
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 January 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.586368
This article is part of the Research Topic New Insights into the Transmission Dynamics and Control of Antimicrobial Resistance to Last-resort Antibiotics View all 22 articles
Mobile colistin-resistant genes (mcr) have become an increasing public health concern. Since the first report of mcr-1 in Thailand in 2016, perspective surveillance was conducted to explore the genomic characteristics of clinical carbapenem-resistant Enterobacterales (CRE) isolates harboring mcr in 2016–2019. Thirteen (0.28%) out of 4,516 CRE isolates were found to carry mcr genes, including 69.2% (9/13) of E. coli and 30.8% (4/13) of K. pneumoniae isolates. Individual mcr-1.1 was detected in eight E. coli (61.5%) isolates, whereas the co-occurrence of mcr-1.1 and mcr-3.5 was seen in only one E. coli isolate (7.7%). No CRE were detected carrying mcr-2, mcr-4, or mcr-5 through to mcr-9. Analysis of plasmid replicon types carrying mcr revealed that IncX4 was the most common (61.5%; 8/13), followed by IncI2 (15.4%; 2/13). The minimum inhibitory concentration values for colistin were in the range of 4–16 μg/ml for all CRE isolates harboring mcr, suggesting they have 100% colistin resistance. Clermont phylotyping of nine mcr-harboring carbapenem-resistant E. coli isolates demonstrated phylogroup C was predominant in ST410. In contrast, ST336 belonged to CC17, and the KL type 25 was predominant in carbapenem-resistant K. pneumoniae isolates. This report provides a comprehensive insight into the prevalence of mcr-carrying CRE from patients in Thailand. The information highlights the importance of strengthening official active surveillance efforts to detect, control, and prevent mcr-harboring CRE and the need for rational drug use in all sectors.
The global spread of carbapenem-resistant Enterobacterales (CRE) has become a leading public health concern. The lack of accessible treatment has resulted in the use of colistin, an outmoded antibiotic, as a last-resort therapeutic drug for human infections by gram-negative bacteria. The widespread use of colistin in humans and animals has led to the emergence of colistin resistance in gram-negative bacteria, and the rates of resistance are continuously increasing (Kempf et al., 2016; Elbediwi et al., 2019). In 2017, the World Health Organization acknowledged that CRE are a serious priority in a published list of globally important of 12 antimicrobial-resistant pathogens (World Health Organization, 2017). Of particular concern is the spread of mcr genes into CRE, which would create strains that are potentially pan-drug resistant (PDR).
A common mechanism of colistin resistance is thought to be associated with chromosomal mediation (Meletis and Skoura, 2018). The discovery of the first plasmid-mediated colistin resistance gene mcr-1 in an Escherichia coli strain isolated from a pig in China led to an increasing number of reports on the identification of mcr in many bacterial species worldwide (Liu et al., 2016; Elbediwi et al., 2019). Thus far, 10 variants of mcr (mcr-1 through to mcr-10) have been reported (Gharaibeh and Shatnawi, 2019; Wang et al., 2020). The mcr gene has been shown to encode a phosphoethanolamine transferase that alters lipid A in the lipopolysaccharide of the bacterial outer membrane by adding a phosphoethanolamine (Gharaibeh and Shatnawi, 2019). This reduces the attachment of colistin to the bacterial outer membrane and, therefore, prevents cell lysis.
The presence of mcr has been reported in different gram-negative bacteria isolated worldwide from animal and human sources (Elbediwi et al., 2019; Gharaibeh and Shatnawi, 2019). In Thailand, the first reported mcr-1-harboring E. coli were isolated from the stool of an asymptomatic person in 2012 (Olaitan et al., 2016). There has since been documentation of mcr- 1-, mcr- 2-, and mcr-3-carrying E. coli and Klebsiella pneumoniae strains isolated from patients (Paveenkittiporn et al., 2017; Srijan et al., 2018; Kamjumphol et al., 2019; Malchione et al., 2019). The current study investigated the genomic characterization of antimicrobial resistance genes and antimicrobial susceptibility of CRE isolated from patients in Thailand during a laboratory-based surveillance program in 2016–2019.
In total, 6,996 multidrug-resistant (MDR) bacterial isolates were collected from individuals during 2016–2019 from a hospital network in 24 provinces throughout Thailand (Figure 1). There were 666, 2,763, 2,618, and 949 MDR isolates surveyed in 2016–2019, respectively. Conventional biochemical tests described elsewhere were used for the species identification of Enterobacterales (Abbott, 2011). The presence of carbapenemase (blaIMP, blaKPC, blaNDM, and blaOXA–48–like) and mcr-1 genes was determined using multiplex PCR (Hatrongjit et al., 2018). The mcr genes (mcr-1 to mcr-9) were detected using PCR, as described elsewhere (Rebelo et al., 2018; Wang et al., 2018; Yang et al., 2018; Yuan et al., 2019). All mcr-positive isolates were subjected to whole-genome sequencing.
Analysis of the minimal inhibitory concentration (MIC) of antimicrobials for mcr-harboring CRE isolates was performed using the Epsilometer test (E test) and interpreted according to the 2020 Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute, 2020), and E. coli ATCC 25922 was used as the control. The E test was based on ampicillin, amoxicillin-clavulanate, ampicillin-sulbactam, piperacillin-tazobactam, cefazolin, cefepime, cefotaxime, cefoxitin, ertapenem, meropenem, imipenem, gentamicin, amikacin, ciprofloxacin, levofloxacin, trimethoprim, fosfomycin, nitrofurantoin, chloramphenicol, tetracycline, aztreonam, and azithromycin. Additionally, a modified carbapenem inactivation method (mCIM) was applied to all CRE isolates according to 2020 CLSI M100-S30.
Broth microdilution was used to determine the MIC of colistin using colistin sulfate (Merck, Germany) at 1, 2, 4, 8, 16, and 32 μg/ml, respectively. According to 2020 CLSI M100-S30, an MIC of ≤2 μg/ml was interpreted as intermediate susceptibility, whereas an MIC of ≥4 μg/ml was considered to indicate resistance.
DNA from 13 mcr-carrying CRE isolates was extracted from nutrient agar plate cultures using a DNeasy blood & tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations, and the concentration was determined using the Qubit dsDNA BR assay kit (Invitrogen, Oregon, United States). Sequence libraries were prepared using a Qiagen QIAseq FX DNA library kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The prepared libraries were sequenced on the Illumina Miseq platform with Illumina MiSeq 2X 250 base paired-end chemistry (Illumina, CA, United States) according to the manufacturer’s instructions. The genomes for each strain were de novo assembled using CLC Genomics Workbench v12.0.2 (Qiagen, Aarhus, Denmark) with mostly default settings, except that the minimum contig size threshold was set to 500 bp.
Analysis of the whole-genome sequence data was performed as described elsewhere (Kerdsin et al., 2019). Briefly, the isolates were identified to species level using KmerFinder 3.1 (Larsen et al., 2014)1. Antimicrobial resistance genes were identified using ResFinder 4.1 (Zankari et al., 2012)2 and the Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al., 2020)3. We investigated chromosome-mediated colistin resistance by analyzing the mgrB, phoPQ, pmrAB, crrB, and rpoN genes. The gene sequences were analyzed using local BLAST + and Clustal W, including K. pneumoniae MGH78578 (GenBank accession number NC_009648.1) and E. coli K12 sub-strain MG1655 (GenBank accession number U00096) genomes as colistin-susceptible references.
Plasmid replicons were analyzed using PlasmidFinder (Carattoli et al., 2014)4. Phylogrouping for E. coli and the KL type of K. pneumoniae were based on analysis using ClermonTyping (Beghain et al., 2018)5 and Kaptive (Wick et al., 2018)6. The virulence genes of E. coli and K. pneumoniae were analyzed using VirulenceFinder 2.0 (Joensen et al., 2014)7 and Institut Pasteur8, respectively.
For multilocus sequence typing (MLST) analysis of the sequence types (STs) of E. coli and K. pneumoniae, we used MLST 2.0 (Larsen et al., 2012)9. The genomic comparison of 13 mcr-harboring CRE isolates was conducted using a modular Single Genome Analysis to search for the genetically closest relatives in the database following the single nucleotide polymorphism (SNP) approach with BacWGSTdb (Feng et al., 2020)10. A phylogenetic tree was constructed using REALPHY and MEGA X via the neighbor-joining method with 500 bootstrap replicates by applying the Tamura three-parameter model (Bertels et al., 2014; Kumar et al., 2018). The tree was visualized and annotated using Interactive Tree of Life (ITOL) V4 (Letunic and Bork, 2016). E. coli K12 substrain MG1655 (accession no. U00096) and K. pneumoniae HS11286 (accession no. CP003200) were used as the reference sequences for SNP analysis. Details of the other genomes used for comparison with our isolates are shown in the Supplementary Materials.
The associations between mcr genes, mcr-harboring E. coli isolates, and non-E. coli isolates were analyzed by calculating the odds ratios (OR) and p-values using the STATA version 14 software package, with p < 0.05 considered to be statistically significant.
The assembled genomic sequences were deposited under the BioProject accession number PRJNA525849 with BioSample accessions: SAMN15497997-SAMN15498009. The accession numbers for each mcr-1-harboring CRE isolate are provided in Table 2.
Of the 6,996 MDR isolates, 4,516 were identified as CRE (64.5%). Of these, 4,235 (93.7%) isolates were classified as carbapenemase-producing Enterobacterales (CPE) and carried carbapenemase genes (blaNDM, blaOXA–48–like, blaIMP, or coexisting carbapenemase genes) according to the mCIM and PCR results. Of all the CPE isolates, 13 (0.3%) carried mcr genes (Table 1). The mcr-carrying rates among carbapenemase-producing E. coli and K. pneumoniae were 1.03 and 0.12%, respectively. Statistical analysis revealed a strong association between mcr and carbapenemase-producing E. coli, with the OR being 11.06 (95% CI, 3.07–49.23) and statistically significant (p < 0.0001) (Table 1).
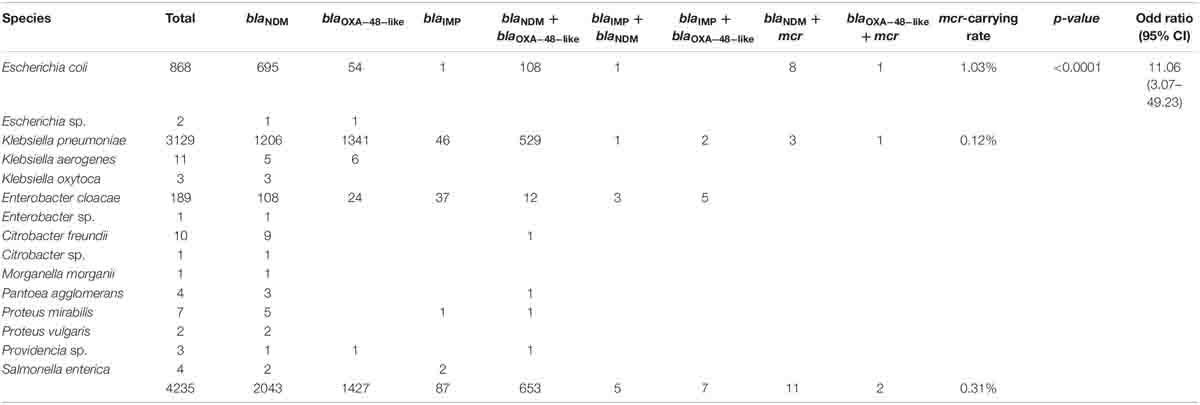
Table 1. Distribution of mcr and carbapenemase genes in carbapenemase-producing Enterobacterales (CPE) during 2016–2019.
The proportions of E. coli and K. pneumoniae isolates showing mcr genes were 69.2% (9/13) and 30.8% (4/13), respectively. Individual mcr-1.1 genes were detected in eight E. coli (61.5%) from 13 mcr-carrying CRE isolates, whereas the co-occurrence of mcr-1.1 and mcr-3.5 was found in only one E. coli isolate (7.7%) (Table 2). However, mcr-2, mcr-4, and mcr-5 through to mcr-9 were not detected. Analysis of the plasmid replicon types carrying mcr revealed that IncX4 was the most common (61.5%; 8/13), followed by IncI2 (15.4%; 2/13), as shown in Table 2. However, three isolates were unidentified. Six carbapenem-resistant E. coli isolates harboring mcr carried IncX4, whereas two isolates contained IncI2. Of the four carbapenem-resistant K. pneumoniae isolates harboring mcr, two carried IncX4 and the other two had unknown plasmid replicon types. The genetic organization of the mcr genes in these 13 isolates is outlined in Figure 2. A common gene found downstream of mcr-1.1 in all isolates encoded the PAP2 family protein, whereas the upstream genes varied. However, a DUF2726-domain-containing protein-encoding gene was commonly found in 8 of the 13 isolates (seven E. coli and one K. pneumoniae). Furthermore, the upstream and downstream genetic organization of mcr-3.5 was quite different from that of mcr-1.1.

Table 2. Distribution of sequence types and antimicrobial resistant genes in carbapenem-resistant E. coli and K. pneumoniae carrying mcr genes.
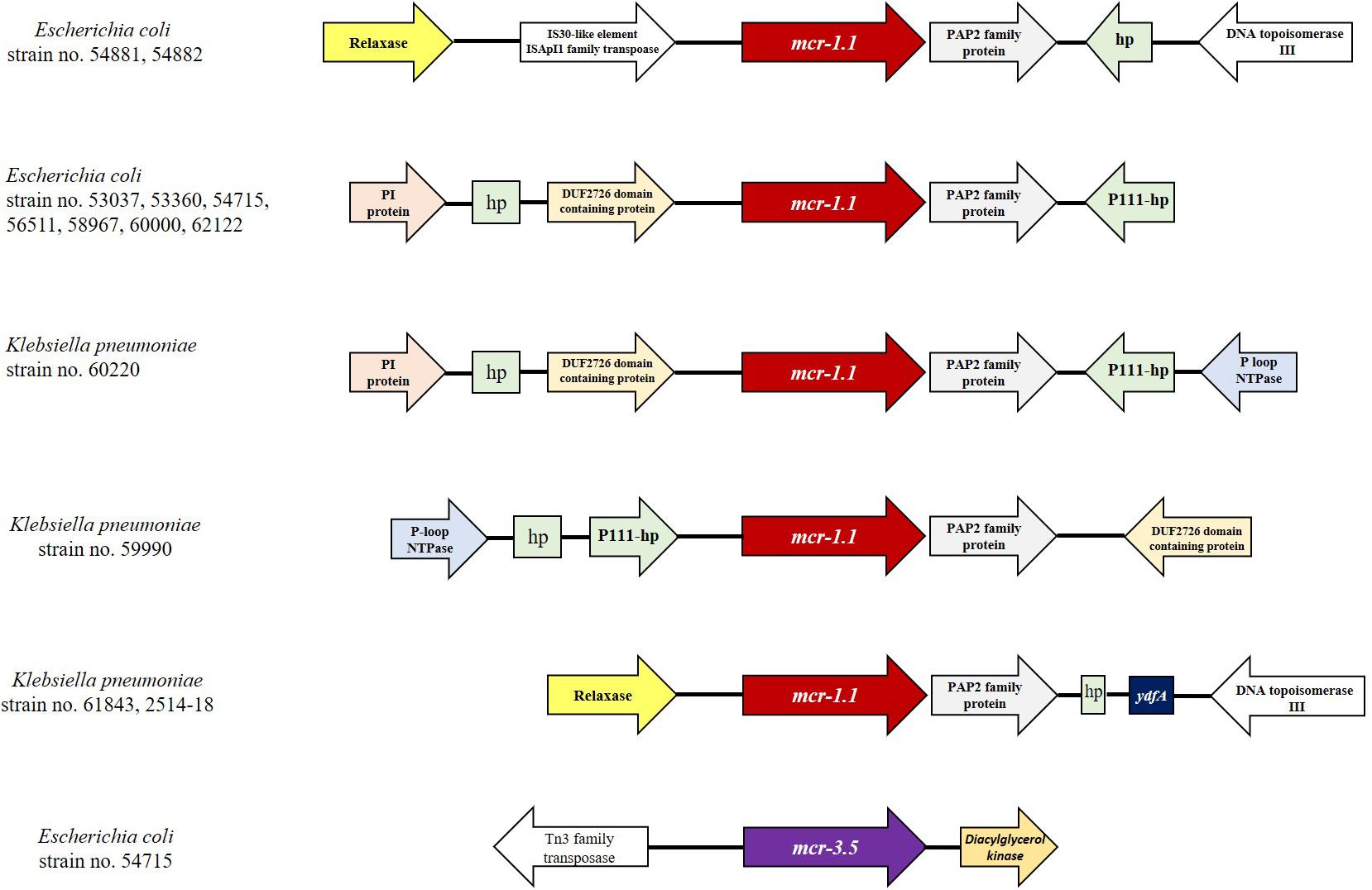
Figure 2. Comparison of the genetic organization of the mcr genes in 13 CRE isolates. The schematic shows the genes flanking the mcr genes in the isolates.
As shown in Table 2, chromosomal-mediated colistin resistance gene mutations, including those in mgrB, pmrAB, phoPQ, crrB, and rpoN, were analyzed. We detected substitutions in pmrAB and rpoN in almost mcr-harboring CRE isolates, whereas phoPQ substitutions were found in three isolates, and no mutations were detected in mgrB. Substitutions were commonly found in the pmrA genes of 12 out of 13 mcr-harboring isolates, while pmrB and rpoN substitutions were detected in 11 isolates. The isolates 54881 and 54882 contained more mutations in the chromosomal-mediated colistin resistance genes than other isolates. Substitution at S29G and E57G in pmrA was predominant in E. coli and K. pneumoniae, respectively. The pmrB substitution at D283G was commonly found in E. coli, whilst all four mcr-1-harboring K. pneumoniae contained a T246A substitution. Only one substitution in phoP (I44L) and one in phoQ (L343V) were detected in mcr-1-carrying E. coli. Substitution at E150D and I165M in rpoN was predominant in E. coli isolates, in contrast to those at F304Y which was commonly found in K. pneumoniae isolates. In addition, one crrB substitution (L296Q) was found in all K. pneumoniae isolates. Insertion or deletion in those described genes was not detected in all mcr-1-harboring CRE isolates.
As shown in Table 2, of the 13 isolates, five and three mcr-harboring E. coli isolates carried blaNDM–1 and blaNDM–5, respectively. Only one E. coli isolate contained blaOXA–48. Three and one mcr-1.1-harboring K. pneumoniae isolates carried blaNDM–1 and blaOXA–181, respectively. Among the β-lactamase genes, blaCTX–M, blaSHV, or blaTEM were detected in almost all isolates (92.3%, 12/13), and only one E. coli isolate had no β-lactamase genes. The predominant blaCTX–M was blaCTX–M–15, which was detected in 53.8% (7/13) of all isolates. The gene blaSHV was found in all K. pneumoniae and one isolate of E. coli (Table 2). Among the ampC β-lactamase genes, blaCMY–2 only was detected in four E. coli isolates (28.6%). The other antimicrobial resistance genes in the mcr-carrying CRE isolates, including those for fluoroquinolones, aminoglycosides, rifampicin, macrolides, chloramphenicol, sulfonamide, tetracycline, fosfomycin, and trimethoprim, are shown in Table 2.
As shown in Table 3, the colistin MIC values for the mcr-harboring isolates were in the range of 4–16 μg/ml. According to the 2020 CLSI M100-S30 guidelines, a microbe with a colistin MIC value of ≥4 μg/ml is resistant, whereas MIC ≤ 2 μg/ml indicates intermediate resistance. Thus, the results indicate that 100% of the mcr-carrying CRE isolated from patients were resistant to colistin. The highest MIC value for colistin (16 μg/ml) was found in two carbapenem-resistant K. pneumoniae isolates. Most of the mcr-harboring E. coli had colistin MIC values of 4 μg/ml (8/9; 88.8%).
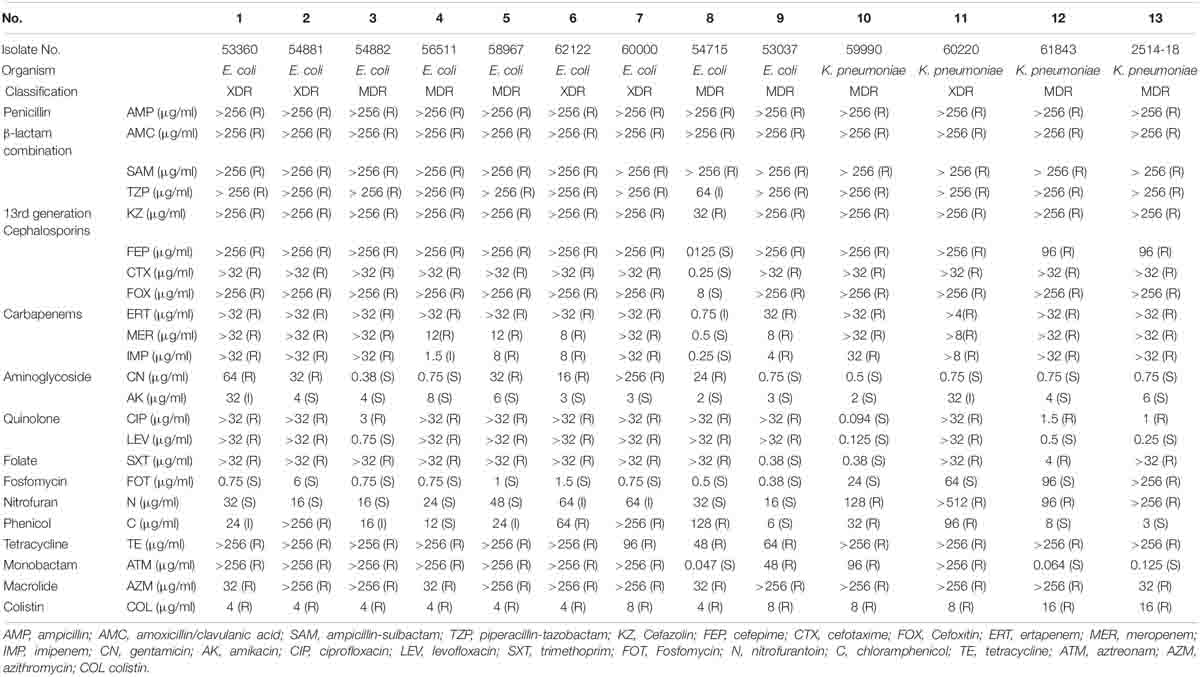
Table 3. Antimicrobial susceptibility of mcr-harboring carbapenem-resistant E. coli and K. pneumoniae.
More than 50% of the mcr-harboring CRE isolates were susceptible to amikacin (11/13), fosfomycin (12/13), and nitrofurantoin (7/13). Of the 13 CRE-harboring mcr isolates, four E. coli isolates and one K. pneumoniae isolate were extensively drug-resistant (XDR; 38.5%). Details of the antimicrobial resistance profiles of the 13 isolates are provided in Table 3. Moreover, 12 of the 13 mcr-harboring CRE isolates were resistant to ciprofloxacin. As shown in Table 2, ciprofloxacin resistance may result from the presence of quinolone resistance genes: oqxA, oqxB, qnrS1, qnrB6, and aac(6′)-Ib-cr.
As summarized in Table 2, Clermont phylotyping of nine mcr-harboring carbapenem-resistant E. coli isolates demonstrated that phylogroup C (5/9; 55.6%) was predominant, followed by phylogroups A (2/9; 22.2%) and D (2/9; 22.2%). The Clermont phylogroup was concordant with clonal complexes (CC). Phylogroup C was concordant with CC23, which contained only ST410. Phylogroup D was concordant with CC38, which consisted of ST3052, whereas phylogroup A was concordant with CC10, which contained either ST10 or ST1287. Four carbapenem-resistant K. pneumoniae isolates harboring mcr were predominantly ST336 (3/4; 75%) and one was ST340 (1/4; 25%). ST336 belonged to CC17 and KL type 25, whereas ST340 belonged to CC258 and KL type 15.
The genetic relationships based on the SNPs of these mcr-harboring isolates are demonstrated in Figures 3, 4. Five E. coli ST410 isolates were widely distributed in several sub-clusters of the ST410 cluster. Strain no. 53360 was closely related to the strain AMA1167 from Denmark, whereas strain no. 56511 clustered with strains from Norway, India, Lebanon, China, and Denmark (Figure 3). Strains no. 58967 and no. 62122 were related to strain KBN10P04869 from South Korea. Strain no. 53037 clustered with strains from Norway, the United States, Brazil, and Germany. Two ST3052 isolates (nos. 54881 and 54882) were closely related to the strain WCHEC020028 from China; they had similar characteristics and were from different individuals in the same hospital ward, indicating that they were likely to have originated from the same source. Strain no. 54715 (ST1287) was related to a strain from the United States (ST617), and these were clustered together with strain no. 6000 (ST10) (Figure 3).

Figure 3. Phylogenetic tree based on single nucleotide polymorphisms (SNP) in E. coli using the neighbor-joining method. The tree was visualized and annotated using Interactive Tree of Life (iToL). The tree is annotated based on sequence types (STs; color highlight), mcr (square), blaNDM (circle), blaKPC (right triangle), and blaOXA–48–like (star). Presentation of the genes is shown by filled symbols. STs 410, 1287, 617, 10, and 3052 are highlighted in yellow, green, brown, pink, and blue, respectively. The isolates used in this study are shown in bold.
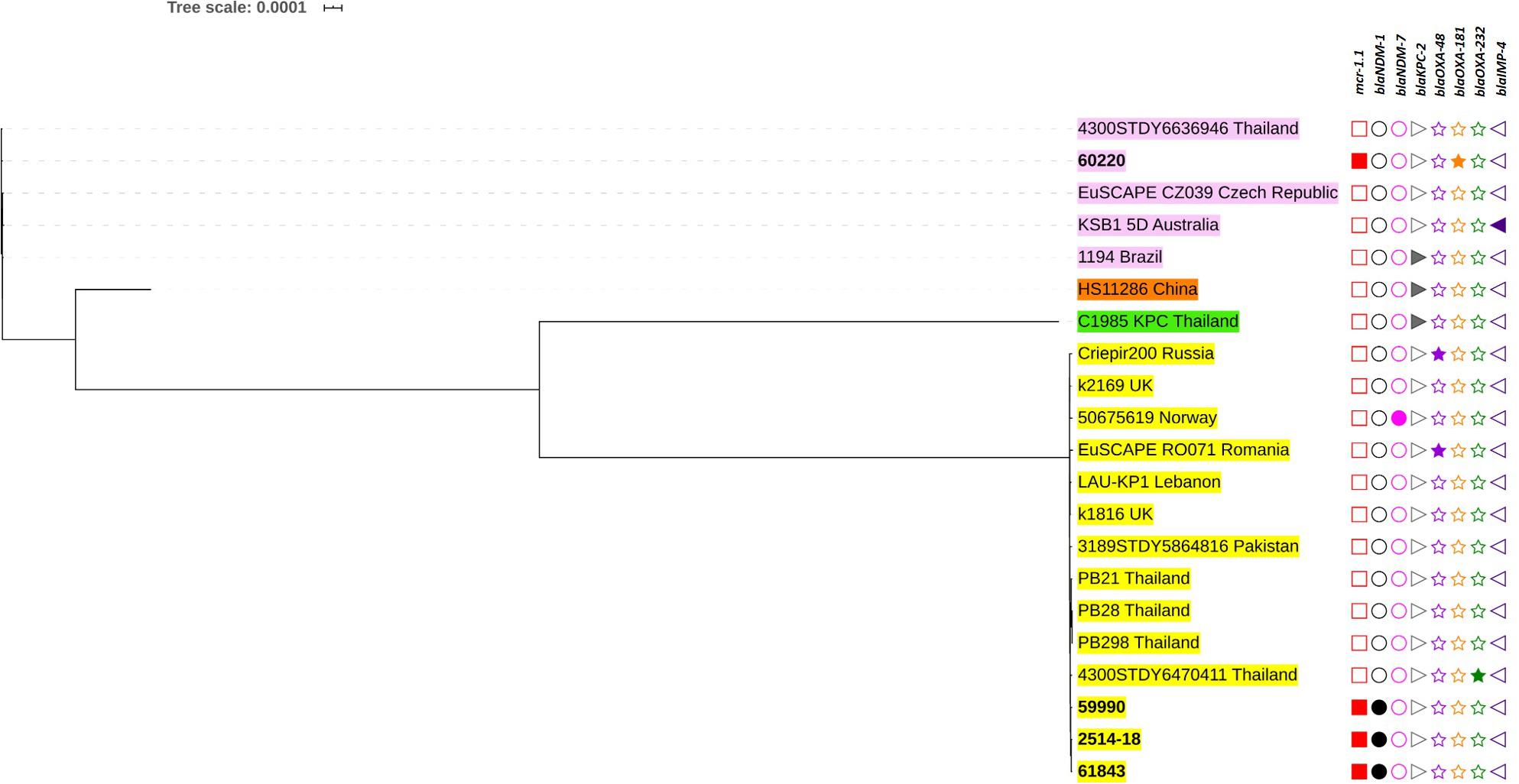
Figure 4. Phylogenetic tree based on single nucleotide polymorphisms (SNP) in K. pneumoniae using the neighbor-joining method. The tree was visualized and annotated using Interactive Tree of Life (iToL). The tree is annotated based on sequence types (STs; color highlight), mcr (square), blaNDM (circle), blaKPC (right triangle), blaOXA–48–like (star), and blaIMP (left triangle). Presentation of the genes is revealed by filled symbols. STs 336, 340, 11, and 4008 are highlighted in yellow, pink, and brown, and green, respectively. The isolates used in this study are shown in bold.
Among the K. pneumoniae isolates, the ST336 isolates were closely related and clustered with other ST336 strains isolated in Thailand (Figure 4). Interestingly, the Thai-ST336 isolates were in a different cluster from that containing the ST336 isolates from other countries. Similarly, isolate no. 60220 (ST340) in this study was closely related to strain 4300STDY6636946 circulating in Thailand (Figure 4).
The discovery that plasmid-mediated colistin resistance is encoded by mcr genes and the high prevalence of human isolates harboring these genes are of global concern. A recent report revealed the overall average prevalence of mcr genes to be 4.7% (0.1–9.3%) in 47 countries across six continents (Elbediwi et al., 2019), and as many as 10 mcr genes (mcr-1 through to mcr-10) have been reported (Gharaibeh and Shatnawi, 2019; Wang et al., 2020). A study of the global prevalence of mcr genes revealed that mcr-1 (4917/5191; 94.7%) is a common gene and has a wider distribution compared with mcr-2 through to mcr-8 (Elbediwi et al., 2019). Human infections with both CRE and non-CRE isolates carrying mcr-1 have been widely reported (Liu et al., 2016; Mediavilla et al., 2016; Paveenkittiporn et al., 2017; Quan et al., 2017; Mendes et al., 2018; Srijan et al., 2018; Zhong et al., 2018; Elbediwi et al., 2019).
The coexistence of mcr and carbapenemase genes, such as blaNDM, blaOXA–48–like, and blaIMP, in CRE isolates has been described in countries worldwide (Mediavilla et al., 2016; Arabaci et al., 2019; Huang et al., 2020; Kananizadeh et al., 2020). The current study found the predominant mcr gene to be mcr-1, which more frequently coexists with blaNDM than blaOXA48–like, highlighting the potential dissemination of mcr-1 and blaNDM among CRE isolates in Thailand. This concurs with a study in China (Huang et al., 2020), where mcr-1 and blaNDM–5 were predominant (78.6%, 11/14). In this study, mcr-1 and blaNDM–1 were the most prevalent resistance genes (61.5%, 8/13). A previous study showed that mcr-3 had a wide distribution in water, animals, food, and human isolates (Elbediwi et al., 2019). We found a 7.7% (1/13) prevalence for mcr-3.5, which co-occurred with mcr-1 and blaOXA–48 in E. coli. The phenomenon of double mcr genes has been reported in isolates from humans, with K. pneumoniae harboring mcr-3 and mcr-8 being isolated from the stool of a healthy individual in Laos (Hadjadj et al., 2019).
Our study revealed that the most common type of plasmid replicon carrying mcr was IncX4. Previous reports have shown IncX4, IncI2, and IncHI2 to be the major plasmid types driving the global dissemination of mcr-1 (Wu et al., 2018). A study in Thailand revealed two predominant plasmid types (IncX4 and IncI2) carrying mcr-1 in CRE (Shanmugakani et al., 2019). This suggests that IncX4 bearing mcr-1 mediates the transmission of CRE and may promote its circulation throughout Thailand. IncX4 and IncI2 acting as vehicles for mcr-1 propagation enhance host fitness and provide a competitive advantage over strains with other plasmid replicon types, resulting in greater plasmid stability (Wu et al., 2018).
On the basis of the Clermont phylotyping scheme, E. coli species can be divided into eight main phylogroups, termed A, B1, B2, C, D, E, F, and G (Clermont et al., 2019). The nine carbapenem-resistant E. coli isolates carrying mcr (55.6%) in this study belonged to phylogroup C, whereas the rest belonged to phylogroups A and D (22.2% each). The E. coli strains responsible for extra-intestinal infection were more likely to be members of phylogroups B2 or D, which show greater pathogenesis than A, B1, or C (Clermont et al., 2013). Strains belonging to phylogroups A, B1, and C are commonly commensal, suggesting that more than half of the E. coli harboring mcr isolated from patients in this study were commensal strains.
Our study revealed seven carbapenem-resistant STs that carry mcr, of which E. coli ST410 (35.7%) and K. pneumoniae ST336 (21.4%) isolates were predominant. Elbediwi et al. (2019) reported that E. coli ST101 carrying mcr-1 have been found in environmental samples, animals, and humans. However, ST10 was the most globally common ST of E. coli carrying mcr-1 (Elbediwi et al., 2019). In Asia, ST116 was found to be the predominant ST that carries mcr-1 isolated from humans, followed by ST117, ST10, ST38, ST101, and ST156 (Elbediwi et al., 2019). E. coli ST410 is internationally considered a new high-risk clone that can cause several types of infection; it is highly resistant and has a global distribution (Roer et al., 2018). This ST has been described in Southeast Asia following multiple introductions through several independent events and differs from clones detected in Europe and North America (Nadimpalli et al., 2019). SNP phylogenetic analysis in this study revealed that the ST410 isolates were diverse or not closely related to other strains. Instead, they were shown to be related to strains from countries other than Thailand. This adds support to the assumption that there have been multiple dissemination events into this area.
Klebsiella pneumoniae ST336 (CC17) is considered an international clone (Rodrigues et al., 2014; Novović et al., 2017; Palmieri et al., 2020) and has been frequently associated with the worldwide spread of blaCTX–M–15 and blaOXA–48–like (Rodrigues et al., 2014; Novović et al., 2017; Palmieri et al., 2020). Interestingly, all ST336 isolates in the current study carried blaNDM–1, but not any blaCTX–M genes, suggesting that they may be from different lineages. To the best of our knowledge, the carbapenem-resistant ST336 isolates in this study are the first to be described as having mcr-1. Previous studies revealed that colistin-resistant ST336 resulted from an mgrB mutation, and no mcr genes have been detected in this ST (Novović et al., 2017; Palmieri et al., 2020). SNP phylogenetic analysis allocated the ST336 isolates to the same cluster as other Thai-ST336 isolates, and this cluster was independent from another ST336 cluster consisting of isolates from other countries. This suggests that Thai-ST336 isolates circulate throughout our country by clonal expansion.
The colistin MIC values (4–16 μg/ml) for our isolates indicated they have 100% resistance. Combinations of mcr and chromosome-mediated colistin resistant genes (pmrAB, phoPQ, crrB, or recN) contributed to the colistin resistance of our isolates. It is interesting that substitutions in pmrAB and crrB in mcr-1-carrying K. pneumoniae are quite different from previous reports (Olaitan et al., 2014a,b; Wright et al., 2015; Cheng et al., 2016). In addition, substitution of phoPQ was not found in our K. pneumoniae isolates comparing to those studies (Olaitan et al., 2014a,b; Wright et al., 2015; Cheng et al., 2016). On the other hand, substitutions of pmrA (G144S), pmrB (H2R, D283G, Y358N), and phoP (I44L) in our mcr-1-harboring E. coli are similar to a study previously described (Choi et al., 2020). In case of rpoN, inactivation of this gene resulting in polymyxin resistance has been observed in Salmonella enterica (Barchiesi et al., 2009). However, polymyxin resistance via rpoN inactivation or substitution is not yet reported in either E. coli or K. pneumoniae. Although our study detected substitution of rpoN, but its role on polymyxin resistance remain to be investigated.
Several studies have shown mcr-1-carrying CRE isolated from humans to have high frequencies of colistin resistance, such as 71.4% in China (Huang et al., 2020), 100% in the United States (Mediavilla et al., 2016), and 100% in Turkey (Arabaci et al., 2019). A previous study in Thailand reported colistin resistance rates of 75.0 and 79.1% in carbapenem-resistant E. coli and K. pneumoniae isolates, respectively (Eiamphungporn et al., 2018). The 13 carbapenem-resistant mcr-1-harboring CRE isolates described here showed a high susceptibility (>50%) to the antibiotics amikacin, fosfomycin, and nitrofurantoin. In contrast, the human mcr-1-harboring CRE isolate from China was reported to be highly susceptible only to tigecycline, amikacin, and aztreonam (Huang et al., 2020), whereas the isolate from the United States was susceptible to more antibiotics, including amikacin, aztreonam, gentamicin, nitrofurantoin, tigecycline, and trimethoprim-sulfamethoxazole (Mediavilla et al., 2016).
Polymyxins, including colistin, were reintroduced into human medical practice by the WHO in 2012 (World Health Organization, 2012). In China, this antibiotic was approved for clinical use in the treatment of bacterial infections in 2017. Since then, the relative prevalence of CRE carrying mcr genes increased from 0.41 to 1.38% (Huang et al., 2020). A study in Singapore revealed that prior exposure to polymyxin (adjusted OR, 21.31; 95% CI, 3.04–150.96) and carbapenem (OR 3.74; CI 1.13–12.44) were independent risk factors for polymyxin-resistant CRE among hospitalized patients (Teo et al., 2019). A study in Thailand demonstrated that chronic kidney diseases (OR 3.95; CI 1.26–12.32) and exposure to antimicrobials for less than 3 months (OR 2.29; CI 0.29–18.21) were risk factors associated with infections by mcr-1-carrying Enterobacteriaceae (Shanmugakani et al., 2019). In patients infected with polymyxin-resistant CRE, the 30-day all-cause in-hospital mortality was 50.0% compared with a 38.1% mortality in patients with polymyxin-susceptible CRE (Teo et al., 2019). Therefore, minimizing the use of polymyxin and carbapenem is strongly recommended.
The findings of the current study provide comprehensive insights into the prevalence of mcr-carrying CRE in patients in Thailand. In general, mcr-1 was present in E. coli and K. pneumoniae isolates. The co-occurrence of two mcr genes was also demonstrated in CRE isolated from patients. To slow the emergence of XDR or PDR strains, priority should be given to strengthening official surveillance, active control, and prevention efforts as well as minimizing the dissemination of mcr genes among CRE isolates in humans.
The datasets generated for this study can be found in the Raw sequencing data were deposited in the Sequence Read Archive (SRA) of NCBI under the BioProject ID PRJNA380676.
The Human Research Ethics Committee of Department of Medical Sciences, Ministry of Public Health, reviewed this study and judged that the protocol constituted routine public health activities and therefore did not involve human subject research. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
WP and AK conceived and designed the study, performed the data analysis, drafted the manuscript, and performed critical revisions of the manuscript for intellectual content. WK performed the laboratory experiments and analyzed the data. RU performed the statistical analysis and critical revision of the manuscript for intellectual content. All the authors read, edited, and approved the final manuscript.
This work was supported by the National Institute of Health, Department of Medical Sciences, Ministry of Public Health, Thailand, and the Division of Global Health Protection, Thailand MoPH–U.S. CDC Collaboration.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. Charatdao Bunthi, Division of Global Health Protection, Thailand MoPH–U.S. CDC Collaboration for critical review of the manuscript and the Kasetsart University Research and Development Institute (KURDI), Bangkok, Thailand for English-editing assistance. We also thank Suzanne Leech, Ph.D., from Edanz Group (https://en-author-services.edanzgroup.com/ac) for editing the draft of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.586368/full#supplementary-material
Abbott, S. (2011). “Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae,” in Manual of Clinical Microbiology, 10th Edn, Vol. 2, eds J. Versalovic, K. C. Carroll, G. Funke, J. H. Jorgensen, M. L. Landry, and D. W. Warnock, (Washington, DC: ASM Press), 639–657.
Alcock, B. P., Raphenya, A. R., Lau, T., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Arabaci, Ç, Dal, T., Başyiǧit, T., Genişel, N., and Durmaz, R. (2019). Investigation of carbapenemase and mcr-1 genes in carbapenem-resistant Klebsiella pneumoniae isolates. J. Infect. Dev. Ctries 13, 504–509. doi: 10.3855/jidc.11048
Barchiesi, J., Espariz, M., Checa, S. K., and Soncini, F. C. (2009). Downregulation of RpoN-controlled genes protects Salmonella cells from killing by the cationic antimicrobial peptide polymyxin B. FEMS Microbiol. Lett. 291, 73–79. doi: 10.1111/j.1574-6968.2008.01437.x
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4:e000192. doi: 10.1099/mgen.0.000192
Bertels, F., Silander, O. K., Pachkov, M., Rainey, P. B., and van Nimwegen, E. (2014). Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 31, 1077–1088. doi: 10.1093/molbev/msu088
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cheng, Y. H., Lin, T. L., Lin, Y. T., and Wang, J. T. (2016). Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 60, 3709–3716. doi: 10.1128/AAC.00009-16
Choi, Y., Lee, J. Y., Lee, H., Park, M., Kang, K., Lim, S. K., et al. (2020). Comparison of fitness cost and virulence in chromosome- and plasmid-mediated colistin-resistant Escherichia coli. Front. Microbiol. 11:798. doi: 10.3389/fmicb.2020.00798
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Clermont, O., Dixit, O. V. A., Vangchhia, B., Condamine, B., Dion, S., Bridier-Nahmias, A., et al. (2019). Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 21, 3107–3117. doi: 10.1111/1462-2920.14713
Clinical and Laboratory Standards Institute, (2020). Performance Standards for Antimicrobial Susceptibility Testing: CLSI Document M100–S30, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Eiamphungporn, W., Yainoy, S., Jumderm, C., Tan-Arsuwongkul, R., Tiengrim, S., and Thamlikitkul, V. (2018). Prevalence of the colistin resistance gene mcr-1 in colistin-resistant Escherichia coli and Klebsiella pneumoniae isolated from humans in Thailand. J. Glob. Antimicrob. Resist. 15, 32–35. doi: 10.1016/j.jgar.2018.06.007
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018). Microorganisms. 7:E461. doi: 10.3390/microorganisms7100461
Feng, Y., Zou, S., Chen, H., Yu, Y., and Ruan, Z. (2020). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 3:gkaa821. doi: 10.1093/nar/gkaa821
Gharaibeh, M. H., and Shatnawi, S. Q. (2019). An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: a review. Vet. World 12, 1735–1746. doi: 10.14202/vetworld.2019
Hadjadj, L., Baron, S. A., Olaitan, A. O., Morand, S., and Rolain, J. M. (2019). Co-occurrence of variants of mcr-3 and mcr-8 Genes in a Klebsiella pneumoniae isolate from Laos. Front. Microbiol. 10:2720. doi: 10.3389/fmicb.2019.02720
Hatrongjit, R., Kerdsin, A., Akeda, Y., and Hamada, S. (2018). Detection of plasmid-mediated colistin-resistant and carbapenem-resistant genes by multiplex PCR. MethodsX 5, 532–536. doi: 10.1016/j.mex.2018.05.016
Huang, H., Dong, N., Shu, L., Lu, J., Sun, Q., Chan, E. W., et al. (2020). Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China, 2014-2019. Emerg. Microbes Infect. 9, 237–245. doi: 10.1080/22221751.2020.1717380
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Kamjumphol, W., Wongboot, W., Suebwongsa, N., Kluabwang, P., Chantaroj, S., and Okada, K. (2019). Draft genome sequence of a colistin-resistant Escherichia coli ST226: a clinical strain harbouring an mcr-1 variant. J. Glob. Antimicrob. Resist. 16, 168–169. doi: 10.1016/j.jgar.2019.01.009
Kananizadeh, P., Oshiro, S., Watanabe, S., Iwata, S., Kuwahara-Arai, K., Shimojima, M., et al. (2020). Emergence of carbapenem-resistant and colistin-susceptible Enterobacter cloacae complex co-harboring blaIMP–1 and mcr-9 in Japan. BMC Infect. Dis. 20:282. doi: 10.1186/s12879-020-05021-7
Kempf, I., Jouy, E., and Chauvin, C. (2016). Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48, 598–606. doi: 10.1016/j.ijantimicag.2016.09.016
Kerdsin, A., Deekae, S., Chayangsu, S., Hatrongjit, R., Chopjitt, P., Takeuchi, D., et al. (2019). Genomic characterization of an emerging blaKPC–2 carrying Enterobacteriaceae clinical isolates in Thailand. Sci. Rep. 9:18521. doi: 10.1038/s41598-019-55008-x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Larsen, M. V., Cosentino, S., Lukjancenko, O., Saputra, D., Rasmussen, S., Hasman, H., et al. (2014). Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539. doi: 10.1128/JCM.02981-13
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Malchione, M. D., Torres, L. M., Hartley, D. M., Koch, M., and Goodman, J. L. (2019). Carbapenem and colistin resistance in Enterobacteriaceae in Southeast Asia: review and mapping of emerging and overlapping challenges. Int. J. Antimicrob. Agents 54, 381–399. doi: 10.1016/j.ijantimicag.2019.07.019
Mediavilla, J. R., Patrawalla, A., Chen, L., Chavda, K. D., Mathema, B., Vinnard, C., et al. (2016). Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM–5, causing a complicated urinary tract infection in a patient from the United States. mBio 7:e01191-16. doi: 10.1128/mBio.01191-16
Meletis, G., and Skoura, L. (2018). Polymyxin resistance mechanisms: from intrinsic resistance to mcr genes. Recent Pat. Antiinfect. Drug Discov. 13, 198–206. doi: 10.2174/1574891X14666181126142704
Mendes, A. C., Novais, Â, Campos, J., Rodrigues, C., Santos, C., Antunes, P., et al. (2018). mcr-1 in carbapenemase-producing Klebsiella pneumoniae with hospitalized patients, Portugal, 2016-2017. Emerg. Infect. Dis. 24, 762–766. doi: 10.3201/eid2404.171787
Nadimpalli, M. L., de Lauzanne, A., Phe, T., Borand, L., Jacobs, J., Fabre, L., et al. (2019). Escherichia coli ST410 among humans and the environment in Southeast Asia. Int. J. Antimicrob. Agents 54, 228–232. doi: 10.1016/j.ijantimicag.2019.05.024
Novović, K., Trudić, A., Brkić, S., Vasiljević, Z., Kojić, M., Medić, D., et al. (2017). Molecular epidemiology of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae in Serbia from 2013 to 2016. Antimicrob. Agents Chemother. 61:e02550-16. doi: 10.1128/AAC.02550-16
Olaitan, A. O., Chabou, S., Okdah, L., Morand, S., and Rolain, J. M. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16:147. doi: 10.1016/S1473-3099(15)00540-X
Olaitan, A. O., Diene, S. M., Kempf, M., Berrazeg, M., Bakour, S., Gupta, S. K., et al. (2014a). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. doi: 10.1016/j.ijantimicag.2014.07.020
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014b). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Palmieri, M., D’Andrea, M. M., Pelegrin, A. C., Mirande, C., Brkic, S., Cirkovic, I., et al. (2020). Genomic epidemiology of carbapenem- and colistin-resistant Klebsiella pneumoniae isolates from Serbia: predominance of ST101 strains carrying a novel OXA-48 plasmid. Front. Microbiol. 11:294. doi: 10.3389/fmicb.2020.00294
Paveenkittiporn, W., Kerdsin, A., Chokngam, S., Bunthi, C., Sangkitporn, S., and Gregory, C. J. (2017). Emergence of plasmid-mediated colistin resistance and New Delhi metallo-β-lactamase genes in extensively drug-resistant Escherichia coli isolated from a patient in Thailand. Diagn. Microbiol. Infect. Dis. 87, 157–159. doi: 10.1016/j.diagmicrobio.2016.11.005
Quan, J., Li, X., Chen, Y., Jiang, Y., Zhou, Z., Zhang, H., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17, 400–410. doi: 10.1016/S1473-3099(16)30528-X
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro. Surveill. 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
Rodrigues, C., Machado, E., Ramos, H., Peixe, L., and Novais, Â (2014). Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: a successful story of international clones (ST15, ST147, ST336) and epidemic plasmids (IncR, IncFIIK). Int. J. Med. Microbiol. 304, 1100–1108. doi: 10.1016/j.ijmm.2014.08.003
Roer, L., Overballe-Petersen, S., Hansen, F., Schønning, K., Wang, M., Røder, B. L., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e00337-18. doi: 10.1128/mSphere.00337-18
Shanmugakani, R. K., Akeda, Y., Sugawara, Y., Laolerd, W., Chaihongsa, N., Sirichot, S., et al. (2019). PCR-dipstick-oriented surveillance and characterization of mcr-1- and carbapenemase-carrying Enterobacteriaceae in a Thai hospital. Front. Microbiol. 10:149. doi: 10.3389/fmicb.2019.00149
Srijan, A., Margulieux, K. R., Ruekit, S., Snesrud, E., Maybank, R., Serichantalergs, O., et al. (2018). Genomic characterization of nonclonal mcr-1-positive multidrug-resistant Klebsiella pneumoniae from clinical samples in Thailand. Microb. Drug Resist. 24, 403–410. doi: 10.1089/mdr.2017.0400
Teo, J. Q., Chang, C. W., Leck, H., Tang, C. Y., Lee, S. J., Cai, Y., et al. (2019). Risk factors and outcomes associated with the isolation of polymyxin B and carbapenem-resistant Enterobacteriaceae spp.: a case-control study. Int. J. Antimicrob. Agents 53, 657–662. doi: 10.1016/j.ijantimicag.2019.03.011
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. doi: 10.1038/s41426-018-0124-z
Wick, R. R., Heinz, E., Holt, K. E., and Wyres, K. L. (2018). Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 56:e00197-18. doi: 10.1128/JCM.00197-18
World Health Organization, (2012). Critically Important Antimicrobials for Human Medicine, 3rd Revision. Geneva: WHO.
World Health Organization, (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and deveLopment of New Antibiotics. Geneva: WHO.
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/AAC.04037-14
Wu, R., Yi, L. X., Yu, L. F., Wang, J., Liu, Y., Chen, X., et al. (2018). Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 9:331. doi: 10.3389/fmicb.2018.00331
Yang, Y. Q., Li, Y. X., Lei, C. W., Zhang, A. Y., and Wang, H. N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1791–1795. doi: 10.1093/jac/dky111
Yuan, Y., Li, Y., Wang, G., Li, C., Xiang, L., She, J., et al. (2019). Coproduction of mcr-9 and NDM-1 by colistin-resistant Enterobacter hormaechei isolated from bloodstream infection. Infect. Drug Resist. 12, 2979–2985. doi: 10.2147/IDR.S217168
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhong, L. L., Phan, H. T. T., Shen, C., Vihta, K. D., Sheppard, A. E., Huang, X., et al. (2018). High rates of human fecal carriage of mcr-1-positive multidrug-resistant Enterobacteriaceae emerge in China in association with successful plasmid families. Clin. Infect. Dis. 66, 676–685. doi: 10.1093/cid/cix885
Keywords: mcr, carbapenem-resistant Enterobacterales, Thailand, colistin, genome
Citation: Paveenkittiporn W, Kamjumphol W, Ungcharoen R and Kerdsin A (2021) Whole-Genome Sequencing of Clinically Isolated Carbapenem-Resistant Enterobacterales Harboring mcr Genes in Thailand, 2016–2019. Front. Microbiol. 11:586368. doi: 10.3389/fmicb.2020.586368
Received: 23 July 2020; Accepted: 16 November 2020;
Published: 11 January 2021.
Edited by:
Shaolin Wang, China Agricultural University, ChinaReviewed by:
Mehmet Demirci, Beykent University, TurkeyCopyright © 2021 Paveenkittiporn, Kamjumphol, Ungcharoen and Kerdsin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anusak Kerdsin, QW51c2FrLmtlQGt1LnRo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.