
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 November 2020
Sec. Virology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.587005
This article is part of the Research Topic Reverse Genetics and Infectious Models for RNA Virus Pathogenesis and Co-Evolution View all 7 articles
 Man Yu1†
Man Yu1† Xinyue Bi1†
Xinyue Bi1† Yuanmin Huang1
Yuanmin Huang1 Yong Chen2
Yong Chen2 Jun Wang2
Jun Wang2 Ruina Zhang2
Ruina Zhang2 Yunkang Lei2
Yunkang Lei2 Zihao Xia1
Zihao Xia1 Mengnan An1*
Mengnan An1* Yuanhua Wu1*
Yuanhua Wu1*In the pathogen infection and host defence equilibrium, plant viruses have evolved to efficiently replicate their genomes, to resist the attack from host defence responses and to avoid causing severe negative effect on growth and metabolism of the hosts. In this study, we generated chimeric tobacco mosaic virus (TMV) variants, in which the coat protein (CP) sequences were substituted with that of cucumber green mottle mosaic virus (CGMMV) or pepper mild mottle virus (PMMoV) to address the role of these in virus infection and host symptomology. The results showed that the chimeric viruses (TMV-CGCP or TMV-PMCP) induce stunting and necrotic symptoms in tobacco plants. We analyzed the transcriptomic changes in tobacco plants after infection of TMV and its chimeras using a high-throughput RNA sequencing approach and found that infection of the chimeric TMV induced significant up-regulation of host defence responsive genes together with salicylic (SA) or abscisic acid (ABA) responsive genes, but down-regulation of auxin (Aux) responsive genes. We further confirmed the increase in the levels of SA and ABA, together with the reduced levels of Aux after infection of chimeric TMV in tobacco plants. These data suggest novel roles of tobamovirus CP in induction of host symptoms and defence responses.
Plant viruses are obligate intracellular parasites and rely on the host machinery for the establishment of their infection cycle (McLeish et al., 2018). Virus-host interaction induces significant molecular alterations in the host plant, which leads to the display of disease symptoms (Lu et al., 2012). Plants usually develop symptoms such as mosaic, stunting, chlorosis or necrosis after virus infection, which may significantly reduce the quality of crops (McLeish et al., 2018; Conti et al., 2017). To combat virus infection, plants mainly employ innate immunity such as pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Gouveia et al., 2016; Calil and Fontes, 2017). In addition, plants have developed RNA silencing, autophagy or other strategies to limit the virus (Ding, 2010; Kushwaha et al., 2019). To counteract host defence, plant viruses also use elaborate ways to suppress or evade host defence machinery to ensure their survival (Wang, 2015; Pallas and Garcia, 2011). In this process, plant viruses hijack host factors for their genome replication, intercellular movement and systemic infection, leading to significant alterations in host gene regulatory network (e.g., genes involved in defence responses and multiple signaling cascades) and metabolic pathway (e.g., biosynthesis of various phytohormones) (Whitham et al., 2003; Collum and Culver, 2016). The high-throughput sequencing techniques, such as RNA sequencing (RNA-Seq) are powerful tools for the identifications of genes whose expression is altered during virus infection in plants (Chen et al., 2017; Li et al., 2017; Sheng et al., 2019).
Tobacco mosaic virus (TMV) belongs to the genus Tobamovirus with a positive single-strand RNA and the genome encodes at least four proteins (Rybicki, 2015). Among them, the 126 and 183 kDa proteins are translated from TMV genomic RNA and play crucial roles in viral genome replication (Ishibashi and Ishikawa, 2016). The 30 kDa movement protein (MP) expressed from sub-genomic RNA facilitates movement of the virus between host cells. The 17.5 kDa coat protein (CP) of TMV is a multifunctional protein responsible for virion formation, viral systemic movement, cross-protection and genomic RNA stability (Callaway et al., 2001; Ivanov and Mäkinen, 2012). It was reported that the sequences of TMV CP are responsible for the differential disease symptomology of the infected plants (Dawson et al., 1988; Culver, 2002). Furthermore, viral CP was also shown to contribute significantly to determining host range of tobamoviruses (Weber and Bujarski, 2015).
The researches of TMV and other tobamovirus species have greatly contributed to the development of virology and viral evolution (Moury et al., 2017). Viruses like cucumber green mottle mosaic virus (CGMMV) and pepper mild mottle virus (PMMoV) are well-investigated tobamoviruses and their genetic structures are similar to that of TMV (Dardick et al., 1999), but only have approximately 45–65% nucleotide sequence similarity. For example, certain viral species have evolved the differential host ranges in which CGMMV mainly infects the family Cucurbitaceae while PMMoV infects pepper plants (Li et al., 2015; Dombrovsky et al., 2017). In contrast, TMV seems to have wider host range than that of CGMMV and PMMoV. To date, the critical viral proteins or the nucleotide sequences involved in the determination of host ranges of tobamoviruses have not been well understood.
The application of homologous recombination technology to construct chimeric viruses provide important tools to unravel the functions of specific viral RNA sequences or the coding proteins during infection (Yelina et al., 2002; Lukhovitskaya et al., 2013; Tuo et al., 2015; Yu et al., 2019). In a previous study, we generated a PMMoV infectious clone and several chimeric clones to clarify critical RNA sequences required for efficient virus infection (Yu et al., 2019).
In this study, we constructed two chimera TMV-CGCP and TMV-PMCP, in which the CP of TMV was substituted with that of CGMMV and PMMoV using the homologous recombination technique. Intriguingly, Nicotiana tabacum inoculated with the chimeric viruses showed significant stunting compared with those inoculated with wild-type TMV. We further investigated the global changes of gene expressions during TMV and the chimeric virus infection in N. tabacum using RNA-seq technology. The results showed that the host resistance genes were significantly up-regulated in leaves infected with TMV-CGCP or TMV-PMCP. Furthermore, infection of the chimeras also induced up-regulation of the genes involved in host defence responses and affected phytohormone biosynthesis or signaling. Specifically, the levels of salicylic acid (SA) and abscisic acid (ABA) were increased, whereas the levels of auxin was decreased by infection of the chimeric viruses.
To construct pCB-TMV-CGCP and pCB-TMV-PMCP that express chimeric TMV encoding heterologous coat protein sequences from CGMMV and PMMoV (Figure 1A), the plasmid pCB-TMV-SY, an infectious clone of TMV Shenyang isolate (MG516107) was used as a template, and TMV-CG + /TMV-CG- and TMV-PM + /TMV-PM- primer pairs were used to amplify two vector DNA fragments with approximately 1,1000 bp in size by TransStart FastPfu PCR SuperMix (TransGen Biotech, Beijing, China). The amplified PCR products were thereafter treated with DMT enzyme (TransGen, Biotech, China) to digest the non-mutated parental plasmid. Two insert DNA fragments were amplified using pCB-CGMMV-LN (KY040049) and pCB-PMMoV-HLD (Yu et al., 2019; Figure 1A) as templates by primer pairs T-CG-CP + /T-CG-CP- and T-PM-CP + /T-PM- CP-, respectively, using high fidelity KOD Plus DNA polymerase (Toyobo, Osaka, Japan). Then the PCR products were purified using an agarose gel DNA Recovery Kit (TIANGEN, Beijing) and ligated with the amplified vector fragments using ClonExpressTM MultiS One Step Cloning Kit (Vazyme, Nanjing, China).
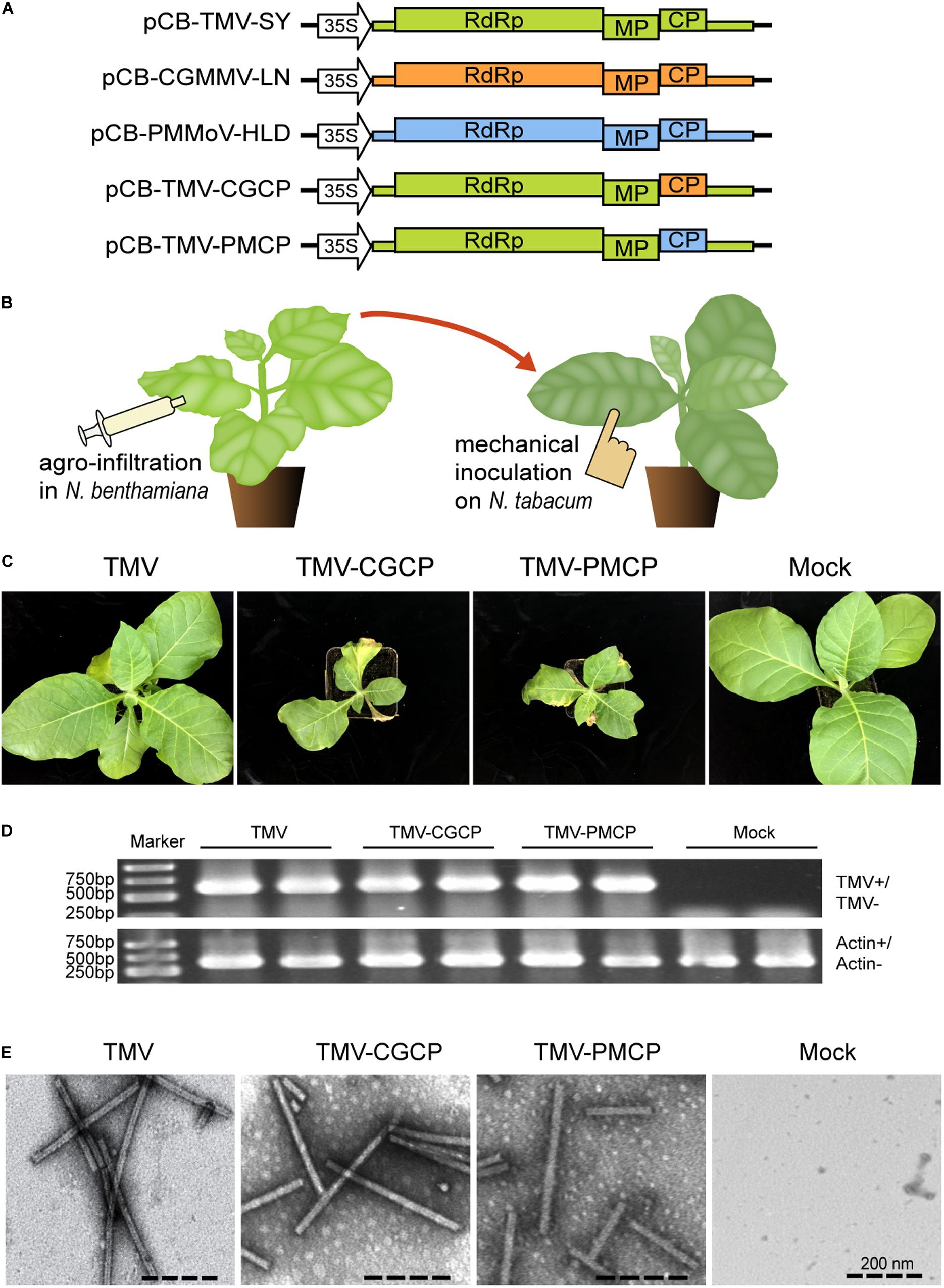
Figure 1. Symptoms of N. tabacum infected by TMV or the chimeric viruses TMV-CGCP, TMV-PMCP. Mock-inoculated plants were used as control treatments (Mock). (A) Schematic representation of the TMV, CGMMV, PMMoV and the chimeras of TMV constructed in this study. (B) Diagram of the virus inoculation methods. Agrobacterium tumefaciens strain GV3101 containing pCB-TMV-SY, pCB-TMV-CGCP or pCB-TMV-PMCP was infiltrated into leaves of N. benthamiana and then the sap of its upper leaves were mechanically inoculated onto leaves of N. tabacum. (C) Symptoms developed by TMV, TMV-CGCP and TMV-PMCP on N. tabacum. Infection of TMV-CGCP or TMV-PMCP induced significant stunting and necrotic symptoms in N. tabacum compared with those infected by wild-type TMV at 12 dpi. (D) RT-PCR analysis of TMV and the chimeras from the systemic leaves of N. tabacum. Host gene actin was used as a control. (E) Electron micrograph of viral particles of TMV and the chimeras isolated from N. tabacum leaves.
Plasmids of pCB-TMV-SY, pCB-CGMMV-LN and pCB-PMMoV-HLD were used as templates to generate 510 bp, 516 bp and 504 bp sized PCR products using primer pairs TMV-CP + /TMV-Myc- CP-, PMMoV-CP + /PMMoV-Myc-CP- and CGMMV-CP + /CGMMV-Myc- CP-, respectively. The three amplified DNA fragments were integrated to the BgIII and HindIII double digested pGD vector using ClonExpressTM MultiS One Step Cloning Kit (TransGen, Biotech, China), respectively, to construct pGD-TMCP-Myc, pGD-CGCP-Myc and pGD-PMCP-Myc.
To construct plasmids of pCB-TMV-fsCP, pCB-TMV-CGfsCP and pCB-TMV-PMfsCP (Supplementary Figure S1A) that cannot express the respective viral coat proteins, pCB-TMV-SY, pCB-CGMMV-LN and pCB-PMMoV-HLD were used as templates and were amplified by primer pairs TMVfsCP + /TMVfsCP-, T-CGCP-fsCP + /T-CGCP- fsCP-, T-PMCP-fsCP + /T-PMCP- fsCP-, respectively. The amplified DNA fragments were treated with DMT enzymes and subjected to self-ligation using a Fast Mutagenesis System kit (TransGen, Biotech, China). The constructs were transformed into Agrobacterium tumefaciens GV3101. All of the PCR primers used in this study are listed in Supplementary Table S1.
The virions of wild-type TMV, TMV-CGCP, and TMV-PMCP were extracted and purified from the corresponding agrobacterium infiltrated N. benthamiana leaves using Gooding’s method (Gooding and Hebert, 1967). The purified virions adjusted to the final concentration of 2 mg/ml were treated by phosphotungstic acid stain method (Quintarelli et al., 1971) and were observed using a transmission electron microscopy HT7700 Exalens (Hitachi, Ltd., Tokyo, Japan).
Plants of N. benthamiana and N. tabacum cv. NC89 were cultivated in a growth climate chamber at 25°C with 16 h/8 h (light/dark) cycles. A. tumefaciens cultures harboring pCB-TMV, pCB-TMV-CGCP or pCB-TMV-PMCP were infiltrated into N. benthamiana leaves at (5–7) leaf stage. The upper leaves of N. benthamiana infected with TMV, TMV-CGCP or TMV-PMCP at 7 days post inoculation (dpi) were harvested and mechanically inoculated on the leaves of N. tabacum variety NC89 for symptom observation and subsequent analysis.
Total RNA was extracted from leaves of N. tabacum that were inoculated with wild-type or chimeric TMV using TRIzon Reagent (TIANGEN, Beijing, China). Three independent biological replicates were used for RNA-seq. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, United States) according to the manufacturer’s instructions and index codes were added to attribute sequences for each sample. The mRNA was purified from total RNA using poly-T oligo-attached magnetic beads and fragmented using divalent cations at elevated temperatures in NEBNext First Strand Synthesis Reaction Buffer (5X). The cDNA was synthesized and purified with AMPure XP system (Beckman Coulter, Beverly, United Statesto select approximately 240 bp fragments. The size-selected, adaptor-ligated cDNA was treated with USER Enzyme (NEB, United States) before PCR, then the purified PCR products were submitted to sequence on an Illumina sequencing platform (HiSeqTM 4000) conducted by the Biomarker Technologies Corporation, Beijing, China.
The raw reads generated by Illumina sequencing were submitted to the Sequence Read Archive database at NCBI (SRA)1, with the SRA BioProject accession number [PRJNA594802 (TMV-inoculated samples): SRR10662682, SRR10662681, SRR10662680; PRJNA594806 (TMV-CGCP-inoculated samples): SRR10662707, SRR10662706, SRR10662705; PRJNA594848 (TMV-PMCP-inoculated samples): SRR10664114, SRR10664113, SRR10664112; PRJNA594821 (Mock-inoculated samples): SRR10662770, SRR10662769, SRR10662768]. The clean reads were mapped to the reference genome sequence using Hisat2 tools soft.2 Differential expression analysis of TMV vs. TMV-CGCP and TMV vs. TMV-PMCP were performed using the DEseq.3 The differentially expressed genes (DEGs) were determined by adjusting P values to <0.01 using the Benjamini and Hochberg’s approach for controlling the false discovery rate <0.05. To further analysis of DEGs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway4 were implemented.
Plant salicylic acid (SA), jasmonic acid (JA), abscisic acid (ABA), and indole-3-acetic acid (IAA) were determined as described with some modifications (Pan et al., 2010). The leaves of N. tabacum inoculated with TMV, TMV-CGCP, TMV-PMCP or buffer control were ground into powder with liquid nitrogen. Approximately 50 mg sample powder were weighed and mixed with working internal standard solution. Then plant hormones were extracted with extraction solvent (Isopropyl alcohol: water: hydrochloric acid = 2:1:0.002). The homogenate was incubated at 4°C on a shaker at a speed of 900 rpm for 30 min. Then trichloromethane (1 ml) was added to each sample and placed on the shaker at a speed of 900 rpm for an additional 30 min at 4°C. Two phases were formed by centrifuging at 14,000 rpm and 4°C for 5 min. Then the solvent from the lower phase (1.2 ml) was transferred to a new tube and concentrated using a nitrogen generator with nitrogen flow. The concentrated samples were resuspended in 0.1 ml methanol and centrifuged for 5 min at 14,000 rpm. Finally, the supernatants were filtered using a syringe and syringe filter and prepared for high-performance liquid chromatography-mass spectrometry (HPLC-MS) analysis.
The RNA samples were reverse-transcribed to synthesize cDNA using a HiScript II Q RT SuperMix for qPCR kit (Vazyme, Nanjing, China). Real-time quantitative PCR (RT- qPCR) was performed using Universal ChamQTM Universal SYBR® qPCR Master Mix (Vazyme, Nanjing, China). Three biological replications were performed for each treatment. Actin was used as a reference for the analysis of gene expression. The primers used for RT-qPCR were shown in Supplementary Table S2.
The leaves of 3-week-old N. benthamiana infiltrated with pGD-TMCP-Myc, pGD-PMCP-Myc or pGD-CGCP-Myc were collected and grinded with plant protein extraction buffer [25 mM Tris, pH 7.5, 10% (v/v) glycerol, 150 mM NaCl, 1 × protease inhibitor cocktail (Roche), and 0.15% (v/v) Non-idet P-40]. After centrifugation at 12,000 rpm for 10 min at 4°C, the supernatant was collected. The total proteins were separated on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels and stained with Coomassie Brilliant Blue, thereafter transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, United Staes). The membrane was treated with Myc-tag antibody (Abcam, Shanghai, China) and alkaline phosphatase (AP)-conjugate goat anti-rabbit secondary antibody (Sangon Biotech, Shanghai, China). The treated membrane was incubated with chemiluminescent substrate CDP-star (Roche Mannheim, Germany) and visualized the target bands by using the Chemical luminous imaging system Tanon 5200 (Tanon, Shanghai, China).
Nicotiana benthamiana is well applied as a model plant in the study of plant physiology or pathology including plant virology. Especially, N. benthamiana is extremely sensitive to virus infection and can be systemically infected by a wide range of viruses (Bally et al., 2018). In this study, we constructed chimeric virus expression vectors to investigate the functions of three tobamovirus coat proteins on the pathogenicity of wild-type and chimeric TMV (Figure 1A). A. tumefaciens GV3101 containing pCB-TMV, pCB-TMV-CGCP or pCB-TMV-PMCP were infiltrated into N. benthamiana leaves and visible shriveled symptoms appeared on the upper non-inoculated leaves at 5 dpi. RNA extracted from upper un-inoculated leaves of 10 agro-infiltrated N. benthamiana plants were reverse transcribed and subjected to sequencing analysis. The results demonstrated that no sequence mutation occurred in the chimeric viruses after their systemic infection in plants (data not shown). Consequently, the leaves of N. benthamiana infected with the wild-type or the chimeras of TMV were extracted and mechanically inoculated onto the leaves of N. tabacum (Figure 1B). The results showed that TMV together with the chimeric viruses can cause chlorosis and mosaic symptoms on the upper un-inoculated leaves of N. tabacum at 12 dpi (Figure 1C). Interestingly, TMV-CGCP and TMV-PMCP also induced stunting in N. tabacum and mosaic symptoms on the upper leaves compared with those inoculated with wild-type TMV (Figure 1C). Specifically, the leaves inoculated by TMV-CGCP and TMV-PMCP typically showed necrotic symptoms (Figure 1C). Furthermore, we confirmed the systemic infection of TMV, TMV-CGCP or TMV-PMCP in N. tabacum by detecting viral RNA from the upper systemic leaves by RT-PCR (Figure 1D). To exclude the effect of CP on virus accumulation and host symptoms, we also further constructed three chimeric viruses TMV-fsCP, TMV-CGfsCP and TMV-PMfsCP that cannot express CP by introducing point mutation at the start codon of the respective CP ORF (Supplementary Figure S1A). The results showed that all of N. tabacum inoculated with the CP defective viruses did not develop observable diseased symptoms. We also confirmed their systemic infection in the upper leaves and relative RNA accumulation by RT-qPCR at 10 dpi (Supplementary Figures S1B,C). The accumulation levels of TMV were much higher than that of the chimeric viruses by RT-qPCR quantification (Supplementary Figure S1C). Furthermore, the accumulation levels of TMV, TMV-CGCP and TMV-PMCP were approximately 11-, 2-, and 3-folds higher than that of TMV-fsCP, TMV-CGfsCP and TMV-PMfsCP (Supplementary Figure S1C). The results also showed no significant differences between the accumulation of TMV-fsCP vs. TMV-CGfsCP and TMV-fsCP vs. TMV-PMfsCP.
Transmission electron microscope was used to observe the virus particles to determine whether the introduced heterologous coat protein can affect the virion morphology of the chimeric viruses. The results indicated that TMV-CGCP and TMV-PMCP can form rod-like virions with approximately 300 nm in length as well as wild-type TMV (Figure 1E) and there was no significant difference in the virion morphology between wild-type TMV and its chimeras, which indicated that the heterologous substituted CP do not affect the virion formation.
To further investigate the critical determinants that modulate the host symptoms among TMV and the chimeras in N. tabacum, genome-wide gene expression analysis was conducted and the upper systemic leaves of TMV or the chimeric viruses inoculated N. tabacum plants were collected for RNA-seq. The mock-inoculated plants treated with PBS only were used as negative controls and each treatment had three biological replicates. A total of 70.28 Gb raw reads were obtained and the clean reads were mapped to the reference genome database using HISAT2 software (Table 1). A total of 4860 DEGs were obtained in the TMV vs. TMV-CGCP, including 3210 up-regulated and 1659 down-regulated genes were identified. In TMV vs. TMV-CGCP, 3715 DEGs were obtained including 2444 up-regulated and 1271 down-regulated genes (Figure 2A and Supplementary Table S3). A total of 2192 DEGs in TMV vs. TMV-CGCP overlap with those in TMV vs. TMV-PMCP (Figure 2B). Differentially regulated genes were identified based on the criteria of significance FDR<0.05 and log2-fold change (log2FC) ≥ 1.0 or ≤ −1.0 (Figure 2C). The DEGs of each treatment are listed in Supplementary Table S3. To further classify the DEGs, GO and KEGG analysis were performed (Supplementary Tables S4, S5). The DEGs were mainly classified into various functions, including transporter activity, signal transducer activity and nucleic acid binding transcription factor activity in GO analysis (Supplementary Figure S4). Total annotation of the DEGs in KEGG pathways were shown (Figure 3 and Supplementary Table S6). The results indicated both TMV vs. TMV-CGCP and TMV vs. TMV-PMCP were significantly enriched in plant-pathogen interaction and plant hormone signal transduction pathways (Figure 3). Then we selected several key DEGs involved in plant-pathogen interaction and plant hormone signal transduction pathways for further investigation.
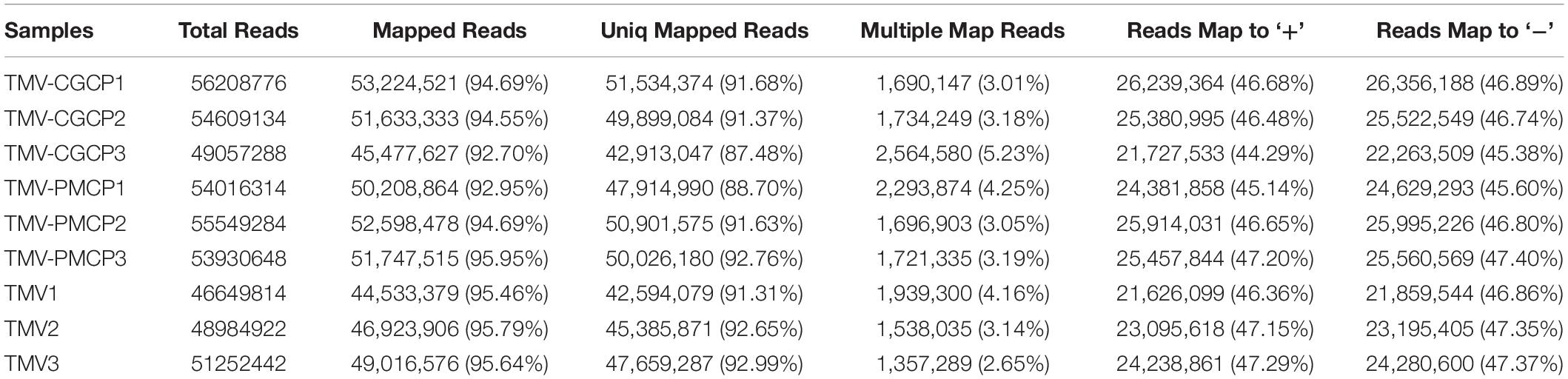
Table 1. Illumina sequencing data and the read numbers aligned onto the N. tabacum reference genome.
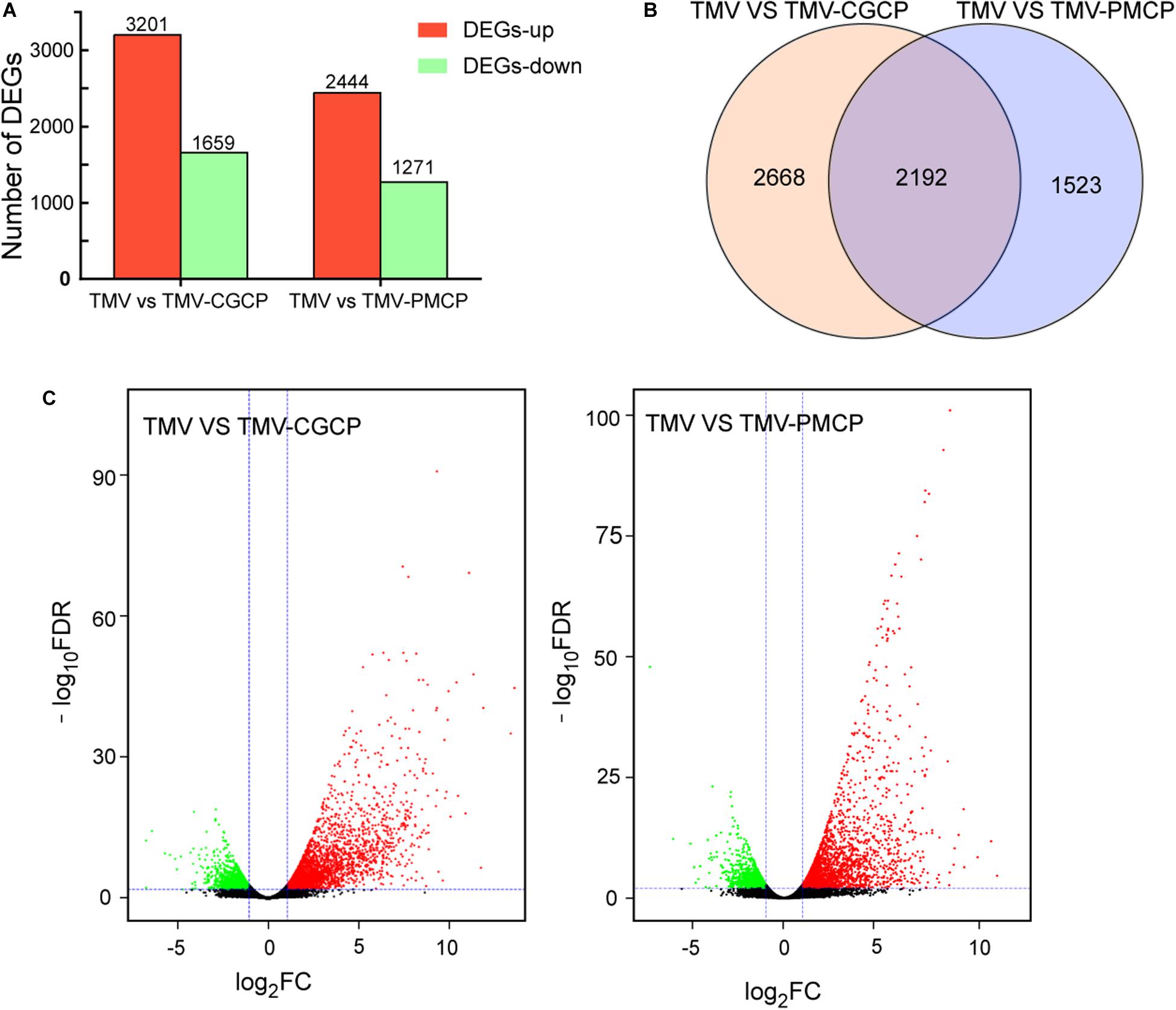
Figure 2. Differential gene expression in virus-infected N. tabacum plants. (A) Numbers of up-regulated (red) and down-regulated (green) gene were shown. (B) Venn diagram of differentially expressed genes (DEGs) between TMV vs. TMV-CGCP and TMV vs. TMV-PMCP. (C) Volcano plots showing DEGs of TMV vs. TMV-CGCP and TMV vs. TMV-PMCP. The red and green colors represent the significantly up- and down-regulated genes, respectively, (FDR<0.05 and |log2 FC| ≥ 1).
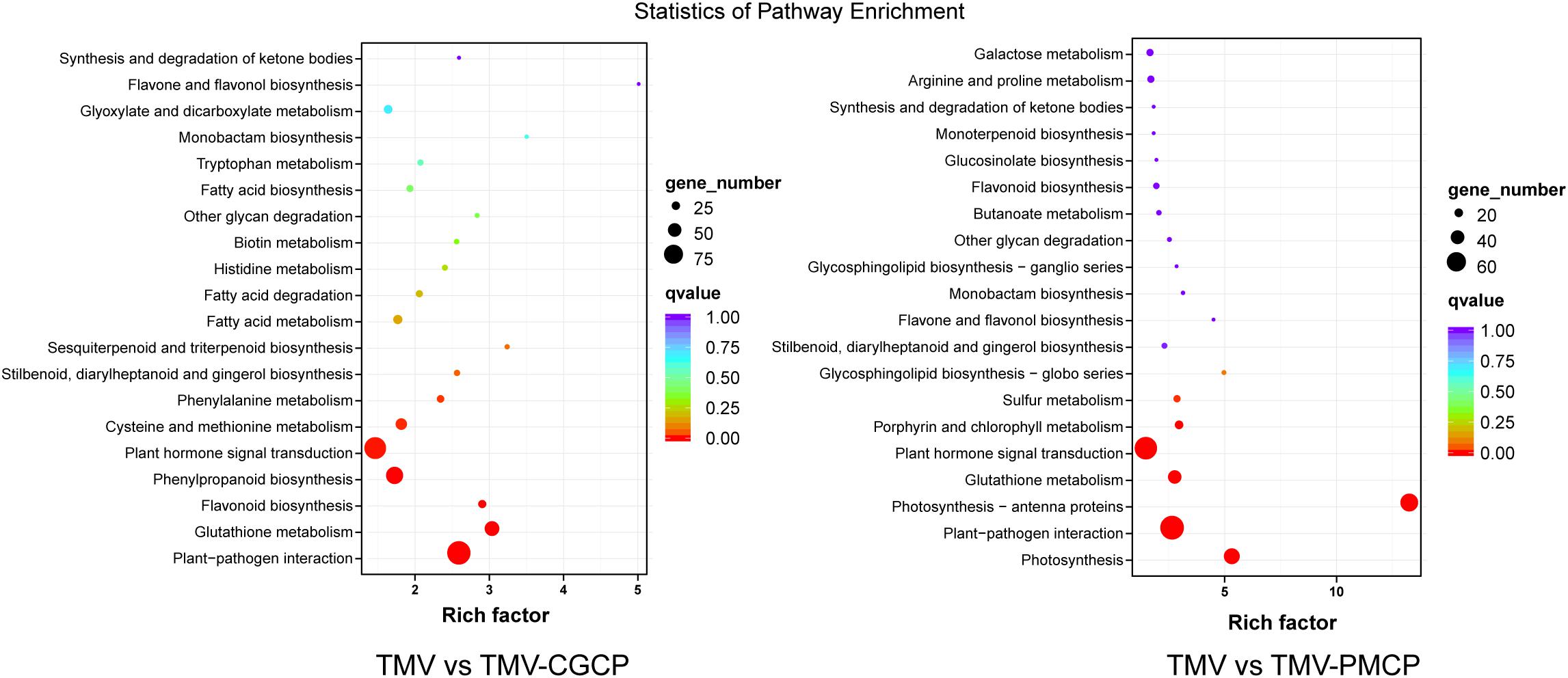
Figure 3. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of TMV vs. TMV-CGCP and TMV vs. TMV-PMCP. The rich factor reflects the degree of enriched DEGs in a given pathway. The number of enriched DEGs in the pathway is indicated by the circle area, and the circle color represents the ranges of the corrected p-value.
To validate the results of RNA-seq, 8 genes responding to TMV or the chimeras infection in N. tabacum were selected for RT-qPCR verification. The selected genes included abscisic acid receptor PYL4 (LOC107778387), auxin-responsive protein IAA14 (LOC107764405), heat shock protein 90 (Hsp90, LOC107816677), WRKY transcription factor 40 (WRKY40, LOC107792337), WRKY transcription factor 50 (WRKY50, LOC107772027), serine/threonine-protein kinase RLK1 (LOC107794887), pathogenesis-related protein-1C (PR1, LOC107763263) and TGACG-sequence-specific DNA-binding protein TGA-2.1 (LOC107782457). The results showed that the expression levels of the auxin responsive genes were significantly decreased, whereas expression of other genes were increased in chimeras infected plants compared with TMV, which were consistent with the results of RNA-seq (Figure 4). Additionally, regression analyses showed that there was a positive correlation between RNA-seq and RT-qPCR data (Supplementary Figure S3).
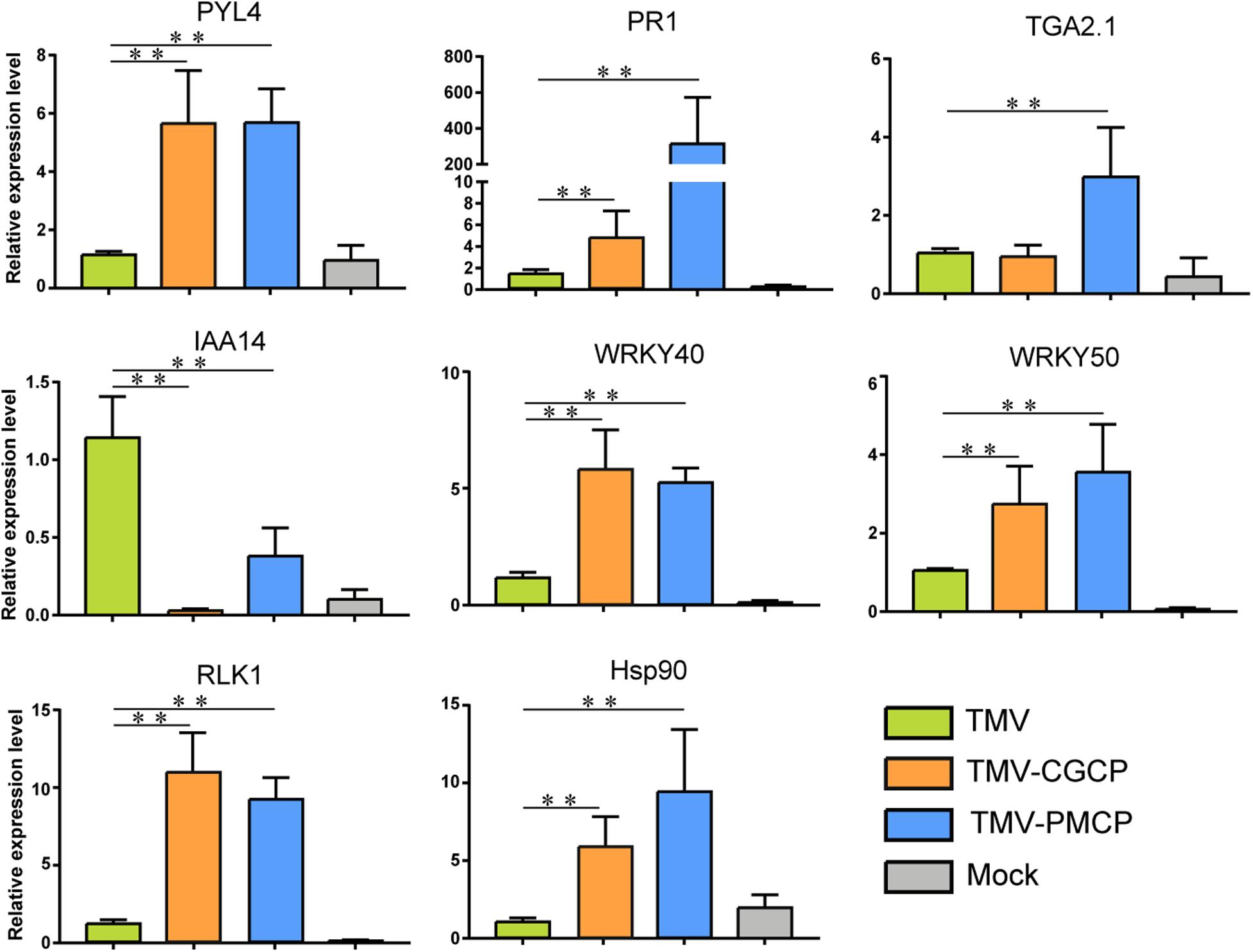
Figure 4. RT-qPCR analysis of the relative expression levels of PYL4, IAA14, Hsp90, WRKY40, WRKY50, RLK1, PR1, and TGA2.1 in the TMV, TMV-CGCP or TMV-PMCP infected samples. Asterisks indicate a statistically significant difference compared with TMV, “∗∗” indicate an extremely significant difference (P < 0.01). Primers used for qPCR were listed in Supplementary Table S6. Information of the genes indicated in the figure were showed in Table 2. Mock-inoculated plants were used as control treatments (Mock).
Plant virus infection often induces host-virus interactions with a strong plant immune response. Leucine-rich repeat LRR domain present in receptor-like kinases (RLKs) plays an important role in pathogen recognition through signaling pathways (Gouveia et al., 2016). In this study, we selected typical genes involved in plant-pathogen interaction and phytohormone signaling from the results of RNA-seq and listed their regulation after the chimeric virus infection (Table 2). We found several differentially expressed genes that belong to RLK gene-family were induced by chimeric virus infection (Table 2 and Supplementary Table S3). In addition, a variety of genes involved in host-pathogen interaction such as FLS2, brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1), enhanced disease susceptibility 1 protein (EDS1) and heat shock proteins (Hsps) were significantly increased (Table 2). Infection of the TMV chimeras also induced differential expression of genes involved in immune signaling transduction such WRKY transcription factors, NAC transcriptional factors and mitogen-activated protein kinase kinase kinase (MAPKKK). Four DEGs including RLK1, Hsp90, WRKY40 and WRKY50 involved in defence response were selected to verify their regulation using RT-qPCR. The results demonstrated that the expression levels of all the above genes were significantly up-regulated by the infection of chimeras of TMV (Table 2). The results of RT-qPCR were consistent with that of RNA-seq (Figure 4 and Table 2).
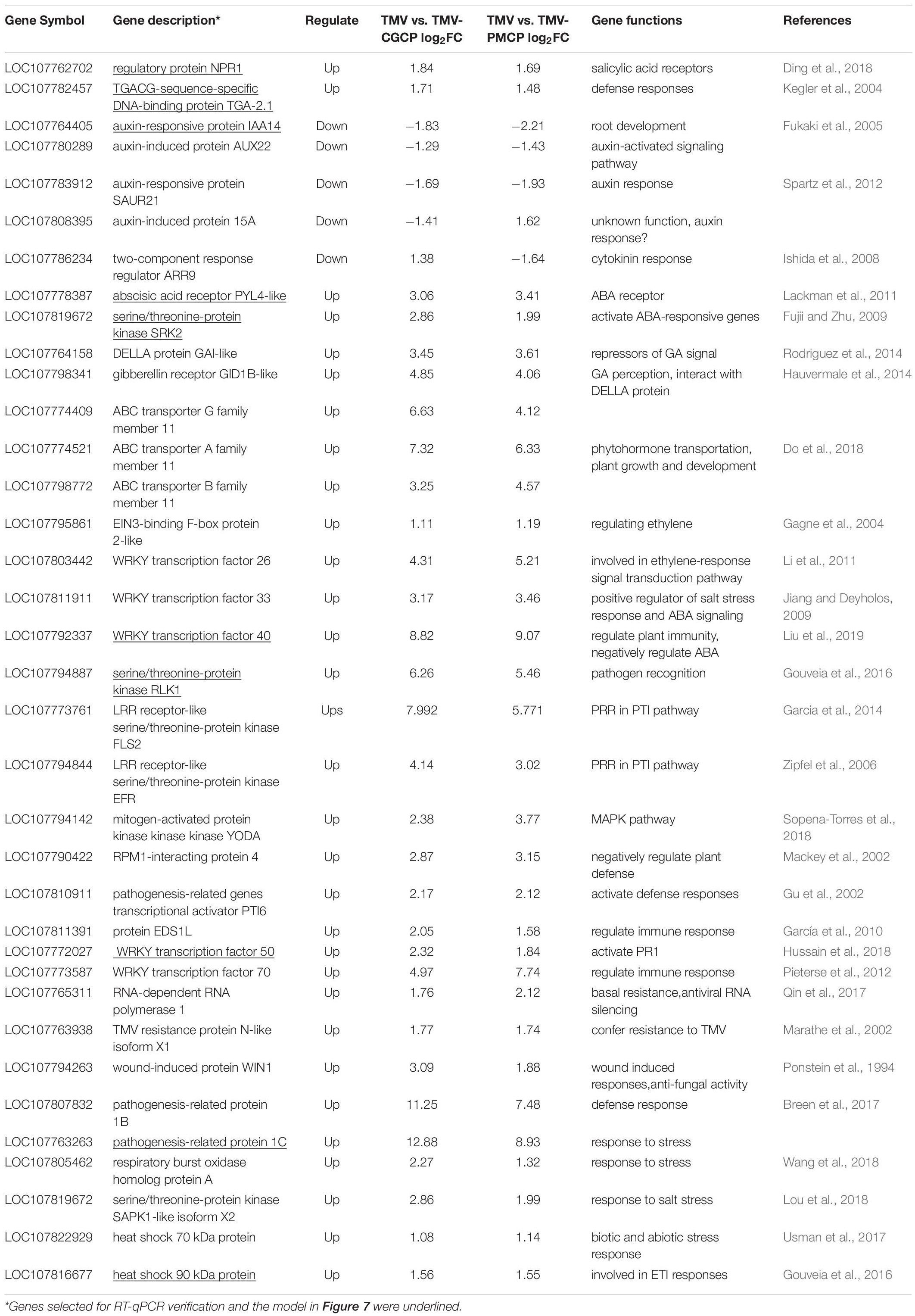
Table 2. A general table showing critical DEGs involved in plant pathogen interaction and phytohormone signaling induced by TMV and the chimeras in N. tabacum.
The change of plant hormones is often accompanied by virus infection. Phytohormones play important roles in plant growth and development as well as host symptoms induced by virus infection (Collum and Culver, 2016). In this study, the transcriptome data indicated that various genes involved in hormone signaling pathways in TMV chimeras infected plants were differentially regulated compared with that of wild-type TMV (Figure 4 and Table 2). For SA signaling pathway, a variety of genes like non-expressor of pathogenesis-related genes 1 (NPR1), TGA Transcription Factor genes and pathogenesis-related protein-1 (PR1) were significantly increased upon chimeric TMV infection. By contrast, the expression of auxin signaling pathway-related genes including auxin transporter AUX1, AUX/IAA family and small auxin-up RNA (SAUR) were significantly decreased after infection of the chimeric viruses. In addition, expressions of ethylene (ET) related genes ethylene insensitive3 (EIN3) and ABA related genes including abscisic acid receptor PYR/PYL family and serine/threonine-protein kinase SRK2 (SNRK2) were induced in TMV-CGCP or TMV-PMCP infected plants. We selected PYL4, PR1, TGA2.1, and IAA14 to verify their regulation using RT-qPCR. The results demonstrated that the expression levels of PR1, TGA2.1, and PYL4 were significantly up-regulated and IAA14 was significantly down-regulated by the infection of chimeric viruses (Figure 4). The results were consistent with that of RNA-seq (Figure 4 and Table 2).
Then, we determined the level of phytohormone including SA, IAA, ABA and JA by HPLC- MS. Mock-inoculated N. tabacum plants were used as control treatments. Our results showed that the SA level of N. tabacum inoculated with chimeras TMV-CGCP or TMV-PMCP was significantly higher than that in TMV or mock-inoculated plants, while the content of IAA in the chimeric virus inoculated plants exhibited obvious decrease than that in wild-type TMV inoculated plants (Figure 5). This result was parallel with the result of the transcriptome. Additionally, ABA level was significantly increased in TMV-CGCP inoculated plants, while ABA content in TMV-PMCP inoculated plants was similar to that of TMV inoculated or mock-inoculated plants (Figure 5). The levels of JA showed no significant difference among all of the virus inoculated plants (Figure 5).
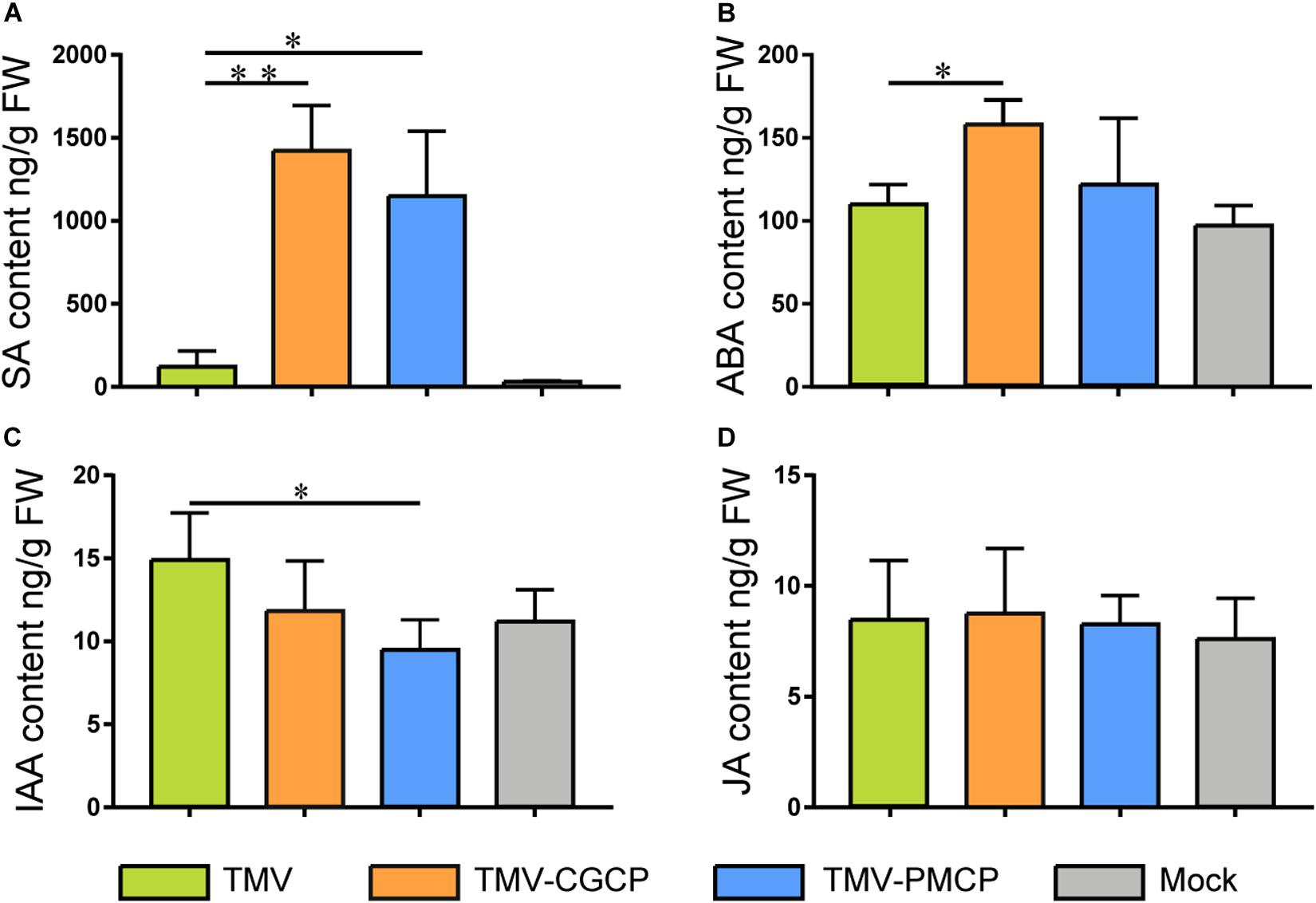
Figure 5. Phytohormone concentrations in N. tabacum infected with TMV, TMV-CGCP or TMV-PMCP. Mock-inoculated plants were used as control treatments (Mock). Contents of (A) SA, (B) IAA, (C) ABA and (D) JA in leaves of N. tabacum were determined at 10 dpi. Asterisks indicate a statistically significant difference compared with TMV, “∗” indicate a significant difference (P < 0.05) and “∗∗” indicate an extremely significant difference (P < 0.01).
To test if the CP from different tobamoviruses can induce differential expression of the defence or phytohormone responsive genes without viral genome RNA, we constructed three constructs (Figure 6A) and transiently express the Myc-tag fused coat protein of TMV (TMCP-Myc), CGMMV (CGCP-Myc), and PMMoV (PMCP-Myc) in N. benthamiana leaves, respectively. The results of western blot analysis showed that the accumulation levels of Myc-tag fused TMV, PMMoV, and CGMMV CP were similar (Figure 6B). Moreover, 8 genes corresponded with defence or phytohormone responses were selected for RT-qPCR verification and the results showed the significant up-regulation of RLK1, Hsp90, PR1, PYL, IAA14, TGA, WRKY40, and WRKY50 induced by transient expression of CGCP-Myc or PMCP-Myc. The regulatory patterns of the gene expression were generally consistent with that induced by infection of TMV-CGCP or TMV-PMCP (Figure 4 and Figure 6C).
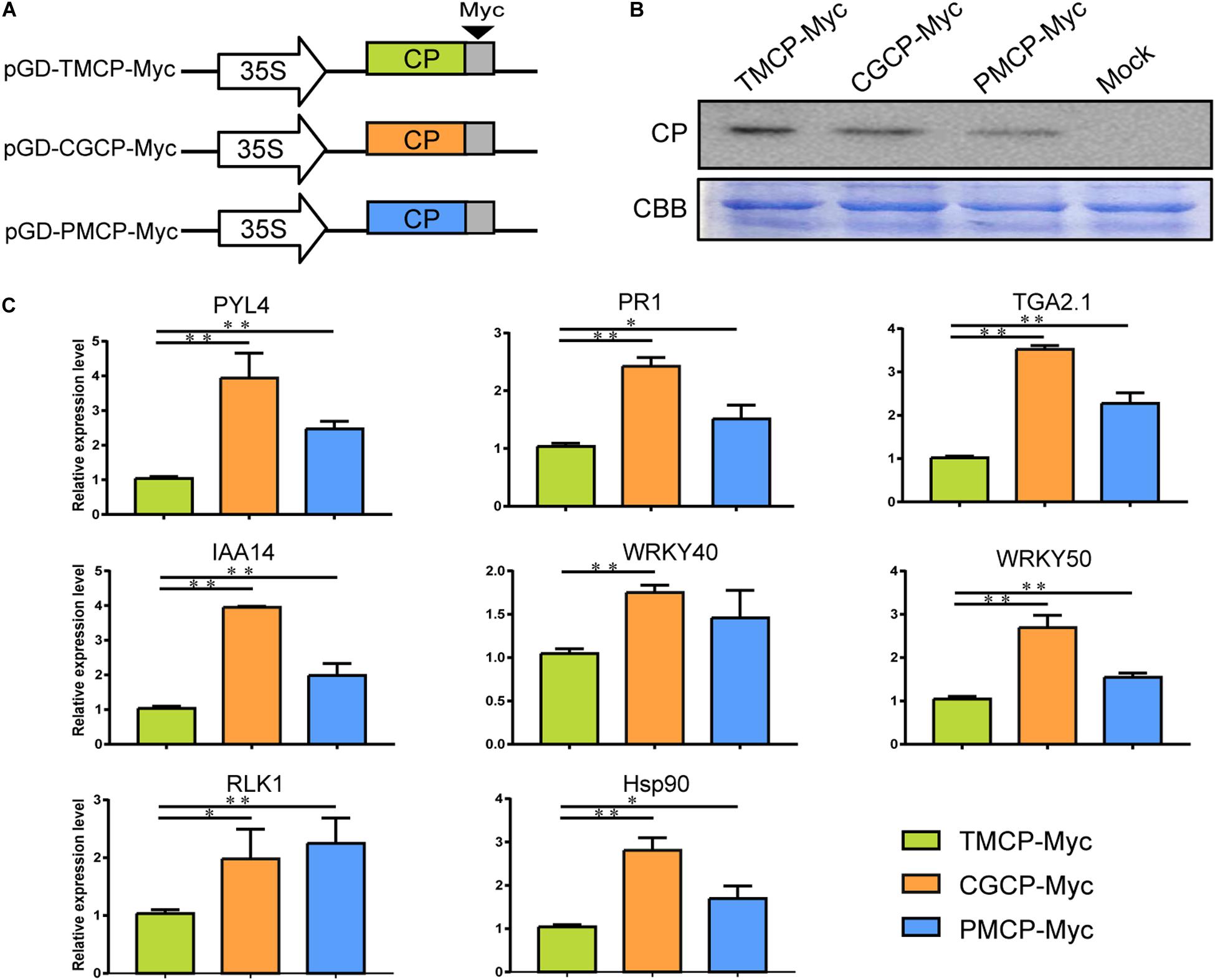
Figure 6. Expression of defence or phytohormone responsive genes regulated by transient expression of CP of TMV, CGMMP or PMMoV with Myc-tag in N. benthamiana. (A) Schematic representation of the transient CP expression vector pGD-TMCP-Myc, pGD-CGCP-Myc and pGD-PMCP-Myc. (B) Western blot analysis of the three tobamovirus CPs in leaves of N. benthamiana infiltrated with pGD-TMCP-Myc, pGD-CGCP-Myc and pGD-PMCP-Myc at 2 dpi. The Coomassie brilliant blue-stained cellular proteins were shown below the western blot as loading controls. (C) RT-qPCR analysis of relative expression levels of PYL4, IAA14, Hsp90, WRKY40, WRKY50, RLK1, PR1, and TGA in N. benthamiana leaves infiltrated with pGD-TMCP-Myc, pGD-CGCP-Myc or pGD-PMCP-Myc at 2 dpi. Information of the genes indicated in the figure were showed in Table 2. Asterisks indicate a statistically significant difference compared with TMV, “∗” indicate a significant difference (P < 0.05) and “∗∗” indicate an extremely significant difference (P < 0.01). For others, refer to the legends of Figure 5. Primers used for qPCR were listed in Supplementary Table S6.
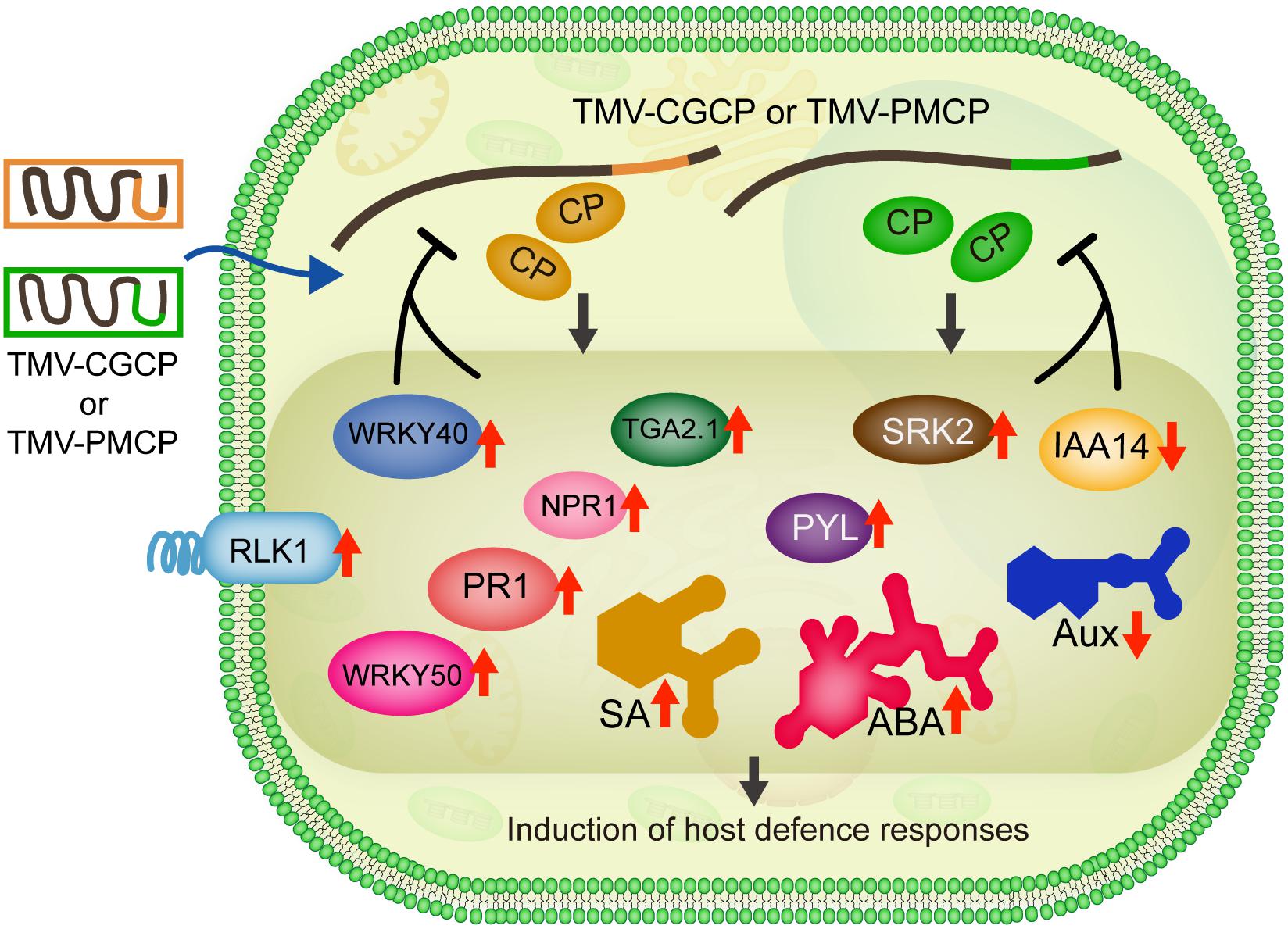
Figure 7. Model for gene and phytohormone regulation involved in induction of host defence responses by infection of chimeric TMV. Infection of TMV-CGCP as well as TMV-PMCP trigger the up-regulation of defence-responsive genes such as RLK1, WRKY40 as well as genes involved in phytohormone signaling pathways (SA: NPR1, TGA2.1, WRKY50 and PR1; ABA: SRK2 and PYL) and induce increase in the levels of SA and ABA, but reduce the expression of IAA14 and levels of Aux, respectively.
Application of chimeric viruses containing heterologous sequence substitution can provide important messages to clarify the critical functions of the RNA sequences or the coding proteins of plant viruses during infection (Yelina et al., 2002; Lukhovitskaya et al., 2013; Tuo et al., 2015; Yu et al., 2019). For example, chimeric barley stripe mosaic virus (BSMV) with exchange in γb protein from poa semilatent virus (PSLV) indicated RNA silencing suppression function of this viral protein. In addition, infection of chimeric TMV and potato virus X (PVX) expressing p12 of chrysanthemum virus B indicated that p12 plays an important role in modulation of host cell morphogenesis (Lukhovitskaya et al., 2013). Furthermore, a series of BSMV chimeras with exchanges in MP from various plant viruses revealed the functions of exogenous MP on the virus-coded transport and host symptoms (Solovyev et al., 1996, 1997, 1999). We previously also constructed various chimeric PMMoV clones to further elucidate the molecular mechanisms for the virus infection (Yu et al., 2019). In this work, chimeric virus TMV-CGCP and TMV-PMCP with heterologous tobamovirus coat proteins were constructed using homologous recombination technique, which greatly shortens the construction procedure as the unique restriction enzyme sites are not required to be introduced into the virus infectious clone.
To combat viral infection, plants have evolved sophisticated innate immune systems, which are generally divided into the PTI and ETI (Gouveia et al., 2016; Calil and Fontes, 2017). After recognition of invading pathogens by pattern recognition receptors (PRRs), the mitogen-activated protein kinases (MAPKs) cascades are rapidly activated and the transduction signal can eventually activate various WRKY transcription factors in plants (Gouveia et al., 2016). RLK1 is one of the well-conserved PRRs being critical for the perception of conserved microbe-associated molecular patterns (MAMPs) to activate PTI in different plant species (Hu et al., 2018). Notably, many studies have shown that WRKY transcription factors from various plant species are induced in response to viral infection (Huang et al., 2016; Huh et al., 2012). In this study, the results indicated that infection of TMV-CGCP or TMV-PMCP can induce rapid host defence responses by markedly up-regulating gene expression of RLK1, WRKY40 and WRKY50 compared with that of wild-type TMV infection. WRKY40 was reported to associate with a transcription factor BZR1 to mediate plant immune signaling (Lozano-Durán et al., 2013). In a previous study, we have also shown that WRKY40 was up-regulated by an anti-viral biological agent Ningnanmycin (An et al., 2019), which also confirmed the function of WRKY40 in plant immune responses. It was revealed that Arabidopsis WRKY50 together with TGA transcription factors synergistically activate expression of PR1 (Hussain et al., 2018), which were in agreement with our results showing that expression of PR1 together with WRKY50 were consistently up-regulated by infection of chimeric TMV. Plants can use elaborate strategies by inducing phytohormone-signaling networks to get through abiotic or biotic stresses, as well as inducing resistance response to pathogen infection (Calil and Fontes, 2017; Verma et al., 2016). In this study, the infection of TMV-CGCP and TMV-PMCP caused stunting in tobacco, which was possibly related to the significant changes in hormone expression.
The prominent contribution of ABA to plant defence response against abiotic stress conditions has long been investigated (Verma et al., 2016). In addition to its roles in development and abiotic stress responses, ABA also plays a multifaceted role in plant immunity (Ton et al., 2009). Additionally, ABA was shown to be involved in enhanced or reduced plant anti-viral responses (Alazem et al., 2017; Xie et al., 2018). ABA induce callose deposition at plasmodesmata (PD) as well as the RNA silencing pathway, which is proved to be involved in antiviral mechanisms (Alazem et al., 2017). Study has also shown a positive correlation between ABA and host resistance against TMV (Chen et al., 2013). In this study, our results showed that infection of the chimeric virus TMV-CGCP and TMV-PMCP induced significant increase of hormone ABA and ABA-responsive genes PYL4 and SRK2 in tobacco compared with that was infected by TMV. It was revealed that PYL4 acts as a receptor for ABA and is involved in plant metabolism and growth (Lackman et al., 2011). SNRK2s are a family of plant-specific protein kinases, which are activated by ABA and play an important role in the activation of ABA responsive genes (Fujii and Zhu, 2009). In this study, TMV-CGCP and TMV-PMCP induced significant increase of hormone ABA and ABA-responsive genes PYL4 and SRK2 in tobacco, which indicated that ABA signaling may be involved in the induced host resistance against the chimeric virus infection.
The increase in the levels of SA plays critical roles for host plants in resistance of a broad spectrum of pathogens such as biotrophic pathogens (or obligate parasites) when they trigger PTI or ETI responses. The SA is also widely mentioned in viral defence immunity (Conti et al., 2017). Exogenous SA treatment reduces TMV RNA accumulation by interfering with TMV replication in mesophyll cells and also inhibits cell to cell movement in epidermal cells (Chivasa et al., 1997; Murphy and Carr, 2002; Zhu et al., 2014). Moreover, an increase in SA levels in the pathogen-infected plant tissues results in the induction of PR1, which was considered as a defence marker gene (Breen et al., 2017). Therefore, the significant induction of PR1 by infection of the chimeric TMV is consistent with the marked increase in SA levels after the chimeric TMV infection in tobacco. In addition, it was reported that TGA2.1 can interact with NPR1 and play crucial roles in SA induction (Thurow et al., 2005). Therefore, the significant induction of PR1 and TGA2.1 is consistent with the marked increase of SA levels after the chimeric virus infection in tobacco.
SA regulates plant basal defences via activating the host systemic acquired resistance (SAR) and hypersensitive responses. Induction of SAR also suppresses a serial of auxin-responsive genes (Kovtun et al., 1998), suggesting that enhanced resistance to diseases would necessitate repression of auxin signaling (Verma et al., 2016). A previous study showed that TMV p126 replicase interacts with a variety of Aux/IAA proteins to influence the expression of a large number of auxin responsive genes and induce specific disease symptoms (Padmanabhan et al., 2005). In general, the SA and JA signaling pathways influence each other through a complex network of antagonistic interactions (Koornneef and Pieterse, 2008). JA, a kind of plant hormone, is mainly responsible for defence against necrotrophic pathogens and herbivorous insects (Wasternack and Hause, 2013). A study had indicated that SA as well as JA are required for systemic response to TMV (Zhu et al., 2014). In this study, SA was rapidly induced by infection of TMV-CGCP or TMV-PMCP than TMV, whereas the JA accumulation remained consistent after TMV or the chimeric TMV infection. The results indicated that the significant increase of SA after the chimeric TMV infection may be responsible for the stunting and necrotic symptoms in tobacco.
Viral CP is crucial for viral RNA encapsidation to form virions, which can protect the virus from nuclease degradation and RNA silencing of the host plants (Ivanov and Mäkinen, 2012). The CP also plays important roles in the infection, proliferation, systemic movement, pathogenicity and transmission of the virus (Bendahmane et al., 2002; Conti et al., 2017). CP is also closely involved in the disease symptomology of the virus-infected plants. For cucumber mosaic virus (CMV), single amino acid substitutions in the CP ORF affected the thylakoid structure of tobacco chloroplasts (Mochizuki and Ohki, 2011). Additionally, the N. benthamiana NbPCIP1 was found to interact with the CP of PVX, which affect infectivity, pathogenicity, and symptom expression of the virus (Park et al., 2009). A previous study showed that TMV with truncate CP induced stunt and necrosis symptoms in tobacco (Kurihara and Watanabe, 2004), which are very similar to the symptoms caused by TMV-CGCP and TMV-PMCP. By comparing the CP amino acid sequences of TMV, CGMMV and PMMoV, we found that the amino acid similarity of TMV CP and CGMMV CP was approximately 35%, whereas the amino acids of PMMoV CP are more closely related with that of TMV with the similarity of 70% (Data not shown). It was reported that an amino acid substitution introduced into CP ORF (CPT42W) can restrict the accumulation of MP and result in strong resistance to TMV infection in plants containing CPT42W (Bendahmane et al., 2002). In this study, infection of TMV-CGCP and TMV-PMCP showed reduced viral RNA accumulation compared with wild-type TMV, which possibly due to the induced resistance of the plants caused by the respective CPs. Furthermore, the transiently expressed CP exhibit similar regulatory effect on host gene expression. Therefore, it is possible that free CP subunit of CGMMV or PMMoV also affect host transcriptome as well as the chimera virions. Our results also indicated that the exogenous substituted CP does not affect the ability of virion assembly of TMV-CGCP and TMV-PMCP despite the low sequence and amino acid similarity. Such results also demonstrated that the CP of CGMMV or PMMoV can recognize RNA sequences of TMV and subsequently perform virion assembly.
In the previous work, we have showed that CP defective virus PMMoV-fsCP can systemically infected N. benthamiana and C. annuum and confirmed the introduced mutation was maintained in the systemic leaves of the inoculated plants (Yu et al., 2019). In this study, the CP defective viruses TMV-fsCP, TMV-CGfsCP and TMV-PMfsCP can systemically infect tobacco but do not induce observable symptoms. These data indicated that CP is dispensable for systemic movement of TMV, but it may contribute to increasing the infection efficiency and play important roles in symptom development.
Normally, viruses do not induce the death of plants in the infection defence equilibrium. Some plant virus even has RNA silencer elements in its genome RNA to suppress the virus from too fast replication (Pogany et al., 2003), which better facilitate the virus to survive and spread. Therefore, the reduction of the adverse effects of the virus infection in host plants is beneficial for the transmission, survival and evolution of the viruses. These chimeric viruses expressing coat proteins from CGMMV or PMMoV may affect host metabolic pathways or affect the interactions between other viral proteins and the host proteins, thus leading to the significant defence responses of tobacco. To clarify the statements, further investigations are required to determine the critical domain or amino acids of coat protein as well as other viral proteins of TMV. We collectively summarized our data in Figure 7. Based on these results, we also speculate that the CP of wild-type TMV may elaborately suppress or evade the host defence machinery of tobacco to reduce the adverse impact on the plant, which should be due to the results of long-period of sophisticated co-evolution between plant viruses and their hosts.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YW conceived the research project. MA, ZX, MY, and XB conceived and designed the experiments. MY, XB, and YH performed the main experiments. YC, JW, RZ, and YL cultivated N. benthamiana plants and N. tabacum cv and NC89 and treated samples for transcriptome sequencing. MA, MY, and XB carried out the transcriptome analysis and analyzed the data. YW, MA, ZX, MY, and XB prepared the reagents and materials and analysis tools. MA and MY prepared the figures and tables. MY wrote the original draft. YW, MA, ZX, and XB reviewed the drafts of the manuscript. All authors reviewed and approved the manuscript.
This research was funded by the National Key R&D Program of China (2017YFD0201104) and Key Scientific and Technological Projects of Sichuan Tobacco Company (2020510600270011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.587005/full#supplementary-material
Supplementary Figure 1 | Construction of chimeras TMV-fsCP, TMV-CGfsCP and TMV-PMfsCP and their symptoms and accumulation in N. tabacum. (A) Schematic representation of genome structure of the construct TMV-fsCP, TMV-CGfsCP and TMV-PMfsCP, site mutation was introduced to the start codon of the CP based on TMV, TMV-CGCP and TMV-PMCP, respectively. (B) Symptoms induced in N. tabacum by TMV-fsCP, TMV-CGfsCP and TMV-PMfsCP at 7 dpi. Mock-inoculated plants were used as control treatments (Mock). (C) Relative accumulation of virus in N. tabacum inoculated with TMV-fsCP, TMV-CGfsCP and TMV-PMfsCP at 7 dp, compared to TMV, TMV-CGMP and TMV-PMCP, respectively. Asterisks indicate a statistically significant difference compared with TMV, “∗” indicate a significant difference (P < 0.05) and “∗∗” indicate an extremely significant difference (P < 0.01).
Alazem, M., He, M. H., Moffett, P., and Lin, N. S. (2017). Abscisic acid induces resistance against bamboo mosaic virus through Argonaute2 and 3. Plant Physiol. 174, 339–355. doi: 10.1104/pp.16.00015
An, M., Zhou, T., Guo, Y., Zhao, X., and Wu, Y. (2019). Molecular regulation of host defense responses mediated by biological anti-TMV agent Ningnanmycin. Viruses 11:815. doi: 10.3390/v11090815
Bally, J., Jung, H., Mortimer, C., Naim, F., Philips, J. G., Hellens, R., et al. (2018). The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annu. Rev. Phytopathol. 56, 405–426. doi: 10.1146/annurev-phyto-080417-050141
Bendahmane, M., Szecsi, J., Chen, I., Berg, R. H., and Beachy, R. N. (2002). Characterization of mutant tobacco mosaic virus coat protein that interferes with virus cell-to-cell movement. Proc. Natl. Acad. Sci. U.S.A. 99, 3645–3650. doi: 10.1073/pnas.062041499
Breen, S., Williams, S. J., Outram, M., Kobe, B., and Solomon, P. S. (2017). Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22, 871–879. doi: 10.1016/j.tplants.2017.06.013
Calil, I. P., and Fontes, E. P. B. (2017). Plant immunity against viruses: antiviral immune receptors in focus. Ann. Bot. 119, 711–723. doi: 10.1093/aob/mcw200
Callaway, A., Giesman-Cookmeyer, D., Gillock, E. T., Sit, T. L., and Lommel, S. A. (2001). The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39, 419–460. doi: 10.1146/annurev.phyto.39.1.419
Chen, L., Fei, C., Zhu, L., Xu, Z., Zou, W., Yang, T., et al. (2017). RNA-seq approach to analysis of gene expression profiles in dark green islands and light green tissues of cucumber mosaic virus-infected Nicotiana tabacum. PLoS One 12:e0175391. doi: 10.1371/journal.pone.0175391
Chen, L., Zhang, L., Li, D., Wang, F., and Yu, D. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, E1963–E1971. doi: 10.1073/pnas.1221347110
Chivasa, S., Murphy, A. M., Naylor, M., and Carr, J. P. (1997). Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 9, 547–557. doi: 10.1105/tpc.9.4.547
Collum, T. D., and Culver, J. N. (2016). The impact of phytohormones on virus infection and disease. Curr. Opin. Virol. 17, 25–31. doi: 10.1016/j.coviro.2015.11.003
Conti, G., Rodriguez, M. C., Venturuzzi, A. L., and Asurmendi, S. (2017). Modulation of host plant immunity by Tobamovirus proteins. Ann. Bot. 119, 737–747. doi: 10.1093/aob/mcw216
Culver, J. N. (2002). Tobacco mosaic virus assembly and disassembly: determinants in pathogenicity and resistance. Annu. Rev. Phytopathol. 40, 287–308. doi: 10.1146/annurev.phyto.40.120301.102400
Dardick, C. D., Taraporewala, Z., Lu, B., and Culver, J. N. (1999). Comparison of tobamovirus coat protein structural features that affect elicitor activity in pepper, eggplant, and tobacco. Mol. Plant Microbe Interact. 12, 247–251. doi: 10.1094/MPMI.1999.12.3.247
Dawson, W., Bubrick, P., and Grantham, G. (1988). Modifications of the tobacco mosaic virus coat protein gene affecting replication, movement, and symptomatology. Phytopathology 78, 783–789. doi: 10.1094/phyto-78-783
Ding, S. W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. doi: 10.1038/nri2824
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant Immunity. Cell 173, 1454–1467.e15. doi: 10.1016/j.cell.2018.03.044
Do, T. H. T., Martinoia, E., and Lee, Y. (2018). Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 41, 32–38. doi: 10.1016/j.pbi.2017.08.003
Dombrovsky, A., Tran-Nguyen, L. T. T., and Jones, R. A. C. (2017). Cucumber green mottle mosaic virus: rapidly increasing global distribution, etiology, epidemiology, and management. Annu. Rev. Phytopathol. 55, 231–256.
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Fukaki, H., Nakao, Y., Okushima, Y., Theologis, A., and Tasaka, M. (2005). Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44, 382–395. doi: 10.1111/j.1365-313X.2005.02537.x
Gagne, J. M., Smalle, J., Gingerich, D. J., Walker, J. M., Yoo, S. D., Yanagisawa, S., et al. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 6803–6808. doi: 10.1073/pnas.0401698101
García, A. V., Blanvillain-Baufumé, S., Huibers, R. P., Wiermer, M., Li, G., Gobbato, E., et al. (2010). Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 6:e1000970. doi: 10.2307/3871462
Garcia, A. V., Charrier, A., Schikora, A., Bigeard, J., Pateyron, S., de Tauzia-Moreau, M. L., et al. (2014). Salmonella enterica flagellin is recognized via FLS2 and activates PAMP-triggered immunity in Arabidopsis thaliana. Mol. Plant 7, 657–674. doi: 10.1093/mp/sst145
Gooding, G. V. Jr., and Hebert, T. T. (1967). A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology 57:1285. doi: 10.1098/rstl.1767.0047
Gouveia, B. C., Calil, I. P., Machado, J. P., Santos, A. A., and Fontes, E. P. (2016). Immune receptors and co-receptors in antiviral innate Immunity in plants. Front. Microbiol. 7:2139. doi: 10.3389/fmicb.2016.02139
Gu, Y. Q., Wildermuth, M. C., Chakravarthy, S., Loh, Y. T., Yang, C., He, X., et al. (2002). Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14, 817–831. doi: 10.1105/tpc.000794
Hauvermale, A. L., Ariizumi, T., and Steber, C. M. (2014). The roles of the GA receptors GID1a, GID1b, and GID1c in sly1-independent GA signaling. Plant Signal. Behav. 9:e28030. doi: 10.4161/psb.28030
Hu, L., Ye, M., Kuai, P., Ye, M., Erb, M., and Lou, Y. (2018). OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol. 219, 1097–1111. doi: 10.1111/nph.15247
Huang, Y., Li, M. Y., Wu, P., Xu, Z. S., Que, F., Wang, F., et al. (2016). Members of WRKY group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genomics 17:788. doi: 10.1186/s12864-016-3123-2
Huh, S. U., Choi, L. M., Lee, G. J., Kim, Y. J., and Paek, K. H. (2012). Capsicum annuum WRKY transcription factor d (CaWRKYd) regulates hypersensitive response and defense response upon tobacco mosaic virus infection. Plant Sci. 197, 50–58. doi: 10.1016/j.plantsci.2012.08.013
Hussain, R. M. F., Sheikh, A. H., Haider, I., Quareshy, M., and Linthorst, H. J. M. (2018). Arabidopsis WRKY50 and TGA transcription factors synergistically activate expression of PR1. Front. Plant Sci. 9:930. doi: 10.3389/fpls.2018.00930
Ishibashi, K., and Ishikawa, M. (2016). Replication of tobamovirus RNA. Annu. Rev. Phytopathol. 54, 55–78. doi: 10.2183/pjab.80.215
Ishida, K., Yamashino, T., and Mizuno, T. (2008). Expression of the cytokinin-induced type-a response regulator gene ARR9 is regulated by the circadian clock in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 72, 3025–3029. doi: 10.1271/bbb.80402
Ivanov, K. I., and Mäkinen, K. (2012). Coat proteins, host factors and plant viral replication. Curr. Opin. Virol. 2, 712–718. doi: 10.1016/j.coviro.2012.10.001
Jiang, Y., and Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Kegler, C., Lenk, I., Krawczyk, S., Scholz, R., and Gatz, C. (2004). Functional characterization of tobacco transcription factor TGA2.1. Plant Mol. Biol. 55, 153–164. doi: 10.1007/s11103-004-0110-9
Koornneef, A., and Pieterse, C. M. (2008). Cross talk in defense signaling. Plant Physiol. 146, 839–844. doi: 10.1104/pp.107.112029
Kovtun, Y., Chiu, W. L., Zeng, W., and Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720. doi: 10.1038/27240
Kurihara, Y., and Watanabe, Y. (2004). A TMV-Cg mutant with a truncated coat protein induces cell death resembling the hypersensitive response in Arabidopsis. Mol. Cells 17, 334–339. doi: 10.1111/j.1365-313X.2004.02142.x
Kushwaha, N. K., Hafrén, A., and Hofius, D. (2019). Autophagy-virus interplay in plants: from antiviral recognition to proviral manipulation. Mol. Plant Pathol. 20, 1211–1216. doi: 10.1111/mpp.12852
Lackman, P., González-Guzmán, M., Tilleman, S., Carqueijeiro, I., Pérez, A. C., Moses, T., et al. (2011). Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci. U.S.A. 108, 5891–5896. doi: 10.1073/pnas.1103010108
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Li, X., An, M., Xia, Z., Bai, X., and Wu, Y. (2017). Transcriptome analysis of watermelon (Citrullus lanatus) fruits in response to cucumber green mottle mosaic virus (CGMMV) infection. Sci. Rep. 7:16747. doi: 10.1038/s41598-017-17140-4
Li, X. D., An, M. N., and Wu, Y. H. (2015). First report of pepper mild mottle virus in Northeast China. Plant Dis. 100:541. doi: 10.1094/PDIS-06-15-0722-PDN
Liu, Z., Shi, L., Weng, Y., Zou, H., Li, X., Yang, S., et al. (2019). ChiIV3 acts as a novel target of WRKY40 to mediate pepper immunity against ralstonia solanacearum infection. Mol. Plant Microbe Interact. 32, 1121–1133. doi: 10.1094/MPMI-11-18-0313-R
Lou, D., Wang, H., and Yu, D. (2018). The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 18:203. doi: 10.1186/s12870-018-1408-0
Lozano-Durán, R., Macho, A. P., Boutrot, F., Segonzac, C., Somssich, I. E., and Zipfel, C. (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2:e00983. doi: 10.7554/eLife.00983
Lu, J., Du, Z. X., Kong, J., Chen, L. N., Qiu, Y. H., Li, G. F., et al. (2012). Transcriptome analysis of Nicotiana tabacum infected by cucumber mosaic virus during systemic symptom development. PLoS One 7:e43447. doi: 10.1371/journal.pone.0043447
Lukhovitskaya, N. I., Solovieva, A. D., Boddeti, S. K., Thaduri, S., Solovyev, A. G., and Savenkov, E. I. (2013). An RNA virus-encoded zinc-finger protein acts as a plant transcription factor and induces a regulator of cell size and proliferation in two tobacco species. Plant Cell 25, 960–973. doi: 10.1105/tpc.112.106476
Mackey, D., Holt, B. F., Wiig, A., and Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. doi: 10.1016/S0092-8674(02)00661-X
Marathe, R., Anandalakshmi, R., Liu, Y., and Dinesh-Kumar, S. P. (2002). The tobacco mosaic virus resistance gene. N. Mol. Plant. Pathol. 3, 167–172. doi: 10.1046/j.1364-3703.2002.00110.x
McLeish, M. J., Fraile, A., and Garcia-Arenal, F. (2018). Ecological complexity in plant virus host range evolution. Adv. Virus Res. 101, 293–339. doi: 10.1016/bs.aivir.2018.02.009
Mochizuki, T., and Ohki, S. T. (2011). Single amino acid substitutions at residue 129 in the coat protein of cucumber mosaic virus affect symptom expression and thylakoid structure. Arch. Virol. 156, 881–886. doi: 10.1007/s00705-010-0910-y
Moury, B., Fabre, F., Hébrard, E., and Froissart, R. (2017). Determinants of host species range in plant viruses. J. Gen. Virol. 98, 862–873. doi: 10.1099/jgv.0.000742
Murphy, A. M., and Carr, J. P. (2002). Salicylic acid has cell-specific effects on tobacco mosaic virus replication and cell-to-cell movement. Plant Physiol. 128, 552–563. doi: 10.1104/pp.010688
Padmanabhan, M. S., Goregaoker, S. P., Golem, S., Shiferaw, H., and Culver, J. N. (2005). Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J. Virol. 79, 2549–2558. doi: 10.1128/JVI.79.4.2549-2558.2005
Pallas, V., and Garcia, J. A. (2011). How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 92, 2691–2705. doi: 10.1099/vir.0.034603-0
Pan, X., Ruth, W., and Wang, X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5, 986–992. doi: 10.1038/nprot.2010.37
Park, M. R., Park, S. H., Cho, S. Y., and Kim, K. H. (2009). Nicotiana benthamiana protein, NbPCIP1, interacting with Potato virus X coat protein plays a role as susceptible factor for viral infection. Virology 386, 257–269. doi: 10.1016/j.virol.2008.12.044
Pieterse, C. M., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pogany, J., Fabian, M. R., White, K. A., and Nagy, P. D. (2003). A replication silencer element in a plus-strand RNA virus. EMBO J. 22, 5602–5611. doi: 10.1093/emboj/cdg523
Ponstein, A. S., Bres-Vloemans, S. A., Sela-Buurlage, M. B., van den Elzen, P. J., Melchers, L. S., and Cornelissen, B. J. (1994). A novel pathogen- and wound-inducible tobacco (Nicotiana tabacum) protein with antifungal activity. Plant Physiol. 104, 109–118. doi: 10.1104/pp.104.1.109
Qin, L., Mo, N., Zhang, Y., Muhammad, T., Zhao, G., Zhang, Y., et al. (2017). CaRDR1, an RNA-dependent RNA polymerase plays a positive role in pepper resistance against TMV. Front. Plant Sci. 8:1068. doi: 10.3389/fpls.2017.01068
Quintarelli, G., Zito, R., and Cifonelli, J. A. (1971). On phosphotungstic acid staining. I. J. Histochem. Cytochem. 19, 641–647. doi: 10.1177/19.11.641
Rodriguez, M. C., Conti, G., Zavallo, D., Manacorda, C. A., and Asurmendi, S. (2014). TMV-Cg coat protein stabilizes DELLA proteins and in turn negatively modulates salicylic acid-mediated defense pathway during Arabidopsis thaliana viral infection. BMC Plant Biol. 14:210. doi: 10.1186/s12870-014-0210-x
Rybicki, E. P. (2015). A Top Ten list for economically important plant viruses. Arch. Virol. 160, 17–20. doi: 10.1007/s00705-014-2295-9
Sheng, Y., Yang, L., Li, C., Wang, Y., and Guo, H. (2019). Transcriptomic changes in Nicotiana tabacum leaves during mosaic virus infection. 3 Biotech 9:220. doi: 10.1007/s13205-019-1740-6
Solovyev, A. G., Savenkov, E. I., Grdzelishvili, V. Z., Kalinina, N. O., Morozov, S. Y., Schiemann, J., et al. (1999). Movement of hordeivirus hybrids with exchanges in the triple gene block. Virology 253, 278–287. doi: 10.1006/viro.1998.9528
Solovyev, A. G., Zelenina, D. A., Savenkov, E. I., Grdzelishvili, V. Z., Morozov, S., Maiss, E., et al. (1997). Host-controlled cell-to-cell movement of a hybrid barley stripe mosaic virus expressing a dianthovirus movement protein. Intervirology 40, 1–6. doi: 10.1159/000150514
Solovyev, A. G., Zelenina, D. A., Savenkov, E. I., Grdzelishvili, V. Z., Morozov, S. Y., Lesemann, D. E., et al. (1996). Movement of a barley stripe mosaic virus chimera with a tobacco mosaic virus movement protein. Virology 217, 435–441. doi: 10.1006/viro.1996.0137
Sopena-Torres, S., Jorda, L., Sanchez-Rodriguez, C., Miedes, E., Escudero, V., Swami, S., et al. (2018). YODA MAP3K kinase regulates plant immune responses conferring broad-spectrum disease resistance. New Phytol. 218, 661–680. doi: 10.1111/nph.15007
Spartz, A. K., Lee, S. H., Wenger, J. P., Gonzalez, N., Itoh, H., Inzé, D., et al. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70, 978–990. doi: 10.1111/j.1365-313X.2012.04946.x
Thurow, C., Schiermeyer, A., Krawczyk, S., Butterbrodt, T., Nickolov, K., and Gatz, C. (2005). Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 44, 100–113. doi: 10.1111/j.1365-313X.2005.02513.x
Ton, J., Flors, V., and Mauch-Mani, B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. doi: 10.1016/j.tplants.2009.03.006
Tuo, D., Shen, W., Yan, P., Li, X., and Zhou, P. (2015). Rapid construction of stable infectious full-length cDNA clone of papaya leaf distortion mosaic virus using In-Fusion cloning. Viruses 7, 6241–6250. doi: 10.3390/v7122935
Usman, M. G., Rafii, M. Y., Martini, M. Y., Yusuff, O. A., Ismail, M. R., and Miah, G. (2017). Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnol. Genet. Eng. Rev. 33, 26–39. doi: 10.1080/02648725.2017.1340546
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wang, A. (2015). Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66. doi: 10.1146/annurev-phyto-080614-120001
Wang, W., Chen, D., Zhang, X., Liu, D., Cheng, Y., and Shen, F. (2018). Role of plant respiratory burst oxidase homologs in stress responses. Free Radic. Res. 52, 826–839. doi: 10.1080/10715762.2018.1473572
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Weber, P. H., and Bujarski, J. J. (2015). Multiple functions of capsid proteins in (+) stranded RNA viruses during plant-virus interactions. Virus Res. 196, 140–149. doi: 10.1016/j.virusres.2014.11.014
Whitham, S. A., Quan, S., Chang, H. S., Cooper, B., Estes, B., Zhu, T., et al. (2003). Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33, 271–283. doi: 10.1046/j.1365-313x.2003.01625.x
Xie, K., Li, L., Zhang, H., Wang, R., Tan, X., He, Y., et al. (2018). Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 41, 2504–2514. doi: 10.1111/pce.13372
Yelina, N. E., Savenkov, E. I., Solovyev, A. G., Morozov, S. Y., and Valkonen, J. P. (2002). Long-distance movement, virulence, and RNA silencing suppression controlled by a single protein in hordei- and potyviruses: complementary functions between virus families. J. Virol. 76, 12981–12991. doi: 10.1128/jvi.76.24.12981-12991.2002
Yu, M., Liu, H., Zheng, H., Yan, F., Zhao, X., Xia, Z., et al. (2019). Viral sequences required for efficient viral infection differ between two chinese pepper mild mottle virus isolates. Virus Res. 267, 9–15. doi: 10.1016/j.virusres.2019.04.007
Zhu, F., Xi, D. H., Yuan, S., Xu, F., Zhang, D. W., and Lin, H. H. (2014). Salicylic acid and jasmonic acid are essential for systemic resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol. Plant Microbe Interact. 27, 567–577. doi: 10.1094/MPMI-11-13-0349-R
Keywords: tobacco mosaic virus, coat protein, transcriptome analysis, chimeric viruses, plant hormones
Citation: Yu M, Bi X, Huang Y, Chen Y, Wang J, Zhang R, Lei Y, Xia Z, An M and Wu Y (2020) Chimeric Tobamoviruses With Coat Protein Exchanges Modulate Symptom Expression and Defence Responses in Nicotiana tabacum. Front. Microbiol. 11:587005. doi: 10.3389/fmicb.2020.587005
Received: 24 July 2020; Accepted: 12 October 2020;
Published: 06 November 2020.
Edited by:
Yi-Ping Li, Sun Yat-sen University, ChinaReviewed by:
Masamichi Nishiguchi, Ehime University, JapanCopyright © 2020 Yu, Bi, Huang, Chen, Wang, Zhang, Lei, Xia, An and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mengnan An, YW5tZW5nbmFuQHN5YXUuZWR1LmNu; Yuanhua Wu, d3V5aDA5QHN5YXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.