
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 November 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.587098
This article is part of the Research Topic (p)ppGpp and Its Homologs: Enzymatic and Mechanistic Diversity Among the Microbes View all 19 articles
During nutrient deprivation, the bacterial cell undergoes a stress response known as the stringent response. This response is characterized by induction of the nucleotide derivative guanosine tetraphosphate (ppGpp) that dramatically modulates the cell’s transcriptome. In Escherichia coli, ppGpp regulates transcription of as many as 750 genes within 5 min of induction by binding directly to RNA polymerase (RNAP) at two sites ~60 Å apart. One proposal for the presence of two sites is that they have different affinities for ppGpp, expanding the dynamic range over which ppGpp acts. We show here, primarily using the Differential Radial Capillary Action of Ligand Assay (DRaCALA), that ppGpp has a similar affinity for each site, contradicting the proposal. Because the ppGpp binding sites are formed by interactions of the β’ subunit of RNAP with two small protein factors, the ω subunit of RNAP which contributes to Site 1 and the transcription factor DksA which contributes to Site 2, variation in the concentrations of ω or DksA potentially could differentially regulate ppGpp occupancy of the two sites. It was shown previously that DksA varies little at different growth rates or growth phases, but little is known about variation of the ω concentration. Therefore, we raised an anti-ω antibody and performed Western blots at different times in growth and during a stringent response. We show here that ω, like DksA, changes little with growth conditions. Together, our data suggest that the two ppGpp binding sites fill in parallel, and occupancy with changing nutritional conditions is determined by variation in the ppGpp concentration, not by variation in ω or DksA.
When nutritional resources change, cells adjust their transcriptional output to match the new environment. In almost all bacterial species, this is accomplished in part by synthesis of the secondary messengers guanosine tetraphosphate (ppGpp; guanosine 5'-diphosphate 3'-diphosphate) and guanosine pentaphosphate (pppGpp; guanosine 5'-triphosphate 3'-diphosphate), respectively, collectively referred to here as ppGpp (reviewed in Hauryliuk et al., 2015; Liu et al., 2015; Irving and Corrigan, 2018). In proteobacteria like Escherichia coli, the basal level of ppGpp (from ~1 to 10 μM) only moderately affects gene expression, but induction of RelA in response to the accumulation of deacylated tRNA(s) increases the ppGpp concentration 100–1,000-fold, dramatically changing gene expression (Ryals et al., 1982; Varik et al., 2017).
In this so-called stringent response, transcription of hundreds of genes, many of which are related to translation, is inhibited within 5 min of ppGpp induction, and transcription of hundreds of other genes, many of which are related to pathways involved in amino acid biosynthesis, is stimulated (Durfee et al., 2008; Traxler et al., 2008; Sanchez-Vazquez et al., 2019). This reprogramming of the transcriptome is accomplished in E. coli by direct binding of ppGpp to RNA polymerase (RNAP; Sanchez-Vazquez et al., 2019). ppGpp also binds directly to many proteins other than RNAP, altering their activities and contributing further to the remodeling of cellular metabolism (Zhang et al., 2018; Wang et al., 2019).
Two proteins that are not essential for the catalytic activity of E. coli RNAP are nevertheless required for the effects of ppGpp on transcription initiation, the 10.2 kDa RNAP subunit ω and the 17.5 kDa transcription factor DksA (Paul et al., 2004, 2005; Vrentas et al., 2005). Genetic and biochemical evidence indicated that ppGpp binds to two sites on RNAP ~60 Å apart (Ross et al., 2013, 2016), with Site 1 at the interface of the ω and β’ subunits of RNAP and Site 2 at the interface of DksA and the secondary channel rim of the β’ subunit (Figure 1). Crystal structures of the RNAP-ppGpp complex are consistent with the models based on the genetic and biochemical studies, and indicate that at Site 1 the ppGpp phosphates are coordinated by residues 2–5 and other residues in ω, as well as by residues R417, K615 in β’. The guanine base is coordinated by several residues in β’, including I619, D622, and R362 (Mechold et al., 2013; Zuo et al., 2013). A strain lacking rpoZ, the gene encoding the RNAP ω subunit, (i.e., lacking Site 1) displays a modest lag in recovering from a downshift from a rich to a minimal medium (Gentry et al., 1991; Ross et al., 2013, 2016), while strains lacking dksA have more pronounced defects in recovery from a downshift and in transcriptional regulation by ppGpp (Paul et al., 2004; Ross et al., 2016; Sanchez-Vazquez et al., 2019).
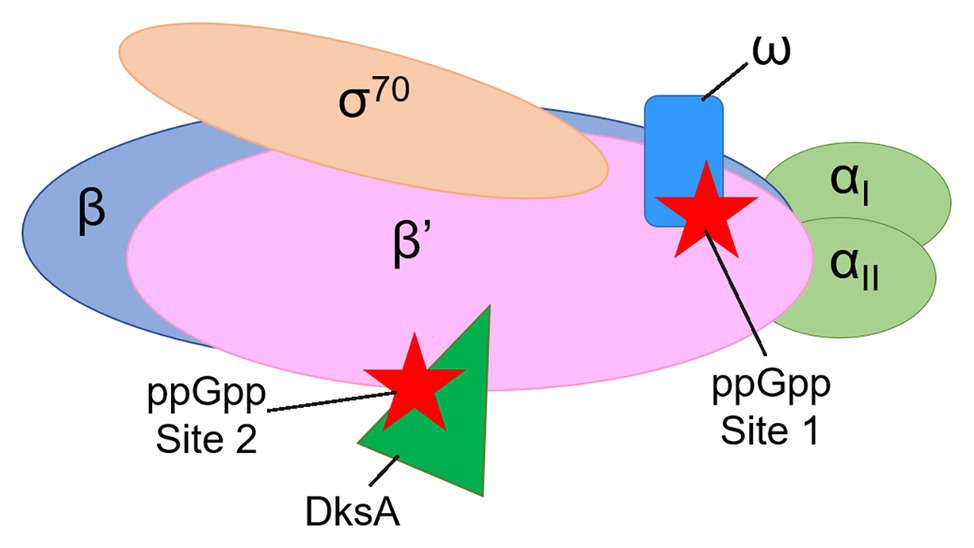
Figure 1. Cartoon representation of Escherichia coli RNA polymerase (RNAP) showing the locations of guanosine tetraphosphate (ppGpp) binding Sites 1 and 2. The αI and αII subunits (a homodimer, encoded by the rpoA gene) are shown in light green. The β subunit (encoded by the rpoB gene) is in blue. The β’ subunit (encoded by the rpoC gene) is in pink. The ω subunit (encoded by the rpoZ gene) is in blue. The σ70 subunit (encoded by the rpoD gene) is in tan. ppGpp (red stars) binds at the interface of the ω and β’ subunits (Site 1) and at the interface of the transcription factor DksA (dark green) and the β’ subunit (Site 2).
The two ppGpp binding sites in RNAP are generally conserved among proteobacteria based on conservation of the residues that contribute to binding (Ross et al., 2013, 2016). However, where it has been investigated, the effects of ppGpp on transcription in some evolutionarily distant bacterial phyla do not involve direct binding of ppGpp to RNAP. For example, in Bacillus subtilis, ppGpp inhibits transcription by binding to protein targets involved in nucleotide metabolism, leading to reduced levels of GTP, the initiating nucleotide for many promoters (Krasny and Gourse, 2004; Liu et al., 2015).
The identification of a second ppGpp-binding pocket (Site 2) in E. coli RNAP provided an explanation for why disruption of Site 1 had only a modest effect on the stringent response. Analysis of the effects of specific mutations in dksA or rpoC (the gene encoding β’) on transcription in vitro, crosslinking of 6-thio-ppGpp and binding of ppGpp to the RNAP-DksA complex suggested that Site 2 is at the interface of DksA and the β’ subunit rim helices at the entrance to the RNAP secondary channel (Ross et al., 2016). These results and a subsequent crystal structure of an RNAP-DksA-ppGpp complex indicated that residues defined by the mutational studies, including K98, R91, and K139 in DksA, coordinate the phosphates of ppGpp (Molodtsov et al., 2018), and the guanosine base is coordinated by two additional residues implicated by the mutational studies, β’ N680 and DksA L95 (Ross et al., 2016; Molodtsov et al., 2018). A recent cryo-EM structure, in which flexible regions of RNAP were not constrained by crystal packing forces, showed that DksA R129 is also in direct contact with ppGpp (Chen, unpublished), consistent with biochemical analysis of effects of DksA substitution variants on ppGpp binding and function at Site 2 (Ross et al., 2016).
Guanosine tetraphosphate binding to Site 1 alone (in the absence of DksA) has a modest inhibitory effect on transcription from the rrnB P1 promoter at saturating ω concentrations (~3-fold), and does not activate transcription from amino acid biosynthesis promoters. ppGpp binding to Site 2 alone (i.e., with a near saturating concentration of DksA but in the absence of the ω subunit) has a larger inhibitory effect on rrnB P1 (~6-fold) and is sufficient for full activation of amino acid biosynthesis promoters (Vrentas et al., 2005; Ross et al., 2016). When both ppGpp binding sites are present, inhibition of rrnB P1 is greater than with either site alone (15–20-fold). These results are consistent with the growth properties of strains lacking ppGpp (Xiao et al., 1991), ω (Gentry and Burgess, 1989; Gentry et al., 1991), or DksA (Paul et al., 2004, 2005), or containing only Site 1 (Ross et al., 2013), only Site 2 (Ross et al., 2016), or both (Ross et al., 2016; Sanchez-Vazquez et al., 2019).
The evolutionary rationale for having two sites is unclear. One model is that the two sites have different affinities for ppGpp, expanding the dynamic range over which ppGpp acts. To test that model, here, we use the Differential Radial Capillary Action of Ligand Assay (DRaCALA; Roelofs et al., 2011), as modified to measure ppGpp binding to RNAP (Ross et al., 2016), to determine the binding affinities of ppGpp for each of the two sites on E. coli RNAP independently as well as together.
We find that both binding sites have similar intrinsic affinities for ppGpp. In addition, we find that the concentrations of ω at different times in cell growth vary only slightly. In conjunction with previous measurements of the concentrations of DksA in vivo (Rutherford et al., 2007), our results indicate that the two binding sites fill with ppGpp in parallel and not sequentially as ppGpp concentrations increase. Furthermore, the binding affinities of RNAP for ppGpp that we determined in vitro are consistent with the reported effects of ppGpp on transcription even in non-stressed conditions in vivo.
Strains and plasmids are listed in Supplementary Table S1 in Supplemental data. To construct the pET23a-His10-SUMO-rpoZ plasmid used to purify ω for antibody development, a gene block (Integrated DNA Technologies) of rpoZ was inserted into pET23a-His10-SUMO (Invitrogen; RLG14235) at the BamH1 and HindIII sites using HiFi DNA Assembly (NEB) to create RLG15371.
Purification of RNAP [wild-type (WT) or mutant; Supplementary Figure S1], the ω subunit of RNAP (Supplementary Figure S2), DksA (WT and variants), GreB, and TraR were as described in Expanded Materials and Methods.
Binding of [32P]-ppGpp to RNAP, RNAP/DksA, RNAP/TraR, or RNAP/GreB complexes was measured by the DRaCALA, adapted from Roelofs et al. (2011). See Expanded Materials and Methods for details.
The most widely used mathematical model for multisite ligand binding to a protein was first proposed by Hill (1910). By plotting fractional binding of the enzyme as a function of ligand concentration, one can calculate the dissociation constant and the “Hill coefficient,” which indicates whether the binding of multiple ligands is positively or negatively cooperative (see Expanded Materials and Methods for details).
All Kd,app values for each set of binding experiments were subjected to a rank sum test (Sigma Plot) to determine if the Kd,app values in each set were statistically different from other values. Statistically different values were then given a p-value.
Polyclonal antibodies were raised by Covance, Inc., following injection of rabbits with purified ω. Quantitative western blots were performed on cells grown in LB as described in Expanded Materials and Methods.
Our previous studies evaluated the relative roles of Sites 1 and 2 on the transcriptional effects of ppGpp, an indirect indicator of ppGpp binding to RNAP (Ross et al., 2013, 2016). These experiments showed that Site 2 had a much larger effect on both inhibition and activation than Site 1, even though the concentration of ppGpp needed for half-maximal effects on transcription appeared similar, ~12–21 μM for Site 1 at saturating ω and ~19 μM for Site 2 at nearly saturating DksA (Ross et al., 2013, 2016). To measure ppGpp binding more directly, we used DRaCALA (Roelofs et al., 2011; Ross et al., 2016) to determine the affinities of ppGpp for each of the two sites on RNAP independently (Figure 2) as well as together (Figure 3). The specificity of this assay for ppGpp was established previously using unlabeled nucleotides as competitors (Supplementary Figure S3A; Ross et al., 2016). Unlabeled ppGpp competed for binding of 32P-ppGpp to each of the two sites, while GDP partially competed, and ATP did not compete at all. As this is a non-equilibrium binding assay, the Kd values are reported here as apparent Kd (Kd,app) values. A table summarizing all the apparent Kd values reported here is provided as Supplementary Table S2.
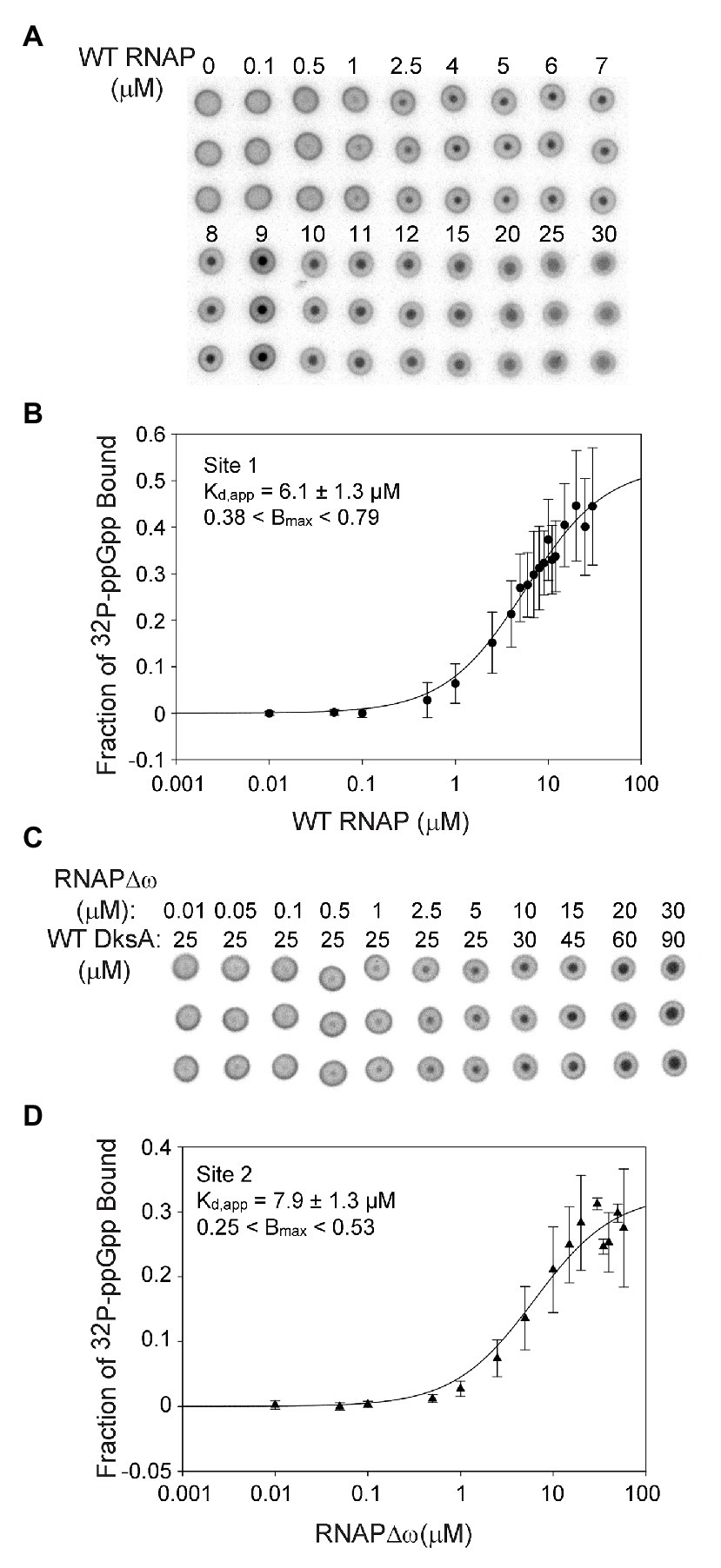
Figure 2. Sites 1 and 2 have similar binding affinities for ppGpp. (A) Differential Radial Capillary Action of Ligand Assay (DRaCALA) analysis of [32P]-ppGpp binding to Site 1. Increasing amounts of wild-type (WT) RNAP (without DksA) were equilibrated with a constant amount of [32P]-ppGpp in DRaCALA buffer and spotted on nitrocellulose filters. Triplicate filters from one representative experiment are shown. (B) The plot shown is a one-site saturation binding curve using averaged [32P]-ppGpp binding data from seven experiments conducted with five separate preparations of wild-type RNAP. Kd,app and Bmax values were determined by fitting each individual experiment to a one-site saturation ligand binding curve. Kd,app and Bmax values and error shown in the inset were averaged from the values for the seven experiments. See Expanded Materials and Methods for further details. (C) Representative DRaCALA analysis of [32P]-ppGpp binding to Site 2. Same as in (A) except RNAP lacked the ω subunit (RNAPΔω), and DksA was included. (D) Same as (B) except the reaction contained RNAPΔω and DksA.
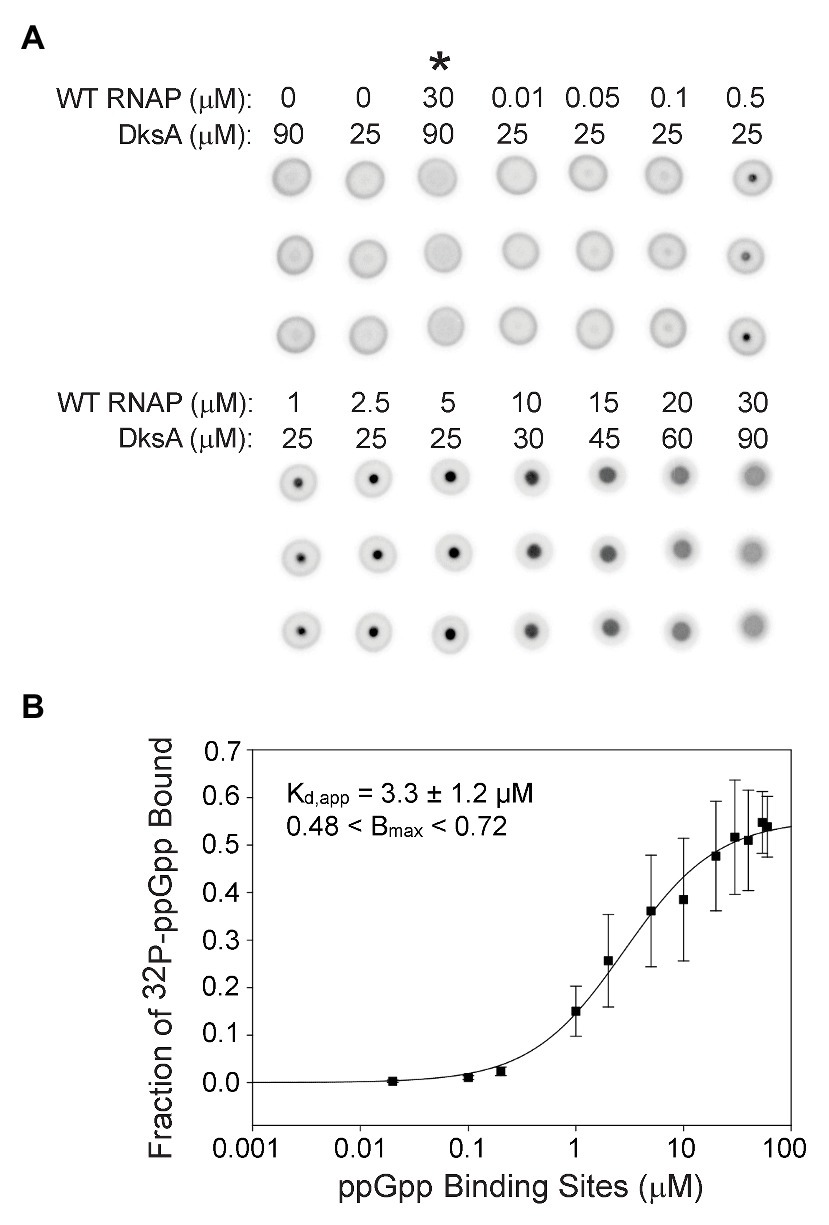
Figure 3. Guanosine tetraphosphate binds with a higher affinity when RNAP contains both ppGpp binding sites. (A) [32P]-ppGpp binding to wild-type RNAP in the presence of DksA as measured by DRaCALA. Triplicate filters from one representative experiment are shown. Wild-type RNAP and DksA concentrations are indicated above each column of three filters. Control reactions are in the first three columns. Asterisk above the third column indicates the presence of unlabeled competitor ppGpp (1 mM) in the reaction. (B) The plot shown is a one-site saturation binding curve for the average [32P]-ppGpp binding data with the error bars representing one SD from the mean from seven independent experiments conducted with five individually purified preparations of WT RNAP, as described in the legend for Figure 2. The Kd,app, Bmax, and error shown in the inset were determined by averaging Kd,app values from each of the individual experiments, as in Figure 2.
For measuring binding of ppGpp to Site 1, wild-type RNAP was purified by concurrent overproduction of the four subunits of core RNAP, α, β, and β’ encoded by one plasmid and overproduction of ω encoded by a second plasmid (see Expanded Materials and Methods). DksA was not included in the binding reactions with wild-type RNAP and ppGpp, eliminating binding to Site 2 (Ross et al., 2016). Increasing amounts of RNAP were combined with a fixed low concentration of [32P]-ppGpp (~5 nM). The observed fraction of [32P]-ppGpp bound at each RNAP concentration from each of seven independent experiments was fit to a one site saturation binding curve (see Expanded Materials and Methods for description of curve fitting). The Kd,app’s for each independent binding curve were then averaged, resulting in a Kd,app of 6.1 ± 1.3 μM for ppGpp binding to Site 1 (Figures 2A,B).
We ensured that the ω subunit was saturating in the RNAP preparations by adding increasing amounts of purified ω to the binding reaction and measuring the fraction of [32P]-ppGpp bound (Supplementary Figure S3B). The additional ω did not increase binding of ppGpp, indicating that ω was already saturating in our RNAP preparation derived from cells overexpressing ω. In contrast, we showed previously that effects of ppGpp on transcription increased slightly when RNAP was purified without concurrent overproduction of ω (Vrentas et al., 2005).
The Site 2 ppGpp binding pocket consists of residues from both DksA and β’ (Ross et al., 2016; Molodtsov et al., 2018). For measuring binding of [32P]-ppGpp to Site 2, independent of ppGpp binding to Site 1, we used logic similar to that described above, but with purified DksA added to the reactions and using an RNAP lacking ω (RNAPΔω) obtained by purification of RNAP from a strain deleted for rpoZ (ΔrpoZ). We showed previously that [32P]-ppGpp does not bind to DksA in the absence of RNAP (Ross et al., 2016), and as described above for Site 1, unlabeled ppGpp competed with [32P]-ppGpp for binding to Site 2 (Supplementary Figure S3A). The data from each of seven separate experiments measuring ppGpp binding to Site 2 were fit to one-site saturation binding curves, and the Kd,app values from each experiment were averaged (Figures 2C,D). The Kd,app for Site 2 was 7.9 ± 1.3 μM, similar to the ppGpp binding affinity for Site 1.
For comparison with the affinities measured above, we measured binding of [32P]-ppGpp to RNAPs that we had shown previously by in vitro transcription did not respond to ppGpp. The Kd,app for an RNAP purified from a strain without ω (ΔrpoZ) to which no DksA was added (and thus lacked Sites 1 and 2) was >100 μM (Supplementary Figure S4A). Similarly, RNAP purified from a strain without ω (ΔrpoZ) overexpressing ωΔ2–5 (RNAPΔω + ωΔ2–5) also had a Kd,app >100 μM (Supplementary Figure S4B). We define the very weak binding of ppGpp to these two RNAPs as non-specific or background binding, more than an order of magnitude weaker than the affinity of RNAP containing Site 1 or Site 2 for ppGpp.
We also tested ppGpp binding to another RNAP variant that previous work had shown responded very poorly to ppGpp in vitro using in vitro transcription as an assay (Ross et al., 2013). “RNAP M7” contains a four residue deletion at the N-terminus of ω, ω (Δ2–5), plus three other substitutions in β’ residues close to or within the Site 1 binding pocket. Surprisingly, this RNAP bound ppGpp with a Kd,app of 16.2 + 4.2 μM, only ~2-fold worse than the RNAPs with wild-type Site 1 or 2 (Supplementary Figure S4C). RNAPs containing a subset of the β’ substitutions present in M7 (e.g., β’K615A/R417A) were very defective in responding to ppGpp when assayed by in vitro transcription and by crosslinking with the zero-length crosslinker 6-thio ppGpp (Ross et al., 2013), yet displayed significant levels of binding in preliminary DRaCALA assays (Ross and Gourse, unpublished data). We suggest that crosslinking and function require very precise positioning of ppGpp in the binding pocket, but DRaCALA assays sometimes can detect binding modes that are non-functional. ppGpp binding to the M7 RNAP may represent such a non-functional binding mode.
Even though Sites 1 and 2 are located ~60 Å apart on RNAP (Ross et al., 2016; Molodtsov et al., 2018) and have similar affinities for ppGpp, it was conceivable that having both sites would alter the overall ppGpp binding affinity. Therefore, we compared [32P]-ppGpp binding to RNAPs containing only Site 1 or Site 2 (Figure 2) with [32P]-ppGpp binding to an RNAP saturated with both ω and DksA (i.e., containing both sites; Figure 3A). For the RNAP with both ppGpp binding sites, we calculated the Kd,app from a plot in which the X-axis indicates the concentration of binding sites (twice the RNAP concentration). The data were fit for each of seven separate experiments, and the Kd,app values were averaged (Figure 3B). For the RNAP with both binding sites, the Kd,app was 3.3 ± 1.2 μM, a significant difference from the RNAPs with only one binding site (6.1 or 7.9 μM; p = 0.006 for both sites compared to Site 1 or p = 0.01 for both sites compared to Site 2). If the nominal concentration of RNAP were used on the X-axis as in Figure 2, rather than with the concentration of ppGpp binding sites, the Kd,app would be even tighter, 1.7 ± 1.2 μM. Thus, when plotted either way, there was a significant difference in the Kd,app of the enzyme for ppGpp with both sites vs. only one site.
The RNAP concentration in the DRaCALA reactions was 100–1,000-fold higher than the ppGpp concentration. Therefore, ppGpp could not fill both binding sites on an RNAP molecule at the same time. The measured binding affinity of the enzyme containing both sites must therefore represent an average of the affinities of ppGpp bound to one site or the other in the population of RNAPs. Nevertheless, we checked for cooperativity by comparing the fits using the equations for one-site and two-site saturation binding curves and the Hill equation. Both resulted in the same Kd,app. The Hill coefficient was ~1, and the curve was not sigmoidal when the data were plotted on a linear scale (Supplementary Figure S5), consistent with a lack of cooperativity.
Although there was no evidence for cooperativity, there was an increase in the affinity of ppGpp for wild-type RNAP containing DksA (both sites) compared to the affinity for RNAP with only one site (3.3 ± 1.2 μM for the RNAP containing both sites, compared to 6.1 ± 1.3 μM or 7.9 ± 1.3 μM for the RNAPs with either Site 1 or Site 2, respectively; Figures 2, 3). Since the DksA concentration was saturating in the DRaCALA reactions measuring binding of ppGpp to RNAP with both sites present, DksA was a candidate to explain the increase in affinity. That is, we hypothesized that DksA binding in the RNAP secondary channel might allosterically alter the binding environment of Site 1. To address this hypothesis, we utilized “separation of function” DksA variants, i.e., variants defective for ppGpp binding but competent for RNAP binding. DksA residues K98 and R129 both contact ppGpp directly (Ross et al., 2016; Molodtsov et al., 2018). DksA variants containing alanine substitutions at either of these positions are still able to bind to RNAP and reduce the lifetime of RNAP-promoter complexes (Ross et al., 2016), but they do not support ppGpp binding to Site 2, and they eliminate the effects of ppGpp on transcription by RNAPΔω (Ross et al., 2016). No binding was detected in DRaCALA experiments with RNAPΔω and either DksA-R129A or DksA-K98A, consistent with predictions for a complex lacking both binding sites for ppGpp (Supplementary Figures S6A,B).
Guanosine tetraphosphate bound to wild-type RNAP in the absence of DksA (i.e., to Site 1) with a Kd,app of 6.1 ± 1.3 μM (Figure 2). ppGpp bound to Site 1 in wild-type RNAP in the presence of DksA-R129A, with a Kd,app of ~1.8 ± 0.5 μM (Figure 4A), ~3-fold more tightly than to Site 1 without DksA (a statistically significant difference with a value of p = 0.001). In the presence of DksA-K98A, ppGpp bound to Site 1 with a Kd,app of ~1.3 ± 0.1 μM (Figure 4B), although the significance of this measurement is less certain because of the smaller number of replicates performed. Nevertheless, together these results indicate that ppGpp binds more tightly to Site 1 when DksA is bound to RNAP.
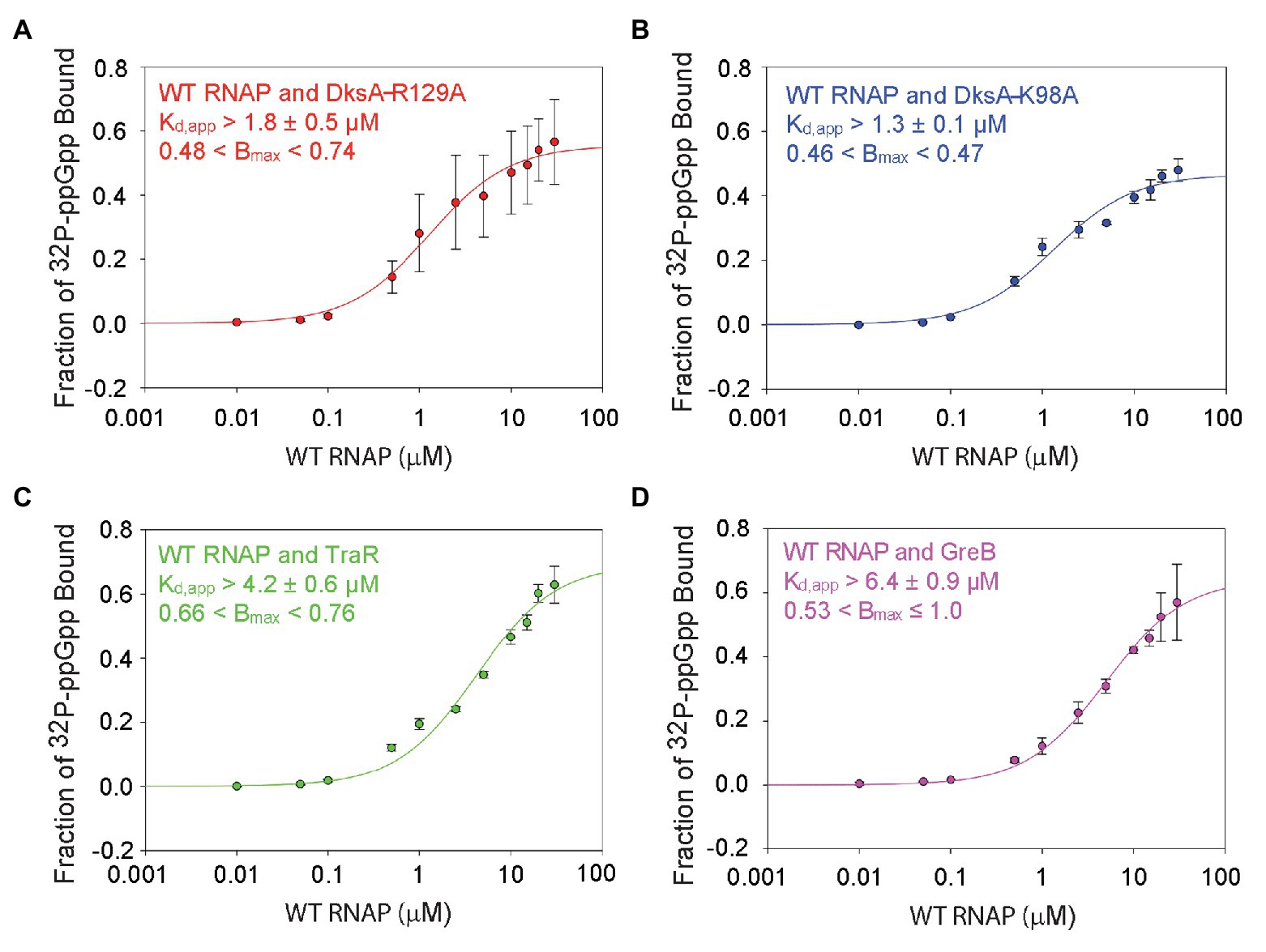
Figure 4. DksA enhances ppGpp binding to Site 1. DRaCALA measurements were performed with wild-type RNAP and the indicated secondary channel binding factors. (A) Wild-type RNAP with DksA-R129A. One-site saturation binding curves with data averaged from four independent experiments. Kd,app and Bmax values, and error shown are averages from the values determined for each independent experiment. (B) Wild-type RNAP with DksA-K98A. Data from two independent experiments, as described in (A). (C) Wild-type RNAP with TraR. Data from three independent experiments, as described in (A). (D) Wild-type RNAP with GreB. Data from three independent experiments, as described in (A).
We also tested whether other secondary channel binding proteins increased the affinity of Site 1 for ppGpp. TraR is a distant homolog of DksA that is encoded by the F element (Blankschien et al., 2009). Although TraR is only half the size of DksA, it has an effect on transcription by itself that is as strong as the effect of DksA and ppGpp together (Gopalkrishnan et al., 2017). However, TraR lacks the residues in DksA that interact with ppGpp, and therefore ppGpp does not bind to RNAPΔω or increase the effect of TraR on transcription (Gopalkrishnan et al., 2017). The Kd,app for ppGpp binding to Site 1 in the wild-type RNAP-TraR complex was at least 4.2 ± 0.6 μM (Figure 4C; the curve did not plateau, so the affinity of ppGpp for Site 1 must be considered a lower estimate) vs. 6.1 ± 1.3 μM in the absence of TraR (Figure 2B). These Kd,app values for binding of ppGpp to Site 1 were not statistically different from each other. The TraR result is discussed further in the next section.
GreB is another secondary channel binding factor that does not function in conjunction with ppGpp (Rutherford et al., 2007; Lee et al., 2012). The Kd,app of ppGpp for the wild-type RNAP-GreB complex (i.e., containing Site 1) was ~6.4 ± 0.9 μM (Figure 4D), very similar to that in the absence of GreB (Figure 2), and ppGpp did not bind to the RNAPΔω-GreB complex (Supplementary Figure S6C). In summary, the mutant DksA proteins increased the affinity of ppGpp for Site 1, and these effects were specific, since other secondary channel binding factors did not increase the affinity of ppGpp for Site 1.
The crystal structures of the RNAP-DksA-ppGpp complex and the RNAP-TraR complex (Molodtsov et al., 2018) provide a potential explanation for the observed effect of DksA, and not TraR, on ppGpp binding to Site 1. In the structures, DksA residues near its coiled coil tip (D64 and N68) are located 6–10 Å from β’ residues K598/K599. β’ K598 and K599 are at the N-terminus of a α-helix that extends to ppGpp binding Site 1 (Figure 5A). β’ residue K615, near the C-terminal end of this α-helix, interacts not only with ppGpp but also with residues in ω that bind to ppGpp. The proximity of DksA to β’-K598/K599 leads us to speculate that a DksA interaction with these residues in β’ might allosterically affect residues in Site 1, explaining the effect of DksA on ppGpp binding to Site 1 (Figure 4). Consistent with a direct interaction between DksA and K598/K599, an RNAP variant that contains alanine substitutions for K598 and K599 was partially resistant to DksA’s ability to shorten the lifetime of RNAP-promoter complexes (Vrentas, 2008).
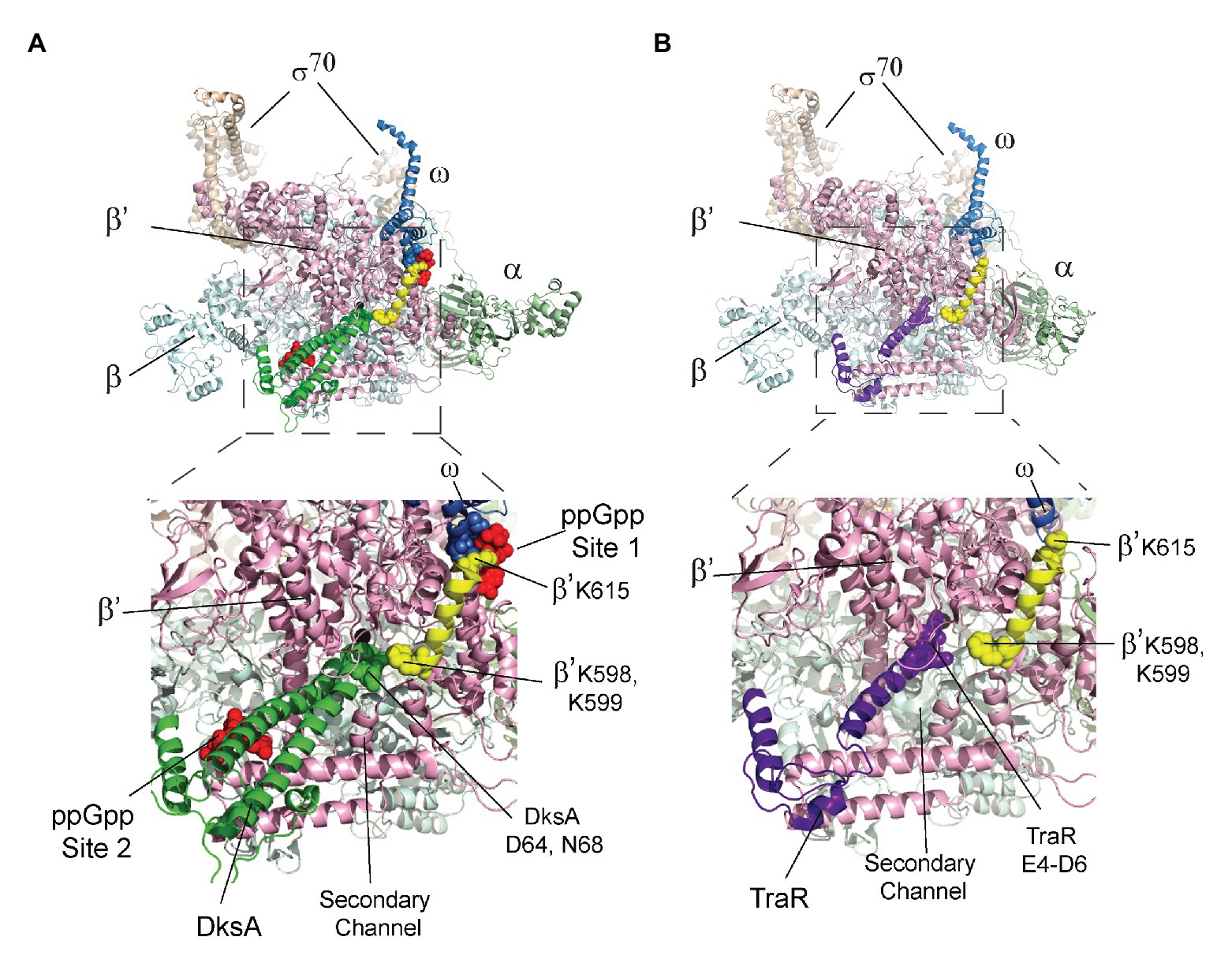
Figure 5. The β’ K598-K615 α-helix connects DksA and Site 1. The top image in each panel shows the crystal structure of RNAP with either (A) DksA or (B) TraR bound in the secondary channel. An expanded view of the boxed region in each panel is shown below the structures of RNAP. (A) X-ray crystal structures of wild-type RNAP holoenzyme with DksA and ppGpp soaked into the crystal (PDB 5VSW; Molodtsov et al., 2018). Coloring: α, green; β, cyan; β’, light pink; ω, gray; σ, tan; and ppGpp, red spheres. The β’ K598-K615 α-helix is yellow with K598, K599, and K615 shown as spheres. DksA is green with D64 and N68 shown as spheres. ω residues A2-T5 are shown as gray spheres. (B) RNAP holoenzyme with TraR (PDB 5W1S; Molodtsov et al., 2018). The β’ K598-K615 α-helix is yellow, and TraR is dark purple with residues E4, D6, and E7 near the TraR N-terminus shown as spheres.
In contrast, in the structure of the RNAP-TraR complex (Figure 5B; Molodtsov et al., 2018 Chen et al., 2019), there is a much greater separation (16–20 Å) between the N-terminus of TraR (corresponding to the coiled-coil tip region of DksA) and β’-K598/K599. We suggest that the increased separation between TraR and K598/K599 might explain the absence of a significant effect of TraR on ppGpp binding to Site 1 (Figure 4C). Thus, the structural information supports the biochemical data suggesting there is an allosteric effect of DksA on binding of ppGpp to Site 1.
DksA concentrations are constant throughout log phase and decrease only slightly in stationary phase (Rutherford et al., 2007). Therefore, changes in Site 2 binding of ppGpp are more likely a function of changes in ppGpp than DksA concentration, at least during exponential growth. In contrast, there is little information about the concentration of ω at different stages in growth or under different nutritional conditions. We reported previously that the magnitude of the effect of ppGpp on transcription in vitro increased slightly when RNAP was purified from cells in which ω was overproduced, suggesting that a small fraction of RNAP lacks ω in cells not overproducing ω (Vrentas et al., 2005).
To determine directly whether ω protein levels vary in vivo, we raised an antibody against ω and examined ω protein levels using Western blots. The antibody reacted with a band of the expected size in a strain with wild-type rpoZ but not one lacking rpoZ (Supplementary Figure S2B, compare lanes 1 and 6). Fortuitously, the anti-ω antibody also reacted with σ70. This cross-reactivity was verified using antibody to σ70 and ω, and Western blots with purified ω and σ70 (Supplementary Figures S2C,D).
As with DksA, the ω concentration remained relatively constant in cells growing exponentially in rich medium (LB), but it declined ~2-fold when cells transitioned to stationary phase (~OD600 = 1; Figures 6A,B). In contrast, the σ70 concentration was relatively constant. It is possible that the small decrease in ω levels during stationary phase could decrease the saturation of RNAP with ω and create a subpopulation of RNAP molecules defective in ppGpp binding to Site 1, but the changes in ω concentration are small and unlikely to be a major determinant of regulation by ppGpp during non-starvation conditions.
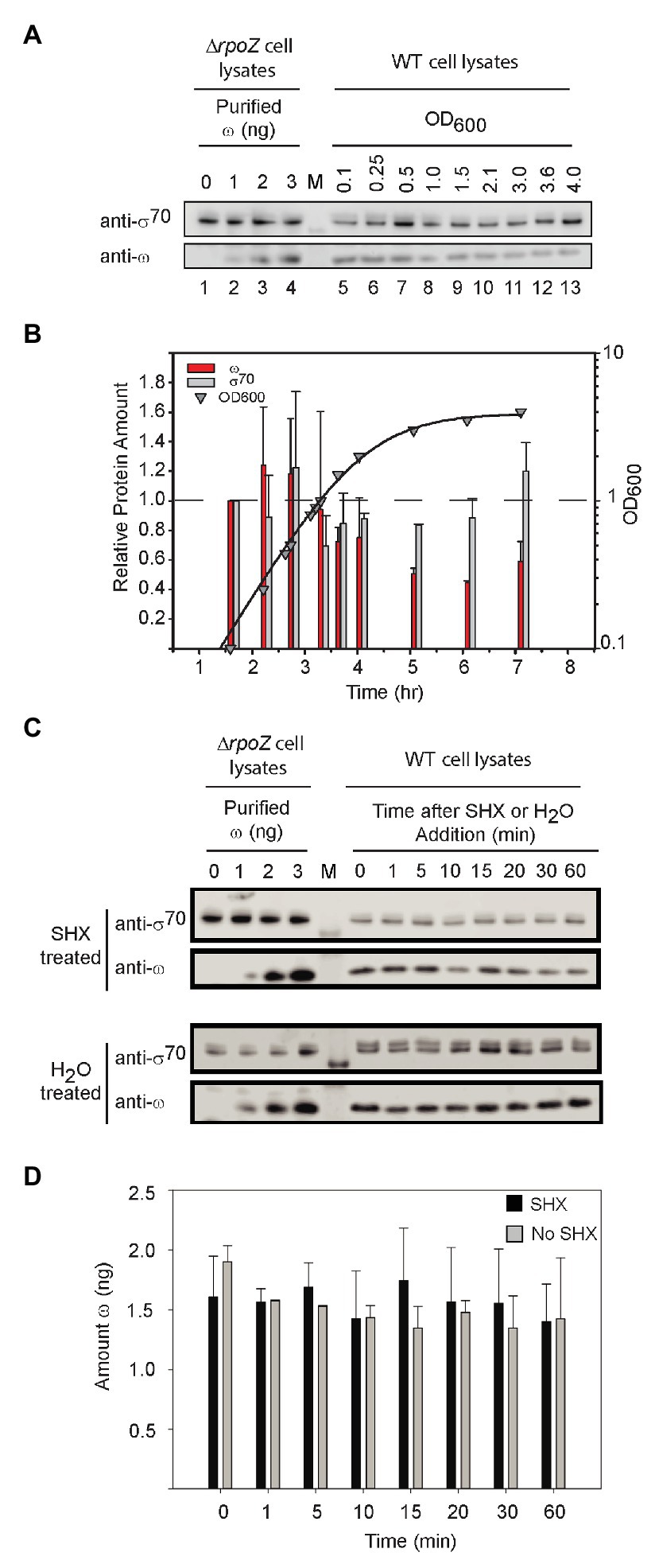
Figure 6. ω levels are relatively constant. (A) Representative Western blots using an anti-ω antibody. Purified ω was added to cell lysates from ΔrpoZ cells (RLG14044) to create a standard curve for estimation of the amount of ω (lanes 1–4). Wild-type cells were grown in LB, and lysates were collected at different OD600 for analysis of ω concentrations using an anti-ω antibody (lanes 5–13). Lane labeled M indicates molecular weight marker. The anti-ω antibody cross-reacted with σ70 (see Supplementary Figure S2). (B) The amounts of ω and σ70 at different times in growth were determined from experiments like that in (A) and are shown relative to the amounts present at an OD600 of 0.1 (first time point, ~1.5 h). Bars indicate the averages of three independent experiments, with error bars representing one SD from the mean. ω bars, red; σ70 bars, gray. ω decreased a maximum of ~2-fold, whereas σ70 (gray bars) remained relatively constant. (C) Representative Western blots with anti-ω antibody and cell lysates following treatment with serine hydroxamate (SHX) which starves cells for serine. Lanes labeled 0–3 indicate amounts of ω added FIGURE 6to cell lysates, lane labeled M indicates molecular weight marker, and lanes at right labeled 0–60 indicate minutes after SHX addition. (D) Average levels of ω in cells treated with SHX (black bars), compared to untreated cells (gray bars). ω bands in each Western blot were quantified using the ω standard curve derived from the same blot. The bars represent the average ω (in ng) for three separate experiments; error bars represent one SD from the mean.
We also examined ω levels when ppGpp was induced to high concentration. After starvation of cells for serine aminoacyl tRNA by addition of serine hydroxamate (SHX; Figure 6C), ω concentrations were stable for at least 60 min, neither increasing nor decreasing well beyond the time needed for a typical stringent response (Figure 6D). We conclude that Site 1 occupancy by ppGpp is determined by changes in ppGpp concentration, not by changes in ω concentration, in both starved and unstarved cultures.
Although the regulatory role of ppGpp during the stringent response has been recognized for more than half a century (Cashel and Gallant, 1969), the mechanistic explanations for the effects of ppGpp on transcription are only now becoming clear. The recent discovery that there are two binding sites for ppGpp on RNAP has shed new light on the mechanisms of transcription regulation by ppGpp (Ross et al., 2016; Molodtsov et al., 2018). Here, we show that the two binding sites on RNAP have very similar intrinsic affinities for ppGpp, contradicting a model in which different affinities of the two sites for ppGpp increase the dynamic range of its effects on transcription during the stringent response. Instead, our data are more consistent with a model in which both binding sites reversibly bind ppGpp, are saturated to approximately the same extent when ppGpp concentrations are low, and become fully occupied at approximately the same time when ppGpp is induced to high levels.
We did not test binding of pppGpp (the pentaphosphate) to the two sites in RNAP. Available data indicate that, in E. coli, pppGpp is less abundant than ppGpp in vivo and that its effects on transcription in vivo and in vitro, particularly at Site 2, are less potent than those of ppGpp (Potrykus and Cashel, 2008; Mechold et al., 2013). Structures of the RNAP/DksA/ppGpp complex indicate that ppGpp at each site is partially solvent exposed (Zuo et al., 2013; Molodtsov et al., 2018), suggesting that each site could accommodate the additional phosphate group in pppGpp. However, it is not known whether the additional phosphate group could alter binding affinity.
Guanosine tetraphosphate binding to the two sites on RNAP can affect at least two different steps in transcription, impacting the kinetics of initiation by multiple mechanisms and resulting in different effects on transcriptional output from different promoters (Ross et al., 2016; Sanchez-Vazquez et al., 2019). For example, ppGpp binding to Site 2 can activate transcription from certain promoters whereas binding of ppGpp to Site 1 cannot (Ross et al., 2016; Gourse et al., 2018). In addition, it was also shown long ago that ppGpp affects transcription elongation in vitro (Kingston et al., 1981). Since those studies were performed without DksA, the effects on elongation were likely a result of binding to Site 1. Further studies will be needed to understand the mechanism(s) responsible for the effects of ppGpp on elongation, and which other factors play roles in these effects (see for example, Singh et al., 2016).
Given the concentration of ppGpp reported for non-starvation conditions in rich medium (~1–10 μM; Ryals et al., 1982), a concentration of RNAP in vivo of ~10 μM (Bremer and Dennis, 2008; Li et al., 2014), and the apparent binding affinities for ppGpp reported here, in a significant fraction of RNAP molecules it is likely that one or the other site would not be saturated with ppGpp. In the lower range of ppGpp concentrations (i.e., non-starvation conditions), when one site or the other is bound by ppGpp, this could result in some stochastic variation in regulation of transcription by ppGpp in subpopulations of RNAP molecules, creating some “bet-hedging” (Rocha et al., 2002). Stochasticity would be less impactful under starvation conditions when high ppGpp concentrations are present, and both ppGpp binding sites on all RNAP molecules would be saturated.
Depending on which step during transcription is affected, ppGpp function requires the presence of ω or DksA. However, since the concentrations of ω and DksA change little during exponential growth in E. coli, and no more than 2-fold in stationary phase, differences in the occupancies of the two ppGpp binding sites over time (and thus the impact of ppGpp on transcription) must result primarily from changes in ppGpp, not ω or DksA concentrations.
Interestingly, our data indicate that DksA increases the affinity of the wild-type RNAP for ppGpp (Kd,app = 3.3 ± 1.2 μM average affinity for the two binding sites on wild-type RNAP versus 6.1 ± 1.3 μM or 7.9 ± 1.3 μM, respectively, for RNAPs containing only Site 1 or 2). In addition, the affinity of ppGpp for the RNAPs in complex with the “separation of function” DksA variants is 1.3 ± 0.1–1.8 ± 0.5 μM. These results suggest that the increased affinity of Site 1 for ppGpp in the presence of DksA accounts for the reduced average affinity of wild-type RNAP for ppGpp relative to the affinity for either Site 1 or Site 2 alone. Thus, DksA allosterically enhances binding of ppGpp to Site 1, even though DksA and Site 1 are separated by ~30 Å at their position of closest approach (Figure 5). Although a limited number of secondary channel binding proteins was tested, we suggest this enhancement of ppGpp affinity for Site 1 is unique to DksA (Figure 4). The physiological consequence, if any, of the enhancement in affinity of Site 1 for ppGpp by DksA remains to be determined.
Finally, the concentrations of ppGpp observed during the stringent response appear to be much higher than needed for full occupancy of the two sites on RNAP. Although the high concentrations undoubtedly evolved in part to increase the kinetics of ppGpp occupancy of RNAP, some 70 proteins in E. coli bind ppGpp, only a subset of which bind ppGpp with an affinity as high as RNAP (Pao and Dyess, 1981; Hou et al., 1999; Kanjee et al., 2011a,b; Zhang et al., 2018; Anderson et al., 2019; Wang et al., 2019). We suggest that the high levels of ppGpp produced during severe starvations could be needed for ppGpp to bind to the proteins with lower affinity ppGpp binding sites.
All datasets presented in this study are included in the article/Supplementary Material.
AM designed, performed, and analyzed experiments and wrote the paper. WR analyzed experiments and wrote the paper. DT performed and analyzed experiments. RG designed and analyzed experiments and wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by a grant to RG from the National Institutes of Health (R01 GM37048). AM was supported in part by predoctoral fellowships from the National Science Foundation and the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank J.-H. Lee for purified GreB, S. Gopalkrishnan for purified TraR, and K. Myers, R. Landick, J. Keck, D. Brow, P. Kiley, and T. Record for helpful comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.587098/full#supplementary-material
Anderson, B. W., Liu, K., Wolak, C., Dubiel, K., She, F., Satyshur, K. A., et al. (2019). Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction. Elife 8:e47534. doi: 10.7554/eLife.47534
Blankschien, M. D., Potrykus, K., Grace, E., Choudhary, A., Vinella, D., Cashel, M., et al. (2009). TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 5:e1000345. doi: 10.1371/journal.pgen.1000345
Bremer, H., and Dennis, P. P. (2008). Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus 3. doi: 10.1128/ecosal.5.2.3
Cashel, M., and Gallant, J. (1969). Two compounds implicated in the function of the RC genes of Escherichia coli. Nature 221, 838–841. doi: 10.1038/221838a0
Chen, J., Gopalkrishnan, S., Chiu, C., Chen, A. Y., Campbell, E. A., Gourse, R. L., et al. (2019). E. coli TraR allosterically regulates transcription initiation by altering RNA polymerase conformation. Elife 8:e49375. doi: 10.7554/eLife.49375
Durfee, T., Hansen, A. M., Zhi, H., Blattner, F. R., and Jin, D. J. (2008). Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 190, 1084–1096. doi: 10.1128/JB.01092-07
Gentry, D. R., and Burgess, R. R. (1989). rpoZ, encoding the omega subunit of Escherichia coli RNA polymerase, is in the same operon as spoT. J. Bacteriol. 171, 1271–1277. doi: 10.1128/jb.171.3.1271-1277.1989
Gentry, D., Xiao, H., Burgess, R. R., and Cashel, M. (1991). The omega subunit of Escherichia coli K-12 RNA polymerase is not required for stringent RNA control in vivo. J. Bacteriol. 173, 3901–3903. doi: 10.1128/jb.173.12.3901-3903.1991
Gopalkrishnan, S., Ross, W., Chen, A. Y., and Gourse, R. L. (2017). TraR directly regulates transcription initiations by mimicking the combined effects of the global regulators DksA and ppGpp. Proc. Natl. Acad. Sci. U. S. A. 114, E5539–E5548. doi: 10.1073/pnas.1704105114
Gourse, R. L., Chen, A. Y., Gopalkrishnan, S., Sanchez-Vazquez, P., Myers, A., and Ross, W. (2018). Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184. doi: 10.1146/annurev-micro-090817-062444
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
Hill, A. V. (1910). The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J. Physiol. 40, iv–vii.
Hou, Z., Cashel, M., Fromm, H. J., and Honzatko, R. B. (1999). Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase. J. Biol. Chem. 274, 17505–17510. doi: 10.1074/jbc.274.25.17505
Irving, S. E., and Corrigan, R. M. (2018). Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology 164, 268–276. doi: 10.1099/mic.0.000621
Kanjee, U., Gutsche, I., Alexopoulos, E., Zhao, B. Y., El Bakkouri, M., Thibault, G., et al. (2011a). Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 30, 931–944. doi: 10.1038/emboj.2011.5
Kanjee, U., Gutsche, I., Ramachandran, S., and Houry, W. A. (2011b). The enzymatic activities of the Escherichia coli basic aliphatic amino acid decarboxylases exhibit a pH zone of inhibition. Biochemistry 50, 9388–9398. doi: 10.1021/bi201161k
Kingston, R. E., Nierman, W. C., and Chamberlin, M. J. (1981). A direct effect of guanosine tetraphosphate on pausing Escherichia coli RNA polymerase during RNA chain elongation. J. Biol. Chem. 256, 2787–2797.
Krasny, L., and Gourse, R. L. (2004). An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23, 4473–4483. doi: 10.1038/sj.emboj.7600423
Lee, J. -H., Lennon, C. W., Ross, W., and Gourse, R. L. (2012). Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J. Mol. Biol. 416, 503–517. doi: 10.1016/j.jmb.2011.12.028
Li, G. -W., Burkhardt, D., Gross, C., and Weissman, J. S. (2014). Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635. doi: 10.1016/j.cell.2014.02.033
Liu, K., Bittner, A. N., and Wang, J. D. (2015). Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 24, 72–79. doi: 10.1016/jmib.2015.01.012
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S., and Cashel, M. (2013). Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189. doi: 10.1093/nar/gkt302
Molodtsov, V., Sineva, E., Zhang, L., Huang, X., Cashel, M., Ades, S. E., et al. (2018). Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA. Mol. Cell 69, 828.e5–839.e5. doi: 10.1016/j.molcel.2018.01.035
Pao, C. C., and Dyess, B. T. (1981). Effect of unusual guanosine nucleotides on the activities of some Escherichia coli cellular enzymes. Biochim. Biophys. Acta 677, 358–362. doi: 10.1016/0304-4165(81)90247-6
Paul, B. J., Barker, M. M., Ross, W., Schneider, D. A., Webb, C., Foster, J. W., et al. (2004). DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322. doi: 10.1016/j.cell.2004.07.009
Paul, B. J., Berkmen, M. B., and Gourse, R. L. (2005). DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. U. S. A. 102, 7823–7828. doi: 10.1073/pnas.0501170102
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Rocha, E. P. C., Matic, I., and Taddei, F. (2002). Over-representation of repeats in stress response genes: a strategy to increase versatility under stress conditions? Nucleic Acids Res. 30, 1886–1894. doi: 10.1093/nar/30.9.1886
Roelofs, K. G., Wang, J., Sintim, H. O., and Lee, V. T. (2011). Differential radial capillary action of ligand assay for high-through put detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U. S. A. 108, 15528–15533. doi: 10.1073/pnas.1018949108
Ross, W., Sanchez-Vazquez, P., Chen, A. Y., Lee, J. -H., Burgos, H. L., and Gourse, R. L. (2016). ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell 62, 811–823. doi: 10.1016/j.molcel.2016.04.029
Ross, W., Vrentas, C. E., Sanchez-Vazquez, P., Gaal, T., and Gourse, R. L. (2013). The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50, 420–429. doi: 10.1016/j.molcel.2013.03.021
Rutherford, S. T., Lemke, J. J., Vrentas, C. E., Gaal, T., Ross, W., and Gourse, R. L. (2007). Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol. 366, 1243–1257. doi: 10.1016/j.jmb.2006.12.013
Ryals, J., Little, R., and Bremer, H. (1982). Control of RNA synthesis in Escherichia coli after a shift to higher temperature. J. Bacteriol. 151, 1425–1432. doi: 10.1128/JB.151.3.1425-1432.1982
Sanchez-Vazquez, P., Dewey, C. N., Kitten, N., Ross, W., and Gourse, R. L. (2019). Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 116, 8310–8319. doi: 10.1073/pnas.1819682116
Singh, N., Bubunenko, M., Smith, C., Abbott, D. M., Stringer, A. M., Shi, R., et al. (2016). SuhB associates with nus factors to facilitate 30S ribosome biogenesis in Escherichia coli. mBio 7:e00114. doi: 10.1128/mBio.00114-16
Traxler, M. F., Summer, S. M., Nguyen, H. T., Zacharia, V. M., Hightower, G. A., Smith, J. T., et al. (2008). The global ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x
Varik, V., Oliveira, S. R. A., Hauryliuk, V., and Tenson, T. (2017). HPLC-based quantification of bactgerial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci. Rep. 7:11022. doi: 10.1038/s41598-017-10988-6
Vrentas, C. E. (2008). A study of the requirements for the response of Escherichia coli RNA polymerase to guanosine tetraphosphate (ppGpp). University of Wisconsin-Madison.
Vrentas, C. E., Gaal, T., Ross, W., Ebright, R. H., and Gourse, R. L. (2005). Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 19, 2378–2387. doi: 10.1101/gad.1340305
Wang, B., Dai, P., Ding, D., Del Rosario, A., Grant, R. A., Pentelute, B. L., et al. (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150. doi: 10.1038/s41589-018-0183-4
Xiao, H., Kalman, M., Ikehara, K., Zemel, S., Glaser, G., and Cashel, M. (1991). Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990.
Zhang, Y., Zbornikova, E., Rejman, D., and Gerdes, K. (2018). Novel (p)ppGpp binding and metabolizing proteins in Escherichia coli. mBio 9, e02188–e02217. doi: 10.1128/mBio.02188-17
Keywords: bacterial transcription, RNA polymerase, ppGpp, DksA, omega subunit, stringent response
Citation: Myers AR, Thistle DP, Ross W and Gourse RL (2020) Guanosine Tetraphosphate Has a Similar Affinity for Each of Its Two Binding Sites on Escherichia coli RNA Polymerase. Front. Microbiol. 11:587098. doi: 10.3389/fmicb.2020.587098
Received: 24 July 2020; Accepted: 06 October 2020;
Published: 05 November 2020.
Edited by:
Gert Bange, University of Marburg, GermanyReviewed by:
Vasili Hauryliuk, Umeå University, SwedenCopyright © 2020 Myers, Thistle, Ross and Gourse. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard L. Gourse, cmdvdXJzZUBiYWN0Lndpc2MuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.