- Molecular Biology of Archaea, Faculty of Biology, Institute of Biology II, University of Freiburg, Freiburg, Germany
In Sulfolobus acidocaldarius, the protein phosphatase PP2A plays important regulatory roles in many cellular processes, including cell growth, cell shape and synthesis of the archaellum. A conserved prokaryotic protein, designated as SaUspA, was identified as an interaction partner of the phosphatase PP2A. SaUspA belongs to the universal stress protein (USP) superfamily, members of which are found in bacteria, archaea, plants and invertebrates. Biochemical analysis showed that SaUspA is a homodimeric ATP-binding protein, which also in vitro binds to PP2A. SaUspA did not hydrolyze ATP, but stimulated the phosphatase activity of PP2A and might in this manner affect many other processes. Interestingly, binding of ATP further enhanced SaUspA’s interaction with PP2A. In contrast to bacterial usp genes, environmental stress conditions including stationary phase, starvation stress, high salinity stress and UV stress did not stimulate expression of saUspA. Deletion of saUspA led to premature production of the archaellin FlaB in S. acidocaldarius although motility was not affected. The ΔsaUspA mutant showed a significant growth defect under high salinity stress and complementation of ATP-binding deficient mutant SaUspAG97A failed to restore this growth defect. Compared with the wild type strain, its growth or survival was not affected under heavy metal stress and UV stress. To date, this is the first study in which the physiological role of USP homologs in archaea have been reported.
Introduction
The universal stress protein (USP) superfamily is highly conserved in bacteria, archaea, plants and other eukaryotes (Vollmer and Bark, 2018). Expression of members of this family is triggered by a multitude of different environmental stress conditions, which suggests an important role of these proteins in general stress responses (Kvint et al., 2003; Vollmer and Bark, 2018). In the past three decades, significant effort has been made to explore the physiological functions of members of the USP superfamily in bacteria and plants. In bacteria, USP proteins were found to be involved in cell growth and survival under stress conditions, motility, biofilm formation, and virulence of pathogen to host (Kvint et al., 2003; Nachin et al., 2005; O’Connor and McClean, 2017; Vollmer and Bark, 2018). In plants, these proteins were critical for the tolerance against abiotic stresses including drought, osmotic stress and anoxia (Gonzali et al., 2015; Gutiérrez-beltrán et al., 2017; Vollmer and Bark, 2018).
UspA was originally discovered in Escherichia coli and was the founding member of the USP superfamily (Nystrdm and Neidhardt, 1992). Later, five paralogs of UspA were identified in E. coli, including UspC, UspD, UspE, UspF, and UspG (Gustavsson et al., 2002). A number of environmental stress conditions, such as nutrient starvation (carbon source, phosphate, sulfur and nitrogen) and osmotic stress, heat shock, UV exposure, and exposure to high salinity or heavy metals induced their expression (Kvint et al., 2003). With the increasing amount of sequenced genomes, more and more USP family members were found in different kinds of organisms such as bacteria, archaea, fungi, plants and invertebrates (Vollmer and Bark, 2018). USP superfamily members are divided into two major groups: non-ATP-binding and ATP-binding members (Kvint et al., 2003; Vollmer and Bark, 2018). UspA from E. coli and Haemophilus influenzae are the representative members of the first group, and the second group is represented by MJ0577 from Methanocaldococcus jannaschii (Zarembinski et al., 1998; Sousa and Mckay, 2001). Notably, ATP-binding is important for the function of the ATP-binding USP proteins. In Mycobacterium tuberculosis, ATP-binding deficient mutants of USP homologs Rv2623 and Rv2624c failed to regulate cell growth and pathogenicity (Drumm et al., 2009). Homodimer formation of E. coli USP proteins and heterodimer formation of different USP proteins with each other were shown to be responsible for the functional diversity in response to different stresses (Vollmer and Bark, 2018). In E. coli, UspA, UspC, and UspD formed homo- and hetero-dimers, and UspF and UspG can form three types of dimers (Nachin et al., 2008). In addition, some members of USP superfamily were phosphorylated in vitro/vivo. Phosphorylation of UspA in E. coli occurred specifically at serine/threonine residues in vivo, which depended on the TypA tyrosine phosphoprotein (Freestone et al., 1997). With cell lysates, the autophosphorylation of UspA occurs in vitro. UspG also showed autophosphorylation activity in vitro (Weber and Jung, 2006). In M. tuberculosis, threonine-237 of Rv2623 was found to be phosphorylated, and mutation to alanine decreased its interaction with a putative ATP binding cassette transporter Rv1747 and abolished its regulatory function in cell growth (Glass et al., 2017).
As described above, USP proteins are rather well studied in bacteria and plants. Although the structures of USP proteins from M. jannaschii and Archaeoglobus fulgidus have been determined (Zarembinski et al., 1998; Tkaczuk et al., 2013), the physiological roles of USP proteins have not been addressed in archaea yet.
In Sulfolobus acidocaldarius, three USP homologs are encoded in the genome and we designated them UspA (Saci_0887), UspB (Saci_1357) and UspC (Saci_1640), which for clarity we will call here SaUspA, SaUspB, and SaUspC, respectively. We recently identified SaUspA as an important interaction partner of PP2A, one of the two phosphatases of S. acidocaldarius (Ye et al., 2020), and here, we focused on the biochemical characterization of SaUspA and investigated its physiological role in S. acidocaldarius. Our results indicate that SaUspA belongs to the ATP-binding USP group and that ATP-binding by SaUspA is important for its interaction with the threonine/serine phosphatase PP2A. Our results also showed that SaUspA plays a role in the adaptation to high salinity stress.
Materials and Methods
Strains and Growth Conditions
S. acidocaldarius strains (MW001 and derived mutants) used in this study (Supplementary Table S1) were cultivated in Brock’ basal medium supplemented with 0.1% (w/v) NZ-amine, 0.2% (w/v) dextrin and 10 μg/mL uracil (Brock et al., 1972; Wagner et al., 2012). For S. acidocaldarius strains containing plasmids, the same medium without uracil was used. E. coli strains (Supplementary Table S1) were grown in Lysogeny broth (LB) medium or on LB agar plates with the appropriate antibiotics (50 μg/mL ampicillin, 25 μg/mL kanamycin or 30 μg/mL chloramphenicol). E. coli Top 10 was used for plasmids propagation. E. coli ER1281 was used to methylate plasmids for S. acidocaldarius transformation. E. coli Rosetta (DE3)/pLysS was used for overexpressing S. acidocaldarius proteins.
Overexpression and Purification of S. acidocaldarius Proteins in E. coli
After the transformation of the expression plasmids (pSVA1037, pSVA1076, pSVA2009, pSVA5134, or pSVA5139) (Supplementary Table S1) into E. coli Rosetta (DE3)/pLysS, 20 mL of overnight culture of E. coli was inoculated in 2 L LB medium supplemented with the appropriate antibiotics and cultivated at 37°C and 140 rpm. At an OD600 of 0.5–0.6, the culture was cooled down and expression was induced with 500 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16°C overnight. On the next day, cells were harvested by centrifugation and re-suspended in lysis buffer (50 mM Tris pH8, 150 mM NaCl) with DNaseI and protease inhibitor. Cells were lysed by using the French press, and the supernatant was collected after centrifugation at 4°C and 48,000 × g for 20 min. To remove E. coli proteins, the supernatant was heated in a water bath to 70°C for 20 min followed by ultracentrifugation at 236,000 × g for 45 min. The supernatant was used for His-tag or Strep-tag purification depending on the isolated protein. For SaUspA and SaUspAG97A, an extra ammonium sulfate precipitation step was performed to obtain nucleotide free protein. All proteins were applied to a Superdex 200 10/300 GL column or Superdex 200 16/600 column (GE Healthcare) equilibrated in lysis buffer before use.
In vitro Protein Interaction Analysis
StrepII tagged SaUspA or SaUspAG97A were used in interaction assays with His6 tagged PP2A. Shortly, 500 μL of each purified protein (0.5 mg/mL) were mixed and incubated at 55°C for 30 min. The mixture was applied to a 1000 μL PureSpeed (PS) tip with 20 μL IMAC resin (Mettler Toledo) as described in the manufacture’s instruction. 50 μL of lysis buffer containing 500 mM imidazole was used in the elution step. To prepare the ATP-bound sample, SaUspA or SaUSPG97A were incubated with 1 mM ATP in lysis buffer containing 5 mM MgCl2 at 25°C for 30 min before incubation with PP2A-his. Fractions from all assays were separated on SDS-PAGE and analyzed by Western blot using α-His and/or α-Strep antibodies.
TNP-ATP Binding Assay
Binding of TNP-ATP [2′-(or-3′)-O-(Trinitrophenyl) Adenosine 5′-Triphosphate] was monitored by fluorescence increase upon binding to the protein using a Fluoromax-4 spectrofluorometer (Horiba). The excitation wavelength was set to 409 nm and the emission was detected at 540 nm. Slit widths for excitation and emission were set to 5 nm and 20 nm, respectively. Binding was measured by titration of 2.5 μM SaUspA or SaUspAG97A with increasing TNP-ATP concentrations (0–30 μM) at 25°C in lysis buffer containing 5 mM MgCl2. Fluorescence was corrected for TNP-ATP fluorescence in the absence of proteins and fitted with the Hill equation: F = [TNP-ATP]n/(K + [TNP-ATP]n), with F = fluorescence, [TNP-ATP] = concentration of TNP-ATP, Kd = dissociation constant and n = hill constant.
In vitro Phosphorylation
ArnC or ArnD were purified and in vitro phosphorylation assays with ArnC or ArnD were set up and performed as described by Reimann et al. (2012) in lysis buffer containing 1 mM MnCl2.
Enzyme Assays of PP2A
Protein serine/threonine phosphatase activity of PP2A was determined by using the artificial p-peptides RRA(pT)VA as the substrate according to manufacturer’s instruction (Promega) (Reimann et al., 2013). The reaction was performed with 100 ng PP2A at 70°C in lysis buffer containing 1 mM MnCl2. The release of free phosphate was determined using the Malachite green assay (Van Veldhoven and Mannaerts, 1987).
RNA Isolation and qRT-PCR
10 mL S. acidocaldarius culture was collected from the respective indicated growth conditions and time points, and total RNA was isolated with TRIzol (Invitrogen). Chromosomal DNA was removed from total RNA samples and cDNA was synthesized according to the standard protocol of the QuantiTect Reverse Transcription Kit (Qiagen). cDNA samples were analyzed with the qPCRBIO SyGreen Mix (PCR Biosystems) and Rotor-GeneQ Real-time PCR cycler (Qiagen). secY was used as the reference to normalize the qRT-PCR data. For analysis, at least three biological replicates and two technical replicates were used.
Construction of Markerless In-Frame Deletion Mutant for saUspA in S. acidocaldarius
The markerless in-frame deletion mutant of saUspA (saci_0887) was constructed as described by Wagner et al. (2012). To construct the deletion mutant plasmid pSVA5113, a PCR was performed to obtain an overlap fragment of 549 bp of the upstream region and of 568 bp of the downstream region using primers 7350/7351 and 7352/7353 (see Supplementary Table S2). The overlap fragment was cloned into pSVA407. After sequencing, the plasmid was methylated in E. coli ER1821 and transformed into S. acidocaldarius MW001. Transformants were selected with first selection gelrite plates lacking uracil [0.6% gelrite (w/v)] and second selection gelrite plates containing uracil (10 μg/mL) and 5-FOA (100 μg/mL). Obtained colonies were screened for the deletion and positive clones were sequenced.
Assays for Cell Survival and Aggregation Under UV Stress
UV treatment of S. acidocaldarius was performed as described by Wolferen et al. (2015). To determine cell survival rates, 10 mL culture (OD600 0.2–0.3) was poured in plastic petri dishes and treated with different doses of UV light (75, 125, and 200 J/m2) (254 nm; Spectroline UV cross-linker). A culture without UV treatment was used as control. All the cultures were serially diluted in medium and spread on solid gelrite plates. Plates were cultivated at 75°C for 3–4 days until colonies were visible. Colonies from UV treated and non-UV treated samples were counted. To quantify the cell survival rates, data from at least three independent experiments was used. For analyzing ups pili dependent cell aggregation, a dose of 75 J/m2 UV light was used. Samples without UV treatment were used as the control. After UV treatment, samples were cultivated at 75°C for 3 h and analyzed with phase-contrast microscopy.
Nutrient Starvation Assays and Western Blots
Cells were grown in nutrient rich medium (Brock’ basal medium with NZ-amine and dextrin) and at an OD600 of 0.4–0.5, cells were harvested by centrifugation at 75°C for 10 min and the supernatant was discarded. Cell pellets were resuspended in an equal volume of pre-warmed Brock’ basal medium without NZ-amine and dextrin and further cultivated at 75°C and 120 rpm. Samples were harvested at indicated time points.
Protein samples were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Roche). The membrane was blocked in blocking buffer with 0.2% I-BlockTM (Thermo Fisher) for 1 h, followed by incubation with the primary antibody α-FlaB (Eurogentec) (Lassak et al., 2012) overnight. After incubation with the secondary anti-rabbit-HRP antibody (Invitrogen) for 1 h, chemiluminescent signals were visualized by ECL Chemocam Imager (INTAS) with Clarity Western ECL blotting substrate (Bio-Rad).
Motility Assays
Motility assays were performed as described by Lassak et al. (2012). The respective S. acidocaldarius cultures were grown overnight till an OD600 of 0.4–0.5. 5 μL of each respective culture was spotted on semi-solid gelrite plates (0.15% gelrite (w/v)). Plates were incubated in closed plastic storage boxes at 75°C for 4–5 days before they were scanned.
Results
SaUspA Interacts With PP2A
SaUspA was recently identified as an interaction partner of the serine/threonine phosphatase PP2A during early nutrient starvation in S. acidocaldarius (Ye et al., 2020). Here, we set-out to further characterize SaUspA and its interaction with PP2A. To that end, C-terminally his6-tagged PP2A (PP2A-his) and C-terminally StrepII-tagged SaUspA (SaUspA-Strep) were purified. Size exclusion chromatography of SaUspA (molecular weight: 16.2 kDa) resulted in a monodisperse peak eluting at a volume corresponding to a molecular weight of ∼ 30 kDa (Figures 1A,B) suggesting that SaUspA forms a dimer. Dimerization of USP proteins occurs via a small C-terminal dimerization domain with a conserved sequence (See Figure 2A; Gutiérrez-beltrán et al., 2017). Overexpression in E. coli of a SaUspA variant which lacked the 5 C-terminal residues forming the dimerization domain (SaUspAΔdim) was not successful, which implied that the conserved dimerization domain is also critical for the stability of SaUspA. To further confirm the previously observed interaction between PP2A and SaUspA, a co-purification assay with PP2A-his and SaUspA-Strep was performed on Ni-NTA resin. SaUspA-Strep did not bind to the Ni-NTA resin by itself (Figure 1C), but SaUspA was identified in the elution fraction in the presence of PP2A-his (Figure 1D), further demonstrating that both proteins interact.
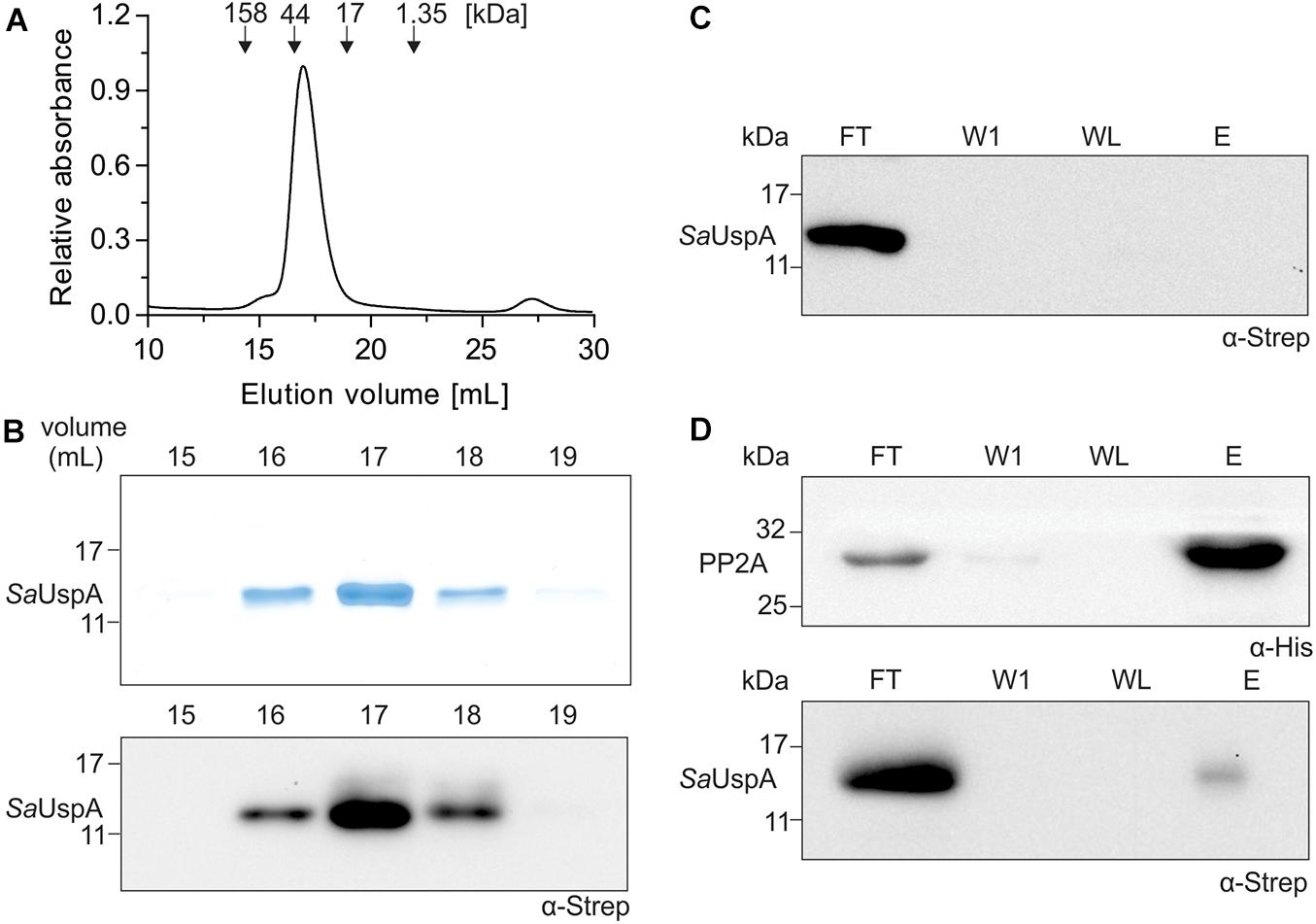
Figure 1. SaUspA is purified as a dimer and interacts with PP2A. (A) Purified Strep II-tagged SaUspA was analyzed on a Superdex 200 10/300 GL column. Molecular weight standard of 158, 44, 17, and 1.35 kDa are indicated. (B) Fractions from (A) were analyzed by SDS-PAGE (upper panel) and Western blot with α-Strep antibody (lower panel). (C) Strep II-tagged SaUspA was applied to Ni-NTA resin, the resin was washed and eluted with buffer containing 10 and 500 μM imidazole, respectively. Fractions were analyzed by Western blot with α-Strep antibody. FT, flowthrough; W1, first wash fraction; WL, last wash fraction; E, elution fraction. (D) In vitro interaction assays for PP2A and SaUspA. His-tagged PP2A and StrepII-tagged SaUspA were mixed and a co-purification was performed as in (C). All experiments were repeated at least three times and representative figures are shown.
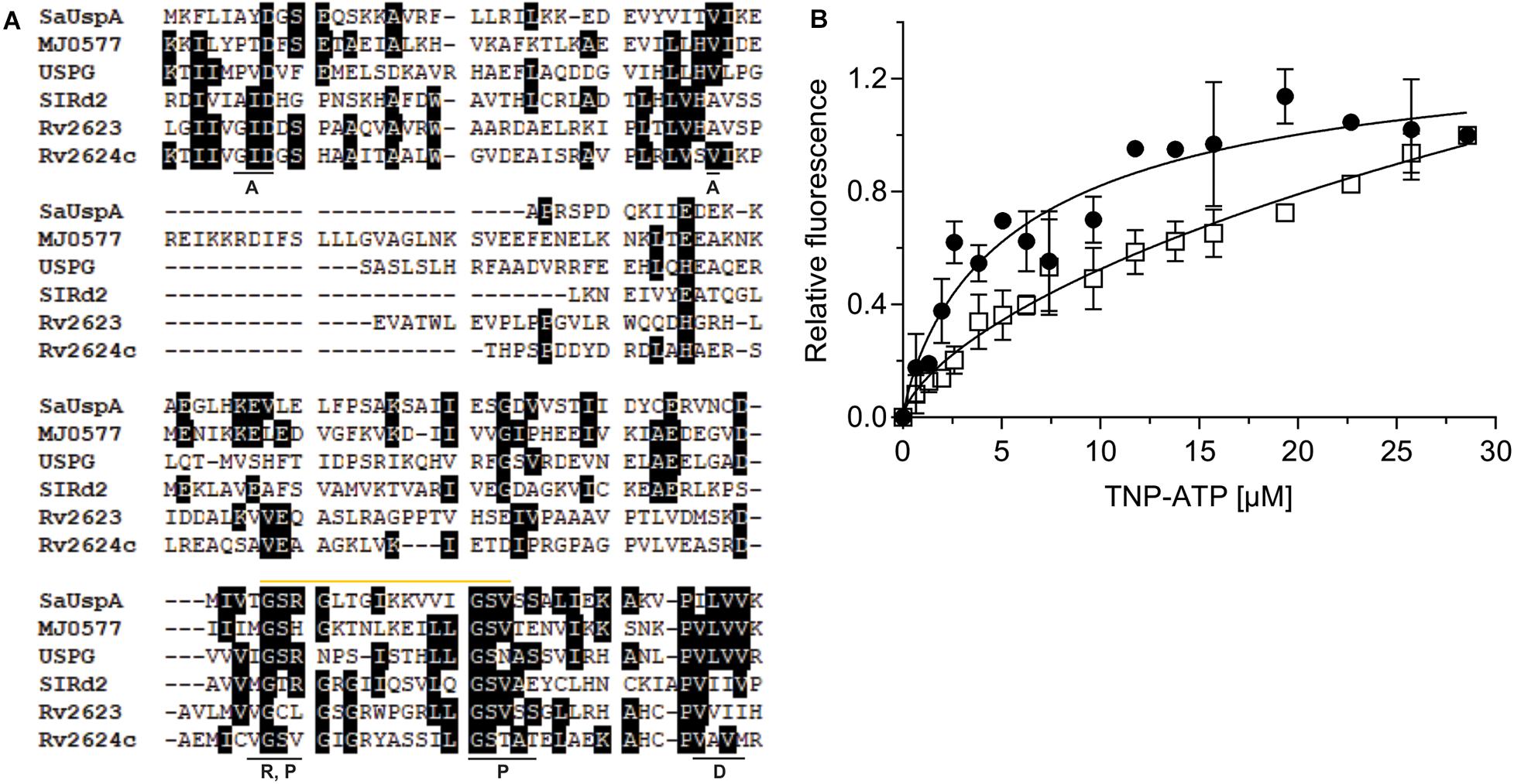
Figure 2. SaUspA is an ATP-binding protein. (A) Multiple sequence alignment of SaUspA and ATP-binding USP proteins including E. coli UspG, M. jannaschii MJ0577, N. benthamiana SlRd2, M. tuberculosis Rv2623 and Rv2624c. Conserved residues were marked with black boxes. Residues underlined with a black bar indicate residues important for dimerization (D) or residues interacting with the adenine (A), phosphate (P) or ribose (R) moieties of ATP are indicated. The Walker or P-loop motif [G-2x-G-9x-G-G(S/T)] responsible for ATP-binding is indicated by the yellow bar. (B) ATP-binding assays of SaUspA. The relative increase in fluorescence upon ATP-binding was determined by titrating 2.5 μM SaUspAWT (WT) (black circle) or SaUspAG97A (G97A) (open square) with increasing concentrations of TNP-ATP. Data was obtained from at least three independent experiments and binding curves were fitted to the Hill equation.
SaUspA Is an ATP-Binding Protein
A multiple protein sequence alignment with five USP proteins that have previously been shown to bind ATP, M. jannaschii MJ0557, E. coli UspG, Nicotiana benthamiana SIRd2, M. tuberculosis Rv2623 and Rv2624c was made (Zarembinski et al., 1998; Drumm et al., 2009; Jia et al., 2016; Gutiérrez-beltrán et al., 2017). This showed that the conserved ATP-binding Walker or P-loop motif [G-2X-G-9X-G(S/T)] and the C-terminus dimerization domain are conserved between SaUspA and these other five USP proteins (Figure 2A), suggesting that SaUspA belongs to the ATP-binding members of the USP family. To investigate whether SaUspA binds ATP, ATP-binding assays were performed using the fluorescent ATP analog TNP-ATP. To determine the binding affinity, 2.5 μM of SaUspA was titrated with 1–30 μM TNP-ATP. Normalized fluorescence intensities were plotted against the TNP-ATP concentration and data was fitted with the Hill equation (Hill coefficient of 0.84) resulting in a dissociation constant of 7 +/− 2 μM (Figure 2B), demonstrating that SaUspA could bind TNP-ATP. SaUspA was also tested for ATP hydrolysis, but ATP hydrolysis was below our detection limits. In M. tuberculosis, The D15E and G117A mutants in Rv2623 showed 34% and 21% ATP binding compared to wild type Rv2623 (Drumm et al., 2009), and mutation of D17 into E and G119 to alanine abolished Rv2624c’s ATP binding ability (Jia et al., 2016). To test whether these two residues are also important for ATP binding in SaUspA, the corresponding amino acids in SaUspA (D8 and G97) were mutated to alanine. No protein could be obtained when SaUspAD8A was expressed in E. coli, suggesting this mutation either destabilized the protein or that SaUspA is stabilized by ATP binding and the mutated protein can no longer bind ATP. However, SaUspAG97A was successfully expressed and purified. SaUspAG97A bound TNP-ATP-binding with a 50-fold lower dissociation constant of around 350 μM (Figure 2B), demonstrating that G97 of SaUspA plays a role in ATP binding.
SaUspA Is Phosphorylated by ArnC
In bacteria and plants, several members of the USP superfamily are phosphorylated, however, only a few kinases responsible for phosphorylation were identified (Freestone et al., 1997; Lenman et al., 2008; Merkouropoulos et al., 2008; Glass et al., 2017; Gutiérrez-beltrán et al., 2017; Potel et al., 2018). In S. acidocaldarius, a number of protein kinases have been characterized and ArnC and ArnD are relatively well studied (Reimann et al., 2012; Hoffmann et al., 2017, 2019; Bischof et al., 2019; Maklad et al., 2020). To investigate whether SaUspA can autophosphorylate or is phosphorylated by ArnC or ArnD in vitro, SaUspA was incubated with or without ArnC or ArnD and [γ−32P] ATP (Figure 3). When SaUspA was incubated with [γ−32P] ATP, no autophosphorylation could be detected, whereas, as described previously (Reimann et al., 2012) autophosphorylation was detected for both ArnC and ArnD. Incubation of SaUspA with ArnC resulted in phosphorylation of SaUspA and loss of phosphorylation of ArnC, while no effects could be observed when SaUspA was incubated with ArnD. Thus, SaUspA did not show autophosphorylation activity in vitro, but could be phosphorylated by ArnC.
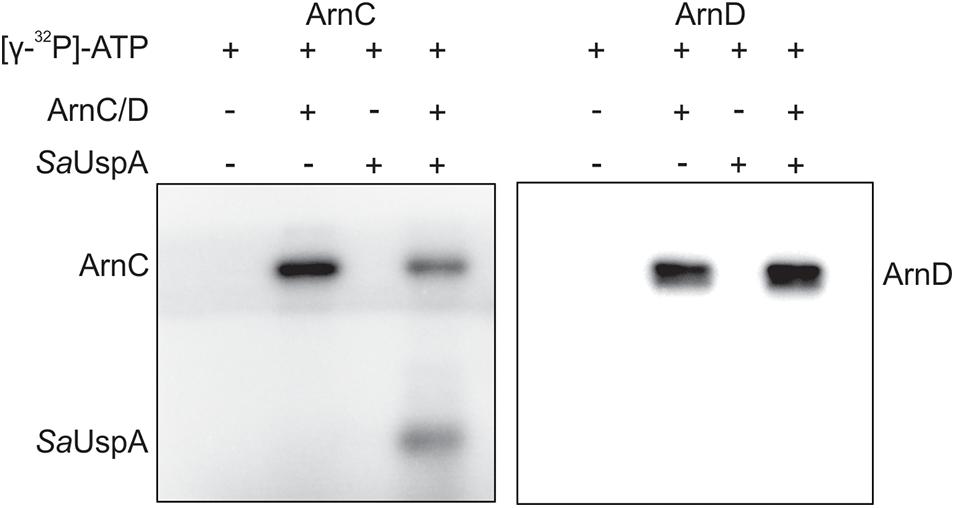
Figure 3. In vitro phosphorylation assays of SaUspA. Purified SaUspA was incubated at 55°C using Mn2+ as cofactor in the presence or absence of the kinases ArnC or ArnD and [γ−32P] ATP.
Binding of ATP by SaUspA Enhances Its Interaction With PP2A
Since SaUspA binds ATP and interacts with PP2A in the in vitro assays, it was tested whether the interaction between PP2A and SaUspA was modified by ATP binding. Upon the addition of ATP, a clear increase of SaUspA was detected in the elution fraction of the interaction assay when compared to the sample without ATP (Figure 4A). This indicated that ATP-binding of SaUspA enhanced its interaction with PP2A. When the assays were performed with the ATP-binding deficient mutant SaUspAG97A (Figure 4B), interaction between SaUspAG97A and PP2A was still observed, but no increase could be observed upon the addition of ATP, further demonstrating that ATP-binding to SaUspA stimulates binding to PP2A.
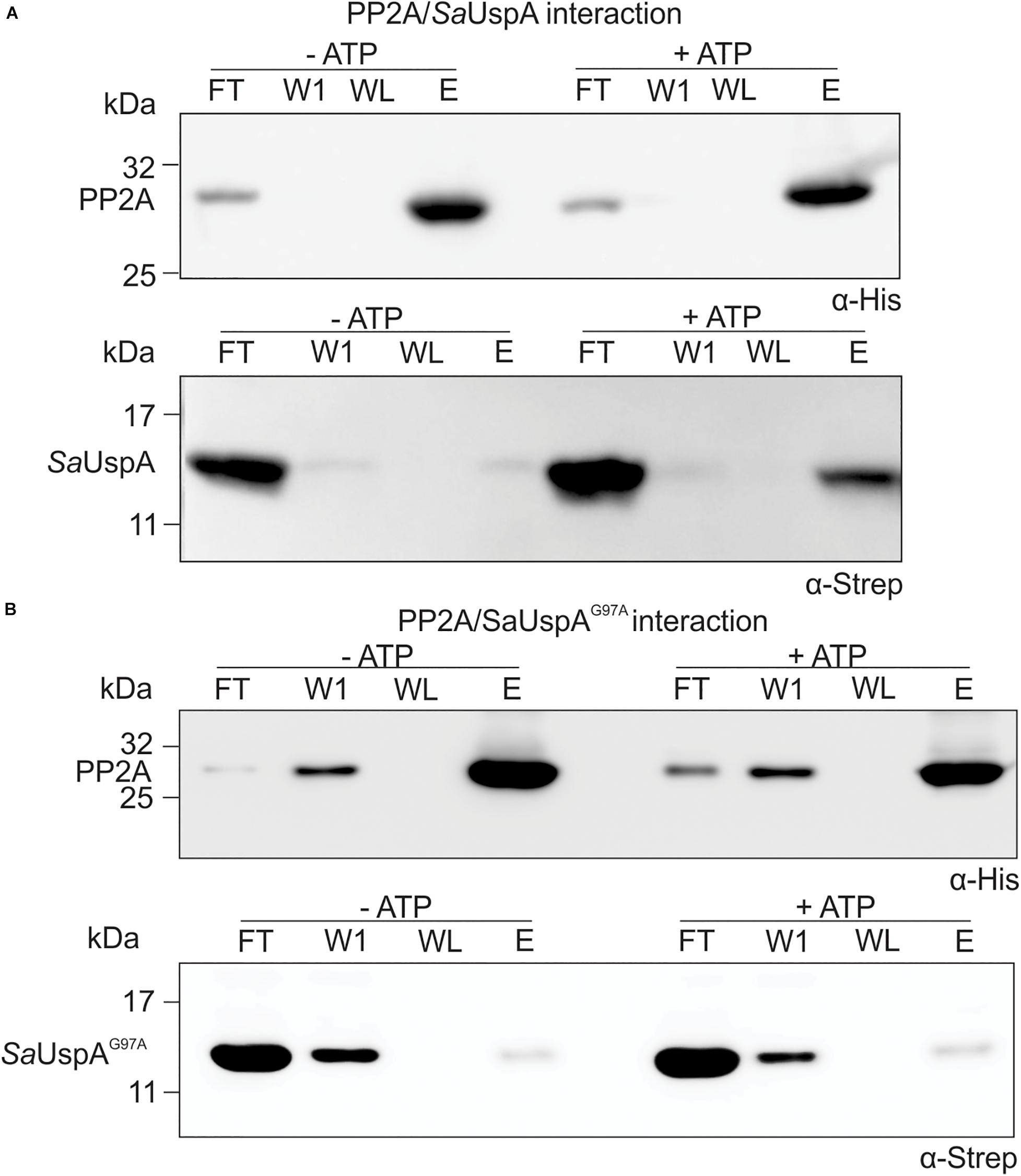
Figure 4. ATP-binding to SaUspA increases the interaction with PP2A. In vitro interaction assays for PP2A and SaUspA. (A) PP2A-his and SaUspA-Strep were mixed and applied to Ni-NTA resin, the resin was washed and eluted with buffer containing 10 and 500 μM imidazole, respectively. Fractions were analyzed by Western blot with α-His and α-Strep antibody. (B) As in (A) but using SaUspAG97A. FT, flowthrough; W1, first wash fraction; WL, last wash fraction; E, elution fraction. All experiments were repeated at least three times and representative figures are shown.
SaUspA Stimulates the Phosphatase Activity of PP2A
Eukaryotic PP2A is a heterotrimeric complex containing a scaffolding, a regulatory and a catalytic subunit (Shi, 2009). The regulatory subunit is involved in regulating the activity of PP2A (Drewes et al., 1993; Gong et al., 1994; Santoro et al., 1998). In addition, a large number of proteins have been identified which interact with one or more subunits or even a specific PP2A, thereby regulating PP2A’s function (Dozier et al., 2004; McConnell et al., 2007). PP2A is the homolog of the eukaryotic PP2A catalytic subunit, but no homologs of the scaffolding or regulatory subunits have been identified in archaea. Thus, it seems likely that proteins like SaUspA, which were previously identified in the pull-down assay (Ye et al., 2020) regulate the activity of PP2A. To investigate whether SaUspA regulates the activity of PP2A, the PP2A phosphatase activity was determined at different concentrations of an artificial phospho-peptide (Figure 5A). As observed previously (Reimann et al., 2013), the phosphatase activity of PP2A had a Km of 84 +/− 16 μM and a Vmax of 274 +/− 16 U/mg, respectively. Remarkably, in the presence of SaUspA, no effect was observed on the Km, but the Vmax increased to 583 +/− 55 U/mg. To our knowledge, this is the first observation of the regulation of the activity of an archaeal phosphatase by another protein. Since ATP enhances the interaction between SaUspA and PP2A, ATP might affect the stimulation of PP2A by SaUspA. However, no effects were observed in the presence of ATP. Most likely PP2A and SaUspA already form a complex at the concentration used in this assay, and ATP-dependent stimulation of the interaction occurs only at lower concentrations. To ensure that the observed effect is specific for the interaction between SaUspA and PP2A, a similar experiment was performed with PP2A and AbfR1 (Figure 5B). AbfR1 is a regulator of biofilm and motility but was not identified as a strong interaction partner in the previous pull-down assays (Li et al., 2017; Ye et al., 2020). When PP2A was assayed in the presence of AbfR1, its phosphatase activity did not show significant differences, which showed that the stimulating effect on PP2A activity was specific to SaUspA.
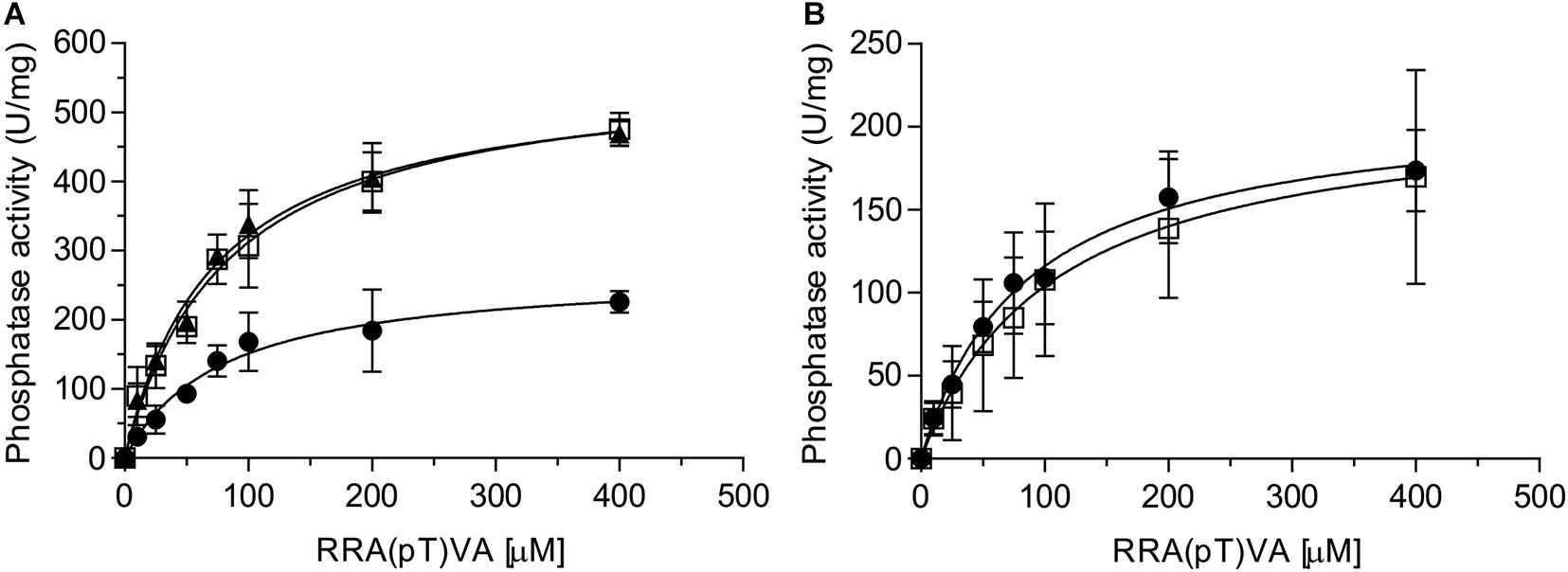
Figure 5. SaUspA stimulates the phosphatase activity of PP2A. The phosphatase activity of PP2A was determined in the absence (black circle) or presence of SaUspA (3 μM) with (black triangle) or without (open square) ATP (A), or AbfR1 (3 μM) (open square) (B). All experiments were performed at 70°C in biological triplicates.
Transcription of saUspA Is Growth Phase Independent and Neither Induced by Nutrient Starvation nor by High Salinity Stress
Universal stress proteins in bacteria and eukarya are important for many different responses to stress, but the role of USP proteins in archaea is currently still unknown. To determine the physiological role of SaUspA, the transcriptional activity of the saUspA gene under different conditions was studied. Cells of S. acidocaldarius MW001 growing in nutrient-rich medium were collected at different OD600 and RNA was isolated and analyzed by qPCR. The transcript levels of saUspA did not change significantly during the different stages of the growth curve and were, in contrast to other studied usp genes (Nystrdm and Neidhardt, 1992; Liu et al., 2007), not induced in the stationary phase (Figure 6A). Also under nutrient depletion (Figure 6B) or high salinity stress (400 mM NaCl) (Figure 6C), no significant up- or down regulation of saUspA was observed. Also previous transcriptome data of S. acidocaldarius (Götz et al., 2007) showed that exposure to 200 J/m2 UV irradiation did not change the transcription of saUspA significantly.
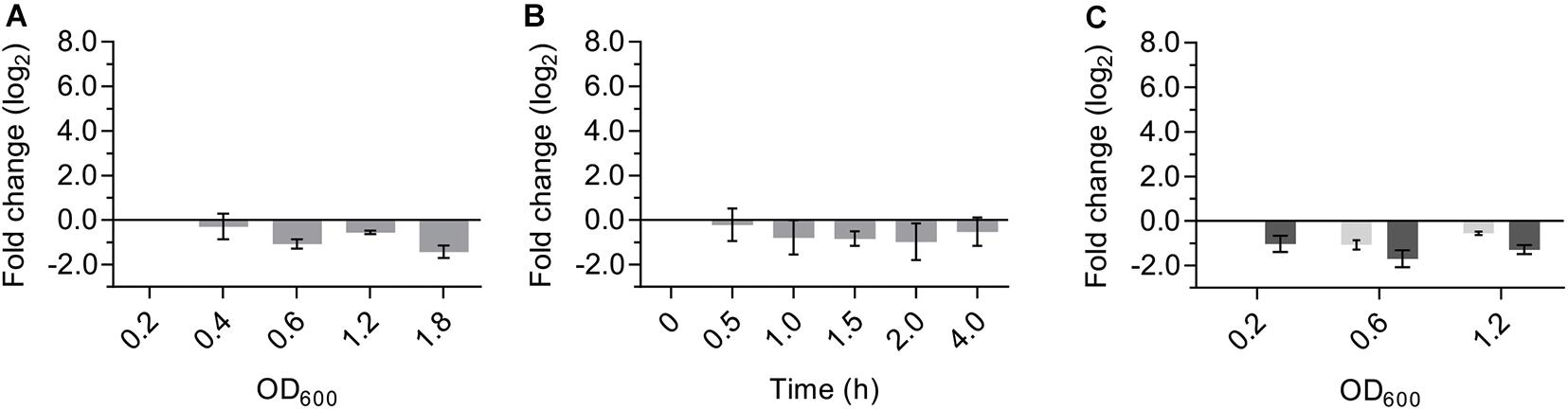
Figure 6. Transcription levels of saUspA do not change in response to different stress conditions. (A) S. acidocaldarius was grown in nutrient-rich medium and cell samples were taken at different OD600. OD600 of 0.2, 0.4, 0.6, and 1.2 belong to the exponential phase of cell growth. OD600 of 1.8 belongs to the stationary phase of cell growth. Samples of OD600 0.2 were used as the control for analysis. (B) S. acidocaldarius was grown in nutrient-depleted medium and cell samples were taken at different time points. Samples taken at t0 were used as the control for analysis. (C) S. acidocaldarius was grown in nutrient-rich medium in the presence (dark gray) and absence (light gray) of NaCl (400 mM), and cell samples were taken at OD600 0.2, 0.6, and 1.2. Samples of OD600 0.2 in the absence of NaCl were used as the control for analysis. Transcription level of saUspA was analyzed by qRT-PCR. Relative transcription level was normalized to secY. The values represent fold changes (mean ± SD) compared with the control from biological triplicates.
Effect of saUspA Deletion on S. acidocaldarius Cells Responding to Different Stress Conditions
To identify a physiological role of SaUspA, a markerless in-frame deletion mutant was constructed. Deletion of saUspA did neither show an effect on normal growth nor any effect on cell survival and growth during UV stress or during growth on heavy metals (Supplementary Figures S1–S3). It has been shown previously that S. acidocaldarius can grow in NaCl concentrations up to 400 mM (Meyer et al., 2011). MW001 and the ΔsaUspA strain were both cultivated in 100 to 400 mM NaCl (Figure 7A). Increasing concentrations of NaCl slowed the growth of both strains and at 400 mM NaCl, the ΔsaUspA mutant grew significantly slower than the wild type strain MW001. Furthermore, the ΔsaUspA mutant showed a clear increase in the lag phase. Microscopic analysis of the strain under these conditions did not reveal any changes in size or morphology of the cells compared to the MW001 strain. Thus the ΔsaUspA stain shows decreased growth under high salinity condition. To test whether this phenotype could be complemented by expression of SaUspA, a plasmid which expressed C-terminally HA-tagged SaUspA, SaUspAΔdim, SaUspAD8A, or SaUspAG97A from the saUspA promotor was constructed and transformed to the S. acidocaldarius ΔsaUspA strain. No expression of the SaUspAΔdim and SaUspAD8A proteins could be detected using Western blots analysis with an anti-HA antibody (Figure 7B), which confirmed that ATP binding and dimerization are important for the stability of this protein. SaUspAG97A which showed reduced ATP binding was also expressed at a lower level than WT SaUspA (Figure 7B). Expression of SaUspA fully complemented the growth defect of the ΔsaUspA mutant whereas the reduced ATP binding mutant SaUspAG97A could only restore the growth defect partially (Figure 7B).
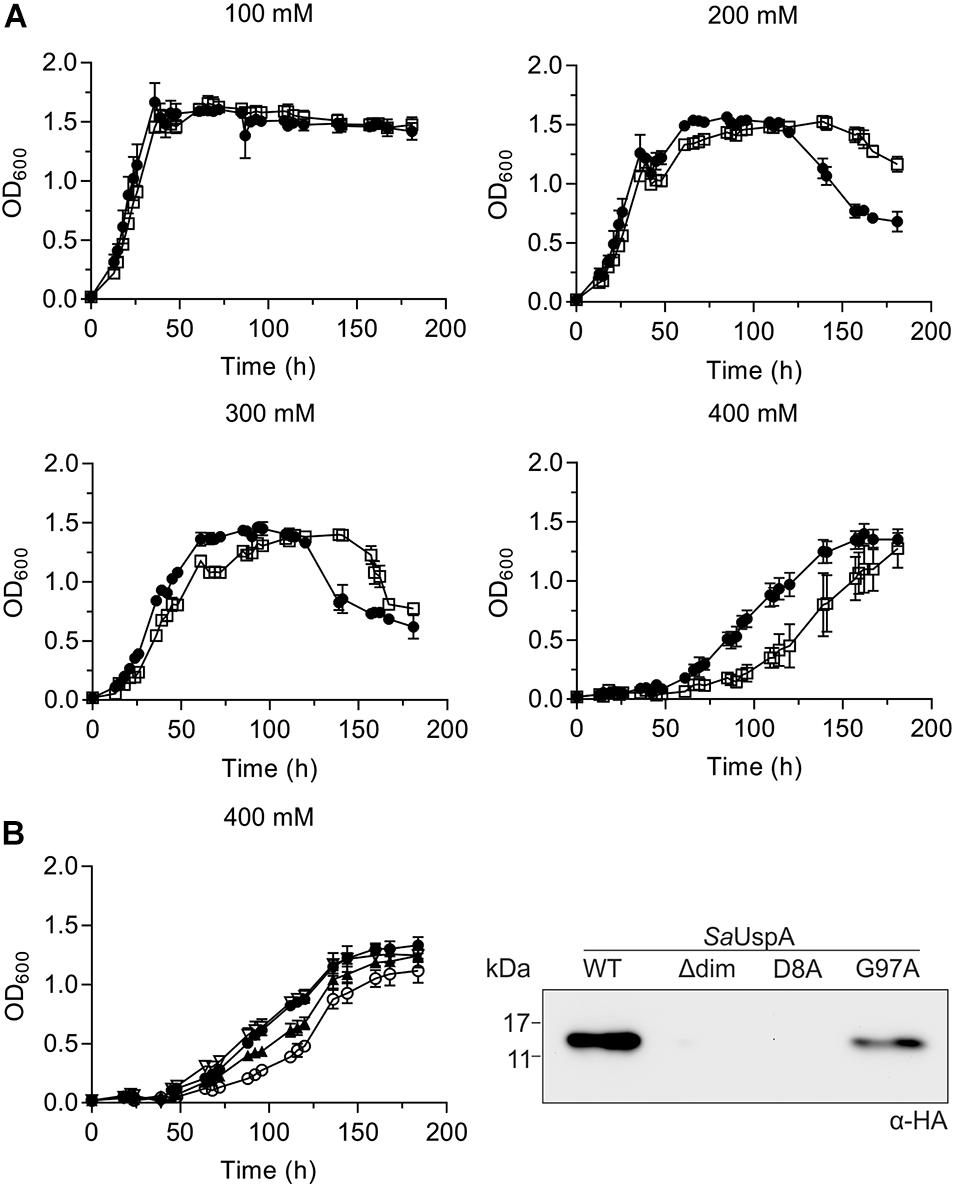
Figure 7. The saUspA deletion mutant is affected during growth on high salt concentrations. (A) Growth curves of S. acidocaldarius MW001 (black circle) and the Δ saUspA mutant (open square) in the presence of 100, 200, 300, and 400 mM NaCl. The growth curves were performed in triplicate and standard deviations are shown. (B) Left panel: complementation of the ΔsaUspA mutant growth defect in presence of high salinity with plasmid-based expressed SaUspA and its ATP-binding deficient mutant from its native promoter. Empty vector (open circle) and plasmids containing saUspAWT (open triangle) and saUspAG97A (black triangle) were used to complement the ΔsaUspA mutant. MW001 containing empty vector (black circle) was used a control. Cells were grown in the presence of 400 mM NaCl. Right panel: plasmid-based expression of HA-tagged SaUspAWT (WT), SaUspAΔdim (Δdim), SaUspAD8A (D8A) and SaUspAG97A (G97A) in the S. acidocaldarius ΔsaUspA mutant was confirmed by western blot analysis. S. acidocaldarius cells were grown in nutrient-rich medium without uracil and cell samples were taken at OD600 of 0.4, which was analyzed by Western blot with α-HA antibody.
Deletion of saUspA Affects the Timing of FlaB Expression
One of the phenotypes of the Δpp2a strain is hyper-motility (Reimann et al., 2013). Motility in archaea is driven by the archaellum (Jarrell and Albers, 2012), which in S. acidocaldarius is expressed under starvation conditions (Lassak et al., 2012). To test whether SaUspA plays a role in the archaellum regulation network in S. acidocaldarius, expression of the archaellin FlaB was analyzed by Western blot analysis during the first 2 h after induction of starvation conditions (Figure 8A and Supplementary Figure S4). FlaB expression increased during nutrient starvation in both the wild type strain and the ΔsaUspA mutant strain. Compared with the WT strain, increased FlaB expression was observed in ΔsaUspA mutant at 0, 0.5, 1.0, and 1.5 h. However, at time point 2.0 h, no significant difference in FlaB expression was observed. Thus, deletion of saUspA leads to early expression of FlaB during nutrient starvation, however, the final expression level of FlaB was not affected. In addition, motility of ΔsaUspA mutant on semi-solid gelrite plates was analyzed (Figure 8B), and as expected from the similar FlaB levels after 2 h, no differences in motility were observed. Thus, deletion of saUspA did not have significant effects on the motility of S. acidocaldarius cells.
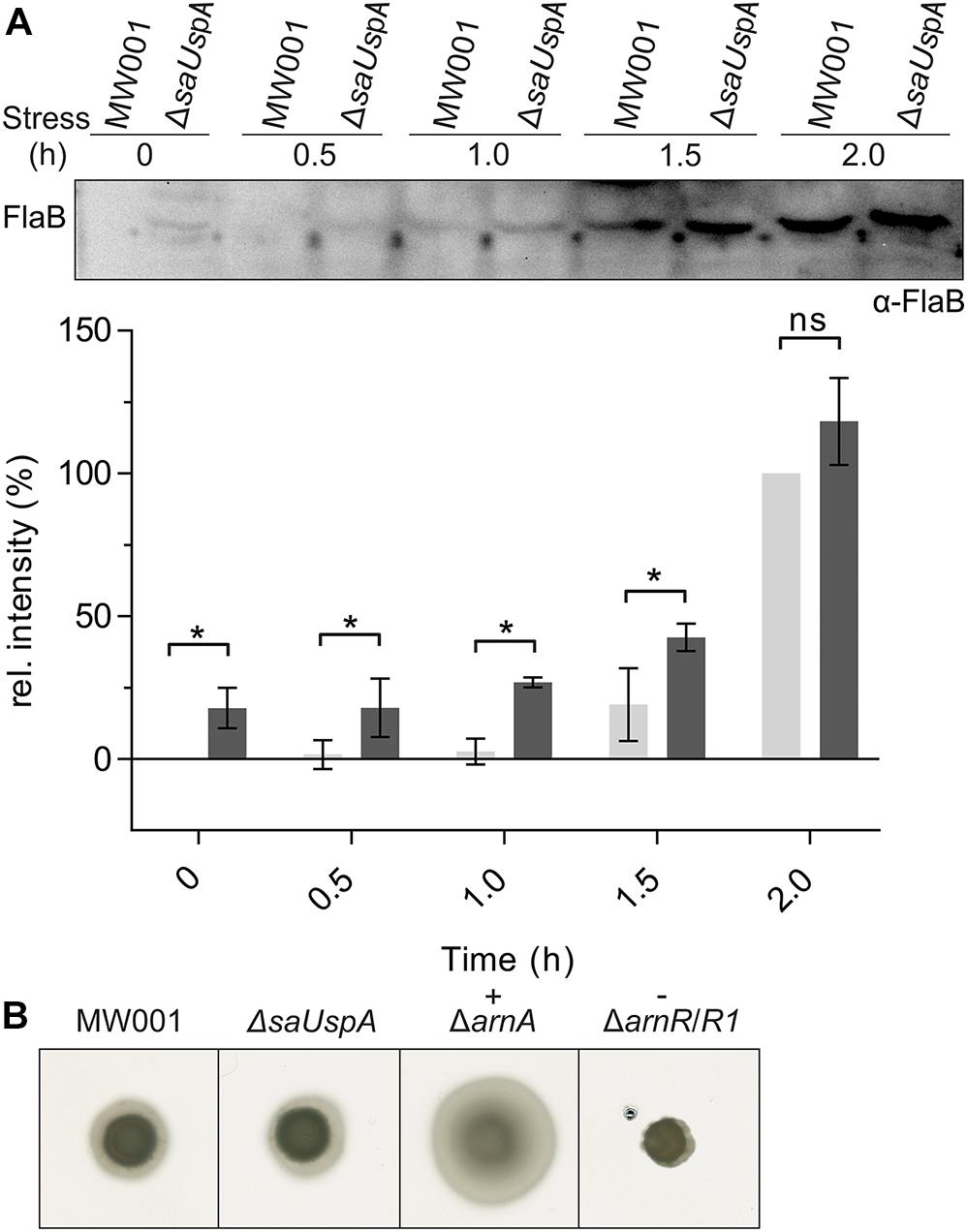
Figure 8. Effect of saUspA deletion on FlaB expression and motility. (A) Time course of FlaB expression during 2 h growth in nutrient-depleted medium. Wild type and ΔsaUspA mutant cells were grown in nutrient-depleted medium for 2 h and cell samples were taken at different time points. The FlaB expression level was analyzed by Western blot with an α-FlaB antibody. A representative Western blot is shown. A quantification of Western blots of three biological replicates is shown for MW001 (light gray) and the ΔsaUSPA mutant (dark gray) below the blot. The expression levels of FlaB at 0 and 2 h for strain MW001 were used as minimal and maximal values for data normalization. Significant differences between MW001 and ΔsaUSPA mutant (p-value < 0.05) are indicated by an asterisk while ns indicates no significance (p-value > 0.05). (B) Motility assay. Same amounts of cells were spotted on semi-solid gelrite plates. After incubating at 75°C for 5 days, the plates were scanned and recorded. ΔarnA and ΔarnR/R1 were used as hyper-motile and non-motile control, respectively. The figures shown here were representative examples of biological triplicates.
Discussion
SaUspA, a member of the universal stress protein family, was recently identified as a binding partner of the serine/threonine phosphatase PP2A (Ye et al., 2020), during the early nutrient starvation response of S. acidocaldarius. PP2A is the homolog of the eukaryotic PP2A catalytic subunit, a heterotrimeric complex containing also a scaffolding, a regulatory and a catalytic subunit (Drewes et al., 1993; Gong et al., 1994; Santoro et al., 1998; Shi, 2009). This heterotrimeric complex interacts with different proteins which interact with one or more subunits or even a specific PP2A, thereby regulating PP2A’s function (Dozier et al., 2004; McConnell et al., 2007). In previous studies, it was proposed that PP2A is involved in the archaellum regulatory network during normal growth and starvation when expression of the archaellum is strongly induced (Reimann et al., 2013). In bacteria, proteins of the USP superfamily were previously shown to be involved in the adaptation to various nutrient starvation conditions and also functioned as regulators in motility (Nystrom and Neidhardt, 1994; Gustavsson et al., 2002; Nachin et al., 2005). Based on these findings, we set out to study whether SaUspA might be involved in nutrient starvation signaling or motility in S. acidocaldarius by modulating the activity of PP2A.
SaUspA Is Stabilized by ATP-Binding and Dimerization
As was previously observed for other USPs, SaUspA formed a stable homodimer. Deletion of the C-terminal dimerization domain resulted in strongly reduced levels of SaUspA, both when expressed in E. coli or in S. acidocaldarius, Indeed, also truncation of six amino acids containing the C-terminal dimerization motif from E coli UspG, abolished dimer formation and resulted in a protein more sensitive to proteolysis (Weber and Jung, 2006). In S. lycopersicum, SlRd2 was still expressed after truncation of the dimerization motif but, its function in LiCl resistance and interaction with SlCipk6 was lost (Gutiérrez-beltrán et al., 2017). A similar observation was made for ATP binding. Previous studies showed that the substitution of the two conserved residues D17 and G109 in the ATP binding of Rv2624c resulted in reduced ATP-binding affinity and reduced thermal stability (Jia et al., 2016). Whereas WT SaUspA could be stably overexpresssed, overexpression of SaUspA with a mutation in D8 could neither be detected in S. acidocaldarius nor when the protein was heterologously expressed in E. coli. Mutagenesis of G97 resulted in reduced expression of SaUspA in both S. acidocaldarius and in E. coli and strongly reduced ATP binding. This shows that for SaUspA both ATP binding and dimerization are important for the stability of the protein. Although protein phosphorylation was found in some members of the USP superfamily (Freestone et al., 1997; Weber and Jung, 2006; Gutiérrez-beltrán et al., 2017; Potel et al., 2018), the physiological role is poorly understood. In M. tuberculosis, Rv2623 was phosphorylated at threonine 237, which was recognized by one FHA domain of Rv1747. The substitution of threonine 237 with a non-phosphorylatable alanine decreased their interaction and abolished their function in regulation growth (Glass et al., 2017). However, although high affinity binding of ATP to SaUspA was observed, neither ATP hydrolysis nor autophosphorylation was observed. In addition, no phosphorylated form of SaUspA could be identified in previous phosphoproteomic studies of S. acidocaldarius (Reimann et al., 2013). In vitro phosphorylation of SaUspA by ArnC but not ArnD was observed, but the relevance of this observation is currently still unknown.
The Interaction Between PP2A and SaUspA Is Enhanced by ATP Binding and Stimulates the Phosphatase Activity of PP2A
Within this study, the previously observed interaction between PP2A and SaUspA (Ye et al., 2020) was confirmed in vitro. Remarkably, this interaction occurred with higher affinity in the presence of ATP. Since SaUspA has no autophosphorylation activity, the increased interaction is caused by binding of ATP to SaUspA and not by autophosphorylation of SaUspA. We have previously proposed that the proteins that interact with PP2A, a conserved putative ATP/GTP binding protein (Saci_1281), the archaellum regulators ArnA and ArnB and SaUspA might regulate the activity of PP2A and thus might regulate different cellular processes (Ye et al., 2020). Here it is demonstrated that binding of SaUspA to PP2A stimulates the phosphatase activity of PP2A. Although members of the USP superfamily have been studied for years, the mechanism by which they function are still largely unknown. We here propose that one possible mechanism by which UPSs influence the stress response is by interacting and modulating the activity of phosphatases.
SaUspA Plays a Role in Nutrient Starvation and High Salinity Stress
To determine in which cellular processes SaUspA plays a role, the effect of SaUspA on different processes was studied. Remarkably, unlike the stress-dependent transcription stimulation of usp genes in bacteria (Vollmer and Bark, 2018), the transcription level of saUspA was constant in S. acidocaldarius during growth in nutrient-rich medium and was not induced by nutrient starvation, high salinity and UV stress.
Deletion of saUspA did also not affect cell survival, growth during UV stress or during growth on heavy metals. A clear effect was observed under nutrient starvation conditions, where ΔsaUspA cells expressed FlaB much earlier even under non-nutrient starvation condition, which indicated that the deletion of saUspA led to the induction of signaling pathways that normally occur during nutrient limitation. Remarkably, this was also observed in the Δpp2a deletion strain (Reimann et al., 2013), suggesting that the UspA-PP2A interaction might be required for the response to nutrient starvation. Furthermore, the ΔsaUspA strain grew significantly slower on high salinity medium. The growth defect could be complemented by the plasmid-based expression of SaUspA, demonstrating that SaUspA also plays a role during salinity stress. Remarkably, the expression levels of SaUspA are constant under these high salt stress conditions, demonstrating that the presence, but not the upregulation of SaUspA is important for the function of SaUspA under these stress conditions. Furthermore, effects of the deletion of saUspA might be compensated by the two other USP superfamily members, SaUspB and SaUspC. These proteins were, however, not identified as interaction partners of PP2A. There are many possible mechanisms by which SaUspA might be involved in resistance to high concentrations (∼400 mM) of NaCl, but the mechanism in S. acidocaldarius is unknown. For example, in E. coli the UspA family protein UspC binds to and stabilizes the two component system KdpD/KdpE which controls the induction of the high-affinity K+-transport system KdpFABC under conditions of salt stress (Heermann et al., 2009). Alternatively, SaUspA might influence proteins involved in growth at high salinity via its interaction with PP2A. In, for example, the halophilic archaeon M. portucalensis FDF1T most proteins participating in uptake and synthesis pathways of the compatible solute betaine were phosphorylated including the glycine betaine BtaABC transporter and enzymes involved in the methionine transmethylation cycle and betaine biosynthesis (Wu et al., 2016).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
XY, CD, and S-VA designed the experiments, analyzed the data, and wrote the manuscript. XY contributed to most of the experiments and figures. CD contributed to in vitro phosphorylation assays using radioactive-labeled ATP. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Chinese Scholarship Council (Ph.D. scholarship to XY) and the German Research Foundation (DFG) to S-VA (Grant Number: AL1206-43).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.598821/full#supplementary-material
References
Bischof, L. F., Haurat, M. F., and Albers, S.-V. (2019). Two membrane-bound transcription factors regulate expression of various type-IV-pili surface structures in Sulfolobus acidocaldarius. PeerJ 7:e6459. doi: 10.7717/peerj.6459
Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972). Sulfolobus?: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 68, 54–68. doi: 10.1007/bf00408082
Dozier, C., Bonyadi, M., Baricault, L., Tonasso, L., and Darbon, J. (2004). Regulation of Chk2 phosphorylation by interaction with protein phosphatase 2A via its B’regulatory subunit. Biol. Cell 96, 509–517. doi: 10.1016/j.biolcel.2004.04.010
Drewes, G., Mandelkow, E.-M., Baumann, K., Goris, J., Merlevede, W., and Mandelkow, E. (1993). Dephosphorylation of tau protein and Alzheimer paired helical filaments by calcmeurin and phosphatase-2A. FEBS Lett. 336, 425–432. doi: 10.1016/0014-5793(93)80850-t
Drumm, J. E., Mi, K., Bilder, P., Sun, M., Lim, J., Bielefeldt-ohmann, H., et al. (2009). Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-Binding?: requirement for establishing chronic persistent infection. PLoS Pathog. 5:e1000460. doi: 10.1371/journal.ppat.1000460
Freestone, P., Nystro, T., Trinei, M., and Norris, V. (1997). The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J. Mol. Biol. 274, 318–324. doi: 10.1006/jmbi.1997.1397
Glass, L. N., Swapna, G., Chavadi, S. S., Tufariello, M., Mi, K., Drumm, J. E., et al. (2017). Mycobacterium tuberculosis universal stress protein Rv2623 interacts with the putative ATP binding cassette (ABC) transporter Rv1747 to regulate mycobacterial growth. PLoS Pathog. 2623:1–25. doi: 10.1371/journal.ppat.1006515
Gong, C.-X., Grundke-Iqbal, I., and Iqbal, K. (1994). Dephosphorylation of Alzheimer’s disease abnormally phosphorylated tau by protein phosphatase-2A. Neuroscience 61, 765–772. doi: 10.1016/0306-4522(94)90400-6
Gonzali, S., Loreti, E., Cardarelli, F., Novi, G., Parlanti, S., Pucciariello, C., et al. (2015). Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 1, 1–9. doi: 10.1038/nplants.2015.151
Götz, D., Paytubi, S., Munro, S., Lundgren, M., Bernander, R., White, M. F., et al. (2007). Open access responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 8:R220. doi: 10.1186/gb-2007-8-10-r220
Gustavsson, N., Diez, A. A., and Nyström, T. (2002). The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43, 107–117. doi: 10.1046/j.1365-2958.2002.02720.x
Gutiérrez-beltrán, E., Personat, J. M., Torre, F., and De Pozo, O. (2017). A universal stress protein involved in oxidative stress is a phosphorylation target for protein kinase CIPK6 1. Plant Physiol. 173, 836–852. doi: 10.1104/pp.16.00949
Heermann, R., Weber, A., Mayer, B., Ott, M., Hauser, E., Gabriel, G., et al. (2009). The universal stress protein UspC scaffolds the KdpD / KdpE signaling cascade of Escherichia coli under salt stress. J. Mol. Biol. 386, 134–148. doi: 10.1016/j.jmb.2008.12.007
Hoffmann, L., Anders, K., Bischof, L. F., Ye, X., Reimann, J., Khadouma, S., et al. (2019). Structure and interactions of the archaeal motility repression module ArnA-ArnB that modulates archaellum gene expression in Sulfolobus acidocaldarius. J. Biol. Chem. 294, 7460–7471. doi: 10.1074/jbc.ra119.007709
Hoffmann, L., Schummer, A., Reimann, J., Haurat, M. F., Wilson, A. J., Beeby, M., et al. (2017). Expanding the archaellum regulatory network – the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius. Microbiologyopen 6:e00414. doi: 10.1002/mbo3.414
Jarrell, K. F., and Albers, S.-V. (2012). The archaellum: an old motility structure with a new name. Trends Microbiol. 20, 307–312. doi: 10.1016/j.tim.2012.04.007
Jia, Q., Hu, X., Shi, D., Zhang, Y., Sun, M., Wang, J., et al. (2016). Universal stress protein Rv2624c alters abundance of arginine and enhances intracellular survival by ATP binding in mycobacteria. Sci. Rep. 6:35462. doi: 10.1038/srep35462
Kvint, K., Nachin, L., Diez, A., and Nystro, T. (2003). The bacterial universal stress protein?: function and regulation. Curr. Opin. Microbiol. 6, 140–145. doi: 10.1016/S1369-5274(03)00025-0
Lassak, K., Neiner, T., Ghosh, A., Klingl, A., Wirth, R., and Albers, S. (2012). Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124. doi: 10.1111/j.1365-2958.2011.07916.x
Lenman, M., Sörensson, C., and Andreasson, E. (2008). Enrichment of phosphoproteins and phosphopeptide derivatization identify universal stress proteins in elicitor-treated arabidopsis. Mol. Plant Microbe Interact. 21, 1275–1284. doi: 10.1094/mpmi-21-10-1275
Li, L., Banerjee, A., Bischof, L. F., Maklad, H. R., Hoffmann, L., Henche, A., et al. (2017). Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius. Mol. Microbiol. 105, 777–793. doi: 10.1111/mmi.13735
Liu, W., Karavolos, M. H., Bulmer, D. M., Allaoui, A., Demarco, R., Hormaeche, C. E., et al. (2007). ARTICLE in press Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb. Pathog. 42, 2–10. doi: 10.1016/j.micpath.2006.09.002
Maklad, H. R., Gutierrez, G. J., Esser, D., Siebers, B., and Peeters, E. (2020). Phosphorylation of the acyl-CoA binding pocket of the FadR transcription regulator in Sulfolobus acidocaldarius. Biochimie 175, 120–124. doi: 10.1016/j.biochi.2020.05.007
McConnell, J. L., Gomez, R. J., McCorvey, L. R. A., Law, B. K., and Wadzinski, B. E. (2007). Identification of a PP2A-interacting protein that functions as a negative regulator of phosphatase activity in the ATM/ATR signaling pathway. Oncogene 26, 6021–6030. doi: 10.1038/sj.onc.1210406
Merkouropoulos, G., Andreasson, E., Hess, D., Boller, T., and Peck, S. C. (2008). An arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP. J. Biol. Chem. 283, 10493–10499. doi: 10.1074/jbc.M800735200
Meyer, B. H., Zolghadr, B., Peyfoon, E., Pabst, M., Panico, M., Morris, H. R., et al. (2011). Sulfoquinovose synthase – an important enzyme in the N - glycosylation pathway of Sulfolobus acidocaldarius. Mol. Microbiol. 82, 1150–1163. doi: 10.1111/j.1365-2958.2011.07875.x.Sulfoquinovose
Nachin, L., Brive, L., Persson, K., Svensson, P., and Nyström, T. (2008). Heterodimer formation within universal stress protein classes revealed by an in silico and experimental approach. J. Mol. Biol. 380, 340–350. doi: 10.1016/j.jmb.2008.04.074
Nachin, L., Nannmark, U., and Nystro, T. (2005). Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187, 6265–6272. doi: 10.1128/JB.187.18.6265
Nystrdm, T., and Neidhardt, F. C. (1992). Cloning, mapping and nucleotide sequencing of a gene encoding a universai stress protein in Eschericha coli. Mol. Microbiol. 6, 3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x
Nystrom, T., and Neidhardt, F. C. (1994). Expression and role of the universal stress protein, UspA, of Escherichia coii during growth arrest. Mol. Microbiol. 11, 537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x
O’Connor, A., and McClean, S. (2017). The role of universal stress proteins in bacterial infections. Curr. Med. Chem. 24, 3970–3979. doi: 10.2174/0929867324666170124145543
Potel, C. M., Lin, M., Heck, A. J. R., and Lemeer, S. (2018). Widespread bacterial protein histidine phosphorylation revealed by mass spectrometry- based proteomics. Nat. Methods 15, 187–190. doi: 10.1038/nmeth.4580
Reimann, J., Esser, D., Orell, A., Amman, F., Pham, T. K., Noirel, J., et al. (2013). Archaeal signal transduction: impact of protein phosphatase deletions on cell size, motility, and energy metabolism in Sulfolobus acidocaldarius. Mol. Cell. Proteomics 12, 3908–3923. doi: 10.1074/mcp.M113.027375
Reimann, J., Lassak, K., Khadouma, S., Ettema, T. J. G., Yang, N., Driessen, A. J. M., et al. (2012). Regulation of archaella expression by the FHA and von Willebrand domain-containing proteins ArnA and ArnB in Sulfolobus acidocaldarius. Mol. Microbiol. 86, 24–36. doi: 10.1111/j.1365-2958.2012.08186.x
Santoro, M. F., Annand, R. R., Robertson, M. M., Peng, Y.-W., Brady, M. J., Mankovich, J. A., et al. (1998). Regulation of protein phosphatase 2A activity by caspase-3 during apoptosis. J. Biol. Chem. 273, 13119–13128. doi: 10.1074/jbc.273.21.13119
Shi, Y. (2009). Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. doi: 10.1016/j.cell.2009.10.006
Sousa, M. C., and Mckay, D. B. (2001). Structure of the universal stress protein of Haemophilus influenzae. Structure 9, 1135–1141. doi: 10.1016/s0969-2126(01)00680-3
Tkaczuk, K. L., Shumilin, I. A., Chruszcz, M., Evdokimova, E., Savchenko, A., and Minor, W. (2013). Structural and functional insight into the universal stress protein family. Evol. Appl. 6, 434–449. doi: 10.1111/eva.12057
Van Veldhoven, P. P., and Mannaerts, G. P. (1987). Inorganic and organic phosphate measurements in the nanomolar range. Anal. Biochem. 161, 45–48. doi: 10.1016/0003-2697(87)90649-x
Vollmer, A. C., and Bark, S. J. (2018). Twenty-Five Years of Investigating the Universal Stress Protein?: Function, Structure, and Applications, 1st Edn. Amsterdam: Elsevier Inc.
Wagner, M., Wolferen, M., Van, Wagner, A., Lassak, K., Meyer, B. H., et al. (2012). Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 3:214. doi: 10.3389/fmicb.2012.00214
Weber, A., and Jung, K. (2006). Biochemical properties of UspG, a universal stress protein of Escherichia coli†. Biochemistry 45, 1620–1628. doi: 10.1021/bi051301u
Wolferen, M., Van, Ma, X., and Albers, S. (2015). DNA processing proteins involved in the UV-Induced stress response of sulfolobales. J. Bacteriol. 197, 2941–2951. doi: 10.1128/JB.00344-15
Wu, W.-L., Lai, S.-J., Yang, J.-T., Chern, J., Liang, S.-Y., Chou, C.-C., et al. (2016). Phosphoproteomic analysis of Methanohalophilus portucalensis FDF1 T identified the role of protein phosphorylation in methanogenesis and osmoregulation. Sci. Rep. 6:29013. doi: 10.1038/srep29013
Ye, X., Vogt, M. S., Van Der Does, C., BildlWolfgang, W., Schulte, U., Essen, L.-O., et al. (2020). The phosphatase PP2A interacts with ArnA and ArnB to regulate the oligomeric state and the stability of the ArnA/B complex. Front. Microbiol. 11:1849. doi: 10.3389/fmicb.2020.01849
Zarembinski, T. I., Hung, L. W., Mueller-Dieckmann, H. J., Kim, K. K., Yokota, H., Kim, R., et al. (1998). Structure-based assignment of the biochemical function of a hypothetical protein?: a test case of structural genomics. Proc. Natl. Acad. Sci. U.S.A. 95, 15189–15193. doi: 10.1073/pnas.95.26.15189
Keywords: Sulfolobus acidocaldarius, protein phosphatase, universal stress protein, ATP binding, archaellin, high salinity stress, archaea
Citation: Ye X, van der Does C and Albers S-V (2020) SaUspA, the Universal Stress Protein of Sulfolobus acidocaldarius Stimulates the Activity of the PP2A Phosphatase and Is Involved in Growth at High Salinity. Front. Microbiol. 11:598821. doi: 10.3389/fmicb.2020.598821
Received: 25 August 2020; Accepted: 23 October 2020;
Published: 13 November 2020.
Edited by:
Jerry Eichler, Ben-Gurion University of the Negev, IsraelReviewed by:
Changyi Zhang, University of Illinois at Urbana–Champaign, United StatesXu Peng, University of Copenhagen, Denmark
Copyright © 2020 Ye, van der Does and Albers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sonja-Verena Albers, c29uamEuYWxiZXJzQGJpb2xvZ2llLnVuaS1mcmVpYnVyZy5kZQ==
 Xing Ye
Xing Ye Sonja-Verena Albers
Sonja-Verena Albers