- Department of Genetics, Microbiology and Statistics, School of Biology, Universitat de Barcelona, Barcelona, Spain
Post-transcriptional regulation mediated by regulatory small RNAs (sRNAs) has risen as a key player in fine-tuning gene expression in response to environmental stimuli. Here, we show that, in Salmonella enterica, the central metabolic regulator CRP-cAMP differentially regulates the sRNAs CsrB and CsrC in a growth phase-dependent manner. While CsrB expression remains unchanged during growth, CsrC displays a growth phase-dependent expression profile, being weakly expressed at the logarithmic growth phase and induced upon entry into stationary phase. We show that CRP-cAMP contributes to the expression pattern of CsrC by repressing its expression during the logarithmic growth phase. The CRP-cAMP mediated repression of CsrC is independent of SirA, a known transcriptional CsrB/CsrC activator. We further show that the sRNA Spot 42, which is derepressed in a Δcrp strain, upregulates CsrC during logarithmic growth. We propose a model where the growth-dependent regulation of CsrC is sustained by the CRP-cAMP-mediated repression of Spot 42. Together, our data point toward a differential regulation of the sRNAs CsrB and CsrC in response to environmental stimuli, leading to fine-tuning of gene expression via the sequestration of the RNA-binding protein CsrA.
Introduction
Bacteria need to adapt rapidly to changing environmental conditions, which is particularly crucial for pathogenic bacteria during the process of infection. While transcription plays a major role in the regulation of gene expression, bacteria display a plethora of post-transcriptional mechanisms that allow the fine-tuning of gene expression in response to environmental cues. Salmonella enterica has been investigated extensively with respect to gene regulation and has become a model for the study of post-transcriptional RNA-mediated regulation.
Among the described post-transcriptional regulation mechanisms, a prominent role has been attributed to sRNAs that modulate gene expression primarily via binding to target mRNAs (Hör et al., 2020). However, sRNAs can also regulate gene expression at the post-translational level, such as the two sRNAs CsrB and CsrC. These sRNAs contain several stem loop structures with GGA motifs in their loop regions, enabling them to bind CsrA. CsrA is a widely conserved RNA-binding protein that inhibits translation by binding to GGA motifs around the ribosome-binding site of target mRNAs (Romeo and Babitzke, 2018). Binding of CsrB and CsrC to CsrA titrates CsrA away from its target mRNAs, thereby counteracting its inhibitory activity (Liu et al., 1997; Weilbacher et al., 2003).
CRP is a transcription factor that acts as a metabolic sensor and becomes active upon binding to the intracellular second messenger cAMP (cyclic adenosine monophosphate) (Görke and Stülke, 2008). Interaction of CRP-cAMP with DNA leads to activation or repression of its target genes. In E. coli, it has been shown that CRP-cAMP represses the expression of both CsrB and CsrC (Pannuri et al., 2016). While repression of CsrB occurs through an indirect mechanism, the repression of CsrC occurs by direct binding of CRP-cAMP to the promoter region of csrC, where it competes with the csrC activator UvrY (Pannuri et al., 2016). In Salmonella, the homolog of the two-component system BarA-UvrY is BarA-SirA (Johnston et al., 1996), which, as in E. coli, it also positively regulates the expression of CsrB and CsrC (Teplitski et al., 2003; Fortune et al., 2006; Martínez et al., 2011). Expression studies using S. enterica cultures on solid media indicates that CRP-cAMP, in contrast to its role described in E. coli, positively regulate the expression of CsrB and CsrC via upregulation of sirA (Teplitski et al., 2006). In S. enterica other sRNAs, such as CyaR and Spot 42, are also regulated by CRP-cAMP, acting as an activator for CyaR and as a repressor for Spot 42 (Papenfort et al., 2008; El Mouali et al., 2018).
In this study we investigated the role of CRP-cAMP in the expression of the sRNAs CsrB and CsrC in the model organism S. enterica using liquid cultures. We describe that CsrC, but not CsrB, display a growth-dependent expression pattern. CsrC expression is silenced during logarithmic growth and highly expressed upon entry into stationary phase, while CsrB expression seems to be constitutive through the growth curve. CRP-cAMP plays a relevant role in this growth-dependent regulatory network. CRP-cAMP differentially regulates the levels of CsrB and CsrC as it does not affect CsrB expression but represses CsrC expression during logarithmic growth. Remarkably, while SirA is required for full expression of CsrC, the CRP-cAMP-mediated repression during logarithmic growth seems to be independent of SirA-mediated regulation. Our data further indicates that Spot 42 contributes to the CRP-cAMP-induced repression of CsrC, suggesting that CRP-cAMP and Spot 42 converge into the differential regulation of CsrB and CsrC in Salmonella.
Results
CRP-cAMP Represses CsrC Levels at the Logarithmic Growth Phase
In order to study the CRP-cAMP mediated regulation of CsrB and CsrC in Salmonella, the expression of both sRNAs was monitored using transcriptional reporter fusions. The regulatory elements controlling CsrB and CsrC expression in Salmonella have been identified in silico and characterized experimentally (Martínez et al., 2014). Accordingly, the described regulatory regions of CsrB (−403, +18) and CsrC (−347, +60) were cloned as lacZ transcriptional fusions in the pQF50 vector (Farinha and Kropinski, 1989), allowing to monitor the levels of CsrB and CsrC during growth. The contribution of CRP-cAMP was assessed by determining the expression level of CsrB and CsrC in wild-type (WT) and in a Δcrp mutant strain, lacking the transcriptional factor CRP. In rich media, no differences in growth rate could be observed in WT and Δcrp strains carrying either csrB-lacZ or csrC-lacZ (Figure 1A). Transcriptional expression was monitored during logarithmic growth (OD600 nm 0.4) and upon entry into stationary phase (OD600 nm 2.0). In the WT, CsrB expression was apparently identical in the two growth phases (Figure 1B). By contrast, CsrC displayed a growth-dependent expression pattern, being less expressed during logarithmic growth and being induced after entering stationary phase (Figure 1B). Remarkably, CRP-cAMP contributes to the CsrC growth-dependent expression pattern. In Δcrp, the expression of CsrC is induced when compared to WT at the logarithmic phase and to a lesser extent at the stationary phase, indicating that CRP-cAMP represses CsrC expression (Figure 1B). By contrast, no effect on CsrB regulation by CRP-cAMP was observed. These results were unexpected since it was previously described that CRP-cAMP acts as an activator of both CsrB and CsrC expression when Salmonella is grown on LB agar media (Teplitski et al., 2006). To discern if the discrepancy could be consequence of differences in the genetic constructs used to monitor gene expression, a similar experiment as in Teplitski et al. (2006) was performed using our strains. The transcriptional expression was monitored after growth on LB agar media. Consistently with the previous report, the transcriptional expression of CsrB and CsrC was strongly diminished in a Δcrp derivative strain compared to WT when Salmonella cells were grown on LB agar media as noted by the white colony phenotype of Δcrp when compared to the blue colony phenotype of WT (Supplementary Image S1; Teplitski et al., 2006). Altogether, CRP-cAMP seem to be required for activation of the expression of CsrB and CsrC in solid media, while it acts as a repressor of CsrC particularly at logarithmic growth phase. This indicates that CRP-cAMP-mediated regulation of CsrB and CsrC is dependent on the growth conditions.
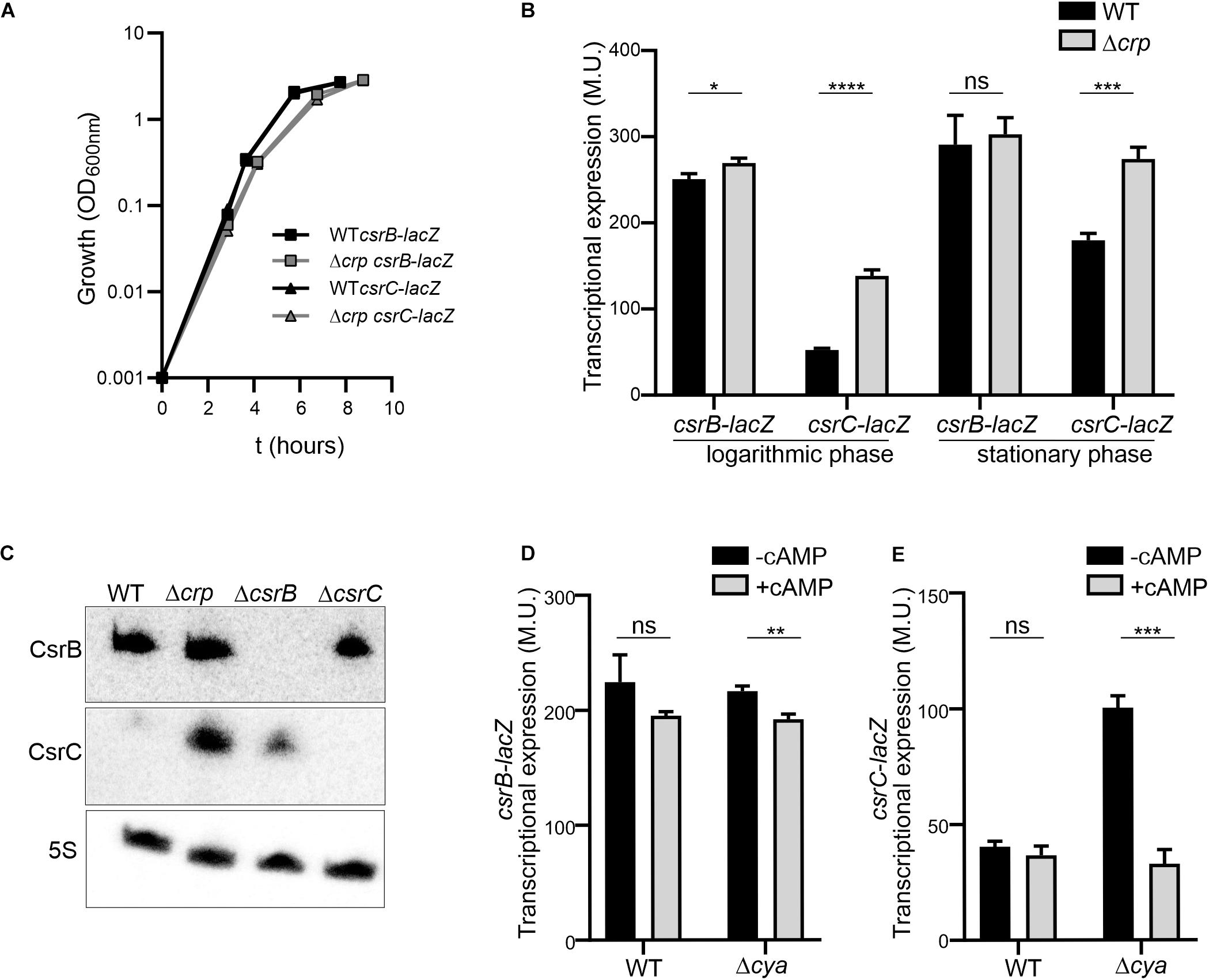
Figure 1. The expression of csrC is repressed at the logarithmic growth phase in a CRP-cAMP mediated manner. (A) Growth curve in LB at 37°C of Salmonella SV5015 strain (WT) and its isogenic Δcrp mutant, carrying either a plasmid fusion of csrB-lacZ or csrC-lacZ. (B) Transcriptional expression of csrB and csrC. Samples from cultures in A were taken at either an OD600 nm of 0.4 or an OD600 nm of 2.0 for determination of the β-galactosidase activity in Miller units (M.U.). (C) Northern blot analyses of the csrB and csrC transcripts. Total RNA samples from cultures of WT (SV5015) and its derivatives Δcrp, ΔcsrB, and ΔcsrC grown in LB up to an OD600 nm of 0.4 were analyzed. Detection of the 5S transcript was used as a control. Transcriptional expression of csrB (D) and csrC (E) in the cya mutant strain and chemical complementation. Cultures of WT, Δcrp and Δcya derivatives were grown in LB either in the absence or in the presence of cAMP (5 mM) at 37°C up to logarithmic phase (OD600 nm of 0.4) and samples were taken for β-galactosidase measurement. In B, C and D; β-galactosidase activity was determined for three independent cultures, average and standard deviation are presented. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant.
The differential regulation of CsrC and CsrB by CRP-cAMP at the logarithmic growth phase was further corroborated by the direct RNA detection of CsrB and CsrC. In the WT, CsrC transcript was barely detected, indicating that CsrC expression is tightly silenced. In the Δcrp strain, high levels of CsrC were detected indicating that CRP-cAMP is involved in the CsrC silencing during logarithmic growth (Figure 1C). Contrasting the CRP-cAMP-dependent changes in CsrC levels, CsrB was not influenced by knockout of Δcrp (Figure 1C), thereby agreeing with the results of our transcriptional fusions (Figure 1B). In addition, deletion of ΔcsrB cause a mild upregulation of CsrC, presumably by affecting the positive feed forward loop that free CsrA protein exerts on its repressors CsrB and CsrC (Romeo and Babitzke, 2018).
CRP becomes active upon binding to cAMP, which is produced by the adenylate cyclase Cya. Therefore, absence of crp or cya should display similar expression profiles. Accordingly, the expression of CsrC in logarithmic growth phase is induced in the Δcya derivative strain when compared to WT while no change was observed in the expression of CsrB (Figure 1D). To further confirm the involvement of CRP-cAMP in CsrC regulation, chemical complementation of Δcya was carried out by ectopic addition of cAMP. The addition of cAMP repressed the expression of CsrC in the Δcya derivative strain while no effect was observed in CsrB expression (Figure 1E). Our data indicate that CRP-cAMP is involved in the growth-dependent regulation of CsrC by repressing its expression.
CRP-cAMP-Mediated Repression of CsrC via a SirA-Independent Pathway
To further characterize the CRP-cAMP mediated regulation of CsrC, a chromosomal csrC-lacZ fusion was generated. In agreement with earlier results, chromosomal csrC-lacZ has a growth dependent expression pattern, where it is lowly expressed at the logarithmic growth phase and induced upon entry into early stationary phase (Figure 2A). Interestingly, we further observed an eightfold induction of the chromosomal csrC-lacZ fusion in the Δcrp background when compared to the WT, which was only true during logarithmic growth (Figure 2A).
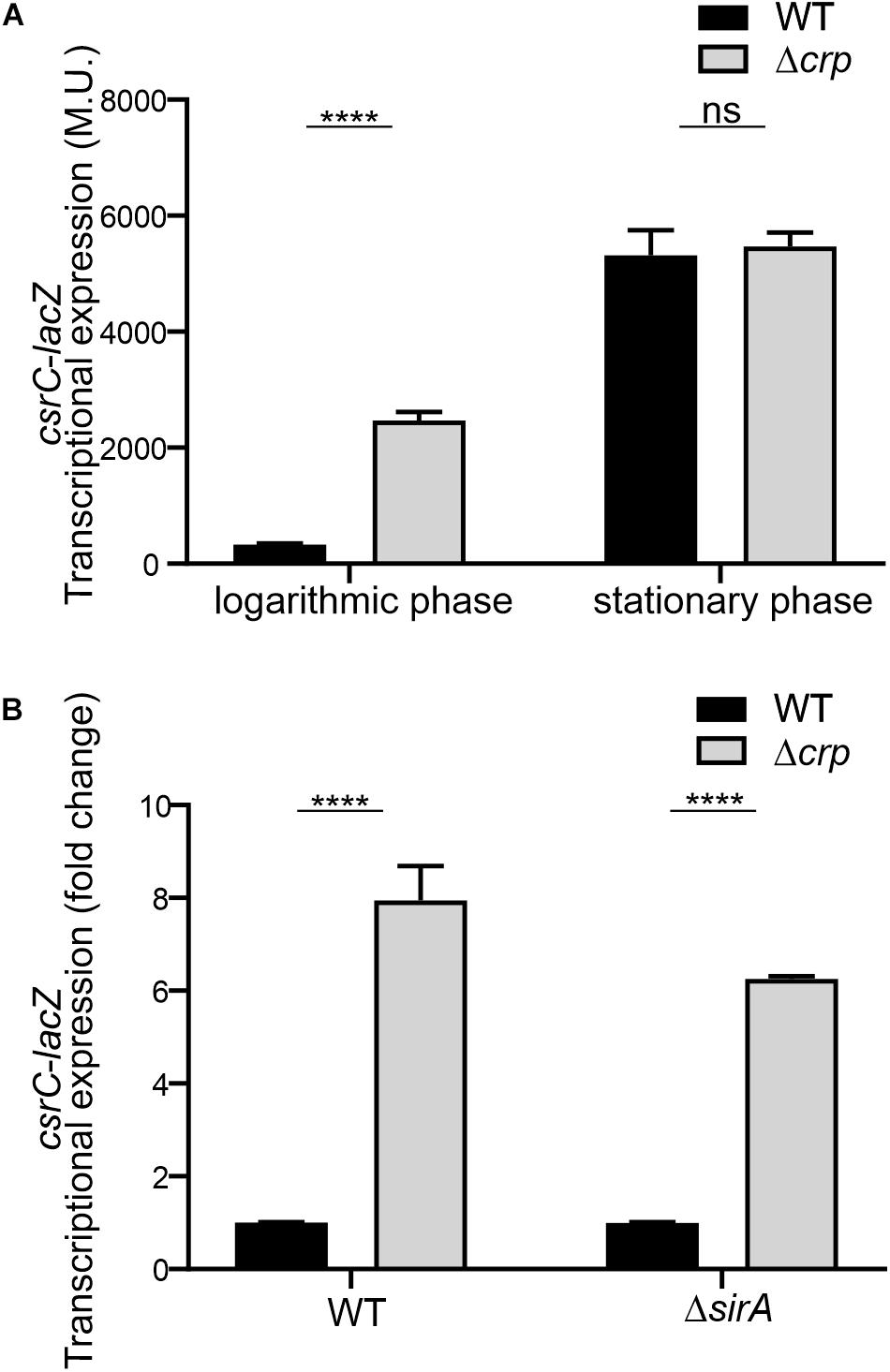
Figure 2. CRP-cAMP mediated repression of csrC is independent of SirA. (A) Transcriptional expression of chromosomal csrC-lacZ fusion was monitored in WT and Δcrp mutant genetic backgrounds. Cultures were grown in LB at 37°C up to an OD600 nm of either 0.4 or 2.0. (B) Transcriptional expression of the chromosomal csrC-lacZ fusion was monitored in WT, Δcrp, ΔsirA, and ΔcrpΔsirA genetic backgrounds. The transcriptional expression is shown in relative values. In each case, the reference (crp+, WT) was set as 1.0. Miller units of the crp+ backgrounds were 308 ± 19 and 28 ± 0.8 for WT and ΔsirA, respectively. β-galactosidase activity was determined for three independent cultures, average and standard deviation is presented. ****p < 0.0001; ns, not significant.
The BarA-SirA two-component system was described to positively regulate the expression of CsrC (Teplitski et al., 2006). The possible involvement of BarA-SirA in the regulation of CsrC by CRP-cAMP was assessed. SirA deletion leads to a decrease of the overall transcriptional expression of the chromosomal csrC-lacZ fusion, 28.2 ± 0.8 Miller units in ΔsirA compared to 308.8 ± 19.3 Miller units in WT. Interestingly, despite the overall lower expression levels of CsrC in the ΔsirA background, the deletion of Δcrp led to a sixfold increase in the transcriptional expression of csrC-lacZ compared to WT (Figure 2B), indicating that CRP-cAMP modulates csrC expression via a SirA-independent mechanism.
In E. coli, it was proposed that CRP-cAMP represses CsrC expression through binding to the upstream region of csrC and competing with UvrY, the SirA homolog (Pannuri et al., 2016). In Salmonella, SirA induces CsrC expression by binding ∼160–168 bp upstream of the csrC promoter (Figure 3A; Martínez et al., 2014). Alignment of the upstream promoter regions of CsrC from E. coli and Salmonella showed that SirA binding site is conserved, whereas the CRP-cAMP binding site displays a lesser extent of conservation (Supplementary Image S2). To characterize if CRP-cAMP binding to the csrC promoter is required for the described regulation, an additional csrC-lacZ fusion was generated where the putative binding sites of these transcriptional factors are not present. As shown in Figure 3A, the pQF50 cloned fragment csrC347-lacZ (−347, +60) maintains SirA/CRP binding sites while csrC91-lacZ (−91, +60) does not. Remarkably, in both csrC347-lacZ and csrC91-lacZ an induction of expression is observed in the Δcrp strain when compared to WT (Figure 3B). These results suggest that CRP-cAMP represses the expression of CsrC during logarithmic growth phase via a mechanism that is independent of the competition between SirA and CRP for the csrC promoter.
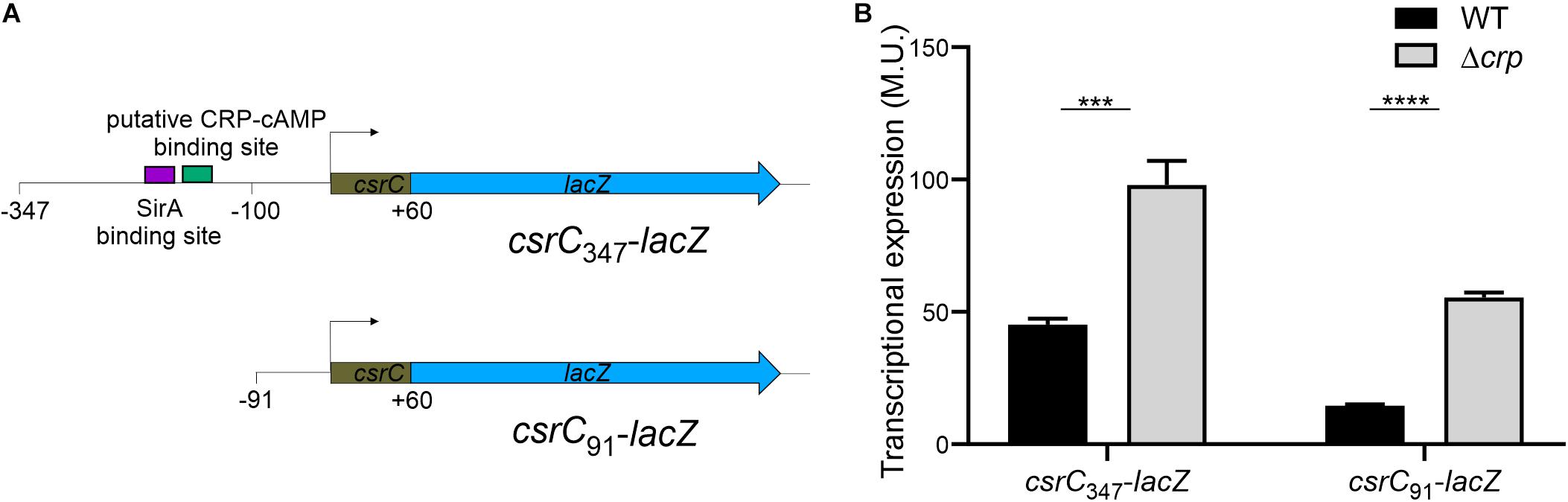
Figure 3. CRP-cAMP represses CsrC in absence of SirA binding site. (A) Diagram of the lacZ transcriptional fusions present in the pQFcsrC and pQFcsrC-91 plasmids. The csrC upstream sequences cloned in pQF50, –347/ + 60 and –91/+60, are depicted. The relative position of the SirA binding site and the putative CRP-cAMP binding site are indicated with a purple and green rectangle. (B) Transcriptional expression of the lacZ fusions described in A in WT and Δcrp genetic backgrounds. In all cases, cultures were grown in LB at 37°C up to an OD600 nm of 0.4. β-galactosidase activity was determined for three independent cultures, average and standard deviation are presented. ***p < 0.001; ****p < 0.0001.
The sRNA Spot 42 Positively Regulates CsrC
In Salmonella, CRP-cAMP represses the expression of the regulatory sRNA Spot 42 in a growth-dependent manner (El Mouali et al., 2018). The similarity of the expression profiles of Spot 42 and CsrC, let us to hypothesize that CRP-cAMP might regulate CsrC via modulation of Spot 42 levels. To this end, we determined CsrC expression during logarithmic growth upon ectopic expression of Spot 42. Remarkably, CsrC transcript levels were strongly induced upon Spot 42 overexpression (pBRSpot 42) when compared to the vector control strain (pBRVC) (Figure 4A). In contrast, no accumulation of CsrB was detected upon overexpression of Spot 42 (Figure 4A). Further supporting our hypothesis, Spot 42 seemed to differentially regulate CsrB and CsrC expression (Figure 4A), similar to the described CRP-cAMP-mediated regulation (Figure 1C). To verify this observation, we overexpressed Spot 42 in the chromosomal csrC-lacZ fusion background, resulting in a threefold induction of CsrC when compared to the vector control strain (Figure 4B). The Spot 42-mediated regulation of CsrC may account for the described repression of CsrC by CRP-cAMP. The contribution of Spot 42 on the induction of CsrC in Δcrp was further assessed. The CsrC derepression in a Δcrp mutant strain drops in absence of Spot 42 (Δspf) as compared to a Spot 42 proficient strain indicating that Spot 42 contributes to the CsrC derepression in a Δcrp background (Figure 4C).
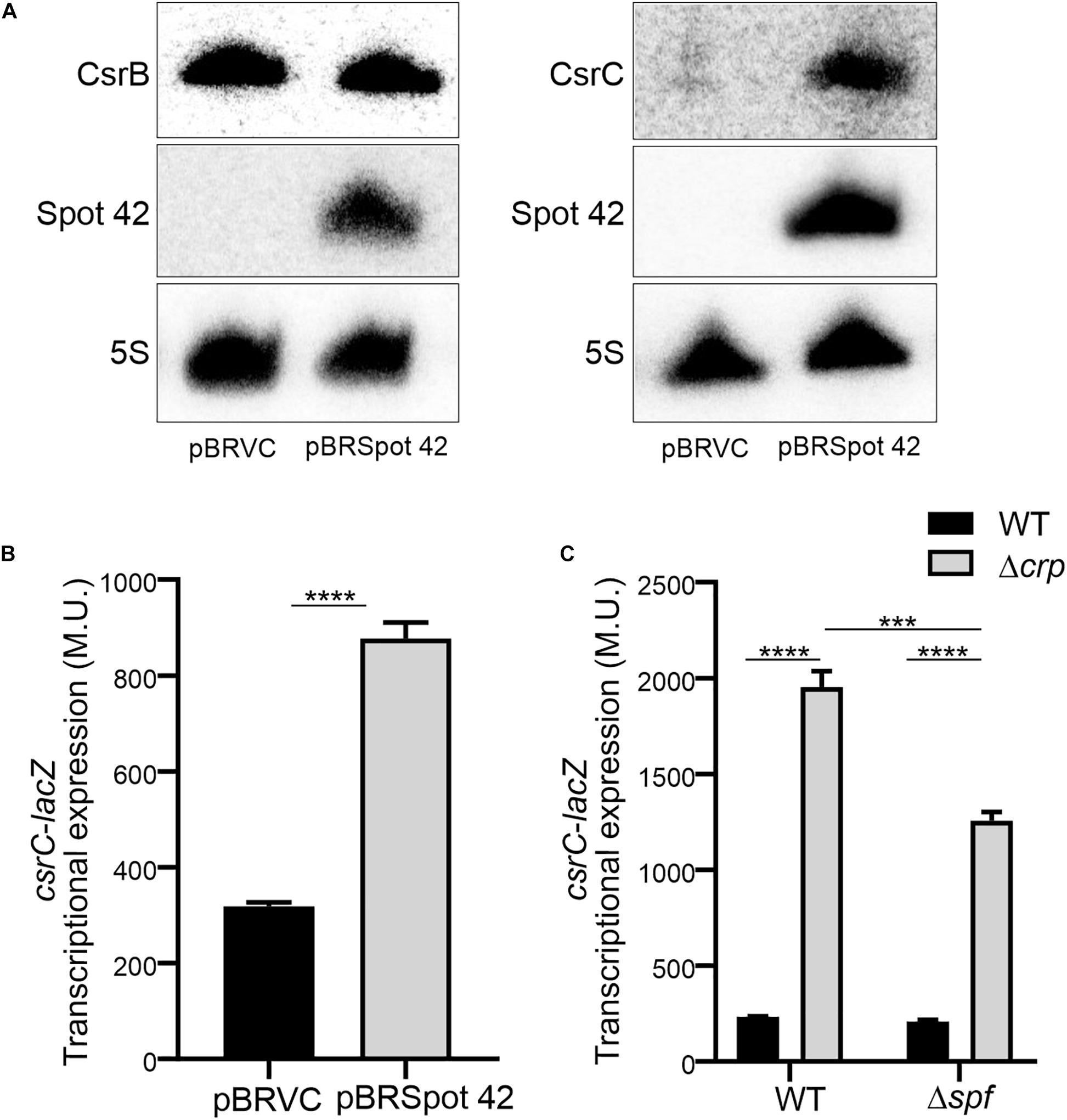
Figure 4. Spot 42 positively regulates the expression of CsrC. (A) Northern blot detection of CsrB, CsrC, and Spot 42 was carried out in strains carrying either the pBRplacVC (control) or pBRplac Spot 42 (overexpressing the sRNA Spot 42). 5S RNA was monitored as loading control. (B) Transcriptional expression of the chromosomal csrC-lacZ fusion upon ectopic expression of the sRNA Spot 42 compared to the strain carrying the control vector (pBRplacVC). (C) Transcriptional expression of the chromosomal csrC-lacZ fusion was monitored in WT, Δspf, Δcrp, and ΔspfΔcrp genetic backgrounds. In all cases cultures were grown in LB at 37°C up to an OD600 nm of 0.4. β-galactosidase activity was determined for three independent cultures, average and standard deviation is presented. ****p < 0.0001, ***p < 0.001.
Interestingly, it has been described that SirA can bind to the promoter region of spf (Zere et al., 2015), suggesting that SirA might be regulating Spot 42. However, transcriptomic data of ΔsirA compared to WT in Salmonella indicates that Spot 42 it is not regulated by SirA (Colgan et al., 2016). The effect of SirA on Spot 42 expression was assessed, using a chromosomal spf-lacZ in the presence and absence of SirA in both WT and Δcrp genetic backgrounds at logarithmic growth phase. As previously shown, Spot 42 expression is induced in the Δcrp mutant compared to WT (El Mouali et al., 2018; Figure 5A). Remarkably, no regulation was observed in the absence of SirA and induction of Spot 42 expression in the Δcrp mutant strain is still observed in absence of SirA (Figure 5A). These results indicate that SirA it is not regulating Spot 42 expression under the studied conditions. Furthermore, SirA was not required for the positive effect of Spot 42 on CsrC expression as concluded from csrC-lacZ expression studies under Spot 42 overexpression in both WT and ΔsirA. In both genetic backgrounds, the overexpression of Spot 42 induces the expression of csrC-lacZ to a similar fold (Figure 5B), consistent with no involvement of SirA in the CRP-cAMP mediated regulation of CsrC.
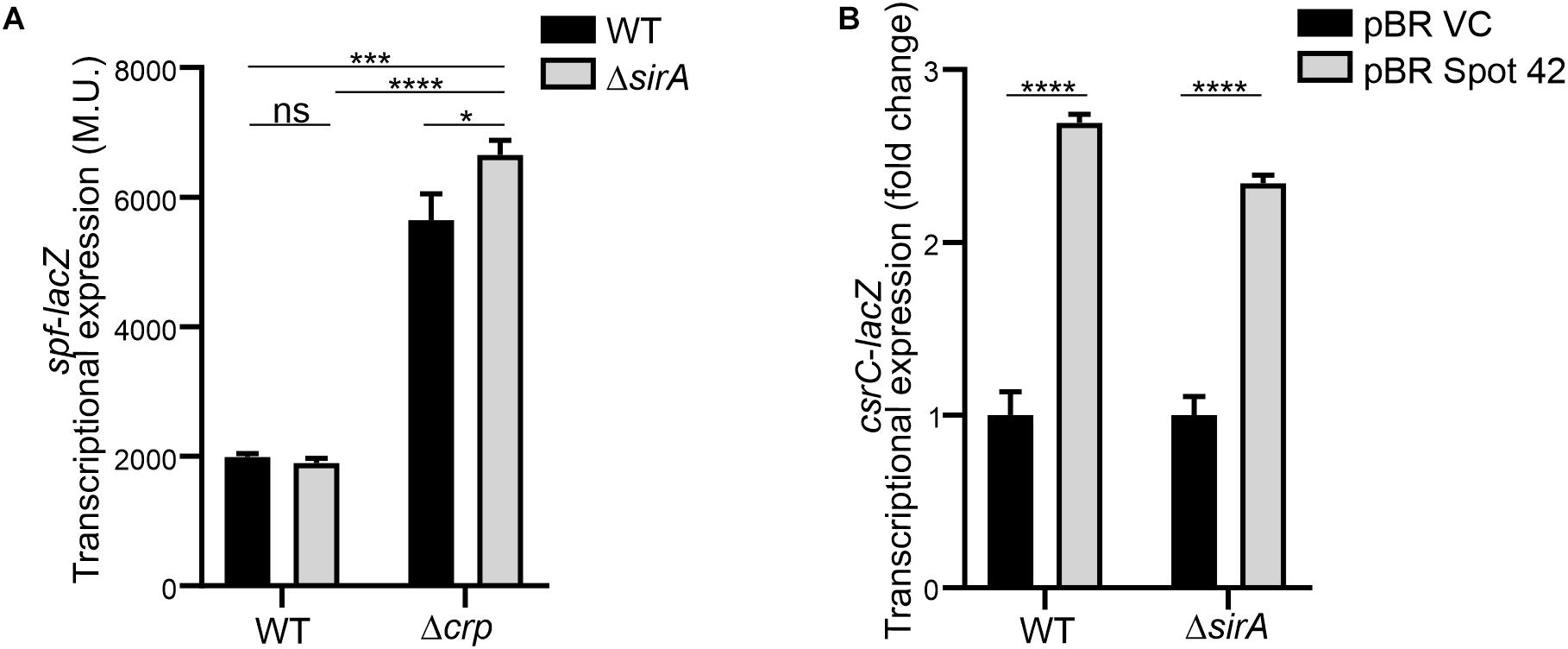
Figure 5. Spot 42 regulates CsrC in a SirA-independent manner. (A) Transcriptional expression of the chromosomal spf-lacZ fusion (Spot 42 expression) was monitored in WT, Δcrp, ΔsirA, and ΔcrpΔsirA genetic backgrounds. (B) Transcriptional expression of the chromosomal csrC-lacZ fusion upon ectopic expression of the sRNA Spot 42 compared to the strain carrying the control vector (pBRplacVC) in a WT and a ΔsirA backgrounds. The transcriptional expression is shown in relative values. In each case, the reference (pBRVC) was set as 1.0. Miller units of the pBRVC backgrounds were 159 ± 21 and 20 ± 2.2 for WT and ΔsirA, respectively. In all cases, cultures were grown in LB at 37°C up to an OD600 nm of 0.4. β-galactosidase activity was determined for three independent cultures, average and standard deviation are presented. *p < 0.05; ***p < 0.001; ****p < 0.0001; ns, not significant.
Altogether, our data indicate that CRP-cAMP differentially regulates CsrB and CsrC at the logarithmic growth phase where it represses specifically CsrC but not CsrB. The CRP-cAMP regulated sRNA Spot 42 arise as new regulator of the Csr regulon in Salmonella.
Discussion
CRP-cAMP is a global regulator initially described to regulate metabolic genes in response to nutrient stimuli. Adding to its more prominent role as transcriptional regulator of mRNAs, CRP-cAMP was also described to regulate the expression of sRNAs such as CyaR, Spot 42, and FnrS (Polayes et al., 1988; Papenfort et al., 2008; De Lay and Gottesman, 2009; Durand and Storz, 2010). CRP-cAMP also modulates the expression of the sRNAs CsrB and CsrC which regulate CsrA protein activity via titration. Previous studies reveal apparent discrepancies in the role of CRP-cAMP on CsrB and CsrC expression. In E. coli, CsrB and CsrC are both repressed by CRP-cAMP, being CsrC expression directly repressed by CRP-cAMP via binding competition with the transcriptional activator UvrY (Pannuri et al., 2016). CRP-cAMP activity is modulated, among others, by the phosphorylation state of the EIIAGlc, which is involved in glucose transport. When phosphorylated, it stimulates cAMP production, promoting the activity of CRP-cAMP. Surprisingly, dephosphorylated EIIAGlc interacts with CsrD and promotes the degradation of CsrB and CsrC by RNAseE (Leng et al., 2016). In other words, conditions that promote the transcriptional derepression of CsrC and CsrB via CRP-cAMP also promote the degradation of these transcripts (Leng et al., 2016; Pannuri et al., 2016). In contrast, in Salmonella CRP-cAMP has been described to play a positive role on CsrB and CsrC expression when it is grown on LB agar media (Teplitski et al., 2006). Although we corroborate these data, we also demonstrate that in LB liquid media, CRP-cAMP differentially regulates CsrB and CsrC. At the logarithmic phase, CsrC but not CsrB is repressed. The alterations in the cell physiology when growing planktonically and within a colony may account for the different regulation described for CsrB and CsrC in Salmonella. In addition of being differentially regulated by CRP-cAMP, CsrB, and CsrC depicted distinct expression profiles during the growth curve. CsrB expression seems to be constitutive, whereas, CsrC expression is silenced during logarithmic growth and induced in early stationary phase. The fact that production of CsrC but not CsrB is growth phase dependent indicates that each RNA responds differently to specific environmental inputs. CRP-cAMP is involved in the growth-dependent regulation of CsrC, suggesting that specific physiological signals that alter CRP-cAMP levels would cause alterations in the levels of CsrC and the concomitant alterations of free CsrA levels that would modulate gene expression to promote adaptation to the new conditions. The differential regulation of CsrB and CsrC, not only by CRP-cAMP, but potentially by additional regulators would provide sensitivity to the Csr system. Far from an ON/OFF state, the Csr regulon would display a scale of grays that would allow the bacteria to fine-tune gene expression in response to environmental stimuli. Interestingly, differential regulation of CsrB and CsrC by CRP-cAMP has been reported in Yersinia pseudotuberculosis, where CRP-cAMP activates the expression of CsrC and represses the expression of CsrB, highlighting complex species-specific regulation of the Csr system (Heroven et al., 2012).
SirA is an activator of CsrB and CsrC (Teplitski et al., 2006; Martínez et al., 2011, 2014. While we observe that SirA is required for full activation of CsrC, our data let us conclude that SirA is not involved in the deregulation of CsrC when CRP-cAMP is depleted. The CsrC deregulation was detected both in absence of SirA and when the SirA binding site was removed from the csrC promoter. Our data indicate that the sRNA Spot 42, which is transcriptionally repressed by CRP-cAMP at the logarithmic growth phase, positively regulates CsrC expression but it does not affect CsrB (Figure 6). This suggests a model where CRP-cAMP differentially regulates CsrB and CsrC via derepression of the trans-encoded sRNA Spot 42. Of note, Spot 42 does not seem to be solely responsible for the derepression of CsrC in absence of crp, as a partial derepression of CsrC is still observed in absence of crp and spf. Suggesting that, CRP-cAMP additionally represses CsrC expression through a Spot 42 independent mechanism (Figure 6).
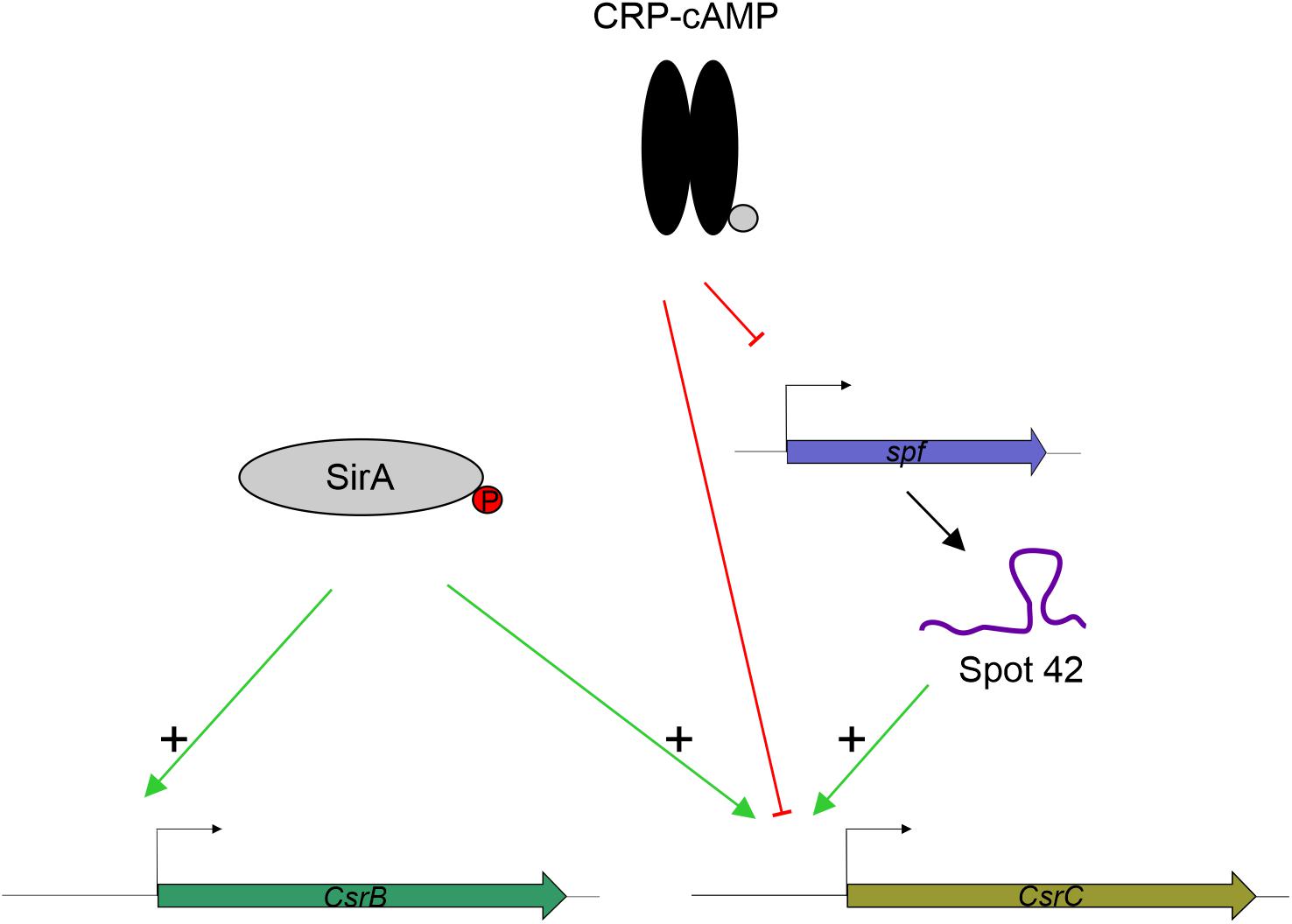
Figure 6. Proposed model for CRP-cAMP mediated repression of CsrC at the logarithmic growth phase. In green, positive regulation is indicated. In red, negative regulation is indicated. SirA, when phosphorylated, it positively regulates CsrB and CsrC. CRP-cAMP represses CsrC and Spot 42. In addition, Spot 42 positively regulates CsrC levels.
The role of Spot 42 in the regulation of CsrC seem to be restricted to logarithmic growth phase and it is not involved in the stationary phase dependent induction. Expression of csrC-lacZ is induced at stationary phase compared to logarithmic growth phase in both the WT and the Δspf backgrounds (Supplementary Image S3). Consistently, transcriptomic data indicates that the expression level of Spot 42 is downregulated 100-fold upon entry into stationary phase when compared to logarithmic growth (Kröger et al., 2013). In agreement, Spot 42 might be responsible to fine-tune the expression of CsrC in conditions where the CRP-cAMP activity is downregulated similarly as it occurs for the Spot 42-regulated hilD mRNA (El Mouali et al., 2018).
In this report, we provide new insights in the link between the CRP-cAMP and Csr regulons in Salmonella. We demonstrate that CRP-cAMP differentially regulates CsrB and CsrC. These two sRNAs are considered functionally redundant but our results suggest that they respond distinctly under specific environmental and physiological conditions in Salmonella. The presence of CsrB alone would affect the free pool of CsrA to a lesser extent than when both CsrB and CsrC sRNAs are present. This fine-tuning of the free CsrA levels would affect the Csr regulon where high affinity mRNA targets will require reduced amounts of free CsrA whereas low affinity targets will require full derepression of CsrA. We show that CRP-cAMP and its target sRNA Spot 42 contribute to the overall levels of CsrB and CsrC. Its contribution allows Salmonella to tightly control the levels of free CsrA in response to environmental stimuli. These features contribute to the versatility of pathogenic bacteria such as the model organism Salmonella enterica.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains, derivatives of Salmonella enterica serovar Typhimurium SL1344, were cultivated in Lysogeny broth (LB; tryptone 10 g/l, yeast extract 5 g/l and sodium chloride 10 g/l). When required, media was supplemented with the antibiotics indicated, ampicillin (Amp) 100 μg/ml, chloramphenicol (Cm) 15 μg/ml or kanamycin (Km) 50 μg/ml. Bacterial cultures were inoculated at an OD600 nm of 0.001, cultures reaching OD600 nm 0.4 were considered logarithmic growth and reaching OD600 nm 2.0 was considered early stationary phase. The bacterial strains and plasmids used in this study are listed in Supplementary Tables S1, S2, respectively.
Genetic Manipulations
Deletion strains were generated by standard gene replacement as previously described (Datsenko and Wanner, 2000). For chromosomal csrC-lacZ transcriptional fusion. The csrC:Cm strain was cured from the antibiotic resistance as previously described (Cherepanov and Wackernagel, 1995). The resulting csrC:frt strain carrying the FRT scar was further used to obtain chromosomal csrC-lacZ by integration of pKG136 as previously described (Ellermeier et al., 2002). Chromosomal spf-lacZ was obtained as for CsrC (El Mouali et al., 2018). Oligonucleotides used for strains construction are listed in Supplementary Table S3.
Plasmidic transcriptional fusions were generated for csrB and csrC. The regulatory regions of interest were PCR amplified, BamHI/HindIII digested and ligated within pQF50 (Farinha and Kropinski, 1989). Spot 42 was cloned in pBRplac (Guillier and Gottesman, 2006; Beisel and Storz, 2011). Spot 42 was PCR amplified, AatII/EcoRI digested and ligated in pBRplac. Spot 42 was expressed constitutively in Salmonella (El Mouali et al., 2018). Oligonucleotides used for cloning are listed in Supplementary Table S3.
β-Galactosidase Activity Assay
Strains of interest were grown to logarithmic growth phase (OD600 nm 0.4) or early stationary phase (OD600 nm 2.0). β-galactosidase activity was measured as described previously (Miller, 1992). Shortly, 100 μl of culture was added to 900 μl of buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 50 mM 2-mercaptoethanol, pH 7) and cells were lysed by the addition of 10 μl of toluene. Reactions were incubated at 28°C and β-galactosidase activity was measured upon addition of 200 μl of ONPG (4 mg/ml). Reactions were stopped by the addition of 500 μl of Na2CO3 (1 M). The OD420 nm and OD550 nm was measured and Miller Units were calculated as previously described (Miller, 1992). β-galactosidase activity determination was performed in technical duplicates for each of three biological replicates.
Total RNA Isolation and Northern Blot
Strains of interest were grown to logarithmic growth phase (OD600 nm 0.4). The biomass of 4 units of OD600 nm was collected, and total RNA extracted by classic hot phenol method. Shortly, the cells were resuspended in 600 μl of TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) containing 0.5 mg/ml lysozyme. Then, 60 μl of 10% SDS (w/v) was added; mixed by inversion and incubated at 64°C for 1–2 min. After the incubation, 66 μl of sodium acetate pH 5.2 (1 M) was added and mixed by inversion. For RNA extraction, 750 μl of Roti-Aqua phenol was added to the samples, mixed by inversion and incubated at 64°C for 6 min. Upon centrifugation (15 min, 13,000 rpm, 4°C), the top aqueous layer was transferred to a fresh 2 ml microcentrifuge tube, and 750 μl of chloroform was added. The samples were mixed and upon centrifugation (12 min, 13,000 rpm, 15°C), the upper aqueous layer was transferred into a new tube and precipitated with 30:1 mix of ethanol and sodium acetate (1 M, pH 6.5). The samples were incubated for 2–3 h or overnight at −20°C. The samples were then centrifuged and precipitated RNA was resuspended in water and concentration measured by NanoDrop. Samples of 10 μg of total RNA were subjected to electrophoretic separation in Tris-Borate-EDTA (TBE) 8% acrylamide gels containing 8.3 M urea. RNAs were transferred to Hybond N + (GE Healthcare) filters by semi-dry TBE based transference. Transcripts of interest were detected by hybridization with 5′ radiolabeled oligos as probes. Images were obtained with the FLA-5100 imaging system (Fujifilm). Oligonucleotides used as probes are listed in Supplementary Table S3.
Statistical Analysis
Graph Pad 8.0 software was used for data analysis. Unpaired two-tailed Student’s t-test were carried out for two groups comparison and p < 0.05 were considered significant.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YE, GE-M, and DG-P contributed to the investigation. YE and CB contributed to the conceptualization, investigation, formal analysis and writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness (grant AGL2013-45339R), the Spanish Ministry of Science, Innovation and Universities (MCIU), State Bureau of Investigation (AIE), and European Regional Development Fund (FEDER) (grant PGC2018-096958-B-I00), and the Catalonian Government (grant 2017SGR499). YE was recipient of an APIF fellowship from the University of Barcelona.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Caroline Tawk (Yale University) and Jens Hör (University of Würzburg) for critical reading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.570536/full#supplementary-material
References
Beisel, C. L., and Storz, G. (2011). The base-pairing RNA Spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell 41, 286–297. doi: 10.1016/j.molcel.2010.12.027
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-a
Colgan, A. M., Kröger, C., Diard, M., Hardt, W. D., Puente, J. L., Sivasankaran, S. K., et al. (2016). The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 12:e1006258. doi: 10.1371/journal.pgen.1006258
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De Lay, N., and Gottesman, S. (2009). The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 191, 461–476. doi: 10.1128/JB.01157-08
Durand, S., and Storz, G. (2010). Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 75, 1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x
El Mouali, Y., Gaviria-Cantin, T., Sánchez-Romero, M. A., Gibert, M., Westermann, A. J., Vogel, J., et al. (2018). CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet. 14:e1007401. doi: 10.1371/journal.pgen.1007401
Ellermeier, C. D., Janakiraman, A., and Slauch, J. M. (2002). Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290, 153–161. doi: 10.1016/s0378-1119(02)00551-6
Farinha, M. A., and Kropinski, A. M. (1989). Construction of broad-host-range vectors for general cloning and promoter selection in Pseudomonas and Escherichia coli. Gene 77, 205–210. doi: 10.1016/0378-1119(89)90068-1
Fortune, D. R., Suyemoto, M., and Altier, C. (2006). Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect. Immun. 74, 331–339. doi: 10.1128/IAI.74.1.331-339.2006
Görke, B., and Stülke, J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624. doi: 10.1038/nrmicro1932
Guillier, M., and Gottesman, S. (2006). Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 59, 231–247. doi: 10.1111/j.1365-2958.2005.04929.x
Heroven, A. K., Sest, M., Pisano, F., Scheb-Wetzel, M., Steinmann, R., Böhme, K., et al. (2012). Crp Induces Switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front. Cell. Infect. Microbiol. 2:158. doi: 10.3389/fcimb.2012.00158
Hör, J., Matera, G., Vogel, J., Gottesman, S., and Storz, G. (2020). Trans-acting small RNAs and their effects on gene expression in Escherichia coli and Salmonella enterica. EcoSal Plus 9. doi: 10.1128/ecosalplus.ESP-0030-2019
Johnston, C., Pegues, D. A., Hueck, C. J., Lee, C. A., and Miller, S. I. (1996). Transcriptional activation of Salmonella Typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22, 715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x
Kröger, C., Colgan, A., Srikumar, S., Händler, K., Sivasankaran, S. K., Hammarlöf, D. L., et al. (2013). An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14, 683–695. doi: 10.1016/j.chom.2013.11.010
Leng, Y., Vakulskas, C. A., Zere, T. R., Pickering, B. S., Watnick, P. I., Babitzke, P., et al. (2016). Regulation of CsrB/C sRNA decay by EIIA(Glc) of the phosphoenolpyruvate: carbohydrate phosphotransferase system. Mol. Microbiol. 99, 627–639. doi: 10.1111/mmi.13259
Liu, M. Y., Gui, G., Wei, B., Preston, J. F., Oakford, L., and Yüksel, Ü, et al. (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272, 17502–17510. doi: 10.1074/jbc.272.28.17502
Martínez, L. C., Martinez-Flores, I., Salgado, H., Fernandez-Mora, M., Medina-Rivera, A., Puente, J. L., et al. (2014). In Silico identification and experimental characterization of regulatory elements controlling the expression of the Salmonella csrB and csrC Genes. J. Bacteriol. 196, 325–336. doi: 10.1128/JB.00806-13
Martínez, L. C., Yakhnin, H., Camacho, M. I., Georgellis, D., Babitzke, P., Puente, J. L., et al. (2011). Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol. Microbiol. 80, 1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x
Miller, J. H. (1992). A Short Course in Bacterial Genetics: A Laboratory Manual. Cold Spring Harbor, NY: CSHL Press.
Pannuri, A., Vakulskas, C. A., Zere, T., McGibbon, L. C., Edwards, A. N., Georgellis, D., et al. (2016). Circuitry linking the catabolite repression and Csr global regulatory systems of Escherichia coli. J. Bacteriol. 198, 3000–3015. doi: 10.1128/JB.00454-416
Papenfort, K., Pfeiffer, V., Lucchini, S., Sonawane, A., Hinton, J. C. D., and Vogel, J. (2008). Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 68, 890–906. doi: 10.1111/j.1365-2958.2008.06189.x
Polayes, D. A., Rice, P. W., Garner, M. M., and Dahlberg, J. E. (1988). Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J. Bacteriol. 170, 3110–3114. doi: 10.1128/jb.170.7.3110-3114.1988
Romeo, T., and Babitzke, P. (2018). Global regulation by CsrA and its RNA antagonists. Microbiol. Spectr. 6:e0009-2017. doi: 10.1128/microbiolspec.rwr-0009-2017
Teplitski, M., Goodier, R. I., and Ahmer, B. M. M. (2003). Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 185, 7257–7265. doi: 10.1128/jb.185.24.7257-7265.2003
Teplitski, M., Goodier, R. I., and Ahmer, B. M. M. (2006). Catabolite repression of the SirA regulatory cascade in Salmonella enterica. Int. J. Med. Microbiol. 296, 449–466. doi: 10.1016/j.ijmm.2006.06.001
Weilbacher, T., Suzuki, K., Dubey, A. K., Wang, X., Gudapaty, S., Morozov, I., et al. (2003). A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48, 657–670. doi: 10.1046/j.1365-2958.2003.03459.x
Keywords: CsrC, post-transcriptional regulation, sRNA, CRP-cAMP, growth phase regulation, Spot 42
Citation: El Mouali Y, Esteva-Martínez G, García-Pedemonte D and Balsalobre C (2020) Differential Regulation of CsrC and CsrB by CRP-cAMP in Salmonella enterica. Front. Microbiol. 11:570536. doi: 10.3389/fmicb.2020.570536
Received: 08 June 2020; Accepted: 17 September 2020;
Published: 14 October 2020.
Edited by:
Omar Orellana, University of Chile, ChileCopyright © 2020 El Mouali, Esteva-Martínez, García-Pedemonte and Balsalobre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youssef El Mouali, eW91c3NlZi5lbG1vdWFsaWJlbm9tYXJAaGVsbWhvbHR6LWhpcmkuZGU=
†Present address: Youssef El Mouali, Helmholtz Institute for RNA-based Infection Research (HIRI), Würzburg, Germany
 Youssef El Mouali
Youssef El Mouali Guillem Esteva-Martínez
Guillem Esteva-Martínez David García-Pedemonte
David García-Pedemonte Carlos Balsalobre
Carlos Balsalobre