
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 05 November 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.590522
This article is part of the Research Topic Bacteriocins and Other Ribosomally Synthesised and Post-translationally Modified Peptides (RiPPs) as Alternatives to Antibiotics View all 13 articles
Due to the rapid emergence of multi-resistant bacterial strains in recent decades, the commercially available effective antibiotics are becoming increasingly limited. On the other hand, widespread antimicrobial peptides (AMPs) such as the lantibiotic nisin has been used worldwide for more than 40 years without the appearance of significant bacterial resistance. Lantibiotics are ribosomally synthesized antimicrobials generated by posttranslational modifications. Their biotechnological production is of particular interest to redesign natural scaffolds improving their pharmaceutical properties, which has great potential for therapeutic use in human medicine and other areas. However, conventional protein engineering methods are limited to 20 canonical amino acids prescribed by the genetic code. Therefore, the expansion of the genetic code as the most advanced approach in Synthetic Biology allows the addition of new amino acid building blocks (non-canonical amino acids, ncAAs) during protein translation. We now have solid proof-of-principle evidence that bioexpression with these novel building blocks enabled lantibiotics with chemical properties transcending those produced by natural evolution. The unique scaffolds with novel structural and functional properties are the result of this bioengineering. Here we will critically examine and evaluate the use of the expanded genetic code and its alternatives in lantibiotics research over the last 7 years. We anticipate that Synthetic Biology, using engineered lantibiotics and even more complex scaffolds will be a promising tool to address an urgent problem of antibiotic resistance, especially in a class of multi-drug resistant microbes known as superbugs.
The use of antibiotics has enormously empowered modern medicine to save human lives and perform medical and surgical treatments under safe conditions (Aslam et al., 2018). In addition, treatment with antibiotics is one of the most important approaches to combat or prevent bacterial infections in animal populations and in the food industry. The widespread use of antibiotics began already during the Second World War, and Alexander Fleming himself was one of the first to recognize how improper use of these compounds could lead to antibiotic-resistant bacteria (Fleming, 1945). The habitats of our planet are saturated with various toxic substances, especially those of anthropogenic origin, which has contributed significantly to the selection of resistant strains (Davies and Davies, 2010). Indeed, the excessive use of antibiotics in agriculture, in the environment, animals (food, pets, aquatic fauna) and human medicine, and their release into the environment through fertilizer/feces is the most plausible cause of antimicrobial resistance (AMR) (Manyi-Loh et al., 2018).
The emergence of pathogens causing enhanced morbidity and mortality due to modifications in various physiological and biochemical mechanisms has led to multi-resistant microbes known also as “superbugs” (Coast et al., 1996). For example, in different microbial strains, resistance has emerged at multiple levels of defense, starting from the entry level (through the limitation of drug permeability and the existence of efflux pumps) to the degradation of the antibiotics via their modification or hydrolysis. Next, lateral gene transfer, alteration of binding through target modification and escape of toxicity through bypass reactions and metabolic shunts were (among others) documented as mechanisms for the development of single or multiple resistances (Yelin and Kishony, 2018). To sum up, due to the rapid appearance of multi-drug resistant bacterial strains in last few decades, the use of commercially available antibiotics has become increasingly limited. A promising alternative could be the use of antimicrobial peptides (AMPs) with many variants already in clinical trials (Table 1). Many bioactive peptides produced from various microorganisms (e.g., actinobacteria, cyanobacteria, bacilli, fungi) have a potential to serve as drugs against numerous pathogens (Greber and Dawgul, 2017; Ongey et al., 2017; Koo and Seo, 2019).
Antimicrobial peptides (AMPs) also known as host defence peptides are one of the most versatile natural products which represent alternatives to traditional antibiotics. In fact, AMPs are a special group of natural products that occur in almost every form of life, namely microorganisms, plants, and animals as an innate immune response (Ageitos et al., 2017). They can be used to treat various infections caused by pathogenic species with better efficacy, selectivity and specificity. Structurally, AMPs are versatile peptides with diverse lengths of almost always less than 80 amino acids in a linear or cyclic structure with a broad set of biotechnological applications, e.g., antibiotics, and inhibitors (Oliva et al., 2018).
Antimicrobial peptides can be classified on the basis of their parent organism, their biosynthetic production pathway (e.g., ribosomal or non-ribosomal origin), their activity spectrum, and their structural features (usually based on predominant secondary structure) or physicochemical properties (e.g., cationic/anionic, hydrophilic/hydrophobic etc.). They are structurally divided into five main classes, including (i) linear α-helical peptides, (ii) β–sheet-containing peptides, (iii) peptides with α- and β-structural elements, and (iv) extended or non-αβ secondary structure elements (e.g., polyproline helices) (Wang, 2015). Furthermore, they also occur in the form of (v) cyclic peptides and other topologically complex scaffolds, which are often ribosomally produced and post-translationally modified peptides (RiPPs) (Koehbach and Craik, 2019). Another classification system divides AMPs into natural, encrypted (Brand et al., 2012), and designed AMPs which are a product of specific genes or as part of larger proteins released after proteolysis, and the artificial ones developed by rational design techniques, respectively (Cardoso et al., 2016; Porto et al., 2017). Figure 1 summarizes the various methods of AMPs classification in the current literature.
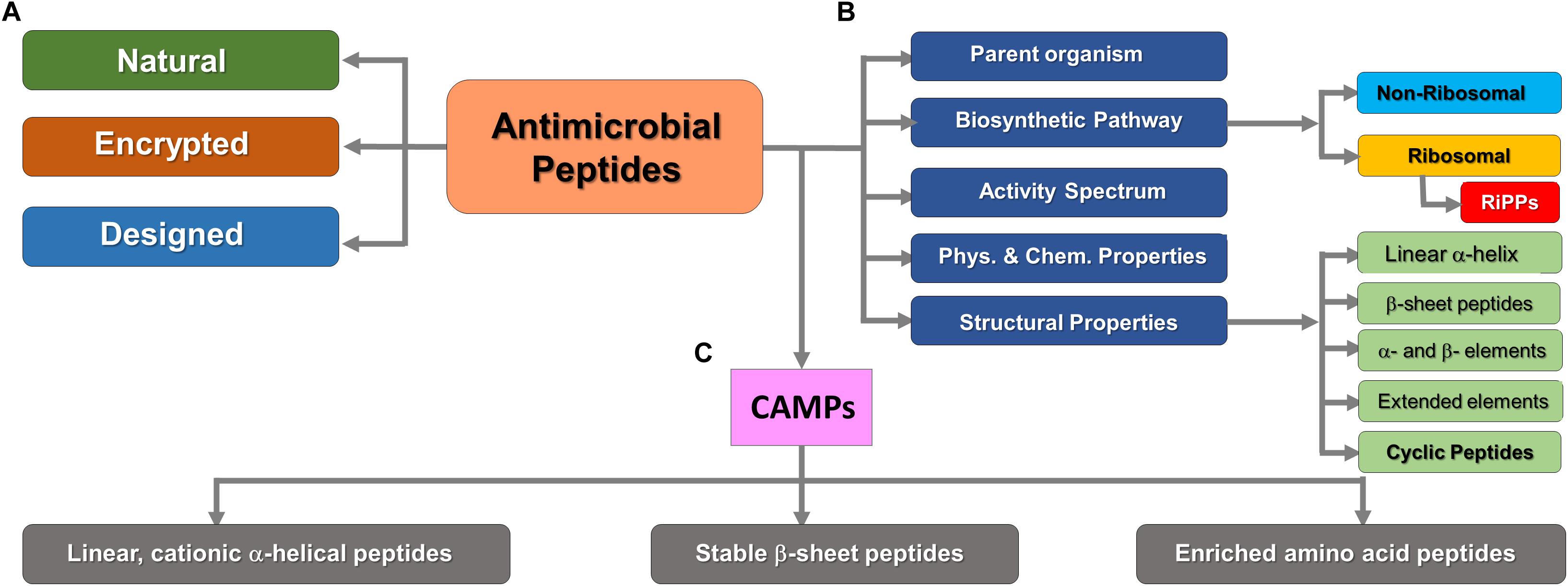
Figure 1. Classification of AMPs on the basis of various criteria. (A) Based on gene encoding pattern (Data-mining strategy). (B) Significant cellular or thermodynamic criteria. (C) Subdivision of positively charged (cationic) AMPs. RiPPs are structurally diverse but exclusively synthesized by ribosomes. This figure was compiled from a variety of sources (Porto et al., 2017; Koehbach and Craik, 2019).
Three models (Carpet/Aggregation/Detergent-like model, Toroidal model, and Barrel-Stave model) have been proposed to explain the mechanism of action of AMPs for the ability to kill bacterial pathogens via membrane permeability (Figure 2). In all these models AMPs act in two sequential steps (Brogden, 2005). In the first step, electrostatic interactions are established between the positively charged amino acids in CAMPs with the negatively charged membrane surface, such as the pyrophosphate group of lipid-II in the cell membrane of microorganism or phosphatidylethanolamines in the outer leaflet of the target cell membrane. In the second step, the formation of pores within the membrane causes the bilayer to break open, which leads to extensive leakages and finally to cell death, particularly in Gram-positive bacteria. Detailed descriptions of detergent-like effects of amphipathic cationic AMPs and other models of membrane disruption by AMPs are described in great detail in the contemporary literature (see e.g., Zweytick et al., 2006). Achievable chemical diversity and variability in the production of peptides together with almost unlimited sequence space for matching their physicochemical properties and generating broad-spectrum antimicrobial activity make them a valid alternative to antibiotics in the effort to combat multi-drug resistant strains and save human lives in the face of future epidemic and other challenges (Ageitos et al., 2017). Nevertheless, several obstacles remain to be overcome in order to develop AMPs for medical use, such as toxicity, stability, and even bacterial resistance. Another obstacle is the lack of standard experimental procedures for quantifying AMP activity and we still do not have a clear picture of several AMP’s mechanisms of action (Torres et al., 2019).
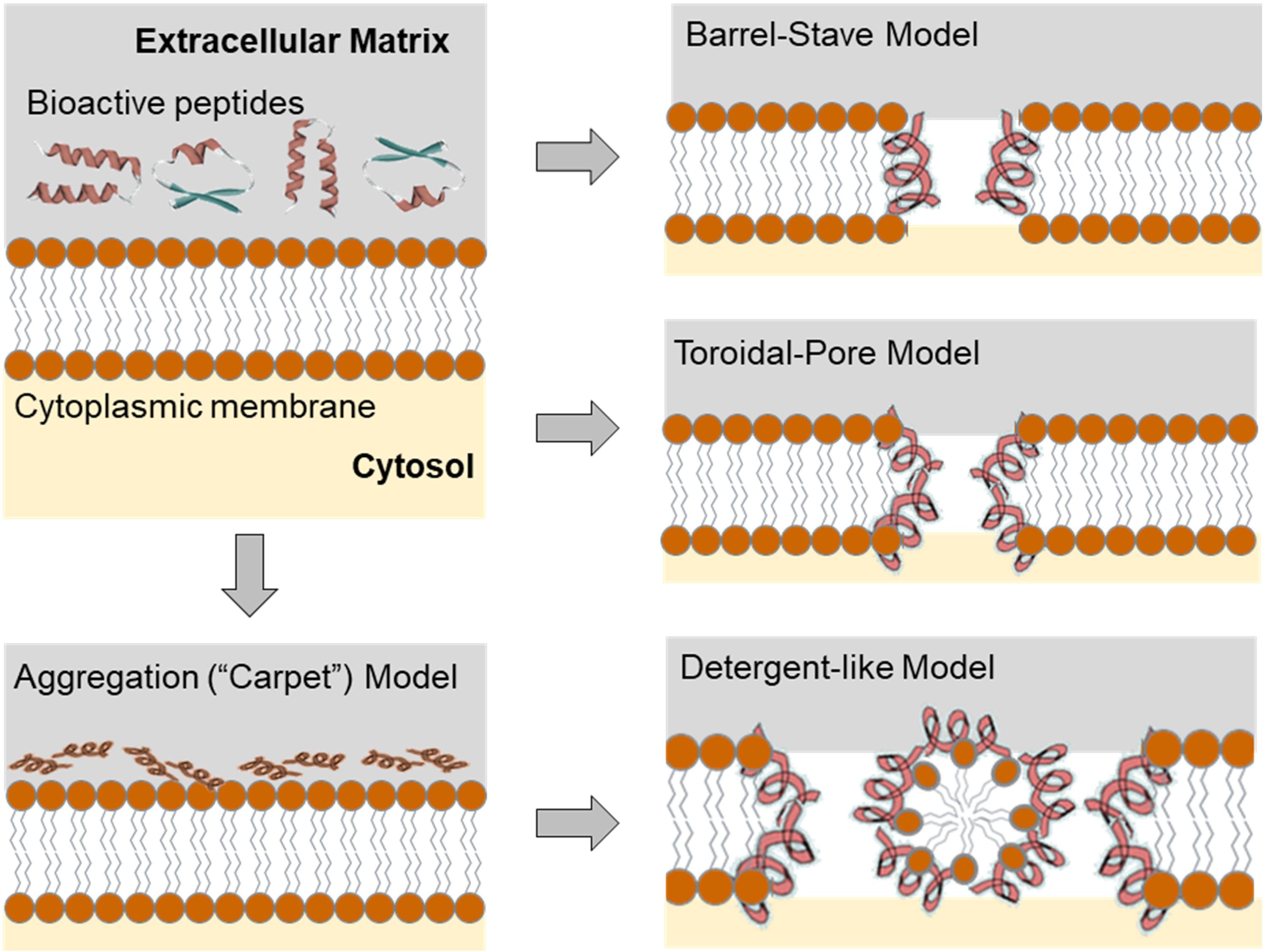
Figure 2. Configuration of the target membrane environment leading to the initial electrostatic and hydrophobic interactions of the AMPs on the bacterial membrane. Initial molecular event of the effect of the AMPs leads to the different scenarios that can be described using transmembrane pore and non-pore models. The carpet model, without pores: AMPs aggregate on the bilayer surface resulting in detergent-like disintegration, whereby the membrane is fragmented into micelles. Conversely, in the “barrel stave” model, membrane pores are formed by interactions between the hydrophobic surface of the pore and the acyl chains of the lipid core of the bilayer. The “toroidal pore” model (wormhole) combines the effects described by both the barrel stave and the carpet model. For a more comprehensive overview see Sun et al. (2018). However, it should be noted that the different mechanisms of AMPs action presented here do not cover the entire range of ribosomally produced and post-translationally modified peptides (RiPPs, vide infra), since not all members of this group damage membranes or form pores, but have different targets and mechanisms of action.
RiPPs are an important class of AMPs, ribosomally synthesized and post-translationally modified bio-active peptides in all three domains of life and show great structural diversity. They are subdivided into more than twenty well-known categories including lanthipeptides, linaridins, proteusins, linear azol(in)e-containing peptides (LAPs), cyanobactins, thiopeptides, bottromycins, microcins, lasso peptides, microviridins, sactipeptides, bacterial head-to-tail cyclized peptides, amatoxins and phallotoxins, cyclotides, orbitides, conopeptides, glycocins, autoinducing peptides (AIPs), pyrroloquinoline quinone (PQQ), pantocin A and thyroid hormones, etc. (Arnison et al., 2013).
In this context, lanthipeptides are classified as one the largest group of RiPPs assigned in class I of bacterial-origin AMPs (bacteriocins). They are transformed into a mature active form by special post-translational modifications (PTMs) carried out by a battery of dedicated enzymes. Class I of bacteriocins include lantibiotics, linear azol(in)e-containing peptides (LAPs), thiopeptides, bottromycins, glycocins, lasso peptides, head-to-tail cyclized bacteriocins, sactibiotics, and lipolanthines (Acedo et al., 2018). Bacterial lanthipeptides exhibit outstanding properties such as conformational rigidity, increased metabolic and chemical stability as well as intense cell permeability that distinguish them from other heat-stable (class-II) and thermo-labile (class-III) bacteriocins (Alvarez-Sieiro et al., 2016). In addition to the four earlier types of lanthipeptides classified based on the biosynthetic mechanisms involved in their structural maturation, the recently discovered modified peptides, including Cocaoidin and Lexapeptide, exhibited novel modification features such as different biosynthetic gene clusters and have been introduced as class V lanthipeptides (Ortiz-López et al., 2020; Xu et al., 2020).
In general, lantibiotics show an efficient ability to kill Gram-positive bacteria and are particularly important for those that exhibit drug resistance such as methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pneumoniae (MRSP), vancomycin intermediate S. aureus (VISA), vancomycin-resistant enterococci (VRE), Clostridioides difficile, etc. Instability and/or insolubility at physiological pH, susceptibility to proteolytic degradation, and a low level of their production are features that mainly limit the use of lantibiotics in the clinic. It should also be noted that the antimicrobial activity of lanthipeptides is well understood and characterized in some RiPPs such as nisin (achieved by lipid-II binding) (Kuipers et al., 1996). Later, the lipid-II binding activity of some lantibiotics was also demonstrated (Wiedemann et al., 2001; Breukink and de Kruijff, 2006), although in some of them the mechanisms of action have yet to be clarified. Nonetheless, low cytotoxicity of lantibiotics to human cells due to the absence of their main target lipid II or negative net charge in eukaryotic membranes (Lagedroste et al., 2019) along with a few examples of naturally occurring resistance (Draper et al., 2015; Khosa et al., 2016a,b; Clemens et al., 2018) make lantibiotics excellent lead compounds for development of new therapeutic options.
A wide range of technologies are at our disposal for producing a peptide such as chemical synthesis, recombinant DNA technologies, and in vitro translation systems. The choice of a strategy for gene-encoded peptide production is largely determined by their size and chemical complexity. Peptides with sophisticated PTMs-generated structures such as RiPPs are preferably produced biosynthetically using host expression cells. Host cells with installed and functional PTM enzymes should be able to express RiPPs in an intact active form with distinctive architecture dominated by lanthionine bridges. In this context, the absence of a widely used large-scale production platform for lantibiotics with consistent quality is one of the main drawbacks of their therapeutic application.
There are generally two options for recombinant expression of lantibiotics, homologous or heterologous expression systems. Homologous expression strategies have limited use in particular due to the difficulty of cultivating the original producers (Maffioli et al., 2015; Mohr et al., 2015) and/or the difficulty of lantibiotics expression induction under laboratory conditions (Wescombe et al., 2011; Garg et al., 2012). Conversely, heterologous biosynthesis with host cells such as Lactococcus lactis, Bacillus subtilis, Bacillus cereus, and E. coli are normally preferred and, in many cases, well established. So far, several genetically manipulatable Streptomyces strains, including Streptomyces lividans (Ahmed et al., 2020), S. albus (Myronovskyi et al., 2018), S. coelicolor M145 (Gomez-Escribano and Bibb, 2011), S. venezuelae ATCC 15439 (Kim et al., 2015), S. avermitilis (Komatsu et al., 2010) have been reported to synthesize natural products from genetically engineered biosynthetic gene clusters. However, Escherichia coli is still the most common and attractive option. A list of successful examples of recombinant expression of different RiPPs families in heterologous hosts such as E. coli and Streptomyces strains were recently compiled by Zhang and colleagues (Zhang et al., 2018). Heterologous production of lantibiotics is generally conducted as inactive form of the antimicrobial peptide (pre-lantibiotic) to prevent any detrimental effects on host cell growth and viability.
In addition, there is great interest to express lantibiotics in microbial host that should facilitate a straightforward and efficient engineering (Kuipers et al., 1996). Numerous conventional protein engineering approaches such as site-directed mutagenesis, directed evolution and various computational tools aid synthetic biologists and biochemists to selectively diversify the properties of expressed therapeutic peptides. These include manipulations of thermodynamic stability, increased bioavailability, reduced aggregation or enhanced specificity and proteolytic stability (Adhikari et al., 2019). In addition, the implementation of in vitro and in vivo approaches in combination with genome mining data and high-throughput screening strategies has opened up unprecedented opportunities to modify and even improve antimicrobial activity, manipulate the physicochemical properties and widening of the antibacterial spectrum in the production of lantibiotics (Field et al., 2015). Recently an attempt to design and biosynthesize a two-lipid II binding motifs-containing lantibiotic, called TL19, is the latest example showing 64-fold stronger activity against Enterococcus faecium than nisin (Zhao et al., 2020).
The potential of classical protein engineering in complex RiPPs is perhaps best illustrated by recombinant nisin, well known for its broad-spectrum antibacterial activity produced in certain strains of L. lactis (Lubelski et al., 2008). Nisin as the most widely used lantibiotic in the food industry over the past 50 years has undergone various methods of bioengineering with an aim to improving its function and/or physicochemical features (Shin et al., 2016). To expand the scope of its activity, several bioengineered variants of nisin have been generated by site-directed mutagenesis and by classical chemical modifications (Ross et al., 2012), including in vitro chemical synthesis (Knerr and van der Donk, 2012; Koopmans et al., 2015). For example, the use of the saturation mutagenesis approach has led to the production of nisin S29 derivatives with increased activity against a number of Gram-positive antibiotic-resistant pathogens. In addition, the increased antimicrobial activity was generated in comparison to the wild type nisin A when tested against Gram-negative food-borne pathogens (Field et al., 2012). Moreover, recombinant production of the nisin Z mutants N20K, M21K, N27K, H31K improved peptide solubility at alkaline pH (Rollema et al., 1995). Next, residue alterations at distinct locations enabled the improvement of antimicrobial activity (Islam et al., 2009; Healy et al., 2013; Geng and Smith, 2018), the enhancement of diffusion through complex polymers (Rouse et al., 2012), and widening effect on some Gram-negative bacteria (Field et al., 2012).
Studies on the effects of the hinge region (NMK) length between rings C and D on antimicrobial activity and host specificity of nisin was also performed (Zhou et al., 2015). Although most variants with shorter or larger hinge length are less active than the wild type, some variants (+2, +1, −1, −2) exhibited higher antimicrobial activity than the wild type nisin A in agar-well-diffusion assays against L. lactis MG1363, Listeria monocytogenes, Enterococcus faecalis VE14089, Bacillus sporothermodurans IC4, and Bacillus cereus 4153. In addition, an extended nisin A variant of the hinge region (20NMKIV24) has been introduced, bypassing the human pathogen’s lantibiotic resistance while showing a slight decrease in antimicrobial activity (Zaschke-Kriesche et al., 2019). In this context, Figure 3 represents different variants of nisin produced by classical protein engineering together with a graphical representation of the mechanism of action of this bioactive peptide.
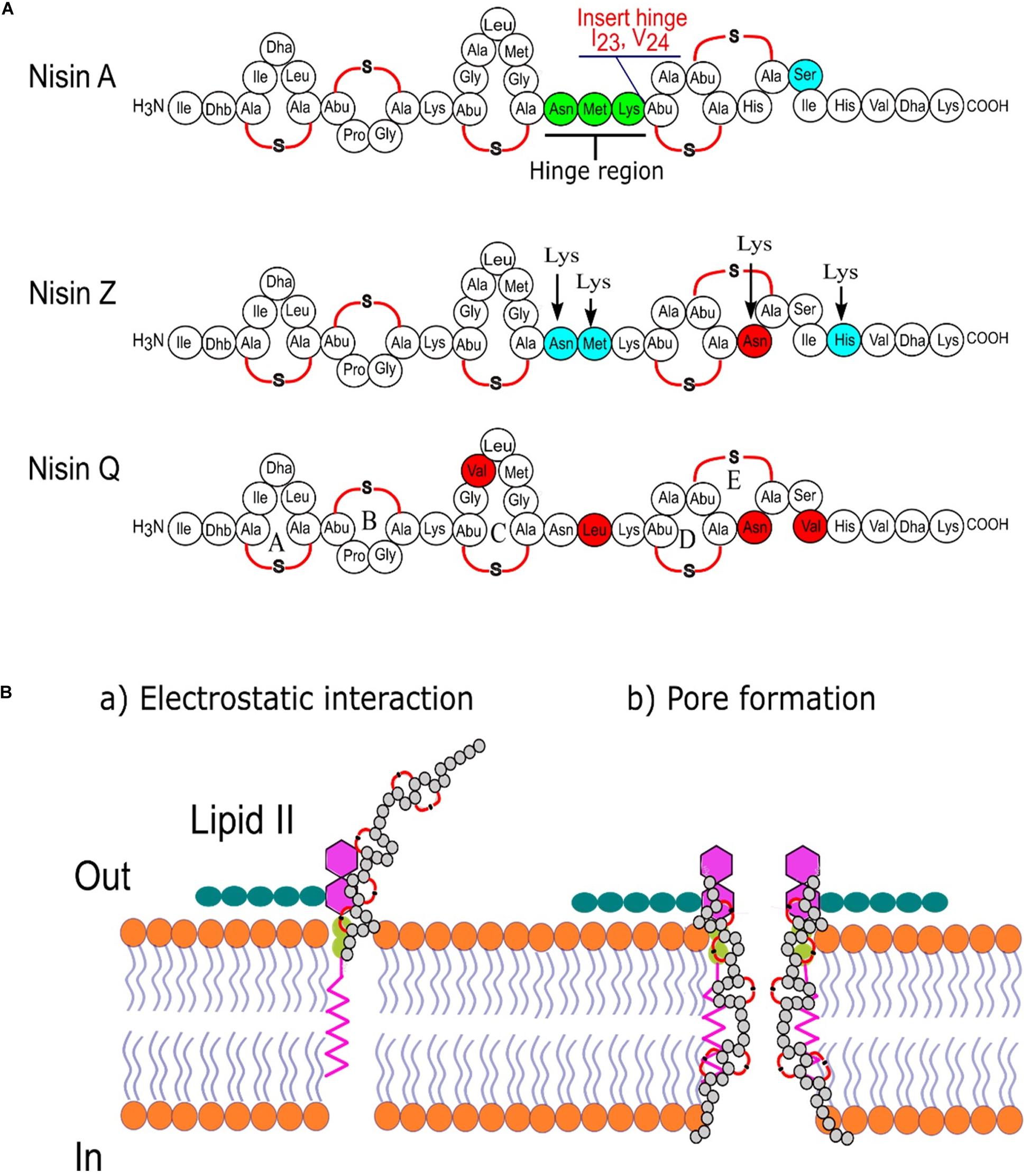
Figure 3. Nisin is one of the best-studied ribosomally synthesized, pore-forming, cationic, antimicrobial peptides. (A) Sequence comparison of three nisin variants from L. lactis and marking the important regions of nisin targeted for bioengineering by classical molecular biology approaches (Chatterjee et al., 2005); (B) two-steps mode of action of nisin.
However, the expression of lantibiotics in popular prokaryotic hosts like E. coli is challenging as these bacteria have no enzymes to perform suitable PTMs to convert pro-peptide into a mature bioactive peptide. Therefore, their recombinant expression has to be coupled with the co-expression of active PTM biosynthetic gene clusters either in vitro or in vivo. These strategies are not in the focus of our study and interested readers are directed to numerous studies and reviews dedicated to this topic (Zhang et al., 2018; Myronovskyi and Luzhetskyy, 2019). We are mainly concerned with the in vivo design strategies that focus to expand the functional scope of AMPs from ribosomal templates beyond the classical protein engineering approaches.
It was argued that chemical and functional diversity delivered by PTMs in AMPs such as lantibiotics can be further supplemented or even expanded by the co-translational insertion of non-canonical amino acids (ncAAs) (Budisa, 2013). Indeed, a limited and conservative set of 20 canonical amino acids used by ribosomes to encode polypeptides in nature usually does not cover enough chemical space required to substantially expand their functional and structural diversity. In nature, this is normally achieved by site-selective PTMs that create special non-canonical amino acid side chains such as dehydroalanine (Dha) and dehydrobutyrine (Dhb) in lantibiotics (Figure 3). To directly mimic these and similar PTMs, chemists used, e.g., rhodium-catalyzed arylations (Key and Miller, 2017), P450-catalyzed cyclopropanations (Gober et al., 2017), and photocatalytic activation (de Bruijn and Roelfes, 2018) in thiopeptides, as well as metalloporphyrin-catalyzed alkylation of methionine in nisin (Maaskant and Roelfes, 2019) as attempts to obtaining novel AMPs with improved properties and/or activities. However, it is difficult to mimic PTM machinery chemically, whereas classical peptide synthetic protocols such as solid-phase peptide synthesis (SPPS) could not cover the chemical complexity of these natural products.
Therefore, the use of recombinant DNA technology that operates with heterologous expression of biosynthetic gene clusters and enriched with reprogrammed protein translation (Figure 4) can provide a reasonable solution to the above-mentioned problems (Hoesl and Budisa, 2012). It enables specific insertion of the biological, chemical, and physical properties delivered by ncAAs that can be accurately defined by the chemist at the bench. Cells equipped with various bioorthogonal chemistries have the potential to perform catalytic transformations that are not found in biology, including the creation of new metabolic and informational pathways (Devaraj, 2018). These modifications can also serve as redox sensors, spectroscopic markers (e.g., spin-labeling) or sensors of protein-ligand interactions. Metal-binding amino acids could lead to new structural, catalytic, or regulatory elements in proteins, while diazirines allow site-specific photo-crosslinking of the target protein to its substrate. Photoactive and photo-isomerizable ncAAs with novel photo-physical properties such as azobenzene side chains can be used to photo-regulate protein activity. Similarly, photocaged ncAAs can be activated by light to turn on enzymatic activity spatiotemporally (for review see Young and Schultz, 2010).
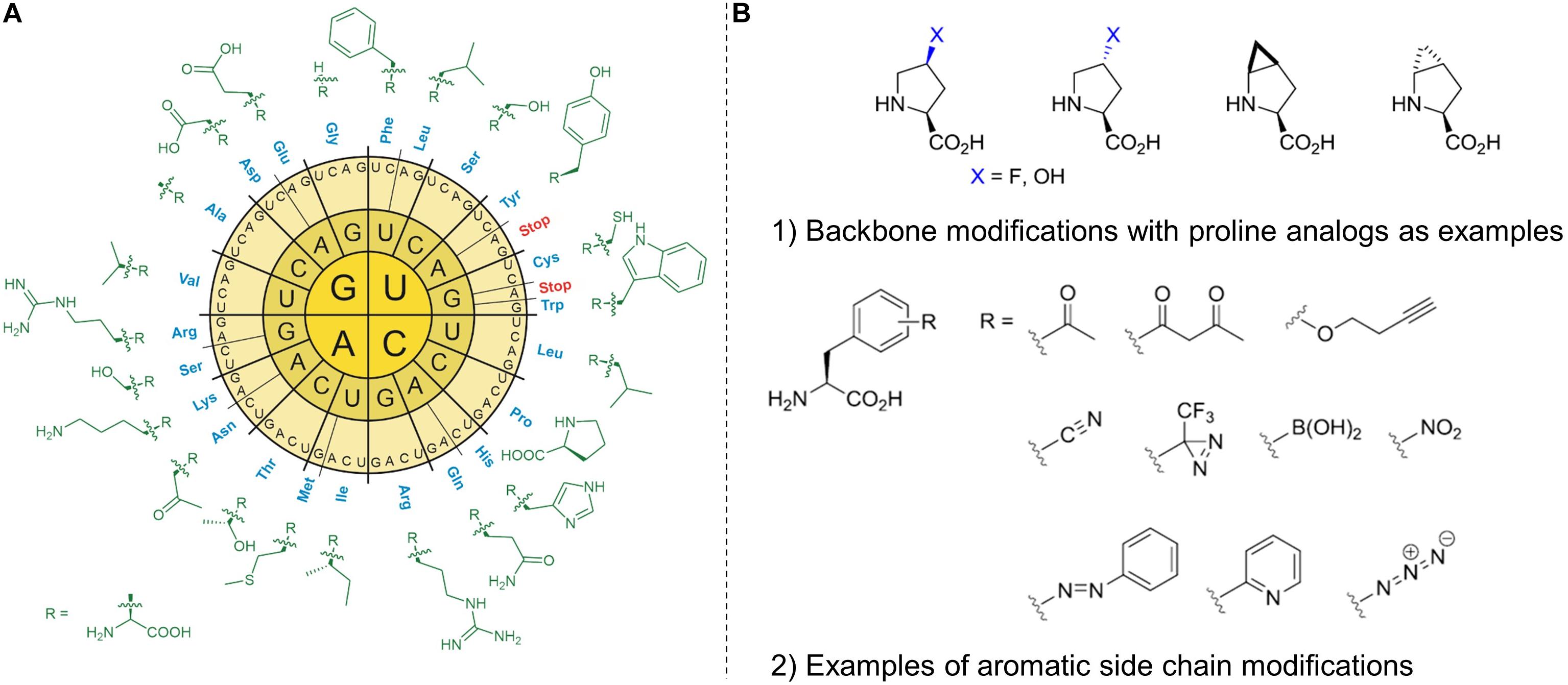
Figure 4. The radial arrangement of the standard genetic code in RNA format (A) with the side chains of 20 canonical amino acids (cAA). The standard amino acid repertoire of the genetic code can be modified or expanded (B) by non-canonical amino acids (ncAAs). By reprogramming protein translation, the ncAAs can be incorporated into recombinant proteins using in vivo and in vitro methods. These insertions offer useful modifications that can be made to the protein main chain (backbone) and the amino acid side chains (aliphatic or aromatic). Modified backbones and side chains often have unique physicochemical properties with the potential to dramatically expand the chemical and functional space of ribosomally synthesized peptides and proteins.
Directed evolution of aminoacyl-tRNA synthase (aaRS) plays a key role in the generation of such molecular tools. Desired ncAAs can be incorporated in a site-directed manner by reading in-frame stop codons, usually amber stop codons (UAG) with a suitable orthogonal pair (o-pair). Such a pair consists of an aaRS enzyme to activate the ncAA and its associated orthogonal tRNA (vide infra). However, the transition to recombinant production with such a possibility by using heterologous expression hosts is not trivial. For that reason, the residue-specific replacement of a particular amino acid at all positions in a protein sequence (known as selective pressure incorporation, vide infra) is a reasonable alternative as it does not require o-pairs (Budisa, 2004). Finally, the capacity of ncAAs incorporation should not interfere with the sequence of posttranslational modifications leading to the formation of lantibiotics and the restoration of their bioactivity.
With such systems in hands we would have very sophisticated tools to rationally engineer RiPPs scaffolds as now they can be redesigned by reprogrammed protein translation which utilize various ncAAs. This approach would have many advantages over the above discussed classical chemical or recombinant approaches as it enables: (i) genetic encoding of desired ncAAs and its sequence positioning; (ii) recombinant production under mild conditions at room temperature and atmospheric pressure; (iii) the targeted functionalization (e.g., various site-directed bioconjugations, light induced cross-linking, metal or cofactor binding, fluorophore or pharmacophore attachment, adhesiveness, etc.) (Cropp and Schultz, 2004). Crucial requirements to expand the scope of protein synthesis with ncAAs should be cellular uptake, their intracellular metabolic stability and translational activity (i.e., incorporation). Finally, it is necessary to reallocate (reassign) coding triplets (codons) in the genetic code to insert ncAAs in target protein/peptide sequences.
The bioexpression of lantibiotics is not fully comparable to the routine recombinant expression of soluble proteins because these polypeptides contain precursor (consisting of leader and core peptide) and recognition sequences. The core peptide is modified by post-translational modification (PTM) enzymes and upon proteolysis and export, transformed into a complex natural product. Unlike non-ribosomal natural products, native RiPPs cannot explore amino acids beyond the canonical 20 proteinogenic amino acids in the synthesis of precursor peptides, limiting their structural diversity at this level of ribosomal synthesis. On the other hand, from a biotechnological point of view, the great advantage of the ribosomal synthesis (coupled efficiently with PTMs) of bioactive compounds would be the possibility of achieving high chemical diversity at low genetic cost and to avoid supply of expensive precursors. The drawback that many unusual building blocks (found in non-ribosomal synthesis) are absent in RiPPs can be efficiently circumvented by genetic code expansion (GCE). The insertion of ncAAs (in different combinations of their numbers and chemistry) into growing peptide represents a novel level of chemical diversification of these sequences (Figure 5). Incorporated ncAAs can either directly interfere with PTMs or occupy sequence positions not affected by the PTM machineries. This provides suitable bio-orthogonal reactive groups (‘handles’) amenable for various chemoselective ligand couplings. In addition, both site- and residue-specific incorporation techniques generally allow for fine chemical manipulations of the amino acid side chains, e.g., of Pro, Trp, Tyr, Met and to visualize them in spectroscopic recordings (Baumann et al., 2018).
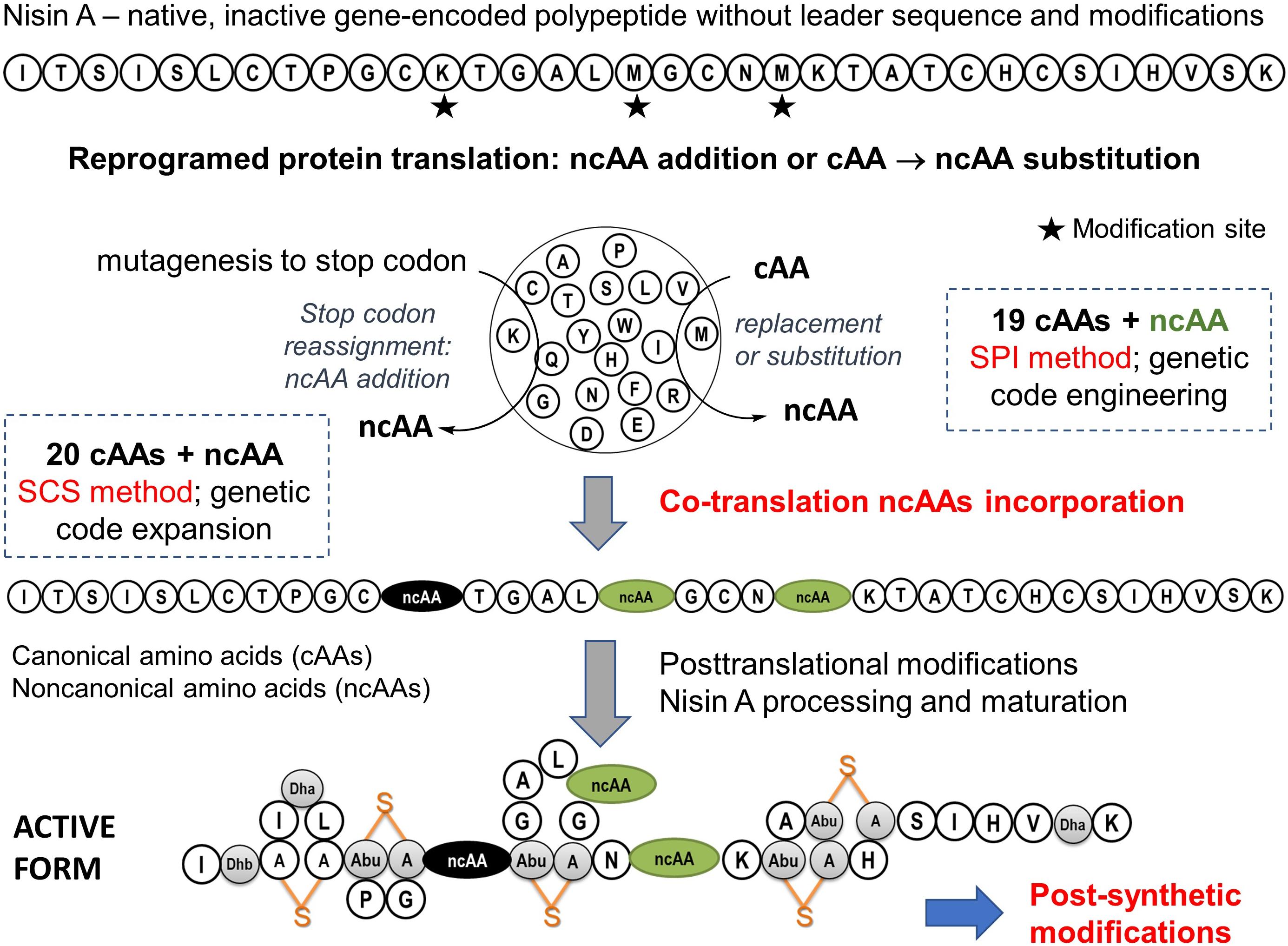
Figure 5. Expansion of scope of the nisin A biosynthesis by ncAAs. The biosynthetic production of active nisin involves the expression of peptide precursors with 20 canonical amino acids as standard building blocks, followed by posttranslational modifications (PTMs). The active form of natural nisin is characterized by the presence of unusual posttranslationally introduced bridge structures known as lanthionines (depicted in orange). It is generally expected that the incorporation of ncAAs (i.e., co-translational modifications) along with post-synthetic modification options (e.g., click chemistry) will create additional functional and structural levels of RiPPs diversification. For example, new variants with improved properties such as stability, specificity, bioavailability, action spectrum and half-life could be created. This is particularly important in view of the fact that nisin is highly effective against a number of human clinical pathogens, including many multi-drug resistant strains (Piper et al., 2009). In recent years it has been clearly demonstrated that several variants of nisin increase activity or stability in different experimental setups (reviewed in Field et al., 2015).
Non-canonical amino acids equipped with specific chemical groups (azides, olefins, ketones and aldehydes, loaded and unloaded alkynes, halogens, oximes, hydrazones, boronic acid esters and acids, etc.) give polypeptide sequences a unique reactivity and chemoselectivity that allow easy and efficient bioconjugations to a variety of ligands (Agostini et al., 2017). The copper(I)-catalyzed Huisgen cycloaddition reaction between azides and alkynes (also known as “click chemistry”) is widely used to functionalize labeled proteins since it takes place under physiological conditions allowing full retention of the protein structure (Kolb et al., 2001). For example, click chemistry proved to be efficient in producing nisin-peptoid hybrids for therapeutic use (Bolt et al., 2018). Further, in vitro metathesis reaction confirmed the capability of the variants for post-biosynthetic modifications such as conjugation reactions with ligands, labels, or tags using bio-orthogonal chemistry. Other alternatives include recently developed very efficient chemoselective methods; mainly copper-click, photoclick, and catalyzed oxime/hydrazone chemistries (Devaraj, 2018). This methodology could provide the RiPPs derivatives with an increase in chemical diversity such as enhanced proteolytic resistance, or increased bioavailability. Finally, bioexpression with various ncAAs combined with the possibilities for further chemical processing of RiPPs after maturation (i.e., semi-synthesis of mature products) makes the number of possible chemical combinations in antibiotic design virtually limitless.
To gain access to such a sophisticated chemistry in recombinant proteins, the incorporation of suitable ncAAs must be established in order to have possibilities for the site- or residue-specific functionalization of e.g., bioorthogonal alkyne- or azide-containing moieties. The simplest strategy for the in vivo insertion of ncAAs is based on the use of auxotrophic microbial systems and is known as selective pressure incorporation (SPI) (Minks et al., 2000). This methodology also exploits natural substrate tolerance (“substrate promiscuity”) of protein translation machinery components toward ncAAs analogs. For example, with a Met-auxotrophic E. coli strain in Met-depleted medium, it was possible to express proteins in which methionine was replaced by different medium-supplemented Met analogs with a terminal azide or alkyne as a side chain (Figure 6). The incorporation protocol typically requires the addition of the ncAA analog in the growing medium, which is taken up by the cell machinery and used in protein translation (Budisa, 2004). Furthermore, the SPI is well known for higher translation yields without the need for extensive host engineering.
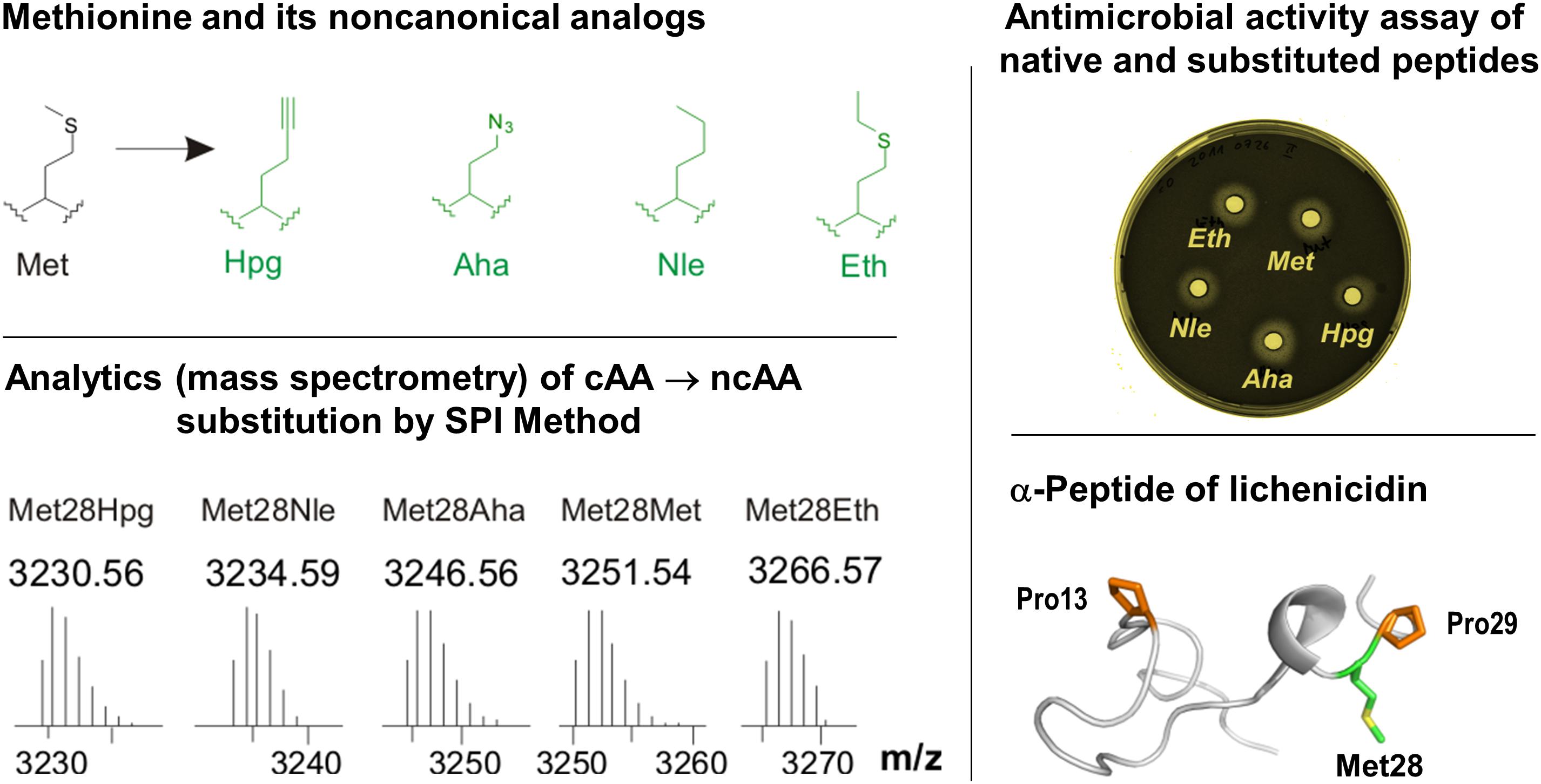
Figure 6. Incorporation of Met analogs into the one subunit of the two-component lantibiotic lichenicidin using the SPI method. Top left: Structures of methionine and its analogs. Met, L-methionine; Aha, L-azidohomoalanine; Hpg, L-homopropargylglycine; Nle, L-norleucine and Eth, L-ethionine. These analogs were used to individually replace Met residues in the precursor peptide of lichenicidin (bottom right), which was also analytically confirmed (bottom left). The substituted α-peptide of lichenicidin underwent PTM-activity, whereby the linear precursor peptide was converted into a polycyclic form that exhibits antimicrobial activity when mixed with the β-peptide (top right). For more details see Oldach et al. (2012). Most recently, Kuipers and co-workers have demonstrated the use of click chemistry in the engineering of nisin by using SPI with the methionine analogs Hpg and Aha (Deng et al., 2020).
The SPI method essentially requires a genetically and metabolically stable auxotrophic expression host with ncAA recognized and activated by endogenous translation apparatus. Efficient and robust protein expression systems with controlled fermentation and expression conditions usually yield target proteins often similar to the wt-counterparts. This was demonstrated in 2012, when the first application of the SPI method for RiPPs was reported (Oldach et al., 2012). In particular, the residue-specific incorporation of various Trp, Met, and Pro analogs into the modular two-component lantibiotic lichenicidin proved to be fully attainable (Figure 6). In this system, the plasmid-encoded SPI-based expression of modular propeptides is coupled with the expression of the fosmid-encoded PTM machinery in the E. coli host cells. It has also been shown that lichenicidin with the Met analog homopropragylglycine (Hpg) can be post-synthetically functionalized by click chemistry using azidofluorescein as a ligand.
However, the fundamental disadvantage of the SPI method is that it allows residue-specific substitution of the canonical amino acid of interest by isosteric analogs, uncontrolled substitutions in the entire proteome, possibly endangering the heterologous PTM machinery and thus damaging the host expression cell. Furthermore, often in this approach the incorporation of ncAA is achieved in a statistical manner, which leads to a heterogeneity of the protein samples. This can be circumvented by using site-directed incorporation methods (Figure 7). However, if a site-directed ncAA incorporation is desired, the protein translation must be orthogonalized. A specific position in the protein to be labeled is mutated to the in-frame stop (usually amber) codon in the target gene. An essential part of this approach is therefore the introduction of in-frame stop codons (UAG) in target mRNAs decoded by the ribosome. In-frame stop codon suppression (i.e., readthrough) is achieved via a specially designed suppressor tRNA charged with ncAA (by a dedicated enzyme) and leads to the ncAA incorporation in a site-specific manner. This expands the amino acid repertoire of the genetic code and provides a basis for the technology known as stop-codon suppression (SCS) also as GCE. GCE generally requires the introduction of orthogonal aminoacyl-tRNA synthetase:tRNA pairs (o-pairs) into recombinant expression systems. Heterologous expression systems operating with orthogonal pairs are also known as orthogonal translation systems (OTS) in the literature. This research area is very well covered by a number of recent reviews (see, for example, Soye et al., 2015; Arranz-Gibert et al., 2018).
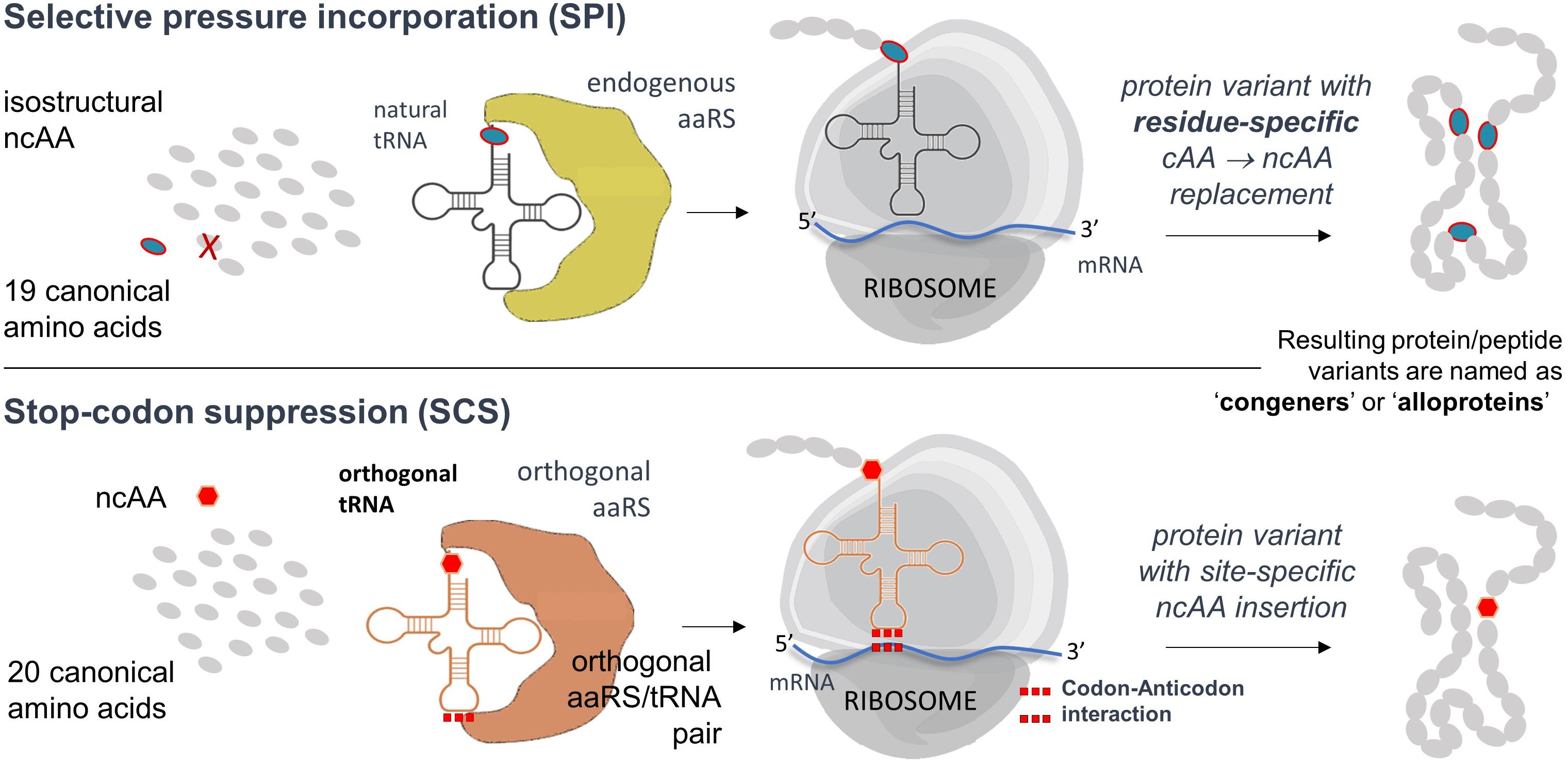
Figure 7. Flow chart of the SPI and SCS methods. Both methods can be performed separately or combined in a single expression experiment (Hoesl and Budisa, 2011). Further details, advantages and disadvantages of both methods are described in the text.
In orthogonal translation, usually a TAG (amber) stop codon is assigned to the ncAA and the orthogonal tRNA anticodon is mutated to CUA for position-specific incorporation. Orthogonal pairs of archaeal origin can be into eubacterial (such as E. coli) or eukaryotic hosts (such as Saccharomyces cerevisiae) and used to affect orthogonal translation (Wang et al., 2006). In particular, to engineer o-pairs, it is necessary to redesign an existing aaRS to activate the desired new ncAA substrate. Commonly used orthogonal pairs are based on Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (MjTyrRS) and Methanosarcina mazei/barkeri pyrrolysyl-tRNA synthetases (MmPylRS or MbPylRS). Compared to E. coli or eukaryotic hosts they show a distant phylogeny (due to their archaeal origin) that allows their implementation as o-pairs (i.e., they do not show cross-reactivity with the endogenous translational apparatus) (Budisa, 2006). Enzyme variants are generated by mutagenesis of the plasmid library and selected iterative positive/negative selection cycles (Bryson et al., 2017). This technology enables the incorporation of ncAAs containing different chemical entities into protein, both at one position (site-directed) and at several positions (multi-site mode). Recently, we have developed a computer-aided approach (Baumann et al., 2019) to create highly diversified libraries of aminoacyl-tRNA synthetases (AARSs) and have demonstrated the engineering of orthogonal translations with up to 50-fold improved insertion efficiency. This was made possible by creating much larger libraries (with > 109 clones). In conventional directed evolution protocols, only about 107 clones are generated.
We and others have already applied and optimized orthogonal pairs for site-specific incorporation of fluorinated amino acids (Völler et al., 2015). Recently, we also have developed a novel and highly efficient orthogonal tyrosyl-tRNA synthetase from Methanocaldococcus jannaschii (MjTyrRS) for photo-activatable L-DOPA derivatives (Hauf et al., 2017). Similarly, we evolved a novel pyrrolysyl-tRNA synthetase from Methanosarcina mazei (MmPylRS), which is able to incorporate S-allylcysteine (Sac) in response to amber stop codons (Exner et al., 2017). This Mm- or Methanosarcina barkeri PylRS (MbPylRS) variants should be an ideal starting enzyme for the evolution of enzyme mutants with high specificity for smaller substrates. These and other advances in the field are recently summarized in a series of specific reviews (Arranz-Gibert et al., 2019; Budisa and Schneider, 2019) and highlights (Mayer, 2019; Yanagisawa et al., 2019).
Without a doubt, GCE will soon become a groundbreaking technology that will allow the catalysis of reactions that are not possible with canonical amino acids alone. More than 200 distinct ncAAs have been incorporated into various peptides and proteins so far (Vargas-Rodriguez et al., 2018). This technology has already yielded new-to-nature RiPPs and proteins in general with improved or novel physicochemical and biological properties which enabled the study of the structure-function relationship of proteins, imaging (Uttamapinant et al., 2015), probing (Chatterjee et al., 2013) and even the development of new therapeutics. Furthermore, it is possible to mask side chain functionalities of amino acids of interest by replacing them site-specifically with e.g., photo-caged ncAA. These masked substrates can then be non-invasively unmasked by UV light irradiation revealing critical functionalities (e.g., photo-caged residues which are substrates for MjTyrRS and Mm or MbPylRS-based orthogonal translation) (Drienovská and Roelfes, 2020). For the incorporation of ncAAs into RiPPs, various methods have been considered so far, ranging from in vitro synthesis reactions to residue and site-specific in vivo incorporation (Bartholomae et al., 2018). To maximize the yield and cost-effective production of lantibiotics, the biological synthesis of these compounds offers a viable alternative strategy with less downstream processing.
The deeper understanding of the molecular mechanisms behind the activities of AMPs, their selectivity and rational manipulation requires the application of novel technologies of peptide modifications. Incorporation of ncAAs is a particularly promising tool, which is able to deliver new chemical functions and specific probes into peptide structures. For example, Nagao et al. reported the first example of heterologous expression of the type II lantibiotic, nukacin ISK-1 in E. coli by co-expression of NukA and the modification enzyme NukM (Nagao et al., 2005). In the following years, this approach was increasingly replaced by the insertion of different ncAAs that mimic different PTMs, such as the photocrosslinker p-Benzoyl-L-Phe (pBpa) in protein and proteome chemistry due to the orthogonal translation rapid development using different vectors and expression systems in E. coli (Miyazaki et al., 2018). In general, the ncAA-insertion provides many advantageous properties when compared with PTMs, such as functional diversity, probing and controllable activity. In this context, Table 2 represents a list of the new-to-nature RiPPs biosynthetically produced via GCE methodology.
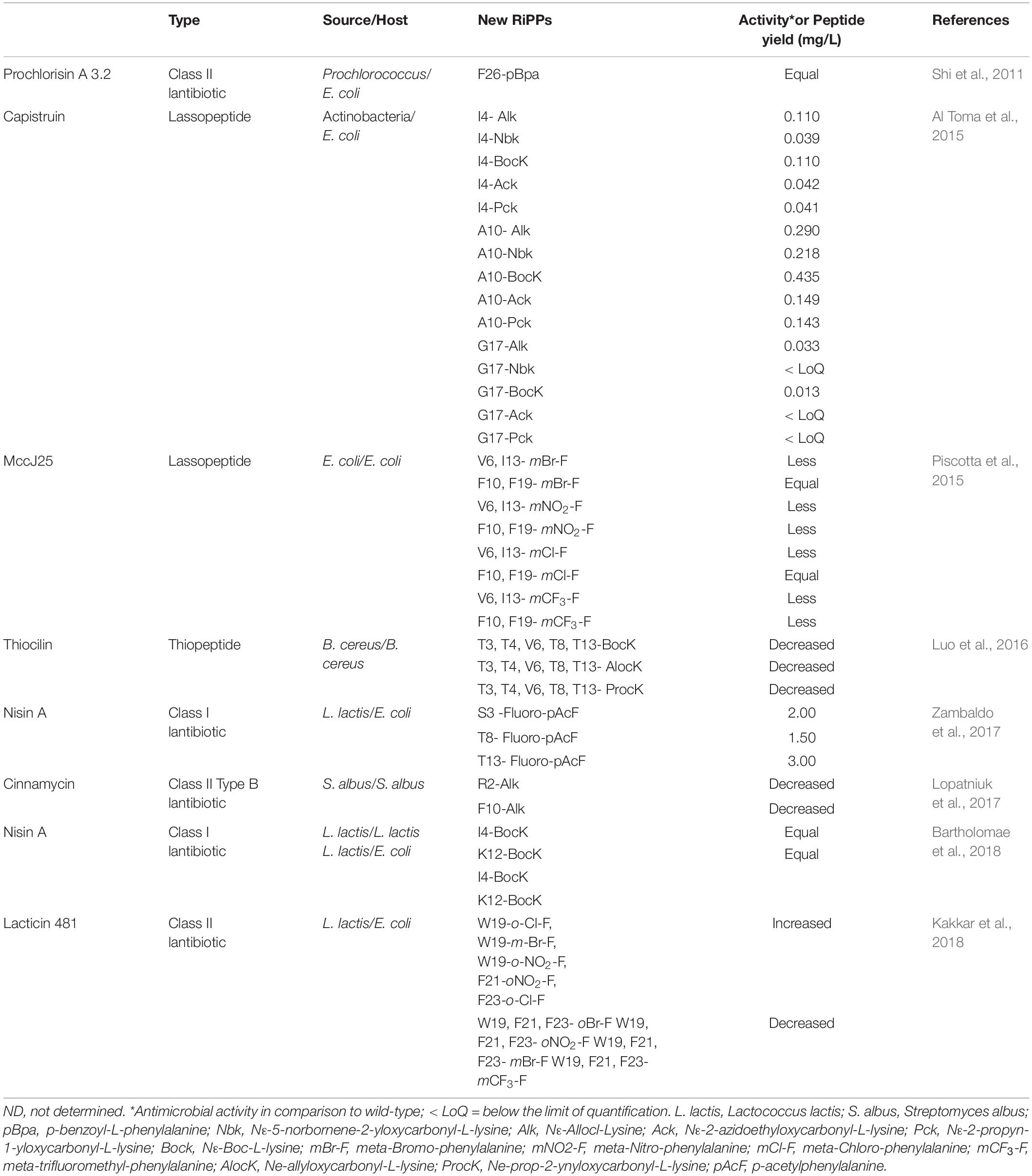
Table 2. List of the new-to-nature RiPPs produced by genetic code expansion technology reported so far.
We define lanthipeptides expressed recombinantly with ncAAs as “new-to-nature” because they are endowed with chemical functionalities that are normally a domain of classical organic chemistry. Most of such genetically encoded modifications introduced in proteins and peptides (in vivo and in vitro) take place on amino acid side chains and very seldom on protein backbones (like e.g., modified proline analogs; see Figure 4; Larregola et al., 2012). For example, D-amino acids, β-, or γ-amino acids, dipeptides and dipeptidomimetic analogs can be inserted into target polypeptide sequences by using engineered in vitro translation systems (Lee et al., 2019). Translation with backbone modification is difficult because ribosomes are being evolved to facilitate the polymerization of α-L-amino acids into polypeptides. Since the majority of the canonical amino acids can be considered to be derivatives of alanine (Kubyshkin and Budisa, 2019), the ncAA insertions (via SCS) or substitutions (via SPI) are mainly additions of new side-chain functionalities- a sort of useful co-translational addition to existing PTMs that should greatly expand the functional performance of modified peptides and proteins.
The engineering of orthogonal aminoacyl-tRNA synthetase/tRNA orthogonal pairs (o-pairs) for the site-specific incorporation of ncAAs is undoubtedly an important tool in protein and peptide design. However, it should be noted that orthogonalization of protein translation is usually associated with a significant decrease in system performance. Therefore, bio-expressions with orthogonal pairs require that the desired ncAA is available in large excess (Völler and Budisa, 2017). In addition, o-pairs based on both MjTyrRS and Mm(Mb)PylRS scaffolds can have advantages and disadvantages in different environments. In bacterial host cells, MjTyrRS often show a higher performance compared with PylRS-based systems (Rauch et al., 2016). In general, orthogonal translation with PylRS-derived o-pairs leads to lower yields of target protein, since this enzyme generally has a low catalytic efficiency (Baumann et al., 2016). While o-pairs based on MjTyrRS allow a higher number of in-frame stop codons that can be suppressed, this is not the case with orthogonal translation based on PylRS (Zhao et al., 2018). Many strategies have been reported to alleviate these limitations, including the computational redesign of both enzymes as discussed elsewhere (Baumann et al., 2019).
The incorporation of ncAAs via SCS into proteins in general and RiPPs in particular is usually associated with reduced expression efficiency since the readthrough of stop codons is context-dependent. In particular, the insertion of in-frame stop codons in target sequences might generate so-called “context effects” with the competition with ribosomal release factors (RFs) for the readthrough (Plotkin and Kudla, 2011). Next, the insertion of a stop codon into a specific sequence is often associated with a high level of interference with natural mRNA folding (Gorochowski et al., 2015). This requires the optimization of mRNA sequences by applying a computer-aided design that should restore or even improve the strength of binding site interaction within the ribosome (Del Campo et al., 2015). For these reasons, “context effects” during the incorporation of ncAA in target sequences were investigated by randomly generating in-frame stop codons along the gene sequence with different proteins (summarized in Agostini et al., 2017).
The screening of an amber stop codon library with lantibiotic genes to study site-specific ncAAs incorporation efficiency could be of great importance to improve and enhance the biosynthesis of RiPPs with ncAAs. For example, we and others recently established a reporter platform for SCS approach using three expression vectors capable of independent control of different groups of genes. Each sense codon in the core peptide region of NisA was replaced by TAG (amber) codon and screening of possibly suitable incorporation positions was assessed upon incorporation of various analogs (Bartholomae et al., 2018). Next, in addition to the feasibility of a position to incorporate ncAAs, the admissibility of target genes that can fold and modify is critical and must always be considered. This is best illustrated by the report of Shi et al. (2011), in which the orthogonal translation was performed with three different types of bioactive peptides, prochlorosins from class II lanthipeptides, two-component lantibiotic haloduracin and the class I lantibiotic, nisin.
As discussed above, the residue-specific replacement of a particular amino acid at all positions in a protein sequence with ncAA (i.e., SPI method) proved to be a reasonable alternative in some cases (Hoesl and Budisa, 2011). Moreover, incorporation of three different tryptophan analogs (5-fluoroTrp, 5-hydroxyTrp, and 5-methylTrp) into different positions of nisin (I1W, I4W, M17W, and V32W), which included the overexpression of tryptophanyl-tRNA synthetase (TrpRS) in L. lactis strain PA1002, showed a reasonable incorporation efficiency and suitable production yield (Zhou et al., 2016). However, in a comparative study between the efficiency of ncAA incorporation in orthogonal translation and by SPI method into the lasso peptide capistruin, the SCS approach showed higher efficiency (Al Toma et al., 2015).
After induction and expression of the target gene, various conventional methods such as immobilized metal affinity chromatography (IMAC) and in rare cases, additional column chromatography were applied for the purification purposes. Due to the lack of a cell wall-anchored protease (NisP), and to avoid the requirement for immunity as well as to achieve higher production yield in E. coli (Montalbán-López et al., 2018), activation of the purified pre-lantibiotics is usually done by using native proteases or some commercial enzymes in vitro. To assess proper maturation of the lantibiotics, MS-analyses and bioactivity assays have been employed. In addition, RP-HPLC is one of the most convenient methods used for the measurement of the concentration before and after the activation by proteolytic cleavage of the leader peptide. Monitoring and quantification of the active lantibiotic by integrating the leader peptide peaks in RP-HPLC might be complicated due to alteration of running properties of the pre-lantibiotics and activated forms during the experiment and make it difficult to distinguish between similar peaks. This strategy has been used to determine the concentration of the activated lantibiotic during the production of genome-mining new lantibiotics as a hybrid containing the leader peptide of nisin (Lagedroste et al., 2019).
Another important issue is incomplete modification of target peptides such as lack of lanthionine ring formation or the defected dehydrations of Ser/Thr residues which lead to the disrupted recognition by the leader peptidases and severe decline of their antimicrobial activities. Although MS analysis of the cleavage products of pre-lantibiotics provides useful information about the dehydration reaction by the decrease of a water molecule mass (−18 Da), the technique cannot indicate the formation of methyl-lanthionine rings. In this regard, use of alkylation agents such as CDAP to bind the free cysteine thiol or peptide digestion approach using tandem MS-MS analysis to determine different patterns between wild type and mutant variants has recently been introduced (Kluskens et al., 2005). Verification of antimicrobial activity of recombinant lantibiotics can also be conducted by growth inhibition assay. Using lantibiotic-sensitive strains such as L. lactis strain NZ9000, Listeria monocytogenes, Clostridioides difficile, etc. have been reported in the literature (Field et al., 2010; Cebrián et al., 2019).
Several techniques have been developed for verification and quantification of nisin, such as turbidity assays (Berridge and Barrett, 1952), colorimetric assays (Wang et al., 2007) and agar diffusion bioassay (Mocquot and Lefebvre, 1956). Agar-based bioassay, as the most widely used method for quantifying nisin activity was implemented based on diffusion through agar gel seeded with nisin-sensitive indicator bacteria (Pongtharangkul and Demirci, 2004). The diameter of the inhibition zone generated by growth inhibition of indicator bacteria is correlated with the concentration of nisin. Factors such as nisin structure, concentration of agar, pH, existence of detergent, number of indicator cells and temperature can affect diffusion of nisin through agar. Several researchers have attempted to improve the accuracy, sensitivity, and reproducibility of conventional agar diffusion bioassays for nisin by optimizing several factors such as concentration of agar and buffering conditions, addition of a detergent like Tween 20, incubation time adjustment, and the temperature of pre-diffusion of the agar plates and even the size of wells on agar (Rogers and Montville, 1991; Wolf and Gibbons, 1996; Bonev et al., 2008; Lalpuria et al., 2013).
A modified form of agar diffusion bioassay was established upon activation of nisin by the membrane-associated protease enzyme (NisP). Antimicrobial activity of precursor lantibiotics and ncAA-modified variants synthesized by native producers or heterologously expressed by host cells could be determined using a sensitive indicator strain harboring appropriate genes for the maturation of the lantibiotic. For example, the nisin-sensitive indicator L. lactis NZ9000 pNZnisPT pIL253 showed reasonable applicability to assess functional and physicochemical properties of modified nisin variants like proline, tryptophan, and lysine incorporated analogs (Bartholomae et al., 2018). In addition, a bioactivity assay for two-component lantibiotics such as lichenicidin which requires the synergistic activity of two peptides was introduced which is called “deferred antagonist bioassay.” Using agar well diffusion plates by mixing the cell-free supernatants of both separately producing strains in suitable proportions against growth of the indicator strain and the measurement of inhibition radii after appropriate incubation at reasonable temperature aid us to easily screen a large library of lantibiotic mutants by avoiding time-consuming purification steps (Barbosa et al., 2019).
Bindman et al. (2015) have demonstrated that co-translational backbone modification α-hydroxy acidic insertion works in RiPPs in response to in-frame amber codons (SCS method). They have shown the possibility of lantibiotic activation after co-translational incorporation of α-hydroxy acids into the precursor peptides in E. coli. The biosynthesis of lacticin 481 or nukacin ISK-1 analogs by incorporating Boc-HO-1, HO-Phe(3-Br)-OH or HO-Tyr(propargyl)-OH at the junction of the leader peptide to the core region of the lantibiotics (position + 1) was successful. This enabled an improved removal step of the leader peptide in a general manner without the need to screen for various proteases (Bindman et al., 2015). Efforts have also been reported to increase the chemical diversity of lasso peptides and even to determine the crystal structure of these complicated cyclic peptides in the presence of bromine atoms (Piscotta et al., 2015). Sixteen possible variants were expressed in E. coli at four positions (Val6, Ile3, Phe10, Phe19). Thereby, four meta-substituted Phe derivatives (m-ClPhe, m-BrPhe, m-NO2Phe and m-CF3Phe) were incorporated into MccJ25 using an engineered “polyspecific” MmPylRS. The results showed that the yield of ncAA-substituted MccJ25 variants intensively depended on the position and the chemical nature of the substitutions.
The use of ncAAs with 1,3- or 1,2-aminothiol reactive groups to promote the cyclization of a downstream target peptide sequence via a C-terminal ligation/ring contraction mechanism has also been reported (Frost et al., 2015). It has been demonstrated to be useful for the formation of macrocyclic side-chain-to-tail peptides in vitro in a pH-controlled manner. This strategy controls the spontaneous cyclization of peptides of variable length and completely random sequences with a wide range of molecular arrangements, namely cyclic, lariat or C-terminal fused to a carrier protein in living bacterial cells. The co-expression of glutamyl-charged tRNAGlu in order to acylate specific Ser/Thr side chains with glutamate prior to the dehydration reaction was proposed by Zambaldo et al. (2017). The main intention was to overcome an incomplete dehydration phenomenon which occurs during incorporation of α-chloroacetamide-containing ncAA to nisin variants having altered macrocyclic topologies and antimicrobial activities. In an attempt to optimize biophysical properties of cinnamycin from Streptococcus albus for medical and industrial applications, three distinct pyrrolysine analogs were incorporated into two distinct positions of the antibiotic in Streptomycetes using the orthogonal pyrrolysyl-tRNA synthetase/tRNAPyl pair from Methanosarcina barkeri. In spite of a low rate of incorporation, the data revealed that the type of ncAA and the position of incorporation are important to achieve suitable amounts of the new deoxycinnamycin derivatives (Lopatniuk et al., 2017). Last but not least, the stereochemical configuration of thioether bridges is an important issue, since recent studies indicate that it is the property of the sequence of the core peptide and not the modification enzyme that determines the stereochemical outcome of ring formation (Garg et al., 2016).
Although most of the reports regarding the site-specific incorporation of ncAAs in RiPPs have shown lower levels of antimicrobial activity of variants than the wild type, some rare reports presented slightly improved activity on analogs (Table 2). Notably, novel modified lantibiotic variants have encrypted potential to show appropriate activity against newly emerged pathogens.
It has been estimated that 1.2 trillion USD are required to cover additional health expenditure per year expected by 2050 due to the rise of various AMRs (World Health Organization, 2020). The World Health Organization (WHO) has announced a global AMR response to coordinate this in collaboration with international partners. According to the WHO report on antimicrobial agents in clinical development stages in 2019, the majority of antibacterial agents are direct-acting small molecules (n = 108, 42.9%), followed by non-traditional approaches (n = 90, 35.7%), then AMPs (n = 27, 10.7%) as the main antibiotic agents (World Health Organization, 2019). It was also argued that the alarming rise in antibiotic resistance rates in the late 20th century is a clear indication that the golden age of antibiotics might be over (Wernicki, 2013). All these figures and projections urge us to reconsider current approaches and anticipate possible future paths to alternatives. In many cases, superbugs are characterized by increased multi-drug resistance, improved transmissibility and virulence, resulting in increased morbidity and mortality.
Multi-drug resistance in bacteria is a very complex problem, deeply rooted in the genetic and biochemical flexibility of bacteria, i.e., a highly pleiotropic phenomenon. Obviously, this problem cannot be solved only by simple solutions such as chemical modifications or variations of known lead substances. Therefore, we believe that the time is coming to use more extensively the most advanced methods of Synthetic Biology such as expanded genetic code to combat superbugs that have continuously evolved in most industrialized nations over the last 60 years. Here we attempted to summarize the efforts that have been made by the help of expanded genetic code to introduce new-to-nature lanthipeptides as promising alternatives to classical chemical agents. We have also tried to present new insights and progress in the field as well as the limitations and challenges which should be overcome in the future. We are firmly convinced that “radical” methods of genetic engineering (Davies and Church, 2019), such as the expansion of the genetic code, can be a useful addition to the struggle against superbugs as a global threat to all of mankind.
In the present medical context and in the current environment of failing antibiotic protection, an obvious functional expansion is a fortification against post-surgery antibiotic infections, for example by coupling therapeutic proteins to antimicrobial modules. New approaches such as genetically encoded chemical conversion (GECCO) has also been recently introduced (Yang et al., 2019) to overcome limitations of translational machinery. This and other newly emerged approaches such as OTS optimization (Dulic et al., 2018), cell-free protein synthesis (Jin and Hong, 2018), genomically recorded strain development (Amiram et al., 2015), artificial codon box division (Iwane et al., 2016), and quadruplet codon and orthogonal ribosome (d’Aquino et al., 2018) are expected to open new avenues for exploiting chemistry in live systems to bioengineering of lanthipeptides. GCE as a research field has now reached the maturity to be efficiently implemented in the bioengineering of lanthipeptides to understand their structural complexity and the behavior of the entire biosynthesis machinery. This should provide a solid basis for expanding the chemical space of recombinant AMPs. The aim is not only improving their therapeutic properties to combat AMR but also to repurpose them functionally for, e.g., anticancer or antiviral activities. The application of co-translational incorporation of ncAAs by more than 200 different chemical entities available to us for rational manipulation of various scaffolds offers unprecedented opportunities to manage the supply of advanced peptide-based antimicrobials and other sophisticated drugs in the future.
HRK-H and NB planned and conceived. All authors read, critically revised, and approved the final manuscript.
This research was supported by SynCrop from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement No 764591 and by Canada Research Chairs Program (Grant No. 950-231971).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
NB would like to thank Prof. Dr. Christian Thomsen, President of the Technical University of Berlin, for his continuous support. We acknowledge support by the Open Access Publication Fund of TU Berlin. We offer our sincere apologies to numerous authors of relevant works who were not included in this review; the omission was unintentional.
Acedo, J. Z., Chiorean, S., Vederas, J. C., and Van Belkum, M. J. (2018). The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol. Rev. 42, 805–828. doi: 10.1093/femsre/fuy033
Adhikari, S., Leissa, J. A., and Karlsson, A. J. (2019). Beyond function: engineering improved peptides for therapeutic applications. AIChE J. 66:e16776. doi: 10.1002/aic.16776
Ageitos, J. M., Sánchez-Pérez, A., Calo-Mata, P., and Villa, T. G. (2017). Antimicrobial peptides (AMPs): ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 133, 117–138. doi: 10.1016/j.bcp.2016.09.018
Agostini, F., Voller, J. S., Koksch, B., Acevedo-Rocha, C. G., Kubyshkin, V., and Budisa, N. (2017). Biocatalysis with unnatural amino acids: enzymology meets xenobiology. Angew. Chem. Int. Ed. 56, 9680–9703. doi: 10.1002/anie.201610129
Ahmed, Y., Rebets, Y., Estevez, M. R., Zapp, J., Myronovskyi, M., and Luzhetskyy, A. (2020). Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microbial. Cell Fact. 19:5. doi: 10.1186/s12934-020-1277-8
Al Toma, R. S., Kuthning, A., Exner, M. P., Denisiuk, A., Ziegler, J., Budisa, N., et al. (2015). Site-directed and global incorporation of orthogonal and isostructural noncanonical amino acids into the ribosomal lasso peptide capistruin. ChemBioChem 16, 503–509. doi: 10.1002/cbic.201402558
Alvarez-Sieiro, P., Montalbán-López, M., Mu, D., and Kuipers, O. P. (2016). Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. doi: 10.1007/s00253-016-7343-9
Amiram, M., Haimovich, A. D., Fan, C., Wang, Y.-S., Aerni, H.-R., Ntai, I., et al. (2015). Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol. 33, 1272–1279. doi: 10.1038/nbt.3372
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/c2np20085f
Arranz-Gibert, P., Patel, J. R., and Isaacs, F. J. (2019). The role of orthogonality in genetic code expansion. Life 9:58. doi: 10.3390/life9030058
Arranz-Gibert, P., Vanderschuren, K., and Isaacs, F. J. (2018). Next-generation genetic code expansion. Curr. Opin. Chem. Biol. 46, 203–211. doi: 10.1016/j.cbpa.2018.07.020
Aslam, B., Wang, W., Arshad, M. I., Khurshid, M., Muzammil, S., Rasool, M. H., et al. (2018). Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 11:1645. doi: 10.2147/IDR.S173867
Barbosa, J., Caetano, T., Mösker, E., Süssmuth, R., and Mendo, S. (2019). Lichenicidin rational site-directed mutagenesis library: a tool to generate bioengineered lantibiotics. Biotechnol. Bioeng. 116, 3053–3062. doi: 10.1002/bit.27130
Bartholomae, M., Baumann, T., Nickling, J. H., Peterhoff, D., Wagner, R., Budisa, N., et al. (2018). Expanding the genetic code of Lactococcus lactis and Escherichia coli to incorporate non-canonical amino acids for production of modified lantibiotics. Front. Microbiol. 9:657. doi: 10.3389/fmicb.2018.00657
Baumann, T., Exner, M., and Budisa, N. (2016). “Orthogonal protein translation using pyrrolysyl-tRNA synthetases for single-and multiple-noncanonical amino acid mutagenesis,” in Synthetic Biology–Metabolic Engineering, eds H. Zhao and A.-P. Zeng (Berlin: Springer), 1–19.
Baumann, T., Hauf, M., Richter, F., Albers, S., Möglich, A., Ignatova, Z., et al. (2019). Computational aminoacyl-tRNA synthetase library design for photocaged tyrosine. Int. J. Mol. Sci. 20:2343.
Baumann, T., Schmitt, F.-J., Pelzer, A., Spiering, V. J., Von Sass, G. J. F., Friedrich, T., et al. (2018). Engineering’golden’fluorescence by selective pressure incorporation of non-canonical amino acids and protein analysis by mass spectrometry and fluorescence. J. Vis. Exp. 134:e57017. doi: 10.3791/57017
Berridge, N., and Barrett, J. (1952). A rapid method for the turbidimetric assay of antibiotics. Microbiology 6, 14–20. doi: 10.1099/00221287-6-1-2-14
Bindman, N. A., Bobeica, S. C., Liu, W. R., and Van Der Donk, W. A. (2015). Facile removal of leader peptides from lanthipeptides by incorporation of a hydroxy acid. J. Am. Chem. Soc. 137, 6975–6978. doi: 10.1021/jacs.5b04681
Boakes, S., and Dawson, M. J. (2014). “Discovery and development of NVB302, a semisynthetic antibiotic for treatment of Clostridium difficile infection,” in Natural Products: Discourse, Diversity, and Design, eds A. Osbourn, R. Goss, and G. T. Carter (Hoboken, NJ: John Wiley & Sons), 455–468. doi: 10.1002/9781118794623.ch24
Boakes, S., Weiss, W. J., Vinson, M., Wadman, S., and Dawson, M. J. (2016). Antibacterial activity of the novel semisynthetic lantibiotic NVB333 in vitro and in experimental infection models. J. Antibiot. 69, 850–857. doi: 10.1038/ja.2016.47
Bolt, H. L., Kleijn, L. H. J., Martin, N. I., and Cobb, S. L. (2018). Synthesis of antibacterial nisin-peptoid hybrids using click methodology. Molecules 23:1566. doi: 10.3390/molecules23071566
Bonev, B., Hooper, J., and Parisot, J. (2008). Principles of assessing bacterial susceptibility to antibiotics using the agar diffusion method. J. Antimicrob. Chemother. 61, 1295–1301. doi: 10.1093/jac/dkn090
Brand, G. D., Magalhães, M. T., Tinoco, M. L., Aragão, F. J., Nicoli, J., Kelly, S. M., et al. (2012). Probing protein sequences as sources for encrypted antimicrobial peptides. PLoS One 7:e45848. doi: 10.1371/journal.pone.0045848
Breukink, E., and de Kruijff, B. (2006). Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5, 321–323. doi: 10.1038/nrd2004
Brogden, K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250. doi: 10.1038/nrmicro1098
Bryson, D. I., Fan, C., Guo, L.-T., Miller, C., Söll, D., and Liu, D. R. (2017). Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 13, 1253–1260. doi: 10.1038/nchembio.2474
Budisa, N. (2004). Prolegomena to future experimental efforts on genetic code engineering by expanding its amino acid repertoire. Angew Chem. Int. Ed. 43, 6426–6463. doi: 10.1002/anie.200300646
Budisa, N. (2006). Engineering the Genetic Code: Expanding the Amino Acid Repertoire for the Design of Novel Proteins. Hoboken, NJ: John Wiley & Sons.
Budisa, N. (2013). Expanded genetic code for the engineering of ribosomally synthetized and post-translationally modified peptide natural products (RiPPs). Curr. Opin. Biotechnol. 24, 591–598. doi: 10.1016/j.copbio.2013.02.026
Budisa, N., and Schneider, T. (2019). Expanding the DOPA universe with genetically encoded, mussel-inspired bioadhesives for material sciences and medicine. ChemBioChem 20, 2163–2190. doi: 10.1002/cbic.201900030
Cardoso, M. H., Ribeiro, S. M., Nolasco, D. O., De La Fuente-Núñez, C., Felício, M. R., Gonçalves, S., et al. (2016). A polyalanine peptide derived from polar fish with anti-infectious activities. Sci. Rep. 6:21385. doi: 10.1038/srep21385
Cebrián, R., Macia-Valero, A., Jati, A. P., and Kuipers, O. P. (2019). Design and expression of specific hybrid lantibiotics active against pathogenic Clostridium spp. Front. Microbiol. 10:2154. doi: 10.3389/fmicb.2019.02154
Chalekson, C. P., Neumeister, M. W., and Jaynes, J. (2002). Improvement in burn wound infection and survival with antimicrobial peptide D2A21 (Demegel). Plast. Reconstr. Surg. 109, 1338–1343. doi: 10.1097/00006534-200204010-00020
Chatterjee, A., Guo, J., Lee, H. S., and Schultz, P. G. (2013). A genetically encoded fluorescent probe in mammalian cells. J. Am. Chem. Soc. 135, 12540–12543. doi: 10.1021/ja4059553
Chatterjee, C., Paul, M., Xie, L., and Van Der Donk, W. A. (2005). Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–684.
Clemens, R., Zaschke-Kriesche, J., Khosa, S., and Smits, S. H. (2018). Insight into two ABC transporter families involved in lantibiotic resistance. Front. Mol. Biosci. 4:91. doi: 10.3389/fmolb.2017.00091
Coast, J., Smith, R. D., and Millar, M. R. (1996). Superbugs: should antimicrobial resistance be included as a cost in economic evaluation? Health Econ. 5, 217–226. doi: 10.1002/(sici)1099-1050(199605)5:3<217::aid-hec200<3.0.co;2-s
Cropp, T. A., and Schultz, P. G. (2004). An expanding genetic code. Trends Genet. 20, 625–630. doi: 10.1016/j.tig.2004.09.013
d’Aquino, A. E., Kim, D. S., and Jewett, M. C. (2018). Engineered Ribosomes for Basic Science and Synthetic Biology. Annu. Rev. Chem. Biomol. Eng. 9, 311–340. doi: 10.1146/annurev-chembioeng-060817-084129
Davies, J., and Davies, D. (2010). Resistance origins and evolution of antibiotic. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Davies, K., and Church, G. M. (2019). Radical technology meets radical application: an interview with George Church. CRISPR J. 2, 346–351. doi: 10.1089/crispr.2019.29074.gch
de Bruijn, A. D., and Roelfes, G. (2018). Chemical modification of dehydrated amino acids in natural antimicrobial peptides by photoredox catalysis. Chem. Eur. J. 24, 11314–11318. doi: 10.1002/chem.201803144
Del Campo, C., Bartholomäus, A., Fedyunin, I., and Ignatova, Z. (2015). Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet. 11:e1005613. doi: 10.1371/journal.pgen.1005613
Deng, J., Viel, J. H., Chen, J., and Kuipers, O. P. (2020). Synthesis and characterization of heterodimers and fluorescent nisin species by incorporation of methionine analogs and subsequent click chemistry. ACS Synth. Biol. 9, 2525–2536. doi: 10.1021/acssynbio.0c00308
Devaraj, N. K. (2018). The future of bioorthogonal chemistry. ACS Cent. Sci. 4, 952–959. doi: 10.1021/acscentsci.8b00251
Draper, L. A., Cotter, P. D., Hill, C., and Ross, R. P. (2015). Lantibiotic resistance. Microbiol. Mol. Biol. Rev. 79, 171–191. doi: 10.1128/mmbr.00051-14
Drienovská, I., and Roelfes, G. (2020). Expanding the enzyme universe with genetically encoded unnatural amino acids. Nat. Catal. 3, 1–10.
Dulic, M., Cvetesic, N., Zivkovic, I., Palencia, A., Cusack, S., Bertosa, B., et al. (2018). Kinetic origin of substrate specificity in post-transfer editing by leucyl-tRNA synthetase. J. Mol. Biol. 430, 1–16. doi: 10.1016/j.jmb.2017.10.024
Exner, M. P., Kuenzl, T., To, T. M., Ouyang, Z., Schwagerus, S., Hoesl, M. G., et al. (2017). Design of S-allylcysteine in situ production and incorporation based on a novel pyrrolysyl-tRNA synthetase variant. ChemBioChem 18, 85–90. doi: 10.1002/cbic.201600537
Feng, Q., Huang, Y., Chen, M., Li, G., and Chen, Y. (2015). Functional synergy of α-helical antimicrobial peptides and traditional antibiotics against Gram-negative and Gram-positive bacteria in vitro and in vivo. Eur. J. Clin. Microbiol. Infect. Dis. 34, 197–204. doi: 10.1007/s10096-014-2219-3
Field, D., Begley, M., O’connor, P. M., Daly, K. M., Hugenholtz, F., Cotter, P. D., et al. (2012). Bioengineered nisin a derivatives with enhanced activity against both Gram positive and Gram negative pathogens. PLoS One 7:e46884. doi: 10.1371/journal.pone.0046884
Field, D., Cotter, P. D., Hill, C., and Ross, R. P. (2015). Bioengineering lantibiotics for therapeutic success. Front. Microbiol. 6:1363. doi: 10.3389/fmicb.2015.01363
Field, D., Quigley, L., O’connor, P. M., Rea, M. C., Daly, K., Cotter, P. D., et al. (2010). Studies with bioengineered Nisin peptides highlight the broad-spectrum potency of Nisin V. Microbial. Biotechnol. 3, 473–486. doi: 10.1111/j.1751-7915.2010.00184.x
Fleming, A. (1945). Penicillin - Nobel Lecture. Retrieved from NobelPrize.org. Nobel Media AB 2020. Available online at: https://www.nobelprize.org/prizes/medicine/1945/fleming/lecture/
Frost, J. R., Jacob, N. T., Papa, L. J., Owens, A. E., and Fasan, R. (2015). Ribosomal synthesis of macrocyclic peptides in Vitro and in Vivo mediated by genetically encoded aminothiol unnatural amino acids. ACS Chem. Biol. 10, 1805–1816. doi: 10.1021/acschembio.5b00119
Garg, N., Goto, Y., Chen, T., and Van Der Donk, W. A. (2016). Characterization of the stereochemical configuration of lanthionines formed by the lanthipeptide synthetase GeoM. Pept. Sci. 106, 834–842. doi: 10.1002/bip.22876
Garg, N., Tang, W., Goto, Y., Nair, S. K., and Van Der Donk, W. A. (2012). Lantibiotics from Geobacillus thermodenitrificans. Proc. Natl. Acad. Sci. U.S.A. 109, 5241–5246. doi: 10.1073/pnas.1116815109
Geng, M., and Smith, L. (2018). Modifying the lantibiotic mutacin 1140 for increased yield, activity, and stability. Appl. Environ. Microbiol. 84, e830–e818. doi: 10.1128/AEM.00830-18
Gober, J. G., Ghodge, S. V., Bogart, J. W., Wever, W. J., Watkins, R. R., Brustad, E. M., et al. (2017). P450-mediated non-natural cyclopropanation of dehydroalanine-containing thiopeptides. ACS Chem. Biol. 12, 1726–1731. doi: 10.1021/acschembio.7b00358
Gomez-Escribano, J. P., and Bibb, M. J. (2011). Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microbial. Biotechnol. 4, 207–215. doi: 10.1111/j.1751-7915.2010.00219.x
Gorochowski, T. E., Ignatova, Z., Bovenberg, R. A., and Roubos, J. A. (2015). Trade-offs between tRNA abundance and mRNA secondary structure support smoothing of translation elongation rate. Nucleic Acids Res. 43, 3022–3032. doi: 10.1093/nar/gkv199
Greber, K. E., and Dawgul, M. (2017). Antimicrobial peptides under clinical trials. Curr. Top. Med. Chem. 17, 620–628. doi: 10.2174/1568026616666160713143331
Håkansson, J., Ringstad, L., Umerska, A., Johansson, J., Andersson, T., Boge, L., et al. (2019). Characterization of the in vitro, ex vivo, and in vivo efficacy of the antimicrobial peptide DPK-060 used for topical treatment. Front. Cell Infect. Microbiol. 9:174. doi: 10.3389/fcimb.2019.00174
Hauf, M., Richter, F., Schneider, T., Faidt, T., Martins, B. M., Baumann, T., et al. (2017). Photoactivatable mussel-based underwater adhesive proteins by an expanded genetic code. ChemBioChem 18, 1819–1823. doi: 10.1002/cbic.201700327
Healy, B., Field, D., O’connor, P. M., Hill, C., Cotter, P. D., and Ross, R. P. (2013). Intensive mutagenesis of the nisin hinge leads to the rational design of enhanced derivatives. PLoS One 8:e79563. doi: 10.1371/journal.pone.0079563
Hoesl, M. G., and Budisa, N. (2011). Expanding and engineering the genetic code in a single expression experiment. ChemBioChem 12, 552–555. doi: 10.1002/cbic.201000586
Hoesl, M. G., and Budisa, N. (2012). Recent advances in genetic code engineering in Escherichia coli. Curr. Opin. Biotechnol. 23, 751–757.
Huo, L., Ökesli, A., Zhao, M., and Van Der Donk, W. A. (2017). Insights into the biosynthesis of duramycin. Appl. Environ. Microbiol. 83:e02698-16. doi: 10.1128/AEM.02698-16
Islam, M. R., Shioya, K., Nagao, J., Nishie, M., Jikuya, H., Zendo, T., et al. (2009). Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol. Microbiol. 72, 1438–1447. doi: 10.1111/j.1365-2958.2009.06733.x
Iwane, Y., Hitomi, A., Murakami, H., Katoh, T., Goto, Y., and Suga, H. (2016). Expanding the amino acid repertoire of ribosomal polypeptide synthesis via the artificial division of codon boxes. Nat. Chem. 8, 317–325. doi: 10.1038/nchem.2446
Jin, X., and Hong, S. H. (2018). Cell-free protein synthesis for producing ‘difficult-to-express’ proteins. Biochem. Eng. J. 138, 156–164. doi: 10.1016/j.bej.2018.07.013
Kakkar, N., Perez, J. G., Liu, W. R., Jewett, M. C., and Van Der Donk, W. A. (2018). Incorporation of nonproteinogenic amino acids in class I and II lantibiotics. ACS Chem. Biol. 13, 951–957. doi: 10.1021/acschembio.7b01024
Kers, J. A., Sharp, R. E., Defusco, A. W., Park, J. H., Xu, J., Pulse, M. E., et al. (2018). Mutacin 1140 lantibiotic variants are efficacious against Clostridium difficile infection. Front. Microbiol. 9:415. doi: 10.3389/fmicb.2018.00415
Key, H. M., and Miller, S. J. (2017). Site-and stereoselective chemical editing of thiostrepton by Rh-catalyzed conjugate arylation: new analogues and collateral enantioselective synthesis of amino acids. J. Am. Chem. Soc. 139, 15460–15466. doi: 10.1021/jacs.7b08775
Khosa, S., Frieg, B., Mulnaes, D., Kleinschrodt, D., Hoeppner, A., Gohlke, H., et al. (2016a). Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Sci. Rep. 6:18679. doi: 10.1038/srep18679
Khosa, S., Hoeppner, A., Gohlke, H., Schmitt, L., and Smits, S. H. (2016b). Structure of the response regulator NsrR from Streptococcus agalactiae, which is involved in lantibiotic resistance. PLoS One 11:e149903. doi: 10.1371/journal.pone.0149903
Kim, E. J., Yang, I., and Yoon, Y. J. (2015). Developing Streptomyces venezuelae as a cell factory for the production of small molecules used in drug discovery. Arch. Pharm. Res. 38, 1606–1616. doi: 10.1007/s12272-015-0638-z
Kluskens, L. D., Kuipers, A., Rink, R., De Boef, E., Fekken, S., Driessen, A. J., et al. (2005). Post-translational modification of therapeutic peptides by NisB, the dehydratase of the lantibiotic nisin. Biochemistry 44, 12827–12834. doi: 10.1021/bi050805p
Knerr, P. J., and van der Donk, W. A. (2012). Chemical synthesis and biological activity of analogues of the lantibiotic epilancin 15X. J. Am. Chem. Soc. 134, 7648–7651. doi: 10.1021/ja302435y
Koehbach, J., and Craik, D. J. (2019). The vast structural diversity of antimicrobial peptides. Trends Pharmacol. Sci. 40, 517–528. doi: 10.1016/j.tips.2019.04.012
Kolb, H. C., Finn, M. G., and Sharpless, K. B. (2001). Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. 40, 2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::aid-anie2004<3.3.co;2-x
Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E., and Ikeda, H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651. doi: 10.1073/pnas.0914833107
Koo, H. B., and Seo, J. (2019). Antimicrobial peptides under clinical investigation. Pept. Sci. 111:e24122. doi: 10.1002/pep2.24122
Koopmans, T., Wood, T. M., ’t Hart, P., Kleijn, L. H., Hendrickx, A. P., Willems, R. J., et al. (2015). Semisynthetic lipopeptides derived from nisin display antibacterial activity and lipid II binding on par with that of the parent compound. J. Am. Chem. Soc. 137, 9382–9389. doi: 10.1021/jacs.5b04501
Kubyshkin, V., and Budisa, N. (2019). Anticipating alien cells with alternative genetic codes: away from the alanine world! Curr. Opin. Biotechnol. 60, 242–249. doi: 10.1016/j.copbio.2019.05.006
Kudrimoti, M., Curtis, A., Azawi, S., Worden, F., Katz, S., Adkins, D., et al. (2017). Dusquetide: reduction in oral mucositis associated with enduring ancillary benefits in tumor resolution and decreased mortality in head and neck cancer patients. Biotechnol. Rep. 15, 24–26. doi: 10.1016/j.btre.2017.05.002
Kuipers, O. P., Bierbaum, G., Ottenwälder, B., Dodd, H. M., Horn, N., Metzger, J., et al. (1996). Protein engineering of lantibiotics. Antonie Van Leeuwenhoek 69, 161–170. doi: 10.1007/BF00399421
Lagedroste, M., Reiners, J., Smits, S. H., and Schmitt, L. (2019). Systematic characterization of position one variants within the lantibiotic nisin. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-018-37532-4
Lalpuria, M., Karwa, V., Anantheswaran, R. C., and Floros, J. D. (2013). Modified agar diffusion bioassay for better quantification of Nisaplin®. J. Appl. Microbiol. 114, 663–671. doi: 10.1111/jam.12078
Larregola, M., Moore, S., and Budisa, N. (2012). Congeneric bio-adhesive mussel foot proteins designed by modified prolines revealed a chiral bias in unnatural translation. Biochem. Biophys. Res. Commun. 421, 646–650. doi: 10.1016/j.bbrc.2012.04.031
Lee, J., Schwieter, K. E., Watkins, A. M., Yu, H., Schwarz, K. J., Lim, J., et al. (2019). Expanding the limits of the second genetic code with ribozymes. Nat. Commun. 10, 1–12. doi: 10.1038/s41467-019-12916-w
Lopatniuk, M., Myronovskyi, M., and Luzhetskyy, A. (2017). Streptomyces albus: a new cell factory for non-canonical amino acids incorporation into ribosomally synthesized natural products. ACS Chem. Biol. 12, 2362–2370. doi: 10.1021/acschembio.7b00359
Lubelski, J., Rink, R., Khusainov, R., Moll, G. N., and Kuipers, O. P. (2008). Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol. Life Sci. 65, 455–476. doi: 10.1007/s00018-007-7171-2
Luo, X., Zambaldo, C., Liu, T., Zhang, Y., Xuan, W., Wang, C., et al. (2016). Recombinant thiopeptides containing noncanonical amino acids. Proc. Natl. Acad. Sci. 113, 3615–3620. doi: 10.1073/pnas.1602733113
Maaskant, R. V., and Roelfes, G. (2019). Bioorthogonal metalloporphyrin-catalyzed selective methionine alkylation in the lanthipeptide nisin. ChemBioChem 20, 57–61. doi: 10.1002/cbic.201800493
Maffioli, S. I., Monciardini, P., Catacchio, B., Mazzetti, C., MüNch, D., Brunati, C., et al. (2015). Family of class I lantibiotics from actinomycetes and improvement of their antibacterial activities. ACS Chem. Biol. 10, 1034–1042. doi: 10.1021/cb500878h
Manyi-Loh, C., Mamphweli, S., Meyer, E., and Okoh, A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:795.
Mayer, C. (2019). Selection, addiction and catalysis: emerging trends for the incorporation of noncanonical amino acids into peptides and proteins in vivo. ChemBioChem 20, 1357–1364. doi: 10.1002/cbic.201800733
Ming, L., and Huang, J. A. (2017). The antibacterial effects of antimicrobial peptides OP-145 against clinically isolated multi-resistant strains. Jpn. J. Infect. Dis. 70, 601–603. doi: 10.7883/yoken.JJID.2017.090
Minks, C., Alefelder, S., Moroder, L., Huber, R., and Budisa, N. (2000). Towards new protein engineering: in vivo building and folding of protein shuttles for drug delivery and targeting by the selective pressure incorporation (SPI) method. Tetrahedron 56, 9431–9442. doi: 10.1016/S0040-4020(00)00827-9
Miyazaki, R., Myougo, N., Mori, H., and Akiyama, Y. (2018). A photo-cross-linking approach to monitor folding and assembly of newly synthesized proteins in a living cell. J. Biol. Chem. 293, 677–686. doi: 10.1074/jbc.M117.817270
Mocquot, G., and Lefebvre, E. (1956). A simple procedure to detect nisin in cheese. J. Appl. Bacteriol. 19, 322–323. doi: 10.1111/j.1365-2672.1956.tb00083.x
Mohr, K. I., Volz, C., Jansen, R., Wray, V., Hoffmann, J., Bernecker, S., et al. (2015). Pinensins: the first antifungal lantibiotics. Angew. Chem. Int. Ed. 54, 11254–11258. doi: 10.1002/anie.201500927
Montalbán-López, M., Deng, J., Van Heel, A. J., and Kuipers, O. P. (2018). Specificity and application of the lantibiotic protease NisP. Front. Microbiol. 9:160. doi: 10.3389/fmicb.2018.00160
Myronovskyi, M., and Luzhetskyy, A. (2019). Heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 36, 1281–1294. doi: 10.1039/C9NP00023B
Myronovskyi, M., Rosenkränzer, B., Nadmid, S., Pujic, P., Normand, P., and Luzhetskyy, A. (2018). Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 49, 316–324. doi: 10.1016/j.ymben.2018.09.004
Nagao, J.-I., Harada, Y., Shioya, K., Aso, Y., Zendo, T., Nakayama, J., et al. (2005). Lanthionine introduction into nukacin ISK-1 prepeptide by co-expression with modification enzyme NukM in Escherichia coli. Biochem. Biophys. Res. Commun. 336, 507–513. doi: 10.1016/j.bbrc.2005.08.125
Nilsson, E., Björn, C., Sjöstrand, V., Lindgren, K., Münnich, M., Mattsby-Baltzer, I., et al. (2009). A novel polypeptide derived from human lactoferrin in sodium hyaluronate prevents postsurgical adhesion formation in the rat. Ann. Surg. 250, 1021–1028. doi: 10.1097/SLA.0b013e3181b246a7
Oldach, F., Al Toma, R., Kuthning, A., Caetano, T., Mendo, S., Budisa, N., et al. (2012). Congeneric lantibiotics from ribosomal in vivo peptide synthesis with noncanonical amino acids. Angew. Chem. Int. Ed. 51, 415–418. doi: 10.1002/anie.201106154
Oliva, R., Chino, M., Pane, K., Pistorio, V., De Santis, A., Pizzo, E., et al. (2018). Exploring the role of unnatural amino acids in antimicrobial peptides. Sci. Rep. 8:8888. doi: 10.1038/s41598-018-27231-5
Ongey, E. L., Yassi, H., Pflugmacher, S., and Neubauer, P. (2017). Pharmacological and pharmacokinetic properties of lanthipeptides undergoing clinical studies. Biotechnol. Lett. 39, 473–482. doi: 10.1007/s10529-016-2279-9
Ortiz-López, F. J., Carretero-Molina, D., Sánchez-Hidalgo, M., Martín, J., González, I., Román-Hurtado, F., et al. (2020). Cacaoidin, first member of the new lanthidin RiPP family. Angew. Chem. Int. Ed. 20:5187. doi: 10.1002/anie.202005187
Piper, C., Draper, L. A., Cotter, P. D., Ross, R. P., and Hill, C. (2009). A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species. J. Antimicrob. Chemother. 64, 546–551. doi: 10.1093/jac/dkp221
Piscotta, F. J., Tharp, J. M., Liu, W. R., and Link, A. J. (2015). Expanding the chemical diversity of lasso peptide MccJ25 with genetically encoded noncanonical amino acids. Chem. Commun. 51, 409–412. doi: 10.1039/c4cc07778d
Plotkin, J. B., and Kudla, G. (2011). Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12, 32–42. doi: 10.1038/nrg2899
Pongtharangkul, T., and Demirci, A. (2004). Evaluation of agar diffusion bioassay for nisin quantification. Appl. Microbiol. Biotechnol. 65, 268–272. doi: 10.1007/s00253-004-1579-5
Porto, W. F., Pires, A. S., and Franco, O. L. (2017). Computational tools for exploring sequence databases as a resource for antimicrobial peptides. Biotechnol. Adv. 35, 337–349. doi: 10.1016/j.biotechadv.2017.02.001
Rauch, B. J., Porter, J. J., Mehl, R. A., and Perona, J. J. (2016). Improved incorporation of noncanonical amino acids by an engineered tRNATyr suppressor. Biochemistry 55, 618–628. doi: 10.1021/acs.biochem.5b01185
Rogers, A. M., and Montville, T. J. (1991). Improved agar diffusion assay for nisin quantification. Food Biotechnol. 5, 161–168. doi: 10.1080/08905439109549799
Rollema, H. S., Kuipers, O. P., Both, P., De Vos, W. M., and Siezen, R. J. (1995). Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl. Environm. Microbiol. 61, 2873–2878.
Ross, A. C., Mckinnie, S. M., and Vederas, J. C. (2012). The synthesis of active and stable diaminopimelate analogues of the lantibiotic peptide lactocin S. J. Am. Chem. Soc. 134, 2008–2011. doi: 10.1021/ja211088m
Rouse, S., Field, D., Daly, K. M., O’connor, P. M., Cotter, P. D., Hill, C., et al. (2012). Bioengineered nisin derivatives with enhanced activity in complex matrices. Microbial. Biotechnol. 5, 501–508.
Shi, Y., Yang, X., Garg, N., and Van Der Donk, W. A. (2011). Production of lantipeptides in Escherichia coli. J. Am. Chem. Soc. 133, 2338–2341. doi: 10.1021/ja109044r
Shin, J. M., Gwak, J. W., Kamarajan, P., Fenno, J. C., Rickard, A. H., and Kapila, Y. L. (2016). Biomedical applications of nisin. J. Appl. Microbiol. 120, 1449–1465. doi: 10.1111/jam.13033
Soye, B. J. D., Patel, J. R., Isaacs, F. J., and Jewett, M. C. (2015). Repurposing the translation apparatus for synthetic biology. Curr. Opin. Chem. Biol. 28, 83–90. doi: 10.1016/j.cbpa.2015.06.008
Srinivas, N., Jetter, P., Ueberbacher, B. J., Werneburg, M., Zerbe, K., Steinmann, J., et al. (2010). Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013. doi: 10.1126/science.1182749
Sun, E., Belanger, C. R., Haney, E. F., and Hancock, R. E. (2018). “Host defense (antimicrobial) peptides,” in Peptide Applications in Biomedicine, Biotechnology and Bioengineering, ed. S. Koutsopoulos (Amsterdam: Elsevier), 253–285.
Torres, M. D., Sothiselvam, S., Lu, T. K., and De La Fuente-Nunez, C. (2019). Peptide design principles for antimicrobial applications. J. Mol. Biol. 431, 3547–3567. doi: 10.1016/j.jmb.2018.12.015
Uttamapinant, C., Howe, J. D., Lang, K., BeraìNek, V. C., Davis, L., Mahesh, M., et al. (2015). Genetic code expansion enables live-cell and super-resolution imaging of site-specifically labeled cellular proteins. J. Am. Chem. Soc. 137, 4602–4605. doi: 10.1021/ja512838z
Vargas-Rodriguez, O., Sevostyanova, A., Söll, D., and Crnkoviæ, A. (2018). Upgrading aminoacyl-tRNA synthetases for genetic code expansion. Curr. Opin. Chem. Biol. 46, 115–122. doi: 10.1016/j.cbpa.2018.07.014
Völler, J., Biava, H., Koksch, B., Hildebrandt, P., and Budisa, N. (2015). Orthogonal translation meets electron transfer: in vivo labeling of cytochrome c for probing local electric fields. ChemBioChem 16, 742–745. doi: 10.1002/cbic.201500022
Völler, J.-S., and Budisa, N. (2017). Coupling genetic code expansion and metabolic engineering for synthetic cells. Curr. Opin. Biotechnol. 48, 1–7. doi: 10.1016/j.copbio.2017.02.002
Wang, F., Cao, L. T., and Hu, S. H. (2007). A rapid and accurate 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide colorimetric assay for quantification of bacteriocins with nisin as an example. J. Zhejiang. Uni. Sci. B 8, 549–554. doi: 10.1631/jzus.2007.B0549
Wang, G. (2015). “Improved methods for classification, prediction, and design of antimicrobial peptides,” in Computational Peptidology, eds P. Zhou and J. Huang (New York, NY: Springer New York), 43–66.
Wang, L., Xie, J., and Schultz, P. G. (2006). Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 35, 225–249. doi: 10.1146/annurev.biophys.35.101105.121507
Wernicki, A. (2013). The End of the Golden Age of Antibiotics? J. Vet. Sci. Anim. Husb. 1:e103. doi: 10.15744/2348-9790.1.e103
Wescombe, P. A., Upton, M., Renault, P., Wirawan, R. E., Power, D., Burton, J. P., et al. (2011). Salivaricin 9, a new lantibiotic produced by Streptococcus salivarius. Microbiology 157, 1290–1299. doi: 10.1099/mic.0.044719-0
Wiedemann, I., Breukink, E., Van Kraaij, C., Kuipers, O. P., Bierbaum, G., De Kruijff, B., et al. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779. doi: 10.1074/jbc.M006770200
Wolf, C., and Gibbons, W. (1996). Improved method for quantification of the bacteriocin nisin. J. Appl. Bacteriol. 80, 453–457. doi: 10.1111/j.1365-2672.1996.tb03242.x
World Health Organization (2019). Antibacterial Agents in Preclinical Development: An Open Access Database. Geneva: WHO.
Xu, M., Zhang, F., Cheng, Z., Bashiri, G., Wang, J., Hong, J., et al. (2020). Functional genome mining reveals a class V lanthipeptide containing a D-amino acid introduced by an F420H2-dependent reductase. Angew. Chem. Int. Ed. 20:8035. doi: 10.1002/anie.202008035
Yanagisawa, T., Kuratani, M., Seki, E., Hino, N., Sakamoto, K., and Yokoyama, S. (2019). Structural basis for genetic-code expansion with bulky lysine derivatives by an engineered pyrrolysyl-tRNA synthetase. Cell Chem. Biol. 26, 936–949. e913. doi: 10.1016/j.chembiol.2019.03.008
Yang, B., Wang, N., Schnier, P. D., Zheng, F., Zhu, H., Polizzi, N. F., et al. (2019). Genetically introducing biochemically reactive amino acids dehydroalanine and dehydrobutyrine in proteins. J. Am. Chem. Soc. 141, 7698–7703. doi: 10.1021/jacs.9b02611
Yelin, I., and Kishony, R. (2018). Antibiotic resistance. Cell 172, 1136–1136. e1131. doi: 10.1016/j.cell.2018.02.018
Young, T. S., and Schultz, P. G. (2010). Beyond the canonical 20 amino acids: expanding the genetic lexicon. J. Biol. Chem. 285, 11039–11044. doi: 10.1074/jbc.R109.091306
Zambaldo, C., Luo, X., Mehta, A. P., and Schultz, P. G. (2017). Recombinant macrocyclic lanthipeptides incorporating non-canonical amino acids. J. Am. Chem. Soc. 139, 11646–11649. doi: 10.1021/jacs.7b04159
Zaschke-Kriesche, J., Reiners, J., Lagedroste, M., and Smits, S. H. (2019). Influence of nisin hinge-region variants on lantibiotic immunity and resistance proteins. Bioorg. Med. Chem. 27, 3947–3953. doi: 10.1016/j.bmc.2019.07.014
Zhang, Y., Chen, M., Bruner, S. D., and Ding, Y. (2018). Heterologous production of microbial ribosomally synthesized and post-translationally modified peptides. Front. Microbiol. 9:1801. doi: 10.3389/fmicb.2018.01801
Zhao, D. L., Wang, D., Tian, X. Y., Cao, F., Li, Y. Q., and Zhang, C. S. (2018). Anti-Phytopathogenic and Cytotoxic Activities of Crude Extracts and Secondary Metabolites of Marine-Derived Fungi. Mar. Drugs 16:36. doi: 10.3390/md16010036
Zhao, X., Yin, Z., Breukink, E., Moll, G. N., and Kuipers, O. P. (2020). An engineered double lipid II binding motifs-containing lantibiotic displays potent and selective antimicrobial activity against Enterococcus faecium. Antimicrob. Agents Chemother. 64:e02050-19. doi: 10.1128/AAC.02050-19
Zhou, L., Shao, J., Li, Q., Van Heel, A. J., De Vries, M. P., Broos, J., et al. (2016). Incorporation of tryptophan analogues into the lantibiotic nisin. Amino Acids 48, 1309–1318. doi: 10.1007/s00726-016-2186-3
Zhou, L., Van Heel, A. J., and Kuipers, O. P. (2015). The length of a lantibiotic hinge region has profound influence on antimicrobial activity and host specificity. Front. Microbiol. 6:11. doi: 10.3389/fmicb.2015.00011
Keywords: genetic code expansion, chemoselectivity, RiPPs, lantibiotics, non-canonical amino acids, nisin, superbugs, synthetic biology
Citation: Karbalaei-Heidari HR and Budisa N (2020) Combating Antimicrobial Resistance With New-To-Nature Lanthipeptides Created by Genetic Code Expansion. Front. Microbiol. 11:590522. doi: 10.3389/fmicb.2020.590522
Received: 01 August 2020; Accepted: 13 October 2020;
Published: 05 November 2020.
Edited by:
Harsh Mathur, Teagasc Food Research Center, IrelandReviewed by:
Sander H. J. Smits, Heinrich Heine University, GermanyCopyright © 2020 Karbalaei-Heidari and Budisa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hamid Reza Karbalaei-Heidari, a2FyYmFsYWVpQHNoaXJhenUuYWMuaXI=; aGFtaWQua2FyYmFsYWVpaGVpZGFyaUB1bWFuaXRvYmEuY2E=; Nediljko Budisa, bmVkaWxqa28uYnVkaXNhQHVtYW5pdG9iYS5jYQ==; bmVkaWxqa28uYnVkaXNhQHR1LWJlcmxpbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.