- 1Institute of Fungus Resources, Department of Ecology, College of Life Sciences, Guizhou University, Guiyang, China
- 2Department of Microbiology, Guiyang College of Traditional Chinese Medicine, Guiyang, China
- 3Key Laboratory of Plant Resource Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), Guizhou University, Guiyang, China
- 4Engineering Research Center of Industrial Microbiology, Ministry of Education, Fujian Normal University, Fuzhou, China
The family Thelebolaceae belongs to the order Thelebolales, class Leotiomycetes, and contains 22 genera. In this study, we introduce a new genus Solomyces gen. nov. in the family Thelebolaceae, which is supported by morphological observation and multilocus-based [internal transcribed spacers (ITS) + LSU and ITS + LSU+ MCM7+ EF1A+ RPB2] phylogenetic analysis. Maximum-likelihood and Bayesian phylogenetic inference analyses indicated that Solomyces is a distinct genus within this family. The new genus is compared against related Thelebolaceae genera, and its description and illustration are provided. This genus comprises one new species and one unnamed species (including two strains). We also report the addition of four new species – Pseudogymnoascus shaanxiensis, Pseudogymnoascus guizhouensis, Pseudogymnoascus sinensis, and Geomyces obovatus – in the family Thelebolaceae and present their morphological and phylogenetic characterizations.
Introduction
The class Leotiomycetes (Pezizomycotina) was erected by Eriksson and Winka (1997) to accommodate inoperculate discomycetes. Fungi in the class Leotiomycetes are ecologically diverse and include mycorrhizas, root and leaf endophytes, plant pathogens, aquatic and aeroaquatic hyphomycetes, mammalian pathogens, and saprobes (Johnston et al., 2019). Leotiomycetes currently comprises 19 orders and order-level clades, 54 families, and 13 family level clades (Ekanayaka et al., 2019; Karunarathna et al., 2020). Previously, however, this class included a wide range of taxa based on traditional morphological taxonomy (Korf, 1973; Spooner, 1987), and the current classification of Leotiomycetes is still largely based on morphologically defined taxa, especially at higher taxonomic levels (Johnston et al., 2019). Nevertheless, sexual or asexual morphs of many Leotiomycetes taxa are not recorded, and a few links between sexual and asexual morphs in Leotiomycetes have been confirmed (Sutton and Hennebert, 1995; Sati and Pathak, 2016; Ekanayaka et al., 2017; Johnston et al., 2019). Thelebolales, an order in the class Leotiomycetes, consists of Thelebolaceae and the Alatospora–Miniancora clade, in which some genera (e.g., Caccobius, Coprobolus, and Leptokalpion) are erected based on their sexual, but not asexual, morphology (Wijayawardene et al., 2017).
Haeckel (1894) introduced the order Thelebolales; however, the taxonomy of this order has long been contentious. Some researchers believed that the order contained only one family (De Hoog et al., 2005), whereas others suggested that at least two families were involved (Ekanayaka et al., 2019; Johnston et al., 2019; Batista et al., 2020). The family Thelebolaceae is important in the order Thelebolales because of several species that can produce antifreeze proteins, ice-binding proteins, and some secondary metabolites with potential application values that offer valuable resources for biotechnological exploitation (Batista et al., 2020); the family was introduced by Eckblad (1968) and typified by the genus Thelebolus with Thelebolus stercoreus as the type species. Members of this family are characterized by absent, apothecial, or cleistothecial ascomata (Van Brummelen, 1985; Stchigel et al., 2001; De Menezes et al., 2017; Ekanayaka et al., 2019). During an extensive diversity survey of Geomyces and allied genera in China, we gathered a collection of fungal isolations. In this study, we introduced their morphological, cultural, and phylogenetic characterization and propose one new genus and five new species.
Materials and Methods
Isolates and Morphology
Soil samples were obtained from Dali City, Yunnan Province; Guiyang City, Guizhou Province; Xi’an and Hanzhong City, Shaanxi Province; and Yichang City, Hubei Province, China. Samples were treated according to the method described by Zhang et al. (2019). Fungi were isolated using a modified baiting technique with chicken feathers as the substrate (Vanbreuseghem, 1952). The feathers were washed, thoroughly rinsed with distilled water, dried, cut into 2-cm fragments, and autoclaved. Plates containing soil material and sterile feathers were incubated at room temperature for 1 month. Fungi were isolated and purified using a conventional dilution technique described by Zhang et al. (2019), as follows: 2 g of soil sample was suspended in 9 ml of distilled water, and the prepared suspension was vortexed, diluted to 1:10,000, and cultured on Sabouraud’s dextrose agar (SDA; 10 g of peptone, 40 g of dextrose, 20 g of agar, 1 L of ddH2O) supplemented with chloramphenicol and cycloheximide. The plates were incubated at 25°C until fungal colony growth was observed. The axenic strains were then transferred to potato dextrose agar (PDA; Shanghai Bio-way Technology Co., Ltd., China) plates for purification. Dried holotypes were deposited in the Mycological Herbarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), or the Institute of Fungus Resources, Guizhou University (GZUIFR, formally the Herbarium of Guizhou Agricultural College; code, GZAC). Ex-type strains and other strains were deposited in the China General Microbiological Culture Collection Center (CGMCC) or the GZUIFR.
The pure strains were incubated on PDA at 25°C for 14 days in the dark to determine the macroscopic characteristics, diameters, and colony colors (surface and reverse). The characterization and measurement of fungal microscopic characteristics were performed in 25% lactic acid. Images were obtained using an optical microscope (OM, DM4 B, Leica, Germany) with differential interference contrast (DIC). The taxonomic descriptions and names of the new taxa were introduced into MycoBank1 and Faces of Fungi2.
DNA Extraction, PCR Amplification, and Sequencing
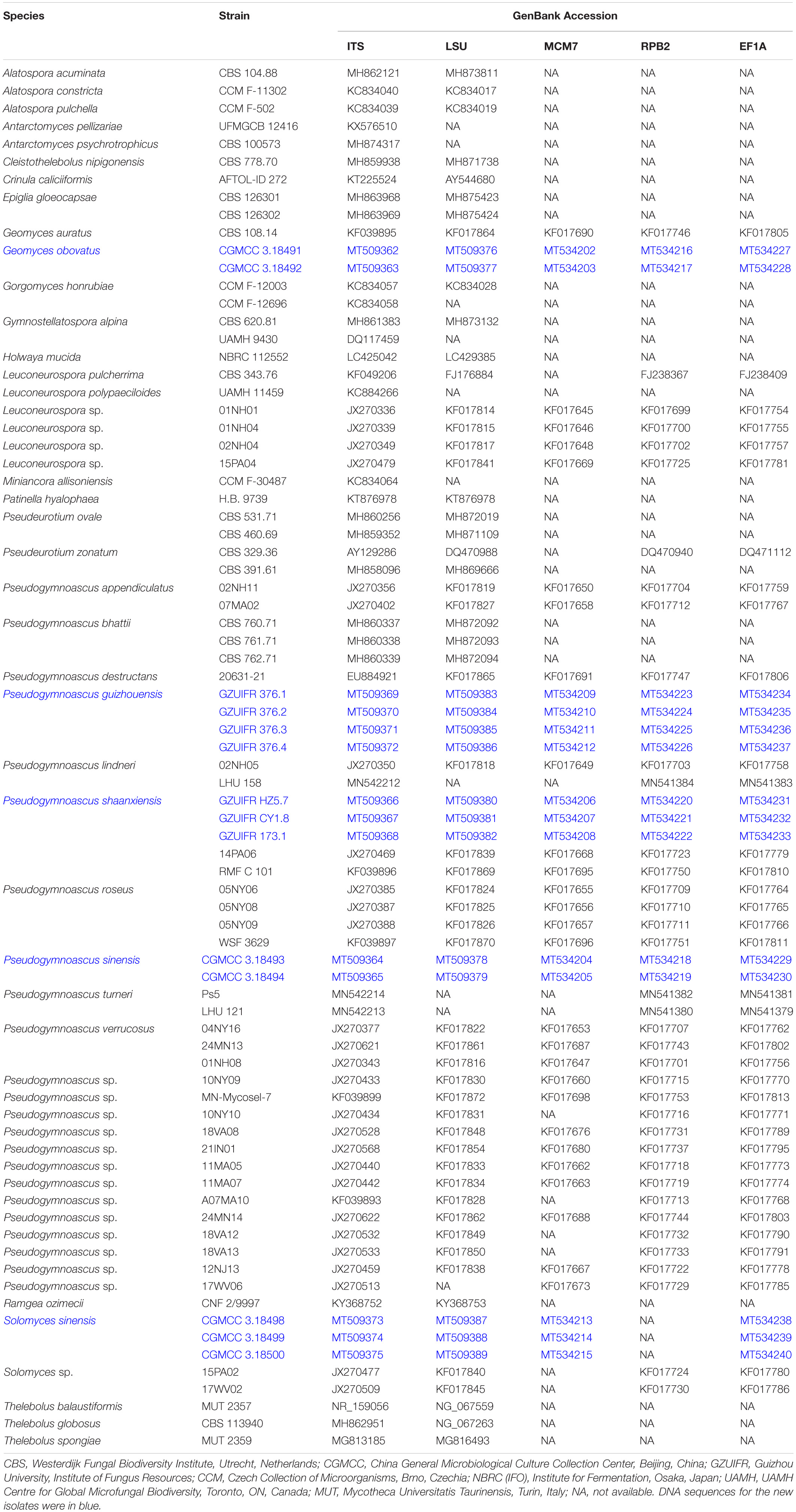
Total DNA was extracted using the BioTeke Fungus Genomic DNA Extraction kit (DP2032, BioTeke, Beijing, China) following the manufacturer’s instructions. Total DNA was used for the amplification and sequencing of five fragments: ribosomal internal transcribed spacers (ITS; primers ITS1/ITS4) (White et al., 1990), the 28S LSU rRNA gene (primers LROR/LR7) (Vilgalys and Hester, 1990; Vilgalys and Sun, 1999), translation elongation factor 1 alpha (EF1A; primers 983F/2218R) (Rehner and Buckley, 2005), RNA polymerase II subunit 2 (RPB2; primers fRPB2-7cF/RPB2-3053b) (Liu et al., 1999; Reeb et al., 2004), and minichromosomal maintenance protein 7 (MCM7; primers MCM7-709/MCM7-1348) (Schoch et al., 2009). The PCR products were sequenced with the abovementioned primers at a commercial sequencing service provider (Shanghai Sangon Biological Engineering Technology & Services Co., Shanghai, China) in an ABI 3730xl DNA Analyzer, using the Sanger method. The consensus sequences were obtained using the SeqMan software v. 7 (DNASTAR Lasergene, Madison, WI, United States). Sequences generated in this study were deposited in GenBank (Table 1).
Phylogenetic Analyses
Sequence data were selected from data previously published by Crous et al. (2019), Ekanayaka et al. (2019), Johnston et al. (2019), and Minnis and Lindner (2013) and then downloaded from GenBank for molecular phylogenetic analyses (Table 1). Multiple sequence alignments for ITS, LSU, MCM7, RPB2, and EF1A were carried out using MAFFT v7.037b (Katoh and Standley, 2013). Sequence editing was performed with MEGA6 (Tamura et al., 2013). The concatenated ITS and LSU, and ITS, LSU, MCM7, RPB2, and EF1A sequences were assembled by SequenceMatrix v.1.7.8 (Vaidya et al., 2011). Gene concordance was assessed with the “hompart” command in PAUP4.0b10 (Swofford, 2002).
Sequence alignment and maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic analyses were performed according to the methodology described by Zhang et al. (2019). Maximum-likelihood (ML) analyses were constructed with IQ-TREE v. 1.6.11 (Nguyen et al., 2015). The best-fit model of substitution for each locus was estimated using IQ-TREE’s ModelFinder function (Kalyaanamoorthy et al., 2017) under the Bayesian information criterion (BIC). Bootstrap analysis was performed using the ultrafast bootstrap approximation (Minh et al., 2013) with 1,000 replicates, and bootstrap support (BS) ≥90% was considered as statistically significant. For Bayesian inference (BI), a Markov chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v.3.2 (Ronquist et al., 2012) for the combined sequence datasets. The selection of the best-fit nucleotide substitution model for each locus was calculated by the Akaike information criterion (AIC) with ModelTest v.3.7 (Posada and Crandall, 1998). After the BI analyses, both runs were examined with Tracer v.1.5 (Drummond and Rambaut, 2007) to determine burn-in and check for convergence.
Two multilocus phylogenies were analyzed to evaluate the various generic placements and establish the phylogenetic relationships in Thelebolaceae. The genus placement was evaluated based on a concatenated ITS and LSU dataset. This phylogeny was constructed to assess if Solomyces is a well-delimited genus. The second multilocus phylogeny was based on a concatenated alignment of ITS, LSU, MCM7, RPB2, and EF1A sequences and included 49 isolates of Geomyces and its allied genera. This analysis was performed to evaluate the generic boundaries and species groupings within the genera Solomyces, Geomyces, and Pseudogymnoascus. All the trees were displayed in FigTree 1.4.23 and edited in Microsoft Paint. DNA alignments have been deposited at TreeBASE.
Results
Phylogenetic Analyses
The first concatenated matrix contains 1,220 nucleotides, i.e., 445 from the ITS and 775 from the LSU. The second concatenated alignment contained 2,808 nucleotides (ITS: 434, LSU: 806, MCM7: 503, RPB2: 437, and EF1A: 628). The best-fit evolutionary model for each locus in the two datasets is listed in Table 2. The tree topology of the Bayesian inference agrees with that of the ML tree (Figures 1, 2). The phylogenies indicated that each genus clusters into a monophyletic clade (Figure 1). In the phylogenetic tree, the new genus Solomyces forms a well-supported (1 BYPP/100 MLBS) clade separated from other genera in Thelebolaceae (Figure 1). Solomyces sinensis represents a separate subclade and is located within the new genus near another unnamed species (includes two strains, 15PA02 and 17WV02, from Hibernacular soil). Furthermore, all the new species of Pseudogymnoascus and Geomyces are placed in a distinct branch.
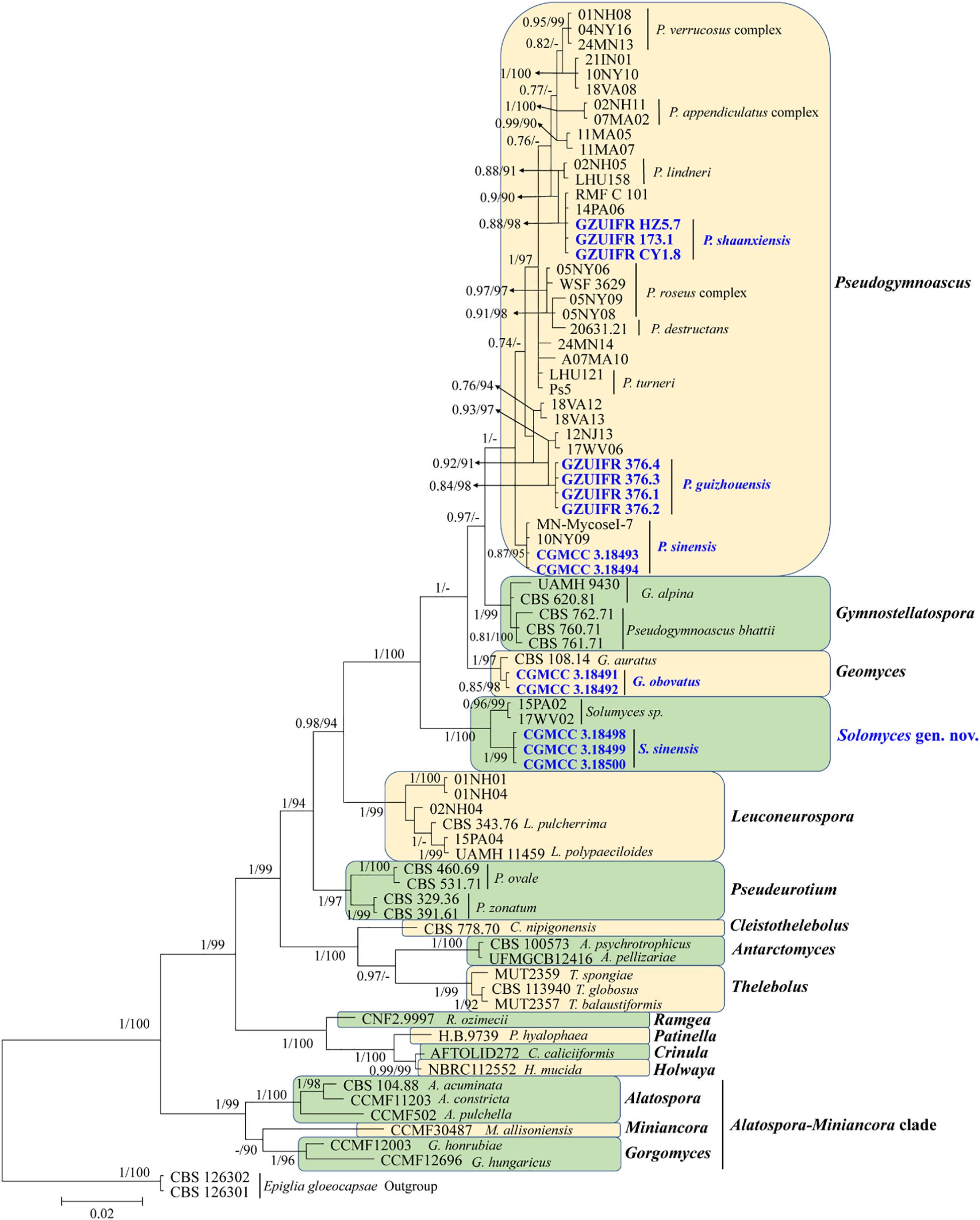
Figure 1. Phylogenetic relationships among the genera in Thelebolales based on Bayesian inference and maximum-likelihood analyses of the concatenated internal transcribed spacers (ITS) and LSU dataset. Posterior probabilities ≥0.7 and maximum-likelihood bootstrap values ≥90% are shown above the internal branches. The new isolates are presented in bold and blue.
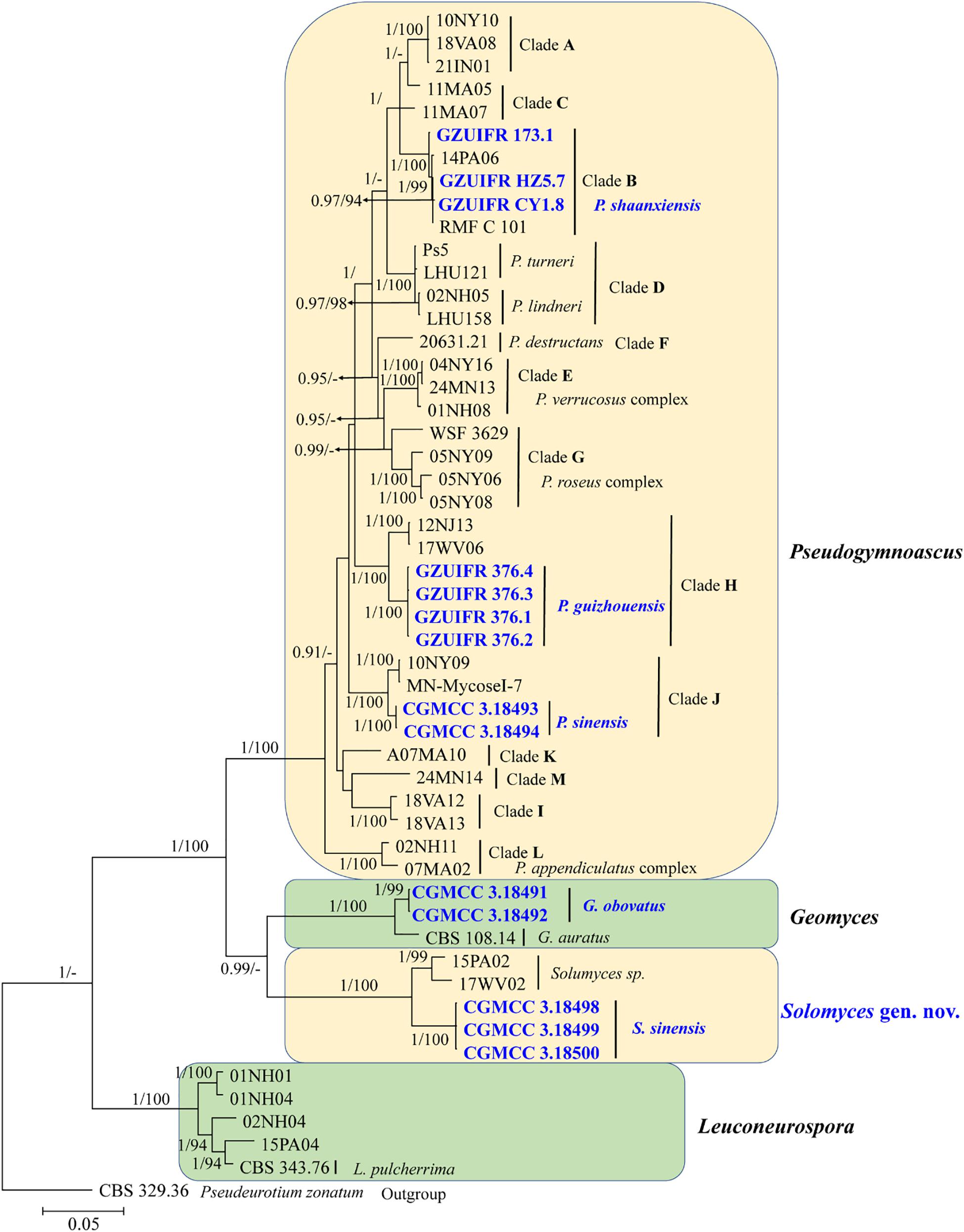
Figure 2. The consensus tree from the Bayesian inference and Geomyces and its allied genera based on the ITS, LSU, MCM7, RPB2, and EF1A concatenated dataset. Posterior probabilities ≥0.7 and maximum-likelihood bootstrap values ≥90% are shown above the internal branches. The new isolates are presented in bold and blue.
Taxonomy
In this section, Solomyces Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, gen. nov. and five new species – S. sinensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.; Geomyces obovatus Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.; Pseudogymnoascus guizhouensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.; Pseudogymnoascus shaanxiensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.; and Pseudogymnoascus sinensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov. – are described and illustrated. We also propose a new combination, Gymnostellatospora bhattii (Samson) Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, comb. nov. based on phylogenetic analyses.
Geomyces obovatus Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.
MycoBank number: MB 835718, Facesoffungi number: FoF 08691 (Figure 3).
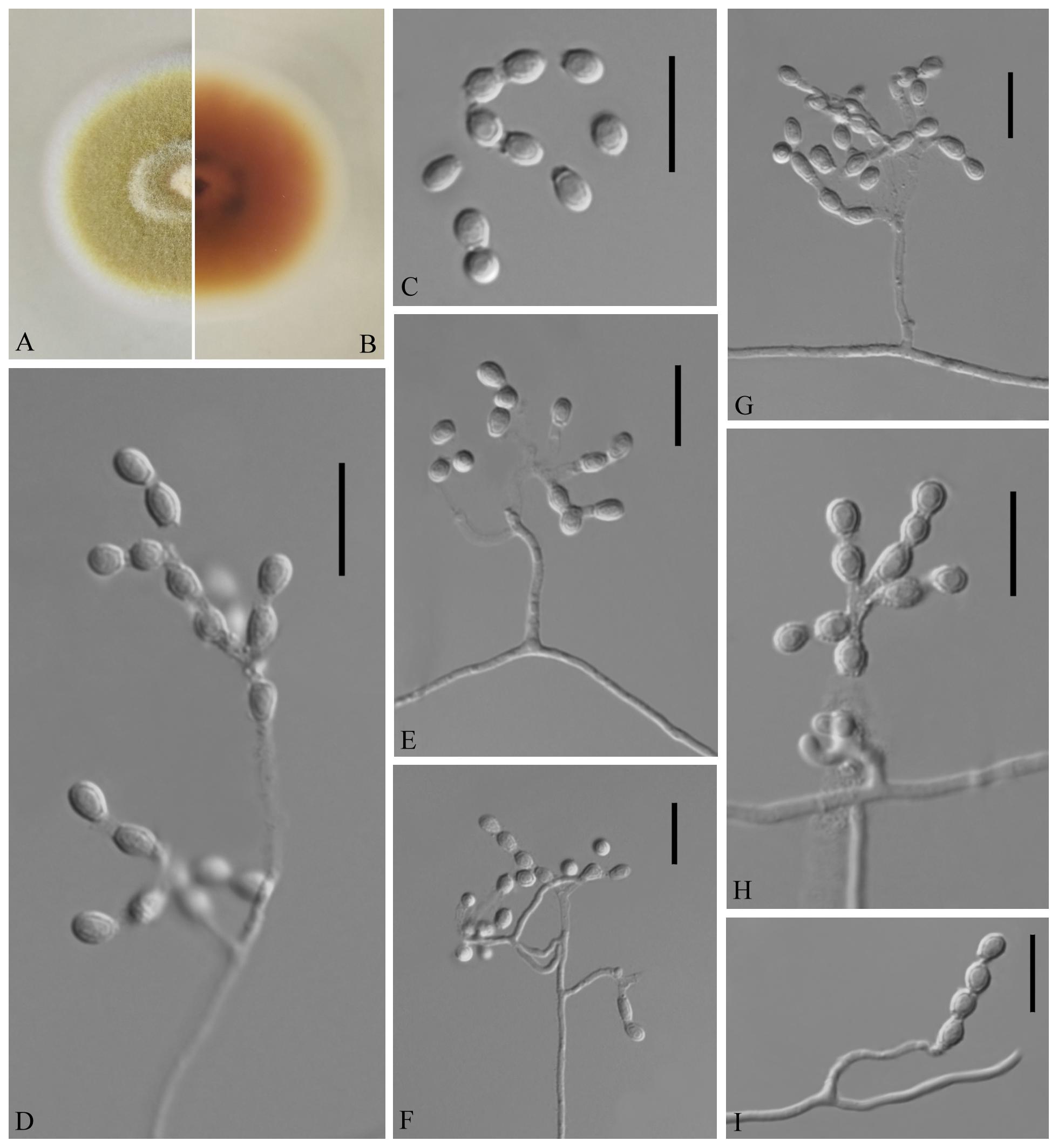
Figure 3. Geomyces obovatus [from ex-holotype China General Microbiological Culture Collection Center (CGMCC) 3.18491]. (A,B) The front and reverse of a G. obovatus colony on potato dextrose agar (PDA) after 14 days at 25°C. (C–I) Conidiophores and conidia. Scale bars = 10 μm.
Etymology: Referring to the obovoid conidia.
Holotype: permanently preserved in a metabolically inactive state, GZAC 20.8.
Description based on GZAC 20.8. Asexual: Colonies on PDA, reaching 28–30 mm in diameter after 14 days at 25°C, slightly felty to floccose, margin identified, khaki at center, and white at edge; reverse hazel. Aerial mycelium abundant, smooth and thin walled, septate, 0.5- to 2-μm wide. Racquet hyphae absent. Conidia abundant, normally terminal and intercalary conidia in a series of 1–4, rarely solitary on short stalk, smooth or echinulate, 3.0–5.0 × 2.5–4.0 μm (av. = 3.5 × 2.5 μm, n = 50). Terminal conidia obovoid, rarely subglobose; intercalary conidia alternate and pyriform, barrel or irregularly shaped.
Sexual morph: not observed.
Geographical distribution: China.
Material examined: China, Guizhou, Guiyang, Qianlingshan Park, 26°60′N, 106°69′E, from soil beside a road, September 2016, Zhi. Y. Zhang (GZAC 20.8 – holotype; CGMCC 3.18491 = GZUIFR QL20.8.1 – ex-type living cultures; ibid., CGMCC 3.18492 = GZUIFR QL20.8.2). The living cultures were kept in sterile 30% glycerol and deposited in a −80°C freezer.
Notes: The Geomyces was introduced by Traaen (1914), and several species have subsequently been described. Currently, the genus contains seven species (Traaen, 1914; Dal Vesco, 1957; Sigler and Carmichael, 1976; Hocking and Pitt, 1988; Li and Cui, 1989; Luo et al., 2016; Chen et al., 2017): Geomyces auratus, Geomyces asperulatus, Geomyces vinaceus, Geomyces pulvereus, Geomyces laevis, Geomyces guiyangensis, and Geomyces fujianensis. G. asperulatus and G. vinaceus likely belong to Pseudogymnoascus (Minnis and Lindner, 2013); G. pulvereus and G. laevis do not have any available sequence data; G. pulvereus lacks intercalary conidia (Hocking and Pitt, 1988), and G. laevis has oblong-elliptical or barrel-shaped intercalary conidia (Li and Cui, 1989). Only ITS sequences are available for G. guiyangensis and G. fujianensis, and the ITS data for CGMCC 3.18491 (the type strain of G. obovatus) show similarity to those of G. guiyangensis (strain G014512) (465/509; 91%, with 4 gaps) and G. fujianensis (strain G242) (464/526; 88%, with 11 gaps); however, the latter two species have no intercalary conidia (Luo et al., 2016; Chen et al., 2017). The proposed new species, G. obovatus, is morphologically and phylogenetically related to G. auratus, but they can be distinguished by their ITS (512/524; 97%, with three gaps), LSU (931/938; 99%, no gaps), MCM7 (592/619; 95%, no gaps), RPB2 (696/717; 97%, no gaps), and EF1A (843/855; 98%, with one gap) sequence data.
Gymnostellatospora bhattii (Samson) Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, comb. nov.
Basionym: Pseudogymnoascus bhattii Samson, Acta Botanica Neerlandica 21: 519. 1972.
Notes: This species was erected by Samson (1972) based on morphological characteristics. However, our phylogenetic analysis based on Thelebolaceae ITS and LSU data indicated that this species was closely related to Gymnostellatospora and separated from Pseudogymnoascus. Based on morphological characteristics, it was difficult to distinguish the closely related species, and even the genera, through traditional taxonomy, and modern phylogenetic methods were a very important adjunct. We therefore transferred P. bhattii to Gymnostellatospora and named the species G. bhattii.
Pseudogymnoascus guizhouensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.
MycoBank number: MB 835716, Facesoffungi number: FoF 08692 (Figure 4).
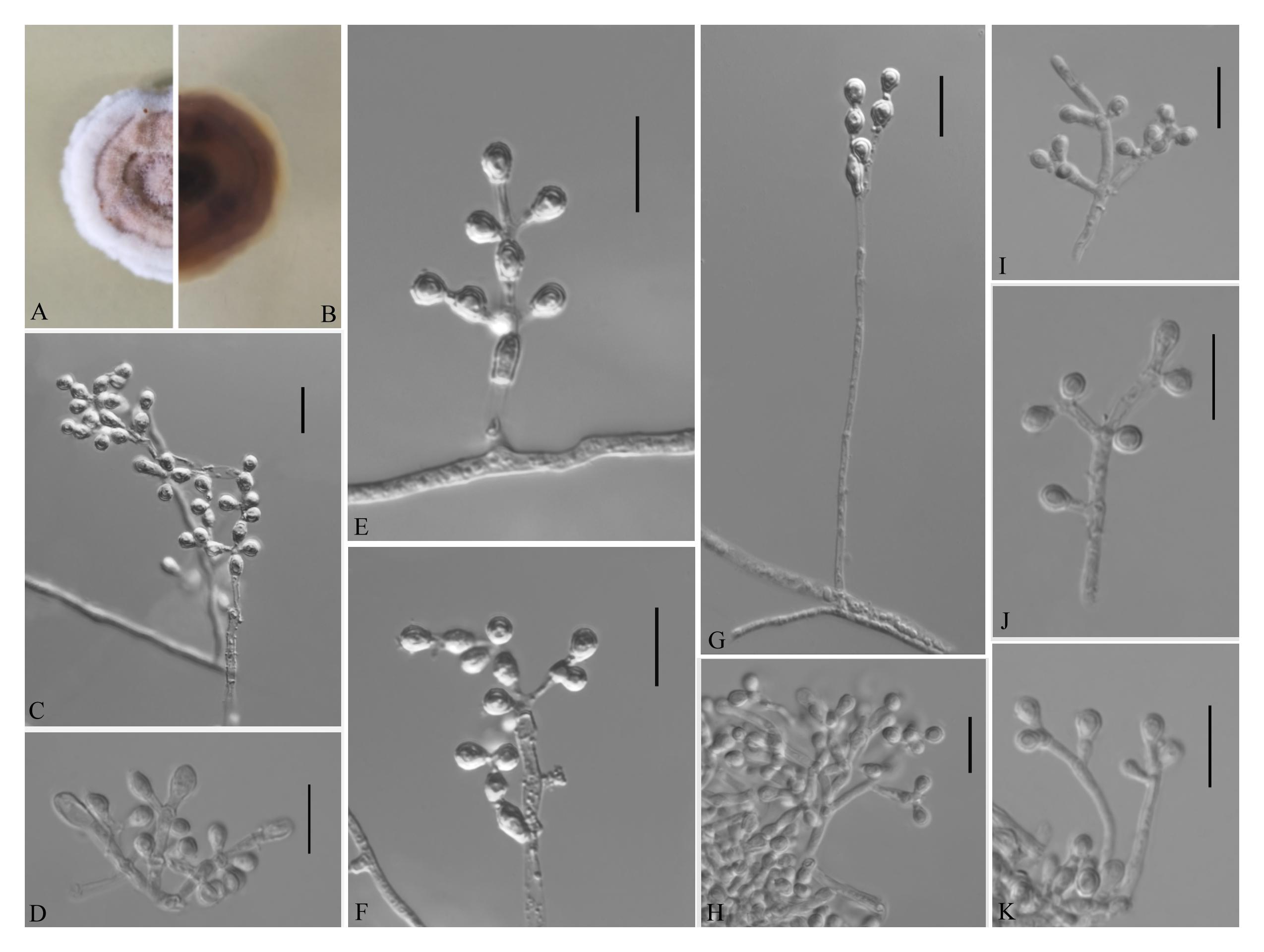
Figure 4. Pseudogymnoascus guizhouensis [from ex-holotype Institute of Fungus Resources, Guizhou University (GZUIFR) 376.1]. (A,B) The front and reverse of a P. guizhouensis colony on PDA after 14 days at 25°C. (C–K) Conidiophores and conidia. Scale bars = 10 μm.
Etymology: Refers to the region from which the fungus was isolated.
Holotype: permanently preserved in a metabolically inactive state, GZAC 376.
Description based on GZAC 376. Asexual: Colonies on PDA, reaching 20–23 mm in diameter after 14 days at 25°C, elevate, powdery, floccose, margin identified, locally indented, pale purple at center and white at the edge, producing a clear exudate; reverse brown. Aerial mycelium abundant, smooth and thin walled, septate, 1.5- to 3.0-μm wide. Racquet hyphae absent. Conidia normally borne on verticillate branches, terminal, intergrading with intercalary conidia, sometimes borne laterally and solitary on hyphae, smooth walled or echinulate, obovoid, pyriform, subglobose or clavate, 3.0–5.5 × 3.0–3.5 μm (av. = 4.0 × 3.0 μm, n = 50). Intercalary conidia are borne on the outer branches of the hyphae or verticillate hyphae, alternate, in series of 1–2, smooth walled or echinulate, cuneiform, barrel shaped, 3.5–6.0 × 3.0–3.5 μm (av. = 4.0 × 3.0 μm, n = 50).
Sexual morph: not observed.
Geographical distribution: China.
Material examined: China, Guizhou, Guiyang, Guizhou University, 26°45′N, 106°67′E, from epiphytic soil of Cinnamomum camphora, June 2019, Zhi. Y. Zhang (GZAC 376 – holotype; GZUIFR 376.1 – ex-type living culture; ibid., GZUIFR 376.2; ibid., GZUIFR 376.3; ibid., GZUIFR 376.4). The living cultures were kept in sterile 30% glycerol and deposited in a −80°C freezer.
Notes: Morphologically, Pseudogymnoascus guizhouensis is similar to Pseudogymnoascus linderi, Pseudogymnoascus turneri (both isolated from sediment), and Pseudogymnoascus destructans (isolated from a wing of a small brown bat, Myotis lucifugus), based on the obovoid and subglobose conidia. However, P. guizhouensis differs from P. linderi and P. turneri as it has cuneiform, barrel-shaped intercalary conidia (the intercalary conidia of P. linderi and P. turneri are globose to truncate) (Crous et al., 2019). P. guizhouensis can be distinguished from P. destructans by the size of conidia and intercalary conidia (4.0 × 3.0 μm vs. 5.0–12.0 × 2.0–3.5 μm, respectively) (Gargas et al., 2009). Phylogenetically, our isolates GZUIFR 376.1, GZUIFR 376.2, GZUIFR 376.3, and GZUIFR 376.4 cluster together very well and form a single clade separated from other Pseudogymnoascus species (Figures 1, 2).
Pseudogymnoascus shaanxiensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.
MycoBank number: MB 835715, Facesoffungi number: FoF 08689 (Figure 5).
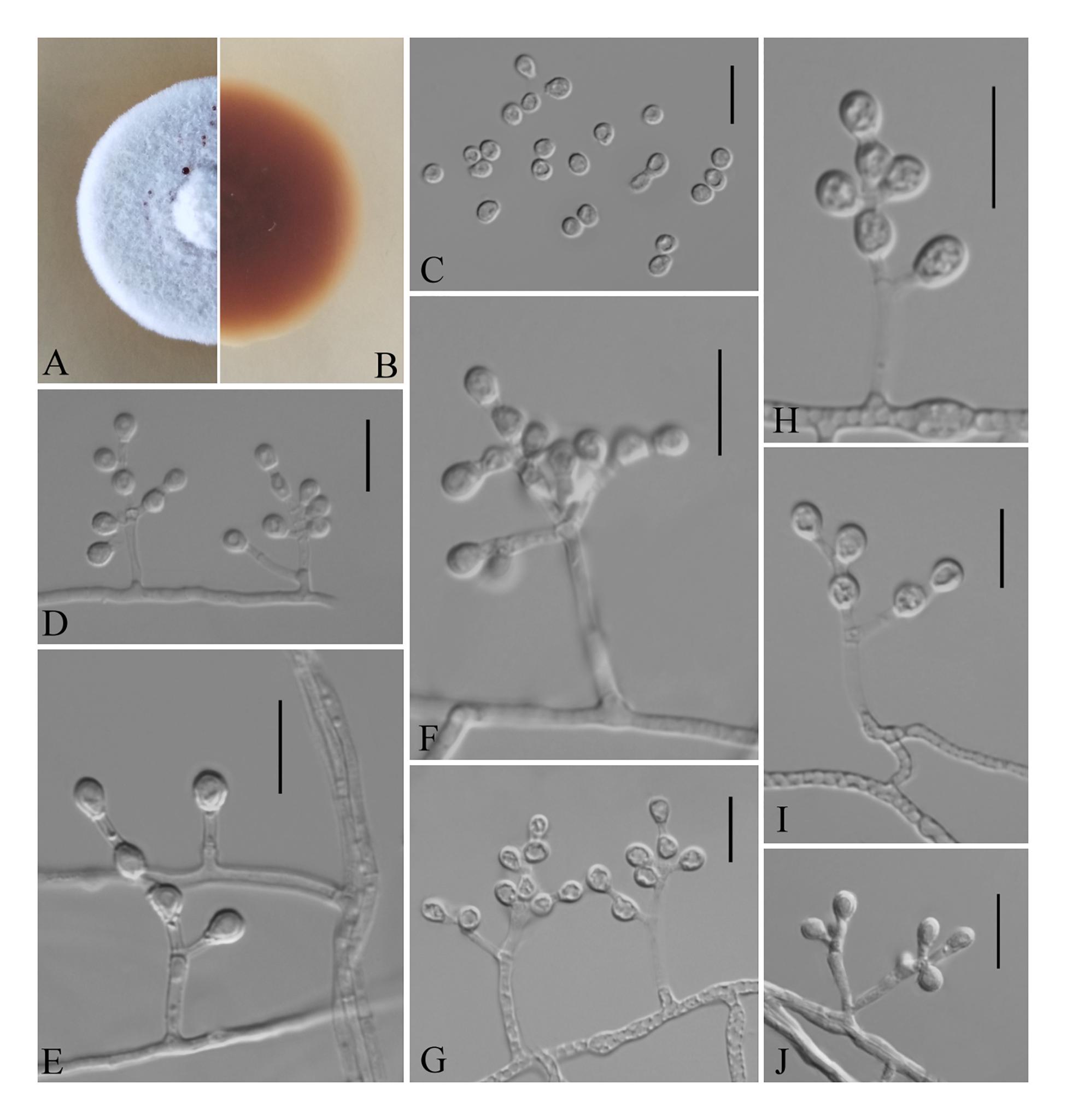
Figure 5. Pseudogymnoascus shaanxiensis (from ex-holotype GZUIFR 173.1). (A,B) The front and reverse of a P. shaanxiensis colony on PDA after 14 days at 25°C. (C) Conidia. (D–J) Conidiophores and conidia. Scale bars = 10 μm.
Etymology: Refers to Shaanxi, the province where the isolate was collected.
Holotype: permanently preserved in a metabolically inactive state, HMAS 255395.
Description based on HMAS 255395. Asexual: Colonies on PDA, reaching 21–23 mm in diameter after 14 days at 25°C, velvety to floccose, margins regular, white, producing a diffusible faint yellow pigment and clear exudates; reverse brown. Hyphae hyaline, smooth-walled, septate, 1.5- to 2.5-μm wide. Racquet hyphae absent. Conidia abundant, pyriform, sometimes subglobose, smooth 3.5–5.0 × 2.5–3.0 μm (av. = 4.0 × 3.0 μm, n = 50). Intercalary conidia subglobose, pyriform, or irregularly shaped, smooth, 3.5–4.0 × 3.0–4.0 μm (av. = 3.5 × 3.0 μm, n = 50). Terminal and lateral conidia on hyphae, short stalk or side branch, solitary, always forming verticillate and opposite branches with an acute angle to the axis near the apex.
Sexual morph: not observed.
Geographical distribution: China and America.
Material examined: China, Shaanxi, Xi’an, Xincheng district, 34°27′N, 108°95′E, from epiphytic soil of Broussonetia papyrifera, September 2018, Zhi. Y. Zhang (HMAS 255395 – holotype; GZUIFR 173.1 – ex-type living culture); Hanzhong, Shaanxi province, from epiphytic soil of Trachycarpus fortunei, September 2018, Zhi. Y. Zhang (GZUIFR HZ5.7); Yichang, Hubei province, from soil beside a park, September 2018, Zhi. Y. Zhang (GZUIFR CY 1.8). The living cultures were kept in sterile 30% glycerol and deposited in a −80°C freezer.
Notes: Morphologically, P. shaanxiensis resembles Pseudogymnoascus appendiculatus and Pseudogymnoascus verrucosus because of the pyriform conidia. However, P. shaanxiensis differs from P. appendiculatus and P. verrucosus based on its subglobose, pyriform intercalary conidia (the intercalary conidia of P. appendiculatus and P. verrucosus are subglobose to elongate) (Rice and Currah, 2006). Phylogenetically, our isolates GZUIFR 173.1, GZUIFR HZ5.7, and GZUIFR CY 1.8 cluster with 14PA06 and RMF C 101 in a distinct subclade and are separated from other clades (Figures 1, 2). Furthermore, since 2013 (Minnis and Lindner, 2013), isolates 14PA06 and RMF C 101 have remained an undescribed species owing to the lack of morphological characteristics. Therefore, we introduce P. shaanxiensis sp. nov. in this study.
Pseudogymnoascus sinensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.
MycoBank number: MB 835717, Facesoffungi number: FoF 08690 (Figure 6).
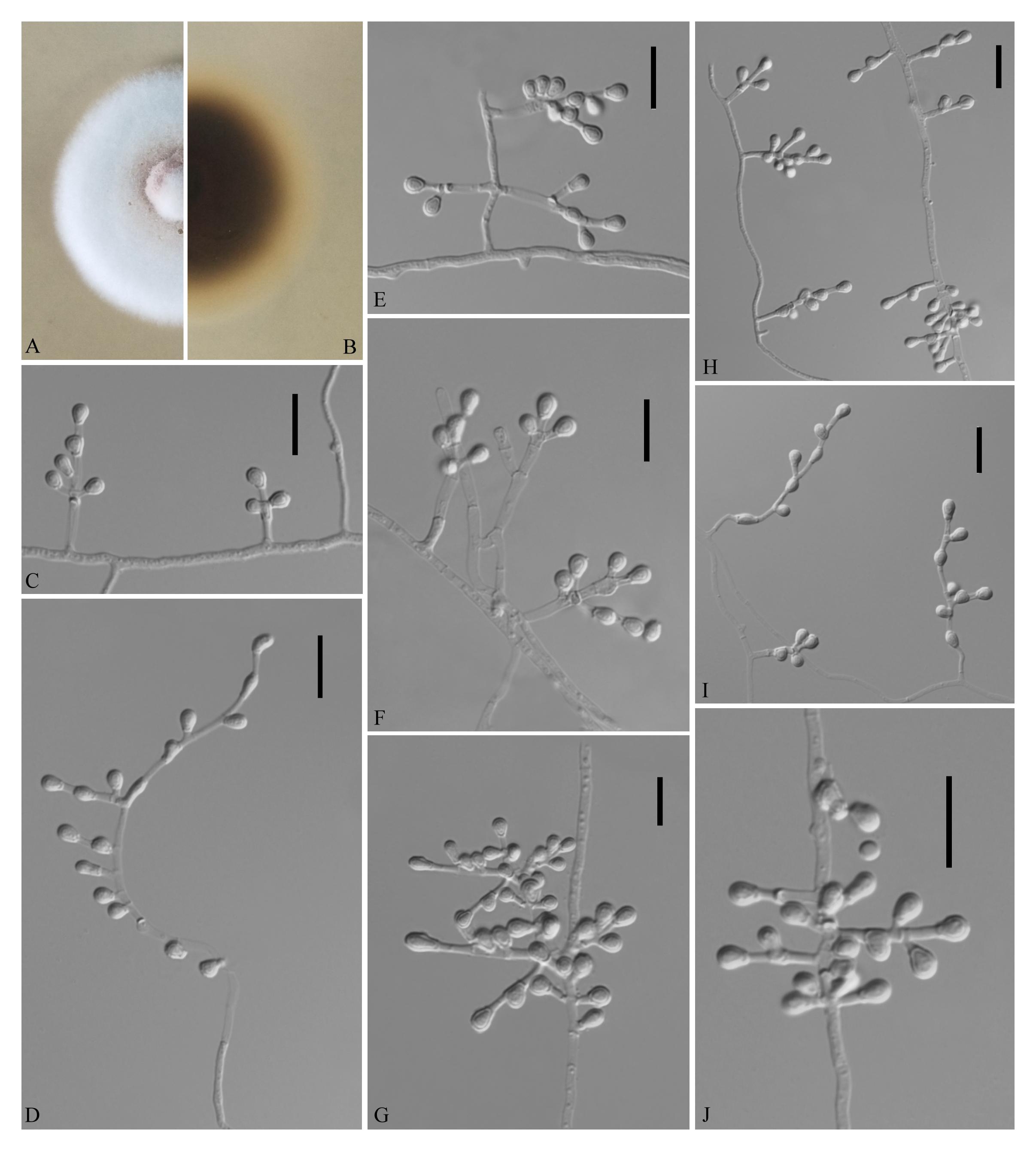
Figure 6. Pseudogymnoascus sinensis (from ex-holotype CGMCC 3.18493). (A,B) The front and reverse of a P. sinensis colony on PDA after 14 days at 25°C. (C–J) Conidiophores and conidia. Scale bars = 10 μm.
Etymology: In reference to China, the country where the type specimen was obtained.
Holotype: permanently preserved in a metabolically inactive state, HMAS 255394.
Description based on HMAS 255394. Asexual: Colonies on PDA, reaching 20–21 mm in diameter after 14 days at 25°C, powdery, floccose, margin identified, light pink at the center and pewter at the edge; reverse brown. Aerial mycelium abundant, smooth and thin walled, septate, 1- to 2-μm wide. Racquet hyphae absent. Conidia abundant, terminal and lateral conidia on hyphae, short stalk or side branches, sometimes forming verticillate and opposite branches with an acute angle to the axis near the apex, solitary, obovoid, 3.0–5.0 × 2.5–3.0 μm (av. = 4.0 × 2.5 μm, n = 50). Intercalary conidia are borne on the outer branches of the hyphae or verticillate hyphae, smooth walled, drum, obovoid, pyriform, or irregularly shaped, 3.0–4.5 × 2.5–5.0 μm (av. = 3 × 3 μm, n = 50).
Sexual morph: not observed.
Geographical distribution: China.
Material examined: China, Guizhou, Guiyang, the Affiliated Hospital of Guizhou Medical University, 26°59′N, 106°71′E, from soil beside a road, September 2016, Zhi. Y. Zhang (HMAS 255394 – holotype; CGMCC 3.18493 = GZUIFR K278.1 – ex-type living cultures; ibid., CGMCC 3.18494 = GZUIFR K278.2). The living cultures were kept in sterile 30% glycerol and deposited in a −80°C freezer.
Notes: Morphologically, P. sinensis is similar to P. linderi and P. turner, based on its obovoid conidia. However, P. sinensis differs from P. linderi and P. turneri as it has drum, obovoid, pyriform, or irregularly shaped intercalary conidia (the intercalary conidia of P. linderi and P. turneri are globose to truncate) (Crous et al., 2019). Phylogenetically, our isolates CGMCC 3.18493 and CGMCC 3.18494 cluster together very well and form a single clade separated from other Pseudogymnoascus species (Figure 1).
Solomyces Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, gen. nov.
MycoBank number: MB 835713; Facesoffungi number: FoF 08688.
Etymology: Solo- (from Solum), in reference to its isolation from soil.
Saprobic on soil. Asexual morph: Conidia: terminal and lateral conidia borne on hyphae, short protrusions, or side branches. Conidia solitary, sometimes 2 in chains, pyriform, sometimes subglobose. Intercalary conidia abundant, olivary, subglobose to globose. Sexual morph: Unknown.
Geographical distribution: China and America.
Type species: Solomyces sinensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang.
Notes: Solomyces is introduced to accommodate Solomyces sinensis and another unnamed species (containing two strains, 15PA02 and 17WV02, from Hibernacular soil in Pennsylvania and West Virginia, respectively; Minnis and Lindner, 2013). The morphology of Solomyces species is similar to that of Geomyces and the asexual morphs of Pseudogymnoascus. However, Geomyces differ in having terminal and lateral conidia borne on hyphae, short protrusions or side branches; intercalary conidia barrel shaped, and conidiophores abundant, always forming verticillate and opposite branches with an acute angle to the axis near the apex (Van Oorschot, 1980; Chen et al., 2017). As can be seen in Figures 1, 2, strains of these genera appear in distinct clades in a phylogeny based on multiple strains, thereby justifying the erection of the new genus Solomyces.
Solomyces sinensis Zhi.Y. Zhang, Y.F. Han and Z.Q. Liang, sp. nov.
MycoBank number: MB 835714, Facesoffungi number: FoF 08687 (Figure 7).
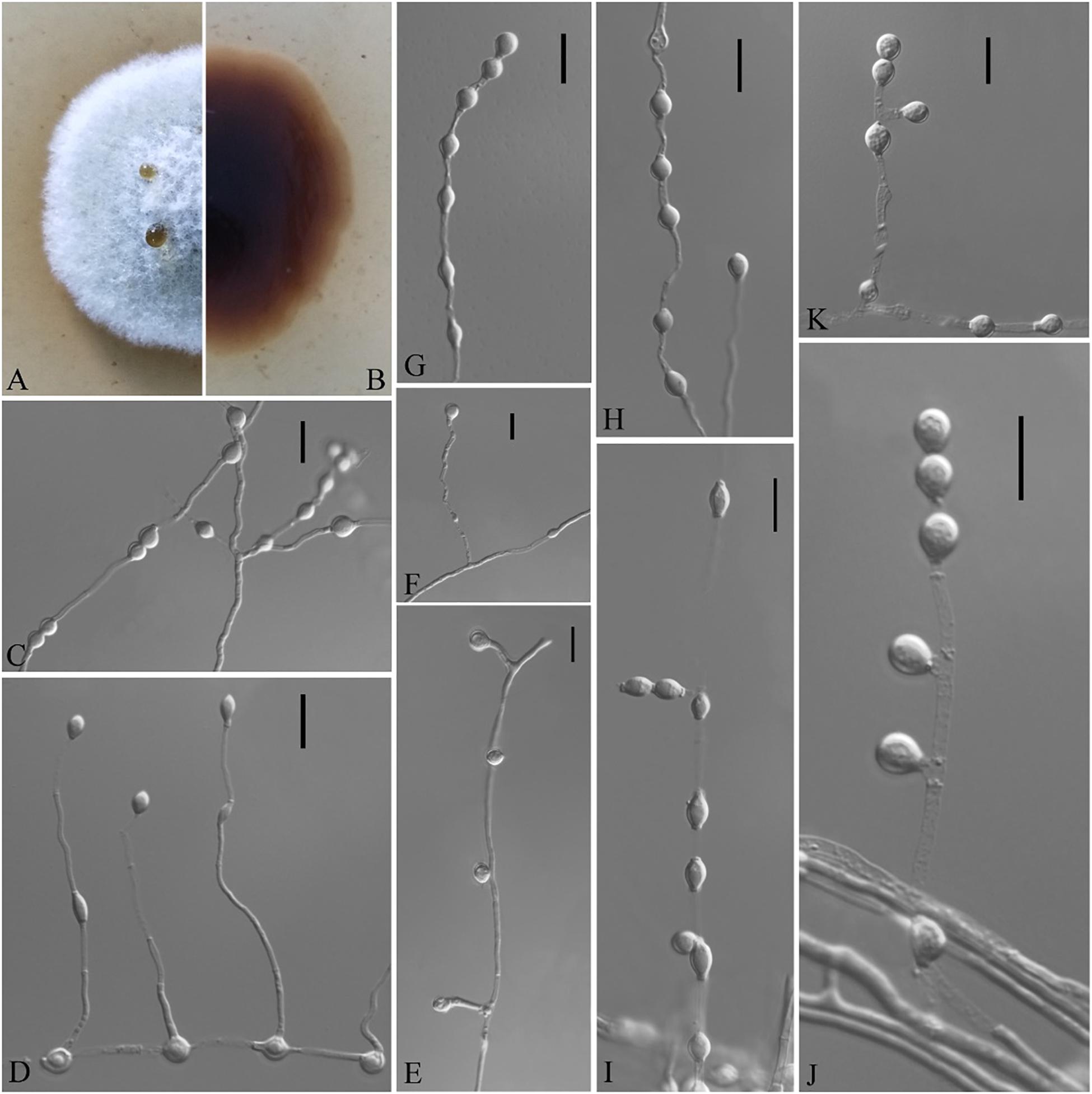
Figure 7. Solomyces sinensis (from ex-holotype CGMCC 3.18498). (A,B) The front and reverse of a S. sinensis colony on PDA after 14 days at 25°C. (C–K) Conidiophores and conidia. Scale bars = 10 μm.
Etymology: In reference to China, the country where the type specimen was obtained.
Holotype: permanently preserved in a metabolically inactive state, HMAS 255397.
Description based on HMAS 255394. Asexual: Colonies on PDA, reaching 16–17 mm in diameter after 14 days at 25°C, elevate at the center, velvety to floccose, margins regular, pewter at the center and white at the edge, producing a diffusible faint yellow pigment and clear exudates; reverse brown. Aerial mycelium abundant, smooth and thin walled, septate, 1- to 2-μm wide. Terminal and lateral conidia borne on hyphae, short protrusions, or side branches. Conidia solitary, sometimes two in chains, pyriform, sometimes subglobose, smooth or rarely rough walled, ecru, 4.0–6.0 × 3.0–5.5 μm (av. = 5.5 × 4.0, n = 50). Intercalary conidia abundant, olivary, subglobose to globose, 4.0–7.0 × 3.0–5.5 μm (av. = 5.0 × 4.0, n = 50).
Sexual morph: not observed.
Geographical distribution: China.
Material examined: China, Guizhou, Guiyang, the Affiliated Hospital of Guizhou Medical University, 26°59′N, 106°71′E, from soil beside a road, September 2016, Zhi. Y. Zhang (HMAS 255397 – holotype; CGMCC 3.18498 = GZUIFR K277.1 – ex-type living cultures; ibid., CGMCC 3.18499 = GZUIFR K277.2; ibid., CGMCC 3.18500 = GZUIFR K277.3). The living cultures were kept in sterile 30% glycerol and deposited in a −80°C freezer.
Notes: Solomyces sinensis was isolated from soil in Guizhou Province, China. We did not compare morphological characteristics between S. sinensis and another two strains within Solomyces owing to the lack of morphological description of these two strains (Minnis and Lindner, 2013). However, S. sinensis is phylogenetically distinct from these strains with high statistical support (1.00 BYPP, 100% MLBS) (Figures 1, 2).
Discussion
In this study, Solomyces gen. nov. is introduced with an asexual morph. Five new species are also described. All the new taxa belong in the order Thelebolales, the members of which are ubiquitous in the environment. Several taxa belonging to this order have been isolated from tropical to arctic regions. They are often coprophilous and frequently isolated from freshwater and saline lakes (De Hoog et al., 2005). They have also been recorded from soils, epiphytic soils in tree holes (Chen et al., 2017), mine sediments (Crous et al., 2019), and sponges (Bovio et al., 2018). Thelebolales have been recorded as saprobic on dead plant material, rarely as plant-parasitic, and also as animal pathogens, e.g., the well-known white-nose disease of bats (Lorch et al., 2011). In addition, several members of Thelebolales are keratinophilic; i.e., they can invade and degrade keratin material (Saxena et al., 2005). All the proposed new taxa in our study were isolated using the baiting technique, a method specifically designed for the isolation of keratinophilic fungi. Consequently, additional studies are needed to assess whether our new taxa can also degrade keratin material.
Recent studies demonstrated that the Thelebolales comprise at least two families or groups (Ekanayaka et al., 2019; Johnston et al., 2019; Batista et al., 2020). Some studies have proposed that the Thelebolales consists of Pseudeurotiaceae and Thelebolaceae based on three distinct pieces of evidence (568 ITS sequences, 15 genes concatenated from 279 species, and a phylogenomic approach from 51 complete genomes) and on a phylogenomic approach, respectively (Johnston et al., 2019; Batista et al., 2020). The Thelebolaceae, represented by the Thelebolus and Antarctomyces, share a common ancestor with the family Pseudeurotiaceae, represented by the genus Pseudogymnoascus (Johnston et al., 2019; Batista et al., 2020). However, a different study indicated that Pseudeurotiaceae was nested within Thelebolaceae based on phylogenetic analysis and synonymized Pseudeurotiaceae under Thelebolaceae (Ekanayaka et al., 2019). The same authors discovered that several genera, previously classified as Leotiaceae and Leotiomycetes genera incertae sedis, clustered within Thelebolales as a sister clade to the Thelebolaceae and defined them as the Alatospora–Miniancora clade (Ekanayaka et al., 2019). The studies by Ekanayaka et al. contained more genus taxa in Thelebolales; therefore, we continued the phylogenetic analysis in Thelebolales based on this study.
Recent reports have indicated that the Thelebolales contained 22 genera. However, because no ITS and LSU sequence data were available for Ascophanus, Ascozonus, Caccobius, Coprobolus, Leptokalpion, Neelakesa, and Pseudascozonus (Ekanayaka et al., 2019), we could not compare the phylogenetic relationships between these genera and Solomyces. In our phylogenetic analysis, our three isolates (CGMCC 3.18498, CGMCC 3.18499, and CGMCC 3.18500; Solomyces sinensis) and two isolates of Minnis and Lindner (2013) (15PA02 and 17WV02, from Hibernacular soil in Pennsylvania and West Virginia, respectively) formed an independent clade with strong statistical support (BYPP 1/MLBS 100%) and were close to Geomyces. Morphologically, the asexual stage of Ascophanus, Ascozonus, Caccobius, Coprobolus, Leptokalpion, Neelakesa, and Pseudascozonus is not recorded in the literature (Wijayawardene et al., 2017). Therefore, no morphological comparison can be done between these genera and Solomyces. However, Solomyces differs from Geomyces by terminal and lateral conidia borne on hyphae, short protrusions or side branches, olivary, subglobose to globose intercalary conidia, and absence of the forming verticillate and opposite branches with an acute angle to the axis near the apex of conidiophores (Sigler and Carmichael, 1976).
Based on morphological characteristics, it was difficult to distinguish closely related species, and even genera, using traditional taxonomy, and modern phylogenetic methods were a very important adjunct. Although Crous et al. (2019) described the new species, P. linderi and P. turneri, based on the similarity of morphological characteristics between these two new species and P. bhattii, they did not compare the phylogenetic relationship. Our phylogenetic analysis indicated that P. bhattii (type strain CBS 760.71) was nested within Gymnostellatospora (Figure 1), and we, therefore, transferred P. bhattii to the Gymnostellatospora and named it G. bhattii.
Data Availability Statement
The sequences generated in this study can be found in GenBank. The accession numbers of the sequences deposited in GenBank are ITS: MT509362–MT509372, LSU: MT509376–MT509386, MCM7: MT534202–MT534212, RPB2: MT534216–MT534226, and EF1A: MT534227–MT534237.
Author Contributions
YH and JH were responsible for conceptualization and funding acquisition. ZZ, CD, WC, QM, and XL were responsible for data acquisition. ZZ, CD, and WC did the formal analysis. ZZ wrote the first draft. YH and ZL wrote, reviewed, and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Key-Area Research and Development Program of Guangdong Province (2018B020205003), the Program of High-level innovative scientific talents of Guizhou Province [(2020)6005],[cpsenter]the Ministry of Science and Technology of China (2013FY110400), the National Natural Science Foundation of China (31460010 and 1860002), the Construction Program of Biology First-Class Discipline in Guizhou [GNYL(2017)009], and the Graduate Innovation Fund Project of Guizhou Province [YJSCXJH(2019)026].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Batista, T. M., Hilário, H. O., de Brito, G. A. M., Moreira, R. G., Furtado, C., de Menezes, G. C. A., et al. (2020). Whole-genome sequencing of the endemic Antarctic fungus Antarctomyces pellizariae reveals an ice-binding protein, a scarce set of secondary metabolites gene clusters and provides insights on Thelebolales phylogeny. Genomics 112, 2915–2921. doi: 10.1016/j.ygeno.2020.05.004
Bovio, E., Garzoli, L., Poli, A., Prigione, V., Firsova, D., McCormack, G. P., et al. (2018). The culturable mycobiota associated with three Atlantic sponges, including two new species: Thelebolus balaustiformis and T. spongiae. Fungal Syst. Evol. 1, 141–167. doi: 10.3114/fuse.2018.01.07
Chen, W. H., Zeng, G. P., Luo, Y., Liang, Z. Q., and Han, Y. F. (2017). Morphological traits and molecular analysis for Geomyces fujianensis sp. nov. from China. Mycosphere 8, 38–43. doi: 10.5943/mycosphere/8/1/5
Crous, P. W., Wingfiled, M. J., Lombard, L., Roets, F., Swart, W. J., Alvarado, P., et al. (2019). Fungal planet description sheets: 951–1041. Persoonia 43, 223–425. doi: 10.3767/persoonia.2019.43.06
Dal Vesco, G. (1957). Geomyces vinaceus” n. sp. forma conidica di “Pseudogymnoascus vinaceus. Raillo. Allionia 3, 1–15.
De Hoog, G. S., Göttlich, E., Platas, G., Genilloud, O., Leotta, G., and van Brummelen, J. (2005). Evolution, taxonomy and ecology of the genus Thelebolus in Antarctica. Stud. Mycol. 51, 33–76.
De Menezes, G. C., Godinho, V. M., Porto, B. A., Gonçalves, V. N., and Rosa, L. H. (2017). Antarctomyces pellizariae sp. nov., a new, endemic, blue, snow resident psychrophilic ascomycete fungus from Antarctica. Extremophiles 21, 259–269. doi: 10.1007/s00792-016-0895-x
Drummond, A., and Rambaut, A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Eckblad, F. E. (1968). The genera of the operculate discomycetes. A re-evaluation of their taxonomy, phylogeny and nomenclature. Norw. J. Bot. 15, 1–191.
Ekanayaka, A., Hyde, K. D., Gentekaki, E., McKenzie, E. H. C., Zhao, Q., Bulgakov, T. S., et al. (2019). Preliminary classification of Leotiomycetes. Mycosphere 10, 310–489. doi: 10.5943/mycosphere/10/1/7
Ekanayaka, A. H., Ariyawansa, H. A., Hyde, K. D., Jones, E. B. G., Daranagama, D. A., Phillips, A. J. L., et al. (2017). Discomycetes: the apothecial representatives of the phylum Ascomycota. Fungal Divers. 87, 237–298. doi: 10.1007/s13225-017-0389-x
Gargas, A., Trest, M. T., Christensen, M., Volk, T. J., and Blehert, D. S. (2009) Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon 108, 147–154. doi: 10.5248/108.147
Haeckel, E. (1894). Systematische Phylogenie. Entwurf eines Natürlichen Systems der Organismen auf Grund ihrer Stammesgeschichte. Theil1, Systematische Phylogenie der Protisten und Pflanzen. (Berlin: G. Reimer), 1–400.
Hocking, A. D., and Pitt, J. I. (1988). Two new species of xerophilic fungi and a further record of Eurotium halophilicum. Mycologia 80, 82–88. doi: 10.2307/3807497
Johnston, P. R., Quijada, L., Smith, C. A., Baral, H.-O., Hosoya, T., Baschien, C., et al. (2019). A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10:1. doi: 10.1186/s43008-019-0002-x
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Karunarathna, A., Peršoh, D., Ekanayaka, A. H., Jayawardena, R. S., Thilini Chethana, K. W., Goonasekara, I. D., et al. (2020). Patellariopsidaceae Fam. Nov. With Sexual-Asexual Connection and a New Host Record for Cheirospora botryospora (Vibrisseaceae, Ascomycota). Front. Microbiol. 11:906. doi: 10.3389/fmicb.2020.00906
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Korf, R. P. (1973). “Discomycetes and Tuberales,” in The Fungi: An Advanced Treatise, Vol. IVA, eds G. C. Ainsworth, F. K. Sparrow, and A. S. Sussman (New York, NY: Academic Press).
Li, Z. Q., and Cui, C. Q. (1989). Study on the Psychrophilic/Psychrotrophic microorganisms. III. Geomyces laevis, a new species of Geomyces. Mycosystema 8, 47–50.
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lorch, J. M., Meteyer, C. U., Behr, M. J., Boyles, J. G., Cryan, P. M., Hicks, A. C., et al. (2011). Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 480, 376–378. doi: 10.1038/nature10590
Luo, Y., Chen, W. H., Wang, Y., Han, Y. F., and Liang, Z. Q. (2016). A new Geomyces species producing melanin in the medium. Mycosystema 35, 123–130. doi: 10.13346/j.mycosystema.140246
Minh, Q., Nguyen, M., and von Haeseler, A. A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Minnis, A. M., and Lindner, D. L. (2013). Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol. 117, 638–649. doi: 10.1016/j.funbio.2013.07.001
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Reeb, V., Lutzoni, F., and Roux, C. (2004). Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol. Phylogenet. Evol. 32, 1036–1060. doi: 10.1016/j.ympev.2004.04.012
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Rice, A. V., and Currah, R. S. (2006). Two new species of Pseudogymnoascus with Geomyces anamorphs and their phylogenetic relationship with Gymnostellatospora. Mycologia 98, 307–318. doi: 10.3852/mycologia.98.2.307
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Samson, R. A. (1972). Notes on Pseudogymnoascus, Gymnoascus and related genera. Acta Bot. Neerlandic. 21, 517–527. doi: 10.1111/j.1438-8677.1972.tb00804.x
Sati, S. C., and Pathak, R. (2016). Anamorph (asexual stage) Teleomorph (sexual stage) Connections in Aquatic hyphomycetes. Int. J. Plant Reprod. Biol. 8, 128–135.
Saxena, P., Kumar, A., and Shrivastava, J. N. (2005). Keratinophilic fungi: a microbial way to manage poultry waste feathers. Indian J. Microbiol. 45, 151–154.
Schoch, C. L., Sung, G. H., López-Giráldez, F., Townsend, J. P., Miadlikowska, J., Hofstetter, V., et al. (2009). The Ascomycota Tree of Life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol. 58, 224–239. doi: 10.1093/sysbio/syp020
Sigler, L., and Carmichael, J. W. (1976). Taxonomy of Malbranchea and some other hyphomycetes with arthroconidia. Mycotaxon 4, 349–488.
Spooner, B. M. (1987). Helotiales of Australasia: geoglossaceae, Orbiliaceae, Scelrotiniaceae, Hyaloscyphaceae. Bibl. Mycol. 116, 1–711.
Stchigel, A. M., Josep, C. A., Mac Cormack, W., and Guarro, J. (2001). Antarctomyces psychrotrophicus gen. et sp. nov., a new ascomycete from Antarctica. Mycol. Res. 105, 377–382. doi: 10.1017/s0953756201003379
Sutton, B. C., and Hennebert, G. L. (1995). “Interconnections amongst anamorphs and their possible contribution to ascomycetes systematics,” in Ascomycete Systematics: Problems and Perspectives in the Nineties, ed. D. L. Hawksworth (New York, NY: Plenum), 77–100.
Swofford, D. L. (2002). PAUP∗ 4.0b10: Phylogenetic Analysis Using Parsimony (∗and other methods). Sunderland, MA: Sinauer.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Traaen, A. E. (1914). Untersuchungen über bodenpilze aus Norwegen. Nytt Mag. Naturvidensk. 52, 19–121.
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Van Brummelen, J. (1985). Pseudoascozonus, a new genus of Pezizales. Proc. Indian Acad. Sci. 94, 363–367.
Van Oorschot, C. A. (1980). A revision of chrysosporium and allied genera (No. 20). Stud. Mycol. 20, 1–89.
Vanbreuseghem, R. (1952). Technique biologique pour l’isolement des dermatophytes du sol. Ann. Soc. Belg. Med. Trop. 32, 175–178.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vilgalys, R., and Sun, B. L. (1999). Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 91, 4599–4603. doi: 10.1073/pnas.91.10.4599
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Hyde, K. D., Rajeshkumar, K. C., Hawksworth, D. L., Madrid, H., Kirk, P. M., et al. (2017). Notes for genera: Ascomycota. Fungal Divers. 86, 1–594. doi: 10.1007/s13225-017-0386-0
Keywords: taxonomy, phylogeny, Thelebolales, new genus, new species
Citation: Zhang Z, Dong C, Chen W, Mou Q, Lu X, Han Y, Huang J and Liang Z (2020) The Enigmatic Thelebolaceae (Thelebolales, Leotiomycetes): One New Genus Solomyces and Five New Species. Front. Microbiol. 11:572596. doi: 10.3389/fmicb.2020.572596
Received: 15 June 2020; Accepted: 29 September 2020;
Published: 29 October 2020.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Hiran A. Ariyawansa, National Taiwan University, TaiwanDonald Henry Pfister, Harvard University, United States
Copyright © 2020 Zhang, Dong, Chen, Mou, Lu, Han, Huang and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfeng Han, c3dhbGxvdzExMjhAMTI2LmNvbQ==
 Zhiyuan Zhang
Zhiyuan Zhang Chunbo Dong1
Chunbo Dong1