jones et al., 2017). Thus, co-expression of toxin–immunity complex followed by co-immunoprecipitation or pulldown assay (Brymora et al., 2004; Kaboord and Perr, 2008; Lin and Lai, 2017a) can lead to the discovery of toxin-interacting proteins. This serves as a straightforward method to identify RS factors that may play a role in toxin entry or activation. In addition, the toxin proteins can be used as a bait in well-established protein–protein interaction platforms such as bacterial two-hybrid (BTH) (Battesti and Bouveret, 2012) or yeast two-hybrid (YTH) (Mehla et al., 2015; Lin and Lai, 2017b) to identify potential RS factors by screening a recipient genomic library.
RS factors that are hijacked to activate toxin activity can be identified based on the knowledge of the toxin’s mode of action. For example, periplasmic disulfide bond-forming protein DsbA that is known to be required for folding or stabilization of proteins located in the periplasm could be critical for activity of periplasmic bacterial toxins such as peptidoglycan hydrolases and phospholipase (Kadokura and Beckwith, 2010). Based on this knowledge, the role of DsbA in T6SS-mediated antibacterial activity of S. marcescens was investigated and found to be required for the activity of the peptidoglycan hydrolase Ssp2 and the periplasmic toxin Ssp4 (Mariano et al., 2018). It is possible that DsbA plays a broader role for toxin activation delivered by multiple antibacterial systems. Besides DsbA, involvement of the ClpAP protease in T6SS susceptibility (Lin et al., 2020) also suggested that various types of proteases may be used for activating toxin activity by either cleaving full-length toxin proteins into more active truncated forms or degrading proteins that may inhibit toxin activity. A recent report showed that self-cleavage at both the N- and C- termini of an Rhs-family T6SS toxin TseI is not required for secretion but critical for its toxin activity (Pei et al., 2020). This finding also suggests that protease cleavage could be a strategy used for toxin activation in the recipient cell. Since the mechanism for N-terminal cleavage of TseI remains unknown, it may be mediated by an unknown protease residing in the recipient cell. Future work to test these potential modifying enzymes in activating antibacterial toxins of various systems shall shed light to understand the molecular basis of toxin action once they are translocated into the recipient cells.
Understanding the mode of action of antibacterial toxins and their target spectra may help us develop novel antibacterial therapies in biomedical and agricultural applications (Sana et al., 2017; Bernal et al., 2018; Trunk et al., 2018; Khakhum et al., 2019; Allsopp et al., 2020). For example, accumulating evidence indicated that T6SSs in commensal bacteria such as Bacteroides fragilis and Pseudomonas protegens play a critical role in the defense against invading bacterial pathogens and impact microbial community in the gut of mammalian and insect, respectively (Chatzidaki-Livanis et al., 2016; Wexler et al., 2016; Vacheron et al., 2019). T6SS is also widespread in plant-associated beneficial bacteria such as Pseudomonas putida and Pseudomonas fluorescens functioning as a biocontrol agent in protecting plants with their antagonistic activity against bacterial and fungal pathogens (Decoin et al., 2014; Bernal et al., 2017). However, these beneficial bacteria are also susceptible to killing by competitor bacteria equipped with antibacterial weapons. Thus, engineering commensal bacteria to protect from or defend against pathogenic bacteria in a polymicrobial community may be beneficial for human and plant health.
Based on the current knowledge, we proposed three strategies for defense against pathogens in a polymicrobial community (Figure 6). One conventional way is engineering strains with specific or arrays of various immunity genes that may offer broad-spectrum protection (Sana et al., 2017; Trunk et al., 2018; Khakhum et al., 2019). With the understanding of the RS factors, alternative approaches could be designed in these commensal bacteria with better survival and competitive capacity. First, engineering the strains with deletion or point mutation in the common RS gene can increase the resistance against killing from various bacteria harboring multiple antibacterial weapons. The common RS factor EF-Tu utilized by both CDI and perhaps T6SS for enhanced killing is a potential RS target. However, since EF-Tu is an essential gene, the detailed molecular mechanisms and amino acid residues critical for toxicity enhancement are required prior to engineer the EF-Tu variant combining proper physiological function and resistance to antibacterial killing. Second, these RS factors can be ideal targets to screen natural products or synthetic chemicals to shut down their expression or ability in enhancing toxin entry or activity. This method offers advantages to bypass genetic modification and more flexibility in temporal and spatial control for such applications.
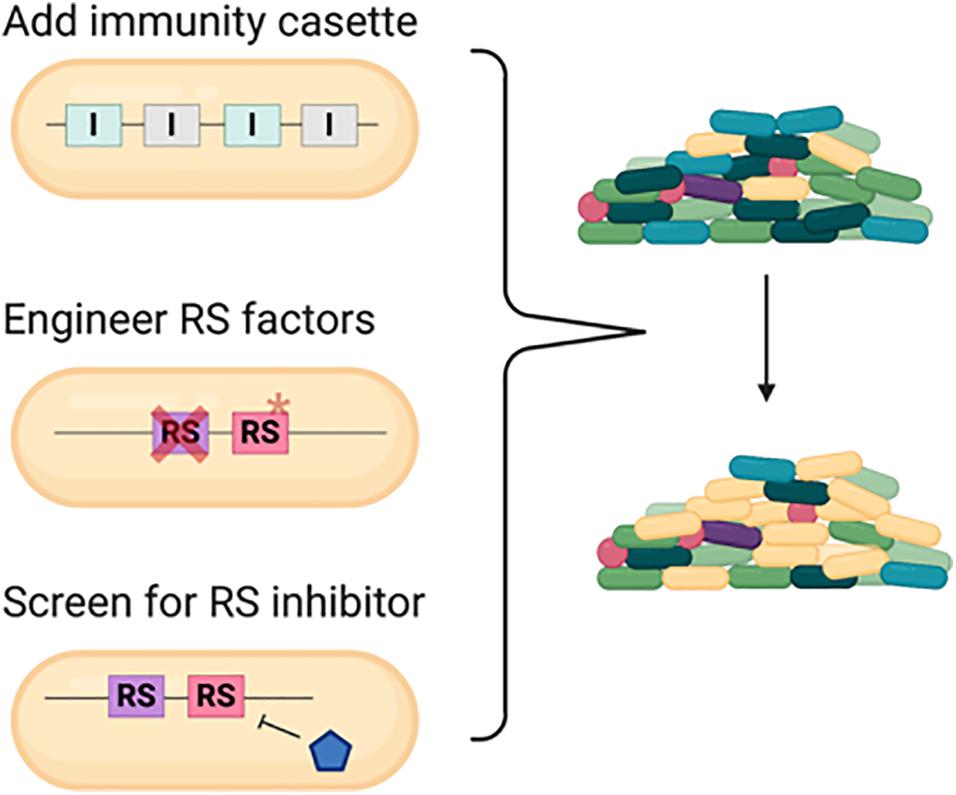
Figure 6. Strategies to engineer commensal bacteria for protection from or defense against pathogens in a polymicrobial community. Conventional strategy is to add an array of immunity gene cassette to the commensal bacteria. With the understanding of the recipient susceptibility (RS) factor, disrupting the RS factor and/or screening for specific inhibitor to conditionally inhibit the RS factor could also serve as novel methods. Cross represents deletion and asterisk represents mutation.
In conclusion, bacteria have deployed versatile bacterial secretion systems as antibacterial weapons for fitness and survival. Similar to the arms race between hosts and pathogens, the bacterial attackers evolve to recognize or hijack recipient cell factors to maximize the antagonism by enhancing the entry or toxicity of bacterial toxins. It is also worth mentioning that some of the recipient proteins are attacking “hotspots.” For example, the OmpC/OmpF OM receptor is the target of both CdiAEC536 and CdiAECL, the IM receptor PstG is the common translocator for multiple CdiA proteins, and the cytoplasmic factor EF-Tu is targeted by multiple CdiA proteins and perhaps Tse6 (Table 1). For receptor-mediated recognition and antibacterial activity at intraspecies levels, different secretion systems tend to target the same or highly similar receptors. We argue that future identification of more RS factors involved in toxins transported by different secretion systems may reveal more toxins targeting “hotspots” to further accelerate the development of novel antibacterial therapies in biomedical and agricultural applications.
H-HL, AF, and E-ML conceived the review. H-HL and E-ML wrote the first draft. All the authors contributed to complete the final version of the manuscript.
Funding for the Lai laboratory and H-HL is Academia Sinica Investigator Award to E-ML (grant no. AS-IA-107-L01). Funding for AF is MRC grant MR/S02316X/1.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All figures were created with BioRender.
Akiyama, Y. (2002). Proton-motive force stimulates the proteolytic activity of FtsH, a membrane-bound ATP- dependent protease in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 8066–8071. doi: 10.1073/pnas.122616899
Allen, J. P., and Hauser, A. R. (2019). Diversity of contact-dependent growth inhibition systems of Pseudomonas aeruginosa. J. Bacteriol. 201:e00776-18.
Allsopp, L. P., Bernal, P., Nolan, L. M., and Filloux, A. (2020). Causalities of war: the connection between type VI secretion system and microbiota. Cell. Microbiol. 22:e13153.
Aoki, S. K., Diner, E. J., De Roodenbeke, C. T. K., Burgess, B. R., Poole, S. J., Braaten, B. A., et al. (2010). A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468:439. doi: 10.1038/nature09490
Aoki, S. K., Malinverni, J. C., Jacoby, K., Thomas, B., Pamma, R., Trinh, B. N., et al. (2008). Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol. Microbiol. 70, 323–340. doi: 10.1111/j.1365-2958.2008.06404.x
Aoki, S. K., Pamma, R., Hernday, A. D., Bickham, J. E., Braaten, B. A., and Low, D. A. (2005). Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248. doi: 10.1126/science.1115109
Aoki, S. K., Webb, J. S., Braaten, B. A., and Low, D. A. (2009). Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J. Bacteriol. 191, 1777–1786. doi: 10.1128/jb.01437-08
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008.
Basler, M., Ho, B. T., and Mekalanos, J. J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. doi: 10.1016/j.cell.2013.01.042
Basler, M., and Mekalanos, J. J. (2012). Type 6 secretion dynamics within and between bacterial cells. Science 337:815. doi: 10.1126/science.1222901
Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J., and Mekalanos, J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. doi: 10.1038/nature10846
Battesti, A., and Bouveret, E. (2012). The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334. doi: 10.1016/j.ymeth.2012.07.018
Beck, C. M., Willett, J. L. E., Cunningham, D. A., Kim, J. J., Low, D. A., and Hayes, C. S. (2016). CdiA effectors from uropathogenic Escherichia coli use heterotrimeric osmoporins as receptors to recognize target bacteria. PLoS Pathog. 12:e1005925. doi: 10.1371/journal.ppat.1005925
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi: 10.1038/ismej.2016.169
Bernal, P., Llamas, M. A., and Filloux, A. (2018). Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 20, 1–15. doi: 10.1111/1462-2920.13956
Bhandari, V., Wong, K. S., Zhou, J. L., Mabanglo, M. F., Batey, R. A., and Houry, W. A. (2018). The role of ClpP protease in bacterial pathogenesis and human diseases. ACS Chem. Biol. 13, 1413–1425. doi: 10.1021/acschembio.8b00124
Bönemann, G., Pietrosiuk, A., Diemand, A., Zentgraf, H., and Mogk, A. (2009). Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325. doi: 10.1038/emboj.2008.269
Bremer, E., Middendorf, A., Martinussen, J., and Valentin-Hansen, P. (1990). Analysis of the tsx gene, which encodes a nucleoside-specific channel-forming protein (Tsx) in the outer membrane of Escherichia coli. Gene 96, 59–65. doi: 10.1016/0378-1119(90)90341-n
Brunet, Y. R., Espinosa, L., Harchouni, S., Mignot, T., and Cascales, E. (2013). Imaging type VI secretion-mediated bacterial killing. Cell Rep. 3, 36–41. doi: 10.1016/j.celrep.2012.11.027
Brunet, Y. R., Zoued, A., Boyer, F., Douzi, B., and Cascales, E. (2015). The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 11:e1005545. doi: 10.1371/journal.pgen.1005545
Brymora, A., Valova, V. A., and Robinson, P. J. (2004). Protein-protein interactions identified by pull-down experiments and mass spectrometry. Curr. Protoc. Cell Biol. 22, 17.5.1–17.5.51.
Cao, Z., Casabona, M. G., Kneuper, H., Chalmers, J. D., and Palmer, T. (2016). The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol. 2:16183.
Cascales, E., Buchanan, S. K., Duché, D., Kleanthous, C., Lloubès, R., Postle, K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229.
Chang, Y.-W., Rettberg, L. A., Ortega, D. R., and Jensen, G. J. (2017). In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep. 18, 1090–1099. doi: 10.15252/embr.201744072
Chatzidaki-Livanis, M., Geva-Zatorsky, N., and Comstock, L. E. (2016). Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. U.S.A. 113, 3627–3632. doi: 10.1073/pnas.1522510113
Christie, P. J. (2019). The rich tapestry of bacterial protein translocation systems. Protein J. 38, 389–408. doi: 10.1007/s10930-019-09862-3
Costa, T. R. D., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Micro. 13, 343–359. doi: 10.1038/nrmicro3456
Coulthurst, S. (2019). The Type VI secretion system: a versatile bacterial weapon. Microbiology 165, 503–515. doi: 10.1099/mic.0.000789
Cui, L., Vigouroux, A., Rousset, F., Varet, H., Khanna, V., and Bikard, D. (2018). A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 9:1912.
De Gregorio, E., Esposito, E. P., Zarrilli, R., and Di Nocera, P. P. (2018). Contact-dependent growth inhibition proteins in Acinetobacter baylyi ADP1. Curr. Microbiol. 75, 1434–1440. doi: 10.1007/s00284-018-1540-y
Decoin, V., Barbey, C., Bergeau, D., Latour, X., Feuilloley, M. G. J., Orange, N., et al. (2014). A type VI secretion system is involved in Pseudomonas fluorescens bacterial competition. PLoS One 9:e89411. doi: 10.1371/journal.pone.0089411
Depardieu, F., and Bikard, D. (2020). Gene silencing with CRISPRi in bacteria and optimization of dCas9 expression levels. Methods 172, 61–75. doi: 10.1016/j.ymeth.2019.07.024
Diner, E. J., Beck, C. M., Webb, J. S., Low, D. A., and Hayes, C. S. (2012). Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI). Genes Dev. 26, 515–525. doi: 10.1101/gad.182345.111
English, G., Trunk, K., Rao, V. A., Srikannathasan, V., Hunter, W. N., and Coulthurst, S. J. (2012). New secreted toxins and immunity proteins encoded within the type VI secretion system gene cluster of Serratia marcescens. Mol. Microbiol. 86, 921–936. doi: 10.1111/mmi.12028
Felisberto-Rodrigues, C., Durand, E., Aschtgen, M.-S., Blangy, S., Ortiz-Lombardia, M., Douzi, B., et al. (2011). Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 7:e1002386. doi: 10.1371/journal.ppat.1002386
Figaj, D., Ambroziak, P., Przepiora, T., and Skorko-Glonek, J. (2019). The role of proteases in the virulence of plant pathogenic bacteria. Int. J. Mol. 20:672. doi: 10.3390/ijms20030672
Filloux, A., and Sagfors, A. (2015). “3 - News and views on protein secretion systems,” in The Comprehensive Sourcebook of Bacterial Protein Toxins (Fourth Edition), eds J. Alouf, D. Ladant, and M. R. Popoff (Boston: Academic Press), 77–108. doi: 10.1016/b978-0-12-800188-2.00003-3
Foster, K. R., and Bell, T. (2012). Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850. doi: 10.1016/j.cub.2012.08.005
Gabor, E., Göhler, A.-K., Kosfeld, A., Staab, A., Kremling, A., and Jahreis, K. (2011). The phosphoenolpyruvate-dependent glucose–phosphotransferase system from Escherichia coli K-12 as the center of a network regulating carbohydrate flux in the cell. Eur. J. Cell Biol. 90, 711–720. doi: 10.1016/j.ejcb.2011.04.002
García-Bayona, L., Gozzi, K., and Laub, M. T. (2019). Mechanisms of resistance to the contact-dependent bacteriocin CdzC/D in Caulobacter crescentus. J. Bacteriol. 201:e00538-18.
García-Bayona, L., Guo, M. S., and Laub, M. T. (2017). Contact-dependent killing by Caulobacter crescentus via cell surface-associated, glycine zipper proteins. eLife 6:e24869.
Ghoul, M., and Mitri, S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. doi: 10.1016/j.tim.2016.06.011
González-Rivera, C., Khara, P., Awad, D., Patel, R., Li, Y. G., Bogisch, M., et al. (2019). Two pKM101-encoded proteins, the pilus-tip protein TraC and Pep, assemble on the Escherichia coli cell surface as adhesins required for efficient conjugative DNA transfer. Mol. Microbiol. 111, 96–117. doi: 10.1111/mmi.14141
Gordon, J. E., Costa, T. R. D., Patel, R. S., Gonzalez-Rivera, C., Sarkar, M. K., Orlova, E. V., et al. (2017). Use of chimeric type IV secretion systems to define contributions of outer membrane subassemblies for contact-dependent translocation. Mol. Microbiol. 105, 273–293. doi: 10.1111/mmi.13700
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4:10.1128. doi: 10.1128/microbiolspec.VMBF-0012-2015
Hersch, S. J., Watanabe, N., Stietz, M. S., Manera, K., Kamal, F., Burkinshaw, B., et al. (2020). Envelope stress responses defend against type six secretion system attacks independently of immunity proteins. Nat. Microbiol. 5, 706–714. doi: 10.1038/s41564-020-0672-6
Ho, B. T., Basler, M., and Mekalanos, J. J. (2013). Type 6 secretion system–mediated immunity to type 4 secretion system–mediated gene transfer. Science 342, 250–253. doi: 10.1126/science.1243745
Hood, R. D., Singh, P., Hsu, F., G’Vener, T. Z. N., Carl, M. A., Trinidad, R. R. S., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Johnson, P. M., Beck, C. M., Morse, R. P., Garza-Sánchez, F., Low, D. A., Hayes, C. S., et al. (2016). Unraveling the essential role of CysK in CDI toxin activation. Proc. Natl. Acad. Sci. U.S.A. 113, 9792–9797. doi: 10.1073/pnas.1607112113
Jones, A. M., Garza-Sánchez, F., So, J., Hayes, C. S., and Low, D. A. (2017). Activation of contact-dependent antibacterial tRNase toxins by translation elongation factors. Proc. Natl. Acad. Sci. U.S.A. 114, E1951–E1957.
Kaboord, B., and Perr, M. (2008). Isolation of proteins and protein complexes by immunoprecipitation. Methods Mol. Biol. 424, 349–364. doi: 10.1007/978-1-60327-064-9_27
Kadokura, H., and Beckwith, J. (2010). Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid. Redox Signal. 13, 1231–1246. doi: 10.1089/ars.2010.3187
Kaundal, S., Uttam, M., and Thakur, K. G. (2016). Dual role of a biosynthetic enzyme, CysK, in contact dependent growth inhibition in bacteria. PLoS One 11:e0159844. doi: 10.1371/journal.pone.0159844
Khakhum, N., Bharaj, P., Myers, J. N., Tapia, D., Kilgore, P. B., Ross, B. N., et al. (2019). Burkholderia pseudomallei ΔtonB Δhcp1 live attenuated vaccine strain elicits full protective immunity against aerosolized melioidosis infection. mSphere 4:e00570-18.
Kiel, N., Amber, S., Wilbur, J. S., Diner, E. J., Aoki, S. K., Poole, S. J., et al. (2012). The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol. Microbiol. 84, 516–529. doi: 10.1111/j.1365-2958.2012.08039.x
Klein, T. A., Ahmad, S., and Whitney, J. C. (2020). Contact-dependent interbacterial antagonism mediated by protein secretion machines. Trends Microbiol. 28, 387–400. doi: 10.1016/j.tim.2020.01.003
Langklotz, S., Baumann, U., and Narberhaus, F. (2012). Structure and function of the bacterial AAA protease FtsH. Biochim. Biophys. Acta 1823, 40–48. doi: 10.1016/j.bbamcr.2011.08.015
Lazzaro, M., Feldman, M. F., and García Véscovi, E. (2017). A transcriptional regulatory mechanism finely tunes the firing of type VI Secretion system in response to bacterial enemies. mBio 8:e00559-17.
Le, N.-H., Peters, K., Espaillat, A., Sheldon, J. R., Gray, J., Di Venanzio, G., et al. (2020). Peptidoglycan editing provides immunity to Acinetobacter baumannii during bacterial warfare. Sci. Adv. 6:eabb5614. doi: 10.1126/sciadv.abb5614
LeRoux, M., De Leon, J. A., Kuwada, N. J., Russell, A. B., Pinto-Santini, D., Hood, R. D., et al. (2012). Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc. Natl. Acad. Sci. U.S.A. 109, 19804–19809. doi: 10.1073/pnas.1213963109
LeRoux, M., Kirkpatrick, R. L., Montauti, E. I., Tran, B. Q., Peterson, S. B., Harding, B. N., et al. (2015). Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife 4:e05701.
Liang, X., Kamal, F., Pei, T.-T., Xu, P., Mekalanos, J. J., and Dong, T. G. (2019). An onboard checking mechanism ensures effector delivery of the type VI secretion system in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 116, 23292–23298. doi: 10.1073/pnas.1914202116
Lien, Y.-W., and Lai, E.-M. (2017). Type VI secretion effectors: methodologies and biology. Front. Cell Infect. Microbiol. 7:254. doi: 10.3389/fcimb.2017.00254
Lin, H.-H., Yu, M., Sriramoju, M. K., Hsu, S.-T. D., Liu, C.-T., and Lai, E.-M. (2020). A high-throughput interbacterial competition screen identifies ClpAP in enhancing recipient susceptibility to type VI secretion system-mediated attack by Agrobacterium tumefaciens. Front. Microbial. 10:3077. doi: 10.3389/fmicb.2019.03077
Lin, J.-S., and Lai, E.-M. (2017a). “Protein–protein interactions: co-immunoprecipitation,” in Bacterial Protein Secretion Systems: Methods and Protocols, eds L. Journet and E. Cascales (New York, NY: Springer), 211–219. doi: 10.1007/978-1-4939-7033-9_17
Lin, J.-S., and Lai, E.-M. (2017b). “Protein–protein interactions: yeast two-hybrid system,” in Bacterial Protein Secretion Systems: Methods and Protocols, eds L. Journet and E. Cascales (New York, NY: Springer), 177–187.
Lossi, N. S., Manoli, E., Förster, A., Dajani, R., Pape, T., Freemont, P., et al. (2013). The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J. Biol. Chem. 288, 7536–7548. doi: 10.1074/jbc.m112.439273
Ma, L.-S., Hachani, A., Lin, J.-S., Filloux, A., and Lai, E.-M. (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104. doi: 10.1016/j.chom.2014.06.002
Ma, L.-S., Lin, J.-S., and Lai, E.-M. (2009). An Icmf family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker A motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191, 4316–4329. doi: 10.1128/jb.00029-09
MacIntyre, D. L., Miyata, S. T., Kitaoka, M., and Pukatzki, S. (2010). The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U.S.A. 107, 19520–19524. doi: 10.1073/pnas.1012931107
Mariano, G., Monlezun, L., and Coulthurst, S. J. (2018). Dual role for DsbA in attacking and targeted bacterial cells during type VI secretion system-mediated competition. Cell Rep. 22, 774–785. doi: 10.1016/j.celrep.2017.12.075
Mehla, J., Caufield, J. H., and Uetz, P. (2015). The yeast two-hybrid system: a tool for mapping protein-protein interactions. Cold Spring Harb. Protoc. 2015, 425–430.
Michalska, K., Gucinski, G. C., Garza-Sánchez, F., Johnson, P. M., Stols, L. M., Eschenfeldt, W. H., et al. (2017). Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs. Nucleic Acids Res. 45, 10306–10320. doi: 10.1093/nar/gkx700
Michalska, K., Quan Nhan, D., Willett, J. L. E., Stols, L. M., Eschenfeldt, W. H., Jones, A. M., et al. (2018). Functional plasticity of antibacterial EndoU toxins. Mol. Microbiol. 109, 509–527. doi: 10.1111/mmi.14007
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Pei, T.-T., Li, H., Liang, X., Wang, Z.-H., Liu, G., Wu, L.-L., et al. (2020). Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system. Nat. Commun. 11:1865.
Pietrosiuk, A., Lenherr, E. D., Falk, S., Bönemann, G., Kopp, J., Zentgraf, H., et al. (2011). Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J. Biol. Chem. 286, 30010–30021. doi: 10.1074/jbc.m111.253377
Poole, S. J., Diner, E. J., Aoki, S. K., Braaten, B. A., T’kint De Roodenbeke, C., Low, D. A., et al. (2011). Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 7:e1002217. doi: 10.1371/journal.pgen.1002217
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513. doi: 10.1073/pnas.0706532104
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Quentin, D., Ahmad, S., Shanthamoorthy, P., Mougous, J. D., Whitney, J. C., and Raunser, S. (2018). Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat. Microbiol. 3, 1142–1152. doi: 10.1038/s41564-018-0238-z
Rapisarda, C., Cherrak, Y., Kooger, R., Schmidt, V., Pellarin, R., Logger, L., et al. (2019). In situ and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex. EMBO J. 38:e100886.
Rigel, N. W., and Silhavy, T. J. (2012). Making a beta-barrel: assembly of outer membrane proteins in Gram-negative bacteria. Curr. Opin. Microbiol. 15, 189–193. doi: 10.1016/j.mib.2011.12.007
Robitaille, S., Trus, E., and Ross, B. D. (2020). Bacterial defense against the type VI secretion system. Trends Microbiol. 165:789.
Ruhe, Z. C., Low, D. A., and Hayes, C. S. (2020). Polymorphic toxins and their immunity proteins: diversity, evolution, and mechanisms of delivery. Annu. Rev. Microbiol. 74, 497–520. doi: 10.1146/annurev-micro-020518-115638
Ruhe, Z. C., Nguyen, J. Y., Beck, C. M., Low, D. A., and Hayes, C. S. (2014). The proton-motive force is required for translocation of CDI toxins across the inner membrane of target bacteria. Mol. Microbiol. 94, 466–481. doi: 10.1111/mmi.12779
Ruhe, Z. C., Nguyen, J. Y., Xiong, J., Koskiniemi, S., Beck, C. M., Perkins, B. R., et al. (2017). CdiA effectors use modular receptor-binding domains to recognize target bacteria. mBio 8:e00290-17.
Ruhe, Z. C., Subramanian, P., Song, K., Nguyen, J. Y., Stevens, T. A., Low, D. A., et al. (2018). Programmed secretion arrest and receptor-triggered toxin export during antibacterial contact-dependent growth inhibition. Cell 175, 921.e14–933.e14.
Ruhe, Z. C., Wallace, A. B., Low, D. A., and Hayes, C. S. (2013). Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. mBio 4:e00480-13.
Sana, T. G., Lugo, K. A., and Monack, D. M. (2017). T6SS: the bacterial “fight club” in the host gut. PLoS Pathog. 13:e1006325. doi: 10.1371/journal.ppat.1006325
Schell, M. A., Ulrich, R. L., Ribot, W. J., Brueggemann, E. E., Hines, H. B., Chen, D., et al. (2007). Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64, 1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x
Schwarz, S., West, T. E., Boyer, F., Chiang, W.-C., Carl, M. A., Hood, R. D., et al. (2010). Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068. doi: 10.1371/journal.ppat.1001068
Souza, D. P., Oka, G. U., Alvarez-Martinez, C. E., Bisson-Filho, A. W., Dunger, G., Hobeika, L., et al. (2015). Bacterial killing via a type IV secretion system. Nat. Commun. 6:6453.
Tikhonova, E. B., and Zgurskaya, H. I. (2004). AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 279, 32116–32124. doi: 10.1074/jbc.m402230200
Toska, J., Ho, B. T., and Mekalanos, J. J. (2018). Exopolysaccharide protects Vibrio cholerae from exogenous attacks by the type 6 secretion system. Proc. Natl. Acad. Sci. U.S.A. 115, 7997–8002. doi: 10.1073/pnas.1808469115
Trunk, K., Peltier, J., Liu, Y.-C., Dill, B. D., Walker, L., Gow, N. A. R., et al. (2018). The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931. doi: 10.1038/s41564-018-0191-x
Vacheron, J., Péchy-Tarr, M., Brochet, S., Heiman, C. M., Stojiljkovic, M., Maurhofer, M., et al. (2019). T6SS contributes to gut microbiome invasion and killing of an herbivorous pest insect by plant-beneficial Pseudomonas protegens. ISME J. 13, 1318–1329. doi: 10.1038/s41396-019-0353-8
Wang, J., Brodmann, M., and Basler, M. (2019). Assembly and subcellular localization of bacterial type VI secretion systems. Annu. Rev. Microbiol. 73, 621–638. doi: 10.1146/annurev-micro-020518-115420
Wexler, A. G., Bao, Y., Whitney, J. C., Bobay, L.-M., Xavier, J. B., Schofield, W. B., et al. (2016). Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. U.S.A. 113, 3639–3644. doi: 10.1073/pnas.1525637113
Whitney, J. C., Quentin, D., Sawai, S., LeRoux, M., Harding, B. N., Ledvina, H. E., et al. (2015). An interbacterial NAD(P)+ glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163, 607–619. doi: 10.1016/j.cell.2015.09.027
Willett, J. L. E., Gucinski, G. C., Fatherree, J. P., Low, D. A., and Hayes, C. S. (2015a). Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc. Natl. Acad. Sci. U.S.A. 112, 11341–11346. doi: 10.1073/pnas.1512124112
Willett, J. L. E., Ruhe, Z. C., Goulding, C. W., Low, D. A., and Hayes, C. S. (2015b). Contact-dependent growth inhibition (CDI) and CdiB/CdiA two-partner secretion proteins. J. Mol. Biol. 427, 3754–3765. doi: 10.1016/j.jmb.2015.09.010
Wu, C.-F., Lien, Y.-W., Bondage, D., Lin, J.-S., Pilhofer, M., Shih, Y.-L., et al. (2020). Effector loading onto the VgrG carrier activates type VI secretion system assembly. EMBO Rep. 21:e47961.
Wu, C.-F., Santos, M. N. M., Cho, S.-T., Chang, H.-H., Tsai, Y.-M., Smith, D. A., et al. (2019). Plant pathogenic Agrobacterium tumefaciens strains have diverse type VI effector-immunity pairs and vary in in planta competitiveness. Mol. Plant Microbe Interact. 32, 961–971. doi: 10.1094/mpmi-01-19-0021-r
Keywords: recipient susceptibility factor, antibacterial activity, bacterial secretion system, CDI, T6SS, effector, Cdz
Citation: Lin H-H, Filloux A and Lai E-M (2020) Role of Recipient Susceptibility Factors During Contact-Dependent Interbacterial Competition. Front. Microbiol. 11:603652. doi: 10.3389/fmicb.2020.603652
Received: 07 September 2020; Accepted: 13 October 2020;
Published: 12 November 2020.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Bruno Yasui Matsuyama, University of São Paulo, BrazilCopyright © 2020 Lin, Filloux and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erh-Min Lai, ZW1sYWlAZ2F0ZS5zaW5pY2EuZWR1LnR3
†Present address: Hsiao-Han Lin, Environmental Genomics and Systems Biology, Lawrence Berkeley National Lab, Berkeley, CA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.