
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 October 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.565349
This article is part of the Research Topic Exposure, Risks, and Drivers of the Mobile Antimicrobial Resistance Genes in the Environment – a Global Perspective View all 17 articles
 Xiuping Zhang1,2†
Xiuping Zhang1,2† Xinxin Li1†
Xinxin Li1† Weihua Wang3†
Weihua Wang3† Jiali Qi1
Jiali Qi1 Dong Wang1
Dong Wang1 Lei Xu4
Lei Xu4 Yong Liu1
Yong Liu1 Yanming Zhang1
Yanming Zhang1 Kangkang Guo1*
Kangkang Guo1*Multiple-drug resistance bacteria containing antimicrobial resistance genes (ARGs) are a concern for public health. Integrons are bacterial genetic elements that can capture, rearrange, and express mobile gene cassettes responsible for the spread of ARGs. Few studies link genotype and phenotype of swine-related ARGs in the context of mobile gene cassette arrays among commensal Escherichia coli (E. coli) in nonclinical livestock isolates from intensive farms. In the present study, a total of 264 isolates were obtained from 330 rectal swabs to determine the prevalence and characteristics of antibiotic-resistant gene being carried by commensal E. coli in the healthy swine from four intensive farms at Anhui, Hebei, Shanxi, and Shaanxi, in China. Antimicrobial resistance phenotypes of the recovered isolates were determined for 19 antimicrobials. The E. coli isolates were commonly nonsusceptible to doxycycline (75.8%), tetracycline (73.5%), sulfamethoxazole-trimethoprim (71.6%), amoxicillin (68.2%), sulfasalazine (67.1%), ampicillin (58.0%), florfenicol (56.1%), and streptomycin (53.0%), but all isolates were susceptible to imipenem (100%). Isolates [184 (69.7%)] exhibited multiple drug resistance with 11 patterns. Moreover, 197 isolates (74.6%) were detected carrying the integron-integrase gene (intI1) of class 1 integrons. A higher incidence of antimicrobial resistance was observed in the intI1-positive E. coli isolates than in the intI1-negative E. coli isolates. Furthermore, there were 17 kinds of gene cassette arrays in the 70 integrons as detected by sequencing amplicons of variable regions, with 66 isolates (94.3%) expressing their gene cassettes encoding for multiple drug resistance phenotypes for streptomycin, neomycin, gentamicin, kanamycin, amikacin, sulfamethoxazole-trimethoprim, sulfasalazine, and florfenicol. Notably, due to harboring multiple, hybrid, and recombination cassettes, complex cassette arrays were attributed to multiple drug resistance patterns than simple arrays. In conclusion, we demonstrated that the prevalence of multiple drug resistance and the incidence of class 1 integrons were 69.7 and 74.6% in commensal E. coli isolated from healthy swine, which were lower in frequency than that previously reported in China.
Antimicrobial-resistant bacteria, especially multiple drug-resistant (MDR) strains have caused many outbreaks of food-borne diseases and infectious diseases worldwide, threatening human and animal health (Paitan, 2018). Integrons located on either chromosome or mobile genetic elements (MGEs), such as plasmids and transposons, are considered responsible for the horizontal gene transfer of antimicrobial resistance (AMR; Stalder et al., 2012). Integrons are natural recombination and expression systems with the ability to acquire gene cassettes (Partridge et al., 2018). Gene cassettes are a major source of the resistance genes found in clinical, commensal, and environmental isolates of bacteria. A gene cassette is a small mobile element (0.5–1 kb) consisting of a single gene (occasionally two) and a recombination site (attC; Partridge et al., 2009). Gene cassettes encode proteins that facilitate interactions with their extracellular environment (Timothy et al., 2019). Several cassettes may be inserted into the same integron forming a tandem array. Class 1 integrons in bacteria ubiquitously reside in gastrointestinal tracts of animals and humans, and their abundance and genetic diversity can readily change in response to environmental pressures (Alonso et al., 2017). The class 1 integrons are diverse and significant players in the spread of AMR. E. coli has been shown to be a significant reservoir of genes encoding for AMR and has been suggested as a useful indicator for resistance in bacterial communities (Ge et al., 2020; Holcomb and Stewart, 2020; Zhang et al., 2020b). Highly dynamic and diverse of E. coli populations exist in the swine intestinal microbiota and in the farm environment throughout the full production cycle, suggesting the potential for carriage of antimicrobial resistance and the presence of clinical integrons (Marchant and Morenoa, 2013; Van den Meersche et al., 2020).
Development of MDR in intestinal flora is closely associated with integrons and their gene cassettes (Olivier et al., 2018). Integrons are genetic platforms for captured gene cassettes, which are regarded as adaptation and evolution of bacteria (José, 2018). They have traditional genetic structure of two conserved segments (5'-CS and 3'-CS) and a variable region (Barraud and Ploy, 2015). At least 130 different (<98% identical) cassettes that carry known or predicted antibiotic resistance genes have been identified, along with many cassettes of unknown function (Partridge et al., 2009). Class 1 integrons play a crucial role in the propagation of ARGs, and have been surveyed in numerous ecosystems and animals. Zhang et al. found the average content of class I integrons as 1.31 × 104 copies/100 ml in drinking water from 71 cities in China (Zhang et al., 2020a). However, the data are still lacking on the epidemiology of class 1 integrons and integron-borne gene cassettes in commensal E. coli among livestock herds, in particular at the source of pork production.
To promote effective antimicrobial stewardship, the government of China issued the National Action Plan to Contain Animal Original Antimicrobial Resistance (2017–2020) on June 22, 2017. This action is in line with the strategic objectives of WHO’s Global Action Plan on antimicrobial resistance. Tracking and surveillance of antimicrobial resistant bacteria in livestock is a critical step toward protecting humans and animals from infections. Anhui, Hebei, Shanxi, and Shaanxi provinces are mainly livestock-raising areas in China, with the amount of fattening swine for market (unit: million heads) of 283.74, 370.96, 81.46, 115.08, with the number of stocks at year-end (unit: million heads) of 135.63, 182.08, 54.95, 83.9, respectively, in 2019 (NBSC, 2019). However, data of antimicrobial resistance in livestock are inadequate in these areas.
The aim of the present study was to obtain the phenotypic and genotypic characterization of selected antimicrobial resistance determinants found in commensal E. coli isolated from intensively farmed swine. We detected the MDR phenotypic profiles, the abundance of class 1 integrons, and gene cassettes of class 1 integrons through antimicrobial susceptibility testing, PCR assay, and DNA sequencing.
From March to May, 2019, a total of 330 rectal sterile swabs were collected from healthy growing-finishing swine of four intensive farms in Anhui, Hebei, Shanxi, and Shaanxi province, China (Table 1). The annual production of the intensive farms was 1,000–1,200 heads (i.e., the breeding stock was 500–600 heads), with growing-finishing swine (mixed sex, same aged from 70 to 185 days) raised in the mode of all-in and all-out. Random sampling was conducted on 90‐ to 180-days growing-finishing swine in 10% of the breeding stock herds, with one swab per swine. Healthy swines were determined by observation indicators of their good physical and mental state, with normal body temperature, appearance, and behavior, feed intake and drinking, and excretion. The Animal Welfare and Research Ethics Committee of Northwest A&F University (Yangling, China) approved the protocol of the experiment (protocol number: NWAFUSM2018005). All the rectal sterile swabs were placed into an ice box and transferred to the laboratory within 6 h for further bacteriological analysis.
Isolation and identification of E. coli were performed with rectal sterile swabs and transferred to sterile culture tubes containing 5 ml of Luria-Bertani (LB) broth and mixed vigorously (200 r/min) at 37°C for 8 h. After enrichment, a loop of LB broth culture was streaked onto eosin-methylene blue medium (EMB) agar and incubated at 37°C for 24 h. Colonies showing a metallic sheen were considered presumptive E. coli isolates, and positive colonies were chosen for further biochemical identification using Gram-negative identification cards of an automated VITEK2 microbial identification system (BioMerieux, France), according to the manufacturers.
All E. coli isolates were tested susceptible to seven classes of antimicrobials by the Kirby-Bauer disk diffusion method on Mueller-Hinton (MH) agar plates following Clinical and Laboratory Standards Institute (CLSI) procedures (CLSI, 2017). A panel of 19 antimicrobial agents (Table 2) are commonly used against clinical infections. In animal husbandry, discs containing streptomycin (STR, 10 μg), neomycin (NEO, 30 μg), gentamicin (GEN, 10 μg), kanamycin (K, 30 μg), amikacin (AK, 30 μg), amoxicillin (AMC, 20 μg), ampicillin (AMP, 10 μg), cephalexin (CL, 30 μg), cefotaxime (CTX, 30 μg), imipenem (IMP, 10 μg), ciprofloxacin (CIP, 5 μg), enrofloxacin (ENR, 10 μg), norfloxacin (NOR, 10 μg), tetracycline (TET, 30 μg), doxycycline (DX, 30 μg), sulfamethoxazole-trimethoprim (SMZ/TMP, 25 μg), sulfasalazine (SIZ, 300 μg), erythromycin (ERY, 15 μg), and florfenicol (FF, 30 μg) were used. The reference strain E. coli ATCC 25922 was used for quality control. Susceptibility decision was described as resistant (R), intermediate (I), or susceptible (S) as the CLSI and the sensitivity criteria for Enterobacteriaceae (Hangzhou Microbiology Co., Ltd., China, 2019). Isolates were considered as nonsusceptible when reported as either intermediate susceptible (I) or resistant (R; Jarlier et al., 2019). E. coli isolates exhibiting MDR was defined as acquired nonsusceptible to ≥1 agent in ≥3 antimicrobial categories (Magiorakos et al., 2012).
All E. coli isolates were screened by PCR for the class 1 integron-integrase (intl1) gene to confirm the presence of class 1 integrons. The primers were intI1 F 5'-ACGAGCGCAAGGTTTCGGT-3' and intI1 R 5'-GAAAGGTCTGGTCATACATG-3' (Bass et al., 1999). The amplification protocol was performed as follows: initial denaturation (94°C for 5 min), followed by 30 cycles of denaturation (94°C for 30 s), annealing (56°C for 30 s), extension (72°C for 2 min), then a final extension (72°C for 10 min). Amplicons were separated by electrophoresis in a 1.5% agarose gel and sequenced (Beijing Qingke Biotech Co., Ltd. China).
The variable regions of the integron-positive isolates were further amplified for gene cassettes by PCR using primers of intI1-V FP 5'-TCATGGCTTGTTATGACTGT-3' and intI-V RP 5'-GTAGGGCTTATTATGCACGC-3' (White et al., 2000). PCR conditions were the same as above except for the annealing temperatures for the fragments. After separated by electrophoresis in 1.5% agarose gel, the gel-recovered product was ligated with pMD19-T vector (TaKaRa Bio Group, Japan) at 16°C for 4 h and then transferred into E. coli DH5α cells (QiaGen Biotech CO., LTD, China). The E. coli DH5α cultures were inoculated on the LB agar plates supplemented with 100 μg/ml ampicillin. The amplified fragments of different sizes were sequenced. The nucleotide sequences were analyzed by using Nucleotide BLAST on NCBI website1 and the integron database INTEGRALL.2 Standard Nucleotide BLAST was set as Search database Nucleotide collection (nr/nt) using Megablast (optimized for highly similar sequences). The general parameters were set by selecting the maximum number of aligned sequences to display (100) and expected threshold (10). Once the nucleotide BLAST results were available, pairwise and CDS features were chosen for alignment view.
Data were analyzed using the Statistical Product and Service Solutions software (SPSS, version 20.0). Chi square test and Fisher’s exact test of analysis of variance (ANOVA) were used to determine the statistical significance of data. Values were considered as having a statistically significant difference if p < 0.05 and as an extremely distinct difference if p < 0.01.
In our study, we investigated the prevalence of antimicrobial resistance in 264 E. coli isolates recovered from rectal swabs. They were resistant to at least one antimicrobial agent (Table 3, left). A high proportion of E. coli isolates were nonsusceptible to doxycycline (75.8%), tetracycline (73.5%), sulfamethoxazole-trimethoprim (71.6%), amoxicillin (68.2%), sulfasalazine (67.1%), ampicillin (58.0%), florfenicol (56.1%), and streptomycin (53.0%). However, all isolates were susceptible to imipenem. A total of 184 (69.7%) E. coli isolates exhibited MDR, with 11 MDR patterns (Figure 1). Importantly, 16 E. coli isolates (6.0%) displayed resistance against 13 antimicrobial agents. The prevalence of E. coli MDR in Anhui, Hebei, Shanxi, and Shaanxi were 78.6, 80, 74.4, and 58.0%, respectively (Table 4).
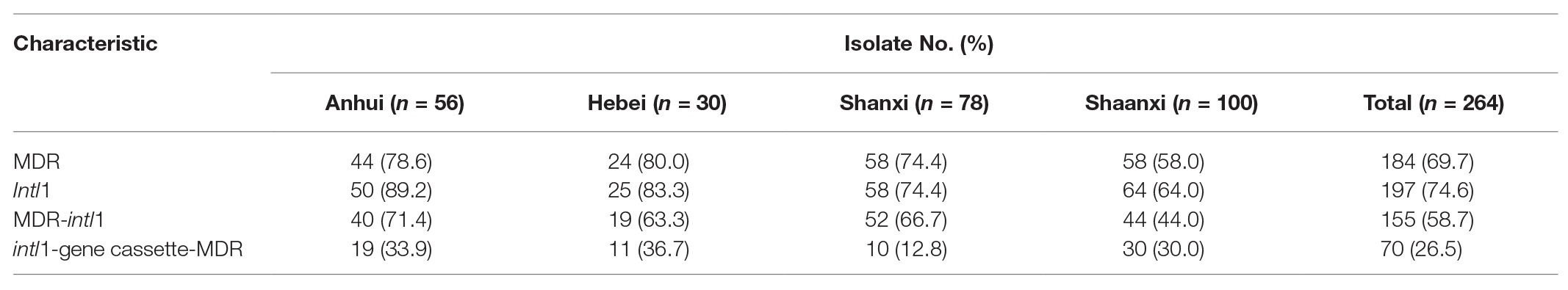
Table 4. Distribution of multiple drug-resistant (MDR), class 1 integrons, and gene cassettes among E. coli isolates in Anhui, Hebei, Shanxi, and Shaanxi.
The IntI1 gene was detected in 197 (74.6%) E. coli isolates, including 155 (78.7%) MDR isolates. IntI1-positive E. coli isolates were most commonly nonsusceptible to doxycycline (70.1%), tetracycline (69.0%), sulfamethoxazole-trimethoprim (67.0%), amoxicillin (66.5%), sulfasalazine (64.5%), florfenicol (54.3%), and ampicillin (53.3%). There were significant differences in AMR between intI1-positive E. coli isolates and intI1-negative E. coli isolates, not only in single antimicrobial resistance but also in multiple drugs resistance (p < 0.01), except sulfasalazine (p = 0.313), kanamycin (p = 0.055), and amikacin (p = 0.128).
After sequencing the variable regions of class 1 integrons, we found 17 gene cassette arrays and 10 gene cassettes (aadA1, aadA2, aadA5, aadA16; dfrA1, dfrA7, dfrA12, dfrA17, cmlA1, and orfF) in 70 E. coli isolates (Table 5). Among them, 66 (85.7%) isolates had gene cassettes that corresponded with respective phenotypic MDR patterns, encoding resistance to streptomycin, neomycin, gentamicin, kanamycin, amikacin, sulfamethoxazole-trimethoprim, sulfasalazine, and florfenicol. However, there were three E. coli isolates that did not express their cassette arrays for MDR patterns at all, including dfrA12-orfF-aadA2/1, aadA1-aadA2, aadA2, and aadA1 (Table 5, marked with lower case c). Different gene cassette arrays, even from the same array showed different phenotypic patterns of MDR. In addition, prevailing arrays were different in the four farms: five arrays in Anhui (n = 19), five arrays in Hebei (n = 11), eight arrays in Shanxi (n = 10), and six arrays in Shaanxi (n = 30).
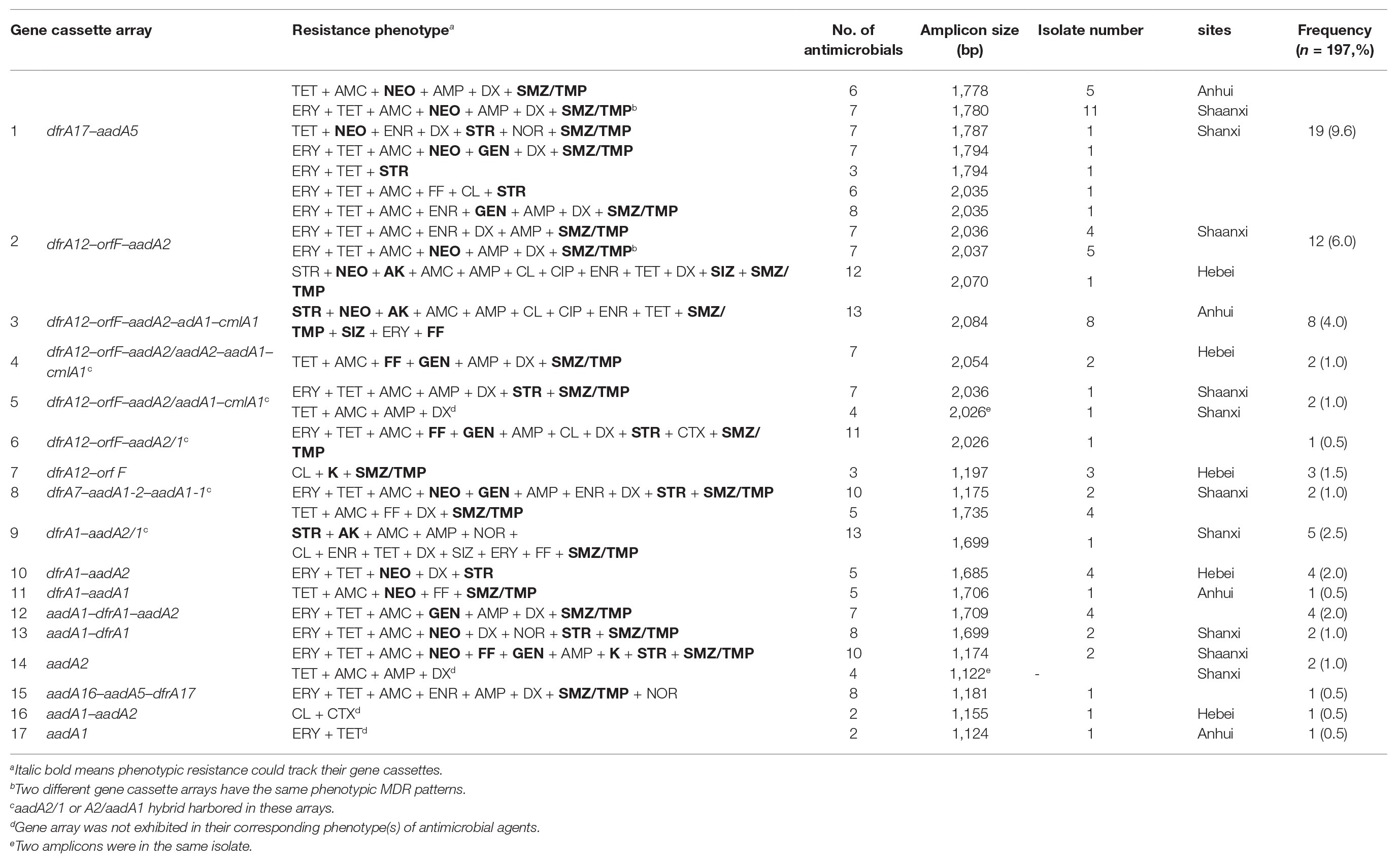
Table 5. Characterization of cassette arrays and integron-associated antibiotic resistance in 70 isolates of E. coli.
Cassette arrays showed the following characteristics. First, the top three cassette arrays were dfrA17–aadA5 (9.6%), dfrA12–orfF–aadA2 (6.0%), and dfrA12–orfF-aadA2–adA1–cmlA1 (4.0%). They were confirmed by the same or similar sizes of amplicons, but they exhibited different patterns of phenotypical MDR. This phenomenon also occurred in dfrA1–aadA2/1 (2.5%), aadA2 (1.5%), and dfrA12–orfF–aadA2/1 (1.0%). Second, in six arrays, E. coli isolates with aadA2/1 and aadA2/aadA2 hybrid gene cassettes exhibited more complex MDR, such as dfrA1–aadA2/1, dfrA7–aadA1-2–aadA1-1, dfrA12–orfF–aadA2/aadA2–aadA1–cmlA1, dfrA12-orfF–aadA2/aadA1–cmlA1, dfrA12–orfF–aadA2/aadA1–cmlA1, and dfrA12–orfF–aadA2/1. Third, E. coli isolates with simple cassette arrays performed poorly on MDR, including dfrA12–orfF, dfrA1–aadA2, dfrA1–aadA1, aadA1–dfrA1, aadA1–aadA2, aadA2, and aadA1. In addition, one isolate yielded two amplicons, 2,026 and 1,122 bp, and they formed into two arrays of dfrA12–orfF–aadA2/aadA1–cmlA1 and aadA2. Another special array, aadA16–aadA5–dfrA17 was found for the first time.
In the present study, we reported the frequency of MDR and the incidence of class 1 integrons from nonclinical swine and were 69.7 and 74.6% in commensal E. coli, which was lower than those that had been reported among swine origin nonclinical E. coli in several provinces of China, such as Sichuan (99.23, 87.69%; Lin et al., 2014), Xiamen (100, 92.2%; Liu et al., 2015), Harbin (100, 91.67%; Zhang et al., 2013), Hulunbeir (77.8, 95.59%; Qiu, 2015), Liaoning (92.31, 71.43%; Zhao et al., 2017), and Yunnan (96.15, 84.62%; Li et al., 2019) between 2010 and 2019. This indicates that there is a clear difference in prevalence between the previous investigated districts and the four provinces. Antimicrobial application is responsible for emergence and spread of MDR intestinal bacteria in animal husbandry (Matjuda and Aiyegoro, 2016). Veterinary antimicrobials always have the most priority treatment, and some of them are often taken as growth promoters for swine fattening (Ochoa et al., 2016). Increasing diversity and abundance of the antimicrobial resistance in E. coli clinical isolates witnessed the extensive and excessive usage of veterinary antimicrobials in swine herds of 18 provinces in China between 2010 and 2019 (Zhang et al., 2017, 2019a; Yang et al., 2019). Swine farming could lead to enhanced concentration levels of various veterinary antimicrobials and ARGs in groundwater and soils (Qiao et al., 2018). The increasing prevalence of MDR E. coli and other antimicrobial-resistant microorganisms causes unpredictable consequences to the environment and humans. In China, alternative strategies have been effectively explored to combat veterinary antimicrobial resistance, such as vaccination, chicken egg yolk antibodies (Zhang et al., 2011), lactic acid bacteria (Yang et al., 2015), antimicrobial peptides (Kang et al., 2017), bacteriocins (Qian et al., 2020), antibiotic adjuvants (Liu et al., 2019), phytocompounds (Chang et al., 2019), and metal-based nanoparticles (Ali et al., 2020). In addition, the implementation of management strategies has been done to reduce antimicrobial usage in animal husbandry, including good hygiene practice and biosecurity measures (Hu and Cowling, 2020). Therefore, it is possible that the frequency of MDR and the incidence of class 1 integrons in the present intensive farms are in the decline process due to good veterinary hygiene managements and reduced antimicrobial usage.
Phenotypic susceptibility testing showed that doxycycline, tetracycline, sulfamethaxazole/trimethoprim, amoxicillin, sulfasalazine, florfenicol, and ampicillin resistance are still challenging intensive farms in Anhui, Hebei, Shanxi, and Shaanxi. Many studies pointed out that E. coli strains from swine farms had formed high resistance to the first‐ or the second-generation antibiotics, and some of the original drugs had lost their antibacterial effect, such as streptomycin, tetracycline, cephalothin, ampicillin, ofloxacin, and sulfamethoxazole (Tang, 2014; Zhang et al., 2017). The evolution of E. coli antimicrobial resistance is closely correlated with the emergence and dissemination of specific ARGs (Jiang et al., 2017a; Divya and Hatha, 2019; Wang et al., 2019) and virulence genes (VGs; Wang et al., 2010; Cheng et al., 2020). This indicates that commensal E. coli in healthy swine, with resistance against doxycycline, tetracycline, sulfamethaxazole/trimethoprim, amoxicillin, sulfasalazine, florfenicol, and ampicillin, may easily obtain antibiotic resistance genes and virulence genes that jeopardize swine health.
Notably, 11 isolates were highly resistant to florfenicol for carrying the cmlA gene (in arrays of dfrA12–orfF–aadA2–adA1–cmlA1, dfrA12–orfF–aadA2/aadA2–aadA1–cmlA1, and dfrA12–orfF–aadA2/aadA1–cmlA1), which encodes a specific chloramphenicol transporter. Florfenicol, a fluorinated chloramphenicol derivative, has been widely used against both Gram-positive and Gram-negative bacteria (Belaynehe et al., 2018). Another notable antimicrobial agent is imipenem, belonging to the carbapenems antibiotics, which has a broader range of activity against Gram-negative and Gram-positive bacteria. The carbapenem-resistant genes (blaOXA-48, blaGES-1, blaKPC-2, and blaNDM-1) were considered to be the most harmful to human health (Yang et al., 2016). In China, KPC-2, NDM, and OXA-48-like carbapenemases were predominant among the carbapenem-resistant Enterobacteriaceae (CRE) isolates from adult and children patients (Han et al., 2020). However, in the present study, all E. coli isolates were susceptible to imipenem, and carbapenem-resistant genes were not detected by PCR. It demonstrates that the carbapenem-resistant genes did not exist or was extremely low in the investigated farms, which is consistent with the carbapenem-resistant pollution that is not severe in the drinking water of China (Xin et al., 2019).
In the present study, the prevalence of MDR E. coli is closely associated with the presence of class 1 integrons. Our findings are in agreement with the previous studies that class 1 integrons are the most ubiquitous classes of integrons in the enteric bacteria (Stokes et al., 2001). According to the study of Yohann Lacotte, in the stress-free environmental settings of E. coli, class 1 integrons live a relaxed life at low-cost structure, which can favor their maintenance and prevalence in cassette networks (Lacotte et al., 2017). In the present study, the healthy swine gut meant a safe environment for the low-cost class 1 integrons, thus 74.6 % (n = 197) integrons were highly prevalent in common E. coli isolates. They can offer a wider platform for the acquisition, rearrangement, and expression of gene cassettes. In addition, isolates (n = 29) without integrons also showed MDR, suggesting that other determinants might contribute to their resistance. Farm animals and manure are a source of food-borne and water-borne human pathogens (Bailey et al., 2011). Being a DNA pollutant (Gillings, 2018), the abundance of intI1 ranged from 3.83 × 10−4 to 4.26 × 100 intI1/cell in eight ecosystems, even in giant pandas (47%) and remote rural area animals (6.7%) in China (Ma et al., 2017; Rehman et al., 2017; Zou et al., 2018). Xia et al. reviewed that E. coli had the highest positive rates (65.4%) of integrons from human patients with Gram-negative bacteria isolates in China during 2000–2014 (Xia et al., 2016). It suggests that MDR E. coli with a high number of class 1 integrons were widely distributed in swine farms, also indicating a higher incidence of lateral gene-transfer events.
In the present study, 17 gene cassette arrays in the commensal E. coli isolates were more diverse than those that had been reported in different animals and their products during the past 10 years in China, Spain, and Australia. There are nine arrays that are summarized from swine farming settings and pork products (dfrA1–aadA1, aadA22, dfr17–aadA5, dfrA12–orfF–aadA2, dfrXII–orfF–aadA2, aadA2, dfrA1–catB3–aacA4, aadB–aadA2, and dfrA12–aadA2–cmlA1–aadA1; Chen, 2013; Wei, 2014; Zhang et al., 2017). Nine arrays were present in E. coli isolates from beef carcasses, including linF–aadA2, dfrA17–aadA5, aadB–blaOXA-10, dfr12–orfF–aadA2, dfrA1–aadA1, dfrA12–aadA2, aadA2, dfr12, and aadB–aadA2 (Chen et al., 2017). Five arrays were in E. coli isolates from waterfowls (dfrA1–orfC, aadA2, aadA1, dfrA1–aadA1, and dfrA1–orfC–aadA1; Zhang et al., 2019b). The different arrangements of gene cassettes of E. coli isolates from healthy swine were not only reported in China but also in other countries. For instance, nine different gene cassette arrays were described in 393 intestinal E. coli isolates in Spain, with dfrA1–aadA1 in a dominant position (Marchant et al., 2013). Ten gene cassette arrays were observed in 103 class 1 integron-positive E. coli from two commercial production facilities in New South Wales, Australia (Reid et al., 2017). These results highlighted the role of pork, beef, and poultry products as a potential source for MDR E. coli strains and the necessity for controlling animal product safety.
In this study, the common gene cassette arrays were still prevalent in MDR E. coli. High frequency of 11 simple gene arrays were found, including dfrA17–aadA5, dfrA12–orfF–aadA2, dfrA1–aadA1, dfrA12–orfF, dfrA1–aadA2, aadA1–dfrA1–aadA2, aadA1–dfrA1, aadA1–aadA2, and aadA2. Then, the complex gene cassette arrays exhibited more MDR patterns through multiple cassettes, hybrid cassettes, and recombination cassettes. Isolates could contain two, three, four, and five cassettes in a single array. The complexity of the cassette array is strongly correlated with MDR patterns and phenotype.
In addition, six isolates were identified carrying hybrid gene cassettes (gene cassettes arrays marked in lowercase letter b, in Table 3). Our results were in agreement with the reported array dfrA12–orfF–aadA2 cassette, in which aadA2 often was replaced by the corresponding part of the aadA1 cassette (Alicia et al., 2005). Moreover, aadA16–aadA5–dfrA17 was a novel array. To the best of our knowledge, this was the first time to report a relative higher incidence of complex cassettes prevailing in intensive-farming swine in China.
We noticed that 12 isolates had a larger variable region containing four or five different gene cassettes, dfrA12–orfF–aadA2–adA1–cmlA1 (n = 8), dfrA12–orfF-aadA2/aadA2–aadA1–cmlA1 (n = 2), dfrA12–orfF–aadA2/aadA1–cmlA1 (n = 1), and dfrA12–orfF–aadA2/1 (n = 1). They were highly consistent with nonclassic integrons that Partridge (Partridge et al., 2009) summarized as the 5'-CS but not the typical 3'-CS or incomplete transposition (tni) region. Similar arrays have been reported in one E. coli isolate from retail meat (pork; Jiang et al., 2017b), two commensal E. coli strains from feces (Moran et al., 2016; Oliva et al., 2018), and three clinical E. coli strains from different sources (samples from patients, dogs, swine, food products, and environment; Antunes et al., 2007; Sa’enz et al., 2010; María et al., 2011; Siqueira et al., 2016). In addition, four gene arrays did not exhibit their corresponding MDR phenotypes. This may due to the fact that many would affect the expression of gene cassettes, such as the strict regulation of integrase expression, the strength of the Pc promoter, the gene cassette arrangement, and the antibiotic concentrations (Vinué et al., 2011). Therefore, our further work will focus on these special arrays and their roles in conferring antibiotic resistance.
In conclusion, this study reported that the prevalence of antimicrobial resistance and the presence of class 1 integrons of commensal E. coli in the present four provinces were lower than previously reported in other regions. A high number of commensal E. coli with class 1 integrons were widely distributed in investigated farms, indicating a higher incidence of lateral resistant gene-transfer events. What is more, a portion of commensal E. coli harboring diverse cassette arrays contributed to their MDR phenotypes. Our finding indicates that the nonclinical swine raised in intensive farm would be a reservoir of MDR E. coli, which is a potential health risk.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
KG and YZ conceived and designed the study. XZ performed the data analyses and wrote the manuscript. XL and JQ performed antimicrobial susceptibility testing and molecular experiments. WW, DW, and YL collected the samples and isolated Escherichia coli. LX reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Key Research and Development Program of Shaanxi Province (Project No. 2018NY-098); The Innovation and Entrepreneurship Training Program for Undergraduates of Northwest A&F University (201810712015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ali, A., Ovais, M., Cui, X., Rui, Y., and Chen, C. (2020). Safety assessment of nanomaterials for antimicrobial applications. Chem. Res. Toxicol. 33, 1082–1109. doi: 10.1021/acs.chemrestox.9b00519
Alicia, M., Gestal, H., Stokes, W., Partridge, S. R., and Hall, R. M. (2005). Recombination between the dfrA12-orfF-aadA2 cassette array and an aadA1 gene cassette creates a hybrid cassette, aadA8b. Appl. Environ. Microbiol. 49, 4771–4774. doi: 10.1128/AAC.49.11.4771-4774.2005
Alonso, C., Zarazaga, M., Ben, S. R., Jouini, A., Ben, S. K., and Torres, C. (2017). Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett. Appl. Microbiol. 64, 318–334. doi: 10.1111/lam.12724
Antunes, P., Machado, J., and Peixe, L. (2007). Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3' conserved sequence region among Salmonella isolates. Antimicrob. Agents Chemother. 51, 1545–1548. doi: 10.1128/AAC.01275-06
Bailey, K. G. B., Gaze, W. H., Zhang, L., Kay, P., Boxall, A., Hawkey, P. M., et al. (2011). Integron prevalence and diversity in manured soil. Appl. Environ. Microbiol. 77, 684–687. doi: 10.1128/AEM.01425-10
Barraud, O., and Ploy, M. C. (2015). Diversity of class 1 integron gene cassette rearrangements selected under antibiotic pressure. J. Bacteriol. 197, 2171–2178. doi: 10.1128/JB.02455-14
Bass, L., Liebert, C. A., Lee, M. D., Summers, A. O., White, D. G., Thayer, S. G., et al. (1999). Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43, 2925–2929. doi: 10.1128/AAC.43.12.2925
Belaynehe, K. M., Shin, S. W., and Yoo, H. S. (2018). Interrelationship between tetracycline resistance determinants, phylogenetic group affiliation and carriage of class 1 integrons in commensal Escherichia coli isolates from cattle farms. BMC Vet. Res. 14:340. doi: 10.1186/s12917-018-1661-3
Chang, F., Shen, S., Shi, P., Zhang, H., Ye, L., Zhou, Q., et al. (2019). Antimicrobial resins with quaternary ammonium salts as a supplement to combat the antibiotic resistome in drinking water treatment plants. Chemosphere 221, 132–140. doi: 10.1016/j.chemosphere.2019.01.047
Chen, C. (2013). Multi-antibiotics resistance and integron gene cassette system analysis of E. coli isolated from pork production chain. Master’s degree dissertation (in Chinese), Sichuan Agricultural University
Chen, C., Ke, S., Li, C., Wu, Y., Chen, T., Lai, C., et al. (2017). High diversity of antimicrobial resistance genes, class 1 integrons, and genotypes of multidrug-resistant Escherichia coli in beef carcasses. Microb. Drug Resist. 23, 915–924. doi: 10.1089/mdr.2016.0223
Cheng, P., Yang, Y., Zhang, J., Li, F., Li, X., Liu, H., et al. (2020). Antimicrobial resistance and virulence profiles of mcr-1-positive Escherichia coli isolated from swine farms in Heilongjiang Province of China. J. Food Prot. doi: 10.4315/jfp-20-190 [Epub ahead of print]
CLSI (2017). Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth-third Informational Supplement M100-S23. Wayne, PA, USA: Clinical and Laboratory Standards Institute.
Divya, S., and Hatha, A. (2019). Screening of tropical estuarine water in south-west coast of India reveals emergence of ARGs-harboring hypervirulent Escherichia coli of global significance. Int. J. Hyg. Environ. Health 222, 235–248. doi: 10.1016/j.ijheh.2018.11.002
Ge, B., Domesle, K., Gaines, S., Lam, C., Bodeis Jones, S., Yang, Q., et al. (2020). Prevalence and antimicrobial susceptibility of Indicator organisms Escherichia coli and Enterococcus spp. isolated from U.S. animal food, 2005-2011. Microorganisms 8:1048. doi: 10.3390/microorganisms8071048
Gillings, M. R. (2018). DNA as a pollutant: the clinical class 1 integron. Curr. Pollut. Rep. 4, 49–55. doi: 10.1007/s40726-018-0076-x
Han, R., Shi, Q., Wu, S., Yin, D., Peng, M., Dong, D., et al. (2020). Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front. Cell. Infect. Microbiol. 10:314. doi: 10.3389/fcimb.2020.00314
Holcomb, D., and Stewart, J. (2020). Microbial indicators of fecal pollution: recent progress and challenges in assessing water quality. Current Environ. Health Rep. 7, 311–324. doi: 10.1007/s40572-020-00278-1
Hu, Y., and Cowling, B. (2020). Reducing antibiotic use in livestock, China. Bull. World Health Organ. 98, 360–361. doi: 10.2471/blt.19.243501
Jarlier, V., Högberg, L. D., Heuer, O. E., Campos, J., and Simonsen, G. S. (2019). Strong correlation between the rates of intrinsically antibiotic-resistant species and the rates of acquired resistance in gram-negative species causing bacteraemia, EU/EEA, 2016. Euro Surveill. 24:1800538. doi: 10.2807/1560-7917.ES.2019.24.33.1800538
Jiang, X., Ellabaan, M., Charusanti, P., Munck, C., Blin, K., Tong, Y., et al. (2017a). Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 8:15784. doi: 10.1038/ncomms15784
Jiang, X., Xu, Y., Li, Y., Zhang, K., Liu, L., Wang, H., et al. (2017b). Characterization and horizontal transfer of qacH-associated class 1 integrons in Escherichia coli isolated from retail meats. Int. J. Food Microbiol. 258, 12–17. doi: 10.1016/j.ijfoodmicro.2017.07.009
José, A. E., Loot, C., and Mazel, D. (2018). “Integrons as adaptive devices” in Molecular mechanisms of microbial evolution. Grand challenges in biology and biotechnology. ed. P. H. Rampelotto (Cham: Springer).
Kang, W., Liu, H., Ma, L., Wang, M., Wei, S., Sun, P., et al. (2017). Effective antimicrobial activity of a peptide mutant Cbf-14-2 against penicillin-resistant bacteria based on its unnatural amino acids. Eur. J. Pharm. Sci. 105, 169–177. doi: 10.1016/j.ejps.2017.05.030
Lacotte, Y., Cécile, P. M., and Sophie, R. (2017). Class 1 integrons are low-cost structures in Escherichia coli. ISME J. 11, 1535–1544. doi: 10.1038/ismej.2017.38
Li, F., Liao, D., Li, N., Miao, H., Song, J., Zhao, W., et al. (2019). Correlations between integron gene cassttes and antimicrobial ressitancen of pathogenic Escherichia coli isolated from pigs. China J. Vet. Sci. 39, 228–234. doi: 10.16303/j.cnki.1005-4545.2019.02.07
Lin, J., Shu, G., Zhang, H., Cao, S., and Wen, X. (2014). Detection of resistance and characterization of integron-cassettes in Escherichia coli from intestinal tract of healthy food-animals, enteric strains. Chinese J. Vet. Sci. 34, 55–60. doi: 10.16303/j.cnki.1005-4545.2014.01.011
Liu, Y., Li, R., Xiao, X., and Wang, Z. (2019). Antibiotic adjuvants: an alternative approach to overcome multi-drug resistant gram-negative bacteria. Crit. Rev. Microbiol. 45, 301–314. doi: 10.1080/1040841x.2019.1599813
Liu, Z., Zhang, Z., Yan, H., Li, J., and Shi, L. (2015). Isolation and molecular characterization of multidrug-resistant Enterobacteriaceae strains from pork and environmental samples in Xiamen, China. J. Food Prot. 78, 78–88. doi: 10.4315/0362-028X.JFP-14-172
Ma, L., Li, A. D., Yin, X. L., and Zhang, T. (2017). The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments. Environ. Sci. Technol. 51, 5721–5728. doi: 10.1021/acs.est.6b05887
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., and Giske, C. G. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marchant, M., and Morenoa, M. A. (2013). Dynamics and diversity of Escherichia coli in animals and system management of the manure on a commercial farrow-to-finish pig farm. Appl. Environ. Microbiol. 79, 853–859. doi: 10.1128/AEM.02866-12
Marchant, M., Vinué, L., Torres, C., and Moreno, M. (2013). Change of integrons over time in Escherichia coli isolates recovered from healthy pigs and chickens. Vet. Microbiol. 163, 124–132. doi: 10.1016/j.vetmic.2012.12.011
María de, T. H., Yolanda, D., Mansilla, E. C., Bezares, B. R., and Marta, G. C. (2011). Genetic characterization of the mechanisms of resistance to amoxicillin/clavulanate and third-generation cephalosporins in Salmonella enterica from three Spanish hospitals. Int. Microbiol. 14, 173–181. doi: 10.2436/20.1501.01.146
Matjuda, D. S., and Aiyegoro, O. A. (2016). Soil bacteriological pollution in pig farm vicinity: assessment of bacterial dynamics and detection of antimicrobial resistance gene. Afr. J. Microbiol. Res. 10, 1625–1636. doi: 10.5897/AJMR2016.8138
Moran, R. A., Holt, K. E., and Hall, R. M. (2016). pCERC3 from a commensal ST95 Escherichia coli: a ColV virulence-multiresistance plasmid carrying a sul3-associated class 1 integron. Plasmid 84, 11–19. doi: 10.1016/j.plasmid.2016.02.002
NBSC (2019). China Statistical Yearbook [Online]. Available: http://www.stats.gov.cn/tjsj/ndsj/2019/indexch.htm (Accessed September 22, 2020).
Ochoa, T. J., Duarte, G., and Oscar, G. (2016). “Antibiotic resistance in Escherichia coli” in Escherichia coli in the Americas. ed. A. G. Torres (Cham: Springer International Publishing).
Oliva, M., Monno, R., Addabbo, P., Pesole, G., Scrascia, M., Calia, C., et al. (2018). IS26 mediated antimicrobial resistance gene shuffling from the chromosome to a mosaic conjugative FII plasmid. Plasmid 100, 22–30. doi: 10.1016/j.plasmid.2018.10.001
Olivier, B., Marianne, P., Elodie, C. D., Delphine, C., Claire, B., Vincent, G., et al. (2018). Antibiotic resistance acquisition in the first week of life. Front. Microbiol. 9:1467. doi: 10.3389/fmicb.2018.01467
Paitan, Y. (2018). Current trends in antimicrobial resistance of Escherichia coli. Curr. Top. Microbiol. Immunol. 416, 181–211. doi: 10.1007/82_2018_110
Partridge, S., Guy, T., Enrico, C., and Jonathan, R. I. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–e00017. doi: 10.1128/CMR.00088-17
Qian, Z., Zhao, D., Yin, Y., Zhu, H., and Chen, D. (2020). Antibacterial activity of lactobacillus strains isolated from Mongolian yogurt against Gardnerella vaginalis. Biomed. Res. Int. 2020:3548618. doi: 10.1155/2020/3548618
Qiao, M., Ying, G., Singer, A. C., and Zhu, Y. (2018). Review of antibiotic resistance in China and its environment. Environ. Int. 110, 160–172. doi: 10.1016/j.envint.2017.10.016
Qiu, J. (2015). “Incidence investigation and prevention of swine colibacillosis in Hulunbeir” in Dissertation for the veterinary doctoral degree (in Chinese). Northeast Agricultural University.
Rehman, M. U., Zhang, H., Huang, S., Iqbal, M. K., Mehmood, K., Luo, H., et al. (2017). Characteristics of integrons and associated gene cassettes in antibiotic-resistant Escherichia coli isolated from free-ranging food animals in China. J. Food Sci. 82, 1902–1907. doi: 10.1111/1750-3841.13795
Reid, C., Wyrsch, E., Roy, C., Zingali, T., Liu, M., Darling, A., et al. (2017). Porcine commensal Escherichia coli: a reservoir for class 1 integrons associated with IS26. Microb. Genom. 3:e000143. doi: 10.1099/mgen.0.000143
Sa’enz, Y., Vinue´, L., Ruiz, E., Somalo, S., Martı´nez, S., Rojo-Bezares, B., et al. (2010). Class 1 integrons lacking qacEΔ1 and sul1 genes in Escherichia coli isolates of food, animal and human origins. Vet. Microbiol. 144, 493–497. doi: 10.1016/j.vetmic.2010.01.026
Siqueira, A. K., Michael, G. B., Domingos, D. F., Ferraz, M. M., Ribeiro, M. G., Schwarz, S., et al. (2016). Diversity of class 1 and 2 integrons detected in Escherichia coli isolates from diseased and apparently healthy dogs. Vet. Microbiol. 194, 79–83. doi: 10.1016/j.vetmic.2016.05.005
Stalder, T., Barraud, O., Casellas, M., Dagot, C., and Ploy, M. (2012). Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 3:119. doi: 10.3389/fmicb.2012.00119
Stokes, H. W., Holmes, A. J., Nield, B. S., Holley, M. P., Nevalainen, H. K. M., Mabbutt, B. C., et al. (2001). Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Eniviron. Microbiol. 67, 5240–5246. doi: 10.1128/AEM.67.11.5240-5246.2001
Tang, Q. (2014). Study on the correlation between the drug-resistance of Escherichia coli and drug use in pig farms. Doctoral dissertation. Hunan Agricultural University.
Timothy, M. G., Jemma, L. G., John, A., and Gillings, M. (2019). High diversity and rapid spatial turnover of integron gene cassettes in soil. Environ. Microbiol. 21, 1567–1574. doi: 10.1111/1462-2920.14551
Van den Meersche, T., Rasschaert, G., Vanden Nest, T., Haesebrouck, F., Herman, L., Van Coillie, E., et al. (2020). Longitudinal screening of antibiotic residues, antibiotic resistance genes and zoonotic bacteria in soils fertilized with pig manure. Environ. Sci. Pollut. Res. Int. 27, 28016–28029. doi: 10.1007/s11356-020-09119-y
Vinué, L., Jové, T., Torres, C., and Ploy, M. C. (2011). Diversity of class 1 integron gene cassette pc promoter variants in clinical Escherichia coli strains and description of a new P2 promoter variant. Int. J. Antimicrob. Agents 38, 526–529. doi: 10.1016/j.ijantimicag.2011.07.007
Wang, X., Jiang, H., Liao, X., Liu, J., Zhang, W., Zhang, H., et al. (2010). Antimicrobial resistance, virulence genes, and phylogenetic background in Escherichia coli isolates from diseased pigs. FEMS Microbiol. Lett. 306, 15–21. doi: 10.1111/j.1574-6968.2010.01917.x
Wang, L., Wang, J., Wang, J., Zhu, L., Yang, L., and Yang, R. (2019). Distribution characteristics of antibiotic resistant bacteria and genes in fresh and composted manures of livestock farms. Sci. Total Environ. 695:133781. doi: 10.1016/j.scitotenv.2019.133781
Wei, W. (2014). The establishment of multiplex PCR for porcine Enterotoxingenitc Escherichia coli and analysis of integron which mediated it multiresistance. Master’s doctoral dissertation. Jiangxi Agricultural University.
White, P. A., Mciver, C. J., Deng, Y. M., and Rawlinson, W. D. (2000). Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182, 265–269. doi: 10.1111/j.1574-6968.2000.tb08906.x
Xia, W., Xu, T., Qin, T., Liu, P., Liu, Y., Kang, H., et al. (2016). Characterization of integrons and novel cassette arrays in bacteria from clinical isloates in China, 2000-2014. J. Biomed. Res. 30, 292–303. doi: 10.7555/jbr.30.20150153
Xin, R., Zhang, K., Wu, N., Zhang, Y., and Niu, Z. (2019). The pollution level of the Bla carbapenemase gene in coastal water and its host bacteria characteristics. Environ. Pollut. 244, 66–71. doi: 10.1016/j.envpol.2018.10.023
Yang, F., Hou, C., Zeng, X., and Qiao, S. (2015). The use of lactic acid bacteria as a probiotic in swine diets. Pathogens 4, 34–45. doi: 10.3390/pathogens4010034
Yang, F., Mao, D., Zhou, H., and Luo, Y. (2016). Prevalence and fate of carbapenemase genes in a wastewater treatment plant in northern China. PLoS One 11:e0156383. doi: 10.1371/journal.pone.0156383
Yang, F., Zhang, K., Zhi, S., Li, J., Tian, X., Gu, Y., et al. (2019). High prevalence and dissemination of β-lactamase genes in swine farms in northern China. Sci. Total Environ. 651, 2507–2513. doi: 10.1016/j.scitotenv.2018.10.144
Zhang, S., Abbas, M., Rehman, M., Huang, Y., Zhou, R., Gong, S., et al. (2020b). Dissemination of antibiotic resistance genes (ARGs) via integrons in Escherichia coli: a risk to human health. Environ. Pollut. 266:115260. doi: 10.1016/j.envpol.2020.115260
Zhang, B., Ku, X., Yu, X., Sun, Q., Wu, H., Chen, F., et al. (2019a). Prevalence and antimicrobial susceptibilities of bacterial pathogens in Chinese pig farms from 2013 to 2017. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-45482-8
Zhang, P., Shen, Z., Zhang, C., Song, L., Wang, B., Shang, J., et al. (2017). Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Vet. Microbiol. 203, 49–55. doi: 10.1016/j.vetmic.2017.02.008
Zhang, S., Wang, S., Li, H., and Li, L. (2011). Vitellogenin, a multivalent sensor and an antimicrobial effector. Int. J. Biochem. Cell Biol. 43, 303–305. doi: 10.1016/j.biocel.2010.11.003
Zhang, X., Wang, L., Xu, G., Yue, J., Su, H., Zhang, L., et al. (2013). Inspection of multidrug resistant Escherichia coli conjuagal plasmid and derived from swine in Harbin. J. Northeast Agric. Univ. 44, 40–45.
Zhang, K., Xin, R., Zhao, Z., Ma, Y., Zhang, Y., and Niu, Z. (2020a). Antibiotic resistance genes in drinking water of China: occurrence, distribution and influencing factors. Ecotoxicol. Environ. Saf. 188:109837. doi: 10.1016/j.ecoenv.2019.109837
Zhang, S., Yang, H., Rehman, M., Yang, K., Dong, M., Yang, J., et al. (2019b). Class 1 integrons as predominant carriers in Escherichia coli isolates from waterfowls in Hainan, China. Ecotoxicol. Environ. Saf. 183:109514. doi: 10.1016/j.ecoenv.2019.109514
Zhao, F., Zhu, Q., Guan, M., ChunJing,, Wang, Z., Cao, D., et al. (2017b). Investigation and analysis of whole carrying integrons of Escherichia coli from swine in Liaoning area. Progress in Veterinary Medicine (Chinese with English abstract).
Keywords: antimicrobial resistance, multiple-drug resistance, class 1 integrons, gene cassette, commensal Escherichia coli, antibiotic-resistant bacteria, antibiotic-resistant genes
Citation: Zhang X, Li X, Wang W, Qi J, Wang D, Xu L, Liu Y, Zhang Y and Guo K (2020) Diverse Gene Cassette Arrays Prevail in Commensal Escherichia coli From Intensive Farming Swine in Four Provinces of China. Front. Microbiol. 11:565349. doi: 10.3389/fmicb.2020.565349
Received: 25 May 2020; Accepted: 01 September 2020;
Published: 14 October 2020.
Edited by:
Lisa Durso, United States Department of Agriculture, United StatesReviewed by:
Dingqiang Chen, Southern Medical University, ChinaCopyright © 2020 Zhang, Li, Wang, Qi, Wang, Xu, Liu, Zhang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kangkang Guo, Z3Vva2syMDA3QG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.