- 1Ministry of Agriculture Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, Institute of Insect Sciences, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 2Entomology Section, Central Cotton Research Institute, Sakrand, Pakistan
- 3Department of Entomology, Faculty of Veterinary and Animal Husbandry, Somali National University, Mogadishu, Somalia
Termitomyces species are wild edible mushrooms that possess high nutritional value and a wide range of medicinal properties. However, the cultivation of these mushrooms is very difficult because of their symbiotic association with termites. In this study, we aimed to examine the differences in physicochemical indices and microbial communities between combs with Termitomyces basidiomes (CF) and combs without Termitomyces basidiomes (CNF). High-performance liquid chromatography (HPLC), inductively coupled plasma optical emission spectrometry (ICP-OES), gas chromatography equipped with a flame ionization detector (GC-FID), some commercial kits, high-throughput sequencing of the 16s RNA, and internal transcribed spacer (ITS) were used. Humidity, pH, and elements, i.e., Al, Ba, Fe, Mn, Ni, S, Ca, and Mg were higher while amino acids particularly alanine, tyrosine, and isoleucine were lower in CF as compared to CNF. The average contents of fatty acids were not significantly different between the two comb categories. The bacterial genera Alistipes, Burkholderia, Sediminibacterium, and Thermus were dominant in all combs. Brevibacterium, Brevundimonas, and Sediminibacterium were significantly more abundant in CF. Basidiomycota and Ascomycota were also identified in combs. Termitomyces clypeatus, Termitomyces sp. Group3, and Termitomyces sp. were the most dominant species in combs. However, any single Termitomyces species was abundantly present in an individual comb.
Introduction
Termitomyces is a rare genus among wild edible mushrooms, most abundantly present in (sub)tropical areas of Southeast Asia and Africa (Aanen et al., 2009; Siddiquee et al., 2015). Termitomyces mushrooms are widely consumed because of their taste, flavor, and medicinal benefits (Zhao et al., 2016; Hsieh and Ju, 2018). A wide spectrum of degrading enzymes is produced by Termitomyces, which could be used in industrial manufacturing such as papermaking (Aanen and Eggleton, 2005; da Costa et al., 2018; Banerjee et al., 2019).
Termitomyces appears only on the nests of fungus-growing termites (Macrotermitine) during rainy seasons. The mutualistic association between Termitomyces and termites started 24–34 million years ago in rainforests of sub-Saharan Africa (Aanen and Eggleton, 2005; Mueller et al., 2005; da Costa et al., 2018). Approximately 330 species of fungus-growing termites have been reported so far, which domesticate 30 described species of Termitomyces (Kirk et al., 2008; Makonde et al., 2013). Individual termite colonies have been found to cultivate only single species of Termitomyces (Makonde et al., 2013). In this symbiotic association, old termite workers forage plant materials from the outside environment and store them inside the colony (Poulsen et al., 2014). Young termite workers ingest this stored plant biomass along with Termitomyces nodules containing abundant asexual spores and excrete lignin-rich excrements to build fresh fungus comb (da Costa et al., 2019a). Termitomyces grow on the fresh comb and decompose it efficiently. After 45 days of growth, the comb is mature. Well-decomposed mature combs are consumed by the older workers (Li et al., 2017). Mature combs produce basidiomes during rainy seasons. The evolution of this intricate relationship makes it extremely difficult to cultivate Termitomyces mushrooms artificially, although their mycelium can grow on several nutritive media and substrates (Tibuhwa, 2012; Ono et al., 2016; da Costa et al., 2019b).
Termitomyces rely on some nutrients from combs to fulfill most parts of their life cycle. Some studies investigated the composition of physicochemical properties such as pH, moisture, and soluble protein in combs and termite guts (Gomathi et al., 2009; Zeleke et al., 2013). Amino acids are important nutritional sources for most fungi. They play central roles both as building blocks of proteins and as intermediates in metabolism (Wu, 2009). The fungus comb nodules are enriched with amino acids (Chiu et al., 2019). Bacterial communities in combs also play significant roles. In recent years, some researchers identified various bacteria and fungi in combs (Otani et al., 2014; Li et al., 2016; Liang et al., 2020). Some strains isolated from combs can stimulate the hyphal growth of Termitomyces (Sawhasan et al., 2012).
In this study, we compared physicochemical parameters including contents in amino acids, elements, sugars, fatty acids, and other basic indices between combs without Termitomyces basidiomes (CNF) and combs with Termitomyces basidiomes (CF) of the fungus-growing termite, Odontotermes formosanus. Microbial communities (bacteria and fungi) were also identified.
Materials and Methods
Collection and Preparation of Fungus Combs
Six healthy fungal combs of O. formosanus were collected from a forested area of Hangzhou City, Zhejiang Province, P. R. China (N 30°18′, E 120°5′). There were three CNF and three CF. Samples were wrapped in plastic bags separately and transported to the laboratory within 6 h of excavation. For each sample, the pH and moisture contents were estimated immediately, and the rest of the combs were stored at −80°C for subsequent experiments.
Basic Chemical Compositions of Fungus Combs
Moisture content and pH were tested as described by Zeleke et al. (2013) with some modifications. Approximately, 1 g of each sample was ground and dissolved with 5 ml distilled water, centrifuged at 8,000 × g for 10 min; the supernatants were used for the determination of pH. Moisture content was estimated by drying the comb to a constant weight. Contents of total soluble protein, total reducing sugar, and polysaccharides including lignin, hemicellulose, and cellulose were determined by using commercial kits (Comin Biotechnology Co., Ltd., Suzhou, China).
Element Content Determination
Inductively coupled plasma optical emission spectrometry (ICP-OES) was used to quantify elements. The procedure for determining the element contents from the samples was similar to that of Toyama-Kato et al. (2003) with minor modifications. Around 0.1 g comb was dried at 40°C, digested, heated to dryness, dissolved with 20 ml HNO3 (1 M), and filtered using a membrane filter (pore size 0.22 μm). Then, 1 ml filtered solution was diluted 10 times with 1 M HNO3 for the detection of calcium and magnesium separately. The operating conditions were shown in Supplementary Table S1.
Determination of Amino Acids
The extraction procedure: Approximately 0.05 g dried comb with 5 ml HCl (6 mol/L, containing 0.1% phenol) was ground and transferred to an Eppendorf tube. After hydrolysis for 20 h at 100°C, 1 ml hydrolysate was collected in a petri dish and cooled down to room temperature and blown to almost dryness using a nitrogen blowing instrument (NBI, Organomation, United States). Then, 1 ml HCl (0.1 mol/L) was added to redissolve, and the turbid liquid was filtered.
Samples and standards derivatization: 200 μl comb extract from the extraction procedure mentioned above and 200 μl standard solution of 17 amino acids were transferred to Eppendorf tubes with 20 μl norleucine as an internal standard. Then, 200 μl triethylamine acetonitrile and 100 μl phenyl isothiocyanate–acetonitrile were added. After mixing, tubes were stable for 1 h at 25°C. Finally, 400 μl n-hexane was added in each tube, left for 10 min after shaking, and then the lower solutions were collected and diluted five times, then filtered (pore size 0.45 μm). Finally, a 10-μl sample was injected for each analysis.
The amino acid analysis was carried out with a high-performance liquid chromatography (HPLC) system (Rigol L3000, Beijing RIGOL Technology Co., Ltd., China) equipped with a UV detector (HPLC-UV), and the column was Kromasil C18 (250 mm × 4.6 mm, 5 μm). The chromatographic conditions were: a flow rate at 1 ml/min, mobile phase A was sodium acetate–acetonitrile solution and phase B was 80% acetonitrile solution. The gradient was: 100% A for 15 min, 90% A with 10% B for 10 min, 70% A with 30% B for 8 min, 55% A with 45% B for 1 s, 100% B for 5 min, and 100% A for the last 7 min.
Volatile Fatty Acid Analysis
Different concentrations of standard solutions (10, 50, 100, 200, and 500 μg/ml) were prepared for formic acid, acetic acid, propionic acid, n-butyric acid, n-valeric acid, n-hexylic acid, and n-heptanoic acid. About 100 μg/ml 2-ethylbutyric acid solution was prepared as an internal reference solution.
Approximately 1 g sample was weighed and homogenized with 5 ml ethyl alcohol for 3 min, then kept at room temperature and shaken occasionally for 10 min. The suspension was transferred into an Eppendorf tube and centrifuged for 20 min at 6,000 rpm. The internal standard (2-ethylbutyric acid solution) was added according to Zhao et al. (2006), and the supernatant was injected into a gas chromatography system equipped with a flame ionization detector (GC-FID) for analysis.
The capillary column (30 m × 320 μm × 1.80 μm) was used, and nitrogen was applied at a flow rate of 15.0 ml/min as the carrier gas. The initial oven temperature was 100°C, maintained for 5 min, then raised to 200°C at 20°C/min and held for 5 min. Hydrogen, air, and nitrogen were the makeup gases at the flow rates of 35, 350, and 30 ml/min, respectively. The volume of the injected sample was 1 μl, and the run time for each solution was 15 min.
Microbial Community Characterizations in Fungus Combs
DNA was extracted from approximately 0.5 g of each comb using the DNeasy PowerSoil kit (Qiagen, GER) for bacteria and omega M5635-02 kit (Omega, United States) for fungi separately, according to manufacturer’s instructions. The primers 341F (5′-CCTACGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) were selected to amplify the V3-V4 variable regions of the bacterial 16S r RNA gene (Otani et al., 2014; Liang et al., 2020). ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify the fungal ITS region (Makonde et al., 2017). The amplification reaction for bacteria was performed in 25 μl final mixture [14.75 μl dd H2O, 2 μl dNTP (2.5 mM), 5 μl 5 × Reaction buffer, 1 μl of each primer (10 μM), 1 μl DNA template, and 0.25 μl Fast pfu DNA Polymerase (NEB, United States)], and the conditions for PCR were 98°C for 5 min, followed by 25 cycles of 98°C for 30 s, 52°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 5 min. The fungal amplification reaction was the same as that for bacteria, and the conditions for PCR were 98°C for 5 min, followed by 28 cycles of 98°C for 30 s, 52°C for 45 s, and 72°C for 45 s, with a final extension at 72°C for 5 min. The final target PCR products were visualized with agarose gel electrophoresis, then extracted and purified from the gel. DNA concentrations were quantified using Quant-iT PicoGreen ds DNA Assay Kit (Invitrogen, CA) and Microplate reader (BioTek, United States). Finally, samples were sequenced commercially (Personalbio Biotech, Shanghai, China) on Illumina MiSeq platform (paired-end) for bacteria and Pacbio Sequel platform (single-molecule real-time) for fungus.
Statistical Analysis
Differences in physicochemical parameters were tested with one-way ANOVA analysis and Student’s t-test using SPSS, v.20.0 at alpha = 0.05. Pyrotag reads were denoised using the DADA2 method according to Benjamini and Hochberg (1995). Raw sequences were quality-filtered according to Koljalg et al. (2013) using the QIIME Release 8.0 software. Silva (Release132)1 with 16S r RNA gene sequences of cockroach (Quast et al., 2013) and UNITE (Release 8.0)2 (Koljalg et al., 2013) were used as reference databases for bacteria and fungi, respectively. Packages (stat, ape, Venn diagram, pheatmap, and ggplot2) of R software3 were used to map and compare the relative abundance of bacteria and fungi at different levels. QIIME2 (2019.4) was used to calculate every index of alpha diversity including Chao1 (Chao, 1984), Observed species, Shannon (Shannon, 1948), Simpson (Simpson, 1949), Pielou’s evenness (Pielou, 1966), and Good’s coverage (Good, 1958). Kruskal–Wallis test (H-test) and Dunn test were applied to test for differences in alpha diversity.
Results
Basic Parameters of Combs
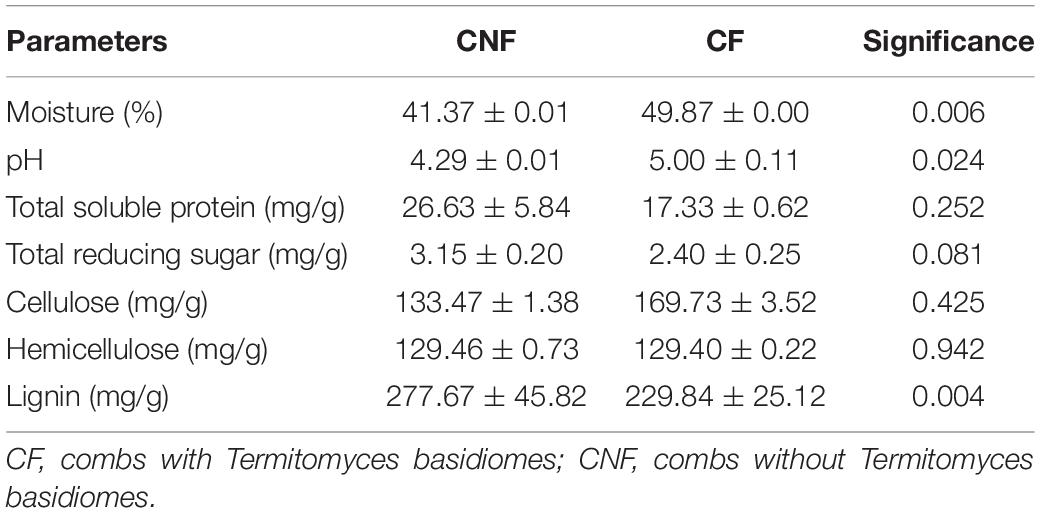
Lignin was significantly (P < 0.01) lower in CF (229.84 ± 25.12) as compared to CNF (277.67 ± 45.82), whereas pH (P < 0.05) and moisture (P < 0.01) were relatively higher in CF as compared to CNF. The contents in total soluble protein, total reducing sugar, hemicellulose, and cellulose were not significantly different between CF and CNF (P > 0.05; Table 1).
Concentration of Elements in Combs
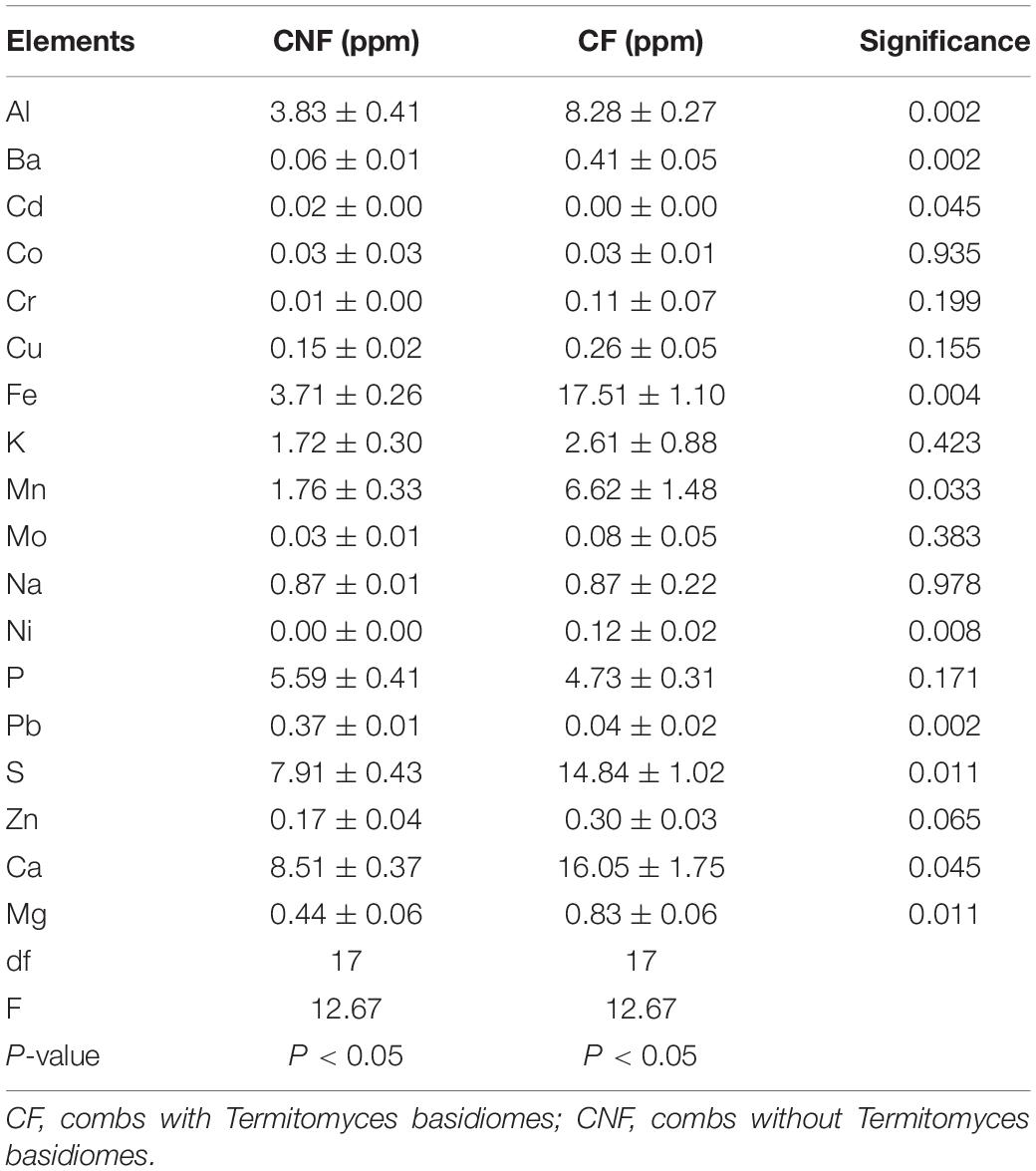
Eighteen elements in each sample were determined by ICP-OES. The average concentration of each element was different between CF and CNF. All elements (except Cd, Co, Na, P, and Pb) were higher in CF as compared to CNF (Table 2). Cd and Pb were lower in CF than in CNF.
Amino Acid Analysis
We compared 17 amino acids between CF and CNF by HPLC-UV (Supplementary Figure S1 and Supplementary Table S2). The amino acid contents were higher in CNF as compared to CF (Figure 1). Among all amino acids, aspartic acid content was highest in both comb types, i.e., CNF (2,107.24 μg/g ± 169.44) and CF (1,910.73 μg/g ± 109.67), followed by glutamic acid (1,864.38 μg/g ± 104.48 and 1,708.38 μg/g ± 175.92, respectively), while average cysteine was lowest in both combs (5.70 μg/g ± 1.93 and 6.05 μg/g ± 1.63, respectively). Amino acid profiles of combs were changed a lot after basidiome formation. Serine and cysteine contents were more abundant in CF (1,030.54 μg/g ± 65.32 and 6.05 μg/g ± 1.63, respectively) than those in CNF (1,004.41 μg/g ± 85.94 and 5.70 μg/g ± 1.93, respectively), while other amino acids were lower in CF. Surprisingly, the relative abundance of alanine, tyrosine, and isoleucine significantly decreased with the appearance of basidiomes (Student’s t-test, P < 0.05).
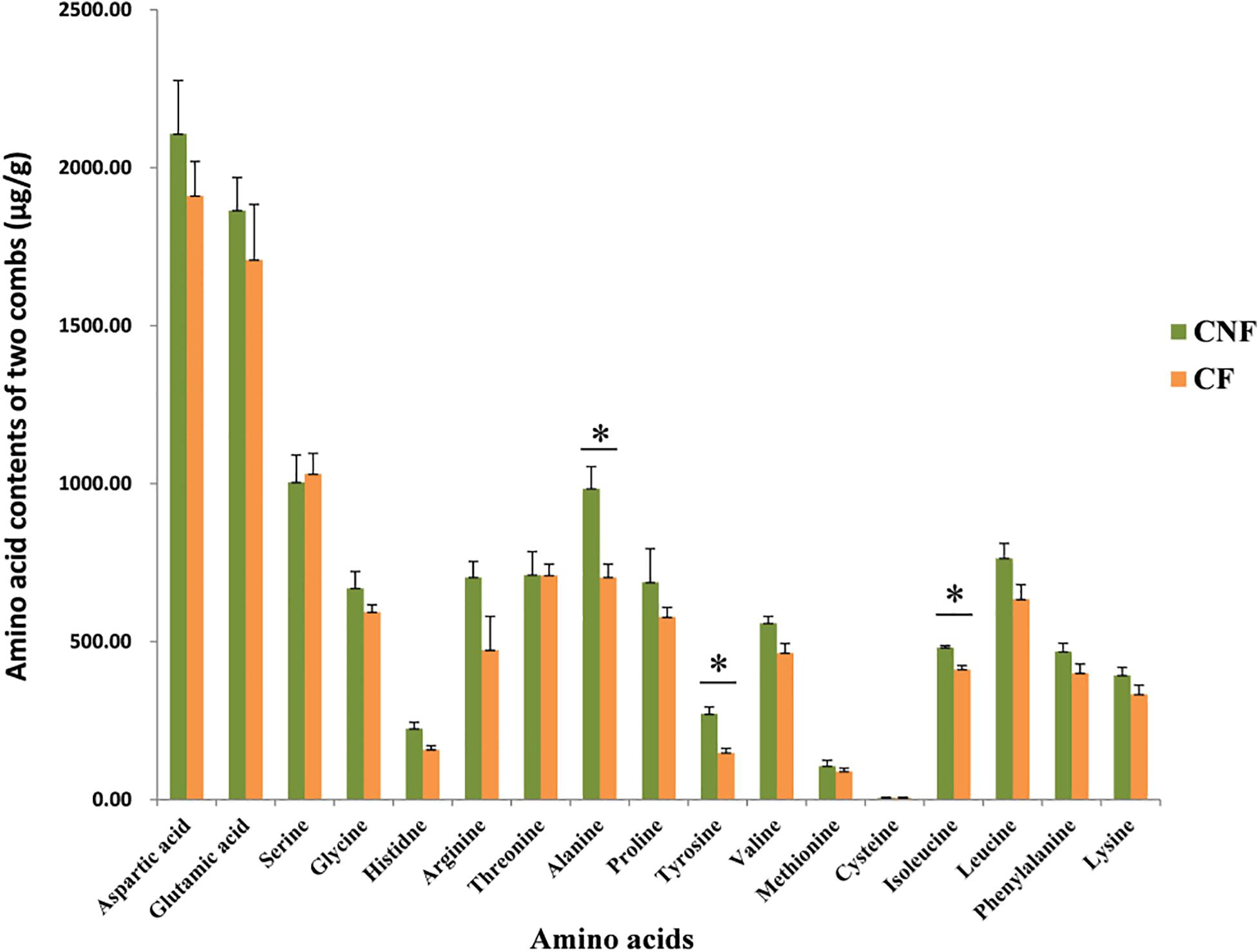
Figure 1. Comparisons of amino acids between combs with Termitomyces basidiomes and combs without Termitomyces basidiomes. ∗ indicates high significance (P < 0.05).
Fatty Acid Analysis
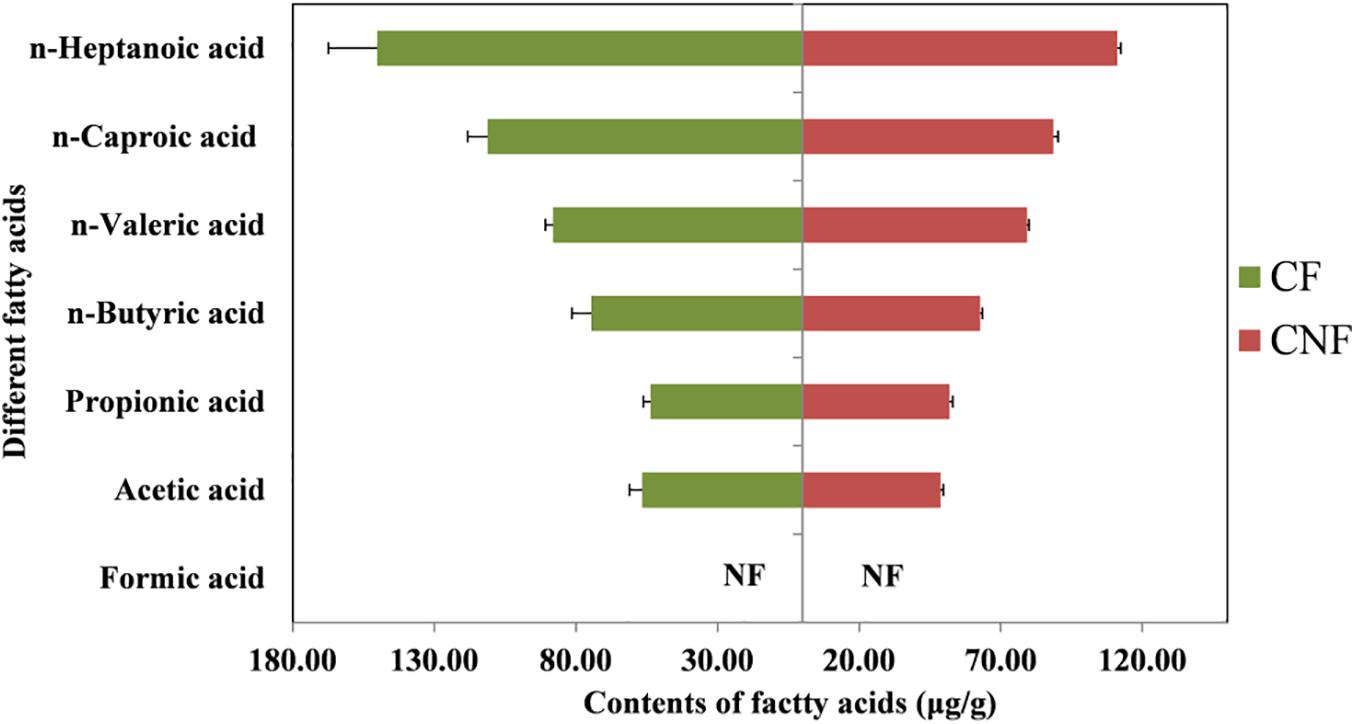
The contents of six fatty acids were measured in CNF and CF (Supplementary Tables S3, S4). The average contents of fatty acids were not significantly different between comb categories. The average contents of caproic acid in both CNF (64.54 μg/g ± 1.20) and CF (94.59 μg/g ± 11.39) were the highest followed by butyric acid (57.80 μg/g ± 0.92 and 74.78 μg/g ± 7.93, respectively) and acetic acid (47.89 μg/g ± 1.21 and 53.86 μg/g ± 4.11, respectively). Whereas the average contents of propionic acid were lowest in CNF (1.09 μg/g ± 0.01) and CF (1.00 μg/g ± 0.04) followed by valeric acid (1.37 μg/g ± 0.02 and 1.20 μg/g ± 0.12, respectively) and heptanoic acid (1.73 μg/g ± 0.05 and 1.63 μg/g ± 0.22, respectively). We did not find formic acid in combs (Figure 2 and Supplementary Table S4).
Bacterial Community Analysis
Between 21,566 and 24,198 high-quality sequences were obtained from each comb. The Good’s coverage for every comb was more than 99.6% (Supplementary Table S5). The microbial diversity indices (Shannon and Simpson), estimated community richness (Chao1 and Observed species), and evenness (Pielou’s evenness) were not statistically different between CF and CNF (Student’s t-test; P > 0.05; Figure 3A). The unweighted pair-group method with arithmetic means (UPGMA) clustering analysis grouped all combs into two distinct groups, i.e., CF and CNF (Figure 3B). The Venn diagram showed that CNF and CF occupied 532 (41.30%) and 441 (31.91%) operational taxonomic units (OTUs), respectively. The OTU 345 (26.79%) was common between both comb categories (Figure 3C).
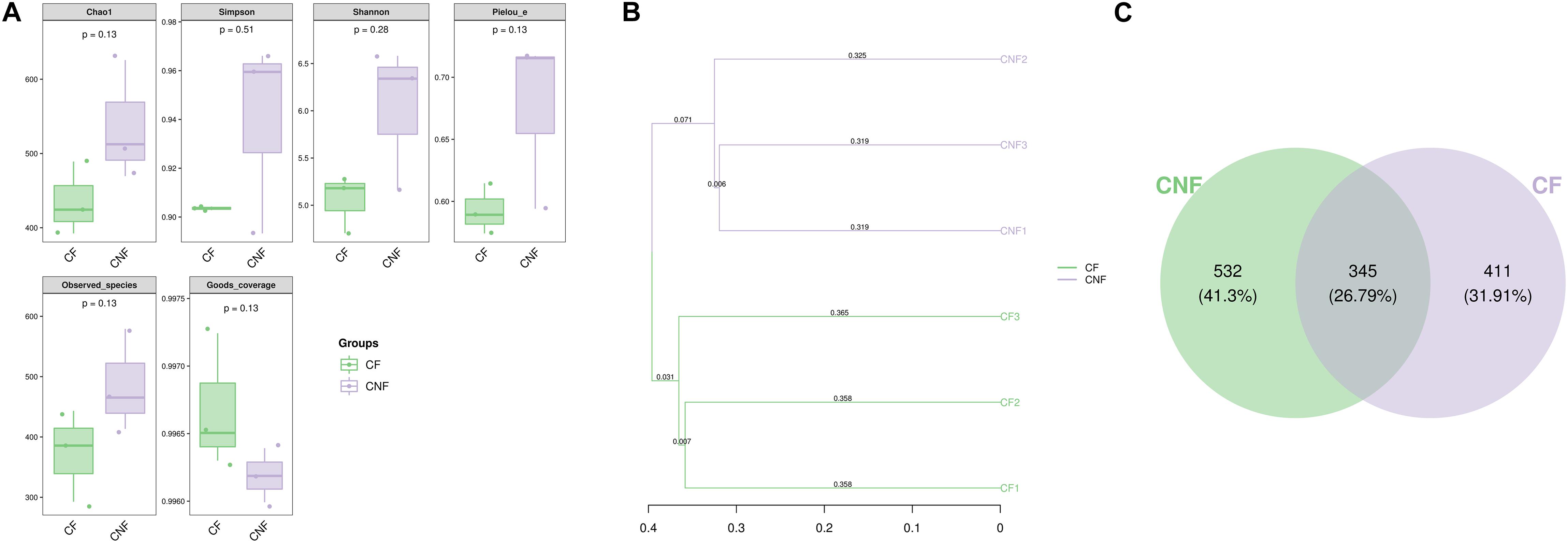
Figure 3. (A) The box plots demonstrated significance among six combs in different indicators, including numbers of sequences, coverage, richness, diversity, and evenness. (B) Clustering tree using unweighted pair-group method with arithmetic means (UPGMA) revealed the bacterial similarities among combs. (C) Venn diagram of OTU between combs without Termitomyces basidiomes (CNF) and combs with Termitomyces basidiomes (CF).
The abundance of various bacterial groups at different taxonomic levels (phylum, family, and genus) was investigated. A total of 20 bacterial phyla were found and compared (Figure 4A) among all combs. Proteobacteria (23.97–68.52%), Bacteroidetes (18.85–43.83%), and Firmicutes (2.45–16.44%) were the dominant identified OTUs among all. With the growth of basidiomes, Actinobacteria and Cyanobacteria increased significantly, while Planctomycetes and Chlorobi decreased (Supplementary Table S6).
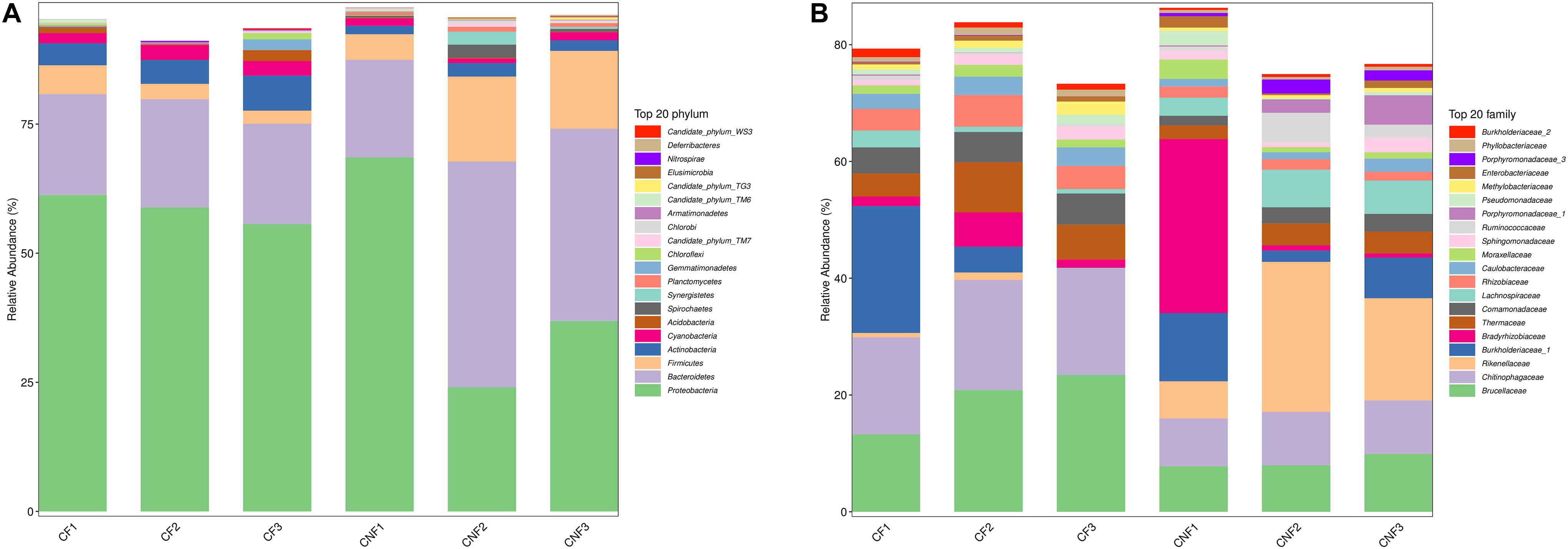
Figure 4. The bacterial community structure in combs with Termitomyces basidiomes (CF) and combs without Termitomyces basidiomes (CNF) combs. (A) The relative abundance of bacteria at the phyla level. (B) The bacterial composition at the family level. CF1, CF2, and CF3 represent three combs with basidiomes, whereas CNF1, CNF2, and CNF3 represent three combs without basidiomes.
At the family level, Brucellaceae (7.82–23.56%), Chitinophagaceae (8.20–18.85%), Rikenellaceae (0.02–25.84%), Burkholderiaceae_1 (0.06–21.96%), and Bradyrhizobiaceae (0.71–29.78%) were dominant in all combs (Figure 4B). Furthermore, Chitinophagaceae (Bacteroidetes), Comamonadaceae (Proteobacteria), Rhizobiaceae (Proteobacteria), Caulobacteraceae (Proteobacteria), and Phyllobacteriaceae (Proteobacteria) were significantly high in CF (Student’s t-test; P < 0.05) (Supplementary Table S7).
At the genus level, Sediminibacterium (8.18–18.80%), Alistipes_II (0.02–18.99%), Burkholderia_1 (0.01–21.40%), and Thermus (2.35–8.53%) were the most abundant bacterial genera in all combs. Sediminibacterium (Chitinophagaceae, Bacteroidetes), Brevundimonas (Caulobacteraceae, Proteobacteria), and Brevibacterium (Brevibacteriaceae, Actinobacteria) were significantly more abundant in combs with basidiomes, while the members of Alistipes IV (Rikenellaceae, Bacteroidetes) were significantly less abundant (Student’s t-test; P < 0.05) (Figure 5 and Supplementary Table S8).
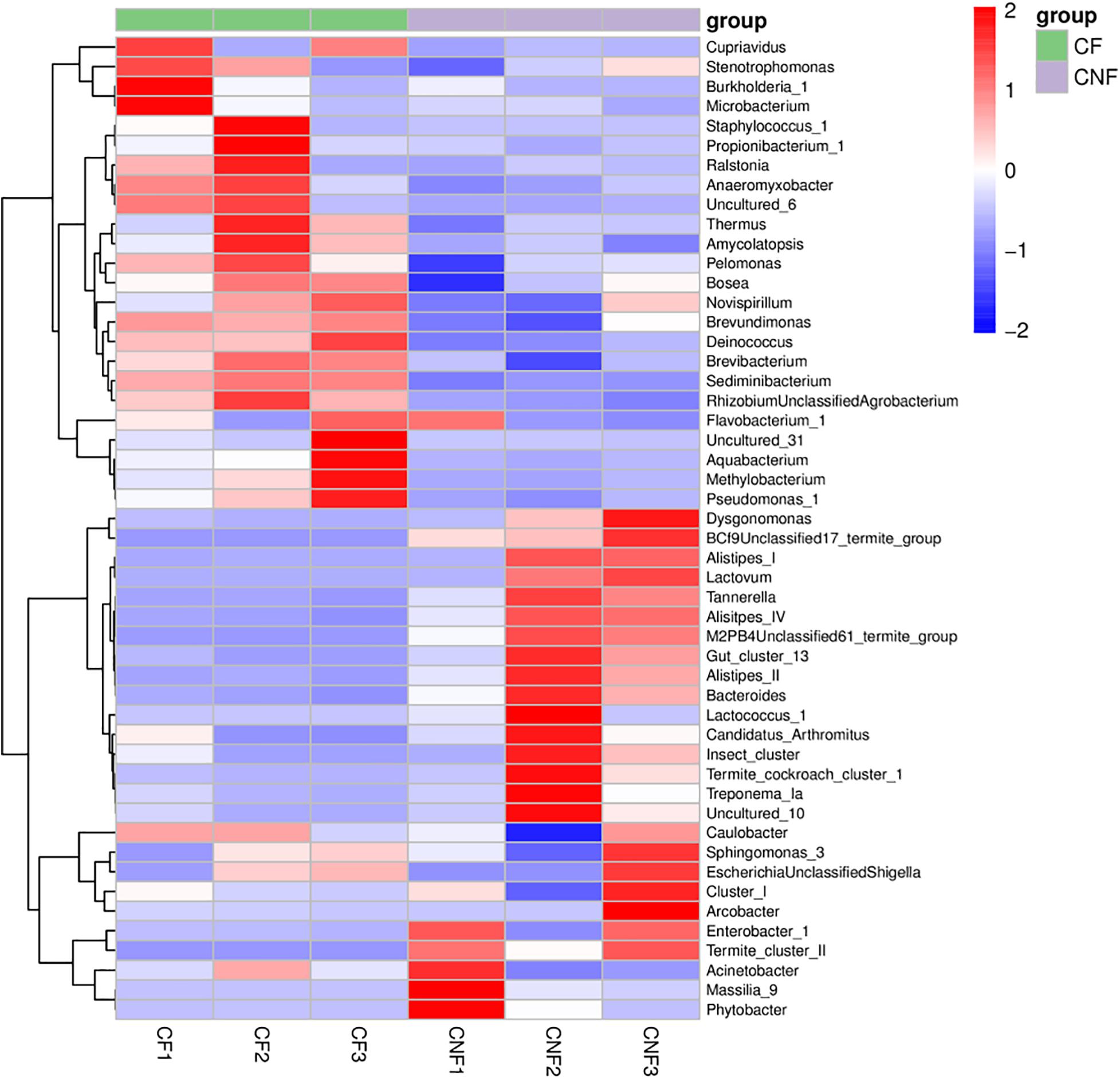
Figure 5. The heat map illustrated the relative abundances of bacteria among combs at the genus level.
Fungal Community Analysis
Between 4,008 and 5,854 high-quality sequences were obtained from every comb. The coverage for every comb was closed to 100% (Supplementary Table S9). Alpha diversity indices (Shannon, Simpson, Chao1, Observed species, and Pielou’s evenness) were not significantly different between comb types (Student’s t-test; P > 0.05).
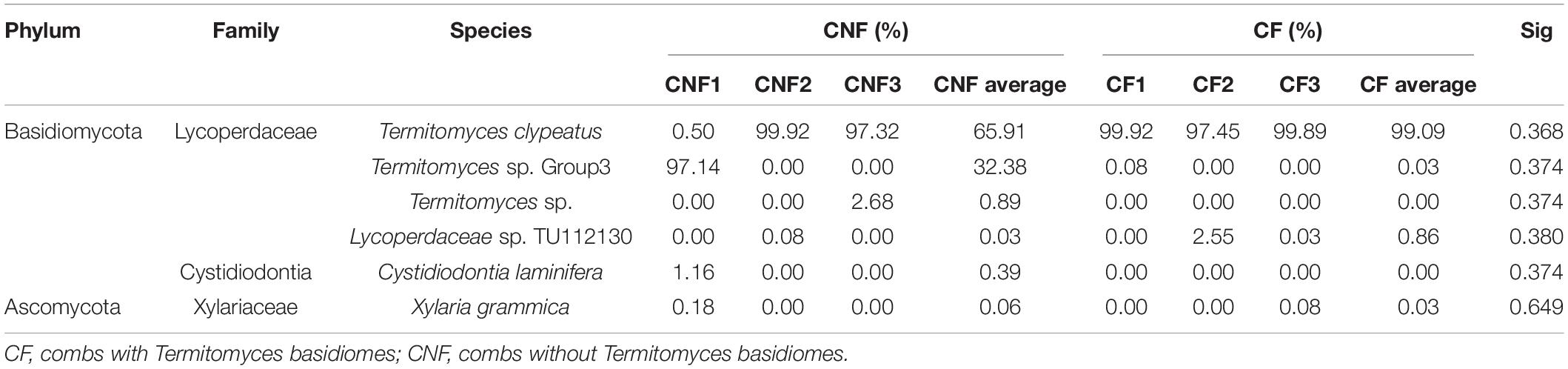
Two phyla, i.e., Basidiomycota and Ascomycota, were found in both types of combs. Basidiomycota was the most prevalent phylum. At the family level, Lycoperdaceae (the family to which Termitomyces belongs), Cystostereaceae, and Xylariaceae were identified. Lycoperdaceae was the most abundant in all combs. The species including Xylaria grammica (0.00–0.18%), Cystidiodontia laminifera (0.00–1.16%), Lycoperdaceae sp. TU112130 (0.00–2.55%), Termitomyces clypeatus (0.50–99.92%), Termitomyces sp. (0.00–2.68%), Termitomyces sp. Group3 (0.00–97.14%), and fungal sp. (0.00–1.02%) were identified. Individual combs were dominated by a single Termitomyces species (Table 3).
Discussion
The present study showed differences in physicochemical properties and microbial communities between CF and CNF. Most fungi grow well at acidic pH and low humidity (Hesse, 1955; Gomathi et al., 2009), and we indeed found low moisture and pH in all combs (Table 1). However, we disagree with the previous study reporting that moisture content decreased after basidiome formation (Sidde Gowda and Rajagopal, 1990). Less lignin was recorded from combs with basidiomes as compared to combs without basidiomes (Table 1), indicating that Termitomyces degrades lignin during the comb maturation period (Hyodo et al., 2000; Bashir et al., 2015).
Our results showed the variation tendency of 18 elements including 16 metal and two non-metal elements in combs (Table 2). Li et al. (2012) also found metal ions in fungus combs. Element contents were relatively higher in CF as compared to CNF. Mushrooms can absorb large amounts of water and minerals from their substrates (Koutrotsios et al., 2020). The bioaccumulation capacity of mushroom is influenced by the substrate, fungal lifestyle, age of the fruiting body, species, type of element, and environment (Campos et al., 2009; Lee et al., 2009; Koutrotsios et al., 2020). We assume that the high concentrations of elements in combs are associated with lignocellulose degradation and fungal growth. Some studies demonstrated that elements are important for fungal metabolic functions related to enzyme production and lignocellulose degradation (Li et al., 2012; Koutrotsios et al., 2020), but less information is available on the significant roles of elements in fungal growth. Cuero et al. (2003) reported that various metal ions caused some phenotypic or genotypic influences in fungus and could stimulate fungal growth. Moreover, some elements play a crucial role in enzymatic reactions regarding nitrogen fixation (Hoffman et al., 2014). For example, molybdenum, iron, and magnesium are essential for the structure formation of metal clusters in nitrogenase and MgATP energy supply (Hoffman et al., 2014), which may strengthen nitrogen fixation.
Amino acids were higher in CNF as compared to CF. This may be due to the uptake of amino acids in basidiomes from combs. It has been reported that amino acids are transported into a fungal cell from the substrate (Whitaker, 1976). Termitomyces mushrooms are capable of transforming amino acids or synthesizing amino acids from non-protein nitrogen (Botha and Eicker, 1992; Deacon, 2006). In termite–fungus association, the amino acids are accumulated in matured fungus combs and fungal nodules (Botha and Eicker, 1992). Chiu et al. (2019) compared 18 amino acids between fungus combs and fungal nodules of Termitomyces. They allocated 7% amino acids to nodules and 93% to combs. Moreover, it has also been reported that amino acids are important for fungal growth and reproduction (Steinberg, 1950; Yang et al., 2020). However, knowledge on the role of amino acids on fungal growth is less satisfactory.
All fatty acids were found in our experiment except formic acid (Figure 2 and Supplementary Table S4). Some short-chain fatty acids such as acetic acid, propionic acid, and butyric acid were also reported previously both in termite guts and fungus combs (Li et al., 2016). It has been reported that fatty acids are produced by microorganisms living in termite guts during the fermentation of fiber and starch (Odelson and Breznak, 1983; Thorburn et al., 2014; Ciarlo et al., 2016).
The bacterial communities in fungus combs are involved in lignocellulose degradation, nutrient supplementation, nitrogen fixation, and antibiotic production (Liang et al., 2020), but whether these bacteria play significant roles in the appearance of basidiomes from the comb remains to be proven. We found that the major bacterial lineages were similar among all combs, but there were marked changes in their abundance (Figures 4, 5 and Supplementary Tables S6–S8). Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, and Proteobacteria were plentifully found in all combs (Figure 3A and Supplementary Table S6). We are consistent with the previous studies on fungus combs (Otani et al., 2014; Liang et al., 2020). It has been reported that some members of Bacteroidetes and Firmicutes can play important roles in the metabolism of carbohydrates (Poulsen et al., 2014; Delmont et al., 2018). Proteobacteria along with Bacteroidetes and Firmicutes are related with nitrogen input, which may help meet the nutritional needs of Termitomyces (Poulsen et al., 2014; Delmont et al., 2018). The nitrogenase reductase nifH genes have been identified in Proteobacteria (Delmont et al., 2018). Liang et al. (2020) suggested that Proteobacteria in fungus comb assist fungus in atmospheric nitrogen fixation. Our findings also indicated that Cyanobacteria and Actinobacteria were more abundant in CF than in CNF (Figure 3A and Supplementary Table S6). Biologists suggested that many species of Cyanobacteria found in fungus combs can fix N2 (Stewart, 1980; Makonde et al., 2015). It has been described that the C/N ratio can maximize fungal growth performance in vitro (Shik et al., 2016). Actinobacteria have also been isolated from fungus combs previously (Visser et al., 2012; Benndorf et al., 2018; Yin et al., 2019). They are generally known as defensive symbionts because they produce antibiotic compounds to suppress invading pathogens (Visser et al., 2012; Benndorf et al., 2018; Yin et al., 2019). Based on these results, we believed that some bacteria from combs are important to create a suitable growth condition for Termitomyces by maintaining the C/N balance or inhibiting the infectious microbes.
Fungal communities of Basidiomycota and Ascomycota were also identified in combs; however, the members of Termitomyces were dominant in all combs (Table 3). These results were the same as reported previously by Shinzato et al. (2005) that Termitomyces was the only visible fungus on the nests of termites, and the individual nest has been found to contain only single species of Termitomyces within the fungus garden.
Conclusion
The current study investigated the differences in physicochemical factors (pH, moisture, total soluble protein, total reducing sugar, cellulose, hemicellulose, lignin, amino acids, metal ions, and fatty acids) and microbial communities (bacteria and fungi) between CF and CNF. A noticeable increase of humidity, pH, and elements (Al, Ba, Fe, Mn, Ni, S, Ca, and Mg) and a decrease of some amino acids (alanine, tyrosine, and isoleucine) were recorded in CF as compared to CNF. This study also revealed a high level of bacterial diversity in fungal combs. The majority of the bacterial communities might assist Termitomyces in lignocellulose degradation and maintenance of the C/N ratio. The possible role of physicochemical characteristics and bacterial communities on the onset of basidiome development by Termitomyces remains unknown, and further in-depth studies are needed.
Data Availability Statement
The datasets were submitted to the Sequence Read Archive of NCBI (http://www.ncbi.nlm.nih.gov; BioProject PRJNA66 2821).
Author Contributions
GY and JM: conceptualization, project administration, and funding acquisition. GY: methodology. GY and FA: software, writing—original draft preparation, and visualization. GY, QZ, and MG: validation. GY and SL: formal analysis and data curation. GY and MG: investigation. GY and QZ: resources. FA, HG, and JM: writing—review and editing. JM: supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31770686) and a project supported by the Scientific Research Fund of Zhejiang Provincial Education Department (Y201941400).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yaer Zhu for technical assistance in GC-FID analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.581219/full#supplementary-material
Abbreviations
CF, combs with Termitomyces basidiomes; CNF, combs without Termitomyces basidiomes; HPLC, high-performance liquid chromatography; ICP-OES, inductively coupled plasma optical emission spectrometry; GC-FID, gas chromatography equipped with a flame ionization detector.
Footnotes
References
Aanen, D. K., de Fine Lichat, H. H., Debets Alfons, J. M., Kerstes Niels, A. G., Hoekstra, R. F., and Boomsma, J. J. (2009). High symbiont relatedness stabilizes mutualistic cooperation in fungus growing termites. Science 326, 1103–1105. doi: 10.1126/science.1173462
Aanen, D. K., and Eggleton, P. (2005). Fungus-growing termites originated in African rain forest. Curr. Biol. 15, 851–855. doi: 10.1016/j.cub.2005.03.043
Banerjee, S., Roy, A., Madhusudhan, M. S., Bairagya, H. R., and Roy, A. (2019). Structural insights of a cellobiose dehydrogenase enzyme from the basidiomycetes fungus Termitomyces clypeatus. Comput. Biol. Chem. 82, 65–73. doi: 10.1016/j.compbiolchem.2019.05.013
Bashir, H., Gangwar, R., and Mishra, S. (2015). Differential production of lignocellulolytic enzymes by a white rot fungus Termitomyces sp. OE147 on cellulose and lactose. Biochim. Biophys. Acta 1854, 1290–1299. doi: 10.1016/j.bbapap.2015.07.005
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Benndorf, R., Guo, H., Sommerwerk, E., Weigel, C., Garcia-Altares, M., Martin, K., et al. (2018). Natural products from actinobacteria associated with fungus-growing termites. Antibiotics 7:83. doi: 10.3390/antibiotics7030083
Botha, W. J., and Eicker, A. (1992). Nutritional value of Termitomyces mycelial protein and growth of mycelium on natural substrates. Mycol. Res. 96, 350–354. doi: 10.1016/S0953-7562(09)80949-0
Campos, J. A., Tejera, N. A., and Sanchez, C. J. (2009). Substrate role in the accumulation of heavy metals in sporocarps of wild fungi. Biometals 22, 835–841. doi: 10.1007/s10534-009-9230-7
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Statist. 11, 265–270.
Chiu, C. I., Ou, J. H., Chen, C.-Y., and Li, H.-F. (2019). Fungal nutrition allocation enhances mutualism with fungus-growing termite. Fungal Ecol. 41, 92–100. doi: 10.1016/j.funeco.2019.04.001
Ciarlo, E., Heinonen, T., Herderschee, J., Fenwick, C., Mombelli, M., Roy, D. L., et al. (2016). Impact of the microbial derived short chain fatty acid propionate on host susceptibility to bacterial and fungal infections in vivo. Sci. Rep. 6:37944. doi: 10.1038/srep37944
Cuero, R., Ouellet, T., Yu, J., and Mogongwa, N. (2003). Metal ion enhancement of fungal growth, gene expression and aflatoxin synthesis in Aspergillus flavus: RT-PCR characterization. J. Appl. Microbiol. 94, 953–961. doi: 10.1046/j.1365-2672.2003.01870.x
da Costa, R. R., Hu, H., Pilgaard, B., Vreeburg Sabine, M. N., Schuckel, J., Pedersen Kristine, S. K., et al. (2018). Enzyme activities at different stages of plant biomass decomposition in three species of fungus-growing termites. Appl. Environ. Microbiol. 84:e01815-17. doi: 10.1128/AEM.01815-17
da Costa, R. R., Hu, H., Li, H., and Poulsen, M. (2019a). Symbiotic plant biomass decomposition in fungus-growing termites. Insects 10:87. doi: 10.3390/insects10040087
da Costa, R. R., Vreeburg Sabine, M. E., Shik, J. Z., Aanen, D. K., and Poulsen, M. (2019b). Can interaction specificity in the fungus-farming termite symbiosis be explained by nutritional requirements of the fungal crop? Fungal Ecol. 38, 54–61. doi: 10.1016/j.funeco.2018.08.009
Delmont, T. O., Quince, C., Shaiber, A., Esen, O. C., Lee, S. T., Rappe, M. S., et al. (2018). Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813. doi: 10.1038/s41564-018-0176-9
Gomathi, V., Esakkiammal, M., Thilagavathi, S. S., and Ramalakshmi, A. (2009). Environmental influence on physico-chemical and biological activities of fungus growing termite (Isoptera: Macrotermitinae). Asia J. Bio Sci. 4, 88–92.
Good, I. J. (1958). The population frequency of species and theestimation of the population parameters. Biometrics 40, 237–246. doi: 10.2307/2333344
Hesse, P. R. (1955). A chemical and physical study of the soils of termite mounds in East Africa. J. Ecol. 43, 449–461.
Hoffman, B. M., Lukoyanov, D., Yang, Z. Y., Dean, D. R., and Seefeldt, L. C. (2014). Mechanism of nitrogen fixation by nitrogenase: the next stage. Chem. Rev. 114, 4041–4062. doi: 10.1021/cr400641x
Hsieh, H. M., and Ju, Y.-M. (2018). Medicinal components in Termitomyces mushrooms. Appl. Microbiol. Biotechnol. 102, 4987–4994. doi: 10.1007/s00253-018-8991-8
Hyodo, F., Inoue, T., Azuma, J. I., Tayasu, I., and Abe, T. (2000). Role of the mutualistic fungus in lignin degradation in the fungus-growing termite Macrotermes gilvus (Isoptera; Macrotermitinae). Soil Biol. Biochem. 32, 653–658. doi: 10.1016/S0038-0717(99)00192-3
Kirk, P. M., Cannon, P. F., Minter, D. W., and Stalpers, J. A. (2008). Dictionary of the Fungi, 10th Edn. Wallingford: CABI.
Koljalg, U., Nilsson, R., Abarenkov, K., Tedersoo, L., Taylor Andy, F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Koutrotsios, G., Danezis, G., Georgiou, C., and Zervakis, G. I. (2020). Elemental content in Pleurotus ostreatus and Cyclocybe cylindracea mushrooms: correlations with concentrations in cultivation substrates and effects on the production process. Molecules 25:2179. doi: 10.3390/molecules25092179
Lee, C. Y., Park, J. E., Kim, B. B., Kim, S. M., and Ro, H. S. (2009). Determination of mineral components in the cultivation substrates of edible mushrooms and their uptake into fruiting bodies. Mycobiology 37, 109–113. doi: 10.4489/MYCO.2009.37.2.109
Li, H.-J., Dietrich, C., Zhu, N., Mikaelyan, A., Ma, B., Pi, R., et al. (2016). Age polyethism drives community structure of the bacterial gut microbiota in the fungus-cultivating termite Odontotermes formosanus. Environ. Microbiol. 18, 1440–1451. doi: 10.1111/1462-2920.13046
Li, H.-J., Sun, J.-Z., Zhao, J.-M., Deng, T.-F., Lu, J.-R., Dong, Y., et al. (2012). Physicochemical conditions and metal ion profiles in the gut of the fungus-growing termite Odontotermes formosanus. J. Insect Physiol. 58, 1368–1375. doi: 10.1016/j.jinsphys.2012.07.012
Li, H.-J., Yelle, D. J., Li, C., Yang, M.-Y., Ke, J., Zhang, R.-J., et al. (2017). Lignocellulose pretreatment in a fungus-cultivating termite. PNAS 114, 4709–4714.
Liang, S.-Y., Wang, C.-P., Ahmad, F., Yin, X.-J., Hu, Y., and Mo, J.-C. (2020). Exploring the effect of plant substrates on bacterial community structure in termite fungus-combs. PLoS One 15:e0232329.
Makonde, H. M., Boga, H. I., Osiemo, Z., Mwirichia, R., Stielow, J. B., Goker, M., et al. (2013). Diversity of Termitomyces associated with fungus-farming termites assessed by cultural and culture-independent methods. PLoS One 8:e56464. doi: 10.1371/journal.pone.0056464
Makonde, H. M., Mwirichia, R., Muwawa, E. M., Klenk, H. P., and Boga, H. I. (2017). Fungal diversity and community structure in gut, mound and surrounding soil of fungus-cultivating termites. Afr. J. Microbiol. Res. 11, 504–515. doi: 10.5897/AJMR2017.8484
Makonde, H. M., Mwirichia, R., Osiemo, Z., Boga, H. I., and Klenk, H. P. (2015). 454 Pyrosequencing-based assessment of bacterial diversity and community structure in termite guts, mounds and surrounding soils. Springerplus 4:471. doi: 10.1186/s40064-015-1262-6
Mueller, U. G., Gerardo, N., Aanen, D. K., Six, D. L., and Schultz, T. R. (2005). The evolution of agriculture in insects. Annu. Rev. Ecol. Evol. Syst. 36, 563–595. doi: 10.1146/annurev.ecolsys.36.102003.152626
Odelson, D. A., and Breznak, J. A. (1983). Volatile fatty acid production by the hindgut microbiota of xylophagous termites. Appl. Environ. Microbiol. 45, 1602–1613.
Ono, K., Hata, T., Yoshimura, T., and Kinjo, K. (2016). Wood decaying properties of the termite mushroom Termitomyces eurrhizus. J. Wood Sci. 63, 83–94. doi: 10.1007/s10086-016-1588-x
Otani, S., Mikaelyan, A., Nobre, T., Hansen, L. H., Kone, N. A., Sorensen, S. J., et al. (2014). Identifying the core microbial community in the gut of fungus-growing termites. Mol. Ecol. 23, 4631–4644. doi: 10.1111/mec.12874
Pielou, E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Poulsen, M., Hu, H., Li, C., Chen, Z., Xu, L., Otani, S., et al. (2014). Complementary symbiont contributions to plant decomposition in a fungus-farming termite. PNAS 111, 14500–14505. doi: 10.5524/100055
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Sawhasan, P., Worapong, J., Flegel, T. W., and Vinijsanun, T. (2012). Fungal partnerships stimulate growth of Termitomyces clypeatus stalk mycelium in vitro. World J. Microbiol. Biotechnol. 28, 2311–2318. doi: 10.1007/s11274-012-1038-x
Shannon, C. E. (1948). A mathematical theory of communication. Bell System Tech. J. 27, 379–423, 623–656. doi: 10.1002/j.1538-7305.1948.tb00917.x
Shik, J. Z., Gormez, E., Kooij, P. W., Santos, J. C., Wcislo, W. T., and Boomsma, J. J. (2016). Nutrition mediates the expression of cultivar-farmer conflict in a fungus-growing ant. PNAS 113, 10121–10127.
Shinzato, N., Muramatsu, M., Watanabe, Y., and Matsui, T. (2005). Termite-regulated fungal monoculture in fungus combs of a macrotermitine termite Odontotermes formosanus. Zool. Sci. 22, 917–922. doi: 10.2108/zsj.22.917
Sidde Gowda, D. K., and Rajagopal, D. (1990). Association of Termitomyces spp. with fungus growing termites. Proc. Indian Acad. Sci. 99, 311–315. doi: 10.1007/BF03186400
Siddiquee, S., Rovina, K., Naher, L., Rodrigues, K. F., and Uzzaman, A. Z. (2015). Phylogenetic relationships of Termitomyces aurantiacus inferred from internal transcribed spacers DNA sequences. Adv. Biosci. Biotechnol. 06, 358–367. doi: 10.4236/abb.2015.65035
Steinberg, R. A. (1950). Growth of fungi in synthetic nutrient solutions. Bot. Rev. 16, 208–228. doi: 10.1007/bf02878505
Stewart, W. D. P. (1980). Some aspects of structure and function in N2-fixing cyanobacteria. Ann. Rev. Microbiol. 34, 497–536. doi: 10.1146/annurev.mi.34.100180.002433
Thorburn, A. N., Macia, L., and Mackay, C. R. (2014). Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 40, 833–842. doi: 10.1016/j.immuni.2014.05.014
Tibuhwa, D. D. (2012). Termitomyces species from Tanzania, their cultural properties and unequalled basidiospores. JBLS 3, 140–159. doi: 10.4172/2157-7463.1000117
Toyama-Kato, Y., Yoshida, K., Fujimori, E., Haraguchi, H., Shimizu, Y., and Kondo, T. (2003). Analysis of metal elements of hydrangea sepals at various growing stages by ICP-AES. Biochem. Eng. J. 14, 237–241. doi: 10.1016/S1369-703X(02)00220-6
Visser, A. A., Nobre, T., Currie, C. R., Aanen, D. K., and Poulsen, M. (2012). Exploring the potential for actinobacteria as defensive symbionts in fungus-growing termites. Microb. Ecol. 63, 975–985. doi: 10.1007/s00248-011-9987-4
Whitaker, A. (1976). Amino acid transport into fungi: an essay. Trans. Br. Mycol. Soc. 67, 365–376. doi: 10.1016/S0007-1536(76)80160-X
Wu, G. (2009). Amino acids: metabolism, functions, and nutrition. Amino Acids 37, 1–17. doi: 10.1007/s00726-009-0269-0
Yang, G.-Y., Ahmad, F., Liang, S.-Y., Fouad, H., Guo, M.-X., Gaal, H. A., et al. (2020). Termitomyces heimii associated with fungus-growing termite produces Volatile Organic Compounds (VOCs) and lignocellulose-degrading enzymes. Appl. Biochem. Biotechnol. 192, 1270–1283. doi: 10.1007/s12010-020-03376-w
Yin, C., Jin, L., Li, S., Xu, X., and Zhang, Y. (2019). Diversity and antagonistic potential of Actinobacteria from the fungus-growing termite Odontotermes formosanus. 3 Biotech. 9:45. doi: 10.1007/s13205-019-1573-3
Zeleke, J., Gessesse, A., and Abate, D. (2013). Substrate-utilization properties of Termitomyces culture isolated from termite mound in the great rift valley region of Ethiopia. J. Nat. Sci. Res. 3, 16–21.
Zhao, G., Nyman, M., and Jonsson, J. A. (2006). Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed. Chromatogr. 20, 674–682. doi: 10.1002/bmc.580
Keywords: Termitomyces, fruiting, bacteria, fungus, cultivation, termite
Citation: Yang G, Ahmad F, Zhou Q, Guo M, Liang S, Gaal HA and Mo J (2021) Investigation of Physicochemical Indices and Microbial Communities in Termite Fungus-Combs. Front. Microbiol. 11:581219. doi: 10.3389/fmicb.2020.581219
Received: 08 July 2020; Accepted: 14 December 2020;
Published: 15 January 2021.
Edited by:
Olivier Raspé, Mae Fah Luang University, ThailandReviewed by:
Samantha Chandranath Karunarathna, Kunming Institute of Botany, ChinaKandikere Ramaiah Sridhar, Mangalore University, India
Copyright © 2021 Yang, Ahmad, Zhou, Guo, Liang, Gaal and Mo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianchu Mo, bW9qaWFuY2h1QHpqdS5lZHUuY24=
 Guiying Yang
Guiying Yang Farhan Ahmad
Farhan Ahmad Qihuan Zhou1
Qihuan Zhou1 Hassan Ahmed Gaal
Hassan Ahmed Gaal