- 1State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing, China
- 2Central Laboratory, Chongqing Public Health Medical Center, Chongqing, China
- 3Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China
Pyrazinamide (PZA) is widely used to treat drug-sensitive or multidrug resistance tuberculosis. However, conventional PZA susceptibility tests of clinical isolates are rather difficult because of the requirement of acid pH. Since resistance to pyrazinamide is primary mediated by mutation of pncA, an alternative way of PZA susceptibility test is to analyze the pyrazinamidase activities of Mycobacterium tuberculosis clinical isolates. Therefore, a database containing the full spectrum of pncA mutations along with pyrazinamidase activities will be beneficial. To characterize mutations of pncA in M. tuberculosis from Chongqing, China, the pncA gene was sequenced and analyzed in 465 clinical isolates. A total of 124 types of mutations were identified in 424 drug-resistant isolates, while no mutation was identified in the 31 pan-susceptible isolates. Ninety-four of the 124 mutations had previously been reported, and 30 new mutations were identified. Based on reported literatures, 294 isolates could be predicted resistant to pyrazinamide. Furthermore, pyrazinamidase activities of the 30 new mutations were tested using the Escherichia coli pncA gene knockout strain. The results showed that 24 of these new mutations (28 isolates) led to loss of pyrazinamidase activity and six (8 isolates) of them did not. Taken together, 322 isolates with pncA mutations could be predicted to be PZA resistant among the 424 drug-resistant isolates tested. Analysis of pncA mutations and their effects on pyrazinamidase activity will not only enrich our knowledge of comprehensive pncA mutations related with PZA resistance but also facilitate rapid molecular diagnosis of pyrazinamide resistance in M. tuberculosis.
Introduction
Nowadays, Mycobacterium tuberculosis (M. tuberculosis) is still one of the most important human pathogens worldwide, which caused about 1.45 million deaths in 2018 (World Health Organization [WHO], 2019). The emergence of multidrug (MDR) and extensively resistant (XDR) M. tuberculosis strains makes it even more difficult to control the disease of tuberculosis (TB). Pyrazinamide (PZA), a synthetic derivative of nicotinamide, was firstly synthesized in 1936, and subsequently used for the treatment of TB since 1952 (Palomino and Martin, 2014). Till present, PZA has been listed as a first-line agent in the modern short-course TB regimen. Due to its unique bactericidal activity against slow growing or metabolically inactive M. tuberculosis that is not killed by other TB drugs, it shortens the time of therapy from 9 to 12 to 6 months and is used in treatment of both drug-susceptible M. tuberculosis and PZA-susceptible MDR M. tuberculosis (Mitchison, 1985; Zhang and Mitchison, 2003).
Although having been used in the treatment of M. tuberculosis for more than 60 years, the accurate mode of action of PZA still remains elusive. The most widely accepted mechanism is that PZA is a prodrug and needs to be converted to its active form of pyrazinoic acid (POA) by the pyrazinamidase (PZase) encoded by the pncA gene. POA as an ionophore in the cytosol is expelled by a weak efflux pump into the acidic conditions. The protonated POA re-enters into cytosol and releases proton. Because of the inefficiency of the efflux pump, repetition of this cycle causes POA accumulation and acidification in cytosol and leads to cellular damage (Zhang et al., 1999). Mutation of the pncA gene aborts or reduces PZase activity and is the major reason why clinical M. tuberculosis strains acquire PZA resistance. However, some clinical PZA-resistant strains with positive PZase do not have pncA gene mutations, suggesting the existence of alternative mechanisms (Zhang and Mitchison, 2003).
So far, the real target of POA has never been identified. Overexpression of fatty acid synthetase I (FAS-I) encoded by eukaryotic-like fas1 gene from M. tuberculosis, M. bovis BCG, or Mycobacterium avium in Mycobacterium smegmatis confers resistance to 5-Cl-PZA, a PZA analog, and FAS-I used to be thought as the target of PZA (Zimhony et al., 2000). However, it was lately realized that POA failed to directly inhibits mycobacterial fatty acid synthase FAS-I, and this enzyme is the target of 5-Cl-PZA, not PZA (Boshoff et al., 2002). The ribosomal protein S1, a vital protein involved in ribosome-sparing process of trans-translation, was also speculated to be the target of POA. Overexpression of RpsA did confer PZA resistance, and three clinical PZA-resistant isolates with no mutation in pncA-harbored RpsA mutations (Shi et al., 2011). However, later research showed that M. tuberculosis RpsA interacts with single-stranded RNA, but not with POA (Dillon et al., 2017), and strains with deletion of trans-translation-relevant genes did not show any change of PZA susceptibility (Personne and Parish, 2014). The aspartate decarboxylase encoded by panD is involved in synthesis of β-alanine, which is a precursor for the biosynthesis of co-enzyme A and pantothenate. Whole genome sequencing analyses demonstrated that PZA-resistant clinical isolates lacking pncA and rpsA mutations were observed with panD mutations (Zhang et al., 2013). On the other hand, although supplementation of precursors of coenzyme A biosynthesis, such as β-alanine, pantothenate, or pantetheine could antagonize the action of PZA, the gene deletion of pantothenate biosynthesis pathway did not antagonize PZA activity. This finding demonstrated that pantothenate biosynthesis pathway including panD is not involved in PZA activity (Dillon et al., 2014). Very recently, researchers found that an ATP-dependent ATPase (ClpC1) involved in protein degradation (Zhang et al., 2017), and a bifunctional enzyme Rv2783 involved in metabolism of RNA (Njire et al., 2017) also confer PZA resistance in clinical isolates. Though evidences have shown that mutations of rpsA, panD, and clpC1 are involved in PZA resistance, contributions of those mutations to clinical resistance of PZA in M. tuberculosis still need further investigation.
Though drug susceptibility testing (DST) is recommended for both first- and second-line anti-tuberculosis drugs (World Health Organization [WHO], 2019), DST for PZA is complicated due to the requirement of acidic pH (Zhang et al., 2002). At present, Bactec MGIT 960 system (Becton Dickinson Biosciences, Sparks, MD) still has unreliable results presented (Chedore et al., 2010; Piersimoni et al., 2013). In some cases, molecular test is rapid, simple, and reliable for prediction of drug resistance. Although the exact mechanism of PZA action is still in research, a couple of genes involved in PZA resistance have been found as mentioned above. One of most important gene is pncA, which accounts for 70–97% of the PZA resistance mutations (Hirano et al., 1997). However, unlike the resistance conferring regions of rpoB to rifampicin, katG to isoniazid, no clear hotspot for pncA mutations has been found. Mutations are highly diverse and scattered throughout the pncA gene, complicating the molecular assessment of PZA resistance and making DNA sequencing the best option so far (Miotto et al., 2014). Moreover, there are reports showing that some specific pncA mutations observed in clinical isolates do not affect the susceptibility to PZA (Xia et al., 2015; Allix-Béguec et al., 2018). As an assistant PZA susceptibility judgment method, classical PZase tests, namely Wayne’s method, used orange-red color change of POA to react with ammonium ferrous sulfate (Wayne, 1974). Subsequent modified Wayne’s method (Zhang et al., 2013; Aono et al., 2018) omitted protein precipitate step which used trichloroacetic acid. However, all the reported Wayne’s methods need large amount of M. tuberculosis culture and hence a long growing cycle is inevitable. In addition, so far, all methods based on Wayne’s work need to be performed in high level biosafety labs (BSL-3) due to the requirement of large amount of bacterial cultures.
Since over 60% MDR TB cases globally are PZA resistant (Whitfield et al., 2015), and the vital role of PZA in the treatment of TB, rapid and easy to perform molecular assays for PZA susceptibility tests are highly desirable. To achieve this, the pncA genes of 465 clinical M. tuberculosis isolates from Chongqing district were sequenced and mutations were analyzed. Subsequently, PZA susceptibilities of the isolates containing previous reported pncA mutations were predicted based on reported literatures. In addition, an Escherichia coli pncA gene knockout strain was used to analyze the PZase activities of the newly identified pncA mutations and hence predict the PZA susceptibilities of the corresponding isolates. The results were reported herein.
Materials and Methods
Clinical M. tuberculosis Isolates
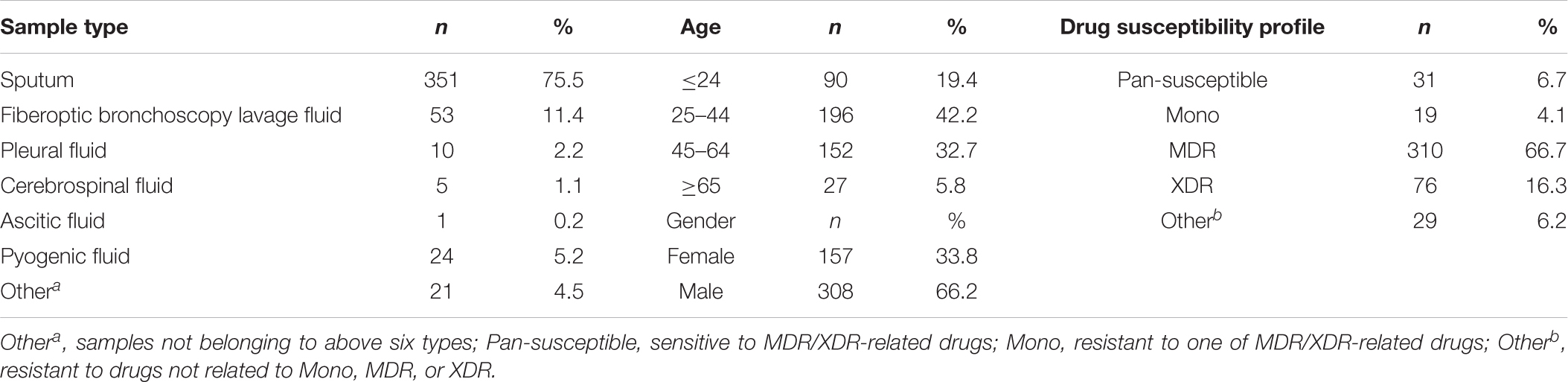
A total of 465 randomly selected clinical samples from different patients (Table 1) mostly came from Chongqing local and surrounding areas between April 2014 and August 2018 in Chongqing Public Health Medical Center (Chongqing, China) were cultured in BACTEC MGIT 960 system (Becton Dickinson, Sparks, MD, United States), according to manufacturer’s instructions (Kruuner et al., 2006). The flagged positive samples were subjected to DST using proportion method on Lowenstein-Jensen (L-J) solid medium (Encode, Zhuhai, China) containing MDR- and XDR-related drugs, isoniazid, rifampicin, ofloxacin, levofloxacin, moxifloxacin, amikacin, and capreomycin. The Mycobacterium species identification were assigned based on sequence polymorphisms in 16S rRNA, hsp65, and rpoB (Kim and Shin, 2018), and on the results of 5 μg/ml TCH and 500 μg/ml PNB on L-J solid medium. Isolates were frozen in 25% glycerol at −70°C refrigerator until use.
Mycobacterial Genomic DNA
Colonies of isolates on L-J solid medium were transferred with several loopfuls into 1.5 ml Eppendorf tubes and suspended in 1 ml deionized water, and then centrifuged for DNA extraction. DNA isolation was conducted using HiPure Mycobacterial DNA Kit (Magen, Guangzhou, China). In brief, the mycobacterial cell pellet was washed twice with deionized water and suspended with 200 μl GTL containing lysozyme (10 μg/ml) and proteinase K (10 μg/ml). The bacterial cells were incubated at 37°C for 20 min, 56°C for 20 min, 95°C for 20 min and then mixed with 600 μl GXP. After centrifugation, the DNA in supernatant was collected on silica membrane, washed with GDW, and resuspended in TE buffer.
pncA Gene Amplification and Sequencing Analysis
The entire open reading frame of pncA flanking 150 bp of upstream putative regulatory sequence was amplified using forward primer pncA-F (5′-GGCCCGATGAAGGTGTCGTA) and reverse primer pncA-R (5′-CGGACGGATTTGTCGCTCA CTAC). The pncA primers were designed according to the M. tuberculosis reference sequence (GenBank accession number AL123456.3).
A S1000 Thermal Cycler (BIO-RAD, Hercules, CA, United States) was used for amplification of pncA with the following procedures: a predegeneration step at 98°C for 10 min, degeneration at 98°C for 15 s, annealing at 61.2°C for 15 s, extension at 72°C for 10 s, and terminated with final extension at 72°C for 5 min. Agarose gel electrophoresis was performed, and the DNA was purified with a Omega Kit (Omega Bio-tek, Norcross, GA, United States) according to the manufacturer’s instructions. The purified DNA products were sequenced in a DNA sequencer (ABI, model 3730XL, Carlsbad, CA, United States) with primers same as the PCR amplification. The sequencing results were compared with the reference sequence using nucleotide BLAST1, and the mutation polymorphisms were depicted according to Sequence Variant Nomenclature2.
PZase Activity Tests in Escherichia coli W3110ΔpncA
Based on Wayne’s method (Wayne, 1974; Zhang et al., 2013; Aono et al., 2018), a modified protocol was used to test the PZase activities of the 30 new mutations identified in this study, by using fast-growing E. coli. Plasmids pKD4, pKD46, pCP20, pCA24N, and E. coli K12 strain W3110 were kindly provided by TB research group in Wuhan Institute of Virology of Chinese Academy of Sciences. E. coli W3110 Δ pncA strain was constructed according to a previous publication (Li et al., 2017). Primers used for construction (pncA-ko-s, pncA-ko-a), verification (pncA-jd-s, pncA-jd-a) of E. coli W3110 Δ pncA, and pncA amplification (pncA-f, pncA-r) from clinical TB strains are listed in Table 2. All the culture mediums and antibiotics were purchased from Merck (Darmstadt, Germany) and restriction endonucleases from TAKARA (Tokyo, Japan).

Table 2. Primers used in construction of E. coli W3110 Δ pncA strain and amplification of M. tuberculosis pncA.
For PZase activity tests, products of pncA in clinical TB strains were amplified, purified, digested using SacII and BglII, and then ligated to the same digested plasmid pCA24N. The verified pCA24N-pncA was transformed into E. coli W3110 Δ pncA. Recombinant strains transformed with pncA genes from different M. tuberculosis clinical isolates were cultured in 37°C incubator to mid-log phase (OD600, 0.7∼1.0), then 10 ml culture of each strain was used and a final concentration of 100 μg/ml pyrazinamide was added. After overnight incubation, final concentration of 2% ammonium ferrous sulfate was added into 2 ml culture supernatant of each strain and optical density at wave length 530 nm (OD530) was measured. The strain contained wild-type pncA from M. tuberculosis H37Rv was set as positive control and with the vector as negative control. Cutoff value was set as the ratio between the sample and the negative control that is more than 2.1 (Yamashita et al., 1995).
Drug Susceptibility Testing of PZA
Isolates stored at −70°C refrigerators were quickly thawed in 37°C water bath and then streaked on 7H10 solid medium (Difco, Becton, Dickinson and company, Sparks, MD, United States) supplemented with 10% (V/V) oleic-albumin-dextrose-catalase (OADC, Difco) and 0.5% (V/V) glycerin. After 3–4 weeks of incubation under 37°C, colonies on 7H10 plates were picked into 7H9 liquid medium (Difco) for another 3–4 weeks of incubation. The 7H10 medium used for PZA DST were adjusted to pH 5.7–5.8 using hydrochloric acid and then steam sterilized at temperature of 121°C for 15 min, afterward OADC and glycerin were added when the medium were cooled to 50°C. PZA (Merck, Germany) powder was dissolved in dimethyl sulfoxide (Merck, Germany) to final concentration of 60 mg/ml. The 7H10 plates containing serial concentrations of PZA (0, 6.25, 12.5, 25, 50, 100, 200, and 400 μg/ml) were prepared. Bacteria cells in mid-log phase cultured in 7H9 medium were adjusted to McFarland concentration of 1.0 and then diluted to 100-fold using physiological saline solution. Aliquot of 10 μl diluents were dropped onto the 7H10 medium with serial concentrations of PZA and incubated at 37°C for 4 weeks. M. tuberculosis H37Rv (ATCC 27294) was set as the control. Minimum inhibitory concentration (MIC) is defined as the lowest drug concentration that inhibits 99% growth of the bacteria.
Results
Sample Characteristics
Characteristics of all the selected M. tuberculosis strains are listed in Table 1. The sputum samples accounted for the largest proportion (75.5%). The age bracket 25–44 made up 42.2% in the total samples, illustrating TB are mainly popular in young adults. The male patients (66.2%) were nearly two times more than the female patients (33.8%). According to the DST results of MDR- and XDR-relevant drugs, in the 465 clinical isolates, there are 310 (66.7%) MDR and 76 (16.3%) XDR TB isolates.
Mutations in the pncA Gene
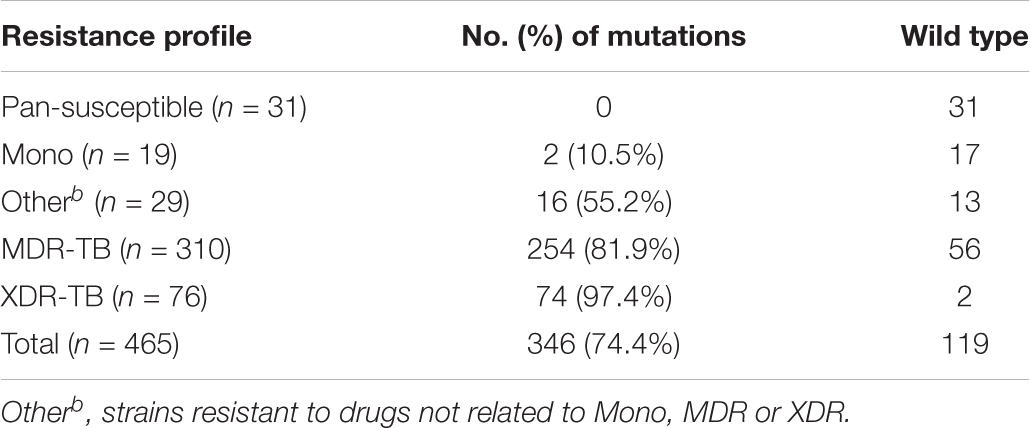
The pncA gene of 465 clinical isolates was sequenced. Of these, 346 (74.4%) isolates harbored at least one mutation compared with the wild-type reference sequence (Table 3). For the 31 isolates pan susceptible to MDR- and XDR-related drugs, no mutations in the pncA coding zone and putative upstream regulatory region were identified. Among the 19 mono (resistant to one of the MDR/XDR related drugs), 29 Other (resistant to drugs not related to MDR/XDR), 310 MDR and 76 XDR TB isolates, 2 (10.5%), 16 (55.2%), 254 (81.9%), and 74 (97.4%) of them harbored mutations in the pncA gene (including its upstream region), respectively (Table 3). Of the 124 types of mutations identified, three mutations at positions −12 and −11 upstream of pncA were identified in 11 isolates (Supplementary Table S1). Except for those three mutations, the remaining 121 mutations were distributed within the pncA open reading frame. Among the mutations observed in this study, 94 have previously been reported, 30 have not (Supplementary Tables S1, S4). We only found one synonymous mutation in the 124 mutational types and the rest of them were non-synonymous mutations (123 of 124). The majority of these mutations were nucleotide substitution (100 of 124) which caused amino acid substitution, 18 types with insertions, and six types with deletions (Supplementary Table S2). Seven types of mutations causing protein translation stop at amino acid positions 10, 41, 68, 103, 119, 122, and 141 were identified in 11 isolates (Supplementary Tables S1, S4).
The most frequent change observed in this study was 407_408 insA, which was found in 51 isolates (Supplementary Tables S1, S4). Meanwhile, 33 isolates contained a point mutation at position 395 and 40 isolates had mutation at position 226. We also observed four isolates had double mutations, 193T > C with 359T > C (1 isolate), 176C > A with 254T > C (1 isolate), 257A > G with 538G > T (1 isolate), and 310A > G with 388G > A (1 isolate). No M. bovis BCG-specific mutation 169C > G was observed in this study. The detailed information of all mutations is listed in Supplementary Table S1.
Correlation of Previously Reported pncA Mutations to PZA Susceptibility and PZase Activity
Due to technical difficulties, PZA MIC tests are not available in most of the clinical hospitals. Therefore, the MICs and PZase activities of isolates with previously reported mutations were collected and are listed in Supplementary Table S4 based on previous publications. Among the 465 samples, 310 contained mutations previously reported. Resistance profile to PZA and PZase activities (negative or positive) were summarized in Table 4. 294 isolates were predicted to be resistant to PZA and 273 were predicted to be PZase negative. There were 211 (68.1%) and 67 (88.2%) isolates predicted to be resistant to PZA in MDR and XDR strains, respectively. Among the 94 types of mutations previously reported, T > G at position 40 and C > T at position 176 were consistently reported susceptible to PZA with the MIC < 100 μg/ml (Stoffels et al., 2012; Xia et al., 2015); A > G at position 35 and 139 and A > C at positions 143 and 403 had literatures showing inconsistent MIC results at a PZA concentration of 100 μg/ml (Morlock et al., 2000; Somoskovi et al., 2007; Juréen et al., 2008; Mphahlele et al., 2008; Stoffels et al., 2012; Xia et al., 2015; Ramirez-Busby et al., 2017; Sheen et al., 2017). Mutation T > C at position 347 and insertion of G between 389 and 390 had inconsistent PZase results in former studies (Aono et al., 2014; Ramirez-Busby and Valafar, 2015). The rest of the mutations showed consistent results related to PZA resistance (Supplementary Table S1). Effects of 30 of the 310 isolates with previously reported mutations on PZase activities had not been determined (Table 4).
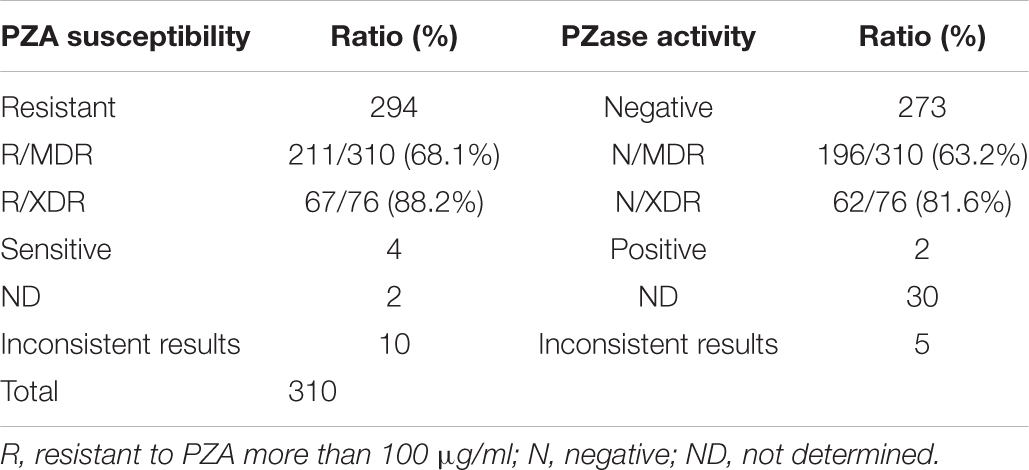
Table 4. Prediction of PZA susceptibilities and PZase activities in 310 isolates by previously reported literatures and their distribution ratios in MDR and XDR TB isolates.
Correlation of Novel pncA Mutations to PZase Activity
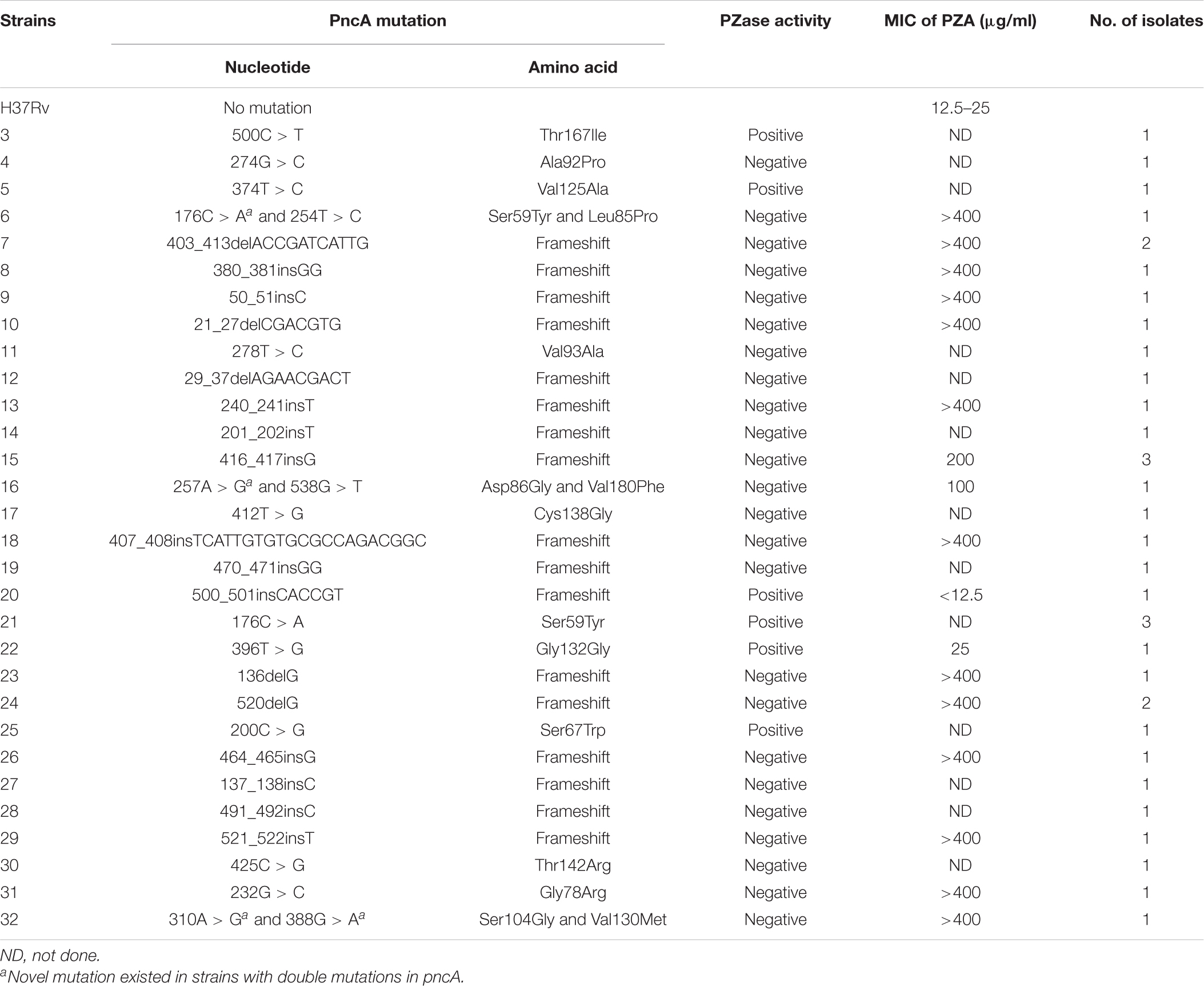
As shown in Figure 1, among the 30 novel mutations, 24 were PZase activity negative and six were positive. Mutations at positions 500C > T, 374T > C, 500_501insCACCGT, 176C > A, 396T > G, and 200C > G maintained their activities. Mutation 396T > G had the highest PZase activity which was even more than that of the positive control. Activity at mutation position 500_501insCACCGT was slightly higher than that of the negative control. All the new mutations without enzyme activity had the values close to that of the negative control. Detailed values of enzyme activity are listed in “Supplementary Dataset S1.”
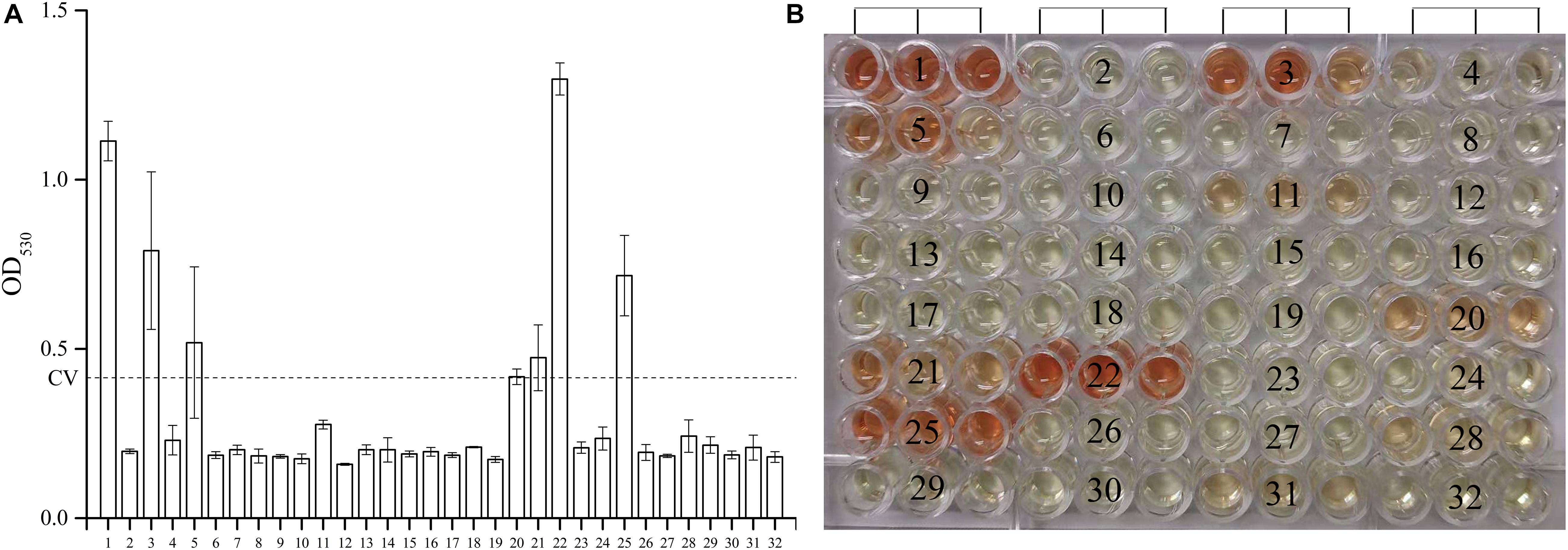
Figure 1. Effects of the 30 new mutations on the enzymatic activity of Mycobacterium tuberculosis PncA. (A) Measurements of OD530 of different wells containing different recombinant E. coli W3110ΔpncA strains. (B) Original color of different wells in 96-well plate. (1) Escherichia coli W3110ΔpncA pCA24N-pncA (positive control); (2) E.coli W3110ΔpncA pCA24N (negative control); (3) 500C > T (positive); (4) 274G > C (negative); (5) 374T > C (positive); (6) 176C > A and 254T > C (negative); (7) 403_413delACCGATCATTG (negative); (8) 380_381insGG (negative); (9) 50_51insC (negative); (10) 21_27delCGACGTG (negative); (11) 278T > C (negative); (12) 29_37delAGAACGACT (negative); (13) 240_241insT (negative); (14) 201_202insT (negative); (15) 416_417insG (negative); (16) 257A > G and 538G > T (negative); (17) 412T > G (negative); (18) 407_408insTCATTGTGTGCGCCAGACGGC (negative); (19) 470_471insGG (negative); (20) 500_501insCACCGT (positive); (21) 176C > A (positive); (22) 396T > G (positive); (23) 136delG (negative); (24) 520delG (negative); (25) 200C > G (positive); (26) 464_465insG (negative); (27) 137_138insC (negative); (28) 491_492insC (negative); (29) 521_522insT (negative); (30) 425C > G (negative); (31) 232G > C (negative); (32) 310A > G and 388G > A (negative). Every result was shown by test using three independent colonies (mean ± SD); OD, optical density; CV, cutoff value (dash line).
Correlation of PZase Activity to PZA Susceptibility
Seventeen isolates (15 PZase negative and 2 positive) with novel pncA mutations were subjected to PZA drug susceptibility test by using H37Rv as control. The PZA MIC of H37Rv was determined to be 12.5–25 μg/ml. The isolates with PZase-negative mutations 176C > A and 254T > C, 403_413delACCGATCATTG, 380_381insGG, 50_51insC, 21_27delCGACGTG, 240_241insT, 416_417insG, 257A > G and 538G > T, 407_408insTCATTGTGTGCGCCAGACGGC, 136delG, 520delG, 464_465insG, 521_522insT, 232G > C, and 310A > G and 388G > A, all showed PZA resistance phenotype. On the contrary, the two isolates with PZase-positive mutations 500_501insCACCGT and 396T > G were all shown to be PZA sensitive. Thus, the PZA DST results of all 17 isolates tested were shown to be well consistent with the PZase activity test using our E. coli gene knockout strain (Table 5).
Discussion
Because of the vital role for the treatment of non-replicative M. tuberculosis and shortening the time course of chemotherapy, PZA is contained in traditional treatment for drug-sensitive TB and in modern therapy to treat MDR and XDR TB. Recently, several clinical studies have been carried out for treatment of drug-sensitive and MDR TB with the combination of PZA, PA824, moxifloxacin (Dawson and Diacon, 2013), and bedaquiline (Diacon et al., 2012). Although PZA has been used for decades, it is shocking that the precise mechanism of PZA action is still obscure. One reason is that the action of PZA relies on acidic environment which results in not so well growing of the bacteria. This acidic condition also leads to unreliable results of PZA susceptibility tests. The inaccurate susceptibility results of PZA complicated the research of its mechanism of action and caused improper treatment of TB and spread of PZA resistance. Presently, bacteria culture-based PZA susceptibility tests are usually not performed in most of the hospitals worldwide. Therefore, alternative methods are required. Given that PZA resistance is frequently related to pncA mutations (Hirano et al., 1997) and PZase activity (Aono et al., 2014), it makes direct sequencing of pncA gene (including its upstream region) and PZase tests good choices. However, not all pncA mutations can cause PZA resistance, which weakens the potential of extensive utilization of direct gene sequencing. On the other hand, all PZase methods based on Wayne’s work require large amount of bacterial cultures and thus have to be performed in BSL-3 labs. Therefore, an in vitro PZase analysis following pncA amplification will be very attractive. Especially, with a continuously updated database containing the full spectrum of pncA mutations and the corresponding pyrazinamidase activities, PZA susceptibility tests may not be that difficult any more.
In order to investigate the prevalence of PZA resistance in M. tuberculosis isolated from Chongqing district, a total of 465 clinical isolates (including 31 pan-susceptible, 19 mono-resistant, 310 MDR, 76 XDR, and 29 other strains) were collected. Subsequently, the pncA gene was amplified, sequenced, and analyzed. The results showed that pncA mutations were identified in 346 isolates. We found that pncA polymorphisms were presented in 254 (81.9%) MDR and 74 (97.4%) XDR isolates (Table 3). The proportions of pncA mutational strains were higher than those from South Africa (70% in MDR, 96% in XDR), Georgia (72% in MDR, 90% in XDR) (Allana et al., 2017), Kazakhstan (75% in MDR) (Akhmetova et al., 2015), and Ukraine (78% in MDR, 88% in XDR) (Daum et al., 2019). We also found that predicted PZA-resistant isolates based on previous literatures in Chongqing district accounts for 68.1% in MDR isolates (Table 4), which was higher than those from Beijing (57.7% in MDR) (Gu et al., 2016), Zhejiang (43.1% in MDR) (Xia et al., 2015), and Taiwan (42.9% in MDR) (Huang et al., 2003). We ascribe this to samples coming from a specific geographic area. Resistance to first-line drugs increased PZA usage. Most of the patients received empirical PZA treatment without the susceptibility tests and increased the possibility of PZA resistance. Otherwise, samples may belong to retreat cases. Former study illustrated that new cases had lower PZA resistance possibility compared with retreat cases (Gu et al., 2016). Additionally, some predictive resistant isolates were borderline resistant at the concentration of 100 μg/ml. The existence of low-level resistance may indicate other mechanism of resistance to PZA, such as an efflux pump involved in the uptake of PZA. Low-level resistance to PZA simultaneously harbored pncA mutations. However, whether the resistance was caused by the mutations is uncertain. Considering the high correlation of PZA resistance and MDR, routine assessment of PZA susceptibility would be necessary.
Among the 346 isolates containing pncA mutations in this study, 294 isolates were predicted to be PZA resistant according to previous publications (Supplementary Table S4).
Except for those mutations previously reported, 30 new mutations were identified in this study (Table 5). Most of them were deletion or insertion of various lengths of nucleotides. The new mutations scattered between nucleotide position 21–521 (Table 5). Some of the substitutional new mutations, 176C > A, 232G > C, 412T > G, and 425C > G occurred at previously reported nucleotide positions. Mutation 176C > T have been shown to be sensitive to PZA at the concentration of 100 μg/ml (Xia et al., 2015). Our results revealed that the PZase activity of the new mutation 176C > A is positive (Figure 1), suggesting the corresponding clinical isolate is sensitive to PZA. Mutations 232G > T (Somoskovi et al., 2007), 232G > A (Ando et al., 2010), 412T > C (Miyagi et al., 2004), and 425C > T (Scorpio et al., 1997) had been shown to cause PZA resistance. Our results also showed that new mutations 232G > C, 412T > G, and 425C > G also led to loss of PZase activity, suggesting the corresponding clinical isolates are resistant to PZA.
Many other new mutations occurred at previously reported different nucleotides but same amino acid positions. We found that strains with mutations in 50_51insC (Gly17), 136delG (Ala46), 201_202insT (Ser67-Trp68), 240_241insT (Asp80-Phe81), 278T > C (Val93Ala), 310A > G (Ser104Gly), 380_381insGG (Glu127), 388G > A (Val130Met), 464_465insG (Val155), 470_471insGG (Val157), 491_492insC (Ser164), 520delG (Glu174), and 521_522insT (Glu174) were all shown to be PZase negative. Previously, strains with mutations at these amino acid positions, Gly17Asp (Daum et al., 2019), Ala46Val (Cheng et al., 2000), Ser67Pro (Hameed et al., 2020), Trp68Arg (Khan et al., 2019), Asp80Asn (Bwalya et al., 2018), Phe81Val (Wu et al., 2019), Val93Met (Daum et al., 2019), Ser104Ile (Hameed et al., 2020), Glu127Lys (Doustdar et al., 2009), Val130Gly (Sengstake et al., 2017), Val155Ala (Daum et al., 2019), Val157Gly (Khan et al., 2019), Ser164Pro (Hameed et al., 2020), and Glu174Gly (Tan et al., 2014), were reported to be resistant to PZA. PZase-positive mutations, 200C > G (Ser67Trp), 374 T > C (Val125Ala), 396T > G (Gly132Gly), 500C > T (Thr167Ile), and 500_501insCACCGT (Thr167) were identified in five isolates (one isolate for each mutation). Among them, 396T > G (Gly132Gly) is a synonymous mutation. Previous studies also showed the presence of mutations (Ser67Pro and Val125) in PZA susceptible isolates (Allix-Béguec et al., 2018).
Among the 30 isolates with novel pncA mutations, 17 of them were successfully recovered and then subjected to PZA DST, and the data showed that the results of PZA DST test were completely consistent to those of the PZase activity tests using the E. coli ΔpncA strain (Figure 1 and Table 5).
We also tried to predict PZA susceptibilities of isolates with novel single mutations using the SUSPECT-PZA webserver3 (Karmakar and Rodrigues, 2020). The prediction results (Supplementary Table S3) were in well consistency with our results, except for mutation 274G > C (sample 4 in Figure 1, Ala92Pro), mutation 278T > C (sample 11, Val93Ala), and mutation 374T > C (sample 5, Val125Ala). From Figure 1, Supplementary Data Sheet S1, and Table 5, we could see that the PZase activity of the mutant 374T > C (Val125Ala) was comparable with that of the 500_501insCACCGT mutant. The PZase activity of 500_501insCACCGT mutant was determined to be positive, and the strain containing the 500_501insCACCGT mutation (K2501) was determined to be sensitive to PZA. However, though the PZase activity of the 278T > C mutant was determined to be negative according to our standard, we did observe residual PZase activity of the mutant (Figure 1 and Supplementary Dataset S1). Unfortunately, the strain with this specific mutation could not be recovered from the refrigerator. We speculated that mutation 278T > C might cause borderline PZA resistance, which needs to be handled with extra caution.
Based on the crystal structure of M. tuberculosis PncA, it has been reported that three major regions (amino acids position 3–17, 61–85, and 132–142) are most commonly associated with PZase activity changes (Mehmood et al., 2019), and other regions (amino acids position 15–20, 50–70, 85–100, 130–140, and 160–180) are related to PncA flexibility (Khan and Malik, 2019). New mutations that caused PZase-negative phenotype in this study, mutations 21_27delCGACGTG (Val7-Val9) and 50_51insC (17Gly), are located in amino acid regions 3 to 17. Mutations 201_202insT (Ser67-Trp68) and 240_241insT (Asp80-Phe81) are located in amino acid regions 61 to 85. Mutations 257A > G (Asp86Gly), 274G > C (Ala92Pro), and 278T > C (Val93Ala) are located in amino acid regions 85–100. Mutation 388G > A (Val130Met) is located in amino acid regions 130–140. Mutations 491_492insC (Ser164), 520delG (Glu174), and 521_522insT (Glu174) are located in regions 160–180. So, it is not surprising to see that those mutations lead to the loss of the PZase activity.
Although researches on the mechanisms of PZA action and resistance are still underway, great improvement have been made. Except for pncA, more genes have been found to be related with PZA resistance, such as rpsA, panD, clpC1, rv1411c, and rv0521 (Shi et al., 2018). However, mutations in pncA still remain the leading cause of PZA resistance. According to previous publications, PZase activity and PZA susceptibility are very closely related. Conventional culture-based methods for PZA susceptibility are often unreliable and time consuming. Therefore, indirect method such as pncA gene sequencing and PZase tests would be better choices. To achieve this, a comprehensive database of pncA mutations with PZase activities is required. In this study, we not only discovered 30 new pncA mutations in M. tuberculosis clinical isolates but also developed a new method for PZAse activity test, which will accelerate the utilization of indirect methods for PZA susceptibility tests.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
This study used archived strains isolated from clinical samples, no human subjects and no identifiable human data were included. The institutional Ethics Committee of Chongqing Public Health Medical Center approved this study (Permit Number: 2017-CQGW-001-KY) and the exemption from written informed consent had been acquired.
Author Contributions
JD and YC conceived the idea of study and designed the methodology. ZY and ML prepared all the isolates. KL and JG carried out the experiments. KL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Science and Technology Major Project of China during the 13th Five-year plan period (2018ZX10302104), the Natural Science Foundation of Chongqing (cstc2018jcyjAX0428), and the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB29020000).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Miss Jinyue Li (Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, People’s Republic of China) for the technical support of PZase activity tests.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.594171/full#supplementary-material
Footnotes
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://varnomen.hgvs.org/
- ^ http://biosig.unimelb.edu.au/suspect_pza/
References
Akhmetova, A., Kozhamkulov, U., Bismilda, V., Chingissova, L., Abildaev, T., Dymova, M., et al. (2015). Mutations in the pncA and rpsA genes among 77 Mycobacterium tuberculosis isolates in Kazakhstan. Int. J. Tuberc. Lung. Dis. 19, 179–184. doi: 10.5588/ijtld.14.0305
Allana, S., Shashkina, E., Mathema, B., Bablishvili, N., Tukvadze, N., Shah, N. S., et al. (2017). pncA gene mutations associated with pyrazinamide resistance in drug-resistant tuberculosis, South Africa and Georgia. Emerg. Infect. Dis. 23, 491–495. doi: 10.3201/eid2303.161034
Allix-Béguec, C., Arandjelovic, I., Bi, L., Beckert, P., Bonnet, M., Bradley, P., et al. (2018). Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N. Engl. J. Med. 379, 1403–1415. doi: 10.1056/NEJMoa1800474
Ando, H., Mitarai, S., Kondo, Y., Suetake, T., Sekiguchi, J. I., Kato, S., et al. (2010). Pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis isolates in Japan. Clin. Microbiol. Infect. 16, 1164–1168. doi: 10.1111/j.1469-0691.2009.03078.x
Aono, A., Chikamatsu, K., Yamada, H., Igarashi, Y., Murase, Y., Takaki, A., et al. (2018). A simplified pyrazinamidase test for pyrazinamide drug susceptibility in Mycobacterium tuberculosis. J. Microbiol. Methods 154, 52–54. doi: 10.1016/j.mimet.2018.09.018
Aono, A., Chikamatsu, K., Yamada, H., Kato, T., and Mitarai, S. (2014). Association between pncA gene mutations, pyrazinamidase activity, and pyrazinamide susceptibility testing in Mycobacterium tuberculosis. Antimicrob, Agents Chemother. 58, 4928–4930. doi: 10.1128/aac.02394-14
Boshoff, H. I., Mizrahi, V., and Barry, C. E. III (2002). Effects of pyrazinamide on fatty acid synthesis by whole mycobacterial cells and purified fatty acid synthase I. J. Bacteriol. 184, 2167–2172.
Bwalya, P., Yamaguchi, T., Mulundu, G., Nakajima, C., Mbulo, G., Solo, E. S., et al. (2018). Genotypic characterization of pyrazinamide resistance in Mycobacterium tuberculosis isolated from Lusaka, Zambia. Tuberculosis 109, 117–122. doi: 10.1016/j.tube.2017.12.007
Chedore, P., Bertucci, L., Wolfe, J., Sharma, M., and Jamieson, F. (2010). Potential for erroneous results indicating resistance when using the bactec MGIT 960 system for testing susceptibility of Mycobacterium tuberculosis to Pyrazinamide. J. Clin. Microbiol. 48, 300–301.
Cheng, S. J., Thibert, L., Sanchez, T., Heifets, L., and Zhang, Y. (2000). pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob. Agents Chemother. 44, 528–532.
Daum, L. T., Konstantynovska, O. S., Solodiankin, O. S., Poteiko, P. I., Bolotin, V. I., Rodriguez, J. D., et al. (2019). Characterization of novel Mycobacterium tuberculosis pncA gene mutations in clinical isolates from the Ukraine. Diagn. Microbiol. Infect. Dis. 93, 334–338. doi: 10.1016/j.diagmicrobio.2018.10.018
Dawson, R., and Diacon, A. (2013). PA-824, moxifloxacin and pyrazinamide combination therapy for tuberculosis. Expert Opin. Investig. Drugs 22, 927–932. doi: 10.1517/13543784.2013.801958
Diacon, A. H., Dawson, R., von Groote-Bidlingmaier, F., Symons, G., Venter, A., Donald, P. R., et al. (2012). 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet 380, 986–993. doi: 10.1016/s0140-6736(12)61080-0
Dillon, N. A., Peterson, N. D., Feaga, H. A., and Keiler, K. C. (2017). Anti-tubercular activity of pyrazinamide is independent of trans-translation and RpsA. Sci. Rep. 7:6135. doi: 10.1038/s41598-017-06415-5
Dillon, N. A., Peterson, N. D., Rosen, B. C., and Baughn, A. D. (2014). Pantothenate and pantetheine antagonize the antitubercular activity of pyrazinamide. Antimicrob. Agents Chemother. 58, 7258–7263. doi: 10.1128/AAC.04028-14
Doustdar, F., Khosravi, A. D., and Farnia, P. (2009). Mycobacterium tuberculosis genotypic diversity in pyrazinamide-resistant isolates of Iran. Microb. Drug Resist. 15, 251–256. doi: 10.1089/mdr.2009.0066
Gu, Y., Yu, X., Jiang, G., Wang, X., Ma, Y., Li, Y., et al. (2016). Pyrazinamide resistance among multidrug-resistant tuberculosis clinical isolates in a national referral center of China and its correlations with pncA, rpsA, and panD gene mutations. Diagn. Microbiol. Infect. Dis. 84, 207–211. doi: 10.1016/j.diagmicrobio.2015.10.017
Hameed, H. A., Tan, Y., Islam, M. M., Lu, Z., Chhotaray, C., and Wang, S. (2020). Detection of novel gene mutations associated with pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis clinical Isolates in Southern China. Infect. Drug Resist. 13, 217–227. doi: 10.2147/idr.s230774
Hirano, K., Takahashi, M., Kazumi, Y., Fukasawa, Y., and Abe, C. (1997). Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber Lung. Dis. 78, 117–122.
Huang, T. S., Lee, S. S., Tu, H. Z., Huang, W. K., Chen, Y. S., Huang, C. K., et al. (2003). Correlation between pyrazinamide activity and pncA mutations in Mycobacterium tuberculosis isolates in Taiwan. Antimicrob. Agents Chemother. 47, 3672–3673. doi: 10.1128/aac.47.11.3672-3673.2003
Juréen, P., Werngren, J., Toro, J. C., and Hoffner, S. (2008). Pyrazinamide resistance and pncA gene mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother, 52, 1852–1854. doi: 10.1128/aac.00110-08
Karmakar, M., and Rodrigues, C. H. M. (2020). Structure guided prediction of Pyrazinamide resistance mutations in pncA. Sci. Rep. 10:1875. doi: 10.1038/s41598-020-58635-x
Khan, M. T., and Malik, S. I. (2019). Structural dynamics behind variants in pyrazinamidase and pyrazinamide resistance. J. Biomol. Struct. Dyn. 38, 3003–3017. doi: 10.1080/07391102.2019.1650113
Khan, M. T., Malik, S. I., Ali, S., Masood, N., Nadeem, T., Khan, A. S., et al. (2019). Pyrazinamide resistance and mutations in pncA among isolates of Mycobacterium tuberculosis from Khyber Pakhtunkhwa. Pakistan. BMC Infect. Dis. 19:116. doi: 10.1186/s12879-019-3764-2
Kim, S. H., and Shin, J. H. (2018). Identification of nontuberculous mycobacteria using multilocous sequence analysis of 16S rRNA, hsp65, and rpoB. J. Clin. Lab Anal. 32:e22184. doi: 10.1002/jcla.22184
Kruuner, A., Yates, M. D., and Drobniewski, F. A. (2006). Evaluation of MGIT 960-based antimicrobial testing and determination of critical concentrations of first- and second-line antimicrobial drugs with drug-resistant clinical strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 44, 811–818. doi: 10.1128/jcm.44.3.811-818.2006
Li, K., Li, T., Yang, S. S., Wang, X. D., Gao, L. X., Wang, R. Q., et al. (2017). Deletion of nudB causes increased susceptibility to antifolates in Escherichia coli and Salmonella enterica. Antimicrob. Agents Chemother. 61:AAC.02378-16. doi: 10.1128/AAC.02378-16
Mehmood, A., Khan, M. T., Kaushik, A. C., Khan, A. S., Irfan, M., and Wei, D. Q. (2019). Structural dynamics behind clinical mutants of PncA-Asp12Ala, Pro54Leu, and His57Pro of Mycobacterium tuberculosis associated with pyrazinamide resistance. Front. Bioeng. Biotechnol. 7:404. doi: 10.3389/fbioe.2019.00404
Miotto, P., Cabibbe, A. M., Feuerriegel, S., Casali, N., Drobniewski, F., Rodionova, Y., et al. (2014). Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. mBio 5:e01819-14. doi: 10.1128/mBio.01819-14
Mitchison, D. A. (1985). The action of antituberculosis drugs in short-course chemotherapy. Tubercle 66, 219–225. doi: 10.1016/0041-3879(85)90040-6
Miyagi, C., Yamane, N., Yogesh, B., Ano, H., and Takashima, T. (2004). Genetic and phenotypic characterization of pyrazinamide-resistant Mycobacterium tuberculosis complex isolates in Japan. Diagn. Microbiol. Infect. Dis. 48, 111–116. doi: 10.1016/j.diagmicrobio.2003.09.013
Morlock, G. P., Crawford, J. T., Butler, W. R., Brim, S. E., Sikes, D., Mazurek, G. H., et al. (2000). Phenotypic characterization of pncA mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44, 2291–2295. doi: 10.1128/aac.44.9.2291-2295.2000
Mphahlele, M., Syre, H., Valvatne, H., Stavrum, R., Mannsåker, T., Muthivhi, T., et al. (2008). Pyrazinamide resistance among South African multidrug-resistant Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 46, 3459–3464. doi: 10.1128/jcm.00973-08
Njire, M., Wang, N., Wang, B., Tan, Y., Cai, X., Liu, Y., et al. (2017). Pyrazinoic acid inhibits a bifunctional enzyme in Mycobacterium tuberculosis. Pathogens 61:e00070-17. doi: 10.1128/aac.00070-17
Palomino, J. C., and Martin, A. (2014). Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics 3, 317–340. doi: 10.3390/antibiotics3030317
Personne, Y., and Parish, T. (2014). Mycobacterium tuberculosis possesses an unusual tmRNA rescue system. Tuberculosis 94, 34–42. doi: 10.1016/j.tube.2013.09.007
Piersimoni, C., Mustazzolu, A., Giannoni, F., Bornigia, S., Gherardi, G., and Fattorini, L. (2013). Prevention of false resistance results obtained in testing the susceptibility of Mycobacterium tuberculosis to pyrazinamide with the Bactec MGIT 960 system using a reduced inoculum. J. Clin. Microbiol. 51, 291–294. doi: 10.1128/jcm.01838-12
Ramirez-Busby, S. M., Rodwell, T. C., Fink, L., and Catanzaro, D. (2017). A multinational analysis of mutations and heterogeneity in PZase, RpsA, and PanD associated with pyrazinamide resistance in M/XDR Mycobacterium tuberculosis. Sci. Rep. 7:3790. doi: 10.1038/s41598-017-03452-y
Ramirez-Busby, S. M., and Valafar, F. (2015). Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 59, 5267–5277. doi: 10.1128/aac.00204-15
Scorpio, A., Lindholm-Levy, P., Heifets, L., Gilman, R., Siddiqi, S., Cynamon, M., et al. (1997). Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41, 540–543.
Sengstake, S., Bergval, I. L., Schuitema, A. R., de Beer, J. L., Phelan, J., de Zwaan, R., et al. (2017). Pyrazinamide resistance-conferring mutations in pncA and the transmission of multidrug resistant TB in Georgia. BMC Infect. Dis. 17:491. doi: 10.1186/s12879-017-2594-3
Sheen, P., Requena, D., Gushiken, E., Gilman, R. H., Antiparra, R., Lucero, B., et al. (2017). A multiple genome analysis of Mycobacterium tuberculosis reveals specific novel genes and mutations associated with pyrazinamide resistance. BMC Genomics 18:769. doi: 10.1186/s12864-017-4146-z
Shi, W., Chen, J., Zhang, S., Zhang, W., and Zhang, Y. (2018). Identification of Novel Mutations in LprG (rv1411c), rv0521, rv3630, rv0010c, ppsC, and cyp128 Associated with Pyrazinoic Acid/Pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 62:e00430-18. doi: 10.1128/aac.00430-18
Shi, W., Zhang, X., Jiang, X., Yuan, H., Lee, J. S., and Barry, C. E. III, et al. (2011). Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333, 1630–1632. doi: 10.1126/science.1208813
Somoskovi, A., Dormandy, J., Parsons, L. M., Kaswa, M., Goh, K. S., Rastogi, N., et al. (2007). Sequencing of the pncA gene in members of the Mycobacterium tuberculosis complex has important diagnostic applications: identification of a species-specific pncA mutation in “Mycobacterium canettii” and the reliable and rapid predictor of pyrazinamide resistance. J. Clin. Microbiol. 45, 595–599. doi: 10.1128/jcm.01454-06
Stoffels, K., Mathys, V., Fauville-Dufaux, M., Wintjens, R., and Bifani, P. (2012). Systematic analysis of pyrazinamide-resistant spontaneous mutants and clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 5186–5193. doi: 10.1128/aac.05385-11
Tan, Y., Hu, Z., Zhang, T., Cai, X., Kuang, H., Liu, Y., et al. (2014). Role of pncA and rpsA gene sequencing in detection of pyrazinamide resistance in Mycobacterium tuberculosis isolates from southern China. J. Clin. Microbiol. 52, 291–297. doi: 10.1128/jcm.01903-13
Wayne, L. G. (1974). Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am. Rev. Respir. Dis. 109, 147–151. doi: 10.1164/arrd.1974.109.1.147
Whitfield, M. G., Soeters, H. M., Warren, R. M., York, T., Sampson, S. L., Streicher, E. M., et al. (2015). A global perspective on pyrazinamide resistance: systematic review and meta-analysis. PLoS One 10:e0133869. doi: 10.1371/journal.pone.0133869
Wu, X., Lu, W., Shao, Y., Song, H., Li, G., Li, Y., et al. (2019). pncA gene mutations in reporting pyrazinamide resistance among the MDR-TB suspects. Infect. Genet. Evol. 72, 147–150. doi: 10.1016/j.meegid.2018.11.012
Xia, Q., Zhao, L. L., Li, F., Fan, Y. M., Chen, Y. Y., Wu, B. B., et al. (2015). Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Zhejiang. China. Antimicrob. Agents Chemother. 59, 1690–1695. doi: 10.1128/aac.04541-14
Yamashita, N., Deguchi, M., Hosotsubo, H., and Asari, S. (1995). [Clinical usefulness of microparticle enzyme immunoassay for HBeAg and HBeAb in chronic hepatitis B patients]. Kansenshogaku Zasshi 69, 1323–1328. doi: 10.11150/kansenshogakuzasshi1970.69.1323
Zhang, S., Chen, J., Shi, W., Cui, P., Zhang, J., Cho, S., et al. (2017). Mutation in clpC1 encoding an ATP-dependent ATPase involved in protein degradation is associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg. Microbes Infect. 6:e8. doi: 10.1038/emi.2017.1
Zhang, S., Chen, J., Shi, W., Liu, W., Zhang, W., and Zhang, Y. (2013). Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg. Microbes Infect. 2:e34. doi: 10.1038/emi.2013.38
Zhang, Y., and Mitchison, D. (2003). The curious characteristics of pyrazinamide: a review. Int. J. Tuberc. Lung. Dis. 7, 6–21.
Zhang, Y., Permar, S., and Sun, Z. (2002). Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J. Med. Microbiol. 51, 42–49.
Zhang, Y., Scorpio, A., Nikaido, H., and Sun, Z. (1999). Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Bacteriol. 181, 2044–2049.
Keywords: tuberculosis, pyrazinamide, pncA mutations, PZase activity, Escherichia coli
Citation: Li K, Yang Z, Gu J, Luo M, Deng J and Chen Y (2021) Characterization of pncA Mutations and Prediction of PZA Resistance in Mycobacterium tuberculosis Clinical Isolates From Chongqing, China. Front. Microbiol. 11:594171. doi: 10.3389/fmicb.2020.594171
Received: 12 August 2020; Accepted: 26 November 2020;
Published: 11 January 2021.
Edited by:
Wladimir Sougakoff, INSERM U1135 Centre d’Immunologie et de Maladies Infectieuses, FranceReviewed by:
Nadine Christine Lemaitre, University Hospital Center (CHU) of Amiens, FranceStephanie Petrella, Université de Paris, France
Copyright © 2021 Li, Yang, Gu, Luo, Deng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiaoyu Deng, ZGVuZ2p5QHdoLmlvdi5jbg==; Yaokai Chen, eWFva2FpY2hlbkBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Kun Li
Kun Li Zhongping Yang2†
Zhongping Yang2† Jiaoyu Deng
Jiaoyu Deng Yaokai Chen
Yaokai Chen