
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 29 October 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.594737
This article is part of the Research Topic Cell Surface Proteins of Gram-positive Pathogenic Bacteria View all 15 articles
Gram-positive bacterial pathogens have an array of proteins on their cell surface that mediate interactions with the host environment. In particular, bacterial cell wall-associated (CWA) proteins play key roles in both colonization and pathogenesis. Furthermore, some CWA proteins promote specialization for host-species or mediate colonization of specific anatomical niches within a host. In this mini review, we provide examples of the many ways by which major pathogens, such as Staphylococci, Streptococci and Listeria monocytogenes, utilize CWA proteins for both host- and niche-specialization. We describe different biological mechanisms mediated by CWA proteins including: the acquisition of iron from hemoglobin in the bloodstream, adherence to and invasion of host cells, and innate immune evasion through binding to the plasma proteins fibrinogen, immunoglobulin G, and complement. We also discuss the limitations of using animal models for understanding the role of specific CWA proteins in host-specialization and how transformative technologies, such as CRISPR-Cas, offer tremendous potential for developing transgenic models that simulate the host environment of interest. Improved understanding of the role of CWA proteins in niche- or host-specificity will allow the design of new therapeutic approaches which target key host–pathogen interactions underpinning Gram-positive bacterial infections.
Bacteria have typically evolved to occupy particular niches within host-species or the environment. For example, Streptococcus uberis and some clones of Staphylococcus aureus are specialized for infection of the bovine mammary gland and Listeria monocytogenes has a tropism for transcytosis of the placenta and intestinal epithelia (Lecuit et al., 2004). Furthermore, bacterial pathogens have adapted to colonize either a single host-species (specialist) or multiple host-species (generalist) (Woolhouse et al., 2001; Bäumler and Fang, 2013). For example, the human pathogen Streptococcus pyogenes, associated with infections such as pharyngitis, skin infections, rheumatic fever and necrotizing fasciitis, and the equine pathogen Streptococcus equi subsp. equi, associated with the respiratory disease strangles, have a very restricted host-specialization. In contrast, the multi-host “generalist” S. aureus is associated with an array of different infections in humans, livestock and wild animal species (Sheppard et al., 2018). However, individual clonal lineages or subtypes of S. aureus have evolved the capacity to infect distinct host-species following a host-switch event (Lowder et al., 2009; Fitzgerald, 2012; Richardson et al., 2018; Haag et al., 2019). Adaptation to a novel niche or host-species involves multiple evolutionary mechanisms including mutation, recombination and horizontal gene transfer. Comparative genomic analysis of S. aureus from different host-species has identified genetic signatures affecting cell surface proteins, marked by gene acquisition, diversification and loss of function events, suggesting a key role for cell surface proteins in host- and niche- specialization (Herron-Olson et al., 2007; Ben Zakour et al., 2008; Lowder et al., 2009; Guinane et al., 2010; Resch et al., 2013; Spoor et al., 2015). Signatures of adaptation affecting the cell envelope may reveal pathways that could be disrupted therapeutically. For instance, S. aureus has undergone several host-switch events from humans into rabbits underpinned by natural adaptive mutations in a gene encoding DltB, a membrane-associated protein involved in D-Ala modification of wall teichoic acids (Viana et al., 2015). These data highlighted the key importance of DltB in host–pathogen interactions, a discovery reinforced by a subsequent study that identified DltB as a novel druggable target (Pasquina et al., 2016).
The aim of this mini review is to highlight the role of cell wall-associated (CWA) proteins of Gram-positive bacteria in overcoming host- and niche-specific barriers to infection. We provide examples of CWA proteins that are either covalently bound to cell wall peptidoglycan, or indirectly attached to the cell wall via non-covalent interactions with wall teichoic acids. Important host- or niche-specific CWA protein interactions underpinning biological mechanisms are described including: acquisition of iron from hemoglobin, adherence to and invasion of host cells, and evasion of the innate immune response (Figure 1). Understanding the mechanisms by which bacteria adapt to different niches or host-species can reveal critical host–pathogen interactions that could potentially be targeted to develop novel therapeutic approaches. However, demonstration of the in vivo importance of a host-specific CWA protein interaction is often challenging outside of the natural host. Here, we also discuss the potential for the design of transgenic models that express host-specialized surface protein receptors and would facilitate studies into the role of host- or niche-specific interactions in bacterial pathogenesis.
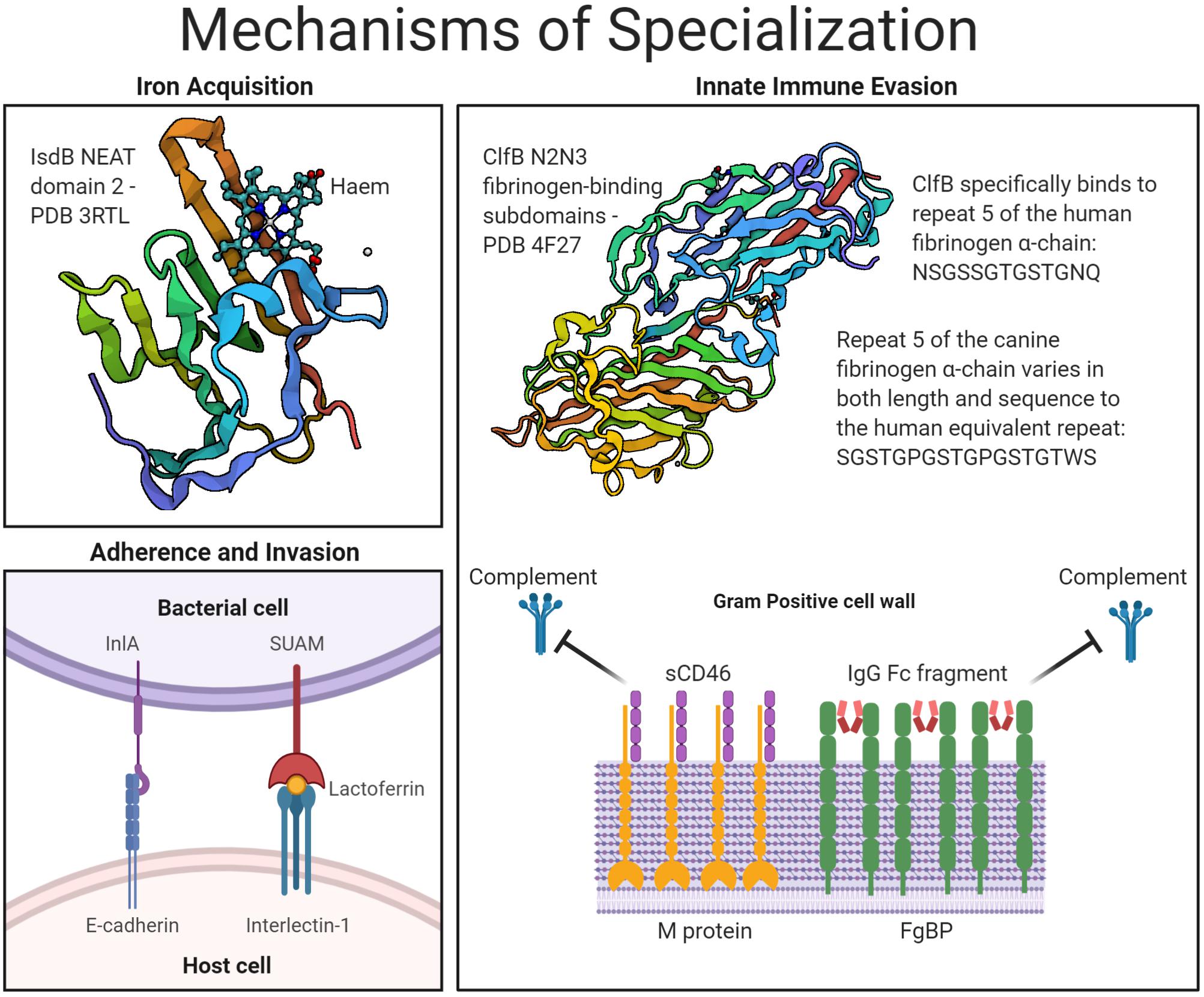
Figure 1. Mechanisms of host- and niche-specialization by Gram-positive surface proteins. An overview of the CWA bacterial proteins described under the three key headings of: iron acquisition, adherence and invasion, and innate immune evasion. Protein structures accessed from PDB files 3RTL, for IsdB in complex with haem (Gaudin et al., 2011), and 4F27, for ClfB in complex with fibrinogen (Xiang et al., 2012).
Bacterial adaptation to a particular host-species or niche often requires modification of surface proteins to allow them to interact with distinct ligands or polymorphic host receptors. Iron is an essential nutrient required as a co-factor for amino acid biosynthesis, the TCA cycle, DNA replication, cellular respiration, and electron transport (Sheldon and Heinrichs, 2015). In the host, iron is often sequestered in molecules such as hemoglobin in the blood and lactoferrin in mucosal secretions and milk. During bacterial infection, iron availability is actively decreased by limiting iron uptake from food digestion in a process termed nutritional immunity (Weinberg, 1974). However, bacteria have evolved numerous mechanisms, some of which involve utilization of surface proteins, to overcome this limitation and to successfully compete with the host for iron.
One such mechanism involves the iron-regulated surface-determinant (Isd) pathway present in a number of Gram-positive bacteria, including S. aureus, S. pyogenes, Bacillus anthracis, and L. monocytogenes (Sheldon and Heinrichs, 2015). In S. aureus, the Isd pathway is involved in acquisition of iron from hemoglobin and myoglobin in the bloodstream, allowing S. aureus to rapidly adapt to low iron conditions (Torres et al., 2006). S. aureus also responds to low iron conditions by altering the expression of metabolic proteins to generate a more acidic environment, promoting the release of iron from host proteins (Friedman et al., 2006). The Isd pathway is represented by multiple cell envelope-associated proteins encoded in an operon, with the near iron transporter (NEAT) domain 1 of the CWA protein IsdB interacting directly with hemoglobin (Torres et al., 2006). Heme is then transferred to NEAT domain 2 of IsdB and along a chain of Isd proteins concluding in heme binding to the IsdE membrane-associated lipoprotein. This allows IsdF-mediated translocation of the heme iron into the bacterial cytoplasm for use in metabolism (Muryoi et al., 2008; Zhu et al., 2008; Bowden et al., 2014; Gianquinto et al., 2019). Notably, IsdB is a host-specialized CWA protein with enhanced binding affinity for human hemoglobin, in comparison to hemoglobin from other mammalian sources (Pishchany et al., 2010). Strikingly, mice expressing a human hemoglobin protein have increased susceptibility to S. aureus infection in comparison to those expressing the native murine hemoglobin (Pishchany et al., 2010). The crystal structure of IsdB, in complex with human hemoglobin, revealed that binding results in conformational changes that displace heme from the β-subunits and subsequently the α-subunits of hemoglobin (Bowden et al., 2018). This IsdB-hemoglobin interface exhibits signatures of positive selection during the evolution of primates, involving both the α- and β-globins, suggesting that the role of IsdB in human-specialization is part of a long evolutionary relationship between S. aureus and hemoglobin (Choby et al., 2018). The specialization of IsdB to human hemoglobin, is similarly observed in the closely related Staphylococcus argenteus and Staphylococcus schweitzeri species, in addition to the more distantly related human-specialized Staphylococcus lugdunensis (Zapotoczna et al., 2012; Choby et al., 2018). Recently, a novel iron acquisition system in S. lugdunensis, dependent on an ECF-type transporter, has also demonstrated host-restriction based on the inability to lyse non-human erythrocytes to release hemoproteins (Jochim et al., 2020). These findings highlight how the co-evolution of bacterium and host can lead to bacterial adaptation to a key nutrient source in a particular host.
In addition to nutrient acquisition, all bacterial pathogens must overcome an array of anatomical, physiological and immunological barriers to colonize and establish infection. Adherence to and invasion of host cells is an important strategy for bacterial colonization, immune evasion, and dissemination. Gram-positive bacteria exploit a range of mechanisms to mediate binding and uptake by different cell types (Ribet and Cossart, 2015; Josse et al., 2017). Such interactions may promote establishment in a particular niche found across multiple host-species or may require diversification, to facilitate binding to tissue or cells that have polymorphic ligands or receptors in a distinct host-species.
A well characterized example of a CWA protein mediating both niche- and host-specialized cellular internalization is the internalin A (InlA) protein of L. monocytogenes, a veterinary pathogen that causes encephalitis in ruminants and a highly virulent foodborne pathogen that can cause miscarriage, stillbirth, or premature labor in pregnant women or meningitis in newborns (Williams et al., 2011; Roulo et al., 2014). InlA interacts with the N-terminal region of E-cadherin expressed by epithelial cells, including those present on the placental villous trophoblast barrier, promoting niche-specialization (Mengaud et al., 1996; Schubert et al., 2002; Lecuit et al., 2004). Primarily, this interaction allows L. monocytogenes, where E-cadherin is exposed, to pass through the intestinal epithelial barrier via transcytosis either on the luminal surface around intestinal goblet cells or intercellular junctions that form as part of normal gut homeostasis (Pentecost et al., 2006; Nikitas et al., 2011). It has been demonstrated that heterologous expression of InlA confers the capacity for internalization to the invasion-deficient Listeria innocua, and that natural premature stop codons in the inlA gene of L. monocytogenes clinical isolates are associated with a reduced capacity to invade intestinal epithelium and an increased infective dose in experimental models (Gaillard et al., 1991; Van Stelten et al., 2011; Su et al., 2019).
Determination of the crystal structure of the InlA interaction with E-cadherin provided evidence for host-specialization with stronger binding observed to human compared to murine E-cadherin (Lecuit et al., 1999; Schubert et al., 2002). Elegant mutational experiments demonstrated that a single proline residue at position 16 of E-cadherin was sufficient to alter host-tropism (Lecuit et al., 1999). Presence of Pro16 in E-cadherin, as naturally occurs in humans, rabbits, and guinea pigs, facilitates strong binding, whereas Glu16, as occurs in rodents, leads to a reduced affinity (Lecuit et al., 1999). Of note, rabbits are considered to be the natural hosts of L. monocytogenes with cows also encoding Pro16 in bovine E-cadherin (Murray et al., 1926; Zimin et al., 2009). InlA of L. monocytogenes has therefore evolved to promote both niche- and host-specialized invasion of intestinal and placental epithelial cells in the natural rabbit host, and conserved residues in human and bovine E-cadherin underpin the capacity for zoonotic infection.
Streptococcus uberis, a leading cause of bovine mastitis, has also evolved a niche- and host-specialized mechanism of adherence and invasion. Lactoferrin is an iron-sequestering glycoprotein with antimicrobial properties that is highly expressed in milk during bovine mastitis (Harmon et al., 1976; Hagiwara et al., 2003; Chaneton et al., 2008). Due to its abundance and importance in host defense, many bacterial pathogens have evolved mechanisms for interacting with lactoferrin (Valenti and Antonini, 2005; Lu et al., 2020). S. uberis encodes two lactoferrin-binding proteins that demonstrate preference for bovine lactoferrin, in comparison to the human variant, and promote evasion of the antimicrobial properties of lactoferrin in the mammary gland niche (Fang and Oliver, 1999; Moshynskyy et al., 2003). In particular, S. uberis adhesion molecule (SUAM) uses lactoferrin as a cross-bridge with host interlectin-1 to promote adherence to and internalization of S. uberis into bovine mammary epithelial cells (Fang et al., 2000; Moshynskyy et al., 2003; Almeida et al., 2006; Patel et al., 2009; Chen et al., 2011). A mutant S. uberis strain deficient in SUAM expression was attenuated for virulence in an experimentally infected mammary gland, suggesting a key role for SUAM in the pathogenesis and niche-specificity of S. uberis (Patel et al., 2009; Almeida et al., 2015).
In addition to the acquisition of key nutrients and the subversion of host cells, overcoming the host immune response is critical for the pathogenesis of Gram-positive bacteria. A wide array of innate immune mechanisms have evolved to prevent bacterial infections including the development of blood clots at wound sites and damaged blood vessels, and the activation of the complement cascade leading to intracellular killing by phagocytic cells such as neutrophils, macrophages, and dendritic cells. In response, Gram-positive bacteria can utilize surface proteins to disrupt these immune mechanisms in a niche or host-specialized manner by interacting with host proteins such as the plasma glycoprotein fibrinogen, to inhibit both hemostasis and phagocytosis, and complement factors such as factor H and CD46, to inhibit complement deposition.
Fibrinogen plays a central role in hemostasis and contains α- β- and γ-chains as a dimer of trimers (Mosesson, 2005). The ubiquity and abundance of fibrinogen has led a wide range of bacterial pathogens to develop ways of subverting fibrinogen for adherence to host cells, abscess formation, and immune evasion (Ko and Flick, 2016; Thomer et al., 2016). Staphylococcal species, including S. aureus, S. lugdunensis, and S. pseudintermedius, commonly adhere to the C-terminal of the fibrinogen γ-chain to provide a generalized but host-specific interaction, which interferes with fibrinogen-mediated coagulation and platelet aggregation (McDevitt et al., 1997; Geoghegan et al., 2010; Pietrocola et al., 2013). In the case of clumping factor A (ClfA) of S. aureus, the fibrinogen γ-chain binding is equivalent for human, canine, feline, murine, and porcine fibrinogen but demonstrates reduced binding to bovine fibrinogen and no detectable ovine fibrinogen-binding, due to a single amino acid substitution in the ovine fibrinogen γ-chain (Geoghegan et al., 2010). This host-specific fibrinogen-interaction of ClfA is essential for S. aureus-mediated sepsis, in murine infection models, as well as mediating phagocytosis inhibition and bacterial aggregation, key innate immune evasion strategies of S. aureus (Higgins et al., 2006; Flick et al., 2013; Claes et al., 2018).
Additional Staphylococcal CWA proteins demonstrate a more host-specialized fibrinogen interaction with the repeat region of the fibrinogen α-chain, which exhibits both inter- and intraspecies variation (Murakawa et al., 1993; Inaba et al., 2008; Pickering et al., 2019). For example, clumping factor B (ClfB) of S. aureus interacts solely with repeat 5 of the human fibrinogen α-chain, promoting human-specialized platelet aggregation, with S. pseudintermedius surface protein L (SpsL) exhibiting canine-specialized high-affinity binding to the repeat region of the canine fibrinogen α-chain (Miajlovic et al., 2007; Walsh et al., 2008; Pickering et al., 2019). SpsL is unique in exhibiting both canine-specific binding to the repeat region of the fibrinogen α-chain and a secondary weaker binding interaction that is also observed with human fibrinogen (Bannoehr et al., 2011; Pickering et al., 2019). This canine-specialized fibrinogen interaction promotes S. pseudintermedius aggregation and phagocytosis inhibition by neutrophils, demonstrating the role of fibrinogen-binding as a host-specialized immune evasion strategy of S. pseudintermedius (Pickering et al., 2019). Adherence to the fibrinogen α-chain is also utilized by the Srr1 and Srr2 glycoproteins of Streptococcus agalactiae for host-specialization, with specific binding to human repeats 6-8, demonstrating that this region of fibrinogen is the target of host-specialization for an array of Gram-positive bacteria (Seo et al., 2013).
Streptococcus pyogenes is a major human-specific pathogen that utilizes a range of innate immune evasion strategies to cause acute pharyngitis and impetigo as well as severe invasive infections such as necrotizing fasciitis (Laabei and Ermert, 2019). The most abundant CWA protein presented on the surface of S. pyogenes is the M protein, with certain M protein serotypes associated with specific niches such as the throat or skin (Fischetti, 1989; Cunningham, 2000). The M protein has an array of host ligands that promote both host cell adherence and immune evasion (Fischetti, 1989). The binding of M protein to complement components such as factor H, C4BP and CD46 all lead to complement inhibition (Horstmann et al., 1988; Thern et al., 1995). In the case of CD46 complement regulatory protein, which aids the cleavage of complement factors C3b and C4b on host cells, M protein has been demonstrated to interact in a human-specific manner (Riley-Vargas et al., 2004; Lövkvist et al., 2008). The interaction of M protein with human cellular CD46 mediates binding to keratinocytes and invasion of lung epithelial cells ultimately leading to cell death (Okada et al., 1995; Rezcallah et al., 2005). During epithelial cell apoptosis, soluble CD46 is shed from the host cell leading to reduced killing of S. pyogenes in whole blood due to its interaction with M protein (Lövkvist et al., 2008). This immune evasion mechanism is human-specialized as evidenced by increased whole blood survival in transgenic mice expressing human CD46 (Lövkvist et al., 2008). These human-expressing CD46 mice were also more susceptible to experimental infection with increased mortality and bacterial levels during a bloodstream infection and the development of necrotizing fasciitis after subcutaneous injection (Lövkvist et al., 2008; Matsui et al., 2009). S. pyogenes has therefore developed a human-specialized immune evasion strategy associated with complement inhibition.
A final example of host-specific immune evasion is the fibrinogen-binding protein (FgBP) of the equine-specific S. equi subsp. equi, the cause of the respiratory disease strangles in horses. FgBP is a CWA protein which was originally characterized as a fibrinogen-binding protein with preference for equine Fg and as a protective antigen in a murine infection model (Meehan et al., 1998, 2000). However, it was later identified as an IgG-binding protein of equine, human, rabbit, porcine, and feline IgG but not of murine, rat, goat, sheep, cow or chicken IgG (Meehan et al., 2001). By interacting with the Fc interdomain region of IgG4 and IgG7, FgBP disrupts complement deposition and antibody-mediated activation of the classical complement pathway, leading to increased survival in equine whole blood, representing a host-specific immune evasion strategy of S. equi subsp. equi (Meehan et al., 2001; Lewis et al., 2008).
The examples provided here include structural, biochemical, and molecular evidence for niche- and host-specialized interactions. However, it is often challenging to validate the importance of these host-pathogen interactions in appropriate infection models, especially when the bacteria exhibit a human tropism (Douam et al., 2015). In some cases, an effective model may not exist and the ethical and/or cost implications of performing experimental infections of large animals is prohibitive. Importantly, small animal models may not have ligands or receptors in common with the natural host and would therefore not support the function of the bacterial surface protein in question. Using knockdown and heterologous expression systems in cell lines in vitro can provide useful information relating to specific host–pathogen interactions but they cannot address the relevance of the interaction in a complex infection setting in vivo. The development of three-dimensional (3D) organoid systems derived from stem cells for infection biology is a highly promising nascent research area that will provide more complex multicellular systems for investigating host–pathogen interactions in vitro (Iakobachvili and Peters, 2017). One such system has been developed from murine mammary gland tissue to create a 3D mammary organoid that can be manipulated to induce lactation or involution (Sumbal et al., 2020). A similar system could be used to generate a bovine mammary organoid allowing the in vitro examination of the lactoferrin-dependent internalization of S. uberis.
Another possibility is to construct transgenic mice that express receptors supporting the function of surface proteins. In addition to the examples already mentioned here for S. aureus IsdB and S. pyogenes M protein (Lövkvist et al., 2008; Matsui et al., 2009; Pishchany et al., 2010), this approach was carried out to examine the role of L. monocytogenes InlA during colonization of the gut (Figure 2). Mice were genetically manipulated to either express human E-cadherin from a specialized promoter in enterocytes or a “humanized” Glu16Pro version of the murine E-cadherin in all E-cadherin-expressing cells (Lecuit et al., 2001; Lecuit and Cossart, 2002; Disson et al., 2008, 2009). Both systems demonstrated the requirement of InlA for intestinal crossing of L. monocytogenes with the chimeric mice also demonstrating that both InlA and internalin B are required for placental invasion during murine pregnancy (Lecuit et al., 2001; Disson et al., 2008). In the reverse scenario, InlA on the L. monocytogenes cell surface has been “murinized” through two single amino acid substitutions, which enhanced binding to murine E-cadherin in vitro and led to infection of the murine intestinal epithelium in vivo (Wollert et al., 2007). This subsequently led to higher bacterial burden in multiple organs including the liver and spleen, demonstrating the importance of this InlA-dependent route of bacterial dissemination (Wollert et al., 2007). Further utilization of this “murinized” L. monocytogenes strain demonstrated that the C57BL/6J mouse cell line has inherent resistance to oral challenge by Listeria with more susceptible murine cell lines exhibiting faster dissemination of the mutated strain as well as increased cytokine production (Bergmann et al., 2013). Advances in genome editing tools, including CRISPR-Cas systems, have made transgenesis of rodents a relatively facile approach, allowing the development of new transgenic models that can facilitate investigations into the importance of host-specialized bacterial interactions in vivo (Qin et al., 2016). For example, recent studies have employed CRISPR-Cas edited mice expressing the human integrin component CD11b to examine the function of S. aureus toxin LukAB in vivo (Boguslawski et al., 2020).
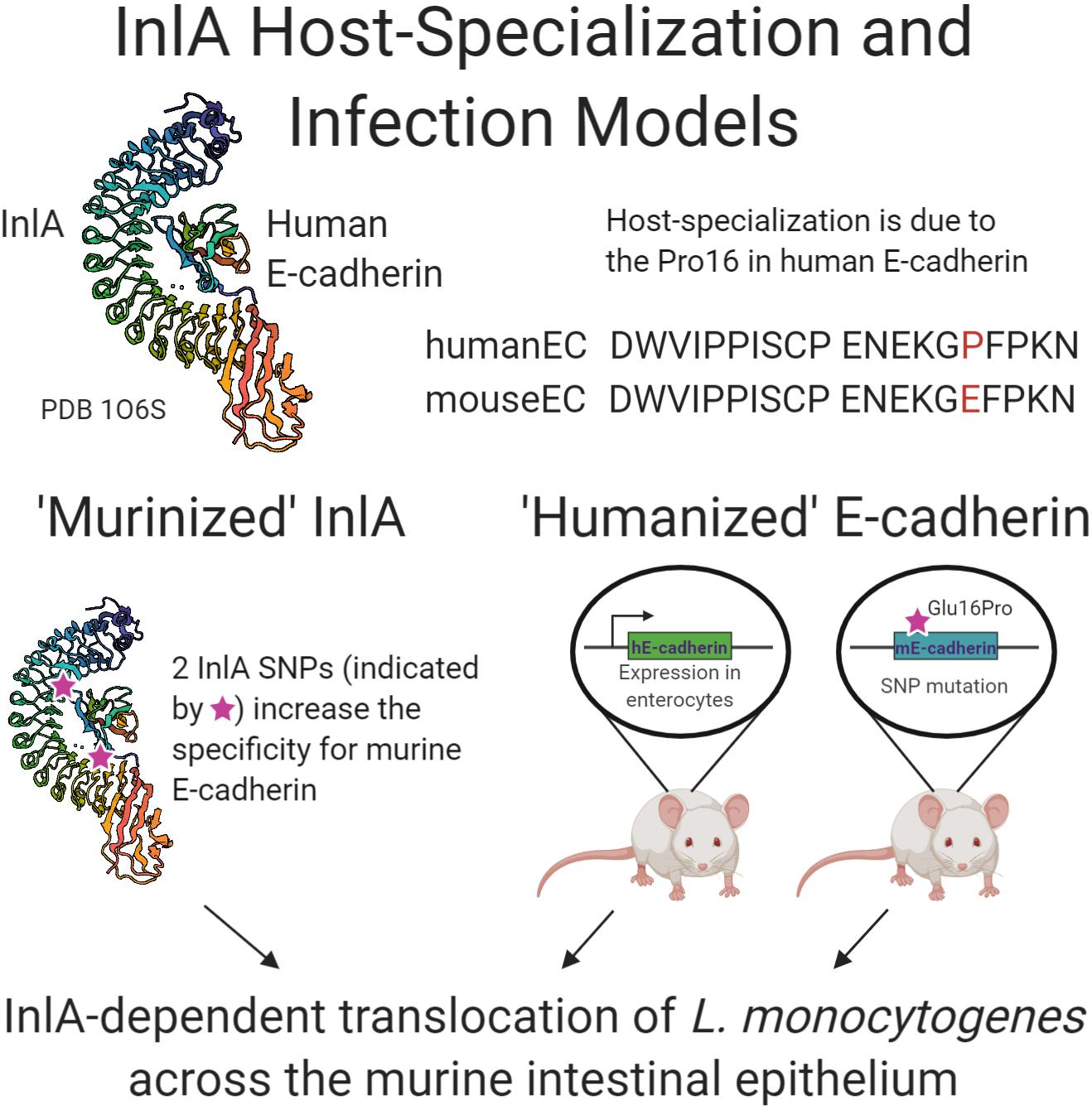
Figure 2. Schematic summary of research examining the interaction of internalin A of L. monocytogenes with human E-cadherin. Crystal structure generation of InlA in complex with human E-cadherin, PDB 1O6S (Schubert et al., 2002), allowed the “murinization” of InlA with enhanced binding to murine E-cadherin. Two transgenic murine models were developed that express the human E-cadherin protein either solely in the intestine or throughout the mouse. All three approaches demonstrated the role of InlA in crossing the intestinal epithelial barrier during L. monocytogenes experimental infection.
Surface proteins of Gram-positive bacteria are essential for an array of different host-pathogen interactions. Accordingly, bacterial adaptation to a new host-species or anatomical niche often requires the acquisition of new proteins or diversification of existing proteins to enable binding to polymorphic ligands or receptors. As highlighted by the examples provided in the current review, important advances have been made regarding our understanding of the molecular basis for such interactions in vitro. In addition, new approaches for transgenesis and mutagenesis offer great promise for the development of new organoid or whole animal models of infection that allow the dissection of the role in pathogenesis of niche and/or host-specialized surface proteins, many of which have multiple binding tropisms involving several different receptor ligands. Development of appropriate experimental models will not only allow the validation of surface proteins that play important roles in vivo but also improve on the current model systems used for the evaluation of novel therapeutic and vaccine candidates. The identification of alternative approaches for control of infection is of particular importance for bacteria that exhibit high levels of antibiotic resistance and are becoming increasingly difficult to treat in a clinical setting.
All authors wrote and edited the manuscript.
JRF is supported by the Biotechnology and Biological Sciences Research Council (UK) Institute Strategic Grant (ISP2) (BBS/E/D/20002173), and a Wellcome Trust collaborative award 201531/Z/16/Z.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank collaborators and members of the Fitzgerald group past and present for their contributions to research on this topic. Figures were created with BioRender.com.
Almeida, R. A., Dego, O. K., Headrick, S. I., Lewis, M. J., and Oliver, S. P. (2015). Role of Streptococcus uberis adhesion molecule in the pathogenesis of Streptococcus uberis mastitis. Vet. Microbiol. 179, 332–335. doi: 10.1016/j.vetmic.2015.07.005
Almeida, R. A., Luther, D. A., Park, H.-M., and Oliver, S. P. (2006). Identification, isolation, and partial characterization of a novel Streptococcus uberis adhesion molecule (SUAM). Vet. Microbiol. 115, 183–191. doi: 10.1016/j.vetmic.2006.02.005
Bannoehr, J., Ben Zakour, N. L., Reglinski, M., Inglis, N. F., Prabhakaran, S., Fossum, E., et al. (2011). Genomic and surface proteomic analysis of the canine pathogen Staphylococcus pseudintermedius reveals proteins that mediate adherence to the extracellular matrix. Infect. Immun. 79, 3074–3086. doi: 10.1128/iai.00137-11
Bäumler, A., and Fang, F. C. (2013). Host specificity of bacterial pathogens. Cold Spring Harb. Perspect. Med. 3:a010041. doi: 10.1101/cshperspect.a010041
Ben Zakour, N. L., Sturdevant, D. E., Even, S., Guinane, C. M., Barbey, C., Alves, P. D., et al. (2008). Genome-wide analysis of ruminant Staphylococcus aureus reveals diversification of the core genome. J. Bacteriol. 190, 6302–6317. doi: 10.1128/jb.01984-07
Bergmann, S., Beard, P. M., Pasche, B., Lienenklaus, S., Weiss, S., Gahan, C. G. M., et al. (2013). Influence of internalin a murinisation on host resistance to orally acquired listeriosis in mice. BMC Microbiol. 13:90. doi: 10.1186/1471-2180-13-90
Boguslawski, K. M., McKeown, A. N., Day, C. J., Lacey, K. A., Tam, K., Vozhilla, N., et al. (2020). Exploiting species specificity to understand the tropism of a human-specific toxin. Sci. Adv. 6:eaax7515. doi: 10.1126/sciadv.aax7515
Bowden, C. F. M., Chan, A. C. K., Li, E. J. W., Arrieta, A. L., Eltis, L. D., and Murphy, M. E. P. (2018). Structure–function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin. J. Biol. Chem. 293, 177–190. doi: 10.1074/jbc.M117.806562
Bowden, C. F. M., Verstraete, M. M., Eltis, L. D., and Murphy, M. E. P. (2014). Hemoglobin binding and catalytic heme extraction by IsdB near iron transporter domains. Biochemistry 53, 2286–2294. doi: 10.1021/bi500230f
Chaneton, L., Tirante, L., Maito, J., Chaves, J., and Bussmann, L. E. (2008). Relationship between milk lactoferrin and etiological agent in the mastitic bovine mammary gland. J. Dairy Sci. 91, 1865–1873. doi: 10.3168/jds.2007-0732
Chen, X., Dego, O. K., Almeida, R. A., Fuller, T. E., Luther, D. A., and Oliver, S. P. (2011). Deletion of sua gene reduces the ability of Streptococcus uberis to adhere to and internalize into bovine mammary epithelial cells. Vet. Microbiol. 147, 426–434. doi: 10.1016/j.vetmic.2010.07.006
Choby, J. E., Buechi, H. B., Farrand, A. J., Skaar, E. P., and Barber, M. F. (2018). Molecular basis for the evolution of species-specific hemoglobin capture by Staphylococcus aureus. mBio 9:e01524-18. doi: 10.1128/mBio.01524-18
Claes, J., Ditkowski, B., Liesenborghs, L., Veloso, T. R., Entenza, J. M., Moreillon, P., et al. (2018). Assessment of the dual role of clumping factor a in S. aureus adhesion to endothelium in absence and presence of plasma. Thromb. Haemost. 118, 1230–1241. doi: 10.1055/s-0038-1660435
Cunningham, M. W. (2000). Pathogenesis of group a streptococcal infections. Clin. Microbiol. Rev. 13, 470–511. doi: 10.1128/cmr.13.3.470-511.2000
Disson, O., Grayo, S., Huillet, E., Nikitas, G., Langa-Vives, F., Dussurget, O., et al. (2008). Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 455, 1114–1118. doi: 10.1038/nature07303
Disson, O., Nikitas, G., Grayo, S., Dussurget, O., Cossart, P., and Lecuit, M. (2009). Modeling human listeriosis in natural and genetically engineered animals. Nat. Protoc. 4, 799–810. doi: 10.1038/nprot.2009.66
Douam, F., Gaska, J. M., Winer, B. Y., Ding, Q., von Schaewen, M., and Ploss, A. (2015). Genetic dissection of the host tropism of human-tropic pathogens. Annu. Rev. Genet. 49, 21–45. doi: 10.1146/annurev-genet-112414-054823
Fang, W., Almeida, R. A., and Oliver, S. P. (2000). Effects of lactoferrin and milk on adherence of Streptococcus uberis to bovine mammary epithelial cells. Am. J. Vet. Res. 61, 275–279. doi: 10.2460/ajvr.2000.61.275
Fang, W., and Oliver, S. P. (1999). Identification of lactoferrin-binding proteins in bovine mastitis-causing Streptococcus uberis. FEMS Microbiol. Lett. 176, 91–96. doi: 10.1111/j.1574-6968.1999.tb13647.x
Fischetti, V. A. (1989). Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2, 285–314. doi: 10.1128/cmr.2.3.285
Fitzgerald, J. R. (2012). Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 20, 192–198. doi: 10.1016/j.tim.2012.01.006
Flick, M. J., Du, X., Prasad, J. M., Raghu, H., Palumbo, J. S., Smeds, E., et al. (2013). Genetic elimination of the binding motif on fibrinogen for the S. aureus virulence factor ClfA improves host survival in septicemia. Blood 121, 1783–1794. doi: 10.1182/blood-2012-09-453894
Friedman, D. B., Stauff, D. L., Pishchany, G., Whitwell, C. W., Torres, V. J., and Skaar, E. P. (2006). Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2:e87. doi: 10.1371/journal.ppat.0020087
Gaillard, J. L., Berche, P., Frehel, C., Gouln, E., and Cossart, P. (1991). Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65, 1127–1141. doi: 10.1016/0092-8674(91)90009-N
Gaudin, C. F. M., Grigg, J. C., Arrieta, A. L., and Murphy, M. E. P. (2011). Unique heme-iron coordination by the hemoglobin receptor IsdB of Staphylococcus aureus. Biochemistry 50, 5443–5452. doi: 10.1021/bi200369p
Geoghegan, J. A., Ganesh, V. K., Smeds, E., Liang, X., Höök, M., and Foster, T. J. (2010). Molecular characterization of the interaction of staphylococcal microbial surface components recognizing adhesive matrix molecules (MSCRAMM) ClfA and Fbl with fibrinogen. J. Biol. Chem. 285, 6208–6216. doi: 10.1074/jbc.M109.062208
Gianquinto, E., Moscetti, I., De Bei, O., Campanini, B., Marchetti, M., Luque, F. J., et al. (2019). Interaction of human hemoglobin and semi-hemoglobins with the Staphylococcus aureus hemophore IsdB: a kinetic and mechanistic insight. Sci. Rep. 9:18629. doi: 10.1038/s41598-019-54970-w
Guinane, C. M., Ben Zakour, N. L., Tormo-Mas, M. A., Weinert, L. A., Lowder, B. V., Cartwright, R. A., et al. (2010). Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2, 454–466. doi: 10.1093/gbe/evq031
Haag, A. F., Fitzgerald, J. R., and Penadés, J. R. (2019). Staphylococcus aureus in animals. Microbiol. Spect. 7, 1–19. doi: 10.1128/microbiolspec.GPP3-0060-2019
Hagiwara, S., Kawai, K., Anri, A., and Nagahata, H. (2003). Lactoferrin concentrations in milk from normal and subclinical mastitic cows. J. Vet. Med. Sci. 65, 319–323. doi: 10.1292/jvms.65.319
Harmon, R. J., Schanbacher, F. L., Ferguson, L. C., and Smith, K. L. (1976). Changes in lactoferrin, immunoglobulin G, bovine serum albumin, and alpha-lactalbumin during acute experimental and natural coliform mastitis in cows. Infect. Immun. 13, 533–542.
Herron-Olson, L., Fitzgerald, J. R., Musser, J. M., and Kapur, V. (2007). Molecular correlates of host specialization in Staphylococcus aureus. PLoS One 2:e1120. doi: 10.1371/journal.pone.0001120
Higgins, J., Loughman, A., Van Kessel, K. P. M., Van Strijp, J. A. G., and Foster, T. J. (2006). Clumping factor A of Staphylococcus aureus inhibits phagocytosis by human polymorphonuclear leucocytes. FEMS Microbiol. Lett. 258, 290–296. doi: 10.1111/j.1574-6968.2006.00229.x
Horstmann, R. D., Sievertsen, H. J., Knobloch, J., and Fischetti, V. A. (1988). Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc. Natl. Acad. Sci. U.S.A. 85, 1657–1661. doi: 10.1073/pnas.85.5.1657
Iakobachvili, N., and Peters, P. J. (2017). Humans in a dish: the potential of organoids in modeling immunity and infectious diseases. Front. Microbiol. 8:2402. doi: 10.3389/fmicb.2017.02402
Inaba, Y., Tsurifune, K., Sasazaki, S., and Mannen, H. (2008). A new tandem repeat in bovine fibrinogen Aα gene. Anim. Biotechnol. 19, 122–126. doi: 10.1080/10495390801929122
Jochim, A., Adolf, L., Belikova, D., Schilling, N. A., Setyawati, I., Chin, D., et al. (2020). An ECF-type transporter scavenges heme to overcome iron-limitation in Staphylococcus lugdunensis. eLife 9:e57322. doi: 10.7554/eLife.57322
Josse, J., Laurent, F., and Diot, A. (2017). Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Front. Microbiol. 8:2433. doi: 10.3389/fmicb.2017.02433
Ko, Y.-P., and Flick, M. J. (2016). Fibrinogen is at the interface of host defense and pathogen virulence in Staphylococcus aureus infection. Semin. Thromb. Hemost. 42, 408–421. doi: 10.1055/s-0036-1579635
Laabei, M., and Ermert, D. (2019). Catch me if you can: Streptococcus pyogenes complement evasion strategies. J. Innate Immun. 11, 3–12. doi: 10.1159/000492944
Lecuit, M., and Cossart, P. (2002). Genetically-modified-animal models for human infections: the Listeria paradigm. Trends Mol. Med. 8, 537–542. doi: 10.1016/S1471-4914(02)02413-9
Lecuit, M., Dramsi, S., Gottardi, C., Fedor-Chaiken, M., Gumbiner, B., and Cossart, P. (1999). A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18, 3956–3963. doi: 10.1093/emboj/18.14.3956
Lecuit, M., Nelson, D. M., Smith, S. D., Khun, H., Huerre, M., Vacher-Lavenu, M.-C., et al. (2004). Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: role of internalin interaction with trophoblast E-cadherin. Proc. Natl. Acad. Sci. U.S.A. 101, 6152–6157. doi: 10.1073/pnas.0401434101
Lecuit, M., Vandormael-Pournin, S., Lefort, J., Huerre, M., Gounon, P., Dupuy, C., et al. (2001). A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292, 1722–1725. doi: 10.1126/science.1059852
Lewis, M. J., Meehan, M., Owen, P., and Woof, J. M. (2008). A common theme in interaction of bacterial immunoglobulin-binding proteins with immunoglobulins illustrated in the equine system. J. Biol. Chem. 283, 17615–17623. doi: 10.1074/jbc.M709844200
Lövkvist, L., Sjölinder, H., Wehelie, R., Aro, H., Norrby-Teglund, A., Plant, L., et al. (2008). CD46 contributes to the severity of group a streptococcal infection. Infect. Immun. 76, 3951–3958. doi: 10.1128/iai.00109-08
Lowder, B. V., Guinane, C. M., Ben Zakour, N. L., Weinert, L. A., Conway-Morris, A., Cartwright, R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550. doi: 10.1073/pnas.0909285106
Lu, J., Francis, J., Doster, R. S., Haley, K. P., Craft, K. M., Moore, R. E., et al. (2020). Lactoferrin: a critical mediator of both host immune response and antimicrobial activity in response to streptococcal infections. ACS Infect. Dis. 6, 1615–1623. doi: 10.1021/acsinfecdis.0c00050
Matsui, H., Sekiya, Y., Nakamura, M., Murayama, S. Y., Yoshida, H., Takahashi, T., et al. (2009). CD46 transgenic mouse model of necrotizing fasciitis caused by Streptococcus pyogenes infection. Infect. Immun. 77, 4806–4814. doi: 10.1128/iai.00577-09
McDevitt, D., Nanavaty, T., House-Pompeo, K., Bell, E., Turner, N., Mcintire, L., et al. (1997). Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur. J. Biochem. 247, 416–424. doi: 10.1111/j.1432-1033.1997.00416.x
Meehan, M., Lynagh, Y., Woods, C., and Owen, P. (2001). The fibrinogen-binding protein (FgBP) of Streptococcus equi subsp. equi additionally binds IgG and contributes to virulence in a mouse model. Microbiology 147, 3311–3322. doi: 10.1099/00221287-147-12-3311
Meehan, M., Muldowney, D. A., Watkins, N. J., and Owen, P. (2000). Localization and characterization of the ligand-binding domain of the fibrinogen-binding protein (FgBP) of Streptococcus equi subsp. equi. Microbiology 146, 1187–1194. doi: 10.1099/00221287-146-5-1187
Meehan, M., Nowlan, P., and Owen, P. (1998). Affinity purification and characterization of a fibrinogen-binding protein complex which protects mice against lethal challenge with Streptococcus equi subsp. equi. Microbiology 144, 993–1003. doi: 10.1099/00221287-144-4-993
Mengaud, J., Ohayon, H., Gounon, P., Mège, R.-M., and Cossart, P. (1996). E-Cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84, 923–932. doi: 10.1016/S0092-8674(00)81070-3
Miajlovic, H., Loughman, A., Brennan, M., Cox, D., and Foster, T. J. (2007). Both complement- and fibrinogen-dependent mechanisms contribute to platelet aggregation mediated by Staphylococcus aureus clumping factor B. Infect. Immun. 75, 3335–3343. doi: 10.1128/iai.01993-06
Mosesson, M. W. (2005). Fibrinogen and fibrin structure and functions. J. Thromb. Haemost. 3, 1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x
Moshynskyy, I., Jiang, M., Fontaine, M. C., Perez-Casal, J., Babiuk, L. A., and Potter, A. A. (2003). Characterization of a bovine lactoferrin binding protein of Streptococcus uberis. Microb. Pathog. 35, 203–215. doi: 10.1016/S0882-4010(03)00150-5
Murakawa, M., Okamura, T., Kamura, T., Shibuya, T., Harada, M., and Niho, Y. (1993). Diversity of primary structures of the carboxy-terminal regions of mammalian fibrinogen A alpha-chains. Characterization of the partial nucleotide and deduced amino acid sequences in five mammalian species; rhesus monkey, pig, dog, mouse and Syrian hamster. Thromb. Haemost. 69, 351–360.
Murray, E. G. D., Webb, R. A., and Swann, M. B. R. (1926). A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes (n.sp.). J. Pathol. Bacteriol. 29, 407–439. doi: 10.1002/path.1700290409
Muryoi, N., Tiedemann, M. T., Pluym, M., Cheung, J., Heinrichs, D. E., and Stillman, M. J. (2008). Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 283, 28125–28136. doi: 10.1074/jbc.M802171200
Nikitas, G., Deschamps, C., Disson, O., Niault, T., Cossart, P., and Lecuit, M. (2011). Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J. Exp. Med. 208, 2263–2277. doi: 10.1084/jem.20110560
Okada, N., Liszewski, M. K., Atkinson, J. P., and Caparon, M. (1995). Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A Streptococcus. Proc. Natl. Acad. Sci. U.S.A. 92, 2489–2493. doi: 10.1073/pnas.92.7.2489
Pasquina, L., Santa Maria, J. P., McKay Wood, B., Moussa, S. H., Matano, L. M., Santiago, M., et al. (2016). A synthetic lethal approach for compound and target identification in Staphylococcus aureus. Nat. Chem. Biol. 12, 40–45. doi: 10.1038/nchembio.1967
Patel, D., Almeida, R. A., Dunlap, J. R., and Oliver, S. P. (2009). Bovine lactoferrin serves as a molecular bridge for internalization of Streptococcus uberis into bovine mammary epithelial cells. Vet. Microbiol. 137, 297–301. doi: 10.1016/j.vetmic.2009.01.013
Pentecost, M., Otto, G., Theriot, J. A., and Amieva, M. R. (2006). Listeria monocytogenes invades the epithelial junctions at sites of cell extrusion. PLoS Pathog. 2:e3. doi: 10.1371/journal.ppat.0020003
Pickering, A. C., Vitry, P., Prystopiuk, V., Garcia, B., Höök, M., Schoenebeck, J., et al. (2019). Host-specialized fibrinogen-binding by a bacterial surface protein promotes biofilm formation and innate immune evasion. PLoS Pathog. 15:e1007816. doi: 10.1371/journal.ppat.1007816
Pietrocola, G., Geoghegan, J. A., Rindi, S., Di Poto, A., Missineo, A., Consalvi, V., et al. (2013). Molecular characterization of the multiple interactions of SpsD, a surface protein from Staphylococcus pseudintermedius, with host extracellular matrix proteins. PLoS One 8:e66901. doi: 10.1371/journal.pone.0066901
Pishchany, G., McCoy, A. L., Torres, V. J., Krause, J. C., Crowe, J. E., Fabry, M. E., et al. (2010). Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe 8, 544–550. doi: 10.1016/j.chom.2010.11.002
Qin, W., Kutny, P. M., Maser, R. S., Dion, S. L., Lamont, J. D., Zhang, Y., et al. (2016). Generating mouse models using CRISPR-Cas9-mediated genome editing. Curr. Protoc. Mouse Biol. 6, 39–66. doi: 10.1002/9780470942390.mo150178
Resch, G., François, P., Morisset, D., Stojanov, M., Bonetti, E. J., Schrenzel, J., et al. (2013). Human-to-bovine jump of Staphylococcus aureus CC8 is associated with the loss of a β-hemolysin converting prophage and the acquisition of a new staphylococcal cassette chromosome. PLoS One 8:e58187. doi: 10.1371/journal.pone.0058187
Rezcallah, M. S., Hodges, K., Gill, D. B., Atkinson, J. P., Wang, B., and Cleary, P. P. (2005). Engagement of CD46 and α5β1 integrin by group A streptococci is required for efficient invasion of epithelial cells. Cell. Microbiol. 7, 645–653. doi: 10.1111/j.1462-5822.2004.00497.x
Ribet, D., and Cossart, P. (2015). How bacterial pathogens colonize their hosts and invade deeper tissues. Microb. Infect. 17, 173–183. doi: 10.1016/j.micinf.2015.01.004
Richardson, E. J., Bacigalupe, R., Harrison, E. M., Weinert, L. A., Lycett, S., Vrieling, M., et al. (2018). Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2, 1468–1478. doi: 10.1038/s41559-018-0617-0
Riley-Vargas, R. C., Gill, D. B., Kemper, C., Liszewski, M. K., and Atkinson, J. P. (2004). CD46: expanding beyond complement regulation. Trends Immunol. 25, 496–503. doi: 10.1016/j.it.2004.07.004
Roulo, R. M., Fishburn, J. D., Amosu, M., Etchison, A. R., and Smith, M. A. (2014). Dose response of Listeria monocytogenes invasion, fetal morbidity, and fetal mortality after oral challenge in pregnant and nonpregnant Mongolian gerbils. Infect. Immun. 82, 4834–4841. doi: 10.1128/IAI.01514-14
Schubert, W.-D., Urbanke, C., Ziehm, T., Beier, V., Machner, M. P., Domann, E., et al. (2002). Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111, 825–836. doi: 10.1016/S0092-8674(02)01136-4
Seo, H. S., Minasov, G., Seepersaud, R., Doran, K. S., Dubrovska, I., Shuvalova, L., et al. (2013). Characterization of fibrinogen binding by glycoproteins Srr1 and Srr2 of Streptococcus agalactiae. J. Biol. Chem. 288, 35982–35996. doi: 10.1074/jbc.M113.513358
Sheldon, J. R., and Heinrichs, D. E. (2015). Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol. Rev. 39, 592–630. doi: 10.1093/femsre/fuv009
Sheppard, S. K., Guttman, D. S., and Fitzgerald, J. R. (2018). Population genomics of bacterial host adaptation. Nat. Rev. Genet. 19, 549–565. doi: 10.1038/s41576-018-0032-z
Spoor, L. E., Richardson, E., Richards, A. C., Wilson, G. J., Mendonca, C., Gupta, R. K., et al. (2015). Recombination-mediated remodelling of host-pathogen interactions during Staphylococcus aureus niche adaptation. Microb. Genom. 1:e000036. doi: 10.1099/mgen.0.000036
Su, X., Cao, G., Zhang, J., Pan, H., Zhang, D., Kuang, D., et al. (2019). Characterization of internalin genes in Listeria monocytogenes from food and humans, and their association with the invasion of Caco-2 cells. Gut Pathog. 11:30. doi: 10.1186/s13099-019-0307-8
Sumbal, J., Chiche, A., Charifou, E., Koledova, Z., and Li, H. (2020). Primary mammary organoid model of lactation and involution. Front. Cell Dev. Biol. 8:68. doi: 10.3389/fcell.2020.00068
Thern, A., Stenberg, L., Dahlbäck, B., and Lindahl, G. (1995). Ig-binding surface proteins of Streptococcus pyogenes also bind human C4b-binding protein (C4BP), a regulatory component of the complement system. J. Immunol. 154, 375–386.
Thomer, L., Schneewind, O., and Missiakas, D. (2016). Pathogenesis of Staphylococcus aureus bloodstream infections. Annu. Rev. Pathol. 11, 343–364. doi: 10.1146/annurev-pathol-012615-044351
Torres, V. J., Pishchany, G., Humayun, M., Schneewind, O., and Skaar, E. P. (2006). Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188, 8421–8429. doi: 10.1128/jb.01335-06
Valenti, P., and Antonini, G. (2005). Lactoferrin: an important host defence against microbial and viral attack. Cell. Mol. Life Sci. 62:2576. doi: 10.1007/s00018-005-5372-0
Van Stelten, A., Simpson, J. M., Chen, Y., Scott, V. N., Whiting, R. C., Ross, W. H., et al. (2011). Significant shift in median guinea pig infectious dose shown by an outbreak-associated Listeria monocytogenes epidemic clone strain and a strain carrying a premature stop codon mutation in inlA. Appl. Environ. Microbiol. 77, 2479–2487. doi: 10.1128/aem.02626-10
Viana, D., Comos, M., McAdam, P. R., Ward, M. J., Selva, L., Guinane, C. M., et al. (2015). A single natural nucleotide mutation alters bacterial pathogen host tropism. Nat. Genet. 47, 361–366. doi: 10.1038/ng.3219
Walsh, E. J., Miajlovic, H., Gorkun, O. V., and Foster, T. J. (2008). Identification of the Staphylococcus aureus MSCRAMM clumping factor B (ClfB) binding site in the αC-domain of human fibrinogen. Microbiology 154, 550–558. doi: 10.1099/mic.0.2007/010868-0
Weinberg, E. D. (1974). Iron and susceptibility to infectious disease. Science 184, 952–956. doi: 10.1126/science.184.4140.952
Williams, D., Dunn, S., Richardson, A., Frank, J. F., and Smith, M. A. (2011). Time course of fetal tissue invasion by Listeria monocytogenes following an oral inoculation in pregnant guinea pigs. J. Food Prot. 74, 248–253. doi: 10.4315/0362-028x.Jfp-10-163
Wollert, T., Pasche, B., Rochon, M., Deppenmeier, S., van den Heuvel, J., Gruber, A. D., et al. (2007). Extending the host range of Listeria monocytogenes by rational protein design. Cell 129, 891–902. doi: 10.1016/j.cell.2007.03.049
Woolhouse, M. E. J., Taylor, L. H., and Haydon, D. T. (2001). Population biology of multihost pathogens. Science 292, 1109–1112. doi: 10.1126/science.1059026
Xiang, H., Feng, Y., Wang, J., Liu, B., Chen, Y., Liu, L., et al. (2012). Crystal structures reveal the multi-ligand binding mechanism of Staphylococcus aureus ClfB. PLoS Pathog. 8:e1002751. doi: 10.1371/journal.ppat.1002751
Zapotoczna, M., Heilbronner, S., Speziale, P., and Foster, T. J. (2012). Iron-regulated surface determinant (Isd) proteins of Staphylococcus lugdunensis. J. Bacteriol. 194, 6453–6467. doi: 10.1128/jb.01195-12
Zhu, H., Xie, G., Liu, M., Olson, J. S., Fabian, M., Dooley, D. M., et al. (2008). Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J. Biol. Chem. 283, 18450–18460. doi: 10.1074/jbc.M801466200
Keywords: specialized, niche, host, surface protein, adaptation, bacteria, interaction
Citation: Pickering AC and Fitzgerald JR (2020) The Role of Gram-Positive Surface Proteins in Bacterial Niche- and Host-Specialization. Front. Microbiol. 11:594737. doi: 10.3389/fmicb.2020.594737
Received: 14 August 2020; Accepted: 05 October 2020;
Published: 29 October 2020.
Edited by:
Timothy J. Foster, Trinity College Dublin, IrelandReviewed by:
Maria Rapala-Kozik, Jagiellonian University, PolandCopyright © 2020 Pickering and Fitzgerald. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Ross Fitzgerald, Um9zcy5GaXR6Z2VyYWxkQGVkLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.