- Department of Plant Pathology and Microbiology, National Taiwan University, Taipei, Taiwan
Fusarium wilt of tomato caused by the ascomycetous fungus Fusarium oxysporum f. sp. lycopersici (Fol) is widespread in most tomato planting areas. Calcineurin is a heterodimeric calcium/calmodulin-dependent protein phosphatase comprised of catalytic (Cna1) and regulatory (Cnb1) subunits. Calcineurin has been studied extensively in human fungal pathogens, but less is known about its roles in plant fungal pathogens. It is known that calcineurin regulates fungal calcium signaling, growth, drug tolerance, and virulence. However, the roles of calcineurin in Fol have not yet been characterized. In this study, we deleted calcineurin CNA1 and CNB1 genes to characterize their roles in conidiation, chlamydospore formation and virulence in Fol. Our results revealed that both cna1 and cnb1 mutants show defects in calcineurin phosphatase activity, vegetative growth and conidiation as compared to the wild type. Furthermore, calcineurin mutants exhibited blunted and swollen hyphae as observed by scanning electron microscopy. Interestingly, we found that Fol calcineurin is critical for chlamydospore formation, a function of calcineurin previously undocumented in the fungal kingdom. According to transcriptome analysis, the expression of 323 and 414 genes was up- and down-regulated, respectively, in both cna1 and cnb1 mutants. Based on the pathogen infection assay, tomato plants inoculated with cna1 or cnb1 mutant have a dramatic reduction in disease severity, indicating that calcineurin has a vital role in Fol virulence. In conclusion, our findings suggest that Fol calcineurin is required, at least in part, for phosphatase activity, vegetative growth, conidiation, chlamydospore formation, and virulence.
Introduction
Calcineurin, a calcium/calmodulin-dependent serine/threonine protein phosphatase, which forms a heterodimer comprised of a catalytic A subunit (Cna) and regulatory B subunit (Cnb) is conserved in eukaryotes (Juvvadi et al., 2014). In the fungal kingdom, calcineurin is responsible for maintaining a diverse range of cellular processes such as ion homeostasis, growth, morphogenesis, stress response, and pathogenicity by activating downstream events (Park et al., 2019). In general, when fungal cells encounter external stress, the plasma membrane or cell compartment Ca2+ influx system will be activated, resulting in an increased intracellular Ca2+ concentration. In the calcineurin cascade, calmodulin (CaM) and regulatory B subunit of calcineurin act as a sensor for Ca2+ signals, then Ca2+/CaM will specifically bind to the catalytic A and regulatory B subunits of calcineurin to form a Ca2+/CaM-calcineurin complex, leading to functional phosphatase activity (Stemmer and Klee, 1994). The activated calcineurin subsequently dephosphorylates the downstream targets, such as Crz1 and Prz1, allowing their nuclear import and inducing the expression of target genes (Juvvadi and Steinbach, 2015).
In baker’s yeast Saccharomyces cerevisiae, calcineurin is required for adaptation and growth under environmental stress and at higher alkaline pH (Cyert, 2003). Previous reports revealed that calcineurin plays roles in stress response and growth at mammalian body temperature (37°C) in Cryptococcus neoformans (Park et al., 2016). Similarly, in Candida species, calcineurin is required for drug tolerance, virulence and survival in serum (Yu et al., 2015; Park et al., 2019). Moreover, calcineurin is essential for proper hyphal growth in the filamentous fungus Aspergillus fumigatus (Juvvadi et al., 2011), and is necessary for cell cycle progression in Aspergillus nidulans (Rasmussen et al., 1994).
By using RNAi strategies, the role of the catalytic subunit Cna1 was characterized in plant fungal pathogen Sclerotinia sclerotiorum and was found to be critical for sclerotial development (Harel et al., 2006). In Magnaporthe oryzae, Cna1 was found to be involved in appressorium formation, conidiation, and hyphal growth (Choi et al., 2009). Meanwhile, Cna1 plays roles in ear gall formation and morphogenesis of Ustilago maydis (Egan et al., 2009). In Ustilago hordei, null mutation in either the catalytic or regulatory subunit resulted in sensitivity to environmental stresses and severely reduced virulence in barley plants (Cervantes-Chavez et al., 2011). In addition, the calcineurin signaling pathway was involved in the morphogenetic differentiation and haustoria formation in Puccinia striiformis f. sp. tritici (Zhang et al., 2012).
However, the roles of calcineurin signaling have not been reported in the important plant fungal pathogen Fusarium oxysporum f. sp. lycopersici, a devastating pathogen that causes tomato wilt. Fusarium wilt of tomato, a soil-borne disease, is one of the limiting factors in tomato yield. Fol can produce three kinds of asexual spores including microconidia, macroconidia and chlamydospore, and these infectious propagules play critical roles in the disease cycle (Agrios, 2005). In Fol, formation of chlamydospores serves as a survival strategy during undesirable conditions. Once in favorable weather, chlamydospores will act as primary inocula to invade tomato roots, clog water flow and nutrient movement by colonizing the vascular bundles, leading to yellowed leaves and wilted plants, and eventually decreased yield of tomato. Meanwhile, the macroconidia and microconidia can be produced from stem surfaces and leaves of Fol infected tomato, which in turn serve as the secondary inocula, thereafter infect neighboring healthy plants by spreading manner (Pietro et al., 2003; Manikandan et al., 2018).
In the present study, we characterized calcineurin function through deletion of the CNA1 and CNB1 genes in Fol. Our results revealed that the calcineurin has vital functions in conidiation, chlamydospore formation, and virulence in Fol.
Materials and Methods
Strains, Media and Chemicals
F. oxysporum f.sp. lycopersici (Fol) 4287 wild type (WT) and calcineurin mutants (Table 1) were grown on potato dextrose agar (PDA) (0.4% potato starch from infusion, 2% dextrose, 1.5% agar) (BioShop, Burlington, ON, Canada). A soil medium made with 200 g horticultural substrate (Kekkila Finnish peat moss), 0.2 g CaCO3, 0.5% glucose and 700 mL ddH2O (filtered after autoclaved) was used for chlamydospore production (Chen et al., 2015). For the pharmacological inhibition test of calcineurin, PDA medium containing calcineurin inhibitor FK506 (Astellas Pharma, Tokyo, Japan) or cyclosporine A (LC Laboratories, Woburn, United States) were used. The growth temperature for all fungal strains was set at 25°C.
Generation of Calcineurin Mutants and Complementary Strains
To disrupt the FolCNA1 gene, 5′ and 3′ non-coding region (NCR) of FolCNA1 were PCR amplified from genomic DNA of Fol using primer sets JC753/JC768 (5′ NCR of FolCNA1, 618 bp) and JC769/JC756 (3′NCR of FolCNA1, 546 bp). Primers JC766/JC767 were used to amplify the hygromycine B resistance gene (hygR) from the plasmid pPK2-hphgfp (Punt et al., 1987), which also contained GFP (gfp) under control of the A. nidulans gpdA promoter (PgpdA) and trpC terminator (TtrpC). These PCR products were treated with ExoSAP-IT (USB Corp., Ohio, United States) to remove additional primers and dNTPs. The FolCNA1 gene disruption cassette (5848 bp) was generated by fusion PCR of these three PCR fragments with primers JC753/JC756 (Supplementary Figure S3A). The same approach was employed to generate FolCNB1 deletion mutants; the 5′ and 3′ non-coding region (NCR) of FolCNB1 were PCR amplified from genomic DNA of Fol using primer sets JC839/JC840 (5′ NCR of FolCNB1, 514 bp) and JC841/JC842 (3′NCR of FolCNB1, 521 bp). The FoCNB1 gene disruption cassette (5654 bp) was generated by fusion PCR of the DNA fragments of 5′ and 3′ NCR of FolCNB1 and hygR using primers JC839/JC842 (Supplementary Figure S3B).
The FolCNA1 and FolCNB1 gene disruption cassettes were introduced into Fol wild type 4287 using the protoplast transformation method to generate cna1 and cnb1 deletion mutants (Moradi et al., 2013). Transformants were confirmed by PCR using primer sets JC788/JC961, JC772/JC773, and JC922/JC827 for cna1 mutants, and JC922/JC961, JC843/JC844, and JC922/JC903 for cnb1 mutants, to identify whether the targeted DNA is successfully recombined and replaced. The observation of GFP fluorescence was also conducted to confirm those mutants (Supplementary Figure S2A). Two independent mutants of cna1 (HFW1 and LHS2) and cnb1 (HFW3 and HWF4) were obtained in this study.
For complementation analysis of the cna1 mutant, a full length of FolCNA1 genomic DNA containing 5′NCR plus CNA1 open reading frame (ORF), and its 3′ NCR were PCR amplified from genomic DNA of Fol by using primer sets JC753/JC838 (5′ NCR+ORF of FolCNA1, 3567 bp) and JC837/JC756 (3′NCR of FolCNA1, 526 bp), respectively. Moreover, primers JC836/JC767 were used to amplify the bleomycin resistance gene (BleoR) from the plasmid pRW1 (Michielse et al., 2009a). These PCR products were treated with ExoSAP-IT (USB Corp., Ohio, United States) to remove additional primers and dNTPs. The reaction of fusion PCR containing three DNA fragments were carried out with primers JC753/JC756, to generate 6655 bp complementation cassette (5′NCR-FolCNA1-BleoR-3′NCR) (Supplementary Figure S4A). The similar approach was used to generate the complementary strain of cnb1 mutant; 5′ NCR plus FolCNB1 ORF and 3′ NCR of FolCNB1 were PCR amplified from genomic DNA of Fol using primer sets JC839/JC925 (5′ NCR+ORF of FolCNB1, 2093 bp) and JC841/JC842 (3′NCR of FolCNB1, 500 bp), respectively. The complementation cassette of cnb1 mutant (5′NCR-FolCNB1-BleoR-3′NCR, 5155 bp; Supplementary Figure S4A) was generated by fusion PCR containing DNA fragments of 5′ NCR+ FolCNB1 and BleoR and the 3′ NCR of FolCNB1 with using primers JC839/JC842. The CNA1 and CNB1 complementation cassettes were then introduced into cna1 and cnb1 mutant, respectively, using the protoplast transformation method. Transformants were confirmed by PCR using primer sets JC1404/JC1405 and JC772/JC773 for cna1:CNA1 (YHH1); JC1404/JC1405 and JC843/JC844 for cnb1:CNB1 (YHH2), to verify the expected DNA recombination in complemented strains (Supplementary Figure S4B).
Colony Morphology Observation
The wild type and calcineurin mutants were streaked out from −80°C stock, cultured on PDA and incubated at 25°C for 2 weeks, and cut 5 mm in diameter with a hole punch. Agar disks containing cultures were put on PDA plates in the presence or absence of 1 μg/mL FK506 or 100 μg/mL cyclosporine A with three replicates. All plates were then incubated at 25°C for 7 days, and the diameters of fungal colonies were measured daily. Data were analyzed by two-way ANOVA and Bonferroni post-test using GraphPad Prism 5 (GraphPad Software, CA, United States).
Chlamydospore Formation
Soil medium was used to trigger Fol strains to produce chlamydospores. Conidial suspension (500 μL) at a concentration of 5 × 105 conidia/mL was inoculated into 100 mL soil medium, and agitated at 25°C for 7 days. Cultures filtered through miracloth, and mycelium-chlamydospore aggregates were resuspended in 20 mL ddH2O and homogenized with beads using a Geno/Grinder machine (SPEX SamplePrep) at 1700 rpm for 15 s.
For the morphological observation of the chlamydospore, mycelium-chlamydospore aggregates were stained with 0.3% Calcofluor white (CFW; Fluorescent Brightener 28; Sigma, St. Louis, United States) and Nile red (NR; Sigma, St. Louis, United States) for 5 min. After staining, chlamydospores were washed twice in sterile water and observed by fluorescence microscopy.
The concentration of chlamydospore was determined via a hemocytometer. Analyses were performed by one-way ANOVA and Dunnett’s post-test using GraphPad Prism 5. The aggregates were dried, and chlamydospores produced per gram of dried mycelia (spores/g) were calculated according to the formula shown below:
Calcineurin Activity Assay
To assay for calcineurin phosphatase activity, 500 μL conidia suspension of Fol wild type and calcineurin mutants at a concentration of 5 × 105 conidia/mL were cultured in PDB medium (BioShop, potato dextrose broth: 0.4% potatoes from infusion) at 25°C, and agitated for 72 h. Mycelia were frozen in liquid nitrogen and ground into powder. Protein crude extracts were prepared by homogenizing the mycelia powder in a buffer containing 50 mM Tris-HCl (pH 7.4), 1 mM EGTA, 0.2% Triton X-100, 1 mM PMSF (Roche, San Francisco, CA, United States) and 1:100 protease inhibitor cocktail was added (Roche, San Francisco, CA, United States). The homogenates were clarified by centrifugation at 5,000 × g for 10 min at 4°C and then at 20,000 × g for 60 min at 4°C. Protein concentration in the extracts were determined by Bradford’s method (Bradford, 1976). Phosphatase activity of calcineurin was assayed according to methods described previously (Perrino et al., 1995; Juvvadi et al., 2011). In brief, the phosphatase activity assay was performed in a total volume of 100 μL containing reaction mixture [25 mM Tris (pH 7.2), 25 mM MES (pH 7.0), 1 mM MnCl2, 50 mM p-nitrophenyl phosphate] and homogenate. After incubation at 30°C for 10 min, the reaction was stopped by addition 10 μL of 13% (w/v) K2HPO4. The absorbance of the samples was measured at 405 nm via microplate spectrophotometer (Spectra MAX 190, Molecular Devices, CA, United States). The difference of absorbance values between the amounts of p-nitrophenol represented the phosphatase activity of calcineurin. The experiments were performed in triplicate, and the data were analyzed by one-way ANOVA and Dunnett’s post-test using GraphPad Prism 5. The calcineurin phosphatase activity was calculated according to the formula shown below:
Microscopy Analysis
For scanning electron microscopy (SEM) observation, Fol strains were grown on PDA and incubated at 25°C for 2 weeks, and cut 5 mm in diameter with a hole punch. The agar disks were fixed with 4% glutaraldehyde for 3 h and treated with 0.1 M cacodylate for 1 h. The fixed mycelia were then dehydrated consecutively with 30, 50, 70, 85, 90, 95, and 100% ethanol for 20 min each. Final treatment was conducted with 100% acetone for 20 min, and the specimens were allowed to dry in critical-point dryer (Hitachi HCP-2, Japan) and then sputter coated with gold by ion-coater (Hitachi E101, Japan). The specimens were observed on a FE-SEM (FEI Inspect S, United States) using an accelerating voltage of 15 kV. These instruments and chemicals were provided by Technology Commons, College of Life Science, National Taiwan University.
For the observation of the hyphal compartment, strains were cultured in PDB and incubated at 25°C for 24 h. Mycelia were collected and stained with 0.3% Calcofluor white (CFW; Fluorescent Brightener 28; Sigma, St. Louis, United States) for 5 min. After staining, mycelia were washed twice in sterile water and observed by fluorescence microscopy. The average length between septum-to-septum was measured by randomly selecting 20 mycelia from each of three technical replicates. Considering of the apical growth of fungi, the hyphal tips and lateral branches were excluded in the measurement and each mycelium were fixed length at 200 μm.
Conidial Germination Rate
In order to examine the germination rate, Fol strains were streaked out from −80°C stock and grown on PDA, incubated at 25°C for 2 weeks. Conidia were collected using ddH2O, filtrated through one-layer miracloth and washed several times. Conidial suspension was adjusted to a concentration of 5 × 105 conidia/mL and incubated at 25°C with shaking at 250 rpm to keep the spores suspended. The germination rate was calculated by randomly counting 50 conidia from each of three replicates over 8 h, and germinated conidia were defined as those containing germ tubes longer than half the length of the conidia.
Plant Infection Assay
The tomato cultivar Solanum lycopersicum Farmer 301 (Known-You Seed Co., Kaohsiung, Taiwan) was used in this study. Two-week-old tomato seedlings were inoculated by submerging roots in a suspension of 5 × 106 conidia/mL of Fol wild type or calcineurin mutants for 30 min, and then transplanting into cultural substrate [vermiculite:perlite:peat moss (1:1:1)]. Tomato plants were grown in a green house under a 16/8 h light-dark photoperiod at 28°C. Disease severity was recorded at 21 dpi. Scales of disease index were graded into five degrees from 0 to 4: 0 = no symptom; 1 = one to three yellowing leaves but not withering; 2 = at least four yellowing leaves but only one to two withering; 3 = at least four yellowing leaves and three withering; and 4 = all vascular bundles are brown, plant either dead, very small or wilted. Ten plants were used for each treatment. Data were analyzed by one-way ANOVA and Dunnett’s post-test using GraphPad Prism 5. The formula of disease severity is shown below:
RNA Sequencing Analysis
Fol wild type, and cna1 (HFW1) and cnb1 (HFW3) mutants were cultured in PDB at 25°C for 14 h. For total RNA extraction, mycelia were collected and frozen in liquid nitrogen, then ground into powder. The powder was transferred to a 1.5 mL centrifuge tube followed by adding an equal volume of TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and 100 μL chloroform, mixed well and centrifuged at 14,000 rpm for 5 min at 4°C. The upper phase was collected and precipitated with isopropanol by centrifuging at 14,000 rpm for 10 min at 4°C. The supernatants were discarded, and the nucleic acid pellets were serially washed in 70 and 100% ethanol. The RNA was then re-suspended with DEPC water.
RNA sequencing was performed by Genomics BioSci &Tech. Co., Ltd, New Taipei City, Taiwan. The RNA quality analysis was conducted using Agilent 2100 Bio-analyzer and Real-Time PCR system, to confirm that the RNA samples had a RIN value ≥ 7. The oligo(dT)-attached beads were used to purify mRNA, which was fragmented for cDNA synthesis. The first strand of cDNA was synthesized by reverse transcriptase with a random primer, and double strand cDNA was synthesized by using dUTP in place of dTTP. The double strand cDNAs were subsequently purified, and single nucleotide adenine was added to the 3′ end. Then, multiple indexing adapters were ligated to both ends of the double strand cDNAs and enriched by PCR amplification. End preparation and three libraries including wild type, HFW1 and HFW3 were generated and sequenced on an Illumina HiSeq platform. To identify differentially expressed genes (DEGs), read counts were analyzed with edgeR v.3.5 package (McCarthy et al., 2012). The RNA sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE1374681 (token: gzqpigoonhgdjoh).
The up- or down-regulated DEGs having a Log2 fold change > 2 or <−2, respectively (P < 0.05), based on Student’s t-test were set as the threshold for significantly differential expression. By protein functional annotation2 and NCBI database search, we chose the genes with known or predicted function in Fol that were correlated with the fungal growth and virulence. For expression confirmation, 2 μg of total RNA was used for reverse transcription with a high-capacity cDNA reverse transcription kit (Applied Biosystems, CA, United States). qRT-PCR was carried out using 10 ng of cDNA as the template with SYBR Green PCR Master Mix (Thermo Fisher Scientific, Vilnius, Lithuania) in a StepOnePlus machine (Applied Biosystems, Foster City, CA, United States). The relative expression levels of PMC1, YVC1, EGL7, SGE1, PSD2, HOG1, CNA1, and CNB1 genes were normalized to the Actin gene of Fol, by using the 2–ΔΔCt method. Primer sequences for each gene are listed in Supplementary Table S1.
Results
Identification of Calcineurin in F. oxysporum f. sp. lycopersici
The nucleotide sequence of calcineurin catalytic subunit A (XP_018243626) and regulatory subunit B (XP_018234243) were derived from the whole genome sequence of Fusarium oxysporum f. sp. lycopersici 4287 in the previous study (Ma et al., 2010). Both CNA1 and CNB1 genes having a single copy in the genome of Fol were verified by BLAST search3. The predicted open reading frame (ORF) of CNA1 (XP_018243626) is 1,701 bp encoding a protein with 566 amino acids, which consists of several sub-domains including catalytic, calmodulin binding, calcineurin B subunit binding, and autoinhibitory domains (Supplementary Figure S1A). The predicted ORF of CNB1 (XP_018234243) is 525 bp encoding a protein with 174 amino acids, which comprises four EF-hand calcium binding domains (Supplementary Figure S1B). Sequence comparison of Fol Cna1/Cnb1 amino acids and other previously reported orthologs in other phytopathogenic fungi and model organisms revealed that Fol Cna1 shares 86% identity with the calcineurin A subunit of Magnaporthe grisea, and 87% identity with Sclerotinia sclerotiorum. The Fol Cnb1 shares 93% identity with both calcineurin B subunit of S. sclerotirum and M. grisea. Phylogenetic analysis using neighbor-joining criteria performed with MEGA software4 showed that Fol Cna1 (Figure 1A) and Cnb1 (Figure 1B) cluster with other filamentous fungi but are distant from those of yeasts.
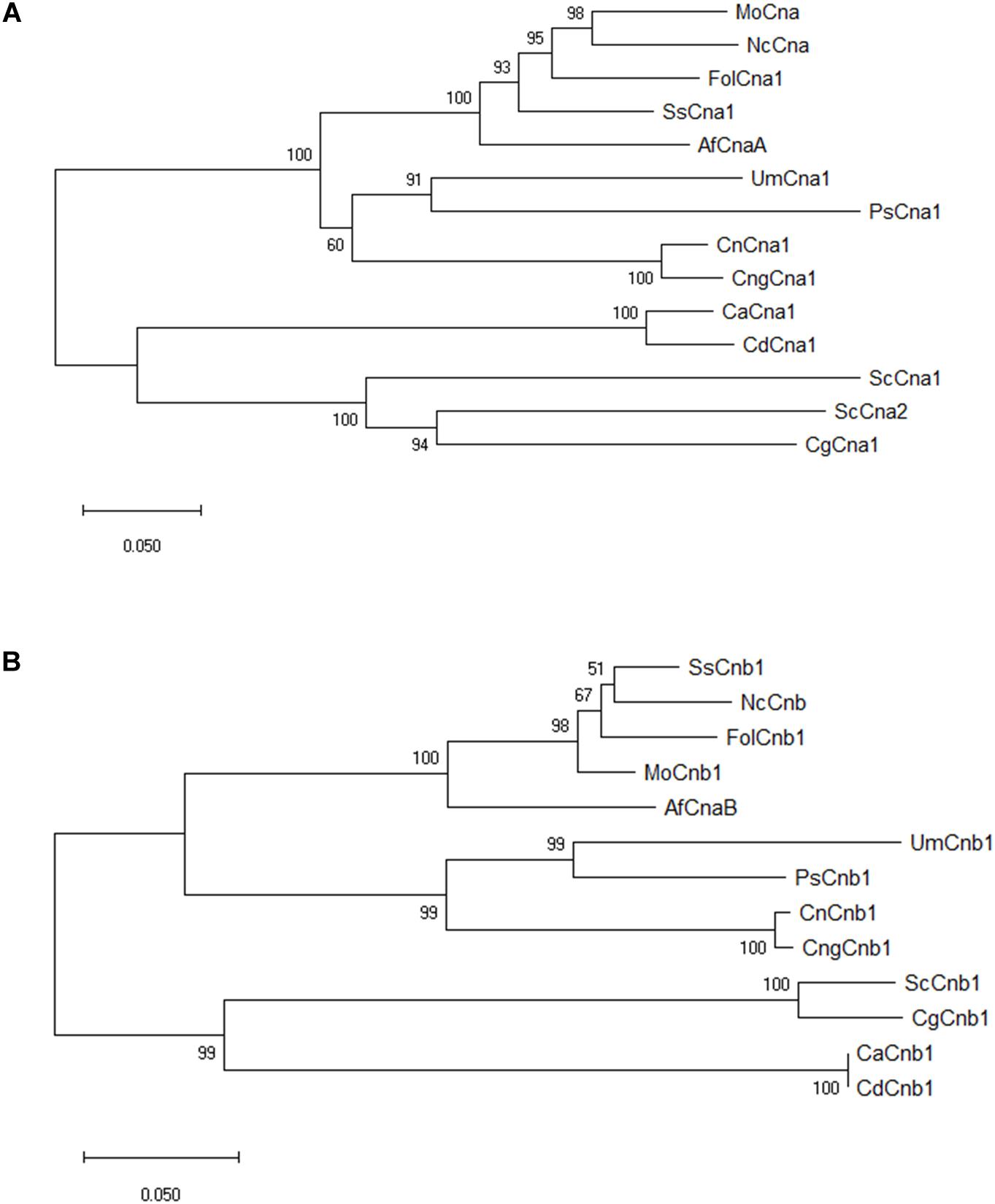
Figure 1. Phylogenetic analyses of F. oxysporum f. sp. lycopersici calcineurin orthologs in filamentous fungi and model organism. By comparing the selected orthologs of calcineurin catalytic Cna1 (A) and regulatory Cnb1 (B) subunits, phylogenic trees were constructed using MEGA software with the neighbor-joining method. The bootstrap confidence value was estimated by 1000 replicates. FolCna1 and FolCnb1 (Fusarium oxysporum f. sp. lycopersici 4287, XP_018243626 and XP_018234243), MoCna and MoCnb (Magnaporthe oryzae 70-15, XP_003711354 and XP_003709672), NcCna and NcCnb (Neurospora crassa OR74A, XP_961193 and CAA73345), SsCna1 and SsCnb1 (Sclerotinia sclerotiorum 1980 UF-70, XP_001597594 and XP_001598128), AfCnaA and AfCnaB (Aspergillus fumigatus Af293, XP_753703 and XP_747624), UmCna1 and UmCnb1 (Ustilago maydis, AAP48999 and EAK82139), PsCna1 and PsCnb1 (Puccinia striiformis f. sp. tritici, AFW98882 and AFW98883), ScCna1 and ScCna2 (Saccharomyces cerevisiae, AAA34465 and AAA34466), ScCnb1 (Saccharomyces cerevisiae, NP_012731), CnCna1 an CnCnb1 (Cryptococcus neoformans var. neoformans, AAD44336 and AAG13937), CngCna1 and CngCnb1 (Cryptococcus neoformans var. gattii WM276, XP_003196247 and XP_003196466), CaCna1 an CaCnb1 (Candida albicans SC5314, XP_718995 and XP_721917), CdCna1 and CdCnb1 (Candida dubliniensis CD36, XP_002416744 and XP_002420770), CgCna1 and CgCnb1 (Candida glabrata, XP_449251 and XP_448800).
Calcineurin Is Required for Phosphatase Activity in F. oxysporum f. sp. lycopersici
In order to understand whether Cna1 and Cnb1 are critical for calcineurin phosphatase activity in Fol, we generated two independent mutants of cna1 and cnb1, respectively, by homologous recombination as described in Materials and Methods. The putative transformants were first screened by drug resistance (HygR) and green fluorescence (GFP) (Supplementary Figure S2A), and finally reconfirmed by PCR analysis (Supplementary Figure S2B). The cna1 mutants (HFW1 and LHS2) and cnb1 mutants (HFW3 and HFW4) were selected for further studies.
To examine whether the calcineurin activity was still exhibited in the mutants, the p-nitrophenyl phosphate (pNPP) was used as substrate for detection of phosphatase activity. Under normal treated condition, we found that phosphatase activity was reduced by approximately 40% in calcineurin mutants compared with the wild type (Figure 2). Concerning that pNPP might not be specific to the serine/threonine phosphatase, additional assays were carried out with or without calcineurin inhibitor (TFP, trifluoperazine) (Gupta et al., 1990), to verify whether the reduction of phosphatase activity in mutants was due to the inactivation of calcineurin. As shown in Figure 2, compared with the wild type under control treatment, enzyme activity had no difference between the wild type and calcineurin mutants in the presence of TFP, suggesting that the reduced phosphatase activity was a result of inactivation of calcineurin. To confirm the calcium dependence of Fol calcineurin activity, the Ca2+ and its chelator EGTA were supplemented into the reaction buffer, respectively. In the wild type, results showed that the Ca2+ and EGTA could elevate and decrease about 16.5 and 19.4% phosphatase activity, respectively, when compared with the untreated control (Figure 2), indicating the critical roles of Ca2+ in calcineurin activity of Fol.
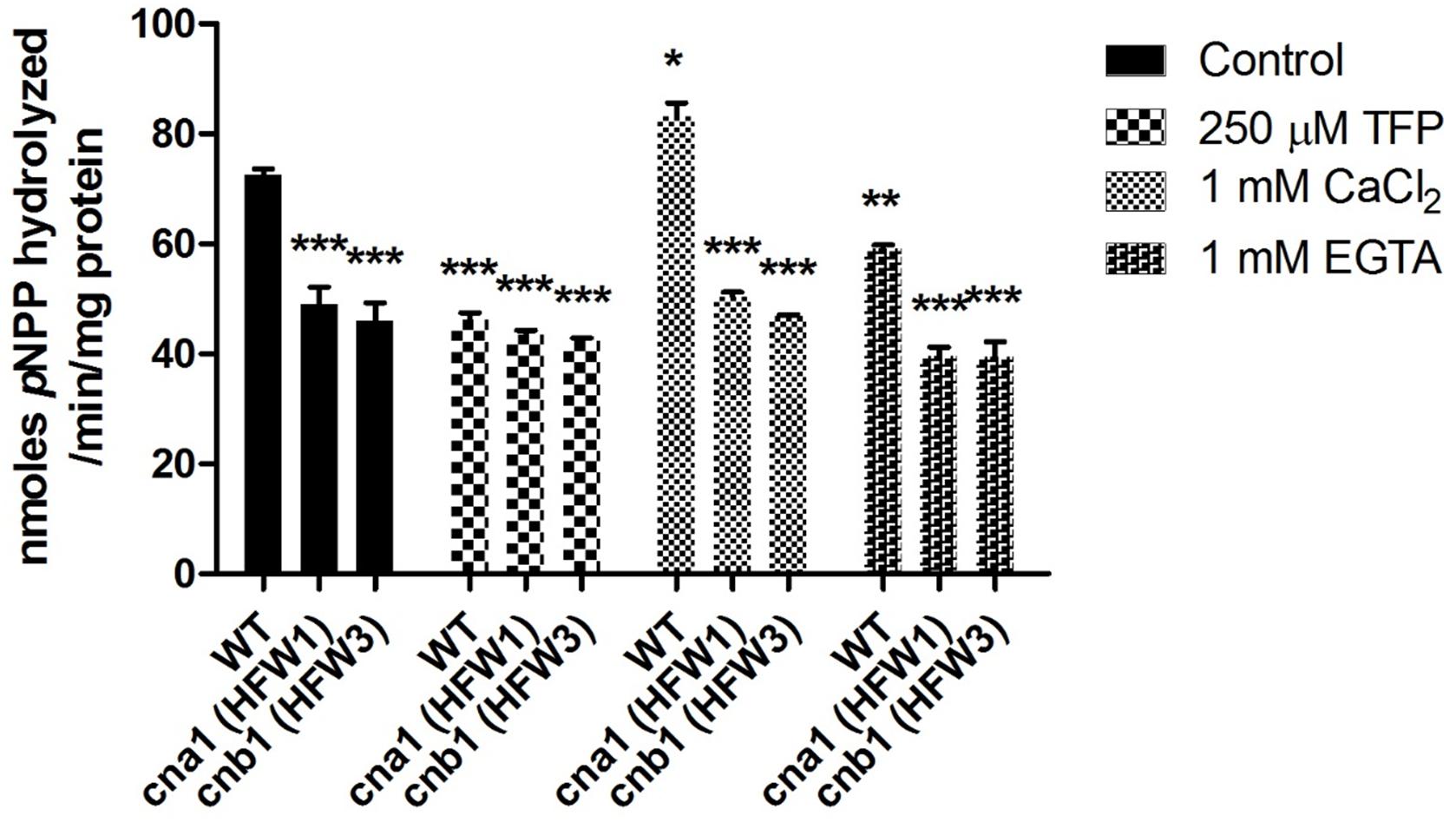
Figure 2. Calcineurin is required for phosphatase activity in Fusarium oxysporum f. sp. lycopersici. Phosphatase activity in the wild type and calcineurin mutants was determined after 72 h growth using p-nitrophenyl phosphate (pNPP) as the substrate. Ca2+ dependence of calcineurin activity was determined by adding 1mM CaCl2 or 1mM EGTA in the reaction. Inhibition of calcineurin activity was determined by adding 250 μM TFP in the reaction. Three technical replicates were performed, and results were analyzed by two-way ANOVA and followed by Bonferroni post-test. Error bars represent standard deviations. ∗P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001 were compared with the WT in the control group.
Calcineurin Governs Vegetative Growth of F. oxysporum f. sp. lycopersici
To investigate the roles of calcineurin in vegetative growth, the colony diameters of wild type and calcineurin mutants were examined daily after inoculation of mycelial agar disks. The wild type showed an extended and radiated mycelial growth on PDA medium. In contrast, the cna1 mutants (HFW1 and LHS2) and cnb1 mutants (HFW3 and HFW4) had compact, wrinkled and sunk colonies (Figure 3A). The average colony diameter of the wild type after 1-week measurement was 72.6 ± 1.0 mm, those of cna1 mutants (HFW1 and LHS2) were 4.6 ± 0.6 and 6.0 ± 0.2 mm; and those of cnb1 mutants (HFW3 and HFW4) were 6.0 ± 0.5 and 6.5 ± 0.2 mm (Figure 3B). The complementary strains (cna1:CNA1 and cnb1:CNB1) showed colony morphology similar to the wild type (Supplementary Figure S4C). Although cnb1:CNB1 grew slower, its growth can be restored as the wile type after slightly extended incubation time, while both cna1 and cnb1 mutants showed retarded growth regardless of extended incubation. These results demonstrated that reduced vegetative growth in cna1 and cnb1 mutants was due to the loss of calcineurin function.
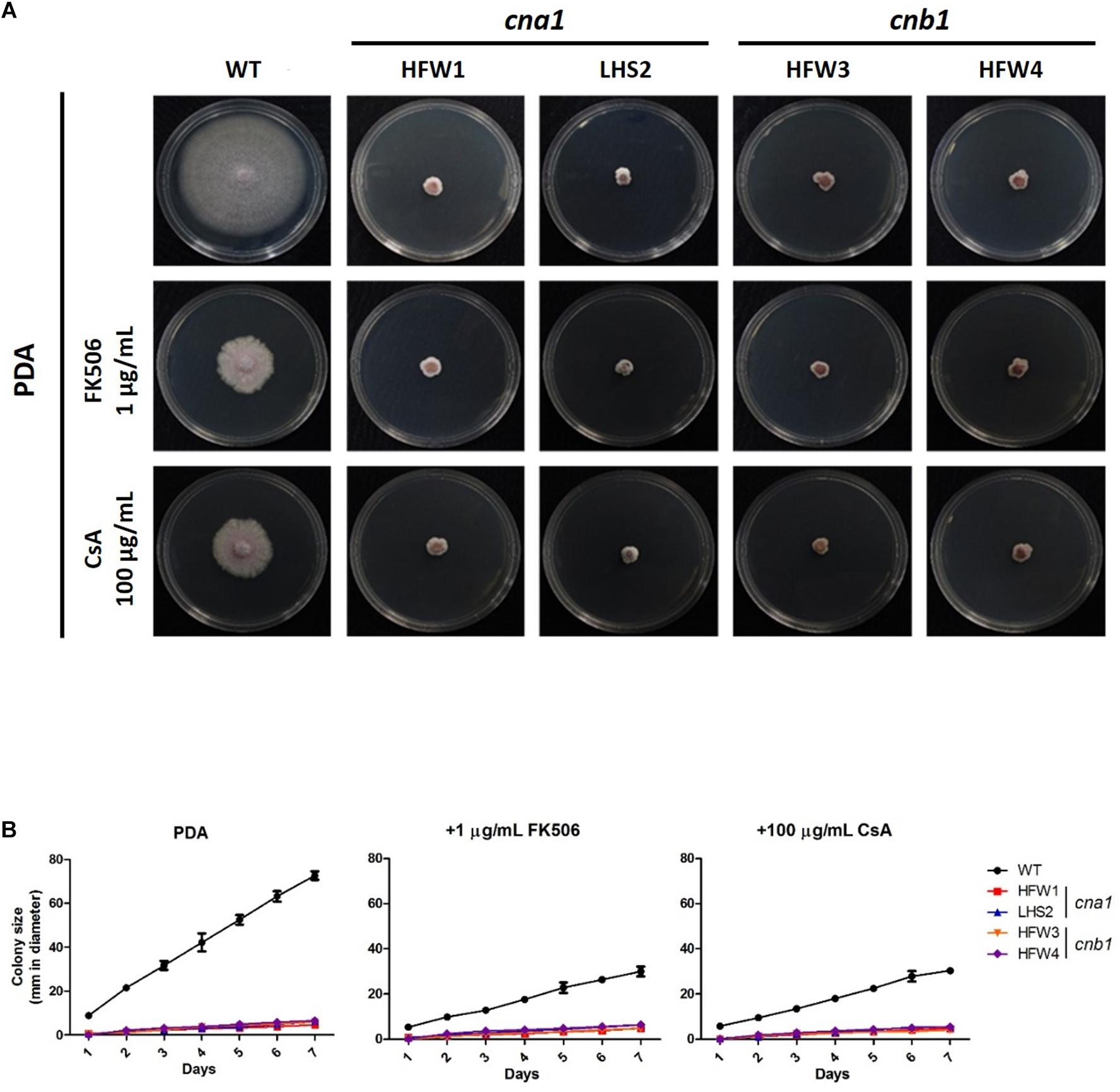
Figure 3. Genetic deletion and pharmacological inhibition of calcineurin cause growth defect in Fusarium oxysporum f. sp. lycopersici. (A) Pharmacological inhibition of calcineurin by calcineurin inhibitors partially mimics genetic deletion of calcineurin in causing growth defect of F. oxysporum f. sp. lycopersici. Vegetative growth of the wild type and calcineurin mutants in the absence or presence of a calcineurin inhibitor. All plates were incubated at 25°C for 1 week. (B) Growth kinetics of the wild type and calcineurin mutants in the presence or absence of a calcineurin inhibitor. Agar discs with the wild type or calcineurin mutants were put onto PDA, PDA + 1 μg/mL FK506, or PDA + 100 μg/mL cyclosporine A (CsA), and colony sizes of each strain were measured in diameter daily for 7 days. Error bars represent standard deviations from three technical replicates.
In order to verify the vegetative growth defects caused by defective calcineurin activity, the calcineurin inhibitor FK506 or cyclosporine A was added to PDA medium. The average diameter of colonies was measured daily after inoculation. As shown in Figure 3, wild type showed a limited vegetative growth on PDA medium containing FK506 (1 μg/mL) or cyclosporine A (100 μg/mL), but was able to extend 29.6 ± 0.9 and 30.0 ± 0.8 mm after 1 week, respectively, whereas the growth of calcineurin mutants was retarded on medium with either inhibitor. In the presence of FK506, the average colony diameters of cna1 mutants were 4.8 ± 0.4 mm (HFW1) and 6.3 ± 0.6mm (LHS2); and those of cnb1 mutants were 4.8 ± 0.1 mm (HFW3) and 6.5 ± 0.5 mm (HFW4). In the presence of cyclosporin A, the average diameter of cna1 and cnb1 colonies was 4.6 ± 0.2 mm (HFW1), 5.5 ± 0.8 mm (LHS2), 4.1 ± 0.7 mm (HFW3), and 5.5 ± 0.5 mm (HFW4), respectively. These results indicate that pharmacological inhibition of calcineurin partially mimics genetic deletion of calcineurin in causing growth defects of Fusarium oxysporum f. sp. lycopersici.
Calcineurin Controls Hyphal Development and Conidiation in Fusarium oxysporum f. sp. lycopersici
To further reveal the details of hyphal compartment in wild type and calcineurin mutants, strains were grown on PDA medium for 2 weeks followed by scanning electron microscope (SEM) analysis. Calcineurin mutants developed irregular, blunted and slightly swollen hyphae compared with the wild type (Figure 4A). Because calcineurin mutants developed abnormal hyphae and grew slower than the wild type, we hypothesized that calcineurin might be involved in hyphal development. To test this hypothesis, conidia suspension of wild type and calcineurin mutants were cultured overnight, and mycelia were harvested for calcofluor white (CFW) staining. Observation under a fluorescent microscope revealed that calcineurin mutants displayed an abnormal distribution of septa (Figure 4A). The average septal distance of wild type was 14.05 ± 4.47 μm, while those of calcineurin mutants were 10.22 ± 3.65 μm (HFW1), 10.48 ± 3.7 μm (LHS2), 10.42 ± 3.49 μm (HFW3), and 10.43 ± 3.2 μm (HFW4), respectively, resulting in a significant difference between the wild type and calcineurin mutants (Figure 4B).
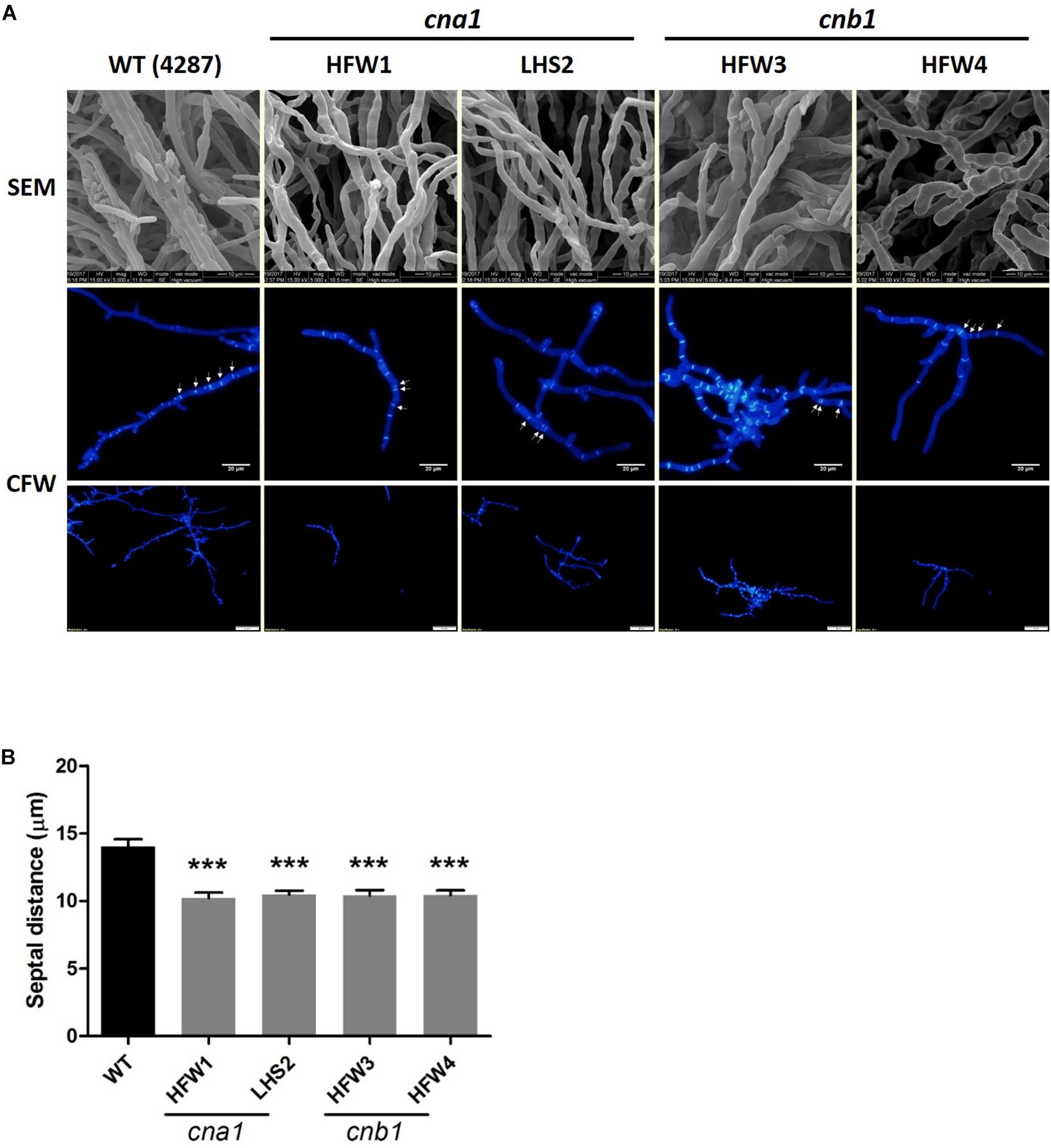
Figure 4. Irregular hyphae and abnormal distribution of septa in calcineurin mutants as revealed by SEM observation and CFW staining, respectively. (A) Hyphal development of 2-week-old wild type and calcineurin mutants was observed under a scanning electron microscope (SEM). Scale bars, 10 μm. Calcofluor white (CFW) staining of hyphae was used to determine the distance of the septa and distribution of hyphal compartment. Scale bars, 50 μm. (B) The average length of septal distance. Data were analyzed by one-way ANOVA. Asterisks indicate significant differences between wild type and calcineurin mutants (P < 0.001). Error bars represent standard errors of mean.
We also monitored the production and germination rate of conidia in wild type and calcineurin mutants. The mycelia agar disks of strains were cultured on the PDA medium for 1 week, conidia were collected and resuspended in ddH2O for number calculation. The results showed that wild type produced more than 108 conidia/mL, whereas cna1 and cnb1 mutants produced less than 105 conidia/mL (Figure 5A), resulting in a larger than 1000-fold difference between the wild type and calcineurin mutants. To examine the rate of germination, conidia (105conidia/mL) derived from the wild type and calcineurin mutants were cultured and germinated in PDB broth for 8 h. The germination rates were calculated at 1-h intervals. As shown in Figure 5B, cna1 and cnb1 mutants displayed a germination rate slightly slower than the wild type at 5 h (P < 0.01 in HFW1; P < 0.001 in LHS2, HFW3, and HFW4), while all strains reached nearly 100% germination rate at 8 h. These results demonstrated that calcineurin plays a vital role in hyphal growth and conidiation, but only slightly affects conidia germination in Fol.
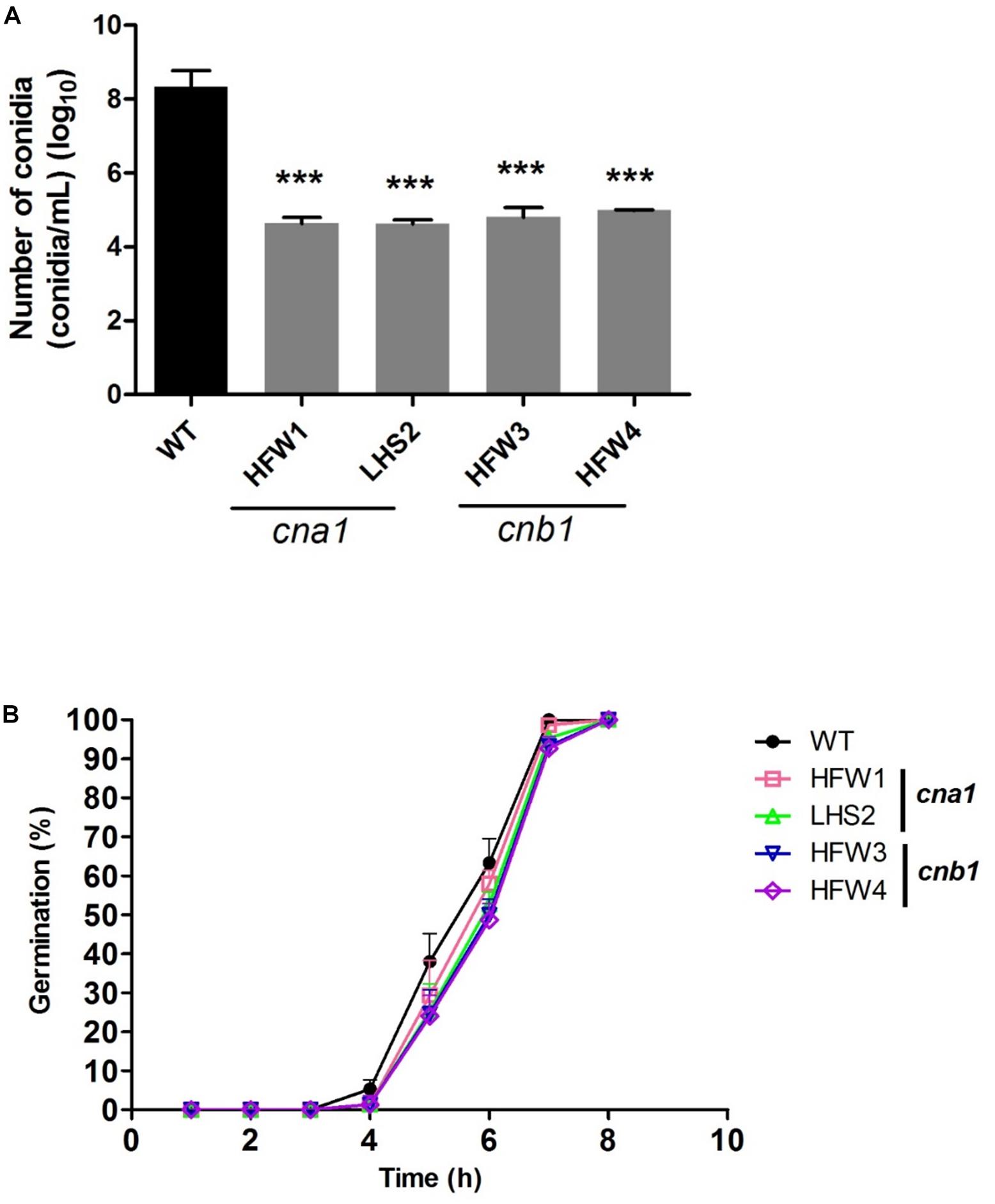
Figure 5. The conidia production and germination rate were reduced in calcineurin mutants. (A) Agar disks of wild type and calcineurin mutants were grown on PDA medium at 25°C for 2 weeks and the number of conidia was counted. Error bars represent standard deviations. Asterisks indicated significant differences (P < 0.001) between wild type and calcineurin mutants. (B) Conidia of the wild type and calcineurin mutants were suspended in PDB and adjusted to 105 conidia/mL. The germination rate was calculated by randomly counting 50 conidia from each of 3 technical replicates over 8 h growth at 25°C.
Calcineurin Mediates Chlamydospore Formation in F. oxysporum f. sp. lycopersici
The role of calcineurin in fungal chlamydospore formation has not yet been reported. To examine this role, the number of chlamydospores were calculated in wild type and calcineurin mutants after incubation for 14 days in chlamydospore-inducing soil medium at 25°C. The number of chlamydospores were defined as the amount of chlamydospore produced per gram of dried mycelia as described in Materials and Methods. Our data showed that many chlamydospores were produced by wild type, but only a few were produced in calcineurin mutants (Figure 6A and Supplementary Figure S5). The chlamydospore production in wild type, cna1 mutants (HFW1 and LHS2), and cnb1 mutants (HFW3 and HFW4) was calculated as [117.5 ± 7.5] × 108, [18.7 ± 6.2] × 108, [15.0 ± 5.0] × 108, [0.75 ± 0.0] × 108, and [0.45 ± 0.35] × 108 (chlamydospores/gram mycelia), respectively, resulting in a significant difference between the wild type and calcineurin mutants (Figure 6B).
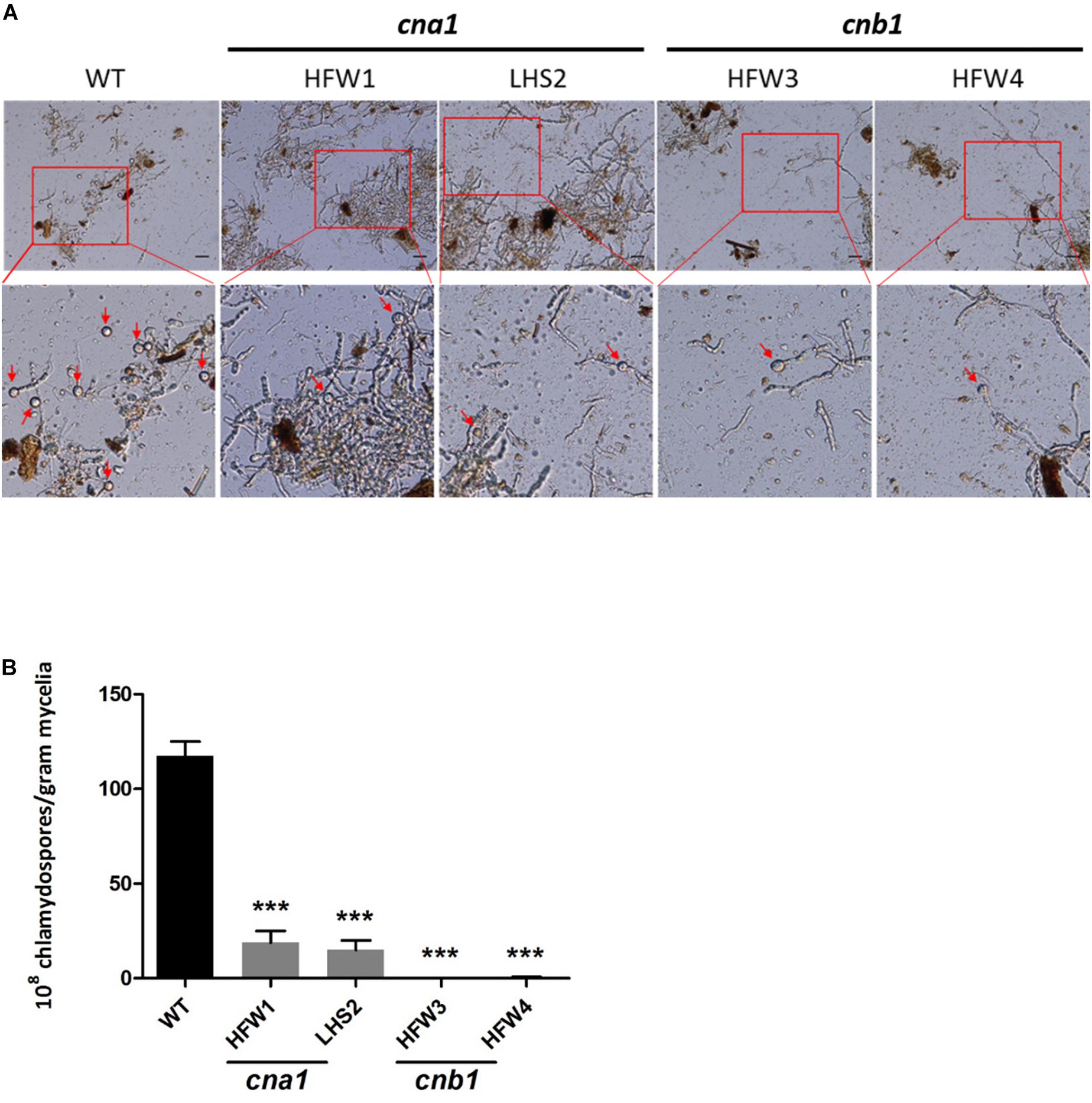
Figure 6. Chlamydospore production was impaired in calcineurin mutants. (A) Conidia from wild type and calcineurin mutants of Fol were inoculated into soil medium to induce chlamydospore formation and then collected as described in section “Materials and Methods.” Arrows indicate chlamydospores. The images in the rectangles of upper panel were enlarged in the lower panel. Scale bar, 50 μm. (B) The average number of chlamydospores produced by mycelia per gram from (A) was analyzed by one-way ANOVA and Bonferroni post-test. Error bars represent standard deviations. Asterisks indicate significant differences (P < 0.001) between wild type and calcineurin mutants. The data were derived from three technical replicates.
Calcineurin Is Required for Virulence of F. oxysporum f. sp. lycopersici on Tomato
To evaluate the virulence of Fol wild type and calcineurin mutants, we conducted a tomato infection assay by the root-dipping inoculation method. Two-week-old tomato seedlings were each inoculated with wild type or a calcineurin mutant, and then grown for 21 days in a green house. The wild type infected tomato plants showed typical Fusarium wilt symptoms, such as yellowing leaves and browning vascular tissues, while only a few leaves displayed yellowing symptoms in calcineurin mutant inoculated seedlings (Figure 7A). All plants wilted and died at 30 days post inoculation (dpi) after wild type infection, whereas plants inoculated with calcineurin mutants revealed a dramatic reduction in disease symptoms and no death was observed at 30 dpi. After 21 dpi, the disease severity of infection with wild type was 100%, and 30, 10, 0 and 0% for the HFW1, LHS2, HFW3 and HFW4, respectively (Figure 7B). Meanwhile, histological observation of tomato infected with wild type or calcineurin mutant was conducted. Cross-section the stem 1 cm above the ground after 21 dpi, as shown in Supplementary Figure S6, the mycelium of Fol wild type could colonize pith, xylem, phloem, and cortex cells of tomato seedlings (Supplementary Figure S6B). Whereas no hyphae can be found in the vascular tissues or cortex cells when tomato seedlings inoculated with cna1 or cnb1 mutants (Supplementary Figures S6C,D). Based on these results, we suggested that calcineurin mutants could invade tomato root but unable to colonize in vascular tissue, indicating ting that calcineurin might be contributed to virulence of Fol on tomato host.
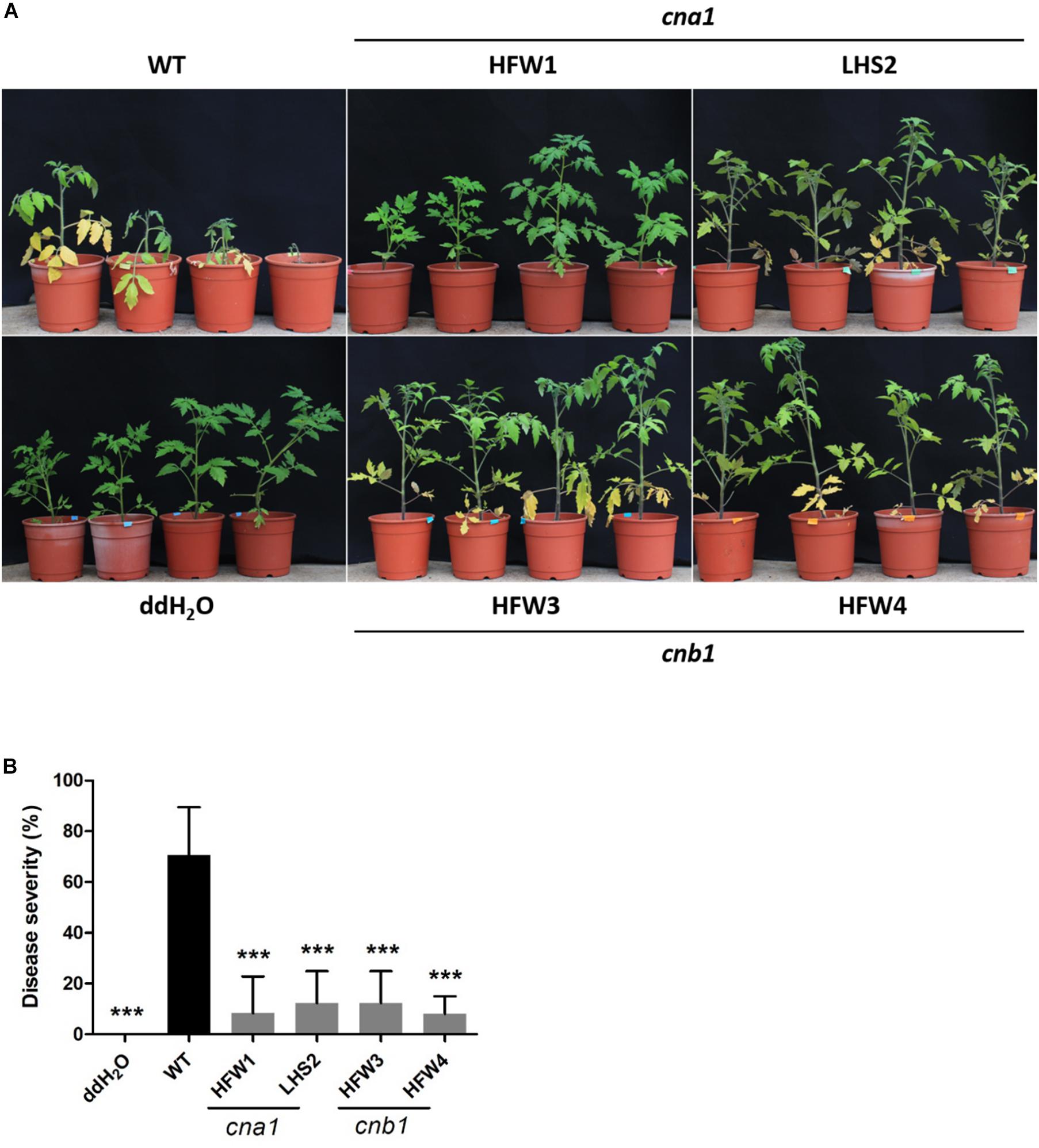
Figure 7. Calcineurin is critical for tomato infection in F. oxysporum f. sp. lycopersici. Tomato seedlings were inoculated by submerging roots in a suspension medium containing 5 × 106 conidia/mL of wild type or calcineurin mutant for 30 min, followed by planting in horticultural substrates, and maintaining in the greenhouse at 28°C. (A) Representative plant phenotypes after 21 days of inoculation. (B) Disease severity was evaluated after 21 days of inoculation. Error bars represented standard deviations. Asterisks indicated significant differences between wild type and calcineurin mutants or mock treatment (P < 0.001).
RNA Sequencing Analysis of Calcineurin-Mediated Genes in F. oxysporum f. sp. lycopersici
To further understand the pleiotropic phenotypes of calcineurin mutants in Fol, we identified calcineurin-mediated genes via RNA sequencing analysis. Strains were grown in PDB at 25°C for 14 h and total RNA was extracted for RNA sequencing analysis. By pairwise analysis, a total of 2139 genes were differentially expressed in wild type and cna1 mutant (HFW1) based on the selection criteria of four-fold change in expression (Log2 fold change > 2 or < -2, and P < 0.05 based on Student’s t-test). Among these, 1079 genes were upregulated and 1060 genes were downregulated in cna1 mutant (Figure 8A). From the comparison of wild type against cnb1 mutant (HFW3), 840 genes were upregulated while 572 genes were downregulated in cnb1 mutant (Figure 8A).
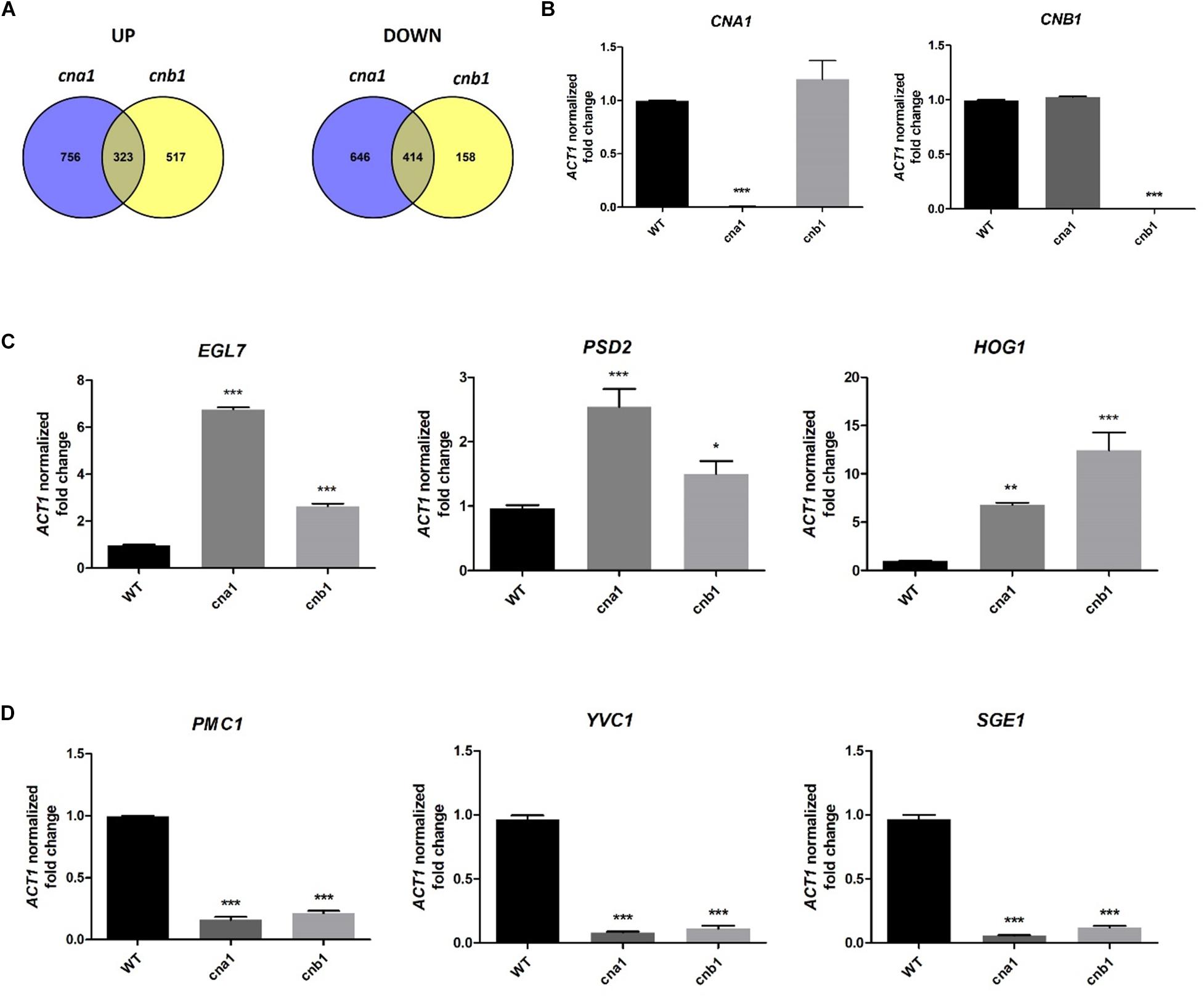
Figure 8. Genes regulated by calcineurin in Fusarium oxysporum f. sp. lycopersici. The 106 conidia/mL of wild type and calcineurin mutants were inoculated in PDB media, and mycelia were collected for RNA extraction after incubation at 25°C for 14 h. (A) Venn diagram illustrating the number of genes expressed differentially between the wild type and cna1 (HFW1) or cnb1 mutant (HFW3) (Log2FC > 2 or < –2, and P < 0.05). A total of 323 genes were upregulated, and 414 genes were downregulated in both cna1 and cnb1 mutants as compared with the wild type. (B) The relative expression levels of CNA1 or CNB1 genes in wild type, cna1 or cnb1 mutant by qRT-PCR analysis. The expression profiles of three upregulated (EGL7, PSD2, HOG1) (C) and three downregulated genes (PMC1, YVC1, SGE1) (D) compared to the wild type were confirmed by qRT-PCR. Error bars represent standard deviation, and results are representative of three technical replicates of one out of three biological replicates, which showed similar results. Asterisks indicated significant differences compared to wild type according to one-way ANOVA and Dunnett’s post-test. *P < 0.05, **P < 0.01, and ***P < 0.001.
Based on the Venn diagram analysis with selection criterion of fourfold change in expression, we identified 737 genes that were differentially expressed coordinately in both cna1 and cnb1 mutants, among which 323 (20% overlapping, 323/1596) and 414 (34% overlapping, 414/1218) genes were up- and down-regulated, respectively (Figure 8A). Moreover, gene ontology (GO) enrichment analysis was performed to better understand the function of those differentially expressed genes (DEGs) that coordinately regulated in both cna1 and cnb1 mutants. The up- and down-regulated genes are functionally correlated with the cellular component, molecular function and biological process, such as those related to cell membrane (GO:0016020, GO:0005886), cell periphery (GO:0071944), and transmembrane transporter (GO:0022891, GO:0022892) (Supplementary Figure S7).
Previous reports demonstrated that calcineurin is involved in ion homeostasis, hyphal growth, cell wall integrity and virulence in filamentous fungi (Juvvadi et al., 2014). We, therefore, screened from those transcripts that overlapped in both cna1 and cnb1 mutants that were associated with the above biological functions. As shown in Table 2 (only showing genes functionally annotated in Fol), out of 737 transcripts, 7 were found to be related to cell wall integrity. Among which 6 genes were upregulated and found to encode endo-β-1,4-glucanase (FolEGLD), endoglucanase (FolEGL4, FolEGL7), chitin synthase (FolCHSD), phosphatidylserine decarboxylase (FolPSD2), and protein kinase (FolHOG1); in addition, one gene encoding chitinase (FolCHS1) was found to be downregulated. As well as cell wall synthesis-related genes, those involved in the transportation of ions and small molecules were also found to be regulated. Three genes were downregulated, P-type Ca2+ ATPase (FolPMC1), Mg2+ translocating P-type ATPase (FolMGTB), and Ca2+ channel (FolYVC1); and one gene encoding Cu2+ exporting ATPase (FolCCC2) was upregulated (Table 2). Interestingly, we found a nuclear protein, SIX gene expression 1 (Sge1), a gene essential for pathogenicity in Fol, was downregulated in both calcineurin mutants.
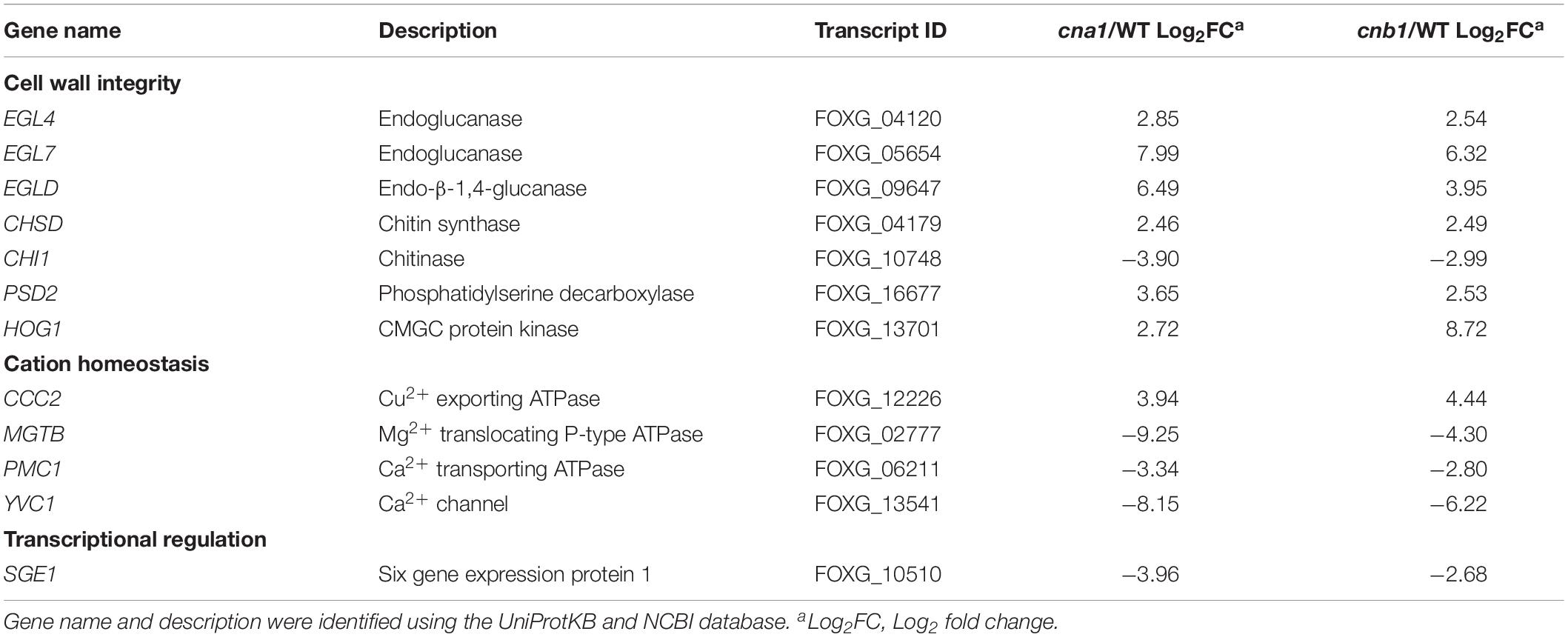
Table 2. Calcineurin-regulated genes are related to cell wall integrity, cation homeostasis, and transcriptional regulation.
To validate the reliability of the candidate gene expression data obtained from RNA sequencing results, the transcription levels of three upregulated genes (EGL7, PSD2, HOG1) and three downregulated genes (PMC1, YVC1, SGE1) were determined by quantitative real-time RT-PCR (qRT-PCR). As shown in Figure 8B, qRT-PCR results confirmed that CNA1 or CNB1 transcription was abolished in cna1 or cnb1 mutant, respectively (Figure 8B). Meanwhile, the relative expression levels of EGL7, PSD2, and HOG1 were increased in both cna1 and cnb1 mutants (Figure 8C), while the expression of PMC1, YVC1, and SGE1 were reduced in both cna1 and cnb1 mutants as compared with the wild type (Figure 8D). These data were consistent with RNA sequencing results.
Discussion
In this study, the biological function of two calcineurin genes, FolCNA1 and FolCNB1, from the tomato wilt fungus F. oxysporum f. sp. lycopersici was characterized. Calcineurin is globally conserved and found in all eukaryotes (Chen et al., 2010; Park et al., 2019). By phylogenetic tree analysis, our results showed that FolCna1 and FolCnb1 were closely related to calcineurin orthologs from other Ascomycete fungi (Figure 1), indicating that functional similarity of calcineurin might be present among the Ascomycetous fungi.
Fol Calcineurin mutants showed severe defects in vegetative and hyphal growth, and a dramatic decrease in virulence. The colony of calcineurin mutants grown on the PDA media were compact and stunted and the radial growth was severely restricted, whereas the wild type displayed longer and dispersed hyphae (Figure 3). These features were similar to the calcineurin A mutant of A. fumigatus and Botrytis cinerea reported in previous studies, which had a tiny, dense, and extremely blunted colony (Steinbach et al., 2006; Juvvadi et al., 2008; Harren et al., 2012). Moreover, blunted hyphae and shorter hyphal compartments could be found in Fol calcineurin mutants under SEM observation and CFW staining (Figure 4). These morphological changes indicate that cell wall integrity might be altered in Fol calcineurin mutants.
Fol calcineurin mutants had a partial reduction in phosphatase activity as compared to the wild type (Figure 2), suggesting that other components or heterodimer partner might contribute to the remaining phosphatase activity. It will be interesting to test whether deletion of both CNA1 and CNB1 genes in Fol will abolish phosphatase activity. Meanwhile, a pull-down assay using either Fol calcineurin subunit as bait might identify a component interacting with calcineurin that has phosphatase activity or calcineurin associated function (Rodríguez et al., 2009; Park et al., 2016).
The soil inhabitant phytopathogenic fungus Fol could survive in plant residues or soil as mycelia or spores during the absence of a host (Agrios, 2005). In the field, Fol mainly survives as a type of chlamydospore, a thick-walled dormant propagule. Extensive studies have shown that chlamydospores play important role in maintaining the Fusarium wilt disease and causing more severe disease symptoms than those of macroconidia or microconidia (Gordon, 2017; Srinivas et al., 2019). In our findings, cna1 and cnb1 mutants reduced conidia production (Figure 5) and impaired chlamydospore formation (Figure 6). Moreover, many of the chlamydospores formed in cna1 or cnb1 mutants have oval-like structure with enlarged suspensor cell instead of typical round structure, indicating that these chlamydospores might be immature or malformed (Supplementary Figure S5) and consequently reduced the disease incidence of tomato Fusarium wilt and affected the life cycle of Fol. However, the conidia germination rates were only slightly altered in cna1 and cnb1 mutants compared with the wild type, indicating that calcineurin plays an important role in the production of conidia and chlamydospore. To the best of our knowledge, calcineurin has not previously been found to affect the formation of chlamydospore in fungi, our findings therefore define a new role for calcineurin in Fol.
Several studies have shown that calcineurin is indispensable for pathogenicity in phytopathogenic fungi. A calcineurin gene MCNA silenced by RNA interference in rice blast fungus Magnaporthe. oryzae resulted in a drastic reduction in mycelial growth, conidiation, appressorium formation, and pathogenicity (Choi et al., 2009). The expression of antisense CNA1 in S. sclerotiorum led to the inhibition of hyphal growth, decreased sclerotia production, and attenuated pathogenicity (Harel et al., 2006). The calcineurin A mutant in gray mold fungus B. cinerea displayed severe defects in hyphae growth which caused a failure to penetrate the bean leaves (Harren et al., 2012). Our studies showed that calcineurin is critical for hyphal growth, conidia production, septal distribution and virulence in Fol. These results revealed that calcineurin plays a vital role in virulence of phytopathogenic fungi.
Due to the importance of calcineurin in fungal growth and virulence, it serves as an ideal antifungal drug. The calcineurin inhibitor such as FK506 or cyclosporin A can bind to FKBP12 or cyclophilin, respectively, and formed a complex with calcineurin and subsequently blocked the phosphatase activity. However, because calcineurin was a conserved phosphatase in eukaryotes, calcineurin inhibitor would also cause immune suppression in mammals. A previous study in our lab revealed that a non-conserved motif within fungal FKBP12 by crystal structure analysis and developed an FK506 analog (APX879), which could attenuate immune suppression in mammalian but enhance antifungal activity in animal therapeutic models (Juvvadi et al., 2019). Moreover, a Serine-Proline Rich Region (SPRR) was found to be a unique domain in filamentous fungal calcineurin, and mutation of the phosphorylated serine residues in the SPRR could block the phosphorylation of CnaA in A. fumigatus (Juvvadi et al., 2013), providing a potential fungal-specific drug target of calcineurin inhibitor.
According to the transcriptome analysis, a few genes related to fungal cell wall integrity were found to be regulated in Fol calcineurin mutants. The cell wall degrading enzymes (EGLD, EGL4, EGL7) and chitin synthase (CHSD) were abundantly expressed in both cna1 and cnb1 mutants (Table 2). In A. fumigatus, it has been shown that β-1,3-glucanase and chitinase plays an important role in hyphal extension by cleavage of the long chain of β-1,3-glucan and broke down the glycosidic bonds of chitin, respectively, both resulting in cell wall remodeling (Hartl et al., 2011; Langner and Gohre, 2016). In addition, the expression of putative HOG1 ortholog, one of MAPK family members, was coordinately regulated in both Fol cna1 and cnb1 mutants. Previous research also showed that the Hog signaling pathways were involved in cell wall integrity of C. neoformans (Bahn and Jung, 2013), indicating that calcineurin might affect cell wall integrity via regulating Hog signaling in Fol. Our study revealed that the colony growth of calcineurin mutants grown on PDA medium containing CFW was similar to those grown on PDA medium only, and the protoplast formation efficiency had no significant difference between wild type and calcineurin mutants (Supplementary Figure S8). As a result, we supposed that the fungal cell wall interference agents (CFW and lysing enzyme) may not have significant effect on Fol calcineurin mutants. Collectively, these results suggested that the biosynthesis of the fungal cell wall might be disturbed in Fol calcineurin mutants, thereby resulting in abnormal hyphal development. We therefore presume that calcineurin might be required for septa formation and proper deposition.
The calcium signaling pathways are critical for virulence, infection, or survival in many pathogenic fungi (Liu et al., 2015). The vacuolar calcium channel YvcA (YVC1 ortholog) in A. fumigatus was responsible for transferring calcium from the cytoplasm to the vacuoles, and the null mutation of YvcA displayed attenuated virulence in the murine model (de Castro et al., 2014). In addition, the P-type ATPase, encoded by PMC1 has been identified as a downstream regulated gene of calcineurin in eukaryotic cells, which is involved in calcium sequestration and regulation. Previous studies in Candida spp. demonstrated that the expression of PMC1 was regulated by calcineurin (Sanglard et al., 2003; Chen et al., 2011), and was required for pathogenesis in C. albicans (Luna-Tapia et al., 2019). The pmcA (PMC1 ortholog) mutant in A. fumigatus was found to be avirulent (Dinamarco et al., 2012). Our results demonstrated that Fol cna1 and cnb1 mutants showed reduced virulence in tomato plants (Figure 7) and downregulated expression of FolPMC1 and FolYVC1 in both calcineurin mutants (Table 2). Additionally, according to our transcriptomic analysis, SIX gene expression 1 (SGE1), a transcription factor in Fol, was downregulated in both cna1 and cnb1 mutants (Table 2). Deletion of FolSGE1 resulted in loss of virulence and inhibited the expression of the effector proteins (secreted in xylem proteins, SIX) during colonization in tomato plant (Lievens et al., 2009; Michielse et al., 2009b). Together, these studies indicate that Fol calcineurin control of virulence is potentially, at least in part, via regulating downstream genes such as YVC1, PMC1, or SGE1. Further experiments to overexpress FolYVC1, FolPMC1, or FolSGE1 in Fol calcineurin mutant might provide further clarification about this influence.
Conclusion
The deletion of CNA1 and CNB1 in Fol resulted in impaired vegetative growth, irregular hyphal development, and the reduction of conidia production and chlamydospore formation. Fol calcineurin mutants also contribute to attenuated virulence in tomato plants. The transcriptome analysis revealed the expressions of cell wall integrity and virulence-related genes were regulated by calcineurin. Taken together, our findings demonstrate that calcineurin signaling plays vital roles in conidiation, chlamydospore formation and virulence of Fol.
Data Availability Statement
The RNA sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE137468 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137468, token: gzqpigoonhgdjoh).
Author Contributions
Y-HH, L-HH, and Y-HL designed the experiments. H-FW and L-HH generated the calcineurin mutants. Y-HH conducted the experiments and wrote the manuscript. Y-LC supervised, designed the experiments, analyzed and interpreted the data, and wrote the manuscript. All the authors read and approved the manuscript.
Funding
This work was financially supported by grants 107-2320-B-002-061-MY3, 106-2923-B-002-001-MY3 from the Ministry of Science & Technology and grant 108AS-1.2.1-ST-aI from the Council of Agriculture, Executive Yuan, Taiwan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. James Swezey and Corby Kistler for providing Fusarium oxysporum f. sp. lycopersici 4287 strain and plasmids pPK2-hphgfp and pRW1. We are grateful to Miranda Loney for language editing, and the Technology Commons, College of Life Science, National Taiwan University for scanning electron microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.539702/full#supplementary-material
Supplementary Figure 1 | Schematic representation of calcineurin domains in F. oxysporum f. sp. lycopersici 4287. (A) The catalytic subunit CNA1 encodes a protein of 566 amino acids, which consists of a catalytic domain, a Cnb binding helix (BBH), a calmodulin-binding domain (CaMBD), and an autoinhibitory domain (AID). (B) The regulatory subunit Cnb1 encodes a protein of 174 amino acids, comprising of four EF-hand Ca2+-binding domains.
Supplementary Figure 2 | Green fluorescence protein observation and PCR analysis of F. oxysporum f. sp. lycopersici wild type and calcineurin mutants. Green fluorescence protein (GFP) fluorescence (A) and PCR analysis (B) were used to confirm the correct disruption of FolCNA1 and FolCNB1. Scale bars, 50 μm. Primers used to confirm wild type and calcineurin mutants are marked with numbers 1 to 6.
Supplementary Figure 3 | Schematic diagram of FolCNA1 and FolCNB1 disruption construct. A diagram showing targeted gene disruption in Fol non-coding region (NCR) flanking 5′ and 3′ ends of the FolCNA1 (A) or FolCNB1 (B) gene was fused with the hygromycin resistance cassette under the control of PgpdA promoter and TtrpC terminator by fusion PCR, and further transformed into the protoplast of the wild type (4287) to carry out homologous recombination. Arrows represent primers used to amplify specific genes or to verify whether successful for targeted DNA replacement.
Supplementary Figure 4 | The growth defects caused by the genetic deletion of CNA1 and CNB1 were restored in the complemented strains. (A) Schematic diagram of cna1 and cnb1 complementation construct. A diagram showing FolCNA1 or FolCNB1 open reading frame (ORF) fused with the bleomycin resistance cassette by fusion PCR, and further transformed into the protoplast of the cna1 or cnb1 mutant, respectively, to carry out homologous recombination. Arrows represent primers used to amplify specific genes or to verify whether successful for targeted DNA replacement. (B) PCR analysis was used to confirm the complementary strains (cna1:CNA1 and cnb1:CNB1). Primers were listed in Supplementary Table S1. (C) Radial growth of the wild type, calcineurin mutants, and complementary strains. All plates were incubated at 25°C for 1 week. (D) Growth kinetics of the wild type and complementary strains. Colony sizes of each strain were measured in diameter daily for 7 days. Error bars represent standard deviations of three technical replicates.
Supplementary Figure 5 | Calcofluor white (CFW) and Nile red (NR) staining of Fol wild type and calcineurin mutants. Conidia from wild type and calcineurin mutants were inoculated into soil extract to induce chlamydospore formation and then collected as described in section “Materials and Methods.” Arrows indicate the chlamydospores. Scale bar, 20 μm.
Supplementary Figure 6 | Histological observation in stem cross sections of tomato seedlings inoculated with the Fol wild type or calcineurin mutant. The 2-week-old tomato seedlings were inoculated with a suspension of 5 × 106 conidia/mL of Fol wild type or calcineurin mutant by submerging roots, and then transplanting into green house for 21 days. Cross section conducted at the stems 1 cm above the ground were examined and photographed. Tomato seedling inoculated with dH2O. Scale bar, 200 μm (A), Fol wild type. Scale bar, 200 μm (B), cna1 mutant. Scale bar, 200 μm (left) and 50 μm (right) (C), or cnb1 mutant. Scale bar, 200 μm (D). Arrows indicate the infected hyphae constitute a cross network between cortex cells and cell surface, while inoculated with Fol wild type (B). Whereas no obvious hyphae can be found on the cortex cells or cell surface, while seedlings inoculated with dH2O or calcineurin mutants (A,C,D). (pi: pith; co: cortex; xy: xylem; ph: phloem).
Supplementary Figure 7 | Gene ontology enrichment analysis of the DEGs that coordinately regulated in both cna1 and cnb1 mutants. Genes are functional correlated with the Cellular component (A), Molecular function (B), and (C) Biological process.
Supplementary Figure 8 | The cell wall integrity analysis of Fol wild type and calcineurin mutants. (A) The cell wall interference agent calcofluor white (CFW) can slightly reduce the colony growth of calcineurin mutants compared with the wild type. Vegetative growth of the wild type and calcineurin mutants was observed in the absence or presence of CFW. All plates were incubated at 25°C for 3 days and photographed. (B) The protoplast formation efficiency has no significant difference between the wild type and calcineurin mutants. The 108 conidia/mL of the wild type and calcineurin mutants were inoculated in PDB media and incubated at 25°C for 16 h, then mycelia were collected and protoplasted with 10 mg/mL lysing enzyme. The protoplasts were counted using a hemocytometer.
Supplementary Table 1 | Primers used in this study.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137468
- ^ https://www.uniprot.org/uploadlists/
- ^ https://www.ncbi.nlm.nih.gov
- ^ https://www.megasoftware.net/
References
Bahn, Y. S., and Jung, K. W. (2013). Stress signaling pathways for the pathogenicity of Cryptococcus. Eukaryotic Cell 12, 1564–1577. doi: 10.1128/ec.00218-13
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cervantes-Chavez, J. A., Ali, S., and Bakkeren, G. (2011). Response to environmental stresses, cell-wall integrity, and virulence are orchestrated through the calcineurin pathway in Ustilago hordei. Mol. Plant Microbe Interact. 24, 219–232. doi: 10.1094/mpmi-09-10-0202
Chen, Y. L., Brand, A., Morrison, E. L., Silao, F. G. S., Bigol, U. G., Malbas, F. F., et al. (2011). Calcineurin controls drug tolerance, hyphal growth, and virulence in Candida dubliniensis. Eukaryotic Cell 10, 803–819. doi: 10.1128/ec.00310-10
Chen, Y. L., Kozubowski, L., Cardenas, M. E., and Heitman, J. (2010). On the roles of calcineurin in fungal growth and pathogenesis. Curr. Fungal Infect. Rep. 4, 244–255. doi: 10.1007/s12281-010-0027-5
Chen, Y. Y., Lin, T. C., Chung, W. H., and Wang, C. L. (2015). Antogonistic microorganisms that inhibit chlamydospore germination of Fusarium oxysporum f. sp. lycopersici. Plant Pathol. Bull. 24, 251–266.
Choi, J. H., Kim, Y., and Lee, Y. H. (2009). Functional analysis of MCNA, a gene encoding a catalytic subunit of calcineurin, in the rice blast fungus Magnaporthe oryzae. J. Microbiol. Biotechnol. 19, 11–16.
Cyert, M. S. (2003). Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311, 1143–1150. doi: 10.1016/s0006-291x(03)01552-3
de Castro, P. A., Chiaratto, J., Winkelströter, L. K., Bom, V. L. P., Ramalho, L. N. Z., Goldman, M. H. S., et al. (2014). The involvement of the Mid1/Cch1/Yvc1 calcium channels in Aspergillus fumigatus virulence. PLoS One 9:e103957. doi: 10.1371/journal.pone.0103957
Dinamarco, T. M., Freitas, F. Z., Almeida, R. S., Brown, N. A., dos Reis, T. F., Ramalho, L. N. Z., et al. (2012). Functional characterization of an Aspergillus fumigatus calcium transporter (PmcA) that is essential for fungal infection. PLoS One 7:e37591. doi: 10.1371/journal.pone.0037591
Egan, J. D., García-Pedrajas, M. D., Andrews, D. L., and Gold, S. E. (2009). Calcineurin is an antagonist to PKA protein phosphorylation required for postmating filamentation and virulence, while PP2A is required for viability in Ustilago maydis. Mol. Plant Microbe Interact. 22, 1293–1301. doi: 10.1094/mpmi-22-10-1293
Gordon, T. R. (2017). Fusarium oxysporum and the Fusarium wilt syndrome. Annu. Rev. Phytopathol. 55, 23–39. doi: 10.1146/annurev-phyto-080615-095919
Gupta, R. C., Khandelwal, R. L., and Sulakhe, P. V. (1990). Effects of sulfhydryl agents, trifluoperazine, phosphatase inhibitors and tryptic proteolysis on calcineurin isolated from bovine cerebral cortex. Mol. Cell Biochem. 97, 43–52.
Harel, A., Bercovich, S., and Yarden, O. (2006). Calcineurin is required for sclerotial development and pathogenicity of Sclerotinia sclerotiorum in an oxalic acid-independent manner. Mol. Plant Microbe Interact. 19, 682–693. doi: 10.1094/mpmi-19-0682
Harren, K., Schumacher, J., and Tudzynski, B. (2012). The Ca2+/calcineurin-dependent signaling pathway in the gray mold Botrytis cinerea: the role of calcipressin in modulating calcineurin activity. PLoS One 7:e41761. doi: 10.1371/journal.pone.0041761
Hartl, L., Gastebois, A., Aimanianda, V., and Latgé, J.-P. (2011). Characterization of the GPI-anchored endo β-1,3-glucanase Eng2 of Aspergillus fumigatus. Fungal Genet. Biol. 48, 185–191. doi: 10.1016/j.fgb.2010.06.011
Juvvadi, P. R., Fortwendel, J. R., Pinchai, N., Perfect, B. Z., Heitman, J., and Steinbach, W. J. (2008). Calcineurin localizes to the hyphal septum in Aspergillus fumigatus: implications for septum formation and conidiophore development. Eukaryot Cell 7, 1606–1610. doi: 10.1128/ec.00200-08
Juvvadi, P. R., Fortwendel, J. R., Rogg, L. E., Burns, K. A., Randell, S. H., and Steinbach, W. J. (2011). Localization and activity of the calcineurin catalytic and regulatory subunit complex at the septum is essential for hyphal elongation and proper septation in Aspergillus fumigatus. Mol. Microbiol. 82, 1235–1259. doi: 10.1111/j.1365-2958.2011.07886.x
Juvvadi, P. R., Fox, D., Bobay, B. G., Hoy, M. J., Gobeil, S. M. C., Venters, R. A., et al. (2019). Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents. Nat. Commun. 10:4275.
Juvvadi, P. R., Gehrke, C., Fortwendel, J. R., Lamoth, F., Soderblom, E. J., Cook, E. C., et al. (2013). Phosphorylation of Calcineurin at a novel serine-proline rich region orchestrates hyphal growth and virulence in Aspergillus fumigatus. PLoS Pathog. 9:e1003564. doi: 10.1371/journal.ppat.1003564
Juvvadi, P. R., Lamoth, F., and Steinbach, W. J. (2014). Calcineurin as a multifunctional regulator: unraveling novel functions in fungal stress responses, hyphal growth, drug resistance, and pathogenesis. Fungal Biol. Rev. 28, 56–69. doi: 10.1016/j.fbr.2014.02.004
Juvvadi, P. R., and Steinbach, W. J. (2015). Calcineurin orchestrates hyphal growth, septation, drug resistance and pathogenesis of Aspergillus fumigatus: where do we go from here? Pathogens 4, 883–893. doi: 10.3390/pathogens4040883
Langner, T., and Gohre, V. (2016). Fungal chitinases: function, regulation, and potential roles in plant/pathogen interactions. Curr. Genet. 62, 243–254. doi: 10.1007/s00294-015-0530-x
Lievens, B., Houterman, P. M., and Rep, M. (2009). Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 300, 201–215. doi: 10.1111/j.1574-6968.2009.01783.x
Liu, S., Hou, Y., Liu, W., Lu, C., Wang, W., and Sun, S. (2015). Components of the calcium-calcineurin signaling pathway in fungal cells and their potential as antifungal targets. Eukaryotic Cell 14, 324–334. doi: 10.1128/ec.00271-14
Luna-Tapia, A., DeJarnette, C., Sansevere, E., Reitler, P., Butts, A., Hevener, K. E., et al. (2019). The vacuolar Ca2+ ATPase pump Pmc1p is required for Candida albicans pathogenesis. mSphere 4:e00715-18.
Ma, L. J., van der Does, H. C., Borkovich, K. A., Coleman, J. J., Daboussi, M.-J., Di Pietro, A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373.
Manikandan, R., Harish, S., Karthikeyan, G., and Raguchander, T. (2018). Comparative proteomic analysis of different isolates of Fusarium oxysporum f.sp. lycopersici to exploit the differentially expressed proteins responsible for virulence on tomato plants. Front. Microbiol. 9:420. doi: 10.3389/fmicb.2018.00420
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Michielse, C. B., van Wijk, R., Reijnen, L., Cornelissen, B. J., and Rep, M. (2009a). Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large-scale insertional mutagenesis. Genome Biol. 10:R4.
Michielse, C. B., van Wijk, R., Reijnen, L., Manders, E. M., Boas, S., Olivain, C., et al. (2009b). The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. PLoS Pathog. 5:e1000637. doi: 10.1371/journal.ppat.1000637
Moradi, S., Sanjarian, F., Safaie, N., Mousavi, A., and Bakhshi Khaniki, G. R. (2013). A modified method for transformation of Fusarium graminearum. J. Crop Protect. 2, 297–304.
Park, H. S., Chow, E. W., Fu, C., Soderblom, E. J., Moseley, M. A., Heitman, J., et al. (2016). Calcineurin targets involved in stress survival and fungal virulence. PLoS Pathog. 12:e1005873. doi: 10.1371/journal.ppat.1005873
Park, H. S., Lee, S. C., Cardenas, M. E., and Heitman, J. (2019). Calcium-calmodulin-calcineurin signaling: a globally conserved virulence cascade in eukaryotic microbial pathogens. Cell Host Microbe 26, 453–462. doi: 10.1016/j.chom.2019.08.004
Perrino, B. A., Ng, L. Y., and Soderling, T. R. (1995). Activation of calcineurin A subunit phosphatase activity by its calcium-binding B subunit. J. Biol. Chem. 270, 340–346. doi: 10.1074/jbc.270.1.340
Pietro, A. D., Madrid, M. P., Caracuel, Z., Delgado-Jarana, J., and Roncero, M. I. G. (2003). Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. doi: 10.1046/j.1364-3703.2003.00180.x
Punt, P. J., Oliver, R. P., Dingemanse, M. A., Pouwels, P. H., and van den Hondel, C. A. (1987). Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56, 117–124. doi: 10.1016/0378-1119(87)90164-8
Rasmussen, C., Garen, C., Brining, S., Kincaid, R. L., Means, R. L., and Means, A. R. (1994). The calmodulin-dependent protein phosphatase catalytic subunit (calcineurin A) is an essential gene in Aspergillus nidulans. EMBO J. 13, 2545–2552. doi: 10.1002/j.1460-2075.1994.tb06544.x
Rodríguez, A., Roy, J., Martínez-Martínez, S., López-Maderuelo, M. D., Niño-Moreno, P., Ortí, L., et al. (2009). A conserved docking surface on calcineurin mediates interaction with substrates and immunosuppressants. Mol. Cell 33, 616–626. doi: 10.1016/j.molcel.2009.01.030
Sanglard, D., Ischer, F., Marchetti, O., Entenza, J., and Bille, J. (2003). Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48, 959–976. doi: 10.1046/j.1365-2958.2003.03495.x
Srinivas, C., Devi, D. N., Murthy, K. N., Mohan, C. D., Lakshmeesha, T. R., Singh, B., et al. (2019). Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: biology to diversity– A review. Saudi J. Biol. Sci. 7, 1315–1324.
Steinbach, W. J., Cramer, R. A., Perfect, B. Z., Asfaw, Y. G., Sauer, T. C., Najvar, L. K., et al. (2006). Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryotic Cell 5, 1091–1103.
Stemmer, P. M., and Klee, C. B. (1994). Dual calcium ion regulation of calcineurin by calmodulin and calcineurin B. Biochemistry 33, 6859–6866.
Yu, S. J., Chang, Y. L., and Chen, Y. L. (2015). Calcineurin signaling: lessons from Candida species. FEMS Yeast Res. 15:fov016.
Keywords: Fusarium oxysporum, calcineurin, conidiation, chlamydospore, virulence
Citation: Hou Y-H, Hsu L-H, Wang H-F, Lai Y-H and Chen Y-L (2020) Calcineurin Regulates Conidiation, Chlamydospore Formation and Virulence in Fusarium oxysporum f. sp. lycopersici. Front. Microbiol. 11:539702. doi: 10.3389/fmicb.2020.539702
Received: 04 March 2020; Accepted: 25 September 2020;
Published: 22 October 2020.
Edited by:
Keiko Yoshioka, University of Toronto, CanadaReviewed by:
Shizhu Zhang, Nanjing Normal University, ChinaNancy Keller, University of Wisconsin–Madison, United States
Nandhitha Venkatesh, University of Wisconsin-Madison in collaboration with reviewer NK
Copyright © 2020 Hou, Hsu, Wang, Lai and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-Lien Chen, ychen28@ntu.edu.tw
 Yi-Hsuan Hou
Yi-Hsuan Hou Ying-Lien Chen
Ying-Lien Chen