- 1Department of Food Science and Nutrition, Illinois Institute of Technology, Bedford Park, IL, United States
- 2Division of Food Processing Science and Technology, U.S. Food and Drug Administration, Bedford Park, IL, United States
- 3Department of Microbiology and Molecular Genetics, University of California, Irvine, Irvine, CA, United States
- 4Department of Food Science and Human Nutrition, University of Illinois at Urbana-Champaign, Urbana, IL, United States
- 5Department of Biology, Illinois Institute of Technology, Chicago, IL, United States
Salmonella enterica subspecies I (ssp 1) is the leading cause of hospitalizations and deaths due to known bacterial foodborne pathogens in the United States and is frequently implicated in foodborne disease outbreaks associated with spices and nuts. However, the underlying mechanisms of this association have not been fully elucidated. In this study, we evaluated the influence of storage temperature (4 or 25°C), relative humidity (20 or 60%), and food surface characteristics on the attachment and survival of five individual strains representing S. enterica ssp 1 serovars Typhimurium, Montevideo, Braenderup, Mbandaka, and Enteritidis on raw in-shell black peppercorns, almonds, and hazelnuts. We observed a direct correlation between the food surface roughness and S. enterica ssp 1 attachment, and detected significant inter-strain difference in survival on the shell surface under various storage conditions. A combination of low relative humidity (20%) and ambient storage temperature (25°C) resulted in the most significant reduction of S. enterica on shell surfaces (p < 0.05). To identify genes potentially associated with S. enterica attachment and survival on shell surfaces, we inoculated a library of 120,000 random transposon insertion mutants of an S. Enteritidis strain on almond shells, and screened for mutant survival after 1, 3, 7, and 14 days of storage at 20% relative humidity and 25°C. Mutants in 155 S. Enteritidis genes which are involved in carbohydrate metabolic pathways, aerobic and anaerobic respiration, inner membrane transport, and glutamine synthesis displayed significant selection on almond shells (p < 0.05). Findings of this study suggest that various food attributes, environmental factors, and an unexpectedly complex metabolic and regulatory network in S. enterica ssp 1 collectively contribute to the bacterial attachment and survival on low moisture shell surface, providing new data for the future development of knowledge-based intervention strategies.
Introduction
Salmonella enterica subspecies I serovars have been implicated in a number of multi-state foodborne disease outbreaks associated with nuts and spices in recent decades in the United States. In 2004, serovar Enteritidis was linked to raw almonds in an outbreak in 12 states, leading to 29 cases of infections (Centers for Disease Control Prevention [CDC], 2004). In 2010, 272 cases of illnesses were reported in an outbreak of S. Montevideo linked to black pepper (Centers for Disease Control Prevention [CDC], 2010). In 2016, pistachios were implicated in an outbreak of S. Montevideo and S. Senftenberg infections, which sickened 11 persons in 9 states (Centers for Disease Control Prevention [CDC], 2018). Other reported salmonellosis outbreaks linked to nuts and nut-associated products included Turkish pine nuts contaminated by S. Enteritidis, nut butter by S. Braenderup, and peanut butter by S. Typhimurium and S. Mbandaka (Scheil et al., 1998; Centers for Disease Control Prevention [CDC], 2009, 2011, 2014). These foodborne outbreaks highlight the need for a thorough examination of the mechanisms of S. enterica survival and persistence on nuts and spices.
Salmonella enterica may contaminate raw nuts and spices at any stage of the food production process, including growing, harvesting, handling, processing, packaging, and storage. Following harvest, nuts and spices may be directly sold as ready-to-eat food without further heat treatment such as roasting; thus, S. enterica potentially residing on the surfaces may not be eradicated prior to packaging and retail. Storage temperature and relative humidity (RH) are considered to be two important environmental factors that influence S. enterica survival on nut and spice surfaces. Harvested raw nuts are usually stored at an RH between 55 and 60% to ensure quality and to limit mold growth (Codex Alimentarius, 2014). Ambient storage (25°C) is often used to sustain flavor quality but may lead to shortened shelf life of raw nuts, compared to storage at 4°C (Senesi et al., 1996; Bruhn et al., 2010).
Once attached to the shells, S. enterica is capable of surviving for long periods of time on low water activity foods, including nuts. For instance, Beuchat and Heaton (1975) demonstrated that S. enterica survive more than 32 weeks at low temperatures (≤ 5°C) on pecans. Blessington et al. (2012) reported that S. enterica survive under ambient storage conditions (23–25°C, 25–35% RH) on walnuts for more than 97 days. Extended survival of S. enterica was also documented by Kimber et al. (2012) on pistachios for at least 12 months at various storage temperatures including −19, 4, and 24°C (Centers for Disease Control Prevention [CDC], 2009).
Despite many previous efforts to assess the extended survival period of S. enterica on various raw nuts, scientific data are scarce on how different food surface characteristics, such as surface roughness, may impact bacterial attachment and survival, and how various S. enterica serovars and strains may differ in their relative ability to attach to and survive on nut shell surface. In addition, the molecular basis underlying the ability of S. enterica to survive on raw in-shell nuts remains largely unknown.
In this study, we assessed the population dynamics of several foodborne outbreak-associated S. enterica serovars on one spice, i.e., black peppercorns, and two different raw nuts, i.e., almonds and hazelnuts, under different storage conditions. We further employed electron microscopy and high-throughput sequencing techniques to explore the physical and molecular mechanisms that S. enterica uses to attach to and survive on the surface of these foods. Findings of this study shed new light on the behavior and physiology of this pathogen on nuts and peppercorns, furthering a better understanding of its dissemination and transmission that results in human foodborne illnesses.
Materials and Methods
Bacterial Strains
Five bacterial strains representing S. enterica subspecies I serovars Typhimurium (strain ATCC 19585), Montevideo (strain FDA691341), Braenderup (strain ATCC BAA-1739), Mbandaka (strain FDA660151), and Enteritidis (strain ATCC 13076) were used in this study (Table 1). Single colonies of each S. enterica stock culture were recovered in Brain Heart Infusion (BHI, Becton Dickinson and Co., Franklin Lakes, NJ) broth at 37°C with shaking at 200 rpm for 24 h. A total of 100 μl cultures were spread onto the surface of BHI agar plates and incubated for 24 h at 37°C. Bacterial cells were then harvested from the BHI agar plates using a spreader and 1 mL of pre-warmed (37°C) phosphate buffered saline (PBS, pH 7.4), spun down, and re-suspended in 1 mL of PBS. Bacterial suspensions were transferred to 15 mL sterile centrifuge tubes and adjusted to a final concentration of 10–11 log CFU/mL with PBS prior to inoculation. The initial inoculum level was determined by serial dilution and enumeration on Xylose Lysine Deoxycholate (XLD, Becton Dickinson and Co.) agar plates.
Food Samples
Whole black peppercorns, raw in-shell almonds, and raw in-shell hazelnuts were selected for this study to assess the attachment and survival of S. enterica. Peppercorn, almond, and hazelnut samples were purchased from local vendors and stored in sterile 500 mL containers at room temperature prior to experimental procedures. The background microbiota on the un-inoculated samples was determined to be below 1 log CFU/g prior to our experiments, and remained largely unchanged throughout the 2-week storage after inoculation with S. enterica.
Microbiological Assessment of S. enterica Population Dynamics on Peppercorns, Almonds, and Hazelnuts
Approximately 20 g of each food was surface-inoculated by mixing each sample with 1 mL of 10–11 log CFU/mL of individual cultures of S. Typhimurium, S. Montevideo, S. Braenderup, S. Mbandaka, and S. Enteritidis in 4 × 8 in whirl-pak bags (Nasco, Fort Atkinson, WI). The bags were then hand-massaged for 10 min. Inoculated samples were dried for 2 h in a biosafety cabinet and then placed in incubators under ambient conditions (i.e., 25°C and 60% RH) to simulate typical commercial storage and packaging conditions. Saturated salt solutions (Supplementary Table 1) were used to maintain the RH for storage as previously reported (Greenspan, 1977). At 0, 1, 3, 7, and 14 days, inoculated samples were transferred to 50 mL sterile centrifuge tubes containing 20 mL PBS for enumeration of viable S. enterica.
We developed a vortex-based separation method to collect S. enterica cells from the surface of artificially inoculated nut samples. At each sampling time point, 20 g of each sample was submerged in 20 mL PBS buffer in a 50 mL conical polypropylene centrifuge tube and vortexed at 300 rpm for 10 min. The bacterial cell suspension was collected. An additional 20 mL PBS buffer was then added to the sample in the same tube. A subsequent vortex at 3000 rpm for 10 min with 3 mm diameter sterile glass beads was performed to remove the remaining surface-attached S. enterica from the shells. The bacterial cell suspension from the subsequent 3000 rpm vortex was collected and mixed with that from the previous vortex at 300 rpm for enumeration. This method allowed maximum removal of surface-attached bacterial cells from shell samples while minimizing potential mechanical cell lysis. Two different culture media, BHI agar and XLD agar, were used to recover bacterial cells and to differentiate healthy versus impaired S. enterica cells. S. enterica was enumerated by serial dilution of the homogenates with PBS, followed by plating onto both of these agars.
An additional storage temperature at 4°C was tested to compare the attachment and survival behaviors of S. Enteritidis and S. Typhimurium on black peppercorn, almond, and hazelnut surfaces to those at 25°C under 20 or 60% relative humidity conditions, respectively.
Measurement of Shell Surface Roughness Using CLSM
The surface roughness of black peppercorns, almond shells, and hazelnut shells was measured using a confocal laser scanning microscopy (CLSM)-based method developed by Fransisca and Feng (2012). Briefly, each shell sample was firmly attached to a microscope slide in twelve replicates. CLSM (Nanofocus, Surf explorer®) was used to capture three-dimensional (3D) images of the surfaces at 20 × magnification with a measurement area of 800 μm × 800 μm. In order to obtain high-quality 3D images, we adjusted width and height based on the size of the samples. Three random spots were measured for each sample. Images were analyzed using the MountainsMap software (Digital Surf, Besançon, France) to convert 3D surface texture and surface morphological traits to ISO 25178-2 3D parameters (ISO, 2012). The surface area roughness parameter Sq is calculated as follows:
where Z2 (x, y) represents the squared distance from individual heights and depths to the mean plane.
Imaging of S. enterica Surface Attachment on Shell Surface Using SEM
Images were captured using scanning electron microscopy (SEM) to illustrate the patterns and morphology of S. enterica during initial attachment to the surface of the samples. Samples were prepared using the methods developed by Wen et al. (2009) and He et al. (2013), with minor modifications. Briefly, shell samples were dissected into small pieces (∼1 cm2) and integral and flat pieces were selected for imaging. Entire shell surfaces were spot-inoculated with 10 μL of 10–11 log CFU/mL of individual cultures of S. Enteritidis and S. Typhimurium and then air-dried for 24 h in a biosafety cabinet at ambient temperature (25°C). Inoculated shell samples were fixed with a primary fixative (2.5% glutaraldehyde in 0.1M cacodylate buffer, pH 7.2) for 2 h followed by a secondary fixative (1% osmium tetroxide aqueous solution in 0.1M cacodylate buffer, pH 7.2) without light for 1 h at room temperature. Fixed samples were dehydrated by sequential washes with electron microscopy-grade ethanol solutions (25, 50, 75, 85, 95, and 100%), and then dried using 100% hexamethyldisilazane (HMDS). Chemical reagents were purchased from Electron Microscopy Sciences (EMS) (Hatfield, PA). Samples were mounted on aluminum studs, sputter coated with Au and Pt to an 8 nm thickness, and examined using a JSM-6320F field emission scanning electron microscope (JEOL USA Inc, Peabody, MA) at an instrument magnification of 10,000 × at the University of Illinois Chicago microscopy core facility.
Transposon Sequencing Analysis of S. enterica Serovar Enteritidis PT4 on Almonds
A transposon insertion library in a fully sequenced S. enterica serovar Enteritidis PT4 strain was utilized, which had been constructed as previously described (de Moraes et al., 2017, 2018; Jayeola et al., 2020). Briefly, the EZ-Tn5 < T7/KAN-2 > promoter insertion kit (Epicentre Technologies Corporation, Madison, WI) was employed, and specific primers were used to add N18 random barcodes to both sides of the EZ-Tn5 transposon for sequencing analysis (Supplementary Table 2). The final transposon insertion library consisted of approximately 120,000 random insertion mutants.
To identify specific genes that were involved in S. enterica survival on almond shells, we adopted a published method (de Moraes et al., 2017; Jayeola et al., 2020), with minor modifications. Briefly, the entire Tn5 mutant library was surface-inoculated on in-shell almonds using the inoculation protocol described above. After 0, 1, 3, 7, and 14 days of storage under ambient conditions, S. enterica cells were collected from almond shells as described above, with minor modifications. Specifically, homogenates collected from the two-step separation were enriched in Luria Bertani (LB; Becton Dickinson and Co.) broth with 50 μg/mL kanamycin at 37°C with shaking for 8 h to reach ∼8 log CFU/mL. Enriched S. enterica cells were pelleted and plated onto LB agar containing kanamycin and incubated at 37°C for 24 h. Genomic DNA was extracted using Generation Capture Columns from Qiagen (Hilden, Germany). The DNA was subsequently used as template for PCR using primers flanking the N18 barcode, and the frequency of each barcode was determined by Illumina sequencing, as previously described (de Moraes et al., 2017, 2018; George et al., 2018; Jayeola et al., 2020).
Mapping and Identification of S. enterica Mutants Under Selection
To map each transposon insertion site on the genome, custom Perl scripts were used to trim the Tn5 priming sites and the corresponding N18 barcodes from the raw sequence reads. Bowtie2 (Version 2.3.0) was used to align the filtered reads to the S. Enteritidis P125109 reference genome. Custom Perl scripts were used to identify each mapped read to its corresponding N18 barcode.
For use in fitness experiments, the overall abundance of the N18 barcodes in each annotated gene was summed in each strand separately, thereby reporting genome features on both strands of each feature; e.g., coding and non-coding strands. Aggregated abundances for the input and output libraries were analyzed for statistical significance with EdgeR (Version 3.5), to identify mutants under positive or negative selection (Robinson et al., 2009; Robinson and McCarthy, 2010).
Statistical Analysis
All experiments were repeated at least three times, each with additional technical duplicates and independent cultures, to ensure data reproducibility for statistical analysis. S. enterica counts collected on selective XLD agar plates were used to minimize false positive counts due to background microflora. D-values were determined with the Bigelow model (Edwards and Berry, 1987). The data were also analyzed using post hoc Tukey’s honest significant difference (HSD) test, student’s t-test (GraphPad Prism software package, Version 5; Microsoft Excel), and analyses of variance (ANOVA), where a p-value of less than 0.05 was considered significant.
Results
S. enterica Inter-Strain Variation in Attachment and Survival on Nuts and Spice
Among hundreds of different pathogenic S. enterica ssp 1 serovars that can cause salmonellosis, only a handful have been implicated in foodborne disease outbreaks associated with raw nuts and spices. We therefore compared the relative ability of different S. enterica ssp 1 serovars to attach to and survive on raw nuts and spices, which serves as an initial step of food contamination. Five S. enterica strains, representing ssp 1 serovars Typhimurium, Montevideo, Braenderup, Mbandaka, and Enteritidis, and three shell surface, black peppercorns, almonds, and hazelnuts, were assessed.
Immediately after inoculation and air-drying, and prior to storage (i.e., at Day 0), we collected and enumerated S. enterica cells attached on black peppercorns, almonds and hazelnuts to evaluate the initial level of bacterial attachment for each strain on each type of shell surface. All five S. enterica strains displayed better initial attachment to black peppercorns and almonds than to hazelnuts; however, significant differences were observed among different strains (Supplementary Table 3). For example, S. Mbandaka displayed the highest initial attachment levels, ranging from 7.42 to 8.18 log CFU/g. In contrast, the attachment of S. Enteritidis on hazelnuts and black pepper was significantly lower than that of all other tested strains on the same surfaces (p < 0.05), suggesting S. enterica strains do differ in their capability to attach to nut or spice surfaces.
Over the course of the 14-day storage period at 25°C and 60% RH, all five S. enterica strains displayed an overall population reduction on different shell surfaces (Figure 1 and Supplementary Table 3). However, the rates of reduction were different among S. enterica strains and surfaces (p < 0.05). For instance, the S. Typhimurium population decreased by less than 1 log CFU/g on black pepper and almonds, whereas the S. Enteritidis population declined rapidly by 3 to 4 log CFU/g on hazelnuts. S. Montevideo, S. Braenderup, and S. Mbandaka showed similar behaviors of survival to that of S. Typhimurium.
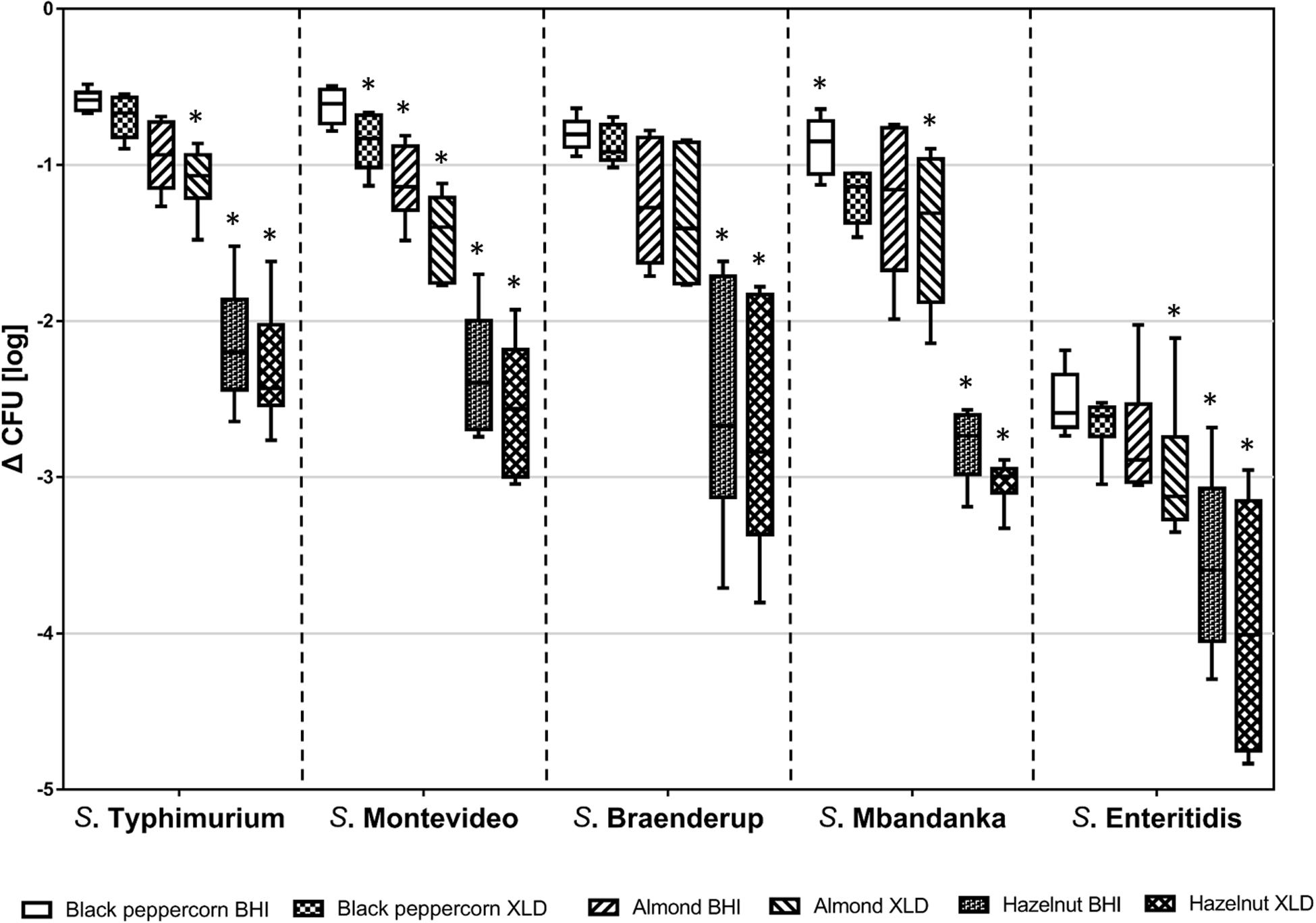
Figure 1. Log reduction of five S. enterica strains on shell surfaces over 14-day storage at 25°C and 60% relative humidity. The box-and-whiskers represent the maximum, 75th percentile, median (center line), 25th percentile, and the minimum values from top to bottom of the box, respectively. * indicates significant log reduction of S. enterica of the same strain (p < 0.05) among different shell surfaces. Error bars indicate standard deviations of means (n = 6).
Counting of surviving CFUs was performed on both a rich medium (BHI) and a more stringent selective medium (XLD). CFUs obtained on XLD were lower, suggesting that some bacterial cells were impaired due to the stresses that S. enterica encountered on the shells. Background microbiota including bacteria other than Salmonella and molds contributed to the higher microbial counts on non-selective BHI. To minimize the false positive plate counts of S. enterica due to the background microbiota, we chose to use XLD counts for further data analysis.
To better illustrate the relative ability of the bacterial strains to survive under the same storage conditions (i.e., 25°C and 60% RH), we calculated the D-values (i.e., time required to reach a 90% population reduction) of different S. enterica strains on the three types of surfaces based on XLD plate counts (Table 2). The majority of tested S. enterica strains survived better on black pepper and almond, with D-values ranging from 12 to 22 days, than on hazelnut, with D-values ranging from 4 to 7 days. Again, S. Enteritidis was substantially less capable of surviving on all three types of surfaces, with D-values that were 2–3 times lower than those of the other S. enterica strains.
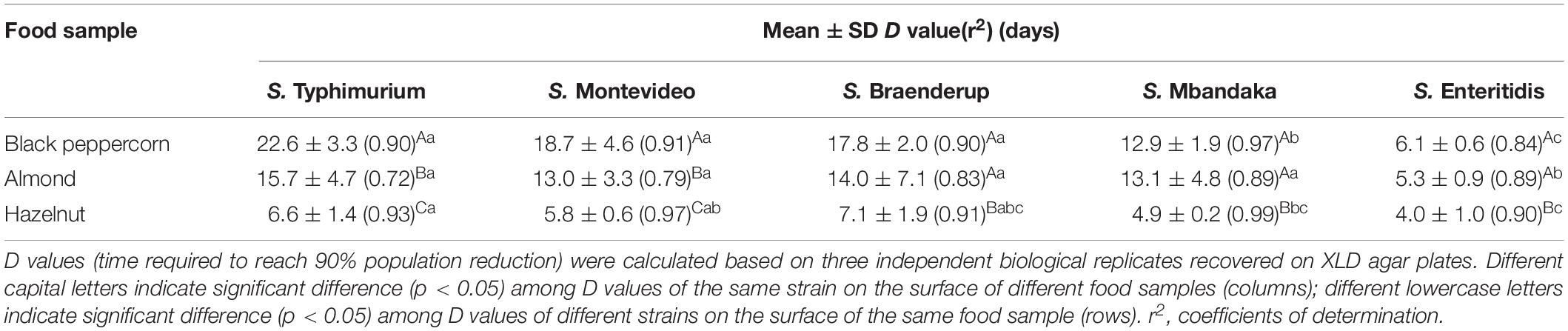
Table 2. D values for different S. enterica strains on the surface of nut samples stored at 25°C and 60% relative humidity.
Effects of Storage Temperature and Relative Humidity on S. enterica Survival on Nuts
Two important environmental factors that have been recognized to have a collective impact on S. enterica survival on food surfaces are storage temperature and relative humidity. In order to evaluate the effect of these factors on S. enterica survival on peppercorns, almonds, and hazelnuts we chose to compare two serovars, S. Enteritidis and S. Typhimurium, which appeared to be substantially different from each other with regards to surface attachment and survival under ambient storage condition, using three additional storage conditions. Figure 2 illustrates the population dynamics of these two strains on the surface of black peppercorn, almond and hazelnut under 4°C/20% RH, 4°C/60% RH, 25°C/20% RH, and 25°C/60% RH, respectively, over a 14-day storage.
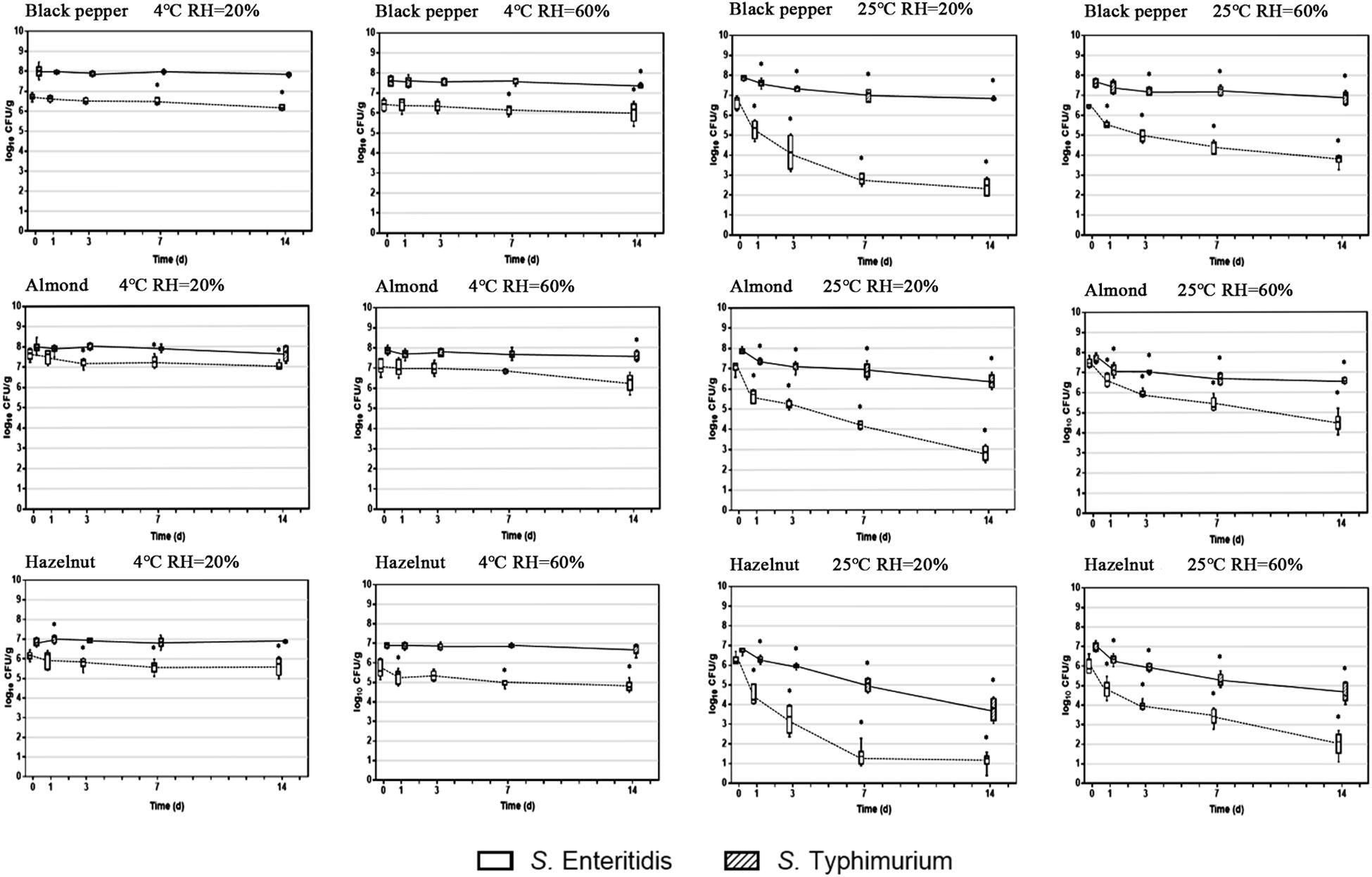
Figure 2. Population dynamics of S. enterica Enteritidis and Typhimurium on black peppercorn, almond, and hazelnut over 14 days of storage. Two storage temperatures (4 and 25°C) and two relative humidity conditions (20 and 60%) were evaluated. The solid lines represent S. Typhimurium and the dotted lines represent S. Enteritidis. The box-and-whiskers represent the maximum, 75th percentile, median (center line), 25th percentile, and the minimum values from top to bottom of the box, respectively. * Indicates significant population changes of the same S. enterica strain (p < 0.05) in comparison to initial inoculation. Error bars indicate standard deviations of means (n = 6).
At 4°C, bacterial populations of both S. Enteritidis and S. Typhimurium did not show a significant reduction on all three types of surfaces, independent from RH. However, at 25°C, both S. enterica strains displayed a population decrease on the three surfaces. For instance, S. Enteritidis showed a reduction of ∼ 5 log CFU/g on hazelnut at 20% RH. Interestingly, ambient storage temperature seemed to be less impactful on S. Typhimurium survival on the surfaces, with an average population reduction of < 2 log CFU/g. Our results show that a combination of ambient temperature (25°C) and low humidity (20%) led to an overall higher bacterial reduction for S. enterica on peppercorns, almonds, and hazelnuts. Furthermore, storage temperature appeared to have a more direct impact on overall S. enterica survival than relative humidity.
Effects of Surface Topology on S. enterica Attachment
To explore the physical mechanisms of S. enterica attachment on raw nut shells and peppercorns, we compared the patterns and morphology of attached bacteria in relation to the roughness of the different surfaces using scanning microscopy. The left column in Figure 3 shows the reconstructed 3-D confocal laser scanning microscopy (CLSM) images of the surfaces of black peppercorns, almonds and hazelnuts. Surface roughness was calculated based on the 3-D images and accompanying areal roughness parameters by Matlab as described (Wang et al., 2009; Fransisca and Feng, 2012). Hazelnuts had a relatively smooth surface with an Sq value of 4.8 ± 1.1 μm, whereas black peppercorns had a rough surface with an Sq value of 20.4 ± 3.2 μm. The numbers of attached S. enterica cells were positively correlated with the surface roughness for all five S. enterica strains on all tested surfaces when stored at 25°C and 60% RH. The right column in Figure 3 shows the scanning electron microscopy (SEM) imaging of surface-attached S. enterica cells, and demonstrated that S. enterica tended to aggregate in the uneven spaces of the surfaces, such as grooves and cavities, with minor morphological changes.
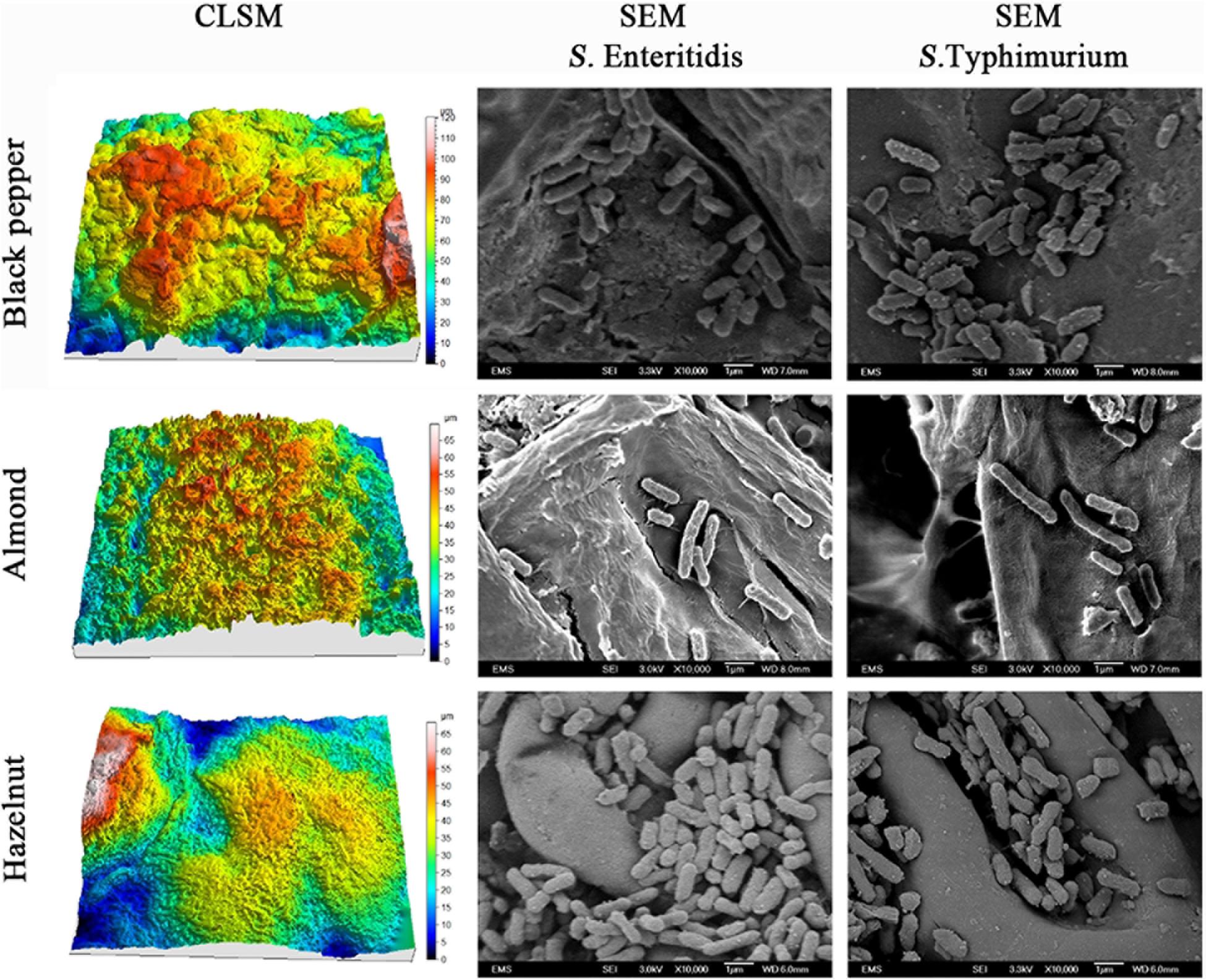
Figure 3. Reconstructed three-dimensional CLSM images showing the surface topology of black peppercorn, almond, and hazelnut at 1,000 × magnifications, and SEM images showing the locations of surface-attached S. enterica Enteritidis and Typhimurium cells on different shell surfaces at 10,000 × magnification.
S. Enteritidis Genes Under Selection on the Surface of Almonds
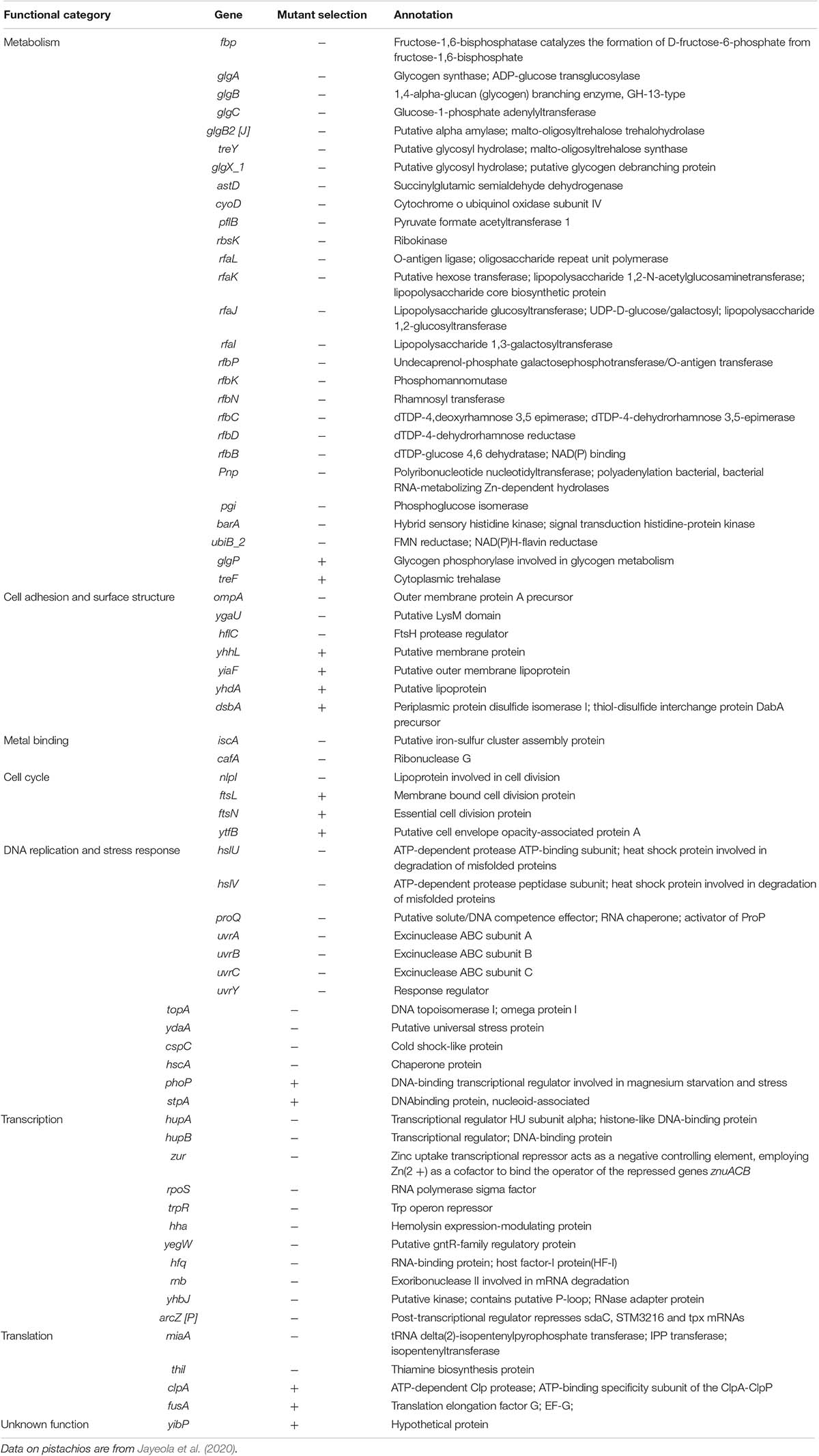
We inoculated a random transposon insertion library of an S. Enteritidis strain on almond shells to screen for S. enterica genes that were either positively or negatively selected during bacterial residence on the shell surface over a 7-day ambient storage (25°C under 60% RH). We monitored the proportion of each transposon mutant at days 0, 1, 3, 7, and 14 (Supplementary Table 6). Using this approach, we identified a total of 102 negatively selected and 53 positively selected S. enterica genes with statistical significance (Supplementary Tables 4, 5), including genes encoding metabolic regulators, pathogenicity island proteins, membrane proteins, fimbrial subunits, transcriptional regulators, and hypothetical proteins. Table 3 lists a subset of S. enterica genes that were under strong selection on almond shells with functional annotations. More than half of the negatively selected genes were involved in various metabolism pathways. Figure 4 illustrates a number of negatively selected genes involved in carbohydrate metabolic pathways, aerobic and anaerobic respiration, inner membrane transport, and glutamine synthesis. In addition, we found that every successfully assayed mutant in the pSEN virulence plasmid was positively selected. As the plasmid has its own origin of replication, this observation was likely due to the copy number of the plasmid being higher relative to the copy number of the genome at later time points.
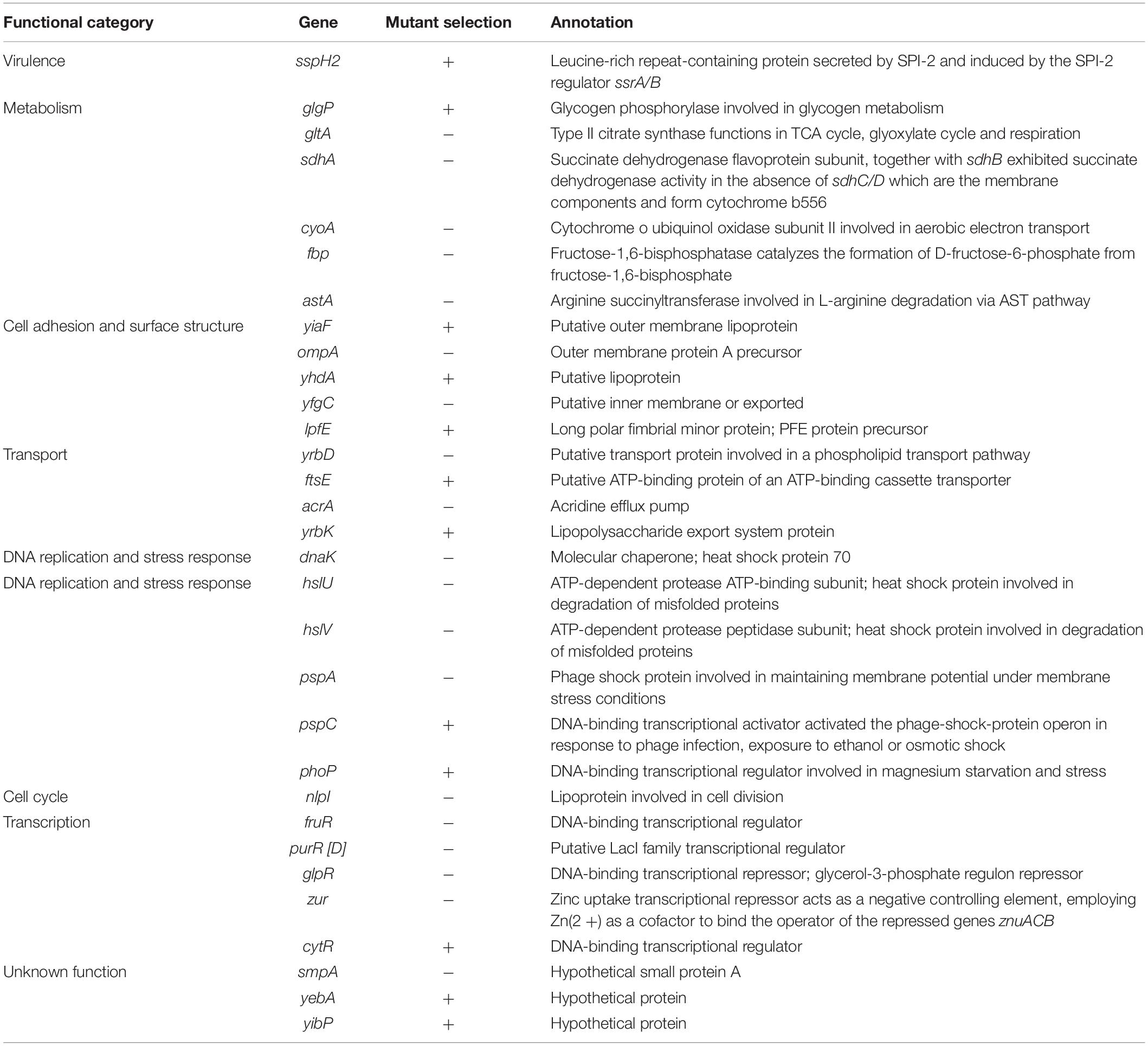
Table 3. Selected S. enterica Enteritidis genes under selection on almonds at 14 days of storage at 25°C and 60% relative humidity (p < 0.05).
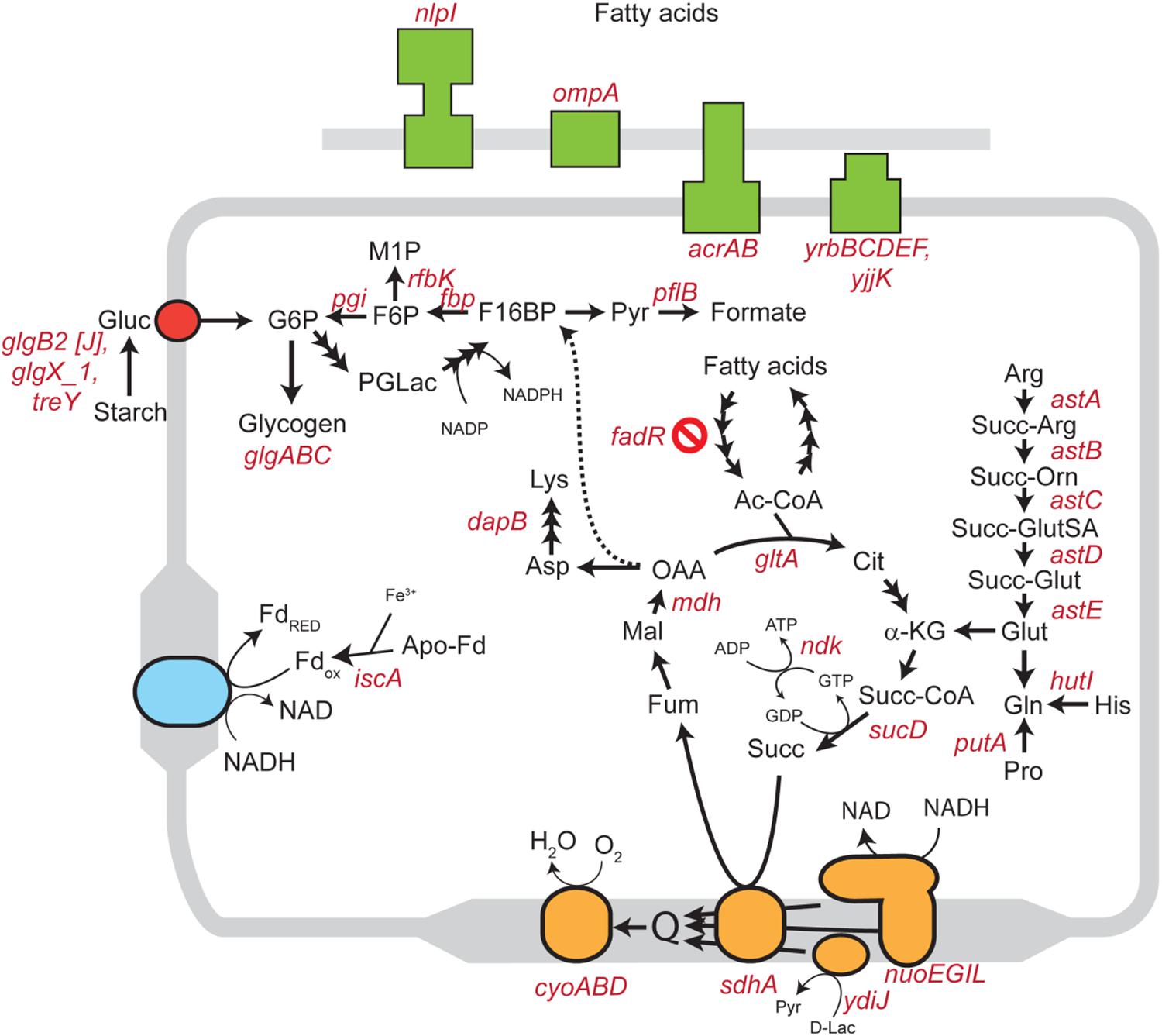
Figure 4. Schematic diagram showing various S. enterica genes involved in carbohydrate, lipid, amino acid, and energy metabolism for which mutants displayed significantly negative selection on almond surfaces after 7-day storage at 25°C. Genes involved in aerobic and anaerobic respiration, cholesterol transport systems, and inner membrane transport systems in S. Enteritidis are highlighted in orange, blue, green and red colors, respectively. Abbreviations: Gluc, glucose; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; M1P, D-mannitol 1-phosphate; F16BP, fructose 1,6-bisphosphate; Pyr, pyruvate; PGLac, phosphogluconolactone; Fd, ferredoxin; Ac-CoA, acetyl coenzyme A; Cit, citrate; α-KG, alpha-ketoglutarate; Succ-CoA, succinyl-CoA; Succ, succinate; Fum, fumarate; Mal, malate; OAA, oxaloacetate; Asp, aspartic acid; Lys, lysine; Arg, arginine; Succ-Arg, N2-succinylarginine; Succ-Orn, N2-succinylornithine; Succ-GluSA, N-succinyl-L-glutamate 5-semialdehyde; Succ-Glut, N-succinyl-L-glutamate; Glut, glutamate; Gln, glutamine; His, histidine; Pro, proline; D-Lac, D-lactate.
Interestingly, a large number of Tn5 mutants in S. Enteritidis which were strongly selected on almonds were also found to be strongly selected in S. enterica on pistachios (Jayeola et al., 2020). Table 4 lists the S. enterica genes that were selected on both almonds and pistachios. Supplementary Table 7 provides a complete list of genes that were either consistently or differentially selected on almonds and pistachios. Among the genes for which mutants were positively selected in both S. Enteritidis on almonds and S. Typhimurium on pistachios was the glgP gene. Strikingly, mutants in glgP were selected in the opposite direction than in glgABC encoded in the same operon. We selected this gene for further analysis. We constructed a deletion mutant of the glgP gene in S. Enteritidis and a complemented strain containing glgP encoded on a plasmid. The constructs displayed no significant difference in growth in BHI at 37°C for 48 h when compared to the wild-type strain (data not shown), suggesting that this gene is not needed in rich media. After 7-days storage on almond shells at 25°C and 60% RH, S. Enteritidis ΔglgP displayed better survival than its complemented strain and the wild-type strain (data not shown). To our knowledge, no positively selected mutant in Salmonella has previously been confirmed using a single gene deletion mutant on a food product.
Discussion
A mechanistic understanding of the attachment and survival of S. enterica on nuts and spices is an important step toward developing effective control and mitigation strategies to reduce or eliminate pathogen contamination in these food products. Yet, this aspect of food safety has not been thoroughly elucidated by previous studies. Our study demonstrated that different S. enterica strains display variable abilities to attach to and survive on raw shell surfaces. For instance, S. Enteritidis was less capable of attaching to nut shells, whereas S. Mbandaka showed a better capability of attachment. In addition, the overall rate of survival of different S. enterica strains attached to shell surface also appeared to be quite variable, suggesting strain- or serovar-specific behavior should be taken into consideration– an important aspect when evaluating and validating a particular mitigation strategy on raw in-shell foods.
It is known that desiccation can inhibit S. enterica proliferation and metabolic functions (Deng et al., 2012). Water activity (aw) on raw shells can range from 0.321 ± 0.20 on peanuts (Fong and Wang, 2016) to 0.585 ± 0.003 on chia seeds. On shell surfaces, S. enterica displayed an overall population reduction over the 14-day period at ambient storage conditions. This reduction was likely caused by the limited water availability for bacterial use (Uesugi et al., 2006; Keller et al., 2012; Blessington et al., 2013; Bowman et al., 2015). We observed that the magnitude of bacterial reduction varied among different S. enterica spp 1 strains. In addition to inhibiting bacterial growth, desiccation can also trigger various stress response mechanisms, enabling the pathogen to persist in low moisture environments for even longer periods of time. Some published studies have shown that once adapted to a desiccated environment, S. enterica can survive for months or even years in orchards and surrounding environments (Beuchat and Heaton, 1975; Blessington et al., 2012; Kimber et al., 2012). A consistent finding of these studies was that the populations of S. enterica initially declined quickly under low moisture stress, followed by a slower population decline and long term persistence (Gurtler et al., 2014), similar to what we observed in this study.
Storage temperature plays a critical role in S. enterica survival on shell surface. In this study, we demonstrated that the rate of reduction of S. enterica stored at ambient temperature (25°C) was significantly higher than that under refrigeration temperature (4°C). Note that the rate of the population reduction of S. Enteritidis on almond shells under ambient storage conditions (0.21 ± 0.03 log CFU/g per day) was higher than the rates observed for S. enterica on other nut products under similar conditions. These conditions included almond kernels (0.25 ± 0.05 log CFU/g per month), pecan halves (0.15 log CFU/g per month), and peanut kernels (0.22 log CFU/g per month) (Uesugi et al., 2006; Brar et al., 2015; Santillana Farakos et al., 2016). Refrigeration temperature, in contrast, was protective for S. enterica on nut shells, consistent with other studies (Uesugi et al., 2006; Brar et al., 2015).
One factor, among possibly many others, which can directly influence the attachment and survival of S. enterica on nuts and peppercorns, is the shell surface roughness (Ra). Our study showed that all tested S. enterica strains survived better on the rougher surface of almond shells and black peppercorns than on the smoother surface of hazelnut shells. A positive correlation between surface roughness and survival of S. enterica on surfaces was observed when samples were stored at 25°C and 60% RH (Figure 1 and Supplementary Table 3). This is consistent with studies of other bacterial pathogens in other food models. For instance, Wang et al. (2009) reported that E. coli O157:H7 displayed a higher attachment rate on the rough cantaloupe surface (Ra = 14.18 ± 0.25 μm) than on a smooth apple surface (Ra = 1.43 ± 0.13 μm). Our SEM analysis further demonstrated that S. enterica tended to aggregate at locations of uneven surface structures, such as grooves and cavities (Figure 3), which could provide a “safe harbor” for the pathogen to attach and escape decontamination.
A number of previous studies suggested that the overall bacterial attachment and survival on various biotic and abiotic surfaces can be mediated by genes encoding transcription factors, regulators, virulence determinants, and cell surface components such as cellulose, flagella, and fimbriae (Barak et al., 2005; Marvasi et al., 2013; Salazar et al., 2013; Tan et al., 2016; de Moraes et al., 2017; Jayeola et al., 2020). We used a library of over 120,000 mutant strains, each with a random Tn5 transposon insertion, to identify the subset of mutants that is under selection during up to 14 days of ambient storage on almond shells. When a Tn5 mutant is lost or less abundant in an “output” pool collected after an experimental treatment, compared to an “input” pool prior to treatment (initial Tn5 library inoculated on almond shells), this “negative selection” suggests a loss of bacterial fitness in a particular environment. This fitness loss indicates an important functional role for the location of the transposon insertion in bacterial survival. Our transposon sequencing analysis identified a few hundred genes in S. Enteritidis from a wide variety of functional groups for which mutants were under selection after 7-day storage on almond shells under ambient conditions (Table 3). We identified a number of S. enterica genes related to the bacterial stress response, DNA repair systems, and cellular metabolisms, for which mutants were negatively selected on almond shells. These included genes involved in carbohydrate metabolic pathways, aerobic and anaerobic respiration, inner membrane transport, and glutamine synthesis (Figure 4).
Two prominent genes that were negatively selected in our screen are dnaK and pspA. The chaperone protein DnaK was previously found to play a role in S. Enteritidis survival in low moisture peanut oil (Deng et al., 2012). In E. coli, this protein can interact with the co-chaperone DnaJ under different stress conditions, thereby preventing the aggregation of stress-denatured proteins (Liberek et al., 1991). Gene pspA encodes the phage shock protein A (PspA) in the pspABCDE operon, which helps to maintain proton-motive force (ΔμH +) of the cell under membrane stress conditions (Kleerebezem et al., 1996). PspA expression is also induced under more general environmental stress conditions, including heat (50°C), ethanol (10%) or hyperosmolarity shock (Model et al., 1997).
Other negatively selected S. enterica genes on almonds included sdhA and the ast operon. Gene sdhA encodes a membrane-bound subunit of the succinate dehydrogenase (SDH) flavoprotein in the tricarboxylic acid (TCA) cycle (Park et al., 1995). The astABCDE operon encodes enzymes in the ammonia-producing arginine succinyltransferase (AST) pathway and controls arginine catabolism during bacterial aerobic exponential growth (Schneider et al., 1998). The negative selection of stress response, DNA repair and metabolism-related genes mentioned above in S. Enteritidis suggested a collective contribution of various pathways in promoting and sustaining S. enterica survival and persistence on almond shells.
We found that a number of genes belonging to cell adhesion and surface structures were under positive selection when mutated by Tn5, suggesting that expression of these genes is disadvantageous for S. enterica survival on almond shells. Notably, mutations in phoP and glgP were positively selected in both S. Enteritidis on almonds and in S. Typhimurium on pistachios. PhoP is part of a two-component system that controls virulence, Mg2+ homeostasis, and resistance to antimicrobial peptides, and controls 9% of genes in Salmonella (Soncini et al., 1996; Walters et al., 2006; Baker et al., 2014; Colgan et al., 2016; Bansal et al., 2017). Gene glgP encodes glycogen phosphorylase, which catalyzes the rate-limiting step in glycogen breakdown by removing glucose units from the polysaccharide (Alonso-Casajus et al., 2006). In E. coli, a close relative of S. enterica, the ability to metabolize glycogen is associated with a rapid response to carbon source starvation (Sekar et al., 2020). That glycogen catabolism is disadvantageous in two different Salmonella serovars in two different nuts is surprising, and warrants additional experimental validation. Another example is a gene encoding the fimbrial adhesin protein LpfE, which was under positive selection. LpfE is part of the lpfABCC’DE fimbrial operon, which encodes long polar fimbriae – a structure that contributes to the attachment of S. Typhimurium to murine small intestine Peyer’s patch cells and participates in the adherence phenotype during EHEC O157:H7 microcolony formation (Baumler et al., 1996; Torres et al., 2002). The lpfABCC’DE fimbrial operon in EHEC O113:H21 has also been shown to increase bacterial adherence to the epithelial cells of Chinese hamster ovary-K1 (CHO-K1) cells (Doughty et al., 2002). The positive selection of lpfE on almonds suggested that expression of this gene is, in contrast, disadvantageous for S. enterica attachment to and/or survival on the almond surface.
Interestingly, a comparison of our S. Enteritidis genes under selection on almonds with genes under selection from a recent study using S. Typhimurium on pistachios (Jayeola et al., 2020), revealed that a majority of these genes were commonly selected on both (Table 4). For example, genes hslU, fbp, and zur were all negatively selected on both almonds and pistachios. Gene hslU encodes an ATP-dependent protease for the degradation of misfolded polypeptides during heat-shock (Rohrwild et al., 1996; Burton et al., 2005). Gene fbp encodes a key enzyme, fructose-1,6-bisphosphatase, which functions in the Calvin cycle and the gluconeogenesis pathway (Nel and Terblanche, 1992; Daie, 1993). This gene is also related to bacterial attachment and biofilm formation in Streptococcus infections (Christie et al., 2002; Barros et al., 2015; Fessia et al., 2019). Zinc uptake transcriptional repressor Zur acts as a negative controlling element when employing Zn2+ as a cofactor to bind the operator of the repressed genes cluster znuACB (Patzer and Hantke, 2000). The strong negative selection of these genes collectively indicated the involvement of their corresponding proteins in the survival of S. enterica on low moisture nut shells.
Some limitations inherent to the nature of our experiments did exist. For instance, we used artificial surface inoculation to create very high bacterial loads on nut shells, a typical approach used in bacterial survival and challenge studies. Such high level of bacterial loads may be unrealistic in the event of natural contamination of nuts and spices. In addition, the relative ability of S. enterica attachment and survival on different shell surface was evaluated by separating bacterial cells from the shells at specific sampling time points and enumerating on BHI and XLD media. S. enterica cells that became severely injured and non-culturable but remained infectious could be underestimated. Several other factors such as bacterial acclimatization during air-drying and recovery can also influence the initial bacterial loads on shell surfaces. Therefore, the initial inoculation levels may not accurately reflect the bacterial ability of attachment and survival on respective shell surfaces, which remains a major technical challenge for this type of studies.
Future studies on the population dynamics of S. enterica over a longer storage period (e.g., 10–40 weeks or more) will provide additional information on how this pathogen behaves on nuts and spices. Use of lower inoculation levels may better reflect contamination events that are likely to occur in foodborne disease outbreaks. While in this study we demonstrated striking concordance in the S. enterica genes under selection in two different serovars on two different nuts (Table 4), studies on other low-moisture foods, using mutant libraries of other S. enterica strains and serovars, will provide further evidence for genes and pathways which are critical for the survival and persistence of this pathogen. Additional experiments may also involve individual gene knockouts (including complementation) for gene functional characterization. These follow-up studies using other S. enterica strains and food models will lead to a better understanding of the survival mechanisms, stress physiology, transmission, and epidemiology of this pathogen in different low-moisture foods.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
YL, MM, and WZ designed the experiments. YL, PD, SP, WC, P-SP, OJ, and MM performed the experiments and analyzed the data. YL wrote the manuscript draft. YL, JS, YH, MT, OJ, HF, MM, SP, and WZ edited and approved the manuscript. WZ and MM secured the funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the United States Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA) Agriculture and Food Research Initiative (AFRI) awards 2014-67017-21707, 2015-67017-23360, 2016-67017-24432, 2017-67015-26085; NIFA Hatch grant CA-D-PLS-2327-H, NIFA-BARD 2017-67017-26180 and by NIH grant AI 139557. JS was supported by the Oak Ridge Institute for Science and Education Research (ORISE) Participation Program to the U.S. Food and Drug Administration (FDA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Bo Zhang, Yang Zhai, Ke Sui, Susanne E. Keller, Leikai Chen, Chunxuan Feng, and Christina Carstens for their technical assistance in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.582202/full#supplementary-material
References
Alonso-Casajus, N., Dauvillee, D., Viale, A. M., Munoz, F. J., Baroja-Fernandez, E., Moran-Zorzano, M. T., et al. (2006). Glycogen phosphorylase, the product of the glgP gene, catalyzes glycogen breakdown by removing glucose units from the nonreducing ends in Escherichia coli. J. Bacteriol. 188, 5266–5272.
Baker, J. J., Johnson, B. K., and Abramovitch, R. B. (2014). Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources. Mol. Microbiol. 94, 56–69.
Bansal, R., Anil Kumar, V., Sevalkar, R. R., Singh, P. R., and Sarkar, D. (2017). Mycobacterium tuberculosis virulence regulator PhoP interacts with alternative sigma factor SigE during acid stress response. Mol. Microbiol. 104, 400–411.
Barak, J. D., Gorski, L., Naraghi-Arani, P., and Charkowski, A. O. (2005). Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 71, 5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005
Barros, E. M., Lemos, M., Souto-Padrón, T., and Giambiagi-deMarval, M. (2015). Phenotypic and genotypic characterization of biofilm formation in Staphylococcus haemolyticus. Curr. Microbiol. 70, 829–834.
Baumler, A. J., Tsolis, R. M., and Heffron, F. (1996). The lpf fimbrial operon mediates adhesion of Salmonella Typhimurium to murine Peyer’s patches. Proc. Natl. Acad. Sci. U.S.A. 93, 279–283.
Beuchat, L. R., and Heaton, E. K. (1975). Salmonella survival on pecans as influenced by processing and storage conditions. Appl. Microbiol. 29, 795–801. doi: 10.1128/aem.29.6.795-801.1975
Blessington, T., Mitcham, E. J., and Harris, L. J. (2012). Survival of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes on inoculated walnut kernels during storage. J. Food Prot. 75, 245–254. doi: 10.4315/0362-028X.JFP-11-278
Blessington, T., Theofel, C. G., Mitcham, E. J., and Harris, L. J. (2013). Survival of foodborne pathogens on inshell walnuts. Int. J. Food Microbiol. 166, 341–348. doi: 10.1016/j.ijfoodmicro.2013.07.016
Bowman, L. S., Waterman, K. M., Williams, R. C., and Ponder, M. A. (2015). Inoculation preparation affects survival of Salmonella enterica on whole black peppercorns and cumin seeds stored at low water activity. J. Food Prot. 78, 1259–1265. doi: 10.4315/0362-028X.JFP-14-483
Brar, P. K., Proano, L. G., Friedrich, L. M., Harris, L. J., and Danyluk, M. D. (2015). Survival of Salmonella, Escherichia coli O157:H7, and Listeria monocytogenes on raw peanut and pecan kernels stored at 224, 4, and 22uC. J. Food Prot. 78, 323–332. doi: 10.4315/0362-028X.JFP-14-327
Bruhn, C., Harris, L. J., Giovanni, M., and Metz, D. (2010). Nuts: Safe Methods for Consumers to Handle, Store, and Enjoy. UCANR Publications. Available online at: https://escholarship.org/uc/item/04f9p7vv (accessed September 18, 2020).
Burton, R. E., Baker, T. A., and Sauer, R. T. (2005). Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat. Struct. Mol. Biol. 12, 245–251.
Centers for Disease Control Prevention [CDC] (2004). Outbreak of Salmonella serotype enteritidis infections associated with raw almonds - United States and Canada, 2003-2004. Morb. Mortal. Wkly. Rep. 53, 484–487.
Centers for Disease Control Prevention [CDC] (2009). Multistate outbreak of Salmonella infections associated with peanut butter and peanut butter–containing products — United States, 2008–2009. MMWR. Morb. Mortal. Wkly. Rep. 58, 85–90.
Centers for Disease Control Prevention [CDC] (2010). Salmonella montevideo infections associated with salami products made with contaminated imported black and red pepper — United States, July 2009–April 2010. MMWR. Morb. Mortal. Wkly. Rep. 59, 1647–1650.
Centers for Disease Control Prevention [CDC] (2011). Multistate Outbreak of Human Salmonella Enteritidis Infections Linked to Turkish Pine Nuts (Final Update). Available online at: https://www.cdc.gov/salmonella/2011/pine-nuts-11-17-2011.html (accessed June 19, 2020).
Centers for Disease Control Prevention [CDC] (2014). Multistate Outbreak of Salmonella Braenderup Infections Linked to Nut Butter Manufactured by nSpired Natural Foods, Inc. (Final Update). Available online at: https://www.cdc.gov/salmonella/braenderup-08-14/index.html (accessed June 19, 2020).
Centers for Disease Control Prevention [CDC] (2018). Multistate Outbreak of Salmonella Montevideo Infections Linked to Raw Sprouts (Final Update). Available online at: https://www.cdc.gov/salmonella/montevideo-01-18/index.html (accessed June 19, 2020).
Christie, J., McNab, R., and Jenkinson, H. F. (2002). Expression of fibronectin-binding protein FbpA modulates adhesion in Streptococcus gordonii The GenBank accession number for the sequence reported in this paper is X65164. Microbiology 148, 1615–1625.
Codex Alimentarius (2014). Code of Hygienic Practice for Spices and Dried Aromatic Herbs. Rome: Food and Agriculture Organization.
Colgan, A. M., Kroger, C., Diard, M., Hardt, W.-D., Puente, J. L., Sivasankaran, S. K., et al. (2016). The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 12:e1006258. doi: 10.1371/journal.pgen.1006258
Daie, J. (1993). Cytosolic fructose-1, 6-bisphosphatase: a key enzyme in the sucrose biosynthetic pathway. Photosynth. Res. 38, 5–14.
de Moraes, M. H., Desai, P., Porwollik, S., Canals, R., Perez, D. R., Chu, W., et al. (2017). Salmonella persistence in tomatoes requires a distinct set of metabolic functions identified by transposon insertion sequencing. Appl. Environ. Microbiol. 83:e03028-16. doi: 10.1128/AEM.03028-3016
de Moraes, M. H., Soto, E. B., González, I. S., Desai, P., Chu, W., Porwollik, S., et al. (2018). Genome-wide comparative functional analyses reveal adaptations of Salmonella sv. newport to a plant colonization lifestyle. Front. Microbiol. 9:877. doi: 10.3389/fmicb.2018.00877
Deng, X., Li, Z., and Zhang, W. (2012). Transcriptome sequencing of Salmonella enterica serovar Enteritidis under desiccation and starvation stress in peanut oil. Food Microbiol. 30, 311–315. doi: 10.1016/j.fm.2011.11.001
Doughty, S., Sloan, J., Bennett-Wood, V., Robertson, M., Robins-Browne, R. M., and Hartland, E. L. (2002). Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70, 6761–6769.
Edwards, D., and Berry, J. J. (1987). The efficiency of simulation-based multiple comparisons. Biometrics 43, 913–928. doi: 10.2307/2531545
Fessia, A. S., Dieser, S. A., Raspanti, C. G., and Odierno, L. M. (2019). Genotyping and study of adherence-related genes of Streptococcus uberis isolates from bovine mastitis. Microb. Pathog. 130, 295–301.
Fong, K., and Wang, S. (2016). Strain-specific survival of Salmonella enterica in peanut oil, peanut shell, and Chia seeds. J. Food Prot. 79, 361–368. doi: 10.4315/0362-028X.JFP-15-419
Fransisca, L., and Feng, H. (2012). Effect of surface roughness on inactivation of Escherichia coli O157:H7 87-23 by new organic acid-surfactant combinations on alfalfa, broccoli, and radish seeds. J. Food Prot. 75, 261–269. doi: 10.4315/0362-028X.JFP-11-279
George, A. S., Cox, C. E., Desai, P., Porwollik, S., Chu, W., de Moraes, M. H., et al. (2018). Interactions of Salmonella enterica serovar Typhimurium and Pectobacterium carotovorum within a tomato soft rot. Appl. Environ. Microbiol. 84:e1913-17. doi: 10.1128/AEM.00927-918
Greenspan, L. (1977). Humidity fixed points of binary saturated aqueous solutions. J. Res. Natl. Bur. Stand Sect. A Phys. Chem. 81, 89–96. doi: 10.6028/jres.081A.011
Gurtler, J. B., Doyle, M. P., and Kornacki, J. L. (2014). The Microbiological Safety of Low Water Activity Foods and Spices. New York, NY: Springer.
He, Y., Li, Y., Salazar, J. K., Yang, J., Tortorello, M. L., and Zhang, W. (2013). Increased water activity reduces the thermal resistance of Salmonella enterica in peanut butter. Appl. Environ. Microbiol. 79, 4763–4767. doi: 10.1128/AEM.01028-1013
ISO (2012). ISO 25178-2” GPS—Surface Texture: Areal—Part 2: Terms, Definitions and Surface Texture Parameters. Geneva: International Organization for Standardization.
Jayeola, V., McClelland, M., Porwollik, S., Chu, W., Farber, J., and Kathariou, S. (2020). Identification of novel genes mediating survival of Salmonella on low-moisture foods via transposon sequencing analysis. Front. Microbiol. 11:726. doi: 10.3389/fmicb.2020.00726
Keller, S. E., Grasso, E. M., Halik, L. A., Fleischman, G. J., Chirtel, S. J., and Grove, S. F. (2012). Effect of growth on the thermal resistance and survival of Salmonella Tennessee and oranienburg in peanut butter, measured by a new thin-layer thermal death time device. J. Food Prot. 75, 1125–1130. doi: 10.4315/0362-028X.JFP-11-477
Kimber, M. A., Kaur, H., Wang, L., Danyluk, M. D., and Harris, L. J. (2012). Survival of Salmonella, Escherichia coli O157:H7, and Listeria monocytogenes on inoculated almonds and pistachios stored at -19, 4, and 24°C. J. Food Prot. 75, 1394–1403. doi: 10.4315/0362-028X.JFP-12-023
Kleerebezem, M., Crielaard, W., and Tommassen, J. (1996). Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15, 162–171.
Liberek, K., Marszalek, J., Ang, D., Georgopoulos, C., and Zylicz, M. (1991). Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878. doi: 10.1073/pnas.88.7.2874
Marvasi, M., Cox, C. E., Xu, Y., Noel, J. T., Giovannoni, J. J., and Teplitski, M. (2013). Differential regulation of Salmonella Typhimurium genes involved in O-antigen capsule production and their role in persistence within tomato fruit. Mol. Plant Microbe Interact. 26, 793–800. doi: 10.1094/MPMI-09-12-0208-R
Model, P., Jovanovic, G., and Dworkin, J. (1997). The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24, 255–261.
Nel, W., and Terblanche, S. E. (1992). Plant fructose-1, 6-bisphosphatases: characteristics and properties. Int. J. Biochem. 24, 1267–1283.
Park, S. J., Tseng, C. P., and Gunsalus, R. P. (1995). Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol. Microbiol. 15, 473–482. doi: 10.1111/j.1365-2958.1995.tb02261.x
Patzer, S. I., and Hantke, K. (2000). The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275, 24321–24332.
Robinson, M. D., and McCarthy, D. J. (2010). edgeR: Differential Expression Analysis of Digital Gene Expression Data. Available online at: https://www.bioconductor.org/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf (accessed September 18, 2020).
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2009). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rohrwild, M., Coux, O., Huang, H. C., Moerschell, R. P., Yoo, S. J., Seol, J. H., et al. (1996). HslV-HslU: a novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc. Natl. Acad. Sci. U.S.A. 93, 5808–5813.
Salazar, J. K., Deng, K., Tortorello, M. L., Brandl, M. T., Wang, H., and Zhang, W. (2013). Genes ycfR, sirA and yigG contribute to the surface attachment of Salmonella enterica Typhimurium and Saintpaul to fresh produce. PLoS One 8:e57272. doi: 10.1371/journal.pone.0057272
Santillana Farakos, S. M., Pouillot, R., Anderson, N., Johnson, R., Son, I., and Van Doren, J. (2016). Modeling the survival kinetics of Salmonella in tree nuts for use in risk assessment. Int. J. Food Microbiol. 227, 41–50. doi: 10.1016/j.ijfoodmicro.2016.03.014
Scheil, W., Cameron, S., Dalton, C., Murray, C., and Wilson, D. (1998). A South Australian Salmonella Mbandaka outbreak investigation using a database to select controls. Aust. N. Z. J. Public Health 22, 536–539. doi: 10.1111/j.1467-842X.1998.tb01434.x
Schneider, B. L., Kiupakis, A. K., and Reitzer, L. J. (1998). Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180, 4278–4286. doi: 10.1128/jb.180.16.4278-4286.1998
Sekar, K., Linker, S. M., Nguyen, J., Grünhagen, A., Stocker, R., and Sauer, U. (2020). Bacterial glycogen provides short-term benefits in changing environments. Appl. Environ. Microbiol. 86:e00049-20. doi: 10.1128/AEM.00049-20
Senesi, E., Rizzolo, A., Colombo, C., and Testoni, A. (1996). Influence of pre-proce- ssing storage conditions on peeled almond quality. Ital. J. Food Sci. 8, 115–125.
Soncini, F. C., Vescovi, E. G., Solomon, F., and Groisman, E. A. (1996). Molecular basis of the magnesium deprivation response in Salmonella Typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178, 5092–5099.
Tan, M. S. F., White, A. P., Rahman, S., and Dykes, G. A. (2016). Role of fimbriae, flagella and cellulose on the attachment of Salmonella Typhimurium ATCC 14028 to plant cell wall models. PLoS One 11:e0158311. doi: 10.1371/journal.pone.0158311
Torres, A. G., Giron, J. A., Perna, N. T., Burland, V., Blattner, F. R., Avelino-Flores, F., et al. (2002). Identification and characterization of lpfABCC’DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157: H7. Infect. Immun. 70, 5416–5427.
Uesugi, A. R., Danyluk, M. D., and Harris, L. J. (2006). Survival of Salmonella Enteritidis phage type 30 on inoculated almonds stored at -20, 4, 23, and 35°C. J. Food Prot. 69, 1851–1857. doi: 10.4315/0362-028X-69.8.1851
Walters, S. B., Dubnau, E., Kolesnikova, I., Laval, F., Daffe, M., and Smith, I. (2006). The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60, 312–330.
Wang, H., Feng, H., Liang, W., Luo, Y., and Malyarchuk, V. (2009). Effect of surface roughness on retention and removal of Escherichia coli O157:H7 on surfaces of selected fruits. J. Food Sci. 74, E8–E15. doi: 10.1111/j.1750-3841.2008.00998.x
Keywords: Salmonella, low-moisture food, transposon sequencing, interstrain difference, storage condition, population dynamics, confocal laser scanning microscopy, gene function analysis
Citation: Li Y, Salazar JK, He Y, Desai P, Porwollik S, Chu W, Paola P-SS, Tortorello ML, Juarez O, Feng H, McClelland M and Zhang W (2020) Mechanisms of Salmonella Attachment and Survival on In-Shell Black Peppercorns, Almonds, and Hazelnuts. Front. Microbiol. 11:582202. doi: 10.3389/fmicb.2020.582202
Received: 10 July 2020; Accepted: 27 August 2020;
Published: 23 October 2020.
Edited by:
Chrysoula C. Tassou, Institute of Technology of Agricultural Products, Hellenic Agricultural Organization, GreeceReviewed by:
Agapi Doulgeraki, Institute of Technology of Agricultural Products, Hellenic Agricultural Organization, GreeceSandeep Tamber, Health Canada, Canada
Copyright © 2020 Li, Salazar, He, Desai, Porwollik, Chu, Paola, Tortorello, Juarez, Feng, McClelland and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael McClelland, bW1jY2xlbGxAdWNpLmVkdQ==; Wei Zhang, emhhbmd3QGlpdC5lZHU=
 Ye Li
Ye Li Joelle K. Salazar
Joelle K. Salazar Yingshu He
Yingshu He Prerak Desai
Prerak Desai Steffen Porwollik
Steffen Porwollik Weiping Chu3
Weiping Chu3 Oscar Juarez
Oscar Juarez Hao Feng
Hao Feng Michael McClelland
Michael McClelland Wei Zhang
Wei Zhang