
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 15 October 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.557819
The classical bordetellae possess several partially characterized virulence mechanisms that are studied in the context of a complete extracellular life cycle in their mammalian hosts. Yet, classical bordetellae have repeatedly been reported within dendritic cells (DCs) and alveolar macrophages in clinical samples, and in vitro experiments convincingly demonstrate that the bacteria can survive intracellularly within mammalian phagocytic cells, an ability that appears to have descended from ancestral progenitor species that lived in the environment and acquired the mechanisms to resist unicellular phagocytic predators. Many pathogens, including Mycobacterium tuberculosis, Salmonella enterica, Francisella tularensis, and Legionella pneumophila, are known to parasitize and multiply inside eukaryotic host cells. This strategy provides protection, nutrients, and the ability to disseminate systemically. While some work has been dedicated at characterizing intracellular survival of Bordetella pertussis, there is limited understanding of how this strategy has evolved within the genus Bordetella and the contributions of this ability to bacterial pathogenicity, evasion of host immunity as well as within and between-host dissemination. Here, we explore the mechanisms that control the metabolic changes accompanying intracellular survival and how these have been acquired and conserved throughout the evolutionary history of the Bordetella genus and discuss the possible implications of this strategy in the persistence and reemergence of B. pertussis in recent years.
The bordetellae are Gram-negative coccobacilli of the class Betaproteobacteria that are known to cause disease in a wide range of animals including small mammals and humans. Despite widespread vaccination, Bordetella pertussis – the etiological agent of whooping cough or pertussis – still affects tens of millions and causes hundreds of thousands of deaths, mostly in children younger than age 5, every year (Yeung et al., 2017). Characterized by paroxysmal cough accompanied by an inspirational whooping sound (hence the name), pertussis can last for months and cause severe respiratory complications and even death, particularly in infants and adults with underlying health conditions. B. pertussis is closely related to Bordetella parapertussis and Bordetella bronchiseptica, and the three species are collectively known as the “classical bordetellae” (Figure 1, Table 1). Multiple lineages appear to be host-restricted, with B. pertussis and B. parapertussis infecting only humans, although another independent lineage, named B. parapertussisov causes pneumonia in sheep (Porter et al., 1994; Park et al., 2012). In contrast, B. bronchiseptica appears to be more closely related to the progenitor of the classical bordetellae and is a respiratory pathogen of diverse mammals, causing a variety of pathologies ranging from chronic and often asymptomatic infection to acute bronchopneumonia and kennel cough in dogs (Goodnow, 1980).
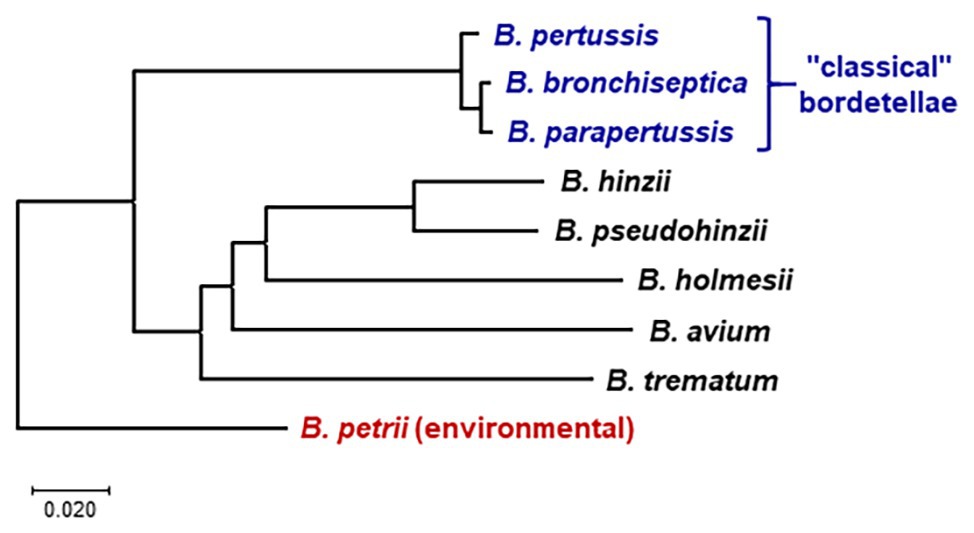
Figure 1. Whole genome phylogeny of nine Bordetella species based on pairwise average nucleotide identities (ANIs). The neighbor-joining tree was constructed from a distance matrix compiled from pairwise ANI between the genomes calculated at https://www.ezbiocloud.net/tools/ani (Yoon et al., 2017). The tree is drawn to scale in MEGA X (Kumar et al., 2018), with branch lengths in the same units as the evolutionary distances used to infer the phylogenetic tree. The tree was rooted according to the previously determined evolutionary relations between Bordetella species (Linz et al., 2016; Hamidou Soumana et al., 2017). Bordetella petrii is the only environmental among the analyzed species; all other species are human or animal pathogens.
In contrast to the closely related classical bordetellae, several Bordetella species with broader genetic diversity have been identified, collectively referred to as “non-classical” Bordetella (Figure 1, Table 1). Some members of the non-classical bordetellae are also known to be host specific animal pathogens. The emerging human pathogen Bordetella holmesii, initially isolated from the blood of septicemic patients (Weyant et al., 1995), has since increasingly been isolated from patients with pertussis-like respiratory infections (Yih et al., 1999; Rodgers et al., 2013). Bordetella avium causes respiratory infections in poultry and wild birds (Kersters et al., 1984). The other “avian” species, Bordetella hinzii, colonizes the respiratory tracts of poultry and was shown to cause disease during experimental infection in turkeys (Vandamme et al., 1995; Register and Kunkle, 2009), and has also been isolated from immunocompromised humans with respiratory disease and septicemia (Cookson et al., 1994; Funke et al., 1996). The closely related Bordetella pseudohinzii, identified as a pathobiont in mouse breeding colonies of commercial vendors (Ivanov et al., 2015, 2016), was found to cause chronic, transmissible otitis media in mice (Dewan et al., 2019). Bordetella trematum, an opportunistic human pathogen, can cause severe skin disease and chronic otitis media (Vandamme et al., 1996). The environmental species Bordetella petrii, originally isolated from an anaerobic bioreactor enriched with river sediment (von Wintzingerode et al., 2001), has subsequently been isolated from soil samples and also from immunocompromised patients (Fry et al., 2005; Nagata et al., 2015). These Bordetella species share many phenotypic characteristics that makes them successful animal pathogens.
Genome sequencing and multilocus sequence typing of the classical bordetellae revealed that B. parapertussis and B. pertussis independently evolved from a B. bronchiseptica-like ancestor (Parkhill et al., 2003; Diavatopoulos et al., 2005). Despite differences in host range and disease, the classical bordetellae are over 98% similar at the DNA sequence level and share many important virulence factors, including the well-known toxins such as adenylate cyclase toxin (ACT), pertussis toxin (PTX), and dermonecrotic toxin, and putative adhesins such as pertactin (PRN; Parkhill et al., 2003). Since these virulence factors are present in B. pertussis, B. parapertussis, and B. bronchiseptica but absent from the non-classical bordetellae, genes encoding these factors are believed to have been acquired before the divergence of the classical bordetellae (Park et al., 2012; Linz et al., 2016). Gain and loss of multiple genes, including those encoding bacterial toxins, protein secretion systems, and other virulence-associated factors, appear to have shaped the diversification and speciation in the genus. Gene loss has been more frequent than gene gain since their divergence, and loss of hundreds of genes was associated with the specialization of several host-restricted species, including the recently evolved human pathogens B. pertussis, B. parapertussis, and B. holmesii (Parkhill et al., 2003; Linz et al., 2016).
Gene loss and acquisition, via horizontal transfer, as a function of speciation and diversification is not restricted to the genus Bordetella and is well-documented in many other species and genera, including Bordetella’s sister genus Achromobacter (Li et al., 2013), other proteobacteria such as Acinetobacter (Chan et al., 2015), as well as bacteria from other phyla such as Streptococcus (Kilian and Tettelin, 2019) and Mycobacterium (Chiner-Oms et al., 2019). For example, several genome studies showed that horizontal transfer in the genus Mycobacterium has been a major player in shaping the species’ pathogenicity, leading to divergence (Ahmed et al., 2007; Saini et al., 2012; Panda et al., 2018). Thus, just like in other microbes, selective gene acquisition and loss has led to niche-specific adaptations that have shaped the metabolic versatility and pathogenicity in the genus Bordetella.
The genus Bordetella has largely been considered host-restricted pathogens with variable host-specificity. However, the recent discovery of several environmental species and 16S meta-analysis studies have revealed that the genus likely arose from an environmental origin (Hamidou Soumana et al., 2017). In addition, B. bronchiseptica was found to infect and persist within the amoeba Dictyostelium discoideum, to utilize its life cycle, translocating to the fruiting bodies and disseminating along with amoeba spores (Taylor-Mulneix et al., 2017a). These observations suggest that amoeba may represent an environmental niche for this, and possibly other animal-pathogenic Bordetella species and their progenitors. Similar to B. bronchiseptica, a wide variety of bacteria has been reported to form endosymbiotic relationships with amoebae, including bacterial genera as diverse as Amoebophilus (phylum Bacteroidetes), Mycobacterium (phylum Actinobacteria), Parachlamydia and Protochlamydia (phylum Chlamydia), Caedibacter (class Alphaproteobacteria), Procabacter (class Betaproteobacteria), and Legionella and Caedibacter (class Gammaproteobacteria; Molmeret et al., 2005; Schmitz-Esser et al., 2008; Samba-Louaka et al., 2018; Dubois et al., 2019). The interaction with amoebae as an environmental reservoir provides protection against external hazards, and possibly a competitive advantage against other bacteria, while enhancing bacterial dissemination along with the amoebic host. During interaction with amoeba, B. bronchiseptica undergoes phenotypic modulation inducing the expression of genes involved in chemotaxis, motility, and growth (e.g., flagella flhD and chemotaxis gene cheZ), while suppressing the expression of virulence factors such as the ACT (encoded by cyaA) and filamentous hemagglutinin (FHA) encoded by fhaB (Taylor-Mulneix et al., 2017a). Thus, in addition to possessing mechanisms to colonize a wide range of mammals including swine, rats, rabbits, sheep, dogs, and cats, B. bronchiseptica is able to establish a successful symbiotic relationship with amoeba in the environment outside a mammalian host.
The ability to adapt to profoundly different environments requires the capacity to sense and respond to changes in the surroundings. Bordetella species have evolved tools to rapidly modulate transcription in order to respond to such changes. Activation of virulence in Bordetella is largely controlled by the BvgAS two-component system, which consists of a sensor protein, BvgS, a transcriptional activator, BvgA, and a transcriptional repressor, BvgR. During growth at temperatures at and below 25°C, the sensor protein is unphosphorylated and inactive, and the bacteria are in the so-called Bvg minus (Bvg−) phase in which transcription of virulence genes is repressed. When receiving inducing signals such as temperature of 37°C, which mimics presence in a mammalian host, the BvgS sensor protein autophosphorylates, goes through a phosphorylation cascade and subsequently transfers a phosphor group to BvgA. Upon phosphorylation by BvgS, BvgA binds to the promoter regions of the Bvg-activated genes and induces the transcription of virulence genes in response to temperature, which results in the expression of virulence factors, such as the type six secretion system (T6SS), type three secretion system (T3SS), PRN, FHA, ACT, PTX, and others (Cummings et al., 2006; Nakamura et al., 2006; Nicholson et al., 2009; Hester et al., 2012).
Under laboratory conditions, the classical bordetellae, including B. bronchiseptica, can respond to different environmental stimuli by switching between two distinct lifestyles. When cultured at 37°C in the so-called Bvg+ phase, in vitro conditions that mimic the infectious phase at temperatures in the mammalian host (Luu et al., 2017), expression of genes associated with colonization and virulence, the so-called virulence activated genes, is upregulated (Moon et al., 2017; Chen and Stibitz, 2019). Activation of the Bvg+ phase is necessary and sufficient to facilitate bacterial colonization during infection. While in this phase, flagella and chemotaxis genes are repressed, and the bacteria are non-motile (Akerley et al., 1992). However, when cultured at 25°C, mimicking environmental conditions, B. bronchiseptica adopts a second lifestyle during which gene expression of virulence-related factors is repressed, while transcription of a large alternative set of genes is activated, the so-called Bvg− phase. The biological significance of the Bvg− phase has long been hypothetical, mainly due to the lack of a clear role for the virulence-repressed state during in vivo studies and lack of knowledge of ex vivo growth. Several authors have speculated that activation of virulence-repressed genes might serve a role during persistence in an environmental reservoir (Cotter and Miller, 1994; Hamidou Soumana et al., 2017; Moon et al., 2017; Taylor-Mulneix et al., 2017b; Linz et al., 2019). Since many of the Bvg− phase transcribed genes are predicted to be metabolic enzymes and transport proteins, they are suspected to enhance acquisition of nutrients, growth, and proliferation in environmental settings. In addition, the transcriptionally active genes in the Bvg− phase include chemotaxis and flagella synthesis genes, suggesting bacterial motility.
Indirect evidence and experimental data support the biological relevance of the Bvg− phase in common ancestors of pathogenic Bordetella. First, in addition to the identification of numerous Bordetella-like bacteria among 16S ribosomal RNA (rRNA) sequences and metagenomes from soil samples (Wang et al., 2007; Hamidou Soumana et al., 2017; Garrido-Sanz et al., 2018), the animal-pathogenic B. hinzii and B. bronchiseptica were shown to grow efficiently in soil extract at 25°C (Hamidou Soumana et al., 2017). Second, using strains engineered to remain locked in a single phenotypic state by a point mutation in bvgS leading to constitutive phosphorylation (Bvg+ phase-locked mutant) or by bvgS gene deletion (Bvg− phase-locked mutant; Cotter and Miller, 1994), the Bvg− phase was shown to mediate B. bronchiseptica interactions with the soil amoeba D. discoideum (Taylor-Mulneix et al., 2017a). Tens of thousands of B. bronchiseptica wild-type and Bvg− phase-locked bacteria were isolated from the amoeba fruiting bodies, in contrast to only 100 Bvg+ phase-locked B. bronchiseptica, indicating that the Bvg− phase contributes to survival inside amoeba trophozoites and fruiting bodies (Taylor-Mulneix et al., 2017a). Interestingly, B. bronchiseptica not only survived but also multiplied inside amoeba fruiting bodies, as indicated by significantly increasing bacterial numbers over time, and were, then, disseminated along with amoeba spores by insects (Taylor-Mulneix et al., 2017a). The ability to switch between the Bvg+ and Bvg− life styles appears to be conserved among the bordetellae (perhaps with the exception of Bordetella ansorpii), as bvgA and bvgS gene homologs have been identified in the genomes of animal-associated species as well as of the environmental B. petrii (Gerlach et al., 2004; Gross et al., 2008, 2010; Linz et al., 2016). Together, these observations suggest the presence of potential environmental reservoirs for many, if not most, animal-pathogenic and human-pathogenic Bordetella species. Although many Bordetella species are adapted to mammals, in which the Bvg+ phase is active, they still conserved the ability to respond to changes such as temperature fluctuations (Coote, 2001) by switching to the ancient Bvg− phase for proliferation in environmental conditions.
Even though B. pertussis can phenotypically modulate between the two Bvg states (Stibitz et al., 1989; Moon et al., 2017; Chen and Stibitz, 2019), to this date, no evidence of an outside-host or environmental niche has been found for this human-restricted pathogen. This has led to the hypothesis that genes induced during the Bvg− phase are likely the vestigial remnants of an important phenotype of ancient Bordetella. But although B. pertussis has lost over 20% of its genome since diverging from B. bronchiseptica, the Bvg regulon in B. pertussis is largely conserved, demonstrating that the system has been under purifying selection. In addition, the ability to switch between life styles seems to be conserved among the bordetellae, as bvgA and bvgS gene homologs have been found in the genomes of animal-associated species as well as of the environmental B. petrii (Gerlach et al., 2004; Gross et al., 2010; Linz et al., 2016, 2019), contradicting the “vestigial” hypothesis.
Recent work has shed some light onto the potential role of the Bvg− phase in B. pertussis. First, Moon and colleagues analyzed B. pertussis gene expression under simulated Bvg− conditions in the laboratory and identified the expression of genes involved in the fatty acid and lipid metabolism, of sugar and amino acid transporters, pyruvate dehydrogenase, genes of the phenylacetic acid catabolic pathway, and of the glycolate oxidation pathway (Moon et al., 2017). Moreover, peptidoglycan and lipopolysaccharide (LPS) synthesis genes as well as capsular polysaccharide biosynthesis loci were actively transcribed in the Bvg− phase. Further, transcription of several genes encoding cold shock proteins was observed in this phase. These findings led the authors to speculate that transcriptional changes mediated by the switch from Bvg+ to Bvg− in response to rapid temperature drops might be an important adaptation that facilitates survival outside the human host during transmission of B. pertussis (Moon et al., 2017). Second, as was shown in the baboon model of B. pertussis infection, transmission of B. pertussis occurs via airflow of aerosolized respiratory droplets (Warfel et al., 2012). Not surprising, this study also found that the distance between the infected host and the naive susceptible individual influences the rate of transmission. Thus, since B. pertussis must survive in airborne respiratory droplets to transmit to a new host, extended B. pertussis survival in cool air outside the host may be important for B. pertussis transmission (Trainor et al., 2015). And third, Bvg− mutants were found to accumulate among a B. pertussis population in the nasopharynx of monkeys, suggesting an active role in facilitating bacterial persistence (Karataev et al., 2016). Taken together, these observations strongly suggest that the Bvg− phase constitutes a conserved strategy that may have contributed to the survival of ancient Bordetella, and even though many Bordetella species have evolved to infect mammalian hosts, the ability to respond and adapt to environmental changes remains active. Thus, it appears likely that the virulence-repressed state indeed plays an active and important role in the biology of host-pathogenic Bordetella species.
In vitro studies have shown that, upon entering human alveolar epithelial cell line A549, a significant portion of intracellular B. pertussis evades phagolysosomal fusion and remains viable in nonacidic compartments by a mechanism that is dependent on microtubule assembly, lipid raft integrity, and the activation of a tyrosine-kinase-mediated signaling (Lamberti et al., 2013). Numbers of viable intracellular bacteria increase from an average of one after 3 h to over five per A549 cell after 24 h. During this period, B. pertussis secretes a wide range of proteins involved in stress response, iron uptake, metabolism, and regulation, which allow the bacteria to reside and persist within host cells (Lamberti et al., 2013). Interestingly, intracellular survival appears to be dependent on the type of host cell, as viable B. pertussis were found to persist for 3 days in human macrophages and epithelial cells (Friedman et al., 1992; Lamberti et al., 2013) but less than 24 h in mouse DCs (Guzman et al., 1994a). B. parapertussis and B. bronchiseptica were also found to persist inside host phagocytes, emphasizing that the ability to survive intracellularly is not unique to B. pertussis (Guzman et al., 1994a; Gorgojo et al., 2012; Bendor et al., 2015).
One aspect commonly overlooked is the role of the Bvg− phase during intracellular persistence of Bordetella spp. inside host phagocytic cells. Spontaneous mutants lacking the bvgS gene, which encodes the sensor component of the BvgAS regulon, and the parental wildtype B. bronchiseptica displayed similar viability in DCs for over 72 h post infection. These data indicated that intracellular survival may be Bvg-independent or perhaps involves genes that are actively transcribed in the Bvg− phase (Guzman et al., 1994a,b). Indeed, assessment of B. bronchiseptica gene expression during intracellular survival in macrophages supported these observations. Upon uptake by macrophages, B. bronchiseptica activates the expression of genes involved in protein repair, DNA repair, oxidative stress response, pH homeostasis, chaperone functions, and activation of specific metabolic pathways (Figure 2). By contrast, the expression of genes involved in bacterial virulence, which is a hallmark of Bvg-based modulation of gene expression in the mammalian host, is suppressed. None of the known virulence factors, including toxins, T6SS and T3SS, and the adhesins PRN and FHA, were transcriptionally active (Rivera et al., 2019), and neither were any of the 205 genes that were previously identified to encode proteins that are secreted under Bvg+ conditions (Luu et al., 2017). A similar expression pattern was recently reported for B. pertussis inside macrophages, where expression of genes encoding virulence factors such as the T3SS, ACT, PTX, PRN, fimbriae 2 (FIM2), tracheal colonization factor (TCF), and the serum resistance protein BrkA was also suppressed (Petrackova et al., 2020). Surprisingly, despite conducting these experiments at 37°C, a temperature known to induce the expression of virulence factors, both B. bronchiseptica and B. pertussis displayed strong suppression of genes associated with virulence. This seems to be a contradiction. However, all tested classical and non-classical Bordetella species with the sole exception of B. avium were able to persist in murine macrophages (Rivera et al., 2019); yet, the presence of virulence factors in Bordetella species was shown to be species-specific (Linz et al., 2016) due to multiple events of gene acquisition and gene loss in the genus. For example, the well-known virulence factors of the classical bordetellae, including PTX, are not present in the non-classical species (Linz et al., 2016).
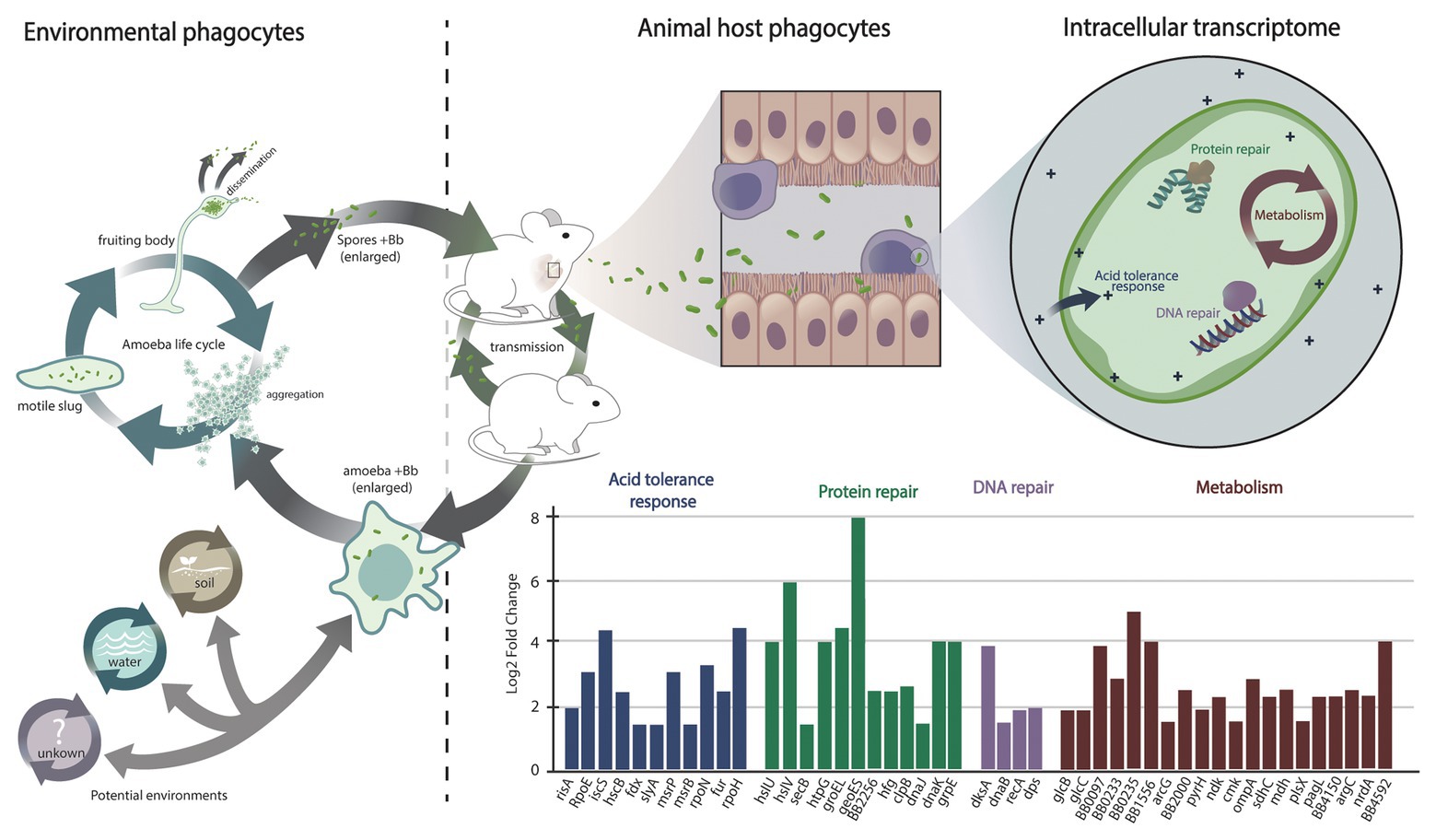
Figure 2. Interaction of Bordetella with environmental and animal host phagocytes and its transcriptional response during intracellular survival. Environmental phagocytes such as amoeba from soil, water, and other possible habitats (indicated by a question mark) represent an environmental reservoir for animal-pathogenic Bordetella species and their progenitors. Bordetella can resist digestion by amoebic phagocytes, translocate to the amoeba fruiting bodies, and disseminate along with amoeba spores. Animals and wind can spread bacteria and spores to new geographic locations, where the bacteria can either stay associated with the amoeba in a stable transmission cycle or infect a new mammalian host. Among the mammalian hosts, the bacteria can establish a transmission cycle that is independent but interconnected with the Bordetella life cycle in the amoebic host. During infection of a mammalian host, the bacteria are attacked by animal host immune cells, including macrophages. Uptake of Bordetella bronchiseptica by macrophages during infection of the mammalian respiratory tract triggers a bacterial SOS response that is marked by the expression of genes involved in protein repair, in DNA repair, and in acid tolerance. In addition, specific metabolic changes are associated with nutrient and oxygen deprivation, including transcriptional upregulation of genes of the glyoxylate cycle and downregulation of genes encoding proteins of the oxidative respiratory chain. In contrast to bacteria from other genera, the expression of genes involved in bacterial virulence is suppressed.
In addition to BvgAS, other regulatory systems have also been implicated in promoting bacterial persistence inside macrophages. A two-component regulatory system named RisAS was found to be required for persistence of B. bronchiseptica inside spleen DC line CB1 and macrophage-like cell line J774A.1 (Jungnitz et al., 1998). Phenotypic exploration of a RisA mutant strain revealed that the RisAS regulon promotes resistance against oxidative stress, production of acid phosphatase, and in vivo persistence. RisA is optimally expressed at 37°C and when the bacteria are contained within eukaryotic phagocytes (Jungnitz et al., 1998). Notably, activation of RisA is regulated independently of BvgAS. Recent transcription analyses of RisA mutants under Bvg+and Bvg− conditions showed that RisA very likely plays a major role during intracellular survival in mammalian phagocytes. The RisA-dependent in vitro transcription profiles (Coutte et al., 2016) were largely congruent with the transcriptional response of B. bronchiseptica and B. pertussis inside macrophages (Rivera et al., 2019; Petrackova et al., 2020). Under Bvg+ conditions, BvgA is phosphorylated and induces transcription of virulence-activated genes as well as of the transcriptional repressor BvgR. BvgR has diguanylate phosphodiesterase activity, which degrades intracellular c-di-GMP to GMP. In the Bvg− phase, and in the absence or at very low levels of c-di-GMP, unphosphorylated RisA activates transcription of a variety of genes, including those involved in osmotic and oxidative stress response, pH homeostasis, and chaperone functions such as groEL (Coutte et al., 2016). At the same time, expression of many iron-regulated genes including tonB is suppressed. At sufficiently high levels of intracellular c-di-GMP, RisA binds to c-di-GMP. Unphosphorylated, c-di-GMP-associated RisA also suppresses expression of iron genes. In contrast, phosphorylated c-di-GMP-binding RisA initiates expression of many virulence-repressed genes, including those for O-antigen and capsule biosynthesis. In addition, phosphorylated and c-di-GMP-bound RisA not only inhibits the expression of the virulence-activated genes but also suppresses the expression of flagellar and chemotaxis genes (Coutte et al., 2016). Thus, the interplay between BvgA and RisA emphasizes the complexity of the regulatory network required for bacterial adaptation to the host intracellular environment.
In addition, the avirulent Bvg− phase has been shown to be required for survival, persistence, and replication of B. bronchiseptica within amoeba (Taylor-Mulneix et al., 2017a). It is plausible that while many pathogenic bacteria respond to stress by inducing the expression of virulence, Bordetella species employ an ancient conserved stress response to cope with the challenges of an intracellular environment. These observations suggest a critical role of the Bvg− phase in modulating gene expression during bacterial interaction within host phagocytes. However, despite these findings, it is important to recognize that these observations only constitute a snapshot of a rather dynamical and complex process. The above studies at the messenger RNA (mRNA) level have shown suppression of virulence upon internalization in macrophages. In contrast, similar analyses of the B. pertussis proteome reported unchanged abundance of virulence proteins PTX and ACT in intracellular B. pertussis compared to in vitro grown under Bvg+ conditions (Lamberti et al., 2016; Valdez et al., 2016), which was interpreted as these proteins contributing to bacterial persistence in macrophages. This apparent contradiction may be due to slow degradation of the already synthesized proteins, which would suggest that the protein levels might decrease at later time points. In this case, the avirulent Bvg− phase would represent the authentic phenotype during intracellular survival.
The ability to survive inside macrophage spans beyond the classical bordetellae (Rivera et al., 2019). Comparative genome analyses between B. bronchiseptica and non-classical Bordetella species revealed the conservation of genes involved in intracellular persistence; approximately 80% of the 318 transcriptionally upregulated B. bronchiseptica genes during intracellular persistence were present in the genomes of non-classical Bordetella spp., in contrast to ~50% of the total of 4,981 evaluated B. bronchiseptica genes. Phenotypic analyses validated the significance of this observation by demonstrating that both the classical and the non-classical Bordetella spp. can persist inside RAW 264.7 macrophages (Rivera et al., 2019). The shared ability for intracellular persistence, and evolutionary conservation of the genes suspected to be involved in it, strongly suggests that intracellular survival represents an ancestral trait, the origin of which precedes speciation in the genus. As such, this feature must have evolved at some point during evolution of Bordetella spp. from environmental bacteria to mammalian respiratory pathogens. B. bronchiseptica and also the sheep-associated B. parapertussisov can resist digestion and utilize the life cycle of the ubiquitous soil amoeba D. discoideum to multiply and disseminate (Taylor-Mulneix et al., 2017a,b). The ancient interaction with these and potentially other environmental phagocytes could have been an important evolutionary milestone on the way to intracellular survival in phagocytic cells. Thus, intracellular persistence in environmental phagocytes may have been a “training ground” – as some authors named it – for the subsequent evolution of bacteria, including Bordetella spp., to animal pathogens (Greub and Raoult, 2004; Molmeret et al., 2005; Salah et al., 2009; Taylor-Mulneix et al., 2017b). While this point of view of an ancient evolutionary adaptation preceding speciation suggests that many, if not all, animal-pathogenic Bordetella species can interact with amoeba, this prediction remains to be evaluated.
Internalization by macrophages triggered a strong general stress response known as SOS response (Simmons et al., 2008) in B. bronchiseptica (Figure 2), characterized by suppression of cell division via downregulation of the fts locus and by upregulation of DNA repair genes, of protein chaperone genes, of oxidative stress response, and of acid tolerance genes (Rivera et al., 2019). As expected under microaerophilic/hypoxic conditions inside macrophages, transcription of the nuo genes that encode the oxidative respiratory chain was strongly suppressed. In contrast, genes of the glyoxylate cycle displayed elevated expression, including the gene glcB encoding malate synthase G and its transcriptional activator glcC, malate dehydrogenase gene mdh, citrate synthase gene gltA, and aconitase gene acnB (Rivera et al., 2019). The glyoxylate cycle is important in the utilization of acetate or fatty acids as the main carbon source and may be essential for nucleotide and amino acid biosynthesis under intracellular conditions (Munoz-Elias and McKinney, 2006). The avian pathogen B. avium is the only animal-adapted Bordetella species that lacks malate synthase transcriptional regulator glcC, and this species was severely impaired in its ability to persist in macrophages. Indeed, deletion of this gene in B. bronchiseptica resulted in a significant reduction in intracellular survival, but complementation of the knockout mutant with a plasmid-borne gene copy restored the wild-type phenotype, confirming a critical role of glcC during intracellular persistence (Rivera et al., 2019). Besides the glyoxylate cycle, several genes of fatty acid synthesis pathways such as 3-oxoacyl-ACP reductase BB4150, long chain fatty acid Co-A ligase BB0233, outer membrane protein OmpA, and ABC transport protein encoded by BB1556 were found to be strongly induced, suggesting that increased membrane biosynthesis. In addition, elevated expression of genes involved in amino acid biosynthesis and transport, of de novo nucleotide biosynthesis as well as of numerous ribosomal protein genes were indicative of general extensive metabolic activity in the bacterial cell in response to internalization by macrophages (Rivera et al., 2019).
Notably, the array of genes expressed during intracellular survival holds similarity to those expressed in response to low pH exposure (pH less than 4) following acid adaptation (pH at 5.5; Fingermann and Hozbor, 2015). Synthesis of ribosomal proteins (ribosomal protein L1, L4, and L5), molecular chaperones (GroEL, HSP 90, and DnaK), proteases (BB3293 and BB1248), metabolic related proteins (succinate-semialdehyde dehydrogenase, aconitate hydratase, and argininesuccinate synthase), and LPS modification have been reported to contribute to B. bronchiseptica acid tolerance response. Furthermore, B. bronchiseptica grown in the avirulent phase or a Bvg− phase-locked strain displayed greater resistance to lethal acid challenge, and thus, increased survival rate in acid. These similarities suggest that upon entry in host macrophages, B. bronchiseptica modulates the transcription of genes involved in the acid tolerance response while suppressing the expression of virulence genes as a mechanism to enhance resistance against stringent acid conditions.
Interestingly, the activation of an acid tolerance response is one of the many features utilized by pathogens known to cause human infection and persist inside host phagocytes, including macrophages and DCs (Thakur et al., 2019). For example, S. enterica serovar Typhimurium can subvert macrophage mediated killing and survive within the acidic environment of macrophages by altering lysosomal pH. Several regulatory proteins involved in reduction of acidic pH and activation of the stress response have been previously identified for intracellular S. enterica. These includes the environmental response regulator protein sigma factor RpoH, OmpR, and ferric uptake regulator protein (Fur), and stress response proteins alkyl hydroperoxide reductase (AhpC), DNA protection during starvation protein (Dps), recombinase A (RecA), and Mg(2+) transport ATPase protein C (MgtC; Imre et al., 2013; Lund et al., 2014). Expression of gene homologs encoding for RpoH, OmpR, Fur, AhpC, Dps, and RecA has been previously identified for intracellular B. bronchiseptica (Rivera et al., 2019), while MgtC has been reported to contribute to B. pertussis persistence inside macrophages (Lamberti et al., 2016; Cafiero et al., 2018).
In many bacteria including Brucella spp., Listeria monocytogenes, S. enterica, Vibrio cholerae, as well as B. pertussis and B. bronchiseptica, another important stress response protein is the RNA-chaperone protein Hfq (Robertson and Roop, 1999; Christiansen et al., 2004; Ding et al., 2004; Sittka et al., 2007; Bibova et al., 2013, 2015; Lamberti et al., 2016; Rivera et al., 2019; Alvarez Hayes et al., 2020). Hfq regulates the binding of non-coding RNAs to mRNAs, which can result either in translational repression and mRNA degradation or in activation of translation. Hfq was found to mediate the response to osmotic and heat stress, as well as to starvation in the stationary growth phase (Robertson and Roop, 1999). Besides stress resistance, Hfq plays a role in the post-transcriptional regulation of virulence factor expression (Christiansen et al., 2004; Sittka et al., 2007). For example, in B. pertussis, an Hfq deletion mutant displayed reduced expression of the TCF and the T3SS (Bibova et al., 2015). In addition, Hfq deletion mutants were found to be severely impaired in their ability to survive inside macrophages (Bibova et al., 2013; Alvarez Hayes et al., 2020), suggesting an important role of the Hfq-mediated stress response during intracellular survival.
Similar to B. bronchiseptica, Francisella tularensis – a facultative intracellular pathogen and the causative agent of tularemia – can infect and proliferate within mammalian phagocytes as well as in amoeba (Abd et al., 2003; El-Etr et al., 2009; Ozanic et al., 2015). Upon infection, intracellular F. tularensis subverts host defenses by escaping the phagosome (Celli and Zahrt, 2013) or by synthesizing proteins that promote persistence inside macrophages, such as macrophage growth locus protein MglA, stringent starvation protein A (SspA), chaperone HtpG, carbamoyl-phosphate synthase (CarA), and iron binding rubredoxin (RubA; Fuller et al., 2008; Charity et al., 2009; Bell et al., 2010; Chong and Celli, 2010; Dai et al., 2010; Bent et al., 2013). Previous work has demonstrated that intracellular B. pertussis can avoid the phagosome-lysosome fusion and replicate inside macrophages within 48 h post infection (Lamberti et al., 2010). In addition, upregulation of genes encoding for MglA (1.7 fold-change), AroG (2.7 fold-change), HtpG (4.2 fold-change), CarA (1.5 fold change), RubA (2.1), and to a lesser degree for SspA (1.2 fold change) has been reported for intracellular B. bronchiseptica at 2 h post infection of RAW 246.7 macrophages (Rivera et al., 2019).
For intracellular pathogens, the ability to persist inside host-phagocytes is contingent upon adjustment to the intracellular environment. Lethal pH, resource starvation, and oxidative burst are among the threats intracellular bacteria will face upon entry in host cells, which often result in damage to protein and DNA integrity and nutrient depravation. Mechanisms commonly employed by intracellular bacteria such as L. monocytogenes, S. enterica, and Clostridium spp. include the synthesis of protein complexes that promote bacterial cell homeostasis under demanding environmental conditions (Lund et al., 2014). Synthesis of molecules such as recombinase RecA, chaperones DnaK, GroEL, GroES, Hsp90, and protease Clp promotes DNA repair and folding of critical enzymes and removal of damage proteins, thus maintains cell integrity upon stress. Like in many intracellular facultative pathogens, the presence of genes encoding for these proteins in the Bordetella genome, and their elevated expression during survival inside macrophages, serves as evidence that the bacteria possess the mechanisms to cope with the hazardous condition often present inside host cells, including professional phagocytic cells (Rivera et al., 2019).
Host response to Bordetella spp. infections has largely been studied in mice challenged with B. pertussis or with its close relative B. bronchiseptica. Upon infection, B. pertussis secretes a wide array of virulence factors that among other functions, mediate adherence to host epithelial cells and promote bacterial survival in the host respiratory track.
For instance, B. pertussis internalized by host cells can persist intracellularly within ciliated epithelial cells and alveolar macrophages. Studies in murine models have shown that innate immune cells and antimicrobial peptides help to control the infection, while complete bacterial clearance requires cellular immunity mediated by T-helper type 1 (Th1) and Th17 cells (Mills et al., 1999). During the early stages of infection, local and innate immune cells including macrophages, DCs, neutrophils, and natural killer (NK) cells are recruited and largely accountable for the control and reduction of B. pertussis (Carbonetti, 2007; Carbonetti et al., 2007; Higgs et al., 2012). Among the effectors that promote bacterial reduction, early secretion of interferon-gamma (INF-γ) by NK cells, DC, and Th1 cells greatly enhance macrophage-mediated killing of B. pertussis; mice lacking INF-γ developed a lethal infection after challenge with B. pertussis (Mahon and Mills, 1999; Higgins et al., 2006).
The adaptive immune system plays a crucial role in clearance of B. pertussis infection. Activation and recruitment of a Th1 and Th17 adaptive immune response mediates bacterial clearance from the lower and upper respiratory track of mice. Modulation of cellular Th1 immunity results in the secretion of IL-12 and INF-γ, which in turn promotes neutrophil recruitment and enhances macrophage phagocytic activity. It is largely recognized that optimal immunity to primary and secondary B. pertussis infections is conferred by the priming and activation of Th1 cells. Similar to convalescent immunity, vaccination with whole cell pertussis (wP) – which is based on heat-killed B. pertussis bacteria – promotes the secretion of Th1 derived cytokines and enhances macrophage activity, resulting in increased killing of phagocytosed bacteria. Protection conferred by wP has been widely studied, and the rapid decline in the number of pertussis cases following its introduction is strong and compelling evidence for its effectiveness. However, while whole cell pertussis vaccines (wP) were highly effective at clearing the infection and providing long-term protection, several cases of vaccine reactogenicity were reported, which led to its replacement in many countries by acellular vaccines (aP) that are composed of three to five purified immunogenic antigens. Even though acellular pertussis vaccine is effective at clearing B. pertussis infection from the lungs and protecting against disease pathology, vaccination with aP, in contrast to immunization with wP vaccine, does not provide protection against bacterial colonization of the upper respiratory tract of mice and fails to prevent transmission among non-human primates (Warfel and Merkel, 2013; Warfel et al., 2014).
Despite wide vaccination coverage, the incidence of pertussis is increasing, which prompted the National Institute of Allergy and Infectious Diseases (NIAID) to add B. pertussis to the list of priority emerging infectious diseases/pathogens in 2015. Multiple factors have been suspected to contribute to the reemergence of pertussis. These include greater awareness and reporting, improved detection methodology, vaccine hesitancy and incomplete vaccine boosters in some areas, antigenic shifts in B. pertussis to escape vaccine-mediated immunity, more rapid waning of aP-induced immunity and decreased long-term protection compared with whole cell vaccines, and a less effective aP-induced immune response that does not prevent colonization and transmission of B. pertussis, resulting in increased carriage and asymptomatic transmission (Cherry, 2012; Klein et al., 2012; Bart et al., 2014; Warfel et al., 2014; Althouse and Scarpino, 2015; Dewan et al., 2020). As outlined above, infection with B. pertussis naturally induces a significant Th1-type T-lymphocyte cytokine response in mice that is characterized by high levels of IL-2, IFN-γ, and TNF-α, a type of immune response that is characteristic of infection by intracellular pathogens (Spellberg and Edwards, 2001; Thakur et al., 2019). While protective immunity generated by wP vaccination also promotes a Th1 immune response, the less efficacious aP vaccines induce a strong Th2 and antibody response. Since the aP vaccine-induced Th2 response does not effectively target intracellular pathogens, we propose that the inability to clear intracellular bacteria could allow B. pertussis to evade host immunity during survival inside phagocytic cells. This would protect the bacteria from antibodies, complement activation, and bactericidal activity, and thus, would result in suboptimal vaccine protection. Thus, given that the number of pertussis cases have been on the rise following the widespread use of aP vaccines, it seems plausible that an inadequate immune response to intracellular B. pertussis may have been contributing to the reemergence of whooping cough.
Whooping cough is caused by B. pertussis and, to a lesser extent, B. parapertussis. Similar to B. pertussis, the incidence of whooping cough-like disease caused by B. parapertussis has been increasing over the past decades. Antibodies elicited against the wP vaccine were shown to also target B. parapertussis, and thus, provide some level of protection, even though less protection than against B. pertussis (Willems et al., 1998; Zhang et al., 2009). In contrast to wP vaccines, aP-elicited antibodies showed no efficacy against B. parapertussis. On the contrary, aP vaccination induced host immunity that somehow interfered with the optimal clearance of B. parapertussis. Therefore, it was speculated that widespread aP vaccination against B. pertussis might increase the risk of acquiring an B. parapertussis infection (Long et al., 2010). Further studies revealed that the B. parapertussis O-antigen inhibited antibody binding to the bacteria (Zhang et al., 2009; Gorgojo et al., 2012). In the absence of opsonizing antibodies, neutrophils were unable to kill B. parapertussis after phagocytic uptake. A high proportion of the engulfed bacteria persisted in nonacidic phagosomes, establishing an intracellular infection (Gorgojo et al., 2012, 2014). Intracellular survival and persistence of B. parapertussis is tempting us to speculate that the inability of the aP vaccine-induced Th2 response to effectively target intracellular pathogens may be among the reasons for the increasing incidence of B. parapertussis.
Pathogenic members of the genus Bordetella have adapted to colonize, replicate, and transmit in animal hosts. The environmental origin of the species suggests that the ability to survive and persist in environmental phagocytes could have protected the bacteria from external dangers while ensuring transmission to novel environments and hosts. The interaction of the bacteria with environmental amoeba may have forced the evolution of schemes that allowed them to interact with mammalian phagocytes, specifically the evolution of tools to avoid and survive phagocytic digestion. Thus, a once critical survival strategy in the environment could potentially affect the success of the bacteria during mammalian host infection. This mechanism to evade the host immune system appears to be contributing to their emergence as important animal and human pathogens. Interestingly, similar to the interaction of B. bronchiseptica with amoebae (Taylor-Mulneix et al., 2017a), the transcriptional response to internalization by mammalian phagocytes occurred in the environmental and avirulent Bvg− phase (Rivera et al., 2019; Petrackova et al., 2020). Despite the interaction with macrophages at 37°C, the mammalian host temperature, and thus, supposedly Bvg+ conditions, transcription of the well-known virulence factors was downregulated. While this seems contradictory, it makes sense because the presence of virulence factors in Bordetella species is largely species-specific (Linz et al., 2016) but both classical and non-classical Bordetella species were able to persist in murine macrophages (Rivera et al., 2019). Evolution and host specialization, such as the evolution of B. pertussis from a B. bronchiseptica-like ancestor and its adaptation to a closed transmission cycle with only a human host (Dewan and Harvill, 2019), were often accompanied by genome reduction. In case of B. pertussis and the human lineage of B. parapertussis, this loss of hundreds of genes ended their ability to utilize amoebae as an environmental reservoir (Taylor-Mulneix et al., 2017b). By contrast, B. pertussis’ and B. parapertussis’ ability to persist in macrophages was preserved, likely because of strong selection for a favorable trait in the mammalian/human host. Careful examination of the mechanisms by which Bordetella reside inside host cells and the role of intracellular persistence during infection can provide valuable insight and new approaches to confront the reemergence of whooping cough.
IR, BL, and EH wrote the review. IR and BL contributed equally to this manuscript and should be considered equally contributing first authors. All authors approved the submitted version.
This work was supported by grants AI104399, GM113681, and AI142678 of the National Institutes of Health to EH. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MR declared a past co-authorship with the author EH to the handling editor.
Abd, H., Johansson, T., Golovliov, I., Sandstrom, G., and Forsman, M. (2003). Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69, 600–606. doi: 10.1128/aem.69.1.600-606.2003
Ahmed, N., Saini, V., Raghuvanshi, S., Khurana, J. P., Tyagi, A. K., Tyagi, A. K., et al. (2007). Molecular analysis of a leprosy immunotherapeutic bacillus provides insights into Mycobacterium evolution. PLoS One 2:e968. doi: 10.1371/journal.pone.0000968
Akerley, B. J., Monack, D. M., Falkow, S., and Miller, J. F. (1992). The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J. Bacteriol. 174, 980–990. doi: 10.1128/jb.174.3.980-990.1992
Althouse, B. M., and Scarpino, S. V. (2015). Asymptomatic transmission and the resurgence of Bordetella pertussis. BMC Med. 13:146. doi: 10.1186/s12916-015-0382-8
Alvarez Hayes, J., Surmann, K., Lamberti, Y., Depke, M., Dhople, V., Blanca, B., et al. (2020). Hfq modulates global protein pattern and stress response in Bordetella pertussis. J. Proteome 211:103559. doi: 10.1016/j.jprot.2019.103559
Bart, M. J., Harris, S. R., Advani, A., Arakawa, Y., Bottero, D., Bouchez, V., et al. (2014). Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. mBio 5:se01074. doi: 10.1128/mBio.01074-14
Bell, B. L., Mohapatra, N. P., and Gunn, J. S. (2010). Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect. Immun. 78, 2189–2198. doi: 10.1128/IAI.00021-10
Bendor, L., Weyrich, L. S., Linz, B., Rolin, O. Y., Taylor, D. L., Goodfield, L. L., et al. (2015). Type six secretion system of Bordetella bronchiseptica and adaptive immune components limit intracellular survival during infection. PLoS One 10:e0140743. doi: 10.1371/journal.pone.0140743
Bent, Z. W., Brazel, D. M., Tran-Gyamfi, M. B., Hamblin, R. Y., VanderNoot, V. A., and Branda, S. S. (2013). Use of a capture-based pathogen transcript enrichment strategy for RNA-Seq analysis of the Francisella tularensis LVS transcriptome during infection of murine macrophages. PLoS One 8:e77834. doi: 10.1371/journal.pone.0077834
Bibova, I., Hot, D., Keidel, K., Amman, F., Slupek, S., Cerny, O., et al. (2015). Transcriptional profiling of Bordetella pertussis reveals requirement of RNA chaperone Hfq for type III secretion system functionality. RNA Biol. 12, 175–185. doi: 10.1080/15476286.2015.1017237
Bibova, I., Skopova, K., Masin, J., Cerny, O., Hot, D., Sebo, P., et al. (2013). The RNA chaperone Hfq is required for virulence of Bordetella pertussis. Infect. Immun. 81, 4081–4090. doi: 10.1128/IAI.00345-13
Cafiero, J. H., Lamberti, Y. A., Surmann, K., Vecerek, B., and Rodriguez, M. E. (2018). A Bordetella pertussis MgtC homolog plays a role in the intracellular survival. PLoS One 13:e0203204. doi: 10.1371/journal.pone.0203204
Carbonetti, N. H. (2007). Immunomodulation in the pathogenesis of Bordetella pertussis infection and disease. Curr. Opin. Pharmacol. 7, 272–278. doi: 10.1016/j.coph.2006.12.004
Carbonetti, N. H., Artamonova, G. V., Van Rooijen, N., and Ayala, V. I. (2007). Pertussis toxin targets airway macrophages to promote Bordetella pertussis infection of the respiratory tract. Infect. Immun. 75, 1713–1720. doi: 10.1128/IAI.01578-06
Celli, J., and Zahrt, T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010314. doi: 10.1101/cshperspect.a010314
Chan, A. P., Sutton, G., DePew, J., Krishnakumar, R., Choi, Y., Huang, X. Z., et al. (2015). A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol. 16:143. doi: 10.1186/s13059-015-0701-6
Charity, J. C., Blalock, L. T., Costante-Hamm, M. M., Kasper, D. L., and Dove, S. L. (2009). Small molecule control of virulence gene expression in Francisella tularensis. PLoS Pathog. 5:e1000641. doi: 10.1371/journal.ppat.1000641
Chen, Q., and Stibitz, S. (2019). The BvgASR virulence regulon of Bordetella pertussis. Curr. Opin. Microbiol. 47, 74–81. doi: 10.1016/j.mib.2019.01.002
Cherry, J. D. (2012). Why do pertussis vaccines fail? Pediatrics 129, 968–970. doi: 10.1542/peds.2011-2594
Chiner-Oms, A., Sanchez-Buso, L., Corander, J., Gagneux, S., Harris, S. R., Young, D., et al. (2019). Genomic determinants of speciation and spread of the Mycobacterium tuberculosis complex. Sci. Adv. 5:eaaw3307. doi: 10.1126/sciadv.aaw3307
Chong, A., and Celli, J. (2010). The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbiol. 1:138. doi: 10.3389/fmicb.2010.00138
Christiansen, J. K., Larsen, M. H., Ingmer, H., Sogaard-Andersen, L., and Kallipolitis, B. H. (2004). The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186, 3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004
Cookson, B. T., Vandamme, P., Carlson, L. C., Larson, A. M., Sheffield, J. V., Kersters, K., et al. (1994). Bacteremia caused by a novel Bordetella species, “B. hinzii.” J. Clin. Microbiol. 32, 2569–2571. doi: 10.1128/JCM.32.10.2569-2571.1994
Coote, J. G. (2001). Environmental sensing mechanisms in Bordetella. Adv. Microb. Physiol. 44, 141–181. doi: 10.1016/s0065-2911(01)44013-6
Cotter, P. A., and Miller, J. F. (1994). BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62, 3381–3390. doi: 10.1128/IAI.62.8.3381-3390.1994
Coutte, L., Huot, L., Antoine, R., Slupek, S., Merkel, T. J., Chen, Q., et al. (2016). The multifaceted RisA regulon of Bordetella pertussis. Sci. Rep. 6:32774. doi: 10.1038/srep32774
Cummings, C. A., Bootsma, H. J., Relman, D. A., and Miller, J. F. (2006). Species‐ and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188, 1775–1785. doi: 10.1128/JB.188.5.1775-1785.2006
Dai, S., Mohapatra, N. P., Schlesinger, L. S., and Gunn, J. S. (2010). Regulation of Francisella tularensis virulence. Front. Microbiol. 1:144. doi: 10.3389/fmicb.2010.00144
Dewan, K. K., and Harvill, E. T. (2019). Did new transmission cycles in anthropogenic, dense, host populations encourage the emergence and speciation of pathogenic Bordetella? PLoS Pathog. 15:e1007600. doi: 10.1371/journal.ppat.1007600
Dewan, K. K., Linz, B., DeRocco, S. E., and Harvill, E. T. (2020). Acellular pertussis vaccine components: today and tomorrow. Vaccines 8:217. doi: 10.3390/vaccines8020217
Dewan, K. K., Taylor-Mulneix, D. L., Campos, L. L., Skarlupka, A. L., Wagner, S. M., Ryman, V. E., et al. (2019). A model of chronic, transmissible otitis media in mice. PLoS Pathog. 15:e1007696. doi: 10.1371/journal.ppat.1007696
Diavatopoulos, D. A., Cummings, C. A., Schouls, L. M., Brinig, M. M., Relman, D. A., and Mooi, F. R. (2005). Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1:e45. doi: 10.1371/journal.ppat.0010045
Ding, Y., Davis, B. M., and Waldor, M. K. (2004). Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53, 345–354. doi: 10.1111/j.1365-2958.2004.04142.x
Dubois, V., Pawlik, A., Bories, A., Le Moigne, V., Sismeiro, O., Legendre, R., et al. (2019). Mycobacterium abscessus virulence traits unraveled by transcriptomic profiling in amoeba and macrophages. PLoS Pathog. 15:e1008069. doi: 10.1371/journal.ppat.1008069
El-Etr, S. H., Margolis, J. J., Monack, D., Robison, R. A., Cohen, M., Moore, E., et al. (2009). Francisella tularensis type A strains cause the rapid encystment of Acanthamoeba castellanii and survive in amoebal cysts for three weeks postinfection. Appl. Environ. Microbiol. 75, 7488–7500. doi: 10.1128/AEM.01829-09
Fingermann, M., and Hozbor, D. (2015). Acid tolerance response of Bordetella bronchiseptica in avirulent phase. Microbiol. Res. 181, 52–60. doi: 10.1016/j.micres.2015.09.001
Friedman, R. L., Nordensson, K., Wilson, L., Akporiaye, E. T., and Yocum, D. E. (1992). Uptake and intracellular survival of Bordetella pertussis in human macrophages. Infect. Immun. 60, 4578–4585. doi: 10.1128/IAI.60.11.4578-4585.1992
Fry, N. K., Duncan, J., Malnick, H., Warner, M., Smith, A. J., Jackson, M. S., et al. (2005). Bordetella petrii clinical isolate. Emerg. Infect. Dis. 11, 1131–1133. doi: 10.3201/eid1107.050046
Fuller, J. R., Craven, R. R., Hall, J. D., Kijek, T. M., Taft-Benz, S., and Kawula, T. H. (2008). RipA, a cytoplasmic membrane protein conserved among Francisella species, is required for intracellular survival. Infect. Immun. 76, 4934–4943. doi: 10.1128/IAI.00475-08
Funke, G., Hess, T., von Graevenitz, A., and Vandamme, P. (1996). Characteristics of Bordetella hinzii strains isolated from a cystic fibrosis patient over a 3-year period. J. Clin. Microbiol. 34, 966–969. doi: 10.1128/JCM.34.4.966-969.1996
Garrido-Sanz, D., Manzano, J., Martin, M., Redondo-Nieto, M., and Rivilla, R. (2018). Metagenomic analysis of a biphenyl-degrading soil bacterial consortium reveals the metabolic roles of specific populations. Front. Microbiol. 9:232. doi: 10.3389/fmicb.2018.00232
Gerlach, G., Janzen, S., Beier, D., and Gross, R. (2004). Functional characterization of the BvgAS two-component system of Bordetella holmesii. Microbiology 150, 3715–3729. doi: 10.1099/mic.0.27432-0
Goodnow, R. A. (1980). Biology of Bordetella bronchiseptica. Microbiol. Rev. 44, 722–738. doi: 10.1128/MMBR.44.4.722-738.1980
Gorgojo, J., Harvill, E. T., and Rodriguez, M. E. (2014). Bordetella parapertussis survives inside human macrophages in lipid raft-enriched phagosomes. Infect. Immun. 82, 5175–5184. doi: 10.1128/IAI.02553-14
Gorgojo, J., Lamberti, Y., Valdez, H., Harvill, E. T., and Rodriguez, M. E. (2012). Bordetella parapertussis survives the innate interaction with human neutrophils by impairing bactericidal trafficking inside the cell through a lipid raft-dependent mechanism mediated by the lipopolysaccharide O antigen. Infect. Immun. 80, 4309–4316. doi: 10.1128/IAI.00662-12
Greub, G., and Raoult, D. (2004). Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433. doi: 10.1128/cmr.17.2.413-433.2004
Gross, R., Guzman, C. A., Sebaihia, M., dos Santos, V. A., Pieper, D. H., Koebnik, R., et al. (2008). The missing link: Bordetella petrii is endowed with both the metabolic versatility of environmental bacteria and virulence traits of pathogenic bordetellae. BMC Genomics 9:449. doi: 10.1186/1471-2164-9-449
Gross, R., Keidel, K., and Schmitt, K. (2010). Resemblance and divergence: the “new” members of the genus Bordetella. Med. Microbiol. Immunol. 199, 155–163. doi: 10.1007/s00430-010-0148-z
Guzman, C. A., Rohde, M., Bock, M., and Timmis, K. N. (1994a). Invasion and intracellular survival of Bordetella bronchiseptica in mouse dendritic cells. Infect. Immun. 62, 5528–5537. doi: 10.1128/IAI.62.12.5528-5537.1994
Guzman, C. A., Rohde, M., and Timmis, K. N. (1994b). Mechanisms involved in uptake of Bordetella bronchiseptica by mouse dendritic cells. Infect. Immun. 62, 5538–5544. doi: 10.1128/IAI.62.12.5538-5544.1994
Hamidou Soumana, I., Linz, B., and Harvill, E. T. (2017). Environmental origin of the genus Bordetella. Front. Microbiol. 8:28. doi: 10.3389/fmicb.2017.00028
Hester, S. E., Lui, M., Nicholson, T., Nowacki, D., and Harvill, E. T. (2012). Identification of a CO2 responsive regulon in Bordetella. PLoS One 7:e47635. doi: 10.1371/journal.pone.0047635
Higgins, S. C., Jarnicki, A. G., Lavelle, E. C., and Mills, K. H. (2006). TLR4 mediates vaccine-induced protective cellular immunity to Bordetella pertussis: role of IL-17-producing T cells. J. Immunol. 177, 7980–7989. doi: 10.4049/jimmunol.177.11.7980
Higgs, R., Higgins, S. C., Ross, P. J., and Mills, K. H. (2012). Immunity to the respiratory pathogen Bordetella pertussis. Mucosal Immunol. 5, 485–500. doi: 10.1038/mi.2012.54
Imre, A., Bukovinszki, A., Lovell, M. A., Li, H., Zhou, X., and Barrow, P. A. (2013). Gene expression analysis of Salmonella enterica SPI in macrophages indicates differences between serovars that induce systemic disease from those normally causing enteritis. Vet. Microbiol. 167, 675–679. doi: 10.1016/j.vetmic.2013.07.034
Ivanov, Y. V., Linz, B., Register, K. B., Newman, J. D., Taylor, D. L., Boschert, K. R., et al. (2016). Identification and taxonomic characterization of Bordetella pseudohinzii sp. nov. isolated from laboratory-raised mice. Int. J. Syst. Evol. Microbiol. 66, 5452–5459. doi: 10.1099/ijsem.0.001540
Ivanov, Y. V., Shariat, N., Register, K. B., Linz, B., Rivera, I., Hu, K., et al. (2015). A newly discovered Bordetella species carries a transcriptionally active CRISPR-Cas with a small Cas9 endonuclease. BMC Genomics 16:863. doi: 10.1186/s12864-015-2028-9
Jungnitz, H., West, N. P., Walker, M. J., Chhatwal, G. S., and Guzman, C. A. (1998). A second two-component regulatory system of Bordetella bronchiseptica required for bacterial resistance to oxidative stress, production of acid phosphatase, and in vivo persistence. Infect. Immun. 66, 4640–4650. doi: 10.1128/IAI.66.10.4640-4650.1998
Karataev, G. I., Sinyashina, L. N., Medkova, A. Y., Semin, E. G., Shevtsova, Z. V., Matua, A. Z., et al. (2016). Insertional inactivation of virulence operon in population of persistent Bordetella pertussis bacteria. Genetika 52, 422–430. doi: 10.1134/S102279541603008X
Kersters, K., Hinz, K. H., Hertle, A., Segers, P., Lievens, A., Siegmann, O., et al. (1984). Bordetella avium sp. nov., isolated from the respiratory tracts of turkeys and other birds. Int. J. Syst. Bacteriol. 34, 56–70. doi: 10.1099/00207713-34-1-56
Kilian, M., and Tettelin, H. (2019). Identification of virulence-associated properties by comparative genome analysis of Streptococcus pneumoniae, S. pseudopneumoniae, S. mitis, three S. oralis subspecies, and S. infantis. mBio 10:e01985-19. doi: 10.1128/mBio.01985-19
Klein, N. P., Bartlett, J., Rowhani-Rahbar, A., Fireman, B., and Baxter, R. (2012). Waning protection after fifth dose of acellular pertussis vaccine in children. N. Engl. J. Med. 367, 1012–1019. doi: 10.1056/NEJMoa1200850
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lamberti, Y., Cafiero, J. H., Surmann, K., Valdez, H., Holubova, J., Vecerek, B., et al. (2016). Proteome analysis of Bordetella pertussis isolated from human macrophages. J. Proteome 136, 55–67. doi: 10.1016/j.jprot.2016.02.002
Lamberti, Y., Gorgojo, J., Massillo, C., and Rodriguez, M. E. (2013). Bordetella pertussis entry into respiratory epithelial cells and intracellular survival. Pathog. Dis. 69, 194–204. doi: 10.1111/2049-632X.12072
Lamberti, Y. A., Hayes, J. A., Perez Vidakovics, M. L., Harvill, E. T., and Rodriguez, M. E. (2010). Intracellular trafficking of Bordetella pertussis in human macrophages. Infect. Immun. 78, 907–913. doi: 10.1128/IAI.01031-09
Li, X., Hu, Y., Gong, J., Zhang, L., and Wang, G. (2013). Comparative genome characterization of Achromobacter members reveals potential genetic determinants facilitating the adaptation to a pathogenic lifestyle. Appl. Microbiol. Biotechnol. 97, 6413–6425. doi: 10.1007/s00253-013-5018-3
Linz, B., Ivanov, Y. V., Preston, A., Brinkac, L., Parkhill, J., Kim, M., et al. (2016). Acquisition and loss of virulence-associated factors during genome evolution and speciation in three clades of Bordetella species. BMC Genomics 17:767. doi: 10.1186/s12864-016-3112-5
Linz, B., Ma, L., Rivera, I., and Harvill, E. T. (2019). Genotypic and phenotypic adaptation of pathogens: lesson from the genus Bordetella. Curr. Opin. Infect. Dis. 32, 223–230. doi: 10.1097/QCO.0000000000000549
Long, G. H., Karanikas, A. T., Harvill, E. T., Read, A. F., and Hudson, P. J. (2010). Acellular pertussis vaccination facilitates Bordetella parapertussis infection in a rodent model of bordetellosis. Proc. Biol. Sci. 277, 2017–2025. doi: 10.1098/rspb.2010.0010
Lund, P., Tramonti, A., and De Biase, D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 38, 1091–1125. doi: 10.1111/1574-6976.12076
Luu, L. D. W., Octavia, S., Zhong, L., Raftery, M., Sintchenko, V., and Lan, R. (2017). Characterisation of the Bordetella pertussis secretome under different media. J. Proteome 158, 43–51. doi: 10.1016/j.jprot.2017.02.010
Mahon, B. P., and Mills, K. H. (1999). Interferon-gamma mediated immune effector mechanisms against Bordetella pertussis. Immunol. Lett. 68, 213–217. doi: 10.1016/s0165-2478(99)00070-x
Mills, K. H., Ryan, M., McGuirk, P., Griffin, F., Murphy, G., and Mahon, B. (1999). The immunology of Bordetella pertussis infection. Biologicals 27:77. doi: 10.1006/biol.1999.0183
Molmeret, M., Horn, M., Wagner, M., Santic, M., and Abu Kwaik, Y. (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71, 20–28. doi: 10.1128/AEM.71.1.20-28.2005
Moon, K., Bonocora, R. P., Kim, D. D., Chen, Q., Wade, J. T., Stibitz, S., et al. (2017). The BvgAS regulon of Bordetella pertussis. mBio 8:e01526-17. doi: 10.1128/mBio.01526-17
Munoz-Elias, E. J., and McKinney, J. D. (2006). Carbon metabolism of intracellular bacteria. Cell. Microbiol. 8, 10–22. doi: 10.1111/j.1462-5822.2005.00648.x
Nagata, J. M., Charville, G. W., Klotz, J. M., Wickremasinghe, W. R., Kann, D. C., Schwenk, H. T., et al. (2015). Bordetella petrii sinusitis in an immunocompromised adolescent. Pediatr. Infect. Dis. J. 34:458. doi: 10.1097/INF.0000000000000564
Nakamura, M. M., Liew, S. Y., Cummings, C. A., Brinig, M. M., Dieterich, C., and Relman, D. A. (2006). Growth phase‐ and nutrient limitation-associated transcript abundance regulation in Bordetella pertussis. Infect. Immun. 74, 5537–5548. doi: 10.1128/IAI.00781-06
Nicholson, T. L., Buboltz, A. M., Harvill, E. T., and Brockmeier, S. L. (2009). Microarray and functional analysis of growth phase-dependent gene regulation in Bordetella bronchiseptica. Infect. Immun. 77, 4221–4231. doi: 10.1128/IAI.00136-09
Ozanic, M., Marecic, V., Abu Kwaik, Y., and Santic, M. (2015). The divergent intracellular lifestyle of Francisella tularensis in evolutionarily distinct host cells. PLoS Pathog. 11:e1005208. doi: 10.1371/journal.ppat.1005208
Panda, A., Drancourt, M., Tuller, T., and Pontarotti, P. (2018). Genome-wide analysis of horizontally acquired genes in the genus Mycobacterium. Sci. Rep. 8:14817. doi: 10.1038/s41598-018-33261-w
Park, J., Zhang, Y., Buboltz, A. M., Zhang, X. Q., Schuster, S. C., Ahuja, U., et al. (2012). Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13:545. doi: 10.1186/1471-2164-13-545
Parkhill, J., Sebaihia, M., Preston, A., Murphy, L. D., Thomson, N., Harris, D. E., et al. (2003). Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35, 32–40. doi: 10.1038/ng1227
Petrackova, D., Farman, M. R., Amman, F., Linhartova, I., Dienstbier, A., Kumar, D., et al. (2020). Transcriptional profiling of human macrophages during infection with Bordetella pertussis. RNA Biol. 17, 731–742. doi: 10.1080/15476286.2020.1727694
Porter, J. F., Connor, K., and Donachie, W. (1994). Isolation and characterization of Bordetella parapertussis-like bacteria from ovine lungs. Microbiology 140, 255–261. doi: 10.1099/13500872-140-2-255
Register, K. B., and Kunkle, R. A. (2009). Strain-specific virulence of Bordetella hinzii in poultry. Avian Dis. 53, 50–54. doi: 10.1637/8388-070108-Reg.1
Rivera, I., Linz, B., Dewan, K. K., Ma, L., Rice, C. A., Kyle, D. E., et al. (2019). Conservation of ancient genetic pathways for intracellular persistence among animal pathogenic bordetellae. Front. Microbiol. 10:2839. doi: 10.3389/fmicb.2019.02839
Robertson, G. T., and Roop, R. M. Jr. (1999). The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34, 690–700. doi: 10.1046/j.1365-2958.1999.01629.x
Rodgers, L., Martin, S. W., Cohn, A., Budd, J., Marcon, M., Terranella, A., et al. (2013). Epidemiologic and laboratory features of a large outbreak of pertussis-like illnesses associated with cocirculating Bordetella holmesii and Bordetella pertussis-Ohio, 2010-2011. Clin. Infect. Dis. 56, 322–331. doi: 10.1093/cid/cis888
Saini, V., Raghuvanshi, S., Khurana, J. P., Ahmed, N., Hasnain, S. E., Tyagi, A. K., et al. (2012). Massive gene acquisitions in Mycobacterium indicus pranii provide a perspective on mycobacterial evolution. Nucleic Acids Res. 40, 10832–10850. doi: 10.1093/nar/gks793
Salah, I. B., Ghigo, E., and Drancourt, M. (2009). Free-living amoebae, a training field for macrophage resistance of mycobacteria. Clin. Microbiol. Infect. 15, 894–905. doi: 10.1111/j.1469-0691.2009.03011.x
Samba-Louaka, A., Robino, E., Cochard, T., Branger, M., Delafont, V., Aucher, W., et al. (2018). Environmental Mycobacterium avium subsp. paratuberculosis hosted by free-living amoebae. Front. Cell. Infect. Microbiol. 8:28. doi: 10.3389/fcimb.2018.00028
Schmitz-Esser, S., Toenshoff, E. R., Haider, S., Heinz, E., Hoenninger, V. M., Wagner, M., et al. (2008). Diversity of bacterial endosymbionts of environmental acanthamoeba isolates. Appl. Environ. Microbiol. 74, 5822–5831. doi: 10.1128/AEM.01093-08
Simmons, L. A., Foti, J. J., Cohen, S. E., and Walker, G. C. (2008). The SOS regulatory network. EcoSal Plus 3, 1–30. doi: 10.1128/ecosalplus.5.4.3
Sittka, A., Pfeiffer, V., Tedin, K., and Vogel, J. (2007). The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63, 193–217. doi: 10.1111/j.1365-2958.2006.05489.x
Spellberg, B., and Edwards, J. E. Jr. (2001). Type 1/type 2 immunity in infectious diseases. Clin. Infect. Dis. 32, 76–102. doi: 10.1086/317537
Stibitz, S., Aaronson, W., Monack, D., and Falkow, S. (1989). Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature 338, 266–269. doi: 10.1038/338266a0
Taylor-Mulneix, D. L., Bendor, L., Linz, B., Rivera, I., Ryman, V. E., Dewan, K. K., et al. (2017a). Bordetella bronchiseptica exploits the complex life cycle of Dictyostelium discoideum as an amplifying transmission vector. PLoS Biol. 15:e2000420. doi: 10.1371/journal.pbio.2000420
Taylor-Mulneix, D. L., Hamidou Soumana, I., Linz, B., and Harvill, E. T. (2017b). Evolution of bordetellae from environmental microbes to human respiratory pathogens: amoebae as a missing link. Front. Cell. Infect. Microbiol. 7:510. doi: 10.3389/fcimb.2017.00510
Thakur, A., Mikkelsen, H., and Jungersen, G. (2019). Intracellular pathogens: host immunity and microbial persistence strategies. J. Immunol. Res. 2019:1356540. doi: 10.1155/2019/1356540
Trainor, E. A., Nicholson, T. L., and Merkel, T. J. (2015). Bordetella pertussis transmission. Pathog. Dis. 73:ftv068. doi: 10.1093/femspd/ftv068
Valdez, H. A., Oviedo, J. M., Gorgojo, J. P., Lamberti, Y., and Rodriguez, M. E. (2016). Bordetella pertussis modulates human macrophage defense gene expression. Pathog. Dis. 74:ftw073. doi: 10.1093/femspd/ftw073
Vandamme, P., Heyndrickx, M., Vancanneyt, M., Hoste, B., De Vos, P., Falsen, E., et al. (1996). Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Ruger and Tan 1983. Int. J. Syst. Bacteriol. 46, 849–858. doi: 10.1099/00207713-46-4-849
Vandamme, P., Hommez, J., Vancanneyt, M., Monsieurs, M., Hoste, B., Cookson, B., et al. (1995). Bordetella hinzii sp. nov., isolated from poultry and humans. Int. J. Syst. Bacteriol. 45, 37–45. doi: 10.1099/00207713-45-1-37
von Wintzingerode, F., Schattke, A., Siddiqui, R. A., Rosick, U., Gobel, U. B., and Gross, R. (2001). Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int. J. Syst. Evol. Microbiol. 51, 1257–1265. doi: 10.1099/00207713-51-4-1257
Wang, F., Grundmann, S., Schmid, M., Dorfler, U., Roherer, S., Munch, J. C., et al. (2007). Isolation and characterization of 1.2,4-trichlorobenzene mineralizing Bordetella sp. and its bioremediation potential in soil. Chemosphere 67, 896–902. doi: 10.1016/j.chemosphere.2006.11.019
Warfel, J. M., Beren, J., and Merkel, T. J. (2012). Airborne transmission of Bordetella pertussis. J. Infect. Dis. 206, 902–906. doi: 10.1093/infdis/jis443
Warfel, J. M., and Merkel, T. J. (2013). Bordetella pertussis infection induces a mucosal IL-17 response and long-lived Th17 and Th1 immune memory cells in nonhuman primates. Mucosal Immunol. 6, 787–796. doi: 10.1038/mi.2012.117
Warfel, J. M., Zimmerman, L. I., and Merkel, T. J. (2014). Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc. Natl. Acad. Sci. U. S. A. 111, 787–792. doi: 10.1073/pnas.1314688110
Weyant, R. S., Hollis, D. G., Weaver, R. E., Amin, M. F., Steigerwalt, A. G., O’Connor, S. P., et al. (1995). Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J. Clin. Microbiol. 33, 1–7. doi: 10.1128/JCM.33.1.1-7.1995
Willems, R. J., Kamerbeek, J., Geuijen, C. A., Top, J., Gielen, H., Gaastra, W., et al. (1998). The efficacy of a whole cell pertussis vaccine and fimbriae against Bordetella pertussis and Bordetella parapertussis infections in a respiratory mouse model. Vaccines 16, 410–416. doi: 10.1016/s0264-410x(97)80919-x
Yeung, K. H. T., Duclos, P., Nelson, E. A. S., and Hutubessy, R. C. W. (2017). An update of the global burden of pertussis in children younger than 5 years: a modelling study. Lancet Infect. Dis. 17, 974–980. doi: 10.1016/S1473-3099(17)30390-0
Yih, W. K., Silva, E. A., Ida, J., Harrington, N., Lett, S. M., and George, H. (1999). Bordetella holmesii-like organisms isolated from Massachusetts patients with pertussis-like symptoms. Emerg. Infect. Dis. 5, 441–443. doi: 10.3201/eid0503.990317
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Keywords: Bordetella, evolution, intracellular survival, phagocytes, transcriptome
Citation: Rivera I, Linz B and Harvill ET (2020) Evolution and Conservation of Bordetella Intracellular Survival in Eukaryotic Host Cells. Front. Microbiol. 11:557819. doi: 10.3389/fmicb.2020.557819
Received: 11 May 2020; Accepted: 28 August 2020;
Published: 15 October 2020.
Edited by:
Awdhesh Kalia, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Vikram Saini, All India Institute of Medical Sciences, IndiaCopyright © 2020 Rivera, Linz and Harvill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Israel Rivera, aXNyYWVscml2ZXJhY29sb25AZ21haWwuY29t; Eric T. Harvill, aGFydmlsbEB1Z2EuZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.