- 1Beijing Center for Disease Prevention and Control, Institute for Nutrition and Food Hygiene, Beijing, China
- 2Beijing Centers for Disease Preventive Medical Research, Beijing, China
- 3Beijing Advanced Innovation Center for Food Nutrition and Human Health, College of Veterinary Medicine, China Agricultural University, Beijing, China
Campylobacter species are zoonotic pathogens and the leading cause of bacterial enteritis worldwide. With the increase of antimicrobial resistance to fluoroquinolones and macrolides, they have been identified by the World Health Organization (WHO) as high-priority antimicrobial-resistant pathogens. There is currently little known about the prevalence and antimicrobial resistance characteristics of Campylobacter species in Beijing. In this study, we performed a 2-year surveillance of Campylobacter in Beijing, China. We used multilocus sequence typing (MLST) and antimicrobial susceptibility testing to analyze 236 Campylobacter isolates recovered from 230 clinical infectious cases in Beijing between 2017 and 2018. The Campylobacter isolation rate in diarrhea patients was 7.81%, with higher isolation rates in male patients than female patients and in autumn compared with other seasons. We identified 125 sequence types (STs) of 23 cloning complexes (CCs) among the 236 isolates, including four new alleles and 19 new STs. The most commonly isolated STs of Campylobacter jejuni were ST-22 and ST-760 (4.50%), and the most commonly isolated ST of Campylobacter coli was ST-9227 (16.67%). We also compared our isolates with clinical Campylobacter isolates from other countries in Asia, CC-353 of Campylobacter coli was found in eight countries, CC-1034 and CC-1287 of Campylobacter coli were found only in China. All C. jejuni isolates were resistant to at least one antimicrobial. C. jejuni showed the highest rate of resistance toward ciprofloxacin (94.50%), followed by tetracycline (93.50%), and nalidixic acid (92.00%), while C. coli showed highest resistance toward ciprofloxacin (94.44%) and tetracycline (94.44%) followed by nalidixic acid (88.89%). The most commonly observed MDR combination of C. jejuni were quinolone, phenicol and tetracycline (11.50%), while the most commonly observed MDR combination of C. coli were macrolide, quinolone, phenicol, tetracycline and lincosamide (30.56%). Surveillance of molecular characterization will provide important information for prevention of Campylobacter infection. This study enhances insight into Campylobacter infections in diarrheal patients, with relevance for treatment regimens in Beijing.
Introduction
Campylobacter species are responsible for the greatest number of bacterial-mediated diarrhea outbreaks in the world (World Health Organization, 2013), with the thermophilic Campylobacter jejuni and Campylobacter coli being the most common pathogens. The European Food Safety Agency (EFSA) reports that C. jejuni has caused the most cases of bacterial foodborne diseases in the European Union for many years (European Food Safety Agency and European centre for Disease Prevention and Control, 2015). According to statistics on the number of foodborne pathogenic microorganisms in the United States by the United States Centers for Disease Control and Prevention (USA-CDC) in 2016, the number of cases caused by C. jejuni ranked highest (Centers for Disease Control and Prevention [CDC], 2017). In addition, Campylobacter infection can lead to extraintestinal complications and campylobacteriosis such as Guillain Barré syndrome, reactive arthritis, and irritable bowel syndrome (Nachamkin et al., 1998; Zia et al., 2003; Connor, 2005). An outbreak of C. jejuni resulted in 32 Guillain Barré syndrome patients in Shuangyang District, Changchun City, Jilin Province, China, during 2007 (Zhang et al., 2010b).
Campylobacter can colonize poultry, swine, and ruminants (Gallay et al., 2007; Elhadidy et al., 2018); therefore, people are easily infected when handling and consuming raw and semi-raw contaminated foods, as well as contaminated water (Klein-Jöbstl et al., 2016; Kovanen et al., 2016). Most Campylobacter infections are mild, self-limiting, and resolve within a few days. However, effective antimicrobial treatment is essential for people with severe or prolonged Campylobacter infections, and the elderly, young, or immunocompromised patients (Reddy and Zishiri, 2017). Clinical treatment of gastroenteritis caused by Campylobacter is usually performed with macrolides and fluoroquinolones; however, occasionally, severe systemic infection requires the use of aminoglycoside antimicrobials, such as gentamicin (Blaser and Engberg, 2008; Pernica et al., 2016). Campylobacter isolates from both developed and developing countries show resistance to several antimicrobials, including aminoglycosides, fluoroquinolones, macrolides, and tetracyclines (Padungton and Kaneene, 2003; Ruiz-Palacios, 2007; Ge et al., 2013; Abdi-Hachesoo et al., 2014; Shobo et al., 2016; Reddy and Zishiri, 2017), which led the World Health Organization in 2017 to list Campylobacter spp. as one of the six high-priority antimicrobial-resistant pathogens (World Health Organization, 2017). Therefore, identifying the molecular characteristics of resistant isolates and estimating their frequency in different populations is important for disease prevention aimed at controlling the emergence of resistant isolates and detecting resistant infections rapidly.
Molecular typing of bacteria plays an important role in the epidemiological investigation of pathogen transmission pathways by identifying outbreaks and diseases (O’Mahony et al., 2011). However, only a few studies have investigated the isolation rate and molecular characterization of clinical Campylobacter spp. in China (Zhang et al., 2010a, 2014; Wei et al., 2015; Pan et al., 2016; Ju et al., 2018). To understand the genotypes and antimicrobial resistance of clinical Campylobacter isolates in Beijing, we combined antimicrobial susceptibility tests and molecular subtyping analysis with epidemiological information.
Materials and Methods
Study Population and Campylobacter Isolation
From 2017 to 2018, Campylobacter isolates were obtained from 2,945 patients with acute diarrhea in 14 hospitals in pilot areas (Xicheng, Fengtai, Shunyi, Fangshan, Yanqing, and Mentougou Districts) of a Campylobacter surveillance program in Beijing. Patients were defined as having ≥3 watery, loose, mucosal, or bloody stools over a 24-h period. Five milligrams of fresh stool sample was collected from each of the diarrheal patients. The samples were maintained in Cary-Blair medium (Oxoid CM0935), at 4°C and transported to the laboratory for bacterial isolation within 24 h. A Campylobacter isolation kit incorporating a membrane filter method (ZC-CAMPY-002, Qingdao Sinova Biotechnology Co., Ltd., Qingdao, China) was used to isolate Campylobacter. Briefly, 1 mL of fecal specimen suspension was transferred to 4 mL of enrichment medium provided in the kit. The main component of the enrichment medium was a modified Preston broth. The enrichment medium was then cultured at 42°C for 48 h in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2). Approximately 300 μL of enrichment medium was spotted on the membrane filter (Type: 0.45 μm) surface of the kit and spread onto Karmali and Columbia agar plates. Five (if less than 5, pick all) or more colonies resembling Campylobacter were picked after 48 h of incubation at 42°C in a microaerobic atmosphere. All isolates were firstly identified using matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry (Bruker, Leipzig, Germany) and further checked by PCR, according to a previously described method (Wang et al., 2002).
Epidemiological Data
Demographic and epidemiological data were managed using WPS Office (Kingsoft, China). All patients were residents of Beijing. Patients were divided according to age into child patients (≤5 years old), adolescent patients (6–17 years old), adult patients (18–64 years old), and elderly patients (≥65 years old). Seasons were classified according to the onset date of symptoms: spring (March, April, May), summer (June, July, August), autumn (September, October, November), and winter (December, January, February).
Multilocus Sequence Typing
MLST was performed by sequencing seven housekeeping loci (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) according to a previously described method (Dingle et al., 2001). Briefly, genomic DNA was extracted using a commercial kit (QIAamp DNA mini kit, Germany). Amplification reactions were performed in a 20 μL mixture containing 10 μL of 2 × PCR master (Sangon Biotec., Shanghai, China), 0.5 μL of forward and reverse primer, 1 μL of DNA template, and 7.5 μL of pure water. PCR was performed as 1 cycle of 5 min at 94°C; 35 cycles of 1 min at 94°C, 2 min at 50°C, and 1 min at 72°C; and a final extension of 5 min at 72°C. After amplification, 2 μL of the PCR product was subjected to electrophoresis through a 1.5% agarose gel. The PCR products were then purified and sequenced (3730xl DNA Analyzer, Applied Biosystems, United States). The alleles of each gene were determined from a comparison of seven alleles in the MLST database1. For each isolate, the number of alleles was assigned and sequence types (STs) and cloning complexes (CCs) were determined. New alleles and STs were submitted to the PubMLST database. A minimum spanning tree (MST) and dendrogram of MLST data was created using BioNumerics v.7.6 (bioMérieux, Marcy-l’Étoile, France).
Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) of all C. jejuni isolates was determined according to the agar dilution method recommended by CLSI using a commercial kit (Zhongchuang Biotechnology Ltd. Corp., Qingdao, China). Six classes of antimicribials were chosen for this study: macrolides, fluoroquinolones, aminoglycosides, chloramphenicols, tetracyclines, and lincosamides. The breakpoints for resistance used in this study were based on standards used in the National Antimicrobial Resistance Monitoring System (NARMS, Page last reviewed: March 15, 2019). The following MIC values were determined for C. jejuni: erythromycin (≥8 μg mL–1), azithromycin (≥0.5 μg mL–1), nalidixic acid (≥32 μg mL–1), ciprofloxacin (≥1 μg mL–1), gentamicin (≥4 μg mL–1), streptomycin (≥16 μg mL–1), chloramphenicol (≥32 μg mL–1), florfenicol (≥8 μg mL–1), tetracycline (≥2 μg mL–1), telithromycin (≥8 μg mL–1), and clindamycin (≥1 μg mL–1). MIC values for C. coli were as follows: erythromycin (≥16 μg mL–1), azithromycin (≥1 μg mL–1), nalidixic acid (≥32 μg mL–1), ciprofloxacin (≥1 μg mL–1), gentamicin (≥4 μg mL–1), streptomycin (≥16 μg mL–1), chloramphenicol (≥32 μg mL–1), florfenicol (≥8 μg mL–1), tetracycline (≥4 μg mL–1), telithromycin (≥8 μg mL–1), and clindamycin (≥2 μg mL–1). Multi-drug resistance (MDR) was defined as resistance to three or more classes of antimicrobials in this study. C. jejuni ATCC 33560 was used as a control.
Statistical Analysis
SPSS software, version 20.0 (SPSS, Inc.) was used to analyze any significant statistical differences in the results. Differences in the frequencies of Campylobacter isolation rate across STs, CCs, and MDR isolates, and other variables, including epidemiological data, were examined using χ2 and Fisher’s exact tests for dichotomous variables; p < 0.05 was considered significant.
Results
Description of Campylobacter Cases Identified and Prevalence of Campylobacter spp. in Beijing
We collected 2,945 stool samples from individual diarrhea patients in 14 hospitals in Beijing during 2017 and 2018; 236 Campylobacter isolates were recovered, comprising 200 isolates of C. jejuni (84.75%, 200/236) and 36 isolates of C. coli (15.25%, 36/236). There were 230 positive cases of Campylobacter infection, including five cases with co-infections. Among these, three cases were co-infected with C. jejuni and C. coli, one case was co-infected with two C. jejuni isolates, and one case was co-infected with two C. jejuni isolates and one C. coli isolate.
The isolation rate of Campylobacter in diarrhea patients was 7.81% (230/2945). Of these patients, 148 were male (64.35%) while 82 were female (35.65%). Patients ranged in age from 3 to 91 years old. Campylobacter isolation rate was higher in autumn than in other seasons (spring, χ2 = 3.4909, P < 0.0682; summer, χ2 = 6.8528, P < 0.0105; winter, χ2 = 6.7567, P < 0.0130), and higher in male than in female (χ2 = 8.6054, P < 0.0037) (Table 1). No statistically significant difference was found in the Campylobacter isolation rate between the child and other age groups (adolescent, χ2 = 0.9406, P < 0.3321; adult, χ2 = 0.3216, P < 0.5707; elderly, χ2 = 0.0012, P < 0.9719).
Multilocus Sequence Typing
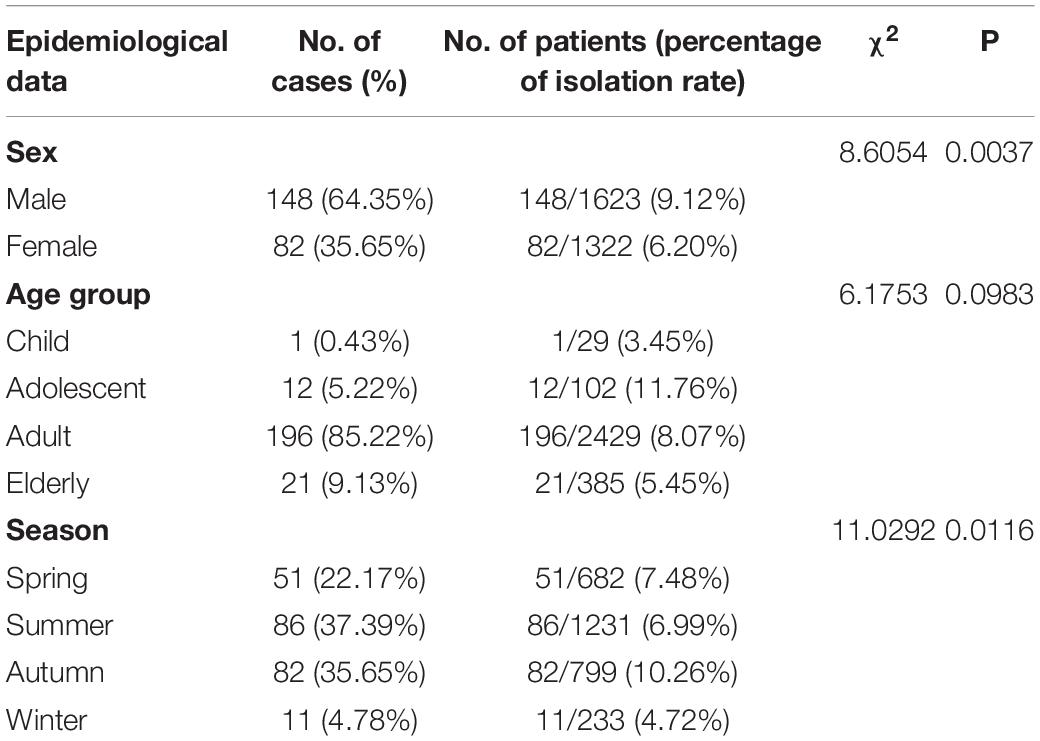
We identified 125 STs of 23 CCs among the 236 isolates, including four new alleles and 19 new STs. The new STs are shown in Supplementary Table S1. Thirty STs from 53 isolates did not belong to any known clonal complex. Among the 200 isolates of C. jejuni, we recovered 103 STs. The most commonly isolated STs were ST-22 and ST-760 (9/200, 4.50%), followed by ST-6500 (7/200, 3.50%), and ST-653 (6/200, 3.00%); 59 STs had only one isolate (Figure 1). The most commonly isolated CC was CC-21 (42/200, 21.00%). Among 36 isolates of C. coli, we recovered 22 STs. The most commonly isolated was ST-9227 (6/36, 16.67%). CC-828 was the only CC found in C. coli (30/36, 83.33%), and six isolates did not belong to any known CC (Figure 2).
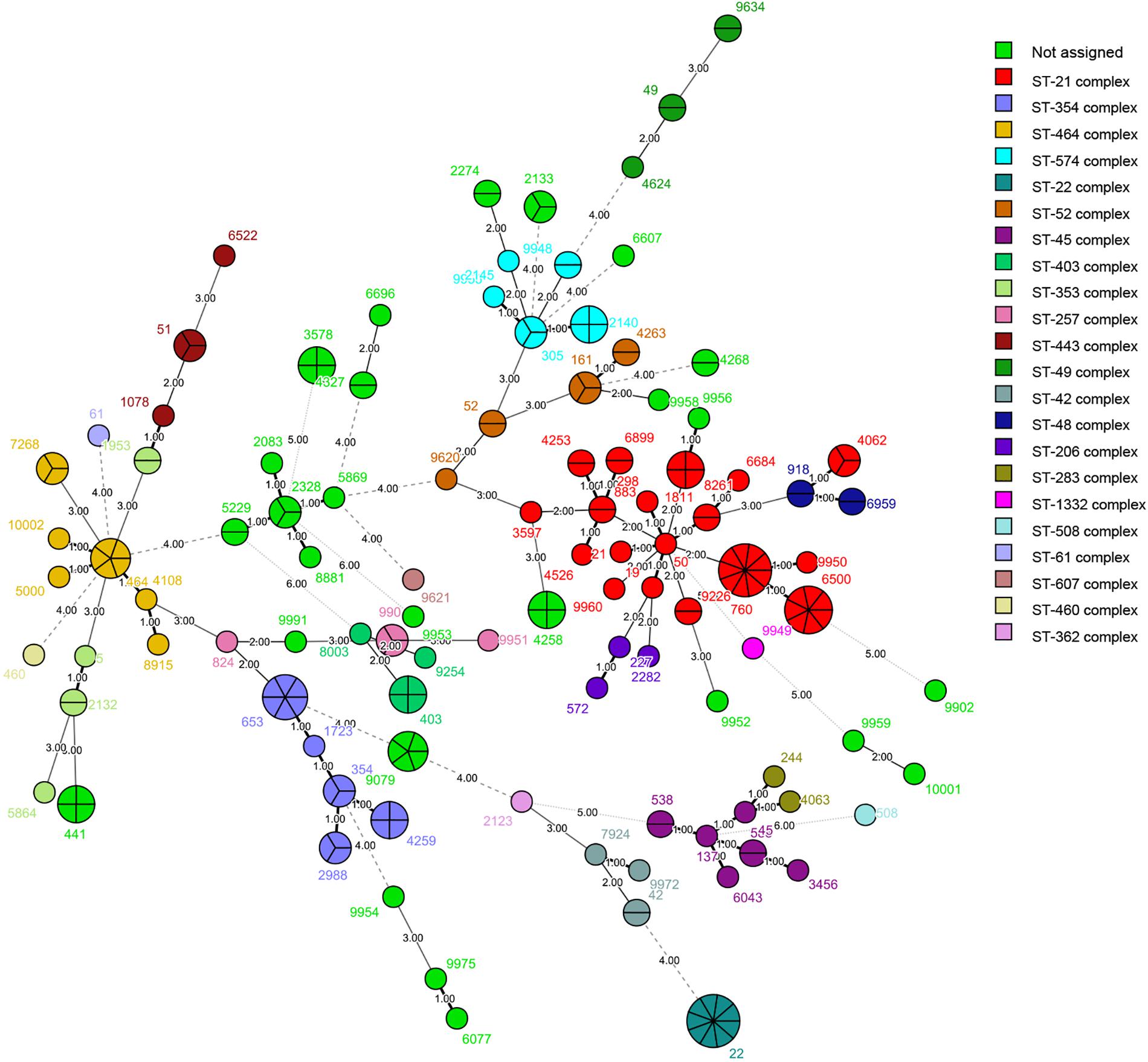
Figure 1. Phylogenetic analysis. Minimum spanning tree (MST) of MLST data of 200 C. jejuni human isolates collected in 2017 and 2018 in Beijing, China. Each color represents one clonal complex (CC). Isolates are represented by circles, and the size of the circle is proportional to the number of isolates. Branches and numbers represent allelic differences between isolates.
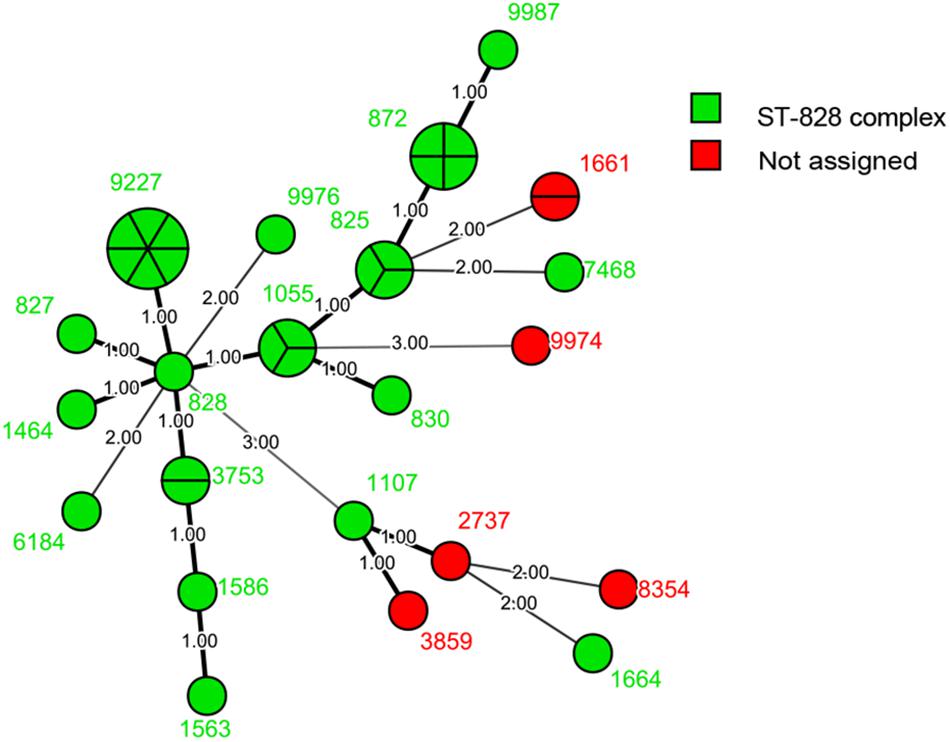
Figure 2. Phylogenetic analysis. Minimum spanning tree (MST) of MLST data of 36 C. coli human isolates collected in 2017 and 2018 in Beijing, China. Green represents the ST-828 complex and red represents isolates that do not belong to any known clonal complex. Isolates are represented by circles, and the size of the circle is proportional to the number of isolates. Branches and numbers represent allelic differences between isolates.
Comparison of Isolates From This Study With Those From Other Districts in China
We compared the distribution of Campylobacter isolates from diarrhea patients from Beijing with those from patients from other districts in China using MLST allelic profiles available from previous reports (Zhang et al., 2010a, 2014, 2015; Wei et al., 2015; Pan et al., 2016; Ju et al., 2018). We constructed dendrograms for C. jejuni (Figure 3) and C. coli (Figure 4) isolates. In total, 187 STs were recovered from 376 isolates of C. jejuni. There were no obvious regional characteristics for these STs: ST-5 (n = 6) was recovered from four districts while ST-22 (n = 14, most common), ST-51 (n = 8), ST-2328 (n = 7), ST-354 (n = 6), ST-2274 (n = 6), ST-1811 (n = 4), and ST-2132 (n = 4) were recovered from three districts of China. A total of 47 STs were recovered from 114 isolates of C. coli. Among these, nine STs were found in both Beijing and Shanghai City. CC-21 (57/376, 15.16%) and CC-828 (95/114, 83.33%) were the most commonly isolated CCs for C. jejuni and C. coli, respectively.
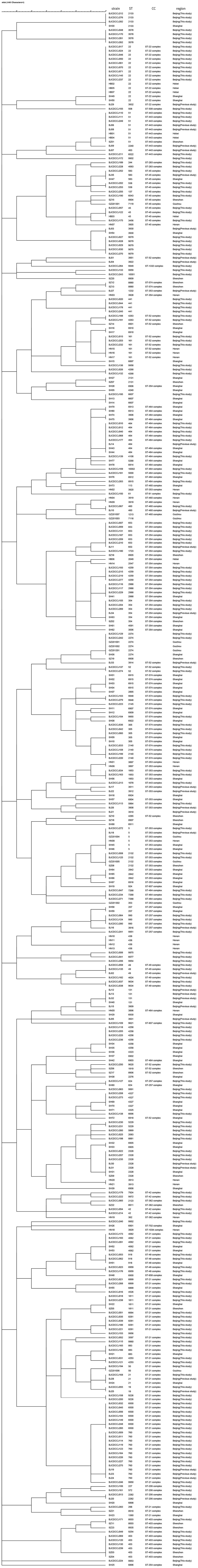
Figure 3. Dendrogram of MLST data of C. jejuni human isolates collected from this study and other district of China, Showed the number of differences in MLST alleles. The similarity coefficient were calculated using the categorical (difference) and complete linkage for the cluster analysis.

Figure 4. Dendrogram of MLST data of C. coli human isolates collected from this study and other district of China, showed the number of differences in MLST alleles. The similarity coefficient were calculated using the categorical (difference) and complete linkage for the cluster analysis.
Comparison of Isolates With Those From Other Countries in Asia
We compared our isolates with clinical Campylobacter isolates from other countries in Asia by selecting 1,454 Campylobacter isolates from patients with diarrhea from the PubMLST database (see text footnote 1) on January 18, 2019: 1336 C. jejuni and 118 C. coli isolates. The 1,536 human C. jejuni isolates were divided into 631 STs and 30 CCs, with 22 of these CCs found in this study. The most prevalent CCs were CC-21 (259/1536, 16.86%), CC-52 (105/1536, 6.84%), CC-353 (104/1536, 6.77%), CC-354 (102/1536, 6.64%), CC-574 (100/1536, 6.51%), CC-464 (67/1536, 4.36%), CC-257 (53/1536, 3.45%), and CC-22 (53/1536, 3.45%). CC-353 was found in eight countries, CC-21, CC-443 (41/1536, 2.67%), CC-45 (37/1536, 2.41%), and CC-574 were found in seven countries, and CC-206 (45/1536, 2.93%) and CC-257 were found in six countries. CC-1034 and CC-1287 were found only in China, CC-692 and CC-177 were found only in Thailand, and CC-41 and CC-446 were found only in Bangladesh and Israel, respectively (Figure 5). The 154 human C. coli isolates were divided into 95 STs and 2 CCs. One hundred and thirty-one isolates belonged to CC-828, 3 isolates from different countries belonged to CC-1150, and the remaining isolates did not belong to any known CC (Figure 6).
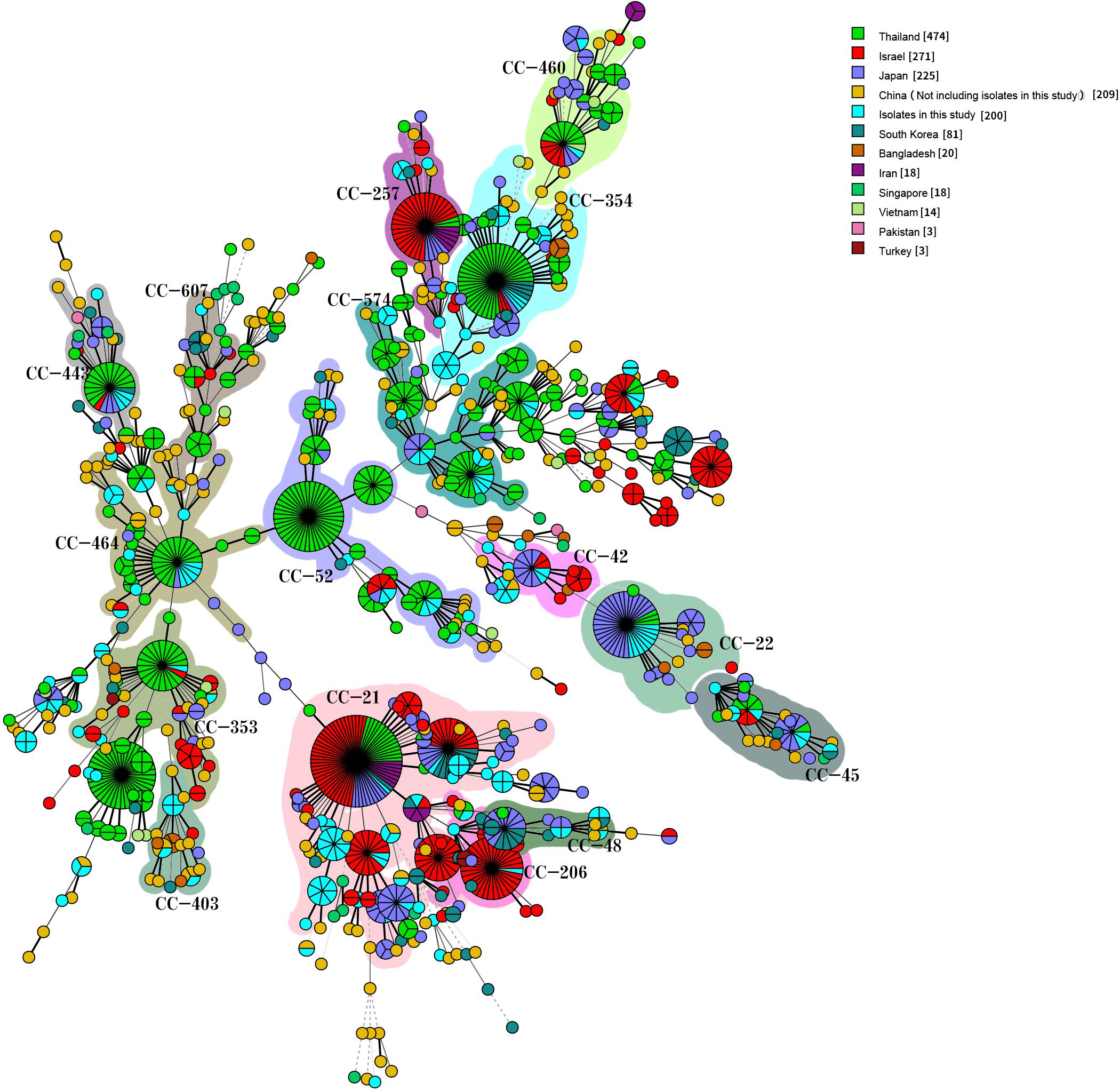
Figure 5. Genetic relationships among the 200 clinical isolates C. jejuni in this study and 1,336 isolates C. jejuni from Asia in the pubmlst database (Data taken time: January 18, 2019). A minimum spanning tree was reconstructed based on CCs from this study and the MLST database. The size of circles is proportional to the number of isolates, and the sources of the isolates are colored as indicated. Shadow zones in different colors represent different clonal complexes.
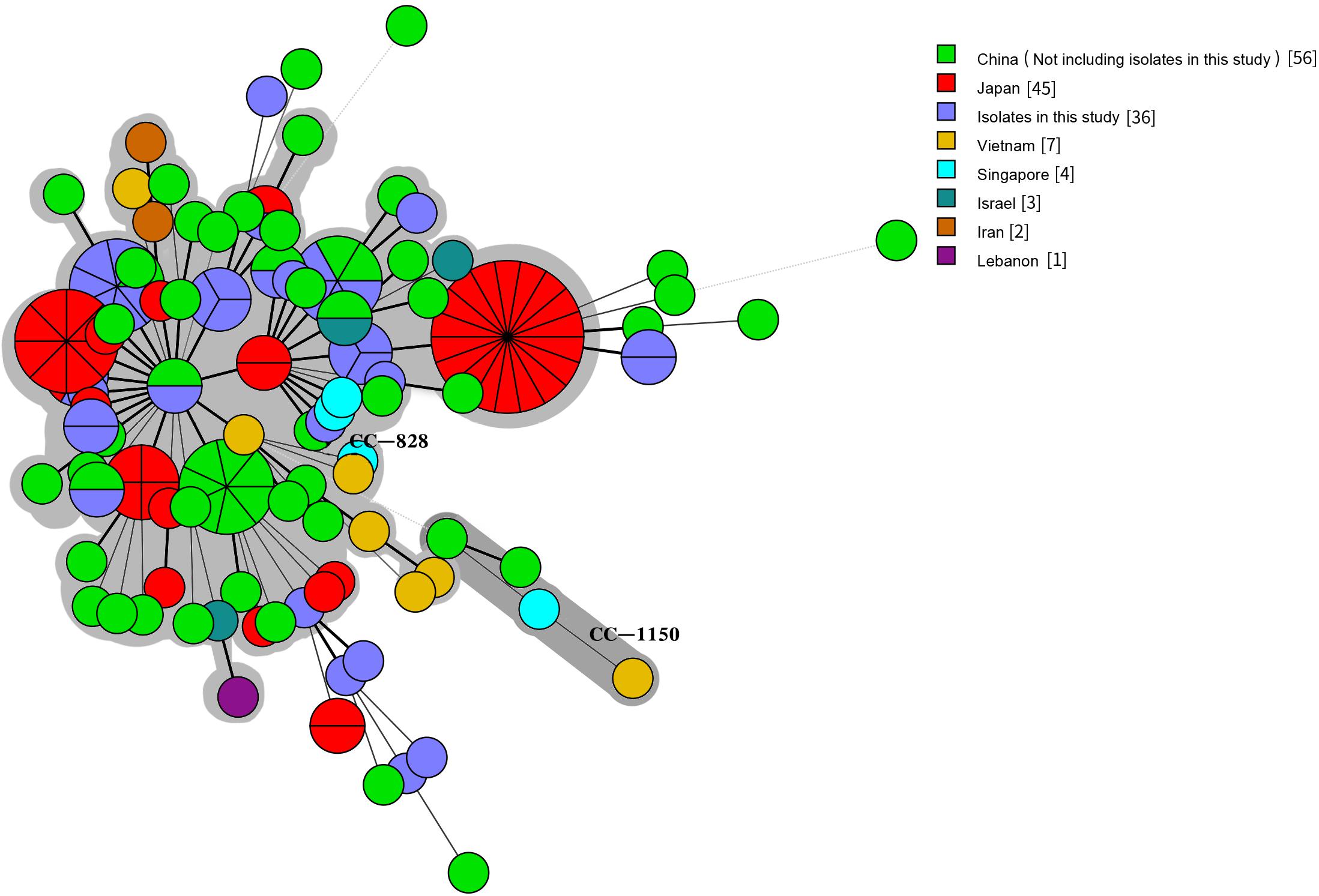
Figure 6. Genetic relationships among the 36 clinical isolates C. coli in this study and 118 isolates C. coli from Asia in the pubmlst database (Data taken time: January 18, 2019). A minimum spanning tree was reconstructed based on CCs from this study and the MLST database. The size of circles is proportional to the number of isolates, and the sources of isolates are colored as indicated. Shadow zones in different color represent different clonal complexes.
Antibacterial Susceptibility Testing
The MICs of the quality control isolate (C. jejuni ATCC 33560) were within the reference quality control range. No C. jejuni isolate was susceptible to all 11 antimicrobials tested; however, 179 C. jejuni isolates (89.50%) were resistant to three or more antimicrobials and two isolates were susceptible to only one antimicrobial (erythromycin and gentamicin, respectively). C. jejuni showed highest resistance to ciprofloxacin (94.50%), followed by tetracycline (93.50%), and nalidixic acid (92.00%) (Table 2). The most prevalent resistance pattern was a combination of nalidixic acid, ciprofloxacin, and tetracycline (33.33%) (Supplementary Figure S1). C. coli showed highest resistance to ciprofloxacin (94.44%) and tetracycline (94.44%), followed by nalidixic acid (88.89%). Thirty-five C. coli isolates (97.22%) were resistant to three or more antimicrobials, three isolates were resistant to all antimicrobials tested except chloramphenicol, and nine isolates were resistant to all antimicrobials except chloramphenicol and flofenicol; this was the most common resistance pattern. No C. coli isolates were resistant to chloramphenicol and one isolate was susceptible to all 11 antimicrobials (Supplementary Figure S2).
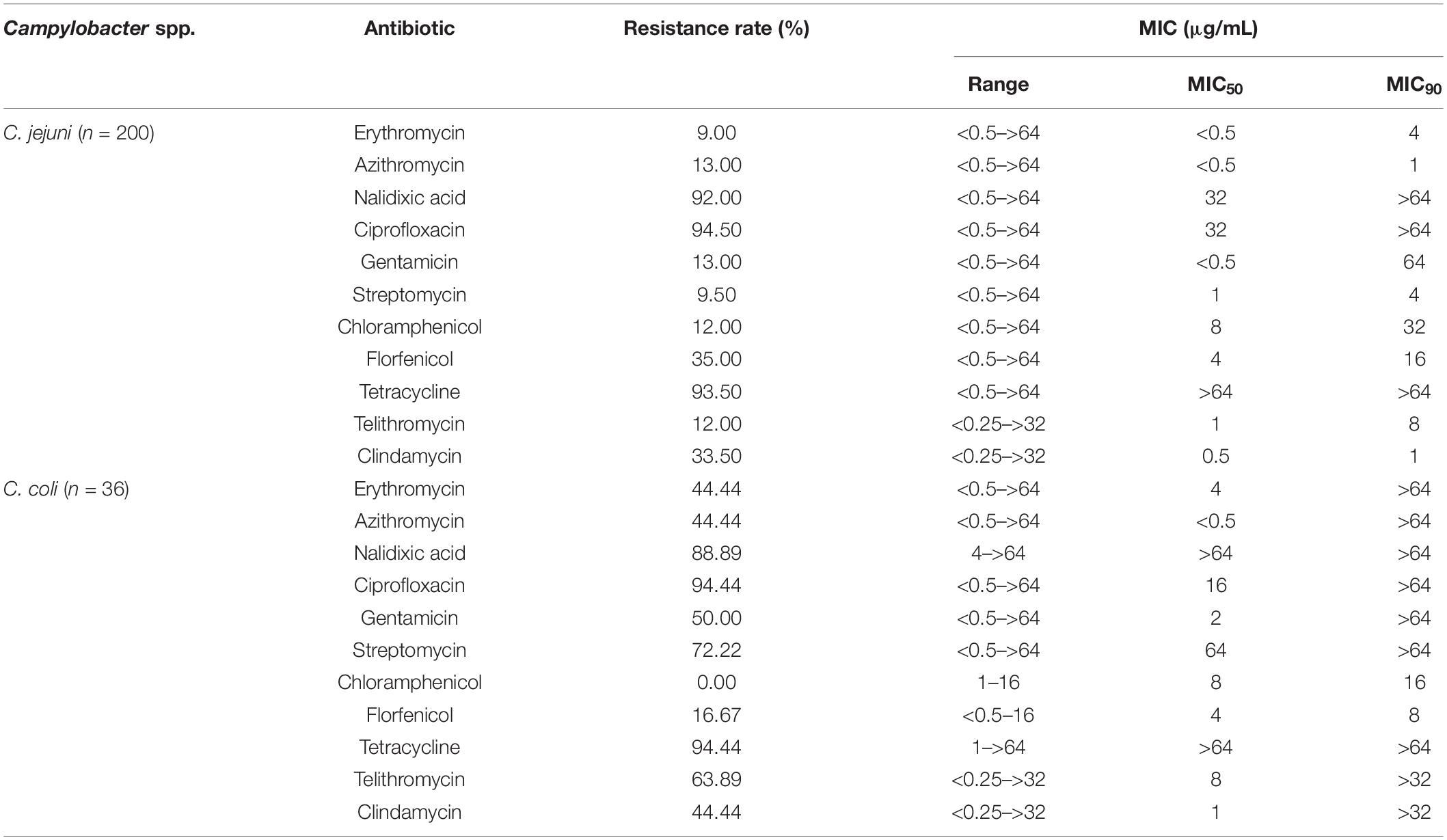
Table 2. Resistance rates (percentages) and minimum inhibitory concentrations (MICs) of human Campylobacter spp. isolates in Beijng.
The MIC50 and MIC90 values for Campylobacter are shown in Table 2. Statistical differences for antibacterial susceptibility between different antimicrobials are shown in Supplementary Table S2. Statistically, C. jejuni exhibited lower resistance rates to erythromycin (χ2 = 31.0821, P < 0.0001), azithromycin (χ2 = 20.6197, P < 0.0001), gentamicin (χ2 = 27.5356, P < 0.0001), streptomycin (χ2 = 77.7753, P < 0.0001), and telithromycin (χ2 = 51.5031, P < 0.0001) than C. coli, while the C. coli resistance rate to florfenicol (χ2 = 4.6967, P = 0.0332) was lower than that of C. jejuni. In addition, among 8–10 antimicrobial-resistant isolates, the isolation rate of C. coli was higher than that of C. jejuni (χ2 = 26.2338, P < 0.0001).
For multi-drug resistance (MDR), 110 C. jejuni isolates (55.00%) were resistant to three or more classes of antimicrobials. Among these, 23 isolates (11.50%) were resistant to quinolone, phenicol and tetracycline, simultaneously 21 isolates (10.50%) were resistant to quinolone, phenicol, tetracycline and lincosamide. Seven isolates (3.50%) were resistant to all six classes of antimicrobials. We also recovered 27 MDR isolates of C. coli (75.00%), of which 11 isolates (30.56%) were resistant to macrolide, quinolone, phenicol, tetracycline, and lincosamide. Three isolates (8.33%) were resistant to all six classes of antimicrobials.
We detected 81 STs representing 19 CCs from 137 MDR isolates. Among these, ST-872, ST-1055, ST-2140, ST-9959, and ST-9976 were resistant to 10 antimicrobials (ST-872, ST-1055, and ST-9976 were sensitive to chloramphenicol. ST-2140 and ST-9959 were sensitive to gentamicin and erythromycin, respectively). Moreover, all ST-9226 isolates (n = 3) were resistant to 8–9 antimicrobials (sensitive to streptomycin, chloramphenicol, and florfenicol). All ST-1811 isolates (n = 2) were resistant to six antimicrobials (both isolates were resistant to nalidixic acid, ciprofloxacin chloramphenicol, florfenicol, and tetracycline, however one isolate was resistant to clindamycin and the other was resistant to gentamicin). Eleven isolates of CC-828 (11/30, 36.67%) were resistant to 8–10 antimicrobials, and three isolates (ST-403) of CC-403 (3/6, 50.00%) were resistant to eight antimicrobials. This suggested that the above STs and CCs are related to MDR of Campylobacter spp.
Discussion
Campylobacter is one of the major causes of gastroenteritis in the world. The transmission chain of Campylobacter spp. is not completely known but chickens are considered to be the major reservoir for transmission to humans. Additional sources of infection are likely to include red meat, unpasteurized milk, and contaminated water (Mylius et al., 2007; Pernica et al., 2016). The present study provides more complete and updated information about the prevalence, antimicrobial resistance, and genetic diversity of Campylobacter isolates during 2017 and 2018 in Beijing. Campylobacter infections can be fatal among very young children (World Health Organization, 2020). Several studies of Campylobacter prevalence in children have been conducted previously. For instance, Zhu et al. (2016) reported that 2.9% of children with diarrhea were infected by Campylobacter in Wuhan (southwest China). Ju et al. (2018) reported that 4.0% of child patients with diarrhea were infected by Campylobacter in Shenzhen (southern China). Our study revealed a Campylobacter prevalence of 3.45% (1/29) in child patients with diarrhea, which was not statistically significantly different from other age groups. Moreover, Wang et al. (2015) reported no Campylobacter infections in child patients with diarrhea in Beijing. The discrepancy in prevalence may be caused by the different choice of surveillance hospitals. Only one of the 19 hospitals selected in this study is a children’s hospital. Our results indicated that the isolation rate of Campylobacter is higher in autumn than in other seasons. We suggest that attention should be paid to food hygiene in this season to reduce the risk of foodborne diseases.
In this study, we also highlighted the higher genetic diversity of Campylobacter isolates circulating in Beijing. We identified a total of 125 STs including 19 new STs, with the new STs reaching ST-10002 in this study. Data from an earlier study involving patient diarrheal isolates of Campylobacter collected in China revealed that the most commonly isolated ST in Hebei province is ST-22, consistent with our study. However, the most commonly isolated STs in Guizhou province, Henan province, Shanghai city, and Shenzhen city are ST-2274, ST-436, ST-6915, and ST-403, respectively (Zhang et al., 2010a; Wei et al., 2015; Pan et al., 2016; Ju et al., 2018). This difference in STs may be due to the low number of isolates. The most commonly isolated CCs in this study were CC-21 (42/200, 21.00%) and CC-828 (30/36, 83.33%) from C. jejuni and C. coli, respectively. Many studies have shown different major CCs of Campylobacter in different countries and regions, but CC-21, CC-45, CC-48, and CC-353 are the major CCs containing the largest number of isolates in many studies (O’Mahony et al., 2011; Klein-Jöbstl et al., 2016; Kovanen et al., 2016; Pernica et al., 2016; Dunn et al., 2018; Rokney et al., 2018). Studies have also shown that CC-21 is the most commonly isolated CC of C. jejuni in China (Zhang et al., 2010a, 2014, 2015; Wei et al., 2015; Pan et al., 2016; Ju et al., 2018), including human and chicken isolates. MLST analyses of Campylobacter in Asian diarrhea patients show that CC-21 is the most commonly isolated CC of C. jejuni in China, Israel, Iran, Japan, and South Korea. CC-1150 of C. coli has been found in Vietnam, Singapore, and China. These data highlight the diversity of Campylobacter spp. in Beijing and confirm previous MLST studies with C. jejuni and C. coli isolates from humans in China.
ST-2274 of C. jejuni and ST-9227 of C. coli caused outbreaks of foodborne diseases resulting in seven and three patients with diarrhea in Beijing, respectively (Qu et al., 2019; Li et al., 2020). ST-2274 was found in Thailand and Guizhou province and Shanghai city of China. ST-9227 has not been reported in the PubMLST database in Asia except in Beijing. An outbreak of Guillain Barré syndrome caused by C. jejuni was previously reported in Changchun City, Jilin Province, China, and the strains isolated from patients were all ST-2993 (Zhang et al., 2010b). However, no other ST-2993 isolates in the PubMLST database were from other country of Asia.
Antimicrobial resistance of Campylobacter is receiving more attention. Infection with antimicrobial-resistant isolates is often associated with longer disease duration, higher risk of invasive disease, and higher healthcare costs (Lopes et al., 2019). The use of antimicrobials in animal agriculture and human medicine can affect the development of resistance in Campylobacter. It is believed that the unrestricted use of antimicrobials, especially in developing countries, has led to increased resistance to Campylobacter spp. (Gibreel and Taylor, 2006; Deng et al., 2015). In some countries, in contrast to therapeutic agents, antibacterial agents are still used as growth promoters (Osterlund et al., 2003; Gibreel and Taylor, 2006; Qin et al., 2011; Wang et al., 2014). Antimicrobial-resistant strains of Campylobacter are a zoonotic pathogens that can be transmitted to humans through contaminated food, water, or milk, seriously jeopardizing treatment (Smith et al., 1999; Piddock et al., 2000; Engberg et al., 2004; Smith and Fratamico, 2010; Schweitzer et al., 2011). Determination of the antimicrobial resistance level of Campylobacter isolates is crucial for the control and prevention of human infection, particularly for cases in which therapy is recommended. Macrolides and fluoroquinolones are the primary choice for treating campylobacteriosis (Chen et al., 2010). However, an increase in erythromycin-resistant Campylobacter in poultry has been observed in China recently (Zhang A. et al., 2016). In our study, 44.44% of C. coli were resistant to erythromycin. We also found that 75.00% of C. coli were MDR isolates and 11 isolates (30.56%) were resistant to macrolide, quinolone, phenicol, tetracycline, and lincosamide. Therefore, the clinical treatment of campylobacteriosis caused by C. coli should be carefully reconsidered. The overall resistance level of C. jejuni is not as high as that of C. coli, but it cannot be ignored. High resistance rates of C. jejuni to quinolone and tetracycline, with a high prevalence of MDR, has been reported in China previously (Zhang T. et al., 2016; Li et al., 2017). In our study, 55.00% of C. jejuni were MDR isolates, with quinolone, phenicols, and tetracycline the most commonly observed MDR combination (11.50%), followed by quinolone, phenicol, tetracycline and lincosamides (10.50%). A significant association between ST-464 and resistance to ciprofloxacin, nalidixic acid, and tetracycline (cip-nal-tet) has been reported (Cha et al., 2016). Five isolates of ST-464 were recovered in our study, all of which were resistant to cip-nal-tet. One study also claims that ST-403 has strong antimicrobial resistance (Ju et al., 2018), which was shown in our study. Some studies, such as in Michigan and Iran (Cha et al., 2016; Divsalar et al., 2019), reported low resistance rate of C. jejuni to florfenicol and clindamycin. However, Higher resistance rates of C. jejuni to these two antimicrobials have been reported in China. Pan et al. (2016) reported 21.8% of C. jejuni isolated from children with diarrhea were resistant to clindamycin. Ju et al. (2018) reported florfenicol resistance rate of C. jejuni close to 70%. A 17-year study of C. jejuni resistance in diarrhea patients of Beijing showed that the resistance rate of florfenicol and clindamycin is increasing (Zhou et al., 2016). Alarming was resistance of C. jejuni to florfenicol (35.00%) and clindamycin (33.50%) in our study. We found 17 ioslates resistant to florfenicol, clindamycin and chloramphenicol. At the same time, all 17 ioslates were resistant to cip-nal-tet, and belong to 17 different STs. The resistance pattern of these antimicrobials limits the choice of suitable antimicrobials for human Campylobacter disease. Thus, we suggest that clinicians should consider multi-drug treatment.
An isolate of C. coli (ST-825, CC-828) sensitive to all antimicrobials was isolated from an adult man who claimed to have eaten seafood at home (unpublished data). We are very curious about this result, because in addition to this isolate, other C. coli isolates were resistant to two or more classes of antimicrobials. Therefore, the detection of resistance genes in C. coli is the focus of our next research.
In China, a few studies have reported the prevalence of Campylobacter in food and animals (Zhang et al., 2015; Han et al., 2016; Zhu et al., 2017). However, descriptions of clinical Campylobacter are very limited. In this study, we described the characteristics of molecular typing and antimicrobial susceptibility profiles of clinical Campylobacter isolates in Beijing, the capital city of China. The clinical isolates analyzed in this study were recovered from a systematic surveillance program, which provided a unique opportunity to characterize Campylobacter spp. in Beijing.
Conclusion
Based on our results, attention should be drawn to the prevalence of isolates with high antimicrobial resistance rates and genetic similarity in Beijing and other districts of China. Food manufacturers should continually assess levels of contamination in food production and processing to effectively decrease Campylobacter infection and dissemination of resistant isolates in humans. Importantly, this study provides detailed insight into the phylogeny and antimicrobial resistance profiles of Campylobacter spp. in Beijing. Further surveillance is necessary to detect emerging resistance patterns and to assess the impact of strategies designed to mitigate antimicrobial resistance.
Data Availability Statement
All relevant data is contained within the article. All datasets generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The protocol was approved by the ethics committee of Beijing Center for Disease Prevention and Control (Beijing CDC). Participants received information on the study’s purpose and of their right to keep information confidential. Written consent was obtained from each participant and children’s parents or their guardians.
Author Contributions
JJ and XM were involved in the collection of isolates and collected the clinical data. YL, XZ, and PZ performed the molecular subtyping and antibiotic susceptibility tests. XZ, JJ, and PZ performed the data analysis. PZ, XZ, ZS, XM, and QC designed the study, drafted, and revised this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (Grant No. 2017YFC1601500) and the Cultivation Fund of Beijing Center for Disease Prevention and Control, Beijing Research Center for Preventive Medicine (Grant No. 2019-BJYJ-04).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.554784/full#supplementary-material
Footnotes
References
Abdi-Hachesoo, B., Khoshbakht, R., Sharifiyazdi, H., Tabatabaei, M., Hosseinzadeh, S., and Asasi, K. (2014). Tetracycline resistance genes in Campylobacter jejuni and C. coli isolated from poultry carcasses. Jundishapur J. Microbiol. 7:e12129. doi: 10.5812/jjm.12129
Blaser, M. J., and Engberg, J. (2008). “Clinical aspects of Campylobacter jejuni and Campylobacter coli infections,” in Campylobacter, 3rd Edn, eds I. Nachamkin, C. Szymanski, and M. Blaser (Washington, DC: American Society of Microbiology), 99–121. doi: 10.1128/9781555815554.ch6
Centers for Disease Control, Prevention [CDC] (2017). Campylobacter, Salmonella led Bacterial Foodborne Illnesses in 2016 (Final Update). Available online at: https://www.cdc.gov/media/releases/2017/p0420-campylobacter-salmonella.html (accessed February 10, 2020).
Cha, W., Mosci, R., Wengert, S. L., Singh, P., Newton, D. W., Salimnia, H., et al. (2016). Antimicrobial susceptibility profiles of human Campylobacter jejuni isolates and association with phylogenetic lineages. Front. Microbiol. 7:589. doi: 10.3389/fmicb.2016.00589
Chen, X., Naren, G. W., Wu, C. M., Wang, Y., Dai, L., Xia, L. N., et al. (2010). Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet. Microbiol. 144, 133–139. doi: 10.1016/j.vetmic.2009.12.035
Connor, B. A. (2005). Sequelae of traveler’s diarrhea: focus on postinfectious irritable bowel syndrome. Clin. Infect. Dis. 41, S577–S586. doi: 10.1086/432956
Deng, F., Shen, J., Zhang, M., Wu, C., Zhang, Q., and Wang, Y. (2015). Constitutive and inducible expression of the rRNA methylase gene erm (B) in Campylobacter. Antimicrob. Agents Chemother. 59, 6661–6664. doi: 10.1128/AAC.01103-15
Dingle, K. E., Colles, F. M., Wareing, D. R., Ure, R., Fox, A. J., Bolton, F. E., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39, 14–23. doi: 10.1128/JCM.39.1.14-23.2001
Divsalar, G., Kaboosi, H., Khoshbakht, R., Shirzad-Aski, H., and Ghadikolaii, F. P. (2019). Antimicrobial resistances, and molecular typing of Campylobacter jejuni isolates, separated from food-producing animals and diarrhea patients in Iran. Comp. Immunol. Microbiol. Infect. Dis. 65, 194–200. doi: 10.1016/j.cimid.2019.06.001
Dunn, S. J., Pascoe, B., Turton, J., Fleming, V., Diggle, M., Sheppard, S. K., et al. (2018). Genomic epidemiology of clinical Campylobacter spp. at a single health trust site. Microb. Genom. 4:e000227. doi: 10.1099/mgen.0.000227
Elhadidy, M., Miller, W. G., Arguello, H., Álvarez-Ordóñez, A., Duarte, A., Dierick, K., et al. (2018). Genetic basis and clonal population structure of antibiotic resistance in Campylobacter jejuni isolated from broiler carcasses in Belgium. Front. Microbiol. 9:1014. doi: 10.3389/fmicb.2018.01014
Engberg, J., Neimann, J., Nielsen, E. M., Aarestrup, F. M., and Fussing, V. (2004). Quinolone-resistant Campylobacter infections: risk factors and clinical consequences. Emerg. Infect. Dis. 10, 1056. doi: 10.3201/eid1006.030669
European Food Safety Agency, and European centre for Disease Prevention and Control (2015). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food–borne outbreaks in 2014. EFSA J. 13:4329. doi: 10.2903/j.efsa.2015.4329
Gallay, A., Prouzet-Mauléon, V., Kempf, I., Lehours, P., Labadi, L., Camou, C., et al. (2007). Campylobacter antimicrobial drug resistance among humans, broiler chickens, and pigs. France. Emerg. Infect. Dis. 13, 259–266. doi: 10.3201/eid1302.060587
Ge, B., Wang, F., Sjolund-Karlsson, M., and McDermott, P. F. (2013). Antimicrobial resistance in campylobacter: susceptibility testing methods and resistance trends. J. Microbiol. Methods 95, 57–67. doi: 10.1016/j.mimet.2013.06.021
Gibreel, A., and Taylor, D. E. (2006). Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 58, 243–255. doi: 10.1093/jac/dkl210
Han, X., Zhu, D., Lai, H., Zeng, H., Zhou, K., Zou, L., et al. (2016). Prevalence, antimicrobial resistance profiling and genetic diversity of Campylobacter jejuni and Campylobacter coli isolated from broilers at slaughter in China. Food Control 69, 160–170. doi: 10.1016/j.foodcont.2016.04.051
Ju, C. Y., Zhang, M. J., Ping, Y., Lu, J. R., Yu, M. H., Hui, C., et al. (2018). Genetic and antibiotic resistance characteristics of Campylobacter jejuni isolated from diarrheal patients, poultry and cattle in Shenzhen. Biomed. Environ. Sci. 31, 579–585. doi: 10.3967/bes2018.079
Klein-Jöbstl, D., Sofka, D., Iwersen, M., Drillich, M., and Hilbert, F. (2016). Multilocus sequence typing and antimicrobial resistance of Campylobacter jejuni isolated from dairy calves in Austria. Front. Microbiol. 7:72. doi: 10.3389/fmicb.2016.00072
Kovanen, S., Kivistö, R., Llarena, A. K., Zhang, J., Kärkkäinen, U. M., Tuuminen, T., et al. (2016). Tracing isolates from domestic human Campylobacter jejuni infections to chicken slaughter batches and swimming water using whole-genome multilocus sequence typing. Int. J. Food Microbiol. 226, 53–60. doi: 10.1016/j.ijfoodmicro.2016.03.009
Li, B., Ma, L., Li, Y., Jia, H., Wei, J., Shao, D., et al. (2017). Antimicrobial resistance of Campylobacter species isolated from broilers in live bird markets in Shanghai. China. Foodborne Pathog. Dis. 14, 96–102. doi: 10.1089/fpd.2016.2186
Li, Y., Gu, Y., Lv, J., Liang, H., Zhang, J., Zhang, S., et al. (2020). Laboratory study on the gastroenteritis outbreak caused by a multidrug-resistant Campylobacter coli in China. Foodborne Pathog. Dis. 17, 187–193. doi: 10.1089/fpd.2019.2681
Lopes, B. S., Strachan, N. J., Ramjee, M., Thomson, A., MacRae, M., Shaw, S., et al. (2019). Nationwide stepwise emergence and evolution of multidrug-resistant Campylobacter jejuni sequence type 5136, United Kingdom. Emerg. Infect. Dis. 25:1320. doi: 10.3201/eid2507.181572
Mylius, S. D., Nauta, M. J., and Havelaar, A. H. (2007). Cross–contamination during food preparation: a mechanistic model applied to chicken–borne Campylobacter. Risk Anal.: Int. J. 27, 803–813. doi: 10.1111/j.1539-6924.2006.00872.x
Nachamkin, I., Allos, B. M., and Ho, T. (1998). Campylobacter species and guillain-barré syndrome. Clin. Microbiol. Rev. 11, 555–567. doi: 10.1128/CMR.11.3.555
O’Mahony, E., Buckley, J. F., Bolton, D., Whyte, P., and Fanning, S. (2011). Molecular epidemiology of Campylobacter isolates from poultry production units in southern Ireland. PLoS One 6:e28490. doi: 10.1371/journal.pone.0028490
Osterlund, A., Hermann, M., and Kahlmeter, G. (2003). Antibiotic resistance among Campylobacter jejuni/coli strains acquired in Sweden and abroad: a longitudinal study. Scand. J. Infect. Dis. 35, 478–481. doi: 10.1080/00365540310010949
Padungton, P., and Kaneene, J. B. (2003). Campylobacter spp. in human, chickens, pigs and their antimicrobial resistance. J. Vet. Med. Sci. 65, 161–170. doi: 10.1292/jvms.65.161
Pan, H., Ge, Y., Xu, H., Zhang, J., Kuang, D., Yang, X., et al. (2016). Molecular characterization, antimicrobial resistance and Caco-2 cell invasion potential of Campylobacter jejuni/coli from young children with diarrhea. Pediatr. Infect. Dis. J. 35, 330–334. doi: 10.1097/INF.0000000000001016
Pernica, J. M., Steenhoff, A. P., Welch, H., Mokomane, M., Quaye, I., Arscott-Mills, T., et al. (2016). Correlation of clinical outcomes with multiplex molecular testing of stool from children admitted to hospital with gastroenteritis in Botswana. J, Pediatric Infect. Dis. Soc. 5, 312–318. doi: 10.1093/jpids/piv028
Piddock, L. J., Ricci, V., Stanley, K., and Jones, K. (2000). Activity of antibiotics used in human medicine for Campylobacter jejuni isolated from farm animals and their environment in Lancashire. UK. J. Antimicrob. Chemother. 46, 303–306. doi: 10.1093/jac/46.2.303
Qin, S. S., Wu, C. M., Wang, Y., Jeon, B., Shen, Z. Q., Wang, Y., et al. (2011). Antimicrobial resistance in Campylobacter coli isolated from pigs in two provinces of China. Int. J. Food Microbiol. 146, 94–98. doi: 10.1016/j.ijfoodmicro.2011.01.035
Qu, M., Zhang, M., Zhang, X., Jia, L., Xu, J., Chu, Y., et al. (2019). Molecular and epidemiologyical analysis of a Campylobacter jejuni outbreak in China, 2018. J. Infect. Dev. Ctries. 13, 1086–1094. doi: 10.3855/jidc.11408
Reddy, S., and Zishiri, O. T. (2017). Detection and prevalence of antimicrobial resistance genes in Campylobacter spp. isolated from chickens and humans. Onderstepoort J. Vet. Res. 84, 1–6. doi: 10.4102/ojvr.v84i1.1411
Rokney, A., Valinsky, L., Moran-Gilad, J., Vranckx, K., Agmon, V., and Weinberger, M. (2018). Genomic epidemiology of Campylobacter jejuni transmission in Israel. Front. Microbiol. 9:2432. doi: 10.3389/fmicb.2018.02432
Ruiz-Palacios, G. M. (2007). The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44, 701–703. doi: 10.1086/509936
Schweitzer, N., Dan, A., Kaszanyitzky, E., Samu, P., Toth, A. G., Varga, J., et al. (2011). Molecular epidemiology and antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli isolates of poultry, swine, and cattle origin collected from slaughterhouses in Hungary. J. Food Prot. 74, 905–911. doi: 10.4315/0362-028X.JFP-10-376
Shobo, C. O., Bester, L. A., Baijnath, S., Somboro, A. M., Peer, A. K., and Essack, S. Y. (2016). Antibiotic resistance profiles of Campylobacter species in the South Africa private health care sector. J. Infect. Dev. Ctries. 10, 1214–1221. doi: 10.3855/jidc.8165
Smith, J. L., and Fratamico, P. M. (2010). Fluoroquinolone resistance in Campylobacter. J. Food Prot. 73, 1141–1152. doi: 10.4315/0362-028X-73.6.1141
Smith, K. E., Besser, J. M., Hedberg, C. W., Leano, F. T., Bender, J. B., Wicklund, J. H., et al. (1999). Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. N. Engl. J. Med. 340, 1525–1532. doi: 10.1056/NEJM199905203402001
Wang, G., Clark, C. G., Taylor, T. M., Pucknell, C., Barton, C., Price, L., et al. (2002). Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40, 4744–4747. doi: 10.1128/JCM.40.12.4744-4747.2002
Wang, X., Wang, J., Sun, H., Xia, S., Duan, R., Liang, J., et al. (2015). Etiology of childhood infectious diarrhea in a developed region of China: compared to childhood diarrhea in a developing region and adult diarrhea in a developed region. PLoS One 10:e142136. doi: 10.1371/journal.pone.0142136
Wang, Y., Zhang, M., Deng, F., Shen, Z., Wu, C., Zhang, J., et al. (2014). Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob. Agents. Chemother. 58, 5405–5412. doi: 10.1128/AAC.03039-14
Wei, X., You, L., Tian, K., and Li, S. (2015). Multilocus sequence typing of Campylobacter jejuni isolates from Guizhou province. Zhonghua Liuxingbingxue Zazhi. 36, 1326–1328. doi: 10.3760/cma.j.issn.0254-6450.2015.11.029
World Health Organization (2013). The Global View of Campylobacteriosis: Report of an Expert Consultation. Utrecht: World Health Organization.
World Health Organization (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics (Final Update). Available online at: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed April 1, 2020).
World Health Organization (2020). Campylobacter. Avaliable at: https://www.who.int/news-room/fact-sheets/detail/campylobacter (accessed August 27, 2020).
Zhang, A., Song, L., Liang, H., Gu, Y., Zhang, C., Liu, X., et al. (2016). Molecular subtyping and erythromycin resistance of Campylobacter in China. J. Appl. Microbiol. 121, 287–293. doi: 10.1111/jam.13135
Zhang, T., Luo, Q., Chen, Y., Li, T., Wen, G., Zhang, R., et al. (2016). Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in Central China. Gut Pathog. 8:48. doi: 10.1186/s13099-016-0132-2
Zhang, G., Zhang, X., Hu, Y., Jiao, X. A., and Huang, J. (2015). Multilocus sequence types of Campylobacter jejuni isolates from different sources in eastern China. Curr. Microbiol. 71, 341–346. doi: 10.1007/s00284-015-0853-3
Zhang, M., Gu, Y., He, L., Ran, L., Xia, S., Han, X., et al. (2010a). Molecular typing and antimicrobial susceptibility profiles of Campylobacter jejuni isolates from north China. J. Med. Microbiol. 59, 1171–1177. doi: 10.1099/jmm.0.022418-0
Zhang, M., Li, Q., He, L., Meng, F., Gu, Y., Zheng, M., et al. (2010b). Association study between an outbreak of Guillain-Barre syndrome in Jilin, China, and preceding Campylobacter jejuni infection. Foodborne Pathog. Dis. 7, 913–919. doi: 10.1089/fpd.2009.0493
Zhang, M., Liu, X., Xu, X., Gu, Y., Tao, X., Yang, X., et al. (2014). Molecular subtyping and antimicrobial susceptibilities of Campylobacter coli isolates from diarrheal patients and food-producing animals in China. Foodborne Pathog. Dis. 11, 610–619. doi: 10.1089/fpd.2013.1721
Zhou, J. Y., Zhang, M. J., Yang, W. N., Fang, Y. Q., Wang, G. Q., and Hou, F. Q. (2016). A seventeen-year observation of the antimicrobial susceptibility of clinical Campylobacter jejuni and the molecular mechanisms of erythromycin-resistant isolates in Beijing. China. Int. J. Infect. Dis. 42, 28–33. doi: 10.1016/j.ijid.2015.11.005
Zhu, J., Yao, B., Song, X., Wang, Y., Cui, S., Xu, H., et al. (2017). Prevalence and quantification of Campylobacter contamination on raw chicken carcasses for retail sale in China. Food Control 75, 196–202. doi: 10.1016/j.foodcont.2016.12.007
Zhu, X. H., Tian, L., Cheng, Z. J., Liu, W. Y., Li, S., Yu, W. T., et al. (2016). Viral and bacterial etiology of acute diarrhea among children under 5 years of age in Wuhan. China. Chin. Med. J. Engl. 129:1939. doi: 10.4103/0366-6999.187852
Keywords: Campylobacter spp., human, MLST, antimicrobial susceptibility, China
Citation: Zhang P, Zhang X, Liu Y, Jiang J, Shen Z, Chen Q and Ma X (2020) Multilocus Sequence Types and Antimicrobial Resistance of Campylobacter jejuni and C. coli Isolates of Human Patients From Beijing, China, 2017–2018. Front. Microbiol. 11:554784. doi: 10.3389/fmicb.2020.554784
Received: 23 April 2020; Accepted: 22 September 2020;
Published: 19 October 2020.
Edited by:
Frederique Pasquali, University of Bologna, ItalyReviewed by:
Yosra A. Helmy, The Ohio State University, United StatesMarja-Liisa Hänninen, University of Helsinki, Finland
Copyright © 2020 Zhang, Zhang, Liu, Jiang, Shen, Chen and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Chen, Y2NoZW5xaWFuQDI2My5uZXQ=; Xiaochen Ma, eGlhb2NoLW1hQDEyNi5jb20=
 Penghang Zhang
Penghang Zhang Xiaoai Zhang
Xiaoai Zhang Yuzhu Liu1,2
Yuzhu Liu1,2 Zhangqi Shen
Zhangqi Shen