- 1Guangdong Key Laboratory of Animal Conservation and Resource Utilization, Guangdong Public Laboratory of Wild Animal Conservation and Utilization, Institute of Zoology, Guangdong Academy of Science, Guangzhou, China
- 2College of Veterinary Medicine, Agricultural University of Hebei, Baoding, China
- 3Zhengzhou Zoo, Zhengzhou, China
- 4Beijing Key Laboratory of Captive Wildlife Technologies, Beijing Zoo, Beijing, China
- 5College of Global Change and Earth System Science, Beijing Normal University, Beijing, China
- 6Department of Infectious Diseases, Zhejiang Provincial Centre for Disease Control and Prevention, Hangzhou, China
- 7South China Institute of Environmental Sciences, Ministry of Ecology and Environment, Guangzhou, China
Due to its drug resistant nature, β-lactamase represents a serious challenge for public health. Extended-spectrum β-lactamase (ESBL) producing Klebsiella pneumoniae clones are increasingly reported worldwide. Little is known about the prevalence and biological characteristics of drug-resistant strains in zoos. During routine surveillance at the Zhengzhou Zoo of China, we found Klebsiella pneumoniae isolate in healthy Red Kangaroos (Macropus Rufus) with severe MDR. The Klebsiella pneumoniae were especially resistant to Cefuroxime Sodium (MIC, > 64 μg/mL), Ceftriaxone (MIC, >8 μg/mL) and Cefepime (MIC, >64 μg/mL), and belonged to ST290. Subsequently, whole genome sequencing (WGS) showed that the Chrome Chr-M297-1 harbored blaDHA–3, blaSHV–1, blaCTX–M–14, fosA5, dfrA3, sul3, etc., and pM297-1.1 [222,864 bp, IncFIB(K)], which carried nine antimicrobial genes including blaCTX–M–14, blaTEM–191, aph(3″)-Ib, aph(6)-Id and qnrS1, etc., and pM297-1.2 [225,763 bp, IncFII(K)] carried 22 antimicrobial genes including blaTEM–1, blaCTX–M–3, aph(3′)-Ia, aac(3)-IIa, aac(6′)-Ib-cr, aadA16, qnrB2, qnrS1, qacEΔ1, mphA, sul1, and dfrA27, etc. A traceability analysis then revealed that these two plasmids were highly similar to those recovered from human clinical samples in some southern cities in Sichuan Province, China (>99%), suggesting that these plasmids are spreading in China. Furthermore, two plasmids harboring conjugal transfer genes facilitated the transmission of antimicrobial genes by conjugation with E. coli J53. Our research shows that the transmission and adaptation of Klebsiella pneumoniae producing ESBLs is occurring in zoo environments, suggesting that zoos may be becoming important potential reservoirs for clinically important drug-resistant genes. It is therefore necessary to monitor the emergence and spread of drug-resistant gene strains in captive wild animals held in zoo environments.
Introduction
The emergence and dissemination of antimicrobial resistance (AMR) in the environment has become a global concern. AMR has become an area of focus over the past two decades and is now recognized as a potential and serious threat to global public health (Tacconelli et al., 2018). AMRs can be disseminated rapidly through various pathways, including foodborne pathogens, insects, wastewater, pet, food-production, or wild animals (Yang Q.E. et al., 2019). There were more than 25 instances of human infectious disease outbreak reported during the period between 1990 and 2000, 11 of which were related to animals in farms, petting zoos, and zoos (Bender and Shulman, 2004). A human may be infected through direct or indirect contact with wild animals during interactive activities in zoos, a situation that is lesser discussed in relation to public health, and there has to date, been little research on the role of wild animals in zoos and the epidemiology of multidrug resistance (MDR). Some previous studies have shown that bacterial isolates from wild animals that live in close proximity to humans have stronger drug resistance compared to wild animals living in remote areas (Rolland et al., 1985; Cole et al., 2005; Skurnik et al., 2006; Kozak et al., 2009; Allen et al., 2011). Captive wild animals in zoos have more close contact with humans and, therefore, are potential natural reservoirs for AMRs and antibiotic-resistant bacteria.
Bacteria can meet the evolutionary challenge of combating antimicrobial chemotherapy by acquiring preexisting resistance determinants from the bacterial gene pool. This is achieved through the concerted activities of mobile genetic elements that can move within or between DNA molecules, which include insertion sequences, transposons, and gene cassettes/integrons, and those that are able to transfer between bacterial cells, such as plasmids and integrative conjugative elements. MDR bacteria from captive wild animals can carry various mobile genetic elements, for example, a study on a Czech zoo found that IncI1 harbored blaCTX–M–1 and qnrS1 in E. coli (Dobiasova et al., 2013), integron (class I and II). Another study found plasmid carrying blaCMY–26,qnr and aac(6′)-Ib-cr in Gram-negative bacterial isolates from a Japanese zoo (Ahmed et al., 2007), and MDR Salmonella Enterobacter has been recovered from captive wild animals in Ohio (Farias et al., 2015). These studies indicate that these environments facilitate the emergence and dissemination of multidrug resistant pathogenic bacteria, posing a serious public health risk due to the interactions between humans and animals in these environments.
To further understand the routes of dissemination of AMRs harboring bacteria in a zoo, this study collected fresh animal feces samples as part of the routine monitoring of bacterial diseases in Zhengzhou zoo, Henan province, China. The Enterobacteriaceae in these samples were isolated, and we discovered an MDR Klebsiella pneumoniae isolate from a Red Kangaroo had severe drug resistance, including second-generation cephalosporins (Cefuroxime Sodium), third-generation cephalosporins (Ceftriaxone) and even, fourth-generation cephalosporins (Cefepime). A whole genome sequencing (WGS) analysis was then conducted to evaluate the relationship between the plasmid, drug-resistant gene related elements, and human clinical isolates.
Materials and Methods
Bacterial Isolates
Enterobacteriaceae was isolated from fresh fecal samples of animals at Zhengzhou Zoo, Henan province, China. Briefly, fecal samples were collocated into an Eppendorf tube with 500 μL sterile saline, which was gently shaken, and allowed to stand for 10 min. The supernatant was used to inoculate onto the MacConkey Agar (Beijing SanYao Science & Technology Development Co, Beijing, China) plate at 35°C for 18 h. The colonies with different morphologies and colors were then stored and subjected to further analysis. Species identification was carried out using a 16S rRNA sequence (TIANYI HUIYUAN, China).
Drug Susceptibility Testing
Drug susceptibility testing was performed by the broth microdilution method (microbial susceptibility kit, BIO-KONT, China), according to the CLSI guidelines (CLSI, 2019). Fifteen antimicrobial drugs were used to screen MDR, including Ampicillin, Ampicillin/Sulbactam, Piperacillin/Tazobactam, Aztreonam, Cefuroxime sodium, Ceftriaxone, Cefepime, Ciprofloxacin, Levofloxacin, Meropenem, Colistin, Chloramphenicol, Trimethoprim/Sulfamethoxazole, Nitrofurantoin, and Amikacin. We tested drug susceptibility to Tetracycline and Doxycycline by the Kirby-Bauer disk diffusion method (OXOID, United Kingdom), and E. coli ATCC25922 was used as a quality-control strain.
Whole-Genome Sequencing and Bioinformatics Analysis
Based on drug susceptibility testing, an MDR Klebsiella pneumoniae isolate (named M297-1) was identified in a Red Kangaroo. This multidrug resistant (MDR) bacteria was subjected to WGS using the Oxford Nanopore Technologies (ONT) MinION platform (Biomarker Technologies, China) (Ashton et al., 2015; Loman et al., 2015). Sequencing was then carried out according to the standard protocol provided by ONT, and high-quality genomic DNA was extracted by NanoDrop, Qubit, and 0.35% agarose gel electrophoresis for purity, concentration, and integrity. Large fragments of DNA were recovered by the BluePippin automatic nucleic acid recovery system. The library was constructed by ligation sequencing kit (SQK-LSK109 Ligation Sequencing Kit, Oxford Nanopore Technologies, United Kingdom), and DNA damage repair and terminal repair magnetic bead purification were used to connect and re-purify, and the Qubit library was quantified and sequenced on the machine.
A phylogenetic tree was constructed based on 16s rRNAs and Klebsiella pneumoniae isolate multilocus sequence typing (MLST) was conducted by using MLST 1.8 or PubMLST1. Plasmid replicon typing, plasmid multilocus sequence typing, and identification of resistance genes were performed using Plasmid Finder2 or pMLST 2.0, and Resfinder 2.0 or CARD, respectively. The comparison of the similarity between plasmids and known plasmids was conducted using PLSDB databases3. The plasmid sequence was annotated with DFAST4, and the genomic structure was compared in EasyFig. The comparative map of the plasmid genome was drawn by the Illustrator for Biological Sequences (IBS) software v1.0. (Liu et al., 2015) and modified manually. Transposons and insertion sequences were determined using ISfinder.
Conjugation Experiments and the Evaluation of Plasmid Stability
To investigate the transferability of the plasmid in Klebsiella pneumoniae M297-l isolate, we performed conjugation assays with sodium-azide resistance E. coli J53 as a recipient strain. Briefly, overnight cultures of MDR Klebsiella pneumoniae M297-l as a donor and the recipient E. coli J53 strain were 1:10 mix and conducted on nitrocellulose membranes on a MacConkey Agar plate by incubation at 35°C for 16–20 h. After incubation, we subsequently diluted the 10-fold serial, mixed the culture in sterile saline, and aliquoted 100 μL of diluted culture onto MacConkey Agar plates supplemented with 20 mg/L of cefotaxime and 200 mg/L of sodium azide. E. coli transconjugants were screened by drug susceptibility testing. To further evaluate the stability of the plasmid of E. coli transconjugants, E. coli transconjugants were passaged continuously in MHB without antibiotics and detected in a McConkey Agar plate containing 20 mg/L cefotaxime according to previous reports (Walsh et al., 2011; Di Luca et al., 2017; Wein et al., 2019).
Similarly, we selected the conjugate strains based on the drug sensitivity test and used the Illumina sequencing platform (BGI, China) for high-throughput sequencing of the whole genome of the transconjugants plasmid.
Accession Numbers
The sequence data and details of the sequenced samples, including the date and location of the collection and source, were submitted to the GenBank. Accession numbers for Chr-M297-1, plasmid pM297-1.1, and pM297-1.2 from Klebsiella pneumoniae M297-1, respectively. Klebsiella pneumoniae M297-1 assembly contigs were deposited under study accession number CP051490, CP051491, and CP051492.
Results
MDR Klebsiella pneumoniae M297-1 Isolates From Red Kangaroo
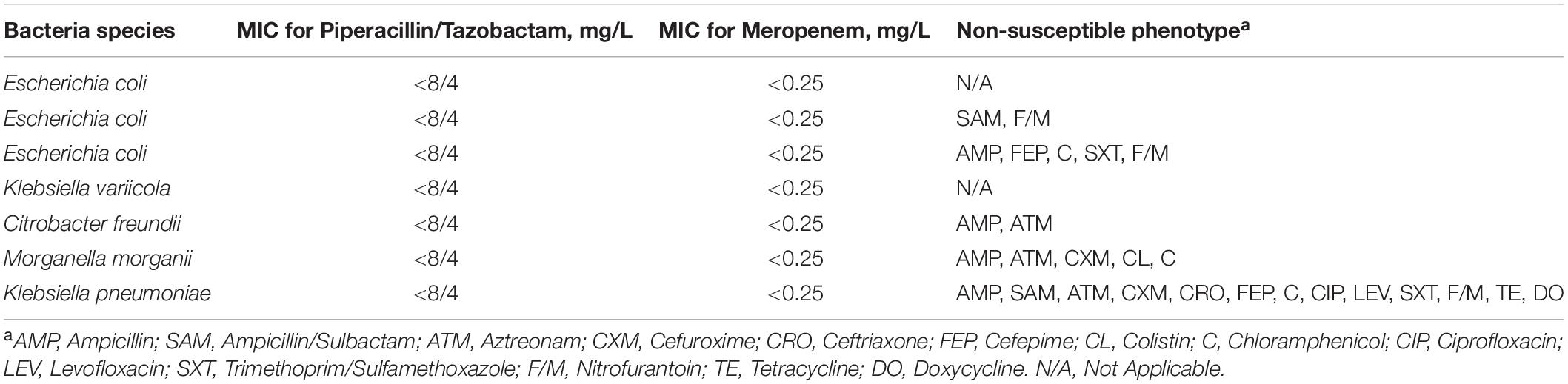
Overall, 24 isolates were isolated from animal fecal samples in Zhengzhou Zoo. These included 33 isolates of Escherichia coli and 1 isolate of Klebsiella pneumoniae. The susceptibility profiles indicated that most of the isolates were susceptible to some of the common antimicrobial drugs in clinical use. The antimicrobial resistance of the isolates from the Red Kangaroo (n = 4) was serious (Table 1) as compared with other samples the isolate of Klebsiella pneumoniae (named as M297-1) had high resistance to major groups of antimicrobial drugs including group A (Ampicillin), group B (Cefuroxime, Cefepime, Ceftriaxone, Ciprofloxacin, Levofloxacin, Trimethoprim/Sulfamethoxazole, Ampicillin/Sulbactam), group C (Aztreonam, Chloramphenicol), and group U (Nitrofurantoin).
Genomic Structure of the Chr-M297-1 Harboring blaDHA–3, blaSHV–1, and blaCTX–M–14
The isolate was subjected to WGS analysis. The chromosomal DNA of M297-1 (named as Chr-M297-1) is 5,750,384 bp in size and exhibits 100% identity with a query coverage of 100% to the K. pneumoniae by Ribosomal Multilocus Sequence Typing (rMLST). Isolate M297-1 belongs to ST 290)5, has the highest homology with Klebsiella pneumoniae isolate (99.93%) from human clinical samples based on phylogenetic tree of 16s rRNA (Supplementary Figure S1). For ST290, only five isolates were recorded in the database, of which one strain was from a dairy cow and the other four strains were from human clinical samples. Chr-M297-1 possesses 9 gene islands and 1 prophage, 169 antimicrobial genes most importantly carrying 3 β-lactamase genes (blaDHA–3, blaSHV–1, and blaCTX–M–14), fosA5, dfrA3, sul3, etc. (Table 2). In addition, the resistance pump was mainly composed of ABC family genes, as well as MFS, SMR, and MATE family genes.
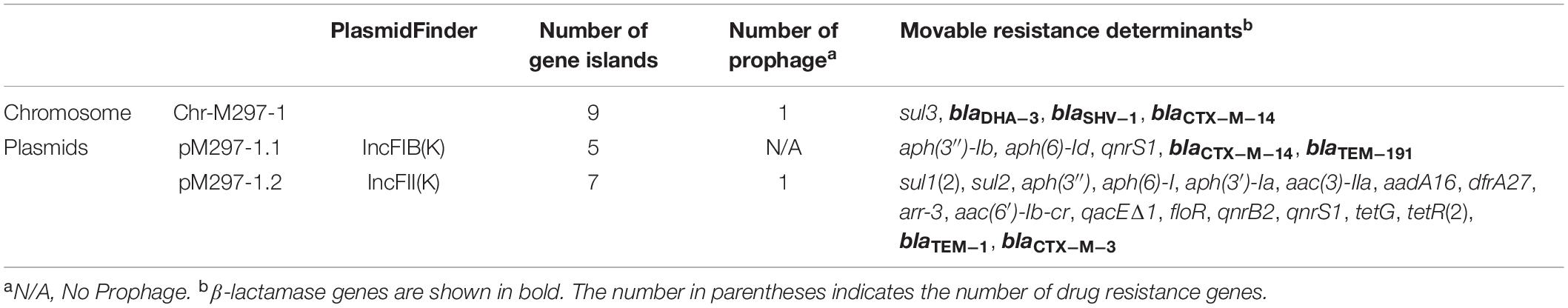
Table 2. Genomic information of Klebsiella pneumoniae isolate M297-1 from Red Kangaroo in Zhengzhou zoo, Henan province.
Genomic Structure of Plasmid Carrying Important Antimicrobial Genes
K. pneumoniae isolate M297-1 contains two plasmids pM297-1.1 (222,864 bp) and pM297-1.2 (225,763 bp), which belong to IncFIB(K) and IncFII(K), respectively. pM297-1.1 contains five Genomic_islands, two β-lactamase genes (blaCTX–M–14 and blaTEM–191), two aminoglycoside resistant genes [aph(3″)-Ib and aph(6)-Id], quinolone resistance gene qnrS1 and others. According to PLSDB database, only three plasmids were similar to pM297-1.1 focusing on the multidrug resistant region, which is tnpA-other-blaCTX–M–14-tnpA/Tn903-blaTEM–191—tnpA/IS2-other-qnrS1-tnpR/Tn552-ISKpn19-tnpR/Tn4653 (Figure 1). More specifically, there were conjugal transfer protein genes (traABCDEGHIKLMNPQTUVX-trbBCEFI-finO), Lactose permease, and two aminoglycoside resistant genes [transposase-aph(3″)-Ib and aph(6)-Id] co-located in the same larger Genomic_island 11 (42,044–101,359 bp).
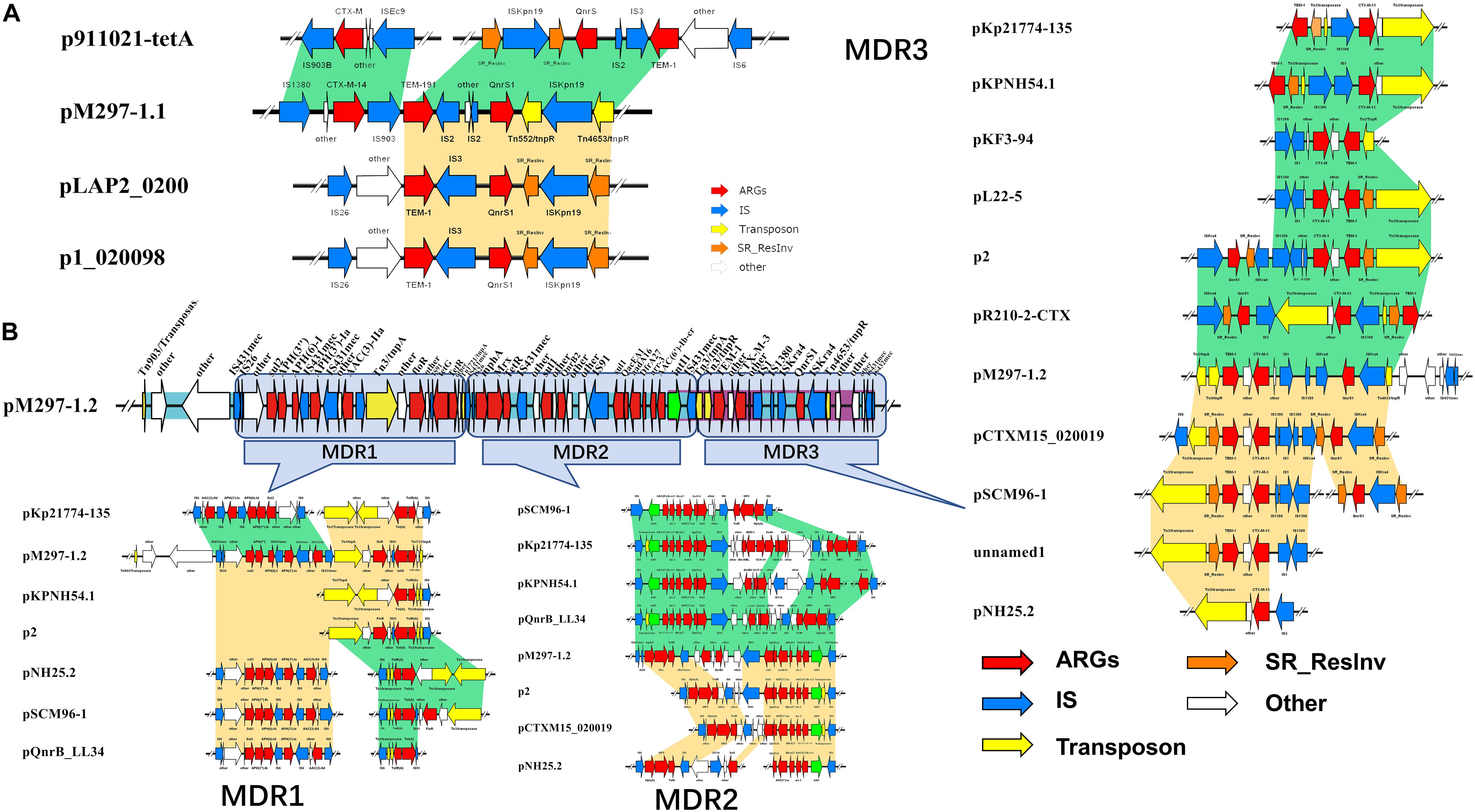
Figure 1. Antimicrobial genes, insertion sequences, and composite transposons in plasmids from MDR K. pneumoniae isolate M297-1. (A) pM297-1.1 and its similar linear property particle characteristics and comparison; (B) pM297-1.2 and its similar linear property particle characteristics and comparison. Dark green and golden shadows represent shared areas with high similarity (>90%), showing the relative positions of genes identified in the complete nucleotide sequences of these bacteria. These genes are marked with arrows, which represent the coding sequence and the associated transcription direction, and the size of the arrow is proportional to the length of the gene. The red shadow represents the antibiotic resistance gene, the blue shadow indicates the insertion sequence, the yellow shadow indicates the transposase gene, the green shadow indicates the integrase gene, and the orange shadow indicates the serine recombinase family protein gene. the blank indicates that it has nothing to do with drug resistance or unknown functional protein genes. Turquoise shadow, gene island; purple shadow, prophage.
pM297-1.2 contains 7 Genomic_islands and 1 prophage. Comparing to pM297-1.1, pM297-1.2 harbors more antimicrobial genes, including 2 extended-spectrum β-lactamase (ESBLs) genes (blaCTX–M–3 and blaTEM–1), 5 aminoglycoside resistance genes [aph(6)-I,aph(3′)-Ia, aph(3)-IIa,aadA16, aph(6′)-Ib-cr], fluoroquinolones resistant genes (qnrB2 and qnrS1), mphA, sul1, qacEΔ1 multidrug exporter, etc. Notably, pM297-1.2 carries chloramphenicol resistant genes (floR), tetracycline resistant genes (tetG), sulfonamide resistance gene (sul1, sul2) (Table 2), but pM297-1.1 does not contain these genes.
The genomic region of pM297-1.1 and pM297-1.2 were divided into different Multidrug Resistance regions according to the composition of antimicrobial genes and related antimicrobial genes. The pM297-1.1 possessed one multidrug resistance gene region (tnpA-other-blaCTX–M–14-insC/Tn903-blaTEM–191-insD-other-insC21-qnrS1-tnpR/Tn552-other-tnpR/Tn4653), which was entirely conserved in p911021-tetA, p1_020098, pLAP2_020009, NZ_CP040176.1 (similarity, >99.86%). This genomic region was flanked by insC/Tn903 and insC21 sequences and co-harbored other genes, encoding ESBLs (blaCTX–M–14 and blaTEM–191, Figure 1A, pM297-1.1), and one gene conferring resistance to fluoroquinolones (qnrS1, Figure 1A, pM297-1.1).
The multidrug resistance region of pM297-1.2 appeared to consist of three parts, which are named MDR1, MDR2, and MDR3. MDR1 was bracketed by derivatives of Tn903, Tn3, and Tn1721(Tn21 subfamily) and harbored resistance genes against aminoglycoside, sulfonamide, and chloramphenicol. MDR2 carried one class I integron (intl1) and co-harbored other genes that have resistance to aminoglycoside, sulfonamide, rifampicin, and fluoroquinolones. Notably, MDR3 was bracketed by derivatives of Tn3 harboring two β-lactamases genes blaCTX–M–3 and blaTEM–1 and Tn4653 gene carrying one gene resistance to fluoroquinolones. Some plasmid, including pKp21774-135, pKPNH54.1, pKF3-94, pL22-5, p2, pR210-2-CTX, pCTXM15_020019, pSCM96-1, pNH25.2, were found to possess the MDR3 by a database search. These two plasmids evolved from other plasmids by inserting, deleting, or replacing.
Traceability Analysis of These Two Plasmids
In the PLSDB database, five plasmids from Klebsiella pneumoniae in human clinical samples had high similarity to plasmid pM297-1.1. pM297-1.2 had high similarity to thirteen plasmids, of which 9 were derived from Klebsiella pneumoniae, and 1 from Klebsiella variicola, Klebsiella quasipneumoniae, Klebsiella sp., and Escherichia coli in human clinical samples from different regions (Supplementary Table S1).
Conjugative Transfer and Stability of Plasmids Harboring MDR Regions
Transconjugants were obtained from mating experiments with Klebsiella pneumoniae M297-1 donors, in E. coli J53 recipients, at rates of 10–5 to 10–6 transconjugants per recipient cell. All of the 13 putative transconjugants that were tested were found to be the recipient background species and to be β-lactamase positive by antimicrobial susceptibility testing. Notably, 8 of the 13 transconjugants were resistant to sulfamethoxazole, chloramphenicol, and tetracycline, which was suggested to be transconjugants carrying with pM297-1.2 (Table 3). Another 4 strains were not resistant to the sulfamethoxazole, chloramphenicol, and tetracycline (Table 3), which was considered to be transconjugants carried pM297-1.1. All positive transconjugant colonies tested, resistance to cefotaxime, and remained detectable throughout the 6-day passage experiment, even in the absence of antibiotic selection.

Table 3. Drug susceptibility testing of E. coli transconjugants from Klebsiella pneumoniae M297-1 as donor and E. coli J53 as a recipient strain.
We sequenced the whole genome of the J53-2 transconjugants plasmid, which is completely sensitive to Chloramphenicol, Trimethoprim/Sulfamethoxazole, Tetracycline, and Doxycycline, and also sequenced the transconjugants plasmid J53-12, which is highly resistant to Chloramphenicol, Trimethoprim/Sulfamethoxazole, Tetracycline, and Doxycycline. The results show that there was a complete genome of plasmid pM297-1.1 in J53-2 and a complete genome of pM297-1.2 in J53-12. This result confirmed that the two plasmids that were carried by strain M297-1 could be transferred to E. coli.
We confirmed that the two plasmids carried by Klebsiella pneumoniae M297-1 had strong transferability and stability in new host bacteria.
Discussion
In this study, we reported firstly that the multidrug-resistant Klebsiella pneumoniae M297-1 was carried by healthy Red Kangaroo, which suggests that an asymptomatic animal host, which is likely to be ignored, may become an important reservoir for drug-resistant pathogens, such as Salmonella entericus (Perron et al., 2008). Salmonella isolated from captive wild animals in Ibadan, western Nigeria, has been observed to be resistant to sulfadiazine and penicillin (Falade and Durojaiye, 1976). Among the 232 isolates of Gram-negative bacteria isolated from mammals, reptiles, and birds raised in an Asakusa Zoo, Hiroshima Prefecture, Japan, 21.1% of Gram-negative bacteria carry at least one drug-resistant gene and have multiple drug-resistant phenotypes (Ahmed et al., 2007). The ESBLs and fluoroquinolone resistance genes detected in Czech zoos were associated with the transmission of specific E. coli clones and plasmids of specific incompatible groups between different animal species (Dobiasova et al., 2013). S. aureus was taken from zoo and wildlife in Germany from 2008 to 2016, and two isolates from juvenile red squirrels showed multiple drug resistant phenotypes (Fessler et al., 2018). The frequency of AMRs detected in animals living in human settlements is significantly higher than that in animals living in the wild (Grall et al., 2015; Kock et al., 2018). The isolates from wild animals show a similar pattern of drug resistance to E. coli from human clinical sources in their study areas (Jobbins and Alexander, 2015). Other studies have indicated that drug-resistant Salmonella isolates identified from zoo environments, including those from animals and their keepers, have been confirmed to be clone-related (Farias et al., 2015; Milton et al., 2018). Therefore, wild animals may be important hosts and storage hosts for the spread of drug-resistant bacteria, and human activities significantly affect the microbial community of captive wild animals in zoo environments, which are not necessarily monitored for such risks.
In the present study, the ST290 sequence types K. pneumoniae M297-1 from a Red Kangaroo were closely related to that found in human clinical isolates, but only 5 cases of this sequence type have been reported worldwide, in the United States, Australia, and China (Figure 2). The plasmid of isolates carrying blaKPC–2, blaIMP–8, blaTEM–1, and blaCTX–M–15 gene transmission have been found in neonatal infections (Jin et al., 2017; Kong et al., 2020). The first case of nosocomial epidemic infection caused by NDM-5 metallo-β-lactamase ST290 isolates were reported in China (Wang et al., 2019). Therefore, our study further showed that zoo-derived MDR bacteria are closely related to human-derived MDR.
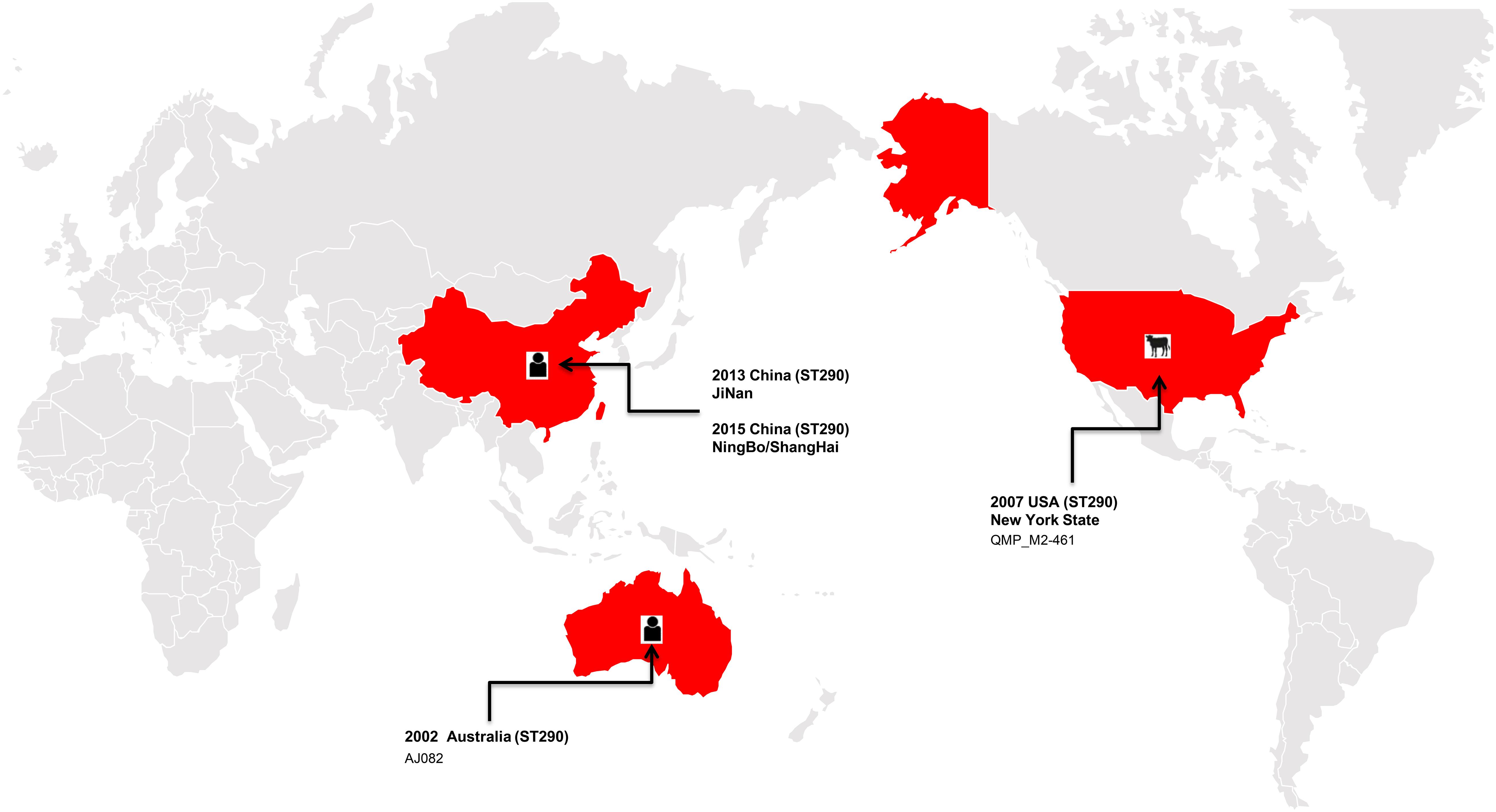
Figure 2. The distribution of the ST290 of Klebsiella pneumoniae isolates in the world, among which three isolates were found in China, one in the United States, and one in Australia.
bGWAS provided comprehensive information about isolate M297-1. The whole genome analysis of M297-1 confirmed that its plasmids carried clinically related ESBLs genes including blaTEM–1, blaTEM–191, blaCTX–M–3, and blaCTX–M–14 (Table 2 and Figure 1). Since the late 1990s, the multidrug-resistant Enterobacteriaceae that produces ESBLs has become an important cause of urinary tracts and bloodstream infections in humans (Pitout and Laupland, 2008; Wyres et al., 2020). This is a rapidly evolving class of β-lactamases, usually from blaTEM–1, blaTEM–2, or blaSHV–1 genes, which can hydrolyze third-generation cephalosporins and aztreonam and can be inhibited by clavulanic acid (Paterson and Bonomo, 2005). Although new members of the ESBLs family are often found, the earlier blaCTX–M–14 and blaCTX–M–15 enzymes are prevalent in the world at large (Bush and Fisher, 2011; Bush and Bradford, 2020), while blaCTX–M–3 enzymes are prevalent mainly in Europe (Canton et al., 2008). Resistance to extended-spectrum cephalosporins such as ceftazidime, cefotaxime, and cefepime were often observed when ESBLs appeared in Klebsiella sp. (Babic et al., 2006). ESBLs were also detected on the M297-1 chromosome, but it also stably carried the ESBLs positive plasmids pM297-1.1 and pM297-1.2, indicating that drug resistance itself is not the only selection criterion for maintaining ESBLs cod plasmids. Other drug resistant genes, such as those of sulfonamides, fluoroquinolones, aminoglycosides, and tetracyclines and found on chromosomes and plasmids, suggest that the findings of this study may have evolved under the pressure of many antibiotics.
Mobile genetic elements, such as insertion sequence, transposon, integron, and prophage, can mobilize antibiotic resistance genes. The detection of blaTEM and blaCTX–M type β-lactamases genes in a variety of genetic backgrounds suggests that their mobilization may involve multiple mechanisms. In this study, for plasmid pM297-1.1, IS1380 is located upstream of blaCTX–M–14, and downstream we detected IS903, which has also located upstream of blaTEM–191 and blaTEM–191. This may also be related to its downstream truncated IS2 gene, and similar structures also exist in similar plasmids retrieved (p911021-tetA, MG288679.1; p1_020098, NZ_CP036307.1; pLAP2_020009, CP038004.1) (Figure 1). This seems to imply that the β-lactamase gene carried by pM297-1.1 may form a tandem structure of the drug resistance gene after multiple homologous recombinations. For the MDR 3 region of plasmid pM297-1.2, the resistance gene box of β-lactamase genes in Tn3-like transposon was tnpA-tnpR-blaTEM–1-other-blaCTX–M–3, and also located on the prophage. We found that a similar plasmid (pKF3-94, pL22-5, p2, pCTXM15_ 020019, pSCM96-1) also had a complete or truncated similar structure (Figure 1). There are usually two IS431mec insertion sequence genes from Staphylococcus aureus upstream and downstream of aminoglycosides, fluoroquinolones, sulfonamides, and tetracyclines in the pM297-1.2 MDR region. The structure of the intI1-gene cassette carried by the plasmid pM297-1.2 supports the concept of mobile elements to transfer antimicrobial genes between different bacteria. The drug resistance gene cassette [aac(6′)-1b-cr-arr-3-dfrA27-aadA16] is derived from the plasmid (ACC_NUCCORE : EU675686) carried by the multi-drug resistance E. coil isolated from the urine of patients in Huashan Hospital (Wei et al., 2009). This structure is completely preserved in pM297-1.2 from Klebsiella pneumoniae M297-1 (Figure 1) and is located on the Genomic Island. The drug resistance gene cassette mainly encodes β-lactamases, acetyltransferases, and nucleoside transferases, which do not require significant cell interaction and can be integrated into the metabolic network. Therefore, the interference from the existing genome is minimal, and it is the best example of a single gene corresponding to a single phenotype (Ghaly et al., 2020). In any case, this genetic factor will help bacteria evolve multiple determinants of antibiotic resistance in different habitats (Edge and Hill, 2005).
Asia is one of the centers of antibiotic resistance, and there are a number of drug-resistant strains including K. pneumoniae, and a large number of acquired gram-negative MDR strains have been found (Jean and Hsueh, 2011). The prevalent STs in Asia include ST15, ST23, ST14, and ST231. Among them, ST15 is very common, ST23 is significantly related to Southeast Asia, while ST14 and ST231 are significantly related to South Asia (Wyres et al., 2020). In this study, most of the plasmids retrieved from the PLSDB database were carried by K. pneumoniae. Among the K. pneumoniae isolates carrying highly similar plasmids to pM297-1.2, only two isolates of ST15 were isolated from Thailand and China, and one strain of ST23 and one strain of ST14 from China. The isolates carrying highly similar plasmids to pM297-1.1 did not detect the above four sequence types (Supplementary Table S1). As a result, the two plasmids in our study belong to IncF, a narrow host spectrum plasmid widely in Enterobacteriaceae, and can carry a variety of AMR genes and play a major role in the spread of specific antimicrobial genes (Carattoli, 2011; Rozwandowicz et al., 2018). Some studies showed that IncFIB and IncFII plasmids are effective vectors of β-lactamases. These plasmids can promote the transfer of antimicrobial genes when exposed to antibiotics (Rooney et al., 2019). blaNDM, blaOXA, and other genes spread in the Enterobacteriaceae flora of medical facilities by relying on these plasmids (Simner et al., 2018; Wu et al., 2019; Strydom et al., 2020), while blaCTX–M can become the dominant gene carried by IncFII plasmids in France and China (Du et al., 2012; Dahmen et al., 2013). It seems that blaCMY–42 is replacing blaCTX–M–15 in some areas (Paul et al., 2020). In particular, some IncFIB cannot only express a high level of drug resistance but also enhance the virulence of K. pneumoniae after conjugation with K. pneumoniae (Yang X. et al., 2019). The results of this suggest that the co-prevalence of plasmid IncFIB and IncFII in the same isolates may lead to serious public health problems.
Similar plasmids were found by Sweden, Thailand, and China, respectively. Except for one isolate in Sweden and two isolates in Thailand, the other isolates carrying similar plasmids are mainly distributed among the southern cities of Sichuan province, China (Figure 3 and Supplementary Table S1). It has been suggested that during the period from 2010 to 2019, the epidemic distribution of this drug-resistant plasmid was gradually spreading in China, and the difference in carrying drug-resistant genes indicates that it is evolving. However, the phylogenetic analysis of the related isolates showed that M297-1 is located on a different branch with other K. pneumoniae isolates carrying similar plasmids (Figure 4). Since strain M297-1 comes from red kangaroo samples, while other strains carrying similar plasmids come from human clinical samples, it is speculated that the reason why M297-1 and other K. pneumoniae carrying similar plasmids are in different branches may be closely related to their hosts.
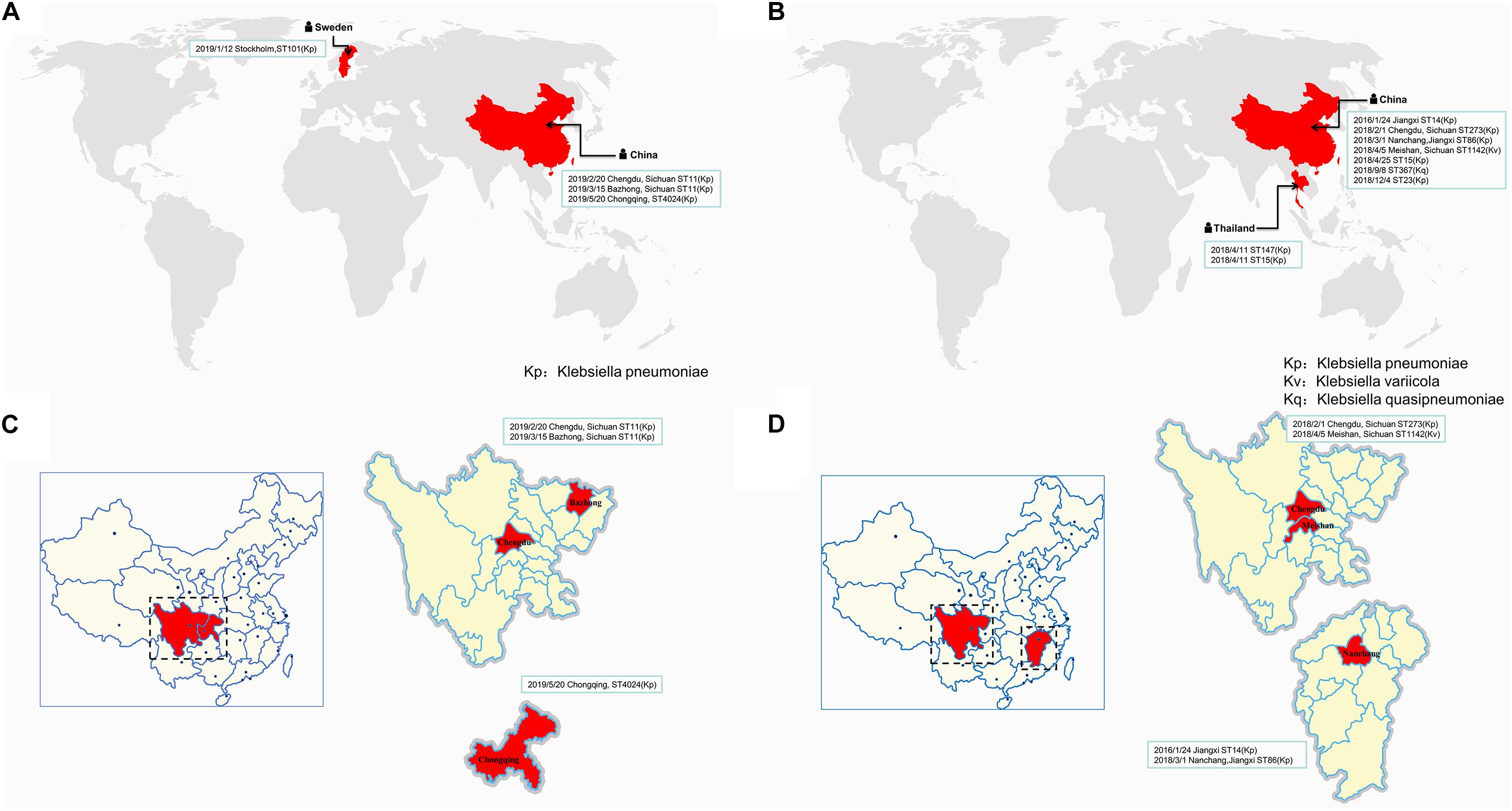
Figure 3. The distribution of different isolates carrying similar plasmids in the world. The distribution of different isolates carrying similar plasmids that are highly similar to plasmids pM297-1.1 (A) and pM297-1.2 (B), while (C,D) indicate the distribution in China. The reporting date, region, and ST typing of the isolates are shown in the box.

Figure 4. Phylogenetic relationship between reference K. pneumoniae isolates carrying similar plasmids and M297-1 based on 16s rRNA sequences. The circle indicates the strain carrying highly similar plasmids to pM297-1.1, the triangle indicates the strain carrying highly similar plasmids to pM297-1.2, and the red arrow points to M297-l. Orange, China; light green, Sweden; dark blue, Thailand; gray, missing information.
Conclusion
In conclusion, this study demonstrated the high resistance of 13 drugs including Ceftriaxone and Cefepime on MDR K. pneumoniae isolate, taken from healthy Red Kangaroos in Zhengzhou zoo, China. This is the first report on a K. pneumoniae M297-1 (ST290) from a wild animal carrying blaDHA–3, blaSHV–1, blaCTX–M–14, blaTEM–191, blaTEM–1, and blaCTX–M–3 in China and two conjugal transferable plasmids that co-harbor other antimicrobial genes: aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, aac(3)-IIa, aac(6′)-Ib-cr, aadA16, qnrB2, qnrS1, qacEΔ1, mphA, sul1, and dfrA27. This research confirmed that there is a close relationship between drug-resistant strains carried by wild animals in zoos and human clinical isolates. This suggests that zoos may be becoming important reservoirs for clinically important MDR isolates, which pose a serious potential public health risk. These potential reservoirs should not be ignored and monitoring these environments is of vital importance in preventing major threats to public health in the future.
Data Availability Statement
The sequence data and details of the sequenced samples, including the date and location of collection and source, were submitted to the GenBank. Accession numbers for Chr-M297-1, plasmid pM297-1.1, and pM297-1.2 from Klebsiella pneumonia M297-1, respectively. Bioproject IDs PRJNA624988 and PRJNA624988, respectively. Klebsiella pneumoniae M297-1 assembly contigs are deposited under study accession number CP051490, CP051491, and CP051492, BioProject ID PRJNA624988.
Ethics Statement
The animal study was reviewed and approved by the Animal Experimental Ethics Committee of Institute of Zoology (No. GIABR20191104).
Author Contributions
CW and JQ designed the study. XW, QK, JZ, ZL, and FJ isolated and identified isolates. JL, JY, CZ, TJ, GD, and SL collected samples and analyzed the data. XW, CW, and JQ wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Introduction of Leading Talents Program of Guangdong Academy of Sciences (No. 2016GDASRC-0205), and was a GDAS Special Project of Science and Technology Development (No. 2018GDASCX-0107), and received support from the Earmarked Fund for the Hebei Dairy Cattle Innovation Team of Modern Agro-industry Technology Research System (No. HBCT2018120205).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.560474/full#supplementary-material
Supplementary Figure 1 | Phylogenetic relationship between Klebsiella pneumoniae isolate M297-1 and the reference strain from human clinical samples based on the 16s rRNA.
Supplementary Table 1 | Comparison between reference plasmid and pM297-1.1 and pM297-1.2 from Klebsiella pneumoniae M297-1 from Red Kangaroo in Zhengzhou zoo, Henan province.
Footnotes
- ^ http://pubmlst.org/
- ^ https://cge.cbs.dtu.dk/services/PlasmidFinder/
- ^ https://ccb-microbe.cs.uni-saarland.de/plsdb/
- ^ https://dfast.nig.ac.jp/
- ^ https://pubmlst.org/rmlst/
References
Ahmed, A. M., Motoi, Y., Sato, M., Maruyama, A., Watanabe, H., Fukumoto, Y., et al. (2007). Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl. Environ. Microbiol. 73, 6686–6690. doi: 10.1128/aem.01054-07
Allen, S. E., Boerlin, P., Janecko, N., Lumsden, J. S., Barker, I. K., Pearl, D. L., et al. (2011). Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in southern Ontario. Canada. Appl. Environ. Microbiol. 77, 882–888. doi: 10.1128/aem.01111-10
Ashton, P. M., Nair, S., Dallman, T., Rubino, S., Rabsch, W., Mwaigwisya, S., et al. (2015). MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 33, 296–300. doi: 10.1038/nbt.3103
Babic, M., Hujer, A. M., and Bonomo, R. A. (2006). What’s new in antibiotic resistance? Focus on beta-lactamases. Drug Resist. Updat. 9, 142–156. doi: 10.1016/j.drup.2006.05.005
Bender, J. B., and Shulman, S. A. (2004). Reports of zoonotic disease outbreaks associated with animal exhibits and availability of recommendations for preventing zoonotic disease transmission from animals to people in such settings. J. Am. Vet. Med. Assoc. 224, 1105–1109. doi: 10.2460/javma.2004.224.1105
Bush, K., and Bradford, P. A. (2020). Epidemiology of beta-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 33, 2–26. doi: 10.1128/cmr.00047-19
Bush, K., and Fisher, J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65, 455–478. doi: 10.1146/annurev-micro-090110-102911
Canton, R., Novais, A., Valverde, A., Machado, E., Peixe, L., Baquero, F., et al. (2008). Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14(Suppl. 1), 144–153. doi: 10.1111/j.1469-0691.2007.01850.x
Carattoli, A. (2011). Plasmids in Gram negatives: molecular typing of resistance plasmids. Int. J. Med. Microbiol. 301, 654–658. doi: 10.1016/j.ijmm.2011.09.003
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing, 29th Edn. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Cole, D., Drum, D. J., Stalknecht, D. E., White, D. G., Lee, M. D., Ayers, S., et al. (2005). Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 11, 935–938. doi: 10.3201/eid1106.040717
Dahmen, S., Haenni, M., Chatre, P., and Madec, J. Y. (2013). Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J. Antimicrob. Chemother. 68, 2797–2801. doi: 10.1093/jac/dkt291
Di Luca, M. C., Sorum, V., Starikova, I., Kloos, J., Hulter, N., Naseer, U., et al. (2017). Low biological cost of carbapenemase-encoding plasmids following transfer from Klebsiella pneumoniae to Escherichia coli. J. Antimicrob. Chemother. 72, 85–89. doi: 10.1093/jac/dkw350
Dobiasova, H., Dolejska, M., Jamborova, I., Brhelova, E., Blazkova, L., Papousek, I., et al. (2013). Extended spectrum beta-lactamase and fluoroquinolone resistance genes and plasmids among Escherichia coli isolates from zoo animals, Czech Republic. FEMS Microbiol. Ecol. 85, 604–611. doi: 10.1111/1574-6941.12149
Du, X. D., Li, D. X., Hu, G. Z., Wang, Y., Shang, Y. H., Wu, C. M., et al. (2012). Tn1548-associated armA is co-located with qnrB2, aac(6’)-Ib-cr and blaCTX-M-3 on an IncFII plasmid in a Salmonella enterica subsp. enterica serovar Paratyphi B strain isolated from chickens in China. J. Antimicrob. Chemother. 67, 246–248. doi: 10.1093/jac/dkr407
Edge, T. A., and Hill, S. (2005). Occurrence of antibiotic resistance in Escherichia coli from surface waters and fecal pollution sources near Hamilton. Ontario. Can. J. Microbiol. 51, 501–505. doi: 10.1139/w05-028
Falade, S., and Durojaiye, O. A. (1976). Salmonellae isolated from captive animals in Ibadan, Western State of Nigeria. J. Wildl. Dis. 12, 464–467. doi: 10.7589/0090-3558-12.3.464
Farias, L. F., Oliveira, C. J., Medardus, J. J., Molla, B. Z., Wolfe, B. A., and Gebreyes, W. A. (2015). Phenotypic and Genotypic Characterization of Salmonella enterica in Captive Wildlife and Exotic Animal Species in Ohio, USA. Zoonoses. Publ. Health 62, 438–444. doi: 10.1111/zph.12170
Fessler, A. T., Thomas, P., Muhldorfer, K., Grobbel, M., Brombach, J., Eichhorn, I., et al. (2018). Phenotypic and genotypic characteristics of Staphylococcus aureus isolates from zoo and wild animals. Vet. Microbiol. 218, 98–103. doi: 10.1016/j.vetmic.2018.03.020
Ghaly, T. M., Geoghegan, J. L., Tetu, S. G., and Gillings, M. R. (2020). The Peril and Promise of Integrons: Beyond Antibiotic Resistance. Trends Microbiol. 28, 455–464. doi: 10.1016/j.tim.2019.12.002
Grall, N., Barraud, O., Wieder, I., Hua, A., Perrier, M., Babosan, A., et al. (2015). Lack of dissemination of acquired resistance to beta-lactams in small wild mammals around an isolated village in the Amazonian forest. Environ. Microbiol. Rep. 7, 698–708. doi: 10.1111/1758-2229.12289
Jean, S. S., and Hsueh, P. R. (2011). High burden of antimicrobial resistance in Asia. Int. J. Antimicrob. Agents 37, 291–295. doi: 10.1016/j.ijantimicag.2011.01.009
Jin, Y., Song, X., Liu, Y., Wang, Y., Zhang, B., Fan, H., et al. (2017). Characteristics of carbapenemase-producing Klebsiella pneumoniae as a cause of neonatal infection in Shandong, China. Exp. Ther. Med. 13, 1117–1126. doi: 10.3892/etm.2017.4070
Jobbins, S. E., and Alexander, K. A. (2015). From Whence They Came–Antibiotic-Resistant Escherichia Coli In African Wildlife. J. Wildl. Dis. 51, 811–820. doi: 10.7589/2014-11-257
Kock, R., Daniels-Haardt, I., Becker, K., Mellmann, A., Friedrich, A. W., Mevius, D., et al. (2018). Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin. Microbiol. Infect. 24, 1241–1250. doi: 10.1016/j.cmi.2018.04.004
Kong, N., Li, G., Yang, C., Wang, X., Cao, M., Zhang, L., et al. (2020). Prevalence of blaKPC-2-harbouring Klebsiella pneumoniae ST290 in a tertiary hospital in China. J. Glob. Antimicrob. Resist. 20, 344–345. doi: 10.1016/j.jgar.2020.01.025
Kozak, G. K., Boerlin, P., Janecko, N., Reid-Smith, R. J., and Jardine, C. (2009). Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario. Canada. Appl. Environ. Microbiol. 75, 559–566. doi: 10.1128/aem.01821-08
Liu, W., Xie, Y., Ma, J., Luo, X., Nie, P., Zuo, Z., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Loman, N. J., Quick, J., and Simpson, J. T. (2015). A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12, 733–735. doi: 10.1038/nmeth.3444
Milton, A. A. P., Agarwal, R. K., Priya, G. B., Athira, C. K., Saminathan, M., Reddy, A., et al. (2018). Occurrence, antimicrobial susceptibility patterns and genotypic relatedness of Salmonella spp. isolates from captive wildlife, their caretakers, feed and water in India. Epidemiol. Infect. 146, 1543–1549. doi: 10.1017/s0950268818001553
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/cmr.18.4.657-686.2005
Paul, D., Babenko, D., and Toleman, M. A. (2020). Human carriage of cefotaxime-resistant Escherichia coli in North-East India: an analysis of STs and associated resistance mechanisms. J. Antimicrob. Chemother. 75, 72–76. doi: 10.1093/jac/dkz416
Perron, G. G., Quessy, S., and Bell, G. (2008). A reservoir of drug-resistant pathogenic bacteria in asymptomatic hosts. PLoS One 3:e3749. doi: 10.1371/journal.pone.0003749
Pitout, J. D., and Laupland, K. B. (2008). Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166. doi: 10.1016/s1473-3099(08)70041-0
Rolland, R. M., Hausfater, G., Marshall, B., and Levy, S. B. (1985). Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl. Environ. Microbiol. 49, 791–794.
Rooney, C. M., Sheppard, A. E., Clark, E., Davies, K., Hubbard, A. T. M., Sebra, R., et al. (2019). Dissemination of multiple carbapenem resistance genes in an in vitro gut model simulating the human colon. J. Antimicrob. Chemother. 74, 1876–1883. doi: 10.1093/jac/dkz106
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Simner, P. J., Antar, A. A. R., Hao, S., Gurtowski, J., Tamma, P. D., Rock, C., et al. (2018). Antibiotic pressure on the acquisition and loss of antibiotic resistance genes in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1796–1803. doi: 10.1093/jac/dky121
Skurnik, D., Ruimy, R., Andremont, A., Amorin, C., Rouquet, P., Picard, B., et al. (2006). Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57, 1215–1219. doi: 10.1093/jac/dkl122
Strydom, K. A., Chen, L., Kock, M. M., Stoltz, A. C., Peirano, G., Nobrega, D. B., et al. (2020). Klebsiella pneumoniae ST307 with OXA-181: threat of a high-risk clone and promiscuous plasmid in a resource-constrained healthcare setting. J. Antimicrob. Chemother. 75, 896–902. doi: 10.1093/jac/dkz550
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/s1473-3099(17)30753-3
Walsh, T. R., Weeks, J., Livermore, D. M., and Toleman, M. A. (2011). Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11, 355–362. doi: 10.1016/s1473-3099(11)70059-7
Wang, Z., Li, M., Shen, X., Wang, L., Liu, L., Hao, Z., et al. (2019). Outbreak of blaNDM-5-Harboring Klebsiella pneumoniae ST290 in a Tertiary Hospital in China. Microb. Drug Resist. 25, 1443–1448. doi: 10.1089/mdr.2019.0046
Wei, Q., Jiang, X., Yang, Z., Chen, N., Chen, X., Li, G., et al. (2009). dfrA27, a new integron-associated trimethoprim resistance gene from Escherichia coli. J. Antimicrob. Chemother. 63, 405–406. doi: 10.1093/jac/dkn474
Wein, T., Hulter, N. F., Mizrahi, I., and Dagan, T. (2019). Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat. Commun. 10:2595. doi: 10.1038/s41467-019-10600-7
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., and Zong, Z. (2019). NDM Metallo-beta-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 32, e00115–8. doi: 10.1128/cmr.00115-18
Wyres, K. L., Nguyen, T. N. T., Lam, M. M. C., Judd, L. M., van Vinh Chau, N., Dance, D. A. B., et al. (2020). Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome. Med. 12:11. doi: 10.1186/s13073-019-0706-y
Yang, Q. E., Tansawai, U., Andrey, D. O., Wang, S., Wang, Y., Sands, K., et al. (2019). Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): An increasing public health risk. Environ. Int. 122, 281–290. doi: 10.1016/j.envint.2018.11.021
Keywords: multidrug-resistance, Klebsiella pneumonia, genomic characteristic, extended-spectrum β-lactamase-producing, public health risk5c
Citation: Wang X, Kang Q, Zhao J, Liu Z, Ji F, Li J, Yang J, Zhang C, Jia T, Dong G, Liu S, Hu G, Qin J and Wang C (2020) Characteristics and Epidemiology of Extended-Spectrum β-Lactamase-Producing Multidrug-Resistant Klebsiella pneumoniae From Red Kangaroo, China. Front. Microbiol. 11:560474. doi: 10.3389/fmicb.2020.560474
Received: 09 May 2020; Accepted: 13 August 2020;
Published: 14 October 2020.
Edited by:
Charlene Renee Jackson, United States Department of Agriculture (USDA), United StatesReviewed by:
Khalid Zerouali, University of Hassan II Casablanca, MoroccoXiaogang Xu, Fudan University, China
Copyright © 2020 Wang, Kang, Zhao, Liu, Ji, Li, Yang, Zhang, Jia, Dong, Liu, Hu, Qin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhua Qin, cWpocXFxQDEyNi5jb20=; Chengmin Wang, d2FuZ2NobUBnaWFici5nZC5jbg==
†These authors have contributed equally to this work
 Xue Wang1,2†
Xue Wang1,2† Shelan Liu
Shelan Liu Chengmin Wang
Chengmin Wang