
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 07 October 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.574736
This article is part of the Research Topic Fungal Primary and Secondary Metabolism and its Importance for Virulence and Biomedical Applications View all 13 articles
Protein acetylation is a universal post-translational modification that fine-tunes the major cellular processes of many life forms. Although the mechanisms regulating protein acetylation have not been fully elucidated, this modification is finely tuned by both enzymatic and non-enzymatic mechanisms. Protein deacetylation is the reverse process of acetylation and is mediated by deacetylases. Together, protein acetylation and deacetylation constitute a reversible regulatory protein acetylation network. The recent application of mass spectrometry-based proteomics has led to accumulating evidence indicating that reversible protein acetylation may be related to fungal virulence because a substantial amount of virulence factors are acetylated. Additionally, the relationship between protein acetylation/deacetylation and fungal drug resistance has also been proven and the potential of deacetylase inhibitors as an anti-infective treatment has attracted attention. This review aimed to summarize the research progress in understanding fungal protein acetylation/deacetylation and discuss the mechanism of its mediation in fungal virulence, providing novel targets for the treatment of fungal infection.
The post-translational modification (PTM) of proteins is a major regulatory mechanism in all life forms. PTM refers to the chemical modification of amino acid residues in proteins by the addition of different chemical groups, which confer new properties on modified proteins, including changes in enzyme activity, subcellular localization, interaction partners, protein stability, and DNA binding (Mann and Jensen, 2003; Verdin and Ott, 2015). Currently, nearly 200 different types of PTMs have been identified, including acetylation, phosphorylation, alkylation, methylation, ubiquitination, and glycosylation (Garavelli, 2004). Protein acetylation, which refers to the covalent binding of an acetyl group to an amino acid residue of a protein, is the most well-known PTM besides phosphorylation (Ali et al., 2018). The most widely studied protein acetylation is that of lysine residues, although acetylation of serine and threonine side chains has also been reported (Tang and Yu, 2019). Thus, unless otherwise specified, in this review, acetylation refers only to that of lysine residues.
The acetyl group can be attached to the α-amino group on the N-terminal end of the protein or the ε-amino group on the side chain of lysine residues; therefore, acetylation can be classified as Nα -acetylation or Nε -acetylation (Hentchel and Escalante-Semerena, 2015). At present, two mechanisms that can regulate acetylation have been identified: one mechanism is mainly regulated by lysine acetyltransferases (KATs), while the other mechanism is non-enzymatic, which can directly introduce lysine residues through the non-enzymatic reaction of acetyl phosphate or acetyl-CoA (Wagner and Hirschey, 2014; Lee et al., 2018). In prokaryotes, acetyl phosphate can modulate bacterial virulence through non-enzymatic acetylation (Ren et al., 2019). The main contributor of non-enzymatic acetylation in eukaryotes may be acetyl-CoA in the mitochondria; however, its role still needs to be studied further and is not the focus of this review (Weinert et al., 2014).
Protein deacetylation is the reverse reaction of acetylation that is catalyzed by lysine deacetylases (KDACs), which consist of two protein families, namely, classical Zn2+-dependent histone deacetylases (HDACs) and NAD+-dependent sirtuins (Table 1; Narita et al., 2019). Although acetylation occurs in an enzymatic or non-enzymatic manner, removal of the acetyl group requires KDACs. KDACs play a vital role in numerous biological processes by allowing chromatin condensation, thereby inhibiting transcription (Rupert et al., 2016). Many eukaryotic KATs and KDACs were initially identified as histone-specific enzymes and were historically named histone acetyltransferases and HDACs. In this review, we uniformly used the more specific terms KATs and KDACs.
A substantial amount of evidence showed that acetylation and deacetylation play essential roles in modifying the chromosome structure and regulating gene expression (Nicolas et al., 2018). Acetylation and deacetylation can also modify many key cellular processes relevant to physiology and disease, such as enzymatic activity, signal transduction, DNA damage repair, cell division, metabolism, autophagy, protein stability, and protein localization and interactions (Eckschlager et al., 2017; Narita et al., 2019). Hence, protein acetylation and deacetylation can interfere with every step in a regulatory process, thereby altering cell fate and function.
Research on acetylation/deacetylation is currently focused on metabolism, tumor treatment, and other aspects, while there is less research on microbial acetylation, especially regarding microbial virulence. The development of proteomics has resulted in accumulating evidence that protein acetylation/deacetylation is related to microbial virulence and drug resistance (Hnisz et al., 2010; Li et al., 2017; Brandão et al., 2018; Ren et al., 2019). The role of acetylation in regulating bacterial virulence was summarized in a review conducted by Ren et al. (2017). However, the relationship between the specific mechanism of acetylation and fungal pathogenicity remains unclear.
Fungal pathogens have a negative impact on the global economy, food security, and human and animal welfare, not only because they have caused pestilence and famine but also because of the difficulty in treating fungal infectious diseases as well as increased resistance to antifungal drugs (Fisher et al., 2016; Motaung et al., 2017). In animals and plants, an unprecedented number of fungal and fungal-like diseases have led to some of the most serious deaths and extinctions in wild species (Fisher et al., 2016). Therefore, the virulence of pathogenic fungi must be explored. This review discusses how acetylation/deacetylation regulates fungal virulence. First, we discussed the widespread distribution of this modification in the fungal community and listed some virulence-related acetylated proteins present in fungi. Then, we highlighted recent examples to illustrate the unexpected role of acetylation/deacetylation in fungal virulence to suggest novel targets for the development of anti-infective drugs and the treatment of infectious diseases.
Protein acetylation is a conserved evolutionary modification that occurs in eukaryotic and prokaryotic proteins and was first discovered in histones (Phillips, 1963; Allfrey et al., 1964). Reversible protein acetylation was studied in the context of the histones until the late 1990s. Recent advancements in high-resolution mass spectrometry and high-affinity purification technology for acetylated lysine peptides revealed that protein acetylation/deacetylation is not restricted to histones, which resulted in detailed studies of the acetylated proteome and its function (Schilling et al., 2019).
Previous studies and database searches revealed that protein acetylation is widespread in fungi. Zhou et al. detected 477 acetylated proteins (5.28%) among all 9,038 proteins of Candida albicans, which was the first study on acetylome in human pathogenic fungi, providing an important initiating point for further study of the functional analysis of acetylated proteins in such fungal pathogens (Zhou X. et al., 2016). The comparative analysis of fungal acetylomes plays an important role in determining the essential role of acetylation in the virulence of human fungal pathogens (Li et al., 2019). Significant differences in the number and sites of acetylated proteins were found according to the stage of human fungal pathogen growth. For example, 2,335 proteins in the mycelium growing stage were identified to be acetylated in Trichophyton rubrum, which was >10 times higher than that in the conidia stage, and may be explained by conidia being in a quiescent state with low metabolic activity (Xu et al., 2018). Further evidence of protein acetylation in human fungal pathogens was observed in Histoplasma capsulatum, Cryptococcus neoformans, and Aspergillus fumigatus (Xie et al., 2016; Brandão et al., 2018; Lin et al., 2020).
In plant pathogenic fungi, Yang et al. identified 1,313 high-confidence acetylation sites in 727 acetylated proteins in Aspergillus flavus, while 577 acetylated sites were reported in 364 different proteins in Fusarium graminearum (Zhou S. et al., 2016; Yang et al., 2019). Several published studies have described the acetylome of different fungal species, including plant pathogenic fungi Phytophthora sojae, Botrytis cinerea, and Magnaporthe oryzae; fungal insect pathogens, such as Beauveria bassiana and Metarhizium anisopliae; and nonpathogenic fungi species, such as Saccharomyces cerevisiae and Yarrowia lipolytica; which are considered as important resources to explore the physiological role of this modification in eukaryotes (Henriksen et al., 2012; Mukherjee et al., 2012; Li et al., 2016; Lv et al., 2016; Wang et al., 2017; Cai et al., 2018c; Liang et al., 2018).
Interestingly, most of the identified acetylated proteins were involved in the regulation of glucose, lipid, and amino acid metabolism (Wang et al., 2017). Important findings regarding the control of metabolism via protein acetylation were reported in prokaryotes (Wang et al., 2010). A large number of metabolic enzymes are also acetylated in S. cerevisiae, which is consistent with the enzymes that regulate central metabolism through reversible acetylation, ensuring that cells respond to environmental changes by rapidly sensing the cellular energy state and flexibly changing rates or direction (Wang et al., 2010; Henriksen et al., 2012). This result can be explained by the central role of acetyl-CoA in intermediary metabolism because acetyl-CoA acts as the acetyl-donor for both enzymatic and non-enzymatic acetylation (Trefely et al., 2019). In other words, the dynamic interplay between cellular metabolism and acetylation plays a key role in epigenetics; however, this is not the focus of this review.
In summary, protein acetylation is widely distributed in fungi. Aside from modifying many key cellular processes, such as enzymatic activity, signal transduction, cell division, and metabolism, it also controls morphological transformation, biofilm formation, acetic acid stress tolerance, and other processes, thereby affecting the entire fungal life cycle (Kim et al., 2015; Cheng et al., 2016; Narita et al., 2019; Lin et al., 2020).
All known bacterial KATs that have been discovered to date belong to the Gcn5-related N-acetyltransferase family; fewer deacetylases are encoded by prokaryotes, which means that acetylation and deacetylation processes in fungi are more complex with a higher proportion of acetylated proteins in eukaryotes (Tables 1, 2; Hentchel and Escalante-Semerena, 2015). KATs play a vital role in the morphogenetic hyphae growth, biofilm formation, drug resistance, and virulence (Kong et al., 2018; Lin et al., 2020). In B. bassiana, deletion of gcn5 led to severe defects in colony growth and loss of cuticle infection (Cai et al., 2018b). In P. sojae, although Δgcn5 mutants had a normal development, their virulence in soybean was significantly reduced (Zhao et al., 2015). KDACs are also necessary for fungal pathogenesis, which were found to be decisive regulators of genes involved in pathogenicity and fungal toxin production, regulating a number of physiological processes, including thermotolerance, capsule formation, melanin synthesis, protease activity, and cell wall integrity (Bauer et al., 2016, 2019; Brandão et al., 2018).
Many fungal phenotypes have shown a specific correlation with virulence, such as biofilm formation, capsule production, melanin formation, and the secretion of various proteins (Staniszewska et al., 2012; Alspaugh, 2015). Additionally, cellular features, such as the cell wall, hyphae formation, stress response and morphological transition, allow the rapid and effective adaptation of fungal pathogens to varying conditions, which is conducive to their survival in the environment and in infected hosts (Wang et al., 2013; Alspaugh, 2015; Kim et al., 2015). Here, we primarily focused on the KATs and KDACs to discuss the role of acetylation and deacetylation in fungal virulence (Table 1), particularly in A. fumigatus, C. neoformans, and C. albicans, which are important clinical and useful research models for studying fatal infectious fungal pathogens in humans.
Generally, pathogens are subjected to various environmental challenges, such as temperature variations, an acidic pH, and oxidative stress. Reversible acetylation has emerged as one of the processes critical to maintaining cellular homeostasis and shaping responses to environmental stimuli (O’Meara et al., 2010; Chang et al., 2015; Wang J.-J. et al., 2018). In C. neoformans, the loss of acetylation gene gcn5 caused a reduction in toxicity in a murine intranasal infection model and growth defects at high temperatures (O’Meara et al., 2010). In B. bassiana, the deletion of gcn5 led to a 97% reduction in the conidiation capacity as well as severe defects in the growth of fungal colonies and conidial thermotolerance (Cai et al., 2018b). Moreover, Mst2, which can specifically acetylate histone H3K14 through cooperation with Gcn5 to regulate global acetylation events in B. bassiana, was found to play an important role in sustaining multiple stress tolerances such as osmotic and oxidative stress tolerance, cell wall perturbing stress tolerance, thermotolerance, and UV-B resistance (Wang J.-J. et al., 2018). Furthermore, the Δgcn5 mutants of S. cerevisiae and Schizosaccharomyces pombe showed defects in the cellular response to many stressors, including elevated temperatures, high salt concentrations, and nutrient deprivation (Chang et al., 2015).
The absence of acetyltransferase Rtt109 in S. cerevisiae not only activated the transcription of stress-responsive genes but also improved the resistance to oxidative stress, which ultimately contributed to the improvement in acetic acid tolerance (Cheng et al., 2016). The KDAC sirtuin 2, played a role in starvation stress resistance in yeasts; the deacetylase gene rpd3 was also considered essential for starvation stress resistance (Fulco et al., 2003; Nakajima et al., 2016). A previous study on B. bassiana suggested that Δrpd3 significantly reduced the conidial tolerance to wet-heat stress at 45°C but increased the conidial resistance to UV-B irradiation, and the fungal virulence was greatly attenuated in the absence of rpd3 (Cai et al., 2018c). Studies have found that the downregulation of RPD3-type deacetylase RpdA leads to avirulence of A. fumigatus in a murine model for pulmonary aspergillosis (Bauer et al., 2019). In addition, KDACs also play a decisive role as virulence factors in the pathogenic fungus Cochliobolus carbonum and B. bassiana (Baidyaroy et al., 2001; Zhang et al., 2020). Although the mechanism of virulence attenuation and adverse environmental tolerance remains unclear, previous evidence suggests that acetylation/deacetylation can control the virulence level of pathogens by regulating their stress response.
Hyphae have a strong ability to adhere and invade the host, making it easy to maintain their colonization and escape attacks from the host immune system, probably through the release of cell type-specific virulence factors, such as adhesins (e.g., Hwp1, Als3, Als10, Fav2, and Pga55), tissue-degrading enzymes (e.g., Sap4, Sap5, and Sap6), and antioxidant defense proteins (e.g., Sod5) (Sudbery, 2011; Noble et al., 2017). Acetylation and deacetylation play critical regulatory roles in regulating the initiation and maintenance of hyphal development (Garnaud et al., 2016; Kong et al., 2018; Lee et al., 2019). Tribus et al. (2010) found that the repression of the promoter of rpdA knockdown strains resulted in distorted and hyperbranched hyphae and a tremendous loss of radial growth of fungal colonies. MoHOS2-mediated histone deacetylation is important for the development of M. oryzae. In the absence of this mechanism, M. oryzae exhibits defects in hyphae formation, thereby impairing its growth ability inside the host plant (Lee et al., 2019). In B. bassiana, the hyphal cells of Δhos2 mutants are significantly longer than those of the wild type strains, which was concurrent with its inability to develop intact nuclei in hyphal cells (Cai et al., 2018a). The hyphal growth defects of four acetyltransferase mutants of F. graminearum, namely, ΔFgGCN5, ΔFgRTT109, ΔFgSAS2, and ΔFgSAS3 mutants in solid medium, have also been reported (Kong et al., 2018).
In C. albicans, deacetylase Rpd31 and Set3C (Set3/Hos2 HDAC complex) are crucial repressors of the yeast-to-hyphae transition in C. albicans (Hnisz et al., 2010; Garnaud et al., 2016). The acetyltransferase activity of nucleosome acetyltransferase of H4 (NuA4) and the deacetylase activity of Hda1 have also been reported as essential for hyphal initiation and maintenance (Wang X. et al., 2018). Furthermore, NuA4 dynamically regulates hyphal growth by merging and separating with the SWR1 complex, which was mediated by the acetylation of Eaf1 at K173 (lysine residue 173) (Wang X. et al., 2018). Gcn5 was also required for the invasive and filamentous growth of C. albicans, while gcn5 mutant impaired the hyphal elongation in sensing serum and attenuated the C. albicans virulence in a mouse systemic infection model (Chang et al., 2015). Further evidence of the association between acetylation and hyphal growth was found in the acetyltransferase Esa1, which belongs to the MYST (Moz, YBF2, Sas2p, and Tip) family. Wang et al. found that Esa1 was not important for the general growth of C. albicans but was important for its filamentous growth and that Esa1 deletion could prevent filament formation under all hyphal induction conditions (Wang et al., 2013). Overall, hyphal initiation, development, and maintenance are complex processes regulated by acetylation and deacetylation in both filamentous fungi and budding yeasts.
One of the key virulence traits of fungi is morphological plasticity (Li et al., 2017). Although some human fungal pathogens mainly exist in the form of budding yeast cells (such as C. neoformans) or filamentous hyphal structures (such as Aspergillus), C. albicans alternates between these and other forms, usually in response to specific environmental cues (Takagi et al., 2019). In addition to yeast-to-hyphae transition, C. albicans can undergo a reversible switch between two morphologies, known as the white and opaque phases. Although the white and opaque cell types share the same genome, white cells caused more severe virulent in toxicity in mouse models (Kvaal et al., 1997). The class II deacetylase Hda1 selectively inhibits white-to-opaque switches, while the class I deacetylase Rpd31 suppresses transitions in both directions (Srikantha et al., 2001). Moreover, the sirtuins Hst3 and Sir2 were repressors of the white-to-opaque switch, whereas Set3C was the key activator (Figure 1; Pérez-Martín et al., 1999; Hnisz et al., 2009; Stevenson and Liu, 2011). Thus, C. albicans requires interaction with KDACs function for its morphological plasticity, which is central to its pathogenesis.
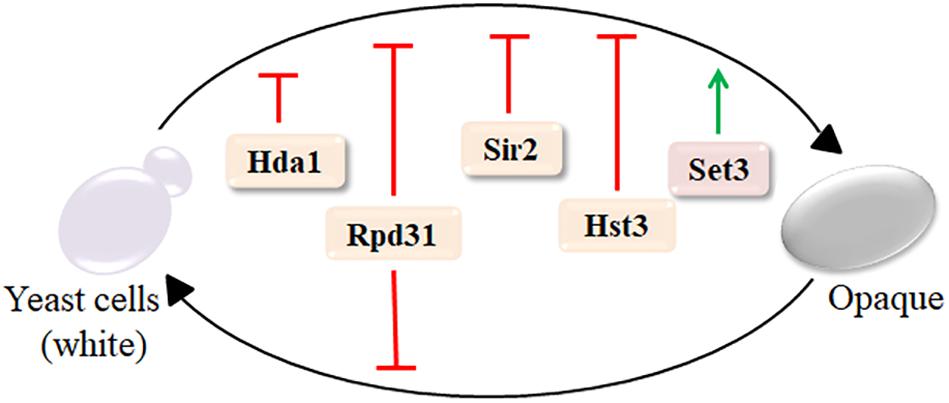
Figure 1. Lysine deacetylases (KDACs) regulate the white-to-opaque switch in C. albicans. Set3 only activates the white-to-opaque switch; Hda1, Sir2, and Hst3 inhibit the progress; and Rpd31 suppresses transitions in both directions. Green arrows indicate activation, while red lines indicate inhibitory functions. Reproduced with the kind permission from Frontiers in Microbiology (Garnaud et al., 2016) with adjusted color scheme.
Biofilm formation on host tissues and indwelling medical devices is highly associated with fungal pathogenicity and drug resistance because the extracellular matrix hinders drug diffusion (Nobile et al., 2014). Fungal adhesion on both biotic and abiotic surfaces is the first phase of biofilm formation, which is closely related to the fungal cell wall and is critical to all later stages of biofilm development (Nett et al., 2011; Lohse et al., 2018). The relationship between biofilm resistance and the cell wall integrity pathway has been confirmed (Nett et al., 2011). In C. albicans, Nobile et al. (2014) found that the deletion of set3 and hos2 in C. albicans reduced biofilm formation and biomass, and these mutants appeared more resistant to yeast dispersion in vivo. Heat shock protein 90 (Hsp90) was a key regulator of biofilm dispersion and drug resistance and could be acetylated on lysine 27 and 270 (Robbins et al., 2012). Compromised Hsp90 function reduced the biofilm formation of C. albicans in vitro and impaired the dispersal of biofilm cells, blocking their capacity to serve as reservoirs of infection (Robbins et al., 2011). Moreover, Hsp90 was involved in the resistance of A. fumigatus biofilms to drugs (Robbins et al., 2011). Another study on A. fumigatus found that acetyltransferase GcnE was also required for biofilm formation (Lin et al., 2020). The list of device-associated infections caused by biofilms is expanding daily. Thus, the urgent determination of the mechanisms whereby acetylation/deacetylation participates in regulating biofilm formation is crucial.
A distinguishing feature of fungi is their ability to produce a variety of small molecules that contribute to their survival and pathogenicity. These substances include compounds such as pigments, which play a role in virulence and protect fungi from environmental damage, and toxins that kill host tissues or hinder competition from other organisms. The absence of deacetylase HdaA in Aspergillus nidulans caused the upregulation of carcinogenic sterigmatocystin (Shwab et al., 2007). In A. fumigatus, ΔhdaA knockout strains had a decreased production of the virulence factor gliotoxin (Lee et al., 2009). In the plant pathogenic fungus Fusarium fujikuroi, the deletion of hda1 or hda2 inhibited the production of red polyketide pigment bikaverin, plant hormone gibberellin, and mycotoxin fumaric acid; however, the deletion of hda1 did not affect the production of mycotoxin fusarins, and the deletion of hda2 did not affect the production of pigment fusarubin. This finding indicated that the impact of acetylation on transcriptional regulation is usually more complex because of the functional complementarity of different KDAC genes (Studt et al., 2013).
AflO, a key enzyme in aflatoxin biosynthesis, was acetylated at lysine 241 and 384 and played a vital role in the pathogenicity of A. flavus (Yang et al., 2019). Six proteins involved in the virulence of B. cinerea were found to be acetylated (BcSak1, Hpt1, Bcchs2, CHSV, PKS, and BOS1) (Lv et al., 2016). In F. graminearum, 10 virulence-related proteins were also acetylated, including Kin4, Sty1, and Gpmk1 (Zhou X. et al., 2016). Deoxynivalenol (DON), a mycotoxin produced by F. graminearum, is a virulence factor that helps fungi colonize and spread within spikes (Kong et al., 2018). The DON production levels of ΔFgSAS3 and ΔFgGCN5 mutants were almost zero compared with that of wild type strain (Kong et al., 2018). Although DON is not a protein, acetylation plays an important role in its metabolism.
Melanin is a pigmented polymer that protects fungal cells against oxidative stress, phagocytosis, and antifungal drugs. It also modifies the host immune responses by reducing the susceptibility of melanized microbes to the host defense mechanisms (Brandão et al., 2015, 2018). Brandão et al. (2018) found that the change in virulence of Δhda1 mutants of C. neoformans might be due to its markedly reduced formation of capsule, melanin, and extracellular proteases, all of which are specifically required for the survival of microbes in the host. Maeda et al. (2017) found that deletion of the HdaA homolog in Magnaporthe orzyae increased the expression of melanin biosynthesis genes. Although the effect of KDAC inhibitors (KDACis) on melanin synthesis has been confirmed, the specific mechanism of acetylation in melanin formation remains unclear (Brandão et al., 2015). The abovementioned studies suggest that protein acetylation/deacetylation can affect the virulence of fungi by participating in the regulation of secondary metabolite biosynthesis.
Invasive infections caused by fungal pathogens are a major public health issue. More than 1.6 million people worldwide develop serious fungal diseases that have a major or fatal impact on their lives (Bongomin et al., 2017). Although the development of new antifungal drugs is an important strategy for the treatment of fungal infections, the urgent development of new infection treatment strategies in view of the uncontrolled increase in the incidence of drug-resistant fungal infections worldwide is crucial (Perlin et al., 2017). Enzymes that control chromatin modification could form a new group of antimicrobial target genes because they are involved in many pathophysiological processes that regulate virulence (Tscherner et al., 2015; Bauer et al., 2016). These results may shed new light on KATs/KDACs as a potential therapeutic target for developing an anti-infection drug (Figure 2).
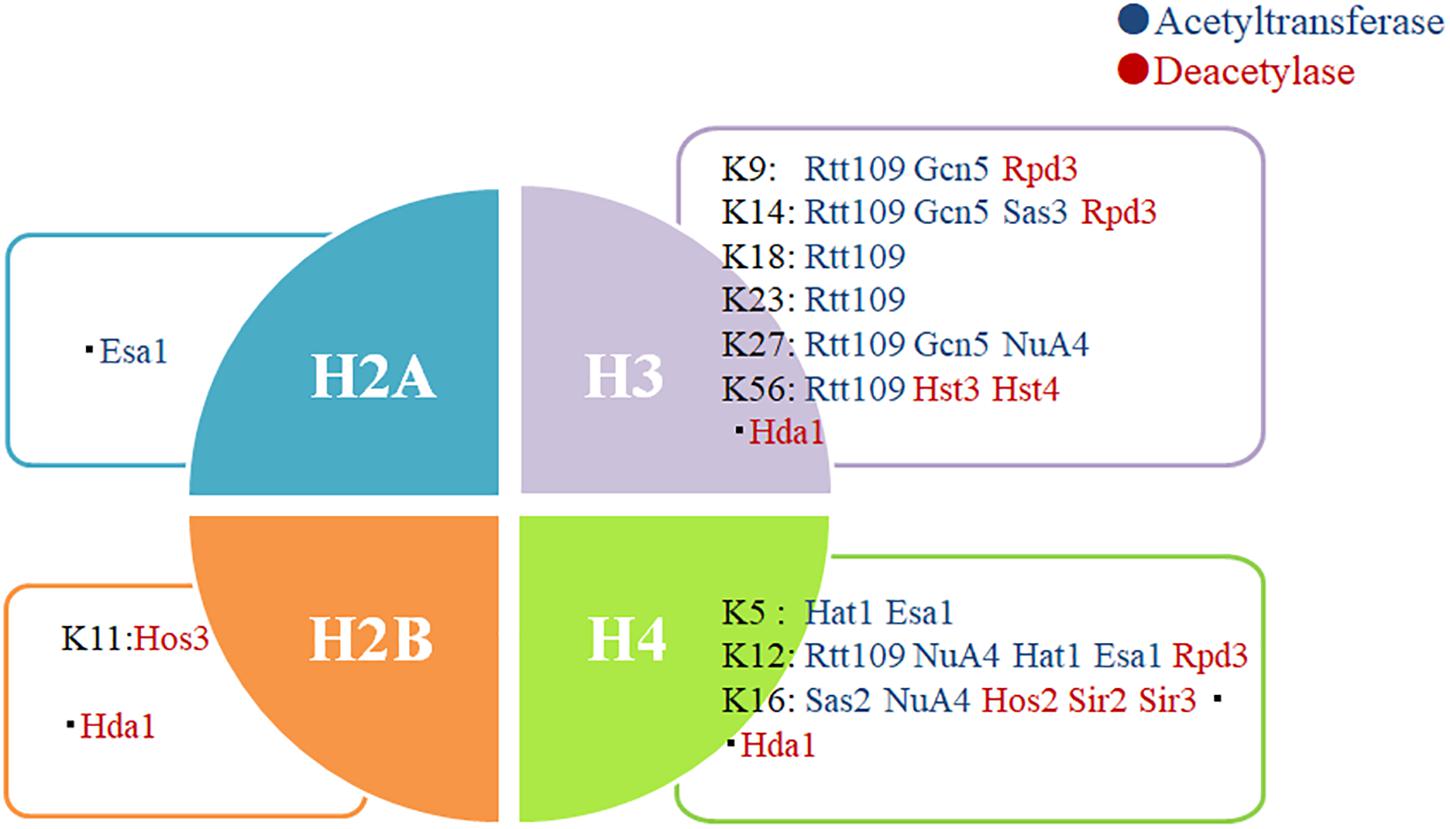
Figure 2. Some important acetylation sites of histones. Protein acetylation on the nucleosomal histones is essential in the regulation of chromatin structure and expression of genes. Some histone acetylation sites that play an important role in fungi have been summarized. A few studies have reported the action sites of Esa1 and Hda1 on histones.
Because KDACis can regulate reversible protein acetylation by inhibiting KDAC activity, altering gene expression; inducing cell cycle arrest, cell differentiation, and apoptosis; reducing angiogenesis; and modulating immune response; they became a new hotspot in the research of tumor-targeted therapy (Spange et al., 2009; Eckschlager et al., 2017). Currently, KDACis are broadly classified into five main groups: hydroxamates, cyclic peptides, benzamides, short-chain fatty acids, and sirtuin inhibitors (Table 3; von Knethen and Brüne, 2019). At present, KDACis are widely used in clinical practice; however their application value in the treatment of antifungal infections requires further investigation.
Targeting fungal deacetylases as a therapeutic strategy has a particular advantage. Inhibiting fungal KDACs may have beneficial and synergistic effects by reducing the virulence and growth of fungi, while also decreasing their tolerance and resistance to the existing antifungal drugs (Pfaller et al., 2015; Zhang and Xu, 2015). Previous studies found that KDACis could inhibit the production of toxic factors in C. neoformans; in S. cerevisiae, the deacetylase inhibitor trichostatin A (TSA) could eliminate its resistance to azole drugs by inhibiting the KDAC activity (Robbins et al., 2012; Brandão et al., 2015). Lys56 acetylation of histone H3 in C. albicans was an ideal target for antifungal therapy, and reduced levels of H3K56ac sensitized C. albicans to genotoxic and antifungal agents (Figure 2; Wurtele et al., 2010). In A. fumigatus, Hsp90 acetylation was also involved in the regulation of drug resistance (Lamoth et al., 2014). Deacetylases Hda1 and Rpd3 can regulate the function of Hsp90 to control fungal drug resistance; therefore, KDACis can block the emergence and maintenance of Hsp90-dependent azole resistance (Robbins et al., 2012). These studies demonstrate that KDACis hold great promise in the treatment of infections resistant to antifungal agents.
Little attention has been paid to acetylase inhibitors because KATs are rarely considered as drug targets. One of the reasons may be the promising application of KDACis in various diseases, and the other is that only a small number of acetylase inhibitors have been identified (Spange et al., 2009). Thus, we mainly discuss the application of KDACis in the treatment of fungal infections using several representative KDACis as examples.
TSA is the best-known broad-spectrum KDACi. It was first isolated from a culture medium of Streptomyces platensis and initially appeared as an antifungal drug to inhibit the growth of Trichophyton and Aspergillus (Tsuji et al., 1976). The inhibition of RpdA activity by TSA resulted in a significant delay in the growth and germination of fungal species, such as A. fumigatus, A. nidulans, Aspergillus terreus, Penicillium chrysogenum, and Neurospora crassa (Bauer et al., 2016). Hnisz et al. (2010) found that TSA was involved in triggering the yeast-to-hyphae conversion of C. albicans by inhibiting Set3C, which controls protein kinase A signaling through Efg1. TSA also increased the susceptibility of Candida sp. to azole antifungals by inhibiting the biosynthesis of ergosterol (Smith and Edlind, 2002). Lamoth et al. (2014) found that the combination of TSA and azole drugs in the treatment of A. fumigatus also showed a promising possibility. Considering instability and rapid metabolism of TSA, the development of highly selective inhibitors is very important for mitigating potential toxicities caused by high doses (Tan and Liu, 2015). Sodium butyrate, apicidin, and suberoylanilide hydroxamic acid are also effective broad-spectrum KDACis; however, their use in antifungal therapy requires further investigation.
MGCD290, a fungal-specific Hos2 inhibitor in Candida sp., displayed moderate activity when used alone (Pfaller et al., 2009, 2015). However, the use of MGCD290 in combination with fluconazole, voriconazole, and posaconazole significantly increased the susceptibility of fungal species in vitro such as azole-resistant Candida, Mucor, and Fusarium sp. (Pfaller et al., 2009; Lamoth et al., 2015). When fluconazole, which had inactive activity against filamentous fungi, was used in combination with MGCD290, there was a distinctly favorable influence of the fluconazole MICs of Aspergillus strains, resulting in a conversion from resistance to susceptibility (Pfaller et al., 2009). Interestingly, Hos2 was a homologous gene of HosA in A. nidulans (Pidroni et al., 2018). However, the deletion of HosA did not affect the efficacy of any antifungal drugs, which contradicted the specificity of MGCD290 (Pidroni et al., 2018). These contradictory results may be explained by the different biological functions of HosA-type proteins in different Aspergillus species, or more likely by the fact that MGCD290 does not specifically act on HosA-type enzymes in filamentous fungi (Pidroni et al., 2018). Therefore, although the specificity of MGCD290 is a debatable issue, MGCD290 has great application prospects in antifungal therapies.
Nicotinamide (NAM), a form of vitamin B3, is a typical non-competitive inhibitor of sirtuins (Orlandi et al., 2017). NAM possesses an antibacterial activity, inhibits cell proliferation and enhances the antiproliferative effect of cytostatic drugs (Xing et al., 2019). NAM’s potential to inhibit the growth of Mycobacterium tuberculosis, Plasmodium falciparum, and HIV has been demonstrated in clinical trials (Murray, 2003; Tcherniuk et al., 2017). Furthermore, NAM displayed broad-spectrum activity against multiple clinical isolates, including C. parapsilosis, C. tropicalis, C. glabrata, C. krusei, C. neoformans, and fluconazole-resistant C. albicans (Xing et al., 2019). NAM also reduced the kidney burden in a mouse model of disseminated candidiasis (Wurtele et al., 2010). Moreover, two different Aspergillus species, A. fumigatus, and A. nidulans, were very sensitive to NAM (Wurtele et al., 2010). NAM also reduced the activity of some enzymes produced by fungi, such as C. albicans, T. rubrum, and Trichophyton mentagrophytes, which supports the use of NAM as an antifungal drug (Ciebiada-Adamiec et al., 2010). In addition to NAM, sirtuin inhibitors include the specific SIRT1 and SIRT2 inhibitors sirtinol, cambinol and EX-527, but their value in the treatment of infection is unclear (Eckschlager et al., 2017).
As a common PTM, protein acetylation plays an essential role in metabolism, virulence, transcription, and translation, among other processes. Acetylation is primarily catalyzed by specific acetyltransferases but can also occur due to the non-enzymatic reactions of acetyl phosphates. At present, only a few studies have reported on the latter, and the relationship between the two different acetylation processes in the regulation of microbial virulence remains unclear. Furthermore, it is unknown whether other non-enzymatic/enzymatic acetylation mechanisms exist and how these (de)acetylation mechanisms cooperate.
Additionally, fungal virulence is a complex phenotype involving multiple factors, making it difficult to explain by analyzing a single type of PTM because there may be multiple PTMs on the same protein, and one protein usually has multiple acetylated lysine residues. How do multiple PTMs cooperate in response to different environmental changes? Besides acetylation, are there other types of acylation that affect the regulation of acetylation, such as malonylation, glutarylation, succinylation, methylation, propionylation, and butyrylation? Do these acetylation modifications have an effect on fungal virulence? These questions need to be addressed in future studies.
At present, research on acetylation and deacetylation mainly focuses on human metabolism, tumors, and other aspects. By contrast, studies on microbial acetylation/deacetylation are limited and mainly focus on a few microbial species, such as Escherichia coli, Salmonella typhimurium, M. tuberculosis, S. cerevisiae, and C. albicans; and studies on human fungal pathogens mainly focus on C. albicans, C. neoformans, and A. fumigatus (Brandão et al., 2018; Bauer et al., 2019; Lin et al., 2020). With the rapid development of protein detection technology, such as high-resolution mass spectrometry, and the broad application of the protein chip, developments in these fields will greatly enrich the investigations on the role of acetylation/deacetylation in regulating microbial physiological process, especially that of microbial pathogenesis and immunity. Therefore, further study of other pathogens is important to reveal the effect of protein acetylation/deacetylation on fungal toxicity and its potential mechanism and may provide some novel potential drug targets for drug development. Finally, in most cases, the effect of the regulation of protein acetylation in host cells by specific pathogens on the quality of immune responses to a broad range of pathogens has not been studied. Future investigations need to be rationally designed to analyze both the pathogen itself and the host’s immune status to avoid excessive damage to the host’s tissues.
All authors contributed to the critical analysis of the collected data and writing of the manuscript. All authors approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (32060040, 31760261, and 31660035), the Science and Technology Research Project of Jiangxi Provincial Education Department (60224), the Key Research and Development Projects of Jiangxi Natural Science Foundation (20192BBG70067, 20202BAB206062, and 20202BAB216045), and the Key Projects of Jiangxi Province Science Foundation for Youths (20192ACBL21042).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Enago (www.enago.cn) for the English language editing.
Ali, I., Conrad, R. J., Verdin, E., and Ott, M. (2018). Lysine acetylation goes global: from epigenetics to metabolism and therapeutics. Chem. Rev. 118, 1216–1252. doi: 10.1021/acs.chemrev.7b00181
Allfrey, V. G., Faulkner, R., and Mirsky, A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794. doi: 10.1073/pnas.51.5.786
Alspaugh, J. A. (2015). Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet. Biol. 78, 55–58. doi: 10.1016/j.fgb.2014.09.004
Baidyaroy, D., Brosch, G., Ahn, J. H., Graessle, S., Wegener, S., Tonukari, N. J., et al. (2001). A gene related to yeast HOS2 histone deacetylase affects extracellular depolymerase expression and virulence in a plant pathogenic fungus. Plant Cell 13, 1609–1624. doi: 10.1105/tpc.010168
Bauer, I., Misslinger, M., Shadkchan, Y., Dietl, A.-M., Petzer, V., Orasch, T., et al. (2019). The Lysine Deacetylase RpdA Is Essential for Virulence in Aspergillus fumigatus. Front. Microbiol. 10:2773. doi: 10.3389/fmicb.2019.02773
Bauer, I., Varadarajan, D., Pidroni, A., Gross, S., Vergeiner, S., Faber, B., et al. (2016). A Class 1 Histone Deacetylase with Potential as an Antifungal Target. mBio 7:e00831-16. doi: 10.1128/mBio.00831-16
Bongomin, F., Gago, S., Oladele, R. O., and Denning, D. W. (2017). Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 3:57. doi: 10.3390/jof3040057
Brandão, F., Esher, S. K., Ost, K. S., Pianalto, K., Nichols, C. B., Fernandes, L., et al. (2018). HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci Rep. 8:5209. doi: 10.1038/s41598-018-21965-y
Brandão, F. A., Derengowski, L. S., Albuquerque, P., Nicola, A. M., Silva-Pereira, I., and Poças-Fonseca, M. J. (2015). Histone deacetylases inhibitors effects on Cryptococcus neoformans major virulence phenotypes. Virulence 6, 618–630. doi: 10.1080/21505594.2015.1038014
Cai, Q., Tong, S.-M., Shao, W., Ying, S.-H., and Feng, M.-G. (2018a). Pleiotropic effects of the histone deacetylase Hos2 linked to H4-K16 deacetylation, H3-K56 acetylation, and H2A-S129 phosphorylation in Beauveria bassiana. Cell Microbiol. 20:e12839. doi: 10.1111/cmi.12839
Cai, Q., Wang, J.-J., Fu, B., Ying, S.-H., and Feng, M.-G. (2018b). Gcn5-dependent histone H3 acetylation and gene activity is required for the asexual development and virulence of Beauveria bassiana. Environ. Microbiol. 20, 1484–1497. doi: 10.1111/1462-2920.14066
Cai, Q., Wang, Z.-K., Shao, W., Ying, S.-H., and Feng, M.-G. (2018c). Essential role of Rpd3-dependent lysine modification in the growth, development and virulence of Beauveria bassiana. Environ. Microbiol. 20, 1590–1606. doi: 10.1111/1462-2920.14100
Chang, P., Fan, X., and Chen, J. (2015). Function and subcellular localization of Gcn5, a histone acetyltransferase in Candida albicans. Fungal Genet. Biol. 81, 132–141. doi: 10.1016/j.fgb.2015.01.011
Cheng, C., Zhao, X., Zhang, M., and Bai, F. (2016). Absence of Rtt109p, a fungal-specific histone acetyltransferase, results in improved acetic acid tolerance of Saccharomyces cerevisiae. FEMS Yeast Res. 16:fow010. doi: 10.1093/femsyr/fow010
Ciebiada-Adamiec, A., Małafiej, E., and Ciebiada, I. (2010). Inhibitory effect of nicotinamide on enzymatic activity of selected fungal strains causing skin infection. Mycoses 53, 204–207. doi: 10.1111/j.1439-0507.2009.01696.x
Dubey, A., Lee, J., Kwon, S., Lee, Y.-H., and Jeon, J. (2019). A MYST family histone acetyltransferase, MoSAS3, is required for development and pathogenicity in the rice blast fungus. Mol. Plant Pathol. 20, 1491–1505. doi: 10.1111/mpp.12856
Eckschlager, T., Plch, J., Stiborova, M., and Hrabeta, J. (2017). Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 18:1414. doi: 10.3390/ijms18071414
Fisher, M. C., Gow, N. A. R., and Gurr, S. J. (2016). Tackling emerging fungal threats to animal health, food security and ecosystem resilience. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 371:20160332. doi: 10.1098/rstb.2016.0332
Fulco, M., Schiltz, R. L., Iezzi, S., King, M. T., Zhao, P., Kashiwaya, Y., et al. (2003). Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 12, 51–62. doi: 10.1016/s1097-2765(03)00226-0
Garavelli, J. S. (2004). The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics 4, 1527–1533. doi: 10.1002/pmic.200300777
Garnaud, C., Champleboux, M., Maubon, D., Cornet, M., and Govin, J. (2016). Histone deacetylases and their inhibition in candida species. Front. Microbiol. 7:1238. doi: 10.3389/fmicb.2016.01238
Graessle, S., Dangl, M., Haas, H., Mair, K., Trojer, P., Brandtner, E. M., et al. (2000). Characterization of two putative histone deacetylase genes from Aspergillus nidulans. Biochim. Biophys. Acta 1492, 120–126. doi: 10.1016/s0167-4781(00)00093-2
Heidinger-Pauli, J. M., Onn, I., and Koshland, D. (2010). Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics 185, 1249–1256. doi: 10.1534/genetics.110.116871
Henriksen, P., Wagner, S. A., Weinert, B. T., Sharma, S., Bacinskaja, G., Rehman, M., et al. (2012). Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11, 1510–1522. doi: 10.1074/mcp.M112.017251
Hentchel, K. L., and Escalante-Semerena, J. C. (2015). Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 79, 321–346. doi: 10.1128/MMBR.00020-15
Hnisz, D., Majer, O., Frohner, I. E., Komnenovic, V., and Kuchler, K. (2010). The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 6:e1000889. doi: 10.1371/journal.ppat.1000889
Hnisz, D., Schwarzmüller, T., and Kuchler, K. (2009). Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol. Microbiol. 74, 1–15. doi: 10.1111/j.1365-2958.2009.06772.x
Itoh, E., Shigemoto, R., Oinuma, K.-I., Shimizu, M., Masuo, S., and Takaya, N. (2017). Sirtuin A regulates secondary metabolite production by Aspergillus nidulans. J. Gen. Appl. Microbiol. 63, 228–235. doi: 10.2323/jgam.2016.11.002
Jacob, T. R., Peres, N. T. A., Martins, M. P., Lang, E. A. S., Sanches, P. R., Rossi, A., et al. (2015). Heat Shock Protein 90 (Hsp90) as a Molecular Target for the Development of Novel Drugs Against the Dermatophyte Trichophyton rubrum. Front. Microbiol. 6:1241. doi: 10.3389/fmicb.2015.01241
Kim, J., Lee, J.-E., and Lee, J.-S. (2015). Histone deacetylase-mediated morphological transition in Candida albicans. J. Microbiol. 53, 805–811. doi: 10.1007/s12275-015-5488-3
Kim, J., Park, S., and Lee, J.-S. (2018). Epigenetic Control of Oxidative Stresses by Histone Acetyltransferases in Candida albicans. J. Microbiol. Biotechnol. 28, 181–189. doi: 10.4014/jmb.1707.07029
Kong, X., van Diepeningen, A. D., van der Lee, T. A. J., Waalwijk, C., Xu, J., Xu, J., et al. (2018). The Histone Acetyltransferases Are Important for Morphogenesis, DON Biosynthesis, and Pathogenicity. Front. Microbiol. 9:654. doi: 10.3389/fmicb.2018.00654
Kvaal, C. A., Srikantha, T., and Soll, D. R. (1997). Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect. Immun. 65, 4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997
Kwon, S., Lee, J., Jeon, J., Kim, S., Park, S.-Y., Jeon, J., et al. (2018). Role of the Histone Acetyltransferase Rtt109 in Development and Pathogenicity of the Rice Blast Fungus. Mol. Plant Microbe Interact. 31, 1200–1210. doi: 10.1094/MPMI-01-18-0015-R
Lamoth, F., Juvvadi, P. R., Soderblom, E. J., Moseley, M. A., Asfaw, Y. G., and Steinbach, W. J. (2014). Identification of a key lysine residue in heat shock protein 90 required for azole and echinocandin resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 58, 1889–1896. doi: 10.1128/AAC.02286-13
Lamoth, F. D. R., Juvvadi, P. R., and Steinbach, W. J. (2015). Histone deacetylase inhibition as an alternative strategy against invasive aspergillosis. Front. Microbiol. 6:96. doi: 10.3389/fmicb.2015.00096
Lee, I., Oh, J.-H., Shwab, E. K., Dagenais, T. R. T., Andes, D., and Keller, N. P. (2009). HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production. Fungal Genet. Biol. 46, 782–790. doi: 10.1016/j.fgb.2009.06.007
Lee, J., Lee, J.-J., and Jeon, J. (2019). A histone deacetylase, MoHOS2 regulates asexual development and virulence in the rice blast fungus. J. Microbiol. 57, 1115–1125. doi: 10.1007/s12275-019-9363-5
Lee, S.-Y., Choi, Y.-S., Kim, E.-H., Cheong, H.-K., Lee, Y.-J., Lee, J.-G., et al. (2018). Nonenzymatic acetylation of ubiquitin Lys side chains is modulated by their neighboring residues. FEBS J. 285, 1277–1289. doi: 10.1111/febs.14404
Li, D., Lv, B., Tan, L., Yang, Q., and Liang, W. (2016). Acetylome analysis reveals the involvement of lysine acetylation in diverse biological processes in Phytophthora sojae. Sci Rep. 6:29897. doi: 10.1038/srep29897
Li, X., Robbins, N., O’Meara, T. R., and Cowen, L. E. (2017). Extensive functional redundancy in the regulation of Candida albicans drug resistance and morphogenesis by lysine deacetylases Hos2, Hda1, Rpd3 and Rpd31. Mol. Microbiol. 103, 635–656. doi: 10.1111/mmi.13578
Li, Y., Li, H., Sui, M., Li, M., Wang, J., Meng, Y., et al. (2019). Fungal acetylome comparative analysis identifies an essential role of acetylation in human fungal pathogen virulence. Commun. Biol. 2:154. doi: 10.1038/s42003-019-0419-1
Li, Y., Wang, C., Liu, W., Wang, G., Kang, Z., Kistler, H. C., et al. (2011). The HDF1 histone deacetylase gene is important for conidiation, sexual reproduction, and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 24, 487–496. doi: 10.1094/MPMI-10-10-0233
Liang, M., Zhang, S., Dong, L., Kou, Y., Lin, C., Dai, W., et al. (2018). Label-Free Quantitative Proteomics of Lysine Acetylome Identifies Substrates of Gcn5 in Magnaporthe oryzae Autophagy and Epigenetic Regulation. mSystems 3, e00270-18. doi: 10.1128/mSystems.00270-18
Lin, C.-J., Hou, Y.-H., and Chen, Y.-L. (2020). The histone acetyltransferase GcnE regulates conidiation and biofilm formation in Aspergillus fumigatus. Med. Mycol. 58, 248–259. doi: 10.1093/mmy/myz043
Lin, Y.-Y., Lu, J.-Y., Zhang, J., Walter, W., Dang, W., Wan, J., et al. (2009). Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136, 1073–1084. doi: 10.1016/j.cell.2009.01.033
Lohse, M. B., Gulati, M., Johnson, A. D., and Nobile, C. J. (2018). Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 16, 19–31. doi: 10.1038/nrmicro.2017.107
Lv, B., Yang, Q., Li, D., Liang, W., and Song, L. (2016). Proteome-wide analysis of lysine acetylation in the plant pathogen Botrytis cinerea. Sci Rep. 6:29313. doi: 10.1038/srep29313
Maeda, K., Izawa, M., Nakajima, Y., Jin, Q., Hirose, T., Nakamura, T., et al. (2017). Increased metabolite production by deletion of an HDA1-type histone deacetylase in the phytopathogenic fungi, Magnaporthe oryzae (Pyricularia oryzae) and Fusarium asiaticum. Lett. Appl. Microbiol. 65, 446–452. doi: 10.1111/lam.12797
Mann, M., and Jensen, O. N. (2003). Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261. doi: 10.1038/nbt0303-255
Motaung, T. E., Saitoh, H., and Tsilo, T. J. (2017). Large-scale molecular genetic analysis in plant-pathogenic fungi: a decade of genome-wide functional analysis. Mol. Plant Pathol. 18, 754–764. doi: 10.1111/mpp.12497
Mukherjee, K., Fischer, R., and Vilcinskas, A. (2012). Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front. Zool. 9:25. doi: 10.1186/1742-9994-9-25
Murray, M. F. (2003). Nicotinamide: an oral antimicrobial agent with activity against both Mycobacterium tuberculosis and human immunodeficiency virus. Clin. Infect. Dis 36, 453–460. doi: 10.1086/367544
Nakajima, E., Shimaji, K., Umegawachi, T., Tomida, S., Yoshida, H., Yoshimoto, N., et al. (2016). The Histone Deacetylase Gene Rpd3 Is Required for Starvation Stress Resistance. PLoS One 11:e0167554. doi: 10.1371/journal.pone.0167554
Narita, T., Weinert, B. T., and Choudhary, C. (2019). Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174. doi: 10.1038/s41580-018-0081-3
Nett, J. E., Sanchez, H., Cain, M. T., Ross, K. M., and Andes, D. R. (2011). Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot. Cell 10, 1660–1669. doi: 10.1128/EC.05126-11
Nicolas, D., Zoller, B., Suter, D. M., and Naef, F. (2018). Modulation of transcriptional burst frequency by histone acetylation. Proc. Natl. Acad. Sci. U.S.A. 115, 7153–7158. doi: 10.1073/pnas.1722330115
Nobile, C. J., Fox, E. P., Hartooni, N., Mitchell, K. F., Hnisz, D., Andes, D. R., et al. (2014). A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. mBio 5, e01201-14. doi: 10.1128/mBio.01201-14
Noble, S. M., Gianetti, B. A., and Witchley, J. N. (2017). Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 15, 96–108. doi: 10.1038/nrmicro.2016.157
O’Meara, T. R., Hay, C., Price, M. S., Giles, S., and Alspaugh, J. A. (2010). Cryptococcus neoformans histone acetyltransferase Gcn5 regulates fungal adaptation to the host. Eukaryot. Cell 9, 1193–1202. doi: 10.1128/EC.00098-10
Orlandi, I., Pellegrino Coppola, D., Strippoli, M., Ronzulli, R., and Vai, M. (2017). Nicotinamide supplementation phenocopies SIR2 inactivation by modulating carbon metabolism and respiration during yeast chronological aging. Mech. Ageing Dev. 161, 277–287. doi: 10.1016/j.mad.2016.06.006
Pérez-Martín, J., Uría, J. A., and Johnson, A. D. (1999). Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 18, 2580–2592. doi: 10.1093/emboj/18.9.2580
Perlin, D. S., Rautemaa-Richardson, R., and Alastruey-Izquierdo, A. (2017). The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect. Dis. 17, e383–e392. doi: 10.1016/S1473-3099(17)30316-X
Pfaller, M. A., Messer, S. A., Georgopapadakou, N., Martell, L. A., Besterman, J. M., and Diekema, D. J. (2009). Activity of MGCD290, a Hos2 histone deacetylase inhibitor, in combination with azole antifungals against opportunistic fungal pathogens. J. Clin. Microbiol. 47, 3797–3804. doi: 10.1128/JCM.00618-09
Pfaller, M. A., Rhomberg, P. R., Messer, S. A., and Castanheira, M. (2015). In vitro activity of a Hos2 deacetylase inhibitor, MGCD290, in combination with echinocandins against echinocandin-resistant Candida species. Diagn. Microbiol. Infect. Dis. 81, 259–263. doi: 10.1016/j.diagmicrobio.2014.11.008
Phillips, D. M. (1963). The presence of acetyl groups of histones. Biochem. J. 87, 258–263. doi: 10.1042/bj0870258
Pidroni, A., Faber, B., Brosch, G., Bauer, I., and Graessle, S. (2018). A Class 1 Histone Deacetylase as Major Regulator of Secondary Metabolite Production in Aspergillus nidulans. Front. Microbiol. 9:2212. doi: 10.3389/fmicb.2018.02212
Ren, J., Sang, Y., Lu, J., and Yao, Y. F. (2017). Protein acetylation and its role in bacterial virulence. Trends Microbiol. 25, 768–779. doi: 10.1016/j.tim.2017.04.001
Ren, J., Sang, Y., Qin, R., Su, Y., Cui, Z., Mang, Z., et al. (2019). Metabolic intermediate acetyl phosphate modulates bacterial virulence via acetylation. Emerg. Microbes Infect. 8, 55–69. doi: 10.1080/22221751.2018.1558963
Reyes-Dominguez, Y., Narendja, F., Berger, H., Gallmetzer, A., Fernandez-Martin, R., Garcia, I., et al. (2008). Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD-prnB bidirectional promoter. Eukaryot. Cell 7, 656–663. doi: 10.1128/EC.00184-07
Robbins, N., Leach, M. D., and Cowen, L. E. (2012). Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Rep. 2, 878–888. doi: 10.1016/j.celrep.2012.08.035
Robbins, N., Uppuluri, P., Nett, J., Rajendran, R., Ramage, G., Lopez-Ribot, J. L., et al. (2011). Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 7:e1002257. doi: 10.1371/journal.ppat.1002257
Rupert, C. B., Heltzel, J. M. H., Taylor, D. J., and Rusche, L. N. (2016). Sporadic Gene Loss After Duplication Is Associated with Functional Divergence of Sirtuin Deacetylases Among Candida Yeast Species. G3 6, 3297–3305. doi: 10.1534/g3.116.033845
Schilling, B., Meyer, J. G., Wei, L., Ott, M., and Verdin, E. (2019). High-resolution mass spectrometry to identify and quantify acetylation protein targets. Methods Mol. Biol. 1983, 3–16. doi: 10.1007/978-1-4939-9434-2_1
Shwab, E. K., Bok, J. W., Tribus, M., Galehr, J., Graessle, S., and Keller, N. P. (2007). Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6, 1656–1664. doi: 10.1128/EC.00186-07
Smith, W. L., and Edlind, T. D. (2002). Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: correlation with reduction in CDR and ERG upregulation. Antimicrob. Agents Chemother. 46, 3532–3539. doi: 10.1128/aac.46.11.3532-3539.2002
Soukup, A. A., Chiang, Y.-M., Bok, J. W., Reyes-Dominguez, Y., Oakley, B. R., Wang, C. C. C., et al. (2012). Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol. 86, 314–330. doi: 10.1111/j.1365-2958.2012.08195.x
Spange, S., Wagner, T., Heinzel, T., and Krämer, O. H. (2009). Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41, 185–198. doi: 10.1016/j.biocel.2008.08.027
Srikantha, T., Tsai, L., Daniels, K., Klar, A. J., and Soll, D. R. (2001). The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol. 183, 4614–4625. doi: 10.1128/JB.183.15.4614-4625.2001
Staniszewska, M., Bondaryk, M., Piłat, J., Siennicka, K., Magda, U., and Kurzatkowski, W. (2012). [Virulence factors of Candida albicans]. Przegl. Epidemiol. 66, 629–633.
Stevenson, J. S., and Liu, H. (2011). Regulation of white and opaque cell-type formation in Candida albicans by Rtt109 and Hst3. Mol. Microbiol. 81, 1078–1091. doi: 10.1111/j.1365-2958.2011.07754.x
Studt, L., Schmidt, F. J., Jahn, L., Sieber, C. M. K., Connolly, L. R., Niehaus, E. M., et al. (2013). Two histone deacetylases, FfHda1 and FfHda2, are important for Fusarium fujikuroi secondary metabolism and virulence. Appl. Environ. Microbiol. 79, 7719–7734. doi: 10.1128/AEM.01557-13
Sudbery, P. E. (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9, 737–748. doi: 10.1038/nrmicro2636
Takagi, J., Singh-Babak, S. D., Lohse, M. B., Dalal, C. K., and Johnson, A. D. (2019). Candida albicans white and opaque cells exhibit distinct spectra of organ colonization in mouse models of infection. PLoS One 14:e0218037. doi: 10.1371/journal.pone.0218037
Tan, S., and Liu, Z.-P. (2015). Natural products as zinc-dependent histone deacetylase inhibitors. ChemMedChem 10, 441–450. doi: 10.1002/cmdc.201402460
Tang, Y., and Yu, W. (2019). SIRT1 and p300/CBP regulate the reversible acetylation of serine-threonine kinase NDR2. Biochem. Biophys. Res. Commun. 518, 396–401. doi: 10.1016/j.bbrc.2019.08.069
Tcherniuk, S. O., Chesnokova, O., Oleinikov, I. V., and Oleinikov, A. V. (2017). Nicotinamide inhibits the growth of P. falciparum and enhances the antimalarial effect of artemisinin, chloroquine and pyrimethamine. Mol. Biochem. Parasitol. 216, 14–20. doi: 10.1016/j.molbiopara.2017.06.004
Trefely, S., Doan, M. T., and Snyder, N. W. (2019). Crosstalk between cellular metabolism and histone acetylation. Methods Enzymol. 626, 1–21. doi: 10.1016/bs.mie.2019.07.013
Tribus, M., Bauer, I., Galehr, J., Rieser, G., Trojer, P., Brosch, G., et al. (2010). A novel motif in fungal class 1 histone deacetylases is essential for growth and development of Aspergillus. Mol. Biol. Cell 21, 345–353. doi: 10.1091/mbc.E09-08-0750
Tribus, M., Galehr, J., Trojer, P., Brosch, G., Loidl, P., Marx, F., et al. (2005). HdaA, a major class 2 histone deacetylase of Aspergillus nidulans, affects growth under conditions of oxidative stress. Eukaryot. Cell 4, 1736–1745. doi: 10.1128/EC.4.10.1736-1745.2005
Tscherner, M., Zwolanek, F., Jenull, S., Sedlazeck, F. J., Petryshyn, A., Frohner, I. E., et al. (2015). The Candida albicans histone acetyltransferase Hat1 regulates stress resistance and virulence via distinct chromatin assembly pathways. PLoS Pathog. 11:e1005218. doi: 10.1371/journal.ppat.1005218
Tsuji, N., Kobayashi, M., Nagashima, K., Wakisaka, Y., and Koizumi, K. (1976). A new antifungal antibiotic, trichostatin. J. Antibiot. 29, 1–6. doi: 10.7164/antibiotics.29.1
Verdin, E., and Ott, M. (2015). 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264. doi: 10.1038/nrm3931
von Knethen, A., and Brüne, B. (2019). Histone deacetylation inhibitors as therapy concept in sepsis. Int. J. Mol. Sci. 20:346. doi: 10.3390/ijms20020346
Wagner, G. R., and Hirschey, M. D. (2014). Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 54, 5–16. doi: 10.1016/j.molcel.2014.03.027
Wang, G., Guo, L., Liang, W., Chi, Z., and Liu, L. (2017). Systematic analysis of the lysine acetylome reveals diverse functions of lysine acetylation in the oleaginous yeast Yarrowia lipolytica. AMB Express 7:94. doi: 10.1186/s13568-017-0393-2
Wang, J.-J., Cai, Q., Qiu, L., Ying, S.-H., and Feng, M.-G. (2018). The histone acetyltransferase Mst2 sustains the biological control potential of a fungal insect pathogen through transcriptional regulation. Appl. Microbiol. Biotechnol. 102, 1343–1355. doi: 10.1007/s00253-017-8703-9
Wang, X., Zhu, W., Chang, P., Wu, H., Liu, H., and Chen, J. (2018). Merge and separation of NuA4 and SWR1 complexes control cell fate plasticity in Candida albicans. Cell Discov. 4:45. doi: 10.1038/s41421-018-0043-0
Wang, Q., Zhang, Y., Yang, C., Xiong, H., Lin, Y., Yao, J., et al. (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007. doi: 10.1126/science.1179687
Wang, X., Chang, P., Ding, J., and Chen, J. (2013). Distinct and redundant roles of the two MYST histone acetyltransferases Esa1 and Sas2 in cell growth and morphogenesis of Candida albicans. Eukaryot. Cell 12, 438–449. doi: 10.1128/EC.00275-12
Wassano, N. S., Leite, A. B., Reichert-Lima, F., Schreiber, A. Z., Moretti, N. S., and Damasio, A. (2020). Lysine acetylation as drug target in fungi: an underexplored potential in Aspergillus spp. Braz. J. Microbiol. 51, 673–683. doi: 10.1007/s42770-020-00253-w
Weinert, B. T., Iesmantavicius, V., Moustafa, T., Schölz, C., Wagner, S. A., Magnes, C., et al. (2014). Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 10:716. doi: 10.1002/msb.134766
Wurtele, H., Tsao, S., Lépine, G., Mullick, A., Tremblay, J., Drogaris, P., et al. (2010). Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat. Med. 16, 774–780. doi: 10.1038/nm.2175
Xie, L., Fang, W., Deng, W., Yu, Z., Li, J., Chen, M., et al. (2016). Global profiling of lysine acetylation in human histoplasmosis pathogen Histoplasma capsulatum. Int. J. Biochem. Cell Biol. 73, 1–10. doi: 10.1016/j.biocel.2016.01.008
Xing, X., Liao, Z., Tan, F., Zhu, Z., Jiang, Y., and Cao, Y. (2019). Effect of nicotinamide against Candida albicans. Front. Microbiol. 10:595. doi: 10.3389/fmicb.2019.00595
Xu, X., Liu, T., Yang, J., Chen, L., Liu, B., Wang, L., et al. (2018). The First Whole-Cell Proteome- and Lysine-Acetylome-Based Comparison between Trichophyton rubrum conidial and mycelial stages. J. Proteome Res. 17, 1436–1451. doi: 10.1021/acs.jproteome.7b00793
Yang, G., Yue, Y., Ren, S., Yang, M., Zhang, Y., Cao, X., et al. (2019). Lysine acetylation contributes to development, aflatoxin biosynthesis and pathogenicity in Aspergillus flavus. Environ. Microbiol. 21, 4792–4807. doi: 10.1111/1462-2920.14825
Yin, Z., Chen, C., Yang, J., Feng, W., Liu, X., Zuo, R., et al. (2019). Histone acetyltransferase MoHat1 acetylates autophagy-related proteins MoAtg3 and MoAtg9 to orchestrate functional appressorium formation and pathogenicity in Magnaporthe oryzae. Autophagy 15, 1234–1257. doi: 10.1080/15548627.2019.1580104
Zhang, L., and Xu, W. (2015). Histone deacetylase inhibitors for enhancing activity of antifungal agent: a patent evaluation of WO2014041424(A1). Expert Opin. Ther. Pat. 25, 237–240. doi: 10.1517/13543776.2014.981256
Zhang, N., Yang, Z., Zhang, Z., and Liang, W. (2020). BcRPD3-Mediated Histone Deacetylation Is Involved in Growth and Pathogenicity of Botrytis cinerea. Front. Microbiol. 11:1832. doi: 10.3389/fmicb.2020.01832
Zhao, W., Wang, T., Liu, S., Chen, Q., and Qi, R. (2015). The histone acetyltransferase PsGcn5 mediates oxidative stress responses and is required for full virulence of Phytophthora sojae. Microb. Pathog. 87, 51–58. doi: 10.1016/j.micpath.2015.07.015
Zhou, S., and Wu, C. (2019). Comparative acetylome analysis reveals the potential roles of lysine acetylation for DON biosynthesis in Fusarium graminearum. BMC Genomics 20:841. doi: 10.1186/s12864-019-6227-7
Zhou, S., Yang, Q., Yin, C., Liu, L., and Liang, W. (2016). Systematic analysis of the lysine acetylome in Fusarium graminearum. BMC Genomics 17:1019. doi: 10.1186/s12864-016-3361-3
Keywords: protein acetylation, protein deacetylation, fungal infection, virulence, KDAC inhibitors
Citation: Chen J, Liu Q, Zeng L and Huang X (2020) Protein Acetylation/Deacetylation: A Potential Strategy for Fungal Infection Control. Front. Microbiol. 11:574736. doi: 10.3389/fmicb.2020.574736
Received: 21 June 2020; Accepted: 17 September 2020;
Published: 07 October 2020.
Edited by:
Laure Ries, University of São Paulo, BrazilReviewed by:
Ingo Bauer, Innsbruck Medical University, AustriaCopyright © 2020 Chen, Liu, Zeng and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaotian Huang, eHRodWFuZ0BuY3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.