- College of Food Science and Engineering, Ocean University of China, Qingdao, China
Antimicrobial peptides (AMPs) are a class of small peptides that widely exist in nature and they are an important part of the innate immune system of different organisms. AMPs have a wide range of inhibitory effects against bacteria, fungi, parasites and viruses. The emergence of antibiotic-resistant microorganisms and the increasing of concerns about the use of antibiotics resulted in the development of AMPs, which have a good application prospect in medicine, food, animal husbandry, agriculture and aquaculture. This review introduces the progress of research on AMPs comprehensively and systematically, including their classification, mechanism of action, design methods, environmental factors affecting their activity, application status, prospects in various fields and problems to be solved. The research progress on antivirus peptides, especially anti-coronavirus (COVID-19) peptides, has been introduced given the COVID-19 pandemic worldwide in 2020.
Introduction
Alexander Fleming discovered lysozyme in 1922, and this discovery marked the birth of modern innate immunity. Since then, antibiotics and antimicrobial peptides (AMPs) have been discovered. A total of 3,240 AMPs have been reported in the antimicrobial peptide database (APD31) updated on August 24, 2020.
Different types of AMPs have the following commonalities: their number of amino acid residues is between 10 and 60 (average: 33.26), and almost all AMPS are cationic (average net charge: 3.32). However, several anionic AMPs also exist, and they have several acidic amino acids like aspartic acid and glutamic acid (Malkoski et al., 2001; Schittek et al., 2001; Lai et al., 2007).
The anti-microbial resistance of microorganisms is becoming increasingly serious with the abuse of antibiotics in medicine, agriculture and animal husbandry, especially in developing countries. Research from Kenya has detected substantial amounts of antibiotic residues in edible meat (Ayukekbong et al., 2017). The prevalence of vancomycin-resistant Enterococcus (VRE) and methicillin-resistant Staphylococcus aureus (MRSA) is increasing in clinical medicine, so the countermeasures are urgently needed to address these bacterial infections. However, from the perspective of pharmaceutical companies, the development of new antibiotic drugs results in low profit. Thus, replacing antibiotics has become a consideration in the pharmaceutical, agricultural, animal husbandry, and food industries.
Research on AMPs is continuously developing and considerable amounts of data on AMPs have been stored in AMP databases. However, the mechanism of AMPs remains incompletely understood, and further work needs to be performed to determine the relationship between different physicochemical properties to obtain low-cost and highly safe AMPs with remarkable antimicrobial effects and the specificity and a high capacity for synergies of AMPs should also be further developed (Lazzaro et al., 2020).
Classification of AMPs
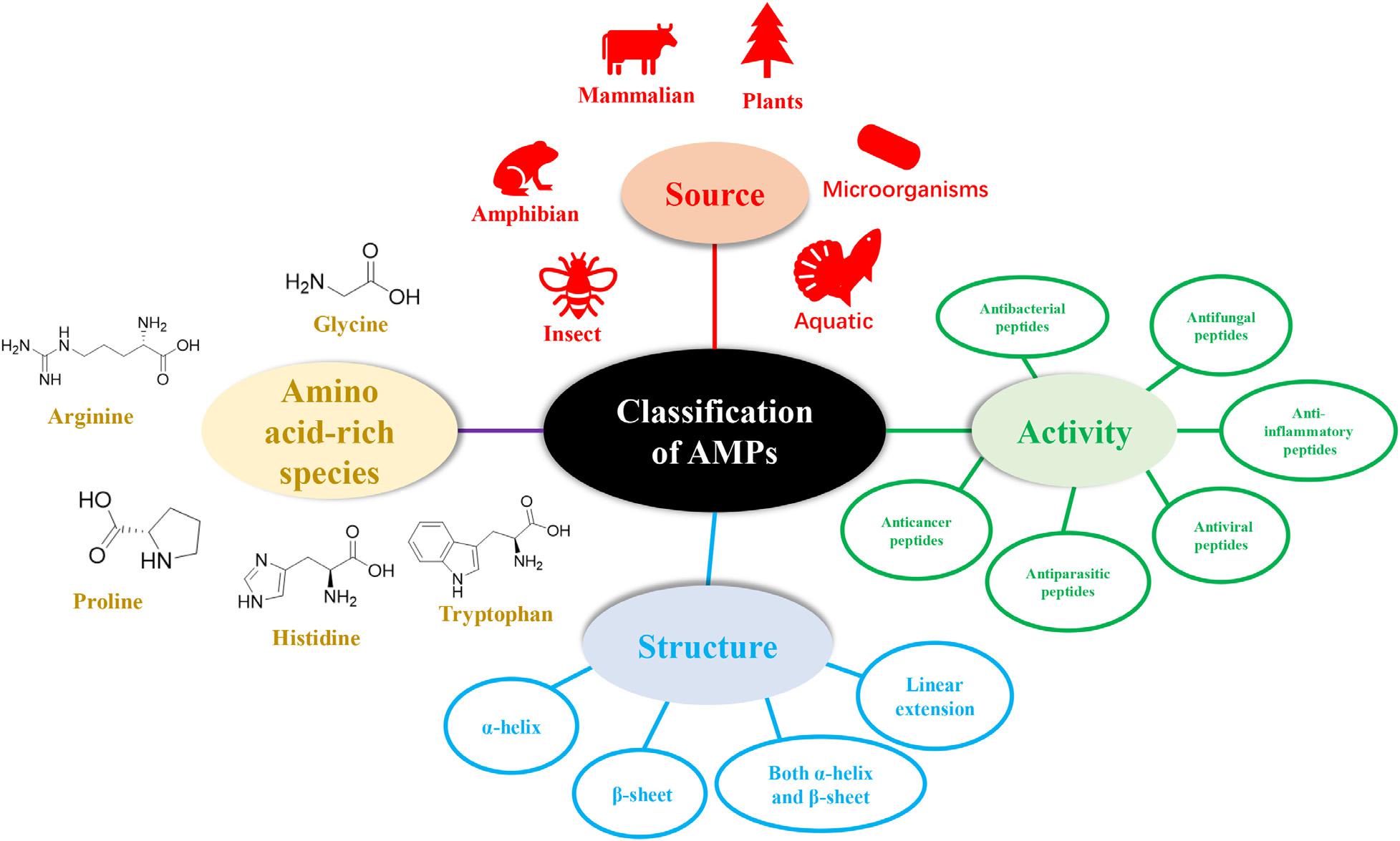
The diversity of natural AMPs causes difficulty in their classification. AMPs are classified based on (1) source, (2) activity, (3) structural characteristics, and (4) amino acid-rich species (Figure 1).
Classification of AMPs Based on Sources
The sources of AMPs can be divided into mammals (human host defense peptides account for a large proportion), amphibians, microorganisms, and insects according to statistical data in APD3. The AMPs found in oceans have also attracted widespread attention.
Mammalian Antimicrobial Peptides
Mammalian antimicrobial peptides are found in human, sheep, cattle, and other vertebrates. Cathelicidins and defensins are the main families of AMPs. Defensins can be divided into α-, β-, and θ-defensins depending on the position of disulfide bonds (Reddy et al., 2004). Human host defense peptides (HDPs) can protect human from microbial infections but show different expressions in every stage of human growth. For example, cathelicidin LL-37, a famous AMP derived from the human body, is usually detected in the skin of newborn infants, whereas human beta-defensin 2 (hBD-2) is often expressed in the elderly instead of the young (Gschwandtner et al., 2014). HDPs can be identified in many parts of the body such as skin, eyes, ears, mouth, respiratory tract, lung, intestine, and urethra. Besides, AMPs in human breast milk also play an important role in breastfeeding because it can decrease the morbidity and mortality of breast-feeding infants (Field, 2005). What’s interesting is that Casein201 (peptide derived from β-Casein 201–220 aa), identified in colostrum, shows different levels in preterm human colostrum and term human colostrums (Zhang et al., 2017). Dairy is an important source of AMPs, which are generated through milk enzymatic hydrolysis. Several AMPs have been identified from α-lactalbumin, β-lactoglobulin, lactoferrin, and casein fractions, and the most famous peptide obtained is lactoferricin B (LfcinB) (Sibel Akalın, 2014). Furthermore, whether the AMPs derived from dairy products can be used for dairy preservation is also an interesting subject to develop.
In addition to antimicrobial activity, HDPs, such as cathelicidins and defensins, also affect immune regulation, apoptosis, and wound healing (Wang, 2014).
Amphibian-Derived Antimicrobial Peptides
Antimicrobial peptides from amphibians play an important role in the protection of amphibians from the pathogens that have induced the global amphibian population decline (Rollins-Smith, 2009). Frogs are the main source of amphibian AMPs and the most famous AMP from frogs is magainin; the skin secretions of frogs from genera Xenopus, Silurana, Hymenochirus, and Pseudhymenochirus under the Pipidae family are rich in AMPs (Conlon and Mechkarska, 2014). Furthermore, cancrin, which has an amino acid sequence of GSAQPYKQLHKVVNWDPYG, has been reported as the first AMP from the sea amphibian Rana cancrivora (Lu et al., 2008). This marks a broader source of AMPs of amphibians.
Insect-Derived Antimicrobial Peptides
Antimicrobial peptides are mainly synthesized in fat bodies and blood cells of insects, which is one of the main reasons for insects’ strong adaptability to survival (Vilcinskas, 2013). Cecropin is the most famous family of AMPs from insects, and it can be found in guppy silkworm, bees, Drosophila. Cecropin A shows activity against different inflammatory diseases and cancers (Dutta et al., 2019). What should be known is that the number of AMPs varies greatly between species, for example, invasive harlequin ladybird (Harmonia axyridis) and black soldier fly (Hermetia illucens) have up to 50 AMPs, while pea aphid (Acyrthosiphon pisum) lacks AMPs (Shelomi et al., 2020). Jellein, a peptide derived from bee royal jelly, shows promising effects on several bacteria and fungi, and its lauric acid-conjugated form can inhibit the parasite Leishmania major (Zahedifard et al., 2020).
Microorganisms-Derived Antimicrobial Peptides
Antimicrobial peptides can be obtained from microorganisms like bacteria and fungi, and some famous peptides are nisin, gramicidin from Lactococcus lactis, Bacillus subtilis, and Bacillus brevis (Cao et al., 2018). Due to the high price of chemical synthesis of AMPs, the biological expression has attracted the increase of attention. Specific yeast species like Pichia pastoris, Saccharomyces cerevisiae, and bacteria like Escherichia coli, B. subtilis, and plants have been used for expression systems (Parachin et al., 2012), but it should be noticed that because of the toxicity, proteolytic degradation, and purification, AMPs are difficult to be produced in E. coli, which is necessary to take advantage of fusion tags (Yu et al., 2015).
Besides, numerous AMPs have also been extracted and isolated from the stems, seeds, and leaves of plants, and they are classified into several groups, including thionins, defensins and snakins (Tang et al., 2018). More marine-derived AMPs have been reported to have given the increasing value allotted by people to marine resources. Although most of the reported marine AMPs have been tested in vitro, several of these AMPs have shown promising results in vivo, for example, As-CATH4 shows an immunity-stimulating effect in vivo and can enhance the anti-infective capability of drugs used in combination with it (Semreen et al., 2018). Myticusin-beta is an immune-related AMP of Mytilus coruscus and a promising alternative to antibiotics (Oh et al., 2020). Moreover, GE33, known as pardaxin, is a marine AMP and the GE33-based vaccine has shown the ability to enhance antitumor immunity in mice (Huang et al., 2013).
Classification Based on Activity
The activity of AMPs can be divided into 18 categories according to the statistics of the ADP3 database. These categories can be summarized as antibacterial, antiviral, antifungal, antiparasitic, anti-human immunodeficiency virus (HIV), and anti-tumor peptides (Figure 2).
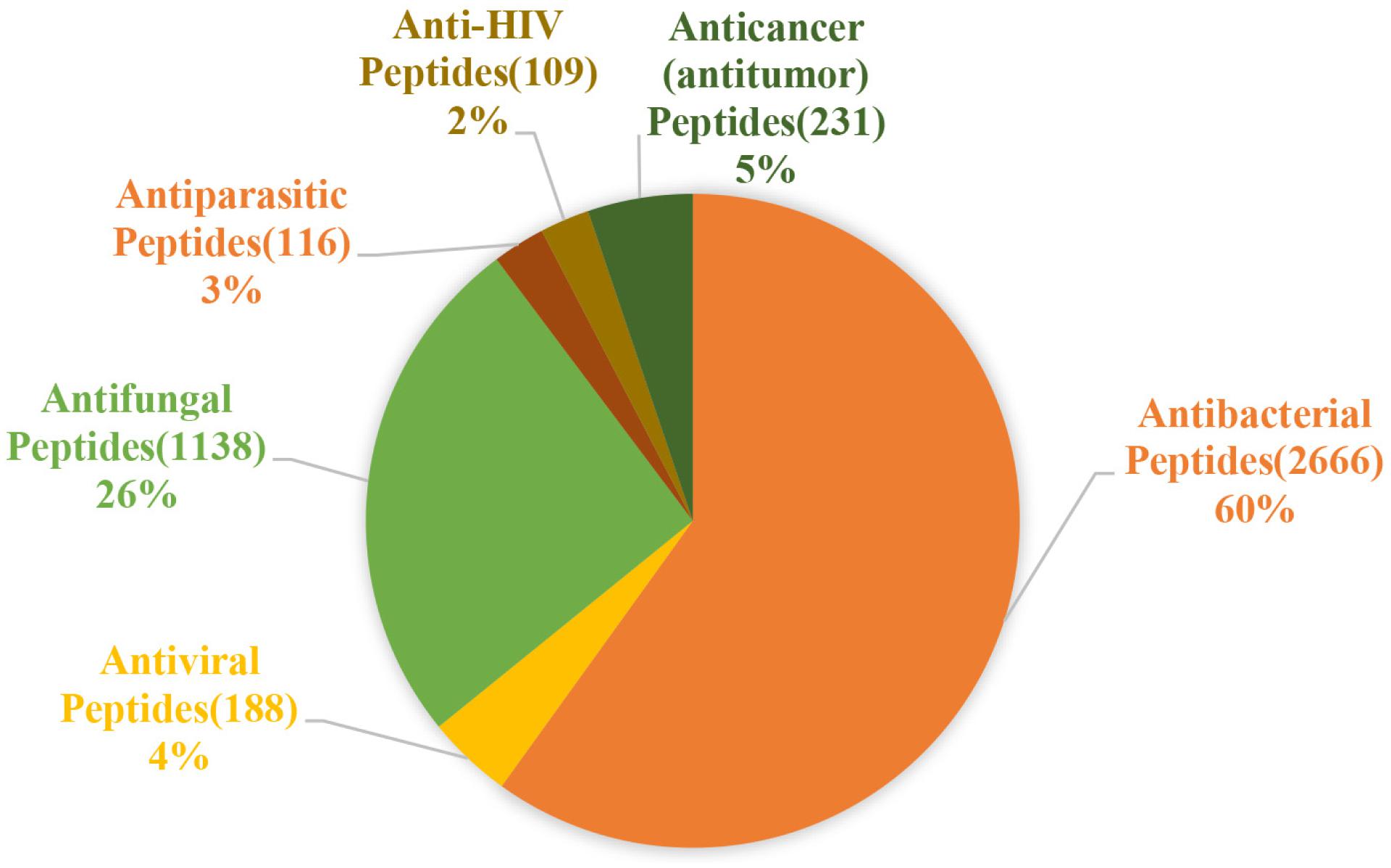
Figure 2. Statistics of the main functions of antimicrobial peptides. Antibacterial peptides account for the largest proportion, approximately 60%, followed by antifungal peptides, which account for 26%, and antiviral, antiparasitic, anticancer, anti-HIV peptides account for almost the same about 2–5% (the figure is drawn based on data in APD3).
Antibacterial Peptides
Antibacterial peptides account for a large part of AMPs and have a broad inhibitory effect on common pathogenic bacteria, such as VRE, Acinetobacter baumannii, and MRSA in clinical medicine and S. aureus, Listeria monocytogenes, E. coli in food and Salmonella, Vibrio parahaemolyticus in aquatic products. Many natural and synthetic AMPs like nisin, cecropins and defensins have shown good inhibition activity to Gram-positive bacteria and Gram-negative bacteria. In recent research, AMPs P5 (YIRKIRRFFKKLKKILKK-NH2) and P9 (SYERKINRHFKTLKKNLKKK-NH2), which are designed based on Aristicluthys nobilia interferon-I, inhibit MRSA and show a low cytotoxicity (Li C. et al., 2019).
Antifungal Peptides (AFPs)
Antifungal peptides are a subclass of AMPs that address fungal infections with enhanced drug resistance. Many AFPs have shown excellent anti-fungal activities against common pathogenic fungi, such as Aspergillus and Candida albicans in clinical medicine, yeast, filamentous fungi (e.g., Aspergillus flavus), mold in food and agriculture. Except for brevinin, ranatuerin, cecropins, many synthetic peptides also show good antifungal activity. For example, AurH1, derived from aurein 1.2, can effectively treat C. albicans infection, which has a lethal rate up to 40% (Madanchi et al., 2020). Aflatoxin, which is a carcinogen produced by A. flavus, is harmful to the human body. Many AFPs can inhibit the growth of A. flavus. For example, an AFP with a sequence of FPSHTGMSVPPP can inhibit the growth of A. flavus MD3. A total of 37 antifungal peptides isolated from Lactobacillus plantarum TE10 and their mixture can reduce A. flavus spore formation in fresh maize seeds (Muhialdin et al., 2020). Moreover, two chemically synthesized radish AMPs show a good inhibitory effect against different yeast species, such as Zygosaccharomyces bailii and Zygosaccharomyces rouxii (Shwaiki et al., 2020).
Antiviral Peptides (AVPs)
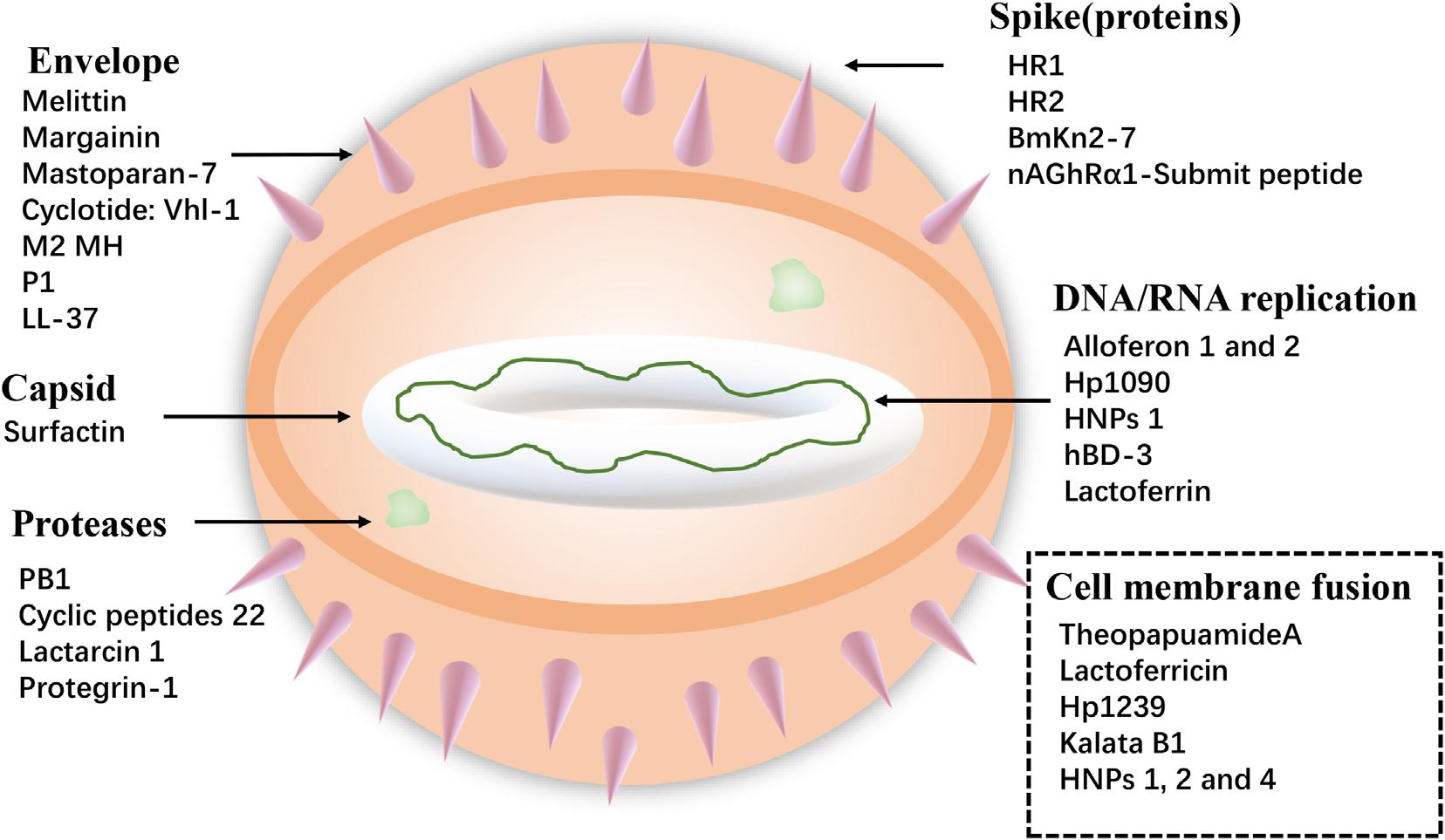
Viruses cause serious harm to human life and huge economic losses to the animal husbandry. The COVID-19, which is the recent outbreak, has caused great loss of lives and properties. Furthermore, foot-and-mouth disease virus, avian influenza virus (AIV), and HIV are long-term threats to human life. So, it is extremely urgent to solve these problems, and antiviral peptides provide new ways. Antiviral peptides show a strong killing effect on viruses mainly by (1) inhibiting virus attachment and virus cell membrane fusion, (2) destroying the virus envelope, or (3) inhibiting virus replication (Jung et al., 2019) (shown in Figure 3). A recent report has shown that AMP Epi-1 mediates the inactivation of virus particles and has good inhibitory activity against foot-and-mouth disease virus (Huang et al., 2018). Moreover, infectious bronchitis virus (IBV) is the pathogen of infectious bronchitis and the inoculation of swine intestinal AMP (SIAMP)–IBV mixed solution remarkably reduced the mortality of chicken embryos compared with the IBV infection group, showing the good inhibitory activity of SIAMP on IBV (Sun et al., 2010). Anti-HIV peptides are a subclass of anti-viral peptides. The most important examples of these peptides include defensins (including α- and β-defensins, which have different mechanisms), LL-37, gramicidin D, caerin 1, maximin 3, magainin 2, dermaseptin-S1, dermaseptin-S4, siamycin-I, siamycin-II, and RP 71955 (Madanchi et al., 2020) and antiviral peptide FuzeonTM (enfuvirtide) has been commercialized as an anti-HIV drug (Ashkenazi et al., 2011).
Due to the global spread of the COVID-19 (Figure 4A), the antiviral peptides against the coronavirus will be discussed in more detail. Coronaviruses (CoVs) belong to the family Coronaviridae; they are enveloped viruses with a positive-sense single-stranded RNA genome and have a helical symmetry (Franks and Galvin, 2014). CoVs, including severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (Mustafa et al., 2018), and the recent outbreak of COVID-19 have caused serious threats to human life and property. CoVs can cause life-threatening respiratory diseases and the viral particle is formed by spike glycoprotein (S), the envelope (E), the membrane (M), and the nucleocapsid (N) (Vilas Boas et al., 2019). It should be noted that their infectivity requires viral spike (S) protein. Fusion inhibitor peptides combine with the S protein to interfere with its folding and prevent infection. Besides, the S2 domain of the SARS-CoV S protein contains heptad repeat HR1 and HR2 sequences. Peptide HR2 (HR2: SLTQINTTLLDLTYEMLSLQQVVKALNESYIDLKEL) and its lipid-binding peptide is highly similar or even identical to the near-membrane portion of S protein ferredoxin, which interferes with refolding into post-fusion fusion-catalyzing domains (FDs) (Du et al., 2009; Park and Gallagher, 2017). According to recent research, the lipopeptide EK1C4, derived from EK1 (SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL), is the most effective fusion inhibitor against COVID-19 S protein-mediated membrane fusion (Xia et al., 2020). Homology modeling and protein-peptide docking showed that temporin has potential therapeutic applications against MERS-CoV (Marimuthu et al., 2019). Two AMPs from the non-structural protein nsp10 of SARS-CoV, K12, and K29, can inhibit SARS-CoV replication (Ke et al., 2012). Furthermore, rhesus theta-defensin 1 (RTD-1) treated animals have a marked reduction in mortality in the presence of SARS-CoV while the peptide alone shows airway inflammation and the one possible mechanism of action for RTD-1 is immunomodulatory (Wohlford-Lenane et al., 2009). In general, AMPs against coronavirus can be roughly classified as i) peptides derived from HR1, HR2 and RBD subunits of the spike protein, ii) peptides derived from other AMPs, iii) Peptides derived from non-structural protein (Mustafa et al., 2018). Furthermore, molecular docking analysis indicated that peptides were employed to disrupt the interaction between COVID-19 and ACE2 (angiotensin-converting enzyme 2) to inhibit COVID-19 entrance in cells (Figure 4B) (Souza et al., 2020). Finally, it should be noted that this therapy lacks clinical trials and the main method of animal experiments is an intranasal administration. This reminds us that nasal drug delivery (NDD) is a potential therapy for AMPs as anti-coronavirus drugs. Besides, the antiviral database AVPdb2 includes numerous antiviral peptides.
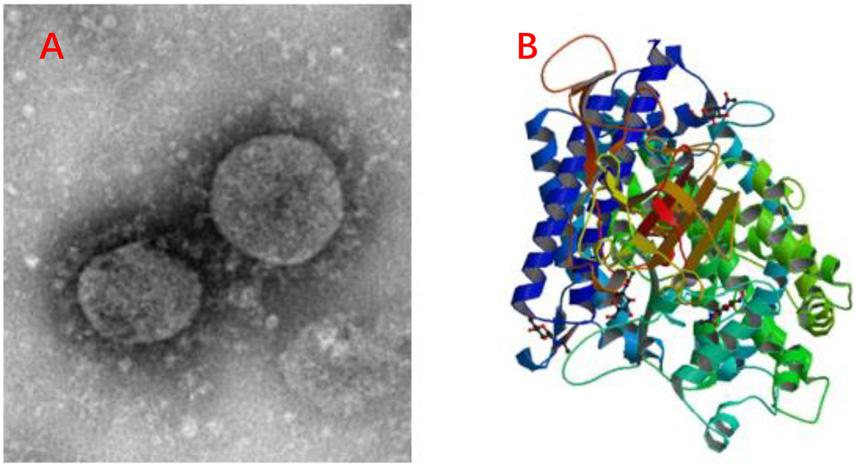
Figure 4. Information of COVID-19. (A) C-F13-nCoV Wuhan strain 02, Strain Number: CHPC 2020.00002; NPRC 2020.00002, Source: National Pathogen Resource Collection Center (National Institute for Viral Disease Control and Prevention under Chinese Center for Disease Control and Prevention). (B) Structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2. (10.2210/pdb6LZG/pdb).
Antiparasitic Peptides
Parasitic protozoa can cause diseases in human and animals through a variety of routes, including animal-to-person or person-to-person contact, water, soil, and food (Chalmers et al., 2020). And with the increase in parasite drug resistance, the need for new treatments has increased. Antiparasitic peptides show their killing effect on parasites which cause diseases such as malaria and leishmaniasis (Mangoni et al., 2005; Rhaiem and Houimel, 2016) and AMPs like cathelicidin, temporins-SHd show high inhibition activity against parasites (Abbassi et al., 2013). In recent research, Epi-1, a marine synthetic AMP, can remarkably inhibit Trichomonas vaginalis by destroying its membrane (Neshani et al., 2019). The peptide Jellein derived from bee royal jelly which has introduced above and 4-amino acid AMP KDEL (lysine, aspartic acid, glutamic acid, and leucine) has shown a significant effect on the Leishmania parasite (Cao et al., 2019; Zahedifard et al., 2020). However, it should be noted that their mechanisms are not the same. Cyanobacterial peptides differ from higher-eukaryote AMPs because their antiparasitic action depends on specific protein targets. Thus, these target parasites can be distinguished accurately even though they belong to the same family or genus (Rivas and Rojas, 2019).
Anticancer Peptides (ACPs)
The ACPs show anticancer mechanisms by (1) recruiting immune cells (such as dendritic cells) to kill tumor cells, (2) inducing the necrosis or apoptosis of cancer cells, (3) inhibiting angiogenesis to eliminate tumor nutrition and prevent metastasis, and (4) activating certain regulatory functional proteins to interfere with the gene transcription and translation of tumor cells (Wu D. et al., 2014; Ma et al., 2020). Tritrpticin and its analogs induce considerable toxicity toward Jurkat cells in vitro, whereas indolicidin and puroindoline A can also act as ACPs (Arias et al., 2020). It should be noted that both net charge and hydrophobicity play important roles in optimizing the anticancer activity of ACPs and they can constrain and influence each other. Thus, achieving a balance between net charge and hydrophobicity is important for better anticancer activity.
Besides the peptide mentioned above, anti-inflammatory, anti-diabetic peptides, spermicidal peptides etc. have been noticed, but they are not the same as antimicrobial peptides. Simply put, anti-inflammatory peptides decrease the release of inflammatory mediators and inflammatory cytokines (nitric oxide, interleukin-6, and interleukin-1β) and some of them also inhibit inflammatory signals like NF-κB, MAPK, and JAK-STAT pathways (Meram and Wu, 2017; Gao et al., 2020). Anti-diabetic peptides play their function by modulating the G protein-coupled receptor kinase (GRK 2/3) or activating glucagon-like peptide-1 (GLP-1), glucagon receptors (Marya et al., 2018; Graham et al., 2020). However, it is not accurate to classify these types of peptides as AMPs and bioactive peptides may be more convincing.
Classification of AMPs Based on Amino Acid-Rich Species
Proline-Rich Peptides (PrAMPs)
Proline is a typical non-polar amino acid. PrAMPs behave differently from other AMPs, that is, they enter bacterial cytoplasm by the inner membrane transporter SbmA instead of killing bacteria through membrane destruction (Mattiuzzo et al., 2007). Once in the cytoplasm, PrAMPs target ribosomes and block the binding of aminoacyl-tRNA to peptidyltransferase center or trap decoding release factors on the ribosome during the termination of translation to interfere with protein synthesis (Seefeldt et al., 2015). For instance, Tur1A, which is an orthologous AMP of bovine PrAMP Bac7 discovered from Tursiops truncatus, interferes with the transition from the initial phase to the extension phase of protein synthesis by binding to ribosomes. In addition, different PrAMPs lack a high sequence similarity but have short motifs containing repeating proline and arginine (Arg) residues (e.g., -PPXR- in Bac5 and -PRPX- in Bac7) (Mardirossian et al., 2018, 2019). Although PrAMPs mainly kill Gram-positive bacteria, pPR-AMP1, a proline-rich AMP identified from crab (Scylla paramamosain), exhibits antimicrobial activity against Gram-positive and Gram-negative bacteria (Imjongjirak et al., 2017). Besides, pieces of research have shown that PrAMPs have immunostimulation activity (Li W. et al., 2016).
Tryptophan- and Arginine-Rich Antimicrobial Peptides
Tryptophan (Trp), as a non-polar amino acid, has a remarkable effect on the interface region of the lipid bilayer, whereas Arg, as a basic amino acid, confers peptide charge and hydrogen bond interactions, which are essential properties to combine with the bacterial membrane’s abundant anionic component. And it seems that Trp residues play the role of natural aromatic activators of Arg-rich AMPs by ion-pair-π interactions (Walrant et al., 2020), thereby promoting enhanced peptide-membrane interactions (Chan et al., 2006). In addition to indolicidin and Triptrpticin which both are famous AMPs that rich in Arg and Trp residues. Octa 2 (RRWWRWWR) is also a typical Trp- and Arg-rich AMP that inhibits Gram-negative E. coli and Pseudomonas aeruginosa and Gram-positive S. aureus. And short Trp- and Arg-rich AMPs designed based on bovine and murine lactoferricin have also shown strong inhibitory action against bacteria (Strøm et al., 2002; Bacalum et al., 2017).
Histidine-Rich Peptides
Histidine is a common basic amino acid, and histidine-rich AMPs show good membrane permeation activity. HV2 is a histidine-rich AMP designed based on RR(XH)2XDPGX(YH)2RR–NH2 (where X represents I, W, V, and F). This peptide increases the permeability of bacterial cell membranes to cause cell membrane rupture and death. In addition, HV2 inhibits bacterial movement in a concentration-dependent manner and shows a strong anti-inflammatory effect by inhibiting the production of tumor necrosis factor α (TNF-α) (Dong et al., 2019). An AMP designed based on Octa 2 has shown good therapeutic potential by replacing its Arg residues with histidine (Bacalum et al., 2017). Furthermore, L4H4, which is designed based on the linear cationic amphiphilic peptide magainin, also shows good antibacterial activity and cell penetration properties by inserting four histidine sequences in leucine and alanine (Lointier et al., 2020).
Glycine-Rich Antimicrobial Peptides
The R group of glycine is generally classified as a non-polar amino acid in biology. Glycine-rich AMPs, such as attacins and diptericins, widely exist in nature (Lee et al., 2001; Kwon et al., 2008). These peptides contain 14% to 22% glycine residues, which have an important effect on the tertiary structure of the peptide chain. A glycine-rich AMP derived from salmonid cathelicidins activates phagocyte-mediated microbicidal mechanisms, which differ from the mechanism of conventional AMPs (D’Este et al., 2016). Furthermore, the glycine-rich central–symmetrical GG3 is an ideal commercial drug candidate against clinical Gram-negative bacteria (Wang et al., 2015).
Classification Based on Antimicrobial Peptide Structures
Antimicrobial peptides can be divided into four categories based on their structures including linear α-helical peptides, β-sheet peptides, linear extension structure, and both α-helix and β-sheet peptides (Figure 5) (Lei et al., 2019). Moreover, progressively cyclic peptides and AMPs with more complex topologies (including lasso peptides and thioether bridged structures) are reported (Koehbach and Craik, 2019).
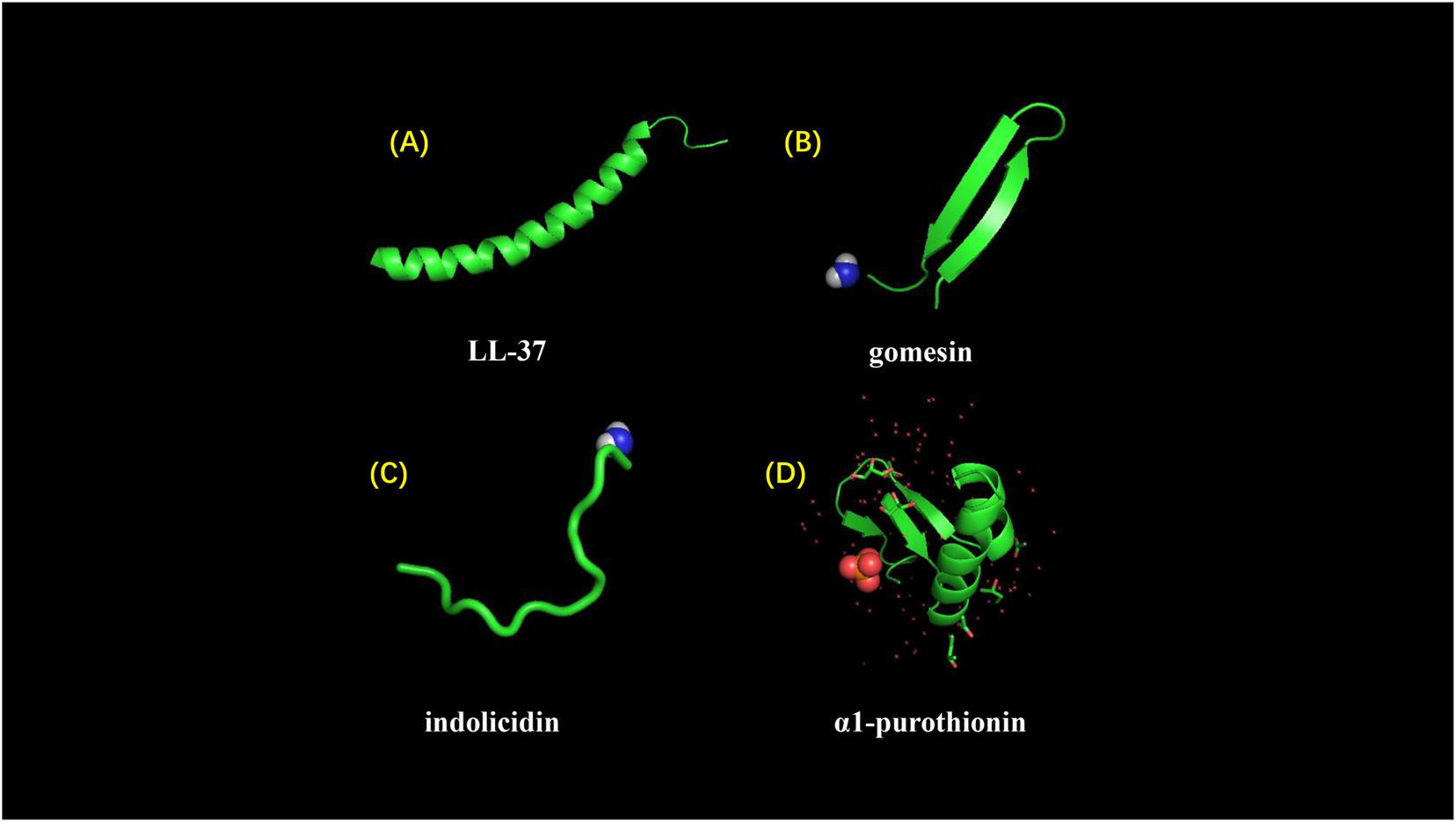
Figure 5. Different structures of AMPs. (A) LL-37 adopts a typical α-helical conformation (10.2210/pdb2K6O/pdb). (B) Gomesin is a β-sheet peptide and stabilized by disulfide bonds (10.2210/pdb1KFP/pdb). (C) Indolicidin is a AMP with linear extension structure instead of well-defined 3D structure (10.2210/pdb1G89/pdb). (D) α1-purothionin adopts both alpha-helix and beta-sheet conformation, and arrows indicate extension direction (10.2210/pdb2plh/pdb).
Antimicrobial Peptide Action Mechanism
Membrane Targeting Mechanism
The membrane-targeting mechanisms of AMPs can be described through models, including the pole and carpet models and the pole model can be further divided into the toroidal pore and barrel-stave models (Figure 6).
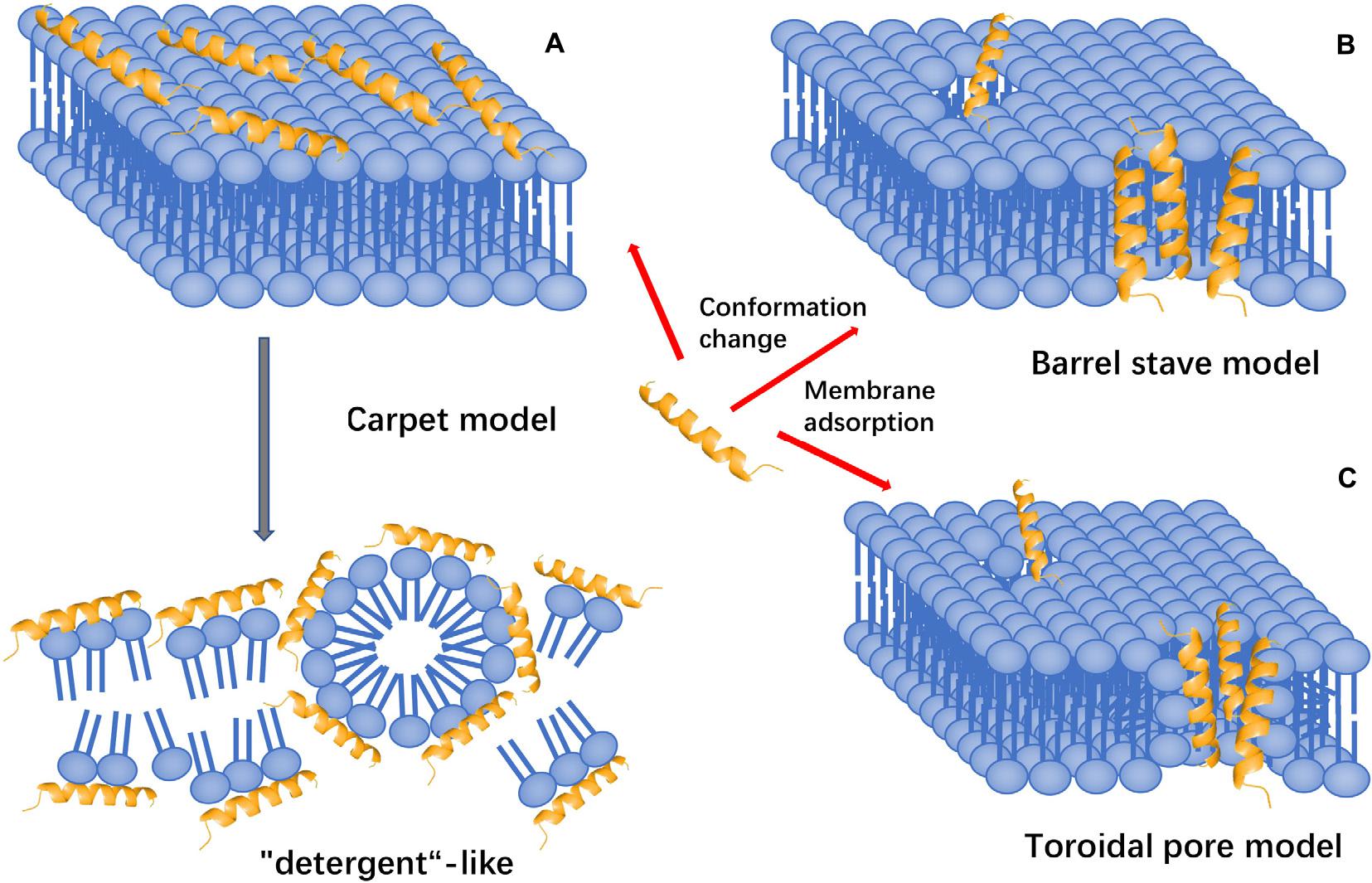
Figure 6. Models of action for extracellular AMP activity. (A) Carpet model: accumulation of AMPs on the surface and then destroy the cell membrane in the manner of “detergent”. (B) Barrel stave model: AMPs aggregate with each other and are inserted into the bilayer of the cell membrane in the form of multimers and arrange parallel to the phospholipids, then form a channel. (C) Toroidal pore model: accumulation of AMPs vertically embed in the cell membrane, and then, bend to form a ring hole.
The Toroidal Pore Model
The toroidal pore model is also known as the wormhole model. In this model, AMPs vertically embedded in the cell membrane accumulate and then bend to form a ring hole with a diameter of 1–2 nm (Matsuzaki et al., 1995, 1996). The typical examples of this model are magainin 2, lacticin Q, and arenicin. Furthermore, cationic peptides, including TC19, TC84, and BP2, compromise the membrane barrier by creating fluid domains (Omardien et al., 2018).
Barrel-Stave Model
Antimicrobial peptides aggregate with each other, penetrate the bilayer of the cell membrane in the form of multimers, and form channels that result in the cytoplasmic outflow. In severe cases, AMPs can induce cell membrane collapse and lead to cell death (Lohner and Prossnigg, 2009). For instance, Alamethicin performs its pore-forming activity by using this model. Besides, hairpin AMP protegrin-1 can form stable octameric β-barrels and tetrameric arcs (half barrels) in implicit and explicit membranes by simulations (Lipkin and Lazaridis, 2015).
Carpet-Like Model
Antimicrobial peptides are arranged parallel to the cell membrane. Their hydrophilic end faces the solution, and their hydrophobic end faces the phospholipid bilayer. AMPs will cover the membrane surface that similar to a carpet and destroy the cell membrane in a ‘detergent’-like manner (Oren and Shai, 1998). However, this pore-forming mechanism requires a certain concentration threshold and the required concentration of AMPs is high. Human cathelicidin LL-37 exhibits its activity through this mechanism, and AMPs with β-sheet structure also play a role in this model (Shenkarev et al., 2011; Corrêa et al., 2019). Polarized light-attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR) was used to study the effect of AMP cecropin P1 on the bacterial cell membrane and found that it was an applied flat on the surface of the pathogen’s cell membrane to destabilize and eventually destroy the cell membrane (Lyu et al., 2019).
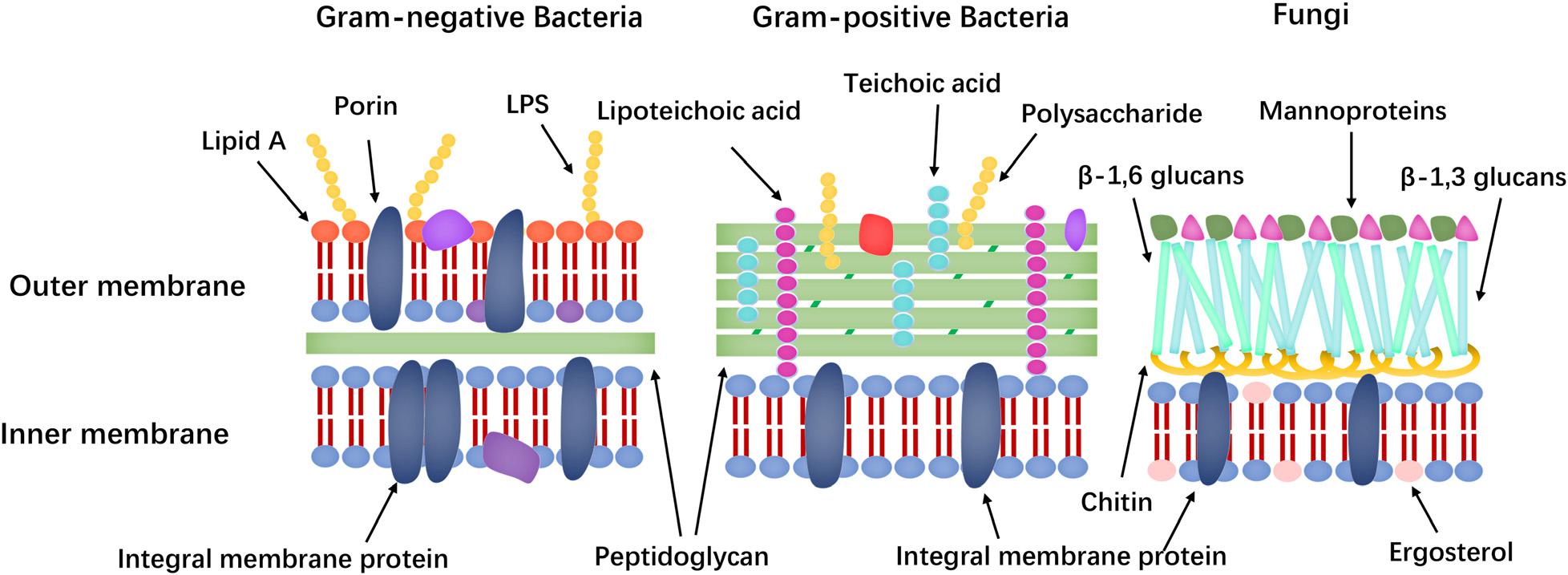
Membrane targeting mechanisms (the cell membrane composition differences of bacteria and fungi shown in Figure 7) can be further refined to address the large differences in the lipid composition of the cell membranes of bacteria, fungi, and mammals. The main lipids in cell membranes include glycerophospholipids (GPLs), lysolipids, sphingolipids, and sterols. Phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL) are the most common anionic lipids in bacteria, whereas phosphatidylcholine (PC), phosphatidylinositol (PI), PE, and phosphatidic acid (PA) are the main GPLs in fungal cell membranes (Ejsing et al., 2009; Singh and Prasad, 2011; Li et al., 2017). Furthermore, fungal cell membranes are more anionic than mammalian cell membranes and have higher PC content. Meanwhile, ergosterol is the sterol found in the plasma membrane of lower eukaryotes, such as fungi, whereas that of animals contains cholesterol (Faruck et al., 2016). Many AMPs take advantage of differences in membrane components to exert their effects.
Antimicrobial peptides are promising to be anti-biofilm agents but it should be noticed that they are different from the cell penetrating peptides (CPPs) which typically comprise 5–30 amino acids and can translocate across the cell membrane. CPPs could be categorized according to physicochemical properties into three classes: Cationic, amphipathic, and hydrophobic, but anti-biofilm peptides have stricter requirements for these physicochemical properties. Anti-biofilm peptides target the biofilms by different mechanisms including (1) degradation of signals within biofilms; (2) permeabilize within cytoplasmic membrane/EPS; (3) modulating EPS production etc. and then can address chronic multi-resistant bacterial infections (Pletzer et al., 2016; Ribeiro et al., 2016; Guidotti et al., 2017; Derakhshankhah and Jafari, 2018; Rajput and Kumar, 2018). For instance, SAAP-148, synthesized based on LL-37, showed activity to prevent biofilm formation by S. aureus and A. baumannii (Crunkhorn, 2018).
Non-membrane Targeting Mechanism
The way of AMPs entering cells is direct penetration or endocytosis. After entering the cytoplasm, AMPs will identify and act on the target. Depending on the target, AMPs can be divided into the following categories.
Inhibition of Protein Biosynthesis
Antimicrobial peptides affect transcription, translation, and assembly into functional peptides through molecular chaperone folding by interfering with related enzymes and effector molecules. For example, Bac7 1–35 targets ribosomes to inhibit protein translation (Mardirossian et al., 2014), whereas Tur1A inhibits protein synthesis in E. coli and Thermus thermophilus by inhibiting the transition from the initial phase to the extension phase. However, the differences between Tur1A and Bac7 also lead to various ways of binding to ribosomes and interacting with the ribosomal peptide exit tunnel (Mardirossian et al., 2018). But some AMPs’ have different targets. For instance, genome-wide transcription shows that the AMP DM3 can affect many important intracellular pathways of protein biosynthesis (Le et al., 2016).
Chaperones are key proteins for correctly folding and assembling newly synthesized proteins and make them have stereoisomerism, which makes AMPs have cell selectivity and can prevent cytotoxicity. According to a previous review: both pyrhocoricin and drosocin can prevent DnaK from refolding misfolded proteins by inducing a permanent closure of the DnaK peptide-binding cavity (Kragol et al., 2001; Le et al., 2017; Wrońska and Boguś, 2020).
Inhibition of Nucleic Acid Biosynthesis
Antimicrobial peptides can affect key enzymes or induce the degradation of nucleic acid molecules to inhibit nucleic acid biosynthesis. Indolicidin, a C-terminal-amidated cationic Trp-rich AMP with 13 amino acids, specifically targets the abasic site of DNA to crosslink single- or double-stranded DNA and it can also inhibit DNA topoisomerase I (Subbalakshmi and Sitaram, 1998). TFP (Tissue factor pathway inhibitor)1-1TC24, which is an AMP from tongues, enters the cytoplasm of target cells after the rupture of cell membrane and then degrades DNA and RNA (He et al., 2017).
Inhibition of Protease Activity
Many AMPs can inhibit various metabolic activities by inhibiting protease activity. For example, histatin 5 has a strong inhibitory effect on the proteases secreted by the host and bacteria. AMPs eNAP-2 and indolicidin inhibit microbial serine proteases, elastase, and chymotrypsin (Le et al., 2017). Cathelicidin-BF is a peptide isolated from the venom of Bungarus fasciatus, it can effectively inhibit thrombin-induced platelet aggregation and further block protease-activated receptor 4 (Shu et al., 2019).
Inhibition of Cell Division
Antimicrobial peptides inhibit cell division by inhibiting DNA replication and DNA damage response (SOS response), blocking the cell cycle or causing the failure of chromosome separation (Lutkenhaus, 1990). For instance, APP (GLARALTRLLRQLTRQLTRA), which is an AMP with 20 amino acid residues, can efficiently kill C. albicans because of its cell-penetrating efficiency, strong DNA-binding affinity, and ability to induce S-phase arrest in intracellular environment (Li L. et al., 2016). MciZ, which has 40 amino acid residues, is an effective inhibitor of bacterial cell division, Z-ring formation, and localization (Cruz et al., 2020).
Moreover, it has been reported that several AFPs have damaging effects on the organelles of fungi. For example, Histintin 5 can interact with mitochondria, causing the production of ROS, and inducing cell death (Helmerhorst et al., 2001).
In addition to intracellular targets, differences in cell wall composition, such as lipopolysaccharide (LPS), lipid A and mannoproteins, are potential targets for AMPs. Specifically, Gram-positive and Gram-negative bacteria are classified based on their bacterial cell wall structure. Gram-positive bacteria have a layer of cross-linked peptidoglycan, whereas Gram-negative bacteria have an additional outer membrane with an inner leaflet containing only phosphatidic acid and an outer leaflet made of LPS. LPS has numerous negatively charged phosphate groups, which combine with a salt bridge with a divalent cation (e.g., Ca2+ and Mg2+) to form an electrostatic network (Nikaido, 2003). This electrostatic zone is the main barrier against hydrophobic antibiotics and causes the low permeability of Gram-negative bacteria. The main components of the fungal cell wall are mannoprotein, β-glucans and chitin (polymers of 1,4-β-N-acetylglucosamine) and the mutations in the relevant genes of the LPS pathway and phospholipid trafficking provide resistance to the AMPs (Cabib, 2009; Spohn et al., 2019). Mannoproteins in fungal cell walls include a variety of proteins, including structural proteins, cell adhesion proteins (floccrin and lectin) and enzymes involved in cell wall synthesis and remodeling (hydrolytic enzymes and transglycosylase). These proteins differ from human cell membrane proteins and are potential targets of AFPs (Rautenbach et al., 2016). Furthermore, teichoic acid and lipoteichoic acid in the cell wall are also potential targets of AMPs and these theories could support the design of AMPs with low cytotoxicity.
Design Methods of Antimicrobial Peptides
Antimicrobial peptides have good application prospects. However, AMPs have the following problems. (1) AMPs damage the cell membrane of eukaryotes and cause hemolytic side effects; (2) rising production costs and technical problems limit their manufacture; (3) their stability is limited at certain pH; (4) AMPs have reduced activity under the presence of iron and certain serum; (5) AMPs are easily hydrolyzed by proteases. Therefore, the ideal AMP should meet the following characteristics: (i) high antimicrobial activity; (ii) low toxicity to mammalian membranes; (iii) high protease and environment stability; (iv) low serum binding capacity and (v) ease of access and low cost production (Li et al., 2017). Therefore, designing AMPs to achieve the desired effect has attracted increasing attention. The rational design of antibacterial peptides should focus on the following five aspects: chain length, secondary structure, net charge, hydrophobicity, and amphiphilicity and these have been mentioned in many studies and this review will focus more on several specific methods of antimicrobial peptide design.
Site-Directed Mutation
Site-directed mutation refers to the redesign of natural antimicrobial peptides by adding, deleting or replacing one, or several amino acid residues (Torres et al., 2019).
De novo Design Peptides
The de novo design of peptides attaches importance to the design of amphiphilic AMPs (Guha et al., 2019). For example, GALA is a well-known de novo-designed AMP. Amphipathic α-helical peptide GALA is created by placing protonatable glutamic acid residues in most positions with the spacing of i to i + 4 (Goormaghtigh et al., 1991). The repeated sequence (XXYY)n, where X1 and X2 are hydrophobic amino acids, Y1 and Y2 are cationic amino acids, and n is the number of repeat units, is designed based on the hydrophobicity cycle that mimics natural α-helical AMPs and successfully designs broad-spectrum α-helical AMPs. Sequences (LKKL)3 and (WKKW)2.5 have the highest selectivity (Khara et al., 2017). Moreover, LlKmW2 model peptides are also de novo-designed peptides. Amphipathic helical properties were conferred by using leucines and lysines, and two tryptophan residues were positioned at the amphipathic interface between the hydrophilic ending side and the hydrophobic starting side. Among the model peptides, L4K5W2 has good anti-MRSA activity (Lee et al., 2011).
Template-Based Design Method
Sequence templates can be obtained by comparing a large number of structurally homologous fragments of natural AMPs (such as HDPs) and extracting conservative patterns based on the type of residue (such as charged, polar, hydrophobic, etc.) (Zelezetsky and Tossi, 2006). Based on the modification, the parameters, such as helix formation tendency, cationic, amphiphilicity and overall hydrophobicity, can be systematically changed. For instance, cecropin, magainin, protegrin, and lactoferrin have all been used as AMP templates (Fjell et al., 2012).
Based on the Self-Assembly of Antimicrobial Peptides
Peptides can form nanostructures, such as micelles, vesicles, nanotubes, nanoparticle nanobelt, and nanofibre nanotube, and can increase or impart antibacterial activity to AMPs during the self-assembly of peptides. For example, KLD-12 (KLD) is a self-assembling peptide with 12 amino acid residues that can adopt nanostructures and are known for their tissue engineering properties. The addition of Arg residues in KLD shows no remarkable change in its β-sheet secondary structure and the self-assembly characteristics of the forming nanostructures (Tripathi et al., 2015). Dimer structure can also be used to enhance the antimicrobial activity of AMPs and reduce toxicity, but membrane-destabilizing effects are reduced after dimer formation (Malekkhaiat Häffner and Malmsten, 2018).
Chemical Modification
Various chemical modifications of AMPs, including residue phosphorylation, the addition of D-amino acids or unnatural amino acids (homoarginine), cyclization, halogenation, acetylation, and peptidomimetics, have been used to improve the stability of peptides against proteases. Given that the enzyme is stereospecific, the incorporation of unnatural D-amino acids into the AMP sequence can reverse the stereochemistry and prevent protease degradation (Zhong et al., 2020). The so-called peptidomimetics, whose main elements mimic the structure of peptides, are usually produced by modifications, such as chain extension or heteroatom incorporation of existing peptides (Patch and Barron, 2002). Ornine, which is an unnatural residue with a positive charge and has a high resistance to protease activity, is also used in non-chemical modification. Replacing Trp residues with family residues, such as β-dihydrophenylalanine, can stabilize secondary structures and improve antibacterial properties (Maurya et al., 2013).
Halogenation
Halogenation is highly related to the activity, specificity, and stability of AMPs. In the latest report, Halogen is introduced into jelleine-I which is a short peptide isolated from the royal jelly of honeybees (Apis mellifera) by replacing phenylalanine with a halogenated phenylalanine analog, increasing the antibacterial activity in vitro and anti-biofilm activity. In addition, the proteolytic stability of jelleine-1 is increased by 10–100 times by halogenation (Jia et al., 2019). The halogenated peptidomimetic α,α-disubstituted β-amino amides are also promising bacteriostatic drugs that have inhibitory effects on more than 30 multi-resistant clinical isolates of Gram-positive and Gram-negative bacteria (Paulsen et al., 2019). Halogenation is also related to the specificity of AMPs. The o-fluorine substitution in phenylalanine residues maintains the activity of temporin L on E. coli but leads to the loss of activity on S. aureus and P. aeruginosa (Setty et al., 2017).
Cyclisation
Three modes of cyclisation, including cyclisation via disulfide bonds, head-to-tail cyclisation and internal bonding between side chains, have been found in natural AMPs. The synthesis of disulfide bonds often complicates the development of synthetic peptides. The circularisation of the main chain of arenicin-1 molecule resulted in increased activity against drug-resistant clinical isolates but caused no substantial effect on cytotoxicity (Orlov et al., 2019). The HDPs tachyplesins I, II, and III and their cyclic analogs cTI, cTII, and cTIII, respectively, have similar structures and activities and can resist bacterial and cancer cells. The cyclisation of the backbone reduces the hemolytic activity and improves the stability of the peptides whilst maintaining effective anticancer and antibacterial activities (Vernen et al., 2019).
Capping
Capping refers to the addition of specific motifs or modifications, such as amidation at the C-terminus and acetylation at the N-terminus, rendering AMPs with more natural peptide characteristics. Post-translational modifications play an important role in the function of AMPs and are the most commonly used in peptide design. The C-terminal Rana box (consisting of a C-terminal cyclic heptapeptide with a conservative disulfide bond) and amide group are important C-terminal capping methods. For example, the C-terminal amide group of maximin H5 can enhance antibacterial efficacy without increasing lytic ability (Dennison et al., 2015). The N-terminal lipidated analog C4VG16KRKP shows enhanced antibacterial activity against various Gram-negative bacteria. The functions of N-terminal lipidation include (i) increasing LPS neutralization, (ii) increasing stability to proteases and peptidases, and (iii) reducing cytotoxicity (Datta et al., 2016). Furthermore, hydrophobic end labeling is a commonly used method to increase the activity of antimicrobial peptides. Acyl lipid peptides have a linear or cyclic structure in which one or more hydrocarbon tails are connected to the N-terminus of a short oligopeptide (Chu-Kung et al., 2010). Lipopeptides have covalently attached hydrophobic moieties, such as sterols or fatty acids. Aromatic amino acid terminal labeling is also the main hydrophobic terminal labeling method. Tryptophan (W) and phenylalanine (F) are the commonly used aromatic amino acids. Their large and polarisable residues have an affinity for the interface, and the W/F tag is also sensitive to the differences between ergosterol and cholesterol and can prevent self-assembly. This condition results in low aggregation numbers and high critical aggregation concentrations (Schmidtchen et al., 2014).
Conjugation
Peptide conjugation has been the goal of most research in recent years to produce active and stable AMPs with high selectivity. Different side chains or AMP fragments can be used aside from the repetition of the same amino acid motifs. For example, conjugating fatty acids with a length of 8–12 carbon atoms to the 4th or 7th side chain of the D-amino acids of Ano-D4,7 improves antibacterial selectivity and anti-biofilm activity. In addition, the new peptide exhibits high stability against trypsin, serum, salt, and different pH environments (Zhong et al., 2020). The conjugation of different AMPs can also be performed. For example, the hybrid peptide (PA2–GNU7) constructed by the addition of PA2 to GNU7 has a high activity and specificity to P. aeruginosa (Kim et al., 2020).
Synthetic Mimics of AMPs (SMAMPs)
SMAMPs include a broad family of molecular entities based on the structure and function of AMPs. However, their backbones are not entirely based on α-amino acids, including β-amino acid oligomers, arylamide oligomers, and phenylene ethynylenes (Michael Henderson and Lee, 2013). For instance, SMAMP10, which is a potential drug for intravenous treatment, causes no drug resistance and has a strong inhibitory effect on MRSA and vancomycin-resistant Enterococcus faecium (Tew et al., 2006).
Peptoids
Peptoids are peptide isomers, in which the side chain is bonded to the main chain nitrogen instead of α-carbon or poly-N-substituted glycine in which the side chain is connected to amide nitrogen instead of the α-carbon on the main chain (Andreev et al., 2018). For example, the cationic peptide SA4 (IOWAGOLFOLFO-NH2) and its poly-N-substituted glycine homolog SPO (nInOnWnAnGnOnLnFnOnLnFnO-NH2) inhibit the planktonic and biofilm formation of A. baumannii strains, which are susceptible to multi-drug resistance (Sharma et al., 2019).
Use of Motifs
Motifs with specific functions have been reported increasingly. These motifs can be repeated units for combining into new antimicrobial peptides, or specific amino acid combination units appearing at the end (such as capping) of or even in the peptide chain.
Motif at the End of the Chain
ATCUN motif
This motif includes two tripeptide structures, including Gly–Gly–His or Val–Ile–His, which are added at the end of the peptide chain. ATCUN-containing AMPs in the presence of hydrogen peroxide and ascorbic acid combine with Cu2+ to induce the valence of copper ions between +2 and +3 oxidation states and form an ATCUN–Cu (II) complex, generating ROS by Fenton-like reactions. Extracellular polymeric substances (EPS) are important for biofilms and can enhance the resistance of cells to antibacterial agents (Flemming, 2016). ATCUN–AMPs have been used to degrade environmental DNA, which is one of the major components of EPS. Several related practical applications have been reported. For example, the biological activity against carbapenem-resistant Enterobacteriaceae is increased by adding this motif to the N-terminus of an alpha-helical AMP (such as CM15). Besides, the Cu–ATCUN derivative of OV-3 containing a C-terminal GGC sequence showed high levels of membrane permeation and lipid peroxidation. The concept of catalytic metal drugs has attracted widespread attention although the concept is still in its infancy because of the role of metal ions (Alexander et al., 2017; Agbale et al., 2019).
Rana box
Rana box: Rana box is a heptapeptide motif (CGLXGLC) from the nigrocin family. Rana box consists of two cysteine residues that are separated by four or five other residues on the side and can form a cyclic disulfide bond. Rana box peptide has shown structural analogies with polymyxin (colistin), and the primary structure of the Rana box motif is important in determining bacteriostatic activity (Kozić et al., 2015). The deletion of the ‘Rana box’ motif will cause the AMP antibacterial effect to disappear, but replacing the natural ‘Rana box’ sequence of AMPs with amidated phenylalanine can expand its efficacy against antibiotic-resistant microorganisms, including MRSA and P. aeruginosa, and reduce cytotoxicity. This phenomenon also shows that the effect of the motif on AMPs needs to be determined based on the specific situation and is not completely beneficial (Bao et al., 2018).
LPS binding motif
The LPS binding motif (G-WKRKRF-G) can produce a broad spectrum of antibacterial activity when introduced into the C-terminus of temporin-1 Ta and temporin-1 Tb (close isoforms of temporin) (Mohanram and Bhattacharjya, 2016).
γ-core motif
Antifungal Peptides have a conserved GXC(X3–9) C γ-core motif (residues 5–14, GKCYKKDNIC; d-isomer) at its N-terminus, which is a cation part of the ring. This conserved motif interferes with the integrity of the plasma membrane of the cell (Yount and Yeaman, 2004). Conserved γ-core motifs are directly involved in protein–membrane interactions and strongly contribute to membrane binding (Utesch et al., 2018).
Motif in the Chain
If replace d-Phe1-Pro2 sequence in peptide chain with d-Phe-2-Abz turn motif (2-Abz is an abbreviation of 2-aminobenzoic acid D-amino acid) in AMP Tyrc A, and nuclear magnetic resonance shows that this change retains the β-hairpin structure. Unlike the traditional β-turn motif, the D-Phe-2-Abz motif can be used as a tool for β-hairpin libraries. The hydrophobic peptide can be formed into the nucleated β-hairpin formation by adding the D-Phe-2-Abz motif. Moreover, the inclusion of this part in two designed cationic amphiphilic peptides can produce broad-spectrum antibacterial activity and low hemolysis rate (Cameron et al., 2017; Cameron et al., 2018).
NGR motif
The NGR motif is composed of Asn–Gly–Arg, and AMPs with this structure have strong cytotoxicity (Table 1). The data indicate that the new AMPs containing NGR may bind to CD13+ or αvβ3+ tumor cells by binding to CD13 or αvβ3, respectively, to exert anti-tumor activity, especially on CD13+ tumor cells (Zhang et al., 2015).
“Glycine zipper” of GxxxG motifs
The central GxxxG motif can induce strong self-assembly and have been already used in the design of AMPs (Brosig and Langosch, 1998; Krauson et al., 2012).
Motif-Based Polyvalent Peptide Synthesis (Dimers, Tetramers, etc.)
Bovine lactoferrin B is an AMP composed of 25 amino acid residues and has antibacterial, antifungal, and antiparasitic activities. The multivalent molecules LfcinB (20–25)2 and LfcinB (20–25)4 contain the LfcinB (20–25) motif (RRWQWR) and show inhibition activity against E. coli, P. aeruginosa, and S. aureus. Chimeric peptide chimera 3 containing two motifs, namely, the RRWQWR of LfcinB (20–25) and the RLLR of BFII (32–35), shows high antibacterial activity against E. coli ATCC 25922 and S. aureus ATCC 25923 (Vargas-Casanova et al., 2019; Pineda-Castañeda et al., 2020).
Computer Design
Computer design includes simple statistical modeling, Structure-activity relationships study (Abdel Monaim et al., 2018), neural networks (Müller et al., 2018), deep learning (Veltri et al., 2018), word embedding (Hamid and Friedberg, 2018) and machine learning. For example, a machine learning method by Matlab is proposed based on the concept of scoring the contribution of each amino acid’s antibacterial activity (Wu X. et al., 2014). The genetic algorithm was used to design the amphiphilic α-helical peptide guavalin 2, which has an uncommon amino acid composition (three tyrosine and three glutamine residues) and interestingly causes membrane hyperpolarization, which is a different mechanism from those of other AMPs (Porto et al., 2018). Two research methods have been developed based on the research background of quantitative structure–activity relationships: prediction method based on AMP therapeutic index and the identification of novel potential AMPs from the expressed sequence tag database based on the principles of the highly conserved signal peptide subclasses related to AMPs (Juretić et al., 2011).
Rational Library Design
In this way, a variety of AMP variants can be obtained. If combined with high-throughput screening, it can effectively obtain the desired AMP. For instance, some new AMPs are designed by the combinatorial peptide library of melittin and show higher activity and lower cytotoxicity (Krauson et al., 2015).
Environmental Factors Affecting the Activity of Antimicrobial Peptides
Metal Ions
Cations, such as Na+ and Mg2+, may affect AMP activity (Zhu et al., 2015). However, the different valences of metal ions have varied effects on AMPs. For example, divalent cations show stronger antagonism to bacteria than monovalent cations with thanatin and s-thanatin, which are insect AMPs (Wu et al., 2008). In the presence of NaCl, the signal response during the association phase remarkably decreased in single-cycle and multi-cycle kinetic experiments, resulting in a decreased association rate. This occurrence may be caused by the shielding effect of NaCl between the cationic peptide and the zwitterionic membrane. Another possible reason is that Na+ can bind to the phospholipid bilayer, where the ions interact with the phosphate and the carbonyl oxygen of lipid head groups (Sabapathy et al., 2020). The reduced activity of synthetic peptide [RLLR]5 under high salt concentration is possibly caused by the destruction of its α-helix structure.
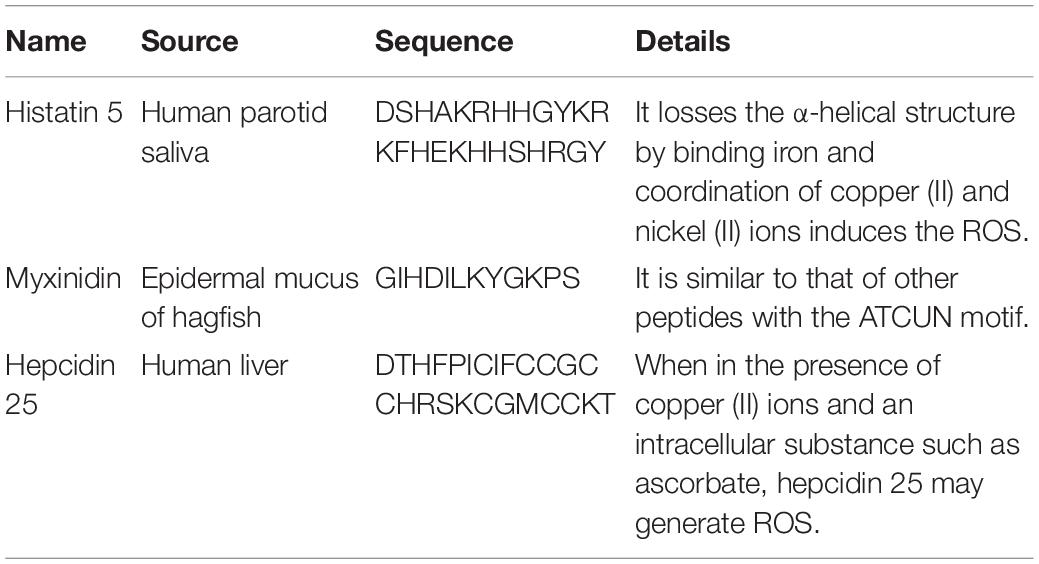
Table 2 shows that several AMPs, including histatin, myxinidin, and hepcidin, contain ATCUN motifs (Amino Terminal Copper and Nickel with XXH sequence). Iron is the most abundant metal ion in human saliva, but the combination with this metal ion results in the loss of the α-helix of histatin 5 and greatly reduces its antifungal activity (Puri et al., 2015). However, the coordination of copper (II) and nickel (II) ions can induce the formation of ROS, which is essential for bactericidal activity (Jeżowska-Bojczuk and Stokowa-Sołtys, 2018).
Anionic AMPs have a large number of negatively charged aspartic and glutamic acid residues (Lakshmaiah Narayana and Chen, 2015). They require zinc as a functional cofactor and the zinc complex shows stronger antibacterial activity (Jiang et al., 2014). Several of these AMPs use metal ions to form cationic salt bridges with the negatively charged components of the microbial membrane to penetrate the membrane. Anionic AMPs may attach to ribosomes or inhibit ribonuclease activity when in the cytoplasm (Jeżowska-Bojczuk and Stokowa-Sołtys, 2018).
Metal ions also affect the self-assembly of peptides. These ions can recognize specific amino acids, such as lysine and glutamic acid, and may form salt bridges between peptide molecules to induce peptide self-assembly. For example, Zn2+ can stabilize the aggregation of peptides on the cell membrane, which results in the enhanced antibacterial effect of DCD-1L in the presence of Zn2+ (Tian et al., 2015).
pH
Many AMPs are stable and retain their antimicrobial activity in a wide pH range. AMPs have enhanced activity at low pH because of their basic properties. This condition is related to the protonation of histidine at acidic pH, which promotes electrostatic interactions with anionic surfaces, including LPS and the anions of phospholipids, and subsequently enhances antibacterial properties. The effect of pH on the antibacterial activity of AMPs varies. For example, thanatin’s activity at neutral pH is slightly higher than that under acidic conditions. By contrast, the activity of xylan on E. coli, Listeria, and C. albicans is remarkably higher at pH 5.5 than at pH 7.4 (Holdbrook et al., 2018). The inactivation of the histidine-containing AMP C18G-His under low pH conditions involves pH-dependent changes in the state of the aggregates in the solution, because the aggregates, which are sensitive to pH and lipid composition, may be affected by binding and conformation. Peptides can also enhance bacterial membrane permeability at low pH (Hitchner et al., 2019). Thrombin-derived C-terminal peptides (TCPs) will also change the mode of CD14 (a protein that is abundant in human plasma) from anti-inflammatory mode to bacterial elimination mode from pH 7.4 to pH 5.5 (Holdbrook et al., 2018). A dimer (e.g., P-113) can be created to provide AMPs with resistance to a higher pH range. The sensitivity of this pH-sensitive AMP can be used to achieve a certain targeting effect in practical applications. In addition, charge interaction is one of the most important factors in peptide self-assembly. pH affects the charge state of amino acid and substituent functional groups. Therefore, adjusting the pH is the most common method for controlling peptide assembly and disassembly (Tian et al., 2015).
Proteases
Proteases have a strong destructive effect on AMPs. For instance, LL-37, which has the strongest inhibitory effect on chlamydial infection, is inhibited by the protease chlamydial protease-like activity factor (CPAF) secreted by Chlamydia (Tang et al., 2015). Studies have been focused on the design of AMP carriers to solve this problem (Lewies et al., 2017; Nordström et al., 2019). The presence of chitosan–silica solid support of KR-12 peptide can protect it be hydrolyzed by α-trypsin, and the degree of protection is increased by 38% compared with the free KR-12 (Diosa et al., 2020). However, several enzymes, such as protease 65, esterase 66 and phosphatase 67, cut the blocking group of the peptide and trigger the self-assembly of the peptide, which positively affects AMPs (Tian et al., 2015).
Current Progress and Application of Antimicrobial Peptides
Medicine
Antimicrobial peptides can regulate pro-inflammatory reactions, recruit cells, stimulate the proliferation of cells, promote wound healing, modify gene expression and kill cancer cells to participate in the immune regulation of human skin, respiratory infections, and inflammatory diseases (de la Fuente-Núñez et al., 2017). For example, α-defensins HNP-1, HNP-2, and HNP-3 showed effective antibacterial activity against adenovirus, human papilloma virus, herpes virus, influenza virus and cytomegalovirus. Pulmonary diseases, such as idiopathic pulmonary fibrosis, alveolar proteinosis, and acute respiratory distress syndrome, show elevated levels of AMPs (Guaní-Guerra et al., 2010). Likewise, AMPs secreted by the Paneth cells in the mammalian gut are important to shape the gut microbiota (Bevins and Salzman, 2011).
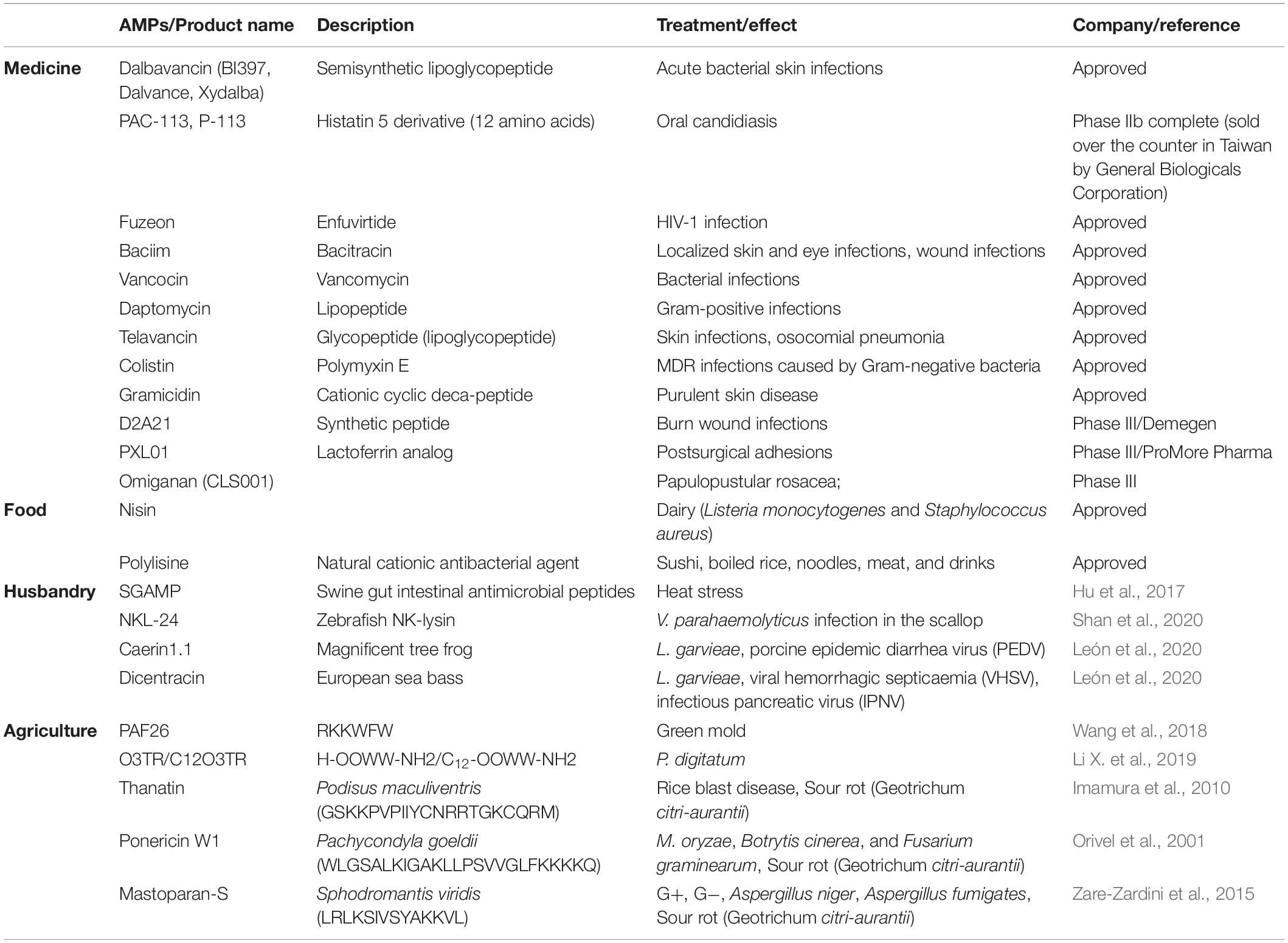
The application of AMPs in medicine, such as dental, surgical infection, wound healing and ophthalmology is developing now. But there are only three AMPs that have been approved by FDA including gramicidin, daptomycin, and colistin.
Dental caries, endodontic infections, candidiasis, and periodontal disease are common diseases in the human oral cavity. Dental caries is a prevalent oral disease and some acidogenic bacteria like Streptococcus sp. are the main caries-associated pathogens (Izadi et al., 2020). Several AMPs have good application potential. For instance, peptide ZXR-2 (FKIGGFIKKLWRSLLA) has shown potent activities against pathogenic bacteria of dental caries, Streptococcus mutans, Streptococcus sobrinus, and Porphyromonas gingivalis and peptide PAC-113 (Clinical trial identifier: NCT00659971) that has been sold over the counter in Taiwan for treating oral candidiasis (Chen L. et al., 2017).
In surgical infection and wound healing: surgical infection occurs after surgery, burns, accidental injury, skin disease, and chronic wound infections have a serious hazard to human life (Thapa et al., 2020). Several AMPs have shown the therapeutic potential of these diseases. For example, AMP PXL150 shows pronounced efficacy as an anti-infective agent in burn wounds in mice and AMP D2A21 has been in the third phase of clinical trials for treating burn wound infections (Björn et al., 2015).
In ophthalmology: Human eyes are prone to be infected by several organisms including bacteria and fungi in which S. aureus, Streptococcus pneumoniae, P. aeruginosa, Aspergillus spp., and C. albicans are the most relevant pathogens (Silva et al., 2013). Although AMPs such as Lactoferricin B, Protegrin-1 exhibited antimicrobial activity against these pathogenic bacteria, their application in the field of ophthalmology is only at the theoretical stage. With the popularity of contact lenses and the increase in cases of related eye infections, antimicrobial peptides have shown good application prospects in ophthalmology (Khan and Lee, 2020).
Additional methods need to be performed for the application of AMPs as drugs in medicine. The main strategies include (1) constructing precursors to reduce cytotoxicity and improve protease stability, (2) using AMPs in combination with existing antibacterial agents, (3) inducing the correct expression of AMPs with appropriate drugs and using engineering probiotics as vectors to express AMPs. For example, in the field of wound repair, different formulation strategies, such as loading AMPs in nanoparticles, hydrogels, creams, gels, ointments, or glutinous rice paper capsules, have been developed to effectively deliver AMPs to the wound (Borro et al., 2020; Thapa et al., 2020). In recent research, the sponges developed from modified starch and HS-PEG-SH are covalently immobilized with AMP showed effective antibacterial activity (Yang et al., 2019).
More technical means, including pheromone-labeled AMPs, local environment-triggered AMPs (enzyme precursor drug release system, pH-activated AMPs, etc.), have been developed to improve the targeting mechanism of AMPs. Furthermore, nanotubes, quantum dots, graphene, and metal nanoparticles have been proposed to be a potential method to enhance drug delivery of AMPs (Magana et al., 2020). Hybrid peptides have also been used to build targeting peptides. For example, PA2, which is a P. aeruginosa-targeting peptide, was combined with GNU7 (a broad-spectrum AMP) to construct a hybrid peptide (PA2–GNU7) that targets OprF protein and has good bactericidal activity and specificity (Kim et al., 2020). Furthermore, some antibiotics, for instance, daptomycin (a lipopeptide), lugdunin which is a 21-membered cyclic peptide consists of 6 amino acid residues plus a thiazolidine moiety and telavancin (a glycopeptide) have been widely used for the clinic (Durand et al., 2019; Lampejo, 2020). Although they are antibiotics, they have provided broader ideas for the design of AMPs.
Food
Food preservatives have potential harm to the human body. Therefore, natural preservatives are being advocated by more people. AMPs have a good inhibitory effect on common bacteria and fungi in food, and many AMPs are resistant to acids, alkalis, and high temperatures are easily hydrolyzed by proteases in the human body. Thus, AMPs are a promising alternative to preservatives. Nisin is a bacteriocin produced by L. lactis subspecies. Lactic acid bacteria have been widely used as food preservatives. Nisin is categorized as generally recognized as safe (GRAS) by the US Food and Drug Administration (FDA) and is used as a food preservative in other countries (Khan and Oh, 2016). However, only nisin and polylysine are currently approved by the FDA as food additives (Santos et al., 2018). Pedocin PA-1, a bacteriocin consisting of 44 amino acids produced by a diplococcus, is also used as a food preservative and is sold on the market under the trade name ALTA 2431. Pedocin PA-1 is used as a food additive to inhibit the growth of L. monocytogenes, which can cause meat deterioration (Settanni and Corsetti, 2008). Enterocin AS-48 is an AMP used to preserve cider, fruit and vegetable juices, and enterocin CCM4231 is used to preserve soy milk (Rai et al., 2016; Santos et al., 2018). Encapsulating bacteriocins into liposomes is a new method used to overcome the problems of AMPs in food applications (such as proteolytic degradation or interaction with food ingredients) (da Silva Malheiros et al., 2010).
Moreover, active packaging by adding AMPs is a novel packaging method that has great potential in the food industry. For instance, ε-poly-L-lysine is used in conjunction with starch biofilms to show good inhibitory effects on Aspergillus parasiticus (aflatoxin producer) and Penicillium expansum and nisin have the potential to be dairy preservative because it is a highly surface-active molecule (Luz et al., 2018).
Animal Husbandry and Aquaculture
The European Union banned the use of animal growth promoters in animal feed in 2006. Thus, a new antibacterial strategy is needed. Many AMPs are the potential to be used in poultry, swine, and ruminants breeding and aquaculture because they can improve production performance (Liu et al., 2008; Bao et al., 2009), immunity and promote intestinal health and some of them have a stronger inhibitory effect on bacterial inflammation if used with antibiotics (Wang et al., 2019; Cote et al., 2020). For example, SIAMP has a good effect on the treatment of IBV in chicken (Sun et al., 2010). By adding swine gut intestinal antimicrobial peptides (SGAMP), broilers showed higher average daily gain and feed efficiency under chronic heat stress conditions (Hu et al., 2017). Frog caerin 1.1, European sea bass dicentracin and NK-lysine peptides (NKLPs) have good inhibitory effects on Nodavirus, Septicaemia haemorrhagic virus, Infectious pancreatic necrosis virus and Spring viremia carp virus, which are devastating to fish farming (León et al., 2020). The AMP in soybean meal fermented by B. subtilis E20 effectively inhibits V. parahaemolyticus and Vibrio alginolyticus and enhances the resistance level of Litopenaeus vannamei against V. parahaemolyticus when added to feeds (Cheng et al., 2017).
Agriculture
For agriculture, the plant pathogenic infection of bacteria and fungi causes the loss of economy, for instance, Aspergillus flavus infection of corn and peanuts, citrus green mold caused by Penicillium digitatum, gray mold disease caused by Botrytis cinerea on strawberries and Geotrichum citri-aurantii infection of citrus fruit all cause great harm to the growth and post-harvest of agricultural products (Liu et al., 2007; Liu et al., 2019). Several AFPs have shown prospect to control these problems. However, the practical application of antimicrobial peptides in the transportation and preservation of agricultural products is still lacking, because the use of antimicrobial peptides will greatly increase the cost in the transportation of fruits and vegetables (application examples of AMPs in these four fields are shown in Table 3).
Conclusion
Antimicrobial peptides constitute a global research hotspot, but many key issues in design and application need to be solved urgently. Several restrictive factors hinder the application of AMPs. The interaction of multidisciplinary subjects, such as biology, materials science, chemistry, bioinformatics, molecular informatics and pharmacy can further develop prospective AMPs. Computer molecular dynamics simulation, cell membrane simulation, and more methods are being applied to study the mechanism of AMPs. How to further understand the correlation between AMPs and various targets instead of conducting one-sided experimental research might improve experimental designs to obtain stronger systemic and scientific demonstrations. On this basis, further animal experiments are required instead of simple cell-level experiments to test the effect of AMPs under complex physiological conditions. Several complicated methods, such as the chemical method of peptidomimetics and non-natural amino acid modifications, have been applied in designing AMPs to solve the problem of protease hydrolysis. Most methods use chemical substrates, but the cost of these methods cannot be ignored in practice. In addition, chemical synthesis and the use of engineered bacteria are currently the mainstream for such procedures. Finding a better biological preparation method, reducing the cost and increasing the yield is important problems in practical application. Furthermore, studying the AMP expression of the organism itself and finding a better expression vector are necessary for mass production in the future as more AMPs in nature are discovered. Further research is needed on the reported AMPs to solve the problem on structure–function relationship. As a branch of peptide drugs, AMPs need to progress with the advancement of medical science against the background of the current low success rate of the clinical application of AMPs. More attention can be focused on food, agriculture, and animal husbandry.
Author Contributions
QK and YH: conceptualization, methodology, writing – original draft preparation, and writing – review and editing. All authors contributed to writing and reviewing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National Key R&D Program of China (2019YFD0901705).
Footnotes
References
Abbassi, F., Raja, Z., Oury, B., Gazanion, E., Piesse, C., Sereno, D., et al. (2013). Antibacterial and leishmanicidal activities of temporin-SHd, a 17-residue long membrane-damaging peptide. Biochimie 95, 388–399. doi: 10.1016/j.biochi.2012.10.015
Abdel Monaim, S. A. H., Jad, Y. E., El-Faham, A., de la Torre, B. G., and Albericio, F. (2018). Teixobactin as a scaffold for unlimited new antimicrobial peptides: SAR study. Bioorgan. Med. Chem. 26, 2788–2796. doi: 10.1016/j.bmc.2017.09.040
Agbale, C. M., Sarfo, J. K., Galyuon, I. K., Juliano, S. A., Silva, G. G. O., Buccini, D. F., et al. (2019). Antimicrobial and antibiofilm activities of helical antimicrobial peptide sequences incorporating metal-binding motifs. Biochemistry 58, 3802–3812. doi: 10.1021/acs.biochem.9b00440
Alexander, J. L., Yu, Z., and Cowan, J. A. (2017). Amino terminal copper and nickel binding motif derivatives of ovispirin-3 display increased antimicrobial activity via lipid oxidation. J. Med. Chem. 60, 10047–10055. doi: 10.1021/acs.jmedchem.7b01117
Andreev, K., Martynowycz, M. W., Huang, M. L., Kuzmenko, I., Bu, W., Kirshenbaum, K., et al. (2018). Hydrophobic interactions modulate antimicrobial peptoid selectivity towards anionic lipid membranes. Biochim. Biophys. Acta Biomembr. 1860, 1414–1423. doi: 10.1016/j.bbamem.2018.03.021
Arias, M., Haney, E. F., Hilchie, A. L., Corcoran, J. A., Hyndman, M. E., Hancock, R. E. W., et al. (2020). Selective anticancer activity of synthetic peptides derived from the host defence peptide tritrpticin. Biochim. Biophys. Acta Biomembr. 1862:183228. doi: 10.1016/j.bbamem.2020.183228
Ashkenazi, A., Wexler-Cohen, Y., and Shai, Y. (2011). Multifaceted action of fuzeon as virus-cell membrane fusion inhibitor. Biochim. Biophys. Acta Biomembr. 1808, 2352–2358. doi: 10.1016/j.bbamem.2011.06.020
Ayukekbong, J. A., Ntemgwa, M., and Atabe, A. N. (2017). The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob. Resist. Infect. Control 6:47. doi: 10.1186/s13756-017-0208-x
Bacalum, M., Janosi, L., Zorila, F., Tepes, A.-M., Ionescu, C., Bogdan, E., et al. (2017). Modulating short tryptophan- and arginine-rich peptides activity by substitution with histidine. Biochim. Biophys. Acta Gen. Subj. 1861, 1844–1854. doi: 10.1016/j.bbagen.2017.03.024
Bao, H., She, R., Liu, T., Zhang, Y., Peng, K. S., Luo, D., et al. (2009). Effects of pig antibacterial peptides on growth performance and intestine mucosal immune of broiler chickens. Poult. Sci. 88, 291–297. doi: 10.3382/ps.2008-00330
Bao, K., Yuan, W., Ma, C., Yu, X., Wang, L., Hong, M., et al. (2018). Modification targeting the “rana box” motif of a novel nigrocin peptide from Hylarana latouchii enhances and broadens its potency against multiple bacteria. Front. Microbiol. 9:2846. doi: 10.3389/fnins.2017.2846
Bevins, C. L., and Salzman, N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368. doi: 10.1038/nrmicro2546
Björn, C., Noppa, L., Näslund Salomonsson, E., Johansson, A.-L., Nilsson, E., Mahlapuu, M., et al. (2015). Efficacy and safety profile of the novel antimicrobial peptide PXL150 in a mouse model of infected burn wounds. Int. J. Antimicrob. Agents 45, 519–524. doi: 10.1016/j.ijantimicag.2014.12.015
Borro, B. C., Nordström, R., and Malmsten, M. (2020). Microgels and hydrogels as delivery systems for antimicrobial peptides. Colloids Surf. B Biointerf. 187:110835. doi: 10.1016/j.colsurfb.2020.110835
Brosig, B., and Langosch, D. (1998). The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. Public. Protein Soc. 7, 1052–1056. doi: 10.1002/pro.5560070423
Cabib, E. (2009). Two novel techniques for determination of polysaccharide cross-links show that Crh1p and Crh2p attach chitin to both β(1-6)- and β(1-3)glucan in the Saccharomyces cerevisiae cell wall. Eukaryot. Cell 8:1626. doi: 10.1128/EC.00228-09
Cameron, A. J., Edwards, P. J. B., Harjes, E., and Sarojini, V. (2017). Tyrocidine A analogues bearing the planar d-Phe-2-Abz turn motif: how conformation impacts bioactivity. J. Med. Chem. 60, 9565–9574. doi: 10.1021/acs.jmedchem.7b00953
Cameron, A. J., Varnava, K. G., Edwards, P. J. B., Harjes, E., and Sarojini, V. (2018). Acyclic peptides incorporating the d-Phe-2-Abz turn motif: investigations on antimicrobial activity and propensity to adopt β-hairpin conformations. J. Pept. Sci. 24:e3094. doi: 10.1002/psc.3094
Cao, J., de la Fuente-Nunez, C., Ou, R. W., Torres, M. D. T., Pande, S. G., Sinskey, A. J., et al. (2018). Yeast-based synthetic biology platform for antimicrobial peptide production. ACS Synthet. Biol. 7, 896–902. doi: 10.1021/acssynbio.7b00396
Cao, L., Jiang, W., Cao, S., Zhao, P., Liu, J., Dong, H., et al. (2019). In vitro leishmanicidal activity of antimicrobial peptide KDEL against Leishmania tarentolae. Acta Biochim. Biophys. Sin. 51, 1286–1292. doi: 10.1093/abbs/gmz128
Chalmers, R. M., Robertson, L. J., Dorny, P., Jordan, S., Kärssin, A., Katzer, F., et al. (2020). Parasite detection in food: current status and future needs for validation. Trends Food Sci. Technol. 99, 337–350. doi: 10.1016/j.tifs.2020.03.011
Chan, D. I., Prenner, E. J., and Vogel, H. J. (2006). Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim. Biophys. Acta Biomembr. 1758, 1184–1202. doi: 10.1016/j.bbamem.2006.04.006
Chen, L., Jia, L., Zhang, Q., Zhou, X., Liu, Z., Li, B., et al. (2017). A novel antimicrobial peptide against dental-caries-associated bacteria. Anaerobe 47, 165–172. doi: 10.1016/j.anaerobe.2017.05.016
Chen, Y., Cai, S., Qiao, X., Wu, M., Guo, Z., Wang, R., et al. (2017). As-CATH1-6, novel cathelicidins with potent antimicrobial and immunomodulatory properties from Alligator sinensis, play pivotal roles in host antimicrobial immune responses. Biochem. J. 474, 2861–2885. doi: 10.1042/bcj20170334
Cheng, A.-C., Lin, H.-L., Shiu, Y.-L., Tyan, Y.-C., and Liu, C.-H. (2017). Isolation and characterization of antimicrobial peptides derived from Bacillus subtilis E20-fermented soybean meal and its use for preventing Vibrio infection in shrimp aquaculture. Fish Shellf. Immunol. 67, 270–279. doi: 10.1016/j.fsi.2017.06.006
Chu-Kung, A. F., Nguyen, R., Bozzelli, K. N., and Tirrell, M. (2010). Chain length dependence of antimicrobial peptide-fatty acid conjugate activity. J. Colloids Interf. Sci. 345, 160–167. doi: 10.1016/j.jcis.2009.11.057
Conlon, J. M., and Mechkarska, M. (2014). Host-defense peptides with therapeutic potential from skin secretions of frogs from the family pipidae. Pharmaceuticals 7, 58–77.
Corrêa, J. A. F., Evangelista, A. G., Nazareth, T. D. M., and Luciano, F. B. (2019). Fundamentals on the molecular mechanism of action of antimicrobial peptides. Materialia 8:100494. doi: 10.1016/j.mtla.2019.100494
Cote, C. K., Blanco, I. I., Hunter, M., Shoe, J. L., Klimko, C. P., Panchal, R. G., et al. (2020). Combinations of early generation antibiotics and antimicrobial peptides are effective against a broad spectrum of bacterial biothreat agents. Microb. Pathog. 142:104050. doi: 10.1016/j.micpath.2020.104050
Crunkhorn, S. (2018). Synthetic peptides eradicate resistant infections. Nat. Rev. Drug Discov. 17:166. doi: 10.1038/nrd.2018.31
Cruz, G. F., de Araujo, I., Torres, M. D. T., de la Fuente-Nunez, C., Oliveira, V. X., Ambrosio, F. N., et al. (2020). Photochemically-generated silver chloride nanoparticles stabilized by a peptide inhibitor of cell division and its antimicrobial properties. J. Inorgan. Organ. Polym. Mater. 30, 2464–2474. doi: 10.1007/s10904-019-01427-2
da Silva Malheiros, P., Daroit, D. J., and Brandelli, A. (2010). Food applications of liposome-encapsulated antimicrobial peptides. Trends Food Sci. Technol. 21, 284–292. doi: 10.1016/j.tifs.2010.03.003
Datta, A., Kundu, P., and Bhunia, A. (2016). Designing potent antimicrobial peptides by disulphide linked dimerization and N-terminal lipidation to increase antimicrobial activity and membrane perturbation: structural insights into lipopolysaccharide binding. J. Colloids Interf. Sci. 461, 335–345. doi: 10.1016/j.jcis.2015.09.036
de la Fuente-Núñez, C., Silva, O. N., Lu, T. K., and Franco, O. L. (2017). Antimicrobial peptides: role in human disease and potential as immunotherapies. Pharmacol. Therap. 178, 132–140. doi: 10.1016/j.pharmthera.2017.04.002
Dennison, S. R., Mura, M., Harris, F., Morton, L. H. G., Zvelindovsky, A., and Phoenix, D. A. (2015). The role of C-terminal amidation in the membrane interactions of the anionic antimicrobial peptide, maximin H5. Biochim. Biophys. Acta Biomembr. 1848, 1111–1118. doi: 10.1016/j.bbamem.2015.01.014
Derakhshankhah, H., and Jafari, S. (2018). Cell penetrating peptides: a concise review with emphasis on biomedical applications. Biomed. Pharmacother. 108, 1090–1096. doi: 10.1016/j.biopha.2018.09.097
D’Este, F., Benincasa, M., Cannone, G., Furlan, M., Scarsini, M., Volpatti, D., et al. (2016). Antimicrobial and host cell-directed activities of Gly/Ser-rich peptides from salmonid cathelicidins. Fish Shellf. Immunol. 59, 456–468. doi: 10.1016/j.fsi.2016.11.004
Diosa, J., Guzman, F., Bernal, C., and Mesa, M. (2020). Formation mechanisms of chitosan-silica hybrid materials and its performance as solid support for KR-12 peptide adsorption: impact on KR-12 antimicrobial activity and proteolytic stability. J. Mater. Res. Technol. 9, 890–901. doi: 10.1016/j.jmrt.2019.11.029
Dong, N., Wang, C., Zhang, T., Zhang, L., Xue, C., Feng, X., et al. (2019). Bioactivity and bactericidal mechanism of histidine-rich β-hairpin peptide against Gram-negative bacteria. Intern. J. Mol. Sci. 20:3954. doi: 10.3390/ijms20163954
Du, L., He, Y., Zhou, Y., Liu, S., Zheng, B. J., and Jiang, S. (2009). The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 7, 226–236. doi: 10.1038/nrmicro2090
Durand, G. A., Raoult, D., and Dubourg, G. (2019). Antibiotic discovery: history, methods and perspectives. Int. J. Antimicrob. Agents 53, 371–382. doi: 10.1016/j.ijantimicag.2018.11.010
Dutta, P., Sahu, R. K., Dey, T., Lahkar, M. D., Manna, P., and Kalita, J. (2019). Beneficial role of insect-derived bioactive components against inflammation and its associated complications (colitis and arthritis) and cancer. Chem. Biol. Interact. 313:108824. doi: 10.1016/j.cbi.2019.108824
Ejsing, C. S., Sampaio, J. L., Surendranath, V., Duchoslav, E., Ekroos, K., Klemm, R. W., et al. (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 2136–2141. doi: 10.1073/pnas.0811700106
Faruck, M. O., Yusof, F., and Chowdhury, S. (2016). An overview of antifungal peptides derived from insect. Peptides 80, 80–88. doi: 10.1016/j.peptides.2015.06.001
Field, C. J. (2005). The immunological components of human milk and their effect on immune development in infants. J. Nutr. 135, 1–4. doi: 10.1093/jn/135.1.1
Fjell, C. D., Hiss, J. A., Hancock, R. E. W., and Schneider, G. (2012). Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11, 37–51. doi: 10.1038/nrd3591
Franks, T. J., and Galvin, J. R. (2014). “Coronavirus,” in Viruses and the Lung, eds A. Fraire, B. Woda, R. Welsh, and R. Kradin (Berlin: Springer), doi: 10.1007/978-3-642-40605-8_13
Fuse, N., Hayashi, Y., Fukata, J., Tominaga, T., Ebisui, O., Satoh, Y., et al. (1993). Purification and characterization of new anti-adrenocorticotropin rabbit neutrophil peptides (defensins). Eur. J. Biochem. 216, 653–659.
Gao, R., Shu, W., Shen, Y., Sun, Q., Bai, F., Wang, J., et al. (2020). Sturgeon protein-derived peptides exert anti-inflammatory effects in LPS-stimulated RAW264.7 macrophages via the MAPK pathway. J. Funct. Foods 72:104044. doi: 10.1016/j.jff.2020.104044
Goormaghtigh, E., De Meutter, J., Szoka, F., Cabiaux, V., Parente, R. A., and Ruysschaert, J. M. (1991). Secondary structure and orientation of the amphipathic peptide GALA in lipid structures: an infrared-spectroscopic approach. Eur. J. Biochem. 195, 421–429. doi: 10.1111/j.1432-1033.1991.tb15721.x
Graham, G. V., Conlon, J. M., Abdel-Wahab, Y. H., and Flatt, P. R. (2020). Glucagon from the phylogenetically ancient paddlefish provides a template for the design of a long-acting peptide with effective anti-diabetic and anti-obesity activities. Eur. J. Pharmacol. 878:173101. doi: 10.1016/j.ejphar.2020.173101
Gschwandtner, M., Zhong, S., Tschachler, A., Mlitz, V., Karner, S., Elbe-Bürger, A., et al. (2014). Fetal human keratinocytes produce large amounts of antimicrobial peptides: involvement of histone-methylation processes. J. Invest. Dermatol. 134, 2192–2201. doi: 10.1038/jid.2014.165
Guaní-Guerra, E., Santos-Mendoza, T., Lugo-Reyes, S. O., and Terán, L. M. (2010). Antimicrobial peptides: general overview and clinical implications in human health and disease. Clin. Immunol. 135, 1–11. doi: 10.1016/j.clim.2009.12.004
Guha, S., Ghimire, J., Wu, E., and Wimley, W. C. (2019). Mechanistic landscape of membrane-permeabilizing peptides. Chem. Rev. 119, 6040–6085. doi: 10.1021/acs.chemrev.8b00520
Guidotti, G., Brambilla, L., and Rossi, D. (2017). Cell-penetrating peptides: from basic research to clinics. Trends Pharmacol. Sci. 38, 406–424. doi: 10.1016/j.tips.2017.01.003
Hamid, M.-N., and Friedberg, I. (2018). Identifying antimicrobial peptides using word embedding with deep recurrent neural networks. Bioinformatics 35, 2009–2016. doi: 10.1093/bioinformatics/bty937
He, S.-W., Zhang, J., Li, N.-Q., Zhou, S., Yue, B., and Zhang, M. (2017). A TFPI-1 peptide that induces degradation of bacterial nucleic acids, and inhibits bacterial and viral infection in half-smooth tongue sole, Cynoglossus semilaevis. Fish Shellf. Immunol. 60, 466–473. doi: 10.1016/j.fsi.2016.11.029
Helmerhorst, E. J., Troxler, R. F., and Oppenheim, F. G. (2001). The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 98, 14637–14642. doi: 10.1073/pnas.141366998
Henderson, J. T., Chopko, A. L., and van Wassenaar, P. D. (1992). Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Archiv. Biochem. Biophys. 295, 5–12. doi: 10.1016/0003-9861(92)90480-K
Hitchner, M. A., Santiago-Ortiz, L. E., Necelis, M. R., Shirley, D. J., Palmer, T. J., Tarnawsky, K. E., et al. (2019). Activity and characterization of a pH-sensitive antimicrobial peptide. Biochim. Biophys. Acta Biomembr. 1861:182984. doi: 10.1016/j.bbamem.2019.05.006
Holdbrook, D. A., Singh, S., Choong, Y. K., Petrlova, J., Malmsten, M., Bond, P. J., et al. (2018). Influence of pH on the activity of thrombin-derived antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 1860, 2374–2384. doi: 10.1016/j.bbamem.2018.06.002
Hu, F., Gao, X., She, R., Chen, J., Mao, J., Xiao, P., et al. (2017). Effects of antimicrobial peptides on growth performance and small intestinal function in broilers under chronic heat stress. Poult. Sci. 96, 798–806. doi: 10.3382/ps/pew379
Huang, H.-N., Pan, C.-Y., and Chen, J.-Y. (2018). Grouper (Epinephelus coioides) antimicrobial peptide epinecidin-1 exhibits antiviral activity against foot-and-mouth disease virus in vitro. Peptides 106, 91–95. doi: 10.1016/j.peptides.2018.07.003
Huang, H.-N., Rajanbabu, V., Pan, C.-Y., Chan, Y.-L., Wu, C.-J., and Chen, J.-Y. (2013). A cancer vaccine based on the marine antimicrobial peptide pardaxin (GE33) for control of bladder-associated tumors. Biomaterials 34, 10151–10159. doi: 10.1016/j.biomaterials.2013.09.041
Imamura, T., Yasuda, M., Kusano, H., Nakashita, H., Ohno, Y., Kamakura, T., et al. (2010). Acquired resistance to the rice blast in transgenic rice accumulating the antimicrobial peptide thanatin. Transg. Res. 19, 415–424. doi: 10.1007/s11248-009-9320-x
Imjongjirak, C., Amphaiphan, P., Charoensapsri, W., and Amparyup, P. (2017). Characterization and antimicrobial evaluation of SpPR-AMP1, a proline-rich antimicrobial peptide from the mud crab Scylla paramamosain. Dev. Compar. Immunol. 74, 209–216. doi: 10.1016/j.dci.2017.05.003
Izadi, N., Keikha, M., Ghazvini, K., and Karbalaei, M. (2020). Oral antimicrobial peptides and new therapeutic strategies for plaque-mediated diseases. Gene Rep. 21:100811. doi: 10.1016/j.genrep.2020.100811
Jeżowska-Bojczuk, M., and Stokowa-Sołtys, K. (2018). Peptides having antimicrobial activity and their complexes with transition metal ions. Eur. J. Med. Chem. 143, 997–1009. doi: 10.1016/j.ejmech.2017.11.086
Jia, F., Zhang, Y., Wang, J., Peng, J., Zhao, P., Zhang, L., et al. (2019). The effect of halogenation on the antimicrobial activity, antibiofilm activity, cytotoxicity and proteolytic stability of the antimicrobial peptide jelleine-I. Peptides 112, 56–66. doi: 10.1016/j.peptides.2018.11.006
Jiang, L., Wang, B., Li, B., Wang, C., and Luo, Y. (2014). Preparation and identification of peptides and their zinc complexes with antimicrobial activities from silver carp (Hypophthalmichthys molitrix) protein hydrolysates. Food Res. Int. 64, 91–98. doi: 10.1016/j.foodres.2014.06.008
Jung, Y., Kong, B., Moon, S., Yu, S.-H., Chung, J., Ban, C., et al. (2019). Envelope-deforming antiviral peptide derived from influenza virus M2 protein. Biochem. Biophys. Res. Commun. 517, 507–512. doi: 10.1016/j.bbrc.2019.07.088
Juretić, D., Vukičević, D., Petrov, D., Novković, M., Bojović, V., Lučić, B., et al. (2011). Knowledge-based computational methods for identifying or designing novel, non-homologous antimicrobial peptides. Eur. Biophys. J. 40, 371–385. doi: 10.1007/s00249-011-0674-7
Ke, M., Chen, Y., Wu, A., Sun, Y., Su, C., Wu, H., et al. (2012). Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2’-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 167, 322–328. doi: 10.1016/j.virusres.2012.05.017
Khan, I., and Oh, D.-H. (2016). Integration of nisin into nanoparticles for application in foods. Innovat. Food Sci. Emerg. Technol. 34, 376–384. doi: 10.1016/j.ifset.2015.12.013
Khan, S. A., and Lee, C.-S. (2020). Recent progress and strategies to develop antimicrobial contact lenses and lens cases for different types of microbial keratitis. Acta Biomater. 113, 101–118. doi: 10.1016/j.actbio.2020.06.039
Khara, J. S., Obuobi, S., Wang, Y., Hamilton, M. S., Robertson, B. D., Newton, S. M., et al. (2017). Disruption of drug-resistant biofilms using de novo designed short α-helical antimicrobial peptides with idealized facial amphiphilicity. Acta Biomater. 57, 103–114. doi: 10.1016/j.actbio.2017.04.032
Kim, H., Jang, J. H., Kim, S. C., and Cho, J. H. (2020). Development of a novel hybrid antimicrobial peptide for targeted killing of Pseudomonas aeruginosa. Eur. J. Med. Chem. 185:111814. doi: 10.1016/j.ejmech.2019.111814
Koehbach, J., and Craik, D. J. (2019). The vast structural diversity of antimicrobial peptides. Trends Pharmacol. Sci. 40, 517–528. doi: 10.1016/j.tips.2019.04.012
Kozić, M., Vukičević, D., Simunić, J., Rončević, T., Antcheva, N., Tossi, A., et al. (2015). Predicting the minimal inhibitory concentration for antimicrobial peptides with rana-box domain. J. Chem. Inform. Model. 55, 2275–2287. doi: 10.1021/acs.jcim.5b00161
Kragol, G., Lovas, S., Varadi, G., Condie, B. A., Hoffmann, R., and Otvos, L. (2001). The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40, 3016–3026. doi: 10.1021/bi002656a
Krauson, A. J., Hall, O. M., Fuselier, T., Starr, C. G., Kauffman, W. B., and Wimley, W. C. (2015). Conformational fine-tuning of pore-forming peptide potency and selectivity. J. Am. Chem. Soc. 137, 16144–16152. doi: 10.1021/jacs.5b10595
Krauson, A. J., He, J., and Wimley, W. C. (2012). Gain-of-function analogues of the pore-forming peptide melittin selected by orthogonal high-throughput screening. J. Am. Chem. Soc. 134, 12732–12741. doi: 10.1021/ja3042004
Kwon, Y. M., Kim, H. J., Kim, Y. I., Kang, Y. J., Lee, I. H., Jin, B. R., et al. (2008). Comparative analysis of two attacin genes from Hyphantria cunea. Compar. Biochem. Physiol. Part B Biochem. Mol. Biol. 151, 213–220. doi: 10.1016/j.cbpb.2008.07.002
Lai, Y., Villaruz, A. E., Li, M., Cha, D. J., Sturdevant, D. E., and Otto, M. (2007). The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in Staphylococci. Mol. Microbiol. 63, 497–506. doi: 10.1111/j.1365-2958.2006.05540.x
Lakshmaiah Narayana, J., and Chen, J.-Y. (2015). Antimicrobial peptides: possible anti-infective agents. Peptides 72, 88–94. doi: 10.1016/j.peptides.2015.05.012
Lampejo, T. (2020). Dalbavancin and telavancin in the treatment of infective endocarditis: a literature review. Int. J. Antimicrob. Agents 56:106072. doi: 10.1016/j.ijantimicag.2020.106072
Lazzaro, B. P., Zasloff, M., and Rolff, J. (2020). Antimicrobial peptides: application informed by evolution. Science 368:eaau5480. doi: 10.1126/science.aau5480
Le, C. F., Gudimella, R., Razali, R., Manikam, R., and Sekaran, S. D. (2016). Transcriptome analysis of Streptococcus pneumoniae treated with the designed antimicrobial peptides, DM3. Sci. Rep. 6:26828. doi: 10.1038/srep26828
Le, C.-F., Fang, C.-M., and Sekaran, S. D. (2017). Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 61:e02340-16. doi: 10.1128/AAC.02340-16
Lee, J. H., Cho, K. S., Lee, J., Yoo, J., Lee, J., and Chung, J. (2001). Diptericin-like protein: an immune response gene regulated by the anti-bacterial gene induction pathway in drosophila. Gene 271, 233–238. doi: 10.1016/S0378-1119(01)00515-7
Lee, S.-H., Kim, S.-J., Lee, Y.-S., Song, M.-D., Kim, I.-H., and Won, H.-S. (2011). De novo generation of short antimicrobial peptides with simple amino acid composition. Regul. Peptid. 166, 36–41. doi: 10.1016/j.regpep.2010.08.010
Lei, J., Sun, L., Huang, S., Zhu, C., Li, P., He, J., et al. (2019). The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 11, 3919–3931.
León, R., Ruiz, M., Valero, Y., Cárdenas, C., Guzman, F., Vila, M., et al. (2020). Exploring small cationic peptides of different origin as potential antimicrobial agents in aquaculture. Fish Shellf. Immunol. 98, 720–727. doi: 10.1016/j.fsi.2019.11.019
Lewies, A., Wentzel, J. F., Jordaan, A., Bezuidenhout, C., and Du Plessis, L. H. (2017). Interactions of the antimicrobial peptide nisin Z with conventional antibiotics and the use of nanostructured lipid carriers to enhance antimicrobial activity. Int. J. Pharm. 526, 244–253. doi: 10.1016/j.ijpharm.2017.04.071
Li, C., Zhu, C., Ren, B., Yin, X., Shim, S. H., Gao, Y., et al. (2019). Two optimized antimicrobial peptides with therapeutic potential for clinical antibiotic-resistant Staphylococcus aureus. Eur. J. Med. Chem. 183:111686. doi: 10.1016/j.ejmech.2019.111686
Li, J., Koh, J.-J., Liu, S., Lakshminarayanan, R., Verma, C. S., and Beuerman, R. W. (2017). Membrane active antimicrobial peptides: translating mechanistic insights to design. Front. Neurosci. 11:73. doi: 10.3389/fnins.2017.00073
Li, L., Sun, J., Xia, S., Tian, X., Cheserek, M. J., and Le, G. (2016). Mechanism of antifungal activity of antimicrobial peptide APP, a cell-penetrating peptide derivative, against Candida albicans: intracellular DNA binding and cell cycle arrest. Appl. Microbiol. Biotechnol. 100, 3245–3253. doi: 10.1007/s00253-015-7265-y
Li, W., Sani, M.-A., Jamasbi, E., Otvos, L., Hossain, M. A., Wade, J. D., et al. (2016). Membrane interactions of proline-rich antimicrobial peptide, Chex1-Arg20, multimers. Biochim. Biophys. Acta Biomembr. 1858, 1236–1243. doi: 10.1016/j.bbamem.2016.02.035
Li, X., Wang, W., Liu, S., Ruan, C., Yi, L., Deng, L., et al. (2019). Effects of the peptide H-OOWW-NH2 and its derived lipopeptide C12-OOWW-NH2 on controlling of citrus postharvest green mold. Postharvest Biol. Technol. 158:110979. doi: 10.1016/j.postharvbio.2019.110979
Lipkin, R. B., and Lazaridis, T. (2015). Implicit membrane investigation of the stability of antimicrobial peptide β-barrels and arcs. J. Membr. Biol. 248, 469–486. doi: 10.1007/s00232-014-9759-4
Liu, S., Wang, W., Deng, L., Ming, J., Yao, S., and Zeng, K. (2019). Control of sour rot in citrus fruit by three insect antimicrobial peptides. Postharvest Biol. Technol. 149, 200–208. doi: 10.1016/j.postharvbio.2018.11.025
Liu, T., She, R., Wang, K., Bao, H., Zhang, Y., Luo, D., et al. (2008). Effects of rabbit sacculus rotundus antimicrobial peptides on the intestinal mucosal immunity in chickens. Poult. Sci. 87, 250–254. doi: 10.3382/ps.2007-00353
Liu, Z., Zeng, M., Dong, S., Xu, J., Song, H., and Zhao, Y. (2007). Effect of an antifungal peptide from oyster enzymatic hydrolysates for control of gray mold (Botrytis cinerea) on harvested strawberries. Postharvest Biol. Technol. 46, 95–98. doi: 10.1016/j.postharvbio.2007.03.013
Lohner, K., and Prossnigg, F. (2009). Biological activity and structural aspects of PGLa interaction with membrane mimetic systems. Biochim. Biophys. Acta Biomembr. 1788, 1656–1666. doi: 10.1016/j.bbamem.2009.05.012
Lointier, M., Aisenbrey, C., Marquette, A., Tan, J. H., Kichler, A., and Bechinger, B. (2020). Membrane pore-formation correlates with the hydrophilic angle of histidine-rich amphipathic peptides with multiple biological activities. Biochim. Biophys. Acta Biomembr. 1862:183212. doi: 10.1016/j.bbamem.2020.183212
Lu, Y., Ma, Y., Wang, X., Liang, J., Zhang, C., Zhang, K., et al. (2008). The first antimicrobial peptide from sea amphibian. Mol. Immunol. 45, 678–681. doi: 10.1016/j.molimm.2007.07.004
Lutkenhaus, J. (1990). Regulation of cell division in E. coli. Trends Genet. 6, 22–25. doi: 10.1016/0168-9525(90)90045-8
Luz, C., Calpe, J., Saladino, F., Luciano, F. B., Fernandez-Franzón, M., Mañes, J., et al. (2018). Antimicrobial packaging based on ε-polylysine bioactive film for the control of mycotoxigenic fungi in vitro and in bread. J. Food Process. Preserv. 42:e13370. doi: 10.1111/jfpp.13370
Lyu, Y., Fitriyanti, M., and Narsimhan, G. (2019). Nucleation and growth of pores in 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) / cholesterol bilayer by antimicrobial peptides melittin, its mutants and cecropin P1. Colloids Surf. B Biointerf. 173, 121–127. doi: 10.1016/j.colsurfb.2018.09.049
Ma, R., Wong, S. W., Ge, L., Shaw, C., Siu, S. W. I., and Kwok, H. F. (2020). In vitro and MD simulation study to explore physicochemical parameters for antibacterial peptide to become potent anticancer peptide. Mol. Ther. Oncolyt. 16, 7–19. doi: 10.1016/j.omto.2019.12.001
Madanchi, H., Shoushtari, M., Kashani, H. H., and Sardari, S. (2020). Antimicrobial peptides of the vaginal innate immunity and their role in the fight against sexually transmitted diseases. New Microb. New Infect. 34:100627. doi: 10.1016/j.nmni.2019.100627
Magana, M., Pushpanathan, M., Santos, A. L., Leanse, L., Fernandez, M., Ioannidis, A., et al. (2020). The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 20, e216–e230. doi: 10.1016/S1473-3099(20)30327-3
Malekkhaiat Häffner, S., and Malmsten, M. (2018). Influence of self-assembly on the performance of antimicrobial peptides. Curr. Opin. Colloids Interf. Sci. 38, 56–79. doi: 10.1016/j.cocis.2018.09.002
Malkoski, M., Dashper, S. G., O’Brien-Simpson, N. M., Talbo, G. H., Macris, M., Cross, K. J., et al. (2001). Kappacin, a novel antibacterial peptide from bovine milk. Antimicrob. Agents Chemother. 45, 2309–2315. doi: 10.1128/AAC.45.8.2309-2315.2001
Mangoni, M. L., Saugar, J. M., Dellisanti, M., Barra, D., Simmaco, M., and Rivas, L. (2005). Temporins, small antimicrobial peptides with leishmanicidal activity. J. Biol. Chem. 280, 984–990. doi: 10.1074/jbc.M410795200
Mardirossian, M., Grzela, R., Giglione, C., Meinnel, T., Gennaro, R., Mergaert, P., et al. (2014). The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem. Biol. 21, 1639–1647. doi: 10.1016/j.chembiol.2014.10.009
Mardirossian, M., Pérébaskine, N., Benincasa, M., Gambato, S., Hofmann, S., Huter, P., et al. (2018). The dolphin proline-rich antimicrobial peptide Tur1A inhibits protein synthesis by targeting the bacterial ribosome. Cell Chem. Biol. 25, 530–539.e537. doi: 10.1016/j.chembiol.2018.02.004
Mardirossian, M., Sola, R., Beckert, B., Collis, D. W. P., Di Stasi, A., Armas, F., et al. (2019). Proline-rich peptides with improved antimicrobial activity against E. coli, K. pneumoniae, and A. baumannii. Chemmedchem 14, 2025–2033. doi: 10.1002/cmdc.201900465
Marimuthu, S. K., Nagarajan, K., Perumal, S. K., Palanisamy, S., and Subbiah, L. (2019). Insilico alpha-helical structural recognition of temporin antimicrobial peptides and its interactions with middle east respiratory syndrome-coronavirus. Int. J. Pept. Res. Ther. 26, 1473–1483. doi: 10.1007/s10989-019-09951-y
Martin, N. I., Sprules, T., Carpenter, M. R., Cotter, P. D., Hill, C., Ross, R. P., et al. (2004). Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry 43, 3049–3056. doi: 10.1021/bi0362065
Marya, K. H., Nabavi, S. M., and Habtemariam, S. (2018). Anti-diabetic potential of peptides: future prospects as therapeutic agents. Life Sci. 193, 153–158. doi: 10.1016/j.lfs.2017.10.025
Matsuzaki, K., Murase, O., Fujii, N., and Miyajima, K. (1995). Translocation of a channel-forming antimicrobial peptide, magainin 2, across lipid bilayers by forming a pore. Biochemistry 34, 6521–6526. doi: 10.1021/bi00019a033
Matsuzaki, K., Murase, O., Fujii, N., and Miyajima, K. (1996). An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 35, 11361–11368. doi: 10.1021/bi960016v
Mattiuzzo, M., Bandiera, A., Gennaro, R., Benincasa, M., Pacor, S., Antcheva, N., et al. (2007). Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 66, 151–163. doi: 10.1111/j.1365-2958.2007.05903.x
Maurya, I. K., Thota, C. K., Sharma, J., Tupe, S. G., Chaudhary, P., Singh, M. K., et al. (2013). Mechanism of action of novel synthetic dodecapeptides against Candida albicans. Biochim. Biophys. Acta Gen. Subj. 1830, 5193–5203. doi: 10.1016/j.bbagen.2013.07.016
Meram, C., and Wu, J. (2017). Anti-inflammatory effects of egg yolk livetins (α, β, and γ-livetin) fraction and its enzymatic hydrolysates in lipopolysaccharide-induced RAW 264.7 macrophages. Food Res. Intern. 100, 449–459. doi: 10.1016/j.foodres.2017.07.032
Michael Henderson, J., and Lee, K. Y. C. (2013). Promising antimicrobial agents designed from natural peptide templates. Curr. Opin. Solid State Mater. Sci. 17, 175–192. doi: 10.1016/j.cossms.2013.08.003
Mohanram, H., and Bhattacharjya, S. (2016). ‘Lollipop’-shaped helical structure of a hybrid antimicrobial peptide of temporin B-lipopolysaccharide binding motif and mapping cationic residues in antibacterial activity. Biochim. Biophys. Acta Gen. Subj. 1860, 1362–1372. doi: 10.1016/j.bbagen.2016.03.025
Muhialdin, B. J., Algboory, H. L., Kadum, H., Mohammed, N. K., Saari, N., Hassan, Z., et al. (2020). Antifungal activity determination for the peptides generated by Lactobacillus plantarum TE10 against Aspergillus flavus in maize seeds. Food Control 109:106898. doi: 10.1016/j.foodcont.2019.106898
Müller, A. T., Hiss, J. A., and Schneider, G. (2018). Recurrent neural network model for constructive peptide design. J. Chem. Inform. Model. 58, 472–479. doi: 10.1021/acs.jcim.7b00414
Mustafa, S., Balkhy, H., and Gabere, M. N. (2018). Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): a review. J. Infect. Public Health 11, 9–17. doi: 10.1016/j.jiph.2017.08.009
Neshani, A., Zare, H., Akbari Eidgahi, M. R., Khaledi, A., and Ghazvini, K. (2019). Epinecidin-1, a highly potent marine antimicrobial peptide with anticancer and immunomodulatory activities. BMC Pharmacol. Toxicol. 20:33. doi: 10.1186/s40360-019-0309-7
Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656. doi: 10.1128/mmbr.67.4.593-656.2003
Nordström, R., Andrén, O. C. J., Singh, S., Malkoch, M., Davoudi, M., Schmidtchen, A., et al. (2019). Degradable dendritic nanogels as carriers for antimicrobial peptides. J. Colloids Interf. Sci. 554, 592–602. doi: 10.1016/j.jcis.2019.07.028
Oh, R., Lee, M. J., Kim, Y.-O., Nam, B.-H., Kong, H. J., Kim, J.-W., et al. (2020). Myticusin-beta, antimicrobial peptide from the marine bivalve, Mytilus coruscus. Fish Shellf. Immunol. 99, 342–352. doi: 10.1016/j.fsi.2020.02.020
Omardien, S., Drijfhout, J. W., Vaz, F. M., Wenzel, M., Hamoen, L. W., Zaat, S. A. J., et al. (2018). Bactericidal activity of amphipathic cationic antimicrobial peptides involves altering the membrane fluidity when interacting with the phospholipid bilayer. Biochim. Biophys. Acta Biomembr. 1860, 2404–2415. doi: 10.1016/j.bbamem.2018.06.004
Oren, Z., and Shai, Y. (1998). Mode of action of linear amphipathic α-helical antimicrobial peptides. Pept. Sci. 47, 451–463.
Orivel, J., Redeker, V., Le Caer, J. P., Krier, F., Revol-Junelles, A. M., Longeon, A., et al. (2001). Ponericins, new antibacterial and insecticidal peptides from the venom of the ant Pachycondyla goeldii. J. Biol. Chem. 276, 17823–17829. doi: 10.1074/jbc.M100216200
Orlov, S. D., Shamova, V. O., Eliseev, E. I., Zharkova, S. M., Chakchir, B. O., Antcheva, N., et al. (2019). Redesigning Arenicin-1, an antimicrobial peptide from the marine polychaeta Arenicola marina, by strand rearrangement or branching, substitution of specific residues, and backbone linearization or cyclization. Mar. Drugs 17:376. doi: 10.3390/md17060376
Parachin, N. S., Mulder, K. C., Viana, A. A. B., Dias, S. C., and Franco, O. L. (2012). Expression systems for heterologous production of antimicrobial peptides. Peptides 38, 446–456. doi: 10.1016/j.peptides.2012.09.020
Park, J.-E., and Gallagher, T. (2017). Lipidation increases antiviral activities of coronavirus fusion-inhibiting peptides. Virology 511, 9–18. doi: 10.1016/j.virol.2017.07.033
Patch, J. A., and Barron, A. E. (2002). Mimicry of bioactive peptides via non-natural, sequence-specific peptidomimetic oligomers. Curr. Opin. Chem. Biol. 6, 872–877. doi: 10.1016/S1367-5931(02)00385-X
Paulsen, M. H., Ausbacher, D., Bayer, A., Engqvist, M., Hansen, T., Haug, T., et al. (2019). Antimicrobial activity of amphipathic α,α-disubstituted β-amino amide derivatives against ESBL - CARBA producing multi-resistant bacteria; effect of halogenation, lipophilicity and cationic character. Eur. J. Med. Chem. 183:111671. doi: 10.1016/j.ejmech.2019.111671
Pineda-Castañeda, H. M., Bonilla-Velásquez, L. D., Leal-Castro, A. L., Fierro-Medina, R., García-Castañeda, J. E., and Rivera-Monroy, Z. J. (2020). Use of click chemistry for obtaining an antimicrobial chimeric peptide containing the LfcinB and buforin II minimal antimicrobial motifs. Chem. Select 5, 1655–1657. doi: 10.1002/slct.201903834
Pletzer, D., Coleman, S. R., and Hancock, R. E. W. (2016). Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr. Opin. Microbiol. 33, 35–40. doi: 10.1016/j.mib.2016.05.016
Porto, W. F., Irazazabal, L., Alves, E. S. F., Ribeiro, S. M., Matos, C. O., Pires, A. S., et al. (2018). In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 9:1490. doi: 10.1038/s41467-018-03746-3
Puri, S., Li, R., Ruszaj, D., Tati, S., and Edgerton, M. (2015). Iron binding modulates candidacidal properties of salivary histatin 5. J. Dent. Res. 94, 201–208. doi: 10.1177/0022034514556709
Rai, M., Pandit, R., Gaikwad, S., and Kövics, G. (2016). Antimicrobial peptides as natural bio-preservative to enhance the shelf-life of food. J. Food Sci. Technol. 53, 3381–3394. doi: 10.1007/s13197-016-2318-2315
Rajput, A., and Kumar, M. (2018). “Anti-biofilm peptides: a new class of quorum quenchers and their prospective therapeutic applications,” in Biotechnological Applications of Quorum Sensing Inhibitors, ed. V. C. Kalia (Singapore: Springer), 87–110.
Rautenbach, M., Troskie, A. M., and Vosloo, J. A. (2016). Antifungal peptides: to be or not to be membrane active. Biochimie 130, 132–145. doi: 10.1016/j.biochi.2016.05.013
Reddy, K. V. R., Yedery, R. D., and Aranha, C. (2004). Antimicrobial peptides: premises and promises. Int. J. Antimicrob. Agents 24, 536–547. doi: 10.1016/j.ijantimicag.2004.09.005
Rhaiem, R. B., and Houimel, M. (2016). Targeting Leishmania major parasite with peptides derived from a combinatorial phage display library. Acta Trop. 159, 11–19. doi: 10.1016/j.actatropica.2016.03.018
Ribeiro, S. M., Felício, M. R., Boas, E. V., Gonçalves, S., Costa, F. F., Samy, R. P., et al. (2016). New frontiers for anti-biofilm drug development. Pharmacol. Therap. 160, 133–144. doi: 10.1016/j.pharmthera.2016.02.006
Rivas, L., and Rojas, V. (2019). Cyanobacterial peptides as a tour de force in the chemical space of antiparasitic agents. Arch. Biochem. Biophys. 664, 24–39. doi: 10.1016/j.abb.2019.01.030
Rollins-Smith, L. A. (2009). The role of amphibian antimicrobial peptides in protection of amphibians from pathogens linked to global amphibian declines. Biochim. Biophys. Acta Biomembr. 1788, 1593–1599. doi: 10.1016/j.bbamem.2009.03.008
Sabapathy, T., Deplazes, E., and Mancera, R. L. (2020). Revisiting the interaction of melittin with phospholipid bilayers: the effects of concentration and ionic strength. Intern. J. Mol. Sci. 21:746. doi: 10.3390/ijms21030746
Santos, J. C. P., Sousa, R. C. S., Otoni, C. G., Moraes, A. R. F., Souza, V. G. L., Medeiros, E. A. A., et al. (2018). Nisin and other antimicrobial peptides: production, mechanisms of action, and application in active food packaging. Innovat. Food Sci. Emerg. Technol. 48, 179–194. doi: 10.1016/j.ifset.2018.06.008
Schittek, B., Hipfel, R., Sauer, B., Bauer, J., Kalbacher, H., Stevanovic, S., et al. (2001). Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat. Immunol. 2, 1133–1137. doi: 10.1038/ni732
Schmidtchen, A., Pasupuleti, M., and Malmsten, M. (2014). Effect of hydrophobic modifications in antimicrobial peptides. Adv. Colloid Interf. Sci. 205, 265–274. doi: 10.1016/j.cis.2013.06.009
Seefeldt, A. C., Nguyen, F., Antunes, S., Pérébaskine, N., Graf, M., Arenz, S., et al. (2015). The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. Biol. 22, 470–475. doi: 10.1038/nsmb.3034
Selsted, M. E., Brown, D. M., DeLange, R. J., Harwig, S. S., and Lehrer, R. I. (1985). Primary structures of six antimicrobial peptides of rabbit peritoneal neutrophils. J. Biol. Chem. 260, 4579–4584.
Semreen, M. H., El-Gamal, M. I., Abdin, S., Alkhazraji, H., Kamal, L., Hammad, S., et al. (2018). Recent updates of marine antimicrobial peptides. Saudi Pharm. J. 26, 396–409. doi: 10.1016/j.jsps.2018.01.001
Settanni, L., and Corsetti, A. (2008). Application of bacteriocins in vegetable food biopreservation. Int. J. Food Microbiol. 121, 123–138. doi: 10.1016/j.ijfoodmicro.2007.09.001
Setty, S. C., Horam, S., Pasupuleti, M., and Haq, W. (2017). Modulating the antimicrobial activity of temporin L through introduction of fluorinated phenylalanine. Intern. J. Peptide Res. Therap. 23, 213–225. doi: 10.1007/s10989-016-9553-5
Shan, Z., Yang, Y., Guan, N., Xia, X., and Liu, W. (2020). NKL-24: a novel antimicrobial peptide derived from zebrafish NK-lysin that inhibits bacterial growth and enhances resistance against Vibrio parahaemolyticus infection in yesso scallop, Patinopecten yessoensis. Fish Shellf. Immunol. 106, 431–440. doi: 10.1016/j.fsi.2020.08.020
Sharma, D., Choudhary, M., Vashistt, J., Shrivastava, R., and Bisht, G. S. (2019). Cationic antimicrobial peptide and its poly-N-substituted glycine congener: antibacterial and antibiofilm potential against A. baumannii. Biochem. Biophys. Res. Commun. 518, 472–478. doi: 10.1016/j.bbrc.2019.08.062
Shelomi, M., Jacobs, C., Vilcinskas, A., and Vogel, H. (2020). The unique antimicrobial peptide repertoire of stick insects. Dev. Compar. Immunol. 103:103471. doi: 10.1016/j.dci.2019.103471
Shenkarev, Z. O., Balandin, S. V., Trunov, K. I., Paramonov, A. S., Sukhanov, S. V., Barsukov, L. I., et al. (2011). Molecular mechanism of action of β-hairpin antimicrobial peptide arenicin: oligomeric structure in dodecylphosphocholine micelles and pore formation in planar lipid bilayers. Biochemistry 50, 6255–6265. doi: 10.1021/bi200746t
Shu, G., Chen, Y., Liu, T., Ren, S., and Kong, Y. (2019). Antimicrobial peptide cathelicidin-BF inhibits platelet aggregation by blocking protease-activated receptor 4. Intern. J. Pept. Res. Therap. 25, 349–358. doi: 10.1007/s10989-018-9677-x
Shwaiki, L. N., Arendt, E. K., and Lynch, K. M. (2020). Anti-yeast activity and characterisation of synthetic radish peptides Rs-AFP1 and Rs-AFP2 against food spoilage yeast. Food Control 113:107178. doi: 10.1016/j.foodcont.2020.107178
Sibel Akalın, A. (2014). Dairy-derived antimicrobial peptides: action mechanisms, pharmaceutical uses and production proposals. Trends Food Sci. Technol. 36, 79–95. doi: 10.1016/j.tifs.2014.01.002
Silva, N. C., Sarmento, B., and Pintado, M. (2013). The importance of antimicrobial peptides and their potential for therapeutic use in ophthalmology. Int. J. Antimicrob. Agents 41, 5–10. doi: 10.1016/j.ijantimicag.2012.07.020
Singh, A., and Prasad, R. (2011). Comparative lipidomics of azole sensitive and resistant clinical isolates of Candida albicans reveals unexpected diversity in molecular lipid imprints. PLoS One 6:e19266. doi: 10.1371/journal.pone.0019266
Souza, P. F. N., Lopes, F. E. S., Amaral, J. L., Freitas, C. D. T., and Oliveira, J. T. A. (2020). A molecular docking study revealed that synthetic peptides induced conformational changes in the structure of SARS-CoV-2 spike glycoprotein, disrupting the interaction with human ACE2 receptor. Int. J. Biol. Macromol. 164, 66–76. doi: 10.1016/j.ijbiomac.2020.07.174
Spohn, R., Daruka, L., Lázár, V., Martins, A., Vidovics, F., Grézal, G., et al. (2019). Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 10:4538. doi: 10.1038/s41467-019-12364-6
Strøm, M. B., Rekdal, Ø, and Svendsen, J. S. (2002). Antimicrobial activity of short arginine- and tryptophan-rich peptides. J. Pept. Sci. 8, 431–437. doi: 10.1002/psc.398
Subbalakshmi, C., and Sitaram, N. (1998). Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 160, 91–96. doi: 10.1111/j.1574-6968.1998.tb12896.x
Sun, Q., Wang, K., She, R., Ma, W., Peng, F., and Jin, H. (2010). Swine intestine antimicrobial peptides inhibit infectious bronchitis virus infectivity in chick embryos. Poult. Sci. 89, 464–469. doi: 10.3382/ps.2009-00461
Tang, L., Chen, J., Zhou, Z., Yu, P., Yang, Z., and Zhong, G. (2015). Chlamydia-secreted protease CPAF degrades host antimicrobial peptides. Microb. Infect. 17, 402–408. doi: 10.1016/j.micinf.2015.02.005
Tang, S.-S., Prodhan, Z. H., Biswas, S. K., Le, C.-F., and Sekaran, S. D. (2018). Antimicrobial peptides from different plant sources: isolation, characterisation, and purification. Phytochemistry 154, 94–105. doi: 10.1016/j.phytochem.2018.07.002
Tew, G. N., Clements, D., Tang, H., Arnt, L., and Scott, R. W. (2006). Antimicrobial activity of an abiotic host defense peptide mimic. Biochim. Biophys. Acta Biomembr. 1758, 1387–1392. doi: 10.1016/j.bbamem.2006.03.001
Thapa, R. K., Diep, D. B., and Tønnesen, H. H. (2020). Topical antimicrobial peptide formulations for wound healing: current developments and future prospects. Acta Biomater. 103, 52–67. doi: 10.1016/j.actbio.2019.12.025
Tian, X., Sun, F., Zhou, X.-R., Luo, S.-Z., and Chen, L. (2015). Role of peptide self-assembly in antimicrobial peptides. J. Pept. Sci. 21, 530–539. doi: 10.1002/psc.2788
Torres, M. D. T., Sothiselvam, S., Lu, T. K., and de la Fuente-Nunez, C. (2019). Peptide design principles for antimicrobial applications. J. Mol. Biol. 431, 3547–3567. doi: 10.1016/j.jmb.2018.12.015
Tripathi, J. K., Pal, S., Awasthi, B., Kumar, A., Tandon, A., Mitra, K., et al. (2015). Variants of self-assembling peptide, KLD-12 that show both rapid fracture healing and antimicrobial properties. Biomaterials 56, 92–103. doi: 10.1016/j.biomaterials.2015.03.046
Utesch, T., de Miguel Catalina, A., Schattenberg, C., Paege, N., Schmieder, P., Krause, E., et al. (2018). A computational modeling approach predicts interaction of the antifungal protein AFP from Aspergillus giganteus with fungal membranes via its γ-core motif. mSphere 3:e00377-18. doi: 10.1128/mSphere.00377-318
Vargas-Casanova, Y., Rodríguez-Mayor, A. V., Cardenas, K. J., Leal-Castro, A. L., Muñoz-Molina, L. C., Fierro-Medina, R., et al. (2019). Synergistic bactericide and antibiotic effects of dimeric, tetrameric, or palindromic peptides containing the RWQWR motif against Gram-positive and Gram-negative strains. RSC Adv. 9, 7239–7245. doi: 10.1039/C9RA00708C
Veltri, D., Kamath, U., and Shehu, A. (2018). Deep learning improves antimicrobial peptide recognition. Bioinformatics 34, 2740–2747. doi: 10.1093/bioinformatics/bty179
Vernen, F., Harvey, J. P., Dias, A. S., Veiga, S. A., Huang, Y.-H., Craik, J. D., et al. (2019). Characterization of tachyplesin peptides and their cyclized analogues to improve antimicrobial and anticancer properties. Intern. J. Mol. Sci. 20:4184. doi: 10.3390/ijms20174184
Vilas Boas, L. C. P., Campos, M. L., Berlanda, R. L. A., de Carvalho Neves, N., and Franco, O. L. (2019). Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 76, 3525–3542. doi: 10.1007/s00018-019-03138-w
Vilcinskas, A. (2013). Evolutionary plasticity of insect immunity. J. Insect. Physiol. 59, 123–129. doi: 10.1016/j.jinsphys.2012.08.018
Walrant, A., Bauzá, A., Girardet, C., Alves, I. D., Lecomte, S., Illien, F., et al. (2020). Ionpair-π interactions favor cell penetration of arginine/tryptophan-rich cell-penetrating peptides. Biochim. Biophys. Acta Biomembr. 1862:183098. doi: 10.1016/j.bbamem.2019.183098
Wang, G. (2014). Human antimicrobial peptides and proteins. Pharmaceuticals 7, 545–594. doi: 10.3390/ph7050545
Wang, J., Chou, S., Xu, L., Zhu, X., Dong, N., Shan, A., et al. (2015). High specific selectivity and membrane-active mechanism of the synthetic centrosymmetric α-helical peptides with Gly-Gly pairs. Sci. Rep. 5:15963. doi: 10.1038/srep15963
Wang, J., Dou, X., Song, J., Lyu, Y., Zhu, X., Xu, L., et al. (2019). Antimicrobial peptides: promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 39, 831–859. doi: 10.1002/med.21542
Wang, W., Deng, L., Yao, S., and Zeng, K. (2018). Control of green and blue mold and sour rot in citrus fruits by the cationic antimicrobial peptide PAF56. Postharvest Biol. Technol. 136, 132–138. doi: 10.1016/j.postharvbio.2017.10.015
Wohlford-Lenane, C. L., Meyerholz, D. K., Perlman, S., Zhou, H., Tran, D., Selsted, M. E., et al. (2009). Rhesus theta-defensin prevents death in a mouse model of severe acute respiratory syndrome coronavirus pulmonary disease. J. Virol. 83, 11385–11390. doi: 10.1128/JVI.01363-09
Wrońska, A. K., and Boguś, M. I. (2020). Heat shock proteins (HSP 90, 70, 60, and 27) in Galleria mellonella (lepidoptera) hemolymph are affected by infection with Conidiobolus coronatus (entomophthorales). PLoS One 15:e0228556. doi: 10.1371/journal.pone.0228556
Wu, D., Gao, Y., Qi, Y., Chen, L., Ma, Y., and Li, Y. (2014). Peptide-based cancer therapy: opportunity and challenge. Cancer Lett. 351, 13–22. doi: 10.1016/j.canlet.2014.05.002
Wu, G., Ding, J., Li, H., Li, L., Zhao, R., Fan, X., et al. (2008). Effects of cations and pH on antimicrobial activity of thanatin and s-thanatin against Escherichia coli ATCC25922 and B. subtilis ATCC 21332. Curr. Microbiol. 57, 552–557. doi: 10.1038/npre.2008.2006.1
Wu, X., Wang, Z., Li, X., Fan, Y., He, G., Wan, Y., et al. (2014). In vitro and in vivo activities of antimicrobial peptides developed using an amino acid-based activity prediction method. Antimicrob. Agents Chemother. 58:5342. doi: 10.1128/AAC.02823-14
Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., et al. (2020). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 30, 343–355. doi: 10.1038/s41422-020-0305-x
Yang, X., Liu, W., Xi, G., Wang, M., Liang, B., Shi, Y., et al. (2019). Fabricating antimicrobial peptide-immobilized starch sponges for hemorrhage control and antibacterial treatment. Carbohyd. Polym. 222:115012. doi: 10.1016/j.carbpol.2019.115012
Yount, N. Y., and Yeaman, M. R. (2004). Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. U.S.A. 101, 7363–7368. doi: 10.1073/pnas.0401567101
Yu, H., Li, H., Gao, D., Gao, C., and Qi, Q. (2015). Secretory production of antimicrobial peptides in Escherichia coli using the catalytic domain of a cellulase as fusion partner. J. Biotechnol. 214, 77–82. doi: 10.1016/j.jbiotec.2015.09.012
Zahedifard, F., Lee, H., No, J. H., Salimi, M., Seyed, N., Asoodeh, A., et al. (2020). Comparative study of different forms of jellein antimicrobial peptide on leishmania parasite. Exp. Parasitol. 209:107823. doi: 10.1016/j.exppara.2019.107823
Zare-Zardini, H., Taheri-Kafrani, A., Ordooei, M., Ebrahimi, L., Tolueinia, B., and Soleimanizadeh, M. (2015). Identification and biochemical characterization of a new antibacterial and antifungal peptide derived from the insect Sphodromantis viridis. Biochem. Biokhimiia 80, 433–440. doi: 10.1134/S0006297915040069
Zelezetsky, I., and Tossi, A. (2006). Alpha-helical antimicrobial peptides—using a sequence template to guide structure-activity relationship studies. Biochim. Biophys. Acta Biomembr. 1758, 1436–1449. doi: 10.1016/j.bbamem.2006.03.021
Zhang, F., Cui, X., Fu, Y., Zhang, J., Zhou, Y., Sun, Y., et al. (2017). Antimicrobial activity and mechanism of the human milk-sourced peptide Casein201. Biochem. Biophys. Res. Commun. 485, 698–704. doi: 10.1016/j.bbrc.2017.02.108
Zhang, Z., Hou, L., Feng, L., Huang, S., Luo, M., Shao, S., et al. (2015). An antimicrobial peptide containing NGR motif has potent antitumor activity against CD13+ and CD13- tumor cells. Tumor Biol. 36, 8167–8175. doi: 10.1007/s13277-015-3402-6
Zhong, C., Zhu, N., Zhu, Y., Liu, T., Gou, S., Xie, J., et al. (2020). Antimicrobial peptides conjugated with fatty acids on the side chain of D-amino acid promises antimicrobial potency against multidrug-resistant bacteria. Eur. J. Pharm. Sci. 141:105123. doi: 10.1016/j.ejps.2019.105123
Zhu, Q. Z., Hu, J., Mulay, S., Esch, F., Shimasaki, S., and Solomon, S. (1988). Isolation and structure of corticostatin peptides from rabbit fetal and adult lung. Proc. Natl. Acad. Sci. U.S.A. 85, 592–596. doi: 10.1073/pnas.85.2.592
Keywords: antimicrobial peptides, classification, coronavirus, mode of action, design, motifs, application
Citation: Huan Y, Kong Q, Mou H and Yi H (2020) Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 11:582779. doi: 10.3389/fmicb.2020.582779
Received: 13 July 2020; Accepted: 23 September 2020;
Published: 16 October 2020.
Edited by:
Giovanna Batoni, University of Pisa, ItalyReviewed by:
Cesar de la Fuente-Nunez, University of Pennsylvania, United StatesPaula Gomes, University of Porto, Portugal
Copyright © 2020 Huan, Kong, Mou and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Kong, kongqing@ouc.edu.cn; kongqingg@163.com
 Yuchen Huan
Yuchen Huan Qing Kong
Qing Kong Haijin Mou
Haijin Mou Huaxi Yi
Huaxi Yi