- 1Institute of Basic Medicine, The First Affiliated Hospital of Shandong First Medical University, Jinan, China
- 2Science and Technology Innovation Center, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, China
The global coronavirus disease 2019 (COVID-19) pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is one of seven human coronaviruses. G-quadruplexes are intrinsic obstacles to genome replication. Whether G-quadruplexes are present in human coronaviruses is unknown. In the current study, we have predicted that all seven human coronaviruses harbor G-quadruplex sequences. Conserved G-quadruplex sequences in SARS-CoV and SARS-CoV-2 were analyzed and verified by circular dichroism (CD) spectroscopy and Thioflavin T fluorescence assay. Similar to SARS-CoV, SARS-CoV-2 encodes an nsP3 protein, which is predicted to associate with G-quadruplexes. Targeting G-quadruplex sequences in the SARS-CoV-2 genome by G-quadruplex ligands could be a new way to conquer COVID-19.
Introduction
Ongoing coronavirus disease 2019 (COVID-19) pandemic has been a major global threat for human health (Lu and Zhang, 2020; Tu et al., 2020), with over 20 million confirmed cases in over 200 countries and regions. COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a betacoronavirus genus of Coronaviridae family. Among seven types of Coronaviridae family of viruses which could infect humans, HCoV-229E, HCoV-HKU1, HCoV-NL63, and HCoV-OC43 are common around the world, but SARS-CoV, MERS-CoV, and SARS-CoV-2 are more recent and rare.
Both SARS-CoV-2 and SARS-CoV are human SARS-related coronavirus (SARSr-CoVs). SARSr-CoVs have positive-stranded RNA genomes of about 30 kb in length, which encodes multiple proteins. One of the most complex tasks for all viruses is to replicate the entire genome. The replication rate of SARS-CoV-2 is higher than SARS-CoV (Chu et al., 2020). However, the underlying mechanism is not clear. There are intrinsic obstacles to genome replication. For instance, the folding of G-rich sequences into G-quadruplex structures is one source of replication stress. G4 structures’ formation requires at least four or more contiguous runs of guanosine nucleotides exist in a short sequence. G-tetrads are formed around K+ ions through four Hoogsteen-type hydrogen bonds, and then the tetrads stack to adopt G4 structures (Figure 1A).
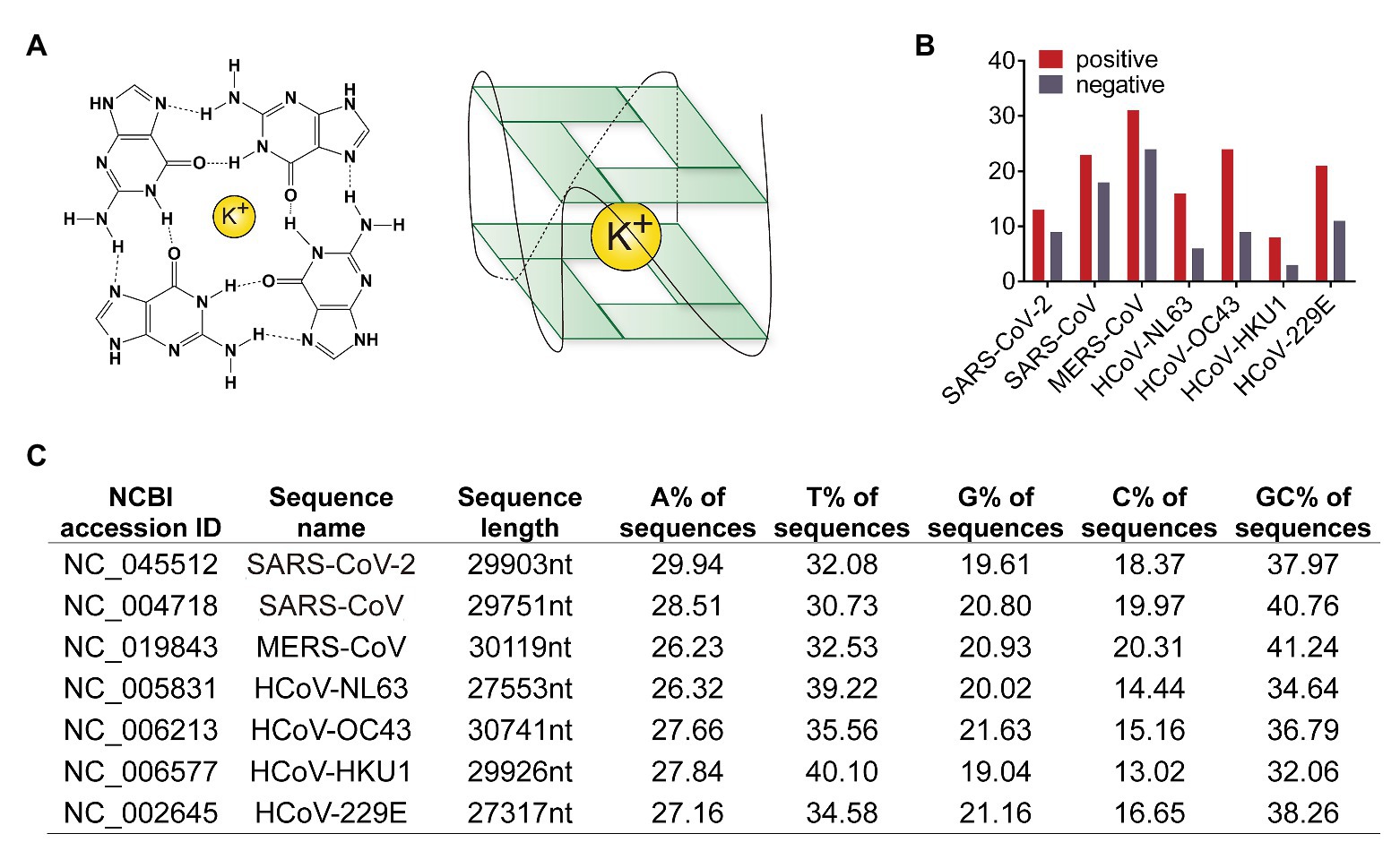
Figure 1. Human coronaviruses genomic RNA possesses G-quadruplexes. (A) Schematic diagram of G-quadruplex structure. Guanines forming G-tetrad structure with a potassium ion inside for stable. (B) Numbers of predicted G-quadruplex forming sites in seven human coronaviruses. The red column represents the G-quadruplex numbers in positive strand, and the gray showed the number in negative strand. (C) Sequence features of human coronavirus genomes. Lane one to three, NCBI accession ID, names, and sequence length of genome reference sequences. Lane four to seven, the content of each base in genome sequences. Lane eight, GC content in genome sequences.
Recently, the important functions for virus G4 structures have been demonstrated. G4 structures in the long terminal repeat promoter of the human immunodeficiency virus (HIV) was critical for promoter activity (Amrane et al., 2014; Perrone et al., 2014). A G4 structure in Epstein-Barr virus (EBV) functioned as a cis-acting regulatory region to translate EBV encoded nuclear antigen 1 (EBNA1) mRNA (Murat et al., 2014). G4 structures have also been observed in human papillomavirus (HPV), hepatitis B virus (HBV), Nipah virus, hepatitis C virus (HCV), Zika virus, and Ebola virus (Ruggiero and Richter, 2020). These studies highlight a critical role for G4 structures in viruses.
Whether G4 structures are present in the genomes of human coronaviruses including SARS-CoV-2 is largely unknown. To address this question, we analyzed human coronavirus RNA genomes and predicted several conserved G4. G4 ligands could be developed as antiviral agents for human coronaviruses, including SARS-CoV-2, causing the current COVID-19 pandemic. Moreover, we identified SARS-CoV-2 contains less G4 than SARS-CoV, which partially explains why SARS-CoV-2 replicated faster than SARS-CoV-2.
Materials and Methods
Sequence Analysis
Genomes of bat SARSr-CoV, SARS-CoV, and SARS-CoV-2 strains were downloaded from the NCBI virus database. Potential G4 forming sequences in human coronavirus genomes were predicted by QGRS mapper1 (Kikin et al., 2006) and Quadbase22 (Parashar and Shantanu, 2016). The parameters were as below: max. length, 30; min. G-group size, 2; and loop size from 0 to 12. Global genome alignment among human coronavirus was conducted using R package DECIPHER with default parameters (Wright, 2015). The results were exported in Fasta format and visualized in MEGA X. Sequence logos were generated by weblogo tools3 (Crooks et al., 2004).
Thioflavin T Fluorescence Assay
Single strand RNA oligmers of conserved G4 sequences in SARS-CoV-2, as well as mutant sequences, were synthesized. The sequences were listed in Table 1. RNA oligmers were desolved in RNase free buffer containing 20 mM Tris-HCl and 40 mM KCl to a final concentration of 2 μM. Then the RNA solution was heated at 90°C for 5 min and slowly cooling down to room temperature. Thioflavin T (ThT) powder were bought from Aladdin Industrial Corporation and dissolved in the buffer above. Oligmers and ThT were mixed at final concentration 2 and 2 μM, respectively. The fluorescence at 495 nm emission was collected after 425 nm excitation using SpectraMax microplate reader.
Circular Dichroism Spectrum
G4 RNA oligmers were dissolved in RNase free buffer containing 20 mM Tris-HCl and 40 mM KCl to a final concentration of 2 μM. Circular dichroism (CD) spectrum was collected by Chirascan V100 from wavelength 200 to 400 nm.
Plasmids Construction and Primer Extension Assay
G4 sequences were inserted after start codon of green fluorescent protein (GFP) sequences and cloned into pCHA vector. Primers used for plasmid construction were below: forward primer for nsP10, 5'-CCGGAATTCATGGGTATGTGGAAAGGTTATGGCGTGAGCAAGGGCGCC-3'; forward primer for Sa, 5'-CCGGAATTCATGGGTTGGACCTTTGGTGCAGGTGTGAGCAAGGGCGCC-3'; forward primer for N, 5'-CCGGAATTCATGGGCTGGCAATGGCGGGTGAGCAAGGGCGCC-3'; and reverse primer for nsP10, Sa, and N, 5'-CGCGGATCCTCACTTGTACAGCTCATCCAT-3'. Primers used in primer extension assay were below: forward primer, 5'-GTGGAGCAATAGCAGAGCTC-3' and reverse primer, TCACTTGTACAGCTCATCCA-3'. These plasmids were used as PCR templates. N,N΄-(9-(4-(Dimethylamino)phenylamino)acridine-3,6-diyl)bis(3-(pyrrolidin-1-yl)propanamide) (BRACO-19) and meso-5,10,15,20-Tetrakis-(N-methyl-4-pyridyl)porphine, Tetratosylate (TMPyP4) at concentrations of 0, 5, 10, 20, and 40 μM were added into PCR reaction mixture. The PCR products were analyzed by electrophoresis in agarose gels stained with Gelred.
Protein Expression Assay
G4-based plasmids expressing GFP constructed above were transfected into 293T cells using PEI transfection reagent. Forty micromolar BRACO-19 and TMPyP4 were added into cells 4 h after transfection and the cell lysis and were collected to detect the expression level of GFP by SDS-PAGE followed by Western blot.
Structure Analysis
The 3D structure of the SARS-CoV-2 nsP3 SARS-unique domain (SUD) was obtained through homology modeling using the swiss-model4 (Waterhouse et al., 2018). The template was downloaded from the PDB database with PDB ID 2W2G (Tan et al., 2009). The superimposed image and atom distances were generated by chimera software (Downloaded from http://www.rbvi.ucsf.edu/chimera; Pettersen et al., 2004).
G4 Ligands
The previously reported G4 ligands with antiviral function were BRACO-19 (Read et al., 1999), PHENdc3 (De Cian et al., 2007), IZCZ-3 (Hu et al., 2018), PIPER (Fedoroff et al., 1998), PDP (Müller et al., 2010), PDS (Rodriguez et al., 2008), TMPyP4 (Parkinson et al., 2007), c-exNDI (Collie et al., 2012), and quarfloxin (Drygin et al., 2009). The chemical structures were drawn using Chemdraw software.
Results
Prediction of G4 Sequences in Human Coronaviruses
Genome sequences of MERS-CoV, SARS-CoV, SARS-CoV-2, HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1 were obtained from the NCBI nucleotide database. Genomes of bat SARSr-CoV as well as SARS-CoV and SARS-CoV-2 strains were downloaded from the NCBI virus database. Potential G4 forming sequences in human coronavirus genomes were predicted by QGRS mapper (Kikin et al., 2006). In both positive and negative strands, SARS-CoV-2 possessed less number of predicated G4 and less GC content than SARS-CoV (Figures 1B,C and Supplementary Tables S1 and S2). SARS-CoV-2 with less G4 sequences replicates faster because of less energy is required to bypass G4 structure.
Global genome alignment among human coronaviruses was conducted and conserved G4 forming sites were analyzed according to the corresponding position in genomes and confirmed manually (Supplementary Table S3). Further, another G4 prediction tool quadbase2 was used to confirm the conserved sequences, and the results were listed in Supplementary Table S4. As shown in Supplementary Table S3, the G4 sequences in ORF1ab were conserved in four coronaviruses, which were SARS-CoV, SARS-CoV-2, HCoV-OC43, and HCoV-229E. The G4 sequences in the S protein-coding region were conserved in five coronaviruses (SARS-CoV, SARS-CoV-2, MERS-CoV, HCoV-NL63, and HCoV-229E), including the three with highest mortality rates. G4 sequences are key RNA secondary structures in the viral genomes. Consequently, these conserved G4 sequences could be interesting targets in developing of innovative drugs against human coronaviruses.
Interestingly, SARS-CoV and SARS-CoV-2 shared the most similar G4 sequences among human coronaviruses. Seven conserved G4 sequences in SARS-CoV and SARS-CoV-2 genomes were listed in Figure 2A, and the schematic diagram of G4 sites were shown in Figure 2B. There were two in the nsP1 coding region and three in S coding region, and the other two were in nsP10 and N coding regions. According to the SARS-CoV-2 genome annotation, we identified the conserved position of G4 sequences (Figure 2A).
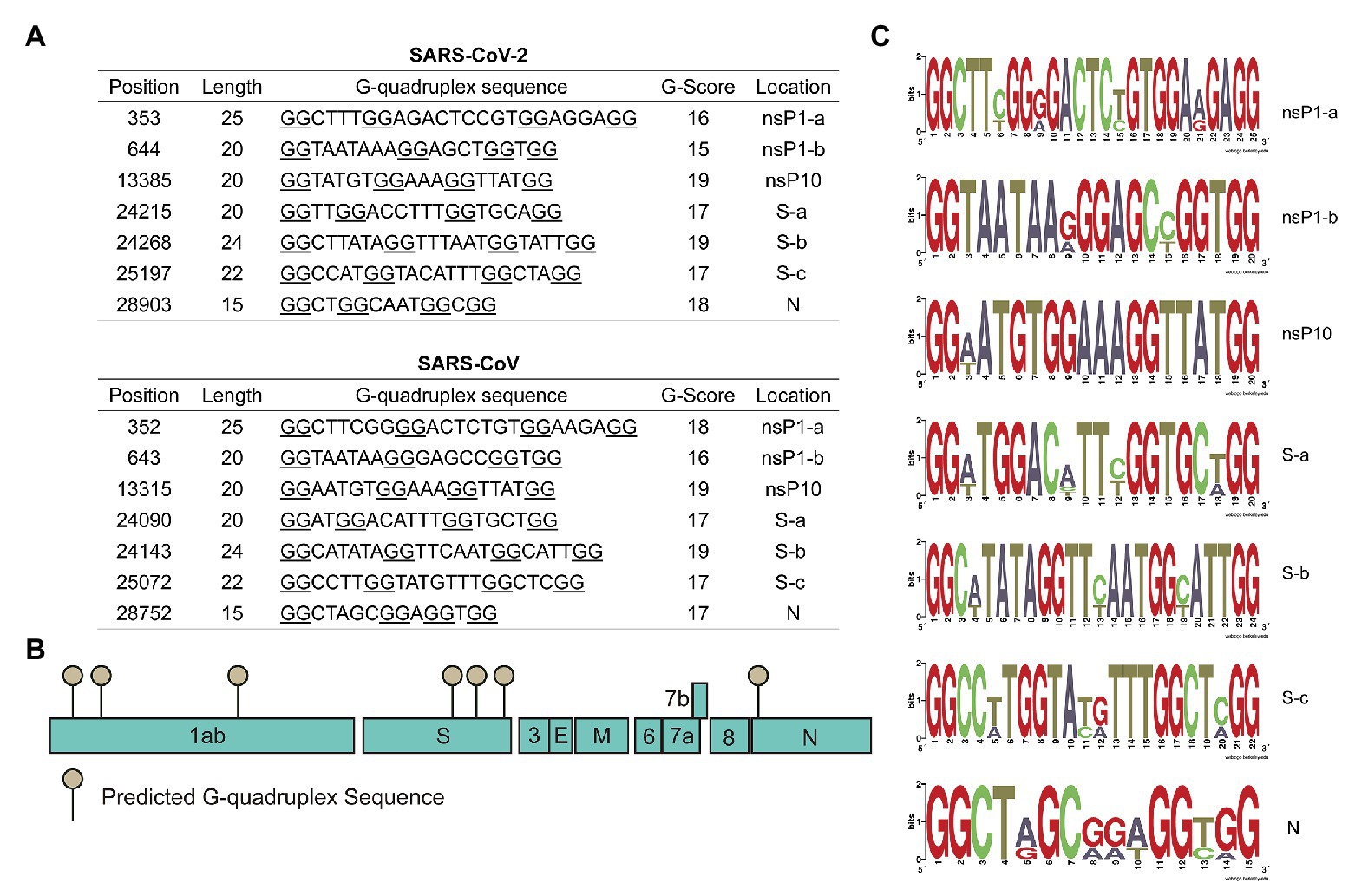
Figure 2. Severe acute respiratory syndrome-related coronavirus (SARSr-CoV) possesses G-quadruplexes. (A) Conserved G-quadruplex sequences in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and SARS-CoV genomes. (B) A model of the conserved G-quadruplex forming sites in SARS-CoV-2 genome. (C) Sequence logos formed by G-quadruplex sequences in typical SARSr-CoV.
Conserved G4 sequences in SARSr-CoV were analyzed, including bat SARS-related coronavirus, SARS-CoV, and SARS-CoV-2. The results showed that the seven G4 sites identified above were also conserved in bat SARSr-CoV (Supplementary Tables S5 and S6). To better visualize the conserved sequences, sequence logos were generated using the weblogo tool. Results in Figure 2C showed that critical G4 sequences were conserved in typical SARSr-CoV. To further explore G4 sequences’ evolution in SARS-CoV-2, all the SARS-CoV-2 strains available on the NCBI database (up to 8th April 2020) from different countries were downloaded and analyzed. Genome alignment was conducted using R package DECIPHER, and it turned out that G4 sites were highly conserved in SARS-CoV-2 strains (Supplementary Table S7 and Supplementary Figure S1). Finally, the conserved G4 sequences in all SARSr-CoV strains were aligned and logos were generated (Supplementary Figure S2), indicating that the G4 sequences were evolutionarily conserved. Our observation of the strong conservation of the G4 sites in SARSr-CoV genomes supports a hypothesis that these sequences are very important for SARSr-CoV.
Characterization of G-Quadruplex Structures
G4 structure-specific binding to ThT to induce its fluorescence (Renaud de la Faverie et al., 2014). Single strand RNA oligomers of these conserved sequences in SARS-CoV-2 were synthesized. ThT fluorescence assay results showed that these conserved sequences were adopted to form G4 structures in SARS-CoV-2 genomes (Figure 3A).
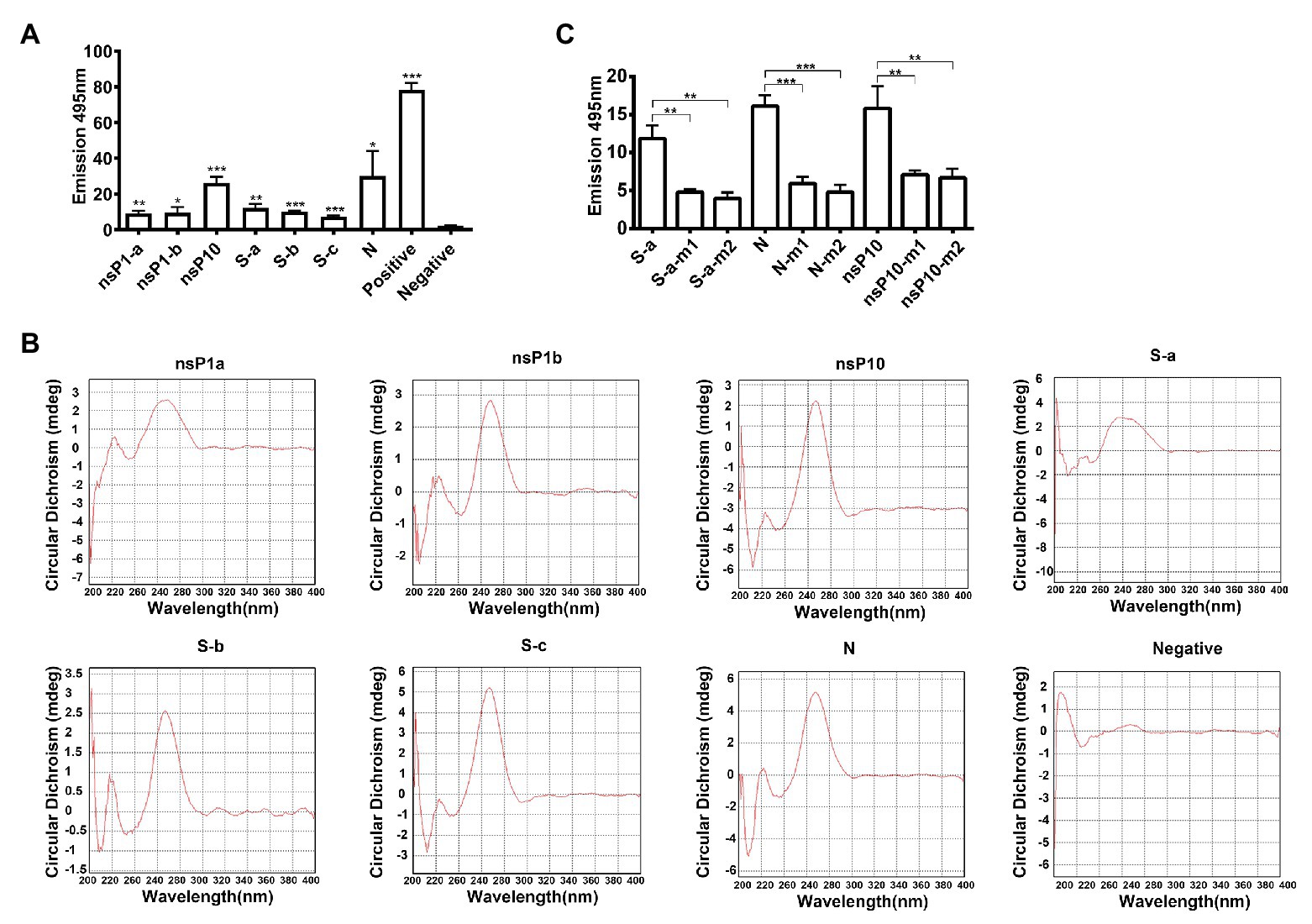
Figure 3. Characterization of G-quadruplex structures. (A) Thioflavin T (ThT) fluorescence assay for each conserved G-quadruplex sequences. *p < 0.05, **p < 0.01, and ***p < 0.001 based on the Student t-test. All results are from three independent experiments. Error bars, SD. (B) Circular dichroism (CD) spectrum of conserved G-quadruplex sequences. (C) ThT fluorescence assay for G-quadruplex single or double mutations. m1, single guanine mutation; m2, double guanines mutation. *p < 0.05, **p < 0.01, and ***p < 0.001 based on the Student t-test. All results are from three independent experiments. Error bars, SD.
Circular dichroism spectrum was employed to further confirm the existence of G4 structure in SARS-CoV-2 genome. All these seven conserved sequences have absorbance at about 264 nm, indicating that G4 structures in SARS-CoV-2 genome were adopted to form parallel-strand topologies (Figure 3B). To analyze the effect of single guanine in the G4 region, single or two nucleotide mutations of nsP10, S-a, and N G4 sequences were designed and oligomers were synthesized. The results in Figure 3C showed that the fluorescence signals of mutation oligomers were decreased significantly compared with that of wide type.
SARS-CoV-2 nsP3 Potentially Associates With G-Quadruplex Sequences
SARSr-CoV encodes an nsP3 protein, which possesses two SUD (M and N) capable of interacting with G4 sequences and potentially essential for unwinding G4 folds in RNA (Figure 4A; Kusov et al., 2015). Stimulated structure of SUD from SARS-CoV-2 nsP3 showed a similar structure to SARS-CoV SUD. Interestingly, an L-Y hydrophobic interaction in SARS-CoV-2 SUD replaced the disulfide bond in SARS-CoV SUD (Figure 4B). The two SUD from SARS-CoV-2 and SARS-CoV are conserved, indicating the co-evolution of nsP3 and G4 sequences (Supplementary Figure S3). The utility of SUD adds additional support for the importance of these genomic G4 folds in SARSr-CoV.
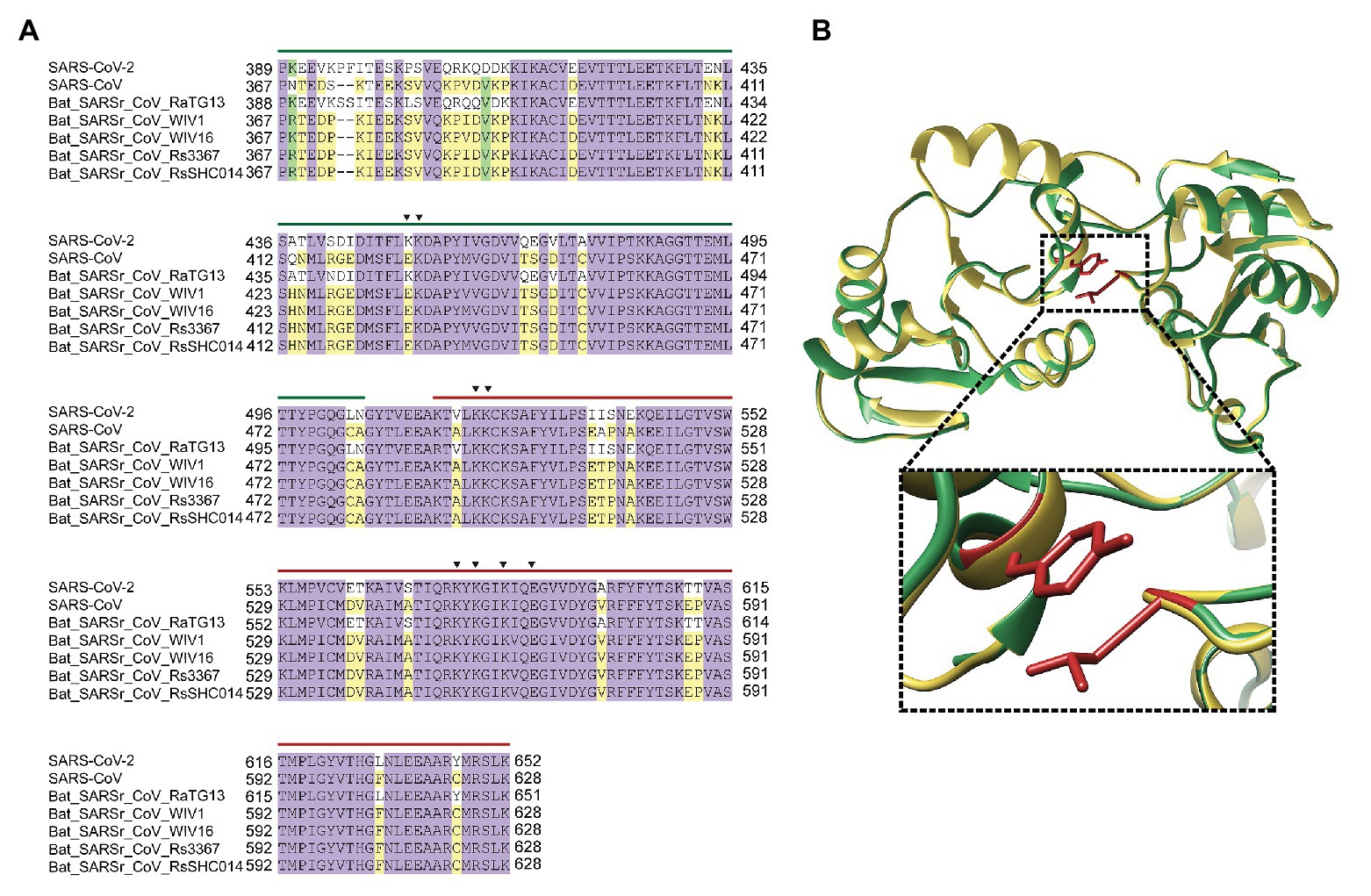
Figure 4. SARS-CoV-2 nsP3 potentially associates with G-quadruplex sequences. (A) Sequence alignment of nsP3 SARS-unique domain (SUD)-M and SUD-N in typical SARSr-CoV. The key amino acids in G-quadruplex binding were indicated in triangle. Green line is SUD-M, while red line is SUD-N. (B) SUD 3D structure superimpose of SARS-CoV and SARS-CoV-2. The enlarged image showed the atom distance between L516 and Y647 in SARS-CoV-2.
G-Quadruplex Ligands Block G4-Based Gene Expression
G4 ligands could stabilize RNA G4 and have been demonstrated to be potential antivirus strategies for HIV, HBV, HCV, and Ebola virus (Ruggiero and Richter, 2018). Figure 5A listed the G4 ligands which have been reported to exert antiviral activities in the past few years. G4 sequences were present in human coronaviruses, and these G4 ligands may be developed as potential drugs against SARS-CoV-2, SARS-CoV, and MERS-CoV.
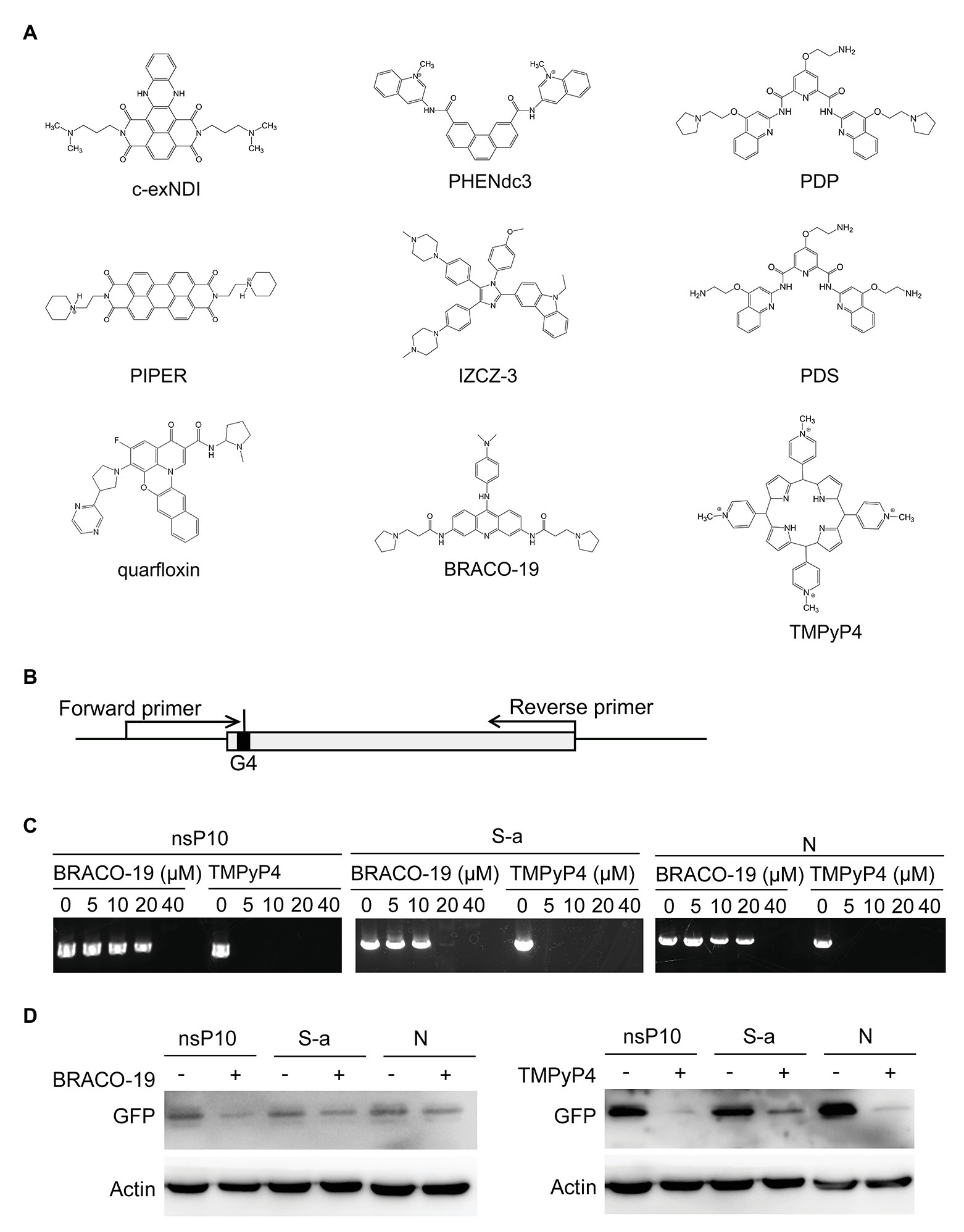
Figure 5. G-quadruplex ligands BRACO-19 and TMPyP4 block G4-based gene expression. (A) G-quadruplex ligands with antiviral roles. (B) Schematic diagram of G-quadruplex containing plasmids construction and primer extension strategies. (C) PCR products of primer extension assay were observed by agarose electrophoresis. The concentrations of G-quadruplex ligands treatment were as indicated. (D) GFP expression level with or without treatment were detected through Western blot.
Primer extension assay were performed to detect whether G4 stabilization affect DNA replication. Plasmids were constructed by inserting the G4 sequences into GFP gene after translation start codon ATG, and two primers were selected to amplify the GFP gene (Figure 5B). Two G4 binding ligands, BRACO-19 and TMPyP4, were used to stabilize the G4 structure. As shown in Figure 5C, with the increase of treatment concentration, the yield of PCR product reduced. To further detect the influence of G4 structure in protein expression, plasmids constructed above were transfected into cells and GFP expression were examined in the presence or absence of G4 ligands. The results showed both BRACO-19 and TMPyP4 treatment decreased the expression of GFP inserted by G4 sequences (Figure 5D).
Discussion
Our study provides a paradigm for assessing G4 functions in viral genomes. Through computational search, different G4-forming sequences were predicted from the human coronaviruses including SARS-CoV-2. The formation of G4s was determined by CD spectroscopy and ThT fluorescence assay. The role of a G4 in gene expression was addressed using primer extension assay and Western blot. Overall, our results point to a potential role for G4s in controlling SARS-CoV-2 viral gene expression. The role of G4s in SARS-CoV-2 viral replication awaits further investigation. We believe that genome-wide analyses of G4s in more viruses will help us to establish a general link between virus life cycle and viral G4s.
G4 structures could hinder gene expression. SARS-CoV-2 contains fewer predicated G4 than SARS-CoV (Figure 1B), which partially explains why SARS-CoV-2 replicates faster than SARS-CoV. G4 sequences are potential antiviral targets. We showed G4 ligands including TMPyP4 and BRACO-19 could inhibit G4 reporter expression (Figure 5D), indicating that G4 ligands could inhibit G4-contaning virus genome replication.
SARS-CoV-2 G4 could be used to develop tools for SARS-CoV-2 studies. G4 has been applied to detect HCV (Luo et al., 2019) and HIV (Sontakke and Srivatsan, 2020). Likewise, G4 could be developed as a potential biosensor for SARS-CoV-2 detection. G4 is applied to set up HCV helicase assay (Leung et al., 2015). In the future, G4 might be used to measure SARS-CoV-2 nsP13 helicase assay.
Many viral proteins associated with virus-encoded G4. HIV-1 nucleocapsid protein NCp7 binds and unfolds the HIV-1 G4 and promotes reverse transcription (Butovskaya et al., 2019). HCV helicase NS3 unwound viral G4 (Leung et al., 2015). SUD domains of SARS-CoV nsP3 were shown to bind to viral G4 (Tan et al., 2009) and play a critical role in viral replication and transcription (Kusov et al., 2015). Based on the similarity of SARS-CoV SUD and SARS-CoV-2 SUD (Figure 4B), nsP3 from SARS-CoV-2 was predicted to associate with viral G4 through SUD domain. Moreover, it is possible that SARS-CoV-2 helicase nsP13 may unwind viral G4 to enhance viral replication.
Viral G4 also associated with host proteins. Cellular nucleolin interacted with viral core G4 to suppress HCV replication (Bian et al., 2019). Nucleolin directly binds to EBV G4 in EBNV1 mRNA sequence to inhibit EBNV1 protein expression (Lista et al., 2017). Nucleolin stabilizes the HIV-1 LTR G4s, and the human ribonucleoprotein A2B1 (HnRNP A2/B1) unwinds the G4s to promote HIV-1 transcription (Scalabrin et al., 2017). Whether host proteins interact with G4 of SARS-CoV-2 remains inconclusive. In the future, we will identify host proteins involved in the function of SARS-CoV-2 G4.
In summary, our results have predicted that all seven human coronaviruses harbor G4 sequences, indicating that G4 structures are crucial elements in the genomes of human coronaviruses. Thus, targeting G4 in viral genomes is a new way to develop antiviral agents. Analysis through genome alignment demonstrated that SARS-CoV and SARS-CoV-2 contained seven conserved G4 sequences. ThT fluorescence assay and CD spectroscopy showed that these conserved G4 sequences in SARS-CoV-2 were able to form G4 folds. Whether these G4s are important for maintaining global genome structure remains an open question. Further studies are needed to better understand these G4s in human coronaviruses and, more specifically, the SARS-CoV-2.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
HC performed the bioinformatics study and experiments. LZ conceived the research. HC and LZ wrote the manuscript and approved the final version for publication.
Funding
This work was supported by the National Key Plan for Research and Development of China (2016YFD0500300), National Major S & T Project for the Prevention and Treatment of Major Infectious Diseases in China (2017ZX10004206-007), and Academic Promotion Program of Shandong First Medical University (2019LJ001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.567317/full#supplementary-material
Footnotes
1. http://bioinformatics.ramapo.edu/QGRS/index.php/analyze.php
2. http://quadbase.igib.res.in/TetraPlexFinder
References
Amrane, S., Kerkour, A., Bedrat, A., Vialet, B., Andreola, M. L., and Mergny, J. L. (2014). Topology of a DNA G-quadruplex structure formed in the HIV-1 promoter: a potential target for anti-HIV drug development. J. Am. Chem. Soc. 136, 5249–5252. doi: 10.1021/ja501500c
Bian, W. X., Xie, Y., Wang, X. N., Xu, G. H., Fu, B. S., Li, S., et al. (2019). Binding of cellular nucleolin with the viral core RNA G-quadruplex structure suppresses HCV replication. Nucleic Acids Res. 47, 56–68. doi: 10.1093/nar/gky1177
Butovskaya, E., Soldà, P., Scalabrin, M., Nadai, M., and Richter, S. N. (2019). HIV-1 nucleocapsid protein unfolds stable RNA G-quadruplexes in the viral genome and is inhibited by G-quadruplex ligands. ACS Infect. Dis. 5, 2127–2135. doi: 10.1021/acsinfecdis.9b00272
Chu, H., Chan, J. F., Wang, Y., Yuen, T. T., Chai, Y., Hou, Y., et al. (2020). Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 71, 1400–1409. doi: 10.1093/cid/ciaa410
Collie, G. W., Promontorio, R., Hampel, S. M., Micco, M., Neidle, S., and Parkinson, G. N. (2012). Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 134, 2723–2731. doi: 10.1021/ja2102423
Crooks, G. E., Hon, G., Chandonia, J. -M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
De Cian, A., Cristofari, G., Reichenbach, P., De Lemos, E., Monchaud, D., Teulade-Fichou, M. P., et al. (2007). Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl. Acad. Sci. U. S. A. 104, 17347–17352. doi: 10.1073/pnas.0707365104
Drygin, D., Siddiqui-Jain, A., O’Brien, S., Schwaebe, M., Lin, A., Bliesath, J., et al. (2009). Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 69, 7653–7661. doi: 10.1158/0008-5472.can-09-1304
Fedoroff, O. Y., Salazar, M., Han, H., Chemeris, V. V., Kerwin, S. M., and Hurley, L. H. (1998). NMR-based model of a telomerase-inhibiting compound bound to G-quadruplex DNA. Biochemistry 37, 12367–12374. doi: 10.1021/bi981330n
Hu, M. H., Wang, Y. Q., Yu, Z. Y., Hu, L. N., and Tan, J. H. (2018). Discovery of a new four-leaf clover-like ligand as a potent c-MYC transcription inhibitor specifically targeting the promoter G-quadruplex. J. Med. Chem. 61, 2447–2459. doi: 10.1021/acs.jmedchem.7b01697
Kikin, O., D’Antonio, L., and Bagga, P. S. (2006). QGRS mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 34, W676–W682. doi: 10.1093/nar/gkl253
Kusov, Y., Tan, J., Alvarez, E., Enjuanes, L., and Hilgenfeld, R. (2015). A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication–transcription complex. Virology 484, 313–322. doi: 10.1016/j.virol.2015.06.016
Leung, K. H., He, H. Z., He, B., Zhong, H. J., Lin, S., Wang, Y. T., et al. (2015). Label-free luminescence switch-on detection of hepatitis C virus NS3 helicase activity using a G-quadruplex-selective probe. Chem. Sci. 6, 2166–2171. doi: 10.1039/c4sc03319a
Lista, M. J., Martins, R. P., Billant, O., Contesse, M. A., Findakly, S., Pochard, P., et al. (2017). Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 8:16043. doi: 10.1038/ncomms16043
Lu, Y., and Zhang, L. (2020). Social media WeChat infers the development trend of COVID-19. J. Infect. 81, e82–e83. doi: 10.1016/j.jinf.2020.03.050
Luo, X., Xue, B., Feng, G., Zhang, J., Lin, B., Zeng, P., et al. (2019). Lighting up the native viral RNA genome with a fluorogenic probe for the live-cell visualization of virus infection. J. Am. Chem. Soc. 141, 5182–5191. doi: 10.1021/jacs.8b10265
Müller, S., Kumari, S., Rodriguez, R., and Balasubramanian, S. (2010). Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. 2, 1095–1098. doi: 10.1038/nchem.842
Murat, P., Zhong, J., Lekieffre, L., Cowieson, N. P., Clancy, J. L., Preiss, T., et al. (2014). G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 10, 358–364. doi: 10.1038/nchembio.1479
Parashar, D., and Shantanu, C. (2016). QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 44, W277–W283. doi: 10.1093/nar/gkw425
Parkinson, G. N., Ghosh, R., and Neidle, S. (2007). Structural basis for binding of porphyrin to human telomeres. Biochemistry 46, 2390–2397. doi: 10.1021/bi062244n
Perrone, R., Butovskaya, E., Daelemans, D., Palù, G., Pannecouque, C., and Richter, S. N. (2014). Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 69, 3248–3258. doi: 10.1093/jac/dku280
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., and Meng, E. C. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Read, M. A., Wood, A. A., Harrison, J. R., Gowan, S. M., Kelland, L. R., Dosanjh, H. S., et al. (1999). Molecular modeling studies on G-quadruplex complexes of telomerase inhibitors: structure-activity relationships. J. Med. Chem. 42, 4538–4546. doi: 10.1021/jm990287e
Renaud de la Faverie, A., Guédin, A., Bedrat, A., Yatsunyk, L. A., and Mergny, J. L. (2014). Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res. 42:e65. doi: 10.1093/nar/gku111
Rodriguez, R., Müller, S., Yeoman, J. A., Trentesaux, C., Riou, J. F., and Balasubramanian, S. (2008). A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 130, 15758–15759. doi: 10.1021/ja805615w
Ruggiero, E., and Richter, S. N. (2018). G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 46, 3270–3283. doi: 10.1093/nar/gky187
Ruggiero, E., and Richter, S. N. (2020). Viral G-quadruplexes: new frontiers in virus pathogenesis and antiviral therapy. Annu. Rep. Med. Chem. 54, 101–131. doi: 10.1016/bs.armc.2020.04.001
Scalabrin, M., Frasson, I., Ruggiero, E., Perrone, R., Tosoni, E., Lago, S., et al. (2017). The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci. Rep. 7:45244. doi: 10.1038/srep45244
Sontakke, V. A., and Srivatsan, S. G. (2020). A dual-app nucleoside probe reports G-quadruplex formation and ligand binding in the long terminal repeat of HIV-1 proviral genome. Bioorg. Med. Chem. Lett. 30:127345. doi: 10.1016/j.bmcl.2020.127345
Tan, J., Vonrhein, C., Smart, O. S., Bricogne, G., Bollati, M., Kusov, Y., et al. (2009). The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 5:e1000428. doi: 10.1371/journal.ppat.1000428
Tu, H., Tu, S., Gao, S., Shao, A., and Sheng, J. (2020). The epidemiological and clinical features of COVID-19 and lessons from this global infectious public health event. J. Inf. Secur. 81, 1–9. doi: 10.1016/j.jinf.2020.04.011
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Keywords: COVID-19, SARS-CoV-2, SARS-CoV, G-quadruplexes, nsP3
Citation: Cui H and Zhang L (2020) G-Quadruplexes Are Present in Human Coronaviruses Including SARS-CoV-2. Front. Microbiol. 11:567317. doi: 10.3389/fmicb.2020.567317
Edited by:
Yufeng Wang, University of Texas at San Antonio, United StatesCopyright © 2020 Cui and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leiliang Zhang, YXJtemhhbmdAaG90bWFpbC5jb20=
 Haoran Cui
Haoran Cui Leiliang Zhang
Leiliang Zhang