
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 03 December 2020
Sec. Biology of Archaea
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.586025
This article is part of the Research TopicProceedings of the International Workshop on Geo-Omics of ArchaeaView all 22 articles
DNA damage response (DDR) in eukaryotes is largely regulated by protein phosphorylation. In archaea, many proteins are phosphorylated, however, it is unclear how the cells respond to DNA damage through global protein phosphorylation. We previously found that Δrio1, a Rio1 kinase homolog deletion strain of Sulfolobus islandicus REY15A, was sensitive to UV irradiation. In this study, we showed that Δrio1 grew faster than the wild type. Quantitative phosphoproteomic analysis of the wild type and Δrio1, untreated and irradiated with UV irradiation, revealed 562 phosphorylated sites (with a Ser/Thr/Tyr ratio of 65.3%/23.8%/10.9%) of 333 proteins in total. The phosphorylation levels of 35 sites of 30 proteins changed with >1.3-fold in the wild type strain upon UV irradiation. Interestingly, more than half of the UV-induced changes in the wild type did not occur in the Δrio1 strain, which were mainly associated with proteins synthesis and turnover. In addition, a protein kinase and several transcriptional regulators were differentially phosphorylated after UV treatment, and some of the changes were dependent on Rio1. Finally, many proteins involved in various cellular metabolisms exhibited Riol-related and UV-independent phosphorylation changes. Our results suggest that Rio1 is involved in the regulation of protein recycling and signal transduction in response to UV irradiation, and plays regulatory roles in multiple cellular processes in S. islandicus.
Protein post-translational modifications (PTMs) are rapid regulatory mechanisms of cells in response to various stresses. In eukaryotes, protein phosphorylation, one of the most important PTMs, mediates multiple cellular processes and signal cascades, including DNA damage response (DDR) and DNA repair (Sirbu and Cortez, 2013; Jette and Lees-Miller, 2015; Blackford and Jackson, 2017).
Transcriptomic studies shown that multiple genes exhibited transcriptional changes in several archaea after UV or γ-irradiation, including the up-regulation of genes involved in DNA repair, and the down-regulation of genes involved in DNA replication and cell division (McCready et al., 2005; Frols et al., 2007; Gotz et al., 2007; Williams et al., 2007). Phosphoproteomic analyses showed that a number of archaeal DDR and DNA repair proteins were regulated by phosphorylation (Aivaliotis et al., 2009; Esser et al., 2012; Reimann et al., 2013). Importantly, some kinases were induced or repressed by DNA damages. For example, Saccharolobus solfataricus Rio1, an atypical protein kinase, was transcriptionally induced at the early stage (30 min) of UV-treatment and the mRNA level of Rio1 increased in γ-irradiated Pyrococcus furiosus cells (Gotz et al., 2007; Williams et al., 2007). Another study showed that a typical (Hanks-type) protein kinase (homolog of SiRe_2030) was repressed in UV-treated S. solfataricus (Frols et al., 2007). In addition, two regulators, Orc1-2 and TFB3, were found to be phosphorylated in S. solfataricus and Sulfolobus acidocaldarius (Esser et al., 2012; Reimann et al., 2013). These observations suggest that protein phosphorylation plays an important role in DDR and/or DNA repair. However, a detailed mechanism of how these processes are regulated is still unknown.
Rio kinases are found in all three domains of life. In eukaryotes, Rio kinases participate in ribosome biogenesis, cell cycle progression, and genome integrity (Angermayr et al., 2002; Ferreira-Cerca et al., 2012; Widmann et al., 2012). Importantly, human RIO1 is overexpressed in different tumor types and promotes tumor growth (Weinberg et al., 2017). Recently, yeast Rio1 interactome containing more than 100 putative interactors has been mapped (Iacovella et al., 2018). Yeast Rio1 was able to regulate the transcription of a number of genes by binding to their promoter regions (Iacovella et al., 2018), indicating that Rio1 plays a global regulatory role in the cell. Although archaeal Rios was suggested to have a conserved role in ribosome biogenesis (Knuppel et al., 2018), their function has not been established thus far. Our previous study revealed that Rio1 phosphorylated the Holliday junction resolvase Hjc in S. islandicus and regulated its nuclease activity (Huang et al., 2019). However, it remains unclear what other role(s) archaeal Rio1 plays in DNA damage-induced regulatory networks. And above this, the effects of DNA damage on the entire phosphoproteome have not been reported in archaea.
Mass spectrometry-based phosphoproteomics is a very powerful tool for identifying and quantifying protein phosphorylation, and has been widely applied in the investigation of eukaryotic DDR and DNA repair (Ajon et al., 2011; Makarova et al., 2015; van Wolferen et al., 2016; Cao et al., 2019). In this study, we performed quantitative phosphoproteomic analysis on S. islandicus REY15A and rio1 deletion strain treated or untreated with UV. We found that both UV-treatment and rio1 deletion resulted in phosphorylation changes of proteins in various cellular processes. Moreover, we found that the phosphorylation changes in Δrio1 were much fewer than those in E233S after UV irradiation, most of which were related to protein metabolism. The UV-independent phosphorylation regulated by Rio1 was also analyzed, indicating that phosphorylation changes of many proteins participate in various cellular metabolic processes, reminiscent of those in yeast.
Sulfolobus islandicus strain REY15A (E233S) (ΔpyrEFΔlacS) (hereafter E233S) and its transformants were cultured as described previously (Deng et al., 2009) with slight modification. In particular, yeast extract was not added to the growth medium. D-arabinose (0.2% w/v) was used for the induction of protein overexpression. Gelrite (0.8% w/v) was used for making culture plates.
Assay of the sensitivity of strains to UV irradiation on plates was performed as described previously (Huang et al., 2019). For UV-treatment of cells in liquid medium, a 30 mL aliquot at an OD600 ∼ 0.2–0.3 was transferred to a petri dish (9 cm in diameter). The cells were irradiated with UV at 100 or 200 J/m2 (Frols et al., 2007) in a CL-1000 UV crosslinker (UVP Bio-imaging systems, CA, United States) or mock-treated, and transferred back to the culture flasks. The cells were then incubated at 75°C in the dark. The OD600 values were measured every 6 or 12 h. Growth curves were drawn based on data from three biological repeats.
The wild type and Δrio1 strains were cultivated at an initial OD600 ∼ 0.04. After growing overnight, the samples (OD600 ∼ 0.2) were taken and observed under a Nikon Eclipse80i Microscope (Nikon corporation, Japan). The sizes of the cells (in diameter) were measured with software NIS-Elements AR 3.1. Samples at different times from early to late log phase were also taken and diluted by 10,000-fold, and 2–10 μl of the diluted samples were plated in triplicate. The plates were incubated at 75°C for 7–10 days before colony counting.
A volume of 250 mL of the wild type or Δrio1 (Δsire_0171) strain was cultivated to OD600 ∼ 0.28. A 50 mL aliquot of each culture was then transferred to petri dish (15 cm in diameter) and put into a CL-1000 Ultraviolet crosslinker. The cells were irradiated with UV at 200 J/m2 or mock-treated and then transferred back to a culture flask. The cells were then incubated at 75°C for 30 min with shaking at 110 rpm in the dark before collected by centrifugation at 6,245 g for 10 min and washed with PBS (phosphate-buffered saline) buffer three times. The cell pellets were stored at −80°C before phosphoproteomics analysis. Each sample was prepared in duplicate. The sample treatment and proteomic and bioinformatic analysis were conducted by BPI (Beijing Protein Innovation Co., Ltd, Beijing, China).
Samples were first ground in liquid nitrogen into a powder and then transferred to a 5 ml centrifuge tube. After that, four volumes (500 μl) of lysis buffer (10 mM dithiothreitol, 1% protease inhibitor cocktail (Merck, Darmstadt, Germany), and 1% phosphatase inhibitor) was added to the cell powder, followed by sonication three times on ice using a high intensity ultrasonic processor (Scientz, Ningbo Xinzhi Ultra-Sonic Technology Co., LTD., Zhejiang, China). An equal volume of Tris-saturated phenol (pH 8.0) was then added, and the mixture was further vortexed for 5 min. After centrifugation (4°C, 5,500 g, 10 min), the upper layer of the phenol phase was transferred to a new centrifuge tube. Proteins were precipitated by adding five volumes (2 mL) of ammonium acetate-saturated methanol, and the samples were then incubated at −20°C overnight. After centrifugation at 5,500 g for 10 min at 4°C, the supernatant was discarded. The remaining precipitate was washed with ice-cold methanol once, followed by ice-cold acetone three times. The proteins were re-dissolved in 8M urea, and the protein concentration was determined with a BCA (Bicinchoninic acid) Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, United States) according to the manufacturer’s instructions.
For digestion, the protein solutions were reduced with 5 mM dithiothreitol for 30 min at 56°C and then alkylated with 11 mM iodoacetamide for 15 min at room temperature in the dark. The urea concentration of the protein sample was then diluted by adding 100 mM triethylamonium bicarbonate (TEAB) to less than 2 M. Finally, trypsin was added at 1:50 trypsin to protein mass ratio for the first digestion at 37°C overnight and a 1:100 trypsin to protein mass ratio for a second 4 h digestion.
After trypsin digestion, peptides were desalted using a Strata X C18 SPE column (Phenomenex, Torrance, CA, United States) and vacuum dried. Peptides were then resuspended in 0.5 M TEAB and processed according to the manufacturer’s protocol for the TMT kit. Briefly, one unit of TMT reagent was thawed and dissolved in 100% acetonitrile. The peptide mixtures were then incubated at room temperature for 2 h and pooled, desalted, and dried by vacuum centrifugation using an Eppendorf Concentrator plus.
The tryptic peptides were then fractionated using high pH reverse-phase HPLC on a Thermo Agilent 300Extend C18 column (5 μm particles, 4.6 mm inner diameter, 250 mm length). Briefly, peptides were first separated with a gradient of 8−32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Then, the peptides were combined into four fractions and dried by vacuum centrifugation for 6 h.
Tryptic peptide mixtures were incubated with an IMAC (immobilized metal affinity chromatography) microsphere suspension with vibration in loading buffer (50% acetonitrile/6% trifluoroacetic acid) and enriched phosphopeptides were then collected by centrifugation. To remove non-specifically adsorbed peptides, the IMAC microspheres were sequentially washed with 50% acetonitrile/6% trifluoroacetic acid and 30% acetonitrile/0.1% trifluoroacetic acid. Elution buffer containing 10% NH4OH was added, and the enriched phosphopeptides were eluted from the IMAC microspheres by vibration. The supernatant containing phosphopeptides was collected and lyophilized by vacuum centrifugation. The samples were desalted according to the instruction of the C18 ZipTips column for LC-MS/MS analysis.
Tryptic peptides were dissolved in 0.1% formic acid (solvent A) and then directly loaded on a homemade reverse-phase analytical column (15 cm length, 75 μm inner diameter). Solvent A contained 0.1% formic acid in 2% acetonitrile. The gradient comprised a range of 4–22% solvent B (0.1% formic acid in 90% acetonitrile) over 40 min, 23–35% over 8 min, climbing to 80% in 4 min then holding at 80% for the last 4 min, all at a constant flow rate of 400 nL/min on an EASY-nLC 1000 UPLC system.
These peptides were subjected to an NSI source followed by tandem mass spectrometry (MS/MS) on a Q ExactiveTM Plus (Thermo Fisher Scientific, Waltham, MA, United States) coupled inline to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350−1800 for the full scan, and intact peptides were detected by the Orbitrap at a resolution of 70,000. The fixed first mass was set as 100 m/z. Peptides were selected for MS/MS using an NCE setting of 28, and the fragments were detected on the Orbitrap at a resolution of 35,000. A data-dependent procedure that alternated between one MS scan followed by 10 MS/MS scans with 30.0 s dynamic exclusion was then applied. The automatic gain control (AGC) was set to 1E5. The signal threshold was set as 20,000 ions/s, and the maximal injection time was set as 100 ms.
The resulting MS/MS data were processed using the Maxquant search engine (v.1.5.2.8). Tandem mass spectra were then searched against the Sulfolobus uniprot database (Uniprot_Sulfolobus_islandicus_REY15A, 3993 sequences). A reverse decoy database was concatenated for calculation of the false positive rate (FPR). Trypsin/P was specified as the cleavage enzyme allowing up to 2 missing cleavages. The minimum length of peptides was set to 7 aa, and the maximum number of modification sites was set to 5. The mass tolerance for precursor ions was set to 20 ppm in the first search and 5 ppm in the main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl on cystine was specified as a fixed modification. Phosphorylation on Ser/Thr/Tyr was specified as variable modifications. The quantitative method was set as TMT-10plex. Protein identification and the FDR of the identified peptide spectrum match (PSM) were adjusted to <1%. The localization probability of site modifications was filtered with a value of >0.75.
The Gene Ontology (GO) annotation proteome was derived from the UniProt-GOA database1. First, the identified protein IDs were converted to UniProt IDs and then mapped to GO IDs. If some identified proteins were not annotated by UniProt-GOA database, InterProScan software was used to annotate the protein’s GO function based on the protein sequence alignment method. Then proteins were classified by Gene Ontology annotation based on three categories: biological process, cellular component, and molecular function. Functional descriptions of the identified protein domains were annotated by InterProScan v.5.14-53.02 based on the protein sequence alignment method, and the InterPro domain database was used. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to annotate protein pathways in this dataset. The KEGG online service tool KAAS was used to annotate each protein’s KEGG database description. Then the annotation results were mapped on the KEGG pathway database using the KEGG online service tools KEGG mapper.
Soft MoMo (motif-x algorithm) was used to search for a motif of sequences derived from amino acids in specific positions of modified-13-mers (6 amino acids upstream and downstream of a site) in all protein sequences. All the database protein sequences were used as the background database parameter. The minimum number of occurrences was set to 20. The emulate original motif-x option was set, and all other parameters were set to the default.
To perform enrichment analysis of GO, KEGG, and protein domain for each category, a two-tailed Fisher’s exact test was employed to test the enrichment of each differentially modified protein against all identified proteins. The GO/pathway/protein domains with a corrected p-value < 0.05 were considered significant.
Sulfolobus islandicus FHA and vWAs were co-expressed in E. coli using the pRSFDuet-1 plasmid (Novagen). The genes encoding FHA and vWAs were amplified with corresponding primers (Supplementary Table S1) and inserted into the NdeI/XhoI and SacI (or BamHI for the vWA2 gene)/SalI, respectively. For co-expression of FHA and vWAs in S. islandicus, the arabinose promoter was cloned into the original multiple cloning site (MCS) of pSeSD (Peng et al., 2012). The gene fragments of Flag-vWAs and His-FHA were amplified and inserted into the XhoI/NheI in MCS1 and NdeI/SalI in MCS2, respectively, of the modified pSeSD, yielding pSeSD-Flag-vWA1-His-FHA and pSeSD-Flag-vWA2-His-FHA.
The vector pET15b was applied for the construction of plasmids expressing N-terminal His-tagged Sis7d (SiRe_0668) and SMC-like protein (SiRe_0508) in E. coli. The genes were amplified using their corresponding primers (Supplementary Table S1). The PCR products were digested with NdeI/SalI and ligated into the same restriction sites in pET15b. The phosphor-mimic mutants, Sis7d-T41E/S47E and SiRe_0508-Y263E/Y266E were constructed by SOE PCR and inserted into the NdeI/SalI sites of pET15b.
The plasmids for protein expression were transformed into E. coli BL21-Codonplus-RIL. E. coli harboring the pET15b plasmids was cultivated in 1 L of LB medium containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) (LAC). Proteins were induced by addition of IPTG and incubation at 37°C for 4 h. The cells were collected by centrifugation at 10,000 g for 3 min and disrupted by sonication in lysis buffer (50 mM Tris-HCl pH 8.0, 200 mM NaCl, 5% glycerol). The proteins were purified with Ni-NTA agarose (Invitrogen) column, pooled and frozen by liquid nitrogen for storage at −80°C.
To detect the interaction between S. islandicus FHA and vWAs in E. coli, pRSFDuet-1 based co-expression plasmids were transformed into E. coli BL21-Codonplus-RIL. Single transformants were picked into 30 ml LAC medium and subsequently inoculated in 500 ml LAC medium. His-tagged FHA and Flag-tagged-vWAs were induced as described above and purified by anti-Flag beads (Sigma) according the standard protocol. Briefly, E. coli cells were resuspended in 40 ml buffer A (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% glycerol) and cell extract was obtained by sonication. The supernatant of heat-treated cell extract was incubated with anti-Flag beads pre-equilibrated with buffer A at 25°C for 1 h. The beads were washed with 9 ml of buffer A and the target proteins were eluted with 3 ml of 0.1 M Gly-HCl buffer. The eluted samples were analyzed by Western blot using anti-His antibody to detect the presence or absence of His-vWAs. pRSFDuet-1 containing the single gene (either FHA or vWA) was used as negative control. Similar procedure was followed for the pull down assay of the interaction between FHA and vWAs.
To analyze the interaction between FHA and vWAs in vivo, pSeSD based co-expression plasmids were transformed into S. islandicus. One hundreds milliliters of S. islandicus cells harboring the co-expression plasmid were cultivated to OD600∼0.2 and the protein expression was induced with 0.2% D-arabinose for 12 h. The cells were collected and resuspended in 10 ml lysis buffer and cell extract was obtained by sonication. Soluble His-tagged FHA was purified with Ni-NTA agarose column and eluted with 2 ml lysis buffer containing 250 mM imidazole. The eluted samples were analyzed by Western blot using anti-Flag antibody to detect the presence or absence of Flag-vWAs. Strains harboring the expression plasmids for single gene, pSeSD-His-FHA, pSeSD-Flag-vWA1, or pSeSD-Flag-vWA2, were used as negative controls. Similar procedure was followed for the pull down assay of the interaction between FHA and vWAs from S. islandicus REY15A.
Electrophoretic mobility shift assay was performed in a 20 μl reaction mixture containing 25 mM Tris-HCl pH 8.0, 25 mM NaCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, FAM-labeled dsDNA (500 bp), and various concentrations of the target protein. The mixture was incubated at 37°C for 30 min and analyzed in 8% native PAGE. The gel was scanned by TyphoonTM 162 FLA 7000 image analyzer (GE Healthcare).
Previous transcriptomic analysis in S. solfataricus revealed that rio1, a kinase gene, was up-regulated by 2.8-fold upon UV irradiation (Gotz et al., 2007), and the rio1 deletion strain (Δrio1) of S. islandicus exhibited higher sensitivity to UV irradiation than the wild type strain E233S (Huang et al., 2019). Here, we confirmed that Δrio1 was more sensitive to a relatively high dosage of UV irradiation than E233S. In liquid medium, both E233S and Δrio1 were sensitive to UV irradiation, but Δrio1 was slightly more sensitive to UV irradiation at 200 J/m2 than E233S (Supplementary Figure S1A). In the spotting assay, Δrio1 grew almost the same as the wild type after UV irradiation at 15 J/m2 on plates, but it grew more slowly than the wild type at 25 J/m2 UV (Supplementary Figure S1B). Interestingly, in liquid culture Δrio1 grew faster (5.3 h/generation) and reached a higher density than E233S (6.3 h/generation) with or without UV treatment (Figure 1A and Supplementary Figure S1A). The mean cell size of Δrio1 (1.21 ± 0.15 μm) was comparable to that of the wild type (1.38 ± 0.22 μm) (Figure 1B). Colony formation assay revealed that Δrio1 contained more live cells than the wild type (Figure 1C). The results indicate that Rio1 negatively regulates cell growth under normal growth conditions.
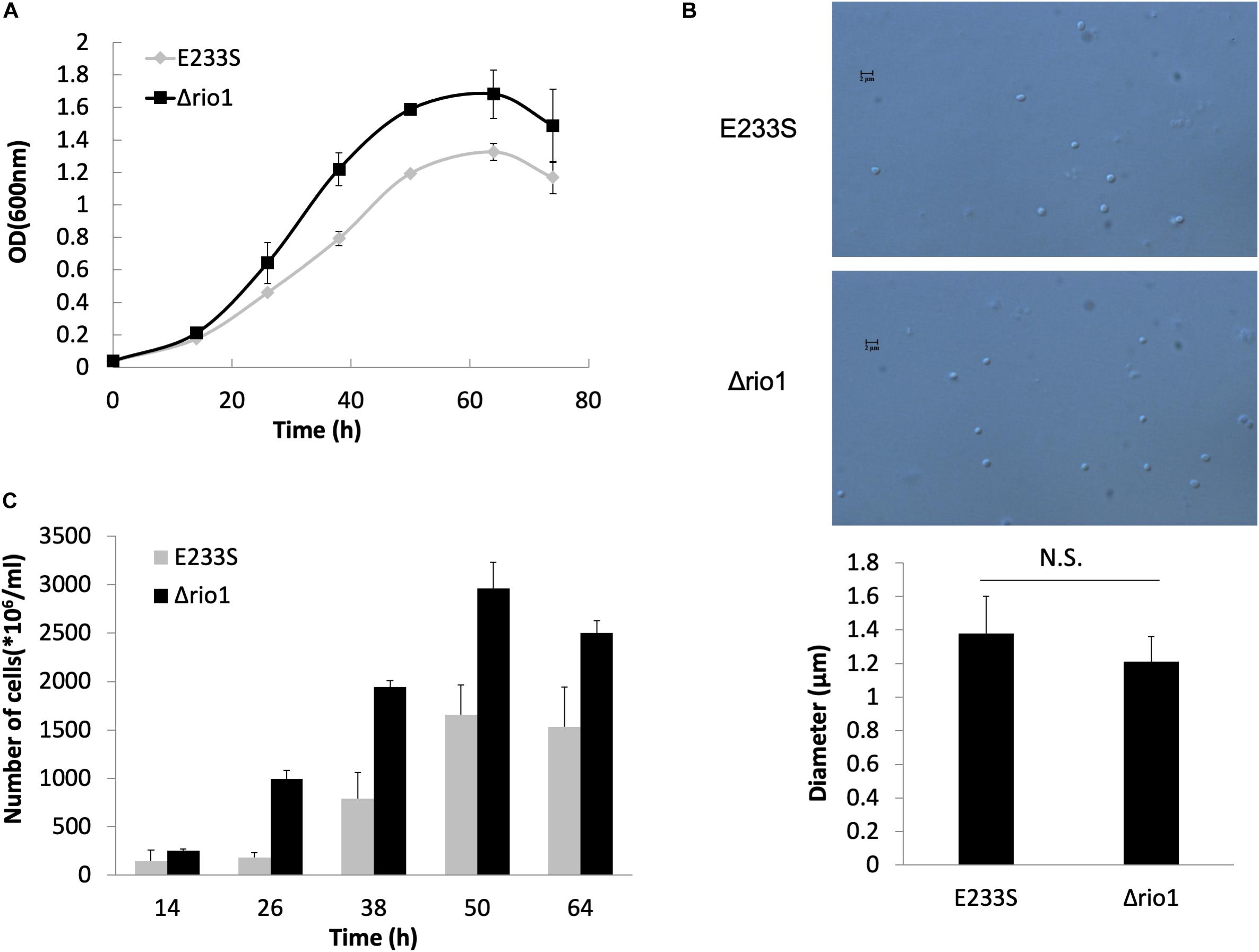
Figure 1. Δrio1 grew faster than the wild type strain E233S. (A) Growth curves of Δrio1 and E233S. The cultures were transferred to fresh medium at a same initial OD600 value. Samples were taken at an interval of 6 or 12 h for measuring ODs until stationary phase. (B) Cell size of Δrio1 and E233S. The samples at early log phase (OD600 ∼ 0.2) were used for microscopy. Diameters of 190–200 cells were measured for each strain and the averages are shown at the bottom. Representative photos are shown. Scale bar (2 μm) is indicated. N.S., non-significant. (C) Colony formation assay of Δrio1 and E233S. Samples of the two strains were taken at same time point during their growth and diluted for plating. Colonies were counted after 7–10 days incubation at 75°C.
To reveal the phosphorylation regulatory network in DDR and Rio1-mediated processes in S. islandicus, we performed quantitative phosphoproteomic analysis on both UV-treated and untreated wild type strain E233S and Δrio1. A schematic of the experimental design is shown in Figure 2. E233S was untreated or treated with UV to investigate the UV phospho-signaling changes. The samples of untreated and UV-treated Δrio1 were compared to determine whether the UV-induced differential phosphorylation events observed in wild type cells were a result of Rio1 signaling. Comparison between E233S and Δrio1 was performed to reveal Rio1-mediated phosphorylation changes. In a previous transcriptomic analysis (Gotz et al., 2007), S. solfataricus (a close relative of S. islandicus) cells were treated with 200 J/m2 UV. Therefore, the same dosage of UV was applied in the analysis in this study. The samples were then incubated for 30 min in the dark after UV irradiation before collection for quantitative phosphoproteomic analysis by TMT labeling. Two biological replicates in the early exponential growth phase (OD600 of 0.28) were analyzed for each sample, and relative standard deviation (RSD) and hierarchical clustering analyses of all differential phosphorylation evens (>1.3-fold, see bellow, Cao et al., 2019) from five comparison data confirmed that these measurements were reliable (Supplementary Figures S2A,B).
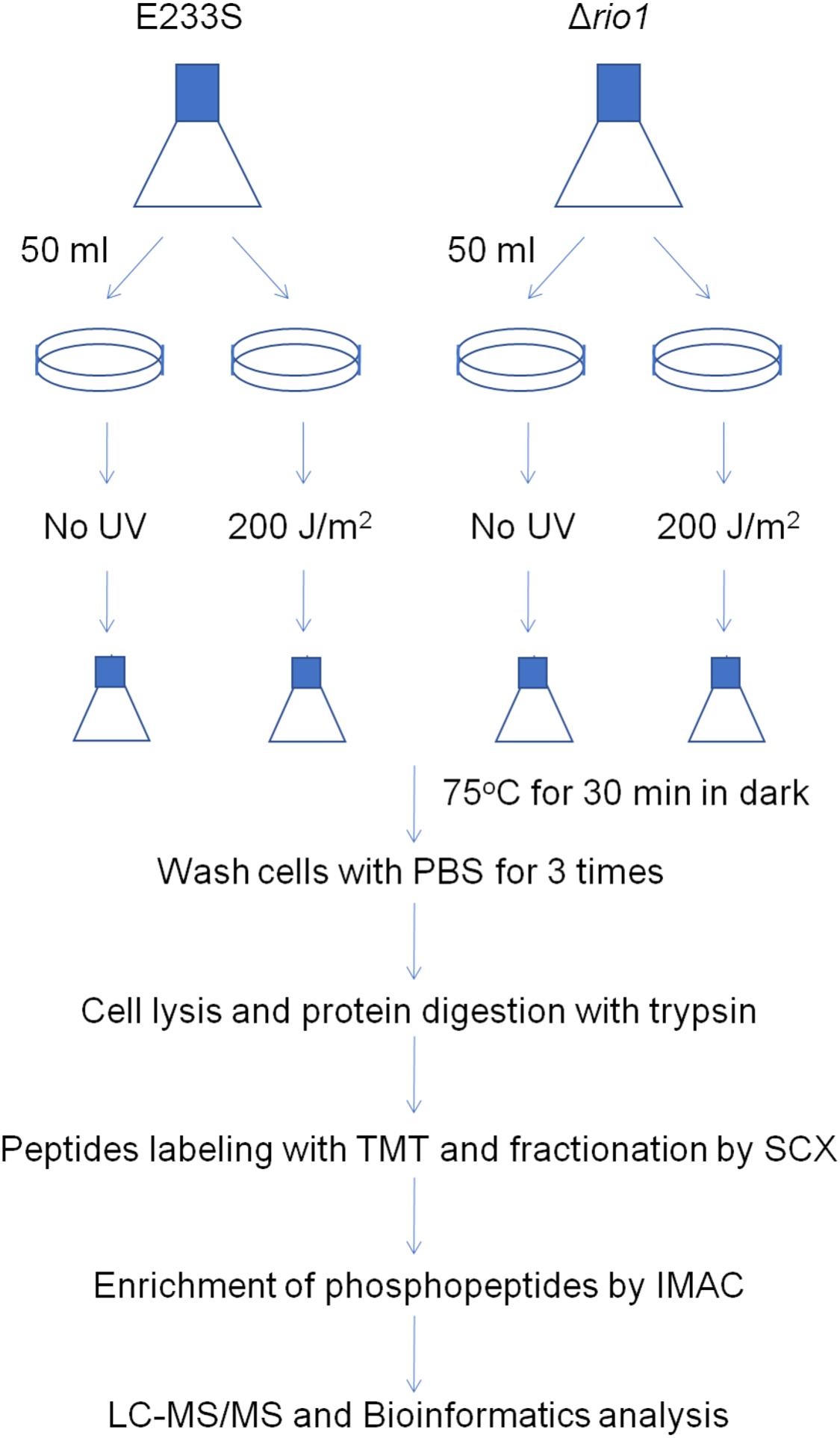
Figure 2. Schematic of sample preparation for phosphoproteomics analysis. A volume of 250 mL of E233S and Δrio1 strain was cultivated to OD600 ∼ 0.28. Aliquots of 50 mL were transferred to petri dishes (15 cm in diameter), which were subsequently put in a CL-1000 Ultraviolet crosslinker. Cells were irradiated with 200 J/m2 UV or mock-treated and transferred back to a culture flask. The flasks were incubated at 75°C for 30 min in the dark. Cells were collected by centrifugation at 6,245 g for 10 min and washed with PBS buffer three times. The cell pellets were ground in liquid nitrogen, and total proteins were digested with trypsin. The peptides were labeled with TMT and fractionated by SCX. The phosphopeptides were enriched by IMAC and subjected to LC-MS/MS and bioinformatics analysis. Two repeats were used for each treatment.
Overall, 1357 peptides were identified, containing 672 phosphorylated sites from 352 proteins. Among these phosphorylated sites, 562 phosphorylated sites from 333 proteins were identified with localization probabilities > 0.75, and 448 unique phosphorylated sites from 284 proteins were further quantified (Supplementary Table S1). A previous phosphoproteomic analysis of S. solfataricus upon change of carbon source identified 690 phosphorylated sites from 540 proteins (Esser et al., 2012), while another phosphoproteomic analysis of two protein phosphatase deletion strains (Δpp2a and Δptp) in S. acidocaldarius showed that numbers of phosphorylated sites increased compared with those from a wild type strain (from 54 to 477 and 715, respectively) (Reimann et al., 2013). Our data are comparable to those from these reports. Among the proteins identified in our study, more than 61% of the phosphorylated proteins contained only one phosphorylated site, while the proteins with two and three phosphorylated sites were 19.0 and 10.2%, respectively. Fewer than 10% of the phosphorylated proteins contained more than three phosphorylated sites (Supplementary Figure S3). Specifically, there were 13 proteins containing five or more phosphorylated sites, indicating those proteins were intensively regulated by phosphorylation. These proteins included nucleoside diphosphate kinase, aspartokinase, phosphoglucosamine mutase, phosphoenolpyruvate synthase, alcohol dehydrogenase GroES domain protein, NADP-dependent isocitrate dehydrogenase, DNA/RNA-binding protein Alba, 30S ribosomal protein S5, translation elongation factor 1α, thermosomes (SiRe_1214 and SiRe_1716), and von Willebrand type A (vWA) domain proteins (SiRe_1008 and SiRe_1011) (Supplementary Table S1). All the identified phosphorylated proteins were classified according to archaeal clusters of orthologous groups (arCOG) functional categories (Makarova et al., 2015). Similar to the previous phosphoproteomic analyses of Sulfolobus species (Esser et al., 2012; Reimann et al., 2013), the phosphorylated proteins belonged to most arCOG categories (19 out of 26 arCOG), confirming that protein phosphorylation plays a global regulatory role in archaeal cells (Supplementary Figure S4).
We found that the ratios of phosphorylated Ser, Thr, and Tyr were 65.3, 23.8, and 10.9%, respectively. The ratio of phosphorylated Tyr in our data was lower than the two previous phosphoproteomic analyses (53.6% in wild type S. solfataricus and 36.2−45.7% in the wild type, Δptp, or Δpp2a of S. acidocaldarius) (Esser et al., 2012; Reimann et al., 2013). These differences may be due to the different phases of the cells used for analysis. A culture in the early log phase (OD600 ∼ 0.28) was used in this study, while in the two previous reports, cells of late exponential growth phase (OD600 ∼ 0.7 − 0.8) were used. It was reported that proteins carrying phosphorylated Tyr residues were significantly more abundant in Hela cells (Sharma et al., 2014). The lower OD600 of the samples in this study might have an effect on protein profiles (probably low abundant proteins), while cultures with a higher OD600 contained more abundant proteins that might have had more phosphorylated Tyr sites. Moreover, the relatively low level of Tyr phosphorylation could have been caused by the Tyr kinase activity versus phosphor-tyrosine phosphatase (PTP) activity (Hunter, 2009). It seems that most Tyr kinases in eukaryotes are only activated in specific circumstances for phosphorylation, whereas phosphorylated Tyr residues usually have a short half-life due to the high activity of PTPs unless they have been protected by binding to SH2 or PTP domain proteins (Sadowski et al., 1986). Thus far, it is unclear whether archaeal Tyr kinases are activated under specific conditions. However, the activity of S. acidocaldarius PTP was much higher than that of PP2A (Reimann et al., 2013), indicating that the half-life of Tyr phosphorylation may be too short to be significantly detected in certain stages of Sulfolobus cells.
For quantification of phosphorylation changes, those with more than 1.3-fold differential phosphorylation in both biological replicates were selected for detailed analysis. The use of >1.3-fold cutoff was because phosphorylated sites with >1.3-fold change comprised 5–10% of the total phosphorylated sites detected and this threshold was used in several proteomics studies in archaea (e.g., Cao et al., 2019). The phosphorylation changes might reflect the increase or decrease of steady state protein in phosphorylated state. However, the methodology used in this study could not exclude that variation in synthesis rates and/or degradation rates of proteins also lead to phosphorylation changes.
To identify UV-dependent protein/site phosphorylation, the phosphorylated proteins and sites were compared between UV-treated E233S and untreated E233S. The phosphorylation of 15 sites on 14 proteins increased after UV-treatment, while the phosphorylation of 20 sites on 18 proteins decreased (Figure 3). These proteins were mainly associated with cellular metabolism (arCOG C, E, G, H); translation, ribosomal structure, and biogenesis (arCOG J); and posttranslational modification, protein turnover, and chaperones (arCOG O) (Figure 4A). This indicated that the processes of both cellular metabolism and protein synthesis changed (probably inhibited for preserving energy after DNA damage) by phosphorylation and PTMs were induced on specific proteins in response to UV irradiation.
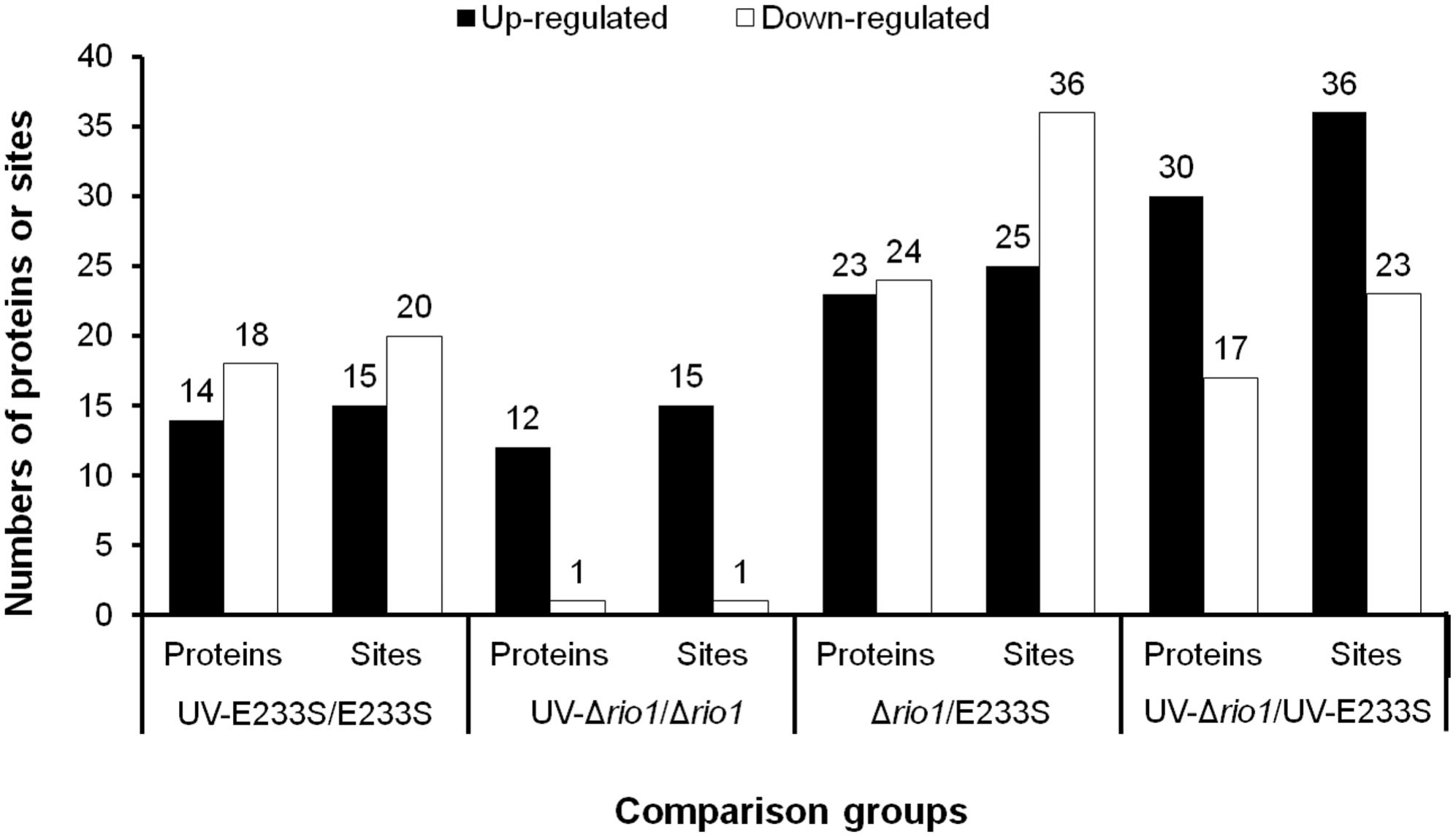
Figure 3. Statistics of differentially phosphorylated protein sites in various comparison groups. The numbers of phosphorylated proteins (sites) with a threshold of >1.3-fold are summarized here. The increased (up-) and decreased (down-) amount of phosphorylated proteins and sites are shown in filled and empty bars, respectively.
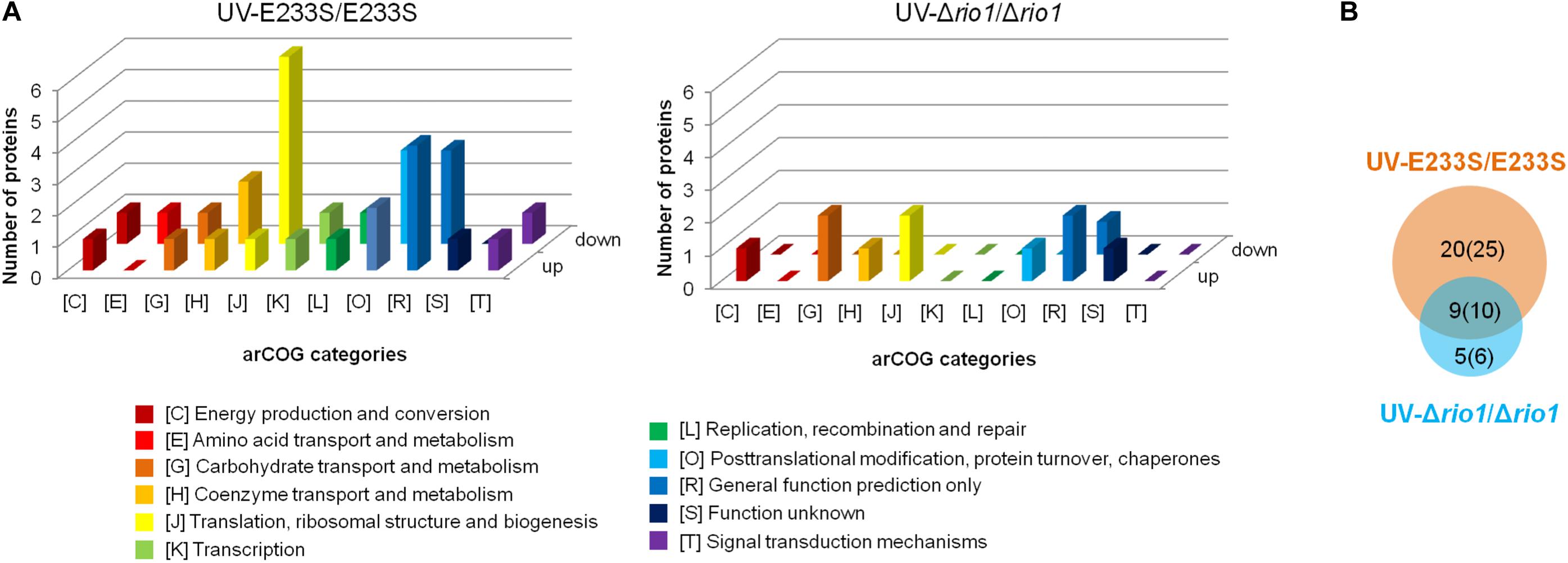
Figure 4. Analysis of differentially phosphorylated proteins in a comparison of UV-treated E233S versus E233S and UV-treated Δrio1 versus Δrio1. (A) COG analysis of differentially phosphorylated proteins in UV-treated E233S versus E233S (left panel) and UV-treated Δrio1 versus Δrio1 (right panel). Both increased (up) and decreased (down) phosphorylated proteins are shown. The categories of arCOG are indicated at the bottom. (B) Overlap analysis of the numbers of differentially phosphorylated proteins (sites) between UV-treated E233S versus E233S (orange) and UV-treated Δrio1 versus Δrio1 (cyan). The numbers in brackets are those of phosphorylated sites.
Interestingly, two and three differentially phosphorylated sites were identified in the Hanks-type protein kinase (SiRe_2056, ePK1) and vWA domain-containing protein SiRe_1011 (vWA2), respectively. The phosphorylation level of ePK1 at T314 decreased by 0.59-fold, whereas that on T592 increased 2.0-fold. The phosphorylation of three residues of vWA2, S304, T314, and T327 decreased by 0.64, 0.68, and 0.74-fold, respectively (Table 1 and Supplementary Table S2). The phosphorylation of different protein sites may correlate with various regulatory functions. Protein kinases would transfer signals and vWA domain proteins may mediate protein-protein interaction (Whittaker and Hynes, 2002). Strikingly, decreased phosphorylation of ePK1 at T314 and vWA2 at S304, T314, and T327 also occurred when rio1 was deleted (Table 1). Moreover, phosphorylation of another vWA domain-containing protein SiRe_1008 (vWA1) at T353 decreased in both UV-treated E233S cells and UV-treated Δrio1, indicative of UV-dependent but Rio1-independent manner (Table 1 and Supplementary Tables S2, S3).
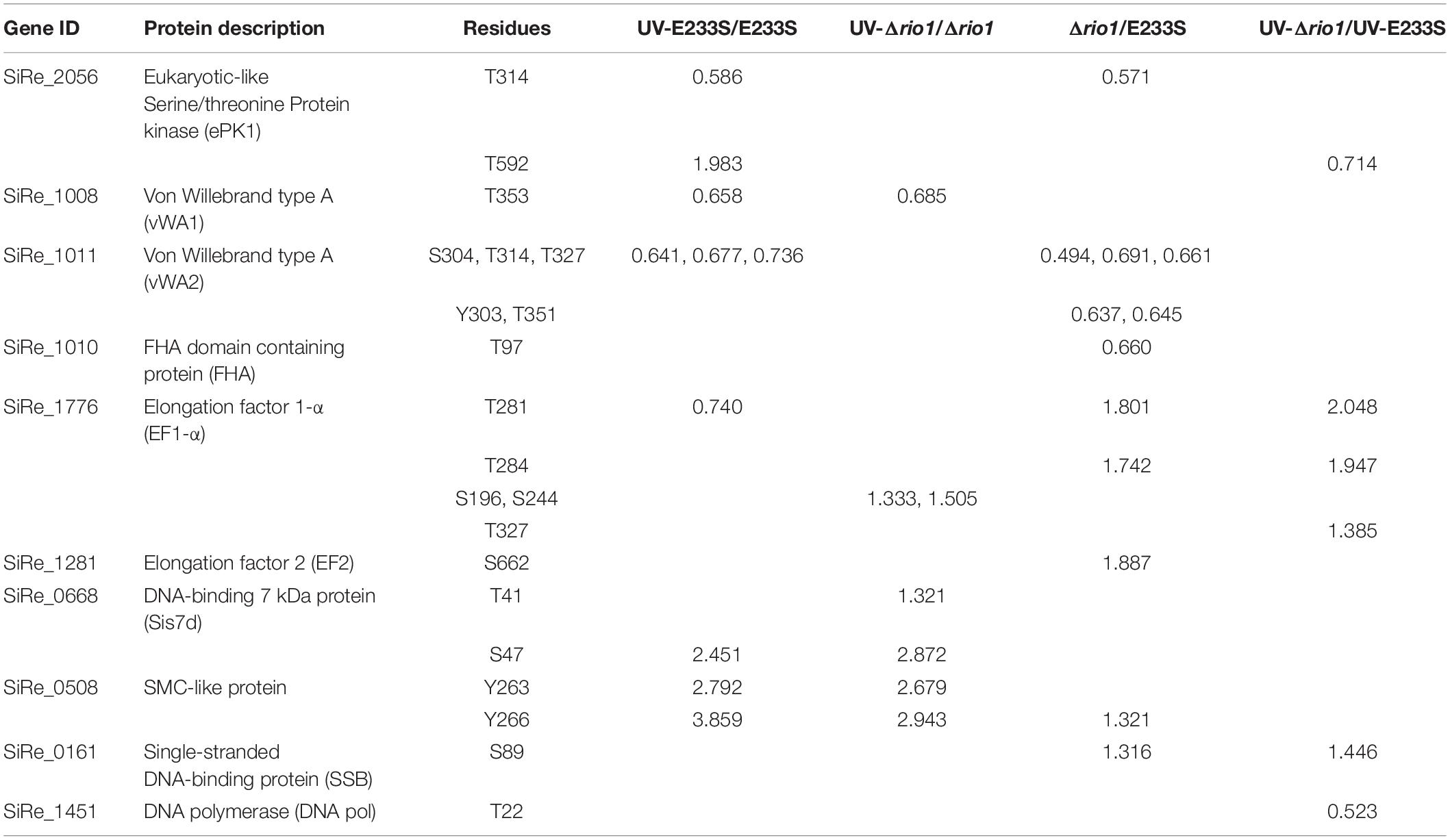
Table 1. Differentially phosphorylated proteins involved in signal transduction, DNA replication/repair, and translation in the four comparisons of UV-E233S/E233S, UV-Δrio1/Δrio1, Δrio1/E233S, and UV-Δrio1/UV-E233S.
In the S. islandicus genome, an FHA domain-containing protein SiRe_1010 is adjacent to the two vWA domain proteins (SiRe_1008 and SiRe_1011) and its phosphorylation at T97 decreased in Δrio1 versus E233S (Table 1 and Supplementary Table S4). In eukaryotes, the FHA domain is often a part of large protein kinases and is involved in DDR, DNA repair, and replication (Durocher and Jackson, 2002; Mohammad and Yaffe, 2009). In bacteria, FHA-containing proteins were able to bind phosphorylated proteins (Alderwick et al., 2006; Niebisch et al., 2006). In S. acidocaldarius, the interaction of FHA and vWA1 was mediated by phosphorylation and the cell motility was regulated by this interaction (Reimann et al., 2012; Albers and Jarrell, 2015; Hoffmann et al., 2019; Ye et al., 2020). Our data suggested that vWA2 might also be phosphorylated and interacted with FHA in vivo. To test this hypothesis, co-expression of FHA and vWA1 (or vWA2) was performed in E. coli and S. islandicus. In vivo pull down was carried out by anti-Flag beads to obtain Flag-FHA and Western blot with His-tag specific antibody to determine the co-exist of His-vWAs in E. coli, or by Ni-NTA purification of His-tagged FHA and Western blot for detection of co-purified vWAs by anti-Flag antibody in S. islandicus. We revealed that vWA2 interacted with FHA (Figures 5B,D). Similar to S. acidocaldarius counterparts, vWA1 was able to interact with FHA both in E. coli and S. islandicus (Figures 5A,C). Interestingly, more proteins were pulled down in S. islandicus cells than in E. coli. We assume that PTM including phosphorylation enhances the protein interaction. Consistently, a recent in vitro study showed that phosphorylation of S. acidocaldarius vWA1 stimulated formation of FHA-vWA complex, while dephosphoryation by PP2A resulted in dissociation of the complex (Ye et al., 2020). Our results indicate that phosphorylation of vWA2 may be also involved in signal transduction together with FHA. Since S. acidocaldarius Δrio1 are less motile (Hoffmann et al., 2017) and our data showed that phosphorylation of FHA and vWA2 was decreased when rio1 was deleted, it is also possible that Rio1 might be involved in cell motility by regulating FHA/vWAs phosphorylation. Further study is needed to investigate whether and how the interactions between FHA and vWA1/vWA2 are regulated, and if these interactions play a broad role in stress responses.
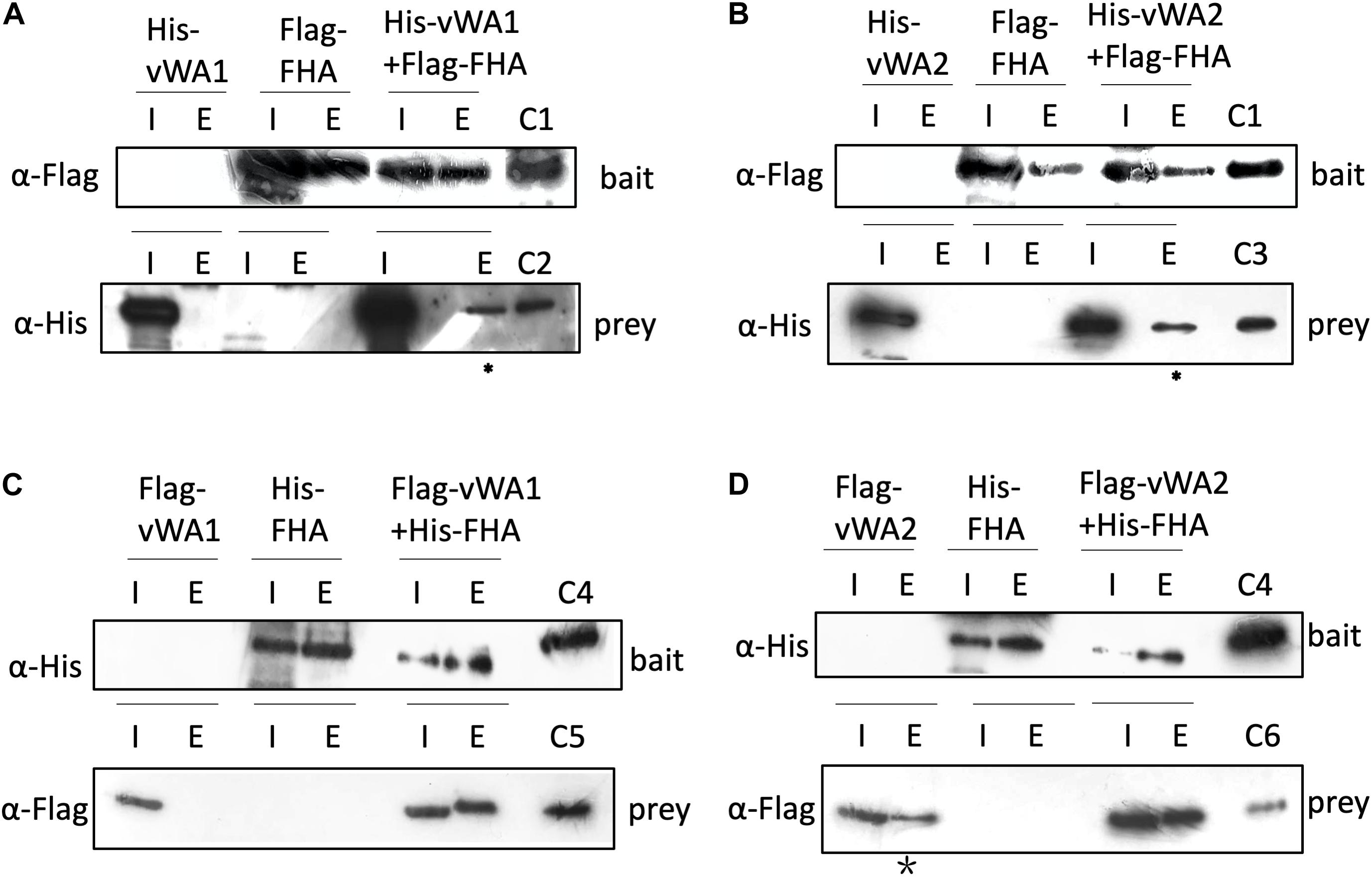
Figure 5. Interaction of FHA and vWAs by pull down. (A,B) Co-purification of His-vWA1(A)/vWA2(B) and Flag-FHA in E. coli. His-vWA1 (or vWA2) and Flag-FHA were co-expressed in E. coli. Five hundred milliliters of the E. coli cells were resuspended in 40 ml buffer A and protein samples were prepared as described in the section “Materials and Methods.” Total soluble protein and the enriched sample (20 μl) from approximately 40 ml supernatant (0.05%) and 9 ml elute (0.22%), respectively, were analyzed by Western blot. C1, control of Flag-FHA purified from E. coli; C2, His-tagged vWA1 purified from E. coli; C3, His-tagged vWA2 purified from E. coli. The asterisks represent samples concentrated by 12 times (2.6% of the total elute) before loading to the gels. (C,D) Co-purification of Flag-vWA1(C)/vWA2(D) and His-FHA in S. islandicus. One hundreds milliliters of S. islandicus cells harboring the co-expression plasmid were cultivated to OD600∼0.2 and protein samples were prepared as described in the section “Materials and Methods.” Total soluble protein and the enriched sample (40 μl) from approximately 10 ml supernatant (0.4%) and 2 ml elute (2.0%), respectively, were analyzed by Western blot. C4, His-tagged FHA purified from E. coli; C5, Flag-vWA1 purified from S. islandicus; C6, Flag-vWA2 purified from S. islandicus. The star indicates that Flag-vWA2 bound to Ni-NTA beads weakly. I, input; E, elute.
We found that the phosphorylation levels of two chromosomal proteins changed with UV irradiation. While previous report showed that the transcription of the chromatin protein Sso7d was repressed upon UV irradiation (Frols et al., 2007; Gotz et al., 2007), the phosphorylation level at T41 and S47 of the counterpart in S. islandicus, Sis7d, increased in our study (Table 2). Sso7d is able to bind DNA so as to increase the thermal stability of packed DNA (Robinson et al., 1998; White and Bell, 2002). Decreased Sso7d/Sis7d level would allow chromatin to be more flexible for DNA repair. T41 and S47 are located at a conserved core loop close to the DNA backbone (41, 42). Therefore, phosphorylation of Sis7d may reduce its DNA binding ability by introducing negative charge. To test the effect of phosphorylation on Sis7d, the wild type and phosphor-mimic mutant proteins (T41E/S47E) were purified and their DNA binding activities were assayed. As expected, DNA binding ability of mutant Sis7d decreased compared with the wild type protein (Figure 6A). In addition, we found that the phosphorylation of an SMC (Structural Maintenance of Chromosome) domain-containing protein (SiRe_0508) at Y263 and Y266 increased after UV treatment (Table 2). Since the SMC family proteins are usually involved in chromosome organization (Diebold-Durand et al., 2017; Wang et al., 2017), SiRe_0508 phosphorylation may inhibit its DNA affinity to impede chromosome remodeling and facilitate DNA repair. As predicted, EMSA revealed that phosphor-mimic mutant SiRe_0508-Y263E/Y266E exhibited lower dsDNA affinity than the wild type enzyme (Figure 6B). Exactly how the phosphorylation of SiRe_0508 regulates its function needs further investigation.
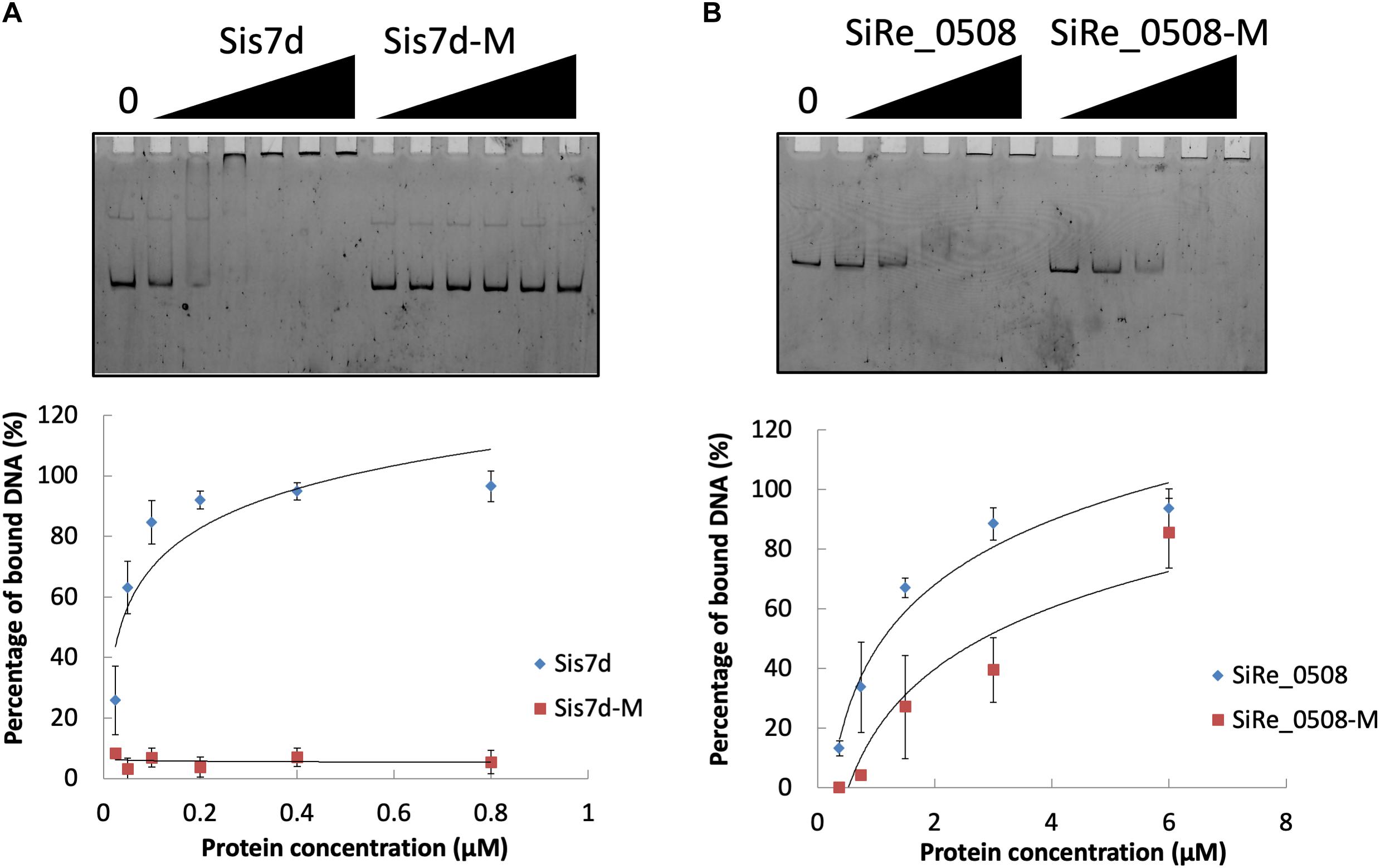
Figure 6. DNA binding ability of chromosome remodeling proteins and their phosphor-mimic mutants. (A) EMSA of Sis7d and its mutants Sis7d-T41E/S47E. The reaction was performed at 37°C for 30 min and analyzed in 8% native PAGE (see section “Materials and Methods”). Protein concentration of 0.025, 0.05, 0.1, 0.2, 0.4, and 0.8 μM was used. (B) EMSA of SiRe_0508 and SiRe_0508-Y263E/Y266E. Protein concentration of 0.38, 0.75, 1.5, 3, and 6 μM was used. Representative gels are shown. The quantitative data was obtained from at least three independent experiments. Error bars indicate standard deviation.
Nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), and homologous recombination repair are supposed to repair UV-induced DNA lesions (Courcelle and Hanawalt, 2001; Goosen and Moolenaar, 2008). In the current study, phosphorylation of multiple DNA replication/repair proteins were identified, including DnaG, DNA Ligase, replicative DNA polymerase B (PolB1, SiRe_1451), DNA topoisomerase, RFC, RadA, SSB, Hjm, XPB1, and XPD (Supplementary Table S1), although the phosphorylation levels of DNA repair proteins altered not always significantly. However, phosphorylation levels of some of the proteins changed 1.2–1.3 times or more in at least one of the biological replicates, such as PolB1, DNA topoisomerase, RadA, and XPB1. Proteins involved in the NER, BER, and MMR pathways have been shown to change significantly in their phosphorylation patterns at 6 h after UV treatment in Hela cells (Edifizi et al., 2017; Xu et al., 2017). This indicated that phosphorylation of DNA replication and repair proteins also changes in response to UV treatment in archaeal cells but to a lesser degree compared with eukaryotes. In addition, we could not rule out that the phosphorylation change of DNA repair proteins may become more apparent at the later stages of response to UV irradiation.
To investigate which pathways induced by UV irradiation were dependent on Rio1, the phosphorylation changes between UV-treated Δrio1 cells and untreated Δrio1 cells (UV-Δrio1/Δrio1) were compared with those between UV-treated E233S cells with untreated E233S cells (UV-E233S/E233S). Our results revealed that phosphorylation of 15 sites on 12 proteins increased in UV-Δrio1/Δrio1 (9 sites overlapped with UV-E233S/E233S), while that of only one site decreased in UV-Δrio1/Δrio1, which was much fewer than the number of decreased phosphorylated sites (20) from a comparison of UV-E233S/E233S (Figures 3, 4B). This suggested that Rio1 was responsible for the reduction (>1.3-fold) of most protein phosphorylation induced by UV irradiation (Figure 4A and Supplementary Table S3). We identified 25 sites on 21 proteins with >1.3-fold differential phosphorylation only in UV-treated E233S cells versus E233S, among which 6 sites on 6 proteins exhibited increased phosphorylation, and 19 sites on 17 proteins had lower phosphorylation levels (Figure 4B and Table 2). In addition, there were 10 sites (9 increased and 1 decreased) on 9 proteins that changed in both groups, representing those phosphorylation events induced by UV but independent of Rio1 (Table 2).
Among the proteins with decreased phosphorylation levels identified only in UV-treated E233S, the majority were associated with translation, ribosomal structure, and biogenesis (arCOG J, including TEF1α, the exosome complex component Rrp41, and three ribosomal proteins) and posttranslational modification, protein turnover, and chaperones (arCOG O, including ePK1, CDC48, and two thermosomes, Figure 3A and Table 2). These indicated that Rio1 mediated protein metabolism after UV irradiation in an indirect manner. In a study of Caenorhabditis elegans, the genes involved in protein synthesis, refolding, and degradation were transcriptionally reduced upon UV-induced DNA damage, suggesting that protein recycling was impaired due to general energy depletion (Edifizi et al., 2017). Our data revealed that these processes were also regulated by phosphorylation after UV treatment, probably by inhibition. It is unknown whether the inhibition of the phosphorylation by Rio1 was due to inactivation of a second protein kinase or enhancement of protein phosphatase. However, we cannot exclude that Rio1 might directly interact with these proteins for inhibition, since the interaction of yeast Rio1 with ribosomal proteins, exosome complex components, and chaperones has been detected by yeast two-hybrid (Iacovella et al., 2018). Moreover, in eukaryotes, Rio1 is a subunit of the pre-40S ribosome and is involved in pre-rRNA processing and small ribosomal subunit maturation via a direct interaction with other pre-40S subunits (Ferreira-Cerca et al., 2014; Turowski et al., 2014; Ameismeier et al., 2018). Most likely, one of the Rio1 roles in response to UV irradiation was inhibition of protein synthesis and turnover to reduce energy consuming.
Several proteins exhibited increased phosphorylation levels in UV-treated E233S but not in UV-treated Δrio1, among which were ePK1 (SiRe_2056), the S-layer protein SlaA (SiRe_1612), and a thermosome (SiRe_1214). Our in vitro kinase assay showed that Rio1 was able to phosphorylate ePK1 (Supplementary Figure S5), conversely, ePK1 could also phosphorylate Rio1 as revealed previously (Huang et al., 2017). The detailed regulatory mechanism between these kinases needs further investigation. It will be very interesting to determine whether ePK1 and SlaA are potential Rio1 substrates in vivo and what roles they play in DDR.
Remarkably, PolB1 phosphorylation decreased in UV-treated Δrio1 cells as compared to UV-treated E233S cells but not significantly different (<1.3-fold) in Δrio1 cells compared to E233S cells, implying that the phosphorylation change were induced by UV and amplified by Rio1, affecting (probably inhibiting) DNA replication (Table 1 and Supplementary Table S1).
Previously, we found that Rio1 phosphorylated the Holliday junction resolvase Hjc, which inhibited its catalytic activity and increased the cell viability in the presence of multiple DNA lesions. We proposed a regulatory role for Rio1 in stalled DNA replication fork resolution (Huang et al., 2019). Phosphorylation of Hjc was not identified in this study, probably due to low protein level, which could become even lower since hjc was transcriptionally down-regulated after UV treatment in S. acidocaldarius (13). Similarly, we did not identify phosphorylation of Orc1-2 or Tfb3 in a recently identified Orc1-2 dependent DDR network in Sulfolobus (Feng et al., 2018; Schult et al., 2018; Sun et al., 2018). The reason could also be due to low abundance of phosphorylated proteins at early stage of UV treatment. In this network, the Orc1-2 is up-regulated in the presence of DNA damage agents NQO or UV. The protein binds to the promoter regions of the gene itself and DDR genes and activates the expression of the Orc1-2, transcription factor TFB3, proteins involved in DNA synthesis (Dpo2), the ups operon and the ced gene transfer system (Ajon et al., 2011; van Wolferen et al., 2016), and represses the expression of genes involved in DNA replication initiation, genome segregation, and cell division (Sun et al., 2018). Since we did not find a conserved DNA motif for Orc1-2 binding to the DDR gene promoters (5′-ANTTTC-3′) in the rio1 promoter region (data not shown), which was bound by Orc1-2 for regulation (Le et al., 2017; Sun et al., 2018), Rio1-mediated network might be independent of the Orc1-2-centered network. In addition, Rio1 was induced earlier than that of Orc1-2 after UV-treatment (peak at 30 min versus 90 min) according to a previous transcriptomic study (Gotz et al., 2007), suggesting that the expression of rio1 after UV irradiation may not in fact be induced by Orc1-2 and the Rio1-mediated network might be independent of the Orc1-2-centered network. Whether and how Rio1 interplays with the Orc1-2- centered network need further investigation.
The comparison of Δrio1 cells versus E233S cells revealed that phosphorylation of 25 sites on 23 proteins increased, while that at 36 sites on 24 proteins decreased (Figure 3 and Supplementary Table S4). The arCOG categories of these proteins revealed that more proteins associated with various cellular metabolisms (arCOG C, E, G, H) appeared in Δrio1 cells versus E233S compared with UV-treated E233S versus untreated E233S cells (Supplementary Figure S6), suggesting that Rio1 also regulates metabolism without DNA damage treatment in addition to protein synthesis/turnover.
To further investigate the impact of differentially phosphorylated proteins (>1.3-fold) on cellular physiological processes, the functional enrichment including KEGG pathway and GO categories was analyzed for each comparison group. For KEGG enrichment, multiple proteins with increased phosphorylation were enriched in Δrio1. These proteins are involved in various metabolic pathways, including galactose metabolism, starch and sucrose metabolism, alanine, aspartate, and glutamate metabolism, monobactam biosynthesis, cysteine and methionine metabolism, glycine, serine, and threonine metabolism, and lysine biosynthesis (Figure 7A). The latter five metabolic pathways were also enriched in the UV-treated Δrio1 versus UV-treated E233S cells, confirming the important roles of Rio1 in cellular metabolism independent of UV irradiation (Figure 7A and Supplementary Table S5). As Δrio1 grew faster than wild type, Rio1 may regulate growth rate via protein synthetic capacity and/or cellular metabolism.

Figure 7. Functional enrichment of the five comparison groups. (A) KEGG pathway. (B) Molecular function. Both increase (up) and decrease (down) of differentially phosphorylated proteins involved in processes or pathways are shown. Red to blue represents the enrichment strength in log10 values.
In addition, GO enrichment based on molecular function showed that rio1 deletion mainly led to increased phosphorylation of proteins with lyase or hydrolase activities, whereas UV irradiation resulted in increased phosphorylation of those with ion or small molecular binding activities, suggesting that these two treatments (rio1 deletion and UV irradiation) led to phosphorylation changes of proteins with different activities for different downstream reactions (Figure 7B).
Remarkably, the phosphorylation of three Pur proteins (SiRe_1379/PurL, SiRe_1382/PurC, and SiRe_1753/PurA) decreased in both Δrio1 cells compared to E233S cells and UV-treated Δrio1 cells compared to UV-treated E233S (Supplementary Tables S4, S5), but not in UV-E233S/E233S, indicating the phosphorylation change is Rio1-dependent but not UV-induced. It has been shown that the gene cluster of nucleotide (purine) biosynthesis (purC/Saci_1607, purS/Saci_1608, and purL/Saci_1610) and a putative purine transporter gene (Saci_0214) were transcriptionally down-regulated in response to UV in S. acidocaldarius (Hoffmann et al., 2017; Schult et al., 2018). These changes suggested that nucleotide synthesis and uptake were both repressed after UV treatment, probably for the purpose of slowing down DNA replication. Consistent with this, the transcripts of the gene cluster (Saci_1607-Saci_1613) and the uptake gene (Saci_0214) have been shown to be higher in S-phase, where a large number of DNA replication events occur (Lundgren and Bernander, 2007). Our results suggest that purine biosynthesis is regulated probably by Rio1-mediated phosphorylation. Strikingly also, phosphorylation of SSB increased after rio1 deletion regardless of UV-treatment (Table 1). Phosphorylation of these proteins could provide another layer of regulation, although it was in a UV-independent manner in certain circumstances.
To understand whether Sulfolobus phosphorylation has a bias toward any amino acid motif, we analyzed the amino acid composition of the identified phosphorylation sites using software MoMo. The frequencies of 6 amino acids surrounding all phosphorylation sites were summarized, and the result showed that Arg appeared at the +3 position of 39 out of 367 phosphorylated Ser sites, which was enriched by 2.7-fold compared with the background of Arg containing peptides from all identified peptides (Figure 8). In addition, Arg was enriched at the +2 position of phosphorylated Thr by 4.6-fold (Figure 8). These results indicate that the positively charged amino acid Arg, which frequently appeared at the +2/3 position downstream of phosphorylated sites, might facilitate the phosphorylation reaction of certain Sulfolobus protein kinases.
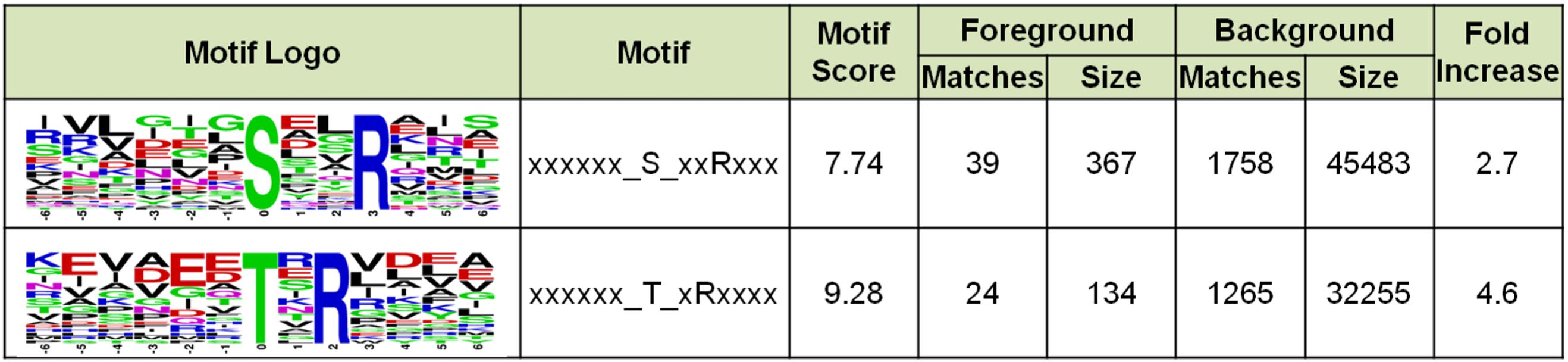
Figure 8. Motif enrichment analysis of the identified phosphorylation Ser and Thr sites in this study. Ser and Thr at specific positions in modified-13-mers (6 amino acids upstream and downstream of phosphorylation site) for all identified protein sequences were collected, and all the database protein sequences were used as the background database parameter. The software package MoMo (motif-x algorithm) was used for motif enrichment analysis. The minimum number of occurrences was set to 20.
Our study revealed a complex network and a number of putative target proteins of Rio1 in early response to DNA damage. Many of the phenomena need further verification and some are worthy further exploration. Nevertheless, Saccharomyces cerevisiae Rio1 was shown to interact with a number of factors involved in ribosome production, protein synthesis and turnover, metabolism, energy production, and cell cycle progress by yeast two-hybrid screen (Iacovella et al., 2018), suggesting that there may be conserved function and mechanism of Rio1 proteins from eukaryotes and archaea. Further study on which subset of proteins is regulated by Rio1-dependent phosphorylation and whether archaeal Rio1 binds certain gene promoters directly for regulation, similar to yeast Rio1, will also be needed. Furthermore, it will be interesting to study on how Rio1 is regulated in response to DNA damage and other stresses.
The datasets generated for this study can be found in the ProteomeXchange, accession: PXD021663. Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD021663.
QH designed the experiments, conducted most of the experiments, analyzed the data, and wrote the manuscript. ZL performed the colony formation assay and in vivo pull down. PW conducted the strain cultivation experiments. JN helped to analyze the data and revised the manuscript. YS conceived the project and revised the manuscript. All the authors approved the final version of the manuscript.
This study was supported by grants from the National Key R&D Program of China (2020YFA0906800), Natural Science Foundation of China (31970546 and 31670061 to YS; 31470200 and 31970119 to JN; 31900055 to QH), and The Fundamental Research Funds of Shandong University (2018HW015 to QH).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank BPI (Beijing, China) for the technical support of phosphoproteomic analysis. We also thank members of the CRISPR and Archaea Biology Research Center for helpful discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.586025/full#supplementary-material
PTM, post-translational modification; DDR, DNA damage response; UV, ultraviolet; TFB3, transcription factor B3; Rio, right open reading frame; MCS, multiple cloning site; TMT, tandem mass tags; SCX, strong cation exchange; IMAC, immobilized metal affinity chromatography; PBS, phosphate-buffered saline; LC-MS/MS, liquid chromatography tandem mass spectrometry; Q-TOF, quadrupole time-of-flight; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; arCOG, archaeal clusters of orthologous groups; RSD, relative standard deviation; vWA, von Willebrand type A; TEF, translation elongation factor; PTP, phosphotyrosine phosphatases; SMC, structural maintenance of chromosome.
Aivaliotis, M., Macek, B., Gnad, F., Reichelt, P., Mann, M., and Oesterhelt, D. (2009). Ser/Thr/Tyr protein phosphorylation in the archaeon Halobacterium salinarum–a representative of the third domain of life. PLoS One 4:e4777. doi: 10.1371/journal.pone.0004777
Ajon, M., Frols, S., van Wolferen, M., Stoecker, K., Teichmann, D., Driessen, A. J., et al. (2011). UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 82, 807–817. doi: 10.1111/j.1365-2958.2011.07861.x
Albers, S. V., and Jarrell, K. F. (2015). The archaellum: how Archaea swim. Front. Microbiol. 6:23. doi: 10.3389/fmicb.2015.00023
Alderwick, L. J., Molle, V., Kremer, L., Cozzone, A. J., Dafforn, T. R., Besra, G. S., et al. (2006). Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 2558–2563. doi: 10.1073/pnas.0507766103
Ameismeier, M., Cheng, J., Berninghausen, O., and Beckmann, R. (2018). Visualizing late states of human 40S ribosomal subunit maturation. Nature 558, 249–253. doi: 10.1038/s41586-018-0193-0
Angermayr, M., Roidl, A., and Bandlow, W. (2002). Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in the control of cell cycle progression. Mol. Microbiol. 44, 309–324. doi: 10.1046/j.1365-2958.2002.02881.x
Blackford, A. N., and Jackson, S. P. (2017). ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817. doi: 10.1016/j.molcel.2017.05.015
Cao, J., Wang, T., Wang, Q., Zheng, X., and Huang, L. (2019). Functional insights into protein acetylation in the hyperthermophilic archaeon Sulfolobus islandicus. Mol. Cell Proteomics 18, 1572–1587. doi: 10.1074/mcp.ra119.001312
Courcelle, J., and Hanawalt, P. C. (2001). Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8196–8202. doi: 10.1073/pnas.121008898
Deng, L., Zhu, H., Chen, Z., Liang, Y. X., and She, Q. (2009). Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13, 735–746. doi: 10.1007/s00792-009-0254-2
Diebold-Durand, M. L., Lee, H., Ruiz Avila, L. B., Noh, H., Shin, H. C., Im, H., et al. (2017). Structure of full-length SMC and rearrangements required for chromosome organization. Mol. Cell 67:e335.
Edifizi, D., Nolte, H., Babu, V., Castells-Roca, L., Mueller, M. M., Brodesser, S., et al. (2017). Multilayered reprogramming in response to persistent DNA damage in C. elegans. Cell Rep. 20, 2026–2043. doi: 10.1016/j.celrep.2017.08.028
Esser, D., Pham, T. K., Reimann, J., Albers, S. V., Siebers, B., and Wright, P. C. (2012). Change of carbon source causes dramatic effects in the phospho-proteome of the archaeon Sulfolobus solfataricus. J. Proteome Res. 11, 4823–4833. doi: 10.1021/pr300190k
Feng, X., Sun, M., Han, W., Liang, Y. X., and She, Q. (2018). A transcriptional factor B paralog functions as an activator to DNA damage-responsive expression in archaea. Nucleic Acids Res. 46:7465. doi: 10.1093/nar/gky302
Ferreira-Cerca, S., Kiburu, I., Thomson, E., LaRonde, N., and Hurt, E. (2014). Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 42, 8635–8647. doi: 10.1093/nar/gku542
Ferreira-Cerca, S., Sagar, V., Schafer, T., Diop, M., Wesseling, A. M., Lu, H., et al. (2012). ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1316–1323. doi: 10.1038/nsmb.2403
Frols, S., Gordon, P. M., Panlilio, M. A., Duggin, I. G., Bell, S. D., Sensen, C. W., et al. (2007). Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 189, 8708–8718. doi: 10.1128/jb.01016-07
Goosen, N., and Moolenaar, G. F. (2008). Repair of UV damage in bacteria. DNA Repair (Amst.) 7, 353–379. doi: 10.1016/j.dnarep.2007.09.002
Gotz, D., Paytubi, S., Munro, S., Lundgren, M., Bernander, R., and White, M. F. (2007). Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 8:R220.
Hoffmann, L., Anders, K., Bischof, L. F., Ye, X., Reimann, J., Khadouma, S., et al. (2019). Structure and interactions of the archaeal motility repression module ArnA-ArnB that modulates archaellum gene expression in Sulfolobus acidocaldarius. J. Biol. Chem. 294, 7460–7471. doi: 10.1074/jbc.ra119.007709
Hoffmann, L., Schummer, A., Reimann, J., Haurat, M. F., Wilson, A. J., Beeby, M., et al. (2017). Expanding the archaellum regulatory network - the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius. Microbiologyopen 6:e00414. doi: 10.1002/mbo3.414
Huang, Q., Mayaka, J. B., Zhong, Q., Zhang, C., Hou, G., Ni, J., et al. (2019). Phosphorylation of the archaeal Holliday junction resolvase Hjc inhibits its catalytic activity and facilitates DNA repair in Sulfolobus islandicus REY15A. Front. Microbiol. 10:1214. doi: 10.3389/fmicb.2019.01214
Huang, Q., Zhong, Q., Mayaka, J. B. A., Ni, J., and Shen, Y. (2017). Autophosphorylation and Cross-phosphorylation of protein kinases from the crenarchaeon Sulfolobus islandicus. Front. Microbiol. 8:2173. doi: 10.3389/fmicb.2017.02173
Hunter, T. (2009). Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 21, 140–146. doi: 10.1016/j.ceb.2009.01.028
Iacovella, M. G., Bremang, M., Basha, O., Giaco, L., Carotenuto, W., Golfieri, C., et al. (2018). Integrating Rio1 activities discloses its nutrient-activated network in Saccharomyces cerevisiae. Nucleic Acids Res. 46, 7586–7611. doi: 10.1093/nar/gky618
Jette, N., and Lees-Miller, S. P. (2015). The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 117, 194–205. doi: 10.1016/j.pbiomolbio.2014.12.003
Knuppel, R., Christensen, R. H., Gray, F. C., Esser, D., Strauss, D., Medenbach, J., et al. (2018). Insights into the evolutionary conserved regulation of Rio ATPase activity. Nucleic Acids Res. 46, 1441–1456. doi: 10.1093/nar/gkx1236
Le, T. N., Wagner, A., and Albers, S. V. (2017). A conserved hexanucleotide motif is important in UV-inducible promoters in Sulfolobus acidocaldarius. Microbiology 163, 778–788. doi: 10.1099/mic.0.000455
Lundgren, M., and Bernander, R. (2007). Genome-wide transcription map of an archaeal cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 2939–2944. doi: 10.1073/pnas.0611333104
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life (Basel) 5, 818–840. doi: 10.3390/life5010818
McCready, S., Muller, J. A., Boubriak, I., Berquist, B. R., Ng, W. L., and DasSarma, S. (2005). UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Syst. 1:3.
Mohammad, D. H., and Yaffe, M. B. (2009). 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair 8, 1009–1017. doi: 10.1016/j.dnarep.2009.04.004
Niebisch, A., Kabus, A., Schultz, C., Weil, B., and Bott, M. (2006). Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J. Biol. Chem. 281, 12300–12307. doi: 10.1074/jbc.m512515200
Peng, N., Deng, L., Mei, Y., Jiang, D., Hu, Y., Awayez, M., et al. (2012). A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 78, 5630–5637. doi: 10.1128/aem.00855-12
Reimann, J., Esser, D., Orell, A., Amman, F., Pham, T. K., Noirel, J., et al. (2013). Archaeal signal transduction: impact of protein phosphatase deletions on cell size, motility, and energy metabolism in Sulfolobus acidocaldarius. Mol. Cell Proteomics 12, 3908–3923. doi: 10.1074/mcp.m113.027375
Reimann, J., Lassak, K., Khadouma, S., Ettema, T. J., Yang, N., Driessen, A. J., et al. (2012). Regulation of archaella expression by the FHA and von Willebrand domain-containing proteins ArnA and ArnB in Sulfolobus acidocaldarius. Mol. Microbiol. 86, 24–36. doi: 10.1111/j.1365-2958.2012.08186.x
Robinson, H., Gao, Y. G., McCrary, B. S., Edmondson, S. P., Shriver, J. W., and Wang, A. H. (1998). The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 392, 202–205. doi: 10.1038/32455
Sadowski, I., Stone, J. C., and Pawson, T. (1986). A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol. Cell Biol. 6, 4396–4408. doi: 10.1128/mcb.6.12.4396
Schult, F., Le, T. N., Albersmeier, A., Rauch, B., Blumenkamp, P., van der Does, C., et al. (2018). Effect of UV irradiation on Sulfolobus acidocaldarius and involvement of the general transcription factor TFB3 in the early UV response. Nucleic Acids Res. 46, 7179–7192. doi: 10.1093/nar/gky527
Sharma, K., D’Souza, R. C., Tyanova, S., Schaab, C., Wisniewski, J. R., Cox, J., et al. (2014). Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594. doi: 10.1016/j.celrep.2014.07.036
Sirbu, B. M., and Cortez, D. (2013). DNA damage response: three levels of DNA repair regulation. Cold Spring Harb. Perspect. Biol. 5:a012724. doi: 10.1101/cshperspect.a012724
Sun, M., Feng, X., Liu, Z., Han, W., Liang, Y. X., and She, Q. (2018). An Orc1/Cdc6 ortholog functions as a key regulator in the DNA damage response in Archaea. Nucleic Acids Res. 46, 6697–6711. doi: 10.1093/nar/gky487
Turowski, T. W., Lebaron, S., Zhang, E., Peil, L., Dudnakova, T., Petfalski, E., et al. (2014). Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 42, 12189–12199. doi: 10.1093/nar/gku878
van Wolferen, M., Wagner, A., van der Does, C., and Albers, S. V. (2016). The archaeal Ced system imports DNA. Proc. Natl. Acad. Sci. U.S.A. 113, 2496–2501. doi: 10.1073/pnas.1513740113
Wang, X., Brandao, H. B., Le, T. B., Laub, M. T., and Rudner, D. Z. (2017). Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355, 524–527. doi: 10.1126/science.aai8982
Weinberg, F., Reischmann, N., Fauth, L., Taromi, S., Mastroianni, J., Kohler, M., et al. (2017). The atypical kinase RIOK1 promotes tumor growth and invasive behavior. EBioMedicine 20, 79–97. doi: 10.1016/j.ebiom.2017.04.015
White, M. F., and Bell, S. D. (2002). Holding it together: chromatin in the Archaea. Trends Genet. 18, 621–626. doi: 10.1016/s0168-9525(02)02808-1
Whittaker, C. A., and Hynes, R. O. (2002). Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387. doi: 10.1091/mbc.e02-05-0259
Widmann, B., Wandrey, F., Badertscher, L., Wyler, E., Pfannstiel, J., Zemp, I., et al. (2012). The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell 23, 22–35. doi: 10.1091/mbc.e11-07-0639
Williams, E., Lowe, T. M., Savas, J., and DiRuggiero, J. (2007). Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles 11, 19–29. doi: 10.1007/s00792-006-0002-9
Xu, H., Chen, X., Ying, N., Wang, M., Xu, X., Shi, R., et al. (2017). Mass spectrometry-based quantification of the cellular response to ultraviolet radiation in HeLa cells. PLoS One 12:e0186806. doi: 10.1371/journal.pone.0186806
Keywords: archaea, Sulfolobus islandicus, phosphoproteome, UV irradiation, Rio, protein phosphorylation
Citation: Huang Q, Lin Z, Wu P, Ni J and Shen Y (2020) Phosphoproteomic Analysis Reveals Rio1-Related Protein Phosphorylation Changes in Response to UV Irradiation in Sulfolobus islandicus REY15A. Front. Microbiol. 11:586025. doi: 10.3389/fmicb.2020.586025
Received: 22 July 2020; Accepted: 09 November 2020;
Published: 03 December 2020.
Edited by:
Sonja-Verena Albers, University of Freiburg, GermanyReviewed by:
Changyi Zhang, University of Illinois at Urbana–Champaign, United StatesCopyright © 2020 Huang, Lin, Wu, Ni and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinfeng Ni, amluZmduaUBzZHUuZWR1LmNu; Yulong Shen, eXVsZ3NoZW5Ac2R1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.