
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 October 2020
Sec. Evolutionary and Genomic Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.581698
This article is part of the Research Topic Genomics and Effectomics of Filamentous Plant Pathogens View all 20 articles
 Dong Liang1,2
Dong Liang1,2 Christian Benjamin Andersen2
Christian Benjamin Andersen2 Ramesh R. Vetukuri2
Ramesh R. Vetukuri2 Daolong Dou1*
Daolong Dou1* Laura J. Grenville-Briggs2*
Laura J. Grenville-Briggs2*Crop protection strategies that are effective but that reduce our reliance on chemical pesticides are urgently needed to meet the UN sustainable development goals for global food security. Mycoparasitic oomycetes such as Pythium oligandrum and Pythium periplocum, have potential for the biological control of plant diseases that threaten crops and have attracted much attention due to their abilities to antagonize plant pathogens and modulate plant immunity. Studies of the molecular and genetic determinants of mycoparasitism in these species have been less well developed than those of their fungal counterparts. Carbohydrate-active enzymes (CAZymes) from P. oligandrum and P. periplocum are predicted to be important components of mycoparasitism, being involved in the degradation of the cell wall of their oomycete and fungal prey species. To explore the evolution of CAZymes of these species we performed an in silico identification and comparison of the full CAZyme complement (CAZyome) of the two mycoparasitic Pythium species (P. oligandrum and P. periplocum), with seven other Pythium species, and four Phytophthora species. Twenty CAZy gene families involved in the degradation of cellulose, hemicellulose, glucan, and chitin were expanded in, or unique to, mycoparasitic Pythium species and several of these genes were expressed during mycoparasitic interactions with either oomycete or fungal prey, as revealed by RNA sequencing and quantitative qRT-PCR. Genes from three of the cellulose and chitin degrading CAZy families (namely AA9, GH5_14, and GH19) were expanded via tandem duplication and predominantly located in gene sparse regions of the genome, suggesting these enzymes are putative pathogenicity factors able to undergo rapid evolution. In addition, five of the CAZy gene families were likely to have been obtained from other microbes by horizontal gene transfer events. The mycoparasitic species are able to utilize complex carbohydrates present in fungal cell walls, namely chitin and N-acetylglucosamine for growth, in contrast to their phytopathogenic counterparts. Nonetheless, a preference for the utilization of simple sugars for growth appears to be a common trait within the oomycete lineage.
The oomycetes are notorious as plant pathogens that cause devastating diseases in crop plants and our natural landscapes. Efficient control of many oomycete diseases relies on the usage of synthetic pesticides, which may have detrimental effects on the environment. The Genus Pythium contains predominantly saprotrophs and necrotrophic pathogens that occupy diverse ecological niches and infect various plant, arthropod, and even, human hosts (Plaats-Niterink, 1981). Some members of this fungal-like eukaryotic lineage are mycoparasites, obtaining nutrients from living fungal or oomycete hosts and are thus of great interest as potential biological control agents as part of an Integrated Pest Management (IPM) system for the control of crop diseases.
Mycoparasitic oomycetes have been less-well studied than their plant pathogenic counterparts, nevertheless, two species, Pythium oligandrum and Pythium periplocum have been investigated in this context. P. oligandrum has been documented as an antagonist and/or mycoparasite of a wide range of hosts including plant pathogenic Pythium spp. (Benhamou and Chet, 1997), Phytophthora spp. (Picard et al., 2000a; Horner et al., 2012), ascomycetes and basidiomycetes (Bradshaw-Smith et al., 1991; Benhamou et al., 1997). As well as an ability to antagonize fungi and oomycetes, these species display several other key features important for biological control of plant diseases. They typically grow faster than their plant pathogenic counterparts, meaning that they can outcompete other species for rhizosphere space and nutrition and they are able to promote both plant growth (Le Floch et al., 2003), and induced resistance in host plants (Takenaka et al., 2011; Ouyang et al., 2015). Whilst these experiments demonstrate their potential as biological control agents and detailed microscopic analysis has revealed the nature of their mycoparasitic interactions with various prey species (Benhamou et al., 1999; Picard et al., 2000b), there have been fewer mechanistic studies investigating the molecular or genetic determinants of their mycoparasitic lifestyle. Several cell wall-degrading enzymes and putative effectors were previously revealed to be expressed by P. oligandrum in the presence of oomycete tissue (Horner et al., 2012) and microarray analysis was recently used to investigate P. oligandrum-plant interactions (Yacoub et al., 2018), which showed significant reprogramming of the Vitus virifera root transcriptome in the presence of P. oligandrum. However, these studies were limited either in their methodology (a small sequencing study of 3,000 cDNA clones) or scope (focus on changes in plant roots), respectively, and thus we still lack a detailed mechanistic understanding of mycoparasitism in the oomycete lineage.
To provide a more complete basis for detailed molecular and genetic analysis of mycoparasitic Pythium species, we have sequenced and assembled the genomes of both P. oligandrum (Kushwaha et al., 2017a) and P. periplocum (Kushwaha et al., 2017b). Two other isolates of P. oligandrum have also been sequenced (Berger et al., 2016; Faure et al., 2020). We have also performed RNA sequencing of selected Pythium oligandrum-prey and Pythium periplocum-prey interactions, a detailed analysis of which will be published elsewhere.
The Carbohydrate-active enzyme complement (CAZyome) is the repertoire of predicted genes coding for enzymes involved in carbohydrate metabolism in an organism (CAZymes), including the synthesis, degradation, and modification of structural components of the cell wall. The CAZymes can be divided into five superfamilies, glycoside hydrolases (GH), glycosyl transferases (GT), polysaccharide lyases (PL), and carbohydrate esterases (CE) based on their activity and sequence similarity (Lombard et al., 2013). Comparative analysis of the mycoparasitic fungi Trichoderma atrovidirde and Trichoderma virens with other closely related fungal species, reveals the expansion of the CAZyome, particularly of genes from the family GH18, which comprises proteins with putative functions as chitinases in the mycoparasitic Trichoderma species. Other CAZy families expanded were those encoding endo-β-N acetylglucosimindases and β-1,3-glucanases from the families GH17, GH55, GH64, and GH81 (Kubicek et al., 2011). Chitin is a major component of the fungal cell wall, and therefore an obvious target for mycoparasitic lytic attack. Carbohydrate binding domains (CBMs) are also more abundant in the chitinase sequences from mycoparasitic Trichoderma species, compared to other fungi. As well as an expansion in the number of genes encoding chitinases, these Trichoderma genomes also contain an expanded number of GH75 chitosanases. It has long been known that the binding and degradation of chitin is important for successful mycoparasitism in these Trichoderma species. T. virens strains with chitinase knock-out mutants show reduced mycoparasitic ability, whilst strains that constitutively overexpress the same gene show enhanced biocontrol capabilities (Baek et al., 1999). The addition of CBMs to chitinases from Trichoderma harzianum has been shown to increase the antifungal activity of this species (Limón et al., 2004). Interestingly, Druzhinina et al. (2018) found nearly half of CAZy families in the mycoparasitic Trichoderma species were obtained by lateral gene transfer from plant-associated filamentous Ascomycete fungi, which has allowed Trichoderma species to expand their nutritional base (Druzhinina et al., 2018).
In the present study, we report the detailed mining of the P. oligandrum and P. periplocum genomes to investigate the presence and role of genes encoding CAZymes. Expression was investigated through analysis of RNA sequencing data from Pythium oligandrum-Phytopthora infestans and Pythium periplocum-Ph. infestans as well as P. periplocum-Botryis cinerea interactions. Transcript abundance of selected genes was confirmed through qRT-PCR analysis of the same parasite-prey interactions. The genomes of our mycoparasitic Pythium species were compared to those of nine other oomycete pathogens with different host and lifestyle ranges, to test the hypothesis that like their fungal counterparts, mycoparasitic oomycetes have expanded CAZyomes and that deployment of these cell wall degrading enzymes is also important for mycoparasitic oomycete-oomycete or oomycete-fungal interactions.
Our findings suggest that an expanded CAZyome may be one hallmark of mycoparasitism in eukaryotic microbes. Several of the CAZy encoding gene families appear to have been acquired by in mycoparasitic oomycetes through horizontal gene transfer. Our data also suggests that some CAZy-encoding genes act as pathogenicity factors, residing in similar genomic locations and with the potential to undergo rapid evolution in a similar manner to effector genes from phytopathogenic oomycetes.
Whilst it is well known that mycoparasitic fungi such as species within the Trichoderma Genus secrete an array of cell wall degrading enzymes during mycoparasitism of their prey, there is more limited information on the molecular mechanisms of mycoparasitism in the oomycete lineage. Previously several CAZyme-encoding genes were found to be expressed by P. oligandrum when interacting with tissue from the oomycete prey species, Ph. infestans (Horner et al., 2012). However, no comprehensive analysis of the CAZyome of P. oligandrum or P. periplocum has so far been carried out. In order to identify the key features and evolution of the mycoparasitic Pythium CAZyome, putative CAZyme encoding genes from P. oligandrum, P. periplocum and 11 other oomycete species were predicted using the dbCAN CAZy annotation pipeline (Yin et al., 2012). We first evaluated genome completeness in terms of the expected gene content of the thirteen species used for the comparison by carrying out a Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis (Seppey et al., 2019). Complete and single copy BUSCO groups account for at least 80% of each genome (Supplementary Figure S1), and thus we concluded that the genome completeness of each organism was comparable. The CAZyme complement of the genomes of the mycoparasitic species Pythium oligandrum (Kushwaha et al., 2017a) and Pythium periplocum (Kushwaha et al., 2017b), was compared to those from the plant pathogens: Pythium ultimum (Lévesque et al., 2010), Pythium aphanodermatum, Pythium arrhenomanes, Pythium iwayamai, Pythium irregulare, Phytopythium vexans (Adhikari et al., 2013), Phytophthora ramorum (Tyler et al., 2006), Phytophthora infestans (Haas et al., 2009), and Phytophthora capsica (Lamour et al., 2012), and the human pathogen Pythium insidiosum (Rujirawat et al., 2015).
We predicted 516 proteins in the CAZyome of P. oligandrum, 431 proteins in that of P. periplocum, and 321–719 proteins in the other oomycete species (Figure 1A). The total number of CAZy families predicted in P. oligandrum was 106, with 102 predicted in P. periplocum. The total number of CAZy families in the other oomycetes ranged from 93 to 110 (Figure 1A). These numbers are in line with previously reported predictions of the oomycete CAZyome (Ospina-Giraldo et al., 2010; Adhikari et al., 2013), indicating that our predictions are reliable. Within the Pythium genus, the total number of CAZy encoding genes was higher on average in the mycoparasitic Pythium species compared to the plant pathogenic Pythium species. However, since phytopathogenic Phytophthora species exhibit higher numbers of total CAZy genes, there is no significant difference within the oomycete lineage in the total numbers of CAZy encoding genes or in the total number of CAZy families per species.
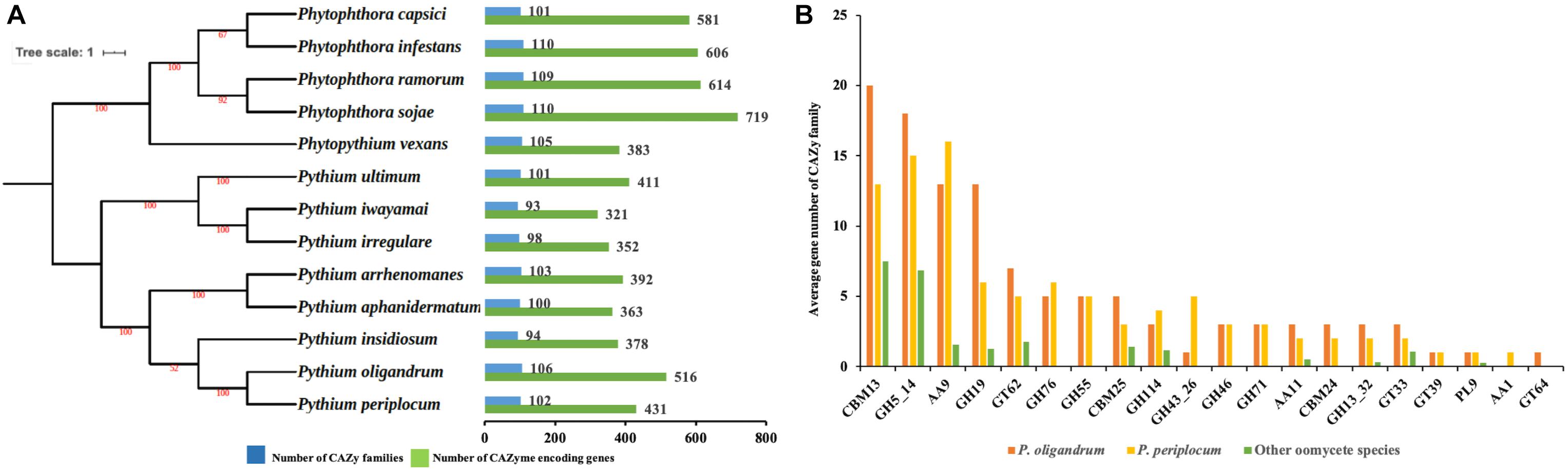
Figure 1. The CAZyme of two mycoparasitic Pythium species. (A) Species tree of oomycetes in this study and distribution of CAZy proteins. The species tree was constructed using the maximum likelihood method with 1,000 bootstraps based on a concatenated alignment of housekeeping genes identified by CEGMA analysis. Green bars indicate the total number of CAZyme encoding genes. Blue bars indicate the number of CAZy families present. (B) Average gene number of CAZy families unique or expanding in mycoparasitic Pythium. CAZy families were identified as unique or expanding in mycoparasitic Pythium species compare to other oomycetes by a T-test analysis.
To identify redundancy within the dataset and to compare CAZymes between the species, a comparison of the average gene member count per CAZy family was performed. Twenty CAZy families were identified as either expanded or unique within the mycoparasitic Pythium genomes when compared to the other species used in the analysis (p < 0.05, T-test; Figure 1B). These families include: three Auxiliary Activity families (AA1, AA11, AA9), three Carbohydrate-Binding Module families (CBM13, CBM24, CBM25), nine Glycoside Hydrolase families (GH114, GH13_32, GH19, GH43_26, GH46, GH5_14, GH55, GH71, GH76), four Glycosyl Transferase families (GT33, GT39, GT62, GT64), and one Polysaccharide Lyase family (PL9).
Alongside these analyses, we also mined RNA sequencing (RNA-Seq) data from the interactions between P. oligandrum with Ph. infestans. P. periplocum with Ph. infestans and from P. periplocum with Botrytis cinerea, to check the expression of these and other CAZy families. As well as the detailed analysis of the CAZy families described below, this also allowed us to identify differentially expressed genes during mycoparasitism of oomycete prey that were not encoded by families expanded in the mycoparasites, but that nonetheless may play a role in parasite-prey interactions (Supplementary Figure S2).
Since CAZy families with a putative role in the degradation of fungal or oomycete cell walls were predominantly expanded in the mycoparasitic oomycetes, we therefore focused the rest of our studies on carbohydrate binding and the metabolism of, cellulose, glucan, chitin and hemicelluloses, the major constituents of the cell walls of these prey species. Thus, eight CAZy families namely, AA9, GH5_14, GH55, GH71, GH19, GH46, GH76, and GH43_26 were chosen for detailed analysis. Among them, representatives of the CAZy families GH55, GH71, GH46, GH76, and GH43_26 were only detected in mycoparasitic Pythium species and were absent in the other oomycete genomes tested (Figure 1B and Supplementary Table S2) and thus appear to be unique components of the mycoparasitic oomycete CAZyme. Moreover, we also performed ortholog clustering using OrthoFinder (Emms and Kelly, 2019), which provides ortholog relationship analysis within each of the CAZy families mentioned above (Supplementary Table S3).
The major structural component of the oomycete cell wall is cellulose (Sietsma et al., 1969) and cellulose has been implicated in the maintenance of correct cell morphology and the production of appressoria (Grenville-Briggs et al., 2008). Penetration of plant tissue also requires active cellulose synthesis in the phytopathogenic oomycete, Ph. infestans (Grenville-Briggs et al., 2008). Homologs of the predicted cellulose synthase encoding genes [the CesA genes from the glycoside hydrolase family 2 (GT2)], previously identified in Ph. infestans (Grenville-Briggs et al., 2008) were identified in both P. oligandrum and P. periplocum, and this family was not expanded in the mycoparasites compared to the phytopathogens, although P. oligandrum contains one extra copy of the gene encoding CesA3 compared to P. periplocum, and the gene encoding CesA1 appears to be absent from P. periplocum (Supplementary Figure S7A). Although the CesA family is not expanded in the mycoparasites, genes encoding all eight CesA proteins identified in the mycoparasitic Pythium species, and a further GT2 protein from P. periplocum, were differentially expressed during interactions of the mycoparasite with Ph. infestans (Supplementary Figures S2A, S7B). Thus, although, it is not known whether appressorium production is also a significant component of oomycete mycoparasitism, the synthesis of cellulose may of general importance for growth during mycoparasitic interactions.
We hypothesize that cellulose degradation enzymes may be important pathogenicity determinants in the mycoparasitic Pythium species, since they are part of the cell wall of oomycete prey. Two families of genes encoding enzymes predicted to be involved in the degradation of cellulose were expanded in the mycoparasitic species, namely AA9 and GH5_14.
The auxiliary activity family 9 (AA9), proteins are lytic polysaccharide monooxygenases that are predominantly found in fungi. The AA9 family was significantly expanded in the mycoparasitic Pythium genomes compared to the other oomycete species studied. 16 AA9 proteins were predicted in P. oligandrum and 17 in P. periplocum. A range of one to five AA9 proteins were identified in plant pathogenic Pythium species, one AA9 protein was identified in the human infecting Pythium, and six AA9 proteins in Phytophthora species. Q86K62, an Auxiliary Activities 10 (AA10) protein in Dictyostelium discoideum, was used as outgroup to produce an AA9 phylogenetic tree, which consists of four clades (Figure 2B). Among clades, 17 mycoparasitic Pythium AA9 proteins are present in clade 1 (eight P. oligandrum and nine P. periplocum). Eight AA9 proteins are present in clade 3 (four from P. oligandrum and four from P. periplocum). Therefore, expansion of AA9 family in mycoparasitic Pythium appears to occur primarily in clade 1 and clade 3.
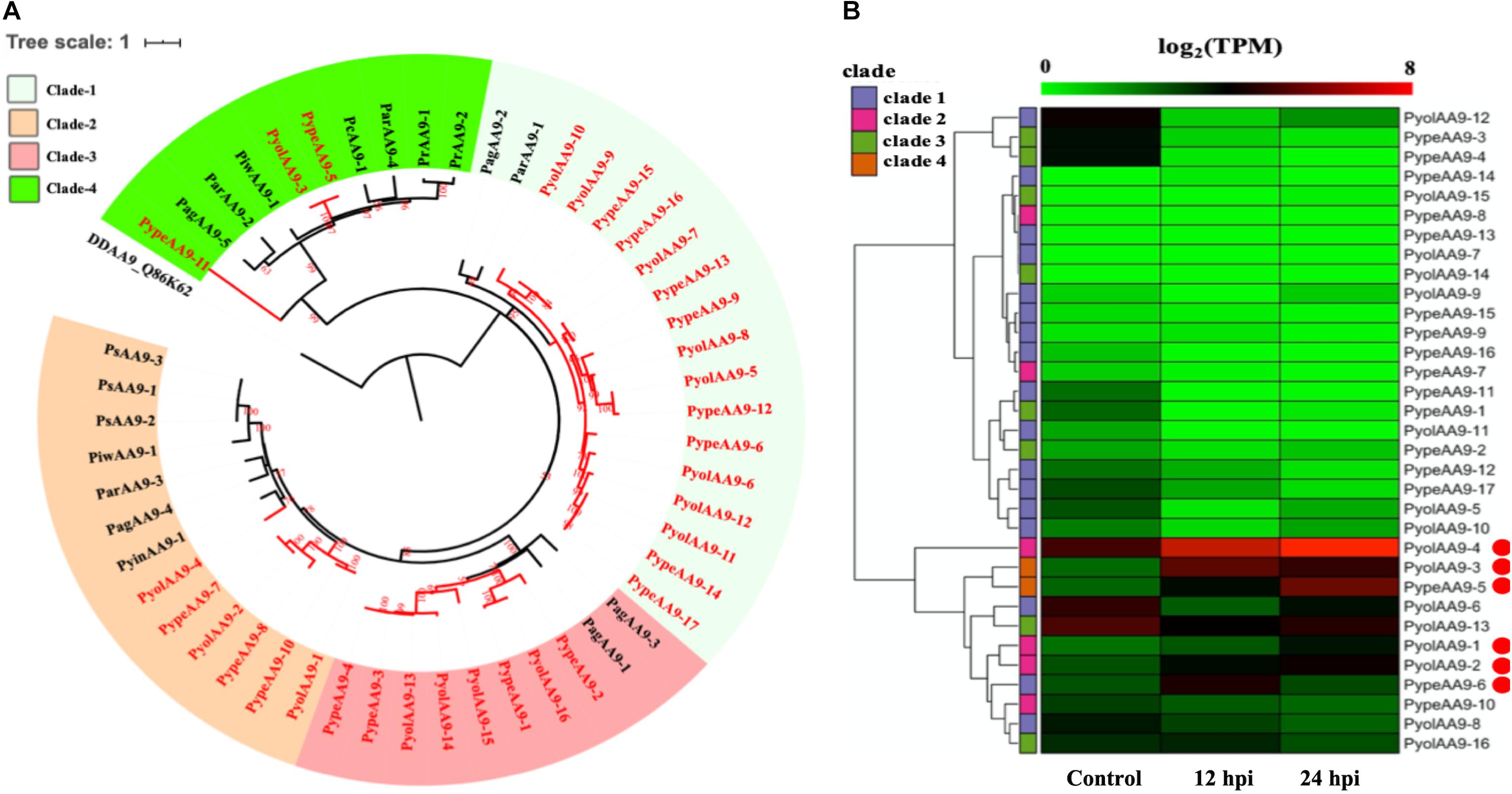
Figure 2. Analysis of auxiliary activity family 9 (AA9). (A) Phylogenetic tree of AA9 proteins identified in oomycete species. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). DDAA9_Q86K62 (an AA10 protein verified in Dictyostelium discoideum) was used as an outgroup. AA9 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype)are marked in red. (B) RNAseq Expression profiles of AA9 genes from P. oligandrum and P. periplocum during interactions with Ph. infestans during in vitro growth (control) or at 12 or 24 h post interaction (hpi). The phylogeny of each protein is marked using colored bars to represent each clade. The genes with the highest expression level during the interactions with Ph. infestans are marked with red circles. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
Previous studies have identified a “two-speed” architecture in the genomes of plant pathogenic oomycetes, in which the gene sparse regions exhibit higher plasticity, and hence drive adaptive evolution, that the gene dense regions (Raffaele and Kamoun, 2012). Pathogenicity determinants such as the RxLR effectors from phytopathogenic oomycetes tend to occupy regions that are gene poor, but repeat rich, facilitating rapid evolution of those regions (Dong et al., 2015). Detailed analysis of the Ph. infestans genome (Haas et al., 2009) postulated that accelerated effector evolution is driven by tandem gene duplication and homologous recombination. Based on gene density analysis, the majority of the P. oligandrum AA9 genes, encoding AA9 proteins assigned to clade 1, are located within the gene sparse regions (Supplementary Figure S3A). AA9 genes encoding clade 3 AA9 proteins, however, are present in the gene dense regions. Genome location analysis of the AA9 genes within the P. oligandrum genome shows that they cluster in two tandem arrays, consisting of AA9 genes encoding AA9 proteins of clade 1 and clade 3, respectively (Supplementary Figure S4A), suggesting that members of the AA9 family may be under rapid evolution in mycoparasitic oomycetes and further, may be important pathogenicity determinants for this group of organisms.
Using normalized TPM read counts from RNA-Seq data of the interaction between either P. oligandrum or P. periplocum and the prey Ph. infestans, we found seven AA9 genes were highly expressed specifically in the interactions between host and prey (Figure 2B). Among them, PyolAA9-2, PyolAA9-3, and PyolAA9-4 were significantly up-regulated at 12 and 24 hpi of the interaction and may be of general importance during parasite-prey interactions. PypeAA9-6 was only significantly up-regulated at 12 hpi, so may act predominantly in the early stages of mycoparasitism and thus could be described as a putative pathogenicity factor. PypeAA9-5 and PyolAA9-1were significantly up-regulated at 24 hpi and therefore play an important role in the later stage of mycoparasitism. PyolAA9-13 was comparatively highly expressed during both in vitro growth of both Pythium species as well as during interactions with oomycete prey and thus may be important for remodeling of the parasite cell wall during both vegetative growth and feeding.
We were able to design specific primers for qRT-PCR amplification of three of the AA9 genes, namely PyolAA9-3, PyolAA9-13, and PyolAA9-16 (Figure 10A). Both genes showed an induction during mycoparasitism of Ph. infestans, with PyolAA9-3 displaying the highest relative expression at 6 h, during the early colonization of Ph. infestans tissue. PyolAA9-13 was induced at 6 h and continued to show an elevated expression relative to in vitro growth levels from 6 to 48 h, with a peak at 36 h. PyolAA9-16 displayed constitutive levels of expression during both vegetative growth and the first 6 hpi which then reduced during the later stages of the interaction. These results suggest that AA9 family proteins have a role in mycoparasitism of Ph. infestans by mycoparasitic Pythium species.
An AA9 gene from the saprotrophic fungus Chaetomium globosum has previously been shown to promote cellulose hydrolysis by GH5_14 family cellulases and also exhibits the same synergistic effect in xylan hydrolysis (Kim et al., 2015, 2016). Since we also found the GH5_14 (predicted enzyme activity as endo-1,4-β-D-glucanases/cellulases (EC3.2.1.4), to be significantly expanded in the mycoparasitic Pythium genomes, compared to their phytopathogenic counterparts, we also checked for the presence of the genes encoding the GH5_14 family in gene sparse regions of the genome and for duplication events. The GH5_14 family comprises 18 proteins in P. oligandrum and 15 in P. periplocum. The phylogenic tree of GH5_14 proteins from all oomycetes in this study can be divided into seven clades (Figure 3A). Two plant GH5_14 proteins, ZmGH5_B6TTA1 and OjGH5_Q8RU06, were used as outgroups. GH5_14 proteins from the mycoparasitic Pythium species are present in clade 1, clade 2, clade 4, and clade 6. Notably, most of the GH5_14 proteins from the mycoparasitic Pythiums are present in clade 1 (10 from P. oligandrum and seven from P. periplocum). Thus, the expansion of the GH5_14 family occurred primarily in clade 1. Furthermore, genes encoding those proteins in clade 1 reside in the gene sparse regions of the genome and are located in tandem arrays, suggesting that these two families of associated genes are undergoing rapid evolution (Supplementary Figures S3B, S4B) and thus may also be important as pathogenicity determinants.
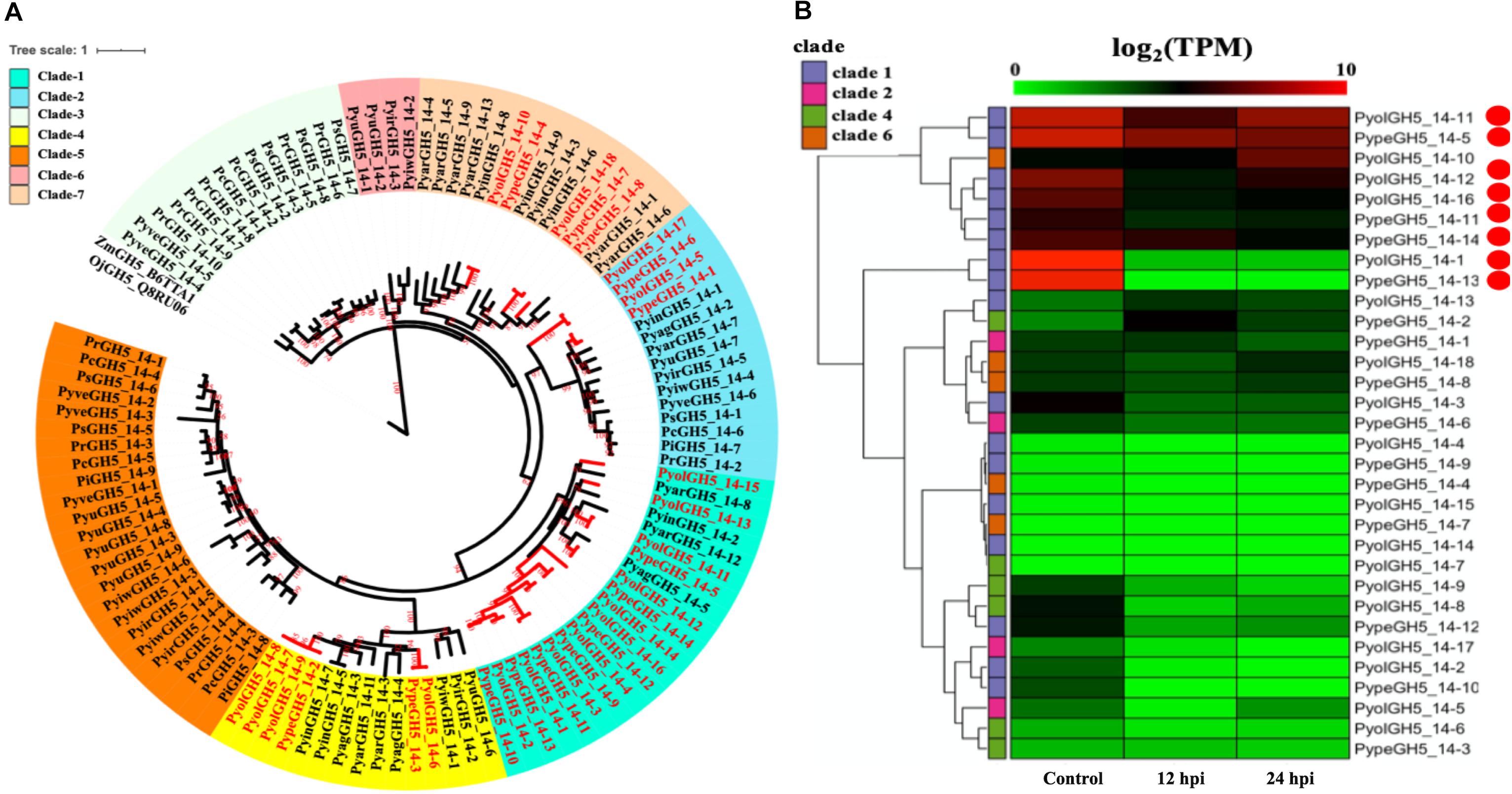
Figure 3. Analysis of glycoside hydrolase family 5_14 (GH5_14). (A) Phylogenetic tree of GH5_14 proteins identified in oomycete species. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). ZmGH5_B6TTA1 and OjGH5_Q8RU06 (GH5_14 proteins verified in plant species) were used as outgroups. GH5_14 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are marked in red. (B) RNAseq Expression profiles of GH5_14 genes from P. oligandrum and P. periplocum during in vitro growth or interactions with Ph. infestans and 12 or 24 h post interaction. The phylogeny of each protein is marked using colored bars to represent each clade. The genes with the highest expression level during the interactions with Ph. infestans are marked with red circles. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
Using normalized TPM read counts from RNA-Seq data of the interaction between either P. oligandrum or P. periplocum and the prey, Ph. infestans, we could determine that eight putative cellulase, GH5_14 genes encoding proteins assigned to clade 1, were highly expressed during mycoparasite-prey interactions, or during in vitro growth (Figure 3B), among them, two genes, PyolGH5_14-11 and PypeGH5_14-5, showed near constitutive expression levels in all conditions tested, suggesting a role in the remodeling of the mycoparasitc cell wall during all phases of growth. Four genes, PypeGH5_14-11, PyolGH5_14-12, PypeGH5_14-14, PypeGH5_14-16, were highly expressed during in vitro growth and slightly down-regulated during interactions with the prey species. PyolGH5_14-1 and PypeGH5_14-13, were highly expressed mainly during in vitro growth. This suggests the latter two genes have a role in the modification of the cell wall of the mycoparasite during vegetative growth, and little to no role in mycoparasitism. The genes encoding GH5_14 proteins in clade 2, clade 4, and clade 6 were all expressed at relatively low levels during in vitro growth, and/or during the interaction with Ph. infestans. However, transcripts from PyolGH5_14-10, assigned to clade 6, were highly abundant in the Ph. infestans interaction at 24 hpi.
Detailed expression analysis of a time course of Ph. infestans infection by P. oligandrum revealed that, of the genes tested, PyolGH5_14-16 was highly expressed during mycoparasitism, peaking at 36 h with a relative expression level greater than 300 times that of the in vitro expression (Figure 10A). Furthermore, data from three independent biological replicates shows that the expression of this gene oscillates between higher and lower expression over the interaction time course. A temporal switching of gene expression may be a response to the availability of new host hyphae, reflecting enzyme production only as and when needed, e.g., through digestion of a single hypha at a time. Alternatively, this could be part of a mechanism to avoid accidental self-damage. The fungus Trichoderma reesei, is a prolific producer of cellulases, and as such is a model organism for industrial production of several CAZy enzymes involved in cell wall degradation. Major cellulase transcription factor genes have been identified that show differential regulation under light or dark conditions and photoreceptors play an important role in regulation of nutritional uptake in this organism (Schmoll, 2018). PyolGH5_14-16 transcript abundance peaks occurred during the night sampling (12 and 36 h) of the interaction time course and this pattern is reminiscent of a circadian rhythm. However, this may also reflect gene induction under the somewhat artificial in vitro conditions under which the experiments were performed and thus investigation of the expression of CWDEs under natural or field conditions would be interesting for the future. PyolGH5_14-1 and PyolGH5_14-12 show a decreased expression during mycoparasitism, relative to in vitro levels, indicating a potential role predominantly during vegetative growth. Neither the GH6 family of putative cellobiohydrolases (EC 3.2.1.91) or the GH5_20 family of endo-β-1,4-glucanases/cellulases (EC 3.2.1.4) were expanded in the mycoparasitic oomycetes, however, five GH6 family genes from P. oligandrum were differentially expressed (significantly upregulated) during the early stages of interactions with Ph. infestans. whilst five GH5_20 genes were down-regulated (Supplementary Figure S2A). Four GH5_20 genes from P. periplocum were also down-regulated at the same time point, as were three GH3 β-glucosidases (EC 3.2.1.21) (Supplementary Figure S2B), indicating these may be used to cleave celluloses from other substrates, or involved in other phases of the mycoparasitic lifecycle. Genes encoding 10 putative GH17 family endo-1,3-β-glucosidases (EC 3.2.1.39) from P. oligandrum were upregulated during the early stages of interactions with Ph. infestans five from P. periplocum were upregulated in the later stages of mycoparasitism (Supplementary Figure S2), indicating that the different mycoparasites may use a progression of different cellulose degrading enzymes at different time points in the mycoparasitic life cycle.
Thus, the metabolism of cellulose is likely to be important for both vegetative oomycete growth and for mycoparasitism of oomycete prey and several of the genes involved in this process display the genomic hallmarks of pathogenicity factors undergoing rapid evolution.
Glucans are structurally related to cellulose and are another important component of the oomycete cell wall. In contrast to cellulose, glucans are largely absent from plant cell walls and thus dissecting the occurrence and evolution of enzymes involved in the degradation of glucans may provide unique insights into the evolution of the mycoparasitic oomycete species. GH55 CAZy family members are predicted to function as exo-β-1,3-glucanases (EC 3.2.1.58 degrading glucans containing β-1,3-linkages as well as participating in the hydrolysis of laminarin, a component of fungal cell walls (Bara et al., 2003). Thus, genes in this class may potentially target both oomycete and fungal prey. Genes assigned to the CAZy family GH55 were found only in the mycoparasitic Pythium and were absent in the other oomycete genomes tested, indicating that they might be important for mycoparasitism. We found genes encoding five GH55 proteins each in P. oligandrum and P. periplocum. Phylogenetic analysis shows that oomycete GH55 proteins are most closely related to those from a variety of fungal species (Figure 4A). It has recently been shown that almost half of the cell wall degrading carbohydrate active enzymes found in mycoparasitic Trichoderma species were obtained via horizontal gene transfer, a process by which genetic material may be transferred between distinct evolutionary lineages, either cross-Kingdom or within Kingdom (Doolittle, 1999; Andersson, 2009), from plant associated filamentous fungi (Druzhinina et al., 2018). Our data supports the hypothesis that P. oligandrum and P. periplocum have developed the ability to be mycoparasitic, in part, through the acquisition of CWDEs from the GH55 family, via horizontal gene transfer from filamentous fungi. The flanking regions of mycoparasitic Pythium GH55 genes show conserved collinearity with regions of the P. ultimum genome (Supplementary Figure S5A), although GH55 genes are absent in P. ultimum. Therefore, mycoparasitic Pythium GH55 genes appear to reside within conserved regions of their genomes and may potentially have been inserted there via horizontal gene transfer from fungi.
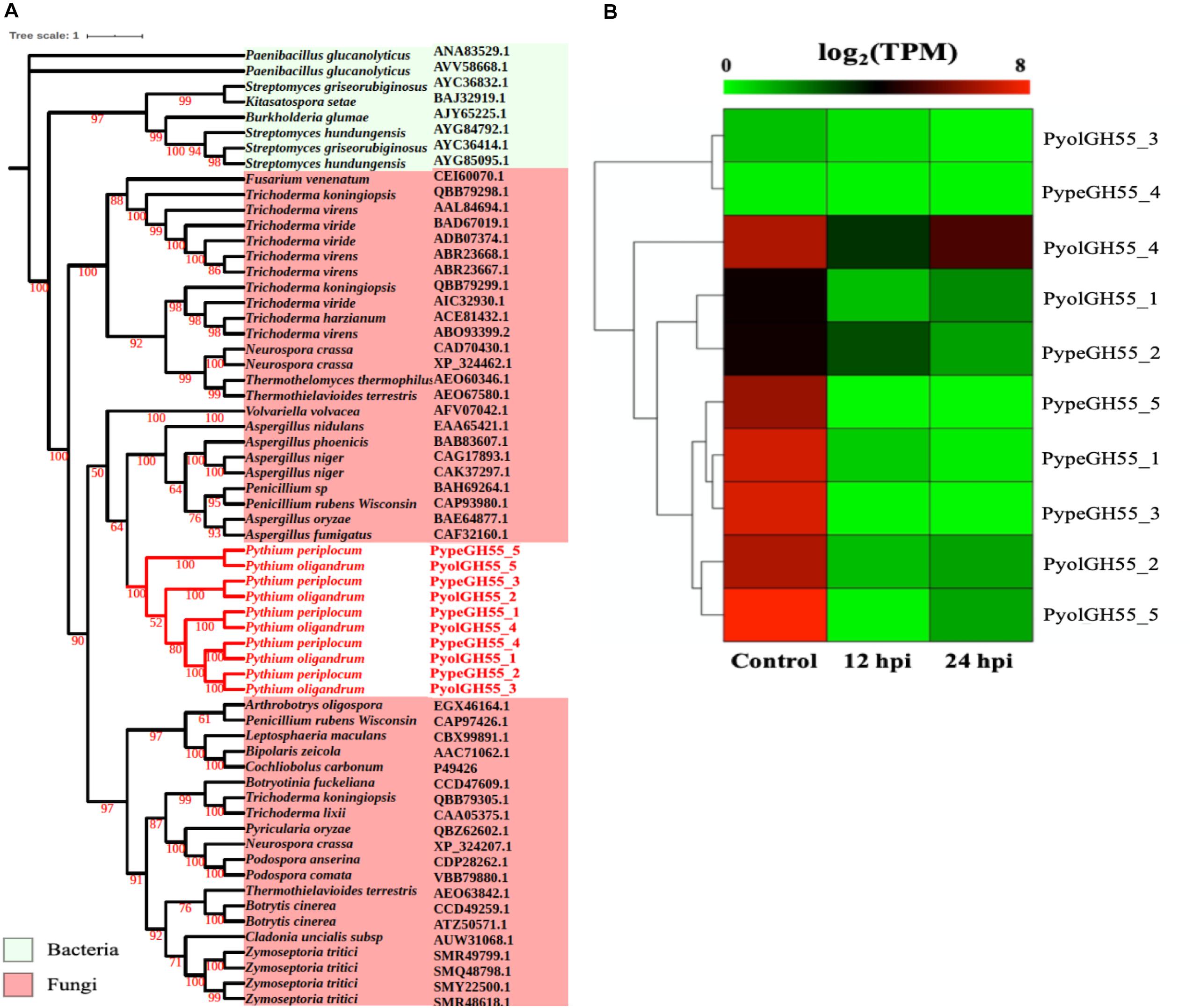
Figure 4. Analysis of glycoside hydrolase family 55 (GH55). (A) Phylogenic tree of GH55 proteins identified in P. oligandrum and P. periplocum. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). GH55 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are shown in red. (B) RNAseq Expression profiles of GH55 genes detected in P. oligandrum and P. periplocum during in vitro growth or interactions with Ph. infestans at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
Analysis of normalized TPM read counts from the RNA-Seq interaction libraries, revealed two different expression patterns. Six GH55 genes, (PyolGH55_4, PypeGH55_5, PypeGH55_1, PypeGH55_3, PyolGH55_2, PyolGH55_5), were highly expressed during in vitro growth of and significantly down-regulated in the interaction stages (Figure 4B). Notably, PyolGH55_4 was highly expressed both during in vitro growth and at the later stage of the interaction between P. oligandrum Ph. infestans. Four GH55 genes (PyolGH55_3, PypeGH55_4, PyolGH55_1, PypeGH55_2), show a low level of expression in all stages tested, suggesting that they do not have a major role during in vitro growth or in the interaction with Ph. infestans as a prey species. They may, however, be expressed more highly in other growth stages or in the interaction with other prey species.
Like the GH55 family, members of GH71 family were only identified in mycoparasitic Pythium among oomycete species in this study. GH71 proteins have a predicted enzymatic activity as α-1,3-glucanases (EC 3.2.1.59). We found three GH71 proteins in each of the mycoparasitic species P. oligandrum and P. periplocum, respectively. Phylogenetic analysis groups these genes within a branch of fungal GH71 proteins (Figure 5A). Thus, this suggests that the GH71 genes from the mycoparasitic Pythium species may also have been obtained via horizontal transfer from fungi. Two of the genes, PyolGH71-1 and PypeGH71-1, are highly expressed during in vitro growth and are significantly down-regulated during the interaction with Ph. infestans (Figure 5B). The remaining members of this family show low levels of expression in the stages tested in this study, which is a similar expression profile to the GH55 glucanase family. PyolGH71-1 showed high relative expression levels at 36 and 48 h of the interaction with Ph. infestans, suggesting either a role in degradation of prey glucans that are exposed later during mycoparasitism or a role in P. oligandrum cell wall rearrangement and/or growth, after nutrient uptake from the prey.
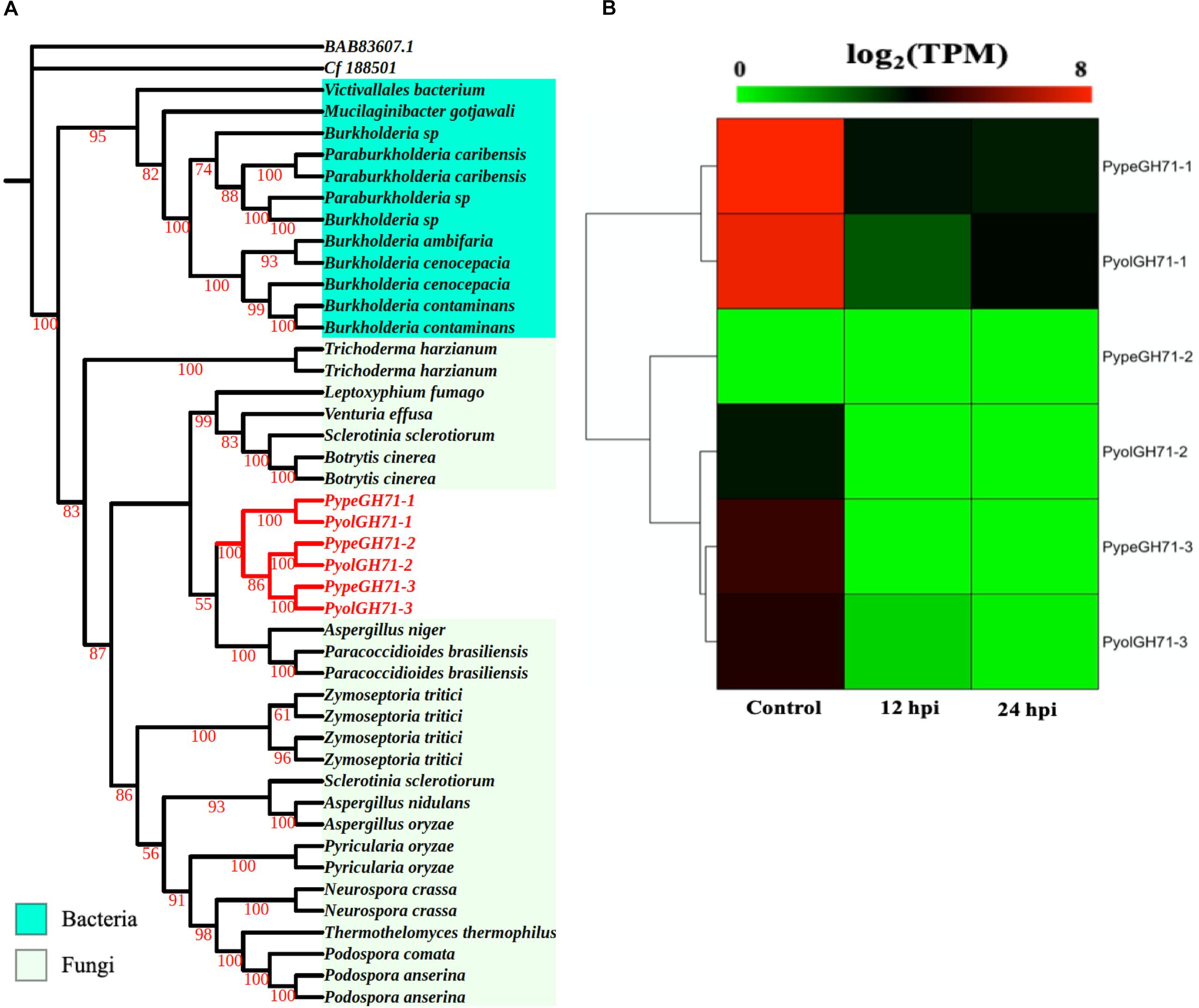
Figure 5. Analysis of glycoside hydrolase family 71 (GH71). (A) Phylogenic tree of GH71proteins identified in P. oligandrum and P. periplocum. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). GH71 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are shown in red. (B) RNAseq Expression profiles of GH71 genes detected in P. oligandrum and P. periplocum during in vitro growth or interactions with Ph. infestans at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
We next used the GH55 and GH71 protein sequences detected in P. oligandrum and P. periplocum in blastp searches against the NR database, excluding sequences from P. oligandrum and P. periplocum themselves. Compiling the resultant data as a phylogenetic tree, further confirms that GH55 and GH71 proteins appear to have been transferred horizontally from Ascomycete fungi to the oomycetes (Supplementary Figure S8). Moreover, we also made Hidden Markov Model (HMM) searches against the genomes of other available oomycetes, namely, Hyaloperonospora arabidopsidis, Albugo laibachii, Saprolegnia parasitica, Saprolegnia diclina, Phytophthora parasitica, and Pythium brassicum. However, we could not find any GH55 and GH71-like genes in these species, and thus conclude that they are absent in other oomycetes, for which we have genome data. Therefore, it appears that horizontal gene transfer of GH55 and GH71 genes occurred within the latest common ancestor of P. oligandrum.
Members of the GH19 family, with predicted activity as chitinases (EC 3.2.1.14), are abundant in mycoparasitic Pythium species (Figure 6A). Here we found 13 GH19 proteins in P. oligandrum and six in P. periplocum. A range of one to four GH19 proteins were found in other oomycetes in this study. As shown in Figure 6A, GH19 proteins can be divided into only two clades. GH19 proteins from the mycoparasitic Pythium species were only present in clade 2, which also includes one GH19 proteins from each of the plant pathogens P. aphanidermatum, and Ph. capsici and three genes from the human and animal pathogen P. insidiosum. Based on gene density analysis, most of the GH19 genes identified, are predicted to be present in the gene sparse regions of the genomes of both P. oligandrum and P. periplocum (Supplementary Figures S3C,D). Moreover, one tandem array, consisting of genes PyolGH19-3 to PyolGH19-10, is present in the genome of P. oligandrum (Supplementary Figure S4C), suggesting that these genes are able to undergo rapid evolutionary change, and may be important pathogenicity factors in fungal mycoparasitism.
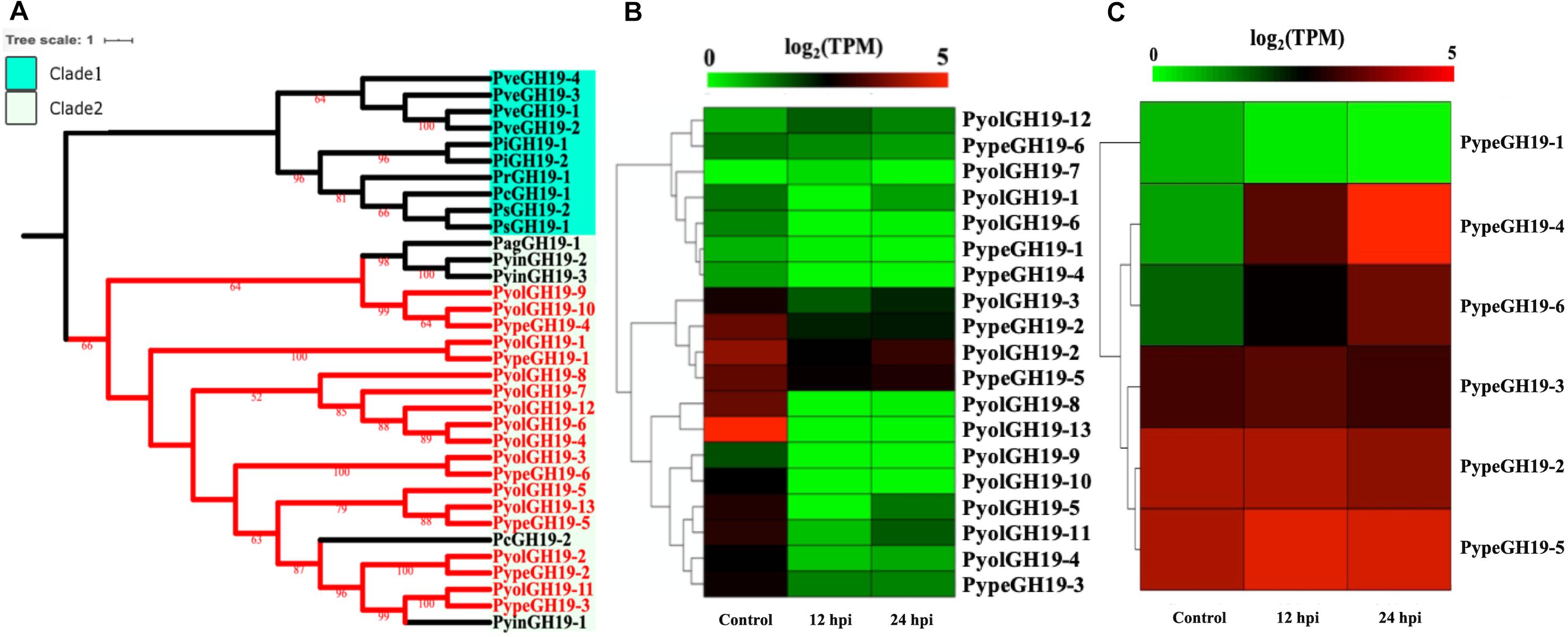
Figure 6. Analysis of glycoside hydrolase family 19 (GH19). (A) Phylogenetic tree of GH19 proteins identified in oomycete species. Maximum-likelihood method with 1,000 bootstraps (values displayed per branch) was used to construct tree based on alignment of GH19 proteins. GH19 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are shown in red. (B) RNAseq Expression profile of GH19 genes detected in P. oligandrum and P. periplocum during in vitro growth or interactions with Ph. infestans at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene. (C) RNAseq Expression profile of GH19 genes detected in P. periplocum during in vitro growth or interactions with B. cinerea at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
Analysis of normalized TPM read counts from our RNA-Seq interaction libraries reveals that GH19 genes (PypeGH19-2, PyolGH19-2, PypeGH19-5, PyolGH19-8, PyolGH19-13), were highly expressed during in vitro growth and significantly down-regulated during the interaction with the prey oomycete Ph. infestans (Figure 6B). Thus, GH19 genes seem to be dispensable for interactions between mycoparasitic Pythium species and Ph. infestans, which has a cellulosic cell wall and no detectable chitin (Sietsma et al., 1969; Mélida et al., 2013). To test the hypothesis that GH19 proteins are used for parasitism of fungal prey, we mined our sequenced transcriptomes of interactions between P. periplocum and the gray mold fungus, Botrytis cinerea. Three P. periplocum GH19 genes (PypeGH19-2, PypeGH19-3, and PypeGH19-5), were highly expressed during both in vitro growth and during the interactions between P. periplocum and B. cinerea. Two P. periplocum GH19 genes (PypeGH19-4, and PypeGH19-6), were highly expressed solely during interactions between P. periplocum and B. cinerea and PypeGH19-1 induction was not detected in any of the conditions we tested (Figure 6C).
To confirm the role of GH19 family members in mycoparasitism of B. cinerea, we performed qRT-PCR analysis of selected GH19 genes during the early stages of parasitism by either P. periplocum or P. oligandrum. Four P. periplocum GH19 genes, (PypeGH19-1, PypeGH19-4, PypeGH19-5, PypeGH19-6), were significantly upregulated (between two and fifteen -fold change) at 12 and 24 h of interactions with B. cinerea. Six P. oligandrum GH19 genes (PyolGH19-1, PyolGH19-2, PyolGH19-3, PyolGH19-8, PyolGH19-9, PyolGH19-11), were significantly induced (7–1,300-fold change) during the interaction with B. cinerea. Thus, chitinase gene expression is likely to be important for mycoparasitism of fungal prey and the mycoparasitic Pythium species are able to sense the cell wall components of their prey and adjust the expression of their CWDEs accordingly, since these genes are not expressed in the presence of a non-chitinous prey species.
The GH46 family (EC.3.2.1.132) is predicted to have chitosanase activity and was detected only in the mycoparasitic Pythium and not in the other oomycete species screened in this study. Chitosan is a cationic polysaccharide composed of β-1,4-linked D-glucosamine (GlcN) that is derived from chitin, with chitosanase being the major substrate-specific enzyme that acts on the β-1,4-glycosidic linkages of chitosan (Sun et al., 2018). We detected three GH46 genes in both P. oligandrum and P. periplocum respectively. To explore the phylogenic relationship of the mycoparasitic Pythium GH46 genes, we retrieved GH46 proteins from a diverse range of species (Viens et al., 2015). As Figure 7A shows, the phylogenic tree of the GH46 family is divided into five clades, as previously reported (Viens et al., 2015). Mycoparasitic Pythium GH46 proteins are present in clade C, which includes GH46 proteins from Chlorella virus. Thus, we inferred that mycoparasitic Pythium may have been obtained GH46 genes by horizontal transferred from a viral donor. Genes neighboring the GH46 gene family in the mycoparasitic Pythium show collinearity with regions of the P. ultimum genome (Supplementary Figure S5B). Therefore, it would appear that the mycoparasitic Pythium GH46 genes were transferred into conserved regions of their genomes.
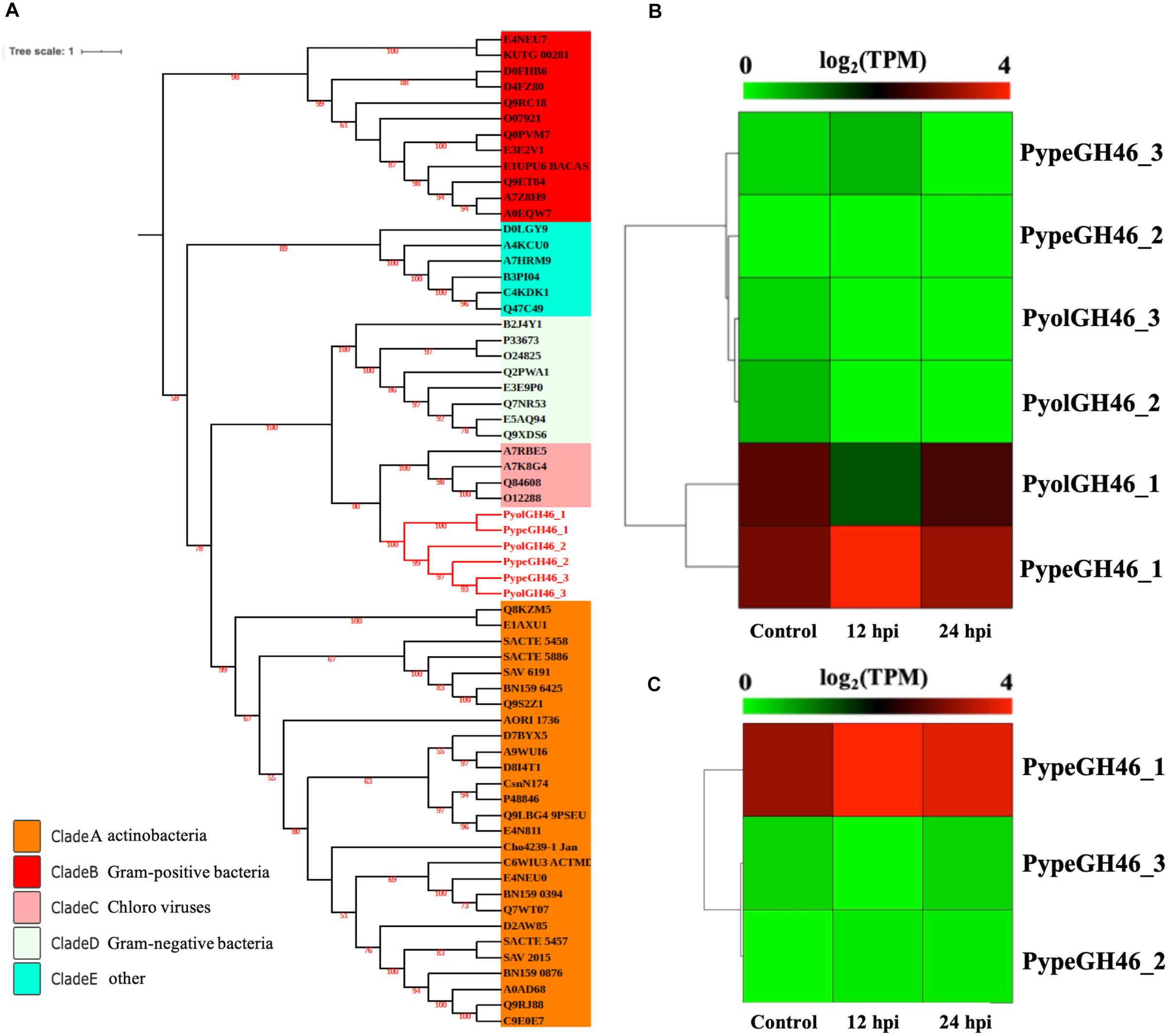
Figure 7. Analysis of glycoside hydrolase family 46 (GH46). (A) Phylogenetic tree of GH46 proteins identified in oomycete species. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). GH46 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are marked in red. (B) RNAseq expression profile of GH46 genes detected in P. oligandrum and P. periplocum during in vitro growth or during interactions with Ph. infestans at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene. (C) RNAseq Expression profile of GH46 genes detected in P. periplocum during in vitro growth or interacting with B. cinerea at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
Interestingly, transcripts of one GH46 gene (PypeGH46-1) were highly abundant, relative to in vitro levels, during the parasitism of both Ph. infestans and B. cinerea by P. periplocum. One gene from P. oligandrumm (PyolGH46-1) was also expressed during parasitism of Ph. infestans (Figures 7B,C). Quantitative RT-PCR assays of GH46 genes during the early stages of interactions of either P. periplocum or P. oligandrum with B. cinerea reveal five genes (PyolGH46-1, PyolGH46-2, PyolGH46-3, PypeGH46-1, and PypeGH46-3), that are induced between 2 and 50-fold during parasitism of this fungus (Figure 10).
It has long been thought that the oomycetes are predominantly cellulosic in contrast to the chitinous fungi with whom they share many ecological niches. However detailed analysis of the structural carbohydrates in the cell wall of several plant and fish pathogenic oomycetes has identified three types of cell wall-based differences in structural carbohydrates within different oomycete Genera (Mélida et al., 2013). Type I contains glucuronic acid and mannose and no N-acetylglucosamine (GlcNAc), type II is characterized by cross linking between cellulose and 1,3,β glucans and up to 5% GlcNAc and the third type contains the highest GlcNAc content along with unusual carbohydrates (Mélida et al., 2013). This analysis did not include any of the mycoparasitic oomycetes or members of the Pythium genus though, so it is not clear which cell wall type P. oligandrum or P. periplocum contain, although based on our data showing that chitin metabolism-linked genes are expressed under in vitro conditions, we hypothesize that the mycoparasitic oomycetes may be modulating related carbohydrates or chito-oligosaccharides within their own cell walls. Alternatively, these genes may be expressed in vitro in preparation for degradation of prey cell walls, or in response to a depletion of nutrients.
Four GH76 family members were found each in both P. oligandrum and P. periplocum although GH76 genes were absent in the other oomycete species screened. GH76 proteins are annotated as α-1,6-mannanases (EC 3.2.1.101), targeting α-1,6-mannosidic linkages and thus participating in the deconstruction of fungal cell walls (Cuskin et al., 2015). The phylogenic analysis, with an exo-β-1,3-glucanase of GH55 proteins (BAB83607.1) as the outgroup, shows that mycoparasitic Pythium GH76 proteins group most closely in the GH76 proteins from bacterial species (Figure 8A). Thus, these genes may have been horizontally transferred to a common mycoparasitic Pythium ancestor from bacteria.
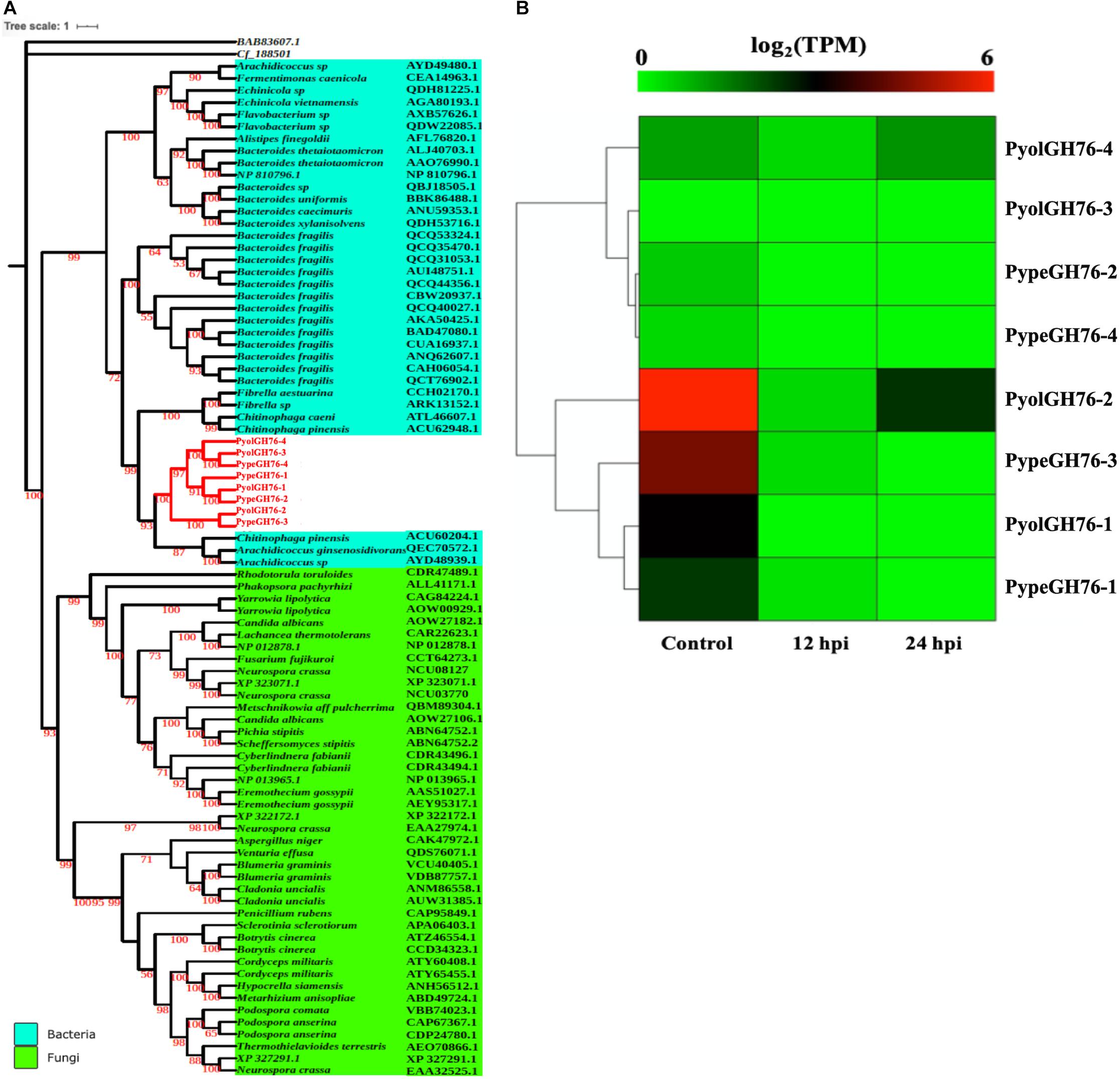
Figure 8. Analysis of glycoside hydrolase family 76 (GH76). (A) Phylogenetic tree of GH76 proteins identified in P. oligandrum and P. periplocum with GH76 proteins of bacteria and fungi. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). GH76 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are marked in red. BAB83607.1 and Cf_188501 (GH55 proteins verified in fungal species) were used as an outgroup. (B) RNAseq Expression profile of GH76 genes detected in P. oligandrum and P. periplocum during in vitro growth or interactions with Ph. infestans at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
Analysis of normalized TPM read counts from our RNA-Seq libraries revealed that one of the P. oligandrum genes (PyolGH76-2) and one of the P. periplocum genes (PypeGH76-3) were highly expressed during in vitro growth (Figure 8B). However, there was little to no detectable differential expression during interactions with the prey species tested, meaning that these genes are dispensable for antagonism of the fungal and oomycete prey used in this study.
GH43_26 genes were only identified in mycoparasitic Pythium among oomycetes in our study. Here we found one GH43_26 protein in P. oligandrum and eight GH43_26 proteins in P. periplocum. These genes have predicted activity as α-L-arabinofuranosidases (EC 3.2.1.55), which hydrolyze terminal non-reducing alpha-L-arabinofuranoside residues in alpha-L-arabinosides. Figure 9A shows that GH43_26 proteins from mycoparasitic Pythiums (red branch) group within a fungal GH43_26 clade (green branch). An endo-β-1,4-xylanase from the GH11 family (AHE13930.1) was used as an outgroup. This suggests that this gene family may have been horizontally transferred from fungi to mycoparasitic Pythium. Three of the GH43_26 genes (PypeGH43_26-7, PypeGH43_26-5, PyolGH43_26-1) were highly expressed both during in vitro growth and 24 hpi of the interaction with Ph. infestans indicating that they may be of some importance for growth of the mycoparasitic species themselves and of a lesser importance during mycoparasitism of oomycete prey (Figure 9B).
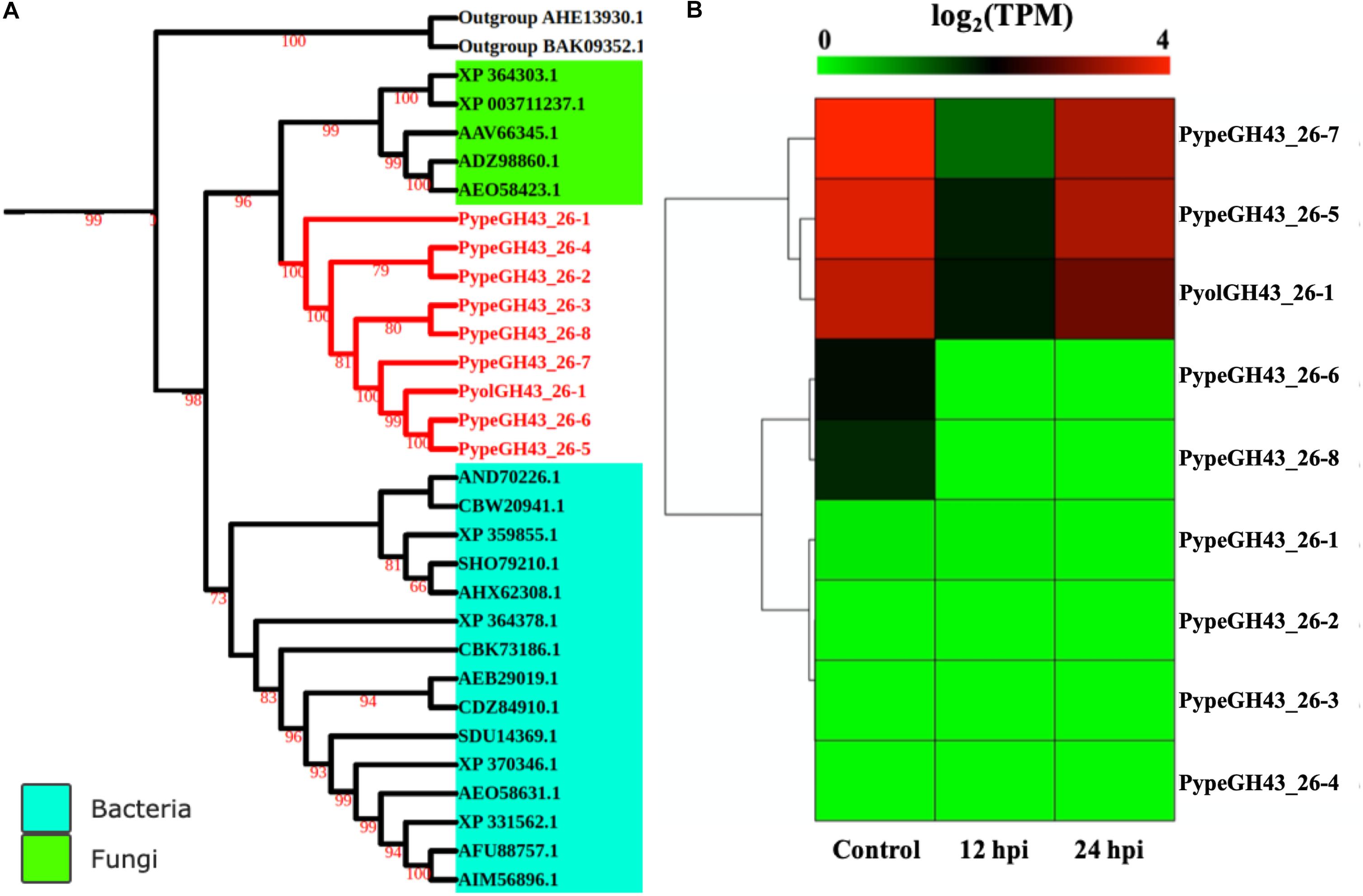
Figure 9. Analysis of glycoside hydrolase family 43_26 (GH43_26). (A) Phylogenetic tree of GH43_26 proteins identified in P. oligandrum and P. periplocum with GH43_26 proteins of bacteria and fungi retrieved from CAZy database. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). GH43_26 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are marked in red. AHE13930.1 and BAK095352.1 (GH11 proteins verified in fungi species) were used as an outgroup. (B) RNAseq Expression profile of GH43_26 genes detected in P. oligandrum and P. periplocum during in vitro growth or at 12 or 24 h post interaction with Ph. infestans. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
The GT71 family encodes genes with activity as α-mannosyltransferases (EC 2.4.1.-), and whilst this family was not expanded, nine GT71 genes were significantly upregulated in P. oligandrum during interactions with Ph. infestans. GH16 family members with predicted xyloglucanase activity (EC 3.2.1.151) were not expanded in the mycoparasitic oomycetes, however, three putative GH16 genes from P. oligandrum were upregulated during interactions with the prey oomycete Ph. infestans (Supplementary Figure S2). The family with the highest number of differentially expressed genes when interacting with the oomycete prey in both P. oligandrum and P. periplocum was CE1. Thirteen genes from P. oligandrum and fifteen genes from P. periplocum were upregulated during the early interactions with Ph. infestans (Supplementary Figure S2) and thus these family members may play a role in the degradation of hemicelluloses of oomycete origin. This Carbohydrate esterase family has diverse esterase functions including as acetyl xylan esterases (EC 3.1.1.72). GH30_1 family endo-β-1,4-xylanases from both mycoparasites were downregulated in the same interactions, indicating that they are not important in the interaction with oomycetes.
Since destruction of cellulose appears to be important to oomycete-oomycete mycoparasitic interactions, and these Pythium species appear able to sense the major carbohydrate component of the prey cell wall and adapt their CWDE gene expression, we next investigated the presence and expression of cellulose binding module containing proteins, with the hypothesis that these may play a role in the sensing or binding of oomycete prey cells.
The cellulose-binding elicitor lectin (CBEL) family was found to contain two cellulose-binding domains (CBDs), belonging to the family 1 carbohydrate binding modules (CBM1). The CBM1 family is almost exclusively found only in fungi and oomycetes. In the phytopathogenic oomycetes, CBEL family members have been shown to have necrosis-inducing activity in host plants, and to bind host cellulose (Mateos et al., 1997; Gaulin et al., 2006). In a previous study of a small cDNA library of P. oligandrum interacting with dead tissue from Ph. infestans, a CBEL gene from P. oligandrum was shown to be upregulated during the interaction (Horner et al., 2012).
In the current study we found six CBEL proteins each in P. oligandrum and P. periplocum. A range of 2–10 CBEL proteins were detected in the other oomycete genomes screened. Supplementary Figure S6A shows that CBEL proteins were divided into four phylogenetic clades with a Trichoderma reesei exoglucanase used as an outgroup. Mycoparasitic Pythium CBEL proteins were only present in clade 4 (Supplementary Figure S6A). Based on domain architecture analysis, CBD (CBM1) domains are present in all putative CBEL proteins and PAN domains are only present in CBEL proteins grouped within clade 4. PAN domains are found in a diverse array of proteins and have been implicated in protein-protein or protein-carbohydrate interactions. Some of the mycoparasitic Pythium CBEL proteins are also predicted to contain other domains, including transglutaminase elicitors (TGase_elicitor), Leucine Rich Repeats (LRR_4 or LRR_8) and elicitin domains.
Analysis of normalized TPM read counts from our RNA-Seq libraries revealed that putative CBEL genes from P. oligandrum (PyolCBEL_2, PyolCBEL_4, PyolCBEL_5), and P. periplocum (PypeCBEL_3, PypeCBEL_6) were highly expressed during in vitro growth. These CBEL proteins are predicted to contain only a PAN domain and carbohydrate binding domain along with a signal peptide. Of these, PyolCBEL_5 and PypeCBEL_3 were also somewhat expressed during interactions with Ph. infestans (Supplementary Figure S6B). The Transcripts from mycoparasitic Pythium CBEL proteins, predicted to contain either a TGase_elicitor or an Elicitin domain, were not highly expressed during the interaction with Ph. infestans under the conditions tested (Figure 6B).
Based on qRT-PCR (Figure 10A), transcripts of PyolCBEL-1, PyolCBEL-2, and PyolCBEL-3 were all more abundant in the interaction with Ph. infestans than during in vitro growth of P. oligandrum, with PyolCBEL-1 and PyolCBEL-3 showing the highest expression levels. PyolCBEL-1 peaked at almost 150-fold the in vitro expression level at 6 hpi compared and PyolCBEL-3 shows a 12-fold expression increase at the same time point. PyolCBEL-2 was expressed at near in vitro levels throughout the infection time course, and PyolCBEL-4 was expressed at extremely low levels throughout the interaction. Three genes containing CMB63 domains from P. periplocum were highly upregulated during the early stages of the interactions with Ph. infestans (Supplementary Figure S2B), but interestingly CBM63 genes were not differentially expressed in the P. oligandrum-Ph. infestans interaction. The CBM63 module from Bacillus subtilis expansin protein EXLX1 has been experimentally shown to bind cellulose (Nikolaos et al., 2011).
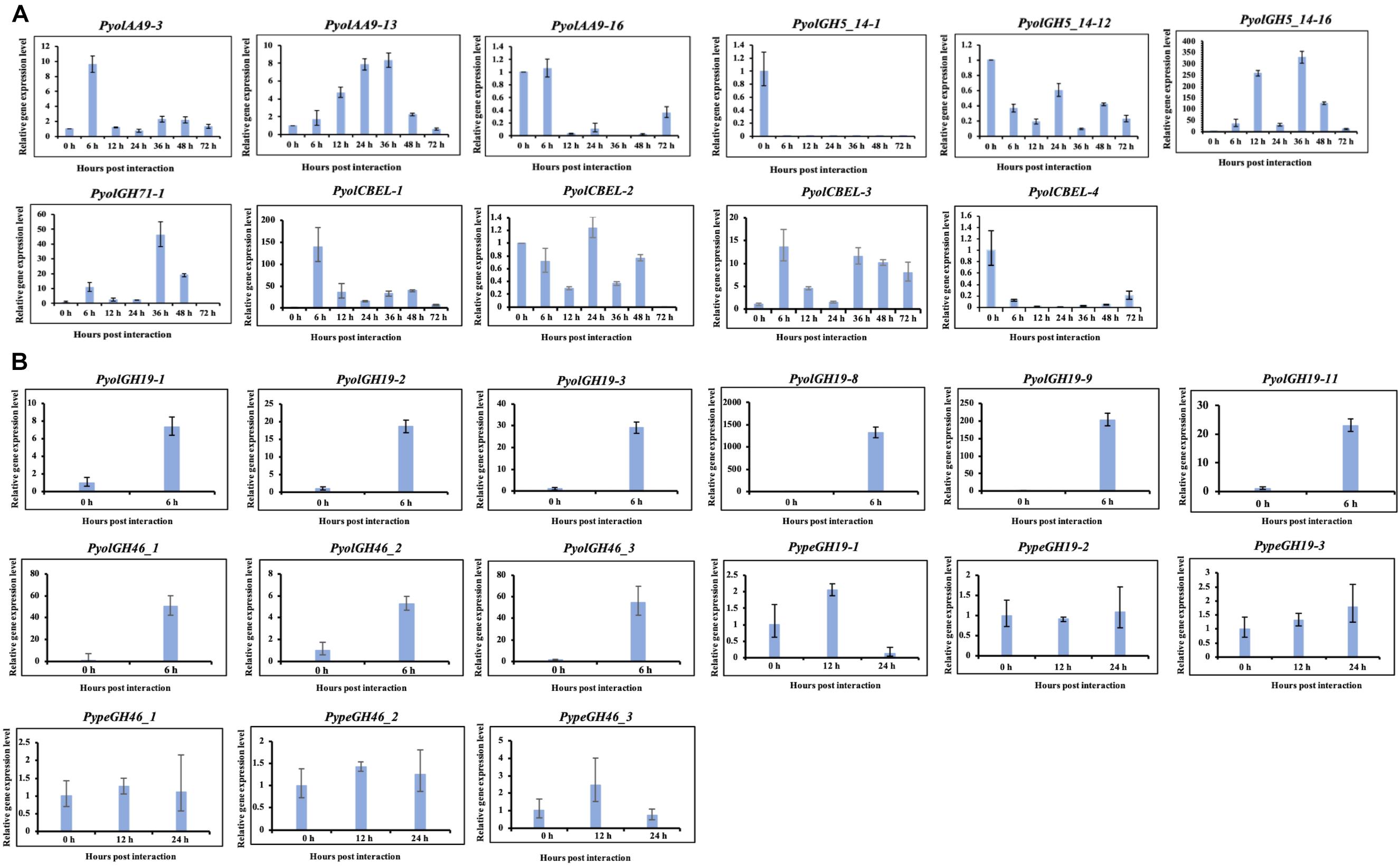
Figure 10. Quantitative Real-Time PCR verification of expression of selected CAZy genes in P. oligandrum and P. periplocum during in vitro growth or during interactions with Ph. infestans or B- cinnerea. In vitro growth at 0 h was used as the reference and normalized to 1.0. The α-tubulin genes of P. oligandrum and P. periplocum (Pyoltua and Pypetua) were used as internal reference genes. Data displayed as the average of three biological replicates and error bars indicate standard deviations. 6, 12, 24, 36, 48, 72 h represent hours post interaction with Ph. infestans or B. cinerea. (A) Quantitative Real-Time PCR verification for CAZy genes of P. oligandrum and P. periplocum during interactions with Ph. infestans. (B) Quantitative Real-Time PCR verification for CAZy genes of P. oligandrum and P. periplocum during interactions with B. cinerea.
Overall, we compared our RNA-seq data and qRT-PCR assay results (Figures 2B, 3B, 5B, 6B,C, 7B,C, 10 and Supplementary Figure S6). From the RNA-seq analysis we concluded that, PyolAA9-3, PyolAA9-13, PyolGH5_14-16, PypeGH46-1, and PyolCBEL-1 were upregulated during interactions with Ph. infestans or B. cinema. PyolAA9-16, PyolGH5_14-1, PyolCBEL-2, and PyolCBEL-4 were downregulated during interaction with Ph. infestans. All the above genes mentioned also show similar expression profiles using qRT-PCR assays. PyolGH5_14-12, PypeGH19-1, PypeGH19-2, PypeGH19-3, PypeGH46-2, PypeGH46-3, PyolCBEL-3 were constitutively expressed. There was generally a good agreement between the RNA-seq and qRT-PCR expression data, showing the reliability of our data. However, PyolGH5_14-12, PypeGH19-1, and PyolCBEL-3 expression using RNA-seq is not so consistent with that shown by qRT-PCR.
To investigate the extent to which the mycoparasitic oomycetes could directly utilize the major complex carbohydrate components of oomycete and fungal cell walls as the sole sources of carbon we performed in vitro growth assays in minimal media amended with either cellulose, chitin or the monomeric unit of chitin, N-acetylglucosamine with glucose or yeast extract amendments used as control conditions. We compared the growth of P. oligandrum and P. periplocum to that of Ph. infestans (Figure 11).
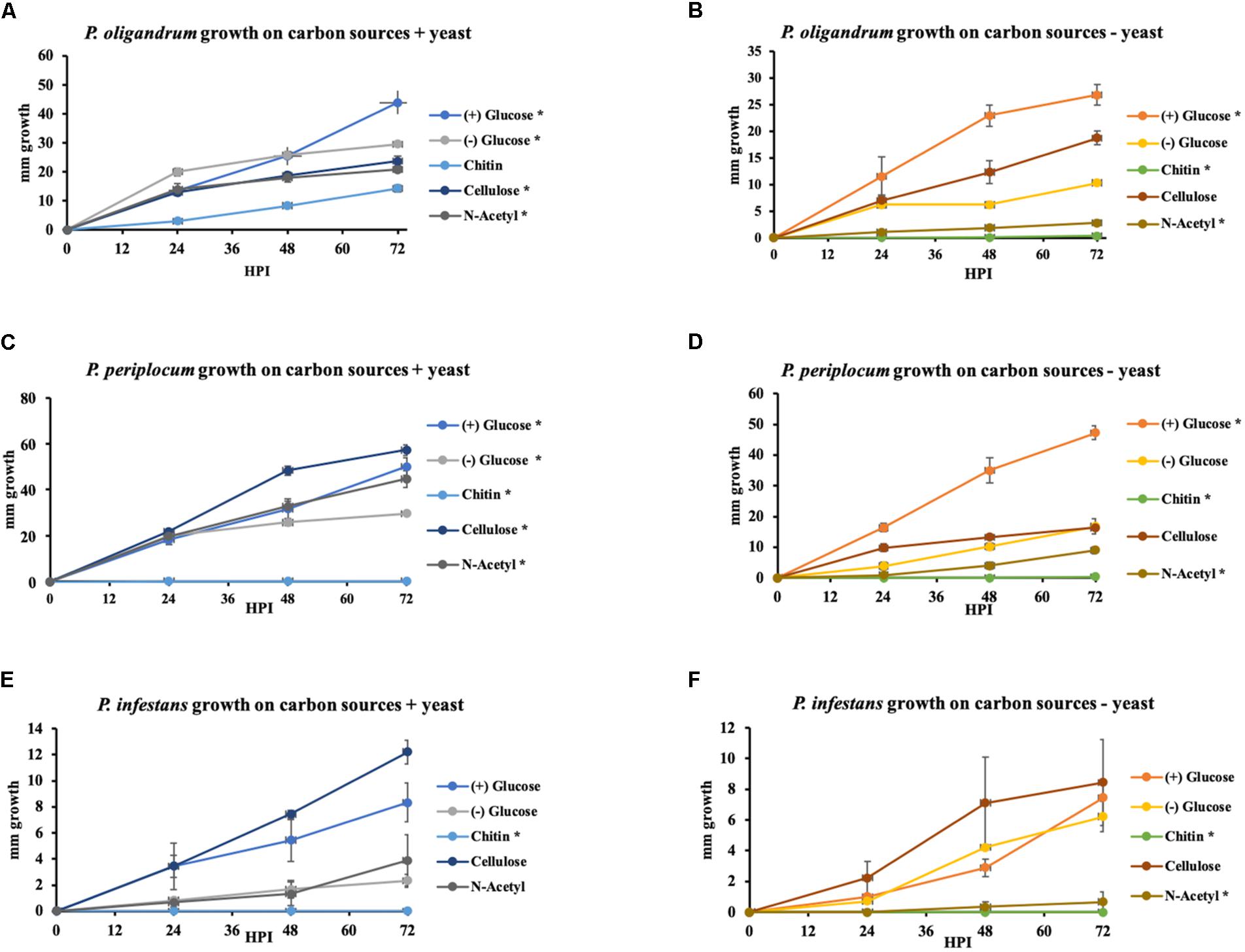
Figure 11. Growth of P. oligandrum, P. periplocum, and Ph. infestants on varying carbon sources representing the major structural carbohydrates in fungal or oomycete cell walls. P. oligandrum, P. periplocum, and Ph. infestans growth on the carbon sources, cellulose, glucose, chitin, and N-Acetyl-glucosamine in modified Plich media plus or minus yeast, denoted by different colored lines. X-axis shows hours post inoculation (hpi). Y-axis displays colony diameter in mm. (A,B) Growth rate of P. oligandrum. (C,D) Growth rate of P. periplocum. (E,F) Growth rate of Ph. infestans. * denotes statistically significant difference from the control (-glucose, -yeast) data at p < 0.05 (t-test).
In either the presence or absence of yeast, P. oligandrum grew largest with glucose as a carbon source and was also able to utilize cellulose, chitin and N-acetylglucosamine as carbon sources. In the presence of yeast, P. periplocum grew equally well in most treatments, but in the absence of yeast, grew best with glucose as the sole carbon source. In the absence of yeast P. periplocum showed a strong preference for growth in glucose media and very reduced growth in the other substrates. In either the presence or absence of yeast, P. infestans grew largest with cellulose as the primary carbon source, and was not able to utilize chitin or N-acetylglucosamine reflecting the adaptation of this species to cellulosic plant hosts. Several CAZy encoded genes that function in the release of the simple sugar glucose from cellulose (e.g., GH6 and GH17 family members) were upregulated during mycoparasitism of the cellulosic host Ph. infestans, and genes for the degradation of the cell wall carbohydrates from fungal and oomycete prey were abundant and expanded in the mycoparasite genomes.
Taken together, these data suggest that degradation of more complex carbohydrate constituents of fungal and oomycete cell walls is only of limited importance to mycoparasitic oomycetes. Instead, destruction of the major components of prey cell walls in order to gain access to simple sugars within the cells appears to be the predominant strategy of these mycoparasites. This is in line with similar strategies used by phytopathogenic Pythium species, where CWDE secretion to allow maceration of the plant cell wall and subsequent uptake of simple sugars from within plant cells appears to be the major mechanism by which these species obtain nutrition (Zerillo et al., 2013). However, due to the expansion of CWDEs within mycoparasitic oomycete genomes they show a clear ability to utilize more complex carbohydrates not found within the plant cell wall when necessary. The mycoparasites also exhibit overall faster growth in vitro than the plant pathogen indicating a more efficient usage of these carbon sources for growth, and potentially allowing these species to outcompete their plant pathogenic or saprotrophic counterparts within the agroecosystem.
P. oligandrum and P. periplocum have long been recognized for their ability to inhibit plant pathogens (Paul, 1999; Rey et al., 2008), although the molecular basis of their interactions remains largely unknown. A small cDNA library sequencing study previously identified the potential role of CWDEs in P. oligandrum mycoparasitism (Horner et al., 2012). Through a detailed in silico and transcriptomic analysis, we have investigated the CAZyome of both P. oligandrum and P. periplocum in relation to plant pathogenic oomycetes. In conclusion, we found that tandem duplication and horizontal gene transfer events have been the major drivers for the formation of the distinctive and expanded CAZyome of mycoparasitic Pythium species.
We identified three major CAZy families from which the member genes are predominantly located in the gene sparse regions of their respective genomes and that have expanded through tandem duplication. These are hallmarks of genes undergoing rapid evolution such as the RxLR effectors of plant pathogenic oomycetes (Haas et al., 2009; Dong et al., 2015). Based on our data, we postulate, for the first time, that mycoparasitic oomycete genomes also display a “two-speed genome” phenomenon in a similar manner to plant pathogenic oomycetes, but that the key pathogenicity determinants, at least those responsible for the onset of infection are CAZyme CWDEs that afford the mycoparasitic oomycetes the opportunity to macerate microbial tissue from which they can extract the sugars and other nutrients they require for growth and reproduction.
We have identified that carbohydrate active enzymes such as cellulose degrading enzymes are potentially important pathogenicity determinates in the mycoparasitic oomycetes. Since both mycoparasite and prey oomycetes all contain such enzymes, we hypothesize that these enzymes must be tightly regulated or compartmentalized to limit or prevent self-degradation and thus the regulation of CAZy gene function in mycoparasitic oomycetes is an important topic for further investigation in the future. Such studies will help to answer one of the most important questions in mycoparasitism; how self, and non-self, recognition occurs.
Furthermore, we have provided evidence for the horizontal gene transfer of five CAZy families from bacteria, fungi or viruses to the mycoparasites. In contrast, mycoparasitic fungi of the Trichoderma species are reported to have obtained almost 50% of their CWDE complement by horizontal, or lateral, gene transfer (Druzhinina et al., 2018). However, Richards et al. (2011) investigated the occurrence of horizontal gene transfer events from fungi to oomycetes and concluded that only a small number of such events could be found, thus our results are in line with these previous findings (Richards et al., 2011).
Taken together our data suggest a possible phytopathogenic ancestral state for the Pythium Genus, with the ability to mycoparasitise other eukaryotic microbes an adaptation derived from expansion of several key CAZyme gene families, and horizontal gene transfer events that occurred in the last common ancestor of P. oligandrum and P. periplocum.
The published genomes of the following oomycetes were used for the comparative analysis of their CAZyme encoding gene complement: P. oligandrum (CBS 530.74) (Kushwaha et al., 2017a), P. periplocum (CBS 532.74) (Kushwaha et al., 2017b), P. ultimum (DAOM BR144) (Lévesque et al., 2010), P. irregulare (DAOM BR486) (Adhikari et al., 2013), P. iwayamai (DAOM BR242034) (Adhikari et al., 2013), P. arrhenomanes (ATCC 12531) (Adhikari et al., 2013), P. aphanidermatum (DAOM BR444) (Adhikari et al., 2013), P. insidiosum (Strain Pi-S) (Rujirawat et al., 2015), Phytopythium. vexans (DAOM BR484) (Adhikari et al., 2013), Ph. infestans (T30-4) (Haas et al., 2009), Ph. sojae (P6497) (Tyler et al., 2006), Ph. ramorum (Pr102) (Tyler et al., 2006), Ph. capsici (LT1534) (Lamour et al., 2012).
To explore overall distribution of CAZymes among the above 11 oomycetes, the genome assemblies, transcript sequences, and protein sequences were downloaded from the NCBI Genome portal1 and the Joint Genome Portal2, along with the gene model annotation files in GFF3 format, which were used for CAZy annotation and synteny analysis.
Benchmarking Universal Single-Copy Orthologs BUSCO v4 (Seppey et al., 2019)3 was used to evaluate the completeness of the genome assemblies of the oomycetes used in this study. Default parameters were used with lineage-specific datasets set to stramenopiles_odb10.
P. oligandrum (CBS 530.74) and P. periplocum (CBS 532.74) were maintained on V8 media amended with CaCO3 as previously described (Horner et al., 2012; Kushwaha et al., 2017a, b) at 18°C in the dark. Ph. infestans (88,069) was maintained on rye sucrose agar at 18°C in the dark as described (Grenville-Briggs et al., 2008). B. cinerea (B05) was maintained on either V8 media or corn meal agar (CMA) at 20°C in the dark. Prior to confrontation, P. oligandrum, P. periplocum and B. cinnerea were grown in liquid V8 broth amended with calcium carbonate and Ph. infestans were grown in liquid pea broth, at 20°C in the dark.
CAZyme-encoding genes were predicted using the dbCAN pipeline (Yin et al., 2012). Briefly, hidden Markov models of all CAZy families were download from CAZy database (Lombard et al., 2013)4. Hidden Markov searches using the predicted proteomes of the oomycete species listed above were performed, with a cut-off value of 1E-03. To predict members of the CBEL family, only candidates containing two CBM_1 domain and a signal peptide were retained.
Transmembrane domain searches were conducted using the TMHMM Server v2.0 (Käll et al., 2005)5. SignalP 3.0 was used to predict the presence of a signal peptide for secretion (Bendtsen et al., 2004)6. Domain architecture predictions were conducted with by searching the Pfam database (El-Gebali et al., 2018)7. For CAZy families with diverse enzyme activity, the exact enzyme activity annotation of members in these families were conducted by using BLASTP search of verified proteins in the ExPASy database (Artimo et al., 2012)8.
Fisher’s exact test, conducted in the R program suite v3.5.3, was used to compare the gene counts of CAZy family members in the mycoparasitic Pythium species with those of the plant pathogenic Pythium, human Pythium, and Phytophthora species, respectively. The CAZy families with p-values higher than 1E-05 were removed. The CAZy families with higher gene counts in mycoparasitic Pythium were thus retained for further analysis.
Protein sequences were aligned by MUSCLE (Edgar, 2004). Phylogenetic analysis of full-length proteins alignments was performed by IQ-TREE (Nguyen et al., 2014), using the Maximum Likelihood approach and 1,000 bootstrap values. The models for phylogenetic analysis were automatically selected by IQ-TREE program. The visualization and modification of phylogenetic trees were performed using the iTOL server (Letunic and Bork, 2006). Branches with the bootstrap values higher than 50 were displayed.
The predicted proteomes of P. oligandrum and P. periplocum were used to search against the predicted proteome of P. ultimum using BLASTP, using the default maximum hits setting with an E-value cut-off of 1E-10. Identification of gene collinearity between mycoparasitic Pythium and P. ultimum was performed using MCScanX (Wang et al., 2012). TBtools (Chen et al., 2018) was employed for visualization of the synteny analysis.
We screened significantly differentially expressed CAZy genes and the CAZy families they belong to. By calculating the average fold change of these CAZy families, with treatment by log2, we could distinguish overall expression profiles of differentially expressed CAZy genes that were assigned to the same CAZy family. We used a log2-treated average fold change approach, to analyze the changes in expression of genes from each family where values equal to, or less than, −1 (to the left of the dashed black line in Supplementary Figure S2) correspond to down regulation of most of the genes within a family and values equal to, or greater than, 1 (to the right of the dashed red line in Supplementary Figure S2) correspond to up-regulation of the majority of the genes in that family. Numbers of genes from each gene family, on which this analysis is based are shown in the left of each panel of Supplementary Figure S2.
For confrontation assays, approximately 5 cm3 of mycelium, washed with sterile dH20, from mycoparasite (P. oligandrum or P. periplocum) and prey (either Ph. infestans or B. cinnerea) were placed at opposite sides of a polycarbonate membrane on V8 agar. The interaction zone (1 cm) was sampled at the point of contact (0 h post interaction, hpi) and then subsequently at 12 and 24 hpi. Control samples of the mycoparasites were prepared using two mycelial plugs from the same organism interacting with each other for 3 days, and mycoparasitism samples were prepared using either the oomycete Ph. infestans or the fungus B. cinnerea as the prey, which was prepared from liquid media samples as described above. Collected samples were snap frozen in liquid nitrogen and used for RNA extraction. Five replicates of each interaction were prepared and three of these replicates were randomly chosen for RNA extraction and sequencing. For detailed quantitative RT-PCR of a time course of mycoparasitism, the interacting mycelium from the confrontations between the mycoparasite and the prey in sterile tap water, was excised at 0, 6, 12, 24, 36, 48, and 72 hpi and immediately snap frozen and ground in liquid nitrogen, prior to RNA extraction.
Approximately 100 mg RNA from each sample was extracted using the RNeasy Plant Mini Kit (#74904 QIAGEN) according to the manufacturers protocol, and treated with RNase-free DNase for 20 min at 37°C (Ambion, TURBO DNA-free Kit). The extracted RNA was qualitatively visualized by agarose gel electrophoresis and a NanoDrop 1,000 spectrophotometer (Thermo Fisher Scientific) was used to quantify the total amount of RNA. Prior to RNA sequencing, the integrity of the samples was corroborated using the ExperionTM Automated Electrophoresis System (Bio-Rad Laboratories, Hercules, United States).
Polyadenylated messenger RNA was captured from 200 ng total RNA per sample using magnetic beads and Illumina adaptors with sample specific barcode sequences were ligated before subsequent library amplification using PCR using the Illumina TruSeq RNA poly-A selection kit. Sequencing of 150 bp paired-end libraries was carried out using the Illumina NovaSeq6000 S4 platform (SciLifeLab, Stockholm). All raw sequencing data in this study have been deposited in National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA637834 and a full analysis of these data will be presented elsewhere. The resulting bcl2fastq demultiplexed FastQ files were de novo mapped to our previously published reference genomes (Kushwaha et al., 2017a, b) and Adaptor cleaning was conducted by Trimmomatic (Bolger et al., 2014). Normalization and quantification of expression levels was performed by Salmon (Patro et al., 2015). The full analysis of differential gene expression during mycoparasitic interactions will be published elsewhere, however, in the current study we have mined these data to investigate the expression levels of selected CAZyme-encoding genes as presented in the results. pheatmap package of R project (Kolde, 2012) was used to produce heatmaps showing the log fold change in gene expression values (TPM counts) for selected CAZyme-encoding genes between the transcriptome samples.
cDNA synthesis was carried out using the Superscript IV Reverse transcriptase cDNA synthesis kit (Thermo Fisher Scientific) using 2 μg template RNA. All cDNA samples were diluted to 20 ng–1 prior to qRT-PCR. The gene expression levels were evaluated using quantitative RT-PCR (Biorad real-time PCR cycler using SYBG as the fluorescent dye). P. oligandrum and P. periplocum α-tubulin genes (Pyoltua; Genbank accessions MT623563 and MT811915; Supplementary Table S1). Primers (listed in Supplementary Table S1) were designed in Primer39 and the NCBI BLASTN web platform was used to check the specificity of the sequences for the genes in question, with the low complexity filter turned off. The internal reference genes list above were used to normalize expression levels of CAZyme candidates from the corresponding species.
To study growth of mycoparasitic Pythium on different carbon substrates, representing the major structural components of oomycete and fungal cell walls, we compared the growth of P. oligandrum, P. periplocum, and Ph. infestans in modified Plich media (van West et al., 1999) amended with, cellulose, glucose, chitin and N-Acetylglucosamine individually at 25 mM∗mL–1 both with and without yeast extract. Modified plich media without glucose or yeast extract served as controls. 7.5 mm plugs from 7-day old liquid cultures of P. oligandrum CBS 530.74 and P. periplocum CBS 532.74 cultivated in V8 media and Ph. infestans 88,069 cultivated in pea-broth were used to investigate the growth rate of the oomycetes on different the carbon sources. Ph. infestans was used as a comparative control. Growth of each organism was quantified as diagonal growth in mm and measured every 24 hpi for three constitutive days. In total 10 treatments were performed. The experiment was repeated three times with three independent biological replicates. Statistical analysis of the growth assays was calculated as the area under the growth curve using the trapezoidal method. The differences among treatments was assessed using student’s T-test, on the means of the area under the curve, assuming two-tailed distribution and two-sample with unequal variance. Significant difference was accepted if (p < 0.05).
RNA-seq data (NCBI accession: PRJNA637834); P. oligandrum internal reference gene (Genbank accession: MT623563); P. periplocum internal reference gene (Genbank accession: MT811915).
LG-B designed the experiments and analysis methods. DL designed and performed the bioinformatic analysis. CA and RV designed and performed the wet lab experiments and analyzed the wet lab experimental data. DL, CA, and LG-B wrote the manuscript. LG-B and DD revised the manuscript. All authors reviewed and approved the final manuscript.
This work was supported by funding to LG-B from The Swedish Foundation for Strategic Research (through the Future Research Leaders Fellowship program, FFL5) and the Swedish Research Council Formas (through a Future Research Leaders Project grant). This project has also received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No. 766048 to LG-B, which supports CA. Funding to DD and DL was provided by the Chinese Fundamental Research Funds for the Central Universities (KYT202001, JCQY201901, and JCQY201904). We gratefully acknowledge support from Science for Life Laboratory (SciLifeLab), the Knut and Alice Wallenberg Foundation, the National Genomics Infrastructure funded by the Swedish Research Council, and Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure and for SciLifeLab National Genomics Project funding.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.581698/full#supplementary-material
Supplementary Figure 1 | BUSCO analysis for evaluation of genome assembly completeness. X-axis represent genome assemblies of organisms mentioned in this article. Y-axis represent number of each category of BUSCO groups.
Supplementary Figure 2 | Comparison of significantly expressed genes assigned to CAZy families that are not unique or expanded in the mycoparasitic Pythium species. Numbers within the heatmaps represent the count of significantly expressed genes assigned to each corresponding CAZy family. Green histograms represent log2-treated average fold change of Pythium CAZy genes significantly expressed from in vitro growth to 12 hpi in the presence of Ph. infestans. Light blue histograms represent log2-treated average fold change of Pythium CAZy genes significantly expressed from 12 hpi in the presence of Ph. infestans to 24 hpi in the same interaction. Black and red dashed lines represent log2-treated average fold change equal to -1 and 1 respectively. (A) overview of significantly expressed CAZy genes of P. oligandrum. (B) overview of significantly expressed CAZy genes of P. periplocum.
Supplementary Figure 3 | Analysis of gene density for selected CAZy gene families. (A) Gene density heatmap of AA9 genes in P. oligandrum. (B) Gene density heatmap of GH5_14 genes in P. oligandrum. Dots with different color represent intergenic distance of encoding genes in different clades according to respective phylogenic tree. (C) Gene density heatmap of GH19 genes in P. oligandrum. (D) Gene density heatmap of GH19 genes in P. periplocum. Dots with black color represent intergenic distance of GH19 encoding genes detected in P. oligandrum and P. periplocum.
Supplementary Figure 4 | Analysis of genome location of selected CAZy genes. (A) Genome location of P. oligandrum AA9 encoding genes from clades 1 and 3. Blue arrows represent the orientation of P. oligandrum AA9 encoding genes from clades 1 and 3. (B) Genome location of P. oligandrum GH5_14 encoding genes encoding from clade 1. Blue arrows represent the orientation of P. oligandrum GH5_14 genes from clade 1. (C) Genome location of P. oligandrum GH19 genes. Blue arrows represent orientation of P. oligandrum GH19 genes.
Supplementary Figure 5 | Synteny analysis. (A) Synteny analysis of GH55 genes between mycoparasitic Pythium species and P. ultimum. The red arrows represent GH55 genes identified in P. oligandrum and P. periplocum. The blue arrows represent genes near to GH55 genes. The gray lines represent syntenic relationships. (B) Synteny analysis of GH46 genes between mycoparasitic Pythium and P. ultimum. The red arrows represent the orientation of GH46 genes identified in P. oligandrum and P. periplocum. The blue arrows represent the orientation of genes neighboring the GH46 genes. The gray lines represent syntenic relationships.
Supplementary Figure 6 | Analysis of the cellulose-binding elicitor lectin (CBEL) family. (A) Phylogenetic tree of CBEL proteins identified in oomycete species. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). CBEL proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are marked in red. TrCBHI_P62694 (an exoglucanase verified in Trichoderma reesei) was used as an outgroup. Domain architecture of CBEL proteins is shown on to the left of the phylogenetic tree. (B) RNAseq Expression profile of CBEL genes detected in P. oligandrum and P. periplocum during in vitro growth or during interactions with Ph. infestans at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
Supplementary Figure 7 | Analysis of the cellulose synthase (CesA) family. (A) Phylogenetic tree of CesA proteins identified in oomycete species. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). CesA3 proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are marked in red. CesA proteins identified in Arabidopsis thaliana (At) were used as outgroup genes. The domain architecture of each of the CesA proteins is shown to the left of the Phylogenic tree. (B) RNAseq Expression profile of CesA genes detected in P. oligandrum and P. periplocum during in vitro growth or during interactions with Ph. infestans at 12 or 24 h post interaction. Expression levels are expressed as the log2 fold change of transcripts per million (TPM), per gene.
Supplementary Figure 8 | Phylogenic analysis of GH55 and GH71 proteins detected in mycoparasite Pythium and their homologous proteins detected by BLAST search. Maximum likelihood tree, with 1,000 bootstraps (values displayed per branch). Proteins identified in P. oligandrum (Pyol) and P. periplocum (Pype) are shown in red.
Supplementary Table 1 | Primers for CAZyme-coding genes verified by qRT-PCR in this study.
Supplementary Table 2 | ID and gene model sequences of CAZyme-coding genes in this study.
Supplementary Table 3 | Ortholog clustering for each CAZy families mentioned in this study.
Adhikari, B. N., Hamilton, J. P., Zerillo, M. M., Tisserat, N., Lévesque, C. A., and Buell, C. R. (2013). Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes. PLoS One 8:e75072. doi: 10.1371/journal.pone.0075072
Andersson, J. O. (2009). “Horizontal gene transfer between microbial eukaryotes,” in Horizontal Gene Transfer, eds M. B. Gogarten., J. P. Gogarten, and L. Olendzenski (Totowa, FL: Humana Press), 473–487. doi: 10.1007/978-1-60327-853-9_27
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucl. Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Baek, J.-M., Howell, C. R., and Kenerley, C. M. (1999). The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr. Genet. 35, 41–50. doi: 10.1007/s002940050431
Bara, M. T. F., Lima, A. L., and Ulhoa, C. J. (2003). Purification and characterization of an exo-β-1, 3-glucanase produced by Trichoderma asperellum. FEMS Microbiol. Lett. 219, 81–85. doi: 10.1016/s0378-1097(02)01191-6
Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Benhamou, N., and Chet, I. (1997). Cellular and molecular mechanisms involved in the interaction between Trichoderma harzianum and Pythium ultimum. Appl. Environ. Microbiol. 63, 2095–2099. doi: 10.1128/aem.63.5.2095-2099.1997
Benhamou, N., Rey, P., Chérif, M., Hockenhull, J., and Tirilly, Y. (1997). Treatment with the mycoparasite Pythium oligandrum triggers induction of defense-related reactions in tomato roots when challenged with Fusarium oxysporum f. sp. radicis-lycopersici. Phytopathology 87, 108–122. doi: 10.1094/phyto.1997.87.1.108
Benhamou, N., Rey, P., Picard, K., and Tirilly, Y. (1999). Ultrastructural and cytochemical aspects of the interaction between the mycoparasite Pythium oligandrum and soilborne plant pathogens. Phytopathology 89, 506–517. doi: 10.1094/phyto.1999.89.6.506
Berger, H., Yacoub, A., Gerbore, J., Grizard, D., Rey, P., Sessitsch, A., et al. (2016). Draft genome sequence of biocontrol agent Pythium oligandrum strain Po37, an oomycota. Genome Announc. 4, 00215–00216e. doi: 10.1128/genomea.00215-16
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bradshaw-Smith, R., Whalley, W., and Craig, G. (1991). Interactions between Pythium oligandrum and the fungal footrot pathogens of peas. Mycol. Res. 95, 861–865. doi: 10.1016/s0953-7562(09)80050-6
Chen, C., Xia, R., Chen, H., and He, Y. (2018). TBtools, a Toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 13:289660. doi: 10.1101/289660
Cuskin, F., Lowe, E. C., Temple, M. J., Zhu, Y., Cameron, E. A., Pudlo, N. A., et al. (2015). Human gut bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165. doi: 10.1038/nature13995
Dong, S., Raffaele, S., and Kamoun, S. (2015). The two-speed genomes of filamentous pathogens: waltz with plants. Curr. Opin. Genet. Dev. 35, 57–65. doi: 10.1016/j.gde.2015.09.001
Doolittle, W. F. (1999). Phylogenetic classification and the universal tree. Science 284, 2124–2128. doi: 10.1126/science.284.5423.2124
Druzhinina, I. S., Chenthamara, K., Zhang, J., Atanasova, L., Yang, D., Miao, Y., et al. (2018). Massive lateral transfer of genes encoding plant cell wall-degrading enzymes to the mycoparasitic fungus Trichoderma from its plant-associated hosts. PLoS Genet. 14:e1007322. doi: 10.1371/journal.pgen.1007322
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2018). The Pfam protein families database in 2019. Nucl. Acids Res. 47, D427–D432. doi: 10.6019/tol.pfam_fams-t.2018.00001.1
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14. doi: 10.1101/466201
Faure, C., Veyssière, M., Boëlle, B., San Clemente, H., Bouchez, O., Lopez-Roques, C., et al. (2020). Long-read genome sequence of the sugar beet rhizosphere mycoparasite Pythium oligandrum. G3 10, 431–436. doi: 10.1534/g3.119.400746
Gaulin, E., Dramé, N., Lafitte, C., Torto-Alalibo, T., Martinez, Y., Ameline-Torregrosa, C., et al. (2006). Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18, 1766–1777. doi: 10.1105/tpc.105.038687
Grenville-Briggs, L. J., Anderson, V. L., Fugelstad, J., Avrova, A. O., Bouzenzana, J., Williams, A., et al. (2008). Cellulose synthesis in Phytophthora infestans is required for normal appressorium formation and successful infection of potato. Plant Cell 20, 720–738. doi: 10.1105/tpc.107.052043
Haas, B. J., Kamoun, S., Zody, M. C., Jiang, R. H., Handsaker, R. E., Cano, L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398. doi: 10.1038/nature08358
Horner, N. R., Grenville-Briggs, L. J., and Van West, P. (2012). The oomycete Pythium oligandrum expresses putative effectors during mycoparasitism of Phytophthora infestans and is amenable to transformation. Fungal Biol. 116, 24–41. doi: 10.1016/j.funbio.2011.09.004
Käll, L., Krogh, A., and Sonnhammer, E. L. (2005). An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21(Suppl._1), i251–i257. doi: 10.1093/bioinformatics/bti1014
Kim, I. J., Nam, K. H., Yun, E. J., Kim, S., Youn, H. J., Lee, H. J., et al. (2015). Optimization of synergism of a recombinant auxiliary activity 9 from Chaetomium globosum with cellulase in cellulose hydrolysis. Appl. Microbiol. Biotechnol. 99, 8537–8547. doi: 10.1007/s00253-015-6592-3
Kim, I. J., Youn, H. J., and Kim, K. H. (2016). Synergism of an auxiliary activity 9 (AA9) from Chaetomium globosum with xylanase on the hydrolysis of xylan and lignocellulose. Proc. Biochem. 51, 1445–1451. doi: 10.1016/j.procbio.2016.06.017
Kubicek, C. P., Herrera-Estrella, A., Seidl-Seiboth, V., Martinez, D. A., Druzhinina, I. S., Thon, M., et al. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12:R40. doi: 10.1007/978-1-4899-0280-1_12
Kushwaha, S. K., Vetukuri, R. R., and Grenville-Briggs, L. J. (2017a). Draft genome sequence of the mycoparasitic oomycete Pythium oligandrum Strain CBS 530.74. Genome Announc. 5, 317–346e. doi: 10.1128/genomea.00346-17
Kushwaha, S. K., Vetukuri, R. R., and Grenville-Briggs, L. J. (2017b). Draft genome sequence of the mycoparasitic oomycete Pythium periplocum strain CBS 532.74. Gen. Announc. 5, 17–57e. doi: 10.1128/genomea.00057-17
Lamour, K. H., Mudge, J., Gobena, D., Hurtado-Gonzales, O. P., Schmutz, J., Kuo, A., et al. (2012). Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol. Plant Microbe Interact. 25, 1350–1360. doi: 10.1094/MPMI-02-12-0028-R
Le Floch, G., Rey, P., Benizri, E., Benhamou, N., and Tirilly, Y. (2003). Impact of auxin-compounds produced by the antagonistic fungus Pythium oligandrum or the minor pathogen Pythium group F on plant growth. Plant Soil 257, 459–470. doi: 10.1023/a:1027330024834
Letunic, I., and Bork, P. (2006). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Lévesque, C. A., Brouwer, H., Cano, L., Hamilton, J. P., Holt, C., Huitema, E., et al. (2010). Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11:R73. doi: 10.1186/gb-2010-11-7-r73
Limón, M., Chacón, M., Mejías, R., Delgado-Jarana, J., Rincón, A., Codón, A., et al. (2004). Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl. Microbiol. Biotechnol. 64, 675–685. doi: 10.1007/s00253-003-1538-6
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2013). The carbohydrate-active enzymes database (CAZy) in 2013. Nucl. Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Mateos, F. V., Rickauer, M., and Esquerré-Tugayé, M.-T. (1997). Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol. Plant Microbe Interact. 10, 1045–1053. doi: 10.1094/mpmi.1997.10.9.1045
Mélida, H., Sandoval-Sierra, J. V., Diéguez-Uribeondo, J., and Bulone, V. (2013). Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell 12, 194–203. doi: 10.1128/ec.00288-12
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nikolaos, G., Akira, T., Nikolas, N., and Daniel, J. G. (2011). Structure-function analysis of the bacterial expansin EXLX1. J. Biol. Chem. 286, 16814–16823. doi: 10.1074/jbc.m111.225037
Ospina-Giraldo, M. D., Griffith, J. G., Laird, E. W., and Mingora, C. (2010). The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genom. 11:525. doi: 10.1186/1471-2164-11-525
Ouyang, Z., Li, X., Huang, L., Hong, Y., Zhang, Y., Zhang, H., et al. (2015). Elicitin−like proteins Oli−D1 and Oli−D2 from Pythium oligandrum trigger hypersensitive response in Nicotiana benthamiana and induce resistance against Botrytis cinerea in tomato. Mol. Plant Pathol. 16, 238–250. doi: 10.1111/mpp.12176
Patro, R., Duggal, G., and Kingsford, C. (2015). Salmon: accurate, versatile and ultrafast quantification from RNA-seq data using lightweight-alignment. BioRxiv 2015:021592. doi: 10.1101/021592
Paul, B. (1999). Pythium periplocum, an aggressive mycoparasite of botrytis cinerea causing the gray mould disease of grape-vine. FEMS Microbiol. Lett. 181, 277–280. doi: 10.1111/j.1574-6968.1999.tb08855.x
Picard, K., Ponchet, M., Blein, J.-P., Rey, P., Tirilly, Y., and Benhamou, N. (2000a). Oligandrin. a proteinaceous molecule produced by the mycoparasite pythium oligandrum induces resistance to phytophthora parasitica infection in tomato plants. Plant Physiol. 124, 379–396. doi: 10.1104/pp.124.1.379
Picard, K., Tirilly, Y., and Benhamou, N. (2000b). Cytological effects of cellulases in the parasitism of Phytophthora parasitica by Pythium oligandrum. Appl. Environ. Microbiol. 66, 4305–4314. doi: 10.1128/aem.66.10.4305-4314.2000
Plaats-Niterink, A. (1981). Monograph of the genus Pythium in studies in mycology. Centraalbureau Voor Schimmelcultures, (Netherlands: Baarn).
Raffaele, S., and Kamoun, S. (2012). Genome evolution in filamentous plant pathogens: why bigger can be better. Nat. Rev. Microbiol. 10, 417–430. doi: 10.1038/nrmicro2790
Rey, P., Le Floch, G., Benhamou, N., and Tirilly, Y. (2008). “Pythium oligandrum biocontrol: its relationships with fungi and plants,” in Plant-microbe interactions, E. Ait Barka and C. Clément, (Kerala: Research Signpost), 43–57.
Richards, T. A., Soanes, D. M., Jones, M. D., Vasieva, O., Leonard, G., Paszkiewicz, K., et al. (2011). Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. U.S.A. 108, 15258–15263. doi: 10.1073/pnas.1105100108
Rujirawat, T., Patumcharoenpol, P., Lohnoo, T., Yingyong, W., Lerksuthirat, T., Tangphatsornruang, S., et al. (2015). Draft genome sequence of the pathogenic oomycete Pythium insidiosum strain Pi-S, isolated from a patient with pythiosis. Genome Announc. 3, 515–574e. doi: 10.1128/genomeA.00574-15
Schmoll, M. (2018). Regulation of plant cell wall degradation by light in Trichoderma. Fungal Biol. Biotechnol. 5, 10. doi: 10.1186/s40694-018-0052-7
Seppey, M., Manni, M., and Zdobnov, E. M. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245. doi: 10.1007/978-1-4939-9173-0_14
Sietsma, J., Eveleigh, D., and Haskins, R. (1969). Cell wall composition and protoplast formation of some oomycete species. Biochim. Biophys. Acta 184, 306–317. doi: 10.1016/0304-4165(69)90033-6
Sun, H., Cao, R., Li, L., Zhao, L., and Liu, Q. (2018). Cloning, purification and characterization of a novel GH46 family chitosanase, Csn-CAP, from Staphylococcus capitis. Proc. Biochem. 75, 146–151. doi: 10.1016/j.procbio.2018.09.021
Takenaka, S., Yamaguchi, K., Masunaka, A., Hase, S., Inoue, T., and Takahashi, H. (2011). Implications of oligomeric forms of POD-1 and POD-2 proteins isolated from cell walls of the biocontrol agent Pythium oligandrum in relation to their ability to induce defense reactions in tomato. J. Plant Physiol. 168, 1972–1979. doi: 10.1016/j.jplph.2011.05.011
Tyler, B. M., Tripathy, S., Zhang, X., Dehal, P., Jiang, R. H., Aerts, A., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266. doi: 10.1126/science.1128796
van West, P., Kamoun, S., Van’t Klooster, J. W., and Govers, F. (1999). Internuclear gene silencing in Phytophthora infestans. Mol. Cell 3, 339–348. doi: 10.1016/s1097-2765(00)80461-x
Viens, P., and Lacombe-Harvey, M. -È, and Brzezinski, R. (2015). Chitosanases from family 46 of glycoside hydrolases: from proteins to phenotypes. Mar. Drugs 13, 6566–6587. doi: 10.3390/md13116566
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl. Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Yacoub, A., Gerbore, J., Magnin, N., Haidar, R., Compant, S., and Rey, P. (2018). Transcriptional analysis of the interaction between the oomycete biocontrol agent. Pythium oligandrum, and the roots of Vitis vinifera L. Biol. Control 120, 26–35. doi: 10.1016/j.biocontrol.2017.02.007
Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F., and Xu, Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucl. Acids Res. 40, W445–W451. doi: 10.1093/nar/gks479
Keywords: mycoparasitism, CAZy, carbohydrate active enzymes, cell wall degrading enzymes, biological control, comparative genomics, oomycete genomics
Citation: Liang D, Andersen CB, Vetukuri RR, Dou D and Grenville-Briggs LJ (2020) Horizontal Gene Transfer and Tandem Duplication Shape the Unique CAZyme Complement of the Mycoparasitic Oomycetes Pythium oligandrum and Pythium periplocum. Front. Microbiol. 11:581698. doi: 10.3389/fmicb.2020.581698
Received: 09 July 2020; Accepted: 28 September 2020;
Published: 29 October 2020.
Edited by:
James Hane, Curtin University, AustraliaReviewed by:
M. Carmen Limon, Seville University, SpainCopyright © 2020 Liang, Andersen, Vetukuri, Dou and Grenville-Briggs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daolong Dou, ZGRvdUBuamF1LmVkdS5jbg==; Laura J. Grenville-Briggs, bGF1cmEuZ3JlbnZpbGxlLmJyaWdnc0BzbHUuc2U=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.