
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 December 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.603578
This article is part of the Research Topic Antibiotic Resistance and its Continuity in the Environmental Niche View all 19 articles
CTX-M-producing Escherichia coli are spreading since 1999 both in clinical and in community settings. Environmental samples such as rivers have also been pointed out as being vectors for ESBL producers. In this report, we have investigated the presence and the diversity of CTX-M-producing E. coli isolates in two samplings of the Seine River (next to Notre Dame), Paris France, performed in June 2016 and 2017. The total number of bacteria growing on the selective ChromID ESBL agar was 3.1 × 105 cfu/L (23.8% of all growing bacteria) in 2016, whereas it was 100-fold lower in 2017 (3 × 103 cfu/L; 8.3% of all growing bacteria). However, among them, the prevalence of ESBL-producing E. coli increased from <0.1 to 1.1% in one-year. ESBLs were exclusively of the CTX-M-type: CTX-M-1 (n = 5), CTX-M-15 (n = 7), CTX-M-14 (n = 1), and CTX-M-27 (n = 2). The isolates belonged to several multi locus sequence types, and a wide diversity of incompatibility groups of plasmids were identified in those E. coli isolates. The occurrence and diversity of E. coli isolates belonging to many clones and producing many CTX-M-variants have been identified in our study. The presence of these bacteria in rivers that are open again for recreational usage (swimming) is worrying as it may contribute to further dissemination of ESBL producers in the community.
Escherichia coli is an ubiquitous human pathogen, most commonly involved in urinary tract infections and bacteremia in humans and animals (Rogers et al., 2011). Plasmid-mediated extended-spectrum β-lactamases (ESBLs) have become predominant in community-onset E. coli infection (Pitout et al., 2005). The first human CTX-M variant (previously named MEN-1) was initially reported in 1991 from a clinical E. coli isolate from France (Bernard et al., 1992). Since, CTX-M-producing E. coli have increasingly spread both in hospitals and in the community (Cantón et al., 2012) and represent now the most prevalent ESBLs worldwide. They are divided into five groups based on amino acid sequence: the CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 groups (Cantón et al., 2012).
The CTX-M-15 variant from the CTX-M-1 group, was first described in 2001 from several enterobacterial isolates from India on large-sized plasmids along with an ISEcp1 insertion sequence upstream of the blaCTX–M–15 gene (Karim et al., 2001). Since 2008 (Coque et al., 2008), it has rapidly become the most prevalent ESBL worldwide in humans, especially linked to an E. coli group B2, serogroup O25b, sequence type 131 (ST131). A collection from eight European countries demonstrated also the presence of ST131, including 6% of ESBL-producing E. coli isolates recovered from companion animals (Ewers et al., 2010). The water environment is conducive to the transfer of resistance genes between species, where ESBL producers from various sources get in contact with a broad range of potential recipients. A previous study, conducted in 2011, reported occurrence and diversity of ESBL-producing Aeromonas spp. in the Seine river (Girlich et al., 2011). However, at this date, no ESBL-producing E. coli isolates were identified in those samples, and the presence of CTX-M E. coli producers in rivers was still an exceptional event as shown by Kim et al. (2008) in Korea in 2008 or by Dhanji et al. (2011) in the United Kingdom in 2011. More recently, the emergence of ESBL-producing E. coli occurred in urban rivers. In Austria, CTX-M-producing E. coli were identified in the River Mur in the center of Graz, Austria’s second largest city (Zarfel et al., 2017). In Guadeloupe, the predominance of CTX-M-producing E. coli has been reported from waste water treatment plant effluents (Guyomard-Rabenirina et al., 2017). In contrast to what was reported from European countries, the occurrence of CTX-M-E. coli producers was high in the Pearl River in China (Ye et al., 2017).
The aim of the present study was to investigate the presence of expanded-spectrum cephalosporin (ESC)-resistant E. coli isolates in the water of the Seine River, Paris, France, sampled in June of two consecutive years (2016 and 2017) at the same centrally located sampling spot (next to the Notre Dame). We report here, the isolation of CTX-M-type ESBL-producing E. coli isolates and the in-depth genomic characterization of 15 of them.
Sampling of the Seine River water, Paris, France, was performed in June of two consecutive years (2016 and 2017) at the same centrally located sampling spot (next to the Notre Dame). Samples were collected c.a. 1 m from the shore and c.a. 20 cm below the water surface using a 1-L sterile plastic bottle connected to a rope. The bottle was immediately closed, transferred on ice to the bacteriology laboratory of the Bicêtre Hospital, Le Kremlin-Bicêtre, France, and directly processed upon arrival. Four hundred milliliters of water was filtrated through a nitrocellulose membrane (0.45 μm, Millipore), and the bacteria were resuspended from the membranes in 2 ml of sterile water. Aliquots (100 μl) were then plated on ChromID ESBL plates (bioMérieux, Marcy l’Etoile, France). Pink-colored colonies growing ChromID ESBL were identified by mass spectrometry (MALDI-TOF, Bruker, France), and the ESBL phenotype was evidenced by a double disk synergy test (Karim et al., 2001). Plasmids, extracted by the Kieser method were electroporated into E. coli Top10, as previously described (Girlich et al., 2011). In case electroporation did not work, mating out assay was performed as previously described (Girlich et al., 2011). Transformants or transconjugants were selected on cefotaxime (0.5 μg/ml) agar. Identification of replicon types of the plasmid incompatibility (Inc) groups was performed by PCR as previously described by Carattoli et al. (Carattoli, 2009). Using this typing scheme, 18 Inc groups may be identified: Hl1, Hl2, I1-Iγ, X, L/M, N, FIA, FIB, W, Y, P, FIC, A/C, T, FIIAs, F, K, and B/O.
NG-Test CTXM-Multi, a rapid Lateral Flow Immuno Assay (LFIA, NG-Biotech, Guipry, France) was used to detect all five CTX-M-groups, as previously described (Bernabeu et al., 2020). Briefly, one colony was resuspended in the extraction buffer, vortexed, and 100 μl was dropped on the LFIA strip. Results were eye read after 15 min of migration.
Whole genome sequencing was performed on 15 selected ESBL-producing E. coli isolates using Illumina technology on a Nextseq 500 sequencer as previously described (Dabos et al., 2019). De novo assembly was performed by CLC Genomics Workbench v7.0.4 (Qiagen, Les Ulis, France) after quality trimming (Qs ≥ 20). The acquired antimicrobial resistance genes were identified using ResFinder (Bortolaia et al., 2020), incompatibility groups of plasmids were determined using Plasmid finder (Clausen et al., 2018), and the sequence type was obtained using the Multi Locus Sequence Typing (MLST) modules of the Center for Genomic Epidemiology with genes adk, fumC, gyrB, icd, mdh, purA, and recA1 (Larsen et al., 2012).
Total bacterial count on Mueller Hinton agar was 1.3 × 106 cfu/L of Seine water samples in 2016, whereas it was 3.5 × 104 cfu/L in 2017. Bacterial count growing on ChromID ESBL agar was 3.1 × 105 cfu/L (23.8% of all growing bacteria) in 2016, whereas it was 100-fold lower in 2017 (3 × 103 cfu/L; 8.3% of all growing bacteria). ESBL-producing E. coli isolates recovered were 11 per 100 ml and 4 per 500 ml of water, in 2016 and 2017, respectively. Among the total bacteria growing on ChromID ESBL agar, <0.1% (2.7 × 102 cfu/L) were ESBL-producing E. coli in the samples from June 2016, whereas 1.1% (33 cfu/L) ESBL-producing E. coli isolates were identified in May 2017. ESBLs produced by the E. coli isolates were exclusively CTX-M enzymes (Table 1). NG-Test CTX-M-gr1, a LFIA specific for group 1 CTX-M-β-lactamases gave positive results for 12/15 ESBL-producing E. coli isolates.
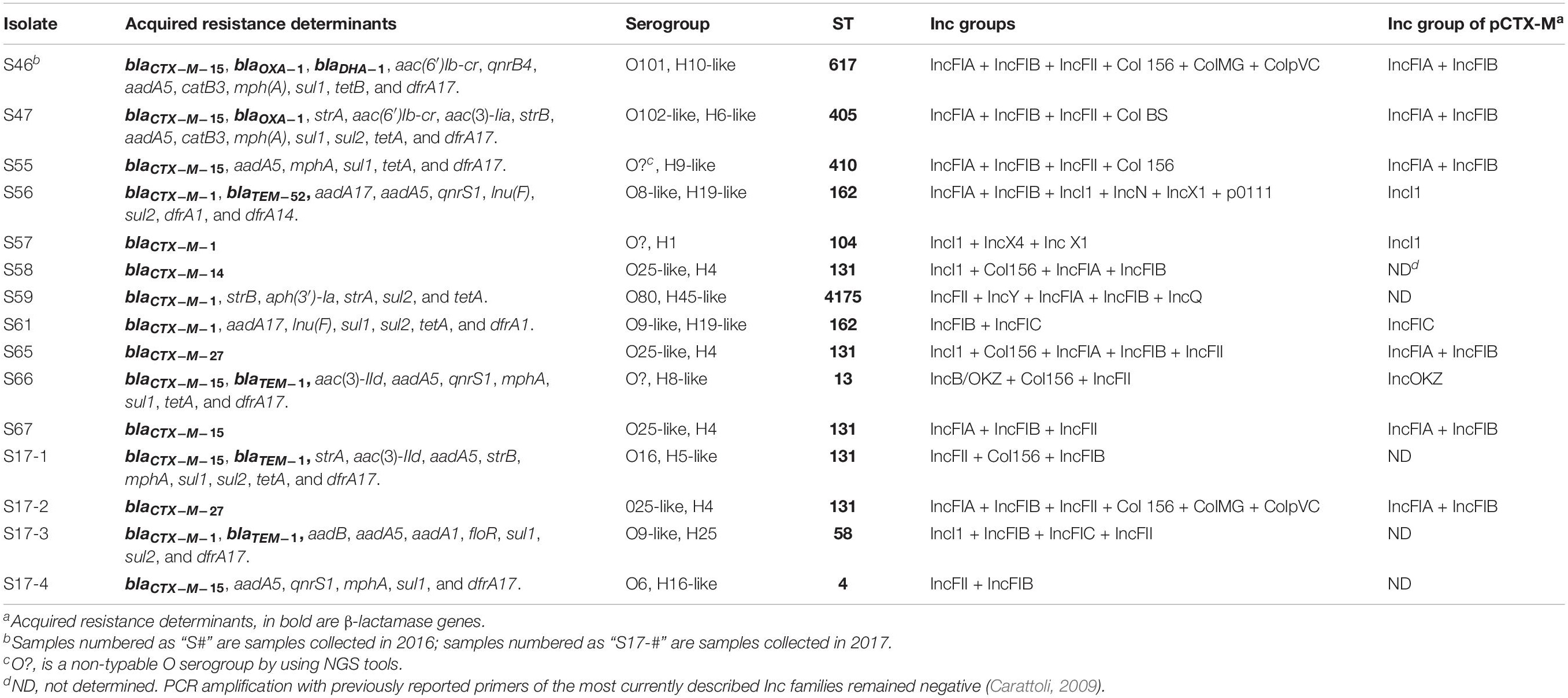
Table 1. Genetic characteristic of ESBL-producing E. coli isolates from the Seine river from 2016 and 2017.
WGS identified blaCTX–M–1 (n = 5), blaCTX–M–15 (n = 7), blaCTX–M–14 (n = 1), blaCTX–M–27 (n = 2), blaTEM–52 (n = 1) ESBL genes, blaDHA–1 (n = 1) cephalosporinase gene, and blaTEM–1 (n = 3) and blaOXA–1 (n = 2) penicillinase genes (Table 1). The results of the WGS were in accordance with those of the NG-Test CTX-M- MULTI LFIA, validating this latest test for the rapid detection of the five groups of CTX-M-producing Enterobacterales, as previously reported (Bernabeu et al., 2020). As observed in other studies, group 1 CTX-M-producing E. coli isolates were dominant in our study (Zarfel et al., 2017; Hooban et al., 2020).
As commonly found in CTX-M-producers, most isolates were multidrug resistant, possessing aminoglycoside-modifying enzyme [e.g., aac(6′)-Ib, aadA1, aadA5, or aph(3′)-Ia], quinolone-resistance genes (qnrS1), tetracycline (tetA, tetB), chloramphenicol (catB3), and trimethoprim/sulfamethoxazole (dfrA14, dfrA17, sul1, and sul2). Those resistance gene are, for most of them, carried by a class 1 integron (Table 1; Cantón et al., 2012).
Clonal relationship of these isolates was initially assessed by MLST and then by WGS SNP analysis. MLST analysis revealed a wide diversity of clonal groups with 10 different STs among the 15 isolates. Noticeably, only two STs were represented with at least two isolates being ST162 (n = 2) and ST131 (n = 5) (Figure 1). WGS-based phylogeny confirmed this diversity but also indicated that the five ST131 isolates can be divided into two subclones (Figure 1). It can be noticed that ESBL distribution did not follow the clonal relationship. For instance, in ST131 isolates, three types of ESBLs were identified: blaCTX–M–14, blaCTX–M–15, and blaCTX–M–27 genes. The wide diversity of clones may reflect the large spread of ESBLs in the community. Indeed, we did not identify a clonal spread of ESBL-producing E. coli but rather unrelated isolates that are present in the Seine River. Among the five ST131 E. coli isolates, four were genetically close (Figure 1). However, these isolates did not share the same resistome indicating that ST131 is widely distributed independent of the ESBL content as previously observed (Pitout and Finn, 2020). Moreover, the two closest ST131 (S65 and S17-2) possessing the blaCTX–M–27 ESBL gene were recovered one-year apart indicating the persistence or continuing contamination by this clone.
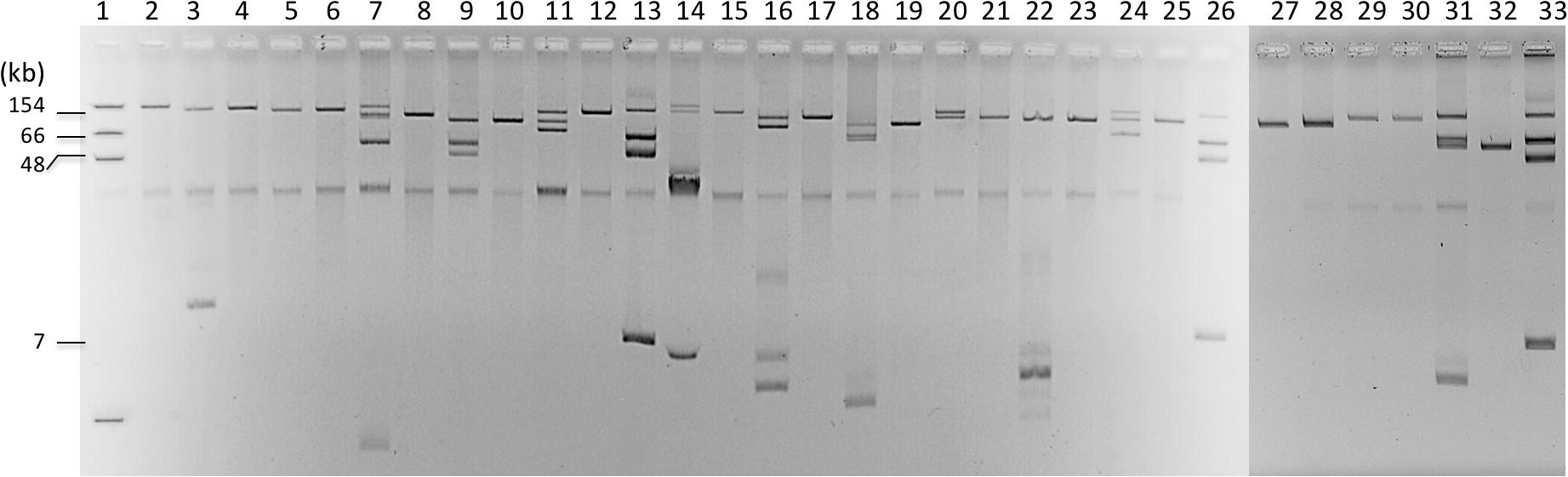
Figure 1. Phylogenetic tree of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from Seine River. The phylogeny was performed using CSIPhylogeny (https://cge.cbs.dtu.dk/services/CSIPhylogeny/). Year of isolation is indicated by colored circles and broad-spectrum β-lactamases by colored pentagons.
The blaCTX–M genes were located on large plasmids of different sizes (Figure 2) belonging to diverse incompatibility groups (Table 1). Several studies have shown that plasmids of the IncF family were the predominant group that carry the blaCTX–M–15 gene, whereas the blaCTX–M–14 gene is carried on a variety of plasmid types, including on IncF, especially in the Far-East, and on IncK, in Western Europe (Bevan et al., 2017). Horizontal transfer of antimicrobial resistance plasmids by conjugation in Enterobacterales occurs in the human gut, animals, and the environment (Bevan et al., 2017; Zarfel et al., 2017). As previously reported, the main ESBL types identified in companion animals were CTX-M-14 (26.8%), CTX-M-15 (24.4%), CTX-M-27 (19.5%), and CTX-M-55 (19.5%) (Kawamura et al., 2017), and the most prevalent STs were ST131 (n = 15, 35.7%), followed by ST38, ST10, and ST410 (Kawamura et al., 2017). For example, among those STs, ST10/CC10 corresponds to an international cluster already identified in humans, wildlife infections, domestic farm animals, companion animals, and commercial chicken meat (Nascimento et al., 2017).
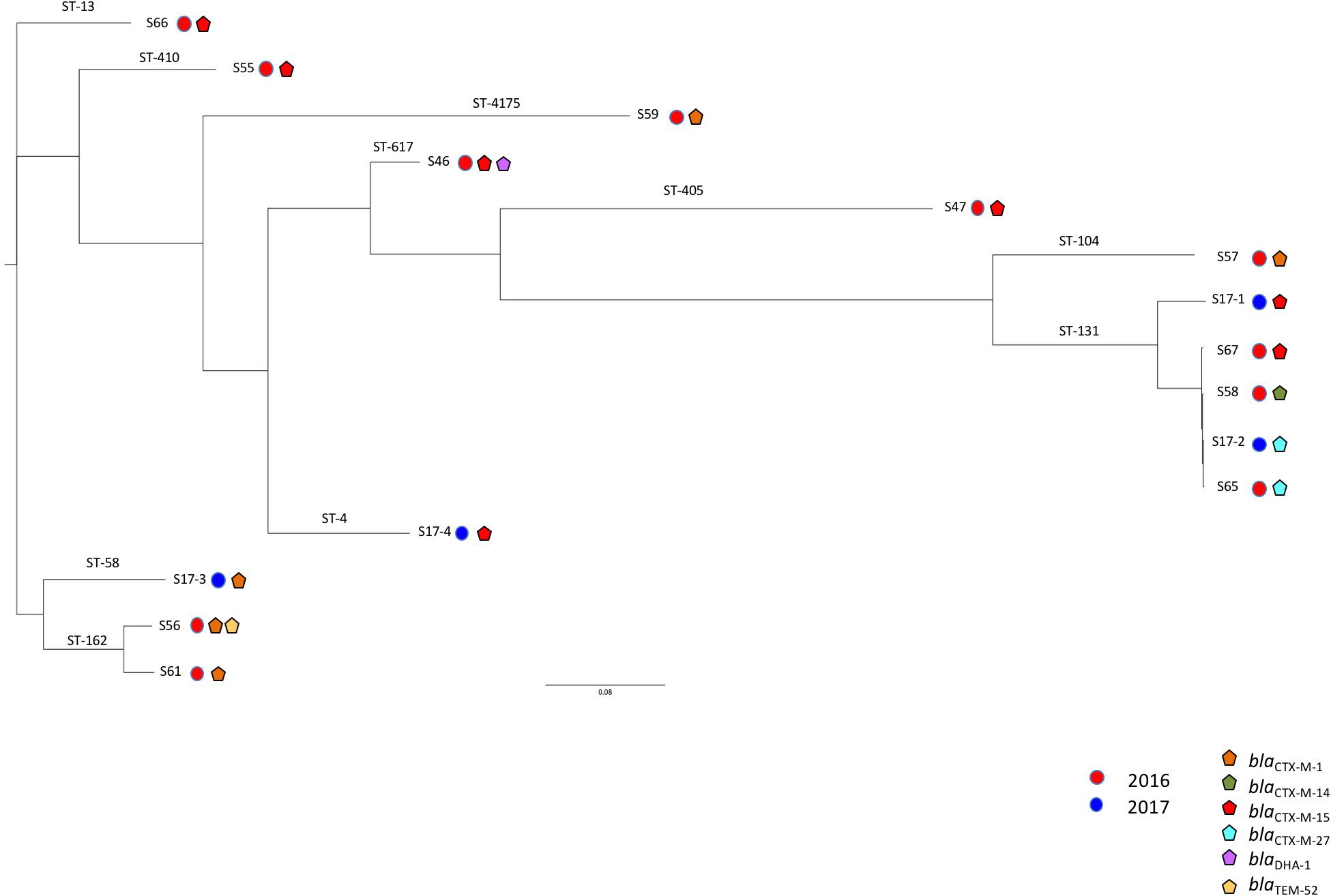
Figure 2. Plasmid extractions from cultures of the different isolates and their transconjugants or transformants. Lanes 1, 3, 5, 7, 9, 11, 14, 16, 18, 20, 22, and 24 correspond to S46, S47, S55, S56, S57, S59, S61, S65, S66, 17.1, S17.2, S17.3, S58, S59, and S17.4; Lanes 2, 4, 6, 8, 10, 12, 15, 17, 19, 21, 23, 25, 28, 30, and 32 correspond to their respective E. coli transformants or transconjugants; Lanes 13, 26, and 33 correspond to E. coli 50192 harboring four plasmids: 7, 48, 66, and 154 kb.
We identified in this study the occurrence of different ESBL-producing E. coli isolates from the Seine River in Paris, France. A wide diversity of clones was identified here. The most prevalent, with 4 isolates out of 15, was ST131. This result is not surprising given its widespread occurrence, but the presence of different CTX-M-variants belonging to different groups of enzymes was unexpected, as ST131 is frequently associated to CTX-M-15. We also identified one ST410 isolate. This clone has recently attracted not only the light by its association with the spread of the carbapenemase OXA-181 (Patiño-Navarrete et al., 2020) but also for its isolation in animals (Yang et al., 2019). Of note, two isolates of ST162 were recovered in this study. This clone has been reported to be associated with the resistance gene in wild avian isolates (Oteo et al., 2018). However, it remains difficult to conclude on the original source of these isolates, which could be of avian/environmental or human sources. Hooban et al. (2020) reported that most of the ESBL producers identified in aquatic environments around the world between 2010 and 2017 expressed blaCTX–M genes (n = 21 among 29 studies), followed by blaTEM (n = 18), and blaSHV (n = 11). Surprisingly, among eight Chinese studies, only three identified CTX-M-producing E. coli isolates in rivers and lakes (Hooban et al., 2020). The prevalence of ESBL producers among waterborne thermo-tolerant coliforms ranged in amount from 11% (Ye et al., 2017) to 17% in Chinese rivers (Chen et al., 2010). Ye et al. (2017) identified only CTX-M-variants as ESBLs, with additional variants: i.e., CTX-M-55 and CTX-M-65 in addition to CTX-M-14 and CTX-M-15. Notably, a previous study, 6 years earlier, reported TEM (37.6%) and SHV (84.1%) as being the most common ESBL among clinical isolates from the same city of Chongqing in 2004 (Chen et al., 2010). In Brazil, four studies reported the presence of CTX-M but also of KPC-2 carbapenemase K. pneumoniae producers in rivers, lakes, and sea water (Hooban et al., 2020). The presence of blaCTX–M genes in water is more and more frequent worldwide, most often associated with highly self-transferable plasmids. In all cases, it is likely that the transfer of these bacteria from the sewage to the rivers occurred. Most worrying, concomitant spread of carbapenemase genes has been witnessed in many countries including Switzerland, Spain, Portugal, Austria, United States, Brazil, India, and China (Hooban et al., 2020).
The epidemic dissemination of CTX-M-encoding genes is largely due to their localizations on mobile genetic elements, such as plasmids, transposons, and integrons, which allow these genes to easily spread among bacterial communities (Cantón et al., 2012). In 2016, the samples were collected a few days after floods that occurred between the end of May and the beginning of June 2016, thus explaining a high prevalence of ESBL-E. coli isolates in the Seine river that likely originated from animal feces that have been drained by the rains. However, the presence and diversity of those isolates one-year later is more worrying, as it indicates a persistent contamination of the Seine river with ESBLs-producing E. coli isolates. This is especially worrying given that the many rivers all over Europe open again for different recreational and sporting activities, including swimming.
The datasets generated for this study can be found in NCBI BioProject, NCBI Accession No. PRJNA662045.
TN: conception, data analysis, writing, and proof-reading. RB: data analysis and proof-reading. DG: experimental work, data analysis, writing, and proof-reading. All authors contributed to the article and approved the submitted version.
This work was supported by the Assistance Publique–Hôpitaux de Paris, by a grant from the Université Paris-Saclay, and by the LabEx LERMIT supported by a grant from the French National Research Agency (ANR-10-LABX-33). This work was also funded in part by a grant from Joint Program Initiative on Antimicrobial Resistance (ANR-14-JAMR-0002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Institut Pasteur PIBNet for WGS of bacterial isolates.
Bernabeu, S., Ratnam, K. C., Boutal, H., Gonzalez, C., Vogel, A., Devilliers, K., et al. (2020). A lateral flow immunoassay for the rapid identification of CTX-M-producing enterobacterales from culture plates and positive blood cultures. Diagnostics 10:E764.
Bernard, H., Tancrede, C., Livrelli, V., Morand, A., Barthelemy, M., and Labia, R. (1992). A novel plasmid-mediated extended-spectrum β-lactamase not derived from TEM- or SHV-type enzymes. J. Antimicrob. Chemother. 29, 590–592. doi: 10.1093/jac/29.5.590
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Cantón, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/aac.01707-08
Chen, H., Shu, W., Chang, X., Chen, J., Guo, Y., and Tan, Y. (2010). The profile of antibiotics resistance and integrons of extended-spectrum beta-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ. Pollut. 158, 2459–2464. doi: 10.1016/j.envpol.2010.03.023
Clausen, P. T. L. C., Aarestrup, F. M., and Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19:307. doi: 10.1186/s12859-018-2336-6
Coque, T. M., Novais, A., Carattoli, A., Poirel, L., Pitout, J., Peixe, L., et al. (2008). Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14, 195–200.
Dabos, L., Patiño-Navarrete, R., Nastro, M., Famiglietti, A., Glaser, P., Rodriguez, C. H., et al. (2019). SME-4-producing Serratia marcescens from Argentina belonging to clade 2 of the S. marcescens phylogeny. J. Antimicrob. Chemother. 74, 1836–1841. doi: 10.1093/jac/dkz115
Dhanji, H., Murphy, N. M., Akhigbe, C., Doumith, M., Hope, R., Livermore, D. M., et al. (2011). Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum β-lactamase from UK river water. J. Antimicrob. Chemother. 66, 512–516. doi: 10.1093/jac/dkq472
Ewers, C., Grobbel, M., Stamm, I., Kopp, P. A., Diehl, I., Semmler, T., et al. (2010). Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 65, 651–660. doi: 10.1093/jac/dkq004
Girlich, D., Poirel, L., and Nordmann, P. (2011). Diversity of clavulanic acid-inhibited extended-spectrum β-lactamases in Aeromonas spp. from the Seine River, Paris, France. Antimicrob. Antimicrob. Agents Chemother. 55, 1256–1261. doi: 10.1128/aac.00921-10
Guyomard-Rabenirina, S., Dartron, C., Falord, M., Sadikalay, S., Ducat, C., Richard, V., et al. (2017). Resistance to antimicrobial drugs in different surface waters and wastewaters of Guadeloupe. PLoS One 12:e0173155. doi: 10.1371/journal.pone.0173155
Hooban, B., Joyce, A., Fitzhenry, K., Chique, C., and Morris, D. (2020). The role of the natural aquatic environment in the dissemination of extended spectrum beta-lactamase and carbapenemase encoding genes: a scoping review. Water Res. 180:115880. doi: 10.1016/j.watres.2020.115880
Karim, A., Poirel, L., Nagarajan, S., and Nordmann, P. (2001). Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201, 237–241. doi: 10.1016/s0378-1097(01)00276-2
Kawamura, K., Sugawara, T., Matsuo, N., Hayashi, K., Norizuki, C., Tamai, K., et al. (2017). Spread of CTX-type extended-spectrum β-lactamase-producing Escherichia coli isolates of epidemic clone B2-O25-ST131 among dogs and cats in Japan. Microb. Drug Resist. 23, 1059–1066. doi: 10.1089/mdr.2016.0246
Kim, J., Kang, H. Y., and Lee, Y. (2008). The identification of CTX-M-14, TEM-52, and CMY-1 enzymes in Escherichia coli isolated from the Han River in Korea. J. Microbiol. Seoul Korea 46, 478–481. doi: 10.1007/s12275-008-0150-y
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/jcm.06094-11
Nascimento, T., Cantamessa, R., Melo, L., Fernandes, M. R., Fraga, E., Dropa, M., et al. (2017). International high-risk clones of Klebsiella pneumoniae KPC-2/CC258 and Escherichia coli CTX-M-15/CC10 in urban lake waters. Sci. Total Environ. 598, 910–915. doi: 10.1016/j.scitotenv.2017.03.207
Oteo, J., Mencía, A., Bautista, V., Pastor, N., Lara, N., González-González, F., et al. (2018). Colonization with Enterobacteriaceae-Producing ESBLs, AmpCs, and OXA-48 in Wild Avian Species, Spain 2015-2016. Microb. Drug Resist. 24, 932–938. doi: 10.1089/mdr.2018.0004
Patiño-Navarrete, R., Rosinski-Chupin, I., Cabanel, N., Gauthier, L., Takissian, J., Madec, J.-Y., et al. (2020). Stepwise evolution and convergent recombination underlie the global dissemination of carbapenemase-producing Escherichia coli. Genome Med. 12:10.
Pitout, J. D. D., and Finn, T. J. (2020). The evolutionary puzzle of Escherichia coli ST131. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 81:104265. doi: 10.1016/j.meegid.2020.104265
Pitout, J. D. D., Nordmann, P., Laupland, K. B., and Poirel, L. (2005). Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56, 52–59. doi: 10.1093/jac/dki166
Rogers, B. A., Sidjabat, H. E., and Paterson, D. L. (2011). Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66, 1–14. doi: 10.1093/jac/dkq415
Yang, H., Rehman, M. U., Zhang, S., Yang, J., Li, Y., Gao, J., et al. (2019). High prevalence of CTX-M belonging to ST410 and ST889 among ESBL producing E. coli isolates from waterfowl birds in China’s tropical island, Hainan. Acta Trop. 194, 30–35. doi: 10.1016/j.actatropica.2019.03.008
Ye, Q., Wu, Q., Zhang, S., Zhang, J., Yang, G., Wang, H., et al. (2017). Antibiotic-resistant extended spectrum ß-lactamase- and plasmid-mediated ampc-producing Enterobacteriaceae Isolated from Retail Food Products and the Pearl River in Guangzhou, China. Front. Microbiol. 8:96. doi: 10.3389/fmicb.2017.00096
Zarfel, G., Lipp, M., Gürtl, E., Folli, B., Baumert, R., and Kittinger, C. (2017). Troubled water under the bridge: screening of river Mur water reveals dominance of CTX-M harboring Escherichia coli and for the first time an environmental VIM-1 producer in Austria. Sci. Total Environ. 593–594, 399–405. doi: 10.1016/j.scitotenv.2017.03.138
Keywords: CTX-M-14, CTX-M-15, Escherichia coli, Seine river, plasmids
Citation: Girlich D, Bonnin RA and Naas T (2020) Occurrence and Diversity of CTX-M-Producing Escherichia coli From the Seine River. Front. Microbiol. 11:603578. doi: 10.3389/fmicb.2020.603578
Received: 07 September 2020; Accepted: 09 November 2020;
Published: 09 December 2020.
Edited by:
Marina Spinu, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Sebastian Guenther, University of Greifswald, GermanyCopyright © 2020 Girlich, Bonnin and Naas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thierry Naas, dGhpZXJyeS5uYWFzQGFwaHAuZnI=; dGhpZXJyeS5uYWFzQGJjdC5hcGhwLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.