
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 20 January 2021
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.602789
This article is part of the Research Topic CRISPR-Cas Systems in Bacteria and Archaea View all 10 articles
Type III CRISPR-Cas systems, which are widespread in both bacteria and archaea, provide immunity against DNA viruses and plasmids in a transcription-dependent manner. Since an unprecedented cyclic oligoadenylate (cOA) signaling pathway was discovered in type III systems in 2017, the cOA signaling has been extensively studied in recent 3 years, which has expanded our understanding of type III systems immune defense and also its counteraction by viruses. In this review, we summarized recent advances in cOA synthesis, cOA-activated effector protein, cOA signaling-mediated immunoprotection, and cOA signaling inhibition, and highlighted the crosstalk between cOA signaling and other cyclic oligonucleotide-mediated immunity discovered very recently.
CRISPR-Cas systems are known to provide adaptive immunity against viruses and plasmids in prokaryotes. Based on the composition of effector complexes, CRISPR-Cas systems were divided into two classes which could be further subdivided into six types (types I–VI) and multiple subtypes (Makarova et al., 2020b). Class 1 systems (including type I, III, and IV), which have multi-subunit effector complex, are widespread in bacteria and archaea; whereas class 2 (including type II, V, and VI), which contain single-subunit effector complex, are almost completely presented in bacteria (Mohanraju et al., 2016). The effector complexes of type I, II, and V (and possibly IV) target double-stranded DNA (dsDNA), while Type VI system targets RNA (Makarova et al., 2020b). Unlike them, type III effector complex targets both RNA and single-stranded DNA (ssDNA) of the invaders (Tamulaitis et al., 2017). The type III system can be further divided into six subtypes (III A–F), in which Type III-A/D system forms a Csm effector complex composed of five subunits (Csm 1–5) and a single CRISPR RNA (crRNA), while Type III-B/C forms a Cmr effector complex consisting of six subunits (Cmr 1–6) and a crRNA (Makarova et al., 2020b). The effector complexes of type III systems exhibit both target RNA cleavage activity and target RNA-activated ssDNA cleavage activity (Elmore et al., 2016; Estrella et al., 2016; Kazlauskiene et al., 2016). Type III systems provide immunity against invaders depending on the target RNA transcription (Deng et al., 2013; Goldberg et al., 2014). The crRNA-guided Csm/Cmr complexes recognize the complementary target RNA and cleave it into 6 nt nucleotide intervals using the multiple copies of Csm3 or Cmr4 subunit (Hale et al., 2009; Estrella et al., 2016; Kazlauskiene et al., 2016). Target RNA binding also activates the cyclic oligoadenylate (cOA) synthesis activity of Cas 10 subunit. More details about the transcription-dependent immunity and the structural basis of type III effector complexes and effector proteins had been reviewed elsewhere (Pyenson and Marraffini, 2017; Tamulaitis et al., 2017; Molina et al., 2020). In this review, we systematically discuss the recent advances in cOA signaling pathway of type III systems.
The Cas 10 subunit of type III effector complex and the ancillary ribonuclease Csm6/Csx1 are two important components involved in cOA signaling. Cas 10 contains an N-terminal histidine-aspartate (HD) domain and two Palm domains with a GGDD motif inserted into the second Palm domain (Tamulaitis et al., 2017; Figure 1A). The HD domain is responsible for ssDNA cleavage activity, while the Palm domains are homologous to nucleotide polymerases and nucleotide cyclase (Makarova et al., 2002, 2011; Zhu and Ye, 2012), and were hypothesized to synthesize cyclic nucleotides like cyclic di-AMP (Burroughs et al., 2015). However, there was no experimental evidence to verify the domains function for a long time (Koonin and Makarova, 2018). Csm6/Csx1 contains an N-terminal CRISPR-associated Rossman fold (CARF) domain which was predicted to sense nucleotide derivative and a C-terminal higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain which often functions as ribonuclease (Anantharaman et al., 2013; Makarova et al., 2014; Figure 1A). In 2017, two independent studies revealed that the two proteins were involved in a cOA signaling pathway, which had never been found in prokaryotes (Kazlauskiene et al., 2017; Niewoehner et al., 2017). It was found that the Palm domains were responsible for cOA synthesis, and the CARF domain of Csm6/Csx1 can sense the corresponding cOA (Kazlauskiene et al., 2017; Niewoehner et al., 2017; Figure 1B). When target RNA is recognized by effector complex, Cas10 subunit can be activated and can generate cOA, which in turn allosterically activates the ribonuclease Csm6/Csx1 through binding the CARF domain, resulting in non-specific RNA degradation (Kazlauskiene et al., 2017; Niewoehner et al., 2017; Figure 1B).
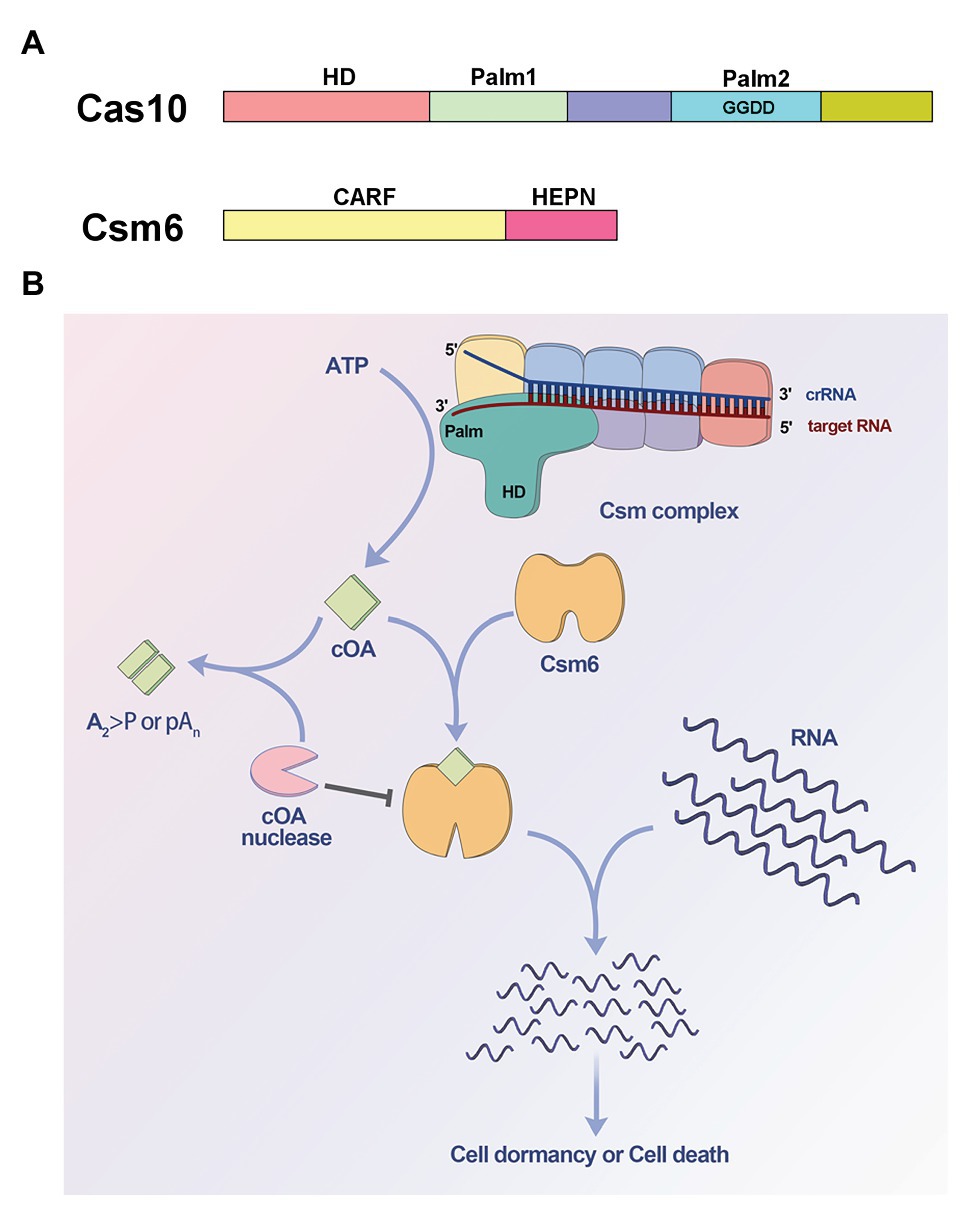
Figure 1. Cyclic oligoadenylate (cOA) signaling-mediated immunity in type III CRISPR-Cas systems. (A) Domain organization of Cas10 and Csm6. Cas10 contains an N-terminal histidine-aspartate (HD) domain and two Palm domains (Palm1 and Palm2), and the GGDD motif is inserted into Palm2 domain. Csm6 contains an N-terminal CRISPR-associated Rossman fold (CARF) domain and a C-terminal higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain. (B) Model for cOA signaling pathway of type III systems. The Palm domains of Cas10 subunit are activated and convert ATP into cOA molecules, when the target RNA is recognized by the CRISPR RNA (crRNA)-guided effector complex. The synthesized cOA allosterically activates Csm6 by binding the CARF domain, which subsequently degrades RNA non-specifically, resulting in host cell dormancy or cell death. On the other hand, cOA nucleases including ring nuclease and membrane-associated DHH-DHHA1 family nuclease can degrade cOA molecules to switch off cOA signaling, thereby acting as off-switch for the systems.
Cas10 is the largest subunit of type III effector complex and is a signature protein of type III systems (Tamulaitis et al., 2017; Koonin and Makarova, 2018). The cOA synthesis activity of Cas10 is subject to tight spatial and temporal control. The Cas10 subunit is activated and converts ATP into cOA molecules only when target RNA is recognized by the crRNA-guided effector complex, and cOA synthesis will be deactivated abruptly following target RNA cleavage and dissociation from the effector complex (Kazlauskiene et al., 2017; Niewoehner et al., 2017; Rouillon et al., 2018). Unlike type I, II, and V CRISPR-Cas systems which distinguish self from non-self DNA in a protospacer adjacent motif (PAM)-dependent manner, type III systems were proposed to rely on the 5'-handle of crRNA (8 nt) and the 3'-flanking sequence of the target RNA to avoid autoimmunity (Marraffini and Sontheimer, 2010; Kazlauskiene et al., 2016; Tamulaitis et al., 2017). Non-complementarity between crRNA 5'-handle and 3'-flanking sequence of the target RNA is essential for Cas10 activation for cOA synthesis (Kazlauskiene et al., 2017). Previous studies showed type III systems were much tolerant of mismatches in target RNA (Pyenson et al., 2017; Goldberg et al., 2018; Rouillon et al., 2018). To some degree, target RNA binding-mediated Cas10 activation is also tolerant of crRNA-target mismatches, but base pairs in direct contact with Cas10 subunit, such as those adjacent to the 3' end of target RNA, are very stringent (Rouillon et al., 2018; Nasef et al., 2019). Base-pairing of 3'-flanking target RNA sequence to the 5'-handle of crRNA affects activation of both ssDNA cleavage and cOA synthesis (Guo et al., 2019; Johnson et al., 2019; Foster et al., 2020). Structural studies on Csm complexes show that the interaction between non-complementary 3'-flanking target RNA sequence and Cas10 subunit is crucial to induce a conformational change of Cas10 subunit for activation of its single-stranded DNase (ssDNase) and cOA synthetase activities (Jia et al., 2019c; You et al., 2019). Moreover, recent studies on the Cmr complex show that a unique stalk loop in Cmr3 is critical for avoiding autoimmunity and triggering Cas10 activation (Guo et al., 2019; Sofos et al., 2020). In addition, the length of the crRNA-target duplex also affects Cas10 activation. Twenty five base pairs or longer crRNA-target duplex are required for efficient activation of ssDNA cleavage and cOA generation (You et al., 2019; Sofos et al., 2020).
Since cOA synthesis was identified in Type III-A system of Streptococcus thermophilus and Enterococcus italicus, respectively (Kazlauskiene et al., 2017; Niewoehner et al., 2017), the Cas10 subunits of effector complexes from various bacteria and archaea, which harbor type III-A/B/D systems were verified to generate various cOA molecules (cOAn, n = 3–6; Han et al., 2018; Rouillon et al., 2018; Grüschow et al., 2019; Nasef et al., 2019; Foster et al., 2020). Notably, the major cOA species produced by effector complex is not always the one that activates the effector ribonuclease in the same system (Kazlauskiene et al., 2017; Rouillon et al., 2019; Smalakyte et al., 2020). That may be because the in vitro cOA synthesis could be affected by reaction conditions, thus the synthesized major cOA species may be different from those in vivo (Smalakyte et al., 2020). Recently, alternative nucleotide signal molecules were found to be synthesized by GDDEF cyclase, cGAS/DncV-like nucleotidyltransferases (CD-NTases), and ppGpp synthetase homolog (Hallberg et al., 2016; Ahmad et al., 2019; Whiteley et al., 2019), leading us to consider the existence of a subfamily of Cas10-like proteins that can synthesize other kinds of cyclic oligonucleotide molecules. Structure studies on Csm effector complexes bound to substrates (AMPPNP and ATP) have shown that each Palm domain has a conserved serine residue (Ser273 and Ser549 in the Streptococcus thermophilus Csm1), which forms hydrogen bonds with base of ATP and confers specificity for ATP (Jia et al., 2019a; You et al., 2019). Moreover, a biochemical study on Cmr effector complexes also shows that two conserved serine residues in the Palm 1 domain of Cmr2 are important for ATP binding and cOA synthesis, and the study further reveals a cooperative substrate binding mechanism for efficient cOA synthesis (Han et al., 2018). Cas10 with substitutions of the conserved serine residues still retains a certain degree of cOA synthesis activities, yet whether the nucleotide specificity is affected remains unclear (Han et al., 2018; You et al., 2019). It will be interesting to investigate the possibility of other cyclic oligonucleotides synthesis from uncharacterized type III effector complexes.
The effector proteins Csm6 and Csx1, representatives of CARF family proteins, can be activated by either cOA4 or cOA6, depending on their preferences (Shah et al., 2019). Very recently, the crystal structures of complexes of Csm6/Csx1 with cOA molecules have been determined (Jia et al., 2019b; Molina et al., 2019; Garcia-Doval et al., 2020). Studies on these structures reveal that one cOA binds to each CARF domains of the symmetrical homodimer of Csm6/Csx1, resulting in conformational change of Csm6/Csx1 and HEPN domain activation (Jia et al., 2019b; Molina et al., 2019; Garcia-Doval et al., 2020). Furthermore, the CARF domain of Csm6 can autoregulate its RNase activity through degrading its cOA activators (Athukoralage et al., 2019; Jia et al., 2019b; Garcia-Doval et al., 2020). However, the CARF domain of Csx1 cannot cleave its cOA activator, suggesting that the degradation of cOA by CARF domains is not a general mechanism for CARF family proteins (Molina et al., 2019).
Since CARF domain is responsible for sensing cOA, other CARF domain-containing proteins may also serve as the effector proteins. Bioinformatics analysis shows that the CARF domain is also fused to various other domains in type III systems, implying cOA signaling may provide immunity through activating various CARF domain proteins not just Csm6/Csx1 (Makarova et al., 2014, 2020a; Koonin and Makarova, 2018; Shah et al., 2019). For examples, CARF domain is fused to other RNase domains such as ribosome-dependent endoribonuclease RelE and PIN, and DNase domains of restriction endonuclease (REase) and HD nuclease, suggesting that RNA and even DNA can be degraded by such cOA-activated CARF domain proteins (Makarova et al., 2014, 2020a; Koonin and Makarova, 2018). Additionally, CARF domains are also fused with domains such as helix-turn-helix (HTH), AAA+ ATPase, or adenosine deaminase, suggesting that RNA transcription can also be regulated by such cOA-activated CARF domain proteins (Makarova et al., 2014, 2020a; Koonin and Makarova, 2018). Very recently, an effector protein containing two CARF domains and one DNA nuclease-like domain (named Can1) and another effector protein containing a Csx1 protein fused to a ring nuclease CRISPR-associated ring nuclease 2 (Crn2) domain (named Csx1-Crn2) are characterized (McMahon et al., 2020; Samolygo et al., 2020). Unlike Csm6/Csx1, Can1 is a monomeric enzyme with DNA nuclease activity (McMahon et al., 2020), while Csx1-Crn2 degrades cOA4 by the Crn2 domain to limit its cOA4-activated ribonuclease activity (Samolygo et al., 2020). These results demonstrated the diversity of CARF domain-containing effectors.
It has been known that activated Cas10 subunit produces cOAs ranging from cOA3 to cOA6 (Kazlauskiene et al., 2017). However, it is unlikely that cOA3 or cOA5 can activate CARF domain proteins like Csm6/Csx1 which assembles as homodimer with 2-fold symmetry, because the two cOAs lack symmetry to fit the dimer interface of CARF domains (Rouillon et al., 2018). Thus, it was questioned why type III effector complex generates cOA3 and cOA5, which are even the predominant products (Kazlauskiene et al., 2017; Smalakyte et al., 2020); and whether there are any other kinds of effector proteins presented in type III systems. Very recently, it is found that a novel CD-NTase produces cOA3, which in turn activates its effector endonuclease NucC to degrade DNA non-specifically to provide immunity against bacteriophage (Lau et al., 2020; Ye et al., 2020). Interestingly, NucC homologs as accessory proteins are also encoded within type III CRISPR/Cas systems and can be strongly activated by cOA3, indicating the existence of effector proteins without CARF domains in type III systems (Lau et al., 2020; Malone et al., 2020). Indeed, many other kinds of accessory proteins have been identified in type III systems (Shah et al., 2019). New cOA-activated effector proteins may still exist and remain to be identified, especially in some type III systems that contain Cas10 but lack any CARF domain proteins (Koonin and Makarova, 2018).
The effector protein Csm6 has been shown to be essential for type III-A CRISPR-Cas systems against phage and plasmid even before the cOA signaling was discovered in 2017 (Hatoum-Aslan et al., 2014; Jiang et al., 2016). Anti-phage activity of Csm6 was demonstrated to be dependent on Cas10 activation and cOA synthesis in vivo at the time of cOA signaling discovery (Niewoehner et al., 2017). Since Csm6/Csx1 cleaves RNA with a preference for only one or two nucleotides (Kazlauskiene et al., 2017; Foster et al., 2019; Jia et al., 2019b; Molina et al., 2019), it is largely sequence non-specific, and degrades RNA of host and invader indiscriminately. Thus, the activated effector Csm6/Csx1 is deleterious to the host, and it was proposed that cOA signaling confers host defense through inducing cell dormancy to arrest infection or inducing programmed host cell death to abort infection (Kazlauskiene et al., 2017). Indeed, it has been observed that Csm6 activation resulted in degradation of both host and plasmid transcripts, and induced growth arrest of the host which was critical for plasmid clearance (Rostøl and Marraffini, 2019). Recently, it was found that a kind of jumbo phages form nucleus-like structures during infection to protect their DNA from DNA-targeting nucleases (Chaikeeratisak et al., 2017; Mendoza et al., 2020). However, a type III system can provide robust immunity against such nucleus-forming jumbo phage (Malone et al., 2020). In this case, the cOA signaling is essential for the type III system against the jumbo phage (Malone et al., 2020). Interestingly, the effector protein involved in the cOA signaling is a NucC-like DNA nuclease but not the ribonuclease (Malone et al., 2020), which is not capable of cleaving the jumbo phage DNA in principle. Thus, it is likely that the cOA signaling confers defense by inducing host dormancy or abortive infection through non-specifically degrading the host genome.
It is worth noting that Csm6/Csx1 activation is crucial for efficient immunity against virus when targets are late-expressed viral genes but not the early-expressed genes (Jiang et al., 2016; Bhoobalan-Chitty et al., 2019). It was suggested that the Cas10 ssDNase is sufficient to clear the invaders when targets are early-expressed genes (Jiang et al., 2016; Bhoobalan-Chitty et al., 2019). In this case, it is not necessary for the host to activate cOA signaling pathway which might also be toxic to the host. Indeed, it was shown that targeting the late-expressed viral gene exhibits a relatively stronger antiviral immunity than targeting the early-expressed viral gene when the Cas10 ssDNase is inactivated, indicating that cOA signaling-mediated immunity may be stronger in targeting the late-expressed viral gene than the early-expressed gene (Bhoobalan-Chitty et al., 2019). Recent studies have shown that the Palm domain of Cas10 can be strongly activated even when the target genes are transcribed at very low levels (Rostøl and Marraffini, 2019; Athukoralage et al., 2020b), so it is unlikely that transcripts from early-expressed genes cannot activate Cas10. One possibility is that the activity of Cas10 Palm domain is inhibited in the early infection stage by some unknown mechanism in host cells, and this inhibition is released in the late infection stage for the activation of Cas10 Palm domain. Such hypothesis could be supported by findings that cellular nucleotides such as dATP, AMP, and ADP can also bind to the adenosine binding sites of Cas10 and affect cOA synthesis (Kazlauskiene et al., 2017). The level of these nucleotides may be decreased during viral replication, promoting activation of cOA synthesis.
Histidine-aspartate domain of Cas10, which is responsible for non-specific ssDNA degradation, is also involved in immunity against viruses and plasmids. It was reported that cOA signaling should be coupled with Cas10 ssDNase activity for efficient clearance of invader genomes (Jiang et al., 2016; Rostøl and Marraffini, 2019; Varble and Marraffini, 2019). However, in some studies, inactivation of HD domain of Cas10 has little effect on immunity against invaders, suggesting that without the assistance of Cas10 ssDNase activity, the type III effector complex with cOA signaling can still provide sufficient immunoprotection (Foster et al., 2019; Liu et al., 2019; Malone et al., 2020). Cas10 ssDNase was previously proposed to be involved in ssDNA cleavage at the transcription bubble, but a recent study argued this mechanism, leaving the real role of the Cas10 ssDNase unclear (Liu et al., 2019). Thus, how cOA signaling cooperates with the Cas10 ssDNase for immune defense remains to be investigated.
Due to its toxicity to the host, extant cOA should be removed after clearance of the invaders to enable host cells to return to normal growth state (Figure 1B). The first cOA nuclease, also named CRISPR-associated ring nuclease 1 (Crn1), was identified from crenarchaeote Sulfolobus solfataricus (Athukoralage et al., 2018). Crn1 is a CARF domain-containing protein that forms a homodimer, and specifically cleaves cOA4 into linear di-adenylate products to switch off the cOA4-activated effector proteins (Athukoralage et al., 2018). Interestingly, the CARF domain of effector protein Csm6 is also found to be capable of degrading cOA, thereby functioning as self-limiting ribonucleases (Athukoralage et al., 2019; Jia et al., 2019b; Garcia-Doval et al., 2020). Notably, very recently, it was found the HEPN domain of Csm6 can also degrade cOA to self-regulate its RNase activity (Smalakyte et al., 2020). Moreover, recent studies report that the widespread CRISPR associated protein Csx3 is a novel ring nuclease, named Crn3 (CRISPR associated ring nuclease 3; Athukoralage et al., 2020a; Brown et al., 2020). Interestingly, an unusual cooperative catalytic mechanism was found in which an active site of Csx3 tetramer is formed by two dimers sandwiching a cOA4 substrate (Athukoralage et al., 2020a). In addition, a metal-dependent and membrane-associated DHH-DHHA1 family nuclease (MAD) from Sulfolobus islandicus has recently been identified as a novel cOA-degrading enzyme (Zhao et al., 2020). MAD can accelerate the clearance of high-level cOA and may cooperate with cellular ring nuclease to remove cOA (Zhao et al., 2020).
Since cOA signaling promotes strong antiviral immunity, conversely, virus can utilize different strategies to restrict cOA signaling for immune evasion. Obviously, cOA degradation is a simple and efficient way for viruses to evade immune response. Indeed, a new family of viral anti-CRISPR (Acr) protein, AcrIII-1, was recently identified as a ring nuclease that specifically degrades cOA4, suggesting that it functions as Acr protein against cOA4-triggered type III CRISPR-Cas immunity (Athukoralage et al., 2020c). AcrIII-1 has a higher activity for cOA4 degradation than Crn1 and Crn3, and is unrelated to the CARF family proteins (Athukoralage et al., 2020a,c). AcrIII-1 homologs are widespread in various prokaryotes, where AcrIII-1 homologs may function as host-encoded ring nuclease like Crn1 and Crn3, thus named Crn2 (Athukoralage et al., 2020c). Very recently, a novel type III CRISPR-Cas inhibitor AcrIIIB1, encoded by Sulfolobus virus, has been identified to inhibit type III-B system immunity by binding to its effector complex to affect cOA signaling, demonstrating another strategy developed by virus to evade cOA signaling-mediated immunity (Bhoobalan-Chitty et al., 2019).
Currently, all of the characterized cOA nucleases (Crn1–3 and MAD) specifically degrade cOA4. However, given that Cas10 synthesizes cOAs ranging from cOA3 to cOA6, it is rational to predict the existence of other cOA-specific nucleases in prokaryotes and viruses. A cOA6-activated Csm6 can cleave cOA6 by its CARF domain (Garcia-Doval et al., 2020), indicating there may be presence of other CARF domain proteins such as Crn1 homologs that can specifically cleave cOA6. Moreover, MAD which is distinct from ring nucleases has a board substrate spectrum including cyclic di-nucleotides and ssRNA, implying MAD could degrade various cOAs (Zhao et al., 2020). Additionally, AcrIIIB1 utilizes a special strategy to inhibit cOA signaling, but its homologous proteins are only found in a few archaeal viruses (Bhoobalan-Chitty et al., 2019). The inhibitory activity of AcrIIIB1 also inspires us to explore other Acr proteins in bacteriophages and more strategies for evasion of cOA signaling such as inhibiting the effector protein activities.
In recent studies, a large family of CD-NTases have been found to produce a wide variety of cyclic di- and trinucleotides including 3'3' cyclic UMP-AMP, 3'3'3' cyclic AMP-AMP-GMP, and 2'3'3' cyclic AMP-AMP-AMP, which had never been reported previously (Whiteley et al., 2019; Lowey et al., 2020). These cyclic di- and trinucleotides can activate the downstream effector proteins, such as patatin-like phospholipases, DNA endonucleases, proteases, and pore-forming transmembrane proteins, to mediate anti-phage immunity by abortive infection (Cohen et al., 2019; Lau et al., 2020; Lowey et al., 2020; Ye et al., 2020), which is similar to the action of effector proteins of cOA signaling in type III systems. This newly discovered anti-bacteriophage defense system is termed cyclic oligonucleotide-based anti-phage signaling system (CBASS; Cohen et al., 2019), which is widespread and diverse in bacteria and crosstalks with cOA signaling. For example, cOA3-activated DNA endonuclease in CBASS is also present in some type III systems and is considered to be an important effector protein of cOA signaling for immunity against phage (Lau et al., 2020; Malone et al., 2020). Very recently, it was found that the major CBASS-associated protein effectors contain a SAVED domain, which is a fusion of two CARF-like domains, but recognize diverse asymmetric cyclic oligonucleotide signals such as 3'3'3'cyclic AMP-AMP-GMP and 3'3'3' cyclic AMP-AMP-AMP (cOA3) which are synthesized by CD-NTases (Lowey et al., 2020). Interestingly, Bioinformatics analysis showed that CD-NTases and SAVED domains fused to protein partners such as Lon protease and pore-forming transmembrane protein, which are occasionally incorporated into type III CRISPR loci (Burroughs et al., 2015; Lowey et al., 2020), which may increase the complexity of the regulation of the cOA signaling and enhance cOA signaling-mediated immunity.
Cyclic oligonucleotide molecules discovered in the past 3 years have greatly expanded our understanding on nucleotide signaling molecules in prokaryotes for decades. Due to the diversity of the effector proteins in cOA signaling and crosstalk between cOA signaling and CBASS, cOA signaling is far from fully elucidated. Future studies of cOA regulation will further expand our understanding of the role of cOA signaling and give us new insights into the cyclic oligonucleotides involved in antiviral defense systems.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31900032, 31670175, and 31870165) and Shenzhen Science and Technology Innovation Fund (JCYJ20170413115637100).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ahmad, S., Wang, B., Walker, M. D., Tran, H. R., Stogios, P. J., Savchenko, A., et al. (2019). An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678. doi: 10.1038/s41586-019-1735-9
Anantharaman, V., Makarova, K. S., Burroughs, A. M., Koonin, E. V., and Aravind, L. (2013). Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 8:15. doi: 10.1186/1745-6150-8-15
Athukoralage, J. S., Graham, S., Grüschow, S., Rouillon, C., and White, M. F. (2019). A type III CRISPR ancillary ribonuclease degrades its cyclic oligoadenylate activator. J. Mol. Biol. 431, 2894–2899. doi: 10.1016/j.jmb.2019.04.041
Athukoralage, J. S., Graham, S., Rouillon, C., Grüschow, S., Czekster, C. M., and White, M. F. (2020b). The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling. Elife 9:e55852. doi: 10.7554/eLife.55852
Athukoralage, J. S., McMahon, S. A., Zhang, C., Grüschow, S., Graham, S., Krupovic, M., et al. (2020c). An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575. doi: 10.1038/s41586-019-1909-5
Athukoralage, J. S., McQuarrie, S., Grüschow, S., Graham, S., Gloster, T. M., and White, M. F. (2020a). Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage. Elife 9:e57627. doi: 10.7554/eLife.57627
Athukoralage, J. S., Rouillon, C., Graham, S., Grüschow, S., and White, M. F. (2018). Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 562, 277–280. doi: 10.1038/s41586-018-0557-5
Bhoobalan-Chitty, Y., Johansen, T. B., Di Cianni, N., and Peng, X. (2019). Inhibition of type III CRISPR-Cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell 179, 448–458. doi: 10.1016/j.cell.2019.09.003
Brown, S., Gauvin, C. C., Charbonneau, A. A., Burman, N., and Lawrence, C. M. (2020). Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type-III CRISPR-Cas that degrades the second messenger cA4. J. Biol. Chem. 295, 14963–14972. doi: 10.1074/jbc.RA120.014099
Burroughs, A. M., Zhang, D., Schäffer, D. E., Iyer, L. M., and Aravind, L. (2015). Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654. doi: 10.1093/nar/gkv1267
Chaikeeratisak, V., Nguyen, K., Khanna, K., Brilot, A. F., Erb, M. L., Coker, J. K., et al. (2017). Assembly of a nucleus-like structure during viral replication in bacteria. Science 355, 194–197. doi: 10.1126/science.aal2130
Cohen, D., Melamed, S., Millman, A., Shulman, G., Oppenheimer-Shaanan, Y., Kacen, A., et al. (2019). Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 574, 691–695. doi: 10.1038/s41586-019-1605-5
Deng, L., Garrett, R. A., Shah, S. A., Peng, X., and She, Q. (2013). A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 87, 1088–1099. doi: 10.1111/mmi.12152
Elmore, J. R., Sheppard, N. F., Ramia, N., Deighan, T., Li, H., Terns, R. M., et al. (2016). Bipartite recognition of target RNAs activates DNA cleavage by the type III-B CRISPR-Cas system. Genes Dev. 30, 447–459. doi: 10.1101/gad.272153.115
Estrella, M. A., Kuo, F. T., and Bailey, S. (2016). RNA-activated DNA cleavage by the type III-B CRISPR-Cas effector complex. Genes Dev. 30, 460–470. doi: 10.1101/gad.273722.115
Foster, K., Grüschow, S., Bailey, S., White, M. F., and Terns, M. P. (2020). Regulation of the RNA and DNA nuclease activities required for Pyrococcus furiosus type III-B CRISPR-Cas immunity. Nucleic Acids Res. 48, 4418–4434. doi: 10.1093/nar/gkaa176
Foster, K., Kalter, J., Woodside, W., Terns, R. M., and Terns, M. P. (2019). The ribonuclease activity of Csm6 is required for anti-plasmid immunity by type III-A CRISPR-Cas systems. RNA Biol. 16, 449–460. doi: 10.1080/15476286.2018.1493334
Garcia-Doval, C., Schwede, F., Berk, C., Rostøl, J. T., Niewoehner, O., Tejero, O., et al. (2020). Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat. Commun. 11:1596. doi: 10.1038/s41467-020-15334-5
Goldberg, G. W., Jiang, W., Bikard, D., and Marraffini, L. A. (2014). Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637. doi: 10.1038/nature13637
Goldberg, G. W., McMillan, E. A., Varble, A., Modell, J. W., Samai, P., Jiang, W., et al. (2018). Incomplete prophage tolerance by type III-A CRISPR-Cas systems reduces the fitness of lysogenic hosts. Nat. Commun. 9:61. doi: 10.1038/s41467-017-02557-2
Grüschow, S., Athukoralage, J. S., Graham, S., Hoogeboom, T., and White, M. F. (2019). Cyclic oligoadenylate signalling mediates Mycobacterium tuberculosis CRISPR defence. Nucleic Acids Res. 47, 9259–9270. doi: 10.1093/nar/gkz676
Guo, T., Zheng, F., Zeng, Z., Yang, Y., Li, Q., She, Q., et al. (2019). Cmr3 regulates the suppression on cyclic oligoadenylate synthesis by tag complementarity in a type III-B CRISPR-Cas system. RNA Biol. 16, 1513–1520. doi: 10.1080/15476286.2019.1642725
Hale, C. R., Zhao, P., Olson, S., Duff, M. O., Graveley, B. R., Wells, L., et al. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956. doi: 10.1016/j.cell.2009.07.040
Hallberg, Z. F., Wang, X. C., Wright, T. A., Nan, B., Ad, O., Yeo, J., et al. (2016). Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3', 3'-cGAMP). Proc. Natl. Acad. Sci. U. S. A. 113, 1790–1795. doi: 10.1073/pnas.1515287113
Han, W., Stella, S., Zhang, Y., Guo, T., Sulek, K., Peng-Lundgren, L., et al. (2018). A type III-B Cmr effector complex catalyzes the synthesis of cyclic oligoadenylate second messengers by cooperative substrate binding. Nucleic Acids Res. 46, 10319–10330. doi: 10.1093/nar/gky844
Hatoum-Aslan, A., Maniv, I., Samai, P., and Marraffini, L. A. (2014). Genetic characterization of antiplasmid immunity through a type III-A CRISPR-Cas system. J. Bacteriol. 196, 310–317. doi: 10.1128/JB.01130-13
Jia, N., Jones, R., Sukenick, G., and Patel, D. J. (2019a). Second messenger cA(4) formation within the composite Csm1 palm pocket of type III-A CRISPR-Cas Csm complex and its release path. Mol. Cell 75, 933–943. doi: 10.1016/j.molcel.2019.06.013
Jia, N., Jones, R., Yang, G., Ouerfelli, O., and Patel, D. J. (2019b). CRISPR-Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA(4) cleavage with ApA>p formation terminating RNase activity. Mol. Cell 75, 944–956. doi: 10.1016/j.molcel.2019.06.014
Jia, N., Mo, C. Y., Wang, C., Eng, E. T., Marraffini, L. A., and Patel, D. J. (2019c). Type III-A CRISPR-Cas Csm complexes: assembly, periodic RNA cleavage, DNase activity regulation, and autoimmunity. Mol. Cell 73, 264–277.e5. doi: 10.1016/j.molcel.2018.11.007
Jiang, W., Samai, P., and Marraffini, L. A. (2016). Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164, 710–721. doi: 10.1016/j.cell.2015.12.053
Johnson, K., Learn, B. A., Estrella, M. A., and Bailey, S. (2019). Target sequence requirements of a type III-B CRISPR-Cas immune system. J. Biol. Chem. 294, 10290–10299. doi: 10.1074/jbc.RA119.008728
Kazlauskiene, M., Kostiuk, G., Venclovas, Č., Tamulaitis, G., and Siksnys, V. (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609. doi: 10.1126/science.aao0100
Kazlauskiene, M., Tamulaitis, G., Kostiuk, G., Venclovas, Č., and Siksnys, V. (2016). Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell 62, 295–306. doi: 10.1016/j.molcel.2016.03.024
Koonin, E. V., and Makarova, K. S. (2018). Discovery of oligonucleotide signaling mediated by CRISPR-associated polymerases solves two puzzles but leaves an enigma. ACS Chem. Biol. 13, 309–312. doi: 10.1021/acschembio.7b00713
Lau, R. K., Ye, Q., Birkholz, E. A., Berg, K. R., Patel, L., Mathews, I. T., et al. (2020). Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell 77, 723–733. doi: 10.1016/j.molcel.2019.12.010
Liu, T. Y., Liu, J. J., Aditham, A. J., Nogales, E., and Doudna, J. A. (2019). Target preference of type III-A CRISPR-Cas complexes at the transcription bubble. Nat. Commun. 10:3001. doi: 10.1038/s41467-019-10780-2
Lowey, B., Whiteley, A. T., Keszei, A. F. A., Morehouse, B. R., Mathews, I. T., Antine, S. P., et al. (2020). CBASS immunity uses CARF-related effectors to sense 3'-5'- and 2'-5'-linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell 182, 38.e17–49.e17. doi: 10.1016/j.cell.2020.05.019
Makarova, K. S., Anantharaman, V., Grishin, N. V., Koonin, E. V., and Aravind, L. (2014). CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 5:102. doi: 10.3389/fgene.2014.00102
Makarova, K. S., Aravind, L., Grishin, N. V., Rogozin, I. B., and Koonin, E. V. (2002). A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 30, 482–496. doi: 10.1093/nar/30.2.482
Makarova, K. S., Aravind, L., Wolf, Y. I., and Koonin, E. V. (2011). Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct 6:38. doi: 10.1186/1745-6150-6-38
Makarova, K. S., Timinskas, A., Wolf, Y. I., Gussow, A. B., Siksnys, V., Venclovas, Č., et al. (2020a). Evolutionary and functional classification of the CARF domain superfamily, key sensors in prokaryotic antivirus defense. Nucleic Acids Res. 48, 8828–8847. doi: 10.1093/nar/gkaa635
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020b). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi: 10.1038/s41579-019-0299-x
Malone, L. M., Warring, S. L., Jackson, S. A., Warnecke, C., Gardner, P. P., Gumy, L. F., et al. (2020). A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat. Microbiol. 5, 48–55. doi: 10.1038/s41564-019-0612-5
Marraffini, L. A., and Sontheimer, E. J. (2010). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. doi: 10.1038/nature08703
McMahon, S. A., Zhu, W., Graham, S., Rambo, R., White, M. F., and Gloster, T. M. (2020). Structure and mechanism of a type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate. Nat. Commun. 11:500. doi: 10.1038/s41467-019-14222-x
Mendoza, S. D., Nieweglowska, E. S., Govindarajan, S., Leon, L. M., Berry, J. D., Tiwari, A., et al. (2020). A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature 577, 244–248. doi: 10.1038/s41586-019-1786-y
Mohanraju, P., Makarova, K. S., Zetsche, B., Zhang, F., Koonin, E. V., and van der Oost, J. (2016). Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353:aad5147. doi: 10.1126/science.aad5147
Molina, R., Sofos, N., and Montoya, G. (2020). Structural basis of CRISPR-Cas type III prokaryotic defence systems. Curr. Opin. Struct. Biol. 65, 119–129. doi: 10.1016/j.sbi.2020.06.010
Molina, R., Stella, S., Feng, M., Sofos, N., Jauniskis, V., Pozdnyakova, I., et al. (2019). Structure of Csx1-cOA4 complex reveals the basis of RNA decay in type III-B CRISPR-Cas. Nat. Commun. 10:4302. doi: 10.1038/s41467-019-12244-z
Nasef, M., Muffly, M. C., Beckman, A. B., Rowe, S. J., Walker, F. C., Hatoum-Aslan, A., et al. (2019). Regulation of cyclic oligoadenylate synthesis by the Staphylococcus epidermidis Cas10-Csm complex. RNA 25, 948–962. doi: 10.1261/rna.070417.119
Niewoehner, O., Garcia-Doval, C., Rostøl, J. T., Berk, C., Schwede, F., Bigler, L., et al. (2017). Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548. doi: 10.1038/nature23467
Pyenson, N. C., Gayvert, K., Varble, A., Elemento, O., and Marraffini, L. A. (2017). Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe 22, 343.e3–353.e3. doi: 10.1016/j.chom.2017.07.016
Pyenson, N. C., and Marraffini, L. A. (2017). Type III CRISPR-Cas systems: when DNA cleavage just isn’t enough. Curr. Opin. Microbiol. 37, 150–154. doi: 10.1016/j.mib.2017.08.003
Rostøl, J. T., and Marraffini, L. A. (2019). Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR-Cas immunity. Nat. Microbiol. 4, 656–662. doi: 10.1038/s41564-018-0353-x
Rouillon, C., Athukoralage, J. S., Graham, S., Grüschow, S., and White, M. F. (2018). Control of cyclic oligoadenylate synthesis in a type III CRISPR system. Elife 7:e36734. doi: 10.7554/eLife.36734
Rouillon, C., Athukoralage, J. S., Graham, S., Grüschow, S., and White, M. F. (2019). Investigation of the cyclic oligoadenylate signaling pathway of type III CRISPR systems. Methods Enzymol. 616, 191–218. doi: 10.1016/bs.mie.2018.10.020
Samolygo, A., Athukoralage, J. S., Graham, S., and White, M. F. (2020). Fuse to defuse: a self-limiting ribonuclease-ring nuclease fusion for type III CRISPR defence. Nucleic Acids Res. 48, 6149–6156. doi: 10.1093/nar/gkaa298
Shah, S. A., Alkhnbashi, O. S., Behler, J., Han, W., She, Q., Hess, W. R., et al. (2019). Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families. RNA Biol. 16, 530–542. doi: 10.1080/15476286.2018.1483685
Smalakyte, D., Kazlauskiene, M., Havelund, J. F., Rukšėnaitė, A., Rimaite, A., Tamulaitiene, G., et al. (2020). Type III-A CRISPR-associated protein Csm6 degrades cyclic hexa-adenylate activator using both CARF and HEPN domains. Nucleic Acids Res. 48, 9204–9217. doi: 10.1093/nar/gkaa634
Sofos, N., Feng, M., Stella, S., Pape, T., Fuglsang, A., Lin, J., et al. (2020). Structures of the Cmr-β complex reveal the regulation of the immunity mechanism of type III-B CRISPR-Cas. Mol. Cell 79, 741.e7–757.e7. doi: 10.1016/j.molcel.2020.07.008
Tamulaitis, G., Venclovas, Č., and Siksnys, V. (2017). Type III CRISPR-Cas immunity: major differences brushed aside. Trends Microbiol. 25, 49–61. doi: 10.1016/j.tim.2016.09.012
Varble, A., and Marraffini, L. A. (2019). Three new Cs for CRISPR: collateral, communicate, cooperate. Trends Genet. 35, 446–456. doi: 10.1016/j.tig.2019.03.009
Whiteley, A. T., Eaglesham, J. B., de Oliveira Mann, C. C., Morehouse, B. R., Lowey, B., Nieminen, E. A., et al. (2019). Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. doi: 10.1038/s41586-019-0953-5
Ye, Q., Lau, R. K., Mathews, I. T., Birkholz, E. A., Watrous, J. D., Azimi, C. S., et al. (2020). HORMA domain proteins and a trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol. Cell 77, 709.e7–722.e7. doi: 10.1016/j.molcel.2019.12.009
You, L., Ma, J., Wang, J., Artamonova, D., Wang, M., Liu, L., et al. (2019). Structure studies of the CRISPR-Csm complex reveal mechanism of co-transcriptional interference. Cell 176, 239–253. doi: 10.1016/j.cell.2018.10.052
Zhao, R., Yang, Y., Zheng, F., Zeng, Z., Feng, W., Jin, X., et al. (2020). A membrane-associated DHH-DHHA1 nuclease degrades type III CRISPR second messenger. Cell Rep. 32:108133. doi: 10.1016/j.celrep.2020.108133
Keywords: cyclic oligonucleotide, type III systems, CRISPR immune defense, CARF domain proteins, Cas10, CD-NTase
Citation: Huang F and Zhu B (2021) The Cyclic Oligoadenylate Signaling Pathway of Type III CRISPR-Cas Systems. Front. Microbiol. 11:602789. doi: 10.3389/fmicb.2020.602789
Received: 04 September 2020; Accepted: 11 November 2020;
Published: 20 January 2021.
Edited by:
Aixin Yan, The University of Hong Kong, Hong KongReviewed by:
Wenyuan Han, Huazhong Agricultural University, ChinaCopyright © 2021 Huang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengtao Huang, aHVhbmdfZmVuZ3Rhb0AxMjYuY29t; Bin Zhu, YmluX3podUBodXN0LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.