
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 October 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.571093
Many Stramenopile species belonging to oomycetes from the genus Saprolegnia infect fish, amphibians, and crustaceans in aquaculture farms and natural ecosystems. Saprolegnia parasitica is one of the most severe fish pathogens, responsible for high losses in the aquaculture industry worldwide. Most of the molecules reported to date for the control of Saprolegnia infections either are inefficient or have negative impacts on the health of the fish hosts or the environment resulting in substantial economic losses. Until now, the whole proteome of S. parasitica has not been explored for a systematic screening of novel inhibitors against the pathogen. The present study was designed to develop a consensus computational framework for the identification of potential target proteins and their inhibitors and subsequent experimental validation of selected compounds. Comparative analysis between the proteomes of Saprolegnia, humans and fish species identified proteins that are specific and essential for the survival of the pathogen. The DrugBank database was exploited to select food and drug administration (FDA)-approved inhibitors whose high binding affinity to their respective protein targets was confirmed by computational modeling. At least six of the identified compounds significantly inhibited the growth of S. parasitica in vitro. Triclosan was found to be most effective with a minimum inhibitory concentration (MIC100) of 4 μg/ml. Optical microscopy showed that the inhibitors affect the morphology of hyphal cells, with hyper-branching being commonly observed. The inhibitory effects of the compounds identified in this study on Saprolegnia’s mycelial growth indicate that they are potentially usable for disease control against this class of oomycete pathogens. Similar approaches can be easily adopted for the identification of potential inhibitors against other plant and animal pathogenic oomycete infections.
Most species from the oomycete genus Saprolegnia are opportunistic parasites (Plumb, 1999) that cause the disease saprolegniasis in various fish species (van West, 2006), amphibians (Blaustein et al., 1994), crustaceans (Diéguez-Uribeondo et al., 1994), and other aquatic animals (Fernandez-Beneitez et al., 2011). The virulent strains of Saprolegnia parasitica infect eggs, juvenile, and adult fish, leading to important losses in fish farms worldwide (van West, 2006). The chemical malachite green was used effectively for the treatment of saprolegniasis (Willoughby and Roberts, 1992) until it was banned in 2002 by the United States and several other countries due to its carcinogenic and mutagenic effects in humans and animals (Srivastava et al., 2004). The aquaculture production is increasing every year, and due to the lack of suitable and effective chemicals to control saprolegniasis, a re-emergence and spread of the disease is observed worldwide. Losses due to S. parasitica are higher in fish farms than in wild environments because the farmed animals are typically held in high densities and exposed to constant stress and various types of pollutants, factors that altogether increase the risk of infection and spread of the disease.
To date, multiple chemicals have been trialed to control saprolegniasis (Table 1), but their negative impact on the environment or the health of fish farmers or the fish hosts typically prevents their practical use in aquaculture. Two main classes of molecules have been investigated that have either general antimicrobial activity or a more targeted mode of action. From the first class, the least harmful compound is sodium chloride, but this salt is efficient only at high concentrations, which makes its use impractical in freshwater (Das et al., 2012). Hydrogen peroxide also has minimal impact on the environment and has been used to efficiently treat Saprolegnia infections in catfish (Howe et al., 1999). More recently, however, its application has been shown to provoke negative side effects, such as reduced fish growth and damage to the gills (Mustafa, 2019). Likewise, while boric acid inhibits germination and colonization of spores as well as mycelial growth in Saprolegnia species (Ali et al., 2014), an excess of boron affects DNA integrity of blood and sperm cells in Nile tilapia (Acar et al., 2018) and adversely impacts the growth of rainbow trout (Öz et al., 2018). Peracetic acid is another nonspecific antimicrobial chemical with proven activity against S. parasitica (Marchand et al., 2012), but its use in fish farms is hindered by the fact that it causes lacrimation at low concentrations as well as extreme discomfort and irritation of the upper respiratory tract in humans (N. R. Council, 2010). Other efficient compounds have adverse effects on the environment. Examples are formaldehyde, sometimes used for the treatment of eggs and larval stages (Gieseker et al., 2006), and copper sulfate, which inhibits Saprolegnia growth by affecting multiple biological functions, including protein and energy biogenesis (Hu et al., 2016).
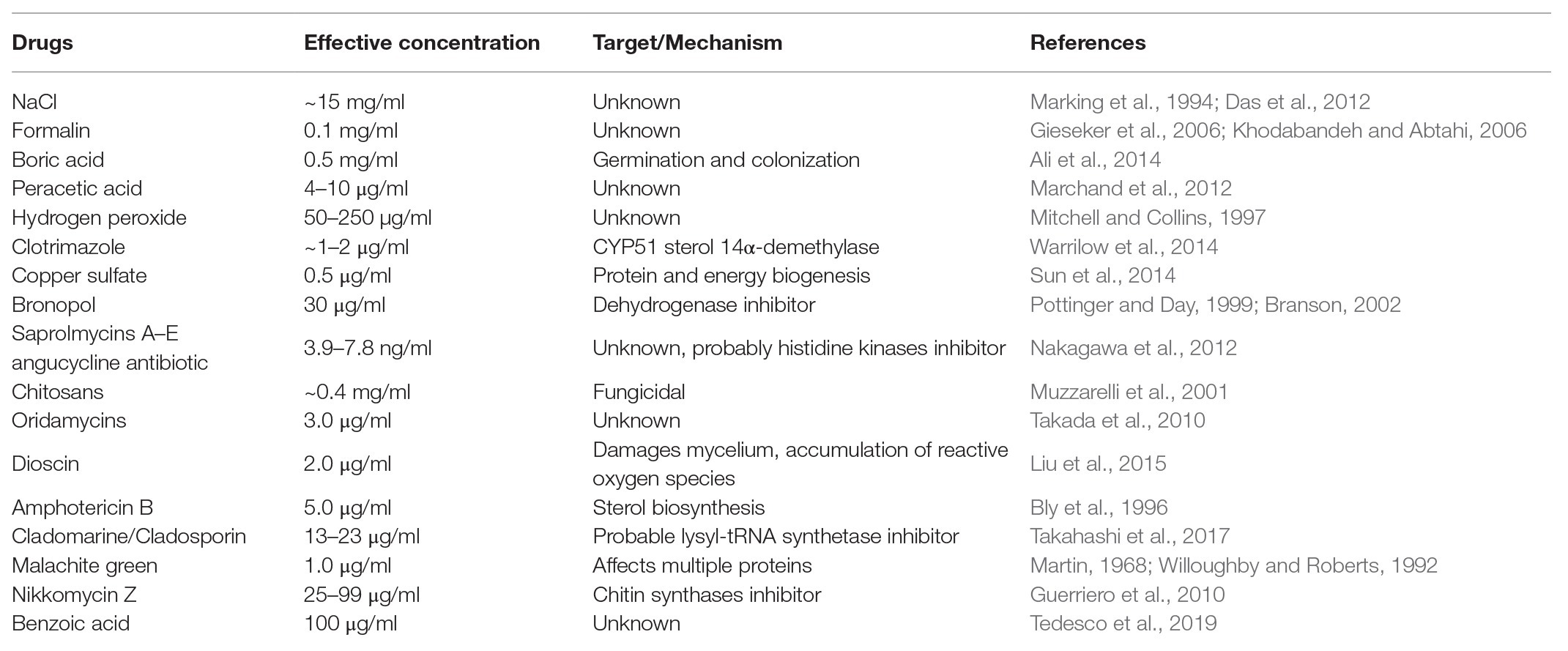
Table 1. List of compounds reported earlier to be effective in controlling the growth and spread of S. parasitica in aquaculture.
Other chemicals with more specific modes of action have been tested against Saprolegnia species. For example, bronopol (2-bromo-2-nitropropane-1,3-diol) and its methyl derivative 2-methyl-4-isothiazolin-3-one oxidize thiol groups in proteins and are, as such, inhibitors of dehydrogenases. These chemicals have been identified as promising compounds for the control of Saprolegnia infections in ova and adult fish (Branson, 2002; Oono and Hatai, 2007), although tolerant strains of Saprolegnia have been reported (Rezinciuc et al., 2014). Further chemicals of the oxyalkylchalcone class have been developed to overcome bronopol resistance. Promising molecules with higher inhibitory activity than bronopol on Saprolegnia growth are 2',4'-dihydroxychalcone and its oxyalkylated derivative produced by chemical synthesis, 2-hydroxy, 4-farnesyloxychalcone (Flores et al., 2016). However, these molecules were tested in vitro only and there are no results that demonstrate both their efficiency in infected animals and innocuousness to the environment. An attractive alternative approach to the chemical synthesis of novel inhibitors is the search for natural molecules that exhibit antimicrobial properties. Relevant examples that have been shown to be effective against S. parasitica with weak or no activity against fungi, bacteria, microalgae, or zooplankton are the angucycline antibiotics saprolmycins A and E from Streptomyces sp. (Nakagawa et al., 2012). Likewise, cladomarine and quellenin, two antibiotics produced by the deep-sea fungus Penicillium coralligerum (YK-247) and Aspergillus sp. (strain YK-76), respectively, were shown to exhibit anti-Saprolegnia activity (Takahashi et al., 2017). However, despite the discovery of these promising natural anti-oomycete compounds, their large-scale production has proven to be challenging and too expensive to be commercially viable.
Despite the numerous efforts mentioned above, we are still lacking compounds that can be used in the aquaculture industry to tackle saprolegniasis. Bioinformatic approaches have allowed the identification of inhibitors of bacterial pathogens of humans (Solanki and Tiwari, 2018; Bhardwaj et al., 2019; Cesur et al., 2020). Inspired by these successful strategies, here, we aim to identify specific protein targets and potential lead inhibitors of S. parasitica using combined in silico subtractive proteomics with a sequence-based approach. Our strategy presented in Figure 1 consisted in the analysis of the whole predicted proteome of S. parasitica and its comparison with fish and human proteomes to identify non-homologous essential proteins from the pathogen. The DrugBank database was subsequently searched to select potential food and drug administration (FDA)-approved inhibitors of these proteins, which were subsequently tested in vitro for their efficacy in arresting the growth of S. parasitica. With this approach, we have identified several inhibitors that exhibit MIC100 as low as 4 μg/ml. Their interactions with their protein targets were also studied by computational modeling, providing further evidence of the specific nature of the observed inhibitory effects. Similar strategy to identify and test novel compounds can be extended to other pathogenic oomycetes.
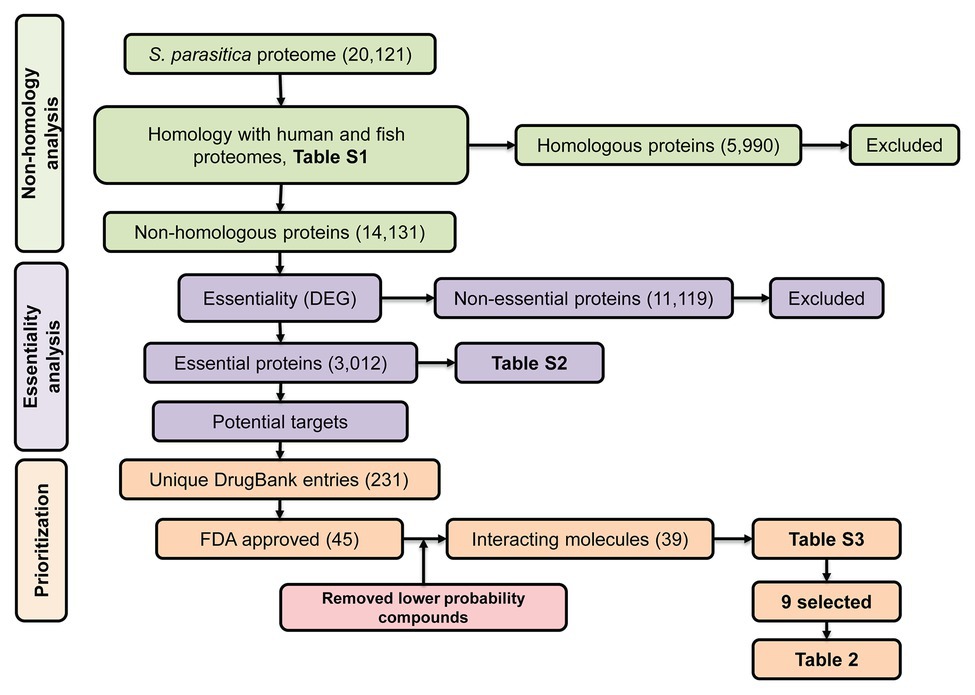
Figure 1. Schematic representation of the subtractive proteomics approach used in the present study. The whole proteome of Saprolegnia parasitica was searched using blastp (E-value < 1.0E-03, bitscore > 100) against human and fish proteomes followed by Database of Essential Genes (DEG) and finally, against DrugBank database for identification of food and drug administration (FDA)-approved inhibitor compounds.
The predicted proteome of S. parasitica (strain CBS 223.65) was retrieved from the NCBI website1 using genome assembly ASM15154v2. Conserved protein domains were predicted using pfam v31.0 (Finn et al., 2016) and conserved domain database (CDD; Marchler-Bauer et al., 2017). Protein sequences deduced from the human (Homo sapiens GRCh38.p11) and 15 fish genomes (Supplementary Table S1) were compared with the sequences from S. parasitica using blastp (BLAST 2.2.31+). The Saprolegnia proteins that did not present any significant sequence similarity (E-value < 1.0E-03, bitscore > 100) with any of the human or fish proteins were considered as non-homologous and used for further analysis. Their sequences were searched against the Database of Essential Genes (DEG v15.2; Luo et al., 2014) using blastp to create a list of essential proteins from S. parasitica. The DEG database comprises genes from archaea, bacteria, and eukaryotes that are known to be vital for the organism considered. As it is generally assumed that proteins essential to one organism are also likely to be essential to other organisms, proteins from S. parasitica similar to proteins from the DEG database, i.e., proteins with an E-value < 1.0E-05 and a bitscore > 100, were considered as potential targets of anti-oomycete compounds and used for further analysis.
An effective way to estimate the druggability of a protein, i.e., its capacity to bind one or several inhibitors, is to identify in the first instance similar proteins that have been shown to interact with these inhibitors (Keller et al., 2006). The DrugBank database (v5.1.1; Wishart et al., 2018) is a unique collection of bioinformatic and cheminformatic resources that combines drug data with information on the corresponding protein targets. The non-homologous essential proteins from S. parasitica identified as described above were searched against the DrugBank database using blastp. The proteins that showed significant scores (E-value < 1.0E-05, bitscore > 100) and their corresponding FDA-approved compounds were further considered.
Homology modeling was performed using the CPHmodels 3.2 Server (Nielsen et al., 2010) that uses profile-profile alignment guided by protein secondary structure and exposure predictions for template identification. The generated models were further minimized using Steepest Descent and the conjugate gradient method in Discovery Studio 2.5 (Biovia, 2018). The models were used for molecular docking with selected lead compounds using AutoDock Vina (Trott and Olson, 2010) in PyRx (Dallakyan, 2008) with default parameters. The 3D structures of the active compounds were retrieved from the DrugBank database and minimized in PyRx before docking. The docked complexes were further visualized using PyMol (Seeliger and de Groot, 2010) and LigPlot+ (Laskowski and Swindells, 2011), and the corresponding binding energies were calculated using AutoDock Vina (Trott and Olson, 2010).
S. parasitica (strain CBS223.65) was maintained on potato dextrose agar (PDA) by inoculating Petri dishes with agar plugs carrying pre-grown mycelium. The cultures were kept at 24°C in the dark, and inoculations were repeated every 15 days. Growth inhibition experiments were conducted in the synthetic medium of Machlis (1953). First, sesame seeds carrying S. parasitica mycelium were prepared by spreading autoclave-sterilized seeds (121°C for 20 min) on PDA plates that contained a central agar plug covered with mycelium. After 5 days at 24°C, the seeds coated with hyphal cells were transferred to 24-well flat-bottom tissue culture plates (one seed per well) containing 1 ml Machlis medium in each well. Dilutions of the selected compounds were used to determine the effective concentration of each inhibitor (Table 2). Copper sulfate (Sun et al., 2014), boric acid (Ali et al., 2014), benzoic acid (Tedesco et al., 2019), and malachite green (Martin, 1968), all known to be effective inhibitors of S. parasitica, were used as positive controls. Solvents of each class of inhibitor, i.e., sterile water, dimethyl sulfoxide (DMSO), ethanol, and acetone, were used as negative controls. Varying concentrations of each solvent (0.01, 0.1, 1.0, and 10.0%) were also tested to determine whether they had any effect on the growth of the mycelium (Supplementary Figures S1A,B). All concentrations were tested in three or more replicates. The plates were incubated at 24°C for 4 days, and the extent of mycelial growth in each well was observed and measured daily (colony diameter). MIC100 were defined as the inhibitor concentrations for which no growth was observed in four independent replicates after 4 days of incubation at 24°C.
Additional growth inhibition experiments were conducted on PDA plates. These were prepared using the MIC100 determined in the Machlis medium and the same positive and negative controls as listed above (Supplementary Figure S2). Mycelial growth was quantified by measuring the diameter of the colonies on the PDA plates pre-inoculated with single agar plugs (5 mm) placed at the center of the plates. Experiments were conducted in triplicates and measurements of colony diameters were made at 0 (day 0), 24 (day 1), 48 (day 2), and 72 h (day 3) of culture at 24°C (Figure 2).
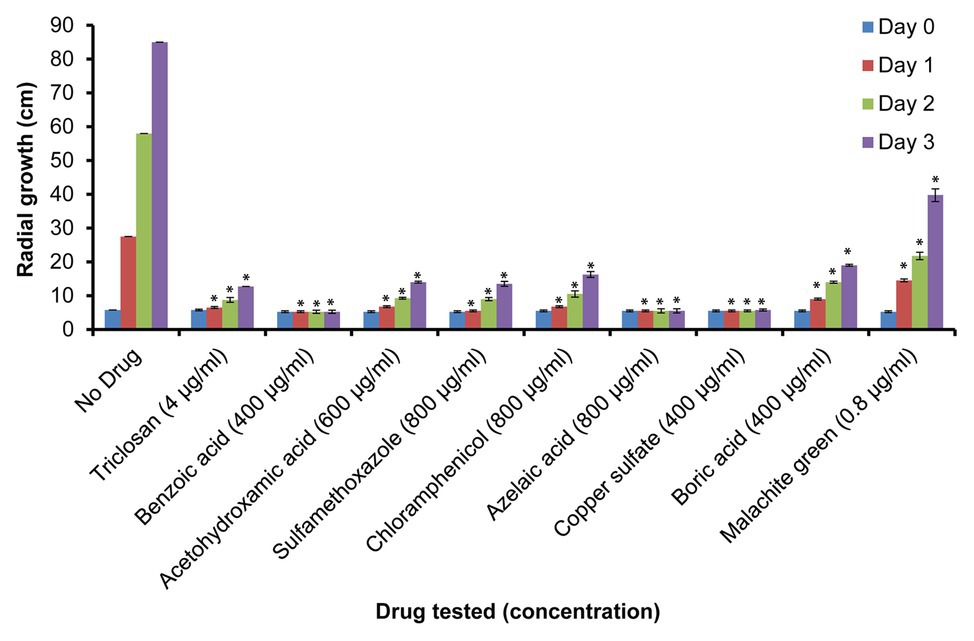
Figure 2. Relative growth of S. parasitica on potato dextrose agar (PDA) plates. Growth of mycelium (measured as diameter in cm) in the presence of sub-inhibitory concentrations of the tested compounds after 0 (day 0), 24 (day 1), 48 (day 2), and 72 h (day 3) of culture at 24°C. Four independent replicates were measured for each drug. A Student’s t-test was also performed to check significant differences between drug treated samples compared to the sample without drug (*p < 0.0001).
The effect of the tested compounds on the morphology of hyphal cells was observed by optical microscopy in 24-well flat-bottom tissue culture plates which contained 1 ml Machlis medium and appropriate dilutions of the lead compounds. Controls were performed in parallel as indicated above. Observations were made using a Leica Microsystems Ltd. DFC295 optical microscope, and images were recorded using the Leica Application Suite software v4.1.0.
To identify essential pathogen-specific proteins and potential specific growth inhibitors of S. parasitica, the sequences of all predicted proteins from the pathogen were compared with human and fish protein sequences (Supplementary Table S1). Among the 20,121 proteins encoded by the genome of S. parasitica, 5,990 were similar to human or fish proteins and were therefore disregarded for further analysis (Figure 1). Out of the remaining 14,131 non-homologous proteins, 3,012 showed significant similarity with essential proteins in the DEG database. These proteins were considered essential for S. parasitica and used for further analysis (Supplementary Table S2). Out of these, 117 were similar to target sequences in the DrugBank database, yielding 231 DrugBank unique entries, of which 45 are FDA approved (Supplementary Table S3). As the DrugBank database enlists not only inhibitors for their respective proteins but also interacting compounds, six of these 45 entries (DB01987, DB03147, DB03247, DB00140, DB09510, and DB09513) were unlikely to be efficient inhibitors. Hence, the list of potential inhibitors of growth was narrowed down to 39 compounds (Supplementary Table S3). These were further subdivided into two groups: the first group gathers compounds that potentially interact with two or more proteins from S. parasitica, whereas the second group consists of molecules that are predicted to bind one protein only (Supplementary Table S3).
This group contains eight compounds that can potentially interact with 29 proteins from Saprolegnia (Supplementary Table S3). Interestingly, among these molecules, three are semisynthetic echinocandin derivatives, namely caspofungin, anidulafungin, and micafungin. Echinocandins are well-characterized antifungal agents (Gil-Alonso et al., 2015; Perfect, 2017) that inhibit 1,3-β-D-Glucan synthases, i.e., the enzymes responsible for the formation of the major cell wall β-glucans in fungi. Earlier work from our group has shown that 1,3-β-D-glucans are major components of the cell wall of S. parasitica and other oomycete species (Bulone et al., 1990; Bulone and Fèvre, 1996; Mélida et al., 2013). 1,3-β-Glucan synthases have no mammalian homologues and are therefore ideal targets for antifungal and anti-oomycete inhibitors (Perfect, 2017). CDD analysis of the S. parasitica proteome led to the identification of at least 10 proteins that belong to the glucan synthase family, including SPRG_05460, which was not identified in our in silico screening as an echinocandin-binding protein. We did test multiple echinocandins in previous work and showed that they do not inhibit the 1,3-β-Glucan synthases of Saprolegnia in vitro and are therefore unlikely to stop the growth of this class of micro-organisms (Bulone, 1992). This probably reflects different properties between the fungal and oomycete enzymes as one would expect given their low level of similarity, which our blast results revealed to be of 29–34%.
Picric acid, citric acid, and artenimol are three other compounds from this group of inhibitors. However, their specificity is predicted to be low, essentially because of their general antimicrobial activity. Indeed, picric acid has been used as an antiseptic for a long time (Tidy, 1915) and is predicted to inhibit at least eight proteins from S. parasitica (Supplementary Table S3). The same applies to citric acid, which we have predicted to interact with at least four proteins from S. parasitica, including two uncharacterized proteins (SPRG_06530, a probable FtsZ protein, and SPRG_11850 and a phosphoribosylaminoimidazole carboxylase), a tryptophan synthase (SPRG_05079) and a cell division protein FtsZ (SPRG_11942). Tryptophan synthase catalyzes the final step of the tryptophan biosynthetic pathway. This enzyme is absent in mammals, which makes it an attractive target for many other pathogens (Shen et al., 2009). The cell division protein FtsZ has been described as a potential drug target in various bacterial pathogens (Shen et al., 2009), and phosphoribosylaminoimidazole carboxylase has been proposed previously as a target for drug therapy against Candida albicans (Donovan et al., 2001). Despite these observations, the diversity of enzymes and non-catalytic proteins to which citric acid potentially binds to suggests that its mode of action is not specific. Similarly, artenimol (dihydroartemisinin) is predicted to interact with at least two different Saprolegnia proteins, namely SPRG_00366, a predicted pyridoxal biosynthesis lyase (PdxS), and SPRG_09913, a negative regulator of GroEL domains. Interestingly, this compound has been shown to be active against a range of organisms as diverse as viruses (Efferth et al., 2008), malarial protozoa (Pull et al., 2019), plant pathogenic fungi (Tang et al., 2000), and even certain tumor cells (Jansen et al., 2011) but its efficacy against oomycetes has not been reported.
Another compound identified in group one is formic acid. It is predicted to interact with at least three chorismate binding enzymes, an anthranilate synthase protein (SPRG_06062), and two hypothetical proteins (SPRG_12737 and SPRG_16397). The antimicrobial activity of formic acid has been reported on various bacterial and yeast pathogens (Plumed-Ferrer and von Wright, 2011; Lastauskiene et al., 2014). However, chronic exposure to this molecule has been shown to have adverse health effects similar to those observed in methanol poisoning, i.e., metabolic acidosis and ocular toxicity (Thompson, 1992).
α-Phosphoribosylpyrophosphoric acid is predicted to interact with two proteins of unknown functions and anthranilate phosphoribosyltransferase, which is involved in the biosynthesis of aromatic amino acids. This compound is a key intermediate of the biosynthesis of nucleotides and likely to interfere with other vital processes of the fish hosts. This suggests that it would not be suitable for the management of saprolegniasis.
In conclusion, the compounds from this first group of inhibitors are expected to lack specificity (e.g., citric acid) or to be inactive against Saprolegnia (e.g., echinocandins), and, in some cases, to have adverse health effects on the fish hosts or humans (e.g., formic acid). Thus, in order to select more specific inhibitors potentially usable against Saprolegnia, we have focused the rest of our study on molecules that are predicted to bind a single essential protein in the pathogen.
This group comprises 31 compounds, the majority being sulfonamides or “sulfa drugs” (18 molecules). Sulfonamides are predicted to interact with the protein SPRG_19504 in S. parasitica, which belongs to the dihydropteroate synthase (DHPS) family and contains a 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase (HPPK) domain and a pterin-binding domain. In several bacteria and lower eukaryotes, HPPK (EC 2.7.6.3) and DHPS (EC 2.5.1.15) enzymes catalyze sequential reactions in the folic acid biosynthetic pathway (Illarionova et al., 2002). Higher eukaryotes obtain folate from dietary sources and lack necessary enzymes for folate biosynthesis (Matherly, 2001), whereas eubacteria and lower eukaryotes are able to synthesize tetrahydrofolate. Thus, the folate pathway is an interesting potential target for anti-infective agents. Indeed, DHPS is a well-known target of the sulfonamide class of antibacterial drugs (Hevener et al., 2010). CDD analysis of the proteome of S. parasitica revealed the presence of two additional proteins involved in the last step of the folic acid biosynthetic pathway and annotated as dihydrofolate reductase (DHFR), i.e., SPRG_10565 and SPRG_17339. These observations suggest that sulfonamides are potential effective inhibitors of the growth of Saprolegnia.
Quaternary ammonium compounds like cetrimonium and didecyldimethylammonium are predicted to interact with SPRG_00820, a hypothetical protein that contains a mycolic acid cyclopropane synthetase domain. An antimycotic activity was reported for cetrimide, in particular, against fungal keratitis caused by Fusarium solani, without any negative effect on eye corneal tissues (Mahmoud, 2007). Similarly, didecyldimethylammonium chloride (DDAC) is able to inactivate photosystem II (PSII) and disintegrate marine phytoplankton species (van Slooten et al., 2015). It was also found effective against Escherichia coli where it induced the formation of blebs in the cell membrane, causing leakage of the intracellular compartment and subsequent cell death (Yoshimatsu and Hiyama, 2007).
Triclosan and triclocarban are commonly used broad-spectrum antibacterial/antifungal agents that block fatty acid biosynthesis by inhibiting enoyl-ACP reductase (Dhillon et al., 2015). Both molecules are predicted to interact with the protein SPRG_20215 in S. parasitica, an uncharacterized protein that is predicted to contain a short-chain dehydrogenase/reductase (SDR) domain. Triclosan has been found to be effective against various Gram-negative and Gram-positive bacteria, as well as Mycobacteria (Schweizer, 2001) and Plasmodium falciparum (Surolia and Surolia, 2001), where it targets dihydrofolate reductase (Bilsland et al., 2018). This inhibitor is also active against the parasite Leishmania panamensis with no toxicity toward mammalian cells at concentrations as high as 200 μg/ml (Otero et al., 2014).
The benzimidazoles albendazole and thiabendazole are predicted to interact with the protein SPRG_07119 which carries a FAD-binding domain and is structurally similar to fumarate reductase 2 (PDB 5GLG) from Saccharomyces cerevisiae. Benzimidazoles are commonly used as fungicides and anthelminthic drugs where they specifically inhibit microtubule assembly (Davidse, 1986).
Two other approved compounds from group 2 are acetohydroxamic acid (Ohta et al., 2001) and ecabet (Shibata et al., 1997), which inhibit ureases. In Saprolegnia, both drugs are predicted to interact with the protein SPRG_06801, which is predicted to be a nickel-dependent amidohydrolase that catalyzes the decomposition of urea into carbamate and ammonia. As such, this catabolic reaction represents an important source of nitrogen in bacteria, fungi, and plants. Urease activity has been reported in Cryptococcus gattii (Feder et al., 2015) and the recently discovered novel systemic fungal pathogen Emergomyces africanus (Lerm et al., 2017). Urease is considered an essential enzyme and is explored as a target for the development of novel antibacterial agents (Follmer, 2010). Our in silico proteome analysis suggests that S. parasitica contains urease and therefore that inhibitors of this enzyme are worth testing against the pathogen.
Azelaic acid is predicted to interact with SPRG_13682, a hypothetical protein with 5'-3' exonuclease activity, and an N-terminal resolvase-like domain. Azelaic acid has been found effective against various Gram-positive and Gram-negative bacterial pathogens by inhibiting several oxidoreductases and interfering with glycolysis (Nazzaro-Porro, 1987).
Chloramphenicol was also identified in group 2. It is a common antibacterial agent that acts by inhibiting protein synthesis (Xaplanteri et al., 2003) and is considered fungistatic (Rooke and Shattock, 1983). It has been reported to exert variable inhibitory activity on the growth of Phytophthora species (Panek and Tomsovsky, 2017). Indeed, resistant strains of Phytophthora have been reported (Ann and Ko, 1992) alongside sensitive strains.
The last three compounds in group 2 are tromethamine, glycerol, and benzoic acid (Supplementary Table S3). Tromethamine is predicted to interact with the hypothetical protein SPRG_04175, which contains a biotin and thiamin synthesis associated domain. Glycerol is predicted to interact with a histidinol dehydrogenase (SPRG_01876) and it has been explored previously for its antibacterial activity (Saegeman et al., 2008). Benzoic acid is a common food preservative used to prevent the growth of yeast and molds (Brown and Swinburne, 1971). It most likely acts by interacting with the putative hydrolase SPRG_02663, a hypothetical Xaa-Pro dipeptidyl-peptidase with a C-terminal non-catalytic domain. This compound showed activity against various stages of fungal development (Amborabé et al., 2002) and it has recently been reported to be effective against S. parasitica (Tedesco et al., 2019), but its mechanism of inhibition was not investigated in these studies.
In conclusion, most of the inhibitors from group 2 have been shown to be effective against other microbial pathogens, which support the validity of our approach, but a large majority of these compounds have never been tested against Saprolegnia. Therefore, the next step of our work involved the use of in vitro assays in the presence of representative chemicals of the different classes from group 2 to determine whether these selected inhibitors affect the growth of S. parasitica and to which extent.
From the above list of DrugBank compounds predicted to interact with single proteins of S. parasitica (group 2), nine representative compounds were tested for their growth inhibitory effect on Saprolegnia mycelium (Table 2; Supplementary Figure S1). In addition, copper sulfate, boric acid, and malachite green were used as positive controls, and the solvents of the different inhibitors were tested as negative controls (see Materials and Methods section). The solvents used for the preparation of stock solutions of the selected compounds, i.e., water, dimethyl sulfoxide (DMSO), ethanol, and acetone, did not inhibit the growth of S. parasitica at concentrations up to 1% v/v (Supplementary Figures S1A,B). These results are in keeping with previous reports (Xue-Gang et al., 2013; Hoskonen et al., 2015). However, some growth inhibition was observed with DMSO and ethanol at concentrations higher than 2% v/v, while growth was not perturbed in acetone at concentrations as high as 10% v/v.
Triclosan and triclocarban are predicted to target the protein SPRG_20215. Triclosan was found to be very effective when tested in liquid Machlis medium, with an MIC100 concentration of 4 μg/ml (Supplementary Figure S1C), and significant levels of inhibition still observed at a lower concentration of 2 μg/ml. Benzoic acid (targeting the hypothetical protein SPRG_02663) and acetohydroxamic acid (targeting the putative urease SPRG_06801) arrested growth completely at 600 and 800 μg/ml, respectively. In addition, significant growth inhibition was still observed at lower concentrations (Supplementary Figures S1D,E). Recently, another group also reported benzoic acid as an effective compound against S. parasitica (Tedesco et al., 2019). However, in our experimental conditions, the 100 μg/ml concentration tested by Tedesco et al. (2019) had no visible effect on Saprolegnia growth (Supplementary Figure S1D). Other compounds such as the sulfonamide sulfamethoxazole (800 μg/ml), chloramphenicol (800 μg/ml), and azelaic acid (800 μg/ml) showed growth inhibition at less than 1,000 μg/ml (Supplementary Figures S1F,G,H). Furthermore, glycerol, albendazole, and thiabendazole did not show any significant inhibitory effect up to a concentration of 1,000 μg/ml (Supplementary Figures S1I,J,K). The compounds that significantly inhibited the growth of S. parasitica at concentrations lower than 1,000 μg/ml were considered effective, and their activity was further analyzed.
As expected and reported earlier, the positive controls, such as copper sulfate (400 μg/ml; Sun et al., 2014), boric acid (400 μg/ml; Ali et al., 2014), and malachite green (2 μg/ml; Martin, 1968; Supplementary Figures S1L,M,N) arrested the growth of mycelium in the liquid medium of Machlis. However, the use of sesame seeds to inoculate the culture medium leads to variable amounts and quality of mycelium per cell culture well (Stueland et al., 2005). This variability of inoculum size and quality is most likely responsible for the discrepancies reported in the literature with regard to the most efficient inhibitory concentrations of the different drugs.
To further evaluate the effect of the selected drugs on the growth of S. parasitica on a solid medium, we prepared PDA plates supplemented with the minimum inhibitory concentrations of each drug determined in the in vitro assays in liquid Machlis medium (Table 2). As no growth was observed for any of the compounds tested, a lower (sub-inhibitory) concentration was used to test the effect of each inhibitor (Table 2; Figure 2; Supplementary Figure S2). As expected, normal growth was observed on PDA plates devoid of inhibitor. Similar to the observations in liquid medium (Supplementary Figure S1), no inhibition was observed in the presence of glycerol, albendazole, and thiabendazole (1,000 μg/ml each) after 72 h culture at 24°C (Supplementary Figures S2K,L,M). Significant levels of inhibition were observed in the presence of triclosan (4 μg/ml), benzoic acid (400 μg/ml), acetohydroxamic acid (600 μg/ml), sulfamethoxazole (800 μg/ml), azelaic acid (1,000 μg/ml), chloramphenicol (800 μg/ml), copper sulfate (400 μg/ml), boric acid (400 μg/ml), and malachite green (0.8 μg/ml; Figure 2; Supplementary Figure S2).
Optical microscopy observations were made to analyze the effect of the selected inhibitors on the morphology of S. parasitica (Figure 3). In the absence of inhibitor, the hyphae appeared as translucent elongated cells. As opposed to the control, hyper-branching was commonly observed in the presence of some of the drugs that arrest the growth of the mycelium (Figure 3). The level of hyper-branching varied for the different compounds and was dose-dependent (Supplementary Figure S3). Pronounced hyper-branching was observed in the presence of triclosan (4 μg/ml), followed by benzoic acid (400 μg/ml), sulfamethoxazole (400 μg/ml), and boric acid (400 μg/ml). In the presence of acetohydroxamic acid, chloramphenicol, azelaic acid, and copper sulfate, low levels of side branching were visible (Figure 3). Interestingly, compared to other strong inhibitors of growth, no pronounced hyper-branching was visible in the presence of malachite green (Figure 3J). Hyper-branching of the mycelium of S. parasitica in the presence of various inhibitors has not been explored previously. Therefore, an in-depth study of the mechanisms underlying the formation of such branching structures would be necessary to understand the effect of the inhibitors on cell development.
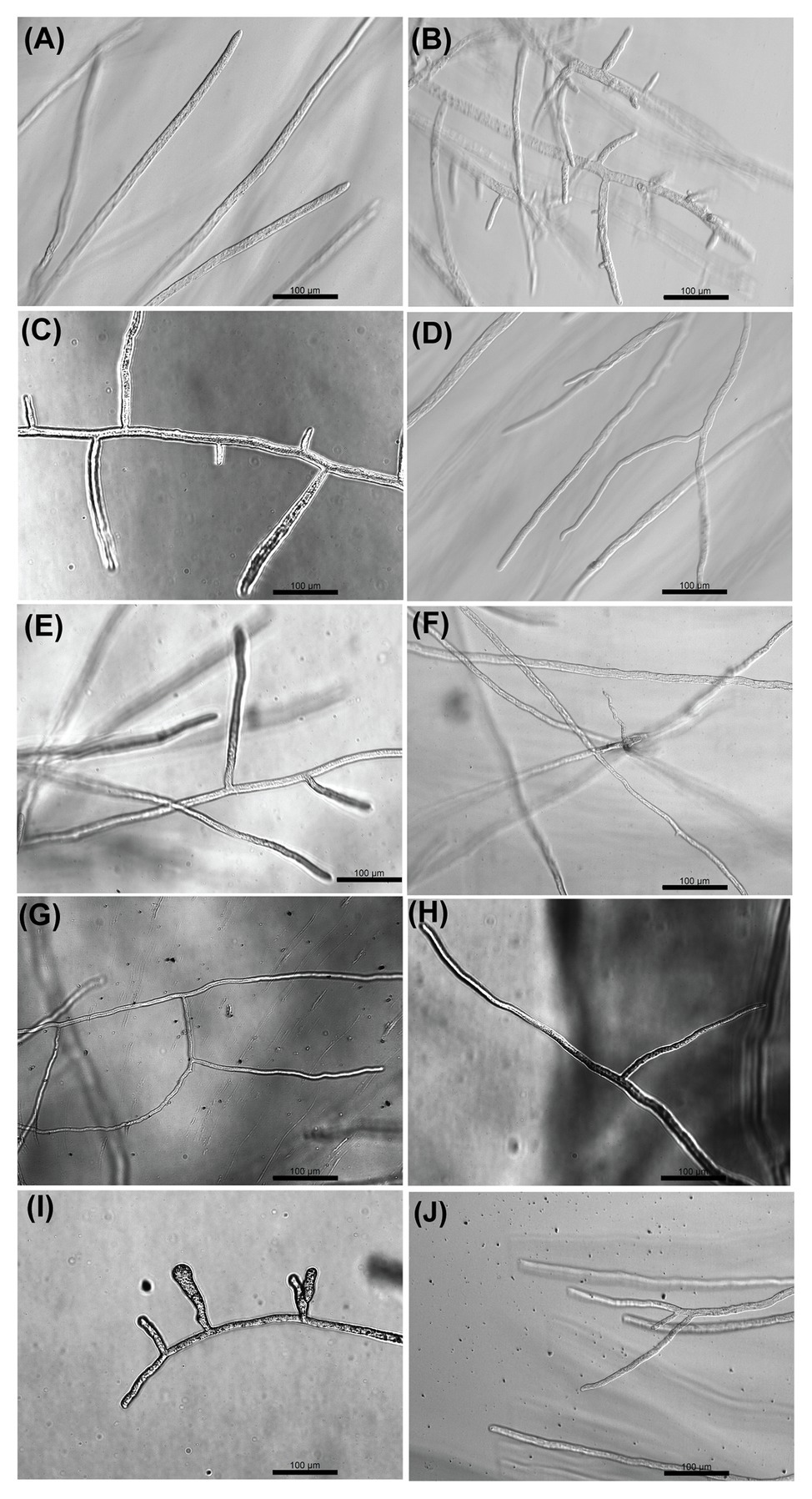
Figure 3. Optical microscopy observation of the effect of selected compounds on the growth of hyphae of S. parasitica after 24 h culture at 24°C in Machlis medium. (A) No Drug, (B) Triclosan (4 μg/ml), (C) Benzoic acid (400 μg/ml), (D) Acetohydroxamic acid (600 μg/ml), (E) Sulfamethoxazole (400 μg/ml), (F) Chloramphenicol (800 μg/ml), (G) Azelaic acid (800 μg/ml), (H) Copper sulfate (200 μg/ml), (I) Boric acid (400 μg/ml), and (J) Malachite green (0.8 μg/ml). Scale bars = 100 μm.
In this work, homology modeling and in silico docking studies were performed to better understand the mode of interaction between proteins and inhibitors. To this end, we used the six compounds shown to be effective in the in vitro assays along with their respective predicted target proteins (Figure 4; Supplementary Table S4). The six target proteins were SPRG_20215, SPRG_02663, SPRG_06801, SPRG_19504, SPRG_35021, and SPRG_13682. The 3D template structure for each of the selected proteins and corresponding percent sequence identity are presented in Supplementary Table S4. All proteins had a significantly high sequence identity of >30%, which allowed the generation of a reliable 3D protein structure, except for SPRG_02663, which has a slightly lower sequence identity (25.5%) with its template protein. The docking results show an approximate binding pattern of the inhibitors with their target proteins. Analysis of the docked complexes (compounds with their respective proteins) showed a considerable number of hydrogen bonding within the protein cavity or the active sites, which supports the specificity of the binding of each inhibitor with its respective protein. Furthermore, all the compounds showed significantly high binding energy toward their target, ranging from −7.7 to −4.4 kcal mol−1, which indicates strong interactions between the ligands and their targets (Figure 4; Supplementary Table S4).
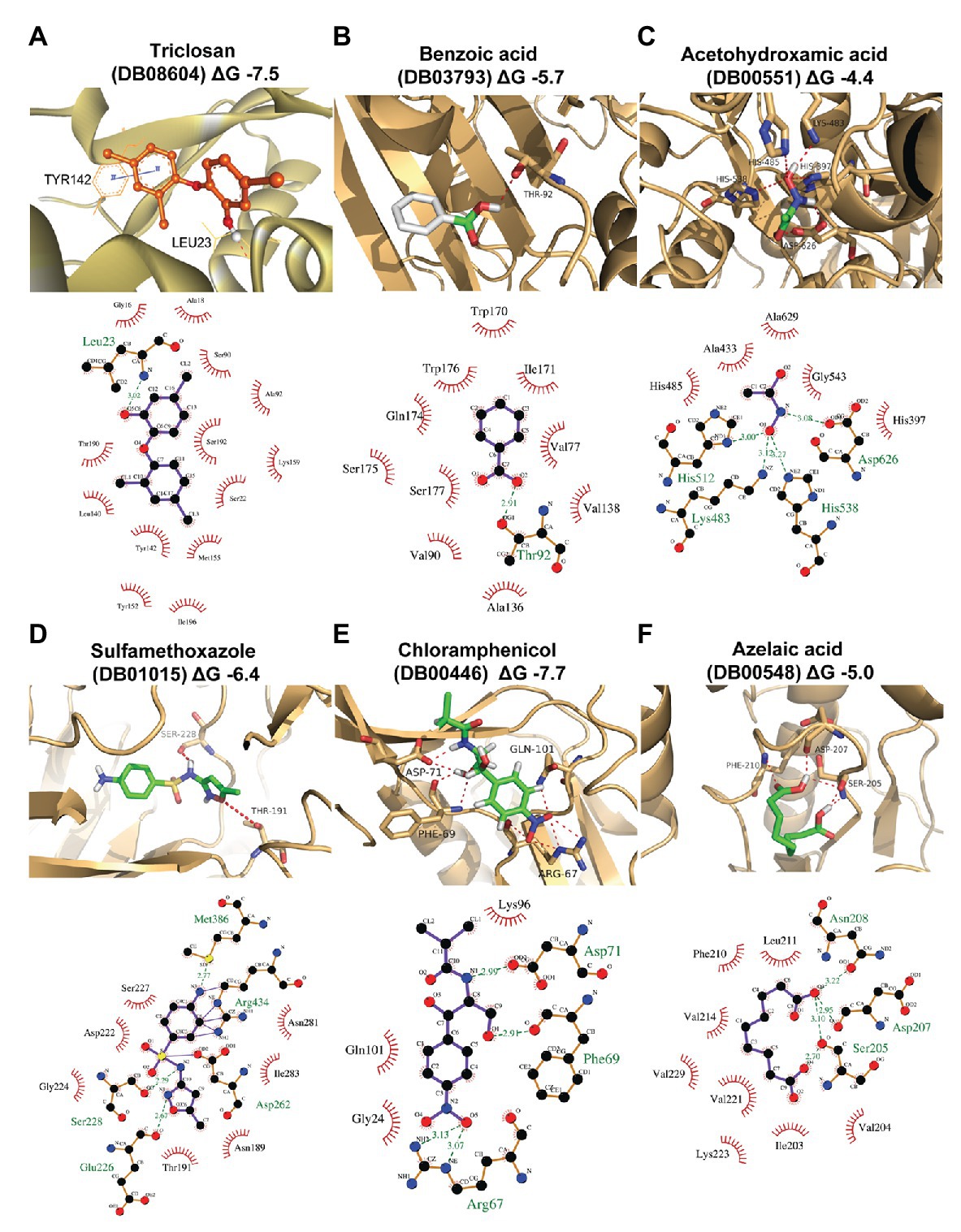
Figure 4. Homology modeling and docking of the tested compounds with their respective target proteins. For each of the protein-ligand complexes, the LigPlot cartoons show the interactions between each inhibitor and the amino acid residues in the active site cleft of the respective proteins. (A) SPRG_20215 with triclosan (DB08604; binding energy −7.5 kcal/mol), (B) SPRG_02663 with benzoic acid (DB03793; binding energy −5.7 kcal/mol), (C) SPRG_06801 (putative urease) with acetohydroxamic acid (DB00551; binding energy −4.4 kcal/mol), (D) SPRG_19504 with sulfamethoxazole (DB01015; binding energy −4 kcal/mol), (E) SPRG_35021 with chloramphenicol (DB00446; binding energy −7.7 kcal/mol), and (F) SPRG_13682 with azelaic acid (DB00548; binding energy −5.0 kcal/mol).
The results of the virtual screening were visualized using LigPlot to show the details of the interactions of the proteins with their respective ligands (Figure 4). Triclosan (DB08604), which is predicted to bind to the uncharacterized protein SPRG_20215, interacts through a single H bond between the O5 atom of the drug and the backbone amino group of Leu23. In addition, there are 13 residues from the protein that contribute to hydrophobic interactions. Similar interacting residues have been reported earlier for an enoyl-acyl carrier protein reductase from Thermus thermophilus (Otero et al., 2012). The binding of benzoic acid (DB03793) to SPRG_02663 involves essentially hydrophobic interactions and a single H bond only. The SPRG_19504 protein complex with sulfamethoxazole (DB01015) involves seven amino acid residues from the protein that interact with the inhibitor through hydrophobic bonds while four residues are involved in H bonds. The interaction between SPRG_13682 and azelaic acid (DB00548) shows that the inhibitor interacts within the core hydrophobic pocket of the protein, with six hydrophobic residues involved in addition to the four H bonds associated with two polar and one negatively charged residues of the protein. Chloramphenicol (DB00446) interacts with SPRG_35021 through four H bonds while three other residues from the protein are engaged in hydrophobic interactions. The smallest inhibitor analyzed, acetohydroxamic acid (DB00551), forms H bonds with charged residues like His, Asp, and Lys, and it also interacts through hydrophobic bonds with five surrounding residues of the putative urease protein SPRG_06801. In summary, the docking study reveals that all complexes analyzed involve H bonding and hydrophobic interactions; it supports the initial screening results and provides initial evidence for the likely mechanisms of action of the six drug-proteins pairs identified in this work.
We have identified several compounds along with their probable target proteins using a subtractive proteomics approach and tested nine of these molecules in vitro for their efficacy against the growth of S. parasitica’s mycelium. Molecular docking studies showed that these compounds exhibit minimum binding energies to their respective targets and present a strong affinity toward the active pockets of the proteins, which further supports the experimentally observed growth inhibition. Considering both the low effective concentration of some of these compounds against Saprolegnia and the fact that they are already used in humans suggest that they would be safe to use in aquaculture. Moreover, the concentration of triclosan effective against Saprolegnia under our in vitro conditions (4 μg/ml) is significantly lower to its “no-observed-adverse-effect level” (NOAEL) in humans (0.3% w/w; Lee et al., 2019). Similarly, acetohydroxamic acid, which is effective against S. parasitica at around 600–800 μg/ml, is used at up to 15 mg/kg/day for the treatment of human urinary infections and stones (Griffith et al., 1981; Williams et al., 1984). Azelaic acid (600–800 μg/ml against S. parasitica) has not shown any toxic effect during oral (≤20 g/day) or topical (20% cream) administration to humans (Fitton and Goa, 1991). Both sulfamethoxazole and chloramphenicol have been widely used as antibacterial agents in humans and animals. Sulfamethoxazole (800 μg/ml against S. parasitica) has been used in combination with trimethoprim for over 30 years, with a recommended dosage of up to 40 mg/kg/12 h (Dudley et al., 1984). Similarly, also effective at 800 μg/ml against S. parasitica, chloramphenicol has a recommended dosage of 25 mg/kg/6 h (Powell and Nahata, 1982). Future work is required to confirm the probable mechanism of action of these promising compounds as well as their efficacy on the different developmental stages of S. parasitica (Srivastava et al., 2018). It may also become possible to increase their efficacy by downstream modification and make them usable for the treatment of saprolegniasis in fish eggs, juvenile, and adult fish. Furthermore, the current integrative approach is not limited to saprolegniasis but it can also be employed for the identification of potential inhibitors of other oomycete pathogens.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SK, VB, and VS designed the experimental work, analyzed the data and wrote the manuscript. RM contributed to the molecular modeling experiments. All authors contributed to the article and approved the submitted version.
This work was supported by grants to VB from the European Commission (Horizon 2020 FET-OPEN Grant Agreement No. 828940) and to VS from the Swedish Research Council FORMAS (grant #2019-00912).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.571093/full#supplementary-material
CDD, Conserved domain database; DDAC, Didecyldimethylammonium chloride; DEG, Database of essential genes; DHFR, Dihydrofolate reductase; DHPS, Dihydropteroate synthase; DMSO, Dimethyl sulfoxide; FDA, Food and drug administration; PDA, Potato dextrose agar; SDR, Short-chain dehydrogenases/reductases.
Acar, Ü., İnanan, B. E., Zemheri, F., Kesbiç, O. S., and Yılmaz, S. (2018). Acute exposure to boron in Nile tilapia (Oreochromis niloticus): median-lethal concentration (LC50), blood parameters, DNA fragmentation of blood and sperm cells. Chemosphere 213, 345–350. doi: 10.1016/j.chemosphere.2018.09.063
Ali, S. E., Thoen, E., Evensen, O., and Skaar, I. (2014). Boric acid inhibits germination and colonization of Saprolegnia spores in vitro and in vivo. PLoS One 9:e91878. doi: 10.1371/journal.pone.0091878
Amborabé, B. -E., Fleurat-Lessard, P., Chollet, J. -F., and Roblin, G. (2002). Antifungal effects of salicylic acid and other benzoic acid derivatives towards Eutypa lata: structure-activity relationship. Plant Physiol. Biochem. 40, 1051–1060. doi: 10.1016/s0981-9428(02)01470-5
Ann, P., and Ko, W. (1992). Survey of antibiotic resistance and dependence in Phytophthora. Mycologia 84, 82–86. doi: 10.2307/3760405
Bhardwaj, T., Haque, S., and Somvanshi, P. (2019). Comparative assessment of the therapeutic drug targets of C. botulinum ATCC 3502 and C. difficile str. 630 using in silico subtractive proteomics approach. J. Cell. Biochem. 120, 16160–16184. doi: 10.1002/jcb.28897
Bilsland, E., van Vliet, L., Williams, K., Feltham, J., Carrasco, M. P., Fotoran, W. L., et al. (2018). Plasmodium dihydrofolate reductase is a second enzyme target for the antimalarial action of triclosan. Sci. Rep. 8:1038. doi: 10.1038/s41598-018-19549-x
Blaustein, A. R., Hokit, D. G., O’Hara, R. K., and Holt, R. A. (1994). Pathogenic fungus contributes to amphibian losses in the Pacific northwest. Biol. Conserv. 67, 251–254. doi: 10.1016/0006-3207(94)90616-5
Bly, J., Quiniou, S., Lawson, L., and Clem, L. (1996). Therapeutic and prophylactic measures for winter saprolegniosis in channel catfish. Dis. Aquat. Org. 24, 25–33. doi: 10.3354/dao024025
Branson, E. (2002). Efficacy of bronopol against infection of rainbow trout (Oncorhynchus mykiss) with the fungus Saprolegnia species. Vet. Rec. 151, 539–541. doi: 10.1136/vr.151.18.539
Brown, A. E., and Swinburne, T. (1971). Benzoic acid: an antifungal compound formed in Bramley’s seedling apple fruits following infection by Nectria galligena Bres. Physiol. Plant Pathol. 1, 469–475. doi: 10.1016/0048-4059(71)90009-9
Bulone, V. (1992). Etude des glycanes pariétaux de Saprolegnia monoica et caractérisation biochimique et immunologique de leurs enzymes de synthèse. PhD thesis. Villeurbanne, France: University of Lyon.
Bulone, V., and Fèvre, M. (1996). A 34-kilodalton polypeptide is associated with 1,3-beta-glucan synthase activity from the fungus Saprolegnia monoica. FEMS Microbiol. Lett. 140, 145–150. doi: 10.1111/j.1574-6968.1996.tb08328.x
Bulone, V., Girard, V., and Fevre, M. (1990). Separation and partial purification of 1,3-beta-glucan and 1,4-β-Glucan synthases from Saprolegnia. Plant Physiol. 94, 1748–1755. doi: 10.1104/pp.94.4.1748
Cesur, M. F., Siraj, B., Uddin, R., Durmuş, S., and Çakır, T. (2020). Network-based metabolism-centered screening of potential drug targets in Klebsiella pneumoniae at genome scale. Front. Cell. Infect. Microbiol. 9:447. doi: 10.3389/fcimb.2019.00447
Das, S. K., Murmu, K., Das, A., Shakuntala, I., Das, R. K., Ngachan, S. V., et al. (2012). Studies on the identification and control of pathogen Saprolegnia in selected Indian major carp fingerlings at mid hill altitude. J. Environ. Biol. 33, 545–549.
Davidse, L. C. (1986). Benzimidazole fungicides: mechanism of action and biological impact. Annu. Rev. Phytopathol. 24, 43–65. doi: 10.1146/annurev.py.24.090186.000355
Dhillon, G. S., Kaur, S., Pulicharla, R., Brar, S. K., Cledon, M., Verma, M., et al. (2015). Triclosan: current status, occurrence, environmental risks and bioaccumulation potential. Int. J. Environ. Res. Public Health 12, 5657–5684. doi: 10.3390/ijerph120505657
Diéguez-Uribeondo, J., Cerenius, L., and Söderhäll, K. (1994). Saprolegnia parasitica and its virulence on three different species of freshwater crayfish. Aquaculture 120, 219–228. doi: 10.1016/0044-8486(94)90080-9
Donovan, M., Schumuke, J. J., Fonzi, W. A., Bonar, S. L., Gheesling-Mullis, K., Jacob, G. S., et al. (2001). Virulence of a phosphoribosylaminoimidazole carboxylase-deficient Candida albicans strain in an immunosuppressed murine model of systemic candidiasis. Infect. Immun. 69, 2542–2548. doi: 10.1128/IAI.69.4.2542-2548.2001
Dudley, M. N., Levitz, R., Quintiliani, R., Hickingbotham, J., and Nightingale, C. (1984). Pharmacokinetics of trimethoprim and sulfamethoxazole in serum and cerebrospinal fluid of adult patients with normal meninges. Antimicrob. Agents Chemother. 26, 811–814. doi: 10.1128/AAC.26.6.811
Efferth, T., Romero, M. R., Wolf, D. G., Stamminger, T., Marin, J. J., and Marschall, M. (2008). The antiviral activities of artemisinin and artesunate. Clin. Infect. Dis. 47, 804–811. doi: 10.1086/591195
Feder, V., Kmetzsch, L., Staats, C. C., Vidal-Figueiredo, N., Ligabue-Braun, R., Carlini, C. R., et al. (2015). Cryptococcus gattii urease as a virulence factor and the relevance of enzymatic activity in cryptococcosis pathogenesis. FEBS J. 282, 1406–1418. doi: 10.1111/febs.13229
Fernandez-Beneitez, M. J., Ortiz-Santaliestra, M. E., Lizana, M., and Dieguez-Uribeondo, J. (2011). Differences in susceptibility to Saprolegnia infections among embryonic stages of two anuran species. Oecologia 165, 819–826. doi: 10.1007/s00442-010-1889-5
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Fitton, A., and Goa, K. L. (1991). Azelaic acid: a review of its pharmacological properties and therapeutic efficacy in acne and hyperpigmentary skin disorders. Drugs 41, 780–798. doi: 10.2165/00003495-199141050-00007
Flores, S., Montenegro, I., Villena, J., Cuellar, M., Werner, E., Godoy, P., et al. (2016). Synthesis and evaluation of novel oxyalkylated derivatives of 2',4'-dihydroxychalcone as anti-oomycete agents against bronopol resistant strains of Saprolegnia sp. Int. J. Mol. Sci. 17:1366. doi: 10.3390/ijms17081366
Follmer, C. (2010). Ureases as a target for the treatment of gastric and urinary infections. J. Clin. Pathol. 63, 424–430. doi: 10.1136/jcp.2009.072595
Gieseker, C., Serfling, S., and Reimschuessel, R. (2006). Formalin treatment to reduce mortality associated with Saprolegnia parasitica in rainbow trout, Oncorhynchus mykiss. Aquaculture 253, 120–129. doi: 10.1016/j.aquaculture.2005.07.039
Gil-Alonso, S., Jauregizar, N., Canton, E., Eraso, E., and Quindos, G. (2015). In vitro fungicidal activities of anidulafungin, caspofungin, and micafungin against Candida glabrata, Candida bracarensis, and Candida nivariensis evaluated by time-kill studies. Antimicrob. Agents Chemother. 59, 3615–3618. doi: 10.1128/AAC.04474-14
Griffith, D. P., Moskowitz, P., and Feldman, S. (1981). “Infection-induced stones: status of clinic trials with Urostat™ (acetohydroxamic acid)” in Urolithiasis. eds. L. H. Smith, W. G. Robertson, and B. Finlayson (Boston, MA: Springer), 199–208.
Guerriero, G., Avino, M., Zhou, Q., Fugelstad, J., Clergeot, P. H., and Bulone, V. (2010). Chitin synthases from Saprolegnia are involved in tip growth and represent a potential target for anti-oomycete drugs. PLoS Pathog. 6:e1001070. doi: 10.1371/journal.ppat.1001070
Hevener, K. E., Yun, M. K., Qi, J., Kerr, I. D., Babaoglu, K., Hurdle, J. G., et al. (2010). Structural studies of pterin-based inhibitors of dihydropteroate synthase. J. Med. Chem. 53, 166–177. doi: 10.1021/jm900861d
Hoskonen, P., Heikkinen, J., Eskelinen, P., and Pirhonen, J. (2015). Efficacy of clove oil and ethanol against Saprolegnia sp. and usability as antifungal agents during incubation of rainbow trout Oncorhynchus mykiss (Walbaum) eggs. Aquac. Res. 46, 581–589. doi: 10.1111/are.12200
Howe, G. E., Gingerich, W. H., Dawson, V. K., and Olson, J. J. (1999). Efficacy of hydrogen peroxide for treating saprolegniasis in channel catfish. J. Aquat. Anim. Health 11, 222–230. doi: 10.1577/1548-8667(1999)011<0222:EOHPFT>2.0.CO;2
Hu, K., Ma, R. R., Cheng, J. M., Ye, X., Sun, Q., Yuan, H. L., et al. (2016). Analysis of Saprolegnia parasitica transcriptome following treatment with copper sulfate. PLoS One 11:e0147445. doi: 10.1371/journal.pone.0147445
Illarionova, V., Eisenreich, W., Fischer, M., Haussmann, C., Romisch, W., Richter, G., et al. (2002). Biosynthesis of tetrahydrofolate. Stereochemistry of dihydroneopterin aldolase. J. Biol. Chem. 277, 28841–28847. doi: 10.1074/jbc.M204046200
Jansen, F. H., Adoubi, I., Comoe, J. C. K., de Cnodder, T., Jansen, N., Tschulakow, A., et al. (2011). First study of oral Artenimol-R in advanced cervical cancer: clinical benefit, tolerability and tumor markers. Anticancer Res. 31, 4417–4422.
Keller, T. H., Pichota, A., and Yin, Z. (2006). A practical view of ‘druggability’. Curr. Opin. Chem. Biol. 10, 357–361. doi: 10.1016/j.cbpa.2006.06.014
Khodabandeh, S., and Abtahi, B. (2006). Effects of sodium chloride, formalin and iodine on the hatching success of common carp, Cyprinus carpio, eggs. J. Appl. Ichthyol. 22, 54–56. doi: 10.1111/j.1439-0426.2006.00662.x
Laskowski, R. A., and Swindells, M. B. (2011). LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786. doi: 10.1021/ci200227u
Lastauskiene, E., Zinkeviciene, A., Girkontaite, I., Kaunietis, A., and Kvedariene, V. (2014). Formic acid and acetic acid induce a programmed cell death in pathogenic Candida species. Curr. Microbiol. 69, 303–310. doi: 10.1007/s00284-014-0585-9
Lee, J. D., Lee, J. Y., Kwack, S. J., Shin, C. Y., Jang, H. J., Kim, H. Y., et al. (2019). Risk assessment of Triclosan, a cosmetic preservative. Toxicol. Res. 35, 137–154. doi: 10.5487/TR.2019.35.2.137
Lerm, B., Kenyon, C., Schwartz, I. S., Kroukamp, H., de Witt, R., Govender, N. P., et al. (2017). First report of urease activity in the novel systemic fungal pathogen Emergomyces africanus: a comparison with the neurotrope Cryptococcus neoformans. FEMS Yeast Res. 17:fox069. doi: 10.1093/femsyr/fox069
Liu, L., Shen, Y. F., Liu, G. L., Ling, F., Liu, X. Y., Hu, K., et al. (2015). Inhibition of dioscin on Saprolegnia in vitro. FEMS Microbiol. Lett. 362:fnv196. doi: 10.1093/femsle/fnv196
Luo, H., Lin, Y., Gao, F., Zhang, C. T., and Zhang, R. (2014). DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 42, D574–D580. doi: 10.1093/nar/gkt1131
Machlis, L. (1953). Growth and nutrition of water molds in the subgenus Euallomyces. II. Optimal composition of the minimal medium. Am. J. Bot. 40, 450–460. doi: 10.1002/j.1537-2197.1953.tb06505.x
Mahmoud, Y. A. (2007). In vitro and in vivo antifungal activity of cetrimide (cetyltrimethyl ammonium bromide) against fungal keratitis caused by Fusarium solani. Mycoses 50, 64–70. doi: 10.1111/j.1439-0507.2006.01313.x
Marchand, P. A., Phan, T. M., Straus, D. L., Farmer, B. D., Stüber, A., and Meinelt, T. (2012). Reduction of in vitro growth in flavobacterium columnare and Saprolegnia parasitica by products containing peracetic acid. Aquac. Res. 43, 1861–1866. doi: 10.1111/j.1365-2109.2011.02995.x
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Marking, L. L., Rach, J. J., and Schreier, T. M. (1994). American fisheries society evaluation of antifungal agents for fish culture. Progress. Fish Cult. 56, 225–231. doi: 10.1577/1548-8640(1994)056<0225:AFSEOA>2.3.CO;2
Matherly, L. H. (2001). Molecular and cellular biology of the human reduced folate carrier. Prog. Nucleic Acid Res. Mol. Biol. 67, 131–162. doi: 10.1016/s0079-6603(01)67027-2
Mélida, H., Sandoval-Sierra, J. V., Dieguez-Uribeondo, J., and Bulone, V. (2013). Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell 12, 194–203. doi: 10.1128/EC.00288-12
Mitchell, A., and Collins, C. (1997). Review of the therapeutic uses of hydrogen peroxide in fish production. Aquac. Mag. 23, 74–79.
Mustafa, S. (2019). Assessment of hydrogen peroxide on histopathology and survival rate in common carp, Cyprinus carpio l. infected with saprolegniasis. Iraqi J. Agri. Sci. 50, 697–704. doi: 10.36103/ijas.v2i50.669
Muzzarelli, R. A., Muzzarelli, C., Tarsi, R., Miliani, M., Gabbanelli, F., and Cartolari, M. (2001). Fungistatic activity of modified chitosans against Saprolegnia parasitica. Biomacromolecules 2, 165–169. doi: 10.1021/bm000091s
Nakagawa, K., Hara, C., Tokuyama, S., Takada, K., and Imamura, N. (2012). Saprolmycins A-E, new angucycline antibiotics active against Saprolegnia parasitica. J. Antibiot. 65, 599–607. doi: 10.1038/ja.2012.86
Nazzaro-Porro, M. (1987). Azelaic acid. J. Am. Acad. Dermatol. 17, 1033–1041. doi: 10.1016/s0190-9622(87)70294-1
Nielsen, M., Lundegaard, C., Lund, O., and Petersen, T. N. (2010). CPHmodels-3.0—remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res. 38, W576–W581. doi: 10.1093/nar/gkq535
N. R. Council (2010). Acute exposure guideline levels for selected airborne chemicals. Peracetic acid, acute exposure guideline levels. Washington: National Academies Press.
Ohta, T., Shibata, H., Kawamori, T., Iimuro, M., Sugimura, T., and Wakabayashi, K. (2001). Marked reduction of helicobacter pylori-induced gastritis by urease inhibitors, acetohydroxamic acid and flurofamide, in Mongolian gerbils. Biochem. Biophys. Res. Commun. 285, 728–733. doi: 10.1006/bbrc.2001.5229
Oono, H., and Hatai, K. (2007). Antifungal activities of bronopol and 2-methyl-4-isothiazolin-3-one (MT) against Saprolegnia. Biocontrol Sci. 12, 145–148. doi: 10.4265/bio.12.145
Otero, J. M., Noel, A. J., Guardado-Calvo, P., Llamas-Saiz, A. L., Wende, W., Schierling, B., et al. (2012). High-resolution structures of Thermus thermophilus enoyl-acyl carrier protein reductase in the apo form, in complex with NAD+ and in complex with NAD+ and triclosan. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 1139–1148. doi: 10.1107/S1744309112033982
Otero, E., Vergara, S., Robledo, S. M., Cardona, W., Carda, M., Velez, I. D., et al. (2014). Synthesis, leishmanicidal and cytotoxic activity of triclosan-chalcone, triclosan-chromone and triclosan-coumarin hybrids. Molecules 19, 13251–13266. doi: 10.3390/molecules190913251
Öz, M., Inanan, B. E., and Dikel, S. (2018). Effect of boric acid in rainbow trout (Oncorhynchus mykiss) growth performance. J. Appl. Anim. Res. 46, 990–993. doi: 10.1080/09712119.2018.1450258
Panek, M., and Tomsovsky, M. (2017). In vitro growth response of Phytophthora cactorum, P. nicotianae and P. x pelgrandis to antibiotics and fungicides. Folia Microbiol. 62, 269–277. doi: 10.1007/s12223-017-0493-z
Perfect, J. R. (2017). The antifungal pipeline: a reality check. Nat. Rev. Drug Discov. 16, 603–616. doi: 10.1038/nrd.2017.46
Plumb, J. A. (1999). Overview of warmwater fish diseases. J. Appl. Aquac. 9, 1–10. doi: 10.1300/j028v09n02_01
Plumed-Ferrer, C., and von Wright, A. (2011). Antimicrobial activity of weak acids in liquid feed fermentations, and its effects on yeasts and lactic acid bacteria. J. Sci. Food Agric. 91, 1032–1040. doi: 10.1002/jsfa.4278
Pottinger, T. G., and Day, J. G. (1999). A Saprolegnia parasitica challenge system for rainbow trout: assessment of Pyceze as an anti-fungal agent for both fish and ova. Dis. Aquat. Org. 36, 129–141. doi: 10.3354/dao036129
Powell, D. A., and Nahata, M. C. (1982). Chloramphenicol: new perspectives on an old drug. Drug Intell. Clin. Pharm. 16, 295–300. doi: 10.1177/106002808201600404
Pull, L., Lupoglazoff, J. M., Beardmore, M., Michel, J. F., Buffet, P., Bouchaud, O., et al. (2019). Artenimol-piperaquine in children with uncomplicated imported falciparum malaria: experience from a prospective cohort. Malar. J. 18:419. doi: 10.1186/s12936-019-3047-9
Rezinciuc, S., Sandoval-Sierra, J. V., and Dieguez-Uribeondo, J. (2014). Molecular identification of a bronopol tolerant strain of Saprolegnia australis causing egg and fry mortality in farmed brown trout, Salmo trutta. Fungal Biol. 118, 591–600. doi: 10.1016/j.funbio.2013.11.011
Rooke, D. M., and Shattock, R. (1983). Effect of chloramphenicol and streptomycin on developmental stages of Phytophthom infestans. Microbiology 129, 3401–3410. doi: 10.1099/00221287-129-11-3401
Saegeman, V. S., Ectors, N. L., Lismont, D., Verduyckt, B., and Verhaegen, J. (2008). Short‐ and long-term bacterial inhibiting effect of high concentrations of glycerol used in the preservation of skin allografts. Burns 34, 205–211. doi: 10.1016/j.burns.2007.02.009
Schweizer, H. P. (2001). Triclosan: a widely used biocide and its link to antibiotics. FEMS Microbiol. Lett. 202, 1–7. doi: 10.1111/j.1574-6968.2001.tb10772.x
Seeliger, D., and de Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 24, 417–422. doi: 10.1007/s10822-010-9352-6
Shen, H., Yang, Y., Wang, F., Zhang, Y., Ye, N., Xu, S., et al. (2009). Characterization of the putative tryptophan synthase beta-subunit from Mycobacterium tuberculosis. Acta Biochim. Biophys. Sin. 41, 379–388. doi: 10.1093/abbs/gmp017
Shibata, K., Kasuga, O., Yasoshima, A., Matsushita, T., and Kawakami, Y. (1997). Bactericidal effect of ecabet sodium on clarithromycin‐ and metronidazole-resistant clinical isolates of Helicobacter pylori. Jpn. J. Antibiot. 50, 525–531.
Solanki, V., and Tiwari, V. (2018). Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 8:9044. doi: 10.1038/s41598-018-26689-7
Srivastava, V., Rezinciuc, S., and Bulone, V. (2018). Quantitative proteomic analysis of four developmental stages of Saprolegnia parasitica. Front. Microbiol. 8:2658. doi: 10.3389/fmicb.2017.02658
Srivastava, S., Sinha, R., and Roy, D. (2004). Toxicological effects of malachite green. Aquat. Toxicol. 66, 319–329. doi: 10.1016/j.aquatox.2003.09.008
Stueland, S., Heier, B. T., and Skaar, I. (2005). A simple in vitro screening method to determine the effects of drugs against growth of Saprolegnia parasitica. Mycol. Prog. 4, 273–279. doi: 10.1007/s11557-006-0131-7
Sun, Q., Hu, K., and Yang, X. L. (2014). The efficacy of copper sulfate in controlling infection of Saprolegnia parasitica. J. World Aquacult. Soc. 45, 220–225. doi: 10.1111/jwas.12113
Surolia, N., and Surolia, A. (2001). Triclosan offers protection against blood stages of malaria by inhibiting enoyl-ACP reductase of Plasmodium falciparum. Nat. Med. 7, 167–173. doi: 10.1038/84612
Takada, K., Kajiwara, H., and Imamura, N. (2010). Oridamycins A and B, anti-Saprolegnia parasitica indolosesquiterpenes isolated from Streptomyces sp. KS84. J. Nat. Prod. 73, 698–701. doi: 10.1021/np1000522
Takahashi, K., Sakai, K., Nagano, Y., Orui Sakaguchi, S., Lima, A. O., Pellizari, V. H., et al. (2017). Cladomarine, a new anti-saprolegniasis compound isolated from the deep-sea fungus, Penicillium coralligerum YK-247. J. Antibiot. 70, 911–914. doi: 10.1038/ja.2017.58
Tang, H. Q., Hu, J., Yang, L., and Tan, R. X. (2000). Terpenoids and flavonoids from Artemisia species. Planta Med. 66, 391–393. doi: 10.1055/s-2000-8538
Tedesco, P., Fioravanti, M. L., and Galuppi, R. (2019). In vitro activity of chemicals and commercial products against Saprolegnia parasitica and Saprolegnia delica strains. J. Fish Dis. 42, 237–248. doi: 10.1111/jfd.12923
Thompson, M. (1992). NTP technical report on the toxicity studies of formic acid (CAS no. 64-18-6) administered by inhalation to F344/N rats and B6C3F1 mice. Toxic. Rep. Ser. 19, D1–D3.
Tidy, H. (1915). The strength of picric acid as an antiseptic. Lancet 186, 604–605. doi: 10.1016/S0140-6736(00)54228-7
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
van Slooten, C., Peperzak, L., and Buma, A. G. (2015). Assessment of didecyldimethylammonium chloride as a ballast water treatment method. Environ. Technol. 36, 435–449. doi: 10.1080/09593330.2014.951401
van West, P. (2006). Saprolegnia parasitica, an oomycete pathogen with a fishy appetite: new challenges for an old problem. Mycologist 20, 99–104. doi: 10.1016/j.mycol.2006.06.004
Warrilow, A. G., Hull, C. M., Rolley, N. J., Parker, J. E., Nes, W. D., Smith, S. N., et al. (2014). Clotrimazole as a potent agent for treating the oomycete fish pathogen Saprolegnia parasitica through inhibition of sterol 14alpha-demethylase (CYP51). Appl. Environ. Microbiol. 80, 6154–6166. doi: 10.1128/AEM.01195-14
Williams, J. J., Rodman, J. S., and Peterson, C. M. (1984). A randomized double-blind study of acetohydroxamic acid in struvite nephrolithiasis. N. Engl. J. Med. 311, 760–764. doi: 10.1056/NEJM198409203111203
Willoughby, L., and Roberts, R. (1992). Towards strategic use of fungicides against Saprolegnia parasitica in salmonid fish hatcheries. J. Fish Dis. 15, 1–13. doi: 10.1111/j.1365-2761.1992.tb00631.x
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082. doi: 10.1093/nar/gkx1037
Xaplanteri, M. A., Andreou, A., Dinos, G. P., and Kalpaxis, D. L. (2003). Effect of polyamines on the inhibition of peptidyltransferase by antibiotics: revisiting the mechanism of chloramphenicol action. Nucleic Acids Res. 31, 5074–5083. doi: 10.1093/nar/gkg686
Xue-Gang, H., Lei, L., Cheng, C., Kun, H., Xian-Le, Y., and Gao-Xue, W. (2013). In vitro screening of Chinese medicinal plants for antifungal activity against Saprolegnia sp. and Achlya klebsiana. N. Am. J. Aquac. 75, 468–473. doi: 10.1080/15222055.2013.808298
Keywords: disease control, fish pathogen, growth inhibitors, oomycete, Saprolegnia parasitica, in silico proteomics
Citation: Kumar S, Mandal RS, Bulone V and Srivastava V (2020) Identification of Growth Inhibitors of the Fish Pathogen Saprolegnia parasitica Using in silico Subtractive Proteomics, Computational Modeling, and Biochemical Validation. Front. Microbiol. 11:571093. doi: 10.3389/fmicb.2020.571093
Received: 09 June 2020; Accepted: 22 September 2020;
Published: 16 October 2020.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Francisco P. Chávez, University of Chile, ChileCopyright © 2020 Kumar, Mandal, Bulone and Srivastava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vaibhav Srivastava, dmFzcmlAa3RoLnNl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.