- 1Tuberculosis Research Section, Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, United States
- 2Drug Discovery Unit, College of Life Sciences, James Black Centre, University of Dundee, Dundee, United Kingdom
- 3Inflammation and Innate Immunity Unit, Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, United States
Mycobacterium tuberculosis resides in the lungs in various lesion types with unique microenvironmental conditions. This diversity is in line with heterogeneous disease progression and divergent drug efficiency. Fluorescent reporter strains can be used to decipher the micromilieu and to guide future treatment regimens. Current reporters using replicating plasmids, however, are not suitable for long-term mouse infections or studies in non-human primates. Using a combination of recombinant DNA and protein optimization techniques, we have developed reporter strains based on integrative plasmids, which exhibit stimulus-response characteristics and fluorescence intensities comparable to those based on replicating plasmids. We successfully applied the concepts by constructing a multi-color reporter strain able to detect simultaneous changes in environmental pH, Mg2+ concentrations, and protein expression levels.
Introduction
Tuberculosis (TB) is the leading cause of death from a single infectious agent worldwide. In 2019 the World Health Organization (WHO) reported that global TB infections resulted in an estimated 1.4 million fatalities (Global Tuberculosis Report, 2020). Mycobacterium tuberculosis (Mtb) mainly infects the lungs, where it leads to very heterogeneous lesion types, that possess a wide range of pathological and immunological characteristics. Representative pathologies of pulmonary disease in humans include air-filled cavities and nodules with a wide range of interior composition, including necrotic, fibrotic, and caseous features, often found simultaneously in infected individuals (Lenaerts et al., 2015). The diverse locations where the bacilli reside are accompanied by unique microenvironmental conditions, such as hypoxic stress (Via et al., 2008; Belton et al., 2016; Prosser et al., 2017), nutrient limitation (Kim et al., 2010; Berney and Berney-Meyer, 2017), ion starvation or toxicity (Wagner et al., 2005; Hood and Skaar, 2012; Marcela Rodriguez and Neyrolles, 2014), and low pH (Kempker et al., 2017; Baker et al., 2019). Metabolic and physiologic adaptations of Mtb bacteria under these circumstances (Ehrt et al., 2018), as well as the lesion architecture itself (Prideaux et al., 2015; Strydom et al., 2019), strongly influence disease progression and drug efficiency. Given the intricacies of the assorted conditions present during infection and the connection to treatment outcome, it is important to develop tools to probe the local bacterial environment in vivo and use this information to guide future treatment regimens.
To date, several fluorescent mycobacterial reporter strains have been successfully applied in mice and have revealed novel insights about the local microenvironment and bacteria-host interactions (Abramovitch, 2018; MacGilvary and Tan, 2018). For instance, Abramovitch et al. (2011) and Tan et al. (2013) developed reporter strains that monitor pH, an environmental cue known to be important for Mtb in vitro growth (Supplementary Figure 1A) and intracellular survival (Vandal et al., 2009). While pH is well studied as a critical signal during host infections, the impact of environmental ions has only recently been studied leading to the development of chloride (Cl–) and potassium (K+)-responsive reporter strains (Tan et al., 2013; MacGilvary et al., 2019). For magnesium (Mg2+) ions, however, no tools are currently available to reliably quantify or read out its concentration. The importance of Mg2+ for mycobacterial growth has been demonstrated in in vitro culture using media with low Mg2+ concentration (Supplementary Figure 1B; Buchmeier et al., 2000; Piddington et al., 2000; Walters et al., 2006; Goodsmith et al., 2015) and inhibitors of Mg2+ homeostasis (Supplementary Figure 2A; Lopez Quezada et al., 2019; Park et al., 2019). In addition, there is accumulating evidence that survival of Mtb within the acidic pH of phagosomes is Mg2+ dependent (Buchmeier et al., 2000; Piddington et al., 2000). Mtb mutants lacking the persistence-associated integral membrane protein (PerM, rv0955) (Goodsmith et al., 2015) or the Mg2+ transporter C (MgtC, rv1811) (Buchmeier et al., 2000) showed Mg2+-dependent growth defects in vitro. In vivo, these gene deletions resulted in attenuation in the chronic phase of infection and loss of virulence, respectively (Buchmeier et al., 2000; Goodsmith et al., 2015). In this context, it has been hypothesized that Mg2+ restriction might be a mechanism to control Mtb infections. Thus, there is a need for new reporter strains for this divalent ion.
The drawback of current reporter strains is the use of replicating plasmids, which are not suitable for long term mouse infections or studies in non-human primates due to the loss of the reporter construct over time (our unpublished data; Abramovitch, 2018). Furthermore, relying on a single plasmid limits the number of responsive elements that can be introduced into one reporter strain, thereby restricting the ability to correlate different environmental factors and gain deeper insights into the complexity of the microenvironment.
To overcome these limitations, we have developed novel strategies to generate reporter strains using exclusively integrative plasmids, which exhibit stimulus-response characteristics and fluorescence intensities (FIs) comparable to those based on replicating plasmids. Screening a library of fluorescent proteins (FPs), designing new multicistronic gene constructs, testing various mycobacterial promoters, and exploiting the flexibility of mRNA-based responsive elements gave us the toolbox to accomplish this goal. We successfully applied these concepts by constructing a multi-color reporter strain able to detect simultaneous changes in environmental pH, Mg2+ concentrations, and protein expression levels.
Materials and Methods
Bacterial Strains, Media, and Culture Conditions
The bacterial strains used in this study are listed in Supplementary Table 1. Escherichia coli (E. coli) NEB5α strains were cultured in Luria-Bertani liquid media (LB; Sigma) or grown on LB agar (Invitrogen). For selection of recombinant E. coli, Carbenicillin (Car), Kanamycin (Kan), and Hygromycin (Hyg) were used at concentrations of 50, 50, and 200 μg/ml, respectively. Mtb bacteria were cultured in Difco Middlebrook 7H9 liquid media (BD) supplemented with ADGNTw (Supplementary Table 2) or grown on Difco Middlebrook 7H11 agar (BD) supplemented with OADGN (Supplementary Table 2). For selection of recombinant Mtb, Kan and Hyg were used at concentrations of 25 and 50 μg/ml, respectively. For acid and Mg2+ stress conditions Mtb bacteria were grown in Sauton’s medium (Supplementary Table 2) adjusted to different pH values and supplemented with desired magnesium sulfate (MgSO4) concentrations.
Formation of New Gene Constructs
Cloning vectors, suicide plasmids, and oligonucleotides used in this study are listed in Supplementary Tables 3, 4, respectively. Promoter and gene sequences are listed in Supplementary Tables 5, 6. The plasmids were constructed using oligo annealing, restriction enzyme-based cloning, and Gibson Assembly.
Plasmid Modifications
The backbone of plasmid pMV361 (Stover et al., 1991) was amplified using primers 1 and 2, digested with NcoI and reannealed, in order to introduce an NcoI restriction site at the translational start site and simplify future cloning steps. The L5 integrase was removed using primers 3 and 4 with terminal BamHI restriction sites. Furthermore, the hsp60 promoter (Stover et al., 1991) was exchanged with Psmyc from pML1357 (Huff et al., 2010) exploiting the XbaI and NcoI restriction sites. The new plasmid was named pL5 Psmyc (Supplementary Figure 3).
pML1357, an integrative plasmid targeted to the mycobacterial Giles integration site (Huff et al., 2010), was purchased from Addgene and modified by exchanging the multiple cloning site and smyc promoter with those from pL5 Psmyc using XbaI and HindIII restriction sites. The Giles integrase of pML1357 was removed by PCR amplification of the plasmid using primers 7 and 8 with XhoI restriction sites and reannealing to form pML1357dI. Primers 9 and 10 were used to introduce a second multiple cloning site into pML1357dI replacing the xylE gene encoding catechol 2,3 dioxygenase. The new plasmid was named pGiles Psmyc (Supplementary Figure 3).
In order to transfer the Giles integrase into a suicide vector, the gene for the integrase together with its promoter was amplified from pML1357 using primers 11 and 12 and inserted into the non-mycobacterial plasmid pUC19 (Norrander et al., 1983). The plasmid was named pUC19-GI.
Library of FP Genes
To enable quantification of protein expression levels via western blot, an N-terminal 6x-His tag was added to all FPs used in this study. A DNA sequence encoding the 6x-His tag was introduced downstream of the smyc promoter (Kaps et al., 2001; Ehrt et al., 2005) in the pL5 Psmyc plasmid via oligo annealing using oligomers 13 and 14. FP genes, purchased from Addgene (Supplementary Table 3), were amplified using the respective primers in Supplementary Table 4 and introduced by restriction enzyme-based cloning, usually NcoI and HindIII, downstream of the Psmyc-His element.
Multicistronic Constructs
The Addgene-sourced mWasabi gene contained an internal NcoI site. To enable NcoI-based cloning, the restriction site was removed by a single point mutation (C354 → G354) using Site-Directed Mutagenesis. The modified mWasabi dN gene was subsequently cloned downstream of the Psmyc-His element using NcoI and HindIII restriction sites. In addition, a synthetic, codon-optimized version of mWasabi was purchased (Eurofins) based on reported mycobacterial codon preferences.1 The COmWasabi synthetic gene was introduced into the NcoI and ClaI sites downstream of Psmyc-His with primer pair 41 and 42 containing the restriction sites BspHI and ClaI. NcoI and BspHI ligation led to destruction of the NcoI site. His-tagged COmWasabi together with the ribosome binding site (RBS) was then amplified using primers 43 and 42 and cloned into the restriction sites HindIII and ClaI downstream of mWasabi dN. The new gene construct was named double green and could be easily transferred using NcoI- and ClaI-based cloning.
In order to generate the double red construct, the mScarlet gene was codon optimized and cloned downstream of the Psmyc-His element using primers 21 and 44 with the restriction sites NcoI and HindIII. Subsequently, His-tagged COmScarlet together with its RBS was transferred into the HindIII restriction site at the 3′ end of the mScarlet gene using primer pair 45 and 44. For the triple red construct, an RBS and the His-tagged gene encoding mRuby3 was introduced downstream of the two mScarlet genes by exploiting the ClaI restriction site. Since the RBS-HismRuby3 DNA element was amplified with a forward primer containing an AclI restriction site, the ClaI site at the 5′ end was deleted while it remained intact at the 3′ end.
Promoter Library
For the promoter library, the smyc promoter upstream of eGFP was exchanged with the promoters Phsp60, PG13 (Barker et al., 1999), Pmsp12 (Chan et al., 2002), PMOP (George et al., 1995), and Pleft (Nesbit et al., 1995), respectively. Plasmids containing Phsp60, PG13, and Pmsp12 were purchased from Addgene. PMOP was obtained from the vector pMH29 (George et al., 1995) and a synthetic DNA sequence of Pleft, identical to the sequence described by Jain et al. (2012), but with an NcoI site at the 3′ end, was ordered from Eurofins. The promoters Phsp60, PG13, Pmsp12, and Pleft were introduced into the XbaI and NcoI restriction sites upstream of the eGFP gene. PMOP was inserted into the same restriction sites, but using primer pair 52 and 53 containing SpeI and NcoI sites.
The Pleft* element was constructed by Gibson Assembly ligating the PCR product of Pleft, amplified using primers 56 and 57, and the XbaI/BsaHI digested pL5 Psmyc-eGFP plasmid. We used the same oligo annealing strategy, described above, to introduce a 6x-His tag downstream of the Pleft* element.
Mono-Responsive Reporter Gene Constructs
pGiles was used as the backbone for constructing the pH-responsive reporter plasmid. The triple red gene construct, reported above, was amplified from the respective pL5 Psmyc-triple red plasmid and transferred into the second multiple cloning site of pGiles carrying the native promoter of rv2390c (Tan et al., 2013) in the restriction sites BmtI and BglII. To accomplish this insertion, we applied Gibson Assembly. The plasmid was digested with the enzymes BglII and NotI and the three genes were amplified together using primers 58 and 59. These primers contained ApaI and BglII intrinsic restriction sites, respectively, which led to a new BglII site downstream of the triple red element and an ApaI site at the 5′ end. Subsequently, using the restriction sites BmtI and ApaI the native promoter of rv2390c was exchanged with a 6x-His-tagged native promoter of the mycobacterial gene rv2395A (Abramovitch et al., 2011) amplified from Mtb H37Rv genomic DNA.
The Mg2+-responsive elements together with their native promoters upstream of the mycobacterial genes rv1535 and rv1806 (Pnp1535 and Pnp1806) were amplified from Mtb genomic DNA using primers 62–65. These DNA fragments were cloned into the first multiple cloning site of pGiles exploiting the restriction sites XbaI and NcoI. A 6x-His tag was inserted into the NcoI site by oligo annealing. mWasabi dN was introduced downstream between NcoI and HindIII sites, using primers 39 and 40.
In order to construct the Psmyc1806 and Pleft1806 fusion products, we applied Gibson Assembly. As vectors, pL5 containing either Psmyc-mWasabi dN or Pleft*-HismClover3 were used. The plasmid backbone was amplified with primers 66 and 67 or 70 and 71. The Mg2+-responsive element was amplified from Pnp1806-mWasabi dN with primers 68, 69, 72, and 73 and ligated with the respective plasmid part. Downstream of Psmyc1806 a 6x-His tag was introduced as described above. Subsequently, the fused elements were transferred into the XbaI and NcoI restriction sites upstream of mWasabi dN in the pGiles plasmid.
For the Mg2+ responsive double green constructs, the His-tagged gene encoding COmWasabi together with its RBS was introduced downstream of the mWasabi dN gene, as described above.
Tri-Responsive Reporter Gene Construct
To reduce the number of plasmids needed for the construction of the Mtb tri-responsive reporter strain, the DNA regions encoding Pnp2395A-triple red and Pleft1806-double green were combined in a single plasmid. The original plasmids containing these reporter constructs were digested with XbaI and ClaI and newly ligated, introducing Pleft1806-double green into the vector containing the pH-responsive element.
The double blue system under the control of the constitutive promoter Psmyc was designed as follows. The gene of mTagBFP2 was codon optimized and introduced into the NcoI and ClaI restriction sites, downstream of the Psmyc-His element in pL5 Psmyc-His, using primer pair 74 and 75 with BspHI and ClaI sites, respectively, thus, deleting the NcoI cloning site. The DNA region encoding His-tagged COmTagBFP2 together with the RBS was amplified using primers 43 and 75 and cloned downstream of Psmyc-HismTagBFP2 in the pL5 Psmyc-HismTagBFP2 plasmid.
Generation of Fluorescent Mtb Strains and Reporter Strains
Plasmids were incorporated into electrocompetent Mtb H37Rv (or HN878 for in vivo assay) cells via electroporation using a Bio-Rad GenePulser Xcell electroporator and 2 mm electroporation cuvettes (Bio-Rad). The conditions for electroporation were 2500 V, 25 μF, and 1000 Ω. All pL5-based plasmids were co-electroporated with a suicide plasmid, pBS-Int (Springer et al., 2001), which encodes for the L5 integrase. The suicide plasmid used for incorporation of pGiles plasmids was pUC19-GI. Transformants were selected on 7H11 agar (BD) containing Kan, Hyg, or both antibiotics.
Western Blot
Mtb H37Rv bacteria expressing the FP constructs were grown in 7H9 ADGNTw media (20 mL) to an OD600 of 0.6. Cultures were centrifuged (3000 × g, RT, 10 min) and resuspended in lysis buffer (Thermo Fisher Scientific; 600 μl) containing 1× protease inhibitor (Roche). Bacterial lysis was performed by bead-beating (Roche MagnaLyzer; 6500 rpm, 45 s, three times with cooling on ice in between) with 0.1 mm Zirconia/Silica beads (BioSpec Products; 400 μl). Lysates were centrifuged (21,130 × g, 4°C, 5 min) and protein concentrations of the supernatants determined by BCA assay (Pierce). Samples were then boiled (95°C, 5 min) with reducing loading SDS-PAGE sample buffer (Bio-Rad) and equivalent protein quantities of each sample loaded onto Any kD Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad; 220 V, 35 min) with a Color Prestained Protein Standard (Broad Range, 10–250 kDa, NEB). Proteins were transferred onto a nitrocellulose membrane (Invitrogen) using the Trans-Blot Turbo Transfer System (Bio-Rad; 25 V, 10 min). The membrane was blocked with BSA-TBST [5% BSA in TBST (1× TBS with 0.1% Tween 20)] for 1 h at RT, washed three times (15 min each) with TBST, cut in half, and incubated with the primary mouse antibodies anti-His6 (Sigma; 1:3000 dilution in BSA-TBST) or anti-GroEL (Abcam, ab20519; 1:2000 dilution in BSA-TBST) overnight at 4°C. After three washing steps for 15 min, each with TBST, the secondary goat anti-mouse horseradish peroxidase (HRP) antibody (Invitrogen; 1:10,000 dilution in BSA-TBST) was applied and the membrane was incubated for 1 h at RT. The blot was washed three times (15 min each) with TBST, developed using Clarity and Clarity Max ECL Western Blotting Substrate (Bio-Rad), and analyzed with the ChemiDoc Imaging System (Bio-Rad). Relative quantifications of protein expression levels were carried out using the Bio-Rad Image Lab Software.
In vitro Assays
Measuring Signal Intensities of Fluorescent Mtb Strains
The various H37Rv strains expressing the FP constructs were grown to mid-logarithmic phase in 7H9 ADGNTw media containing the required antibiotic. A total of 200 μl each were transferred into a clear-bottomed black 96 well plate (Costar) and the plate was placed in a CLARIOstar Plus microplate reader (BMG LABTECH). Fluorescence measurements were taken using the optimal excitation and emission wavelengths of each FP and values were normalized to the respective optical densities (OD600) of culture in each well.
Detection of FI Changes Based on Environmental Conditions
Reporter strains, grown in 7H9 ADGNTw medium, were adapted for one week to Sauton’s medium (pH 7.0, 500 μM MgSO4) starting with an initial OD600 of 0.01. Subsequently, 1 ml of culture (OD600: 0.8) was centrifuged (20,293 × g, RT, 2 min), the pellet washed once with Sauton’s medium without MgSO4, and finally resuspended in 800 μl of the same Sauton’s medium (pH 7.0, no MgSO4) leading to an OD600 of 1.0. A total of 24 well plates were assembled with Sauton’s media containing the required antibiotic, pH values and Mg2+ concentrations (1 ml/well), and the reporter strain suspension was added. Starting ODs were adjusted to the bacterial growth characteristics in the respective medium, e.g., for a 4 days time point the OD600 in medium with a pH of 7.0 and 500 μM MgSO4 was set to 0.025, while OD600s in a media with low pH (5.0) or low Mg2+ concentrations (10 μM) were set to 0.1. After the respective incubation time at 37°C, 200 μl of each well was transferred to a clear-bottomed black 96 well plate (Costar) and FIs determined as described above. The same experimental set up was applied to analyze the impact of other divalent ions (Ca2+, Co2+, Cu2+, Mn2+, Ni2+, Zn2+) or the CorA inhibitor pyrimidinetrione amide analog 10 (PAA10) (Park et al., 2019) on riboswitch-based fluorescence response. In the case of the divalent ions, a MgSO4 concentration of 10 μM was used and a divalent ion concentration around fourfold lower than the determined minimal inhibitory concentrations (MICs). The influence of PAA10 was tested using six different concentrations (0, 5, 25, 50, 100, 250 nM).
Determination of Minimal Inhibitory Concentrations
The various divalent ions (Ca2+, Co2+, Cu2+, Mn2+, Ni2+, Zn2+) and the molecule PAA10 were serially (1:2) diluted in Sauton’s medium (pH 7.0, 500 μM MgSO4) in 96-well round-bottom plates (Thermo Fisher Scientific; 50 μl/well). The starting concentrations of the divalent ions and PAA10 were 5 mM and 4 μM, respectively. Mtb H37Rv wild-type (WT) bacteria were grown in Sauton’s medium (pH 7.0, 500 μM MgSO4) to an OD600 of 0.2. The culture was diluted (1:1000) in the same medium and 50 μl was added to each well. Plates were analyzed after 14 days of incubation at 37°C. The MIC was determined as the lowest concentration of the tested substance that showed complete inhibition of visible bacterial growth.
In vivo Assay
In vivo Detection of Pleft* FP Strains by Flow Cytometry
For in vivo infections, WT B6.SJL (CD45.1/1) mice were intrapharyngeally inoculated with 50 CFU of Mtb HN878 (WT, L5 attB:Pleft* mWasabi, L5 attB:Pleft* mCherry, or L5 attB:Pleft* mScarlet). Lungs were harvested 37 days post-infection and dissociated via GentleMACS and Lung Cell Isolation Buffer (Miltenyi Biotec). Digested lungs were passed through a 100 μm cell strainer and cells washed and purified with 37% Percoll/RPMI. Cells for flow cytometry were washed, counted and subsequently fixed overnight using an Intracellular Fixation & Permeabilization Buffer (eBioscience/Thermo Fisher Scientific). Samples were acquired on a X50 Symphony flow cytometer (BD Biosciences) and analyzed using FlowJo software (BD Biosciences).
In vivo Detection of Mtb HN878 (L5 attB:Pleft* mScarlet) by Confocal Microscopy
C3HeB/FeJ mice were infected with Mtb HN878 (L5 attB:Pleft* mScarlet) by aerosol (50–100 CFU/lung). Mice were treated with standard TB drugs at 5 weeks post-infection for 4 weeks and followed up for 4 weeks prior to disease reactivation. At 13 weeks post-infection, lungs were harvested after perfusion with 4% paraformaldehyde and incubated in a fixation and permeabilization solution (BD Biosciences) for overnight at 4°C followed by washing and dehydration in 30% sucrose. Lungs were embedded in OCT compound (Sakura) and stored at −80°C until used. A total of 20 μm sections of frozen lung tissue were incubated in a blocking buffer (1% BSA, 0.3% Triton X-100, 1% Fc block in 1× PBS) for 2 h at RT followed by staining with anti-mouse CD68 antibody (FA11; Biolegend) overnight at 4°C. Tissues were imaged on a Leica SP8 confocal microscope and images were analyzed with Imaris software (Bitplane).
Results
We sought to develop fluorescent reporter strains based on stable integrative plasmids that would be detectable in vivo at equivalent bacterial densities as those achieved currently with episomal, multi-copy constructs. To determine the brightness difference between episomal multicopy and chromosomal single copy expression, we first measured the signal intensity of mycobacteria expressing eGFP either from the replicating plasmid pOLYG or from a gene construct integrated into the L5 attB site of the mycobacterial chromosome, in vitro. In both strains, expression of the FP was under control of the same mycobacterial promoter, Psmyc. The FI of Mtb (pOLYG Psmyc eGFP) was approximately 10 times higher than that of Mtb (L5 attB:Psmyc eGFP), carrying a single copy of the gene (Figure 1A).
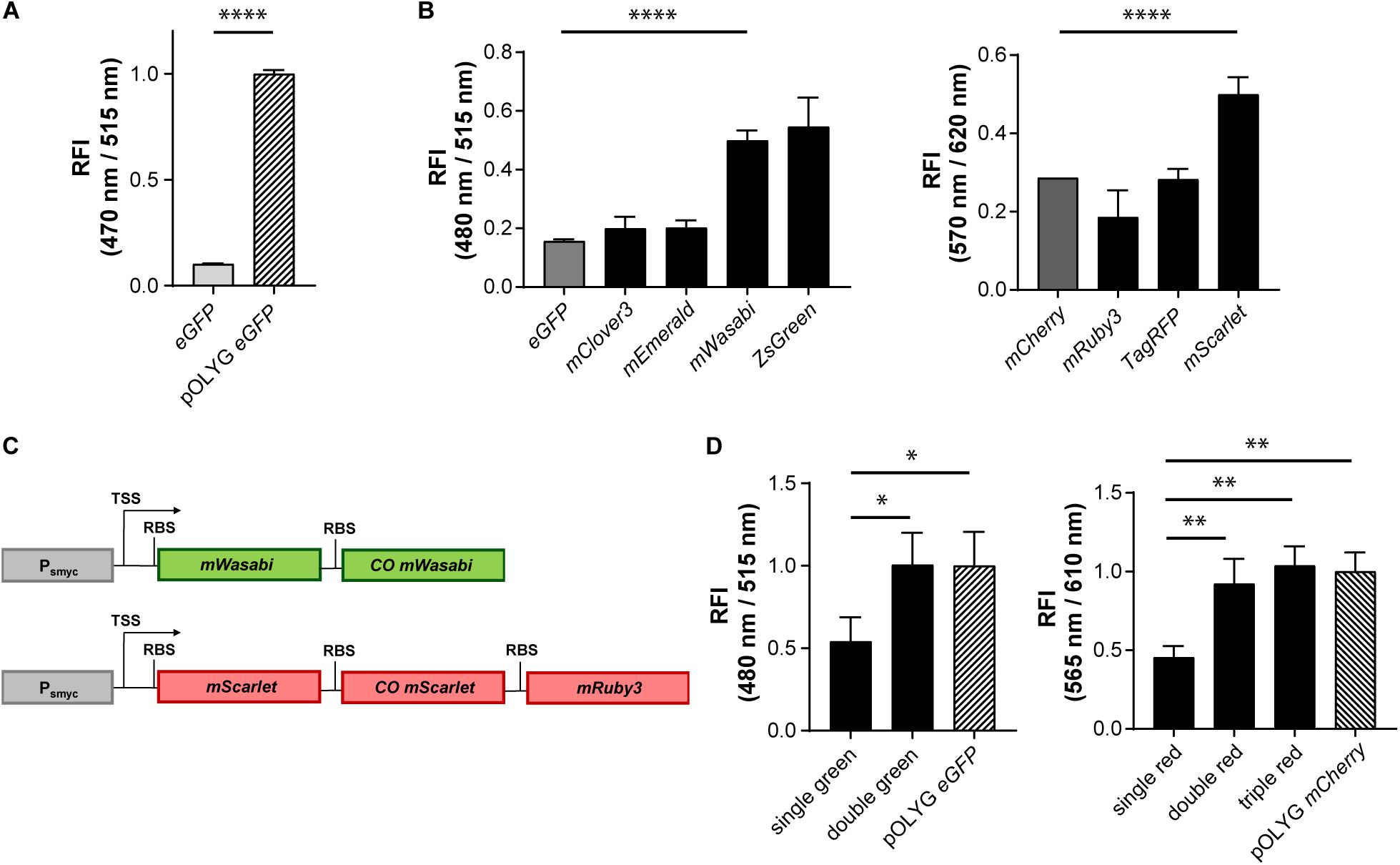
Figure 1. Increase in relative fluorescence intensity using different fluorescent proteins and multicistronic constructs. (A) Relative fluorescence intensity (RFI) of Mtb H37Rv strains expressing eGFP either from an integrated gene construct or from the replicating plasmid pOLYG, and (B) Mtb H37Rv strains expressing different green (left) or red (right) fluorescent proteins. (C) Design of double green and triple red gene constructs. TSS indicates transcriptional start site; RBS indicates ribosome binding site. (D) RFIs of Mtb H37Rv strains containing the double or triple gene constructs chromosomally integrated compared to the replicating plasmids pOLYG Psmyc-eGFP and pOLYG Psmyc-mCherry, respectively. All genes were under the control of the smyc promoter. FIs were detected during the logarithmic growth phase of the strains, analyzed relative to their optical densities at 600 nm, and normalized with FIs of Mtb (pOLYG Psmyc eGFP) and Mtb (pOLYG Psmyc mCherry) equal to 1. Data are representative of at least three independent experiments; the error bars indicate standard deviation; for statistical analysis groups were compared by Student’s unpaired, two-tailed t-tests, p-values: ****p < 0.0001, ∗∗p < 0.01, ∗p < 0.05.
To increase the brightness of Mtb strains carrying the gene construct chromosomally, we screened a library of FPs with excitation and emission wavelengths comparable to those of the commonly used green and red FPs eGFP and mCherry, constructed multicistronic FP constructs, and tested expression strengths of various mycobacterial promoters.
Increase of Signal Intensity Using Different FPs
Genes encoding five different green FPs (eGFP, mClover3, mEmerald, mWasabi, ZsGreen) and four red FPs (mCherry, mRuby3, TagRFP, mScarlet) were cloned downstream of the mycobacterial smyc promoter within an integrative plasmid. Mtb bacteria were transformed with these constructs and FIs relative to the OD600s of the strains were measured. All mycobacterial strains expressing alternative green FPs showed higher FIs compared to the eGFP expressing reference strain. The brightest signals were observed for Mtb (L5 attB:Psmyc mWasabi) and Mtb (L5 attB:Psmyc ZsGreen) (Figure 1B) with a threefold increase above that of Mtb (L5 attB:Psmyc eGFP). For Mtb (L5 attB:Psmyc mWasabi) a comparable 3.5 times higher protein expression level was detected by western blot. In contrast, Mtb (L5 attB:Psmyc ZsGreen) had a similar expression level to that of Mtb (L5 attB:Psmyc eGFP) (Supplementary Figure 4A). In the group of red-fluorescent Mtb strains, only the one expressing mScarlet showed a significantly higher FI compared to the mCherry expressing strain (Figure 1B). Conversely, the protein expression level of Mtb (L5 attB:Psmyc mScarlet) was 10-fold lower than Mtb (L5 attB:Psmyc mCherry) as quantified by western blot (Supplementary Figure 4B).
Increase of Signal Intensity Using Multicistronic FP Constructs
Introducing different FPs resulted in an increase in FI of the stably integrated single copy constructs, however brightness levels were still significantly below those of Mtb (pOLYG Psmyc eGFP) and Mtb (pOLYG Psmyc mCherry). To enhance the FI of the single copy mWasabi construct, a second, codon optimized, allele encoding mWasabi, along with an RBS, was cloned downstream of the Psmyc-gene region. Codon optimization was performed in order to increase expression levels as well as limit homologous recombination and thus the loss of gene constructs over time. A similar bicistronic gene sequence was designed for the red FPs including mScarlet and codon optimized mScarlet (COmScarlet). Furthermore, a tricistronic construct was developed containing the gene for mRuby3 downstream of the two mScarlet genes (Figure 1C). The new plasmids were transformed into Mtb resulting in the strains Mtb (L5 attB:Psmyc double green), Mtb (L5 attB:Psmyc double red), and Mtb (L5 attB:Psmyc triple red). Comparison of the FIs showed a twofold signal increase for Mtb (L5 attB:Psmyc double green) as well as Mtb (L5 attB:Psmyc double red) compared to the equivalent Mtb strain containing only one gene of the FP. The fluorescence readout signal of Mtb (L5 attB:Psmyc double green) was equivalent to that achieved by Mtb (pOLYG Psmyc eGFP). Mtb (L5 attB:Psmyc triple red) showed a slightly higher FI than Mtb (L5 attB:Psmyc double red), which was comparable to that of Mtb (pOLYG Psmyc mCherry) (Figure 1D).
Increase of Signal Intensity Using Different Mycobacterial Promoters
As an alternative strategy to increase FIs, we cloned a variety of mycobacterial promoters (Psmyc, Phsp60, PG13, Pmsp12, PMOP, and Pleft) upstream of the gene encoding eGFP in a chromosomally integrating plasmid (Figure 2A). The strongest eGFP expression level was detected for Mtb (L5 attB:PMOP eGFP), with a FI fivefold higher compared to the Mtb strain containing the commonly used hsp60 promoter and less than 1.5 times brighter compared to Mtb (L5 attB:Psmyc eGFP). The lowest fluorescence signal was measured for Mtb (L5 attB:Pmsp12 eGFP). Mtb (L5 attB:Phsp60 eGFP), Mtb (L5 attB:PG13 eGFP), and Mtb (L5 attB:Pleft eGFP) had similar brightness levels. Analysis of the Pleft and Psmyc promoter-RBS sequences (Supplementary Table 5) revealed that Psmyc contained an RBS (AGGAGG) similar to the consensus RBS of Mtb (DeJesus et al., 2013; Newton-Foot and Gey van Pittius, 2013), while the putative RBS for Pleft was identified as GGGAGA. Under the assumption that the consensus RBS is associated with stronger expression levels, we exchanged the RBS downstream of the left promoter to that of Psmyc to generate a new promoter-RBS element, which was named Pleft*. Pleft* was cloned upstream of the gene encoding eGFP and subsequently transformed into Mtb. When the FI of Mtb (L5 attB:Pleft* eGFP) was compared to the other mycobacterial promoters previously tested, there was a 25-fold increase in expression strength compared to Mtb (L5 attB:Phsp60 eGFP) and a sixfold increase in expression over the Psmyc containing Mtb strain (Figure 2B).
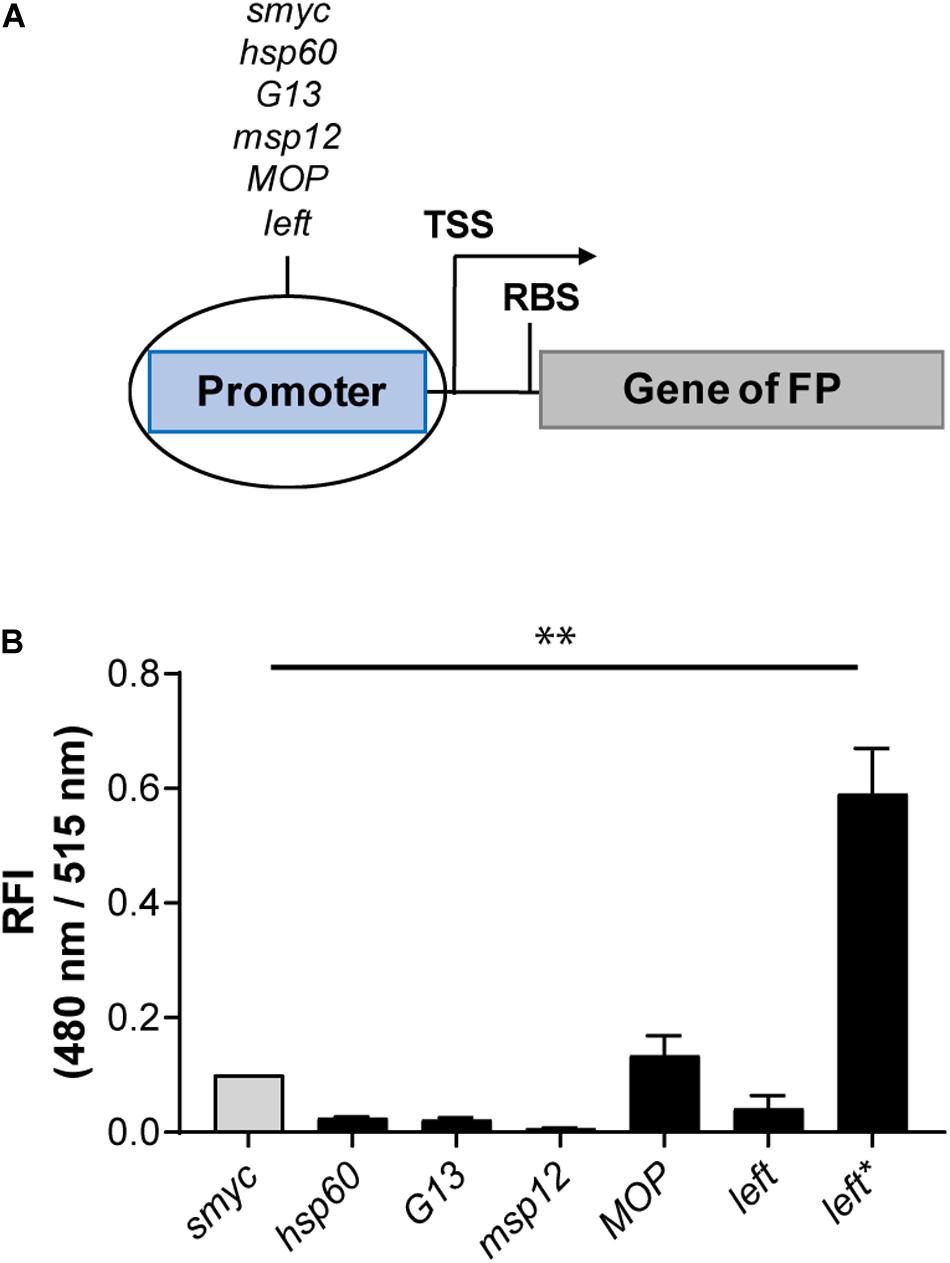
Figure 2. Introduction of the Pleft* element led to a strong fluorescence increase. (A) Design of the promoter-gene constructs. TSS indicates transcriptional start site; RBS indicates ribosome binding site. (B) RFI of Mtb H37Rv strains expressing eGFP under the control of different constitutive promoters. All gene constructs were chromosomally integrated. FIs were detected during the logarithmic growth phase of the strains, analyzed relative to their optical densities at 600 nm, and normalized with FI of Mtb (L5 attB:Psmyc eGFP) equal to 0.1. Data are representative of at least two independent experiments; the error bars indicate standard deviation; for statistical analysis groups were compared by Student’s unpaired, two-tailed t-tests, p-value: ∗∗p < 0.01.
Combination of Bright Protein Candidates and the New Pleft* Construct
After identification of Pleft* as a strong promoter-RBS element, we combined it with the alternative green and red FP candidates described previously. Pleft* was cloned upstream of the genes encoding mWasabi and mScarlet in chromosomally-integrating plasmids, and transformed into Mtb. Mtb (L5 attB:Pleft* mWasabi) showed fivefold higher FI compared to the Mtb (L5 attB:Psmyc double green), and Mtb (pOLYG Psmyc eGFP) strains. The signal intensity of the Mtb (L5 attB:Pleft* mScarlet) strain was about 15 times higher than the Mtb (L5 attB:Psmyc triple red), and the Mtb (pOLYG Psmyc mCherry) strains (Figure 3A). In addition, Mtb (L5 attB:Pleft* mWasabi) and Mtb (L5 attB:Pleft* mScarlet) colonies appear visibly green and pink, respectively, when grown on agar plates (Figure 3B). Additional agar plate images of Mtb strains expressing other FPs under the control of the left∗ promoter are depicted in Supplementary Figure 5. Despite their brightness, Mtb (L5 attB:Pleft* mScarlet) had very similar growth characteristics and Mtb (L5 attB:Pleft* mWasabi) only minimal growth differences on agar plates and in liquid media (Supplementary Figure 6) in comparison to a WT strain.

Figure 3. Fluorescence intensities were strongly increased when combining the Pleft* element with genes expressing mWasabi or mScarlet. (A) Fluorescence comparison between Mtb H37Rv strains containing either the replicating plasmids pOLYG Psmyc-eGFP and pOLYG Psmyc-mCherry, respectively, or the chromosomally integrated gene constructs Psmyc-double green/-triple red, and Pleft*-HismWasabi/-HismScarlet. RFI were determined during the logarithmic growth phase of the strains, analyzed relative to their optical densities at 600 nm, and normalized with FIs of Mtb (pOLYG Psmyc eGFP) and Mtb (pOLYG Psmyc mCherry) equal to 1. Data are representative of at least two independent experiments; the error bars indicate standard deviation; for statistical analysis groups were compared by Student’s unpaired, two-tailed t-tests, p-values: ****p < 0.0001. (B) Mtb (L5 attB:Pleft* mWasabi) and Mtb (L5 attB:Pleft* mScarlet) colonies grown on 7H11 agar. (C) Flow cytometric analysis of Mtb HN878 (L5 attB:Pleft* mScarlet) at 610 nm in lung single-cell suspensions from WT B6.SJL (CD45.1/1) 37 days post-infection with 50 CFU. Data are representative of one experiment with three mice. (D) Lung tissue image of C3HeB/FeJ mice 13 weeks post-infection with Mtb HN878 (L5 attB:Pleft* mScarlet). mScarlet expressing bacilli (red) were detected intracellularly in macrophages (CD68+, blue). Scale bar indicates 20 μm.
Detection of Pleft* FP Strains in Lungs of WT Mice
The new, bright Pleft* FP strains described above represent a useful starting point for the construction of more advanced reporter strains. Based on their strong fluorescence profile, these strains also provide an attractive alternative to commonly used fluorescent Mtb strains for in vivo imaging. This was tested by infecting WT B6.SJL (CD45.1/1) mice with Mtb HN878 (WT), Mtb HN878 (L5 attB:Pleft* mCherry), Mtb HN878 (L5 attB:Pleft* mScarlet), and Mtb HN878 (L5 attB:Pleft* mWasabi) and analyzing single-cell suspensions of mouse lungs after fixation 5 weeks post infection by flow cytometry. Cells infected with the fluorescent Pleft* strains could clearly be distinguished from uninfected cells (Figure 3C and Supplementary Figure 7). In addition, Mtb HN878 (L5 attB:Pleft* mScarlet) bacteria were successfully visualized in lungs of C3HeB/FeJ mice 13 weeks post-infection using confocal microscopy (Figure 3D), indicating long-term expression of the FP during mouse infections.
Exploiting Multicistronic FP Constructs to Generate a Bright pH Reporter
To apply these new findings to the optimization of more complex, chromosomally-integrated reporter constructs, we first exchanged the smyc promoter in the triple red gene construct for the previously reported pH-responsive promoter of the Mtb gene rv2395A (Abramovitch et al., 2011; Figure 4A). After successful transformation of Mtb, FIs of the new reporter strain Mtb (Giles attB:Pnp2395A triple red) were measured in vitro as a function of pH and time. In agreement with the results described previously using a multi-copy reporter construct (Abramovitch et al., 2011), our multicistronic, integrated, pH reporter displayed a threefold increase in fluorescence after 3 days incubation at acidic pH (5.5) compared to neutral pH (7.0). Importantly, the detected fluorescence readout at pH 5.5 reached the level of Mtb (pOLYG Psmyc mCherry) (Figure 4B). We further investigated the reporter’s response under a range of physiologically relevant pH values. While no significant difference in FI was detected at pH 6.5 compared to pH 7.0, a pH-dependent increase was apparent from pH 6.0 to pH 4.5, with signal intensities twofold higher at pH 5.0 and 4.5 compared to the reference strain Mtb (pOLYG Psmyc mCherry) (Figure 4C and Supplementary Figure 8).

Figure 4. Mtb (Giles attB:Pnp2395A triple red) showed pH-dependent fluorescence with high signal intensity despite chromosomal integration. (A) Design of the pH-responsive gene construct. TSS indicates transcriptional start site; RBS indicates ribosome binding site. RFI of Mtb (Giles attB:Pnp2395A triple red) was analyzed as a function of (B) time and (C) pH dependency at a Mg2+ concentration of 500 μM after 6 days of incubation. FIs were detected during the logarithmic growth phase of the strain, analyzed relative to its optical densities at 600 nm, and normalized with FI of Mtb (pOLYG Psmyc mCherry) equal to 1. Data are representative of at least three biological replicates; the error bars indicate standard deviation; for statistical analysis groups were compared by Student’s unpaired, two-tailed t-tests, p-values: ****p < 0.0001, ns: not significant.
Generation of Novel Mg2+ Reporter Strains Using mRNA-Based Responsive Elements
Conformational changes of the untranslated region (UTR) of the mRNA can be induced by a variety of small molecules, e.g., metabolites or drugs, and ion concentrations, leading to changes in expression levels of the downstream gene (Sinumvayo et al., 2018). These unique responsive oligonucleotides are termed riboswitches and represent a promising tool for reporter strain construction. A number of riboswitches have been identified in Mtb bacteria by sequence homology, including two homologous motifs upstream of the Mtb genes rv1535 and rv1806. Based on transcriptomic profiling under ion starvation, these riboswitches were classified as Mg2+-responsive elements (Walters et al., 2006). We cloned both riboswitches with their native promoters upstream of the gene encoding mWasabi (Figure 5A) and integrated the new reporter constructs into the Giles attachment site of the Mtb chromosome. The new reporter strains, Mtb (Giles attB:Pnp1535 mWasabi) and Mtb (Giles attB:Pnp1806 mWasabi), showed Mg2+-dependent fluorescence responses in vitro inversely correlated to the concentration of the divalent ion. Grown under normal conditions with high levels of Mg2+ these strains were much less fluorescent than the episomally encoded pOLYG eGFP control (Figure 5B). While the FI of Mtb (Giles attB:Pnp1535 mWasabi) was two times greater at the lower (10 μM) compared to the higher Mg2+ concentration (500 μM), Mtb (Giles attB:Pnp1806 mWasabi) displayed fluorescence differences of sevenfold under these conditions (Figure 5C). The Mg2+ response was time dependent reaching maximal levels after approximately 4–7 days of incubation (Figure 5D).
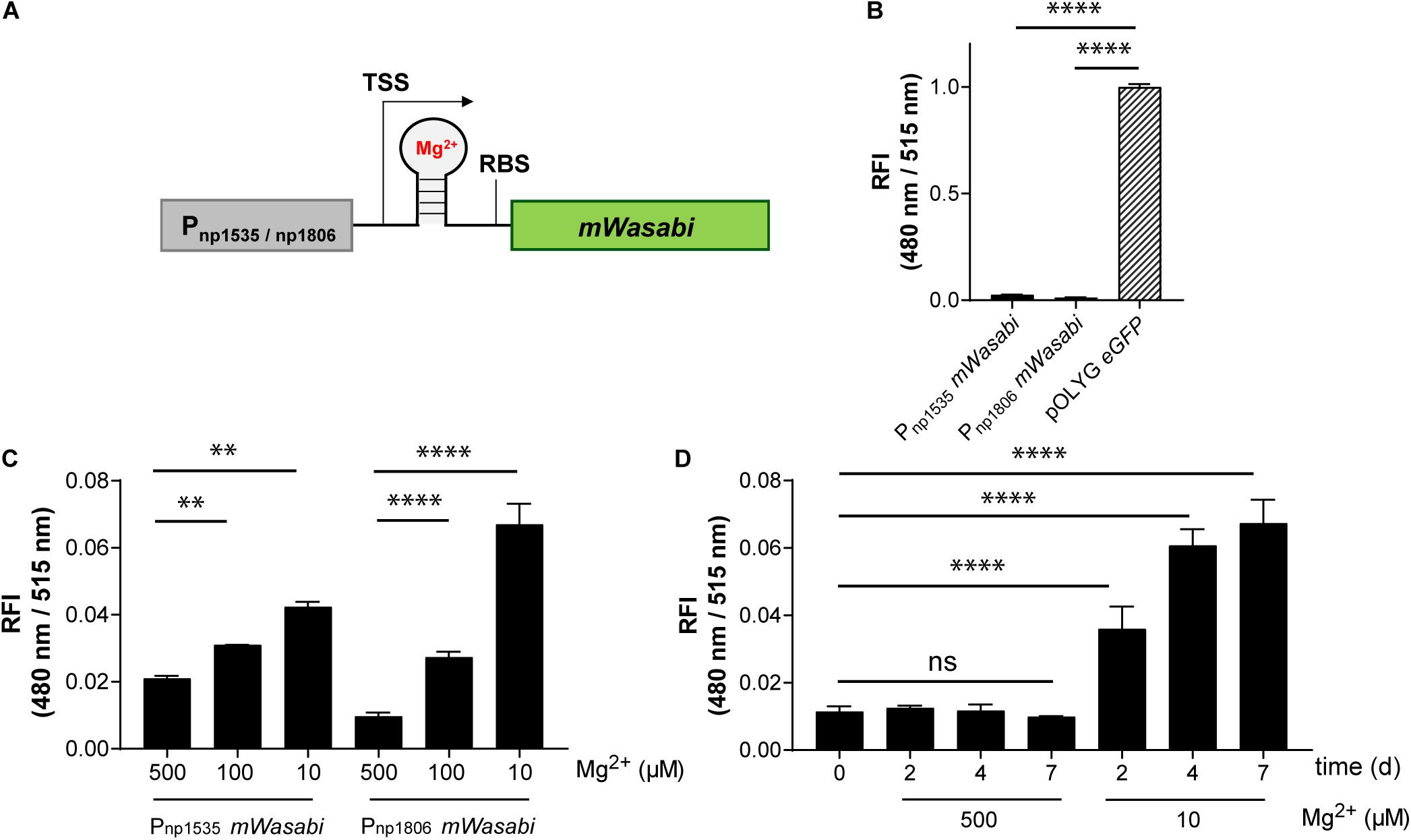
Figure 5. Mtb reporter strains carrying the native promoter regions upstream of the genes rv1535 or rv1806 showed Mg2+-dependent readout signals, but with low fluorescence intensities. (A) Design of Mg2+-responsive gene constructs. TSS indicates transcriptional start site; RBS indicates ribosome binding site. (B) Fluorescence comparison between Mtb bacteria expressing eGFP from the replicating plasmid pOLYG and the reporter strains Mtb (Giles attB:Pnp1535 mWasabi) and Mtb (Giles attB:Pnp1806 mWasabi) at a Mg2+ concentration of 500 μM and pH 7.0. (C) Mg2+ dependence of the RFI of the reporter strains at pH 7.0 after 7 days of incubation. (D) Time dependency of the fluorescence increase of Mtb (Giles attB:Pnp1806 mWasabi) when exposed to low Mg2+ levels (10 μM) at pH 7.0. FIs were detected during the logarithmic growth phase of the strains, analyzed relative to their optical densities at 600 nm, and normalized with FI of Mtb (pOLYG Psmyc eGFP) equal to 1. Data are representative of at least three biological replicates; the error bars indicate standard deviation; for statistical analysis groups were compared by Student’s unpaired, two-tailed t-tests, p-values: ****p < 0.0001, ∗∗p < 0.01, ns: not significant.
Increasing the FI of the Riboswitch-Based Mtb Reporter Strain
While the data above confirmed the utility of the np1535 and np1806 riboswitch elements in generating Mg2+-responsive reporter systems, the overall FIs of the chromosomally-integrated constructs were 40- and 86-fold, respectively, lower than the corresponding signal achieved by the constitutively active reference strain Mtb (pOLYG Psmyc eGFP) (Figure 5B). In order to improve the readout signal of the Mg2+ responsive reporter, we applied the strategies described above, including (a) promoter exchange and (b) formation of multicistronic FP constructs. The essential DNA element representing the responsive riboswitch was identified based on published transcriptional start site (TSS) data (Cortes et al., 2013) and by sequence alignment of the DNA regions upstream of the Mtb genes rv1535 and rv1806. An oligonucleotide region of around 250 base pairs was determined, which showed 70% sequence homology (Supplementary Table 5). The predicted RBS, AGGAGG, and AGGAGA, of those genes closely resembled the consensus RBS of Mtb (DeJesus et al., 2013; Newton-Foot and Gey van Pittius, 2013), which suggests that they were suitable elements for strong gene expression systems without further modifications. Based on this analysis, we fused the putative core riboswitch of np1806, along with the native RBS, to either the smyc or left promoter. Furthermore, these new promoter-riboswitch constructs were placed upstream of either mWasabi or double green (Figure 6A). To determine the basal fluorescence of the Mtb strains carrying the newly designed plasmids, we measured signal intensity at a high Mg2+ concentration of 500 μM. While Psmyc1806 and the native promoter led to similar FIs, Mtb (Giles attB:Pleft1806 mWasabi) showed a fivefold increased brightness, which correlates with the fold-change we observed previously comparing the promoter elements Psmyc and Pleft* (Figures 2B, 6B and Supplementary Figure 9). When testing the FIs of the Mtb strains containing two gene copies of mWasabi, the results displayed a fluorescent signal output between seven and nine times higher than the reporter strains harboring a single mWasabi gene (Figure 6B and Supplementary Figure 10). Importantly, the Mg2+ responsiveness of the new Mtb reporter strains remained intact despite manipulations of the promoter and gene regions. Mtb (Giles attB:Pleft1806 double green) showed a fivefold fluorescence enhancement at low Mg2+ concentration (10 μM) compared to high cation levels (500 μM), achieving a similar brightness to Mtb (pOLYG Psmyc eGFP) (Figure 6C).
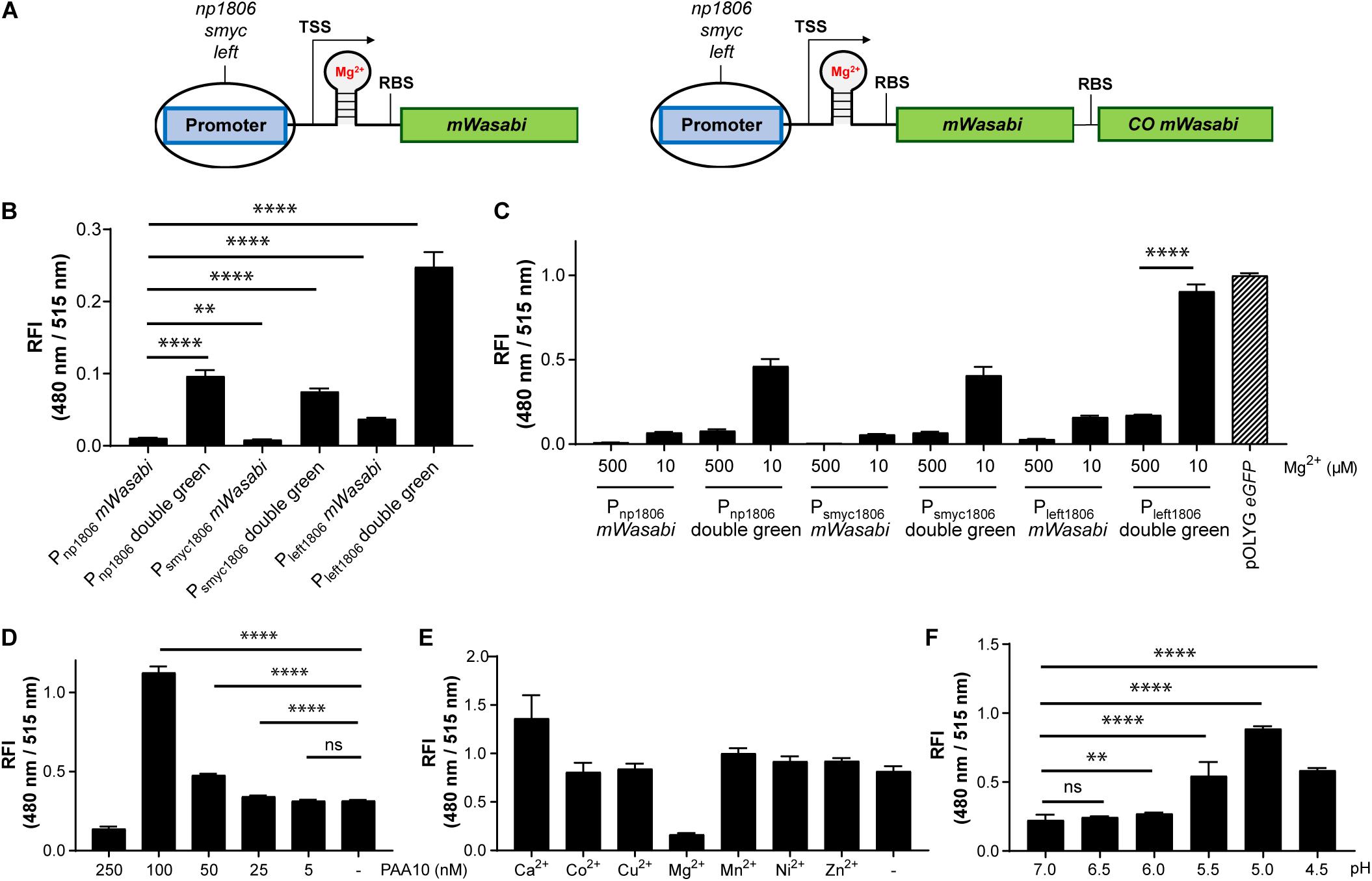
Figure 6. Using protein-riboswitch fusions and multicistronic constructs yielded increased fluorescence intensity while maintaining a Mg2+-specific response. (A) Design of Mg2+-responsive gene constructs. TSS indicates transcriptional start site; RBS indicates ribosome binding site. Influence of protein-riboswitch fusions and multicistronic constructs on (B) the relative fluorescence intensity (RFI) at a Mg2+ concentration of 500 μM and pH 7.0, and (C) Mg2+-dependent fluorescence increase at pH 7.0 after 7 days of incubation. RFIs of Mtb (Giles attB:Pleft1806 double green) were detected (D) at a Mg2+ concentration of 500 μM and pH 7.0 when exposed for 4 days to different concentrations of the CorA inhibitor pyrimidinetrione amide analog 10 (PAA10), (E) RFI after 6 days coincubation at a Mg2+ concentration of 10 μM and pH 7.0 with the addition of calcium chloride (CaCl2; 1 mM), cobalt(II) sulfate (CoSO4; 100 μM), copper(II) sulfate (CuSO4; 40 μM), magnesium sulfate (MgSO4; 1 mM), manganese(II) sulfate (MnSO4; 150 μM), nickel(II) sulfate (NiSO4; 300 μM), or zinc sulfate (ZnSO4; 1 mM), (F) RFI after 6 days incubation in media with Mg2+ concentrations of 500 μM and different pH values. FIs were analyzed relative to the optical densities at 600 nm and normalized with FI of Mtb (pOLYG Psmyc eGFP) equal to 1. Data are representative of at least four biological replicates; the error bars indicate standard deviation; for statistical analysis groups were compared by Student’s unpaired, two-tailed t-tests, p-values: ****p < 0.0001, ∗∗p < 0.01, ns: not significant.
The Mtb (Giles attB:Pleft1806 Double Green) Reporter Strain Selectively Responds to Mg2+
The fluorescence response of the novel riboswitch-based reporter strain was further analyzed across a broader range of physiologically relevant Mg2+ conditions and demonstrated a robust concentration and time dependency (Supplementary Figure 10). To verify Mg2+ specificity the influence of a known inhibitor (PAA10) of the mycobacterial Mg2+ transporter CorA (Park et al., 2019) was examined. A significant fluorescence increase was detected from an inhibitor concentration of 25 nM, increasing to a maximum at inhibitor concentrations close to its MIC (250 nM) (Supplementary Figure 2A). At a concentration of 100 nM, the detected FI of the reporter strain suggests that intracellular Mg2+ levels are lower than those obtained at external media MgSO4 concentrations of 10 μM (Figure 6D and Supplementary Figure 2B). To exclude promiscuity towards other divalent ions, the response profile of the novel reporter strain Mtb (Giles attB:Pleft1806 double green) was explored under low magnesium (10 μM) and high divalent ion (Ca2+, Co2+, Cu2+, Mn2+, Ni2+, Zn2+) concentrations, close to their MIC values or at 1 mM if the MIC was greater than 2.5 mM (Supplementary Figure 11). None of the tested ions had a significant impact on the FI induced by the absence of Mg2+ ions, indicating high specificity of the riboswitch (Figure 6E and Supplementary Figure 11).
Influence of Environmental pH on the Fluorescence Response of Mtb (Giles attB:Pleft1806 Double Green)
Based on previous data showing that lower pH values influence the Mg2+ levels required for mycobacterial growth (Buchmeier et al., 2000; Piddington et al., 2000), we predicted that acidic pH would impact the fluorescence signal of Mtb (Giles attB:Pleft1806 double green). Screening the reporter’s response at high Mg2+ concentrations (500 μM) and a range of pH values supported this assumption. At pH 5.5, there was a 2.5-fold increase in the fluorescence signal compared to pH 7.0. When the pH dropped to 5.0, a fourfold increase in FI was observed and a threefold increase was measured for pH 4.5. These data imply that the intrabacterial Mg2+ levels are 50 times lower than environmental ion concentrations at pH 5.0 (Figure 6F and Supplementary Figure 12), suggesting that the Mg2+ uptake of Mtb is dependent on extracellular pH.
These results show that to fully understand the micromilieu, both pH and Mg2+ must be evaluated in parallel. Therefore, a reporter with the ability to simultaneously respond to pH and Mg2+ had to be constructed. In addition, this strain had to contain an expression system under the control of a constitutive promoter to exclude false positive or negative results based on fluctuations in protein expression levels.
Expanding the FP Panel for Mtb Bacteria
For construction of a triple reporter, we took advantage of the variety of available FPs, in order to track multiple conditions simultaneously without spectral crosstalk. Eleven different genes (mTagBFP2, mT-Sapphire, mTurquoise2, mTFP1, LSSmOrange, CyOFP1, Ypet, mPapaya, mOrange2, mKate2, mCardinal) encoding FPs with a large variety of excitation and emission wavelengths were cloned downstream of the Psmyc-His element within pL5 Psmyc-His. Signal intensities of the new fluorescent Mtb strains were screened under different wavelength pairings and possible combinations with minimal spectral overlap were identified. In addition to Mtb (L5 attB:Psmyc mWasabi) and Mtb (L5 attB:Psmyc mScarlet), four other Mtb strains (L5 attB:Psmyc mTagBFP2, LSSmOrange, mPapaya, mCardinal) exhibited fluorescence characteristics which allowed signal detection in parallel (Supplementary Figures 13, 14).
Generation of a Three-Color Fluorescent Strain to Simultaneously Monitor pH, Mg2+, and Protein Expression
We exploited the compatibility of mWasabi, mScarlet, and mTagBFP2 to generate the intended triple reporter strain detecting pH, Mg2+ concentration, and protein expression levels. The newly identified reporter constructs Pnp2395A-triple red and Pleft1806-double green were introduced into different multiple cloning sites within the same plasmid, which integrates into the Giles attB site of the mycobacterial chromosome. In addition, the constitutive smyc promoter was cloned upstream of mTagBFP2 and COmTagBFP2 and introduced into a separate plasmid integrating into the L5 attB site (Figure 7A). The FIs of the new triple reporter Mtb strain (protein expression: blue, λex:400 nm/λem:450 nm; Mg2+ concentration: green, λex:480 nm/λem:515 nm; pH: red, λex:565 nm/λem:610 nm), carrying both plasmids integrated at different sites of the chromosome, were compared to those harboring the individual reporter constructs separately. No significant differences in signal intensities were detected (Figure 7B). Furthermore, the new tri-responsive reporter strain had identical growth characteristics in 7H9 medium compared to the strains containing the single multicistronic gene constructs under control of the smyc promoter or a WT strain, respectively (Supplementary Figure 15).
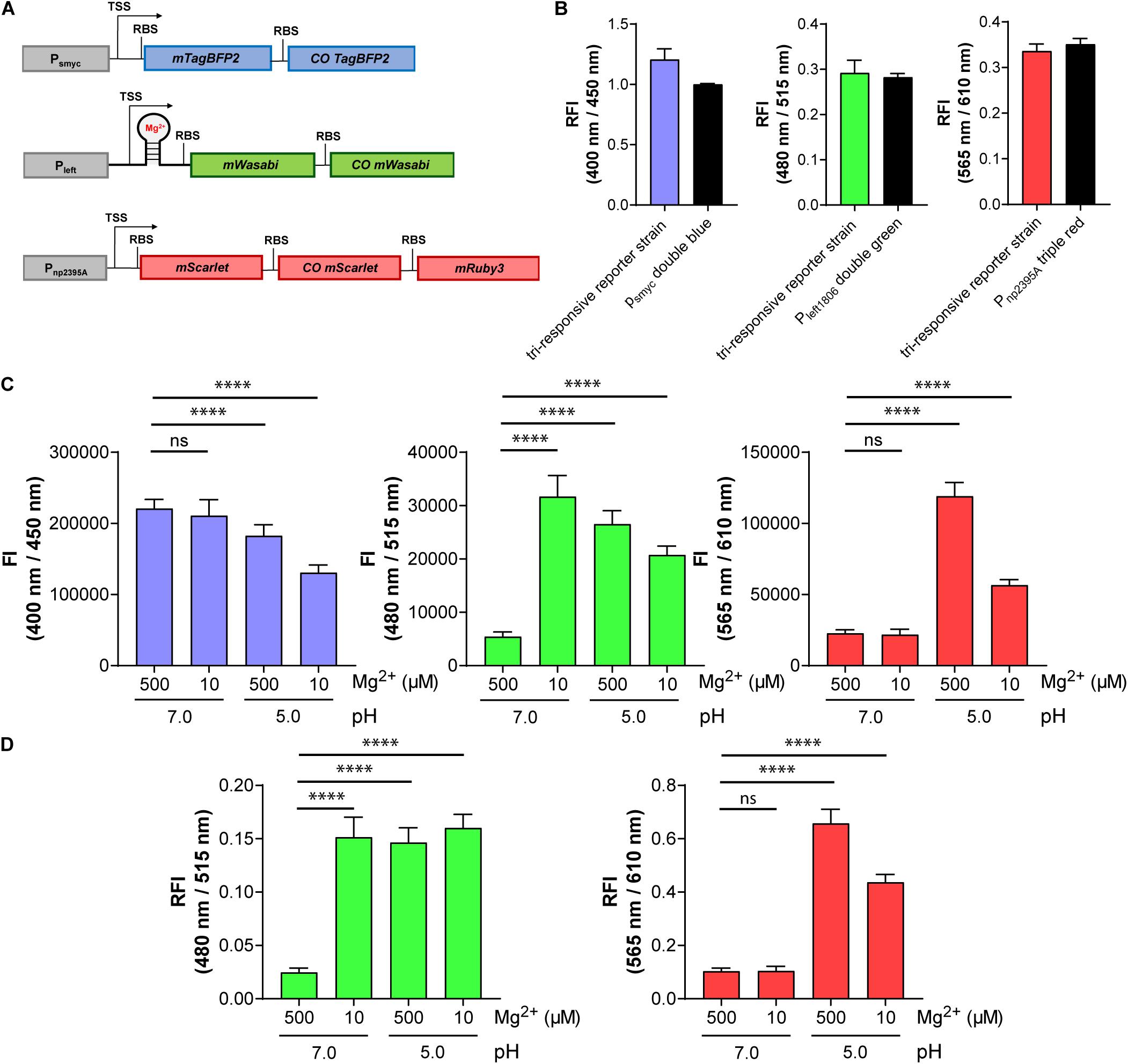
Figure 7. The triple reporter strain enables simultaneous visualization of three conditions in parallel using fluorescence signals at three different wavelengths: protein expression (blue), Mg2+ concentration (green), and pH (red). (A) Design of the gene constructs included in the tri-responsive reporter strain. TSS indicates transcriptional start site; RBS indicates ribosome binding site. (B) RFI of the triple reporter strain in comparison with the single reporter strains Mtb (L5 attB:Psmyc double blue), Mtb (Giles attB:Pleft1806 double green), and Mtb (Giles attB:Pnp2395A triple red) detected at their respective optimal excitation and emission wavelengths in media with pH 7.0 and a Mg2+ concentration of 500 μM. Data were normalized with FIs of Mtb (L5 attB:Psmyc double blue), Mtb (pOLYG Psmyc eGFP), and Mtb (pOLYG Psmyc mCherry) equal to 1. (C) FIs of the three fluorescent proteins detected after 6 days of Mtb (tri-responsive reporter) growth in neutral (pH 7.0) and acidic pH (pH 5.0), as well as high (500 μM) and low (10 μM) Mg2+ concentrations. (D) FIs at 480/515 nm (green) and 565/610 nm (red) depicted relative to changes in protein expression levels (400/450 nm, blue). All FIs were detected during the logarithmic growth phase of the strain and analyzed relative to its optical densities at 600 nm. Data are representative of at least three biological replicates; the error bars indicate standard deviation; for statistical analysis groups were compared by Student’s unpaired, two-tailed t-tests, p-values: ****p < 0.0001, ns: not significant.
Testing Response Characteristics of the Tri-Responsive Reporter Strain
The triple reporter strain was cultured under a range of physiologically relevant pH values and Mg2+ concentrations and the OD600-normalized FIs were measured. Red fluorescence increased significantly when the bacteria were cultured in media at pH 5.0, versus pH 7.0, as observed previously with the individual pH-responsive reporter strain, and the FI at the low pH was twofold greater for high (500 μM) compared to low (10 μM) Mg2+ concentration (Figure 7C, right panel). Green FI increased to a similar extent when the strain was grown under either low Mg2+ concentrations (10 μM), acidic pH (5.0), or a combination of the two conditions (Figure 7C, middle panel). A sixfold difference in green FI was measured when comparing 10 to 500 μM Mg2+ levels at pH 7.0. The green fluorescence signal output was higher in low Mg2+ and neutral pH compared to low or high Mg2+ and acidic pH. A decrease in blue FI was observed under acidic conditions, with a stronger reduction in media containing 10 μM Mg2+ (Figure 7C, left panel). Plotting the red and green FIs relative to the blue signal (in effect normalizing for protein expression levels; Figure 7D) led to alterations in the data, particularly evident in the reduced variation between the Mg2+ (green) responses across all of the tested “activating” conditions. In all cases, the response of the triple reporter strain was time dependent (Supplementary Figures 16–19).
Discussion
Using a combination of recombinant DNA and protein optimization techniques, we have developed a novel reporter strain which, despite chromosomal integration, exhibits high FIs. This triple reporter strain is able to monitor protein expression, pH, and, for the first time, Mg2+ concentration simultaneously. One method to accomplish the brightness of the new strain was to exploit the variety of available FPs. Although several green and red FPs are described to have higher molecular brightness than eGFP and mCherry, those two FPs are still most frequently used for the development of Mtb reporter strains (Abramovitch, 2018; MacGilvary and Tan, 2018). In this study, our green FP of choice was mWasabi, an FP derived from mTFP1 that exhibits high photostability, preferable narrow excitation and emission spectra, and improved molecular brightness relative to eGFP (Ai et al., 2008). While mWasabi has previously been employed in various mycobacterial model systems (Takaki et al., 2013; Ganji et al., 2015; Bernut et al., 2016; Sukheja et al., 2017), to the best of our knowledge it has never been quantitatively compared to eGFP as a reporter protein in this genus. Here, we show that Mtb bacteria expressing mWasabi had a promising threefold higher FI compared to Mtb expressing eGFP from the same promoter, supporting its use as an improved mycobacterial reporter gene. One potential disadvantage of mWasabi is its sensitivity towards acidic pH, similar to that of eGFP (Zhou et al., 2012; Tanida et al., 2014). However, for our purpose the pH dependence was not of great concern, since Mtb is known to maintain its intra-bacterial pH between 6.8 and 7.5, even when exposed to acid environmental conditions (Vandal et al., 2008). Besides mWasabi, ZsGreen, a GFP-like protein isolated from a Zoanthus species (Matz et al., 1999), led to a comparable high fluorescence signal when expressed in Mtb bacteria. The protein expression level of ZsGreen was 3.5-fold lower compared to mWasabi, which could be an advantage regarding energy cost and possible toxicity associated with excessive protein production. Despite these benefits, we decided to proceed with mWasabi, since this green FP had already been successfully used for in vivo imaging of Mycobacterium marinum (Stirling et al., 2020). Future studies will explore the utility of mNeonGreen, the brightest monomeric green FP yet described (Shaner et al., 2013). While molecular brightness, calculated as the product of extinction coefficient and quantum yield, might be used as a first indication for the applicability of FPs in reporter strain development, there was no correlation between molecular brightness and final signal intensity of fluorescent Mtb bacteria, in this study (Supplementary Figure 20). The discrepancy between molecular brightness, protein expression level, and FI, might be explained by proper intra-bacterial folding of FPs (Kremers et al., 2007).
While a variety of red FPs have been previously screened as alternatives to mCherry, so far only the tandem dimer tdTomato was identified to exceed the brightness of mCherry when expressed in mycobacteria (Carroll et al., 2010; Kong et al., 2016). Recently, mScarlet was engineered with the highest ever calculated molecular brightness and the longest fluorescence lifetime in the red FP spectral class. Furthermore, it shows high photostability (t1/2 = 277 s) and acid tolerance (Bindels et al., 2017). Based on these data, we introduced mScarlet into Mtb and observed a higher FI for Mtb (L5 attB:Psmyc mScarlet) compared to Mtb (L5 attB:Psmyc mCherry). The high readout signal in combination with the low expression level of mScarlet is promising for reporter strain development. In addition, its monomeric character is favorable over the tandem dimer of tdTomato for the construction of multicistronic FP systems.
In order to monitor three conditions in parallel, we had to introduce an additional FP with excitation and emission wavelengths varying from those of mWasabi and mScarlet. Four FPs, mTagBFP2 (Subach et al., 2011), LSSmOrange (Shcherbakova et al., 2012), mPapaya (Hoi et al., 2013), and mCardinal (Chu et al., 2014), were identified as potential candidates. Comparing the fluorescence output signals at the optimal wavelengths for each FP identified Mtb (L5 attB:Psmyc mTagBFP2) as the brightest strain with around 100-fold higher FI compared to Mtb (L5 attB:Psmyc mCardinal) (Supplementary Figure 21). Based on the low signal intensity detected for Mtb bacteria expressing mCardinal, we excluded this FP for reporter strain development. However, with excitation and emission wavelengths with maxima at 604 and 659 nm, it might still be a promising candidate for future in vivo studies. We also did not choose mPapaya, since spectral cross talk with mScarlet and mWasabi are more likely to occur with this yellow FP compared to LSSmOrange and mTagBFP2. Both LSSmOrange (data not shown) and mTagBFP2 were successfully used to construct analogous triple reporter strains. In both cases, expressing all three FPs in a single strain did not impact the FIs and response characteristics of each reporter system. However, microscopes are commonly equipped with filter sets suitable for detection of this blue FP, but not LSSmOrange, which has excitation and emission wavelengths at 437 and 572 nm. Furthermore, photoconvertible characteristics were described for this long stokes shift (LSS) FP. Irradiation with a strong 400 nm laser resulted in a change in the absorption peak of LSSmOrange from 437 to 553 nm and thereby in the loss of the LSS (Fron et al., 2015). While the laser intensity of confocal microscopes is most likely not strong enough to induce this photoconversion, we decided not to continue with LSSmOrange. Additional benefits of mTagBFP2 are its high photostability (Subach et al., 2011), as well as a low sensitivity to pH and hypoxia (Tu et al., 2014), two environmental conditions Mtb bacteria are often exposed to within the lungs.
To increase brightness all of the reporter constructs contained at least two genes encoding FPs with similar excitation and emission wavelengths. Insertion of a second gene had the intended additive effect resulting in a twofold fluorescence increase. Construction of a three gene system led only to a slight signal enhancement compared to the two gene element. These results are in line with studies from Lim et al. (2011), showing that protein expression levels are anti-proportional to the distance of the gene location to the TSS.
Introducing the left promoter with a modified RBS was another successful method to obtain increased FIs of mycobacterial strains. Pleft was first identified as a promoter in the genome of the mycobacteriophage L5 (Nesbit et al., 1995). An initial phage-based study using a Pleft-mVenus expression system suggested 100-fold greater FI compared to the analogous hsp60 construct (Jain et al., 2012). However, following reports introducing Pleft directly into Mtb via replicating plasmids detected only a threefold increase, leading to FIs similar to those of Psmyc-gene constructs (Ehrt et al., 2005; Kanno et al., 2016; Kong et al., 2016). In our hands, Mtb bacteria containing Pleft-eGFP showed comparable FIs to Mtb (L5 attB:Phsp60 eGFP). The only difference between our Pleft and the one used by Jain et al. (2012) and Kanno et al. (2016) were three nucleotides in front of the start codon (cat → tcc), which were exchanged in order to construct an NcoI cloning site. The apparent discrepancies in expression levels may be due to effects on RNA folding and thereby protein production (Horbal et al., 2018). Expression from the left promoter was further increased by replacement of its RBS with the one from Psmyc creating Pleft*, which allowed us to generate integrated fluorescent strains with FIs exceeding those obtained using episomal expression from pOLYG by several fold.
In order to exploit the strength of Pleft for reporter strain development, the responsive element had to be outside of the promoter region. In this study, we utilized riboswitches. Their high ligand affinity and specificity, as well as a precise dose-dependence, makes them ideal candidates for reporter strain development (Sherwood and Henkin, 2016; McCown et al., 2017; Sinumvayo et al., 2018). While a number of riboswitches, including cyclic-di-AMP-, cobalamin-, tRNA-, and Mg2+-sensing RNA elements, have been predicted in Mtb using sequence homology (Arnvig and Young, 2012; Nawrocki et al., 2015; Schwenk and Arnvig, 2018), only a single B12-dependent riboswitch upstream of the metE gene has been experimentally validated to date (Warner et al., 2007). The first synthetic promoter-riboswitch fusion was introduced in 2012 into mycobacteria by using a theophylline-responsive riboswitch, which enabled the construction of a new conditional gene knockdown system (Seeliger et al., 2012). To date, however, riboswitches have not been exploited to design novel reporter strains in mycobacteria.
In this study, we applied two Mg2+ responsive elements, also termed ykok leader or Mbox, for this purpose, which were described to be located upstream of the genes rv1535 and rv1806. Transcription of both of these genes was known to be strongly upregulated 13.2- and 5.7-fold, respectively, in Mtb bacteria when Mg2+-starved for 48 h (Walters et al., 2006). In line with this, we detected a fluorescence increase for both Mtb strains carrying the native promoter elements upstream of mWasabi when MgSO4 concentration in culture was reduced from 500 to 10 μM. The signal enhancement was 3.5 times higher for Mtb (Giles attB:Pnp1806 mWasabi) compared to Mtb (Giles attB:Pnp1535 mWasabi).
Construction of a Pleft-riboswitch(1806) fusion led to a fivefold signal increase compared to the native promoter element, while keeping Mg2+-response characteristics intact. The new Mtb (Giles attB:Pleft1806 double green) reporter strain exclusively responded to Mg2+ but not to other divalent ions and thus will enable specific monitoring of Mg2+ concentrations in vivo, even in the presence of Cu2+, and Zn2+ ions, which were previously described to reach high μM concentrations in Mtb-containing phagosomes (Wagner et al., 2005).
While other divalent ions did not interfere with the reporter response, pH values of 5.5 or lower led to an increase of the fluorescence signal compared to neutral pH, even in media with high Mg2+ concentrations (500 μM). The pH-dependence of rv1806 expression has already been described (Rohde et al., 2007). The mechanism of this pH response, however, is still poorly understood. Destruction of a DNA region, controversially expressing either a potential nucleic acid binding protein AprA (rv2395A) or the small non-coding RNA (ncRNA) mcr7 (Abramovitch et al., 2011; Solans et al., 2014), led to reduced expression of rv1806 (Abramovitch et al., 2011). While aprA/mcr7 is under control of the pH-dependent PhoPR two component system, rv1806 is expressed at WT levels in a PhoP transposon mutant (Abramovitch et al., 2011). More confusingly, the transcriptomic profiling strongly differs between PhoP mutants and Mg2+-starved WT bacteria. However, disruption of PhoPR prevents growth in low-Mg2+ media and addition of excess Mg2+ during infection can partially overcome the growth defect of PhoP mutants in THP-1 macrophages (Walters et al., 2006). Interestingly, in our study, Mtb (Giles attB:Pnp2395A triple red) indeed showed a reduced output signal when Mg2+ levels were lowered at acidic pH, and this could not be fully explained by reduction of total protein expression levels. Walters et al. (2006) predicted the pH and Mg2+ correlative effects to be based on cell wall remodeling. However, this has still to be confirmed. Nevertheless, it underlines the necessity to introduce a pH-responsive gene system into the Mg2+-dependent Mtb reporter strain in order to enable differentiation between fluorescence increase based on acid environmental conditions or low Mg2+ concentrations.
Our novel triple reporter strain, detecting Mg2+ levels, pH and protein expression levels, will be used in the future to study the importance of Mg2+ ions in vivo and the potential of Mg2+ transporters as drug targets. One potential candidate is the mycobacterial protein of the proline-glutamate family PE20, whose expression is controlled by the riboswitch(1806), used in this study. PE20 in combination with the proline-proline-glutamate containing protein 31 (PPE31) was shown to be responsible for Mg2+ uptake across the outer membrane of Mtb in a phthiocerol dimycocerosate (PDIM)-dependent manner (Wang et al., 2020).
The transporter, predicted to be responsible for Mg2+ uptake across the inner membrane of Mtb, is the transmembrane protein CorA. Recently, two groups have identified novel anti-mycobacterial molecules, whose efficacy was Mg2+-dependent. Mutations of resistant strains exclusively mapped to the gene rv1239c, which encodes CorA (Lopez Quezada et al., 2019; Park et al., 2019). We explored one of the compounds from these studies, PAA10 (Park et al., 2019). Treatment of the triple reporter strain with this compound showed an increased fluorescence signal at concentrations close to its known MIC value. When the reporter was cultured in high (500 μM) Mg2+ levels, the FI, detected at a PAA10 concentration of 100 nM, exceeded that measured at 10 μM environmental MgSO4, confirming the proposed mechanism of action of intracellular Mg2+ starvation. Our new reporter strain will be used to identify Mg2+ concentrations of different Mtb lesions within the lungs which may address whether the lack of in vivo efficacy of PAA10 (Park et al., 2019) was due to poor exposure to the compound at the site of infection or due to high concentrations of Mg2+ in the Mtb microenvironment.
Future applications of the reporter strains described in this study will invariably require more physiologically relevant model systems than homogenous bacterial cultures, and studies to gauge their utility in cell culture macrophage and animal infection models are ongoing. It is unknown, for example, how reporter gene expression will be affected by less discrete, and temporally fluctuating, changes in pH or ion concentrations as expected to be encountered in more complex biological systems. Similarly, while we could isolate live fluorescent bacteria from mice 13 weeks post infection, it is not clear what effect transgene overexpression may have on long-term fitness and pathogenicity of the organism. Furthermore, while the analytical methods used here (fluorescence plate reader) emphasized population averages, future work will utilize confocal microscopy to analyze individual bacterial responses within spatially and temporally defined regions. Inter-bacterial heterogeneity at the single cell level will provide important information about the heterogeneity of the local milieu. While mindful of these caveats that arise from migrating technical assays from in vitro to in vivo, we are encouraged by prior work in the application of engineered mycobacterial reporter strains to study in vivo phenomena (Tan et al., 2013; Sukumar et al., 2014).
Besides the Mg2+-dependent riboswitch used in this study, other responsive RNA elements might be attractive tools for reporter strain development in the future. Only a few riboswitches have been identified in mycobacteria to date. However, it is likely that the mycobacterial genome contains additional ones, which still need to be discovered (McCown et al., 2017). Furthermore, it should be possible to exploit riboswitches of other species, since the response mechanism is solely RNA-based and independent of species-specific regulatory proteins. In addition, riboswitches allow for the possibility of de novo design to any ligand. Indeed, the first de novo designed riboswitch was recently developed to the second-line anti-TB drug ciprofloxacin (Groher et al., 2018). Using antibiotic-dependent riboswitches for Mtb reporter strain development will allow the direct monitoring of drug concentration across diverse lesion types in vivo and offers a promising approach to get deeper insights into treatment efficiency.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by NIAID Animal Care and Use Committee.
Author Contributions
KK, GP, HB, and CB designed the experiments. KK, AB, GP, MA, H-JY, HF, and SG conducted the experiments. KK, AB, GP, MA, H-JY, KM-B, HB, and CB wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Disease, National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.591866/full#supplementary-material
Footnotes
References
Abramovitch, R. B. (2018). Mycobacterium tuberculosis reporter strains as tools for drug discovery and development. IUBMB Life 70, 818–825. doi: 10.1002/iub.1862
Abramovitch, R. B., Rohde, K. H., Hsu, F. F., and Russell, D. G. (2011). aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol. Microbiol. 80, 678–694. doi: 10.1111/j.1365-2958.2011.07601.x
Ai, H. W., Olenych, S. G., Wong, P., Davidson, M. W., and Campbell, R. E. (2008). Hue-shifted monomeric variants of i fluorescent protein: identification of the molecular determinants of color and applications in fluorescence imaging. BMC Biol. 6:13. doi: 10.1186/1741-7007-6-13
Arnvig, K., and Young, D. (2012). Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol. 9, 427–436. doi: 10.4161/rna.20105
Baker, J. J., Dechow, S. J., and Abramovitch, R. B. (2019). Acid fasting: modulation of Mycobacterium tuberculosis metabolism at acidic pH. Trends Microbiol. 27, 942–953. doi: 10.1016/j.tim.2019.06.005
Barker, L. P., Porcella, S. F., Wyatt, R. G., and Small, P. L. (1999). The Mycobacterium marinum G13 promoter is a strong sigma 70-like promoter that is expressed in Escherichia coli and mycobacteria species. FEMS Microbiol. Lett. 175, 79–85. doi: 10.1111/j.1574-6968.1999.tb13604.x
Belton, M., Brilha, S., Manavaki, R., Mauri, F., Nijran, K., Hong, Y. T., et al. (2016). Hypoxia and tissue destruction in pulmonary TB. Thorax 71, 1145–1153. doi: 10.1136/thoraxjnl-2015-207402
Berney, M., and Berney-Meyer, L. (2017). Mycobacterium tuberculosis in the face of host-imposed nutrient limitation. Microbiol. Spectr. 5:e0030-2016.
Bernut, A., Nguyen-Chi, M., Halloum, I., Herrmann, J. L., Lutfalla, G., and Kremer, L. (2016). Mycobacterium abscessus-induced granuloma formation is strictly dependent on TNF signaling and neutrophil trafficking. PLoS Pathog. 12:e1005986. doi: 10.1371/journal.ppat.1005986
Bindels, D. S., Haarbosch, L., van Weeren, L., Postma, M., Wiese, K. E., Mastop, M., et al. (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 14, 53–56. doi: 10.1038/nmeth.4074
Buchmeier, N., Blanc-Potard, A., Ehrt, S., Piddington, D., Riley, L., and Groisman, E. A. (2000). A parallel intraphagosomal survival strategy shared by mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35, 1375–1382. doi: 10.1046/j.1365-2958.2000.01797.x
Carroll, P., Schreuder, L. J., Muwanguzi-Karugaba, J., Wiles, S., Robertson, B. D., Ripoll, J., et al. (2010). Sensitive detection of gene expression in mycobacteria under replicating and non-replicating conditions using optimized far-red reporters. PLoS One 5:e9823. doi: 10.1371/journal.pone.0009823
Chan, K., Knaak, T., Satkamp, L., Humbert, O., Falkow, S., and Ramakrishnan, L. (2002). Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc. Natl. Acad. Sci. U.S.A. 99, 3920–3925. doi: 10.1073/pnas.002024599
Chu, J., Haynes, R. D., Corbel, S. Y., Li, P., González-González, E., Burg, J. S., et al. (2014). Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat. Methods 11, 572–578. doi: 10.1038/nmeth.2888
Cortes, T., Schubert, O. T., Rose, G., Arnvig, K. B., Comas, I., Aebersold, R., et al. (2013). Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 5, 1121–1131. doi: 10.1016/j.celrep.2013.10.031
DeJesus, M. A., Sacchettini, J. C., and Ioerger, T. R. (2013). Reannotation of translational start sites in the genome of Mycobacterium tuberculosis. Tuberculosis 93, 18–25. doi: 10.1016/j.tube.2012.11.012
Ehrt, S., Guo, X. V., Hickey, C. M., Ryou, M., Monteleone, M., Riley, L. W., et al. (2005). Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. doi: 10.1093/nar/gni013
Ehrt, S., Schnappinger, D., and Rhee, K. Y. (2018). Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 496–507. doi: 10.1038/s41579-018-0013-4
Fron, E., De Keersmaecker, H., Rocha, S., Baeten, Y., Lu, G., Uji-i, H., et al. (2015). Mechanism behind the apparent large stokes shift in LSSmOrange investigated by time-resolved spectroscopy. J. Phys. Chem. B 119, 14880–14891. doi: 10.1021/acs.jpcb.5b09189
Ganji, R., Dhali, S., Rizvi, A., Sankati, S., Vemula, M. H., Mahajan, G., et al. (2015). Proteomics approach to understand reduced clearance of mycobacteria and high viral titers during HIV-mycobacteria co-infection. Cell Microbiol. 18, 355–368. doi: 10.1111/cmi.12516
George, K. M., Yuan, Y., Sherman, D. R., and Barry, C. E. III (1995). The biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Identification and functional analysis of CMAS-2. J. Biol. Chem. 270, 27292–27298. doi: 10.1074/jbc.270.45.27292
Global Tuberculosis Report (2020). World Health Organization, 29th Edn. Available online at: https://apps.who.int/iris/bitstream/handle/10665/336069/9789240013131-eng.pdf (accessed November 21, 2020).
Goodsmith, N., Guo, X. V., Vandal, O. H., Vaubourgeix, J., Wang, R., Botella, H., et al. (2015). Disruption of an M. tuberculosis membrane protein causes a magnesium-dependent cell division defect and failure to persist in mice. PLoS Pathog. 11:e1004645. doi: 10.1371/journal.ppat.1004645
Groher, F., Bofill-Bosch, C., Schneider, C., Braun, J., Jager, S., Geißler, K., et al. (2018). Riboswitching with ciprofloxacin-development and characterization of a novel RNA regulator. Nucleic Acids Res. 46, 2121–2132. doi: 10.1093/nar/gkx1319
Hoi, H., Howe, E. S., Ding, Y., Zhang, W., Baird, M. A., Sell, B. R., et al. (2013). An engineered monomeric Zoanthus sp. yellow fluorescent protein. Chem. Biol. 20, 1296–1304. doi: 10.1016/j.chembiol.2013.08.008
Hood, M. I., and Skaar, E. P. (2012). Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525–537. doi: 10.1038/nrmicro2836
Horbal, L., Siegl, T., and Luzhetskyy, A. (2018). A set of synthetic versatile genetic control elements for the efficient expression of genes in Actinobacteria. Sci. Rep. 8:491. doi: 10.1038/s41598-017-18846-1
Huff, J., Czyz, A., Landick, R., and Niederweis, M. (2010). Taking phage integration to the next level as a genetic tool for mycobacteria. Gene 468, 8–19. doi: 10.1016/j.gene.2010.07.012
Jain, P., Hartman, T. E., Eisenberg, N., O’Donnell, M. R., Kriakov, J., Govender, K., et al. (2012). φ(2)GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J. Clin. Microbiol. 50, 1362–1369. doi: 10.1128/jcm.06192-11
Kanno, A. I., Goulart, C., Rofatto, H. K., Oliveira, S. C., Leite, L. C. C., and McFadden, J. (2016). New Recombinant Mycobacterium bovis BCG expression vectors: improving genetic control over mycobacterial promoters. Appl. Environ. Microbiol. 82, 2240–2246. doi: 10.1128/aem.03677-15
Kaps, I., Ehrt, S., Seeber, S., Schnappinger, D., Martin, C., Riley, L. W., et al. (2001). Energy transfer between fluorescent proteins using a co-expression system in Mycobacterium smegmatis. Gene 278, 115–124. doi: 10.1016/s0378-1119(01)00712-0
Kempker, R. R., Heinrichs, M. T., Nikolaishvili, K., Sabulua, I., Bablishvili, N., Gogishvili, S., et al. (2017). Lung tissue concentrations of pyrazinamide among patients with drug-resistant pulmonary tuberculosis. Antimicrob. Agents Chemother. 61:e00226-17. doi: 10.1128/AAC.00226-17
Kim, M. J., Wainwright, H. C., Locketz, M., Bekker, L. G., Walther, G. B., Dittrich, C., et al. (2010). Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol. Med. 2, 258–274. doi: 10.1002/emmm.201000079
Kong, Y., Yang, D., Cirillo, S. L., Li, S., Akin, A., Francis, K. P., et al. (2016). Application of fluorescent protein expressing strains to evaluation of anti-tuberculosis therapeutic efficacy In Vitro and In Vivo. PLoS One 11:e0149972. doi: 10.1371/journal.pone.0149972
Kremers, G.-J., Goedhart, J., van den Heuvel, D. J., Gerritsen, H. C., and Gadella, T. W. J. Jr. (2007). Improved green and blue fluorescent proteins for expression in bacteria and mammalian cells. Biochemistry 46, 3775–3783. doi: 10.1021/bi0622874
Lenaerts, A., Barry, C. E. III, and Dartois, V. (2015). Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol. Rev. 264, 288–307. doi: 10.1111/imr.12252
Lim, H. N., Lee, Y., and Hussein, R. (2011). Fundamental relationship between operon organization and gene expression. Proc. Natl. Acad. Sci. U.S.A. 108, 10626–10631. doi: 10.1073/pnas.1105692108
Lopez Quezada, L., Silve, S., Kelinske, M., Liba, A., Diaz Gonzalez, C., Kotev, M., et al. (2019). Bactericidal disruption of magnesium metallostasis in Mycobacterium tuberculosis is counteracted by mutations in the metal ion transporter CorA. mBio 10:e-1405-19. doi: 10.1128/mBio.01405-19
MacGilvary, N. J., Kevorkian, Y. L., and Tan, S. (2019). Potassium response and homeostasis in Mycobacterium tuberculosis modulates environmental adaptation and is important for host colonization. PLoS Pathog. 15:e1007591. doi: 10.1371/journal.ppat.1007591
MacGilvary, N. J., and Tan, S. (2018). Fluorescent Mycobacterium tuberculosis reporters: illuminating host-pathogen interactions. Pathog. Dis. 76:fty017. doi: 10.1093/femspd/fty017
Marcela Rodriguez, G., and Neyrolles, O. (2014). Metallobiology of tuberculosis. Microbiol. Spectr. 2:e0012-2013. doi: 10.1128/microbiolspec.MGM2-0012-2013
Matz, M. V., Fradkov, A. F., Labas, Y. A., Savitsky, A. P., Zaraisky, A. G., Markelov, M. L., et al. (1999). Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17, 969–973. doi: 10.1038/13657
McCown, P. J., Corbino, K. A., Stav, S., Sherlock, M. E., and Breaker, R. R. (2017). Riboswitch diversity and distribution. RNA 23, 995–1011. doi: 10.1261/rna.061234.117
Nawrocki, E. P., Burge, S. W., Bateman, A., Daub, J., Eberhardt, R. Y., Eddy, S. R., et al. (2015). Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137.
Nesbit, C. E., Levin, M. E., Donnelly-Wu, M. K., and Hatfull, G. F. (1995). Transcriptional regulation of repressor synthesis in mycobacteriophage L5. Mol. Microbiol. 17, 1045–1056. doi: 10.1111/j.1365-2958.1995.mmi_17061045.x
Newton-Foot, M., and Gey van Pittius, N. C. (2013). The complex architecture of mycobacterial promoters. Tuberculosis 93, 60–74. doi: 10.1016/j.tube.2012.08.003
Norrander, J., Kempe, T., and Messing, J. (1983). Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26, 101–106. doi: 10.1016/0378-1119(83)90040-9
Park, Y., Ahn, Y. M., Jonnala, S., Oh, S., Fisher, J. M., Goodwin, M. B., et al. (2019). Inhibition of CorA-Dependent Magnesium Homeostasis Is Cidal in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 63:e01006-19. doi: 10.1128/AAC.01006-19
Piddington, D. L., Kashkouli, A., and Buchmeier, N. A. (2000). Growth of Mycobacterium tuberculosis in a defined medium is very restricted by Acid pH and Mg2+ levels. Infect. Immun. 68, 4518–4522. doi: 10.1128/iai.68.8.4518-4522.2000
Prideaux, B., Via, L. E., Zimmerman, M. D., Eum, S., Sarathy, J., O’Brien, P., et al. (2015). The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat. Med. 21, 1223–1227. doi: 10.1038/nm.3937
Prosser, G., Brandenburg, J., Reiling, N., Barry, C. E., Wilkinson, R. J., and Wilkinson, K. A. (2017). The bacillary and macrophage response to hypoxia in tuberculosis and the consequences for T cell antigen recognition. Microbes Infect. 19, 177–192. doi: 10.1016/j.micinf.2016.10.001
Rohde, K. H., Abramovitch, R. B., and Russell, D. G. (2007). Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2, 352–364. doi: 10.1016/j.chom.2007.09.006
Schwenk, S., and Arnvig, K. B. (2018). Regulatory RNA in Mycobacterium tuberculosis, back to basics. Pathog. Dis. 76:fty035. doi: 10.1093/femspd/fty035
Seeliger, J. C., Topp, S., Sogi, K. M., Previti, M. L., Gallivan, J. P., and Bertozzi, C. R. (2012). A riboswitch-based inducible gene expression system for mycobacteria. PLoS One 7:e29266. doi: 10.1371/journal.pone.0029266
Shaner, N. C., Lambert, G. G., Chammas, A., Ni, Y., Cranfill, P. J., Baird, M. A., et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409. doi: 10.1038/nmeth.2413
Shcherbakova, D. M., Hink, M. A., Joosen, L., Gadella, T. W., and Verkhusha, V. V. (2012). An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J. Am. Chem. Soc. 134, 7913–7923. doi: 10.1021/ja3018972
Sherwood, A. V., and Henkin, T. M. (2016). Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374. doi: 10.1146/annurev-micro-091014-104306
Sinumvayo, J. P., Zhao, C., and Tuyishime, P. (2018). Recent advances and future trends of riboswitches: attractive regulatory tools. World J. Microbiol. Biotechnol. 34:171.
Solans, L., Gonzalo-Asensio, J., Sala, C., Benjak, A., Uplekar, S., Rougemont, J., et al. (2014). The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 10:e1004183. doi: 10.1371/journal.ppat.1004183
Springer, B., Sander, P., Sedlacek, L., Ellrott, K., and Böttger, E. C. (2001). Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int. J. Med. Microbiol. 290, 669–675. doi: 10.1016/s1438-4221(01)80004-7
Stirling, D. R., Suleyman, O., Gil, E., Elks, P. M., Torraca, V., Noursadeghi, M., et al. (2020). Analysis tools to quantify dissemination of pathology in zebrafish larvae. Sci. Rep. 10:3149. doi: 10.1038/s41598-020-59932-1
Stover, C. K., de la Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T., et al. (1991). New use of BCG for recombinant vaccines. Nature 351, 456–460.
Strydom, N., Gupta, S. V., Fox, W. S., Via, L. E., Bang, H., Lee, M., et al. (2019). Tuberculosis drugs’ distribution and emergence of resistance in patient’s lung lesions: a mechanistic model and tool for regimen and dose optimization. PLoS Med. 16:e1002773. doi: 10.1371/journal.pmed.1002773
Subach, O. M., Cranfill, P. J., Davidson, M. W., and Verkhusha, V. V. (2011). An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One 6:e28674. doi: 10.1371/journal.pone.0028674
Sukheja, P., Kumar, P., Mittal, N., Li, S.-G., Singleton, E., Russo, R., et al. (2017). A novel small-molecule inhibitor of the Mycobacterium tuberculosis demethylmenaquinone methyltransferase MenG is bactericidal to both growing and nutritionally deprived persister cells. mBio 8:e02022-16. doi: 10.1128/mBio.02022-16
Sukumar, N., Tan, S., Aldridge, B. B., and Russell, D. G. (2014). Exploitation of Mycobacterium tuberculosis reporter strains to probe the impact of vaccination at sites of infection. PLoS Pathog. 10:e1004394. doi: 10.1371/journal.ppat.1004394
Takaki, K., Davis, J. M., Winglee, K., and Ramakrishnan, L. (2013). Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat. Protoc. 8, 1114–1124. doi: 10.1038/nprot.2013.068
Tan, S., Sukumar, N., Abramovitch, R. B., Parish, T., and Russell, D. G. (2013). Mycobacterium tuberculosis responds to chloride and pH as synergistic cues to the immune status of its host cell. PLoS Pathog. 9:e1003282. doi: 10.1371/journal.ppat.1003282
Tanida, I., Ueno, T., and Uchiyama, Y. (2014). A super-ecliptic, pHluorin-mKate2, tandem fluorescent protein-tagged human LC3 for the monitoring of mammalian autophagy. PLoS One 9:e110600. doi: 10.1371/journal.pone.0110600
Tu, J. M., Chang, M. C., Huang, L. L., Chang, C. D., Huang, H. J., Lee, R. H., et al. (2014). The blue fluorescent protein from Vibrio vulnificus CKM-1 is a useful reporter for plant research. Bot. Stud. 55:79. doi: 10.1186/s40529-014-0079-x
Vandal, O. H., Nathan, C. F., and Ehrt, S. (2009). Acid resistance in Mycobacterium tuberculosis. J. Bacteriol. 191, 4714–4721. doi: 10.1128/jb.00305-09
Vandal, O. H., Pierini, L. M., Schnappinger, D., Nathan, C. F., and Ehrt, S. (2008). A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat. Med. 14, 849–854. doi: 10.1038/nm.1795
Via, L. E., Lin, P. L., Ray, S. M., Carrillo, J., Allen, S. S., Eum, S. Y., et al. (2008). Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76, 2333–2340. doi: 10.1128/iai.01515-07
Wagner, D., Maser, J., Lai, B., Cai, Z., Barry, C. E. III, Höner Zu Bentrup, K., et al. (2005). Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J. Immunol. 174, 1491–1500. doi: 10.4049/jimmunol.174.3.1491
Walters, S. B., Dubnau, E., Kolesnikova, I., Lava, L. F., Daffe, M., and Smith, I. (2006). The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60, 312–330. doi: 10.1111/j.1365-2958.2006.05102.x
Wang, Q., Boshoff, H. I. M., Harrison, J. R., Ray, P. C., Green, S. R., Wyatt, P. G., et al. (2020). PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science 367, 1147–1151. doi: 10.1126/science.aav5912
Warner, D. F., Savvi, S., Mizrahi, V., and Dawes, S. S. (2007). A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J. Bacteriol. 189, 3655–3659. doi: 10.1128/jb.00040-07
Keywords: Mycobacterium tuberculosis, fluorescent protein, reporter strain, riboswitch, pH, magnesium
Citation: Kolbe K, Bell AC, Prosser GA, Assmann M, Yang H-J, Forbes HE, Gallucci S, Mayer-Barber KD, Boshoff HI and Barry CE III (2020) Development and Optimization of Chromosomally-Integrated Fluorescent Mycobacterium tuberculosis Reporter Constructs. Front. Microbiol. 11:591866. doi: 10.3389/fmicb.2020.591866
Received: 05 August 2020; Accepted: 13 November 2020;
Published: 09 December 2020.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Ying Kong, University of Tennessee Health Science Center (UTHSC), United StatesWilliam Jacobs, Albert Einstein College of Medicine, United States
Copyright © 2020 Kolbe, Bell, Prosser, Assmann, Yang, Forbes, Gallucci, Mayer-Barber, Boshoff and Barry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Helena I. Boshoff, SEJPU0hPRkZAbmlhaWQubmloLmdvdg==; Clifton E. Barry III, Q0JBUlJZQG5pYWlkLm5paC5nb3Y=
†These authors have contributed equally to this work
 Katharina Kolbe
Katharina Kolbe Alice C. Bell1†
Alice C. Bell1† Gareth A. Prosser
Gareth A. Prosser Maike Assmann
Maike Assmann He Eun Forbes
He Eun Forbes Katrin D. Mayer-Barber
Katrin D. Mayer-Barber Helena I. Boshoff
Helena I. Boshoff Clifton E. Barry III
Clifton E. Barry III