- Department of Life Sciences and Systems Biology, School of Nature Sciences, Università degli Studi di Torino, Turin, Italy
Successful inter-kingdom relationships are based upon a dynamic balance between defense and cooperation. A certain degree of competition is necessary to guarantee life spread and development. On the other hand, cooperation is a powerful tool to ensure a long lasting adaptation to changing environmental conditions and to support evolution to a higher level of complexity. Bacteria can interact with their (true or potential) parasites (i.e., phages) and with their multicellular hosts. In these model interactions, bacteria learnt how to cope with their inner and outer host, transforming dangerous signals into opportunities and modulating responses in order to achieve an agreement that is beneficial for the overall participants, thus giving rise to a more complex “organism” or ecosystem. In this review, particular attention will be addressed to underline the minimal energy expenditure required for these successful interactions [e.g., moonlighting proteins, post-translational modifications (PTMs), and multitasking signals] and the systemic vision of these processes and ways of life in which the system proves to be more than the sum of the single components. Using an inside-out perspective, I will examine the possibility of multilevel interactions, in which viruses help bacteria to cope with the animal host and bacteria support the human immune system to counteract viral infection in a circular vision. In this sophisticated network, bacteria represent the precious link that insures system stability with relative low energy expenditure.
Introduction
The existence of symbiotic relationships between bacteria and organisms belonging to other kingdoms has long been recognized, although only recently, thanks to the advancement of molecular and omic techniques, in-depth insights into their reciprocal interaction at a molecular level have been achieved (Qin et al., 2010; Mendes and Raaijmakers, 2015). The fundamental role that bacteria have played during higher organisms’ evolution, and the huge impact that they continue to have in host development, self-recognition, physiology and adaptation to changing environments have been extensively discussed (Zilber-Rosenberg and Rosenberg, 2008; Schnorr, 2015). Paradigmatic for describing these interactions are the examples concerning root (Ramírez-Puebla et al., 2013) and the animal gut (Ley et al., 2008) microbiota where both specificity and biodiversity have supported the establishment and evolution of what is now called holobiont, the unit on which evolutionary selection acts (Rosenberg and Zilber-Rosenberg, 2016).
In parallel to this multicellular organism-targeted relationship, one of the oldest and most important challenges for bacteria is the interaction with bacteriophages, viruses infecting prokaryotic cells. At a first sight, phages are parasites/predators that, after exploiting host biosynthetic machinery, kill the cell. From this standpoint, they represent a danger to be absolutely avoided by bacteria that have set-up complex mechanisms of phage resistance (Mirzaei and Maurice, 2017) that have in turn generated mutations in bacteriophages both to overcome resistance and to adapt to new bacterial species (Mirzaei and Maurice, 2017). The latter phage strategy promotes “host jumps” that provide a host-range expansion and ensures virus survival (De Sordi et al., 2019a). However, not only bacteria provide bacteriophage diversification, but also phages support bacterial diversification as shown by the high variability of phage receptors in the same bacterial species in a certain ecological niche (De Sordi et al., 2019b). These reciprocal dynamics, by creating a high selective pressure, have deeply contributed to the evolution of both partners resulting in the overall expansion of their genetic diversity, as suggested by Allen and Abedon (2013). This co-evolution has been recently confirmed to be very fast (just 8 days) by Wandro (2019) who observed that the interaction between Efv-phi1 lytic phage and its host Enterococcus faecium brought to mutations in the surface bacterial structures and RNA polymerase as well as in the tail proteins of the virus. Beside these surface modifications, bacteria can employ sophisticated strategies aimed to escape phage infection after nucleic acid injection. Among these, are worth mentioning the R-M system and the CRISPR (cluster regularly interspaced short palindromic repeats) sequences. The former involves restriction modification systems providing degradation of the phage genome (De Sordi et al., 2019b). The latter is consistent with abundant phage spacers that are identical to a sequence named the proto-spacer found in the genome of the infecting virulent phage. Therefore, this system counteracts nucleic acid invaders (both DNA and RNA) via a sequence specific strategy (Deveau et al., 2010). However, even in this case resistance mechanisms set up by viruses render both these mechanisms uneffective (Samson et al., 2013). In the case of CRISPR-based bacterial phage resistance, phage mutants exist that carry a single point mutation or a deletion in their proto-spacer and therefore they can successfully complete their lytic cycles (Labrie et al., 2010). The CRISPR system and the phage adaptive response represent one the best examples of co-evolution between bacteria and phages that shape the structure of microbial communities (Andersson and Banfield, 2008). On the other hand, bacteria have learnt how to cope with phage infection renegotiating this stressing event to arrange benefits for both contenders, and how to adapt to mammalian host immune system. This ability to convert a negative affair into an opportunity is part of the strategies referred in 2001 by Hoagland et al. (2001) that described the 16th rules of the living, among which three are worth mentioning: a) different assembling of simple modules to create diversity at low-cost, b) high information exchange to optimize instead that maximize, c) interconnection and integration of new aspects to change a bad event into an opportunity (i.e., changing predation/parasitism into symbiosis) These aspects are particularly relevant in the fast-growing and fast-evolving bacterial populations, however, they constitute a model also for guiding human behavior in a period (climate change and scarcity of resources) that reveals our overall fragility.
When considering bacterial interactions with both host and phage our traditional learning approach seems sometimes inadequate to fully understand the reality. The huge number of interactions and feed-backs, the difficulty to establish a clear cause/effect model, and the overall complexity of the network suggest that a circular and systemic point of view is best fitting for elucidating the whole dynamic. In the present review, I will try to explore, although partly in a reductionist way, the molecular mechanisms underlying this Russian doll model of multilevel interactions, considering on one side the molecular effectors at the host-bacteria interface and on the other side the bacterial advantages linked to phage infection. Finally, some aspects of the complex, systemic and holistic ecosystem made up by host, bacteria and phages will be discussed.
The Interactive Biochemistry at the Bacteria Animal Host Interface
Bacteria–host interaction is an unpredictable relationship. What is clear analyzing the literature is that in a symbiosis the whole is more than the sum of the parts and therefore we cannot advance without a systems biology perspective (Relman, 2008). Often, the relationship becomes so intimate that both partners lose some genes and become strongly dependent on the presence of the other partner like in the inter-kingdom syntrophy [for example, lactic acid bacteria (LAB) that cannot synthesize amino acids and need milk proteins or animal inability to synthesize vitamins that are provided by the gut bacteria] (Ellers et al., 2012). This co-evolution has been in some cases so drastic that bacteria lost their identity to become organelles (Margulis’ theory) giving origin to a system of higher complexity (Dyall et al., 2004). Therefore, the apparent involution of each partner is sometimes the door to have access to a new life experience.
In humans, the presence of a commensal microbiota increases the host fitness providing new genes, thus conferring new metabolic capabilities, higher flexibility and adaptation to new ecosystems, like a pen-drive that enhances the capacity of a computer without dramatically altering the hard disk (Webster, 2014). However, besides genetics, several host factors (age, circadian rhythms, hormones, diet, and drugs) can affect symbiont bacteria diversity and single cell level phenotypic pattern (Zdziarski et al., 2010), therefore, the relationship should be considered dynamic and often unpredictable. This is the major challenge when exogenous microbiota supply is planned to treat gut dysbiosis (De Vrieze, 2013) or enhance plant fitness using eco-friendly agriculture (Berg et al., 2016). In this scenario, molecular communication and co-metabolism are crucial factors in host-microbe interactions.
Cross-signaling between bacteria and their host is a well-established concern (Mazzoli and Pessione, 2016). From one side, the host can modulate microbiota releasing compounds (top-down control) and, on the other side, bacteria can use their signaling molecules to modify host biology (bottom-up control) also responding to host stimuli (Cryan and Dinan, 2012). Actually, it has been experimentally proven that this reciprocal interaction modifies the gene expression profiles of both partners (Zdziarski et al., 2010). Host-derived factors besides modulating microbiota composition (Hevia et al., 2015) can impact bacterial genomics, transcriptomic and proteomics at a single cell level, altering the virulence profiles (Denou et al., 2008; Sandrini et al., 2014) and thus conditioning microbial evolution and diversification. As an example the expression of virulence genes is often elicited by host factors such as stress-induced catecholamines (Cogan et al., 2007) toward whom bacteria have evolved specific receptors (Clarke et al., 2006). Compounds of exogenous origin such as antibiotics should be included among modulating host-supplied factors. Actually, low-levels of antibiotics (like those acquired from the diet) can induce expression of virulence genes and favor DNA exchanges between bacteria (Goh et al., 2002). In parallel, microbial factors, both surface-bound and secreted, influence host physiology by regulating several functions as better described in the next section (Liu et al., 2020). Although secondary metabolites are specifically produced by microbes to interact with their environment and therefore with organisms sharing the same ecological niche (social adaptation) (O’Brien and Wright, 2011), frequently simple molecules derived from central carbon catabolism also play a role in cross-communication.
Bacterial-Derived Multitasking Signaling Compounds
Several bacterial-produced metabolites deeply affect the overall host physiological status, including metabolism (Bäckhed et al., 2004), immunity (Segain et al., 2000), and mental health (Cryan and Dinan, 2012). The three main families of biochemical compounds (lipids, sugars, and proteins) all contribute to bacteria signaling toward the host. Their multitasking roles, targeting bacteria and host, are reported in Table 1.
Short-chain fatty acids (SCFA), namely acetate, lactate, propionate and butyrate, are generally end-products of bacterial carbohydrate catabolism, mainly produced following a fiber-rich diet. By acting as histone deacetylase inhibitors they can induce epigenetic modifications that modulate host functions such as energy storage and immune homeostasis (Davie, 2003). Among SCFA, acetate displays an interesting role in controlling articular diseases by inducing faster resolution of the neutrophil-mediate inflammatory response (Vieira et al., 2017). Butyrate (in concentrations lower than 2 mM) can improve the gut epithelial barrier by enhancing the production and secretion of mucin by goblet cells (Burger-van Paassen et al., 2009) and the expression of tight-junction proteins (Wang et al., 2018) besides stimulating the production of anti-inflammatory cytokines, such as interleukin (IL)-10 (Vipperla and O’keefe, 2012). Butyrate also helps to regulate the balance between proliferation, differentiation, and apoptosis of colonocytes and it can be found in higher quantities in the feces of healthy individuals compared with individuals with colorectal cancer (Serban, 2014).
Exopolysaccharides (EPS), besides being important constituents of the biofilm extracellular polymeric substance and acting as protective shield for preventing cell desiccation and osmotic stress (Pessione, 2012) also have a role as signaling molecules, behaving as growth control agents and immune modulators for the host. Lactobacillus acidophilus EPS can inhibit the growth of Caco-2 colon cancer cells (Deepak et al., 2016) while EPS from L. gasseri can induce apoptosis in cervical tumor cells, also decreasing the production of TNF-α and increasing the IL-10 production thus controlling inflammation (Sungur et al., 2017). Also Bifidobacteria-derived EPS support important functions such as modulation of the composition of the gut microbiota and of communication processes with the host. In this genus the G + C content of the genes encoding EPS synthetic enzymes is different from the one of the whole genome suggesting horizontal gene transfer (HGT) events that underline the importance of acquiring such capability (Hidalgo-Cantabrana et al., 2014).
However, the central role is played by nitrogen compounds such as amino acid (and their metabolic products) peptides and proteins. Glutamate, glutamine, and their derivative gamma aminobutyric acid (GABA), especially produced by LAB, are involved in neuromodulation and in controlling neuro-muscular junctions as well as vascular tension and blood pressure (Mazzoli et al., 2010). The tryptophan-derivative indole can control tight junctions (Bansal et al., 2010). Other amino acids derivatives such as histamine, serotonin, nor-epinephrine as well as inorganic gaseous molecules such as NO and H2S (generated by bacterial conversion of diet compounds) are responsible for controlling enteric neurotransmission (for exhaustive reading, see Mazzoli and Pessione, 2016).
It is interesting to underline that bacteria produce SCFA and EPS for their own benefit and also amines have a specific function in both pH buffering and energy gain increase for the prokaryote partner (Pessione, 2012). Therefore, this host-targeted effect seems to be an additional and cheap function of an otherwise useful molecule.
A further example concerns quorum sensing (QS) autoinducers. In Pseudomonas 3-oxo-C12 homoserine lactone can control different host metabolic pathways including tight-junction proteins modifications, resulting in an altered gut barrier (Vikström et al., 2010), pro- and anti-inflammatory cytokine ratio (Glucksam-Galnoy et al., 2013) and apoptosis (Tateda et al., 2003). Similarly, Pseudomonas quinolone signal can downregulate innate immune responses (Kim et al., 2010) and control in a concentration-dependent manner host neutrophil chemotaxis (Hänsch et al., 2014). A review article by Dobson et al. (2012) elegantly illustrates the multiple roles that Gram-positive secreted peptides can have: first, they behave as QS signals at low concentrations and as bacteriocins at higher concentrations by inducing their own biosynthesis when a cell density threshold is reached (Kleerebezem, 2004). Secondarily, at least in L. plantarum, they can behave as bacteriocins and immune modulating agents altering cytokine profiles of both monocytes (Van Hemert et al., 2010) and dendritic cells (Meijerink et al., 2010). All these examples underline the sustainability of these multitasking molecules that support energy saving for bacteria that use the same compound to achieve many different functions. It is evident that host cells must have evolved receptors for sensing such “non-self” compounds, but it is even smarter the fact that bacteria can use a single molecule to orchestrate relationships with both their siblings and their host, with low energy expenditure, and using the same “language.”
Alongside these small molecules, important mediators for bacteria–host interaction are surface-bound and secreted proteins. Surface-layer proteins and glycoproteins can control bacterial adhesion (Beganoviæ et al., 2011) and some secreted proteins like p40 and p75 can cooperate to the gut epithelium physiology by protecting tight junctions and enhancing IgA production (Wang et al., 2017). However, the emblematic example of a cheap bacterial strategy to fulfill a successful interaction with the animal host is the existence of moonlighting proteins (MPs).
Moonlighting Proteins
Moonlighting proteins are proteins that perform different functions in different cellular compartments (Jeffery, 1999). Some exceptions concern MP having a double task in the same cellular compartment such as surface-linked proteases which also behave as adhesins (Kainulainen and Korhonen, 2014). They are ubiquitous in all living kingdoms and their existence has finally overcome the concept one gene-one function, thus providing a clear explanation for the low number of genes present in most living organisms’ genomes (Henderson and Martin, 2014). In bacteria, MPs represent a very interesting example of economy that exploits the same macromolecule to achieve housekeeping functions inside the cell and to interact with the host when secreted (Table 1). Most of them are central metabolism enzymes (especially glycolytic or TCA cycle enzymes) elongation factors (EF-Tu and Ef-G) and stress-induced proteins (chaperones such as GroEL and DNAk) that after secretion can interact with the animal host favoring bacterial adhesion to mucosal surfaces (when surface-bound) (Jeffery, 2018) and regulating the host immune system (when released) (Henderson et al., 2008). Although it has been recognized that these proteins use different domains to fulfill their intracellular and moonlighting roles, in some cases the two functions are performed by the same molecular region as described for the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) whose adhesive function is inhibited by the co-factor NAD+ that hinders the catalytic site (Kinoshita et al., 2008).
Moonlighting proteins lack any classical export signal sequence related to the most common Sec or TAT secretion machineries and any surface attachment domain (Jeffery, 2018). Although the secretion mechanism for most MPs is not known, some possible non-canonical export ways have been described. Transient post-translational modifications (PTM) have been hypothesized to occur, acting as a signal for partitioning some proteins (among the pool of a certain metabolic enzyme) outside the cell. Enolase, a glycolytic enzyme, can be spontaneously modified by its substrate (2-phosphoglicerate) at the level of lysine341, thus becoming suitable for extracellular release. Catalytically active mutants of enolase bearing glutamine, glutamate, arginine, or alanine in the residue341, cannot be extruded (Boël et al., 2004). A second hypothesis suggests that secondary and tertiary structures could be important for recognition and secretion: in Bacillus subtilis, a hydrophobic alpha-helical domain within enolase that contributes to its secretion has been identified (Yang et al., 2011). Since secreted and intracellular MPs share the same physical structure, the existence of chaperones inducing transient rare conformations that are competent for secretion has been hypothesized but not yet proven (Amblee and Jeffery, 2015). Finally, bacterial membrane vesicles (MVs), better described in the next section, can be exploited to release MP (Henderson and Martin, 2014) as previously demonstrated for similar structures present in human cells (Lancaster and Febbraio, 2005).
Regarding surface binding, classical anchoring motifs are lacking in MPs. Some authors ascribe the capability to be retained on the cell surface to specific conformational states of each protein. It has been demonstrated that GroEL1 is prevalently released into the external environment (Cehovin et al., 2010) whereas GroEL2 is a capsule associated protein (Hickey et al., 2010) in Mycobacteria. While the latter favors adhesion to macrophages, the former is the best fitting in inducing cytokine release by monocytes (Lewthwaite et al., 2001). According to others, environmental factors such as pH, also play a role in the cell-anchoring or release of a specific proteins (Antikainen et al., 2007). Because of their different biological tasks, cell-anchored and released MPs both cooperate to achieve a coordinated interaction with the host. Curiously, the MPs released by a bacterial species can re-associate on the surface of a different one, suggesting that the communication occurs among more than two partners (Oliveira et al., 2012).
As far as adhesion is concerned, there is evidence that bacteria displaying surface-bound MPs can auto-aggregate (a pre-requisite for mucosal adhesion) (Waśko et al., 2014) and also co-aggregate with other bacterial species (a feature that potentially quenches pathogens) (Bergonzelli et al., 2006). Adhesion targets in the host are mucus components (Kinoshita et al., 2008), gut epithelial cells (Granato et al., 2004) or extracellular matrix proteins (collagen, laminin, and fibronectin) (Castaldo et al., 2009; Genovese et al., 2013). A peculiar type of interaction has been described between two MPs, the mammalian MP Hsp60 and the bacterial (Listeria monocytogenes) MP alcohol-acetaldheyde dehydrogenase, both acting inside the cell as cell stress proteins (Wampler et al., 2004). Actually, in multicellular organisms, membrane exposed MPs act as receptors for bacteria (Jin et al., 2003) and viruses (Reyes-del Valle et al., 2005).
Adhesion ability represents an advantage when microbes live in symbiosis with higher organisms, favoring tissues colonization. It is interesting to underline that, since pathogens and commensal bacteria share most of the moonlighting adhesins, beneficial bacteria can act as competitors for gut epithelium adhesion thus supporting surface exclusion of the pathogens (Servin, 2004). In LAB that are paradigmatic examples of probiotic organisms, MPs localize on lipoteichoic acids and in cell division sites with ionic interactions (Kainulainen and Korhonen, 2014). Their extracellular location (surface-attached or released into the environment) depends upon pH: under acidic conditions (pH lower than their isoelectric point, i.e., 5) some positively charged MPs (like enolase and GAPDH) can interact with the negatively charged lipoteichoic acids, whereas at higher pH the molecules, negatively charged, are released from the cell surface (Antikainen et al., 2007). These considerations, however, are not true for some pathogenic bacteria that retain the MPs cell-anchored even in neutral and alkaline conditions (Jagadeesan et al., 2010). Hence, since cell-wall anchored MPs act as adhesins, competitive displacement of pathogens on host mucosal surfaces by beneficial microbes like LAB can only occur in acidic conditions, underlining the huge importance of the environment in balancing commensal–pathogens–host interactions. In this respect extracellular and envelope proteomic data suggested that a selenium supply in the growth medium can enhance the abundance of some adhesive MPs (alpha-enolase, EF-Tu) in L. reuteri Lb2BMDSM16143 (Mangiapane et al., 2014).
Released MPs prevalently act as immune modulating factors. Immune modulation is especially elicited by moonlighting bacterial chaperones. i) GroEL/Hsp60 acts activating human monocytes (Maguire et al., 2002) and stimulates the secretion of IL-8 in human macrophages (Bergonzelli et al., 2006). ii) Bacterial intracellular survival into macrophages is provided by Hsp20 (Schnappinger et al., 2003). iii) A cell-stress protein peptidyl-prolyl isomerase (PPI) also promotes survival inside macrophages (Henderson and Martin, 2011). iv) DNAk stimulates dendritic cells maturation (Floto et al., 2006). Although the last three examples refer to pathogenic bacteria, nevertheless released MPs seem to play an important role in controlling prokaryote–host interaction at multiple levels.
Additional activities of secreted bacterial MPs include i) induction of apoptosis in gastric epithelial cells by PPI (Basak et al., 2005) and ii) the role of GroEL/Hsp60 from Enterobacter aerogenes as toxin available for the insect host to paralyze its prey (Yoshida et al., 2001). Finally, according to Henderson et al. (2008) we can conclude that among MPs, stress chaperones, having evolved specifically to interact with other proteins, may have cell signaling functions similar to cytokines, sometimes creating a network of interaction with host-released MPs.
The number of MPs identified by mass spectrometry (MS) and reported to be displayed on the bacterial cell surface is increasing every day (Wang and Jeffery, 2016). Interestingly, bacterial MPs besides interfacing with the mammalian host can also favor symbiotic relationships between bacteria and yeasts (Katakura et al., 2010) suggesting that they double role has evolved to promote whatever inter-kingdom interactions.
PTMs and Cross-Talk
A further fascinating aspect of the bacteria–host cross-talk is the role of PTMs on proteins. At the end of the last century, once achieved exhaustive information about the majority of plant, animal and microbial genomes an unexpected low number of genes was found. If in Eukarya the presence of introns can partially account for this finding, in prokaryotes it cannot. An important step to elucidate this point is the huge presence of PTMs on proteins. Although for long time bacteria were considered unable to post-translationally modify proteins, it is now evident that the paradigm one gene-one protein is wrong even for prokaryotes (Cain et al., 2014).
The concept of protein species (Jungblut et al., 2008), firstly introduced to avoid confusion with the term “isozyme” (that mean the product of a different gene encoding a protein with analogous function, e.g., catalyzing the same reaction with different efficacy and sometimes different direction), refers to different PT modified proteins and is now well accepted. There is an increasing evidence that PTMs have a huge biological significance because they determine peculiar protein properties like half-life, interaction with other proteins, cellular localization (Kobir et al., 2011) as well as diversified protein functions such as redox signaling, enzyme activity, chemotaxis, and metabolic flux tuning (Dykstra et al., 2013). Actually, each protein species (for instance a protein bearing glycosylation) can solve a specific physiological function not performed by the same gene product that has been processed differently (for instance the same protein when phosphorylated). As an example, methylations or acetylations of cellulosome scaffold proteins have critical functional significance, by impacting differently cellulose utilization efficiency (Dykstra et al., 2013). In this way, only one gene can support several tasks thus guarantee economy of genes and of energy required for transcription and translation (Figure 1).
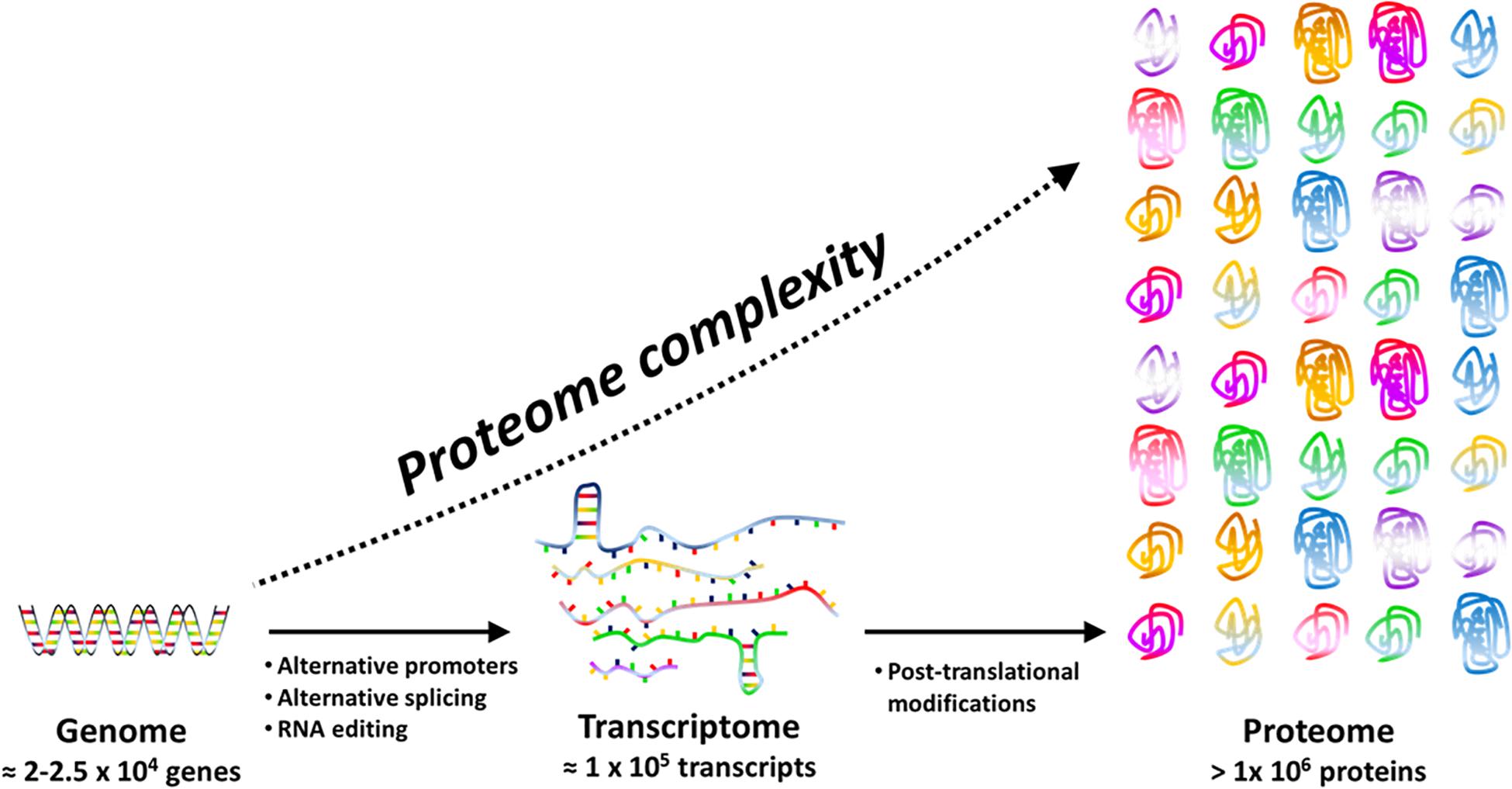
Figure 1. Proteome complexity is the result of transcriptional (e.g. alternative promoters), post-transcriptional (e.g. RNA editing and splicing) and post-translational events. Post-translational events include a large panel of reversible or irreversible chemical modifications that may affect structure, function and cellular location of proteins.
An interesting concern of PTMs is their regulatory/interactive role. Cellular sensing, adjusting and responding to changes in the external environment, is often mediated by reversible PTMs and post-translational regulatory networks in which one PTM can compete with or, on the contrary, facilitate further PTMs by inducing conformational changes on the targeted protein (Soufi et al., 2012) (for comprehensive information on the evolution and functional significance of PTMs see the review by Beltrao et al., 2013). In the specific case of bacteria, the majority of PTMs are transient and occur to ensure rapid adaptation to changing conditions (Grangeasse et al., 2015). Furthermore, in the case of commensal/pathogenic bacteria PTMs on key extracellular proteins can support interaction with the host: serine-glycosylated proteins are important in bacterial adhesion and colonization of host mucosal tissues (Zhou and Wu, 2009), acetylated proteins play a role in virulence (Ren et al., 2017) and multiple PTMs are used by bacteria to alter their surface antigens thus escaping host immune response (Cain et al., 2014).
However, the most intriguing feature is the ability of bacteria to post-translationally modify host proteins (Ribet and Cossart, 2010). This can be achieved by either direct PTM of surface exposed host proteins or by interfering with the host PTM machinery by means of secreted molecules able to penetrate inside the host cell. The preferential targets of these bacterial PTMs are host regulatory proteins like Rho GTPases, since with a single modification on them, bacteria can activate or deactivate the control of a complex network of signals inside the host cell. For instance, RhoGTPases can be either activated by deamidation and/or polyamination or rather inactivated by glycosylation and/or AMPylation thus controlling crucial events such as innate and adaptive immune responses (Ribet and Cossart, 2010). Bacteria can also impact host gene transcription, as occurs by histone-targeted PTMs (Hamon and Cossart, 2008). It is evident that discriminating between “simple” PTMs and epigenetic effects when considering histones is artificial, so the conclusion that bacteria can deeply influence, by epigenetic effects, host gene expression is not so nonsense. Considering that other bacterial modifications on non-proteinaceous molecules have been reported (Pizarro-Cerdá and Cossart, 2004), also nucleic acid modification cannot be totally excluded. As an example, commensal gut bacteria can methylate the DNA of the TLR4 gene thus regulating intestinal inflammation and contributing to the maintenance of intestinal symbiosis (Takahashi et al., 2011) and several pathogens can induce histone acetylation and DNA methylation in the host during infection (Schmeck et al., 2005; Bierne et al., 2012). This opens a new area of investigation on the complex interaction network between bacteria and their hosts. Further information in this field, especially concerning the epigenetic modulation of host gene expression through PTM of histones exerted by commensal/symbiont bacteria, will provide added value to the knowledge of the mechanisms involved in bacteria–host reciprocal control and in supporting the theory of a superorganism constituting the holobiome selection unit (Rosenberg and Zilber-Rosenberg, 2016).
The Inner Host: Bacteria-Phage Contract, a Hidden Opportunity
Bacteria–phage interaction can result in the well known lytic (phage replication and cell lysis) and lysogenic (phage integration into bacterial host DNA) cycles. Occasionally, in particular conditions also pseudolysogeny (phage enters and remains in the cytoplasm without integrating into the bacterial DNA and without replicating) (Cenens et al., 2013) and the so-called chronic cycle (phages particles are continuously released without causing cell lysis) (Mirzaei and Maurice, 2017), can occur. The former evolves toward lytic or lysogenic cycles whereas the latter can cause a lower growth rate in the host bacterial cell because part of the cell energy is directed to phage functions (Munson-McGee et al., 2018).
The phage lytic cycle involving bacterial killing is frequently observed in open environments (soils and waters) where the number of viral particles is about 50 fold higher than the number of living cells (Srinivasiah et al., 2008). Besides promoting antagonistic co-evolution (De Sordi et al., 2019a,b) the lytic cycle is responsible of nutrient cycling among bacterial populations, mineralization of organic bacterial matter and ultimately enrichment of soils (Allen and Abedon, 2013). However, lytic cycle is not the rule in well-established ecosystems such as the microbial biofilm (Jayaraman, 2008) and the human gut (Mirzaei and Maurice, 2017) where lysogenic events mediated by temperate bacteriophages are prevalent because of several mechanisms of resistance set up by bacteria against virulent viruses (Knowles et al., 2017). Independently from the final outcome (lytic, chronic, pseudolysogenic, or lysogenic cycle) and from the stress that phage attack causes to the bacterial cell, phage infection seems to have ensured during evolution some advantages to the bacterial populations.
Possible Advantages Related to Phage Infection
Bacterial proteins that are involved in nutrient uptake (such as porins present on the outer membrane of Gram-negative bacteria), are often used by phages as receptors. In Escherichia coli, as an example, the receptor for the tails of the phages T5, TI, and phi-80 (the ton A/ton B proteins) is used for ferric iron (ferrichrome) uptake (Braun et al., 1976) and in Salmonella the T5-like phage receptor is the uptake protein for vitamin B12 (Kim and Ryu, 2011). In E. coli, it has long been established that the lambda-phage receptor (LamB protein) is a trimeric maltoporin which facilitates maltose uptake (Szmelcman and Hofnung, 1975), although the specific binding sites for phage and for the disaccharide are located on different amino acid residues on this protein (Charbit et al., 1988) (Figure 2). It seems therefore that the risk of phage entry is the price to be paid by the bacterial cell to gain food in nutrient-poor environments. Nevertheless, a question arises: why bacteria (whose ability to resist to phage attack with an arsenal of resistance mechanisms is well-known, see for example Mirzaei and Maurice, 2017) continue to harbor a protein that facilitates phage infection, without modifying it to render it suitable only for nutrient uptake? Evidently, the benefits of obtaining essential nutrients overwhelm the negative effects of phage attack, otherwise this system would be lost or modified. It is tempting to speculate that phage attack to the receptor can induce conformational changes on the protein so as to render it most effective in nutrient capturing. However, at present there are not experimental data supporting this hypothesis. Alternatively, these proteins could have been conserved since phage attack, because of the numerous benefits described in the next sections, cannot be seen only/always as a negative event.
A peculiar phenomenon whose origin is still controversial is the presence of viral-like structures inside prokaryotes (Figure 3). These protein-based particles, similar to phage capsid (but lacking viral nucleic acid), represent a sort of a primitive compartimentalization strategy allowing the formation of micro-compartments or organelles in bacteria. These protein shells confer evolutionary advantage to the bacterial host because they isolate from the rest of the cytosol peculiar metabolic pathways generating toxic compounds such as aldehydes. It is well known that 3-hydroxypropionaldehyde (produced during the conversion of glycerol to 1,3-propanediol) is toxic for several Gram-negative bacteria such as Klebsiella, Enterobacter, and Citrobacter (Barbirato et al., 1998) and it is even used for food preservation being active also toward Listeria monocytogenes and E. coli O157:H7 (Vollenweider and Lacroix, 2004). In L. reuteri the 3-hydroxypropionaldehyde is named reuterin and, when suitably converted into acrolein, is used as bacteriocin-like weapon conferring ecological advantage to this bacterial species toward surrounding bacterial populations (Engels et al., 2016). In Salmonella the enzymes for the metabolization of 1,2 propanediol, generating toxic propionaldehyde are encapsulated in microcompartments (Sampson and Bobik, 2008) that in their aspect and architectural organization (polyhedral protein shells) are very similar to phage particles, especially phage-head. The possible viral origin of these particles is very likely, however, at present, only in Archea the structure (named encapsulin) reveals a common ancestry with the capsid protein of a virus (Sutter et al., 2008). On the other hand, the Gene Transfer Agents are viral-like particles from defective phages that harbor in the capsid bacterial DNA fragments that can be transferred to suitable recipient bacteria sustaining recombination (Lang et al., 2012). These particles do not contain phage nucleic acids, but the bacterial host harbors a prophage in its genome.
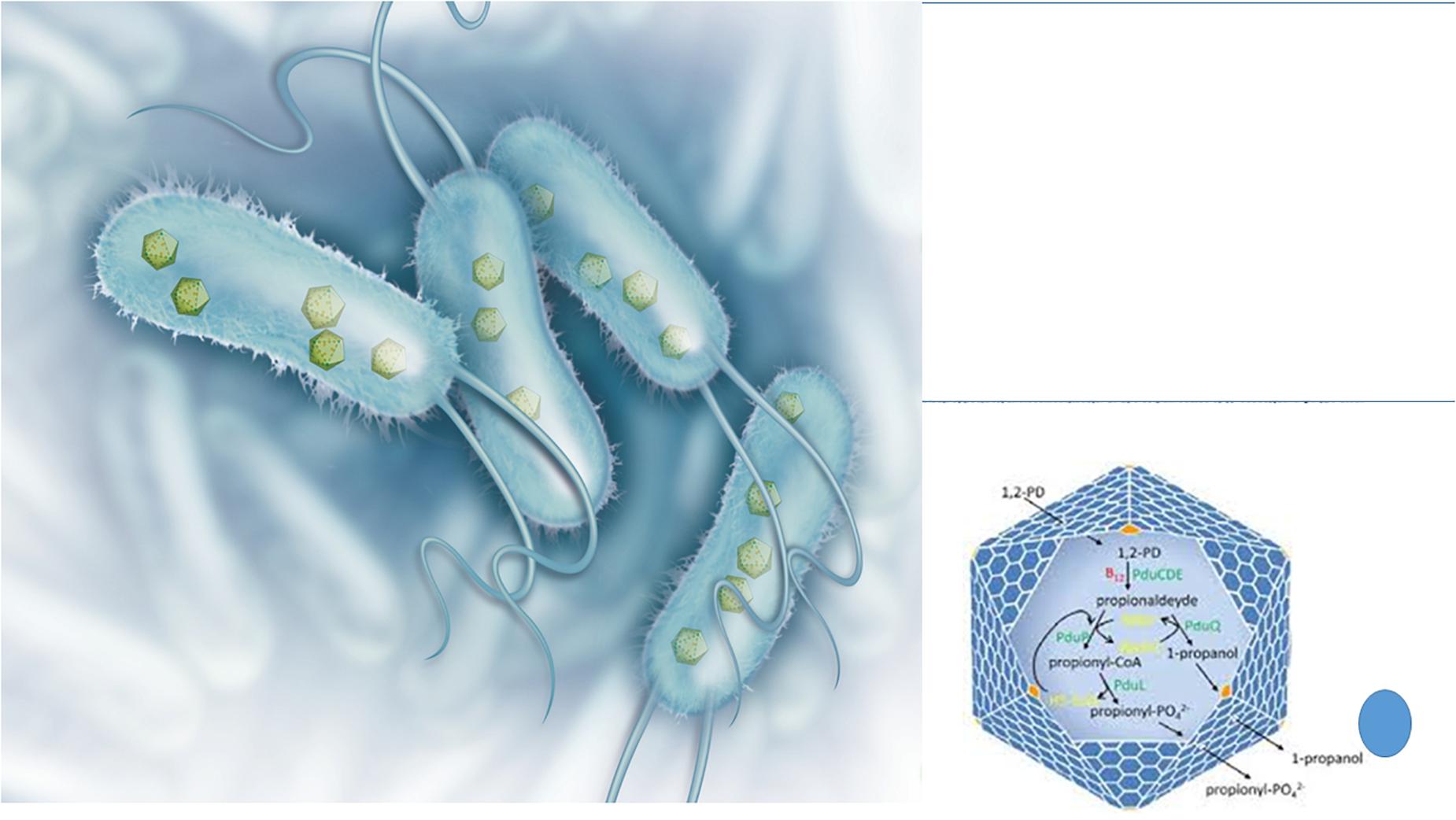
Figure 3. Bacterial structures showing similarities with phages: protein shell microcompartments and the phage head. Microcompartments are used for isolating metabolic pathways generating toxic compounds such as 3-hydroxypropionaldehyde (on the left: illustration originally printed in The Scientist by Thom Graves; on the right: from Lundin et al., 2020).
Finally, a possible viral remnant present in a certain number of Gram-negative bacteria is the type VI secretion system (T6SS). This is a phage-like structure very similar to phage T4 contractile nanomachine (Basler et al., 2012). Actually, T6SS can be illustrated as an inverted phage tail on the surface of the bacterial cell, used for secreting proteins in the extracellular environment or for injecting them directly into target (bacterial or host) cells (Records, 2011) (Figure 4). Structural homologies are common in biology and also the type II secretion system proved to be similar to filamentous phages (Leiman et al., 2009). Nevertheless, besides structural homologies, in the T6SS sequence similarities to phage T4 tail tube are also present (Leiman et al., 2009). Since diversification of functions from one original structural model can occur during evolution, establishing whether the phage tail preceeds the secretion system or vice-versa is hard matter. However, according to Records (2011) it is likely that some Gram-negative bacteria have utilized for their own benefit proteins encoded by phage genes integrated into their genomes. This secretion system offers to bacteria numerous advantages. Actually, among the seven known ways that bacteria evolved to export proteins (type I–VII secretion systems) T6SS is one of the most interesting: it provides a very efficient mean (several proteins can be delivered simultaneously into the target cell) to translocate completely folded proteins across inner and outer membrane of the secreting bacterium and also across other bacteria’s or eukaryotic host membranes (Basler and Mekalanos, 2012). Sometimes protein transfer is finalized to adjacent bacterial cell killing and acquisition of DNA (that can be used as nitrogen/phosphorus source but also for gene recombination) from the prey (Ringel et al., 2017). This strategy, although initially costly in ATP consumption, supports competition between bacteria as well as competence and HGT (Basler, 2015). Furthermore, proteins secreted by T6SS are exchanged among cells and reused for T6SS assembly thus providing recycling (sustainable and shared use of resources) and promoting cooperation between strains to kill competitors (successful interaction) (Vettiger and Basler, 2016). Furthermore, regarding bacteria–host interaction, it has been shown that mutant strains lacking a T6SS orthologous gene trigger stronger inflammatory responses than usual, suggesting that T6SS may function as modulator of acute inflammation thus promoting long-term interaction between bacteria and host (Chow and Mazmanian, 2010). Finally, since genes for T6SS are also conserved in free-living bacteria, it reasonable to assume that besides being involved in interbacteria and host interaction, T6SS also improves the environmental fitness of the bacteria possessing it.
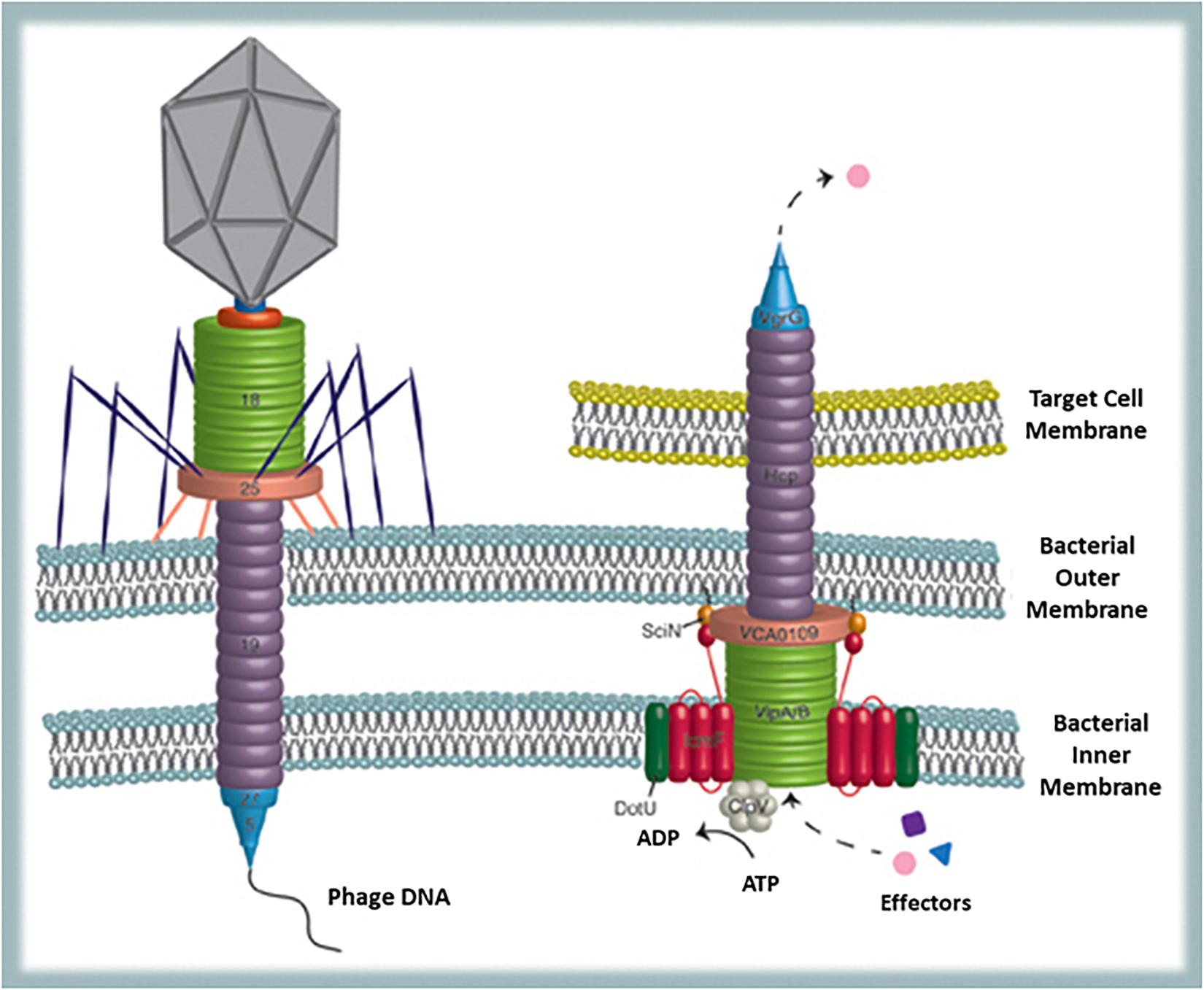
Figure 4. Bacterial structures showing similarities with phages: T6SS and the phage tail. T6SS is used to translocate completely folded proteins across inner and outer membrane of the secreting bacterium and across other bacteria’s or eukaryotic host membranes (from Records, 2011).
Lysogeny and Its Numerous Benefits
Lysogeny cannot be considered just as a simple harboring of “selfish DNA” since it offers some basic advantages to the infected bacterium in whatever habitat, such as the immunity toward further viral infections (Richter et al., 2012) and new functions related to stress resistance (Bondy-Denomy and Davidson, 2014). In specific conditions, other advantages (best fitness) are related to the ecological niche. In open aquatic environments, although the lytic cycle seems to be the general pattern (affecting periodical fluctuations in the bacterial population biodiversity) phage islands controlling genes involved in photosynthesis and light detection have been found in Cyanobacteria genome (Lindell et al., 2007). Ultimately, this results in a better photosynthetic fitness of the bacterial host.
However, it is in complex ecosystems (biofilms, animal-, and vegetal-associated microbiota) that lysogeny reveals its huge potential in driving evolution, suggesting multilevel effects on different partner cells. In the best-studied ecological niche, the human gut, most phages bear integrase genes allowing the prophage lifestyle (Minot et al., 2011). On the other hand, the transition lysogenic-lytic is sometimes dependent also upon QS autoinducers produced by the bacterial host (Silpe and Bassler, 2019). Temperate phages act as vectors for HGT (resulting from encapsidation of bacterial genes by a prophage when the environmental conditions favor a lytic cycles) frequently occurring when bacterial cells are in close proximity. The gene recombination that occurs in a certain ecosystem, favors the exchange of characters crucial for enhancing the metabolic and colonization capabilities of bacteria, meanwhile supporting population diversity and ultimately survival (Harrison and Brockhurst, 2017). However, a recent evaluation, based on a bioinformatics approach, excludes a real role of phages in the propagation of genes involved in antibiotic resistance: actually, the phage genome rarely encodes antibiotic resistance genes and transduction events, being based on erroneous encapsidation of non-viral DNA, seldom target these genes (Enault et al., 2017).
In parallel to the advantages of HGT, phage themselves act as a reservoir of genes important for the host fitness and especially for their social life and interkingdom relationships. Actually, integrated prophages, whose existence depends upon bacterial host survival, can alter virulence gene expression and generally account for a huge number of opportunities offered to their bacterial host (Wandro, 2019). The toxin-antitoxin modules (TA) present in the prokaryote genome have been described as originated from defective viral sources (Villarreal, 2012). They can encode several items. Among these, bacteriocins and immunity factors toward the self-produced bacteriocin (Tikhonenko et al., 1975) can offer advantage to a specific bacterial subpopulation in the fight against other bacteria. The bacteriocin producers therefore, soon become “permanent holder” of the viral genetic element conferring this benefit. Some TA genes such as mazEF induce individual cell death in E. coli during early phases of phage P1 infection (before new viral particles are produced and released) to prevent destruction of the overall bacterial population (Hazan and Engelberg-Kulka, 2004). When we consider a biofilm, a well-structured and “intelligent” microbial community, this behavior of programmed and altruistic cell death is related to the so-called sacrificial cells: the presence of this phage-derived module ensures that these cells set up an autolytic strategy to supply nutrients to the rest of the population. A second type of behavior frequent in a biofilm is persistence or dormancy: the bacterial cells are slow growing or in steady-state to prevent virus entry (Thingstad et al., 2015) or to counteract nutrient starvation. Even in this case the TA modules play a central role (Lewis, 2012). Therefore, it seems evident that from one side biofilms ensure perfect ecological niche for prophage maintenance and evolution and from the other side phages seem to have contributed to bacterial evolution toward a community lifestyle.
An interesting and peculiar mechanism recently discovered, i.e., explosive cell lysis (Turnbull et al., 2016) underlines the role played by prophages in the formation and release of bacterial MVs, previously described both in Gram-negative (Schwechheimer and Kuehn, 2015) and in Gram-positive (Lee et al., 2009) bacteria. These MVs (50–800 nm, average size 250 nm) can contain different cargo-molecules such as viral genomes (Gaudin et al., 2014), cytoplasmic MPs (Henderson and Martin, 2014), DNA, RNA (Zavan et al., 2020), and are involved in important functions, e.g., bacterial communication through hydrophobic QS signals, HGT, sharing of nutrients (especially evident in the biofilm) (Toyofuku et al., 2017b, 2020), also protecting the cells from some antibiotics and phages by acting as decoys (Manning and Kuehn, 2011).
In Pseudomonas aeruginosa a bacteriophage-associated endolysin, encoded into the pyocin gene cluster and coding for a peptidoglycan hydrolase, triggers explosive cell lysis resulting in MVs biogenesis by curling and self-annealing of shattered membrane fragments. Although the pyocin structural genes are not involved, peptidoglycan hydrolase seems not to be the only enzyme responsible for lysis since, at least in E. coli, also holins (facilitating translocation of cell wall hydrolases across the inner membrane) participate to the phenomenon. The inductors of the lytic cycle and vesicle formation can be multiple: light, mutagens, antibiotics and other exogenous stressors, however, endogenous events cannot be excluded (Turnbull et al., 2016).
The described model is not the only one acting on the basis of a phage-mediated stimulus. Toyofuku et al. (2017b) reported that prophages can also induce MVs formation on Gram-positive bacteria, although with a different strategy not implying explosive cell lysis. In B. subtilis a holin-endolysin mediated mechanism occurs. First, holin causes the formation of pores in the cytoplasmic membrane thus facilitating endolysin hydrolysis of peptidoglycan. Once achieved the disruption of peptidoglycan, the cytoplasmic membrane is extruded and the vesicles released, while the bacterial cell dies because membrane integrity is lost. Phage-related proteins have been demonstrated within the vesicles by proteomic studies in both B. subtilis (Kim et al., 2016) and Streptococcus pneumoniae (Resch et al., 2016).
When considering both mechanisms, it is interesting to highlight that only a part of the overall population dies: in P. aeruginosa not all cells harbor the pyocin-prophage insert and in the Gram-positive model the damage caused to the cytoplasmic membrane cannot always be lethal (Toyofuku et al., 2017a). In general, it can be stated that the generation of vesicles is beneficial not exactly for the individual cell but for the microbial community (e.g., the biofilm) in which nutrient exchange and HGT are favored. Moreover, MVs being similar to intact cells and bearing phage receptors can act as decoys attracting antibiotics and phages, thus ensuring an escape lane to living cells (Manning and Kuehn, 2011). Furthermore, also phages can beneficiate of some advantages: since MVs harbor phage receptors, when they fuse with membranes of phage resistant bacterial cells these not-target bacteria become sensitive to phage attack thus allowing the phage population extending the number of hosts and increasing their diffusion among bacterial species. Therefore, in a circular way, also bacterial communities can beneficiate of further HGT by phage transduction. This ultimately results in enhanced fitness of microbial populations as suggested by Nanda et al. (2015). Finally, it is tempting to speculate that MVs can be one of the possible structures giving origin to the lipid envelope present in some phage families. Curiously, the dsRNA phage phi6 (belonging to the Cystoviridae family of rare enveloped phages) displaying a lytic behavior on the target Gram-negative P. aeruginosa acquires the envelope from the bacterial cytoplasmic membrane inside the host cytoplasm (Laurinavičius et al., 2004). Experiments, recently reported by Lyytinen et al. (2019), highlight that the phage-encoded protein P9 can induce in E. coli the formation of intracellular MVs where the protein (possessing a transmembrane domain) is partially incorporated. The presence of internal organelles in prokaryotes are a very rare event observed in Cyanobacteria (thylakoids) and magnetotactic bacteria only. Here again, although the advantages for the host of such a system are still to be fully elucidated, the interaction between bacteria and viruses can give rise to novel opportunities of evolution.
Bacteria–Phage–Host: A Russian-Doll Model for an Inter-Kingdom Coordinated Project
The complex multilevel communication occurring among bacteria, phages and their animal host have driven evolutionary strategies that are worth to be examined. A huge number of inter-kingdom interactions exists in the complex ecosystem of the human gut where many actors are playing on the stage, including viruses, bacteria, archaea, protozoa, and fungi. However, a significant degree of complexity can be observed even when limiting our attention to the central role of bacteria that cross-talk with their inner (phage) and outer (human) host(s), in a Russian doll model represented in Figure 5. First there are some analogies in the behavior that phages and bacteria entertain with their respective host. Similar features characterize phages and bacteria in their ability to set up a form of dormancy (pseudolysogeny in phages and persister cells in bacteria) that allows them waiting for better conditions to activate their reproductive cycle (Łoś and Węgrzyn, 2012). A second analogy is that phage infection is dependent upon the metabolic state of the bacterial-host (De Sordi et al., 2019b) as well as bacterial colonization is dependent upon host metabolism, nutrient availability and circadian rhythms (Rosselot et al., 2016). For instance, planktonic actively duplicating bacterial cells allow to phages high infection efficiency, whereas biofilm-embedded cells are protected from phage (Vidakovic et al., 2018) and viral particle assembly is slow-down during stationary phase (Bryan et al., 2016). In addition, the environment in which bacteria live favors or hinders phage establishment (van Belleghem et al., 2019) like the environment in which the human host lives (hormones, diet, drugs, pollutants, etc…) can favor or exclude certain bacterial populations. Although the mucus layer can harbor a reservoir of bacteria that is maintained thanks to the mucin-derived glycans, regardless of food intake, the lumen microbiota is deeply affected by diet (Donaldson et al., 2016). Actually, the crucial role of diet in selecting saccharolytic or proteolytic bacterial populations has long been established (Gibson and Roberfroid, 1995). Similarly, certain host-environmental conditions can induce production of capsules that mask phage receptors on the bacterial surface of certain species compelling phages to find another target (Roach et al., 2013). Even in this case, however, the arms race between bacteria and viruses favors the appearance of mutant phages that possess enzymes hydrolyzing capsules and ensuring phage access to the receptor (Samson et al., 2013).
On the other hand, it is worth underlining that this multilevel relationship deeply affects the fitness of the three partners finally resulting in a dynamic fluctuation generally driven toward homeostasis (Chaturongakul and Ounjai, 2014), although some possible drifts can occur. It has been experimentally detected that healthy humans display a higher bacterial biodiversity, whereas the phage diversity is increased in inflammatory bowel disease (Norman et al., 2014). Nevertheless, filtered fecal transplantation, although lacking bacterial cells, provides benefits in overcoming Clostridium difficile colitis, suggesting that bacteriophages play a very important role in the human gut homeostasis (Ott et al., 2017).
Partnership Agreement Between Bacteria and the Human Host
The partnership agreement between bacteria and human host to control several aspects of reciprocal well-being is a well-established concern that also includes bacteriophage control. Actually, humans and other animals have evolved a sophisticated immune system not only toward pathogenic bacteria but against viruses as well. Since host immune system does not discriminate between human-targeting viruses and phages, it is likely that immune defenses can control phage populations thus protecting commensal bacteria. At this purpose, it has been demonstrated that viral nucleic acids are recognized by specific toll-like receptors (TLR) namely DNA by TLR9, dsRNA by TLR3 and ssRNA by TLR7 and TLR8, that after sensing the nucleic acid, trigger production of IFNs and other pro-inflammatory cytokines (De Paepe et al., 2014). More specifically, phages have been demonstrated to interact with the animal immune system, by means of head proteins, inducing immunological responses, even if sometimes this interaction can result in a negative modulation of both humoral and cell-mediated immunity probably inducing tolerance toward phages and their bacterial hosts (Da̧browska et al., 2006).
Partnership Agreement Between Bacteria and Phages
In a second model, bacteria and phages can set up a partnership agreement that ensures phage survival and better bacterial fitness toward the mammalian host. The well-known lysogenic conversion support bacteria in producing toxic molecules (such as cholera, diphtheria, and Shiga toxins) that protect them from the host immune reaction and this also ensures temperate phage stabilization into the bacterial genome (Fortier and Sekulovic, 2013). The severe epidemic outbreak of Shiga-toxin producing E. coli O104:H4 occurred in 2011 in Germany was linked to a toxin-encoding phage (Muniesa et al., 2012). An additional example is P. aeruginosa PAO1 bearing a prophage, which although less fitting in swimming, swarming and twitching motility, enhances production of the virulence factor pyocianin and increases resistance to macrophages internalization (Hosseinidoust et al., 2013). In the same bacterial model, the phage-like type VI secretion system allows bacterial translocation of virulence factors (Basler et al., 2012). However, virulence factors like toxins can sometimes have also a beneficial action on the animal host such as carcinogenesis suppression as reported by Doulberis et al. (2015).
Besides helping to produce virulence factors, prophages can also modify surface antigenicity of bacteria thus favoring their host in evading the mammalian immune system (De Paepe et al., 2014). In Gram-negative bacteria, modifications in the O-antigen domain of the outer membrane have been described (Davies et al., 2013). A curious example of surface modification involving a three-partner interaction, has been reported by Mitchell et al. (2007): in this case prophage induction occurs and the production of holins provides enhanced cell permeability and secretion of some parts of the phage “body,” namely, tail fiber proteins that are exposed on the bacterial (Streptococcus mitis) cell surface of not fully lysed cells where they act as adhesins for platelet cells. Furthermore, there is growing evidence that harboring a prophage, besides enhancing aggressiveness toward the host, is a winning strategy for bacteria to improve their resilience to harsh conditions such as presence of bile salts and oxidants, not so rare in the human gut (Wang et al., 2010).
In all the reported examples, phage-bacterial host relationship can be viewed as mutualistic as suggested by Beckett and Williams (2013). Actually, phages provide competitiveness to their bacterial-host in a way that probably proved useless in an abiotic ecological niche (where lysogeny is less observed, see above). On its side, the bacterium continues to cultivate this successful interaction by harboring the virus as a prophage to defend itself from the mammalian host. Therefore, the human/animal host represents a crucial factor for maintaining phage lysogeny. On the other hand, this has been demonstrated also by the abundance of integrase genes detected by metagenomics in the genome of phages living in the gut community (Minot et al., 2011).
Partnership Agreement Between Phages and the Human Host
A third scenario is possible in which the partnership agreement can be signed also between phages and human host. Phages are the most represented entities in the biosphere. It has been established that the phage population in the human gut (where the number of phages reaches 109 per gram versus 1011 bacteria) (Kim et al., 2011) is higher than that of human-targeting viruses (De Paepe et al., 2014). However, it is difficult to establish which families of viruses are present because of the lack of molecular genomic markers similar to the bacterial 16S ribosomal RNA genes, but also because extracting the genetic content from viral particles is still challenging (Roux et al., 2013). According to Manrique et al. (2017) the most represented families are the dsDNA Myoviridae, Siphoviridae and Podoviride and also the ss DNA Microviridae that represent the healthy gut phageome. In some cases, phages, due to their killing activity (lytic cycle) on bacteria can behave as antimicrobial agents controlling bacterial populations and preventing bacterial infection: this has been related to the high number of bacteriophages present at the gut level on mucosal surfaces and interacting with the mucus layer by means of an immunoglobulin-like capsid domain (Barr et al., 2013). According to these authors, host mucosal surfaces and phage coevolve to maintain phage adherence. From one side, this benefits the host since phage adherence to mucus provides a non–host-derived antimicrobial defense, limiting the number of mucosal bacteria and contributing to the maintenance of a selected commensal microbiota. Actually, based on the model kill-the-winner (De Paepe et al., 2014), phage can especially control excessive growth of a specific bacterial population, as occurs during a sudden invasion by an exogenous pathogen, but also when dysbiosis favors a particular endogenous lineage. On the other side, it offers to the phage the opportunity to encounter bacteria thus potentially increasing their replicative success (Barr et al., 2013). Alongside this model that represents a true metazoan-phage symbiosis, indirect effects positively affecting host fitness are reported: phage head and tail proteins can stimulate the immune system sometimes inducing antibodies and pro-inflammatory cytokines (Łusiak-Szelachowska et al., 2014) sometimes reducing inflammation caused by bacterial LPS (Miernikiewicz et al., 2016). A recent and very interesting article suggests that dietary fructose and bacterial-derived SCFA can trigger phage transition from lysogenic to lytic (Oh et al., 2019). This is not the only example in which human host-derived molecules such as component of the western diet, food additives ad nicotine can affect phage populations by controlling their bacterial hosts (Mirzaei and Maurice, 2017). Furthermore, sub-inhibitory doses of antibiotics can trigger prophage excision from bacterial DNA and lytic cycle establishment, thus enhancing bacterial killing, both in E. coli (Zhang et al., 2000) and in pathogenic Gram-positive models resulting in phage expansion and bacterial infection control (Matos et al., 2013). However, it has been reported that most stressors (oxidants, smoke, and antibiotics) mainly induce prophage lytic cycle on beneficial microbes rather than on opportunistic pathogens thus triggering gut dysbiosis (Mills et al., 2013). Therefore, it seems reasonable to assume that the partnership agreement to be beneficial should occur in specific physiological conditions.
Three-Partner Agreement
Finally, it is of interest considering a three-partner agreement: the one mediated by the relatively recently discovered MVs. As discussed in a previous section, MVs exist because of an integrated prophage into a bacterial genome. Besides being important for phage propagation and bacteria–bacteria interaction, MVs can also play a role in the mammalian host physiology, in pathological states and in the reciprocal interactions occurring between host and bacteria. Actually, it has been demonstrated that MVs release can be induced by animal host signals such as changes in temperature, pH, presence of antimicrobial peptides (Deatherage and Cookson, 2012).
As far as the control of host physiology is concerned, MVs can stimulate protective immunity controlling both innate and adaptive immune responses (Deatherage and Cookson, 2012) and also regulate inflammatory pathways (Alaniz et al., 2007). Moreover, MVs from Listeria monocytogenes can inhibit autophagy (Vdovikova et al., 2017) and it has been reported that MVs from Akkermansia muciniphyla (but not those from E. coli) can regulate tight junctions thus controlling intestinal barrier integrity and gut permeability with consequences on the overall inflammatory state of the host organism (Chelakkot et al., 2018).
Modulation of pathological states of the host, such as cancer, by MVs has also been described. Vdovikova et al. (2018) recently demonstrated that MVs from pathogenic (but not from commensal) bacteria such as Vibrio cholerae, can impact gene expression on colon carcinoma cells, by enhancing the transcription of genes involved in cell differentiation. This effect, probably involving epigenetic changes and chromatin accessibility, do not require direct contact between bacteria and cancer cells (Vdovikova et al., 2018). If other cellular structures are involved in the interaction with the bacterial MVs remains to be elucidated. Furthermore, MVs from V. cholerae display the ability to induce higher expression of the nuclear receptor for the Vitamin D that promotes differentiation of colon carcinoma cells (Pálmer et al., 2001).
It is evident from all these examples that the phenotypic effect, finally resulting after MVs signaling to the host, strongly depends upon the cargo transported by the MV (toxin, immunomodulating molecule, and non-human-targeted compound). The contact between MV and the host cell can occur either by endocytosis or by membrane fusion (O’Donoghue and Krachler, 2016) however, in both cases this strategy potently enhances the efficiency by which the active compound (transported and protected) is sensed by the target cell.
Membrane vesicles can also shape the pathogen–host interaction, sometimes favoring sometimes attenuating pathogenesis. Two examples are worth mentioning. In the Salmonella enterica serovar Thypy model, the Cly A cytotoxin is eight fold more active in causing host cell lysis when embedded in MVs than in the free form. The fusion of MVs with the cell membrane of the eukaryotic host can facilitate the toxic action (Wai et al., 2003). On the contrary, in the Helicobacter pylori model the MVs-associated toxin VacA is less active than the soluble form, suggesting that the use of MVs is a strategy used by bacteria to modulate the virulence impact during infection (Ricci et al., 2005). This ambiguity still remains considering the fact that MVs from one side can act as decoys attracting the antibody response, thus allowing the producing bacterium to escape the host immune system, from the other side, they can represent a first signal that allows early detection of the bacterial population by the host immunity that can thus organize and react accordingly (Deatherage and Cookson, 2012). All these data underline that the complex interplay between phages, bacteria and the host is far to be fully understood.
Conclusion
From the analysis of the intricate relationships established between bacteria and their inner and outer hosts, two essential aspects emerge: i) bacteria carry out strategies finalized to communication but also to earn energy and make the cheapest choice, ii) bacteria integrate the “enemy” in view of a broader advantage on the long period and in a larger context. Both behaviors are sustainable. From one hand, they take into account the possibility to use simple building blocks to perform multiple duties (i.e., multitasking signaling compounds and MPs), sometimes using module differentiation (PTMs). From the other hand, the logic of sharing and reuse, together with the conflict mediation has brought during evolution to a shift from a negative event (phage attack) into big opportunities of survival in systemic communities, which ultimately promote a more complex level of organization such as social life and the establishment of a successful integrated lifestyle.
The overall reported data of the last section highlight that a holistic viewpoint can better gave us a framework of what really happens in the phage–bacteria–animal host multilevel community. Phages are the software of bacteria like symbiont bacteria are the software of humans, in a Russian doll-like model. Bacteria are not the same without their prophage(s) and humans are not the same without their tailor-made microbiota. Both can be removed from their respective hosts, however, the hosts pay a cost in reducing its fitness (less resistant/performant bacteria and dysbiosis). Despite their bad reputation due to the fact that both phage and bacteria can kill their host, phages can provide bacteria with new genes, supplying them new opportunities and commensal bacteria can offer to the human hosts new metabolic and signaling pathways not supported by host genome. An agreement often occurs to ensure that the war can be changed into a reciprocally profitable event. Every level sustains intricate and complex relationships with the other levels (and other gut populating organisms as well) and, although more than expected interactions also occur between phages and human host, bacteria are in the center. In this sometimes intricate network of possibilities they prove to be efficient, intelligent in decision-making, and able to respond to both sides. A good lesson also for humans.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work has been supported by Ricerca locale (ex 60%) of Turin University. I am grateful to Anna Luganini for helpful discussion and to Roberto Mazzoli for figures improvement.
References
Alaniz, R. C., Deatherage, B. L., Lara, J. C., and Cookson, B. T. (2007). Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J. Immunol. 179, 7692–7701. doi: 10.4049/jimmunol.179.11.7692
Allen, H. K., and Abedon, S. T. (2013). That’s disturbing! An exploration of the bacteriophage biology of change. Front. Microbiol. 4:295. doi: 10.3389/fmicb.2013.00295
Amblee, V., and Jeffery, C. J. (2015). Physical features of intracellular proteins that moonlight on the cell surface. PLoS One 10:e0130575. doi: 10.1371/journal.pone.0130575
Andersson, A. F., and Banfield, J. F. (2008). Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320, 1047–1050. doi: 10.1126/science.1157358
Antikainen, J., Kupannen, V., Lähteenmäki, K., and Korhonen, T. K. (2007). pH-dependent association of enolase and glyceraldehyde-3-phosphate dehydrogenase of Lactobacillus crispatus with the cell wall and lipoteichoic acids. J. Bacteriol. 189, 4539–4543. doi: 10.1128/jb.00378-07
Bäckhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101, 15718–15723. doi: 10.1073/pnas.0407076101
Bansal, T., Alaniz, R. C., Wood, T. K., and Jayaraman, A. (2010). The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 228–233. doi: 10.1073/pnas.0906112107
Barbirato, F., Soucaille, P., Camarasa, C., and Bories, A. (1998). Uncoupled glycerol distribution as the origin of the accumulation of 3-hydroxypropionaldehyde during the fermentation of glycerol by Enterobacter agglomerans CNCM 1210. Biotechnol. Bioeng. 58, 303–305. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<303::aid-bit28>3.0.co;2-b
Barr, J. J., Auro, R., Furlan, M., Whiteson, K. L., Erb, M. L., Pogliano, J., et al. (2013). Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 10771–10776. doi: 10.1073/pnas.1305923110
Basak, C., Pathak, S. K., Bhattacharyya, A., Pathak, S., Basu, J., and Kundu, M. (2005). The secreted peptidyl prolyl cis, trans-isomerase HP0175 of Helicobacter pylori induces apoptosis of gastric epithelial cells in a TLR4-and apoptosis signal-regulating kinase 1-dependent manner. J. Immunol. 174, 5672–5680. doi: 10.4049/jimmunol.174.9.5672
Basler, Á., and Mekalanos, J. J. (2012). Type 6 secretion dynamics within and between bacterial cells. Science 337, 815–815. doi: 10.1126/science.1222901
Basler, Á., Pilhofer, Á., Henderson, G. P., Jensen, G. J., and Mekalanos, J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. doi: 10.1038/nature10846
Basler, M. (2015). Type VI secretion system: secretion by a contractile nanomachine. Philos. Trans. R. Soc. B Biol. Sci. 370:20150021. doi: 10.1098/rstb.2015.0021
Beckett, S. J., and Williams, H. T. (2013). Coevolutionary diversification creates nested-modular structure in phage–bacteria interaction networks. Interface Focus 3:20130033. doi: 10.1098/rsfs.2013.0033
Beganović, J., Frece, J., Kos, B., Pavunc, A. L., Habjaniè, K., and Šušković, J. (2011). Functionality of the S-layer protein from the probiotic strain Lactobacillus helveticus M92. Antonie Van Leeuwenhoek 100, 43–53. doi: 10.1007/s10482-011-9563-4
Beltrao, P., Bork, P., Krogan, N. J., and van Noort, V. (2013). Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. 9:714. doi: 10.1002/msb.201304521
Berg, G., Rybakova, D., Grube, M., and Köberl, M. (2016). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. doi: 10.1093/jxb/erv466
Bergonzelli, G. E., Granato, D., Pridmore, R. D., Marvin-Guy, L. F., Donnicola, D., and Corthésy-Theulaz, I. E. (2006). GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect. Immun. 74, 425–434. doi: 10.1128/iai.74.1.425-434.2006
Bierne, H., Hamon, M., and Cossart, P. (2012). Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2:a010272. doi: 10.1101/cshperspect.a010272
Boël, G., Pichereau, V., Mijakovic, I., Mazé, A., Poncet, S., Gillet, S., et al. (2004). Is 2-phosphoglycerate-dependent automodification of bacterial enolases implicated in their export? J. Mol. Biol. 337, 485–496. doi: 10.1016/j.jmb.2003.12.082
Bondy-Denomy, J., and Davidson, A. R. (2014). When a virus is not a parasite: the beneficial effects of prophages on bacterial fitness. J. Microbiol. 52, 235–242. doi: 10.1007/s12275-014-4083-3
Braun, V., Hancock, R. E., Hantke, K., and Hartmann, A. (1976). Fuctional organization of the outer membrane of Escherichia coli: phage and colicin receptors as components of iron uptake systems. J. Supramol. Struct. 5, 37–58. doi: 10.1002/jss.400050105
Bryan, D., El-Shibiny, A., Hobbs, Z., Porter, J., and Kutter, E. M. (2016). Bacteriophage T4 infection of stationary phase E. coli: life after log from a phage perspective. Front. Microbiol. 7:1391. doi: 10.3389/fmicb.2016.01391
Burger-van Paassen, N., Vincent, A., Puiman, P. J., Van Der Sluis, M., Bouma, J., Boehm, G., et al. (2009). The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem. J. 420, 211–219. doi: 10.1042/bj20082222
Cain, J. A., Solis, N., and Cordwell, S. J. (2014). Beyond gene expression: the impact of protein post-translational modifications in bacteria. J. Proteomics 97, 265–286. doi: 10.1016/j.jprot.2013.08.012
Castaldo, C., Vastano, V., Siciliano, R. A., Candela, M., Vici, M., Muscariello, L., et al. (2009). Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb. Cell Fact. 8:14. doi: 10.1186/1475-2859-8-14
Cehovin, A., Coates, A. R., Hu, Y., Riffo-Vasquez, Y., Tormay, P., Botanch, C., et al. (2010). Comparison of the moonlighting actions of the two highly homologous chaperonin 60 proteins of Mycobacterium tuberculosis. Infect. Immun. 78, 3196–3206. doi: 10.1128/iai.01379-09
Cenens, W., Makumi, A., Mebrhatu, M. T., Lavigne, R., and Aertsen, A. (2013). Phage–host interactions during pseudolysogeny: lessons from the Pid/dgo interaction. Bacteriophage 3:e1003269.
Charbit, A., Gehring, K., Nikaido, H., Ferenci, T., and Hofnung, M. (1988). Maltose transport and starch binding in phage-resistant point mutants of maltoporin: functional and topological implications. J. Mol. Biol. 201, 487–493. doi: 10.1016/0022-2836(88)90630-4
Chaturongakul, S., and Ounjai, P. (2014). Phage–host interplay: examples from tailed phages and Gram-negative bacterial pathogens. Front. Microbiol. 5:442. doi: 10.3389/fmicb.2014.00442
Chelakkot, C., Choi, Y., Kim, D. K., Park, H. T., Ghim, J., Kwon, Y., et al. (2018). Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 50:e450. doi: 10.1038/emm.2017.282
Chow, J., and Mazmanian, S. K. (2010). A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7, 265–276. doi: 10.1016/j.chom.2010.03.004
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/pnas.0604343103
Cogan, T. A., Thomas, A. O., Rees, L. E., Taylor, A. H., Jepson, M. A., Williams, P. H., et al. (2007). Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut 56, 1060–1065. doi: 10.1136/gut.2006.114926
Cryan, J. F., and Dinan, T. G. (2012). Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712. doi: 10.1038/nrn3346
Da̧browska, K., świtała-Jeleń, K., Opolski, A., and Górski, A. (2006). Possible association between phages, Hoc protein, and the immune system. Arch. Virol. 151, 209–215. doi: 10.1007/s00705-005-0641-7
Davie, J. R. (2003). Inhibition of histone deacetylase activity by butyrate. J. Nutr. 133, 2485S–2493S.
Davies, M. R., Broadbent, S. E., Harris, S. R., Thomson, N. R., and van der Woude, M. W. (2013). Horizontally acquired glycosyltransferase operons drive Salmonellae lipopolysaccharide diversity. PLoS Genet. 9:e1003568. doi: 10.1371/journal.pgen.1003568
De Paepe, M., Leclerc, M., Tinsley, C. R., and Petit, M. A. (2014). Bacteriophages: an underestimated role in human and animal health? Front. Cell. Infect. Microbiol. 4:39. doi: 10.3389/fcimb.2014.00039
De Sordi, L., Lourenço, M., and Debarbieux, L. (2019a). “I will survive”: a tale of bacteriophage-bacteria coevolution in the gut. Gut Microbes 10, 92–99. doi: 10.1080/19490976.2018.1474322
De Sordi, L., Lourenço, M., and Debarbieux, L. (2019b). The battle within: interactions of bacteriophages and bacteria in the gastrointestinal tract. Cell Host Microbe 25, 210–218. doi: 10.1016/j.chom.2019.01.018
Deatherage, B. L., and Cookson, B. T. (2012). Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 80, 1948–1957. doi: 10.1128/iai.06014-11
Deepak, V., Ramachandran, S., Balahmar, R. M., Pandian, S. R. K., Sivasubramaniam, S. D., Nellaiah, H., et al. (2016). In vitro evaluation of anticancer properties of exopolysaccharides from Lactobacillus acidophilus in colon cancer cell lines. In Vitro Cell. Dev. Biol. Anim. 52, 163–173. doi: 10.1007/s11626-015-9970-3
Denou, E., Pridmore, R. D., Berger, B., Panoff, J. M., Arigoni, F., and Brüssow, H. (2008). Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190, 3161–3168. doi: 10.1128/jb.01637-07
Deveau, H., Garneau, J. E., and Moineau, S. (2010). CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 64, 475–493. doi: 10.1146/annurev.micro.112408.134123
Dobson, A., Cotter, P. D., Ross, R. P., and Hill, C. (2012). Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78, 1–6. doi: 10.1128/aem.05576-11
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Doulberis, M., Angelopoulou, K., Kaldrymidou, E., Tsingotjidou, A., Abas, Z., Erdman, S. E., et al. (2015). Cholera-toxin suppresses carcinogenesis in a mouse model of inflammation-driven sporadic colon cancer. Carcinogenesis 36, 280–290. doi: 10.1093/carcin/bgu325
Dyall, S. D., Brown, M. T., and Johnson, P. J. (2004). Ancient invasions: from endosymbionts to organelles. Science 304, 253–257. doi: 10.1126/science.1094884
Dykstra, A. B., Rodriguez, M. Jr., Raman, B., Cook, K. D., and Hettich, R. L. (2013). Characterizing the range of extracellular protein post-translational modifications in a cellulose-degrading bacteria using a multiple proteolyic digestion/peptide fragmentation approach. Anal. Chem. 85, 3144–3151. doi: 10.1021/ac3032838
Ellers, J., Toby Kiers, E., Currie, C. R., McDonald, B. R., and Visser, B. (2012). Ecological interactions drive evolutionary loss of traits. Ecol. Lett. 15, 1071–1082. doi: 10.1111/j.1461-0248.2012.01830.x
Enault, F., Briet, A., Bouteille, L., Roux, S., Sullivan, M. B., and Petit, M. A. (2017). Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 11, 237–247. doi: 10.1038/ismej.2016.90
Engels, C., Schwab, C., Zhang, J., Stevens, M. J., Bieri, C., Ebert, M. O., et al. (2016). Acrolein contributes strongly to antimicrobial and heterocyclic amine transformation activities of reuterin. Sci. Rep. 6:36246.
Floto, R. A., MacAry, P. A., Boname, J. M., Mien, T. S., Kampmann, B., Hair, J. R., et al. (2006). Dendritic cell stimulation by mycobacterial Hsp70 is mediated through CCR5. Science 314, 454–458. doi: 10.1126/science.1133515
Fortier, L. C., and Sekulovic, O. (2013). Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4, 354–365. doi: 10.4161/viru.24498
Gaudin, M., Krupovic, M., Marguet, E., Gauliard, E., Cvirkaite-Krupovic, V., Le Cam, E., et al. (2014). Extracellular membrane vesicles harbouring viral genomes. Environ. Microbiol. 16, 1167–1175. doi: 10.1111/1462-2920.12235
Genovese, F., Coïsson, J. D., Majumder, A., Pessione, A., Svensson, B., Jacobsen, S., et al. (2013). An exoproteome approach to monitor safety of a cheese-isolated Lactococcus lactis. Food Res. Int. 54, 1072–1079. doi: 10.1016/j.foodres.2012.12.017
Gibson, G. R., and Roberfroid, M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412. doi: 10.1093/jn/125.6.1401
Glucksam-Galnoy, Y., Sananes, R., Silberstein, N., Krief, P., Kravchenko, V. V., Meijler, M. M., et al. (2013). The bacterial quorum-sensing signal molecule N-3-oxo-dodecanoyl-L-homoserine lactone reciprocally modulates pro-and anti-inflammatory cytokines in activated macrophages. J. Immunol. 191, 337–344. doi: 10.4049/jimmunol.1300368
Goh, E. B., Yim, G., Tsui, W., McClure, J., Surette, M. G., and Davies, J. (2002). Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. U.S.A. 99, 17025–17030. doi: 10.1073/pnas.252607699
Granato, D., Bergonzelli, G. E., Pridmore, R. D., Marvin, L., Rouvet, M., and Corthésy-Theulaz, I. E. (2004). Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 72, 2160–2169. doi: 10.1128/iai.72.4.2160-2169.2004
Grangeasse, C., Stülke, J., and Mijakovic, I. (2015). Regulatory potential of post-translational modifications in bacteria. Front. Microbiol. 6:500.
Hamon, M. A., and Cossart, P. (2008). Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe 4, 100–109. doi: 10.1016/j.chom.2008.07.009
Hänsch, G. M., Prior, B., Brenner-Weiss, G., Obst, U., and Overhage, J. (2014). The Pseudomonas quinolone signal (PQS) stimulates chemotaxis of polymorphonuclear neutrophils. J. Appl. Biomater. Funct. Mater. 12, 21–26. doi: 10.5301/jabfm.5000204
Harrison, E., and Brockhurst, M. A. (2017). Ecological and evolutionary benefits of temperate phage: what does or doesn’t kill you makes you stronger. Bioessays 39:1700112. doi: 10.1002/bies.201700112
Hazan, R., and Engelberg-Kulka, H. (2004). Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272, 227–234. doi: 10.1007/s00438-004-1048-y
Henderson, B., Derek, J. G., and Chadwick, J. (2008). “Cell stress proteins as modulators of bacteria-host interactions,” in Novartis Foundation Symposium, Vol. 291, eds D. Chadwick and J. A. Goode (Chichester: John Wiley), 141. doi: 10.1002/9780470754030.ch11
Henderson, B., and Martin, A. (2011). Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491. doi: 10.1128/iai.00179-11
Henderson, B., and Martin, A. C. (2014). Protein moonlighting: a new factor in biology and medicine. Biochem. Soc. Trans. 42, 1671–1678. doi: 10.1042/bst20140273
Hevia, A., Delgado, S., Sánchez, B., and Margolles, A. (2015). Molecular players involved in the interaction between beneficial bacteria and the immune system. Front. Microbiol. 6:1285.
Hickey, T. B., Ziltener, H. J., Speert, D. P., and Stokes, R. W. (2010). Mycobacterium tuberculosis employs Cpn60. 2 as an adhesin that binds CD43 on the macrophage surface. Cell. Microbiol. 12, 1634–1647. doi: 10.1111/j.1462-5822.2010.01496.x
Hidalgo-Cantabrana, C., Sánchez, B., Milani, C., Ventura, M., Margolles, A., and Ruas-Madiedo, P. (2014). Genomic overview, and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl. Environ. Microbiol. 80, 9–18. doi: 10.1128/AEM.02977-13
Hoagland, M. B., Dodson, B., and Hauch, J. (2001). Exploring the Way Life Works: The Science of Biology. Burlington, MA: Jones and Bartlett Learning.
Hosseinidoust, Z., Tufenkji, N., and Van De Ven, T. G. (2013). Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 79, 2862–2871. doi: 10.1128/aem.03817-12
Jagadeesan, B., Koo, O. K., Kim, K. P., Burkholder, K. M., Mishra, K. K., Aroonnual, A., et al. (2010). LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria, promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology 156, 2782–2795. doi: 10.1099/mic.0.036509-0
Jayaraman, R. (2008). Bacterial persistence: some new insights into an old phenomenon. J. Biosci. 33, 795–805. doi: 10.1007/s12038-008-0099-3
Jeffery, C. (2018). Intracellular proteins moonlighting as bacterial adhesion factors. AIMS Microbiol. 4, 362–376. doi: 10.3934/microbiol.2018.2.362
Jin, S., Song, Y. C., Emili, A., Sherman, P. M., and Chan, V. L. (2003). JlpA of Campylobacter jejuni interacts with surface exposed heat shock protein 90α and triggers signalling pathways leading to the activation of NF-κB and p38 MAP kinase in epithelial cells. Cell. Microbiol. 5, 165–174. doi: 10.1046/j.1462-5822.2003.00265.x
Jungblut, P. R., Holzhütter, H. G., Apweiler, R., and Schlüter, H. (2008). The speciation of the proteome. Chem. Cent. J. 2, 1–10.
Kainulainen, V., and Korhonen, T. K. (2014). Dancing to another tune—adhesive moonlighting proteins in bacteria. Biology 3, 178–204. doi: 10.3390/biology3010178
Katakura, Y., Sano, R., Hashimoto, T., Ninomiya, K., and Shioya, S. (2010). Lactic acid bacteria display on the cell surface cytosolic proteins that recognize yeast mannan. Appl. Microbiol. Biotechnol. 86, 319–326. doi: 10.1007/s00253-009-2295-y
Kim, K., Kim, Y. U., Koh, B. H., Hwang, S. S., Kim, S. H., Lépine, F., et al. (2010). HHQ and PQS, two Pseudomonas aeruginosa quorum-sensing molecules, down-regulate the innate immune responses through the nuclear factor-κB pathway. Immunology 129, 578–588. doi: 10.1111/j.1365-2567.2009.03160.x
Kim, M., and Ryu, S. (2011). Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 77, 2042–2050. doi: 10.1128/aem.02504-10
Kim, M. S., Park, E. J., Roh, S. W., and Bae, J. W. (2011). Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77, 8062–8070. doi: 10.1128/aem.06331-11
Kim, Y., Edwards, N., and Fenselau, C. (2016). Extracellular vesicle proteomes reflect developmental phases of Bacillus subtilis. Clin. Proteomics 13:6.
Kinoshita, H., Uchida, H., Kawai, Y., Kawasaki, T., Wakahara, N., Matsuo, H., et al. (2008). Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J. Appl. Microbiol. 104, 1667–1674. doi: 10.1111/j.1365-2672.2007.03679.x
Kleerebezem, M. (2004). Quorum sensing control of lantibiotic production: nisin and subtilin autoregulate their own biosynthesis. Peptides 25, 1405–1414. doi: 10.1016/j.peptides.2003.10.021
Knowles, B., Bailey, B., Boling, L., Breitbart, M., Cobián-Güemes, A., Del Campo, J., et al. (2017). Variability and host density independence in inductions-based estimates of environmental lysogeny. Nat. Microbiol. 2:17064.
Kobir, A., Shi, L., Boskovic, A., Grangeasse, C., Franjevic, D., and Mijakovic, I. (2011). Protein phosphorylation in bacterial signal transduction. Biochim. Biophys. Acta 1810, 989–994.
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Lancaster, G. I., and Febbraio, M. A. (2005). Exosome-dependent trafficking of HSP70 A novel secretory pathway for cellular stress proteins. J. Biol. Chem. 280, 23349–23355. doi: 10.1074/jbc.m502017200
Lang, A. S., Zhaxybayeva, O., and Beatty, J. T. (2012). Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10, 472–482. doi: 10.1038/nrmicro2802
Laurinavičius, S., Käkelä, R., Bamford, D. H., and Somerharju, P. (2004). The origin of phospholipids of the enveloped bacteriophage phi6. Virology 326, 182–190. doi: 10.1016/j.virol.2004.05.021
Lee, E. Y., Choi, D. Y., Kim, D. K., Kim, J. W., Park, J. O., Kim, S., et al. (2009). Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9, 5425–5436. doi: 10.1002/pmic.200900338
Leiman, P. G., Basler, M., Ramagopal, U. A., Bonanno, J. B., Sauder, J. M., Pukatzki, S., et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159. doi: 10.1073/pnas.0813360106
Lewis, K. (2012). “Persister cells: molecular mechanisms related to antibiotic tolerance,” in Antibiotic Resistance, ed. A. R. M. Coates (Heidelberg: Springer), 121–133. doi: 10.1007/978-3-642-28951-4_8
Lewthwaite, J. C., Coates, A. R., Tormay, P., Singh, M., Mascagni, P., Poole, S., et al. (2001). Mycobacterium tuberculosis Chaperonin 60.1 Is a More Potent Cytokine Stimulator than Chaperonin 60.2 (Hsp 65) and Contains a CD14-Binding Domain. Infect. Immun. 69, 7349–7355. doi: 10.1128/iai.69.12.7349-7355.2001
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R., and Gordon, J. I. (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. doi: 10.1038/nrmicro1978
Lindell, D., Jaffe, J. D., Coleman, M. L., Futschik, M. E., Axmann, I. M., Rector, T., et al. (2007). Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449, 83–86. doi: 10.1038/nature06130
Liu, Q., Yu, Z., Tian, F., Zhao, J., Zhang, H., Zhai, Q., et al. (2020). Surface components and metabolites of probiotics for regulation of intestinal epithelial barrier. Microb. Cell Fact. 19:23.
Łoś, M., and Węgrzyn, G. (2012). Pseudolysogeny. Adv. Virus Res. 82, 339–349. doi: 10.1016/b978-0-12-394621-8.00019-4
Lundin, A. P., Stewart, K. L., Stewart, A. M., Herring, T. I., Chowdhury, C., and Bobik, T. A. (2020). Genetic characterization of a glycyl radical microcompartment used for 1, 2-propanediol fermentation by uropathogenic Escherichia coli CFT073. J. Bacteriol. 202. doi: 10.1128/JB.00017-20
Łusiak-Szelachowska, M., Żaczek, M., Weber-Da̧browska, B., Miêdzybrodzki, R., Kłak, M., Fortuna, W., et al. (2014). Phage neutralization by sera of patients receiving phage therapy. Viral Immunol. 27, 295–304. doi: 10.1089/vim.2013.0128
Lyytinen, O. L., Starkova, D., and Poranen, M. M. (2019). Microbial production of lipid-protein vesicles using enveloped bacteriophage phi6. Microb. Cell Fact. 18:29. doi: 10.1186/s12934-019-1079-z
Maguire, M., Coates, A. R., and Henderson, B. (2002). Chaperonin 60 unfolds its secrets of cellular communication. Cell Stress Chaperones 7, 317–329. doi: 10.1379/1466-1268(2002)007<0317:cuisoc>2.0.co;2
Mangiapane, E., Lamberti, C., Pessione, A., Galano, E., Amoresano, A., and Pessione, E. (2014). Selenium effects on the metabolism of a Se-metabolizing Lactobacillus reuteri: analysis of envelope-enriched and extracellular proteomes. Mol. Biosyst. 10, 1272–1280. doi: 10.1039/c3mb70557a
Manning, A. J., and Kuehn, M. J. (2011). Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11:258. doi: 10.1186/1471-2180-11-258
Manrique, P., Dills, M., and Young, M. J. (2017). The human gut phage community and its implications for health and disease. Viruses 9:141. doi: 10.3390/v9060141
Matos, R. C., Lapaque, N., Rigottier-Gois, L., Debarbieux, L., Meylheuc, T., Gonzalez-Zorn, B., et al. (2013). Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 9:e1003539. doi: 10.1371/journal.pgen.1003539
Mazzoli, R., and Pessione, E. (2016). The neuro-endocrinological role of microbial glutamate and GABA signaling. Front. Microbiol. 7:1934. doi: 10.3389/fmicb.2016.01934
Mazzoli, R., Pessione, E., Dufour, M., Laroute, V., Giuffrida, M. G., Giunta, C., et al. (2010). Glutamate-induced metabolic changes in Lactococcus lactis NCDO 2118 during GABA production: combined transcriptomic and proteomic analysis. Amino Acids 39, 727–737. doi: 10.1007/s00726-010-0507-5
Meijerink, M., Van Hemert, S., Taverne, N., Wels, M., De Vos, P., Bron, P. A., et al. (2010). Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One 5:e10632. doi: 10.1371/journal.pone.0010632
Mendes, R., and Raaijmakers, J. M. (2015). Cross-kingdom similarities in microbiome functions. ISME J. 9, 1905–1907. doi: 10.1038/ismej.2015.7
Miernikiewicz, P., Kłopot, A., Soluch, R., Szkuta, P., Kêska, W., Hodyra-Stefaniak, K., et al. (2016). T4 phage tail adhesin gp12 counteracts LPS-induced inflammation in vivo. Front. Microbiol. 7:1112. doi: 10.3389/fmicb.2016.01112
Mills, S., Shanahan, F., Stanton, C., Hill, C., Coffey, A., and Ross, R. P. (2013). Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 4, 4–16. doi: 10.4161/gmic.22371
Minot, S., Sinha, R., Chen, J., Li, H., Keilbaugh, S. A., Wu, G. D., et al. (2011). The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625. doi: 10.1101/gr.122705.111
Mirzaei, M. K., and Maurice, C. F. (2017). Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat. Rev. Microbiol. 15, 397–408. doi: 10.1038/nrmicro.2017.30
Mitchell, J., Siboo, I. R., Takamatsu, D., Chambers, H. F., and Sullam, P. M. (2007). Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol. Microbiol. 64, 844–857. doi: 10.1111/j.1365-2958.2007.05703.x
Muniesa, M., Hammerl, J. A., Hertwig, S., Appel, B., and Brüssow, H. (2012). Shiga toxin-producing Escherichia coli O104: H4: a new challenge for microbiology. Appl. Environ. Microbiol. 78, 4065–4073. doi: 10.1128/aem.00217-12
Munson-McGee, J. H., Snyder, J. C., and Young, M. J. (2018). Archaeal viruses from high-temperature environments. Genes 9:128. doi: 10.3390/genes9030128
Nanda, A. M., Thormann, K., and Frunzke, J. (2015). Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 197, 410–419. doi: 10.1128/jb.02230-14
Norman, J. M., Handley, S. A., and Virgin, H. W. (2014). Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology 146, 1459–1469. doi: 10.1053/j.gastro.2014.02.001
O’Brien, J., and Wright, G. D. (2011). An ecological perspective of microbial secondary metabolism. Curr. Opin. Biotechnol. 22, 552–558. doi: 10.1016/j.copbio.2011.03.010
O’Donoghue, E. J., and Krachler, A. M. (2016). Mechanisms of outer membrane vesicle entry into host cells. Cell. Microbiol. 18, 1508–1517. doi: 10.1111/cmi.12655
Oh, J. H., Alexander, L. M., Pan, M., Schueler, K. L., Keller, M. P., Attie, A. D., et al. (2019). Dietary fructose and microbiota-derived short-chain fatty acids promote bacteriophage production in the gut symbiont Lactobacillus reuteri. Cell Host Microbe 25, 273–284. doi: 10.1016/j.chom.2018.11.016
Oliveira, L., Madureira, P., Andrade, E. B., Bouaboud, A., Morello, E., Ferreira, P., et al. (2012). Group B streptococcus GAPDH is released upon cell lysis, associates with bacterial surface, and induces apoptosis in murine macrophages. PLoS One 7:e29963. doi: 10.1371/journal.pone.0029963
Ott, S. J., Waetzig, G. H., Rehman, A., Moltzau-Anderson, J., Bharti, R., Grasis, J. A., et al. (2017). Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 152, 799–811. doi: 10.1053/j.gastro.2016.11.010
Pálmer, H. G., González-Sancho, J. M., Espada, J., Berciano, M. T., Puig, I., Baulida, J., et al. (2001). Vitamin D3 promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of β-catenin signaling. J. Cell Biol. 154, 369–388. doi: 10.1083/jcb.200102028
Pessione, E. (2012). Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front. Cell. Infect. Microbiol. 2:86. doi: 10.3389/fcimb.2012.00086
Pizarro-Cerdá, J., and Cossart, P. (2004). Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat. Cell Biol. 6, 1026–1033. doi: 10.1038/ncb1104-1026
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65.
Ramírez-Puebla, S. T., Servín-Garcidueñas, L. E., Jiménez-Marín, B., Bolaños, L. M., Rosenblueth, M., Martínez, J., et al. (2013). Gut and root microbiota commonalities. Appl. Environ. Microbiol. 79, 2–9. doi: 10.1128/aem.02553-12
Records, A. R. (2011). The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol. Plant Microbe Interact. 24, 751–757. doi: 10.1094/mpmi-11-10-0262
Relman, D. A. (2008). ‘Til death do us part’: coming to terms with symbiotic relationships. Nat. Rev. Microbiol. 6, 721–724. doi: 10.1038/nrmicro1990
Ren, J., Sang, Y., Lu, J., and Yao, Y. F. (2017). Protein acetylation and its role in bacterial virulence. Trends Microbiol. 25, 768–779. doi: 10.1016/j.tim.2017.04.001
Resch, U., Tsatsaronis, J. A., Le Rhun, A., Stübiger, G., Rohde, M., Kasvandik, S., et al. (2016). A two-component regulatory system impacts extracellular membrane-derived vesicle production in group A Streptococcus. mBio 7:e00207-16.
Reyes-del Valle, J., Chávez-Salinas, S., Medina, F., and Del Angel, R. M. (2005). Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J. Virol. 79, 4557–4567. doi: 10.1128/jvi.79.8.4557-4567.2005
Ribet, D., and Cossart, P. (2010). Pathogen-mediated posttranslational modifications: a re-emerging field. Cell 143, 694–702. doi: 10.1016/j.cell.2010.11.019
Ricci, V., Chiozzi, V., Necchi, V., Oldani, A., Romano, M., Solcia, E., et al. (2005). Free-soluble and outer membrane vesicle-associated VacA from Helicobacter pylori: two forms of release, a different activity. Biochem. Biophys. Res. Commun. 337, 173–178. doi: 10.1016/j.bbrc.2005.09.035
Richter, C., Chang, J. T., and Fineran, P. C. (2012). Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) systems. Viruses 4, 2291–2311. doi: 10.3390/v4102291
Ringel, P. D., Hu, D., and Basler, M. (2017). The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep. 21, 3927–3940. doi: 10.1016/j.celrep.2017.12.020
Roach, D. R., Sjaarda, D. R., Castle, A. J., and Svircev, A. M. (2013). Host exopolysaccharide quantity and composition impacts bacteriophage pathogenesis of Erwinia amylovora. Appl. Environ. Microbiol. 79, 3249–3256. doi: 10.1128/aem.00067-13
Rosenberg, E., and Zilber-Rosenberg, I. (2016). Microbes drive evolution of animals and plants: the hologenome concept. mBio 7:e01395.
Rosselot, A. E., Hong, C. I., and Moore, S. R. (2016). Rhythm and bugs: circadian clocks, gut microbiota, and enteric infections. Curr. Opin. Gastroenterol. 32, 7–11. doi: 10.1097/mog.0000000000000227
Roux, S., Krupovic, M., Debroas, D., Forterre, P., and Enault, F. (2013). Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences. Open Biol. 3:130160. doi: 10.1098/rsob.130160
Sampson, E. M., and Bobik, T. A. (2008). Microcompartments for B12-dependent 1, 2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J. Bacteriol. 190, 2966–2971. doi: 10.1128/jb.01925-07
Samson, J. E., Magadán, A. H., Sabri, M., and Moineau, S. (2013). Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687. doi: 10.1038/nrmicro3096
Sandrini, S., Alghofaili, F., Freestone, P., and Yesilkaya, H. (2014). Host stress hormone norepinephrine stimulates pneumococcal growth, biofilm formation and virulence gene expression. BMC Microbiol. 14:180. doi: 10.1186/1471-2180-14-180
Schmeck, B., Beermann, W., van Laak, V., Zahlten, J., Opitz, B., Witzenrath, M., et al. (2005). Intracellular bacteria differentially regulated endothelial cytokine release by MAPK-dependent histone modification. J. Immunol. 175, 2843–2850. doi: 10.4049/jimmunol.175.5.2843
Schnappinger, D., Ehrt, S., Voskuil, M. I., Liu, Y., Mangan, J. A., Monahan, I. M., et al. (2003). Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198, 693–704. doi: 10.1084/jem.20030846
Schwechheimer, C., and Kuehn, M. J. (2015). Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619. doi: 10.1038/nrmicro3525
Segain, J. P., De La Blétiere, D. R., Bourreille, A., Leray, V., Gervois, N., Rosales, C., et al. (2000). Butyrate inhibits inflammatory responses through NFκB inhibition: implications for Crohn’s disease. Gut 47, 397–403. doi: 10.1136/gut.47.3.397
Serban, D. E. (2014). Gastrointestinal cancers: influence of gut microbiota, probiotics and prebi-otics. Cancer Lett. 345, 258–270. doi: 10.1016/j.canlet.2013.08.013
Servin, A. L. (2004). Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28, 405–440. doi: 10.1016/j.femsre.2004.01.003
Silpe, J. E., and Bassler, B. L. (2019). A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell 176, 268–280. doi: 10.1016/j.cell.2018.10.059
Soufi, B., Soares, N. C., Ravikumar, V., and Macek, B. (2012). Proteomics reveals evidence of cross-talk between protein modifications in bacteria: focus on acetylation and phosphorylation. Curr. Opin. Microbiol. 15, 357–363. doi: 10.1016/j.mib.2012.05.003
Srinivasiah, S., Bhavsar, J., Thapar, K., Liles, M., Schoenfeld, T., and Wommack, K. E. (2008). Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res. Microbiol. 159, 349–357. doi: 10.1016/j.resmic.2008.04.010
Sungur, T., Aslim, B., Karaaslan, C., and Aktas, B. (2017). Impact of Exopolysaccharides (EPSs) of Lactobacillus gasseri strains isolated from human vagina on cervical tumor cells (HeLa). Anaerobe 47, 137–144. doi: 10.1016/j.anaerobe.2017.05.013
Sutter, M., Boehringer, D., Gutmann, S., Günther, S., Prangishvili, D., Loessner, M. J., et al. (2008). Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 15, 939–947. doi: 10.1038/nsmb.1473
Szmelcman, S., and Hofnung, M. (1975). Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J. Bacteriol. 124, 112–118. doi: 10.1128/jb.124.1.112-118.1975
Takahashi, K., Sugi, Y., Nakano, K., Tsuda, M., Kurihara, K., Hosono, A., et al. (2011). Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J. Biol. Chem. 286, 35755–35762. doi: 10.1074/jbc.m111.271007
Tateda, K., Ishii, Y., Horikawa, M., Matsumoto, T., Miyairi, S., Pechere, J. C., et al. (2003). The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect. Immun. 71, 5785–5793. doi: 10.1128/iai.71.10.5785-5793.2003
Thingstad, T. F., Pree, B., Giske, J., and Våge, S. (2015). What difference does it make if viruses are strain-, rather than species-specific? Front. Microbiol. 6:320. doi: 10.3389/fmicb.2015.00320
Tikhonenko, A. S., Belyaeva, N. N., and Ivanovics, G. (1975). Electron microscopy of phages liberated by megacin A producing lysogenic Bacillus megaterium strains. Acta Microbiol. Acad. Sci. Hung. 22, 58–59.
Toyofuku, M., Cárcamo-Oyarce, G., Yamamoto, T., Eisenstein, F., Hsiao, C. C., Kurosawa, M., et al. (2017a). Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 8:481.
Toyofuku, M., Morinaga, K., Hashimoto, Y., Uhl, J., Shimamura, H., Inaba, H., et al. (2017b). Membrane vesicle-mediated bacterial communication. ISME J. 11, 1504–1509. doi: 10.1038/ismej.2017.13
Toyofuku, M., Tashiro, Y., Nomura, N., and Eberl, L. (2020). “Functions of MVs in Inter-bacterial communication,” in Bacterial Membrane Vesicles, eds M. Kaparakis-Liaskos and T. Kufer (Cham: Springer), 101–117. doi: 10.1007/978-3-030-36331-4_5
Turnbull, L., Toyofuku, M., Hynen, A. L., Kurosawa, M., Pessi, G., Petty, N. K., et al. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7:11220.
van Belleghem, J. D., Da̧browska, K., Vaneechoutte, M., Barr, J. J., and Bollyky, P. L. (2019). Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses 11:10. doi: 10.3390/v11010010
Van Hemert, S., Meijerink, M., Molenaar, D., Bron, P. A., de Vos, P., Kleerebezem, M., et al. (2010). Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol. 10:293. doi: 10.1186/1471-2180-10-293
Vdovikova, S., Gilfillan, S., Wang, S., Dongre, M., Wai, S. N., and Hurtado, A. (2018). Modulation of gene transcription and epigenetics of colon carcinoma cells by bacterial membrane vesicles. Sci. Rep. 8:7434.
Vdovikova, S., Luhr, M., Szalai, P., Nygård Skalman, L., Francis, M. K., Lundmark, R., et al. (2017). A novel role of Listeria monocytogenes membrane vesicles in inhibition of autophagy and cell death. Front. Cell. Infect. Microbiol. 7:154. doi: 10.3389/fcimb.2017.00154
Vettiger, A., and Basler, M. (2016). Type VI secretion system substrates are transferred and reused among sister cells. Cell 167, 99–110. doi: 10.1016/j.cell.2016.08.023
Vidakovic, L., Singh, P. K., Hartmann, R., Nadell, C. D., and Drescher, K. (2018). Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat. Microbiol. 3, 26–31. doi: 10.1038/s41564-017-0050-1
Vieira, A. T., Galvão, I., Macia, L. M., Sernaglia, E. M., Vinolo, M. A. R., Garcia, C. C., et al. (2017). Dietary fiber and the short-chain fatty acid acetate promote resolution of neutrophilic inflammation in a model of gout in mice. J. Leukoc. Biol. 101, 275–284. doi: 10.1189/jlb.3a1015-453rrr
Vikström, E., Bui, L., Konradsson, P., and Magnusson, K. E. (2010). Role of calcium signalling and phosphorylations in disruption of the epithelial junctions by Pseudomonas aeruginosa quorum sensing molecule. Eur. J. Cell Biol. 89, 584–597. doi: 10.1016/j.ejcb.2010.03.002
Villarreal, L. P. (2012). “The addiction module as a social force,” in Viruses: Essential Agents of Life, ed. G. Witzany (Dordrecht: Springer), 107–145. doi: 10.1007/978-94-007-4899-6_6
Vipperla, K., and O’keefe, S. J. (2012). The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr. Clin. Pract. 27, 624–635. doi: 10.1177/0884533612452012
Vollenweider, S., and Lacroix, C. (2004). 3-Hydroxypropionaldehyde: applications and perspectives of biotechnological production. Appl. Microbiol. Biotechnol. 64, 16–27. doi: 10.1007/s00253-003-1497-y
Wai, S. N., Lindmark, B., Söderblom, T., Takade, A., Westermark, M., Oscarsson, J., et al. (2003). Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115, 25–35. doi: 10.1016/s0092-8674(03)00754-2
Wampler, J. L., Kim, K. P., Jaradat, Z., and Bhunia, A. K. (2004). Heat shock protein 60 acts as a receptor for the Listeria adhesion protein in Caco-2 cells. Infect. Immun. 72, 931–936. doi: 10.1128/iai.72.2.931-936.2004
Wandro, S. J. (2019). Evolution of Bacteriophages Infecting Enterococcus from the Human Microbiome. Ph.D. dissertation, University of California Irvine, Irvine, CA.
Wang, J., Ji, H., Wang, S., Liu, H., Zhang, W., Zhang, D., et al. (2018). Probiotic Lactobacillus plantarum promotes intestinal barrier function by strengthening the epithelium and modulating gut microbiota. Front. Microbiol. 9:1953. doi: 10.3389/fmicb.2018.01953
Wang, W., and Jeffery, C. J. (2016). An analysis of surface proteomics results reveals novel candidates for intracellular/surface moonlighting proteins in bacteria. Mol. Biosyst. 12, 1420–1431. doi: 10.1039/c5mb00550g
Wang, X., Kim, Y., Ma, Q., Hong, S. H., Pokusaeva, K., Sturino, J. M., et al. (2010). Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1:147.
Wang, Y., Wu, Y., Wang, Y., Xu, H., Mei, X., Yu, D., et al. (2017). Antioxidant properties of probiotic bacteria. Nutrients 9:521. doi: 10.3390/nu9050521
Waśko, A., Polak-Berecka, M., Paduch, R., and Jóźwiak, K. (2014). The effect of moonlighting proteins on the adhesion and aggregation ability of Lactobacillus helveticus. Anaerobe 30, 161–168. doi: 10.1016/j.anaerobe.2014.10.002
Webster, N. S. (2014). Cooperation, communication, and co-evolution: grand challenges in microbial symbiosis research. Front. Microbiol. 5:164. doi: 10.3389/fmicb.2014.00164
Yang, C. K., Ewis, H. E., Zhang, X., Lu, C. D., Hu, H. J., Pan, Y., et al. (2011). Nonclassical protein secretion by Bacillus subtilis in the stationary phase is not due to cell lysis. J. Bacteriol. 193, 5607–5615. doi: 10.1128/jb.05897-11
Yoshida, N., Oeda, K., Watanabe, E., Mikami, T., Fukita, Y., Nishimura, K., et al. (2001). Chaperonin turned insect toxin. Nature 411, 44–44. doi: 10.1038/35075148
Zavan, L., Bitto, N. J., and Kaparakis-Liaskos, M. (2020). “Introduction, History, and Discovery of Bacterial Membrane Vesicles,” in Bacterial Membrane Vesicles: Biogenesis, Functions and Applications, eds M. Kaparakis-Liaskos and T. A. Kufer (Cham: Springer), 1–21. doi: 10.1007/978-3-030-36331-4_1
Zdziarski, J., Brzuszkiewicz, E., Wullt, B., Liesegang, H., Biran, D., Voigt, B., et al. (2010). Host imprints on bacterial genomes—rapid, divergent evolution in individual patients. PLoS Pathog. 6:e1001078. doi: 10.1371/journal.ppat.1001078
Zhang, X., McDaniel, A. D., Wolf, L. E., Keusch, G. T., Waldor, M. K., and Acheson, D. W. (2000). Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181, 664–670. doi: 10.1086/315239
Zhou, M., and Wu, H. (2009). Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 155, 317–327. doi: 10.1099/mic.0.025221-0
Keywords: phage–bacteria–human host, multitasking signals, evolution, partnership agreement, viral-like particles, moonlight proteins, PTMs (post-translational modifications), MVs (membrane vesicles)
Citation: Pessione E (2020) The Russian Doll Model: How Bacteria Shape Successful and Sustainable Inter-Kingdom Relationships. Front. Microbiol. 11:573759. doi: 10.3389/fmicb.2020.573759
Received: 17 June 2020; Accepted: 31 August 2020;
Published: 20 October 2020.
Edited by:
Shyam Sundar Paul, Central Institute for Research on Buffaloes (ICAR), IndiaReviewed by:
Lorena Ruiz, Institute of Dairy Products of Asturias (IPLA), SpainChangyi Zhang, University of Illinois at Urbana–Champaign, United States
Copyright © 2020 Pessione. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Enrica Pessione, ZW5yaWNhLnBlc3Npb25lQHVuaXRvLml0
 Enrica Pessione
Enrica Pessione